- 1Graduate School of Biotechnology and Crop Biotech Institute, Kyung Hee University, Yongin, South Korea
- 2Institute for Molecular Physiology, Heinrich-Heine-Universität Düsseldorf, Düsseldorf, Germany
- 3Max Planck Institute for Plant Breeding Research, Cologne, Germany
Many scientific findings have been reported on the beneficial function of reactive oxygen species (ROS) in various cellular processes, showing that they are not just toxic byproducts. The double-edged role of ROS shows the importance of the regulation of ROS level. We report a gene, rrsRLK (required for ROS-scavenging receptor-like kinase), that encodes a cytoplasmic RLK belonging to the non-RD kinase family. The gene was identified by screening rice RLK mutant lines infected with Xanthomonas oryzae pv. oryzae (Xoo), an agent of bacterial leaf blight of rice. The mutant (ΔrrsRLK) lacking the Os01g02290 gene was strongly resistant to many Xoo strains, but not to the fungal pathogen Magnaporthe grisea. ΔrrsRLK showed significantly higher expression of OsPR1a, OsPR1b, OsLOX, RBBTI4, and jasmonic acid-related genes than wild type. We showed that rrsRLK protein interacts with OsVOZ1 (vascular one zinc-finger 1) and OsPEX11 (peroxisomal biogenesis factor 11). In the further experiments, abnormal biogenesis of peroxisomes, hydrogen peroxide (H2O2) accumulation, and reduction of activity of ROS-scavenging enzymes were investigated in ΔrrsRLK. These results suggest that the enhanced resistance in ΔrrsRLK is due to H2O2 accumulation caused by irregular ROS-scavenging mechanism, and rrsRLK is most likely a key regulator required for ROS homeostasis in rice.
Introduction
Despite the massive efforts of international organizations and scientists to solve food problems, 108 million people world-wide face serious food insecurities (Arthur, 2008). Furthermore, the growth rate of the global population demands continued efforts to supply more food to humans. There are diverse causes that reduce the productivity of crops, and losses by biotic invaders are serious. These situations illustrate the importance of research on crop defense systems against small pathogens such as viruses, bacteria, and fungi.
Since the gene-for-gene hypothesis was proposed (Flor, 1971), plant defense systems have been largely concentrated in two types. Both are known to be triggered by interactions between pathogen elicitors and host plant receptors. The first one is rapidly triggered by interactions between pathogen-associated molecular patterns (PAMPs, e.g., chitin, flagellin, and elongation factor-Tu) from pathogens and pattern recognition receptors (PRRs, e.g., chitin elicitor receptor kinase 1, flagella sensing 2, elongation factor-Tu receptor, and Rpg1) of host plants (Brueggeman et al., 2002; Kunze et al., 2004; Chinchilla et al., 2006; Miya et al., 2007). The defense system is called PAMP-Triggered Immunity (PTI), and the interactions mostly occur extracellularly. PAMP-PRR interactions typically support the activation of mitogen-activated protein kinases, the rapid production of reactive oxygen species (ROS), and the induction of pathogenesis-related (PR) gene expression (Pitzschke et al., 2006). Plants also have cytoplasmic receptors that sense pathogenic effectors delivered through type three secretion systems (Jones and Dangl, 2006). We call the effectors and receptors avirulence (Avr) factors and disease resistance (R) proteins, respectively. The second type of defense system (called Effector-Triggered Immunity, ETI) usually causes programmed cell death (Cunnac et al., 2009) and induces much stronger and strain-specific responses compared to PTI (Jones and Dangl, 2006). ETI is often triggered indirectly. R protein senses interaction of pathogen effector and host protein (called a guardee) and triggers ETI (Block et al., 2008). In response to the defense mechanisms of host plants, pathogens have evolved to avoid recognition by receptors for PTI/ETI through either lost or altered effector (Thomma et al., 2011), but host plants have also evolved to detect such changes in pathogens (Almagro et al., 2009).
Although the pathogen molecules recognized by the plant receptors in ETI or PTI are diverse and their interactions with plant receptors are very specific, receptors responsive to PTI or ETI show high similarity in protein structure. The majority of R proteins in ETI typically possess a nucleotide binding site-leucine rich repeat harboring a variable N-terminus such as coiled-coil or toll/interleukin 1 receptor like domains (Boller and Felix, 2009). PRRs typically contain a receptor domain, frequently followed by a protein kinase (Braun and Walker, 1996). Alternatively, the receptor has a protein kinase as a signaling partner (Medzhitov, 2001). Concerning PRRs, members of the interleukin-1 receptor-associated kinase (IRAK) family involved in immune responses of animals and plants are distinguishable from members unrelated to defense (Dardick and Ronald, 2006). Dardick and Ronald (2006) suggested that most known or predicted PRRs have a kinase domain that belongs to a non-arginine (R)-aspartate (D) kinase group. It is well-known that conserved R residue immediately preceding the conserved catalytic D residue in the catalytic loop is critical for phosphorylation processes in a group of kinases (so called RD kinases) (Nolen et al., 2004). However, most kinases associated with PTI replace the positively charged R in the catalytic loop with uncharged residues such as Cys, Gly, Phe, or Leu with an exception of Arabidopsis chitin elicitor receptor kinase (CERK1) (Miya et al., 2007). In the rice genome, 371 of over 1,000 IRAKs (Zhou et al., 2004) have the non-RD motif (Dardick and Ronald, 2006), although only a small number of these have been functionally characterized.
We carried out inoculation tests with an IRAK gene mutant rice population (suppression or activation) carrying the non-RD motif using a pathogenic bacterium, Xanthomonas oryzae pv. oryzae (Xoo). Xoo, an agent of leaf blight in rice, is a major contributor to serious reduction of rice production in Asia and Africa (Khan et al., 2014). In the test, we selected genes that are most likely involved in rice defense. We report an interesting gene in this study, named rrsRLK (required for ROS-scavenging receptor-like kinase), that regulates ROS levels in rice and plays a negative role in defense against Xoo infection.
Materials and Methods
Plants, Bacteria, and Fungi and Growth Conditions
An IRAK gene mutant rice population (generated with Oryza sativa var. japonica cv. Dongjin or Hwayoung) (Jeon et al., 2000) carrying the non-RD motif was used for inoculation tests with Xoo strains and resulted in selection of 3A-10392 (a knock-out line of Os01g02290 in Dongjin, ΔrrsRLKdj), 4A-01523 (an activation line of Os01g02290 in Dongjin, rrsRLKAct1), 2A-50012 (another activation line of Os01g02290 in Dongjin, rrsRLKAct2), and 3B-00367 (a knock-out line of Os01g02290 in Hwayoung, ΔrrsRLKhy) mutant rice plants. Additional knock-out mutant lines that carry a T-DNA insertion at Os01g54930 encoding OsVOZ1 (rice vascular one zinc finger protein 1, K-05631) in Kittake (O. sativa var. japonica) and at Os03g02590 encoding OsPEX11 (rice peroxisomal biogenesis factor 11, 1B-07040) in Dongjin were selected and used. All mutant lines, including wild types (Dongjin and Hwayoung) as controls, are listed in Supplementary Table S1, and the genotyping method to determine segregation family is described in the Supplementary Materials.
Seeds were germinated on petri dishes containing water-drenched filter paper at 28°C for three days, transferred to soil, and then grown in a green house or paddy field before pathogen inoculation. All the inoculation experiments were carried out with 6-week-old rice or 7-week-old rice plants (in the winter season). Inoculation tests were carried out in a restricted chamber with conditions of 28°C with 85% humidity for 14 h during the day and 25°C and 80% humidity for 10 h at night.
Xoo strains (PXO99A, Philippine strain 6, compatible with Dongjin, PXO99 in this study; HB01009, Korean strain 3a, compatible to Hwayoung) and Magnaporthe grisea (M. grisea, PO6-6, a Philippine isolate) strains used in this study are listed in Supplementary Table S1. Xoo strains were cultured at 28°C for three days on plates of peptone sucrose agar (PSA) medium (10 g/L of peptone, 10 g/L of sucrose, 1 g/L of glutamic acid, 16 g/L agar, pH 7.0) and used for inoculation tests. M. grisea, PO6-6 was inoculated on potato dextrose (PDA) medium (24 g/L of potato starch, dextrose, 15 g/L agar) and cultured at 28°C for three days.
Pathogen Inoculation and Disease Evaluation
Inoculation of Xoo strains and M. grisea was carried out using the clipping method (Kauffman et al., 1973) and the leaf punch inoculation method (Lee et al., 2009), respectively. All the details including pathogen preparation for inoculation and scoring lesion lengths on rice leaves are described in the Supplementary Materials.
RT-PCR and Quantitative RT-PCR
RT-PCR and qRT-PCR were carried out to investigate expression of target genes such as Os01g02290, Os01g54930, Os03g02590, and PR genes in each mutant line. Total RNA extraction and cDNA synthesis were carried out using the method provided by the respective company, Takara (Japan) and Clontech Laboratories (United States). RT- and qRT-PCR analyses were repeated more than three times with three biological replicates. Thermo cycling conditions for RT- and qRT-PCR reactions are described in the Supplementary Materials, and used primers are listed in Supplementary Tables S2–S4.
Sub-Cellular Localization of rrsRLK Protein
Full-length cDNA of Os01g02290.2 was amplified by PCR using a cDNA clone (accession number: AK241355, KOME number: J065150L24) supplied from KOME1 and specific primers (Supplementary Table S3) containing recognition sequences of SmaI and SpeI restriction enzymes. The PCR product was inserted into the pGA3452 vector (Kim S.R. et al., 2009) after cutting with SmaI and SpeI. The pGA3452 vector is designed for expression of GFP fusion proteins under control of a maize ubiquitin promoter. The construct was then introduced into protoplast mesophyll and OC cells using the PEG transformation method (Choi et al., 2014) for transient expression of the rrsRLK-sGFP gene. Additional constructs, previously generated to express plasma membrane (PM)-mRFP (Kim et al., 2009) and nuclear localization signal (NLS)-mRFP (Kim et al., 2009) driven by 35S promoters, were also introduced into the protoplasts as markers. Signals of rrsRLK-sGFP, PM-mRFP, and NLS-mRFP were then examined on a fluorescence microscope Axioplan 2 (LSM 510 META; Zeiss, Germany) equipped with filter sets for GFP (excitation wavelength/dichroic transition: 488/543 nm) and RFP (excitation wavelength/dichroic transition: 561/575 nm). Methods for protoplast isolation (Yang et al., 2013) and transformation (Hong et al., 2012) were used for this study with minor modifications (see the Supplementary Materials).
Yeast Two-Hybrid Assay
A Gal4-based system with Gateway technology (Invitrogen, United States) was used for a yeast two-hybrid assay. DNA fragment (nt 859–1197, rrsRLKk) corresponding to the kinase domain of rrsRLK protein was amplified by PCR with specific primers containing attB1 and attB2 sites and KOME full-length cDNA of Os01g02290.2 as a template. The PCR product was cloned into pDONR222 (Invitrogen, United States) by BP recombination to generate the entry clone. Afterward, rrsRLKk was transferred to the yeast destination bait plasmid pDEST32 (Invitrogen, United States) by LR recombination, resulting in pDEST32rrsRLKk (pD32rrsRLKk). To construct a Dongjin cDNA library, cDNA of approximately 0.5–3 kbp was cloned into pDONR222 and subsequently into pDEST22 by LR recombination. This yielded pD22Lib (pDEST22 containing the 0.5–3 kb fragment of rice cDNA Library, AmpR). pD32rrsRLKk contains the DNA-binding domain of Gal4 and the leucine selection marker gene LEU2. pD22Lib contains the GAL4 transcription activation domain and the tryptophan selection marker gene TRP1. All constructs were checked by restriction enzyme analysis and confirmed by DNA sequencing. pD32rrsRLKk and pD22Lib were co-transformed into yeast strain YD116 cells according to the manufacturer’s protocol (Invitrogen, United States). The transformants were cultured on synthetic complete medium, lacking leucine (-Leu) and tryptophan (-Trp). After 72 h, the colonies were picked and mixed with 100 μL of sterile water. Then, 10 μL of the cell suspension was spotted onto selection plates to screen for expression of the three reporter genes (HIS3, URA3, and lacZ). Growth of the yeast was assessed on SC-Leu-Trp-His supplemented with SC-Leu-Trp-Ura and 0-50 mM 3-amino-1, 2, 4-triazole (3AT) as a histidine inhibitor. A change in the blue color of the transformants was monitored for the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). pD32rrsRLKk, pD22OsVOZ1, and pD22OsPEX11 were combined with pDEST32 or pDEST22 and tested for auto-activation of the reporter genes.
In Vivo Coimmunoprecipitation Assays
A coimmunoprecipitation assay was carried out using the previously reported method with minor modifications (Yang et al., 2013). Constructs were generated for transient expression of HA-tagged rrsRLK, Myc-tagged OsVOZ1, and Myc-tagged PEX11. Full-length rrsRLK without the stop codon was amplified by PCR using the primer sets listed in Supplementary Table S2 and inserted into the HpaI/KpnI sites of the HA-tagged vector pGA3698. The pGA3698 vector contains the maize Ubiquitin1 promoter and the 3x HA coding region. pGA3697 vectors carrying full-length OsVOZ1 and OsPEX11 were constructed using HpaI. The pGA3697 vector carried the maize Ubiquitin1 promoter and the 4x Myc coding region. To verify the interaction of rrsRLK-OsVOZ1 and rrsRLK-OsPEX11, a combination of the constructs was transformed into the protoplasts isolated from rice mesophyll or OC cells using PEG-mediated transformation (see Supplementary Materials). The protoplasts were re-suspended in IP buffer [50 mM Tris–HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% (v/v) Triton X-100, 1 mM DTT, 2 mM NaF, 50 μM MG132, and an adequate amount of protease inhibitor cocktail (Roche, United States)]. After vortexing briefly, the samples were centrifuged at 12,000 rpm for 10 min. Next, 10 μL each of protein A and G conjugated to agarose beads (Millipore, United States) were added to the supernatant for pre-clearing to prevent non-specific binding. We used 10% of the pre-cleared extracts as input controls, while the rest was precipitated with an anti-HA monoclonal antibody (Roche, United States) and incubated overnight with gentle shaking. The incubated sample was then mixed with 10 μL each of protein A and G conjugated to agarose beads and incubated for 2 h. After being washed five times with IP buffer, the proteins bound to the beads were eluted with 20 μL of SDS sample buffer. The eluents were separated with electrophoresis on a 10% SDS-polyacrylamide gel and transferred to a PVDF membrane (Millipore, United States). The experiment for monitoring the interaction between rrsRLK, OsVOZ1, and OsPEX11 was performed multiple times using the anti-Myc monoclonal antibody for precipitation and an HRP-conjugated anti-HA monoclonal antibody (Cell Signaling Technology, United States) for detection. Proteins that bound to the membrane were incubated with an HRP-conjugated anti-Myc monoclonal antibody (Cell Signaling technology, United States) and detected using an ECL prime western blotting detection reagent (GE Healthcare, United Kingdom) in LAS-4000 (GE Healthcare, United Kingdom).
H2O2 Measurement by Enzymatic Assay
The roots, stems, and leaves of Se-WTdj and ΔrrsRLKdj (1g each) were homogenized in 100 mM sodium phosphate buffer (pH 6.8). The homogenate was filtered through four layers of gauze and centrifuged at 17,000 × g for 25 min at 4°C. The supernatant was collected for measurement of H2O2 contents. Contents of H2O2 in rice tissues were determined by a modified method of Bernt and Bergmeyer (1974) using peroxidase. To initiate the enzyme reaction, an aliquot of 0.5 mL of supernatant was mixed with 2.5 mL of peroxide reagent consisting of 83 mM sodium phosphate (pH 7.0), 0.005% (w/v) o-dianisidine, and 40 μg/mL of peroxidase (Sigma-Aldrich, United States). After 15 min incubation at room temperature, the reactions were stopped by adding 0.5 mL of 1 N perchloric acid and centrifuged at 5,000 × g for 5 min. The absorbance of the sample was measured at 436 nm and was compared to the extinction of a H2O2 standard for quantitative analysis.
Analysis of Antioxidant Enzyme Activity in ΔrrsRLKdj
To extract total protein from leaves of WT and ΔrrsRLKdj, 1 g leaves was homogenized in 100 mM potassium phosphate buffer (pH 7.8) containing 0.1 mM EDTA, 1% (w/v) PVP (Polyvinylpyrrolidone), and 0.5% (v/v) Triton X-100 at 4°C. The homogenate was filtered through four layers of cheesecloth and centrifuged at 17,000 × g for 25 min at 4°C. The supernatant was collected for measurement of the enzymatic activities of superoxide dismutase (SOD), ascorbate peroxidase (APX), catalase (CAT), and peroxidase (POX). SOD activity was determined based on the method described by Beyer and Fridovich (1987) with a minor modification. The reaction mixture (1.06 mL) was composed of 50 mM potassium phosphate (pH 7.8), 9.9 mM methionine, 57 μM nitro blue tetrazolium, 0.025% triton X-100, 0.4 μM riboflavin, and the 40 μL extract. The mixture was incubated with irradiation of light for 7 min, and absorbance was monitored at A560 nm. To measure APX activity, the protein extract was added to reaction mixture containing 0.5 mM ascorbate and 0.2 mM H2O2 in 990 μL of 50 mM phosphate buffer (pH 7.0), and absorbance was monitored at 290 nm for 10 min. CAT activity was determined spectrophotometrically. Protein extract (1.92 mL) was mixed with 80 μL of 50 mM phosphate buffer (pH 7.0) containing 30 mM H2O2 as a substrate, and absorbance at 240 nm was monitored for 10 min. POX activity was determined by monitoring the formation of guaiacol dehydrogenation product (extinction coefficient 6.39 mM/L cm-1) at 436 nm. Reaction mixture contained 100 mM potassium phosphate (pH 7.0), 0.3 mM guaiacol, and 50 μL of the protein extract. The reaction was initiated by adding 0.1 mM H2O2, and A436 nm was monitored for 10 min.
Results
Lack of the Os01g02290 Gene Induces Strong Resistance Against Xoo
We selected 68 mutant lines from a T-DNA rice mutant population (Jeon et al., 2000). The mutant lines have T-DNA insertion into IRAK genes carrying a non-RD kinase, resulting in suppression or activation of the gene. Inoculation tests using the mutant lines and PXO99 resulted in the selection of a few mutant lines showing shorter lesion lengths than Dongjin rice. One of these is a mutant line (3A-10392, named ΔrrsRLKdj in this study) where Os01g02290 gene expression was suppressed by T-DNA insertion into the first exon (nt 480) (Figures 1A,B). Lesion length on the leaves of the mutant lines was 0.5 ± 0.27 cm and was shorter than that on the leaves of segregate wild type (Se-WTdj) (9.6 ± 1.04 cm) (Figures 1C,D). The population of bacteria extracted from the inoculated leaves was significantly reduced in the mutant rice leaves compared to that extracted from the inoculated Se-WTdj leaves (Figure 1E). These results indicate that rice lacking the Os01g02290 gene shows strong resistance against PXO99. The enhanced resistance against PXO99 was confirmed in the other mutant line, 3B-00367 (named ΔrrsRLKhy in this study). Expression of the gene was inhibited by T-DNA insertion at the first exon (nt 581) of Os01g02290 in Hwayoung rice (Supplementary Figures S1A,B). From the inoculation test with HB01009 (Xoo Korean strain 3a, compatible to Hwayoung), lesion lengths on leaves of the ΔrrsRLKhy rice were reduced about 71.8% (3.13 ± 1.37 cm) compared to those on the leaves of segregate wild type (Se-WThy) (11.11 ± 1.04 cm) (Supplementary Figures S1C,D). We also found gene activation lines (4A-01523 and 2A-50012) in which expression of Os01g02290 was enhanced by T-DNA insertion at nt 6877 (named rrsRLKact1) and nt 4068 (named rrsRLKact2), respectively (Supplementary Figures S1E,F). However, lesion lengths on the leaves of the activation lines were not significantly different from those of Se-WTdj (Supplementary Figures S1G,H).
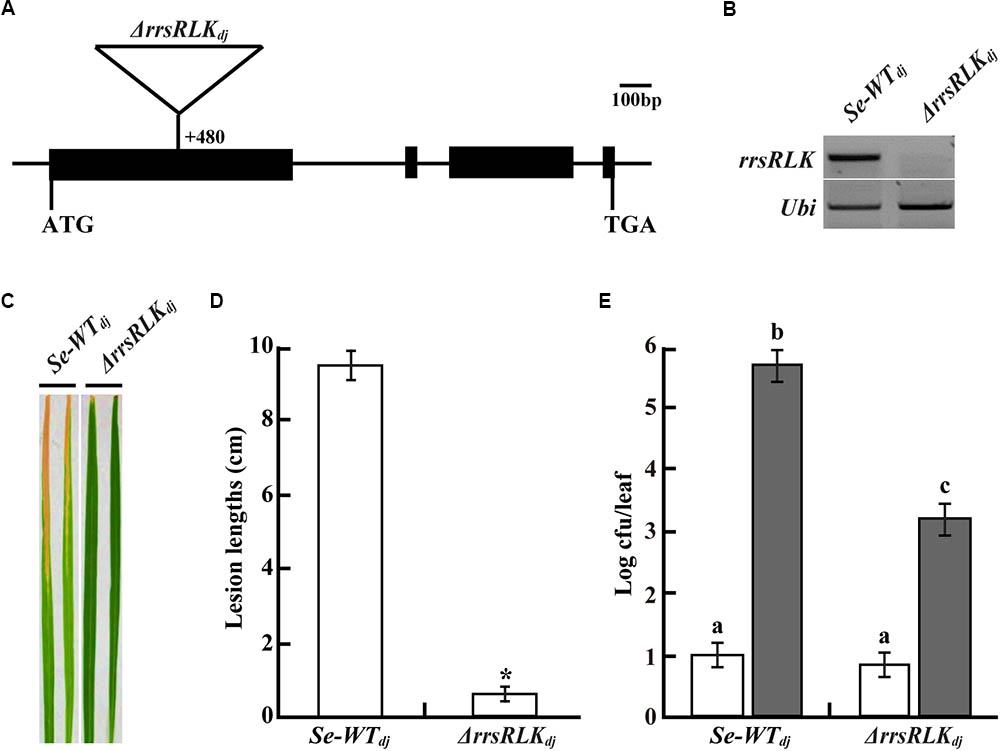
FIGURE 1. (A) Schematic representation of T-DNA insertion (+480 bp from the translational start site) at Os01g02290 encoding rrsRLK in Dongjin (ΔrrsRLKdj). ATG start codon and TGA stop codon are indicated. Os01g02290 consists of four exons (black boxes) and three introns (line between the black boxes). (B) Expression analysis of the rrsRLK gene in SE-WTdj (segregated wild type) and ΔrrsRLKdj mutant lines using RT-PCR with rrsRLK primers. Ubi gene was used as an internal control. The experiment was repeated three times with consistent results. (C) Comparison of lesion development on leaves of SE-WTdj and homozygous mutant lines of ΔrrsRLKdj. Rice plants grown for 6 weeks were inoculated with PXO99 and monitored for lesion development for 14 DAI. The results were consistent until the 6th generation of the mutant, and each experiment was carried out with more than 40 leaves. (D) Lesion lengths scored at 14 DAI on leaves of SE-WTdj and ΔrrsRLKdj mutant lines. The averages and error ranges of each sample were calculated from 40 leaves. Asterisk indicates P < 0.05 (Student’s t-test). Data in (C,D) are results of experiments carried out using the 3rd generation of ΔrrsRLKdj mutant lines. (E) Bacterial population extracted from inoculated leaves of SE-WTdj and ΔrrsRLKdj mutant lines at 0 and 14 DAI. Xoo was extracted from inoculated leaves and incubated on PSA plates after serial dilution for colony counting. Each data point represents the average and standard deviation of three biological replicates. The experiment was repeated three times with similar results. Different letters above bars indicate statistically significant differences as determined by one-way analysis of variance (ANOVA: ∗, a, b, c), P < 0.05.
ΔrrsRLKdj Has a Broad Range of Resistance Against Xoo Strains, but Not to the Fungal Pathogen M. grisea
To test if the resistance of the mutant is specific to the PXO99 strain, another Philippine strain, PXO86, and Korean strains (HB01009, HB01013, HB01014, HB01015) were inoculated on leaves of the ΔrrsRLKdj and Se-WTdj lines. All five strains are known to be compatible with Dongjin. Inoculation of each strain caused long lesion development on the leaves of Se-WTdj rice, while lesion lengths on the leaves of ΔrrsRLKdj were dramatically reduced (Supplementary Figure S2A). The results suggest that absence of the Os01g02290 gene in rice induces a broad range of resistance to Xoo strains. To study the possible resistance of ΔrrsRLKdj against fungal pathogens, an inoculation test with M. grisea (compatible with Dongjin) was carried out. In the test, leaves of the ΔrrsRLKdj and Se-WTdj rice showed similar lesion lengths (Supplementary Figure S2B), indicating that knock-out of the Os01g02290 gene does not induce resistance against the fungal pathogen M. grisea.
Suppression of Os01g02290 Gene Expression Induces Semi-Dwarfism and Lesion Mimic Phenotypes
Morphological changes of ΔrrsRLKdj were monitored through the sixth generation. We found that ΔrrsRLKdj displayed semi-dwarfism. At the 8-leaf stage, the ΔrrsRLKdj showed a 10% reduction in rice height compared to Se-WTdj in paddy fields (Figures 2A,B). The seeds of ΔrrsRLKdj were smaller in panicle length, weight, and height to those of Se-WTdj (Figures 2A,B). The semi-dwarfism was also shown in roots (Figure 2A). In particular, ΔrrsRLKdj had about a 75% decrease in grain numbers compared to Se-WTdj (Figure 2B). Additional changes include reddish brown spot lesions scattered across the entire surface of the leaves, with severity increasing at the tips (Figure 2A). However, the lesion-mimic phenotype on the mutant leaves appeared conditionally; it only appeared on rice grown in the paddy field during the summer season. All of the changes in rice height and seed size that were observed in ΔrrsRLKdj were similarly observed with ΔrrsRLKhy, including the lesion-mimic. These results suggest that the rrsRLK protein from the Os01g02290 gene contributes to both vegetative and reproductive stages in rice. In addition, the lesion-mimic on the mutant leaves, which only appeared during the summer, leads us to suspect that the protein may have a function associated with tolerance under the conditions.
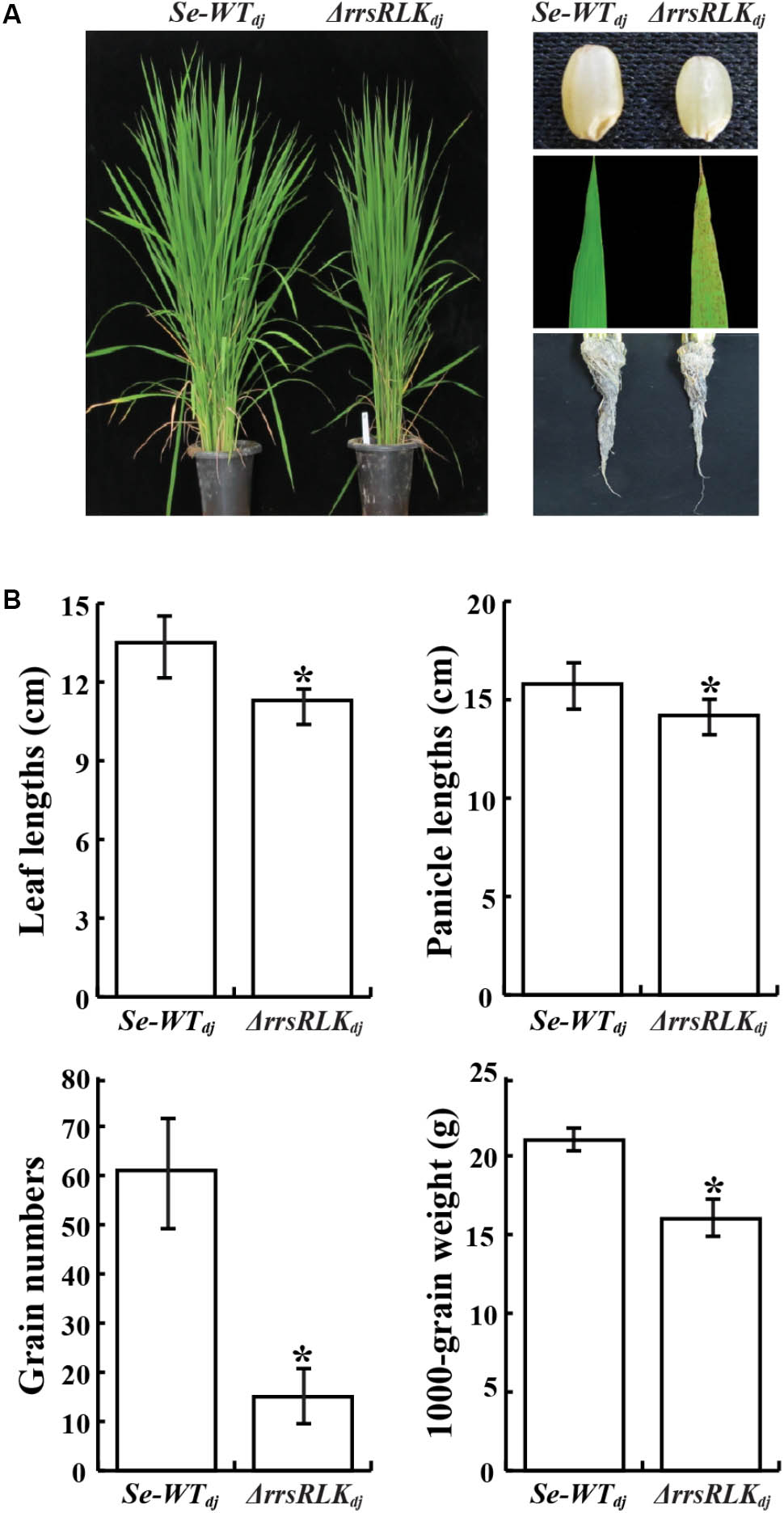
FIGURE 2. (A) Comparison of morphology between SE-WTdj and homozygous lines of ΔrrsRLKdj. Leaf lesions mimic symptoms and grain phenotypes of SE-WTdj and ΔrrsRLKdj (Right). (B) Phenotypic data compared between SE-WTdj and ΔrrsRLKdj (leaf and panicle lengths, grain numbers and weight) from the T3 generation grown in a paddy field were collected in 2015. Asterisks indicate P < 0.05 (Student’s t-test).
Os01g02290.2 Encoding a Tyr Receptor-Like Kinase Is Only Expressed in Rice
According to the rice genome database2, two genetic forms of Os01g02290 can be expressed by alternative splicing. Per annotation of the database, Os01g02290.1 encodes a Ser/Thr receptor-like kinase, and the alternative form Os01g02290.2 encodes a Tyr receptor-like kinase. To determine the expression of both forms, RT-PCR was carried out with specific primers (Supplementary Table S4) and RNAs extracted from each leaf stage of Dongjin and Hwayoung. As shown in Supplementary Figure S3A, the alternative splicing form 1 (Os01g02290.1) did not amplify at any stage, while Os01g02290.2 was expressed at all stages in both rice lines, though it was slightly decreased after the six-leaf stage in both lines. From additional RT-PCR to test tissue specificity of the gene expression, Os01g02290.1 did not appear in any tissue, while Os01g02290.2 was expressed in all tissues. The highest expression was seen in the root of both wild types (Supplementary Figure S3B), while the seed showed very slight expression. These results suggest that only Os01g02290.2 annotated to encode a Tyr receptor-like kinase is functional in rice, and the protein dominantly contributes at the vegetative phase and in the root.
Os01g02290.2 consists of four exons split by three introns. The domain structure of the protein deduced from the DNA sequence of Os01g02290.2 is predicted to have a signal peptide (aa 1–21), a transmembrane (aa 263–285), and a tyrosine kinase (aa 325–393) (Sakai et al., 20133). To determine if the kinase domain phosphorylates tyrosine residue in a substrate protein, a recombinant protein with the kinase domain tagged with 6x Histidine at the N-terminus was generated (Supplementary Figure S3C) and used for the tyrosine kinase activity assay. As shown in Supplementary Figure S3D, the recombinant protein showed significant tyrosine kinase activity, as seen in the control, c-Src (Proto-oncogene tyrosine-protein kinase), while the protein extract from Escherichia coli carrying only the pET28-a vector did not. This result reveals that the domain of the protein encoded by the Os01g02290.2 gene has kinase activity that phosphorylates a tyrosine(s) of a target protein.
Lack of rrsRLK Induces Expressional Changes of PR and Phytohormone-Related Genes
Many pathogenesis-related (PR) and phytohormone-related genes involved in plant defense have been reported (Agrawal et al., 2000; Bari and Jones, 2009). To test if absence of the rrsRLK gene has an effect on expression of PR and hormone-related genes, qRT-PCR analysis was carried out for 20 PR and six hormone-synthesis genes (listed in Supplementary Table S3) in Se-WTdj and ΔrrsRLKdj. From the analyses, we found that expression of OsPR1a, OsPR1b, OsLOX, and RBBTI4 was significantly increased in ΔrrsRLKdj, and OsPR10a was decreased in ΔrrsRLKdj (Figure 3A). The RBBTI4 gene encodes a trypsin inhibitor and is associated with immunity in animals, plants, and microbes (Farmer and Ryan, 1990; Song et al., 1999; Meyskens et al., 2001). Although it was not tested using purified RBBTI4 protein, inhibitory activity against trypsin in total protein extract from ΔrrsRLKdj was significantly increased compared to that from Se-WTdj (Supplementary Figure S4).
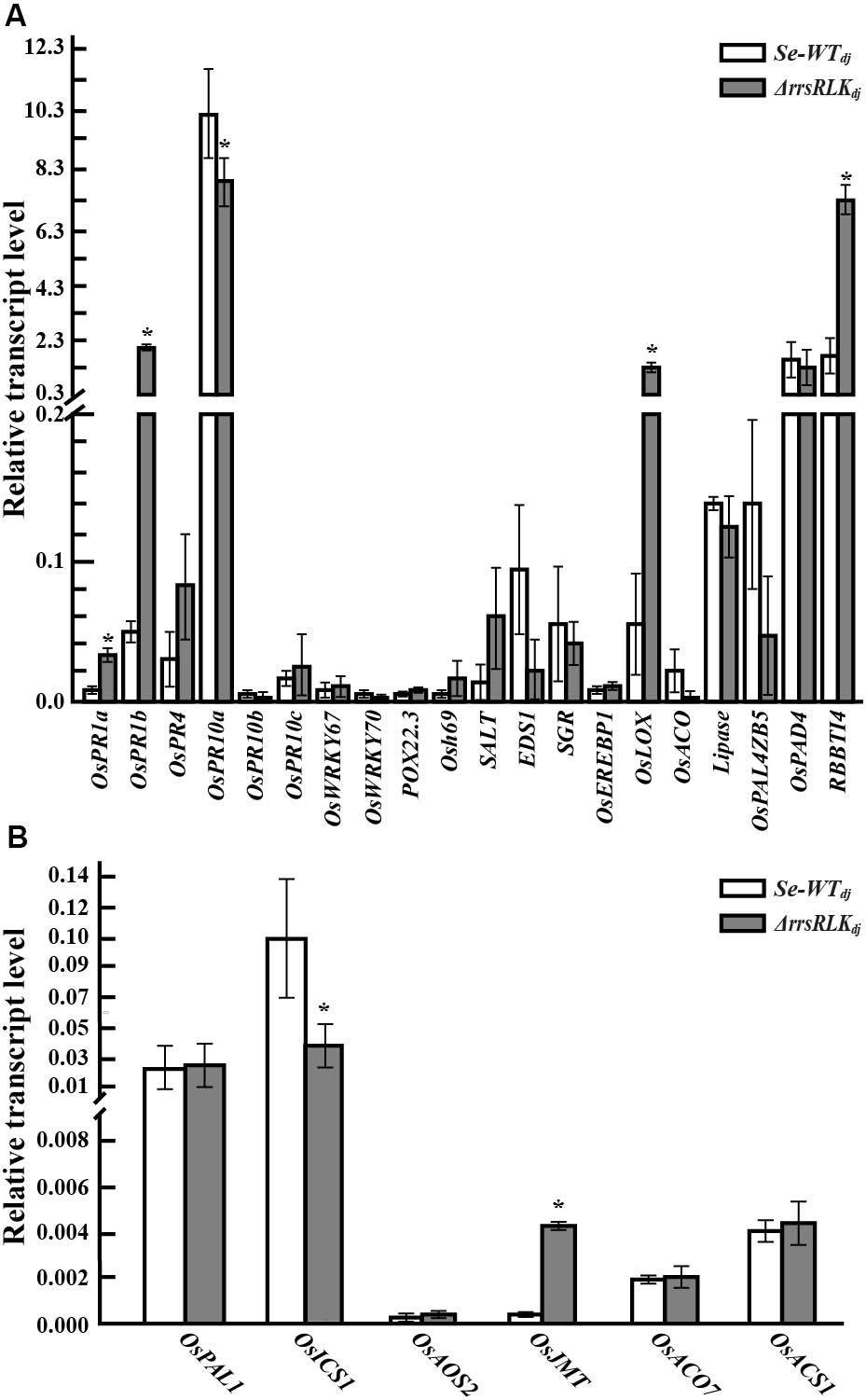
FIGURE 3. The expression analysis of pathogen-related (A) and hormone-related genes (B) in rice plants grown for 6 weeks. RNA for qRT-PCR was extracted from leaves of 6-week-old SE-WTdj and ΔrrsRLKdj mutant lines. Data represent mean ± SD of three independent measurements. Asterisks indicate statistical significance according to Student’s test; ∗P < 0.05.
In the test with phytohormone-related genes involved in salicylic acid (SA), jasmonic acid or methyl-jasmonic acid (JA or MeJA), and ethylene (ET) synthesis, expression of the OsICS1 gene in ΔrrsRLKdj was significantly decreased, and expression of the methyl-jasmonate biosynthesis (OsJMT) gene in ΔrrsRLKdj was significantly increased (Figure 3B). In ET-related genes, we did not observe any change in expression. These results reveal that deficiency of rrsRLK induces PR gene expression including OsPR1a, OsPR1b, OsLOX, and RBBTI4, as well as JA/MeJA synthesis.
The rrsRLK Protein Localizes in Cytoplasm, Not in the Plasma Membrane or Nuclei
The domain structure of rrsRLK protein suggests that it might be localized in the plasma membrane. To determine the subcellular localization of the protein, a construct to generate rrsRLK protein fused by GFP at the C-terminus was introduced in OC cells, and the GFP signal was monitored (Figure 4). In this transient expression assay, the rrsRLK-GFP signal was only detected in cytoplasm and was not merged with any signal from the nuclear marker NLS-mRFP or the plasma membrane marker PM-mRFP (Figure 4). This result indicates that rrsRLK is a cytoplasmic protein, not localized in the plasma membrane or nuclei.
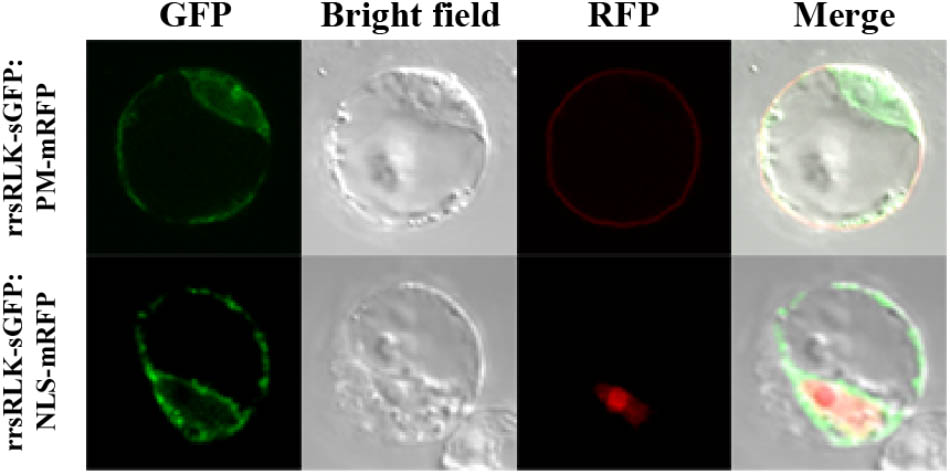
FIGURE 4. Sub-cellular localization of the rrsRLK protein in rice cells. The rrsRLK-GFP fusion protein was transiently expressed in OC cells and monitored with a confocal microscope equipped with filter sets for GFP (excitation wavelength/dichroic transition: 488/543 nm) and RFP (excitation wavelength/dichroic transition: 561/575 nm). GFP, OC cells showing rrsRLK-GFP green fluorescence in cytosol; RFP, the same cell showing plasma membrane and nuclear localization using PM-mRFP and NLS-mRFP markers (Kim et al., 2009); Bright field, a differential interference contrast image; Merged, the merged image of GFP and RFP.
The rrsRLK Protein Interacts With OsPEX11 and OsVOZ1
To identify interacting protein(s) downstream of rrsRLK-mediated signaling, we carried out a yeast two-hybrid assay using GAL4-rrsRLKk (rrsRLK kinase domain, aa 325–393) as a bait protein and a rice cDNA library. We found eight interacting candidates: O. sativa vascular plant one zinc finger protein 1 (OsVOZ1: Os01g0753000), O. sativa peroxisomal biogenesis factor 11 (OsPEX11: Os03g0117100), UMP synthase (Uridine 5′-monophosphate synthase), Chlorophyll A-B binding protein, Dehydrogenase, and three drought-induced proteins. In order to confirm the binding abilities of these candidate proteins, X-gal blue/white selection was carried out (Figure 5A). Only two of the proteins, OsVOZ1 and OsPEX11, showed positive signals. The interactions between full-length rrsRLK and the two proteins were confirmed by a coimmunoprecipitation assay in which the former was tagged with HA (rrsRLK-HA) and the latter with Myc (OsVOZ1-Myc and OsPEX11-Myc) (Figure 5B). The tagged proteins were co-expressed in rice mesophyll protoplasts using the maize Ubiquitin1 promoter. Analysis with anti-HA and anti-Myc antibodies showed that rrsRLK truly interacts with OsVOZ1 and OsPEX11, respectively.
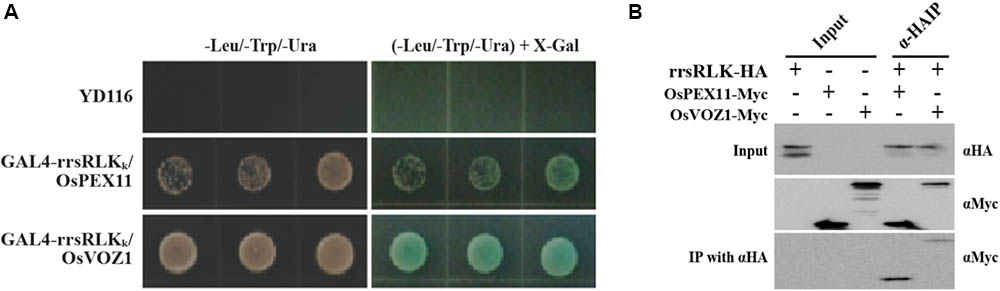
FIGURE 5. Interaction between rrsRLK, OsPEX11, and OsVOZ1. (A) Yeast two-hybrid assay carried out with rrsRLKk as a bait protein. The plasmids (pD32rrsRLKk and pD22Lib) were co-transformed into the yeast strain YD116. The transformants were grown on selective minimal medium without Leu, Trp, and Ura. The interaction was confirmed by measuring the activity of β-galactosidase in yeast. (B) Co-immunoprecipitation assay of rrsRLK, OsVOZ1, and OsPEX11. A combination of the constructs was transformed into rice protoplasts for transient expression and immunoprecipitated with anti-HA (αHA) or anti-Myc (αMyc) antibodies. The immunoprecipitates were analyzed with a Western blot with anti-HA or anti-Myc antibodies. 10% of the pre-cleared extracts were used as input controls.
Lack of OsVOZ1 and OsPEX11 Induced Resistance Against Xoo in Rice
Identification of the two interacting proteins led us to hypothesize that OsVOZ1 and/or OsPEX11 might be signaling partners, and that signaling is negative for Xoo-resistance. To test this hypothesis, we selected T-DNA insertion mutants for OsPEX11 (1B-07040 and 1B-03689) and OsVOZ1 (K-05631) from the rice T-DNA mutant population. 1B-07040 has a T-DNA insertion at the first exon of Os03g0117100 encoding OsPEX11 in Dongjin (ΔOsPEX11dj), and the 1B-03689 line has a T-DNA insertion 100 bp upstream from the end of the sixth exon of the gene in Hwayoung (ΔOsPEX11hy) (Supplementary Figure S5A). In K-05631, T-DNA is inserted 259 bp upstream from the start codon of Os01g0753000 encoding OsVOZ1 in Kittake (ΔOsVOZ1ki) (Supplementary Figure S5B). Expression of the OsPEX11 in 1B-07040 and 1B-03689 and OsVOZ1 in K-05631 was tested with RT-PCR (Supplementary Figures S5C–E) and T2 generation of the mutant rice plants inoculated by Xoo strains [PXO99 for ΔOsPEX11dj and ΔOsVOZ1ki, and HB01009 for ΔOsPEX11hy]. All lesion lengths scored at 14 days after inoculation were dramatically shortened compared to each wild type (Figures 6A,B). Lesion lengths on leaves of the ΔOsPEX11hy (4.8 ± 1.50 cm) and ΔOsPEX11dj (4.6 ± 1.59 cm) strains were reduced by 64.7 and 79.58% compared to the ones of Hwayoung (23.5 ± 1.96 cm) and Dongjin (14.8 ± 2.78 cm), respectively. Lesions on the leaves of the ΔOsVOZ1ki (3.9 ± 1.25 cm) line were reduced by about 65% compared to the ones of Kittake (15.85 ± 2.19cm). These shorter lesion lengths reveal that signals caused by interactions between rrsRLK and the two proteins have negative effects on resistance to Xoo in rice.
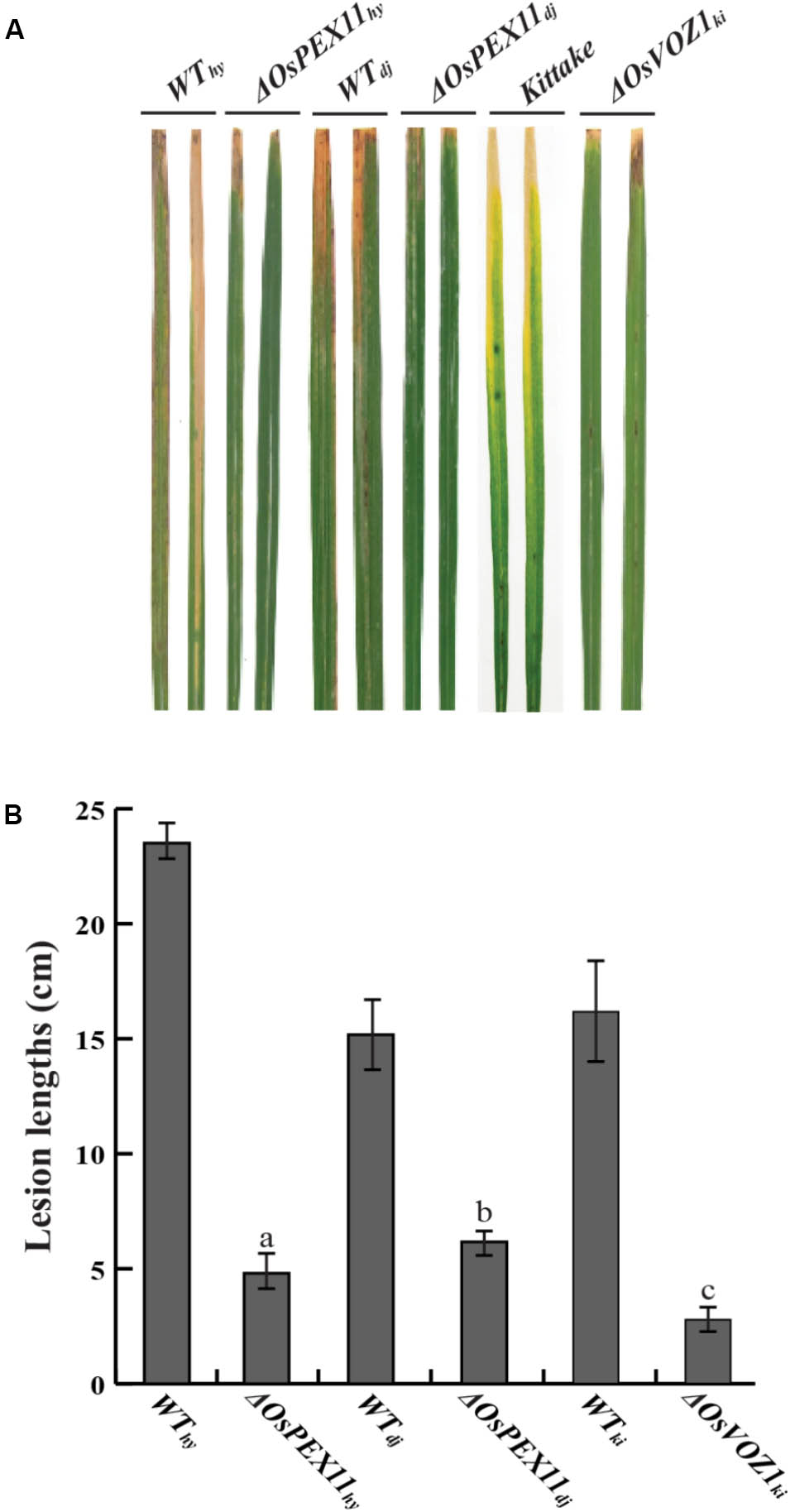
FIGURE 6. Inoculation assay with OsPEX11 (ΔOsPEX11hy and ΔOsPEX11dj) and OsVOZ1 (ΔOsVOZ1ki) gene knock-out mutant lines. (A) Xoo-inoculated leaves of wild type rice (Hwayoung, Dongjin, and Kittake) and homozygous line of OsPEX11 and OsVOZ1. Each line was inoculated with Xoo strains (PXO99 for Dongjin and Kittake and HB01009 for Hwayoung) using a clipping method, and the lesion development on the leaves was monitored for 14 DAI. (B) Lesion lengths of the wild type and mutant rice leaves. The lesion lengths were scored at 14 DAI. The averages and error ranges of each data set were calculated from 40 leaves. The experiment was repeated three times with consistent results. Different letters above bars indicate statistically significant differences as determined by one-way analysis of variance (ANOVA: a, b, c), P < 0.05.
H2O2 Is Accumulated in ΔrrsRLK Rice Due to a Decrease in Activities of ROS-Scavenging Enzymes
PEX11 is an essential protein for peroxisome multiplication (Erdmann and Blobel, 1995; Marshall et al., 1995), and peroxisomes are responsible for detoxification of ROS. The results described above suggest that ΔrrsRLK might have abnormal ROS contents. We investigated H2O2 contents in the roots, stems, and leaves of ΔrrsRLKdj and compared to those of wild type. H2O2 contents were increased in all of the tissues of ΔrrsRLK, particularly in the roots (Figure 7A). Accumulation of H2O2 in ΔrrsRLKdj was also investigated in an experiment using DAB staining. As shown in Supplementary Figure S6A, accumulation of H2O2 in the leaf, stem, and root of the ΔrrsRLKdj was clearly detected. Using a higher magnification than in Supplementary Figure S6A, we noted that the accumulation of H2O2 was higher in cells of ΔrrsRLKdj leaf. In addition, ΔrrsRLKdj cells had giant spots that seem to be peroxisomes, while the wild type has scattered small spots (Supplementary Figure S6B). These results suggest that rrsRLK contributes to regulation of the amount of H2O2 in rice. Furthermore, we tested gene expression of burst oxidase homologs (RBOHs), because additional ROS production in apoplast occurs through the protein (Kadota et al., 2014). However, rice RBOH genes showed no changes in expression (Supplementary Figure S7). Based on the results, we finally carried out experiments to test gene expression against most of available genes (around 80 genes) encoding ROS-scavenging enzymes and to measure the activity of ROS-scavenging enzymes, SOD, CAT, APX, and POX. Among the genes that we tested, Os03g11960 encoding Cu-Zn SOD, Os02g02400 encoding Cat, Os06g37150 encoding L-APX, and Os07g48020 encoding POX were representatively reduced in expression as shown in Supplementary Figure S8. The activity of SOD, CAT, POX, and APX in protein extract from the mutant rice leaves were significantly decreased in ΔrrsRLKdj compared to Dongjin (Figure 7B).
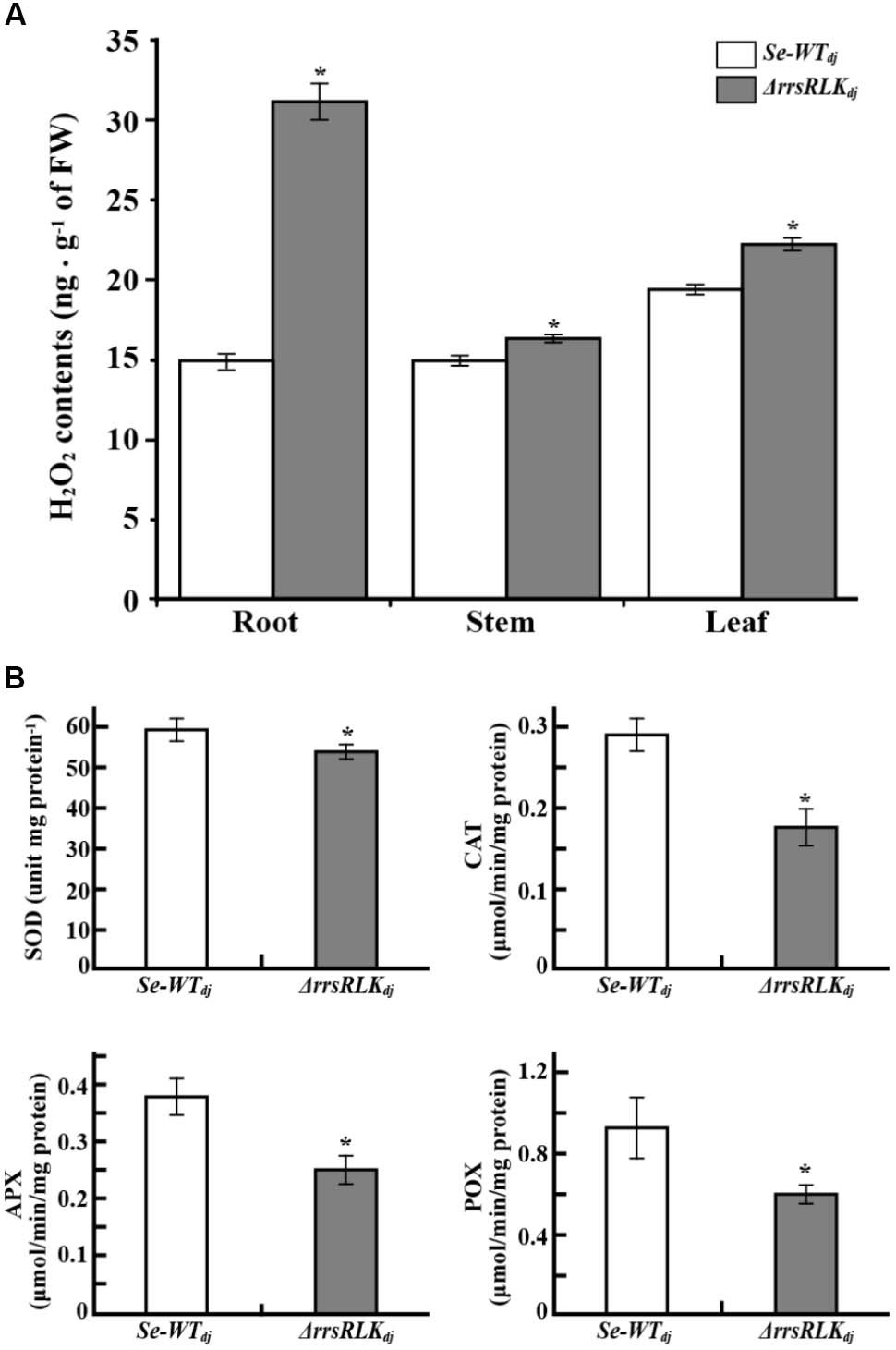
FIGURE 7. (A) H2O2 content in tissues of SE-WTdj and ΔrrsRLKdj. Content of H2O2 in 1 g of tissue (root, stem, and leaf) of SE-WTdj and ΔrrsRLKdj was spectrophotometrically determined at 436 nm by the modified method of Bernt and Bergmeyer (1974) using peroxidase enzyme. (B) Activity assay of superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), and peroxidase (POX) in leaves of SE-WTdj and ΔrrsRLKdj. Total protein was extracted from 1 g of leaves from SE-WTdj and ΔrrsRLKdj rice, and the activity of ROS-scavenging enzymes was spectrophotometrically monitored at A560nm (SOD), A240nm (CAT), A290nm (APX), and A436nm (POX) using a modified method of Beyer and Fridovich (Beyer and Fridovich, 1987). All assays were repeated more than three times, and the error bars of each sample denote the mean ± SD (n = 9). The asterisks indicate that a significant difference (P < 0.05) in the H2O2 was detected between transgenic plants and wild type.
Discussion
In higher plants as well as in animals, ROS (e.g., O2⋅-, H2O2, OH⋅, 1O2) have highly versatile properties. In early studies, ROS were regarded as toxic byproducts in plants, continuously produced through aerobic metabolism such as respiration and photosynthesis. It is now understood that ROS play beneficial roles in most cellular processes. The differentiation of plant tissues is characterized by ROS homeostasis, especially in roots. UPB1, a member of the basic/helix loop-helix (bHLH) TF family, is a well-known factor of root differentiation acting by controlling peroxidases expression (Tsukagoshi et al., 2010). ROS have been also identified as essential partners of auxin-signaling for gravitropism of maize root (Joo et al., 2001) and participate in rice seed germination associated with ABA (Ye et al., 2011). In addition to the evidence that ROS contribute to plant development, it has been suggested that they also play a role as rapidly generated signal molecules in response to stresses (Stael et al., 2015). Perception of PAMPs by RLKs immediately induces additional ROS production (Husi et al., 2000; Mersmann et al., 2010; Petutschnig et al., 2010) in apoplasts by nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (often called respiratory burst oxidase homologs, RBOHs) as a part of the PRR complex (Kadota et al., 2014). Both expression of Rboh genes and activation of the proteins are triggered by PAMP recognition (Ogasawara et al., 2008; Boudsocq et al., 2010; Morales et al., 2016). The additional ROS in apoplasts enter the cell via aquaporin to alter cellular processes such as activation of defense responses, regulation of photosynthesis, modulation of hormonal responses, and inhibition of growth and development (Mittler and Blumwald, 2015). These alterations are eventually terminated by reduction of ROS to appropriate levels by scavenging enzymes. APX, SOD, POX, and CAT are the enzymes responsible for ROS homeostasis (Mittler et al., 2004). Our results showed that H2O2 content is significantly increased in ΔrrsRLK regardless of pathogen infection (Figure 7A and Supplementary Figure S6). H2O2 accumulation in apoplasts of the ΔrrsRLKdj leaves was also observed (Supplementary Figure S6B). However, H2O2 accumulation in the tissues and apoplasts of ΔrrsRLK is not related to additional production by RBOHs because no change of Rboh (A, B, C, D, E, F, G, I) gene expression was observed in the mutant (Supplementary Figure S7). Although we cannot explain why more H2O2 was observed in the apoplasts of ΔrrsRLK, it is clear that abnormal ROS levels conferring broad spectrum resistance to Xoo are due to irregularity of the scavenging system caused by a reduction of scavenging-enzyme activity in the ΔrrsRLK lines (Figure 7B). However, we suspect that H2O2 level in ΔrrsRLK rice is not sufficient to enhance resistance to M. grisea, although it is resistant to Xoo. Around 10 times higher ROS amount by inhibition of H2O2 degradation in Bsr-d1 knock-out rice than wild type confers blast resistance (Li et al., 2017). Compared to the mutant, ΔrrsRLK rice possesses maximal two times higher H2O2 in root, and the increases in stem and leaf were smaller than that in root. Under consideration of H2O2-sensitivity of bacteria and fungi (Hong et al., 2013), this possibly explains why ΔrrsRLK rice has resistance to only the bacterial pathogen.
In our experiments, yeast two-hybrid assays and coimmunoprecipitation tests resulted in identification of two interacting proteins (Figure 5). The knock-out mutants of genes encoding the interacting proteins showed strong resistance to Xoo (Figure 6). One of the two proteins, OsPEX11, is essential in plants for peroxisome multiplication. Peroxisomes are derived from the endoplasmic reticulum and are created by division of pre-existing peroxisomes (Tabak et al., 2006). One of the important roles of plant peroxisomes is detoxification of cells by decreasing H2O2 levels produced by chloroplasts and mitochondria in photorespiration. Since the first peroxisome division protein, Sc-PEX11, was identified in yeast (Erdmann and Blobel, 1995; Marshall et al., 1995), homologs of Sc-PEX11 as well as many proteins involved in peroxisome biogenesis have been reported in Arabidopsis (McCartney et al., 2005). Ectopic expression of Sc-PEX11 caused organelle elongation/tubulation for multiplication, and the null mutant of the gene showed one or two giant peroxisomes in cells (Erdmann and Blobel, 1995; Marshall et al., 1995), as shown in our results (Supplementary Figure S6B). Lesion mimic under summer conditions (Figure 2) can be explained by this. Vigorous H2O2 production under the summer conditions is beyond the detoxifying ability in ΔrrsRLK mutant rice. These changes in ΔrrsRLK clearly illustrate the rrsRLK function in rice biology as a regulator for peroxisome multiplication and ROS homeostasis.
The VOZ protein was reported as a novel transcription factor in A. thaliana (Mitsuda et al., 2004). AtVOZ interacts with phytochrome B and accelerates flowering time in A. thaliana (Yasui et al., 2012). AtVOZ2 is controlled by light quality in a phytochrome-dependent manner (Yasui et al., 2012). Loss of function in voz genes enhanced resistance to freezing, cold, and drought, suggesting that it acts as a negative regulator for resistance against abiotic stresses. VOZs also act as a positive regulator for resistance to the fungal pathogen because the voz1voz2 mutant showed increased susceptibility for Colletotrichum higginsianum (Nakai et al., 2013). Rice cultivar Dongjin has two orthologs of AtVOZ2, OsVOZ1, and OsVOZ2, with 60.4% identity in amino acid sequences. Interestingly, OsVOZ2 knock-out mutant rice showed enhanced resistance (Cheong et al., 2013). In the report, OsVOZ2 is suggested as a target protein of the Xoo type-three effector XopNKX085, and the interaction between OsVOZ2 and XopNKX085 is important for Xoo virulence. This means that OsVOZ2 is a negative regulator for gene expression responsible for defense against Xoo. Although the report concluded that OsVOZ1 is not a target of XopNKX085, our results demonstrate that OsVOZ1 negatively functions in the expression of defense-related genes in rice (Figure 6) and is regulated by interaction with rrsRLK.
Over the past several decades, PR proteins have been reported in a wide variety of plant species (Linthorst and Van Loon, 1991). In our experiments, ΔrrsRLKdj mutant lines showed expression changes of various PR genes. Expression of OsPR1a, OsPR1b, OsLOX, and RBBTI4 was increased in ΔrrsRLKdj, while that of OsPR10a was decreased (Figure 3A). PR1 genes are often used as a marker for studying resistance in plants. In rice, 32 PR1 genes are predicted in the rice genome (van Loon et al., 2006). Among them, OsPR1a and OsPR1b are relatively well studied. These two PR1 genes are known to respond to fungal infection (Agrawal et al., 2001), environmental stresses (Agrawal et al., 2000, 2001), and chemical treatment (Malamy et al., 1990). The expression of OsPR1 genes is generally implicated in the antagonistic relationship between SA and JA (Mitsuhara et al., 2008), and SA-JA antagonism is likely conserved in rice (Yuan et al., 2007). However, the signaling pathways of SA and JA are often documented as synergistic (Xie et al., 2011). OsLOX is a JA biosynthesis gene in rice (Liu et al., 2012). JA biosynthesis requires three rounds of peroxisomal β-oxidation for the conversion of OPDA (12-Oxo-Phytodienoic Acid) to JA (Vick and Zimmerman, 1984; Dave and Graham, 2012). In our experiments, deficiency of rrsRLK induced gene expression of OsLOX and OsJMT (Figure 3). This suggests that rrsRLK or a signal mediated by rrsRLK has a negative effect on JA or MeJA synthesis. Expression of the OsPR10a gene is induced by pathogens including Xoo (Ryu et al., 2006) and diverse stimuli such as SA, JA, ethephone, NaCl, and ABA (Hwang et al., 2008). Although our test results demonstrated that SA and JA have an antagonistic relationship in ΔrrsRLKdj (Figure 3B), it is possible that OsPR10a expression is dominantly affected by the SA signaling in rice. RBBTI genes encode a cysteine-rich serine protease inhibitor in rice (Pang et al., 2013). There is a lot of evidence that protease inhibitors are inducible in pathogen attack (Roby et al., 1987; Rickauer et al., 1989; Cordero et al., 1994; McGurl et al., 1994; Kim and Choi, 2000; Pernas et al., 2000) and contribute to resistance against pathogenic proteases by inhibition (Peng and Black, 1976; Yamaleev et al., 1980; Brown and Adikaram, 1983). Bacterial proteases are one of the (a)virulence factors targeting host proteins. For instances, YopJ of Yersinia pestis and XopD of Xanthomonas campestris pv. vesicatoria are cysteine proteases that alter the sumoylation process by targeting SUMO (small ubiquitin-like modifier) proteins to inhibit multiple signaling (Orth et al., 2000; Hotson et al., 2003). On the other hand, host plants have developed appropriate defense measures, such as BBTI, to suppress the activity of proteases from pathogens. Since the first identification of soybean BBTI proteins (Bowman, 1946; Birk, 1961; Birk et al., 1963), homologs have been identified in many plants and their functions in the biology of plants and their interactions with microbial invaders studied (Haq et al., 2004; Yan et al., 2009). In rice, overexpression of RBBI2-3 and BBTI4 confers resistance to rice blast and bacterial leaf blight (Pang et al., 2013), respectively. Our results agree with the previous report: induced RBBTI4 expression in ΔrrsRLK mutant lines (Figure 3) showed resistance against Xoo strains. Additionally, our results confirm that expression of the RBBTI4 gene is also related to hormone signaling (Rakwal and Komatsu, 2000). Exogenous JA treatment results in reduction of ribulose-1, 5-bisphosphate carboxylase/oxygenase subunits and induction of novel proteins, including RBBTI4. This suggests that induction of RBBTI4 expression is positively regulated by JA signaling. Together, these findings suggest that the expressional change of PR genes in ΔrrsRLK is due to change of phytohormones synthesis, particularly of JA induced by H2O2 accumulation in ΔrrsRLK rice. ROS signals interact with several other signaling pathways such as nitric oxide and the stress hormones salicylic acid, jasmonic acid, and ethylene (Gechev et al., 2006). Their inter-relationships are very complex, and many questions about these relationships have yet to be answered. Hu et al. (2003) reported that H2O2 stimulates JA accumulation, but JA has no effect on H2O2 generation in suspension-cultured cells of Panax ginseng. Zhao and Sakai (2003) observed that H2O2 treatment stimulates activity of LOX, important for the biosynthesis of jasmonate in Cupressus Lusitanica cell cultures. Furthermore, a conditional oxidative stress-signaling mutant of Arabidopsis, which shows H2O2-activated oxidation and glutathione accumulation, accumulates transcripts of the four genes LOX3, OPR3, JAZ10, and VSP2 involved in JA synthesis and signaling, suggesting that intracellular oxidative stress activates JA signaling (Han et al., 2013).
Based on our data, deficiency of rrsRLK in rice causes H2O2 accumulation by abnormal peroxisome multiplication and reduction of activity of ROS-scavenging enzymes. This leads to an increase in a phytohormone such as JA (MeJA) and an induction of PR gene expression. Although questions remain to be answered, if rrsRLK interacts with an effector from Xoo and/or recognizes an internal molecule as a signal to modulate ROS level in rice, we conclude that rrsRLK is one of key regulators for ROS homeostasis and plays a negative role in rice defense against the bacterial pathogen Xoo.
Author Contributions
S-WL conceived and designed the experiments. YY, J-CP, JY, C-YK, M-HC, K-HJ, J-SJ, and GA performed the experiments and conducted bioinformatics analyses. YY, J-CP, and S-WL analyzed the data and wrote the manuscript.
Funding
This work was supported by a grant (20150738) from Kyung Hee University, Yongin, South Korea, and partially supported by a grant from the Mid-Career Researcher Program (NRF-2017R1A2B4004035), and National Research Foundation of Korea.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00577/full#supplementary-material
Footnotes
References
Agrawal, G. K., Jwa, N.-S., and Rakwal, R. (2000). A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphatase inhibitors. Biochem. Biophys. Res. Commun. 274, 157–165. doi: 10.1006/bbrc.2000.3114
Agrawal, G. K., Rakwal, R., and Jwa, N.-S. (2001). Differential induction of three pathogenesis-related genes, PR10, PR1b and PR5 by the ethylene generator ethephon under light and dark in rice (Oryza sativa L.) seedlings. J. Plant Physiol. 158, 133–137. doi: 10.1078/0176-1617-00186
Almagro, L., Gómez Ros, L. V., Belchi-Navarro, S., Bru, R., Ros Barceló, A., and Pedreño, M. A. (2009). Class III peroxidases in plant defence reactions. J. Exp. Bot. 60, 377–390. doi: 10.1093/jxb/ern277
Arthur, J. A. (2008). The African diaspora in the United States and Europe: the Ghanaian Experience. Abingdon: Routledge.
Bari, R., and Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Bernt, E., and Bergmeyer, H. (1974). “Inorganic peroxides,” in Methods of Enzymatic Analysis, ed. H. U. Bergmeyer, (New York, N.Y: Academic Press), 2246–2248. doi: 10.1016/B978-0-12-091304-6.50090-2
Beyer, W. F., and Fridovich, I. (1987). Assaying for superoxide dismutase activity: some large consequences of minor changes in conditions. Anal. Biochem. 161, 559–566. doi: 10.1016/0003-2697(87)90489-1
Bindschedler, L. V., Dewdney, J., Blee, K. A., Stone, J. M., Asai, T., Plotnikov, J., et al. (2006). Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 47, 851–863. doi: 10.1111/j.1365-313X.2006.02837.x
Birk, Y. (1961). Purification and some properties of a highly active inhibitor of trypsin and α-chymotrypsin from soybeans. Biochim. Biophys. Acta 54, 378–381. doi: 10.1016/0006-3002(61)90387-0
Birk, Y., Gertler, A., and Khalef, S. (1963). A pure trypsin inhibitor from soya beans. Biochem. J. 87, 281–284. doi: 10.1042/bj0870281
Block, A., Li, G., Fu, Z. Q., and Alfano, J. R. (2008). Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11, 396–403. doi: 10.1016/j.pbi.2008.06.007
Bogdanove, A. J., Koebnik, R., Lu, H., Furutani, A., Angiuoli, S. V., Patil, P. B., et al. (2011). Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 193, 5450–5464. doi: 10.1128/JB.05262-11
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Boudsocq, M., Willmann, M. R., McCormack, M., Lee, H., Shan, L., He, P., et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422. doi: 10.1038/nature08794
Bowman, D. E. (1946). Differentiation of soy bean antitryptic factors. Proc. Soc. Exp. Biol. Med. 63, 547–550. doi: 10.3181/00379727-63-15668
Braun, D. M., and Walker, J. C. (1996). Plant transmembrane receptors: new pieces in the signaling puzzle. Trends Biochem. Sci. 21, 70–73. doi: 10.1016/S0968-0004(96)80185-X
Brown, A., and Adikaram, N. (1983). A role for pectinase and protease inhibitors in fungal rot development in tomato fruits. J. Phytopathol. 106, 239–251. doi: 10.1111/j.1439-0434.1983.tb00049.x
Brueggeman, R., Rostoks, N., Kudrna, D., Kilian, A., Han, F., Chen, J., et al. (2002). The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 99, 9328–9333. doi: 10.1073/pnas.142284999
Cheong, H., Kim, C.-Y., Jeon, J.-S., Lee, B.-M., Moon, J. S., and Hwang, I. (2013). Xanthomonas oryzae pv. oryzae type III effector XopN targets OsVOZ2 and a putative thiamine synthase as a virulence factor in rice. PLoS One 8:e73346. doi: 10.1371/journal.pone.0073346
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476. doi: 10.1105/tpc.105.036574
Choi, S. C., Lee, S., Kim, S.-R., Lee, Y.-S., Liu, C., Cao, X., et al. (2014). Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3. Plant Physiol. 164, 1326–1337. doi: 10.1104/pp.113.228049
Cordero, M. J., Raventós, D., and Segundo, B. (1994). Expression of a maize proteinase inhibitor gene is induced in response to wounding and fungal infection: systemic wound-response of a monocot gene. Plant J. 6, 141–150. doi: 10.1046/j.1365-313X.1994.6020141.x
Cunnac, S., Lindeberg, M., and Collmer, A. (2009). Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12, 53–60. doi: 10.1016/j.mib.2008.12.003
Dardick, C., and Ronald, P. (2006). Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog. 2:e2. doi: 10.1371/journal.ppat.0020002
Dave, A., and Graham, I. A. (2012). Oxylipin signaling: a distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA). Front. Plant Sci. 3:42. doi: 10.3389/fpls.2012.00042
Erdmann, R., and Blobel, G. (1995). Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 128, 509–523. doi: 10.1083/jcb.128.4.509
Farmer, E. E., and Ryan, C. A. (1990). Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci. U.S.A. 87, 7713–7716. doi: 10.1073/pnas.87.19.7713
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev-phyto-072910-095339
Gechev, T. S., Van Breusegem, F., Stone, J. M., Denev, I., and Laloi, C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28, 1091–1101. doi: 10.1002/bies.20493
Han, Y., Mhamdi, A., Chaouch, S., and Noctor, G. (2013). Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ. 36, 1135–1146. doi: 10.1111/pce.12048
Haq, S. K., Atif, S. M., and Khan, R. H. (2004). Protein proteinase inhibitor genes in combat against insects, pests, and pathogens: natural and engineered phytoprotection. Arch. Biochem. Biophys. 431, 145–159. doi: 10.1016/j.abb.2004.07.022
Hong, J. K., Kang, S. R., Kim, Y. H., Yoon, D. J., Kim, D. H., Kim, H. J., et al. (2013). Hydrogen peroxide- and nitric oxide-mediated disease control of bacteria wilt in tomato plants. Plant Pathol. J. 29, 386–396. doi: 10.5423/PPJ.OA.04.2013.0043
Hong, S.-Y., Seo, P. J., Cho, S.-H., and Park, C.-M. (2012). Preparation of leaf mesophyll protoplasts for transient gene expression in Brachypodium distachyon. J. Plant Biol. 55, 390–397. doi: 10.1007/s12374-012-0159-y
Hotson, A., Chosed, R., Shu, H., Orth, K., and Mudgett, M. B. (2003). Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50, 377–389. doi: 10.1046/j.1365-2958.2003.03730.x
Hu, X., Neill, S., Cai, W., and Tang, Z. (2003). Hydrogen peroxide and jasmonic acid mediate oligogalacturonic acid-induced saponin accumulation in suspension-cultured cells of Panax ginseng. Physiol. Plant. 118, 414–421. doi: 10.1034/j.1399-3054.2003.00124.x
Husi, H., Ward, M. A., Choudhary, J. S., Blackstock, W. P., and Grant, S. G. N. (2000). Proteomic analysis of NMDA receptor–adhesion protein signaling complexes. Nat. Neurosci. 3, 661–669. doi: 10.1038/76615
Hwang, S.-H., Lee, I. A., Yie, S. W., and Hwang, D.-J. (2008). Identification of an OsPR10a promoter region responsive to salicylic acid. Planta 227, 1141–1150. doi: 10.1007/s00425-007-0687-8
Jain, M., Nijhawan, A., Tyagi, A. K., and Khurana, J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651. doi: 10.1016/j.bbrc.2006.04.140
Jeon, J. S., Lee, S., Jung, K. H., Jun, S. H., Jeong, D. H., Lee, J., et al. (2000). T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22, 561–570. doi: 10.1046/j.1365-313x.2000.00767.x
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Joo, J. H., Bae, Y. S., and Lee, J. S. (2001). Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol. 126, 1055–1060. doi: 10.1104/pp.126.3.1055
Kadota, Y., Sklenar, J., Derbyshire, P., Stransfeld, L., Asai, S., Ntoukakis, V., et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55. doi: 10.1016/j.molcel.2014.02.021
Kauffman, H. E., Reddy, A. P. K., Hsieh, S. P. Y., and Merca, S. D. (1973). An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541.
Khan, M. A., Naeem, M., and Iqbal, M. (2014). Breeding approaches for bacterial leaf blight resistance in rice (Oryza sativa L.), current status and future directions. Eur. J. Plant Pathol. 139, 27–37. doi: 10.1007/s10658-014-0377-x
Kim, S.-H., and Choi, N.-S. (2000). Purification and characterization of subtilisin DJ-4 secreted by Bacillus sp. strain DJ-4 screened from Doen-Jang. Biosci. Biotechnol. Biochem. 64, 1722–1725. doi: 10.1271/bbb.64.1722
Kim, S.-R., Lee, D.-Y., Yang, J.-I., Moon, S., and An, G. (2009). Cloning vectors for rice. J. Plant Biol. 52, 73–78. doi: 10.1007/s12374-008-9008-4
Kunze, G., Zipfel, C., Robatzek, S., Niehaus, K., Boller, T., and Felix, G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16, 3496–3507. doi: 10.1105/tpc.104.026765
Lee, B.-M., Park, Y.-J., Park, D.-S., Kang, H.-W., Kim, J.-G., Song, E.-S., et al. (2005). The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33, 577–586. doi: 10.1093/nar/gki206
Lee, S.-K., Song, M.-Y., Seo, Y.-S., Kim, H.-K., Ko, S., Cao, P.-J., et al. (2009). Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil–nucleotide-binding–leucine-rich repeat genes. Genetics 181, 1627–1638. doi: 10.1534/genetics.108.099226
Li, W., Zhu, Z., Chern, M., Yin, J., Yang, C., Ran, L., et al. (2017). A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170, 114–126. doi: 10.1016/j.cell.2017.06.008
Linthorst, H. J., and Van Loon, L. (1991). Pathogenesis-related proteins of plants. Crit. Rev. Plant Sci. 10, 123–150. doi: 10.1080/07352689109382309
Liu, X., Li, F., Tang, J., Wang, W., Zhang, F., Wang, G., et al. (2012). Activation of the jasmonic acid pathway by depletion of the hydroperoxide lyase OsHPL3 reveals crosstalk between the HPL and AOS branches of the oxylipin pathway in rice. PLoS One 7:e50089. doi: 10.1371/journal.pone.0050089
Malamy, J., Carr, J. P., Klessig, D. F., and Raskin, I. (1990). Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004. doi: 10.1126/science.250.4983.1002
Marshall, P. A., Krimkevich, Y. I., Lark, R. H., Dyer, J. M., Veenhuis, M., and Goodman, J. M. (1995). Pmp27 promotes peroxisomal proliferation. J. Cell Biol. 129, 345–355. doi: 10.1083/jcb.129.2.345
McCartney, A. W., Greenwood, J. S., Fabian, M. R., White, K. A., and Mullen, R. T. (2005). Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17, 3513–3531. doi: 10.1105/tpc.105.036350
McGurl, B., Orozco-Cardenas, M., Pearce, G., and Ryan, C. A. (1994). Overexpression of the prosystemin gene in transgenic tomato plants generates a systemic signal that constitutively induces proteinase inhibitor synthesis. Proc. Natl. Acad. Sci. U.S.A. 91, 9799–9802. doi: 10.1073/pnas.91.21.9799
Medzhitov, R. (2001). Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1, 135–145. doi: 10.1038/35100529
Mersmann, S., Bourdais, G., Rietz, S., and Robatzek, S. (2010). Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 154, 391–400. doi: 10.1104/pp.110.154567
Meyskens, F. L. Jr., Farmer, P., and Fruehauf, J. P. (2001). Redox regulation in human melanocytes and melanoma. Pigment Cell Res. 14, 148–154. doi: 10.1034/j.1600-0749.2001.140303.x
Mitsuda, N., Hisabori, T., Takeyasu, K., and Sato, M. H. (2004). VOZ; isolation and characterization of novel vascular plant transcription factors with a one-zinc finger from Arabidopsis thaliana. Plant Cell Physiol. 45, 845–854. doi: 10.1093/pcp/pch101
Mitsuhara, I., Iwai, T., Seo, S., Yanagawa, Y., Kawahigasi, H., Hirose, S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genomics 279, 415–427. doi: 10.1007/s00438-008-0322-9
Mittler, R., and Blumwald, E. (2015). The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27, 64–70. doi: 10.1105/tpc.114.133090
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Morales, J., Kadota, Y., Zipfel, C., Molina, A., and Torres, M.-A. (2016). The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 67, 1663–1676. doi: 10.1093/jxb/erv558
Nakai, Y., Nakahira, Y., Sumida, H., Takebayashi, K., Nagasawa, Y., Yamasaki, K., et al. (2013). Vascular plant one-zinc-finger protein 1/2 transcription factors regulate abiotic and biotic stress responses in Arabidopsis. Plant J. 73, 761–775. doi: 10.1111/tpj.12069
Nolen, B., Taylor, S., and Ghosh, G. (2004). Regulation of protein kinases: controlling activity through activation segment conformation. Mol. Cell 15, 661–675. doi: 10.1016/j.molcel.2004.08.024
Ogasawara, Y., Kaya, H., Hiraoka, G., Yumoto, F., Kimura, S., Kadota, Y., et al. (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 283, 8885–8892. doi: 10.1074/jbc.M708106200
Orth, K., Xu, Z., Mudgett, M. B., Bao, Z. Q., Palmer, L. E., Bliska, J. B., et al. (2000). Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290, 1594–1597. doi: 10.1126/science.290.5496.1594
Pang, Z., Zhou, Z., Yin, D., Lv, Q., Wang, L., Xu, X., et al. (2013). Transgenic rice plants overexpressing BBTI4 confer partial but broad-spectrum bacterial blight resistance. J. Plant Biol. 56, 383–390. doi: 10.1007/s12374-013-0277-1
Peng, J., and Black, L. (1976). Increased proteinase inhibitor activity in response to infection of resistant tomato plants by Phytophthora infestans. Phytopathology 66, 958–963.
Pernas, M., Sánchez-Monge, R., and Salcedo, G. (2000). Biotic and abiotic stress can induce cystatin expression in chestnut. FEBS Lett. 467, 206–210. doi: 10.1016/S0014-5793(00)01157-1
Petutschnig, E. K., Jones, A. M., Serazetdinova, L., Lipka, U., and Lipka, V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285, 28902–28911. doi: 10.1074/jbc.M110.116657
Pitzschke, A., Forzani, C., and Hirt, H. (2006). Reactive oxygen species signaling in plants. Antioxid. Redox Signal. 8, 1757–1764. doi: 10.1089/ars.2006.8.1757
Rakwal, R., and Komatsu, S. (2000). Role of jasmonate in the rice (Oryza sativa L.) self-defense mechanism using proteome analysis. Electrophoresis 21, 2492–2500. doi: 10.1002/1522-2683(20000701)21:12<2492::AID-ELPS2492>3.0.CO;2-2
Rickauer, M., Fournier, J., and Esquerré-Tugayé, M.-T. (1989). Induction of proteinase inhibitors in tobacco cell suspension culture by elicitors of Phytophthora parasitica var. nicotianae. Plant Physiol. 90, 1065–1070. doi: 10.1104/pp.90.3.1065
Roby, D., Toppan, A., and Esquerré-Tugayé, M.-T. (1987). Cell surfaces in plant micro-organism interactions. VIII. Increased proteinase inhibitor activity in melon plants in response to infection by Colletotrichum lagenarium or to treatment with an elicitor fraction from this fungus. Physiol. Mol. Plant Pathol. 30, 453–460. doi: 10.1016/0885-5765(87)90024-5
Ryu, H.-S., Han, M., Lee, S.-K., Cho, J.-I., Ryoo, N., Heu, S., et al. (2006). A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 25, 836–847. doi: 10.1007/s00299-006-0138-1
Sakai, H., Lee, S. S., Tanaka, T., Numa, H., Kim, J., Kawahara, Y., et al. (2013). Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 54:e6. doi: 10.1093/pcp/pcs183
Song, H. K., Kim, Y. S., Yang, J. K., Moon, J., Lee, J. Y., and Suh, S. W. (1999). Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 Å resolution. J. Mol. Biol. 293, 1133–1144. doi: 10.1006/jmbi.1999.3239
Stael, S., Kmiecik, P., Willems, P., Van Der Kelen, K., Coll, N. S., Teige, M., et al. (2015). Plant innate immunity–sunny side up? Trends Plant Sci. 20, 3–11. doi: 10.1016/j.tplants.2014.10.002
Tabak, H. F., Hoepfner, D., Zand, A. V., Geuze, H. J., Braakman, I., and Huynen, M. A. (2006). Formation of peroxisomes: present and past. Biochim. Biophys. Acta 1763, 1647–1654. doi: 10.1016/j.bbamcr.2006.08.045
Thomma, B. P., Nürnberger, T., and Joosten, M. H. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15. doi: 10.1105/tpc.110.082602
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Tsukagoshi, H., Busch, W., and Benfey, P. N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616. doi: 10.1016/j.cell.2010.10.020
van Loon, L. C., Rep, M., and Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Vick, B. A., and Zimmerman, D. C. (1984). Biosynthesis of jasmonic acid by several plant species. Plant Physiol. 75, 458–461. doi: 10.1104/pp.75.2.458
Xie, X.-Z., Xue, Y.-J., Zhou, J.-J., Zhang, B., Chang, H., and Takano, M. (2011). Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol. Plant 4, 688–696. doi: 10.1093/mp/ssr005
Yamaleev, A., Mukhsinov, V. K., Isaev, R., Yamaleeva, A., and Krivchenko, V. (1980). Activity of protease inhibitors and resistance of wheat to the causal agent of hard smut. S-kh. Biol. 15, 143–144.
Yan, K. M., Chang, T., Soon, S. A., and Huang, F. Y. (2009). Purification and characterization of bowman-birk protease inhibitor from rice coleoptiles. J. Chin. Chem. Soc. 56, 949–960. doi: 10.1002/jccs.200900139
Yang, J., Lee, S., Hang, R., Kim, S. R., Lee, Y. S., Cao, X., et al. (2013). OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 73, 566–578. doi: 10.1111/tpj.12057
Yasui, Y., Mukougawa, K., Uemoto, M., Yokofuji, A., Suzuri, R., Nishitani, A., et al. (2012). The phytochrome-interacting VASCULAR PLANT ONE–ZINC FINGER1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell 24, 3248–3263. doi: 10.1105/tpc.112.101915
Ye, N., Zhu, G., Liu, Y., Zhang, A., Li, Y., Liu, R., et al. (2011). Ascorbic acid and reactive oxygen species are involved in the inhibition of seed germination by abscisic acid in rice seeds. J. Exp. Bot. 63, 1809–1822. doi: 10.1093/jxb/err336
Yuan, Y., Zhong, S., Li, Q., Zhu, Z., Lou, Y., Wang, L., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5, 313–324. doi: 10.1111/j.1467-7652.2007.00243.x
Zhao, J., and Sakai, K. (2003). Multiple signalling pathways mediate fungal elicitor-induced b-thujaplicin biosynthesis in Cupressus lusitanica cell cultures. J. Exp. Bot. 54, 647–656. doi: 10.1093/jxb/erg062
Keywords: peroxisomal biogenesis factor 11, vascular one zinc-finger 1, plant defense, reactive oxygen species, ROS-homeostasis, Xanthomonas oryzae pv. oryzae
Citation: Yoo Y, Park J-C, Cho M-H, Yang J, Kim C-Y, Jung K-H, Jeon J-S, An G and Lee S-W (2018) Lack of a Cytoplasmic RLK, Required for ROS Homeostasis, Induces Strong Resistance to Bacterial Leaf Blight in Rice. Front. Plant Sci. 9:577. doi: 10.3389/fpls.2018.00577
Received: 12 February 2018; Accepted: 13 April 2018;
Published: 18 May 2018.
Edited by:
Víctor Flors, Jaume I University, SpainReviewed by:
Lucía Jordá, Universidad Politécnica de Madrid (UPM), SpainKaijun Zhao, Institute of Crop Science, Chinese Academy of Agricultural Sciences, China
Copyright © 2018 Yoo, Park, Cho, Yang, Kim, Jung, Jeon, An and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Won Lee, c3dsZWU2ODAzQGtodS5hYy5rcg==
†These authors have contributed equally to this work.
‡Present address: Jungil Yang, Institute for Molecular Physiology, Heinrich-Heine-Universität Düsseldorf, Düsseldorf, Germany Chi-Yeol Kim, Department of Agricultural Biotechnology, College of Agriculture and Life Sciences, Seoul National University, Seoul, South Korea
 Youngchul Yoo
Youngchul Yoo Jong-Chan Park
Jong-Chan Park Man-Ho Cho
Man-Ho Cho Jungil Yang
Jungil Yang Chi-Yeol Kim
Chi-Yeol Kim Ki-Hong Jung
Ki-Hong Jung Jong-Seong Jeon
Jong-Seong Jeon Gynheung An
Gynheung An Sang-Won Lee
Sang-Won Lee