- State Key Laboratory of Crop Genetics and Germplasm Enhancement, Key Laboratory of Biology and Germplasm Enhancement of Horticultural Crops in East China, Ministry of Agriculture, College of Horticulture, Nanjing Agricultural University, Nanjing, China
MADS-box genes form a large gene family in plants and are involved in multiple biological processes, such as flowering. However, the regulation mechanism of MADS-box genes in flowering remains unresolved, especially under short-term cold conditions. In the present study, we isolated BcMAF1, a Pak-choi (Brassica rapa ssp. Chinensis) MADS AFFECTING FLOWERING (MAF), as a floral repressor and functionally characterized BcMAF1 in Arabidopsis and Pak-choi. Subcellular localization and sequence analysis indicated that BcMAF1 was a nuclear protein and contained a conserved MADS-box domain. Expression analysis revealed that BcMAF1 had higher expression levels in leaves, stems, and petals, and could be induced by short-term cold conditions in Pak-choi. Overexpressing BcMAF1 in Arabidopsis showed that BcMAF1 had a negative function in regulating flowering, which was further confirmed by silencing endogenous BcMAF1 in Pak-choi. In addition, qPCR results showed that AtAP3 expression was reduced and AtMAF2 expression was induced in BcMAF1-overexpressing Arabidopsis. Meanwhile, BcAP3 transcript was up-regulated and BcMAF2 transcript was down-regulated in BcMAF1-silencing Pak-choi. Yeast one-hybrid and dual luciferase transient assays showed that BcMAF1 could bind to the promoters of BcAP3 and BcMAF2. These results indicated that BcAP3 and BcMAF2 might be the targets of BcMAF1. Taken together, our results suggested that BcMAF1 could negatively regulate flowering by directly activating BcMAF2 and repressing BcAP3.
Introduction
The switch from vegetative to reproductive growth, which is called floral transition, is a vital developmental transition in flowering plants for reproductive success. The shoot apical meristem only produces leaf primordia during the vegetative phase, yet the shoot apical meristem can produce floral primordia after the transition. This process is regulated by multiple environmental and endogenous factors, such as temperature and daylength (Simpson and Dean, 2002). To maximize reproductive success, it is important to fulfill this transition at the correct time. Vernalization is a process that promotes plants to acquire flowering competence during long periods of winter cold and coordinating floral development with the seasons until winter has finished (Searle et al., 2006). The mechanisms by which plants regulate flowering time in the vernalization pathway, particularly during short-term cold conditions, and still coordinate floral development is a meaningful area of research.
MADS-box genes regulate multiple and important biological processes in plants, such as floral transition. MADS-box genes act in determining floral organ identity to control floral transition. Mutations of multiple MADS-box genes can change floral organs to leaves and overexpression of these genes can change leaves to floral organs. MADS-box proteins all have an MIKC structure and contain a highly conserved DNA-binding MADS domain in the N terminal, most of which can bind to a minimal consensus motif called the CArG box (Shore and Sharrocks, 1995). Some MADS-box genes are temperature responsive and regulate response to cold conditions. For example, FLOWERING LOCUS C (FLC) plays a key role in vernalization-induced flowering. In Arabidopsis, FLC functions as a floral repressor by directly repressing downstream genes, which promote flowering, such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Deng et al., 2011). The expression of FLC is stably reduced and triggered by vernalization on the epigenetic level and remains low even when returned to warm conditions. This repression can be “remembered” and reactivated only in the next generation, allowing rapid flowering in spring (Sheldon et al., 2000).
The genus Brassica has a close relationship with the model species Arabidopsis, both of which belong to the Brassicaceae family. B. rapa is a member of the genus Brassica whose subgenomes have evolved by genome fractionation from Arabidopsis (Wang et al., 2011). Flowering time is a key developmental trait and wide variation exists among B. rapa accessions. Several major flowering candidate genes, such as FLC and FT, have been identified in B. rapa based on previous studies of flowering time regulation. Six copies of FT have been mapped in B. napus, three of which were related to two major quantitative trait loci clusters for flowering time (Zhao et al., 2009). In addition, four FLC paralogs have been cloned in B. rapa (Schranz et al., 2002). The investigation of FLC expression and vernalization response in B. napus (Zou et al., 2012) and Chinese cabbage (Kim et al., 2007), together with co-localization of FLC paralogs with quantitative trait loci for flowering time in B. rapa (Lou et al., 2007; Xiao et al., 2013) suggests that the Brassica FLC genes and Arabidopsis FLC gene have similar functions. However, the cold-sensing mechanism of FLC is not clear and genetic manipulation of the vernalization trait in B. rapa has not been successfully studied.
MADS AFFECTING FLOWERING, which are FLC-related genes, also act as floral repressors in Arabidopsis (Ratcliffe et al., 2001). Although the functions of MAFs in Arabidopsis have been studied, there is no report on MAFs in Pak-choi. Pak-choi (Brassica rapa ssp. Chinensis), belonging to B. rapa family, is a major vegetable crop that is widely cultivated in Asia (Tian et al., 2004). The vernalization response shows difference among the different cultivars in Pak-choi. Wuyueman was used as the main material in the present study, which requires vernalization and flowers later than other cultivars. We isolated and functionally characterized BcMAF1, a Pak-choi MAF gene, as a floral repressor in Arabidopsis and Pak-choi. Expression profiles in different tissues and during the process of vernalization in Pak-choi were demonstrated. The results showed that BcMAF1 was highly expressed in leaves, stems, and petals, and was induced by short-term cold conditions. Overexpressing BcMAF1 in Arabidopsis caused late flowering and silencing endogenous BcMAF1 in Pak-choi showed early flowering. Yeast one-hybrid assay, dual luciferase transient assay, and qPCR results showed that BcMAF1 could directly repress BcAP3 and activate BcMAF2. In summary, the present study suggested that BcMAF1 played a negative role in regulating flowering by directly repressing BcAP3 and activating BcMAF2 in Pak-choi.
Materials and Methods
Plant Materials
Plants of Pak-choi cultivars Wuyueman and 49caixin were grown in a greenhouse under long day (LD) conditions (16 h light/8 h dark, 22°C/18°C). For the vernalization treatment, 1-month-old seedlings of Pak-choi cultivar Wuyueman were transferred to a new chamber exposure condition of 4°C for 5 weeks. Seedlings grown in the greenhouse without vernalization treatment were the control group. Leaves of vernalized and non-vernalized (control) seedlings were collected after treatment at 0, 1, 2, 3, 4, and 5 weeks; frozen immediately in liquid nitrogen; and stored at -80°C. To analyze the expression of BcMAF1 in different tissues, the leaves, stems, roots, styles, stamens, petals and sepals of flowering Pak-choi cultivar Wuyueman were harvested and frozen in liquid nitrogen. Three biological replications were performed for each sample. The Pak-choi cultivar 49caixin do not require a vernalization treatment and grow faster than other cultivars; therefore, it was only used for virus-induced gene silencing (VIGS).
Arabidopsis overexpressing BcMAF1 seedlings were Col-0 ecotype background and grown in a greenhouse under LD conditions (16 h light/8 h dark, 22°C/18°C). Seeds of 35S:GFP and 35S:BcMAF1-GFP T3 lines were grown on Murashige and Skoog (MS) medium with 35 mg/L hygromycin for expression analysis of downstream genes. Whole seedlings were collected after 15 days for RNA extraction. Both Pak-choi and Arabidopsis samples used for qPCR were harvested at Zeitgeber time (ZT) 16 under LD conditions, in which FT might show relatively higher expression (Osnato et al., 2012). For extracting mesophyll protoplasts, 1-month-old Arabidopsis Col-0 seedlings were grown under short day (SD) conditions (8 h light/16 h dark photoperiod).
Cloning and Sequence Analysis
To clone BcMAF genes, degenerate primers (5′-RATYGAGARCAARAGYAGTNGACAA-3′, Y = C/T, R = A/G, N = A/G/C/T) and 5′-Oligo(dT)20MN-3′(M = A/G/C, N = A/G/C/T) were first designed to amplify the conserved regions of the MAF orthologs based on sequence information from the A. thaliana MAF gene family in TAIR101 and the Chinese cabbage chiifu genome in BRAD2 according to the previous report (Duan et al., 2015). Based on the sequencing results of PCR products from the conserved region of each MAF orthologs and the full length sequences of Chinese cabbage MAF orthologs (data not shown), we designed gene-specific primers (Supplementary Table S1) and performed 5′-and 3′-RACE (Smart RACE cDNA amplification kit; Clontech, Mountain View, CA, United States) to amplify the full-length cDNA sequences of BcMAFs in the cDNA library of Pak-choi cultivar Wuyueman leaves. Total RNA extraction, cDNA synthesis, and amplification by PCR were conducted using the same methods described in our previous report (Huang et al., 2016). The open reading frames (ORFs) of BcMAF1 and BcMAF2 (accession numbers: MG964044 and MG964045) were amplified by BcMAF1-S and BcMAF1-A, and BcMAF2-S and BcMAF2-A primers, respectively, and then inserted into the PMD18-T Vector (Takara, Beijing, China) before sequencing. The ORFs of BcSOC1, BcFT1, BcFT2, and BcAP3 (accession numbers: MG964046, MG964047, MG964048, and MG964049) were also amplified using the same methods. The genomic sequences of BcAP3 and BcMAF2 were amplified by two pairs of primers, BcAP3-S and BcAP3-A, and BcMAF2-S and BcMAF2-A, respectively, from genomic DNA, which was isolated using the Plant Genomic DNA Kit (Tiangen, Beijing, China). The promoters of BcAP3 and BcMAF2 were obtained using corresponding SP1, SP2, and SP3 primers, which were designed based on the genomic sequences of BcAP3 and BcMAF2 by self-formed adaptor PCR with a KX Genome Walking Kit (Zoman Biotechnology, Beijing, China) according to the manufacturer’s instructions (Wang et al., 2007). The promoters were then amplified by two pairs of primers, Y1 and Y2, and Y3 and Y4. All primers used are listed in Supplementary Table S1. Multiple sequence alignment and phylogenetic analysis were performed based on the procedure outlined in our previous report (Huang et al., 2016). The phylogenetic tree was generated with full-length protein sequences using the Neighbor-Joining method by MEGA 6. Bootstrap values were estimated with 1000 replicates. The CArG boxes that existed in promoters were analyzed by the Softberry software program3.
Subcellular Localization of 35S:BcMAF1-GFP Protein
The protein coding region of BcMAF1 without the termination codon was amplified by primers, O1 and O2, and then cloned into the pCambia1302 vector in fusion with the green fluorescent protein (GFP) under the CaMV35S promoter, generating a novel fusion vector (35S:BcMAF1-GFP). The empty vector (35S:GFP) was used as the control. 35S:BcMAF1-GFP and 35S:GFP plasmids were transformed into the Agrobacterium tumefaciens (strain GV3101) using the freeze-thaw method. The obtained Ag. tumefaciens were injected into tobacco leaves based on methods described previously (Zhang et al., 2012). Tobacco leaves were also stained with DAPI (nucleus specific dye) to confirm nuclear localization. After incubation at 25°C for 48 h, GFP in tobacco leaves was observed by confocal microscopy (Leica, TCS SP2, Wetzlar, Germany).
Generation of BcMAF1 Overexpressing Lines
The 35S:BcMAF1-GFP construct was transformed into Arabidopsis Col-0 seedlings by the floral-dip method (Clough and Bent, 1998). Seeds of T0, T1, and T2 transgenic plants were selected on MS medium containing 35 mg/L hygromycin. To confirm the positive transgenic plants, PCR was used with a pair of specific primers (O1 and O2). The transgenic plants transformed with 35S:GFP were used as the control. Western blot analysis was performed based on methods described in a previous study (Jie et al., 2017). The GFP fluorescence in transgenic plants was also detected based on the above method. Of the six independent transgenic lines selected, three T3 homozygous lines (#8, #16, and #23), confirmed by PCR, western blot, and GFP fluorescence observation, were used for all analyses. Flowering time was counted from sowing time to the opening time of the first flower. Total leaf number was counted until the time of bolting. Each measurement was calculated with 30 plants. Values were expressed as means ± standard deviation of mean (SEM). Analysis of variance was used for statistical analysis. Differences between lines were separated using the least significant difference test at P < 0.01.
VIGS-Mediated Silencing of BcMAF1 in Pak-choi
For VIGS assay, a 40 bp specific fragment of BcMAF1 was derived from its coding sequence. The 40 bp specific fragment and its antisense version were synthesized by the GenScript company (China) to form the self-hybridized palindromic oligonucleotide. Then, the self-hybridized palindromic oligonucleotide was inserted into the pTY-S (pTY) vector of the turnip yellow mosaic virus-induced gene silencing (TYMV-VIGS) system to form the BcMAF1-silencing construct before sequencing (Pflieger et al., 2008). The pTY empty vector and the pTY carrying the self-hybridized palindromic oligonucleotide of BcPDS served as negative and positive controls, respectively. The sequences of oligonucleotides used for VIGS are listed in Supplementary Table S2. Two-week-old Pak-choi cultivar 49caixin plants, which usually bolted at 8-weeks-old, were used for VIGS. The pTY, pTY-BcPDS, and pTY-BcMAF1 plasmids (5 μg) coated onto gold particles were bombarded into plants by particle gun bombardment (Bio-Rad, PDS1000/He) based on the previous protocol with some modifications (Hamada et al., 2017). In each experiment, four plants were bombarded with each plasmid in three biological replicates. Three weeks later, leaves showing virus symptoms were sampled to analyze the levels of predicted downstream genes and silencing efficiency. BcMAF1-silencing Pak-choi plants were confirmed by qPCR. Two positive plants, pTY-BcMAF1-1 and pTY-BcMAF1-5, were used for all experiments. Days from sowing to bolting time were counted.
Expression Analysis in Pak-choi and Arabidopsis
qPCR analysis was performed based on methods described in our previous report (Huang et al., 2016). AtACT and BcACT were used as the internal controls in Arabidopsis and Pak-choi, respectively. To evaluate amplification specificity, melting curves were generated for each reaction. Three biological replicates were used for each experiment. The results were analyzed by the 2-ΔΔCt method (Livak and Schmittgen, 2012). Primers for qPCR were designed by Premier 5 and are presented in Supplementary Table S1.
Yeast One-Hybrid Assay
We performed the yeast one-hybrid assay with the Matchmaker® Gold Yeast One-Hybrid System following the standard protocol (Clontech Laboratories Inc., Palo Alto, CA, United States). The promoter fragments of BcMAF2 and BcAP3 were cloned upstream of the reporter AUR1-C gene, which conferred resistance to Aureobasidin A (AbA), into the pAbAi vector at the KpnI and XhoI sites, respectively, and were then integrated into the yeast genome (strain Y1H GOLD). The information of the promoters of BcMAF2 and BcAP3 is shown in Supplementary Table S3. Yeast genomic PCR was used to verify bait strains by promoter-specific primers. The coding sequence of BcMAF1 was inserted into pGADT7 with EcoRI and SacI restriction sites to generate the prey vector. After self-activation test, the prey vector was transformed into the yeast cells containing the promoter fragments on SD/-Leu medium. pGADT7 was used as the negative control. The survival colonies were picked and then spotted onto the SD/-Leu medium containing 300 ng/mL AbA at 30°C for 3 days.
Transient Dual Luciferase Assay in Arabidopsis Protoplasts
To generate the reporter fusion construct, the putative promoter regions of BcMAF2 and BcAP3 were introduced into the pGreenII 0800-LUC vector using KpnI and XhoI restriction sites, in which the Firefly luciferase gene (FLUC) was controlled by the above cloned promoters. Protoplasts were subsequently co-transfected with 20 μg of 35S:BcMAF1-GFP, 20 μg of the recombinant pGreenII 0800-LUC vector, and 2 μg of Renilla luciferase (RLUC) reporter plasmid (as an internal control), and then incubated in darkness for 18 h. Protoplasts co-transfected with the recombinant pGreenII 0800-LUC plasmid and Renilla luciferase reporter plasmid were used as the negative control. Protoplasts isolation and transfection were performed following the described methods (Kang et al., 1998; Yoo et al., 2007) with some modifications. FLUC and RLUC activities were separately quantified by the Dual-Luciferase Reporter Assay System (Promega). The relative FLUC/RLUC activity was used to measure the promoter activity. All assays were repeated three times. Values are expressed as means ± SEM. Differences between treatments were separated using the least significant difference test at P < 0.01.
Results
Isolation and Expression Analysis of BcMAF1
We isolated two MAF genes in Pak-choi and named them as BcMAF1 and BcMAF2. Multiple sequences alignment revealed that both BcMAF1 and BcMAF2 contained a typical MADS domain at the N-terminus (Figure 1A). Phylogenetic analysis of BcMAF1 and BcMAF2 with Arabidopsis MAFs were constructed (Figure 1B), which indicated that BcMAF1 and BcMAF2 showed greater similarity to AtMAF1, AtMAF2, and AtMAF3.
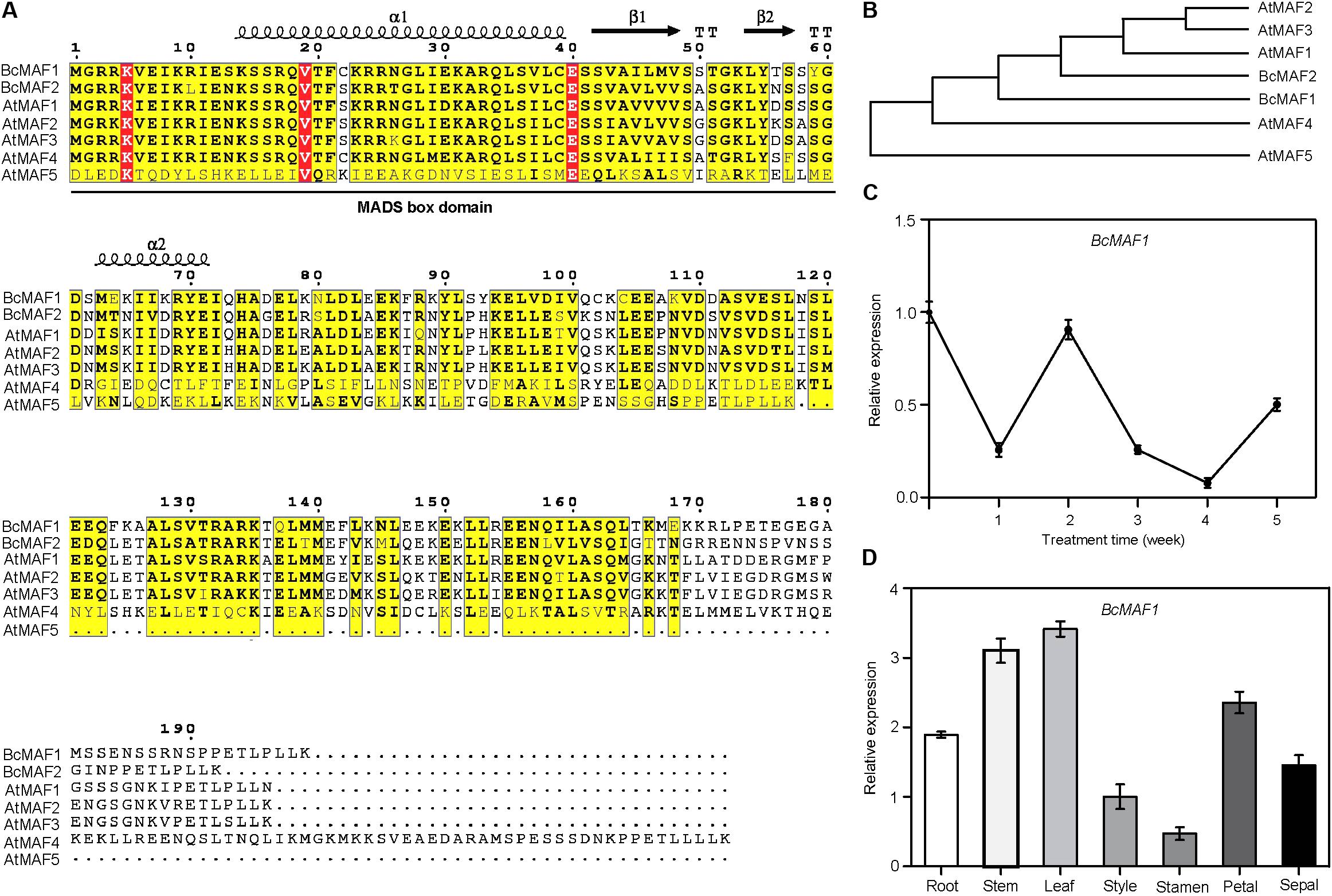
FIGURE 1. BcMAF1 is a short-term cold response gene encoding an MADS protein. (A) Sequence alignment of BcMAF1, BcMAF2, and AtMAFs. Conserved and similar residues are boxed with red and yellow, respectively. The MADS-box domain is indicated by a straight line. (B) The phylogenic tree of BcMAF1, BcMAF2, and AtMAFs. The gene accession numbers are as follows: AtMAF1 (AT1G77080), AtMAF2 (AT5G65050), AtMAF3 (AT5G65060), AtMAF4 (AT5G65070), AtMAF1 (AT5G65080), BcMAF1 (MG964044), and BcMAF2 (MG964045). (C) The expression of BcMAF1 during the process of vernalization in Pak-choi by qPCR. (D) The expression of BcMAF1 in different tissues of Pak-choi by qPCR. Data shown are means ± SEM of three independent experiments.
To determine whether the BcMAF1 or BcMAF2 expression was a response to vernalization in Pak-choi, we performed qPCR using total RNA from leaves of Pak-choi cultivar Wuyueman plants under cold treatment. Specifically, in the time-course analysis of treated samples, BcMAF1 expression in leaves decreased after 1 week of cold treatment and reverted to its original value after 2 weeks, which was more rapid than the expression of BcMAF2 (Figure 1C and Supplementary Figure S1). The abundance of BcMAF1 transcript strongly decreased at 3 weeks, reached the minimum value at 4 weeks, and then increased. The results indicated that BcMAF1 responded to vernalization and might play a more important role than BcMAF2 in premature flowering under short-term cold conditions. We further analyzed the expression level of BcMAF1 in different Pak-choi tissues. The highest BcMAF1 transcript was found in leaves, followed by stems, petals, roots, sepals, styles, and stamens (Figure 1D).
Subcellular Localization of BcMAF1 Protein
We detected the subcellular localization of 35S:BcMAF1-GFP fusion (Figure 2A) by transiently overexpressing it in tobacco leaves using the Agrobacterium infiltration methodology. GFP fluorescence of the 35S:BcMAF1-GFP fusion protein was only observed in the nucleus, which was also confirmed by DAPI staining. However, the fluorescence of 35S:GFP was observed in both nucleus and cytoplasm (Figure 2B). This strongly suggested that BcMAF1 was targeted to the nucleus and might act as a transcription factor.
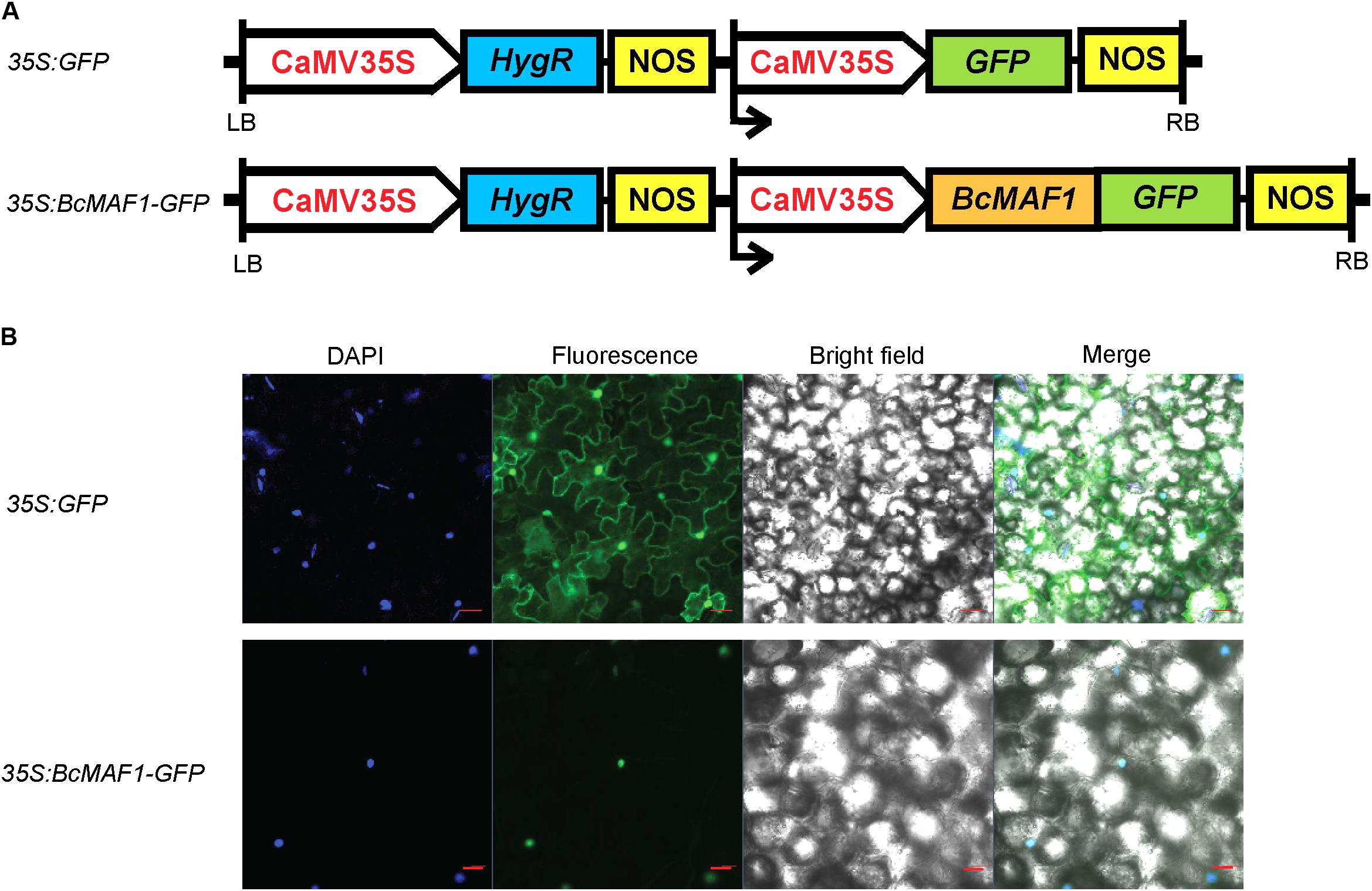
FIGURE 2. Subcellular localization of BcMAF1 protein. (A) 35S:GFP and 35S:BcMAF1-GFP construct. (B) Transient expression of 35S:GFP (Scale bars = 50 μm) and 35S:BcMAF1-GFP fusion protein (Scale bars = 20 μm) in tobacco leaves.
Overexpression of BcMAF1 in Transgenic Arabidopsis Caused Late Flowering
We first overexpressed BcMAF1 in Arabidopsis Col-0 background to investigate its roles in flowering regulation. The PCR products of 597 bp (BcMAF1 coding sequence without termination codon) were amplified in six T3 transgenic lines (#1, #4, #8, #11, #16, and #23), indicating that BcMAF1 had been transformed and expressed in these six lines (Supplementary Figure S2A). The BcMAF1-GFP fusion protein of approximately 50 kDa was detected in four lines (#8, #11, #16, and #23), with the band of #11 weak (Supplementary Figure S2B). Meanwhile, the GFP fluorescence of three T3 lines (#8, #16, and #23) seedlings was also observed (Supplementary Figure S2C). These three T3 lines were used for further characterization. Transgenic plants that overexpressed BcMAF1 showed markedly late flowering than control plants (Figure 3A). An increase in rosette leaves numbers (until the time of bolting) was observed in the three T3 lines (Figure 3B). Flowering time (days to opening of the first flower) was also delayed in the transgenic lines compared to the control line (Figure 3C). These results suggested that BcMAF1 might function as a floral repressor.
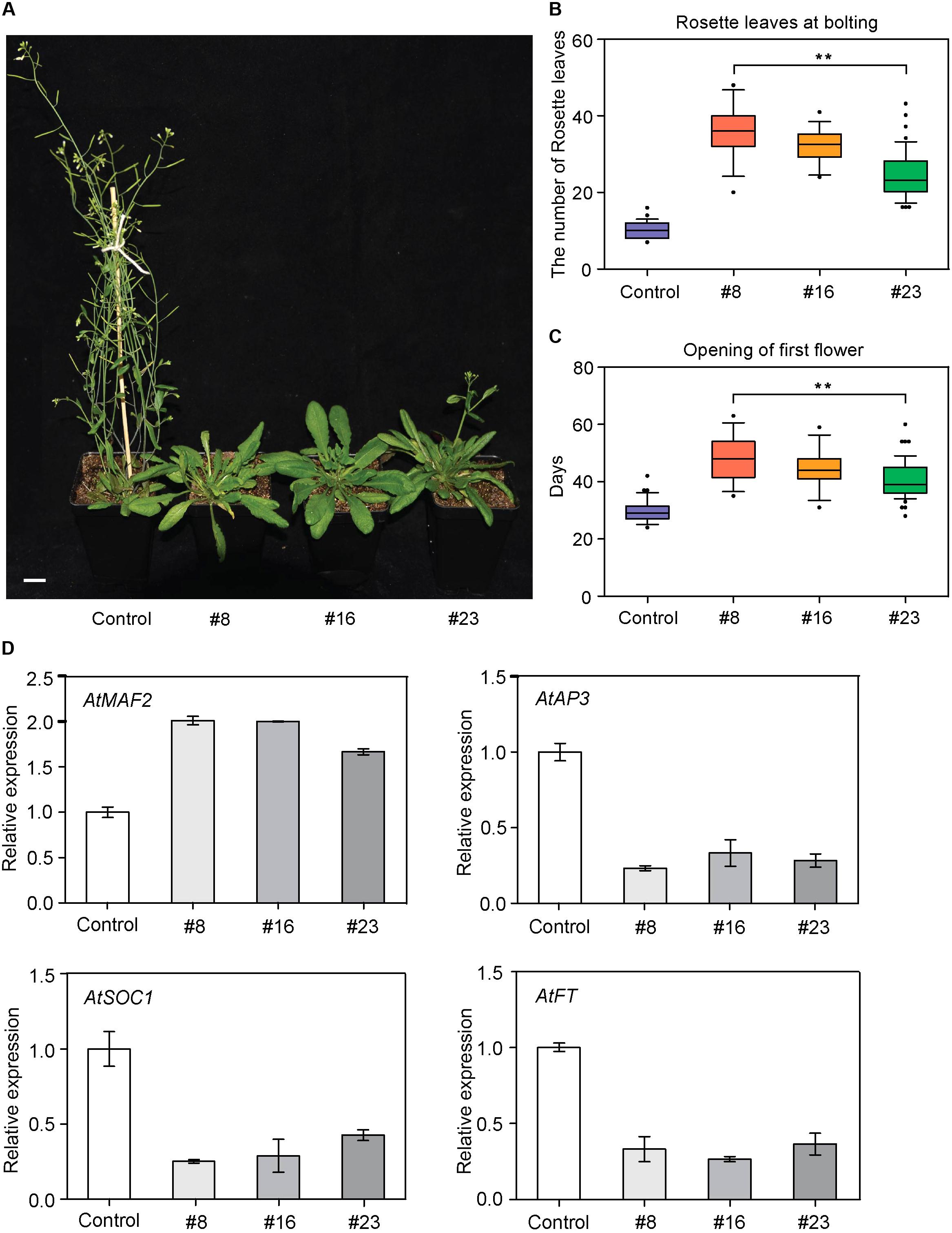
FIGURE 3. Overexpression of BcMAF1 in Arabidopsis. (A) Late flowering phenotype of the transgenic plants overexpressed BcMAF1. The 35S:GFP and 35S:BcMAF1-GFP#8, #16, and #23 plants in a chamber (LD, 16 h light/8 h dark photoperiod at 22°C/18°C). Scale bars = 1.5 cm. Rosette leaf number at bolting (B) Days of opening of first flower (C) in the 35S:GFP and 35S:BcMAF1-GFP#8, #16, and #23 plants. Error bars represent standard deviation of the mean number of 30 plants for each line. ∗∗ indicate significant differences from the control (P < 0.01). (D) Expression analysis of predicted downstream genes in the 35S:GFP and 35S:BcMAF1-GFP#8, #16, and #23 plants.
To elucidate the molecular mechanism of BcMAF1 in contributing to flowering repression, the expressions of four important genes involved in flowering were investigated. As shown in Figure 3D, the transcript level of AtAP3, which promoted flowering and was involved in specifying petal and stamen identities, decreased to less than 33% of the control in the transgenic lines. The transcript level of an important flowering repressor, AtMAF2, was also detected. The expression of AtMAF2 showed an increase in the two-fold transcript of the control in transgenic lines. AtSOC1 and AtFT (genes that promote flowering) expressions were approximately 57% lower in transgenic lines than in the control line. These results suggested that late flowering caused by overexpressing BcMAF1 might be due to the inhibition of AtAP3, AtSOC1, and AtFT, and the activation of AtMAF2 expression.
Virus-Induced BcMAF1 Silencing Caused Early Flowering
To further functionally characterize the role of BcMAF1 in Pak-choi flowering regulation, we silenced BcMAF1 using TYMV-based VIGS. Three weeks after Pak-choi plants underwent particle gun bombardment, the photobleaching or mosaic leaf phenotype typical of Phytoene desaturase (PDS) deficiency or TYMV was observed on the upper leaves of pTY-BcPDS, pTY-BcMAF1, or pTY (control) plants, which indicated that TYMV-mediated gene silencing was effective in Pak-choi. Total RNA was extracted from the upper leaves of the positive plants. To confirm the efficiency of silencing, the expression levels of BcMAF1 and BcPDS in the positive plants were evaluated with qPCR. BcMAF1 expression was significantly reduced by at least 62% in BcMAF1-silencing plants, whereas BcPDS expression was significantly reduced by approximately 52% in BcPDS-silencing plants (Figures 4A,B). As expected, silencing BcMAF1 accelerated bolting by 11–13 days (Supplementary Table S4) and promoted flowering (Figure 4C) in comparison to the control. We then detected the transcript levels of the predicted downstream genes. The levels of BcAP3, BcFT1, BcFT2, and BcSOC1 were higher whereas the level of BcMAF2 was lower in the BcMAF1-silencing plants than in the control plant (Figure 4D). This suggested that BcAP3, BcMAF2, BcFT1, BcFT2, and BcSOC1 might participate in BcMAF1-mediated flowering regulation.
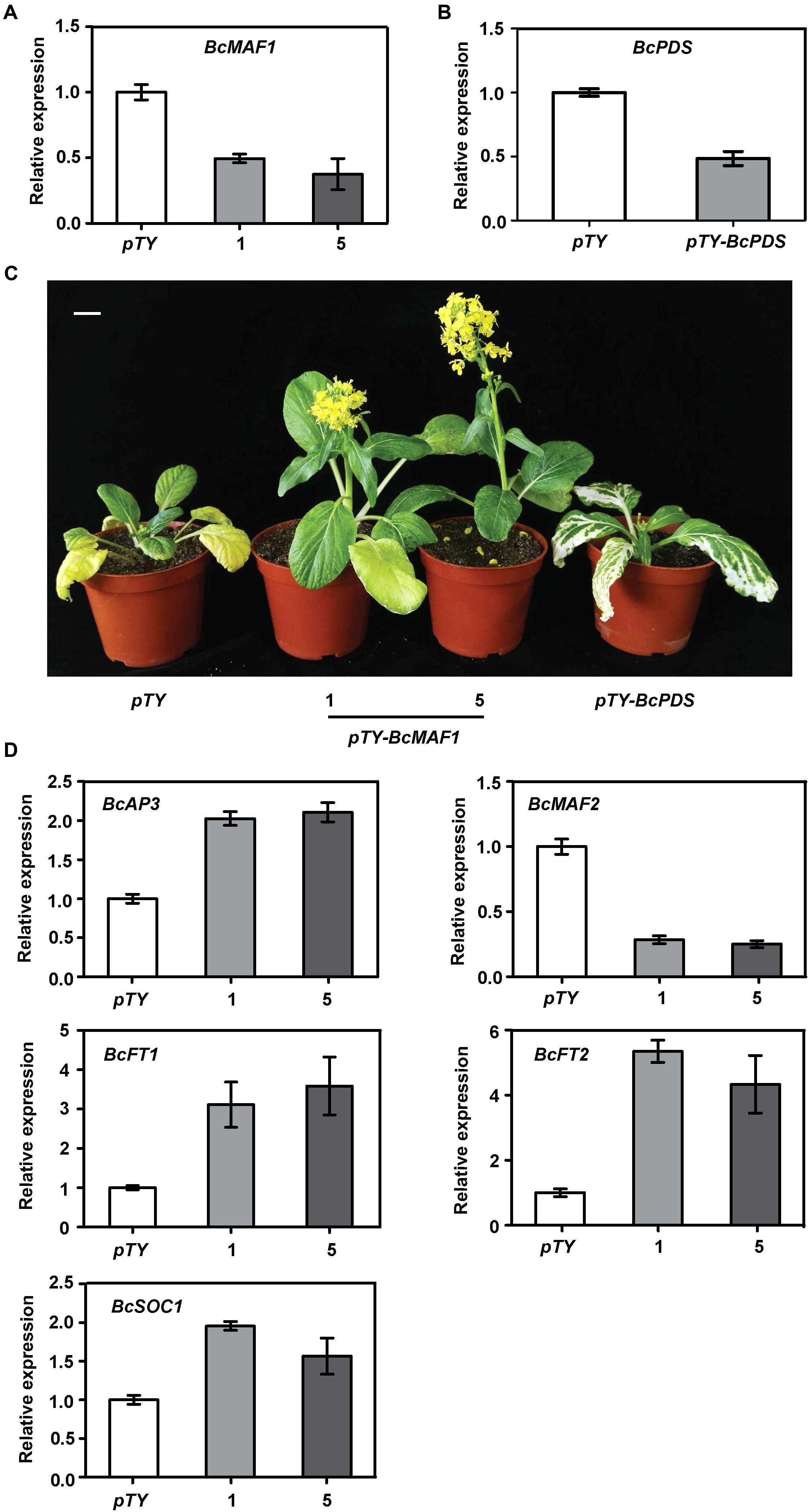
FIGURE 4. Silencing BcMAF1 in Pak-choi. (A) Expression analysis of BcMAF1 in Pak-choi seedlings bombarded with pTY and pTY-BcMAF1 plasmid. (B) Expression analysis of BcPDS in Pak-choi seedlings bombarded with pTY and pTY-BcPDS plasmid. (C) Early flowering phenotype in Pak-choi seedlings silenced BcMAF1. Scale bars = 2.5 cm. (D) Expression analysis of predicted downstream genes in the pTY, pTY-BcMAF1-1, and pTY-BcMAF1-5 plants.
Direct Binding of BcMAF1 to the Promoters of BcMAF2 and BcAP3
A previous study showed that SOC1 and FT were repressed by MAFs (Shen et al., 2014); therefore, we studied the relationships among BcMAF1, BcAP3, and BcMAF2. Some members of the MADS-box family can recognize and bind to the CArG box, a consensus sequence that has a core motif CC(A/T)6GG, in the promoters of their targets (Shore and Sharrocks, 1995). On account of BcMAF1 belonging to the MADS-box family, we speculated that BcMAF1 could bind to promoters owning the CArG box. Transcription regulatory elements analysis indicated that BcMAF2 and BcAP3 promoters possessed at least one CArG box (Supplementary Table S3). Meanwhile, the expression levels of AtMAF2 and AtAP3 were significantly changed in BcMAF1-overexpressing Arabidopsis (Figure 3D), whereas the expression levels of BcMAF2 and BcAP3 were significantly changed in the BcMAF1-silencing Pak-choi (Figure 4D). This suggested that BcMAF2 and BcAP3 might be the targets of BcMAF1 in Pak-choi. To test and verify this hypothesis, we performed the yeast one-hybrid assay. Yeast cells harboring the promoter fragments of BcAP3 or BcMAF2 together with pGADT7-BcMAF1 were all obtained on SD/-Leu medium containing 300 ng/mL AbA, except for the control (Figure 5). The results implied that the BcMAF1 protein had DNA binding activity and could directly bind to the promoters of BcMAF2 and BcAP3.
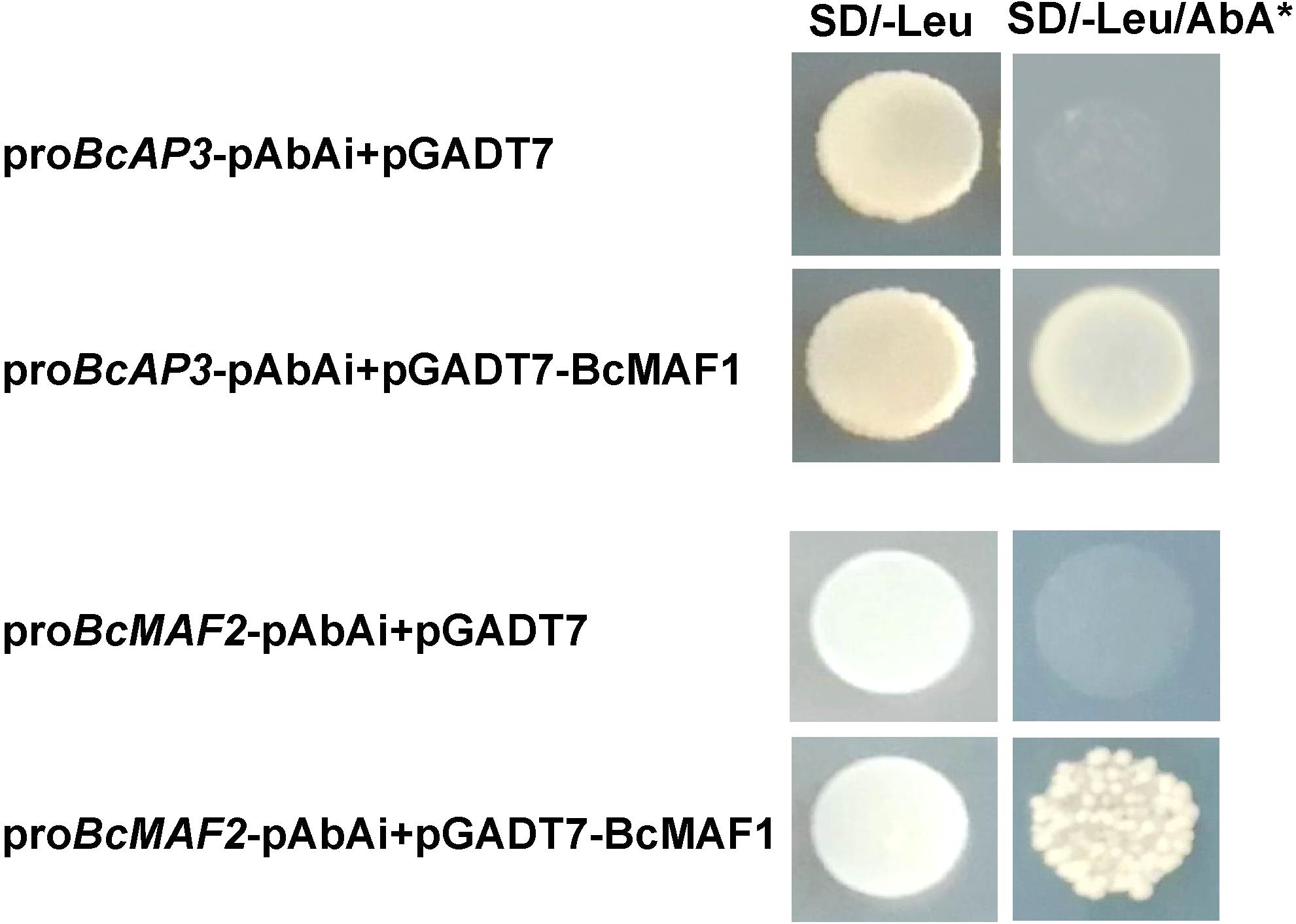
FIGURE 5. Binding activities of BcMAF1 protein with the promoters of BcMAF2 and BcAP3 detected by yeast one-hybrid assays. The yeast cells were grown on SD/-Leu medium plate supplemented with or without 300 ng/mL AbA.
To further confirm the binding activity of BcMAF1 with proBcMAF2 and proBcAP3, we conducted a transient expression experiment using the dual luciferase assay. When 35S:BcMAF1-GFP was co-transfected with proBcMAF2-LUC, the value of FLUC/RLUC was approximately 2-fold higher than the control. In contrast, the value of FLUC/RLUC was approximately 50% lower than the control when 35S:BcMAF1-GFP was co-transfected with proBcAP3-LUC. Together, our results proved that BcMAF2 was activated by BcMAF1 directly binding to its promoter, yet BcAP3 was inhibited by BcMAF1 (Figure 6).
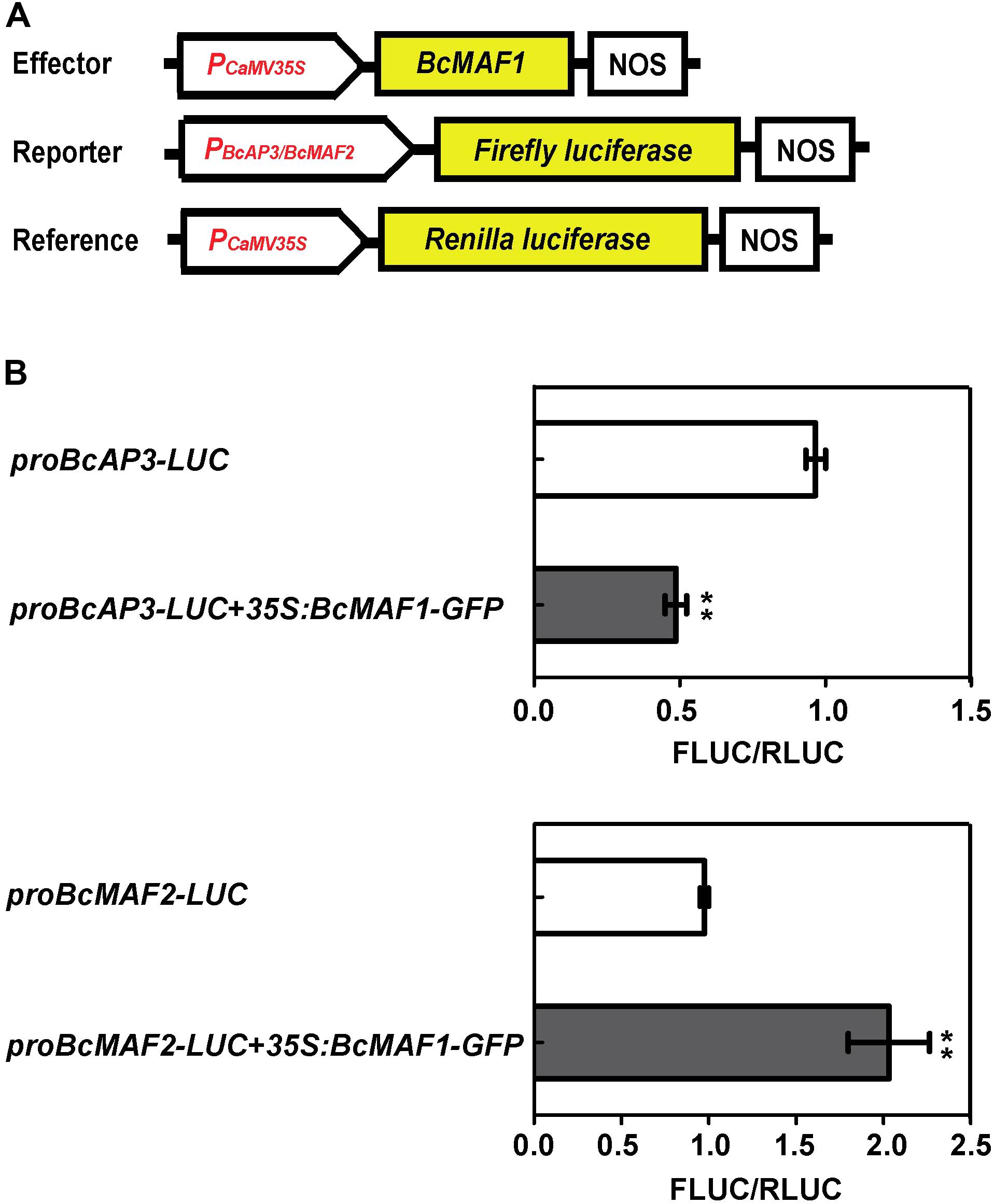
FIGURE 6. Activation or repression of BcMAF2 or BcAP3 promoter by overexpressing BcMAF1 in the dual luciferase transient assay. (A) Diagram of the effector, reporter, and reference plasmids used in the dual luciferase transient assay. (B) The relative FLUC/RLUC activities of BcMAF2 and BcAP3 promoters activated and repressed by recombinant 35S:BcMAF1-GFP in Arabidopsis protoplasts were detected by Dual-Luciferase® Reporter Assay System. Bars represent the mean ± SEM. The relative FLUC/RLUC activity of the negative control was set at 1. ∗∗ indicate a significant difference at P < 0.01.
Discussion
In Arabidopsis, there are five MAF proteins that are highly related to FLC (with 53–87% identity), which show temperature-dependent changes in expression (De Bodt et al., 2003). They also repress floral transition and their expressions are regulated by vernalization (Ratcliffe et al., 2003; Sung et al., 2006). FLC, MAF3, and the other three MAF genes (MAF1, MAF2, and MAF4) can directly interact with each other and form complexes to regulate the genes related to floral transition, such as FT (Gu et al., 2013). However, the molecular mechanisms of MAFs in flowering regulation under short-term cold conditions are still not clear, especially in Pak-choi. In the present study, we successfully isolated a MAF gene and named it as BcMAF1 from Pak-choi cultivar Wuyueman. We found that BcMAF1 was mainly expressed in leaves, stems, and petals, and responded to vernalization. The sharply up-regulated expression of BcMAF1 after 2 weeks of vernalization treatment suggested that BcMAF1 might function in preventing premature flowering. Thus, we further investigated BcMAF1 function by overexpressing it in Arabidopsis and silencing it in Pak-choi, as well as investigated the targets of BcMAF1 in Pak-choi.
Late flowering in BcMAF1-overexpressing Arabidopsis and early flowering in BcMAF1-silencing Pak-choi plants were found (Figure 3A, Figure 4C), which suggested that BcMAF1 could repress flowering. MADS-box transcription factors could bind to the CArG box and two CArG boxes in the BcAP3 promoter were also found (Supplementary Table S3). Additionally, BcMAF1 was highly expressed in petals, whereas AP3 was highly expressed in stamens and petals (Theissen et al., 2000). Therefore, we predicted that BcMAF1 might directly regulate BcAP3 to control the floral transition time, which was in agreement with the down-regulation of AtAP3 in transgenic Arabidopsis and up-regulation of BcAP3 in BcMAF1-silencing Pak-choi (Figures 3D, 4D). Yeast one-hybrid and dual luciferase assays showed that BcMAF1 could directly bind to the BcAP3 promoter (Figures 5, 6).
Floral organ identity is controlled by the combinatorial activity of five classes of floral homeotic genes in Arabidopsis, according to the ABCDE model (Theissen and Saedler, 2001). AP3, a B class gene, is a transcription factor of the MADS-box family and regulates many plant developmental processes, such as floral development and flowering (Irish and Kramer, 1998; Theissen et al., 2000). Mutations in AP3 caused the conversions of petals to sepals and stamens to carpels in Arabidopsis (Bowman et al., 1989; Jack et al., 1992). Additionally, overexpressing poplar AP3 in tobacco plants showed an earlier flowering phenotype (An et al., 2011). AP3 had a positive feedback loop to maintain its own expression (Jack et al., 1992; Goto and Meyerowitz, 1994). AP3, PI, and AP1 could form a heterodimer and then bind to the CArG box in vitro (Honma and Goto, 2001). AP3 is also required for the regulation of female flower development by directly activating ETR1 in the cucumber (Sun et al., 2016). Therefore, we further detected AP1 and ETR1 expressions in the transgenic Arabidopsis and BcMAF1-silencing Pak-choi. AtAP1 and AtETR1 transcript levels were lower in transgenic Arabidopsis, whereas BcAP1-1, BcAP1-2, and BcETR1 transcript levels were higher in silencing Pak-choi (Supplementary Figure S3). These findings suggested that BcMAF1 could also reduce BcETR1, BcAP1-1, and BcAP1-2 by directly inhibiting BcAP3 to regulate not only flowering time but also floral development in Pak-choi. The possible role of BcMAF1 in floral development still requires further investigation.
Previous studies have shown that MAF2, which is encoded a floral repressor, can prevent premature vernalization under short periods of cold. The maf2 mutant shows flower earlier than wild type after short periods of cold; however, it retains a normal vernalization response. This protection process is likely to be independent of FLC because FLC expression is not significantly decreased after short periods of cold (Ratcliffe et al., 2003). Thus, we predicted that BcMAF2, a MAF2 homolog, might also play a key role in short-term cold conditions in Pak-choi, which was confirmed by qPCR (Supplementary Figure S1). The relationship between BcMAF1 and BcMAF2 was further investigated. Yeast one-hybrid and dual luciferase assays demonstrated that BcMAF2 might be another target of BcMAF1 (Figures 5, 6). This explained the up-regulation of AtMAF2 by overexpressing BcMAF1 in transgenic Arabidopsis and the down-regulation of BcMAF2 by silencing endogenous BcMAF1 in Pak-choi (Figures 3D, 4D). Thus, we theorized that BcMAF1 could prevent premature flowering by directly activating BcMAF2 in Pak-choi.
We also examined the temporal expression pattern of BcMAF1 in Pak-choi under SD and LD conditions (Supplementary Figure S4). The abundance of BcMAF1 was low in light conditions and increased 8 h before dusk, with a peak in transcript at dusk under SDs. However, the abundance of BcMAF1 was not significantly changed under LD conditions. Meanwhile, BcMAF1 expression was lower in Pak-choi plants grown under LD conditions than those grown under SD conditions, indicating that LDs may inhibit BcMAF1 transcript to promote flowering. Overall, BcMAF1 expression showed a circadian oscillation under SD conditions. Further research is required to study the role of BcMAF1 under SD conditions.
Taken together, BcMAF1 was a floral repressor and prevented premature flowering in Pak-choi. BcMAF1 acted as a nuclear transcription factor and regulated the expressions of BcAP3 and BcMAF2 by directly binding to their promoters. This is the first study on the functional analysis of the MAF gene in Pak-choi flowering regulation. This research will help further clarify the regulatory mechanisms of flowering not only in Pak-choi but in other Brassicaceae species as well.
Author Contributions
FH: Performed the experiments and wrote the paper. FH, TL, and XH: Manuscript revision and approval. XH: Contributed to the interpretation of the results and coordinated the study. All authors read and approved the final manuscript.
Funding
This work was supported by grants from Integrated Innovation Center of Industrial Technology System of Modern Agriculture (vegetables) of Jiangsu (SXGC[2017]273), the Fundamental Research Funds for the Central Universities (Y0201700179), and Major Program of National Key Research and Development of China (2017YFD0101803).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Isabelle Jupin for providing the plasmid pTY-S.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00290/full#supplementary-material
FIGURE S1 | The expression of BcMAF2 during the process of vernalization in Pak-choi by qPCR.
FIGURE S2 | Identification of T3 transgenic Arabidopsis plants. (A) PCR analysis of T3 transgenic Arabidopsis plants overexpressing empty vector (control) and BcMAF1 (#1, #4, #8, #11, #16, and #23). The amplified fragments were the BcMAF1 coding sequence without termination codon (597 bp). (B) Western blot detection of the BcMAF1-GFP fusion protein in the transgenic Arabidopsis plants. The fusion protein is approximately 50 kDa. The band of marker is 50 kDa. (C) GFP fluorescence observation of 35S:BcMAF1-GFP#8, #16, and #23 seedlings.
FIGURE S3 | Expression analysis of predicted downstream genes of BcAP3 in BcMAF1-overexpressing Arabidopsis (A) and BcMAF1-silencing Pak-choi (B).
FIGURE S4 | Changes in BcMAF1 transcript in response to LD and SD were monitored in Pak-choi by qPCR. Data shown are means ± SEM of three independent experiments.
TABLE S1 | Primers used in the paper.
TABLE S2 | Sequences of oligonucleotides used for VIGS.
TABLE S3 | Information of the promoters of BcMAF2 and BcAP3.
TABLE S4 | Days from sowing to the time of bolting in the silenced Pak-choi plants.
Footnotes
References
An, X., Ye, M., Wang, D., Wang, Z., Cao, G., Zheng, H., et al. (2011). Ectopic expression of a poplar APETALA3 -like gene in tobacco causes early flowering and fast growth. Biotechnol. Lett. 33, 1239–1247. doi: 10.1007/s10529-011-0545-4
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. doi: 10.1105/tpc.1.1.37
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
De Bodt, S., Raes, J., Van De Peer, Y., and Theissen, G. (2003). And then there were many: MADS goes genomic. Trends Plant Sci. 8, 475–483. doi: 10.1016/j.tplants.2003.09.006
Deng, W., Ying, H., Helliwell, C. A., Taylor, J. M., Peacock, W. J., and Dennis, E. S. (2011). FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 6680–6685. doi: 10.1073/pnas.1103175108
Duan, W., Song, X., Liu, T., Huang, Z., Ren, J., Hou, X., et al. (2015). Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol. Genet. Genomics 290, 239–255. doi: 10.1007/s00438-014-0912-7
Goto, K., and Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8:1548. doi: 10.1101/gad.8.13.1548
Gu, X., Le, C., Wang, Y., Li, Z., Jiang, D., Wang, Y., et al. (2013). Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4:1947. doi: 10.1038/ncomms2947
Hamada, H., Linghu, Q., Nagira, Y., Miki, R., Taoka, N., and Imai, R. (2017). An in planta biolistic method for stable wheat transformation. Sci. Rep. 7:11443. doi: 10.1038/s41598-017-11936-0
Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. doi: 10.1038/35054083
Huang, F., Tang, J., and Hou, X. (2016). Molecular cloning and characterization of BcCSP1, a Pak-choi (Brassica rapa ssp. Chinensis) cold shock protein gene highly co-expressed under ABA and cold stimulation. Acta Physiol. Plant. 38:47. doi: 10.1007/s11738-015-2058-6
Irish, V. F., and Kramer, E. M. (1998). Genetic and molecular analysis of angiosperm flower development. Adv. Bot. Res. 28, 197–230. doi: 10.1016/S0065-2296(08)60297-9
Jack, T., Brockman, L. L., and Meyerowitz, E. M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. doi: 10.1016/0092-8674(92)90144-2
Jie, W., Yao, W., Lei, W., Ma, F., Tong, W., Chen, W., et al. (2017). Overexpression of VpEIFP1, a novel F-box/Kelch-repeat protein from wild Chinese Vitis pseudoreticulata, confers higher tolerance to powdery mildew by inducing thioredoxin z proteolysis. Plant Sci. 263, 142–155. doi: 10.1016/j.plantsci.2017.07.004
Kang, S. G., Jin, J. B., Piao, H. L., Pih, K. T., Jang, H. J., Lim, J. H., et al. (1998). Molecular cloning of an Arabidopsis cDNA encoding a dynamin-like protein that is localized to plastids. Plant Mol. Biol. 38, 437–447. doi: 10.1023/A:1006099718761
Kim, S. Y., Park, B. S., Kwon, S. J., Kim, J., Lim, M. H., Park, Y. D., et al. (2007). Delayed flowering time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Cell Rep. 26, 327–336. doi: 10.1007/s00299-006-0243-1
Livak, K. J., and Schmittgen, T. D. (2012). Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔ C T method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lou, P., Zhao, J., Kim, J. S., Shen, S., Del Carpio, D. P., Song, X., et al. (2007). Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J. Exp. Bot. 58, 4005–4016. doi: 10.1093/jxb/erm255
Osnato, M., Castillejo, C., Matias-Hernandez, L., and Pelaz, S. (2012). TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 3:808. doi: 10.1038/ncomms1810
Pflieger, S., Blanchet, S., Camborde, L., Drugeon, G., Rousseau, A., Noizet, M., et al. (2008). Efficient virus-induced gene silencing in Arabidopsis using a ’one-step’ TYMV-derived vector. Plant J. 56, 678–690. doi: 10.1111/j.1365-313X.2008.03620.x
Ratcliffe, O. J., Kumimoto, R. W., Wong, B. J., and Riechmann, J. L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169. doi: 10.1105/tpc.009506
Ratcliffe, O. J., Nadzan, G. C., Reuber, T. L., and Riechmann, J. L. (2001). Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126, 122–132. doi: 10.1104/pp.126.1.122
Schranz, M. E., Quijada, P., Sung, S. B., Lukens, L., Amasino, R., and Osborn, T. C. (2002). Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162, 1457–1468.
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Krober, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., and Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U.S.A. 97, 3753–3758. doi: 10.1073/pnas.97.7.3753
Shen, L., Thong, Z., Gong, X., Shen, Q., Gan, Y., and Yu, H. (2014). The putative PRC1 RING-finger protein AtRING1A regulates flowering through repressing MADS AFFECTING FLOWERING genes in Arabidopsis. Development 141, 1303–1312. doi: 10.1242/dev.104513
Shore, P., and Sharrocks, A. D. (1995). The MADS-Box Family of Transcription Factors. Berlin: Springer. doi: 10.1007/978-3-642-85252-7_7
Simpson, G. G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289.
Sun, J. J., Li, F., Wang, D. H., Liu, X. F., Li, X., Liu, N., et al. (2016). CsAP3: a cucumber homolog to Arabidopsis APETALA3 with novel characteristics. Front. Plant Sci. 7:1181. doi: 10.3389/fpls.2016.01181
Sung, S., Schmitz, R. J., and Amasino, R. M. (2006). A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev. 20, 3244–3248. doi: 10.1101/gad.1493306
Theissen, G., Becker, A., Di, R. A., Kanno, A., Kim, J. T., Münster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. doi: 10.1023/A:1006332105728
Theissen, G., and Saedler, H. (2001). Plant biology. Floral quartets. Nature 409, 469–471. doi: 10.1038/35054172
Tian, C., Wan, P., Sun, S., Li, J., and Chen, M. (2004). Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54, 519–532. doi: 10.1023/B:PLAN.0000038256.89809.57
Wang, S., He, J., Cui, Z., and Li, S. (2007). Self-formed adaptor PCR: a simple and efficient method for chromosome walking. Appl. Environ. Microbiol. 73, 5048–5051. doi: 10.1128/AEM.02973-06
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Xiao, D., Zhao, J. J., Hou, X. L., Basnet, R. K., Carpio, D. P., Zhang, N. W., et al. (2013). The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks. J. Exp. Bot. 64, 4503–4516. doi: 10.1093/jxb/ert264
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, J. Y., Qiao, Y. S., Lv, D., Gao, Z. H., Qu, S. C., and Zhang, Z. (2012). Malus hupehensis NPR1 induces pathogenesis-related protein gene expression in transgenic tobacco. Plant Biol. 14, 46–56. doi: 10.1111/j.1438-8677.2011.00483.x
Zhao, J., Shi, L., Jiang, C., Liu, J., Wu, B., Long, Y., et al. (2009). The evolution of Brassica napus FLOWERING LOCUST paralogues in the context of inverted chromosomal duplication blocks. BMC Evol. Biol. 9:271. doi: 10.1186/1471-2148-9-271
Keywords: BcAP3, BcMAF1, BcMAF2, late flowering, Pak-choi (Brassica rapa ssp. Chinensis)
Citation: Huang F, Liu T and Hou X (2018) Isolation and Functional Characterization of a Floral Repressor, BcMAF1, From Pak-choi (Brassica rapa ssp. Chinensis). Front. Plant Sci. 9:290. doi: 10.3389/fpls.2018.00290
Received: 30 November 2017; Accepted: 19 February 2018;
Published: 06 March 2018.
Edited by:
Stefan de Folter, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional (CINVESTAV-IPN), MexicoCopyright © 2018 Huang, Liu and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xilin Hou, aHhsQG5qYXUuZWR1LmNu
 Feiyi Huang
Feiyi Huang Tongkun Liu
Tongkun Liu Xilin Hou
Xilin Hou