- 1Department of Plant Science and Technology, Chung-Ang University, Anseong, South Korea
- 2Department of Systems Biotechnology, Chung-Ang University, Anseong, South Korea
Xanthomonas axonopodis pv. glycines (Xag) is a Gram-negative bacterium that causes bacterial pustule disease in soybean. To acclimate to new environments, the expression of genes in bacteria is controlled directly or indirectly by diverse transcriptional factors. Among them, LysR type transcriptional regulators are well-characterized and abundant in bacteria. In a previous study, comparative proteomic analysis revealed that LysR type carbohydrate-related transcriptional regulator in Xag (LcrX) was more abundant in XVM2, which is a minimal medium, compared with a rich medium. However, the functions of LcrX in Xag have not been characterized. In this study, we generated an LcrX-overexpressing strain, Xag(LcrX), and the knockout mutant strain, XagΔlcrX(EV), to elucidate the functions of LcrX. Bacterial multiplication of Xag(LcrX) in soybean was significantly impaired, indicating that LcrX is related to virulence. Comparative proteomic analysis revealed that LcrX is mainly involved in carbohydrate metabolism/transport and inorganic ion transport/metabolism. Based on the results of proteomics analysis, diverse phenotypic assays were carried out. A gel electrophoresis mobility shift assay demonstrated that LcrX specifically bound to the putative promoter regions of genes encoding putative fructose 1,6-bisphosphatase and protease. Through a 96-well plate assay under various conditions, we confirmed that the growth of Xag(LcrX) was dramatically affected in the presence of various carbon sources, while the growth of XagΔlcrX(EV) was only slightly changed. Biofilm formation activity was reduced in Xag(LcrX) but enhanced in XagΔlcrX(EV). The production of siderophores was also decreased in Xag(LcrX) but not altered in XagΔlcrX(EV). In contrast, LcrX was not associated with exopolysaccharide production, protease activity, or bacterial motility. These findings provide new insights into the functions of a carbohydrate-related transcriptional regulator in Xag.
Introduction
Xanthomonas axonopodis pv. glycines (Xag) is a Gram-negative plant pathogenic bacterium that causes bacterial pustule disease, one of the most devastating and economically important diseases in soybeans (Athinuwat et al., 2009). Xag infection occurs through natural openings such as the stomata and wounds on soybean leaves, and Xag multiplies within the intercellular spaces of mesophyll cells (Chatnaparat et al., 2012). Initial symptoms are tiny green spots observed at infection sites on leaves, which spread to the leaf surface and appear as a brown or yellow color. Finally, pustules on the leaves appear with rugged surfaces and chlorotic halos. This disease reduced the size and quantity of seeds (Narvel et al., 2001; Darrasse et al., 2013).
Transcription factors (TFs) are trans-acting proteins that can bind to specific cis-acting elements in promoter regions and regulate the initiation of transcription (Balleza et al., 2009). TFs directly or indirectly regulate expression of not only one or several genes but also numerous genes, eventually leading to the global regulation of gene expression (Browning and Busby, 2004; Geertz and Maerkl, 2010). It has been reported that approximately 51% of genes in Escherichia coli are directly or indirectly modulated by seven transcriptional regulators (Lrp, NarL, ArcA, FIS, IHF, FNR, and CRP) under specific conditions (Riley, 1993; Martinez-Antonio and Collado-Vides, 2003). Each TF regulates the expression of at least 50 genes, and some genes are co-regulated by different TFs. The genes regulated by the 7 TFs are involved in at least 15 biological mechanisms, including iron and carbon utilization (Zhang et al., 2005). Specifically, CRP is associated with global regulation of gene expression in E. coli by controlling 22 different TFs. Thus, global gene expression directly or indirectly regulated by TFs can control bacterial behaviors and phenotypic alterations.
Most TFs have two distinct functional domains: a DNA-binding domain that directly recognizes specific DNA sequences such as helix-turn-helix (HTH) domains or zinc fingers domains and an activation domain that is responsible for initiating transcription (Ohlendorf et al., 1983; Blau et al., 1996; Bouhouche et al., 2000). These types of TFs have been historically classified based on their amino acid composition in prokaryotes and eukaryotes (Mitchell and Tjian, 1989; Rivera-Gomez et al., 2011). Among them, the LysR type transcriptional regulator family (LTTR) is most abundant in prokaryotic organisms (Minezaki et al., 2005). Originally, LTTRs were known as transcriptional activators subject to negative auto-regulation, but they were recently described as global transcriptional regulators that control single or operonic gene expression as activators and repressors (Maddocks and Oyston, 2008). LTTRs contain an HTH motif in their DNA-binding domain and generally bind to the LTTR box sequence, T-N11-A, in the promoter regions of their regulons (Schell, 1993; Parsek et al., 1994; Aravind et al., 2005).
Some LTTRs are closely associated with the regulation of virulence-related genes. For example, MvfR regulates the expression of pqs operons, which are known to be regulated by quorum sensing in Pseudomonas aeruginosa (Deziel et al., 2005). Additionally, the virulence of P. aeruginosa lacking MvfR was impaired, while the mutant could still produce the quorum sensing factor N-acyl-L-homoserine lactone. One of the non-classical LTTRs, PA2206, is required for tolerance to oxidative stresses and contributes to global regulation in P. aeruginosa; a PA2206-knockout strain was found to be less virulent compared to the wild-type strain (Reen et al., 2013). In Xanthomonas oryzae pv. oryzae (Xoo), a classical LTTR, GamR, is involved in galactose metabolism and positively regulates hrp expression by binding to the promoters of hrpG and hrpX (Rashid et al., 2016). However, the functions of LysR type transcriptional regulators in Xanthomonas spp. including Xag remain unclear.
Previously, we reported differentially (2-fold) abundant proteins of Xag in rich medium and XVM2, which is a hrp gene inducing medium and has been widely used for identifying virulence-related genes/proteins (Wengelnik et al., 1996; Kim et al., 2003), using label-free comparative proteomic analysis (Park et al., 2017). Among them, LcrX (LysR type carbohydrate-related transcriptional regulator in Xag; Accession No. AOY61799) was abundantly detected in XVM2 compared with a rich medium, but its functions have not been reported. In this study, we generated a knockout mutant, XagΔlcrX(EV), and LcrX-overexpressing strain, Xag(LcrX), to investigate the functions of LcrX in Xag. To predict the biological mechanisms associated with LcrX, a label-free shotgun comparative proteomic analysis and clusters of orthologous groups (COGs) were used. We also confirmed the binding of LcrX through an electrophoretic mobility shift assay (EMSA) and tested the functions of LcrX using diverse phenotypic assays. Proteomic and phenotypic analyses were performed to investigate the role of LcrX in virulence, carbon utilization, iron-uptake, biofilm formation in Xag.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Xanthomonas axonopodis pv. glycines str. 8ra (Seong et al., 2016) and mutant cells of Xag were grown at 28°C in tryptic soy (TS) medium (tryptic soy broth, soybean-casein digested: 30 g/L) or XVM2 medium (0.32 mM K2HPO4, 0.16 mM KH2PO4, 1 mM CaCl2, 5 mM MgSO4, 10 mM (NH4) 2SO4, 20 mM NaCl, 0.01 mM FeSO4, 10 mM fructose, 10 mM sucrose, 0.03% cassamino acid) (Wengelnik and Bonas, 1996). Escherichia coli strains DH5α and BL21 were used as the hosts for cloning and protein expression, respectively, and were routinely cultivated in Luria Bertani medium at 37°C. Antibiotics were used for the selection of bacterial cells at final concentrations of 50 µg/ml kanamycin, 10 µg/ml gentamicin, 100 µg/ml ampicillin, 30 µg/ml cephalexin, and 100 µg/ml rifampicin for E. coli and Xanthomonas strains.
Generation of LcrX Mutant Strains
All primers used to generate plasmids and mutants are listed in Supplementary Table S2. A 1301-bp DNA fragment possessing the open reading frame of lcrX was amplified by PCR from Xag genomic DNA using specific primers. The amplicons were cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) to generate pGEM-lcrX, and then this sequence was confirmed by Sanger sequencing. The middle region of pGEM-lcrX was digested with the restriction enzyme AfeI, and the kanamycin cassette from pUC4K was inserted, producing pGEM-lcrX::KM. The construct was introduced into the wild-type strain, Xag 8ra, by electroporation using a Bio Rad Micropulser™ (Bio-Rad, Hercules, CA, USA). The lcrX knockout mutant (Xag∆lcrX) was selected on TS agar plates containing kanamycin and confirmed by PCR using specific primers. The pBBR1MCS-5 vector (Kovach et al., 1995) was transferred into the Xag∆lcrX mutant, generating Xag∆lcrX(EV). To obtain an LcrX-overexpressing strain and complemented mutant, the open reading frame (903 bp) of lcrX was amplified by PCR with primer pairs containing restriction sites for SalI and BamHI. The amplicon was inserted into the pGEM-T Easy vector, generating pGEMLcrX-OE, which was confirmed by Sanger sequencing. To generate the overexpressing strain, SalI and BamHI were used to cut the lcrX fragment, and then cloned into pBBR1-MCS5, producing pBBR1LcrX. The expression of lcrX was driven by the lac promoter in pBBR1-MCS5. To eliminate the effect of the lacZ promoter in pBBR1-MCS5, we generated a promoter-less vector, pBBR1-MCS5P, in which 76 bp of the lacZ promoter region was deleted. For the complemented strain, the LcrX fragment was cloned, including the 388-bp upstream region possessing a putative promoter of lcrX. The fragment was cut by SalI and BamHI and cloned into the pBBR1MCS-5P vector to generate the pBBR1LcrXP construct. For the overexpressing and complemented strains, the pBBR1LcrX and pBBR1-MCS5P plasmids were transferred into the wild-type and Xag∆lcrX mutant by electroporation, creating Xag(LcrX) and Xag∆lcrX(LcrXP), respectively. The transformants were confirmed by PCR using specific primers (MCS5-F and MCS5-R) (Supplementary Table S7).
Virulence Assay
Virulence assays were performed by infiltration and spray methods in soybean plants, and the susceptible soybean cultivar Jinju 1 was used for testing virulence (Han et al., 2007). In the spray method, Xag strains were cultured in TSB medium for 48 h, harvested, and suspended in 10 mM MgCl2 buffer to an OD600 of 0.3. Soybean cultivar Jinju 1 plants were grown in a greenhouse for 15–18 days, and bacterial cell suspensions were sprayed onto fully expanded trifoliate leaves. Five soybean plants were used for each strain. After inoculation, the infected plants were kept in the humidity chamber (Relative humidity: over 90%) for the first two days. The bacterial population was measured at 0, 3, 6, and 9 days after inoculation. Infected leaves were harvested and ground using mortars and pestles with ten mM MgCl2. After serial dilution, the bacterial population was determined using a colony counting method onto TS agar medium containing the appropriate antibiotics. For infiltration, bacterial suspensions were prepared in 10 mM MgCl2 buffer to an OD600 of 0.3 and diluted to 1:1000. Fully expanded trifoliate leaves were inoculated using needleless syringes. The infected leaf discs were obtained using a cork borer (4 mm in diameter) at intervals of three days after inoculation. The leaf discs were ground with sterile water, and the bacterial population was measured by a colony counting method.
Proteins/Peptide Preparation
Proteomic analysis with two sets, Xag(EV) vs. Xag(LcrX) and Xag(EV) vs. Xag∆lcrX(EV), were independently performed by label-free shotgun comparative proteomic analysis. Previously established protocols were used for protein extraction and peptide quantification (Park et al., 2017). Bacterial cells were grown in XVM2 medium and grown again to a final concentration, an OD600 of 0.5 and harvested by centrifugation at 7300 ×g for 15 min at 28°C. After removing the supernatants, the pellet was washed twice with 50 mM Tris-HCl (pH 7.8), re-suspended in lysis buffer (6 M Guanidine-HCl, 10 mM DTT, 50 mM Tris-HCl, pH 7.8), and disrupted with an Ultrasonic Processor (Colo Parmer, Vernon Hills, IL, USA). After sonication, the samples were centrifuged at 8,000 × g for 20 min at 4°C, and then the supernatants were collected. Protein concentration was determined with a BCA assay kit (Thermo Fisher Scientific, Waltham, MA, USA). The alkylation process was carried out in the presence of 100 mM iodoacetamide. Total proteins were precipitated using trichloroacetic acid, washed with acetone, and then re-suspended in 50 mM ammonium bicarbonate (pH 7.8). To digest the proteins, 5 µg of trypsin was used with 300 µg of total proteins. The digested samples were cleaned up using the Sep-Pak Vac 1cc tC18 cartridge (Waters, Milford, MA, USA). The digested protein samples were quantified with a BCA protein assay kit.
Liquid Chromatography and Tandem Mass Spectrometry
Two microgram samples of digested peptides from three biological replicates in each strain were analyzed by split-free nano liquid chromatography (EASY-nLC II; Thermo Fisher Scientific) linked to an LTQ Velos Pro instrument (Thermo Fisher Scientific). To separate the peptides, we used a 7.5-cm column packed with MAGIG C18AQ 200A (5 µm) material (Michrom BioResources, Auburn, CA, USA). The samples were injected over 420 min with a gradient and flow rate that was maintained at 300 nL/min with water/acetonitrile gradient (solvent A: 100% H2O + 0.1% formic acid; solvent B: 100% acetonitrile + 0.1% formic acid; 7% B for 5 min, 35% B for 380 min, 80% B for 10 min, and final at 7% B for 25 min). To obtain the full mass spectra, six data-dependent scans were carried out over m/z 300–2,000 mass ranges. Charge state selection was allowed for 2+ and 3+ ions. Dynamic exclusion was permitted in one repeat count, 0.5 min repeat duration, and 3 min exclusion duration. The six most intense ions were sequentially selected from each full mass scan. This analysis was repeated using three biological replicates for each sample.
The Thermo proteome discoverer (ver. 1.3.0.399) combined with the SEQUEST search algorithm was used to evaluate the MS/MS spectra using the Xag strain 8ra database from the National Center for Biotechnology Information database. The target-decoy strategy was also used in this analysis to improve the confidence of identification (Elias and Gygi, 2007). Trypsin was assigned as the enzyme, and two missed cleavages were allowed. For all peptides, 100 ppm was used for mass accuracy, and 0.01 was used for the false discovery rate, with a probability score >20. The Scaffold 4 program (Proteome Software, Portland, OR, USA) was used to identify proteins that matched a minimum of two unique peptides. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD016274. Comparative analyses were carried out using peptide spectrum matches (PSMs) (Choi et al., 2008) and proteins commonly detected in the three biological replicates were selected for comparison. PSMs from selected proteins were normalized to the total PSMs from all proteins in each sample. As a comparison parameter, the average value of the PSMs from three biological replicates was used to identify differentially abundant proteins (>2-fold) in Xag(EV) vs. Xag(LcrX) or Xag(EV) vs. Xag∆lcrX(EV). The Student’s t-test was used for statistical analysis. Differentially abundant proteins were classified by clustering the orthologous group (COG) analysis (Lu et al., 2009).
Electrophoretic Mobility Shift Assay
For expression and purification of LcrX, the pOPINM vector containing maltose-binding protein (MBP) (Berrow et al., 2007) was used. The open reading frame of lcrX was amplified using a specific primer set (Supplementary Table S2) with a Lamp Tag (Biofact, Daejeon, Korea), and the amplified fragment was cloned in pOPINM using In-Fusion Cloning technology (Takara Bio, Shiga, Japan) to prepare pOPINM-LcrX. The construct was transformed into E. coli BL21(DE3) cells to generate the BL21(MBP-LcrX) strain. BL21(MBP-LcrX) cells were grown in LB medium at 37°C for 16 h and then transferred into fresh LB (1:50 dilution). When the OD600 reached 0.6, 1 mM isopropyl-b-D-thiogalactoside was added, and the cells were incubated again for 6 h at 28°C. The cells were harvested and re-suspended in lysis buffer (20 mM imidazole, 500 mM sodium chloride, 20 mM sodium phosphate, pH 7.4). Cell lysis was conducted by adding lysozyme (a final concentration, 1 mg/ml) followed by sonication with an Ultrasonic Processor (Colo Parmer, Vernon Hills, IL, USA) 10 times (3 s on/10 s off) on ice. The total lysates were centrifuged at 12,000 × g for 10 min at 4°C. MBP-LcrX protein in the supernatant was purified using His GraviTrap™ columns (GE Healthcare, Little Chalfont, UK) according to the manufacturer’s protocol. The eluates were cleaned with Zeba™ spin desalting columns by the manufacturer’s protocol (Thermo Fisher Scientific) and ultrapure water. The purified MBP-LcrtX was visualized by staining with Coomassie brilliant blue R-250 (Sigma-Aldrich, St. Louis, MO, USA), and immunoblot results were confirmed using anti-His (QIAGEN, Hilden, Germany) and goat anti-rabbit IgG antibodies (Merck Millipore, Burlington, MA, USA).
To test the DNA binding activity of recombinant LcrX, electrophoretic mobility shifting assay (EMSA) was performed as previously described with minor modification (Imperi et al., 2010). The DNA probes for EMSA were generated by PCR using the specific primers listed in Supplementary Table S2. All probes were designed to amplify 210 bp preceding the start codon The binding reaction with the specific DNA probe was carried out in binding buffer (20 mM Tris-HCl (pH 7.5), 1 mM DTT, 0.1 mM EDTA, 10% glycerol, 10 mg/ml bovine serum albumin, 50 mM KCl, 10 mM MgCl2, 5 μg/ml poly di-dc). The reaction mixtures containing recombinant LcrX and 32P-end labeled double-stranded probe DNA were incubated at room temperature for 20 min, and the samples were subjected into 6% native-polyacrylamide gel electrophoresis that had been pre-electrophoresed for 30 min in 1X TBE (89 mM Tris, 89 mM borate, and 2 mM EDTA) at room temperature. The DNA-protein complex was separated by electrophoresis at room temperature and 130 V for 1 h. The gel was dried and exposed to a phosphor screen (PerkinElmer, Waltham, MA, USA), and bands were detected with a Packard Cyclone Phosphor Imager (PerkinElmer).
Quantitative Reverse Transcription PCR
Gene expression was evaluated by quantitative reverse transcription PCR (q-RT-PCR). All oligonucleotide primers used for q-RT-PCR are shown in Supplementary Table S2. Bacterial cells were grown in media and harvested at an OD of 0.6. The total RNA was extracted with a High Pure RNA Isolation Kit (Roche, Mannheim, Germany), and cDNA synthesis was performed following the protocol of the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The 16S ribosomal RNA was employed as a reference gene for normalization. q-RT-PCR was performed using an IQ™ SYBR Green Supermix (Bio-Rad) on a CFX connect™ (Bio-Rad). The experiment was repeated at least twice with three replicates. The 2−ΔΔCt method was used to calculate gene expression levels.
Extracellular Protease Activity
Extracellular protease activity was examined as previously described (Sole et al., 2015) with slight modifications. Xag strains were grown in TSB medium, washed with sterile water, and then adjusted to an OD600 of 1.0. Three microliters of bacterial suspension were dropped onto NYG plates containing 1% skim milk (BD Biosciences, Franklin Lakes, NJ, USA) and 1% agar, and then incubated at 28°C for 7 days. The clear halo zone was measured.
GEN III Microplate Assay
The growth ability of the Xag strain under 94 different conditions was investigated in a GEN III microplate (Biolog™, Hayward, CA, USA) using the provided protocol with slight modifications. The GEN III microplate is a part of a phenotype microarray system consisting of panels of 96 wells containing diverse carbon sources and chemical substrates in each well (94 different conditions). Xag strains were grown in TSB and washed three times with sterile water. Bacterial cells were adjusted to an OD590 of 0.02 in incubating fluid (Biolog™). One hundred microliters of bacterial suspensions were mixed in each well and incubated at 28°C. Growth was then determined by measuring the absorbance at 590 nm after 24 and 48 h using a Spectramax 190 microplate reader.
Bacteria Growth in Rich and Minimal Medium
To determine of Bacteria growth in rich and minimal condition, TSB and XVM2 medium were used. Xag strains were incubated at 28°C and then adjusted to an OD600 of 0.3. After washing twice, bacterial suspensions were diluted to an OD600 of 0.003 with TSB and XVM2. Growth was measured with a UV spectrophotometer at 600 nm at 24-h intervals for 72 or 120h.
Bacteria Growth in Different Sugar Sources
To confirm the cell growth ability in the presence of sugar sources as the sole carbon source, M9 medium was used in the supplement of 0.4% glucose, sucrose, and fructose. Xag cells were grown in TSB, washed twice with sterile water, and re-suspended in M9 medium. The bacterial suspension was adjusted to an OD600 of 0.001 in M9 medium, and growth was measured with a UV spectrophotometer at 600 nm at 24-h intervals for 96 h.
Biofilm Formation and Exopolysaccharide Production Assay
Biofilm formation was determined in XVM2 liquid medium as described previously with slight modifications (Park et al., 2014). Xag strains were grown for 2 days in TSB medium and washed using sterile water. The bacterial suspension was adjusted to an OD600 of 0.3 and diluted to 1:1000 in XVM2 liquid media. The bacterial suspension was incubated in a 96-well PVC (polyvinyl chloride) plate at 28°C for 8 days. After incubation, the supernatant was carefully eliminated, and attached bacterial cells were stained with 0.1% crystal violet. The stained cells were solubilized in 95% ethanol for 20 min, and absorbance was measured at 590 nm with a Spectramax 190 microplate reader (Molecular Devices, Sunnyvale, CA, USA) with 14 biological replicates.
Exopolysaccharide (EPS) production was evaluated as previously described with slight modifications (Bae et al., 2018b). Xag strains were grown to an OD600 of 0.1 in 5 ml XVM2 for 5 days at 28°C. The bacteria suspension was centrifuged at 10,500 ×g for 3 min at 28°C, and 400 µl of the supernatant was added in 1.2 ml of 95% ethanol and stored overnight at −20°C. The samples were centrifuged, and then the pellets were dried at room temperature. The pellets were added to 1 ml sterile water and mixed with 1 ml of 5% aqueous phenol and 5 ml of H2SO4 and EPS was quantified by measuring the absorbance of the mixtures at 488 nm.
Swimming and Swarming Motility Assays
The swimming and swarming motility assays were performed as described previously with slight modifications (Bae et al., 2018a). Swarming and swimming and motilities were determined on an XVM2 plate containing 0.3% agar. Xag strains were grown for 2 days in TSB medium and washed with sterile water. The bacterial suspension was adjusted to an OD600 of 0.3, and 3 µl of each bacterial cell suspension was spotted on the surface for swarming and placed into in the middle of XVM2 plates for swimming by stabbing using tips onto the center and incubated at 28°C for 3 days. The diameter of the colony occupied by each strain was measured.
Chrome Azurol S (CAS) Assay
Siderophore production was assessed by performing a chrome azurol S assay as described previously with slight modifications (Park et al., 2019). XVM2-CAS-agar was used to create an iron-rich condition. For the iron-limited condition, 2, 2′-bipyrudyl (BP) was added at a final concentration of 100 μM (XVM2-CAS-BP). Xag strains were grown on TS agar medium for 2 days, harvested, washed three times with sterile water, and adjusted to an OD600 of 0.3. Three microliters of bacterial suspension were dropped onto the XVM2-CAS or XVM2-CAS-BP plate. The diameters of the halo zone and colony sizes observed on the plates were measured after incubation at 3 days.
In Silico Modeling in LcrX
To predict the structure of LcrX, the deduced amino acid (aa) sequence of LcrX was evaluated with the I-TASSER server (Roy et al., 2010), and the PDB file for LcrX was obtained. PyMOL software (Molecular Graphics System, San Carlos, CA, USA) was used to generate the predicted three-dimensional structure of LcrX.
Statistical Analysis
The statistical significance of quantitative data was identified by Student’s t-test and one-way analysis of variance with Tukey’s multiple comparison using SPSS 12.0K software (SPSS, Inc., Chicago, IL, USA). A P-value of less than 0.05 was considered to indicate a significant difference.
Results
LcrX Is a LysR Type Transcriptional Regulator
LysR type transcriptional regulators (LTTRs) possess two domains, a DNA-binding domain showing HTH motifs and an activation domain for transcription initiation (Johnson et al., 1993; Yura et al., 1993). Analysis of the deduced amino acid sequence of LcrX revealed two conserved domains, a DNA-binding domain (HTH; 8–67 aa) and an activation domain (96–262 aa) (Figure 1A), indicating that LcrX belongs to the LTTR family. The predicted 3D structure of LcrX by I-TASSER (Roy et al., 2010) and PyMOL clearly revealed two distinct domains (Figure 1B), supporting that LcrX is a putative LTTR. Next, we compared the sequence of LcrX at the amino acid level with putative LTTRs in other bacteria (Figure 1C). High homology was observed between LcrX and CAJ25335 (297/300, 99%), AAM38319 (300/300 100%), and AXQ50657 (211/300, 70%) in X. campestris, X. citri, and Stenotrophomonas rhizophila, respectively (Figure 1C), indicating that LcrX is conserved in Xanthomonas spp. and closely related genera. Finally, the deduced amino acid sequence of LcrX was compared to those of other LTTRs whose functions have been characterized (Lamblin and Fuchs, 1994; Wei et al., 2012). However, LcrX exhibited low homology with CynR related to the cyanate metabolism (78/300, 26%) (Lamblin and Fuchs, 1994), OxyR involved in an oxidative stress (62/300, 20%), (Ochsner et al., 2000), and CcmR associated with the inorganic carbon concentrating mechanism (67/300, 22%) (Woodger et al., 2007) from E. coli, P. aeruginosa, and Synechococcus, respectively (Figure 1D), indicating that LcrX has distinct functions from the three LTTRs.
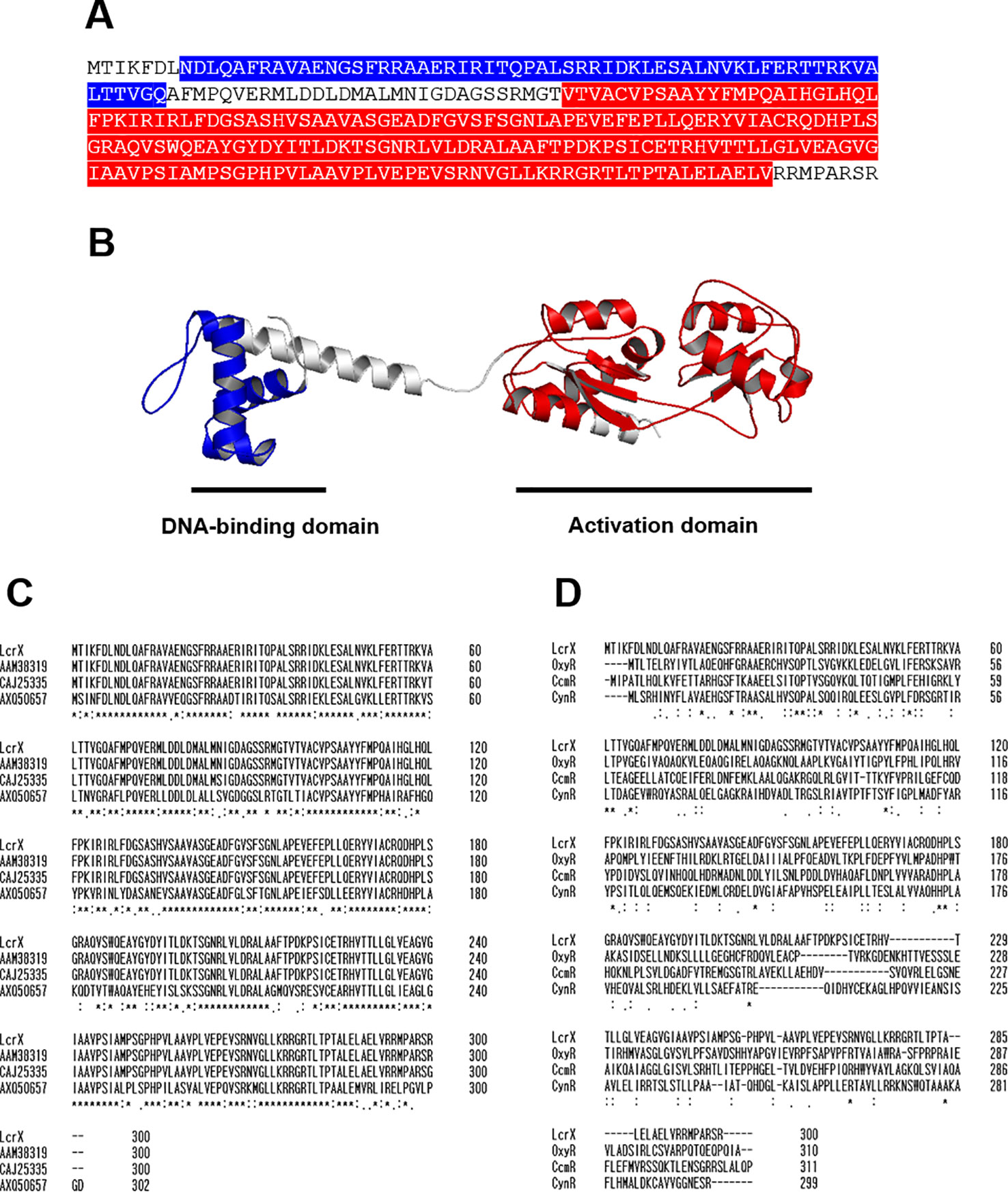
Figure 1 Predicted three-dimensional structure and sequence alignment of LcrX. (A) Deduced amino acid sequence of LcrX where blue and red boxes indicate DNA-binding domain and LysR activation domain, respectively. (B) Predicted 3D structure obtained using the protein modeling server I-TASSER and PyMOL program. (C) Comparison of amino acid sequences of LcrX with its homologs in Xanthomonas spp. and Stenotrophomonas spp. using the ClustalOmega program. CAJ25335 from X. campestris pv. vesicatoria str. 85-10, AAM38319 from X. citri pv. citri str. 306, AXQ50657 from Stenotrophomonas rhizophila str. GA1 (D) Comparison of amino acid sequences of LcrX with LysR type transcriptional regulators whose functions were previously characterized. OxyR from Pseudomonas aeruginosa str. PAO1, CcmR from Synechococcus sp. str. PCC 7002, and CynR from Escherichia coli str. K-12. ‘*’, ‘.’, and ‘:’ indicate most conserved residues, semi-conserved sequence, respectively.
Overexpression of LcrX Reduced the Virulence of Xag
Because LcrX was abundantly detected in XVM2 medium (Park et al., 2017), we examined the effect of LcrX on the virulence of Xag in susceptible soybean using the wild-type strain carrying an empty vector Xag(EV), LcrX-overexpressing strain Xag(LcrX), lcrX-knockout mutant carrying an empty vector Xag∆lcrX(EV), and complemented strain Xag∆lcrX(LcrXP). Expression of LcrX in Xag(LcrX) and Xag∆lcrX(LcrXP) was driven by the Lac promoter and putative native promoter of lcrX, respectively. The virulence of the four strains was examined using two different methods, spray inoculation (Figure 2A) and infiltration with needleless syringes on the leaves (Figure 2B). In both methods, bacterial growth in the leaves of soybean showed similar patterns. In the spray method (Figure 2A), the population of Xag(LcrX) was significantly lower than that of Xag(EV) at 3 (29-fold), 6 (>500-fold), and 9 (> 140-fold) days after inoculation. However, the bacterial growth of Xag∆lcrX(EV) and Xag∆lcrX(LcrXP) was not significantly different from that of Xag(EV). The disease symptoms by Xag(LcrX) were significantly reduced in infected leaves compared with Xag(EV) and Xag∆lcrX(EV) (Supplementary Figure S1A), but Xag(LcrX) showed typical pustule symptoms like other three strains (Supplementary Figure S1B). The difference in population between Xag(LcrX) and Xag(EV) in the infiltration method was lower than that in spray inoculation. Xag(LcrX) also showed a reduced population at 3 (5-fold), 6 (4-fold), 9 (8-fold), and 12 days after inoculation (5-fold) compared to Xag(EV). Xag∆lcrX(EV) and Xag∆lcrX(LcrXP) showed similar growth patterns as Xag(EV). However, disease symptoms caused by the infiltration were not very different among Xag strains (Supplementary Figure S1C). These results indicate that overexpression of LcrX negatively affects the virulence of Xag in soybean, regardless of the inoculation method.
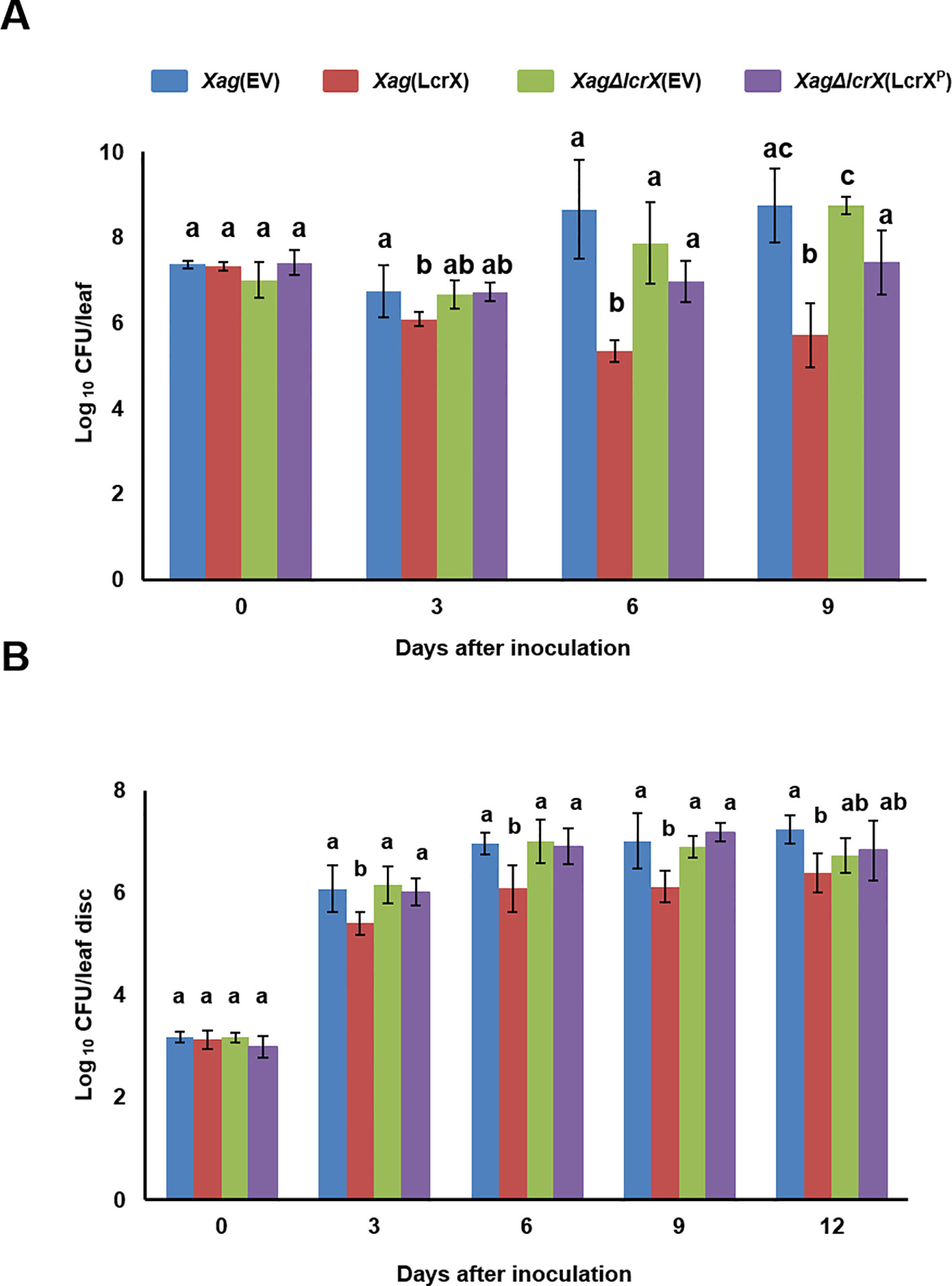
Figure 2 Growth test of Xag(EV), Xag(LcrX), XagΔlcrX(EV), and XagΔlcrX(LcrXP) using two different inoculation methods on soybean leaves. Leaves of soybean cv. Jin-ju were inoculated by (A) the spray method and (B) infiltration. In the spray method, bacterial suspensions were adjusted to an OD600 of 0.3 in 10 mM MgCl2 and sprayed onto fully expanded trifoliate leaves. In the infiltration test, bacterial suspensions were adjusted to an OD600 of 0.3 and serially (10−3) diluted, and then infiltrated with needleless syringes. Bacterial populations were determined using the colony counting method at 3-day intervals. Error bars represent the standard deviation of five biological replicates. The different letters on the bars represent significant differences by one-way ANOVA (p < 0.05). This experiment was repeated at least three times.
Comparative Proteomic Analysis of LcrX
In a previous study, we confirmed that LcrX was abundantly in XVM2 medium rather than in TSB. Therefore, we carried out q-RT-PCR to evaluate the expression of the LcrX transcript. As a result, lcrX expression was much higher (9.8-fold) in XVM2 compared to in TSB (Supplementary Figure S2). Because overexpression of LcrX clearly contributes to reduced virulence, we predicted the biological mechanisms related to LcrX in Xag using a label-free shotgun comparative proteomic analysis combined with COG analysis. In the comparison of Xag(EV) and Xag(LcrX), a total of 1178 and 1119 proteins were commonly detected in the three biological replicates, respectively (Supplementary Table S3). These proteins were subjected to comparative analysis, which showed that 75 and 78 proteins were differentially (over 2-fold) abundant in Xag(EV) and Xag(LcrX), respectively (Supplementary Tables S4 and S5). The LcrX protein was mainly detected in Xag(LcrX), indicating that Xag(LcrX) is indeed an LcrX-overexpressing strain (Supplementary Table S5). COG classification was used to group these differentially abundant proteins by similar functions (Figure 3). Proteins belonging to the G (carbohydrate metabolism and transport), K (transcription), L (replication, recombination, and repair), M (cell wall/membrane/envelope biogenesis), and T (signal transduction mechanisms) groups were abundantly detected in Xag(LcrX) (Figure 3A). Additionally, proteins categorized in the H (coenzyme transport and metabolism), J (translation), O (post-translational modification, protein turnover, and chaperones), and P (inorganic ion transport and metabolism) groups were highly detected in Xag(EV) (Figure 3A). Notably, the N (cell motility) group was not found in Xag(LcrX). Interestingly, at least 8 proteins associated with carbohydrate metabolisms, such as two glycerophosphodiester phosphodiesterases, xylosidase, mannosidase, galactonate dehydratase, galactose dehydrogenase, sorbosone dehydrogenase, and alcohol dehydrogenase, were abundantly detected in Xag(LcrX) (Supplementary Table S5). Moreover, five proteins involved in iron uptake (bacterioferritin, ferrous iron transporter B, and three TonB-dependent receptors) were more abundant in Xag(EV) than in Xag(LcrX) (Supplementary Table S4 and S5). Collectively, these results suggest that LcrX is closely involved in carbohydrate metabolism and iron uptake within Xag.
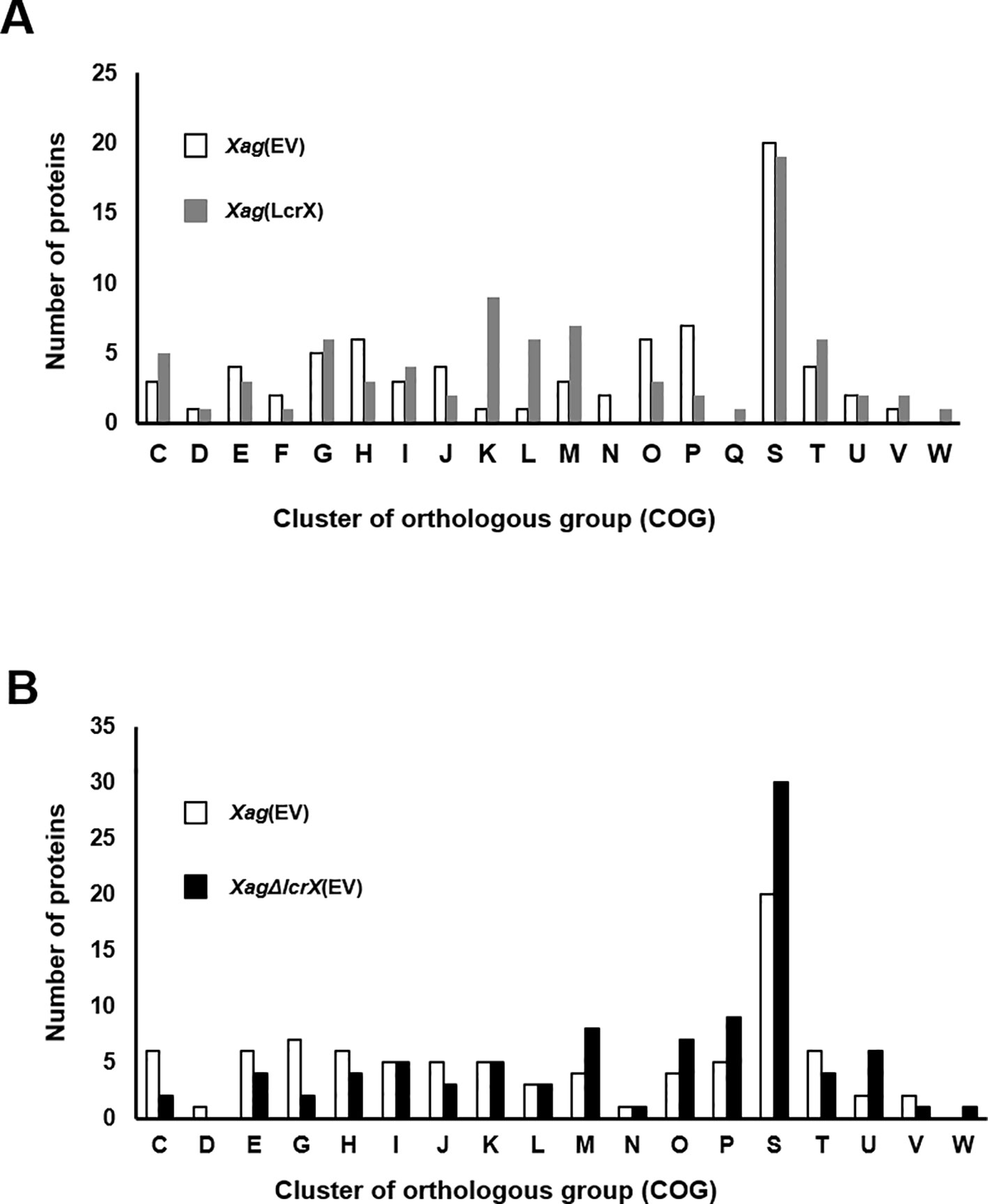
Figure 3 Clusters of orthologous groups (COG) of proteins whose expression is regulated by LcrX. Bar graphs indicate (A) the COG category of 75 and 78 proteins, which were more (over 2-fold) abundant in Xag(EV) and Xag(LcrX), respectively and (B) COG category of 84 and 90 proteins, which were more (over 2-fold) abundant in Xag(EV) and XagΔlcrX(EV), respectively. C, Energy production and conversion; D, Cell cycle control and mitosis; E, Amino acid metabolism and transport; F, Nucleotide metabolism and transport; G, Carbohydrate metabolism and transport; H, Coenzyme metabolism; I, Lipid metabolism; J, Translation; K, Transcription; L, Replication and repair; M, Cell wall/membrane/envelop biogenesis, N, Cell motility; O, Post-translational modification, protein turnover, chaperone functions; P, Inorganic ion transport and metabolism; Q, Secondary structure; R, General functional prediction only; S, Function unknown; T, Signal transduction; U, Intracellular trafficking and secretion; V, Defense mechanisms; W, Extracellular structures.
Next, we compared proteins from Xag(EV) to those from Xag∆lcrX(EV). This experiment was carried out independently of the comparison of Xag(EV) and Xag(LcrX). In LC-MS/MS analysis, 1091 and 1097 proteins were shared in the three biological replicates of Xag(EV) and Xag∆lcrX(EV), respectively (Supplementary Table S3). A total of 174 proteins were differentially expressed between Xag(EV) and Xag∆lcrX(EV), and 84 and 90 proteins were more abundant (over 2-fold) in Xag(EV) and Xag∆lcrX(EV), respectively (Supplementary Table S6 and S7). COG analysis showed that the expression of proteins categorized by the M (cell wall/membrane/envelop biogenesis), O (post-translational modification, protein turnover, and chaperones), P (inorganic ion transport and metabolism), and U (intracellular trafficking, secretion, and vesicular transport) groups was highly affected in Xag∆lcrX(EV) (Figure 3B). Proteins belonging to the C (energy production and conversion) and G (carbohydrate metabolism and transport) groups were abundant in Xag(EV). Similar to in the Xag(EV) vs Xag(LcrX) comparison, seven proteins involved in carbohydrate metabolism (succinate dehydrogenase, pyruvate dehydrogenase, alcohol dehydrogenase, glycosyl hydrolase, glycogen debranching enzyme, glucosidase, and amylase), and six proteins involved in iron uptake (ferrochelatase and five TonB-dependent receptors) were detected (Supplementary Table S6 and S7).
LcrX Directly Bound to Putative Promoters of Fructose 1,6-Bisphosphatase and Protease
If LcrX is a transcriptional regulator, it may regulate the expression of genes by binding to the promoters of its target genes. To examine whether LcrX and putative promoters directly interact, an EMSA was carried out. Despite several attempts, successful purification of the native and soluble LcrX in E. coli BL21 was not possible because expressed LcrX formed inclusion bodies. To increase solubility, we generated LcrX fused to MBP and successfully purified native and soluble MBP-LcrX with a size of approximately 75 kDa (Supplementary Figure S3). Next, we selected putative targets of LcrX in the EMSA using the following categories: I) abundance of proteins in comparative proteomics, II) COG classification combined with phenotypic alteration, and III) the numbers of PSMs detected from LC-MS/MS analysis. As a result, we selected eight genes, AOY61777 (monothiol glutaredoxin and Grx4 family), AOY60939 (fructose 1,6-bisphosphatase, FBP), AOY62387 (TonB-dependent receptor), AOY62229 (hypothetical protein), AOY62452 (protease), AOY64564 (bacterioferritin), AOY64093 (glycine C-acetyltransferase), and LcrX. We selected LcrX because some LTTRs are auto-regulated (Lindquist et al., 1989; Schell, 1993). For the EMSA, we amplified the putative promoter region, which is 210 bp from the start codon, from each gene using specific primer sets. Among the eight candidates, recombinant LcrX bound to the putative promoters of fructose 1,6-bisphosphatase and protease, but not to the other six genes, including LcrX (Supplementary Figure S4). Thus, LcrX is not auto-regulated. To confirm whether LcrX specifically bound to the two promoter regions, unlabeled probes were used as competitors (Figure 4). As the amounts of unlabeled probes increased, probe-LcrX bands gradually decreased in both cases (Figure 4), demonstrating that LcrX binds directly to the putative promoters of FBP and protease. Therefore, we also evaluated the transcripts of both fructose 1,6-bisphosphatase and protease by q-RT-PCR. As expected, the gene expression of FBP and protease was consistent with the pattern of protein abundance (Supplementary Figure S5). The transcripts of both genes were lower and higher in Xag(LcrX) and Xag∆lcrX(EV), respectively, compared to in Xag(EV). Taken together, these data indicate that LcrX negatively regulates the expression of FBP and protease.
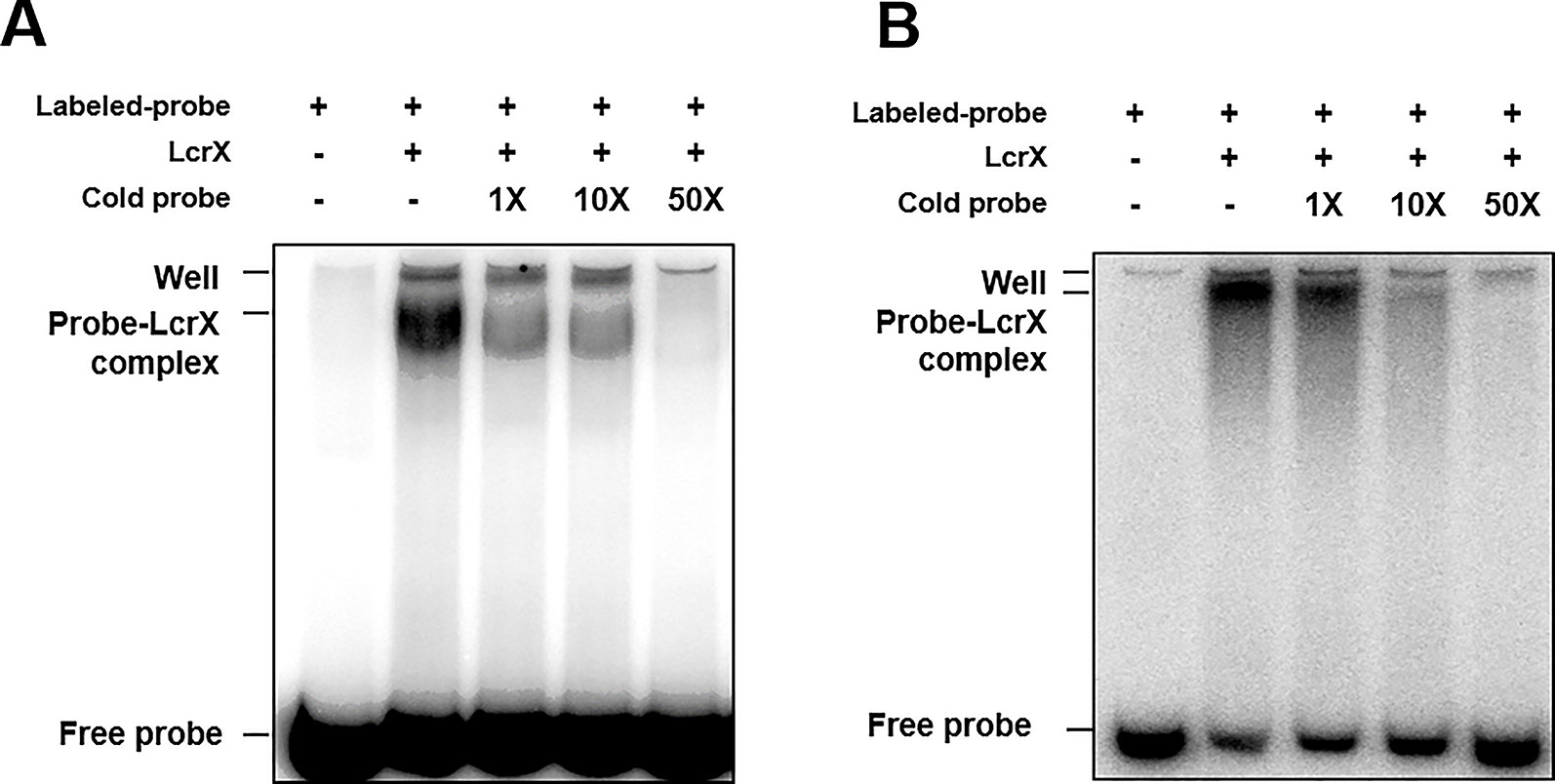
Figure 4 Electrophoretic mobility shift assay of LcrX. EMSA was performed to determine between LcrX and putative promoter probes from (A) AOY60939 (fructose 1,6-bisphosphatase) and (B) AOY62452 (protease) labeled with [γ -32P]. The binding reaction was performed using 0.5 μM of recombinant LcrX with 20 ng of labeled probes. Cold probes, 1X (20 ng), 10X (200 ng), and 50 X (1,000 ng), were added for a competition assay.
LcrX Is Involved in the Growth of Xag Under Diverse Conditions
Because deduced amino acid sequences of protease reveal that the proteins is not a cytoplasmic protein, we examined an extracellular protease activity in four strains, but the activity did not differ under the conditions used (Supplementary Figure S6). Therefore, we focused on the growth ability of Xag under various conditions including in the presence of diverse sugar sources. We compared the growth ability of Xag(EV), Xag(LcrX), and Xag∆lcrX(EV) using a GEN III plate containing 71 different carbon sources, and 23 different chemicals or conditions (Figure 5). After incubation for 24 h, the growth of Xag(LcrX) was significantly different from that of Xag(EV) under 38 different conditions. Among the 38 different conditions, 21 were carbon sources and 17 were chemical substrates (Figure 5A). However, Xag∆lcrX(EV) showed differences in growth under only 7 conditions, one carbon source, and 6 chemical substrates. After incubation for 48 h in the GEN III plate, Xag(LcrX) showed significantly different growth in 18 carbon sources but not in any chemical substrates (Figure 5B). Interestingly, the growth of Xag∆lcrX(EV) was not significantly different from that of Xag(EV) under all conditions after incubation for 48 h.
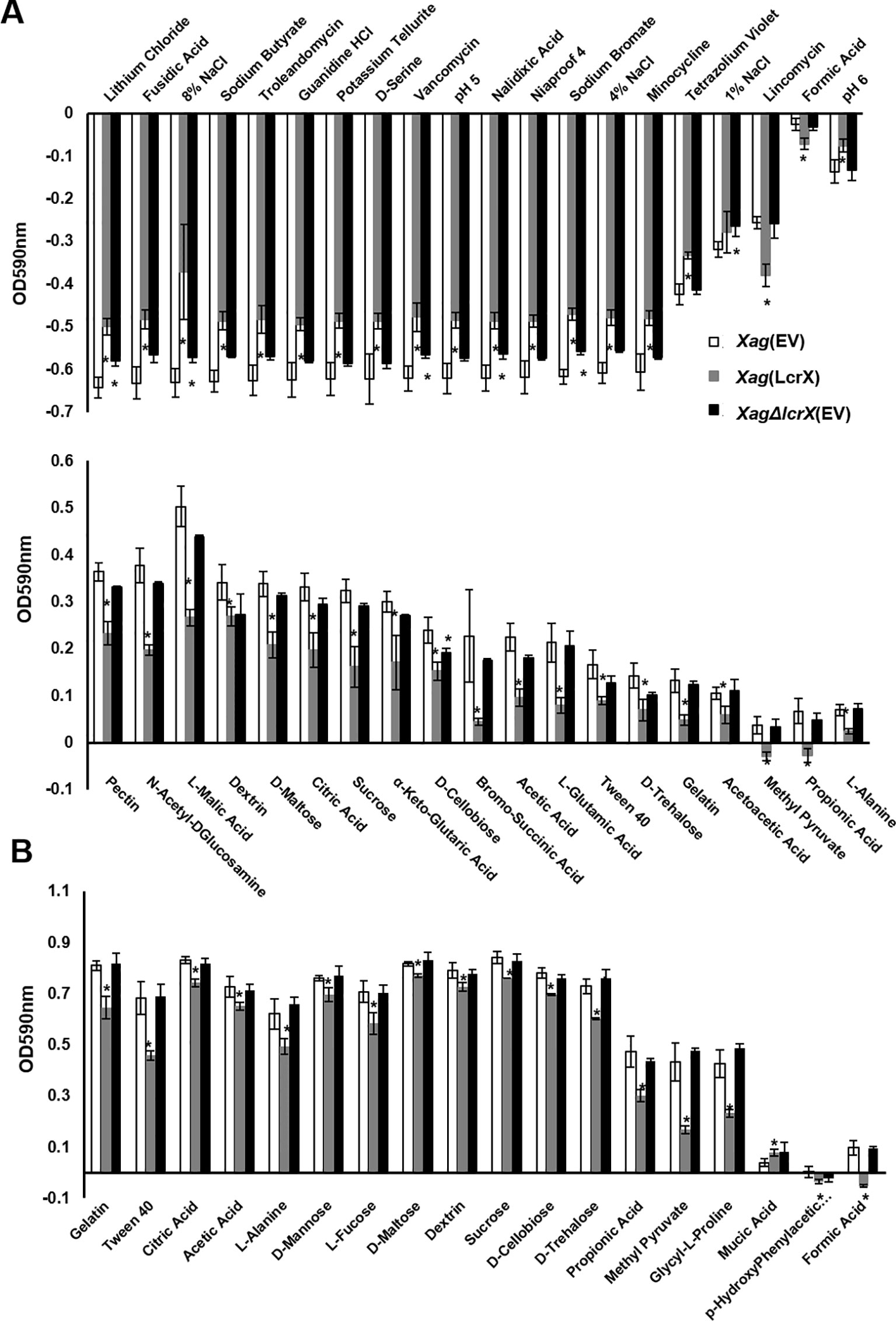
Figure 5 Comparison of growth of Xag(EV), Xag(LcrX), and XagΔlcrX(EV) under 94 different conditions. The GEN III plate, which contains 71 different carbon sources and 23 different chemicals was used for this assay. The bacterial suspensions were adjusted at OD590 of 0.02, incubated in GEN III plate for 2 days, and then measured at 590 nm. The graph represents different sources in which the growth of Xag(LcrX) and/or XagΔlcrX(EV) was different from that of Xag(EV) at (A) 24 h and (B) 48 h after incubation. White, gray, and black indicate Xag(EV), Xag(LcrX), and XagΔlcrX(EV), respectively. The plus and minus values indicate more and less growth compared with the controls, respectively. Bars in the graph represent the means of three biological replicates with the standard deviation and asterisks indicate significant differences (t test, p < 0.05).
LcrX Controls the Growth of Xag in the Presence of Diverse Sugar Sources
Proteomic analysis using the EMSA and GENIII plate assay suggested that LcrX directly regulates the expression of FBP and is closely related to the utilization of different types of sugar sources. We further investigated the growth of four strains in the presence of fructose, glucose, or sucrose as the sole carbon source. In TSB, there was no difference in the growth of the four strains (Figure 6A), indicating that LcrX is not associated with bacterial multiplication in the nutrient-rich condition. However, Xag(LcrX) showed slow growth compared to Xag(EV), while XagΔlcrX(EV) and Xag∆lcrX(LcrXP) displayed similar patterns as Xag(EV) when supplemented with fructose, glucose, or sucrose (Figures 6B–D). In the presence of glucose and fructose, the population of Xag(LcrX) was reduced compared to the other strains at 24, 48, and 72 h after incubation (Figure 6B and 6C). In sucrose, the growth of Xag(LcrX) was significantly lower than those of the three strains (Figure 6D). These data indicate that the functions of LcrX are associated with bacterial growth in the presence of sugar sources.
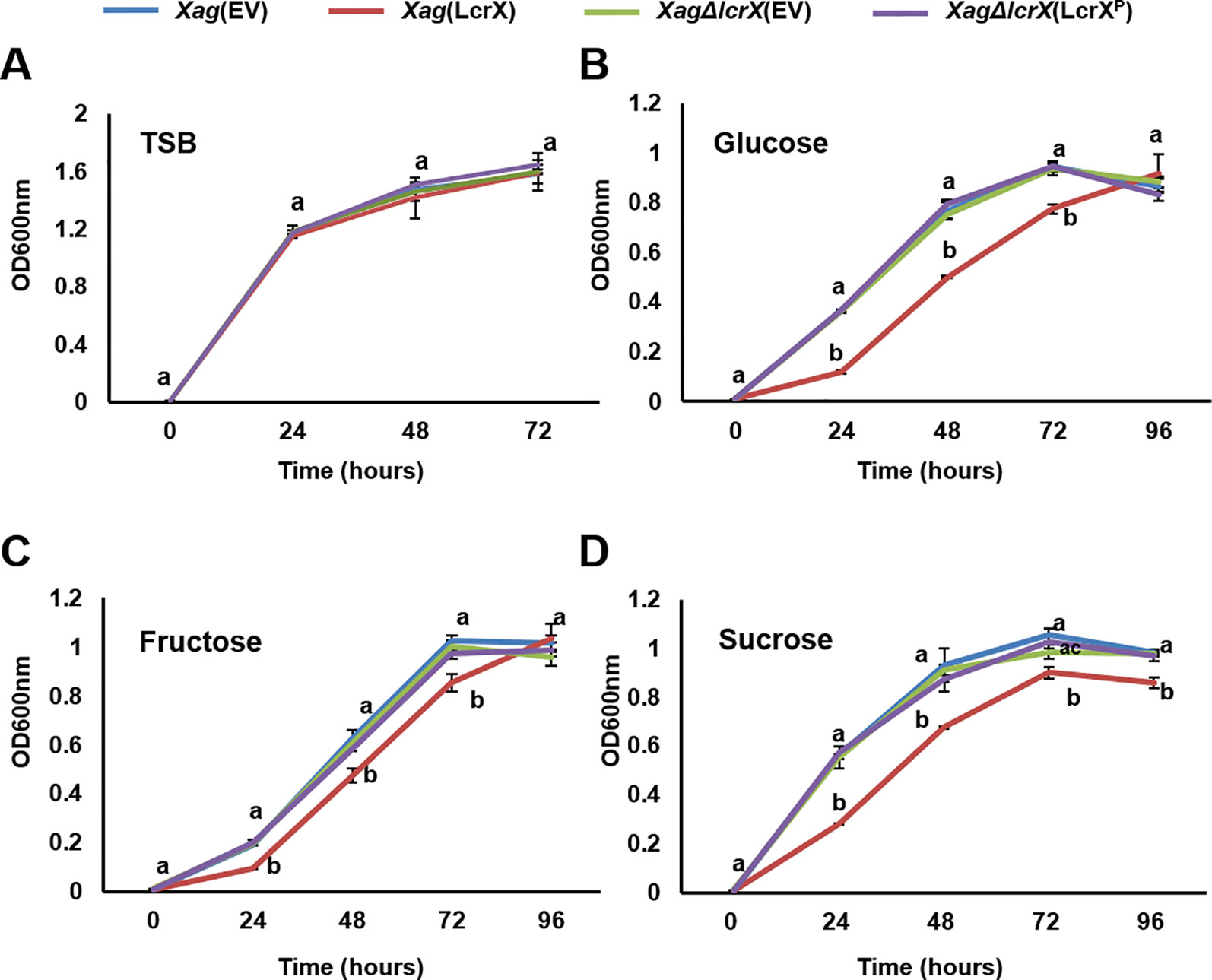
Figure 6 Growth of Xag strains on TSB and M9 minimal medium containing different sugar sources. Xag strains were grown at 28°C in TSB (A), and modified M9 medium with 0.4% glucose (B), fructose (C), or sucrose (D). The bacterial cells were measured at 24-h intervals at an OD of 600 nm. Different letters on error bar graph indicate significant differences by one-way ANOVA (p < 0.05). This experiment was repeated at least three times.
LcrX Is Related to Biofilm Formation But Not EPS Production and Motility
Biofilm formation is an important virulence factor by which bacteria protect themselves against diverse environmental stresses (Costerton et al., 1995). Polysaccharides and proteins are the main components in bacterial biofilms (Nielsen et al., 1997). Proteomic analyses revealed diverse proteins associated with carbohydrate metabolism in both sets [Xag(EV) vs. Xag(LcrX)] and [Xag(EV) vs. Xag∆lcrX(EV)] (Supplementary Tables S4–S7 and Figure 3) and LcrX was involved in carbon utilization of Xag (Figure 6). Therefore, we investigated the biofilm formation abilities and EPS production of Xag(EV), Xag(LcrX), Xag∆lcrX(EV), and Xag∆lcrX(LcrXP). Because we used XVM2 medium for the assays, the growth patterns of the four strains were measured in the medium (Supplementary Figure S7). The four strains showed similar growth patterns, although Xag(LcrX) was slightly low in only 48 h after incubation. Biofilm formation was measured in a 96-well PVC plate assay. The biofilm formation abilities significantly differed between Xag(LcrX) and Xag∆lcrX(EV) (Figure 7A). Biofilm formation by Xag(LcrX) was reduced (1.3-fold), while that by Xag∆lcrX(EV) was significantly increased (1.8-fold) compared to that of Xag(EV). Biofilm formation of the complemented strain, Xag∆lcrX(LcrXP), was restored to the level of Xag(EV), indicating that Xag∆lcrX was functionally complemented by LcrXP. Because bacterial cells produce EPS, a component of biofilm (Sutherland, 2001), we examined whether the change in biofilm formation was affected by EPS production. However, unlike the biofilm formation patterns (Figure 7A), which were reduced in Xag(LcrX) and increased in Xag∆lcrX(EV), there were no significant differences in EPS production between the four strains (Figure 7B). These results indicate that LcrX is involved in biofilm formation but not in EPS production in Xag. Additionally, bacterial motility is known to be closely involved in biofilm formation because flagella contribute to the surface attachment (Lemon et al., 2007). Therefore, we examined bacterial motility using two different assays (Figure 8). The swarming motility in Xag(LcrX) and Xag∆lcrX(EV) was not different from this in Xag(EV) (Figures 8A and 8B). However, the swimming halo in Xag(LcrX) was reduced compared with Xag(EV) (Figures 8C and 8D). XagΔlcrX(EV) tended to enhance the swimming motility compared with Xag(EV). The complemented strain was comparable with Xag(EV). These data suggest that LcrX affects the flagella motility in liquid or inside low-viscosity conditions.
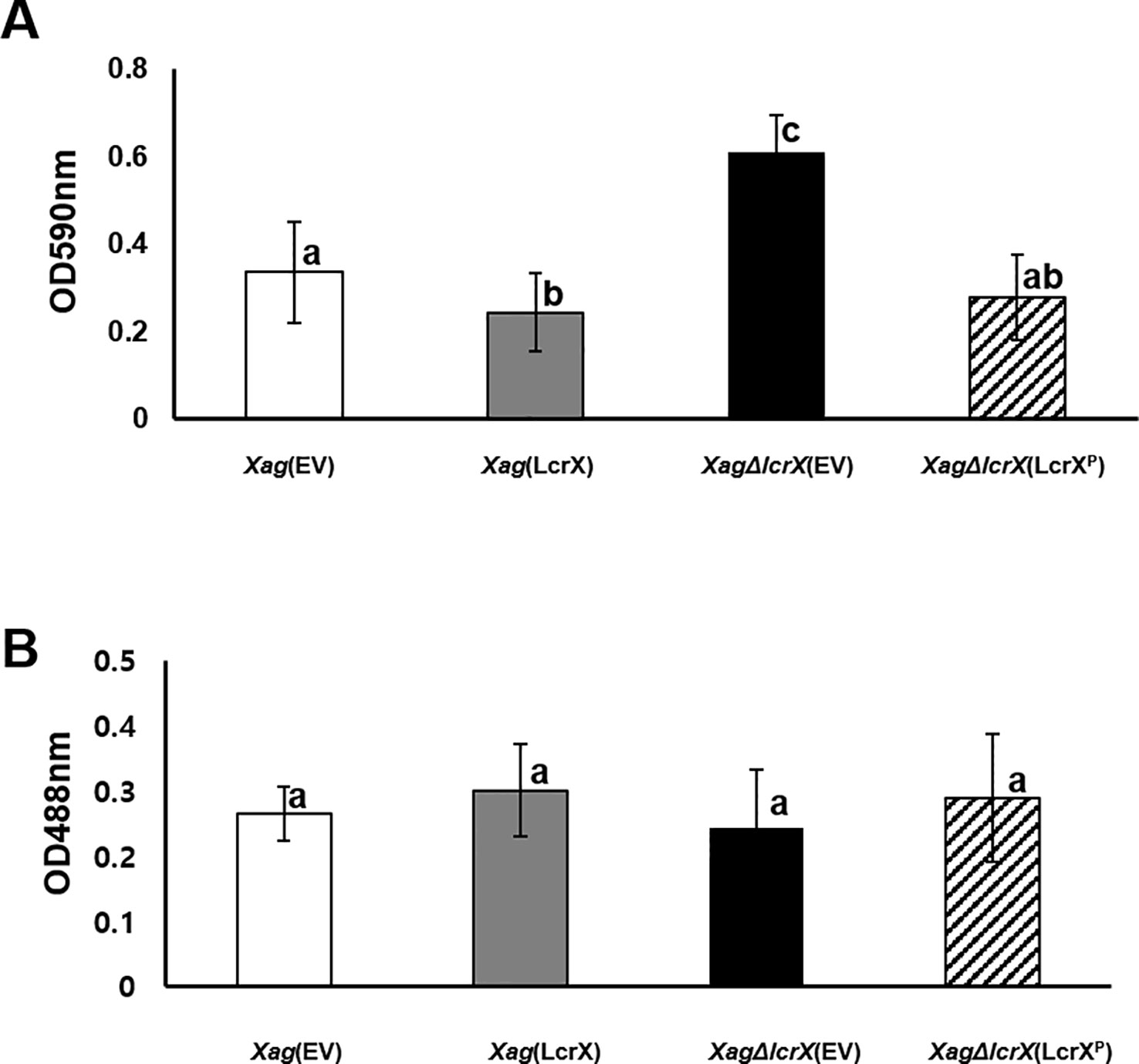
Figure 7 Biofilm formation and exopolysaccharide (EPS) production in Xag(EV), Xag(LcrX), XagΔlcrX(EV), and XagΔlcrX(LcrXP). (A) Biofilm formation ability in Xag strains was evaluated by polyvinyl chloride plate (PVC) plate assay. Xag strains were incubated in the PVC plates for eight days and cells attached to the surface were stained with crystal violet and resolved in 95% ethanol. Biofilm formation was enumerated using a spectrophotometer at 590 nm. Error bars in the graph indicate the means of 14 biological replicates with the standard deviation. (B) EPS production by Xag strains was quantified by the phenol-sulfuric acid method. Xag strains were incubated in TSB for 2 days and in XVM2 for 4 days, and then EPS was quantified by a phenol-sulfuric acid method. EPS was quantified with a spectrophotometer at 488 nm. Bars indicate the means of three biological replicates with the standard deviation. Different letters represent significant differences by one-way ANOVA (p < 0.05). This experiment was repeated at least three times.
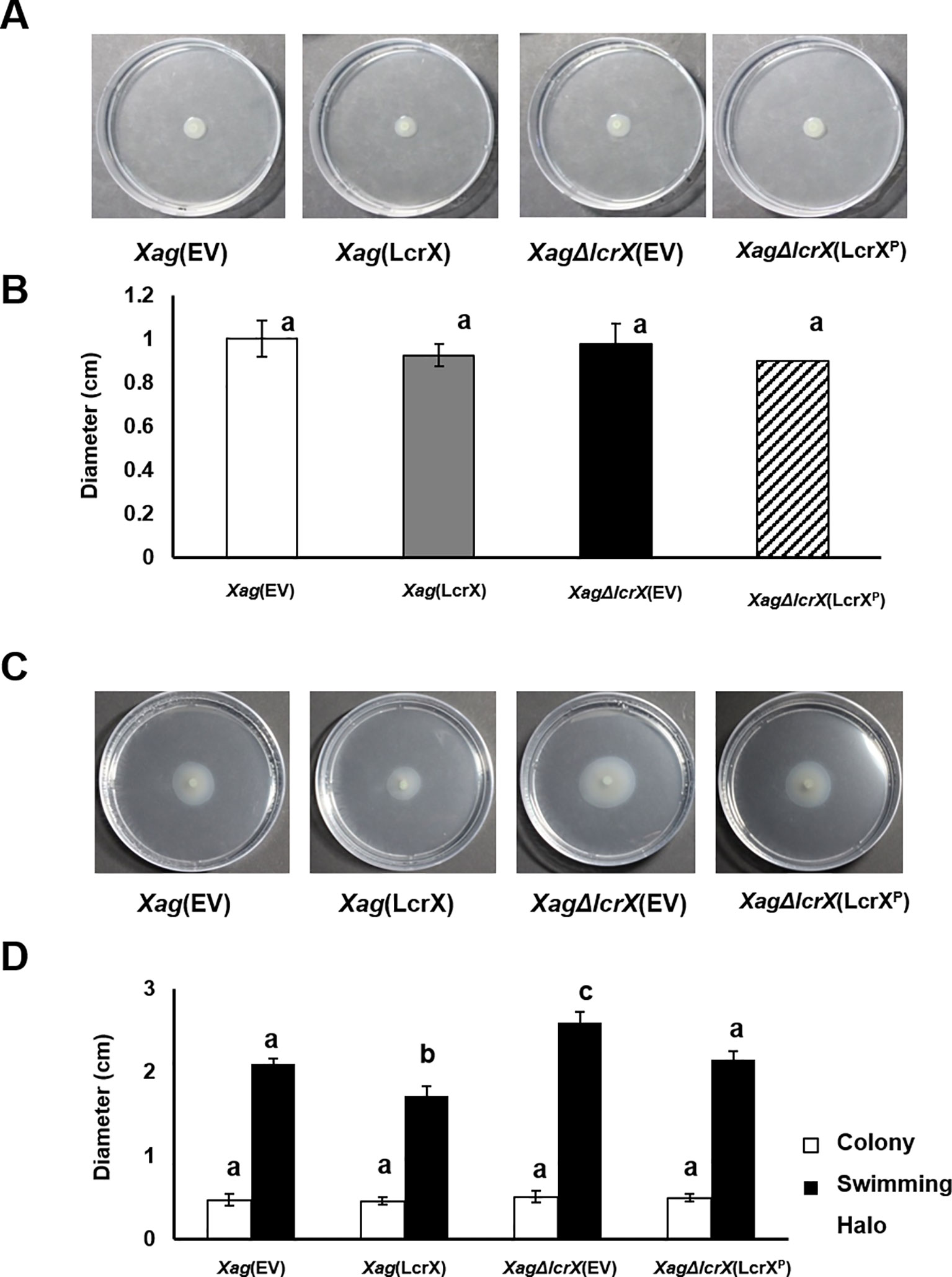
Figure 8 Swimming and swarming motility of Xag strains. Three microliters of bacterial suspension (OD600 of 0.3) were spotted onto the semi-solid medium (0.3% agar) for swarming (A and B) or positioned in to the middle of the medium by stabbing (C and D). Photographs of the motilities (A and C) were taken, and the expansion of colonies and colony halos (C and D) was measured at 3 days after incubation. Bars in the graph indicate the means of five biological replicates with the standard deviation. Different letters represent significant differences by one-way ANOVA (p < 0.05). This experiment was repeated at least five times.
Overexpression of LcrX Reduced Siderophore Secretion
Proteomic analysis showed that diverse iron-related proteins, including TonB-dependent receptors, were less abundant in Xag(LcrX) compared to in Xag(EV) (Supplementary Tables S4 and S5). TonB-dependent receptors are part of the iron-uptake system, which helps bacteria absorb this mineral under iron-limited conditions (Schauer et al., 2008; Noinaj et al., 2010). Therefore, a CAS assay was performed to evaluate siderophore secretion. As shown in Figures 9A and 9B, no obvious difference was found between the four strains under the iron-rich condition. However, under the iron-limited condition, the diameter of the yellow halo, which represents siderophore secretion, was reduced (approximately 70%) in Xag(LcrX) compared with Xag(EV) (Figures 9C and 9D). These data indicate that overexpression of LcrX in Xag diminished siderophore production under the iron-limited condition, but not under the iron-rich condition.
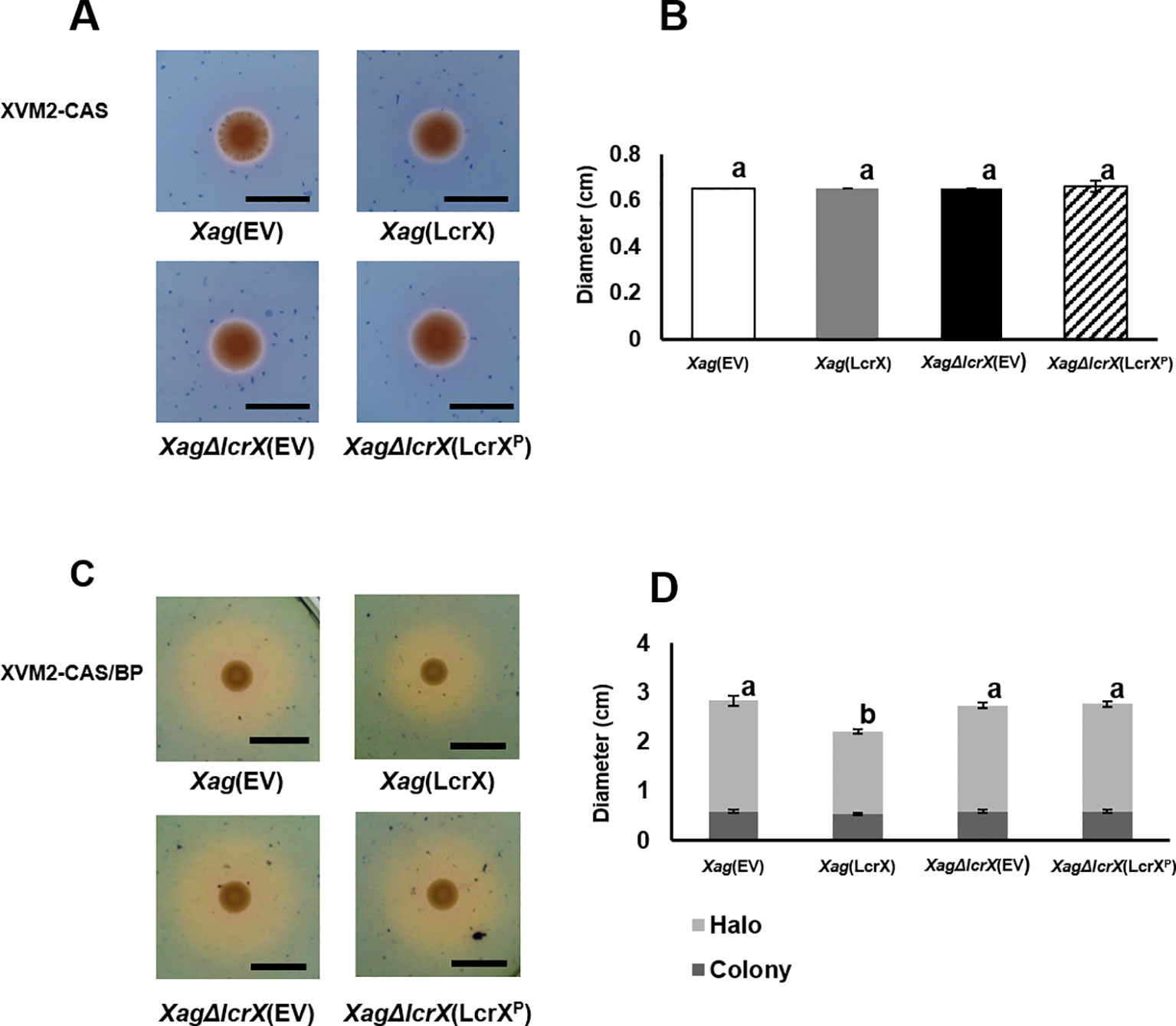
Figure 9 Chrome azurol S (CAS) assay of Xag(EV), Xag(LcrX), XagΔlcrX(EV), and XagΔlcrX(LcrXP). Three microliters of a bacterial suspension at OD600 of 0.3 were dropped on (A and B) the iron-rich medium, XVM2-CAS, and (C and D) iron-limited medium, XVM2-CAS-BP which contained 2,2’-bipyrudyl (BP). (A and C) Photographs of colony and halos were taken (B and D), and their diameters were measured at 3 days after incubation. Black bars in the photographs represent 1 cm. Error bars indicate the means of five biological replicates with the standard deviation. Different letters on the error bar graph represent significant differences by one-way ANOVA (p < 0.05). This experiment was repeated at least three times.
Discussion
In many prokaryotes, gene expression is controlled by diverse TFs to acclimate to new environments. Among the TF families, the LTTR family is highly conserved in bacteria (Rivera-Gomez et al., 2011). A previous study reported that the N-terminus of LTTRs contains an HTH motif as a DNA-binding domain, and the C-terminus possesses an activation domain (Schell, 1993; Zaim and Kierzek, 2003). The predicted 3D structure of LcrX also shows two domains, a DNA-binding domain, and LysR activation domain, suggesting that LcrX belongs to the LTTR family. The low homology with OxyR, CcmR, and CynR (Figure 1C) suggests that LcrX is involved in different mechanisms from those transcriptional regulators. LTTR is the largest family and found not only in Xanthomonas spp., but also in other bacteria and even in fungi (Minezaki et al., 2005). Previous studies reported that the LTTRs are mainly involved in amino acid metabolisms such as IlvY for isoleucine/valine biosynthesis and ArgP for arginine transport in E. coli. (Wek and Hatfield, 1988; Marbaniang and Gowrishankar, 2011). In addition to amino acid metabolism, LTTRs are also associated with carbohydrate metabolism, quorum sensing, cell division, and virulence (Sheehan and Dorman, 1998; Cao et al., 2001; Jones et al., 2003). Therefore, LTTRs may not be restricted to one biological mechanism in bacteria and likely control diverse cellular and metabolic processes, contributing to their phenotypic alteration ability and virulence.
In the EMSA, we found that LcrX directly bound to the putative promoter of protease (AOY62452), which was more (over 3-fold) abundant in XagΔlcrX(EV) compared to in Xag(EV) and in Xag(EV) compared with Xag(LcrX). These data suggest that protease was negatively regulated by LcrX. However, extracellular protease activity was not changed in the mutants. The protease, AOY62452 showed high homology with EcpA (92%, 335/362), which encodes an extracellular protease in Xoc (Zou et al., 2012). Xoc lacking EcpA did not show the extracellular protease activity. Interestingly, extracellular protease activity by EcpA was observed in Xoc, whereas this activity was not observed in Xoo, although the two pathovars infect the same host. A comparison of EcpA from Xoo and Xoc revealed that 1–282 aa are nearly identical, while 283–366 aa are completely different. Furthermore, the protease activity was observed when the N-terminal of Xoo EcpA was fused with the C-terminal of Xoc EcpA, but not vice versa (Zou et al., 2012), indicating that the C-terminal regions are indispensable for protease activity. Comparison of the amino acid sequence between AOY62452 and EcpA of Xoc revealed that the C-terminal region residues (264–296 aa) of AOY62452 differed from those of Xoc EcpA (data not shown). Therefore, the functions of AOY62452 may differ from those of EcpA, and the extracellular protease activity of Xag(LcrX) was not changed, although LcrX negatively regulates AOY62452 expression. Additionally, Xag possesses more than 26 proteins, including 5 proteases, 10 serine proteases, 4 ATP-dependent proteases, 5 metalloproteases, a zinc-dependent protease, a cysteine protease, and over 90 peptidases. The extracellular protease activity of Xag under the given condition may have been affected by other proteases and peptidases.
Interestingly, overexpression of LcrX led to reduced virulence and biofilm formation. Biofilm formation is a major virulence factor because it allows bacteria to protect themselves under stressful conditions, including antimicrobial activity from the host (Costerton et al., 1999; Donlan and Costerton, 2002; Luppens et al., 2002). Escherichia coli and Vibrio cholerae strains with impaired biofilm formation abilities were less virulent compared with their wild-type strains (Zhu et al., 2002; Naves et al., 2008; Huang et al., 2013). In agreement with these previous studies, the reduced biofilm formation of Xag(LcrX) may contribute to virulence in Xag. In the proteomic analysis, the expression of proteins related to carbohydrate metabolism was affected by LcrX. Carbohydrates are one of the main components of biofilm. For example, GlpG is a glycogen phosphorylase involved in glycogen metabolism. An Azospirillum brasilense strain deficient in GlpG had a different monosaccharide composition in extracted EPS and significantly reduced biofilm formation ability compared to the wild-type strain (Lerner et al., 2009). This study demonstrates that glycogen regulation is involved in biofilm formation and sugar metabolic pathways. Therefore, LcrX may be involved in biofilm formation by regulating carbohydrate metabolism. In addition, proteomic data showed that proteins related to signal transduction mechanisms, including histidine kinases, response regulators, and c-di-GMP phosphodiesterase were abundantly in Xag(EV) vs. Xag(LcrX). It is very well-known that these types of proteins function to control biofilm formation. For example, in Streptococcus mutans, which lacks histidine kinase and response regulators, biofilm formation and acid tolerance were reduced (Li et al., 2002). Similarly, the histidine kinase BinK negatively regulates biofilm formation in Vibrio fischeri (Brooks and Mandel, 2016). Additionally, overexpression of RbdA, which regulates c-di-GMP phosphodiesterase, resulted in reduced biofilm formation, whereas biofilm formation was significantly increased in P. aeruginosa cells lacking RdbA (An et al., 2010). LcrX may be a transcriptional regulator that directly or indirectly controls the expression of proteins related to carbohydrate metabolism and signal transduction pathways and regulates biofilm formation as well as virulence. Additionally, it is known that the swimming motility is closely related to an attachment of surfaces in a high humidity condition, biofilm formation, and virulence (Pratt and Kolter, 1998; Tans-Kersten et al., 2004; Zhu et al., 2019). We also showed that swimming motility in Xag(LcrX) was decreased. Therefore, it can be postulated that the reduction of the swimming motility is one of the reasons for the low biofilm in Xag(LcrX), which may contribute to virulence in Xag.
Particularly, iron-associated proteins such as bacterioferritin, ferrous iron transporter B, and TonB-dependent receptors, which are outer membrane proteins and siderophore receptors (Turner et al., 2001), were abundant in Xag(EV) compared with Xag(LcrX). The iron uptake system is important in bacteria because iron is essential for energy generation and cell growth (Skaar, 2010). Therefore, bacteria must produce iron-chelators, called siderophores, to absorb iron (Fe3+) from the environment (Neilands, 1995). According to recent reports, the roles of siderophores in virulence differ in Xanthomonas spp. For example, siderophores in Xoo, the causal agent of bacterial leaf blight disease on rice, which mainly colonizes the xylem vessel and is not essential for virulence, but siderophores from X. oryzae pv. oryzicola (Xoc), which causes bacterial streak disease on rice and colonizes the intercellular space in the mesophyll, are required for optimum virulence (Pandey and Sonti, 2010; Rai et al., 2015; Pandey et al., 2016). In addition, the virulence of X. campestris pv. campestris, which is a causal agent of black rot disease in crucifers, was reduced when the siderophore synthesis gene (xssA) was deficient (Pandey et al., 2017). In this study, we showed that secretion of siderophores from the LcrX-overexpressing strain was dramatically reduced compared to that from the wild-type strain, suggesting that overexpression of LcrX negatively regulates siderophore production. This may explain the reduced virulence of Xag(LcrX).
Proteomic analysis revealed that many proteins in G (carbohydrate metabolism and transport) were differentially abundant between Xag(EV) and Xag(LcrX), and the overexpression of LcrX led reduced biofilm formation consisting of polysaccharides and other biomolecules. Based on these results, we hypothesized that LcrX is involved in regulating carbohydrate metabolism. Indeed, LcrX directly bound to the putative promoter of FBP, which possesses putative LTTR boxes (Supplementary Figure S8). FBP was more (over 5.3-fold) abundant in Xag(EV) compared to in Xag(LcrX), suggesting that LcrX negatively regulates FBP expression. Previous studies reported that FBP and fructose-bisphosphate aldolases, whose substrates are fructose-bisphosphate, are involved in the use of carbon sources and virulence. Mycobacterium marinum, lacking GlpX that encodes a fructose 1, 6-bisphosphatase, showed reduced growth in the presence of the gluconeogenic carbon source and was less virulent in Zebrafish (Tong et al., 2016). Additionally, fructose-bisphosphate aldolase-deficient mutants of Xoc and Francisella novicida displayed growth retardation in various carbon sources and showed reduced virulence in their hosts (Guo et al., 2012; Ziveri et al., 2017).
Proteomic analysis revealed that six and two proteins belonging to the group G (carbohydrate metabolism) were more abundant in Xag(LcrX) and XagΔlcrX(EV), respectively, compared to in Xag(EV) (Supplementary Tables S5 and S7). In addition, the GEN III plate assay at 48 h after of incubation showed that the growth of Xag(LcrX) was reduced under 18 different conditions, which contained different carbon sources and not chemical substances, compared to the growth of Xag(EV), suggesting that LcrX is related to the regulation of carbohydrate metabolism Furthermore, the proteomic analysis and EMSA results support the roles of LcrX in regulating carbohydrate metabolism. Therefore, overexpression of LcrX may trigger growth retardation by negatively regulating carbohydrate metabolism. Bacterial growth patterns in the presence of sugar sources as a sole carbon source showed a similar pattern as the GEN III plate assay. The growth of Xag(LcrX), but not XagΔlcrX(EV), was reduced compared to that of Xag(EV). Collectively, these data demonstrate that under the solo carbon condition, overexpressed LcrX negatively regulates bacterial growth and causes growth retardation in Xag. Finally, these different abilities to use diverse carbon sources by Xag(LcrX) and XagΔlcrX(EV) may explain why Xag(LcrX) was less virulent, while XagΔlcrX(EV) was not.
In this study, the functions of LcrX in Xag were postulated by using the predicted 3D structure and label-free shotgun comparative proteomics combined with COG categorization. We further characterized the functions of LcrX through diverse phenotypic assays, including the GEN III plate assay. Taken together, LcrX is involved in siderophore production, biofilm formation, and carbohydrate metabolism, but not in EPS production and motility, which may contribute to its virulence in Xag. Finally, we demonstrated that LcrX binds to specific putative promoters containing LTTR boxes. This study provides fundamental and valuable information regarding the biological functions of a previously uncharacterized transcriptional regulator in plant pathogenic bacteria.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD016274.
Author Contributions
S-WH conceived the study. S-WH and HP designed the experiments. HP, MK, H-JP, and JL conducted the experiments. ED conducted EMSA. HP and S-WH analyzed the data and prepared the manuscript. All authors reviewed the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Next-Generation BioGreen 21 Program (PJ01328901) of the Rural Development Administration and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1D1A1B07045724), Republic of Korea (to S-WH).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01657/full#supplementary-material
References
An, S., Wu, J., Zhang, L. H. (2010). Modulation of Pseudomonas aeruginosabiofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl. Environ. Microbiol. 76, 8160–8173. doi: 10.1128/AEM.01233-10
Aravind, L., Anantharaman, V., Balaji, S., Babu, M. M., Iyer, L. M. (2005). The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29, 231–262. doi: 10.1016/j.fmrre.2004.12.008
Athinuwat, D., Prathuangwong, S., Cursino, L., Burr, T. (2009). Xanthomonas axonopodispv. glycinessoybean cultivar virulence specificity is determined by avrBs3 homolog avrXg1. Phytopathology 99, 996–1004. doi: 10.1094/PHYTO-99-8-0996
Bae, N., Park, H. J., Park, H., Kim, M., Do, E., Han, S. W. (2018a). Elucidating Functions of FleQ in Xanthomonas oryzae pv. oryzae by comparative proteomic and phenotypic Analyses. Int. J. Mol. Sci. 19, e3038. doi: 10.3390/ijms19103038
Bae, N., Park, H. J., Park, H., Kim, M., Han, S. W. (2018b). Deciphering the functions of the outer membrane porin OprBXo involved in virulence, motility, exopolysaccharide production, biofilm formation and stress tolerance in Xanthomonas oryzae pv. oryzae. Mol. Plant Pathol. 19, 2527–2542. doi: 10.1111/mpp.12727
Balleza, E., Lopez-Bojorquez, L. N., Martinez-Antonio, A., Resendis-Antonio, O., Lozada-Chavez, I., Balderas-Martinez, Y. I., et al. (2009). Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol. Rev. 33, 133–151. doi: 10.1111/j.1574-6976.2008.00145.x
Berrow, N. S., Alderton, D., Sainsbury, S., Nettleship, J., Assenberg, R., Rahman, N., et al. (2007). A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45. doi: 10.1093/nar/gkm047
Blau, J., Xiao, H., Mccracken, S., Ohare, P., Greenblatt, J., Bentley, D. (1996). Three functional classes of transcriptional activation domains. Mol. Cell. Biol. 16, 2044–2055. doi: 10.1128/MCB.16.5.2044
Bouhouche, N., Syvanen, M., Kado, C. I. (2000). The origin of prokaryotic C2H2 zinc finger regulators. Trends Microbiol. 8, 77–81. doi: 10.1016/S0966-842X(99)01679-0
Brooks, J. F., Mandel, M. J. (2016). The histidine kinase BinK is a negative regulator of biofilm formation and squid colonization. J. Bacteriol. 198, 2596–2607. doi: 10.1128/JB.00037-16
Browning, D. F., Busby, S. J. (2004). The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2, 57–65. doi: 10.1038/nrmicro787
Cao, H., Krishnan, G., Goumnerov, B., Tsongalis, J., Tompkins, R., Rahme, L. G. (2001). A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. U.S.A. 98, 14613–14618. doi: 10.1073/pnas.251465298
Chatnaparat, T., Prathuangwong, S., Ionescu, M., Lindow, S. E. (2012). XagR, a LuxR homolog, contributes to the virulence of Xanthomonas axonopodis pv. glycines to soybean. Mol. Plant Microbe Interact. 25, 1104–1117. doi: 10.1094/MPMI-01-12-0008-R
Choi, H., Fermin, D., Nesvizhskii, A. I. (2008). Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell. Proteom. 7, 2373–2385. doi: 10.1074/mcp.M800203-MCP200
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R., Lappin-Scott, H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. doi: 10.1146/annurev.mi.49.100195.003431
Costerton, J. W., Stewart, P. S., Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Darrasse, A., Bolot, S., Serres-Giardi, L., Charbit, E., Boureau, T., Fisher-Le Saux, M., et al. (2013). High-quality draft genome sequences of Xanthomonas axonopodis pv. glycines strains CFBP 2526 and CFBP 7119. Genome Announc. 1, e01036. doi: 10.1128/genomeA.01036-13
Deziel, E., Gopalan, S., Tampakaki, A. P., Lepine, F., Padfield, K. E., Saucier, M., et al. (2005). The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 55, 998–1014. doi: 10.1111/j.1365-2958.2004.04448.x
Donlan, R. M., Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/cmr.15.2.167-193.2002
Elias, J. E., Gygi, S. P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214. doi: 10.1038/nmeth1019
Geertz, M., Maerkl, S. J. (2010). Experimental strategies for studying transcription factor-DNA binding specificities. Brief Funct. Genomics 9, 362–373. doi: 10.1093/bfgp/elq023
Guo, W., Zou, L. F., Li, Y. R., Cui, Y. P., Ji, Z. Y., Cai, L. L., et al. (2012). Fructose-bisphophate aldolase exhibits functional roles between carbon metabolism and the hrp system in rice pathogen Xanthomonas oryzae pv. oryzicola. PloS One 7, e31855. doi: 10.1371/journal.pone.0031855
Han, S. W., Choi, M. S., Lee, S. H., Hwang, D., Hwang, B. K., Heu, S. (2007). Characterization of a novel necrotic response of glycine max line “PI96188” to Xanthomonas axonopodis pv. glycines. Plant Patho. J. 23, 193–202. doi: 10.5423/PPJ.2007.23.3.193
Huang, T. P., Lu, K. M., Chen, Y. H. (2013). A novel two-component response regulator links rpf with biofilm formation and virulence of Xanthomonas axonopodis pv. citri. PloS One 8, e62824. doi: 10.1371/journal.pone.0062824
Imperi, F., Tiburzi, F., Fimia, G. M., Visca, P. (2010). Transcriptional control of the pvdS iron starvation sigma factor gene by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ. Microbiol. 12, 1630–1642. doi: 10.1111/j.1462-2920.2010.02210.x
Johnson, P. F., Sterneck, E., Williams, S. C. (1993). Activation domains of transcriptional regulatory proteins. J. Nutr. Biochem. 4, 386–398. doi: 10.1016/0955-2863(93)90069-9
Jones, R. M., Britt-Compton, B., Williams, P. A. (2003). The naphthalene catabolic (nag) genes of Ralstonia sp. strain U2 are an operon that is regulated by NagR, a LysR-type transcriptional regulator. J. Bacteriol. 185, 5847–5853. doi: 10.1128/JB.185.19.5847-5853.2003
Kim, J. G., Park, B. K., Yoo, C. H., Jeon, E., Oh, J., Hwang, I. (2003). Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185, 3155–3166. doi: 10.1128/jb.185.10.3155-3166.2003
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Lamblin, A. F. J., Fuchs, J. A. (1994). Functional analysis of the Escherichia coli K12 Cyn operon transcriptional regulation. J. Bacteriol. 176, 6613–6622. doi: 10.1128/jb.176.21.6613-6622.1994
Lemon, K. P., Higgins, D. E., Kolter, R. (2007). Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189, 4418–4424. doi: 10.1128/JB.01967-06
Lerner, A., Castro-Sowinski, S., Lerner, H., Okon, Y., Burdman, S. (2009). Glycogen phosphorylase is involved in stress endurance and biofilm formation in Azospirillum brasilense Sp7. FEMS Microbiol. Let. 300, 75–82. doi: 10.1111/j.1574-6968.2009.01773.x
Li, Y. H., Lau, P. C., Tang, N., Svensater, G., Ellen, R. P., Cvitkovitch, D. G. (2002). Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184, 6333–6342. doi: 10.1128/jb.184.22.6333-6342.2002
Lindquist, S., Lindberg, F., Normark, S. (1989). Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC beta-lactamase gene. J. Bacteriol. 171, 3746–3753. doi: 10.1128/jb.171.7.3746-3753.1989
Lu, G. T., Tang, Y. Q., Li, C. Y., Li, R. F., An, S. Q., Feng, J. X., et al. (2009). An adenosine kinase exists in Xanthomonas campestris pathovar campestris and is involved in extracellular polysaccharide production, cell motility, and virulence. J. Bacteriol. 191, 3639–3648. doi: 10.1128/JB.00009-09
Luppens, S. B., Rombouts, F. M., Abee, T. (2002). The effect of the growth phase of Staphylococcus aureus on resistance to disinfectants in a suspension test. J. Food Prot. 65, 124–129. doi: 10.4315/0362-028X-65.1.124
Maddocks, S. E., Oyston, P. C. (2008). Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154, 3609–3623. doi: 10.1099/mic.0.2008/022772-0
Marbaniang, C. N., Gowrishankar, J. (2011). Role of ArgP (IciA) in lysine-mediated repression in Escherichia coli. J. Bacteriol. 193, 5985–5996. doi: 10.1128/JB.05869-11
Martinez-Antonio, A., Collado-Vides, J. (2003). Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6, 482–489. doi: 10.1016/j.mib.2003.09.002
Minezaki, Y., Homma, K., Nishikawa, K. (2005). Genome-wide survey of transcription factors in prokaryotes reveals many bacteria-specific families not found in archaea. DNA Res. 12, 269–280. doi: 10.1093/dnares/dsi016
Mitchell, P. J., Tjian, R. (1989). Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245, 371–378. doi: 10.1126/science.2667136
Narvel, J. M., Jakkula, L. R., Phillips, D. V., Wang, T., Lee, S. H., Boerma, H. R. (2001). Molecular mapping of Rxp conditioning reaction to bacterial pustule in soybean. J. Hered. 92, 267–270. doi: 10.1093/jhered/92.3.267
Naves, P., Del Prado, G., Huelves, L., Gracia, M., Ruiz, V., Blanco, J., et al. (2008). Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb. Pathog. 45, 86–91. doi: 10.1016/j.micpath.2008.03.003
Neilands, J. B. (1995). Siderophores - structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–26726. doi: 10.1074/jbc.270.45.26723
Nielsen, P. H., Jahn, A., Palmgren, R. (1997). Conceptual model for production and composition of exopolymers in biofilms. Water Sci. Technol. 36, 11–19. doi: 10.1016/S0273-1223(97)00318-1
Noinaj, N., Guillier, M., Barnard, T. J., Buchanan, S. K. (2010). TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60. doi: 10.1146/annurev.micro.112408.134247
Ochsner, U. A., Vasil, M. L., Alsabbagh, E., Parvatiyar, K., Hassett, D. J. (2000). Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182, 4533–4544. doi: 10.1128/jb.182.16.4533-4544.2000
Ohlendorf, D. H., Anderson, W. F., Matthews, B. W. (1983). Many gene-regulatory proteins appear to have a similar alpha-helical fold that binds DNA and evolved from a common precursor. J. Mol. Evol. 19, 109–114. doi: 10.1007/bf02300748
Pandey, A., Sonti, R. V. (2010). Role of the FeoB protein and siderophore in promoting virulence of Xanthomonas oryzae pv. oryzae on rice. J. Bacteriol. 192, 3187–3203. doi: 10.1128/JB.01558-09
Pandey, S. S., Patnana, P. K., Lomada, S. K., Tomar, A., Chatterjee, S. (2016). Co-regulation of iron metabolism and virulence associated functions by iron and XibR, a novel iron binding transcription factor, in the plant pathogen Xanthomonas. PloS Pathog. 12, e1006019. doi: 10.1371/journal.ppat.1006019
Pandey, S. S., Patnana, P. K., Rai, R., Chatterjee, S. (2017). Xanthoferrin, the alpha-hydroxycarboxylate-type siderophore of Xanthomonas campestris pv. campestris, is required for optimum virulence and growth inside cabbage. Mol. Plant Pathol. 18, 949–962. doi: 10.1111/mpp.12451
Park, H. J., Jung, H. W., Han, S. W. (2014). Functional and proteomic analyses reveal that wxcB is involved in virulence, motility, detergent tolerance, and biofilm formation in Xanthomonas campestris pv. vesicatoria. Biochem. Biophys. Res. Commun. 452, 389–394. doi: 10.1016/j.bbrc.2014.08.076
Park, H. J., Bae, N., Park, H., Kim, D. W., Han, S. W. (2017). Comparative proteomic analysis of three Xanthomonas spp. cultured in minimal and rich media. Proteomics 17, 1700142. doi: 10.1002/pmic.201700142
Park, H. J., Jung, B., Lee, J., Han, S. W. (2019). Functional characterization of a putative DNA methyltransferase, EadM, in Xanthomonas axonopodis pv. glycines by proteomic and phenotypic analyses. Sci. Rep. 9, 2446. doi: 10.1038/s41598-019-38650-3
Parsek, M. R., Ye, R. W., Pun, P., Chakrabarty, A. M. (1994). Critical nucleotides in the interaction of a LysR-type regulator with its target promoter region. catBC promoter activation by CatR. J. Biol. Chem. 269, 11279–11284.
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450. doi: 10.1093/nar/gky1106
Pratt, L. A., Kolter, R. (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30, 285–293. doi: 10.1046/j.1365-2958.1998.01061.x
Rai, R., Javvadi, S., Chatterjee, S. (2015). Cell-cell signalling promotes ferric iron uptake in Xanthomonas oryzae pv. oryzicola that contribute to its virulence and growth inside rice. Mol. Microbiol. 96, 708–727. doi: 10.1111/mmi.12965
Rashid, M. M., Ikawa, Y., Tsuge, S. (2016). GamR, the LysR-type galactose metabolism regulator, regulates hrp gene expression via transcriptional activation of two key hrp regulators, HrpG and HrpX, in Xanthomonas oryzae pv. oryzae. Appl. Environ. Microbiol. 82, 3947–3958. doi: 10.1128/AEM.00513-16
Reen, F. J., Haynes, J. M., Mooij, M. J., O’gara, F. (2013). A non-classical LysR-type transcriptional regulator PA2206 is required for an effective oxidative stress response in Pseudomonas aeruginosa. PloS One 8, e54479. doi: 10.1371/journal.pone.0054479
Rivera-Gomez, N., Segovia, L., Perez-Rueda, E. (2011). Diversity and distribution of transcription factors: their partner domains play an important role in regulatory plasticity in bacteria. Microbiology 157, 2308–2318. doi: 10.1099/mic.0.050617-0
Roy, A., Kucukural, A., Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. doi: 10.1038/nprot.2010.5
Schauer, K., Rodionov, D. A., De Reuse, H. (2008). New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33, 330–338. doi: 10.1016/j.tibs.2008.04.012
Schell, M. A. (1993). Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47, 597–626. doi: 10.1146/annurev.mi.47.100193.003121
Seong, H. J., Park, H. J., Hong, E., Lee, S. C., Sul, W. J., Han, S. W. (2016). Methylome analysis of two Xanthomonas spp. using single-molecule real-time sequencing. Plant Pathol. J. 32, 500–507. doi: 10.5423/PPJ.FT.10.2016.0216
Sheehan, B. J., Dorman, C. J. (1998). In vivo analysis of the interactions of the LysR-like regulator SpvR with the operator sequences of the spvA and spvR virulence genes of Salmonella typhimurium. Mol. Microbiol. 30, 91–105. doi: 10.1046/j.1365-2958.1998.01041.x
Skaar, E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PloS Pathog. 6, e1000949. doi: 10.1371/journal.ppat.1000949
Sole, M., Scheibner, F., Hoffmeister, A. K., Hartmann, N., Hause, G., Rother, A., et al. (2015). Xanthomonas campestris pv. vesicatoria secretes proteases and xylanases via the Xps type II secretion system and outer membrane vesicles. J. Bacteriol. 197, 2879–2893. doi: 10.1128/JB.00322-15
Sutherland, I. W. (2001). Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147, 3–9. doi: 10.1099/00221287-147-1-3
Tans-Kersten, J., Brown, D., Allen, C. (2004). Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol. Plant Microbe Interact. 17, 686–695. doi: 10.1094/MPMI.2004.17.6.686
Tong, J. F., Meng, L., Wang, X. W., Liu, L. X., Lyu, L. D., Wang, C., et al. (2016). The FBPase encoding gene glpX is required for gluconeogenesis, bacterial proliferation and division in vivo of Mycobacterium marinum. PloS One 11, e0156663. doi: 10.1371/journal.pone.0156663
Turner, P. C., Thomas, C. E., Stojiljkovic, I., Elkins, C., Kizel, G., Ala’aldeen, D. A., et al. (2001). Neisserial TonB-dependent outer-membrane proteins: detection, regulation and distribution of three putative candidates identified from the genome sequences. Microbiology 147, 1277–1290. doi: 10.1099/00221287-147-5-1277
Wei, Q., Phu, N. L. M., Dotsch, A., Hildebrand, F., Panmanee, W., Elfarash, A., et al. (2012). Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 40, 4320–4333. doi: 10.1093/nar/gks017
Wek, R. C., Hatfield, G. W. (1988). Transcriptional activation at adjacent operators in the divergent overlapping Ilvy and Ilvc promoters of Escherichia coli. J. Mol. Biol. 203, 643–663. doi: 10.1016/0022-2836(88)90199-4
Wengelnik, K., Bonas, U. (1996). HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178, 3462–3469. doi: 10.1128/jb.178.12.3462-3469.1996
Wengelnik, K., Van Den Ackerveken, G., Bonas, U. (1996). HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol. Plant Microbe Interact. 9, 704–712. doi: 10.1094/mpmi-9-0704
Woodger, F. J., Bryant, D. A., Price, G. D. (2007). Transcriptional regulation of the CO2-concentrating mechanism in a euryhaline, coastal marine cyanobacterium, Synechococcus sp strain PCC 7002: Role of NdhR/CcmR. J. Bacteriol. 189, 3335–3347. doi: 10.1128/JB.01745-06
Yura, K., Tomoda, S., Go, M. (1993). Repeat of a helix-turn-helix module in DNA-binding proteins. Protein Eng. 6, 621–628. doi: 10.1093/protein/6.6.621
Zaim, J., Kierzek, A. M. (2003). The structure of full-length LysR-type transcriptional regulators. Modeling of the full-length OxyR transcription factor dimer. Nucleic Acids Res. 31, 1444–1454. doi: 10.1093/nar/gkg234
Zhang, Z. G., Gosset, G., Barabote, R., Gonzalez, C. S., Cuevas, W. A., Saier, M. H. (2005). Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J. Bacteriol. 187, 980–990. doi: 10.1128/JB.187.3.980-990.2005
Zhu, J., Miller, M. B., Vance, R. E., Dziejman, M., Bassler, B. L., Mekalanos, J. J. (2002). Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 99, 3129–3134. doi: 10.1073/pnas.052694299
Zhu, Y. L., Hou, H. M., Zhang, G. L., Wang, Y. F., Hao, H. S. (2019). AHLs regulate biofilm formation and swimming motility of Hafnia alvei H4. Front. Microbiol. 10, 1330. doi: 10.3389/fmicb.2019.01330
Ziveri, J., Tros, F., Guerrera, I. C., Chhuon, C., Audry, M., Dupuis, M., et al. (2017). The metabolic enzyme fructose-1,6-bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella. Nat. Commun. 8, 853. doi: 10.1038/s41467-017-00889-7
Keywords: Xanthomonas axonopodis pv. glycines, LysR type transcriptional regulators, soybean, proteomics, virulence
Citation: Park H, Do E, Kim M, Park H-J, Lee J and Han S-W (2020) A LysR-Type Transcriptional Regulator LcrX Is Involved in Virulence, Biofilm Formation, Swimming Motility, Siderophore Secretion, and Growth in Sugar Sources in Xanthomonas axonopodis Pv. glycines. Front. Plant Sci. 10:1657. doi: 10.3389/fpls.2019.01657
Received: 03 June 2019; Accepted: 25 November 2019;
Published: 10 January 2020.
Edited by:
Brigitte Mauch-Mani, Université de Neuchâtel, SwitzerlandReviewed by:
Seiji Tsuge, Kyoto Prefectural University, JapanYong-Qiang He, Guangxi University, China
Copyright © 2020 Park, Do, Kim, Park, Lee and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Wook Han, c3doYW5AY2F1LmFjLmty
 Hanbi Park
Hanbi Park Eunsoo Do
Eunsoo Do Minyoung Kim
Minyoung Kim Hye-Jee Park
Hye-Jee Park Jongchan Lee
Jongchan Lee Sang-Wook Han
Sang-Wook Han