- 1Department of Biology, University of South Dakota, Vermillion, SD, United States
- 2Department of Biochemistry and Molecular Biology, University of Nevada, Reno, Reno, NV, United States
This perspective paper explores the utilization of abiotic stress-responsive transcription factors (TFs) from crassulacean acid metabolism (CAM) plants to improve abiotic stress tolerance in crop plants. CAM is a specialized type of photosynthetic adaptation that enhances water-use efficiency (WUE) by shifting CO2 uptake to all or part of the nighttime when evaporative water losses are minimal. Recent studies have shown that TF-based genetic engineering could be a useful approach for improving plant abiotic stress tolerance because of the role of TFs as master regulators of clusters of stress-responsive genes. Here, we explore the use of abiotic stress-responsive TFs from CAM plants to improve abiotic stress tolerance and WUE in crops by controlling the expression of gene cohorts that mediate drought-responsive adaptations. Recent research has revealed several TF families including AP2/ERF, MYB, WRKY, NAC, NF-Y, and bZIP that might regulate water-deficit stress responses and CAM in the inducible CAM plant Mesembryanthemum crystallinum under water-deficit stress-induced CAM and in the obligate CAM plant Kalanchoe fedtschenkoi. Overexpression of genes from these families in Arabidopsis thaliana can improve abiotic stress tolerance in A. thaliana in some instances. Therefore, we propose that TF-based genetic engineering with a small number of CAM abiotic stress-responsive TFs will be a promising strategy for improving abiotic stress tolerance and WUE in crop plants in a projected hotter and drier landscape in the 21st-century and beyond.
Introduction
Formidable challenges facing humankind include a burgeoning global human population (Godfray et al., 2010; Gerland et al., 2014) and the increasing frequency and intensity of droughts related to global warming (Cook et al., 2014; Singh et al., 2015). In addition, abiotic stresses including high salinity, temperature extremes, increased UV radiation, heavy metals, and high light intensities are and will continue to be major constraints for global crop production and food security (Lesk et al., 2016). Among these abiotic stresses, drought is of major concern as it has dire effects on crop productivity (Fahad et al., 2017), plant growth, and development (Yordanov et al., 2000). By the end of the 21st century, rapid changes in the global climate will likely increase the frequencies of drought by more than 20% compared to current rates (Lobell et al., 2011; Cook et al., 2014). Indeed, Daryanto et al. (2016) showed that an approximately 40% decrease in water availability can decrease wheat (Triticum aestivum L.) and maize (Zea mays L.) yields by 20.6% and 39.3%, respectively. High salinity, another abiotic stress that is harmful to crop production, affects 20% of total cultivated and 33% of irrigated agricultural lands worldwide (Shrivastava and Kumar, 2015). Similarly, low temperatures and heatwaves cause significant reduction in crop yields across the world (Sanghera et al., 2011; Challinor et al., 2014; Hatfield and Prueger, 2015). Thus, novel approaches to mitigate the negative impacts of abiotic stresses on crop yields must be explored and developed to avoid socio-economic collapse due to climate change.
Approaches to enhance sustainable bioenergy production by engineering the CAM pathway into C3 crops to enhance their water-use efficiency (WUE) on marginal lands are already underway (Borland et al., 2014, 2015; Yang et al., 2015; Liu et al., 2018). Another approach to enhance abiotic stress tolerance is to modulate the expression of transcription factors (TFs) or the functions of abiotic stress-adaptive genes that might already be present, but that are not normally expressed in unstressed or C3 plants (Hussain et al., 2011; Rabara et al., 2014; Joshi et al., 2016; Wang et al., 2016; Bechtold, 2018). This approach would involve bioengineering a small number of regulatory genes with potentially global effects made possible by the role of TFs in gene regulation (Rabara et al., 2014; Joshi et al., 2016).
Transcription factor-based genetic engineering could direct such regulatory TFs to modulate a large number of downstream abiotic stress-responsive genes (Rabara et al., 2014). Stress tolerance in plants is generally under polygenic control (Tran et al., 2010) and some of the genes regulating stress-tolerance responses happen to code for TFs (Villalobos et al., 2004; Qiu and Yu, 2009; Zhang et al., 2011; Yang et al., 2012; Cai et al., 2014; Chen et al., 2014; Swain et al., 2017). Therefore, TFs might be ideal candidate regulators for improving abiotic stress tolerance in crop plants. To date, TF-based genetic engineering has mainly repurposed TFs from Arabidopsis thaliana, Glycine max, Oryza sativa, and T. aestivum (Rabara et al., 2014; Joshi et al., 2016; Wang et al., 2016). As far as we know, no reports have analyzed the effects of overexpressing TFs from crassulacean acid metabolism (CAM) plants which have greater abiotic stress tolerances than mesophytes. Most CAM plants are naturally adapted to low-water environments and many other abiotic stresses compared to the agronomically important C3 plants (Borland et al., 2009). Here, we consider using a “next-generation TF-based” approach to exploit abiotic stress-responsive TFs from CAM plants to improve abiotic stress tolerance in crop plants. The current technologies for TF-based approaches to improve plant abiotic stress tolerance have been extensively discussed and reviewed by Rabara et al. (2014), Joshi et al. (2016), and Wang et al. (2016), and those details are therefore only briefly summarized below.
Transcription Factor-Based Approach
Plants are sessile organisms that exhibit various biochemical, physiological, and molecular adaptations to extreme environments (Joshi et al., 2016). For instance, water-deficit stress activates the expression of stress-responsive genes encoding enzymes that synthesize compatible protective sugars, antioxidants, and proteins, including heat shock proteins and some classes of late embryogenesis abundant (LEA) proteins (Tran et al., 2010; Joshi et al., 2016; Wang et al., 2016). In addition to the stress-induced upregulation of the above proteins, the expression of various regulatory proteins including TFs, protein kinases, and protein phosphatases is also activated (Wang et al., 2016).
Transcription factors are master regulators of many cellular processes and can also interact with other transcriptional regulators (Joshi et al., 2016). Importantly, they play a pivotal role in different abiotic stress responses by binding to the upstream cis-regions of promoters in many stress-responsive genes (Yamaguchi-Shinozaki and Shinozaki, 2006). Many studies have been conducted to identify and characterize families of TFs including AP2/ERFBP, MYB, WRKY, NAC, NF-Y, and bZIP that are involved in abiotic stress responses (Umezawa et al., 2006; Golldack et al., 2011; Leyva-González et al., 2012; Wang et al., 2016; Swain et al., 2017; Zanetti et al., 2017). Several TFs have already been overexpressed in crop plants and A. thaliana to improve abiotic stress tolerance (Qiu and Yu, 2009; Zhang et al., 2011; Yang et al., 2012; Cai et al., 2014; Chen et al., 2014). For example, the NAC family is one of the largest TF families in plants and is involved not only in plant growth and development, but also in transcriptional reprogramming associated with plant stress responses (Tran et al., 2010; Nakashima et al., 2012). Mao et al. (2012) reported that overexpression of the TaNAC2 gene from wheat can enhance tolerance to drought, salt, and freezing stresses in A. thaliana. In addition, functional characterization of the NAC045 (Zheng et al., 2009) and SNAC1 genes from O. sativa enhanced drought and salt tolerance in rice (Hu et al., 2006). Furthermore, overexpression of either GmMYB76 or GmMYB177 from soybean significantly enhanced salt and freezing tolerance in A. thaliana (Liao et al., 2008).
A relatively less explored yet high-potential approach is to discover novel abiotic stress-adaptive regulatory genes in extremophytes (i.e., CAM xerophytes and halophytes, desiccation-tolerant plants, or resurrection plants) to use for bioengineering abiotic stress tolerance in crop plants (Inan et al., 2004; Shi et al., 2013; Joshi et al., 2016; Bechtold, 2018). For example, the overexpression of the TF CpMYB10 from the resurrection plant Craterostigma plantagineum in A. thaliana led to desiccation and salt tolerance in transgenic lines (Villalobos et al., 2004).
Crassulacean Acid Metabolism and Abiotic Stress Tolerance
CAM plants have evolved a specialized type of photosynthetic adaptation that allows them to live under conditions of severe water deficit and in semi-arid and arid regions of the world including deserts. These plants have shifted all or part of their primary CO2 uptake and fixation to the nighttime, when evaporative water losses are minimal, and perform C3 carboxylation reactions when stomata are closed during the daytime. This temporal separation of carbon fixation leads to the formation of the four-carbon organic acid malate, which is stored in the vacuole during the night and subsequently undergoes decarboxylation to release CO2 for re-fixation during the day to produce carbohydrates (Borland et al., 2009). Because of this temporal separation of carbon fixation and inverted stomatal behavior, CAM plants can reduce water loss due to transpiration. These characteristics also allow CAM plants to fix net CO2 15% more efficiently than C3 plants (Nobel, 1991) resulting in increased biomass of CAM plants while using less water than C3 plants. Additionally, CAM plants can produce similar amounts of biomass using 80% less water in comparison to C3 plants (Nobel, 1996; Borland et al., 2009). Thus, CAM plants have between 3- and 6-fold higher WUE than C4 and C3 plants, respectively (Garcia et al., 2014; Yang et al., 2015).
In addition to their higher WUE and associated drought tolerance (Yang et al., 2017), CAM plants can tolerate high temperature up to 70°C, whereas C3 plants can tolerate only 50–55°C (Borland et al., 2009). CAM halophytes can also adapt to high salinity, as during the induction of CAM by salt stress in Mesembryanthemum crystallinum (Winter and Holtum, 2014). Moreover, CAM plants can better tolerate higher light intensities (>1000 μmol m-2 s-1) and UV-B irradiation levels than can agronomically important C3 plants (Borland et al., 2009). Furthermore, CAM plants can increase daily net CO2 uptake under increased atmospheric CO2 concentrations, which might be advantageous in global climate change scenarios (Nobel, 1996). Some CAM plants such as Agave salmiana, Opuntia ficus-indica, and Stenocereus queretaroensis can also survive in subzero temperatures, and Agave utahensis Engelm. can tolerate temperatures as low as -18°C (Nobel, 1996).
The abiotic stress-adaptive characteristics of CAM plants will be particularly beneficial for adapting to the consequences of anthropogenic climate change, such as droughts and heatwaves, high soil salinity, temperature extremes, and high light or UV-B irradiation. Many of the stress-adaptive responses involve abscisic acid (ABA)-dependent and -independent response pathways (Song et al., 2016). ABA-dependent and independent signaling events likely participate in the stress-activation of CAM in M. crystallinum (Chu et al., 1990; Taybi and Cushman, 1999, 2002; Abdin et al., 2002; Cushman and Borland, 2002), suggesting that the CAM pathway likely has evolved in response to abiotic stress (Reyes-García and Andrade, 2009; Heyduk et al., 2018; Yin et al., 2018).
Crassulacean acid metabolism is thought to have evolved independently multiple times from ancestral C3 plants (Silvera et al., 2010) because no unique metabolic pathways are required, although some CAM-specific variant enzymes apparently evolved in some instances (Ermolova et al., 2003; Gehrig et al., 2005; Vaasen et al., 2006). However, temporal changes in gene expression of the CAM enzyme variants likely occurred because of alterations in their regulation compared to their orthologs in C3 plants (Hermans and Westhoff, 1990; Lepiniec et al., 1993; Cushman et al., 2008; Heyduk et al., 2018; Yin et al., 2018). Furthermore, the ABA-dependent stress response pathway is involved in CAM activation not only in M. crystallinum (Taybi and Cushman, 2002), but also in other CAM species (Taybi et al., 1995; Rodrigues et al., 2016; Yin et al., 2018). Although CAM is found in over 36 families of vascular plants (Silvera et al., 2010), we rely on only a few major CAM species such as pineapple (Ananas comosus), Agave, and Opuntia as agricultural crops to provide food, forage, fiber, and biofuels (Cushman et al., 2015). Well characterized CAM model species also provide abundant resources for the identification and selection of candidate TFs involved in abiotic stress adaptations (Hartwell et al., 2016). Hence, identification of candidate CAM pathway regulators (i.e., TFs) that are expressed or activated under water-deficit stress or CAM should be prioritized to exploit the molecular and regulatory machinery of abiotic stress adaptation in CAM plants as a vital resource for applications in C3 crop species (Yang et al., 2015; Fernie, 2016; Yin et al., 2018).
Cam Abiotic Stress-Responsive TF-Based Approach
Bioengineering a TF that can confer desirable traits such as increased drought tolerance (Villalobos et al., 2004) or increased biomass (Lim et al., 2018) into C3 A. thaliana will be crucial as a proof of concept for the CAM abiotic stress-responsive TF-based approach to increase abiotic stress tolerance in C3 plants. Fortunately, genetic resources (i.e., genome and transcriptome sequences) for CAM plants are now available for Agave (Abraham et al., 2016), Kalanchoe spp. (Yang et al., 2017), pineapple (Ming et al., 2015, 2016; Wai et al., 2017), and M. crystallinum (Chiang et al., 2016). Although genetic resources for CAM plants are becoming readily available, the underlying regulatory basis of CAM is still not completely understood. Many TFs of unknown function have been identified during recent genome and transcriptome sequencing efforts; thus, there are now many opportunities to analyze the functions of TFs involved in water-deficit-stress response or CAM function and to exploit the potential of bioengineering using CAM plant TFs to improve abiotic stress tolerance in crop plants. Indeed, candidate CAM TFs involved in C3 to CAM transition in obligate CAM species of Agave (Heyduk et al., 2018; Huang et al., 2018; Yin et al., 2018), Kanlanchoe (Moseley et al., 2018), and Manfreda (Heyduk et al., 2018), and weak CAM species of Polianthes and Beschorneria (Heyduk et al., 2018), or the induction of CAM in Tralinum triangulare (Brilhaus et al., 2016) have been identified. Not surprisingly, a number of these candidate TFs are involved in the ABA stress responsive pathway (Heyduk et al., 2018; Yin et al., 2018). More importantly though, many of these candidate TFs are involved in the rewiring of the phase shift from C3 to CAM transition in the evolution of CAM photosynthesis (Heyduk et al., 2018; Moseley et al., 2018; Yin et al., 2018). Although it would be interesting to attempt to reprogram a C3 plant such that it becomes CAM performing, we are not suggesting to shift gene expression patterns of CAM pathway genes that might be present in extant C3 plants, or to regulate the engineered CAM pathway in C3 plants (Yang et al., 2015; Fernie, 2016; Heyduk et al., 2018; Yin et al., 2018), but rather identify and exploit the TFs involved in abiotic stress responses from obligate and inducible CAM plants to modulate the expression of appropriate genes in C3 plants to improve their abiotic stress tolerance.
Eight most abundant candidate TFs under water-deficit stress diel and zeitgeber time have been identified that might regulate the CAM state, water-deficit stress response, or both in M. crystallinum (Garcia et al., 2014; Cushman, unpubl. data; Table 1). Mesembryanthemum crystallinum switches from C3 to CAM when salt or water-deficit stressed (Cushman, 2001; Cushman and Borland, 2002; Taybi and Cushman, 2002). Kalanchoe fedtschenkoi orthologs of these top candidate TFs were also highly expressed during CAM induction in older leaf pairs of K. fedtschenkoi plants (Garcia et al., 2014; Cushman, unpubl. data). It is known that young leaves are C3 performing, whereas mature leaves are CAM performing in K. fedtschenkoi (Cushman, 2001). These TFs share a base mean expression level of >100 FPKM and at least two-fold induction during a transition from C3 to CAM or imposition of water-deficit stress in K. fedtschenkoi and M. crystallinum, respectively (Garcia et al., 2014; Cushman, unpubl. data). These candidate CAM TFs also belong to the families of TFs reported to be involved in abiotic stress responses (Rabara et al., 2014; Joshi et al., 2016; Roy, 2016; Wang et al., 2016). Orthologs of these candidate CAM TFs in A. thaliana also have several reported functions in plant development and abiotic stress tolerance (Zheng et al., 2009; Zhang et al., 2011; Yang et al., 2012; Chen et al., 2014; Swain et al., 2017; Zanetti et al., 2017). Intriguingly, recent functional characterization of two putative CAM regulators of water-deficit stress response or CAM activation via overexpression in A. thaliana strongly suggest that CAM TFs have high potential to increase tolerance to drought and other abiotic stresses in C3 plants.
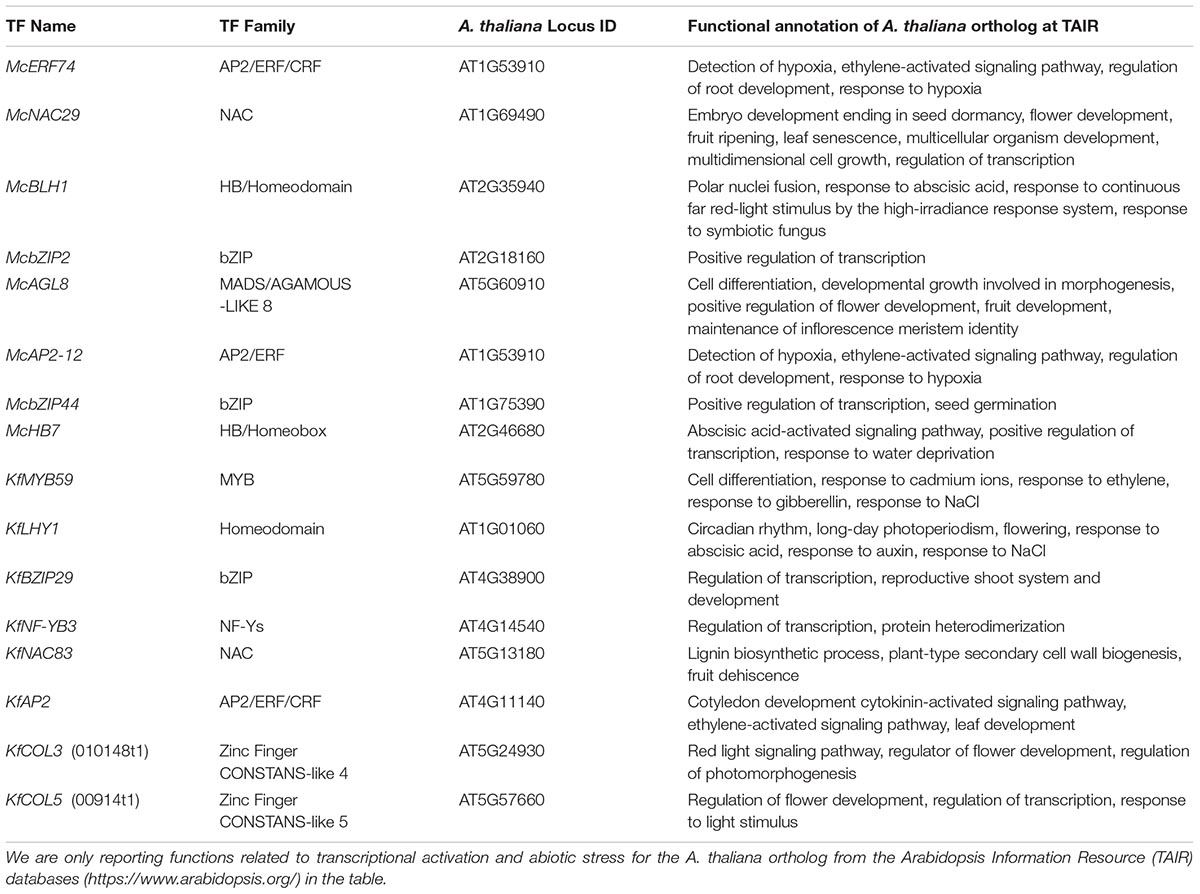
Table 1. List of the top eight candidate transcription factors (TFs) from the inducible CAM plant Mesembryanthemum crystallinum and top eight candidate TFs from the obligate CAM plant Kalanchoe fedtschenkoi hypothesized to regulate the CAM state or water-deficit stress responses in CAM plants and their corresponding orthologs in Arabidopsis thaliana.
One of these candidate CAM TFs is a myeloblastosis (MYB59, closest ortholog in Arabidopsis) TF whose transcripts are 20-fold more abundant in CAM-performing older leaf pairs relative to C3-performing younger leaves in K. fedtschenkoi (Hartwell et al. unpubl. data). Results from four, third-generation (T3) transgenic lines carrying the KlMYB59 indicate increased rosette size and biomass at 4-week-old juvenile stage, and increased shoot length at 8-week-old mature stage in transgenic plants compared to WT (Table 2; Wone et al., unpubl. data; full results being presented in a separate publication). However, transgenic lines show delayed flowering in long-day photoperiod compared to WT plants (16 h light/8 h dark). In addition, these transgenic lines exhibit increased integrated WUE compared to WT plants. Furthermore, transgenic lines have longer primary roots despite exposure to 50 μM selenium compared to WT plants. In addition to MYB59, transcripts of the NAC83 TF (closest ortholog in Arabidopsis) were also more highly expressed in CAM-performing leaves of K. laxiflora and K. fedtschenkoi relative to C3-performing leaves (Cushman et al., unpubl. data). The function of this K. fedtschenkoi NAC83 TF (KfNAC83) is not known in CAM- or C3-performing CAM plants, but its A. thaliana ortholog suggests roles in abiotic stress responses and development (Table 1). Functional characterization of KfNAC83 shows enhanced water-deficit stress tolerance and increased integrated WUE in four independent transgenic T3-generation A. thaliana lines compared to WT plants (Table 2; Wone et al., unpubl. data; full results being presented in a separate publication). Furthermore, KfNAC83-overexpressing lines show significantly increased rosette size, leaves in the mature rosette, shoot biomass, number of siliques, and lateral roots compared to WT. Interestingly, these transgenic lines also showed tolerance to 150 mM NaCl. Collectively, our characterization results strongly suggest that at least two of these candidate CAM TFs have functions in abiotic stress responses and CAM photosynthesis.

Table 2. Results from T3 transgenic A. thaliana lines overexpressing candidate CAM TFs from the obligate CAM plant K. fedtschenkoi hypothesized to regulate water-deficit stress response or CAM activation.
Conclusion
The abiotic stress-adaptive features of CAM plants provide a wealth of genetic resources, specifically TFs, that are now available for functional testing and possible improvement of WUE and abiotic stress responses in C3 photosynthesis plants. Our recent findings strongly suggest that a bioengineering approach using CAM abiotic stress-responsive TFs has the potential to increase abiotic stress tolerance in A. thaliana and possibly in C3 crop plants. Our results indicate that CAM abiotic stress-responsive gene expression can be modulated by the appropriate CAM TFs to generate stress-adaptive phenotypes in A. thaliana and likely other C3 plants because these CAM abiotic stress-responsive genes are apparently conserved and present in C3 plants (Heyduk et al., 2018). Furthermore, although K. fedtschenkoi is distantly related to A. thaliana, transgenic A. thaliana lines carrying the obligate CAM plants’ TFs showed favorable features for translational applications. We are optimistic that overexpressing TFs from the inducible CAM halophyte, M. crystallinum will have similar favorable responses in A. thaliana lines. Co-overexpression of only a small number of obligate and/or inducible CAM plant abiotic stress-responsive TFs with demonstrated abiotic stress-adaptive or -responsive functions would provide a facile approach for bioengineering desirable responses to abiotic stress (Song et al., 2016). Such an approach could open the door to potentially transformative applications to ensure long-term sustainable food, fiber, feed, and fuel production in a projected hotter and drier landscape in the 21st century and beyond.
Author Contributions
BWMW conceived the CAM abiotic stress-responsive TF-based approach to improve C3 crop plant abiotic stress responses. AA, KR, and BWMW wrote the manuscript. AA, KR, and BW conducted the experiments and provided the CAM plant TF overexpression data. WY, TG, and JC provided the CAM plant TF sequences. All authors reviewed the final manuscript.
Funding
This work was funded by South Dakota Board of Regents Competitive Research Grant #A18-0015-001 to BWMW and was supported, in part, by the Department of Energy, Office of Science, Genomic Science Program under Award Number DE-SC0008834 to JC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mary Ann Cushman for her helpful and clarifying comments on the manuscript. We also thank Austin Hacck and Savannah Aanderud for their assistance in maintaining the transgenic lines of Arabidopsis plants.
References
Abdin, M., Rehman, R., Israr, M., Srivastava, P. S., and Bansal, K. C. (2002). Abiotic stress related genes and their role in conferring resistance in plants. Indian J. Biotech. 1, 225–244.
Abraham, P. E., Yin, H., Borland, A. M., Weighill, D., Lim, S. D., De Paoli, H. C., et al. (2016). Transcript, protein and metabolite temporal dynamics in the CAM plant agave. Nat. Plants 2:16178. doi: 10.1038/nplants.2016.178
Bechtold, U. (2018). Plant life in extreme environments: how do you improve drought tolerance? Front. Plant Sci. 9:543. doi: 10.3389/fpls.2018.00543
Borland, A. M., Griffiths, H., Hartwell, J., and Smith, J. A. C. (2009). Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands. J. Exp. Bot. 60, 2879–2896. doi: 10.1093/jxb/erp118
Borland, A. M., Hartwell, J., Weston, D. J., Schlauch, K. A., Tschaplinski, T. J., Tuskan, G. A., et al. (2014). Engineering crassulacean acid metabolism to improve water-use efficiency. Trends Plant Sci. 19, 327–338. doi: 10.1016/j.tplants.2014.01.006
Borland, A. M., Wullschleger, S. D., Weston, D. J., Tuskan, G. A., Hartwell, J., Yang, X., et al. (2015). Climate-resilient agroforestry: physiological responses to climate change and engineering of crassulacean acid metabolism (CAM) as a mitigation strategy. Plant Cell Environ. 38, 1833–1849. doi: 10.1111/pce.12479
Brilhaus, D., Bräutigam, A., Mettler-Altmann, T., Winter, K., and Weber, A. P. M. (2016). Reversible burst of transcriptional changes during induction of crassulacean acid metabolism in Talinum triangulare. Plant Physiol. 170, 102–122. doi: 10.1104/pp.15.01076
Cai, R., Zhao, Y., Wang, Y., Lin, Y., Peng, X., Li, Q., et al. (2014). Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell Tissue Organ. Cult. 119, 565–577. doi: 10.1007/s11240-014-0556-7
Challinor, A. J., Watson, J., Lobell, D. B., Howden, S. M., Smith, D. R., and Chhetri, N. (2014). A meta-analysis of crop yield under climate change and adaptation. Nat. Clim. Chang. 4, 287–291. doi: 10.1038/nclimate2153
Chen, X., Wang, Y., Lv, B., Li, J., Luo, L., Lu, S., et al. (2014). The NAC family transcription factor OsNAP confers abiotic stress response through the aba pathway. Plant Cell Physiol. 55, 604–619. doi: 10.1093/pcp/pct204
Chiang, C.-P., Yim, W. C., Sun, Y.-H., Ohnishi, M., Mimura, T., Cushman, J. C. et al. (2016). Identification of ice plant (Mesembryanthemum crystallinum L.) MicroRNAs using RNA-Seq and their putative roles in high salinity responses in seedlings. Front. Plant Sci. 7:1143. doi: 10.3389/fpls.2016.01143
Chu, C., Dai, Z., Ku, M. S. B., and Edwards, G. E. (1990). Induction of crassulacean acid metabolism in the facultative halophyte Mesembryanthemum crystallinum by abscisic acid. Plant Physiol. 93, 1253–1260. doi: 10.1104/pp.93.3.1253
Cook, B. I., Smerdon, J. E., Seager, R., and Coats, S. (2014). Global warming and 21st century drying. Clim. Dyn. 43, 2607–2627. doi: 10.1007/s00382-014-2075-y
Cushman, J. C. (2001). Crassulacean acid metabolism. A plastic photosynthetic adaptation to arid environments. Plant Physiol. 127, 1439–1448. doi: 10.1104/pp.010818
Cushman, J. C., Agarie, S., Albion, R. L., Elliot, S. M., Taybi, T., and Borland, A. M. (2008). Isolation and characterization of mutants of common ice plant deficient in crassulacean acid metabolism. Plant Physiol. 147, 228-238. doi: 10.1104/pp.108.116889
Cushman, J. C., and Borland, A. M. (2002). Induction of Crassulacean acid metabolism by water limitation. Plant Cell Environ. 25, 295–310. doi: 10.1046/j.0016-8025.2001.00760.x
Cushman, J. C., Davis, S. C., Yang, X., and Borland, A. M. (2015). Development and use of bioenergy feedstocks for semi-arid and arid lands. J. Exp. Bot. 66, 4177–4193. doi: 10.1093/jxb/erv087
Daryanto, S., Wang, L., and Jacinthe, P. A. (2016). Global synthesis of drought effects on maize and wheat production. PLoS One 11:e0156362. doi: 10.1371/journal.pone.0156362
Ermolova, N. V., Ann Cushman, M., Taybi, T., Condon, S. A., Cushman, J. C., and Chollet, R. (2003). Expression, purification, and initial characterization of a recombinant form of plant PEP-carboxylase kinase from CAM-induced Mesembryanthemum crystallinum with enhanced solubility in Escherichia coli. Protein Expr. Purif. 29, 123–131. doi: 10.1016/S1046-5928(03)00014-7
Fahad, S., Bajwa, A. A., Nazir, U., Anjum, S. A., Farooq, A., Zohaib, A., et al. (2017). Crop production under drought and heat stress: plant responses and management options. Front. Plant Sci. 8:1147. doi: 10.3389/fpls.2017.01147
Fernie, A. R. (2016). Systems biology: a new CAM era. Nat. Plants 2:16190. doi: 10.1038/nplants.2016.190
Garcia, T. M., Heyduk, K., Kuzmick, E., and Mayer, J. A. (2014). Crassulacean acid metabolism biology. New Phytol. 204, 738–740. doi: 10.1111/nph.13127
Gehrig, H. H., Wood, J., Ann Cushman, M., Virgo, A. C., Cushman, J., and Winter, K. (2005). Research note: large gene family of phosphoenolpyruvate carboxylase in the crassulacean acid metabolism plant Kalanchoe pinnata (Crassulaceae) characterised by partial cDNA sequence analysis. Funct. Plant Biol. 32, 467–472. doi: 10.1071/FP05079
Gerland, P., Raftery, A. E., Ševèíková, H., Li, N., Gu, D., Spoorenberg, T., et al. (2014). World population stabilization unlikely this century. Science 346, 234–237. doi: 10.1126/science.1257469
Godfray, H. C. J., Beddington, J. R., Crute, I. R., Haddad, L., Lawrence, D., Muir, J. F., et al. (2010). Food security: the challenge of feeding 9 billion people. Science 327, 812–818. doi: 10.1126/science.1185383
Golldack, D., Luking, I., and Yang, O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30, 1383–1391. doi: 10.1007/s00299-011-1068-0
Hartwell, J., Dever, L. V., and Boxall, S. F. (2016). Emerging model systems for functional genomics analysis of crassulacean acid metabolism. Curr. Opin. Plant Biol. 31, 100–108. doi: 10.1016/j.pbi.2016.03.019
Hatfield, J. L., and Prueger, J. H. (2015). Temperature extremes: effect on plant growth and development. Weather Clim. Extrem. 10, 4–10. doi: 10.1016/j.wace.2015.08.001
Hermans, J., and Westhoff, P. (1990). Analysis of expression and evolutionary relationships of phosphoenol-pyruvate carboxylase genes in Flaveria trinervia (C4) and F. pringlei (C3). Mol. Gen. Genet. 224, 459–468. doi: 10.1007/BF00262441
Heyduk, K., Ray, J. N., Ayyampalayam, S., and Leebens-Mack, J. (2018). Shifts in gene expression profiles are associated with weak and strong crassulacean acid metabolism. Am. J. Bot. 105, 587–601. doi: 10.1002/ajb2.1017
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 12987–12992. doi: 10.1073/pnas.0604882103
Huang, X., Wang, B., Xi, J., Zhang, Y., He, C., Zheng, J., et al. (2018). Transcriptome comparison reveals distinct selection patterns in domesticated and wild agave species, the important cam plants. Int. J. Genomics 2018, 1–12. doi: 10.1155/2018/5716518
Hussain, S. S., Kayani, M. A., and Amjad, M. (2011). Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol. Prog. 27, 297–306. doi: 10.1002/btpr.514
Inan, G., Zhang, Q., Li, P., Wang, Z., Cao, Z., Zhang, H., et al. (2004). Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol. 135, 1718–1737. doi: 10.1104/pp.104.041723
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7:1029. doi: 10.3389/fpls.2016.01029
Lepiniec, L., Keryer, E., Philippe, H., Gadal, P., and Cretin, C. (1993). Sorghum phosphoenolpyruvate carboxylase gene family: structure, function and molecular evolution. Plant Mol. Biol. 21, 487–502. doi: 10.1007/BF00028806
Lesk, C., Rowhani, P., and Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Leyva-González, M. A., Ibarra-Laclette, E., Cruz-Ramírez, A., and Herrera-Estrella, L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One 7:e48138. doi: 10.1371/journal.pone.0048138
Liao, Y., Zou, H. F., Wang, H. W., Zhang, W. K., Ma, B., Zhang, J. S., et al. (2008). Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 18, 1047–1060. doi: 10.1038/cr.2008.280
Lim, S. D., Yim, W. C., Liu, D., Hu, R., Yang, X., and Cushman, J. C. (2018). A Vitis vinifera basic helix-loop-helix transcription factor enhances plant cell size, vegetative biomass and reproductive yield. Plant Biotechnol. J. 16, 1595–1615. doi: 10.1111/pbi.12898
Liu, D., Palla, K. J., Hu, R., Moseley, R. C., Mendoza, C., Chen, M., et al. (2018). Perspectives on the basic and applied aspects of crassulacean acid metabolism (CAM) research. Plant Sci. 274, 394–401. doi: 10.1016/j.plantsci.2018.06.012
Lobell, D. B., Schlenker, W., and Costa-Roberts, J. (2011). Climate trends and global crop production since 1980. Science 333, 616–620. doi: 10.1126/science.1204531
Mao, X., Zhang, H., Qian, X., Li, A., Zhao, G., and Jing, R. (2012). TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63, 2933–2946. doi: 10.1093/jxb/err462
Ming, R., VanBuren, R., Wai, C. M., Tang, H., Schatz, M. C., Bowers, J. E., et al. (2015). The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 47, 1435–1442. doi: 10.1038/ng.3435
Ming, R., Wai, C. M., and Guyot, R. (2016). Pineapple genome: a reference for monocots and CAM photosynthesis. Trends Genet. 32, 690–696. doi: 10.1016/j.tig.2016.08.008
Moseley, R. C., Mewalal, R., Motta, F., Tuskan, G. A., Haase, S., and Yang, X. (2018). Conservation and diversification of circadian rhythmicity between a model crassulacean acid metabolism plant Kalanchoë fedtschenkoi and a model C3 photosynthesis plant Arabidopsis thaliana. Front. Plant Sci. 9:1757. doi: 10.3389/fpls.2018.01757
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nobel, P. S. (1991). Achievable productivities of certain CAM plants: basis for high values compared with C3 and C4 plants. New Phytol. 119, 183–205. doi: 10.1111/j.1469-8137.1991.tb01022.x
Nobel, P. S. (1996). Responses of some North American CAM plants to freezing temperatures and doubled CO2 concentrations: implications of global climate change for extending cultivation. J. Arid Environ. 34, 187–196. doi: 10.1006/jare.1996.0100
Qiu, Y., and Yu, D. (2009). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exper. Bot. 65, 35–47. doi: 10.1016/j.envexpbot.2008.07.002
Rabara, R. C., Tripathi, P., and Rushton, P. J. (2014). The potential of transcription factor-based genetic engineering in improving crop tolerance to drought. Omics 18, 601–614. doi: 10.1089/omi.2013.0177
Reyes-García, C., and Andrade, J. (2009). Crassulacean acid metabolism under global climate change. New Phytol. 181, 754–757. doi: 10.1111/j.1469-8137.2009.02762.x
Rodrigues, M. A., Hamachi, L., Mioto, P. T., Purgatto, E., and Mercier, H. (2016). Implications of leaf ontogeny on drought-induced gradients of CAM expression and ABA levels in rosettes of the epiphytic tank bromeliad Guzmania monostachia. Plant Physiol. Biochem. 1, 400–411. doi: 10.1016/j.plaphy.2016.08.010
Roy, S. (2016). Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 11:e1117723. doi: 10.1080/15592324.2015.1117723
Sanghera, G. S., Wani, S. H., Hussain, W., and Singh, N. B. (2011). Engineering cold stress tolerance in crop plants. Curr. Genomics 12, 30–43. doi: 10.2174/138920211794520178
Shi, Y., Yan, X., Zhao, P., Yin, H., Zhao, X., Xiao, H., et al. (2013). transcriptomic analysis of a tertiary relict plant, extreme xerophyte Reaumuria soongorica to identify genes related to drought adaptation. PLoS One 8:e63993. doi: 10.1371/journal.pone.0063993
Shrivastava, P., and Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Silvera, K., Neubig, K., Whitten, W., Williams, N., Winter, K., and Cushman, J. C. (2010). Evolution along the crassulacean acid metabolism continuum. Funct. Plant Biol. 37, 995–1010. doi: 10.1071/FP10084
Singh, B., Bohra, A., Mishra, S., Joshi, R., and Pandey, S. (2015). Embracing new-generation ‘omics’ tools to improve drought tolerance in cereal and food-legume crops. Biol. Plant 59, 413–428. doi: 10.1007/s10535-015-0515-0
Song, L., Huang, S. C., Wise, A., Castanon, R., Nery, J. R., Chen, H., et al. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354, aag1550. doi: 10.1126/science.aag1550
Swain, S., Myers, Z. A., Siriwardana, C. L., and Holt, B. F. III (2017). The multifaceted roles of NUCLEAR FACTOR-Y in Arabidopsis thaliana development and stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 636–644. doi: 10.1016/j.bbagrm.2016.10.012
Taybi, T., and Cushman, J. C. (1999). Signaling events leading to crassulacean acid metabolism (CAM) induction in the common ice plant, Mesembryanthemum crystallinum. Plant Physiol. 121, 545–555. doi: 10.1104/pp.121.2.545
Taybi, T., and Cushman, J. C. (2002). Abscisic acid signaling and protein synthesis requirements for phosphoenolpyruvate carboxylase transcript induction in the common ice plant. J. Plant Physiol. 159, 1235–1243. doi: 10.1078/0176-1617-00834
Taybi, T., Sotta, B., Gehrig, H., Güclü, S., Kluge, M., and Brulfert, J. (1995). Differential effects of abscisic acid on phosphoenolpyruvate carboxylase and CAM operation in Kalanchoë blossfeldiana. Botanica Acta 108, 240–246. doi: 10.1111/j.1438-8677.1995.tb00856.x
Tran, L. S., Nishiyama, R., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2010). Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1, 32–39. doi: 10.4161/gmcr.1.1.10569
Umezawa, T., Fujita, M., Fujita, Y., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17, 113–122. doi: 10.1016/j.copbio.2006.02.002
Vaasen, A., Begerow, D., and Hampp, R. (2006). Phosphoenolpyruvate carboxylase genes in C3, crassulacean acid metabolism (CAM) and C3/CAM intermediate species of the genus Clusia: rapid reversible C3/CAM switches are based on the C3 housekeeping gene. Plant Cell Environ. 29, 2113–2123. doi: 10.1111/j.1365-3040.2006.01583.x
Villalobos, M. A., Bartels, D., and Iturriaga, G. (2004). Stress tolerance and glucose insensitive phenotypes in Arabidopsis overexpressing the CpMYB10 transcription factor gene. Plant Physiol. 135, 309–324. doi: 10.1104/pp.103.034199
Wai, C. M., VanBuren, R., Zhang, J., Huang, L., Miao, W., Edger, P. P., et al. (2017). Temporal and spatial transcriptomic and microRNA dynamics of CAM photosynthesis in pineapple. Plant J. 92, 19–30. doi: 10.1111/tpj.13630
Wang, H., Wang, H., Shao, H., and Tang, X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7:67. doi: 10.3389/fpls.2016.00067
Winter, K., and Holtum, J. A. (2014). Facultative crassulacean acid metabolism (CAM) plants: powerful tools for unravelling the functional elements of CAM photosynthesis. J. Exp. Bot. 65, 3425–3441. doi: 10.1093/jxb/eru063
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, A., Dai, X., and Zhang, W.-H. (2012). A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. 63, 2541–2556. doi: 10.1093/jxb/err431
Yang, X., Cushman, J. C., Borland, A. M., Edwards, E. J., Wullschleger, S. D., Tuskan, G. A., et al. (2015). A roadmap for research on crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world. New Phytol. 207, 491–504. doi: 10.1111/nph.13393
Yang, X., Hu, R., Yin, H., Jenkins, J., Shu, S., Tang, H., et al. (2017). The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat. Commun. 8:1899. doi: 10.1038/s41467-017-01491-7
Yin, H., Guo, H. B., Weston, D. J., Borland, A. M., Ranjan, P., Abraham, P. E., et al. (2018). Diel rewiring and positive selection of ancient plant proteins enabled evolution of CAM photosynthesis in Agave. BMC Genomics 19:588. doi: 10.1186/s12864-018-4964-7
Yordanov, I., Velikova, V., and Tsonev, T. (2000). Plant responses to drought, acclimation, and stress tolerance. Photosynthetica 38, 171–186. doi: 10.1023/a:1007201411474
Zanetti, M. E., Ripodas, C., and Niebel, A. (2017). Plant NF-Y transcription factors: key players in plant-microbe interactions, root development and adaptation to stress. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 645–654. doi: 10.1016/j.bbagrm.2016.11.007
Zhang, X., Wang, L., Meng, H., Wen, H., Fan, Y., and Zhao, J. (2011). Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 75, 365–378. doi: 10.1007/s11103-011-9732-x
Keywords: abiotic stress response, crassulacean acid metabolism, drought tolerance, extremophytes, genetic engineering, transcription factor
Citation: Amin AB, Rathnayake KN, Yim WC, Garcia TM, Wone B, Cushman JC and Wone BWM (2019) Crassulacean Acid Metabolism Abiotic Stress-Responsive Transcription Factors: a Potential Genetic Engineering Approach for Improving Crop Tolerance to Abiotic Stress. Front. Plant Sci. 10:129. doi: 10.3389/fpls.2019.00129
Received: 29 September 2018; Accepted: 25 January 2019;
Published: 22 February 2019.
Edited by:
Wolfram Weckwerth, University of Vienna, AustriaReviewed by:
Dong-Ha Oh, Louisiana State University, United StatesDouglas S. Domingues, São Paulo State University (UNESP), Brazil
Copyright © 2019 Amin, Rathnayake, Yim, Garcia, Wone, Cushman and Wone. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bernard W. M. Wone, YmVybmllLndvbmVAdXNkLmVkdQ==
†Co-first authors
 Atia B. Amin
Atia B. Amin Kumudu N. Rathnayake
Kumudu N. Rathnayake Won C. Yim
Won C. Yim Travis M. Garcia2
Travis M. Garcia2 John C. Cushman
John C. Cushman Bernard W. M. Wone
Bernard W. M. Wone