- 1Southern Regional Collaborative Innovation Center for Grain and Oil Crops, Hunan Agricultural University, Changsha, China
- 2Key Laboratory of Crop Physiology, Ecology and Genetic Breeding, Ministry of Education, College of Agronomy, Jiangxi Agricultural University, Nanchang, China
- 3State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, China
Leaf senescence is one of the most common factors that affects the growth and yield of rice. Although numerous genes affecting leaf senescence have been identified, few involved in cuticular wax synthesis have been described for rice premature leaf senescence. Here, we cloned and characterized Premature Leaf Senescence 4 (PLS4) in rice (Oryza sativa), which encodes a putative 3-oxoacyl-reductase in the fatty acid biosynthetic pathway. Subcellular localization of OsPLS4 was observed in the chloroplast. A single nucleotide substitution in OsPLS4 reduced leaf cuticular wax, and the expression levels of most wax biosynthesis-associated genes were downregulated. TEM showed chloroplast development were defective in the pls4 mutant. Further investigation revealed that the chlorophyll (Chl) content was reduced in the pls4 mutant compared with the WT and that the photosynthesis rate was lower, which caused ROS dramatic accumulation at the heading stage. These results confirmed premature leaf senescence in pls4 plants. Cold treatment indicated that the mutant was more sensitive than the WT was to cold stress. Together, all the above results indicate that the OsPLS4 mutation affects cuticular wax biosynthesis and chloroplast development in rice, causing reduced cuticular wax and premature leaf senescence.
Introduction
Rice (Oryza sativa) is one of the most important cereal crop species worldwide and is consumed by more than half of the global population. Leaf senescence is the final process of leaf development, during which time intracellular organelles and macromolecules are actively destabilized to relocate nutrients into developing tissues or storage organs (Sakuraba et al., 2016). Premature leaf senescence affects rice yields and quality by reducing photosynthetic efficiency and the accumulation of nutrients (Yang et al., 2016; Mao et al., 2017). Hence, understanding the process of premature leaf senescence and the relevant molecular mechanisms could be beneficial for crop breeding.
A large number of senescence-associated genes (SAGs) have been identified, and these genes seem to function in various biological processes, such as the degradation of chloroplasts, the accumulation of reactive oxygen species (ROS), and biotic and abiotic stress responses (Jiao et al., 2012; Liang et al., 2014; Sakuraba et al., 2015; Yang et al., 2016; Leng et al., 2017a). Chlorophyll (Chl) is an essential molecule for the harvest of light energy during photosynthesis. A large number of genes involved in either Chl biosynthesis or degradation have been verified to respond to leaf senescence (Horie et al., 2009; Sato et al., 2009; Yamatani et al., 2013; Yasuhito et al., 2013). Moreover, Chl degradation accelerates leaf yellowing, which is the most notable sign of leaf senescence. Several genes are associated with Chl degradation OsNOL and OsNYC1 (Kusaba et al., 2007; Sato et al., 2009), which encode two short-chain dehydrogenases/reductases that serve as Chl b reductases; OsNYC3 (Morita et al., 2010), which encodes an α/β hydrolase-fold family protein; OsNYC4 (Yamatani et al., 2013), whose product is involved in thylakoid formation and chloroplast precursors; OsPAO, which encodes a pheophorbide oxygenase; and OsRCCR1 (Tang et al., 2011), which encodes a reductase of Chl-like catabolites. Several leaf color-associated mutations have been reported in rice, such as, GATA transcription factor 1 has been shown to regulate chloroplast biogenesis positively (Hudson et al., 2013). The pale green leaf encodes an oxygenase required for the biosynthesis of Chl, which has also been shown to play key roles in rice yields and quality (Yang et al., 2016). Additionally, premature leaf senescence is related with excess ROS overproduction. If ROS accumulation is not reduced within interior plant cells in a timely and effective manner, the cell membranes can disrupted, protein and chloroplast degradation can be promoted, ultimately resulting in early leaf senescence. For example, the LTS1 gene encodes a nicotinate phosphoribosyltransferase (NaPRT1), which is involved in NAD biosynthesis, and a point mutation in OsNaPRT1 leads to hydrogen peroxide (H2O2) accumulation in leaves and triggers premature leaf senescence (Wu et al., 2016). The pls3 mutant presents a reduced Chl content and an increased H2O2 content, the OsMTS1 encodes a methyltransferase, is required for melatonin biosynthesis in rice, and disruption of OsMTS1 can trigger premature leaf senescence (Hong et al., 2018).
Plant cuticular wax serves as an essential barrier for preventing abiotic and biotic stress damage, such as exposure to dust, pathogens and high/low temperature (Aharoni et al., 2004; Monica et al., 2005; Weidenbach et al., 2014). Cuticular wax is mainly composed of mainly very-long-chain fatty acids (VLCFAs; chain length ranging from C20 to C34 carbons) and their derivatives (including aldehydes, alcohols, alkanes, ketones, and esters) (Samuels et al., 2008; Haslam and Kunst, 2013). Cuticular wax biosynthesis is very complex and begins with de novo-synthesized C16 and C18 fatty acids within the plastids. C16 and C18 fatty acids are then elongated to VLCFAs by the fatty acid elongase (FAE) complex, which comprises β-ketoacyl-CoA synthase (KCS), β-ketoacyl-CoA reductase (KCR), β-hydroxy acyl-CoA dehydratase, and enoyl-CoA reductase, in the endoplasmic reticulum (ER). VLCFA elongation involves a four-step reaction cycle. First, the condensation of C16 and C18 acyl-CoA is catalyzed by KCS, yielding β-ketoacyl-CoA; second, the reduction of β-ketoacyl-CoA is catalyzed by KCR; third, the resulting β-hydroxy acyl-CoA is dehydrated by β-hydroxy acyl-CoA dehydratase (HCD); and fourth, the enoyl acyl-CoA is reduced by enoyl-CoA reductase (ECR) (Benning, 2009; Lippold et al., 2012; Li et al., 2016). 3-oxoacyl-(acyl carrier protein[ACP]) reductase (OAR), which catalyzes the first reduction step in each cycle of fatty acid elongation in plants (Slabas et al., 1992; Winter et al., 1997; O’Hara et al., 2007), belongs to a very large family of enzymes, the short-chain alcohol dehydrogenase/reductase (SDR) family, whose members carry out a series of reduction and dehydrogenation reactions via NADH or NADPH (Persson et al., 2003; Wickramasinghe et al., 2006; Kavanagh et al., 2008). The functions of SDR encompass many aspects of primary (lipid synthesis and Chl biosynthesis or degradation) and secondary (terpenoids, steroids, phenolics, and alkaloids) metabolism (Moummou et al., 2012). Several wax biosynthesis genes have previously been identified in rice by the characterization of wax crystal-sparse leaf (wsl) mutants. OsWSL1 encodes a KCS that is involved in the biosynthesis of cuticular waxes on rice leaves (Yu et al., 2008). OsWSL2 encodes a homolog of the Arabidopsis thaliana genes (CER3/WAX2/YRE/FLP1) and the maize gene GL1, which is associated with the elongation of VLCFAs (Mao et al., 2012). OsWSL3 encodes a KCR that affects cuticular wax biosynthesis in rice (Gan et al., 2016). OsWSL4 is predicted to encode a KCS that is homologous to the product of Arabidopsis CER6 (Wang et al., 2017). OsWSL5 is predicted to encode a cytochrome P450 family member CYP96B5, which is involved in the formation of epidermal wax crystals on rice leaf affecting drought sensitivity (Zhang et al., 2020). OsWS1 is involved in wax biosynthesis and is regulated by osa-miR1848 (Xia et al., 2015). Although several wax biosynthesis-related genes have been cloned in rice, few wax related mutants have been reported to be associated with early leaf senescence. In particular, some enzymes related to the wax synthesis pathway regulate plant senescence, which has rarely been studied in rice. However, in Arabidopsis thaliana, two genes (PES1 and PES2) belong to the esterase/lipase/thioesterase family of acyltransferases, which are involved in fatty acid phytyl ester synthesis in chloroplasts, a process involved in maintaining the integrity of the photosynthetic membrane during abiotic stress and senescence (Felix et al., 2012). This study indicated these genes may maintain the integrity of the biological membrane to defense stress and senescence in plants.
In this study, we isolated and characterized a rice premature leaf senescence 4 (pls4) mutant. Using map-based cloning strategy, we found that a single nucleotide substitution in OsPLS4 was responsible for the phenotypic variation, and the OsPLS4 mutation resulted in cuticular wax decrease. OsPLS4 is predicted to encode a 3-oxoacyl-ACP reductase, which participates in the reduction reaction of the first step of fatty acid biosynthesis in chloroplasts. We performed detailed functional analyses to explore the role of OsPLS4 in leaf senescence in rice on the basis genetic, physiological and biochemical approaches. Our results demonstrated that OsPLS4 plays an essential role in cuticular wax synthesis chilling stress in rice.
Materials and Methods
Plant Materials and Growth Conditions
We obtained a premature leaf senescence (pls4) mutant from a mutant population generated by EMS treatment of ZH11 (O. sativa japonica). An F2 mapping population was generated from a cross between the pls4 mutant and rice variety TN1(O. sativa indica). All plants were grown in a paddy field at Jiangxi Agricultural University, Nanchang, Jiangxi Province, China, and at Sanya, Hainan Province, China.
Fine Mapping and Isolation of PLS4
We crossed pls4 with TN1 and used the F2 population for gene mapping. For primary mapping, BSA was performed with DNA pools from 30 F2 individuals with the pls4 phenotype and from 30 F2 individuals with the wild type phenotype. The initial localization was determined via 182 SSR markers from the 12 chromosomes of rice1. Afterward, 985 recessive individuals with the premature leaf senescence(the mutant phenotype) were selected from the F2 population to fine map the PLS4 locus. In the PLS4 primary mapped region, we developed InDel markers on the basis of sequence comparisons between the genomic sequences of Nipponbare and 9311 to narrow down. All primer sequences used for gene mapping are shown in Supplementary Table S2. The products were subsequently separated on 8% polyacrylamide gels.
Functional Complementation of the pls4 Mutant
For the complementation of the pls4 mutant, a 5221-bp genomic DNA fragment that included the PLS4 coding region along with the upstream and downstream sequence were introduced into a pCAMBIA1300 binary vector to generate the transformation construct, yielding pCAMBIA1300-OsPLS4. pCAMBIA1300-OsPLS4 was transformed into the calli generated from the seed embryos of the pls4 mutant by Agrobacterium-mediated transformation. The primers used are listed in Supplementary Table S1.
Scanning and Transmission Electron Microscopy
Tissue sections from the pls4 mutant and the same region in WT were selected at the heading stage for SEM and TEM analysis.
SEM was performed as previously described. The detached leaves were pre-fixed in fixation buffer (2.5% glutaraldehyde-sodium in 100 mM phosphate buffer) at room temperature and post-fixed in 1% OsO4 at 4°C. The samples were dehydrated in a series of ethanol and dried with a critical point dryer. Subsequently, the prepared samples were coated with platinum by a sputtering instrument and examined by SEM (S-4800, Hitachi, Japan) at an accelerating voltage of 10 kV.
For TEM analysis, the pre-fixed tissue segments were processed as previously described, embedded in paraffin wax, mounted, and observed under an H-7650 transmission electron microscope (Hitachi, Japan) (Broun et al., 2004).
Quantitative Real-Time PCR and Promoter-GUS Analysis
Total RNA was extracted from rice tissues using a Takara Plant MiniBEST RNA Extraction Kit (Takara, Japan), and cDNA was synthesized by PrimeScriptTM II reverse transcriptase (Takara, Japan) according to the manufacturer’s instructions. qRT-PCR was performed using a 2 × SYBR Green PCR Master Mix (Applied Biosystems, United States) on a 7500 Real-Time PCR System (Applied Biosystems, United States) in a reaction volume of 20 μL. The OsUBQ (Ubiquitin) gene was used as a control.
For the promoter-GUS assay, the 2040-bp genomic DNA fragment was amplified from PLS4 promoter region upstream of WT genomic DNA via primers (Supplementary Table S2), the resulting fragment was introduced into a pCAMBIA1305-GUS vector upstream of the GUS reporter gene. Rice transformation and histochemical analysis were performed as previously described (Tian et al., 2013).
Subcellular Localization and Promoter Fusions
To produce the subcellular localization of PLS4, the PLS4 coding DNA sequence without a termination codon was amplified and cloned into a subcellular localization pCaMV35S-GFP vector to fuse PLS4 to an enhanced green fluorescent protein (eGFP) (Bottanelli et al., 2012). The control construct (p35S:GFP) and the fusion constructs (p35S:PLS4-GFP) were subsequently transiently expressed into tobacco (Nicotiana benthamiana) epidermal leaf cells as described previously (Batoko et al., 2000). The cells were then examined under a confocal fluorescence microscope (Leica TCS SP5, Germany) after 48 h of incubation. The GFP and Chl fluorescence at 522 and 680 nm was recorded respectively.
To assess the subcellular localization of PLS4, its coding sequences were subcloned into a SpeI- and BamHI-digested pAN580 vector to produce a C-terminal GFP fusion construct driven by the constitutive 35S promoter. The constructs were then cotransformed together with a chloroplast marker fusion construct into rice protoplasts as described previously (Aggarwal et al., 2014). The transformed cells were detected by confocal microscopy (Olympus FV1000 MPE) and images were captured at 488 nm for GFP excitation and at 534 nm for red fluorescent protein (RFP) excitation. The primers used are listed in Supplementary Table S2.
Leaf Cuticular Wax Components Analysis
Cuticular waxes were extracted from mature expanded and the same region blade leaves at the heading stage and measured as described previously (Mao et al., 2012). Briefly, by immersing leaves from the flag at the heading stage into 30 mL of n-hexane at 67°C for 30 s, with 50 μg of n-tetracosane used as an internal standard. The n-hexane was then evaporated under gaseous N2 and the residue was derivatized with 100 μL of bis-N,N-(trimethylsilyl) trifluoroacetamide (BSTFA, Sigma, St. Louis, MO, United States) and 100 μL of pyridine for 60 min at 70°C. All wax samples were analyzed with an Agilent (Santa Clara, CA, United States) 7000C gas chromatography-mass spectrometry (GC-MS) device on a 30 m HP-1MS column. The column was operated with helium as the carrier gas and with splitless injection at 250°C. The oven temperature was increased from 50 to 200°C at 20°C min–1, maintained for 2 min at 200°C, increased at 2°C min–1 to 320°C, and then maintained at 320°C for 14 min. The total amount of cuticular wax was expressed per unit area of the leaf surface. The leaf area was measured with an LI-3000C Portable Area Meter (LI-COR Biosciences, Lincoln, NE, United States). All experiments consisted of five biological replicates.
Measurement of the Chl Content and Photosynthetic Rate
Chl was extracted from the fully expanded leaves of rice plants at four development stages and at the heading stage in two independent complementation lines measured according to previous methods (Porra et al., 1994). Briefly, 0.1 g of leaf tissue was immersed in 25 mL of 95% alcohol for 24 h in darkness. Afterward, the Chl content was measured with a DU800 visible spectrophotometer (Beckman, CA, United States) at 649 and 665 nm.
The photosynthetic rates were measured at the heading stage of WT and pls4 mutant from 10:00–12:00 h on a sunny day by the portable photosynthetic measurement device LI-6400 (LI-COR Biosciences, Lincoln, NE, United States). All experiments consisted of three biological replicates.
Rate of Water Loss and Chl Efflux
The leaf water-loss rate was measured for detached leaves of plants at the heading stage of WT and pls4 mutant. The same parts of the flag leaves were cut into 5cm lengths pieces and placed in a Petri dish. In the dark, the leaves were weighted every 1 h. With respect to Chl leakage, 0.1 g of leaf blades tissue of plants at the heading stage was immersed in 25 mL of 95% alcohol. A series of 100 μL aliquots were collected at 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10 h and subjected to spectrophotometry (absorption measured at 647 and 664 nm) to quantify the amount of Chl that was leached. All experiments consisted of three biological replicates.
Histochemical Analysis, Determination of H2O2 and MDA Contents, and Detection of CAT Activity
The accumulation of the O2– was measured via NBT (0.5 mg mL–1 in 10 mM potassium phosphate buffer, pH 7.6). The staining and bleaching of the leaves were performed as previously described (Yang et al., 2016). The H2O2 and MDA contents and CAT activity were determined by the use of Suzhou Comin Biotechnology kits via spectrophotometry with respect to the H2O2 content, H2O2 and titanium sulfate form a yellow titanium peroxide complex with characteristic absorption at 415 nm (Code: H2O2-1-Y, Comin); with respect to the MDA contents, the MDA is combined with thiobarbituric acid (TBA) to form a red product, which has maximum absorption peak at 532 nm (Code: MDA-2-Y, Comin); and the CAT activity was measured normally (Code: CAT-1-Y, Comin). All experiments consisted of three biological replicates.
Chilling Treatment
To test chilling tolerance, the seedlings of WT and pls4 were treated at 4°C for 72 h. Afterward, they were moved to a temperature-controlled greenhouse that had a 30°C/25°C day/night cycle for recovery for 7 days. After treatment at 4°C, leaves of the seeding were removed every 3 h to measure the expression of genes related to cold tolerance.
Investigation of Agronomic Traits
The plant height and tiller number of different rice lines were measured under field conditions. When the rice plants grew to maturity in the field, their height, tiller number, panicle length, number of grains per panicle, seed setting rate and 1000-grain weight were measured via conventional methods, with five biological replicates per WT and mutant plants.
Results
Characterization of the pls4 Mutant
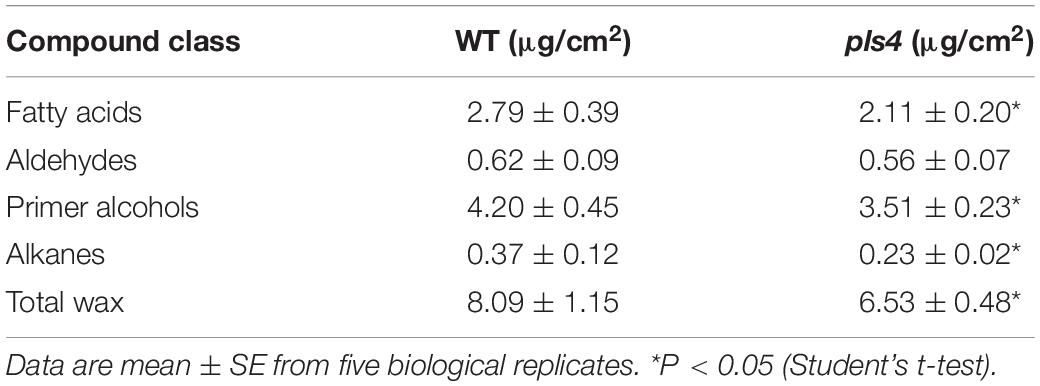
We isolated a premature leaf senescence mutant described as pls4 from a mutant library of the japonica rice variety Zhong Hua 11 (ZH11), which was generated by ethyl methyl sulfonate (EMS) treatment. The pls4 mutant exhibited multiple different phenotypes from wild-type (WT) ZH11. At the tillering stage, compared with those of the WT, the plant height and size of the pls4 mutant were reduced, but there was no obvious difference in leaf color (Figure 1A); compared with that in the WT, the Chl content in the mutant moderately decreased, but the difference was not significant (P > 0.05) (Figure 1D). However, after heading, the leaves color changed from green to yellow and some brown spots were visible (Figures 1B,C), and the Chl content rapidly decreased in pls4 (Figure 1D). Which are typical phenomena associated with leaf senescence.
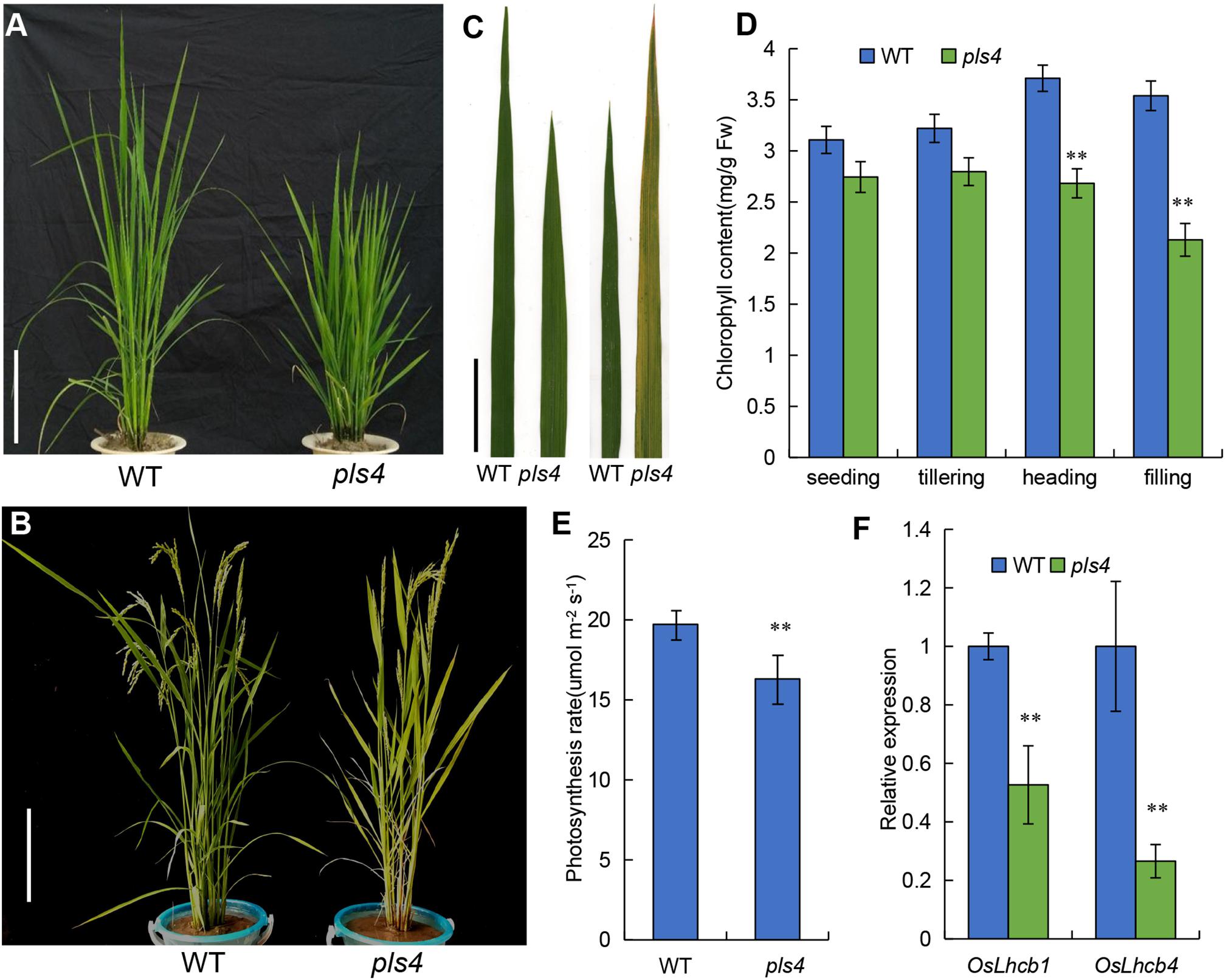
Figure 1. Characteristics of the WT and pls4 mutant. (A,B) Phenotypes of the WT and pls4 mutant at the tillering and heading stages. Bars = 20 cm. (C) The leaves of the WT and pls4 mutant at the tillering and heading stages. Bars = 5 cm. (D) Chl content in the leaves of pls4 and the WT at four growth stages. Fw, fresh weight. (E) Determination of the photosynthesis rate of the WT and pls4 mutant at the heading stage. (F) Expression levels of OsLhcb1 and OsLhcb4; OsLhcb1, light-harvesting Chl a/b-binding protein of photosystem II (PSII) (LOC_Os10g41780); OsLhcb4 (LOC_Os07g37240). The data presented are the means ± SDs of three biological replicates. **P < 0.01 (Student’s t-test).
We found that the photosynthetic rate was significantly lower in the flowering leaf of the pls4 mutant than in the WT (Figure 1E). OsLhcb1 and OsLhcb4 are essential to the photosynthetic system of rice, and their expression was also detected. The qRT-PCR results showed that the expression of both OsLhcb1 and OsLhcb4 was strongly suppressed in the pls4 mutant compared with the WT; the OsLhcb1 and OsLhcb4 expression in the pls4 mutants was only approximately one-half and one-quarter of that in the WT plants, respectively (Figure 1F).
Because photosynthesis and leaf senescence in the late stage strongly impact on yield formation, several agronomic traits, including plant height, tiller number, panicle length, grain number per panicle, seed setting rate and 1000-grain weight, were investigated. The results showed that plant height, tiller number, panicle length and grain number per panicle were moderately affected in the pls4 plants, while the seed setting rate and 1000-grain weight significantly decreased. Both the seed setting rate and 1000-grain weight in the mutant were only approximately 70% of those in the WT (Supplementary Figure S2 and Supplementary Table S3).
Map-Based Cloning of OsPLS4
To identify whether the premature-senescence phenotype of pls4 is caused by a single gene or multiple genes, reciprocal crosses of pls4 with common cultivars (including Nip, TN1 and 93-11) were carried out. The obtained F1 plants from all the crosses displayed a normal phenotype, and none of them showed a Chl degradation-type phenotype (Supplementary Figure S1). Segregation of the traits was observed in all F2 populations, and the ratio of WT plants to plants with a premature leaf senescence phenotype was close to 3:1 according to a χ2 test (Supplementary Table S1). These results indicated that the premature leaf senescence phenotype of pls4 is a qualitative trait that is controlled by a single recessive gene.
To isolate OsPLS4, the F2 population from the cross between the pls4 mutant and TN1 were selected for gene mapping. Bulked segregant analysis (BSA) was used for linkage analysis and primary mapping with 182 simple sequence repeat (SSR) markers. The results revealed that several SSR markers on chromosome 4 were co-segregated with the pls4 phenotype, and the locus was mapped between markers M3 and M4 (Figure 2A). Fine mapping was subsequently performed with InDel markers according to the difference in the primary markers between Nipponbare and 93-11, and the location of the OsPLS4 locus was narrowed to a 52-kb interval between C4 and C7, which included 7 predicted open reading frames (ORFs) (Figure 2A). DNA sequencing analysis of the whole region revealed that a single nucleotide substitution (G-A) was present within the ninth exon of the fourth ORF, LOC_Os04g30760 (Figure 2B). For the remainder of this paper, pls4 will continue to be used as the mutant line name, and OsPLS4 will be used as the gene name. The mutation of LOC_Os04g30760 within pls4 leads to an amino acid change from alanine (Ala) to threonine (Thr) at the 254th amino acid residue (Figure 2B). These results suggested that the mutation of LOC_Os04g30760 was possibly responsible for the abnormal phenotype of pls4.
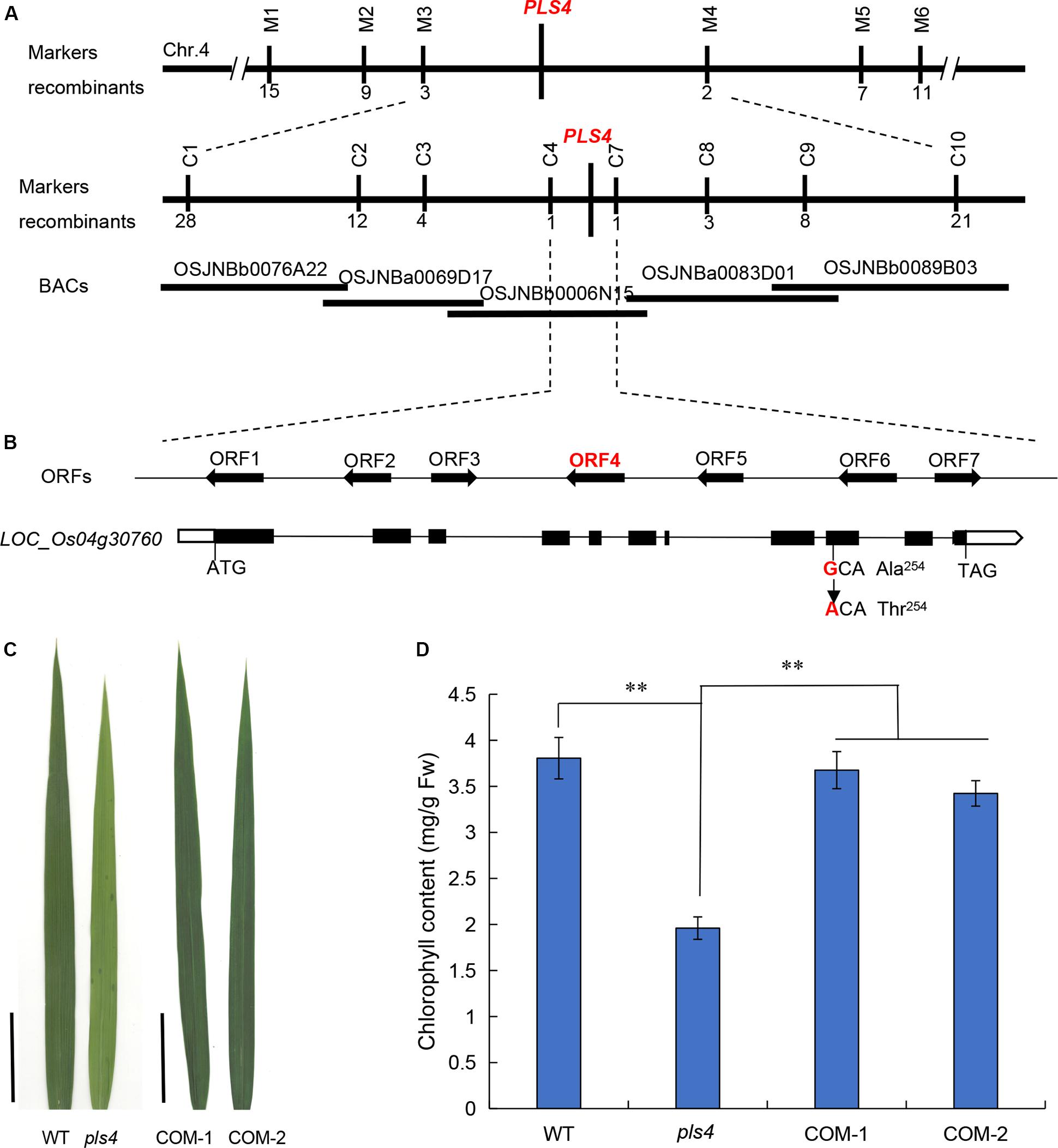
Figure 2. Map-based cloning of the OsPLS4 gene. (A) Fine mapping of PLS4 further localized the mutation to a 52-kb genomic region between C4 and C7 on chromosome 4. The number of recombination events between marker and pls4 phenotype. The target region contains 7 candidate ORFs. (B) Schematic of PLS4. The black rectangles represent exons, and the position of the SNP is indicated in the ninth exon of LOC_Os04g30760; the SNP resulted in the mutation of the encoded protein at position 254 from alanine (Ala) to threonine (Thr). (C) The phenotype of the flag leaves in the transplanted lines, COM-1,-2: two independent complementation plants; Bars = 5 cm. (D) The flag leaf Chl content of the WT, pls4 and two COM independent lines at the heading stage. The data presented are the means ± SDs of three biological replicates. **P < 0.01 (Student’s t-test).
To investigate the linkage between the pls4 phenotype and the mutation of LOC_Os04g30760, we transformed the mutant with the pCAMBIA1300 binary vector (COM), which contained a genomic fragment encompassing the WT gene, including its native promoter and a 957-bp downstream sequence. Twenty-three independent T0 transformants were obtained, and the leaf premature-senescence phenotype was completely restored in the transgenic plants (Figure 2C and Supplementary Figure S3). Moreover, the COM plants were nearly the same as the WT plants in terms of their Chl content and OsPLS4 expression level (Figure 2D and Supplementary Figure S4). In summary, these results indicated that the cloned candidate gene LOC_Os04g30760 was indeed responsible for the pls4 phenotype.
The ORF of OsPLS4 is predicted to encode a protein of 319 amino acids composing 3-oxoacyl-reductase. Compared with homologous proteins from other species (sorghum [Sorghum bicolor], maize [Zea mays], Brachypodium [Brachypodium Beauv], and Arabidopsis), nearly 86.69% of amino acids throughout the protein coned by OsPLS4 are conserved, as revealed by ClustalW alignment (Supplementary Figure S5). Additionally, the mutated amino acid residue within OsPLS4 is truly conserved in monocotyledons (i.e., sorghum, maize and Brachypodium) (Supplementary Figure S5). Molecular phylogenetic analysis was performed with homologous protein sequences of OsPLS4 from various plant species, which revealed that monocotyledons are closely related (Supplementary Figure S6). The results of the BLAST sequence and phylogenetic analyses demonstrate that OsPLS4 is important and conservative in monocotyledonous plants and that the Ala mutation in this study is very important for OsPLS4 function in rice.
Expression Pattern and Subcellular Localization of OsPLS4
To detect the expression pattern of OsPLS4, we used qRT-PCR to analyze its transcriptional level in several tissues, including the roots, culms, leaves, sheaths and panicles, at several stages. The results indicated that OsPLS4 was widely expressed in different rice tissues. OsPLS4 was expressed at the lowest level in the roots and was expressed at the highest level in green tissues. Moreover, even in the leaves, the transcript level of OsPLS4 was different at the different developmental stages. OsPLS4 had the highest expression level at the tillering stage approximately 30 times that in the roots and more than 6 times that in the leaves at other stages (Figure 3A). Analysis of OsPLS4 expression in the tillering leaves of WT and pls4 mutant, the result showed no significant difference (Supplementary Figure S4). The OsPLS4 gene expression pattern was further confirmed by β-glucuronidase (GUS) assays. GUS expression was detected in the roots, leaf blades, sheaths, stems, inflorescences, glumes, and anthers, but not in the stigma papillae, which is consistent with the results of the RT-PCR and qPCR analyses (Figure 3B). Strong GUS signals were detected in the tillering leaves (Figure 3Bb). OsPLS4 was highly expressed in green tissues, indicating that OsPLS4 plays an essential roles at the tillering stage in rice.
Figure 3. Expression patterns and subcellular localization. (A) Expression pattern of OsPLS4. Expression in various organs, including the leaves, roots, culms, leaf sheaths, and panicles at the heading stage in the WT. The data presented are the means ± SDs of three biological replicates. (B) Spatial expression pattern of the OsPLS4 gene in transgenic rice plants harboring the OsPLS4 promoter fused to the GUS gene. (a) germinating seed; (b) spikelet; (c) stem; (d) leaf sheath; (e) tillering leaf; (f) anther. Bars = 1 mm. (C) Subcellular localization of OsPLS4-GFP fusion constructs. The transient expression of OsPLS4-GFP fusion constructs in rice protoplasts and epidermal cells of N. benthamiana leaves was imaged by a confocal microscope. Bars = 5 nm.
The subcellular localization of OsPLS4 was also explored to further study its functions. We fused GFP to the C-terminus of OsPLS4 and then transiently expressed the construct into both rice protoplasts and tobacco leaf epidermal cells. The same result was obtained in both rice and tobacco, and the GFP signal of the fused protein overlapped with that of chloroplast auto-fluorescence. We found that OsPLS4 was located specifically in the chloroplast (Figure 3C). This result indicated that OsPLS4 is involved in fatty acid biosynthesis in the chloroplast.
OsPLS4 Regulates Leaf Cuticular Wax Synthesis
OsPLS4 encodes a putative 3-oxoacyl-reductase, which is a key enzyme involved in fatty acid biosynthesis found in many plant and bacterial species. In plants, fatty acids are a major component of cuticular wax. Therefore, leaf cuticular wax in both WT and pls4 was evaluated. Scanning electron microscopy (SEM) assay revealed that the platelet-like wax crystals were distributed at a high density on the surface of WT leaves (Figures 4A,C), whereas wax crystals were significantly reduced on the surface of the mutant leaves and were sparsely distributed (Figures 4B,D). Moreover, the contents of all wax components were measured with gas chromatography-mass spectrometry (GC-MS). Compared with those of WT leaves, the total wax loads of pls4 leaves were significantly reduced by 19.3% (P < 0.05, Table 1). Further analysis revealed that the contents of the four major components (fatty acids, aldehydes, primer alcohols, and alkanes) were reduced in the mutant; fatty acids, primer alcohols and alkanes were significantly reduced (P < 0.05) by 24.4, 16.4, and 37.8%, respectively (Figure 4H and Table 1).
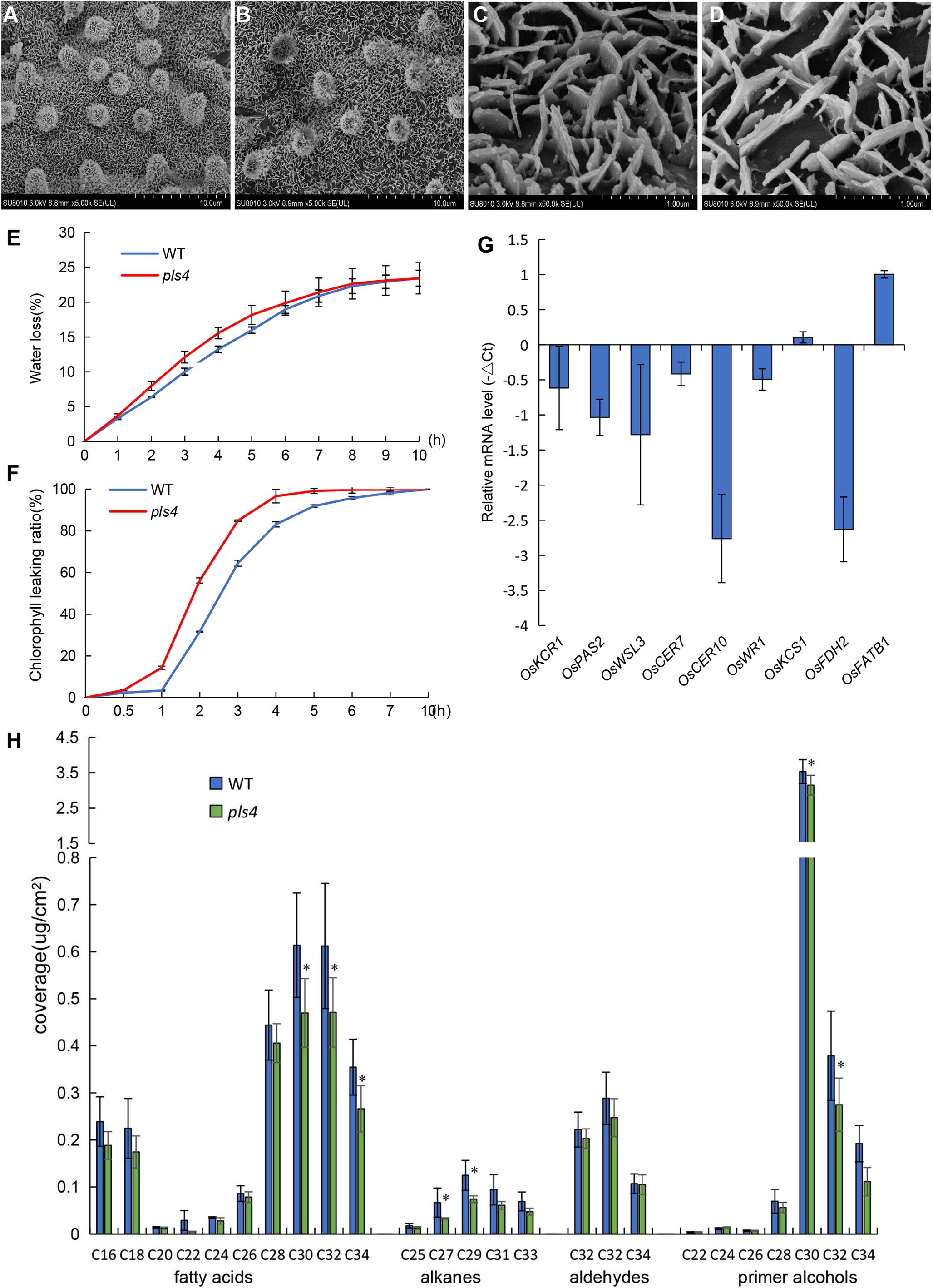
Figure 4. Analysis of cuticular waxes in the leaves of the WT and pls4 mutant. (A–D) Cuticular wax crystals from the leaves of the WT (A,C) and pls4 mutant (B,D) at the heading stage by SEM. Bars = 50 μm (A,B), 5 μm (C,D). (E) Rate of water loss from detached leaves of the WT and pls4 mutant. (F) Chl leakage assays of leaves of the WT and pls4 mutant. (G) Relative expression of a set of genes associated with wax synthesis in the leaves of WT and pls4 mutant at the tillering stage. The data presented are the means ± SDs of three biological replicates. (H) Cuticular wax composition on the leaf surfaces of the WT and pls4 mutant analyzed by GC-MS. Asterisks denote significant differences from the WT. The data presented are the mean ± SD from five biological replicates. *P < 0.05 (Student’s t-test).
Leaf permeability is strongly influenced by the quantity of cuticular wax. Thus, the rate of water loss and Chl leakage were measured in pls4. The water loss rate from detached leaves was faster in the pls4 mutant than in the WT (Figure 4E), and the Chl leakage assay showed that Chl was more readily detected from pls4 leaves than from WT leaves (Figure 4F). These results indicate that the cuticle of pls4 is more permeable than that of the WT due to a scarcity of wax.
To investigate the molecular basis for the sparse crystals on the leaves, we measured the expression levels of wax synthesis-related genes (i.e., OsKCR1, OsPAS2, OsWSL3, OsCER7, OsCER10, OsWR1, OsKCS1, OsFDH2, and OsFATB1). Compared with that in the WT, with the exception of the expression of OsKCS1 and OsFATB1, the expression of these genes in pls4 was downregulated (Figure 4G). The expression of OsCER10 and OsFDH2 was strongly suppressed in pls4, and their expression levels were up to 6 times lower than those in the WT (Figure 4G). OsKCS1 and OsFATB1 encode a β-ketoacyl-CoA synthase and a fatty acyl-ACP thioesterase B, which are both involved in VLCFAs synthesis. As to why the expression of OsKCS1 and OsFATB1 genes upregulated, the regulatory mechanism is still unclear. These results demonstrated that OsPLS4 functions in cuticular wax biosynthesis, not only as an enzyme but also as a regulator of other enzymes involved this process.
Leaf Senescence Is Exacerbated in the pls4 Mutant
The pls4 plants exhibited obvious early leaf senescence at the heading stage (Figures 1B,C). Compared with those in the WT plants, TEM revealed that the number and size of chloroplasts were dramatically reduced in the flag leaves of the pls4 during the heading stage (Figures 5Aa,d). Moreover, in the pls4 mutant, the granum thylakoids (GTKs) were arranged disorderly in the chloroplast, and thylakoid stacking was indistinct (Figures 5Ac,f), while abundant osmium particles also accumulated in the chloroplasts of the pls4 mutant (Figures 5Ab,e).
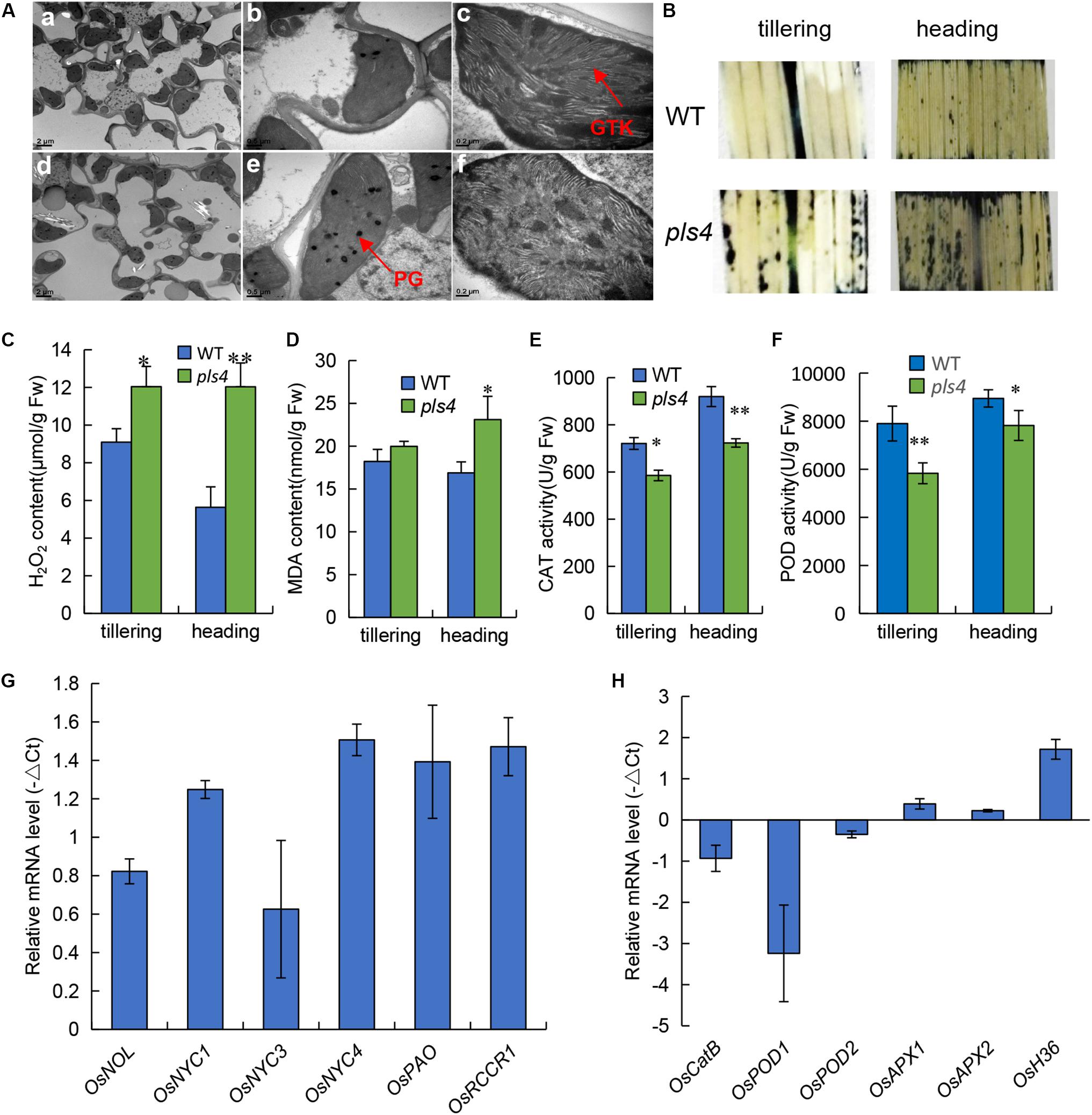
Figure 5. Leaves of pls4 exhibit a severe senescence phenotype under natural conditions. (A) TEM analysis of the flag leaves of the WT and pls4 mutant at the heading stage. GTK, granum thylakoid; PG, plastoglobule. Bars = 2 μm (a,d), 0.5 μm (b,e), 0.2 μm (c,f). (B) Accumulation of O2– radicals in naturally senescence leaves, visualized by staining with NBT. (C–F) H2O2 and MDA contents, CAT and POD activity in the WT and pls4 mutant at the tillering and heading stages. The data presented are the means ± SDs of three biological replicates. *P < 0.05; **P < 0.01 (Student’s t-test). FW, fresh weight. (G) Changes in transcript levels of Chl degradation-associated genes in the leaves of the WT and pls4 mutant at the tillering stage. The genes are as follows: OsNOL (LOC_Os03g45194) and OsNYC1 (LOC_Os01g12710), two short-chain dehydrogenase/reductases, represent Chl b reductases; OsNYC3, which encodes an α/β hydrolase-fold family protein (LOC_Os06g24730); OsNYC4, which encodes thylakoid formation 1, and is involved with chloroplast precursors (LOC_Os07g37250); OsPAO, which encodes a pheophorbide oxygenase (LOC_Os03g05310); and OsRCCR1, which encodes reductase of Chl-like catabolites (LOC_Os10g25030). (H) Relative expression of a set of genes associated with senescence in the WT and the pls4 mutant. The genes include the following: OsCatB, catalase (LOC_Os06g51150); OsPOD1 (LOC_Os01g22370) and OsPOD2 (LOC_Os03g22010), two peroxidases; OsAPX1 (LOC_Os03g17690) and OsAPX2 (LOC_Os07g49400), two ascorbate peroxidases; and OsH36, aminotransferase, senescence-induced protein (LOC_Os05g39770). The data presented are the means ± SDs of three biological replicates.
Leaf variegation and necrotic lesions have been reported to result from ROS accumulation and usually result in cell apoptosis (Jiang et al., 2011). To further confirm leaf senescence by physiological and biochemical detection methods. ROS production in the leaves at the tillering and heading stages was measured via nitro blue tetrazolium (NBT) histochemical analysis. In the WT leaves, ROS were barely detected at both the tillering and heading stages; however, more stained areas appeared in pls4 flag leaves than in WT flag leaves, especially at the heading stage (Figure 5B). These results are in accordance with the different degrees of the phenotypic differences during the two stages. The CAT and POD activity and concentrations of senescence-related substances, such as H2O2 and MDA, were subsequently measured. At both the tillering and heading stages, the concentrations of both H2O2 and MDA were higher in the psl4 leaves than in the WT leaves (Figures 5C,D), but the activity of CAT and POD were lower in pls4 mutant (Figures 5E,F). All of these indicators significantly differed at the heading stage. Therefore, these results suggested that the pls4 mutants exhibits a more notable senescence phenotype than do the WT plants.
To understand the molecular basis for premature leaf senescence in pls4 further, we assayed the expression levels of genes related to Chl degradation (i.e., OsNOL, OsNYC1, OsNYC3, OsNYC4, OsPAO, and OsRCCR1) and ROS scavenging (i.e., OsCatB, OsPOD1, OsPOD2, OsAPX1, and OsAPX2) in both WT and pls4. The expression of all genes related to Chl degradation was upregulated in pls4 (Figure 5G). OsCatB, OsPOD1, OsPOD2, OsAPX1, and OsAPX2 are genes associated with ROS scavenging. Unlike the expression levels of OsAPX1 and OsAPX2, compared with those in the WT plants, the OsCatB, OsPOD1, and OsPOD2 expression levels in the pls4 mutants were downregulated. In particular, the OsPOD1 transcriptional level in the pls4 mutants was approximately 10 times less than that in the WT plants (Figure 5H). OsAPX1 and OsAPX2 are two ascorbate peroxidases genes, APX is a antioxidant enzyme of ROS scavenging in plant, O2– and H2O2 were higher in the pls4 leaves than the WT leaves (Figures 5B,C), which induced the OsAPX gene to be slightly upregulated in pls4 mutant. Additionally, the expression of the common senescence marker OsH36 (Lee et al., 2001), sharply increased in pls4, indicating that more severe senescence occurred in the mutant.
To determine which pathways were influenced in pls4, RNA sequence was performed by the tillering stage leaves, and the differentially expressed genes (DEGs) between pls4 and WT were assayed. In total, we obtained 5867 DEGs, including 3091 upregulated and 2776 downregulated ones in pls4 compared with WT, and the screening conditions were a false discovery rate (FDR) < 0.05 and a |log2FC| > 1. There were 485 DEGs, so pathway annotation was performed (Supplementary Table S5). Interestingly, DEGs between pls4 and WT were mainly enriched in the lipid metabolic and senescence-associated pathway (Supplementary Table S6 and Supplementary Figure S7). These results indicated that the mutation of OsPLS4 may affect wax biosynthesis and premature leaf senescence in the pls4 mutant.
OsPLS4 Is Involved in Chilling Tolerance
It is well known that plant cuticular wax provides an essential barrier for preventing abiotic and biotic damage, and stresses are important factors leading to senescence. In this study, we tested the impact of wax scarcity on the cold response. Three-leaf-stage seedlings of both pls4 and WT were treated at 4°C for 72 h, and the leaves of the mutant became rolled and withered (Figure 6A). The treated seedlings were subsequently incubated at 30°C for 7 days for recovery. Eighty-two percent of the WT seedlings survived, while only a few pls4 seedlings turned green, and most of them withered (Figure 6B). This result indicated that pls4 is more sensitive than WT to cold stress. To explore the molecular mechanism by which OsPLS4 regulates chilling tolerance further, the expression of several key cold tolerance-associated genes was measured every 3 h in treated seedlings. As such, the expression of COLD1, Ctb1 and OsOBF1 was measured. The expression of all three genes was obviously suppressed in pls4, as their expression was hardly detected. After low-temperature treatment, the transcript levels of COLD1, Ctb1 and OsOBF1 peaked at 6 or 9 h after chilling but then decreased in the WT plants (Figures 6D–F). According to the chilling process, the OsPLS4 expression level was similar to that of cold tolerance-associated genes and was higher in the WT than in pls4 (Figure 6C). The results indicated that OsPLS4 responded to low-temperature stress. Furthermore, the mutation of OsPLS4 reduced fatty acid synthesis in the pls4 plants and affect membrane fluidity in plant cells, which is also a key factor in low temperature sensitivity.
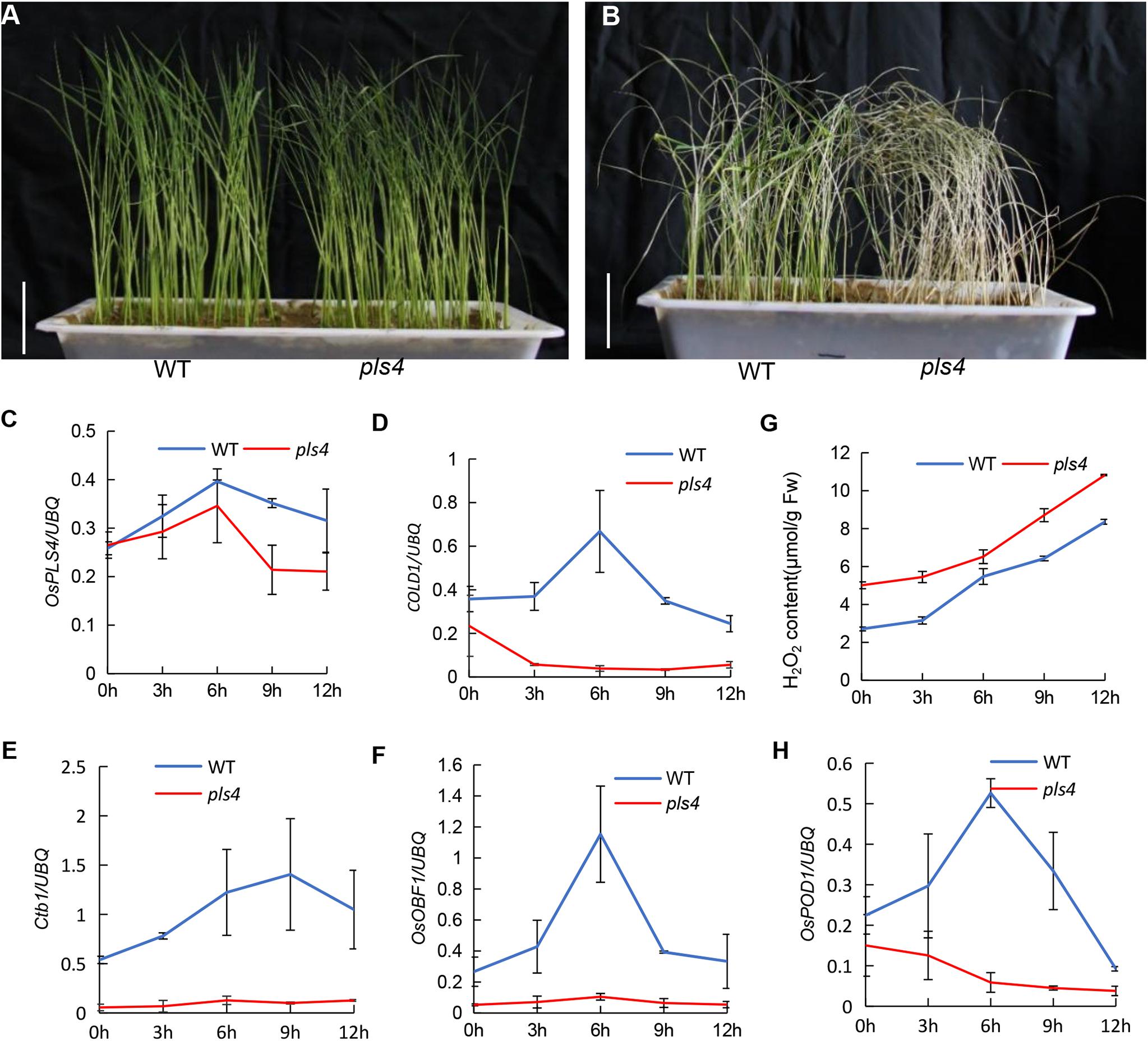
Figure 6. Chilling stress at the seedling stage and related gene expression analysis. (A) The WT and pls4 mutant treated at 4°C for 72 h. Bar = 5 cm. (B) The WT and pls4 mutant treated at 4°C for 72 h, followed by recovery at 30°C/25°C day/night for 7 days. Bar = 5 cm. (C–F) Relative expression of OsPLS4 and genes associated with cold tolerance at different chilling treatment times for the WT and pls4 mutant. The genes included COLD1 (LOC_Os04g51180), Ctb1 (LOC_Os04g52830) and OsOBF (LOC_Os12g37410). (G) H2O2 content in the WT and pls4 mutant at different chilling treatment times. (H) Relative expression of OsPOD1 (peroxidases) in the WT and pls4 mutant at different chilling treatment times. The data presented are the means ± SDs from three biological replicates.
In addition, the more H2O2 accumulated in the pls4 mutant than in the WT in all cold treatments, and as time progressed, the content of H2O2 also increased. Moreover, the expression of OsPOD1, which is essential to scavenging H2O2, increased after the first decrease, and in pls4, this gene expression level was always low (Figures 6G,H). The results again proved that the pls4 plants were more sensitive to chilling stress than the WT plants.
Discussion
To data, we propose that epicuticular wax crystals were significantly reduced on the mutant leaves by one base substitution in OsPLS4. However, the destruction of the cuticle synthesis pathway causes a senescence phenotype at the later growth stage. Leaf yellowing was the most obvious phenotype at the heading stage. This phenotype was confirmed by the disintegration of chloroplasts, reduction in Chl content and photosynthetic rate, the higher accumulation of ROS and MDA and up-regulation of SAGs. In addition, cuticular wax defect caused that the pls4 plants were more sensitive to chilling stress than the WT plants.
Premature leaf senescence severely affects the growth and yield of rice, and leaf cells undergo different metabolic changes during senescence (Hörtensteiner, 2006; Schippers et al., 2015). In the last decade, a series of leaf SAGs have been identified and characterized in rice by using different leaf senescence mutants (Jiao et al., 2012; Liang et al., 2014; Sakuraba et al., 2015; Hong et al., 2018). However, the molecular mechanism controlling leaf senescence remains poorly understood. In this study, we have identified OsPLS4 gene from the pls4 mutant via map-based cloning and a genomic complementation assay. The pls4 mutant differed from previously reported early senescence mutants (Liang et al., 2014; Yang et al., 2016; Leng et al., 2017a). The change of leaves color and reduction of Chl content mainly occurred during the heading stage (Figures 1B,D). TEM revealed that the number and size of chloroplasts were dramatically reduced in yellow leaves of the pls4 during the heading stage, and the granum thylakoids (GTKs) were arranged disorderly in the chloroplast of the pls4 mutant (Figures 5Aa–f). These results indicated that the ultrastructure of chloroplasts was disrupted in the pls4 mutant. In previous studies, SAGs were involved mainly in chloroplast development and degradation, protein synthesis, degradation and transport pathways, the hormone pathway and the programmed cell death pathway among others (Leng et al., 2017b; Hong et al., 2018). However, we isolated and cloned a rice OsPLS4 gene (Figure 2), encoding a putative 3-oxoacyl-ACP reductase, which is a key enzyme involved in fatty acid biosynthesis found in many plant chloroplasts (Joyard et al., 1998; Heredia-Martínez et al., 2018). Moreover, fatty acids are synthesized mainly in thylakoid membranes and are major components of membrane lipids (Joyard et al., 1998; Li et al., 2016). Hence, affected fatty acid synthesis pathways can result in membrane lipids degradation, altering membrane composition, and disrupting membrane fluidity and permeability in chloroplast (Dietz et al., 2016; Li et al., 2016; Zhang et al., 2016). Interesting, our results showed that the OsPLS4 protein was located in the chloroplast, which is consistent with its putative biochemical function (Figure 3C). TEM showed abnormal chloroplast development, with indistinct thylakoid stacking and abundant accumulation of osmium particles in the pls4 mutant (Figure 5A). The Chl content and photosynthetic rate dramatically decreased in pls4 (Figures 1D,E), which both are indicator of premature leaf senescence. Taken together, these results demonstrate that the mutation of OsPLS4 indirectly affected chloroplast normal development and further caused leaf senescence.
Some important changes are observed in premature leaf senescence. For instance, leaf color, degradation of chloroplasts, reduction in Chl content and photosynthesis, accumulation of ROS, reduction of yield-related attributes (Rani et al., 2002; Khanna-chopra, 2012; Huang et al., 2016; Leng et al., 2017b). In addition, cuticle is strongly linked with the release of ROS, Under conditions where the cuticular barrier is broken, ROS are induced (L’Haridon et al., 2011). In our study, ROS accumulation was obvious increased in the pls4 mutant with reduced surface cuticle. NBT staining exhibited that H2O2 production in leaf segments occurred at higher levels in the pls4 mutant than in the WT (Figure 5B), and the concentrations of both H2O2 and MDA increased significantly in the leaves of pls4, whereas the scavenging enzymes CAT and POD activity decreased (Figures 5C–F). These results suggested that the pls4 plants responded actively to the H2O2 and MDA accumulation but the scavenger CAT and POD activity did not increase and execution of senescence phenotype in the pls4 mutant leaves. However, high ROS accumulation cannot been eliminated effectively in plant cell, which damages to thylakoid membranes and other cellular components. Compared with the tillering stage, all these senescence indicators have reached the extremely significant difference at the heading stage (∗∗P < 0.01) (Figures 5B–D), whose high ROS accumulation can explain why the pls4 plants exhibited yellow leaf and Chl content rapidly decreased after heading, but not at the previous stages. According to the expression pattern of OsPLS4, the OsPLS4 expression level was relatively low in the leaves before the tillering stage, and the OsPLS4 expression level was the highest in the leaves at the tillering stage (Figure 3A), which indicated the OsPLS4 gene may play an important role in the tillering stage. Moreover, RNA-seq analysis showed that DEGs were mainly distributed in lipid metabolism and senescence related pathway, which may cause lipid metabolic disorders, altering the cuticle synthesis and leaf senescence of the pls4 mutant. Taken together, these factors may be a reason why the plant height and size of the pls4 mutant reduced compared with WT at the tillering stage and yellow leaf after the flowering. 3-oxoacyl-reductase activity and the molecular regulation mechanism of the OsPLS4 will be further studied to better explain why the mutants exhibit leaf senescence at the heading stage.
The plant cuticular wax biosynthesis process is very complex and begins with fatty acid synthesis in the plastid, followed by assembly in the ER (Kunst and Samuels, 2003; Karki et al., 2019). Several genes have been identified in rice by the characterization of mutants whose leaves are sparsely covered with wax crystals, which are involved in VLCFA elongation of the fatty acid synthesis pathway in the ER, and the subcellular localization of these proteins was mostly in the ER (Yu et al., 2008; Mao et al., 2012; Gan et al., 2016; Wang et al., 2017). In this study, SEM and GC-MS analysis showed leaves surface wax content significant decrease in the pls4 mutant (Figures 4A–D,G and Table 1). However, the subcellular localization of the PLS4 protein was in the chloroplast (Figure 3C), and we speculated that OsPLS4 was involved in the first step of fatty acid synthesis and indirectly affects cuticular wax synthesis. To verify the OsPLS4 function, we determined that the expression levels of wax synthesis-related genes were downregulated in pls4 (Figure 4G), with the exception of OsKCS1 and OsFATB1, which may be due to the OsPLS4 mutation leading to a reduction in intermediate products during the initial stage of fatty acid biosynthesis. In addition, analysis of the water-loss rate and Chl leakage ratio indicated that the leaf surface of the pls4 mutant had less wax than did the leaf surface of the WT (Figures 4E,F). Taken together, these results demonstrate that OsPLS4 is involved in fatty acid synthesis and regulates cuticular wax synthesis in rice.
Plant cuticular wax is the initial protective barrier against the external environment, with a key role in protecting plants from abiotic and biotic damage (Kunst and Samuels, 2003). In plants, the fluidity and stability of membranes is important to sustain the functional activity of membrane proteins and the membranes themselves, which are closely related to temperature; cold-tolerant plants contain a greater abundance of unsaturated fatty acids (Yu et al., 2009; Tovuu et al., 2016; Wang et al., 2019). Moreover, chilling stress mediates a series of physiological and metabolite changes, such as alterations in chlorophyll fluorescence, H2O2, ROS, lipid peroxides, and other metabolites (Wang et al., 2010; Liu et al., 2018). In the present study, we found that the temperature remained low for several days, and leaf senescence was more obvious in the pls4 mutant than in the WT in the field. Moreover, we subjected plants at the seedling stage to low temperature. The results indicated that the pls4 plants were more sensitive to chilling stress than were the WT plants (Figures 6A,B). Furthermore, the H2O2 contents in the pls4 mutant were higher than those in the WT under chilling stress (Figure 6G), which further confirmed this conclusion. The variation trend of OsPLS4 was similar to that of the chilling tolerance-associated genes (Figures 6C–F), confirming that OsPLS4 plays an important role in the clod tolerance of rice. Thus, we demonstrate that OsPLS4 responds to chilling stress by regulating fatty acid synthesis. However, the molecular mechanism controlling chilling stress needs to be further examined with the use of OsPLS4 over-expression plants.
Previous reports showed that decreased photosynthesis leads to a lack of biomass production in plant leaves, which can cause an inadequate supply of nutrients for the rice grain at the filling stage; premature senescent leaves have a substantial influence on grain filling and further affect rice yield. In this study, leaf senescence and low photosynthetic rate of the pls4 mutant affects normal plant growth and the grain-filling rate, eventually resulting in decreased rice grain production and relatively low grain quality (Supplementary Tables S3, S4), which is consistent with the previous results of reports (Yang et al., 2016; Hong et al., 2018). In addition, our results showed that pollen fertility and the seed setting rate obviously decreased in pls4 mutant (Supplementary Figure S2 and Supplementary Table S3). These altered agronomic traits ultimately affected the grain yield. Overall, these results indicated that cuticular wax is not only essential for protecting rice plants from external damage but also necessary for plant growth and development.
Conclusion
A simple mechanism was proposed to explain OsPLS4-mediated cuticular wax biosynthesis and leaf senescence in rice (Figure 7). We confirmed that OsPLS4 was involved in fatty acid synthesis in the chloroplast, and amino acid change of OsPLS4 inhibits fatty acid synthesis to caused chloroplast defective development and weaken photosynthesis, ultimately leading to premature leaf senescence in the pls4 mutant. This senescence mechanism differs from previously reported mechanisms based on genes directly involved in chloroplast synthesis and degradation. Furthermore, decreases in cuticular wax caused the pls4 mutant plants to be more sensitive to chilling stress than the WT plants. Therefore, our study provides insight into new molecular mechanisms involved in premature leaf senescence and theoretical basis for plant stress resistance.
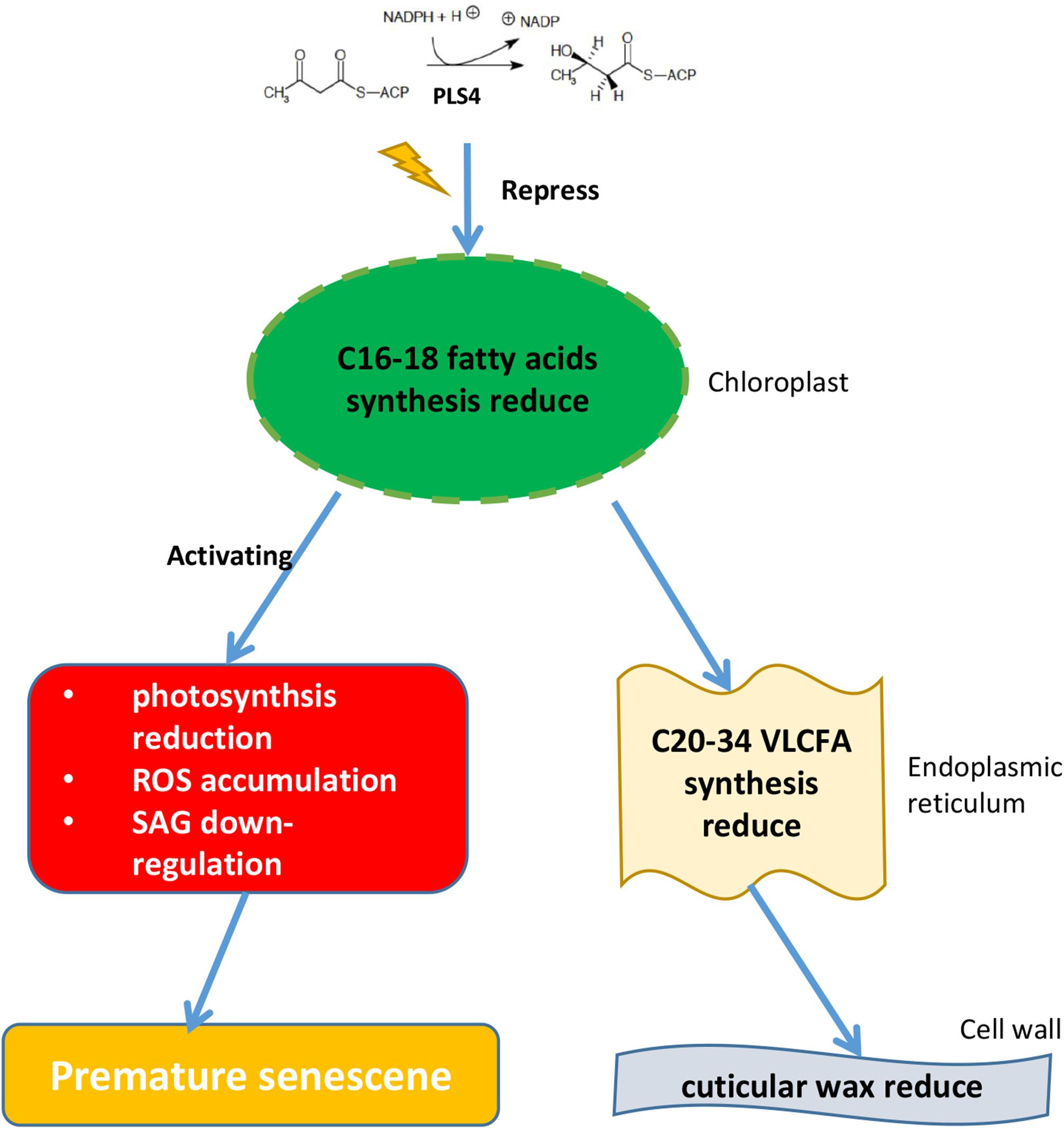
Figure 7. Proposed model for the function of OsPLS4 in cuticular wax formation and leaf senescence. OsPLS4 gene mutation would lead to blocked fatty acid synthesis in chloroplast, which caused the VLCFA synthesis and cuticular wax decrease in pls4 mutant. However, abnormal chloroplast development affected the photosynthetic rate reduction, ROS accumulation and the expression level of the SAGs, ultimately leading to premature leaf senescence in the pls4 mutant.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
HH, WT, and JX conceived and designed the research. TL, YY, LO, and LP carried out the physiological characterization of the mutants and map-based cloning of OsPLS4. DZ, ZQ, JF, and XL performed the plant physiological and chilling treatment experiment. DZ, JX, and ZJ carried out the RNA-seq, confocal microscopy, and bioinformatics data analyses. DZ and JX wrote the manuscript. CZ, XP, and JB supervised the experiments. All authors reviewed the manuscript.
Funding
This study was supported by funding the National Natural Science Foundation of China (Grant Nos. 31960403, 31860373, and 31501286).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the confocal microscopy support provided by Prof. Liu Bo and Yuh-Ru Julie Lee in University of California, Davis, United States.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00782/full#supplementary-material
Footnotes
References
Aggarwal, C., Banas, A. K., Kasprowicz-Maluski, A., Borghetti, C., Labuz, J., Dobrucki, J., et al. (2014). Blue-light-activated phototropin2 trafficking from the cytoplasm to Golgi/post-Golgi vesicles. J. Exp. Bot. 65, 3263–3276. doi: 10.1093/jxb/eru172
Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16, 2463–2480. doi: 10.1105/tpc.104.022897
Batoko, H., Zheng, H. Q., Hawes, C., and Moore, I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12, 2201–2218. doi: 10.1105/tpc.12.11.2201
Benning, C. (2009). Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25, 71–91. doi: 10.1146/annurev.cellbio.042308.113414
Bottanelli, F., Gershlick, D. C., and Denecke, J. (2012). Evidence for sequential action of Rab5 and Rab7 GTPases in prevacuolar organelle partitioning. Traffic 13, 338–354. doi: 10.1111/j.1600-0854.2011.01303.x
Broun, P., Poindexter, P., Osborne, E., Jiang, C. Z., and Riechmann, J. L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 4706–4711. doi: 10.1073/pnas.0305574101
Dietz, K. J., Turkan, I., and Krieger-Liszkay, A. (2016). Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol. 171, 1541–1550. doi: 10.1104/pp.16.00375
Felix, L., Katharina, V. D., Marion, A., Georg, H. L., Vera, W., Jenny Lindberg, Y., et al. (2012). Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. Plant Cell 24, 2001–2014. doi: 10.1105/tpc.112.095588
Gan, L., Wang, X. L., Cheng, Z. J., Liu, L. L., Wang, J. L., Zhang, Z., et al. (2016). Wax crystal-sparse leaf 3 encoding a β-ketoacyl-CoA reductase is involved in cuticular wax biosynthesis in rice. Plant Cell Rep. 35, 1687–1698. doi: 10.1007/s00299-016-1983-1981
Haslam, T. M., and Kunst, L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107. doi: 10.1016/j.plantsci.2013.05.008
Heredia-Martínez, L. G., Andrés-Garrido, A., Martínez-Force, E., Pérez-Pérez, M. E., and Crespo, J. L. (2018). Chloroplast damage induced by the inhibition of fatty acid synthesis triggers autophagy in chlamydomonas. Plant Physiol. 178, 1112–1129. doi: 10.1104/pp.18.00630
Hong, Y., Zhang, Y., Sinumporn, S., Yu, N., Zhan, X., Shen, X., et al. (2018). Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice. Plant J. 95, 877–891. doi: 10.1111/tpj.13995
Horie, Y., Ito, H., Kusaba, M., Tanaka, R., and Tanaka, A. (2009). Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-Protein complexes in Arabidopsis. J. Biol. Chem. 284, 17449–17456. doi: 10.1074/jbc.M109.008912
Hörtensteiner, S. (2006). Chlorophyll degradation during senescence. Annu. Rev. Plant. Biol. 50, 55–77. doi: 10.1146/annurev.arplant.57.032905.105212
Huang, Q. N., Shi, Y. F., Zhang, X. B., Song, L. X., Feng, B. H., Wang, H. M., et al. (2016). Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J. Integr. Plant Biol. 58, 12–28. doi: 10.1111/jipb.12372
Hudson, D., Guevara, D. R., Hand, A. J., Xu, Z., Hao, L., Chen, X., et al. (2013). Rice cytokinin GATA transcription factor1 regulates chloroplast development and plant architecture. Plant Physiol. 162, 132–144. doi: 10.1104/pp.113.217265
Jiang, H., Chen, Y., Li, M., Xu, X., and Wu, G. (2011). Overexpression of SGR results in oxidative stress and lesion-mimic cell death in rice seedlings. J. Integr. Plant Biol. 53, 375–387. doi: 10.1111/j.1744-7909.2011.01037.x
Jiao, B. B., Wang, J. J., Zhu, X. D., Zeng, L. J., Li, Q., and He, Z. H. (2012). A Novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol. Plant. 5, 205–217. doi: 10.1093/mp/ssr081
Joyard, J., Teyssier, E., Miège, C., Berny-Seigneurin, D., Maréchal, E., Block, M. A., et al. (1998). The biochemical machinery of plastid envelope membranes. Plant Physiol. 118, 715–723. doi: 10.1104/pp.118.3.715
Karki, N., Johnson, B. S., and Bates, P. D. (2019). Metabolically distinct pools of phosphatidylcholine are involved in trafficking of fatty acids out of and into the chloroplast for membrane production. Plant Cell 31, 2768–2788.
Kavanagh, K. L., Jornvall, H., Persson, B., and Oppermann, U. (2008). Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol. Life. Sci 65, 3895–3906. doi: 10.1105/tpc.19.00121
Khanna-chopra, R. (2012). Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249, 469–481. doi: 10.1007/s00709-011-0308-z
Kunst, L., and Samuels, A. L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid. Res. 42, 51–80. doi: 10.1016/s0163-7827(02)00045-40
Kusaba, M., Ito, H., Morita, R., Iida, S., Sato, Y., Fujimoto, M., et al. (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex ii and grana degradation during leaf senescence. Plant Cell 19, 1362–1375. doi: 10.1105/tpc.106.042911
Lee, R. H., Wang, C. H., Huang, L. T., and Chen, S. C. (2001). Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 52, 1117–1121. doi: 10.1093/jexbot/52.358.1117
Leng, Y. J., Yang, Y. L., Ren, D. Y., Huang, L. C., Dai, L. P., Wang, Y. Q., et al. (2017a). A rice PECTATE LYASE-like gene is required for plant growth and leaf senescence. Plant Physiol. 174, 1151–1166. doi: 10.1104/pp.16.01625
Leng, Y. J., Ye, G. Y., and Zeng, D. L. (2017b). Genetic dissection of leaf senescence in rice. Int. J. Mol. Sci. 18:2686. doi: 10.3390/ijms18122686
L’Haridon, F., Besson-Bard, A., Binda, M., Serrano, M., Abou-Mansour, E., Balet, F., et al. (2011). A permeable cuticle is associated with the release of reactive oxygen species and induction of innate immunity. PLoS Pathog. 7:e1002148. doi: 10.1371/journal.ppat.1002148
Li, N. N., Xu, C. C., Li-Beisson, Y., and Philippar, K. (2016). Fatty acid and lipid transport in plant cells. Trends Plant Sci. 21, 145–158. doi: 10.1016/j.tplants.2015.10.011
Liang, C. Z., Wang, Y. Q., Zhu, Y. N., Tang, J. Y., Hu, B., Liu, L. C., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Lippold, F., Vom Dorp, K., Abraham, M., Holzl, G., Wewer, V., Yilmaz, J. L., et al. (2012). Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. Plant Cell 24, 2001–2014. doi: 10.1105/tpc.112.095588
Liu, C. T., Wang, W., Mao, B. G., and Chu, C. C. (2018). Cold stress tolerance in rice: physiological changes, molecular mechanism, and future prospects. Yi Chun. 40, 171–185. doi: 10.16288/j.yczz.18-17
Mao, B. G., Cheng, Z. J., Lei, C. L., Xu, F. H., Gao, S. W., Ren, Y. L., et al. (2012). Wax crystal-sparse leaf 2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta 235, 39–52. doi: 10.1007/s00425-011-1481-1481
Mao, C. J., Lu, S. C., Lv, B., Zhang, B., Shen, J. B., He, J. M., et al. (2017). A Rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 174, 1747–1763. doi: 10.1104/pp.17.00542
Monica, S., Hans, H., Elmon, S., Riccardo, V., Francesco, S., and Mario, M. (2005). Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol. 138, 478–489. doi: 10.1104/pp.104.058164
Morita, R., Sato, Y., Masuda, Y., Nishimura, M., and Kusaba, M. (2010). Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 59, 940–952. doi: 10.1111/j.1365-313X.2009.03919.x
Moummou, H., Kallberg, Y., Tonfack, L. B., Persson, B., and van der Rest, B. (2012). The plant short-chain dehydrogenase (SDR) superfamily: genome-wide inventory and diversification patterns. BMC Plant Biol. 12:219. doi: 10.1186/1471-2229-12-219
O’Hara, P., Slabas, A. R., and Fawcett, T. (2007). Antisense expression of 3-oxoacyl-ACP reductase affects whole plant productivity and causes collateral changes in activity of fatty acid synthase components. Plant Cell Physiol. 48, 736–744. doi: 10.1093/pcp/pcm041
Persson, B., Kallberg, Y., Oppermann, U., and Jörnvall, H. (2003). Coenzyme-based functional assignments of short-chain dehydrogenases/reductases (SDRs). Chem. Biol. Interact. 143, 271–278. doi: 10.1016/s0009-2797(02)00223-5
Porra, R. J., Schafer, W., Cmiel, E., Katheder, I., and Scheer, H. (1994). The derivation of the formyl-group oxygen of chlorophyll b in higher plants from molecular oxygen. Eur. J. Biochem. 219, 671–679. doi: 10.1111/j.1432-1033.1994.tb19983.x
Rani, M. H., Liu, Q. E., Yu, N., Zhang, Y. X., Wang, B. F., Cao, Y. R., et al. (2002). ES5 is involved in the regulation of phosphatidylserine synthesis and impacts on early senescence in rice (Oryza sativa L.). Plant Mol Biol. 102, 501–515. doi: 10.1007/s11103-019-00961-964
Sakuraba, Y., Han, S. H., Yang, H. J., Piao, W., and Paek, N. C. (2016). Mutation of rice early flowering3.1 (OsELF3.1) delays leaf senescence in rice. Plant Mol. Biol. 92, 223–234. doi: 10.1007/s11103-016-0507-502
Sakuraba, Y., Piao, W., Lim, J. H., Han, S. H., Kim, Y. S., An, G., et al. (2015). Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 56, 2325–2339. doi: 10.1093/pcp/pcv144
Samuels, L., Kunst, L., and Jetter, R. (2008). Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59, 683–707. doi: 10.1146/annurev.arplant.59.103006.093219
Sato, Y., Morita, R., Katsuma, S., Nishimura, M., Tanaka, A., and Kusaba, M. (2009). Two short-chain dehydrogenase/reductases, NON-Yellow Coloring 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 57, 120–131. doi: 10.1111/j.1365-313X.2008.03670.x
Schippers, J. H. M., Romy, S., Carol, W., and Hai-Chun, J. (2015). Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiol. 169, 914–930. doi: 10.1104/pp.15.00498
Slabas, A. R., Chase, D., Nishida, I., Murata, N., Sidebottom, C., Safford, R., et al. (1992). Molecular cloning of higher-plant 3-oxoacyl-(acyl carrier protein) reductase. Sequence identities with the nodG-gene product of the nitrogen-fixing soil bacterium Rhizobium meliloti. Biochem. J. 283, 321–326. doi: 10.1042/bj2830321
Tang, Y. Y., Li, M. R., Chen, Y., Wu, P. Z., Wu, G. J., and Jiang, H. W. (2011). Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. Plant Physiol. 168, 1952–1959. doi: 10.1016/j.jplph.2011.05.026
Tian, L. H., Dai, L. L., Yin, Z. J., Fukuda, M., Kumamaru, T., Dong, X. B., et al. (2013). Small GTPase Sar1 is crucial for proglutelin and α-globulin export from the endoplasmic reticulum in rice endosperm. J. Exp. Bot. 64, 2831–2845. doi: 10.1093/jxb/ert128
Tovuu, A., Zulfugarov, I. S., Wu, G., Kang, I. S., Kim, C., Moon, B. Y., et al. (2016). Rice mutants deficient in ω-3 fatty acid desaturase (FAD8) fail to acclimate to cold temperatures. Plant Physiol. Biochem. 109, 525–535. doi: 10.1016/j.plaphy.2016.11.001
Wang, H. S., Yu, C., Tang, X. F., Wang, L. Y., Dong, X. C., and Meng, Q. W. (2010). Antisense-mediated depletion of tomato endoplasmic reticulum omega-3 fatty acid desaturase enhances thermal tolerance. J. Integr. Plant Biol. 52, 568–577. doi: 10.1111/j.1744-7909.2010.00957.x
Wang, X., Yu, C., Liu, Y., Yang, L., Li, Y., Yao, W., et al. (2019). GmFAD3A, A ω-3 Fatty acid desaturase gene, enhances cold tolerance and seed germination Rate under low temperature in rice. Int. J. Mol. Sci. 20:3796. doi: 10.3390/ijms20153796
Wang, X. C., Guan, Y. Y., Zhang, D., Dong, X. B., Tian, L. H., and Qu, L. Q. (2017). A β-Ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiol. 173, 944–955. doi: 10.1104/pp.16.01527
Weidenbach, D., Jansen, M., Franke, R. B., Hensel, G., Weissgerber, W., Ulferts, S., et al. (2014). Evolutionary conserved function of barley and Arabidopsis 3-KETOACYL-CoA SYNTHASES in providing wax signals for germination of powdery mildew fungi. Plant Physiol. 166, 1621–1633. doi: 10.1104/pp.114.246348
Wickramasinghe, S. R., Inglis, K. A., Urch, J. E., Muller, S., van Aalten, D. M., and Fairlamb, A. H. (2006). Kinetic, inhibition and structural studies on 3-oxoacyl-ACP reductase from Plasmodium falciparum, a key enzyme in fatty acid biosynthesis. Biochem. J. 393, 447–457. doi: 10.1042/BJ20050832
Winter, E., Brummel, M., Schuch, R., and Spener, F. (1997). Decarboxylation of malonyl-(acyl carrier protein) by 3-oxoacyl-(acyl carrier protein) synthases in plant fatty acid biosynthesis. Biochem. J. 321, 313–318. doi: 10.1042/bj3210313
Wu, L. W., Ren, D. Y., Hu, S. K., Li, G. M., Dong, G. J., Jiang, L., et al. (2016). Mutation of OsNaPRT1 in the NAD salvage pathway leads to withered leaf tips in rice. Plant Physiol. 171:1898. doi: 10.1104/pp.15.01898
Xia, K. F., Ou, X. J., Gao, C. Z., Tang, H. D., Jia, Y. X., Deng, R. F., et al. (2015). OsWS1 involved in cuticular wax biosynthesis is regulated byosa-miR1848. Plant Cell Environ. 38, 2662–2673. doi: 10.1111/pce.12576
Yamatani, H., Sato, Y., Masuda, Y., Kato, Y., Morita, R., Fukunaga, K., et al. (2013). NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll - protein complexes during leaf senescence. Plant J. 74, 652–662. doi: 10.1111/tpj.12154
Yang, Y. L., Xu, J., Huang, L. C., Leng, Y. J., Dai, L. P., Rao, Y. C., et al. (2016). PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J. Exp. Bot. 67, 1297–1310. doi: 10.1093/jxb/erv529
Yasuhito, S., Md Lutfor, R., Sung-Hwan, C., Ye-Sol, K., Hee-Jong, K., Soo-Cheul, Y., et al. (2013). The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J. 74, 122–133. doi: 10.1111/tpj.12110
Yu, C., Wang, H. S., Yang, S., Tang, X. F., Duan, M., and Meng, Q. W. (2009). Overexpression of endoplasmic reticulum omega-3 fatty acid desaturase gene improves chilling tolerance in tomato. Plant Physiol. Bioch. 47, 1102–1112. doi: 10.1016/j.plaphy.2009.07.008
Yu, D., Ranathunge, K., Huang, H., Pei, Z., Franke, R., Schreiber, L., et al. (2008). Wax Crystal-Sparse Leaf1 encodes a β-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 228, 675–685. doi: 10.1007/s00425-008-0770-779
Zhang, D., Yang, H., Wang, X., Qiu, Y., Tian, L., Qi, X., et al. (2020). Cytochrome P450 family member CYP96B5 hydroxylates alkanes to primary alcohols and is involved in rice leaf cuticular wax synthesis. New Phytol. 225, 2094–2107. doi: 10.1111/nph.16267
Keywords: chloroplast, leaf senescence, cuticular wax, fatty acid synthesis, chilling stress, Oryza sativa
Citation: Zhou D, Li T, Yang Y, Qu Z, Ouyang L, Jiang Z, Lin X, Zhu C, Peng L, Fu J, Peng X, Bian J, Tang W, Xu J and He H (2020) OsPLS4 Is Involved in Cuticular Wax Biosynthesis and Affects Leaf Senescence in Rice. Front. Plant Sci. 11:782. doi: 10.3389/fpls.2020.00782
Received: 06 February 2020; Accepted: 18 May 2020;
Published: 11 June 2020.
Edited by:
Andy Pereira, University of Arkansas, United StatesReviewed by:
Lei Wang, Chinese Academy of Sciences, ChinaKurniawan Rudi Trijatmiko, International Rice Research Institute, Philippines
Copyright © 2020 Zhou, Li, Yang, Qu, Ouyang, Jiang, Lin, Zhu, Peng, Fu, Peng, Bian, Tang, Xu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenbang Tang, dGFuZ3dlbmJhbmdAMTYzLmNvbQ==; Jie Xu, eHVqaWUxOTg2MTVAZm94bWFpbC5jb20=; Haohua He, aGhodWE2NEAxNjMuY29t
†These authors have contributed equally to this work
 Dahu Zhou
Dahu Zhou Ting Li
Ting Li Yaolong Yang3
Yaolong Yang3 Jianmin Bian
Jianmin Bian