- Molecular Biology of Vegetable Laboratory, College of Horticulture, Shenyang Agricultural University, Shenyang, China
The rapid spread of clubroot disease, which is caused by Plasmodiophora brassicae, threatens Brassicaceae crop production worldwide. Breeding plants that have broad-spectrum disease resistance is one of the best ways to prevent clubroot. In the present study, eight Chinese cabbage germplasms were screened using published clubroot-resistant (CR) loci-/gene-linked markers. A CR gene Crr3 potential carrier “85-74” was detected which linked to marker BRSTS61; however, “85-74” shows different responses to local pathogens “LAB-19,” “LNND-2,” and “LAB-10” from “CR-73” which harbors Crr3. We used a next-generation sequencing-based bulked segregant analysis approach combined with genetic mapping to detect CR genes in an F2 segregant population generated from a cross between the Chinese cabbage inbred lines “85-74” (CR) and “BJN3-1” (clubroot susceptible). The “85-74” line showed resistance to a local pathogen “LAB-19” which was identified as race 4; a genetic analysis revealed that the resistance was conferred by a single dominant gene. The CR gene which we named CRd was mapped to a 60 kb (1 cM) region between markers yau389 and yau376 on chromosome A03. CRd is located upstream of Crr3 which was confirmed based on the physical positions of Crr3 linked markers. The identification of CRd linked markers can be applied to marker-assisted selection in the breeding of new CR cultivars of Chinese cabbage and other Brassica crops.
Introduction
Chinese cabbage (Brassica rapa ssp. pekinensis) is one of the most important leafy head vegetables cultivated in China, Korea, and Japan. Its production has been undermined by the rapid spread of clubroot disease, resulting in major economic losses. The soil-borne obligate plant pathogen Plasmodiophora brassicae Woronin causes clubroot in Brassica crops, blocking nutrient and water transport (Voorrips et al., 2003). The life cycle of P. brassicae has not been well understood until now. The P. brassicae infection starts from primary zoospores releasing and causes root hair infection and then primary zoospore or secondary zoospores induce cortical infection leading to the formation of galls on the roots (McDonald et al., 2014). Crop rotation and application of fluazinam and cyazofamid can effectively reduce the viability of resting P. brassicae spores and prevent infection (Ransom et al., 1991; Suzuki et al., 1995; Wallenhammar, 1996; Mitani et al., 2003; Townley and Fox, 2003; Miller et al., 2007; Kutcher et al., 2013; Peng et al., 2014). Although these approaches alleviate the symptoms of clubroot, they do not eradicate the disease. Breeding of clubroot-resistant (CR) cultivars is a desirable strategy for controlling clubroot owing to its advantages such as low cost and environment friendliness.
CRa, CRb, CRc, CRk, Crr1, Crr2, Crr3, Crr4, PbBa3.1, PbBa3.3, and QS_B3.1 genes have been identified in European fodder turnips (Matsumoto et al., 1998; Suwabe et al., 2003, 2006; Hirai et al., 2004; Piao et al., 2004; Sakamoto et al., 2008; Chen et al., 2013; Pang et al., 2014). Most of these genes are from different genetic resources and are associated with distinct P. brassicae pathotypes. CRa and CRb were resistant to race 2 and race 4, respectively. Crr1, Crr2, and Crr4 from Siloga were resistant to Ano-01 and Wakayama-01. CRc and CRk from Debra were resistant to isolates M85 and K04. Crr3 was detected from Milan White which was resistant to isolate Ano-01. PbBa3.1 and PbBa3.3 were detected from ECD04 conferring resistance to Pb2 and Pb7, respectively. Several CR genes were also recently mapped in B. rapa (Yu et al., 2016; Huang et al., 2017). Some of the genes and loci including CRa, CRb, and QS_B3.1, Crr3, CRk, and PbBa3.3 were clustered in a proximal region of chromosome A03 in B. rapa; whether they represent a single or multiple genes remains to be determined. CRa and Crr1a have been cloned and are known to encode Toll-interleukin-1 receptor-like domain-nucleotide binding site-leucine-rich repeat (TIR-NBS-LRR) proteins. It was reported that CRb is the same as CRa (Hatakeyama et al., 2017); however, the identities of the remaining CR genes require confirmation.
Molecular markers linked to CR loci or genes are essential for pyramiding several CR genes into one cultivar through marker-assisted selection (MAS). MAS has been successfully used for transforming CR genes into Chinese cabbage (Yoshikawa, 1981; Zhang et al., 2012). However, B. rapa, Brassica oleracea, and Brassica napus plants known to harbor genes conferring specific resistance to P. brassicae (Rocherieux et al., 2004; Werner et al., 2008; Chen et al., 2013) have all exhibited loss of resistance within a few years (Tjallingii, 1965; Kuginuki et al., 1999). Therefore, identifying novel CR genes or alleles associated with resistance to different pathotypes is essential for overcoming the challenges of co-existing pathotypes and the rapid mutation rate of P. brassicae in the field.
Next-generation sequencing (NGS)-based bulked segregant analysis (BSA) is a powerful tool for mapping disease resistance gene/genes that has been applied to Arabidopsis, rice (Oryza sativa), sorghum (Sorghum bicolor), soybean (Gycin emax), wheat (Triticum turgidum), and cotton (Gossypium; Trick et al., 2012; Yang et al., 2013; Uchida et al., 2014; Han et al., 2015; Song et al., 2017; Zhu et al., 2017). In the present study, we found the Chinese cabbage inbred line “85-74,” which exhibited resistance to the local pathogen “LAB-19” (race 4) and was distinct from CR germplasms harboring CRa, Crr1, and Crr3. We employed NGS-based BSA to identify the CR gene/genes in “85-74.” Our results can provide a basis for breeding new CR cultivars of Chinese cabbage.
Materials and Methods
Plant Materials
Eight Chinese cabbage germplasms including “CR Shinki,” “CR-77,” “CR-20,” “CR-75,” “CR-26,” “CR-73,” “85-74,” and “BJN3-1” were used in this study. “CR Shinki” harbors the CRa and CRb gene, and “CR-77,” “CR-20,” “CR-75,” “CR-26,” “CR-73,” and “85-74” were genotyped using published CR loci-/gene-linked markers (Saito et al., 2006; Suwabe et al., 2006; Sakamoto et al., 2008; Ueno et al., 2012; Zhang et al., 2012; Chen et al., 2013; Hatakeyama et al., 2013; Pang et al., 2014). To evaluate the resistance of CR resources against different pathotypes of P. brassicae, eight CR inbred lines were inoculated with 11 local pathogens. Their pathotypes were classified based on Williams’ clubroot differential set (Williams, 1966), which includes Jersey Queen, Badger shipper, Laurentian, and Wilhelmsburger. The clubroot-susceptible inbred line of Chinese cabbage “BJN3-1” served as a negative control.
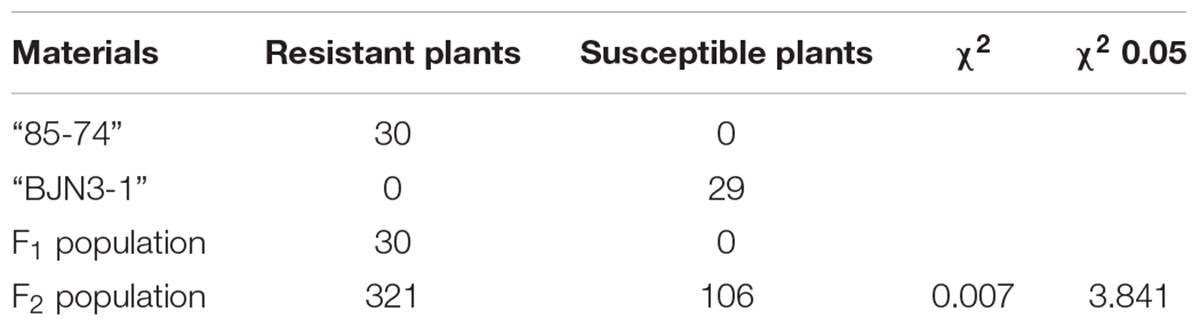
An analysis of CR loci-/gene-linked markers revealed that “85-74” and “CR-73” harbored the Crr3 gene, but showed distinct responses to three local pathogen isolates. We therefore crossed “85-74” and “BJN3-1” and then self-pollinated the offspring to produce an F2 population; 432 F2 individuals were used for genetic analysis and 127 were self-pollinated to generate an F3 population that was used for CR tests.
Pathogen Inoculation and CR Tests
A total of 11 field isolates were collected from infected Chinese cabbage or canola from different province of China and maintained on the roots of a susceptible Chinese cabbage “91-12” and stored in -20°C until required. These pathogens were classified according to Williams’ clubroot differential set and used to evaluate the resistance of eight Chinese cabbage germplasms; additionally, 24 seeds from each of eight CR Chinese cabbage inbred lines along with Williams’s differential set were grown for pathogen screening in the spring of 2014.
A total of 36 seeds of F1 individuals and 432 seeds of F2 individuals of “85-74” and “BJN3-1” were grown and inoculated with “LAB-19” in 2015; 36 seeds from each of 127 F3 populations were grown for the “LAB-19” inoculation test in 2016. All plants were grown in 72-well multipots and maintained in a greenhouse at 20–25°C under a 16-h photoperiod. Resting spores were prepared and inoculation was performed as previously described (Pang et al., 2014). For inoculation, 1 ml of spore suspension was applied to the bottom of the stem base of each 7-days-old seedling; disease resistance was evaluated 6 weeks later.
DNA Extraction and Pool Construction
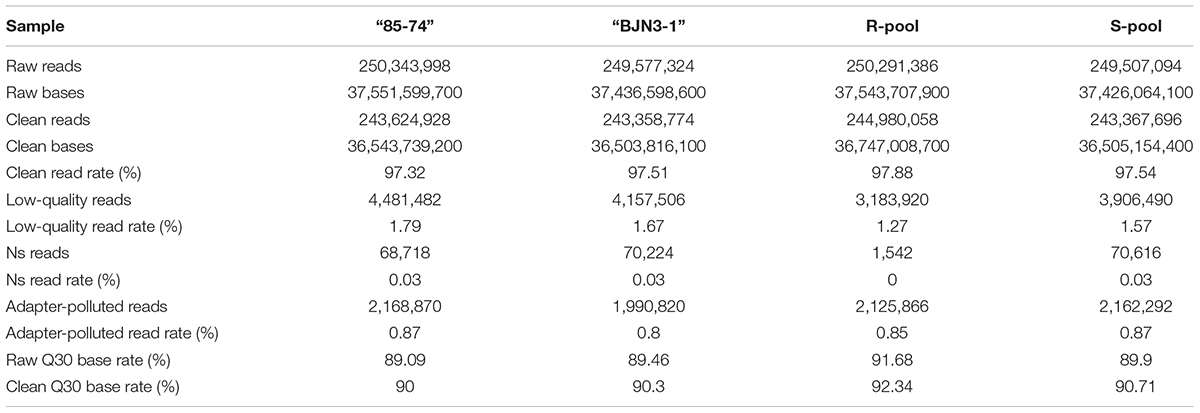
Young leaves from eight Chinese cabbage germplasm were collected and used for characterization of known CR loci/genes. Young leaves from 127 F2 individuals were sampled and used for genome sequencing and gene mapping. The genomic DNA was extracted according to the cetyl trimethylammonium bromide method (Li et al., 2010), with minor modifications. DNA concentration was determined with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). Highly resistant and susceptible F3 families were selected and equal amounts of DNA from selected F2 individuals were mixed together to form resistant and susceptible pools (R- and S-pool, respectively) that were sequenced along with “85-74” and “BJN3-1” with the HiSeq 2500 system (Illumina, San Diego, CA, United States).
Analysis of Markers Linked to Previously Identified CR Genes
Chinese cabbage germplasms were genotyped for known CR loci/genes identification using linked markers (Saito et al., 2006; Suwabe et al., 2006; Sakamoto et al., 2008; Ueno et al., 2012; Chen et al., 2013; Hatakeyama et al., 2013; Pang et al., 2014; Zhang et al., 2014). DNA fragments were amplified by PCR and the susceptible “BJN3-1” line was used as a control. PCR products were separated on a standard agarose (2%) or polyacrylamide (6%) gel and their sizes were compared to that of known resistance genes.
Sequencing and Bioinformatics Analysis
Genome sequencing of the two parental lines and R- and S-pools and statistical analysis of bioinformatics data were carried out by Annoroad Co.1 A library of 300–500-bp insert size was constructed and paired-end sequenced on an Illumina HiSeq 2500 platform. Raw data were processed with Perl scripts to ensure data quality for subsequent analyses. The adopted filtering criteria are as follows: (1) remove the adaptor-polluted reads (reads containing more than five adapter-polluted bases were regarded as adaptor-polluted reads and would be filtered out); (2) remove the low-quality reads; reads with the number of low-quality bases (phred Quality value less than 19) accounting for more than 50% of total bases are regarded as low-quality reads; and (3) remove reads with number of N bases accounting for more than 5%. As for paired-end sequencing data, both reads would be filtered out if any read of the paired-end reads are adaptor-polluted. The obtained Clean Data after filtering will be carried out on statistics analyses on its quantity and quality, including Q30, data quantity, base content statistics, etc. Clean reads were aligned to the reference genome sequence “Chiifu-401-42” from the Ensemble Genome database2 using Burrows–Wheeler Aligner v.0.7.12 (Li and Durbin, 2009). Samtools v.1.2 (Li et al., 2009) was used to sort the reads, and duplicate reads obtained by PCR were removed using the MarkDuplicates command of Picard tools v.1.133. Reads mapped to two or more sites were filtered out. Statistical analyses were carried out using an in-house Perl script.
The Genome Analysis Toolkit (GATK; McKenna et al., 2010) HaplotypeCaller function was used for single nucleotide polymorphism (SNP) and insertion-deletion (InDel) calling. The SNPs and InDels were filtered with the GATK VariantFiltration protocol before further analysis with the following settings: QD < 2.0, MQ < 40, DP < 4, MQRankSum < -12.5. Annotation was performed using ANNOVAR (Wang et al., 2010) for all qualified variants based on the GFF file.
BSA Mapping Using Sequencing Data
To detect candidate loci associated with CR, the SNP index was calculated for all variants (Takagi et al., 2013). To reduce the impact of sequencing and alignment errors, we filtered out variations that met any of the following conditions: (1) loci in parents were heterozygous; (2) depths of variation were <10 or positions were not covered in parents or bulks; (3) variations in SNP index in both bulks were <0.3 or >0.7; or (4) variations were not on chromosomes (e.g., they were on a scaffold). All remaining variants were retained for further analysis. We slid along the genome with a 1-Mb window at a step size of 100 kb to calculate the mean SNP index, and subtracted the SNP index value of the R-pool from that of the S-pool to obtain the ΔSNP index (Takagi et al., 2013). Confidence intervals at 0.1, 0.5, and 0.01 levels were determined by computer simulation; the threshold was set at a 0.01 confidence level to identify candidate quantitative trait loci. The SNPs and InDels in the confidence region were selected and validated through Sanger sequencing.
CRd Mapping and Candidate Genes Analysis
The reference genome sequence from the Ensemble Genome database was downloaded and used for marker development in the CRd candidate region. Simple sequence repeat (SSR) markers were developed using SSR Hunter v.1.3 (Li and Wan, 2005). The two parental lines along with R- and S-pools were used to develop markers linked to the CR gene. A genetic map was constructed with the developed and previously published markers using JoinMap v.4.0 (Stam, 1993; Van Ooijen, 2006). CRd closely linked markers were validated in natural population. The candidate genes in the CRd region were compared with 244 resistance genes in B. rapa4.
Semi-Quantitative RT-PCR Analysis
Total RNA was isolated from 0 day, 7 days, 10, and 13 days after inoculation (DAI) of P. brassicae of “85-74” and “BJN3-1” root tissue using an Easy-BLUETM Total RNA Extraction Kit (Invitrogen, United States). The total RNA from each plant sample amounting to 5 μg was combined with random hexamer primers in a Super Script first-strand cDNA synthesis system according to the manufacturer’s instructions (Invitrogen, United States). Complementary DNA was diluted 10-fold, and 1 μl of the diluted cDNA was used in each 20 μl PCR mixture. Sequence information from B. rapa was used for RT-PCR primers design. Standard PCR was performed, with 5 min denaturation at 94°C followed by 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s. The PCR products were analyzed following electrophoresis on a 1% agarose gel.
Results
Characterization of Chinese Cabbage Germplasm
To characterize CR resources of Chinese cabbage, eight inbred lines of CR Chinese cabbage were genotyped with known CR gene linked markers and infected with 11 different local isolates of P. brassicae. “CR-77” and “CR-20” were found to carry Crr1 and CRa; “CR-75” and “CR Shinki” harbored CRa and CRb; “CR-73” harbored CRk and Crr3. “85-74” was a potential carrier of Crr3, which was linked to BRSTS61. “BJN3-1” had no CR loci, and “CR-26” carried unknown CR gene/genes (Table 1). Based on Williams’ classification system, local pathogens “AHXC-68,” “LAB-16,” “LNXM-1,” “AHHS-62,” “LAB-19,” and “LNND-2” were identified as pathotype 4; “LAB-7” and “LAB-10” were pathotype 2; “HBLC-31” and “AHHS-65” were pathotype 7; and “AHHS-65” was pathotype 11. CR inbred lines showed variable resistance depending on the presence of infection with these pathogens. The negative control “BJN3-1” was infected by all pathogens. “CR-77” and “CR-20” showed resistance to all 11 pathogens; “85-74” was susceptible to “LNND-2” and “LAB-10,” but resistant to the other pathogens tested in this study; “CR Shinki” was susceptible to “LNXM-1” and “AHHS-62,” but resistance to the other pathogens; “CR-75” and “CR-26” were susceptible to “LNXM-1” and “AHHS-62,” respectively; and “CR-73” was susceptible to “LAB-19” (Table 1). “CR-73” and “85-74” showed distinct resistance responses to “LAB-19,” “LNND-2,” and “LAB-10.” This suggests that “85-74” and “CR-73” harbor different CR genes or alleles. Based on these results, “LAB-19” was selected to phenotype F2 and F3 populations derived from the “85-74” and “BJN3-1” cross.
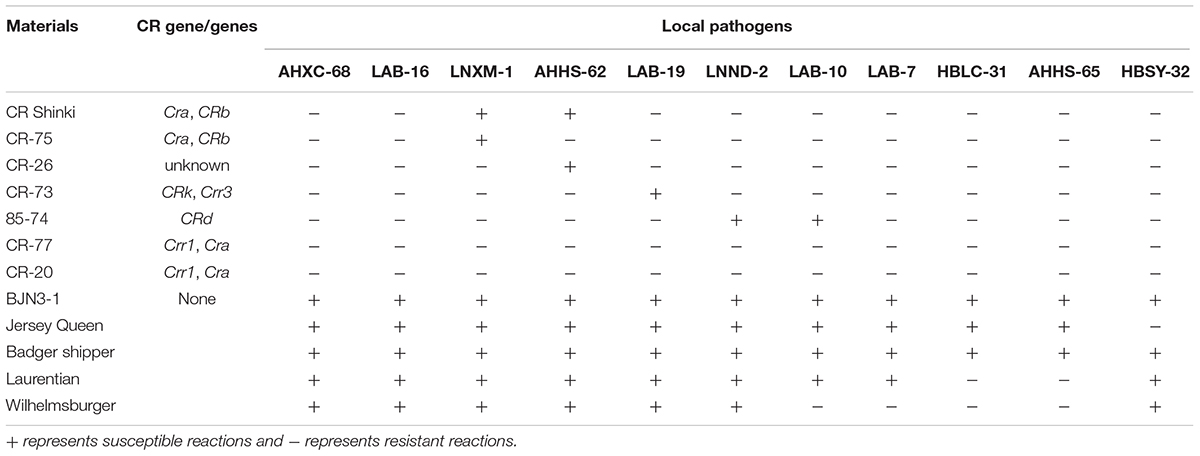
TABLE 1. Clubroot disease resistance test of Chinese cabbage germplasms and pathotypes identification using Williams’ clubroot differential set.
Phenotype Evaluation and R- and S-Pool Construction
To investigate the inheritance of the resistance to local pathogen “LAB-19,” parental lines, F1, and 432 F2 individuals were inoculated with 1 × 107 spores⋅ml-1. The “85-74” and F1 lines were highly resistant whereas “BJN3-1” was susceptible. Among 432 F2 individuals, 321 and 106 were resistant and susceptible, respectively, and exhibited a 3:1 segregation ratio at a 0.05 level of probability (Table 2). These results indicated that CR is controlled by a single dominant gene in “85-74.” Parental lines with 127 F3 families were then inoculated with “LAB-19” in 2016. R- and S-pools were constructed by selecting 19 highly CR and 16 susceptible F2 individuals depending on the F3 family phenotype.
Sequencing Data Analysis
Sequencing data were generated with Illumina HiSeq 2500 with an average insert size of about 350 bp. A total of 250,343,998, 249,577,324, 250,291,386, and 249,507,094 raw reads were obtained from “85-74,” “BJN3-1,” and R- and S-pools (SRA accession: SRP136862), respectively. Clean data were obtained after removing adapter-polluted and low-quality reads and unknown bases (N > 5%), yielding minimum and maximum clean Q30 base rates of 90% and 92.34%, respectively (Table 3). Genome coverage ranged from 90.24 to 91.63%, and average depths were 86.26×, 84.22×, 83.49×, and 85.34× for “85-74,” “BJN3-1,” and R- and S-pools, respectively.
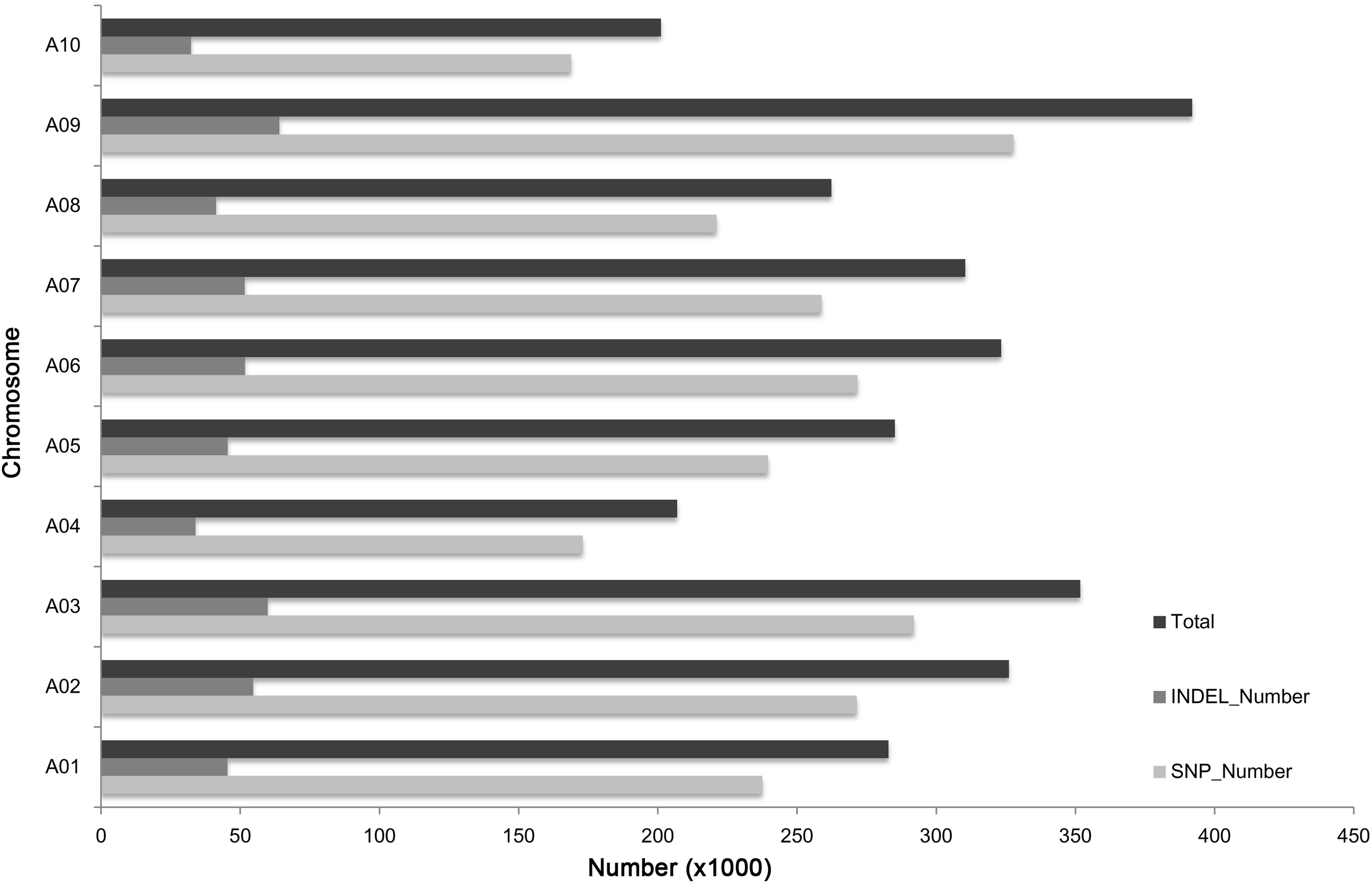
In total, 2,941,775 SNPs and InDels were detected between “85-74” and “BJN3-1” (Figure 1). The average number of sequence variations on the 10 chromosomes was 294,177, with chromosomes A09 and A10 having the highest and lowest number of variations, respectively. Chromosome A03 had a comparatively high number and density of variations in a specific region.
Association Analysis
To calculate SNP index, we filtered out sequence variations that met the above-described conditions. Of the 2,941,775 variations, 599,797 were to calculate SNP index (Figure 2A). According to the ΔSNP index value, a 3.94-Mb candidate region from 13.57 to 17.51 Mb was identified on chromosome A03 at a 0.01 confidence level (Figure 2B). A total of 19,664 SNPs and 4450 InDels were found in the candidate region; 1953 out of 5861 variations in exons caused changes of amino acid sequence (Supplementary Table S1). Twenty pairs of primers were designed for InDels validation (Supplementary Table S2). Seven pairs of primers were not amplified PCR production and the rest of 13 primers produced single band. The PCR products were sequenced and shown exactly same with our BSA-sequencing data (Supplementary Figure S1). The candidate genes in the CRd region were compared with 244 resistance genes in B. rapa5. Four resistance genes were identified which encode TIR-NBS-LRR protein, including Bra001160, Bra001161, Bra001162, and Bra001175 with 20, 4, 42, and 81 sequence variations, respectively, in the exons.
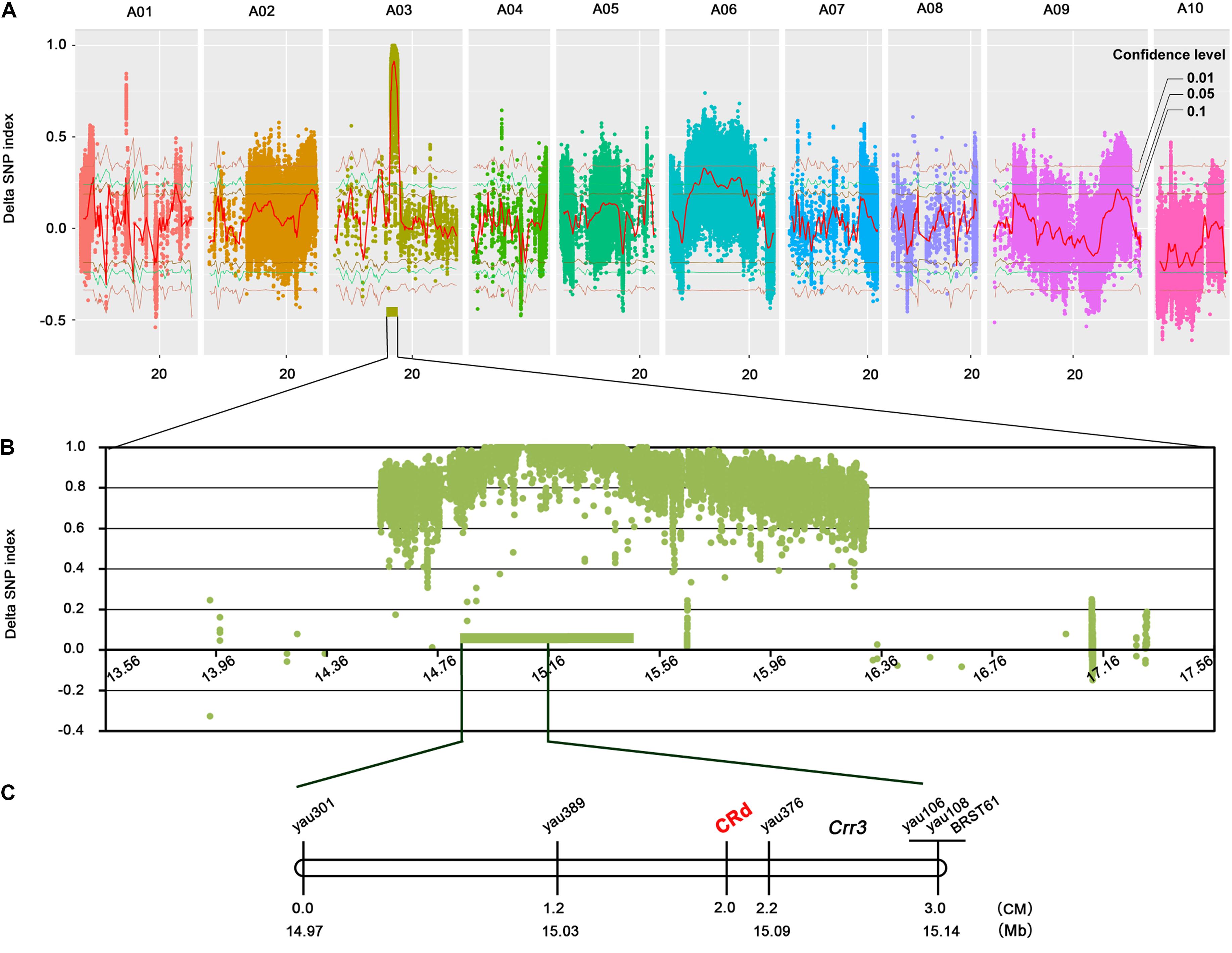
FIGURE 2. CRd mapping by NGS-based BSA and genetic mapping approaches. (A) Genome-wide ΔSNP index Manhattan plots and marker-trait association with 0.1, 0.05, and 0.01 confidence levels. (B) ΔSNP index Manhattan plots in candidate region; the green rectangle indicates the core region. (C) Genetic/physical map of the region harboring CRd on chromosome 3.
CRd Mapping and Candidate Genes Analysis
A core region of 577 kb was found on chromosome A03 with an extremely high average ΔSNP index of 0.9631 (Figure 2B). We designed eight SSR primer pairs within this region and screened for polymorphisms between the two parent lines. Five polymorphic markers were identified including yau301, yau389, yau376, yau106, and yau108; these were used to genotype 127 F2 individuals (Figure 2C and Supplementary Table S3). The Crr3 linked marker BRSTS61 showing polymorphism between “85-74” and “BJN3-1” was used to compare the mapping locations of CRd and Crr3.
A genetic map of the region surrounding the CRd gene was constructed based on the genotypes of seven markers. CRd was mapped to a 1 cM region with the flanking markers yau389 and yau376 (Figure 2C). Alignment of marker sequences to the reference genome sequence of B. rapa revealed a physical distance between yau389 and yau376 of about 60 kb. CRd is located upstream of Crr3 was confirmed based on the physical position of Crr3 linked markers. CRd closely linked markers yau389 and yau376 were validated in natural population and the cultivars which harbor CRd gene all showed resistant to isolate “LAB-19” (Supplementary Figure S2 and Supplementary Table S4). Total four genes Bra001160, Bra001161, Bra001162, and Bra001175 which encode TIR-NBS-LRR protein were identified in the CRd candidate region.
Semi-Quantitative RT-PCR Analysis
Total four genes Bra001160, Bra001161, Bra001162, and Bra001175 which encode TIR-NBS-LRR protein were identified in the CRd candidate region. To examine the expression characteristics of these four genes from “85-74” and “BJN3-1,” we performed RT-PCR analysis with a common primer set (18S) and resistance gene-specific primer (Supplementary Table S5). As shown in Figure 3, 18S expressed in “85-74” and “BJN3-1” from 0 DAI to 13 DAI. Bra001160, Bra001161, and Bra001175 were more highly expressed in “85-74” at 13 DAI. Bra001162 was more highly expressed in “BJN3-1” at 7 DAI, 10 DAI, and 13 DAI.
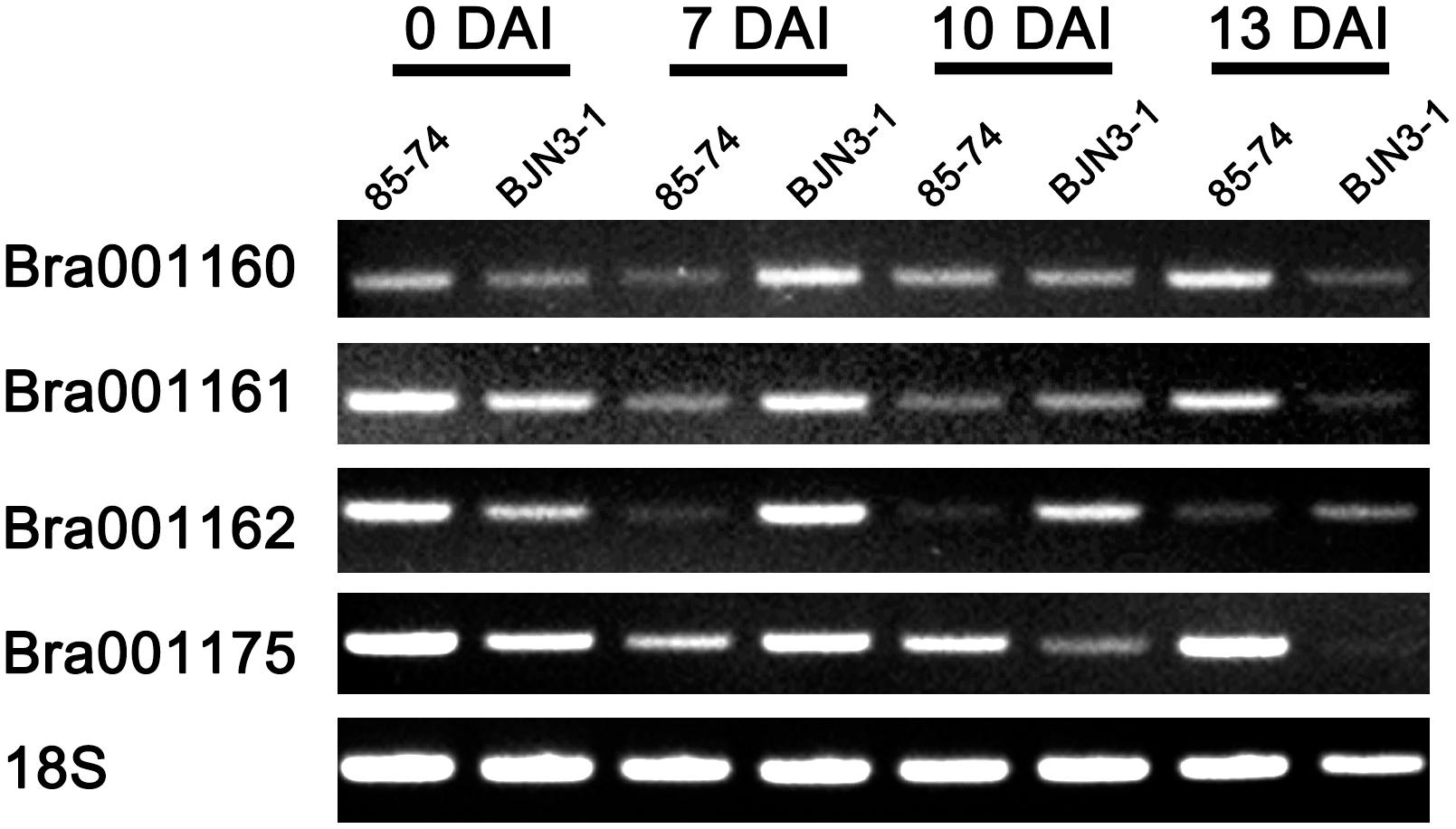
FIGURE 3. Expression levels of Bra001160, Bra001161, Bra001162, and Bra001175 genes in roots of “85-74” and “BJN3-1.”
Discussion
In this study, we used an NGS-based BSA strategy to map the novel CR gene CRd in an F2 population of B. rapa. Previously identified CR loci or genes in B. rapa have been associated with resistance to specific pathotypes of P. brassicae. For instance, the CRa gene confers resistance to P. brassicae isolate M85, which belongs to race 2 according to Williams’s classification (Williams, 1966; Ueno et al., 2012), whereas CRb (Piao et al., 2004) and Crr1 (Hatakeyama et al., 2013) confer resistance to race 4 and Ano-01 (an unknown pathotype), respectively. In the present study, “85-74” showed resistance to all pathotypes of race 2, 4, 7, and 11, except for “LAB-10” (race 2) and “LNND-2” (race 4). “CR-73” showed resistance to all tested isolates except to “LAB-19” (race 4). The variable responses to “LAB-19,” “LAB-10,” and “LNND-2” indicate that “85-74” and “CR-73” have distinct genetic backgrounds. Moreover, “CR-75” was identified carrying CRa and CRb using published CR loci-/gene linked markers and “CR Shinki” harbored CRa and CRb also. However, “CR-75” showed resistant to “AHHS-62” while “CR Shinki” not which indicates that “CR-75” is highly possible carrying other unknown CR gene/genes need to explore through genetic mapping. A given pathotype identified according to Williams’s classification system is expected to produce the same response in hosts; however, pathotype 4 had different infectivity in the Chinese cabbage germplasm inoculation tests. “LAB-7” and “LAB-10” were identified as pathotype 2; however, only the latter infected “85-74” successfully. These results indicate that the Williams’s classification system has a limited capacity for distinguishing co-existing isolates.
Clubroot disease is threating all of the brassica crops. To reduce the economic losses, CR genes have been investigated in canola (Rcr1 and Rcr4, Yu et al., 2016, 2017), cabbage (Anju1, Anju2, Anju4, and GC1, Tomita et al., 2013), and Chinese cabbage (CRa, Crr1, and CRb, Ueno et al., 2012; Hatakeyama et al., 2013; Zhang et al., 2014.) These CR genes and their closely linked markers exploration in brassica crops have been greatly improved the CR breading through MAS strategy. Breeding of CR cultivars is the most environment friendliness and economically effective strategy for controlling clubroot. Plant disease resistance genes are abundant and are clustered together in the genome (Michelmore and Meyers, 1998; Wang et al., 2011). Most of the CR loci or resistance genes reported to date in B. rapa are clustered on chromosome A03 in two specific genomic regions (CRa, CRb, and QS_B3.1 at one locus and CRk and Crr3 at another). CRd is in the same genomic region as CRk and Crr3. The CRk linked marker HC688 was not polymorphic in our population. The sequences of the Crr3 linked markers BrSTS78 and BrSTS33 were searched in the Ensemble Genome database to further distinguish between CRd and Crr3, and were found to be located at 15.091 and 15.331 Mb, respectively, on chromosome A03. Meanwhile, CRd was mapped to between yau389 (15.029 Mb) and yau376 (15.089 Mb). These results confirm that CRd is located upstream of Crr3. Moreover, “85-74” and “CR-73” (harbor Crr3 resistant gene) showed different responses to “LAB-19,” “LAB-10,” and “LNND-2.” Thus, CRd is possible to be a novel CR gene distinct from those previously identified on chromosome A03 of B. rapa.
Most of the disease resistance genes encoding NBS-LRR proteins confer pathogen race-specific resistance (Flor, 1956; Meyers et al., 2003). It was previously reported that CRa and Crr1 encode TIR-NBS-LRR protein. In the present study, Bra001162 was more highly expressed in “BJN3-1” at 7 DAI, 10 DAI, and 13 DAI indicated that Bra001162 may not associate with CR, while Bra001160, Bra001161, and Bra001175 were more highly expressed in “85-74” than “BJN3-1” at 13 DAI (Figure 3). Therefore, Bra001160, Bra001161, and Bra001175 are highly possible to be candidate genes of CRd. These results will be helpful for the CRd gene cloning and validation of transgenic lines in future study.
CRd was mapped into a 1 cM region on chromosome A03 of the B. rapa genome in a small F2 segregant population. However, it was found to be anchored to a relatively short 60 kb region based on the reference genome of B. rapa, indicating the presence of a recombination hotspot at this location. Previously studies have shown that such hotspots in Arabidopsis thaliana are accession-specific or vary depending on the cross (Drouaud et al., 2006, 2007; Kim et al., 2007; Salomé et al., 2012). It is also possible that “85-74” harbors a large insertion that is not present in the reference genome.
Conclusion
We identified the CRd gene in a CR population of B. rapa. Our findings may be useful for breeding cultivars of Chinese cabbage and other Brassica crops with broad-spectrum resistance to multiple P. brassicae pathotypes.
Author Contributions
WP analyzed the data and drafted the manuscript. PF performed the experiments and data analysis. XL and ZZ helped in the data analysis and experiments. ZP conceived the study, participated in its coordination, and helped to draft the manuscript. All authors have read and approved the final manuscript.
Funding
This study was supported by grants from the National Natural Scientific Foundation of China (Project No. 31471882), the National Key Research and Development Program of China (Grant No. 2016YFD0100202-19), and the earmarked fund for China Agriculture Research System (CARS-12).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00653/full#supplementary-material
FIGURE S1 | Agarose gel of PCR product of InDels validation primer set.
FIGURE S2 | Validation of CRd closely linked marker (A) yau376 and (B) yau389 using natural population. Lanes 1–25 are materials that have been listed in Supplementary Table S4. Black arrow indicates the clubroot-resistant band.
TABLE S1 | Details of SNP index and annotation for 1953 SNPs and InDels that can cause changes in the amino acid sequence of the candidate region.
TABLE S2 | Primers designed for InDels validation and PCR product sequences.
TABLE S3 | Markers used for genetic map construction in this study.
TABLE S4 | Materials used for natural validation in this study.
TABLE S5 | Primer set used for semi-quantitative RT-PCR analysis.
Footnotes
- ^ http://www.annoroad.com/
- ^ ftp://ftp.ensemblgenomes.org/pub/release-32/plants/fasta/brassica_rapa/dna/Brassica_rapa.IVFCAASv1.dna.toplevel.fa.gz
- ^ http://broadinstitute.github.io/picard/
- ^ http://brassicadb.org/brad/
- ^ http://brassicadb.org/brad/
References
Chen, J., Jing, J., Zhan, Z., Zhang, T., Zhang, C., and Piao, Z. (2013). Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS One 8:e85307. doi: 10.1371/journal.pone.0085307
Drouaud, J., Camilleri, C., Bourguignon, P. Y., Canaguier, A., and Bérard, A. (2006). Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 16, 106–114. doi: 10.1101/gr.4319006
Drouaud, J., Mercier, R., Chelysheva, L., Bérard, A., and Falque, M. (2007). Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet. 3:e106. doi: 10.1371/journal.pgen.0030106
Flor, H. H. (1956). The complementary genic system in flax and flax rust. Adv. Genet. 8, 29–54. doi: 10.1016/S0065-2660(08)60498-8
Han, Y., Lv, P., Hou, S., Li, S., and Ji, G. (2015). Combining next generation sequencing with bulked segregant analysis to fine map a stem moisture locus in sorghum (Sorghum bicolor L. Moench). PLoS One 10:e0127065. doi: 10.1371/journal.pone.0127065
Hatakeyama, K., Niwa, T., Kato, T., Ohara, T., Kakizaki, T., and Matsumoto, S. (2017). The tandem repeated organization of NB-LRR genes in the clubroot-resistant CRb, locus in Brassica rapa, L. Mol. Genet. Genomics 292, 397–405. doi: 10.1007/s00438-016-1281-1
Hatakeyama, K., Suwabe, K., Tomita, R. N., Kato, T., Nunome, T., Fukuoka, H., et al. (2013). Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS One 8:e54745. doi: 10.1371/journal.pone.0054745
Hirai, M., Harada, T., Kubo, N., Tsukada, M., Suwabe, K., and Matsumoto, S. (2004). A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor. Appl. Genet. 108, 639–643. doi: 10.1007/s00122-003-1475-x
Huang, Z., Peng, G., Liu, X., Deora, A., and Falk, K. C. (2017). Fine mapping of a clubroot resistance gene in Chinese cabbage using SNP markers identified from bulked segregant RNA sequencing. Front. Plant Sci. 8:1448. doi: 10.3389/fpls.2017.01448
Kim, S., Plagnol, V., Hu, T. T., Toomajian, C., and Clark, R. M. (2007). Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 39, 1151–1155. doi: 10.1038/ng2115
Kuginuki, Y., Yoshigawa, H., and Hirai, M. (1999). Variation in virulence of Plasmodiophora brassicae in Japan tested with clubroot resistant cultivars of Chinese cabbage. Eur. J. Plant Pathol. 105, 327–332. doi: 10.1023/A:1008705413127
Kutcher, H. R., Brandt, S. A., Smith, E. G., Ulrich, D., Malhi, S. S., and Johnston, A. M. (2013). Blackleg disease of canola mitigated by resistant cultivars and four-year crop rotations in western Canada. Can. J. Plant Pathol. 35, 209–221. doi: 10.1080/07060661.2013.775600
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, Q., and Wan, J. (2005). SSRHunter: development of local searching software for SSR sites. Hereditas 27, 808–810.
Li, X., Ramchiary, N., Choi, S. R., Van Nguyen, D., Hossain, M. J., Yang, H. K., et al. (2010). Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa genome sequencing project. Genome 53, 939–947. doi: 10.1139/G10-054
Matsumoto, E., Yasui, C., Ohi, M., and Tsukada, M. (1998). Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage. Euphytica 104, 79–86. doi: 10.1023/A:1018370418201
McDonald, M. R., Sharma, K., Gossen, B. D., Deora, A., Feng, J., and Hwang, S. F. (2014). The role of primary and secondary infection in host response to Plasmodiophora brassicae. Phytopathology 104, 1078–1087. doi: 10.1094/PHYTO-07-13-0189-R
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., and Kernytsky, A. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R. W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308
Michelmore, R. W., and Meyers, B. C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. doi: 10.1101/gr.8.11.1113
Miller, S. A., Mera, J. R., and Baysal, F. (2007). Evaluation of Fungicides for the Control of Clubroot of Broccoli, 2006. Plant Disease Management Reports (online). Report 1, V004. St. Paul, MN: The American Phytopathological Society. doi: 10.1094/PDMR01
Mitani, S., Sugimoto, K., Hayashi, H., Takii, Y., Ohshima, T., and Matsuo, N. (2003). Effects of cyazofamid against Plasmodiophora brassicae Woronin on Chinese cabbage. Pest Manag. Sci. 59, 287–293. doi: 10.1002/ps.627
Pang, W., Liang, S., Li, X., Li, P., Yu, S., Lim, Y. P., et al. (2014). Genetic detection of clubroot resistance loci in a new population of Brassica rapa. Hortic. Environ. Biotechnol. 55, 540–547. doi: 10.1007/s13580-014-0079-5
Peng, G., Lahlali, R., Hwang, S. F., Pageau, D., and Hynes, R. K. (2014). Crop rotation, cultivar resistance, and fungicides/biofungicides for managing clubroot (Plasmodiophora brassicae) on canola. Can. J. Plant Pathol. 36(Suppl. 1), 99–112. doi: 10.1080/07060661.2013.860398
Piao, Z., Deng, Y., Choi, S., Park, Y., and Lim, Y. P. (2004). SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 108, 1458–1465. doi: 10.1007/s00122-003-1577-5
Ransom, L. M., Collins, D., Taylor, P. A., and Baker, A. (1991). “A new weapon to beat clubroot,” in Proceedings of the Australasian Plant Pathology Society 8th Biennial Conference, Sydney, NSW.
Rocherieux, J., Glory, P., Giboulot, A., Boury, S., Barbeyron, G., Thomas, G., et al. (2004). Isolate-specific and broad-spectrum QTL are involved in the control of clubroot in Brassica oleracea. Theor. Appl. Genet. 108, 1555–1563. doi: 10.1007/s00122-003-1580-x
Sakamoto, K., Saito, A., Hayashida, N., Taguchi, G., and Matsumoto, E. (2008). Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theor. Appl. Genet. 117, 759–767. doi: 10.1007/s00122-008-0817-0
Saito, M., Kubo, N., Matsumoto, S., Suwabe, K., Tsukada, M., and Hirai, M. (2006). Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor. Appl. Genet. 114, 81–91. doi: 10.1007/s00122-006-0412-1
Salomé, P. A., Bomblies, K., Fitz, J., Laitinen, R. A., and Warthmann, N. (2012). The recombination landscape in Arabidopsis thaliana F2 populations. Heredity 108, 447–455. doi: 10.1038/hdy.2011.95
Song, J., Li, Z., Liu, Z., Guo, Y., and Qiu, L. J. (2017). Next-generation sequencing from bulked-segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean. Front. Plant Sci. 8:919. doi: 10.3389/fpls.2017.00919
Stam, P. (1993). Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 3, 739–744. doi: 10.1111/j.1365-313X.1993.00739.x
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., Fujimura, M., Nunome, T., et al. (2003). Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor. Appl. Genet. 107, 997–1002. doi: 10.1007/s00122-003-1309-x
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., and Kondo, M. (2006). Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics 173, 309–319. doi: 10.1534/genetics.104.038968
Suzuki, K., Sugimoto, K., Hayshi, H., and Komyoji, T. (1995). Biological mode of action of fluazinam, a new fungicide, for Chinese cabbage clubroot. Jpn. J. Phytopathol. 61, 395–398. doi: 10.3186/jjphytopath.61.395
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., and Natsume, S. (2013). QTLseq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. Cell Mol. Biol. 74, 174–183. doi: 10.1111/tpj.12105
Tjallingii, F. (1965). Testing clubroot-resistance of turnips in the Netherlands and the physiologic specialization of Plasmodiophora brassicae. Euphytica 14, 1–22.
Tomita, H., Shimizu, M., Doullah, A. U., Fujimoto, R., and Okazaki, K. (2013). Accumulation of quantitative trait loci conferring broad-spectrum clubroot resistance in Brassica oleracea. Mol. Breed. 32, 889–900. doi: 10.1007/s11032-013-9918-9
Townley, D., and Fox, T. V. (2003). “Control of clubroot disease using cyazofamid and fluazinam fungicides,” in Proceedings of the 8th International Congress for Plant Pathology, Christchurch, 72.
Trick, M., Adamski, N. M., Mugford, S. G., Jiang, C. C., Febrer, M., and Uauy, C. (2012). Combining ANP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol. 12:14. doi: 10.1186/1471-2229-12-14
Uchida, N., Sakamoto, T., Tasaka, M., and Kurata, T. (2014). Identification of EMS-Induced Causal Mutations in Arabidopsis thaliana by Next-Generation Sequencing. Arabidopsis Protocols. New York, NY: Humana Press.
Ueno, H., Matsumoto, E., Aruga, D., Kitagawa, S., Matsumura, H., and Hayashida, N. (2012). Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol. Biol. 80, 621–629. doi: 10.1007/s11103-012-9971-5
Van Ooijen, J. W. (2006). JoinMap 4. Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen: Kyazma B.V.
Voorrips, R. E., Jongerius, M. C., and Kanne, H. J. (2003). Quantitative trait loci for clubroot resistance in Brassica oleracea. Biotechnol. Agric. For. 52, 87–104. doi: 10.1007/978-3-662-05036-1_6
Wallenhammar, A. C. (1996). Prevalance of Plasmodiophora brassicae in a spring oilseed rape growing area in central Sweden and factors influencing soil infestation levels. Plant Pathol. 45, 710–719. doi: 10.1046/j.1365-3059.1996.d01-173.x
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164. doi: 10.1093/nar/gkq603
Wang, X., Wang, H., Wang, J., Sun, R., and Wu, J. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Werner, S., Diederichsen, E., Frauen, M., Schondelmaier, J., and Jung, C. (2008). Genetic mapping of clubroot resistance genes in oilseed rape. Theor. Appl. Genet. 116, 363–372. doi: 10.1007/s00122-007-0674-2
Williams, P. H. (1966). A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology 56, 624–626.
Yang, Z., Huang, D., Tang, W., Zheng, Y., and Liang, K. (2013). Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS One 8:e68433. doi: 10.1371/journal.pone.0068433
Yoshikawa, H. (1981). “Breeding for clubroot resistance in Chinese cabbage,” in Proceedings of the 1st International Symposium, eds N. S. Talekar and T. D. Griggs, Tsukuba, 405–413.
Yu, F., Zhang, X., Peng, G., Falk, K. C., Strelkov, S. E., and Gossen, B. D. (2017). Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 7:4516. doi: 10.1038/s41598-017-04903-2
Yu, F., Zhang, X., Zhen, H., Chu, M., and Tao, S. (2016). Identification of genome-wide variants and discovery of variants associated with Brassica rapa clubroot resistance gene rcr1 through bulked segregant RNA sequencing. PLoS One 11:e0153218. doi: 10.1371/journal.pone.0153218
Zhang, T., Wu, D., Zhao, Z., Wang, Z., and Piao, Z. Y. (2012). Development of near isogenic lines for clubroot resistance in Chinese cabbage and their assessment. Mol. Plant Breed. 6, 722–730.
Zhang, T., Zhao, Z., Zhang, C., Pang, W., Choi, S. R., and Lim, Y. P. (2014). Fine genetic and physical mapping of the CRb gene conferring resistance to clubroot disease in Brassica rapa. Mol. Breed. 34, 1173–1183. doi: 10.1007/s11032-014-0108-1
Keywords: Chinese cabbage, Plasmodiophora brassicae, clubroot disease, bulked segregant analysis, pathogen resistance gene, CR cultivars
Citation: Pang W, Fu P, Li X, Zhan Z, Yu S and Piao Z (2018) Identification and Mapping of the Clubroot Resistance Gene CRd in Chinese Cabbage (Brassica rapa ssp. pekinensis). Front. Plant Sci. 9:653. doi: 10.3389/fpls.2018.00653
Received: 24 January 2018; Accepted: 27 April 2018;
Published: 18 May 2018.
Edited by:
Maoteng Li, Huazhong University of Science and Technology, ChinaReviewed by:
Xiaowu Wang, Biotechnology Research Institute (CAAS), ChinaKun Lu, Southwest University, China
Copyright © 2018 Pang, Fu, Li, Zhan, Yu and Piao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongyun Piao, enlwaWFvQHN5YXUuZWR1LmNu; enlwaWFvc2F1QDE2My5jb20=
†These authors have contributed equally to this work.
 Wenxing Pang
Wenxing Pang Pengyu Fu†
Pengyu Fu† Zhongyun Piao
Zhongyun Piao