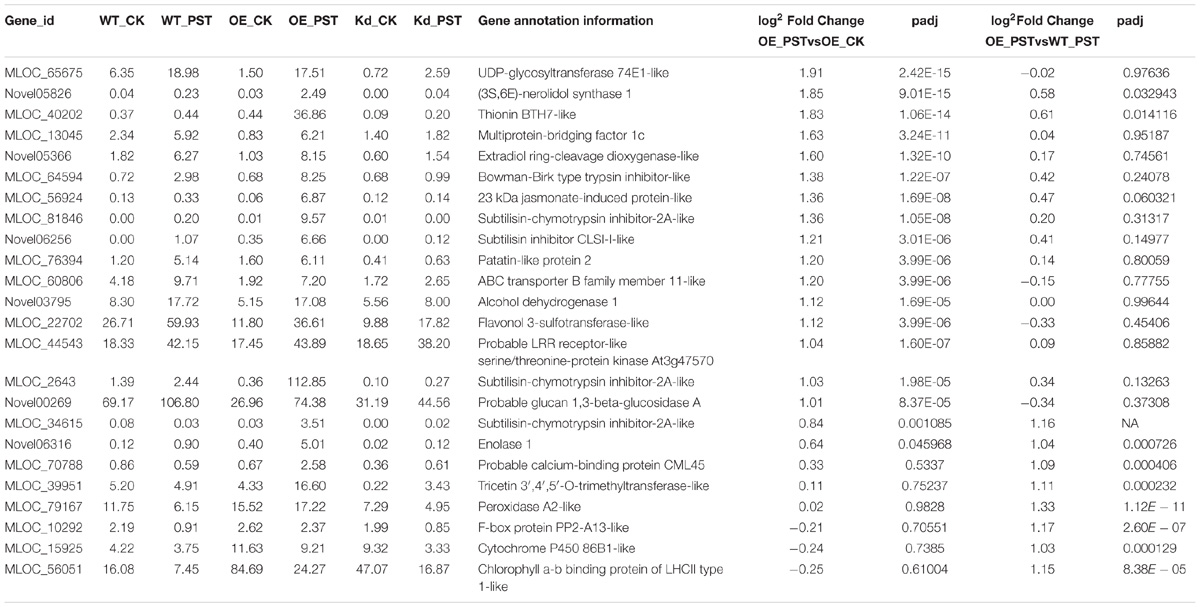
- 1College of Plant Protection, Biological Control Center for Plant Diseases and Plant Pests of Hebei, Hebei Agricultural University, Baoding, China
- 2College of Life Sciences, Hebei Agricultural University, Baoding, China
- 3Graduate School of Chinese Academy of Agricultural Sciences, Beijing, China
Systemic acquired resistance (SAR) in Arabidopsis is established beyond the initial pathogenic infection or is directly induced by treatment with salicylic acid or its functional analogs (SA/INA/BTH). NPR1 protein and WRKY transcription factors are considered the master regulators of SAR. Our previous study showed that NPR1 homologs in wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) regulated the expression of genes encoding pathogenesis-related (PR) proteins during acquired resistance (AR) triggered by Pseudomonas syringae pv. tomato DC3000. In the present examination, AR induced by P. syringae DC3000 was also found to effectively improve wheat resistance to Puccinia triticina (Pt). However, with more complex genomes, genes associated with this SAR-like response in wheat and barley are largely unknown and no specific WRKYs has been reported to be involved in this biological process. In our subsequent analysis, barley transgenic line overexpressing wheat wNPR1 (wNPR1-OE) showed enhanced resistance to Magnaporthe oryzae isolate Guy11, whereas AR to Guy11 was suppressed in a barley transgenic line with knocked-down barley HvNPR1 (HvNPR1-Kd). We performed RNA-seq to reveal the genes that were differentially expressed among these transgenic lines and the wild-type barley plants during the AR. Several PR and BTH-induced (BCI) genes were designated as downstream genes of NPR1. The expression of few WRKYs was significantly associated with NPR1 expression during the AR events. The transient expression of three WRKY genes, including HvWRKY6, HvWRKY40, and HvWRKY70, in wheat leaves by Agrobacterium-mediated infiltration enhanced the resistance to Pt. In conclusion, a profile of genes associated with NPR1-mediated AR in barley was drafted and WRKYs discovered in the current study showed a substantial potential for improving wheat resistance to Pt.
Introduction
Systemic acquired resistance (SAR) is an inducible form of plant defense that confers broad-spectrum immunity to secondary infections beyond the initial infection site. In Arabidopsis, SAR is associated with accumulation of the plant hormone salicylic acid (SA) and transcriptional activation of pathogenesis-related (PR) genes (Zheng and Dong, 2013). The Arabidopsis NPR1 protein (Non-expresser of PR genes 1, also known as NIM1 and SAI1) is a master regulator required for SAR. Upon pathogen infection or treatment with SA or its functional analogs 2,6-dichloroisonicotinic acid (INA) and benzothiadiazole (BTH), NPR1 translocates from the cytoplasm into the nucleus, where it interacts with the TGA2 transcription factor to promote expression of multiple PR genes (Cao et al., 1994; Delaney et al., 1995; Ryals et al., 1997; Shah et al., 1997; Mou et al., 2003). Overexpression of Arabidopsis NPR1 (AtNPR1) in other plant species (e.g., rice and wheat) enhances their resistance against multiple pathogens (Chern et al., 2001; Makandar et al., 2006; Quilis et al., 2008; Gao et al., 2013; Xu et al., 2017). In Arabidopsis and rice, several WRKY transcription factors (TFs) have been suggested to play important roles in the NPR1-mediated SAR. A previous genomic approach has identified several WRKYs, including AtWRKY18, AtWRKY58, and AtWRKY70, as regulatory nodes in the transcriptional network of SAR in Arabidopsis (Wang et al., 2006). On the other hand, OsWRKY3 and OsWRKY71 in rice were reported as upstream genes of the rice NPR1 homolog (NH1) (Liu et al., 2005, 2007). Another WRKY transcription factor in rice, OsWRKY45, was established as an independent regulator in the SA/BTH signaling pathway (Shimono et al., 2007; Nakayama et al., 2013).
Our previous analysis showed that the wheat NPR1 homolog (wNPR1) interacts with four members of the basic-region leucine zipper (bZIP) transcription factor family (also known as TGA) (Cantu et al., 2013). The interactions between NPR1 and TGAs are critical for NPR1 functioning in Arabidopsis and rice (Chern et al., 2001; Després et al., 2003). However, SAR in wheat and barley somewhat differs from that described in model plants of Arabidopsis and rice (Wang et al., 2018). In early investigations, SA/INA/BTH treatment of wheat and barley induced a SAR-like response, BTH-induced resistance (BIR), to various pathogens, including Blumeria graminis and Puccinia triticina (Pt) (Görlach et al., 1996; Beßer et al., 2000; Hafez et al., 2014). Other studies established that most of the PR genes were not induced by such treatments (Vallélianbindschedler et al., 1998; Molina et al., 1999). Moreover, another group of BTH-inducible genes, such as the wheat chemical-induced (WCI) and barley chemical-induced (BCI) genes, may be responsible for the enhanced resistance to various pathogens during BIR.
In another study, Pseudomonas syringae pv. japonica (Psj) or Xanthomonas translucens pv. cerealis (Xtc) induced systemic immunity (SI) against secondary infection of Xtc in uninfected barley leaves (Dey et al., 2014). The findings of the same study also indicated that SI was not associated with barley HvNPR1, or local or systemic accumulation of SA, but with several WRKY and ERF transcription factors.
As a third form of SAR-like response in barley, acquired resistance (AR) to the secondary pathogen Magnaporthe oryzae (Mo) was induced in the area adjacent to the initial infection of P. syringae pv. tomato DC3000 in a PR genes-induced manner, but such resistance was not systemic (Colebrook et al., 2012). The levels of both free and conjugated SA were significantly upregulated in barley leaves infiltrated with P. syringae pv. syringae (Vallélianbindschedler et al., 1998). In our previous research, the induction of several barley PR genes, including HvPR1b, HvPR2, HvPR3_Chit2a, and HvPR5_TLP6, was significantly associated with the expression level of NPR1 in transgenic lines overexpressing wheat NPR1 (wNPR1-OE) or suppressing barley NPR1 (HvNPR1-Kd) during the P. syringae DC3000-triggered AR (Wang et al., 2016). However, genes associated with these biological processes are largely unknown and no specific WRKYs has been reported to be involved in this SAR-like response in barley.
In the present study, we established that AR induced by P. syringae DC3000 also improved wheat resistance to P. triticina (Pt). In addition, the AR responses of the barley transgenic lines wNPR1-OE and HvNPR1-Kd to the Mo isolate Guy11 were evaluated. Further, transcriptome analysis of these transgenic lines during AR response was carried out. The downstream genes of NPR1 were identified based on their expression profiles. The inductions of several transcription factors displayed significant association with the expression of NPR1 during AR. Three differentially expressed WRKY genes, identified in our transcriptome database, showed a considerable potential for improving wheat resistance to Pt.
Materials and Methods
Plants and Pathogens
A transgenic line of barley overexpressing wheat wNPR1 (wNPR1-OE) and a transgenic line with suppressed barley HvNPR1 (HvNPR1-Kd) under maize Ubiquitin promoter in the background of cultivar “Golden Promise” were derived from previous studies (Dey et al., 2014; Wang et al., 2016). The wild-type plants severed as control. The fully expanded third leaves from the experimental plants were used for P. syringae pv. tomato DC3000 infiltration and subsequent M. oryzae (Mo) inoculation. Briefly, P. syringae DC3000 was grown in KB medium with Rif antibiotics and was then diluted to OD600 = 0.5 in sterile water. Third leaves were inoculated with a 1-mL needless syringe by pressure infiltration of bacterial suspensions through the leaf abaxial surface. The borders of the infiltrated region were marked using a marker pen. Control seedlings were infiltrated in the same way with sterile water. After bacterial inoculation, seedlings were maintained at a constant temperature of 23°C to facilitate bacterial growth. Samples for RNA extraction were collected from regions adjacent to the infiltration from the transgenic lines and the wild-type plants 48 h post-inoculation (hpi) after a clear cell death phenotype triggered by P. syringae DC3000 infection was observed (Colebrook et al., 2012; Wang et al., 2016).
The same adjacent regions were also used for Mo inoculation to evaluate the degree of acquired resistance in the transgenic lines and the wild-type plants. Mo isolate Guy11 was grown on complete medium for 11 days at 25°C under a 16/8-h light-dark cycle. Ten microliters of the conidia suspension (5.0 × 105 spores per milliliter) containing 0.05% Tween-20 was dropped to the press-injured spots on the adjacent region and then wrapped with cellophane tape. The plants were kept in a mist chamber at 25°C in the dark for the first 24 hpi and then transferred to a growth chamber at 23°C and 80% humidity under a 16/8-h (light/dark) photoperiod. The inoculated leaves were photographed 5 days later. The size of the lesion caused by Mo on each leaf was measured. The assay for each treatment and phenotype combination consisted of at least six biological replicates. Data were transformed to restore normality and Dunnett’s test was performed using SAS software v9.4 (SAS Institute, Cary, NC, United States).
A similar approach was employed to induce AR in wheat using P. syringae DC3000 infiltration. Water infiltration served as negative control. Fully expanded secondary leaves from seedlings of the wheat susceptible line “Thatcher” were used in the experiments. Urediniospores of highly virulent Pt pathotype THTT were sprayed on the region adjacent to the P. syringae DC3000 infection area 2 days post-infiltration. Inoculated wheat plants were maintained in a moist chamber at 18°C for 16 h in the dark and were next transferred into a growth chamber with 16 h light at 23°C and 8 h darkness at 18°C. The phenotype of leaf rust was recorded at 10 dpi. The percentage of Pt sporulation area in the corresponding region for each leaf was determined using ASSESS (version 2.0) image analysis software for plant disease quantification from the American Phytopathology Society (Lamari, 2008; Zhang et al., 2017). The whole experiment was repeated twice and each repeat consisted of 11–14 biological replicates. The data were transformed to restore normality and general linearized model (GLM) ANOVA was conducted using SAS software version 9.4 (SAS Institute, Cary, NC, United States).
RNA Isolation and qRT-PCR
RNA samples for RNA-seq and qRT-PCR assays were isolated using a plant total RNA extraction kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The first-strand cDNA was synthesized using a reverse transcription kit (Clontech, Mountain View, CA, United States). Then, gene expression was quantified as described before (Wang et al., 2016), using the barley elongation factor 1-a (HvEF1a, GenBank accession number Z50789) and actin (HvActin, GenBank accession number AK362208) as internal references. The qRT-PCR primers, designed for the selected PR genes (HvPR1b, HvPR2, and HvPR3_Chit2a, derived from our previous study), BCI genes (HvBCI1, HvBCI3, and HvBCI7), and NPR1 gene, are listed in Supplementary Table S1. Further, the amplification efficiency for each pair of primers was calculated using five fourfold cDNA dilutions (1:1, 1:4, 1:16, 1:64, and 1:256). To ensure amplification specificity, dissociation curves for the temperature range from 60 to 94°C were generated for each reaction. The threshold values (Ct) generated from the Roche LightCycler 96 were used to quantify the relative gene expression using the Delta-Ct method as described earlier (Wang et al., 2016). Two independent transgenic lines for each of the wNPR1-OE and HvNPR1-Kd were utilized. Each experiment consisting of 4–11 biological replicates was considered as a block. Calculations of the mean and standard error were performed using Microsoft Excel (Microsoft, Redmond, WA, United States). The data were transformed to restore normality and GLM ANOVA was conducted using SAS software version 9.4 (SAS Institute, Cary, NC, United States).
RNA-seq
Library preparation and RNA-seq were performed as described in the Illumina TruSeq RNA Sample Preparation Version 2 Guide, the Illumina HiSeq 1000 System User Guide (Illumina), and the KAPA Library Quantification Kit-Illumina/ABI Prism User Guide (Kapa Biosystems) by Novogene Co., Ltd. The sequencing run was performed on a HiSeq 1000 instrument. Then, the sequence reads were mapped on the Ensembl Genomes Hordeum vulgare genome sequence (International Barley Genome Sequencing Consortium, 2012) using TopHat 2.0.8 (Trapnell et al., 2009) with default parameter settings and an expected mean insert size of 150 bp. The assembled contigs that were not aligned with the reference genome were annotated as “Novel” transcripts. Further, HTSeq was used to assemble the mapped RNA-seq reads into transcripts to quantify their relative abundance (Trapnell et al., 2010). Differentially expressed genes were identified using the default settings of DESeq2 (Love et al., 2014) and filtered for an FDR-adjusted P < 0.05. Statistically significant over-representation of GO categories within differentially expressed genes was determined using the GOseq package (Young et al., 2010). Statistically significant GO terms tested by conditional hypergeometric tests (P < 0.05) were considered enriched. Next, heatmaps were generated by MeV software using FPKM values from the RNA-seq database. Hierarchical clustering analysis was performed by the MeV software, based on which the genes with similar expression patterns were clustered. A summary of GO annotation categories was generated using the GOLevel2 Counter in the TBtools software.
Transient Expression Assay
RNA isolated from barley leaves was utilized for cDNA synthesis using the reverse transcription kit (Clontech, Mountain View, CA, United States). Subsequently, the cDNA sequences of 10 selected WRKY genes were PCR-amplified using the primers listed in Supplementary Table S1. Initially, the PCR products were cloned into a pGEM-T easy vector (Promega, Madison, WI, United States) and then into a wheat transgenic vector pLGY02, which contained the maize (Zea mays) ubiquitin 1 promoter and T-DNA insertion site. The recombinant constructs were transformed into the Agrobacterium strain AGL1, and the wheat leaves were subjected to transient gene expression assays as previously described (Lu et al., 2016). Fresh Agrobacterium was grown overnight in yeast extract broth (YEB) medium supplemented with Rifampicin and Kanamycin. The bacterial pellets obtained after centrifugation were resuspended in an infiltration buffer containing 10 mM MES, 10 mM MgCl2, and 400 μM acetosyringone to an optical density of OD600 = 2.0. For wheat infiltration, the fully expanded secondary leaf of wheat seedlings was infiltrated using a 1-mL syringe without a needle. The border of the infiltration area was marked with a mark pen. Urediniospores of highly virulent Pt pathotype THTT were spray-inoculated 4 days post-infiltration. The inoculated leaves were photographed at 10 days post-inoculation. The percentage of Pt sporulation area in the infiltration region for each leaf was calculated using ASSESS software v2.0 (American Phytopathology Society) (Lamari, 2008; Zhang et al., 2017). The assay for each gene was repeated at least twice, and each repeat consisted of 5–18 biological replicates. Data were transformed to restore normality and Dunnett’s test was carried out using SAS software v9.4 (SAS Institute, Cary, NC, United States). Raw data for all these conducted experiments were archived in Supplementary File S1.
Results
AR Triggered by P. syringae DC3000 in Wheat Reduces the Severity of Pt
AR triggered by P. syringae DC3000 in barley was considered as a SAR-like response providing broad-spectrum protection against subsequent pathogen challenge (Colebrook et al., 2012). To elucidate whether such AR can be utilized to improve the resistance of other Triticeae crops, we examined its effect in the defense reaction of wheat to leaf rust, which is a severe fungal disease in wheat. A highly virulent Pt pathotype THTT was used to inoculate the region adjacent to P. syringae DC3000 or water infiltration area in the wheat leaves of the susceptible line “Thatcher.” Cell death triggered by P. syringae DC3000 was observed 2 days post-infiltration. The susceptible phenotypes of leaf rust were identified in both P. syringae DC3000 and water mock treatments 10 days post-inoculation (Figure 1). The percentage of Pt sporulation area for each leaf was calculated using ASSESS software. Significantly more (P < 0.0001) promoted resistance to Pt was observed in the region adjacent to the P. syringae DC3000 infection area than in the mock control (Figure 1).

FIGURE 1. AR triggered by P. syringae DC3000 in wheat reduces the severity of Pt. A highly virulent Pt pathotype THTT was inoculated in the region adjacent to either P. syringae DC3000 or water infiltration area in wheat leaves of the susceptible line “Thatcher.” The susceptible phenotypes of leaf rust were observed in the region adjacent to both P. syringae DC3000 and water mock infiltration area at 10 dpi. The numbers below the images of the leaves represent the average percentages of the Pt sporulation areas in the corresponding leaf regions (n = 25). Compared with the mock control, a significantly (∗∗∗P < 0.0001) more enhanced resistance to Pt was observed in the region adjacent to the P. syringae DC3000 infection area. The whole experiment was repeated twice, and each repeat, consisting of 11–14 biological replicates, was considered as a block. The data obtained were transformed to restore normality, and general linearized model (GLM) ANOVA was conducted using SAS software version 9.4.
Barley AR to Mo Isolate Guy11 Is Mediated by NPR1
The Mo isolate Guy11 was used to determine the extent of AR triggered by P. syringae DC3000 in a barley transgenic line overexpressing wheat wNPR1 (wNPR1-OE), a transgenic line with knocked-down barley HvNPR1 (HvNPR1-Kd), and wild-type plants (Figure 2). The third leaf of barley plants was infected by infiltration with P. syringae DC3000 or treated with sterile water as a mock control. Mo isolate Guy11 was inoculated in the region adjacent to P. syringae DC3000 infiltration area when a cell death was observed 2 days post-infiltration (dpi). Strong AR against Mo isolate Guy11 was established in the region adjacent to P. syringae DC3000 infection area in the wild-type plants (Figure 2A). The size of the lesions caused by the Mo isolate Guy11 in the region adjacent to P. syringae DC3000 infection area was significantly (P < 0.05) lower than that in the mock control (Figure 2B). The wNPR1-OE transgenic line had more pronounced resistance to Mo infection than the wild-type plants, even in the mock control (Figure 2). The AR triggered by P. syringae DC3000 in the HvNPR1-Kd transgenic line, was suppressed but not fully eliminated (Figure 2), possibly because NPR1 was not completely removed from the HvNPR1-Kd line. These results indicate that the AR to the Mo isolate Guy11, triggered by P. syringae DC3000, is mediated by NPR1.
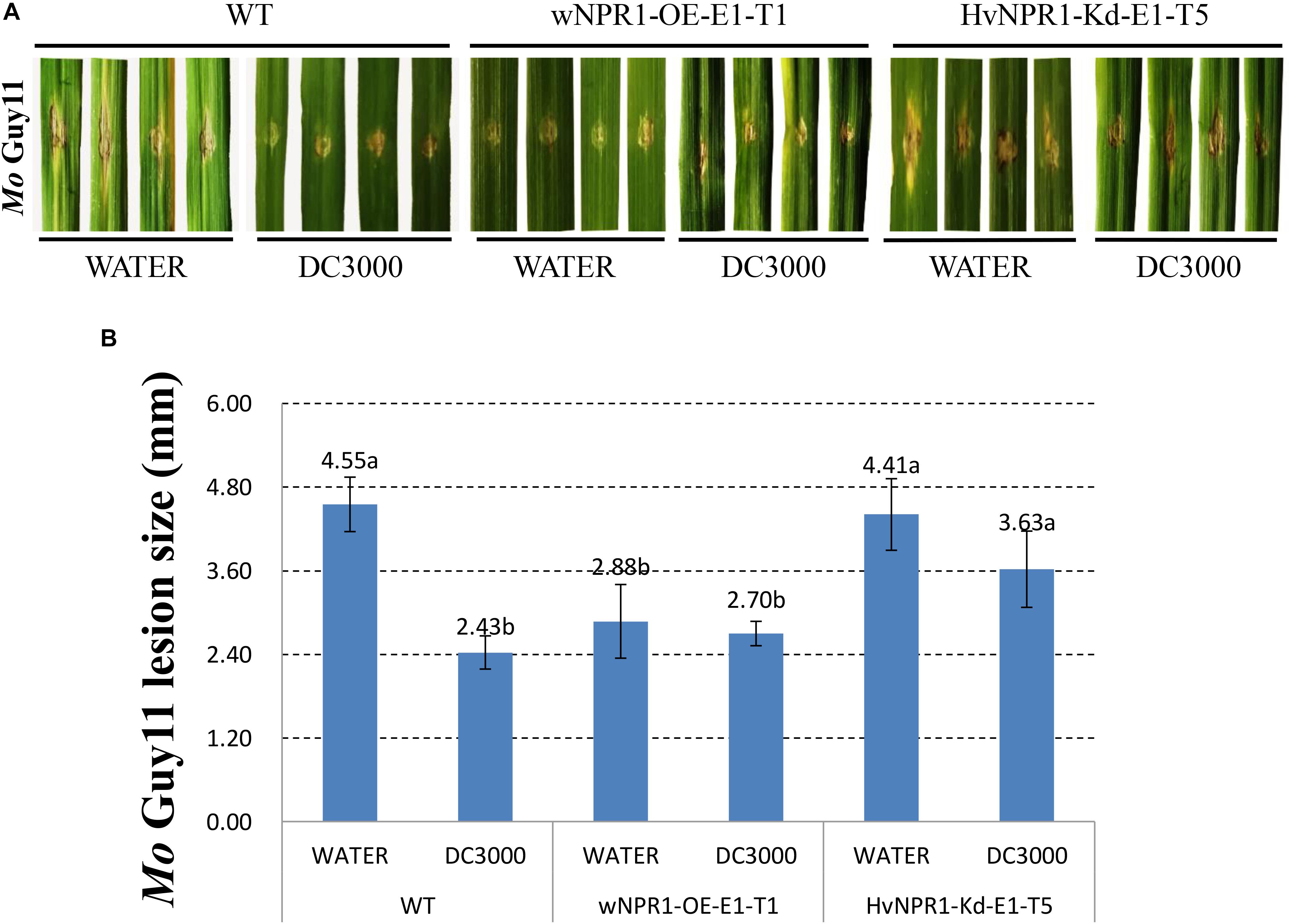
FIGURE 2. Barley AR to Mo isolate Guy11 is mediated by NPR1. (A) Symptoms of M. oryzae isolate Guy11 after inoculation in the region adjacent to P. syringae DC3000 infiltration area in the wNPR1-OE and HvNPR1-Kd barley transgenic lines and the wild-type plants. Distilled water was mock-infiltrated to serve as a control. (B) Quantification of M. oryzae lesion size. The assay for each treatment and phenotype combination consisted of at least six biological replicates. The data were transformed to restore normality, and Dunnett’s test (P < 0.05) was conducted using SAS software v9.4.
RNA-Seq Analysis Was Applied on wNPR1-OE and HvNPR1-Kd Barley Transgenic Lines During AR Triggered by P. syringae DC3000
To explore the gene regulation network during the NPR1-mediated AR in barley, we performed RNA-seq analysis on samples collected from the region adjacent to either P. syringae DC3000 or water infiltration area in the wNPR1-OE and HvNPR1-Kd barley transgenic lines and the wild-type plants. A number of 4–7 biological replicates for each treatment and genotype combination were sent for 6-Gb RNA sequencing (Supplementary Table S2). At least 46 million reads were sequenced (150-bp pair-end) from each sample and mapped on the Ensembl Genomes H. vulgare genome sequence with a total of 31,794 genes, including 7,583 “Novel” transcripts. A clear correlation between the gene expression levels of the biological replicates was observed (R2 > 0.92, Supplementary Figure S1). The abundance of accumulated transcripts was estimated using the fragments per kilobase of transcript per million mapped reads (FPKM) value. All raw data were uploaded to NCBI in BioProject PRJNA431836.
Expression Profiles of PR and BCI Genes in the NPR1-Mediated AR in Barley
Since many of the PR genes were reported as downstream genes of NPR1 during the P. syringae DC3000-triggered AR in our previous study (Wang et al., 2016), in the present investigation, we initially checked the expression levels of all the PR gene families in our RNA-seq database (Figure 3). We noticed that the transcript levels of HvPR1, HvPR2, HvPR3_Chit2a, HvPR5 (TLP6, TLP7, and TLP8), HvPR9, and HvPR13, were significantly associated with the expression of NPR1 as either significantly (P < 0.05) higher induced in the wNPR1-OE transgenic line or lower induced in the HvNPR1-Kd transgenic line (Figure 3). In contrast, the expression levels of PR5 (TLP1), PR15, PR16, PR17a, and PR17b showed induction by P. syringae DC3000 but in a NPR1-independent manner. Since a group of BCI genes were previously reported to be responsible for the BTH-induced resistance in barley (Beßer et al., 2000), we examined the expression patterns of all barley BCI genes in our database and found that HvBCI2 (the same gene as HvPR13) and HvBCI7 were regulated by NPR1 during the P. syringae DC3000-triggered AR (Figure 3). qRT-PCR assay was carried out to validate the gene expression data obtained from the RNA-seq database. Two independent transgenic lines for each of the wNPR1-OE and HvNPR1-Kd were used and the barley elongation factor HvEF1a (GenBank accession number Z50789) were employed as a reference gene. We found that the expression level of NPR1 in the wNPR1-OE and HvNPR1-Kd transgenic lines was significantly (P < 0.01) higher and lower, respectively, than that in the wild-type plants (Supplementary Figure S2). In addition, the expression levels of six selected genes, including HvPR1, HvPR2, HvPR3_Chit2a, HvBCI1, HvBCI3, and HvBCI7, were measured by qRT-PCR. In the wild-type plants, significant (P < 0.01) inductions of HvPR1, HvPR2, HvPR3_Chit2a, and HvBCI1, were observed upon the P. syringae DC3000 treatment (Figure 4). Furthermore, the expression levels of HvPR1, HvPR2, HvPR3_Chit2a, and HvBCI7, were significantly (P < 0.05) upregulated in the wNPR1-OE transgenic lines by the P. syringae DC3000 treatment (Figure 4), which indicated their roles as downstream components of NPR1 during the AR response. On the other hand, the inductions of such downstream genes during the P. syringae DC3000-triggered AR were suppressed in the HvNPR1-Kd transgenic lines (Figure 5). Additionally, we used another reference gene, HvActin (GenBank accession number AK362208), to validate the results of our qRT-PCR assay for HvPR1 and found similar expression patterns (Supplementary Figure S3).
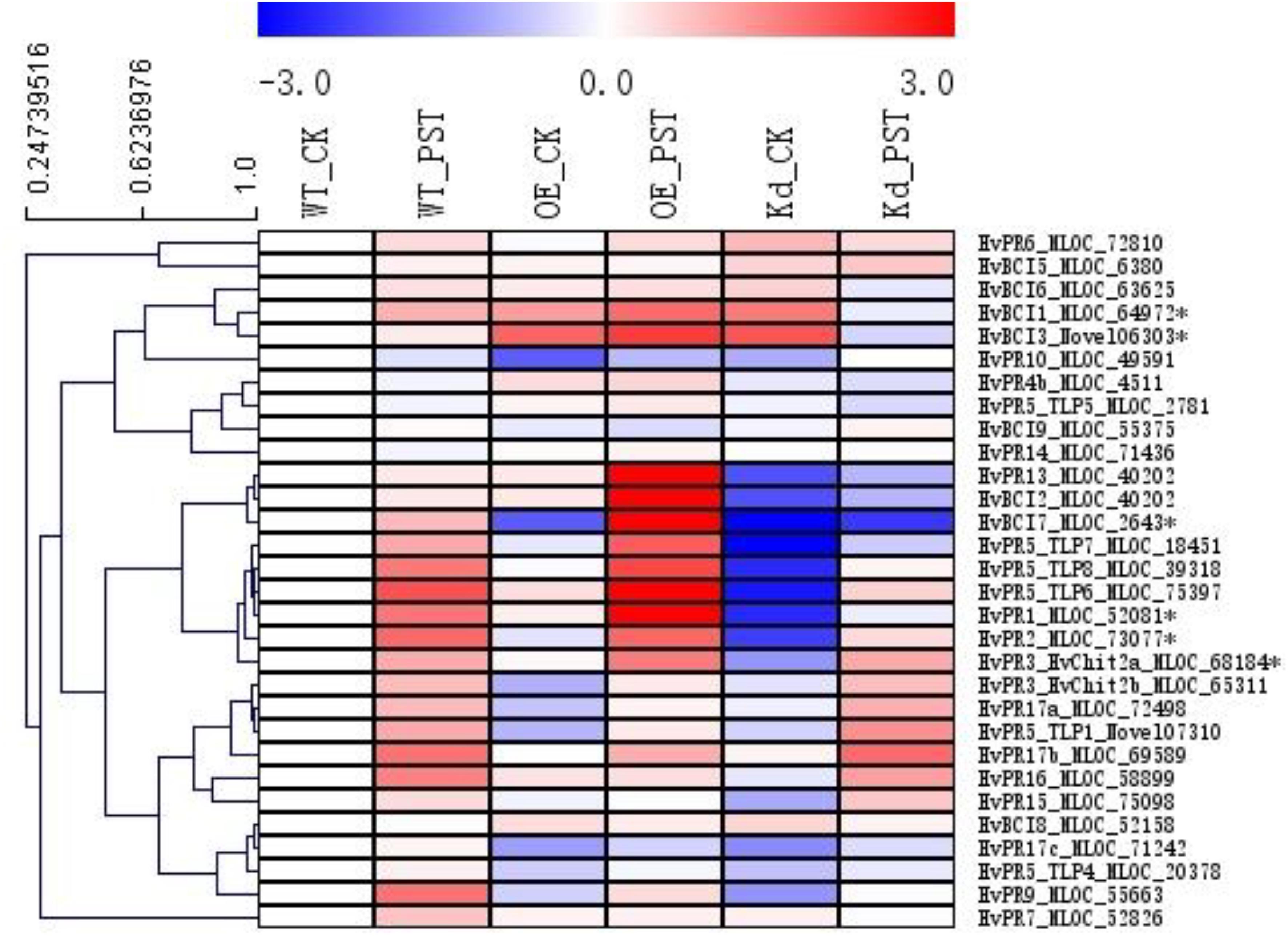
FIGURE 3. Expression profiles of PR and BCI genes during the NPR1-mediated AR. A heatmap was generated by MeV software using the FPKM values of PR and BCI genes from the RNA-seq database. Genes with similar expression patterns were clustered using the “Hierarchical Clustering” function of the MeV software. The expression levels of six genes (∗asterisk-labeled) were further validated by qRT-PCR assay.
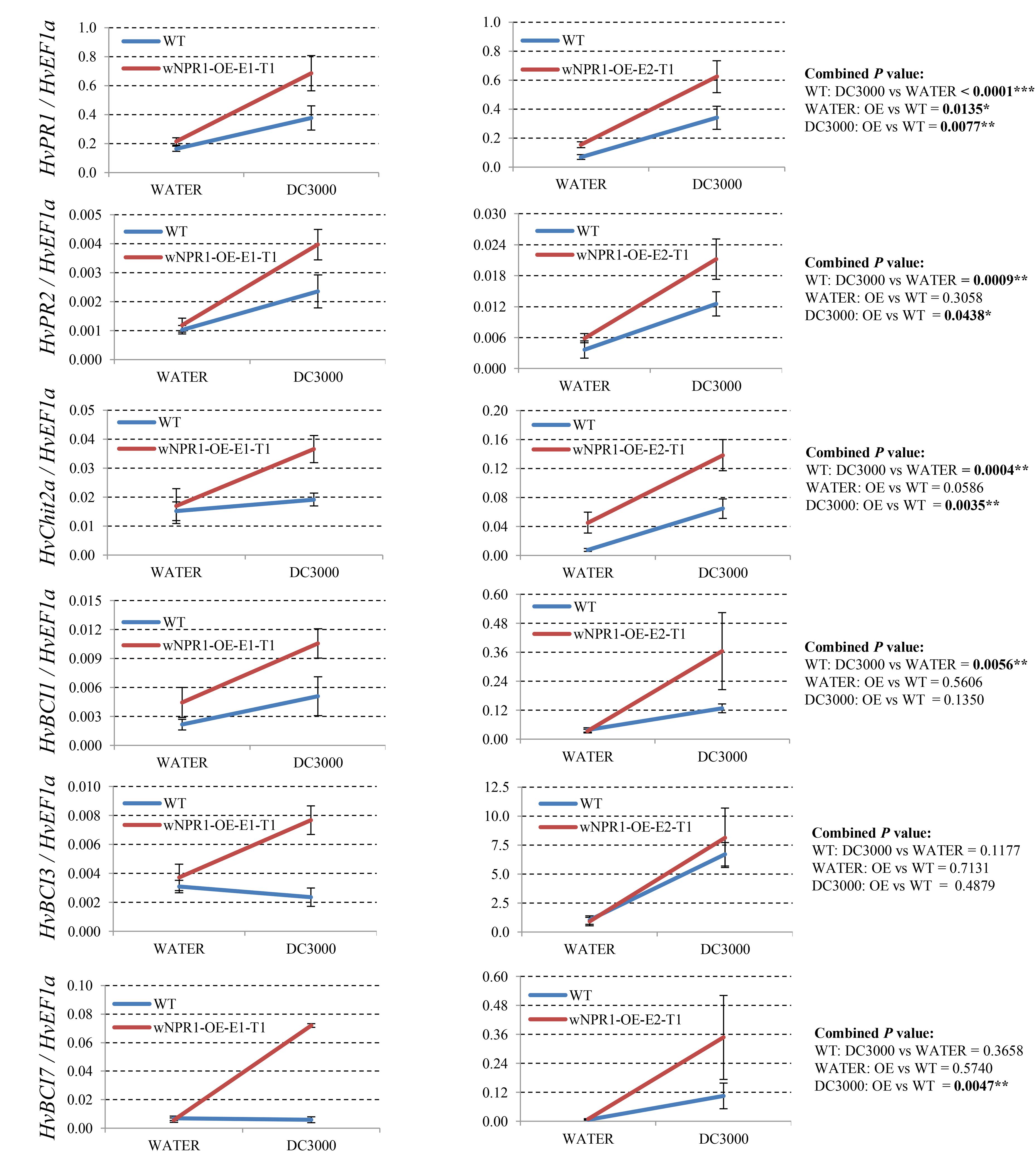
FIGURE 4. Transcript levels of selected PR and BCI genes in the wNPR1-OE transgenic lines. Third leaves of wNPR1-OE barley transgenic lines and wild-type plants were infiltrated with water (control) or P. syringae pv. tomato DC3000. Samples for qRT-PCR assays were collected from the leaf region adjacent to the infection 48 h after inoculation, after a cell death phenotype observed. Using the 2−ΔCT method, the transcript levels were expressed relative to those of the endogenous control HvEF1a. Two independent transgenic lines for the wNPR1-OE were used. Each experiment, consisting of 4–11 biological replicates, was considered as a block. Calculations of the mean and standard error were performed using Microsoft Excel software. Data were transformed to restore normality and general linearized model (GLM) ANOVA (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.0001) was conducted using SAS software version 9.4.
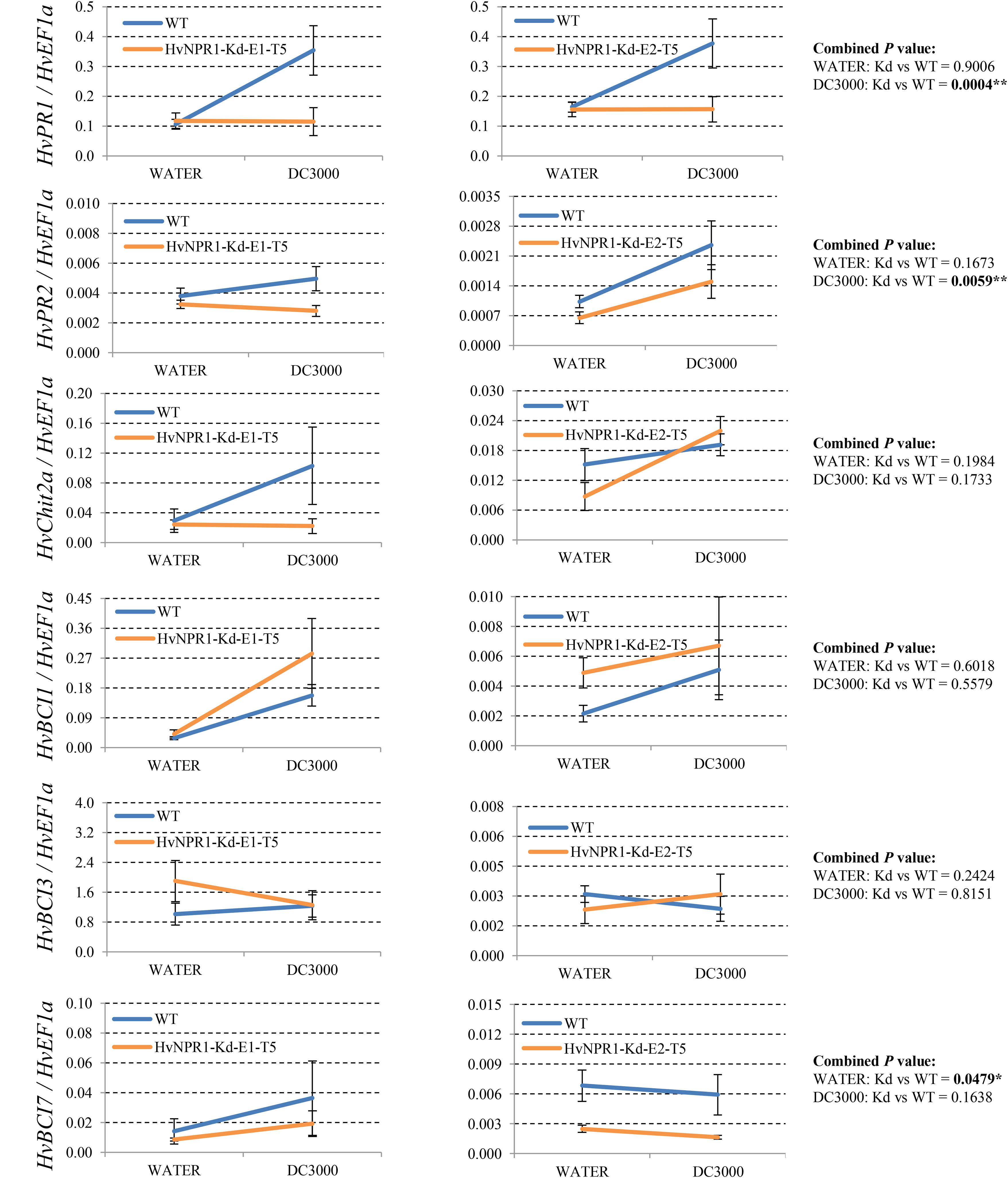
FIGURE 5. Transcript levels of selected PR and BCI genes in the HvNPR1-Kd transgenic lines. Third leaves of HvNPR1-Kd barley transgenic lines and wild-type plants were infiltrated with water (control) or P. syringae pv. tomato DC3000. Then, samples for qRT-PCR assays were collected from the leaf region adjacent to the infection 48 h after inoculation, after a cell death phenotype observed. The transcript levels are expressed relative to those of the endogenous control HvEF1a using the 2−ΔCT method. Two independent transgenic lines for the HvNPR1-Kd were used, and, each experiment, consisting of 4–11 biological replicates, was considered a block. Calculations of the mean and standard error were performed using Microsoft Excel software. Data were transformed to restore normality and general linearized model (GLM) ANOVA (∗P < 0.05, ∗∗P < 0.01) was conducted using SAS software version 9.4.
Differentially Expressed Genes (DEGs) Associated With NPR1 Expression During AR
DEGs in different comparisons, including “WT_PST vs. WT_CK,” “OE_PST vs. OE_CK,” “OE_PST vs. WT_PST,” “Kd_PST vs. Kd_CK,” and “Kd_PST vs. WT_PST,” were identified by DESeq2 (q-value < 0.05 and | log2foldchange| > 1, with gene annotation). Three types of DEGs were manually classified based on their possible roles in the NPR1-mediated AR (Supplementary Figure S4). Type I DEGs were highly upregulated genes in the wNPR1-OE transgenic line after P. syringae DC3000 induction. A total of 24 Type I DEGs were designated from significantly upregulated genes based on the comparisons “OE_PST vs. OE_CK” and “OE_PST vs. WT_PST” (Supplementary Figure S4 and Table 1). Several PR and BCI genes were annotated as Type I DEGs, including probable glucan 1,3-beta-glucosidase A (HvPR2), peroxidase A2-like (HvPR9), and thionin BTH7-like (HvBCI7). GO annotation for the Type I DEGs showed that the majority of these possible downstream genes of NPR1 during AR were annotated with “binding” and “catalytic activity” in the molecular function category, and with “metabolic process” and “response to stimulus” in the biological process category (Supplementary Figure S5A).
Type II DEGs were obtained and categorized into groups from significantly upregulated genes based on the comparisons “Kd_PST vs. Kd_CK” and “Kd_PST vs. WT_PST” (Supplementary Figure S4 and Supplementary Table S3). A total of 64 genes were classified into this group, including several genes encoding transcription factors, such as transcription factor JUNGBRUNNEN 1-like, probable WRKY transcription factor 48, and probable WRKY transcription factor 50. Compared with GO annotations for the Type I DEGs, more genes in this group were annotated with “cellular process” but not “response to stimulus” (Supplementary Figure S5B). A third group of Type III DEGs, including only nine genes, were shared DEGs from Type I and Type II groups (Supplementary Figures S4, S5C and Supplementary Table S4).
The Transient Expression of HvWRKY6, HvWRKY40, and HvWRKY70 Enhanced Wheat Resistance to Pt
A total of 46 WRKY genes were annotated in our RNA-seq database, and, based on their expression patterns, 10 of them were found to be involved in the NPR1-mediated AR (Figure 6). Since the naming system for the WRKY genes in the genera of Triticeae was confusing, a polygenetic tree was generated using sequences of all these 10 WRKY proteins and their homologs from Hordeum vulgare (Hv), Aegilops tauschii (At), Brachypodium distachyon (Bd), Triticum aestivum (Ta), Triticum urartu (Tu), and Sorghum bicolor (Sb) from the GenBank nr database (Supplementary Figure S6). All WRKYs were temporally designated according to their closest homologs in relative plant species, such as HvWRKY4, HvWRKY6, HvWRKY17, HvWRKY19, HvWRKY20, HvWRKY31, HvWRKY40, HvWRKY64, HvWRKY70, and HvWRKY76.
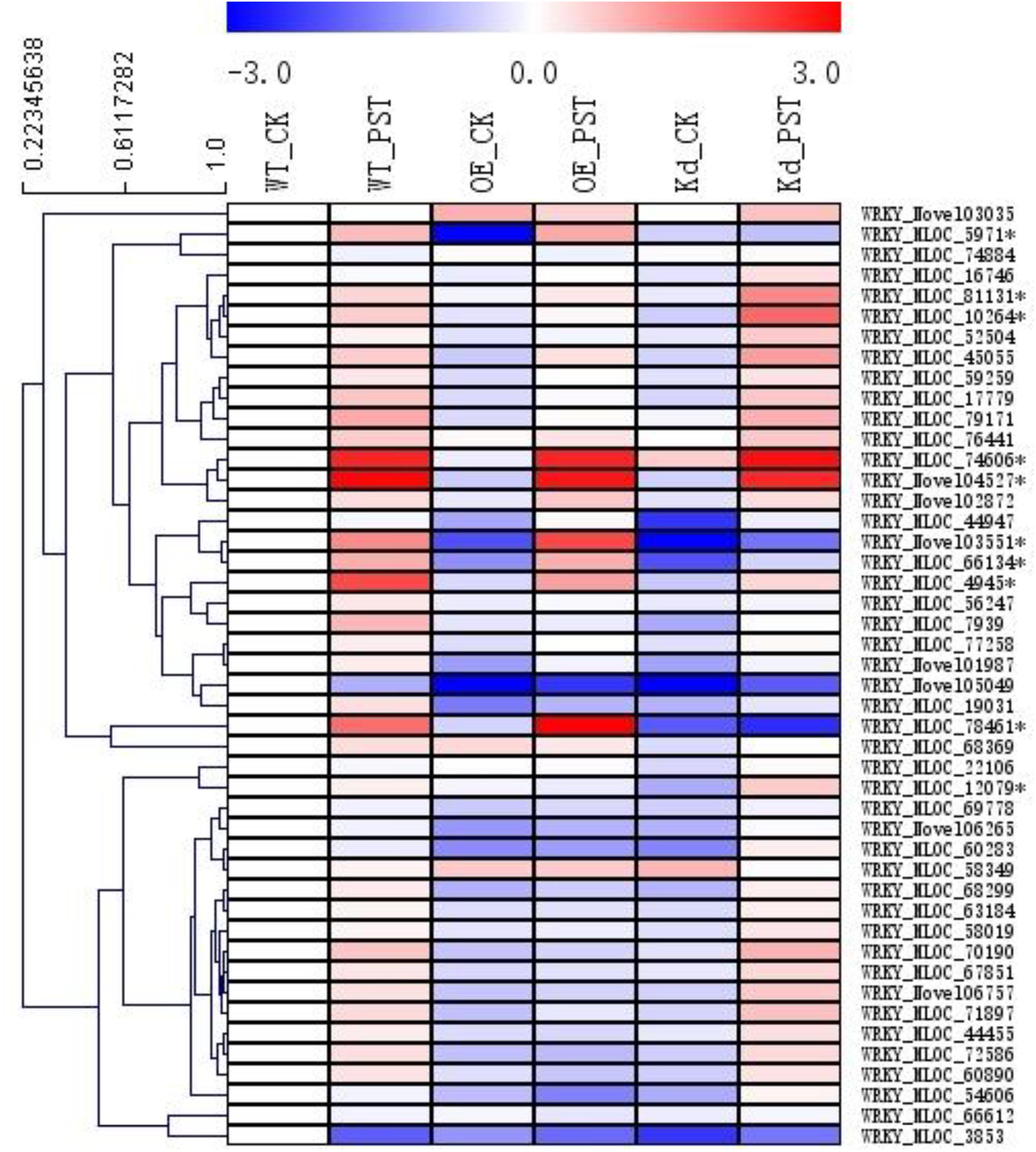
FIGURE 6. Expression patterns of WRKY transcription factor gene families during the NPR1-mediated AR. A heatmap was generated by MeV software using the FPKM values of the genes encoding WRKY transcription factors from the RNA-seq database. Ten differentially expressed WRKYs were selected (∗asterisk-labeled) for further functional characterization. Genes with similar expression patterns were clustered using the “Hierarchical Clustering” function of the MeV software.
To test the potential of the differentially expressed WRKYs for improving wheat resistance to Pt, the open reading frame (ORF) of each WRKY gene was cloned and incorporated into a pLGY02 vector (Ubiquitin promoter, with T-DNA insertion site). The recombinant vector was transformed into the Agrobacterium strain AGL1. The transformed Agrobacterium was infiltrated into the secondary leaves of wheat seedlings of the susceptible line “Thatcher,” and the infiltration area was marked with a mark pen. Urediniospores of the highly virulent Pt pathotype THTT were spray-inoculated 4 days post-infiltration. The phenotype of leaf rust was recorded 10 days post-inoculation (Figure 7). The percentage of Pt sporulation area in the infiltration region of each leaf was calculated using ASSESS software. Wheat with transient expression of TaPR1b (GenBank accession number HQ541962) was utilized as a positive control, whereas the untransformed Agrobacterium strain AGL1 was employed as a negative control. Although susceptible phenotypes were observed in all treatments, Pt sporulation in the leaves of the wheat with transient expression of TaPR1b and three WRKYs, including HvWRKY6, HvWRKY40, and HvWRKY70, were significantly (P < 0.001) reduced or delayed (Figure 7 and Supplementary Table S5). The transient expression of the other seven WRKY genes, including HvWRKY4, HvWRKY17, HvWRKY19, HvWRKY20, HvWRKY31, HvWRKY64, and HvWRKY76, in wheat leaves exerted no positive effect on wheat resistance to Pt (Supplementary Table S5).

FIGURE 7. The transient expression of HvWRKY6, HvWRKY40, and HvWRKY70, enhanced the resistance of wheat to Pt. A total of 10 differentially expressed WRKY genes were transiently expressed in wheat leaves using Agrobacterium. Urediniospores of the highly virulent Pt pathotype THTT were spray-inoculated 4 days post-infiltration. Wheat transiently expressing TaPR1b was used as a positive control, whereas the untransformed Agrobacterium strain AGL1 was employed as a mock control. The phenotypes of leaf rust were recorded 10 days post-inoculation. The susceptible phenotypes were observed in all treatments. The numbers below the images of the leaves represent the average percentages of the Pt sporulation areas in the infiltration regions in each of the treatments. Asterisk indicates the significance of the differences between the treatment and mock established by using Dunnett’s test (∗∗P < 0.001, ∗∗∗P < 0.0001). The Pt sporulation in the wheat leaves transiently expressing TaPR1b and three WRKYs, including HvWRKY6, HvWRKY40, and HvWRKY70, was significantly reduced or delayed. Assay for each gene was performed at least twice and each repeat consisted of 5–18 biological replicates (Supplementary Table S5).
Discussion
NPR1 has been reported as the master regulator of SAR in model plants of Arabidopsis and rice. Recent studies on NPR1 homologs in wheat and barley have provided the initial clue to understand the mechanism of SAR in these two plant species (Cantu et al., 2013; Dey et al., 2014; Wang et al., 2016). Compared with SAR in Arabidopsis, three different SAR-like responses can be induced by either pathogens or SA/INA/BTH treatment in wheat and barley (Wang et al., 2018). In the current study, the observed beneficial effect of acquired resistance, which led to the reduction of Pt infection in wheat (Figure 1), suggested a potential use of NPR1-mediated AR in improving the resistance of Triticeae crops. The diminished AR to M. oryzae observed in the HvNPR1-Kd transgenic line indicated a key role of NPR1 during the AR triggered by P. syringae DC3000 infection (Figure 2). Finally, the resistance of the wNPR1-OE transgenic line to Mo was enhanced (Figure 2), providing valuable evidence that NPR1 can be utilized in improving barley resistance to Mo, possibly also to the recently emerged wheat blast disease (Inoue et al., 2017).
So far, a total number of 18 PR gene families have been designated from plant species, many of which showed involvement in wheat and barley resistance to various pathogens (Loon et al., 2006; Wang et al., 2018). Several barley PR genes, including HvPR1, HvPR2, HvPR3_Chit2a, HvPR4b, and HvPR5_TLP6, were validated as downstream genes of NPR1 during the P. syringae DC3000-triggered AR (Wang et al., 2016). Another group of BCI genes seems to be responsible for the enhanced resistance in barley induced by BTH treatment (Beßer et al., 2000). In the present study, the expression profiles of all the PR and BCI genes were generated using FPKM values from the RNA-seq assay. The NPR1-regulated genes, including HvPR1, HvPR2, HvPR3_Chit2a, HvPR5_TLP6/7/8, HvPR13/BCI2, and HvBCI7, during the P. syringae DC3000-triggered AR, were established (Figures 3, 8). Further studies on HvPR13/BCI2 and HvBCI7 may provide initial evidence for understanding the relationship between AR and BIR. Interestingly, the expression levels of several PR genes, including HvPR3_Chit2b, HvPR5_TLP1, HvPR15, HvPR16, HvPR17a, and HvPR17b, were induced in a NPR1-independent manner, which indicated that other unknown regulators were functioning during the AR triggered by P. syringae DC3000.
In this investigation, we classified three DEG groups based on their expression patterns. Several PR and BCI genes were categorized into Type I and Type III DEGs (Table 1 and Supplementary Table S4). The possible downstream genes of NPR1 during the P. syringae DC3000-triggered AR were mainly identified as Type I DEGs. Interestingly, we also discovered a large group of Type II DEGs (Supplementary Table S3) which were highly induced only in the HvNPR1-Kd transgenic line after P. syringae DC3000 infection. We speculated that some of them were negative regulators or upstream genes of NPR1 during the AR. Furthermore, the regulator network of NPR1 in barley during the P. syringae DC3000-triggered AR was initially established in our investigation (Figure 8).
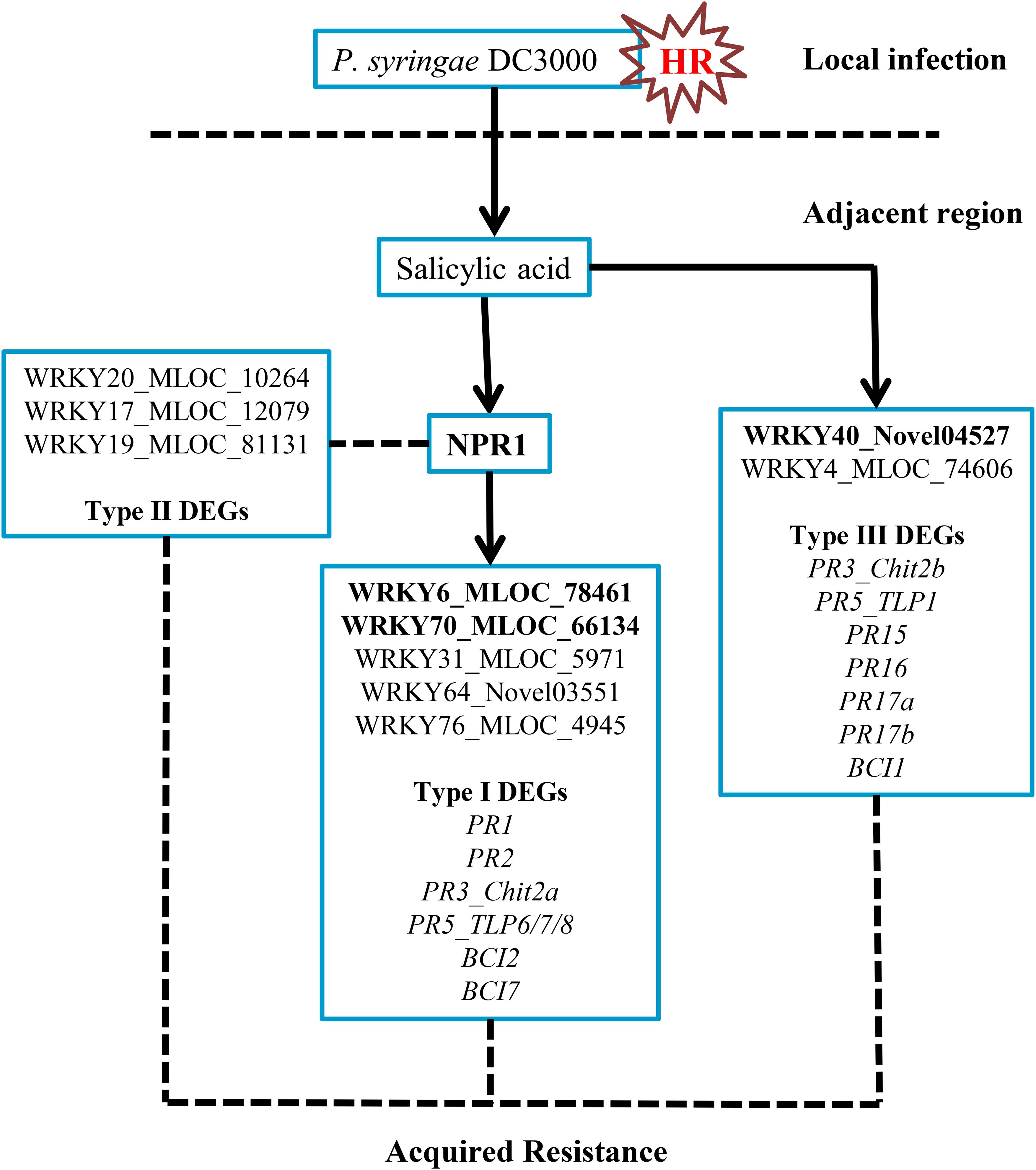
FIGURE 8. Possible regulatory network of the NPR1-mediated AR in barley. The transient expression of three barley WRKY genes, including HvWRKY6, HvWRKY40, and HvWRKY70 (bold-labeled), in wheat leaves by Agrobacterium-mediated infiltration enhanced the resistance to Pt.
In addition, we checked the involvement of barley WRKY transcription factors in the NPR1-mediated AR. In model plants of Arabidopsis and rice, several WRKYs, including AtWRKY18, AtWRKY58, AtWRKY70, OsWRKY03, OsWRKY71, and OsWRKY45, have been suggested to play important roles in the SAR (Liu et al., 2005, 2007; Wang et al., 2006; Shimono et al., 2007; Nakayama et al., 2013). A total of 171 TaWRKYs were identified in wheat, whereas 45 HvWRKYs were detected in barley (Christer et al., 2008; Pan et al., 2017). It was rather difficult to determine the key WRKY genes during SAR in these two crop species. By checking the expression patterns of all the 46 WRKYs identified in our RNA-seq assay (Figure 6), a total of 10 HvWRKY genes with relatively higher induction in either the wNPR1-OE or HvNPR1-Kd transgenic line were selected for further functional validation.
Three differentially expressed HvWRKYs, including HvWRKY6, HvWRKY40, and HvWRKY70, exerted positive effects on wheat resistance to Pt (Figure 7). HvWRKY6_MLOC_78461 was a barley homolog of AtWRKY6_XP_020181741 (Supplementary Figure S6), which had been reported to be associated with both senescence- and defense-related processes (Robatzek and Somssich, 2001). HvWRKY40_NOVEL04527 was clustered with AtWRKY40_EMT21551 in our polygenetic analysis (Supplementary Figure S6). In an earlier study, AtWRKY40 was found to be a repressor of antimycin A-induced mitochondrial retrograde expression and high-light-induced signaling (Aken et al., 2013). In addition, our data evidenced that HvWRKY70_MLOC_66134 was clustered with Arabidopsis AtWRKY70_XP_020165252 (Supplementary Figure S6), which exerted dual roles as negative regulators of SA biosynthesis and positive regulators of SA-mediated gene expression and resistance in Arabidopsis (Wang et al., 2006). We speculated that these three HvWRKYs were key regulators during the NPR1-mediated AR and may represent valuable transgenic resources for resistance improvement of wheat plants.
Conclusion
In conclusion, NPR1 acts as a key regulator during the P. syringae DC3000-triggered AR in barley (Figure 8). In the present study, we have identified genes associated with the NPR1-mediated AR. Several PR and BCI genes were confirmed to be the downstream genes of NPR1. The expression levels of several WRKY transcription factors were significantly associated with NPR1 expression, which might be key components in the NPR1-mediated AR. Furthermore, it is noteworthy that three differentially expressed WRKYs identified in the current study showed potential for resistance improvement of wheat plants.
Author Contributions
XW conceived the original screening and research plans. XW and DL supervised the experiments. JG and WB performed most of the experiments. HL, JW, and XY provided technical assistance to JG. XW designed the experiments and analyzed the data. XW conceived the project and wrote the article with contributions of all the authors. DL supervised and complemented the writing.
Funding
This work was supported by the National Natural Science Foundation of China (31701776 and 31301649), the Provincial Natural Science Foundation of Hebei for Excellent Young Scholar (C2018204091), the Young Talents Project of Hebei Education Department (BJ2016028), and Agricultural Talents Project of Chinese Academy of Agricultural Sciences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Jorge Dubcovsky (University of California, Davis, United States) for advising on the manuscript, Dr. Jinrong Xu (Northwest Agriculture and Forestry University, China) for sharing Mo isolate Guy11, and Dr. C. A. Volt (Helmholtz Zentrum Muenchen, Germany) for distributing the RNAi transgenic barley plants with reduced expression of HvNPR1.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01486/full#supplementary-material
References
Aken, O. V., Zhang, B., Law, S., Narsai, R., and Whelan, J. (2013). AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol. 162, 254–271. doi: 10.1104/pp.113.215996
Beßer, K., Jarosch, B., Langen, G., and Kogel, K.-H. (2000). Expression analysis of genes induced in barley after chemical activation reveals distinct disease resistance pathways. Mol. Plant Pathol. 1, 277–286. doi: 10.1046/j.1364-3703.2000.00031.x
Cantu, D., Yang, B., Ruan, R., Li, K., Menzo, V., Fu, D., et al. (2013). Comparative analysis of protein-protein interactions in the defense response of rice and wheat. BMC Genomics 14:166. doi: 10.1186/1471-2164-14-166
Cao, H., Bowling, S. A., Gordon, A. S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. doi: 10.1105/tpc.6.11.1583
Chern, M. S., Fitzgerald, H. A., Yadav, R. C., Canlas, P. E., Dong, X., and Ronald, P. C. (2001). Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. Cell Mol. Biol. 27, 101–113. doi: 10.1046/j.1365-313x.2001.01070.x
Christer, J., Mangelsen, E., Kilian, J., Berendzen, K. W., Kolukisaoglu, U. H., and Harter, K. (2008). Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9:194. doi: 10.1186/1471-2164-9-194
Colebrook, E. H., Creissen, G., Mcgrann, G. R., Dreos, R., Lamb, C., and Boyd, L. A. (2012). Broad-spectrum acquired resistance in barley induced by the Pseudomonas pathosystem shares transcriptional components with Arabidopsis systemic acquired resistance. Mol. Plant Microbe Interact. 25, 658–667. doi: 10.1094/MPMI-09-11-0246
Delaney, T. P., Friedrich, L., and Ryals, J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92, 6602–6606. doi: 10.1073/pnas.92.14.6602
Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., et al. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15, 2181–2191. doi: 10.1105/tpc.012849
Dey, S., Wenig, M., Langen, G., Sharma, S., Kugler, K. G., Knappe, C., et al. (2014). Bacteria-triggered systemic immunity in barley is associated with WRKY and ETHYLENE RESPONSIVE FACTORs but not with salicylic acid. Plant Physiol. 166, 2133–2151. doi: 10.1104/pp.114.249276
Gao, C., Kou, X., Li, H., Zhang, J., Saad, A., and Liao, Y. (2013). Inverse effects of Arabidopsis NPR1 gene on fusarium seedling blight and fusarium head blight in transgenic wheat. Plant Pathol. 62, 383–392. doi: 10.1111/j.1365-3059.2012.02656.x
Görlach, J., Volrath, S., Knaufbeiter, G., Hengy, G., Beckhove, U., Kogel, K. H., et al. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8, 629–643. doi: 10.1105/tpc.8.4.629
Hafez, Y. M., Soliman, N. K., Saber, M. M., Imbabi, I. A., and Abdelaziz, A. S. (2014). Induced resistance against Puccinia triticina, the causal agent of wheat leaf rust by chemical inducers. Egypt. J. Pest Control 24, 173–181.
Inoue, Y., Vy, T. T. P., Yoshida, K., Asano, H., Mitsuoka, C., Asuke, S., et al. (2017). Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 357, 80–83. doi: 10.1126/science.aam9654
International Barley Genome Sequencing Consortium (2012). A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716. doi: 10.1038/nature11543
Lamari, L. (2008). Assess 2.0 : Image Analysis Software for Plant Disease Quantification. St. Paul, MN: The American Phytopathological Society Press. doi: 10.1094/assess2.0
Liu, X., Bai, X., Wang, X., and Chu, C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979. doi: 10.1016/j.jplph.2006.07.006
Liu, X. Q., Bai, X. Q., Qian, Q., Wang, X. J., Chen, M. S., and Chu, C. C. (2005). OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 15, 593–603. doi: 10.1038/sj.cr.7290329
Loon, L. C. V., Rep, M., and Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, X., Kracher, B., Saur, I. M., Bauer, S., Ellwood, S. R., Wise, R., et al. (2016). Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc. Natl. Acad. Sci. U.S.A. 113, E6486–E6495. doi: 10.1073/pnas.1612947113
Makandar, R., Essig, J. S., Schapaugh, M. A., Trick, H. N., and Shah, J. (2006). Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant Microbe Interact. 19, 123–129. doi: 10.1094/MPMI-19-0123
Molina, A., Görlach, J., Volrath, S., and Ryals, J. (1999). Wheat genes encoding two types of PR-1 proteins are pathogen inducible, but do not respond to activators of systemic acquired resistance. Mol. Plant Microbe Interact. 12, 53–58. doi: 10.1094/MPMI.1999.12.1.53
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
Nakayama, A., Fukushima, S., Goto, S., Matsushita, A., Shimono, M., Sugano, S., et al. (2013). Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biol. 13:150. doi: 10.1186/1471-2229-13-150
Pan, N., Liu, C., Kang, J., and Lv, J. (2017). Genome-wide analysis of WRKY transcription factors in wheat (Triticum aestivum L.) and differential expression under water deficit condition. PeerJ 5:e3232. doi: 10.7717/peerj.3232
Quilis, J., Peñas, G., Messeguer, J., Brugidou, C., and Segundo, B. S. (2008). The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol. Plant Microbe Interact. 21, 1215–1231. doi: 10.1094/MPMI-21-9-1215
Robatzek, S., and Somssich, I. E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 28, 123–133. doi: 10.1046/j.1365-313X.2001.01131.x
Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H. Y., et al. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9, 425–439.
Shah, J., Tsui, F., and Klessig, D. F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10, 69–78. doi: 10.1094/MPMI.1997.10.1.69
Shimono, M., Sugano, S., Nakayama, A., Jiang, C. J., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. doi: 10.1105/tpc.106.046250
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Vallélianbindschedler, L., Métraux, J. P., and Schweizer, P. (1998). Salicylic acid accumulation in barley is pathogen specific but not required for defense-gene activation. Mol. Plant Microbe Interact. 11, 702–705. doi: 10.1094/MPMI.1998.11.7.702
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123. doi: 10.1371/journal.ppat.0020123
Wang, X., Bi, W. S., Gao, J., Yu, X., Wang, H., and Liu, D. (2018). Systemic acquired resistance, NPR1, and pathogenesis-related genes in wheat and barley. J. Integr. Agric. 17, 60345–60347. doi: 10.1016/S2095-3119(17)61852-5
Wang, X., Yang, B., Li, K., Kang, Z., Cantu, D., and Dubcovsky, J. (2016). A conserved Puccinia striiformis protein interacts with wheat NPR1 and reduces induction of pathogenesis-related genes in response to pathogens. Mol. Plant Microbe Interact. 29, 977–989. doi: 10.1094/MPMI-10-16-0207-R
Xu, G., Yuan, M., Ai, C., Liu, L., Zhuang, E., Karapetyan, S., et al. (2017). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494. doi: 10.1038/nature22372
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Zhang, W., Chen, S., Abatea, Z., Nirmala, J., Rouse, M. N., and Dubcovsky, J. (2017). “Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group,” in Proceedings of the National Academy of Sciences of the United States of America Early Edition, Washington, DC.
Keywords: WRKY transcription factors, NPR1, acquired resistance, barley, wheat, Puccinia triticina
Citation: Gao J, Bi W, Li H, Wu J, Yu X, Liu D and Wang X (2018) WRKY Transcription Factors Associated With NPR1-Mediated Acquired Resistance in Barley Are Potential Resources to Improve Wheat Resistance to Puccinia triticina. Front. Plant Sci. 9:1486. doi: 10.3389/fpls.2018.01486
Received: 18 April 2018; Accepted: 25 September 2018;
Published: 17 October 2018.
Edited by:
Víctor Flors, Universitat Jaume I, SpainReviewed by:
Zhonglin Mou, University of Florida, United StatesMickael Malnoy, Fondazione Edmund Mach, Italy
Copyright © 2018 Gao, Bi, Li, Wu, Yu, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodong Wang, emhid3hkQGhlYmF1LmVkdS5jbg== Daqun Liu, bGl1ZGFxdW5AY2Fhcy5jbg==
†These authors have contributed equally to this work
 Jing Gao1†
Jing Gao1† Xiaodong Wang
Xiaodong Wang