- 1Key Laboratory of Northern Japonica Rice Genetics and Breeding, Ministry of Education and Liaoning Province – Key Laboratory of Northeast Rice Biology and Genetics and Breeding, Ministry of Agriculture, Rice Research Institute, Shenyang Agricultural University, Shenyang, China
- 2Shenyang Product Quality Supervision and Inspection Institute, Shenyang, China
Autophagy (self-eating), a conserved pathway in eukaryotes, which is designed to handle cytoplasmic material in bulk and plays an important role in the remobilization of nutrient, such as nitrogen (N) under suboptimal nutrient conditions. Here, we identified a core component of an autophagy gene in rice (Oryza sativa), OsATG8a, with increased expression levels under N starvation conditions. Overexpression of OsATG8a significantly enhanced the level of autophagy and the number of effective tillers in the transgenic rice. In addition, the transgenic lines accumulated more N in grains than in the dry remains and the yield was significantly increased under normal N conditions. Further N allocation studies revealed that the nitrogen uptake efficiency (NUpE) and nitrogen use efficiency (NUE) significantly increased. Otherwise, under suboptimal N conditions, overexpression of OsATG8a did not seem to have any effect on yield and NUE, but NUpE was still improved significantly. Based on our findings, we consider OsATG8a to be a great candidate gene to increase NUE and yield.
Introduction
Nitrogen (N) is one of the most important nutrient elements for plants and is required for growth and development. N is also a vital component of amino acids, proteins, nucleic acids, chlorophyll, and plant hormones (Kraiser et al., 2011). In plants, especially cereal crops, the yield and quality of the grain depend not only on the N absorbed prior to flowering, but also on the mobilization of reused N from mature leaves during senescence (Kichey et al., 2007; Chardon et al., 2012; Havé et al., 2017). The circulation process for organic nitrogen transfer from aging tissues to seeds in plants is an important determinant of productivity and yield, especially under nitrogen deficiency stress (Masclaux-Daubresse et al., 2010). Although the pathways of protein metabolism have not been thoroughly studied in senescing leaves, the following three important pathways are currently considered: plastid resident proteases, senescence-associated vacuoles (SAVs), and autophagy (Otegui et al., 2005; Ishida et al., 2008; Wada et al., 2009; Roberts et al., 2012).
Autophagy, a conserved vacuolar degradation pathway to remove cellular debris, appears to be active at a basal level in plant cells during all developmental stages, (Marshall and Vierstra, 2018). First, autophagy transfers the “cargo” (cytoplasmic components) to “degradation organs” (vacuoles in yeast and plants). Second, the cargo is degraded (Li and Vierstra, 2012). In response to stress or nutrient starvation, autophagy is enhanced to facilitate the degradation of increasing levels of toxic and damaged components and nutrients mobilized from this recycled cell material are then used to maintain cellular processes and in adaptation to stress (Yang and Bassham, 2015). Through studies on a variety of organisms, the mechanism of the autophagic machinery has emerged. The first AuTophaGy-related gene (ATG gene) was found in yeast (Miki and Yoshinori, 1993). Many orthologs of yeast ATG genes have been characterized in different species, including mammals and plants (Wang et al., 2015; Li et al., 2016; Zhai et al., 2016). To date, almost 40 ATG proteins have been identified as evolutionarily conserved among mammals, yeasts, and plant; these proteins form the core autophagic mechanism (Liu and Bassham, 2012).
A study of Arabidopsis thaliana atg mutants showed that autophagy controls the remobilization of N from leaves to seeds (Guiboileau et al., 2012). NUE decreased significantly to about 50% in the seeds of atg mutants compared to the WT under N-limited conditions (Guiboileau et al., 2013). Similar results were found in crops, such as in the rice osatg7-1 mutant, where biomass and NUE during the vegetative growth stage were significantly reduced and the mutant was unable to mobilize N in aging leaves (Wada et al., 2015). Recent research further indicated that the metabolism and endogenous levels of gibberellins and cytokinin in anthers of the osatg7 mutant were defective, which is the reason for defects in pollen maturation of autophagy-deficient plants (Kurusu et al., 2017). In maize (Zea mays), 15N pulse-chase analysis revealed that the amount of recycled N was reduced by two-fold in the atg12 mutant compared with WT (Li F. et al., 2015). A facile multi-omics strategy in maize showed significant effects of autophagy on the formation of eukaryotic proteins and membranes under well-fed growth conditions and nutrient stress (Marshall and Vierstra, 2018). In addition, a study on the model plant A. thaliana also revealed a shorter life span, premature senescence of leaves, metabolome changes in cells, reallocated resources and protein accumulation, and impaired tolerance to biological and abiotic stress in atg mutants (Di et al., 2018).
Among the many plant ATG proteins, ATG8 is an important protein involved in formation of the autophagosome and recruitment of particular types of cargo (Kellner et al., 2016). The ATG8 protein is ubiquitin-like and conjugated to phosphatidylethanolamine (PE) on the autophagic membrane, which is located on the autophagosome membrane (Nakatogawa et al., 2007; Xie et al., 2008; Klionsky and Schulman, 2014). ATG8 is a central player in the autophagy process because it provides a docking site for ATG8 interaction motif (AIM)-containing autophagic receptors that selectively recruit cargo (Kellner et al., 2016). Seven ATG8 isoforms (OsATGa to OsATGg) have been identified in the rice genome. OsATG8a through OsATG8c have high levels of identity with the deduced amino acid sequence and OsATG8d has similarities to AtATG8i (Chung et al., 2009; Xia et al., 2011); OsATG8e, however, has a gene locus but a lack of supporting expressed sequence tag (EST) data (Chung et al., 2009); OsATG8f only has a gene locus ID in RAP-DB but has no corresponding full-length cDNA; and OsATG8i has not been mapped to the rice genome (Xia et al., 2011). Several studies indicate that overexpression of ATG8 homologs promotes plant growth and increases tolerance to N starvation in plants (Slavikova et al., 2008; Xia et al., 2012; Li W. et al., 2015, Li et al., 2016; Kellner et al., 2016; Wang et al., 2016).
In this article, some studies were done to identify the potential roles of OsATG8a (LOC_Os07g32800) on yield and N uptake and reuse in rice. The OsATG8a protein was predominantly localized to the cytoplasm, with autophagosome-like structures seen as fluorescence puncta. Compared to the control plants, the independent 35S-OsATG8a transgenic lines had more effective tillering and produced more panicles and seeds, thus increasing yield. At the same time, autophagy levels were significantly enhanced in the transgenic lines under both N sufficient and deficient conditions. N allocation studies showed that overexpression of OsATG8a significantly increased N allocation to seeds and enhanced nitrogen uptake efficiency (NUpE) and nitrogen use efficiency (NUE). Based on these results, OsATG8a is an important candidate gene for synergistically enhanced N remobilization and better yield in rice.
Materials and Methods
Plant Materials and Growth Conditions
The japonica rice variety Shennong9816 (SN9816) was used in this study. For short term hydroponic culture, rice seedlings were grown in half Hoagland’s solution (with 3 mM KNO3 and 0.25 mM (NH4)2SO4, pH = 5.7) as previously described (Xia et al., 2012) in a growth chamber (28°C/25°C and 10 h light/14 h dark). To analyze the response of OsATG8a expression to N limitation in rice, rice seedlings were cultured with half Hoagland’s solution as sufficient N (NS, with 3.5 mM N) for 14 days, then transferred to the same NS (3.5 mM) solution as control; modified half Hoagland’s solution (with 0.6 mM KNO3 and 0.1 mM (NH4)2SO4, pH = 5.7) for low N (NL, with 0.8 mM N); and N-free solution for deficient N (ND, with 0 mM N); the lack of K+ was replaced with KCl in NL (0.8 mM) and ND (0 mM) solution. After 1 and 3 days of cultivation in NS (3.5 mM), NL (0.8 mM), or ND (0 mM) conditions described above, 10 rice seedlings (SN9816) of each N level were collected as a sample, and the leaves and roots were harvested separately.
Total RNA Extraction and Gene Expression Analysis
Total RNA extraction was conducted using the Eastep Super Total RNA Extraction Kit (Promega) and first strand cDNA was synthesized with the PrimeScript RT Master Mix (TaKaRa). Real-time RT-PCR was performed as described previously (Xu et al., 2011) using SYBR Premix Ex TaqII (TaKaRa) on an Applied Biosystems 7500 Real Time PCR System. The following standard thermal profile was used for all PCRs: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 34 s. All reactions were done at least in triplicate. OsActin1 was used as an internal control. The primers used for RT-PCR analysis are listed in Supplementary Table S1.
Binary Vector Construction and Rice Transformation
The OsATG8a gene has a 360-bp coding sequence (CDS) and encodes 120 amino acids. The complete coding region of OsATG8a was amplified by PCR using SN9816 cDNA as a template and ligated into binary vector pCAMBIA1301, resulting in the 35S-OsATG8a fusion gene. For the ProOsATG8a-GUS construct, a 1405-bp promoter fragment of OsATG8a was amplified by PCR using SN9816 genomic DNA as templates and cloned into vector pCAMBIA1301. The 35S-eGFP-OsATG8a (enhanced green fluorescent protein) fusion constructs were produced by inserting the complete coding region of OsATG8a into the pCAMBIA1300 vector. All constructs were confirmed by DNA sequencing. Sequences of all the primers used in cloning are listed in Supplemental Table S1. The plant expression vector was transformed into SN9816 by Agrobacterium-mediated transformation as described previously (Hiei et al., 1994). 14-day-old seedlings of 35S-OsATG8a homozygous T3 lines and SN9816 grown hydroponically were sampled to confirm the transcript level of OsATG8a.
Protein Isolation and Western Blot Analysis
Fourteen-day-old seedlings of transgenic lines and SN9816 cultured hydroponically in NS (3.5 mM) solution were transferred to NS (3.5 mM) or ND (0 mM) solution for 24 h to analyze the autophagic flux. Total protein extraction was conducted using the MinuteTM Total Protein Extraction Kit for plants (Invent Biotechnologies, Inc.), and total membrane protein extraction was conducted using the MinuteTM Membrane Protein Isolation Kit for plants (Invent Biotechnologies, Inc.). Protein concentration was determined with the BCA Protein Assay Kit (TaKaRa), and equal amount of protein was loaded in each lane. Immunoblotting was done as described previously (Chung et al., 2009; Xia et al., 2012). The obtained proteins were loaded into a 12% SDS-PAGE with 6 M urea gel. After electrophoresis separation, the proteins were transferred to a PVDF membrane for protein profile analysis or immunoblot analysis. Antibodies against Arabidopsis ATG8a and NBR1, α-Tubulin (Agrisera) were used as primary antibodies (1: 1000); goat anti-rabbit (IgG) (Sigma-Aldrich) was used as the secondary antibody (1: 10000). The reactions were detected using the Pro-light HRP Chemiluminescent Kit (TIANGEN). Quantification analysis of the protein band intensities from immunoblots was performed by software Image J. For total protein content of leaves and seeds analysis, proteins were extracted from 100 mg fresh weight of leaves at grain filling stage, 5 mg dry mass of seeds after harvested, and the same volume was loaded in each lane. Then, the total protein content was determined with the BCA Protein Assay Kit (TaKaRa) and Coomassie Brilliant Blue staining. Each experiment was repeated at least three times, and one representative result was shown.
MDC Staining for Detecting Autophagosome Activity
Twenty one-day-old seedlings of transgenic lines and SN9816 cultured with NS (3.5 mM) solution were transferred to NS (3.5 mM) or ND (0 mM) liquid medium for 24 h, then the roots were excised for detecting autophagosome activity by monodansylcadaverine (MDC) staining. MDC staining was conducted as described (Wang et al., 2013), confocal images were acquired using an inverted Zeiss LSM 780 laser scanning microscope.
Pot Trials of Rice
Rice plants were cultivated in pots under natural growth conditions from for three different years at Shenyang Agricultural University (Shenyang, China; Longitude: 123.34°E Latitude: 41.49°N), with each pot containing 14 kg of air-dried soil collected from an experimental farm at Shenyang Agricultural University. The growing season is from April to October every year. Field tests for rice using the T3 generation of 35S-OsATG8a homozygous transgenic rice in the SN9816 background were carried out under natural growth conditions. There were three N levels in this experiment, N225 (225 kg ⋅ ha–1, 2.79 g ⋅ pot–1), N75 (75 kg ⋅ ha–1, 0.93 g ⋅ pot–1), and N0 (without N fertilizer). Urea was used as the only nitrogen source for N225 and N75 treatments and was applied three times during the whole growth period. For N225 or N75 treatment, the rice plants were applied with 81 kg ⋅ ha–1 or 26 kg ⋅ ha–1 urea as basal fertilizer, then 54 kg ⋅ ha–1 or 18 kg ⋅ ha–1 as tillering fertilizer, finally 90 kg ⋅ ha–1 or 30 kg ⋅ ha–1 as panicle fertilizer. Superphosphate and potassium chloride were used as phosphate and potassium fertilizers for each N level, superphosphate was supplied as basal fertilizer, and potassium chloride was supplied as basal fertilizer and twice as panicle fertilizer. There were three separate plots each with 3 N levels, each transgenic line or SN9816 planted ten pots for each N level per plot as replicates, and four plants were grown in a pot.
GUS Histochemical Staining and Fluorometric Assay for GUS Activity
For temporal and spatial expression pattern analysis, tissues from the culm, leaf sheath, leaf blade, and developing seeds of ProOsATG8a-GUS transgenic rice cultivated in pots under N225 conditions, and roots of 14-day-old transgenic seedlings cultured with NS (3.5 mM) solution were sampled for histochemical detection of GUS expression. For the N stress, 14-day-old seedlings of ProOsATG8a-GUS transgenic rice cultured with NS (3.5 mM) solution were transferred to ND (0 mM) liquid medium for 24 h, then GUS histochemical staining was conducted as described previously (Xu et al., 2011), and the fluorimetric assay for GUS activity was performed as described previously (Jefferson, 1989).
Subcellular Localization Assay
The 35S-eGFP-OsATG8a fusion constructs were used to investigate the subcellular localization of OsATG8a. OsMCA1-RFP was used as a cell membrane marker (Kurusu et al., 2012). Rice protoplasts were isolated and transformed according to published methods (Zhang et al., 2011). The fluorescence images were captured via an inverted Zeiss LSM 780 laser scanning microscope.
Measurement of SPAD and Agronomic Traits Analysis
Leaves of transgenic lines and SN9816 at the tillering stage were used to measure the relative chlorophyll content determined by a soil plant analysis development (SPAD) value measured using a SPAD-502 Plus chlorophyll meter (Konica Minolta, Japan). The agronomic traits including plant height, tiller number, seed-setting rate, thousand grain weight, panicle number, and grain yield were analyzed at ripening stage according to methods described previously (Bin et al., 2015). Three random pots with four replicates were chosen to be measured and the agronomic traits were all measured on a single-plant basis.
Biomass, Total N Content, NUpE, and NUE Calculations
Rice plants were sampled at the ripening stage by separating them into culm, leaf sheath, leaf blade, and panicle at harvest, then all samples were kept in a dry oven at 80°C for 7 days, and dry masses were weighed as DWDr (dry weight of culm, leaf sheath, leaf blade, and panicle) and DWSEEDs (dry weight of total seeds). Then the dry mass of these tissue organs was ground to powder and total N concentrations (N%) were measured using an Elemental Analyzer (Elementar, Vario MAX C, Germany), as N% = mg N/100 mg DW. Biomass was calculated as DWDr + DWSEEDs. The harvest index (HI) was calculated as DWSEEDs/(DWSEEDs+DWDr) ratio and was used as an indicator of yield. The N harvest index (NHI) was calculated as (N%SEEDs × DWSEEDs)/(N%SEEDs × DWSEEDs + N%Dr × DWDr). The NUpE was calculated as (total aboveground N accumulation in N225 or N75 pot) – (total aboveground N accumulation in N0 pot)/N supply, and the NUE was calculated as (grain yieid in N225 or N75 pot) – (grain yield in N0 pot)/N supply.
Statistical Analysis
The experiments were repeated in three separate plots as three replicates, and the representative results were shown in the figures. The statistically significant differences of all data were analyzed based on Student’s t-test with the comparisons between two groups of data (transgenic lines vs. SN9816) at significance levels of *P < 0.05 and **P < 0.01.
Results
Subcellular Localization and Temporal and Spatial Expression Patterns of OsATG8a in Rice
ATG8 is a reliable marker for autophagosomes and autophagic activity when fused with a fluorescent protein in plants and animals (Ichimura et al., 2000; Yoshimoto et al., 2004; Contento et al., 2005; Avin-Wittenberg et al., 2012; Izumi et al., 2015). Subcellular localization analysis with rice protoplasts showed that the eGFP-OsATG8a fusion protein was predominantly localized to the cytoplasm, with autophagosome-like structures seen as fluorescence puncta in rice protoplasts (Figure 1A), while the colocalization assay with a cell membrane marker OsMCA1-RFP (Kurusu et al., 2012) showed that the OsATG8a was not localized to cell membrane (not co-localized with RFP). To examine the temporal and spatial expression patterns of OsATG8a, we performed histochemical analysis of the promoter-GUS fusion report systems in ProOsATG8a-GUS transgenic rice. Histochemical staining indicated that GUS activity could be detected in the root, leaf sheath, leaf blade, culm, and the developing seeds of rice (Figure 1B). When the ProOsATG8a-GUS transgenic rice seedlings were subjected to nitrogen deficiency conditions, GUS activity significantly increased (Supplementary Figures S1A,B). To confirm whether the expression of OsATG8a responded to N limitation, we further examined the transcript level of OsATG8a using real-time RT-PCR. When rice seedlings of SN9816 were placed under N stress for 1 or 3 days, the expression levels of OsATG8a in leaves and roots were significantly increased with an increasing degree of N stress compared with NS conditions (Supplementary Figures S1C,D). These results suggest that expression of OsATG8a was induced by N stress.
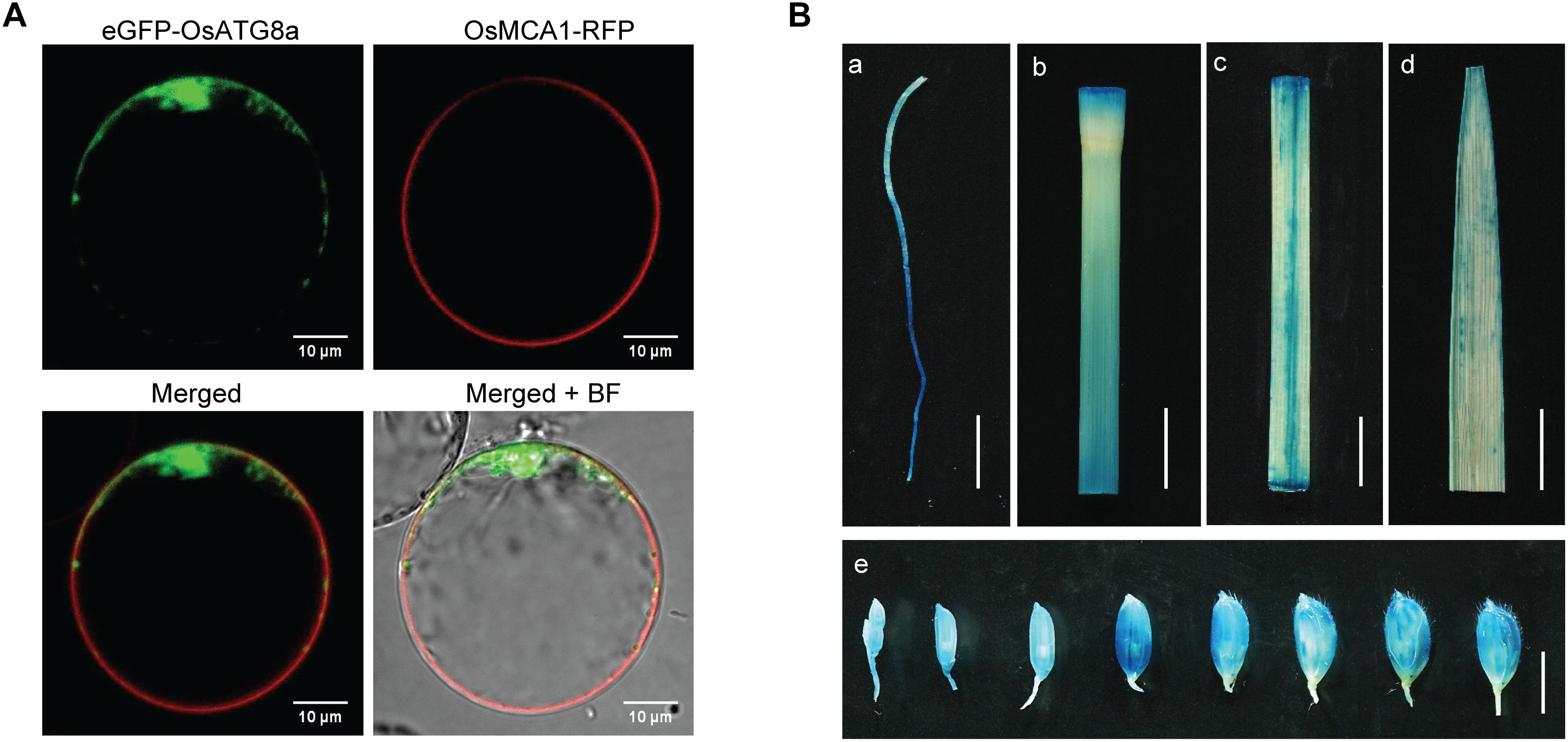
Figure 1. Protein subcellular localization of OsATG8a and expression patterns of OsATG8a gene. (A) Subcellular localization of OsATG8a in rice protoplasts. Panels showed the image of eGFP-OsATG8a, the cell membrane marker OsMCA1-RFP, merged of eGFP-OsATG8a and OsMCA1-RFP, and overlap image of mergerd and bright-field (BF), respectively. Scale bars, 10 μm. (B) GUS histochemical staining of ProOsATG8a-GUS transgenic rice, tissues used for GUS staining including root from 14-day-old seedlings cultured hydroponically (a), culm (b), leaf sheath (c), leaf blade (d), and developing seeds (e). Scale bars, 1 cm in (a–d), 5 mm in (e).
Overexpression of OsATG8a Increases the Number of Tillers and Reduces Plant Height in Transgenic Rice
To analyze the physiological function of OsATG8a in developmental processes in rice, we obtained more than 10 independent transgenic lines. The transcript level of OsATG8a was confirmed in two homozygous transgenic lines using real-time RT-PCR (Figure 2A). Lines 4 and 10 (OE-4, OE-10) are the over-expressed lines. Under both normal growth (N225) and low N (N75) conditions, the tiller number in the transgenic lines was significantly higher than that of the control plant (SN9816) from tillering to ripening stage and the transgenic lines were more robust and vigorous than the control (Figures 2B–D). In the ripening stage, transgenic plants were shorter than control plants (Figure 2E), especially under N75 conditions. Additionally, the relative chlorophyll content SPAD value in flag leaves of transgenic lines was higher than in the control (Figure 2F), which indicated that leaf senescence was slightly delayed in the OsATG8a-overexpressing rice. These results suggest that overexpression of OsATG8a plays a positive role in promoting rice tillering, delaying senescence, and prolonging efficient photosynthetic activities.
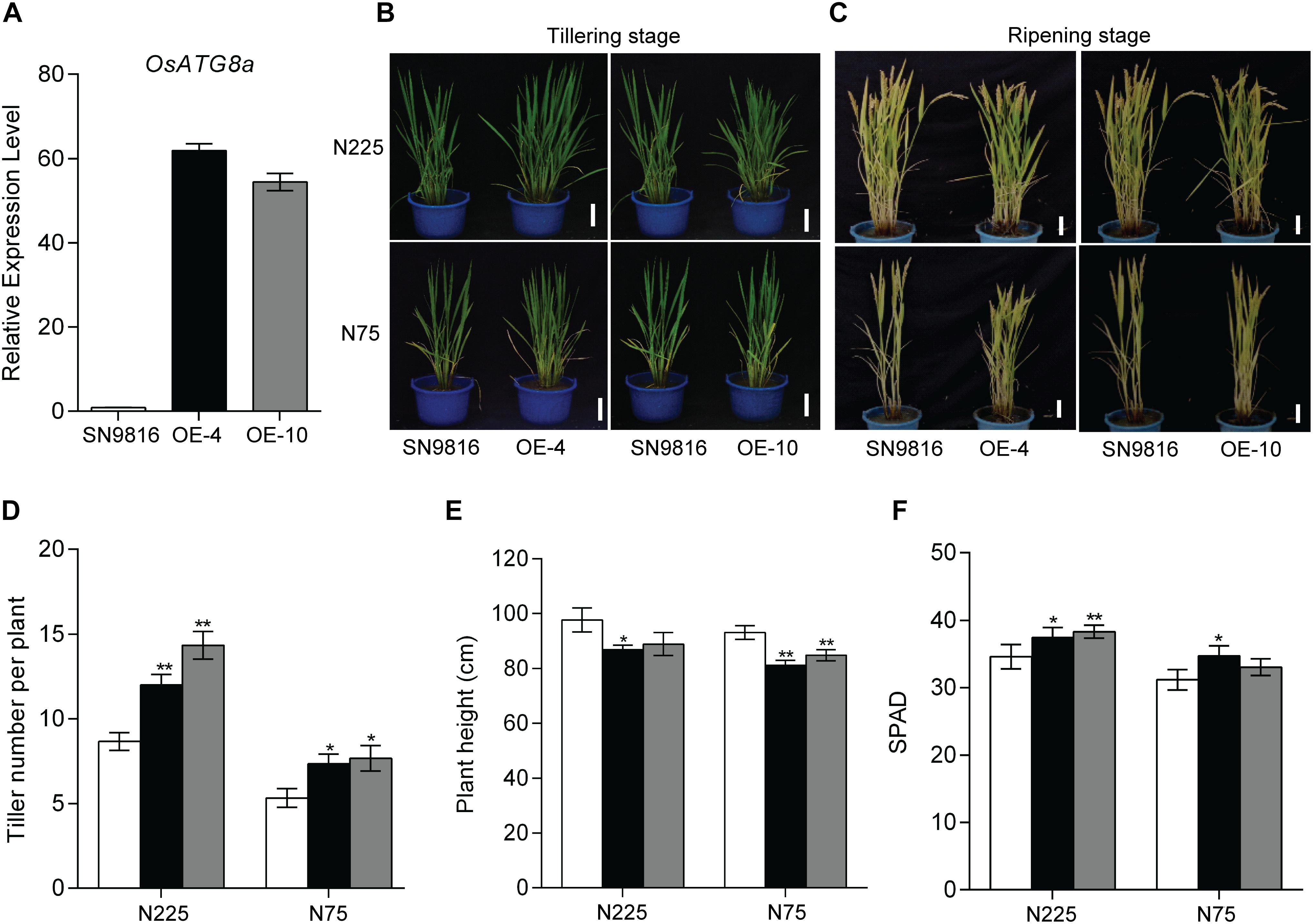
Figure 2. Characterization of OsATG8a-overexpressing transgenic rice. (A) Expression level of OsATG8a in 14-day-old seedlings of SN9816 and transgenic lines (OE-4 and OE-10). OsActin1 was used as an internal control. Values are means ± SD (n = 3). Phenotypes of the whole rice plants under sufficient N (N225) or low N (N75) conditions at tillering stage (B) and ripening stage (C). Scale bars, 10 cm. (D) Tiller number per plant, (E) plant height per plant and (F) SPAD value per plant of SN9816 and OsATG8a-overexpressing lines (OE-4 and OE-10) under N225 or N75 conditions at ripening stage. Values are means ± SD of one representative biological replicate (n = 12) out of three, *P < 0.05, **P < 0.01 (t-test).
Overexpression of OsATG8a Increases Panicle Number and Yield and Promotes NUE in Transgenic Rice
We further investigated the effect of OsATG8a on yield and found that the transgenic lines produced significantly more panicles and had increased yield per plant (Figures 3A,C). Although there was little difference in grain number per panicle compared to the control group, the thousand grain weight and the seed setting rate in transgenic lines were lower than those in the control plants (Figures 3B,D,E). However, the transgenic lines exhibited a significantly higher panicle number under the two N conditions (Figure 3F). These results were consistent with an increased total seed number and yield per plant compared to the control under N225, although the yield per plant was not significantly different under N75 (Figure 3G). We also measured the length and width of seeds, which showed no difference between the transgenic lines and SN9816 (Supplementary Figure S2). Given the increased yield in transgenic lines, we further analyzed plant biomass. The results showed that the biomass of transgenic lines increased by 22.29 or 10.62% under N225 or N75 conditions, respectively (Figure 4A). Harvest index (HI) and NHI are important indicators of seed production that also highlight the efficiency of N allocation to seeds. As shown in Figure 4B, the NHI was significantly higher in the transgenic lines compared to that in the control under N225 condition. The ratio of NHI to HI was used to indicate the variation of NUE as described previously (Guiboileau et al., 2012). Under both N225 and N75 conditions, the HI and the NHI/HI were not different between the transgenic lines and SN9816 (Figures 4C,D). We then analyzed the N content in seeds and dry remains, and compared to the control plant, the transgenic lines accumulated more N in seeds but had lower N in dry remains (Figures 4E,F). Since the N concentration in seeds as well as biomass were both significantly increased in transgenic lines, the NUpE and NUE were consequently increased especially under N225 condition (Figures 4G,H), suggesting that OsATG8a might be essential for both seed yield and NUE. Under N75 condition, the NUpE of transgenic lines was higher than that of control, but NUE was not. These results clarify that overexpression of OsATG8a effectively redeploys N allocation from the leaves to seeds and increases the NUE of rice under N optimal conditions.
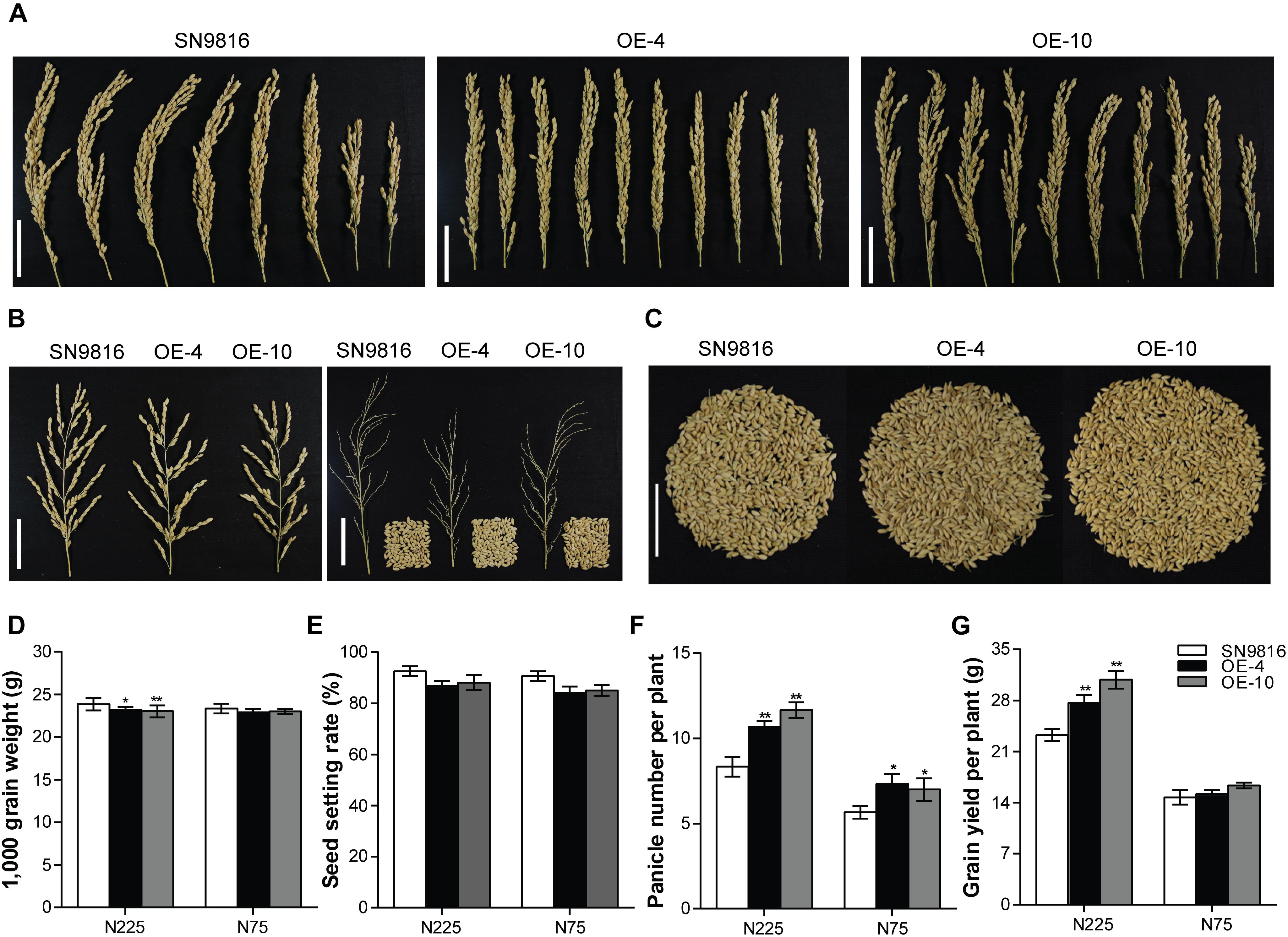
Figure 3. Comparison of yield-related characteristics between SN9816 and 35S-OsATG8a transgenic rice. Phenotypes of (A) panicle number per plant, (B) panicle and grain number per panicle, and (C) grain number per plant in SN9816 and transgenic lines (OE-4 and OE-10) under N225 condition. Scale bars, 5 cm. (D) Thousand grain weight, (E) seed setting rate, (F) panicle number per plant, and (G) grain number per plant of SN9816 and transgenic lines (OE-4 and OE-10) under N225 or N75 conditions. Values are means ± SD of one representative biological replicate (n = 12) out of three, *P < 0.05, **P < 0.01 (t-test).
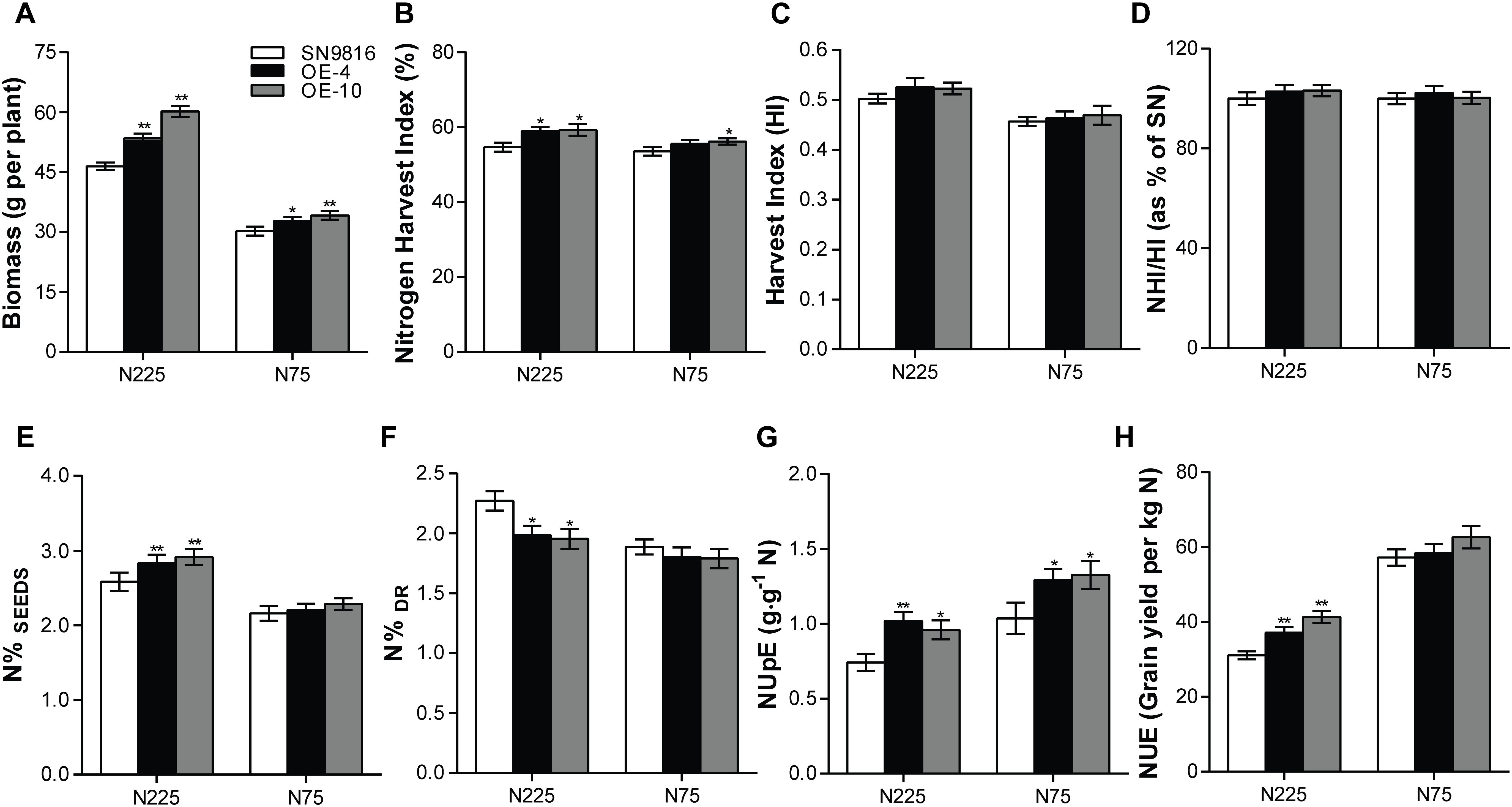
Figure 4. Evaluation of biomass, NUpE, and NUE in transgenic rice. (A) Biomass accumulation as measured by dry weight of the whole plant, (B) Nitrogen harvest index (NHI), (C) Harvest index (HI), (D) the ration of NHI/HI, (E) N concentration in seeds (N%SEEDs) and (F) in dry remains (N%Dr), (G) Nitrogen uptake efficiency (NUpE) and (H) nitrogen use efficiency (NUE) in SN9816 and transgenic lines (OE-4 and OE-10) under N225 or N75 conditions. Values are means ± SD of one representative biological replicate (n = 12) out of three, *P < 0.05, **P < 0.01 (t-test).
Overexpression of OsATG8a Enhances Autophagy Levels in the Transgenic Rice
The autophagy core protein ATG8a usually acts as the autophagic flux marker protein following previous reports (Klionsky and Schulman, 2014; Hafrén et al., 2017; Luo et al., 2017; Marshall and Vierstra, 2018). We first ascertained that the OsATG8a-overexpressing transgenic rice accumulated more ATG8a protein than control plants used ATG8a antibody of Arabidopsis under either NS (3.5 mM) or ND (0 mM) conditions (Figure 5A). Since the Neighbor of BRCA1 (NBR1) in Arabidopsis is an autophagy substrate degraded in the vacuole, the degradation level of NBR1 protein can be used to measure the selective autophagic flux in plants (Steingrim et al., 2011; Hafrén et al., 2017). Thus, we examined the autophagy receptor NBR1 by western blotting. NBR1 protein accumulation was reduced in transgenic lines compared to control plants, especially after N deficiency treatment, and thus autophagic flux increased (Figure 5A). To further analysis the autophagy flux, we monitored the lipidation of ATG8 by detected the ATG8 and lipidated ATG8 (ATG8-PE) adducts by immunoblot of the membrane protein. The ATG8-PE complex was an important component of autophagosome. The result showed that overexpression of OsATG8a enhanced the lipidation of ATG8, especially after N deficiency treatment, as the ATG8-PE as the level of ATG8-PE was higher in transgenic lines membranes (Figure 5A). Except that, we further used MDC staining to examine the number of autophagome in transgenic rice. MDC, an acidophilic dye widely used in mammals and plants can be used as a probe to detect autophagosome (Contento et al., 2005). There was observed more MDC-positive autophagic structures in the root cells of the transgenic lines, and the fluorescence signal intensity was higher than that in the control, especially after N deficiency treatment (Figure 5B). These results showed that overexpression of OsATG8a significantly increases autophagy activity and autophagic flux in transgenic rice.
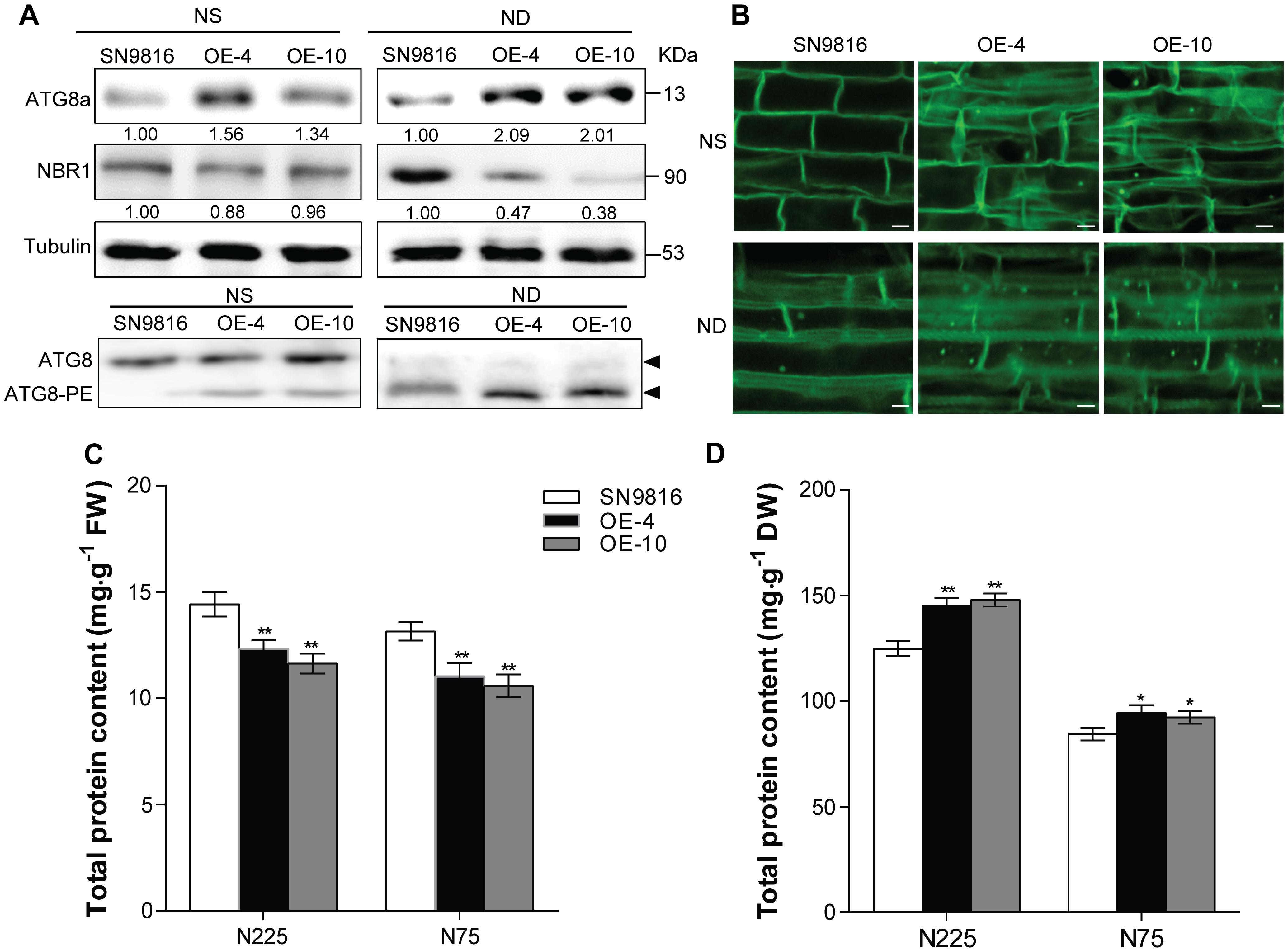
Figure 5. Overexpression of OsATG8a enhances the autophagy levels in rice. 14-day-old seedlings of SN9816 and transgenic lines (OE-4 and OE-10) cultured hydroponically in NS (3.5 mM) solution were transferred to NS (3.5 mM) or ND (0 mM) solution for 24 h (n = 10). (A) Immunoblot analysis of the extracts with anti-Arabidopsis ATG8a and NBR1 antibodies, near equal protein loads were confirmed by immunoblot analysis with an α-tubulin antibody. Numbers below the blot indicate intensity ratios between the NBR1 protein and the α-tubulin band in each line, and the intensity ratios in each line were normalized to SN9816 (value set to 1) under NS (3.5 mM) or ND (0 mM) conditions. The membrane fraction was used to detect the level of lipidated (ATG8-PE) and free ATG8 in the leaves. The ATG8-PE conjugates had a faster migration than ATG8 in the membrane fraction, so the band of ATG8-PE was below the band of ATG8 during SDS-PAGE in the presence of 6 M urea. (B) MDC-stained autophagosomes in roots were observed by confocal microscopy. Protein contents of leaves (at grain filling stage) (C) and seeds (after harvest) (D) extracts in SN9816 and transgenic lines (OE-4 and OE-10) under N225 and N75 conditions. The weight of each sample in (C) or (D) was the same, and the protein content was determined by BCA method as mg protein per gram fresh weight in leaves or mg protein per gram dry weight in seeds. Values are means ± SD (n = 3), *P < 0.05, **P < 0.01 (t-test).
Previous studies showed that autophagy is important for maintaining normal protein profiles in plants (Li F. et al., 2015; Marshall and Vierstra, 2018). To study whether overexpression of OsATG8a affected the protein profiles of leaves and seeds in rice, we compared the protein content in leaves or seeds of transgenic lines and control plants under N225 and N75 conditions. In leaf samples of the same quality, the protein content of leaves at the grain filling stage in transgenic lines was lower than in controls under both N conditions (Figure 5C and Supplementary Figure S3A). The protein content of seed was also higher. Corresponding to that was the protein content of seeds in transgenic lines was much higher than that of SN9816 (Figure 5D and Supplementary Figure S3B), which was consistent with the higher N content (N%SEEDs) in seeds of transgenic lines. These results were consistent with previous studies showing that leaf protein content in rice is affected by N stress (Wada et al., 2015), and the remobilization and reuse of N also increased due to the high autophagy level, so that most of the proteins in the leaves were transferred into the seeds, especially under N stress.
Discussion
Autophagy appears to express at a basal level in plants under normal conditions for maintaining cell homeostasis. In plant cells, especially when nutrients are scarce, autophagy regulates the recycling and reuse of nutrients, and provides nutrients for plants by degrading and clearing macromolecules and organelles that are damaged to improve cell vitality, regulate immune response, and in adaptation to nutrient starvation. Thus, autophagy would be more important under nutrient scarcity (Bassham et al., 2006; Lenz et al., 2011; Li and Vierstra, 2012; Guiboileau et al., 2013; Michaeli et al., 2016; Hafrén et al., 2017; Minina et al., 2018). Therefore, these advantages of autophagy can be used to adjust important agricultural traits, such as increase the tolerance to abiotic and biotic stress, modulate the immune response to pathogen infection, and mitigate the effects of depleted nutrients. Nutrition limitation induces autophagy in plants (Moriyasu and Ohsumi, 1996; Thompson et al., 2005). The transcription of many plant ATG genes is significantly up-regulated similar to high autophagy activity under nutrient-limited conditions (Thompson et al., 2005; Chung et al., 2009; Xia et al., 2011; Wang et al., 2013). In our study, the transcript level of rice OsATG8a was significantly increased under N deficiency (Supplementary Figures S1C,D). Furthermore, the promoter activity of OsATG8a was enhanced under N-starvation conditions in the ProOsATG8a-GUS transgenic rice (Supplementary Figures S1A,B).
We further demonstrated the role of OsATG8a in plant growth, yield, and nitrogen utilization. The 35S-OsATG8a transgenic lines performed better than control SN9816 under normal growth conditions. The increased SPAD in the leaves (Figure 2F) reflected delayed leaf senescence and high nitrogen content in the “source” in transgenic rice and suggests that these plants had higher accumulation capacity of photosynthetic products during the vegetative growth stage. This in turn would provide more nutrients, especially N, for reproductive growth and yield. In this study, overexpression of OsATG8a significantly promoted rice tillering under both normal N and N limited conditions (Figure 2D), consistent with increased yield, especially under normal N conditions (Figure 3G). In the OsATG8a-overexpressing transgenic rice lines, although the number of grains per panicle did not change, the number of panicles was significantly higher than the control, which increased the yield per plant (Figures 3A,F). The increased yield was due to the increased number of effective tillers and panicles in the 35S-OsATG8a transgenic rice. Meanwhile, the GUS activity of OsATG8a promoter was detected in the developing seeds (Figure 1Be), indicating that the OsATG8a gene may also participate in seed formation in rice. Interestingly, the OsATG8a-overexpressing rice was shorter (Figures 2C,E). In agronomy, shorter plant height combined with increased tiller percentage is beneficial for ideal plant type and increasing crop yield. It was shown that overexpressing the soybean GmATG8c gene, AtATG5 or AtATG7 gene in model plants Arabidopsis could increase seed yields and tolerance to abiotic and biotic stress (Xia et al., 2012; Minina et al., 2018). Consistent with these conclusions, it was also proved that OsATG8a plays a positive role in rice, meaning that it may be a feasible candidate gene for agricultural breeding improvement. In addition, using autophagy to change the nutrient cycle could produce more beneficial agronomic traits.
Autophagy plays an important role in nutrient cycling in all eukaryotes, and in plants it is especially important for remobilization and transfer of N from the leaf to the grain (Masclaux-Daubresse et al., 2017). Several atg deletion mutants showed phenotypes such as diminished growth and development, accelerated senescence, reduced yield, severe inhibition of growth phenotype during carbon or N deficiency, the process of N remobilization is hampered due to the lack of autophagy in leaves (Doelling et al., 2002; Thompson et al., 2005; Chung et al., 2010; Anongpat et al., 2011; Guiboileau et al., 2012, 2013; Li F. et al., 2015; Wada et al., 2015; Eguchi et al., 2017). With defective autophagy, the atg mutants had a defect of nitrogen remobilization, N in the cells cannot be effectively recovered or utilized, leading to the accumulation of undigested soluble proteins in their leaves. All of these could inhibit plant growth and reduce NUE of plants (Ren et al., 2014). Changing the autophagy process of crops could improve NUE and increase crop yield (Avin-Wittenberg et al., 2018). The high consumption of NBR1, increased lipidation of ATG8a (Figure 5A), and enhanced number of autophagosomes in transgenic rice especially after N deficiency treatment (Figure 5B) all suggested that the autophagy level was significantly stimulated in the OsATG8a-overexpressing transgenic rice. We hypothesized that the increased yield in these lines may be due to the enhanced autophagy, which promotes the remobilization of N from the leaves to seeds. Corresponding to this was that the protein content of leaves decreased while the protein content of seeds increased (Figures 5C,D), which also supports this conclusion. Compared with the control, the N concentration in dry remains was much lower in transgenic lines, but significantly higher in seeds under adequate N (Figures 4E,F), these results revealed that our transgenic rice made more efficient use of available N, transferring more N to seeds, and wasting less in non-seed areas. NHI, HI, NHI/HI and seed N concentration (N%SEEDs) are major indicators of NUE and seed nutritional quality (Masclaux-Daubresse et al., 2010). Overexpression of OsATG8a in transgenic rice resulted in higher NHI, N%SEEDs, and NUE (Figures 4B,E,H), indicating that OsATG8a plays a vital role in N allocation to seeds and NUE. Similarly, the NUpE of transgenic rice was higher under two N conditions. Under N deficiency conditions, although the NUpE was higher than the control plants in the overexpressed lines, the NUE and NHI/HI did not change, which explained why the grain yield per plant of the overexpressed lines was not significantly different from that of the control group (Figures 3G, 4D,H). At the same time, this may be the reason that N%SEEDs and NHI were not significantly different (Figures 4B,E).
Constitutive expression of some ATG8 genes in the transgenic plants not only led to better tolerance to N starvation, but also accelerated growth under optimal growth conditions (Slavikova et al., 2008; Xia et al., 2012; Li W. et al., 2015, Li et al., 2016; Kellner et al., 2016; Wang et al., 2016). Recent research from Masclaux-Daubresse’s group found that overexpression AtATG8a, AtATG8e, AtATG8f, or AtATG8g in Arabidopsis could not only significantly increase the number of autophagosomes, but also improved N%SEEDs and at the same time reduced N loss to dry biomass under full nitrate conditions. Additionally, the N remobilization efficiency was significantly increased in ATG8 overexpressors relative to the control line but the yield and biomass were not affected (Chen et al., 2018). Although we came to similar conclusions, in our hands, the OsATG8a gene powerfully affected the growth and development of rice, such that the overexpression of OsATG8a significantly increased effective tiller number under two N conditions, and also increased grain yield and biomass under normal growth conditions. Another point worth mentioning is that both NUE and NUpE were significantly improved in overexpressing OsATG8a transgenic rice than control plant.
Taken together, our results suggest that OsATG8a has a positive role in the maintenance of plant fitness. These results have also linked autophagy to plant nitrogen distribution/homeostasis, as well as increased yield in rice. We concluded that OsATG8a is an important candidate gene for promoting plant growth and development and increasing yield and NUE in rice.
Author Contributions
FX designed the study and provided experimental materials. JY performed the GUS staining, total RNA extraction, real time RT-PCR experiments, field trials of rice, phenotypic analysis, measurement of chlorophyll and SPAD, and agronomic traits analysis. XZ performed the binary vector construction, MDC staining, protein isolation, western blot analysis and the subcellular localization assay. XL performed the rice genetic transformation and the NUpE and NUE calculations. NL performed the measurement of biomass and total N content. JY and XZ analyzed the results and prepared the figures and tables. FX and XZ wrote the manuscript. All authors discussed the results and commented on the manuscript, read and approved the final manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program of China (Grant No. 2018YFD0300300).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00584/full#supplementary-material
References
Anongpat, S., Faqiang, L., Taijoon, C., and Vierstra, R. D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23, 3761–3779. doi: 10.1105/tpc.111.090993
Avin-Wittenberg, T., Baluška, F., Bozhkov, P. V., Elander, P. H., Fernie, A. R., Galili, G., et al. (2018). Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J. Exp. Bot. 69, 1335–1353. doi: 10.1093/jxb/ery069
Avin-Wittenberg, T., Honig, A., and Galili, G. (2012). Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 249, 285–299. doi: 10.1007/s00709-011-0296-z
Bassham, D. C., Laporte, M., Marty, F., Moriyasu, Y., Ohsumi, Y., Olsen, L. J., et al. (2006). Autophagy in development and stress responses of plants. Autophagy 2, 2–11. doi: 10.4161/auto.2092
Bin, H., Wei, W., Shujun, O., Jiuyou, T., Hua, L., Ronghui, C., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Chardon, F., Noël, V., and Masclaux-Daubresse, C. (2012). Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. J. Exp. Bot. 63:3401. doi: 10.1093/jxb/err353
Chen, Q., Soulay, F., Saudemont, B., Elmayan, T., Marmagne, A., and Masclaux-Daubresse, C. (2018). Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 60, 343–352. doi: 10.1093/pcp/pcy214
Chung, T., Phillips, A. R., and Vierstra, R. D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62, 483–493. doi: 10.1111/j.1365-313X.2010.04166.x
Chung, T., Suttangkakul, A., and Vierstra, R. D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149, 220–234. doi: 10.1104/pp.108.126714
Contento, A. L., Xiong, Y., and Bassham, D. C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42, 598–608. doi: 10.1111/j.1365-313x.2005.02396.x
Di, B. J., Marmagne, A., Berger, A., Yoshimoto, K., Cueff, G., Chardon, F., et al. (2018). Autophagy controls resource allocations and protein storage accumulation in Arabidopsis seeds. J. Exp. Bot. 69, 1403–1414. doi: 10.1093/jxb/ery012
Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R., and Vierstra, R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277:33105. doi: 10.1074/jbc.m204630200
Eguchi, M., Kimura, K., Makino, A., and Ishida, H. (2017). Autophagy is induced under Zn limitation and contributes to Zn-limited stress tolerance in Arabidopsis (Arabidopsis thaliana). Soil Sci. Plant Nutr. 63, 1–9.
Guiboileau, A., Avila-Ospina, L., Yoshimoto, K., Soulay, F., Azzopardi, M., Marmagne, A., et al. (2013). Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 199, 683–694. doi: 10.1111/nph.12307
Guiboileau, A., Yoshimoto, K., Soulay, F., Bataillé, M. P., Avice, J. C., and Masclaux-Daubresse, C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194, 732–740. doi: 10.1111/j.1469-8137.2012.04084.x
Hafrén, A., Macia, J.-L., Love, A. J., Milner, J. J., Drucker, M., and Hofius, D. (2017). Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. 114, E2026–E2035. doi: 10.1073/pnas.1610687114
Havé, M., Marmagne, A., Chardon, F., and Masclaux-Daubresse, C. (2017). Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. J. Exp. Bot. 68, 2513–2529. doi: 10.1093/jxb/erw365
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313x.1994.6020271.x
Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492. doi: 10.1038/35044114
Ishida, H., Yoshimoto, K., Izumi, M., Reisen, D., Yano, Y., Makino, A., et al. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148, 142–155. doi: 10.1104/pp.108.122770
Izumi, M., Hidema, J., Wada, S., Kondo, E., Kurusu, T., Kuchitsu, K., et al. (2015). Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol. 167, 1307–1320. doi: 10.1104/pp.114.254078
Kellner, R., Jc, D. L. C., Maqbool, A., Kamoun, S., and Dagdas, Y. F. (2016). ATG8 expansion: a driver of selective autophagy diversification? Trends Plant Sci. 22:204. doi: 10.1016/j.tplants.2016.11.015
Kichey, T., Hirel, B., Heumez, E., Dubois, F., and Gouis, J. L. (2007). In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crops Res. 102, 22–32. doi: 10.1016/j.fcr.2007.01.002
Klionsky, D. J., and Schulman, B. A. (2014). Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 21:336. doi: 10.1038/nsmb.2787
Kraiser, T., Gras, D. E., Gutierrez, A. G., Gonzalez, B., and Gutierrez, R. A. (2011). A holistic view of nitrogen acquisition in plants. J. Exp. Bot. 62, 1455–1466. doi: 10.1093/jxb/erq425
Kurusu, T., Koyano, T., Kitahata, N., Kojima, M., Hanamata, S., Sakakibara, H., et al. (2017). Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant Signal. Behav. 12:e1365211. doi: 10.1080/15592324.2017.1365211
Kurusu, T., Nishikawa, D., Yamazaki, Y., Gotoh, M., Nakano, M., Hamada, H., et al. (2012). Plasma membrane protein OsMCA1 is involved in regulation of hypo-osmotic shock-induced Ca2+ influx and modulates generation of reactive oxygen species in cultured rice cells. BMC Plant Biol. 12:11. doi: 10.1186/1471-2229-12-11
Lenz, H. D., Eva, H., Eric, M., Karina, K., Karl, W., Mark, S., et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66, 818–830. doi: 10.1111/j.1365-313X.2011.04546.x
Li, F., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389–1408. doi: 10.1105/tpc.15.00158
Li, W., Chen, M., Zhong, L., Liu, J. M., Xu, Z. S., Li, L. C., et al. (2015). Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 468, 800–806. doi: 10.1016/j.bbrc.2015.11.035
Li, F., and Vierstra, R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17, 526–537. doi: 10.1016/j.tplants.2012.05.006
Li, W., Chen, M., Wang, E., Hu, L., Hawkesford, M. J., Zhong, L., et al. (2016). Genome-wide analysis of autophagy-associated genes in foxtail millet (Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice. BMC Genomics 17:797. doi: 10.1186/s12864-016-3113-4
Liu, Y., and Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. doi: 10.1146/annurev-arplant-042811-105441
Luo, L., Zhang, P., Zhu, R., Fu, J., Su, J., Zheng, J., et al. (2017). Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front. Plant Sci. 8:1459. doi: 10.3389/fpls.2017.01459
Marshall, R. S., and Vierstra, R. D. (2018). Autophagy: the master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208. doi: 10.1146/annurev-arplant-042817-040606
Masclaux-Daubresse, C., Chen, Q., and Havé, M. (2017). Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol. 39, 8–17. doi: 10.1016/j.pbi.2017.05.001
Masclaux-Daubresse, C., Danielvedele, F., Dechorgnat, J., Chardon, F., Gaufichon, L., and Suzuki, A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. doi: 10.1093/aob/mcq028
Michaeli, S., Galili, G., Genschik, P., Fernie, A. R., and Avin-Wittenberg, T. (2016). Autophagy in plants – what’s new on the menu? Trends Plant Sci. 21, 134–144. doi: 10.1016/j.tplants.2015.10.008
Miki, T., and Yoshinori, O. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174. doi: 10.1016/0014-5793(93)80398-e
Minina, E. A., Moschou, P. N., Vetukuri, R. R., Sanchez-Vera, V., Cardoso, C., Liu, Q., et al. (2018). Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 69, 1415–1432. doi: 10.1093/jxb/ery010
Moriyasu, Y., and Ohsumi, Y. (1996). Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 111, 1233–1241. doi: 10.1104/pp.111.4.1233
Nakatogawa, H., Ichimura, Y., and Ohsumi, Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178. doi: 10.1016/j.cell.2007.05.021
Otegui, M. S., Noh, Y. S., Martínez, D. E., Vila Petroff, M. G., Staehelin, L. A., Amasino, R. M., et al. (2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41, 831–844. doi: 10.1111/j.1365-313x.2005.02346.x
Ren, C., Liu, J., and Gong, Q. (2014). Functions of autophagy in plant carbon and nitrogen metabolism. Front. Plant Sci. 5:301. doi: 10.3389/fpls.2014.00301
Roberts, I. N., Caputo, C., Criado, M. V., and Funk, C. (2012). Senescence-associated proteases in plants. Physiol. Plant. 145, 130–139. doi: 10.1111/j.1399-3054.2012.01574.x
Slavikova, S., Ufaz, S., Avinwittenberg, T., Levanony, H., and Galili, G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59:4029. doi: 10.1093/jxb/ern244
Steingrim, S., Trond, L., Kirsten, K., and Terje, J. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993–1010. doi: 10.4161/auto.7.9.16389
Thompson, A. R., Doelling, J. H., Suttangkakul, A., and Vierstra, R. D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110. doi: 10.1104/pp.105.060673
Wada, S., Hayashida, Y., Izumi, M., Kurusu, T., Hanamata, S., Kanno, K., et al. (2015). Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 168, 60–73. doi: 10.1104/pp.15.00242
Wada, S., Ishida, H., Izumi, M., Yoshimoto, K., Ohsumi, Y., Mae, T., et al. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149, 885–893. doi: 10.1104/pp.108.130013
Wang, P., Sun, X., Jia, X., Wang, N., Gong, X., and Ma, F. (2016). Characterization of an autophagy-related gene MdATG8i from apple. Front. Plant Sci. 7:720. doi: 10.3389/fpls.2016.00720
Wang, Y., Yu, B., Zhao, J., Guo, J., Li, Y., Han, S., et al. (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25, 1383–1399. doi: 10.1105/tpc.112.108993
Wang, Y., Zheng, X., Yu, B., Han, S., Guo, J., Tang, H., et al. (2015). Disruption of microtubules in plants suppresses macroautophagy and triggers starch excess-associated chloroplast autophagy. Autophagy 11, 2259–2274. doi: 10.1080/15548627.2015.1113365
Xia, K., Liu, T., Ouyang, J., Wang, R., Fan, T., and Zhang, M. (2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 18, 363–377. doi: 10.1093/dnares/dsr024
Xia, T., Xiao, D., Liu, D., Chai, W., Gong, Q., and Wang, N. N. (2012). Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS One 7:e37217. doi: 10.1371/journal.pone.0037217
Xie, Z., Nair, U., and Klionsky, D. J. (2008). An in cytes from MBC selection: Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19:3290. doi: 10.1091/mbc.e07-12-1292
Xu, F., Meng, T., Li, P., Yu, Y., Cui, Y., Wang, Y., et al. (2011). A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol. 157, 2131–2153. doi: 10.1104/pp.111.182899
Yang, X., and Bassham, D. C. (2015). New insight into the mechanism and function of autophagy in plant cells. Int. Rev. Cell Mol. Biol. 320, 1–40. doi: 10.1016/bs.ircmb.2015.07.005
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. doi: 10.1105/tpc.104.025395
Zhai, Y., Guo, M., Wang, H., Lu, J., Liu, J., Zhang, C., et al. (2016). Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum L. Front. Plant Sci. 7:131. doi: 10.3389/fpls.2016.00131
Keywords: autophagy, OsATG8a, nitrogen use efficiency, yield, rice
Citation: Yu J, Zhen X, Li X, Li N and Xu F (2019) Increased Autophagy of Rice Can Increase Yield and Nitrogen Use Efficiency (NUE). Front. Plant Sci. 10:584. doi: 10.3389/fpls.2019.00584
Received: 18 January 2019; Accepted: 18 April 2019;
Published: 07 May 2019.
Edited by:
Goetz Hensel, Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK), GermanyReviewed by:
Celine Masclaux-Daubresse, INRA Centre Versailles-Grignon, FranceRosemary White, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia
Copyright © 2019 Yu, Zhen, Li, Li and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fan Xu, Y2VsaWFmYW5mYW5AMTYzLmNvbQ==
†These authors have contributed equally to this work
 Jinlei Yu1†
Jinlei Yu1† Xiaoxi Zhen
Xiaoxi Zhen Fan Xu
Fan Xu