- State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Nanjing University, Nanjing, China
The ERECTA family genes (ERfs) have been found to play diverse functions in Arabidopsis, including controlling cell proliferation and cell growth, regulating stomata patterning, and responding to various stresses. This wide range of functions has rendered them as a potential candidate for crop improvement. However, information on their functional roles, particularly their morphological impact, in crop genomes, such as rice, is limited. Here, through evolutionary prediction, we first depict the evolutionary trajectory of the ER family, and show that the ER family is actually highly conserved across different species, suggesting that most of their functions may also be observed in other plant species. We then take advantage of the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats–associated nuclease 9) system to assess their morphological impact on one of the most important crops, rice. Loss-of-function mutants of OsER1 and OsER2 display shortened plant stature and reduced panicle size, suggesting they possibly also functioned in regulating cell proliferation and cell growth in rice. In addition to functions similar to that in Arabidopsis, we also find clues that rice ERfs may play unique functional roles. The OsER2 displayed more severe phenotypic changes than OsER1, indicating putative differentiation in their functions. The OsERL might be of essential in its function, and the proper function of all three rice ER genes might be dependent of their genetic background. Future investigations relating to these functions are key to exploiting ERfs in crop development.
Introduction
The ERECTA (ER) gene has been extensively studied in Arabidopsis thaliana for its famous Landsberg erecta (Ler) ecotype (Torii et al., 1996; van Zanten et al., 2009; Shpak, 2013). Arabidopsis plants with homozygous er alleles have very short inflorescence stems, siliques, and pedicels (Torii et al., 1996). The ER gene encodes a leucine-rich repeat receptor-like kinase (LRR-RLK) that has functional Ser/Thr kinase activity, which in turn plays a regulatory role in plant development. During Arabidopsis organogenesis, the ER gene, together with other RLK members, plays a critical role in controlling cell proliferation and cell growth (Shpak et al., 2003). The ER pathway could regulate shoot apical meristem (SAM) size and floral meristem identity in parallel to the class III HD-ZIP (homeodomain-leucine-zipper) and CLV (CLAVATA) pathways (Mandel et al., 2014, 2016). Natural allelic variations in the Arabidopsis ER locus have been associated with variations in petal shape, suggesting its function in determining petal shape (Abraham et al., 2013). In addition to these morphological functions, ER protein also functions as plant a physiological regulator. The ER gene was found to regulate transpiration efficiency in Arabidopsis (Masle et al., 2005), overexpression of ER significantly increases the thermotolerance in Arabidopsis, rice, and tomato (Qi et al., 2004; Shen et al., 2015). The ER gene has also been reported to be a contributor to shade-avoidance syndrome, possibly working in a background-dependent manner (Kasulin et al., 2013). Finally, being a member of the LRR family, which is best described in resistance (R) genes, ER also affects the resistance to various biotic and abiotic factors (Godiard et al., 2003; Shpak, 2013; Jordá et al., 2016; Takahashi et al., 2016).
Generally, the ER gene functions together with its family members. There are two paralogs of ER in Arabidopsis, namely, ER-LIKE1 (ERL1) and ER-LIKE2 (ERL2). Throughout the secondary growth of Arabidopsis hypocotyl, ER and ERL1 redundantly prevent premature progression of sequential events (Ikematsu et al., 2017). Three ERECTA family genes (ERfs) could also interact synergistically in regulating stomatal patterning (Shpak et al., 2005) and mediating morphological alterations (Uchida et al., 2011). Therefore, the ER family acts cooperatively or redundantly in regulating almost every aspect of plant development (Shpak, 2013; Kosentka et al., 2017; Tameshige et al., 2017; Qu et al., 2017).
Due to its function in regulating plant development and thermotolerance, the ER family may be potentially utilized in agriculture. Rice (Oryza sativa) is the most important food resource in Asian countries, and there is currently a need to increase its yield to feed continuously growing human populations. It is thus essential to identify and characterize functional genes in rice that may assist in improving yield, quality, and resistance. There are several ERfs present in the rice genome (referred to as OsERfs hereafter). The overexpression of OsER1 increases the heat tolerance of transgenic rice, whereas OsER1 plants carrying the T-DNA null allele died quickly when grown at high temperatures (Shen et al., 2015), thereby suggesting that the ER family plays a critical functional role in rice, and has the potential to be used to breed thermotolerant crops.
However, current knowledge on ERfs has been mostly derived from Arabidopsis. Functional studies on ERfs are lagged behind in other crops, such as rice, particularly relating to morphological impacts, thus hindering their application to agricultural programs. The OsER1 gene has only been investigated in terms of its role in thermotolerance (Shen et al., 2015), whereas its phenotypic influences remain unclear. Furthermore, studies on the functional role in response to stresses of OsERL, which also known as OsSIK1, are conflicting. Previous studies have shown that the overexpression of OsERL increases the antioxidant capacity of rice, indicating that it plays an important role in responding to salt and drought stress, possibly by suppressing stomatal development in rice leaves (Ouyang et al., 2010). However, a more recent study did not observe any difference in heat tolerance between OsERL mutants and the wild-type control (Shen et al., 2015), and thus OsERL’s regulatory role in stomatal development remains elusive. Whether these discrepancies among studies were due to different treatments, i.e., salt and drought stresses used by Ouyang et al. (2010) compared to heat stress in Shen et al. (2015), or other factors remain unclear. To our knowledge, no functional assessment of OsER2 has been conducted to date, which is partly due to an incorrect gene structure as well as gene symbol given to OsER2 in public rice databases (see section “Discussion” for details).
The poor understanding of rice ERfs is also attributable to the difficulty in obtaining desired mutants with specific genetic backgrounds. The CRISPR/Cas9 system has emerged as a revolutionary genome editing tool that allows targeted mutagenesis at base pair (bp) resolution. In this study, we investigated: (1) whether all of the ERfs in rice have similar functions as that in Arabidopsis; and (2) whether three OsERfs have unique features that differ from those in Arabidopsis. To answer these, we first predicted the putative functions of OsERfs based on evolutionary analyses, and then we knocked out three OsERfs in the rice genome via the CRISPR/Cas9 system to confirm their functional impact on plant phenotypic characteristics. Our results show that ERfs are highly conserved across the plant kingdom, hence some of their functions could be extrapolated from Arabidopsis to rice. However, different OsERfs also indicate addition functions other than that reported in Arabidopsis. We also reveal the putative influence of genetic background on OsERf function that has not been described in previous reports. These results pave the way for further utilization of ERfs in modern breeding programs.
Materials and Methods
Identification of ERfs and Construction of Phylogenetic Tree
Gene sequences of 56 selected plant species were collected from JGI Phytozome database v12.1 (Goodstein et al., 2012), NCBI GenBank, and individual genome project websites (Supplementary Table S1). The protein sequence of Arabidopsis ER gene was used as a seed to search for ERfs of all selected plants through NCBI tblastn (Altschul et al., 1990) with E-value cutoff set to 0.01. Only blast hits with identity ≥50% and coverage ≥50% were defined as ERfs to distinguish from other LRR-RLK members (Supplementary Table S2). The protein sequences of all identified ERfs were aligned by MUSCLE (Edgar, 2004) with default parameters. The aligned protein sequences were further used to guide the alignments of CDS. A phylogenetic tree was constructed using the Neighbor-Joining (NJ) method with 1,000 replicates of bootstrap test through MEGA 6 (Tamura et al., 2013). Each identified gene was classified as ER or ERL-LIKE (ERL) based on its similarity to Arabidopsis ER gene and was further confirmed by its position in the phylogenetic tree.
Estimation of Genetic Parameters of ERfs
Sequence similarity and Ka, Ks values of ER genes and ERL genes across selected plant species were calculated using MEGA 6 (Supplementary Table S3). For within-population analysis, 247 high-coverage re-sequenced rice cultivars (Supplementary Table S4) were collected from the 3,000 rice genomes project (The 3,000 Rice Genomes Project, 2014). Four groups were sampled, including indica (IND), aus (AUS), temperate japonica (TEJ), and tropical japonica (TRJ), the aromatic (ARO) group was not included due to too limited sample size. The cleaned reads were mapped to the Nipponbare reference genome Os-Nipponbare-Reference-IRGSP-1.0 (Kawahara et al., 2013) using BWA-MEM v0.7.10-r789 (Li, 2013) with default parameters. Variants were called using GATK HaplotypeCaller module (McKenna et al., 2010), and were filtered by removing variant calls with a quality <50. The population-level phylogenetic tree was constructed using variant loci with a minor allele frequency (MAF) ≥0.05. The average nucleotide diversities within different rice subspecies were calculated using public available PERL script “calc_vcf_diversity.pl”1 as described in Wang et al. (2016).
Putative effects of variants in rice ERfs were predicted using SNPeffect 4.0 (De Baets et al., 2012) against MSU Rice Genome Annotation Project (RGAP) Release 7 database (Kawahara et al., 2013). The results for OsER2 were further examined manually based on NCBI model. Tajima’s D-value was estimated using DnaSP v5 (Librado and Rozas, 2009).
Generate of Mutant Plants of Rice ERfs by CRISPR/Cas9
Spacers were designed according to Shan et al. (2014) for three rice ERfs. The complementary oligonucleotides of each spacer were inserted into BsaI restriction site of plasmid of single-guide RNAs (sgRNAs). Then sgRNA were incorporated into the Cas9 vector, which contained Cas9 driven by the maize ubiquitin promoter with Gateway recombination method (Katzen, 2007). The vectors were then transformed into two rice cultivars Oryza sativa L. ssp. indica cultivar Kasalath and Oryza sativa L. ssp. japonica cultivar Wuyungeng24. Kasalath was a traditional variety with high tolerance to unsatisfactory growing conditions such as drought and phosphate deficiency (Sakai et al., 2014). It has been extensively used in rice functional characterization and the genome sequences were available (Sakai et al., 2014). Wuyungeng24 was chose as a representative cultivar of japonica rice for its good fertility.
Verification of Mutagenesis
Genomic DNA of transgenic lines was extracted using protocols suggested by Monna et al. (2002). To identify mutations in regenerated plants, genomic DNA surrounding the targeted regions of sgRNAs was amplified by PCR and afterwards sequenced by Sanger et al. (1977) method. As OsER1 and OsER2 share a high similarity, both genes were verified in regenerated plants of either OsER1 or OsER2 to ensure no off-targeted editing. Plant lines with mutated alleles as well as WT were selected for phenotypic analyses.
Results
ER Family Emerged Early in Plants and Is Highly Conserved in Terms of Copy Number and Gene Structure Across Different Species
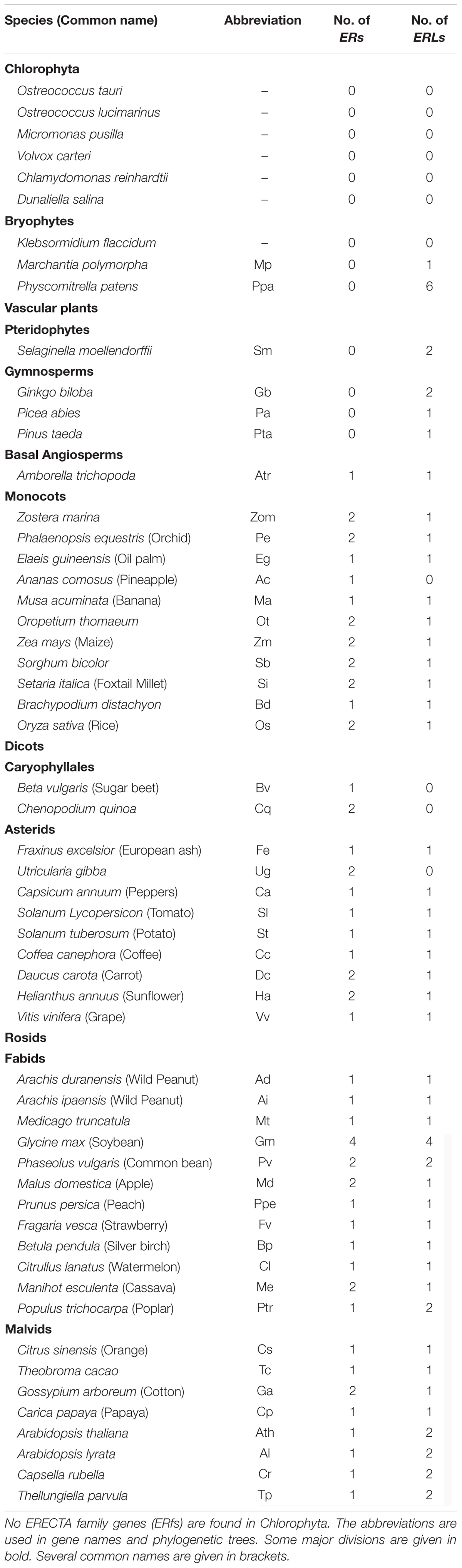
The protein sequence of the A. thaliana ER gene was used as query sequence to search for its homologs in all of the collected genomes (see section “Materials and Methods” for details). No homologs with protein identity ≥50% were found in all of the seven algae (Table 1, BLAST in six species from Chlorophyta and one species Klebsormidium flaccidum from Charophyta). The most ancestral ER family with ERL members only was found in bryophytes, while the first ER gene was found in basal angiosperms (Figure 1, Supplementary Figure S1, and Table 1), suggesting that the ER family only appeared in early land plants (i.e., Embryophyta). All of the collected ERfs were clearly separated into two large clades, which we denoted as the ER clade and ERL clade based on their relationship to the Arabidopsis ER and ERL genes (Figure 1A and Supplementary Figure S1).
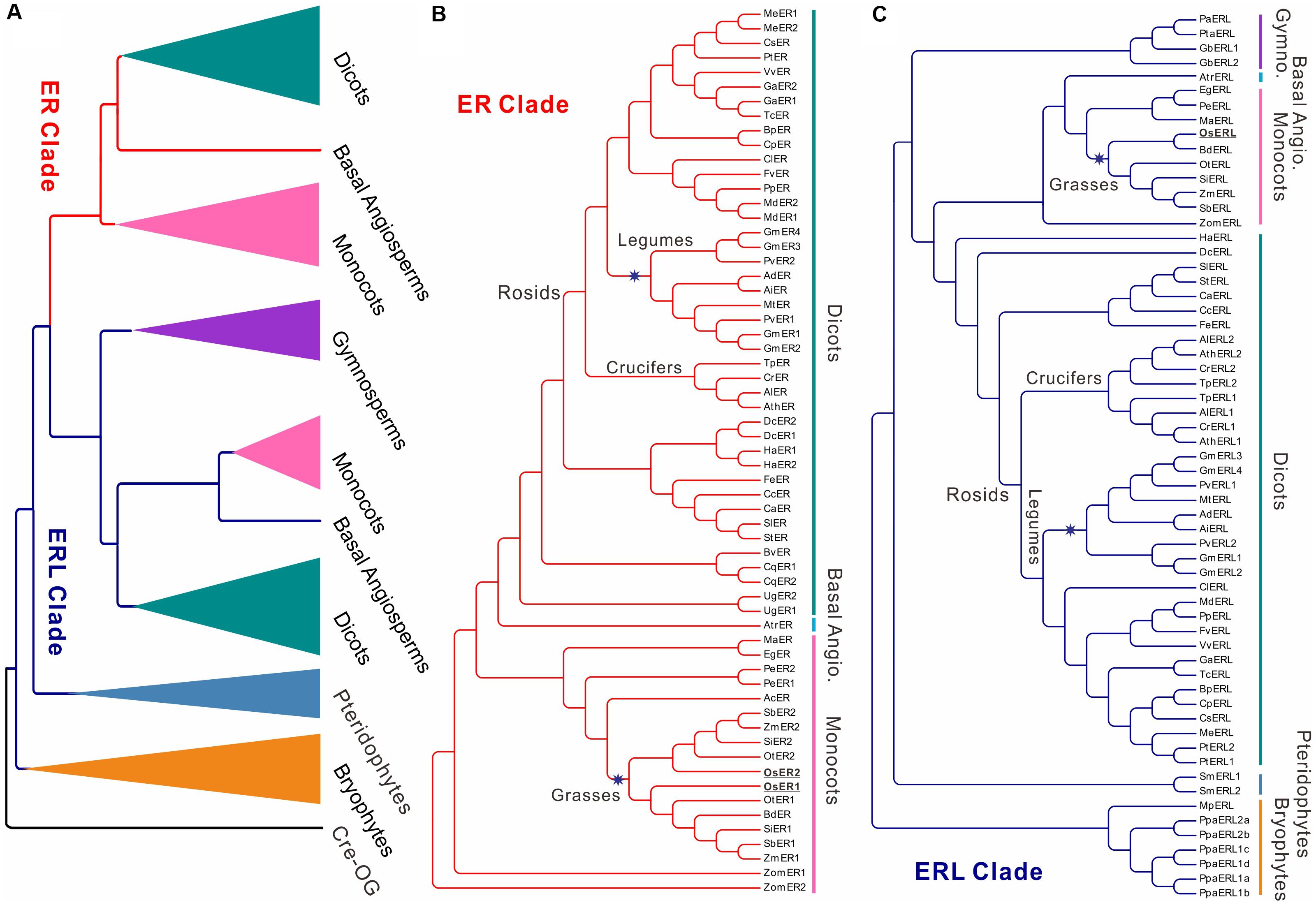
FIGURE 1. Cladogram of ERECTA family genes (ERfs) among 49 plant species. The cladogram was constructed using protein sequences of all identified ERfs. One LRR protein Cre17.g741150.t2.1 (Cre-OG) from Chlamydomonas reinhardtii was picked as an outgroup. Two major clades, ERECTA (ER) clade and ER-LIKE (ERL) clade were identified based on their relationship to Arabidopsis ER and ERL proteins. (A) Simplified cladogram of ER and ERL proteins across major divisions (e.g., kingdoms or sub-kingdoms). Each clade and group are indicated by different colors; (B) Cladogram of all ER proteins across 49 plant species; (C) Cladogram of all ERL proteins across 49 plant species. Major divisions are shown on the right side with the same color schemes as used in (A); two whole-genome duplication (WGD) events specific to grasses and legumes are indicated by blue stars; the rice ER and ERL proteins were highlighted are underlined. The abbreviations for each species were given in Table 1. A full phylogenetic tree with bootstrap values was given in Supplementary Figure S1. Abbreviation: Gymno., Gymnosperms; Basal Angio., Basal Angiosperms.
It is noteworthy that the ERfs of gymnosperms exclusively belong to the ERL clade (Figure 1C), suggesting that the ER clade only emerged from the basal angiosperms. The number of ER genes in each species remains steady across all of the subsequent species with only one or two copies, roughly corresponding to the whole-genome duplication (WGD) events that occurred in each of the species (Figure 1B). The soybean genome possesses the largest ER family with four ER genes and four ERL genes, which were possibly contributed by multiple WGD events (Schmutz et al., 2010). However, the first expansion of the ER family in the soybean genome most likely occurred prior to its divergence from common bean (also containing two copies of ER and ERL), but after their divergence from other legumes (only having one ER and ERL), thus not due to the legume-specific WGD (Figures 1B,C and Table 1).
We have identified three ERfs in the rice genome, including two ER genes, OsER1 (LOC_Os06g10230/Os06g0203800) and OsER2 (LOC_Os02g53720/Os02g0777400), and one ERL gene OsERL (LOC_Os06g03970/Os06g0130100) (Figure 1 and Supplementary Figure S1). Unlike legumes, OsER1 and OsER2 were possibly produced due to the grass-specific WGD (Paterson et al., 2009; Zhang et al., 2012), as most other grass species also contain two copies of the ER genes (Figure 1B and Table 1). Compared to the ER genes, the ERL genes seemed to have an independent evolutionary path, as suggested by the asynchronous copy number variations compared to the ER genes in most species (Figure 1C and Table 1). Besides a possible different evolutionary history of ERs and ERLs, the whole ER family seemed to be highly conserved with no rapid loss or expansion found both across and within species.
In addition to conservation in gene number, the gene structure of ERfs also remained unchanged among most species (Supplementary Table S2). All of the ER genes surveyed here have 22∼29 introns, whereas approximately 90% of the ERL genes contain 25∼27 introns (Supplementary Table S2).
Functional Constraints of ERfs as Revealed by Signatures of Negative Selection Between and Within Species
Molecular signatures of selection have been proven to be a powerful tool in generating functional inferences (Nielsen, 2005). Therefore, we investigated whether the ER family has undergone any selection forces due to its pleiotropic functions. We found a relatively high similarity of ER proteins (78.0% in average) and ERL genes (76.3% in average) across all of the angiosperms, despite a long evolutionary history (Supplementary Table S3). We then compared the number of non-synonymous changes per non-synonymous site (Ka) and the corresponding number of synonymous changes per synonymous site (Ks) values across all of the selected species for each of the ER and ERL genes. Both ER genes as well as the ERL gene displayed 10-fold lower Ka values compared to their Ks values (Supplementary Table S3), indicating strong negative selections. This agrees with the observed conserved function role of the ER family.
A similar signature of negative selection was also detected within rice populations. Among 247 rice cultivars analyzed (Supplementary Table S4), only 12, 4, and 7 single nucleotide polymorphisms (SNPs) with MAF ≥0.05 were detected within the coding sequences (CDS) of OsER1, OsER2, and OsERL, respectively (Supplementary Table S5). OsER1 and OsER2 both harbor only two non-synonymous changes, whereas OsERL contains four polymorphism sites that could alter the encoded amino acids. No insertions/deletions (indels) were found with a MAF ≥0.05 within the coding regions of those genes.
All three genes have limited diversities and small Ka/Ks ratios (Supplementary Table S5), suggesting putative negative selections as observed across species, although this signature was not significant in Tajima’s D-test (Supplementary Table S5). However, one thing that seemed to be clear is that the allele distributions were subjected to the fine structure of the rice population (Supplementary Figure S2A). The population structure of Asian cultivated rice has been extensively reshaped by strong artificial selections, which could be divided into two major subspecies, i.e., indica and japonica, and further subdivided in to five groups: indica (IND), aus (AUS), aromatic (ARO), temperate japonica (TEJ), and tropical japonica (TRJ) (Garris et al., 2005). The selected 247 rice accessions contain four major groups, consisting 120 IND and 27 AUS, which belong to indica subspecies, and 52 TEJ and 48 TRJ, which belong to japonica subspecies (Supplementary Table S4).
Four major haplotype groups (Hap1∼4) of OsER1 were identified based on the linkages of 12 SNP sites (Supplementary Figure S2B). Hap1 showed the highest frequency (∼68%) and was detected in all four of the rice groups (Supplementary Figure S2 and Supplementary Table S6), thus this haplotype could be viewed as the ancestral haplotype that was present before the sub-differentiation of cultivated rice. The Hap2 differed from Hap1 only on one SNP, but was mainly found in TEJ and TRJ from japonica subspecies (Supplementary Figure S2). The two other haplotypes, Hap3 and Hap4, which both distinct from Hap2, could only be found in indica groups (Supplementary Figure S2). There were as few as four SNPs between Hap3 and Hap4 (Supplementary Figure S2). Therefore, those three haplotypes stand for latterly generated polymorphisms during the sub-speciation of indica and japonica rice, and their distances from the ancestral haplotype Hap1 (Supplementary Figure S2) coincided with an earlier origin of japonica rice, which is predated to indica rice (Huang et al., 2012; Yuan et al., 2017).
Unlike OsER1, OsER2 lacks an ancestral haplotype shared by all four of the groups (Supplementary Figure S2), indicating two genes possibly diverged after their derivation from the WGD event in grasses. The subsequent evolution of OsER2 generated three major haplotypes (Hap1 and Hap2 with a frequency over 30%, and Hap3 with a frequency of only 3%) with very limited variants (Supplementary Figure S2B and Supplementary Table S6), and it also followed the differentiation of two subspecies with Hap1, corresponding to indica, and Hap2 and Hap3, corresponding to japonica rice (Supplementary Figure S2A).
In contrast to OsER1 and OsER2, OsERL showed a moderate admixture of different groups (Supplementary Figure S2A), again suggesting an independent evolutionary path of OsERL compared to the OsER genes. Nonetheless, genetic polymorphisms are limited among all of the major haplotypes of OsERL (Supplementary Figure S2B).
OsERfs Have Similar Expression Patterns as Arabidopsis ER Genes
The expression level of the Arabidopsis ER genes is directly linked to their regulatory functions (Shpak, 2013); therefore, we further investigated the expression patterns of OsERfs. Three OsERfs, especially OsER1 and OsERL, were found to share a similar expression level across different organs (Supplementary Figures S3A,C). Three OsERfs showed the highest expression in inflorescences, e.g., spikelet, panicles, and stamens, etc. A high to medium expression level was observed in leaves, shoots, and stems. The roots showed the lowest expression levels. This expression pattern was largely the same as that observed in Arabidopsis (Torii et al., 1996; Supplementary Figure S3C).
The expression levels among different developmental stages were more variable in rice, but the general trend did not differed much from that of Arabidopsis (Supplementary Figures S3B,D). The highest expression level was found in the early stage of vegetative growth, and decreased to an intermediate level at the later phase of the reproductive stage.
In addition to these observed similarities, unlike OsER1 and OsERL, OsER2 also displayed distinct expression patterns, as compared to Arabidopsis. In sperm cells, OsER2 showed extremely high expression levels, whereas OsER1, OsERL, and Arabidopsis ERfs were not expressed (Supplementary Figures S3A,C). OsER2 was also upregulated during heading stage, whereas the other genes were not (Supplementary Figure S3B). These distinct expression patterns indicate that OsER2 possibly plays a distinct role compared to the other rice ER genes.
Generation of OsERf Mutants Using the CRISPR/Cas9 System
Three sgRNAs (spacers) were designed to target each ERf for precise mutagenesis. To minimize the possibility of off-targeted editing, the spacer sequence was designed with no other similar matches (BLAST hits allow no more than five mismatches) except for the target regions. The spacer for OsER1 and OsER2 was designed to target the front-end of their 3rd and 25th exons, respectively (Figures 2A,B), whereas the spacer for OsERL was designed at the 1st exon (Figure 2C). Two rice varieties, O. sativa L. ssp. indica cultivar Kasalath (KA) and O. sativa L. ssp. japonica cultivar Wuyungeng24 (WU), were selected as the recipients for their transformation efficiency. Through Agrobacterium-mediated transformation, the CRISPR/Cas9 construct was introduced into rice embryogenic calli, resulting in 13 regenerated transgenic plants (T0 generation) of KA background and 47 regenerated transgenic plants of WU background. PCR amplification followed by Sanger sequencing was used to confirm whether the vectors were successfully transformed into the transgenic plants (Supplementary Table S7). Since OsER1 and OsER2 share a high similarity, when testing the targeted region in regenerated plants of OsER1, we also verified its best-matched region in OsER2. No mutation was found within the best-matched region of OsER2, suggesting no off-target effect. This was also true for the regenerated plants of OsER2.
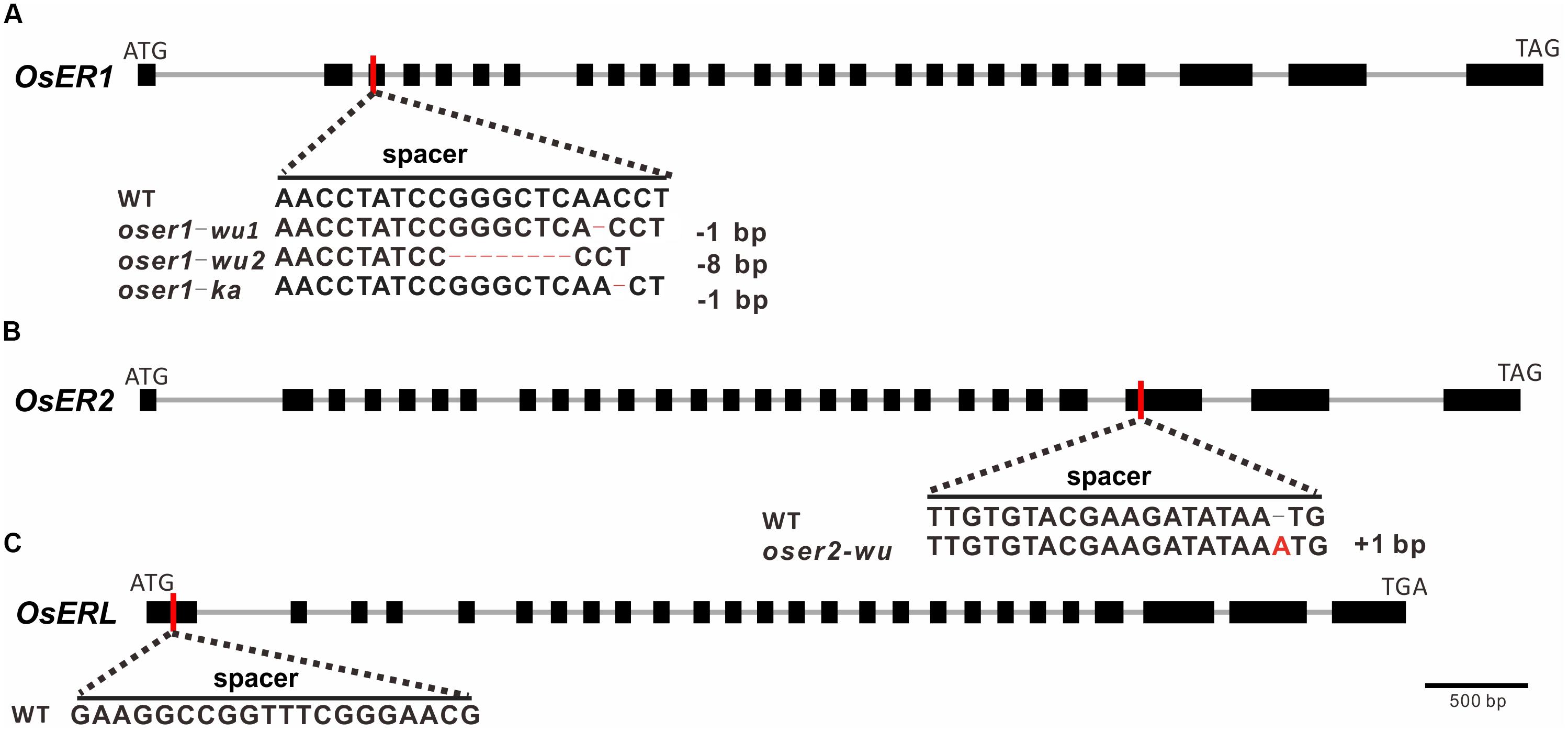
FIGURE 2. Targeted sites, designed spacers, and obtained mutant types for (A) OsER1, (B) OsER2, and (C) OsERL gene. Red vertical lines represent the targeted sites of designed spacers within the coding sequence. Bases edited by the CRISPR/Cas9 system are highlighted with colored bases or dash lines. No mutants were obtained for OsERL after two independent transformation trails.
Of the 13 KA-T0 plants, there were five regenerated plants for OsER1 and eight for OsERL, but no successfully regenerated plants were obtained for OsER2 after three independent transformation trials. Two of the five OsER1-KA plants were successfully mutated at the designed sites (a 40% mutation rate), whereas no mutations were found in the targeted regions of all eight of the OsERL-KA plants compared to the wild type (WT) line. Of the 47 WU-T0 plants, the mutagenesis efficiency was generally higher, with all seven of the OsER1-WU plants and all 20 of the OsER2-WU plants exhibiting successful gene knockout (a 100% mutation rate). Unfortunately, still no mutations were found in the designed regions of all 20 of the OsERL-WU plants. Finally, we failed to generate null mutants for OsERL after another round of independent transformation trials. This raised the possibility that the OsERL functioned as an essential gene that would be lethal when totally lost. In sum, we have obtained nine oser1 mutant plants, with two with a KA background and seven with a WU background, and 20 oser2 mutant plants, all with the WU background.
DNA analysis revealed two mutant types for OsER1-WU plants, one mutant type for OsER1-KA and OsER2-WU plants (Figures 2A,B). Both oser1-wu1 and oser1-ka mutant type contain -1 bp (at position +227 bp and +228 bp of CDS, respectively) frameshift mutations, whereas the oser1-wu2 type has an 8 bp (at position +220 bp) deletion, and oser2-wu harbors a 1 bp (at position +1,940 bp) insertion. All of those frameshift mutations would introduce premature stop codons at protein position 88, 92, 88, and 651 in the oser1-wu1, oser1-wu2, oser1-ka, and oser2-wu mutants, respectively.
OsER Mutants Have Shortened Statures, Reduced Panicle Size, and Seed Setting Rate Compared to the WT
To assess the phenotypic impact of the oser mutants, the T1 generation of three oser1-wu1, six oser1-wu2, six oser1-ka, and 10 oser2-wu plants were obtained through selfing of the T0 plants. During the maturing stage, no obvious phenotypic changes were observed in the oser1-ka mutants. In contrast, all of the oser mutants with the WU background, i.e., oser1-wu1, oser1-wu2, and oser2-wu, showed significantly shortened plant stature as well as reduced panicle size compared to the WT (Figures 3A,B and Supplementary Figures S4A,B). Since no obvious difference was observed between oser1-wu1 and oser1-wu2 mutants, we only focused on oser1-wu1 mutants hereafter to avoid repetition. The oser2-wu mutants exhibited a more severe dwarf phenotype compared to oser1-wu1 (Figures 3A,B and Supplementary Figures S4A,B). Not only the plant stature but also the panicle size of oser2-wu mutants were significantly shorter than oser1-wu1 (two-tailed Students’ t-test, P = 0.00163 for plant height and P = 2.55 ×10-5 for panicle size).
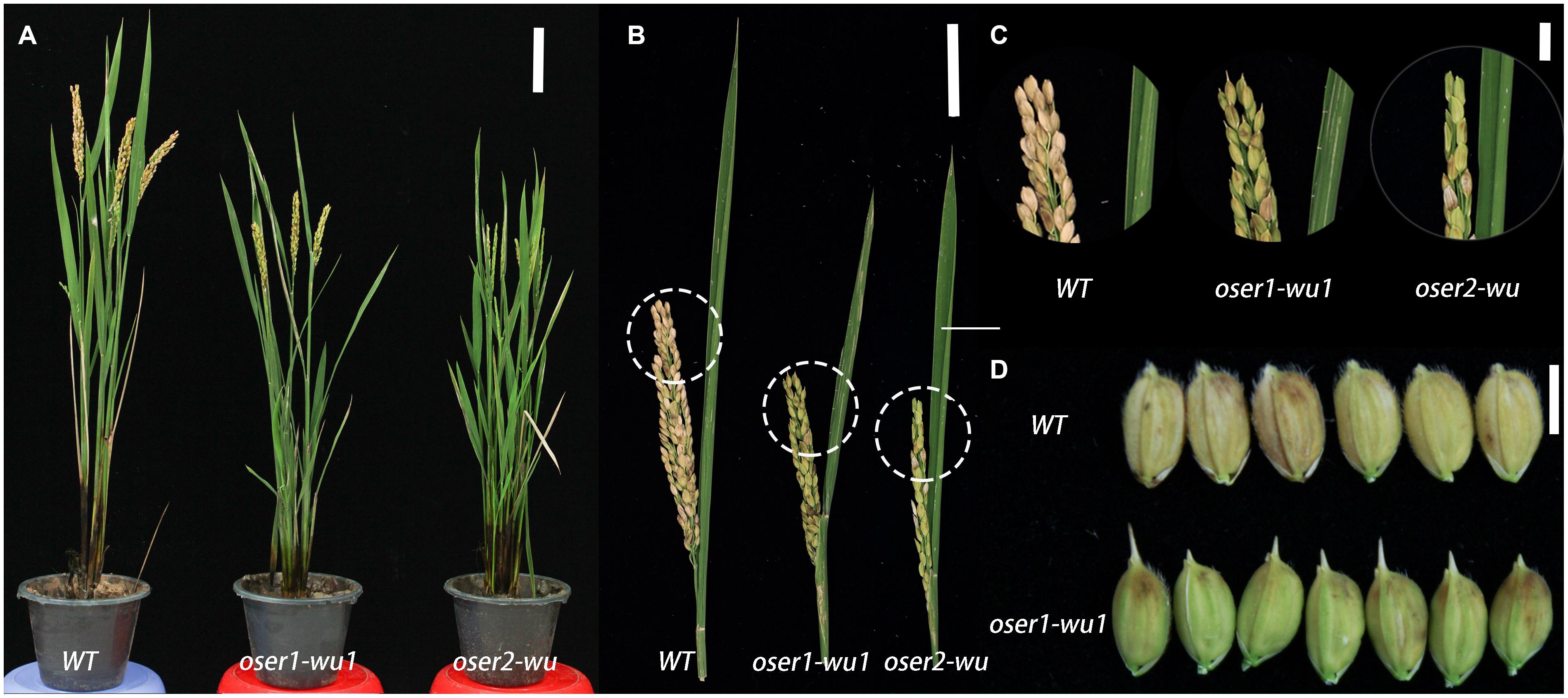
FIGURE 3. Phenotypic analyses of mutant plants for OsER1 and OsER2. (A) Phenotypes of mature plants of oser1-wu1 and oser2-wu mutants. (B) Changes in panicle size in the oser1-wu1 and oser2-wu mutants compared to the WT; (C) Enlarged view of the panicles of each plant as circled in (B); (D) Morphology of seeds of oser1-wu1 compared to the WT, no detectable phenotypic changes observed in seeds of oser2-wu. Bars = 10 cm (A), 5 cm (B), 1 cm (C), and 5 mm (D).
The oser1-wu1 mutant also have slightly elongated awns of seeds compared to WT (Figure 3C), while the similar short awns were not observed in oser2-wu. On the contrary, there was no significant difference of seed setting rate between oser1-wu1 and WT, but a significant impaired seed setting rate was found in oser2-wu (Figure 3D and Supplementary Figure S4C). In conclusion, both OsER genes played a role in control plant height and panicle size of rice WU plants, while OsER2 might be also involved in regulation of reproductive process.
Discussion
The Evolutionary Trajectory of the ER Family
The ER family has been shown to play pleotropic functional roles during various development stages of Arabidopsis (Shpak, 2013); however, whether those functions also occur in other plant species remains unclear. We hypothesize that, owing to the important roles of the ER family, many of its functions are preserved in most plant species. The present study provides four pieces of evidence that support this hypothesis.
First, a gene’s function usually could constraint the evolution route of itself, and in turn, an overview of its phylogeny could provide clues on its function (Eisen, 1998). In this study, we investigated the evolutionary trajectory of the ER family among diverse plant species. A previous study had surveyed 12 plant species, from which the ERfs were found to be present in a very early stage during land-plant evolution (Villagarcia et al., 2012). Here, we expanded the phylogenetic analysis to include a total of 56 plant species (Figure 1, Supplementary Figure S1, Table 1, and Supplementary Table S1), encompassing almost every node of the Viridiplantae, thereby offering a picture with unprecedented high resolution for the evolutionary history of the ER family. We find the ERfs, especially ERs, were rarely lost after their emergence, indicating a functional constraint on those genes.
Second, all of the ERfs have an intact gene structure, which contains multiple introns. In Arabidopsis, the presence of multiple introns in the ER gene was supposed to be essential for its expression, and those introns increase ER expression in an additive manner (Karve et al., 2011). Therefore, the structure of multiple introns or splicing patterns could be a key character of ERfs. The intact gene structure is also found in rice genome, suggesting a preserved ER and ERL gene structure across different plant species, coinciding with its importance in regulating its expression.
However, we did find several species that contain fewer introns within ERLs (Supplementary Table S2), which could be due to two possible reasons. One is that the ERLs, unlike ERs, might face less constraint in their expressions, as suggested by the independent evolution path. Another is that some of these annotated ERLs in public databases were incomplete due to imperfect gene predictions. Here, we prefer the later answer. Even in the well-annotated rice genome, we found that the two most often used database, i.e., MSU and RAP-DB, both predicted a truncated version of OsER2 (Supplementary Figure S5A). Only the NCBI model captures the correct structure of OsER2, which is also supported by the RNA-seq data (Supplementary Figure S5B). Although RAP-DB assigned the “OsER2” symbol to gene Os06g0130100, phylogenetic analysis reveals that this gene actually belongs to the ERL clade, and the bona fide OsER2 gene is the complete version of LOC_Os02g53720/Os02g0777400 as given by the NCBI model (LOC4330905). Therefore, we suggest that caution be exercised in using OsER2 and OsERL from public databases.
The third piece of evidence came from the within-species diversities of rice population. There were only limited polymorphisms among OsERfs, and most diversities were established before rice sub-speciation as implied in OsER1. The sub-speciation process leads to the fixation of differentiated polymorphisms in the OsER1 and OsER2, but not in OsERL, which is in line with their independent evolutionary trajectory. All of the three genes displayed limited variations owing to their functional constraints.
Finally, besides the conservation witnessed at the variants level, an unchanged expression pattern again hints that their functions could be largely preserved in rice compared to Arabidopsis. The expression pattern in Arabidopsis reflects their functional roles in regulating cell proliferation and expansion in vegetative and reproductive organs (Shpak, 2013). Therefore, the observed similar expression patterns between rice and Arabidopsis suggest that most of these functions may also be applicable to rice. In conclusion, all the above evidences suggest that the ER family is one of the most conserved subfamilies in LRR-RLK genes (Liu et al., 2017).
From all above evidences, we could now delineate a picture of how ERfs evolved across different species. Given the ER family’s functional role in stomatal patterning, is it possible that the emergence of the ER family coincided with the emergence of stomata in land plants? However, a contrasting fact is that one ERL gene was found in the liverwort Marchantia polymorpha (Figure 1C and Table 1), which actually lacks stomata (Bowman et al., 2017). Further evidence was found in the genome of seagrass Zostera marina (Figure 1 and Table 1). Z. marina is a marine angiosperm that has lost its stomata as well as all the genes involved in stomatal differentiation (Olsen et al., 2016), whereas the ER family was preserved (Figure 1 and Table 1). These findings suggest that the early ER family may have functioned in other aspects and was only later involved in the regulation networks of stomatal patterning.
The ERLs emerged prior to ERs, and the first ERL appeared with the emergence of the whole LRR-RLK family, which was also absent in algal species (Liu et al., 2017). As mentioned above, the early ERLs may have not differed much from other LRR-RLK members and possibly did not possess those functions specific to ERfs. Since the first ER was only found in Amborella, suggesting the true origin of ERs could be basal angiosperms. Besides a different time of origin, ERs and ERLs also evolved via two independent routes. The expansion of ERs followed well with the WGD events that occurred in different clades, whereas this pattern was rarely observed in the ERLs. At the level of rice populations, the evolution of the ERs consisted of sub-speciation process, whereas the ERLs did not. The independent evolutionary path of the ERs and ERLs may explain the observed distinct functions between ERs and ERLs in some species (Shen et al., 2015).
Combined with the cross-species analyses, we could also delineate the whole history of the three OsERfs. OsER1, and OsER2 originated from a duplication of their common ancestor during the grass-specific WGD, and were both involved in the sub-speciation process of rice population. In contrast, OsERL might has lost one copy after WGD, and its subsequent evolution was less subjected to the rice sub-speciation progress.
The Phenotypic Impacts of Rice ER Genes
The Arabidopsis ER gene was first recognized by its functional roles in contributing to a shortened inflorescence stems, siliques, and pedicels (Torii et al., 1996). A similar phenomenon was observed here in rice with a reduced panicle size in oser1-wu and oser2-wu mutants. In Arabidopsis, three ERfs interacted synergistically in controlling plant organ size, where ERL1 and ERL2 functioned redundantly, and the loss of all ERfs leads to severed dwarfism in plants (Shpak et al., 2004). In rice, the two ERs also functioned with a certain level of redundancy, while the loss of either gene could cause a shortened stature in rice plants. Whether this change in plant height reflected a similar function of rice ERs in cell proliferation and cell growth as Arabidopsis (Shpak et al., 2003) still needs more evidences on cellular level, though. The similarity in the phenotypic impacts of the ER genes in controlling plant architecture between Arabidopsis and rice again emphasizes the conserved functional characters of the ER genes, and suggests that many of their functions as well as mechanisms could be readily to migrate to other plant species, as also indicated by their regulatory roles in thermotolerance among various species (Qi et al., 2004; Shen et al., 2015).
However, some caveats are still necessary when migrating those functions. First, the functions of ERs seemed to be dependent on their genetic background in rice. Distinct from the significantly changed morphological traits in oser1-wu and oser2-wu mutants, the oser1-ka mutation conferred no detectable phenotype. Sequence analysis revealed only one SNP in the penultimate exon between the OsER1-KA allele (belonging to the Hap1 in Supplementary Figure S2) and the OsER1-WU allele (belonging to the Hap2 in Supplementary Figure S2). Another two small indels (≤3 bp) and three SNPs were found between two cultivars within the intron and promoter regions (defined as 3 kb apart from the start codon) of OsER1, respectively. Similarly, the OsER2 differed in only four SNPs between KA (belonging to the Hap1 in Supplementary Figure S2) and WU (belonging to the Hap2 in Supplementary Figure S2), of which two were non-synonymous changes. These limited differences of OsER1 and OsER2 might could not fully explain the distinct phenotypes between oser1-ka and oser1-wu. Since the two selected cultivars belong to different subspecies of rice, this suggests the putative functional diversification of OsERs between indica and japonica rice, which is also depicted in their differentiated evolutionary paths as discussed earlier. It is also curious to note that all of the previous investigation on OsER1 and OsERL were conducted exclusively in japonica rice (Ouyang et al., 2010; Shen et al., 2015). Whether this was simply due to a greater convenience of transformation in japonica rice as compared to indica rice, or was indeed subjected to its genetic backgrounds awaits further confirmation.
Second, although different oser mutants shared many phenotypical changes, the oser2 mutants always have more severed impacts than oser1, indicating unique functional characters for each of the OsER genes besides of their redundant functions. This was more pronounced in their influences on seed setting rate, which was only observed in the mutants of OsER2. The functional role of OsER2 in rice reproduction is in line with its uniquely elevated expression level in sperm cells and during the heading stage (Supplementary Figures S3A,B). Therefore, although OsER1 and OsER2 still share many characters in common, there could have been substantial functional divergence between two genes during grass evolution.
Finally, although we failed to assess the functions of OsERL, some evidence suggests different roles for OsERL compared to OsERs and Arabidopsis ERfs. One piece of evidence was reported previously when dissecting OsER1’s function in thermotolerance, which showed that OsERL is not involved in thermos-response in rice using T-DNA insertional mutant (Shen et al., 2015). Although Shen et al. (2015) also obtained a T-DNA mutant for OsERL and confirmed that its expression level was abolished, the T-DNA was actually inserted into the 24th exon, which was quite near its terminus. The mutants used by Ouyang et al. (2010) were also near the end of the gene, with a Tos17 insertion in the 19th exon and the 23rd intron of OsERL, but they observed that the mutants performed differently under salt and drought treatments. Our OsERL spacer was designed at the very beginning of the gene (Figure 2), which did not successfully generate mutant plants. Therefore, we propose that the basic function of OsERL is essential which could not bear a completely loss, but part of their function, such as stress-tolerance, relies on how many exons were preserved, similar to that observed in Arabidopsis (Karve et al., 2011). Much like the functional roles of OsERL, evolutionary analysis shows that the ERLs underwent an independent evolutionary trajectory compared to ERs, and rice only contains one copy of ERL whereas Arabidopsis has two, thereby resulting in different functional constraints.
Notwithstanding most of these suggestions need to be further tested, the presence of several unique functional characters in rice ERfs compared to Arabidopsis seems to be true. Future exploiting of ERfs into crop development should be aware of these differences.
Author Contributions
LW and JH designed the project. YZ, LW, SL, and SX performed the experiments and analyzed the data. LW wrote the paper. YZ prepared the figures and tables. JH and SY revised the paper. All authors read and approved the final article.
Funding
This work was supported by the National Major Special Project on New Varieties Cultivation for Transgenic Organisms (No. 2016ZX08009001-003), the National Natural Science Foundation of China (31601041, 91631104, and 31671322), and China Postdoctoral Science Foundation (2016M601772).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00473/full#supplementary-material
Footnotes
References
Abraham, M. C., Metheetrairut, C., and Irish, V. F. (2013). Natural variation identifies multiple loci controlling petal shape and size in Arabidopsis thaliana. PLoS One 8:e56743. doi: 10.1371/journal.pone.0056743
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15. doi: 10.1016/j.cell.2017.09.030
De Baets, G., Van Durme, J., Reumers, J., Maurer-Stroh, S., Vanhee, P., Dopazo, J., et al. (2012). SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 40, D935–D939. doi: 10.1093/nar/gkr996
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Eisen, J. A. (1998). Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 8, 163–167. doi: 10.1101/gr.8.3.163
Garris, A. J., Tai, T. H., Coburn, J., Kresovich, S., and McCouch, S. (2005). Genetic structure and diversity in Oryza sativa L. Genetics 169, 1631–1638. doi: 10.1534/genetics.104.035642
Godiard, L., Sauviac, L., Torii, K. U., Grenon, O., Mangin, B., Grimsley, N. H., et al. (2003). ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 36, 353–365. doi: 10.1046/j.1365-313X.2003.01877.x
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Huang, X., Kurata, N., Wei, X., Wang, Z.-X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Ikematsu, S., Tasaka, M., Torii, K. U., and Uchida, N. (2017). ERECTA-family receptor kinase genes redundantly prevent premature progression of secondary growth in the Arabidopsis hypocotyl. New Phytol. 213, 1697–1709. doi: 10.1111/nph.14335
Jordá, L., Sopeña-Torres, S., Escudero, V., Nuñez-Corcuera, B., Delgado-Cerezo, M., Torii, K. U., et al. (2016). ERECTA and BAK1 receptor like kinases interact to regulate immune responses in Arabidopsis. Front. Plant Sci. 7:897. doi: 10.3389/fpls.2016.00897
Karve, R., Liu, W., Willet, S. G., Torii, K. U., and Shpak, E. D. (2011). The presence of multiple introns is essential for ERECTA expression in Arabidopsis. RNA 17, 1907–1921. doi: 10.1261/rna.2825811
Kasulin, L., Agrofoglio, Y., and Botto, J. F. (2013). The receptor-like kinase ERECTA contributes to the shade-avoidance syndrome in a background-dependent manner. Ann. Bot. 111, 811–819. doi: 10.1093/aob/mct038
Katzen, F. (2007). Gateway® recombinational cloning: a biological operating system. Expert Opin. Drug Discov. 2, 571–589. doi: 10.1517/17460441.2.4.571
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. doi: 10.1186/1939-8433-6-4
Kosentka, P. Z., Zhang, L., Simon, Y. A., Satpathy, B., Maradiaga, R., Mitoubsi, O., et al. (2017). Identification of critical functional residues of receptor-like kinase ERECTA. J. Exp. Bot. 68, 1507–1518. doi: 10.1093/jxb/erx022
Li, H. (2013). Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. ArXiv13033997 Q-Bio. Available at: http://arxiv.org/abs/1303.3997 [Accessed May 9, 2016].
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, P.-L., Du, L., Huang, Y., Gao, S.-M., and Yu, M. (2017). Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 17:47. doi: 10.1186/s12862-017-0891-5
Mandel, T., Candela, H., Landau, U., Asis, L., Zelinger, E., Carles, C. C., et al. (2016). Differential regulation of meristem size, morphology and organization by the ERECTA, CLAVATA and class III HD-ZIP pathways. Development 143, 1612–1622. doi: 10.1242/dev.129973
Mandel, T., Moreau, F., Kutsher, Y., Fletcher, J. C., Carles, C. C., and Williams, L. E. (2014). The ERECTA receptor kinase regulates Arabidopsis shoot apical meristem size, phyllotaxy and floral meristem identity. Development 141, 830–841. doi: 10.1242/dev.104687
Masle, J., Gilmore, S. R., and Farquhar, G. D. (2005). The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436, 866–870. doi: 10.1038/nature03835
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Monna, L., Kitazawa, N., Yoshino, R., Suzuki, J., Masuda, H., Maehara, Y., et al. (2002). Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 9, 11–17. doi: 10.1093/dnares/9.1.11
Nielsen, R. (2005). Molecular signatures of natural selection. Annu. Rev. Genet. 39, 197–218. doi: 10.1146/annurev.genet.39.073003.112420
Olsen, J. L., Rouzé, P., Verhelst, B., Lin, Y.-C., Bayer, T., Collen, J., et al. (2016). The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530, 331–335. doi: 10.1038/nature16548
Ouyang, S.-Q., Liu, Y.-F., Liu, P., Lei, G., He, S.-J., Ma, B., et al. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62, 316–329. doi: 10.1111/j.1365-313X.2010.04146.x
Paterson, A. H., Bowers, J. E., Bruggmann, R., Dubchak, I., Grimwood, J., Gundlach, H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556. doi: 10.1038/nature07723
Qi, Y., Sun, Y., Xu, L., Xu, Y., and Huang, H. (2004). ERECTA is required for protection against heat-stress in the AS1/AS2 pathway to regulate adaxial–abaxial leaf polarity in Arabidopsis. Planta 219, 270–276. doi: 10.1007/s00425-004-1248-z
Qu, X., Zhao, Z., and Tian, Z. (2017). ERECTA regulates cell elongation by activating auxin biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 8:1688. doi: 10.3389/fpls.2017.01688
Sakai, H., Kanamori, H., Arai-Kichise, Y., Shibata-Hatta, M., Ebana, K., Oono, Y., et al. (2014). Construction of pseudomolecule sequences of the aus rice cultivar kasalath for comparative genomics of Asian cultivated rice. DNA Res. 21, 397–405. doi: 10.1093/dnares/dsu006
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467.
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Shan, Q., Wang, Y., Li, J., and Gao, C. (2014). Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9, 2395–2410. doi: 10.1038/nprot.2014.157
Shen, H., Zhong, X., Zhao, F., Wang, Y., Yan, B., Li, Q., et al. (2015). Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato. Nat. Biotechnol. 33, 996–1003. doi: 10.1038/nbt.3321
Shpak, E. D. (2013). Diverse roles of ERECTA family genes in plant development. J. Integr. Plant Biol. 55, 1238–1250. doi: 10.1111/jipb.12108
Shpak, E. D., Berthiaume, C. T., Hill, E. J., and Torii, K. U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131, 1491–1501. doi: 10.1242/dev.01028
Shpak, E. D., Lakeman, M. B., and Torii, K. U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor–like kinase signaling pathway that regulates organ shape. Plant Cell 15, 1095–1110. doi: 10.1105/tpc.010413
Shpak, E. D., McAbee, J. M., Pillitteri, L. J., and Torii, K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290–293. doi: 10.1126/science.1109710
Takahashi, T., Shibuya, H., and Ishikawa, A. (2016). ERECTA contributes to non-host resistance to Magnaporthe oryzae in Arabidopsis. Biosci. Biotechnol. Biochem. 80, 1390–1392. doi: 10.1080/09168451.2016.1151345
Tameshige, T., Ikematsu, S., Torii, K. U., and Uchida, N. (2017). Stem development through vascular tissues: EPFL–ERECTA family signaling that bounces in and out of phloem. J. Exp. Bot. 68, 45–53. doi: 10.1093/jxb/erw447
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
The 3,000 Rice Genomes Project (2014). The 3,000 rice genomes project. Gigascience 3:7. doi: 10.1186/2047-217X-3-7
Torii, K. U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R. F., et al. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. doi: 10.1105/tpc.8.4.735
Uchida, N., Igari, K., Bogenschutz, N. L., Torii, K. U., and Tasaka, M. (2011). Arabidopsis ERECTA-family receptor kinases mediate morphological alterations stimulated by activation of NB-LRR-type UNI proteins. Plant Cell Physiol. 52, 804–814. doi: 10.1093/pcp/pcr032
van Zanten, M., Snoek, L. B., Proveniers, M. C. G., and Peeters, A. J. M. (2009). The many functions of ERECTA. Trends Plant Sci. 14, 214–218. doi: 10.1016/j.tplants.2009.01.010
Villagarcia, H., Morin, A.-C., Shpak, E. D., and Khodakovskaya, M. V. (2012). Modification of tomato growth by expression of truncated ERECTA protein from Arabidopsis thaliana. J. Exp. Bot. 63, 6493–6504. doi: 10.1093/jxb/ers305
Wang, L., Zhang, Y., Qin, C., Tian, D., Yang, S., and Hurst, L. D. (2016). Mutation rate analysis via parent–progeny sequencing of the perennial peach. II. No evidence for recombination-associated mutation. Proc. R. Soc. B 283:20161785. doi: 10.1098/rspb.2016.1785
Yuan, Y., Zhang, Q., Zeng, S., Gu, L., Si, W., Zhang, X., et al. (2017). Selective sweep with significant positive selection serves as the driving force for the differentiation of japonica and indica rice cultivars. BMC Genomics 18:307. doi: 10.1186/s12864-017-3702-x
Keywords: ERECTA gene, phylogeny, homologs, rice population genetics, gene function, CRISPR/Cas9
Citation: Zhang Y, Li S, Xue S, Yang S, Huang J and Wang L (2018) Phylogenetic and CRISPR/Cas9 Studies in Deciphering the Evolutionary Trajectory and Phenotypic Impacts of Rice ERECTA Genes. Front. Plant Sci. 9:473. doi: 10.3389/fpls.2018.00473
Received: 03 February 2018; Accepted: 26 March 2018;
Published: 10 April 2018.
Edited by:
Renchao Zhou, Sun Yat-sen University, ChinaReviewed by:
Jing Ding, Nanjing Agricultural University, ChinaJinfeng Chen, University of California, Riverside, United States
Copyright © 2018 Zhang, Li, Xue, Yang, Huang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ju Huang, cG9ueWh1YW5nQG5qdS5lZHUuY24= Long Wang, d2FuZ2xvbmdAbmp1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Yanchun Zhang†
Yanchun Zhang† Sihai Yang
Sihai Yang Long Wang
Long Wang