- Department of Biology, KU Leuven, Leuven, Belgium
Functional conservation of RNAs between different species is a key argument for their importance. While few long non-coding RNAs are conserved at the sequence level, many long non-coding RNAs have been identified that only share a position relative to other genes. It remains largely unknown whether and how these lncRNAs are conserved beyond their position. In Arabidopsis thaliana, the lncRNA COOLAIR is transcribed antisense from FLOWERING LOCUS C (FLC) in response to cold. Despite relatively low sequence similarity, the COOLAIR expression pattern and in vitro RNA secondary structure are highly conserved across the family Brassicaceae, which originated some 50 mya. It is unclear, however, whether COOLAIR functions in distantly related species such as monocots, which diverged some 150 mya. Here, we identified antisense lncRNAs from FLC homologs in various monocot species that share no sequence similarity with A. thaliana COOLAIR. Yet similar to COOLAIR, we found that BdODDSOC1 antisense (BdCOOLAIR1) and BdODDSOC2 antisense (BdCOOLAIR2) are induced by cold in a Brachypodium distachyon winter accession. Across B. distachyon accessions, the sequences of BdCOOLAIR1 and BdCOOLAIR2 are less conserved than exons but more conserved than flanking regions, suggesting a function for the transcript itself. Knock down of the BdODDSOC2 non-overlapping BdCOOLAIR2 transcript did not show a morphological phenotype, but did result in significantly higher BdODDSOC2 expression during cold, indicating that BdCOOLAIR2 performs a role in cis in the rate of BdODDSOC2 silencing. This functional similarity between eudicot and monocot species reveals ancient conservation or convergent evolution of FLC antisense transcription. Either scenario supports its functional importance.
Introduction
Thousands of long non-coding RNAs (lncRNAs) have been identified and new lncRNAs are continuously being discovered. Yet, the functional importance of these transcripts remains debated. One reason is that lncRNAs evolve much faster than protein coding genes, which has been observed frequently by comparing genome and transcriptome sequences between various species (Necsulea et al., 2014; Hezroni et al., 2015). While sharing low primary sequence conservation, a few better studied lncRNAs, such as XIST (Yildirim et al., 2013), HOTAIR (Somarowthu et al., 2015), TUNA (Lin et al., 2014) and cyrano (Ulitsky et al., 2011), have been experimentally demonstrated to have conserved secondary structures or functions between species. Across different plant species, only hundreds of lncRNAs are conserved at the sequence level, while many more are only positionally conserved (Mohammadin et al., 2015; Wang et al., 2015). To date, little has been done to explore the function of these positionally conserved lncRNAs.
COOLAIR is a set of lncRNAs transcribed antisense from FLOWERING LOCUS C (FLC) in Arabidopsis thaliana (Swiezewski et al., 2009). Despite the relatively low nucleotide sequence identity, COOLAIR shows high expression and secondary structure conservation across the Brassicaceae (Castaings et al., 2014; Hawkes et al., 2016; Li et al., 2016). FLC is a MADS box gene and one of the most intensively studied plant genes. It is best known for its function as a repressor of flowering (Michaels and Amasino, 1999; Bloomer and Dean, 2017; Whittaker and Dean, 2017). To allow flowering in spring, FLC needs to be repressed by the autonomous pathway and the vernalization pathway. In the autonomous pathway, FLC is promoted by FRIGIDA (FRI), while repressed by FCA. In the vernalization pathway, FLC expression is reduced by prolonged cold and the locus is epigenetically silenced. COOLAIR is involved in both of these pathways. In the autonomous pathway, FCA, FY, CstF64, and CstF77 promote the use of the proximal splice site of COOLAIR to represses FLC through an FLD-dependent reduction in H3K4me2 (Liu et al., 2010). In response to vernalization, COOLAIR is transiently induced by prolonged cold, reaching a maximum expression level after 2 weeks (Swiezewski et al., 2009). Although COOLAIR is not absolutely required for the silencing of FLC during laboratory vernalization (Helliwell et al., 2011; Csorba et al., 2014), FLC terminator/COOLAIR promoter exchange lines result in the slowing down of the FLC silencing rate, through the switch in chromatin state by erasing H3K36me3 (Csorba et al., 2014). In the slowly vernalizing accession Var2-6, FLC repression is slower due to splicing of the distally polyadenylated COOLAIR (Li et al., 2015), indicating the role of COOLAIR in natural variation. The stable silencing of FLC is associated with two more lncRNAs, COLDWRAP and COLDAIR, which recruit PHD-PRC2 to a specific chromatin region (Heo and Sung, 2011; Kim and Sung, 2017; Kim et al., 2017). However, COOLAIR has not been identified beyond the Brassicaceae, both COLDWRAP and COLDAIR have not even been found beyond A. thaliana (Castaings et al., 2014; Li et al., 2016).
FLOWERING LOCUS C has been identified in monocots through synteny and phylogenetic analysis (Ruelens et al., 2013). FLC homologs in barley, wheat and Brachypodium distachyon, a model for the temperate cereals, are repressed during vernalization, showing a similar expression behavior as FLC in A. thaliana (Greenup et al., 2010; Sharma et al., 2017). In B. distachyon, three FLC homologs have been found: BdODDSOC2, BdODDSOC1, and BdMADS37 (Ruelens et al., 2013). BdODDSOC2 is epigenetically silenced during vernalization in the winter accession BdTR3C and overexpression of BdODDSOC2 in the facultative accession Bd21-3 delays flowering time (Sharma et al., 2017). Besides temperate monocots, there are FLC homologs in subtropical monocots, such as OsMADS51 in Oryza sativa, which is proposed to be involved in promoting flowering in short days (Kim et al., 2007; Ruelens et al., 2013). In contrast to the extensively studied mechanisms of FLC in A. thaliana, whether and how lncRNAs function in the regulation of FLC homologs in distantly related monocots is unknown.
Here we show that the antisense transcript COOLAIR is positionally conserved in FLC homologs across distantly related monocot species. We found that in B. distachyon accession BdTR3C, BdCOOLAIR2 is strongly induced by prolonged cold and represses its neighboring coding gene BdODDSOC2. The functional similarity of FLC/COOLAIR sense-antisense pairs between eudicots and monocots highlights the functional importance of antisense long non-coding RNA transcription.
Materials and Methods
Plant Growth Conditions
Brachypodium distachyon seeds were overnight incubated on wet filter paper in petri dishes, then sown in root trainers (soil:vermiculite = 2:1). For qPCR, root trainers were putted in 8 h/16 h light/dark at 23°C for 3 weeks, then 8 h/16 h light/dark at 4°C for 2 weeks, and finally transferred to 8 h/16 h light/dark at 23°C for 1 week. For phenotyping, root trainers were placed in 8 h/16 h light/dark at 23°C for 3 weeks, then transferred to 8 h/16 h light/dark at 4°C for 2 weeks (Bd21-3) and 6 weeks (BdTR3C), respectively, then transferred to 20h/4h light/dark at 23–25°C until flowering.
RNA Extraction, cDNA Synthesis, and qPCR Analysis
RNA extraction was performed following standard TRIsure protocol. DNA was removed using TURBO DNA free kit (Ambion-Applied), first single strand cDNA was synthesized by SensiFast cDNA Synthesis Kit of Bioline (GC Biotech BIO-65054). qPCR was performed with SensiFast SYBR Hi-Rox Kit (GC Biotech: BIO-92020), at least three biological replicates and two technical replicates were used for each time point.
Genomic Sequence Diversity
Brachypodium distachyon pan-genome sequences were downloaded from the COGE database. Alignment was performed by CLC sequence viewer 7.7, with gap open cost 10.0, gap extension cost 1.0. DNA polymorphisms were calculated by DNASP, sites with alignment gaps were excluded. The figure was drawn with a 50-sites sliding window and 25 sites step size.
RNAi Lines
For RNAi mediated knockdown of BdCOOLAIR2, around 200 bp of BdCOOLAIR2 was first cloned into a pENTR2B vector (Invitrogen), with BamHI (Promega) and EcoRV (Promega) restriction enzymes. Then, the BdCOOLAR2 sequence was inserted into a binary destination vector pIPK007 with the Zea mays UBIQUITIN promoter (Himmelbach et al., 2007) by using GatewayTM LR ClonaseTM Enzyme mix (Invitrogen). The primers for BdCOOLAIR2 cloning are as follows: BdCOOLAIR2_RNAi_F: TGGGTC GGATCCGTCCGGAGGCACACAAAT and BdCOOLAIR2_RNAi_R: CAAACTGATATCGGGACCTGAAGAACACGAGA. B. distachyon transformation was performed according to the Agrobacterium-mediated transformation protocol (Himmelbach et al., 2007; Alves et al., 2009). The primers for genotyping are listed in Supplemental Table 1. Phenotyping was done with T1 plants in a growth chamber, null sibling plants were used as controls (NC).
Calculation of Maximal Information Coefficient
We made use of the Minerva package in R to calculate the MIC of the data (Albanese et al., 2013). To test for significance, we generated 250000 permuted datasets and calculated the MIC for each of these and calculated a p-value as the fraction of datasets with an MIC higher than the MIC for the observed data.
Results
FLC Homologs in B. distachyon and Other Grasses Generate Antisense LncRNAs
To investigate whether there are non-coding RNAs generated from FLC loci in monocots, we first analyzed publicly available datasets. In B. distachyon, we found multiple Expressed Sequence Tags (ESTs) behind the stop codon of BdODDSOC1 and BdODDSOC2, based on the RNA-seq and EST data in the phytozome browser (Goodstein et al., 2012) (Figure 1A). The direction of these ESTs is antisense to BdODDSOC1 and BdODDSOC2 coding transcripts. They are annotated as lncRNAs in GreeNC, a database of plant lncRNAs (Goodstein et al., 2012; Paytuví Gallart et al., 2015). For BdODDSOC1, there are 7 annotated antisense ESTs, while only one (Bradi2g59124.5) has high confidence with a full-length of 2581 bp. For BdODDSOC2, there are 2 annotated antisense ESTs, both with high confidence. The length of those is 475 bp (Bradi2g59186.1) and 372 bp (Bradi2g59186.2), respectively. The coding potential for these three antisense transcripts is very low, –1.130 (Bradi2g59124.5), -1.034 (Bradi2g59186.1) and –1.023 (Bradi2g59186.2), further suggesting that they could function as non-coding transcripts.
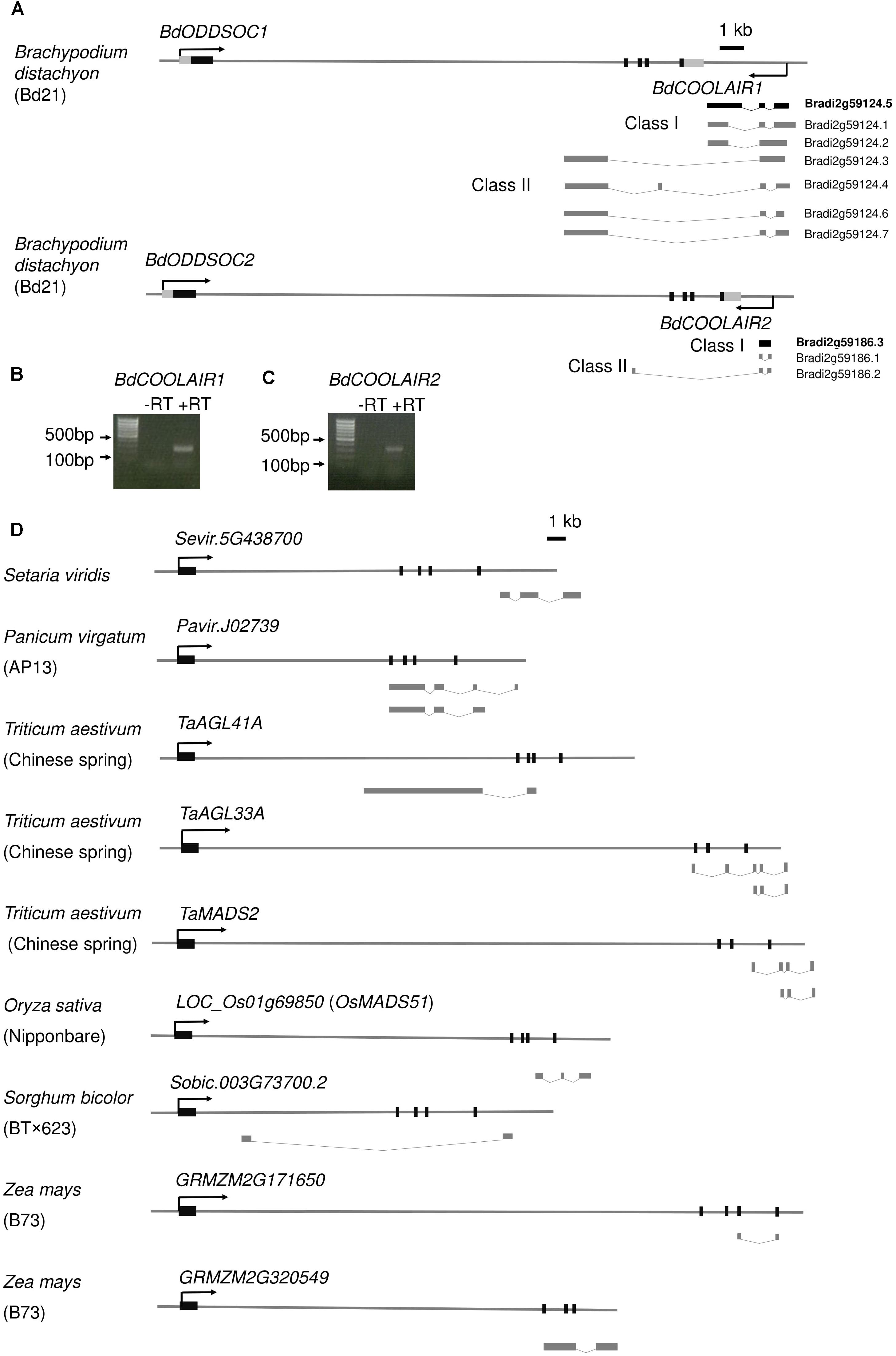
Figure 1. FLC homologs in B. distachyon and other monocots generate antisense long non-coding RNAs. (A) Schematic representation of annotated FLC homolog genomic loci (BdODDSOC1 and BdODDSOC2) and antisense transcripts in B. distachyon. For BdODDSOC1 and BdODDSOC2 genomic loci, the black boxes indicate exons, the black lines indicate introns and flanking region. The directions of sense transcripts (black arrow), Class I antisense transcripts and Class II antisense transcripts are shown. Transcripts in bold (Bradi2g59124.5 and Bradi2g59186.3) are confirmed by RT-PCR. (B) and (C) Agarose gel for BdCOOLAIR1 and BdCOOLAIR2, respectively after RT-PCR. A DNA ladder is shown. (D) Schematic representation of FLC homolog genomic loci and antisense transcripts in various grasses. For FLC homologs, the black boxes indicate exons, the black lines indicate introns and flanking regions. The direction of sense transcripts (black arrow) is shown.
We further confirmed the existence of these antisense transcripts by RT-PCR and Sanger sequencing. Our results are not fully consistent with the EST data, because we could only amplify the high-confidence proximal antisense of BdODDSOC1 and BdODDSOC2 (Figures 1B,C). The RT-PCR confirmed that the antisense transcripts are not overlapping with BdODDSOC1 and BdODDSOC2 (Supplementary Figure S1), which is different from A. thaliana (Swiezewski et al., 2009). Class II lncRNAs are also annotated from EST data while we could not amplify Class II BdCOOLAIR2. We designed primers specific for Class II, but amplified nothing. For Class I BdCOOLAIR2, we got two bands, the bands of Bradi2g59186.3 is much stronger than Bradi2g59186.1 (Figure 1A). Both Bradi2g59186.3 and Bradi2g59186.1 are Class I BdCOOLAIR2. Even though we could not amplify distal antisense transcripts, we cannot exclude their existence because they could be expressed in other accessions or under specific conditions which have not been tested in our study.
To know whether FLC antisense transcripts are also present in other monocots, we further analyzed the public gene expression data available for other grass species. Interestingly, we found that FLC homologs in at least 6 other grasses produce antisense transcripts, including the model plant Setaria viridis, the energy plant Panicum virgatum, the temperate cereal Triticum aestivum and the tropical cereals Oryza sativa, Sorghum bicolor and Zea mays (Figure 1D). Thus, we concluded that FLC antisense transcription is present in various grass species.
To know whether these antisense transcripts are COOLAIR homologs, we analyzed their sequence conservation. The sequence has fully diverged and no sequence conservation could be detected between these antisense transcripts and COOLAIR in A. thaliana (Supplementary Figure S2). Even between grass species, the antisense transcripts share no sequence conservation (Supplementary Figure S3). Because they are all located at the end of FLC homologs, we propose that these antisense transcripts are positionally conserved COOLAIR homologs in grasses. For further study, we named the antisense of BdODDSOC1 and BdODDSOC2 as BdCOOLAIR1 and BdCOOLAIR2, respectively.
ODDSOC2 Antisense Transcription Is Strongly Induced by Cold in a B. distachyon Winter Accession
To investigate the expression patterns of these positionally conserved lncRNAs, we performed quantitative RT-PCR in three accessions. We have previously shown that in the winter accession BdTR3C, BdODDSOC2 is epigenetically silenced by prolonged cold (Figure 2A), while in the spring accession Bd21, BdODDSOC2 is repressed by cold but not stably silenced. In the facultative accession Bd21-3, BdODDSOC2 does not respond to cold (Sharma et al., 2017). Interestingly, we found that in BdTR3C, BdODDSOC2 and BdCOOLAIR2 show opposite expression patterns during early stage cold treatment (Figures 2B,D and Supplementary Figure S4). BdODDSOC2 goes down, while BdCOOLAIR2 is significantly upregulated (approximately 40 folds) and reaches a maximal level after 3 days of cold. To investigate whether BdCOOLAIR2 is specifically induced by cold, we compared its expression to non-cold treatment control conditions. Indeed, without cold, BdODDSOC2 antisense expression is not induced (Figure 2D). To verify whether the relationship between BdCOOLAIR2 and BdODDSOC2 expression holds in a facultative or a spring accession, we checked the expression of BdCOOLAIR2 in Bd21-3 and Bd21. BdCOOLAIR2 is only slightly induced in Bd21-3 (approximately two folds) after 1 day (Figure 2E), and BdCOOLAIR2 is not induced significantly in Bd21 (Figure 2F).
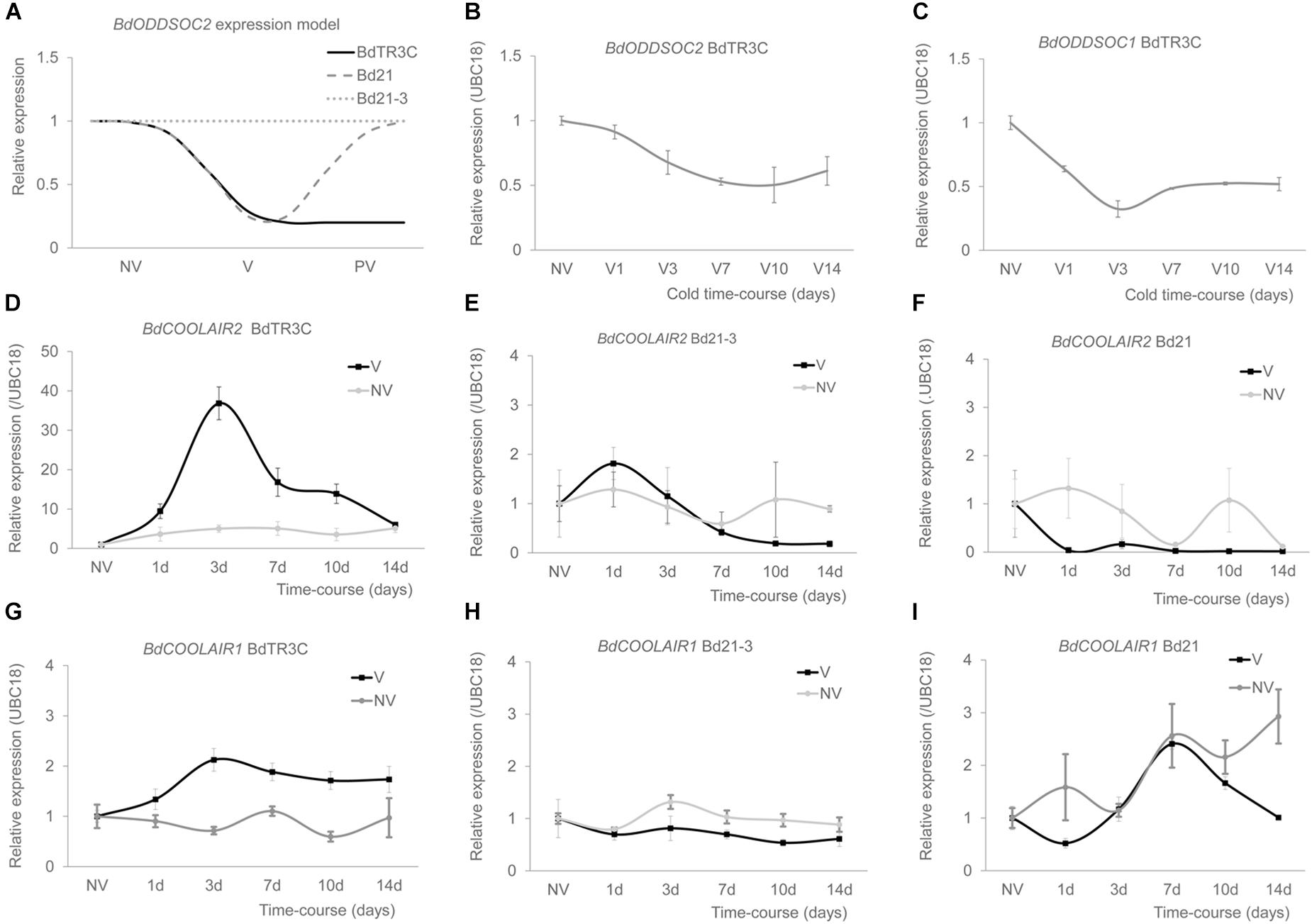
Figure 2. BdCOOLAIR2 and BdCOOLAIR1 are upregulated by vernalization. (A) Model representation of expression patterns of BdODDSOC2 in three accessions: BdTR3C, Bd21 and Bd21-3. NV is non-vernalized, V is vernalized, PV is post-vernalized. In BdTR3C, BdODDSOC2 is epigenetically silenced in vernalization (V) and stays down in post-vernalization (PV); in Bd21, BdODDSOC2 is repressed in vernalization, while it comes back up post-vernalization; in Bd21-3, BdODDSOC2 expression is not affected during vernalization and post-vernalization. (B) BdODDSOC2 is repressed during vernalization. (C) BdODDSOC1 is repressed during vernalization. (D) BdCOOLAIR2 in BdTR3C is induced by vernalization (black). Non-vernalization control is shown (gray). (E) BdCOOLAIR2 expression in Bd21-3, vernalization (black) and non-vernalization (gray) are shown. (F) BdCOOLAIR2 expression in Bd21, vernalization (black) and non-vernalization (gray) are shown. (G) BdCOOLAIR1 in BdTR3C is induced by vernalization (black). Non-vernalization control is shown (gray). (H) BdCOOLAIR1 expression in Bd21-3, vernalization (black) and non-vernalization (gray) are shown. (I) BdCOOLAIR1 expression in Bd21, vernalization (black) and non-vernalization (gray) are shown. Values are means ± SEM of three or four biological replicates, two technical replicates.
For BdODDSOC1, BdCOOLAIR1 also shows an opposite expression pattern in BdTR3C (Figures 2C,G). BdODDSOC1 goes down, while BdCOOLAIR1 goes up, again reaching a maximum level after 3 days of cold. Here only an approximately two-fold induction level is reached. Similar to BdCOOLAIR2, BdCOOLAIR1 is induced by cold (Figure 2G). Similar to BdCOOLAIR2, BdCOOLAIR1 is not induced in Bd21-3 (Figure 2H) or Bd21 (Figure 2I).
BdODDSOC1 and BdODDSOC2 Intronic Transcripts Are Not Induced by Prolonged Cold
Similar to the A. thaliana FLC locus, the BdODDSOC1 and BdODDSOC2 loci have a very large first intron, suggesting that this may play a role in gene regulation. To investigate whether intronic sense lncRNA transcription, similar to COLDAIR, is present in B. distachyon, we investigated RNA-seq data as no ESTs were annotated for the introns. We found that there are multiple peaks located in the long first intron of BdODDSOC1 and BdODDSOC2 and considered that these peaks might point to intronic long non-coding RNAs in these regions. To verify this idea in BdTR3C, we designed primers spanning the entire first intron of BdODDSOC1 and BdODDSOC2 (Supplementary Tables S2, S3). After RT-PCR amplification, we observed a series of bands along the first intron of BdODDSOC1 and BdODDSOC2 (Supplementary Figures S5, S6). However, this did not generate a clear idea about the number of different lncRNAs transcribed from the first intron, nor of the size of possible lncRNAs.
To know whether the transcripts observed in the first intron are induced by prolonged cold, we performed quantitative RT-PCR. They showed a similar expression pattern as sense coding transcripts, and are downregulated during prolonged cold (Supplementary Figures S7, S8). But because long non-coding RNAs may be only induced at a specific time and under specific conditions, which may have gone unnoticed in our experiments, we cannot conclude whether they function similar to COLDAIR.
The BdCOOLAIR Sequence Is Relatively Conserved in B. distachyon
To know how the antisense sequence evolved, we analyzed the DNA sequences using the released B. distachyon pan-genome sequencing data (Gordon et al., 2017). We downloaded the genomic DNA sequence of BdODDSOC2 and BdODDSOC1 for 54 of the genome sequenced B. distachyon accessions and conducted a polymorphism analysis (Figure 3).
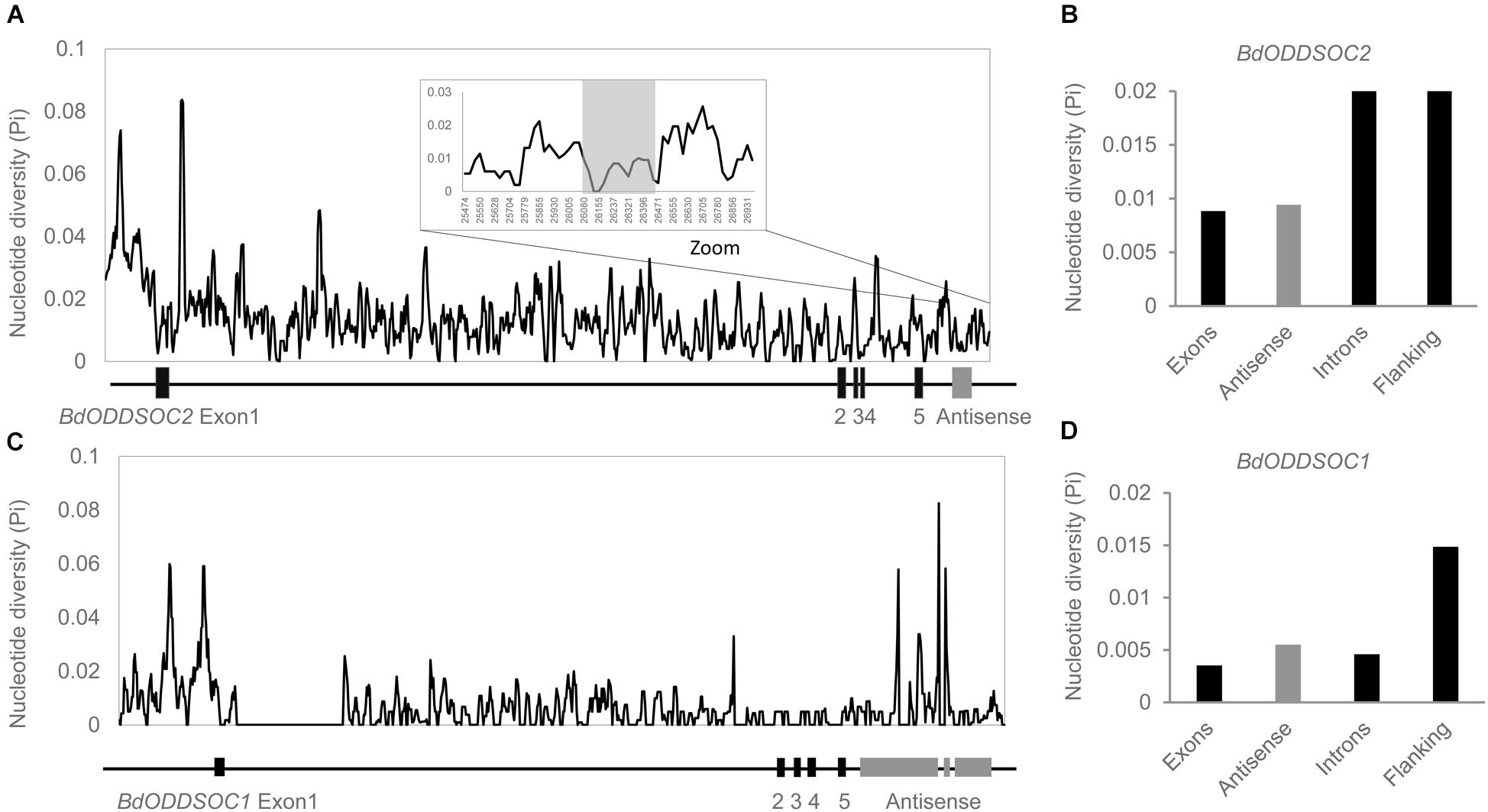
Figure 3. BdCOOLAIR2 sequence is more conserved than intronic and flanking regions. (A) Sliding window plot representation of BdODDSOC2 genomic diversity, with a window size of 28 kb. (B) Bar chart of sequence diversity of different regions of BdODDSOC2. (C) Sliding window plot representation of BdODDSOC1 genomic diversity, with a window size of 23 kb. (D) Bar chart of sequence diversity of different regions of BdODDSOC1. Black indicates exons, introns and flanking regions, gray indicates antisense.
BdODDSOC2 contains five exons, which are most conserved with a nucleotide diversity value (NDV) of 0.008. The intronic and gene flanking region NDV is 0.0209 and 0.0217, respectively. The antisense value (0.009) is higher than the value for exons, but lower than introns and flanking regions. Thus, in BdODDSOC2, the antisense region evolved slightly faster than exons but much more slowly than regions under more neutral selection (introns and flanking regions). This suggests a function for the RNA transcript and not just for the act of transcription (Kopp and Mendell, 2018). BdODDSOC1 shows a comparable result, where the antisense sequence (NDV is 0.005) evolved faster than exons (NDV is 0.0035) and introns (NDV is 0.0045), but more slowly than flanking regions (NDV is 0.015). Therefore, we wanted to test whether BdCOOLAIR2 performs a function at the transcript level rather than testing whether the action of transcription is important.
Silencing of BdODDSOC2 Is Delayed in BdCOOLAIR2 Knock-Down Lines
We have shown that the position of BdCOOLAIR2 does not overlap with BdODDSOC2 (Figure 1A and Supplementary Figure S1), and its sequence evolution suggests that BdCOOLAIR2 has a function at the RNA level (Figures 3A,B). To understand the functional importance of BdCOOLAIR2, we used RNA interference to knock down BdCOOLAIR2 in two accessions: BdTR3C and Bd21-3. We chose BdCOOLAIR2 rather than BdCOOLAIR1, because in BdTR3C, BdODDSOC2 is epigenetically silenced by cold and BdCOOLAIR2 is much more strongly induced than BdCOOLAIR1. We generated 10 RNAi lines for accession BdTR3C and 18 lines for accession Bd21-3.
To investigate whether we successfully knocked down BdCOOLAIR2 using RNAi, we first checked its expression level in RNAi T0 and T1 lines. We confirmed that in induced conditions, BdCOOLAIR2 expression is lower in 5 RNAi lines compared to null sibling controls (Figure 4A and Supplementary Table S4). To study whether BdCOOLAIR2 works in cis, we investigated the expression level of its neighboring coding transcript BdODDSOC2. Interestingly, in BdTR3C, there is a significant difference in BdODDSOC2 expression level between RNAi and null siblings at three days of cold treatment (t(16) = 4.3258, p < 0.001, Figure 4B, Supplementary Figure S9 and Supplementary Tables S5, S6). However, in non-vernalization, 6 weeks of vernalization and post-vernalization conditions, we did not observe a significant difference. Three days of cold treatment is the time point when BdCOOLAIR2 reached its maximum expression level, supporting a role for BdCOOLAIR2 in regulating BdODDSOC2 during cold treatment. Although BdODDSOC2 is still repressed in these RNAi lines, the silencing rate is slower compared with control lines (Figure 4B and Supplementary Figure S11A). BdODDSOC2 and BdODDSOC1 share a lot of sequence similarity. To know whether BdODDSOC1 can be induced in BdCOOLAIR2 RNAi lines, we also tested the expression of BdODDSOC1. We found that after three days of cold treatment, BdODDSOC1 is not induced in BdCOOLAIR2 RNAi lines, which is different with BdODDSOC2 (Supplementary Figure S10). We conclude that like COOLAIR in A. thaliana, BdCOOLAIR2 has a function associated with the rate of BdODDSOC2 silencing in a winter accession (Figure 4C and Supplementary Figure S11B) (Csorba et al., 2014). We showed previously that ectopic expression of BdODDSOC2 delays flowering in Bd21-3. As a result, we would expect an effect of silencing of BdCOOLAIR2 on flowering time because BdODDSOC2 is silenced more slowly. We used 10 BdTR3C RNAi lines and 11 Bd21-3 RNAi lines to phenotype flowering time and leaf number. However, we did not observe a significant difference between these lines and controls (Supplementary Figure S12). This suggests that the effect on expression of BdODDSOC2 is not strong enough to result in a significant phenotype.
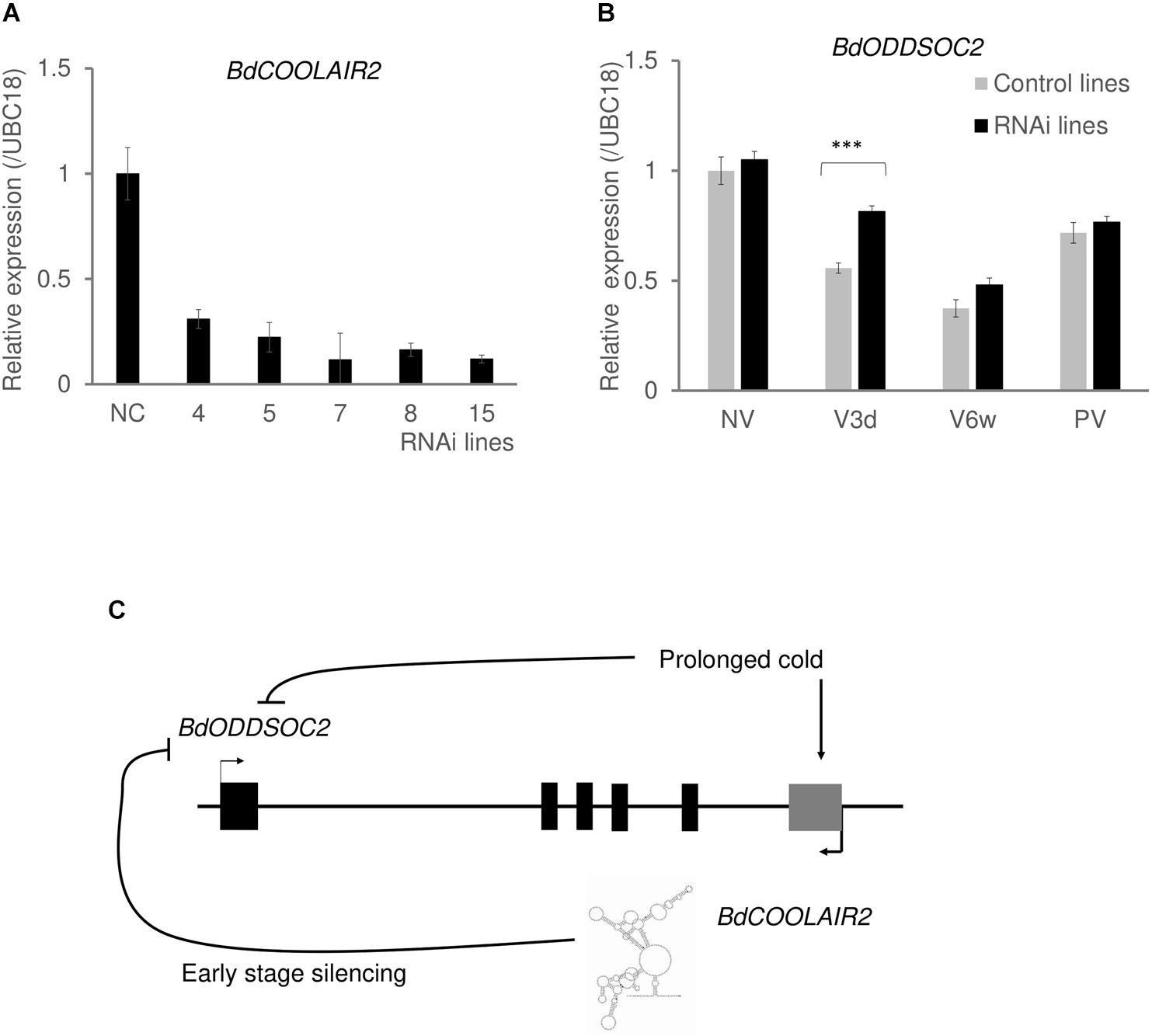
Figure 4. BdCOOLAIR2 function in the silencing of BdODDSOC2. (A) Expression of BdCOOLAIR2 in BdTR3C RNAi lines is significantly lower than in null sibling control lines (NC). Values are means ± SEM of three biological replicates. (B) Silencing of BdODDSOC2 is delayed in BdCOOLAIR2 knock-down lines. BdODDSOC2 expression was tested in BdCOOLAIR2 RNAi lines, in non-vernalization (NV), three days of vernalization (V3d), 6 weeks of vernalization (V6w) and 1 week post-vernalization (PV). Gray bars indicate null-sibling control lines, black bars indicate RNAi lines. Values for control lines are means ± SEM of three biological replicates, while values for RNAi lines are means ± SEM of twelve to fifteen biological replicates, pooled of all RNAi lines. The values of each RNAi line were separately shown in Supplementary Figure S9. Asterisks indicate statistically significant differences with the student’s t-test (∗∗∗p < 0.001). (C) In response to vernalization, the BdCOOLAIR2 transcript has a function associated with silencing of BdODDSOC2.
To further test the antagonistic relationship between BdODDSOC2 and BdCOOLAIR2 (Rosa et al., 2016), we investigated their gene expression levels using qRT-PCR in 74 three-week old individual plants belonging to different accessions (Supplementary Table S7). We observed that in plants with low BdCOOLAIR2 expression, BdODDSOC2 expression levels can be high and in plants with high BdCOOLAIR2 expression, BdODDSOC2 expression levels are almost always low (Supplementary Figure S13). This suggests a non-coexistence relationship, in which either transcript can be present, but they are not expressed together. To formally test such a non-linear association, we made use of the maximal information coefficient (MIC), which measures linear or non-linear correlation between paired variables (Reshef et al., 2011). We found that the calculated MIC for the data (MIC = 0.3752117) is significantly higher than the MICs calculated for a null distribution of permuted datasets of equal size (empirical p value = 0.0127). This further confirms the antagonistic relationship between BdCOOLAIR2 and BdODDSOC2.
Discussion
The expression of COOLAIR and its structure is conserved across closely related Brassicaceae (Castaings et al., 2014; Hawkes et al., 2016; Li et al., 2016). In our study, we demonstrate that beyond the Brassicaceae, COOLAIR is positionally conserved in distantly related monocot species. However, its sequence is unrecognizable. In B. distachyon, we find that BdCOOLAIR2 is strongly induced by prolonged cold in a vernalization requiring accession but not in spring accessions. We further demonstrate that BdCOOLAIR2 sequence diversity is lower than intronic and gene flanking regions, suggesting a function for the transcript itself. We indeed found that the BdCOOLAIR2 transcript plays a role in the repression of BdODDSOC2.
Sense-antisense pairs are pervasive in bacteria, budding yeast, rice, maize and mammalian genomes (Pelechano and Steinmetz, 2013). LncRNA sequences evolve rapidly, and antisense lncRNAs are more positionally conserved than sequence conserved (Necsulea et al., 2014; Hezroni et al., 2015; Mohammadin et al., 2015; Wang et al., 2015). However, only a few studies have been performed to compare the function of lncRNAs across distantly related species (Ulitsky et al., 2011; Yildirim et al., 2013; Lin et al., 2014; Somarowthu et al., 2015). Our data provide more evidence that positionally conserved antisense long non-coding RNAs can be deeply functionally conserved or alternatively, can originate through convergent evolution to fulfill a similar role in gene regulation. The first hypothesis appears more likely as the lack of sequence similarity can be a consequence of sampling density and not enough sequence data is available to fully explore the rapid sequence evolution of lncRNAs over long evolutionary distances. Indeed, FLC homologs in intermediate species such as Theobroma cacao and Vitis vinifera also have annotated antisense transcripts (Supplementary Figure S14).
In A. thaliana, alternative polyadenylation is important for COOLAIR function (Liu et al., 2010; Li et al., 2015) and proximal COOLAIR represses FLC while distal COOLAIR is not involved in repression. In B. distachyon, we only amplified proximal antisense lncRNAs, which is consistent with Arabis alpina, in which proximal antisense is much more conserved than distal antisense (Castaings et al., 2014). However, in Brassica rapa, it was shown that the distal antisense is strongly induced by cold (Li et al., 2016). Therefore, it remains somewhat unclear whether alternative polyadenylation is a conserved aspect of COOLAIR function. In contrast to the antagonistic relationship in a single cell between FLC and COOLAIR (Rosa et al., 2016), it has been recently reported that FRI-containing supercomplex and FRI partners upregulate both FLC and Class II COOLAIR, by promoting the formation of a chromatin loop around FLC locus (Li et al., 2018). In addition, elimination of Class II RNAs in the FLC+MAF2-T and FLC+NOS-T lines is associated with a reduction in FLC expression. Thus, the mechanism behind FLC/COOLAIR antagonistic expression and COOLAIR guided FLC silencing under different pathways needs to be further studied.
We tried to identify sense intronic lncRNAs, but we did not reach a conclusion. The VRE region (promoter of COLDAIR) has evolved rapidly in Brassicaceae (Castaings et al., 2014; Li et al., 2016), with very low sequence conservation. Thus, so far COLDAIR has not been found beyond A. thaliana. We found some transcripts based on RNA-Seq, EST and our RT-PCR data. These transcripts show a similar expression pattern as sense coding transcripts, and they are downregulated during prolonged cold. We propose that these transcripts might be unspliced sense FLC transcripts. However, we cannot exclude the existence of non-coding transcripts in the introns. The reason might be that we did not use suitable accessions or conditions that would induce lncRNAs expression.
In A. thaliana, the sequence of COOLAIR overlaps with FLC, which makes the use of RNAi limited to understand its function. Therefore, FLC terminator/COOLAIR promotor exchange lines were created to truncate the expression of COOLAIR (Wang et al., 2014). In the case of B. distachyon, BdCOOLAIR2 does not overlap with BdODDSOC2 and we generated RNAi lines specifically targeting the antisense lncRNA. In no-cold and post-cold conditions, we did not see a clear effect on BdODDSOC2, however at 3 days of cold, BdODDSOC2 expression is higher in antisense knockdown lines compared with controls. Thus, we demonstrated that BdCOOLAIR2 functions in cis, and at the RNA level. BdCOOLAIR2 knock down does not affect the overall expression of BdODDSOC2 but slows down the repression process. This effect apparently was not strong enough to affect flowering time. However, considering the slow repression of FLC caused by splicing of the distally polyadenylated COOLAIR in the slowly vernalizing accession Var2-6 (Li et al., 2015), it has been proposed that COOLAIR works more importantly in natural conditions in which temperature and other factors fluctuate widely (Whittaker and Dean, 2017). It would be very interesting to verify this hypothesis also in B. distachyon.
Author Contributions
KG and FJ designed the experiments. FJ, KP, and MM carried out the experiments and analyzed the data. ND designed the primers for BdODDSOC1 and BdODDSOC2 introns. FJ and KG wrote the manuscript.
Funding
FJ was supported by the China Scholarship Council (CSC) for 4 years at KU Leuven. ND was supported by a FWO SB fellowship SB131327. The Geuten Lab was supported by KU Leuven grant C24/17/37 and FWO grant G065713.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Anja Vandeperre for technical assistance and other members of the Geuten laboratory for discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00072/full#supplementary-material
References
Albanese, D., Filosi, M., Visintainer, R., Riccadonna, S., Jurman, G., and Furlanello, C. (2013). Minerva and minepy: a C engine for the MINE suite and its R, Python and MATLAB wrappers. Bioinformatics 29, 407–408. doi: 10.1093/bioinformatics/bts707
Alves, S. C., Worland, B., Thole, V., Snape, J. W., Bevan, M. W., and Vain, P. (2009). A protocol for Agrobacterium-mediated transformation of Brachypodium distachyon community standard line Bd21. Nat. Protoc. 4, 638–649. doi: 10.1038/nprot.2009.30
Bloomer, R. H., and Dean, C. (2017). Fine-tuning timing: natural variation informs the mechanistic basis of the switch to flowering in Arabidopsis thaliana. J. Exp. Bot. 68, 5439–5452. doi: 10.1093/jxb/erx270
Castaings, L., Bergonzi, S., Albani, M. C., Kemi, U., Savolainen, O., and Coupland, G. (2014). Evolutionary conservation of cold-induced antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana perennial relatives. Nat. Commun. 5:4457. doi: 10.1038/ncomms5457
Csorba, T., Questa, J. I., Sun, Q., and Dean, C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 111, 16160–16165. doi: 10.1073/pnas.1419030111
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Gordon, S. P., Contreras-Moreira, B., Woods, D. P., Des Marais, D. L., Burgess, D., Shu, S., et al. (2017). Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 8:2184. doi: 10.1038/s41467-017-02292-8
Greenup, A. G., Sasani, S., Oliver, S. N., Talbot, M. J., Dennis, E. S., Hemming, M. N., et al. (2010). ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol. 153, 1062–1073. doi: 10.1104/pp.109.152488
Hawkes, E. J., Hennelly, S. P., Novikova, I. V., Irwin, J. A., Dean, C., and Sanbonmatsu, K. Y. (2016). COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 16, 3087–3096. doi: 10.1016/j.celrep.2016.08.045
Helliwell, C. A., Robertson, M., Finnegan, E. J., Buzas, D. M., and Dennis, E. S. (2011). Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS One 6:e21513. doi: 10.1371/journal.pone.0021513
Heo, J. B., and Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Hezroni, H., Koppstein, D., Schwartz, M. G., Avrutin, A., Bartel, D. P., and Ulitsky, I. (2015). Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 11, 1110–1122. doi: 10.1016/j.celrep.2015.04.023
Himmelbach, A., Zierold, U., Hensel, G., Riechen, J., Douchkov, D., Schweizer, P., et al. (2007). A set of modular binary vectors for transformation of cereals. Plant Physiol. 145, 1192–1200. doi: 10.1104/pp.107.111575
Kim, D.-H., and Sung, S. (2017). Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell 40, 302.e4–312.e4. doi: 10.1016/j.devcel.2016.12.021
Kim, D.-H., Xi, Y., and Sung, S. (2017). Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 13:e1006939. doi: 10.1371/journal.pgen.1006939
Kim, S. L., Lee, S., Kim, H. J., Nam, H. G., and An, G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145, 1484–1494. doi: 10.1104/pp.107.103291
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi: 10.1016/j.cell.2018.01.011
Li, P., Tao, Z., and Dean, C. (2015). Phenotypic evolution through variation in splicing of the noncoding RNACOOLAIR. Genes Dev. 29, 696–701. doi: 10.1101/gad.258814.115
Li, X., Zhang, S., Bai, J., and He, Y. (2016). Tuning growth cycles of Brassica crops via natural antisense transcripts of BrFLC. Plant Biotechnol. J. 14, 905–914. doi: 10.1111/pbi.12443
Li, Z., Jiang, D., and He, Y. (2018). FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat. Plants 4, 836–846. doi: 10.1038/s41477-018-0250-6
Lin, N., Chang, K.-Y., Li, Z., Gates, K., Rana, Z. A., Dang, J., et al. (2014). An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 53, 1005–1019. doi: 10.1016/j.molcel.2014.01.021
Liu, F., Marquardt, S., Lister, C., Swiezewski, S., and Dean, C. (2010). Targeted 3’ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97. doi: 10.1126/science.1180278
Michaels, S. D., and Amasino, R. M. (1999). Flowering locus C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Mohammadin, S., Edger, P. P., Pires, J. C., and Schranz, M. E. (2015). Positionally-conserved but sequence-diverged: identification of long non-coding RNAs in the Brassicaceae and Cleomaceae. BMC Plant Biol. 15:217. doi: 10.1186/s12870-015-0603-5
Necsulea, A., Soumillon, M., Warnefors, M., Liechti, A., Daish, T., Zeller, U., et al. (2014). The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640. doi: 10.1038/nature12943
Paytuví Gallart, A., Gallart, A. P., Pulido, A. H., de Lagrán, I. A. M., Sanseverino, W., and Cigliano, R. A. (2015). GREENC: a Wiki-based database of plant lncRNAs. Nucleic Acids Res. 44, D1161–D1166. doi: 10.1093/nar/gkv1215
Pelechano, V., and Steinmetz, L. M. (2013). Gene regulation by antisense transcription. Nat. Rev. Genet. 14, 880–893. doi: 10.1038/nrg3594
Reshef, D. N., Reshef, Y. A., Finucane, H. K., Grossman, S. R., McVean, G., Turnbaugh, P. J., et al. (2011). Detecting novel associations in large data sets. Science 334, 1518–1524. doi: 10.1126/science.1205438
Rosa, S., Duncan, S., and Dean, C. (2016). Mutually exclusive sense-antisense transcription at FLC facilitates environmentally induced gene repression. Nat. Commun. 7:13031. doi: 10.1038/ncomms13031
Ruelens, P., de Maagd, R. A., Proost, S., Theißen, G., Geuten, K., and Kaufmann, K. (2013). Flowering locus C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 4:2280. doi: 10.1038/ncomms3280
Sharma, N., Ruelens, P., D’hauw, M., Maggen, T., Dochy, N., Torfs, S., et al. (2017). A flowering locus c homolog is a vernalization-regulated repressor in brachypodium and is cold regulated in wheat. Plant Physiol. 173, 1301–1315. doi: 10.1104/pp.16.01161
Somarowthu, S., Legiewicz, M., Chillón, I., Marcia, M., Liu, F., and Pyle, A. M. (2015). HOTAIR forms an intricate and modular secondary structure. Mol. Cell 58, 353–361. doi: 10.1016/j.molcel.2015.03.006
Swiezewski, S., Liu, F., Magusin, A., and Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462, 799–802. doi: 10.1038/nature08618
Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H., and Bartel, D. P. (2011). Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550. doi: 10.1016/j.cell.2011.11.055
Wang, H., Niu, Q.-W., Wu, H.-W., Liu, J., Ye, J., Yu, N., et al. (2015). Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 84, 404–416. doi: 10.1111/tpj.13018
Wang, Z.-W., Wu, Z., Raitskin, O., Sun, Q., and Dean, C. (2014). Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc. Natl. Acad. Sci. U.S.A. 111, 7468–7473. doi: 10.1073/pnas.1406635111
Whittaker, C., and Dean, C. (2017). The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33, 555–575. doi: 10.1146/annurev-cellbio-100616-060546
Keywords: long non-coding RNA, lncRNA, B. distachyon, in cis, vernalization, FLC, positionally conserved, COOLAIR
Citation: Jiao F, Pahwa K, Manning M, Dochy N and Geuten K (2019) Cold Induced Antisense Transcription of FLOWERING LOCUS C in Distant Grasses. Front. Plant Sci. 10:72. doi: 10.3389/fpls.2019.00072
Received: 19 September 2018; Accepted: 17 January 2019;
Published: 01 February 2019.
Edited by:
Elena M. Kramer, Harvard University, United StatesReviewed by:
Rainer Melzer, University College Dublin, IrelandSibum Sung, University of Texas at Austin, United States
Liang Wu, Zhejiang University, China
Copyright © 2019 Jiao, Pahwa, Manning, Dochy and Geuten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koen Geuten, a29lbi5nZXV0ZW5Aa3VsZXV2ZW4uYmU=; a29lbi5nZXV0ZW5AYmlvLmt1bGV1dmVuLmJl
 Fuchao Jiao
Fuchao Jiao Kanchan Pahwa
Kanchan Pahwa Murray Manning
Murray Manning Koen Geuten
Koen Geuten