- 1Shandong Provincial Key Laboratory of Plant Stress, College of Life Sciences, Shandong Normal University, Ji’nan, China
- 2Maize Research Institute, Shandong Academy of Agricultural Sciences, Jinan, China
The Arabidopsis thaliana WD40-repeat protein TRANSPARENT TESTA GLABRA1 (TTG1) controls epidermis development, playing opposite roles in trichome differentiation and root hair formation. We isolated and characterized LbTTG1 (encoding a WD40-repeat protein with high sequence similarity to TTG1) from the recretohalophyte Limonium bicolor, which actively excretes absorbed salt via a salt gland. The complete open reading frame of LbTTG1 was 1,095 bp, encoding a protein of 364 amino acids, and showed highest expression during the salt gland initiation stage. We heterologously expressed LbTTG1 in wild type and ttg1-13 Arabidopsis plants to verify the protein’s function, and the copies of LbTTG1 were identified in transgenic strains using southern blotting. Trichomes were extremely induced on the first true leaves of plants heterologously expressing LbTTG1, whereas no trichomes were produced by ttg1-13 plants. Conversely, plants heterologously expressing LbTTG1 produced fewer root hairs than ttg1-13 plants. In plants heterologously expressing LbTTG1 compared to controls, epidermis differentiation genes (GLABRA1 and GLABRA3) were up-regulated while genes encoding negative regulators of trichome development (TRIPTYCHON and CAPRICE) were down-regulated. Under increased NaCl concentrations, both of the transgenic lines showed enhanced germination and root length, and accumulated less malondialdehyde (MDA) and Na+ and produced more proline, soluble sugar, and higher glutathione S-transferase activity, compared with the ttg1-13 mutant. These results indicate that LbTTG1 participates in epidermis development in Arabidopsis, similarly to other WD40-repeat proteins, and specifically increases salt tolerance of transgenic Arabidopsis by reducing ion accumulation and increasing osmolyte levels.
Introduction
Soil salinization inhibits the growth and production of crops worldwide. Currently, 800 million hectares of land is affected by elevated salinity, and poorly designed irrigation approaches are causing rapid expansion of saline farmland worldwide (Munns and Tester, 2008). Finding ways to make use of large areas of saline land is urgent as a means to expand available arable land (Song et al., 2017). Few crops can survive in salt-affected regions, which leads to substantially reduced production and often results in soil degradation and desertification (Flowers and Colmer, 2008; Song and Wang, 2015). Thus, it is necessary to improve the salt tolerance of non-halophytic plants by transforming them with genes conferring salt tolerance (Yuan et al., 2015).
Halophytes that survive and complete their life cycles in environments with ≥200 mM NaCl can provide a broad bank of genes for improving the salt tolerance of non-halophytes. Two key points of focus should be identifying suitable halophytes and choosing key salt-tolerance genes. A highly efficient Agrobacterium-mediated transformation system has been established for Limonium bicolor, a recretohalophyte that can actively excrete absorbed salt to the outside via a salt gland (Ding et al., 2010; Deng et al., 2015). Salt glands are specific and visible epidermal structures that make recretohalophytes distinct from all non-halophytes and other types of halophytes. A large number of mutants involved in salt gland development and salt secretion were screened by an efficient autofluorescence method (Yuan et al., 2013), and the distribution patterns of salt glands have been demonstrated (Leng et al., 2018). Five distinct stages of epidermis differentiation have been discerned in leaves (Yuan et al., 2015), and the ultrastructures of salt glands have also been observed (Feng et al., 2014; Feng et al., 2015). Given this existing foundation, L. bicolor represents a good model plant for studying salt tolerance and development of the salt gland.
Our previous studies illustrated a series of genes that may participate in salt gland development (Yuan et al., 2015) and salt secretion (Yuan et al., 2016). Surprisingly, genes reported to be involved in trichome initiation differentiation are found in L. bicolor, such as those with homology to GLABRA1 (GL1), TRANSPARENT TESTA GLABRA1 (TTG), GLABRA3 (GL3), TRIPTYCHON (TRY), and CAPRICE (CPC) (Yuan et al., 2015). Given that no trichomes are observed in L. bicolor (Leng et al., 2018), we speculate that salt glands of L. bicolor may evolve from a trichome-like structure under the control of similar regulatory genes.
In Arabidopsis, trichomes act as a physical barrier against biotic stress, and their differentiation network has been clearly illustrated. TTG1, encoding a WD40-repeat protein, participates in trichome initiation, flavonoid production, seed coat mucilage production, and seed oil and storage protein accumulation, and negatively regulates root hair development (Walker et al., 1999). AtTTG1 and AtGL1 combine with AtGL3 or ENHANCER OF GLABRA3 (AtEGL3) to form a TTG1-GL3/EGL3-GL1 complex, which directly regulates AtGL2 and AtTTG2 in the positive regulation of trichome initiation (Zhao et al., 2008). Negative regulators of trichome development are mainly MYB proteins, including TRY, CPC, ENHANCER OF TRY AND CPC1 (ETC1), and ETC2 (Kirik et al., 2004; Tominaga-Wada and Nukumizu, 2012; Tominaga-Wada and Wada, 2016). The functions of homologous genes associated with trichome development in other species have been verified using heterologous expression in Arabidopsis. For example, the transgenic expression of CsTTG1 in cucumber (Cucumis sativa) and Arabidopsis enhances trichome number (Chen et al., 2016), and the cucumber CsGL1 gene mutant csgl1 shows abnormal trichomes on leaves, stems, flowers, and fruits, with expression of papillae instead (Li et al., 2015; Zhao et al., 2015). Therefore, heterologous expression is a useful tool for investigating gene function.
Here, we identified a gene encoding a WD40-repeat protein with high sequence similarity to TTG1 of Arabidopsis by comparing transcriptome data of L. bicolor (Yuan et al., 2015) with expression data for all homologous genes involved in trichome differentiation. WD40-repeat proteins (also known as WD or beta-transducin repeats) are short ∼40 amino acid motifs, often terminating in a Trp-Asp (W-D) dipeptide, and are involved in negative regulation of root hairs and positive of trichomes (Payne et al., 2000; Zhang et al., 2003). This gene, named LbTTG1 in L. bicolor, may play an important role in salt gland development due to its high expression during salt gland differentiation. LbTTG1 played an important role in epidermis formation and can enhance the salt tolerance of Arabidopsis, and the possible reason was also illustrated.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of L. bicolor were collected from a saline, inland environment (N37°20′; E118°36′) in the Yellow River Delta, Shandong, China. Dry seeds were stored in a refrigerator at 4°C for 6 months before use. Seeds were surface-sterilized in 70% ethanol for 5 min, followed by 6% (v/v) sodium hypochlorite (239305, Sigma, USA) with vigorous shaking for 15–20 min, and then washed thoroughly with sterile distilled water before being germinated on Murashige and Skoog (1962) (MS) basal medium containing 3% (w/v) sucrose and 0.9% (w/v) agar, adjusted to pH 5.8 with KOH before autoclaving. Seeds were cultured at 28 ± 3°C/23 ± 3°C (day/night) at a light intensity of 600 µmol/m2/s (15-h photoperiod) and 70% relative humidity. The first true leaves were separately collected at different leaf developmental stages—stage A (undifferentiation, 4–5 days after sowing), stage B (salt gland differentiation, 6–7 days), stage C (stomata differentiation, 8–10 days), stage D (epidermis differentiation, 11–16 days), and stage E (mature, more than 17 days)—for RT-PCR and gene cloning according to (Yuan et al., 2015).
The Arabidopsis thaliana ecotype Col-0 (Columbia-0) was used as a control. The homozygous Arabidopsis AT5G24520 mutant ttg1-13 (CS67772), obtained by fast neutron mutagenesis, was ordered from the Arabidopsis Biological Resource Center. Seeds of Col-0 and CS67772 were sterilized three times using 75% ethanol for 3 min and then three times using 95% ethanol for 1 min, and washed five times with distilled water. Seeds were sown on sterile half-strength (1/2) MS medium containing 0.8% (w/v) sucrose and 0.8% (w/v) agar (pH 5.8) for germination. After 2 days of vernalization at 4°C, seeds were cultured at 22°C/18°C (day/night) under a 16 h/8 h light/dark cycle with a light level of 150 µmol/m2/s and 70% relative humidity (Sui et al., 2017). After culture for 1 week, seedlings were transplanted to pots (10 cm in diameter and 8 cm in height) containing well-mixed soil (soil: vermiculite: perlite, 3:1:1) for further flowering and transformation.
Full-Length Clone of LbTTG1 From L. bicolor
According to (Yuan et al., 2015), approximately 5,000 leaves from stage A, 4,000 from stage B, 2,000 from stage C, 1,000 from stage D, and 1,000 from stage E were separately dissected at the same time of day (8:00 a.m. to 10:00 a.m.), and all leaves were immediately frozen in liquid nitrogen and stored –80°C until use. Total RNA from different samples was extracted using a Total Plant RNA Extraction kit (Karroten, Beijing, China) and purified using ethanol and 3 mol/L sodium acetate to precipitate polysaccharides. Purified total RNA was quantified using a NanoDrop ND-2000 spectrophotometer (NanoDrop products, Wilmington, DE, USA); the concentration of total RNA was greater than 700 ng/µl with optical density (OD)260/280 2.0–2.2 and OD260/230 ≥ 1.0. After electrophoresis to measure integrity, cDNA was obtained using a ReverTra Ace® quantitative PCR (qPCR) RT kit (TOYOBO Co., Ltd, Japan) according to the manufacturer’s instructions.
Lb106501, homologous to the Arabidopsis gene AT5G24520 (AtTTG1) and named LbTTG1, was identified after mapping the leaf developmental transcriptome of L. bicolor to the Uniref database (http://www.uniprot.org/) by applying Blastx with an E-value threshold of 0.005 (Yuan et al., 2015). The assembled sequence of LbTTG1, as a reference for the full-length clone, was obtained from previous transcriptome results (Yuan et al., 2015). All primers were designed using Primer Premier 5.0 (Table S1). A specific intermediate fragment of LbTTG1 was first obtained using cDNA of stage A leaves as template with primers LbTTG1-S and LbTTG1-A; primers for 3′ RACE amplification were designed according to this intermediate fragment. cDNA for 3′ RACE amplification was reverse transcribed using QT primer instead of the primer mix. The 3′ RACE sequence was obtained after continuous 3′-end amplification using primer Q0 paired with LbTTG13′-GSP1 and primer Q1 paired with LbTTG13′-GSP2. The 5′-end sequence of LbTTG1 was obtained by 5′ RACE using a 5′ RACE kit (5′ RACE System for Rapid Amplification of cDNA Ends, Version 2.0, Invitrogen) with primers LbTTG15′-GSP1 and LbTTG15′-GSP2. The full-length sequence of LbTTG1 was determined by combining the sequencing results, and the full-length cDNA of LbTTG1 was obtained using primers LbTTG1 CDS-S and LbTTG1 CDS-A.
Bioinformatics Analysis of LbTTG1 in L. bicolor
The conserved domain of LbTTG1 was predicted using the online tool SMART (http://smart.embl-heidelberg.de/). A phylogenetic tree was reconstructed using the neighbor-joining method with MEGA5.1 software (www.megasoftware.net) and ClustalX software, with statistical support for nodes obtained from at least 1,000 trials.
Subcellular Localization of LbTTG1 by Transient Expression
The open reading frame (ORF) region of LbTTG1 was cloned into the pCAMBIA1300 vector, containing a CaMV 35S promoter, hygromycin resistance gene, and GFP reporter gene, by double digestion with KpnI and BamHI after TA cloning using pEASY-T3 vectors with primers LbTTG1 OE-S and LbTTG1 OE-A (Table S1). The resulting p1300-LbTTG1 vector was transformed into onion epidermis cells using Agrobacterium tumefaciens strain GV3101 (Sun et al., 2007). Fluorescence signals of labeled LbTTG1 were detected by microscopy (TCS S8 MP two-photon laser scanning confocal microscope, Leica, Germany).
qPCR Analysis at Different Developmental Stages
Total RNAs of A-E stage leaves, stems, roots, flowers, and mature leaves under 200 mM NaCl treatment for 24 h were extracted for real-time PCR. qPCR primers were designed using Beacon Designer™ Free Edition software (version 7.8). The amplification procedure consisted of 94°C for 5 min, followed by 35 cycles of 30 s at 94°C, 1 min at 51°C, and 1 min at 72°C. SYBR Green qPCR was carried out in a fluorometric thermal cycler (Bio-Rad CFX96™ Realtime PCR System) using SYBR Premix Ex Taq™ (TaKaRa, Mountain View, CA) in 20 µl reaction mixtures containing 10 µl of 2×SYBR Premix Ex Taq™ (TaKaRa), 50 ng cDNA, and 0.2 µM each primer (Table S1, Lbtubulin RT and LbTTG1 RT) using Lbtubulin as an internal control. The PCR thermal cycle was as follows: denaturation at 95°C for 5 min and 40 cycles at 94°C for 20 s, 58°C for 15 s, and 65°C for 15 s. Three replicate biological experiments were performed. Relative expression levels were calculated using the formula 2–ΔΔC(T).
Vector Construction and Transformation of Arabidopsis Col-0 and ttg1-13
The LbTTG1 ORF was cloned into the pCAMBIA3301 vector under the control of the CaMV 35S promoter to generate p35S::LbTTG1, using primers LbTTG1 OEAt-S and LbTTG1 OEAt-A (Table S1) according to the instructions of the In-Fusion® HD Cloning kit (Clontech Laboratories, Inc.). The p35S::LbTTG1 vector was introduced into A. tumefaciens strain EHA105, which was then transformed into A. thaliana Col-0 and ttg1-13 by the Agrobacterium-mediated floral dip method (Clough and Bent, 2010). After screening with herbicide for three consecutive generations, homozygous Col 35S::LbTTG1 and ttg 35S::LbTTG1 lines were retained for qPCR.
Plants heterologously expressing LbTTG1 were identified by PCR using primers LbTTG1V-S and LbTTG1V-A on genomic DNA. Total RNA of several strains of Col 35S::LbTTG1 and ttg 35S::LbTTG1 was then extracted using a FastPure Plant Total RNA Isolation kit (Vazyme) according to the manufacturer’s instructions. qPCR was conducted to evaluate the expression level of LbTTG1 in Col 35S::LbTTG1 and ttg 35S::LbTTG1 using LbTTG1RT-S and LbTTG1RT-A primers. Amplification of the actin gene of Arabidopsis was used as an internal control (primers AtactinRT-S and AtactinRT-A). Three replications were carried out for each transgenic line. Three lines with high, medium, and low LbTTG1 expression levels, respectively, of Col 35S::LbTTG1 and ttg 35S::LbTTG1 were used for further experiments.
Southern Hybridization of LbTTG1 in L. Bicolor and Transgenic Lines of Arabidopsis
Genomic DNA was extracted from 2-week-old seedlings of L. bicolor and transgenic lines of Arabidopsis using cetyltrimethylammonium bromide (CTAB) method (Guo et al., 2015), then 10 µg was digested with HindIII. After electrophoresis in a 0.7% agarose gel at 25 V and 4°C overnight, positive control (pGBKT7-LbTTG1 digested with NdeI to obtain LbTTG1) and all samples were transferred to a HyBond N+ membrane (Amersham, UK) using upward capillary method for 20 h. Digested DNA was fixed to the membrane at 80°C for 2 h. The probe was synthesized by primers LbTTG1P-S and LbTTG1P-A (Table S1), and was labeled with Digoxigenin-11-dUTP (PCR DIG Probe Synthesis Kit, Roche). DNA gel blot hybridization was performed by Zoonbio Biotechnology Co., Ltd (Nanjing, China) using DIG DNA labeling and detection kit (Roche). After pre-hybridization at 37°C for 2 h and probe denaturation, hybridization was incubated at 37°C overnight. Membrane was washed two times using 2×SSC/0.1%SDS for 5 min at room temperature, then twice for 15 min in 1×SSC/0.1%SDS buffer at 65°C. After blocking for 30 min, the membrane was immersed in antibody solution (Anti-Digoxigenin-AP, Roche) for 30 min. Hybridized membrane was detected in the presence of CSPD (C18H20ClNa2O7P), and the hybridized signals were visualized on X-ray film after 1 h.
Phenotypic Observation of Trichome and Root Hair Development in Col 35S::LbTTG1 and ttg 35S::LbTTG1
Phenotypes of transgenic plants were observed in the T3 generation, and photographs of 1-week-old homozygous seedlings of the T3 generation were taken under a dissecting microscope (Nikon, Japan). The number of trichomes on the first true leaf was calculated for 10 plants of each line. Root hair length and number were also examined in 5-day-old Col 35S::LbTTG1 and ttg 35S::LbTTG1 seedlings; three replicates of 20 plants from each line were performed. The same root position (region from 2 to 3 cm from the root tip) was chosen for calculating root hair number and length. The numbers of trichomes and root hairs were calculated using ImageJ software (http://rsb.info.nih.gov/ij/).
Effect of NaCl Concentration on Salt Tolerance of Different Transgenic Lines: Percentage Germination
Three Col 35S::LbTTG1 and three ttg 35S::LbTTG1 lines with high, medium, and low LbTTG1 expression levels, respectively, were used for salt treatment. All seeds were sowed on 1/2 MS medium with different NaCl concentrations (0, 50, 100, and 150 mM). After 24 h, seed germination percentage was calculated based on radicles breaking the seed coats for more than 1 mm. Germination percentage (%) = number of germinated seeds/total number of seeds × 100%. Fifty seeds of each line were sowed in each treatment, and three replicates were performed.
Effect of NaCl Concentration on Salt Tolerance of Different Transgenic Lines: Root Length and Physiological Indicators
All seeds were germinated on 1/2 MS medium for 24 h before uniform seedlings were transplanted to medium with different NaCl concentrations (0, 50, 100, and 150 mM) to observe the effect of NaCl on root growth. Five-day-old seedlings were photographed for root length measurement using ImageJ software. Twenty seedlings were measured for each line.
Four-day-old, uniform seedlings grown on 1/2 MS medium were transplanted into soil for gradient NaCl treatment (0, 50, 100, and 150 mM). Two-week-old seedlings under different NaCl treatments were pooled to 0.5 g for the measurement of the contents of Na+ and K+, MDA, proline, and soluble sugar according to (Han et al., 2019) and (Guo et al., 2018). Ion concentrations were determined using a flame photometer (M410, Sherwood, UK). Glutathione S-transferase (GST) activity was determined according to (Dinler et al., 2014) that the formation of the conjugate1-chloro-2,4-dinitrobenzene (CDNB) using reduced glutathione resulted in the increase of absorbance at 340 nm. Five replicates were performed for each line.
RT-qPCR of Trichome Formation and Stress-Related Genes
Five-day-old seedlings of all lines grown on 1/2 MS medium were collected for RNA extraction. RT-qPCR was performed using primers targeting genes related to trichome differentiation, including AtTTG1, AtGL1, AtGL3, AtEGL3, AtCPC, and AtTRY, listed as genename-S and genename-A (e.g. AtTTG1-S and AtTTG1-A) in Table S1.
Two-week-old seedlings of all lines grown on 1/2 MS medium were transferred to 1/2 MS liquid medium supplied with 100 mM NaCl solution for 0 and 3 h. Seven stress-related marker genes were selected for RT-qPCR: AtSOS1, AtSOS2, AtSOS3, AtP5CS1, AtP5CS2, AtGSTU5, and AtAREB1 (Table S1). Three replicate biological experiments were performed. Relative expression levels were calculated using the formula 2–ΔΔC(T). Atactin (AtactinRT-S and AtactinRT-A) was used as an internal control.
Statistical Analysis
Statistical analysis was performed using SPSS at P = 0.05 (Duncan’s multiple range tests). ANOVA with orthogonal contrasts and mean comparison procedures was used to detect differences between treatments.
Results
LbTTG1 Encoded WD40-Repeat Protein
The LbTTG1 gene contained a complete ORF of 1,095 bp encoding a protein comprising 364 amino acids with a molecular mass of 39.7 kDa (C1761H2712N462O561S10) (Figure 1A). High identity was illustrated between LbTTG1 and AtTTG1 (Figure 1B). LbTTG1 contained four WD-repeat domains located at 86∼132, 139∼184, 187∼225, and 276∼316 amino acids (Figure 1C), representing regions highly conserved with other TTG1 proteins. To investigate the evolutionary relationships among TTG1 proteins in plants, we reconstructed a phylogenetic tree using full-length amino acid sequences by the neighbor-joining method. LbTTG1 shared the highest evolutionary relationship with a protein from Tartary buckwheat (Fagopyrum tataricum) (Figure 1D).
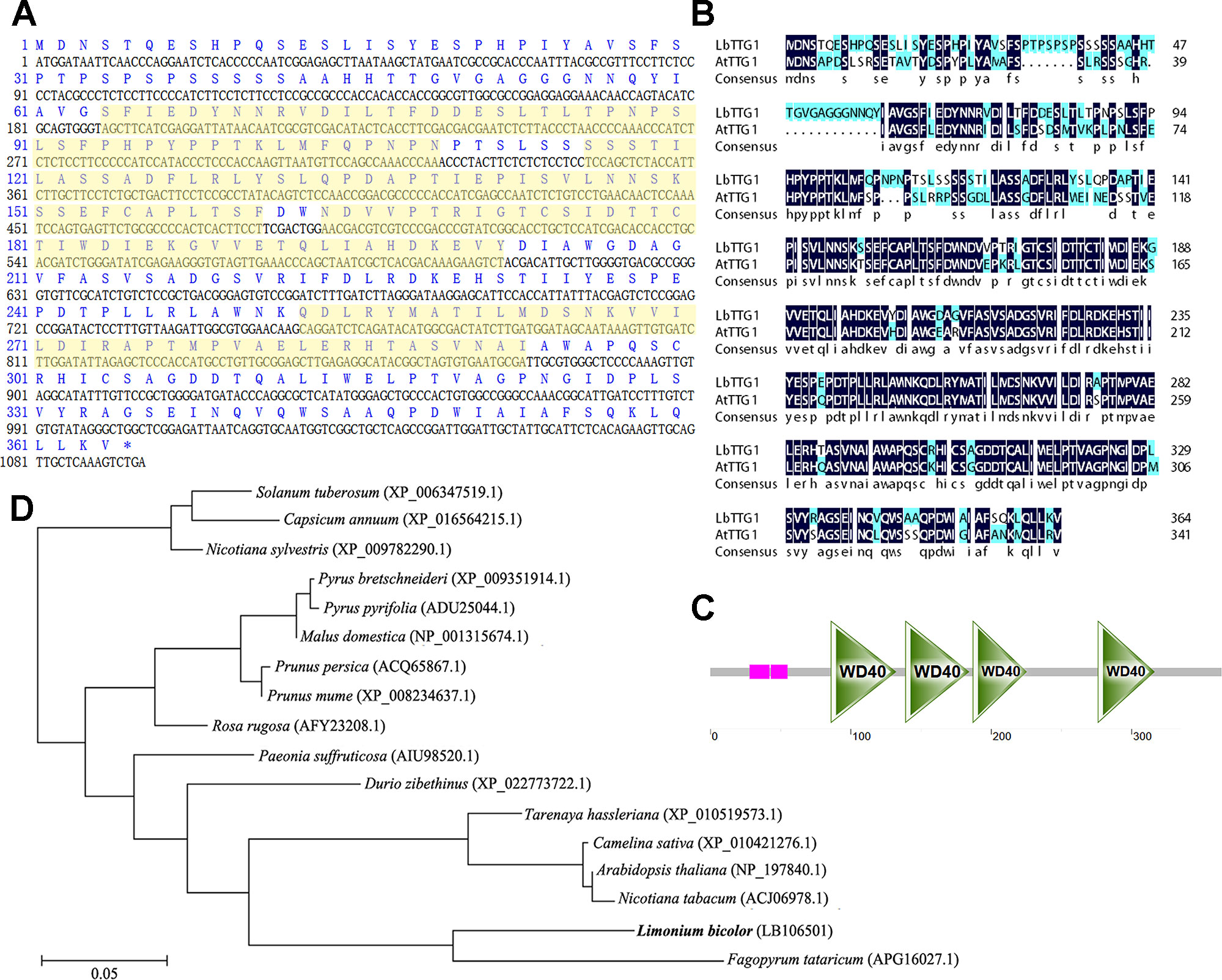
Figure 1 LbTTG1 encoded WD40 repeat protein. (A) Nucleotide and amino acid sequences of LbTTG1 analyzed with DNAman. The yellow shadows indicated four WD-repeat domains. (B) The sequence alignment of TTG1 between Limonium bicolor and Arabidopsis. The identity is 73.9%. (C) Four conserved WD-repeat domains of LbTTG1, located in 86∼132, 139∼184, 187∼225, and 276∼316 amino acids; pink mean the low complexity domain, drawn with SMART. (D) Phylogenetic relationships of different plants based on amino acid residues of LbTTG1, reconstructed by the neighbor-joining method using MEGA and ClustalX software. Bar, 0.05 substitutions per amino acid position.
Subcellular Localization and Expression of LbTTG1 at Different Leaf Developmental Stages
We transformed onion epidermis cells with Agrobacterium containing the p1300-LbTTG1 vector and detected expression of the GFP-LbTTG1 fusion protein in both the nucleus and plasma membrane, while the empty vector only positioned in the membrane (Figure 2).
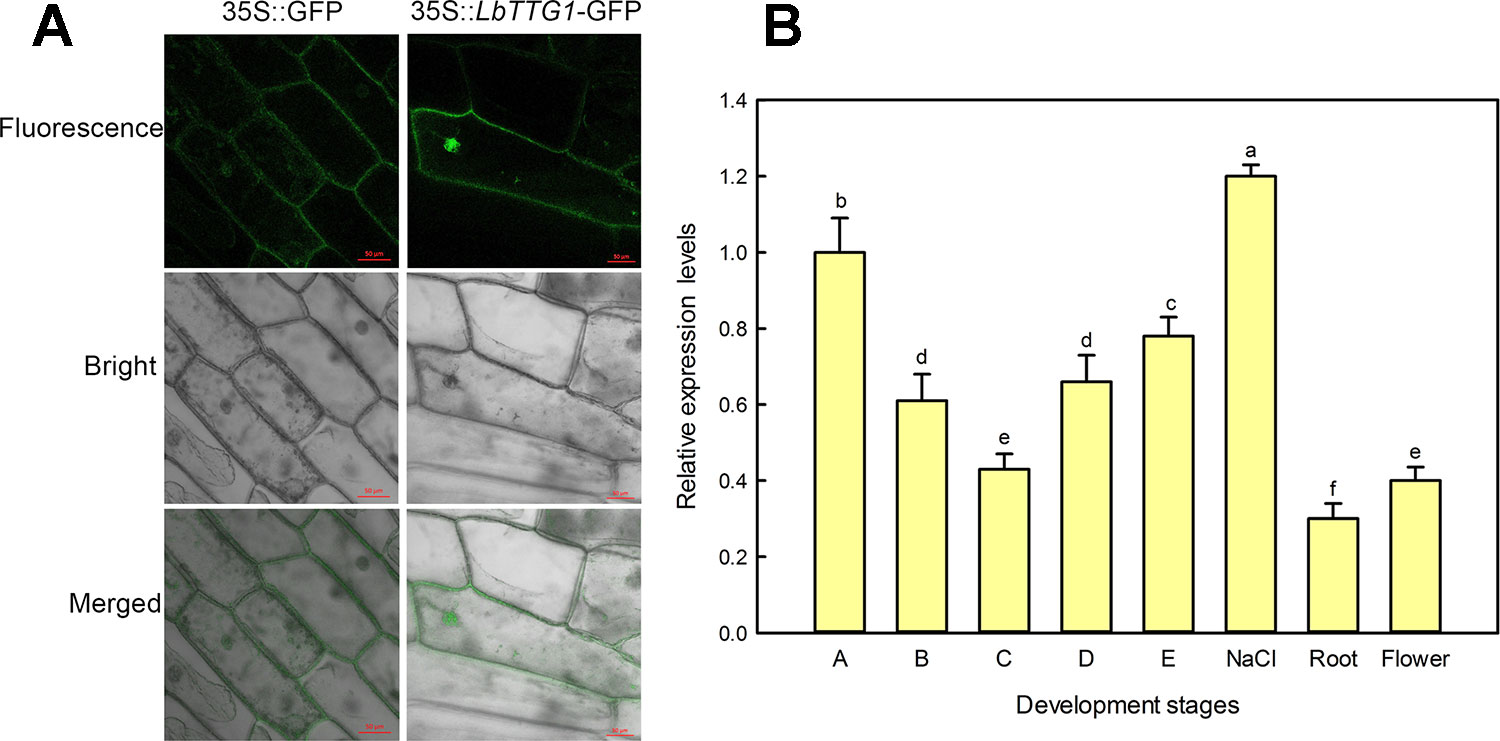
Figure 2 Subcellular localization by onion epidermis transformation and expression level of LbTTG1 at different development stages and conditions. (A) Subcellular localization of LbTTG1 after transformation of onion epidermis. GFP-LbTTG1 fusion protein was expressed in both the nucleus and cytoplasm. 35S::GFP is the empty control vector, and 35S::LbTTG1-GFP is the experiment group. Bars = 50 µm. (B) Expression levels of LbTTG1 at different developmental stages and conditions. Stages A-E were the continues leaf development stage. Stage A, undifferentiation, 4–5 days after sowing; stage B, salt gland differentiation, 6–7 days; stage C, stomata differentiation, 8–10 days; stage D, epidermis differentiation, 11–16 days; stage E, mature, ≥17 days after sowing. NaCl means the mature leaves isolated from the seedlings at stage E under 200 mM NaCl for 24 h. Roots were taken from the seedlings of stage E. Flower was the mixture of flower buds and complete flowers.
What is the level of LbTTG1 expression in different organs, different leaf development, and conditions? qPCR was applied to evaluate the effects at different developmental stages and conditions (Figure 2). During leaf development (A–E), the highest LbTTG1 expression levels were identified at stage A and the lowest at stage C (Figure 2). The highest expression of LbTTG1 is showed under 200 mM NaCl treatment. Basal expression level was observed in roots and flowers.
LbTTG1 Participated in Epidermis Development
To clarify the relationship between LbTTG1 and AtTTG1 and their roles in trichome formation, we heterologously expressed LbTTG1 in Col-0 (Figure 3A) and ttg1-13 (Figure 3B). We identified nine transgenic Col 35S::LbTTG1 lines with high expression of LbTTG1, L2, L3, L4, L9, L11, L16, L18, L22, and L26, whereas there was no expression in wild type plants (Figure 3C). Among the ttg 35S::LbTTG1 lines, distinctly high expression levels were observed in CL3, CL5, CL6, and CL9, whereas the mutant ttg1-13 showed no LbTTG1 expression (Figure 3C).
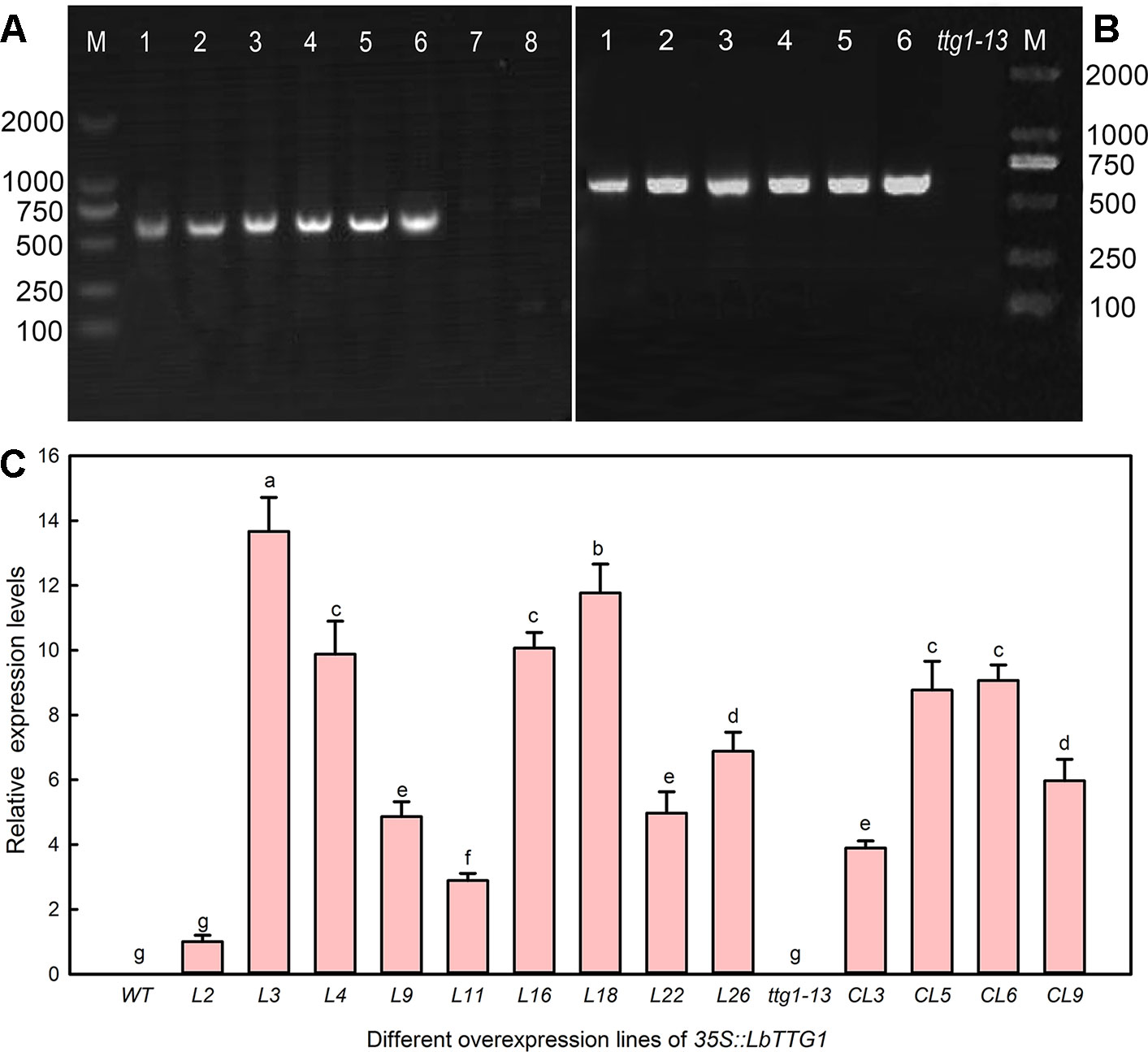
Figure 3 Identification of Arabidopsis lines heterologously expressing LbTTG1. (A) Genomic DNA PCR of Col 35S::LbTTG1 lines; lanes 1–6, different transgenic lines; lane 7, blank control with ddH2O as template; lane 8, negative control with wild type DNA as template. (B) Genomic DNA PCR of ttg 35S::LbTTG1 lines (lanes 1–6). (C) Expression levels of LbTTG1 examined using quantitative PCR in Col 35S::LbTTG1 and ttg 35S::LbTTG1. L numbers represent lines of Col 35S::LbTTG1, e.g. L2 and L3; CL numbers represent complementation lines of ttg 35S::LbTTG1. Data are means of three replicates ± SD; different letters indicate significant differences at P = 0.05 according to Duncan’s multiple range test.
AtTTG1 participates in development of the epidermis, including trichomes and root hairs. We therefore chose three lines harboring each transgene construct with high, medium, and low expression, respectively (L3, L16, and L2 of Col 35S::LbTTG1 and CL6, CL9, and CL3 of ttg 35S::LbTTG1), to compare the effect of the heterologous expression of LbTTG1 on the development of trichomes and root hairs. In order to determine the copies of LbTTG1 in the L. bicolor and transgenic Arabidopsis genome, Southern blotting was first investigated (Figure 4). Two hybridization bands were detected in the genomic of L. bicolor. In transgenic lines of Arabidopsis, L3 of Col 35S::LbTTG1 showed single copy insertion though expression was the highest. L16, L2 of Col 35S::LbTTG1, and CL6, CL9 of ttg 35S::LbTTG1, exhibited two copies. The above Southern blot analysis confirmed that L3, L16, L2, CL6, and CL9 were stably transformed with LbTTG1. However, no bands were detected in line CL3 by southern blotting while PCR verification was positive (Figure 3) for LbTTG1 gene, which may be not stable in transgenic Arabidopsis.
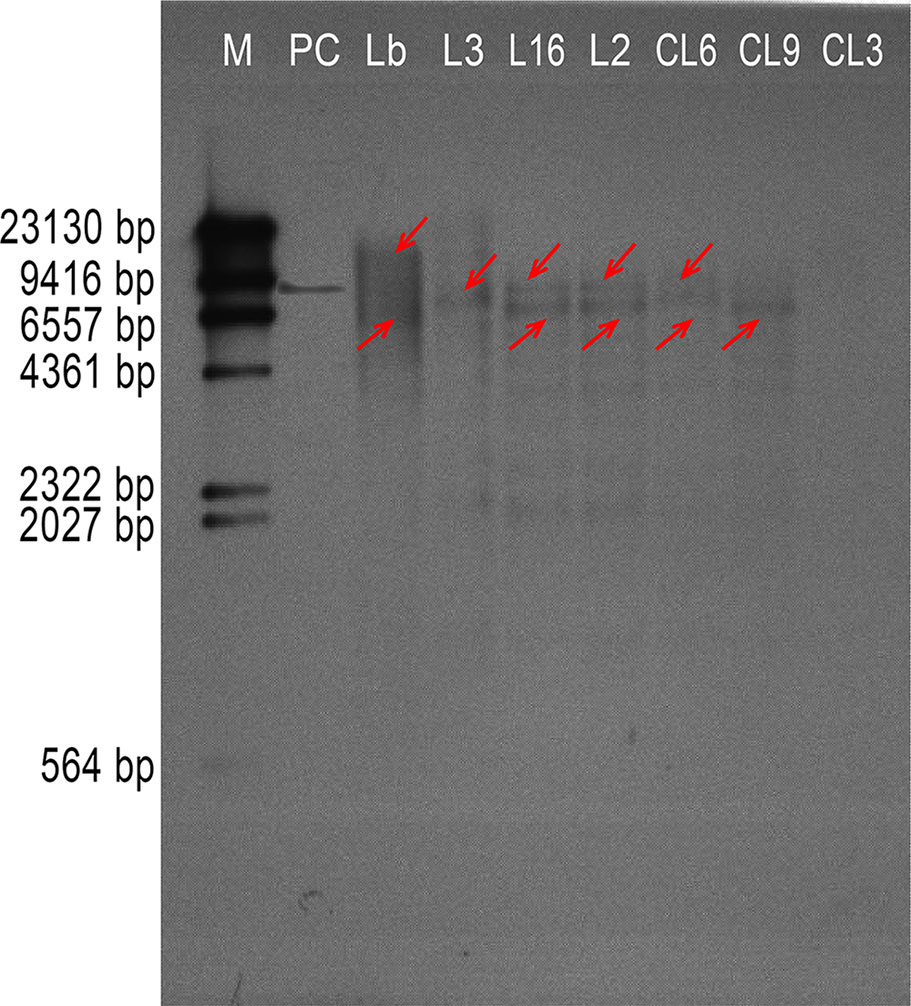
Figure 4 Southern hybridization of LbTTG1 in Limonium bicolor and transgenic lines of Arabidopsis. Lane M, DIG-labeled DNA molecular-weight marker; lane PC, positive control; lane Lb, genomic from L. bicolor; lane L3, L16, and L2, Arabidopsis Col 35S::LbTTG1; lane CL6, CL9, and CL3, Arabidopsis ttg 35S::LbTTG1. The red arrow indicated the possible gene copy.
We compared the number of trichomes per leaf in wild type, ttg 1-13, Col 35S::LbTTG1, and ttg 35S::LbTTG1 plants (Figure 5). Lines heterologously expressing LbTTG1 in a wild type background showed enhanced trichome formation; however, the expression level of LbTTG1 did not have a dose effect on trichome formation. The ttg1-13 mutant showed no trichome development, while heterologous expression of LbTTG1 in these mutants returned trichome number to wild type levels. Line CL9 showed more trichome differentiation than the wild type; however, no significant expression level dose effect was observed. In addition, heterologous expression of LbTTG1 had no effect on the number of trichome branches.
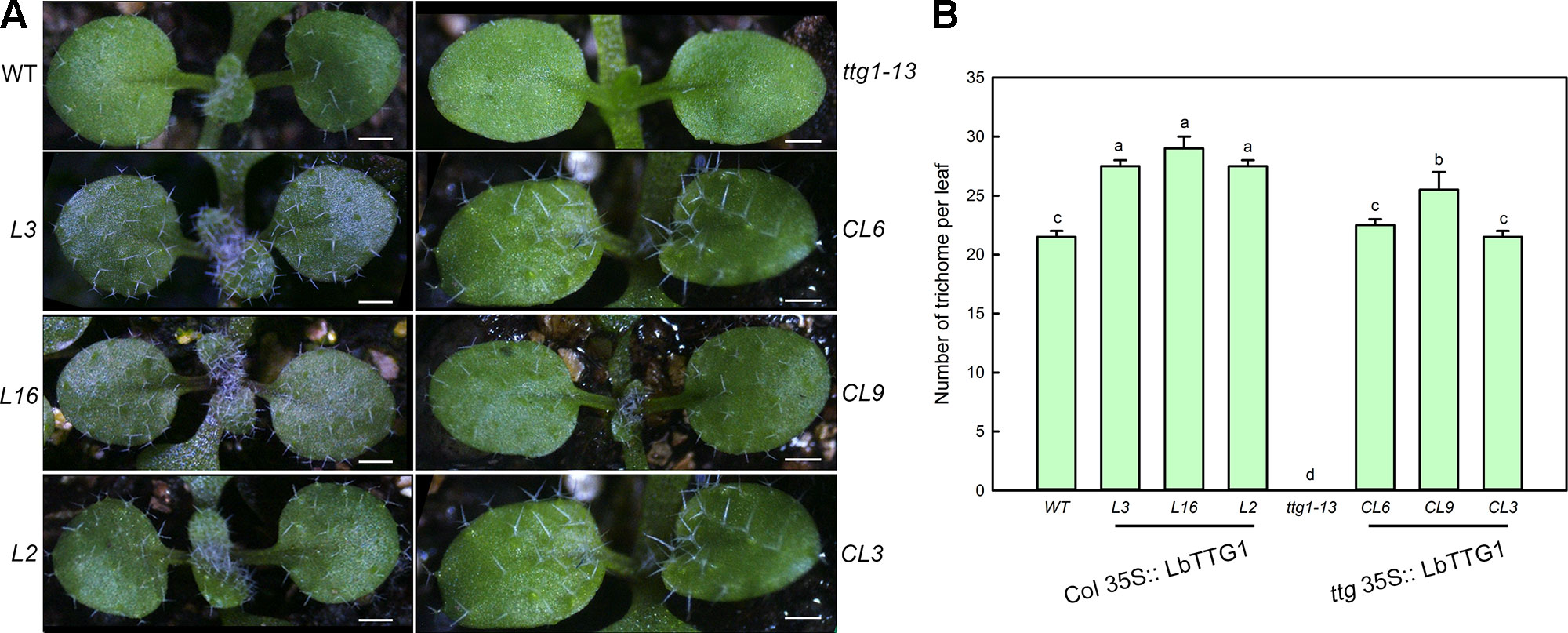
Figure 5 Trichome development was enhanced after transformation of LbTTG1. (A) Trichomes on the first two rosette leaves of Col-0 wild type (WT), ttg1-13, Col 35S::LbTTG1 (L3, L16, and L2), and ttg 35S::LbTTG1 (CL6, CL9, and CL3). Photographs show 2-week-old soil-grown seedlings. (B) Trichome density on the first two rosette leaves of WT, ttg1-13, Col 35S::LbTTG1, and ttg 35S::LbTTG1. Data are mean ± SD of 10 plants; different letters indicate significant differences at P = 0.05 according to Duncan’s multiple range test.
We also measured the development of root hairs in the LbTTG1 expression lines and complementation lines (Figure 6). The TTG1 mutant ttg1-13 had considerably more root hairs than the wild type, whereas all Col 35S::LbTTG1 lines (L3, L16, L2) had fewer. The numbers of root hairs in CL6, CL9, and CL3 ttg 35S::LbTTG1 plants were similar to that in the control (Figure 6). Root hair length showed a similar trend; root hairs were longest in ttg1-13 plants and similar in the complementation lines and wild type. Root hair development was significantly affected by heterologous expression of LbTTG1 in both the wild type and ttg1-13 mutant.
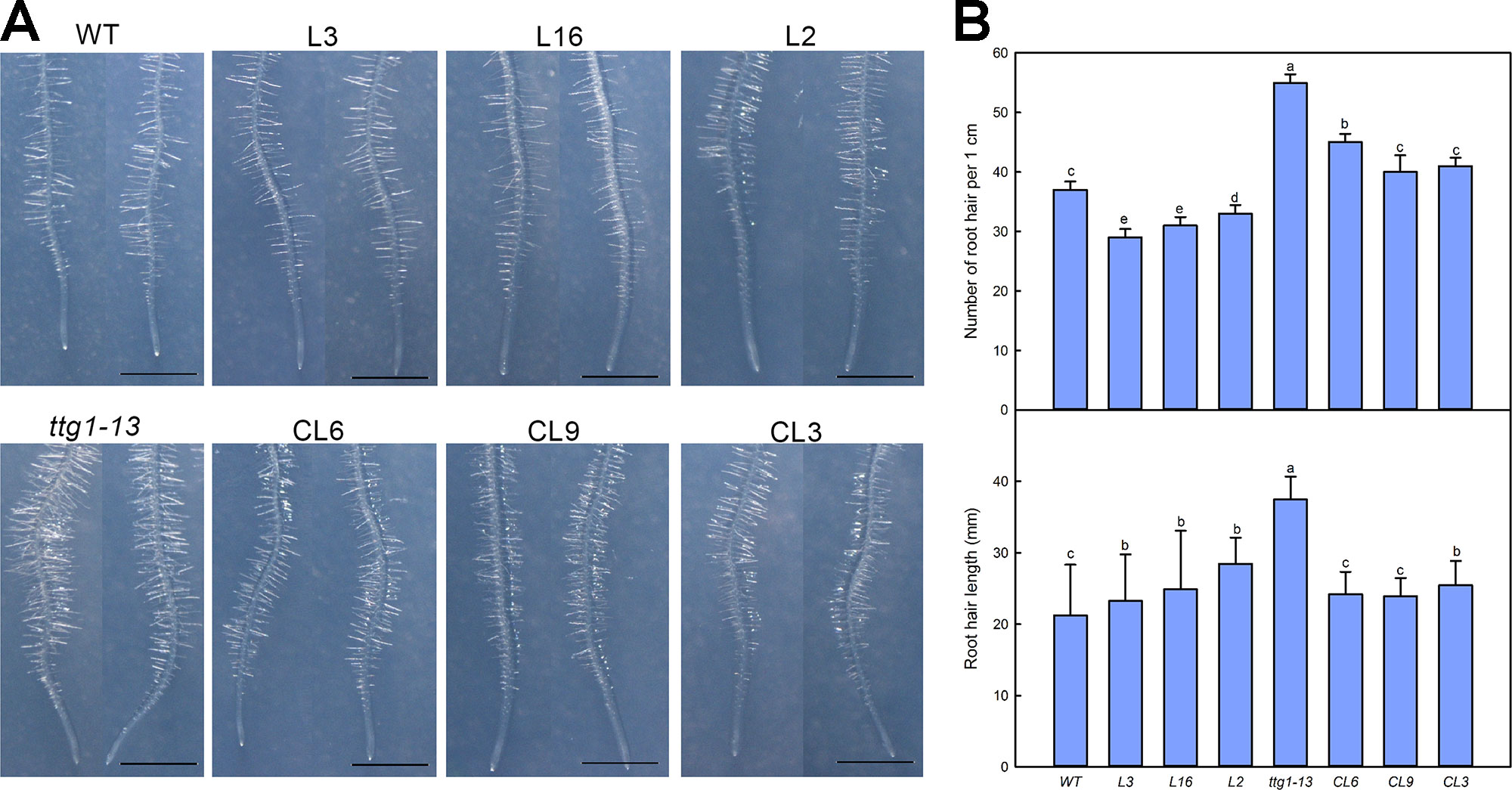
Figure 6 Root hair development of Col 35S::LbTTG1 and ttg 35S::LbTTG1. (A) Phenotypes of root hairs of WT, ttg1-13, Col 35S::LbTTG1 (L3, L16, and L2), and ttg 35S::LbTTG1 (CL6, CL9, and CL3) after culture for 5 days on MS medium. (B) The same position of each root (region from 2 cm to 3 cm from root tip) was chosen for calculating root hair number and length from 10 or 20 plants from each line. Data for root hair number are mean ± SD of 10 plants. Data for root hair length are mean ± SD of 20 plants; different letters indicate significant differences at P = 0.05 according to Duncan’s multiple range test.
Transgenic Lines of LbTTG1 Behave Well in Both Germination Stage and Seedling Stage Under NaCl Treatment
Given that root hairs of transgenic lines were significantly affected, seed germination was further measured using different lines of Col 35S::LbTTG1 and ttg 35S::LbTTG1 (Figure 7). Seeds of different lines were treated with different concentrations of NaCl (0, 50, 100, and 150 mM) to analyze the effect of NaCl on germination at 5 days after sowing (Figure 7). With increasing NaCl concentration, seeds of all lines showed decreased germination. The Col 35S::LbTTG1 lines (L3, L16, L2) showed similar germination trends to the wild type. The mutant ttg1-13 showed much poorer germination than the wild type under all treatments, while the ttg 35S::LbTTG1 complementation lines (CL6, CL9, CL3) showed higher germination than ttg1-13 but still poorer germination than the wild type. Root length showed similar trends under the different NaCl treatments (Figures 7A, B).
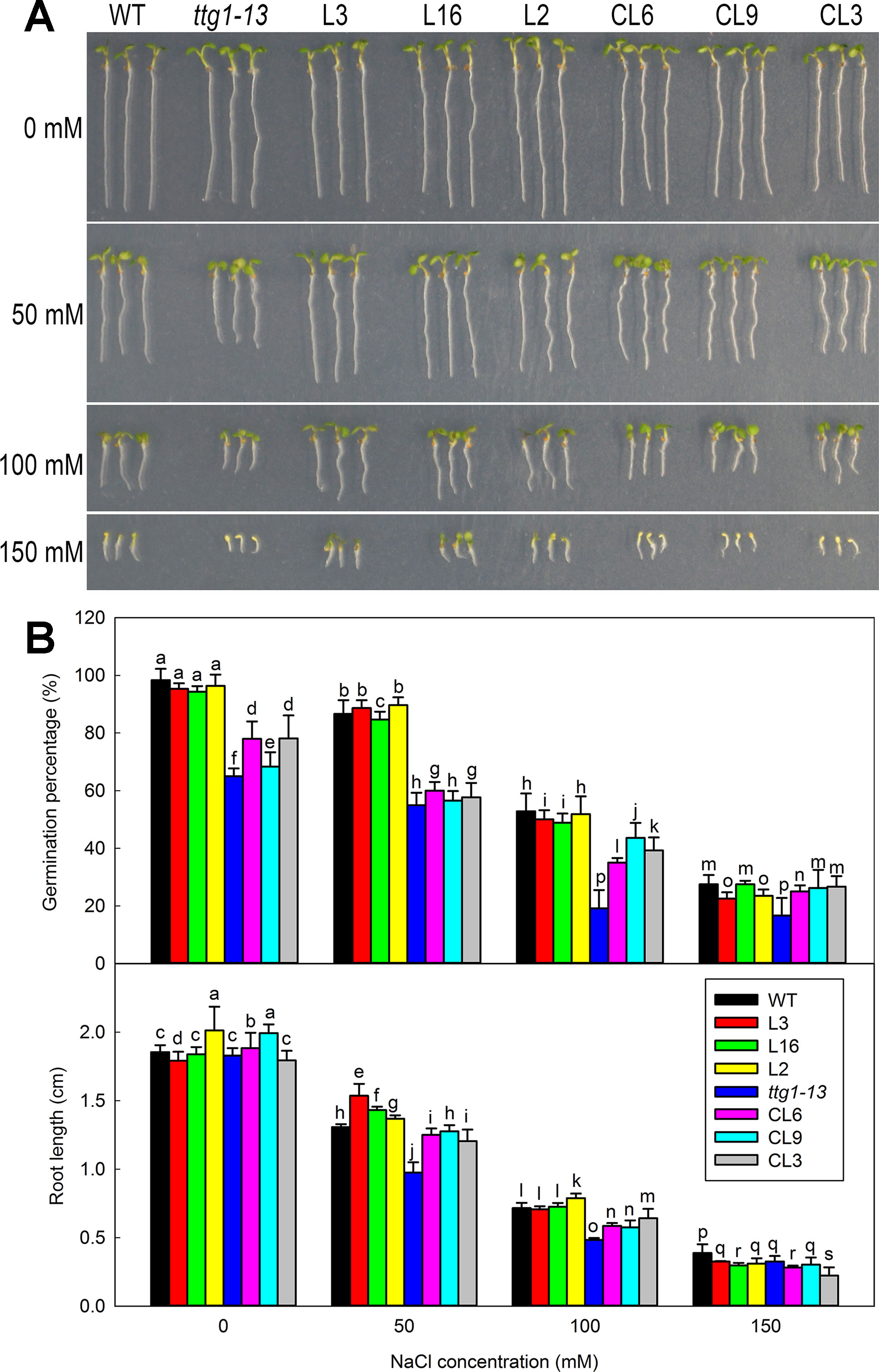
Figure 7 Germination of Col 35S::LbTTG1 and ttg 35S::LbTTG1 seeds under different NaCl concentrations. (A) Uniform, 1-day-old seedlings were transplanted to medium containing different NaCl concentrations (0, 50, 100, and 150 mM). Phenotypes of WT, ttg1-13, Col 35S::LbTTG1 (L3, L16, and L2), and ttg 35S::LbTTG1 (CL6, CL9, and CL3) in 5-day-old seedlings. (B) Germination percentage calculated 24 h after sowing on medium containing different NaCl concentrations (0, 50, 100, and 150 mM). Fifty seeds of each line were sowed in each treatment, and three replicates were performed. Data for germination percentage are mean ± SD. Root length of five-day-old seedlings was calculated using ImageJ software. Data for root length are mean ± SD of 20 plants per line; different letters indicate significant differences at P = 0.05 according to Duncan’s multiple range test.
There was no significant difference in the growth of all lines under no-salt conditions, but growth of ttg1-13 showed a distinct decrease with increasing NaCl concentration (Figure 8) and no true leaf was seen under 150 mM treatment. Different from ttg1-13, seedlings of Col 35S::LbTTG1 and ttg 35S::LbTTG1 performed better, with L16 and L2 especially producing greater biomass than the wild type under 150 mM NaCl (Figure 8). This indicated that the LbTTG1 transgenic lines had enhanced salt tolerance. Better growth (Figure 8) and increased biomass (Figure 8) of complementation lines compared with ttg1-13 also supported this conclusion. Given that 100 mM NaCl treatment dramatically inhibited the growth and biomass of all lines, we chose to use the 0 and 100 mM NaCl conditions for further in-depth measurement of physiological indicators, including Na+ and K+, proline, MDA, GST activity, and soluble sugar (Figure 9). No distinct differences were observed among different lines under control conditions; however, under 100 mM NaCl treatment, ttg1-13 showed the highest Na+ content and MDA content and the lowest K+, proline, GST activity, and soluble sugar contents. By contrast, the transgenic lines had the lowest Na+ content and MDA content and the highest K+, proline, GST activity, and soluble sugar contents. These results indicated that the highly salt-tolerant transgenic lines benefited from accumulation of high levels of organic osmotic adjustment substances, low ion toxicity, and high GST activity.
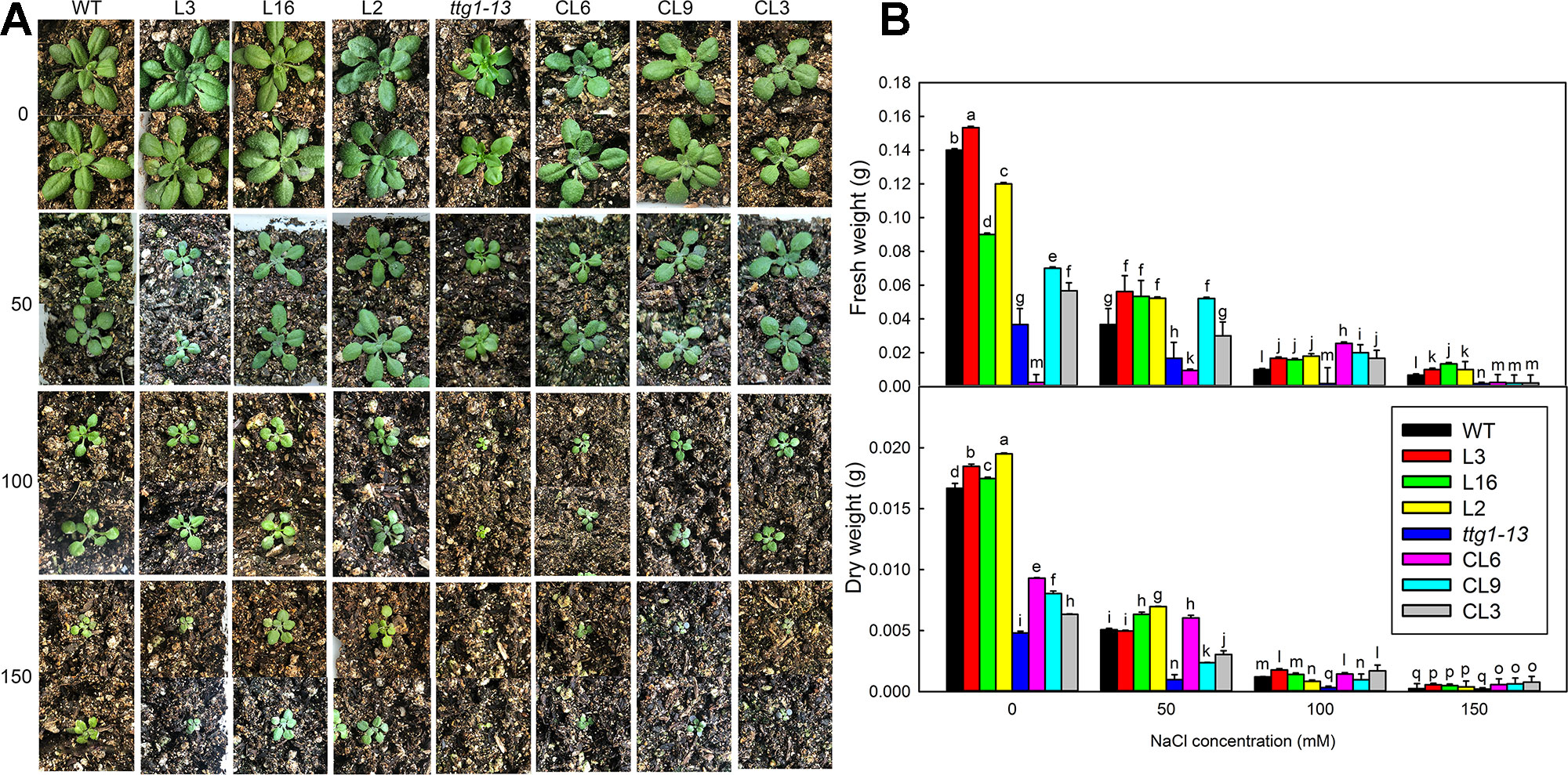
Figure 8 The growth and biomass of different strains under different NaCl treatments. (A) Two-week-old seedlings of WT, ttg1-13, Col 35S::LbTTG1 (L3, L16, and L2), and ttg 35S::LbTTG1 (CL6, CL9, and CL3) under different NaCl treatments (0, 50, 100, and 150 mM). (B) Fresh weight and dry weight of intact seedlings under different NaCl treatments. Data are means ± SD of five replicates; different letters indicate significant differences at P = 0.05 according to Duncan’s multiple range test.
Expression Levels of Epidermis Differentiation and Stress-Related Marker Genes
Transformation of LbTTG1 conferred high heterologous expression levels of LbTTG1 in Col 35S::LbTTG1 and ttg 35S::LbTTG1 plants (Figure 3). Considering that trichome differentiation was enhanced (Figure 5) and root hair development was inhibited (Figure 6) in transgenic lines, we examined key genes involved in epidermis differentiation: AtTTG1, AtGL3, AtEGL3, AtGL1, AtCPC, and AtTRY (Figure 10). LbTTG1 transformation did not alter the expression of AtTTG1 from that of the wild type or ttg1-13. No significant differences were observed in the expression of AtGL3, AtCPC, or AtTRY among different lines. Two genes (AtEGL3 and AtGL1) showed their highest expression in transgenic lines; these proteins may directly combine with LbTTG1 to promote trichome development and inhibit root hair differentiation.
To investigate why transformation of LbTTG1 significantly improved the salt tolerance of Arabidopsis, we chose seven stress-related marker genes for preliminary investigation (Figures 10B, C): AtSOS1 (Salt overly sensitive 1), AtSOS2 (Salt overly sensitive 2), AtSOS3 (Salt overly sensitive 3), AtP5CS1 (Δ1-pyrroline-5-carboxylate synthetase 1), AtP5CS2 (Δ1-pyrroline-5-carboxylate synthetase 2), AtGSTU5 (GST class tau 5), and AtAREB1 (abscisic acid-responsive element binding protein 1). Under short-term 100 mM NaCl treatment, high expression levels of AtSOS1, AtSOS2, AtSOS3, AtP5CS1, AtP5CS2, and AtGSTU5 were seen in the transgenic expression lines in the wild type background (L3, L16, and L2), while the lowest expression levels occurred in ttg1-13, and expression levels in the complementation lines (CL6, CL9, and CL3) were intermediate between the two (Figures 10B, C). A different trend in expression was seen for the abscisic acid-related gene AREB1, with highest expression in ttg1-13 and relatively lower expression in transgenic lines (Figure 10C).
Discussion
LbTTG1 of L. bicolor contains the similar WD40-repeat protein domains as TTG1 proteins from other species (Brueggemann et al., 2010), which may determine epidermis differentiation into trichomes and root hairs (Schellmann and Hulskamp, 2004). Heterologous expression of LbTTG1 in Arabidopsis increased trichome number (Figure 5) and decreased root hair number (Figure 6). Moreover, we observed no dose effect of LbTTG1 expression on trichome and root hair development, indicating that LbTTG1 can enhance the function of AtTTG1 in the positive regulation of trichome formation and negative regulation of root hair development, and a low level of LbTTG1 is sufficient for the formation of the TTG1-EGL3-GL1 complex with EGL3 and GL1 to initiate trichome and root hair differentiation. We used AtTTG1 deletion mutants of Arabidopsis for the heterologous expression of LbTTG1 to illustrate the homology between LbTTG1 and AtTTG1 in epidermis differentiation. Although different complementation lines showed different LbTTG1 expression levels and gene copies, we observed distinct complementation phenotypes similar to those of the wild type, with more trichomes (Figure 5) and fewer root hairs (Figure 6) than in the mutant. All these results indicate that, similar to TTG1 proteins from other species, LbTTG1 and AtTTG1 have homologous functions in epidermis differentiation, probably due to their common WD40 domain (Ben-Simhon et al., 2011; Schaart et al., 2013; Dressel and Hemleben, 2009).
Besides Arabidopsis, the function of TTG1 protein is well illustrated in Setaria italica, Artemisia annua, and others. Similar to what we observed here, SiTTG1 of S. italica (Liu et al., 2017) and AaWD40 from A. annua (Wang et al., 2016) can complement the trichome phenotype of ttg1-13 to restore the wild type phenotype of Arabidopsis, and the latter can enhance the synthesis of anthocyanin. Similar results of enhanced trichome formation were seen in transgenic lines of CsWD40 from Camellia sinensis (Liu et al., 2018). TTG1 genes in different species thus show high conservation in controlling epidermis differentiation. The WD40-repeat domain may be responsible for this, but further transformation with WD40-repeat domain deletions of LbTTG1 will allow deeper understanding of the mechanism. How does LbTTG1 participate in epidermis differentiation? Two key genes (AtEGL3 and AtGL1) involved in epidermis development were highly expressed in transgenic lines (Figure 10). (Zhao et al., 2008) reported that AtTTG1 combines with EGL3 and GL1 to initiate trichome formation. The current study using heterologous expression in Arabidopsis suggested that LbTTG1 may also combine with EGL3 and GL1 to promote trichome formation and inhibit root hair differentiation. Transcriptome and in vitro interaction analyses will provide details of how the downstream and interacting proteins function in the future.
In addition to its function in epidermis differentiation, which is similar to that of its homologs in other species, a specific function of salt tolerance was seen in LbTTG1 transgenic lines in both wild type and ttg1-13 backgrounds. Interestingly, ttg1-13 showed sensitivity to salt, which may be due to increased root hair development (Figure 6), whereas the complementation lines showed increased germination over that of ttg1-13 under increasing NaCl treatment (Figure 7). Similar trends were seen in physiological indicators, including MDA and ion contents. This may be explained in two ways. On the one hand, we can obtain clues from ion contents under different treatments (Figure 9). More root hairs lead to more Na+ being absorbed into the root, so the ttg1-13 mutant showed salt sensitivity due to accumulated Na+ and ionic homeostasis disorder; transgenic lines had fewer root hairs, so these plants showed salt tolerance. On the other hand, in addition to the WD40-repeat domain common to all TTG1 proteins, LbTTG1 was also speculated to have a specific domain (maybe the low complexity domain in Figure 1C) that may determine the specific function of salt tolerance in this halophyte, which had highest expression level under NaCl treatment (Figure 2). We examined expression levels of marker genes for salt tolerance in LbTTG1 transgenic lines (Figures 10B, C), including Na+ efflux, osmotic adjustment, active oxygen scavenging system, and abscisic acid synthesis. SOS1, SOS2, and SOS3 was up-regulated in transgenic lines (Figure 10); SOS3 showed the most significant increase among the three, which may combine with SOS2, and then active SOS1 (a Na+/H+ antiporter located in the plasma membrane) to actively transport Na+ to the outside of the cell, reducing harm to protoplasts (Shi et al., 2000; Qiu et al., 2002; Zhu, 2002). Increased proline accumulation in transgenic lines compared with the wild type and ttg1-13 (Figure 9) may be due to high expression levels of AtP5CS1 and AtP5CS2 (Figure 10), which both controlled the synthesis of proline (Mattioli et al., 2010). Organic osmolytes such as proline are synthesized for osmotic adjustment under salt stress (Székely, 2004), so transgenic lines of LbTTG1 performed well under NaCl treatment. In addition to ion stress and osmotic stress, oxidative stress also causes damage under salt stress conditions (Khan and Ashraf, 2008). GST, one of the indicators of antioxidant enzyme system activity (Wagner et al., 2002), showed high expression in transgenic lines (Figure 9), which was likely due to the high expression level of AtGSTU5 (Figure 10), thus enhanced the salt tolerance of the transgenic lines. Salt stress often induces synthesis of abscisic acid and up-regulation of AREB1 (Fujita et al., 2005; Narusaka et al., 2003). Under salt treatment, ttg1-13 showed higher AREB1 expression levels than transgenic lines, which might also explain the high salt tolerance of transgenic lines.
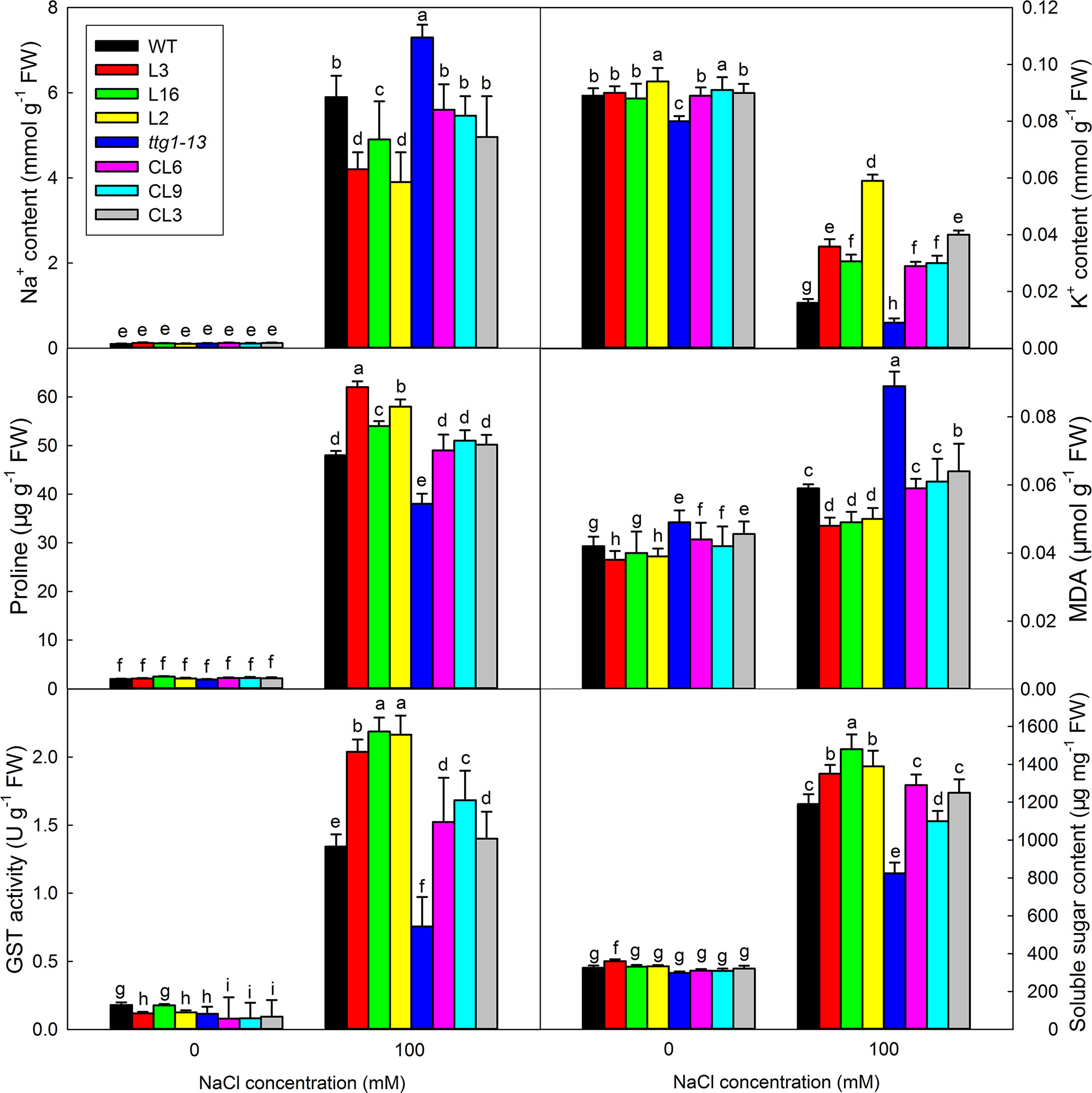
Figure 9 Ion contents, proline, MDA, GST activity, and soluble sugar contents of seedlings under different NaCl treatments. Seedlings under 0 and 100 mM NaCl treatments were pooled for measurement of Na+ and K+ contents, proline, MDA, GST activity, and soluble sugar contents. Data are means ± SD of five replicates; different letters indicate significant differences at P = 0.05 according to Duncan’s multiple range test.
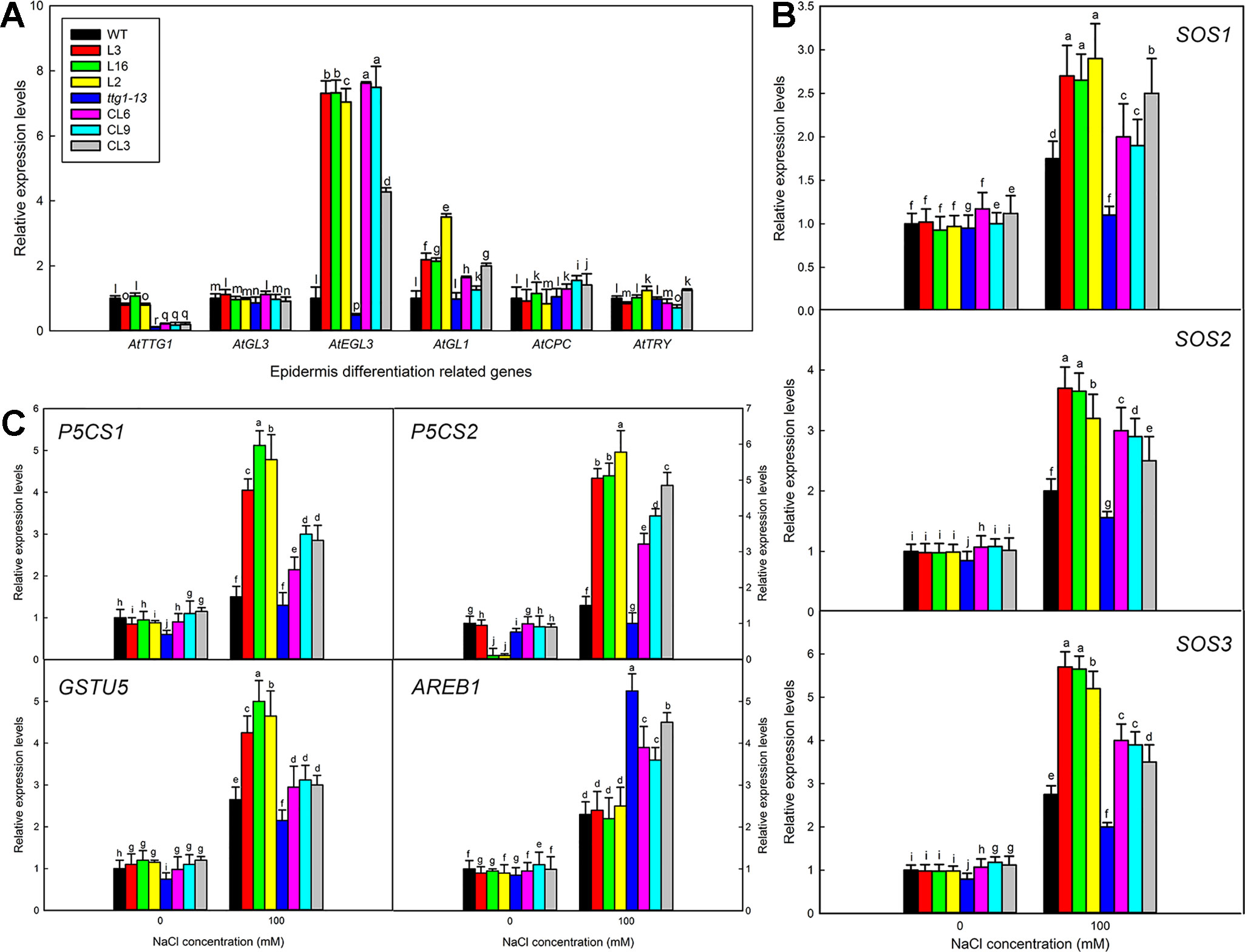
Figure 10 Expression levels of trichome formation and stress-related marker genes. (A) Relative expression levels of AtTTG1, AtGL3, AtEGL3, AtGL1, AtCPC, and AtTRY in 5-day-old seedlings. (B) Relative expression levels of AtSOS1, AtSOS2, AtSOS3, AtAREB1, AtP5CS1, AtP5CS2, and AtGSTU5. Two-week-old seedlings of all lines germinated on MS medium were transferred to 1/2 MS liquid medium containing 100 mM NaCl for 0 and 3 h. Expression of each gene was measured in three replicate biological experiments; different letters indicate significant differences at P = 0.05 according to Duncan’s multiple range test.
In summary, transgenic lines of Arabidopsis heterologously expressing LbTTG1 showed improved salt tolerance at both the germination and seedling stages. The epidermis of transformed plants displayed enhanced trichome and reduced root hair development. A specific domain of LbTTG1 and the common WD40-repeat domain may be responsible for these phenotypes. Given that a transformation system is available for L. bicolor, further studies using the CRISPR-cas9 system will be performed to verify the function of LbTTG1.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
FY and BW designed research. BL, HZ, and XW performed research. BL and GH analyzed data. FY wrote the paper. BW revised the paper.
Funding
This work was supported by the NSFC (National Natural Science Research Foundation of China, project nos. 31600200; 31570251; 31770288), Shandong Province key research and development plan (2015ZDJS03002; 2017CXGC0313), the Natural Science Research Foundation of Shandong Province (ZR2014CZ002), and the Higher Educational Science and Technology Program of Shandong Province (J15LE08).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01456/full#supplementary-material
References
Ben-Simhon, Z., Judeinstein, S., Nadler-Hassar, T., Trainin, T., Bar-Ya’akov, I., Borochov-Neori, H., et al. (2011). A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 234 (5), 865–881. doi: 10.1007/s00425-011-1438-4
Brueggemann, J., Weisshaar, B., Sagasser, M. (2010). A WD40-repeat gene from Malus×domestica is a functional homologue of Arabidopsis thaliana TRANSPARENT TESTA GLABRA1. Plant Cell Rep. 29 (3), 285–294. doi: 10.1007/s00299-010-0821-0
Chen, C., Yin, S., Liu, X., Liu, B., Yang, S., Xue, S., et al. (2016). CsTTG1 Encodes a WD-repeat Protein That Regulates Fruit Wart Formation in Cucumis sativus through Interaction with the Homeodomain-leucine Zipper I Protein Mict. Plant Physiol. 171(2), 1156–1168. doi: 10.1104/pp.16.00112
Clough, S. J., Bent, A. F. (2010). Floral dip: a simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 16 (6), 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Deng, Y., Feng, Z., Yuan, F., Guo, J., Suo, S., Wang, B. (2015). Identification and Functional Analysis of the Autofluorescent Substance in Limonium bicolor Salt Glands. Plant Physiol. Biochem. 97, 20–27. doi: 10.1016/j.plaphy.2015.09.007
Ding, F., Chen, M., Sui, N., Wang, B. S. (2010). Ca2+ significantly enhanced development and salt-secretion rate of salt glands of Limonium bicolor under NaCl treatment. South Afr. J. Bot. 76 (1), 95–101. doi: 10.1016/j.sajb.2009.09.001
Dinler, B. S., Antoniou, C., Fotopoulos, V. (2014). Interplay between GST and nitric oxide in the early response of soybean (Glycine max L.) plants to salinity stress. J. Plant Physiol. 171 (18), 1740–1747. doi: 10.1016/j.jplph.2014.07.026
Dressel, A., Hemleben, V. (2009). Transparent Testa Glabra 1 (TTG1) and TTG1-like genes in Matthiola incana R. Br. and related Brassicaceae and mutation in the WD-40 motif. Plant Biol. 11 (2), 204–212. doi: 10.1111/j.1438-8677.2008.00099.x
Feng, Z., Deng, Y., Zhang, S., Liang, X., Yuan, F., Hao, J., et al. (2015). K+ accumulation in the cytoplasm and nucleus of the salt gland cells of Limonium bicolor accompanies increased rates of salt secretion under NaCl treatment using NanoSIMS. Plant Sci. 238, 286–296. doi: 10.1016/j.plantsci.2015.06.021
Feng, Z., Sun, Q., Deng, Y., Sun, S., Zhang, J., Wang, B. (2014). Study on pathway and characteristics of ion secretion of salt glands of Limonium bicolor. Acta Physiologiae Plantarum 36 (10), 2729–2741. doi: 10.1007/s11738-014-1644-3
Flowers, T. J., Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytol. 179 (4), 945–963. doi: 10.1111/j.1469-8137.2008.02531.x
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 (12), 3470–3488. doi: 10.1105/tpc.105.035659
Guo, J., Li, Y., Han, G., Song, J., Wang, B. S. (2018). NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Funct. Plant Biol. 44 (3), 350–361. doi: 10.1071/FP17181
Guo, J., Wei, Z., Lu, Z., Hao, L., Li, H., Feng, G. (2015). Isolation and Functional Analysis of Chalcone Isomerase Gene from Purple-Fleshed Sweet Potato. Plant Mol. Biol. Rep. 33 (5), 1451–1463. doi: 10.1007/s11105-014-0842-x
Han, G., Yuan, F., Guo, J., Zhang, Y., Sui, N., Wang, B. (2019). AtSIZ1 improves salt tolerance by maintaining ionic homeostasis and osmotic balance in Arabidopsis. Plant Sci. 285, 55–67. doi: 10.1016/j.plantsci.2019.05.002
Khan, A., Ashraf, M. (2008). Exogenously applied ascorbic acid alleviates salt-induced oxidative stress in wheat. Environ. Exp. Bot. 63 (1-3), 224–231. doi: 10.1016/j.envexpbot.2007.10.018
Kirik, V., Simon, M., Huelskamp, M., Schiefelbein, J. (2004). The enhancer of try and cpc1 gene acts redundantly with triptychon and caprice in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268 (2), 506–513. doi: 10.1016/j.ydbio.2003.12.037
Leng, B. Y., Yuan, F., Dong, X. X., Wang, J., Wang, B. S. (2018). Distribution pattern and salt excretion rate of salt glands in two recretohalophyte species of Limonium (Plumbaginaceae). South Afr. J. Of Bot. 115, 74–80. doi: 10.1016/j.sajb.2018.01.002
Li, Q., Cao, C., Zhang, C., Zheng, S., Wang, Z., Wang, L., et al. (2015). The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J. Exp. Bot. 66 (9), 2515–2526. doi: 10.1093/jxb/erv046
Liu, K., Qi, S., Li, D., Jin, C., Gao, C., Duan, S., et al. (2017). TRANSPARENT TESTA GLABRA 1 ubiquitously regulates plant growth and development from Arabidopsis to foxtail millet (Setaria italica). Plant Sci. 254, 60–69. doi: 10.1016/j.plantsci.2016.10.010
Liu, Y., Hou, H., Jiang, X., Wang, P., Dai, X., Chen, W., et al. (2018). A WD40 Repeat Protein from Camellia sinensis Regulates Anthocyanin and Proanthocyanidin Accumulation through the Formation of MYB-bHLH-WD40 Ternary Complexes. Int. J. Mol. Sci. 19 (6), 1686. doi: 10.3390/ijms19061686
Mattioli, R., Falasca, G., Sabatini, S., Altamura, M. M., Costantino, P., Trovato, M. (2010). The proline biosynthetic genes P5CS1 and P5CS2 play overlapping roles in Arabidopsis flower transition but not in embryo development. Physiol. Plant 137 (1), 72–85. doi: 10.1111/j.1399-3054.2009.01261.x
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34 (2), 137–148. doi: 10.1046/j.1365-313X.2003.01708.x
Payne, C. T., Zhang, F., Lloyd, A. M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156 (3), 1349–1362.
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. 99 (12), 8436–8441. doi: 10.1073/pnas.122224699
Schaart, J. G., Dubos, C., Romero De La Fuente, I., Van Houwelingen, A. M., De Vos, R. C., Jonker, H. H., et al. (2013). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria×ananassa) fruits. New Phytol. 197 (2), 454–467. doi: 10.1111/nph.12017
Schellmann, S., Hulskamp, M. (2004). Epidermal differentiation: trichomes in Arabidopsis as a model system. Int. J. Dev. Biol. 49 (5–6), 579–584. doi: 10.1387/ijdb.051983ss
Shi, H. Z., Ishitani, M., Kim, C., Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. 97 (12), 6896–6901. doi: 10.1073/pnas.120170197
Song, J., Shi, W., Liu, R., Xu, Y., Sui, N., Zhou, J., et al. (2017). The role of the seed coat in adaptation of dimorphic seeds of the euhalophyte Suaeda salsa to salinity. Plant Species Biol. 32 (2), 107–114. doi: 10.1111/1442-1984.12132
Song, J., Wang, B. (2015). Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 194, 541–553. doi: 10.1093/aob/mcu194
Sui, N., Tian, S., Wang, W., Wang, M., Fan, H. (2017). Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front. Plant Sci. 8, 1337. doi: 10.3389/fpls.2017.01337
Sun, W., Cao, Z., Li, Y., Zhao, Y., Zhang, H. (2007). A simple and effective method for protein subcellular localization using Agrobacterium-mediated transformation of onion epidermal cells. Biologia 62 (5), 529–532. doi: 10.2478/s11756-007-0104-6
Székely, G. (2004). The role of proline in Arabidopsis thaliana osmotic stress response. Acta Biologica Szegediensis 48 (1-4), 81–81.
Tominaga-Wada, R., Nukumizu, Y. (2012). Expression analysis of an R3-Type MYB transcription factor CPC-LIKE MYB4 (TRICHOMELESS2) and CPL4-related transcripts in Arabidopsis. Int. J. Mol. Sci. 13 (3), 3478–3491. doi: 10.3390/ijms13033478
Tominaga-Wada, R., Wada, T. (2016). The Arabidopsis CAPRICE protein fused to the VP16 transcriptional activation domain alters root hair and trichome development. Plant Biotechnol. 16, 0506. doi: 10.5511/plantbiotechnology.16.0506a
Wagner, U., Edwards, R., Dixon, D. P., Mauch, F. (2002). Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 49 (5), 515–532. doi: 10.1023/A:1015557300450
Walker, A. R., Davison, P. A., Bolognesi-Winfield, A. C., James, C. M., Srinivasan, N., Blundell, T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 (7), 1337–1349. doi: 10.1105/tpc.11.7.1337
Wang, W., Zhang, Q., Guo, D. (2016). An Artemisia WD40-Repeat Gene Regulates Multiple Cellular Functions in Arabidopsis. J. Biosci. Medicines 4 (05), 30. doi: 10.4236/jbm.2016.45003
Yuan, F., Chen, M., Leng, B. Y., Wang, B. (2013). An efficient autofluorescence method for screening Limonium bicolor mutants for abnormal salt gland density and salt secretion. South Afr. J. Bot. 88, 110–117. doi: 10.1016/j.sajb.2013.06.007
Yuan, F., Lyu, M. J. A., Leng, B. Y., Zhu, X. G., Wang, B. S. (2016). The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol. Biol. 91 (3), 241–256. doi: 10.1007/s11103-016-0460-0
Yuan, F., Lyv, M. J., Leng, B. Y., Zheng, G. Y., Feng, Z. T., Li, P. H., et al. (2015). Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 38, 1637–1657. doi: 10.1111/pce.12514
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130 (20), 4859–4869. doi: 10.1242/dev.00681
Zhao, J.-L., Wang, Y.-L., Yao, D.-Q., Zhu, W.-Y., Chen, L., He, H.-L., et al. (2015). Transcriptome profiling of trichome-less reveals genes associated with multicellular trichome development in Cucumis sativus. Mol. Genet. Genomics 290 (5), 2007–2018. doi: 10.1007/s00438-015-1057-z
Zhao, M., Morohashi, K., Hatlestad, G., Grotewold, E., Lloyd, A. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135 (11), 1991–1999. doi: 10.1242/dev.016873
Keywords: Arabidopsis, heterologous expression, Limonium bicolor, root hair, salt stress, trichome, WD40-repeat protein
Citation: Yuan F, Leng B, Zhang H, Wang X, Han G and Wang B (2019) A WD40-Repeat Protein From the Recretohalophyte Limonium bicolor Enhances Trichome Formation and Salt Tolerance in Arabidopsis. Front. Plant Sci. 10:1456. doi: 10.3389/fpls.2019.01456
Received: 30 July 2019; Accepted: 18 October 2019;
Published: 12 November 2019.
Edited by:
Oscar Vicente, Polytechnic University of Valencia, SpainReviewed by:
Yucheng Wang, Northeast Forestry University, ChinaMingxun Chen, Northwest A&F University, China
Copyright © 2019 Yuan, Leng, Zhang, Wang, Han and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Yuan, eXVhbmZhbmdAc2RudS5lZHUuY24=; Baoshan Wang, YnN3YW5nQHNkbnUuZWR1LmNu
†These authors have contributed equally to this work
 Fang Yuan
Fang Yuan Bingying Leng
Bingying Leng Haonan Zhang
Haonan Zhang Xi Wang1
Xi Wang1 Guoliang Han
Guoliang Han Baoshan Wang
Baoshan Wang