- 1College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2Department of Biology, Saint Mary’s University, Halifax, NS, Canada
- 3School of Veterinary and Life Sciences, Murdoch University, Murdoch, WA, Australia
- 4Hubei Collaborative Innovation Centre for Grain Industry, Yangtze University, Jingzhou, China
Grain size and weight are crucial components of barley yield and quality and are the target characteristics of domestication and modern breeding. Despite this, little is known about the genetic and molecular mechanisms of grain size and weight in barley. Here, we evaluated nine traits determining grain size and weight, including thousand grain weight (Tgw), grain length (Gl), grain width (Gw), grain length-width ratio (Lwr), grain area (Ga), grain perimeter (Gp), grain diameter (Gd), grain roundness (Gr), and factor form density (Ffd), in a double haploid (DH) population for three consecutive years. Using five mapping methods, we successfully identified 60 reliable QTLs and 27 hotspot regions that distributed on all chromosomes except 6H which controls the nine traits of grain size and weight. Moreover, we also identified 164 barley orthologs of 112 grain size/weight genes from rice, maize, wheat and 38 barley genes that affect grain yield. A total of 45 barley genes or orthologs were identified as potential candidate genes for barley grain size and weight, including 12, 20, 9, and 4 genes or orthologs for barley, rice, maize, and wheat, respectively. Importantly, 20 of them were located in the 14 QTL hotspot regions on chromosome 1H, 2H, 3H, 5H, and 7H, which controls barley grain size and weight. These results indicated that grain size/weight genes of other cereal species might have the same or similar functions in barley. Our findings provide new insights into the understanding of the genetic basis of grain size and weight in barley, and new information to facilitate high-yield breeding in barley. The function of these potential candidate genes identified in this study are worth exploring and studying in detail.
Introduction
Since domestication about 10,000 years ago in the Fertile Crescent, barley (Hordeum vulgare L.) has become one of the most important cereal crops cultivated around the world, and is widely used as animal feed, potential healthy food products and is a major raw material for malting and brewing industries (Salamini et al., 2002; Collins et al., 2010; Ullrich, 2010). With the rapid growth of the global population and the continuous reduction of arable land worldwide, improving barley yield remains a major challenge for the barley breeding program in the present context of climate change (Fischer and Edmeades, 2010; Feng et al., 2016).
Grain size and weight, which is determined by its three-dimensional structure (length, width, and thickness) and the degree of grain filling, are two crucial components that affect barley yield and malt quality (Zhang X. et al., 2012). In the context of crop domestication and artificial breeding, grain size and weight have always been important agronomic traits for human care and selection. Despite this, little is known about the genetic and molecular mechanisms of grain size and weight in barley. Archaeological evidence suggests that barley grains increased in size starting in the Pre-Pottery Neolithic A (PPNA; 9700–8700 BC) and earliest Pre-Pottery Neolithic B (PPNB; 8700–6200 BC) (Fuller, 2007). Compared to their progenitors, modern barley varieties have larger grains that not only have a favorable effect on seedling vigor and yield, but are also favored by the malting and feed industries as they can increase malt yields and feed production capacity (Gan and Stobbe, 1996; Walker and Panozzo, 2011). Therefore, untangling the genetic factors controlling grain size and weight is crucial for improving barley yield and quality as well as understanding the domestication process that has occurred in barley.
In recent years, the rapid advance of functional genomics research has promoted our understanding of the genetic basis and developmental mechanisms of grain size and weight, many QTLs or genes associated with grain size and weight have been mapped or characterized in detail in rice (Zuo and Li, 2014; Li and Yang, 2017; Liu et al., 2017a, 2018; Wu et al., 2017; Li et al., 2018; Sun et al., 2018; Ying et al., 2018; Zhao et al., 2018), maize (Liu et al., 2015, 2017b; Chen et al., 2016; Zhou et al., 2017), and wheat (Zhang L. et al., 2012; Hu et al., 2016; Ma L. et al., 2016; Ma M. et al., 2016; Geng et al., 2017; Hou et al., 2017; Sajjad et al., 2017; Zhai et al., 2018; Zhang et al., 2018). However, in comparison to the relatively extensive research that has been conducted in other cereal species mentioned above, only limited molecular information is available to understand the biological developmental processes and formation mechanism of barley grain size. To date, some attempts have been made to clarify the genetic basis of barley grain size and weight. For example, Ayoub et al. (2002) detected QTLs for grain size and shape characteristics on all seven linkage groups. Walker et al. (2013) identified 232 QTLs for 11 grain traits across the three environments in a DH population. Zhou et al. (2016) mapped two major QTLs for grain length in a recombinant inbred line (RIL) population. Xu et al. (2018) identified 29 QTL hotspots distributed on all seven chromosomes for grain size and weight. Additionally, several genes affecting barley grain size or weight have been characterized using mutant materials, including Nud (Taketa et al., 2008), Vrs1 (Komatsuda et al., 2007; Sakuma et al., 2017), Vrs2 (Youssef et al., 2017), Vrs3 (Bull et al., 2017; Van Esse et al., 2017), Vrs4 (Koppolu et al., 2013), and Int-c (Ramsay et al., 2011). However, these genes that have been characterized in the past all indirectly affect barley grain size or weight, while genes that directly control yield components have not yet been identified in barley.
Comparative genomic approaches have provided an effective strategy for identifying genes with conserved functions across genomes and species (Su et al., 2011; Liu et al., 2017b), such as wheat powdery mildew resistance gene TmMla1 and barley powdery mildew resistance gene HvMLA1, which were identified as orthologous (Jordan et al., 2011), and the Int-c gene regulating lateral spikelet fertility in barley which was identified as an ortholog of the maize domestication gene ZmTB1 (Ramsay et al., 2011). It has been reported that many of the genes affecting grain size/weight generally have conserved functions, but their functional specificity may be divergent among species (Zhang et al., 2015, 2018; Liu et al., 2017b; Sajjad et al., 2017; Zhai et al., 2018). For example, the GW2 gene that encodes a RING-type E3 ubiquitin ligase to regulate grain weight in rice (Song et al., 2007), its orthologous in maize (Li et al., 2010), wheat (Su et al., 2011; Bednarek et al., 2012; Yang et al., 2012; Hong et al., 2014; Jaiswal et al., 2015; Simmonds et al., 2016; Geng et al., 2017; Zhai et al., 2018; Zhang et al., 2018), and sorghum (Tao et al., 2017) were also found to be involved in the control of grain weight, but with different mechanisms. Similarly, the orthologous genes of rice GS5 gene (Li et al., 2011) in maize (Liu et al., 2015) and wheat (Wang et al., 2015, Ma L. et al., 2016; Wang S. et al., 2016) also control similar phenotypes in grain size and weight as well as in rice. At present, a considerable number of genes associated with grain size/weight have been characterized in rice, maize, and wheat, but whether the orthologs of these genes in barley have the same or similar function remains an open question. Therefore, it is necessary to characterize the orthologs of these grain size/weight genes in barley to provide insight into the genetic mechanisms of barley grain size and weight.
In this study, we performed a QTL mapping of nine grain size and weight traits in a DH population, using an SNP-based high density genetic map and identified 164 barley orthologs of 112 grain size/weight genes from rice, maize, and wheat in the barley genome. The objectives of this study were to identify reliable QTL and QTL hotspots affecting barley grain traits, and to explore the genetic correspondence between the QTLs identified here and grain size/weight genes in other cereal species. The results of this study will enhance our understanding of the genetic basis of grain size and weight in barley and may provide new information to facilitate high-yield breeding in barley.
Materials and Methods
Plant Materials and Field Trials
A doubled haploid (DH) population containing 122 lines derived from Huaai 11 (six-rowed and naked) and Huadamai 6 (two-rowed and hulled) was employed to identify QTLs that control barley grain size and weight. Details of the DH population and their parents can be found in our earlier studies (Ren et al., 2010; Wang et al., 2017). The DH lines and parents were evaluated in natural field conditions over three crop seasons (2015 to 2016, 2016 to 2017 and 2017 to 2018 seasons) in the experimental farm of Huazhong Agricultural University, Wuhan, China (30°48’N, 114°36’E), in a randomized complete block design with three replicates. In each replicate, each line was planted in a two-row plot of 1.5 m length with the spacing of 0.1 m between plants and 0.2 m between rows. Field management, including irrigation, fertilization, weeding and pest control, followed the standard agricultural practices in barley production.
Evaluation of Barley Grain Size and Weight
At maturity, kernels of six uniform plants in the middle of each plot were bulk-harvested and sun-dried for phenotypic evaluation. Then, 200 to 300 fully filled grains of each line were used to measure thousand grain weight (Tgw, g), grain length (Gl, mm), grain width (Gw, mm), grain length-width ratio (Lwr), grain area (Ga, mm2), grain perimeter (Gp, mm), grain diameter (Gd, mm) and grain roundness (Gr) using a camera-assisted phenotyping system (SC-G, Wanshen Detection Technology Co., Ltd., Hangzhou, China) (Yin et al., 2015). Ga and Gp were defined as the actual area and length of the seed projection outline, respectively. Gd was calculated by Gd = , and Gr was calculated by Gr = , where the major axis is the major axis length of the seed fitted ellipse. In addition, in order to assess difference in grain density, we calculated the factor form density (Ffd, g/mm2) according to the following formula: Ffd = (Giura and Saulescu, 1996).
Statistical Analysis
The best linear unbiased prediction (BLUP) value of the three replicate measurements for each year were used for statistical analysis and QTL mapping. The calculations of descriptive statistics, Student’s t-test, normality test (Shapiro–Wilk), correlation analysis and analysis of variance (ANOVA) were performed using SPSS v24.0 (IBM SPSS Statistics, Chicago, IL, United States). Broad sense heritability (H2) estimates were calculated from ANOVA using the following formula: H2 = 1 − MS2/MS1, where MS1 and MS2 are the mean squares of genotype and genotype × environment, respectively (Knapp et al., 1985). Frequency distribution and QTL-likelihood maps for the grain size and weight were drawn using the Origin programs (OriginLab, Northampton, MA, United States).
QTL Analysis
The high-density genetic linkage map for “Huaai 11 × Huadamai 6” population used in this study was constructed previously (Ren et al., 2016), which included 1962 markers on all seven chromosomes, comprising 1894 SNP markers and 68 SSR markers. It spanned 1375.80 cM of the whole-genome with an average marker distance of 0.7 cM. Grain size and weight QTLs detection were performed using the inclusive composite interval mapping (ICIM) algorithm in QTL IciMapping v4.1 software (Meng et al., 2015). Single-environment QTL and Multi-Environment Trials (MET) analyses were performed using the ICIM-ADD (additive and dominance effects) mapping method in “BIP (QTL mapping in biparental populations)” module and “MET (QTL mapping for multi-environment traits)” module, respectively. The scanning step size was 1.0 cM, and the probability in stepwise regression (PIN) was 0.001. The LOD threshold was determined by a 1000 times the permutation test, with a Type 1 error of 0.05. The narrow sense heritability of each MET QTL was estimated from the MET analysis using the following formula: h2 = , where PVE(A) and PVE(E) are the additive genetic (A) and environmental (E) components of the multi-environment variance. Moreover, to overcome the interference of row type (Rt) and caryopsis type (Ct), we used Rt and Ct as a covariate, respectively, and conducted QTL mapping with a covariance analysis. The covariate QTL analysis was performed using software QTL.gCIMapping from the R website (Feng et al., 2018); the critical LOD scores for a significant QTL was set at 3.0, and the walking speed for the genome-wide scan was set at 1 cM. The naming of QTLs followed the nomenclature proposed by Mccouch et al. (1997). If QTLs for different traits were located in the same marker interval or their 1.5-LOD confidence intervals overlapped, the corresponding loci were defined as a pleiotropic or tightly linked QTLs. For the same trait, QTLs repeatedly detected in more than one year were defined as a stable QTLs, QTLs repeatedly detected in at least two years environment and in multiple mapping methods were considered to be a reliable QTLs. The orthologs of rice, wheat and maize genes in the barley reference genome (Mascher et al., 2017) were identified using the Ensembl Plant Database1. BARLEYMAP pipeline (Cantalapiedra et al., 2015)2 was used to compare the marker information and identify potential candidate genes based on the barley physical map. According to the marker density of the genetic map, any barley gene or ortholog located within ± 5 Mb on either side of the QTL peak was identified as a candidate gene for QTL.
Results
Phenotypic Variation and Correlation Analysis
We evaluated nine grain size and weight traits in the DH population and their parents, for three consecutive years. Descriptive statistics for those grain size and weight traits are presented in Table 1. The phenotypic difference in the grain of the two parental lines, Huadamai 6 and Huaai 11, are shown in Figure 1A. The t-test showed that two parents were significantly different (p < 0.05) on all investigated grain size and weight traits (Table 1). Huadamai 6 showed higher values for Tgw, Gl, Gw, Lwr, Ga, Gp, and Gd in all three years than Huaai 11, while Huaai 11 had more Gr and Ffd than Huadamai 6 (Table 1 and Figure 1B).
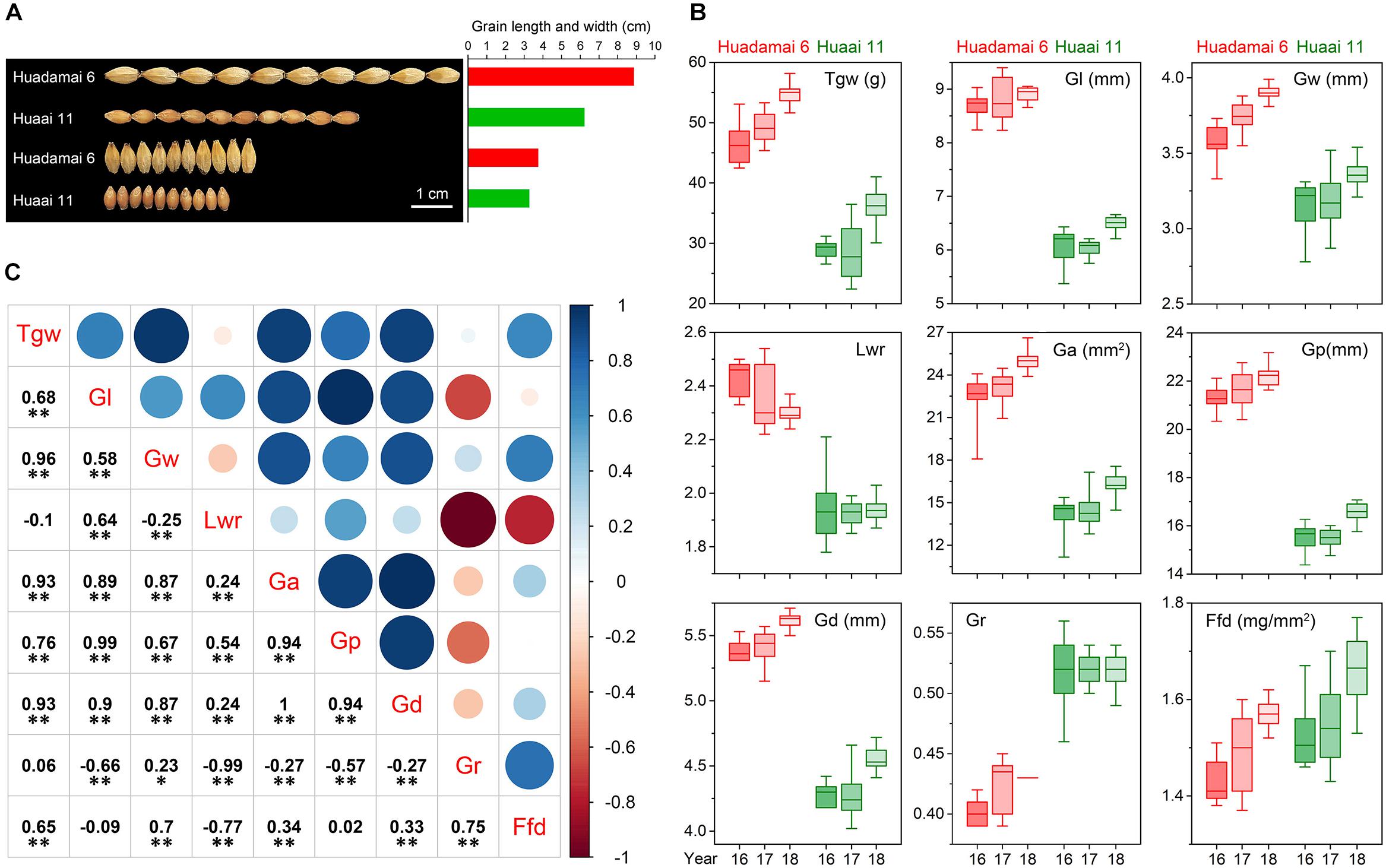
Figure 1. (A) Grain phenotypes of the two parents Huadamai 6 and Huaai 11. (B) Box diagram of nine grain size and weight traits for two parents in three years. (C) Pearson correlation coefficients among nine characteristics of barley grain size across the three years. The two-tailed t-test was applied to test the significance of correlation coefficients (∗p < 0.05, ∗∗ p < 0.01).
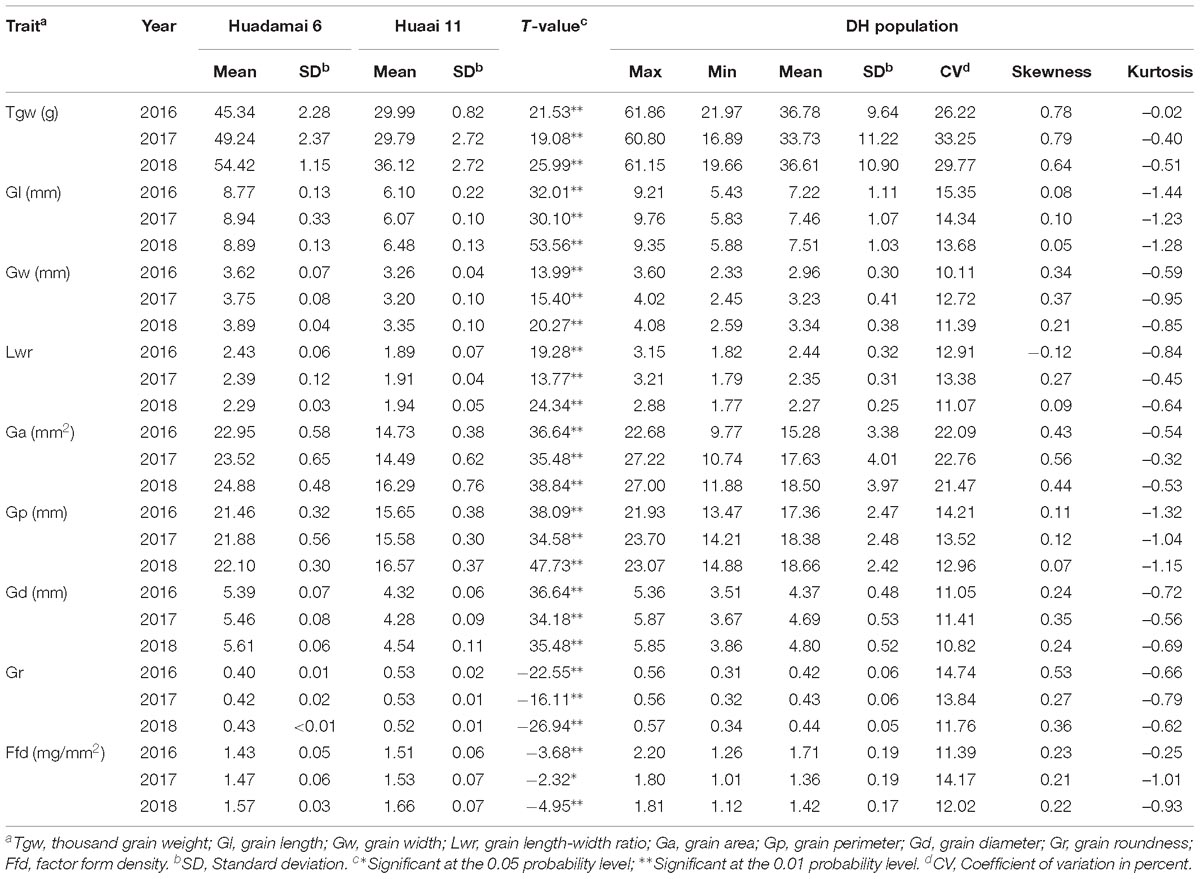
Table 1. Phenotypic performance for the nine grain size and weight traits of the DH population and their parents.
The nine grain size and weight traits in the DH population showed highly phenotypic variation and transgressive segregation (values more extreme than the parental phenotypes) in all years. The phenotypic variation coefficient ranged from 10.11 to 33.25% (Table 1). Variance analysis indicated that the effects of genotype, year and genotype × year interactions were all significant (Table 2). All grain size and weight traits had broad-sense heritability over 95%, which confirmed that genetic effects are the major determinant of the phenotypic variance on grain size and weight in barley. The frequency distributions of nine grain size and weight traits showed continuous variation in all years, indicating the polygenic inheritance (Supplementary Figure S1). Shapiro–Wilk test indicated that some of the grain size and weight traits displayed normal distributions, including Lwr (2016–2018) and Ffd (2016).
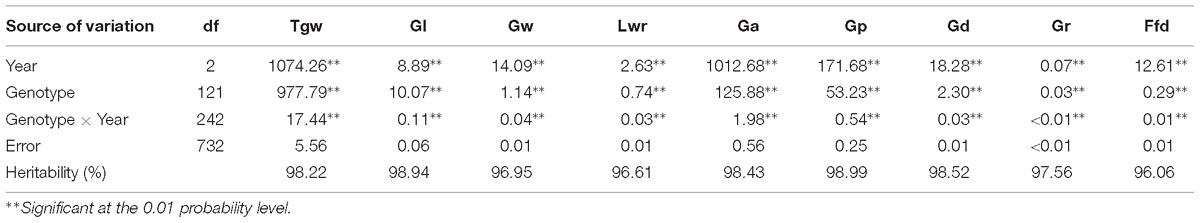
Table 2. Mean squares of ANOVA and heritability for grain size and weight of the DH population in three years.
Additionally, we also calculated the correlation coefficient among the nine grain size and weight traits in the DH population based on the mean value of the three-year data (Figure 1C). The results showed that Tgw had a significantly positive correlation with all grain size and weight traits except Gr and Lwr. Ffd was highly positively correlated with Tgw, Gw, Ga, Gd and Gr, and was negatively correlated with Lwr. Other seven grain size and weight traits (Gl, Gw, Lwr, Ga, Gp, Gd and Gr) also showed a highly significant correlation with each other.
QTL Analysis of Barley Grain Size and Weight Trait
Using the ICIM BIP module for single-environment QTL analysis, we identified 168 QTLs distributed on all chromosomes of barley for nine grain size and weight traits, including 45, 63 and 60 QTLs in 2016, 2017 and 2018, respectively (Supplementary Table S1 and Figure 2A). Out of the 168 identified QTLs, 62 (36.9%) major QTLs (QTLs that can explaining more than 10% of the phenotypic variation) were identified, including 3, 8, 5, 9, 8, 9, 8, 8, and 4 QTLs for Tgw, Gl, Gw, Lwr, Ga, Gp, Gd, Gr, and Ffd, respectively (Supplementary Table S1). Importantly, 3, 3, 5, 4, 5, 3, 3, 4, and 3 stable QTLs (QTLs repeatedly detected in more than one year for the same trait) were identified for Tgw, Gl, Gw, Lwr, Ga, Gp, Gd, Gr, and Ffd, respectively (Supplementary Table S2). Nineteen stable QTLs each explained more than 10% of the phenotypic variation (mean value from all years). The QTL for Gw around 125 cM on chromosomal 2H, identified in all three years, is shown in Figure 3A as an example. Additionally, we also identified 23 pleiotropic or tightly linked QTLs that influenced at least two traits, such as the QTL at 79.5–80.5 cM (511.81–514.43 Mb) on chromosomal 7H simultaneously affected Lwr, Gr, and Ffd (Supplementary Table S3 and Figure 3B).
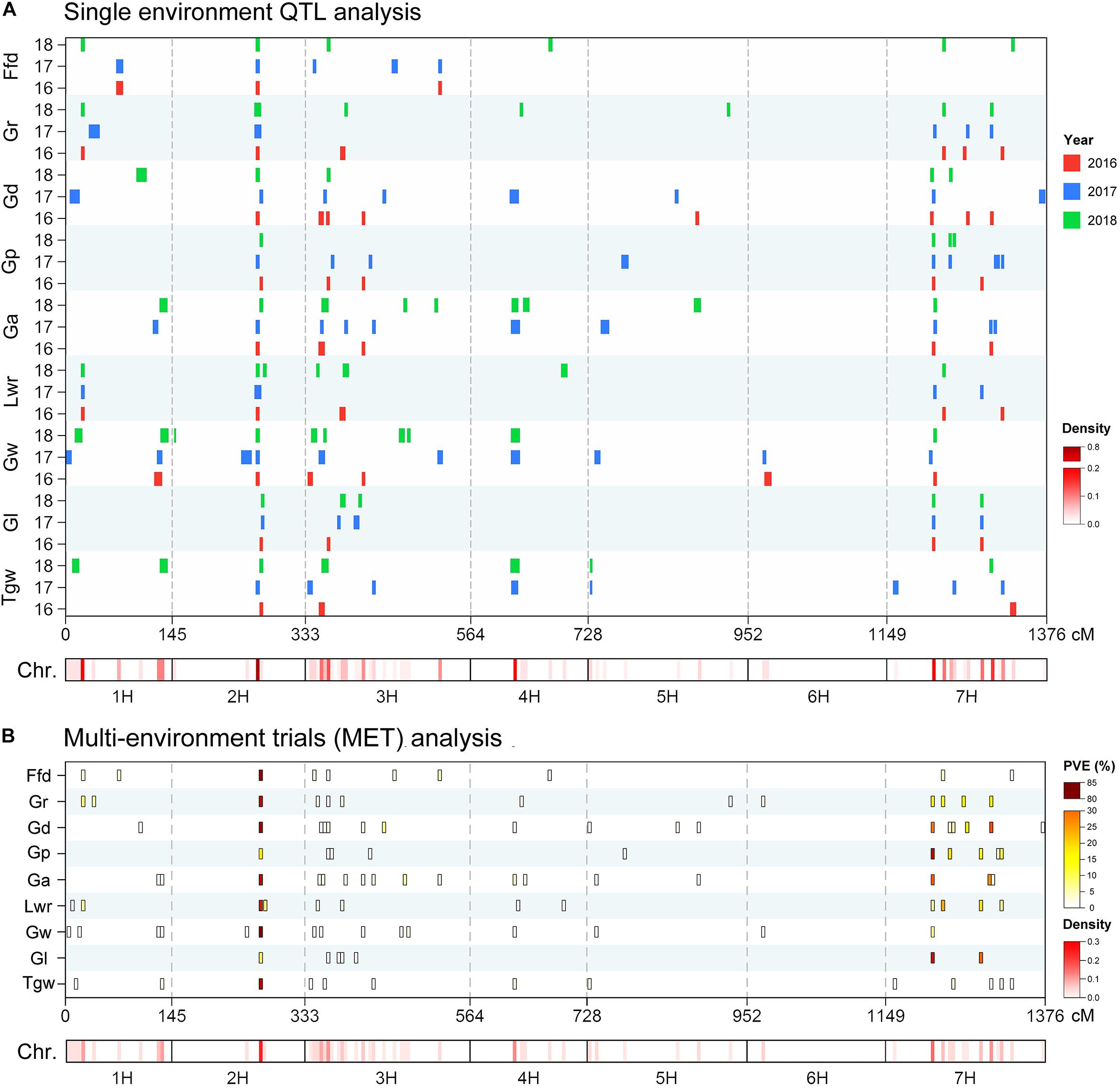
Figure 2. Chromosome distribution of QTLs associated with nine grain size and weight traits identified in the (A) single-environment QTL analysis and (B) multi-environment trials (MET) analysis. Heat map under the X-axis showed the density of QTLs for nine grain size and weight traits across the genome. The window size was 5 cM. QTL bars in single-environment QTL analysis represented the 1.5-LOD support intervals from ICIM mapping.
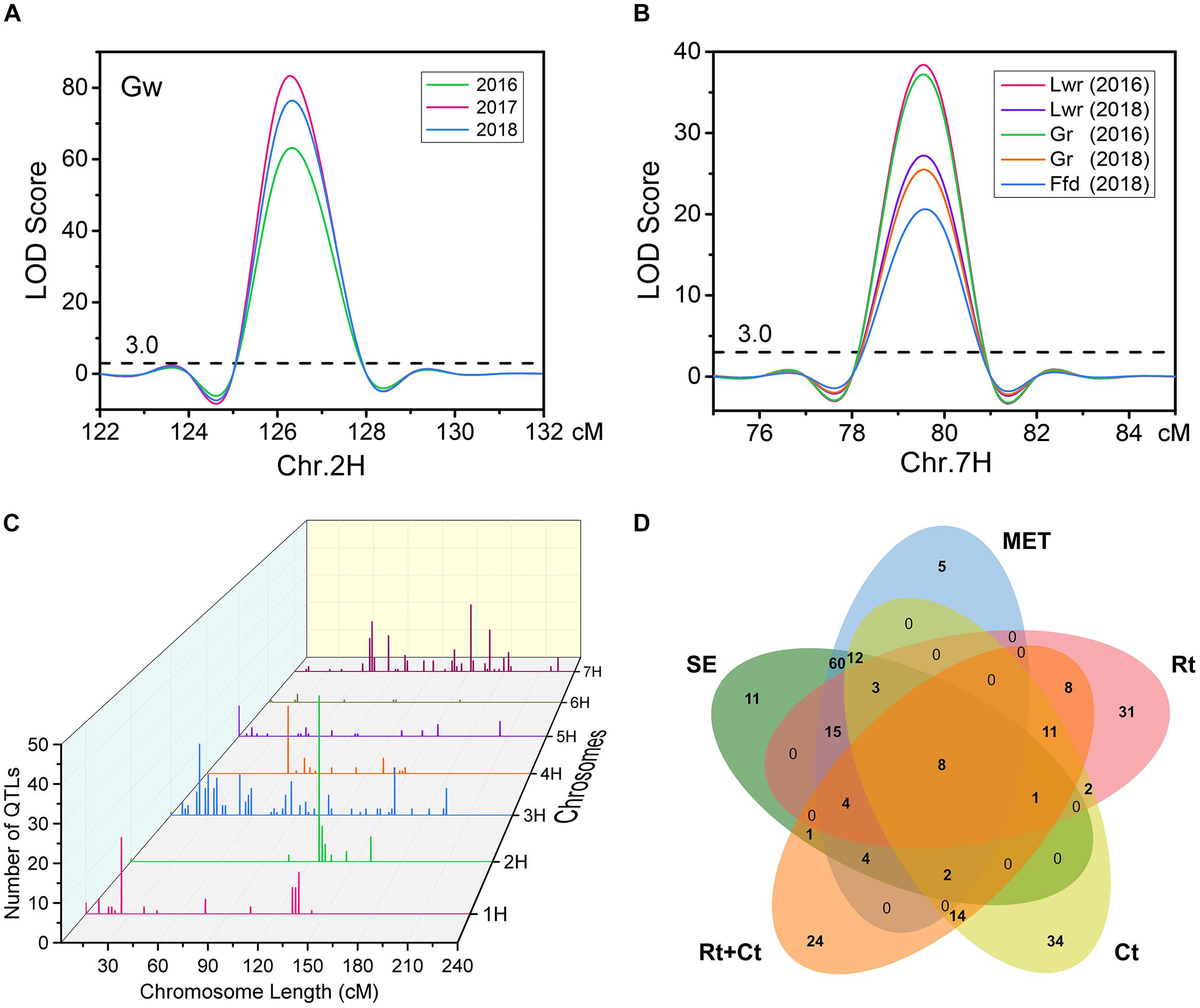
Figure 3. (A) A stable QTL on chromosome 2H for Gw was identified in all three years using single environmental QTL analysis. (B) A pleiotropic or tightly linked QTL on chromosome 7H was identified for Lwr, Gr, and Ffd in single environmental QTL analysis. (C) Distribution characteristics of QTLs for nine grain size and weight traits detected in the five mapping methods. (D) Venn diagram of QTLs identified for nine grain size and weight traits in five mapping methods. SE, single-environment QTL analysis; MET, multi-environment trials (MET) analysis; Rt, covariate QTL analysis using row type as a covariate; Ct, covariate QTL analysis using caryopsis type as a covariate; Rt+Ct, covariate QTL analysis using row type and caryopsis type as a covariate.
We also performed MET analysis using the ICIM MET module. A total of 12, 7, 17, 10, 17, 11, 17, 12, and 10 MET QTLs were identified for Tgw, Gl, Gw, Lwr, Ga, Gp, Gd, Gr, and Ffd, respectively (Supplementary Table S4 and Figure 2B). Of these, 25 (22.1%) MET QTLs each explained more than 10% of the phenotypic variation. The narrow sense heritability of each MET QTL ranged from 24.99 to 99.98%. In addition, we also identified 23 pleiotropic or tightly linked QTLs that affected two or more traits simultaneously (Supplementary Table S5). Notably, a considerable number of loci were repeatedly identified by single-environment QTL analysis and MET analysis. A total of 109 QTLs (96.5%) detected in MET analysis were also identified in a single-environment QTL analysis (Supplementary Table S4). Similarly, 159 QTLs (94.6%) detected in single-environment QTL analysis were also identified in the MET analysis (Supplementary Table S1). In addition, we also found four QTLs were only identified in MET analysis with a low level of explanation of phenotypic variation (Supplementary Table S4).
Furthermore, to eliminate the potential confounding effect of row type (Rt) and caryopsis type (Ct) on grain size and weight traits, we used these two traits as a covariate, and performed covariate QTL analysis using genome-wide composite interval mapping (GCIM) methods. A total of 312 covariate QTLs distributed on all seven chromosomes were successfully identified, including 108, 117, and 87 covariate QTLs for using Rt, Ct, and Rt+Ct as a covariate, respectively (Supplementary Table S6). Among these 312 covariate QTLs, 123 covariate QTLs corresponded to main or MET QTLs detected in single-environment QTL analysis or MET analysis, and the remaining 189 (60.6%) were new QTLs that were not identified in either single-environment QTL analysis or MET analysis (Supplementary Table S6). We integrated all QTLs detected by the five mapping methods and found that most QTLs were distributed on chromosomes 1H, 2H, 3H, and 7H (Figure 3C). Figure 3D shows the Venn diagram of the QTLs for nine grain size and weight traits identified by five mapping methods. Importantly, we identified 60 reliable QTLs (QTLs of the same trait repeatedly detected in at least two years environment and in multiple mapping methods), including 7, 8, 9, 7, 9, 6, 6, 4, and 4 reliable QTLs for Tgw, Gl, Gw, Lwr, Ga, Gp, Gd, Gr, and Ffd, respectively (Table 3). The phenotypic variation explained by each reliable QTL ranged from 0.75 to 70.03% (mean value from all QTLs), with a LOD value ranging from 3.42 to 80.98 (mean value from all QTLs). Among these reliable QTLs, most reliable QTLs were identified in single-environment QTL analysis or MET analysis, only 8 reliable QTLs were only identified in covariance QTL analysis. Importantly, 21 of reliable QTLs had major effects on their respective target traits, of which 20 QTLs were also major QTLs detected in single-environment and multi-environmental.
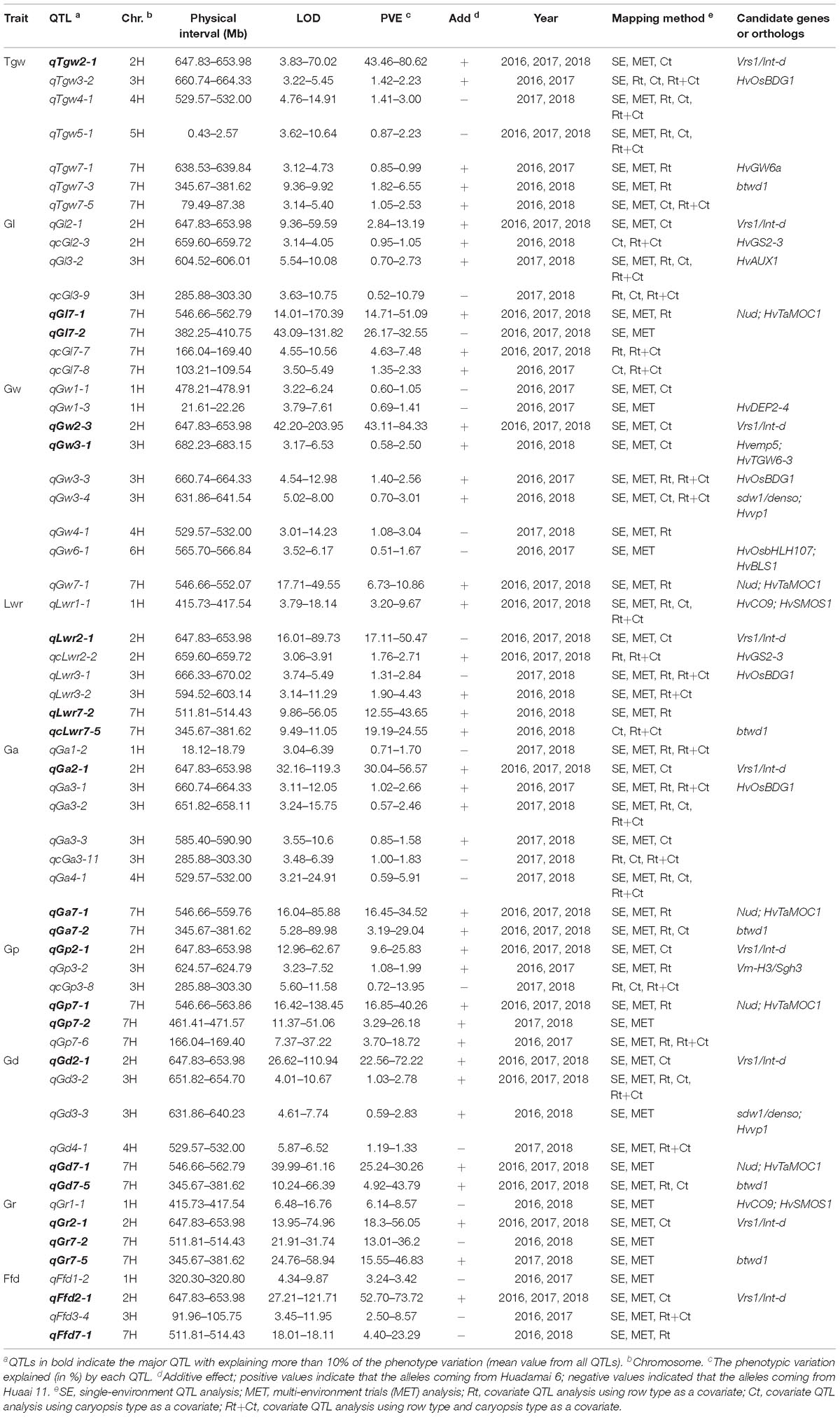
Table 3. Reliable QTLs identified for nine grain size and weight traits in two or more year using multiple mapping method.
QTL Hotspots of Barley Grain Size and Weight Trait
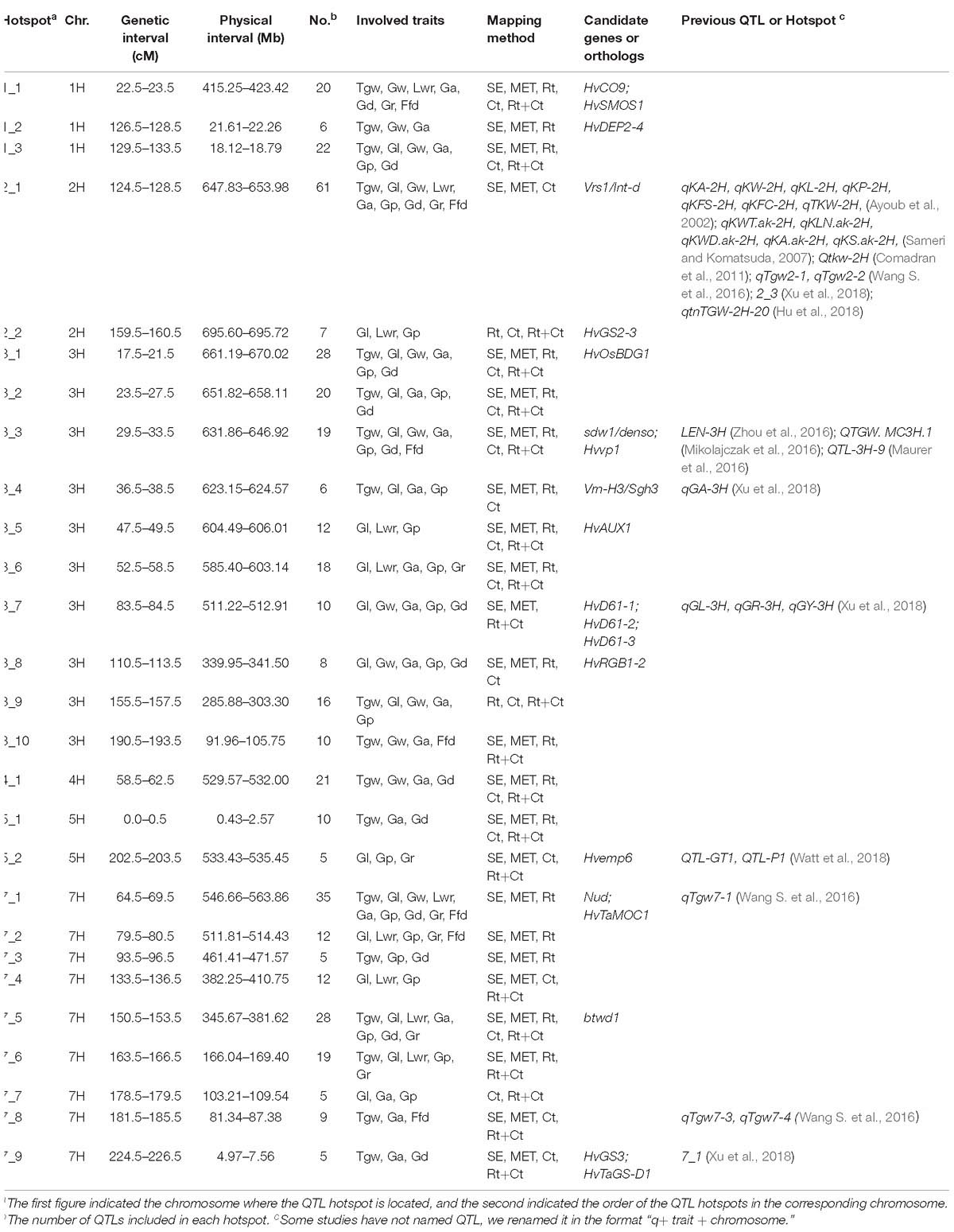
In this study, we found a considerable number of overlapping QTLs for different traits. By integrating all the QTLs identified using the five mapping methods, we found 27 hotspots on six chromosomes involving 421 QTLs, including 3, 2, 10, 1, 2, and 9 hotspots for 1H, 2H, 3H, 4H, 5H, and 7H, respectively (Table 4). For instance, the significant QTL hotspots on chromosome 2H at 124.5–128.5 cM (647.83–653.98 Mb) and on chromosome 7H at 64.5–69.5 cM (546.66–563.86 Mb), included 61 QTLs and 35 QTLs, each influenced all nine grain size and weight traits simultaneously. Another significant QTL hotspot including 28 QTLs, detected on chromosome 7H at 150.5–153.5 cM (345.67–381.62 Mb), was related to Tgw, Gl, Lwr, Ga, Gp, Gd, and Gr. Similarly, the rest of QTL hotspots each affected more than three or more grain traits.
Genetic Correspondence in Diverse Cereals
To identify candidate genes for QTLs and their correspondence to grain size/weight related genes in other cereal species, we collected 38 barley genes and 148 other cereal crops genes (including 94 rice genes, 40 maize genes, and 14 wheat genes) associated with grain size/weight or yield (Supplementary Tables S7, S8). Based on the Ensembl Plant Database (see footnote 1), we identified 164 barley orthologs from 112 other cereal species genes, including 102, 48, and 14 barley orthologs for 70 rice genes, 29 maize genes, and 13 wheat genes, respectively, while no corresponding ortholog was found for 36 genes (Supplementary Table S8 and Supplementary Figure S2). Out of these 112 genes, 93, 7, 5, 2, 1, 1, 2, and 1 gene have one, two, three, four, five, six, seven, and nine orthologs, respectively. We mapped these 38 barley genes and 164 barley orthologs to the barley physical map and aligned these against our QTLs, and identified 45 barley genes or orthologs that were located within ±5 Mb on either side of the QTL peak, including 12, 20, 9, and 4 genes or orthologs for barley, rice, maize, and wheat, respectively (Table 5 and Figure 4). Importantly, 20 of them were located in the 14 QTL hotspot regions on chromosome 1H, 2H, 3H, 5H, and 7H.
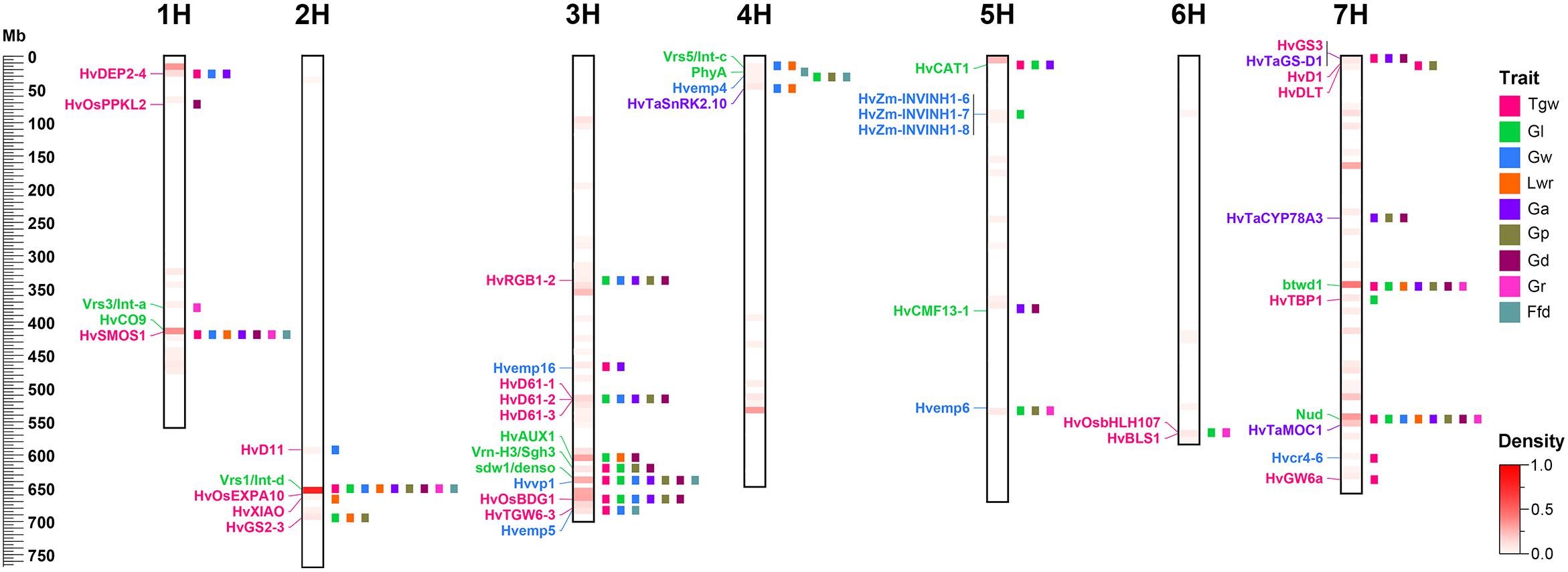
Figure 4. Comparative analysis of QTLs detected in this study with barley yield-associated genes and barley orthologs of grain size or weight genes from rice, wheat, and maize. A total of 20 rice orthologs (shown in red), 4 wheat orthologs (shown in blueviolet), 9 maize orthologs (shown in dodgerblue), and 12 barley genes (shown in lime) were shown in the whole barley genome. The heat map in the chromosome region illustrated the density of QTLs for nine grain size and weight traits. The window size was 10 Mb.
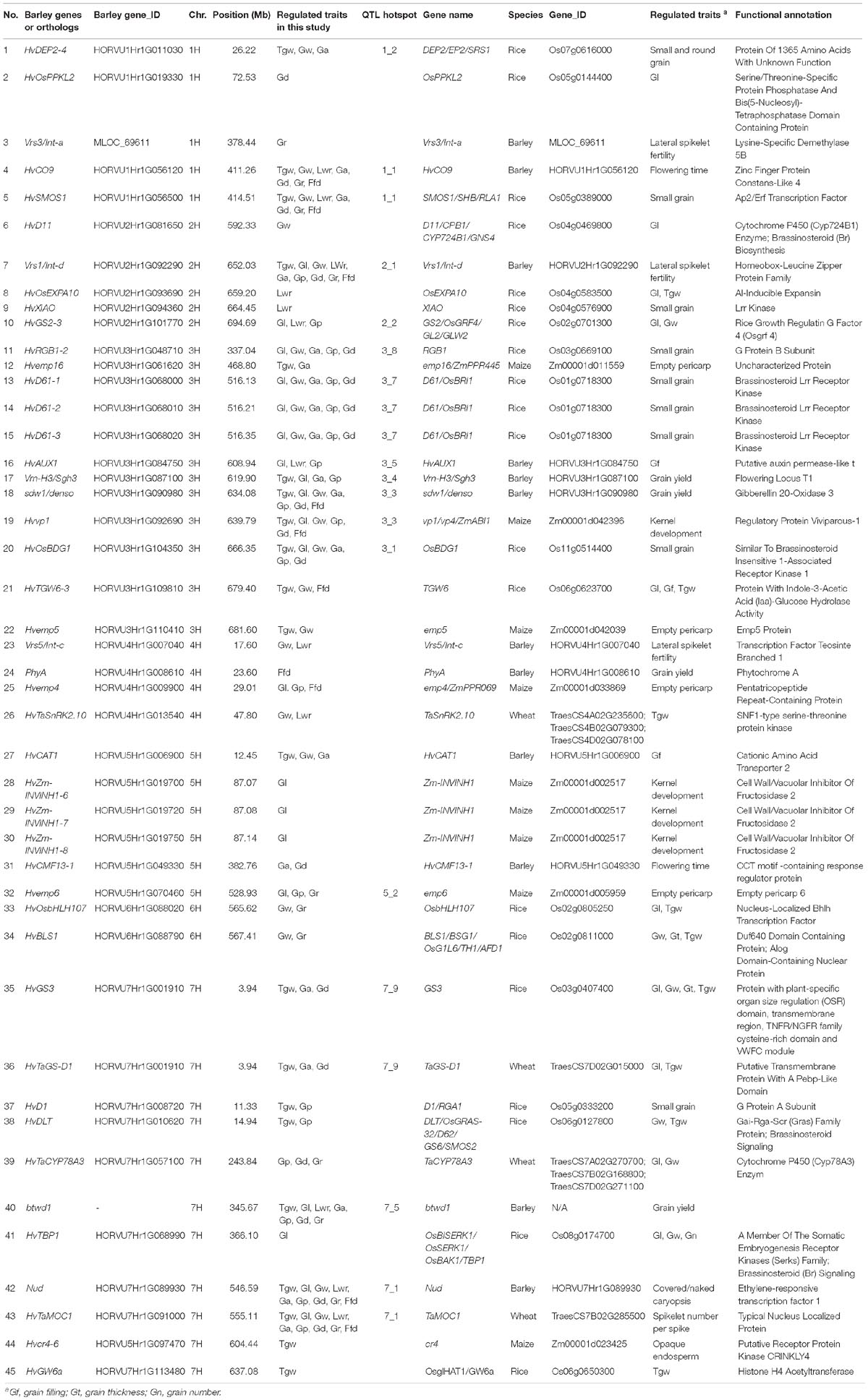
Table 5. Correspondence between QTL of barley grain size and weight and known genes of other cereal grain traits.
Discussion
Improving barley yield has always been an important objective of barley genetic research and breeding programs. Grain size is one of the key factors determining barley yield. Although some QTLs related to grain size and weight have been identified, and several genes affecting grain size or weight have been characterized, our knowledge on the genetic and molecular mechanisms that regulate grain size in barley remain largely unknown. In this study, we have identified 593 QTLs for nine barley grain size and weight traits using five mapping methods. A total of 45 potential candidate genes were identified, providing important insight into the genetic basis of barley grain size and weight.
QTLs for Grain Size and Weight
QTL analysis of thousand grain weight (Tgw) has been performed previously (Ayoub et al., 2002; Coventry et al., 2003; Chen et al., 2004; Li et al., 2005, 2006; Sameri and Komatsuda, 2007; Comadran et al., 2011; Kalladan et al., 2013; Walker et al., 2013; Maurer et al., 2016; Mikolajczak et al., 2016; Wang J. et al., 2016; Xu et al., 2018), but some important information is still missed for the QTL of grain size in barley. To date, QTLs for grain length (Gl) (Ayoub et al., 2002; Sameri and Komatsuda, 2007; Kalladan et al., 2013; Walker et al., 2013; Zhou et al., 2016; Watt et al., 2018; Xu et al., 2018), grain width (Gw) (Ayoub et al., 2002; Sameri and Komatsuda, 2007; Kalladan et al., 2013; Walker et al., 2013; Cu et al., 2016; Xu et al., 2018), grain length-width ratio (Lwr) (Sameri and Komatsuda, 2007; Kalladan et al., 2013), grain area (Ga) (Ayoub et al., 2002; Sameri and Komatsuda, 2007; Xu et al., 2018), grain diameter (Gd) (Cu et al., 2016), and grain roundness (Gr) (Ayoub et al., 2002) have been mapped on almost all seven linkage groups, while QTLs conferring grain perimeter (Gp) and factor form density (Ffd) were rarely reported previously in barley.
In the present study, we successfully identified 60 reliable QTLs for the nine traits of grain size and weight and found 27 hotspot regions that distributed on chromosome 1H, 2H, 3H, 4H, 5H, and 7H (Tables 3, 4). Comparing our QTLs with published results by using BARLEYMAP pipeline (Cantalapiedra et al., 2015), we found some QTLs identified here appeared to coincide to the QTLs described previously. For example, 61 QTLs encompassing all nine traits were clustered in the interval of 647.83–653.98 Mb on chromosome 2H, which corresponds to the QTLs reported previously (Ayoub et al., 2002; Sameri and Komatsuda, 2007; Comadran et al., 2011; Wang J. et al., 2016; Xu et al., 2018), indicating its stability and major effects. By using genome-wide association analysis, Xu et al. (2018) detected a QTL hotspot 7_1 for grain size and weight traits near the barley ortholog of rice Gs3 gene on chromosome 7H (HORVU7Hr1G001910), which may be the same to the QTL hotspot 7_9 (7H: 4.97–7.56 Mb) identified in our study. The QTL hotspot 3_3 consisting of 19 QTLs for Tgw, Gl, Gw, Ga, Gp, Gd, and Ffd on chromosome 3H at 29.5–33.5 cM (631.86–646.92 Mb) identified here may share the same location with the QTL LEN-3H for Gl described by Zhou et al. (2016), the QTL QTGW. MC3H.1 for Tgw detected by Mikolajczak et al. (2016), and the QTL QTL-3H-9 for Tgw found by Maurer et al. (2016).
In addition, some novel QTLs were also identified in our study. For example, QTL hotspot 4_1 for Tgw, Gw, Ga, and Gd on chromosome 4H at 529.57–532.00 Mb identified here are different from the QTLs or QTL hotspots for grain size and weight described on chromosome 4H in previous reports (Maurer et al., 2016; Mikolajczak et al., 2016; Zhou et al., 2016; Xu et al., 2018). Similarly, QTL hotspot 7_2 (511.81–514.43 Mb) and 7_5 (345.67–381.62 Mb) on chromosome 7H contained 12 QTLs and 28 QTLs, respectively, which are different from those QTLs for grain size and weight detected on chromosome 7H in previous studies (Mikolajczak et al., 2016; Zhou et al., 2016; Xu et al., 2018).
Some Barley Yield-Related Genes Were Associated With Grain Size and Weight
Many genes have been proved to have pleiotropic effects. For example, semi-dwarf gene sdw1/denso controls plant height, the number of tillers, grain yield, and grain size (Forster, 2001; Kuczyńska et al., 2013; Kuczyńska et al., 2014). Photoperiod response gene Ppd-H1/Eam1/HvPRR37 has pleiotropic effects on flowering time, leaf size, and yield components (Li et al., 2005; Digel et al., 2016). Thus, we investigated 38 barley genes, including 37 genes that were previously described to influence grain yield and a novel dwarf gene btwd1 (Ren et al., 2016) previously identified in our DH population, to explore whether they also affect barley grain size and weight (Supplementary Table S7). Among these 38 barley genes, 12 were identified as potential candidate genes affecting barley grain size and weight (Table 5). Importantly, seven of them were identified in the QTL hotspot region controlling barley grain size and weight.
The most important QTL hotspot 2_1 on chromosome 2H at 124.5–128.5 cM (647.83–653.98 Mb) contained 61 QTLs for all grain size and weight traits. The vrs1/int-d gene (HORVU2Hr1G092290) was also mapped to this region, which has previously been reported to affect row type, grain size and Tgw (Ayoub et al., 2002; Komatsuda et al., 2007; Sakuma et al., 2017; Hu et al., 2018; Xu et al., 2018). Another significant QTL hotspot region underlying all grain size and weight traits on chromosome 7H at 64.5–69.5 cM (546.66–563.86 Mb) was physically close to nud gene (HORVU7Hr1G089930) that determines the hulled/naked caryopsis phenotype (Taketa et al., 2008). Several studies have previously reported that yield-related QTLs, including Tgw, were tightly linked to nud gene (Barabaschi et al., 2012; Gong et al., 2016). Since our population was derived from a cross between naked six-rowed barley and hulled two-rowed barley, the effects of these two genes on grain traits were in line with our expectations. Due to large effect of these two genes in the population used here, the effect of other QTLs on grain traits was relatively weak and difficult to detect. Therefore, to reduce the confounding effects of these two genes on grain traits, we carried out covariate QTL analysis to find more QTLs association with grain traits. In fact, our strategy was successful because we found more than 60% (189 QTLs) of the covariate QTLs were new QTLs that were not detected in either single-environment QTL analysis or MET analysis (Supplementary Table S6). Although these QTLs had relatively weak effects on the grain traits in this population due to the large influence of the vrs1/int-d and nud genes, they played an important role in revealing the genetic basis of barley grain traits. For mapping barley grain traits, it is better to use parents with the same row type and caryopsis type to construct mapping population to eliminate the influence of these two genes, vrs1/int-d and nud, over other QTLs.
Except for vrs1/int-d and nud genes, the vernalization gene Vrn-H3/Sgh3/HvFT1 (HORVU3Hr1G087100) and flowering gene HvCO9/HvCMF11 (HORVU1Hr1G056120), which are associated with barley yield, were physically close to QTL hotspot regions 1_1 (415.25–423.42 Mb) and 3_4 (623.15–624.57 Mb), respectively (Table 5). Moreover, we also found that semi-dwarf gene sdw1/denso (HORVU3Hr1G090980) and dwarf gene btwd1 [close to SNP marker 7HL_6335336 (7H: 345673515–345673602 bp)] were located in the QTL hotspots region of 3_3 (631.86–646.92 Mb) and 7_5 (345.67–381.62 Mb), respectively (Table 5). These two dwarf or semi-dwarf genes have previously been described to be closely related to yield, and the sdw1/denso semi-dwarf gene has also been reported to have pleiotropic or tightly linkage effects on Tgw and grain size (Forster, 2001; Coventry et al., 2003; Kuczyńska et al., 2013; Maurer et al., 2016; Ren et al., 2016). We predicted 20 candidate genes for btwd1 that were located within ±5 Mb on either side of the nearest SNP marker 7HL_6335336 of the btwd1 gene (Supplementary Table S9). Among them, the most promising candidate gene HORVU7Hr1G066930 (7H: 346181113–346195444 bp) whose annotation information is WD-40 repeat protein-like isoform 1, is an ortholog of rice gene OsTPL/ASP1/OsLIS-L1. The rice OsTPL/ASP1/OsLIS-L1 gene encoding a lissencephaly type-1-like protein and containing the WD40 motif has previously been confirmed to regulate the first internode elongation of rice, resulting in a dwarf phenotype (Gao et al., 2012). These results provided evidence that many of the yield-related genes might have pleiotropic or tightly linkage effects on barley grain size and weight and contributed to phenotypic diversity in barley grain size and weight.
Some Grain Size/Weight Related Genes in Other Cereals May Have Conserved Functions in Barley
Comparative genomics has demonstrated that orthologs from common ancestors generally have conserved functions, which provides an effective strategy for the discovery of barley genes (Devos, 2005; Su et al., 2011; Murat et al., 2017). For example, the rice gene DEP1 that encodes a highly cysteine-rich G protein gamma subunit to regulate grain yield (Kunihiro et al., 2013), its orthologs gene HvDep1 in barley has also been found to have similar functions as in rice (Bélanger et al., 2014; Wendt et al., 2016). At present, a considerable number of genes associated with grain size/weight have been characterized in rice, maize, and wheat. Thus, identification of orthologs for these genes in the barley genome, using comparative genomic approaches, may provide more insights into the genetic mechanisms of barley grain size and weight.
In this study, a total of 32 barley orthologs were identified as potential candidate genes that determine barley grain size or weight, including 20 (one of them is the same as in wheat), 9 and 4 genes from rice, maize, and wheat, respectively (Table 5). The barley ortholog of the rice OsBDG1 gene is on chromosome 3H at 666.35 Mb (HORVU3Hr1G104350), which encodes the leucine-rich repeat receptor-like protein kinase family protein and is nearby the QTL hotspot 3_1 (661.19–670.02 Mb) identified in this study (Table 5). The rice OsBDG1 gene encoding a small protein with short leucine-rich-repeats possessing cell elongation activity, has previously been proven to positively regulate grain size in rice (Jang and Li, 2017). Hence, we believe that HORVU3Hr1G104350 should be a reliable candidate gene affecting grain size as the function of the OsBDG1 gene. The barley ortholog of the rice GS3 gene, HORVU7Hr1G001910, located on chromosome 7H at 3.94 Mb, is close to the QTL hotspot 7_9 (7H: 4.97–7.56 Mb) for Tgw, Ga, and Gd, encoding a grain length protein (Table 5). The rice GS3 gene encoding a membrane protein with several conserved domains including the plant-specific organ size regulation (OSR) domain, is a negative regulator of grain size and organ size (Mao et al., 2010). Many previous studies have confirmed that the GS3 gene can regulate the grain size of rice (Fan et al., 2006; Takano-Kai et al., 2009, 2013; Mao et al., 2010; Nan et al., 2018). Moreover, the GS3 gene also has a similar function in wheat, and its wheat ortholog TaGS-D1 has been reported to be associated with grain weight and grain length in wheat (Zhang et al., 2014). Therefore, we concluded that HORVU7Hr1G001910 is a reliable candidate gene to regulate the grain size or weight of barley. Similarly, the traits regulated by other potential candidate genes also showed phenotypes consistent or partially consistent with the traits contained in their corresponding QTL hotspots. These findings might imply that grain size/weight genes of other cereal species have same or similar functions in barley. These barley orthologs of grain size/weight related genes identified from rice, maize, and wheat in our study, provide promising candidate genes for barley grain size and weight.
Conclusion
In summary, in this study, we identified 60 reliable QTLs and 27 QTL hotspots for the traits of grain size and weight in barley, using a single-environment QTL analysis, MET analysis, and covariate QTL analysis. Moreover, we also systematically explored the genetic correspondence between the QTLs identified in this study and known yield-related genes in barley and grain size/weight related genes in other cereal species. A total of 45 barley genes or orthologs were identified as promising candidate genes for barley grain size and weight, 20 of which were located in the QTL hotspot region underlying barley grain size and weight. These potential candidate genes are worth exploring and studying in detail. Our findings will enhance our understanding of the genetic basis of barley grain size and weight and may provide new information to facilitate high-yield breeding in barley.
Author Contributions
DS and GS conceived this study and designed the experiments. QW, BD, YC, and YW conducted the experiments and phenotyping measurements. QW performed the statistical analysis and wrote the manuscript. XR coordinated the experiments and oversaw the data analysis. DS and GS modified the manuscript. CL produced the Huaai 11 and Huadamai 6 DH population. All authors read and approved the final version of the manuscript.
Funding
This project was supported by the earmarked fund for China Agriculture Research System (CARS-5).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00469/full#supplementary-material
Footnotes
References
Ayoub, M., Symons, J., Edney, J., and Mather, E. (2002). QTLs affecting kernel size and shape in a two-rowed by six-rowed barley cross. Theor. Appl. Genet. 105, 237–247. doi: 10.1007/s00122-002-0941-1
Barabaschi, D., Francia, E., Tondelli, A., Gianinetti, A., Stanca, A. M., and Pecchioni, N. (2012). Effect of the nud gene on grain yield in barley. Czech J. Genet. Plant Breed. 48, 10–22.
Bednarek, J., Boulaflous, A., Girousse, C., Ravel, C., Tassy, C., Barret, P., et al. (2012). Down-regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. J. Exp. Bot. 63, 5945–5955. doi: 10.1093/jxb/ers249
Bélanger, S., Gauthier, M., Jean, M., Sato, K., and Belzile, F. (2014). Genomic characterization of the Hordeum vulgare DEP1 (HvDEP1) gene and its diversity in a collection of barley accessions. Euphytica 198, 29–41. doi: 10.1007/s10681-014-1089-1
Bull, H., Casao, M. C., Zwirek, M., Flavell, A. J., Thomas, W. T. B., Guo, W. B., et al. (2017). Barley SIX-ROWED SPIKE3 encodes a putative Jumonji C-type H3K9me2/me3 demethylase that represses lateral spikelet fertility. Nat. Commun. 8:936. doi: 10.1038/s41467-017-00940-7
Cantalapiedra, C. P., Boudiar, R., Casas, A. M., Igartua, E., and Contreras-Moreira, B. (2015). BARLEYMAP: physical and genetic mapping of nucleotide sequences and annotation of surrounding loci in barley. Mol. Breed. 35:13. doi: 10.1007/s11032-015-0253-1
Chen, G., Suprunova, T., Krugman, T., Fahima, T., and Nevo, E. (2004). Ecogeographic and genetic determinants of kernel weight and colour of wild barley (Hordeum spontaneum) populations in Israel. Seed. Sci. Res. 14, 137–146. doi: 10.1079/ssr2004163
Chen, L., Li, Y. X., Li, C., Wu, X., Qin, W., Li, X., et al. (2016). Fine-mapping of qGW4.05, a major QTL for kernel weight and size in maize. BMC Plant Biol. 16:81. doi: 10.1186/s12870-016-0768-6
Collins, H. M., Burton, R. A., Topping, D. L., Liao, M. L., Bacic, A., and Fincher, G. B. (2010). Variability in fine structures of noncellulosic cell wall polysaccharides from cereal grains: potential importance in human health and nutrition. Cereal Chem. 87, 272–282. doi: 10.1094/Cchem-87-4-0272
Comadran, J., Russell, J. R., Booth, A., Pswarayi, A., Ceccarelli, S., Grando, S., et al. (2011). Mixed model association scans of multi-environmental trial data reveal major loci controlling yield and yield related traits in Hordeum vulgare in mediterranean environments. Theor. Appl. Genet. 122, 1363–1373. doi: 10.1007/s00122-011-1537-4
Coventry, S. J., Barr, A. R., Eglinton, J. K., and Mcdonald, G. K. (2003). The determinants and genome locations influencing grain weight and size in barley (Hordeum vulgare L.). Aust. J. Agric. Res. 54, 1103–1115. doi: 10.1071/Ar02194
Cu, S. T., March, T. J., Stewart, S., Degner, S., Coventry, S., Box, A., et al. (2016). Genetic analysis of grain and malt quality in an elite barley population. Mol. Breed. 36:129. doi: 10.1007/s11032-016-0554-z
Devos, K. M. (2005). Updating the ’crop circle’. Curr. Opin. Plant Biol. 8, 155–162. doi: 10.1016/j.pbi.2005.01.005
Digel, B., Tavakol, E., Verderio, G., Tondelli, A., Xu, X., Cattivelli, L., et al. (2016). Photoperiod1 (Ppd-H1) controls leaf size. Plant Physiol. 172, 405–415. doi: 10.1104/pp.16.00977
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Feng, J. Y., Zhang, J., Wen, Y. J., Zhang, Y. M., Zhang, Y. W., and Dunwell, J. M. (2018). An efficient multi-locus mixed model framework for the detection of small and linked QTLs in F2. Brief. Bioinform doi: 10.1093/bib/bby058 [Epub ahead of print].
Feng, Z., Wu, C., Wang, C., Roh, J., Zhang, L., Chen, J., et al. (2016). SLG controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. J. Exp. Bot. 67, 4241–4253. doi: 10.1093/jxb/erw204
Fischer, R. A., and Edmeades, G. O. (2010). Breeding and cereal yield progress. Crop Sci. 50, S85–S98. doi: 10.2135/cropsci2009.10.0564
Forster, B. P. (2001). Mutation genetics of salt tolerance in barley: an assessment of golden promise and other semi-dwarf mutants. Euphytica 120, 317–328. doi: 10.1023/A:1017592618298
Fuller, D. Q. (2007). Contrasting patterns in crop domestication and domestication rates: recent archaeobotanical insights from the Old World. Ann. Bot. 100, 903–924. doi: 10.1093/aob/mcm048
Gan, Y., and Stobbe, E. H. (1996). Seedling vigor and grain yield of ‘Roblin’ wheat affected by seed size. Agron. J. 88, 456–460. doi: 10.2134/agronj1996.00021962008800030016x
Gao, X., Chen, Z., Zhang, J., Li, X., Chen, G., Li, X., et al. (2012). OsLIS-L1 encoding a lissencephaly type-1-like protein with WD40 repeats is required for plant height and male gametophyte formation in rice. Planta 235, 713–727. doi: 10.1007/s00425-011-1532-7
Geng, J., Li, L., Lv, Q., Zhao, Y., Liu, Y., Zhang, L., et al. (2017). TaGW2-6A allelic variation contributes to grain size possibly by regulating the expression of cytokinins and starch-related genes in wheat. Planta 246, 1153–1163. doi: 10.1007/s00425-017-2759-8
Giura, A., and Saulescu, N. N. (1996). Chromosomal location of genes controlling grain size in a large grained selection of wheat (Triticum aestivum L). Euphytica 89, 77–80. doi: 10.1007/Bf00015722
Gong, X., Wheeler, R., Bovill, W. D., and Mcdonald, G. K. (2016). QTL mapping of grain yield and phosphorus efficiency in barley in a mediterranean-like environment. Theor. Appl. Genet. 129, 1657–1672. doi: 10.1007/s00122-016-2729-8
Hong, Y., Chen, L., Du, L. P., Su, Z., Wang, J., Ye, X., et al. (2014). Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Funct. Integr. Genomics 14, 341–349. doi: 10.1007/s10142-014-0380-5
Hou, J., Li, T., Wang, Y., Hao, C., Liu, H., and Zhang, X. (2017). ADP-glucose pyrophosphorylase genes, associated with kernel weight, underwent selection during wheat domestication and breeding. Plant Biotechnol. J. 15, 1533–1543. doi: 10.1111/pbi.12735
Hu, M. J., Zhang, H. P., Liu, K., Cao, J. J., Wang, S. X., Jiang, H., et al. (2016). Cloning and characterization of TaTGW-7A gene associated with grain weight in wheat via SLAF-seq-BSA. Front. Plant Sci. 7:1902. doi: 10.3389/fpls.2016.01902
Hu, X., Zuo, J., Wang, J., Liu, L., Sun, G., Li, C., et al. (2018). Multi-locus genome-wide association studies for 14 Main agronomic traits in barley. Front. Plant Sci. 9:1683. doi: 10.3389/fpls.2018.01683
Jaiswal, V., Gahlaut, V., Mathur, S., Agarwal, P., Khandelwal, M. K., Khurana, J. P., et al. (2015). Identification of novel SNP in promoter sequence of TaGW2-6A associated with grain weight and other agronomic traits in wheat (Triticum aestivum L.). PLoS One 10:e0129400. doi: 10.1371/journal.pone.0129400
Jang, S., and Li, H. Y. (2017). Oryza sativa BRASSINOSTEROID UPREGULATED1 LIKE1 induces the expression of a gene encoding a small leucine-rich-repeat protein to positively regulate lamina inclination and grain size in rice. Front. Plant Sci. 8:1253. doi: 10.3389/fpls.2017.01253
Jordan, T., Seeholzer, S., Schwizer, S., Töller, A., Somssich, I. E., and Keller, B. (2011). The wheat Mla homologue TmMla1 exhibits an evolutionarily conserved function against powdery mildew in both wheat and barley. Plant J. 65, 610–621. doi: 10.1111/j.1365-313X.2010.04445.x
Kalladan, R., Worch, S., Rolletschek, H., Harshavardhan, V. T., Kuntze, L., Seiler, C., et al. (2013). Identification of quantitative trait loci contributing to yield and seed quality parameters under terminal drought in barley advanced backcross lines. Mol. Breed. 32, 71–90. doi: 10.1007/s11032-013-9853-9
Knapp, S. J., Stroup, W. W., and Ross, W. M. (1985). Exact confidence intervals for heritability on a progeny mean basis. Crop Sci. 25, 192–194. doi: 10.2135/cropsci1985.0011183X002500010046x
Komatsuda, T., Pourkheirandish, M., He, C. F., Azhaguvel, P., Kanamori, H., Perovic, D., et al. (2007). Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. U.S.A. 104, 1424–1429. doi: 10.1073/pnas.0608580104
Koppolu, R., Anwar, N., Sakuma, S., Tagiri, A., Lundqvist, U., Pourkheirandish, M., et al. (2013). Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc. Natl. Acad. Sci. U.S.A. 110, 13198–13203. doi: 10.1073/pnas.1221950110
Kuczyńska, A., Mikołajczak, K., and Æwiek, H. (2014). Pleiotropic effects of the sdw1 locus in barley populations representing different rounds of recombination. Electron. J. Biotechnol. 17, 217–223. doi: 10.1016/j.ejbt.2014.07.005
Kuczyńska, A., Surma, M., Adamski, T., Mikołajczak, K., Krystkowiak, K., and Ogrodowicz, P. (2013). Effects of the semi-dwarfing sdw1/denso gene in barley. J. Appl. Genet. 54, 381–390. doi: 10.1007/s13353-013-0165-x
Kunihiro, S., Saito, T., Matsuda, T., Inoue, M., Kuramata, M., Taguchi-Shiobara, F., et al. (2013). Rice DEP1, encoding a highly cysteine-rich G protein γ subunit, confers cadmium tolerance on yeast cells and plants. J. Exp. Bot. 64, 4517–4527. doi: 10.1093/jxb/ert267
Li, J. Z., Huang, X. Q., Heinrichs, F., Ganal, M. W., and Röder, M. S. (2005). Analysis of QTLs for yield, yield components, and malting quality in a BC3-DH population of spring barley. Theor. Appl. Genet. 110, 356–363. doi: 10.1007/s00122-004-1847-x
Li, J. Z., Huang, X. Q., Heinrichs, F., Ganal, M. W., and Röder, M. S. (2006). Analysis of QTLs for yield components, agronomic traits, and disease resistance in an advanced backcross population of spring barley. Genome 49, 454–466. doi: 10.1139/g05-128
Li, N., Xu, R., Duan, P., and Li, Y. (2018). Control of grain size in rice. Plant Reprod. 31, 237–251. doi: 10.1007/s00497-018-0333-6
Li, Q., Li, L., Yang, X., Warburton, M. L., Bai, G., Dai, J., et al. (2010). Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 10:143. doi: 10.1186/1471-2229-10-143
Li, W., and Yang, B. (2017). Translational genomics of grain size regulation in wheat. Theor. Appl. Genet. 130, 1765–1771. doi: 10.1007/s00122-017-2953-x
Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi: 10.1038/ng.977
Liu, J., Chen, J., Zheng, X., Wu, F., Lin, Q., Heng, Y., et al. (2017a). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3:17043. doi: 10.1038/nplants.2017.43
Liu, J., Huang, J., Guo, H., Lan, L., Wang, H., Xu, Y., et al. (2017b). The conserved and unique genetic architecture of kernel size and weight in maize and rice. Plant Physiol. 175, 774–785. doi: 10.1104/pp.17.00708
Liu, J., Deng, M., Guo, H., Raihan, S., Luo, J., Xu, Y., et al. (2015). Maize orthologs of rice GS5 and their trans-regulator are associated with kernel development. J. Integr. Plant Biol. 57, 943–953. doi: 10.1111/jipb.12421
Liu, Q., Han, R., Wu, K., Zhang, J., Ye, Y., Wang, S., et al. (2018). G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 9:852. doi: 10.1038/s41467-018-03047-9
Ma, L., Li, T., Hao, C., Wang, Y., Chen, X., and Zhang, X. (2016). TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol. J. 14, 1269–1280. doi: 10.1111/pbi.12492
Ma, M., Zhao, H., Li, Z., Hu, S., Song, W., and Liu, X. (2016). TaCYP78A5 regulates seed size in wheat (Triticum aestivum). J. Exp. Bot. 67, 1397–1410. doi: 10.1093/jxb/erv542
Mao, H., Sun, S., Yao, J., Wang, C., Yu, S., Xu, C., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U.S.A. 107, 19579–19584. doi: 10.1073/pnas.1014419107
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
Maurer, A., Draba, V., and Pillen, K. (2016). Genomic dissection of plant development and its impact on thousand grain weight in barley through nested association mapping. J. Exp. Bot. 67, 2507–2518. doi: 10.1093/jxb/erw070
Mccouch, S., Cho, Y., Yano, M., Paule, E., Blinstrub, M., Morishima, H., et al. (1997). Report on QTL nomenclature. Rice Genet. Newsl. 14, 11–13. doi: 10.1007/s10142-013-0328-1
Meng, L., Li, H., Zhang, L., and Wang, J. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Mikolajczak, K., Ogrodowicz, P., Gudyś, K., Krystkowiak, K., Sawikowska, A., Frohmberg, W., et al. (2016). Quantitative trait loci for yield and yield-related traits in spring barley populations derived from crosses between European and Syrian cultivars. PLoS One 11:e0155938. doi: 10.1371/journal.pone.0155938
Murat, F., Armero, A., Pont, C., Klopp, C., and Salse, J. (2017). Reconstructing the genome of the most recent common ancestor of flowering plants. Nat. Genet. 49, 490–496. doi: 10.1038/ng.3813
Nan, J., Feng, X., Wang, C., Zhang, X., Wang, R., Liu, J., et al. (2018). Improving rice grain length through updating the GS3 locus of an elite variety Kongyu 131. Rice 11:21. doi: 10.1186/s12284-018-0217-2
Ramsay, L., Comadran, J., Druka, A., Marshall, D. F., Thomas, W. T. B., Macaulay, M., et al. (2011). INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 43, 169–172. doi: 10.1038/ng.745
Ren, X., Sun, D., Guan, W., Sun, G., and Li, C. (2010). Inheritance and identification of molecular markers associated with a novel dwarfing gene in barley. BMC Genet. 11:89. doi: 10.1186/1471-2156-11-89
Ren, X., Wang, J., Liu, L., Sun, G., Li, C., Luo, H., et al. (2016). SNP-based high density genetic map and mapping of btwd1 dwarfing gene in barley. Sci. Rep. 6:31741. doi: 10.1038/srep31741
Sajjad, M., Ma, X., Habibullah Khan, S., Shoaib, M., Song, Y., Yang, W., et al. (2017). TaFlo2-A1, an ortholog of rice Flo2, is associated with thousand grain weight in bread wheat (Triticum aestivum L.). BMC Plant Biol. 17:164. doi: 10.1186/s12870-017-1114-3
Sakuma, S., Lundqvist, U., Kakei, Y., Thirulogachandar, V., Suzuki, T., Hori, K., et al. (2017). Extreme suppression of lateral floret development by a single amino acid change in the VRS1 transcription factor. Plant Physiol. 175, 1720–1731. doi: 10.1104/pp.17.01149
Salamini, F., Özkan, H., Brandolini, A., Schäfer-Pregl, R., and Martin, W. (2002). Genetics and geography of wild cereal domestication in the near east. Nat. Rev. Genet. 3, 429–441. doi: 10.1038/nrg817
Sameri, M., and Komatsuda, T. (2007). Localization of quantitative trait loci for yield components in a cross oriental × occidental barley cultivar (Hordeum vulgare L.). Jpn. Agric. Res. Q. 41, 195–199. doi: 10.6090/jarq.41.195
Simmonds, J., Scott, P., Brinton, J., Mestre, T. C., Bush, M., Del Blanco, A., et al. (2016). A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. Appl. Genet. 129, 1099–1112. doi: 10.1007/s00122-016-2686-2
Song, X. J., Huang, W., Shi, M., Zhu, M. Z., and Lin, H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Su, Z., Hao, C., Wang, L., Dong, Y., and Zhang, X. (2011). Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 122, 211–223. doi: 10.1007/s00122-010-1437-z
Sun, S., Wang, L., Mao, H., Shao, L., Li, X., Xiao, J., et al. (2018). A G-protein pathway determines grain size in rice. Nat. Commun. 9:851. doi: 10.1038/s41467-018-03141-y
Takano-Kai, N., Jiang, H., Kubo, T., Sweeney, M., Matsumoto, T., Kanamori, H., et al. (2009). Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182, 1323–1334. doi: 10.1534/genetics.108.103002
Takano-Kai, N., Jiang, H., Powell, A., Mccouch, S., Takamure, I., Furuya, N., et al. (2013). Multiple and independent origins of short seeded alleles of GS3 in rice. Breed. Sci. 63, 77–85. doi: 10.1270/jsbbs.63.77
Taketa, S., Amano, S., Tsujino, Y., Sato, T., Saisho, D., Kakeda, K., et al. (2008). Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 4062–4067. doi: 10.1073/pnas.0711034105
Tao, Y., Mace, E. S., Tai, S., Cruickshank, A., Campbell, B. C., Zhao, X., et al. (2017). Whole-genome analysis of candidate genes associated with seed size and weight in Sorghum bicolor reveals signatures of artificial selection and insights into parallel domestication in cereal crops. Front. Plant Sci. 8:1237. doi: 10.3389/fp/s.2017.01237
Van Esse, G. W., Walla, A., Finke, A., Koornneef, M., Pecinka, A., and Von Korff, M. (2017). Six-rowed spike 3 (VRS3) is a histone demethylase that controls lateral spikelet development in barley. Plant Physiol. 174, 2397–2408. doi: 10.1104/pp.17.00108
Walker, C. K., Ford, R., Muñoz-Amatriaín, M., and Panozzo, J. F. (2013). The detection of QTLs in barley associated with endosperm hardness, grain density, grain size and malting quality using rapid phenotyping tools. Theor. Appl. Genet. 126, 2533–2551. doi: 10.1007/s00122-013-2153-2
Walker, C. K., and Panozzo, J. F. (2011). Development of a small scale method to determine volume and density of individual barley kernels, and the relationship between grain density and endosperm hardness. J. Cereal Sci. 54, 311–316. doi: 10.1016/j.jcs.2011.06.008
Wang, J., Sun, G., Ren, X., Li, C., Liu, L., Wang, Q., et al. (2016). QTL underlying some agronomic traits in barley detected by SNP markers. BMC Genet. 17:103. doi: 10.1186/s12863-016-0409-y
Wang, S., Yan, X., Wang, Y., Liu, H., Cui, D., and Chen, F. (2016). Haplotypes of the TaGS5-A1gene are associated with thousand-kernel weight in Chinese bread wheat. Front. Plant Sci. 7:783. doi: 10.3389/fpls.2016.00783
Wang, Q., Sun, G., Ren, X., Wang, J., Du, B., Li, C., et al. (2017). Detection of QTLs for seedling characteristics in barley (Hordeum vulgare L.) grown under hydroponic culture condition. BMC Genet. 18:94. doi: 10.1186/s12863-017-0562-y
Wang, S., Zhang, X., Chen, F., and Cui, D. (2015). A single-nucleotide polymorphism of TaGS5 gene revealed its association with kernel weight in Chinese bread wheat. Front. Plant Sci. 6:1166. doi: 10.3389/fpls.2015.01166
Watt, C., Zhou, G., Mcfawn, L. A., Chalmers, K. J., and Li, C. (2018). Fine mapping of qGL5H, a major grain length locus in barley (Hordeum vulgare L.). Theor. Appl. Genet. 132, 883–893. doi: 10.1007/s00122-018-3243-y
Wendt, T., Holme, I., Dockter, C., Preuß, A., Thomas, W., Druka, A., et al. (2016). HvDep1 is a positive regulator of culm elongation and grain size in barley and impacts yield in an environment-dependent manner. PLoS One 11:e0168924. doi: 10.1371/journal.pone.0168924
Wu, W., Liu, X., Wang, M., Meyer, R. S., Luo, X., Ndjiondjop, M. N., et al. (2017). A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants 3:17064. doi: 10.1038/nplants.2017.64
Xu, X., Sharma, R., Tondelli, A., Russell, J., Comadran, J., Schnaithmann, F., et al. (2018). Genome-wide association analysis of grain yield-associated traits in a pan-European barley cultivar collection. Plant Genome 11:170073. doi: 10.3835/plantgenome2017.08.0073
Yang, Z., Bai, Z., Li, X., Wang, P., Wu, Q., Yang, L., et al. (2012). SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor. Appl. Genet. 125, 1057–1068. doi: 10.1007/s00122-012-1895-6
Yin, C., Li, H., Li, S., Xu, L., Zhao, Z., and Wang, J. (2015). Genetic dissection on rice grain shape by the two-dimensional image analysis in one japonica × indica population consisting of recombinant inbred lines. Theor. Appl. Genet. 128, 1969–1986. doi: 10.1007/s00122-015-2560-7
Ying, J. Z., Ma, M., Bai, C., Huang, X. H., Liu, J. L., Fan, Y. Y., et al. (2018). TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol. Plant 11, 750–753. doi: 10.1016/j.molp.2018.03.007
Youssef, H. M., Eggert, K., Kopplu, R., Alqudah, A. M., Poursarebani, N., Fazeli, A., et al. (2017). VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 49, 157–161. doi: 10.1038/ng.3717
Zhai, H., Feng, Z., Du, X., Song, Y., Liu, X., Qi, Z., et al. (2018). A novel allele of TaGW2-A1 is located in a finely mapped QTL that increases grain weight but decreases grain number in wheat (Triticum aestivum L.). Theor. Appl. Genet. 131, 539–553. doi: 10.1007/s00122-017-3017-y
Zhang, B., Liu, X., Xu, W., Chang, J., Li, A., Mao, X., et al. (2015). Novel function of a putative MOC1 ortholog associated with spikelet number per spike in common wheat. Sci. Rep. 5:12211. doi: 10.1038/srep12211
Zhang, L., Zhao, Y. L., Gao, L. F., Zhao, G. Y., Zhou, R. H., Zhang, B. S., et al. (2012). TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 195, 574–584. doi: 10.1111/j.1469-8137.2012.04194.x
Zhang, X., Wang, J., Huang, J., Lan, H., Wang, C., Yin, C., et al. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U.S.A. 109, 21534–21539. doi: 10.1073/pnas.1219776110
Zhang, Y., Li, D., Zhang, D., Zhao, X., Cao, X., Dong, L., et al. (2018). Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 94, 857–866. doi: 10.1111/tpj.13903
Zhang, Y., Liu, J., Xia, X., and He, Z. (2014). TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Mol. Breed. 34, 1097–1107. doi: 10.1007/s11032-014-0102-7
Zhao, D. S., Li, Q. F., Zhang, C. Q., Zhang, C., Yang, Q. Q., Pan, L. X., et al. (2018). GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9:1240. doi: 10.1038/s41467-018-03616-y
Zhou, H., Liu, S., Liu, Y., Liu, Y., You, J., Deng, M., et al. (2016). Mapping and validation of major quantitative trait loci for kernel length in wild barley (Hordeum vulgare ssp. spontaneum). BMC Genet. 17:130. doi: 10.1186/s12863-016-0438-6
Zhou, Q., Dong, Y., Shi, Q., Zhang, L., Chen, H., Hu, C., et al. (2017). Verification and fine mapping of qGW1.05, a major QTL for grain weight in maize (Zea mays L.). Mol. Genet. Genomics 292, 871–881. doi: 10.1007/s00438-017-1318-0
Keywords: barley (Hordeum vulgare L.), grain size and weight, doubled haploid population, comparative genomics, QTL
Citation: Wang Q, Sun G, Ren X, Du B, Cheng Y, Wang Y, Li C and Sun D (2019) Dissecting the Genetic Basis of Grain Size and Weight in Barley (Hordeum vulgare L.) by QTL and Comparative Genetic Analyses. Front. Plant Sci. 10:469. doi: 10.3389/fpls.2019.00469
Received: 19 December 2018; Accepted: 28 March 2019;
Published: 24 April 2019.
Edited by:
Marion S. Röder, Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK), GermanyReviewed by:
Ahmad M. Alqudah, Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK), GermanyZhongfu Ni, China Agricultural University, China
Zhenyu Gao, China National Rice Research Institute (CAAS), China
Meixue Zhou, University of Tasmania, Australia
Copyright © 2019 Wang, Sun, Ren, Du, Cheng, Wang, Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongfa Sun, c3VuZG9uZ2ZhMUBtYWlsLmh6YXUuZWR1LmNu
 Qifei Wang
Qifei Wang Genlou Sun
Genlou Sun Xifeng Ren1
Xifeng Ren1 Binbin Du
Binbin Du Chengdao Li
Chengdao Li Dongfa Sun
Dongfa Sun