- 1Key Laboratory for Silviculture and Conservation of Ministry of Education, Beijing Forestry University, Beijing, China
- 2State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 3College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
- 4GEOMAR Helmholtz Centre for Ocean Research Kiel, Kiel, Germany
Cytospora canker is a destructive disease of numerous hosts and causes serious economic losses with a worldwide distribution. Identification of Cytospora species is difficult due to insufficient phylogenetic understanding and overlapped morphological characteristics. In this study, we provide an assessment of 23 Cytospora spp., which covered nine genera of Rosaceae, and focus on 13 species associated with symptomatic branch or twig canker and dieback disease in China. Through morphological observation and multilocus phylogeny of internal transcribed spacer (ITS), large nuclear ribosomal RNA subunit (LSU), actin (act), RNA polymerase II subunit (rpb2), translation elongation factor 1-α (tef1-α), and beta-tubulin (tub2) gene regions, the results indicate 13 distinct lineages with high branch support. These include 10 new Cytospora species, i.e., C. cinnamomea, C. cotoneastricola, C. mali-spectabilis, C. ochracea, C. olivacea, C. pruni-mume, C. rosicola, C. sorbina, C. tibetensis, and C. xinjiangensis and three known taxa including Cytospora erumpens, C. leucostoma, and C. parasitica. This study provides an initial understanding of the taxonomy of Cytospora associated with canker and dieback disease of Rosaceae in China.
Introduction
Many species of Rosaceae plants have economic value, which are important parts of the greening of urban parks and widely cultivated around the world. Many species are also famous fresh and dried fruits with excellent varieties. However, most Rosaceae plants are under serious disease of dieback and stem canker caused by Cytospora species, which have contributed to a severe reduction in yield and quality. A study to clarify the pathogens causing stem canker and dieback of Rosaceae plants is urgently needed.
Cytospora was first introduced by Ehrenberg (1818), which is one of the most important pathogenic fungi of hardwoods and coniferous trees in the world (Adams et al., 2005; Fan et al., 2020). About 150 species epithets of Cytospora are associated with dieback and stem canker on over 130 species of woody hosts (Spielman, 1985; Adams et al., 2005; Kirk et al., 2008; Fan et al., 2020). Over 660 species epithets of Cytospora have been described in Index Fungorum (2020). However, most of these were regarded as synonyms, and most descriptions were unable to identify them accurately (Adams et al., 2005). Leucostoma, Valsa, Valsella, and Valseutypella have been treated as the synonyms of Cytospora (Rossman et al., 2015). The traditional identification of Cytospora species was based heavily on their host affiliations; nevertheless, a single species of Cytospora may occur on a different host, and more than one Cytospora species may be isolated from a single host (Adams et al., 2005; Wang et al., 2011; Ariyawansa et al., 2015; Fan et al., 2015a, b; Hyde et al., 2016; Lawrence et al., 2018). Accurate identification needs additional informative morphological observation and multilocus phylogeny to test the relationship among species (Adams and Taylor, 1993; Harrington and Rizzo, 1999; Adams et al., 2002). Adams et al. (2005) introduced 28 species of Cytospora from Eucalyptus by morphology and phylogeny using ITS sequence. A total of 144 strains from Iran represented 20 species of Cytospora based on ITS phylogeny (Fotouhifar et al., 2010). Later, many species were described based on multilocus phylogeny in recent studies (Fan et al., 2014a,b, 2015a,b; Yang et al., 2015; Lawrence et al., 2017, 2018; Zhu et al., 2018, 2020; Jiang et al., 2020; Shang et al., 2020). Fan et al. (2020) provided an assessment of 52 species of Cytospora in China using a six-locus phylogeny [internal transcribed spacer (ITS), large nuclear ribosomal RNA subunit (LSU), actin (act), RNA polymerase II subunit (rpb2), translation elongation factor 1-α (tef1-α), and beta-tubulin (tub2)]. However, most boundaries of known Cytospora species are tentative and indistinct due to the overlapped morphological characteristics, poor condition of multilocus phylogeny (only ITS is available for most species), and the shortage of fresh collected specimens. Thus, a geography- or host-centered strategy to define species of Cytospora using multiphase approaches has been proposed (Fan et al., 2020).
In this study, a total of 29 strains of Cytospora were isolated from symptomatic hosts of Rosaceae in China. The objectives were to (1) define the species of Cytospora associated with canker and dieback disease of Rosaceae, with illustrations and descriptions; (2) supplement a multi-gene DNA dataset of Cytospora, including ITS, LSU, act, rpb2, tef1-α, and tub2.
Materials and Methods
Sample Collection and Isolation
Fresh specimens of Cytospora canker disease symptoms were collected from infected branches or twigs of five host genera of Rosaceae (11 host species) during collecting trips in China (Supplementary Table S1). The symptoms of Cytospora dieback disease included wilting and killing of twigs and branches, which commenced at the tips and progressed downward to the larger branches, inducing wood lesions and canker formation. Cytospora canker disease represents slightly sunken and discolored areas in the bark, diseased inner bark, and the bark above the infected cambium may appear yellow, brown, reddish brown, gray, or black, becoming watery and odorous as the tissues deteriorate. Several prominent dark sporocarps immersed in the bark, erumpent through the surface of bark when mature (Figures 1, 2). The occurrence of canker diseases of Cytospora in Rosaceae is widespread, which could cause a large area of death in several apple orchards (Figure 2). A total of 29 strains were isolated by removing a mucoid spore mass from conidiomata and/or ascomata, spreading the suspension on the surface of 1.8% potato dextrose agar (PDA) in a Petri dish, and incubating at 25°C for up to 24 h. Single-germinating conidia were transferred onto fresh PDA plates. All specimens are deposited at the Museum of the Beijing Forestry University (BJFC) and the working Collection of X.L. Fan (CF) housed at the Beijing Forestry University. Living cultures are deposited at the China Forestry Culture Collection Centre (CFCC).

Figure 1. Disease symptoms associated with Cytospora species. (A,B) Rosa sp. (C) Prunus serrulata. (D,E) Spiraea salicifolia.
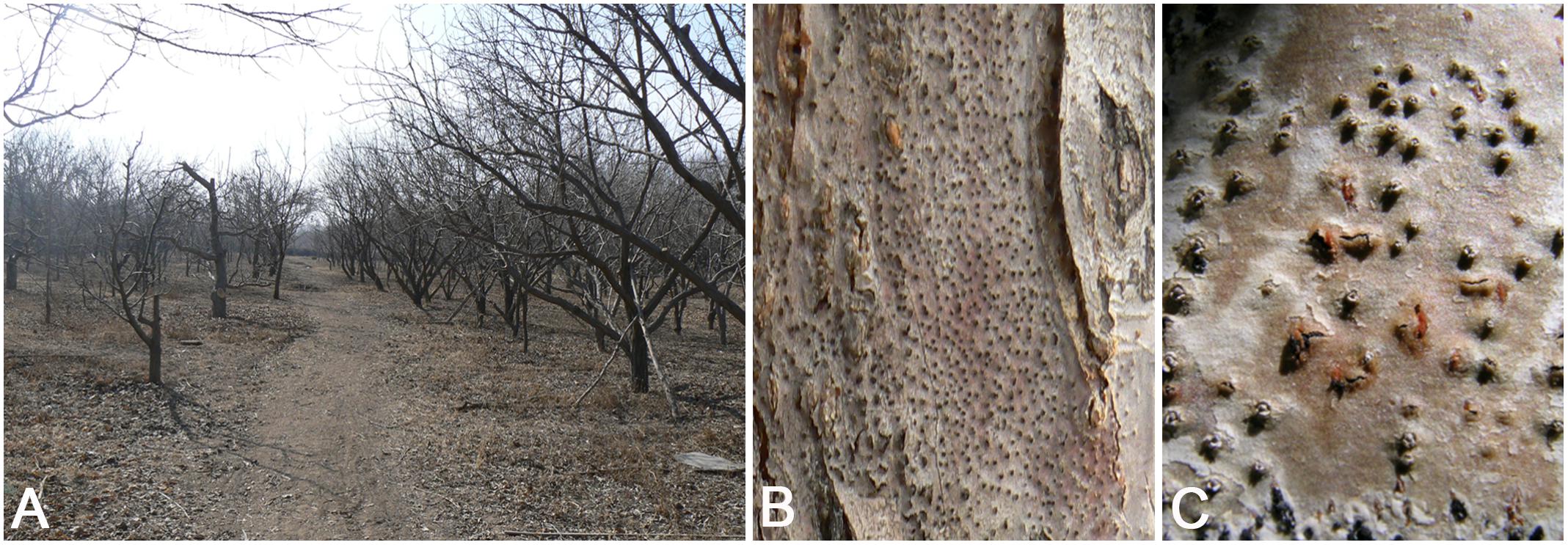
Figure 2. Disease symptoms associated with Cytospora species on Malus sp. (A) Death of the apple trees caused by Cytospora in the orchards. (B,C) Conidiomata on a naturally infected stem in the field.
Morphology
Observation and description of Cytospora species were based on morphological characteristics of the fruiting bodies produced on infected host materials including arrangement and size of stromata; presence and absence of a conceptacle; size, color, and shape of discs; and number and diameter of ostioles per disc. The morphological characteristics were determined under a Leica stereomicroscope (M205). Micro-morphological observations include size and shape of conidiophores and conidia determined under a Nikon Eclipse 80i microscope. Over 30 conidiomata were sectioned, and 50 conidia were selected randomly for measurement. Incubation was done on PDA at 25°C in darkness, and colony diameters and colors were recorded and described after 1 or 2 weeks according to the color charts of Rayner (1970). Adobe Bridge CS v.6 and Adobe Photoshop CS v.5 were used for the manual editing. Taxonomic novelties were deposited in MycoBank (Crous et al., 2004).
DNA Extraction and PCR Amplification
Genomic DNA was extracted from mycelium cultured on PDA with cellophane for 3 days using the modified CTAB method (Doyle and Doyle, 1990). The extracted DNA was estimated visually by electrophoresis in 1% agarose gels, and band intensity was compared with a DNA maker, 1 kbp (Takara Biotech). The qualities of DNA were measured with NanoDropTM 2000 (Thermo, United States). The internal transcribed spacer (ITS) region was amplified with primers ITS1 and ITS4 (White et al., 1990). The large nuclear ribosomal RNA subunit (LSU) region was amplified with primers LROR and LR7 (Vilgalys and Hester, 1990). The actin (act) region was amplified with primers ACT-512F and ACT-783R (Carbone and Kohn, 1999). The RNA polymerase II subunit (rpb2) region was amplified with primers RPB2-5F and fRPB2-7cR (Liu et al., 1999). The translation elongation factor 1-α (tef1-α) gene was amplified with primers EF-688F and EF-1251R (Carbone and Kohn, 1999). The beta-tubulin (tub2) gene was amplified with Bt-2a and Bt-2b (Glass and Donaldson, 1995). The PCR amplicons were electrophoresed in 2% agarose gels. DNA sequencing was carried out using an ABI PRISM® 3730XL DNA Analyzer with BigDye® Terminater Kit v.3.1 (Invitrogen) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China). DNA sequences generated by the forward and reverse primers were used to obtain consensus sequences using Seqman v.9.0.4 (DNASTAR Inc., Madison, WI, United States).
Phylogenetic Analyses
To infer a preliminary phylogenetic relationship for the new sequences, the first alignment based on ITS sequence data was performed using MAFFT v.6 (Katoh and Standley, 2013) and edited manually using MEGA v.6.0 (Tamura et al., 2013). Some characters were excluded from both ends of the alignments to approximate the size of our sequences to those included in the dataset. A second alignment was performed based on a combined six concatenate sequences (ITS, LSU, act, rpb2, tef1-α, and tub2). For individual datasets, sequences were aligned using MAFFT v.6 and edited manually using MEGA v.6.0 and some characters were excluded from both ends of the alignments. A partition homogeneity test (PHT) with heuristic search and 1,000 homogeneity replicates was performed using PAUP v.4.0b10 to test the discrepancy among the six-gene dataset in reconstructing phylogenetic trees. The sequences of Diaporthe vaccinii (CBS 160.32) was included as outgroup in all analyses. The phylogenetic analyses for all the datasets were run using PAUP v.4.0b10 for maximum parsimony (MP) (Swofford, 2003), MrBayes v.3.1.2 for Bayesian inference (BI) (Ronquist and Huelsenbeck, 2003), and RAxML-NG v.0.9.0 for maximum likelihood (ML) (Kozlov et al., 2019). Trees were visualized using FigTree v.1.3.1 (Rambaut and Drummond, 2010).
MP analysis was performed using a heuristic search (1,000 bootstraps) (Hillis and Bull, 1993), with random sequence addition as option to stepwise addition (1,000 replicates and one tree held at each addition step), and maxtrees limited to 200 by replicate. The tree bisection and reconnection (TBR) algorithm was selected (Swofford, 2003). The branches of zero length were collapsed using the command minbrlen, and all equally most parsimonious trees were saved. Other parsimony scores such as tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency (RC) were calculated to describe tree statistics (Swofford, 2003). The branch supports of MP were evaluated with a bootstrapping (BS) method of 1,000 replicates (Hillis and Bull, 1993). For ML and BI analyses, the best-fit evolutionary models for each partitioned locus were estimated by MrModeltest v.2.3 following the Akaike Information Criterion (AIC) (Posada and Crandall, 1998). ML analysis was performed with RAxML-NG1 (Kozlov et al., 2019). The bootstrap was used with 100 replicates and the appropriate models for each gene. BI analysis was done by a Markov chain Monte Carlo (MCMC) algorithm with Bayesian posterior probabilities (BPP) (Rannala and Yang, 1996). Two MCMC chains were run from random trees for 10 million generations, and trees were sampled each 100th generation. The first 25% of the trees were discarded as the burn-in phase of each analysis; branches with significant BPP were calculated to assess the remaining trees (Rannala and Yang, 1996). Phylograms are shown using Figtree v.1.3.1 (Rambaut and Drummond, 2010). All sequences from this study data were deposited in GenBank. The ITS and multi-gene sequence alignment files were deposited in TreeBASE2 (accession number: S25903).
Results
A total of 29 Cytospora isolates from Rosaceae hosts were collected in China. Following alignment, the ITS sequence data comprised 248 Cytospora in group taxa with a total of 629 characters including gaps, of which 365 characters were constant, 63 variable characters were parsimony uninformative, and 201 characters were variable and parsimony informative. A heuristic search generated 200 equally parsimonious trees each with similar clade topologies, and one of which is presented in Supplementary Figure S1 (TL = 1,223, CI = 0.343, RI = 0.855, RC = 0.294). ML and Bayesian results do not significantly differ from the MP tree.
To clarify the phylogenetic position of these Cytospora species, a multi-locus analysis (ITS, LSU, act, rpb2, tef1-α, and tub2) is presented in Figure 3. The final analysis combined sequence data of six genes composed of 218 Cytospora ingroup taxa with a total of 3,712 characters including gaps, of which 2,037 characters were constant, 202 variable characters were parsimony uninformative, and 1,473 characters were variable and parsimony informative. MP analysis generated 200 equally parsimonious trees each with similar clade topologies, and one of which is presented in Figure 3 (TL = 9,782, CI = 0.303, RI = 0.803, RC = 0.244). For ML and BI analyses, the best-fit model of nucleotide evolution was deduced on the AIC (ITS and LSU: GTR, act: TVM, rpb2 and tef1-α: TrN, and tub: HKY). ML method and Bayesian analyses were in agreement and no difference from the MP tree. The MP bootstrap supports (MP-BS) and ML bootstrap (ML-BS) equal to or above 70% were shown in branches in Figure 3. The branches with significant Bayesian posterior probabilities (BPP) equal to or above 0.95 are thickened in the phylograms.
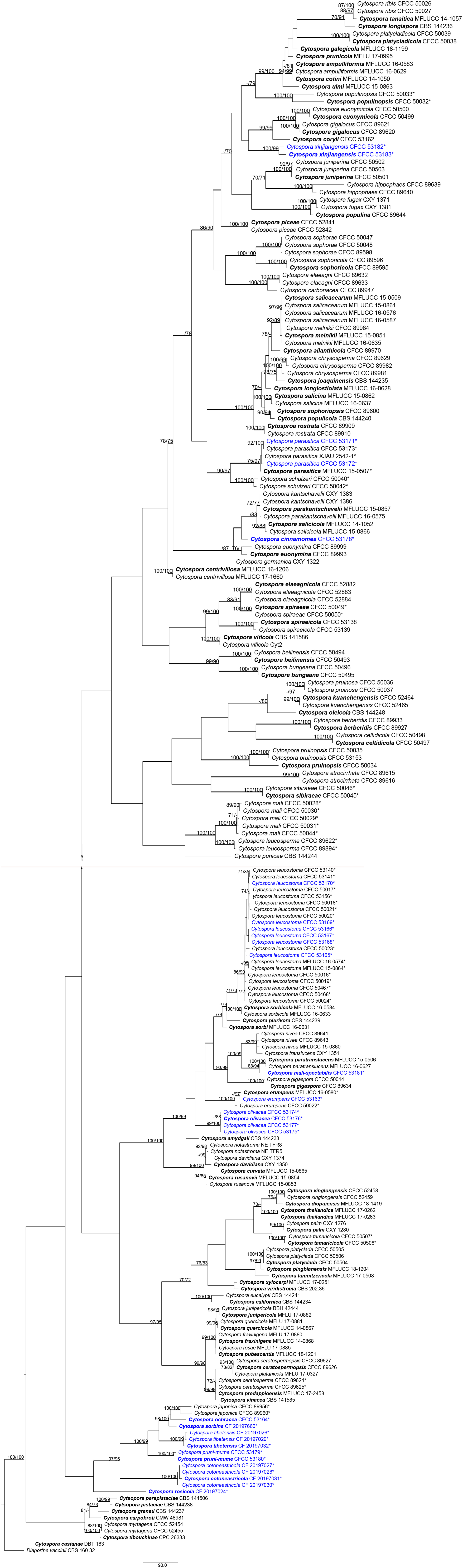
Figure 3. Phylogram of Cytospora based on combined internal transcribed spacer (ITS), large nuclear ribosomal RNA subunit (LSU), actin (act), RNA polymerase II subunit (rpb2), translation elongation factor 1-α (tef1-α), and beta-tubulin (tub2) genes. Maximum parsimony (MP) and maximum likelihood (ML) bootstrap support values above 70% are shown at the first and second positions. Thickened branches represent posterior probabilities above 0.95 from Bayesian inference (BI). Ex-type strains are in bold. Strains in current study are in blue. All the Cytospora species listed from Rosaceae plants in China are marked with *.
The current 29 strains clustered in 13 clades were equivalent to 13 Cytospora species in Figure 3, including three known species (Cytospora erumpens, C. leucostoma, and C. parasitica) and 10 new clades, distinct from all known taxa, are herein described as C. cinnamomea, C. cotoneastricola, C. mali-spectabilis, C. ochracea, C. olivacea, C. pruni-mume, C. rosicola, C. sorbina, C. tibetensis, and C. xinjiangensis. All detailed descriptions and notes are below.
Taxonomy
Cytospora cinnamomea M. Pan & X.L. Fan, sp. nov. (Figure 4)
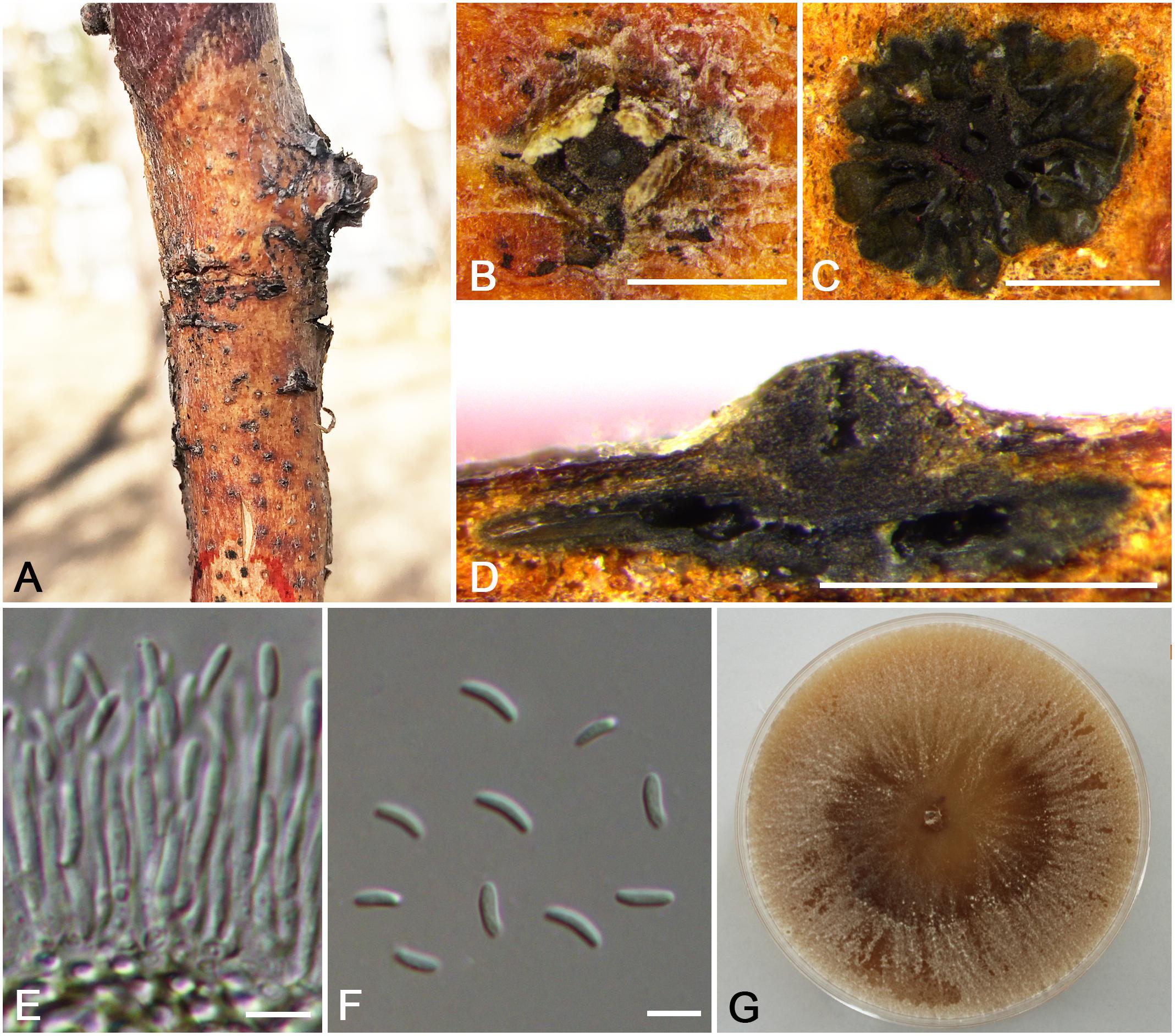
Figure 4. Morphology of Cytospora cinnamomea from Prunus armeniaca (CF 20197654). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F) Conidia. (G) Colonies on potato dextrose agar (PDA) for 2 weeks. Bars: (B–D) = 500 μm; (E) = 10 μm; (F) = 5 μm.
MycoBank MB 834846
Typification: China. Xinjiang Uygur Autonomous Region: Bole Mongol Autonomous Prefecture, 44°46′14.88′′N, 81°13′58.64′′E, from branches of Prunus armeniaca, 20 July 2017, C.M. Tian & X.L. Fan (holotype CF 20197654), ex-type living culture CFCC 53178.
Etymology: Named after the distinctive cinnamon culture color.
Descriptions: Symptoms appeared as circular to ovoid, brown or dark, raised, dehiscent lesions on the twigs or branches, with slightly discolored bark above the infected cambium. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, erumpent, discoid, with multiple locules, 785–1,070 μm ( = 935 μm, n = 30) in diam. Conceptacle absent. Ectostromatic disc dark-brown, circular to ovoid, disc black, 280–560 μm ( = 425 μm, n = 30) in diam. Ostiole conspicuous, circular, gray to brown at the same level as the disc surface, 50–63 μm ( = 57 μm, n = 30) in diam. Locules numerous, irregular arrangement with individual walls. Conidiophores hyaline, unbranched at base or occasionally branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, smooth-walled, elongate-allantoid, unicellular, (4.0–)4.5–6.0(–6.5) × 1–1.5 μm ( = = 5.3 × 1.3 μm, n = 50). Sexual morph: not observed.
Culture characteristics: Cultures on PDA are initially white, fast growing, and covering the 9-cm Petri dish after 3 days, becoming cinnamon to fawn after 30 days. The colonies are flat and with a uniform texture, conidiomata sparse and distributed irregularly on the medium surface.
Habitat and distribution: Known only on Prunus armeniaca from the type locality.
Notes: Cytospora cinnamomea is associated with canker disease of Prunus armeniaca in China. In the combined analysis, the most closely related species to Cytospora cinnamomea are C. kantschavelii, C. parakantschavelii, C. salicicola, and C. euonymina (Figure 3). Cytospora cinnamomea can be distinguished from C. salicicola and C. euonymina by the smaller conidia (4.5–6.0 × 1–1.5 vs. 6.9–7.6 × 1.4–1.5, 6.5–7.5 × 1.5–2 μm) (Saccardo, 1892; Norphanphoun et al., 2017). Cytospora cinnamomea is morphologically similar to C. kantschavelii and C. parakantschavelii, whereas the former species differs from them by having distinct discoid conidiomata and producing cinnamon color in culture media. As for its size of conidia, it is similar with C. parakantschavelii (5.3 × 1.3 vs. 5.3 × 1.4 μm), but different from C. kantschavelii (5.3 × 1.3 vs. 4–5 × 1.2 μm) (Gvritishvili, 1973; Norphanphoun et al., 2017).
Cytospora cotoneastricola M. Pan & X.L. Fan, sp. nov. (Figure 5)

Figure 5. Morphology of Cytospora cotoneastricola from Cotoneaster sp. (CF 20197031). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F) Conidia. Bars: (B–D) = 500 μm; (E,F) = 10 μm.
MycoBank MB 834847
Typification: China. The Tibet Autonomous Region: Nyingchi City, Chayu County, Pineapple Village, 28°56′47.34′′N, 97°25′31.36′′E, from branches of Cotoneaster sp., 30 July 2016, C.M. Tian & X.L. Fan (holotype CF 20197031).
Etymology: Named after the host genus on which it was collected, Cotoneaster sp.
Descriptions: Symptoms appeared as circular to ovoid, yellow or light brown, raised lesions with a distinct black area at the center on the twigs or branches. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, solitary, scattered, discoid to conical, with multi-locules, 1,200–1,480 μm ( = 1,350 μm, n = 30) in diam. Conceptacle conspicuous. Ectostromatic disc yellow to light brown, circular to ovoid, with one ostiole per disc, 165–310 μm ( = 240 μm, n = 30) in diam. Ostiole conspicuous, flask-shaped to conical, gray to black in the center of the disc at the same level as the disc surface, 110–165 μm ( = 131 μm, n = 30) in diam., a column lenticular tissue in the center. Locules dark brown, arranged circularly to triangularly or ellipse with independent walls, ovoid to spherical. Conidiophores hyaline, unbranched at base or occasionally branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, smooth, allantoid, eguttulate, aseptate, thin-walled, (12.5–)13.0–14.0(–14.5) × 2.5–3 μm ( = 13.6 × 2.8 μm, n = 50). Sexual morph: not observed.
Habitat and distribution: Known on Cotoneaster sp. from the type locality and an additional locality in the Tibet Autonomous Region.
Additional materials examined: China. The Tibet Autonomous Region: Nyingchi City, Chayu County, Pineapple Village, 28°56′47.34′′N, 97°25′31.36′′E, from branches of Cotoneaster sp., 30 July 2016, C.M. Tian & X.L. Fan (CF 20197030); Nyingchi City, Chayu County, Pineapple Village, 28°56′47.33′′N, 97°25′31.44′′E, from branches of Cotoneaster sp., 30 July 2016, C.M. Tian & X.L. Fan (CF 20197027); ibid. CF 20197028.
Notes: Cytospora cotoneastricola is described as being associated with canker disease of Cotoneaster sp. in China. Cytospora tibetensis and C. ochracea are associated with the same host. Morphologically, C. cotoneastricola is distinguished from C. tibetensis by having multi-locules and larger size of conidia (13.0–14.0 × 2.5–3 vs. 5.0–5.5 × 1.5–2 μm). Cytospora cotoneastricola also has obvious central column and larger conidia than C. ochracea (13.5–14.0 × 2.5–3 vs. 8.5–9.0 × 1.5–2.5 μm). This species needs to be re-collected from Cotoneaster sp. in Tibet of China, as presently no living culture is available.
Cytospora erumpens Norph. et al., Mycosphere 8: 64, 2017.
Descriptions: see Norphanphoun et al. (2017).
Habitat and distribution: Known on Salix fragilis and Prunus padus. This fungus has been reported from Russia and China.
Material examined: China. Xinjiang Uygur Autonomous Region: Ili Kazak Autonomous Prefecture, 46°01′17.82′′N, 82°45′08.94′′E, from branches of Prunus padus, 16 July 2017, C.M. Tian & X.L. Fan (CF 20197563), living culture CFCC 53163.
Notes: Cytospora erumpens was introduced to cause canker and dieback disease of Salix in Russia (Norphanphoun et al., 2017). Afterward, it was reported on Prunus padus in China (Fan et al., 2020). This fungus can be identified by its black-discoid conidiomata with long ostiolar necks, producing elongate-allantoid conidia (6.4–6.7 × 1.3–1.4 μm) (Norphanphoun et al., 2017). Combined morphology and the DNA sequence data of our strain, which was collected from dead branches of Prunus padus belongs to this species.
Cytospora leucostoma (Pers.) Sacc., Michelia 2: 264, 1881.
Synonyms: Sphaeria leucostoma Pers., Ann. Bot. 11: 23, 1794.
Valsa leucostoma (Pers.) Fr., Summa Veg. Scand., Section Post. (Stockholm): 411, 1849.
Valsa persoonii Nitschke, Pyrenomyc. Germ. 2: 222, 1870.
Leucostoma persoonii (Nitschke) Höhn., Mitt. Bot. Inst. Tech. Hochsch. Wien 5: 78, 1928.
Cytospora donetzica Norphanph et al., Mycosphere 8: 62, 2017.
Valsa ambiens (Pers.) Fr., Summa Veg. Scand., Sectio Post. (Stockholm): 412, 1849.
Cytospora ambiens Sacc., Michelia 1(5): 519, 1879.
Descriptions: see Fan et al. (2020).
Habitat and distribution: Known from mainly Roseaceae, especially Prunoideae. This fungus has been reported around the world.
Materials examined: China. Xinjiang Uygur Autonomous Region: Bole Mongol Autonomous Prefecture, 44°46′12.65′′N, 81°14′02.62′′E, from branches of Sorbus tianschanica, 20 July 2017, C.M. Tian & X.L. Fan (CF 20197672), living culture CFCC 53165; Ili Kazak Autonomous Prefecture, 44°27′39.68′′N, 80°21′12.86′′E, from branches of Prunus armeniaca, 22 July 2017, C.M. Tian & X.L. Fan (CF 20197710), living culture CFCC 53166; ibid. CF 20197711, living culture CFCC 53167; Ili Kazak Autonomous Prefecture, 46°20′55.49′′N, 83°53′55.94′E, from branches of Prunus pseudocerasus, 14 July 2017, C.M. Tian & X.L. Fan (CF 20197513), living culture CFCC 53168. Beijing: Songshan National Nature, 40°30′25.07′′N, 115°48′44.36′′E, from branches of Prunus persica, 20 July 2017, C.M. Tian & X.L. Fan (CF 20191281), living culture CFCC 53169; ibid. CF 20191284, living culture CFCC 53170.
Notes: Cytospora leucostoma is a common species associated with stem canker diseases of woody plants of Rosaceae in China (Fan et al., 2020). This species has obvious black conceptacle, numerous locules, which were subdivided frequently by invaginations with independent walls, and hyaline, allantoid, aseptate conidia with the size of 4.5–5.5 × 1–1.5 μm. In a recent study, Cytospora donetzica has been treated as the synonym of C. leucostoma based on Fan et al. (2020).
Cytospora mali-spectabilis M. Pan & X.L. Fan, sp. nov. (Figure 6)
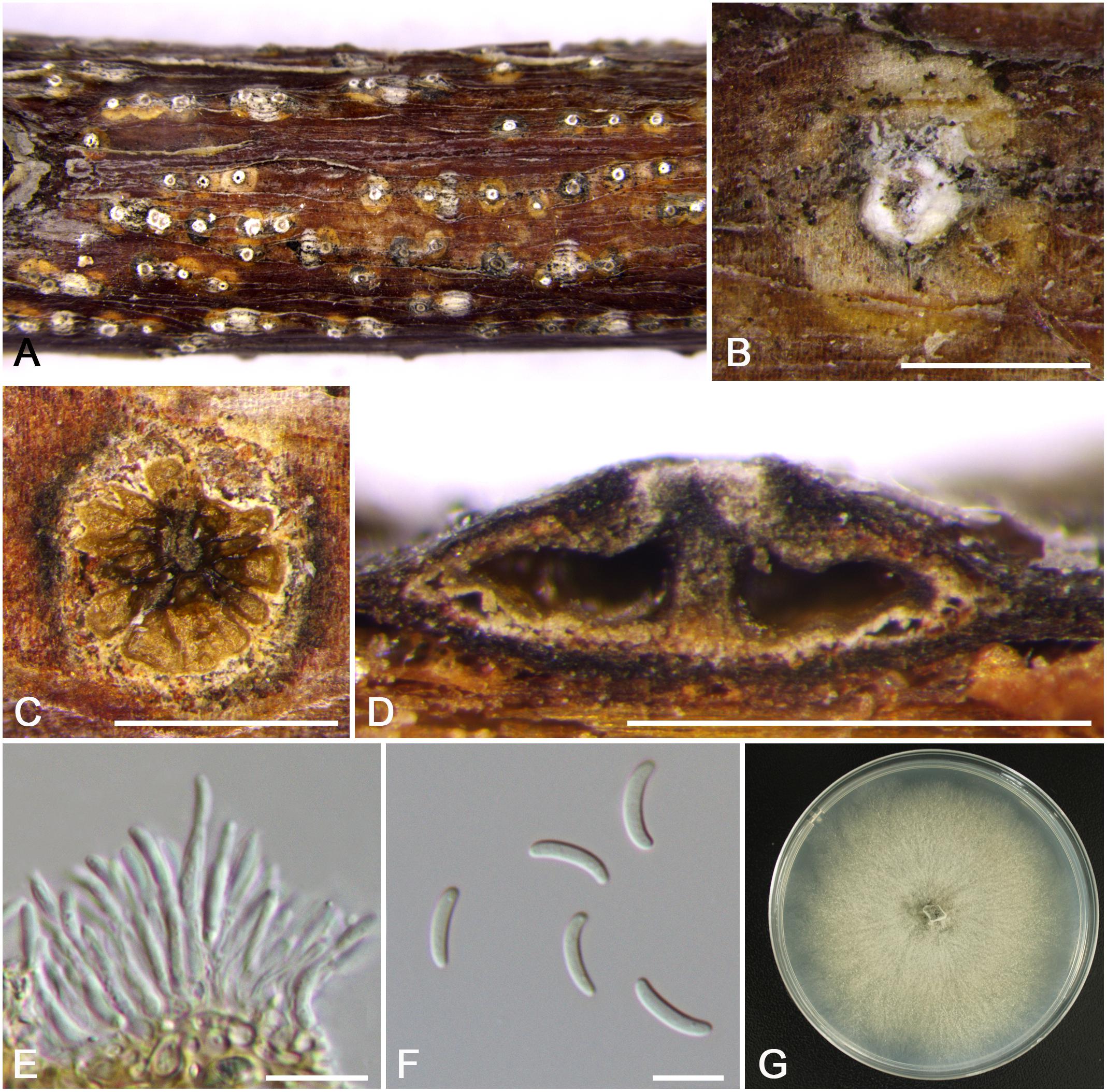
Figure 6. Morphology of Cytospora mali-spectabilis from Malus spectabilis ‘Royalty’ (CF 20197665). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F) Conidia. (G) Colonies on PDA for 2 weeks. Bars: (B–D) = 500 μm; (E,F) = 10 μm.
MycoBank MB 834848
Typification: China. Xinjiang Uygur Autonomous Region: Bole Mongol Autonomous Prefecture, 44°46′10.79′′N, 81°13′56.60′′E, from branches of Malus spectabilis ‘Royalty’, 20 July 2017, C.M. Tian & X.L. Fan (holotype CF 20197665), ex-type living culture CFCC 53181.
Etymology: Named after the host genus on which it was collected, Malus spectabilis ‘Royalty’.
Descriptions: Symptoms appeared as circular, white or ashen, raised lesions with a distinct brown or dark area at the center on the twigs or branches, with discolored bark above the infected cambium. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, erumpent, discoid to conical, 580–675 μm ( = 630 μm, n = 30) in diam., with multi-locule, a column lenticular tissue in the center. Conceptacle absent. Ectostromatic disc white to brown, circular to ovoid, disc dark yellow to brown, 240–350 μm ( = 295 μm, n = 30) in diam. Ostiole conspicuous, circular to ovoid, gray to black at the same level as the disc surface, 60–84 μm ( = 69 μm, n = 30) in diam. Locules complex with wild shapes, subdivided by invaginations with common walls. Conidiophores hyaline, unbranched at base or occasionally branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, unicellular, eguttulate, elongate-allantoid, (8.0–)9.0–10.0(–11.0) × 1.5–2 μm ( = 9.5 × 1.8 μm, n = 50). Sexual morph: not observed.
Culture characteristics: Cultures on PDA are initially white, growing up to 6.5 cm after 3 days, entirely covering the 9-cm Petri dish and becoming buff after 7 days. The colonies ultimately are gray olivaceous and flat with a uniform texture. Conidiomata are randomly distributed on medium surface.
Habitat and distribution: Known only on Malus spectabilis ‘Royalty’ from the type locality.
Notes: Cytospora mali-spectabilis is associated with canker disease of Malus spectabilis ‘Royalty’. In the phylogenetic analyses, C. mali-spectabilis clusters with C. paratranslucens and C. nivea with high bootstrap support (MP/ML/BI = 100/99/1). However, it can be distinguished from C. paratranslucens and C. nivea by larger conidia (9.0–10.0 × 1.5–2 vs. 6.5–7.3 × 1.3–1.5, 7.4–8.8 × 1.5–1.6 μm), and the distinct central column of conidia (Adams et al., 2006; Norphanphoun et al., 2017). Furthermore, C. mali-spectabilis has multiloculate conidiomata sharing a smaller single ostiole (60–84 vs. 70–150 μm) than C. paratranslucens (Norphanphoun et al., 2017). Cytospora mali-spectabilis has absent conceptacle, whereas C. nivea owned black conceptacle surrounding the asexual stroma, usually presenting a huge black ectostromatic disc on the bark surface (Adams et al., 2005; Fan et al., 2014b).
Cytospora ochracea M. Pan & X.L. Fan, sp. nov. (Figure 7)
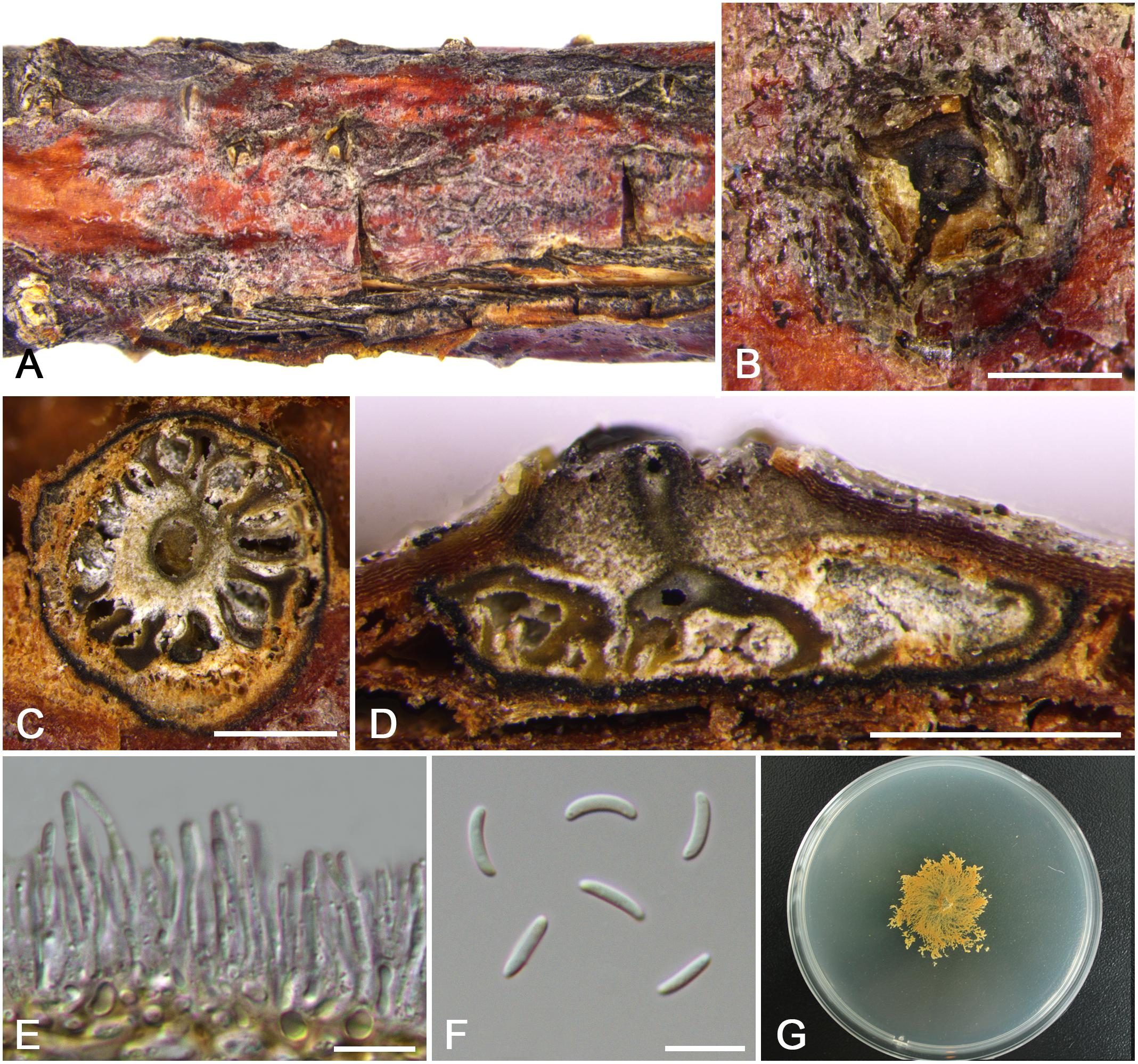
Figure 7. Morphology of Cytospora ochracea from Cotoneaster sp. (CF 20197684). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F) Conidia. (G) Colonies on PDA for 2 weeks. Bars: (B–D) = 500 μm; (E,F) = 10 μm.
MycoBank MB 834849
Typification: China. Xinjiang Uygur Autonomous Region: Bole Mongol Autonomous Prefecture, 44°46′26.42′′N, 81°11′19.28′′E, from branches of Cotoneaster sp., 20 July 2017, C.M. Tian & X.L. Fan (holotype CF 20197684), ex-type living culture CFCC 53164.
Etymology: Named after the distinctive ochreous culture color.
Descriptions: Symptoms appeared as elongate, circular to ovoid, raised, dehiscent lesions surrounded by a black circle on the twigs or branches, with slightly discolored bark above the infected cambium. Sometimes lesions may split along the canker margin. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, erumpent, discoid to conical, 1,150–1,325 μm ( = 1,230 μm, n = 30) in diam. Conceptacle dark. Ectostromatic disc dark brown, circular to ovoid, disc gray white to dark brown, 630–850 μm ( = 740 μm, n = 30) in diam. Ostiole conspicuous, circular, dark-brown to black, 180–260 μm ( = 235 μm, n = 30) in diam. Locules numerous, subdivided frequently by invaginations, irregular arrangement with individual walls. Conidiophores hyaline, unbranched at base or occasionally branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, eguttulate, smooth-walled, elongate-allantoid, aseptate, (7.5–)8.5–9.0(–10.0) × 1.5–2.5 μm ( = 8.8 × 1.9 μm, n = 50). Sexual morph: not observed.
Culture characteristics: Cultures on PDA are initially white, growing up to 5.5 cm after 3 days, becoming ochreous in center after 7 days, deepened in later stage gradually. Colonies are tight, thin with a uniform texture, lacking aerial mycelium. Conidiomata are randomly distributed on medium surface.
Habitat and distribution: Known only on Cotoneaster sp. from the type locality.
Notes: Cytospora ochracea is associated with canker disease of Cotoneaster sp. In the phylogenetic analyses, C. ochracea clusters with C. japonica and C. sorbina with high bootstrap support (MP/ML/BI = 98/100/1). However, it can be distinguished from C. japonica and C. sorbina by larger conidia (8.5–9.0 × 1.5–2.5 vs. 6.5–8.5 × 1.5–2, 4.5–5.5 × 1–1.5 μm) (Fan et al., 2020). Furthermore, C. ochracea has multiloculate conidiomata with individual walls, whereas C. sorbina owned locules with the common walls. Also C. ochracea differs from C. japonica in culture characteristics of its color and growth rate.
Cytospora olivacea M. Pan & X.L. Fan, sp. nov. (Figure 8)

Figure 8. Morphology of Cytospora olivacea from Sorbus tianschanica (CF 20197670). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F) Conidia. (G) Colonies on PDA for 2 weeks. Bars: (B–D) = 500 μm; (E) = 10 μm; (F) = 5 μm.
MycoBank MB 834850
Typification: China. Xinjiang Uygur Autonomous Region: Bole Mongol Autonomous Prefecture, 44°46′10.79′′N, 81°13′56.60′′E, from branches of Sorbus tianschanica, 20 July 2017, C.M. Tian & X.L. Fan (holotype CF 20197670), ex-type living culture CFCC 53176.
Etymology: Named after the distinctive olivaceous culture color.
Descriptions: Symptoms appeared as circular to ovoid, raised, dehiscent lesions on the twigs or branches, with slightly discolored bark above the infected cambium. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, erumpent, flask shaped to conical, with multiple locules, 1,285–1,535 μm ( = 1,415 μm, n = 30) in diam. Conceptacle dark. Ectostromatic disc yellow to light brown, circular to ovoid, disc dark brown, 1,050–1,450 μm ( = 1,250 μm, n = 30) in diam. Ostiole conspicuous, circular, gray to brown at the same level as the disc surface, 330–465 μm ( = 400 μm, n = 30) in diam. Locules numerous, irregular arrangement with independent walls. Conidiophores hyaline, unbranched at base or occasionally branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, smooth-walled, elongate-allantoid, unicellular, 4.0–5.0(–5.5) × 1–1.5 μm ( = 4.4 × 1.3 μm, n = 50). Sexual morph: not observed.
Culture characteristics: Cultures on PDA are initially white and become olivaceous buff, growing fast and entirely covering the 9-cm Petri dish after 3 days, becoming olivaceous gray and slight helical after 30 days. The colonies are flat and with a uniform texture. Conidiomata are randomly distributed on medium surface, extruding a pale white conidial mass.
Habitat and distribution: Known on Cotoneaster sp. and Prunus spp. from the type locality and an additional locality in Xinjiang Uygur Autonomous Region.
Additional materials examined: China. Xinjiang Uygur Autonomous Region: Ili Kazak Autonomous Prefecture, 45°54′18.17′′N, 83°20′45.35′′E, from branches of Prunus dulcis, 15 July 2017, C.M. Tian & X.L. Fan (CF 20197556), living culture CFCC 53175; Bole Mongol Autonomous Prefecture, 44°46′13.44″N, 81°13′58.72″E, from branches of Prunus virginiana, 18 July 2017, C.M. Tian & X.L. Fan (CF 20197601), living culture CFCC 53177; Bole Mongol Autonomous Prefecture, 44°46′13.78″N, 81°13′57.78″E, from branches of Prunus cerasifera, 20 July 2017, C.M. Tian & X.L. Fan (CF 20197652), living culture CFCC 53174.
Notes: Cytospora olivacea is associated with canker disease of Prunus spp. in China. It has multiple locules with black conceptacle, which is commonly discovered in Cytospora spp., while the molecular phylogenies show a clearly different position from all other strains included in this study. Therefore, we describe this species as novel based on morphology and combined sequence data of six genes.
Cytospora parasitica Norph. et al., Fung. Diversity 75: 146, 2015. (Figure 9)
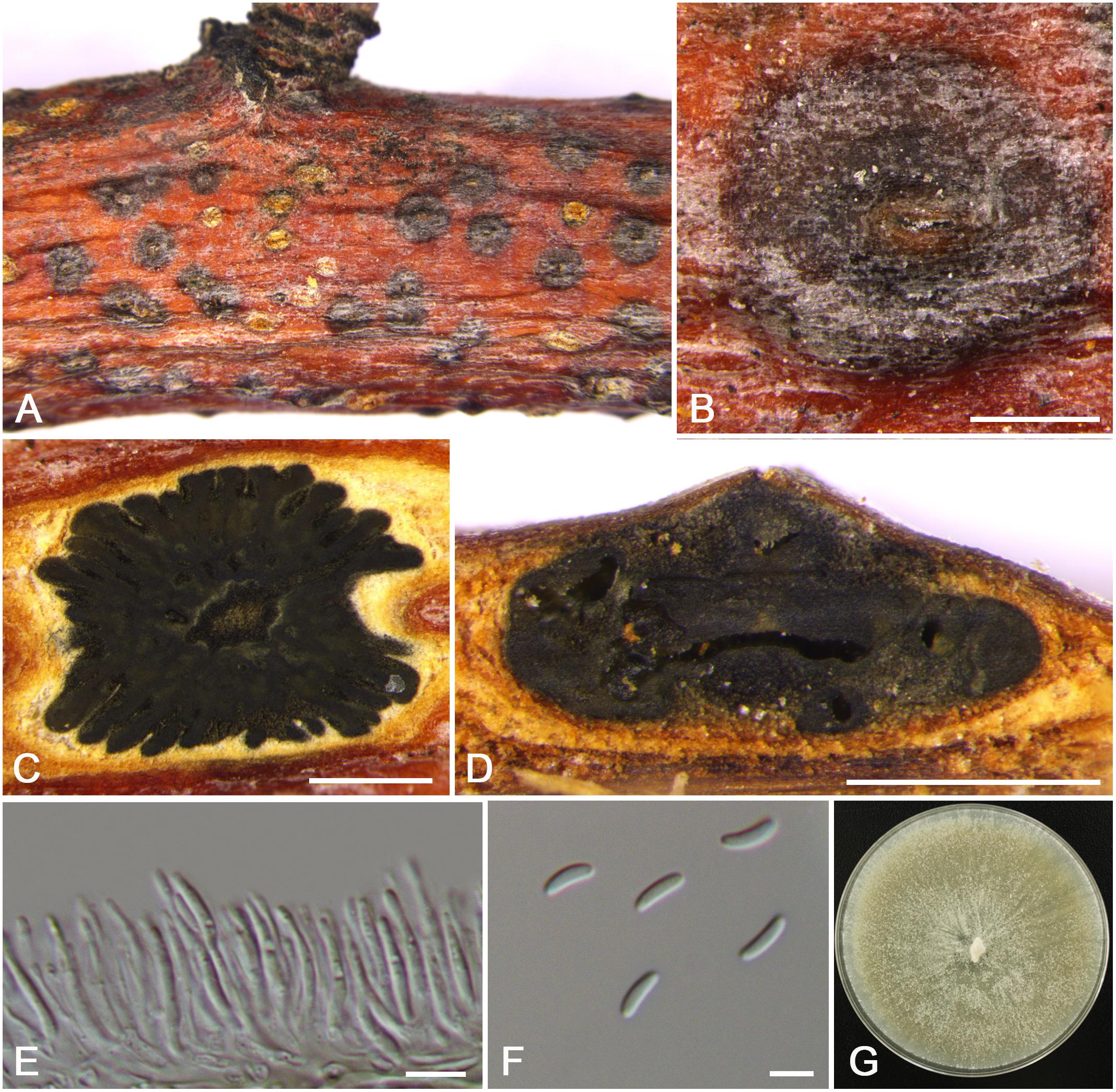
Figure 9. Morphology of Cytospora parasitica from Malus pumila (CF 20197714). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F) Conidia. (G) Colonies on PDA for 2 weeks. Bars: (B–D) = 500 μm; (E) = 10 μm; (F) = 5 μm.
Descriptions: Symptoms appeared as circular, dark brown or dark, slightly sunken, dehiscent lesions with a light brown area at the center on the twigs or branches, with discolored bark above the infected cambium. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, erumpent, flask-shaped to conical, with multiple locules, 1,190–1,650 μm ( = 1,420 μm, n = 30) in diam. Conceptacle absent. Ectostromatic disc dark brown, circular to ovoid, disc gray to black, 1,190–1,480 μm ( = 1,310 μm, n = 30) in diam. Ostiole conspicuous, circular, dark brown to black, 160–210 μm ( = 185 μm, n = 30) in diam. Locules complex multi-loculed irregular arrangement subdivided frequently by invaginations, sharing common walls. Conidiophores hyaline, unbranched at base or occasionally branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, smooth-walled, elongate-allantoid, aseptate, (5.0–)5.5–6.0(–6.5) × 1–2 μm ( = 5.7 × 1.5 μm, n = 50). Sexual morph: not observed.
Culture characteristics: Cultures on PDA are initially white, growing up to 8.5 cm after 3 days and entirely covering the 9-cm Petri dish after 7 days, becoming buff but white mostly. The colonies are flat with a uniform texture, becoming effuse on the surface, without aerial mycelium, conidiomata are randomly distributed on medium surface.
Habitat and distribution: Known from only Malus sp. in China and Russia.
Materials examined: China. Xinjiang Uygur Autonomous Region: Ili Kazak Autonomous Prefecture, 44°16′0.36″N, 80°24′55.53″E, from branches of Malus pumila, 22 July 2017, C.M. Tian & X.L. Fan (CF 20197714), living culture CFCC 53172; Ili Kazak Autonomous Prefecture, 45°56′49.20″N, 82°40′09.56″E, from branches of Malus pumila, 15 July 2017, C.M. Tian & X.L. Fan (CF 20197528), living culture CFCC 53171.
Notes: Cytospora parasitica was introduced by Ariyawansa et al. (2015) relating to canker disease of Malus pumila. Morphologically, our isolates are similar to C. parasitica in having multi-loculate pycnidial conidiomata, producing black area on bark, having smooth-walled, elongate-allantoid, aseptate conidia, whereas the size of our conidia differs from those isolates (5.5–6.0 × 1–2 vs. 6.5–8.0 × 1.3–1.5 μm) (Ariyawansa et al., 2015). Ma et al. (2018) reported this species from the same host plant Malus pumila in Xinjiang of China, which is similar with C. parasitica in the current study.
Cytospora pruni-mume M. Pan & X.L. Fan, sp. nov. (Figure 10)
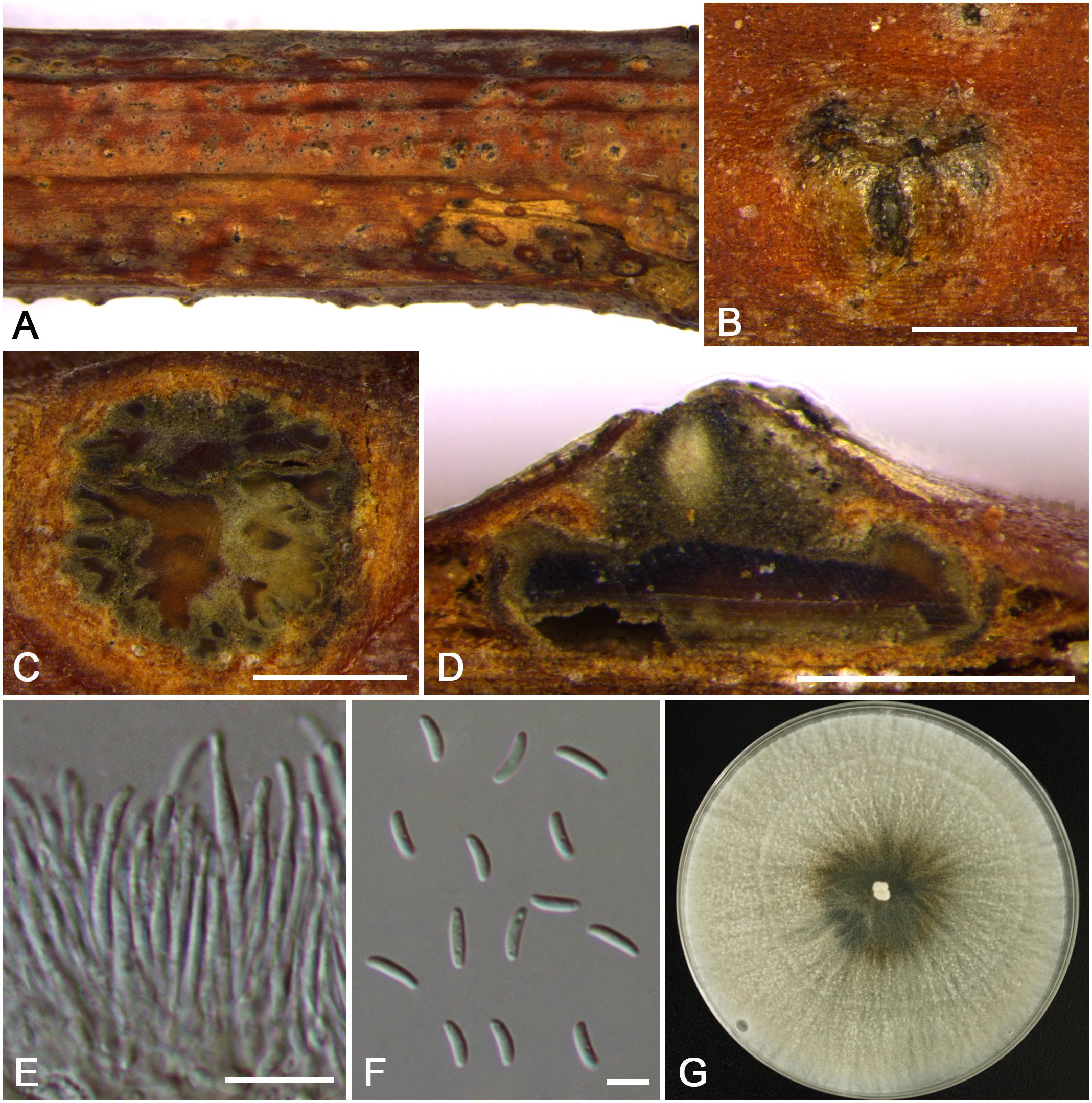
Figure 10. Morphology of Cytospora pruni-mume from Prunus mume (CF 20197515). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F) Conidia. (G) Colonies on PDA for 2 weeks. Bars: (B–D) = 500 μm; (E) = 10 μm; (F) = 5 μm.
MycoBank MB 834851
Typification: China. Xinjiang Uygur Autonomous Region: Ili Kazak Autonomous Prefecture, 46°20′55.51″N, 83°53′55.96″E, from branches of Prunus mume, 14 July 2017, C.M. Tian & X.L. Fan (holotype CF 20197515), ex-type living culture CFCC 53180.
Etymology: Named after the host genus on which it was collected, Prunus mume.
Descriptions: Symptoms appeared as circular, raised, dehiscent lesions on the twigs or branches, with slightly discolored bark above the infected cambium. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, erumpent, discoid, with multiple locules, 870–1,050 μm ( = 920 μm, n = 30) in diam. Conceptacle absent. Ectostromatic disc brown to dark brown, nearly hemispherical, disc dark brown to black, 510–780 μm ( = 630 μm, n = 30) in diam. Ostiole inconspicuous, circular to ovoid, gray to black at the same level as the disc surface. Locule multiple and complex, irregular distribution, subdivided by invaginations with common walls. Conidiophores hyaline, branched at base or occasionally not branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, eguttulate, aseptate, smooth-walled, elongate-allantoid, (5.0–)5.5–6.5(–7.0) × 1.5–2 μm ( = 5.8 × 1.7 μm, n = 50). Sexual morph: not observed.
Culture characteristics: Cultures on PDA are initially white, growing fast and entirely covering the 9-cm Petri dish after 3 days, becoming pale yellow after 7 days. The colonies are flat with a uniform texture, conidiomata were randomly distributed on medium surface.
Habitat and distribution: Known on Prunus mume and Prunus armeniaca from the type locality.
Additional material examined: China. Xinjiang Uygur Autonomous Region: Ili Kazak Autonomous Prefecture, 46°20′54.76″N, 83°53′58.14″E, from branches of Prunus armeniaca, 14 July 2017, C.M. Tian & X.L. Fan (CF 20197512), living culture CFCC 53179.
Notes: Cytospora pruni-mume is associated with canker disease of Prunus mume. The molecular phylogenies show a position clearly distinct from all other strains included in this study (Figure 3). Therefore, we describe this species as a new species.
Cytospora rosicola M. Pan & X.L. Fan, sp. nov. (Figure 11)
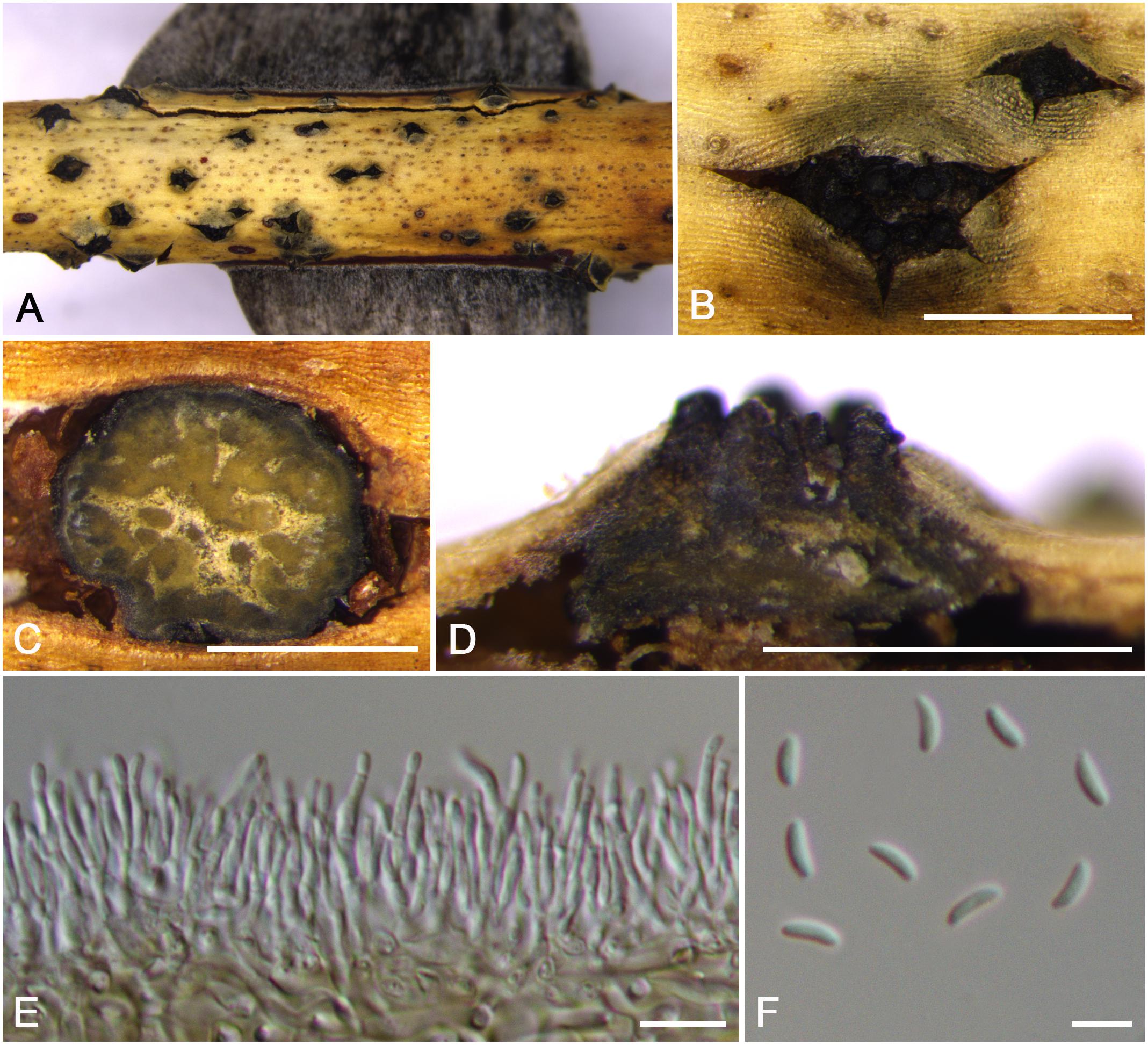
Figure 11. Morphology of Cytospora rosicola from Rosa sp. (CF 20197024). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F) Conidia. Bars: (B–D) = 500 μm; (E) = 10 μm; (F) = 5 μm.
MycoBank MB 834853
Typification: China. The Tibet Autonomous Region: Nyingchi City, Nyingchi County, Nishi Village, 29°45′38.17″N, 94°16′16.71″E, from branches of Rosa sp., 23 July 2016, C.M. Tian & X.L. Fan (holotype CF 20197024).
Etymology: Named after the host genus on which it was collected, Rosa sp.
Descriptions: Symptoms appeared as circular to ovoid, gray or dark brown, raised, dehiscent lesions on the twigs or branches, with discolored bark above the infected cambium. Sometimes lesions may split along the canker margin. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, solitary, scattered, breaking through the outer branch. Locules multiple, circular to ovoid, arranged vesicularly with common walls, 620–710 μm ( = 660 μm, n = 30) in diam. Conceptacle conspicuous. Ectostromatic disc brown to black, circular, disc dark brown, 380–660 μm ( = 520 μm, n = 30) in diam. Ostiole conspicuous, circular to ovoid, dark brown to black at the same level as the disc surface, 280–350 μm ( = 290 μm, n = 30) in diam. Conidiophores hyaline, branched at base or occasionally not branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, allantoid, eguttulate, aseptate, thin-walled, (4.0–)4.5–5.0(–5.5) × 1–2 μm ( = 4.8 × 1.6 μm, n = 50). Sexual morph: not observed.
Habitat and distribution: Known only on Rosa sp. from the type locality.
Notes: Cytospora species associated with Rosa sp. were reported in previous studies such as C. cincta and C. sacculus (Zambettakis and Dzagania, 1986; Fotouhifar et al., 2010). In this study, Cytospora rosicola was also associated with canker disease of Rosa sp. in Tibet. Cytospora rosicola can be distinguished from C. cincta by having smaller and wider conidia (4.5–5.0 × 1–2 vs. 4.5–6.7 × 0.9–1.2), the conspicuous conceptacle, and flask-shaped conidiomata (Mehrabi et al., 2011). Furthermore, this species differs from C. xinjiangensis by conidia size (4.5–5.0 × 1–2 vs. 4.0–4.5 × 1–1.5 μm). Phylogenetically, we treat this species as new, which formed a separate branch. This species needs to be re-collected from Rosa sp. in Tibet of China, as presently no living culture is available.
Cytospora sorbina M. Pan & X.L. Fan, sp. nov. (Figure 12)
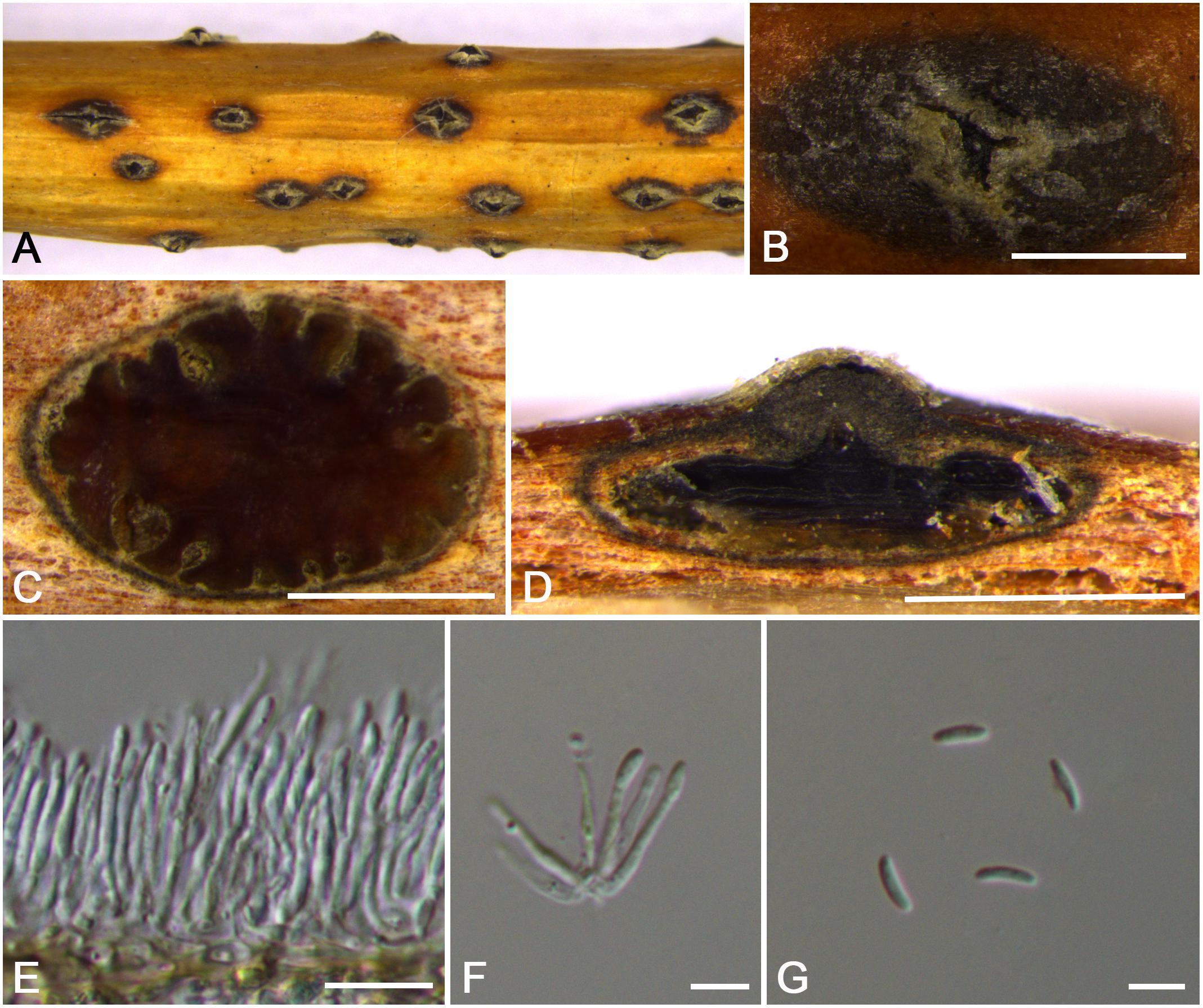
Figure 12. Morphology of Cytospora sorbina from Sorbus tianschanica (CF 20197660). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F,G) Conidia. Bars: (B–D) = 500 μm; (E) = 10 μm; (F,G) = 5 μm.
MycoBank MB 834854
Typification: China. Xinjiang Uygur Autonomous Region: Bole Mongol Autonomous Prefecture, 44°46′13.44′′N, 81°13′58.72′′E, from branches of Sorbus tianschanica, 20 July 2017, C.M. Tian & X.L. Fan (holotype CF 20197660).
Etymology: Named after the host genus on which it was collected, Sorbus tianschanica.
Descriptions: Symptoms appeared as elongate and ovoid, orange or dark brown, raised, dehiscent lesions on the twigs or branches, with slightly discolored bark above the infected cambium. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, erumpent, discoid to conical, with multiple locules, 740–1,120 μm ( = 910 μm, n = 30) in diam. Conceptacle dark. Ectostromatic disc yellow to orange, circular to ovoid, disc gray white to dark brown, 610–1,050 μm ( = 940 μm, n = 30) in diam. Ostiole conspicuous, circular, dark brown to black, 125–280 μm ( = 170 μm, n = 30) in diam. Locules numerous, irregular arrangement with common walls. Conidiophores hyaline, unbranched at base or occasionally branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, smooth-walled, elongate-allantoid, aseptate, (4.0–)4.5–5.5 × 1–1.5 μm ( = 5.0 × 1.3 μm, n = 50). Sexual morph: not observed.
Habitat and distribution: Known only on Sorbus tianschanica from the type locality.
Notes: Cytospora species associated with Sorbus sp. were reported in previous studies such as C. ampolliformis, C. leucostoma, C. populinopsis, C. sorbi, and C. sorbicola (Norphanphoun et al., 2017; Fan et al., 2020). In this study, C. sorbina and C. olivacea are also reported from Sorbus sp. Morphologically, C. sorbina can be distinguished from C. olivacea by the common walls of its locules, as well as the larger conidia (4.5–5.5 × 1–1.5 vs. 4.0–5.0 × 1–1.5 μm). Based on phylogenetic analyses, this species forms separate lineages within the genus Cytospora and sister clade to C. ochreae and C. japonica (Supplementary Figure S1 and Figure 3).
Cytospora tibetensis M. Pan & X.L. Fan, sp. nov. (Figure 13)
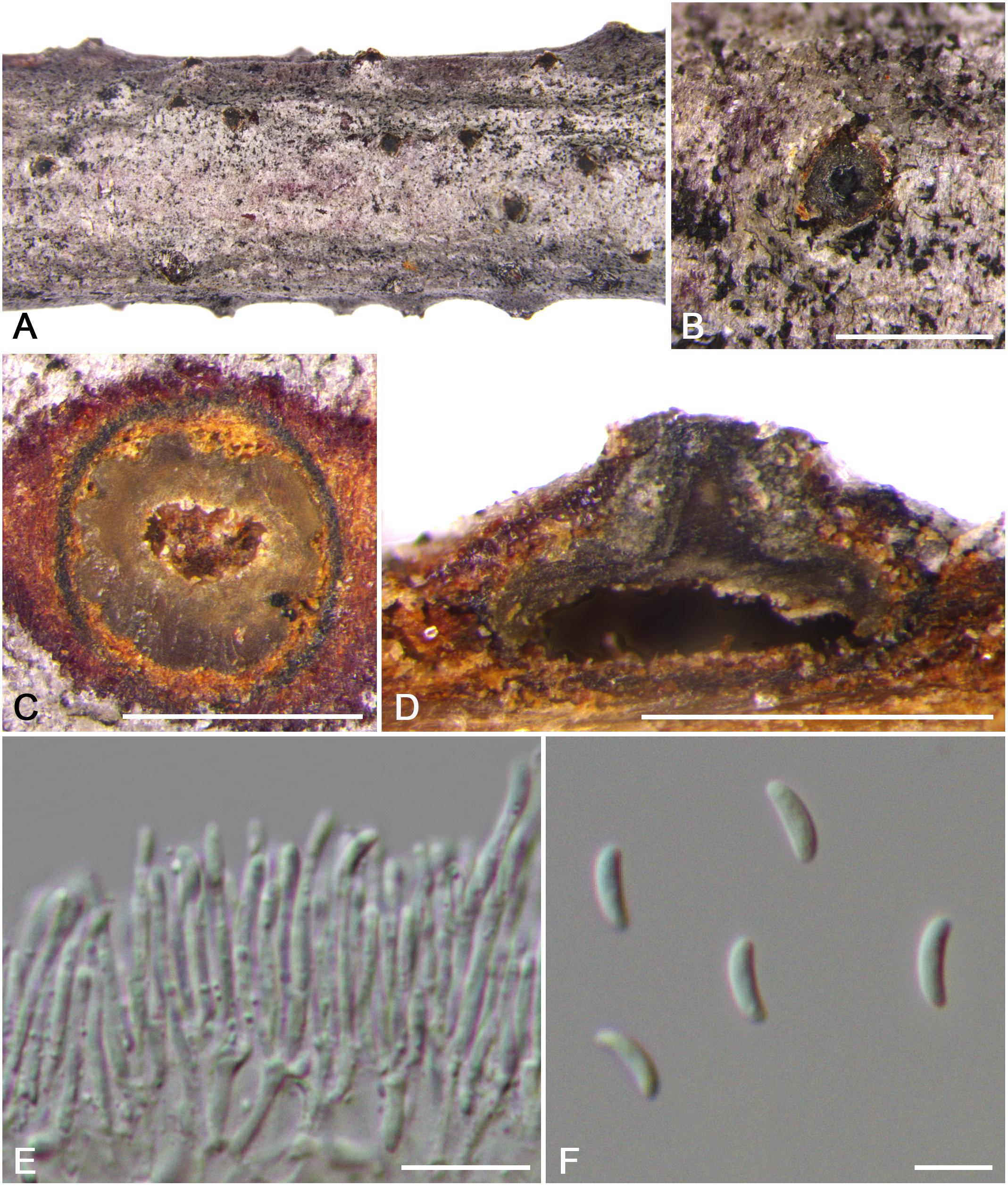
Figure 13. Morphology of Cytospora tibetensis from Cotoneaster sp. (CF 20197032). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F) Conidia. Bars: (B–D) = 500 μm; (E) = 10 μm; (F) = 5 μm.
MycoBank MB 834855
Typification: China. The Tibet Autonomous Region: Nyingchi City, Chayu County, Pineapple Village, 28°56′47.57″N, 97°25′31.93″E, from branches of Cotoneaster sp., 30 July 2016, C.M. Tian & X.L. Fan (holotype CF 20197032).
Etymology: Named after the geographical origin of the type strain, Tibet.
Descriptions: Symptoms appeared as circular and ovoid, dark brown or dark, raised, dehiscent lesions on the twigs or branches. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, erumpent, discoid to conical, 585–650 μm ( = 610 μm, n = 30) in diam. Conceptacle conspicuous. Ectostromatic disc dark brown to gray, circular to ovoid, disc yellow to brown, 270–320 μm ( = 300 μm, n = 30) in diam. Ostiole conspicuous, one ostiole per disc, circular to ovoid, dark brown at the same level as the disc surface, 90–120 μm ( = 105 μm, n = 30) in diam. Locule undivided, circular to ovoid. Conidiophores hyaline, branched at base or occasionally not branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, eguttulate, elongate-allantoid, aseptate, 5.0–5.5(–6.0) × 1.5–2 μm ( = 5.3 × 1.6 μm, n = 50). Sexual morph: not observed.
Habitat and distribution: Known only on Cotoneaster sp. from the type locality.
Additional materials examined: China. The Tibet Autonomous Region: Nyingchi City, Chayu County, Pineapple Village, 28°57′12.91″N, 97°25′20.26″E, from branches of Cotoneaster sp., 30 July 2016, C.M. Tian & X.L. Fan (CF 20197026); Nyingchi City, Chayu County, Pineapple Village, 28°56′47.46″N, 97°25′32.02″E, from branches of Cotoneaster sp., 30 July 2016, C.M. Tian & X.L. Fan (CF 20197029).
Notes: Cytospora tibetensis and C. cotoneastricola were founded on Cotoneaster sp., with the character of black conceptacle and single ostiole. However, C. tibetensis (5.0–5.5 × 1.5–2 μm) differs from C. cotoneastricola (13.0–14.0 × 2.5–3 μm) in having undivided locule with smaller conidia. Phylogenetic analyses based of combined six sequences data indicates that these species form two single lineages, separate from each other with high bootstrap support (Supplementary Figure S1 and Figure 3). Cytospora tibetensis is thus here considered as a novel species. This species needs to be re-collected from Cotoneaster sp. in Tibet of China, as presently no living culture is available.
Cytospora xinjiangensis M. Pan & X.L. Fan, sp. nov. (Figure 14)
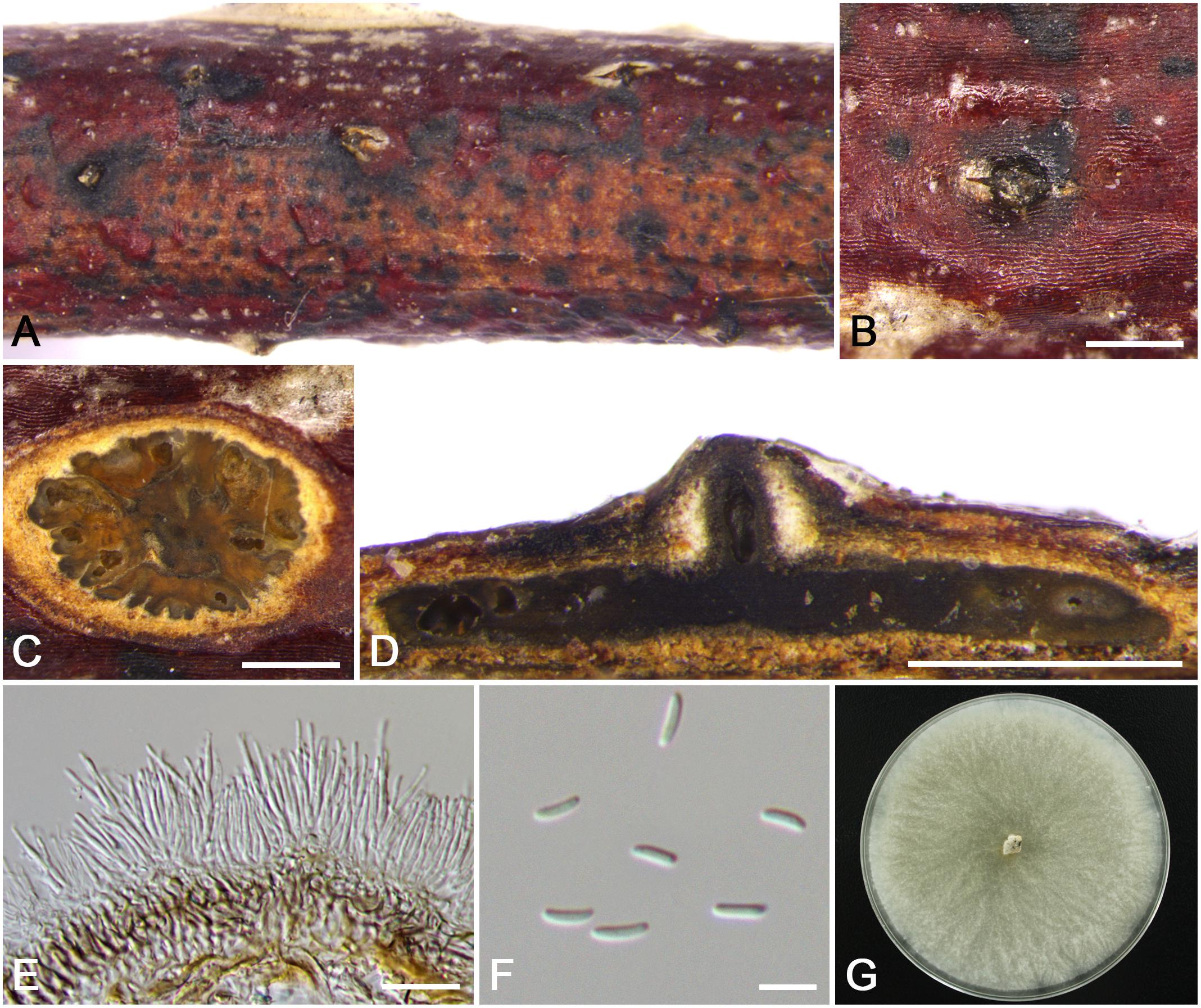
Figure 14. Morphology of Cytospora xinjiangensis from Rosa sp. (CF 20197520). (A,B) Habit of conidiomata on twig. (C) Transverse section of conidioma. (D) Longitudinal section through conidioma. (E) Conidiophores and conidiogenous cells. (F) Conidia. (G) Colonies on PDA for 2 weeks. Bars: (B–D) = 500 μm; (E) = 10 μm; (F) = 5 μm.
MycoBank MB 834852
Typification: China. Xinjiang Uygur Autonomous Region: Ili Kazak Autonomous Prefecture, 45°98′54.00″N, 82°67′01.51″E, from branches of Rosa sp., 15 July 2017, C.M. Tian & X.L. Fan (holotype CF 20197520), ex-type living culture CFCC 53183.
Etymology: Named after the geographical origin of the type strain, Xinjiang.
Descriptions: Symptoms appeared as elongate or circular, dark, slightly sunken, dehiscent lesions on the twigs or branches, with discolored bark above the infected cambium. Asexual morph: Conidiomata pycnidial, immersed in bark, erumpent through the surface of bark when mature, erumpent, discoid, with large multiple locules, 965–1,410 μm ( = 1,145 μm, n = 30) in diam. Conceptacle absent. Ectostromatic disc dark brown to black, circular to ovoid, disc dark brown, 340–610 μm ( = 480 μm, n = 30) in diam. Ostiole unconspicuous, circular to ovoid, gray to black at the same level as the disc surface. Locule multiple and complex, subdivided by invaginations with common walls. Conidiophores hyaline, branched at base or occasionally not branched. Conidiogenous cells enteroblastic, phialidic, sub-cylindrical to cylindrical. Conidia hyaline, eguttulate, elongate-allantoid, aseptate, (3.5–)4.0–4.5(–5.0) × 1–1.5 ( = 4.3 × 1.3 μm, n = 50). Sexual morph: not observed.
Culture characteristics: Cultures on PDA are initially white, growing up to 7.5 cm after 3 days, becoming pale yellow after 14 days, while white are main. Colonies are flat with a uniform texture and becoming helcoid after 30 days. Sterile.
Habitat and distribution: Known only on Rosa sp. from the type locality and an additional locality in Xinjiang Uygur Autonomous Region.
Additional material examined: China. Xinjiang Uygur Autonomous Region: Bole Mongol Autonomous Prefecture, 44°48′47.29′′N, 81°06′22.07′′E, from branches of Rosa sp., 20 July 2017, C.M. Tian & X.L. Fan (CF 20197643), living culture CFCC 53182.
Notes: Cytospora xinjiangensis is associated with canker disease of Rosa sp., which has the same host with C. leucostoma and C. rosicola. Morphologically, C. xinjiangensis has unconspicuous ostiole with small conidia (4.0–4.5 × 1–1.5 μm), compared to the larger conidia of C. leucostoma (5.4–6.4 × 1.2–1.4 μm) (Norphanphoun et al., 2017) and wider conidia of C. rosicola (4.5–5.0 × 1–2 μm). The multigene phylogenetic analyses supported this species as a new species with high support values (MP/ML/BI = 100/99/1) (Figure 3).
Other Species Reported From China
Cytospora ceratosperma (Tode) G.C. Adams & Rossman, IMA Fungus, 6: 147, 2015.
Basionym: Sphaeria ceratosperma Tode, Fung. Mecklenb. Sel. (Lüneburg)2: 53. 1791.
Synonyms: Sphaeria sacculus Schwein., Schr. Naturf. Ges. Leipzig 1: 26. 1822.
Valsa ceratosperma (Tode) Maire, Publ. Inst. Bot. 3(4): 20. 1937.
Cytospora sacculus (Schwein.) Gvrit., Mikol. Fitopatol. 3: 207. 1969.
Notes: Cytospora ceratosperma has been reported in China from twigs and branches of Malus pumila and Pyrus sp. by Wang et al. (2007), which is regarded as a distinct species with multi-ostioles per disc, independent locule walls and grayish to yellow-brown and brownish gray colonies (Adams et al., 2005).
Cytospora japonica (Miyabe & Hemmi) X.L. Fan, Persoonia, 45: 1–45, 2020.
Basionym: Valsa japonica Miyabe & Hemmi, J. Coll. Agric., Imp. Univ. Sapporo 7(4): 296, 1917.
Notes: Cytospora japonica has been reported in China from twigs and branches of Prunus cerasifera by Fan et al. (2020). This species was introduced as a common pathogen in Rosaceae host (Tai, 1979). It is characterized by discoid to conoid conidiomata with hyaline, allantoid, aseptate conidia (6.5–8.5 × 1.5–2 μm) as well as numerous locules, which arranged circularly or irregularly with common walls (Fan et al., 2020).
Cytospora leucosperma (Pers.) Fr., Syst. Mycol. 2: 543, 1823.
Basionym: Naemaspora leucosperma Pers., Observ. Mycol. 1: 81, 1796.
Synonyms: Sphaeria ambiens Pers., Syn. Meth. Fung. 1: 44, 1801.
Valsa ambiens (Pers.) Fr., Summa Veg. Scand., Sectio Post. (Stockholm): 412, 1849.
Cytospora ambiens Sacc., Michelia 1(5): 519, 1879.
Notes: Cytospora leucosperma was chiefly isolated and recorded from Pyrus spp. in China (Teng, 1963; Tai, 1979; Zhuang, 2005). Infected branches collected from Tilia were regarded as the neotype (Urban, 1957; Spielman, 1985), but no living culture and DNA sequence data are available at present. C. leucosperma is similar to C. mali from Malus spp., leading to confusion in both morphology and molecular data (Wang et al., 2011).
Cytospora mali Grove, British Stem- and Leaf-Fungi (Coelomycetes) (Cambridge) 1: 279, 1935.
Synonyms: Valsa mali Miyabe & G. Yamada, M. Miura Agr. Exp. Stn Bull. 4: 17, 1915.
Notes: Cytospora mali was chiefly isolated and discovered from apple (Teng, 1963; Tai, 1979; Wei, 1979; Zhuang, 2005; Wang et al., 2011). It has a similar morphology and close position in phylogeny with C. leucosperma (Fan et al., 2020). Cytospora mali can be distinguished from C. leucosperma by smaller conidiophores (7.5–15 × 1.5 vs. 17–25 × 2–2.5 μm). Furthermore, C. leucosperma is mostly isolated from Pyrus spp. (Fan et al., 2020).
Cytospora populinopsis X.L. Fan & C.M. Tian, Persoonia, 45: 1–45, 2020.
Notes: Cytospora populinopsis was described by Fan et al. (2020) associated with canker disease of Prunus salicina and Sorbus aucuparia in China. It is characterized by having asci with four ascospores, which was similar with C. populina regarded as the pathogen for poplar canker (Fan et al., 2015b). Cytospora populinopsis differs from C. populina based on larger ascospores (13–21 × 2.5–5 vs. 12–13 × 3–4 μm) (Fan et al., 2015b, 2020).
Cytospora rhodophila Sacc., Syll. Fung. (Abellini) 3: 253, 1884.
Notes: Cytospora rhodophila was recorded from Rosa sp. in China (Teng, 1963; Zhuang, 2005).
Cytospora schulzeri Sacc. & P. Syd., Syll. Fung. (Abellini) 14(2): 918, 1899.
Synonyms: Cytospora capitata Schulzer & Sacc., Hedwigia 23: 109, 1884, non.
Cytospora capitata Fuckel, Reisen nach dem Nordpolarmeer 2: 34, 1874.
Valsa malicola Z. Urb., Èeská Mykol. 10: 209, 1956.
Notes: Cytospora schulzeri infected apple trees in China (Teng, 1963; Tai, 1979; Wei, 1979; Zhuang, 2005; Wang et al., 2011). This species differs from C. mali, which was also chiefly isolated and discovered from apple by having numerous ostioles and larger conidia (4.5–6.5 × 1–1.5 vs. 4–5 × 1 μm). In addition, C. schulzeri is distinguished from C. parasitica which is a common species associated with apple by its smaller conidia (4.5–6.5 × 1–1.5 vs. 6.5–8.0 × 1.3–1.5 μm) (Ariyawansa et al., 2015).
Cytospora sibiraeae C.M. Tian et al., Fungal Diversity, 72: 44, 2015.
Notes: Cytospora sibiraeae is regarded as the pathogen responsible for the canker disease of Sibiraea angustata in China, which was reported by Liu et al. (2015). Pathogenic fungi in Sibiraea sp. are rarely reported.
Cytospora spiraeae Fan, Phytotaxa, 338: 57, 2018.
Notes: Cytospora spiraeae was isolated from infected branches or twigs of Spiraea salicifolia. This species is similar with C. schulzeri, which has numerous ostioles and locules with common walls, but it can be distinguished by having smaller locules with a cenetr column compared with C. schulzeri (950–1,100 vs. 1,400–1,500 μm) (Zhu et al., 2018).
Cytospora tamaricicola X.L. Fan & C.M. Tian, Persoonia, 45: 1–45, 2020.
Notes: Cytospora tamaricicola was described by Fan et al. (2020) associated with canker disease of Rosa multiflora and Tamarix chinensis in China. It is characterized by flask shaped to spherical perithecia with biseriate, elongate-allantoid, hyaline, aseptate ascospores (9–11.5 × 2–2.5 μm), pycnidia with multiple locules and thin-walled conidia (5.5–6 × 1–1.5 μm) (Fan et al., 2020).
Key to Cytospora Species on Prunus spp. in China.
1 Asexual morph present…………………………………………………….. 2
1 Asexual morph absent………………………………… C. populinopsis
2 Pycnidium without conceptacle……………………………………….. 3
2 Pycnidium with conceptacle…………………………………………….. 4
3 Locules with the common walls………………….. C. pruni-mume
3 Locules with the independent walls………………………………….. 5
4 Locules with the common walls…………………………. C. japonica
4 Locules with the independent walls………………………………….. 6
5 Size of conidia less than 6 μm…………………….. C. cinnamomea
5 Size of conidia more than 6 μm……………………….. C. erumpens
6 Sexual morph absent…………………………………………. C. olivacea
6 Sexual morph present…………………………………… C. leucostoma
Key to Cytospora species on Malus spp. in China
1 Sexual morph absent………………………………………………………… 2
1 Sexual and asexual morph present……………………………………. 3
2 Size of conidia less than 7 μm…………………………. C. parasitica
2 Size of conidia more than 7 μm……………… C. mali-spectabilis
3 Pycnidial stromata with single ostiole………………………………. 4
3 Pycnidial stromata with numerous ostioles………. C. schulzeri
4 Locules with the common walls………………………………. C. mali
4 Locules with the independent walls…………….. C. ceratosperma
Discussion
In this study, we accepted 23 species of Cytospora from infected plants of Rosaceae in China, including 10 new species (Cytospora cinnamomea, C. cotoneastricola, C. ochracea, C. olivacea, C. pruni-mume, C. rosicola, C. mali-spectabilis, C. sorbina, C. tibetensis, and C. xinjiangensis), and 13 known taxa (Cytospora ceratosperma, C. erumpens, C. japonica, C. leucosperma, C. leucostoma, C. mali, C. parasitica, C. populinopsis, C. rhodophila, C. schulzeri, C. sibiraeae, C. spiraeae, and C. tamaricicola). The current study revealed the attempt to clarify the taxonomy of Cytospora species and extensive host distribution of Rosaceae in China.
The plants of Rosaceae are important ecological and economic tree species in China. However, the current study indicates that the incidence of Cytospora species is serious and have different symptoms in various hosts, including 20 host species of nine genera in Rosaceae, i.e., Cotoneaster, Crataegus, Malus, Prunus, Pyrus, Rosa, Sibiraea, Sorbus, and Spiraea. The result coincides with previous reports that widely extended Cytospora species have been identified to occur in many host species (Adams et al., 2005; Fan et al., 2014a, 2015a, b; Ariyawansa et al., 2015; Liu et al., 2015; Maharachchikumbura et al., 2015, 2016; Hyde et al., 2016; Li et al., 2016) in the current study. Six Cytospora species recovered from diverse Prunus species in California was reported by Lawrence et al. (2018), which are Cytospora amygdali, C. californica, C. eucalypti, C. longispora, C. plurivora, and C. sorbicola. The current results also supplement seven different Cytospora species afflicted Prunus host plants in China. The comparison shows that the species occurrence may be related by geographical and environmental factors, rather than the taxa actually being host specific. Cytospora species and accumulation of DNA dataset are required to expand our understanding of their host range and distribution. Furthermore, Cytospora cotoneastricola, C. ochracea, and C. tibetensis were all collected from Cotoneaster sp., which indicates that the same host could be infected by more than one species. Stevens (1919) summarized the symptoms and species of Rosaceae infected by Cytospora, whereas these reports lacked molecular data. Only a few relative taxonomic studies of Cytospora canker or dieback disease from the plants of Rosaceae were reported, such as Cytospora chrysosperma, C. cincta, C. leucostoma, and C. schulzeri (Mehrabi et al., 2011). Moreover, the host specificity and pathogenicity of many Cytospora species are poorly known. In the current study, Cytospora leucostoma is a common species associated with stem canker diseases of woody plants of Rosaceae, mainly Prunoideae host plants, and C. mali, C. parastica, and C. schulzeri are the common species collected from apple trees.
In China, Cytospora species from cankered apple and pear bark were examined and compared with morphology and ITS sequence data (Wang et al., 2007, 2011). The species identity of the pathogen of Valsa (now Cytospora) canker on pear tree was determined through a combined study of ITS sequence data and cultural characteristics of isolates from apple trees and pear trees in China (Zhang et al., 2007). Ma et al. (2018) clarified and illustrated C. parasitica from the Malus sp. using the ITS, LSU, and tef1-α regions. Fan et al. (2020) summarized 52 species of Cytospora and recommended the dataset of ITS, LSU act, rpb2, tef1-α, and tub2 gene regions. At present, China is a hot place to study these taxa as many species of Cytospora are isolated from important hosts such as Rosaceae. Thus, further studies are required to discover the species of Cytospora in China.
Data Availability Statement
The datasets generated for this study can be found in the MK672943–MK672956, MK672958–MK672985, MK672987–MK673009, MK673011–MK673039, MK673041–MK673069, MK673071–MK673099, and MK673101– MK673111.
Author Contributions
All authors contributed extensively to the work presented in the manuscript. XF and CT conceived and designed the experiments. MP and HZ performed the experiments. MP, HZ, and GB analyzed the data. GB polished the language. MP wrote the manuscript. XF revised and approved the final version of the manuscript.
Funding
This study was financed by the Fundamental Research Funds for the Central Universities (2019ZY23), the National Natural Science Foundation of China (31670647), and College Student Research and Career-Creation Program of Beijing (S201910022007).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00690/full#supplementary-material
Footnotes
References
Adams, G. C., Roux, J., and Wingfield, M. J. (2006). Cytospora species (Ascomycota, Diaporthales, Valsaceae): introduced and native pathogens of trees in South Africa. Australas. Plant Pathol. 35, 521–548. doi: 10.1071/AP06058
Adams, G. C., Roux, J., Wingfield, M. J., and Common, R. (2005). Phylogenetic relationships and morphology of Cytospora species and related teleomorphs (Ascomycota, Diaporthales, Valsaceae) from Eucalyptus. Stud. Mycol. 52, 1–149.
Adams, G. C., Surve-Iyer, R. S., and Iezzoni, A. (2002). Ribosomal DNA sequence divergence and group I introns within Leucostoma species, L. cinctum, L. persoonii and L. parapersoonii sp. nov., ascomycetes that cause Cytospora canker of fruit trees. Mycologia 94, 947–967. doi: 10.1080/15572536.2003.11833153
Adams, G. C., and Taylor, J. W. (1993). Phylogenetic utility of the internal transcribed spacer of nuclear ribosomal DNA in Leucostoma and Valsa. Inoculum 44:1.
Ariyawansa, H. A., Hyde, K. D., Jayasiri, S. C., Buyck, B., Chethana, K. W. T., Dai, D. Q., et al. (2015). Fungal diversity notes 111–252: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 75, 27–274. doi: 10.1007/s13225-015-0346-5
Carbone, I., and Kohn, L. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.2307/3761358
Crous, P. W., Gams, W., Stalpers, J. A., Robert, V., and Stegehuis, G. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Stud. Mycol. 50, 19–22.
Fan, X. L., Bezerra, J. D. P., Tian, C. M., and Crous, P. W. (2020). Cytospora (Diaporthales) in China. Persoonia 45, 1–45. doi: 10.3767/persoonia.2020.45.01
Fan, X. L., Hyde, K. D., Liu, M., Liang, Y. M., and Tian, C. M. (2015a). Cytospora species associated with walnut canker disease in China, with description of a new species C. gigalocus. Fungal Biol. 119, 310–319. doi: 10.1016/j.funbio.2014.12.011
Fan, X. L., Hyde, K. D., Yang, Q., Liang, Y. M., Ma, R., and Tian, C. M. (2015b). Cytospora species associated with canker disease of three anti-desertification plants in northwestern China. Phytotaxa 197, 227–244. doi: 10.11646/phytotaxa.197.4.1
Fan, X. L., Liang, Y. M., Ma, R., and Tian, C. M. (2014a). Morphological and phylogenetic studies of Cytospora (Valsaceae, Diaporthales) isolates from Chinese scholar tree, with description of a new species. Mycoscience 55, 252–259. doi: 10.1016/j.myc.2013.10.001
Fan, X. L., Tian, C. M., Yang, Q., Liang, Y. M., You, C. J., and Zhang, Y. B. (2014b). Cytospora from Salix in northern China. Mycotaxon 129, 303–315. doi: 10.5248/129.303
Fotouhifar, K. B., Hedjaroude, G. A., and Leuchtmann, A. (2010). ITS rDNA phylogeny of Iranian strains of Cytospora and associated teleomorphs. Mycologia 102, 1369–1382. doi: 10.2307/27920441
Glass, N. L., and Donaldson, G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995
Gvritishvili, M. N. (1973). New species of fungi belonging to genus Cytospora Fr. II. Mikol. Fitopatol. 7, 544–549.
Harrington, T. C., and Rizzo, D. M. (1999). “Structure and dynamics of fungal populations,” in Defining Species in the Fungi, ed. J. J. Worrall (Dordrecht: Springer), 43–71.
Hillis, D. M., and Bull, J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analyses. Syst. Biol. 42, 182–192. doi: 10.1093/sysbio/42.2.182
Hyde, K. D., Hongsanan, S., Jeewon, R., Bhat, D. J., McKenzie, E. H. C., Gareth Jones, E. B., et al. (2016). Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 80, 1–270. doi: 10.1007/s13225-016-0373-x
Jiang, N., Yang, Q., Fan, X. L., and Tian, C. M. (2020). Identification of six Cytospora species on Chinese chestnut in China. Mycokeys 62, 1–25. doi: 10.3897/mycokeys.62.47425
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kirk, P. M., Canoon, P. F., Minter, D. W., and Stalpers, J. A. (2008). Ainsworth & Bisby’s Dictionary of the Fungi, 10th Edn. Wallingford: CABI. doi: 10.1079/9780851998268.0000
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Lawrence, D. P., Holland, L. A., Nouri, M. T., Travadon, R., Abramians, A., Michailides, T. J., et al. (2018). Molecular phylogeny of Cytospora species associated with canker diseases of fruit and nut crops in California, with the descriptions of ten new species and one new combination. IMA Fungus 9, 333–370. doi: 10.5598/imafungus.2018.09.02.07
Lawrence, D. P., Travadon, R., Pouzoulet, J., Rolshausen, P. E., Wilcox, W. F., and Baumgartner, K. (2017). Characterization of Cytospora isolates from wood cankers of declining grapevine in North America, with the descriptions of two new Cytospora species. Plant Pathol. 5, 713–725. doi: 10.1111/ppa.12621
Li, G. J., Hyde, K. D., Zhao, R. L., Hongsanan, S., Abdel-Aziz, F. A., Abdel-Wahab, M. A., et al. (2016). Fungal Divers notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 78, 1–237. doi: 10.1007/s13225-016-0366-9
Liu, J. K., Hyde, K. D., Jones, E. B. G., Ariyawansa, H. A., Bhat, D. J., Boonmee, S., et al. (2015). Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 72, 1–197. doi: 10.1007/s13225-015-0324-y
Liu, Y. L., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Ma, R., Liu, Y. M., Yin, Y. X., and Tian, C. M. (2018). A canker disease of apple caused by Cytospora parasitica recorded in China. For. Pathol. 48:e12416. doi: 10.1111/efp.12416
Maharachchikumbura, S. S. N., Hyde, K. D., Jones, E. B. G., McKenzie, E. H. C., Bhat, J. D., Dayarathne, M. C., et al. (2016). Families of Sordariomycetes. Fungal Divers. 72, 1–317. doi: 10.1007/s13225-016-0369-6
Maharachchikumbura, S. S. N., Hyde, K. D., Jones, E. B. G., McKenzie, E. H. C., Huang, S. K., Abdel-Wahab, M. A., et al. (2015). Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 72, 199–301. doi: 10.1007/s13225-015-0331-z
Mehrabi, M., Mohammadi, G. E., and Fotouhifar, K. B. (2011). Studies on Cytospora canker disease of apple trees in Semirom region of Iran. J. Agric. Technol. 7, 967–982.
Norphanphoun, C., Doilom, M., Daranagama, D. A., Phookamsak, R., Wen, T. C., Bulgakov, T. S., et al. (2017). Revisiting the genus Cytospora and allied species. Mycosphere 8, 51–97. doi: 10.5943/mycosphere/8/1/7
Posada, D., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Rambaut, A., and Drummond, A. (2010). FigTree v.1.3.1. Institute of Evolutionary Biology. Edinburgh: University of Edinburgh.
Rannala, B., and Yang, Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rossman, A. Y., Adams, G. C., Cannon, P. F., Castlebury, L. A., Crous, P. W., Gryzenhout, M., et al. (2015). Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 6, 145–154. doi: 10.5598/imafungus.2015.06.01.09
Shang, Q. J., Hyde, K. D., Camporesi, E., Maharachchikumbura, S. S. N., Norphanphoun, C., Brooks, S., et al. (2020). Additions to the genus Cytospora with sexual morph in Cytosporaceae. Mycosphere 11, 189–224. doi: 10.5943/mycosphere/11/1/2
Spielman, L. J. (1985). A monograph of Valsa on hardwoods in North America. Can. J. Bot. 63, 1355–1378. doi: 10.1139/b85-190
Stevens, F. L. (1919). An Apple Canker Due to Cytospora, Vol. 217. Champaign, IL: University of Illinois Agricultural Experiment Station, 367–377.
Swofford, D. L. (2003). PAUP∗: Phylogenetic Analysis Using Parsimony (∗and Other Methods) Version 4.0b10. Sunderland: Sinauer Associates.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Urban, Z. (1957). Vorlaufige mitteilung der ergebnisse einer revision der gattungen Valsa und Valsella. Preslia 29, 394–395.
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wang, X. L., Kang, Z. S., Huang, L. L., and Wei, J. (2011). Re-evaluation of pathogens causing Valsa canker on apple in China. Mycologia 103, 317–324. doi: 10.3852/09-165
Wang, X. L., Kang, Z. S., Huang, L. L., and Yang, P. (2007). Pathogen identification of Valsa canker on pear tree: evidences from rDNA-ITS sequences and cultural characteristics. Mycosystema 26, 517–527.
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Yang, Q., Fan, X. L., Crous, P. W., Liang, Y. M., and Tian, C. M. (2015). Cytospora from Ulmus pumila in northern china. Mycol. Prog. 14, 1–12. doi: 10.1007/s11557-015-1096-1
Zambettakis, C., and Dzagania, A. (1986). Comparaison des caracteres morphologiques entre micromycetes phytopathogenes des regions subtropicales (Georgie) et Mediterraneennes. Bull. Trimest. Soc. Mycol. France 102, 141–154.
Zhang, X. Y., Chen, H. Y., Liang, J., Tian, C. M., and Lu, Q. (2007). Cultural morphology and vegetative compatibility of Cytospora chrysosperma isolates. Bibliogr. Inf. 35, 99–105.
Zhu, H. Y., Pan, M., Bezerra, J. D. P., Tian, C. M., and Fan, X. L. (2020). Discovery of Cytospora species associated with canker disease of tree hosts from Mount Dongling of China. Mycokeys 62, 97–121. doi: 10.3897/mycokeys.62.47854
Zhu, H. Y., Tian, C. M., and Fan, X. L. (2018). Multigene phylogeny and morphology reveal Cytospora spiraeae sp. nov. (Diaporthales, Ascomycota) in China. Phytotaxa 338, 49–62. doi: 10.11646/phytotaxa.338.1.4
Keywords: Cytosporaceae, Diaporthales, phylogeny, taxonomy, 10 new taxa
Citation: Pan M, Zhu H, Bonthond G, Tian C and Fan X (2020) High Diversity of Cytospora Associated With Canker and Dieback of Rosaceae in China, With 10 New Species Described. Front. Plant Sci. 11:690. doi: 10.3389/fpls.2020.00690
Received: 07 March 2020; Accepted: 01 May 2020;
Published: 03 July 2020.
Edited by:
Sajeewa S. N. Maharachchikumbura, University of Electronic Science and Technology of China, ChinaReviewed by:
Chada Norphanphoun, Mae Fah Luang University, ThailandDan Lawrence, University of California, Davis, United States
Copyright © 2020 Pan, Zhu, Bonthond, Tian and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinlei Fan, eGlubGVpZmFuQGJqZnUuZWR1LmNu
 Meng Pan
Meng Pan Haiyan Zhu1,2,3
Haiyan Zhu1,2,3 Guido Bonthond
Guido Bonthond