- 1Molecular Plant Pathology, Swammerdam Institute for Life Sciences (SILS), University of Amsterdam, Amsterdam, Netherlands
- 2Laboratory of Biochemistry, Wageningen University, Wageningen, Netherlands
- 3Keygene N.V., Wageningen, Netherlands
Geminiviruses are plant-infecting DNA viruses that reshape the intracellular environment of their host in order to create favorable conditions for viral replication and propagation. Viral manipulation is largely mediated via interactions between viral and host proteins. Identification of this protein network helps us to understand how these viruses manipulate their host and therefore provides us potentially with novel leads for resistance against this class of pathogens, as genetic variation in the corresponding plant genes could subvert viral manipulation. Different studies have already yielded a list of host proteins that interact with one of the geminiviral proteins. Here, we use affinity purification followed by mass spectrometry (AP-MS) to further expand this list of interacting proteins, focusing on an important host (tomato) and the Replication initiator protein (Rep, AL1, C1) from Tomato yellow leaf curl virus (TYLCV). Rep is the only geminiviral protein proven to be essential for geminiviral replication and it forms an integral part of viral replisomes, a protein complex that consists of plant and viral proteins that allows for viral DNA replication. Using AP-MS, fifty-four ‘high confidence’ tomato proteins were identified that specifically co-purified with Rep. For two of them, an unknown EWS-like RNA-binding protein (called Geminivirus Rep interacting EWS-like protein 1 or GRIEP1) and an isoform of the THO complex subunit 4A (ALY1), we were able to confirm this interaction with Rep in planta using a second method, bimolecular fluorescence complementation (BiFC). The THO subunit 4 is part of the THO/TREX (TRanscription-EXport) complex, which controls RNA splicing and nuclear export of mRNA to the cytoplasm and is also connected to plant disease resistance. This work represents the first step towards characterization of novel host factors with a putative role in the life cycle of TYLCV and possibly other geminiviruses.
Introduction
During the past twenty years, geminiviruses (family Geminiviridae) have emerged as one of the most destructive plant-infecting virus families (Rojas et al., 2005). They infect numerous plant species causing significant yield losses in many economically important crops, both monocots and dicots (Mansoor et al., 2003; Mansoor et al., 2006). Geminiviruses are transmitted between host plants by insect vectors, for example leafhoppers, treehoppers and whiteflies. Based on these vectors and their genome organization, they are currently classified into nine genera (Zerbini et al., 2017). Viral transmission of the largest genus, Begomoviruses (>380 species), is mediated by the whitefly Bemisia tabaci, a highly polyphagous and worldwide spread insect (Zhao et al., 2019). Correspondingly, Begomoviruses have a wide host range, infecting many plant species found in the (sub)tropical regions and in more temperate climate zones (Navas-Castillo et al., 2011). Traditionally, begomovirus infestation was (to some extent effectively) controlled in the field by a combination of (a) insecticide rotation to control whitefly populations and (b) agricultural practices that reduce virus reservoirs in weeds. The success rate of these combined measures varies a lot between crops, cropping systems, and geographical regions (Rojas et al., 2018). To support these aforementioned control measures, they need to be complemented with genetic resistance in crops. So far, few resistance traits against the Tomato yellow leaf curl virus (TYLCV) have been identified and introduced in cultivated crops. Among them were the allelic genes Ty-1 and Ty-3, which encode an RNA-dependent RNA polymerase (Verlaan et al., 2013). Introgression of these genes into tomato cultivars conferred resistance to TYLCV by increasing cytosine DNA methylation of the viral genome, thus acting through viral transcriptional gene silencing (Caro et al., 2015). Another example of a TYLCV resistance locus is Ty-2, which was genetically linked to the gene TYNBS1 that encodes a NB-LRR (Nucleotide-Binding domain and Leucine-Rich Repeat-containing) type plant immune receptor (Yamaguchi et al., 2018). However, following the introduction of the resistance genes Ty-1/3 and Ty2 in tomato cultivars, resistance-breaking strains of TYLCV have emerged in recent years under field conditions, suggesting that these genes do not confer durable resistance against TYLCV (Verlaan et al., 2013; Butterbach et al., 2014; Belabess et al., 2016; Ohnishi et al., 2016). To increase our arsenal of genetic resources for breeding and to help reduce insecticide usage, new genetic strategies are thus needed. One strategy is to screen for resistance based on natural variation in Susceptibility (S) genes (van Schie and Takken, 2014), in this particular case for host genes that are critical for geminiviruses to complete their life cycle in plant cells.
Geminiviruses are typified by their capsid composed of a twinned icosahedral particle that encapsidates the viral genome composed of circular single-stranded DNA (ssDNA) molecule. Within the genus Begomovirus, this genome can either be monopartite (~2.8 kb) encoding six proteins that jointly control viral replication, movement, transmission, and pathogenesis, or bipartite with an ‘A component’ that encodes five or six viral proteins and a ‘B component’ that encodes for two additional proteins (both components being 2.5–2.8 kb in size) (Jeske, 2009). Geminivirus infections start foremost in terminally differentiated cells, i.e. plant cells that are in the G1 phase of the mitotic cell cycle or in the G phase of the endocycle—a variation of the cell cycle characterized by increased ploidy levels and cell expansion without cell division. In order to orchestrate replication of its viral genome, the virus thus needs to reprogram the host cell cycle to promote progression into the S phase of the cell cycle (Hanley-Bowdoin et al., 2004; Hanley-Bowdoin et al., 2013). Only one viral protein, the Replication initiator protein (Rep), is strictly required for viral replication to occur. In fact, Rep is the most conserved geminiviral protein and it is critical for assembly of viral replisomes, a complex of viral and host proteins that forms a DNA replication fork at viral DNA (Rizvi et al., 2015; Ruhel and Chakraborty, 2019). Numerous studies have been attempted to characterize the composition and mode-of-action of this viral replisome. At least one other viral protein, REn (Replication enhancer protein), is known to promote viral replication and as such it is likely also part of viral replisomes (Sunter et al., 1990).
Previous studies support that both Rep and REn interact with host factors in order to create a cellular environment favorable for virus replication, e.g. by up-regulating expression of several host DNA polymerases and DNA replication accessory proteins (Gutierrez, 2000a; Gutierrez, 2000b). One critical component of viral replisomes is the protein PCNA (Proliferating cell nuclear antigen), which acts as a processivity factor of eukaryotic DNA polymerases and forms a DNA clamp at replication forks (Castillo et al., 2003; Morilla et al., 2006). PCNA is a highly conserved protein in eukaryotes that controls the cell cycle, DNA replication and DNA repair pathways (Choe and Moldovan, 2017). Different studies have shown that both Rep and REn from different geminiviruses (TYLCV, Tomato yellow leaf curl Sardinia virus [TYLCSV], Tomato Golden Mosaic Virus [TGMV], Indian mung bean yellow mosaic virus [IMYMV]) interact with PCNA (Castillo et al., 2003; Bagewadi et al., 2004; Settlage et al., 2005) and that Rep from two different Begomoviruses (TYLCV and TGMV) manipulates the SUMOylation status of PCNA—a conserved protein modification of PCNA that directly modulates PCNA function (Arroyo-Mateos et al., 2018). Other host factors that are recruited by members of the Rep protein family to viral replisomes include among others the Replication factor C, RFC (Luque et al., 2002), Replication protein A32, RPA32 (Singh et al., 2007), the recombination and DNA repair related proteins Rad51 and Rad54 (Kaliappan et al., 2012; Suyal et al., 2013a) and the Minichromosome maintenance protein 2, MCM2 (Suyal et al., 2013b). Moreover, Rep also reprograms the host cell cycle by binding transcriptional regulators of the cell cycle like the Retinoblastoma-related protein (RBR) (Xie et al., 1996; Kong et al., 2000) and several members of the NAC family of transcription factors (GRAB1, GRAB2, NAC083) (Suyal et al., 2014) and by interacting with proteins involved in post-translational modification mechanisms, such as ubiquitination and sumoylation (Castillo et al., 2004; Sánchez-Durán et al., 2011; Kushwaha et al., 2017). Besides, Rep from TGMV was also shown to suppress expression of certain viral genes by binding to the viral DNA (Eagle et al., 1994; Shung and Sunter, 2007). Finally, Rep from different geminiviruses was shown to suppress methylation‐mediated transcriptional gene silencing (TGS) by reducing the expression of the responsible DNA methyltransferases (MET1 and CMT3) in the plant species Nicotiana benthamiana and Arabidopsis thaliana (Rodríguez-Negrete et al., 2013).
Clearly, the geminivirus-plant interaction is intrinsically complex involving a multitude of cellular pathways (Hanley-Bowdoin et al., 2013). To better understand how geminiviruses redirect these pathways, we need to characterize in more detail the protein network of the viral proteins with host proteins. A detailed characterization of this network will aid in the identification of key steps needed for viral replication and allows exploitation of this knowledge to attain durable and broad resistance to these viruses e.g. via mutagenesis or natural variation of the underlying genes to ultimately disrupt the viral life cycle. Affinity-based purification followed by mass spectrometry-based protein identification (AP-MS) forms a well-suited technique to characterize the composition of these protein complexes under native conditions (Gavin et al., 2011; Dunham et al., 2012). In the context of plant–virus interactions, AP-MS has been used for example to define the interactome of the NIa protein from the potyvirus Tobacco etch virus (Martínez et al., 2016) and to characterize the global landscape of interactions between TYLCV and one of its host plants, N. benthamiana (Wang et al., 2017a).
Here we expand this list of potential host-interacting proteins by using an AP-MS approach to compose a list of ‘high-confidence’ tomato proteins that co-purify with TYLCV Rep in combination with tomato PCNA. For a small subset of these tomato proteins, (i) an EWS (Ewing Sarcoma protein)-like RNA-binding protein (hereafter named Geminivirus Rep-interacting EWS-like protein 1 [GRIEP1]), and (ii) the THO/TREX (TRanscription-EXport) subunit 4A (ALY1), we confirmed that they interact with Rep and PCNA in planta using an independent method, i.e. the bimolecular fluorescence complementation (BiFC). The THO/TREX complex is involved in mRNA splicing and subsequent export of mRNAs to the cytoplasm and several lines of evidence indicate that nucleocytosolic mRNA transport contributes to the regulation of plant immunity (Ehrnsberger et al., 2019). Moreover, ALY proteins have been shown to play an important role during viral infections (Xu et al., 2020). This work thus extends our inventory of Rep interactions and represents a first step towards characterization of these host factors.
Materials and Methods
Tomato Protoplast Isolation and Transfection
Protoplasts were isolated from in vitro shoot cultures of tomato as described (Shahin, 1985; Tan et al., 1987). A total of 4 × 107 protoplasts per sample were subjected to Polyethylene-glycol (PEG4000) mediated transformation (Negrutiu et al., 1987) with the following plasmids: (i) pK7FWG2 (Karimi et al., 2002) containing Rep from TYLCV (Genbank ID: FJ956702.1) fused to enhanced GFP (EGFP) (referred to as Rep-GFP), (ii) Rep-GFP + pJL-TRBO (Lindbo, 2007) containing tomato (Sl)PCNA fused to the FLAG tag at its N-terminus, referred to as FLAG-PCNA (positive control for interaction), and (iii) FLAG-PCNA as a negative control. Transfected protoplasts were incubated in Gamborg B5 (Duchefa Biochemie) liquid medium at 25°C overnight in dark conditions. Three independent protoplasts isolations and DNA transfections were performed on three different days. The next day the protoplasts were collected in 1.5 ml reaction vials by centrifugation (5 min at 85 g) and frozen at −80°C till further usage.
Protoplasts Protein Extraction and Immunoprecipitation
Protoplasts were defrosted and resuspended in 1 ml per 107 protoplasts of Triton X-buffer (20 mM Tris–HCl pH 7.5, 10 mM KCl, 10% glycerol, 1 mM DTT, 10 mM MgCl2, 2 mM EDTA, 1% Triton X-100, 1 mM PMSF, and 1 mM NaF). Protoplast mixtures were incubated on ice for 30 min and sonicated twice for 15 s. NaCl 2M solution was added to the protoplast suspension at a final concentration of 420 mM and tubes were incubated on ice for 1 h with occasional mixing. All protein sample extracts were then centrifuged at 10,000g for 30 min at 4°C and the supernatant, containing the extracted proteins, was transferred into a fresh tube. Expression of FLAG-PCNA was confirmed with immunoblotting as previously described (Arroyo-Mateos et al., 2018). For mass spectrometry-based identification of the proteins that co-purify with Rep-GFP, 1 ml of total protein extract from each sample was incubated for 1 h with 25 μl (50% slurry) of GFP-Trap_M® beads (Chromotek) at 4°C. After incubation, the beads were captured with a magnetic rack and washed three times in 0.5 ml of washing buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1 mM PMSF protease inhibitor). Antibodies used were anti-Flag (F7425; Sigma-Aldrich) and anti-GFP (3H9; Chromotek).
Tryptic Digestion of the Immunopurified Proteins
The affinity purified proteins were subjected to on-bead tryptic digestion. Briefly, the GFP-Trap_M beads were washed twice with 400 μl of 50 mM ammonium bicarbonate buffer pH 8 (ABC), after which the beads were resuspended in 10 μl 50 mM dithiothreitol (DTT) in 50 mM ABC and incubated at 60°C for 1 h. Subsequentely, 12 μl of 50 mM iodoacetamide in 50 mM ABC was added and the sample incubated at room temperature in the dark for 1 h. Then, 14 μl 50 mM cysteine in ABC was added and the bound proteins were digested overnight at 20°C after adding 1 μl of a Trypsin (Roche, sequencing grade) solution (0.5 μg/μl in 1 mM HCl). Peptide digestion was terminated by acidification to pH3 by adding 1 μl 10% trifluoroacetic acid. The tryptic peptides were concentrated and cleaned using home-made μColumns (Lu et al., 2011). These μColumns were prepared by stacking in a 200 μl tip two 3M™ Empore™ C18 extraction disks and 4 μl of a LiChroprep® RP-18 50% (Merck Millipor) slurry in methanol. The μColumns were washed with 100% methanol and equilibrated with 100 μl of 0.1% formic acid in water. Protein samples were added and eluted through the μColumns. The μColumns were washed with 0.1% formic acid in water and the bound peptides were eluted by adding 1:1 mixture of acetonitrile and 0.1% formic acid in water. The acetonitrile content of the samples was reduced by putting the samples in a centrifuge concentrator (Speed-Vac) at 45°C for 2 h and redissolved into 50 μl 1 ml/l formic acid in water.
Mass Spectrometry Analysis and Data Processing
Digested peptides were analyzed using a nano LC-MS/MS LTQ-Orbitrap XL (Thermo Fisher), as previously described (Gawehns et al., 2015). All tryptic digests were sequentially run as one batch of samples on the nLC-MS machine to minimize technical variation. The MaxQuant software 1.5.2.8 (Cox and Mann, 2008; Hubner et al., 2010) was used to analyze the raw data from the LTQ-Orbitrap (Thermo Fisher) for protein identification and label-free quantification (LFQ). The Uniprot proteome database of tomato (UP000004994) and an in-house made database of contaminants (Peng et al., 2012) were included in the Andromeda search engine (Cox et al., 2014). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD018011.
Data filtering from the MaxQuant output was carried out with Perseus 1.5.5.3 (http://www.perseus-framework.org). Only the LFQ values of the proteins that were identified with at least two tryptic peptides, of which one should be unique and one unmodified, were log10 transformed for further analysis. Proteins that showed a ΔLFQ (log10 LFQ in the sample – log10 LFQ in the control) equal or higher than the ΔLFQ of FLAG-PCNA (internal control) in at least one out of three biological replicates were annotated as ‘putative interactors’ of Rep and included for further analysis.
Computational Analysis
Gene ontology (GO) terms for biological process and cellular component were assigned to the co-purifying tomato proteins using Panther (Mi et al., 2017) and QuickGO (EMBL-EBI, https://www.ebi.ac.uk/QuickGO) tools. GO term enrichment was represented for every sample in a bar graph using Prism 7.0v (GraphPad) software. Venn diagrams were drawn using InteractiVenn (http://www.interactivenn.net/) (Heberle et al., 2015). A manually curated protein–protein interaction network was constructed with Cytoscape (Shannon et al., 2003). For that, the interaction network of every putative host interactor and the known Rep-interactors, as reported by Ruhel and Chakraborty (2019) were retrieved from the STRING protein database (https://string-db.org/). The resulting nodes (proteins) and edges (interactions) were arranged according to the force-directed layout. The protein sequence of the Arabidopsis gene model At4g28990 (closest Arabidopsis homologue of GRIEP1) was analyzed using the InterPro database (https://www.ebi.ac.uk/interpro/) to identify known protein domains in GRIEP1.
Construction of Clones Used for Confocal Microscopy
All molecular DNA cloning techniques were performed using standard methods using (Sambrook et al., 2001). The E. coli strain DH5α was used for subcloning. The CDS of EF1A (XM_004251106; Solyc11g069700.1.1), Nucleolin-like 2 (NM_001319854.1; Solyc02g014310.2.1), Rep-interacting protein 1 (XM_004239224; Solyc05g018340.2.1) and THO complex subunit 4A (a.k.a. SlALY1) (Bruhn et al., 1997) (NM_001347950.1; Solyc10g086400.1.1) were amplified from tomato cDNA by PCR using Phusion DNA polymerase (Thermo Fisher) and the primers listed in Table S1 that contained attB1 and attB2 recombination sites for Gateway-directed cloning. The resulting PCR products were recombined with the Gateway vector pDONR207 (Thermo Fisher) using BP Clonase II (Thermo Fisher) and the resulting pENTR plasmids confirmed by DNA sequencing. The cDNA clones were then introduced into the destination vectors pGWB452 (Nakamura et al., 2010) and pDEST-SCYNEGW (Gehl et al., 2009) using LR Clonase II (Thermo Fisher). All plasmids generated and used in this work are listed in Table S2.
Transient Expression in N. benthamiana by Agroinfiltration and Confocal Microscopy
For transient expression in N. benthamiana plants, the binary constructs were introduced in Agrobacterium tumefaciens GV3101, as previously described (Maio et al., 2019). Briefly, single colonies of A. tumefaciens were grown overnight to an OD600 of 0.8–1.5 in low salt (2.5 g/l NaCl) LB medium and resuspended in infiltration medium (1× MS [Murashige and Skoog] salts (Duchefa), 10 mM MES pH 5.6, 2% w/v sucrose, 200 μM acetosyringone) and incubated at room temperature for at least 2 h. Fully expanded leaves of 4-week old N. benthamiana plants were then syringe-infiltrated with these A. tumefaciens cell suspensions at an OD600 = 1 for all constructs. When two cultures were co-infiltrated for BiFC analysis, they were mixed at a ratio 1:1 to a final OD600 = 1. A. tumefaciens strain carrying the pBIN61 binary vector to express the P19 silencing suppressor (referred to as pBIN61:P19) of Tomato bushy stunt virus (TBSV) was added to every sample at a final OD600 = 0.5. Three days post-infiltration N. benthamiana leaf material was collected for microscopy. Subcellular localization of the GFP-tagged proteins and the reconstituted SCFP fluorophores was detected using a confocal laser scanning microscopy (Zeiss LSM510) with a c-Apochromat 40× 1.2 water-immersion Korr objective. The following beam/filter settings were used: GFP-excitation at 488 nm (argon laser), primary beam-splitting mirrors 405/488, secondary beam splitter 490 nm, band filter BP 505–550 nm; SCFP-excitation at 458 nm (argon laser), primary beam-splitting mirrors 458/514, secondary beam splitter 515 nm, band filter BP 470–500 nm. The experiments were repeated three times with a similar result.
Virus-Induced Gene Silencing in N. benthamiana
For virus-induced gene silencing (VIGS), PCR-amplicons of 300 nucleotides length were designed (https://vigs.solgenomics.net/) (Fernandez-Pozo et al., 2015) and amplified from N. benthamiana cDNA for the closest homologues of the tomato candidate genes GRIEP1 (Nbv6.1trP35301) and SlALY1 (Nbv6.1trP36214) using the primers given in Table S2. These fragments were cloned into the SmaI site of pYL156 (Liu et al., 2002). The resulting vectors TRV2::NbALY1 and TRV2::NbGRIEP1 were confirmed by sequencing and transformed to A. tumefaciens GV3101. The different VIGS constructs were mixed in a 1:1 ratio with A. tumefaciens strain containing pTRV1 at a final OD600 of 0.8 per construct, as described (Liu et al., 2002). This bacterial suspension was syringe-infiltrated in the two first true leaves of two-week old transgenic N. benthamiana plants that contain the 2IR-35S:GFP viral reporter cassette for monitoring TYLCV (isolate Almeria; AJ489258) replication and systemic movement (Maio et al., 2019). As negative control, a TRV2 construct was used that targets the non-plant gene E. coli β-glucuronidase (TRV2::GUS) (Tameling and Baulcombe, 2007), as the latter clone is less aggressive than TRV2 without insert. The VIGS experiment was repeated twice with 14 plants per silencing construct.
Measurement of Viral Replication Activity of Rep in 2IR-GFP Plants
Two weeks post agroinoculation, one half of a fully expended systemic leaf was infiltrated with REPTYLCV-RFP, while the other half was infiltrated with KtoA triple mutant of REPTYLCV-RFP (K65A K69AK99A) at an OD600 of 1.0 (Maio et al., 2019). This latter KtoA triple mutant can no longer replicate the 2IR-GFP cassette (negative control). The replication efficiency of Rep of the viral 2IR-GFP cassette was then estimated indirectly using a radiometric method in which the ratio of RFP fluorescence (Rep levels) over the GFP fluorescence (GFP protein expression from extrachromosomal replisomes) was determined. To this end, in total eight leaf discs (each from a different plant) with a diameter of 5 mm were taken and placed in a 96-wells plate (Perkin-Elmer Optiplate™ 96) containing 100 µl double-distilled water three days post A. tumefaciens infiltration. Fluorescence was measured using microplate reader (BioTek Synergy™ H1 Hybrid multi-mode). GFP fluorescence was excited at 485 nm and the emission was measured at 528 nm (BP16 nm monochromator), while RFP fluorescence was excited at 555 nm and the emission measured at 583 nm (BP 16 nm monochromator). The data was compared by using a student t-test. The experiment was repeated twice with eight plants per silencing construct.
Gene Expression Analysis Using Real Time PCR
In order to quantify the transcript levels of NbALY1 or NbGRIEP1, a total of 100 mg of systemic leaf tissue was collected from 4-week old N. benthamiana plants (2-week post TRV inoculation). Following tissue grinding (Qiagen Tissuelyser II bead mill), total RNA was extracted using TRIzol LS (Thermo Fisher). RNA concentrations were determined measuring the absorbance at 260 nm (A260) using a NanoDrop (Thermo Fisher). A total of 1 µg of RNA was used for cDNA synthesis using RevertAid H reverse transcriptase (Thermo Fisher) following the manufacturer’s instructions. Real time PCR was performed with a QuantStudio™ 3 system (Thermo Fisher) using the EvaGreen kit (Biotium) according to the supplier’s instructions. Melting curves were analyzed to ensure amplification specificity and absence of primer-dimer formation. Primer PCR efficiencies were determined using a serial dilution of a mixed cDNA sample (1:2, 1:4, 1:8 & 1:16) and the qPCR reactions were conducted in duplicate (technical replicate) for four samples (biological replicates). Water and no-template controls were used as negative controls for each primer set. Cycle threshold (Ct) values were calculated using the auto baseline function (Quant studio software). Duplicates for which the Ct value different more than 0.5 were not considered and removed from the analysis. Finally, relative gene expression levels were calculated using the obtained Ct values using qBase+ (Biogazelle, Belgium) applying a method that corrects for primer efficiencies. Finally, the gene expression data was normalized using the N. benthamiana APR as reference gene (Liu et al., 2012). The VIGS experiment was repeated three times with a similar result. The 2IR-GFP extrachromosomical molecules (ECMs) were quantified as described previously (Maio et al., 2019). Primers for the ECMs and the internal reference gene, 25S rRNA, are given in Table S2. The qPCR and data analyses were performed using qBase+ software as described above.
N. benthamiana Protein Extraction and Immunoblotting
Two leaf disks (approximately 50 mg) of N. benthamiana leaf material were harvested, snap frozen in liquid nitrogen and homogenized with plastic pestles. Laemmli buffer (0.1 M Tris pH 6.8, 20% glycerol, 4% SDS, 100 mM DTT, 0.001% Bromophenol blue) was added to each sample (100 μl of buffer per sample). Tubes were vortexed vigorously and heated for 10 min at 96°C. The extracts were then centrifugated at 14,000 rpm at 4°C for 5 min. A total of 10 μl of the protein extract was separated on 10% SDS-PAGE gels and subsequently transferred onto a PVDF membrane. Immunodetection of the proteins was performed according to standard protocols using anti-RFP antibody (Chromotek 6G6; 1:1,000) to detect the Rep-RFP fusion proteins, anti-GFP (Chromotek 3H9; 1:1,000) antibody for GFP and GFP-fusions as primary antibodies and anti-Rat (Pierce 31470; 1:10,000) or anti-Mouse (Pierce 31430; 1:10,000) as secondary antibodies. The labeled proteins were visualized using enhanced chemiluminescence (ECL, 0.1 M Tris–HCl pH 8.5, 1.25 mM luminol [Sigma-Aldrich 09253] in DMSO, 0.2 mM p-Coumaric acid [Sigma C9008] in DMSO, 0.01% H2O2) and detected using MXBE Kodak films (Carestream). Equal loading of the extracted proteins was confirmed by estimating the total amount of Rubisco in each sample using Ponceau S or Coomassie Brilliant Blue staining of the membrane.
Results
Identification of Novel Tomato Proteins That Interact With TYLCV Rep
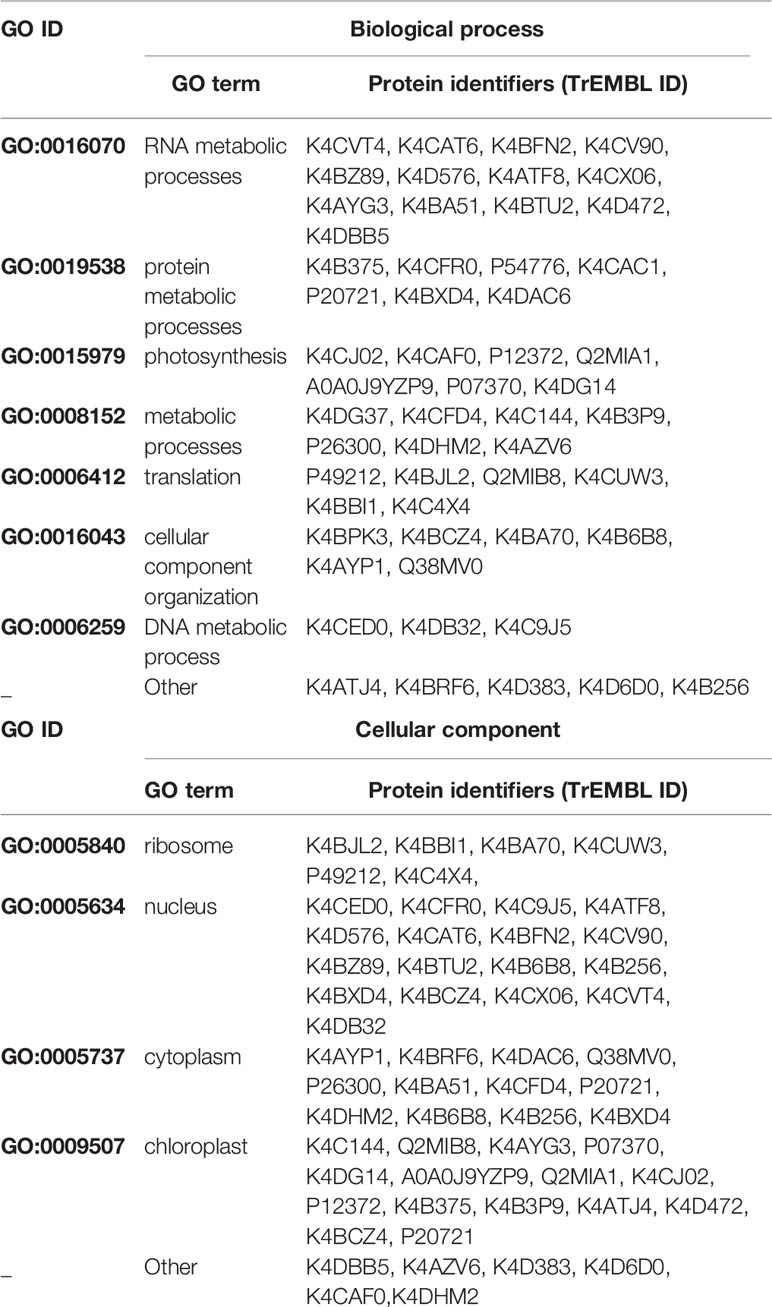
In order to identify novel host proteins that interact with TYLCV Rep, we isolated tomato protoplasts from in vitro cultured plants and transfected them with plasmid DNA to express Rep-GFP alone or in combination with FLAG-PCNA (Figure 1). PCNA was co-expressed together with Rep to have an internal positive control for a Rep interacting protein, but also to promote assembly of viral replisomes. Accumulation of Rep-GFP was visually inspected in every sample by using fluorescence microscopy prior to the affinity purification, while FLAG-PCNA accumulation was confirmed by immunoblotting (Figure S1). The total protein fraction was extracted from three experimental replicates and subjected to affinity purification using anti-GFP resin followed by tryptic digestion in combination with a nano LC–MS/MS analysis. Identification and quantification of the (co)purifying proteins was then performed using MaxQuant. Rep was readily detected in every sample expressing Rep, while PCNA was enriched in the Rep-PCNA samples in comparison to the negative control (PCNA alone) in two out of three biological replicates. In total we identified 427 candidate interactors. To obtain a set of high-confidence interacting proteins, we used as selection criterion that their abundance after the pull-down should be similar or higher than PCNA in one of the replicates. This yielded a total of 54 tomato proteins (Table 1): 27 proteins in the sample expressing Rep-GFP alone, 40 in the sample in which Rep-GFP was expressed together with FLAG-PCNA, and 13 proteins were in present in both samples (Figure 2A). For eight of these interacting proteins, a close homolog was previously identified in a related pull-down experiment for TYLCV Rep in N. benthamiana (Wang et al., 2017a) (Figure 2B and Table 2).
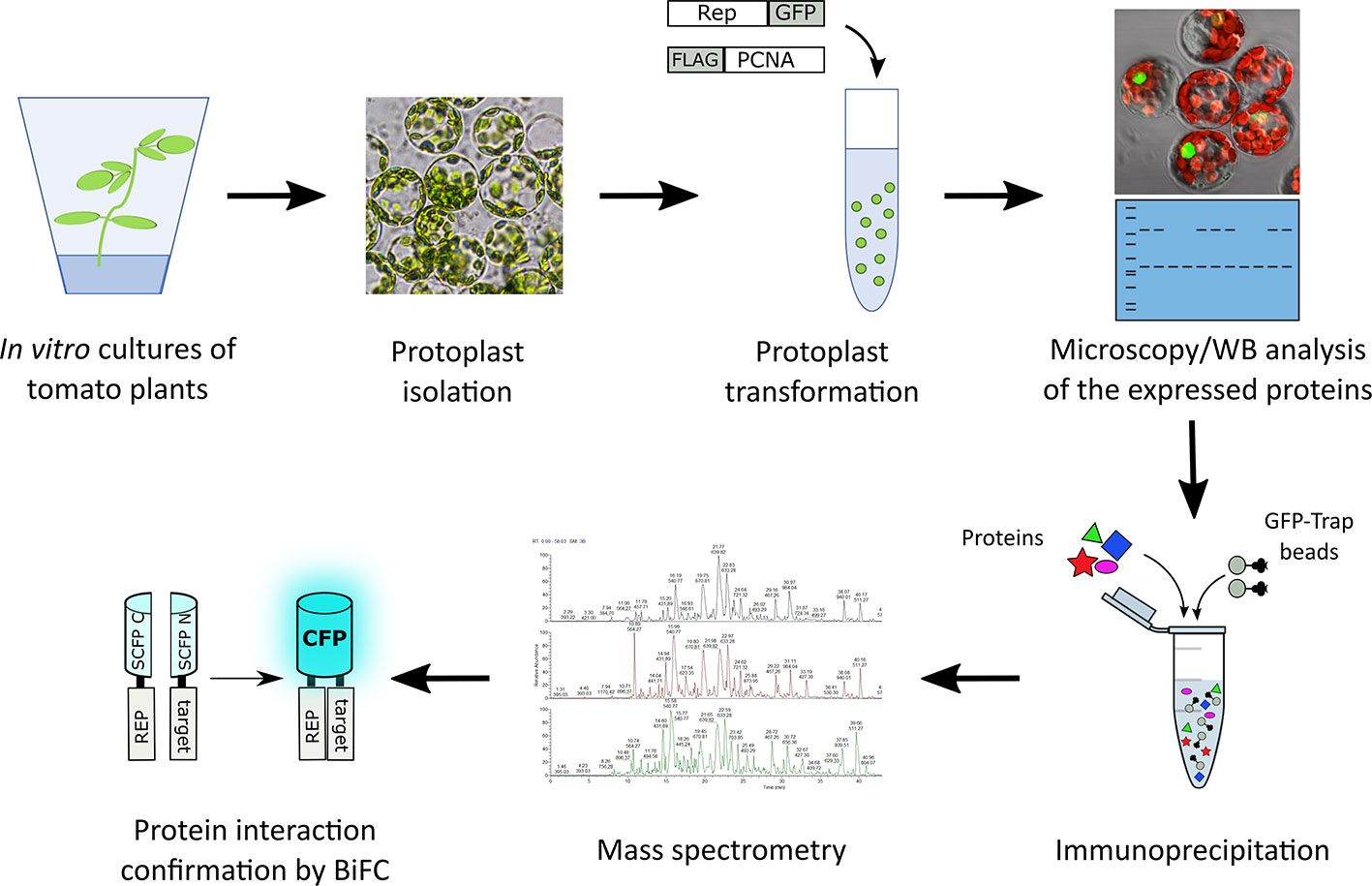
Figure 1 Schematic representation of the strategy followed to identify novel interactors of TYLCV Rep using affinity purification followed by mass spectrometry. Rep fused to GFP was expressed in the presence and absence of FLAG tagged PCNA in tomato protoplasts. The next day, the transfected protoplasts were inspected for a GFP signal using fluorescence microscopy and the total protein fraction was extracted, analyzed for the presence of the overexpressed proteins using immunoblotting (WB) and then subjected to affinity purification using anti-GFP resin (in three independent experiments). Co-purifying proteins were identified after tryptic digestion by tandem mass spectrometry (MS/MS). For a subset of interactors their interaction with Rep was confirmed using bimolecular fluorescence complementation (BiFC). The tryptic digests were prepared in parallel and the resulting peptide samples were analyzed as one batch of runs on the LC-MS.

Table 1 List of the putative interactors of Rep identified by affinity purifications/MS analysis. Proteins in bold were selected for further protein–protein interaction studies.
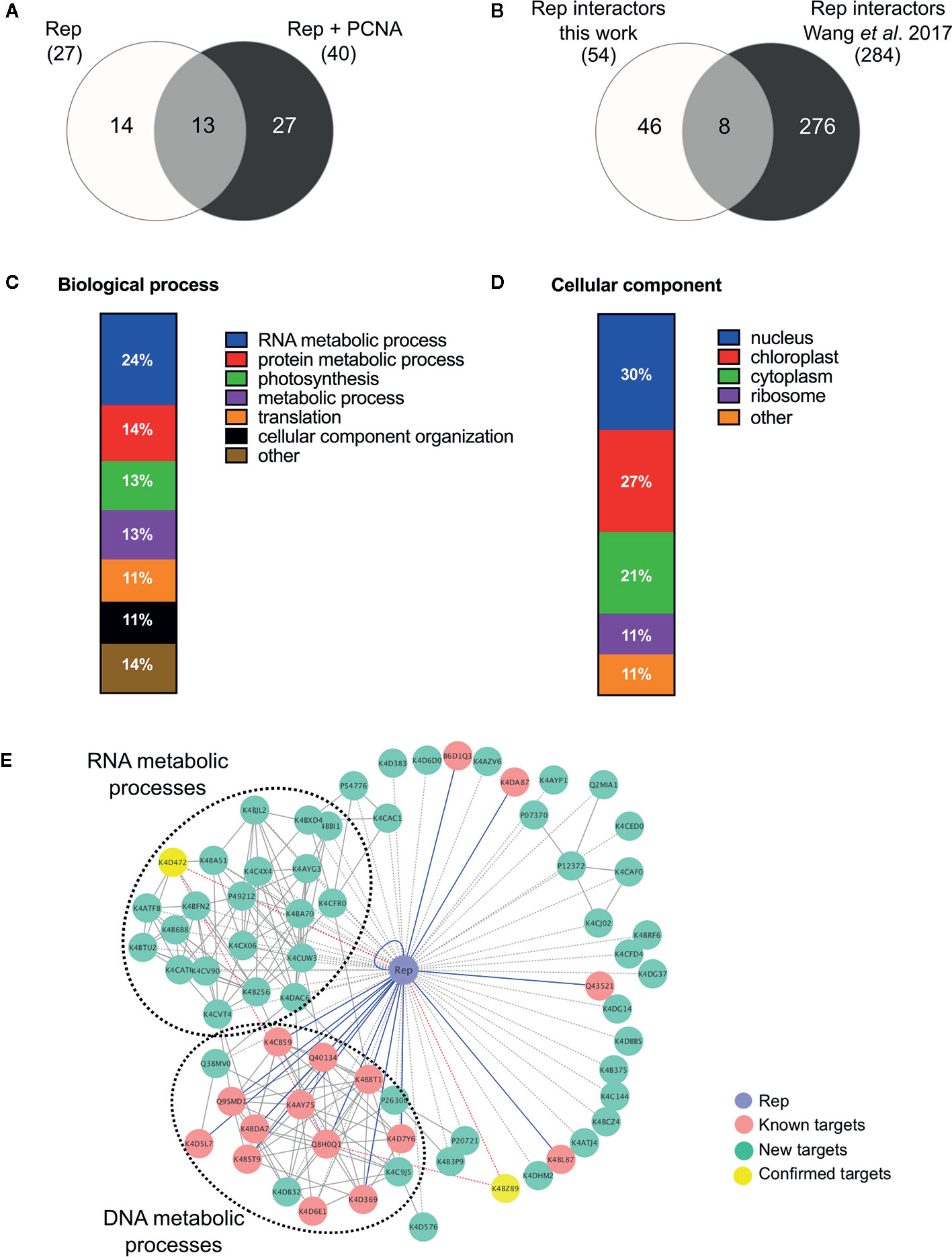
Figure 2 Functional annotation of the tomato interactomes of ‘Rep’ and ‘Rep+PCNA’. (A) Venn diagram depicting the number of interacting proteins that were enriched in the samples expressing ‘Rep’ alone and/or ‘Rep + PCNA’. (B) Venn diagram representing the number of Rep interactors that overlapped with a similar approach taken by Wang et al. (2017a), who purified TYLCV Rep from N. benthamiana cell extracts. (C) Bar chart depicting the proportion of proteins (%) in a subclass of the GO term “Biological process” for the tomato interactors of Rep, combining the two lists of interactors. (D) Bar chart showing the proportion of proteins (%) in a subclass of the GO term “Cellular component”, combining the two lists of interactors. (E) Protein network of the Rep tomato interactome identified in this work (green nodes) and retrieved from the literature (pink nodes). Yellow nodes represent targets whose interaction with Rep was confirmed in this work. Blue lines, known interactions with host proteins; dotted lines, putative interactions with host proteins; red lines, novel confirmed interactions; grey solid lines, protein-protein interactions between plant proteins.
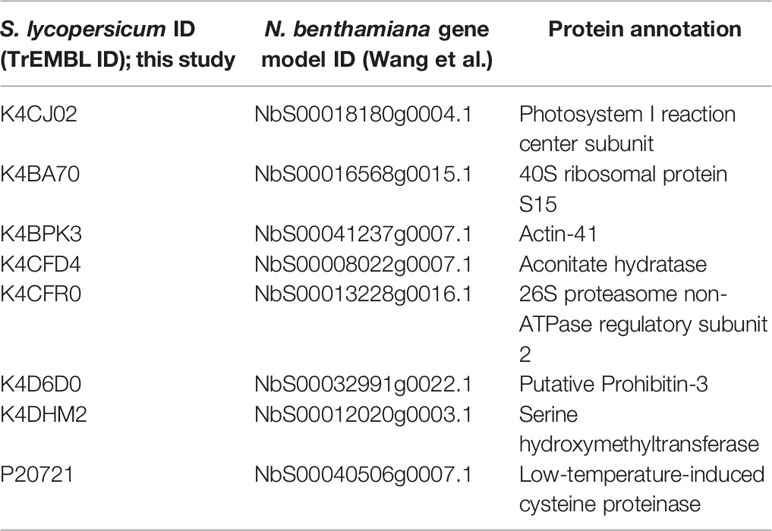
Table 2 List of tomato interactors of TYLCV Rep here found that overlap with the study by Wang et al. (2017a).
Functional Annotation and Protein Network Analysis of the Rep Interactome
In order to gain insight into the cellular processes affected by overexpression of TYLCV Rep in tomato protoplasts, a gene ontology (GO) analysis was conducted assigning functional categories to each interactor. The GO terms “Biological process” and “Cellular component” were assigned and the proportion of plant proteins per GO category (%) was calculated (Table 3). The largest GO category for “Biological process” was formed by proteins belonging to “RNA metabolism”, in particular the subclasses “involved in spliceosome and ribosome assembly”, “regulation of RNA polymerase” and “mRNA/rRNA processing” (Figure 2C). These categories were followed by “protein metabolic processes”, “photosynthesis’, “metabolic processes”, “translation” and “cellular component organization”. Only two GO categories were significantly overrepresented (p-value <0.05), i.e. “RNA metabolic processes” and “metabolic processes”. In total 30% of the identified proteins was assigned to be nucleus based on their annotation (Figure 2D). In order to visualize the Rep-interactome, we plotted the 54 tomato proteins in a protein interaction network (Figure 2E) with the nodes representing (i) the newly identified host proteins (green and yellow), (ii) 16 Rep-interactors derived from previous studies (pink) (reviewed by Ruhel and Chakraborty, 2019) and (iii) Rep (blue). In total, seventy known interactions with Rep (Ruhel and Chakraborty, 2019) were included in this protein network (blue edges and dotted edges) and a total of 148 plant protein interactions (grey edges) were retrieved from the STRING database (Table S3). This network contained two major clusters. The first cluster comprises (mostly) known Rep interactors involved in DNA metabolism, such as DNA replication and repair, while the second cluster is primarily formed by newly-identified candidates implicated in RNA metabolic processes, which hints to that Rep potentially regulates RNA biogenesis in an unknown manner.
Examples of Novel Rep Interactors Linked to Viral DNA Replication and RNA Metabolic Processes
Our list of TYLCV Rep interactors contained several proteins linked to DNA replication and pathogenesis. For example, Elongation factor 1-alpha (EF1A), found in the sample Rep-PCNA, participates in cellular functions like translation, nuclear export, transcription but also apoptosis in virus-infected cells (reviewed in Sasikumar et al., 2012). EF1A is also involved in the propagation of certain positive-strand RNA viruses, as it has been found to interact with viral RNA and certain virus-encoded RNA-dependent RNA polymerases (Abbas et al., 2015).
Another putative novel Rep interactor detected in the AP-MS with Rep-PCNA is Nucleolin-like 2, a major nucleolar protein that regulates the rDNA chromatin structure, rRNA gene expression, pre-rRNA processing and assembly of ribosome particles (Durut and Sáez-Vásquez, 2015). Interestingly, Nucleolin interacts also with PCNA and the Replication protein A 32 (RPA32) during replication stress in human cell lines (Kawamura et al., 2019). These authors also provided evidence that Nucleolin is recruited to damaged replication forks and that recruitment of Nucleolin controls the activation of homologous recombination, a process that is also promoted by geminiviruses in infected cells (Richter et al., 2014). These observations argue for a functional link between Rep and Nucleolin-like 2. Possibly related, the coat protein (CP) of TYLCV was shown to be recruited to the nucleolus in the absence of the virus, while in the presence of the virus it was recruited to discrete nuclear foci (Rojas et al., 2001; Wang et al., 2017b). Expression of Rep was already sufficient to exclude CP from the nucleolus, but its localization in foci was only seen during a TYLCV infection (Wang et al., 2017b). In fact, a plethora of animal and plant viruses but also virus-encoded proteins reside in the nucleolus where they interact with nucleolar proteins and several viral proteins have been shown to co-localize with, reorganize and re-distribute nucleolar proteins such as Nucleolin (Hiscox, 2002; Kim et al., 2004; Chaudhry et al., 2018).
A third interactor again coming from the list of proteins co-purifying with Rep-PCNA and implicated in host-virus interactions is the THO subunit 4A, which is part of the THO core complex of the TREX (TRanscription-EXport) complex. This complex is conserved across eukaryotes and is linked to nuclear export of mRNA (Heath et al., 2016; Sørensen et al., 2017). The THO/TREX complex is targeted by various viral proteins to redirect and control viral mRNA translocation (Schneider and Wolff, 2009; Yarbrough et al., 2014). The closest Arabidopsis homologue of the tomato interactor here identified (THO subunit 4A) is the mRNA export adaptor protein ALY1 (AT5G59950). Once bound to mRNA, the ALY proteins interact with the nuclear export receptor, which then facilitates export of the bound mRNA. Interestingly, compromised ALY1 gene function in Arabidopsis results in loss of RNA-dependent DNA methylation (RdDM) due to deficient export of Argonaute6 mRNA (Choudury et al., 2019). In line with this connection, Rodríguez‐Negrete and co-workers (2013) demonstrated that TYLCSV Rep displays transcriptional gene silencing (TGS) suppressor activity by interfering with the host DNA methylation machinery during infection. In particular, Rep down regulates the transcript levels of key enzymes that maintain this modification, namely the DNA methyltransferases MET1 and CMT3. Arguably, TYLCV Rep might thus inhibit tomato ALY1 function to modulate RdDM activity.
Finally, our list of candidate interactors that interacted with Rep in the absence of PCNA overexpression contained an uncharacterized protein, here called Geminivirus Rep interacting EWS-like protein 1 (GRIEP1), that shows sequence homology to the human RNA-binding protein EWS (Ewing Sarcoma protein). This mammalian protein acts as transcriptional repressor that regulates pri-miRNA processing and plays a role in tumorigenic processes (Ohno et al., 1994; Ouyang et al., 2017). The closest homolog of GRIEP1 in Arabidopsis is At4g28990, a predicted 395 amino acid protein with a predicted Zinc finger domain of the RanBP2-type that groups with the multifunctional TET protein family (TAF15/EWS/TLS; Interpro IPR034870) (Law et al., 2006). This protein family is implicated in transcription, (alternative) splicing, mRNA transport but also DNA repair (Kovar, 2011). To our knowledge this protein has not been studied in any plant species yet. Based on their potential role on DNA replication and/or Rep activity, we selected these four proteins to corroborate their interaction in planta (bold in Table 1).
Confirmation of the Interaction Between Rep and the Putative Interactors
To assess in an independent assay the specificity of the interactions, we used bimolecular fluorescence complementation (BiFC), as it also provides information on where the interaction occurs in the cell. First, we assessed their subcellular localization pattern in N. benthamiana expressing GFP-labelled variants of these proteins. As expected, GFP-EF1A localized to the cytoplasm, while GFP-Nucleolin-like 2 localized exclusively to the nucleolus (Figure 3A). SlALY1 was uniformly distributed in the nucleoplasm with occasionally some small nuclear speckles. GRIEP1 resided, however, in large nuclear bodies (or aggregates) of unknown function or origin. Using immunoblotting, we confirmed that these GFP-fusions accumulated at the expected protein mass in N. benthamiana (Figure S2). We then fused the N-terminal half of the super Cyan fluorescent protein (SCFPN) to the N-terminus of the candidate proteins and the resulting fusion proteins were expressed in N. benthamiana epidermal cells together with Rep or PCNA fused to the C-terminal half of SCFP (SCFPC) (Figure 3B). As negative control, the SCFPN fusion proteins were co-expressed with a non-plant protein (SCFPC-tagged β-Glucuronidase from E. coli). We detected no BiFC signal for any of these four interactors in combination with GUS in any cellular compartment. For SCFPN-EF1A and SCFPN-Nucleolin-like 2, only a faint SCFP signal was observed in some nuclei in combination with TYLCV Rep or PCNA. In contrast, the BiFC couple GRIEP1-Rep showed a strong and specific CFP fluorescence signal in distinct (large) nuclear bodies (NBs) and, to a minor extent, in the cytoplasm. Also, the combination of GRIEP1 and PCNA yielded a strong CFP fluorescence signal in the nucleus with some speckles and a weaker fluorescence signal in the cytoplasm (Figures 3B, C). Finally, SlALY1 interacted with Rep in the nucleoplasm and with PCNA in both the cytoplasm and nucleoplasm. These BiFC experiments thus corroborate our MS data that GRIEP1 and SlALY1 interact specifically and directly with Rep.
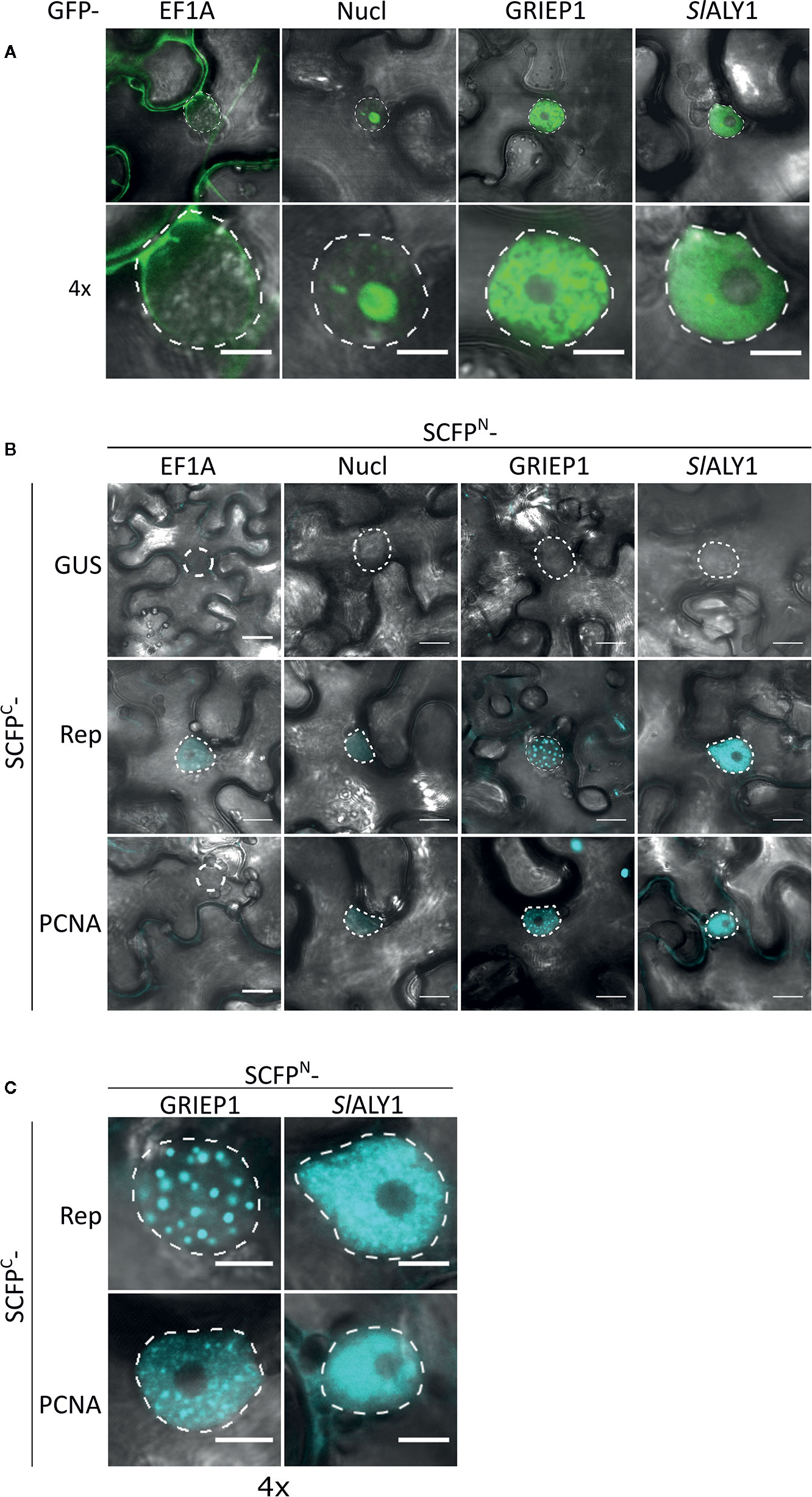
Figure 3 Cellular localization and BiFC protein interaction assay of the Rep-interacting proteins EF1A, Nucleolin-like 2, GRIEP1 and THO. (A) Subcellular localization of GFP-tagged tomato EF1A, Nucleolin-like 2 (Nucl), GRIEP1 and SlALY1 proteins in N. benthamiana epidermal cells upon transient overexpression. Image shown represents a typical cell (top) and a 4× zoom showing only its nucleus (bottom). (B) In a BiFC assay, EF1A and Nucleolin-like 2 do not seem to interact with TYLCV Rep nor PCNA since only a faint background fluorescence signal was detected, whereas GRIEP1 and SlALY1 show strong reconstitution of SCFP fluorophore in combination with Rep and PCNA in nuclei of the transfected cells. (C) 4× zoom of the nuclei of the positive BiFC interaction pairs shown in (B). Scale bar is 5 μm for (A, C) and 20 μm for (B). Dotted lines outline the nucleus. These experiments were repeated three times independently; for each combination, a minimum of 50 cells was analyzed in each experiment.
Next, we used VIGS to assess whether reduced transcript levels of these two confirmed Rep-interactors (GRIEP1 and SlALY) would alter DNA replication activity of Rep. To this end, we took advantage of N. benthamiana expressing a 2IR-GFP reporter cassette (Maio et al., 2019), which allows monitoring of viral replication in situ as a result of TYLCV Rep expressio,. The VIGS constructs caused indeed effective gene silencing of SlALY1 and GRIEP1 without negatively impacting the plant development (Figures S3A, B). Two weeks post TRV inoculation, Rep-RFP was transiently expressed in a fully expanded leaf and 3 days later replication of the viral reporter was (indirectly) quantified by comparing the signal intensities of the GFP and RFP fluorescence (Figure S3C). Plants silenced for GRIEP1 or SlALY1 did not exhibit any reduced Rep replication activity, as measured either by accumulation extrachromosomal circular DNA molecules (ECMs) that derive from the 2IR-GFP cassette or GFP fluorescence (Figures S3E, F). In fact, silencing of GRIEP1 did significantly increase ECM accumulation compared to GUS silenced plants (p-value = 0.0137), and silencing of ALY1 resulted in significantly increased GFP intensity (nearly 2.5-fold increase). These data thus argue that both GRIEP1 and ALY1 might act as inhibitors of Rep DNA replication activity, but we cannot exclude that they impact functions of Rep other than replication activity. Future work should reveal if overexpression of GRIEP1 and ALY1 can repress Rep activity.
Discussion
This work describes an AP-MS proteomics approach to identify novel host proteins that (in)directly interact with TYLCV Rep and to explore their role for geminivirus replication. This yielded 54 putative interactors of TYLCV Rep of which 40 were detected in cells overexpressing Rep together with tomato PCNA and two were confirmed to interact with both Rep and PCNA in the BiFC assay. The majority of the interactors are involved in RNA biogenesis, such as spliceosomal complex assembly, regulation of DNA-dependent RNA polymerase, rRNA expression and maturation and RNA binding, or in protein metabolic processes like catabolic activity, enzyme regulation and proteasome complex. These findings align in part with the proteomics results obtained by Wang and co-workers (2017a), who found that their list of TYLCV Rep interactors in N. benthamiana was enriched for the GO terms belonging to ‘protein catabolic processes’ but not for ‘RNA metabolism’. Moreover, for eight of the Rep interactors here identified a close homolog was found to interact with TYLCV Rep in N. benthamiana (Wang et al., 2017a). Several arguments support the validity of our findings. First, we were able to specifically co-purify PCNA with Rep in two out of three of the independent affinity purifications compared to the negative control, demonstrating that the pulldown conditions were suitable to identify interactors of Rep. In addition, some of the AP-MS hits found were previously found to interact with Rep, e.g. the Histones 2A and 2B (Kong and Hanley-Bowdoin, 2002). Second, a substantial proportion of the putative Rep-interactors were predicted to be nuclear localized in silico. This agrees with the known subcellular localization of Rep and the viral replisome (Kushwaha et al., 2017; Maio et al., 2019). Third, we were able to confirm the interaction between Rep and two selected candidates in planta using the BiFC system and to detect formation of this complex in the nucleus. These two proteins also interacted with PCNA in our BiFC, suggesting that they might be part of viral replisomes or transiently interact with them. GRIEP1 is apparently not required for the Rep-mediated viral DNA replication activity, as the latter remained unchanged in GRIEP1-silenced plants. As well, it was not possible from our experiment to unambiguously determine whether Rep activity was altered as a result of SlALY1-silencing in N. benthamiana. Future studies should reveal if this latter interaction is really involved in viral replication or suppression of transcriptional gene silencing, as reported for other viral proteins (Xu et al., 2020).
Despite the identification of novel interactors of TYLCV Rep in tomato, several limitations apply to our approach. First, we also detected chloroplastic proteins implicated in photosynthesis and proteins involved in ribosome assembly and biogenesis. As a connection is lacking between Rep and photosynthesis and given the difference in compartmentalization of these processes, these proteins likely represent false positives. In fact, photosynthesis-related and ribosomal proteins are highly abundant plant proteins and as such common contaminants picked up in proteomics methods such as AP-MS (Smaczniak et al., 2012). At the same time, many true interactors could easily have been missed due to the stringency of our selection criterium, low protein levels in non-dividing mesophyll cells, weak or transient interactions with Rep or the presence of the tag on Rep, which all could prevent detection of protein-protein interactions. The identified host proteins may also indirectly bind to TYLCV Rep. For example, for Elongation factor 1A and Nucleolin-like 2, we were not able to confirm their interaction using BiFC. This notion of an indirect interaction is corroborated by the observation that, in our interaction network, EF1A is connected to two known interactors of Rep, Ubiquitin conjugation enzyme 2 (UBC2) and Histone 3 (H3). The last aspect to consider is that our experimental results derive from the analysis of epidermal tomato protoplasts that express Rep in the absence of a viral infection. To obtain a more comprehensive picture of the host proteins that interact with Rep, the interactions should also be studied in the context of a virus-infection in phloem cells.
Interestingly, two RNA-binding proteins were confirmed as interacting proteins of both Rep and PCNA. The first one, GRIEP1, is an unknown conserved protein with an EWS-like RNA-binding protein that contains a Zinc finger domain of the RanBP2 type. Interestingly, in Arabidopsis GRIEP1 is mostly expressed in the shoot apex and floral tissues (data available on ePlant, http://bar.utoronto.ca/eplant/), where meristematic cells are dividing, suggesting that the Arabidopsis homolog has a potential role in cell division or differentiation. Moreover, the BiFC couple between Rep and this EWS-like protein resided in nuclear bodies (NBs). We reported previously that the BiFC pair Rep-SCE1 (SUMO E2 conjugating enzyme 1) also aggregates in NBs (Maio et al., 2019). Co-localization studies with marker proteins of different NBs should elucidate the nature and biological function of these different NBs.
The other confirmed interactor is a subunit of the THO complex, SlALY1. The biological function of this THO/TREX complex is well characterized in eukaryotes, where it is recruited to nascent mRNA, where it connects transcriptional elongation to export of mature mRNA (Jimeno et al., 2002; Strässer et al., 2002). This complex is also essential for the nuclear export of the Kaposi’s sarcoma-associated herpes virus mRNAs and for viral DNA replication (Boyne et al., 2008). In plants, the THO/TREX complex has been shown to influence the production of trans-acting small interfering RNAs (Yelina et al., 2010) and a component of this complex appears to be involved in plant disease resistance against powdery mildew pathogen (Pan et al., 2012). As Rep is known to act as suppressor of RNA silencing (Liu et al., 2014), additional studies are needed to elucidate the mechanism and biological consequence of this Rep-THO/TREX interaction.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD018011.
Author Contributions
FM, HB, and MP designed the experiments. FM performed the protoplast isolations and transfections, and prepared the protein samples for nLC-MS/MS measurements. SB performed the nLC-MS/MS measurements. FM and SB analyzed the MS/MS data. FM, TH, MW, and MA-M performed the cloning, agroinfiltration, microscopy analysis, silencing, and qPCR analysis. FM, HB, and MP wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The Topsector T&U program Better Plants for Demands (grant 1409-036 to HB), including the partnering breeding companies, supported this work. FM was financially supported by Keygene N.V. (The Netherlands).
Conflict of Interest
Author MP is employed by the company Keygene N.V.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to L. Tikovsky and H. Lemereis for taking care of our plants; LCAM-FNWI (Molecular Cytology, SILS, University of Amsterdam) for maintenance and support with confocal microscopy. We thank R. Sevenier (Keygene N.V.) for the protoplast isolation and transfection protocol; M. de Vos (Keygene N.V.) for helping with the MS analysis; F. de Lamo for critical review of the manuscript. We thank E. Bejarano (Univeristy of Malaga), D. Baulcome (University of Cambridge, UK), J. Kudla (WWU Muenster, Germany), T. Nagakawa (Shimane University, Japan), J Lindbo for sharing plasmids.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01069/full#supplementary-material
References
Abbas, W., Kumar, A., Herbein, G. (2015). The eEF1A Proteins: At the crossroads of oncogenesis, apoptosis, and viral infections. Front. Oncol. 5, 75. doi: 10.3389/fonc.2015.00075
Arroyo-Mateos, M., Sabarit, B., Maio, F., Sánchez-Durán, M. A., Rosas-Díaz, T., Prins, M., et al. (2018). Geminivirus Replication Protein impairs SUMO Conjugation of PCNA at two acceptor sites. J. Virol. 92, e00611–e00618. doi: 10.1101/305789
Bagewadi, B., Chen, S., Lal, S. K., Choudhury, N. R., Mukherjee, S. K. (2004). PCNA interacts with Indian mung bean yellow mosaic virus rep and downregulates Rep activity. J. Virol. 78, 11890–11903. doi: 10.1128/JVI.78.21.11890-11903.2004
Belabess, Z., Peterschmitt, M., Granier, M., Tahiri, A., Blenzar, A., Urbino, C. (2016). The non-canonical tomato yellow leaf curl virus recombinant that displaced its parental viruses in southern Morocco exhibits a high selective advantage in experimental conditions. J. Gen. Virol. 97, 3433–3445. doi: 10.1099/jgv.0.000633
Boyne, J. R., Colgan, K. J., Whitehouse, A. (2008). Recruitment of the complete hTREX Complex is required for Kaposi’s Sarcoma-Associated Herpesvirus intronless mRNA Nuclear Export and vVirus replication. PLoS Pathog. 4, e1000194. doi: 10.1371/journal.ppat.1000194
Bruhn, L., Munnerlyn, A., Grosschedl, R. (1997). ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 11, 640–653. doi: 10.1101/gad.11.5.640
Butterbach, P., Verlaan, M. G., Dullemans, A., Lohuis, D., Visser, R. G. F., Bai, Y., et al. (2014). Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. Proc. Natl. Acad. Sci. U. S. A. 111, 12942–12947. doi: 10.1073/pnas.1400894111
Caro, M., Verlaan, M. G., Julián, O., Finkers, R., Wolters, A. M. A., Hutton, S. F., et al. (2015). Assessing the genetic variation of Ty-1 and Ty-3 alleles conferring resistance to tomato yellow leaf curl virus in a broad tomato germplasm. Mol. Breed. 35, 132. doi: 10.1007/s11032-015-0329-y
Castillo, A. G., Collinet, D., Deret, S., Kashoggi, A., Bejarano, E. R. (2003). Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 312, 381–394. doi: 10.1016/S0042-6822(03)00234-4
Castillo, A. G., Kong, L. J., Hanley-Bowdoin, L., Bejarano, E. R. (2004). Interaction between a geminivirus replication protein and the plant sumoylation system. J. Virol. 78, 2758–2769. doi: 10.1128/JVI.78.6.2758-2769.2004
Chaudhry, U., Malik, D. A., Saleem, N., Malik, M. T. (2018). Nucleolin: Role in Bacterial and Viral Infections. EC Microbiol. 14, 631–640.
Choe, K. N., Moldovan, G. L. (2017). Forging Ahead through Darkness: PCNA, Still the Principal Conductor at the Replication Fork. Mol. Cell 65, 380–392. doi: 10.1016/j.molcel.2016.12.020
Choudury, S. G., Shahid, S., Cuerda-Gil, D., Panda, K., Cullen, A., Ashraf, Q., et al. (2019). The RNA export factor ALY1 enables genome-wide RNA-directed DNA methylation. Plant Cell 31, 759–774. doi: 10.1105/tpc.18.00624
Cox, J., Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Cox, J., Hein, M. Y., Luber, C. A., Paron, I., Nagaraj, N., Mann, M. (2014). Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell Proteomics 13, 2513–2526. doi: 10.1074/mcp.M113.031591
Dunham, W. H., Mullin, M., Gingras, A. C. (2012). Affinity-purification coupled to mass spectrometry: Basic principles and strategies. Proteomics 12, 1576–1590. doi: 10.1002/pmic.201100523
Durut, N., Sáez-Vásquez, J. (2015). Nucleolin: dual roles in rDNA chromatin transcription. Gene 556, 7–12. doi: 10.1016/j.gene.2014.09.023
Eagle, P. A., Orozco, B. M., Hanley-Bowdoin, L. (1994). A DNA sequence required for geminivirus replication also mediates transcriptional regulation. Plant Cell 6, 1157–1170. doi: 10.1105/tpc.6.8.1157
Ehrnsberger, H. F., Grasser, M., Grasser, K. D. (2019). Nucleocytosolic mRNA transport in plants: export factors and their influence on growth and development. J. Exp. Bot. 70, 3757–3763. doi: 10.1093/jxb/erz173
Fernandez-Pozo, N., Rosli, H. G., Martin, G. B., Mueller, L. A. (2015). The SGN VIGS Tool: User-Friendly Software to Design Virus-Induced Gene Silencing (VIGS) Constructs for Functional Genomics. Mol. Plant 8, 486–488. doi: 10.1016/j.molp.2014.11.024
Gavin, A. C., Maeda, K., Kuhner, S. (2011). Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr. Opin. Biotechnol. 22, 42–49. doi: 10.1016/j.copbio.2010.09.007
Gawehns, F., Ma, L., Bruning, O., Houterman, P. M., Boeren, S., Cornelissen, B. J. C., et al. (2015). The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Front. Plant Sci. 6, 967. doi: 10.3389/fpls.2015.00967
Gehl, C., Waadt, R., Kudla, J., Mendel, R. R., Hänsch, R. (2009). New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2, 1051–1058. doi: 10.1093/mp/ssp040
Gutierrez, C. (2000a). DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J. 19, 792–799. doi: 10.1093/emboj/19.5.792
Gutierrez, C. (2000b). Geminiviruses and the plant cell cycle. Plant Mol. Biol. 43, 763–772. doi: 10.1023/A:1006462028363
Hanley-Bowdoin, L., Settlage, S. B., Robertson, D. (2004). Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol. Plant Pathol. 5, 149–156. doi: 10.1111/j.1364-3703.2004.00214.x
Hanley-Bowdoin, L., Bejarano, E. R., Robertson, D., Mansoor, S. (2013). Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788. doi: 10.1038/nrmicro3117
Heath, C. G., Viphakone, N., Wilson, S. A. (2016). The role of TREX in gene expression and disease. Biochem. J. 473, 2911–2935. doi: 10.1042/BCJ20160010
Heberle, H., Meirelles, G. V., da Silva, F. R., Telles, G. P., Minghim, R. (2015). InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf. 16, 169. doi: 10.1186/s12859-015-0611-3
Hiscox, J. A. (2002). The nucleolus - a gateway to viral infection? Arch. Virol 147, 1077–1089. doi: 10.1007/s00705-001-0792-0
Hubner, N. C., Bird, A. W., Cox, J., Splettstoesser, B., Bandilla, P., Poser, I., et al. (2010). Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754. doi: 10.1083/jcb.200911091
Jeske, H. (2009). Geminiviruses. Curr. Top. Microbiol. Immunol. 331, 185–226. doi: 10.1007/978-3-540-70972-5_11
Jimeno, S., Rondón, A. G., Luna, R., Aguilera, A. (2002). The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 21, 3526–3535. doi: 10.1093/emboj/cdf335
Kaliappan, K., Choudhury, N. R., Suyal, G., Mukherjee, S. K. (2012). A novel role for RAD54: this host protein modulates geminiviral DNA replication. FASEB J. 26, 1142–1160. doi: 10.1096/fj.11-188508
Karimi, M., Inzé, D., Depicker, A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Kawamura, K., Qi, F., Meng, Q., Hayashi, I., Kobayashi, J. (2019). Nucleolar protein nucleolin functions in replication stress–induced DNA damage responses. J. Radiat. Res. 60, 281–288. doi: 10.1093/jrr/rry114
Kim, S. H., Ryabov, E. V., Brown, J. W. S., Taliansky, M. (2004). Involvement of the nucleolus in plant virus systemic infection. Biochem. Soc. Trans. 32, 557–560. doi: 10.1042/BST0320557
Kong, L. J., Hanley-Bowdoin, L. (2002). A Geminivirus Replication Protein Interacts with a Protein Kinase and a Motor Protein That Display Different Expression Patterns during Plant Development and Infection. Plant Cell 14, 1817–1832. doi: 10.1105/tpc.003681
Kong, L. J., Orozco, B. M., Roe, J. L., Nagar, S., Ou, S., Feiler, H. S., et al. (2000). A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19, 3485–3495. doi: 10.1093/emboj/19.13.3485
Kovar, H. (2011). Dr. Jekyll and Mr. Hyde: The Two Faces of the FUS/EWS/TAF15 Protein Family. Sarcoma 2011, 837474. doi: 10.1155/2011/837474
Kushwaha, N. K., Mansi, Chakraborty, S. (2017). The replication initiator protein of a geminivirus interacts with host monoubiquitination machinery and stimulates transcription of the viral genome. PLoS Pathog. 13, e1006587. doi: 10.1371/journal.ppat.1006587
Law, W. J., Cann, K. L., Hicks, G. G. (2006). TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct. Genomic Proteomic 5, 8–14. doi: 10.1093/bfgp/ell015
Lindbo, J. A. (2007). TRBO: a high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 145, 1232–1240. doi: 10.1104/pp.107.106377
Liu, Y., Schiff, M., Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Liu, D., Shi, L., Han, C., Yu, J., Li, D., Zhang, Y. (2012). Validation of Reference Genes for Gene Expression Studies in Virus-Infected Nicotiana benthamiana Using Quantitative Real-Time PCR. PLoS One 7, e46451. doi: 10.1371/journal.pone.0046451
Liu, Y., Jin, W., Wang, L., Wang, X. F. (2014). Replication-associated proteins encoded by Wheat dwarf virus act as RNA silencing suppressors. Virus Res. 190, 34–39. doi: 10.1016/j.virusres.2014.06.014
Lu, J., Boeren, S., de Vries, S. C., van Valenberg, H. J. F., Vervoort, J., Hettinga, K. (2011). Filter-aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins. J. Proteomics 75, 34–43. doi: 10.1016/j.jprot.2011.07.031
Luque, A., Sanz-Burgos, A. P., Ramirez-Parra, E., Castellano, M. M., Gutierrez, C. (2002). Interaction of geminivirus Rep protein with replication factor C and its potential role during geminivirus DNA replication. Virology 302, 83–94. doi: 10.1006/viro.2002.1599
Maio, F., Arroyo-Mateos, M., Bobay, B. G., Bejarano, E. R., Prins, M., van den Burg, H. A. (2019). A lysine residue essential for geminivirus replication also controls nuclear localization of the TYLCV Rep protein. J. Virol 93, e01910–e01918. doi: 10.1128/JVI.01910-18
Mansoor, S., Briddon, R. W., Zafar, Y., Stanley, J. (2003). Geminivirus disease complexes: an emerging threat. Trends Plant Sci. 8, 128–134. doi: 10.1016/S1360-1385(03)00007-4
Mansoor, S., Zafar, Y., Briddon, R. W. (2006). Geminivirus disease complexes: the threat is spreading. Trends Plant Sci. 11, 209–212. doi: 10.1016/j.tplants.2006.03.003
Martínez, F., Rodrigo, G., Aragonés, V., Ruiz, M., Lodewijk, I., Fernández, U., et al. (2016). Interaction network of tobacco etch potyvirus NIa protein with the host proteome during infection. BMC Genomics 17, 87. doi: 10.1186/s12864-016-2394-y
Mi, H., Huang, X., Muruganujan, A., Tang, H., Mills, C., Kang, D., et al. (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189. doi: 10.1093/nar/gkw1138
Morilla, G., Castillo, A. G., Preiss, W., Jeske, H., Bejarano, E. R. (2006). A versatile transreplication-based system to identify cellular proteins involved in geminivirus replication. J. Virol 80, 3624–3633. doi: 10.1128/JVI.80.7.3624-3633.2006
Nakamura, S., Mano, S., Tanaka, Y., Ohnishi, M., Nakamori, C., Araki, M., et al. (2010). Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci. Biotechnol. Biochem. 74, 1315–1319. doi: 10.1271/bbb.100184
Navas-Castillo, J., Fiallo-Olivé, E., Sánchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Negrutiu, I., Shillito, R., Potrykus, I., Biasini, G., Sala, F. (1987). Hybrid genes in the analysis of transformation conditions. Plant Mol. Biol. 8, 363–373. doi: 10.1007/BF00015814
Ohnishi, J., Yamaguchi, H., Saito, A. (2016). Analysis of the Mild strain of tomato yellow leaf curl virus, which overcomes Ty-2 gene–mediated resistance in tomato line H24. Arch. Virol 161, 2207–2217. doi: 10.1007/s00705-016-2898-4
Ohno, T., Ouchida, M., Lee, L., Gatalica, Z., Rao, V. N., Reddy, E. S. (1994). The Ews Gene, Involved in Ewing Family of Tumors, Malignant-Melanoma of Soft Parts and Desmoplastic Small Round-Cell Tumors, Codes for an Rna-Binding Protein with Novel Regulatory Domains. Oncogene 9, 3087–3097.
Ouyang, H., Zhang, K., Fox-Walsh, K., Yang, Y., Zhang, C., Huang, J., et al. (2017). The RNA binding protein EWS is broadly involved in the regulation of pri-miRNA processing in mammalian cells. Nucleic Acids Res. 45, 12481–12495. doi: 10.1093/nar/gkx912
Pan, H. R., Liu, S. M., Tang, D. (2012). HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J. 69, 831–843. doi: 10.1111/j.1365-313X.2011.04835.x
Peng, K., van Lent, J. W. M., Boeren, S., Fang, M., Theilmann, D. A., Erlandson, M. A., et al. (2012). Characterization of Novel Components of the Baculovirus Per Os Infectivity Factor Complex. J. Virol 86, 4981–4988. doi: 10.1128/JVI.06801-11
Richter, K. S., Kleinow, T., Jeske, H. (2014). Somatic homologous recombination in plants is promoted by a geminivirus in a tissue-selective manner. Virology 452–453, 287–296. doi: 10.1016/j.virol.2014.01.024
Rizvi, I., Choudhury, N. R., Tuteja, N. (2015). Insights into the functional characteristics of geminivirus rolling-circle replication initiator protein and its interaction with host factors affecting viral DNA replication. Arch. Virol. 160, 375–387. doi: 10.1007/s00705-014-2297-7
Rodríguez-Negrete, E., Lozano-Durán, R., Piedra-Aguilera, A., Cruzado, L., Bejarano, E. R., Castillo, A. G. (2013). Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 199, 464–475. doi: 10.1111/nph.12286
Rojas, M. R., Jiang, H., Salati, R., Xoconostle-Cázares, B., Sudarshana, M. R., Lucas, W. J., et al. (2001). Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus‘. Virology 291, 110–125. doi: 10.1006/viro.2001.1194
Rojas, M. R., Hagen, C., Lucas, W. J., Gilbertson, R. L. (2005). Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43, 361–394. doi: 10.1146/annurev.phyto.43.040204.135939
Rojas, M. R., Macedo, M. A., Maliano, M. R., Soto-Aguilar, M., Souza, J. O., Briddon, R. W., et al. (2018). World Management of Geminiviruses. Annu. Rev. Phytopathol. 56, 637–677. doi: 10.1146/annurev-phyto-080615-100327
Ruhel, R., Chakraborty, S. (2019). Multifunctional roles of geminivirus encoded replication initiator protein. VirusDis 30, 66–73. doi: 10.1007/s13337-018-0458-0
Sánchez-Durán, M. A., Dallas, M. B., Ascencio-Ibañez, J. T., Reyes, M., II, Arroyo-Mateos, M., Ruiz-Albert, J., et al. (2011). Interaction between geminivirus replication protein and the SUMO-conjugating enzyme is required for viral infection. J. Virol. 85, 9789–9800. doi: 10.1128/JVI.02566-10
Sørensen, B. B., Ehrnsberger, H. F., Esposito, S., Pfab, A., Bruckmann, A., Hauptmann, J., et al. (2017). The Arabidopsis THO/TREX component TEX1 functionally interacts with MOS11 and modulates mRNA export and alternative splicing events. Plant Mol. Biol. 93, 283–298. doi: 10.1007/s11103-016-0561-9
Sambrook, J., Russel, David, W., Maniatis, T. (2001). Molecular cloning Vol. vol. 1-3 (New York: Cold Spring Habour Laboratory Press).
Sasikumar, A. N., Perez, W. B., Kinzy, T. G. (2012). The many roles of the eukaryotic elongation factor 1 complex. WIREs RNA 3, 543–555. doi: 10.1002/wrna.1118
Schneider, J., Wolff, T. (2009). Nuclear functions of the influenza A and B viruses NS1 proteins: do they play a role in viral mRNA export? Vaccine 27, 6312–6316. doi: 10.1016/j.vaccine.2009.01.015
Settlage, S. B., See, R. G., Hanley-Bowdoin, L. (2005). Geminivirus C3 protein: replication enhancement and protein interactions. J. Virol. 79, 9885–9895. doi: 10.1128/JVI.79.15.9885-9895.2005
Shahin, E. A. (1985). Totipotency of Tomato Protoplasts. Theor. Appl. Genet. 69, 235–240. doi: 10.1007/BF00662431
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shung, C. Y., Sunter, G. (2007). AL1-dependent repression of transcription enhances expression of Tomato golden mosaic virus AL2 and AL3. Virology 364, 112–122. doi: 10.1016/j.virol.2007.03.006
Singh, D. K., Islam, M. N., Choudhury, N. R., Karjee, S., Mukherjee, S. K. (2007). The 32 kDa subunit of replication protein A (RPA) participates in the DNA replication of Mung bean yellow mosaic India virus (MYMIV) by interacting with the viral Rep protein. Nucleic Acids Res. 35, 755–770. doi: 10.1093/nar/gkl1088
Smaczniak, C., Li, N., Boeren, S., America, T., van Dongen, W., Goerdayal, S. S., et al. (2012). Proteomics-based identification of low-abundance signaling and regulatory protein complexes in native plant tissues. Nat. Protoc. 7, 2144–2158. doi: 10.1038/nprot.2012.129
Strässer, K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., et al. (2002). TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417, 304–308. doi: 10.1038/nature746
Sunter, G., Hartitz, M. D., Hormuzdi, S. G., Brough, C. L., Bisaro, D. M. (1990). Genetic-Analysis of Tomato Golden Mosaic-Virus: ORF AL2 Is Required for Coat Protein Accumulation While ORF AL3 Is Necessary for Efficient DNA Replication. Virology 179, 69–77. doi: 10.1016/0042-6822(90)90275-V
Suyal, G., Mukherjee, S. K., Choudhury, N. R. (2013a). The host factor RAD51 is involved in mungbean yellow mosaic India virus (MYMIV) DNA replication. Arch. Virol. 158, 1931–1941. doi: 10.1007/s00705-013-1675-x
Suyal, G., Mukherjee, S. K., Srivastava, P. S., Choudhury, N. R. (2013b). Arabidopsis thaliana MCM2 plays role(s) in mungbean yellow mosaic India virus (MYMIV) DNA replication. Arch. Virol. 158, 981–992. doi: 10.1007/s00705-012-1563-9
Suyal, G., Rana, V. S., Mukherjee, S. K., Wajid, S., Choudhury, N. R. (2014). Arabidopsis thaliana NAC083 protein interacts with Mungbean yellow mosaic India virus (MYMIV) Rep protein. Virus Genes 48, 486–493. doi: 10.1007/s11262-013-1028-6
Tameling, W., II, Baulcombe, D. C. (2007). Physical association of the NB-LRR resistance protein Rx with a Ran GTPase–activating protein is required for extreme resistance to Potato virus X‘. Plant Cell 19, 1682–1694. doi: 10.1105/tpc.107.050880
Tan, M. L. M. C., Rietveld, E. M., Vanmarrewijk, G. A. M., Kool, A. J. (1987). Regeneration of Leaf Mesophyll Protoplasts of Tomato Cultivars (L. esculentum) Factors Important for Efficient Protoplast Culture and Plant Regeneration. Plant Cell Rep. 6, 172–175. doi: 10.1007/BF00268470
van Schie, C. C. N., Takken, F. L. W. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Verlaan, M. G., Hutton, S. F., Ibrahem, R. M., Kormelink, R., Visser, R. G. F., Scott, J. W., et al. (2013). The Tomato Yellow Leaf Curl Virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet. 9, e1003399. doi: 10.1371/journal.pgen.1003399
Vizcaino, J. A., Csordas, A., Del-Toro, N., Dianes, J. A., Griss, J., Lavidas, I., et al. (2016). 2016 update of the pride database and its related tools. Nucleic Acids Res. 44, D447–D456. doi: 10.1093/nar/gkw880
Wang, L., Ding, X., Xiao, J., Jiménez-Gόngora, T., Liu, R. Y., Lozano-Durán, R. (2017a). Inference of a Geminivirus-Host Protein-Protein Interaction Network through Affinity Purification and Mass Spectrometry Analysis. Viruses 9, 275. doi: 10.3390/v9100275
Wang, L., Tan, H., Wu, M., Jimenez-Gongora, T., Tan, L., Lozano-Duran, R. (2017b). Dynamic Virus-Dependent Subnuclear Localization of the Capsid Protein from a Geminivirus. Front. Plant Sci. 8, 2165. doi: 10.3389/fpls.2017.02165
Xie, Q., Sanz-Burgos, A. P., Hannon, G. J., Gutiérrez, C. (1996). Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 15, 4900–4908. doi: 10.1002/j.1460-2075.1996.tb00870.x
Xu, M., Mazur, M. J., Tao, X., Kormelink, R. (2020). Cellular RNA Hubs: Friends and Foes of Plant Viruses. Mol. Plant Microbe Interact. 33, 40–54. doi: 10.1094/MPMI-06-19-0161-FI
Yamaguchi, H., Ohnishi, J., Saito, A., Ohyama, A., Nunome, T., Miyatake, K., et al. (2018). An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 131, 1345–1362. doi: 10.1007/s00122-018-3082-x
Yarbrough, M. L., Mata, M. A., Sakthivel, R., Fontoura, B. M. A. (2014). Viral subversion of nucleocytoplasmic trafficking. Traffic 15, 127–140. doi: 10.1111/tra.12137
Yelina, N. E., Smith, L. M., Jones, A. M. E., Patel, K., Kelly, K. A., Baulcombe, D. C. (2010). Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 107, 13948–13953. doi: 10.1073/pnas.0911341107
Zerbini, F. M., Briddon, R. W., Idris, A., Martin, D. P., Moriones, E., Navas-Castillo, J., et al. (2017). ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 98, 131–133. doi: 10.1099/jgv.0.000738
Keywords: geminivirus, Tomato yellow leaf curl virus, viral replication, proteomics, PCNA, Rep, interactome
Citation: Maio F, Helderman TA, Arroyo-Mateos M, van der Wolf M, Boeren S, Prins M and van den Burg HA (2020) Identification of Tomato Proteins That Interact With Replication Initiator Protein (Rep) of the Geminivirus TYLCV. Front. Plant Sci. 11:1069. doi: 10.3389/fpls.2020.01069
Received: 28 February 2020; Accepted: 29 June 2020;
Published: 15 July 2020.
Edited by:
Araceli G. Castillo, Institute of Subtropical and Mediterranean Horticulture La Mayora, SpainReviewed by:
Björn Krenz, German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), GermanyChristina Wege, University of Stuttgart, Germany
Linda Hanley-Bowdoin, North Carolina State University, United States
Copyright © 2020 Maio, Helderman, Arroyo-Mateos, van der Wolf, Boeren, Prins and van den Burg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Harrold A. van den Burg, SC5BLnZhbmRlbkJ1cmdAdXZhLm5s
†Present address: Francesca Maio, Spark Genetics B.V., Utrecht, Netherlands
 Francesca Maio
Francesca Maio Tieme A. Helderman
Tieme A. Helderman Manuel Arroyo-Mateos1
Manuel Arroyo-Mateos1 Sjef Boeren
Sjef Boeren Harrold A. van den Burg
Harrold A. van den Burg