- 1Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture and Rural Affairs, Oil Crops Research Institute of Chinese Academy of Agricultural Sciences, Wuhan, China
- 2College of Life Sciences, Henan Agricultural University, Zhengzhou, China
- 3Key Laboratory of Crop Gene Resources and Germplasm Enhancement in Southern China, Ministry of Agriculture and Rural Affairs, Danzhou, China
- 4Tropical Crops Genetic Resources Institute, Chinese Academy of Tropical Agricultural Sciences, Danzhou, China
The peanut (Arachis hypogaea L.) is an important oilseed crop worldwide. Compared to other common edible vegetable oils, peanut oil contains a higher content of saturated fatty acids (SFAs), approximately 20–40% of which are very long chain fatty acids (VLCFAs). To understand the basis for this oil profile, we interrogated genes for peanut β-ketoacyl-CoA synthase (KCS), which is known to be a key enzyme in VLCFA biosynthesis. A total of 30 AhKCS genes were identified in the assembled genome of the peanut. Based on transcriptome data, nine AhKCS genes with high expression levels in developing seeds were cloned and expressed in yeast. All these AhKCSs could produce VLCFAs but result in different profiles, indicating that the AhKCSs catalyzed fatty acid elongation with different substrate specificities. Expression level analysis of these nine AhKCS genes was performed in developing seeds from six peanut germplasm lines with different VLCFA contents. Among these genes, the expression levels of AhKCS1 or AhKCS28 were, 4–10-fold higher than that of any other AhKCS. However, only the expression levels of AhKCS1 and AhKCS28 were significantly and positively correlated with the VLCFA content, suggesting that AhKCS1 and AhKCS28 were involved in the regulation of VLCFA content in the peanut seed. Further subcellular localization analysis indicated that AhKCS1 and AhKCS28 were located at the endoplasmic reticulum (ER). Overexpression of AhKCS1 or AhKCS28 in Arabidopsis increased the contents of VLCFAs in the seed, especially for very long chain saturated fatty acids (VLCSFAs). Taken together, this study suggests that AhKCS1 and AhKCS28 could be key genes in regulating VLCFA biosynthesis in the seed, which could be applied to improve the health-promoting and nutritional qualities of the peanut.
Introduction
The peanut (Arachis hypogaea L.) is a widely cultivated oilseed crop in tropical and subtropical regions, which provides a significant source of protein, folate, tocopherol, phytosterols, polyphenolics such as resveratrol, fiber, and edible oil (Bertioli et al., 2015; Han, 2016; Bertioli et al., 2019). Fatty acid composition is the main factor in determining the nutritional value and application purpose of vegetable oil (Napier and Graham, 2010; Napier et al., 2014). Peanut oil contains ∼20% saturated fatty acids (SFAs), which is higher than that in many commercial vegetable oils (Wang et al., 2013; Giakoumis, 2018). Limiting the intake of dietary SFAs is recommended by most dietary guidelines, because SFAs promote increasing undesirable low-density lipoprotein (LDL) cholesterol in the blood, leading to a high risk of cardiovascular disease (List, 2004; USDA, 2010; Briggs et al., 2017). In the peanut, SFAs are enriched in VLCSFAs, including arachidic acid (C20:0), behenic acid (C22:0), and lignoceric acid (C24:0) (Wang et al., 2013; Giakoumis, 2018). VLCFAs are fatty acids longer than 18 atoms (Haslam and Kunst, 2013; Giakoumis, 2018). Hence, reducing VLCFA content in the peanut is a prime target to improve the nutritional value of peanuts and peanut oil.
In plants, it has been proved that VLCFA biosynthesis is controlled by a condensing enzyme: β-ketoacyl-CoA synthase (KCS) (Todd et al., 1999; Sassa and Kihara, 2014; Huai et al., 2015; Usher et al., 2017). KCS catalyzes the first step of elongation with the substrate and tissue specificities (Blacklock and Jaworski, 2002, 2006; Haslam and Kunst, 2013; Huai et al., 2015). The expression level and substrate preference of KCS determine the ultimate chain length and contents of VLCFAs (James et al., 1995; Katavic et al., 2001; Denic and Weissman, 2007; Sun et al., 2013; Huai et al., 2015; Shi et al., 2017; Ozseyhan et al., 2018). For example, the KCS from Lunaria annua has been introduced into camelina (Camelina sativa) to produce nervonic acid (C24:1) (Guo et al., 2009; Huai et al., 2015). The fatty acid elongation 1 (FAE1) from Brassica napus is overexpressed in the rapeseed to increase the content of erucic acid (C22:1) (Nath et al., 2009; Li et al., 2012). In contrast, the silencing of FAE1 is used to decrease the contents of VLCFAs (Wang et al., 2010; Shi et al., 2017; Ozseyhan et al., 2018).
According to sequence similarity, KCS genes in higher plants are divided into two gene families, namely FAE1-type and elongation-type (ELO-type) (Haslam and Kunst, 2013; Guo et al., 2016). A total of 21 FAE1-type KCSs were identified in the Arabidopsis genome (Joubes et al., 2008). As the expression patterns and substrate specificities of KCSs are different, they are assigned to play different roles in seed oil biosynthesis (FAE1/KCS18) (James and Dooner, 1990; Lemieux et al., 1990; Kunst et al., 1992; James et al., 1995), development of epidermis (FDH/KCS10, HIC/KCS13) (Gray et al., 2000; Pruitt et al., 2000), suberin metabolism (KCS2/DAISY, KCS9, KCS20) (Franke et al., 2009; Lee et al., 2009; Kim J. et al., 2013), and cuticular lipid metabolism (CER6/CUT1/KCS6, KCS1, KCS16) (Todd et al., 1999; Fiebig et al., 2000; Hegebarth et al., 2017). The ELO-type family is homologous to animal condensing enzymes but shares little homology with the plant KCS enzymes (Leonard et al., 2004; Haslam and Kunst, 2013). Four ELO genes are characterized in Arabidopsis (Quist et al., 2009). Only HOS3 is reported to be involved in the biosynthesis of sphingolipid (Quist et al., 2009), while the functions of other plant ELO homologs remain to be investigated (Haslam and Kunst, 2013).
To reduce the contents of VLCFAs in the peanut, it is necessary to identify which KCS genes contribute to this trait. In this report, we identified AhKCS genes in the peanut, and investigated the expression profiles of these genes. The coding sequences (CDS) and expression levels of AhKCS genes in developing seeds were analyzed in peanut lines with different VLCFA contents. Correlation analysis between the AhKCS gene expression and VLCFA content identified the major genes contributing to VLCFA content. The identified genes were heterologously expressed in yeast and Arabidopsis to confirm their function and substrate specificities. Finally, the candidate AhKCSs for the seed VLCFA content were identified which could be applied in the improvement of peanut oil.
Materials and Methods
Plant Materials and Growth Conditions
Peanut (Arachis hypogaea L.) cultivar Zhonghua16, germplasm lines C-34, C-119, C-140, C-178, C-224, and C-296 were maintained in our lab. Peanut plants were sowed in the Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences (OCRI-CAAS) experimental field in Wuhan, China. Plants were grown under field conditions in a randomized block experimental design with three replications.
Arabidopsis (Arabidopsis thaliana) double mutant fae1/fad2 (Smith et al., 2003) was obtained from Dr. Edgar Cahoon’s Lab at the University of Nebraska-Lincoln, United States. Arabidopsis plants were grown in controlled-environment chambers with 16 h light (21°), 8 h dark (18°) cycle, 100 mE light intensity, and 60% relative humidity. Double mutant fae1/fad2 as control plants were included in each flat area to minimize any spatial aspects of the growth chamber.
Identification of AhKCS Genes
Datasets of the genome sequence were downloaded from the following sources. Arachis hypogaea: PeanutBase1 (Bertioli et al., 2019); Arabidopsis thaliana: TAIR2 (Lamesch et al., 2011).
Arabidopsis KCS genes were downloaded in TAIR, whose Pfam domains were retrieved from Pfam Database3. Standalone similarity searches for peptide sequences were performed through BLASTP under BLAST + executable suite (Camacho et al., 2009). In addition, an HMM search was performed with the “trusted cutoff” as the threshold for detecting the domains. The results of the two rounds of searches were merged, which was subject to another search against a library of Pfam-A families.
Genomic Distribution of AhKCSs
The chromosomal location and syntenic gene pairs information of KCS genes were downloaded from PeanutBase. The syntenic diagram illustrating the detailed genomic distribution of KCS genes was created by Circos4 (Krzywinski et al., 2009).
Alignment and Phylogeny Inference
Multiple sequence alignments were performed using the Clustal W program (Thompson et al., 1994). A phylogenetic tree was constructed using the neighbor-joining (NJ) method in the MEGA X software with 1,000 bootstrap replicates (Kumar et al., 2018).
Transcriptomic Analysis
Raw transcriptome data of 22 different tissues collected from cultivated peanut cv. Tifrunner were downloaded from NCBI (BioSample IDs: SAMN03944933-SAMN03944990) (Clevenger et al., 2016). Reads were mapped to the assembled genome of A. hypogaea which were deposited in-house. Read mapping and calculation of fragments per kilobase per million reads mapped (FPKM) were performed using TopHat2 (Kim D. et al., 2013). A heatmap was generated using pheatmap, an R package5.
RNA Extraction
For investigation of the expression profile in developing seeds, seeds of Zhonghua 16 from five specific stages were collected without pericarp: I – white and flat embryo [approximately 20 days after pollination (DAP)]; II – white and teardrop-shaped embryo (30 DAP); III – white and torpedo to round shaped embryo (40 DAP); IV – light pink and round embryo (50 DAP); V – dark pink, large and round embryo (60 DAP). For association analysis between KCS expression level and VLCFA contents, developing seeds were harvested at stage III from germplasm lines C-34, C-119, C-140, C-178, C-224, and C-296. Total RNA was extracted using TRIzol reagent (Sigma)6 according to the manufacturer’s instructions. Reverse transcription was performed using SuperScript IV First-Strand Synthesis System as described by the manufacturer (Invitrogen)7.
Quantitative Real-Time PCR (qRT-PCR) Analysis
Primers for qRT-PCR were designed using the IDT DNA Real Time PCR primer design tool8 (Supplementary Table S1). Real-time PCRs were performed using paired samples with three technical replicates on a Bio-Rad CFX96 Real-Time system (Bio-Rad)9 and DBI Bioscience Bestar-Real Time PCR Master Mix kit following the manufacturer’s instructions. The data were analyzed with LINREG as previously described (Huai et al., 2015). The experiment was repeated using at least three independent biological replicates, with three technical replicates for each biological sample.
Expression of AhKCS Genes in Yeast
The nine AhKCS genes were individually amplified by PCR from sequenced TA-clones using the primers in Supplementary Table S2, with different restriction sites. The PCR products were digested with corresponding restriction enzymes to the added sites in primers, respectively. The digested fragments were ligated into the vector pYX242, respectively (Supplementary Figure S1). Based on the inserted gene, the resulting plasmids were designated as pYX242-AhKCS1, pYX242-AhKCS4, pYX242-AhKCS10, pYX242-AhKCS13, pYX242-AhKCS17, pYX242-AhKCS23, pYX242-AhKCS25, pYX242-AhyKCS28, and pYX242-AhKCS29.
The vectors with AhKCS genes were introduced into Saccharomyces cerevisiae strain INVSc1 (Invitrogen) using the S.c. EasyComp TM transformation Kit (Invitrogen). Yeast cells transformed with a pYX242 empty vector were used as the control. The transformed yeast cells were selected on minimal agar plates lacking leucine. Transformants were first grown in SC-Leu (synthetic complete minus leucine) medium at 28°C overnight, suspended in SC-Leu medium and grown at 28°C for 2 days; then the transformants were selected and grown as described previously (Guo et al., 2009).
Yeast cells were grown in 50 ml SC-Leu medium at 28°C overnight to OD600 of 1.4, the cells were spun to form a pellet and used for biochemical analysis. Cell pellets were lyophilized and then transferred into glass tubes. Fatty acid methyl esters (FAMEs) were prepared with 2.5% (v/v) sulfuric acid/methanol for gas-chromatography analysis as previously described (Huai et al., 2015).
Cloning of AhKCS Genes From Peanut Plants
The AhKCS genes were cloned from developing seeds at stage III of peanut germplasm lines C-34, C-119, C-140, C-178, C-224, and C-296. Primers were designed based on the sequences of Arahy.IFJ1V3, Arahy.T9PLK1, Arahy.1FMC3R, Arahy.WQ1I1V, Arahy.TIY3DH, Arahy.3ATP19, Arahy.XI5WK7, Arahy.BGR17W, and Arahy.YW30D2. All the cloning primers are listed in Supplementary Table S3.
Subcellular Localization of AhKCS Genes
The AhKCS1 and AhKCS28 genes without a stop codon were amplified by PCR from sequenced TA-clones using the following primers: 5′- CATGGGTACCATGGCTGATGCAAAAGCA-3′ (KpnI) and 5′-CATGGGATCCTGATGGCAGATACCCTTGGA-3′ (BamHI) (the added restriction sites are underlined). The PCR products were digested with KpnI and BamHI, and the digested fragments were separately ligated into the vector pHBT (GenBank accession No. EF090408). As a result, AhKCS genes were inserted between the cauliflower mosaic virus (CaMV) 35S promoter and the green fluorescent protein (GFP) gene. The resulting plasmids were designated as pHBT-AhKCS1-GFP and pHBT-AhKCS28-GFP. The empty vector pHBT-GFP was analyzed as a control. The BnaA.FAE1 from rapeseed was fused with the red fluorescent protein (RFP) gene. The p35S:BnaA.FAE1-RFP construct was used as an endoplasmic reticulum (ER) marker (Wang et al., 2010; Haslam and Kunst, 2013).
The pHBT-GFP, pHBT-AhKCS1-GFP, and pHBT-AhKCS28-GFP were each transiently co-expressed with the ER marker in Arabidopsis protoplasts by PEG transformation, respectively (Nelson et al., 2007). The protoplast cells were isolated from 15-days-old seedlings of wild-type Arabidopsis. The leaf tissues were incubated in a solution containing 1.5% cellulase and 0.75% macerozyme for 4 h with gentle agitation. The protoplasts were collected by centrifuging at 100 g at 4°C for 2 min and resuspended in a solution of 0.4 M mannitol, 15 mM MgCl2, and 4 mM MES (pH 5.7). Then, 100 μl of the protoplast solution with 10 μg plasmid DNA was used for PEG-mediated transformation. After 10 h of incubation in the dark, fluorescence was examined under a laser-scanning confocal microscope (Olympus FV10-ASW).
Expression of AhKCS Genes in Arabidopsis
The AhKCS1 and AhKCS28 genes were amplified by PCR from sequenced TA-clones using the following primers with added NotI restriction sites: 5′-CATGGCGGCCGCATGGCTGATGCAAAAGCA-3′ and 5′-CATGGCGGCCGCTCAGATGGCAGATACCCTTGGA-3′ (the added restriction sites are underlined). The NotI digested fragments were separately inserted into the vector pKMS2 (Huai et al., 2015) between the seed-specific soybean oleosin-1 promoter and 3′UTR, to create pKMS2-AhKCS1 and pKMS2-AhKCS28. The cassettes comprising the oleosin promoter and 3′UTR flanking AhKCS1 or AhKCS28 gene were excised using AscI and respectively inserted into the binary vector pBinGlyRed2 containing a DsRed marker gene (Huai et al., 2015) to generate pBinGlyRed2-AhKCS1 and pBinGlyRed2-AhKCS28 (Supplementary Figure S2).
The constructs pBinGlyRed2-AhKCS1 and pBinGlyRed2-AhKCS28 were introduced into Agrobacterium tumefaciens GV3101 by electro-transformation. The Arabidopsis fae1/fad2 mutant was transformed according to the previously described method (Zhang et al., 2015). DsRed-positive seeds were identified using a green LED flashlight with a red camera filter lens (Huai et al., 2015).
Gas Chromatographic Analysis of Fatty Acid Compositions
For Arabidopsis, the DsRed positive mature seeds from each line were analyzed as a sample. For mature peanut seeds, 20 seeds from each line were collected and grinded as a sample. For developing peanut seeds, 5–10 seeds from each line were collected, lyophilized and grinded as a sample.
FAMEs were prepared from 25 mg finely grounded seeds with 2.5% (v/v) sulfuric acid/methanol as previously described (Huai et al., 2015). Fatty acids were transmethylated at 90°C for 1 h. After cooling to room temperature, 1 ml of aqueous 0.9% NaCl was added, and FAMEs were recovered by three sequential extractions with 1 ml of hexane. FAMEs were analyzed using an Agilent 7890B gas chromatograph with flame ionization detection and the DB-23 column. Fatty acids were identified by retention time according to previous studies (Huai et al., 2018; Liu et al., 2019).
Statistical Analysis
For correlation analysis on expression levels of AhKCS genes and VLCFA contents, the expression level data of each AhKCS gene and the contents of VLCFAs were collected at the same developing stage. For association analysis between AhKCS expression level and VLCFA content in different lines, the expression level data of each AhKCS gene at stage III was correlated with the VLCFA content in mature seeds. Correlation coefficients were estimated using the IBM SPSS Statistics software. For the comparison of multiple means, the test for statistical significance was performed with ANOVA and Fisher’s least significant difference (LSD) multiple-comparison test, using the same software. In all the analyses, only P < 0.05 was considered to be significant.
Results
Identification of AhKCS Gene Family Members in the Peanut Genome
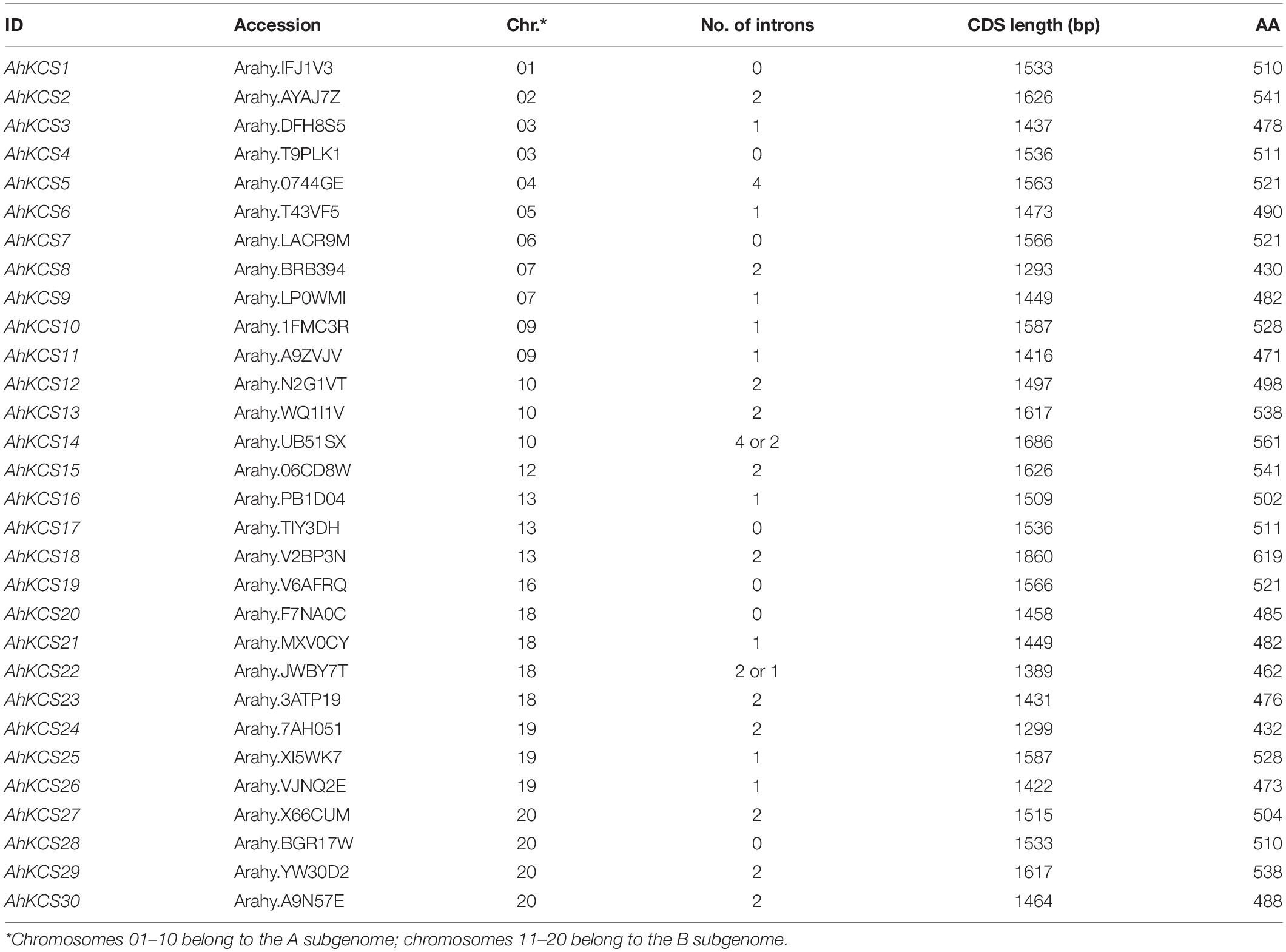
To identify all the AhKCS genes in the genome of the peanut, HMMER and BLAST searches were performed using 21 AtKCS genes from Arabidopsis as the query (Supplementary Table S4). A total of 30 putative AhKCSs were identified in the peanut, and were numbered based on their chromosomal locations, respectively (Table 1). None of these putative AhKCSs showed significant homology to AtELO genes (Supplementary Table S5), indicating that all the identified AhKCS genes were FAE1-type. All AhKCSs contained two domains, a 3-Oxoacyl-[acyl-carrier-protein (ACP)] synthase III C terminal domain (ACP_syn_III_C) and a FAE1/Type III polyketide synthase-like protein domain (FAE1_CUT1_RppA) (Figure 1). The putative AhKCS genes encoded proteins ranging from 432 amino acids to 619 amino acids. Most of these genes contained less than four introns, while seven AhKCS genes harbored no intron throughout their whole open reading frames. The alternatively spliced transcripts were identified in AhKCS14 and AhKCS22 genes (Table 1), implying that different isoforms may play different roles in the development of the peanut.
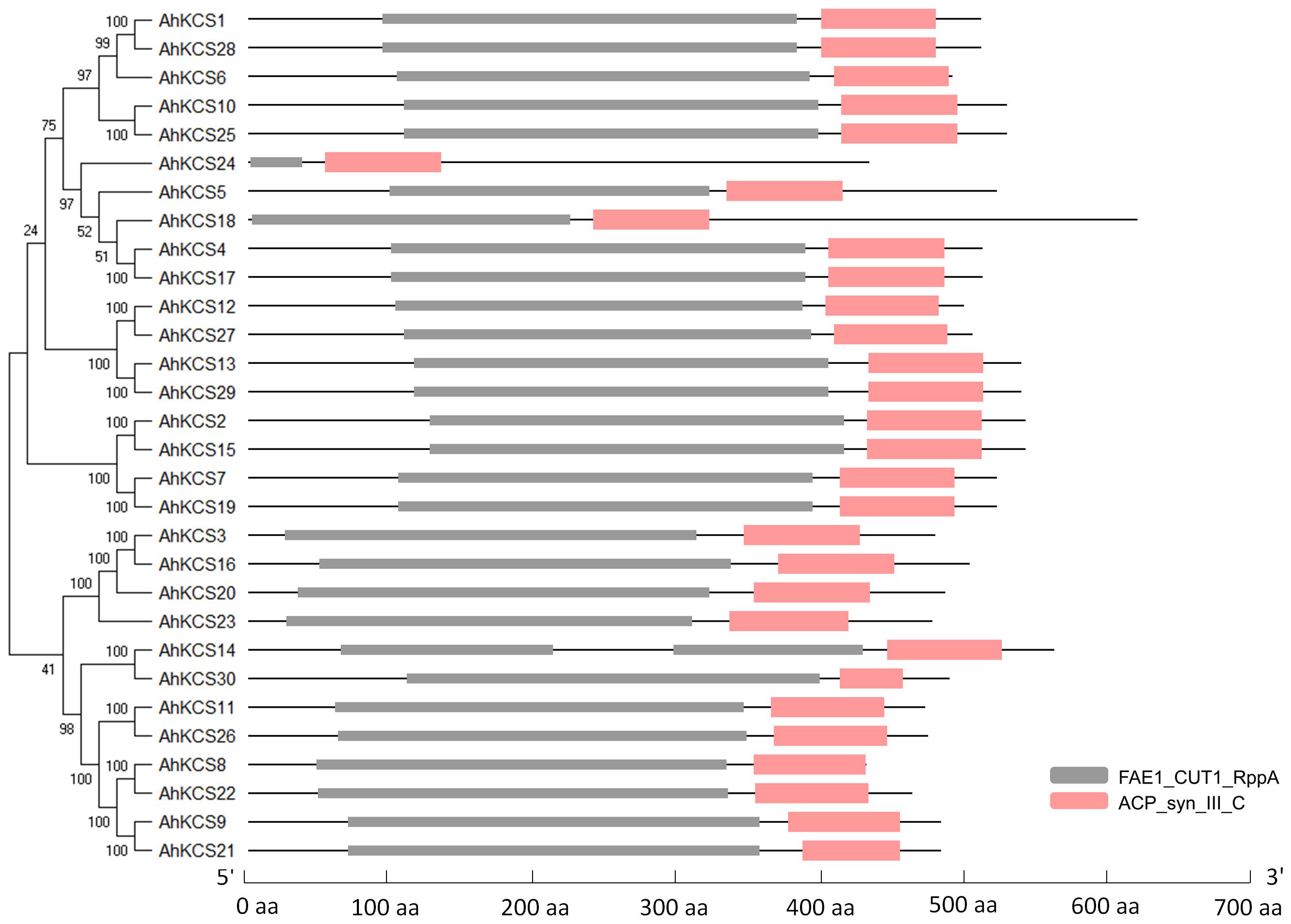
Figure 1. The conserved domains of AhKCSs from peanut. FAE1_CUT1_RppA: FAE1/Type III polyketide synthase-like protein domain (PF08392); ACP_syn_III_C: 3-Oxoacyl-[acyl-carrier-protein (ACP)] synthase III C terminal domain (PF08541).
AhKCS genes were widely distributed throughout genomes but were uneven among chromosomes. There were 14 AhKCS genes (AhKCS1 – AhKCS14) located on nine chromosomes in subgenome A, and 16 AhKCS genes (AhKCS15-AhKCS30) on six chromosomes in subgenome B (Table 1 and Figure 2). No AhKCS gene was detected on chromosome 08 in subgenome A, as well as chromosomes 11, 14, 15, and 17 in subgenome B (Table 1 and Figure 2). More than two AhKCS genes were identified on chromosomes 10 (3), 13 (3), 18 (4), 19 (3), and 20 (4) (Table 1 and Figure 2). Three chromosomes, 03, 07, and 09 each harbored two AhKCS genes, while seven chromosomes, 01, 02, 04, 05, 06, 12, and 16 each had just one AhKCS gene (Table 1 and Figure 2).
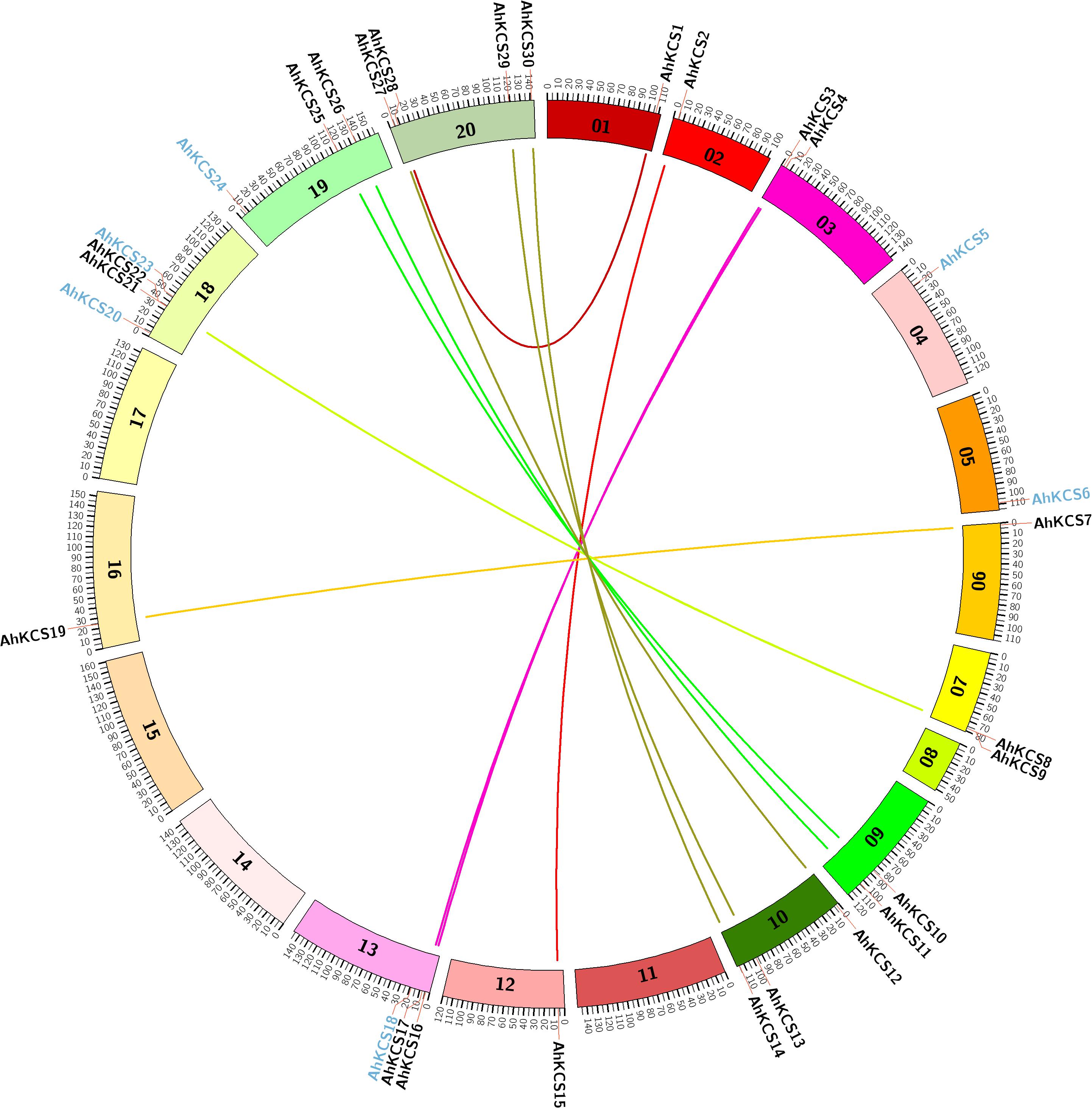
Figure 2. Genome-wide syntenic analysis of AhKCS genes on the chromosomes in peanut. 01–10, chromosomes from the A subgenome; 11–20, chromosomes from the B subgenome. The colored lines indicate the syntenic gene pairs between A and B subgenomes. The blue character represents the non-syntenic KCS genes.
Syntenic gene pairs are illustrated with a Circos diagram (Figure 2). Twelve AhKCS genes in subgenome A were paired with AhKCS genes in subgenome B and shared over 90% homology (Figure 2). AhKCS genes on chromosomes 02, 03, 06, 09, and 10 were syntenic to AhKCS genes on corresponding chromosomes 12, 13, 16, 19, and 20. However, AhKCS1 on chromosome 01 was paired with AhKCS28 on chromosome 20. Similarly, AhKCS8 and AhKCS9 on chromosome 07 were paired with AhKCS21 and AhKCS22 on chromosome 18 (Figure 2). AhKCS5 and AhKCS6 in subgenome A were not paired, as well as four AhKCS genes in subgenome B (AhKCS18, AhKCS20, AhKCS23, and AhKCS24) (Figure 2).
Phylogenetic Analysis of AhKCSs From Peanut Plants
An un-rooted phylogenetic tree was constructed in MEGA X based on the protein sequences of AhKCSs from the peanut, plus the Arabidopsis AtKCSs. The KCS proteins were divided into nine groups: α, β, γ, δ, ε, ζ, η, θ, and ι. No AhKCS was classified into Group ε, η, and ι, while no AtKCS was detected in Group α (Figure 3). These results indicate that AhKCS genes in the peanut might have undergone gene duplication and ensuing sub-functionalization and/or neo-functionalization, generating more complicated functions in the peanut than in Arabidopsis.
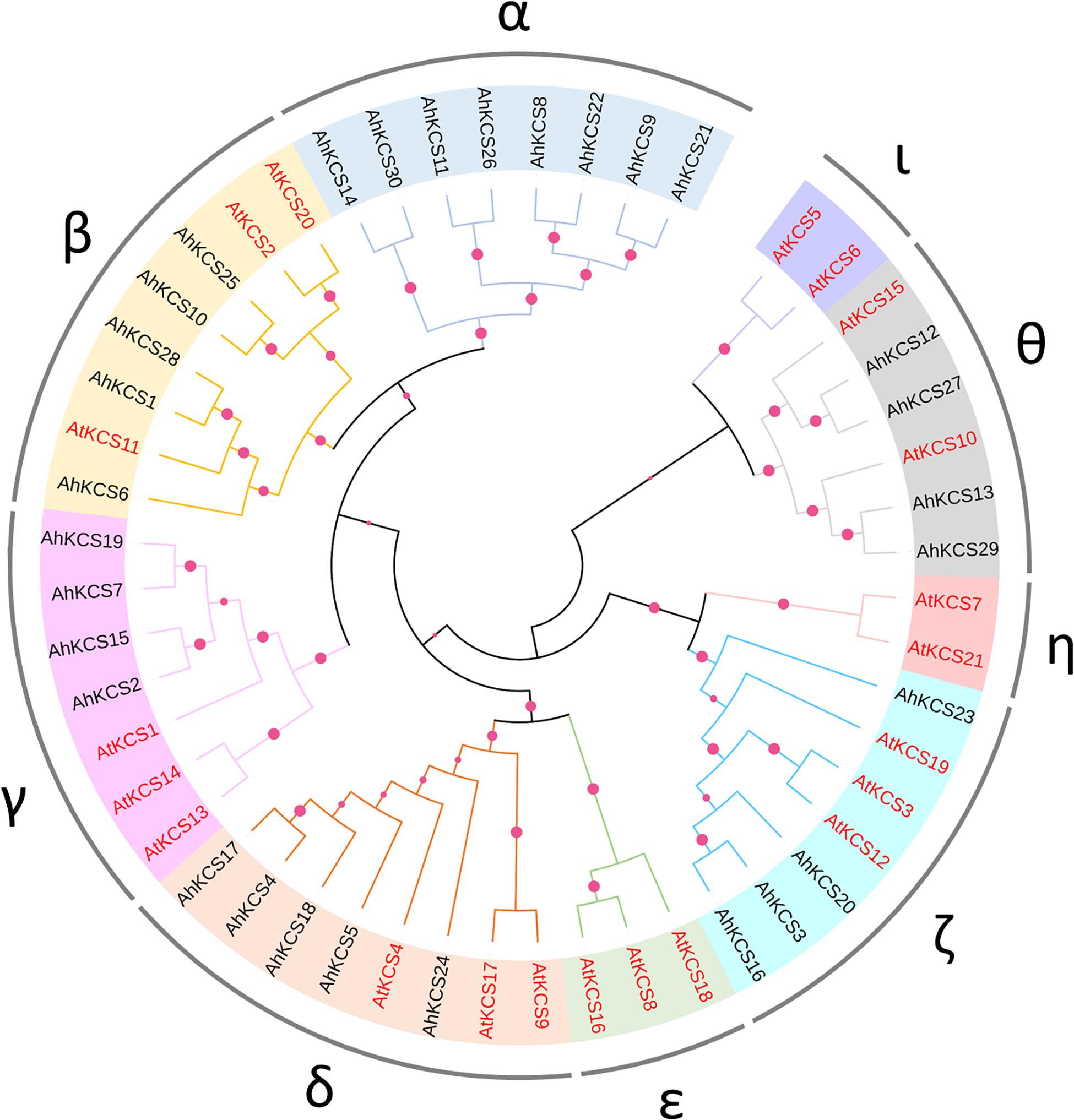
Figure 3. Phylogenetic analysis of AhKCS family members in peanut. A neighbor-joining phylogenetic tree was generated by MEGA X with full-length KCS sequences from the peanut and Arabidopsis. The colored circles at each node represent the bootstrap support (percentage).
Group α contained eight AhKCSs: AhKCS8, AhKCS9, AhKCS11, AhKCS14, AhKCS21, AhKCS22, AhKCS26, and AhKCS30, which were not grouped with any AtKCS. Group β harbored five AhKCSs (AhKCS1, AhKCS6, AhKCS10, AhKCS25, and AhKCS28) and three AtKCSs including the AtKCS2/DAISY AtKCS11 and AtKCS20. There were four AhKCSs (AhKCS2, AhKCS7, AhKCS15, and AhKCS19) and three AtKCSs (AtKCS1, AtKCS13, and AtKCS14) in Group γ. Five AhKCSs (AhKCS4, AhKCS5, AhKCS17, AhKCS18, and AhKCS24) and three AtKCSs (AtKCS4, AtKCS9, and AtKCS17) were classified into Group δ. In Group ζ, 4 AhKCSs (AhKCS3, AhKCS16, AhKCS20, and AhKCS23) were grouped with three AtKCSs (AtKCS3, AtKCS12, and AtKCS19). In Group θ, four AhKCSs (AhKCS12, AhKCS13, AhKCS27, and AhKCS29) were closely related to AtKCS10/FDH and AtKCS15 (Figure 3).
It has been proved that the seed specific expressed AtKCS18/FAE1 controls the VLCFA content in seed of Arabidopsis (James et al., 1995), so the peanut ortholog which was grouped with AtKCS18/FAE1 was of interest. However, there was no AhKCS classified into Group ε with AtKCS18/FAE1 (Figure 3). This might imply that there was no seed-specific AhKCS that regulated VLCFA content only in the seed.
Expression Patterns of AhKCS Genes in Peanut Plants
The expression levels of AhKCS genes in 22 tissues of the peanut were investigated using transcriptome data to identify those with the highest expression in the developing seed (Clevenger et al., 2016). The FPKM values of AhKCS genes were calculated by mapping reads to the peanut genome. A heat-map of AhKCS genes was created to demonstrate their expression profile (Figure 4).
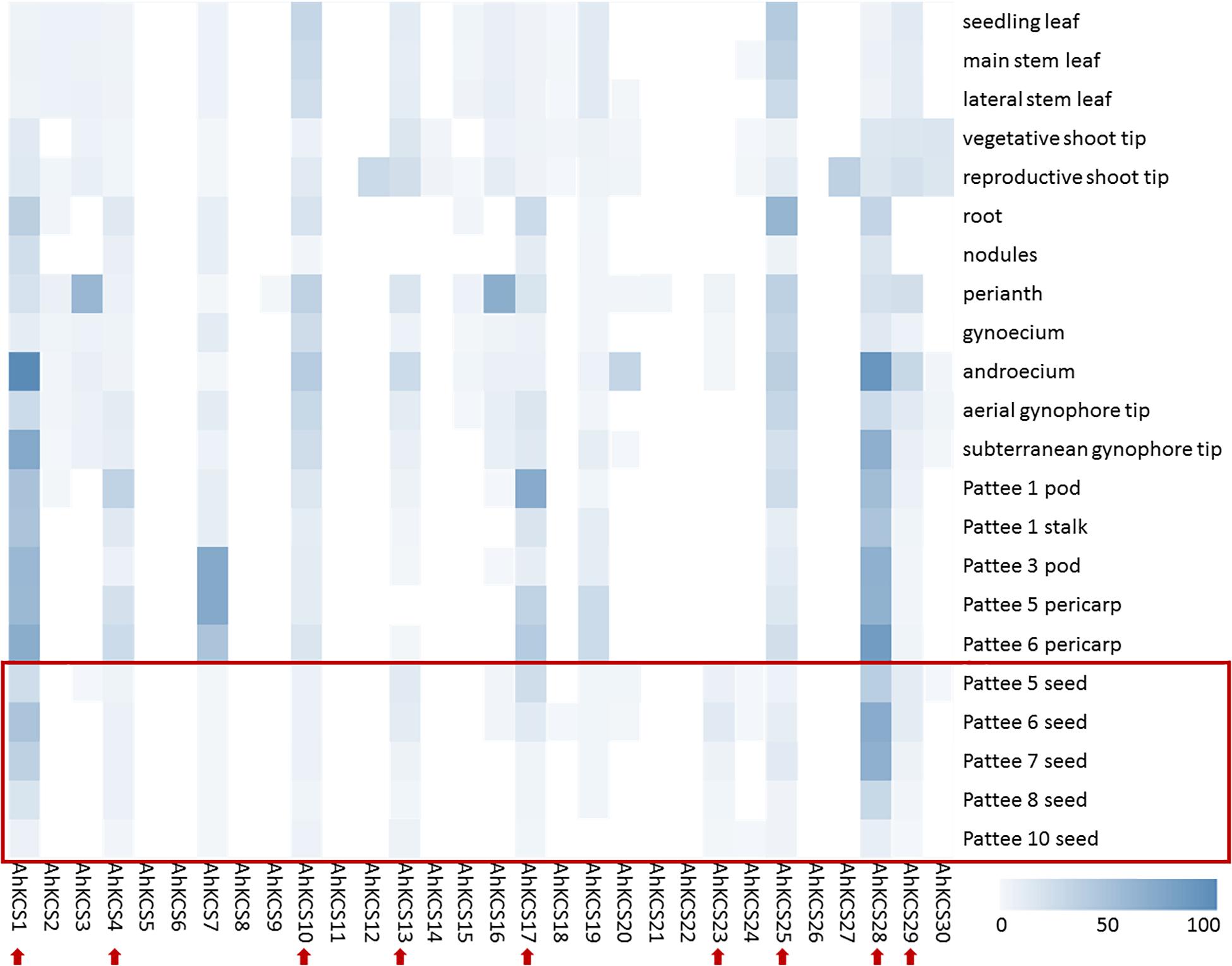
Figure 4. Heat map showing the expression patterns of AhKCS genes in 22 tissues of peanut. Seedling leaves and roots were collected 10 days post emergence; nodules were collected 25 days post emergence; main stem leaves, lateral leaves, vegetative shoot tips from main stem, and reproductive shoot tips from first lateral were collected when the first flower appeared; perianth, gynoecia and androecia were collected when flowers were blooming in the morning of anthesis; aerial gynophore tips were collected at elongating pegs prior to soil penetration; subterranean gynophore tips were collected 24 h after soil penetration; Pattee 1 pod, whole pod at pod swelling; Pattee 1 stalk, gynophore stalk at pod swelling; Pattee 3 pod, pericarp very watery, embryo very small and not easily removed; Pattee 5 pericarp, pericarp soft, not as watery, inner pericarp without cracks; Pattee 5 seed, embryo flat, white or just turning pink at one end; Pattee 6 pericarp, inner pericarp tissue beginning to show cracks or cottony; Pattee 6 seed, torpedo shaped, generally pink at embryonic-axis end of kernels; Pattee 7 seed, torpedo to round shaped, embryonic axis end of kernel pink, other end white to light pink; Pattee 8 seed, round, light pink all over; Pattee 10 seed, large, generally dark pink all over, seed coat beginning to dry out. Red arrows indicate AhKCS genes with some expression level in developing seed. The data was downloaded from NCBI (Clevenger et al., 2016).
In total, the expression of 22 AhKCS genes was detected in at least one tissue of the peanut (Figure 4). Six AhKCS genes (AhKCS5, AhKCS6, AhKCS8, AhKCS11, AhKCS22, and AhKCS26) were not expressed in any of the tested tissues from which the transcriptome data were derived (Figure 4). Eleven AhKCS genes (AhKCS1, AhKCS2, AhKCS4, AhKCS7, AhKCS10, AhKCS13, AhKCS17, AhKCS19, AhKCS25, AhKCS28, and AhKCS29) were constitutively expressed in the 22 tissues (Figure 4). AhKCS12 and AhKCS27 were specifically expressed in the leaf and stem, while AhKCS3, AhKCS16, and AhKCS20 were specifically expressed in the flower (Figure 4). No AhKCS gene was specifically expressed in the roots or gynophores (Figure 4). None of the AhKCS genes showed seed-specific expression, though AhKCS23 was dominantly expressed in both developing seeds and flowers (Figure 4).
Nine AhKCS genes, namely, AhKCS1, AhKCS4, AhKCS10, AhKCS13, AhKCS17, AhKCS23, AhKCS25, AhKCS28, and AhKCS29, were expressed in developing seeds (Figure 4). Except for AhKCS23, the other genes were constitutively expressed (Figure 4). From these results, candidate genes that might regulate VLCFA content in the developing peanut seed were selected for further characterization.
Correlation Analyses on Expression Levels of AhKCS and Contents of VLCFAs
Analysis of VLCFA content in developing seeds of Zhonghua16 were conducted at 20, 30, 40, 50, and 60 DAP. During the development of the seed, no decrease was observed in the accumulation of VLCFAs, except for C22:0 (Figure 5B). The contents of C20:0 and C24:0 continuously increased during the tested developmental stages (Figure 5B). The content of C20:1 soared during 20–40 DAP, plateaued during 40–50 DAP, and then sharply increased during 50–60 DAP (Figure 5B). However, the content of C22:0 sharply increased during 20–40 DAP, and then slightly decreased during 40-50 DAP followed by a moderate increase in the last 10 days (Figure 5B). The total content of VLCFA increased during 20–40 DAP, plateaued during 40–50 DAP, and then continued to increase (Figure 5B).
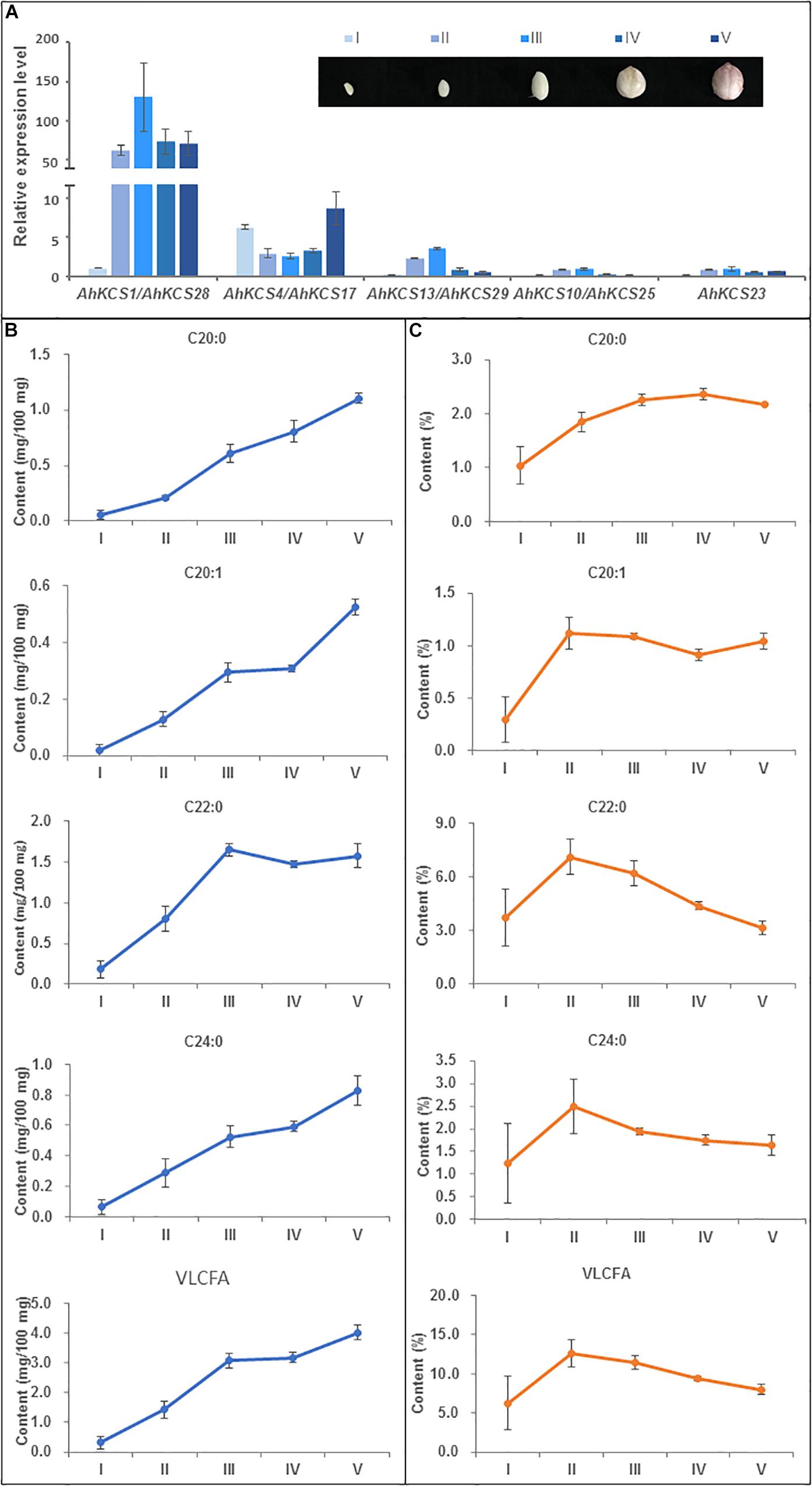
Figure 5. Relationship between AhKCS gene expression and the accumulation of VLCFAs during peanut seed development. (A) qRT-PCR analysis of AhKCS genes in developing peanut seeds. (B) Absolute contents of VLCFAs during peanut seed development. (C) Relative contents of VLCFAs during peanut seed development. Developing peanut seeds were harvested at five specific stages: I – white and flat embryo [approximately 20 days after pollination (DAP)]; II – white and teardrop-shaped embryo (30 DAP); III – white and torpedo to round shaped embryo (40 DAP); IV – light pink and round embryo (50 DAP); V – dark pink, large, and round embryo (60 DAP). AhACTIN gene expression level was used as a constitutive control. Data are presented as means with standard error bars SD, which were calculated based on 3–4 independent replicates. Samples from each replicate were measured using 5–10 developing seeds. Statistical significance was evaluated by ANOVA and Fisher’s least significant difference (LSD) multiple comparison.
Nevertheless, the relative contents of VLCFA first increased and then decreased during seed development (Figure 5C). The relative content of C20:0 gradually increased to a peak at 50 DAP, while the relative content of other VLCFAs, including total content of VLCFA, dramatically rose to a peak at 30 DAP (Figure 5C), suggesting that the relative contents of VLCFAs in the mature seed were lower.
AhKCS gene expression levels in developing peanut seeds were confirmed by qPCR at the same developing stages. Except for AhKCS4 and AhKCS17, the expression levels of other AhKCS genes rapidly increased during 20–40 DAP, and then slightly decreased during 40–60 DAP (Figure 5A). In contrast, the expression level of AhKCS4 and AhKCS17 first decreased during 20–40 DAP, then increased during 40–60 DAP (Figure 5A). The expression level of AhKCS1 and AhKCS28 were 5–10-fold higher than that of any other AhKCS gene in all developing stages except for stage I (Figure 5A). In later stages of development, no significant difference among the expression levels of the remaining AhKCS genes was detected, except for AhKCS1 and AhKCS28 (Figure 5A). These results confirmed that the expression patterns of AhKCS genes by qPCR analysis (Figure 5A) were consistent with transcriptome data (Figure 4).
The correlation analysis on expression levels of AhKCS genes and VLCFA contents was conducted. Only the expression levels of AhKCS1 and AhKCS28 were significantly and positively correlated with the content of C22:0 (R2 = 0.90, p < 0.05) (Supplementary Table S6). Unfortunately, the expression levels of other AhKCS genes were not correlated with any content of VLCFA (Supplementary Table S6). These results implied that among these nine AhKCS genes, AhKCS1, and AhKCS28 are the best candidate contributors to the accumulation of VLCFA in the peanut seed.
Heterologous Expression of AhKCS Genes in Yeast
To analyze the functions of the proteins encoded by the AhKCS genes, the coding regions of each gene was inserted to the vector pYX242 and transformed into yeast, along with BnaA.FAE1 and BnaC.FAE1 genes from rapeseed (Brassic napus L.) (Huai et al., 2018) as controls. Fatty acid profiles of the induced yeast cell were obtained as the evidence of substrate preference.
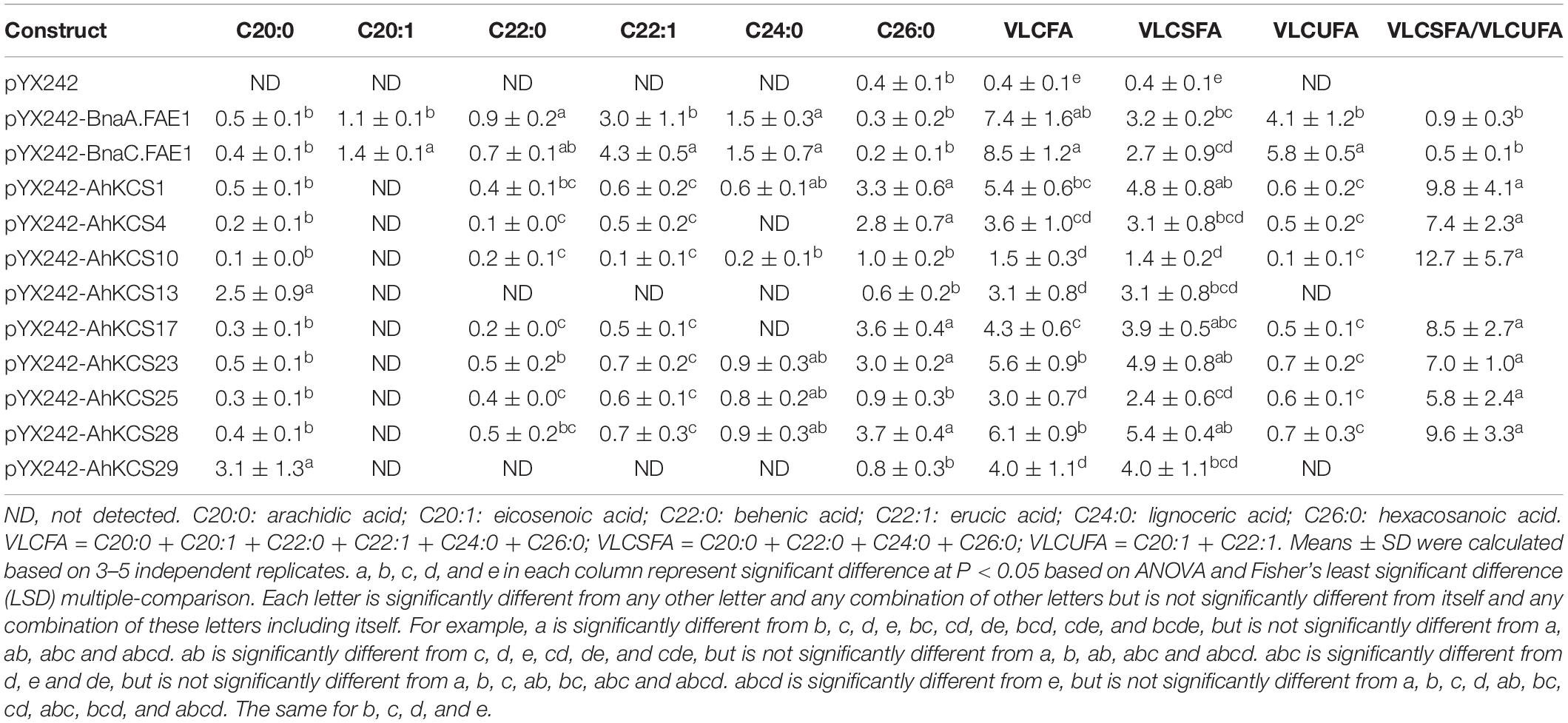
New peaks of saturated and monounsaturated VLCFAs, which are normally present in low abundance in yeast, were detected in yeast cells expressing AhKCS genes, and the VLCFA content in these cells was significantly increased (Figure 6 and Table 2). This result indicated that all the tested AhKCSs possessed fatty acid elongase activities. However, the VLCFA compositions in yeast cells with AhKCS genes were not the same. Five peaks for C20:0, C22:0, C22:1, C24:0, and C26:0 were detected in yeast cells with AhKCS1, AhKCS10, AhKCS23, AhKCS25, and AhKCS28 genes, while only two peaks for C20:0 and C26:0 were detected in yeast cells with AhKCS13 and AhKCS29 (Table 2). There were four peaks for C20:0, C22:0, C22:1, and C26:0 identified in yeast cells with AhKCS4 and AhKCS17 (Table 2). These results suggested that the substrate specificities of AhKCSs differed: AhKCS13 and AhKCS29 tended to accumulate C20:0; AhKCS4 and AhKCS17 preferentially stored C26 fatty acids; AhKCS1, AhKCS10, AhKCS23, AhKCS25, and AhKCS28 actively produced a variety of VLCFAs.
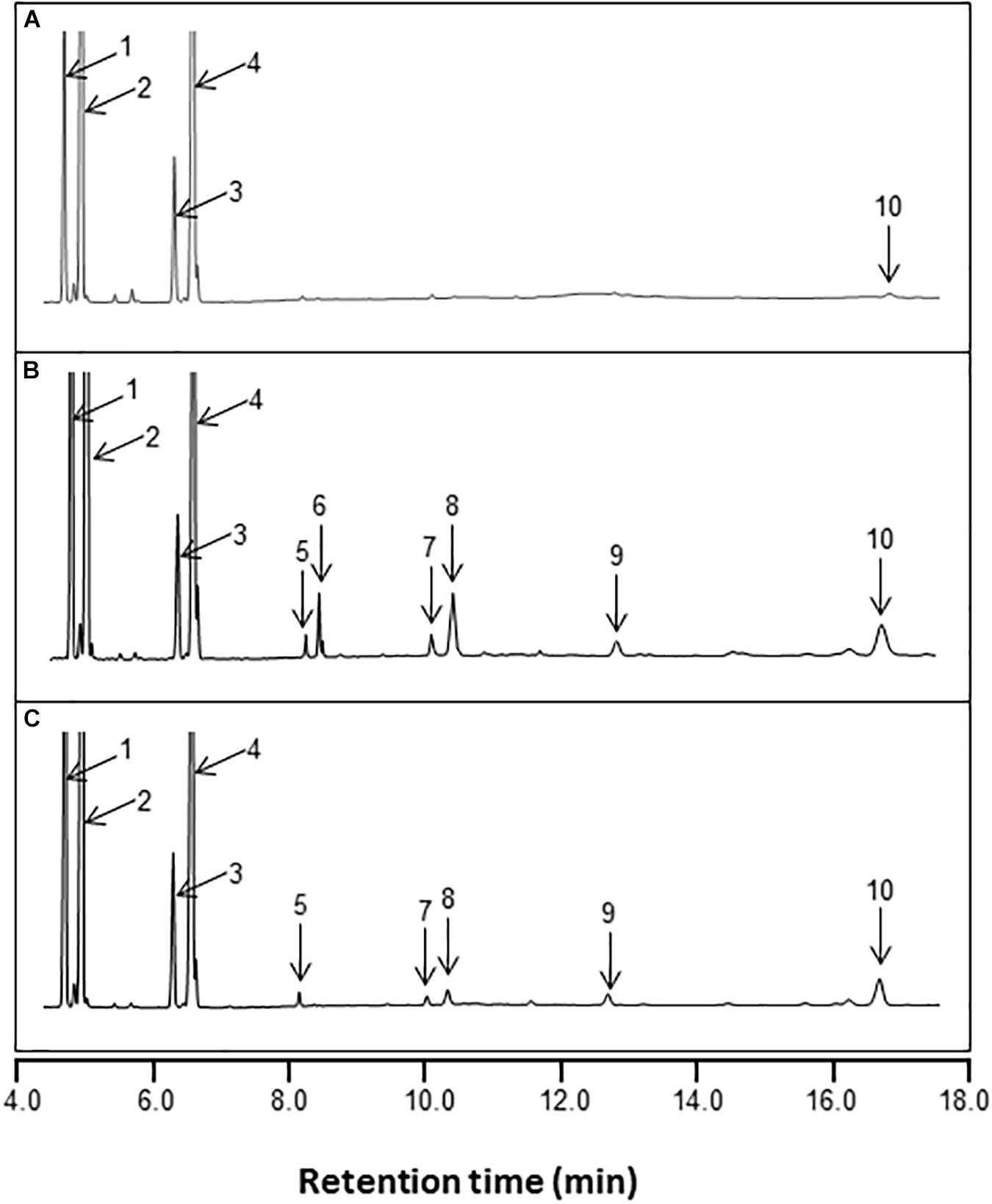
Figure 6. GC analysis of fatty acid methyl esters (FAMEs) from the total lipid fraction of yeast transformed with pYX242 (A), pYX242-BnaA.FAE1 (B), and pYX242-AhKCS1 (C). Fatty acid peak identifies are: 1 = C16:0; 2 = C16:1Δ9; 3 = C18:0; 4 = C18:1Δ9 and C18:1Δ11; 5 = C20:0; 6 = C20:1Δ11; 7 = C22:0; 8 = C22:1Δ13; 9 = C24:0; 10 = C26:0. C16:0, palmitic acid; C16:1, palmitoleic acid; C18:0, stearic acid; C18:1, oleic acid; C20:0, arachidic acid; C20:1, eicosenoic acid; C22:0, behenic acid; C22:1, erucic acid; C24:0, lignoceric acid; C26:0, hexacosanoic acid.
Compared to BnaA.FAE1 and BnaC.FAE1 from rapeseed, AhKCSs exhibited higher preference for C16 and/or C18 SFAs as substrates. In yeast cells expressing BnaA.FAE1 or BnaC.FAE1, C20:1 and C22:1 were detected but only C22:1 was found in AhKCSs-expressing cells (Figure 6 and Table 2). The content of very long-chain unsaturated fatty acids (VLCUFAs; C20:1 + C22:1) was significantly higher than that of very long-chain saturated fatty acids (VLCSFAs; C20:0 + C22:0 + C24:0 + C26:0) in yeast cells expressing BnaC.FAE1 (P < 0.05), though no obvious difference was observed in yeast cells expressing BnaA.FAE1. Conversely, the content of VLCUFA was significantly lower than that of VLCSFA in all yeast cells expressing any AhKCS gene (P < 0.05). Additionally, the ratio of VLCSFA to VLCUFA in yeast cells expressing BnaA.FAE1 or BnaC.FAE1 were 0.9 and 0.5, respectively, while the ratios in all yeast cells expressing any AhKCS gene were greater than 5.0 (Table 2). These results indicated that AhKCSs were more specific to saturated fatty acid substrates than Bna.FAE1s.
Analyses of AhKCS Genes in Peanut Lines With Different Contents of VLCFA
To confirm the roles of AhKCSs in regulating the VLCFA content of the peanut, these nine AhKCS genes were isolated and sequenced from six lines with different VLCFA contents in their seeds. The VLCFA contents in C-34, C-119, and C-140 were 4.6, 4.3, and 4.7%, while the VLCFA contents in C-178, C-296, and C-224 were 9.4, 9.6, and 9.8% (Figure 7B and Supplementary Table S7). However, no single nucleotide polymorphism (SNP) was detected in the coding sequences of AhKCS1, AhKCS4, AhKCS10, AhKCS13, AhKCS25, AhKCS28, and AhKCS29 among these six lines. One nucleotide substitution (C:G → A:T) at position 39 was identified in AhKCS17 of C-119, with no amino acid alternation (Supplementary Figure S3A). Another nucleotide substation (T:A → C:G) at position 1330 was identified in AhKCS23 of C-34 without changing the amino acid (Supplementary Figure S3B). None of the SNPs were therefore associated with VLCFA content.
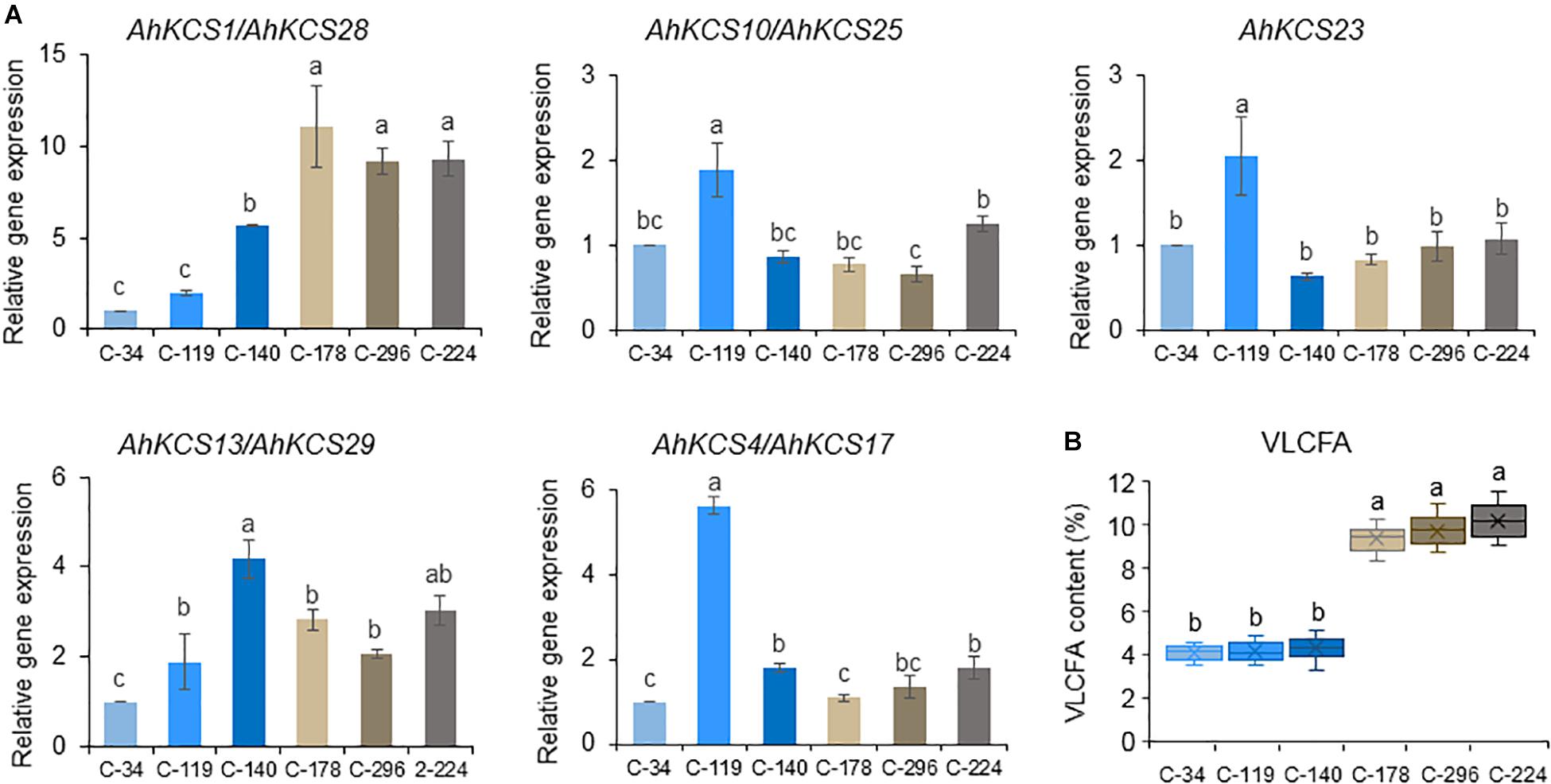
Figure 7. Expression levels of AhKCS genes in developing seeds from peanut lines with different VLCFA contents. (A) qRT-PCR analysis of AhKCS genes in developing seeds from peanut lines with different VLCFA contents. (B) VLCFA contents in mature seeds harvested from C-34, C-119, C-140, C-178, C-296, and C-224. The relative expression values of genes were measured on developing seeds harvested at 50 days after pollination. AhACTIN gene expression level was used as a constitutive control. Values are the means and SD of three independent biological replicates. a, b, and c represent significant difference at P < 0.05 based on ANOVA and Fisher’s least significant difference (LSD) multiple-comparison. Each letter is significantly different from any other letter and any combination of other letters but is not significantly different from itself and any combination of these letters including itself. For example, a is significantly different from b, c and bc, but is not significantly different from a and ab. b is significantly different from a and c, but is not significantly different from b, ab, and bc. c is significantly different from a, b, and ab, but is not significantly different from c and bc. ab is significantly different from c, but is not significantly different from a and b. bc is significantly different from a, but is not significantly different from b and c.
The expression levels of AhKCS genes were further investigated in the developing seeds from these six lines (Figure 7). All nine AhKCS genes were expressed, but only the expression levels of AhKCS1 and AhKCS28 were positively correlated with the VLCFA content (R2 = 0.93, p < 0.05) (Supplementary Table S8). The expression levels of AhKCS1 and AhKCS28 in C-34, C-119, and C-140 were at least twice as low as those in C-178, C-296, and C-224 (Figure 7A). There was no significant correlation between the expression levels of other AhKCS genes and the seed VLCFA content (Figure 7 and Supplementary Table S8). Based on these results, AhKCS1 and AhKCS28 were proposed to regulate the VLCFA content in peanut.
Subcellular Localization of AhKCS1 and AhKCS28
To examine the subcellular localization of AhKCS1 and AhKCS28, they were separately fused with GFP, and then co-expressed with the ER marker in Arabidopsis protoplasts by PEG induction. As expected, the green signal of the empty vector spread throughout the whole cell (Figure 8A). But the green signal of AhKCS1 (Figure 8B) and AhKCS28 (Figure 8C) were completely co-localized with the red ER marker, respectively. No green signal was observed in the nucleus (Figures 8B,C). These results indicated that AhKCS1 and AhKCS28 were located on the ER, where fatty acid elongase complex functions (Haslam and Kunst, 2013).
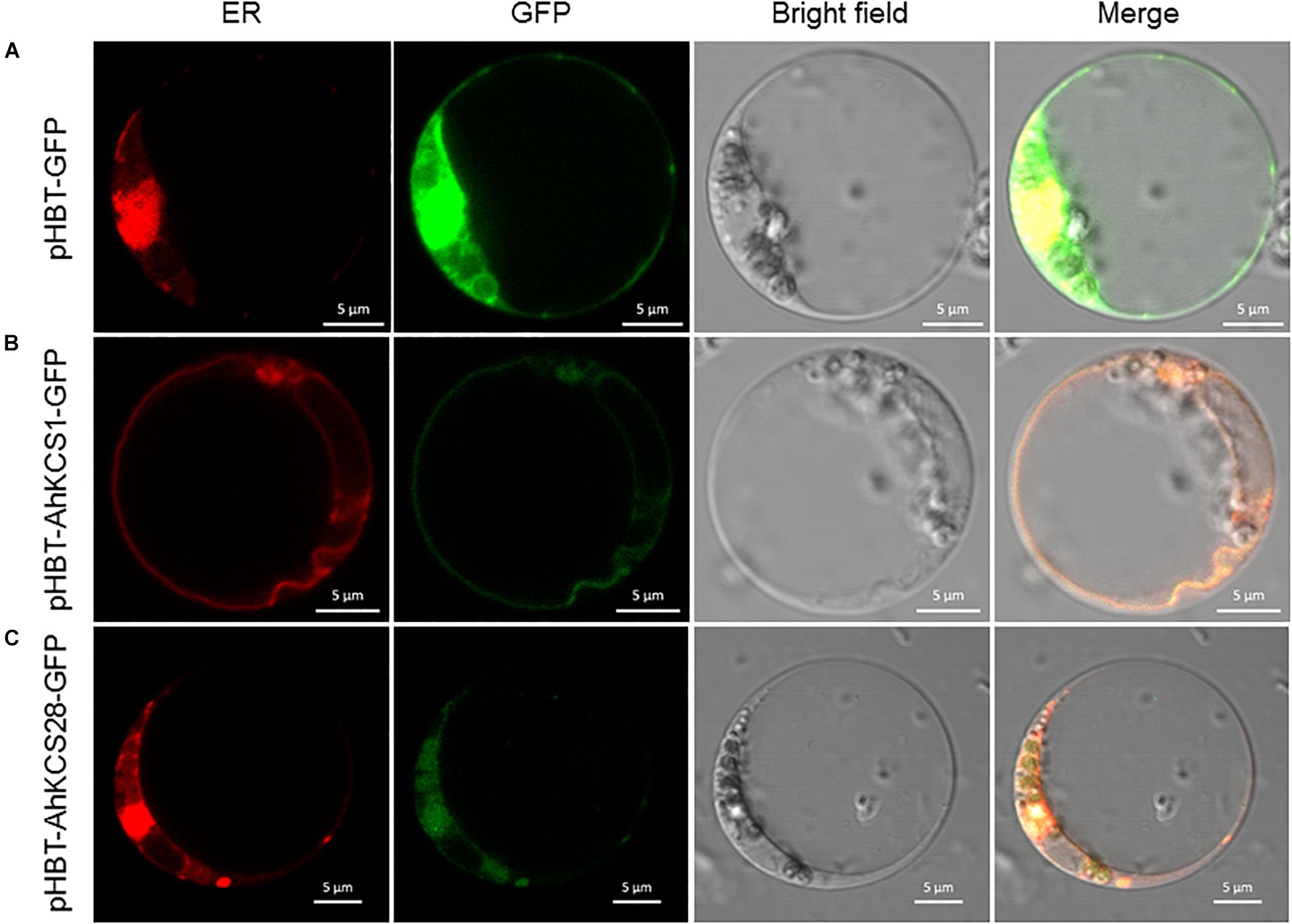
Figure 8. Subcellular location of AhKCS1 and AhKCS28 in the Arabidopsis protoplast cell. (A) An empty GFP vector served as the control. (B) AhKCS1-GFP (green) merged with endoplasmic reticulum (ER) marker (red). (C) AhKCS28-GFP (green) merged with ER marker (red).
Heterologous Expression of AhKCS1 and AhKCS28 Genes in Arabidopsis
To further confirm the function of AhKCS1 and AhKCS28 in plants, the AhKCS1 and AhKCS28 genes were heterologous expressed in the Arabidopsis fae1/fad2 double mutant. Eight AhKCS1-expressing T1 lines and five AhKCS28-expressing T1 lines were obtained, respectively (Supplementary Table S9). The fatty acid composition of the DsRed positive seeds from each line was determined. The VLCFA contents in seeds from the Arabidopsis fae1/fad2 double mutant with BnaA.FAE1 or BnaC.FAE1 (Huai et al., 2018) were also used as controls to compare the substrate specificity.
In seeds of the Arabidopsis fae1/fad2 double mutant, there were 3.2% of C20:0 and 2.2% of C20:1, without other VLCFAs (Figure 9A and Supplementary Table S9). In the seeds from AhKCS1- or AhKCS28-expressing lines, an extra VLCFA C22:0 was detected, and the contents of C20:0 and C20:1 were significantly increased. Moreover, the total VLCFA content was significantly increased from 5.5 to 25.8% and 28.4% in the AhKCS1- or AhKCS28-expressing lines, respectively (Figure 9A and Supplementary Table S9). These results suggested that AhKCS1 and AhKCS28 catalyzed fatty acid elongase activity in plants.
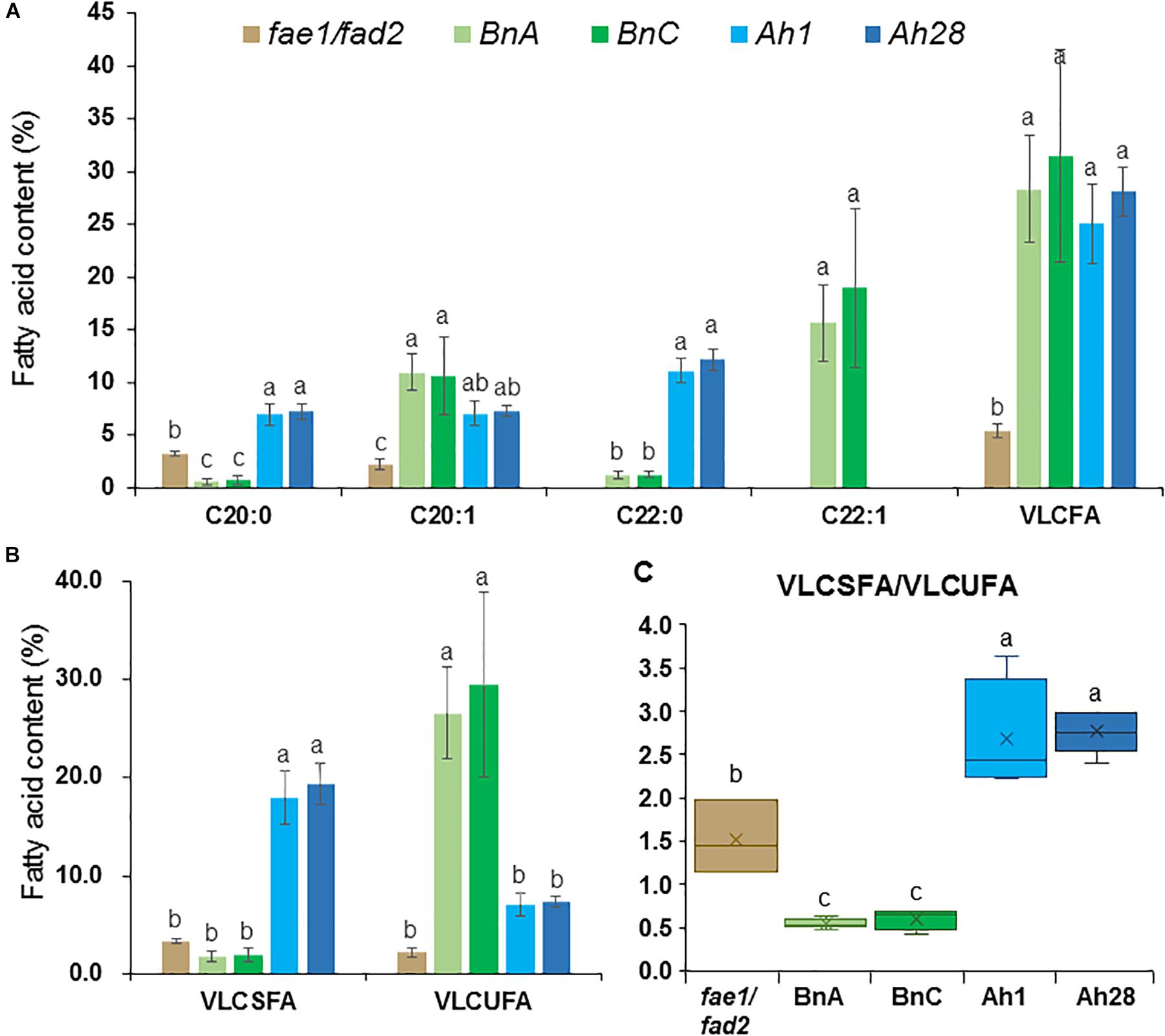
Figure 9. The VLCFA content in mature seeds harvested from Arabidopsis fae1/fad2 double mutant and T2 transgenic lines. (A) The contents of C20:0, C20:1, C22:0, C22:1 and VLCFA. (B) The contents of very long chain saturated fatty acid (VLCSFA) and very long chain unsaturated fatty acid (VLCUFA). (C) The ratio of VLCSFA to VLCUFA. VLCFA = C20:0 + C20:1 + C22:0 + C22:1; VLCSFA = C20:0 + C22:0; VLCUFA = C20:1 + C22:1. BnA, T2 lines transformed with BnaA.FAE1; BnC, T2 lines transformed with BnaC.FAE1; Ah1, T2 lines transformed with AhKCS1; Ah28, T2 lines transformed with AhKCS28. At least three replicates were used for each sample. Samples from each replicate were measured using 25 mg mature seeds. Different letters indicated statistically different at P < 0.05 based on ANOVA and Fisher’s least significant difference (LSD) multiple-comparison. The data was shown in Supplementary Table S9.
The compositions of VLCFAs in the seeds from AhKCS1- or AhKCS28-expressing lines were different to those in the seeds from BnaA.FAE1- and BnaC.FAE1-expressing lines (Figure 9A and Supplementary Table S9). In the BnaA.FAE1- and BnaC.FAE1-expressing lines, two extra VLCFAs were presented such as C22:0 and C22:1, compared to the VLCFAs in the fae1/fad2 double mutant. C22:1 was absent in the seeds from the AhKCS1- or AhKCS28-expressing lines (Figure 9A). The C20:0 content was significantly increased from 3.2 to 6.9% in AhKCS1- and 7.2% in AhKCS28-expressing lines, while it was significantly decreased to 0.6% in BnaA.FAE1- and 0.7% in BnaC.FAE1-expressing lines (0.7%) (Figure 9A). In addition, there were more VLCSFAs and less VLCUFA accumulated in AhKCS1- or AhKCS28-expressing lines compared to BnaA.FAE1- or BnaC.FAE1-expressing lines (Figure 9B). Furthermore, the VLCSFA/VLCUFA ratio in AhKCS1- and AhKCS28-expressing lines were 2.6 and 2.8, but the ratios in BnaA.FAE1- and in BnaC.FAE1-expressing lines were 0.5 and 0.6 (Figure 9C). These results again confirmed that AhKCS1 and AhKCS28 were inclined to use saturated fatty acids as the substrate.
Discussion
The high content of SFAs (15.5–27.5%) is a major negative quality factor in peanut oil. In the peanut, VLCSFA content varies from 2.5 to 8.5%, which accounts for 20–40% of the SFA content. Therefore, decreasing the VLCSFA content is an effective way to reduce SFA total content. It has been proved that the VLCFA content is mainly controlled by KCS in plants (James et al., 1995; Joubes et al., 2008; Huai et al., 2015), so we identified 30 AhKCS genes in the peanut genome (Table 1). There were approximately 20-30 members of the KCS gene family in angiosperms, such as 21 in Arabidopsis, 27 in soybean, and 19 in Medicago truncatula (Guo et al., 2016) – consistent with the large numbers of AhKCS family members in the peanut. All the identified AhKCSs contained two domains: an ACP_syn_III_C domain and a FAE1_CUT1_RppA domain, which have been proven to be necessary for KCSs (Figure 1; Sagar et al., 2015; Guo et al., 2016; Xiao et al., 2016). The ACP_syn_III_C domain is found in 3-Oxoacyl-[acyl-carrier-protein (ACP)] synthase III (EC:2.3.1.41), and is responsible for initiating the chain of fatty acid synthase reactions in plants and bacteria (Abbadi et al., 2000). KCS members harboring the FAE1_CUT1_RppA domain are described as 3-ketoacyl-CoA synthases, type III polyketide synthases, fatty acid elongases or fatty acid condensing enzymes, and are found in both prokaryotic and eukaryotic (mainly plant) species. This domain contains active site residues, as well as motifs involved in substrate binding (Nobutaka et al., 2002). The characteristics of ACP_syn_III_C and FAE1_CUT1_RppA domains supported that the genes identified in this study belong to KCS gene families.
Based on the phylogenetic analysis, all the AtKCS proteins were classified into eight groups (β, γ, δ, ε, ζ, η, θ, and ι) (Figure 3), which is consistent with previous studies (Joubes et al., 2008). All the identified AhKCS proteins were grouped into six groups: α, β, γ, δ, ζ, and θ (Figure 3). The AhKCSs in Group α were not grouped with any AtKCS (Figure 3), suggesting that these AhKCSs might be specific and acquired a different function in the peanut. The AhKCSs in group β were closely related to AtKCS2 and AtKCS20 (Figure 3), which were involved in the biosynthesis of suberin (Lee et al., 2009). The AhKCSs in group θ were grouped with AtKCS10 and AtKCS15 (Figure 3), which were reported to have participated in the biosynthesis of VLCFA in the epidermal cell (Pruitt et al., 2000). The AhKCSs in these two groups might also be involved in the development of the epidermis. Group ι was composed of AtKCS5/CER60 and AtKCS6/Cer6 only (Figure 3), which were responsible for the biosynthesis of cuticular wax (Fiebig et al., 2000). It hinted that the original homolog of AtKCS5/CER60 and AtKCS6/Cer6 might be deleted or functionally diverged in the peanut.
It has been shown that the VLCFA content in the seed is controlled by seed-specific expressed KCS genes, such as AtFAE1 in Arabidopsis (James et al., 1995), BnFAE1 in B. napus (Wang et al., 2010), and CaFAE1 in Crambe abyssinica (Mietkiewska et al., 2007). However, no seed-specific expressing AhKCS gene was found in the peanut (Figure 4). Furthermore, no AhKCS was grouped with AtKCS18/FAE1 (Figure 3). Perhaps because the VLCFAs in the peanut are not as abundant as those in Brassicaceae, the seed-specific AhKCS genes eliminated during evolution.
AhKCS1 and AhKCS28 were considered to be the genes that regulate the content of VLCFA in the peanut for three reasons. First, the expression level of these two genes was the highest in developing seed (Figure 5A), suggesting that these two genes could be one of the main factors to regulate VLCFA biosynthesis in the peanut seed. Second, AhKCS1 and AhKCS28 were the only two genes, whose expression level was significantly and positively correlated with the VLCFA content (Figure 7A and Supplementary Table S8), indicating that the expression level of these two genes might affect the VLCFA content in the peanut seed. Third, AhKCS1 and AhKCS28 exhibited substrate preference for SFA, which were different from BnFAE1s (Table 2 and Figure 9). The substrate specificities are in accordance with the features of VLCFA composition in the peanut and rapeseed, showing that the substrate specificities of these two AhKCSs were involved in determining the VLCFA compositions in the peanut seed. In summary, AhKCS1 and AhKCS28 could be candidate genes for decreasing VLCFA content in the peanut seed.
Due to the high identity of the syntenic AhKCS pairs, the expression level detected in this study is the sum of both genes. For example, the expression level of AhKCS1/AhKCS28 represented the total amount of expression levels of AhKCS1 and AhKCS28. According to the expression profile of AhKCS1 and AhKCS28, both of them were expressed in the developing seed (Figure 4). But the homology between AhKCS1and AhKCS28 was 97.8%, and there were only 14 SNPs between these two genes (Supplementary Figure S4). It is difficult to distinguish the expression levels from each other by qPCR. We designed five pairs of SNP specific primers, but we still failed to separate the expression levels between these two genes (data not shown). Although we could not figure out whose expression level decreased between AhKCS1 and AhKCS28, we still proved that both were involved in the biosynthesis of VLCFAs in the peanut, since both AhKCS1 and AhKCS28 were highly expressed in the developing seed and possessed the fatty acid elongase activities.
Many studies have demonstrated that knocking out the KCS genes with a high expression level in seeds could significantly reduce the VLCFAs. For example, the VLCFA contents were dramatically decreased from >20% to <1% in the Arabidopsis fae1 mutant (James et al., 1995), and from >40% to <2% in canola, which is the fae1 mutant of B. napus (Wang et al., 2010). The fae1 mutants of Camelina sativa were created by the CRISPR/Cas9 technology, and the VLCFA content was reduced from 22 to <2% (Ozseyhan et al., 2018). Suppressing the expression level of KCS genes in seeds could also decrease the VLCFA content. When the expression of FAE1 was silenced by RNAi in seeds of B.napus, the erucic acid was undetectable (Peng et al., 2010; Tian et al., 2011; Shi et al., 2017). Down-regulation of FAE1 genes in Crambe abyssinica significantly reduced the erucic acid content (Li et al., 2016). Therefore, knockout or silencing of the AhKCS1 and AhKCS28 genes in the peanut might be an effective way to decrease the VLCFA content and improve the health-promoting and nutritional qualities of the peanut.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
DH, YLei, and BL conceived and designed the experiments. HJ supplied the peanut lines. XX, JL, LY, YC, XW, NL, YK, and ZW performed the experiments. DH, YLi, PW, and YH analyzed the data. DH wrote the manuscript. DH, HJ, YLei, and BL revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Hubei Province (2017CFB161), the National Natural Science Foundation of China (31671734 and 31871662), the National Key R&D Program of China (2018YFD1000901) and the Fundamental Research Funds for Central Non-profit Scientific Institution (Y2018PT52). The funders had no role in experiment design, data analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Edgar B. Cahoon (Center for Plant Science Innovation and Department of Biochemistry, University of Nebraska-Lincoln) for supplying the vectors and editing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00406/full#supplementary-material
Footnotes
- ^ https://peanutbase.org/
- ^ https://www.arabidopsis.org/
- ^ https://sanger.ac.uk/
- ^ http://circos.ca/
- ^ https://www.r-project.org/
- ^ http://www.sigmaaldrich.com/
- ^ http://www.invitrogen.com/
- ^ http://www.idtdna.com/scitools/Applications/RealTimePCR
- ^ http://www.bio-rad.com
References
Abbadi, A., Brummel, M., Schütt, B. S., Slabaugh, M. B., Schuch, R., and Spener, F. (2000). Reaction mechanism of recombinant 3-oxoacyl-(acyl-carrier-protein) synthase III from Cuphea wrightii embryo, a fatty acid synthase type II condensing enzyme. Biochem. J. 345, 153–160.
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K., et al. (2015). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 47, 438–446.
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884.
Blacklock, B. J., and Jaworski, J. G. (2002). Studies into factors contributing to substrate specificity of membrane-bound 3-ketoacyl-CoA synthases. Eur. J. Biochem. 269, 4789–4798.
Blacklock, B. J., and Jaworski, J. G. (2006). Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem. Biophys. Res. Commun. 346, 583–590.
Briggs, M., Petersen, K., and Kris-Etherton, P. (2017). Saturated fatty acids and cardiovascular disease: replacements for saturated fat to reduce cardiovascular risk. Healthcare 5:29. doi: 10.3390/healthcare5020029
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421.
Clevenger, J., Chu, Y., Scheffler, B., and Ozias-Akins, P. (2016). A developmental transcriptome map for allotetraploid Arachis hypogaea. Fron. Plant Sci. 7:1446. doi: 10.3389/fpls.2016.01446
Denic, V., and Weissman, J. S. (2007). A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130, 663–677.
Fiebig, A., Mayfield, J. A., Miley, N. L., Chau, S., Fischer, R. L., and Preuss, D. (2000). Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12, 2001–2008.
Franke, R., Höfer, R., Briesen, I., Emsermann, M., Efremova, N., Yephremov, A., et al. (2009). The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J. 57, 80–95. doi: 10.1111/j.1365-313X.2008.03674.x
Giakoumis, E. G. (2018). Analysis of 22 vegetable oils’ physico-chemical properties and fatty acid composition on a statistical basis, and correlation with the degree of unsaturation. Renewab. Energy 126, 403–419.
Gray, J. E., Holroyd, G. H., Van Der Lee, F. M., Bahrami, A. R., Sijmons, P. C., Woodward, F. I., et al. (2000). The HIC signalling pathway links CO2 perception to stomatal development. Nature 408, 713–716.
Guo, H.-S., Zhang, Y.-M., Sun, X.-Q., Li, M.-M., Hang, Y.-Y., and Xue, J.-Y. (2016). Evolution of the KCS gene family in plants: the history of gene duplication, sub/neofunctionalization and redundancy. Mol. Genet. Genomics 291, 739–752. doi: 10.1007/s00438-015-1142-3
Guo, Y., Mietkiewska, E., Francis, T., Katavic, V., Brost, J. M., Giblin, M., et al. (2009). Increase in nervonic acid content in transformed yeast and transgenic plants by introduction of a Lunaria annua L. 3-ketoacyl-CoA synthase (KCS) gene. Plant Mol. Biol. 69, 565–575. doi: 10.1007/s11103-008-9439-9
Haslam, T. M., and Kunst, L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107. doi: 10.1016/j.plantsci.2013.05.008
Hegebarth, D., Buschhaus, C., Joubès, J., Thoraval, D., Bird, D., and Jetter, R. (2017). Arabidopsis ketoacyl-CoA synthase 16 (KCS16) forms C36/C38 acyl precursors for leaf trichome and pavement surface wax. Plant Cell Environ. 40, 1761–1776. doi: 10.1111/pce.12981
Huai, D., Zhang, Y., Zhang, C., Cahoon, E. B., and Zhou, Y. (2015). Combinatorial effects of fatty acid elongase enzymes on nervonic acid production in Camelina sativa. Plos One 10:e0131755. doi: 10.1371/journal.pone.0131755
Huai, D., Zhang, Y., Zhang, C., Cahoon, E. B., and Zhou, Y. (2018). Substrate specificities of fatty acid elongae BnaA.FAE1 and BnaC.FAE1 from rapeseed (Brassica napus L.). Chin. J. Oil Crop Sci. 40, 624–632.
James, D. W. Jr., and Dooner, H. K. (1990). Isolation of EMS-induced mutants in Arabidopsis altered in seed fatty acid composition. Theoret. Appl. Genet. 80, 241–245. doi: 10.1007/BF00224393
James, D. W., Lim, E., Keller, J., Plooy, I., Ralston, E., and Dooner, H. K. (1995). Directed Tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) Gene with the maize transposon activator. Plant Cell 7, 309–319.
Joubes, J. M., Raffaele, S., Bourdenx, B., Garcia, C., Laroche-Traineau, J., Moreau, P., et al. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67, 547–566. doi: 10.1007/s11103-008-9339-z
Katavic, V., Friesen, W., Barton, D. L., Gossen, K. K., Giblin, E. M., Luciw, T., et al. (2001). Improving erucic acid content in rapeseed through biotechnology: what can the Arabidopsis FAE1 and the yeast SLC1-1 genes contribute? Crop Sci. 41, 739–747.
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Kim, J., Jung, J. H., Lee, S. B., Go, Y. S., Kim, H. J., Cahoon, R., et al. (2013). Arabidopsis 3-ketoacyl-coenzyme A synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 162, 567–580. doi: 10.1104/pp.112.210450
Krzywinski, M. I., Schein, J. E., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kunst, L., Taylor, D. C., and Underhill, E. W. (1992). Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol. Biochem. 30, 425–434.
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2011). The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Lee, S. B., Jung, S. J., Go, Y. S., Kim, H. U., Kim, J. K., Cho, H. J., et al. (2009). Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 60, 462–475.
Lemieux, B., Miquel, M., Somerville, C., and Browse, J. (1990). Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theoret. Appl. Genet. 80, 234–240. doi: 10.1007/BF00224392
Leonard, A. E., Pereira, S. L., Sprecher, H., and Huang, Y.-S. (2004). Elongation of long-chain fatty acids. Prog. Lipid Res. 43, 36–54.
Li, X., Mei, D., Liu, Q., Fan, J., Singh, S., Green, A., et al. (2016). Down-regulation of crambe fatty acid desaturase and elongase in Arabidopsis and crambe resulted in significantly increased oleic acid content in seed oil. Plant Biotechnol. J. 14, 323–331. doi: 10.1111/pbi.12386
Li, X., Van Loo, E. N., Gruber, J., Fan, J., Guan, R., Frentzen, M., et al. (2012). Development of ultra-high erucic acid oil in the industrial oil crop Crambe abyssinica. Plant Biotechnol. J. 10, 862–870. doi: 10.1111/j.1467-7652.2012.00709.x
List, G. R. (2004). Decreasing trans and saturated fatty acid content in food oils. Food Technol. 1, 23–20.
Liu, N., Chen, H., Huai, D., Xia, F., Huang, L., Chen, W., et al. (2019). Four QTL clusters containing major and stable QTLs for saturated fatty acid contents in a dense genetic map of cultivated peanut (Arachis hypogaea L.). Mol. Breed. 39:23.
Mietkiewska, E., Brost, J. M., Giblin, E. M., Barton, D. L., and Taylor, D. C. (2007). Cloning and functional characterization of the fatty acid elongase 1 (FAE1) gene from high erucic Crambe abyssinica cv. Prophet. Plant Biotechnol. J. 5, 636–645.
Napier, J. A., and Graham, I. A. (2010). Tailoring plant lipid composition: designer oilseeds come of age. Curr. Opin. Plant Biol. 13, 329–336. doi: 10.1016/j.pbi.2010.01.008
Napier, J. A., Haslam, R. P., Beaudoin, F., and Cahoon, E. B. (2014). Understanding and manipulating plant lipid composition: metabolic engineering leads the way. Curr. Opin. Plant Biol. 19, 68–75. doi: 10.1016/j.pbi.2014.04.001
Nath, U. K., Wilmer, J. A., Wallington, E. J., Becker, H. C., and Möllers, C. (2009). Increasing erucic acid content through combination of endogenous low polyunsaturated fatty acids alleles with Ld-LPAAT+ Bn-fae1 transgenes in rapeseed (Brassica napus L.). Theoret. Appl. Genet. 118, 765–773. doi: 10.1007/s00122-008-0936-7
Nelson, B. K., Cai, X., and Nebenführ, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136.
Nobutaka, F., Ohnishi, Y., Ebizuka, Y., and Horinouchi, S. (2002). Alteration of reaction and substrate specificity of a bacterial type III polyketide synthase by site-directed mutagenesis. Biochem. J. 367, 781–789.
Ozseyhan, M. E., Kang, J., Mu, X., and Lu, C. (2018). Mutagenesis of the FAE1 genes significantly changes fatty acid composition in seeds of Camelina sativa. Plant Physiol. Biochem. 123, 1–7. doi: 10.1016/j.plaphy.2017.11.021
Peng, Q., Hu, Y., Wei, R., Zhang, Y., Guan, C., Ruan, Y., et al. (2010). Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in Brassica napus seeds. Plant Cell Rep. 29, 317–325. doi: 10.1007/s00299-010-0823-y
Pruitt, R. E., Vielle-Calzada, J.-P., Ploense, S. E., Grossniklaus, U., and Lolle, S. J. (2000). FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. U.S.A. 97, 1311–1316.
Quist, T. M., Sokolchik, I., Shi, H., Joly, R. J., Bressan, R. A., Maggio, A., et al. (2009). HOS3, an ELO-Like Gene, inhibits effects of ABA and implicates a S-1-P/ceramide control system for abiotic stress responses in Arabidopsis thaliana. Mol. Plant 2, 138–151. doi: 10.1093/mp/ssn085
Sagar, M., Pandey, N., Qamar, N., Singh, B., and Shukla, A. (2015). Domain analysis of 3 Keto Acyl-CoA synthase for structural variations in Vitis vinifera and Oryza brachyantha using comparative modelling. Interdiscip. Sci. 7, 7–20. doi: 10.1007/s12539-013-0017-8
Sassa, T., and Kihara, A. (2014). Metabolism of very long-chain fatty acids: genes and pathophysiology. Biomol. Ther. 22, 83–92. doi: 10.4062/biomolther.2014.017
Shi, J., Lang, C., Wang, F., Wu, X., Liu, R., Zheng, T., et al. (2017). Depressed expression of FAE1 and FAD2 genes modifies fatty acid profiles and storage compounds accumulation in Brassica napus seeds. Plant Sci. 263, 177–182. doi: 10.1016/j.plantsci.2017.07.014
Smith, M. A., Moon, H., Chowrira, G., and Kunst, L. (2003). Heterologous expression of a fatty acid hydroxylase gene in developing seeds of Arabidopsis thaliana. Planta 217, 507–516.
Sun, X., Pang, H., Li, M., Peng, B., Guo, H., Yan, Q., et al. (2013). Evolutionary pattern of the FAE1 gene in Brassicaceae and its correlation with the erucic acid trait. Plos One 8:e83535. doi: 10.1371/journal.pone.0083535
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Tian, B., Wei, F., Shu, H., Zhang, Q., Zang, X., and Lian, Y. (2011). Decreasing erucic acid level by RNAi-mediated silencing of fatty acid elongase 1 (BnFAE1. 1) in rapeseeds (Brassica napus L.). Afr. J. Biotechnol. 10, 13194–13201.
Todd, J., Post-Beittenmiller, D., and Jaworski, J. G. (1999). KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17, 119–130.
USDA (2010). US Department of Health and Human Services. Dietary Guidelines for Americans. Washington, DC: US Government Printing Office.
Usher, S., Han, L., Haslam, R. P., Michaelson, L. V., Sturtevant, D., Aziz, M., et al. (2017). Tailoring seed oil composition in the real world: optimising omega-3 long chain polyunsaturated fatty acid accumulation in transgenic Camelina sativa. Sci. Rep. 7:6570. doi: 10.1038/s41598-017-06838-0
Wang, M. L., Chen, C. Y., Tonnis, B., Barkley, N. A., Pinnow, D. L., Pittman, R. N., et al. (2013). Oil, fatty acid, flavonoid, and resveratrol content variability and FAD2A functional SNP genotypes in the US peanut mini-core collection. J. Agricul. Food Chem. 61, 2875–2882.
Wang, N., Shi, L., Tian, F., Ning, H., Wu, X., Long, Y., et al. (2010). Assessment of FAE1 polymorphisms in three Brassica species using EcoTILLING and their association with differences in seed erucic acid contents. BMC Plant Biol. 10:137. doi: 10.1186/1471-2229-10-137
Xiao, G. H., Wang, K., Huang, G., and Zhu, Y. X. (2016). Genome-scale analysis of the cotton KCS gene family revealed a binary mode of action for gibberellin A regulated fiber growth. J. Integrat. Plant Biol. 58, 577–589. doi: 10.1111/jipb.12429
Keywords: peanut, β-ketoacyl-CoA synthase (KCS), very long chain fatty acid (VLCFA) biosynthesis, expression profiling, function analysis
Citation: Huai D, Xue X, Li Y, Wang P, Li J, Yan L, Chen Y, Wang X, Liu N, Kang Y, Wang Z, Huang Y, Jiang H, Lei Y and Liao B (2020) Genome-Wide Identification of Peanut KCS Genes Reveals That AhKCS1 and AhKCS28 Are Involved in Regulating VLCFA Contents in Seeds. Front. Plant Sci. 11:406. doi: 10.3389/fpls.2020.00406
Received: 15 November 2019; Accepted: 20 March 2020;
Published: 07 May 2020.
Edited by:
Xue-Rong Zhou, Agriculture and Food, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaReviewed by:
Joaquín J. Salas, Instituto de la Grasa (IG), SpainBenjamin Pouvreau, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia
Copyright © 2020 Huai, Xue, Li, Wang, Li, Yan, Chen, Wang, Liu, Kang, Wang, Huang, Jiang, Lei and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Lei, bGVpeW9uZ0BjYWFzLmNu; Boshou Liao, bGJvc2hvdUBob3RtYWlsLmNvbQ==
 Dongxin Huai
Dongxin Huai Xiaomeng Xue1
Xiaomeng Xue1 Jianguo Li
Jianguo Li Xin Wang
Xin Wang Zhihui Wang
Zhihui Wang Yi Huang
Yi Huang Huifang Jiang
Huifang Jiang Yong Lei
Yong Lei Boshou Liao
Boshou Liao