- 1College of Bioscience and Biotechnology, Shenyang Agricultural University, Shenyang, China
- 2College of Horticulture, Shenyang Agricultural University, Shenyang, China
- 3Vegetable Research Institute, Liaoning Academy of Agricultural Sciences, Shenyang, China
- 4Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang Agricultural University, Shenyang, China
Pathogen stress often significantly decreases cucumber production. However, knowledge regarding the molecular mechanism and signals of cucumber disease resistance is far from complete. Here, we report two translationally controlled tumor protein genes, CsTCTP1 and CsTCTP2, that are both negative modulators in the Cucumis sativus defense response to Sphaerotheca fuliginea. Subcellular localization analysis showed that CsTCTP1 and CsTCTP2 were both localized in the cytoplasm. Expression analysis indicated that the transcript levels of CsTCTP1 and CsTCTP2 were linked to the degree of cucumber resistance to S. fuliginea. Transient overexpression of either CsTCTP1 or CsTCTP2 in cucumber cotyledons impaired resistance to S. fuliginea, whereas silencing of either CsTCTP1 or CsTCTP2 enhanced cucumber resistance to S. fuliginea. The relationship of several defense-related genes and ABA and target of rapamycin (TOR) signaling pathway-related genes to the overexpressing and silencing of CsTCTP1/CsTCTP2 in non-infested cucumber plants was investigated. The results indicated that CsTCTP1 participates in the defense response to S. fuliginea by regulating the expression of certain defense-associated genes and/or ABA signaling pathway-associated genes, and CsTCTP2 participates through regulating the expression of TOR signaling pathway-associated genes. Our findings will guide enhancing the resistance of cucumber to powdery mildew.
Introduction
Powdery mildew (PM) caused by Sphaerotheca fuliginea occurs worldwide and is one of the most destructive cucumber diseases in many mild and damp regions (Schulze-Lefert and Vogel, 2000). PM fungi are biotrophic parasites that spread through the air and greatly depress yields worldwide. After a conidial spore absorbs on the host surface, the PM fungus builds one or two germ tubes. An appressorial germ tube develops an appressorium and then penetrates the host cell wall (Hückelhoven, 2005). After the first barrier (the host cell wall) is defeated, the PM fungus develops a haustorium and takes nutrients from the host. A hypha simultaneously continues to spread on the host epidermis. The host defense that inhibits nutrient uptake is thus considered the second barrier.
Increasing amounts of plant data indicate that similar to other eukaryotic phyla, the target of rapamycin (TOR) acts as a master regulator of nutrient, hormone and stress signaling networks (Robaglia et al., 2012; Xiong and Sheen, 2015). Unlike the TOR kinases of animals and yeasts, the plant TOR kinase is present in a TORC1 protein complex, which consists of the TOR, the regulatory-associated protein of TOR (Raptor) and lethal with SEC13 protein 8 (LST8) (Anderson et al., 2005; Laplante and Sabatini, 2012; Moreau et al., 2012; Maegawa et al., 2015). TOR is a conserved Ser/Thr protein kinase, and its N-terminal region interacts with Raptor WD repeats (Adami et al., 2007). Raptor may be involved in substrate recruitment for phosphorylation by TOR, and LST8 might stabilize the complex (Hara et al., 2002; Turck et al., 2004). Evidence also suggests that possible TOR substrates, such as ribosome protein S6 kinase (S6K) and the type 2A-phosphatase-associated protein of 46 kDa (TAP46, a regulator of PP2A phosphatases), are well conserved in plants (Nojima et al., 2003; Ahn et al., 2014). In Arabidopsis, TOR signaling is inhibited upon the interaction of AtRaptor1 and the AtS6K1 N-terminal 44-amino acid region (Son et al., 2016). Cucumber only harbors one S6K gene, and this gene is very similar to AtS6K1 and AtS6K2 (approximately 70% identity; Pu et al., 2017). In animals, TORC1 is inactivated upon phosphorylation of the Raptor protein by AMP-activated kinase (AMPK). Sucrose non-fermenting 1-related kinase 1 (SnRK1), the plant homolog of AMPK, is deduced to have similar functions in inactivating plant TORC1 (Gwinn et al., 2008; Dobrenel et al., 2016). Under stress conditions, SnRK1 is activated to suppress energy-demanding processes, which are positive regulated by TOR, and activate stress-dependent energy-generating processes, thus maintaining energy homeostasis (Baena-González and Sheen, 2008; Jossier et al., 2009).
Plants can escape from pathogen assault owing to a plethora of factors. The expression of defense-associated genes is also an important part in plant–pathogen interactions. For instance, pathogenesis-related protein-1a (PR-1a) and 14-3-3 genes are also thought to be involved in the response of cucumber plants to S. fuliginea (Liu, 2008; Zhang, 2016). Additionally, chitinase, a secondary hydrolytic enzyme, can inhibit hyphal growth by hydrolyzing chitin in the cell wall of mycelial growth tips and thus display resistance against fungal pathogens in plants (Luo et al., 2015). The cucumber pathogen-induced protein CuPi1, a phloem lectin protein, was more abundant in response to S. fuliginea invasion in susceptible cucumber lines than resistant lines (Meng et al., 2016).
The translationally controlled tumor protein (TCTP) is a highly conserved multifunctional protein in Eukarya. Published data revealed that animal TCTP is implicated in an amazing number of cellular events, including cell growth, differentiation, development of organ size, apoptosis, signaling, and stimulus and immune responses (Yang et al., 2005; Hsu et al., 2007; Schmidt et al., 2007; Susini et al., 2008; Vonakis et al., 2008). Great progress has been made in understanding TCTP functions in animals and yeasts, but knowledge of this gene in plants has lagged. The Arabidopsis TCTP gene rescues the corresponding Drosophila TCTP mutant (Brioudes et al., 2010), which underscores the similarity of its function with those of plant and non-plant homologs. Emerging evidence suggests a role of plant TCTP in growth regulation and programmed cell death control (Berkowitz et al., 2008; Toscano-morales et al., 2015; Chou et al., 2016). Plant TCTP was also recently found to respond to pathogen stress (Li et al., 2010; Hinojosamoya et al., 2013; Bruckner et al., 2017). In our previous work, we found that TCTP (XP-004134215) was significantly differentially expressed between two sister cucumber lines (genotypes that were either highly resistant or highly susceptible to S. fuliginea) by 2-D gel electrophoresis (Fan et al., 2014). Cucumber has 2 TCTP genes, CsTCTP1 (XP-004134215) and CsTCTP2 (XP-004135602). Though CsTCTP1 and CsTCTP2 share a high degree of homology, the functions of CsTCTPs to different abiotic stresses may differ (Meng et al., 2017). The molecular and biochemical mechanisms by which TCTP functions in response to S. fuliginea have not been completely established.
TCTP, which is similar to members of the mammalian/dominant suppressor of sec4 (MSS4/DSS4) protein family, contains a domain that putatively binds Rab GTPase. TCTP is also a vital component of the TOR signaling network (Berkowitz et al., 2008; John et al., 2011). Indeed, Arabidopsis TCTP can bind to four AtRab GTPases (AtRABA4a, AtRABA4b, AtRABF1, and AtRABF2b) and interact with Drosophila dRheb GTPases (Brioudes et al., 2010). Additionally, the interaction between TCTP and Rheb is regulated by 14-3-3 (Le et al., 2016). However, the specific relationships between CsTCTP and the TOR signaling pathway in response to S. fuliginea remain unknown.
CsTCTP1 and CsTCTP2 are both regulated by exogenous abscisic acid (ABA) treatment (Meng et al., 2017). Moreover, AtTCTP overexpression enhanced drought tolerance in Arabidopsis and played a role in ABA-mediated stomatal movement (Kim et al., 2012). Accumulating evidence indicates that plant TCTP is related to ABA signal transduction. ABA is also a critical signaling molecule that mediates plant defense responses against pathogen stress (Cao et al., 2011). The ABA signal transduction core components of plants contain ABA receptors (PYR/PYL/RCARs), group A type 2C protein phosphatases (PP2Cs) and subclass III SNF 1-related protein kinases 2 (SnRK2s). The ABA-insensitive (ABI5) gene is thought to be a downstream target of the ABA signaling pathway. In cucumbers, PYL2, PP2C2, and SnRK2.2 are reportedly involved in transducing ABA signals (Wang et al., 2012). The specific relationships between CsTCTP and the TOR signaling pathway in response to S. fuliginea are similarly poorly understood.
In this study, the subcellular localization and expression patterns of CsTCTP1 and CsTCTP2 were determined. Investigation of their defense roles via transient CsTCTP1/CsTCTP2-overexpression and CsTCTP1/CsTCTP2-silencing cucumber plants elucidated that CsTCTP1 and CsTCTP2 both function as negative regulators in host resistance response to S. fuliginea. Furthermore, CsTCTP1 might participate in the defense response to S. fuliginea through regulating the expression of certain defense-associated genes and/or ABA signaling pathway-associated genes, and CsTCTP2 may act by regulating the expression of TOR signaling pathway-associated genes.
Materials and Methods
Plant Materials and Treatments
The cucumber lines/cultivars used in this study were B21-a-2-2-2, XinTaiMiCi, JingYan4, GY14, and B21-a-2-1-2. B21-a-2-2-2 (highly susceptible to S. fuliginea) and B21-a-2-1-2 (highly resistant to S. fuliginea) are two sister cucumber lines that were obtained from the Liaoning Academy of Agricultural Sciences. XinTaiMiCi is susceptible to S. fuliginea and was selected for gene transformation. GY14, an important processing cultivar in North America, is provided by Shanghai Jiao Tong University. JingYan4 is an important planting variety in China that is resistant to downy mildew, PM, and so on.
Cucumber plants were grown in a growth chamber with day/night temperatures of 25/22°C and a 16-h light photoperiod. For rapamycin treatment, XinTaiMiCi cucumbers were grown in a liquid culture to which 10 μM rapamycin had been added. Seven-day-old seedlings of five cucumber lines/cultivars were inoculated with S. fuliginea by spore spray as described previously (Ren, 2011). The cotyledons collected at day 7 of S. fuliginea treatment were placed in liquid nitrogen and stored at -80°C until future study.
Subcellular Localization
The full-length CsTCTP1 and CsTCTP2 ORFs without their stop codons were separately amplified by PCRs and ligated to the 3′ end of the green fluorescent protein (GFP) coding region, yielding p35S:GFP-CsTCTP1 and p35S:GFP-CsTCTP2. The resulting p35S:GFP-CsTCTP1, p35S:GFP-CsTCTP2, and control p35S:GFP plasmids were separately injected into Nicotiana benthamiana leaves and Arabidopsis thaliana protoplasts following the approach of Zhang et al. (2017) and Yoo et al. (2007). The plants were incubated at 22°C for 48 h after injection, at which point GFP signals were observed and photographed using a laser confocal florescence microscope (Leica, TSC SP8, Germany) with an excitation wavelength of 488 nm and a 505–530 nm bandpass emission filter.
TRV-Mediated CsTCTP1 and CsTCTP2 Gene Silencing in Cucumber Cotyledons
A 318 bp sequence of CsTCTP1 (from nucleotides 1 to 318 in the CsTCTP1 cDNA sequence) or a 318 bp sequence of CsTCTP2 (from nucleotides 1 to 318 in the CsTCTP2 cDNA sequence) was subcloned in an antisense orientation into the plasmid pTRV2, yielding pTRV2-CsTCTP1 and pTRV2-CsTCTP2, respectively, which were then introduced into different Agrobacterium tumefaciens EHA105 aliquots. Cotyledons of 8-day-old cucumber seedlings were injected with a mixture (1:1, ratio, v/v) of Agrobacterium cultures containing pTRV1 and a pTRV2, pTRV2-CsTCTP1, or pTRV2-CsTCTP2 plasmid using a needleless syringe. The cotyledons at 7 days post-injection with TRV:00, TRV:CsTCTP1, or TRV:CsTCTP2 were photographed to record their phenotypes. Cotyledons at 14 days post-injection were collected for RNA extraction and molecular detection experiments. Cotyledons at 7 days post-injection were used for S. fuliginea inoculation and fungal biomass detection.
CsTCTP1 and CsTCTP2 Overexpression Transformation Vector and Cucumber Transient Transformation
The cDNAs of CsTCTP1 and CsTCTP2 were subcloned into separate pCAMBIA3301 binary vectors modified with Xba I and Pst I, where they were placed under the control of the 35S promoter. The vector contains a luciferase reporter gene. LUC:00, LUC:CsTCTP1, and LUC:CsTCTP2 vectors were individually transferred into A. tumefaciens strain EHA105.
Agrobacterium tumefaciens EHA105 cells harboring the LUC:00, LUC:CsTCTP1, or LUC:CsTCTP2 vector were cultured to OD600 = 0.8 in induction medium (10 mM MES, 10 mM MgCl2, and 200 mM acetosyringone) and then diluted to OD600 = 0.4. The diluted culture was injected into 8-day-old cucumber cotyledons using a syringe without a needle (Shang et al., 2014). For luciferase assays, 7-day post-injection cotyledons were punched into 1.5-cm diameter round pieces, soaked in 0.3125 mg mL-1 D-luciferin for 45 min and then photographed by an in vivo Plant Imaging System (Berthold, Lb985). For molecular detection experiments, cotyledons with LUC:00, LUC:CsTCTP1, or LUC:CsTCTP2 were collected at 5 days post-injection, placed in liquid nitrogen and stored at -80°C until further studied. Cotyledons with LUC:00, LUC:CsTCTP1 or LUC:CsTCTP2 at 1 day post-injection were used for S. fuliginea treatment for fungal biomass detection.
PCR and qRT-PCR Analysis
Total RNA was isolated using an RNAprep pure Plant Kit(DP432, TIANGEN, China), and cDNA was synthesized by a QuantScript RT Kit (KR103-04, TIANGEN, China). The presence of the introduced CsTCTP1 or CsTCTP2 gene in the transformed cucumber plants was detected by PCRs using a specific primer pair. The two forward primers were CsTCTP1-F, targeting part of the CsTCTP1 gene region, and CsTCTP2-F, targeting part of the CsTCTP2 gene region, and the reverse primer for both cases was LUC-R, targeting part of the luciferase sequence region of the transformation vector. PCRs were conducted in a 20 μL volume containing 1 μL of genomic DNA, 2 μL of Taq DNA Polymerase (TIANGEN), 2 μL of 10 × Taq buffer, 0.5 μL of each primer (10 μM), and 12.5 μL of doubly distilled water.
The relative expression levels of CsTCTP1, CsTCTP2, defense-related genes, and TOR- and ABA-signaling-related genes were analyzed by qRT-PCR. The qRT-PCRs were performed on a SYBR Green I 96-I system (Roche fluorescence quantitative PCR instrument, Basel). The cucumber actin gene, CsActin, was used as an internal reference. The gene-specific primers that were employed are shown in Supplementary Table S1. The expression patterns of the target genes were calculated using the 2-ΔΔCT method. Three independent biological replicates and three technical replicates for each biological replicate were performed, and significance was determined by t-test using SPSS statistical software (P < 0.05).
Coomassie Brilliant Blue R250 Staining and Microscopy
Coomassie brilliant blue R250 staining was used for observing the S. fuliginea hyphae on the cotyledons of control, LUC:00, LUC:CsTCTP1, LUC:CsTCTP2, TRV:00, TRV:CsTCTP1, and TRV:CsTCTP2 transgenic plants at day 7 with S. fuliginea. Control and transgenic infected cotyledons were immersed in destaining solution (containing 22.5 mg of trichloroacetic acid, 15 mL of absolute ethanol, and 5 mL of trichloromethane) for 1 h at 70°C. The samples were then stained with Coomassie brilliant blue R250 solution (containing 3 mg of trichloroacetic acid, 20 mL of H2O, 120 mg of Coomassie brilliant blue R250, and 20 mL of methanol) for 15 min at room temperature. After the samples had been washed with sterile water, they were immersed in 20% glycerol and photographed. The fungus was visualized under a light microscope (Nikon).
Disease Index
The typical symptoms of pathogen whiteness were measured in six disease severity ratings from 0 to 9, where 0 = no symptoms, 1 = 5% of inoculated leaves were whitish, 3 = 5–30% of inoculated leaves were whitish, 5 = 30–50% of inoculated leaves were whitish, 7 = 50–75% of inoculated leaves were whitish, 9 = 75–100% of the inoculated leaves were whitish, and plant death. A Disease Index (DI) was calculated using the following formula: DI (%) = [Σ (N × D)/(H × T)] × 100, where N = number of plants with the respective disease rating; D = the disease rating; H = the highest disease rating; and T = the total number of observed plants. No fewer than 30 cucumbers were surveyed for DI evaluation.
Statistical Analysis
Primer design and sequence alignment were conducted in Primer 5 software. Standard errors of deviation were assessed by Excel. Statistical significance was analyzed by Student’s t-test (P < 0.05) using SPSS software.
Results
Molecular Characterization and Expression Patterns of CsTCTP1 and CsTCTP2
Cucumber harbors two TCTP paralogs with 77% nucleic acid identity, CsTCTP1 (XP-004134215), and CsTCTP2 (XP-004135602), both of which encoded 168 amino acids. Sequence analysis showed that both CsTCTP1 and CsTCTP2 proteins contained conserved TCTP motifs, such as a domain that putatively binds Rab GTPase and TCTP1 (45–55) and TCTP2 (125–147) domains. To explore the subcellular localization of CsTCTP1 and CsTCTP2, we separately infiltrated and transiently transformed p35S:GFP-CsTCTP1, p35S:GFP-CsTCTP2, and control p35S:GFP vectors into N. benthamiana leaves and A. thaliana protoplasts. Confocal microscope examination indicated the exclusive localization of p35S:GFP-CsTCTP1 and p35S:GFP-CsTCTP2 in the cytoplasm, whereas only GFP alone was found in multiple subcellular compartments, including the cytoplasm and nucleus (Figure 1). The results are in accordance with in silico analysis data predicting that CsTCTP1 and CsTCTP2 are both localized in cytoplasm (Chen, 2017).
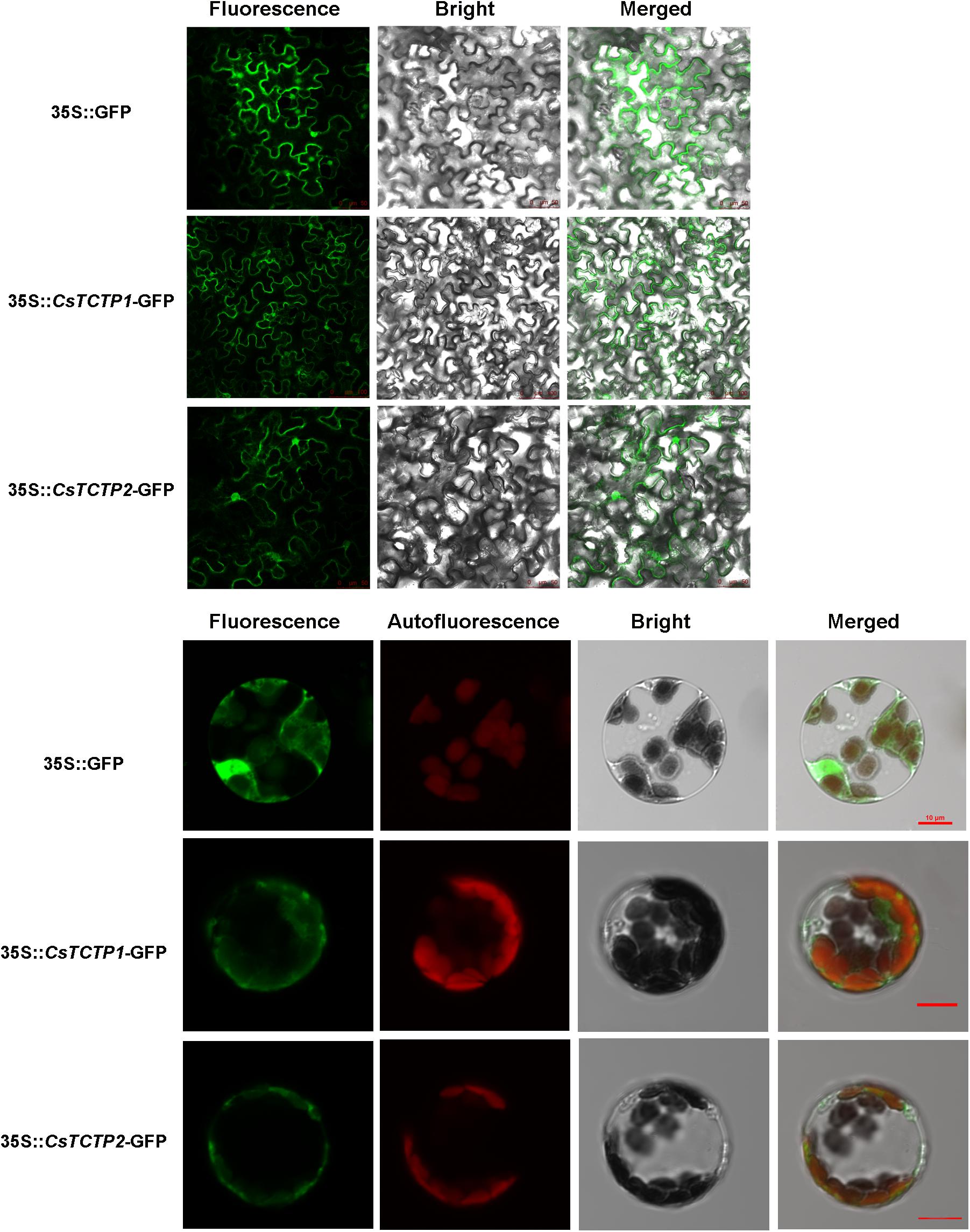
FIGURE 1. Subcellular localization of GFP:CsTCTP1 and GFP:CsTCTP2 fusion proteins in N. benthamiana cells and Arabidopsis protoplasts. Fluorescence, chloroplast autofluorescence, bright field, and merged image were obtained using a Leica confocal microscope.
The patterns of CsTCTP1 and CsTCTP2 expression were investigated in five cucumber lines with different degrees of resistance, including the highly susceptible line B21-a-2-2-2, the susceptible line XinTaiMiCi, the moderately resistant lines JinYan 4 and GY14, and the highly resistant line B21-a-2-1-2, with or without S. fuliginea inoculation for 7 days (Supplementary Figure S1). Without S. fuliginea inoculation, the expression levels of CsTCTP1 and CsTCTP2 were not significantly different in the five cucumber lines. With S. fuliginea infestation, CsTCTP1 and CsTCTP2 transcription increased in susceptible lines and declined in resistant lines. The maximum expression of CsTCTP1 and CsTCTP2 occurred in the B21-a-2-2-2 line with S. fuliginea treatment. The results demonstrated that the different expression patterns of CsTCTP1 and CsTCTP2 in S. fuliginea-susceptible and S. fuliginea-resistant plants after pathogen inoculation are closely linked to the magnitude of cucumber resistance to S. fuliginea and that CsTCTP1 and CsTCTP2 may be involved in the cucumber defense response to S. fuliginea.
CsTCTP1 or CsTCTP2 Silencing Improves Resistance to S. fuliginea in Transgenic Cucumber
To elucidate whether CsTCTP1 or CsTCTP2 was required for cucumber resistance to S. fuliginea, we used tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) to silence CsTCTP1/CsTCTP2 transcripts in the susceptible line XinTaiMiCi. A 5′-terminal fragment specific to CsTCTP1/CsTCTP2 was inserted in an antisense orientation into vector pTRV2, yielding the pTRV2-CsTCTP1/CsTCTP2 recombinant plasmid (Figure 2A). When either the TRV:CsTCTP1 or TRV:CsTCTP2 virus was injected with TRV:00 for 7 days, the chlorotic mosaic symptoms of TRV emerged in the cotyledons of transgenic cucumber plants; however, no symptoms appeared in the control and EHA105-injected cucumbers, suggesting that TRV successfully invaded the cucumber plants injected with TRV:00 and either TRV:CsTCTP1 or TRV:CsTCTP2 (Figure 2B). The transcript levels of CsTCTP1 in TRV:CsTCTP1 transgenic plants and CsTCTP2 in TRV:CsTCTP2 transgenic plants were markedly lower than those of CsTCTP1 and CsTCTP2, respectively, in TRV:00-injected plants (Figure 2C). These results indicated that CsTCTP1/CsTCTP2-silenced plants were successfully generated.
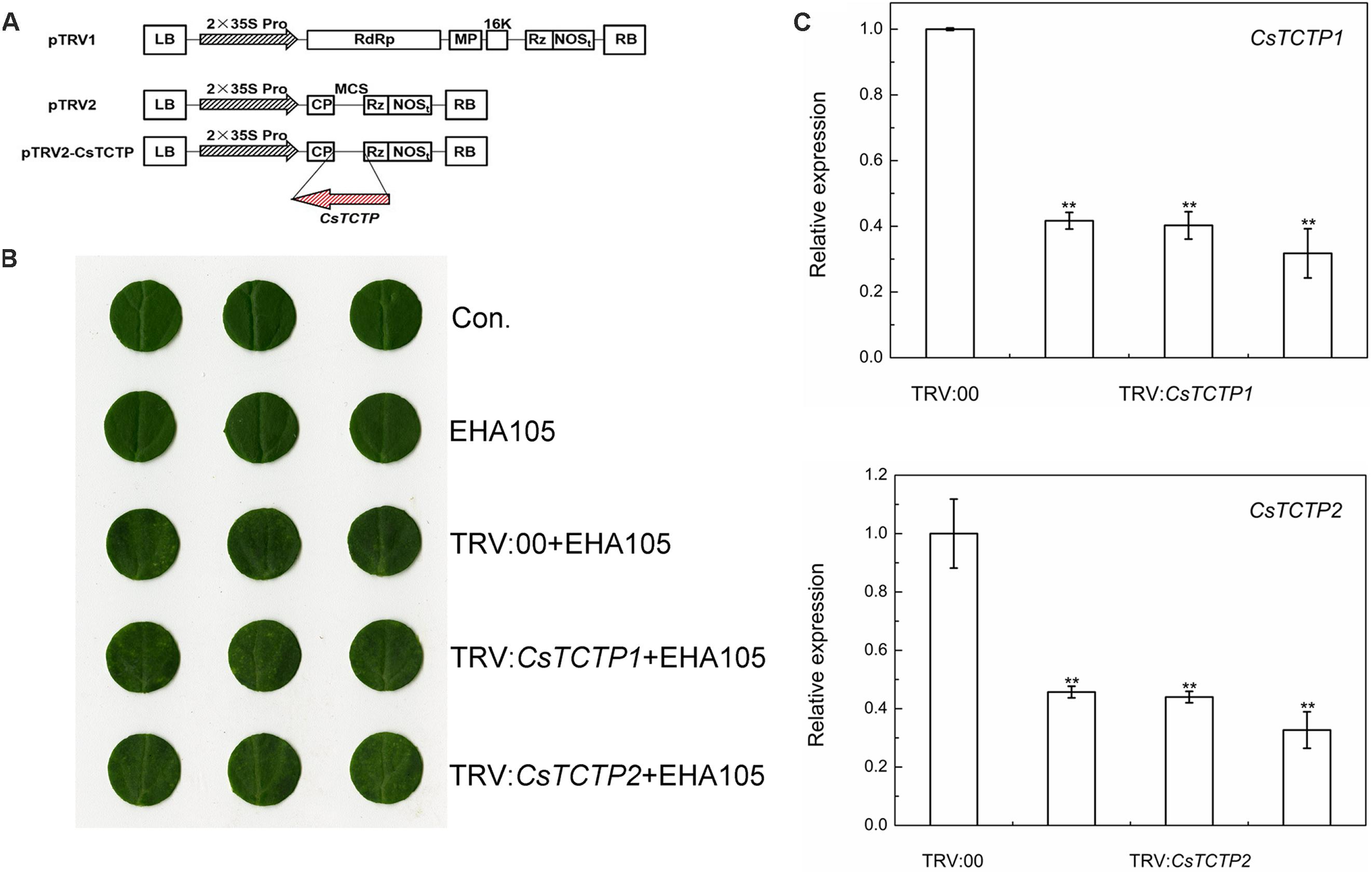
FIGURE 2. Identification of CsTCTP-silenced cucumber plants. (A) CsTCTP1- and CsTCTP2-silencing transformation vectors pTRV-CsTCTP1 and pTRV-CsTCTP2. The arrows show the direction of the inserted fragments. (B) Symptoms in detached cotyledons of silenced cucumber plants. Con. indicates non-injected plants, and EHA105 indicates EHA105-injected plants. (C) qRT-PCR analyses of the expression levels of CsTCTP1 in cucumber plants injected with TRV:00 or TRV:CsTCTP1 and the expression levels of CsTCTP2 in cucumber plants injected with TRV:00 or TRV:CsTCTP2. Statistically significant differences were identified based on three technical replicates (t-test; ∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean values, and data are presented as the mean value ± SE of three biological replicates.
These CsTCTP1-silenced cucumbers and CsTCTP2-silenced cucumbers both exhibited significantly enhanced resistance to S. fuliginea after 7 days of infection (Figure 3A). Many hyphae and branch spores invaded control and TRV:00-injected plants, but only a few initial germ tubes and appressoria were observed in CsTCTP1/CsTCTP2-silenced cucumbers, suggesting that CsTCTP1/CsTCTP2-silencing increases resistance to hyphal development of S. fuliginea (Figure 3A). Furthermore, the DI of the CsTCTP1-silenced plants that had been inoculated with S. fuliginea was 14.81, and the DI of the inoculated CsTCTP2-silenced plants was 16.86, whereas the DIs of the control and TRV:00-injected plants were 27.35 and 25.40, respectively (Figure 3A). These results indicated that the silencing of CsTCTP1 or CsTCTP2 enhanced plant resistance to S. fuliginea.
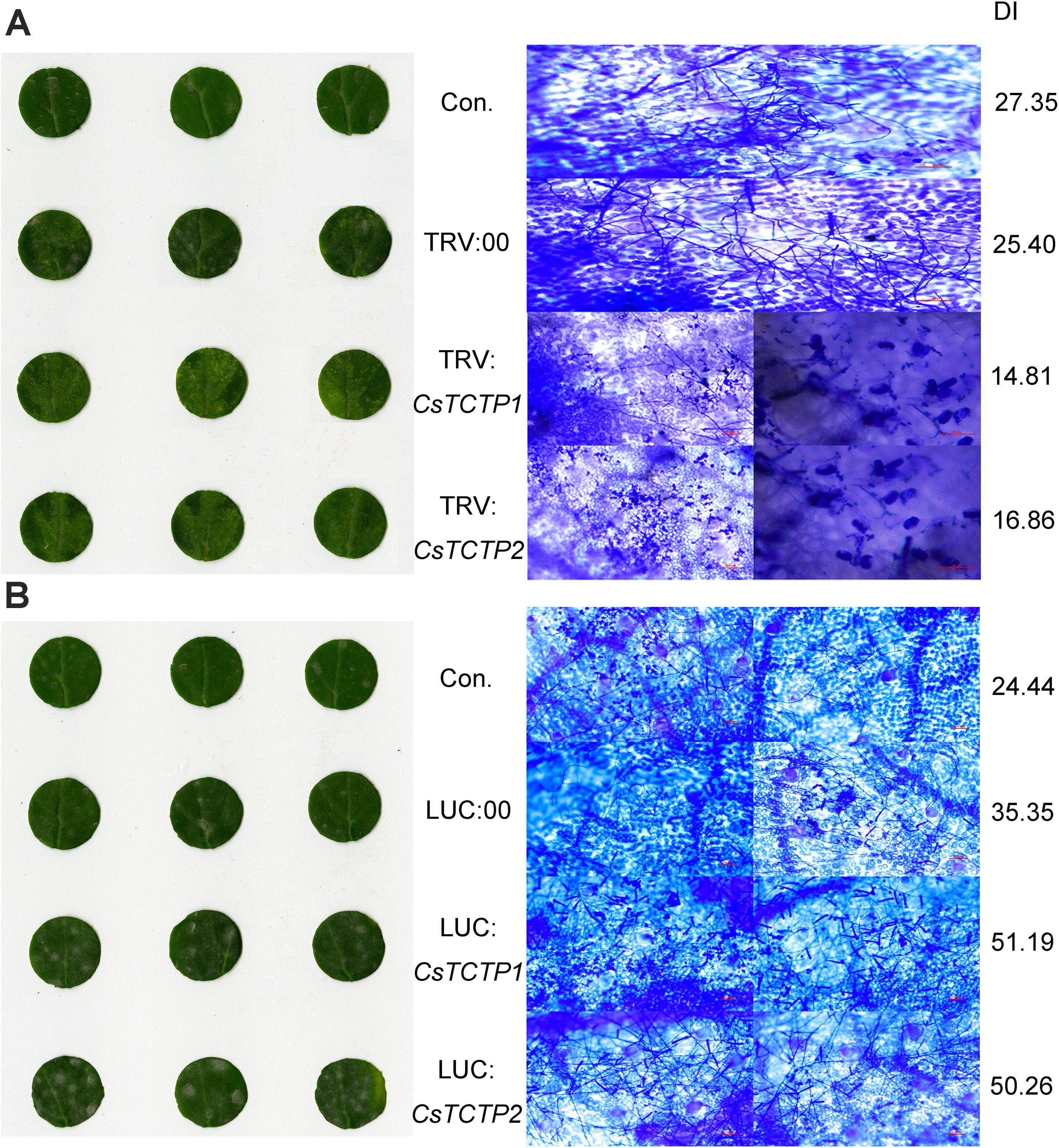
FIGURE 3. Silencing of either CsTCTP1 or CsTCTP2 enhanced cucumber resistance to S. fuliginea, whereas transient overexpression of either CsTCTP1 or CsTCTP2 in cucumber cotyledons impaired resistance to S. fuliginea. (A) Disease symptoms of detached cotyledons of control and CsTCTP1/CsTCTP2-silenced transgenic cucumbers at 7 dpi with S. fuliginea, and Coomassie brilliant blue staining for the examination of the S. fuliginea hyphae of cotyledons of control and CsTCTP1/CsTCTP2-silenced transgenic cucumbers at 7 dpi with S. fuliginea. (B) Disease symptoms of detached cotyledons of control and CsTCTP1/CsTCTP2-overexpressed transgenic cucumbers at 7 dpi with S. fuliginea, and Coomassie brilliant blue staining for the examination of the S. fuliginea hyphae of cotyledons of control and CsTCTP1/CsTCTP2-overexpressed transgenic cucumbers at 7 dpi with S. fuliginea. DI indicates the disease index of PM.
Transient Overexpression of CsTCTP1 or CsTCTP2 in Cucumber Cotyledons Impairs Resistance to S. fuliginea
To gain further insights into the response of CsTCTP1/CsTCTP2 function to S. fuliginea, we constructed the overexpression transformation plasmids LUC:CsTCTP1 and LUC:CsTCTP2, and LUC:00 (Figure 4A), LUC:CsTCTP1 and LUC:CsTCTP2 were separately transformed into and transiently expressed in cucumber XinTaiMiCi cotyledons. Luminescence indicated the expression of LUC in the LUC:00-, LUC:CsTCTP1-, and LUC:CsTCTP2-injected cucumber plants, but the non-injected (control) and EHA105-injected plants did not luminesce (Figure 4B). PCR analysis using the primers CsTCTP1-F/CsTCTP2-F and LUC-R showed that the introduced transgene was only found in CsTCTP1/CsTCTP2-overexpressing plants (Figure 4C). Then, qRT-PCR results showed that the transcript levels of CsTCTP1 in LUC:CsTCTP1 and CsTCTP2 in LUC:CsTCTP2 transgenic cucumbers were markedly higher than those of LUC:00-injected plants (Figure 4D). These results indicated that CsTCTP1 and CsTCTP2 genes had been successfully transiently overexpressed in the transgenic plants.
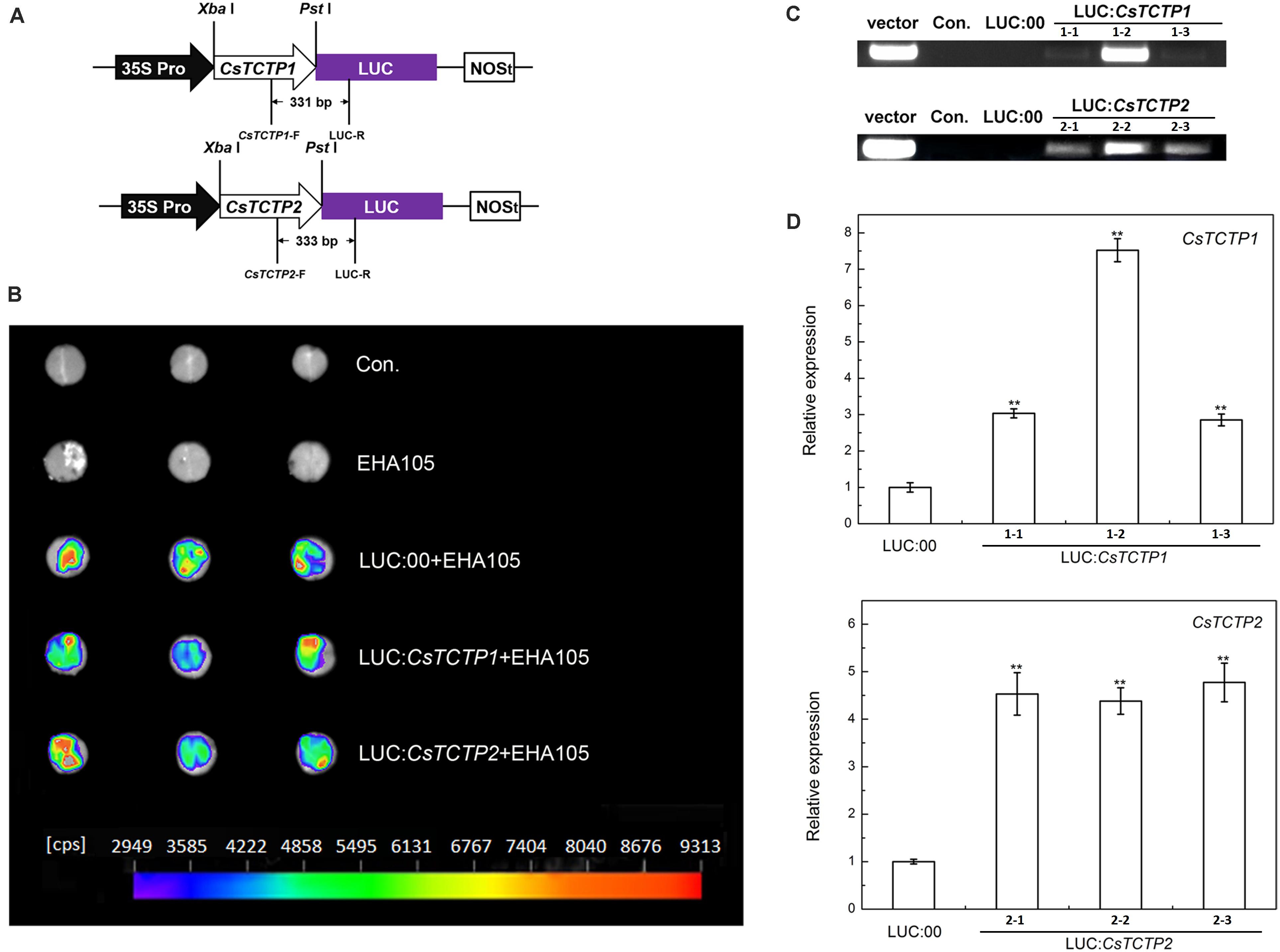
FIGURE 4. Identification of CsTCTP-overexpressing cucumber plants. (A) CsTCTP1- and CsTCTP2-overexpressing transformation vectors LUC:CsTCTP1 and LUC:CsTCTP2. The arrows show the fragment that was amplified in the PCR detection of the recombinant plasmids. (B) Luminescence of detached cotyledons of transient overexpression cucumber plants. Con. indicates non-injected plants, EHA105 indicates EHA105-injected plants, and indicates the number of counts per second. (C) PCR identification of LUC:CsTCTP1-injected and LUC:00-injected cucumbers using primers specific to the CsTCTP1-LUC cassette, and PCR identification of LUC:CsTCTP2-injected and LUC:00-injected cucumbers using primers specific to the CsTCTP2-LUC cassette. Con. indicates non-injected cucumber plants, and vector indicates the transformation vector. (D) qRT-PCR analyses of the expression levels of the CsTCTP1 gene in cucumber plants injected with LUC:00 or LUC:CsTCTP1 and the expression levels of the CsTCTP2 gene in cucumber plants injected with LUC:00 or LUC:CsTCTP2. Statistically significant differences were identified based on three technical replicates (t-test; ∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean values, and data are presented as the mean values ± SE of three biological replicates.
At day 6 with S. fuliginea, the cotyledons of CsTCTP1/-CsTCTP2-overexpressing plants exhibited the whitish appearance typical of PM disease, but control and LUC:00-injected plants demonstrated this appearance to a lesser extent. Microscope observations showed that the hyphal abundances of S. fuliginea were greater on the infested leaves of the CsTCTP1/CsTCTP2-overexpressing plants than on those of control and LUC:00-injected plants. Moreover, the DI of CsTCTP1-overexpressing plants was 51.19, and the DI of CsTCTP2-overexpressing plants was 50.26, while the DIs of the control and LUC:00-injected plants were 24.44 and 35.35, respectively (Figure 3B). These results suggested that overexpression of CsTCTP1 or CsTCTP2 in XinTaiMiCi plants impaired their resistance to S. fuliginea and that CsTCTP1 and CsTCTP2 are required for the cucumber defense response to S. fuliginea infections.
CsTCTP1 and CsTCTP2 Transgenic Plants Modulate the Expression Levels of Defense-Associated Genes
To elucidate whether CsTCTP1/CsTCTP2-mediated resistance against S. fuliginea is related to pathogen-associated genes, we examined the transcriptional responses of four known S. fuliginea-related defense genes, including 14-3-3 (XM-004136782.2), chitinase (HM015248), PR-1a (AF475286), and CuPi1 (U93586.1), to silencing and overexpression of CsTCTP1/CsTCTP2 in non-infested cucumber plants by qRT-PCR (Figure 5). CuPi1, a phloem lectin protein, was more abundant in B21-a-2-2-2 (an S. fuliginea-susceptible genotype) than in B21-a-2-1-2 (Meng et al., 2017). The level of chitinase transcripts was significantly higher in CsTCTP1-silenced plants than in TRV:00-injected plants, whereas it was significantly lower in CsTCTP1-overexpressing cucumbers than in LUC:00-injected cucumbers. Additionally, the level of 14-3-3 transcripts was lower in CsTCTP1-overexpressing plants than LUC:00-injected plants, and PR-1a transcripts were more abundant in CsTCTP1-silenced plants than in TRV:00-injected plants. The transcript level of CuPi1 was higher in CsTCTP1-overexpressing plants than LUC:00-injected plants. The 14-3-3, chitinase and PR-1a expression levels were upregulated in CsTCTP2-overexpressing plants from their values in LUC:00-injected plants and downregulated in CsTCTP2-silenced plants from their levels in TRV:00-injected plants. However, the level of CuPi1 transcripts exhibited in the opposite trend. These findings indicated that the CsTCTP1-induced defense pathway may be linked to defense-associated genes, especially chitinase, whereas the CsTCTP2-induced defense pathway probably worked in another manner.
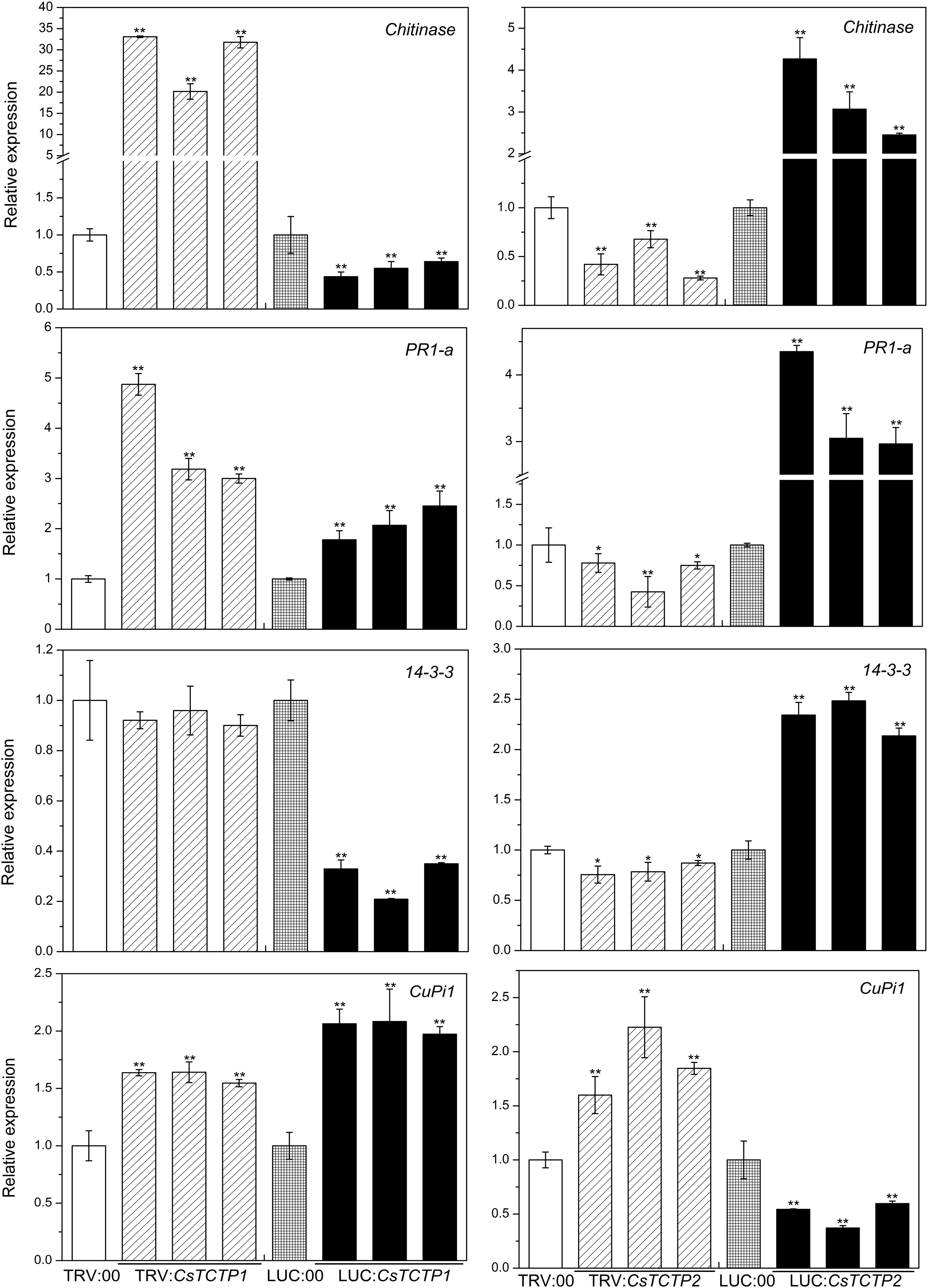
FIGURE 5. Relative expression levels of four genes (chitinase, PR-1a, 14-3-3, and CuPi1) related to S. fuliginea resistance in cucumber. The reported transcription levels of the tested genes in the CsTCTP1/CsTCTP2-silenced and CsTCTP1/CsTCTP2-overexpressing plants are relative to those in the TRV:00- and LUC:00-injected plants, respectively. Statistically significant differences were identified based on three technical replicates (t-test; ∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean, and data are presented as the mean values ± SE of three biological replicates.
CsTCTP1 and CsTCTP2 Transgenic Plants Modulate the Expression Levels of TOR Signaling- and ABA Signaling-Related Genes
To explore whether CsTCTP1/CsTCTP2 functions in the resistance of transgenic cucumber through the TOR signaling pathway, we conducted the following experiments. TOR (XM_011660561.1), Raptor1 (XM_004149881.2), S6K (XM_004138089.2), and SnRK1 (NM_001305710.1) gene expression was quantified in CsTCTP1/CsTCTP2-silenced plants and TRV:00-injected plants, as well as in CsTCTP1/CsTCTP2-overexpressing plants and LUC:00-injected plants (Figure 6). TOR expression levels exhibited no significant difference between CsTCTP1-silenced plants and TRV:00-injected plants, nor does S6K between CsTCTP1-overexpressing plants and LUC:00-injected plants. The transcript levels of SnRK1 was higher in both CsTCTP1-overexpressing plants and CsTCTP1-silenced plants. These results indicated that the expression of TOR, SnRK1, and S6K may be regulated not only by CsTCTP1. The transcript level of Raptor 1 was at its lowest in CsTCTP1-overexpressing plants but was at its highest in CsTCTP1-silenced plants. As shown in Figure 7, TOR and SnRK1 expression levels were lower in CsTCTP2-silenced plants than in TRV:00-injected plants but were higher in CsTCTP2-overexpressing plants, and S6K performed in the opposite manner. The Raptor 1 gene was slightly upregulated in both CsTCTP2-overexpressing plants and CsTCTP2-silenced plants. After treatment with rapamycin (a potential inhibitor to suppress the TOR signaling pathway), cucumbers exhibited remarkably enhanced resistance to S. fuliginea (Supplementary Figure S2). These results implied that CsTCTP2 accumulation is associated with TOR signaling for resistance against S. fuliginea.
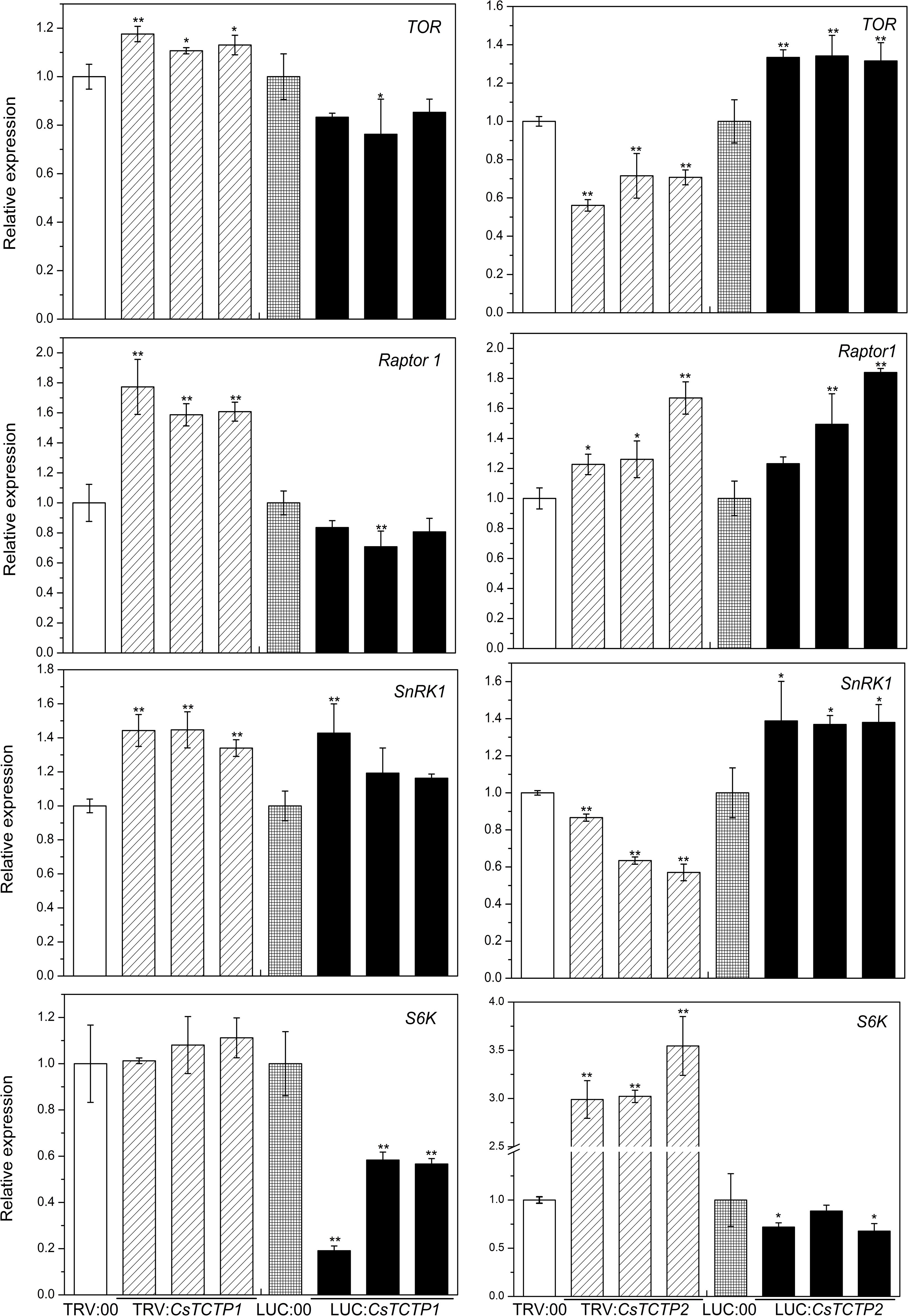
FIGURE 6. Relative expression levels of four genes (TOR, SnRK1, S6K, and Raptor 1) related to the TOR signaling pathway in cucumber. The reported transcription levels of the tested genes in the CsTCTP1/CsTCTP2-silenced and CsTCTP1/CsTCTP2-overexpressing plants are relative to those in the TRV:00- and LUC:00-injected plants, respectively. Statistically significant differences were identified based on three technical replicates (t-test; ∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean values, and data are presented as the mean values ± SE of three biological replicates.
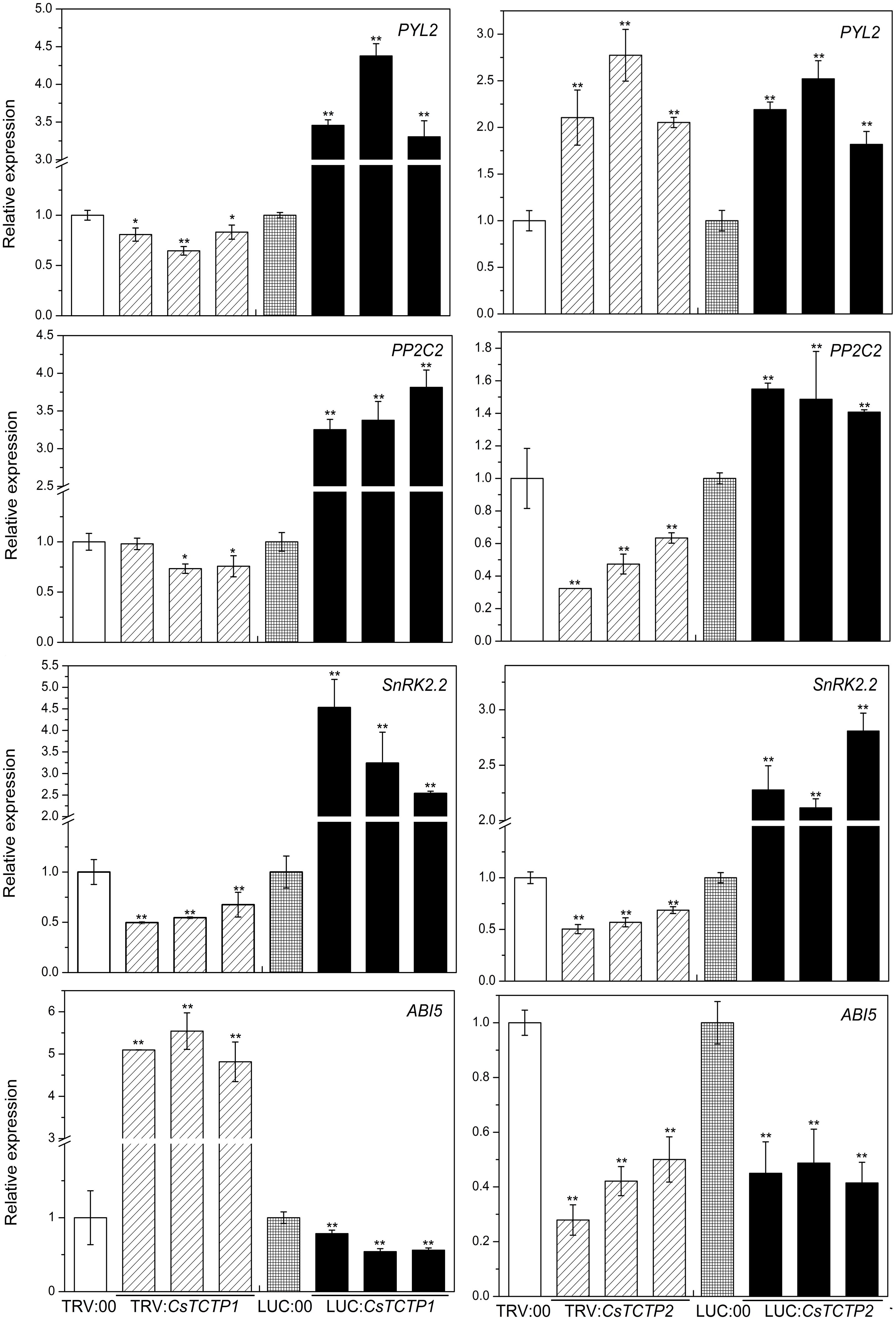
FIGURE 7. Relative expression levels of four genes (PYL2, PP2C2, SnRK2.2, and ABI5) related to the ABA signaling pathway in cucumber. The reported transcription of the tested genes in the CsTCTP1/CsTCTP2-silenced and CsTCTP1/CsTCTP2-overexpressing plants are relative to those in the TRV:00- and LUC:00-injected plants, respectively. Statistically significant differences were identified based on three technical replicates (t-test; ∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean, and data are presented as the mean ± SE of three biological replicates.
Abscisic acid is also an important defense signaling molecule. We next examined the possible molecular mode of action of CsTCTP1 and CsTCTP2 on the expression of ABA signaling genes (Figure 7). The transcript levels of PYL2 (JF789830), PP2C2 (JN566067) and SnRK2.2 (JN566071) genes were significantly higher in the CsTCTP1-overexpressing plants than the control plants and lower in the CsTCTP1-silenced plants. In contrast, ABI5 (XM_004149176.2) gene transcription performed in the opposite manner. The relative expression level of the PYL2 gene was greater in CsTCTP2-silenced/overexpressing plants than TRV:00/LUC:00-injected plants, whereas ABI5 gene expression was downregulated in CsTCTP2 transgenic plants. The transcript levels of PP2C2 and SnRK2.2 genes were higher in CsTCTP2-overexpressing plants than LUC:00-injected plants and lower in the CsTCTP2-silenced plants than TRV:00-injected plants. These results suggested that CsTCTP1 positively modulates the expression of PYL2, PP2C2, and SnRK2.2 and negatively modulates the expression of ABI5. Moreover, CsTCTP2 may positively modulate the expression of PP2C2 and SnRK2.2, whereas the expression of PYL2 and ABI5 may be regulated not only by CsTCTP2 but also by other proteins. These data are in line with the ABA content data (Supplementary Figure S3) showing that ABA signaling genes is regulated by CsTCTP1.
Discussion
TCTP is a highly conserved and multifunctional protein in eukaryotic phyla. Increasing amounts of data suggest that TCTP is regulated by pathogen stress. For instance, TaTCTP (Triticum aestivum) functions under Puccinia striiformis and Erysiphe graminis stresses (Li et al., 2010; Wang, 2010). Comparative proteomic studies showed that CsTCTP1 was differentially expressed by two cucumber sister lines (highly resistant and susceptible lines to S. fuliginea) subjected to S. fuliginea infection (Fan et al., 2014). Here, CsTCTP1 and CsTCTP2 both localized to the cytoplasm, which is in accordance with the in silico analysis. This report is the first to determine the subcellular localization of TCTP in cucumber. This study undoubtedly broadens the understanding of the molecular characterization of TCTPs in plant species. Additionally, the different expression patterns of CsTCTP1 and CsTCTP2 after pathogen inoculation were associated with the resistance degree in five different cucumber cultivars. We thus deduced that CsTCTP1 and CsTCTP2 might related to S. fuliginea stress response.
Although TCTP might be involved in responses to S. fuliginea, its biological functions remain to be identified. The functional characterization of TCTP in cucumber–pathogen interactions has been hampered until recently by the lack of a gene transformation method in cucumber plants. However, Shang et al. (2014) developed a cucumber cotyledon transient agro-infiltration expression system. In this study, we successfully obtained cucumbers that transiently silenced and overexpressed CsTCTP1/CsTCTP2. Function analysis revealed that CsTCTP1/CsTCTP2-silencing markedly enhanced the resistance of these transgenic cucumbers to S. fuliginea. In contrast, CsTCTP1/CsTCTP2-overexpression significantly compromised resistance to S. fuliginea. Inoculation with S. fuliginea resulted in the typical whitish appearance of PM disease on CsTCTP1/CsTCTP2-overexpressing cucumber plants and also caused them to exhibit greater hyphal abundances and a higher DI than control plants. These data indicated that both CsTCTP1 and CsTCTP2 act as negative modulators in the cucumber defense response to S. fuliginea. Notably, overexpression of CsTCTP1 reduced the tolerance to high temperature stress, while overproduction of CsTCTP2 enhanced the resistance to high temperature stress (Meng et al., 2017). Since the function of CsTCTP1 and CsTCTP2 genes may be similar but do not overlap completely, further study of CsTCTP1 and CsTCTP2 regulatory mechanisms is necessary. Studies on TCTP in plants have been conducted by gene transient transformation, mostly in Arabidopsis and tobacco. This work is the first to decipher the functions of CsTCTP1 and CsTCTP2 in cucumber defense against S. fuliginea. This work undoubtedly provides new insights into TCTP biological functions in cucumber.
According to our results, the cucumber TCTP family gene CsTCTP1 may activate defense-associated genes in the cucumber defense response to S. fuliginea. The transcript level of chitinase was more than 20-fold greater in CsTCTP1-silenced cucumbers than TRV:00-injected cucumbers, whereas it was significantly lower in CsTCTP1-overexpressing cucumbers than LUC:00-injected cucumbers. Another cucumber TCTP family gene, CsTCTP2, contributed resistance against S. fuliginea but possibly by other means (probably the TOR signaling pathway). The transcript levels of 14-3-3, chitinase, and PR-1a were greater in CsTCTP2-overexpressing plants than LUC:00-injected plants and lower in CsTCTP2-silencing plants than TRV:00-injected plants. Although CsTCTP1 and CsTCTP2 belong to the same family and are highly similar, they function in the cucumber defense response to pathogens by different mechanisms. Despite having different regulatory mechanisms, the two genes showed the same negative effects on PM.
TOR signaling plays a vital role in balancing cell growth and survival (Aramburu et al., 2014; Hardie, 2015; Schepetilnikov and Ryabova, 2018). Rapamycin is a specific inhibitor of the TOR kinase and produced by Streptomyces hygroscopicus (Molnár et al., 1996). However, understanding the mechanisms of TOR signaling has been limited by the absence of FK506 binding protein 12 (FKBP12) in plant cells. Rapamycin can effectively inhibit Arabidopsis TOR activity when 10 μM rapamycin is added to liquid cultures of seedlings (Xiong and Sheen, 2014). We applied 10 μM rapamycin to liquid cultures, which resulted in significantly enhanced resistance to S. fuliginea and confirmed that the TOR complex negatively regulates pathogen resistance in cucumber (Supplementary Figure S2). Furthermore, diminished expression of the CsTCTP2 gene in cucumber cotyledons results in increased resistance to S. fuliginea. The TORC1 complex, including TOR and Raptor 1, is regulated by CsTCTP2, and a downstream target of TOR signaling, S6K, is also regulated by CsTCTP2. These results revealed that the response of CsTCTP2 to S. fuliginea was associated with TOR signaling. Extant data indicate that SnRK1 is crucial for maintaining energy homeostasis in plant cells under stress conditions (Hulsmans et al., 2016; Wurzinger et al., 2018). The transcription level of SnRK1 was lower here in CsTCTP2-silenced plants than TRV:00-injected plants but was higher in CsTCTP2-overexpressing plants, suggesting that CsTCTP2 plays an important role in maintaining energy homeostasis in cucumber under S. fuliginea stress.
Abscisic Acid is also an important signaling molecule in plant–pathogen interactions (Audenaert et al., 2002; Sánchez-Vallet et al., 2012; Zhou et al., 2013; Ulferts et al., 2015; Sivakumaran et al., 2016). In this study, CsTCTP1 positively modulated the expression of PYL2, PP2C2 and SnRK2.2 and negatively modulated the expression of ABI5. These results revealed that CsTCTP1 may also be involved in the ABA signaling pathway. The exogenous application of ABA suppresses the resistance of plants to pathogens (Cao et al., 2011). The transcription level of CsTCTP1 significantly upregulated after treatment with ABA (Meng et al., 2017). Furthermore, augmented expression of the CsTCTP1 gene in cucumber cotyledons results in decreased resistance to S. fuliginea. These results are in accordance with previous evidence showing that ABA levels negatively correlate with resistance to pathogen stress. In a previous study, ABI5 was expressed more in a resistant line than a susceptible line after S. fuliginea infestation (Chen, 2017). In the current study, the silencing of CsTCTP1 in cucumber cotyledons upregulated ABI5 transcription and simultaneously enhanced resistance to S. fuliginea. These results are also consistent with the response of CsTCTP1 to S. fuliginea being associated with the ABA signaling pathway.
Conclusion
CsTCTP1 and CsTCTP2 are negative modulators in the cucumber defense response to S. fuliginea stress. CsTCTP1 participates in the defense response to S. fuliginea through regulating the expression of certain defense-associated genes and/or ABA signaling pathway-associated genes, and CsTCTP2 participates by regulating the expression of TOR signaling pathway-associated genes. Our findings provide new insights into TCTP function in cucumber defense responses to PM and will guide enhancing the resistance of cucumber to pathogens.
Author Contributions
HF conceived and designed the research. XM analyzed the data and wrote the manuscript. XM, YaY, JZ, NC, TS, and YuY conducted the experiments. All authors read and approved the manuscript.
Funding
This study was funded by National Natural Science Foundation of China (31772314), Natural Science Foundation of Liaoning Province of China (20170540802), and Program for Science and Technology Plan of Shenyang (171246).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Liaoning Academy of Agricultural Sciences for the breeding technology support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00544/full#supplementary-material
References
Adami, A., García-Alvarez, B., Arias-Palomo, E., Barford, D., and Llorca, O. (2007). Structure of TOR and its complex with KOG1. Mol. Cell 27, 509–516. doi: 10.1016/j.molcel.2007.05.040
Ahn, C. S., Ahn, H. K., and Pai, H. S. (2014). Overexpression of the PP2A regulatory subunit Tap46 leads to enhanced plant growth through stimulation of the TOR signalling pathway. J. Exp. Bot. 66, 827–840. doi: 10.1093/jxb/eru438
Anderson, G. H., Veit, B., and Hanson, M. (2005). The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 3:12. doi: 10.1186/1741-7007-3-12
Aramburu, J., Ortells, M. C., Tejedor, S., Buxadé, M., and López-Rodríguez, C. (2014). Transcriptional regulation of the stress response by mTOR. Sci. Signal. 7:re2. doi: 10.1126/scisignal.2005326
Audenaert, K., De Meyer, G. B., and Höfte, M. M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128, 491–501. doi: 10.1104/pp.010605
Baena-González, E., and Sheen, J. (2008). Convergent energy and stress signaling. Trends Plant Sci. 13, 474–482. doi: 10.1016/j.tplants.2008.06.006
Berkowitz, O., Jost, R., Pollmann, S., and Masle, J. (2008). Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell 20, 3430–3447. doi: 10.1105/tpc.108.061010
Brioudes, F., Thierry, A. M., Chambrier, P., Mollereau, B., and Bendahmane, M. (2010). Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc. Natl. Acad. Sci. U.S.A. 107, 16384–16389. doi: 10.1073/pnas.1007926107
Bruckner, F. P., Xavier, A. D. S., Cascardo, R. S., Otoni, W. C., Zerbini, F. M., and Alfenas-Zerbini, P. (2017). Translationally controlled tumour protein (TCTP) from tomato and Nicotiana benthamiana is necessary for successful infection by a potyvirus. Mol. Plant Pathol. 18, 672–683. doi: 10.1111/mpp.12426
Cao, F., Yoshioka, K., and Desveaux, D. (2011). The roles of ABA in plant-pathogen interactions. J. Plant Res. 124, 489–499. doi: 10.1007/s10265-011-0409-y
Chen, Q. (2017). Analysis of CsTCTP Regulated Signal Pathway and Construction of Yeast Two-hybrid System in Cucumber. Master’s thesis, Shenyang Agricultural University, Shenyang.
Chou, M., Xia, C., Feng, Z., Sun, Y., Zhang, D., Zhang, M., et al. (2016). A translationally controlled tumor protein gene Rpf41 is required for the nodulation of Robinia pseudoacacia. Plant Mol. Biol. 90, 389–402. doi: 10.1007/s11103-015-0424-9
Dobrenel, T., Caldana, C., Hanson, J., Robaglia, C., Vincentz, M., Veit, B., et al. (2016). TOR signaling and nutrient sensing. Annu. Rev. Plant Biol. 67, 261–285. doi: 10.1146/annurev-arplant-043014-114648
Fan, H., Ren, L., Meng, X., Song, T., Meng, K., and Yang, Y. (2014). Proteome-level investigation of Cucumis sativus derived resistance to Sphaerotheca fuliginea. Acta Physiol. Plant. 36, 1781–1791. doi: 10.1007/s11738-014-1552-6
Gwinn, D. M., Shackelford, D. B., Egan, D. F., Mihaylova, M. M., Mery, A., Vasquez, D. S., et al. (2008). AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226. doi: 10.1016/j.molcel.2008.03.003
Hara, K., Maruki, Y., Long, X., Yoshino, K., Oshiro, N., Hidaya, S., et al. (2002). Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189. doi: 10.1016/S0092-8674(02)00833-4
Hardie, D. G. (2015). AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr. Opin. Cell Biol. 33, 1–7. doi: 10.1016/j.ceb.2014.09.004
Hinojosamoya, J. J., Xoconostlecázares, B., Toscanomorales, R., Ramírezortega, F., Cabreraponce, J. L., and Ruizmedrano, R. (2013). Characterization of the pumpkin translationally-controlled tumor protein CmTCTP. Plant Signal. Behav. 8:e26477. doi: 10.4161/psb.26477
Hsu, Y., Chern, J. J., Cai, Y., Liu, M., and Choi, K. W. (2007). Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 445, 785–788. doi: 10.1038/nature05528
Hückelhoven, R. (2005). Powdery mildew susceptibility and biotrophic infection strategies. FEMS Microbiol. Lett. 245, 9–17. doi: 10.1016/j.femsle.2005.03.001
Hulsmans, S., Rodriguez, M., de Coninck, B., and Rolland, F. (2016). The SnRK1 energy sensor in plant biotic interactions. Trends Plant Sci. 21, 648–661. doi: 10.1016/j.tplants.2016.04.008
John, F., Roffler, S., Wicker, T., and Ringli, C. (2011). Plant TOR signaling components. Plant Signal. Behav. 6, 1700–1705. doi: 10.4161/psb.6.11.17662
Jossier, M., Bouly, J. P., Meimoun, P., Arjmand, A., Lessard, P., Hawley, S., et al. (2009). SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 59, 316–328. doi: 10.1111/j.1365-313X.2009.03871.x
Kim, Y. M., Han, Y. J., Hwang, O. J., Lee, S. S., Shin, A. Y., Kim, S. Y., et al. (2012). Overexpression of Arabidopsis translationally controlled tumor protein gene AtTCTP enhances drought tolerance with rapid ABA-induced stomatal closure. Mol. Cells 33, 617–626. doi: 10.1007/s10059-012-0080-8
Laplante, M., and Sabatini, D. M. (2012). mTOR signaling in growth control and disease. Cell 149, 274–293. doi: 10.1016/j.cell.2012.03.017
Le, T. P., Thuong, V. L., Kim, A. R., Hsu, Y. C., and Choikw, K. W. (2016). 14-3-3 proteins regulate Tctp–Rheb interaction for organ growth in Drosophila. Nat. Commun. 7:11501. doi: 10.1038/ncomms11501
Li, G., Liu, X. Y., Li, X. P., and Wang, Z. Y. (2010). Cloning of a TCTP gene in wheat and its expression induced by Erysiphe graminis. Biotechnol. Bull. 30, 441–447.
Liu, L. (2008). Inheritance Analysis and QTL Mapping of Powdery Mildew Resistance in Cucumber (Cucumis sativus L.). Doctor’s thesis, Shanghai Jiao Tong University, Shanghai.
Luo, J., Zhang, R., Qi, X., Xu, Q., and Chen, X. (2015). A preliminary study on cloning of chitinase gene and relationships to resistance of powdery mildew in cucumber (Cucumis sativus L.). Mol. Plant Breed. 13, 1584–1591.
Maegawa, K., Takii, R., Ushimaru, T., and Kozaki, A. (2015). Evolutionary conservation of TORC1 components, TOR, Raptor, and LST8, between rice and yeast. Mol. Genet. Genomics 290, 2019–2030. doi: 10.1007/s00438-015-1056-0
Meng, X., Chen, Q., Fan, H., Song, T., Cui, N., Zhao, J., et al. (2017). Molecular characterization, expression analysis and heterologous expression of two translationally controlled tumor protein genes from Cucumis sativus. PLoS One 12:e0184872. doi: 10.1371/journal.pone.0184872
Meng, X., Song, T., Fan, H., Yu, Y., Cui, N., Zhao, J., et al. (2016). A comparative cell wall proteomic analysis of cucumber leaves under Sphaerotheca fuliginea stress. Acta Physiol. Plant. 38:260. doi: 10.1007/s11738-016-2266-8
Molnár, I., Aparicio, J. F., Haydock, S. F., Khaw, L. E., Schwecke, T., König, A., et al. (1996). Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene 169, 1–7. doi: 10.1016/0378-1119(95)00800-4
Moreau, M., Azzopardi, M., Clément, G., Dobrenel, T., Marchive, C., Renne, C., et al. (2012). Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24, 463–481. doi: 10.1105/tpc.111.091306
Nojima, H., Tokunaga, C., Eguchi, S., Oshiro, N., Hidayat, S., Yoshino, K. I., et al. (2003). The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278, 15461–15464. doi: 10.1074/jbc.C200665200
Pu, Y., Luo, X., and Bassham, D. C. (2017). TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front. Plant Sci. 8:1204. doi: 10.3389/fpls.2017.01204
Ren, H. (2011). Inhibitive Mechanisms of Chrysophanol against Sphaerotheca fuliginea. Doctor’s thesis, Agricultural University of Hebei, Baoding.
Robaglia, C., Thomas, M., and Meyer, C. (2012). Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 15, 301–307. doi: 10.1016/j.pbi.2012.01.012
Sánchez-Vallet, A., López, G., Ramos, B., Delgado-Cerezo, M., Riviere, M. P., Llorente, F., et al. (2012). Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant Physiol. 160, 2109–2124. doi: 10.1104/pp.112.200154
Schepetilnikov, M., and Ryabova, L. A. (2018). Recent discoveries on the role of TOR (Target of Rapamycin) signaling in translation in plants. Plant Physiol. 176, 1095–1105. doi: 10.1104/pp.17.01243
Schmidt, I., Fähling, M., Nafz, B., Skalweit, A., and Thiele, B. J. (2007). Induction of translationally controlled tumor protein (TCTP) by transcriptional and post-transcriptional mechanisms. FEBS J. 274, 5416–5424. doi: 10.1111/j.1742-4658.2007.06069.x
Schulze-Lefert, P., and Vogel, J. (2000). Closing the ranks to attack by powdery mildew. Trends Plant Sci. 5, 343–348. doi: 10.1016/S1360-1385(00)01683-6
Shang, Y., Ma, Y., Zhou, Y., Zhang, H., Duan, L., Chen, H., et al. (2014). Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346, 1084–1088. doi: 10.1126/science.1259215
Sivakumaran, A., Akinyemi, A., Mandon, J., Cristescu, S. M., Hall, M. A., Harren, F. J., et al. (2016). ABA suppresses Botrytis cinerea elicited NO production in tomato to influence H2O2 generation and increase host susceptibility. Front. Plant Sci. 7:709. doi: 10.3389/fpls.2016.00709
Son, O., Kim, S., Hur, Y. S., and Cheon, C. I. (2016). Identification of the Raptor-binding motif on Arabidopsis S6 kinase and its use as a TOR signaling suppressor. Biochem. Biophys. Res. Commun. 472, 83–87. doi: 10.1016/j.bbrc.2016.02.068
Susini, L., Besse, S., Duflaut, D., Lespagnol, A., Beekman, C., Fiucci, G., et al. (2008). TCTP protects from apoptotic cell death by antagonizing bax function. Cell Death Differ. 15, 1211–1220. doi: 10.1038/cdd.2008.18
Toscano-morales, R., Xoconostle-Cázares, B., Cabrera-Ponce, J. L., Hinojosa-Moya, J., Ruiz-Salas, J. L., Galván-Gordillo, S. V., et al. (2015). AtTCTP2, an Arabidopsis thaliana homolog of translationally controlled tumor protein, enhances in vitro plant regeneration. Front. Plant Sci. 6:468. doi: 10.3389/fpls.2015.00468
Turck, F., Zilbermann, F., Kozma, S. C., Thomas, G., and Nagy, F. (2004). Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol. 134, 1527–1535. doi: 10.1104/pp.103.035873
Ulferts, S., Delventhal, R., Splivallo, R., Karlovsky, P., and Schaffrath, U. (2015). Abscisic acid negatively interferes with basal defence of barley against Magnaporthe oryzae. BMC Plant Biol. 15:7. doi: 10.1186/s12870-014-0409-x
Vonakis, B. M., MacGlashan, D. W., Vilariño, N., Langdon, J. M., Scott, R. S., and MacDonald, S. M. (2008). Distinct characteristics of signal transduction events by histamine-releasing factor/translationally controlled tumor protein (HRF/TCTP)-induced priming and activation of human basophils. Blood 111, 1789–1796. doi: 10.1182/blood-2007-07-104364
Wang, X. (2010). Defence Responses Including Oxidative Burst and Defenses Gene Expression in the Interaction between Wheat and Stripe Rust and Identification and Functional Characterization of Resistance-related Genes. Doctor’s thesis, Northwest Agriculture and Forestry University, Xi’an.
Wang, Y., Wu, Y., Duan, C., Chen, P., Li, Q., Dai, S., et al. (2012). The expression profiling of the CsPYL, CsPP2C and CsSnRK2 gene families during fruit development and drought stress in cucumber. J. Plant Physiol. 169, 1874–1882. doi: 10.1016/j.jplph.2012.07.017
Wurzinger, B., Nukarinen, E., Nägele, T., Weckwerth, W., and Teige, M. (2018). The SnRK1 kinase as central mediator of energy signaling between different organelles. Plant Physiol. 176, 1085–1094. doi: 10.1104/pp.17.01404
Xiong, Y., and Sheen, J. (2014). The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 164, 499–512. doi: 10.1104/pp.113.229948
Xiong, Y., and Sheen, J. (2015). Novel links in the plant TOR kinase signaling network. Curr. Opin. Plant Biol. 28, 83–91. doi: 10.1016/j.pbi.2015.09.006
Yang, Y., Yang, F., Xiong, Z., Yan, Y., Wang, X., Nishino, M., et al. (2005). An N-terminal region of translationally controlled tumor protein is required for its antiapoptotic activity. Oncogene 24, 4778–4788. doi: 10.1038/sj.onc.1208666
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, C., Chen, H., Cai, T., Deng, T., Zhuang, R., Zhang, N., et al. (2017). Overexpression of a novel peanut NBS-LRR gene AhRRS5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol. J. 15, 39–55. doi: 10.1111/pbi.12589
Zhang, Y. (2016). Research of Cucumber 14-3-3 Proteins in Response to Powdery Mildew Infection. Master’s thesis, Shenyang Agricultural University, Shenyang.
Keywords: ABA signaling pathway, Cucumis sativus, Sphaerotheca fuliginea, translationally controlled tumor protein, transient transformation, TOR signaling pathway
Citation: Meng X, Yu Y, Zhao J, Cui N, Song T, Yang Y and Fan H (2018) The Two Translationally Controlled Tumor Protein Genes, CsTCTP1 and CsTCTP2, Are Negative Modulators in the Cucumis sativus Defense Response to Sphaerotheca fuliginea. Front. Plant Sci. 9:544. doi: 10.3389/fpls.2018.00544
Received: 24 January 2018; Accepted: 09 April 2018;
Published: 25 April 2018.
Edited by:
Essaid Ait Barka, Université de Reims Champagne-Ardenne, FranceReviewed by:
Isabel Diaz, Universidad Politécnica de Madrid (UPM), SpainMostafa Abdelwahed Abdelrahman, Tohoku University, Japan
Copyright © 2018 Meng, Yu, Zhao, Cui, Song, Yang and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyan Fan, SHlmYW43NEAxNjMuY29t
 Xiangnan Meng
Xiangnan Meng Yang Yu1
Yang Yu1 Junyue Zhao
Junyue Zhao Na Cui
Na Cui Haiyan Fan
Haiyan Fan