- 1Co-Innovation Center for Sustainable Forestry in Southern China, Key Laboratory of Forest Genetics and Biotechnology, Ministry of Education, College of Biology and the Environment, Nanjing Forestry University, Nanjing, China
- 2Jiangsu Provincial Key Construction Laboratory of Special Biomass Resource Utilization, Nanjing Xiaozhuang University, Nanjing, China
Increased expression of the insect control protein genes of Bacillus thuringiensis in Populus has been critical to the development of genetically improved plants with agronomically acceptable levels of insect resistance. Bacillus thuringiensis (Cry1Ah1) proteins with highly specific toxicity against Hyphantria cunea were screened using an indoor bioactivity assay to obtain hyper-resistant transgenic poplars. Then, the Cry1Ah1 sequence was optimized and transformed according to the optimal codon in poplar using software of our own design (http://120.79.60.226:8080/u/chen/w/codonpoplar). A vector was constructed to transform poplar NL895. The Cry1Ah1 gene was transformed to poplar NL895 and six transgenic lines were obtained. The expression and insecticidal effect of the Cry1Ah1 gene in transgenic poplar were evaluated by PCR and ELISA, and the specific indoor activity and field insecticidal activity against H. cunea were compared with a control. We concluded that the insecticidal activity of the transgenic NL895 was significantly better against lower instar larvae of H. cunea than against higher instar larvae. The mortality and pupation rates clearly differed among the various instar larvae and between transgenic and non-transgenic poplar. We obtained poplar seedlings with hyper-resistance to H. cunea by screening Bt genes and optimizing their genetic sequence.
Introduction
Poplar is an important fast-growing forest tree. The total area planted with poplar in China is among the largest in the world (Hu et al., 2017; Wang et al., 2018), at about eight million hectares (Lu, 2008; Tun et al., 2018). However, large-scale mono-species planting of poplar can lead to serious pest problems, especially from lepidopteran and coleopteran insects. Lepidopteran pests include Hyphantria cunea Drury, Lymantria dispar Linnaeus, Apocheima cinerarius Erschoff, Malacosoma neustria Motschulsky, and Limacodidae and Notodontidae moths (Wang et al., 2018). Chemical agents and bio-insecticides are not only costly but also tend to result in serious pollution. Therefore, breeding insect-resistant varieties and improving the insect resistance of forest trees have become inevitable choices when cultivating new varieties of poplar. Of all the insect-resistance genes, Bt has been the most widely studied in poplar (Peña and Séguin, 2001). Bt transgenic poplar with insecticidal effects has been developed (McCown et al., 1991). In 1993, China also reported transgenic Populus nigra transformed with the Bt CrylAc gene; the mortality rate of gypsy moth increased to 80–90% (Tian et al., 1993). In 2002, the State Forestry Administration approved a strain of transgenic poplar for commercialization in China; this was the world’s first commercialized transgenic tree species (Wang et al., 2018). During this period, insect-resistant transgenic P. nigra (Hu et al., 2001), P. deltoides (Ramachandran et al., 1993) P. euramericana (Wang et al., 1997), NL-80106 (Populus deltoides × Populus simonii) (Guo et al., 2004) and 741 poplar (Tian et al., 2000; Yang et al., 2003) were successfully cultivated, and there are now resources for continuously introducing new insect-resistant poplar varieties (Zuo et al., 2018).
The fall webworm Hyphantria cunea is a destructive pest that affects a number of ornamental trees and shrubs and several agricultural crops native to North America, where two morphs (red- and black-headed) are found (Cao et al., 2016). The black-headed morph was inadvertently introduced to Asia in 1945 and to Europe in 1946. Currently, it has spread to more than 32 countries worldwide (Sullivan et al., 2012). The extreme polyphagy of the fall webworm (Firidin et al., 2008), including high fecundity, short generation time, and high starvation resistance, facilitate its spread and potential to damage crops. The webworm is a polyphagous pest that inflicts US$1 billion in crop damage annually in China (Subramanian and Mohankumar, 2006). There are more than seven million hectares of poplar plantations in China. The large areas of pure forest and foreign tree species provide conditions for H. cunea to breed and spread, and in recent years it has become increasingly problematic.
Bacillus thuringiensis (Bt) isolates are being screened worldwide in search of new insecticidal genes. More than 320 cry genes grouped into 40 families have been isolated from Bt strains, with an insecticidal spectrum that extends over several invertebrate orders (Crickmore et al., 1998). Because the products of cry1A genes are reported to be toxic to lepidopteran pests, to improve insecticide expression 23 protoxin forms of Bt toxins were selected to test as toxins that act on H. cunea.
To increase gene expression to promote increased insecticide expression, H. cunea (Cry1Ah1) proteins with high toxicity were first screened using an indoor bioactivity assay. Then, our custom software was used to examine the coding sequence and produce synthetic genes, without changing the amino acid sequences. The demonstration of the utility of these genes for providing protection from insects has far-reaching implications for the future of insect-resistant woody plants and for the application of high-expression heterogonous gene design in woody plants (Perlak et al., 1991; Figure 1). Of particular importance and convenience is the highly efficient genetic transformation system coupled with efficient regeneration of poplar that is unsurpassed in other tree crops.
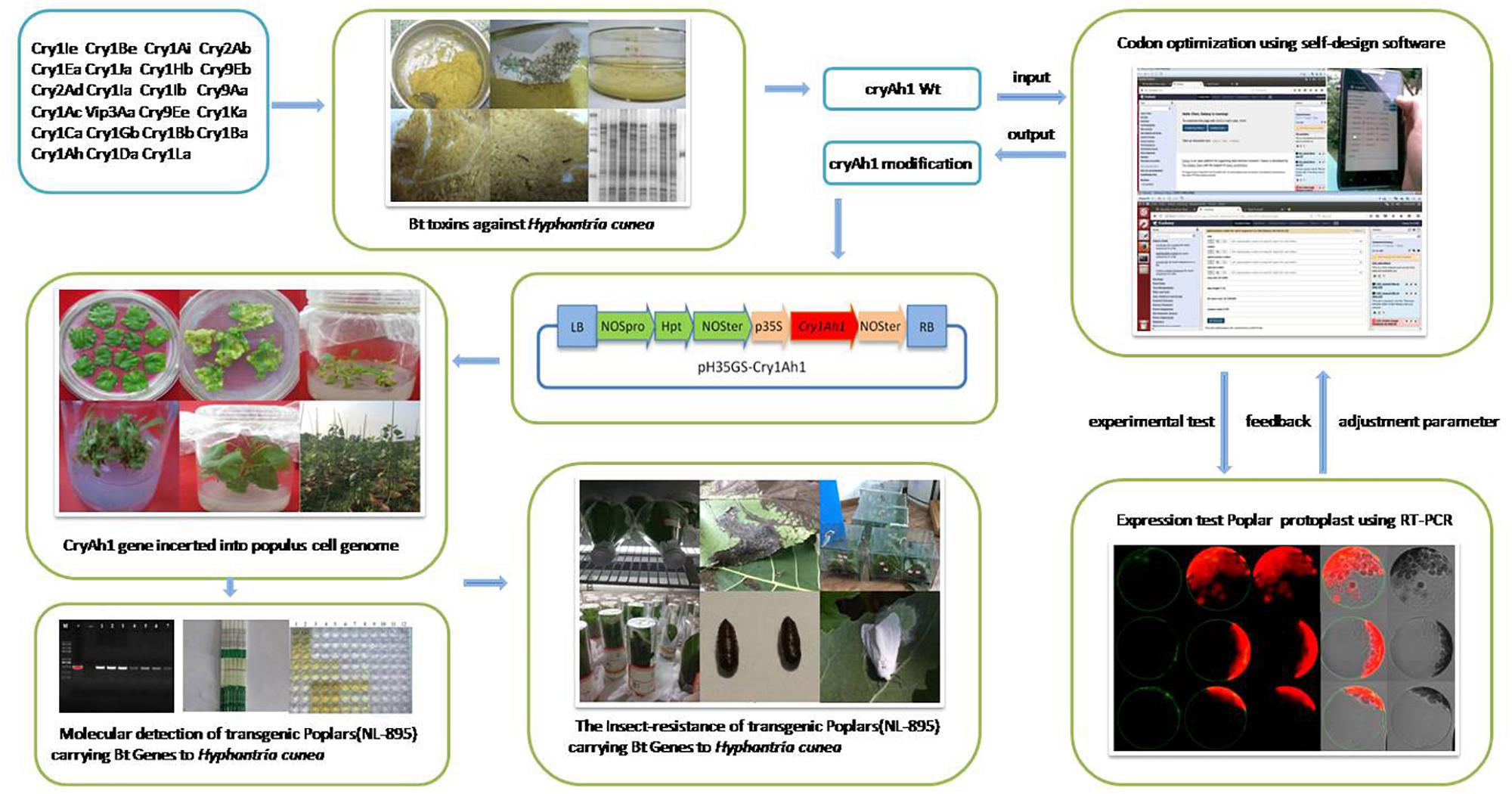
Figure 1. Flow chart of the transformation of transgenic poplars hyper-resistant to Hyphantria cunea using an optimized cry1Ah1 sequence.
Materials and Methods
Plant Materials and Growth Conditions
Leaf disks were prepared from young Populus deltoides × Populus euramericana “Nanlin895” seedlings and pre-cultured for transformation (Zhang et al., 2015). The tested poplars were cultured by the leaf disc method with leaf explants. The seedlings were transplanted after 5 months of growth.
Insect Strains
Insect bioassays were conducted to observe the responses of H. cunea larvae on the leaves of transgenic and control poplars. The first instar larvae of H. cunea were provided by the Research Institute of Forest Ecology, Environment and Protection, of the Chinese Academy of Forestry.
Bt Screening Strains and BT Toxin Extract
Protoxin inclusion bodies were purified from transformed Escherichia coli from the general microbial collection of the Biotechnology and Biological Control Unit and randomly chosen to evaluate their insecticidal activity against worm larvae. The protoxin forms of Bt toxins were used in strains of Bt: Cry1Ie, Cry1Be, Cry1Ai, Cry2Ab, Cry1Ea, Cry1Ja, Cry1Hb, Cry9Eb, Cry2Ad, Cry1Ia, Cry1Ib, Cry9Aa, Cry1Ac, Vip3Aa, Cry9Ee, Cry1Ka, Cry1Ca, Cry1Gb, Cry1Bb, Cry1Ba, Cry1Ah, Cry1Da, and Cry1La1. For all experiments, the strains were provided by the Jie Zhang laboratory at the Institute of Plant Protection of the Chinese Academy of Agricultural Sciences (Gao et al., 2010; Li et al., 2014). Bt strains were routinely cultured on lysogeny broth medium. Protoxins were purified and solubilized as previously described (Lee et al., 1992). Solubilized protoxins were dialyzed against 50 mM Na2CO3 (pH 10). Protoxin concentrations were estimated by measuring total protein using the Bradford (1976) method, with bovine serum (BSA) as the standard, and percent toxin compositions were determined after the proteins had been separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. All protoxin suspensions were stored at 20°C until use.
Insect Bioassay
Bioassays were carried out with H. cunea using the diet-incorporation method with three replicates of 20 insects, each with appropriate controls (Meena et al., 2001). An aliquot of toxin solution was mixed with 15 g of diet at various concentrations. Neonates were fed an artificial diet and exposed to Bt protoxin (Gao et al., 2010). The toxin contents were based on the protein contents. The treated diet was then divided into three replicates and fed to neonates separately. Artificial diet without Bt protoxin was used as a control. Three replicates were set for each concentration, and 20 neonates were used for each replicate. The neonates were allowed to feed freely. Larval mortality was observed until 96 h. The mortality data were used to estimate the 50% lethal concentration (LC50) in terms of μg.g-1 diet as per the maximum-likelihood program.
Codon Optimization Software Construction
We independently developed improved genetic modification software that was used to redesign the cry1Ah1 sequence. The software, CodonPoplar (Supplementary Figure S1), is implemented in C with command-line options for the various parameters and is available from the author. It can be used online at http://120.79.60.226:8080/u/chen/w/codonpoplar. The redesigned sequences include localized regions of A+T richness resembling plant introns, potential plant polyadenylation signal sequences, ATTTA sequences, which have been shown to destabilize mRNA in other systems, and plant codon optimization. This software was used to synthesize the full cry1Ah1 gene as follows:
(i) codon usage was altered to match certain features of host genes according to the optimal poplar codon;
(ii) the GC content was altered to match host genes;
(iii) known RNA instability motifs were avoided;
(1) mRNA degradation signals were removed (ATTTA and ATTAA);
(2) phylogenetic cleavage signals were removed (AATAAA, AATAAT, AATTAA, AACCAA, ATTA, ATTTA, ATAAAA, ATGAAA, AAGCAT, ATATAA, AATCAA, ATACTA, ATACAT, AAAATA, ATTAAA, AATTAA, AATACA, and CATAAA);
(3) polyadenylation sequences were removed (AATAAT type, AATCAA type, AATGAA type, ATGGAA type, AATTAA type, TATAAA type, other types including ATGTAA, TGTGAA, AATGCT, GATATG, ATGCAA, AATGTG, AAAGAT, ATTAA, AATAAA, and AATAAT);
(4) the intronic cleavage sequence (CATTG) contained in the coding region sequence was removed; and
(5) continuous AT enrichment areas greater than four were modified;
(iv) potential RNA secondary structures near the translational start site were removed.
Plasmid Construction and Transformation
The synthetic full-length sequences of the cry1Ah1 genes were cloned into the Gateway entry vector pENTR/D-TOPO or pCR8/GW/TOPO (Invitrogen, Carlsbad, CA, United States), and transferred to the destination vector pH35GS (Kubo et al., 2005) by the LR reaction using LR Clonase II (Invitrogen). In the resulting plasmids, cry1Ah1 was expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The plasmids were electroporated into Agrobacterium tumefaciens strain LB4404 and confirmed by agarose gel electrophoresis.
Putative transgenic plant lines were selected and propagated for PCR and real time (RT)-PCR. Following multiplex selection of transgenic poplar, the plantlets were grown on agar medium for 4 weeks in a confined culture room and moved to a greenhouse when there were four to five leaves on the top bud of the plantlet. After 3 days of acclimatization, the plantlets were transplanted into soil. The soil was then mixed with sterilized peat and perlite (2:1). During the first 2 weeks, water and vinyl membranes were used to maintain humidity.
RT-PCR and Quantitative Real-Time Quantitative PCR (qPCR)
RT-PCR analysis of transgenic plants was performed by synthesis of first-strand cDNA with an enhanced avian RT-PCR kit using 5 μg of total RNA purified from a transgenic plant according to the manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO, United States). The relative quantities of cry1Ah1 transcripts in transgenic poplar plants were analyzed by qPCR performed using a StepOne real-time PCR system (Applied Biosystems, Foster City, CA, United States) and a QuantiFast SYBR green PCR kit (QIAGEN, Hilden, Germany). The poplar β-actin gene (GenBank Accession No. U60482) was used as an endogenous control in all RT-PCR assays. The nucleotide sequences of the forward and reverse primers for the cry1Ah1 and β-actin genes were 5′-TCACTTCCCAAGCACATC-3′ and 5′ATCCTTCTCGGACAGACAA-3′, and 5’CTTCTCCTGTCGGTTTGTCG-3′ and 5′-TGGCAAATTTGAGGAGGTTC-3′, respectively, generating amplicons of 178 and 169 base pairs (bp), respectively. Total RNA extracted from 100 mg of leaf tissue was reverse transcribed into cDNA and used as a template in RT-PCR assays with cry1Ah and β-actin gene-specific primers. Reverse transcription was performed at 50°C for 10 min with an initial denaturation at 95°C for 5 min (for activation of hot-start Taq polymerase), followed by 40 amplification cycles comprised of 10 s denaturation at 95°C and a combined annealing and extension step at 60°C for 30 s in a 25-μl reaction mixture, according to the manufacturer’s instructions (QIAGEN). The relative values obtained from the quantitation of mRNA were expressed as 2-ΔΔCt, where ΔCt represents the difference between the cycle threshold (Ct) values of a target and the endogenous control (β-actin) in the same sample and ΔΔCt is the difference between the ΔCt value of a particular sample and that of the reference sample. The quantitative RT-PCR data represent mean values with standard error of three independent experiments with three replicates of the transgenic plant.
Enzyme-Linked Immunosorbent Assay (ELISA)
Non-transgenic (CK) and transgenic Bt poplar samples were randomly selected. The procedures were performed using a Bt-cry1Ab ELISA kit (Agdia, Elkhart, IN, United States) with 0.5 g of each sample according to the manufacturer’s instructions. Total plant protein was measured by Coomassie blue staining of protein gels and each sample was measured five times.
Insect Bioassay Testing
Six transformed transgenic poplar lines that showed relatively high expression levels of the target genes were selected for the insect-feeding experiment in the laboratory. Bt transgenic poplars and their corresponding non-Bt versions were exposed to first through sixth larval instar H. cunea to determine percent mortalities and times to pupation. The six strains and the non-transgenic poplar NL895 control were all set in the greenhouse in April. In June of that year, the first generation of H. cunea was placed in an artificial climate chamber at 28°C with 14 h of light. Uniform hatching and screening of healthy larvae were performed, and larvae of different ages were grouped and raised. The newly hatched larvae were raised to the test-insect age on leaves of the control poplar to obtain experimental insects at various ages, including 1-day-old larvae, which were used as test insects by raising them on the leaves of the control poplar for 1 day.
Using the indoor artificial group breeding method, from July to September second- to third-generation H. cunea and the third to fifth expanded leaves from the tops of the tested plants were collected and placed in 100-mL triangular flasks. Ten larvae were tested each day, using three flasks and three replicates. Since the larvae can climb, the flasks’ openings were sealed with gas-permeable gauze and then the flasks were inverted in an artificial climate chamber to ensure that the larvae could feed on the leaves normally. The leaves were replaced every 2 days until pupation, and larval growth and mortality were recorded.
Insect Feeding on Transgenic Lines
Transgenic poplars were planted in the spring at a forest station nursery in Xuzhou, Jiangsu Province. Our transgenic poplars were granted a license for field experiments by the Biology Genetic Engineering Security Council of State Forestry Administration and were planted in controlled regions in March 2017. The wild goat cage experiment began in early September. In the natural case, a 50-cm tube of dense nylon mesh with a radius of 15 cm was placed on an upper branch of the plant. With indoor feeding, 1-day-old larvae were obtained as a test source. Each treatment started with 20 1-day-old larvae and 60 buds, and three replicates were performed. After the larvae were added, the ends of the nylon-mesh tube were closed and sealed, and larval growth status and mortality were recorded. The corrected mortality of the larvae at 6 and 12 days was determined. The SPSS statistics package was used to analyze the data. Mortality was corrected using Abbott’s formula:
Results
Bt-Screened H. cunea With High Protein Toxicity Using the Indoor Bioactivity Assay
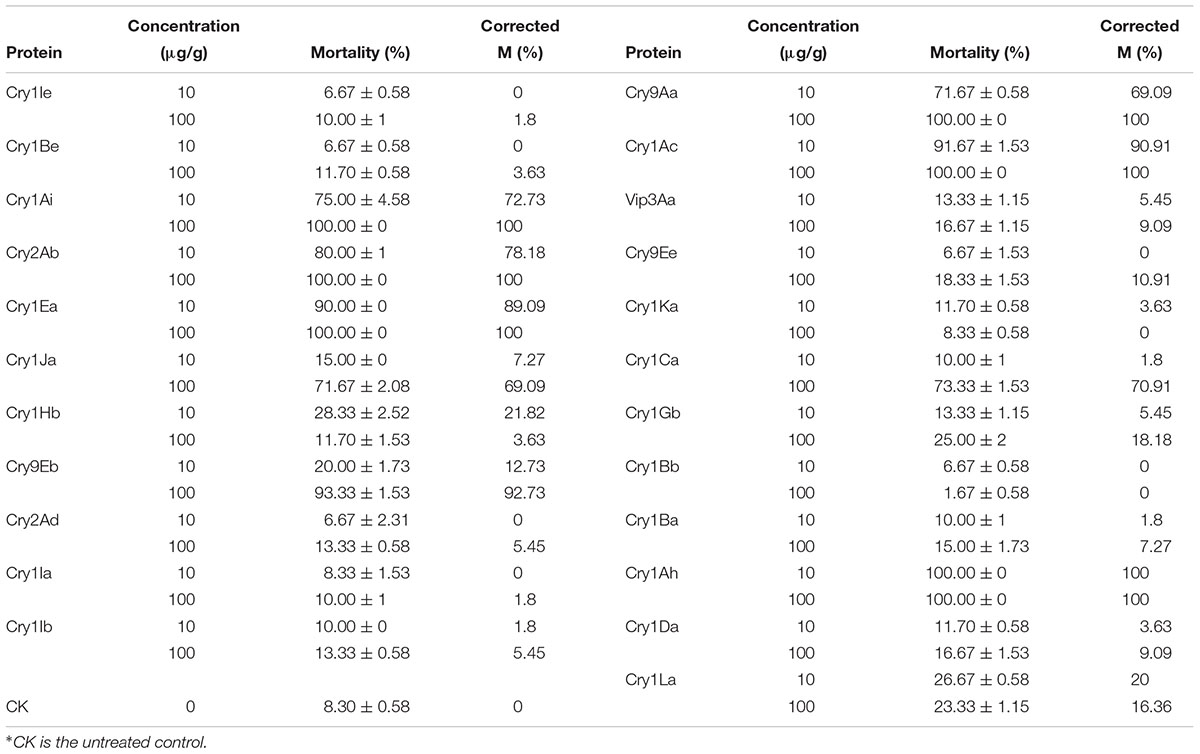
The bioassay results for each protein against the H. cunea larvae are shown in Table 1. When treated with 10 μg/g, the corrected mortality with Cry1Ah1 reached 100%. With Cry1Ea and Cry1Ac, the protein toxicity against the American larvae was significantly lower and the corrected mortality reached 90%. Therefore, Cry1Ah1 was selected as the target gene. In the control group, the sides of the larval bodies were light yellow, and the larvae were very active at spinning and netting. The bodies of larvae fed Cry1Ah1 protein were dark, small, and stiff, and appeared dehydrated (Figure 2).
Figure 2. The larvae after Bt treatment with Cry1Ah. (a) The sides of the larval bodies in the control group were light yellow, and the larvae were very active at spinning and netting. (b) The larvae fed Cry1Ah1 protein were dark, small, and stiff, and appeared dehydrated.
Gene Synthesis
Increased expression of insect-control protein genes of B. thuringiensis in Populus has been critical to the development of genetically improved plants with agronomically acceptable levels of insect resistance.
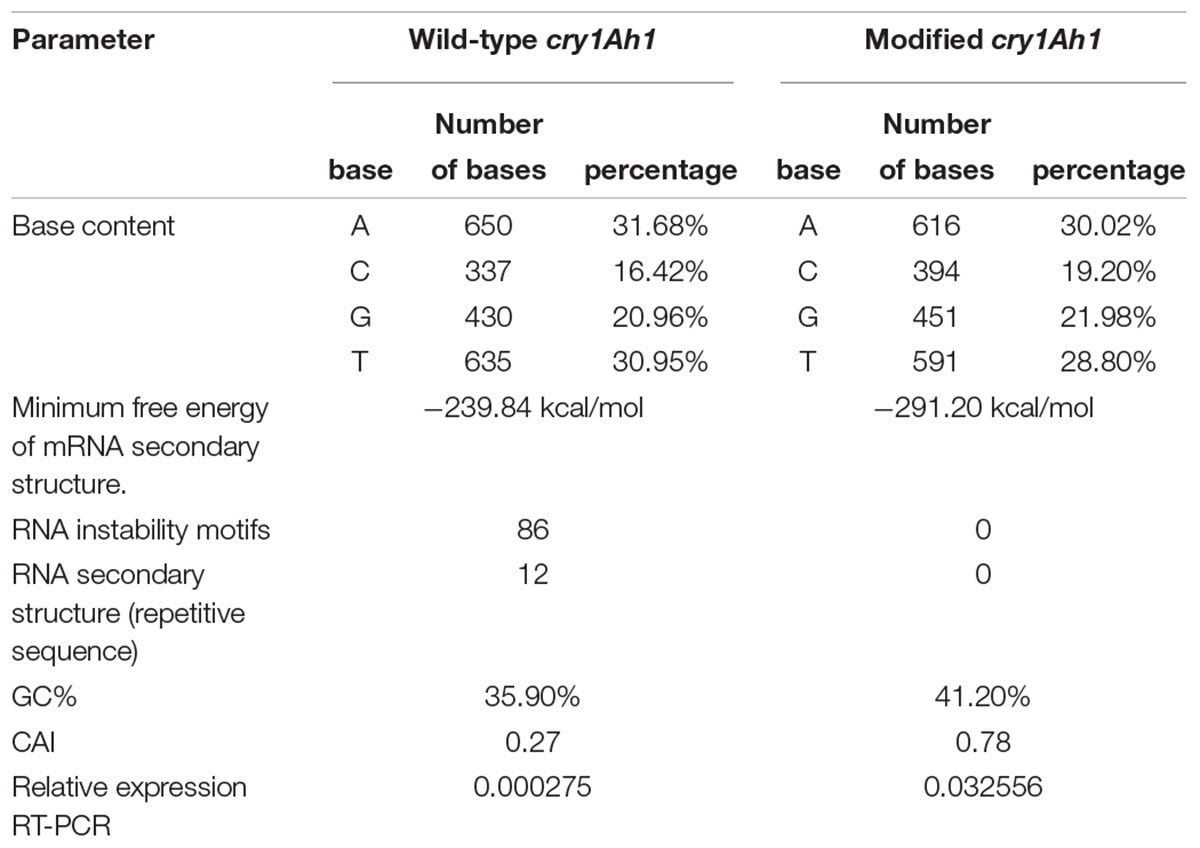
The gene encoding cry1Ah1 was chemically synthesized and the codon was optimized to the full sequence of poplar cells. The synthetic gene sequence is shown (Supplementary Table S1). The gene was synthesized with 41.2% GC content and a 0.78 codon adaptation index (Table 2). The RNA secondary structure was removed and RNA instability motifs were avoided. There was no significant change in the GC contents of the two modified genes, and the minimum free energy of the RNA secondary structure of the modified genes was reduced from -239.84 to -291.20 kcal/mol, which made the transcribed mRNA more stable. Codon usage was adapted to the codon bias of the optimal poplar codon. The vector pH35GS-Cry1Ah1 (Figure 3) was constructed and used to transform poplar clones (Figure 4).
Figure 3. Schematic representation of the T-DNA region in the plasmid vector pH35GS-cry1Ah. LB T-DNA, left border repeat; p35sl CaMV35S promoter; NOSter, NOS terminator; NOSpro, NOS promoter; Hpt, hygromycin resistance gene; RB, T-DNA right border repeat.

Figure 4. Agrobacterium-mediated transformation of poplar. (a) A typical pre-selection plate. (b) Tiny transgenic calli and shoot buds. (c) Transformants with healthy development of shoots and leaves. (d) Transgenic plantlets grown in vitro.
Results of Molecular Detection
The synthetic cry1Ah1 poplar gene fragments were screened by PCR in six independent transgenic lines (Figure 5). PCR primers were used to amplify cry1Ah1. In transgenic plants, the primer pair amplified a band of 561 bp. RT-PCR showed that the modified genes provided enhanced expression (Supplementary Table S2). Expression of the new construct was greater than that of the wild-type gene, and the results showed improvement with respect to RNA stability and expression. The ELISA test of Bt protein expression in transgenic NL895 (Table 3) showed that the total protein content of all lines, including the controls, remained between 24.013 and 25.847 mg/g. Strain No. 6 had the highest toxic protein content, which reached 11500.01 ng/g, accounting for 0.0478% of the total protein expressed by this strain. The results showed that expression of the insecticidal protein was high. Toxic protein as a percentage of total protein content in all lines was at least 0.01%. In general, the higher the expression levels of toxic protein and the higher the percentage of total protein expression, the easier it is to make accumulation of toxins in the body of larvae play a role in killing them.
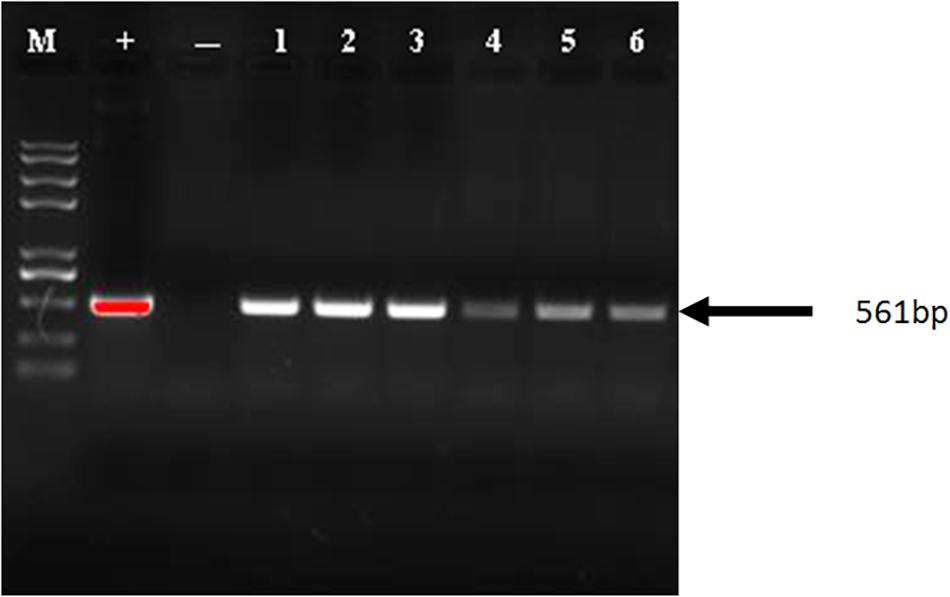
Figure 5. PCR analysis of the Cry1Ah1 gene in transgenic plants. M, marker; +, plasmid; - indicate non-transgenic plant; L1-L6, transgenic plants.
Poplar Leaf Bioassay Based on Feeding by H. cunea Larvae
As shown in Table 4, the insecticidal activity of the transgenic NL895 was significantly better against lower instar larvae of H. cunea than against higher instar larvae in all poplar strains. The corrected mortality of the first to third instars increased gradually, was highest in the third instar larvae, and then gradually decreased in the fourth and fifth instars (Supplementary Figure S2). The corrected mortality of the first, second, and third instar larvae feeding on clone No. 6 was 54.2, 60.9, and 82.6%, respectively, and differed significantly from CK. The pupation rates of the first, second, and third instar larvae feeding on clone No. 6 were 7.4, 14.8, and 15.9%, respectively, and differed significantly from CK. The other five strains of transgenic poplar NL895 also differed from CK (Figure 6).
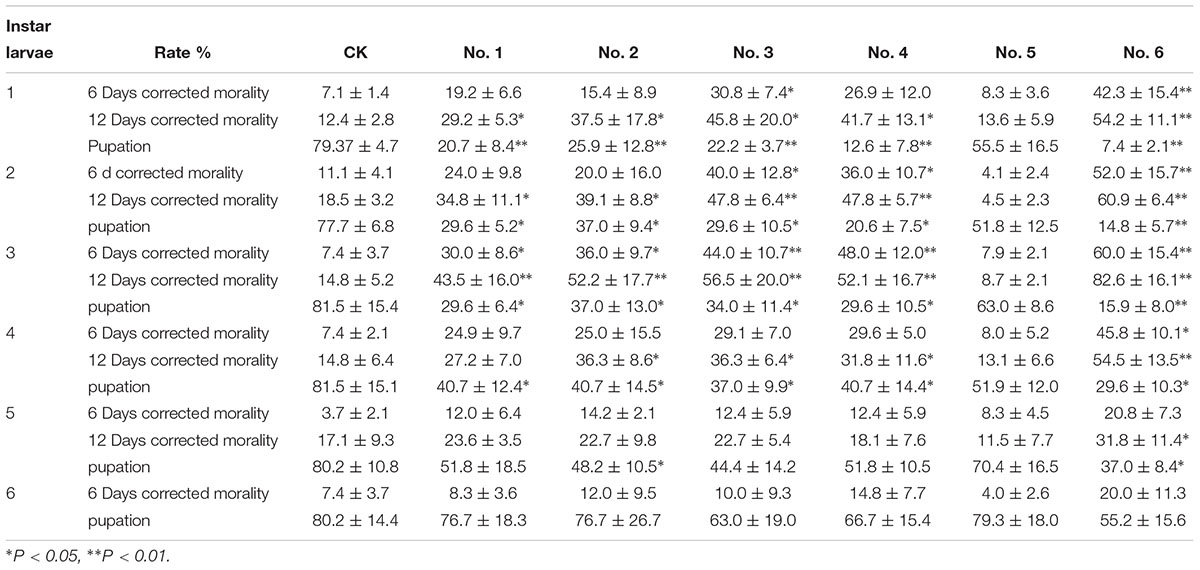
Table 4. Insecticidal activity of transgenic poplars with Bt on different instars of Hyphantria cunea.
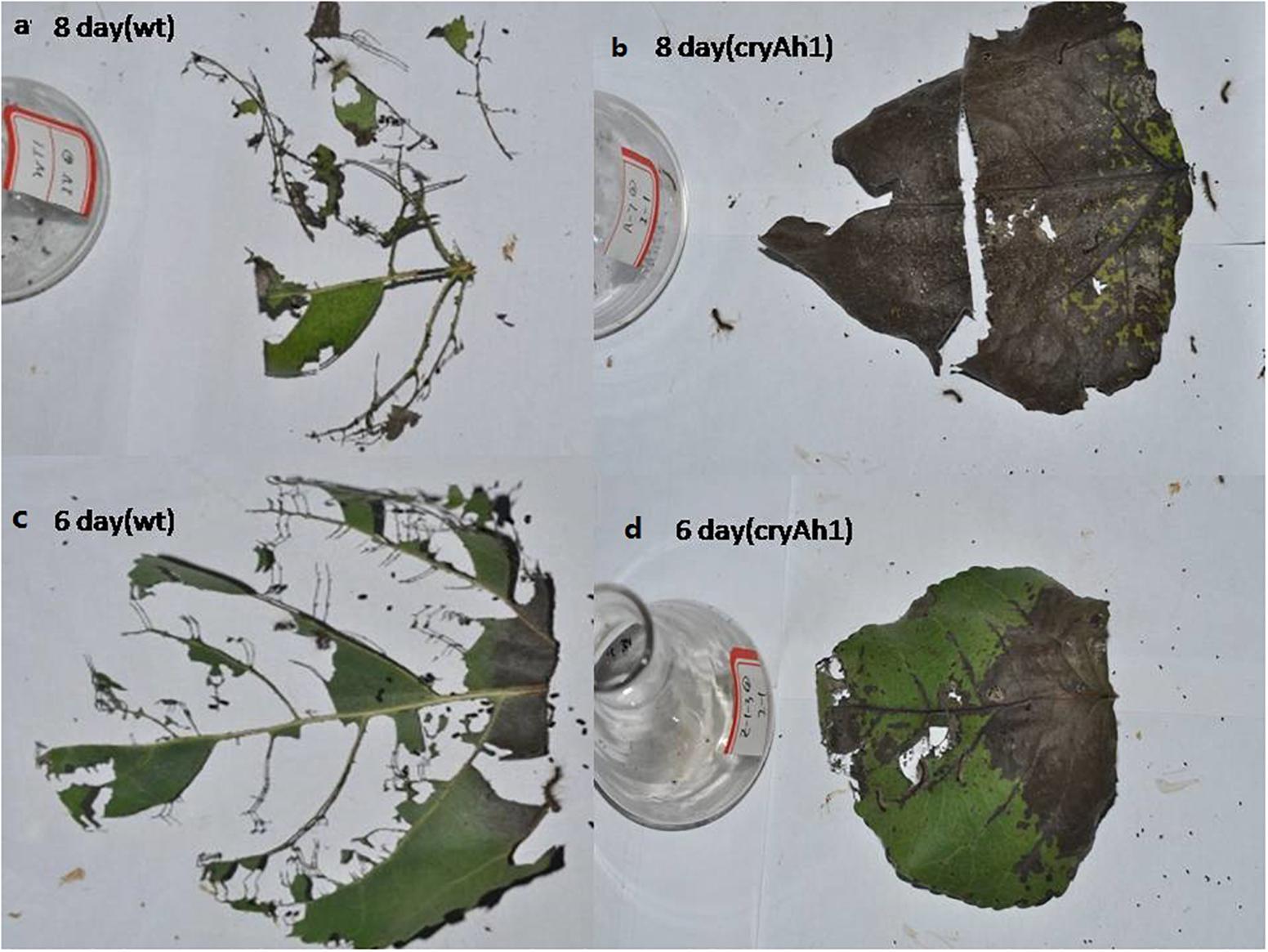
Figure 6. Poplar leaf bioassay of larvae after feeding for 6 or 8 days. Larvae fed (a) non-transgenic and (b) new Cry1Ah1 transgenic poplar leaves for 8 days. Larvae fed (c) non-transgenic and (d) new Cry1Ah1 transgenic poplar leaves for 6 days.
Conversely, the pupation rates of the first to sixth instar larvae increased with age. The corrected mortalities of first to third instar larvae at 6 and 12 days increased with age, and was highest at the third instar, while the corrected mortalities of the third to sixth instar larvae decreased with increasing age. The insecticidal effect also decreased; when the sixth instar larvae were fed each strain of poplar, the insecticidal effect was no longer significant.
Table 5 shows the effects of transgenic Bt poplar on the development of H. cunea. Before 8 days of age, larvae fed on all strains successfully, however, the amounts consumed, growth rates, and pupal weights of the larvae fed on each strain differed. Strain No. 6 showed a significant difference (P < 0.05) at 6 days after feeding and 77 mg was consumed over 8 days of feeding (P < 0.01), compared with the control group, which consumed 183 mg. After 4 and 6 days of feeding on the leaves of No. 6, the body weights of the H. cunea larvae differed significantly from those of the control group (P < 0.05 and P < 0.01, respectively). The larvae fed on line No. 6 developed slowly, with an average body-weight gain of 3.3%, which differed significantly from the control (19.0%) (P < 0.01). The final pupal weight of those larvae that pupated also differed significantly compared with the control. In addition, the feed intakes and body weights of larvae fed on strains 2 and 4 began to show significant differences (P < 0.05) after 6 days, and the rates of weight gain and pupa weights also differed significantly (P < 0.05). Larvae fed on strains 2 and 4 consumed between 91.8 and 99.2 mg (96.3–104.0% of their body weight) over 8 days. There was a significant difference in the food intake (103.5 mg) over 8 days feeding on strain No. 3 by the white moth larvae. Additionally, strain No. 3 did not have a significant impact on the body-weight gain of the larvae. However, strain No. 1, which performed well in previous experiments, did not significantly affect the feeding intake or body-weight gain of insects in this experiment. Strains 6, 2, and 4 all inhibited development to varying degrees; the effect of strain 6 on the development of H. cunea was particularly evident.
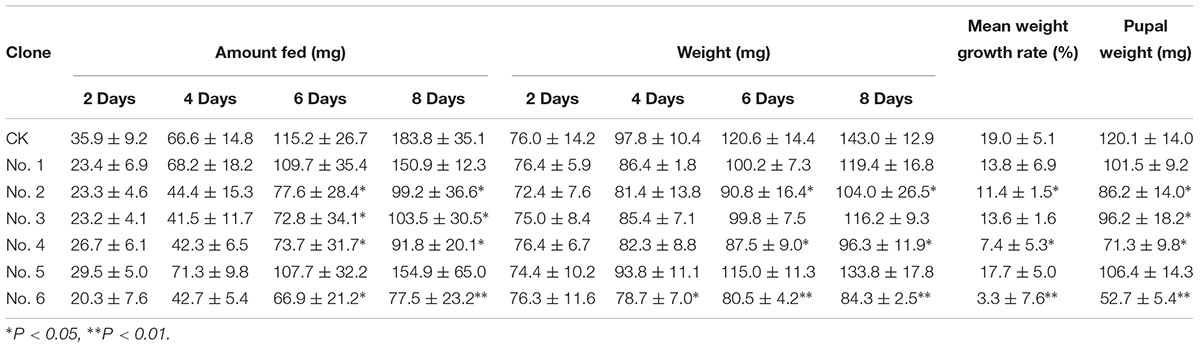
Table 5. Insecticidal activity of transgenic poplars with Bt on the development of Hyphantria cunea.
Insecticidal Activity of Genetically Modified Poplars Against H. cunea in the Field
Results from the specific field-cage experiments carried out at the beginning of July are shown in Table 6. Strain 6 showed the strongest effect, with 99.4% mortality of fifth instar insects. The cumulative mortality by the sixth instar was 99.8% and the pupation rate was 0. Strain 4 had the next strongest effect with 92.5% mortality of fifth instar insects and 99.8% mortality of sixth instar insects. There was no mortality of first instar larvae in the control group, in which the pupation rate was 78%. The pupation rate was less than 15% and mortality was greater than 60% in the third instar for all of the transgenic poplars. The insecticidal activity on H. cunea of genetically modified poplars was much higher in the field than in the laboratory. Field experiments were carried out directly on the leaves of transgenic poplar trees in bagging experiments. Indoor experiments mainly involved picking leaves and feeding them to larvae in an indoor constant-temperature incubator. Perhaps as a result, the leaves were dehydrated and Bt gene expression was weakened indoor.
Discussion
With the development of gene-design and gene-synthesis technology, software can now be used to design genes with greater expression. For the most part, gene-design software is simple. However, it is very difficult to apply to real plants. For example, it is important to remove the phylogenetic cleavage signals for plant transgenes, but most software does not do this. Therefore, we designed new gene-modification and synthesis software called CodonPoplar for poplars. Preliminary research to examine levels of heterogeneous gene expression in poplar was conducted.
The results of the mortality tests using wild larvae at different ages revealed that six lines of Bt-transgenic poplars had highly toxic effects on H. cunea under field conditions, and leaf-feeding by H. cunea decreased significantly. Moreover, the mean rate of pupation was below 10%. The third instar larvae had the highest resistance. H. cunea appears to be highly resistant to hunger. After eating the transgenic leaves, they significantly reduced their food intake, although they did not die immediately.
Kleiner et al. (1995) transferred the Cry1Aa gene to NC5339 poplar and fed transgenic leaves to gypsy moth and Malacosoma larvae. The mortality rates of the first- to third-instar larvae were significantly higher than those of the control group, but larval mortality after the third instar was not remarkable. Rao et al. (2000) fed gypsy moths Bt gene NL-80106 poplar leaves and found that the mortality of the first instar larvae was higher than that of the controls. We also found that the insecticidal effects of transgenic poplars on H. cunea gradually weakened with the growth of the larvae. However, a study of H. cunea larvae at different stages found that the mortality of the first instar larvae with effective transgenic lines was between 29.2 and 54.2%, while the corrected mortality of the third instar larvae was between 43.5 and 82.6%. We speculate that death was caused mainly by accumulation of toxic protein in the larvae, starvation, and other factors. Compared with second and third instar larvae, first instar larvae eat less, which may slow the accumulation of toxins in the larvae. Continuous feeding on leaves leads to accumulation of toxins in the larvae and is the direct cause of larval death. These larvae develop very slowly, and it takes 5–6 days to develop to the second instar. The amount of genetically modified leaves eaten was small. The Bt toxin protein content of the modified leaves was less than that of the Bt toxin extract used for Bt screen strains. In addition, larvae can withstand starvation for up to 10 days. Consequently, this led to a difference in results using the toxin extract for Bt screening strains (results not shown). The H. cunea larvae and pupae fed normal poplar leaves were significantly heavier than those fed transgenic Bt poplar leaves, and mortality was much higher on transgenic Bt poplar leaves than on normal poplar leaves.
Robison et al. (1994) fed gypsy moth larvae transgenic Bt gene NC5339 leaves and found that the larvae ate less than the controls and weight gain was severely delayed. Guo et al. (2011) fed transgenic Bt gene 741 leaves to H. cunea; after 21 days, the control group of larvae began to enter the sixth instar, while the larvae fed the transgenic leaves stayed at the second instar without molting; mean body weight and length were much lower than in the control group (Guo et al., 2011). Their study found that transgenic poplar also had a significant impact on the development of H. cunea. Our paper does not discuss other factors, such as the relationship between insect resistance and the age of transgenic Populus (Ren et al., 2018) or the stability of the toxin in plant tissues (Ren et al., 2017).
Many studies have proven that transgenic poplar has high resistance to H. cunea. The lethal effect of a transgenic single Bt gene line on H. cunea is 80–100% (Tian et al., 2000; Génissel et al., 2003; Ding et al., 2017, 2018; Wang et al., 2018). Although insect resistance has differed significantly among the various transgenic poplar varieties and strains, a positive correlation between toxic protein expression and the mortality of H. cunea larvae was reported (Zhang et al., 2016). In our study, we used a single Bt strain that had a remarkable insecticidal effect on H. cunea. Although expression of modified Bt genes in plants has improved greatly, with the advent of large-scale planting of transgenic plants the evolution of insect resistance cannot be ignored. To slow this process, previous researchers have used a multi-gene strategy (Yang et al., 2003, 2016; Klocko et al., 2014; Zhang et al., 2016; Ding et al., 2017). Additionally, an H. cunea population on transgenic poplar has been systematically surveyed for periods of 3 (Zhang et al., 2015) and 5 years (Zuo et al., 2018). A previous study found that the transformation of Cry1Ac–Cry3A–NTHK1 genes in Populus euramericana “Neva” (Liu et al., 2016) led to mortality exceeding 60% for H. cunea and 100% for first instar larvae of the coleopteran Plagiodera versicolora; the author speculated that this was due to gene interactions (Liu et al., 2016).
In this study, a newly designed Cry1Ah1 gene was successfully transferred into poplar and transgenic poplar lines with hyper-resistance to H. cunea were obtained. In the future, we may use the same method to select and modify new genes for transgenic poplar.
Author Contributions
CX carried out the experimental work and prepared the first draft of the manuscript and figures. HW and LW provided critical inputs for the study as well as during preparation of the manuscript. QZ, CX, and TY designed the research, analyzed the results, and edited the manuscript and figures.
Funding
This work was supported by the National Key Program on Transgenic Research (2018ZX08020002), the National Science Foundation of China (31400567), the State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University: K2013207), and the China Postdoctoral Science Foundation (148825).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00335/full#supplementary-material).
FIGURE S1 | The poplar gene optimization software CodonPoplar interface.
FIGURE S2 | Mortality and pupation rates with Cry1Ah1 for different instar larvae of Hyphantria cunea.
TABLE S1 | Cry1Ah1-U is a codon optimization new sequence and Cry1Ah1-0 is wild sequence.
TABLE S2 | The target gene relative transcript levels for RT-PCR.
Footnote
References
Génissel, A., Leplé, J.-C., Millet, N., Augustin, S., Jouanin, L., and Pilate, G. (2003). High tolerance against Chrysomela tremulae of transgenic poplar plants expressing a synthetic cry3aa gene from Bacillus thuringiensis ssp. tenebrionis. Mol. Breed. 11, 103–110. doi: 10.1023/A:1022453220496
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cao, L., Wei, S., Hoffmann, A. A., Wen, J., and Chen, M. (2016). Rapid genetic structuring of populations of the invasive fall webworm in relation to spatial expansion and control campaigns. Divers. Distrib. 22, 1276–1287. doi: 10.1111/ddi.12486
Crickmore, N., Zeigler, D. R., Feitelson, J., Schnepf, E., Van Rie, J., Lereclus, D., et al. (1998). Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62, 807–813.
Ding, L., Chen, Y., Wang, H., and Wei, J. (2018). Efficacy of Cry1Ac protein against gypsy moth and fall webworm in transgenic poplar (Populus davidiana × Populus bolleana) by bioassay. Can. J. Plant Sci. 98, 844–850. doi: 10.1139/cjps-2017-0322
Ding, L., Chen, Y., Wei, X., Ni, M., Zhang, J., Wang, H., et al. (2017). Laboratory evaluation of transgenic Populus davidiana × Populus bolleana expressing Cry1Ac+ SCK, Cry1Ah3, and Cry9Aa3 genes against gypsy moth and fall webworm. PLoS One 12:e0178754. doi: 10.1371/journal.pone.0178754
Firidin, B., Bílgener, B. M., Yanar, O., Altun, N., and Ince, A. (2008). The effect of nutritional quality of some plant’s leaf on the feeding and development of Hyphantria cunea (Drury) (Lepidoptera: Arctiidae). IJONAS 2,61–68.
Gao, Y., Hu, Y., Fu, Q., Zhang, J., Oppert, B., Lai, F., et al. (2010). Screen of Bacillus thuringiensis toxins for transgenic rice to control Sesamia inferens and Chilo suppressalis. J. Invertebr. Pathol. 105, 11–15. doi: 10.1016/j.jip.2010.05.002
Guo, P. X., Zhang, J., Liu, Z. H., and Pang, X. B. (2011). Investigation of arthropod community and the stability of insect-resistance of the triploid Chinese white poplar transformed with two insect-resistant genes. Hebei J. For. Orchard Res. 1:017.
Guo, T., Ji, B., Zhu, G., and Huang, M. (2004). A study on the insect-resistance of transgenic poplars (NL-80106) carrying bt genes to Micromelalopha troglodyta. J. Nanjing For. Univ. 28, 5–9.
Hu, J. J., Tian, Y. C., Han, Y. F., Li, L., and Zhang, B. E. (2001). Field evaluation of insect-resistant transgenic Populus nigra trees. Euphytica 121, 123–127. doi: 10.1023/A:1012015709363
Hu, J., Zhang, J., Chen, X., Lv, J., Jia, H., Zhao, S., et al. (2017). An empirical assessment of transgene flow from a bt transgenic poplar plantation. PLoS One 12:e0170201. doi: 10.1371/journal.pone.0170201
Kleiner, K. W., Ellis, D. D., McCown, B. H., and Raffa, K. F. (1995). Field evaluation of transgenic poplar expressing a Bacillus thuringiensis cry1A (∞) d-endotoxin gene against forest tent caterpillar (Lepidoptera: Lasiocampidae) and gypsy moth (Lepidoptera: Lymantriidae) following winter dormancy. Environ. Entomol. 24, 1358–1364. doi: 10.1093/ee/24.5.1358
Klocko, A. L., Meilan, R., James, R. R., Viswanath, V., Ma, C., Payne, P., et al. (2014). Bt-cry3aa transgene expression reduces insect damage and improves growth in field-grown hybrid poplar. Can. J. For. Res. 44, 28–35. doi: 10.1139/cjfr-2013-0270
Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. doi: 10.1101/gad.1331305
Lee, M. K., Milne, R. E., Ge, A. Z., and Dean, D. H. (1992). Location of a Bombyx mori receptor binding region on a Bacillus thuringiensis delta-endotoxin. J. Biol. Chem. 267, 3115–3121.
Li, Y., Shu, C., Zhang, X., Crickmore, N., Liang, G., Jiang, X., et al. (2014). Mining rare and ubiquitous toxin genes from a large collection of Bacillus thuringiensis strains. J. Invertebr. Pathol. 122, 6–9. doi: 10.1016/j.jip.2014.07.006
Liu, D., Zhang, J., Dong, Y., Zhang, X., Yang, M., and Gao, B. (2016). Genetic transformation and expression of Cry1Ac–Cry3A–NTHK1 genes in Populus×euramericana “neva”. Acta Physiol. Plant. 38:177. doi: 10.1007/s11738-016-2195-6
Lu, M.Z. (2008) “Advance in transgenic research and its biosafety of forest trees in China,” in Proceeding of the 8th Symposium in Agricultural Biochemistry and Molecular Biology of the Chinese Society of Biochemistry and Molecular Biology, Guiyang, 1–2
McCown, B. H., McCabe, D. E., Russell, D. R., Robison, D. J., Barton, K. A., and Raffa, K. F. (1991). Stable transformation of populus and incorporation of pest resistance by electric discharge particle acceleration. Plant Cell Rep. 9, 590–594. doi: 10.1007/BF00232339
Meena, R. K., Gunda, G. K., Govind, A., Gujar, T., and Kaur, S. (2001). Screening of Bacillus thuringiensis isolates recovered from diverse habitats in India for the presence of cry1a-type genes and cloning of a cry1ac33 gene toxic to Helicoverpa armigera (American bollworm). Asian J. Biotechnol. 4, 53–69.
Peña, L., and Séguin, A. (2001). Recent advances in the genetic transformation of trees. Trends Biotechnol. 19, 500–506. doi: 10.1016/S0167-7799(01)01815-7
Perlak, F. J., Fuchs, R. L., Dean, D. A., McPherson, S. L., and Fischhoff, D. A. (1991). Modification of the coding sequence enhances plant expression of insect control protein genes. Proc. Natl. Acad. Sci. U.S.A. 88, 3324–3328. doi: 10.1073/pnas.88.8.3324
Ramachandran, R., Raffa, K. F., Bradley, D., Miller, M., Ellis, D., and McCown, B. (1993). Activity of an insecticidal protein from Bacillus thuringiensis subsp. thuringiensis HD-290-1 strain to coleopteran and lepidopteran defoliators of poplars. Environ. Entomol. 22, 190–196. doi: 10.1093/ee/22.1.190
Rao, H., Chen, Y., Huang, M., Wang, M., Wu, N., and Fan, Y. (2000). Genetic transformation of poplar NL-80106 transferred by bt gene and its insect-resistance. J. Plant Res. Environ. 9, 1–5.
Ren, Y., Zhang, J., Liang, H., Wang, J., and Yang, M. (2017). Inheritance and expression stability of exogenous genes in insect-resistant transgenic poplar. Plant Cell Tissue Org. Cul. 130, 567–576. doi: 10.1007/s11240-017-1247-y
Ren, Y., Zhang, J., Wang, G., Liu, X., Li, L., Wang, J., et al. (2018). The relationship between insect resistance and tree age of transgenic triploid Populus tomentosa plants. Front. Plant Sci. 9:53. doi: 10.3389/fpls.2018.00053
Robison, D. J., McCown, B. H., and Raffa, K. F. (1994). Responses of gypsy moth (Lepidoptera: Lymantriidae) and forest tent caterpillar (Lepidoptera: Lasiocampidae) to transgenic poplar, Populus spp., containing a Bacillus thuringiensis d-endotoxin gene. Environ. Entomol. 23, 1030–1041. doi: 10.1093/ee/23.4.1030
Subramanian, S., and Mohankumar, S. (2006). Genetic variability of the bollworm, Helicoverpa armigera, occurring on different host plants. J. Insect Sci. 6, 1–8. doi: 10.1673/2006_06_26.1
Sullivan, G. T., Karaca, I., Ozman-Sullivan, S. K., and Kara, K. (2012). Tachinid (Diptera: Tachinidae) parasitoids of overwintered Hyphantria cunea (Drury) (Lepidoptera: Arctiidae) pupae in hazelnut plantations in samsun province, Turkey. J. Entomol. Res. Soc. 14, 21–30.
Tian, Y. C., Han, Y. F., and Li, T. Y. (1993). Studies on insect-resistant transgenic (Populus nigra) plants. Chin. J. Biotechnol. 9, 291–297.
Tian, Y. C., Zheng, J. B., Yu, H. M., Liang, H. Y., Li, C. Q., and Wang, J. M. (2000). Studies of transgenic hybrid poplar 741 carrying two insect resistant genes. Acta Bot. Sin. 42, 263–268. doi: 10.3321/j.issn:1672-9072.2000.03.009
Tun, T. N., Guo, J., Fang, S., and Tian, Y. (2018). Planting spacing affects canopy structure, biomass production and stem roundness in poplar plantations. Scand. J. For. Res. 33, 464–474. doi: 10.1080/02827581.2018.1457711
Wang, G., Dong, Y., Liu, X., Yao, G., Yu, X., and Yang, M. (2018). The current status and development of insect-resistant genetically engineered poplar in China. Front. Plant Sci. 9:1048. doi: 10.3389/fpls.2018.01408
Wang, X., Han, Y., Dai, L., Li, L., and Tian, Y. C. (1997). Studies on insect-resistant transgenic (P. x Euramericana) plants. Sci. Silvae Sin. 33, 74–81.
Yang, M. S., Liang, H. Y., Gao, B. J., Wang, J. M., and Zheng, J. B. (2003). Insecticidal activity and transgene expression stability of transgenic hybrid poplar clone 741 carrying two insect-resistant genes. Silvae Genet. 52,197–201.
Yang, R. L., Wang, A. X., Zhang, J., Dong, Y., Yang, M. S., and Wang, J. M. (2016). Genetic transformation and expression of transgenic lines of Populus x euramericana with insect-resistance and salt-tolerance genes. Genet. Mol. Res. 15:gmr8635 doi: 10.4238/gmr.15028635
Zhang, D. J., Liu, J. X., Lu, Z. Y., Li, C. L., Comada, E., and Yang, M. S. (2015). Impacts of transgenic poplar-cotton agro-ecosystems upon target pests and non-target insects under field conditions. Genet. Mol. Res. 14, 8125–8136. doi: 10.4238/2015.July.27.1
Zhang, Y., Zhang, J., Lan, J., Wang, J., Liu, J., and Yang, M. (2016). Temporal and spatial changes in Bt toxin expression in bt-transgenic poplar and insect resistance in field tests. J. For. Res. 27, 1249–1256. doi: 10.1007/s11676-016-0254-x
Keywords: Bacillus thuringiensis, codon optimization, Hyphantria cunea, poplar, transgenic plant
Citation: Xu C, Wei H, Wang L, Yin T and Zhuge Q (2019) Optimization of the cry1Ah1 Sequence Enhances the Hyper-Resistance of Transgenic Poplars to Hyphantria cunea. Front. Plant Sci. 10:335. doi: 10.3389/fpls.2019.00335
Received: 20 August 2018; Accepted: 04 March 2019;
Published: 26 March 2019.
Edited by:
Isabel Allona, Polytechnic University of Madrid, SpainReviewed by:
Minsheng Yang, Agricultural University of Hebei, ChinaFernando Ponz, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), Spain
Copyright © 2019 Xu, Wei, Wang, Yin and Zhuge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Zhuge, cXpodWdlQG5qZnUuZWR1LmNu
 Chen Xu
Chen Xu Hui Wei1
Hui Wei1 Like Wang
Like Wang Tongming Yin
Tongming Yin Qiang Zhuge
Qiang Zhuge