- 1Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige, Italy
- 2Ethiopian Institute of Agricultural Research, Addis Ababa, Ethiopia
- 3Department of Life Sciences, University of Trieste, Trieste, Italy
- 4CREA Research Centre for Viticulture and Enology, Conegliano, Italy
The development of new resistant varieties to the oomycete Plasmopara viticola (Berk.& Curt) is a promising way to combat downy mildew (DM), one of the major diseases threatening the cultivated grapevine (Vitis vinifera L.). Taking advantage of a segregating population derived from “Merzling” (a mid-resistant hybrid) and “Teroldego” (a susceptible landrace), 136 F1 individuals were characterized by combining genetic, phenotypic, and gene expression data to elucidate the genetic basis of DM resistance and polyphenol biosynthesis upon P. viticola infection. An improved consensus linkage map was obtained by scoring 192 microsatellite markers. The progeny were screened for DM resistance and production of 42 polyphenols. QTL mapping showed that DM resistance is associated with the herein named Rpv3-3 specific haplotype and it identified 46 novel metabolic QTLs linked to 30 phenolics-related parameters. A list of the 95 most relevant candidate genes was generated by specifically exploring the stilbenoid-associated QTLs. Expression analysis of 11 genes in Rpv3-3+/− genotypes displaying disparity in DM resistance level and stilbenoid accumulation revealed significant new candidates for the genetic control of stilbenoid biosynthesis and oligomerization. These overall findings emphasized that DM resistance is likely mediated by the major Rpv3-3 haplotype and stilbenoid induction.
Introduction
Around 68,000 tons of fungicides per year are used in Europe to manage grape diseases, i.e., 65% of all fungicides used in agriculture although viticulture encompasses only 4% of the EU arable land (Eurostat, 2007). A useful strategy to reduce the impact of pesticides on humans, animals and environment is based on genetic improvement, in particular on the introgression of resistance traits from ancestral species into domesticated varieties. The cultivated grapevine (Vitis vinifera L.) is highly susceptible to downy mildew (DM), caused by the biotrophic oomycete Plasmopara viticola (Berk. & M. A. Curtis) Berl. & De Toni, the major disease of temperate-humid climate among various pathogen threats.
A total of 27 Quantitative Trait Loci (QTLs) associated with DM resistance in different genetic backgrounds are known and described (VIVC, 2018). In particular, the major Rpv loci originated from Muscadinia rotundifolia (Merdinoglu et al., 2003), Vitis riparia (Marguerit et al., 2009; Moreira et al., 2011), V. amurensis (Blasi et al., 2011; Schwander et al., 2012; Venuti et al., 2013), V. cinerea (Ochssner et al., 2016), and V. rupestris (Divilov et al., 2018). To close the list, the Rpv3 locus is a major determinant of grapevine DM resistance. Seven conserved Rpv3 haplotypes were identified in five descent groups of resistant varieties and traced back to their founders, which belong to V. rupestris, V. lincecumii, V. riparia, and V. labrusca (Di Gaspero et al., 2012). Until now only two haplotypes at this locus were validated in segregating populations derived from different DM resistance donors (Welter et al., 2007; Zyprian et al., 2016).
The related resistance mechanisms are partially known and are due to gene-for-gene recognition, thus signal cascade and finally defense response. A widespread hot spot of NBS-LRR genes was identified within the genomic region where the Rpv3 locus resides, providing a distinctive advantage for the adaptation of native North American grapevines to resist to P. viticola (Moroldo et al., 2008). The defense response also involves the synthesis of secondary metabolites including the stilbenoids. In fact, besides necrosis (e.g., Boso Alonso and Kassemeyer, 2008; Peressotti et al., 2010) and callose deposition (e.g., Gindro et al., 2003), DM resistance can be accompanied by stilbene accumulation (e.g., Alonso-Villaverde et al., 2011; Malacarne et al., 2011; Mattivi et al., 2011) as activated defense mechanisms. To date no study has investigated the genetic basis of polyphenol, in particular stilbenoid, synthesis or its variation upon P. viticola infection. QTLs associated with synthesis of most polyphenols are not known; only a few research studies on proanthocyanidins, anthocyanins, and flavonol berry composition have recently been attempted in grapevine (Fournier-Level et al., 2009, 2011; Huang et al., 2012, 2013, 2014; Viana et al., 2013; Ban et al., 2014; Azuma et al., 2015; Costantini et al., 2015; Guo et al., 2015; Malacarne et al., 2015).
In this work, we aim to characterize the locus conferring resistance against P. viticola and to identify new polyphenol-related loci in order to shed light on the DM resistance mechanisms in an interspecific segregating population derived from the source “Merzling.'
Materials and Methods
Segregating Population
An interspecific segregating population of 136 putative full-sib individuals derived from the cross between the complex Vitis hybrid “Merzling,” descending from V. vinifera, V. rupestris, and V. lincecumii, and the V. vinifera cv “Teroldego” was studied. This cross between “Merzling,” mid-resistant to DM and high-stilbenoid producer, and “Teroldego,” susceptible to DM and low-stilbenoid producer, was performed at FEM (Edmund Mach Foundation, San Michele all'Adige, Italy) in 1989. During the 2012 growing season the progeny were propagated as grafted plants in 1L-pots filled with soil:sand:peat:vermiculite (3:1:3:3, v/v) in a greenhouse at 25/20°C day/night temperature, with a 16 h photoperiod and relative humidity (RH) of 70 ± 10%. The propagation of the two parental lines was carried out during 2013.
Phenotyping
DM Resistance Assessment
To collect sufficient fresh inoculum, P. viticola propagation was performed on leaves of the susceptible V. vinifera cv “Pinot gris” following the protocol by Vezzulli et al. (2018). Six potted plants (PP) per each progeny individual were maintained in two different growth chambers as three biological replicates for P. viticola- and mock-inoculation, respectively. Firstly, in June 2012 fully expanded leaves of the 10 week-old PP were P. viticola-inoculated (PI) by spraying a suspension of 1 × 105 sporangia/ml onto the abaxial leaf surface and were kept overnight in the dark in a growth chamber at 24°C with 80% RH; mock-inoculated (MI) samples were obtained by analogously spraying distilled water on PP kept under equal conditions. Secondly, in August 2012 the 4–5th leaves from the MI-PP apex of each progeny individual were collected to generate leaf disks (LD) that were inoculated according to Vezzulli et al. (2018). In June and August 2013 the same experiments were respectively repeated on PP and LD of 11 representative progeny individuals along with “Merzling” and “Teroldego” parental lines. Finally, the DM response was evaluated at 8 days post-inoculation (dpi) on PP and at 4, 5, and 6 dpi on LD by means of three parameters evaluated by visual inspection: disease severity (percentage of the disc area showing symptoms of sporulation) and disease incidence (number of discs with sporulation/total number of discs), according to OEPP/EPPO (2001), along with the descriptors OIV 452 for PP or OIV452-1 for LD (overall degree of resistance), recommended by the Organization Internationale de la Vigne et du Vin (OIV, 2009) and adapted according to Bellin et al. (2009) (Table S1A). The latter descriptors, ranking from 1 to 9, are positively correlated with the magnitude of plant response and inversely correlated with the severity of DM symptoms. All percentage data were Arcsin transformed in view of following statistical analysis.
Polyphenol Content Measurement
The 2nd−3rd leaves from the PI- and MI-PP apex were collected at 6 dpi and analyzed for the content of 42 phenolics (18 different stilbenoids) by targeted metabolomics (Vrhovsek et al., 2012), according to Chitarrini et al. (2017) with some modifications. For each metabolite, missing values (indicating concentrations below the quantification limit) were imputed by random sampling from a uniform distribution taking values in the range from 0 to the metabolite specific detection limit. Genotype specific levels were estimated as the median value of each metabolite across three biological replicates. To compensate for the expected non-normal distribution of metabolite concentration, genotype-specific data were subjected to logarithmic transformation (ln). Metabolomic data were used to calculate 22 sum/ratio parameters (Table S1B). In all analyses, the difference (delta) between the PI and MI genotype specific values were considered.
Genotyping and Map Construction
Genomic DNA from the overall 138 studied genotypes was isolated from young leaves using DNeasy Plant Mini Kit (Qiagen, The Netherlands). The 136 putative full-sib individuals and the two parental lines were characterized at the genotypic level by means of 192 microsatellite (Simple Sequence Repeat, SSR) markers, corresponding to an average of ca. 10 SSRs well-scattered along each of the 19 grapevine chromosomes. The microsatellites were chosen based on their polymorphism information in “Merzling” and/or “Teroldego” reported by Salmaso et al. (2008), as well as their unique position and physical distance along the reference genome (http://www.genoscope.cns.fr) (Table S2). The applied mid-throughput genotyping strategy was as reported in Peressotti et al. (2015). Prior to building the “Merzling” × “Teroldego” (M×T) genetic map, SSR markers were tested against the expected segregation ratio using a χ2 goodness-of-fit implemented in JoinMap v.4.1 (JM, Van Ooijen, 2006). Highly distorted (p > 0.05) markers were discarded, while the others (p ≤ 0.05) were used for linkage analysis unless they affected the order of neighboring loci. Molecular markers were grouped and ordered along linkage groups (LGs) using the Kosambi mapping function implemented in JM. Mapping parameters were set at a logarithm of odds (LOD) value of 8 and at a recombination frequency of 0.45. LG number was assigned according to Adam-Blondon et al. (2004) and the linkage map was visually represented with MapChart v.2.2 (Voorrips, 2002).
QTL Analysis and Candidate Gene Selection
Genetic map data were integrated with phenotypic data and QTL mapping was performed separately per each experiment by using the simple Interval Mapping algorithm in MapQTL v.6.0 (Van Ooijen, 2009). QTLs were declared significant if the maximum LOD exceeded the LG-specific LOD threshold (calculated using 1,000 permutations) and mean error rate was <0.05. Linkage groups and QTLs were visualized with MapChart2.2 (Voorrips, 2002) and a R ad-hoc script (R Core Team, 2018). Through the associated markers, each reliable QTL interval was further anchored and aligned on the assembled version of the grapevine reference genome (Jaillon et al., 2007). For the ease of calculation, base pairs (bp) were converted into centiMorgans (cM) by dividing by 433,989, which is the mean physical distance corresponding to 1 cM derived in this mapping work.
In order to characterize these genomic regions, the 12X PN40024 reference genome (http://genomes.cribi.unipd.it) was exploited to extract version 2 (V2) of the gene predictions (GPs) underlying QTLs. The gene annotation adopted was the one reported by Vitulo et al. (2014) and candidate genes (CGs) were selected adopting the following criteria:
i. Proximity to LOD peak offset (in case of large genomic intervals);
ii. Involvement in trait regulation based on literature (reference genes);
iii. Assignment to significantly over-represented functional categories. In particular, a Fisher's exact test was applied to evaluate the over-representation of specific functional categories within QTLs controlling a given parameter. Reference to our analysis was the distribution in the same categories of 31,922 12Xv2 GPs assigned to chromosomes (Vitulo et al., 2014). We considered GPs annotated at the third level, with the exception of GPs annotated at the first and second level if not characterized at a deeper level and significantly represented in the genome. Adjusted p-values by the Benjamini and Hochberg (1995) method for multiple-testing correction were considered significant when ≤ 0.05. For the functional categories significantly enriched a fold enrichment (Fold-change), as the ratio between the frequency of a category in the QTLs controlling a given parameter vs. the one in the genome, was calculated. The functional categories that passed the Fisher's test and having a Fold-change > 0 were considered as over-represented;
iv. Involvement in functional categories of interest.
Quantitative RT-PCR Expression Analysis
Total RNA was isolated from 60 to 80 mg of ground leaves, collected at 6 dpi from each biological replicate (the same powder used for biochemical analysis stored at −80°C), using Spectrum Plant Total RNA Kit (Sigma-Aldrich) according to manufacturer's instructions. cDNAs were synthetized using the SuperscriptVILO™ cDNA Synthesis Kit from 1.5 μg of DNAseI-treated RNA (Thermo Fisher Scientific). Primer sequences were derived from literature or designed on the reference CG sequence using Primer3 v.4.0 software (http://bioinfo.ut.ee/primer3-0.4.0/) according to MIQE guidelines suggestions (Bustin et al., 2009) (Table S3). Due to the high sequence homology among different family members, it was not always possible to design gene-specific primers. Each primer pair was tested by semi-quantitative RT-PCR on cDNAs from one F1 individual, “Merzling” and “Teroldego” parental lines. The corresponding amplicons were checked by electrophoresis and, prior purification by ExoSAP (Euroclone), were Sanger sequenced to verify the specificity of the primer pair. Forward and reverse reads were aligned to the reference CG sequences using Staden Package software (http://staden.sourceforge.net/) in order to confirm their uniqueness.
Finally, the primer pairs that passed this step were employed in qRT-PCR analyses, carried out using the Platinum SYBR Green qPCR SuperMix-UDG in a ViiA™7 thermocycler (Thermo Fisher Scientific). The 384-well plates were set up according to the sample maximization strategy proposed in Hellemans et al. (2007). Each sample was examined in three technical replicates, and dissociation curves were analyzed to verify the specificity of each amplification reaction. Reaction conditions and the analysis protocol were the same as adopted in Malacarne et al. (2015). Six housekeeping genes (VvACT, VvATP16, VvEF1α, VvGAPDH, VvSAND, and VvUBIQ; Table S3) were tested for their stability using GeNorm software (Vandesompele et al., 2002). Normalized relative quantities (NRQs) were then calculated by dividing the RQ by a normalization factor, based on the expression of the two most stable reference genes (VvATP16 and VvGAPDH) (Reid et al., 2006).
Statistical Analysis
Statistical analyses applied to phenotypic data were performed in R (R Core Team, 2018) equipped with tidyverse (Wickham, 2017) https://CRAN.R-project.org/package=tidyverse and ggplot2 (Wickham, 2016) packages. The reproducibility between years was assessed by checking (t-test, p ≤ 0.05) for the presence of a significant difference in the values of the three phenotypic parameters in a selected group of genotypes. In the case of the 64 polyphenol-related parameters, the reproducibility between years was assessed by testing the difference between PI and MI with a non-parametric Wilcoxon test (p ≤ 0.05, corrected for multiplicity applying a Bonferroni correction). The same test was applied to evaluate the presence of a significant induction of the 64 parameters in the PI vs. MI samples.
Phylogenetic Analysis
A total of 90 GPs, including isoforms, encoding peroxidases (IPR000823) were identified in the 12Xv2 PN40024 reference genome (http://genomes.cribi.unipd.it/DATA/V2; Table S6), while the 76 peroxidases from Arabidopsis (Tognolli et al., 2002) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). Codon-based alignments of all the coding gene sequences were made using Macse program (Ranwez et al., 2011). Automatically produced alignments of individual genes were loaded into the Seaview alignment editor and manually edited in amino acid mode to discard obviously misaligned regions. The final selection of alignment columns was saved to produce a 206 pos. long curated alignment containing 167 aminoacid peroxidase sequences. Phylogenetic analyses were performed with the help of Iq-Tree multicore version 1.6. beta4 (Nguyen et al., 2015). All sequences in alignment passed χ2 test of compositional heterogeneity (p < 0.05). The optimal substitution model for the observed alignment was automatically selected under the Bayesian Information Criterion (BIC, Schwarz, 1978) among a set of 468 substitution models. The model assumed LG (Le and Gascuel, 2008) amino-acid replacement matrix with across sites rate heterogeneity modeled via FreeRate model (Soubrier et al., 2012) assuming six rate classes. Branch support values were inferred based on 1,000 bootstrap replicates employing an ultrafast bootstrap approximation (Hoang et al., 2018) as implemented in the IQ-TREE program. Branches were named as suggested by the Super-Nomenclature Committee for Grape Gene Annotation (sNCGGa) (Grimplet et al., 2014). A bootstrap value of 70 (recommended by the Committee) distinguished the genes within the majority of the classes. Whenever a branch containing both Vitis and Arabidopsis gene/genes was found in a subtree, a subclass was defined. The remaining Vitis genes were named independently. Moreover, different members of a subclass were distinguished by a letter and different splicing variants had the same name but followed by a number. In the few cases in which the direct hortolog from Arabidopsis was found, both names were retained.
Results
Downy Mildew Resistance and Polyphenol Content
P. viticola inoculation was performed on the entire progeny (2012) and on the two parental lines along with 11 repeated F1 individuals (2013). The reproducibility of both LD and PP experiments between the 2 years was verified by t-test on the three phenotypic parameters (Figures S1A,B) and it confirmed an overall coherence between 2012 and 2013 infection both for LD and PP. Altogether the phenotypic results indicated an approximately normal distribution of severity, incidence and OIV452(−1) in the progeny and confirmed a mid-resistant and susceptible phenotype for “Merzling” and “Teroldego,” respectively. It is relevant to underline that several F1 individuals were transgressive with respect to the resistance donor “Merzling” exhibiting a higher level of resistance, while none resulted more susceptible than the parental “Teroldego” (Figure 1).
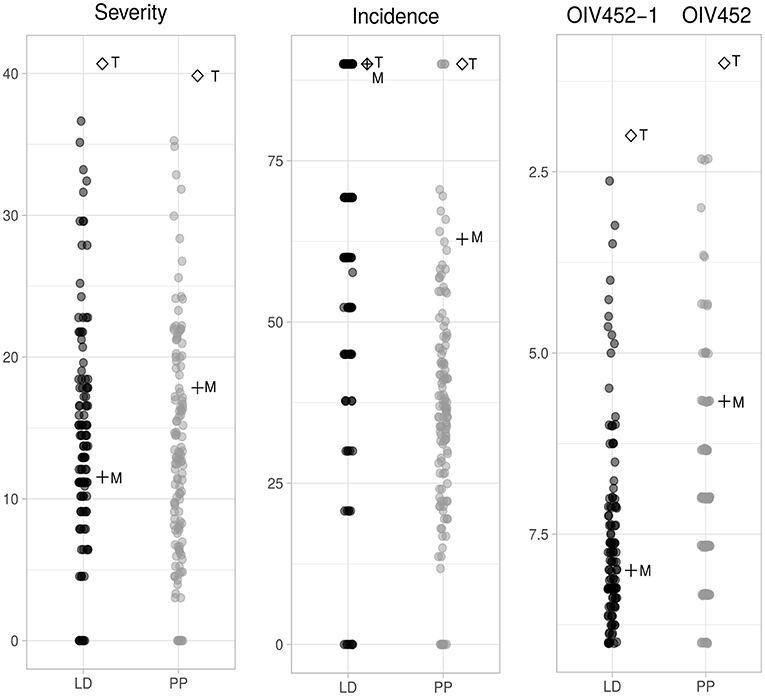
Figure 1. Phenotypic distribution of the three different parameters associated with resistance to P. viticola in the M×T progeny. Severity and incidence are expressed in percentage while the OIV descriptor follows a discrete scale from 1 to 9. These parameters were scored at 6 days post-inoculation (dpi) for leaf disks (LD) and at 8 dpi for potted plants (PP). The data were collected in June and August 2012 for the segregating population and in June and August 2013 for the two parental lines. The reproducibility of the two experiments was tested (p ≤ 0.05) on data collected in August 2012 and 2013 for the same genotypes as shown in Figure S1. M, “Merzling”; T, “Teroldego”.
Analogously to the DM resistance, the polyphenol-related data—the measurement of 64 parameters on PP of 11 repeated F1 individuals (Figure S2)—performed between 2012 and 2013 were found to be reproducible (p ≤ 0.05, corrected for multiplicity applying a Bonferroni correction). The differences turned out to be significant (p ≤ 0.05) in 13 out of 64 cases, exhibiting good reproducibility overall. Moreover, in many cases (e.g., 2,6-DHBA, fertaric acid, cis-piceid, cis-resveratrol, pterorostilbene, cis-ε-viniferin) the range of metabolic content identified in the progeny was greater than in the parents, suggesting a transgressive segregation (Figure S3). A significant delta value between PI and MI plants (p ≤ 0.05, corrected for multiplicity applying a Bonferroni correction) was detected for 32 different value distributions, each representing variation in the content of polyphenols: 22 of stilbenoid class, out of which seven monomeric (trans-resveratrol, piceatannol, pterostilbene, cis-piceid, astringin, isorhamnetin, and the sum of monomeric stilbenoids), five dimeric (trans-ε-viniferin, cis/trans ω-viniferin, pallidol, ampelopsin D+quadrangularin A, and the sum of dimers), four trimeric (α-viniferin, E-cis-miyabenol C, Z-miyabenol C, and the sum of trimers), three tetrameric (isohopeaphenol, ampelopsin H + vaticanol C-like isomer, and the sum of tetramers), the ratio between tetramers and monomers, the sum of polymers and all stilbenoids; two of benzoic acid class (2,6-DHBA and the sum of benzoic acids); the coumarin fraxin; the t-coutaric hydroxycinnamic acid; the flavanone naringenin; four flavonols (quercetin-3-glucoside, rutin, quercetin-3-glucuronide, and the sum of flavonols); and finally the sum of all polyphenols (Figure 2).
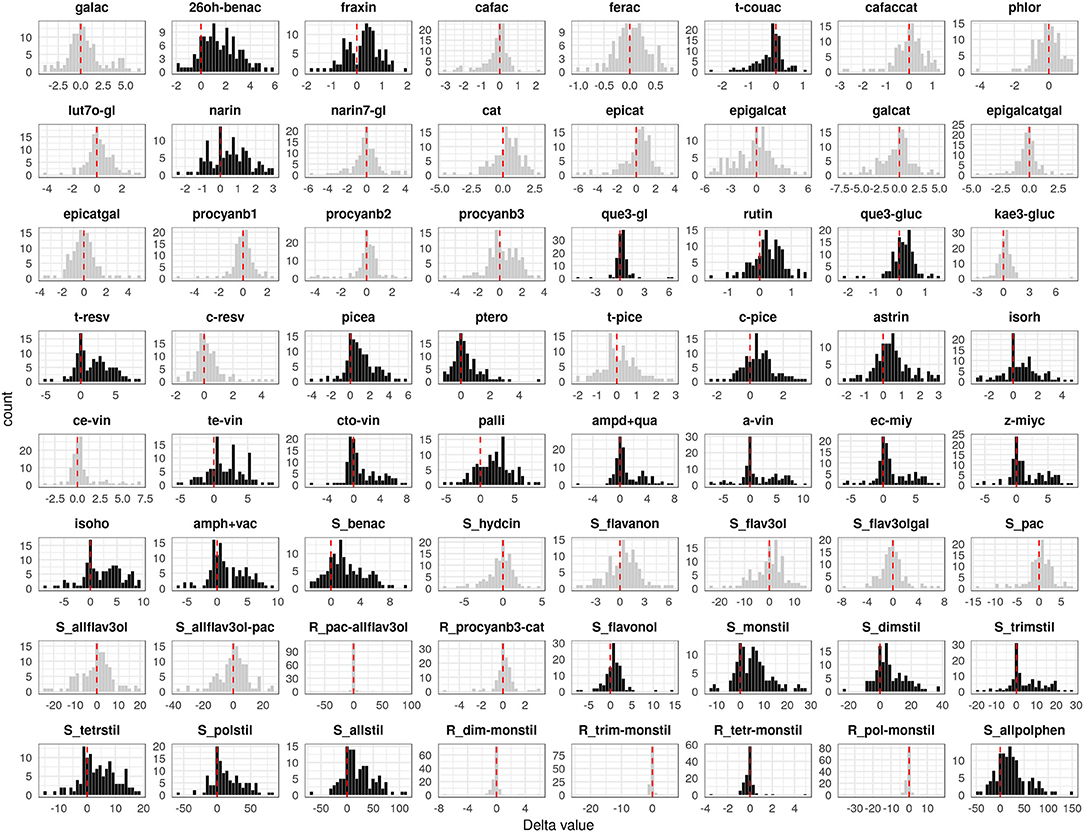
Figure 2. Distribution of the difference between P. viticola-inoculated (PI) and mock-inoculated (MI) values (delta) associated with 64 polyphenol-related parameters recorded in the M×T progeny. Black distributions highlight significantly modulated compounds. The meaning of the parameter abbreviations is reported in Table S1B.
QTL Mapping
The M×T Linkage Map
Out of 136 initial putative full-sib individuals, 129 resulted to be true-to-type F1 individuals upon the genotyping analysis. Of these, three were discarded because they presented >20% missing data (progeny information in Table S4). Out of the 192 scored, 181 markers were ordered into 19 LGs allowing the construction of the M×T consensus map. Marker order was generally consistent between parental and consensus homolog LGs, with local inversion of tightly linked markers, and reflected the backbone of previous published maps (Salmaso et al., 2008; Vezzulli et al., 2008). The remaining 11 markers consisted of one unlinked and 10 showing distorted segregation ratios with a probability p ≤ 0.05. The distribution of the 181 mapped markers into different segregation types showed that 91.1% allowed discrimination between paternal and maternal inherited allele. The total length of the consensus map was 1,162.7 cM with a mean distance between adjacent markers of 6.4 cM (Figure S4). The overall linkage map statistics are reported in Table S4. This map was finally employed in a QTL mapping survey to identify putative genomic regions involved in the genetic control of DM resistance and polyphenol variation upon P. viticola inoculation; a total of 49 significant QTLs associated with 33 different parameters were detected on 12 LGs (Figure 3, Table 1).
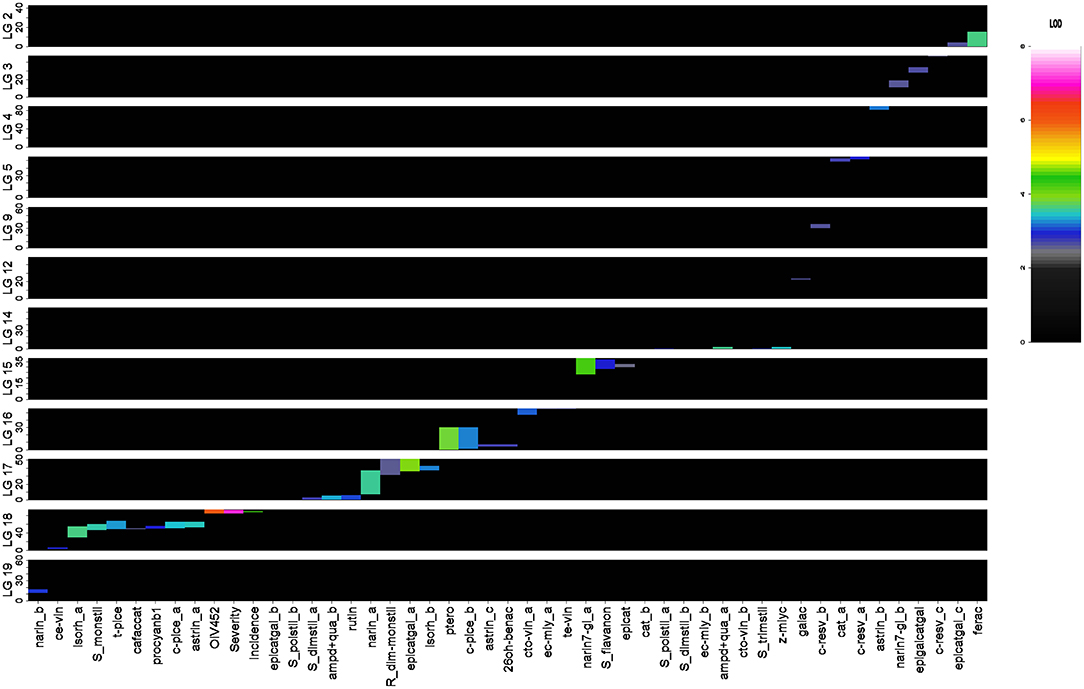
Figure 3. QTL distribution across the 19 M × T LGs. The meaning of the parameter abbreviations is reported in Table S1.
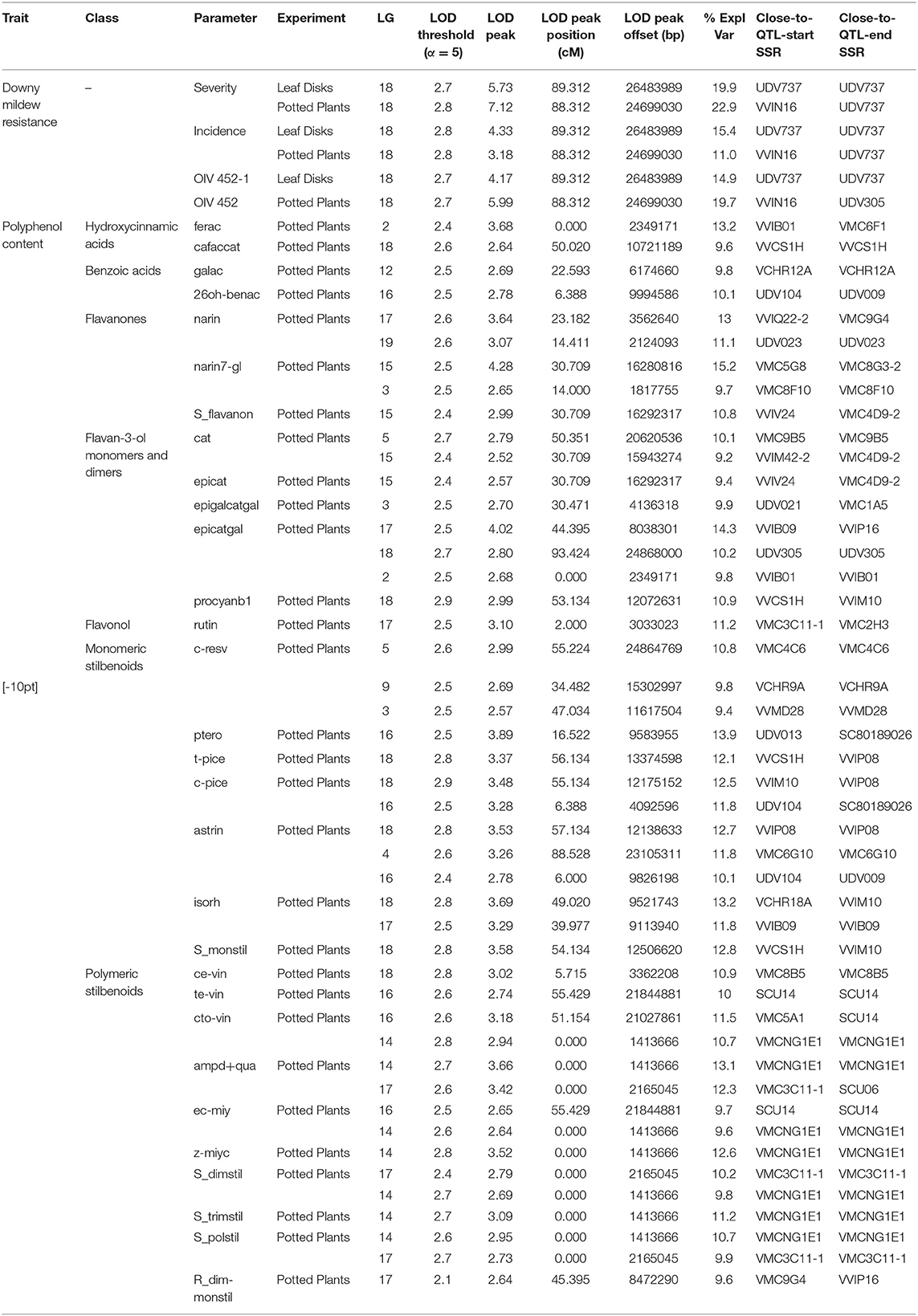
Table 1. List of the 49 identified significant QTLs associated to three DM resistance and 30 polyphenol-related parameters.
The DM Resistance QTLs
Regarding the phenotypic data associated with DM resistance recorded on PP at 6 dpi, the first QTL identified was related to the severity parameter, showing a maximum LOD value of 7.12 (22.9% of explained variance), and located along the distal arm of chromosome 18 (88.3 cM). A co-localized QTL resulted associated with the OIV 452 descriptor with an LOD peak of 5.99 (19.7% of explained variance). Concerning the incidence parameter, the detected QTL was slightly shifted and showed a maximum LOD value of 3.18 (11% of explained variance). These results based on PP phenotypic data were confirmed by the results obtained on LD where a lower LOD peak was calculated per each parameter, except for disease incidence, which is more sensitive to the intra-plant leaf variability (Table 1, Figure 3). This overall genomic interval, ranging from 22.6 to 27.8 Mb of LG 18, co-localizes with the known Rpv3 locus which is characterized by the flanking UDV305 and UDV737 SSR markers (Di Gaspero et al., 2012). Given their genetic profile in this work, the haplotype Rpv3 null−271 was identified as associated with DM resistance in “Merzling” and named Rpv3-3, according to the rules established by the international grapevine research community (www.vitaceae.org; http://www.vivc.de/). Not reliable, confirmed twice between PP and LD experiments, minor QTLs were also identified (data not show).
The Polyphenol-Related QTLs
As concerning polyphenol variation, significant QTLs were newly detected for a total of 30 parameters, including both the original and the derived ones. LOD peak values spanned from 2.52 to 4.28 (Table 1, Figure 3).
Overall, considering the QTL physical distribution, four clusters were located on LGs 18, 17, 16, and 15 by abundance order. In particular, a comprehensive cluster of QTLs positioned on LG 18 was associated with caffeic acid+catechin condensation, five parameters related to monomeric stilbenoids, cis-ε-viniferin, epicatechin gallate and procyanidin B1, and did not result overlapping with the distal region associated with DM resistance (Figure 3).
Regarding hydroxycinnamic acids, one QTL associated with fertaric acid (LOD peak value of 3.68) and one to caffeic acid+catechin condensation (LOD peak value of 2.64) were located on LGs 2 and 18, explaining, respectively 13.2 and 9.6% of the total phenotypic variance. In terms of benzoic acids, gallic acid, and 2,6-DHBA were correspondingly mapped on LGs 12 and 16, with 9.8 and 10.1% of explained variance.
Within the group of monomeric stilbenoids, three QTLs were found to be associated with cis-resveratrol on LGs 5, 9, and 3, explaining 10.8, 9.8, and 9.4% of total variance, respectively. A QTL for pterostilbene was located on LG 16 with a LOD peak value of 3.89, which corresponded to 13.9% of explained variance. Two QTLs, positioned on LGs 16 and 18, were related to the cis-piceid content (11.8 and 12.5% of explained variance), while trans-piceid was mapped only on the LG 18 region which explained a similar variance. Astringin was under control of three genomic regions (LGs 16, 4, and 18) which corresponded to a range from 10.1 to 12.7% of the total phenotypic variance. Two QTLs associated with isorhapontin were positioned on LGs 18 and 17, with 13.2 and 11.8% of explained variance. Finally, a QTL corresponding to the region controlling the content of all the monomeric stilbenoids mentioned above was found on LG 18 for the sum of monomeric stilbenoids. Concerning polymeric stilbenoids, cis-ε-viniferin was mapped on LG 18, in a region upstream to the one associated with monomeric stilbenoids and encompassing 10.9% of the total phenotypic variance; indeed, a QTL related to the trans-ε-viniferin content, explaining a highly similar variance, was detected on LG 16 in a region far from the one associated with the monomeric stilbenoids and coincident with the one explaining the 9.7% of the variance of E-cis-miyabenol content described below. Cis+trans-ω-viniferin was under control of two genomic regions (LGs 16 and 14) which corresponded to the 11.5 and 10.7% of the total phenotypic variance. In particular, 85% of the LG 16 region was specifically associated with this specific dimer. Two genomic regions associated with ampelopsin D+quadrangularin A were identified on LGs 14 and 17; these QTLs showed a LOD peak value of 3.66 and 3.42, with 13.1 and 12.3% of explained variance, respectively. Two QTLs, positioned on LGs 16 and 14, were related to the E-cis-miyabenol content (9.7 and 9.6% of explained variance), while Z-miyabenol C was mapped only on the LG 14 region which explained 12.6% of the total phenotypic variance. Considering derived parameters, two QTLs were identified on LGs 17 and 14 for the sum of dimeric stilbenoids (a mean of 10% of explained variance), whereas the sum of trimeric stilbenoids was associated only with the QTL on LG 14, explaining 11.2% of the total phenotypic variance. For total polymeric stilbenoids, including tetrameric stilbenoids, the two QTLs identified on LGs 14 and 17 were confirmed, with 10.7 and 9.9% of explained variance. Finally, a QTL was detected as associated with the ratio of dimeric to monomeric stilbenoids on a different region of LG 17, explaining 9.6% of the total variance. This region was also controlling the isorhapontin content as previously highlighted. Contrary to LG17, on LG14 the region associated with all the parameters previously described was coincident.
Within the flavanon group, two QTLs related to the naringenin content were found on LGs 17 and 19 (LOD peak value of 3.64 for the major QTL), with 13 and 11.1% of explained variance, respectively. The 56% of the first and the 100% of the second region were specifically associated with naringenin. Indeed, naringenin-7-glucoside was mapped on LGs 15 and 3 (LOD peak value of 4.28 for the major QTL), which respectively explained 15.2 and 9.7 of the total phenotypic variance. In particular, the 72% of the region on LG 13 was specific for this flavanon. For total flavanones, only the QTL located on LG 15 was shared, with 10.8% of explained variance. Regarding flavan-3-ol monomers and dimers, for catechin two QTLs were detected on LGs 5 and 15—of which the first one was private—explaining 10.1 and 9.1% of the total phenotypic variance, respectively. Two QTLs, the first one positioned on LGs 15 coincidently with the region associated with catechin and the second one on LG 3, were respectively related to the epicatechin and epigallocatechin gallate content, explaining a similar percentage of the total phenotypic variance. Three genomic regions associated with epicatechin gallate were identified on LGs 17, 18, and 2; these QTLs spanned from a LOD peak value of 4.02 to 2.68, with a corresponding range of explained phenotypic variance from 14.3 to 9.8%. Procyanidin B1 was mapped on LG 18 with 10.9% of explained variance. Finally, a QTL was associated with the flavonol rutin on LG 17, which explain 11.2% of the total phenotypic variance.
Relationship Among Genetic Background, Resistance Level, and Polyphenol Induction
Of the 30 polyphenolic compounds with an associated QTL, infection significantly affected production of 23. Out of these, synthesis of four compounds was induced in Rpv3-3+ genotypes only, synthesis of another four significantly decreased in Rpv3-3− genotypes only, and one (ampelopsin D+quadrangularin A) was significantly induced and repressed in both Rpv3-3+ and Rpv3-3− genotypes, respectively (Figure 4A).
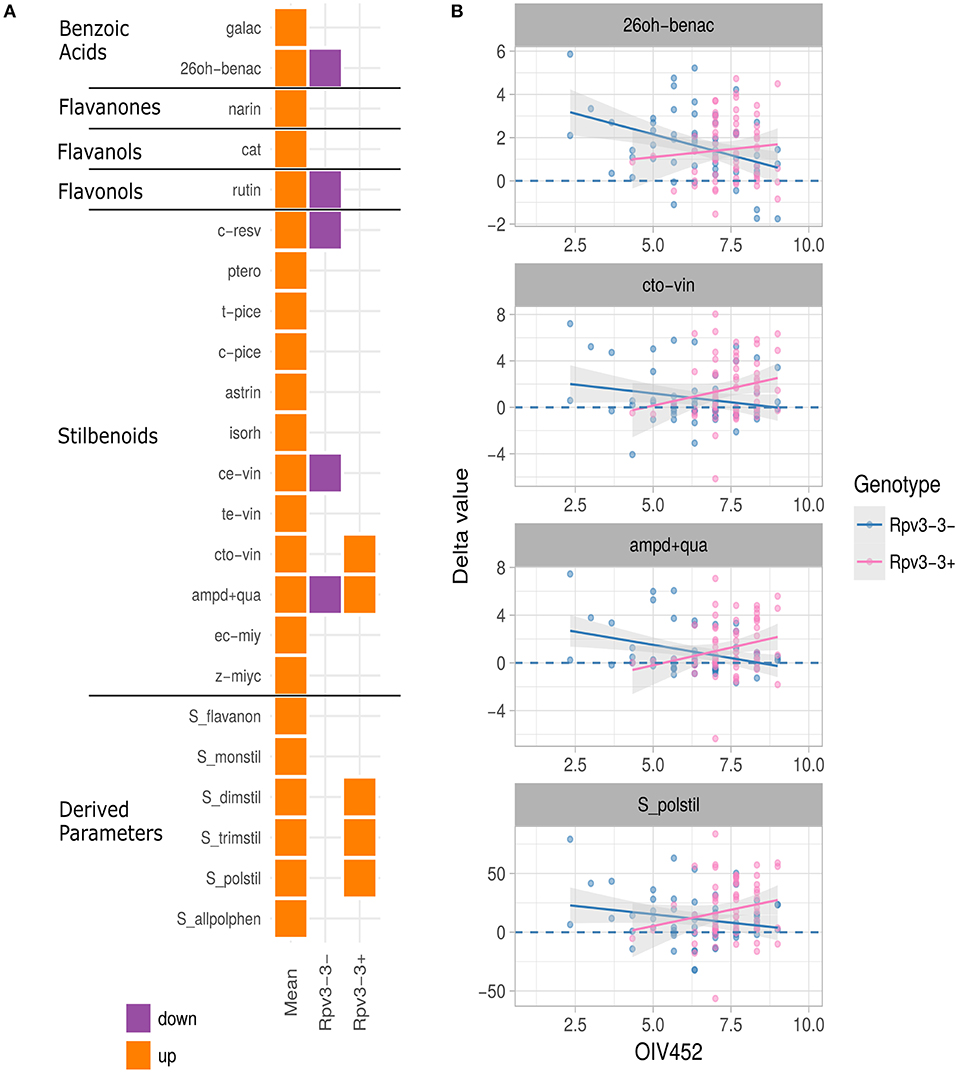
Figure 4. (A) Summary of univariate analysis performed on the 30 polyphenol-related parameters—with a QTL region associated—measured in the M×T progeny. The plot summarizes the outcomes of a series of regression analyses performed to assess the presence of a significant induction (p ≤ 0.05) and its dependence upon the resistance level (measured with the OIV 452 descriptor) for Rpv3-3+ and Rpv3-3− genotypes. Orange and violet colors highlight up and down regulation, respectively. The first column (Mean) indicates the average value of the parameter distribution in the M×T progeny. (B) Examples of the univariate regression analyses. The meaning of the parameter abbreviations is reported in Table S1B.
Considering the Rpv3-3 haplotype, the OIV 452 parameter and the polyphenol delta values, a relationship was highlighted exclusively for the stilbenoids. For instance, cis/trans-ω-viniferin and polymeric stilbenoids, on average induced in the progeny, resulted significantly induced in Rpv3-3+ genotypes showing a high OIV 452 value. By contrast, ampelopsin D+quadrangularin A, on average induced in the progeny as well, showed an opposite profile between Rpv3-3+ and Rpv3-3− genotypes with high OIV 452 values. In addition, the induction of 2,6-DHBA, observed on average in the progeny, was very low in Rpv3-3− genotypes with a high OIV 452 value (Figure 4B). In the Rpv3-3− resistant genotypes no compound synthesis was significantly induced by the fungus (data not shown).
Candidate Gene Identification and Characterization
Candidate Genes Underlying DM Resistance and Polyphenol-Related QTLs
The number of genes identified within each QTL region was extremely variable from a minimum of 5 (LG 18) to a maximum of 984 (LG 16). Due to the high number of GPs underlying the QTLs associated with the 33 assessed parameters, four different main criteria were adopted to select CGs for trait regulation (e.g., Figure S5 for enrichment analysis results). Upon this selection (Table S5), a refined list of the 95 most relevant unique CGs was generated, of which a few were previously characterized as involved in DM response and in the regulation of polyphenol synthesis (the so called “reference genes”), while the majority were newly identified. CGs mainly referred to the functional categories of signaling, secondary metabolism, regulation of transcription, and response to abiotic and biotic stimulus (Table 2).
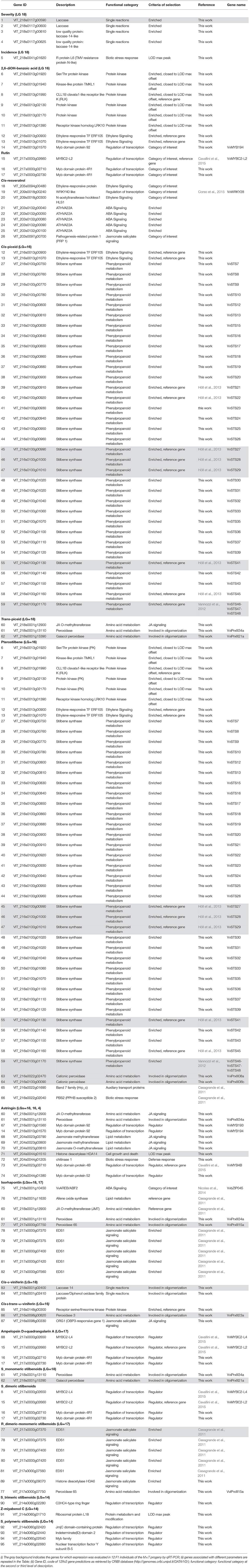
Table 2. List of the 95 most relevant candidate genes associated with DM resistance and polyphenol content.
Besides disease-related (NBS-LRR) genes alone representing a significant part of the grapevine genome (Malacarne et al., 2012) and found also in high numbers within the major QTL on LG 18 associated with DM resistance parameters, four laccases (VIT_218s0117g00590, VIT_218s0117g00600, VIT_218s0117g00610, VIT_218s0117g00625), which can be part of the defense response due to pathogen recognition mediated by the LRR domain, drew our attention (Table S5).
Regarding the polyphenol content trait, we focused on CGs (i) underlying QTLs associated with polyphenols induced by infection and (ii) showing disparity between Rpv3-3+ and Rpv3-3− genotypes characterized by a different level of DM resistance. Within the category of signaling, many selected genes belong to the kinase protein family, as well as to ethylene, ABA and JA signaling pathways. Two genes appear to be of special interest, one coding for the LRK10 Receptor kinase homolog (VIT_216s0013g01390) and the other encoding CLL1B clavata1-like receptor S/T protein kinase (VIT_216s0013g01990) located within the QTL region on LG16 linked to 2,6-DHBA and to pterostilbene. In addition, a gene coding for a Receptor serine/threonine kinase (VIT_216s0148g00300) resulted associated with cis-ω viniferin, as well as two genes encoding the Ethylene-responsive transcription factor ERF105 (VIT_16s0013g00900 and VIT_16s0013g01070) were linked to 2,6-DHBA, pterostilene, and cis-piceid content. In addition, five genes (VIT_203s0132g00040, VIT_203s0132g00050, VIT_203s0132g00080, VIT_203s0132g00090, VIT_203s0132g00100), coding for a cluster of HVA22-like abscisic acid-induced proteins, were linked to cis-resveratrol content and the gene encoding the bZIP factor VvAREB/ABF2 (VIT_218s0001g10450) was exclusively associated to isorhapontin. Three JA O-methyltransferases (VIT_204s0023g03790, VIT_204s0023g03800, VIT_204s0023g03810) were associated with astringin, an additional JA O-methyltransferase (VIT_218s0001g12900) was related to astringin and isorhapontin, five genes (VIT_217s0000g07370, VIT_217s0000g07375, VIT_217s0000g07400, VIT_217s0000g07420, VIT_217s0000g07560) coding for EDS1 were linked to isorhapontin and the ratio between dimeric and monomeric stilbenoids, and finally one gene (VIT_216s0098g00330) encoding ORG1 was associated with the regulation of cis+trans-ω-viniferin content (Table 2).
A cluster of stilbene synthase genes, mostly not specifically related to DM response previously, was found in the QTL intervals associated with cis-piceid and pterostilbene. In addition, six peroxidase genes (herein named VviPrxIII08a, VviPrxIII08b, VviPrxIII15a, VviPrxIII21a, VviPrxIII23a, VviPrxIII34a, Figure 6) underlied monomeric and oligomeric stilbenoid-related QTL regions and two laccase genes (VIT_218s0001g02400 and VIT_218s0001g02410) were associated with ε-viniferin.
Two of the genes recently identified as encoding a set of R2R3-MYB C2 repressors of phenylpropanoid levels (Cavallini et al., 2015) were linked to the regulation of ampelopsin D+quadrangularin A, the sum of dimeric stilbenoids and rutin (VvMYBC2-L2) and to the regulation of trans-piceid and astringin content (VvMYB4B). Another seven MYB genes were identified: VIT_216s0013g01560 (VvMYB193 in Wong et al., 2016) and VIT_216s0013g01570 (VvMYB194 in Wong et al., 2016) in the region controlling both 2,6-DHBA and astringin content, VIT_204s0044g01380 in the region controlling astringin content, VIT_217s0000g02710 and VIT_217s0000g02730 as associated with ampelopsin D+quadrangularin A, the sum of dimeric stilbenoids and rutin content, and finally VIT_214s0060g02640 related to the sum of polymeric stilbenoids. Finally, a WRKY factor (precisely VvWRKY28 in Wang et al., 2014) was associated to cis-resveratrol content.
Newly Identified Genes Candidate to Stilbenoid Oligomerization
Among all CGs, further investigation was dedicated to a set of 11 genes by assessing their transcript level in a set of 12 F1 individuals. The objective was to get further evidence of the association between the identified genomic regions and the traits under investigation. These genotypes exhibited disparity in pathogen resistance level, stilbenoid-related parameters with an mQTL associated and haplotype status at the Rpv3 locus (Figure S6). These genes encoded: (i) five out of six peroxidases identified within monomeric and oligomeric stilbenoid-related QTL regions (VviPrxIII08a, VviPrxIII08b, VviPrxIII15a, VviPrxIII21a, VviPrxIII23a); (ii) two of the six laccases described above (VIT_218s0117g00590 and VIT_218s0001g02400), the first associated with disease severity and the other with ε-viniferin and therefore putatively involved in its oligomerization; (iii) three stilbene synthases (VvSTS27-8-9, VvSTS41, and VvSTS48) already associated with DM response (Vannozzi et al., 2012; Höll et al., 2013) and here related to the regulation of pterostilbene and cis-piceid content; (iv) one Histone deacetylase HDA14 (VIT_204s0044g01510) found in correspondence of the LOD peak offset of the QTL on LG 4 associated with astringin.
The gene expression study revealed a significant correlation between (i) the content of cis-piceid and pterostilbene and the expression level both of VviPrxIII08a and VviPrxIII08b, (ii) the content of ω-viniferin and the expression level of VviPrxIII23a, and finally (iii) the content of cis-piceid and pterostilbene and the expression level both of VvSTS41 and VvSTS48 at 6 dpi (Figure 5).
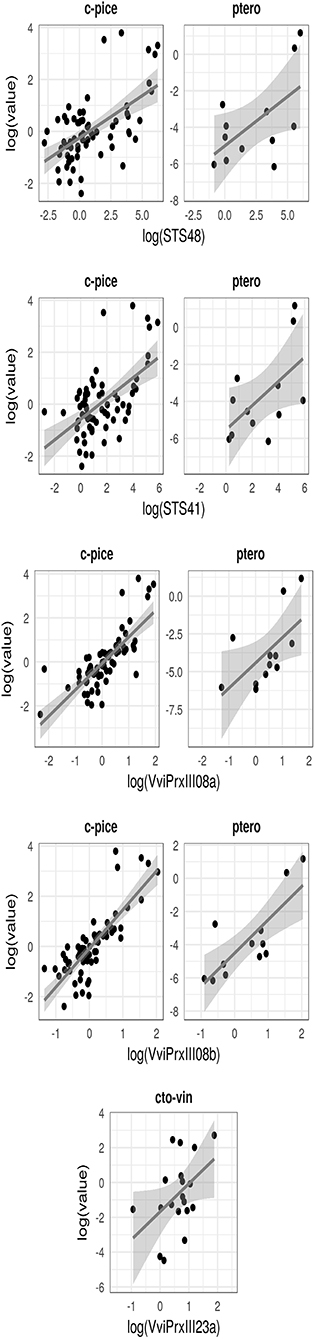
Figure 5. Regression analysis which highlights the association between metabolic induction and normalized relative expression of selected candidate genes in Rpv3-3+ and Rpv3-3− selected genotypes. c-pice, cis-piceide; ptero, pterostilbene; cto-vin, cis + trans-ω-viniferin.
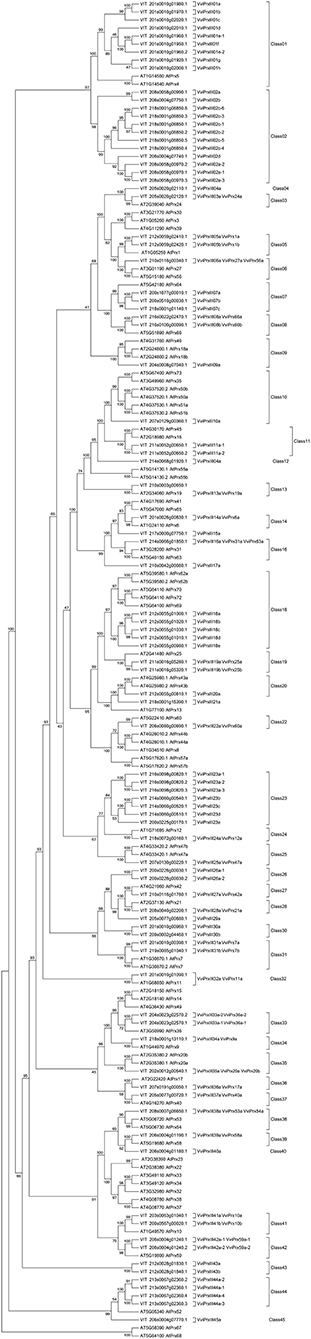
In order to gather additional information about the peroxidases found in the QTL and correlated with stilbenoid induction upon P. viticola infection we performed a phylogenetic analysis of the whole grapevine peroxidase gene family, which allowed us to attribute the 90 GPs to 45 different classes. The six peroxidases herein associated to stilbenoid metabolism are part of different classes: VviPrxIII08a and VviPrxIII08b belong to class 08 and are the putative orthologs of AtPrx66, VviPrxIII15a and VviPrxIII21a are the unique members of class 15 and 23, respectively, VviPrxIII23a has three different isoforms which belong to class 23 together with another four members, and finally VviPrxIII34a belongs to class 34 and is the putative ortholog of AtPrx09 (Figure 6; Table S6).
Discussion
Characterization of the Rpv3-3 haplotype
The M×T genetic map built in this study is an improved version in terms of progeny individuals and LG number as well as marker number/order compared to the one by Salmaso et al. (2008). The high percentage of markers (68%) that was mapped in both parents is close to the 62% of markers positioned into the reference grapevine genetic map, while the 5% of distorted markers is about half of the value previously observed (Adam-Blondon et al., 2004). This upgrading should result in visible improvement in the parental and consensus genetic maps to be suitable for further applications as the association to new phenotypes.
Herein, at the hypervariable microsatellite markers UDV305 and UDV737, we identified the haplotype null-271 as conferring resistance against P. viticola; this genetic variant is located on chromosome 18 and it derives on the “Merzling” side from the grand-parent “Seyval.” This haplotype was named Rpv3-3 according to the VIVC nomenclature, given its co-localization with the Rpv3 locus, which was subjected to selective sweep during grapevine breeding activities. In fact, a seminal study identified seven conserved haplotypes, which are overrepresented in grapevine breeding lines historically selected for DM resistance compared to their wild relatives, and are absent from the susceptible V. vinifera varieties (Di Gaspero et al., 2012). These genetic variants may carry Rpv3 alleles or adjacent Rpv3 paralogs that constantly remained linked with the diagnostic markers. Upon this reported association analysis, nowadays only two wild relative Rpv3 haplotypes have been characterized in segregating populations. The Rpv3-1 haplotype—preserved in the “Seibel 4614” lineage—was firstly identified in the German hybrid “Regent” (Welter et al., 2007; van Heerden et al., 2014) and in the Hungarian hybrid “Bianca” (Bellin et al., 2009) through QTL analyses. The second of these resistance haplotypes—named Rpv3-2, conserved in the “Munson” lineage—has recently been confirmed by QTL mapping (Zyprian et al., 2016). In this work, we have validated the Rpv3-3 haplotype—derived from the descent group founder “Noah”—by means of a QTL analysis on the M×T progeny.
Given this outcome, we attempted the characterization of several genotypes related to Merzling (data not shown). Unlike the comprehensive Rpv3 survey reported by Di Gaspero et al. (2012), we detected the Rpv3-3 haplotype also in the “Merzling” offspring “Solaris,” so far known to carry only the Rpv10 locus derived from V. amurensis. This was due to the masking effect of a different Rpv3 haplotype (Rpv3-1) derived from the second resistance donor Gf.Ga-52-42 in the QTL study (Schwander et al., 2012). Analogously, besides the Rpv 10 locus, we found that “Bronner” and “Cabernet Cortis” unexpectedly inherited the Rpv3-3 haplotype from their parent “Merzling” and “Solaris,” respectively; all these genotypes used as parental lines demonstrated to transmit this acquired haplotype to their respective progenies (Vezzulli S., personal communication). Finally, herein this specific genetic variant was detected also in the related Baron, Prior, and Cabernet Cantor (data not shown). These findings confirmed that grapevine breeders traditionally selected genotypes with a certain degree of pyramiding, namely accumulation of more than one R-locus (Töpfer et al., 2011), suggesting a reinforcement role of the Rpv3-3 haplotype on other R-locus effects. For this reason our investigation can be considered a “genetic” upgrade of the pioneer Rpv3 study by Di Gaspero et al. (2012).
Genes belonging to the NBS-LRR superfamily were detected in the regions underlying QTLs associated with all disease resistance parameters, in agreement with the first studied Rpv3 resistance haplotype encoding NB-LRR and LRR-kinase receptors (Di Gaspero and Foria, 2015). According to the Effector-Triggered Immunity model, R gene products sense the pathogen effectors and activate signal transduction pathways (Cui et al., 2015). In grapevine, Rpv3-dependent resistance follows this model of gene-for-gene interaction (Casagrande et al., 2011). In previous studies, the resistance haplotype was revealed to be necessary and sufficient to trigger a hypersensitive response (HR) leading to cell death in the proximity of sites infected by P. viticola (Bellin et al., 2009; Zyprian et al., 2016). In the current study, weakened or delayed HR was observed on LD of Rpv3-3+ genotypes (data not shown) with lower OIV452-1 values; this overall phenomenon can be due to the combination of different factors—high inoculum concentration and RH—leading to shorter incubation time, more infection sites and faster hyphal growth under highly conducive conditions (Gessler et al., 2011). In addition, it is relevant to highlight that the phenotypic distribution (ranges: OIV 452 from 1.6 to 9, Severity from 0 to 33.3%, Incidence from 0 to 100%) on PP revealed an average mid-resistance level of Rpv3-3+ genotypes, in agreement with the recent study by Foria et al. (2018) which shed light on various Rpv3 haplotypes responsible for different disease resistance degrees. Finally, unlike the expectation in the case of an R-locus, the non-binomial distribution of each DM resistance parameter suggests that this trait is controlled by multiple factors in this genetic background. The M×T QTL study refers to the concept of Advanced Backcross-QTL (AB-QTL)—recently recovered in grapevine—which combines QTL analysis and variety development by designing a mapping/breeding scheme for the simultaneous identification and introgression of wild haplotypes. AB-QTL relies on segregating populations in which most of the wild-parent genome that donates the trait of interest has been purged in early segregating generations by phenotypic selection (Tanksley and Nelson, 1996). This is relevant to guarantee QTL stability once the associated markers are screened in derived breeding materials. In fact, favorable QTL alleles identified in early generations often disappear in later back-cross generations, once the modifier genes that have epistatic interactions with the beneficial QTL alleles are removed from highly V. vinifera genetic backgrounds (Di Gaspero and Foria, 2015). Herein, we did not actually face cases of susceptible individuals carrying the Rpv3-3+ haplotype, whereas we found a few Rpv3-3− individuals displaying DM resistance. Since unreliable minor QTLs were identified in both the M×T consensus and the maternal genetic map (data not shown), and no intra-locus recombination was detected, this phenomenon has still to be elucidated.
Survey of Stilbenoid-Associated Regions Among the Discovered Polyphenol-Related QTLs
In the present study 46 novel metabolic (m)QTLs associated with 30 phenolics-related parameters were discovered. Among the new mQTLs, more than half were associated with stilbenoid-related parameters (monomers, dimers, and trimers), two with hydroxycinnamic acids, two with benzoic acids, five with flavanones, and eight with flavan-3-ol monomers and dimers. Pleiotropic effects, i.e., single gene producing multiple effects on various traits, were recorded. Except for few cases encompassing different classes (e.g., ratio between dimeric and monomeric stilbenoids and naringenin), pleiotropy was detected along genomic intervals associated with parameters falling into the same class, such as monomeric stilbenoids and flavanones. Moreover, epistatic effects have recently been considered in many studies as relevant for complex traits, such as polyphenol biosynthesis. Epistasis, i.e., an additive-by-additive interaction between QTLs, assayed in populations segregating for an entire genome, has been found at a frequency close to that expected by chance alone (Bocianowski, 2013). Therefore, we cannot exclude epistatic effects in the case of characters controlled by more than one region, such as cis-piceid, astringin, and naringenin.
Although few research studies on proanthocyanidin and flavonol berry composition have recently been attempted in grapevine (Huang et al., 2012; Malacarne et al., 2015), most polyphenols do not have a known QTL associated. The mQTLs here identified fill this gap and reveal polyphenols with central role in P. viticola-grapevine interaction. In particular, this analysis allowed the identification of mQTLs associated with 17 different stilbenoid-related parameters, therefore representing a thorough characterization of stilbenoid regulation upon P. viticola infection on leaves.
A cluster of stilbene synthase genes, mostly not specifically related to DM response previously, was identified in the QTL intervals associated with cis-piceid and pterostilbene. Previous works revealed different patterns of transcript accumulation between the different VvSTS family members (Dai et al., 2012; Vannozzi et al., 2012; Höll et al., 2013; Shi et al., 2014) depending on the high variability in their regulatory regions (Chialva et al., 2018). Moreover, six interesting peroxidase and two laccase genes were identified as associated with stilbenoids therefore representing good candidates for ROS-mediated stilbene oligomerization (Calderón et al., 1994; Barceló et al., 2003; Pezet et al., 2004b). Indeed, there is some evidence supporting the enzymatic biotransformation of stilbenes by plant peroxidases (Takaya et al., 2005; Wan et al., 2011) and/or fungal laccases (e.g., Pezet et al., 1991; Breuil et al., 1998, 1999). It is still a debate if the oligomerization is driven by plant or fungal laccases, which catalyze the degradation of stilbene monomers allowing the fungus to escape from the action of grapevine phytoalexins. By phylogenetic reconstruction of the entire grapevine peroxidase gene family, we determined that VviPrxIII08a and VviPrxIII08b are the putative orthologs of AtPrx66 involved in lignification (Tokunaga et al., 2009), VviPrxIII34a is the putative ortholog of AtPrx09 which is closed to Arabidopsis members involved in lignin biosynthesis (Tognolli et al., 2002; Herrero et al., 2013), while VviPrxIII15a, VviPrxIII21a, and VviPrxIII23a have no orthologs in Arabidopsis. A validation of the results obtained at the metabolic level came from a transcriptional investigation in a set of 12 F1 individuals of the progeny, which highlighted a significant association between some monomeric and dimeric stilbenoids and the transcript level of three newly identified peroxidases besides known stilbene synthases.
The time dependent regulation of different inducible stilbenes upon abiotic and biotic stresses was reported to be at least transcriptional (e.g., Vannozzi et al., 2012; Höll et al., 2013; Wong et al., 2016) and coordinated by the action of both MYB and WRKY transcription factors (Malacarne et al., 2018; Vannozzi et al., 2018; Jeandet et al., 2019; Jiang et al., 2019). In the present work additional MYB genes were associated with stilbenoid formation; in particular, the known R2R3-MYB C2 repressors of phenylpropanoid levels (Cavallini et al., 2015) were located along the regions on LG4 and LG17 controlling some monomeric and dimeric stilbenoids, respectively. In addition, it is worth noting that the WRKY factor VvWRKY28 already found co-expressed with VvSTS transcripts in root and leaves (Corso et al., 2015) was here associated with cis-resveratrol content extending the list of WRKY factors identified as involved in the regulation of stilbenoid metabolism. It has recently been shown that the expression level of several members of the stilbene synthase gene family and genes responsible of the oxidative polymerization of phenolic compounds in the phenylpropanoid pathway is highly influenced by the “location” variable in a G×E interaction study (Dal Santo et al., 2018).
Our results showed that ABA and SA/JA signaling play an important role in the regulation of 2,6-DHBA, reported to be the de-activated form of SA (Bellés et al., 2006; Campos et al., 2014; Nawrocka et al., 2018), and of stilbenoid synthesis. A cluster of five genes encoding HVA22-like abscisic acid-induced proteins, belonging to a class of ABA- and stress-inducible proteins (Chen et al., 2002), was identified on LG3 associated with cis-resveratrol. Moreover, the transcriptional regulator VvbZIP045, recently characterized by Nicolas et al. (2014) as a key factor activating down-stream genes of the ABA signaling cascade during the grape berry ripening process, was here associated with the regulation of isorhapontin content. It should be noted that (Wang et al., 2018) have recently showed that Muscadinia rotundifolia “Noble” defense response to P. viticola infection is mediated by stilbene accumulation induced by ABA and SA phytohormones. Moreover, two genes (VIT_216s0013g00900 and VIT_216s0013g01070) encoding Ethylene-responsive transcription factor ERF105, known to regulate the plant response to abiotic and biotic stress (Mizoi et al., 2012; Mishra et al., 2015), were selected within the 2,6 DHBA, cis-piceid, and pterostilbene associated regions. Remarkable was also the significant enrichment of protein kinases in the region on LG16 associated both with 2,6 DHBA and pterostilbene: two genes encoding respectively for CLV1 and LRK10, which are receptor-like kinases (RLK) previously related to plant-microbe interaction and stress responses (Shiu and Bleecker, 2001), and four calcium-dependent protein kinases involved in the translation of pathogen signal-induced changes in the Ca2+ concentration during plant defense reactions (Schulz et al., 2013).
Combination of Rpv3-3 Haplotype, Stilbenoid Induction, and DM Resistance
In our previous study we found evidence of a putative involvement of stilbenoids in conferring DM resistance in the M×T segregating population (Malacarne et al., 2011), without taking into account the genetic background. To shed light into the found Rpv3-mediated DM resistance mechanism we have investigated the genetic bases of this mechanism. Although no overalapping QTLs controlling both DM resistance and stilbenoid-related traits were detected, we could highlight a significant induction of most stilbenoids, especially the polymeric ones, in a high fraction of the individuals. To date several studies have reported on (i) stilbenoid induction upon P. viticola infection (e.g., Langcake and Pryce, 1976; Alonso-Villaverde et al., 2011), (ii) a more rapid and extensive accumulation of stilbenoids in DM-resistant vs. DM-susceptible genotypes (V. vinifera cultivars) (Chitarrini et al., 2017), (iii) an antifungal activity carried out by stilbenoids on P. viticola and other fungi (e.g., Pezet et al., 2004a; Adrian and Jeandet, 2012; Gabaston et al., 2017). What still remains to be elucidated is the defense mechanism underlying DM resistance conferred by different Rpv loci. This is crucial for pyramiding various DM resistance mechanisms, derived from different sources, in the process of genetic improvement.
A deeper look into the Rpv3-3 haplotype within the M×T population highlighted a clear link among genetic background, DM resistance, and stilbenoid induction upon P. viticola infection. Unlike the Rpv3-3− genotypes, the Rpv3-3+ showed on average a significant induction of two dimeric and one trimeric stilbenoids and a positive correlation between the extent of induction and DM resistance. A clear explanation of the resistance in the Rpv3-3− individuals should be further elucidated, since in the present work we could not find any minor reliable DM resistance QTLs, as well as any metabolite exclusively induced in this group of genotypes.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
SV conceptualized the project, carried out genetic mapping and QTL analysis, and drafted the manuscript. GM conceptualized the project, performed the candidate gene selection with related expression analysis, and drafted the manuscript. DM and ZHM carried out the metabolite analysis. UV supported the metabolite analysis. AV and LZ performed the phenotyping assays. MS supported the phenotyping assays. CD and EB carried out the genotyping analysis. RV supported the genotyping analysis. VG performed the phylogenetic study and contributed to the manuscript revision. PF performed the statistical analysis. CM conceptualized as well as coordinated the project, and contributed to the discussion of the results. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully thank Jerome Grimplet (INRA) for the support on gene annotation and nomenclature, Luca Cappellin (FEM) for informatics help, and Susanna Micheli (FEM) for lab assistance. The authors are also grateful to the Grapevine Breeding platform personnel (FEM) for greenhouse and field assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00234/full#supplementary-material
References
Adam-Blondon, A. F., Roux, C., Claux, D., Butterlin, G., Merdinoglu, D., and This, P. (2004). Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor. Appl. Genet. 109, 1017–1027. doi: 10.1007/s00122-004-1704-y
Adrian, M., and Jeandet, P. (2012). Effects of resveratrol on the ultrastructure of Botrytis cinereaconidia and biological significance in plant/pathogen interactions. Fitoterapia 83, 1345–1350. doi: 10.1016/j.fitote.2012.04.004
Alonso-Villaverde, V., Voinesco, F., Viret, O., Spring, J.-L., and Gindro, K. (2011). The effectiveness of stilbenes in resistant vitaceae: ultrastructural and biochemical events during Plasmopara viticola infection process. Plant Physiol. Biochem. 49, 265–274. doi: 10.1016/j.plaphy.2010.12.010
Azuma, A., Ban, Y., Sato, A., Kono, A., Shiraishi, M., Yakushiji, H., et al. (2015). MYB diplotypes at the color locus affect the ratios of tri/di-hydroxylated and methylated/non-methylated anthocyanins in grape berry skin. Tree Genet. Genomes 11:31. doi: 10.1007/s11295-015-0855-0
Ban, Y., Mitani, N., Hayashi, T., Sato, A., Azuma, A., Kono, A., et al. (2014). Exploring quantitative trait loci for anthocyanin content in interspecific hybrid grape (Vitis labruscana × Vitis vinifera). Euphytica 198, 101–114. doi: 10.1007/s10681-014-1087-3
Barceló, A. R., Pomar, F., López-Serrano, M., and Pedreño, M. A. (2003). Peroxidase: a multifunctional enzyme in grapevines. Funct. Plant Biol. 30, 577–591. doi: 10.1071/FP02096
Bellés, J. M., Garro, R., Pallás, V., Fayos, J., Rodrigo, I., and Conejero, V. (2006). Accumulation of gentisic acid as associated with systemic infections but not with the hypersensitive response in plant-pathogen interactions. Planta 223, 500–511. doi: 10.1007/s00425-005-0109-8
Bellin, D., Peressotti, E., Merdinoglu, D., Wiedemann-Merdinoglu, S., Adam-Blondon, A. F., Cipriani, G., et al. (2009). Resistance to Plasmopara viticola in grapevine “Bianca” is controlled by a major dominant gene causing localised necrosis at the infection site. Theor. Appl. Genet. 120, 163–176. doi: 10.1007/s00122-009-1167-2
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Blasi, P., Blanc, S., Wiedemann-Merdinoglu, S., Prado, E., Rühl, E. H., Mestre, P., et al. (2011). Construction of a reference linkage map of Vitis amurensis and genetic mapping of Rpv8, a locus conferring resistance to grapevine downy mildew. Theor. Appl. Genet. 123, 43–53. doi: 10.1007/s00122-011-1565-0
Bocianowski, J. (2013). Epistasis interaction of QTL effects as a genetic parameter influencing estimation of the genetic additive effect. Genet. Mol. Biol. 36, 93–100. doi: 10.1590/S1415-47572013000100013
Boso Alonso, S., and Kassemeyer, H. H. (2008). Different Susceptibility of European Grapevine Cultivars for Downy Mildew. Available online at: https://digital.csic.es/handle/10261/43170 (Accessed October 12, 2018).
Breuil, A. C., Adrian, M., Pirio, N., Meunier, P., Bessis, R., and Jeandet, P. (1998). Metabolism of stilbene phytoalexins by Botrytis cinerea: characterization of a resveratrol dehydrodimer. Tetrahedron Lett. 39, 537–540. doi: 10.1016/S0040-4039(97)10622-0
Breuil, A. C., Jeandet, P., Chopin, F., Adrian, M., Pirio, N., Meunier, P., et al. (1999). Characterization of a pterostilbene dehydrodimer produced by laccase of Botrytis cinerea. Phytopathology 89, 298–302. doi: 10.1094/PHYTO.1999.89.4.298
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Calderón, A. A., Zapata, J. M., and Ros Barceló, A. (1994). Differential expression of a cell wall-localized peroxidase isoenzyme capable of oxidizing 4-hydroxystilbenes during the cell culture of grapevine (Vitis vinifera cv. Airen and Monastrell). Plant Cell Tissue Organ Cult. 37, 121–127. doi: 10.1007/BF00043605
Campos, L., Granell, P., Tárraga, S., López-Gresa, P., Conejero, V., Bellés, J. M., et al. (2014). Salicylic acid and gentisic acid induce RNA silencing-related genes and plant resistance to RNA pathogens. Plant Physiol. Biochem. 77, 35–43. doi: 10.1016/j.plaphy.2014.01.016
Casagrande, K., Falginella, L., Castellarin, S. D., Testolin, R., and Di Gaspero, G. (2011). Defence responses in Rpv3-dependent resistance to grapevine downy mildew. Planta 234, 1097–1109. doi: 10.1007/s00425-011-1461-5
Cavallini, E., Matus, J. T., Finezzo, L., Zenoni, S., Loyola, R., Guzzo, F., et al. (2015). The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine1. Plant Physiol. 167, 1448–1470. doi: 10.1104/pp.114.256172
Chen, C. N., Chu, C. C., Zentella, R., Pan, S. M., and Ho, T. H. D. (2002). AtHVA22 gene family in Arabidopsis: phylogenetic relationship, ABA and stress regulation, and tissue-specific expression. Plant Mol. Biol. 49, 633–644. doi: 10.1023/A:1015593715144
Chialva, C., Muñoz, C., Miccono, M., Eichler, E., Calderón, L., Prieto, H., et al. (2018). Differential expression patterns within the grapevine stilbene synthase gene family revealed through their regulatory regions. Plant Mol. Biol. Rep. 36, 225–238. doi: 10.1007/s11105-018-1073-3
Chitarrini, G., Soini, E., Riccadonna, S., Franceschi, P., Zulini, L., Masuero, D., et al. (2017). Identification of biomarkers for defense response to Plasmopara viticola in a resistant grape variety. Front. Plant Sci. 8:1524. doi: 10.3389/fpls.2017.01524
Corso, M., Vannozzi, A., Maza, E., Vitulo, N., Meggio, F., Pitacco, A., et al. (2015). Comprehensive transcript profiling of two grapevine rootstock genotypes contrasting in drought susceptibility links the phenypropanoid pathway to enhanced tolerance. J. Exp. Bot. 66, 5739–5752. doi: 10.1093/jxb/erv274
Costantini, L., Malacarne, G., Lorenzi, S., Troggio, M., Mattivi, F., Moser, C., et al. (2015). New candidate genes for the fine regulation of the colour of grapes. J. Exp. Bot. 66, 4427–4440. doi: 10.1093/jxb/erv159
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Dai, R., Ge, H., Howard, S., and Qiu, W. (2012). Transcriptional expression of Stilbene synthase genes are regulated developmentally and differentially in response to powdery mildew in norton and cabernet sauvignon grapevine. Plant Sci. 197, 70–76. doi: 10.1016/j.plantsci.2012.09.004
Dal Santo, S., Zenoni, S., Sandri, M., De Lorenzis, G., Magris, G., De Paoli, E., et al. (2018). Grapevine field experiments reveal the contribution of genotype, the influence of environment and the effect of their interaction (G × E) on the berry transcriptome. Plant J. Cell Mol. Biol. 93, 1143–1159. doi: 10.1111/tpj.13834
Di Gaspero, G., Copetti, D., Coleman, C., Castellarin, S. D., Eibach, R., Kozma, P., et al. (2012). Selective sweep at the Rpv3 locus during grapevine breeding for downy mildew resistance. Theor. Appl. Genet. 124, 277–286. doi: 10.1007/s00122-011-1703-8
Di Gaspero, G., and Foria, S. (2015). “Molecular grapevine breeding techniques,” in Grapevine Breeding Programs for the Wine Industry ed A. Reynolds (Cambridge, UK: Woodhead Publishing), 23–37.
Divilov, K., Barba, P., Cadle-Davidson, L., and Reisch, B. I. (2018). Single and multiple phenotype QTL analyses of downy mildew resistance in interspecific grapevines. Theor. Appl. Genet. 131, 1133–1143. doi: 10.1007/s00122-018-3065-y
Eurostat (2007). European Statistics. Available online at: https://ec.europa.eu/eurostat
Foria, S., Magris, G., Morgante, M., and Gaspero, G. D. (2018). The genetic background modulates the intensity of Rpv3-dependent downy mildew resistance in grapevine. Plant Breed. 137, 220–228. doi: 10.1111/pbr.12564
Fournier-Level, A., Hugueney, P., Verriès, C., This, P., and Ageorges, A. (2011). Genetic mechanisms underlying the methylation level of anthocyanins in grape (Vitis vinifera L.). BMC Plant Biol. 11:179. doi: 10.1186/1471-2229-11-179
Fournier-Level, A., Le Cunff, L., Gomez, C., Doligez, A., Ageorges, A., Roux, C., et al. (2009). Quantitative genetic bases of anthocyanin variation in grape (Vitis vinifera L. ssp. sativa) berry: a quantitative trait locus to quantitative trait nucleotide integrated study. Genetics 183, 1127–1139. doi: 10.1534/genetics.109.103929
Gabaston, J., Cantos-Villar, E., Biais, B., Waffo-Teguo, P., Renouf, E., Corio-Costet, M.-F., et al. (2017). Stilbenes from Vitis vinifera L. waste: a sustainable tool for controlling Plasmopara viticola. J. Agric. Food Chem. 65, 2711–2718. doi: 10.1021/acs.jafc.7b00241
Gessler, C., Pertot, I., and Perazzolli, M. (2011). Plasmopara viticola : a review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 50, 3-44–44. doi: 10.14601/Phytopathol_Mediterr-9360
Gindro, K., Pezet, R., and Viret, O. (2003). Histological study of the responses of two Vitis vinifera cultivars (resistant and susceptible) to Plasmopara viticola infections. Plant Physiol. Biochem. 41, 846–853. doi: 10.1016/S0981-9428(03)00124-4
Grimplet, J., Adam-Blondon, A. F., Bert, P. F., Bitz, O., Cantu, D., Davies, C., et al. (2014). The grapevine gene nomenclature system. BMC Genomics 15:1077. doi: 10.1186/1471-2164-15-1077
Guo, Y., Xue, R., Lin, H., Su, K., Zhao, Y., Zhendong, L., et al. (2015). Genetic analysis and QTL mapping for fruit skin anthocyanidin in grape (Vitis vinifera). Pak. J. Bot. 47, 1765–1771.
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., and Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. doi: 10.1186/gb-2007-8-2-r19
Herrero, J., Esteban-Carrasco, A., and Zapata, J. M. (2013). Looking for Arabidopsis thaliana peroxidases involved in lignin biosynthesis. Plant Physiol. Biochem. 67, 77–86. doi: 10.1016/j.plaphy.2013.02.019
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Höll, J., Vannozzi, A., Czemmel, S., D'Onofrio, C., Walker, A. R., Rausch, T., et al. (2013). The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25, 4135–4149. doi: 10.1105/tpc.113.117127
Huang, Y. F., Bertrand, Y., Guiraud, J. L., Vialet, S., Launay, A., Cheynier, V., et al. (2013). Expression QTL mapping in grapevine–revisiting the genetic determinism of grape skin colour. Plant Sci. 207, 18–24. doi: 10.1016/j.plantsci.2013.02.011
Huang, Y. F., Doligez, A., Fournier-Level, A., Le Cunff, L., Bertrand, Y., Canaguier, A., et al. (2012). Dissecting genetic architecture of grape proanthocyanidin composition through quantitative trait locus mapping. BMC Plant Biol. 12:30. doi: 10.1186/1471-2229-12-30
Huang, Y. F., Vialet, S., Guiraud, J. L., Torregrosa, L., Bertrand, Y., Cheynier, V., et al. (2014). A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 201, 795–809. doi: 10.1111/nph.12557
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Jeandet, P., Clément, C., and Cordelier, S. (2019). Regulation of resveratrol biosynthesis in grapevine: new approaches for disease resistance. J. Exp. Bot. 70, 375–378. doi: 10.1093/jxb/ery446
Jiang, J., Xi, H., Dai, Z., Lecourieux, F., Yuan, L., Liu, X., et al. (2019). VvWRKY8 represses stilbene synthase genes through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J. Exp. Bot. 70, 715–729. doi: 10.1093/jxb/ery401
Langcake, P., and Pryce, R. J. (1976). The production of resveratrol by Vitis vinifera and other members of the Vitaceae as a response to infection or injury. Physiol. Plant Pathol. 9, 77–86. doi: 10.1016/0048-4059(76)90077-1
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Malacarne, G., Costantini, L., Coller, E., Battilana, J., Velasco, R., Vrhovsek, U., et al. (2015). Regulation of flavonol content and composition in (syrah×pinot noir) mature grapes: integration of transcriptional profiling and metabolic quantitative trait locus analyses. J. Exp. Bot. 66, 4441–4453. doi: 10.1093/jxb/erv243
Malacarne, G., Perazzolli, M., Cestaro, A., Sterck, L., Fontana, P., Van de Peer, Y., et al. (2012). Deconstruction of the (paleo)polyploid grapevine genome based on the analysis of transposition events involving NBS resistance genes. PLoS ONE 7:e29762. doi: 10.1371/journal.pone.0029762
Malacarne, G., Pilati, S., Valentini, S., Asnicar, F., Moretto, M., Sonego, P., et al. (2018). Discovering causal relationships in grapevine expression data to expand gene networks. a case study: four networks related to climate change. Front. Plant Sci. 9:1385. doi: 10.3389/fpls.2018.01385
Malacarne, G., Vrhovsek, U., Zulini, L., Cestaro, A., Stefanini, M., Mattivi, F., et al. (2011). Resistance to Plasmopara viticola in a grapevine segregating population is associated with stilbenoid accumulation and with specific host transcriptional responses. BMC Plant Biol. 11:114. doi: 10.1186/1471-2229-11-114
Marguerit, E., Boury, C., Manicki, A., Donnart, M., Butterlin, G., Némorin, A., et al. (2009). Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine. Theor. Appl. Genet. 118, 1261–1278. doi: 10.1007/s00122-009-0979-4
Mattivi, F., Vrhovsek, U., Malacarne, G., Masuero, D., Zulini, L., Stefanini, M., et al. (2011). Profiling of resveratrol oligomers, important stress metabolites, accumulating in the leaves of hybrid Vitis vinifera (merzling × teroldego) genotypes infected with Plasmopara viticola. J. Agric. Food Chem. 59, 5364–5375. doi: 10.1021/jf200771y
Merdinoglu, D., Wiedeman-Merdinoglu, S., Coste, P., Dumas, V., Haetty, S., Butterlin, G., et al. (2003). Genetic analysis of downy mildew resistance derived from Muscadinia rotundifolia. Acta Hortic. 603, 451–456. doi: 10.17660/ActaHortic.2003.603.57
Mishra, S., Phukan, U. J., Tripathi, V., Singh, D. K., Luqman, S., and Shukla, R. K. (2015). PsAP2 an AP2/ERF family transcription factor from Papaver somniferum enhances abiotic and biotic stress tolerance in transgenic tobacco. Plant Mol. Biol. 89, 173–186. doi: 10.1007/s11103-015-0361-7
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Moreira, F. M., Madini, A., Marino, R., Zulini, L., Stefanini, M., Velasco, R., et al. (2011). Genetic linkage maps of two interspecific grape crosses (Vitis spp.) used to localize quantitative trait loci for downy mildew resistance. Tree Genet. Genomes 7, 153–167. doi: 10.1007/s11295-010-0322-x
Moroldo, M., Paillard, S., Marconi, R., Fabrice, L., Canaguier, A., Cruaud, C., et al. (2008). A physical map of the heterozygous grapevine “Cabernet Sauvignon” allows mapping candidate genes for disease resistance. BMC Plant Biol. 8:66. doi: 10.1186/1471-2229-8-66
Nawrocka, J., Małolepsza, U., Szymczak, K., and Szczech, M. (2018). Involvement of metabolic components, volatile compounds, PR proteins, and mechanical strengthening in multilayer protection of cucumber plants against Rhizoctonia solani activated by Trichoderma atroviride TRS25. Protoplasma 255, 359–373. doi: 10.1007/s00709-017-1157-1
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nicolas, P., Lecourieux, D., Kappel, C., Cluzet, S., Cramer, G., Delrot, S., et al. (2014). The basic leucine zipper transcription factor ABSCISIC ACID RESPONSE ELEMENT-BINDING FACTOR2 is an important transcriptional regulator of abscisic acid-dependent grape berry ripening processes. Plant Physiol. 164, 365–383. doi: 10.1104/pp.113.231977
Ochssner, I., Hausmann, L., and Töpfer, R. (2016). “Rpv14”, a new genetic source for “Plasmopara viticola” resistance conferred by “Vitis cinerea.” Vitis J. Grapevine Res. 55, 79–81. doi: 10.5073/vitis.2016.55.79-81
OEPP/EPPO (2001). Guidelines for the efficacy evaluation of fungicides. OEPP/EPPO Bull. 31, 313–317.
OIV (2009). Descriptor List for Grape Varieties and Vitis Species, 2nd Edn. Office International de la Vigne et du Vin. Available online at: http://www.oiv.org
Peressotti, E., Dolzani, C., Poles, L., Banchi, E., Stefanini, M., Salamini, F., et al. (2015). A first pedigree-based analysis (PBA) approach for the dissection of disease resistance traits in grapevine hybrids. Acta Hortic. 1082, 113–122. doi: 10.17660/ActaHortic.2015.1082.15
Peressotti, E., Wiedemann-Merdinoglu, S., Delmotte, F., Bellin, D., Di Gaspero, G., Testolin, R., et al. (2010). Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol. 10:147. doi: 10.1186/1471-2229-10-147
Pezet, R., Gindro, K., Viret, O., and Richter, H. (2004a). Effects of resveratrol, viniferins and pterostilbene on Plasmopara viticola zoospore mobility and disease development. Vitis J. Grapevine Res. 43, 145–148.
Pezet, R., Gindro, K., Viret, O., and Spring, J. L. (2004b). Glycosylation and oxidative dimerization of resveratrol are respectively associated to sensitivity and resistance of grapevine cultivars to downy mildew. Physiol. Mol. Plant Pathol. 65, 297–303. doi: 10.1016/j.pmpp.2005.03.002
Pezet, R., Pont, V., and Hoang-Van, K. (1991). Evidence for oxidative detoxication of pterostilbene and resveratrol by a laccase-like stilbene oxidase produced by Botrytis cinerea. Physiol. Mol. Plant Pathol. 39, 441–450. doi: 10.1016/0885-5765(91)90010-F
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Ranwez, V., Harispe, S., Delsuc, F., and Douzery, E. J. P. (2011). MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS ONE 6:e22594. doi: 10.1371/journal.pone.0022594
Reid, K. E., Olsson, N., Schlosser, J., Peng, F., and Lund, S. T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6:27. doi: 10.1186/1471-2229-6-27
Salmaso, M., Malacarne, G., Troggio, M., Faes, G., Stefanini, M., Grando, M. S., et al. (2008). A grapevine (Vitis vinifera L.) genetic map integrating the position of 139 expressed genes. Theor. Appl. Genet. 116, 1129–1143. doi: 10.1007/s00122-008-0741-3
Schulz, P., Herde, M., and Romeis, T. (2013). Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol. 163, 523–530. doi: 10.1104/pp.113.222539
Schwander, F., Eibach, R., Fechter, I., Hausmann, L., Zyprian, E., and Töpfer, R. (2012). Rpv10: a new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor. Appl. Genet. 124, 163–176. doi: 10.1007/s00122-011-1695-4
Schwarz, G. (1978). Estimating the dimension of a model. Ann. Statist. 6, 461–464. doi: 10.1214/aos/1176344136
Shi, J., He, M., Cao, J., Wang, H., Ding, J., Jiao, Y., et al. (2014). The comparative analysis of the potential relationship between resveratrol and stilbene synthase gene family in the development stages of grapes (Vitis quinquangularis and Vitis vinifera). Plant Physiol. Biochem. 74, 24–32. doi: 10.1016/j.plaphy.2013.10.021
Shiu, S. H., and Bleecker, A. B. (2001). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE. 2001:re22. doi: 10.1126/stke.2001.113.re22
Soubrier, J., Steel, M., Lee, M. S. Y., Der Sarkissian, C., Guindon, S., Ho, S. Y. W., et al. (2012). The influence of rate heterogeneity among sites on the time dependence of molecular rates. Mol. Biol. Evol. 29, 3345–3358. doi: 10.1093/molbev/mss140
Takaya, Y., Terashima, K., Ito, J., He, Y.-H., Tateoka, M., Yamaguchi, N., et al. (2005). Biomimic transformation of resveratrol. Tetrahedron 61, 10285–10290. doi: 10.1016/j.tet.2005.08.023
Tanksley, S. D., and Nelson, J. C. (1996). Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 92, 191–203. doi: 10.1007/BF00223376
Tognolli, M., Penel, C., Greppin, H., and Simon, P. (2002). Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288, 129–138. doi: 10.1016/S0378-1119(02)00465-1
Tokunaga, N., Kaneta, T., Sato, S., and Sato, Y. (2009). Analysis of expression profiles of three peroxidase genes associated with lignification in Arabidopsis thaliana. Physiol. Plant. 136, 237–249. doi: 10.1111/j.1399-3054.2009.01233.x
Töpfer, R., Hausmann, L., and Eibach, R. (2011). “Molecular breeding,” in Genetics, genomics and breeding of grapes, eds A. F. Adam-Blondon, J. M. Martínez-Zapater, and C. Kole (Boca Raton, FL: CRC Press), 160–185.
van Heerden, C. J., Burger, P., Vermeulen, A., and Prins, R. (2014). Detection of downy and powdery mildew resistance QTL in a “Regent” × “RedGlobe” population. Euphytica 200, 281–295. doi: 10.1007/s10681-014-1167-4
Van Ooijen, J. (2009). Van Ooijen JW (2009) MapQTL 6, Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species. Wageningen: Kyazma B.V.
Van Ooijen, J. W. (2006). JoinMap.4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen: Kyazma BV.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Vannozzi, A., Dry, I. B., Fasoli, M., Zenoni, S., and Lucchin, M. (2012). Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol. 12:130. doi: 10.1186/1471-2229-12-130
Vannozzi, A., Wong, D. C. J., Höll, J., Hmmam, I., Matus, J. T., Bogs, J., et al. (2018). Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant Cell Physiol. 59, 1043–1059. doi: 10.1093/pcp/pcy045
Venuti, S., Copetti, D., Foria, S., Falginella, L., Hoffmann, S., Bellin, D., et al. (2013). Historical introgression of the downy mildew resistance gene Rpv12 from the Asian species Vitis amurensis into grapevine varieties. PLoS ONE 8:e61228. doi: 10.1371/journal.pone.0061228
Vezzulli, S., Troggio, M., Coppola, G., Jermakow, A., Cartwright, D., Zharkikh, A., et al. (2008). A reference integrated map for cultivated grapevine (Vitis vinifera L.) from three crosses, based on 283 SSR and 501 SNP-based markers. Theor. Appl. Genet. 117, 499–511. doi: 10.1007/s00122-008-0794-3
Vezzulli, S., Vecchione, A., Stefanini, M., and Zulini, L. (2018). Downy mildew resistance evaluation in 28 grapevine hybrids promising for breeding programs in Trentino region (Italy). Eur. J. Plant Pathol. 150, 485–495. doi: 10.1007/s10658-017-1298-2
Viana, A. P., Riaz, S., and Walker, M. A. (2013). Genetic dissection of agronomic traits within a segregating population of breeding table grapes. Genet. Mol. Res. 12, 951–964. doi: 10.4238/2013.April.2.11
Vitulo, N., Forcato, C., Carpinelli, E. C., Telatin, A., Campagna, D., D'Angelo, M., et al. (2014). A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 14:99. doi: 10.1186/1471-2229-14-99
VIVC (2018). Vitis International Variety Catalogue. Available online at: http://www.vivc.de/
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Vrhovsek, U., Masuero, D., Gasperotti, M., Franceschi, P., Caputi, L., Viola, R., et al. (2012). A versatile targeted metabolomics method for the rapid quantification of multiple classes of phenolics in fruits and beverages. J. Agric. Food Chem. 60, 8831–8840. doi: 10.1021/jf2051569
Wan, X., Wang, X. B., Yang, M. H., Wang, J. S., and Kong, L. Y. (2011). Dimerization of piceatannol by Momordicacharantia peroxidase and α-glucosidase inhibitory activity of the biotransformation products. Bioorg. Med. Chem. 19, 5085–5092. doi: 10.1016/j.bmc.2011.07.032
Wang, C., Wu, J., Zhang, Y., and Lu, J. (2018). Muscadinia rotundifolia “Noble” defense response to Plasmopara viticola inoculation by inducing phytohormone-mediated stilbene accumulation. Protoplasma 255, 95–107. doi: 10.1007/s00709-017-1118-8
Wang, M., Vannozzi, A., Wang, G., Liang, Y.-H., Tornielli, G. B., Zenoni, S., et al. (2014). Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 1:14016. doi: 10.1038/hortres.2014.16
Welter, L. J., Göktürk-Baydar, N., Akkurt, M., Maul, E., Eibach, R., Töpfer, R., et al. (2007). Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol. Breed. 20, 359–374. doi: 10.1007/s11032-007-9097-7
Wickham, H. (2017). tidyverse: Easily Install and Load the “Tidyverse”. R package version 1.2.1. Available online at: https://CRAN.R-project.org/package=tidyverse
Wong, D. C. J., Schlechter, R., Vannozzi, A., Höll, J., Hmmam, I., Bogs, J., et al. (2016). A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res. 23, 451–466 doi: 10.1093/dnares/dsw028
Keywords: disease symptom phenotyping, “Merzling” Plasmopara viticola, peroxidase, polyphenols, QTL analysis, stilbenes, Vitis spp.
Citation: Vezzulli S, Malacarne G, Masuero D, Vecchione A, Dolzani C, Goremykin V, Mehari ZH, Banchi E, Velasco R, Stefanini M, Vrhovsek U, Zulini L, Franceschi P and Moser C (2019) The Rpv3-3 Haplotype and Stilbenoid Induction Mediate Downy Mildew Resistance in a Grapevine Interspecific Population. Front. Plant Sci. 10:234. doi: 10.3389/fpls.2019.00234
Received: 09 November 2018; Accepted: 12 February 2019;
Published: 06 March 2019.
Edited by:
Aziz Aziz, Université de Reims Champagne-Ardenne, FranceReviewed by:
Courtney Coleman, University of Missouri, United StatesPhilippe Jeandet, Université de Reims Champagne-Ardenne, France
Copyright © 2019 Vezzulli, Malacarne, Masuero, Vecchione, Dolzani, Goremykin, Mehari, Banchi, Velasco, Stefanini, Vrhovsek, Zulini, Franceschi and Moser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giulia Malacarne, Z2l1bGlhLm1hbGFjYXJuZUBmbWFjaC5pdA==
†These authors have contributed equally to this work
 Silvia Vezzulli
Silvia Vezzulli Giulia Malacarne
Giulia Malacarne Domenico Masuero
Domenico Masuero Antonella Vecchione1
Antonella Vecchione1 Zeraye Haile Mehari
Zeraye Haile Mehari Urska Vrhovsek
Urska Vrhovsek Luca Zulini
Luca Zulini Pietro Franceschi
Pietro Franceschi Claudio Moser
Claudio Moser