- 1Department of Biotechnology, Delhi Technological University, New Delhi, India
- 2Division of Biochemistry, Indian Agricultural Research Institute, New Delhi, India
Soybean is one of the leading oilseed crop in the world and is showing a remarkable surge in its utilization in formulating animal feeds and supplements. Its dietary consumption, however, is incongruent with its existing industrial demand due to the presence of anti-nutritional factors in sufficiently large amounts. Phytic acid in particular raises concern as it causes a concomitant loss of indigestible complexed minerals and charged proteins in the waste and results in reduced mineral bioavailability in both livestock and humans. Reducing the seed phytate level thus seems indispensable to overcome the nutritional menace associated with soy grain consumption. In order to conceive our objective we designed and expressed a inositol polyphosphate 6-/3-/5-kinase gene-specific RNAi construct in the seeds of Pusa-16 soybean cultivar. We subsequently conducted a genotypic, phenotypic and biochemical analysis of the developed putative transgenic populations and found very low phytic acid levels, moderate accumulation of inorganic phosphate and elevated mineral content in some lines. These low phytic acid lines did not show any reduction in seedling emergence and displayed an overall good agronomic performance.
Introduction
For the last 50 years, world population multiplied more rapidly than ever before, and is expected to grow over a third by 2050. Therefore, one of the major challenges agriculture will face in the coming decades is to meet the food demand of the growing population. Scientific community and farmers worldwide are seeking an inexpensive source of energy dense and nutrient rich alternative to serve the purpose. Soybean crop due to its unique nutrient profile provide as a promising option to ensure food security in future. United States Department of Agriculture (USDA) estimated that the global soybean production 2017–2018 will be 348.04 million metric tons. Despite such huge production, the presence of natural anti-nutrients, such as inositol hexakisphosphate (Phytic acid, PA), protease inhibitors, lectins, saponins amongst few others, has limited its consumption (Jiang et al., 2013). PA particularly summon attention as it accounts for over 75% of the total seed phosphorous (Raboy et al., 1984) as well as chelate many divalent cations to form insoluble salts at the biological pH (Halsted et al., 1974; Jacobsen and Slotfeldt-Ellingsen, 1983). Its consumption, however, is inevitable as it is associated with protein bodies found in the cotyledon of the soybean seeds (Lott, 1984; Lott et al., 1995; Wada and Lott, 1997). Therefore, feeding on high-phytate soy-based diets are often feared to exacerbate mineral and protein malnutrition. In addition, phytate excreted by livestock grazing on soy-rich forage contribute significantly to the environmental phosphorus load (Brinch-Pedersen et al., 2000). These problems have provided us with a strong impetus to develop low phytic acid (lpa) soybean grains which could contribute in its biofortification.
The first generation of lpa crops were developed using classical breeding. With advancement in technology these efforts were augmented by the aid of forward genetics. Mutations that block the synthesis or accumulation of PA during seed development have been isolated in the past in a variety of crop species (Maize, Raboy et al., 2000; Shi et al., 2005; Wheat, Guttieri et al., 2004; Barley, Larson et al., 1998; Rasmussen and Hatzack, 1998; and Rice, Larson et al., 2000; Liu et al., 2007). Lpa mutant lines CX1834 (Wilcox et al., 2000) and LR33 (Hitz et al., 2002) were developed in soybean and made available for public breeding efforts. Unfortunately, the crops were unsuccessful due to their poor agronomic performance which called in for a different approach.
Research in the past has evidenced plant genetic engineering techniques to hold unprecedented potential for crop improvement. The first transgenic strategy that was followed to develop lpa soybean was the accumulation of microbial phytases in their seeds (Li et al., 1997; Denbow et al., 1998). However, the cost and the labor incurred for its processing prior to consumption made it an economically unviable alternative. Successively, a sustainable approach directing expression of soybean phytase during embryo development at the site of PA synthesis/storage was followed (Hegeman and Grabau, 2001; Chiera et al., 2004). But only a maximum reduction of 25% in PA level was attained by this method which is significantly less than what has been achieved previously. Therefore, to further reduce the PA level albeit optimally, we thought of genetically engineering soybean by exploring RNA interference (RNAi) technique of reverse genetics, which has recently been used to generate lpa rice (Ali et al., 2013a,b) and wheat (Bhati et al., 2016). Nunes et al., 2006 achieved upto 95% reduction in soybean seed PA by RNAi but at the cost of embryo abortion in the progeny zygotes. The observed effect can partly be accounted for by firstly, the choice of gene and secondly, the promoter used for expression of the transgene. Nunes in order to manipulate PA biosynthesis, followed inhibition of the first step which is suggested to be the most effective strategy. However, suppressing myo-inositol-1-phosphate synthase (GmMIPS) expression may lead to critical alterations in seed PA biosynthesis (Kuwano et al., 2009), subsequently disturbing the cellular phosphorus and inositol homeostasis. Therefore, in our study we decided to chose one of the late PA pathway enzyme, inositol polyphosphate 6-/3-/5-kinase (GmIPK2) as the target gene. Due to multiple specificity of this enzyme, participating in sequential phosphorylation of 1D-myo-inositol-1,4,5-trisphosphate to 1D-myo-inositol-1,3,4,5,6-pentakis-phosphate and its strategic position in the PA biosynthesis pathway we hypothesize that it shall be successful in achieving a greater but optimal level of PA reduction. Further, Nunes et al. (2006) used a constitutive promoter CaMV35S which resulted in a strong expression in vegetative tissues other than developing seeds. From this we conclude that the promoter we use for expression should be active only in developing seeds, the storage site of PA. Also, we kept in mind that the promoter we use should have the same temporal and spatial activity in the seed as GmIPK2 to achieve a critical level of suppression. With regard to this purpose, we selected the promoter of reserve protein vicilin, located in protein bodies which is the same site as GmIPK2 gene expression to drive our RNAi construct.
So far there is no report on successful recovery of fertile lpa transgenics of Indian soybean cultivars. In this study, we were able to generate lpa lines of Glycine max Pusa 16 that along with reduction in phytate levels displayed an increase in available phosphorous and few other important mineral elements assayed. In addition, these lpa transgenics that we developed can provide a valuable system to study seed PA synthesis and can aid in developing markers for the lpa trait in future.
Materials and Methods
Plant Material
Glycine max [L.] Merr. cv. Pusa-16 procured from Division of Genetics, IARI, New Delhi, India was used for isolation of GmIPK2 gene and as the recipient genotype for subsequent genetic modification. Quantitative characteristics of this cultivar grown primarily in northern plain and hill zones of India have been described in studies made by Karnwal and Singh (2009) and Ramteke et al. (2010; Ramteke and Murlidharan, 2012).
Construction of RNAi Expression Vector
Based on the transcript sequence of soybean IPK2 gene available in the plant comparative genomics portal Phytozome v9.1 (Glyma.12G240900) we designed primers to amplify and clone a 305 bp fragment including 250 bp of its 3′ end coding sequence and a conserved sequence of 55 bp from its 3′ untranslated region. This nucleotide fragment was used to amplify and directionally clone sense (GmIPK2_S; FP 5′-CGCGGATCCGCGTTGCAGAAGCTCAAG-3′ and RP 5′-TCCCCGCGGGG AGCGACACTAATTCAAG-3′) and anti-sense (GmIPK2_As; FP 5′-CCGCTCGAGCGGAACGTC TTCGAGTTC-3′ and RP 5′-CCATCGATGGCGCTGTGATTAAGTTCGTA-3′) strands around a 395 bp spacer of soybean fatty acid oley1Δ12 desaturase gene intron (GmFad2-1; FP 5′-TCCCCG CGGGGAAGGTCTGTCTTATTTTGAATC-3′ and RP 5′-CCATCGATGGTATACCGCACTAGT AAACCAC-3′) cloned in pCR2.1-TOPO cloning vector to form a hairpin (ihp) structure which was confirmed by automated sequencing. The binary vector pCWAK that will carry this silencing cassette was constructed inhouse in a previous work by ligating fragments from the expression vector pCW66 and binary vector pAKVS both kindly obtained from Dr. Craig Atkins Laboratory, University of Western Australia, Australia. The final RNAi construct pCWAK-ipk2 was generated by restriction cloning (BamHI/XhoI) of ihp cassette in pCWAK under the control of soybean seed specific vicilin gene promoter and terminator (Figure 1). The plasmid pCWAK-ipk2 also bears phosphinothricin acetyltransferase (PAT) encoding marker gene bar within the T-borders driven by the cauliflower mosaic virus (CaMV) 35S promoter to confer tolerance to the herbicide glufosinate for transgenic selection. After extensive verification of pCWAK-ipk2 by restriction digestion, we subsequently mobilized the binary construct into disarmed Agrobacterium tumefaciens strain EHA105 by triparental mating using Escherichia coli DH5α (pRK2013) helper strain for further use in preliminary transformation experiments.
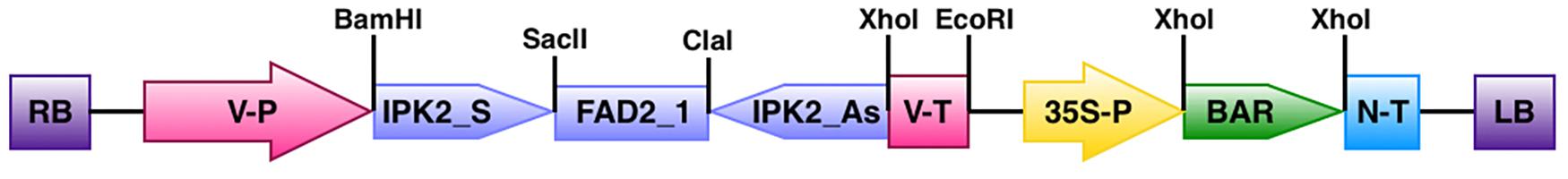
FIGURE 1. Map showing the T-DNA region of binary vector pCWAK-ipk2. RB: right border; V-P: vicilin promoter; IPK2_S: IPK2 sense fragment; FAD2_1: fatty acid oley1 Δ12 desaturase gene intron; IPK2_AS: IPK2 antisense fragment; V-T: vicilin terminator; 35S-P: cauliflower mosaic virus 35S promoter; BAR: bialaphos resistance gene; N-T: nopaline synthase terminator; LB: left border.
Generation of Transgenic Plants Expressing RNAi Construct
Soybean transformation was performed following Agrobacterium-mediated cotyledonary-node method described by Zhang et al., 1999 and Paz et al., 2004 with minor modifications. Briefly, sterilized soybean seeds germinated on half B5 medium were excised to derive two cotyledonary explants per seed and infected with A. tumefaciens harboring the RNAi construct (Figure 2A). Following 3 days of co-cultivation in dark in the presence of thiol compounds (3.3 mM L-cysteine, 1 mM dithiothreitol, 1 mM sodium thiosulfate) to improve cell transformation efficiency (Olhoft and Somers, 2001; Olhoft et al., 2001), the explants were sub-cultured twice for 12–14 days on full B5 medium (shoot induction medium, SIM) supplemented with 4 mg/l glufosinate for selective shoot induction (Figures 2B,C). Four weeks later, healthy explants were transferred to full MS medium (shoot elongation medium, SEM) containing 5 mg/l glufosinate for continued selection and sub-cultured in fresh medium every 2 weeks until shootlets approximately 2–3 cm high were developed (Figures 2D,E). Elongated shootlets were subsequently induced to develop roots by the application of 2mg/l Indole-3-butyric acid (IBA; auxin phytohormone) supplemented in half MS medium (rooting medium, RM) (Figure 2F). Well rooted plantlets were transplanted to soil pots for hardening and grown to maturity under 16 h/8 h light/dark regime at National Phytotron Facility, IARI, Delhi, India (Figures 2G,H).
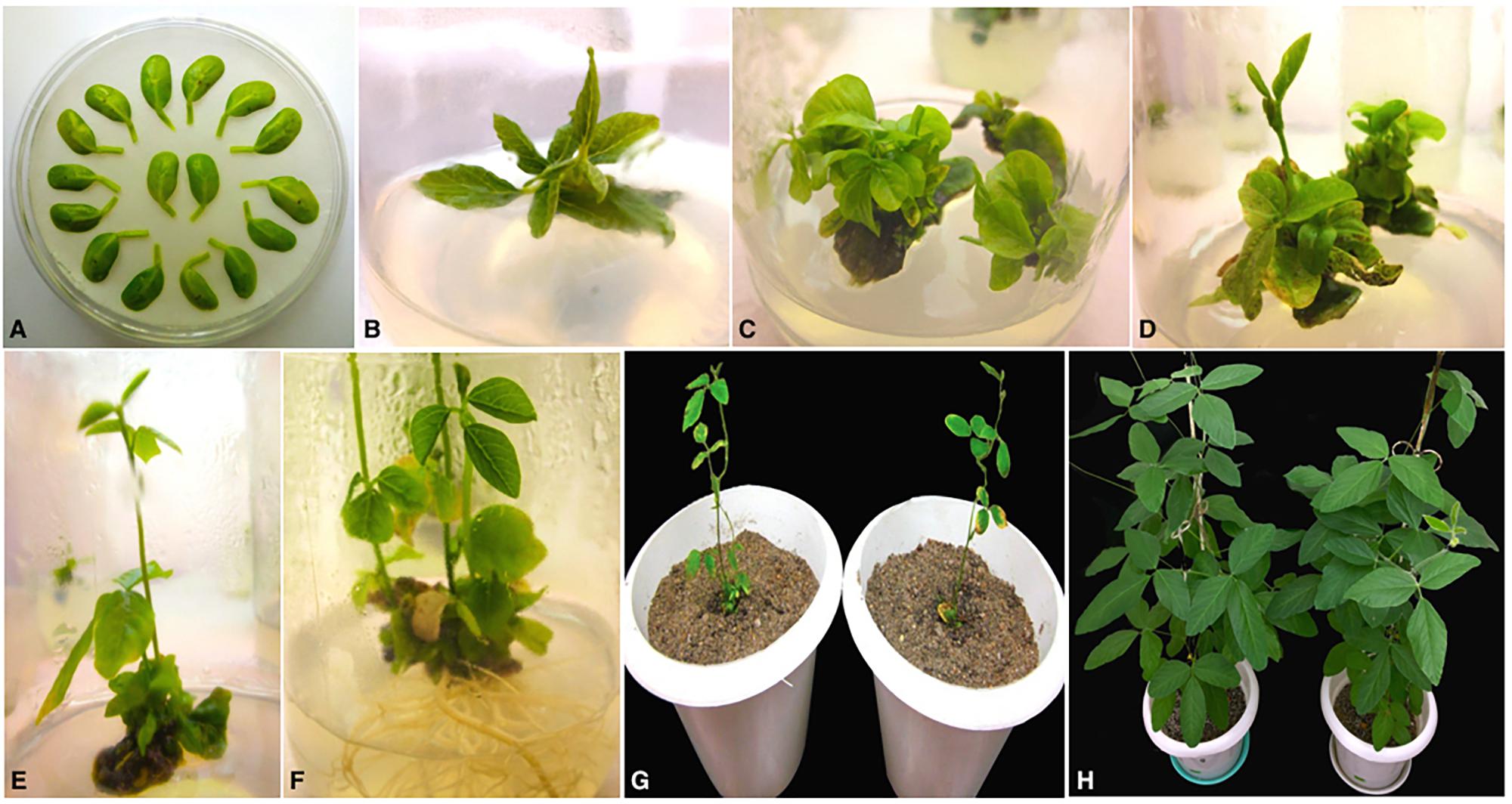
FIGURE 2. Different stages of Agrobacterium-mediated cotyledonary-node transformation of soybean cv. Pusa 16. (A) Co-cultured wounded cotyledonary-node explants (B) shoot induction from axillary meristem on SIM after 2 weeks; (C) shoot development with 4 mg/l glufosinate selection on SIM after 4 weeks of culture; (D) shoot elongation with 5 mg/l glufosinate selection on SEM after 6 weeks of culture; (E) elongated, glufosinate-resistant shoot ready for transfer to RM after 9 weeks of culture; (F) root development on RM; (G) acclimatization of plantlets in sterilized pot mix; (H) mature T0 transgenic plants.
Transgene Integration Analysis
PCR Examination
The putative transformants (T0) that survived hardening were selected for primary screening of transgene integration through PCR analysis. Total genomic DNA was isolated from their leaves and the leaves of non-transformed control plants using genomic DNA mini kit (plant) following manufacturer’s (Geneaid, Taiwan) protocol. GmIPK2_S forward (FP 5′-CGCGGATCCGCGTTGC AGAAGCTCAAG-3′) and GmFAD2-1 reverse (RP 5′-CCATCGATGGTATACCGCACTAGTAA ACCAC-3′) primers were used to amplify a 700 bp fragment using thermal cycling conditions of 1 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 62°C, 30 s at 72°C; and a final extension of 10 min at 72°C. The amplicon generated was purified and sequenced for confirmation.
Segregation Analysis
Segregation analysis of the transgene was performed by studying bar gene expression in the T1 progenies of each independent transformation events characterized. Seeds from these events were germinated in pots filled with Vermiculite, Cocopeat and Sand mixture (1:1:1). Leaf tissues were collected from 30 days old seedlings for genomic DNA isolation and PCR amplification was carried out using bar specific primers (FP 5′-GAACGACGCCCGGCCGACAT-3′ and RP 5′-GTCCAGC TGCCAGAAACCCAC-3′) under thermal cycling conditions of 1 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 65°C, 1 min at 72°C; and a final extension of 3 min at 72°C, to amplify a 500 bp fragment.
Analysis by Southern Blotting
Southern hybridization analysis was carried out on plants cultivated from seeds of PCR positive T0 lines to confirm stable T-DNA integration and to estimate the number of copies inserted. Twenty micrograms of genomic DNA from each sample was digested with PstI which does not cut the T-DNA region. XhoI digested pCWAK-IPK2 plasmid DNA was used as the positive control. The restricted samples were separated by electrophoresis on 0.8% agarose gel and transferred onto positively charged nylon membrane (Axiva, India) by capillary blotting following the method described by Sambrook et al. (1989). The membrane was UV cross-linked and subsequently hybridized with bar-specific probe at 42°C for 16 h, biotin labeled using Biotin DecaLabel DNA Labeling Kit (Thermo Scientific, United States). The hybridized membrane was washed and detected using Biotin Chromogenic Detection Kit according to the protocol described by the manufacturers (Thermo Scientific, United States).
Transcript Analysis by Quantitative Real-Time PCR
Southern positive T2 plants were further analyzed by semiquantitative RT-PCR and quantitative real-time PCR (qRT-PCR) to estimate IPK2 transcript levels. Total RNA was isolated from fresh soybean seed (8–10 mm) tissue of T2 transgenic and non-transgenic plants using TRI Reagent (Sigma-Aldrich, United States). First-strand cDNA was synthesized from 5 μg of DNaseI-treated total RNA using M-MLV Reverse Transcriptase (Promega, United States) following manufacturer’s instructions. qRT-PCR analysis was subsequently performed following SYBR Green (DyNAmo Flash SYBER Green qPCR Kit, Thermo Scientific, United States) chemistry on PikoReal 96 Real-Time PCR platform (Thermo Scientific, United States). Gene-specific primers were designed to amplify a 124 bp fragment from its conserved region (qIPK1F 5′-GGAGCGCTTGCAGAAGC-3′, qIPK1R5′-GACCAGAGGGTTGGT AGC-3′). To normalize the variance among samples, house-keeping gene phosphoenolpyruvate carboxylase (PEPCo) (qPEPCoF5′-CATGCACCAAAGGGTGTTTT-3′, qPEPCoR 5′-TTTTGCG GCAGCTATCTCTC-3′) was used as endogenous control. Amplification was achieved by a two-step PCR reaction with an initial denaturation at 94°C for 4 min followed by 40 cycles of 94°C for 30 s and annealing/extension at 60°C for 30 s. Triplicate quantitative assays were performed on biological triplate corresponding to each sample. Relative mRNA abundance was calculated using the 2-ΔΔCT method described by Livak and Schmittgen, 2001. The specificity of each unique amplification product was determined by melting curve analysis.
Estimation of Seed Phosphorus Levels
Total phosphorous in the seeds was determined colorimetrically by a method described by Chen et al. (1956). Samples (transgenic and non-transgenic control) for the assay were prepared by wet ashing dried ground seeds (250 mg) in 2 ml of concentrated sulfuric acid (Larson et al., 2000; Raboy et al., 2000).
For the analysis of inorganic phosphorus (Pi) levels, we followed a protocol described by Al-Amery et al. (2015) which uses single seed chips (1–2 mg) derived from cotyledonary segment opposite to the embryonic axis. Briefly, seed samples were extracted using 50 μl buffer (25 mM magnesium chloride and 12.5% trichloroacetic acid) for 14–16 h at 37°C with gentle shaking. 10 μl subsample of extracts were loaded in triplicates onto a 96 well plate and diluted with 90 μl water. Each sample was incubated with 100 μl of Chen’s reagent at 37°C for 1 h and the absorbance was read at 882 nm on GloMax®-Multi Detection System (Promega, United States).
Analysis of Phytic Acid Concentration by HPLC
Phytic acid estimation in transgenic seeds was performed by reversed-phase high-performance liquid chromatography (RP-HPLC) as described by Lehrfeld (1989) with some modifications (Pandey et al., 2016). 500 mg dried seed tissue was homogenized in a pestle-mortar, hexane-defatted and subsequently extracted with 0.78 M HCl by sonication (3 min) and mechanical agitation for 1 h at 250 rpm. The extract was centrifuged and one part of clear supernatant was mixed with four parts of HPLC grade water. The diluted sample was passed through conditioned SAX column (Hypersep, Thermo Scientific, United States) connected to a vacuum manifold (Millipore, United States) and eluted with 2 M HCl. The filtrate was evaporated in a vacuum rotary evaporator (Hei-VAP Value Digital G3, Heidolph, Germany), re-dissolved in 1 ml of mobile phase (acetonitrile, formic acid and tetrabutylammonium hydroxide, 4.8:5.1:0.1, v/v/v) and filtered using a 0.2 micron PVDF syringe filter (Millipore, United States). For HPLC analysis, sample was injected onto the C18 RP-column (250 × 4.6 mm, 5 μ; Shimadzu, Japan) equilibrated with isocratic mobile phase at a flow rate of 1.0 ml min-1. PA signals were monitored with a UV-VIS photodiode array detector (SPDM-20Avp, Shimadzu, Japan) at a wavelength of 197 nm. The PA concentration was calculated using the calibration curve prepared with a PA dodecasodium salt standard (Sigma-Aldrich). The software used for analysis was LC-Solutions (version 1.25).
In Vitro Bioavailability Assay
Mature transgenic and non-transgenic seeds were milled (Wiley, Thomas Scientific, United States), weighed (1 g) and digested in triplicates under simulated gastrointestinal conditions following method described by Kiers et al., 2000 to determine the bioavailability of iron (Fe), zinc (Zn), and calcium (Ca). The digest was centrifuged at 9000 rpm for 15 min and supernatant obtained was passed through a 0.45 mm filter. The filtrate was analyzed for Fe, Zn, and Ca using an Atomic Absorption Spectrophotometer (AAnalyst 200, Perkin Elmer, United States). A blank consisting of distilled water was processed in a similar manner and used for sample correction.
Agronomic Evaluation of Transgenic Plants
Germination Assay for Seed Viability
The germination capacity of T3 transgenic seeds as compared to non-transgenic control was assessed by performing controlled germination test (CGT) described by Campion et al., 2009. The seeds were immersed under water for 8 h at 28°C and then transferred to fresh water for an additional 12 h. Following incubation, seeds were rinsed three to four times in distilled water and subsequently germinated in petri dishes lined with filter paper soaked in distilled water under a photoperiod of 28°C 8-h dark and 30°C 16-h light. We also monitored seed vigor by using the standard accelerated aging test (AAT) test. In this we subjected unimbibed seeds to conditions of high temperature (42°C) and high relative humidity (90%) for 72 h and then removed them from the stress conditions and placed under optimum germination conditions.
Phenotypic Analysis
Different morphological traits of transgenic plants were evaluated under controlled conditions to capitulate the influence of transgene integration on phenotype. Mature transgenic plants were harvested and evaluated with respect to the non-transgenic control for plant height, number of pods, number of seeds, seed dry weight, stem length and root length. The height of individual plant was measured as the distance from the soil surface to the tip of the plant. On maturity, upto 100 seeds were harvested and their mean dry weight (SDW) was recorded after air-drying them in oven at 60°C for 72–96 h. Five randomly chosen plants from each transgenic line were evaluated for each parameter studied.
Statistical Analysis
Every experiment was carried out in biological and experimental replications (three to six) for each non-transgenic control and transgenic sample and represented as mean ± standard error. All statistical evaluations were performed using SAS software (version 9.2). Segregation patterns analyzed with chi-square (χ2) goodness of fit were tested at 5% level of significance. We also conducted ANOVA for agronomic traits of transgenics and non-transgenic control and compared the group means by Tukey’s test at 5% level of significance.
Results
Transgene Integration and Segregation Analysis
A preliminary screening was conducted on the T0 plants to identify putative lpa transgenic lines by PCR amplification of ∼700 bp fragment of ihp construct. The assay characterized eight independent transformation events each showing the expected amplicon size which was absent in the case of untransformed control plants (Figure 3).
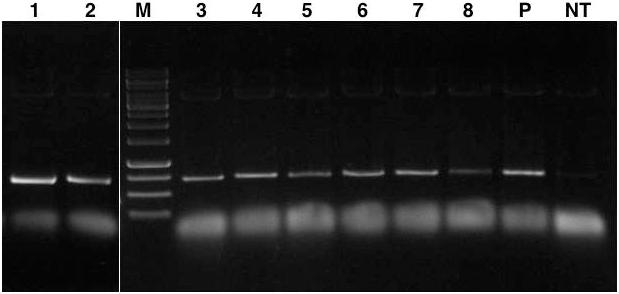
FIGURE 3. PCR amplification of ∼700 bp GmIPK2_S gene plus GmFAD2-1 intron fragment from genomic DNA of T0 transgenic plants. Lanes, M: 1 kb DNA ladder; 1–8: genomic DNA from each transformation event characterized; P: pCWAK-ipk2 plasmid DNA (positive control); NT: genomic DNA from non-transformed plant (negative control). Representative lanes were spliced from the original gel (Supplementary Figure S1).
Seeds from each independent event identified were grown successfully under containment conditions to obtain T1 progeny plants by self pollination. All the derived progenies appeared phenotypically normal and fertile. We performed segregation analysis on them by studying bar gene expression and found that five of the events (P2, P4, P5, P6, and P8) held a good fit to the stable Mendelian inheritance of a single locus (3:1) (Table 1) whilst three (P1, P3, and P7) of them showed a segregation ratio of two transgene loci (15:1).
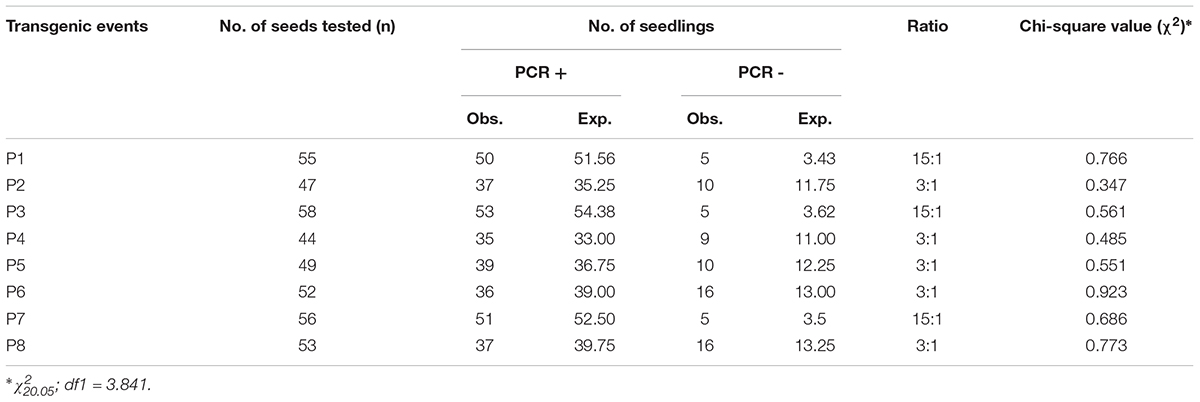
TABLE 1. Segregation of bar gene amongst T1 progenies of eight independent transformation events characterized in soybean cv. Pusa 16.
We further confirmed integration of the transgene cassette by southern blot analysis using bar gene probe (Figure 4). The genomic DNA was digested with PstI endonuclease which is not present within the T-DNA of the construct. Upon detection, all the transgenic lines analyzed showed a distinct pattern of seperation which indicate that each plant originated from an independent transformation event. The blot showed that all the fragments were above 3 kb in size which suggest that the transgenic lines carry intact copies of T-DNA because the shortest fragments in each line were longer than the T-DNA region (∼2.8 kb). The blot also confirmed single copy integration of target gene into the genome of transgenic events P2, P4, P5, P6, and P8. Since single copy insertions are always desirable, T2 generation plants were cultivated from seeds of these five characterized events and PCR screened to select homozygous lines. A positive amplification in all the tested T2 plants derived from the T1 parents, P2-45 and P6-39, suggest their homozygous nature and thus seeds collected from only these parents were selected to characterize lpa trait by further analysis.
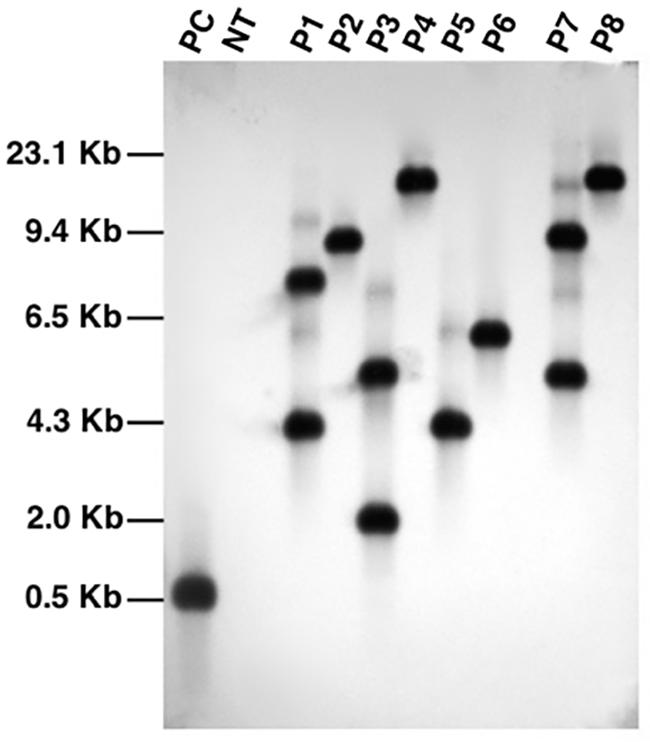
FIGURE 4. Southern blot analysis of PCR characterized transgenic events using bar gene specific probe. Lanes, PC: plasmid pCWAK-ipk2 (positive control); NT: genomic DNA from non-transformed plant (negative control); P1-P8: genomic DNA of independent transgenic events showing hybridization signals indicating transgene integration.
Expression Analysis of Transgenic Plants
To assess the level of reduction in GmIPK2 expression in transgenic seeds with respect to non-transgenic control, we carried out RT-PCR and qRT-PCR analysis. From RT-PCR results, we observed a variable reduction in the expression of GmIPK2 gene with no variation in PEPCo transcript expression between the non-transgenic and transgenic plants thus supporting its use as a stable endogenous reference (Figure 5A). We further quantified the variations in GmIPK2 transcript levels by qRT-PCR and found a maximum reduction of 2.79-fold and 2.56-fold in T3 developing seeds from the transgenic lines P2-45-8 and P6-39-10, respectively (Figure 5B) at P ≥ 0.05. We also studied GmIPK2 expression in other plant tissues of these two lines and found no variation in its transcript levels with respect to the control plants (Supplementary Figures S3, S4) which suggest its successful silencing only in the seeds.
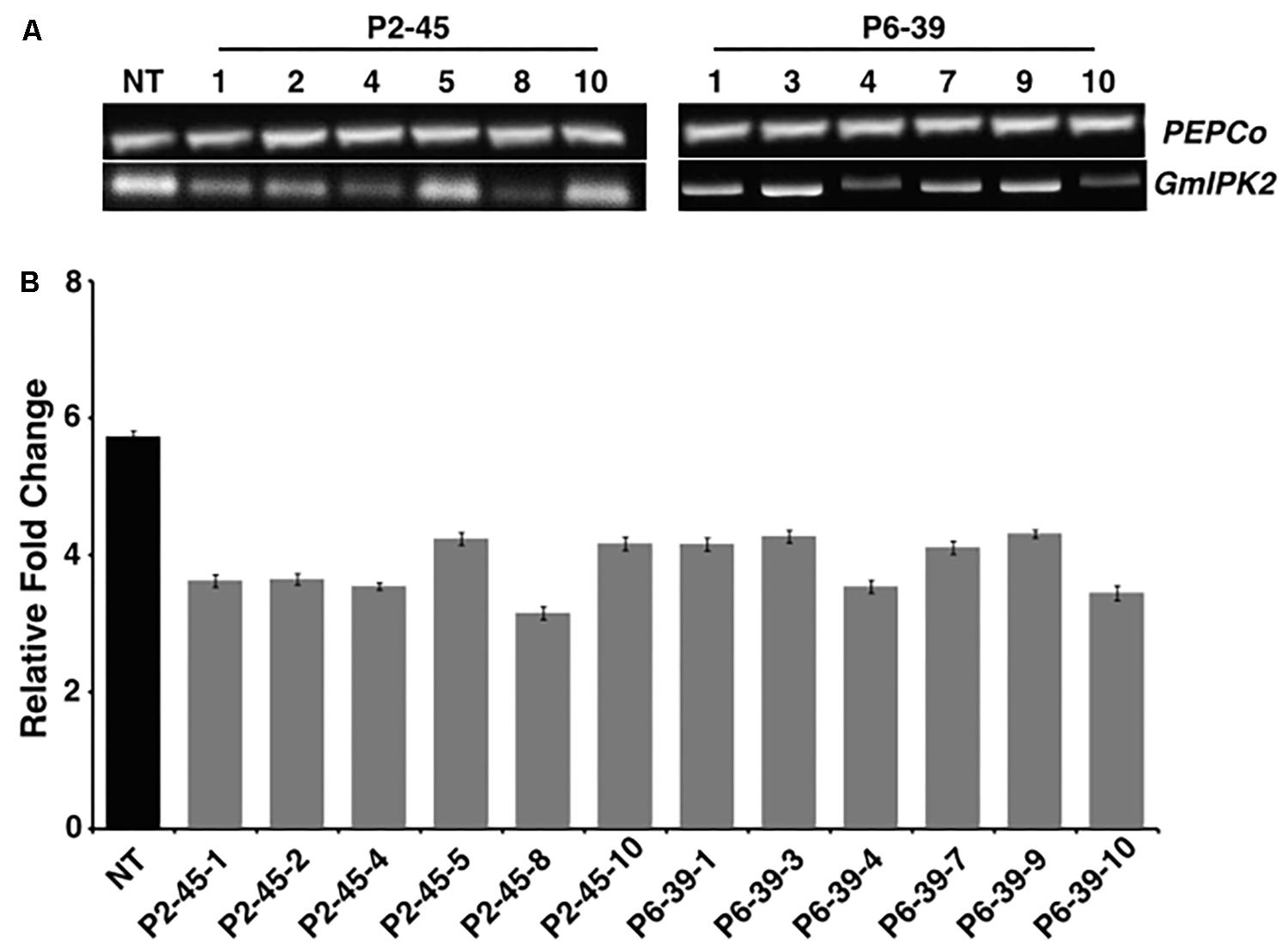
FIGURE 5. Expression analysis of transgenic soybean seeds. (A) RT-PCR amplification showing variation in GmIPK2 transcripts in T3 seeds of P2-45 and P6-39 compared to the PEPCo internal control (NT: Non-transformed plant). Representative lanes were spliced from the original gel (Supplementary Figure S2). (B) Relative fold change measured by qRT-PCR in the above samples indicating varied levels of silencing with maximum reduction observed in P2-45-8. The data presented is mean of technical triplicates corresponding to each biological replicate (n = 3) with error bars indicating standard deviation (SD).
Seed Phytic Acid Quantification
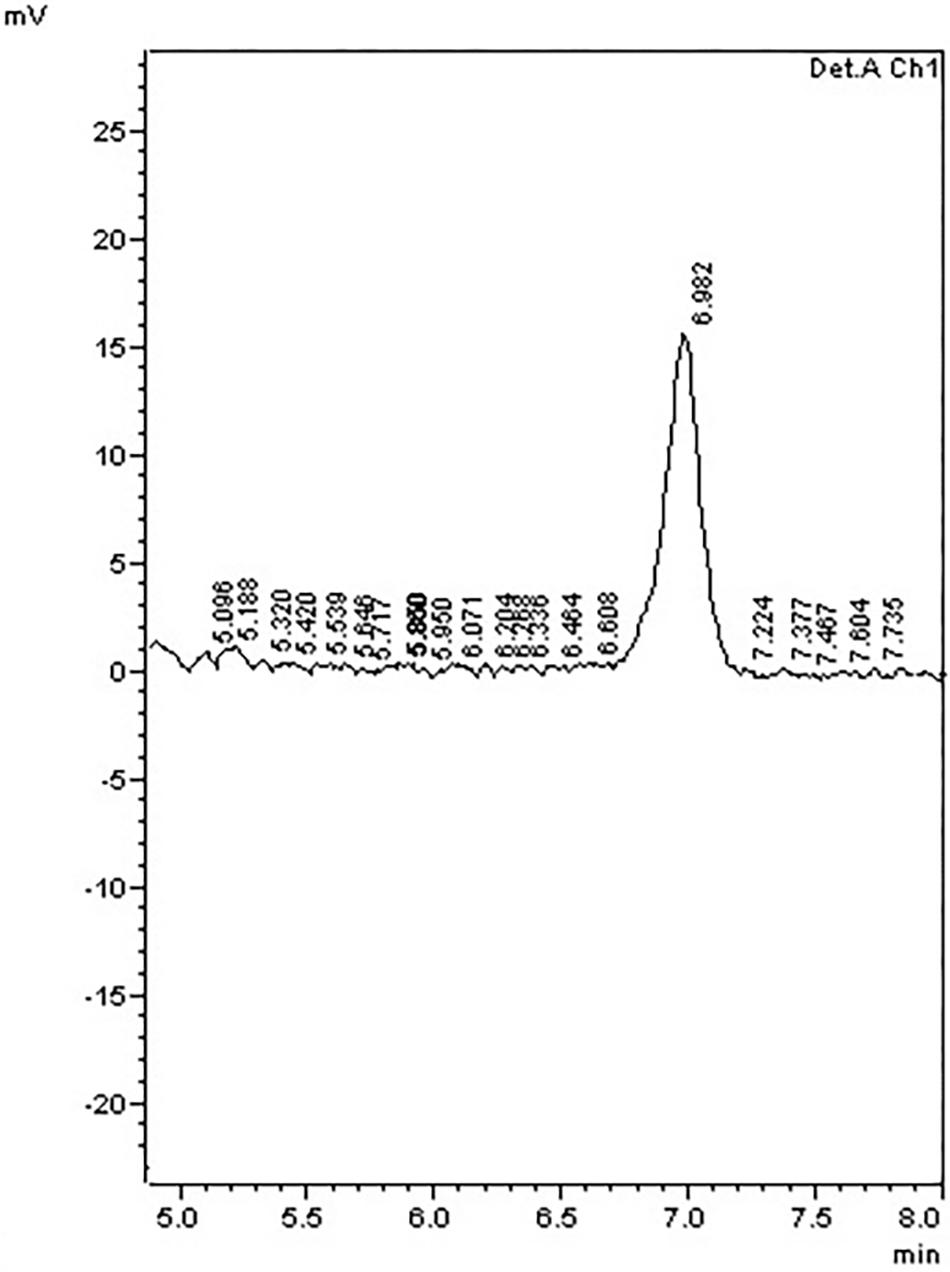
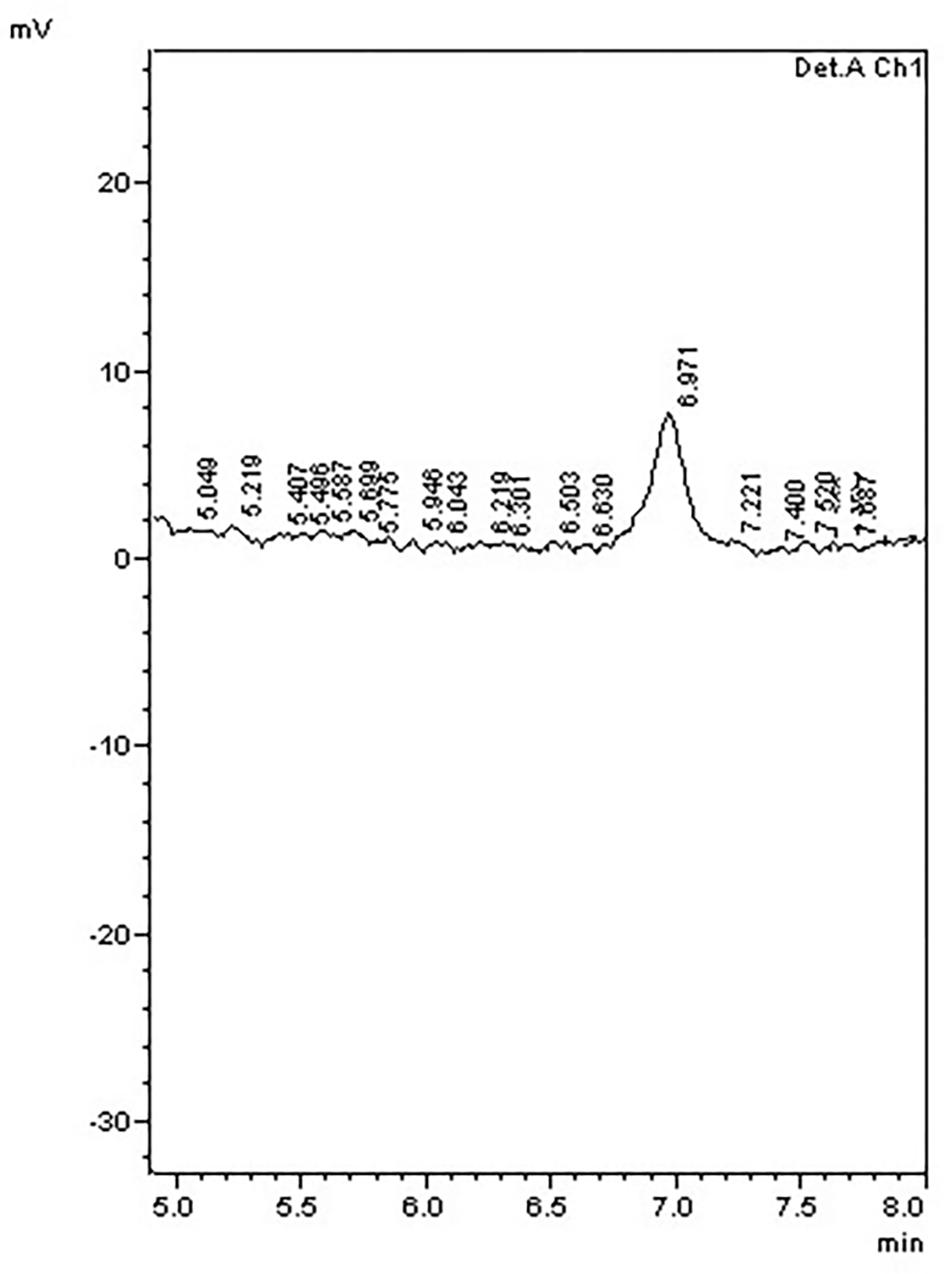
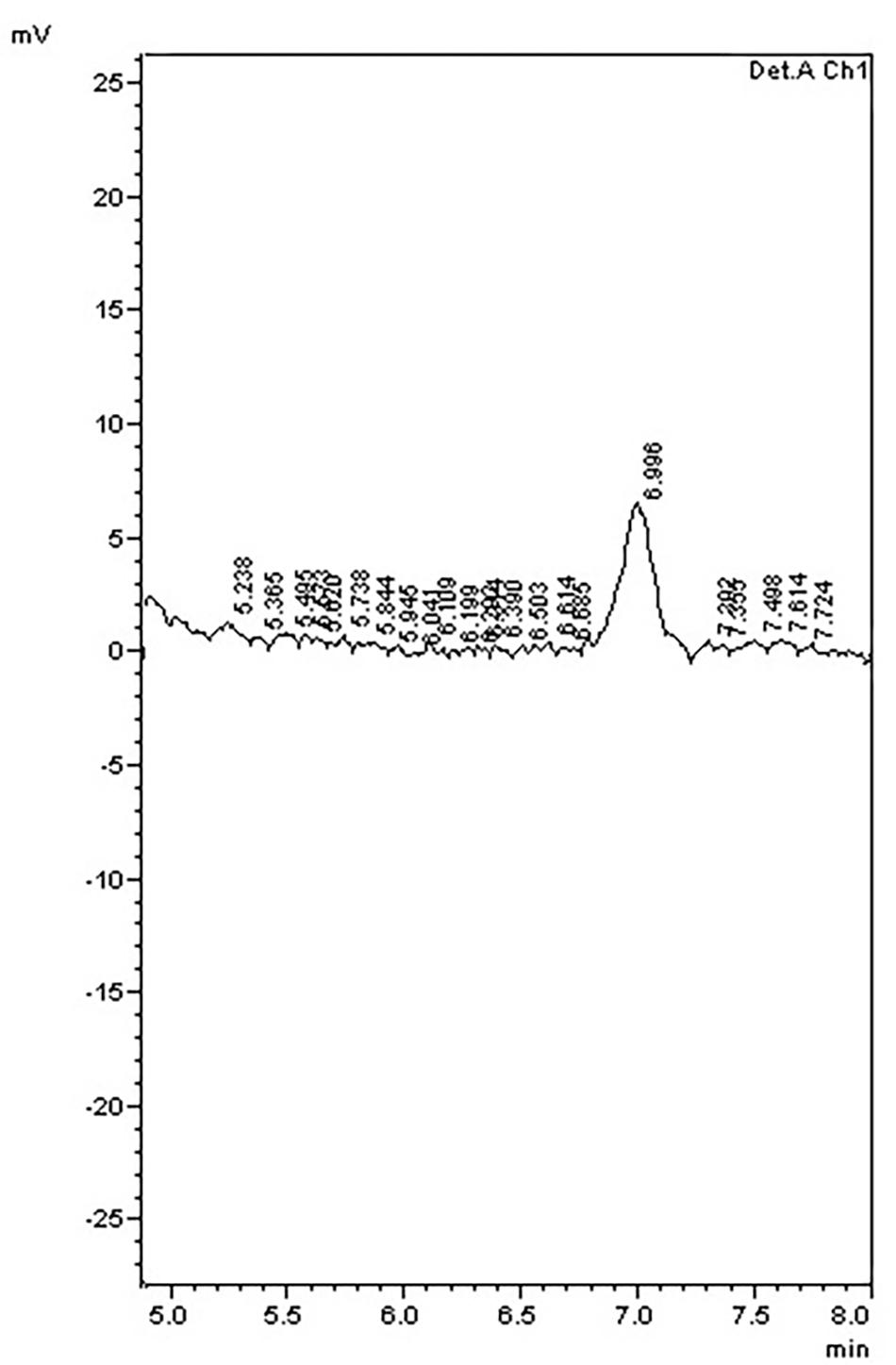
To further examine the effect of GmIPK2 silencing on PA synthesis, we quantified PA content in mature grain extracts of above two lines and non-transgenic samples by HPLC analysis. The chromatogram obtained by scanning the samples at 197 nm displayed PA retention peaks around 7.006 ± 0.09 min, the area under which was used to compute the level of PA. A decrease in the PA content was observed in T3 seeds compared to control which displayed larger peak area indicating its higher concentration in the seeds (Figures 6–8). The mean PA content was 3.48 g 100g-1 for non-transgenic seeds against 1.91 g 100 g-1 and 2.02 g 100 g-1 for the seeds from transgenic lines P2-45-8 and P6-39-10, showing a maximal reduction of 45 and 42%, respectively (Figure 9).
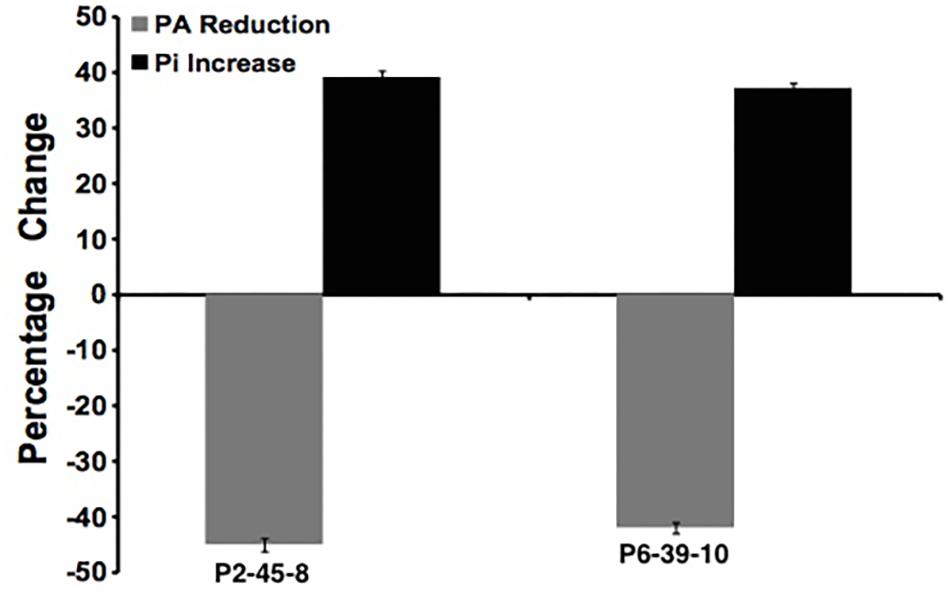
FIGURE 9. Percentage reduction in phytate (gray column) and increase in free phosphate (black column) in P2-45-8 and P6-39-10 transgenic lines.
Analysis of Seed Phosphorus Levels
We estimated the total and free phosphorus (Pi) levels in transgenic as well as non-transgenic seeds to correlate it with the reduction in PA content. No significant difference in total seed phosphorus (P) was observed for the RNAi lines as compared to the control. The average total phosphorus content of T3 seeds was 5.82 mg g-1 (P2-45-8) and 5.75 mg g-1 (P6-39-10), which was observed closely similar to that of non-transgenic seeds, 5.89 mg g-1. However, a significant increase of 39 and 37% in the concentration of Pi was recorded in the T3 seeds obtained from transgenic lines P2-45-8 and P6-39-10, respectively, which gave a dark blue reaction with chen’s reagent, confirming a reduction in the content of its principal storage form PA (Figure 9).
Quantification of Mineral Content in Seeds
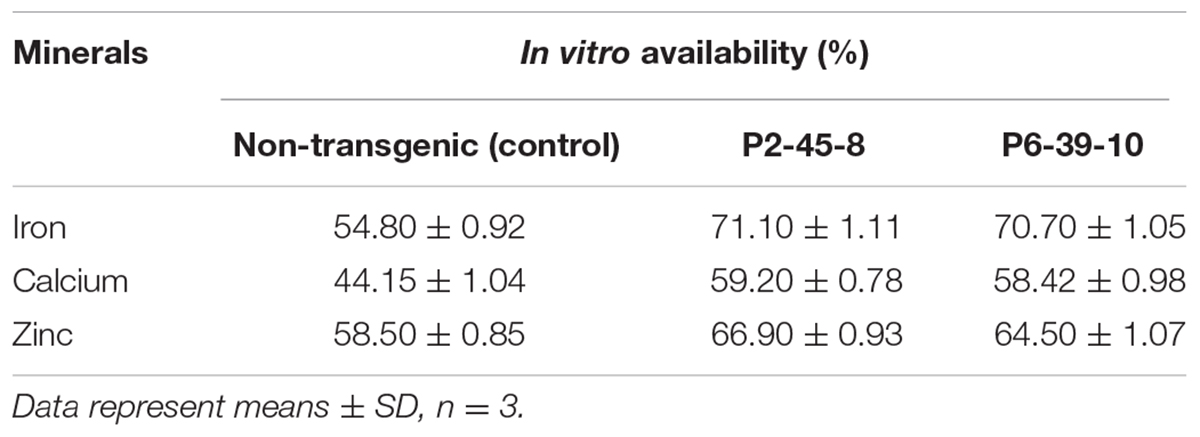
In order to determine whether mitigation of PA improved the mineral bioavailability in soy grains, we made a quantitative estimation of micronutrients in transgenic seeds compared to non-transgenic control by following in vitro digestion method. From the observations made by atomic absorption spectrophotometer, we noticed an increase in the in vitro Fe, Ca, and Zn availability from 54.8, 44.15 and 58.50% for non-transgenic control seeds to 71.1, 59.2, 66.9% for P2-45-8 and 70.7, 58.42, 64.50% for P6-39-10 seeds (Table 2). As mentioned before, PA binding with minerals result in formation of insoluble salts with poor bioavailability, our result suggests that the mineral bioavailability can significantly be improved by reduction of PA in lpa transgenic lines.
Seed Germination and Morphological Trait Analysis of Transgenic Plants
Decrease in seed PA concentration can be accompanied by adverse agronomic consequences. Therefore, we conducted germination tests on the lpa seeds collected from our transgenic lines. The CGT test so conducted confirmed that 98% of the seeds possessed good germination potential whereas AAT test established their high vigor (Supplementary Figure S5A). Also, the morphological analysis of seed germination in each of the tests revealed similar phenotypes in both transgenic and control seeds (Supplementary Figure S5B). Besides, since PA also regulates the growth and development of seedlings, we carried out phenotyping on 75 days old plants. The growth of transgenic plants was compared with the growth of control soybean plants under field growth conditions. Interestingly, P2 and P6 plants exhibited an increase in leaf size which could possibly be explained due to increase in available seed P content (P < 0.05) (Tubana and George, 1997). No significant differences were observed between wild type and transgenic plants in other agronomic traits investigated (P > 0.05) (Table 3). This data confirms that GmIPK2 silenced, lpa soybean seeds are capable of generating plants showing normal morphologies and agronomic traits.
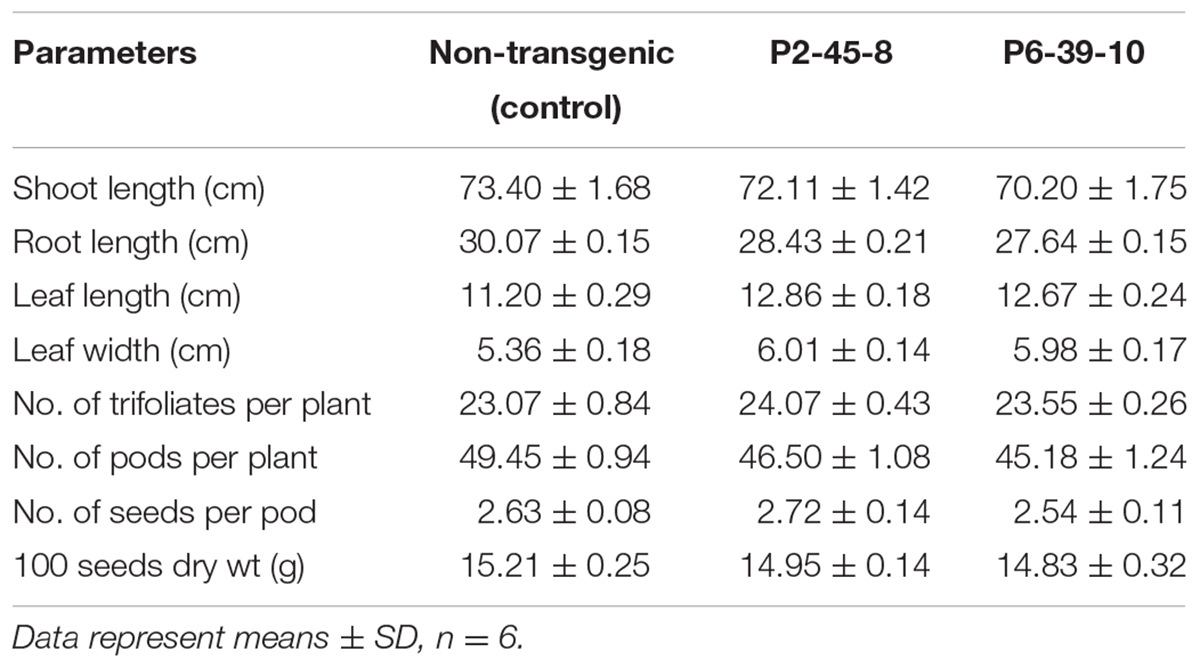
TABLE 3. Morphological and yield contributing characters of T2 transgenic plants raised in green-house.
Discussion
Several mutational and transgenic strategies have been explored to reduce PA in soybean but with undesirable aspects (Meis et al., 2003; Yuan et al., 2007, 2009; Anderson and Fehr, 2008; Maupin and Rainey, 2011; Maupin et al., 2011). We may attribute these negative effects primarily to the constitutive suppression of PA biosynthesis genes which result in an indirect impact on several metabolic processes and signal transduction pathways. Prattley and Stanley (1982) established that the cotyledon of soybean contains 90% of the phytate of the seed, we therefore, expressed our RNAi construct under the control of a cotyledon-specific vicilin promoter (Higgins et al., 1988; Rosche et al., 2002) with the aim to restrict PA perturbations to the seed. As expected, initial characterization of the transgenic genotypes displayed stable integration and inheritance of the transforming DNA with no significant impact on whole plant performance and yield. Segregation patterns observed in the T1 progenies of distinguished lineages demonstrate that both homozygous and heterozygous plants were obtained from self-pollinated seeds of most of the events, suggesting that T0 plants were uniformly transformed and were heterozygous for the transformed genotype. Transgenic plants containing single copy or double copies of the transforming gene were also characterized by segregation analysis. Hobbs et al., 1993 through their studies established that single copy transgenic plants are always desirable to avoid the possibility of gene silencing during introgression of the transgene into the parental line(s) of hybrids, or into improved open-pollinated varieties, we therefore confirmed these single copy events by performing southern blot before proceeding any further analysis.
We used qPCR to check GmIPK2 expression in T3 seeds as well as other plant tissues. From our results, we observed a varied level of reduction of GmIPK2 transcripts in seeds of lines P2-45 and P6-39. These variations can be explained by several factors viz. differences in the site of integration on the chromosomal DNA, possible promoter methylation etc. (Day et al., 2000; Van Leeuwen et al., 2001). On the other hand, no GmIPK2 reduction was detected in other tissues of transgenic and control plants demonstrating seed-specific regulation. We further confirmed the implications of GmIPK2 silencing by testing the extracts from T3 generation seeds for both PA and Pi. A reduction of 45% in the PA content and a 39% increase in Pi levels was found in transgenic line P2-45-8, suggesting a strict correspondence between seed PA and Pi which is in accordance with the previous reports by Larson et al., 2000. Moreover, despite the variations in PA and Pi levels it was found that the mean total P content in the transgenic seeds was unaffected, which suggest that these altered genotypes influence P partitioning to strike a balance for supporting P-related mechanisms in the seed.
Phytic acid represents a major sink for mineral deposition due to its natural strong chelation ability. Research has previously reported the beneficial effects of cereal lpa mutants in improved mineral cations bioavailability (Mendoza et al., 1998; Adams et al., 2002; Hambidge et al., 2004, 2005; Linares et al., 2006; Mazariegos et al., 2006; Veum et al., 2007). We therefore analyzed the mineral concentration of transgenic seeds and found that disruption of GmIPK2 expression caused the Fe, Ca, and Zn contents of seeds to increase. We must also highlight that amongst the three different minerals analyzed, a maximum increase was observed in the levels of Fe followed by Ca and Zn, respectively, supporting the findings of Persson et al., 2009 who suggest that Fe in the seeds is mainly associated with PA.
Phytate reduction in a plant is often negatively correlated with its agronomic performance (Bregitzer and Raboy, 2006) which is initially observed as a direct affect on its seed germination ability. We therefore conducted germination trials on our lpa transgenics and found that they show a good response, with seedling emergence almost identical to the non-transgenic parent type. Our results seem to be in agreement with the recent reports of Shi et al., 2007 and Spear and Fehr, 2007 who suggest that high levels of PA are not an absolute requirement for efficient seed germination or emergence. Lastly, we studied the morphological traits of each transgenic line and found that the lines generated from events P1 and P4 produced seeds with reduced size and weight. The lower seed yield may be attributed to reduction in starch accumulation resulting from indirect affect of disturbed Pi homeostasis on a key regulatory enzyme in starch biosynthesis pathway (Plaxton and Preiss, 1987).
From our research, we can assert that GmIPK2 gene is an appropriate candidate for targeted phytate reduction without any pleotropic effects. Despite our results, there is a need to conduct additional field trials under a variety of conditions, across generations and perform their in depth phenotypic and genotypic analysis in order to conclude the effect of lpa trait. Further studies are therefore underway in our laboratory in an effort to establish the same.
Author Contributions
MP contributed in conception and designing of the study, performed the experiments, acquired the data, performed its analysis and interpretation, and drafted the final manuscript. NB contributed in designing the study, revising the manuscript critically for important intellectual content, and gave final approval of the version to be published. MJ contributed in data acquisition, revising the manuscript critically for important intellectual content, and gave final approval of the version to be published. AD contributed in interpretation of data, revising the manuscript critically for important intellectual content, and gave final approval of the version to be published. AS contributed in conception and designing of the study, revising the manuscript critically for important intellectual content, and gave final approval of the version to be published.
Funding
This work was supported by the National Funds for Basic, Strategic and Frontier Application Research in Agriculture (Grant No. NFBSFARA/RNAi -2011/2011-12), ICAR, Government of India.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00804/full#supplementary-material
References
Adams, C. L., Hambidge, M., Raboy, V., Dorsch, J. A., Sian, L., Westcott, J. L., et al. (2002). Zinc absorption from a low-phytic acid maize. Am. J. Clin. Nutr. 76, 556–559. doi: 10.1093/ajcn/76.3.556
Ali, N., Paul, S., Gayen, D., Sarkar, S. N., Datta, K., and Datta, S. K. (2013a). Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene (IPK1). PLoS One 8:e68161. doi: 10.1371/journal.pone.0068161
Ali, N., Paul, S., Gayen, D., Sarkar, S. N., Datta, S. K., and Datta, K. (2013b). RNAi mediated down regulation of myo-inositol-3-phosphate synthase to generate low phytate rice. Rice 6:12. doi: 10.1186/1939-8433-6-12
Al-Amery, M., Fukushige, H., and Hildebrand, D. (2015). Single seed selection for low phytate lines. J. Am. Oil Chem. Soc. 92, 1119–1123. doi: 10.1007/s11746-015-2681-9
Anderson, B. P., and Fehr, W. R. (2008). Seed source affects field emergence of low-phytate soybean lines. Crop Sci. 48, 929–932. doi: 10.2135/cropsci2007.09.0510
Bhati, K. K., Alok, A., Kumar, A., Kaur, J., Tiwari, S., and Pandey, A. K. (2016). Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 67, 4379–4389. doi: 10.1093/jxb/erw224
Bregitzer, P., and Raboy, V. (2006). Effects of four independent low-phytate mutations on barley agronomic performance. Crop Sci. 46, 1318–1322. doi: 10.2135/cropsci2005.09-0301
Brinch-Pedersen, H., Olesen, A., Rasmussen, S. K., and Holm, P. B. (2000). Generation of transgenic wheat (Triticum aestivum L.) for constitutive accumulation of an Aspergillus phytase. Mol. Breed. 6, 195–206. doi: 10.1023/A:1009690730620
Campion, B., Sparvoli, F., Doria, E., Tagliabue, G., Galasso, I., Fileppi, M., et al. (2009). Isolation and characterisation of an lpa (low phytic acid) mutant in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 118, 1211–1221. doi: 10.1007/s00122-009-0975-8
Chen, P. S., Toribara, T. Y., and Warner, H. (1956). Microdetermination of phosphorus. Anal. Chem. 28, 1756–1758. doi: 10.1021/ac60119a033
Chiera, J. M., Finer, J. J., and Grabau, E. A. (2004). Ectopic expression of a soybean phytase in developing seeds of Glycine max to improve phosphorus availability. Plant Mol. Biol. 56, 895–904. doi: 10.1007/s11103-004-5293-6
Day, C. D., Lee, E., Kobayashi, J., Holappa, L. D., Albert, H., and Ow, D. W. (2000). Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev. 14, 2869–2880. doi: 10.1101/gad.849600
Denbow, D. M., Grabau, E. A., Lacy, G. H., Kornegay, E. T., Russell, D. R., and Umbeck, P. (1998). Soybeans transformed with a fungal phytase gene improve phosphorus availability for broilers. Poultry Sci. 77, 878–881. doi: 10.1093/ps/77.6.878
Guttieri, M., Bowen, D., Dorsch, J. A., Raboy, V., and Souza, E. (2004). Identification and characterization of a low phytic acid wheat. Crop Sci. 44, 418–424. doi: 10.2135/cropsci2004.4180
Halsted, J. C., Smith, J. C. Jr., and Irwin, M. I. (1974). A conspectus of research on zinc requirements of man. J. Nutr. 104, 345–350. doi: 10.1093/jn/104.3.345
Hambidge, K. M., HuVer, J. W., Raboy, V., Grunwald, G. K., Westcott, J. L., Sian, L., et al. (2004). Zinc absorption from low-phytate hybrids of maize and their wild-type isohybrids. Am. J. Clin. Nutr. 79, 1053–1059. doi: 10.1093/ajcn/79.6.1053
Hambidge, K. M., Krebs, N. F., Westcott, J. L., Sian, L., Miller, L. V., Peterson, K. L., et al. (2005). Absorption of calcium from tortilla meals prepared from low phytate maize. Am. J. Clin. Nutr. 82, 84–87. doi: 10.1093/ajcn/82.1.84
Hegeman, C. E., and Grabau, E. A. (2001). A novel phytase with sequence similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiol. 126, 1598–1608. doi: 10.1104/pp.126.4.1598
Higgins, T. J. V., Newbigin, E. J., Stransky, H., Llewellyn, D. J., and Craig, S. (1988). The sequence of a pea vicilin gene and its expression in transgenic tobacco plants. Plant Mol. Biol. 11, 683–695. doi: 10.1007/BF00017468
Hitz, W., Carlson, T. J., Kerr, P. S., and Sebastian, S. A. (2002). Biochemical and molecular characterization of a mutation that confers a decreased raffinosaccharide and phytic acid phenotype on soybean seeds. Plant Physiol. 128, 650–660. doi: 10.1104/pp.010585
Hobbs, S. L., Warkentin, T. D., and Delong, C. M. O. (1993). Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol. Biol. 21, 17–26. doi: 10.1007/BF00039614
Jacobsen, T., and Slotfeldt-Ellingsen, K. D. (1983). Phytic acid and metal availability: a study of Ca and Cu binding foodstuffs. Cereal Chem. 60, 392–395.
Jiang, S., Cai, W., and Xu, B. (2013). Food quality improvement of soy milk made from short-time germinated soybeans. Foods 2, 198–212. doi: 10.3390/foods2020198
Karnwal, M. K., and Singh, K. (2009). Studies on genetic variability, character association and path coefficient for seed yield and its contributing traits in soybean [Glycine max (L.) Merrill]. Legume Res. 32, 70–73.
Kiers, J. L., Nout, M. J. R., and Rombouts, F. M. (2000). In vitro digestibility of processed and fermented soya bean, cowpea and maize. J. Sci. Food Agric. 80, 1325–1331. doi: 10.1002/1097-0010(200007)80:9<1325::AID-JSFA648>3.0.CO;2-K
Kuwano, M., Mimura, T., Takaiwa, F., and Yoshida, K. T. (2009). Generation of stable ‘low phytic acid’ transgenic rice through antisense repression of the 1D-myo-inositol 3-phosphate synthase gene (RINO1) using the 18-kDa oleosin promoter. Plant Biotechnol. J. 7, 96–105. doi: 10.1111/j.1467-7652.2008.00375.x
Larson, S. R., Rutger, J. N., Young, K. A., and Raboy, V. (2000). Isolation and genetic mapping of a non-lethal rice (Oryza sativa L.) low phytic acid mutation. Crop Sci. 40, 1397–1405. doi: 10.2135/cropsci2000.4051397x
Larson, S. R., Young, K. A., Cook, A., Blake, T. K., and Raboy, V. (1998). Linkage mapping of two mutations that reduce phytic acid content of barley grain. Theor. AppI. Genet. 97, 141–146. doi: 10.1007/s001220050878
Lehrfeld, J. (1989). High-performance liquid chromatography analysis of phytic acid on a pH-stable, macroporous polymer column. Cereal Chem. 66, 510–515.
Li, J., Hegeman, C. E., Hanlon, R. W., Lacy, G. H., Denbow, D. M., and Grabau, E. A. (1997). Secretion of active recombinant phytase from soybean cell-suspension cultures. Plant Physiol. 114, 1103–1111. doi: 10.1104/pp.114.3.1103
Linares, L. B., Broomhead, J. N., Guaiume, E. A., Ledoux, D. R., Veum, T. L., and Raboy, V. (2006). Effects of low phytate barley (Hordeum vulgare L.) on zinc utilization in young broiler chicks. Poult. Sci. 86, 299–308. doi: 10.1093/ps/86.2.299
Liu, Q. L., Xu, X. H., Ren, X. L., Fu, H. W., Wu, D. X., and Shu, Q. Y. (2007). Generation and characterization of low phytic acid germplasm in rice (Oryza sativa L.). Theor. AppI. Genet. 114, 803–814. doi: 10.1007/s00122-006-0478-9
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-Δ Δ CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lott, J. N. A. (1984). “Accumulation of seed reserves of phosphorus and other minerals,” in Seed Physiology, Development, Vol. 1, ed. D. R. Murray (Sydney, NSW: Academic Press), 139–166.
Lott, J. N. A., Greenwood, J. S., and Batten, G. D. (1995). “Mechanisms and regulation of mineral nutrient storage during seed development,” in Seed Development and Germination, eds J. Kigel and G. Galili (New York, NY: Marcel Dekker), 215–235.
Maupin, L. M., and Rainey, K. M. (2011). Improving emergence of modified phosphorus composition soybeans: genotypes, germplasm, environments, and selection. Crop Sci. 51, 1946–1955. doi: 10.2135/cropsci2010.10.0585
Maupin, L. M., Rosso, M. L., and Rainey, K. M. (2011). Environmental effects on soybean with modified phosphorus and sugar composition. Crop Sci. 51, 642–650. doi: 10.2135/cropsci2010.07.0396
Mazariegos, M., Hambidge, K. M., Krebs, N. F., Westcott, J. E., Lei, S., Grunwald, G. K., et al. (2006). Zinc absorption in Guatemalan school children fed normal or low-phytate maize. Am. J. Clin. Nutr. 83, 59–64. doi: 10.1093/ajcn/83.1.59
Meis, S. J., Fehr, W. R., and Schnebly, S. R. (2003). Seed source effect on field emergence of soybean lines with reduced phytate and raffinose saccharides. Crop Sci. 43, 1336–1339. doi: 10.2135/cropsci2003.1336
Mendoza, C., Viteri, F. E., Lonnerdal, B., Young, K. A., Raboy, V., and Brown, K. H. (1998). Effect of genetically modified, low-phytic acid maize on absorption of iron from tortillas. Am. J. Clin. Nutr. 68, 1123–1128. doi: 10.1093/ajcn/68.5.1123
Nunes, A., Vianna, G., Cuneo, F., Amaya-Farfán, J., de Capdeville, G., Rech, E., et al. (2006). RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta 224, 125–132. doi: 10.1007/s00425-005-0201-0
Olhoft, P. M., Lin, K., Galbraith, J., Nielsen, N. C., and Somers, D. A. (2001). The role of thiol compounds in increasing Agrobacterium-mediated transformation of soybean cotyledonary-node cells. Plant Cell Rep. 20, 731–737. doi: 10.1007/s002990100388
Olhoft, P. M., and Somers, D. A. (2001). L-Cysteine increases Agrobacterium-mediated T-DNA delivery into soybean cotyledonary node cells. Plant Cell Rep. 20, 706–711. doi: 10.1007/s002990100379
Pandey, V., Krishnan, V., Basak, N., Hada, A., Punjabi, M., Jolly, M., et al. (2016). Phytic acid dynamics during seed development and it’s composition in yellow and black indian soybean (Glycine max L.) genotypes through a modified extraction and HPLC method. J. Plant Biochem. Biotechnol. 25, 367–374. doi: 10.1007/s13562-015-0348-0
Paz, M. M., Shou, H. X., Guo, Z. B., Zhang, Z. Y., Banerjee, A. K., and Wang, K. (2004). Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica 136, 167–179. doi: 10.1023/B:EUPH.0000030670.36730.a4
Persson, D. P., Hansen, T. H., Laursen, K. H., Schjoerring, J. K., and Husted, S. (2009). Simultaneous iron, zinc, sulfur and phosphorus speciation analysis of barley grain tissues using SEC-ICP-MS and IP-ICP-MS. Metallomics 1, 418–426. doi: 10.1039/b905688b
Plaxton, W. C., and Preiss, J. (1987). Purification and properties of nonproteolytic degraded ADP glucose pyrophosphorylase from maize endosperm. Plant Physiol. 83, 105–112. doi: 10.1104/pp.83.1.105
Prattley, C. A., and Stanley, D. W. (1982). Protein-phytate interactions in soybeans. I. Localization of phytate in protein bodies and globoids. J. Food Biochem. 6, 243–253. doi: 10.1111/j.1745-4514.1982.tb00305.x
Raboy, V., Dickinson, D. B., and Below, F. E. (1984). Variation in seed total phosphorus, phytic acid, zinc, calcium, magnesium, and protein among lines of Glycine max and G. soja. Crop Sci. 24, 431–434. doi: 10.2135/cropsci1984.0011183X002400030001x
Raboy, V., Gerbasi, P. F., Young, K. A., Stoneberg, S. D., Pickett, S. G., Bauman, A. T., et al. (2000). Origin and seed phenotype of maize low phytic acid 1–1 and low phytic acid 2–1. Plant Physiol. 124, 355–368. doi: 10.1104/pp.124.1.355
Ramteke, R., Kumar, V., Murlidharan, P., and Agarwal, D. K. (2010). Study on genetic variability and traits interrelationship among released soybean varieties of India [Glycine max (L.) Merrill]. Electron. J. Plant Breed. 1, 1483–1487.
Ramteke, R., and Murlidharan, P. (2012). Characterization of soybean (Glycine max) varieties as per DUS guidelines. Indian J. Agric. Sci. 82, 572–577.
Rasmussen, S., and Hatzack, F. (1998). Identification of two low-phytate barley (Hordeum vulgare L.) grain mutants by TLC and genetic analysis. Hereditas 129, 107–112. doi: 10.1111/j.1601-5223.1998.00107.x
Rosche, E., Blackmore, D., Tegeder, M., Richardson, T., Schroeder, H., Higgins, T. J., et al. (2002). Seed-specific overexpression of a potato sucrose transporter increases sucrose uptake and growth rates of developing pea cotyledons. Plant J. 30, 165–175. doi: 10.1046/j.1365-313X.2002.01282.x
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Shi, J., Wang, H., Hazebroek, J., Ertl, D. S., and Harp, T. (2005). The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 42, 708–719. doi: 10.1111/j.1365-313X.2005.02412.x
Shi, J., Wang, H., Schellin, K., Li, B., Faller, M., Stoop, J. M., et al. (2007). Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 25, 930–937. doi: 10.1038/nbt1322
Spear, J. D., and Fehr, W. R. (2007). Genetic improvement of seedling emergence of soybean lines with low phytate. Crop Sci. 47, 1354–1360. doi: 10.2135/cropsci2006.09.0600
Tubana, B. S., and George, T. (1997). Seed Phosphorus Fertilizer Effect on the Early Growth of Soybean Under Low Phosphorus Supply. Los Baños: International Rice Research Institute.
Van Leeuwen, W., Ruttink, T., Borst-Vrenssen, A. W., van der Plas, L. H., and van der Krol, A. R. (2001). Characterization of position-induced spatial and temporal regulation of transgene promoter activity in plants. J. Exp. Bot. 52, 949–959. doi: 10.1093/jexbot/52.358.949
Veum, T. L., Ledoux, D. R., and Raboy, V. (2007). Low-phytate barley cultivars improve the utilization of phosphorus, calcium, nitrogen, energy, and dry matter in diets fed to young swine. J. Anim. Sci. 85, 961–971. doi: 10.2527/jas.2006-453
Wada, T., and Lott, J. N. A. (1997). Light and electron microscopic and energy dispersive x-ray microanalysis studies of globoids in protein bodies of embryo tissues and the aleurone layer of rice (Oryza sativa L.) grains. Can. J. Bot. 75, 1137–1147. doi: 10.1139/b97-125
Wilcox, J. R., Premachandra, G. S., Young, K. A., and Raboy, V. (2000). Isolation of high seed inorganic P, low-phytate soybean mutants. Crop Sci. 40, 1601–1605. doi: 10.2135/cropsci2000.4061601x
Yuan, F. J., Zhao, H. J., Ren, X. L., Zhu, S. L., Fu, X. J., and Shu, Q. Y. (2007). Generation and characterization of two novel low phytate mutations in soybean (Glycine max L. Merr.). Theor. Appl. Genet. 115, 945–957. doi: 10.1007/s00122-007-0621-2
Yuan, F. J., Zhu, D. H., Deng, B., Fu, X. J., Dong, D. K., Zhu, S. L., et al. (2009). Effects of two low phytic acid mutations on seed quality and nutritional traits in soybean (Glycine max L. Merr). J. Agric. Food Chem. 57, 3632–3638. doi: 10.1021/jf803862a
Keywords: soybean, low phytic acid, inositol polyphosphate 6-/3-/5-kinase, RNAi silencing, seed-specific, Agrobacterium-mediated transformation
Citation: Punjabi M, Bharadvaja N, Jolly M, Dahuja A and Sachdev A (2018) Development and Evaluation of Low Phytic Acid Soybean by siRNA Triggered Seed Specific Silencing of Inositol Polyphosphate 6-/3-/5-Kinase Gene. Front. Plant Sci. 9:804. doi: 10.3389/fpls.2018.00804
Received: 11 September 2017; Accepted: 25 May 2018;
Published: 14 June 2018.
Edited by:
James Lloyd, Stellenbosch University, South AfricaReviewed by:
Francesca Sparvoli, Istituto di Biologia e Biotecnologia Agraria (IBBA), ItalyAjay Kumar Pandey, National Agri-Food Biotechnology Institute, India
Copyright © 2018 Punjabi, Bharadvaja, Jolly, Dahuja and Sachdev. This is an openaccess article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Archana Sachdev, YXJjc19iaW9AeWFob28uY29t
 Mansi Punjabi
Mansi Punjabi Navneeta Bharadvaja
Navneeta Bharadvaja Monica Jolly
Monica Jolly Anil Dahuja
Anil Dahuja Archana Sachdev
Archana Sachdev