- State Key Laboratory for Hybrid Rice, College of Life Sciences, Wuhan University, Wuhan, China
Glycoside hydrolase 3 (GH3) gene family belongs to auxin-responsive gene families and is tightly linked with hormone homeostasis and signaling pathways. However, our knowledge about the evolutionary dynamic of GH3 genes in Gramineae crops is limited. In this study, a comparative genomic and transcriptomic analysis was conducted to study evolutionary patterns and the driving selective forces of GH3 gene family in six representative Gramineae crops, namely, Setaria italica (Si), Zea mays (Zm), Sorghum bicolor (Sb), Hordeum vulgare (Hv), Brachypodium distachyon (Bd), and Oryza sativa ssp. japonica (Os). A total of 17, 13, 11, 9, 8, and 11 GH3 proteins (GH3s) were identified in Si, Zm, Sb, Hv, Bd, and Os, respectively. Phylogenetic, conserved motif, and gene structural analyses could divide all GH3s into two groups (I and II), and all GH3s consisted of seven orthogroups (Ors) on the basis of Or identification result. We further found that genes in the same Or showed similar sequence and structural features, whereas genes in the same groups exhibited intrinsic differences in exon numbers and intron lengths. These results revealed GH3 genes in the same groups have been differentiated. Obvious differences in total numbers of GH3 genes, Ors, and duplication events among these six tested Gramineae crops reflected lineage-specific expansions and homologous gene loss/gain of GH3 gene family during the evolutionary process. In addition, selective force and expression analyses indicated that all GH3 genes were constrained by strong purifying selection, and GH3 genes in conserved Ors showed higher expression levels than that in unconserved Ors. The current study highlighted different evolutionary patterns of GH3 genes in Gramineae crops resulted from different evolutionary rates and duplication events and provided a vital insight into the functional divergence of GH3 genes.
Introduction
Auxin, one of the most important hormones in plants, plays essential roles in embryogenesis, vascular differentiation, phototropism, and plant morphology (Jain et al., 2006; Terol et al., 2006). Glycoside hydrolase 3 (GH3) gene family belongs to auxin-responsive gene families and directly influences the homeostasis of different plant hormones (Vielba, 2018; Kong et al., 2019a). Previous studies demonstrated that GH3 proteins can catalyze various reactions using indole-3-acetic acid (IAA), jasmonic acid (JA), benzoates, and salicylic acid (SA) as substrates (Staswick et al., 2002; Okrent and Wildermuth, 2011; Kong et al., 2019a). In plants, the number of GH3 protein members varies greatly, namely, 19 in Arabidopsis thaliana (Okrent et al., 2009), 13 in Oryza sativa ssp. japonica (Jain et al., 2006; Terol et al., 2006; Fu et al., 2011; Kong et al., 2019a), 13 in Zea mays (Feng et al., 2015), 15 in Solanum lycopersicum (Kumar et al., 2012), two in Physcomitrella patens (Zhang et al., 2018), and 18 in Selaginella moellendorffii (Zhang et al., 2018).
At present, GH3 proteins have been divided into three groups based on evolutionary analysis, sequence similarity, and substrate specificities: group I with JA and/or SA–amido synthetase, group II with IAA–amido synthetase activity, and group III with unknown synthetase activity (Staswick et al., 2002; Okrent et al., 2009). Plant GH3 proteins played important roles in signaling pathways, organ developments, and plant architecture (Fu et al., 2011; Singh et al., 2015; Cano et al., 2018). Among them, OsGH3-1, OsGH3-2, OsGH3-8, and OsGH3-13 are associated with crosstalk modulation between the IAA, JA, and SA signaling pathways under biotic or/and abiotic stresses (Ding et al., 2008; Domingo et al., 2009; Zhang et al., 2009). OsGH3-2 participates in drought and cold tolerances via regulating the levels of auxin and abscisic acid (ABA) (Ding et al., 2008; Du et al., 2012). OsGH3-8 was reportedly associated with rice floret fertility and disease resistance (Ding et al., 2008; Jian et al., 2010; Yadav et al., 2011). Another study reported that “auxin-miR167-ARF8-OsGH3-2” signaling pathway responses to exogenous auxin (Yang et al., 2006). Additionally, “miR156f-OsSPL7-OsGH3-8” signaling pathway modulates the plant architecture (Dai et al., 2018).
As we all know, Gramineae is a large and nearly ubiquitous family of monocotyledonous flowering plants known as grasses and contains many important cereal crops with high economic value, well-characterized phylogeny, and numerous genetic resources for genetic engineering. Gramineae therefore is an exceptional model system to study short-term evolutionary dynamics of gene families in the plants. In this study, six important Gramineae crops (Brachypodium distachyon, Hordeum vulgare, Setaria italica, Sorghum bicolor, Z. mays, and O. sativa ssp. japonica) were selected to study expansion and evolutionary patterns of GH3 genes at the genome-wide scale and to systematically analyze GH3 genes’ phylogenetic relationship, chromosomal location, duplication events, orthogroups (Ors), selective force, gene structure, protein motifs, cis-elements in promoters, and expression profiles. Our results could provide a valuable foundation for further investigation of gene expansion, evolutionary patterns, and functional differentiation in the Gramineae GH3 gene family.
Materials and Methods
Identification and Phylogenetic Analysis of GH3 Genes
Genome datasets of B. distachyon (v3.0), H. vulgare (IBSC_v2), S. italica (v2.0), S. bicolor (NCBIv3), Z. mays (B73_RefGen_v4), and O. sativa ssp. japonica (MSU 7.0) were downloaded from Ensembl Plants (http://plants.ensembl.org/index.html) and TIGR Database (http://rice.plantbiology.msu.edu), respectively. The HMM (Hidden Markox Model) profile of the GH3 auxin-responsive promoter (PF03321) was obtained from Pfam (http://pfam.xfam.org/). All GH3 proteins were separately searched by HMMER 3.2.1 (with default parameters) and BLASTP (E-value of e−5) methods (Kong et al., 2019a). Subsequently, all candidate sequences were subjected to SMART (http://smart.embl-heidelberg.de/) and Pfam (http://pfam.xfam.org/search/sequence) for key conserved domain checks (Singh et al., 2015; Deng et al., 2019), and the candidate GH3 protein sequences without the GH3 auxin-responsive promoter (PF03321) were filtered out (Kong et al., 2018).
All identified GH3 proteins were aligned by ClustalW, and a phylogeny tree was generated in MEGA 6.0 using the neighbor-joining method with 1,000 bootstrap replicates (Zhang et al., 2018). Names of putative GH3 genes were assigned principally based on chromosomal order in each genome in accordance with previous rice GH3 gene study (Jain et al., 2006; Terol et al., 2006; Kong et al., 2019a).
Chromosomal Locations, Gene Structure, and Conserved Motifs
The data about chromosomal locations and gene structures of GH3 genes in all tested species were obtained from GFF3 files and shown by TBtools v0.665 (Chen et al., 2018). Conserved motifs of all GH3 proteins were investigated by MEME Suite 5.0.2 (http://meme-suite.org/tools/meme) with the maximum number of motif sets at 20, the optional width of motifs from 6 to 100 amino acids, and other default parameters (Zhang et al., 2018; Kong et al., 2019b). Next, all identified motifs were annotated by Pfam and InterProScan (http://www.ebi.ac.uk/interpro/search/sequence-search) databases (Mitchell et al., 2018).
Gene Duplication Events, Orthogroups Identification, and Selective Forces
Gene duplication events of the GH3 gene family were analyzed and classified by the “duplicate_gene_classifier” script in MCScanX with an E-value of 1e−5 in BlastP search (Kong et al., 2019b). In general, whole genome duplication (WGD) or segmental duplication gene pairs were defined when the pairs of genes within two segmental regions have collinearity. When two duplicate genes were consecutive, we considered the gene pairs as tandem duplications. When two duplicate genes were separated by a maximum of 20 gene loci, we considered the gene pairs as proximal duplications. Within two duplicate genes from one duplication event, the synonymous (Ks)/nonsynonymous (Ka) substitution (Ka/Ks) rate was analyzed by DnaSP 5.0 (http://www.ub.edu/dnasp/) (Librado and Rozas, 2009), and divergence time was estimated by T = Ks/(2 × 9.1 × 10−9) × 10−6 Mya program (Deng et al., 2019; Kong et al., 2019a).
In this study, OrthoFinder software was used to identify GH3 Ors (Emms and Kelly, 2015). An all-vs-all BlastP search was conducted by diamond software (Buchfink et al. 2015) (https://ab.inf.uni-tuebingen.de/software/) with the following parameters: E-value of 1e−3 as the input file for OrthoFinder software, and Ors then were identified according to the published methods (Emms and Kelly, 2015). Then, Tajima’s D values of all Ors were calculated by DnaSP 5.0 for selective forces analysis among Ors (Librado and Rozas, 2009).
cis-Elements and Expression Analyses of GH3 Genes
Two-Kbp upstream promoter sequences from the start codon (ATG) of all GH3 genes in these six tested species were submitted to the PLANTCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) for the analysis of cis-acting regulatory elements (cis-elements) (Lescot et al., 2002; Liu et al., 2016).
Raw datasets of rice seven essential tissues (DRX000661, DRX000663, DRX000665, DRX000667, DRX000669, DRX000671, and DRX000673) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) and analyzed according to previously published article (Kong et al., 2019a). The fragment per kilobase of exon per million fragments mapped (FPKM) method was adopted to calculate gene expression levels (Kong et al., 2018). In addition, expression date of maize 79 tissues at different development stages was obtained from maize eFP Browser (Hoopes et al., 2018) (data source: Hoopes et al. Atlas, http://bar.utoronto.ca/efp_maize/cgi-bin/efpWeb.cgi). In order to detect the response of GH3 genes to biotic and abiotic stresses, the expression data of maize GH3 genes under salt, cold, heat, and Colletotrichum graminicola was downloaded from maize eFP Browser (Hoopes et al., 2018) (Data Source: Hoopes et al. Stress).
Results
Identification and Classification of GH3 Genes in Gramineae Crops
We investigated a total of 69 nonredundant GH3 genes from the six studied species, with 17 in S. italica, 13 in Z. mays, 11 in S. bicolor, 9 in H. vulgare, 8 in B. distachyon, and 11 in O. sativa ssp. japonica (Table S1). S. italica had more GH3 genes than other tested species, namely, H. vulgare and B. distachyon (Figure 1, Table S1). To reveal the number difference of GH3 genes in these species, group classification and Or identification were conducted. Results showed that all GH3 genes grouped into two groups (I and II) consisted of seven Ors (Figures 1 and 2, Table S2). Further studies indicated that the number of Ors varies greatly among these six species. Or1 and Or2 were existed in all tested species, while Or5, Or3, Or4, Or6, and Or7 were lineage-specific, whereas Or5 was only in O. sativa ssp. japonica, and Or6 and Or7 were only in S. italica (Figure 1). The Or1 and Or2 have more genes than other Ors, and these two Ors existed in all tested crops. We thus defined these two Ors as conserved Ors, whereas others as unconserved Ors. What’s more, we further noticed that gene numbers of these two Ors were different between different crops. For example, Or1 showed four to six genes in S. italica, Z. mays, and S. bicolor, whereas only two to three genes in H. vulgare, B. distachyon, and O. sativa ssp. japonica. These results indicated that the GH3 gene family has lineage-specific expansions and homologous gene loss/gain in Gramineae crops during the evolutionary process.
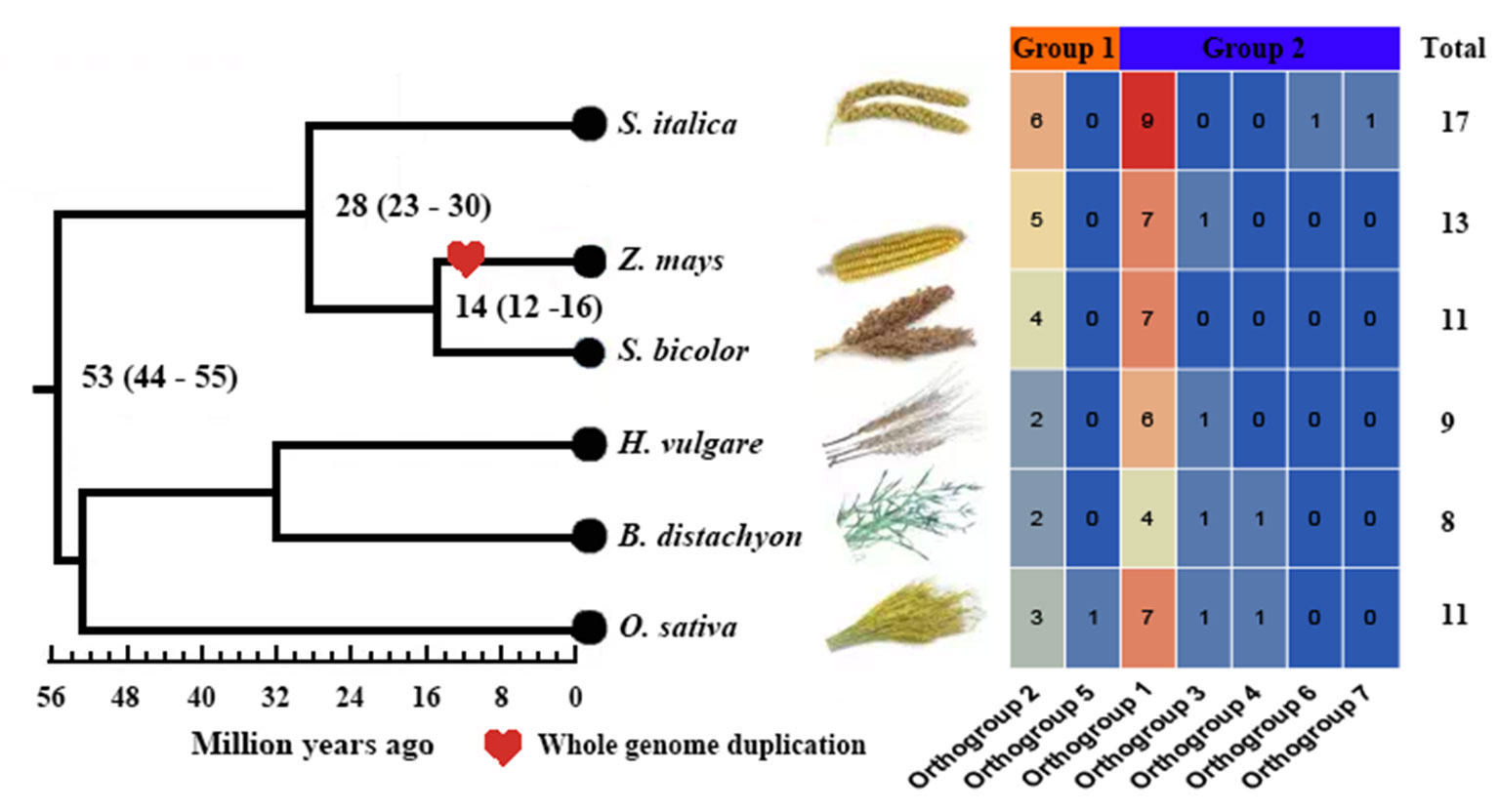
Figure 1 Comparison of the number of GH3 genes in six Gramineae crops. Groups I and II are displayed in different colored boxes. The numbers of orthogroups are shown by heat map: blue means low, and red means high.
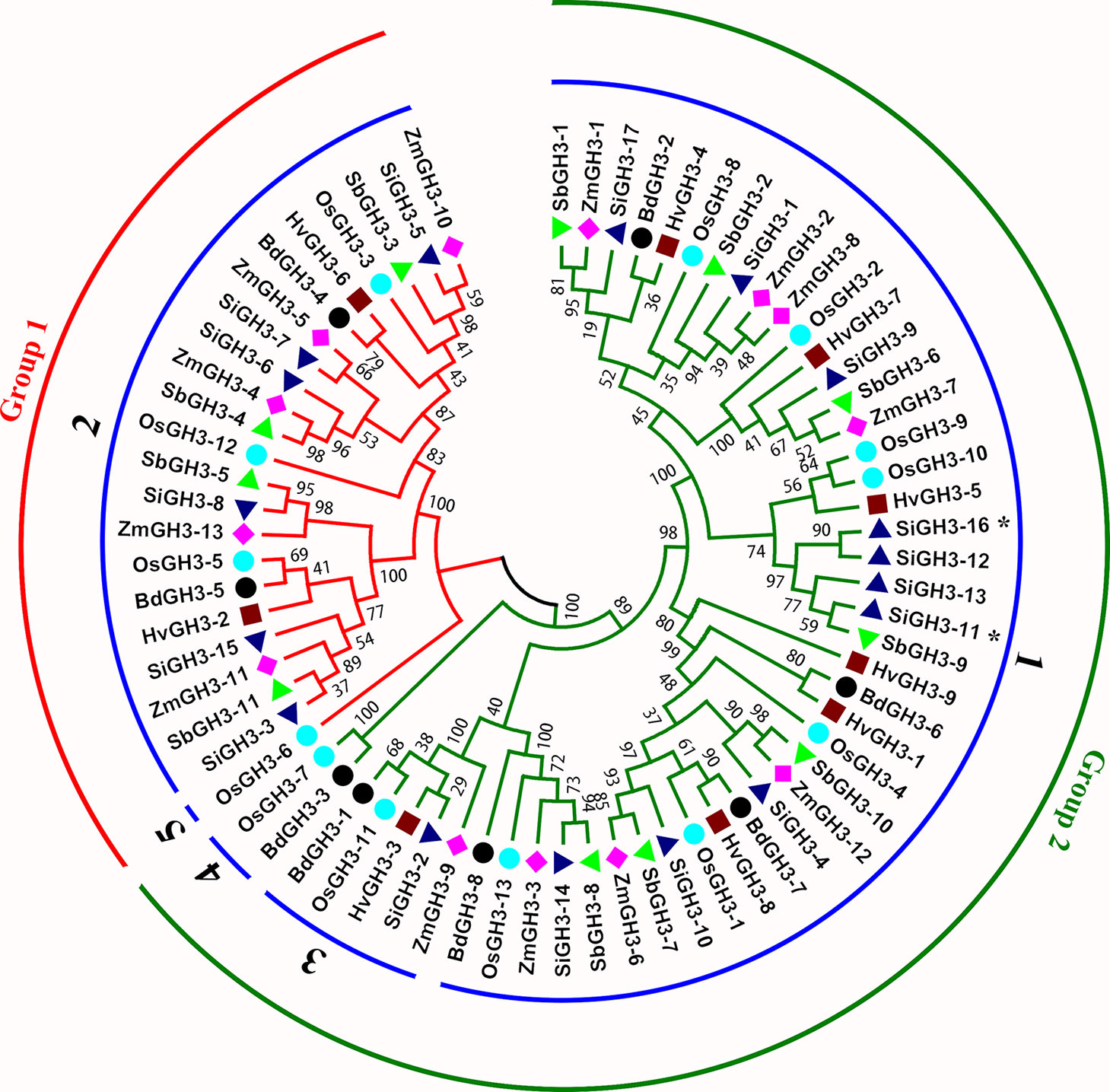
Figure 2 A phylogeny tree of GH3 protein sequences from H. vulgare, B. distachyon, O. sativa ssp. japonica, S. italica, S. bicolor, and Z. mays. Different colors of circles represent different groups. The numbers on blue circles represent different orthogroups, such as 1 means Orthogroup1. The different species are displayed by different shaped markers. SiGH3-11 * and SiGH3-16 belonged to Orthogroup6 and Orthogroup7, respectively.
Chromosomal Locations and Gene Duplication Events
To understand the expansion mechanism of paralogues, we investigated gene locations and duplication modes within each species. Of all tested species, GH3 genes were unevenly distributed on all chromosomes (Chrs) (Figure 3). For example, there were three, four, and one GH3 gene in Chr1, Chr2, and Chr4, respectively, while no GH3 gene in Chr3 and Chr5 (Figure 3A). Interestingly, barley GH3 genes are localized at the proximal regions of the chromosomes, which are the dynamic parts of the chromosomes and very prone to segmental or tandem duplications (Ablazov and Tombuloglu, 2016; Bostancioglu et al., 2018; Tombuloglu, 2018; Tombuloglu et al., 2019). Additionally, WGD or segmental duplication events were found in B. distachyon, S. bicolor, S. italica, Z. mays, and O. sativa ssp. japonica, while tandem duplication, proximal, and dispersed duplication events were not found among these crops (Figures 3A–F). This result suggested WGD or segmental duplication events were the main gene expansion mode of GH3 gene family in Gramineae crops. Interestingly, WGD or segmental duplication events existed only in Or1 (seven pairs) and Or2 (two pairs) (Figure 3), indicating that gene expansions had Or specificity.
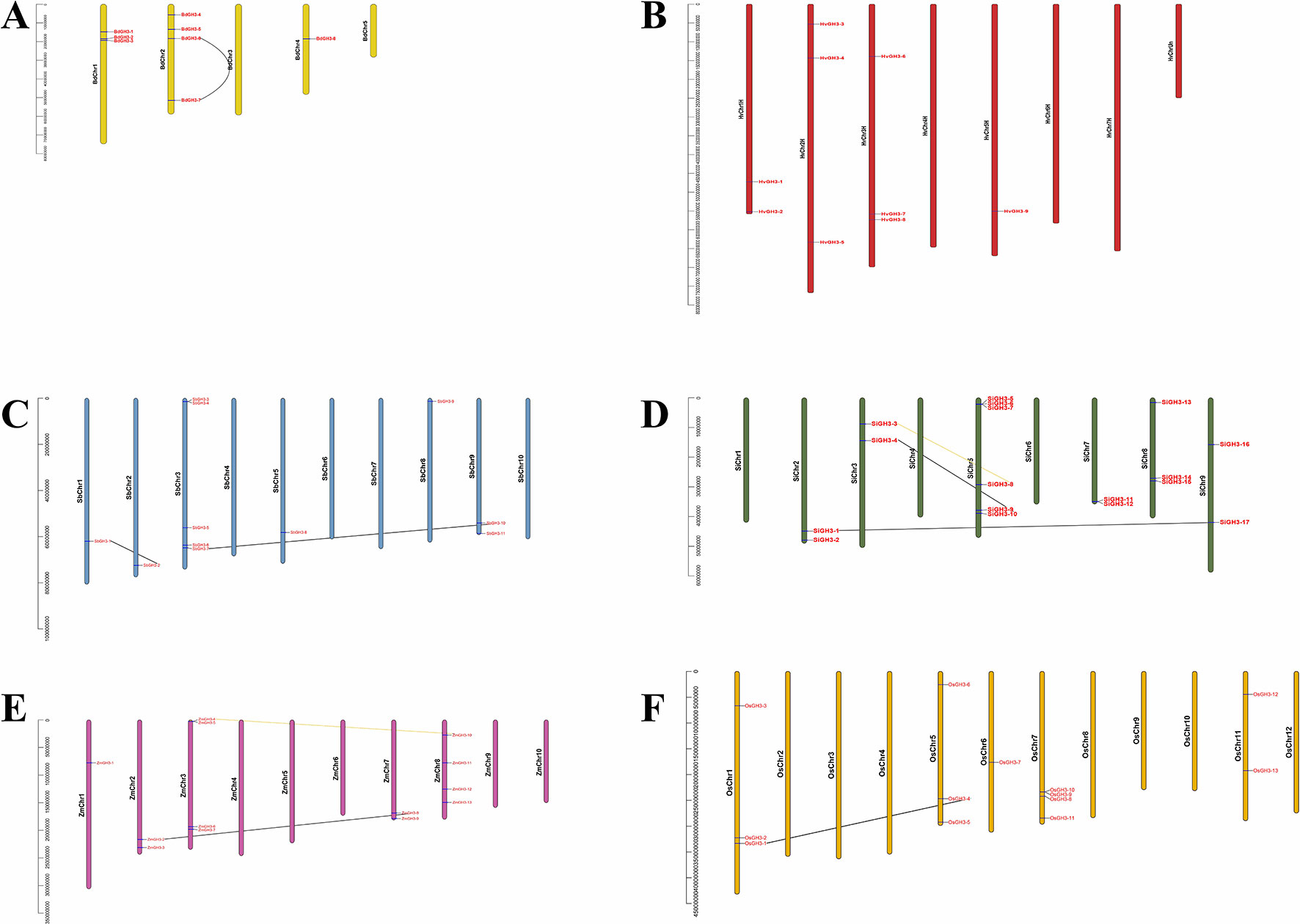
Figure 3 The chromosome location and duplication events of GH3 genes in six species, namely, B. distachyon(A), H. vulgare(B), S. bicolor(C), S. italica(D), Z. mays(E), and O. sativa ssp. japonica(F). The yellow and black lines represent WGD or segmental duplication events in Orthogroup2 (Or2) and Orthogroup1 (Or1), respectively.
Analysis of the Ka/Ks rates of duplicate gene pairs (Table S3) revealed that Ka/Ks ratios of all duplicate gene pairs had less than 1, suggesting that they underwent purifying selection according to the neutral theory. Divergence times of all duplicate gene pairs ranged from 5.43 to 27.70 Mya, except for SiGH3-3 and SiGH3-8 (61.43 Mya) (Table S3). These results indicated that duplication events happen in different stages and play essential roles of GH3 gene family expansions.
Sequence Characteristics and Intron Number Analyses
In this study, 20 conserved motifs (Figure 4, Table S4) were found using MEME suite. We observed that all GH3 proteins showed the similar motifs arrangement, and most identified conserved motifs were parts of GH3 auxin-responsive promoter domain (Table S4), implying that protein sequences of GH3 gene family members are very conservative among different groups/Ors in these tested Gramineae crops. Compared to motifs arrangement, intron/exon structures of all GH3 genes were obviously different between different groups, even in Ors. Phylogenetic tree, exon/intron numbers in CDS, and identified Ors could divide all GH3 genes into obvious classification. Genes in Or2, Or3, and Or4 contained more exons in CDS than Or1 and Or5. These results revealed that GH3 genes from different Ors have been differentiated.
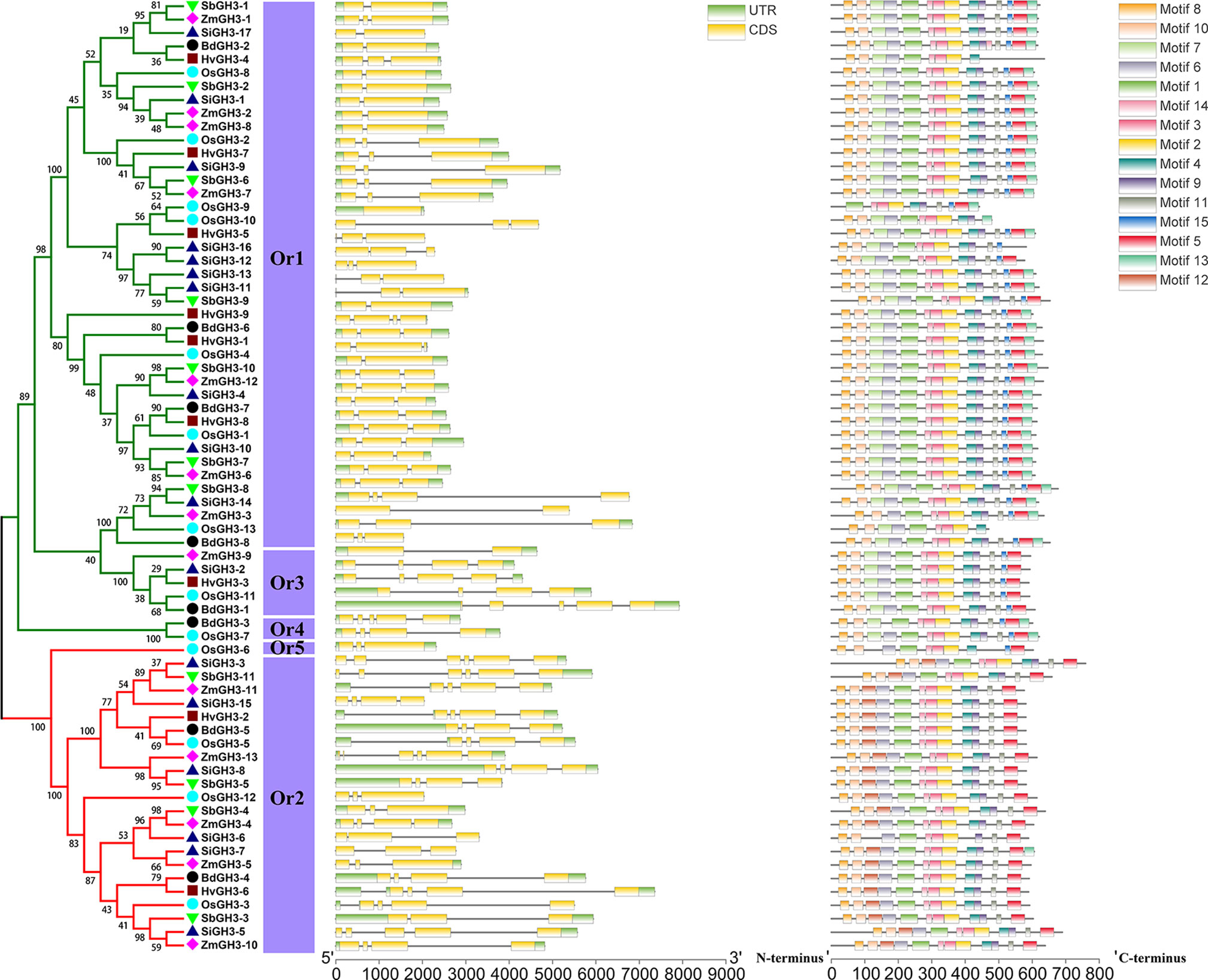
Figure 4 The phylogenetic tree, exon/intron structure, and conserved motif compositions of the GH3 genes in these six species. A phylogeny tree of GH3 protein sequences from H. vulgare, B. distachyon, O. sativa ssp. japonica, S. italica, S. bicolor, and Z. mays. Different colors of branches represent different groups. The different species are indicated by different shaped markers, and Or means orthogroup. The relative lengths of genes and proteins are shown by the widths of the gray bars. The exons and introns are displayed by yellow boxes and gray lines, respectively.
Selective Forces Analysis, Expression, and Function Divergences
To elucidate the evolutionary forces and diverged patterns of different GH3 Ors among Gramineae crops, Tajima’s D and average expression values of Ors were calculated. We found all Ors were under strong purifying selection and ranged from −0.3 to −0.9049 (Table S5). These results pointed out that genes in different Ors faced different evolutionary rates and purifying selection pressures during the evolutionary process.
Expression patterns of seven essential tissues showed that conserved Ors (Or1 and Or2) had higher average expression values than unconserved Ors (Or3, Or4, and Or5) (Table S5) in rice and maize. Further studies showed that OsGH3-2, OsGH3-4, and OsGH3-8 (all in Or1) had similar expression with high expression levels in callus, panicle, and mature seed, while OsGH3-3 and OsGH3-5 (both in Or2) showed completely different expression profiles (Figure 5). This result indicated that genes in Or1 may be redundant, whereas genes in Or2 have functional differentiation. Interestingly, we detected the expression patterns of one WGD or segmental duplication event (OsGH3-1 and OsGH3-4). OsGH3-1 was not expressed in any tissue (Figure 5), indicating this gene had undergone pseudogenization after the duplication event. Differently, in maize, ZmGH3-2 and ZmGH3-8 (one WGD or segmental duplication event) showed similar expression patterns (Figure S1 and Table S6), implying that these two genes are functionally redundant. In addition, we found that the remaining maize GH3 genes showed obviously different expression patterns. For example, ZmGH3-10 showed a high expression level in leaf; ZmGH3-12 was highly expressed at meiotic tassel V18; ZmGH3-7 was strongly expressed at anthers R1.
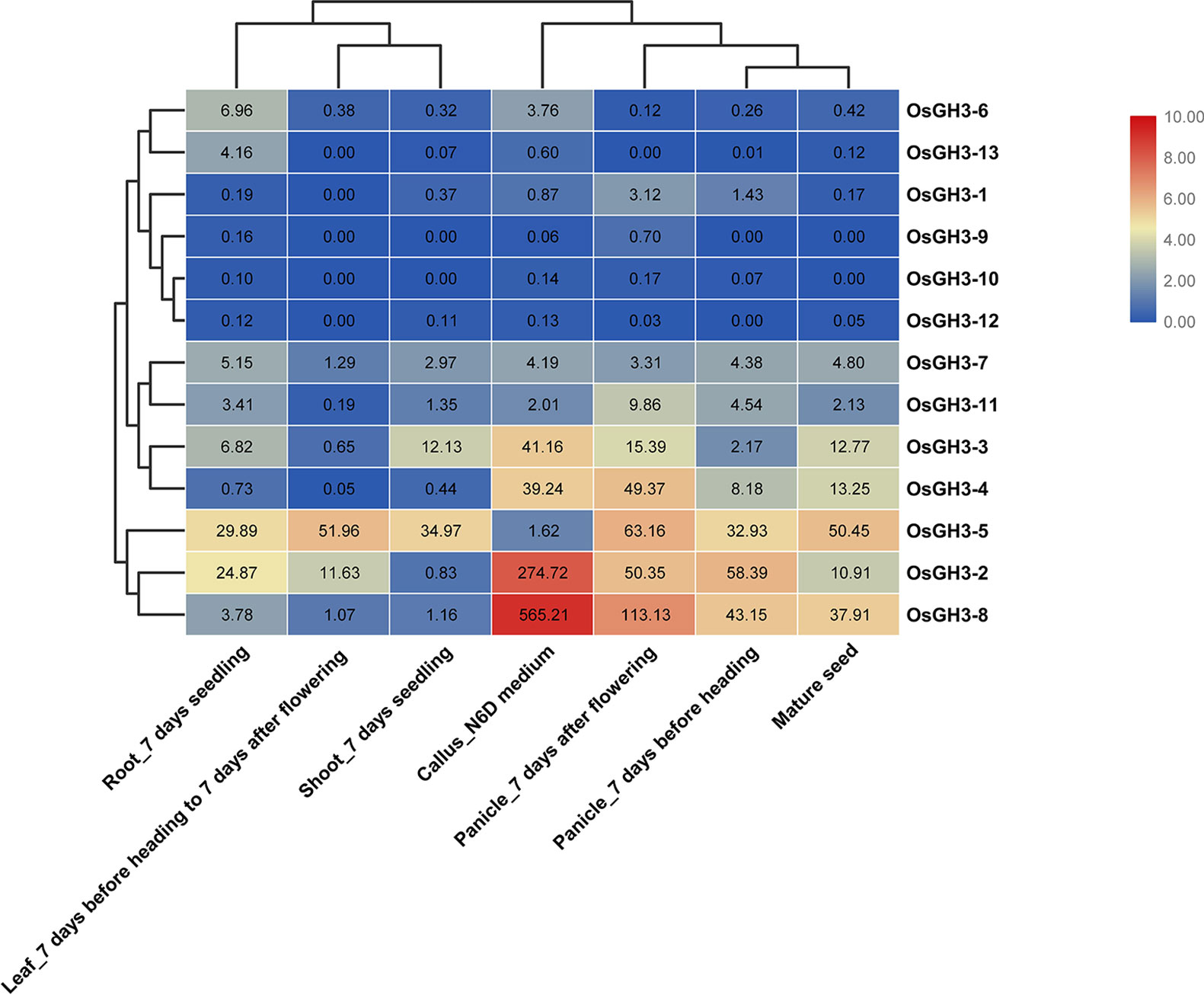
Figure 5 Expression profiles of OsGH3 genes in different tissues. The values in the color scale represent log2FPKM: red/green indicates high level/low level of transcript abundance.
Previous studies reported that GH3 genes play important roles in biotic and abiotic stresses (Fu et al., 2011; Zhang et al., 2018; Kong et al., 2019a). We thus examined the expression changes of maize GH3 gene under salt stress, cold stress, heat stress, and after C. graminicola infection (Figure 6 and Table S7). Under salt, cold, and heat stresses, most maize GH3 genes showed obvious down-regulation. However, ZmGH3-1 showed high up-regulation under cold stress; ZmGH3-9 was highly up-regulated under heat stress; ZmGH3-2 showed clear up-regulation under salt stress; ZmGH3-8 showed slight up-regulation under cold and heat stresses. Similarly, most maize GH3 genes were also down-regulated after C. graminicola infection. Only two maize GH3 genes (ZmGH3-2 and ZmGH3-8) have quite high up-regulation (>30).
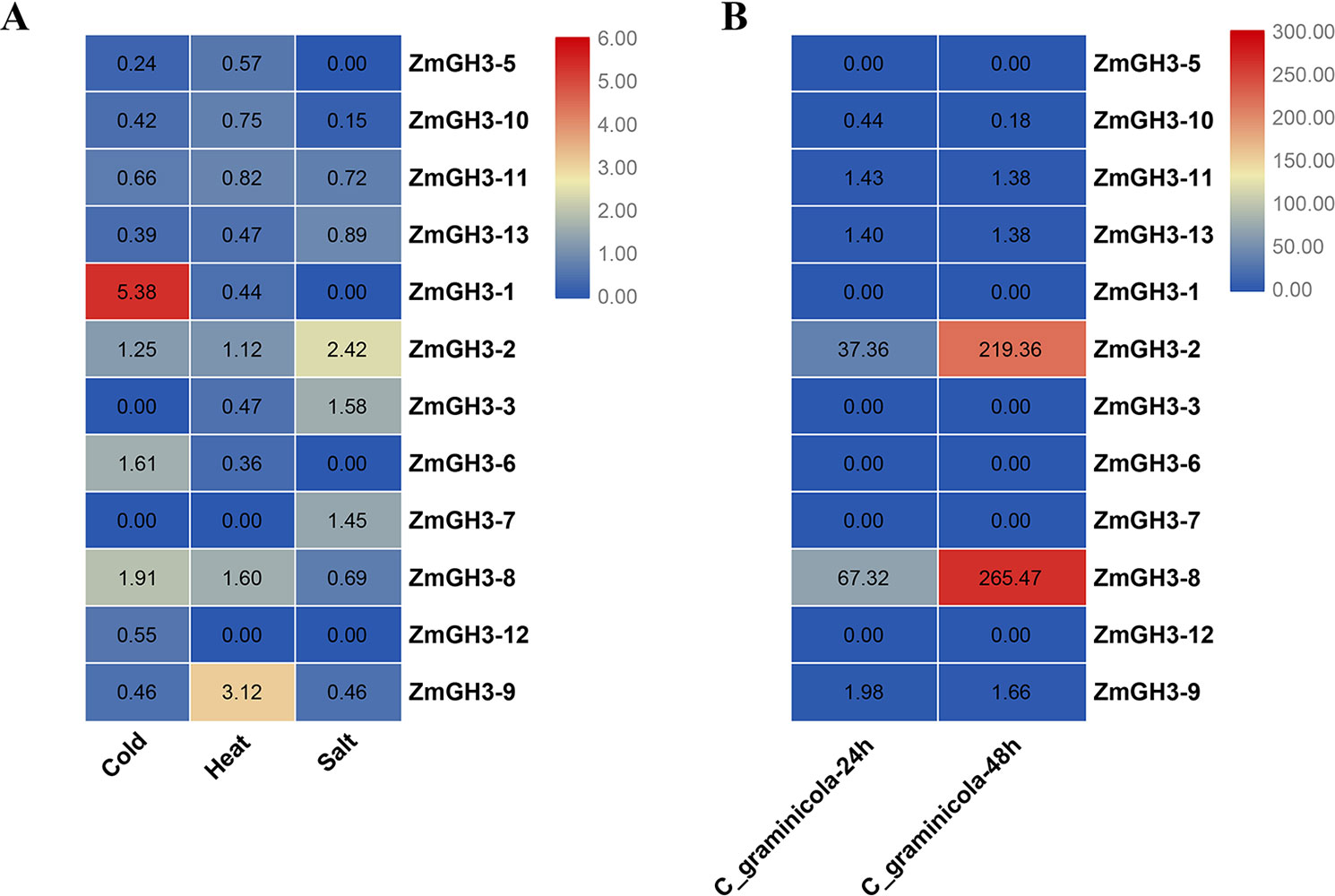
Figure 6 Expression change folds of GH3 genes under salt, cold, and heat (A), as well as after C. graminicola infection (B). Expression change fold = expression value of treated sample/expression value of control sample. The values in the color scale represent change folds from lower (blue color) to higher (red color).
The cis-elements in the gene promoter regions can regulate gene expression, which provides an important insight into gene function prediction. A total of 21 cis-elements were identified through the PLANTCARE website (Lescot et al., 2002) and grouped into four categories, namely, light responsive, growth and development, stress responsive, and phytohormone response (Figure 7). The positioning result of cis-elements showed that cis-elements were unevenly distributed on promoters of all GH3 genes (Figure 7A). We found that 46% of elements belonged to phytohormone response category and were involved in auxin responsiveness (TGA-element and AuxRR element), ABA responsiveness (ABRE), MeJA responsiveness (CGTCA motif and TGACG motif), SA responsiveness (TCA element), and gibberellin responsiveness (P-box, TATC box, and GARE) motifs (Figure 7B). Twenty percent of elements belonged to stress response category associated with multiple stresses, such as drought (MBS) and low temperature (LTR) (Figure 7B). In addition, G-box, Sp1, and GT1 motif were in light responsive category and covered 30% of all elements (Figure 7B). Furthermore, four elements (RY-element, GCN4 motif, MAS-like, and HD-Zip1) in growth and development category related to several processes (Figure 7B). Considering these results, we proposed that GH3 genes participate in light responsive, phytohormone response, stress response, and plant growth and development.
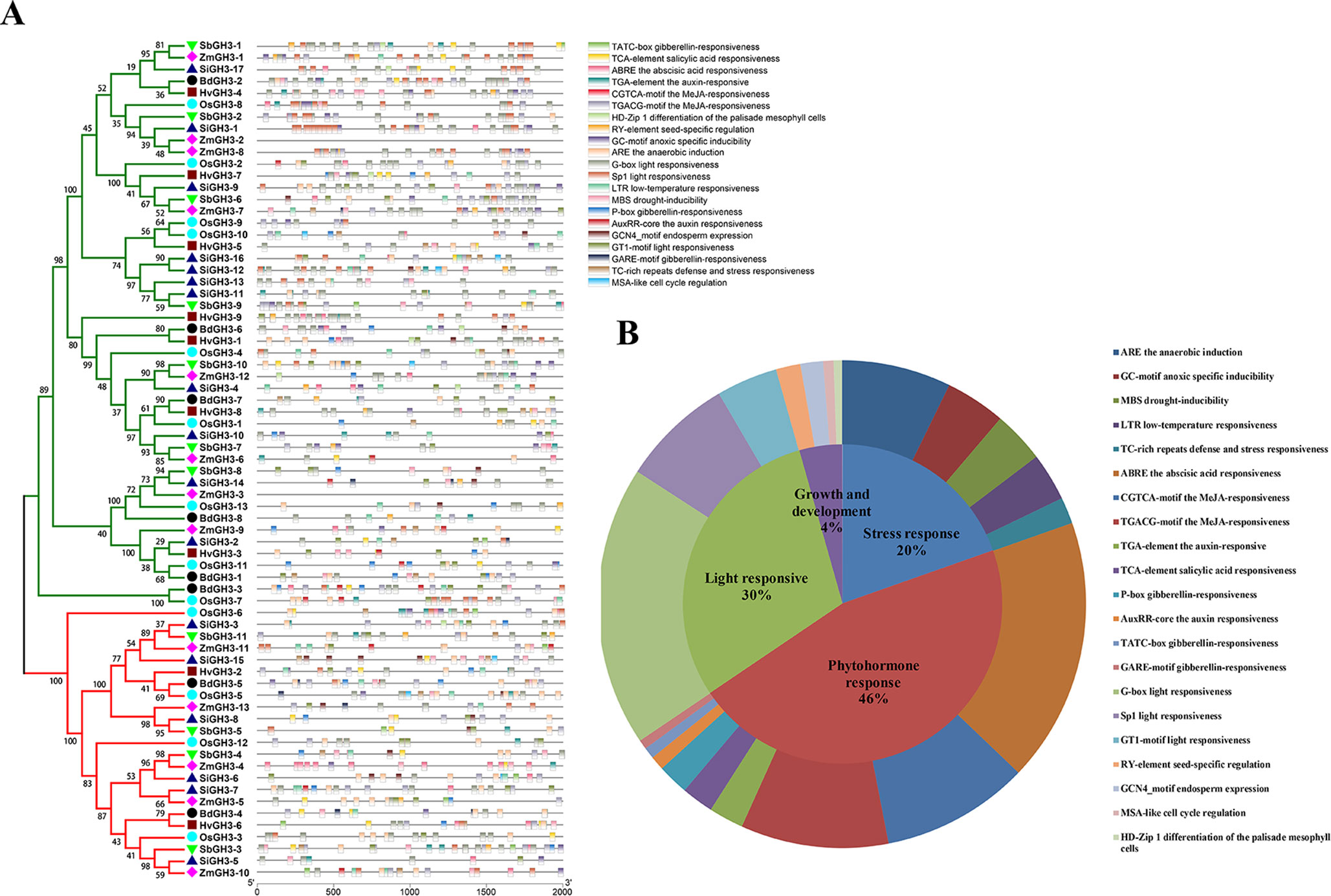
Figure 7 cis-Acting elements in all GH3 genes in Gramineae crops. (A)cis-Acting elements of all GH3 genes showed according to a phylogenetic tree. The different colored boxes indicate different promoter elements in each GH3 gene. (B) Pie charts of different sizes represent the ratio of each promoter element in each category.
Discussion
Evolution of GH3 Genes in Gramineae Crops
In the current study, 69 GH3 genes from six Gramineae crops were identified. We found no direct relevance between genome sizes and the number of GH3 genes. For example, there were 17 GH3 genes in S. italica (genome size: 490 Mbp), while there were nine genes in H. vulgare (genome size: 4.79 Gbp). In addition, there was no direct relevance between WGDs and the number of GH3 genes. Previous studies reported that Z. mays underwent one specific WGD more than other Gramineae plants (Swigoňová et al., 2004). However, Z. mays had fewer GH3 genes than S. italica in this study. In addition, numbers and sizes of Ors vary greatly among these tested Gramineae crops, indicating that the GH3 gene family has undergone different evolutionary approaches in different species, and this conclusion is also supported by our Or identification and selective force results. Our results supported theoretical models of gene family evolution that gene families continuously undergo stochastic gain and loss events (Zimmer et al., 1980; Reed and Hughes, 2004; Hahn et al., 2005). It is well known that gene tandem duplication, segmental duplication, transpositions, and WGDs have significant roles in biological evolution (Xu et al., 2012). In our study, we observed that duplicate gene pairs developed through WGD or segmental duplication, while no gene pair was observed from other mode duplications, specifying that WGD or segmental duplication has played an important role in the expansion of GH3 gene family in some Gramineae species. Our earlier study of GH3 gene family evolution in Oryza genus also verified this conclusion (Kong et al., 2019a). Moreover, we noticed that the divergence times of all duplicate gene pairs ranged from 5.43 to 61.43 Mya, including OsGH3-1 and OsGH3-4 (23.48 Mya). Our previous study in Oryza genus also showed the divergence time of GH3-1 and OsGH3-4 homologous gene pair, which ranged from 23.07 to 31.01 Mya (Kong et al., 2019a). These results indicated that WGD or segmental duplication events play a more important role in B. distachyon, S. italica, Z. mays, and S. bicolor than H. vulgare and O. sativa ssp. japonica.
Functional Diversity in GH3 Genes
The GH3 genes regulate diverse biological processes (Cano et al., 2018) and govern many fundamental aspects of plant growth and development (Liu et al., 2016; Vielba, 2018; Zhang et al., 2018; Kong et al., 2019a). In this study, many elements of light responsive, growth and development, stress responsive, and phytohormone response were identified and supported that GH3 genes have the potential to regulate diverse plant growth and development processes. Selective forces (Tajima’s D) and public data gene expression analysis might help us to be informed on the functional diversity of GH3 genes. We found different Ors were under differential selective forces values, which may lead to functional differentiation between these Ors. As expected, different Ors showed different expression patterns: conserved Ors showed higher expression levels and broader expression tissues than unconserved Ors. These expression patterns confirmed the functional differentiation of different Ors. In addition, conserved Ors showed obvious quantity expansion than unconserved Ors, such as Or2 versus Or5, and Or1 versus Or3. We thus speculate that expansions of Ors are related to plant adaptation to the environment. Current hypotheses proposed that duplicate gene pairs face four fates accompanied by function change after gene duplication events (Lynch and Force, 2000; He and Zhang, 2005; Conant and Wolfe, 2008): loss functions (pseudogenization), new functions (neofunctionalization), partitioning of the original functions (subfunctionalization), and subfunctionalization followed by neofunctionalization (subneofunctionalization). We found gene redundancy (genes in Or1), subfunctionalization (genes in Or2), and pseudogenization (OsGH3-1 and OsGH3-4) in GH3 gene family and supported functional diversity in GH3 genes.
GH3 Genes Play Essential Roles in Plant Growth, Biotic, and Abiotic Stress Responses
Salt, cold, and heat are the major abiotic stresses that frequently affect the growth and development of plants under various natural conditions (Feng et al., 2015; Kong et al., 2019a; Kong et al., 2019b). Auxin-related transcriptional regulation is important for plants to survive and adapt to adverse environmental challenges (Feng et al., 2015; Kong et al., 2019a). For example, OsGH3-2 can enhance drought and cold tolerances via regulating the levels of ABA (Ding et al., 2008; Du et al., 2012). Our earlier study also showed that OsGH3-2 and OsGH3-8 showed obvious up-regulation under salt stress and play important roles in rice salt tolerance (Kong et al., 2019a). In this study, our finding showed that ZmGH3-1, ZmGH3-9, ZmGH3-2, and ZmGH3-8 responded to cold, heat, salt, and cold and salt stresses. These results evidenced that GH3 genes are tightly associated with various abiotic stresses. In addition, our expression result also showed that ZmGH3-2 and ZmGH3-8 showed high up-regulation after C. graminicola infection (>30 fold), indicating these two genes play essential roles in C. graminicola response and tolerance. Overall, ZmGH3-2 and ZmGH3-8 may be good candidate genes for genetic engineering breeding against various biotic and abiotic stresses.
Conclusions
In summary, diverged patterns of GH3 genes in six Gramineae crops were observed from many aspects, including phylogenetic relationship, chromosomal location, duplication events, Ors, selective force, gene structure, protein motifs, cis-elements, and expression patterns. These results revealed that lineage-specific expansions and homologous gene loss/gain occurred during the evolutionary process. GH3 gene family in Gramineae crops was constrained by strong purifying selection. Different selective values of Ors might be related to functional diversity. Taken together, our results will interpret a comprehensive understanding of the evolution of the GH3 gene family in Gramineae crops.
Data Availability Statement
All datasets for this study are included in the manuscript/Supplementary files.
Author Contributions
WK designed and performed the experiments. YZ, XD, SL, and CZ analyzed parts of the data or prepared parts of the figures and tables. YL provided guidance on the whole manuscript. All authors reviewed and approved the final submission.
Funding
This study was funded by the National Key Research and Development Program of China (grant 2016YFD0100400) and the National Special Key Project for Transgenic Breeding (grant 2016ZX08001001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers and editor for their careful reading and helpful comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01297/full#supplementary-material
Table S1 | Summary of information on GH3 genes in six Gramineae crops.
Table S2 | Orthogroups among in six Gramineae crops.
Table S3 | Ka/Ks ratios and divergence times of duplicate gene pairs.
Table S4 | Annotation of conserved motifs in all GH3 proteins.
Table S5 | Tajima’s D and average expression values of Ors. (A) Tajima’s D of each Or was calculated by DnaSP 5.0 (Librado and Rozas, 2009). (B) Average expression values (average FPKM value = total FPKM value of all genes in Or/gene number in Or) of all rice Ors. (C) Average expression values (the same calculation method to Figure B) of all maize Ors.
Table S6 | The expression values of maize GH3 genes in 79 tissues at different stages.
Table S7 | The expression values of maize GH3 genes under salt, cold, and heat, as well as after C. graminicola infection.
Figure S1 | Expression profiles of ZmGH3 genes in various tissues at different stages. The values in the color scale represent expression values: red/green indicates high level/low level of transcript abundance.
References
Ablazov, A., Tombuloglu, H. (2016). Genome-wide identification of the mildew resistance locus O (MLO) gene family in novel cereal model species Brachypodium distachyon. Eur. J. Plant Pathol. 145 (2), 239–253. doi: 10.1007/s10658-015-0833-2
Bostancioglu, S. M., Tombuloglu, G., Tombuloglu, H. (2018). Genome-wide identification of barley MCs (metacaspases) and their possible roles in boron-induced programmed cell death. Mol. Biol. Rep. 45 (9), 1–15. doi: 10.1007/s11033-018-4154-3
Buchfink, B., Xie, C., Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12 (1), 59. doi: 10.1038/nmeth.3176
Cano, A., Sanchez-Garcia, A. B., Albacete, A., Gonzalez-Bayon, R., Justamante, M. S., Ibanez, S., et al. (2018). Enhanced conjugation of auxin by GH3 enzymes leads to poor adventitious rooting in carnation stem cuttings. Front. Plant Sci. 9, 566. doi: 10.3389/Fpls.2018.00566
Chen, C., Xia, R., Chen, H., He, Y. (2018). TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv [Preprint]. doi: 10.1101/289660
Conant, G. C., Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9 (12), 938. doi: 10.1038/nrg2482
Dai, Z., Wang, J., Yang, X., Lu, H., Miao, X., Shi, Z. (2018). Modulation of plant architecture by the miR156f–OsSPL7–OsGH3.8 pathway in rice. J. Exp. Bot. 69 (21), 5117–5130. doi: 10.1093/jxb/ery273
Deng, X., An, B., Zhong, H., Yang, J., Kong, W., Li, Y. (2019). A novel insight into functional divergence of the MST gene family in rice based on comprehensive expression patterns. Genes 10 (3), 239. doi: 10.3390/genes10030239
Ding, X., Cao, Y., Huang, L., Zhao, J., Xu, C., Li, X., et al. (2008). Activation of the indole-3-acetic acid–amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20 (1), 228–240. doi: 10.1105/tpc.107.055657
Domingo, C. F., Tharreau, D., Iglesias, D. J., Talon, M. (2009). Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant-Microbe Interact. 22 (2), 201–210. doi: 10.1094/MPMI-22-2-0201
Du, H., Wu, N., Fu, J., Wang, S. P., Li, X. H., Xiao, J. H., et al. (2012). A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 63 (18), 6467–6480. doi: 10.1093/jxb/ers300
Emms, D. M., Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16 (1), 157. doi: 10.1186/s13059-015-0721-2
Feng, S., Yue, R., Tao, S., Yang, Y., Zhang, L., Xu, M., et al. (2015). Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J. Integr. Plant Biol. 57 (9), 783–795. doi: 10.1111/jipb.12327
Fu, J., Yu, H., Li, X., Xiao, J., Wang, S. (2011). Rice GH3 gene family: regulators of growth and development. Plant Signal. Behav. 6 (4), 570–574. doi: 10.4161/psb.6.4.14947
Hahn, M. W., De Bie, T., Stajich, J. E., Nguyen, C., Cristianini, N. (2005). Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 15 (8), 1153–1160. doi: 10.1101/gr.3567505
He, X. L., Zhang, J. Z. (2005). Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169 (2), 1157–1164. doi: 10.1534/genetics.104.037051
Hoopes, G. M., Hamilton, J. P., Wood, J. C., Esteban, E., Buell, C. R. (2018). An updated gene atlas for maize reveals organ-specific and stress-induced genes. Plant J. 97 (6):1154–1167. doi: 10.1111/tpj.14184
Jain, M., Kaur, N., Tyagi, A. K., Khurana, J. P. (2006). The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genomics 6 (1), 36–46. doi: 10.1007/s10142-005-0142-5
Jian, Z., Nallamilli, B. R., Hana, M., Zhaohua, P. (2010). OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J. 64 (4), 604–617. doi: 10.1111/j.1365-313X.2010.04354.x
Kong, W., Bendahmane, M., Fu, X. (2018). Genome-wide identification and characterization of aquaporins and their role in the flower opening processes in carnation (Dianthus caryophyllus). Molecules 23 (8), 1895. doi: 10.3390/molecules23081895
Kong, W., Gong, Z., Zhong, H., Zhang, Y., Zhao, G., Gautam, M., et al. (2019b). Expansion and evolutionary patterns of glycosyltransferase family 8 in Gramineae crop genomes and their expression under salt and cold stresses in Oryza sativa ssp. japonica. Biomolecules 9 (5), 188. doi: 10.3390/biom9050188
Kong, W., Zhong, H., Deng, X., Gautam, M., Gong, Z., Zhang, Y., et al. (2019a). Evolutionary analysis of GH3 genes in six Oryza species/subspecies and their expression under salinity stress in Oryza sativa ssp. japonica. Plants 8 (2), 30. doi: 10.3390/plants8020030
Kumar, R., Agarwal, P., Tyagi, A. K., Sharma, A. K. (2012). Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Mol. Genet. Genomics 287 (3), 221–235. doi: 10.1007/s00438-011-0672-6
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/Nar/30.1.325
Librado, P., Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 (11), 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, K. D., Wang, J. X., Li, H. L., Zhong, J. D., Feng, S. X., Pan, Y. L., et al. (2016). Identification, expression and IAA-amide synthetase activity analysis of Gretchen Hagen 3 in papaya fruit (Carica papaya L.) during postharvest process. Front. Plant Sci. 7, 1555. doi: 10.3389/Fpls.2016.01555
Lynch, M., Force, A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154 (1), 459. doi: 10.1023/A:1004191625603
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2018). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47 (D1), D351–D360. doi: 10.1093/nar/gky1100
Okrent, R. A., Brooks, M. D., Wildermuth, M. C. (2009). Arabidopsis GH3. 12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 284 (15), 9742–9754. doi: 10.1074/jbc.M806662200
Okrent, R. A., Wildermuth, M. C. (2011). Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Mol. Biol. 76 (6), 489–505. doi: 10.1007/s11103-011-9776-y
Reed, W. J., Hughes, B. D. (2004). A model explaining the size distribution of gene and protein families. Math. Biosci. 189 (1), 97–102. doi: 10.1016/j.mbs.2003.11.002
Singh, V. K., Jain, M., Garg, R. (2015). Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front. Plant Sci. 5, 789. doi: 10.3389/fpls.2014.00789
Staswick, P. E., Tiryaki, I., Rowe, M. L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14 (6), 1405–1415. doi: 10.1105/tpc.000885
Swigoňová, Z., Lai, J., Ma, J., Ramakrishna, W., Llaca, V., Bennetzen, J. L., et al. (2004). Close split of sorghum and maize genome progenitors. Genome Res. 14 (10a), 1916–1923. doi: 10.1101/gr.2332504
Terol, J., Domingo, C., Talon, M. (2006). The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371 (2), 279–290. doi: 10.1016/j.gene.2005.12.014
Tombuloglu, G., Tombuloglu, H., Cevik, E., Sabit, H. (2019). Genome-wide identification of lysin-motif receptor-like kinase (LysM-RLK) gene family in Brachypodium distachyon and docking analysis of chitin/LYK binding. Physiol. Mol. Plant Pathol. 106, 217–225. doi: 10.1016/j.pmpp.2019.03.002
Tombuloglu, H. (2018). Genome-wide analysis of the auxin response factors (ARF) gene family in barley (Hordeum vulgare L.). J. Plant Biochem. Biotechnol. (1), 1–11. doi: 10.1007/s13562-018-0458-6
Vielba, J. M. (2018). Identification and initial characterization of a new subgroup in the GH3 gene family in woody plants. J. Plant Biochem. Biotechnol. 1–11. doi: 10.1007/s13562-018-0477-3
Xu, G., Guo, C., Shan, H., Kong, H. (2012). Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. 109 (4), 1187–1192. doi: 10.1073/pnas.1109047109
Yadav, S. R., Khanday, I., Majhi, B. B., Veluthambi, K., Vijayraghavan, U. (2011). Auxin-responsive OsMGH3, a common downstream target of OsMADS1 and OsMADS6, controls rice floret fertility. Plant Cell Physiol. 52 (52), 2123–2135. doi: 10.1093/pcp/pcr142
Yang, J. H., Han, S. J., Yoon, E. K., Lee, W. S. (2006). Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 34 (6), 1892–1899. doi: 10.1093/nar/gkl118
Zhang, C., Zhang, L., Wang, D., Ma, H., Liu, B., Shi, Z., et al. (2018). Evolutionary history of the glycoside hydrolase 3 (GH3) family based on the sequenced genomes of 48 plants and identification of jasmonic acid–related GH3 proteins in Solanum tuberosum. Int. J. Mol. Sci. 19 (7), 1850. doi: 10.3390/ijms19071850
Zhang, S., Li, C., Cao, J., Zhang, Y., Zhang, S., Xia, Y., et al. (2009). Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 151 (4), 1889–1901. doi: 10.1104/pp.109.146803
Keywords: Glycoside hydrolase 3 (GH3), Gramineae crops, evolutionary patterns, evolutionary dynamic, gene duplication events
Citation: Kong W, Zhang Y, Deng X, Li S, Zhang C and Li Y (2019) Comparative Genomic and Transcriptomic Analysis Suggests the Evolutionary Dynamic of GH3 Genes in Gramineae Crops. Front. Plant Sci. 10:1297. doi: 10.3389/fpls.2019.01297
Received: 17 April 2019; Accepted: 18 September 2019;
Published: 15 October 2019.
Edited by:
Atsushi Fukushima, RIKEN, JapanReviewed by:
Huseyin Tombuloglu, Imam Abdulrahman Bin Faisal University, Saudi ArabiaDong-Wen Lv, University of Florida, United States
Xiangjia Min, Youngstown State University, United States
Copyright © 2019 Kong, Zhang, Deng, Li, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yangsheng Li, bHlzaDIwMDFAd2h1LmVkdS5jbg==
 Weilong Kong
Weilong Kong Yue Zhang
Yue Zhang