- 1Beijing Advanced Innovation Center of Tree Breeding by Molecular Design, Beijing Forestry University, Beijing, China
- 2College of Biological Sciences and Technology, Beijing Forestry University, Beijing, China
Plant hexokinases (HXKs) are a class of multifunctional proteins that not only act as the enzymes required for hexose phosphorylation but also serve as sugar sensors that repress the expression of some photosynthetic genes when internal glucose level increases and regulators of cell metabolism and some sugar-related signaling pathways independent on their catalytic actives. The HXKs have been studied in many plants; however, limited information is available on HXKs of moso bamboo (Phyllostachys edulis). In this study, we identified and characterized 12 hexokinase genes in moso bamboo. Phylogenetic analysis revealed that the moso bamboo hexokinases (PeHXKs) were classifiable into five subfamilies which represented the three types of hexokinases in plants. Gene structure and conserved motif analysis showed that the PeHXK genes contained diverse numbers of introns and exons and that the encoded proteins showed similar motif organization within each subfamily. Multiple sequence alignment revealed that the PeHXK proteins contained conserved domains, such as phosphate 1 (P1), phosphate 2 (P2), adenosine, and a sugar-binding domain. Evolutionary divergence analysis indicated that the PeHXK, OsHXK, and BdHXK families underwent negative selection and experienced a large-scale duplication event approximately 19–319 million years ago. Expression analysis of the PeHXK genes in the leaf, stem, root, and rhizome of moso bamboo seedlings indicated that the PeHXKs perform pivotal functions in the development of moso bamboo. A protein subcellular localization assay showed that PeHXK5a, PeHXK8, and PeHXK3b were predominantly localized in mitochondria, and PeHXK8 protein was also detected in the nucleus. The HXK activity of the PeHXK5a, PeHXK8, and PeHXK3b was verified by a functional complementation assay using the HXK-deficient triple-mutant yeast strain YSH7.4-3C (hxk1, hxk2, and glk1), and the results showed that the three PeHXKs had the plant HXK-specific enzyme traits. The present findings would provide a foundation for further functional analysis of the PeHXK gene family.
Introduction
In plants, sugars are produced predominantly in the leaves during photosynthesis. As one type of sugar, sucrose serves as the core primary metabolite for sink tissues, and as a signaling molecule that regulates plant growth and development (Aguilera-Alvarado and Sanchez-Nieto, 2017). Sucrose is mainly catalyzed by invertase or sucrose synthase into hexoses, including glucose and fructose, both of which must be phosphorylated by hexose-phosphorylating enzymes before further metabolism (Granot, 2008; David-Schwartz et al., 2013). In plants, only two kinds of hexose phosphorylating enzymes, hexokinases (HXKs) and fructokinases (FRKs), have been found (Dai et al., 1999). While HXKs phosphorylate a broad spectrum of hexoses such as glucose and fructose, FRKs specifically phosphorylate fructose (Dai et al., 2002). However, for HXKs in tomato, their affinities to glucose are much higher than that to fructose, and meanwhile, their affinity to fructose is much lower than that of plant FRKs, indicating plant HXKs are actually glucose phosphorylating enzymes (Granot, 2007; Aguilera-Alvarado and Sanchez-Nieto, 2017).
Using biochemical, genetic, or bioinformatics methods, plant HXK gene families have been identified in many species (Karve et al., 2010; Aguilera-Alvarado and Sanchez-Nieto, 2017), including Arabidopsis thaliana (Karve et al., 2008), Physcomitrella patens (Olsson et al., 2003), Lycopersicon esculentum (Damari-Weissler et al., 2006; Granot, 2007), Solanum lycopersicum (Kandel-Kfir et al., 2006), Nicotiana tabacum (Giese et al., 2005), Oryza sativa (Cho et al., 2006), Solanum tuberosum (Jon et al., 2002), Vitis vinifera (Fei et al., 2013), Zea mays (Zhang et al., 2014), Manihot esculenta Grantz (Geng et al., 2017) Camellia sinensis (Li et al., 2017), Brassica napus (Wang et al., 2018), and pear (Pyrus × bretschneideri) (Zhao et al., 2019). On the basis of N-terminal amino acid sequences, the hexokinases are classified into two main types (type A and type B), and two rare types (type C and type D) (Aguilera-Alvarado and Sanchez-Nieto, 2017). The type A HXKs, such as AtHXK3, OsHXK4, and NtHXK2, contain plastid-signaling peptides that indicate their potential localization to plastids (Giese et al., 2005; Cho et al., 2006; Karve et al., 2008, 2010). Somewhat surprising is that type A HXKs have not been predicted in maize or sorghum (both C4 grasses) (Karve et al., 2010). Unlike type A, type B HXKs generally have a highly hydrophobic transmembrane helix associated with mitochondria (Aguilera-Alvarado and Sanchez-Nieto, 2017). However, some type B HXKs may also be localized in the nucleus because a nuclear localization signal is present close to the transmembrane helices (Cho et al., 2009). Type C HXKs, which lack plastid-signaling peptides and membrane-anchored domains, have presently been identified only in Physcomitrella patens and monocotyledonous plants (Cheng et al., 2011; Nilsson et al., 2011).
As ancient and conserved hexokinases, the plant HXKs catalyze hexose phosphorylation, sense glucose level, and correlate with multiple signaling pathways influencing plant growth and development (Rolland et al., 2006; Claeyssen and Rivoal, 2007; Aguilera-Alvarado and Sanchez-Nieto, 2017). In Arabidopsis, AtHXK1 is the best-characterized glucose sensor and is involved in plant development and stress response (Moore et al., 2003; Kim et al., 2006; Rolland et al., 2006; Sarowar et al., 2008). Moreover, AtHXK1 forms a glucose signaling complex with VHA-B1 and RPT5B that directly modulates the transcription of specific target genes (Cho et al., 2007). Additional functions of HXK1 have been elucidated in many other species. For example, overexpression of AtHXK1 in guard cells affected stomatal closure in citrus (Kelly et al., 2013; Lugassi et al., 2015). In tomato, overexpression of AtHXK1 resulted in reduced photosynthesis, slower growth, and induction of senescence, which is indicative of its function in photosynthetic tissues (Dai et al., 1999). Overexpression of pear PbHXK1 in tomato modulated the sugar content and affected plant growth (Zhao et al., 2019). In Nicotiana benthamiana, virus-induced gene silencing of NbHXK1 caused cell death, implying that HXK1 plays a role in the process of cell death (Kim et al., 2006). In apple, MdHXK1 may phosphorylate MdbHLH3 to regulate anthocyanin biosynthesis, and MdNHX1 improves salt tolerance (Hu et al., 2016; Sun et al., 2018). In sunflower (Helianthus annuus L.), HaHXK1 transcript levels were higher in developing seeds than in photosynthetic tissues and the highest HXK activity was detected in the early stages of accumulation of reserve compounds, lipids, and proteins in the seed (Troncoso-Ponce et al., 2011). In addition to HXK1, the functions of some of the other HXK members have been revealed. In rice, OsHXK5 and OsHXK6 function as glucose sensors and overexpression of OsHXK5 and OsHXK6 caused growth retardation in response to glucose treatment (Cho et al., 2009). Reduced OsHXK10 expression in rice resulted in abnormal dehiscence of the anthers in some flowers (Xu et al., 2008). Moreover, plant HXKs also play a regulatory role in interactions between sugars (nutrition) and phytohormones (León and Sheen, 2003; Moore et al., 2003).
Moso bamboo (Phyllostachys edulis) is a member of the Gramineae family and is widely distributed in China and the world with high economic and ecological value (Peng et al., 2013; Zhang et al., 2018). Similar to many other bamboo species, moso bamboo is a perennial with a large genome and complex phenotypes, which is possibly a cause of the slow progress in research on biological functions in the species. In recent years, the publication of the moso bamboo genome (Peng et al., 2013; Wang et al., 2018) and rapid development of plant omics technology have presented the opportunity to explore genetic functions in bamboo, and some gene families have been uncovered (Liu et al., 2017; Cheng et al., 2018; Gao et al., 2018; Hou et al., 2018; Li et al., 2018). With aspect to all the traits of bamboo, the most attractive one is the growth rate that the shoot can grow as fast as one meter per day at peak growth. In order to understand the fast growth, some previous studies have characterized the shoot growth and anatomy that provide insight into the fast-growth bamboo (Lin et al., 2002; Li et al., 2018; Wei et al., 2018). However, the molecular mechanism underlying the fast-growth of bamboo shoot is still unclear. HXKs, which act as catalysts of the first essential step in glucose metabolism, have emerged as important enzymes that mediate sugar sensing and related signals in plant growth (Claeyssen and Rivoal, 2007; Aguilera-Alvarado and Sanchez-Nieto, 2017). With a view that HXKs -related pathways are important for sugar signal transduction which depends on phosphorylated hexoses and intermediate glycolytic products (Aguilera-Alvarado and Sanchez-Nieto, 2017), the plant HXKs are considered to have the function of regulating plant growth and the potential of promoting biomass. In the present study, we identified and characterized PeHXKs in moso bamboo. First, we searched against the bamboo genome database using HXK protein sequences of Arabidopsis and identified hexokinase genes by means of subsequent bioinformatic analysis and experimental verification. We characterized the PeHXKs by investigation of phylogenetic relationships, gene structure, motif analysis, physicochemical properties of the proteins, and subcellular protein localization. Quantitative real-time PCR (qRT-PCR) was used to analyze gene expression patterns and functional complementation in an HXK-deficient yeast strain (YSH7.4-3C) was used to verify the hexokinase activity of PeHXKs. In total, we identified 12 PeHXKs, for which we assessed the conservation in gene and protein structure and the expression profiles in moso bamboo tissues. Also, the HXK activity of the PeHXK5a, PeHXK8, and PeHXK3b was verified. The results presented herein provide a reference for future studies of the PeHXK gene family, especially for the regulation of fast growth and rapid-accumulated biomass in moso bamboo.
Materials and Methods
Identification of HXKs in Moso Bamboo
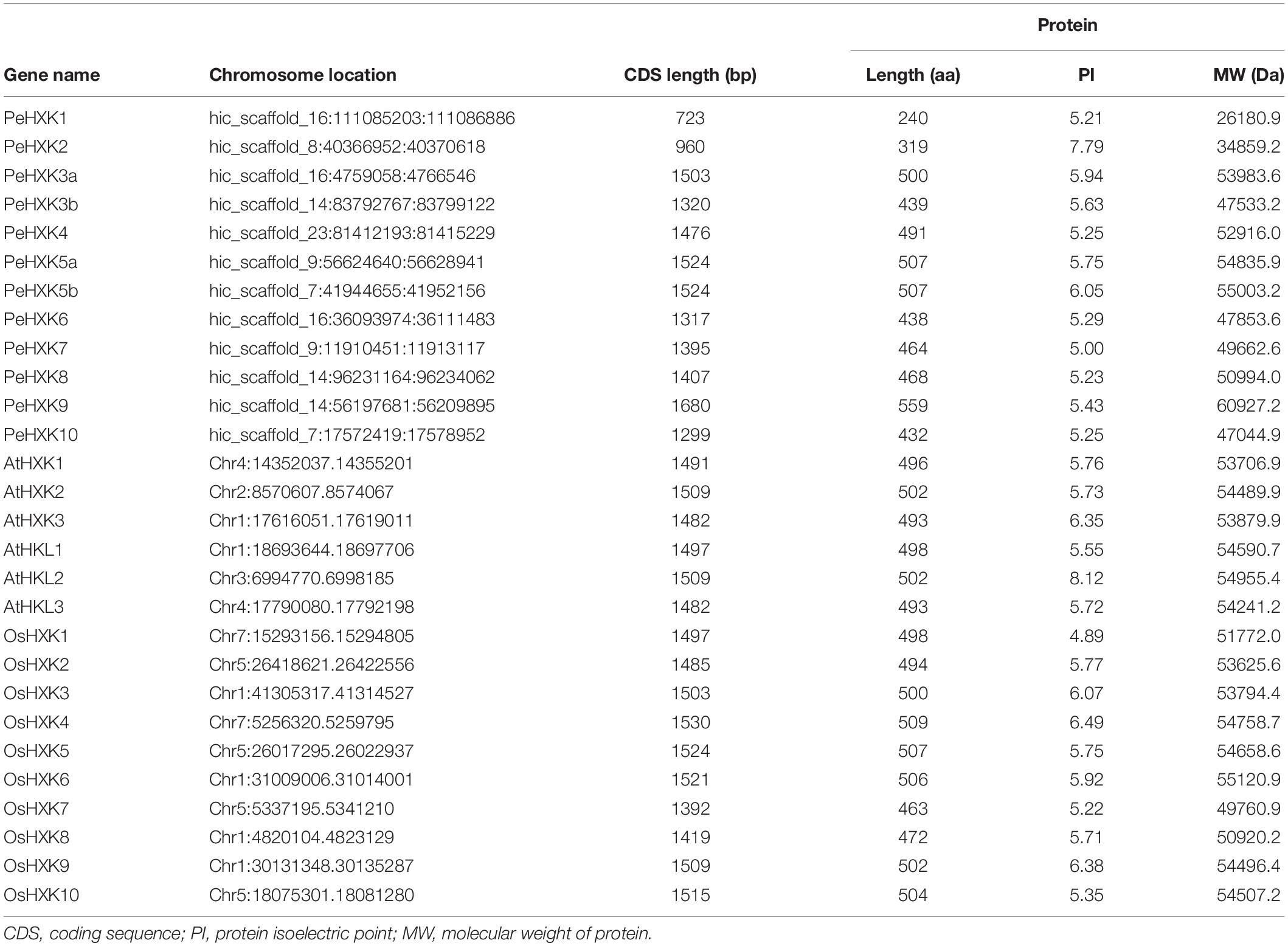
To identify HXK genes of moso bamboo, we used Arabidopsis HXK protein sequences (TAIR1) as query sequences for a BLAST search of the moso bamboo database (Zhao et al., 20182) using a threshold P-value < 10–5. Fifteen candidate HXK sequences were obtained. Three sequences were discarded as a result of a search for conserved domains within the amino acid sequences of the National Center for Biotechnology Information (NCBI) Conserved Domains database3 and the EMBL-EBI Pfam database4. The number of amino acids and length of the coding sequence (CDS) were characterized, and the theoretical isoelectric point (pI) and molecular weight of amino acids were calculated using the ProtParam online tool5 (Gasteiger et al., 2003).
Using the genome and CDS sequences of candidate PeHXKs from the moso bamboo genome database, we mapped the genetic structure with the Gene Structure Display Server 2.0 (GSDS tool6) (Guo et al., 2007). Motif analysis was carried out using MEME7 with a maximum number of 10 motifs and using other default settings (Liu et al., 2018). In the bamboo genome database, we first determined the relative position of each gene on each chromosome, then the location of the PeHXK gene on the chromosome was mapped using MapDraw (Liu and Meng, 2003).
Sequence Alignment and Phylogenetic Tree Construction
We downloaded protein sequences of Arabidopsis HXKs from the TAIR database1, rice HXKs from the NCBI database8, and Populus trichocarpa and Brachypodium distachyon HXKs from the Phytozome database9. The sequences were aligned using MUSCLE, and evolutionary trees were constructed using the neighbor-joining method with MEGA (Liu et al., 2018). The reliability of the tree topology was assessed by means of a bootstrap analysis with 1000 replications. Multiple amino acid sequences of several conserved binding domains were aligned by using SeaView 4 software (Gouy et al., 2010).
Identification of Orthologs and Calculation of Ka and Ks
On the basis of previous reports, orthologs were identified by pairwise alignment (Blanc and Wolfe, 2004). The synonymous substitution rate (Ks) and non-synonymous substitution rate (Ka) were calculated using DnaSP 5 software (Rozas, 2009). We calculated the evolutionary divergence time using the formula T = Ks/2λ (λ = 6.5 × 10–9) as reported previously (Peng et al., 2013). In general, Ka/Ks > 1 indicates the positive selection, Ka/Ks = 1 indicates the neutral selection, and Ka/Ks < 1 suggests negative or stabilizing selection (Juretic et al., 2005).
Plant Material, RNA Extraction, and Quantitative Real-Time PCR (qRT-PCR)
Moso bamboo seeds were collected from Gongcheng Yao Autonomous County, Guangxi Zhuang Autonomous Region, China. The seeds were heated for 1 day in a water bath at 42°C in 200 mg/L gibberellic acid (GA) solution before sowing. The seeds were sown by lightly covering with a mixture of vermiculite and vegetative soil (1:1) and then covered with plastic wrap, and were incubated in a greenhouse at 26°C under a 16 h/8 h (light/dark) photoperiod. The seeds germinated after approximately 1 week and grew to about 10 cm in height after 3 months. The uppermost four leaves (from the shoot apex), stems (including the internodal region and nodes of the whole seedling), roots, and rhizomes were used for the extraction of total RNAs. Tissue samples were collected and immediately placed in liquid nitrogen, and then stored at −80°C until use. Total RNAs were extracted from the samples using the RNAprep Pure Plant Kit (TIANGEN – DP419, Beijing, China). The first-strand cDNA was synthesized using the TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen – AT311, Beijing, China) in accordance with the manufacturer’s instructions. The first-strand cDNA was used as the template in qRT-PCR analyses. The primers (Supplementary Table S2) used for qRT-PCR analyses were designed using the NCBI Primer-BLAST tool10 with a PCR product size of 150–250 bp. An intrinsic membrane protein-encoding gene, Tonoplast Intrinsic Protein 41 (TIP41), was used as the internal reference gene (Fan et al., 2013). The qRT-PCR protocol was 94°C for 30 s, then 40 cycles of 94°C for 5 s and 60°C for 30 s, and used the TransStart® Top Green qPCR SuperMix (TransGen – AQ131, Beijing, China). The values of expression were calculated from three independent biological repeats, each of which was the average of three technical repeats.
Subcellular Localization of PeHXK Proteins
The full-length CDS region of individual PeHXK genes was integrated into the pCambia2300 vector [containing the green fluorescent protein (GFP) sequence]. The full-length CDS region of individual PeHXK genes was amplified by PCR using primers LP (PeHXK5a: CAGACAGTGATGGGGAAGGC; PeHXK8: ATGGCCGCAGCTGCGGTCGCAAT; PeHXK3b: ATGGTCGTTGAGATGCACGC) and RP (PeHXK5a: GTCGAC CTCGGCATACTGAGAGTGC; PeHXK8: CTGTTGCTC AACATACTTGTACTG; PeHXK3b: TATGGAACCTCCTTG TTGCTGTCTA). The GFP sequence was attached to the C-terminus of the target PeHXK protein. The pCambia2300-GFP plasmid was then introduced instantaneously into tobacco leaves by transformation mediated by Agrobacterium tumefaciens strain GV3101. After 3 days, transformed tobacco leaves were observed with a confocal laser scanning microscope. Before the observation of GFP signals, the mitochondria were stained with 500 nM MitoTracker® Red dye by incubation at 37°C for approximately 50 min.
Yeast Complementation Experiments
The yeast triple mutant YSH7.4-3C is deficient in HXK1, HXK2, and GLK1 (Geng et al., 2017) and was kindly provided by Professor Xinwen Hu. The full-length CDS of individual PeHXK genes was integrated into the pDR195 vector (including URA3 as a selection marker) by homologous recombination (Geng et al., 2017). The full-length CDS region of individual PeHXK genes was amplified by PCR using primers LP (PeHXK5a: CAGACAGTGATGGGGAAGGC; PeHXK8: ATGGCCGCAGCTGCGGTCGCAAT; PeHXK3b: ATGGTCGTTGAGATGCACGC) and RP (PeHXK5a: GTCGAC CTCGGCATACTGAGAGTGC; PeHXK8: CTGTTGCTCAACA TACTTGTACTG; PeHXK3b: TATGGAACCTCCTTGTTGCTG TCTA). Given that the yeast triple mutant was unable to utilize glucose or fructose as carbon sources, the reproducible medium was composed of 2% peptone, 1% yeast extract, 2% galactose, and 1.5% agar, and the screening medium for selection of transformed colonies included 0.67% YNB and 2% of a carbon source (D-glucose or D-fructose), supplemented with the appropriate amino acids lacking uracil. As a negative control, the plasmid without the PeHXK gene was transformed into the yeast mutant cultured on medium containing 0.67% YNB and 2% of a carbon source (D-glucose or D-fructose).
Results
Identification and Characterization of Hexokinase Genes in Moso Bamboo
To identify HXK genes in moso bamboo, HXK protein sequences of Arabidopsis were used as query sequences to search the moso bamboo genome. A total of 15 candidate sequences were obtained and three sequences were subsequently discarded after analysis of conserved domains (Table 1 and Supplementary File S3). The molecular weight (MW) of PeHXK proteins and CDS length of the PeHXK genes showed substantial variation. The CDS length ranged from 723 bp (PeHXK1) to 1680 bp (PeHXK9). The encoded protein length ranged from 240 amino acids (aa) (PeHXK1) to 559 aa (PeHXK9). The protein molecular weight ranged from 26.18 kDa (PeHXK1) to 60.93 kDa (PeHXK9). More details for the individual PeHXKs, AtHXKs, and OsHXKs, including additional characteristics such as pI, and scaffold location, were given in Table 1.
Phylogenetic Relationships and Multiple Alignments
Previous studies have identified HXK genes in many plant species. To explore the evolutionary relationships among PeHXKs and HXKs in other plant species, 44 full-length HXK protein sequences, comprising six protein sequences from Arabidopsis, 10 protein sequences from rice, six protein sequences from poplar (Populus trichocarpa), 10 protein sequences from Brachypodium distachyon, and the 12 protein sequences from moso bamboo were used to construct a phylogenetic tree. The accession numbers of the sequences are listed in Supplementary Table S1. The evolutionary tree was approximately divisible into eight subfamilies, of which PeHXK members were distributed among five subfamilies (Figure 1). On the basis of the phylogenetic tree, no homologous sequences of AtHXK1, AtHXK2, and AtHKL3 were present in monocotyledonous plants. Among the PeHXK proteins, PeHXK8, PeHXK1, and PeHXK7 were represented as most similar to three type C OsHXKs (OsHXK1, OsHXK7, and OsHXK8), which contain neither chloroplast transport peptides nor membrane-anchored domains, and PeHXK4 was clustered with OsHXK4, which contains the type A-specific chloroplast transit peptide (Karve et al., 2010). The remaining PeHXK proteins were grouped with type B OsHXKs.
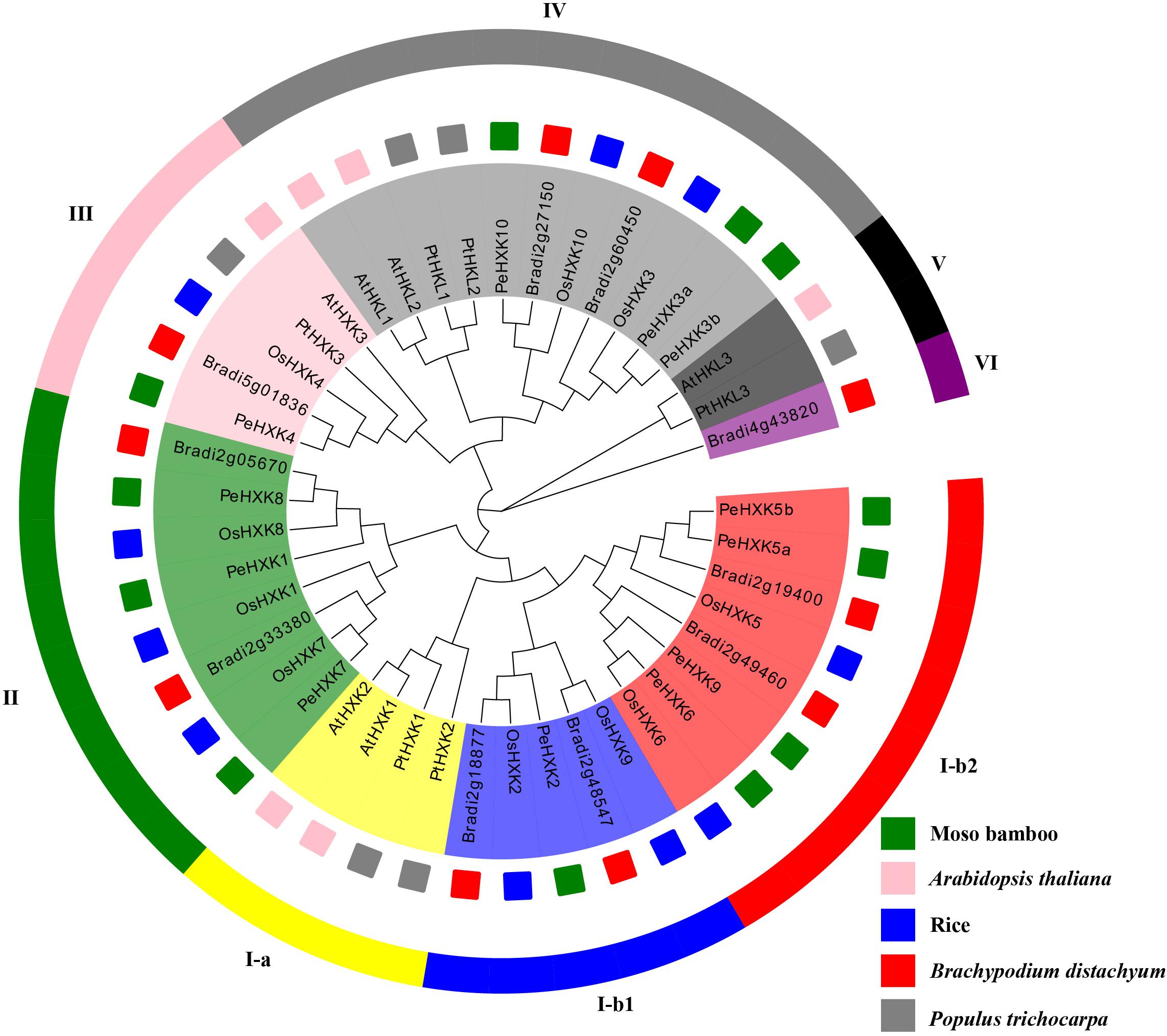
Figure 1. Phylogenetic tree of PeHXK proteins from moso bamboo, Arabidopsis thaliana, rice, Brachypodium distachyon, and Populus trichocarpa. The tree was generated using the Neighbor-Joining (N-J) method with 1000 replicates. The fragments with different colors in the outer ring represent different subfamilies. The small squares with colors inside indicate different species [pink, A. thaliana; blue, O. sativa (rice); red, B. distachyon; green, moso bamboo].
For further characterization of the PeHXK proteins, the protein sequences were aligned using SeaView 4 software. Prediction of conserved sequences was based on previous reports of HXK2 in Saccharomyces cerevisiae (Bork et al., 1992; Katz et al., 2000; Aguilera-Alvarado et al., 2019). The majority of PeHXK proteins contained two conserved domains: an ATP-binding domain with phosphate 1 (P1), phosphate 2 (P2), and adenosine, and a sugar-binding domain. A number of loops and connects were detected in the protein structure. Exceptionally, both PeHXK1 and PeHXK2 had no complete loops. Moreover, PeHXK1 lacked the P1 (in the ATP-binding domain) and sugar-binding core while PeHXK2 had no connect 2, adenosine, and truncated P1 (Figure 2). A more detailed protein sequence comparison between Arabidopsis thaliana, rice, and moso bamboo was shown in Supplementary Figure S1.
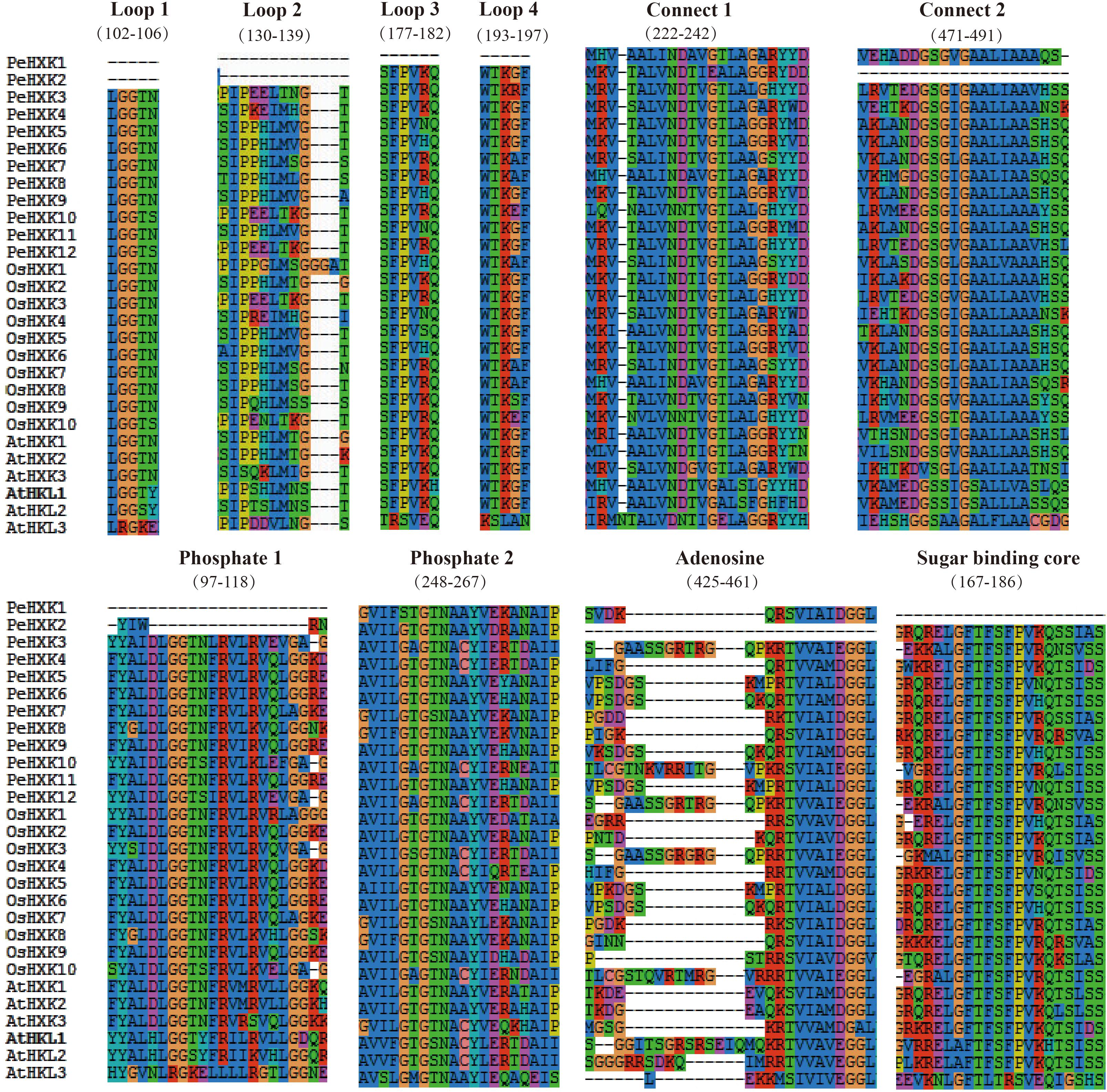
Figure 2. Gene structure of HXKs in moso bamboo, Arabidopsis thaliana, and rice. The HXKs were divided into five distinct sub-groups marked as left. It was specially noted that PeHXK6 and PeHXK9 genomic sequences contain much longer introns, while only OsHXK1 has no intron in the genome at all. The exons, introns, and UTR sections are indicated by yellow rectangles, black lines, and blue rectangles, respectively.
Gene Structure, Chromosomal Location, and Motif Distribution
On the basis of the genome and CDS sequences of the candidate PeHXKs (Supplementary Files S1, S2), the exon–intron structure was predicted using the GSDS tool. The PeHXKs from different subfamilies showed differences in gene structure. The majority of PeHXK genes contained at least nine exons and eight introns, whereas PeHXK1 contained only five exons and four introns. Five genes (PeHXK5b, PeHXK6, PeHXK7, PeHXK8, and PeHXK4) contained both 5′–and 3′- untranslated regions (UTRs), three genes (PeHXK5a, PeHXK3a, and PeHXK3b) contained a 5′-UTR only, and the remaining four genes (PeHXK9, PeHXK2, PeHXK1, and PeHXK10) contained no UTR region (Figure 3).
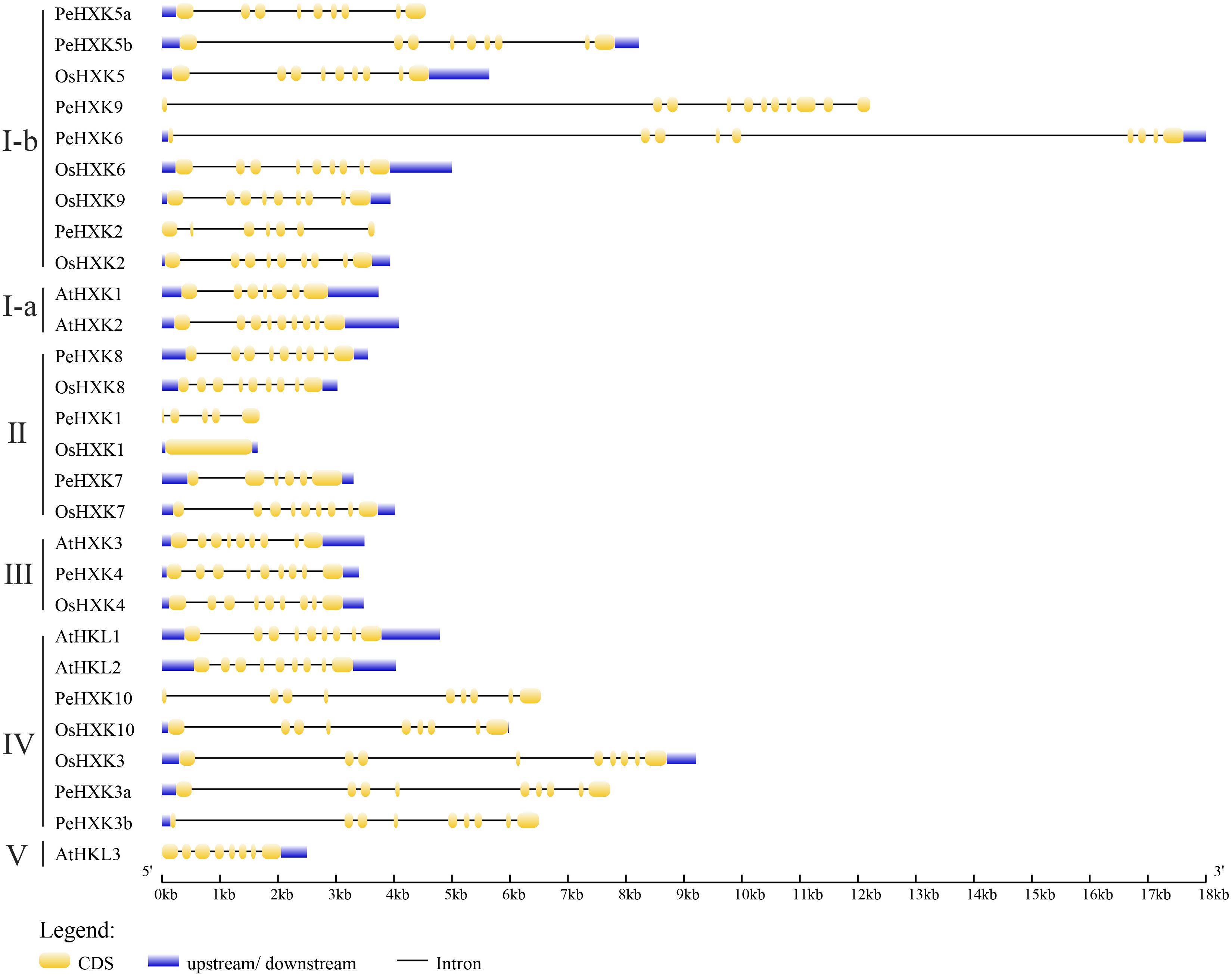
Figure 3. Conserved domain alignment for predicted moso bamboo family proteins. The sequences from moso bamboo, Arabidopsis thaliana, and rice were aligned using MUSCLE. Annotations are based on regions homologous to yeast HXK2 (Kuser et al., 2000). P1, phosphate 1; P2, phosphate 2. The alignment was performed using SeaView4 software (Gouy et al., 2010).
To determine the distribution of PeHXK genes in the moso bamboo genome, we mapped the PeHXK genes to individual chromosomes. The PeHXK genes were located on six chromosomes, of which chromosomes 14 and 16 each carried three PeHXK genes, chromosomes 7 and 9 each carried two PeHXK genes, and one PeHXK was located on each of chromosomes 23 and 8 (Table 1 and Supplementary Figure S2).
To analyze PeHXK protein motif characteristics, we used the MEME tool to predict conserved motifs (Figure 4). In total, 10 conserved motifs in the PeHXK protein sequences were predicted. The majority of PeHXK proteins contained at least nine conserved domains, whereas PeHXK2 and PeHXK1 contained five and four motifs, respectively. It was noteworthy that 10 of the 12 HXK proteins contained motif 1, 3, and 8, which were all components of the ATP-binding domain.

Figure 4. Motif distribution in HXKs from moso bamboo, Arabidopsis thaliana, and rice. Motifs were analyzed using the MEME web server (https://www.swissmodel.expasy.org/). The motifs are represented by different colors. The length of each box in the figure does not represent the actual motif size.
Evolutionary Patterns Among Moso Bamboo, Rice, and Brachypodium distachyon
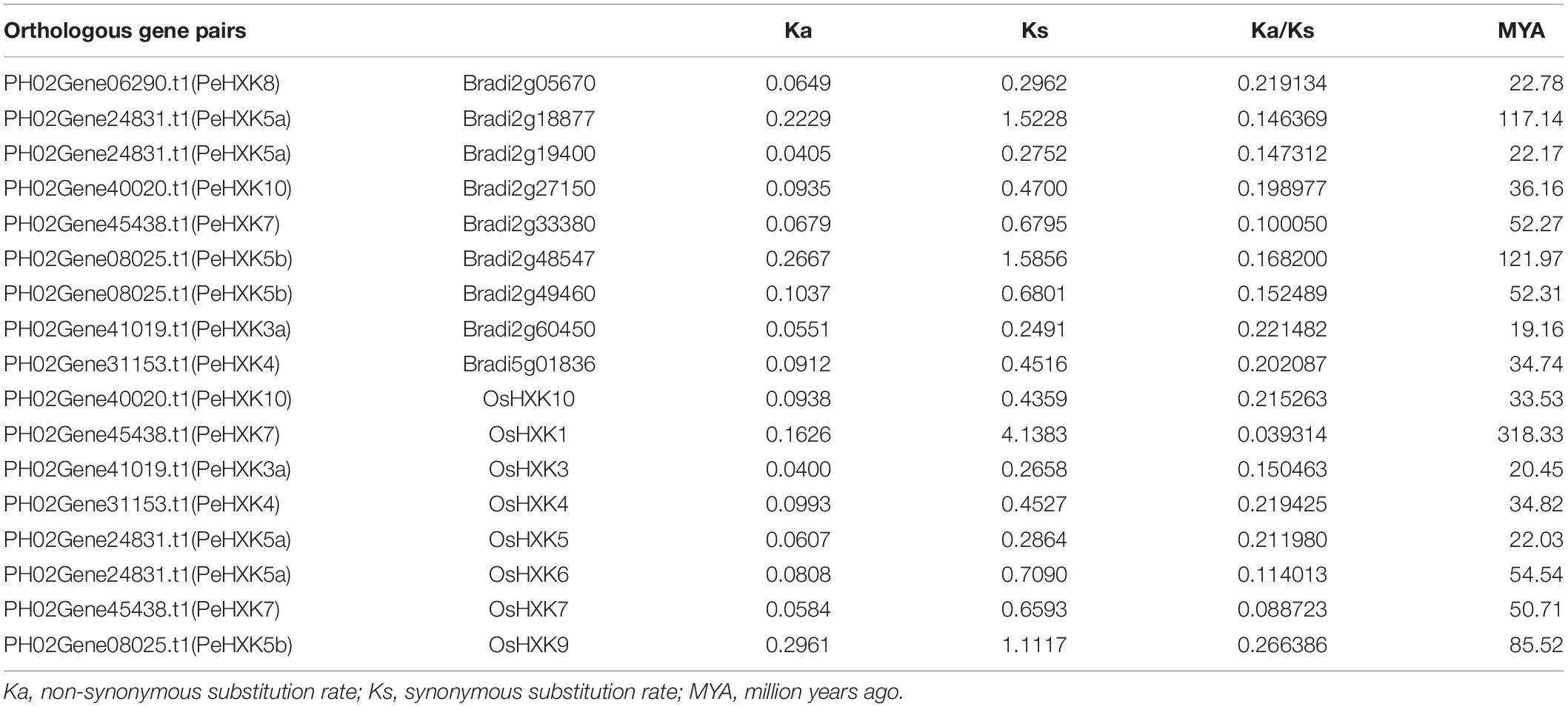
Selection pressure refers to the evolutionary pressure that nature places on organisms to enable those who adapt to the natural environment to survive and reproduce. To investigate the selection pressure on HXK genes in moso bamboo, rice, and Brachypodium distachyon, we identified eight orthologs (Pe-Os) between moso bamboo and rice, and nine orthologs (Pe-Br) between moso bamboo and Brachypodium distachyon using bidirectional best-hit methods (Wu et al., 2016). All gene pairs are listed in Table 2. A previous study (Juretic et al., 2005) showed that the Ka/Ks ratio can be applied as a measure of selection pressure. Accordingly, we calculated the values of Ka, Ks, and Ka/Ks. The value of Ka and Ks respectively represent the non-synonymous substitution rate and synonymous substitution rate. The Ka/Ks ratio was widely used to calculate selection pressure. Generally, the value of Ka/Ks greater than 1, equal to 1, and less than 1 respectively represents positive selection, neutral selection, and negative selection (Juretic et al., 2005). The Ka/Ks values for the Pe-Os and Pe-Br orthologous pairs ranged from 0.039 to 0.266 and 0.100 to 0.22, respectively. The Ks values of Pe-Os pairs ranged from 0.26 to 4.1, suggesting that the orthologs diverged 20 million years ago (MYA). The Ks values of the nine Pe-Br pairs ranged from 0.24 and 1.59, indicating that these orthologs diverged prior to 19 MYA.
Expression Profiles of Moso Bamboo HXK Family Genes
To explore the tissue-specific expression patterns of PeHXK genes, their transcript levels were measured by qRT-PCR in four tissues of moso bamboo seedlings, namely the leaf, stem, root, and rhizome. Five genes (PeHXK5a, PeHXK8, PeHXK5b, PeHXK9, and PeHXK2) were extensively expressed in the tissues, whereas three genes (PeHXK1, PeHXK4, and PeHXK10) were detected at low levels in all four tissues. PeHXK6, PeHXK3a, and PeHXK3b showed a relatively higher transcript level in the stem than in leaf, rhizome, and root. Except for PeHXK1, PeHXK4, and PeHXK10, the remaining PeHXK genes were actively expressed in the stem. Notably, the level of PeHXK2 transcripts was considerably higher in the rhizome (Figure 5).
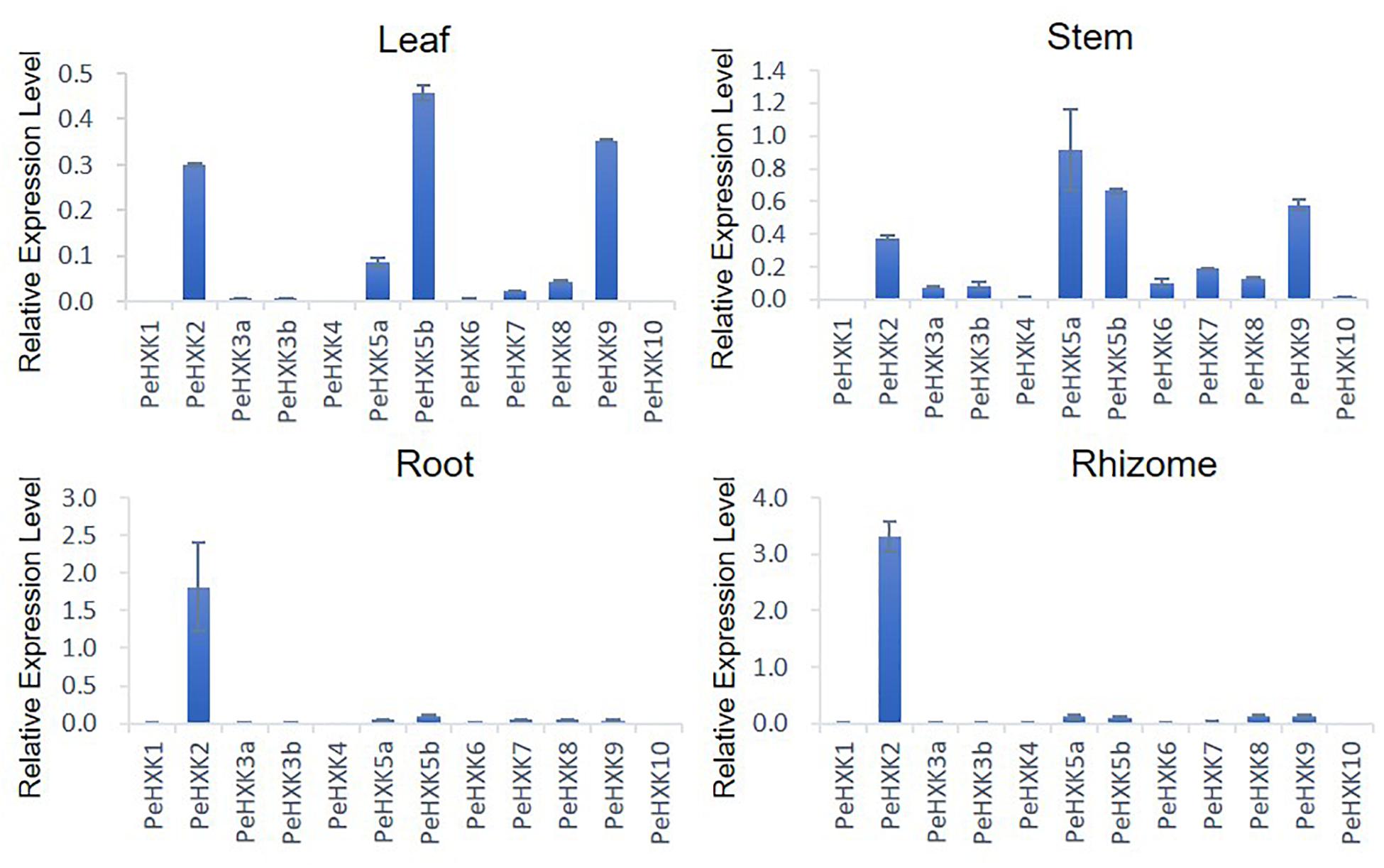
Figure 5. Expression patterns of 12 PeHXK genes in four different tissues. Samples were collected from the leaf, stem, root, and rhizome. The Y-axis indicates relative expression levels of PeHXK genes to that of TIP41, which is used as an inner control (Fan et al., 2013).
Subcellular Localization of Bamboo HXK Family Genes
To determine the subcellular localization of PeHXK proteins, the CDS of three PeHXKs (PeHXK5a, PeHXK8, and PeHXK3b) belonging to different subgroups were fused to the GFP sequence and was transiently expressed in tobacco leaf epidermal cells. The subcellular localization of AtHXK1 and empty plasmid were separately used as a positive and negative control. The fluorescence signals for PeHXK5a-GFP, PeHXK8-GFP, and PeHXK3b-GFP were co-localized with the MitoTracker® Red signal. In addition, PeHXK8-GFP signal was also detected in the nucleus (Figure 6).
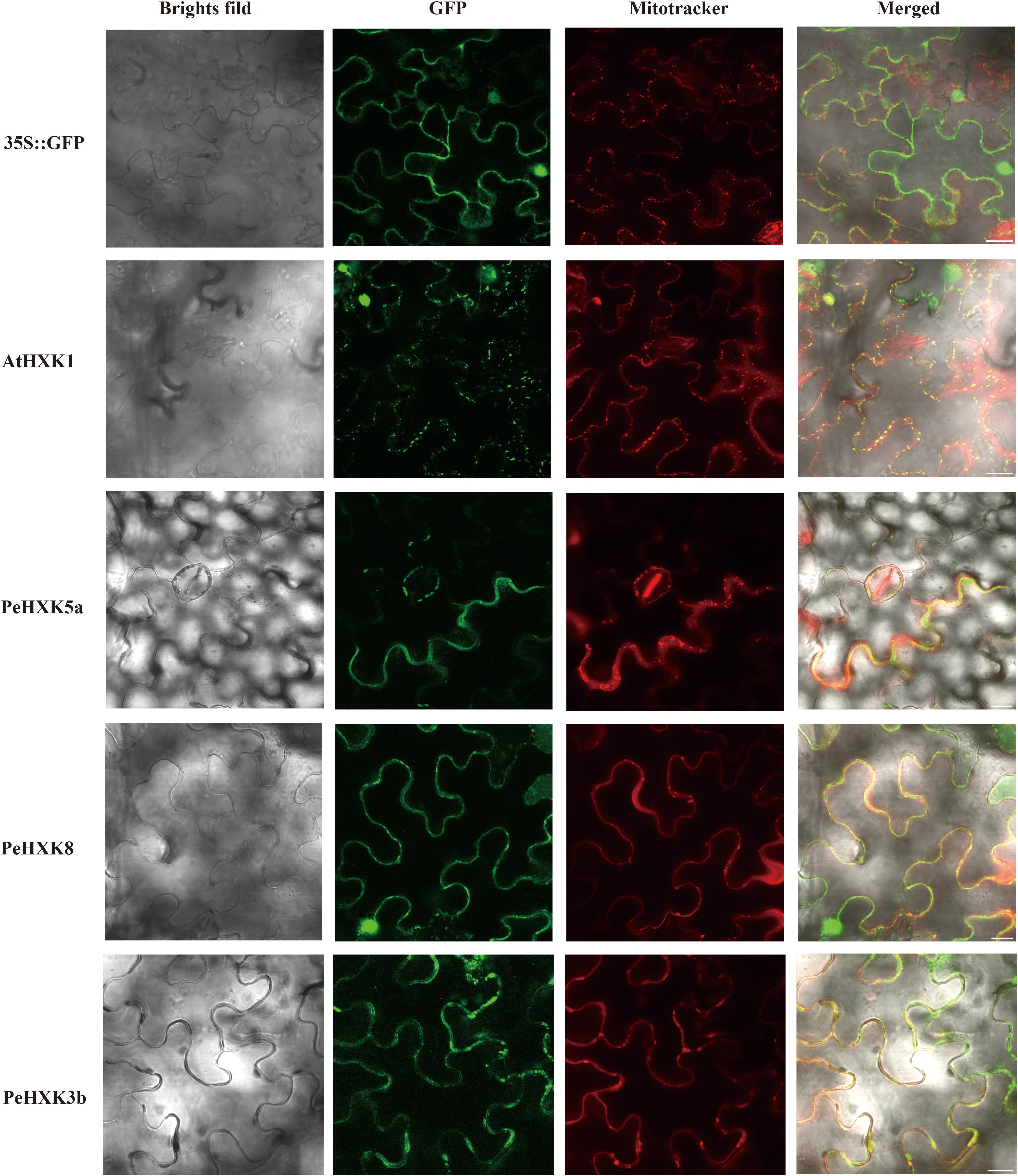
Figure 6. Subcellular location of the PeHXK5a-GFP, PeHXK8-GFP, and PeHXK3b-GFP fusion proteins in tobacco leaves. AtHKX1-GFP and empty vector (pCambia2300GFP) are used as the positive and negative control, respectively. The fluorescence signals were collected by confocal scanning microscopy before pretreatment of dyeing by Mitotracker (red). The merged image was a superposition signals of bright field, GFP and mitotracker signal. Bar = 20 μm.
Complementation of Yeast Mutant
To evaluate the HXK activity of the PeHXKs, we cloned the full-length CDS of PeHXK5a, PeHXK8, and PeHXK3b into the pDR195 yeast expression vector. The yeast triple-mutant strain YSH7.4-3C was transformed with the recombinant vectors and cultured on screening media. Medium in which galactose was the sole carbon source (SGal-URA) was used as a control. The yeast transformed with the empty plasmid, or with plasmids harboring the PeHXK genes (PeHXK5a, PeHXK8, or PeHXK3b) grew normally on Sgal-URA medium. However, on medium in which glucose was the sole source of carbon (SGlu-URA), only yeasts transformed with PeHXK5a, PeHXK8, and PeHXK3b could grow. On medium that contained fructose as the sole carbon source (SFru-URA), yeast transformed with the three PeHXK genes grew better than the control (Figure 7).
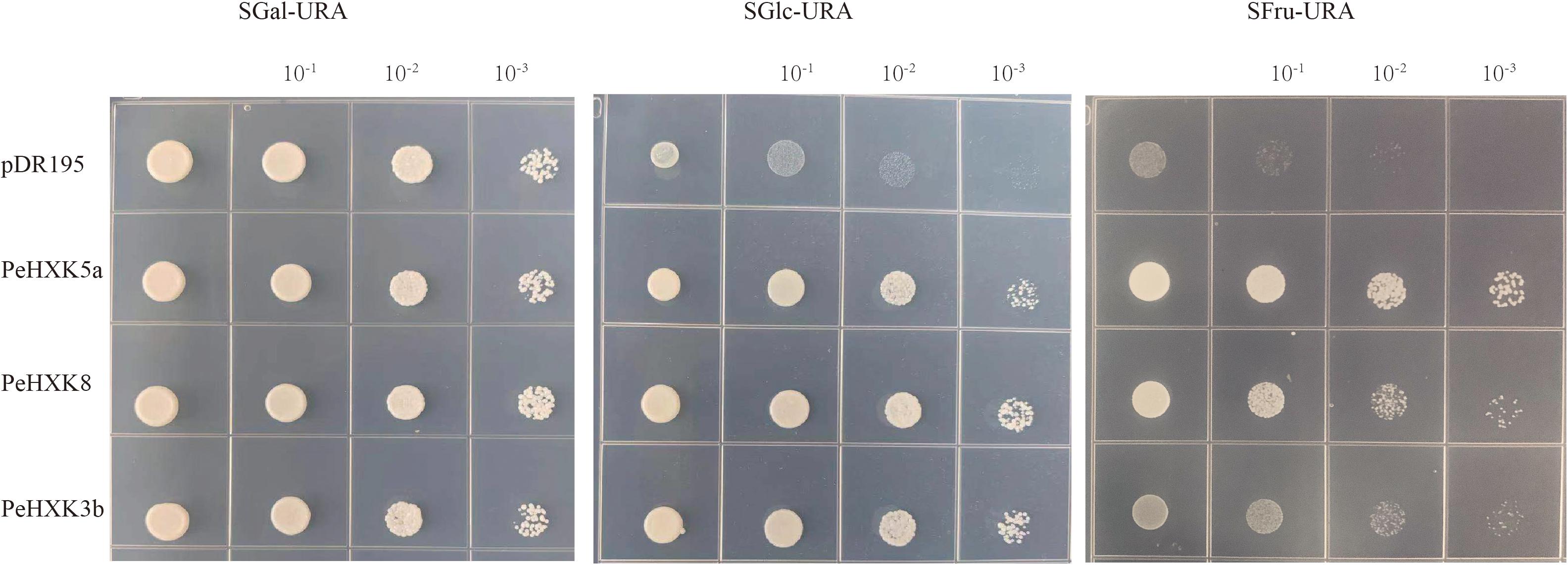
Figure 7. Complementation of the yeast mutant YSH7.4-3C with PeHXK genes. pDR195: the empty pDR195 vector was transformed into YSH7.4-3C as negative control; PeHXK3b, PeHXK5a, and PeHXK8: the recombinant vectors (including the three PeHXK CDS) were transformed into YSH7.4-3C, respectively. SGal-URA, the medium that contained galactose as the sole carbon source and lacked uracil; SGlc-URA, the medium that contained glucose as the sole carbon source and lacked uracil; SFru-URA, the medium that contained fructose as the sole carbon source and lacked uracil.
Discussion
Identification and Characterization of PeHXK Genes
In addition to acting as signaling molecules, hexose sugars are the main carbon source required for the energy supply and storage to fuel and maintain cell life. Moreover, they are an important source for the synthesis of polysaccharide in the plant cell wall. Before being utilized, hexoses need to be phosphorylated by hexose phosphorylating enzymes, including HXKs (Granot et al., 2013). In many plant species, HXKs have been identified as hexokinases phosphorylating glucose and fructose with diversified functions-sensing sugar, controlling gene expression, hormonal interactions and finally regulating plant growth (Granot et al., 2013; Aguilera-Alvarado and Sanchez-Nieto, 2017). However, no information was previously available on members of the HXK gene family in the Bambusoideae, a distinctive subfamily of the Gramineae. As a representative bamboo species, fast-growth moso bamboo, which is distributed widely in China and the world, is an important source of non-timber forest products in human traditional life and thus is of substantial economic and environmental value (Peng et al., 2013; Zhao et al., 2018). Furthermore, it may be considered as a good source of carbohydrate for biofuel production for its environmental benefits and higher annual biomass yield (He et al., 2014). To accelerate the research of bamboo HXKs function, especially regard to the potential of growth-regulation and biomass-increase, here we carried out a genome-wide analysis of HXK genes in moso bamboo. In the present work, 12 HXKs were identified in the moso bamboo genome and characterized with regard to gene and protein structure, conserved motifs, gene evolution, expression pattern, protein subcellular localization, and HXK enzyme activity. A comparison of HXK gene families among multiple plant species revealed an inconsistency in the numbers of PeHXK genes and the genome size of moso bamboo (Figures 3, 4). For example, Arabidopsis, which has an extremely small genome size (125 Mb), contains three HXK genes and three HXK-Like genes, the gramineous plant rice with a genome size of 466 Mb has 10 HXK genes, whereas moso bamboo with a huge genome size of 2.075 Gb contains 12 HXK genes. Apparently, gene duplication events in moso bamboo did not promote an increase in the number of PeHXKs, which indicates that their functions are highly conserved in the species.
High similarity in the protein sequence is commonly associated with function conservation. According to the well-studied tertiary structure of HXKs from yeast and mammals, HXK proteins contain a large and a small domain which is resided largely by the sugar-binding site/core with four additional peptide segments (Loops 1–4) (Bork et al., 1992; Kuser et al., 2000). It also indicates that most of the conserved amino acid residues exist at the fissure of the two domains and generates the glucose and ATP binding sites (Kuser et al., 2000). In our present work, multiple alignments of amino acid sequences revealed that the majority of HXK homologous proteins of bamboo contained complete conserved regions similar to the reported HXKs, including motifs designated phosphate 1, connect 1, phosphate 2, adenosine, and connect 2 (Karve et al., 2010). Exceptions were observed in PeHXK1 and PeHXK2. PeHXK1 lacks L1-L4 loops, and phosphate 1 (P1), and PeHXK2 has no L1-L2 loops, connect 2, and adenosine. Otherwise, PeHXK2 also possesses a big-scale deletion in the P1 motif (Figure 2 and Supplementary Figure S1). For the reasons that the P1 domain is high conserved and required for HXK activity and the loops are the essential component of one of the HXK major domains responsible for glucose binding (Kuser et al., 2000; Karve et al., 2010), the HXK activity consequently would be dismissed in the two HXK proteins in all probability. From a functional point of view, plant HXK family members are classifiable into HXK and HXK-like (HKL) proteins based on whether the protein having the activity to phosphorylate glucose (Moore et al., 2003; Karve et al., 2008; Karve and Moore, 2009). Factually, the functional diversification is considerable in plants. Taking Arabidopsis HXK proteins as an example, both AtHXK1 and AtHXK2 can phosphorylate glucose, sense sugar level, and interact with signaling pathways; AtHKL1, AtHKL2, and AtHKL3 are thought to have just regulatory activity like antagonizing AtHXK1/2; and AtHXK3, as a plastid enzyme, is likely only a catalytic protein (Jang et al., 1997; Granot, 2008; Karve et al., 2010). Similar functional diversification also seems to occur in the rice HXK family (Yu and Chiang, 2008; Cho et al., 2009), thus it is possible that HXK genes in rice and other monocot plants are classified into the two groups. By inspection, we classified the 12 PeHXK genes into four groups (Figure 1). In the Group I-b2, PeHXK5a, PeHXK5b, PeHXK6, and PeHXK9 are located together with OsHXK5 and OsHXK6, while in the Group I-b1, only PeHXK2 shares the same branch with OsHXK2 and OsHXK9; In Group II, the branch possesses OsHXK7, OsHXK8 and their homologous genes PeHXK7, PeHXK8, and PeHXK1; In Group III, PeHXK4 is together with OsHXK4 and AtHXK3; In Group IV, PeHXK10, PeHXK3a, and PeHXK3b are clustered with OsHXK10 and OsHXK3. According to the previous study (Karve et al., 2010; Aguilera-Alvarado and Sanchez-Nieto, 2017), we speculated that PeHXK1, PeHXK2, PeHXK5a, PeHXK5b, PeHXK6, PeHXK9, PeHXK7, and PeHXK8 belong to HXK-protein clade, and PeHXK3a, PeHXK3b, PeHXK10 are HKL proteins. PeHXK4 in Group IV is putative plastid-localization like AtHXK3 and OsHXK4. It is noted that the loss of key motifs in PeHXK1 and PeHXK2 might lead to lost of either catalytic or regulatory function as HXK genes. Furthermore, in regard to the homologous relationships with OsHXK5, OsHXK6, and AtHXK1, we also infer that PeHXK5a, PeHXK5b, and PeHXK6 are candidate glucose sensor in moso bamboo.
Evolutionary Patterns Among HXK Genes of Moso Bamboo, Rice, and Brachypodium distachyon
Duplication events are an important source of novelty in genome evolution. The new gene copies that result from replication may lead to families of genes that evolve novel functions (Moore and Purugganan, 2003). Taking HXK genes as a reference, we estimated that a large-scale duplication event occurred approximately 22–121 and 20–319 MYA on the basis of the estimated divergence times for orthologous gene pairs (Pe-Os and Pe-Br) (Table 2). It was previously estimated that PHD-finger gene families diverged approximately 19–55 and 22–60 MYA between moso bamboo and rice, and between moso bamboo and Brachypodium distachyon, respectively (Liu et al., 2018). These results indicate that different gene families may have been duplicated at different evolutionary time points.
Expression Patterns, Subcellular Localization, and HXK Activity
The expression level of proteins in different tissues and information on subcellular localization of proteins provides a foundation to understand their function in plant growth and development. In Arabidopsis, most HXK genes are expressed extensively in all tissues, while AtHKL3 is only detected in flower (Karve et al., 2008). In rice, the OsHXK family shows the similarly wide range of expression profiles with the exception that OsHXK10 is only actively transcribed in flower and OsHXK1 is not detected in any tissue (Cho et al., 2006). With the exception of AtHKL3, OsHXK1, and OsHXK10, transcripts of all HXK genes in Arabidopsis and rice were detectable in main tissues, suggesting that each plant HXK has either a unique or redundant function in various tissues or organs. In the present study, the expression patterns of PeHXK genes were examined in the leaf, stem, rhizome, and root of moso bamboo seedlings and the results showed that PeHXK family genes have some different profiles (Figure 5). For example, not like AtHXK or OsHXK genes, most PeHXK family members showed a very low expression level in root and rhizome. However, some similar patterns to rice are also observed in PeHXK gene expression. As an example, PeHXK1 and PeHXK10 were almost not detectable in all four tissues tested, like OsHXK1 and OsHXK10, respectively. Moreover, PeHXK5a, PeHXK5b, PeHXK9 showed relatively higher expression levels in leaf and stem, just like their homologous ones in rice, OsHXK5, OsHXK6, and OsHXK9, respectively. Additionally, what is interesting is that the expression levels of PeHXK2, which is mentioned above as inactive with incomplete HXK motifs, showed high transcriptional activity in all four tissues detected, especially in root and rhizome, in both which PeHXK2 is the only one showing high expression, indicating that the PeHXK2 might perform a unique function like as a negative regulator for the other HXKs (Karve and Moore, 2009; Aguilera-Alvarado and Sanchez-Nieto, 2017).
Plants have four HXK types, types A–D, based on their subcellular localization, and differences in subcellular localization result in functional divergence (Karve et al., 2010; Aguilera-Alvarado and Sanchez-Nieto, 2017). To date, except for type D HXKs, which are mitochondrial proteins, have only been identified in the moss Physcomitrella patens (Nilsson et al., 2011), the other three types of HXKs are widely distributed in higher plants. Type A HXKs harboring a chloroplast signal at N-terminus commonly localized at chloroplasts and have been found in moss Physcomitrella patens and higher plants such as Arabidopsis thaliana, Nicotina tabacum, Oryza sativa, Solanum lycopersicum, Vitis vinifera, Camellia sinensis, Brassica napus, pear (Pyrus × bretschneideri), and Spinacia oleracea (Olsson et al., 2003; Cho et al., 2006; Kandel-Kfir et al., 2006; Karve et al., 2008, 2010; Nilsson et al., 2011; Wang et al., 2018; Zhao et al., 2019). Type B HXKs, accounting for most members of HXK family in plants, have a highly hydrophobic helix and associates with mitochondria. Also, some type B HXKs with a nuclear translocation sequence adjacent to the membrane anchor domain can be translocated to the nucleus (Heazlewood et al., 2004; Cho et al., 2006, 2009; Camacho-Pereira et al., 2009; Cheng et al., 2011). Type C HXKs have no signal peptides or membrane anchors and are thought to be cytosolic proteins. They seem to be only present in moss and monocotyledonous plants such as rice and maize (Karve et al., 2010; Cheng et al., 2011; Nilsson et al., 2011). In the present work, we identified one type A PeHXK (PeHXK4), eight type B PeHXKs (PeHXK2, PeHXK3a, PeHXK3b, PeHXK5a, PeHXK5b, PeHXK6, PeHXK9, PeHXK10), and three type C HXKs (PeHXK1, PeHXK7, PeHXK8) based on phylogenetic analysis. To confirm previous reports about the localization of different types of PeHXK proteins, we originally planned to choose three PeHXKs from corresponding three types of PeHXKs. But we failed because the PeHXK4 was not cloned successfully. Thus, finally, we chose PeHXK3b, PeHXK5a, and PeHXK8 as targets to analyze PeHXK protein localization (Figure 6). Our results showed that PeHXK3b and PeHXK5a localized at mitochondria of tobacco leaf epidermal cells, in consistence with that of OsHXK5 and OsHXK6 (Cho et al., 2009). Unlike cytoplasmic protein OsHXK7 which shares the same branch in the phylogenetic tree (Cho et al., 2006), PeHXK8 was detected in both mitochondria and nucleus of leaf epidermal cells. These results suggest that PeHXK3b and PeHXK5a are relatively conserved in their subcellular localization; However, differences in the subcellular localization of PeHXK8 is suggestive of functional variation.
The activity of HXK can be readily determined by means of a yeast complementation assay in vitro (Geng et al., 2017). In the current research, we cloned three PeHXK genes and verified that the three PeHXKs showed HXK activity (Figure 7). It indicated that PeHXK5a and PeHXK8 had an equal ability for phosphorylating glucose and fructose, while PeHXK3b showed favor for glucose as the carbon source to phosphorylate. These results confirmed the two traits of plant HXKs: their low selectivity for substrates, and their preference for glucose over fructose (Karve et al., 2010; Aguilera-Alvarado and Sanchez-Nieto, 2017).
Conclusion
In this study, 12 HXK family genes in moso bamboo were identified and characterized by analysis of phylogenetic relationships, protein and gene structure, structural domains, and estimation of divergence times in evolutionary history. Expression profile analysis implied that these genes were expressed extensively in moso bamboo tissues and may play pivotal roles in plant growth and development. The localization analysis showed that PeHXK3b and PeHX5a were associated with mitochondria while PeHXK8 was localized to both mitochondria and nucleus. An HXK activity assay using the yeast triple-mutant strain YSH7.4-3C verified that the three PeHXKs showed HXK activity with the plant HXK-specific enzyme traits. The present work lays a foundation for further investigation of HXKs in moso bamboo and would accelerate the future cloning and functional analyses of PeHXK genes in moso bamboo.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
LD, SC, and CL conceived and designed the experiments. WZ, YZ, QZ, RW, XW, and SF performed the experiments and interpreted the data. WZ and LD drafted and revised the manuscript.
Funding
This study was financially supported by the National Key Research and Development Program of China (2018 YFD0600101) and the Fundamental Research Funds for the Central Universities (BLX201418), Ministry of Science and Technology of the People’s Republic of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Lianfeng Gu, from the Fujian Agriculture and Forestry University, for kindly providing the seeds of moso bamboo. We thank Xinwen Hu, from Hainan University, for kindly providing the yeast YSH7.4-3C triple mutant and pDR195 vector. We thank Kaihua Jia and Jianfeng Mao, from the Beijing Forestry University, for providing technical support. We also thank Robert McKenzie, Ph.D., from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00600/full#supplementary-material
FIGURE S1 | Amino acids alignment of proteins form moso bamboo, Arabidopsis thaliana, and rice.
FIGURE S2 | The chromosomal distribution of PeHXK genes.
TABLE S1 | List of genes used in this study.
TABLE S2 | Primers used in this study.
FILE S1 | Genomic sequences of PeHXK genes.
FILE S2 | CDS of PeHXK genes.
FILE S3 | Protein sequences of PeHXKs.
Footnotes
- ^ https://www.arabidopsis.org/
- ^ ftp://parrot.genomics.cn/gigadb/pub/10.5524/100001_101000/100498/
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ http://pfam.xfam.org/search#tabview=tab1
- ^ https://web.expasy.org/protparam/
- ^ http://gsds.cbi.pku.edu.cn/
- ^ http://meme-suite.org/tools/meme
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome
References
Aguilera-Alvarado, G. P., Guevara-Garcia, A. A., Estrada-Antolin, S. A., and Sanchez-Nieto, S. (2019). Biochemical properties and subcellular localization of six members of the HXK family in maize and its metabolic contribution to embryo germination. BMC Plant Biol. 19:27. doi: 10.1186/s12870-018-1605-x
Aguilera-Alvarado, G. P., and Sanchez-Nieto, S. (2017). Plant Hexokinases are Multifaceted Proteins. Plant Cell Physiol. 58, 1151–1160. doi: 10.1093/pcp/pcx062
Blanc, G., and Wolfe, K. H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678. doi: 10.1016/j.livsci.2009.01.009
Bork, P., Sander, C., and Valencia, A. (1992). An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U.S.A. 89, 7290–7294. doi: 10.1073/pnas.89.16.7290
Camacho-Pereira, J., Meyer, L. E., Machado, L. B., Oliveira, M. F., and Galina, A. (2009). Reactive oxygen species production by potato tuber mitochondria is modulated by mitochondrially bound hexokinase activity. Plant Physiol. 149, 1099–1110. doi: 10.1104/pp.108.129247
Cheng, W., Zhang, H., Zhou, X., Liu, H., Liu, Y., Li, J., et al. (2011). Subcellular localization of rice hexokinase (OsHXK) family members in the mesophyll protoplasts of tobacco. Biol. Plantarum 55, 173–177. doi: 10.1007/s10535-011-0025-7
Cheng, X., Xiong, R., Liu, H., Wu, M., Chen, F., Hanwei, Y., et al. (2018). Basic helix-loop-helix gene family: genome wide identification, phylogeny, and expression in Moso bamboo. Plant Physiol. Biochem. 132, 104–119. doi: 10.1016/j.plaphy.2018.08.036
Cho, J. I., Ryoo, N., Eom, J. S., Lee, D. W., Kim, H. B., Jeong, S. W., et al. (2009). Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 149, 745–759. doi: 10.1104/pp.108.131227
Cho, J. I., Ryoo, N., Ko, S., Lee, S. K., Lee, J., Jung, K. H., et al. (2006). Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 224, 598–611. doi: 10.2307/23389438
Cho, Y. H., Yoo, S. D., and Sheen, J. (2007). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127, 579–589. doi: 10.1016/j.cell.2006.09.028
Claeyssen, E., and Rivoal, J. (2007). Isozymes of plant hexokinase: occurrence, properties and functions. Phytochemistry 68, 709–731. doi: 10.1016/j.phytochem.2006.12.001
Dai, N., Kandel-Kfir, M., Petreikov, M., Hanael, R., Levin, I., Ricard, B., et al. (2002). The tomato hexokinase LeHXK1 cloning, mapping, expression pattern and phylogenetic relationships. Plant Sci. 163, 581–590. doi: 10.1016/S0168-9452(02)00166-8
Dai, N., Schaffer, A., Petreikov, M., Shahak, Y., Giller, Y., Ratner, K., et al. (1999). Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 11, 1253–1266. doi: 10.1105/tpc.11.7.1253
Damari-Weissler, H., Kandel-Kfir, M., Gidoni, D., Mett, A., Belausov, E., and Granot, D. (2006). Evidence for intracellular spatial separation of hexokinases and fructokinases in tomato plants. Planta 224, 1495–1502. doi: 10.1007/s00425-006-0387-9
David-Schwartz, R., Weintraub, L., Vidavski, R., Zemach, H., Murakhovsky, L., Swartzberg, D., et al. (2013). The SlFRK4 promoter is active only during late stages of pollen and anther development. Plant Sci. 199-200, 61–70. doi: 10.1016/j.plantsci.2012.09.016
Fan, C., Ma, J., Guo, Q., Li, X., Hui, W., and Lu, M. (2013). Selection of reference genes for quantitative real-time pcr in bamboo (Phyllostachys edulis). PLoS One 8:e56573. doi: 10.1371/journal.pone.0056573
Fei, Y., Li, M. L., Pei, P. Y., and Xiu, Q. W. (2013). Hexokinase from grape berries: its prokaryotic expression, polyclonal antibody preparation and biochemical property analyses. J. Plant Biochem. Biot. 22, 324–334. doi: 10.1007/s13562-012-0163-9
Gao, Y., Liu, H., Wang, Y., Li, F., and Xiang, Y. (2018). Genome-wide identification of PHD-finger genes and expression pattern analysis under various treatments in moso bamboo (Phyllostachys edulis). Plant Physiol. Biochem. 123, 378–391. doi: 10.1016/j.plaphy.2017.12.034
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Geng, M. T., Yao, Y., Wang, Y. L., Wu, X. H., Sun, C., Li, R. M., et al. (2017). Structure, expression, and functional analysis of the hexokinase gene family in cassava. Int. J. Mol. Sci. 18:1041. doi: 10.3390/ijms18112398
Giese, J. O., Herbers, K., Hoffmann, M., Klösgen, R. B., and Sonnewald, U. (2005). Isolation and functional characterization of a novel plastidic hexokinase from Nicotiana tabacum. Febs Lett. 579, 827–831. doi: 10.1016/j.febslet.2004.12.071
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Granot, D. (2007). Role of tomato hexose kinases. Funct. Plant Biol. 34, 564–570. doi: 10.1071/FP06207
Granot, D. (2008). Putting plant hexokinases in their proper place. Phytochemistry 69, 2649–2654. doi: 10.1016/j.phytochem.2008.08.026
Granot, D., David-Schwartz, R., and Kelly, G. (2013). Hexose kinases and their role in sugar-sensing and plant development. Front. Plant Sci. 4:44. doi: 10.3389/fpls.2013.00044
Guo, A. Y., Zhu, Q. H., and Xin, C. (2007). GSDS:a gene structure display server. Hereditas 29, 1023–1026. doi: 10.1360/yc-007-1023
He, M. X., Wang, J. L., Qin, H., Shui, Z. X., Zhu, Q. L., Wu, B., et al. (2014). Bamboo: a new source of carbohydrate for biorefinery. Carbohydr. Polym. 111, 645–654. doi: 10.1016/j.carbpol.2014.05.025
Heazlewood, J. L., Tonti-Filippini, J. S., Gout, A. M., Day, D. A., James, W., and Millar, A. H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16, 241–256. doi: 10.1105/tpc.016055
Hou, D., Cheng, Z., Xie, L., Li, X., Li, J., Mu, S., et al. (2018). The R2R3MYB gene family in Phyllostachys edulis: genome-wide analysis and identification of stress or development-related R2R3MYBs. Front. Plant Sci. 9:738. doi: 10.3389/fpls.2018.00738
Hu, D. G., Sun, C. H., Zhang, Q. Y., An, J. P., You, C. X., and Hao, Y. J. (2016). Glucose sensor MdHXK1 phosphorylates and stabilizes MdbHLH3 to promote anthocyanin biosynthesis in apple. PLoS Genet. 12:e1006273. doi: 10.1371/journal.pgen.1006273
Jang, J., León, P., Zhou, L., and Sheen, J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9, 5–19. doi: 10.1105/tpc.9.1.5
Jon, V., Fernie, A. R., Andrea, L., Lothar, W., and Trethewey, R. N. (2002). Potato hexokinase 2 complements transgenic Arabidopsis plants deficient in hexokinase 1 but does not play a key role in tuber carbohydrate metabolism. Plant Mol. Biol. 49, 491–501. doi: 10.1023/a:1015528014562
Juretic, N., Hoen, D. R., Huynh, M. L., Harrison, P. M., and Bureau, T. E. (2005). The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 15, 1292–1297. doi: 10.1101/gr.4064205
Kandel-Kfir, M., Damari-Weissler, H., German, M. A., Gidoni, D., Mett, A., Belausov, E., et al. (2006). Two newly identified membrane-associated and plastidic tomato HXKs: characteristics, predicted structure and intracellular localization. Planta 224, 1341–1352. doi: 10.1007/s00425-006-0318-9
Karve, A., and Moore, B. D. (2009). Function of Arabidopsis hexokinase-like1 as a negative regulator of plant growth. J. Exp. Bot. 60, 4137–4149. doi: 10.1093/jxb/erp252
Karve, A., Rauh, B. L., Xia, X., Kandasamy, M., Meagher, R. B., Sheen, J., et al. (2008). Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 228, 411–425. doi: 10.1007/s00425-008-0746-9
Karve, R., Lauria, M., Virnig, A., Xia, X., Rauh, B. L., and Bd, M. (2010). Evolutionary lineages and functional diversification of plant hexokinases. Mol. Plant 3, 334–346. doi: 10.1093/mp/ssq003
Katz, M. E., Masoumi, A., Burrows, S. R., Shirtliff, C. G., and Cheetham, B. F. (2000). The Aspergillus nidulans xprF gene encodes a hexokinase-like protein involved in the regulation of extracellular proteases. Genetics 156:1559.
Kelly, G., Moshelion, M., David-Schwartz, R., Halperin, O., Wallach, R., Attia, Z., et al. (2013). Hexokinase mediates stomatal closure. Plant J. 75, 977–988. doi: 10.1111/tpj.12258
Kim, M., Lim, J. H., Ahn, C. S., Park, K., Kim, G. T., Kim, W. T., et al. (2006). Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 18, 2341–2355. doi: 10.1105/tpc.106.041509
Kuser, P. R., Krauchenco, S., Antunes, O. A. C., and Polikarpov, I. (2000). The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action. J. Biol. Chem. 275, 20814–20821. doi: 10.1074/jbc.M910412199
León, P., and Sheen, J. (2003). Sugar and hormone connections. Trends Plant Sci. 8, 110–116. doi: 10.1016/s1360-1385(03)00011-6
Li, F., Wu, M., Liu, H., Gao, Y., and Xiang, Y. (2018). Systematic identification and expression pattern analysis of the Aux/IAA and ARF gene families in moso bamboo (Phyllostachys edulis). Plant Physiol. Biochem. 130, 431–444. doi: 10.1016/j.plaphy.2018.07.033
Li, N. N., Qian, W. J., Wang, L., Cao, H. L., Hao, X. Y., Yang, Y. J., et al. (2017). Isolation and expression features of hexose kinase genes under various abiotic stresses in the tea plant (Camellia sinensis). J. Plant Physiol. 209, 95–104. doi: 10.1016/j.jplph.2016.11.007
Lin, J., He, X., Hu, Y., Kuang, T., and Ceulemans, R. (2002). Lignification and lignin heterogeneity for various age classes of bamboo (Phyllostachys pubescens) stems. Physiol. Plant. 114, 296–302. doi: 10.1034/j.1399-3054.2002.1140216.x
Liu, H., Wu, M., Zhu, D., Pan, F., Wang, Y., Wang, Y., et al. (2017). Genome-Wide analysis of the AAAP gene family in moso bamboo (Phyllostachys edulis). BMC Plant Biol. 17:29. doi: 10.1186/s12870-017-0980-z
Liu, H. L., Wu, M., Li, F., Gao, Y. M., Chen, F., and Xiang, Y. (2018). TCP transcription factors in moso bamboo (Phyllostachys edulis): genome-wide identification and expression analysis. Front. Plant Sci. 9:1263. doi: 10.3389/fpls.2018.01263
Liu, R. H., and Meng, J. L. (2003). MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25, 317–321. doi: 10.3321/j.issn:0253-9772.2003.03.019
Lugassi, N., Kelly, G., Fidel, L., Yaniv, Y., Attia, Z., Levi, A., et al. (2015). Expression of Arabidopsis hexokinase in citrus guard cells controls stomatal aperture and reduces transpiration. Front. Plant Sci. 6:1114. doi: 10.3389/fpls.2015.01114
Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W. H., Liu, Y. X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. doi: 10.1126/science.1080585
Moore, R. C., and Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 15682–15687. doi: 10.1073/pnas.2535513100
Nilsson, A., Olsson, T., Ulfstedt, M., Thelander, M., and Ronne, H. (2011). Two novel types of hexokinases in the moss Physcomitrella patens. BMC Plant Biol. 11:32. doi: 10.1186/1471-2229-11-32
Olsson, T., Thelander, M., and Ronne, H. (2003). A novel type of chloroplast stromal hexokinase is the major glucose-phosphorylating enzyme in the moss Physcomitrella patens. J. Biol. Chem. 278, 44439–44447. doi: 10.1074/jbc.M306265200
Peng, Z., Lu, Y., Li, L., Zhao, Q., Feng, Q., Gao, Z., et al. (2013). The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 45, 456–461. doi: 10.1038/ng.2569
Rolland, F., Baenagonzalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. doi: 10.1146/annurev.arplant.57.032905.105441
Rozas, J. (2009). DNA sequence polymorphism analysis Using DnaSP. Methods Mol. Biol. 537:337. doi: 10.1007/978-1-59745-251-9_17
Sarowar, S., Lee, J. Y., Ahn, E. R., and Pai, H. S. (2008). A role of hexokinases in plant resistance to oxidative stress and pathogen infection. J. Plant Biol. 51, 341–346. doi: 10.1007/BF03036136
Sun, M. H., Ma, Q. J., Hu, D. G., Zhu, X. P., You, C. X., Shu, H. R., et al. (2018). The glucose sensor MdHXK1 phosphorylates a tonoplast Na+/H+ exchanger to improve salt tolerance. Plant Physiol. 176, 2977–2990. doi: 10.1104/pp.17.01472
Troncoso-Ponce, M. A., Rivoal, J., Dorion, S., Moisan, M. C., Garces, R., and Martinez-Force, E. (2011). Cloning, biochemical characterization and expression of a sunflower (Helianthus annuus L.) hexokinase associated with seed storage compounds accumulation. J. Plant Physiol. 168, 299–308. doi: 10.1016/j.jplph.2010.07.018
Wang, J., Wang, X., Geng, S., Singh, S. K., Wang, Y., Pattanaik, S., et al. (2018). Genome-wide identification of hexokinase gene family in Brassica napus: structure, phylogenetic analysis, expression, and functional characterization. Planta 248, 171–182. doi: 10.1007/s00425-018-2895-9
Wei, Q., Jiao, C., Ding, Y., Gao, S., Guo, L., Chen, M., et al. (2018). Cellular and molecular characterizations of a slow-growth variant provide insights into the fast growth of bamboo. Tree Physiol. 38, 641–654. doi: 10.1111/pbi.12750
Wu, M., Li, Y., Chen, D., Liu, H., Zhu, D., and Xiang, Y. (2016). Genome-wide identification and expression analysis of the IQD gene family in moso bamboo (Phyllostachys edulis). Sci. Rep. 6:24520. doi: 10.1038/srep24520
Xu, F. Q., Li, X. R., and Ruan, Y. L. (2008). RNAi-mediated suppression of hexokinase gene OsHXK10 in rice leads to non-dehiscent anther and reduction of pollen germination. Plant Sci. 175, 674–684. doi: 10.1016/j.plantsci.2008.07.002
Yu, S. M., and Chiang, C. M. (2008). Distinct hexokinases (HXKs) act as positive and negative regulators in sugar signaling pathways. Plant Biol. 15, 60–61.
Zhang, H., Ying, Y., Wang, J., Zhao, X., Zeng, W., Beahan, C., et al. (2018). Transcriptome analysis provides insights into xylogenesis formation in Moso bamboo (Phyllostachys edulis) shoot. Sci. Rep. 8:3951. doi: 10.1038/s41598-018-21766-3
Zhang, Z., Zhang, J., Chen, Y., Li, R., Wang, H., Ding, L., et al. (2014). Isolation, structural analysis, and expression characteristics of the maize (Zea mays L.) hexokinase gene family. Mol. Biol. Rep. 41, 6157–6166. doi: 10.1007/s11033-014-3495-9
Zhao, B., Qi, K., Yi, X., Chen, G., Xing, L., Qi, X., et al. (2019). Identification of hexokinase family members in pear (Pyrus x bretschneideri) and functional exploration of PbHXK1 in modulating sugar content and plant growth. Gene 711:143932. doi: 10.1016/j.gene.2019.06.022
Keywords: moso bamboo, hexokinase, sequence analysis, expression pattern, subcellular localization, HXK activity
Citation: Zheng W, Zhang Y, Zhang Q, Wu R, Wang X, Feng S, Chen S, Lu C and Du L (2020) Genome-Wide Identification and Characterization of Hexokinase Genes in Moso Bamboo (Phyllostachys edulis). Front. Plant Sci. 11:600. doi: 10.3389/fpls.2020.00600
Received: 17 September 2019; Accepted: 20 April 2020;
Published: 19 May 2020.
Edited by:
Deyu Xie, North Carolina State University, United StatesReviewed by:
Long Li, Northwest A&F University, ChinaGilor Kelly, Agricultural Research Organization (ARO), Israel
Copyright © 2020 Zheng, Zhang, Zhang, Wu, Wang, Feng, Chen, Lu and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Du, ZHVsaWFuZ0BiamZ1LmVkdS5jbg==
 Wenqing Zheng1,2
Wenqing Zheng1,2 Yuan Zhang
Yuan Zhang Shaoliang Chen
Shaoliang Chen Cunfu Lu
Cunfu Lu Liang Du
Liang Du