- 1Rice Research Institute, Sichuan Agricultural University, Sichuan, China
- 2Agriculture and Food, Commonwealth Scientific and Industrial Research Organization (CSIRO), Canberra, ACT, Australia
Lesion mimic mutants display spontaneous cell death, and thus are valuable for understanding the molecular mechanism of cell death and disease resistance. Although a lot of such mutants have been characterized in rice, the relationship between lesion formation and abscisic acid (ABA) synthesis pathway is not reported. In the present study, we identified a rice mutant, lesion mimic mutant 9150 (lmm9150), exhibiting spontaneous cell death, pre-harvest sprouting, enhanced growth, and resistance to rice bacterial and blast diseases. Cell death in the mutant was accompanied with excessive accumulation of H2O2. Enhanced disease resistance was associated with cell death and upregulation of defense-related genes. Map-based cloning identified a G-to-A point mutation resulting in a D-to-N substitution at the amino acid position 110 of OsABA2 (LOC_Os03g59610) in lmm9150. Knock-out of OsABA2 through CRISPR/Cas9 led to phenotypes similar to those of lmm9150. Consistent with the function of OsABA2 in ABA biosynthesis, ABA level in the lmm9150 mutant was significantly reduced. Moreover, exogenous application of ABA could rescue all the mutant phenotypes of lmm9150. Taken together, our data linked ABA deficiency to cell death and provided insight into the role of ABA in rice disease resistance.
Introduction
Abscisic acid (ABA) is one of the multi-functional phytohormones that is involved in many essential physiological processes during growth and development in plants, such as seed maturation, seed desiccation, seed dormancy, germination, and stress-induced responses (Rock and Quatrano, 1995; Leung and Giraudat, 1998; Maia et al., 2014). In rice, ABA modulates seed dormancy mainly through the balance of ABA and GA (ABA/GA ratio) (Liu et al., 2010, 2014; Shu et al., 2013). In different plant–pathogen interaction systems, it is well-established that ABA negatively regulates plant defense. On one hand, ABA-deficient mutants, such as abscisic acid2-1 (aba2-1) and aba3-1 in Arabidopsis, sitiens (sit) in tomato, display enhanced resistance to Golovinomyces cichoracearum and Erwinia chrysanthemi, respectively (Asselbergh et al., 2008; Xiao et al., 2017). Conversely, ABA-increased mutants in Arabidopsis become more susceptible (Gao et al., 2016). On the other hand, exogenous application of ABA increases susceptibility to different pathogens in rice, Arabidopsis and tomato (Asselbergh et al., 2008; Xu et al., 2013; Xiao et al., 2017). ABA-treatment on rice plants leads to enhanced disease symptoms on both susceptible and resistant accessions (Koga et al., 2004; Jiang et al., 2010). However, to our knowledge there is no report of an ABA-deficient mutant associated with disease resistance in rice.
The biosynthesis of ABA has two pathways: (1) the direct pathway via mevalonate pathway in Fungi (Hirai et al., 2000; Inomata et al., 2004) and (2) the indirect pathway via carotenoid pathway that is the main biosynthesis pathway in higher plants. The indirect pathway contains three stages (Milborrow, 2001; Schwartz et al., 2003). First, zeaxanthin is synthesized by zeaxanthin epoxidase (ZEP) in plastids (Marin et al., 1996; Agrawal et al., 2001). Second, zeaxanthin is converted into 9-cis-violaxanthin and 9-cis-neoxanthin by neoxanthin synthase that is encoded by ATABA4 (North et al., 2007). They are dissociated to xanthoxin (Xan) by 9-cis-epoxycarotenoid dioxygenase (NCED) (Iuchi et al., 2001; Riahi et al., 2013; Zhang et al., 2014). Third, Xan is transferred to the cytosol and converted to abscisyl aldehyde (ABAld) by xanthoxin dehydrogenase (XanDH) (Miguel et al., 2002; Endo et al., 2014). Then, ABAld is catalyzed into ABA by Abscisic Aldehyde Oxidase (AAO) (Seo et al., 2000).
XanDH belongs to a short-chain dehydrogenase/reductase (SDR) superfamily that is encoded by ABA2. AtABA2 is a single copy gene in Arabidopsis thaliana and is constitutively expressed. Expression of AtABA2 activates the glucose signal, antagonizes the ethylene signal and promotes the synthesis of ABA (Cheng et al., 2002; Miguel et al., 2002). ZmABA2 is the homolog of AtABA2 in maize. ZmABA2 interacts with ZmMPK5 and coordinately regulates the ABA level in maize (Ma et al., 2016). OsABA2, a rice ABA2 homolog, could restore the Arabidopsis ataba2 mutant phenotypes (Endo et al., 2014), indicating the conservation of ABA biosynthesis function of OsABA2. However, the phenotypic changes in rice resulted from mutation in OsABA2 has not been described.
Previous studies have described mutants that have disrupted functions of genes in ABA biosynthetic pathway in different plants, such as sitiens (sit) and notalilis (not) in tomato (Burbidge et al., 1999; Okamoto et al., 2002), aba1, aba2, and aba3 in Arabidopsis (Léon-Kloosterziel et al., 1996; Schwartz et al., 1997a; Xiong et al., 2002) and pre-harvest sprouting (phs) and viviparous (vp) mutants (vp10, vp13, and vp14) in maize (Tan and Mccarty, 1997; Schwartz et al., 1997b, 2003). The grains of those phs mutants easily germinate in the ear or panicle before harvest under wet conditions. In rice, the osaba1 mutant has a mutation in the gene encoding zeaxanthin epoxidase that involves in ABA synthesis in rice (Agrawal et al., 2001). The genes of phytoene desaturase (OsPSD), ζ-carotene desaturase (OsZDS), carotenoid isomerase (OsCRTISO), and lycopene β-cyclase (β-OsLCY) encode essential enzymes in different steps of the carotenoid synthetic pathway and the disruption of carotenoid biosynthesis leads to PHS trait (Fang et al., 2008). How other genes involve in ABA biosynthesis modulate pre-harvest sprouting remains to be tested.
The lesion mimic mutants (LMMs) display cell death, similar to hypersensitive response (HR), and are highly valuable in the investigation of the molecular mechanism of programmed cell death and defense responses (Greenberg, 1997; Moeder and Yoshioka, 2008; Tamiru et al., 2016). A number of LMMs have been isolated and characterized in Arabidopsis (Cao et al., 1994), maize (Hu et al., 1998), barley (Persson et al., 2009), and rice (Kiyosawa, 1970; Liu et al., 2017). In rice, more than 15 LMM-related genes have been isolated and characterized, which encode proteins of distinct functions, such as OsCUL3a that is the prominent component of cullin 3-based RING E3 ubiquitin ligases (Liu et al., 2017), an AAA-type ATPase (Fekih et al., 2015; Zhu et al., 2016), an eEF1A-like protein (Wang et al., 2017), a mitogen-activated protein kinase kinase kinase (Wang S. et al., 2015), a heat stress transcription factor protein (Utako et al., 2002), a cytochrome P450 monooxygenase (Tadashi et al., 2010), an U-box/armadillo repeat protein (Zeng et al., 2004), a clathrin-associated adaptor protein comlex1 (Qiao et al., 2009), a splicing factor 3b subunit 3 protein (Chen et al., 2012), and a thylakoid-bound protein (Li et al., 2010). Therefore, the molecular mechanism of lesion mimic formation in plant is regulated by a complicated regulatory network. Although a variety of LMM-related genes have been identified, whether ABA synthesis pathway is involved in lesion formation and how ABA modulates disease response in rice remains to be answered.
Here, a new rice lesion mimic mutant 9150, named lmm9150, was isolated and characterized. The lmm9150 mutant exhibited spontaneous cell death on leaves, pre-harvest sprouting under field condition, and enhanced growth of leaves and stems. The lesions in lmm9150 were associated with typical defense responses, such as accumulation of H2O2 and enhanced resistance to bacterial blight and rice blast diseases. Using a map-based cloning approach, we detected a point mutation in LOC_Os03g59610, which encodes OsABA2, a short-chain alcohol dehydrogenase. Knocking-out of OsABA2 with CRISPR/Cas9 led to lesion mimic spots, enhanced disease resistance and growth, and PHS traits. Consistence with the function of OsABA2 in ABA synthesis, the lesion mimic spots and other phenotypes of lmm9150 could be rescued by exogenous application of ABA. Therefore, our results provide further insights into the molecular function of OsABA2 in lesion mimic formation, disease-resistance, seed dormancy, and development in rice.
Materials and Methods
Plant Materials and Growth Conditions
The lmm9150 mutant was generated by ethyl methane sulfonate (EMS) treatment of a Chinese indica cultivar Yixiang1B, which is one of elite backbone parents in hybrid rice breeding programs in China. The wild type (WT) Yixiang1B and lmm9150 were grown, respectively, in the paddy field in Chengdu city (N30.67°E104.06°), Sichuan Province and in Lingshui county (N18.47°E110.04°), Hainan Province, China.
Histochemical Analysis
Leaves from the lmm9150 mutant, with obvious lesion mimic spots, at the seedling stage and the WT at the same growth stage were collected for histochemical analysis. Trypan blue staining was performed to detect cell death as previously described (Zhu et al., 2016). In brief, samples were submerged in lactic acid-phenol-trypan blue solution (2.5 mg/ml trypan blue, 25% (w/v) lactic acid, 23% water-saturated phenol, and 25% glycerol in H2O) and kept at room temperature for 6–12 h, followed by de-staining with solution containing 30% (w/v) chloral hydrate for 5 days. Then, the samples were equilibrated with 50% glycerol for 1 day for taking photos. For detection of hydrogen peroxide (H2O2) accumulation, the 3,3′-diaminobenzidine (DAB) staining was performed as described previously (Zhu et al., 2016).
Chlorophyll Content Measurement
Leaves were soaked in 20 ml of 95% alcohol for 48 h in dark, until the leaves became colorless. The absorbance values were examined by a spectrophotometer at 665 and 649 nm. Then, the content of chlorophylls was calculated as follows:
Chlorophyll (Chl) a absorbance value = 13.95 × OD665-6.88 × OD649
Chlorophyll (Chl) b absorbance value = 24.96 × OD649-7.32 × OD665
Chlorophyll content (mg/g) = (C × V × D)/1000 × W
C: chl a or chl b absorbance value; V: volume of extracting solution; D: dilution index; W: weight of sample.
Chlorophyll content was measured in three biological repeats. Statistical analysis was performed using Student’s t-test.
Inoculation of Pathogens and Disease Resistance Assay
The resistance assay to bacterial blight disease was performed as described previously. In brief, 70-days-old (at tillering stage) plants of the mutant lmm9150 and the WT were used for inoculation of the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo). The Xoo strains, P6, P3, 8248, and X004, which are compatible with the WT, were used for inoculation. Xoo bacterial suspensions with 0.5 of OD600 were used to inoculate by using the scissors-dip method (Zuo et al., 2014). Disease lesion lengths were measured at 15 days post-inoculation (dpi).
For resistance assay to rice blast disease, 60-days-old (at tillering stage) plants of the lmm9150 mutant and the WT were used for inoculation with the fungal pathogen Magnaporthe oryzae (M. oryzae). The M. oryzae strains, ZhongI, and Tetep, which are compatible with the WT, were used for inoculation following a previous report (Park et al., 2012). The disease lesions were observed at 5 dpi.
DNA Extraction and PCR, RNA Extraction, and qRT-PCR
Genomic DNA was extracted from leaves using the cetyltrimethylammonium bromide (CTAB) method (Murray et al., 1980). The PCR mixture was mixed with 2 μl DNA (10–50 ng/μl), 2 μl primers (10 μmol/μl), 0.3 μl dNTPs (10 mmol/L), 0.2 μl Taq (5 U/μl), and 13.5 μl H2O. The running procedure of PCR was performed as the following: pre-denaturation at 95°C for 5 min followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 1 min, with a final extension at 72°C for 10 min. The PCR products were separated by electrophoresis in a 3% agarose gels, stained with ethidium bromide (EB) and photographed.
Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, United States) following the procedures of the manufacturer. The mRNA was digested with DNase I according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, United States) and was subjected to reverse transcription to synthesize first-stand cDNA. Oligo (dT) was used as primer and SuperScript II (Invitrogen, Carlsbad, CA, United States) was used as reverse transcription enzyme.
The qRT-PCR was performed using a Bio-Rad CFX96 Real-Time System coupled to a C1000 Thermal Cycler (Bio-Rad, Hercules, CA, United States). The housekeeping gene Ubiquitin5 (Ubq5) was used as the internal control. The sequences of the primers were listed in Supplementary Table S1.
Genetic Analysis and Map-Based Cloning
Two F1 and two F2 populations derived from the crosses of Yixiang1B × lmm9150 and lmm9150 × Yixiang1B were used for genetic analysis. The F2 population derived from the cross of 02428 ×lmm9150 was used for mapping of the mutant gene. For the bulk segregation analysis (BSA) (Michelmore et al., 1991), equal amount of leaf blades from 10 F2 plants with the lesion mimic phenotype and 10 F2 plants with the WT phenotype were collected for DNA extraction to construct the mutant and the WT DNA pools, respectively. The physical linkage map was then constructed using molecular markers nearby the lmm9150 gene.
The SSR primers were synthesized according to the information of Gramene database1. InDel markers were developed based on the alignment results of the reference 93–11, an indica rice2 and the Nipponbare, a japonica rice2 genome sequence at the candidate region. Primers were designed using Primer3 web version4.0.03. The specificity of each primer in the rice genome was confirmed by BLAST4 and PCR analysis. The sequences of SSR and InDel markers were listed in Supplementary Table S2.
For analysis of the PCR products, amplified products were separated by electrophoresis at 3.0% agarose gel in 0.5 × TBE buffer, and visualized and photographed under UV light.
For whole-genome re-sequencing, the lmm9150 was backcrossed with the WT and self-crossed to generate BC1F2 population. The equal total DNA of 20 BC1F2 plants with lesion mimic spots, was mixed and re-sequenced at Novogene Corporation (Beijing, China). At the same time, the WT genome DNA was also sequenced as control.
Sequence Analysis
Gene prediction was performed using the Rice Genome Annotation Project database5. Sequence alignments were performed using the software DNAMAN Version 6.0. Alignments of amino acid were performed using the software Clustax2.
Construction of Knockout Lines by CRISPR/Cas9
Vector construction was performed to knockout the candidate gene as previously described (Ma et al., 2015). In brief, two targets (Supplementary Table S3), named lmm9150-Y and lmm9150-B, were designed in the coding sequence of conservative region. The two target sequences were amplified by PCR. Using endonuclease, Eco311 and T4 ligase, the lmm9150-Y was inserted into the pBWA (V) H-cas9i2 and the lmm9150-B was inserted into the pBWD (LB) DNAi. Finally, Pbwa (V) H-cas9i2-lmm9150-Y and pBWD (LB) DNAi-lmm9150-B were assembled into the final vector pBWA (V) H-cas9i2-lmm9150 using endonuclease SapI and T4 ligase. The constructs were verified by sequencing and then introduced into the WT by Agrobacterium-mediated transformation as described previously (Jeon et al., 2000). The sequences near the editing region were verified by extracting genomic DNA from T1 transgenic plants (sequencing primer in Supplementary Table S3).
ABA, GA, and Water Loss Assay
Determination of ABA and GA content was carried out using enzyme-linked immunosorbent assay (ELISA) method as previously described (Teng et al., 2006). To test the water-loss rate, seedlings were grown in a paddy field and leaves (300 mg from 10 seedlings) of 40-days-old seedlings were kept on dish at room temperature (28°C). The weight of samples was measured at 20 min interval until 120 min.
Germination Assay
Seeds from the lmm9150 mutant and the WT at 30 days after flowering were used for germination test. Seeds were cultured in a growth chamber at 30°C,14 h light and 24°C,10 h dark cycle for 8 days.
Results
Isolation and Characterization of the lmm9150 Mutant
To explore the molecular connection of lesion mimic and disease resistance in rice, we carried out an extensive forward genetic screening for lesion mimic mutants from an EMS-mutagenized population of indica cultivar Yixiang1B. Subsequently, more than 20 mutants were identified. One of these mutants, named lmm9150, was chosen for further characterization because of its multiple phenotypes in development. The 3-week-old seedlings of the mutant lmm9150 begun to develop brown lesions on the top part of the third leaf in the paddy field in both Chengdu (N: 30.67°) and Hainan (N: 18.47°) (Figure 1A). At tillering stage, the apical part of older leaves also displayed lesion mimic spots (Supplementary Figure S1A). At mature stage, the spots emerged to the top part of flag leaf (Supplementary Figure S1B). In addition, lmm9150 mutant displayed obvious pre-harvest sprouting at 30 days after flowering under natural condition (Figures 1B,C).
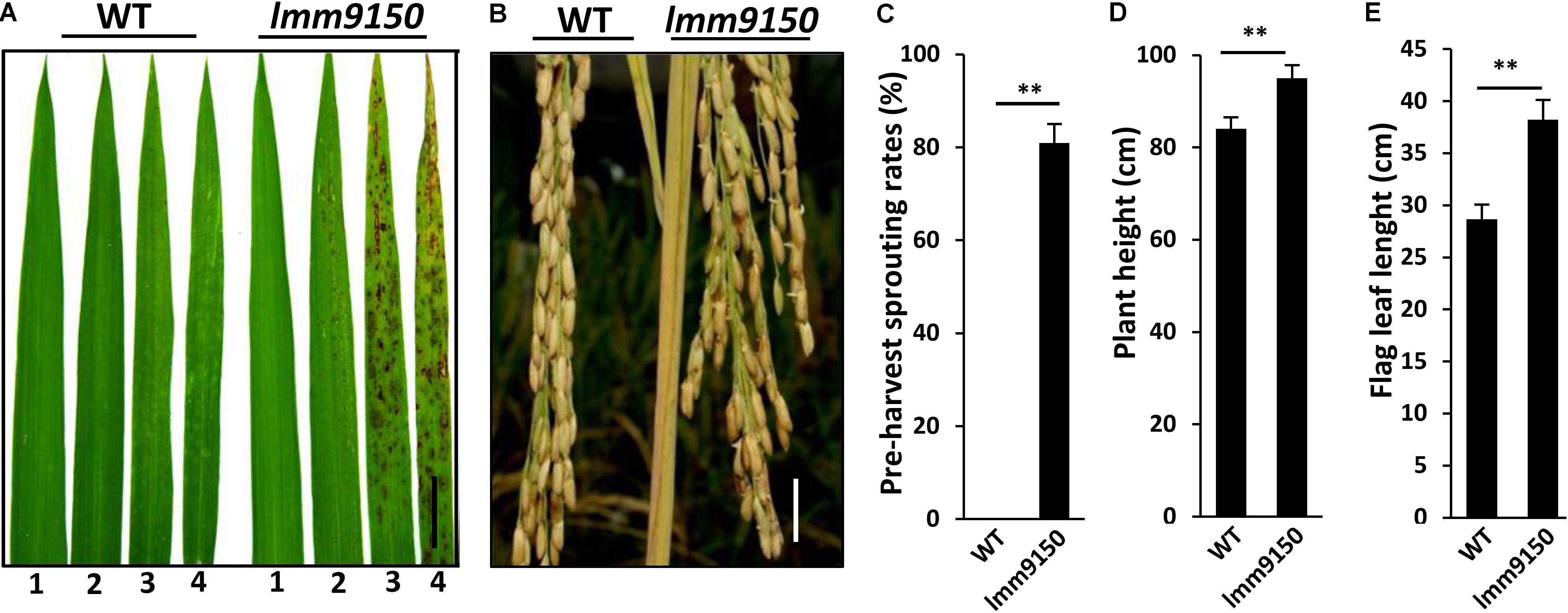
FIGURE 1. Phenotypic characterizations of lmm9150 mutant. (A) Comparison of four leaves between wild type (WT) and lmm9150 mutant shows the lesion mimic spots of lmm9150 at seedling stage. The numbers 1, 2, 3, and 4 represent the first top leaf, the second top leaf, the third top leaf, and the fourth top leaf, respectively. (B) The lmm9150 displays pre-harvest sprouting at yellow mature stage in the paddy field. (C–E) Quantification analyses on pre-harvest sprouting rates (C), plant height (D), and flag leaf length (E), respectively. Data were obtained from 10 main panicles of plants (C), 10 plants (D), and 10 flag leaves of main panicles (E). Statistical analysis was performed using Student’s t-test, ∗∗ indicates P < 0.01. Scale bar: 2 cm in (A,B).
However, apart from plant height and the flag leaf of lmm9150 that were significantly larger than WT (Figures 1D,E), the yield-related agronomic traits, including the number of tillers, seed-setting rate, grain weight per plant, and 1000-grains weight, had no significant difference from those of WT (Supplementary Figures S1C–F).
To characterize the lesions in lmm9150, we performed a trypan blue staining assay to examine cell death. Compared with WT, a lot of blue spots were observed on the leaf of lmm9150 after staining (Figure 2B), indicating existence of dead cells. Because cell death is often associated with the production of reactive oxygen species (ROS) such as H2O2, we further examined the accumulation of H2O2 in lmm9150 by DAB staining assay. Our data indicated that DAB-stained was observed in the mutant but not WT leaf blade, whereas there were rarely observed in the WT (Figure 2C).
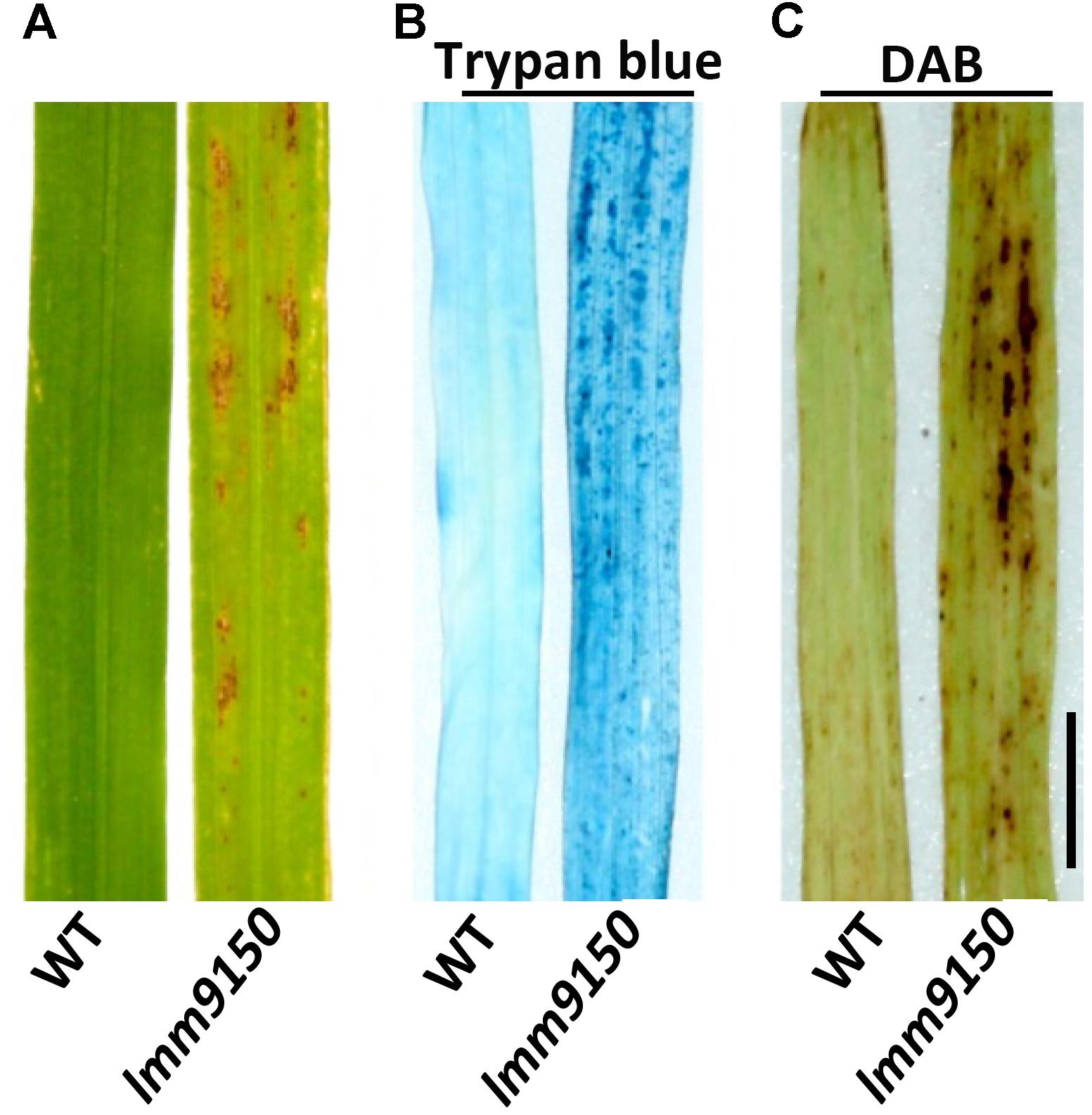
FIGURE 2. Observation of cell death in wild type (WT) and lmm9150 mutant. (A–C) Representative leaf sections of WT and lmm9150 mutant (MT) show lesions (A), dead cells revealed by trypan blue staining (B), and H2O2 accumulation revealed by 3,3′-diaminobenzidine (DAB) staining (C) in the mutant, respectively. The lmm9150 mutant and WT leaf samples were collected at 30 days after sowing. Scale bar: 1 cm.
Next, we examined the content of chlorophylls. Small pieces from cognate positions of flag leaves of WT and lmm9150 at flowering stage were collected for measurement of chlorophylls. The results showed that the content of both chlorophyll a and chlorophyll b in lmm9150 was significantly lower than that of WT (Figures 3A,B), implying that the formation of lesion mimic spots might attributable to the decrease of photosynthesis pigments in the lmm9150 mutant.
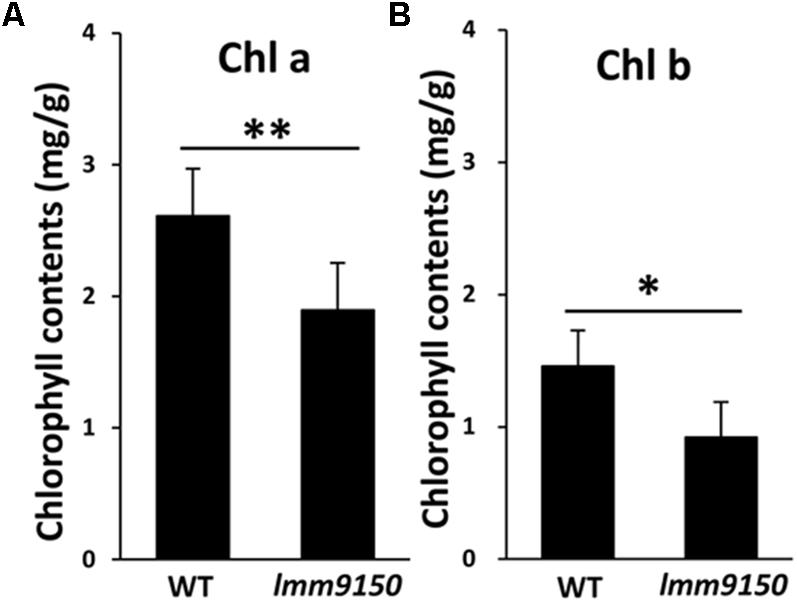
FIGURE 3. Comparison of the chlorophylls content between wild type (WT) and lmm9150 mutant. (A,B) The content of Chlorophyll a (chl a) and chlorophyll b (chl b) shows reduction in lmm9150 in comparison with WT. Mean and SD were obtained from three measurements. Statistical analysis was performed using Student’s t-test, ∗ and ∗∗ represents P < 0.05 and P < 0.01, respectively.
In summary, this mutant began to exhibit lesions as red brown spots from the top part of the third leaf at seedling stage, which eventually extended to the flag leaf at mature stage. Cell death was detected in the mutant, which was accompanied with excessive accumulation of H2O2. In addition, the mutant also exhibited higher plant height, longer flag leaf, and pre-harvest sprouting of seeds at mature stage.
Enhanced Resistance to Bacterial Blight and Rice Blast Pathogens by lmm9150
Many lesion mimic mutants exhibit enhanced defense responses in plants (Wang J. et al., 2015; Zhu et al., 2016). We speculated that the lmm9150 mutant may also become resistant to diseases in rice. To confirm this speculation, we first analyzed the expression of Pathogenesis-Related (PR) genes using quantitative reverse-transcription PCR (qRT-PCR), including PR1a, PR1b, PR10 (Nahar et al., 2011; Sang et al., 2011). The results demonstrated that all the examined PR genes were significantly upregulated in lmm9150 (Figure 4A), implying that lmm9150 was a potential auto-immune mutant. Then, we tested disease resistance of lmm9150 by inoculating Xanthomonas oryzae pv. oryzae (Xoo) and Magnaporthe oryzae at tillering stage, respectively. The results showed that the inoculated lmm9150 leaves exhibited disease lesions obviously shorter than that of WT leaves at 15 days after inoculation of Xoo (Figures 4B–E and Supplementary Figures S2A–C), indicating enhanced resistance. Quantification of the lesion lengths on leaves revealed that the average disease lesion length in lmm9150 was significantly shorter than that of WT. Intriguingly, the lesions on flag leaves were also significantly longer than those on the second leaves in lmm9150 (Figure 4C and Supplementary Figure S2D). In addition, lesions caused by Magnaporthe oryzae on the lmm9150 leaves were remarkably smaller than those of WT leaves (Figure 4D and Supplementary Figure S2E), indicating enhanced resistance to rice blast disease.
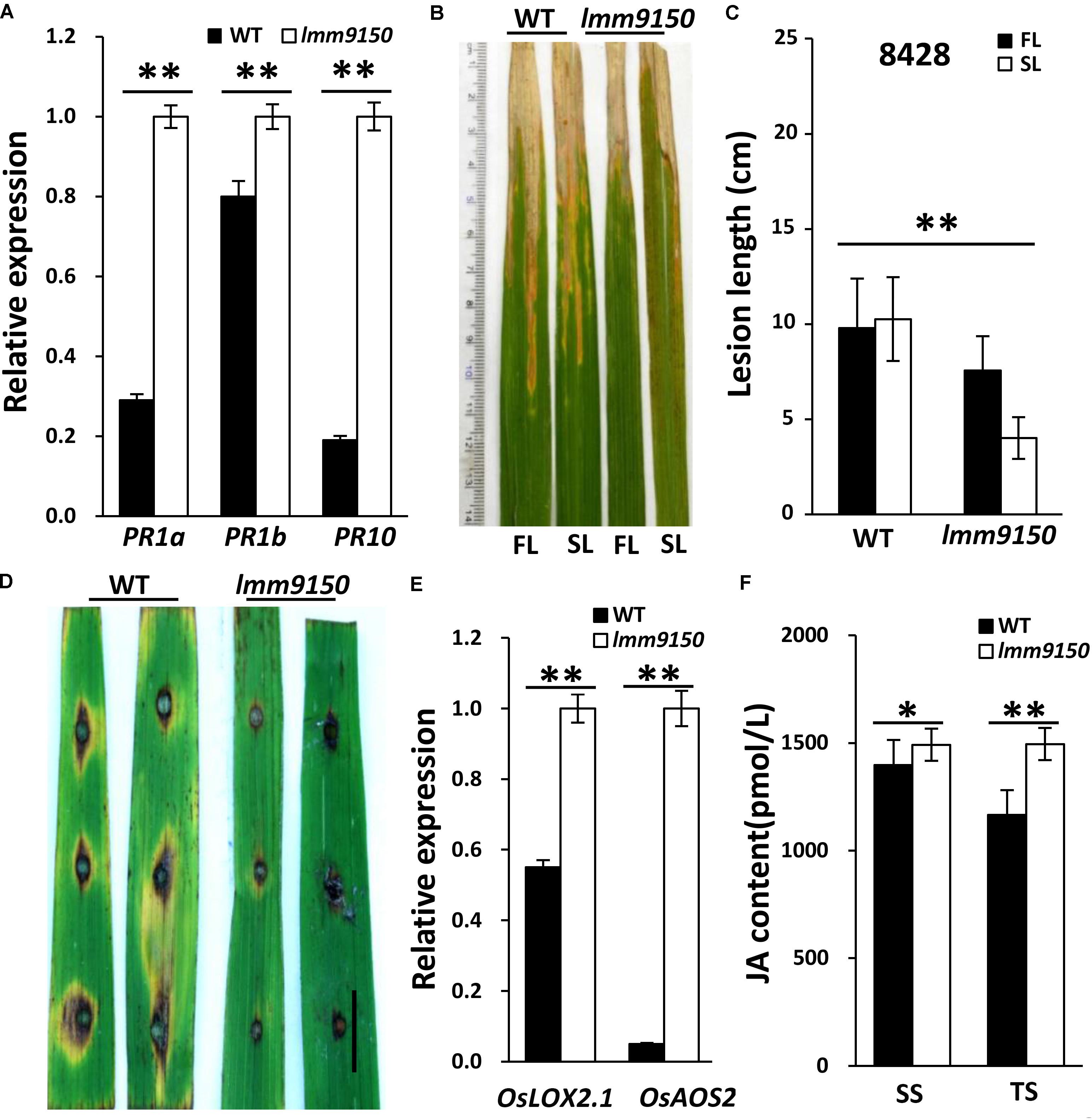
FIGURE 4. Determination of wild type (WT) and lmm9150 plants resistance to bacterial blight and rice blast diseases. (A) Quantitative reverse-transcription PCR (qRT-PCR) data show the relative expression levels of pathogenesis-related (PR) genes in lmm9150 and WT. Mean and SD were obtained from three measurements. (B,C) The disease lesion leaves of WT and lmm9150 mutant were inoculated with Xanthomonas oryzae pv. oryzae (Xoo) strain, 8428, at 15 days post-inoculation. The image (C) shows that the disease lesions of WT are longer than that of lmm9150 and the flag leaf disease lesions of lmm9150 are longer than that of the second leaf of mutant. FL: the flag leaf; SL: the second leaf. Data were obtained from 20 leaves of main panicles. (D) The disease lesions of leaves of WT and lmm9150 mutant were inoculated 5 days post-inoculation with Magnaporthe oryzae (M. oryzae) strain, ZhongI. The experiments were repeated twice with similar results. (E) Quantitative reverse-transcription PCR (qRT-PCR) data show the relative expression levels of jasmonic acid (JA) biosynthesis-related genes in lmm9150 and WT. (F) Comparison of JA content in leaf of WT and lmm9150 mutant. The content of hormone was tested by enzyme-linked immunosorbent assay (ELISA) method. Mean and SD were obtained from three measurements (E,F). The housekeeping gene Ubiquitin5 (Ubq5) was used as control. Statistical analysis was performed using Student’s t-test, ∗ and ∗∗ indicate P < 0.05 and P < 0.01, respectively. Scale bar: 1 cm in (D).
To explore the possible hormone pathway of defense responses activated in lmm9150, we examined the expression of jasmonic acid (JA) and salicylic acid (SA)-related genes. The lipoxygenase2.1 (OsLOX2.1) and allene oxide synthase2 (OsAOS2) are key genes of JA synthesis pathway (Liu et al., 2012). The phytoalexin deficient4 (OsPAD4) and lipase1 (OsEDS1) are key genes of SA signaling pathway (Pieterse et al., 2012). The results indicated that the expression of OsLOX2.1 and OsAOS2 was significantly higher in lmm9150 than that in WT (Figure 4E), indicating increase of JA synthesis. In contrast, the expression of OsPAD4 and OsEDS1 had marginal difference between lmm9150 and WT (Supplementary Figure S2F), implying that SA signaling pathway may not be activated in lmm9150. To address why SA signaling was not activated and confirm JA synthesis was increased, we examined SA level and JA level in leaf blades of WT and lmm9150 at seedling stage and tillering stage. The results showed that JA level of lmm9150 in leaf blades was significantly higher than that of WT at both stages, whereas SA level of lmm9150 in leaf blades had no difference from that of WT (Figure 4F and Supplementary Figure S2G). These data indicated that JA biosynthesis is increased and thus JA-signaling pathway may be associated with the defense responses activated in lmm9150.
Identification of lmm9150 as a Mutant of OsABA2
The two F1 and two F2 populations derived from the crosses of Yixiang1B (WT) ×lmm9150 and lmm9150 × Yixiang1B were used for the genetic analysis. We found that all F1 plants were not observed lesion mimic spots on leaf blades and the segregation between WT and lmm9150 in F2 population fitted 3:1 (Supplementary Table S4). Thus, lmm9150 lesion mimic phenotype was controlled by a single recessive nuclear gene.
To isolate the candidate gene that was responsible for the phenotypes of lmm9150, a mapping population was constructed by crossing lmm9150 with a japonica cultivar 02428. Bulked segregation analysis (BSA) found that eight molecular markers on the end of chromosome 3 co-segregated with the mutant phenotypes of lmm9150. Linkage analysis showed that the mutant gene was mapped to a 488-kb interval between the InDel marker I403.3 and the SSR marker RM3684, co-segregated with I403.2 (Figures 5A,B). Next, the whole-genome was re-sequenced by using the DNA sample bulked from 20 BC1F2 individuals with the lesion mimic phenotype. The genomic DNA of the WT was also sequenced as a control. A single base mutation, which SNP index was one, was found by comparing sequences between the bulked mutants and WT in this interval. Because this SNP localized in the second exon of LOC_Os03g59610, we sequenced the gene in lmm9150 and the WT. As expected, a G-to-A single base substitution was detected at the 1487th base in lmm9150, which resulted in a change from Aspartic acid (D) to Asparagine (N) at the 110th amino acid (Figure 5C and Supplementary Figure S3). These results indicated that lmm9150 was likely arisen from the single base substitution in the LOC_Os03g59610. This gene encodes XanDH, which is orthologous of ABA2 in Arabidopsis (Cheng et al., 2002; Miguel et al., 2002) and had been named OsABA2 in rice (Endo et al., 2014).
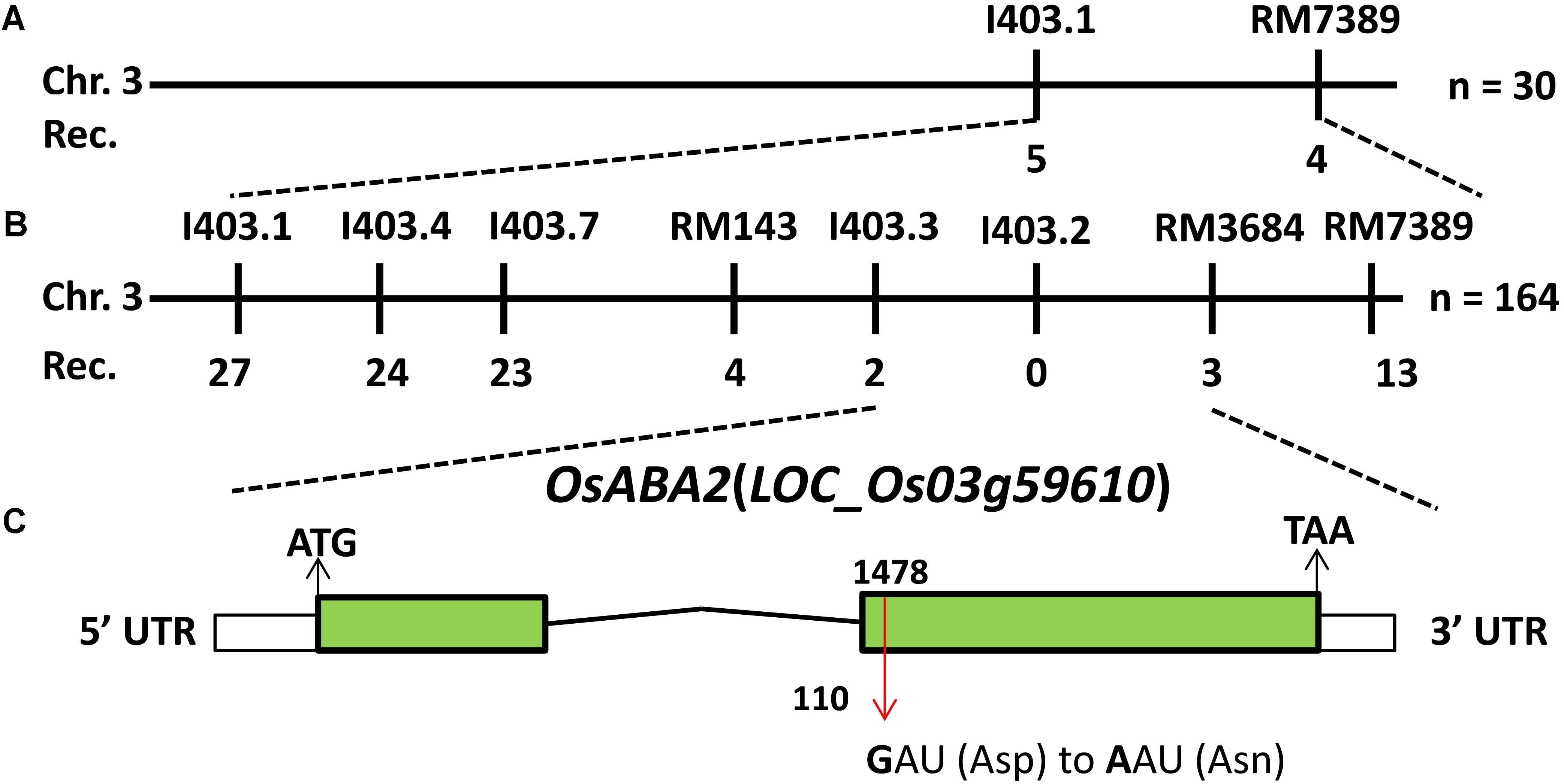
FIGURE 5. Positional cloning of OsABA2. (A) The OsABA2 gene is located on chromosome 3 between InDel marker I403.1 and SSR marker RM7389. (B) The OsABA2 gene is delimited to the region between I403.3 and RM3684 using 164 F2 mutant individuals. (C) The gene structure of OsABA2. Two exons and one intron are indicated by green rectangles and black lines, respectively; a G-to-A base substitution at the 1487th base was detected in the second exon (red arrow) leading to the change of Aspartic acid (Asp) to Asparagine (Asn) at the 110th amino acid.
Sequence alignment of LOC_Os03g59610 between Nipponbare and Yixiang1B revealed a 9-bp deletion in the first exon resulting in deletion of 3 amino acid residue in Yixiang1 B (GenBank accession number: MG334011), which is the WT of lmm9150 (Supplementary Figure S4).
To confirm that the phenotypes of lmm9150 were specifically associated with a mutation in OsABA2, we performed a knockout experiment in the WT background by using CRISPR/Cas9 editing. Five transgenic lines with either deletions or insertions in OsABA2 were obtained (Figure 6A and Supplementary Figure S5). All of these transgenic lines displayed lesion mimic spots on leaves (Figure 6B). Furthermore, gene-edited lines showed higher plant heights (Figure 6C), longer flag leaves (Figure 6D), pre-harvest sprouting phenotype (Figures 6E,F), and enhanced resistance to bacterial blight and rice blast diseases (Figures 6G–I and Supplementary Figures S6A–G). Taken together, these data confirmed that the loss-of-function of OsABA2 led to the lmm9150 phenotypes.
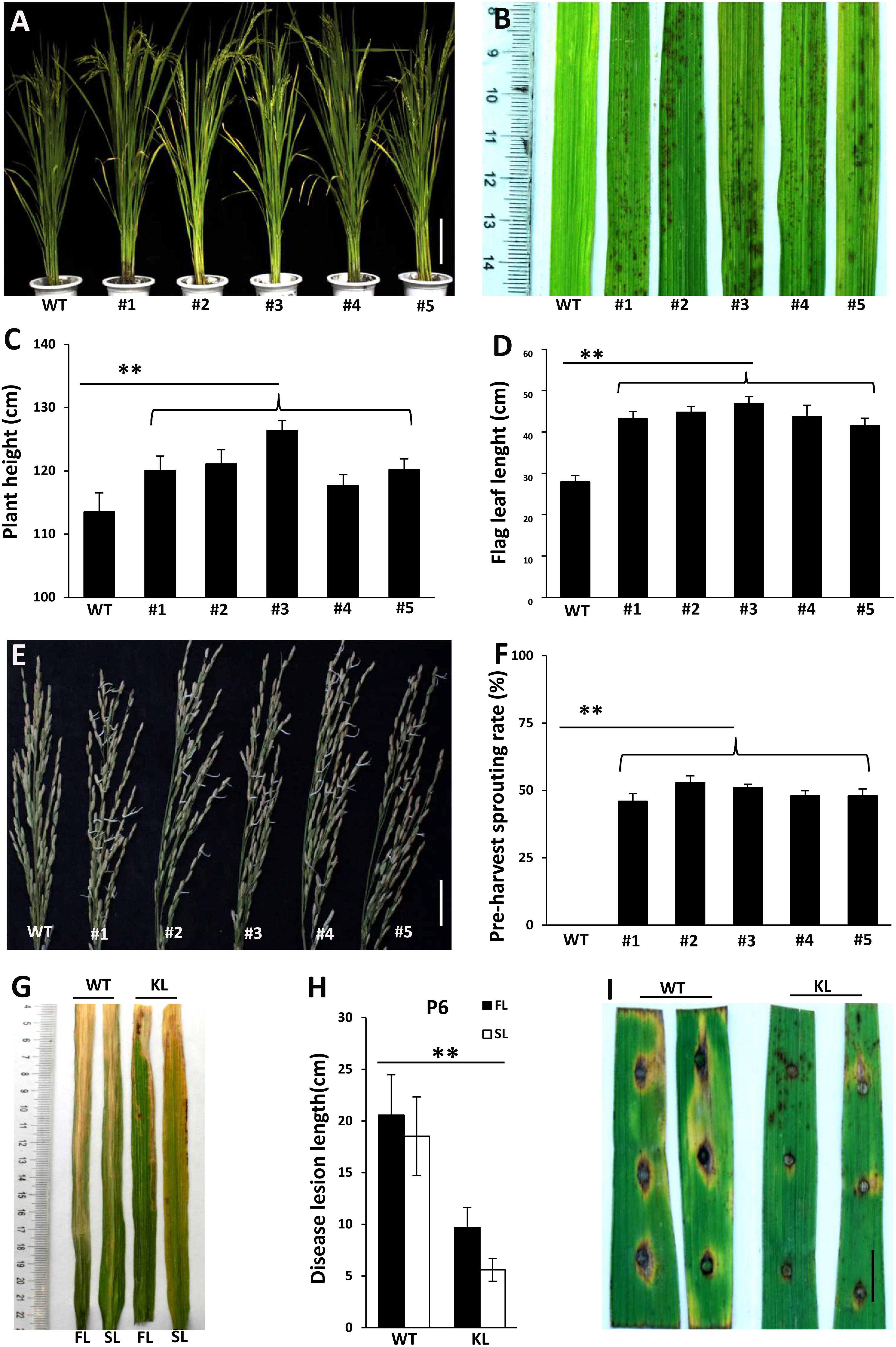
FIGURE 6. Phenotypic characterizations of T1 knockout lines. (A) The plant of the wild type (WT) and 5 T1 knockout lines. The mutations of #1, #3, #4, and #5 knockout lines are deletions. The mutation of #2 knockout line is insertion. (B) 5 knockout lines displays lesion mimic spots on leaves at tillering stage. (C) Comparison of plant height between WT and 5 knockout lines. (D) Comparison of flag leaf length between WT and 5 knockout lines. (E) The panicles for 30 days after flowering were collected and cultivated 3 days, at 28°C. All knockout lines display pre-harvest spouting. (F) Comparison of the pre-harvest sprouting rates between WT and 5 knockout lines 30 days after flowering under natural condition. (G,H) The disease lesion of leaves of WT and #3 knockout line were inoculated with Xanthomonas oryzae pv. oryzae (Xoo) strain, P6, at 15 days post-inoculation. KL: knockout lines; FL: the flag leaf; SL: the second leaf. Data were obtained from 10 plants (C), 10 flag leaves of main panicles (D), 15 main panicles (F), and 20 leaves of main panicles (H). (I) The disease lesions of leaves of WT and #3 knockout line was inoculated 5 days post-inoculation with Magnaporthe oryzae (M. oryzae) strain, ZhongI. The experiments were repeated twice with similar results. Scale bar: 15 cm in (A), 5 cm in (E), 1 cm in (I). Statistical analysis was performed using Student’s t-test, ∗∗ indicates P < 0.01.
The Function of OsABA2 in the Formation of Lesion Mimic Spots, Plant Development, and Seed Dormancy
OsABA2 encodes XanDH, an enzyme involves in ABA biosynthesis pathway (Endo et al., 2014). We hypothesized that mutation in this gene should influence the activity of XanDH and ABA level. To this end, we examined the activity of XanDH in leaf blades at three developmental stages. The results indicated that XanDH activity in lmm9150 was significantly lower than that in WT at all the tested stages (Figure 7A). Next, we examined the ABA level in leaf blades, seeds, and stems of lmm9150. Consistent with the lower activity of XanDH in lmm9150, ABA content in leaves, seeds, and stems were all significantly lower than that in the WT at all the tested developmental stages (Figures 7B–D). By contrast, the GA content in leaf blades, seeds and stems were all significantly higher than that of WT (Supplementary Figures S7A–C). Consistently, ABA/GA ratio was dramatically decreased in leaf blades, seeds and stems, respectively, which may explain why the seeds of lmm9150 showed lack of dormancy (Figures 7E,F and Supplementary Figures S7D,E). Taken together, we speculated that ABA deficiency likely resulted in the phenotypes of lmm9150.
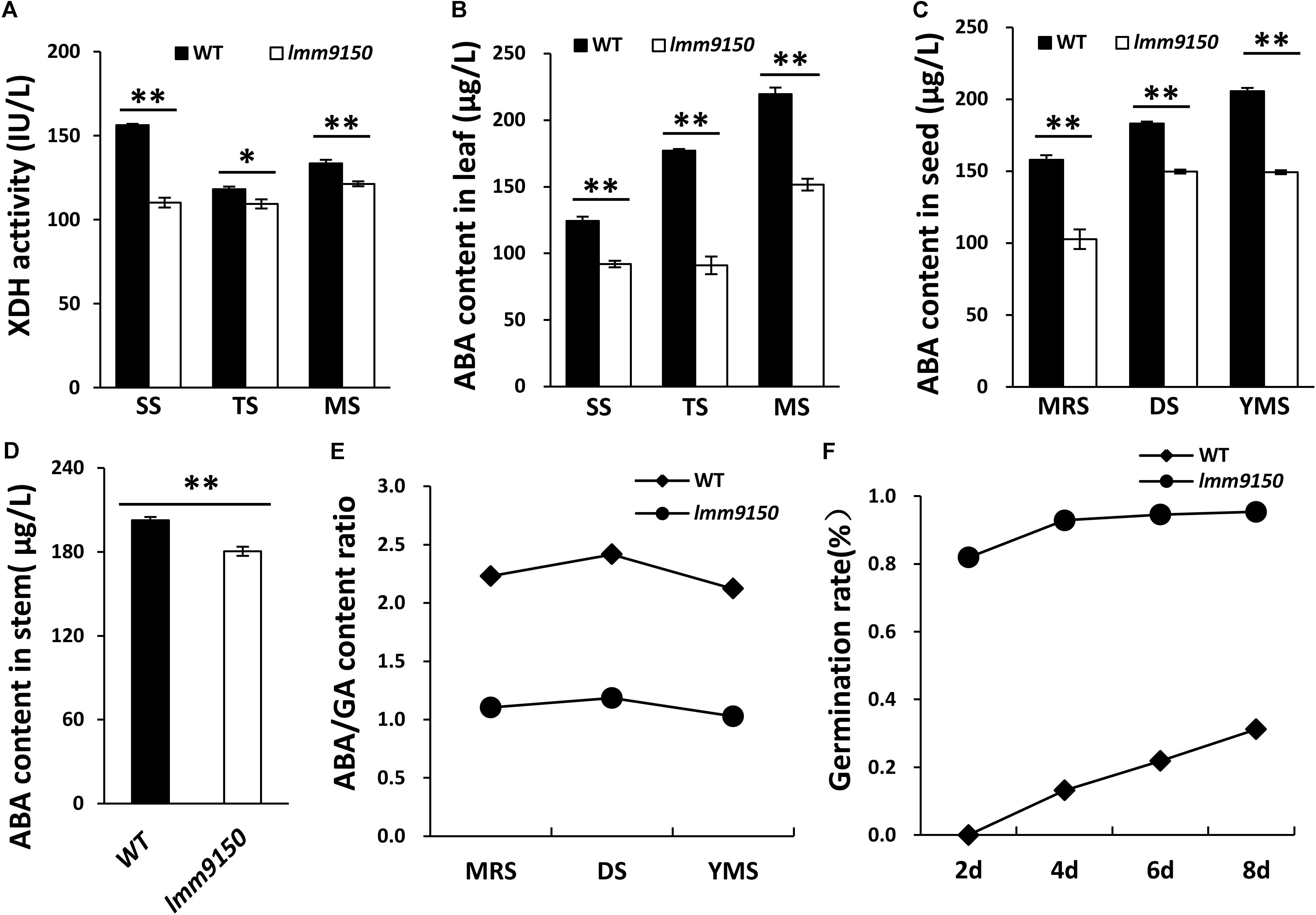
FIGURE 7. Comparison of xanthoxin dehydrogenase (XanDH) activity and abscisic acid (ABA) contents between wild type (WT) and lmm9150 mutant. (A) Changes of XanDH activity in the leaf of WT and the lmm9150 mutant. SS: seedling stage; TS: tillering stage; MS: mature stage. (B–D) Comparison of ABA contents in the leaves (B), seeds (C), and stems (D) between WT and the lmm9150 mutant at different developmental stages. Hormone content was tested using enzyme-linked immunosorbent assay (ELISA) method. MRS: milk ripe stage; DS: dough stage; YMS: yellow mature stage. (E) The ratio of ABA/GA content in seed of WT and the lmm9150 mutant 30 days after flowering. (F) The germination rate of lmm9150 mutant and WT 30 days after flowering. Mean and SD were obtained from three replicates. Statistical analysis was performed using Student’s t-test, ∗ and ∗∗ indicates P < 0.05 and P < 0.01, respectively.
Next, we applied ABA onto the seedlings of lmm9150 and WT. Such ABA-treatment led to three interesting changes of phenotypes in lmm9150. (1) A total of 20 days after treatment, the lmm9150 leaves in mock treatment displayed red brown spots from the older leaves (Figures 8A,B). In contrast, the lmm9150 leaves in ABA-treatment grew as healthy as that of the WT leaves (Figures 8C,D). (2) A total of 20 days after treatment, the plant height of lmm9150 was significantly higher than that of WT in mock treatment (Figures 8E,F). In contrast, the plant height of lmm9150 after ABA-treatment was similar to that of WT at seedling stage (Figures 8G,H). (3) The water loss rate of lmm9150 was obviously higher than that of WT plants at seedling stage (Supplementary Figure S8A). However, after ABA-treatment, the water loss rate of lmm9150 was similar to that of WT (Supplementary Figure S8B).
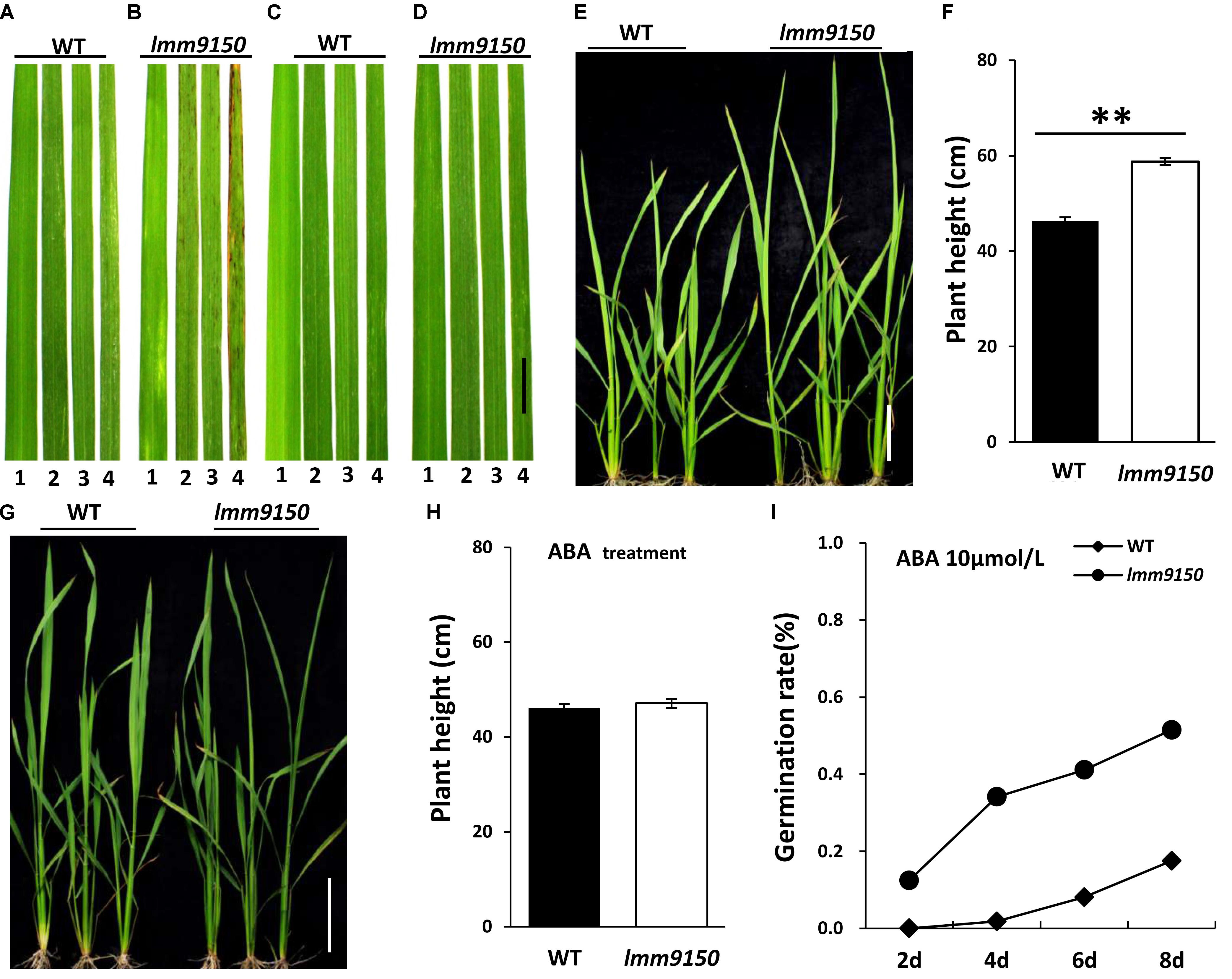
FIGURE 8. Analysis of relationship between abscisic acid (ABA) and plant development in lmm9150 mutant. (A–D) Phenotypic changes in the leaf blades of wild type (WT) and lmm9150 after ABA-treatment. The lmm9150 shows lesion mimic spots without ABA-treatment (A,B) and phenotype of the lmm9150 mutant is similar to WT when lmm9150 was treated with 100 μmol/L ABA for 15 days (C,D). The numbers 1–4 represent the first top leaf, the second top leaf, the third top leaf, and the fourth top leaf, respectively. (E–H) Change of plant height of lmm9150 mutant and WT 20 days after ABA-treatment at seedling stage. Images show that the plant height of lmm9150 and WT after water treatment (E,F) and ABA-treatment (G,H), respectively. The experiments were repeated twice with similar results (A–D,E,G). Data were obtained from 10 seedlings (F,H). (I) Comparison of seed germination rate in WT and lmm9150 mutant with 10 μmol/L ABA. Mean was obtained from three replicates. Statistical analysis was performed using Student’s t-test, ∗∗ indicate P < 0.01.
In addition, we tested the effect of different concentration of ABA on germination of seeds in ABA-containing media. The germination rate of lmm9150 seeds decreased when the concentration of ABA increased in a certain range, reflecting exogenous ABA could effectively inhibit seed germination (Figure 8I and Supplementary Figures S8C,D). Both the seeds of lmm9150 and WT could not germination in media containing 100 μmol/L ABA (Supplementary Figure S8E), indicating that this concentration may be the lethal dosage.
Therefore, all the examined phenotypes of lmm9150, such as lesion mimic spots, plant height, water loss rate, and seed germination, could be rescued through the exogenous application of ABA.
Discussion
Lesion mimic mutants are an ideal tool to investigate the association between PCD and defense responses in plants. In the present study, we isolated a rice lesion mimic mutant, lmm9150, from EMS-mutagenized indica cultivar Yixiang1B. The lmm9150 mutant showed ABA deficiency which likely resulted in some novel and some expected phenotypes, such as spontaneous cell death, pre-harvest sprouting and enhanced growth of stem and leaf (Figures 1, 2, 4). Map-based cloning and CRISPR/Cas9-aided knocking-out identified a point mutation at the LOC_Os03g59610 (OsABA2) locus encoding the xanthoxin dehydrogenase (XanDH) that catalyzes Xan into ABAld in the ABA biosynthesis pathway (Endo et al., 2014). Previously, OsABA2 is reported to be able to restore the ataba2 mutant in Arabidopsis (Endo et al., 2014), indicating that the function of ABA2 in ABA biosynthesis pathway may be conserved in monocot and dicot plants.
The lmm9150 mutant displayed spontaneous cell death on the third leaves starting from the seedling stage to the flag leaf at the yellow mature stage (Figure 1A and Supplementary Figures S1A,B), which was accompanied with excessive accumulation of H2O2 in the mutant (Figure 2C), similar to other rice lesion mimic mutants (Zhu et al., 2016; Liu et al., 2017; Wang et al., 2017). H2O2 is a major by-product of β-oxidation and acts as a signal molecule in the promotion of cell death (Ren et al., 2002; Zhang et al., 2003; Gechev et al., 2005). Thus, the elevated H2O2 in lmm9150 may have contributed to the formation of lesions. It has been reported that ABA suppresses PCD through increasing activities of ROS scavenging enzymes in barley (Bethke et al., 1999; Fath et al., 2001). In maize, loss of ABA synthesis, such as the ABA-deficient mutant viviparous9 (vp9), leads to the early onset of endosperm cell death (Young and Gallie, 2000). Here, we identified that lmm9150 was an ABA-deficient mutant (Figures 7B–D) and exhibited cell death lesions on leaves (Figure 1A and Supplementary Figures S1A,B). Exogenous application of ABA could inhibit the formation of lesions in lmm9150 (Figures 8A–D), confirming that deficiency in ABA biosynthesis can lead to formation of lesion mimics in rice. Nevertheless, the other rice ABA-deficient mutants, osaba1, phs1, phs2, phs3, and phs4, did not exhibit lesion mimic spots. H2O2 level of these mutants was not detected and the superoxide accumulation (O2-) significantly increased in phs3 mutant. It is unknown why these ABA-deficient mutants do not exhibit lesion mimic phenotypes in rice (Agrawal et al., 2001; Fang et al., 2008).
During the lesion mimic formation, disease resistance responses are often auto-activated in LMMs, leading to enhanced resistance to pathogens in rice (Zhu et al., 2016; Liu et al., 2017; Wang et al., 2017). Consistent with this, lmm9150 displayed enhanced resistance to both bacterial blight and rice blast diseases (Figure 4 and Supplementary Figure S2), similar to the enhanced disease resistance in the other ABA-deficient mutants, such as aba2-1, aba3-1 in Arabidopsis and sit in tomato (Xiao et al., 2017). Intriguingly, we noticed that the lesions of bacterial disease on flag leaves were longer than those on the second leaves in lmm9150 (Figure 4C and Supplementary Figure S2D). This could be due to the earlier formation of lesions on the second leaf than that on the flag leaf, as the formation of lesions features the activation of defense against pathogens. Consistent with the enhanced resistance of lmm9150 to pathogens, the expression of three PR genes, PR1a, PR1b, and PR10 were significantly upregulated in lmm9150 (Figure 4A). Moreover, the expression of two marker genes of JA-biosynthesis pathway was upregulated in the mutant, whereas the expression of two marker genes of SA signaling pathway was not obviously changed (Figure 4E and Supplementary Figure S2F). Consistent with this, JA level in lmm9150 was significantly increased, but SA level was the same as the WT (Figure 4F and Supplementary Figure S2G). The hormones, including ABA, JA, and SA, are secondary signal molecules involved in defense responses (Zeevaart and Creelman, 1988; Creelman and Mullet, 1997; Dempsey et al., 1997). It has been reported that the antagonistic interactions between ABA and JA–ET signaling pathway regulate disease resistance in Arabidopsis (Anderson et al., 2004). However, JA signaling and SA signaling can be mutually antagonistic or synergistic in defense response (Takahashi et al., 2004; Bari and Jones, 2009; Francisco et al., 2016). Therefore, the cross-talk between JA and SA is very complicated (Proietti et al., 2013). In addition, SA signaling is usually associated with the upregulation of PR genes (Vallad and Goodman, 2004; Zarate et al., 2007; Jiang et al., 2009). We also detected significantly increased expression of PR genes in lmm9150, although SA level had no obvious change (Figure 4A and Supplementary Figure S2G), which could be due to the activation of defense responses by JA signaling, because JA could also activate the expression of PR genes (Zarate et al., 2007). Collectively, our results indicated that the mutation of OsABA2 may alter ABA biosynthesis, which in turn, promote JA signaling pathway to activate defense against different pathogens. However, the exact mechanisms of the decreased level of ABA in lmm9150 that enhances PR gene expression, JA level, and defense against pathogens are yet to be determined in the future.
Seed dormancy and germination are mainly governed by ABA and GA (Shu et al., 2016). Generally, ABA maintains seed dormancy, whereas GA releases dormancy and promotes germination (Bewley, 1997; Shu et al., 2013; Liu et al., 2014), suggesting that the ratio of ABA/GA content is a major regulator for dormancy and germination (Fang et al., 2008; Finkelstein et al., 2008). Consistent with the previous literatures, our results showed that the ABA content was decreased and the GA content was increased, which resulted in a decreased ratio of ABA/GA and pre-harvest sprouting in the seeds of lmm9150 (Figures 7B,E and Supplementary Figure S7B). These findings were consistent with the observations in the other ABA-deficient mutants in rice that are loss of seed dormancy and exhibit pre-harvest sprouting, including osaba1, phs1, phs2, phs3, and phs4 (Agrawal et al., 2001; Fang et al., 2008). On the other hand, GA is a key regulator of release dormancy and promotion germination in plants (Peng and Harberd, 2002; Ye et al., 2015; Xiong et al., 2017). The balance of ABA/GA determines seed dormancy and germination (White et al., 2000; Fang et al., 2008; Finkelstein et al., 2008). It was possible that the dysfunction of ABA biosynthesis pathway can enhance GA biosynthesis indirectly through a feedback mechanism, as geranylgeranyl pyrophosphate (GGPP) is the common precursor for ABA and GA biosynthesis (Fray et al., 1995; Rodríguezconcepción et al., 2001) and the ABA deficiency may lead to more supply of GGPP for GA production in the mutant. Therefore, it is explainable that the lower ABA/GA ratio in lmm9150 contributed to its pre-harvest sprouting phenotype (Figures 7C,E and Supplementary Figure S7B).
In Arabidopsis, ataba2 displays dwarfism and small size leaf (Cheng et al., 2002; Miguel et al., 2002). In contrast, we found that the plant height of lmm9150 was higher than that of WT and the flag leaf of lmm9150 was longer than that of WT (Figures 1D,E), suggesting that the function of ABA2 in regulating growth in rice is different from that in Arabidopsis, although both genes are involved in ABA biosynthesis. We showed that mutation in OsABA2 led to decreased level of ABA content (Figures 7B–D), but increased level of GA content (Supplementary Figures S7A–C), and exogenous application of ABA could rescue the plant height phenotype of the lmm9150 mutant (Figures 8E–H), suggesting that OsABA2 negatively regulates plant height and leaf length through modulation of ABA and GA levels.
Together, our data demonstrated that novel functions of OsABA2 in modulating plant growth, cell death, and disease resistance, in addition to the expected function in seed dormancy and add new information of the roles of ABA in different biological processes in plant.
Author Contributions
XW designed and performed the project. WW and ML directed the research. YL, QB, PX, and TW performed the analysis of agronomic traits, histochemical staining, shading treatment, and assessed rice disease resistance. YL and QB carried out genetic analysis, map-based cloning, and germination assay. DG and YP performed the mutation analysis and sequence analysis. XD, HZ, XC, and AA assisted in the data analysis. YL, XW, and WW wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program of China (Grant No. 2016YFD0100406), the Special Scientific Research Project of Agricultural Public Welfare of MOA, China (Grant No. 201403002-3), and the National Natural Science Foundation of China (Grant No. 31430072).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00405/full#supplementary-material
FIGURE S1 | Phenotypic characterizations of lmm9150 mutant. (A,B) Comparison of four leaves between wild type (WT) and lmm9150 mutant exhibits the lesion mimic spots of lmm9150 at tillering stage (A) and mature stage (B). The numbers 1, 2, 3 and 4 represent the first top leaf, the second top leaf, the third top leaf and the fourth top leaf, respectively. (C–F) Quantification analysis of tiller numbers (C), grains setting rates (D), grains weight per plant (E), 1000-grains weight (F), respectively. Data were obtained from 10 plants of wild type and mutant (C–E). Mean of 1000-grains weight obtained from 10 replicates. Statistical analysis was performed using Student’s t-test, ∗ and ∗∗ indicates P < 0.05 and P < 0.01, respectively. Scale bar: 2 cm in (A,B).
FIGURE S2 | Determination of wild type (WT) and lmm9150 plants resistance to bacterial blight and rice blast diseases. (A–D) The disease lesion leaves of the wild type (WT) and lmm9150 mutant were inoculated with Xanthomonas oryzae pv. oryzae (Xoo) strains, X004, P6 and P3, at 15 days post-inoculation (dip). The image (D) shows that the disease lesions of WT are longer than that of lmm9150 and the flag leaf disease lesions of lmm9150 are longer than that of the second leaf of mutant. FL: the flag leaf; SL: the second leaf. Data were obtained from 20 leaves of main panicles. (E) The disease lesions of leaves of the wild type (WT) and lmm9150 mutant were inoculated 5 days with Magnaporthe oryzae (M. oryzae) strain, Tetep. The experiments were repeated twice times with similar results. (F) Quantitative reverse-transcription PCR (qRT-PCR) data show the relative expression levels of salicylic acid (SA) signaling-related genes in lmm9150 and wild type (WT). (G) Comparison of SA content in leaf of wild type (WT) and lmm9150. SS: seedling stage; TS: tillering stage. The content of hormones was tested by enzyme-linked immunosorbent assay (ELISA) method. Mean and standard deviation were obtained from three measurements (F,G). The housekeeping gene Ubiquitin5 (Ubq5) was used as control. Statistical analysis was performed using Student’s t-test, ∗and∗∗ indicate P < 0.05 and P < 0.01, respectively. Scale bar: 1 cm in (E).
FIGURE S3 | Comparison of the amino acid sequences of xanthoxin dehydrogenases. Box I indicates cofactor binding site and box II indicates catalytic activity site. The arrow indicates the mutation site of OsABA2 in lmm9150 mutant. BRADI_1g04320 in Brachypodium, GRMZM2G332976 in Zea mays, LOC8057083 in Sorghum bicolor, AT1G52340.1 in Arabidopsis, LOC102577811 in Solanum tuberosum. These symbols describe conversation degree, “∗, : and. ” indicates strong, middle and weak respectively.
FIGURE S4 | Comparison of candidate gene sequence. LOC_Os03g59610 is the reference sequence and comes from a japonica rice Nipponbare. Yixiang1B-CDS sequence was cloned from an indica cv. Yixiang1B, which is the wild type of lmm9150 mutant. The arrow indicates that 9-bp deletion is found in Yixiang1B.
FIGURE S5 | Mutational types of 5 knockout lines. Mutational types of #1, #3, #4 and #5 knockout lines are deletion. Mutational type of #2 knockout line is insertion. The black triangles indicate the mutational sites.
FIGURE S6 | Determination of wild type (WT) and a knockout line plants resistance to bacterial blight and rice blast diseases. (A–F) The disease lesion of leaves of the wild type (WT) and one of knockout lines were inoculated with Xanthomonas oryzae pv. oryzae (Xoo) strains, 8428, P3 and X004, at 15 days post-inoculation. KL: knockout lines; FL: the flag leaf; SL, the second leaf. Data were obtained from 20 leaves of main panicles. (G) The disease lesions of leaves of the wild type (WT) and one of knockout lines was inoculated 5 days with Magnaporthe oryzae (M. oryzae) strain, Tetep. The experiments were repeated twice times with similar results. Scale bar: 1cm in (G). Statistical analysis was performed using Student’s t-test, ∗∗ indicates P < 0.01.
FIGURE S7 | Comparison of gibberellin (GA) and ABA/GA content ratio in the wild type (WT) and lmm9150 mutant. (A–C) Comparison of gibberellin (GA) content in leaves (A), seeds (B) and stems (C) between the wild type (WT) and lmm9150 mutant at different developmental stages. SS: seedling stage; TS: tillering stage; MS: mature stage; MRS: milk ripe stage; DS: dough stage; YMS: yellow mature stage. GA content was tested by enzyme-linked immunosorbent assay (ELISA) method. (D,E) The ratio of ABA/GA content in leaves (D) and stems (E) of the wild type (WT) and the lmm9150 mutant. Mean and standard deviation were obtained from three measurements. Statistical analysis was performed using Student’s t-test, ∗ and ∗∗ indicates P < 0.05 and P < 0.01, respectively.
FIGURE S8 | Effects of ABA on water loss rate and germination in lmm9150 mutant. (A,B) Water loss assays for the leaves of the wild type (WT) and lmm9150 mutant were examined at seedling stage. (C–E) Investigation of germination rate in lmm9150 mutant and wild type seeds with different concentration of ABA (0.1, 1 and 100 μmol/L). Mean were obtained from three measurements.
TABLE S1 | List of primers for qRT-PCR.
TABLE S2 | List of polymorphic molecular markers for mapping.
TABLE S3 | List of targets sequences for PCR.
TABLE S4 | Segregation ratio of F2 populations.
Footnotes
- ^ http://www.gramene.org/microsat
- ^ http://rise2.genomics.org.cn/page/rice/index.jsp
- ^ http://primer3.ut.ee/
- ^ http://ensembl.gramene.org/Tools/Blast?db=core
- ^ http://rice.plantbiology.msu.edu/
References
Agrawal, G. K., Yamazaki, M., Kobayashi, M., Hirochika, R., Miyao, A., and Hirochika, H. (2001). Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol. 125, 1248–1257. doi: 10.1104/pp.125.3.1248
Anderson, J., Badruzsaufari, E., Schenk, P. M., Manners, J. M., Desmond, O. J., and Ehlert, C. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16, 3460–3479. doi: 10.1105/tpc.104.025833
Asselbergh, B., Achuo, A., Höfte, M., and Van Frederique, G. (2008). Abscisic acid deficiency leads to rapid activation of tomato defense responses upon infection with Erwinia chrysanthemi. Mol. Plant Pathol. 9, 11–24. doi: 10.1111/J.1364-3703.2007.00437.X
Bari, R., and Jones, J. G. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Bethke, P. C., Lonsdale, J. E., Fath, A., and Jones, R. L. (1999). Hormonally regulated programmed cell death in barley aleurone cells. Plant Cell 11, 1033–1046. doi: 10.2307/3870796
Bewley, J. D. (1997). Seed germination and dormancy. Plant Cell 9, 1055–1066. doi: 10.1105/tpc.9.7.1055
Burbidge, A., Grieve, T. M., Jackson, A., Thompson, A., Mccarty, D. R., and Taylor, I. (1999). Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 17, 427–431. doi: 10.1046/j.1365-313X.1999.00386.x
Cao, H., Bowling, S. A., Gordon, A. S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. doi: 10.1105/tpc.6.11.1583
Chen, X., Hao, L., Pan, J., Zheng, X., Jiang, G., and Jin, Y. (2012). SPL5, a cell death and defense-related gene, encodes a putative splicing factor 3b subunit 3 (SF3b3) in rice. Mol. Breed. 30, 939–949. doi: 10.1007/s11032-011-9677-4
Cheng, W., Akria, E., Li, Z., Jessica, P., Hueichi, C., and Analilia, A. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14, 2723–2743. doi: 10.1105/tpc.006494
Creelman, R. A., and Mullet, J. E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. 48, 355–381. doi: 10.1146/annurev.arplant.48.1.355
Dempsey, D. A., Shah, J., and Klessig, D. F. (1997). Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18, 547–575. doi: 10.1080/07352689991309397
Endo, A., Nelson, K. M., Thoms, K., Abrams, S. R., Nambara, E., and Sato, Y. (2014). Functional characterization of xanthoxin dehydrogenase in rice. Plant Physiol. 171, 1231–1240. doi: 10.1016/j.jplph.2014.05.003
Fang, J., Chai, C., Qian, Q., Li, C., Tang, J., and Sun, L. (2008). Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 54, 177–189. doi: 10.1111/j.1365-313X.2008.03411.x
Fath, A., Bethke, P. C., and Jones, R. L. (2001). Enzymes that scavenge reactive oxygen species are down-regulated prior to gibberellic acid-induced programmed cell death in barley aleurone. Plant Physiol. 126, 156–166. doi: 10.1104/pp.126.1.156
Fekih, R., Tamiru, M., Kanzaki, H., Abe, A., Yoshida, K., and Kanzaki, E. (2015). The rice (Oryza sativa L.) LESION MIMIC RESEMBLING, which encodes an AAA-type ATPase, is implicated in defense response. Mol. Genet. Genomics 290, 611–622. doi: 10.1007/s00438-014-0944-z
Finkelstein, R., Reeves, W., Ariizumi, T., and Steber, C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. doi: 10.1146/annurev.arplant.59.032607.092740
Francisco, A. R., José, G. G., José, G., Rosario, B. P., Antonio, M. M., and Oswaldo, T. (2016). Partial activation of SA- and JA-defensive pathways in strawberry upon Colletotrichum acutatum interaction. Front. Plant Sci. 7:1036. doi: 10.3389/fpls.2016.01036
Fray, R. G., Wallace, A., Fraser, P. D., Valero, D., Hedden, P., and Bramley, P. M. (1995). Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J. 8, 693–701. doi: 10.1046/j.1365-313X.1995.08050693.x
Gao, S., Guo, W., Feng, W., Liu, L., Song, X., and Chen, J. (2016). LTP3 contributes to disease susceptibility in Arabidopsis by enhancing abscisic acid (ABA) biosynthesis. Mol. Plant Pathol. 17, 412–426. doi: 10.1111/mpp.1229
Gechev, T. S., Minkov, I. N., and Hille, J. (2005). Hydrogen peroxide-induced cell death in Arabidopsis: transcriptional and mutant analysis reveals a role of an oxoglutarate-dependent dioxygenase gene in the cell death process. IUBMB Life 57, 181–188. doi: 10.1080/15216540500090793
Greenberg, J. T. (1997). Programmed cell death in plant-pathogen interactions. Annu. Rev. Plant Physiol. 48, 525–545. doi: 10.1146/annurev.arplant.48.1.525
Hirai, N., Yoshida, R., Todoroki, Y., and Ohigashi, H. (2000). Biosynthesis of abscisic acid by the non-mevalonate pathway in plants, and by the mevalonate pathway in fungi. Biosci. Biotechnol. Biochem. 64, 1448–1458. doi: 10.1271/bbb.64.1448
Hu, G., Yalpani, N., Briggs, S. P., and Johal, G. S. (1998). A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10, 1095–1105. doi: 10.2307/3870714
Inomata, M., Hirai, N., Yoshida, R., and Ohigashi, H. (2004). Biosynthesis of abscisic acid by the direct pathway via ionylideneethane in a fungus, Cercospora cruenta. Biosci. Biotechnol. Biochem. 68, 2571–2580. doi: 10.1271/bbb.68.2571
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., and Kato, T. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Jeon, J. S., Lee, S., Jung, K. H., Jun, S. H., Jeong, D. H., and Lee, J. (2000). T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22, 561–570. doi: 10.1046/j.1365-313x.2000.00767.x
Jiang, C., Shimono, M., Sugano, S., Kojima, M., Yazawa, K., and Yoshida, R. (2010). Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice–Magnaporthe grisea interaction. Mol. Plant Microbe Interact. 23, 791–798. doi: 10.1094/MPMI-23-6-0791
Jiang, C. J., Shimono, M., Maeda, S., Inoue, H., Mori, M., and Hasegawa, M. (2009). Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol. Plant Microbe Interact. 22, 820–829. doi: 10.1094/MPMI-22-7-0820
Kiyosawa, S. (1970). Inheritance of a particular sensitivity of the rice variety, Sekiguchi Asahi, to pathogens and chemicals, and linkage relationship with blast resistance genes. Tokyo Nat. Inst. Agric. Sci. Bull. 21, 61–72.
Koga, H., Dohi, K., and Mori, M. (2004). Abscisic acid and low temperatures suppress the whole plant-specific resistance reaction of rice plants to the infection of Magnaporthe grisea. Physiol. Mol. Plant Pathol. 65, 3–9. doi: 10.1016/j.pmpp.2004.11.002
Léon-Kloosterziel, K. M., Gil, M. A., Ruijs, G. J., Jacobsen, S. E., Olszewski, N. E., and Schwartz, S. H. (1996). Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 10, 655–661. doi: 10.1046/j.1365-313X.1996.10040655.x
Leung, J., and Giraudat, J. (1998). Abscisic acid signals transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. doi: 10.1146/annurev.arplant.49.1.199
Li, J., Pandeya, D., Nath, K., Zulfugarov, I. S., Yoo, S., and Zhang, H. (2010). ZEBRA-NECROSIS, a thylakoid-bound protein, is critical for the photoprotection of developing chloroplasts during early leaf development. Plant J. 62, 713–725. doi: 10.1111/j.1365-313X.2010.04183.x
Liu, Q., Ning, Y., Zhang, Y., Yu, N., Zhao, C., and Zhan, X. (2017). OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell 29, 345–359. doi: 10.1105/tpc.16.00650
Liu, X., Li, F., Tang, J., Wang, W., Zhang, F., and Wang, G. (2012). Activation of the jasmonic acid pathway by depletion of the hydroperoxide lyase OsHPL3 reveals crosstalk between the HPL and AOS branches of the oxylipin pathway in rice. PLoS One 7:e50089. doi: 10.1371/journal.pone.0050089
Liu, Y., Fang, J., Xu, F., Chu, J., Yan, C., and Ppi, M. R. S. (2014). Expression patterns of ABA and GA metabolism genes and-hormone levels during rice seed development and imbibition: a comparison of dormant and non-dormant rice cultivars. J. Genet. Genomics 41, 327–338. doi: 10.1016/j.jgg.2014.04.004
Liu, Y., Ye, N., Rui, L., Chen, M., and Zhang, J. (2010). H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J. Exp. Bot. 61, 2979–2990. doi: 10.1093/jxb/erq125
Ma, F., Ni, L., Liu, L., Li, X., Zhang, H., and Zhang, A. (2016). ZmABA2, an interacting protein of ZmMPK5, is involved in abscisic acid biosynthesis and functions. Plant Biotechnol. J. 14, 771–782. doi: 10.1111/pbi.12427
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., and Qiu, R. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Maia, J., Dekkers, B. J. W., Dolle, M. J., Ligterink, W., and Hilhorst, H. W. M. (2014). Abscisic acid (ABA) sensitivity regulates desiccation tolerance in germinated Arabidopsis seeds. New Phytol. 203, 81–93. doi: 10.1111/nph.12785
Marin, E., Nussaume, L., Quesada, A., Gonneau, M., Sotta, B., and Hugueney, P. (1996). Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15, 2331–2342.
Michelmore, R., Paran, I., and Kesseli, R. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Miguel, G., Apostolova, N., Bellés, J. M., Barrero, J. M., Pedro Piqueras, B., and Ponce, M. R. (2002). The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14, 1833–1846. doi: 10.1105/tpc.002477
Milborrow, B. V. (2001). The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 52, 1145–1164. doi: 10.1093/jexbot/52.359.1145
Moeder, W., and Yoshioka, K. (2008). Lesion mimic mutants: a classical, yet still fundamental approach to study programmed cell death. Plant Signal. Behav. 3, 764–767. doi: 10.1146/annurev.arplant.48.1.525
Murray, M. G., Thompson, W. F., Thomson, W. F., Thompsom, W., Murry, M. G., and Tompson, W. (1980). Rapid isolation of high-molecular-weight plant of high-molecular-weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Nahar, K., Kyndt, T., De, V. D., Höfte, M., and Gheysen, G. (2011). The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 157, 305–316. doi: 10.1104/pp.111.177576
North, H. M., De, A. A., Boutin, J. P., Frey, A., To, A., and Botran, L. (2007). The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 50, 810–824. doi: 10.1111/j.1365-313X.2007.03094.x
Okamoto, M., Min, X., Seo, M., Nakabayashi, K., Kamiya, Y., and Nambara, E. (2002). Complementation of a tomato ABA-deficient sitiens mutant by an Arabidopsis aldehyde oxidase gene, AAO3. Plant Cell Physiol. 43, S42–S42.
Park, C., Chen, S., Shirsekar, G. S., Zhou, B. Z., Khang, C. H., and Pattavipha Songkumarn, A. (2012). The Magnaporthe oryzae effector avrPiz-t targets the ring E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Peng, J., and Harberd, N. P. (2002). The role of GA-mediated signalling in the control of seed germination. Curr. Opin. Plant Biol. 5, 376–381. doi: 10.1016/S1369-5266(02)00279-0
Persson, M., Falk, A., and Dixelius, C. (2009). Studies on the mechanism of resistance to Bipolaris sorokiniana in the barley lesion mimic mutant bst1. Mol. Plant Pathol. 10, 587–598. doi: 10.1111/J.1364-3703.2009.00555.X
Pieterse, C., Van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. C. M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Proietti, S., Bertini, L., Timperio, A. M., Zolla, L., Caporale, C., and Caruso, C. (2013). Crosstalk between salicylic acid and jasmonate in Arabidopsis investigated by an integrated proteomic and transcriptomic approach. Mol. Biosyst. 9, 1169–1187. doi: 10.1039/c3mb25569g
Qiao, Y., Jiang, W., Lee, J., Park, B., Choi, M., and Piao, R. (2009). SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 185, 258–274. doi: 10.1111/j.1469-8137.2009.03047.x
Ren, D., Yang, H., and Zhang, S. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277, 559–565. doi: 10.1074/jbc.M109495200
Riahi, L., Zoghlami, N., Dereeper, A., Laucou, V., Mliki, A., and This, P. (2013). Molecular characterization and evolutionary pattern of the 9-cis-epoxycarotenoid dioxygenase NCED1 gene in grapevine. Mol. Breed. 32, 253–266. doi: 10.1007/s11032-013-9866-4
Rock, C. D., and Quatrano, R. S. (1995). The Role of Hormones during Seed Development. Berlin: Springer. doi: 10.1007/978-94-011-0473-9_31
Rodríguezconcepción, M., Ahumada, I., Diezjuez, E., Sauretgüeto, S., Lois, L. M., and Gallego, F. (2001). 1-Deoxy-D-xylulose 5-phosphate reductoisomerase and plastid isoprenoid biosynthesis during tomato fruit ripening. Plant J. 27, 213–222. doi: 10.1046/j.1365-313x.2001.01089.x
Sang, G. K., Sun, T. K., Wang, Y., Yu, S., Choi, I. S., and Yong, C. K. (2011). The RNase activity of rice probenazole-induced protein1 (PBZ1) plays a key role in cell death in plants. Mol. Cells 31, 25–31. doi: 10.1007/s10059-011-0004-z
Schwartz, S., Léon-Kloosterziel, K. M., and Koornneef, M. (1997a). Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 114, 161–166. doi: 10.1104/pp.114.1.161
Schwartz, S., Tan, B., Gage, D., Zeevaart, J. A., and McCartney, C. (1997b). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. doi: 10.1126/science.276.5320.1872
Schwartz, S. H., Qin, X., and Zeevaart, J. A. (2003). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131, 1591–1601. doi: 10.1104/pp.102.017921
Seo, M., Peeters, A. J. M., Koiwai, H., Oritani, T., Marion-Poll, A., and Zeevaart, J. A. D. (2000). The Arabidopsis aldehyde oxidase (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. U.S.A. 97, 12908–12913. doi: 10.1073/pnas.220426197
Shu, K., Liu, X. D., Xie, Q., and He, Z. H. (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45. doi: 10.1016/j.molp.2015.08.010
Shu, K., Zhang, H., Wang, S., Chen, M., Wu, Y., and Tang, S. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of Abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9:e1003577. doi: 10.1371/journal.pgen.1003577
Tadashi, F., Sylvie, M., Masayuki, I., Masaharu, M., Letian, C., and Hann Ling, W. (2010). Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in Rice. J. Biol. Chem. 285, 11308–11313. doi: 10.1074/jbc.M109.091371
Takahashi, H., Kanayama, Y., Zheng, M. S., Kusano, T., Shu, H., and Ikegami, M. (2004). Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant Cell Physiol. 45, 803–809. doi: 10.1093/pcp/pch085
Tamiru, M., Takagi, H., Abe, A., Yokota, T., Kanzaki, H., and Okamoto, H. (2016). A chloroplast-localized protein LESION AND LAMINA BENDING affects defense and growth responses in rice. New Phytol. 210, 1282–1297. doi: 10.1111/nph.13864
Tan, B. C., and Mccarty, D. R. (1997). Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. U.S.A. 94, 12235–12240. doi: 10.1073/pnas.94.22.12235
Teng, N., Wang, J., Chen, T., Wu, X., Wang, Y., and Lin, J. (2006). Elevated CO2 induces physiological, biochemical and structural changes in leaves of Arabidopsis thaliana. New Phytol. 172, 92–103. doi: 10.1111/j.1469-8137.2006.01818.x
Utako, Y., Masahiro, Y., Hongxuan, L., Motoyuki, A., and Kyoji, Y. (2002). A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. U.S.A. 99, 7530–7535. doi: 10.1073/pnas.112209199
Vallad, G. E., and Goodman, R. M. (2004). Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci. 44, 1920–1934. doi: 10.2135/cropsci2004.1920
Wang, J., Ye, B., Yin, J., Yuan, C., Zhou, X., and Li, W. (2015). Characterization and fine mapping of a light-dependent leaf lesion mimic mutant1 in rice. Plant Physiol. Biochem. 97, 44–51. doi: 10.1016/j.plaphy.2015.09.001
Wang, S., Lei, C., Wang, J., Ma, J., Tang, S., and Wang, C. (2017). SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J. Exp. Bot. 68, 899–913. doi: 10.1093/jxb/erx001
Wang, S., Lim, J., Kim, S., Cho, S., Yoo, S., and Koh, H. (2015). Mutation of SPOTTED LEAF3 (SPL3) impairs abscisic acid-responsive signalling and delays leaf senescence in rice. J. Exp. Bot. 66, 7045–7059. doi: 10.1093/jxb/erv401
White, C. N., Proebsting, W. M., Hedden, P., and Rivin, C. J. (2000). Gibberellins and seed development in maize. I: evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol. 122, 1081–1088. doi: 10.1104/pp.122.4.1081
Xiao, X., Cheng, X., Yin, K., Li, H., and Qiu, J. (2017). Abscisic acid negatively regulates post-penetration resistance of Arabidopsis to the biotrophic powdery mildew fungus. Sci. China Life Sci. 60, 891–901. doi: 10.1007/s11427-017-9036-2
Xiong, L., Lee, H., Ishitani, M., and Zhu, J. K. (2002). Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277, 8588–8596. doi: 10.1074/jbc.M109275200
Xiong, W., Ye, T., Yao, X., Liu, X., Ma, S., and Chen, X. (2017). The dioxygenase GIM2 functions in seed germination by altering gibberellin production in Arabidopsis. J. Integr. Plant Biol. doi: 10.1111/jipb.12619 [Epub ahead of print].
Xu, J., Audenaert, K., Hofte, M., and Vleesschauwer, D. D. (2013). Abscisic acid promotes susceptibility to the rice leaf blight pathogen Xanthomonas oryzae pv. oryzae by suppressing salicylic acid-mediated defenses. PLoS One 8:e67413. doi: 10.1371/journal.pone.0067413
Ye, H., Feng, J., Zhang, L., Zhang, J., Mispan, M. S., and Cao, Z. (2015). Map-based cloning of seed dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 169, 2152–2165. doi: 10.1104/pp.15.01202
Young, T. E., and Gallie, D. R. (2000). Regulation of programmed cell death in maize endosperm by abscisic acid. Plant Mol. Biol. 42, 397–414. doi: 10.1023/A:1006333103342
Zarate, S. I., Kempema, L. A., and Walling, L. L. (2007). Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 143, 866–875. doi: 10.1104/pp.106.090035
Zeevaart, J. A. D., and Creelman, R. A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. 39, 439–473. doi: 10.1146/annurev.pp.39.060188.002255
Zeng, L., Shaohong, Q., Alicia, B., Chengwei, Y., Marietta, B., and Hongyan, Y. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16, 2795–2808. doi: 10.1105/tpc.104.025171
Zhang, S. J., Song, G. Q., Li, Y. L., Gao, J., Liu, J. J., and Fan, Q. Q. (2014). Cloning of 9-cis-epoxycarotenoid dioxygenase gene (TaNCED1) from wheat and its heterologous expression in tobacco. Biol. Plant 58, 89–98. doi: 10.1007/s10535-013-0373-6
Zhang, W., Wang, C., Qin, C., Wood, T., Olafsdottir, G., and Welti, R. (2003). The oleate-stimulated phospholipase D, PLDdelta, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15, 2285–2295. doi: 10.1105/tpc.013961
Zhu, X., Yin, J., Liang, S., Liang, R., Zhou, X., and Chen, Z. (2016). The multivesicular bodies (MVBs)-localized AAA ATPase LRD6-6 inhibits immunity and cell death likely through regulating MVBs-mediated vesicular trafficking in rice. PLoS Genet. 12:e1006311. doi: 10.1371/journal.pgen.1006311
Keywords: abscisic acid, xanthoxin dehydrogenase, gibberellin, ABA/GA ratio, lesion mimic mutant, pre-harvest sprouting, defense responses, Oryza sativa L.
Citation: Liao Y, Bai Q, Xu P, Wu T, Guo D, Peng Y, Zhang H, Deng X, Chen X, Luo M, Ali A, Wang W and Wu X (2018) Mutation in Rice Abscisic Acid2 Results in Cell Death, Enhanced Disease-Resistance, Altered Seed Dormancy and Development. Front. Plant Sci. 9:405. doi: 10.3389/fpls.2018.00405
Received: 09 November 2017; Accepted: 14 March 2018;
Published: 28 March 2018.
Edited by:
Zhengqing Fu, University of South Carolina, United StatesReviewed by:
Aardra Kachroo, University of Kentucky, United StatesMarina Gavilanes-Ruiz, Universidad Nacional Autónoma de México, Mexico
Copyright © 2018 Liao, Bai, Xu, Wu, Guo, Peng, Zhang, Deng, Chen, Luo, Ali, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenming Wang, ajMxNndlbm1pbmd3YW5nQDE2My5jb20= Xianjun Wu, d3V4akBzaWNhdS5lZHUuY24=
†These authors have contributed equally to this work.
 Yongxiang Liao
Yongxiang Liao Que Bai1†
Que Bai1† Daiming Guo
Daiming Guo Hongyu Zhang
Hongyu Zhang Asif Ali
Asif Ali