- 1Departments of Chemistry and Biology, Emory University, Atlanta, GA, United States
- 2Infectious Diseases and Genomic Medicine Group, J Craig Venter Institute, Rockville, MD, United States
- 3Department of Biology, University of Richmond, Richmond, VA, United States
- 4Department of Biology, University of Pennsylvania, Philadelphia, PA, United States
The rich collection of microbes colonizing the plant root making up the rhizosphere function as a multigenomic organ for nutrient distribution. The extent to which its dynamic mutualistic cellular order depends on morphogenic signaling, while likely, remains unknown. We have shown that reaction-diffusion chemical networks constructed with model plant and bacterial metabolites can mimic processes ranging from oxidative burst kinetics to traveling waves and extracellular stationary state reaction-diffusion networks for spatiotemporal ordering of the rhizosphere. Plant parasites and pathogens can be limited by host attachment require dynamic informational networks and continue to provide insight into what controls the rhizosphere. Here we take advantage of Agrobacterium tumefaciens, a plant pathogen with a gated receptor that requires simultaneous perception of two plant metabolites. Genetic manipulations have created receptors allowing each metabolite concentration to be correlated with pathogen behavior. The development of the florescent strains used here provide initial maps of the reaction-diffusion dynamics existing in the rhizosphere, revealing significant differences in the signaling landscape of host and non-host plants before and after wounding, specifically highlighting networks that may inform rhizosphere organization.
Introduction
The rhizosphere, that narrow zone of soil along plant root surfaces containing bacteria, viruses, fungi, and numerous metabolites, functions as an “external metabolome” (Bais et al., 2006; Badri and Vivanco, 2009; Mendes et al., 2011; Philippot et al., 2013; Huang et al., 2014; Glasser et al., 2017). As with any mutualistic multicellular network, organization is likely critical for metabolome function (Dietrich et al., 2008; Stacy et al., 2014; Stacy et al., 2015; Whiteley et al., 2017). The plant root provides reduced carbon and dioxygen to the rhizosphere (Walker et al., 2003), setting up both ends of life’s universal redox network. Indeed, redox-active phenols, quinones, flavins, and phenazines (Bais et al., 2006; Tomilov et al., 2006; Uteau et al., 2015; Rasmann and Turlings, 2016) are prevalent in the rhizosphere where redox active processes have been highlighted in biofilm biogeography (Stacy et al., 2014), allelopathy (Tomilov et al., 2006), quorum sensing (Fuller et al., 2017), and most notably, in the critical spatiotemporal dynamics of semagenesis ensuring success of the parasitic plants (Keyes et al., 2007; Liang et al., 2016; Fuller et al., 2017). The initial discoveries of the reaction diffusion dynamics of semagenesis (Chang et al., 1986; Fate et al., 1990; Smith et al., 1990) motivated further explorations of model reaction-diffusion systems in the rhizosphere (Taran et al., 2019). Given that the developed models have now greatly expanded how reaction-diffusion networks might contribute to the richly dynamic cellular architecture of the rhizosphere, we sought to develop specific probes to map the processes within and on the surfaces of plant tissues with and without environmental perturbation, specifically focusing on plant wounding.
Virulent strains of Agrobacterium tumefaciens, more recently classified as Rhizobium radiobacter, genetically transform many dicotyledonous angiosperms and gymnosperms resulting in the formation of tumors at the site of infection (McCullen and Binns, 2006; Nester, 2014; Binns and Zhao, 2020). These soil bacteria transition from a saprophytic state to a pathogenic state in a process that requires (1) identification of and attachment to host cells that are competent to be transformed and (2) the expression of a series of ‘virulence’ (vir) genes located on their tumor inducing (Ti) plasmids. The latter process is initiated by multiple small molecule signals, specifically low pH, phenols, and sugars, which appear to be characteristic of host wound environments (McCullen and Binns, 2006; Binns and Zhao, 2020). Simple sugars and phenols from the host then serve as xenognostic signals that initiate activation of the virulence genes through the histidine kinase VirA via an integrating AND gate mechanism (Fang et al., 2015).
Fluorescent protein reporters and variants of VirA with altered capacities to recognize the xenognostic signals (McCullen and Binns, 2006; Binns and Zhao, 2020) enable the use of Agrobacterium as a dynamic probe of the extracellular milieu surrounding the host tissues. We accordingly have engineered the signal input modules in Agrobacterium strains carrying gfp under the control of virulence gene promoters to create biosensors. Experiments with these reporter strains provide evidence that Agrobacterium cells accumulate around viable host cells at the wound surface. Different tissues of the same plant, including stem, midrib, and leaf, vary in their wound-induced accumulation, consistent with expected phenol content. This wound-induced signaling response is enhanced in mature plants, consistent with the lower susceptibility of seedlings to tumorigenesis (Robbs et al., 1991). Using a VirA mutant strain that responds to sugar independent of phenol (Fang et al., 2015), we show that wound-induced phenol levels accumulate in host tissues and not in non-host controls. In contrast, wound-induced sugar exudation is similar in host and non-host tissues. Taken together, tumor formation appears uniquely dependent on xenognostic phenol release. Indeed, a strain of Agrobacterium that is hypersensitive to sugar initiates vir gene production, but induces tumors poorly when compared to wildtype Agrobacterium, highlighting this critical role of phenols for pathogenesis. These Agrobacterium strains then serve as valuable probes of the reaction-diffusion networks at plant wound sites, and most importantly, open strategies for similar constructs in other mutualistic microbes to more broadly define the organization necessary for a functioning rhizosphere.
Materials and Methods
Plasmid Construction and Agrobacterium Strains
E. coli strain XL1-Blue was used for plasmid construction, and a list of all plasmids and strains are found in Table 1. pYL355, which contains virA (Y293F) and genes flanking both sides of virA for specific complementation to allow double crossover, was generated by ligating the KpnI fragment from pQF431 into the KpnI-digested pAW162 (Liu, 2012). The BamHI/EcoRI fragment was released from pYL355 and ligated with the BamHI/EcoRI digested pAW190, a derivative of pK18mobsacB, to generate pYL356. Agrobacterium strains with virA(Y293F) substitution on the Ti plasmid were developed by pK18mobsacB-mediated homologous recombination (Schäfer et al., 1994). To generate YHL300, pYL356 was transformed into 358mxGFP, an engineered A348 strain containing virE::GFP as the virulence reporter (Goulian and van der Woude, 2006). The first crossover for virA(Y293F) incorporation was selected by kanamycin resistance, and a single colony was chosen and cultivated overnight in LB at 28°C. The saturated culture was 1:200, 1:1,000, and 1:5,000 diluted with fresh LB, and 100 µl of each dilution was spread onto LB plates containing 10% sucrose. Successful incorporation of virA(Y293F) was verified by sequencing (Beckman Coulter Genomics), and resulted in YHL300. YHL301 was generated by a single transformation of YHL300 with pMP7605, a plasmid containing tac-driven m-Cherry. YHL320 was generated similarly with the method described above, but a transformation of pYL356 into AB520, an A348 derivative with a deletion of the sugar transporter MmsB (Zhao and Binns, 2011; Liu, 2012). After the selection on sucrose plates, successful creation of YHL320 was verified by sequencing, and a dual transformation of pMP7605 and pRG182, a plasmid containing VirB-GFP as the virulence reporter, gave rise to YHL324s+.
Culturing, Inoculation, and Co-Cultivation
All Agrobacterium inoculums were prepared by growth overnight in LB liquid medium with 100 µg/ml gentamicin for AB650wt/YHL301s+, or 15 µg/ml kanamycin and 100 µg/ml gentamicin for YHL324s+ (Liu, 2012). The overnight bacterial culture is pelleted and resuspended to OD0.2 in half strength Murashige and Skoog medium (0.5X MS) buffered to pH5.5 with 50 mM 2-(N-morpholino) ethanesulfonic acid (MES) and mixed with 0.005% triton, and then incubated with plant material for 10 min (mature Nt or Zm), 5 min (young Nt seedlings), or 1 min (Sa seedlings) for inoculation. The bacterial suspension was washed off and the plant material was placed on three types of co-cultivation medium in petri dishes: glucose plates, AS plates and glycerol plates, which contain 0.5X MS medium (pH5.5) supplemented with 56 mM glucose, 100 µM AS, or 100 mM glycerol, respectively. Glucose plates are used for measuring AS equivalents released from plant material, AS plates are for measuring glucose equivalents, and glycerol plates are for measuring a combination of the signals released from the plant material (induction potential). The AS dose responses and glucose dose responses were performed in triplicate in 0.5X MS (pH5.5) liquid medium supplemented with either 56 mM glucose or 100 mM glycerol for measuring AS dose response, or supplemented with 100 µM AS or no AS for measuring the glucose dose response. The range of concentration titrated was 0, 0.1, 1, 10, 100, 300 µM AS or 0, 0.01, 0.1, 1, 10, 100 mM glucose. In all treatments, 15 µg/ml or 50 µg/ml of kanamycin was added to liquid culture or agar plates of YHL324 s+ incubation to maintain the GFP containing plasmid pRG182. Plasmid maintenance was secure in culture, but less stable in planta.
Preparation of Plant Material and Growth Media
Seeds of tobacco (Nicotiana tabacum cv. Havana 38) and Zea mays were purchased from LEHLE Seeds. Prior to germination, seeds were surface sterilized with 50% bleach and 0.1% triton for 10 min, and then washed with excess sterile water for 4–6 times. Seedlings were grown on 0.5X MS + 1% sucrose solid medium (containing filtered sucrose to avoid glucose contamination through autoclaving) until two true leaves emerged and expanded. Some seedlings (>3) were inoculated for examination and the others (>3) were transplanted to soil for obtaining mature plants. Seeds of Striga asiatica were received from USDA under quarantine, surface sterilized with 3% chromic acid, 50% bleach and 0.1% triton for 10 min, and 70% ethanol for 2 min, being rinsed thoroughly with sterile water between each treatment. The seeds were then incubated in sterile water at 28°C for 7–10 days before germination was induced hydroponically in 0.1 mM KCl solution with 10 nM Strigol. Haustorium development was induced hydroponically in 0.1 mM KCl solution with 10 µM DMBQ (Chang and Lynn, 1986).
Wounding mature tobacco or two-month-old maize plants was done by cutting stem/midrib explants with razor blades, or leaf explants with a cork borer (Figure S1). Wounded seedlings of tobacco or S. asiatica were created by pinching hypocotyl/root with pointed tweezers, or cutting the cotyledon with small scissors. Wounding of Kalanchoe diagremontiana was performed with a wooden dowel scraping the cuticle of the leaf 3–4 times.
All plant-Agrobacterium co-cultivation took place on 0.5X MS plates at pH5.5. The pH is buffered to pH5.5 because we found that without buffer the plant tissues changed the pH of the growth media and caused sporadic and/or large fluctuations in the virulence response (data not shown).
Flow Cytometry
A population of Agrobacteria were washed off the plant tissue by vortexing the co-cultivated plant tissue in 0.5X MS liquid medium (pH5.5) with confocal confirmation of effective bacterial removal. The GFP fluorescence of each bacterium was measured by flow cytometry and a population of 10,000 bacteria was counted from each sample with each condition being performed in triplicate. The average GFP fluorescence per bacterium was plotted. GFP fluorescence was excited by a blue sapphire laser at 488 nm laser with emission BP at 525/50 using the long pass 505 nm filter. m-Cherry fluorescence was excited by a green laser at 532 nm with the emission BP at 610/20 using the long pass 600 nm filter.
Fluorescence Imaging
A Zeiss Laser Confocal Scanning Microscope LSM 510 META was used for fluorescence imaging. Argon/2 laser at 488 nm was used to excite GFP and a filter of 505–530 nm was applied for detecting GFP emission. Helium Neon later at 543 nm was used to excite m-Cherry and a filter of 585–615 nm was applied to detect m-Cherry emission. DIC was applied to acquire bright field images. All representative images are presented as a merge of GFP, m-Cherry, and DIC channels.
Results
Construction of Agrobacterium as a Biosensor
We constructed three Agrobacterium tumefaciens strains as biosensors to evaluate the spatiotemporal distribution of phenols and sugars at plant wound sites (Table 2). We began by obtaining a previous strain, 358mxGFP (Goulian and van der Woude, 2006), that contains the gene for green fluorescent protein (GFP) as a replacement for the virulence gene virE2 in strain A348wt. This strain functions as a nonvirulent fluorescent reporter of plant wound signal reception (Liu, 2012). Additionally, the gene encoding mCherry fluorescence protein under the constitutive Tac promoter located in a separate plasmid, pMP7605 (Lagendijk et al., 2010), was added to this strain to give AB650wt. The discovery of a signal integration node in VirA (Fang et al., 2015) led to a simple mutation that separated the co-dependence on sugar and phenol for induction of virulence signals. Insertion of VirAY293F for wild-type VirA in AB650wt generated sugar-sensitive strain YHL301ss, which can be induced by sugar or phenol alone. A third strain, YHL324s+, was prepared by inserting VirAY293F in place of wild-type VirA in AB520. AB520 is a strain carrying a deletion of the multiple monosaccharide transporter B (ΔmmsB) (Zhao and Binns, 2011), which enhances the sensitivity of Agrobacterium to vir-inducing sugars by increasing the relative sugar concentration in the periplasm (Hu et al., 2012). A separate plasmid, pRG182, carrying VirB-GFP was added into YHL324s+ to enable sugar sensing independent of phenols, but with significantly higher sensitivity than YHL301ss. Although YHL324s+ has a different GFP source than AB650wt and YHL301ss and has therefore been left out of any quantitative comparisons between AB650wt and YHL301ss, it has proved useful as a tool for reporting bacterial position due to its signal hypersensitivity. Taken together, strains AB650wt, YHL301ss, and YHL324s+ provide the resources for evaluating the relative levels of sugars and phenols at the wound site of hosts and non-host plants.
The fluorescence of the AB650wt and YHL301ss strains in response to acetosyringone (AS) and glucose was measured independently [100 µM AS or 56 mM (1%) glucose] in liquid media (Figure 1). While we obtained similar data with the YHL324s+ strain (Figure S1), we recognize that this is an imperfect comparison due to the different GFP sources and have therefore focused on the AB650wt and YHL301ss strains. A summary of the sensitivity of these two strains is reported as the ED50 in Table 3. While AB650wt requires AS independent of the concentration of glucose (Figure 1A), YHL301ss is strongly induced by glucose alone (Figure 1B) and displays enhanced sensitivity to phenols (Figure 1C). Maximal induction and sensitivity to AS in both strains are enhanced in the presence of glucose (Figure 1B vs D).
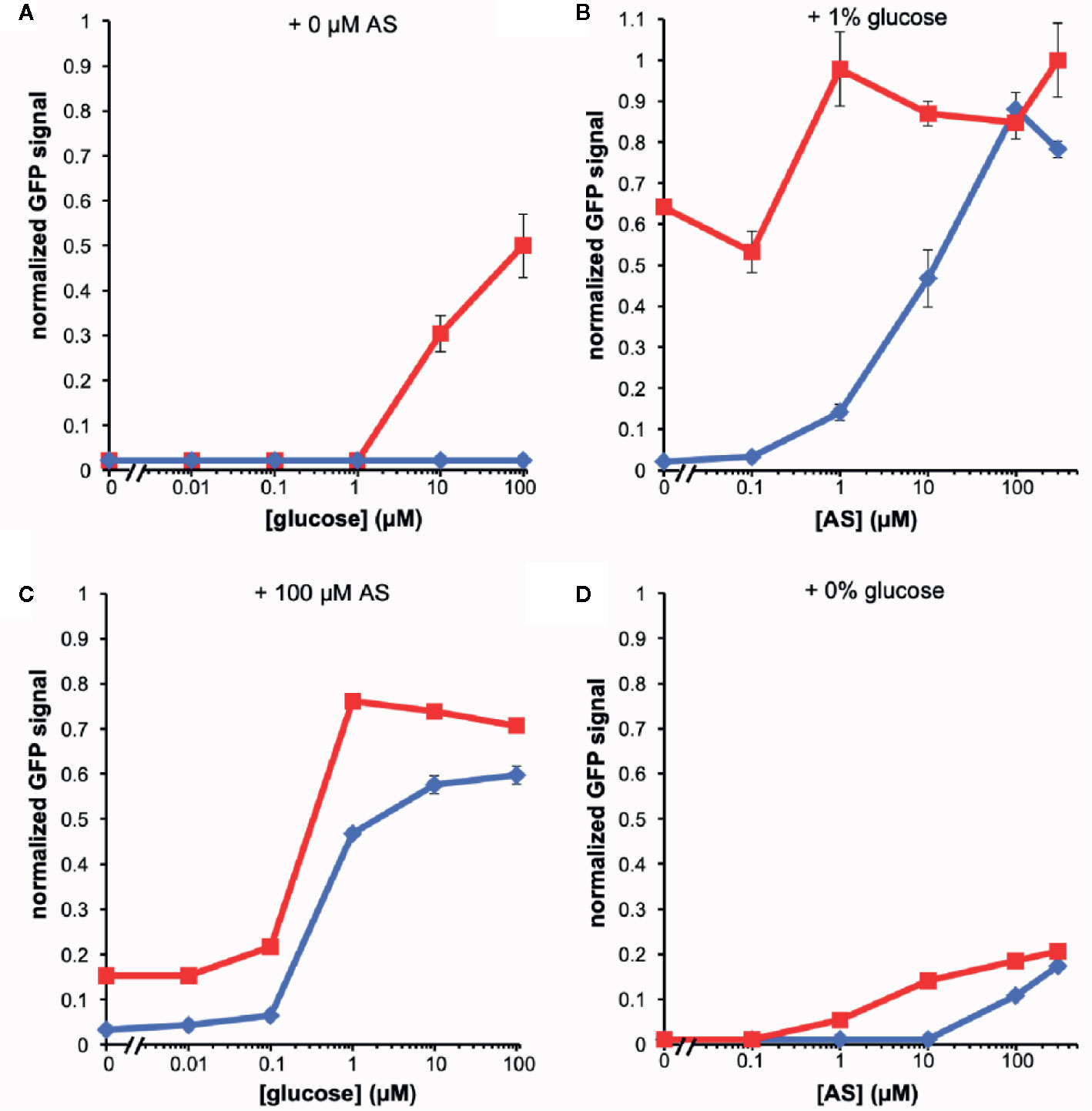
Figure 1 Dose responses of A. tumefaciens fluorescent strains to acetosyringone (AS) or glucose. Upon addition of AS or glucose, virE::gfp [AB650wt (blue diamonds), YHL301ss (red squares)] signal was measured and normalized to the maximal value of YHL301ss GFP signal. Glucose dose responses with no AS (A) or 100 µM AS (C) in the growth medium and AS dose responses with 1% glucose (B) or 1% glycerol (D) in the growth medium. Glycerol is used as a non-inducing carbon source in the absence of glucose. Each data point represents the average of three biological replicates and error bars show ± SE.
Agrobacterium Cells Accumulate in the Apoplast of N. tabacum at Wound Sites
Agrobacterium will naturally colonize plants and form benign bio-films on root surfaces (Heindl et al., 2014) and it is generally accepted that antimicrobial substances released at the wound can limit colonization (Akinsulire et al., 2008; Taye et al., 2011; Meij et al., 2018). We show here that Agrobacterium cells are able to colonize wounded mature tobacco explants (Figure 2A). Unfortunately, without selection either or both markers can be lost in a portion of our bacteria and we cannot use quantitative expression under these conditions; nevertheless, co-incubation with our YHL324s+ strain on 0.5X MS plates confirms the presence of bacteria in optical sections 60-80 µm below the apical surface, corresponding to several cell layers into the wound (Figure 2B). Wildtype Agrobacterium cells (AB650wt) are not detected in unwounded two-week-old tobacco seedlings (Figure 2C), but appear in the apoplast of wounded tobacco seedlings (Figure 2D). In general, young tobacco seedlings do not induce GFP production similarly to mature tobacco plants, which we explore further. These analyses suggest that a wounding event either lowers a physical barrier and/or produces chemical attractants such as sugars or phenols for Agrobacterium colonization of viable cells (Loake et al., 1988).
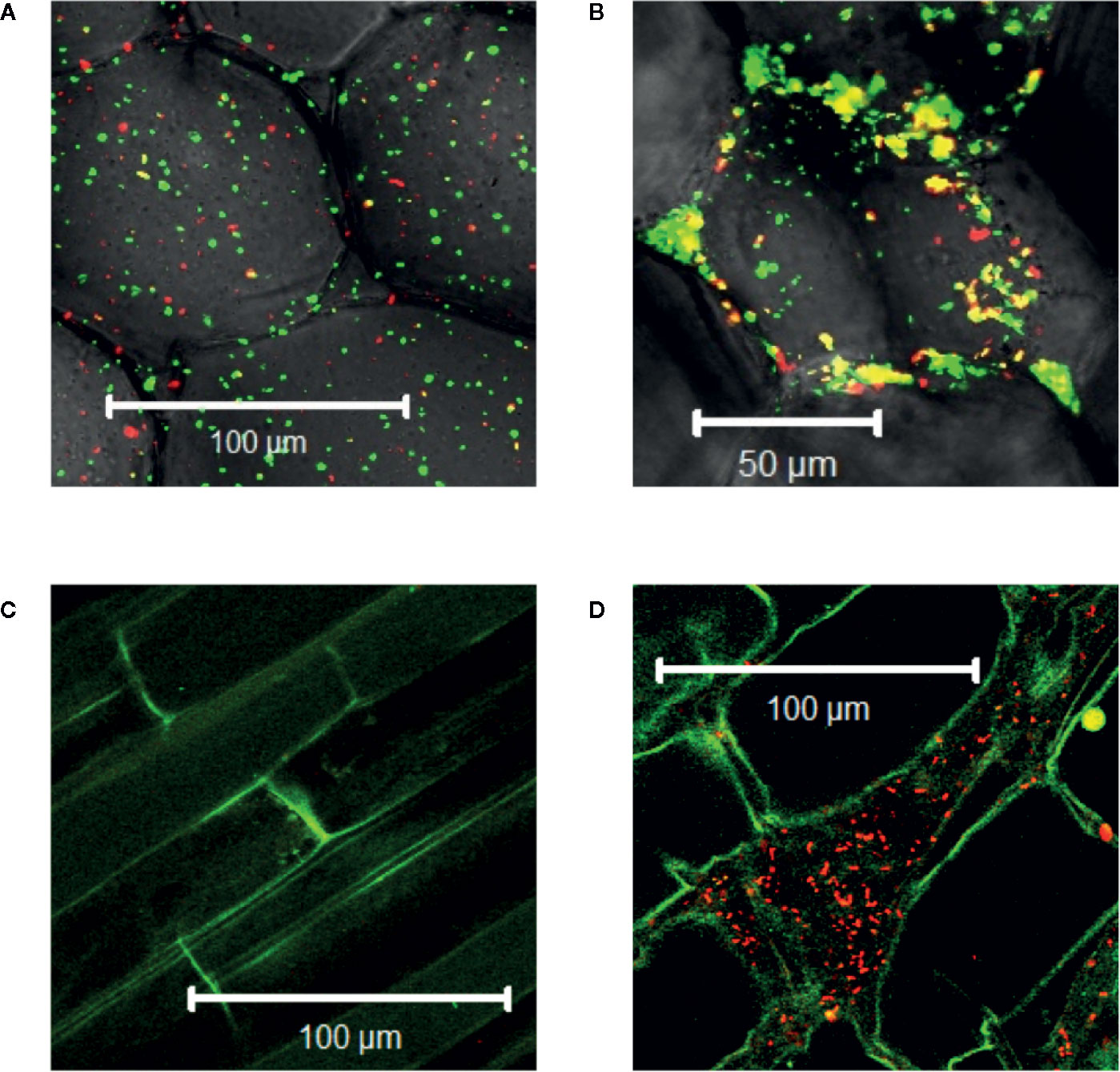
Figure 2 Agrobacterium cells colonizing plants localize to the apoplast. YHL324s+ colonizing the pith tissue of tobacco stem explants on the surface (A) or 60 µm below the surface (B). Unfortunately, co-expression of both markers was not uniform in the absence of selection, limiting quantitative assessment. Nevertheless, unwounded (C) and wounded (D) tobacco seedlings co-cultivated with AB650wt reveal distinct bacterial colonization in the wounded plant. The cells walls of the tobacco seedlings auto-fluoresce, allowing for clear delineation of plant cell structure.
Virulence-Inducing Phenols Accumulate at Living Cells in a Wound
Agrobacterium chemotaxis has been recognized and suggested to be required for functional colonization and infection (Chesnokova et al., 1997; Merritt et al., 2007). Micromolar concentrations of sugars and nanomolar amount of wound phenolics have been reported to attract Agrobacterium to the plant wounds (Ashby et al., 1988; Loake et al., 1988; Winans, 1992). As phenolic monomers and acidic pH are characteristic of plant cell walls and vacuoles, it is possible that phenols and sugars can be released from breached cells, however, Agrobacterium needs to target living cells for transformation. We compared Agrobacterium localization between vascular tissue (comprised of a majority of dead cells) and the adjacent pith or cortex tissue (comprised of living cells) at a cut site in the tobacco stem. Using YHL324s+, we observed colonization and induction on the surface of the stem explants (Figure 3A), however the bacteria were not detected in the vascular channels even though these wide conduits are readily accessible to the bacteria. Instead, accumulation occurred 50–60 μm below the surface in the adjacent cortex tissue within two days of co-cultivation (Figure 3B, Figure S3). While this was a reproducible result, there remains the possibility that these observations are due to alternative differences between these cell types, including the transmittance efficiency, and this will need to be addressed with further experimentation. Population in the apoplast of the living tissue in the cortex is necessary for successful transformation, but how that behavior may be mediated by xenognostic phenol/sugar signals or other molecules impacting Agrobacterium localization will now need to be determined.
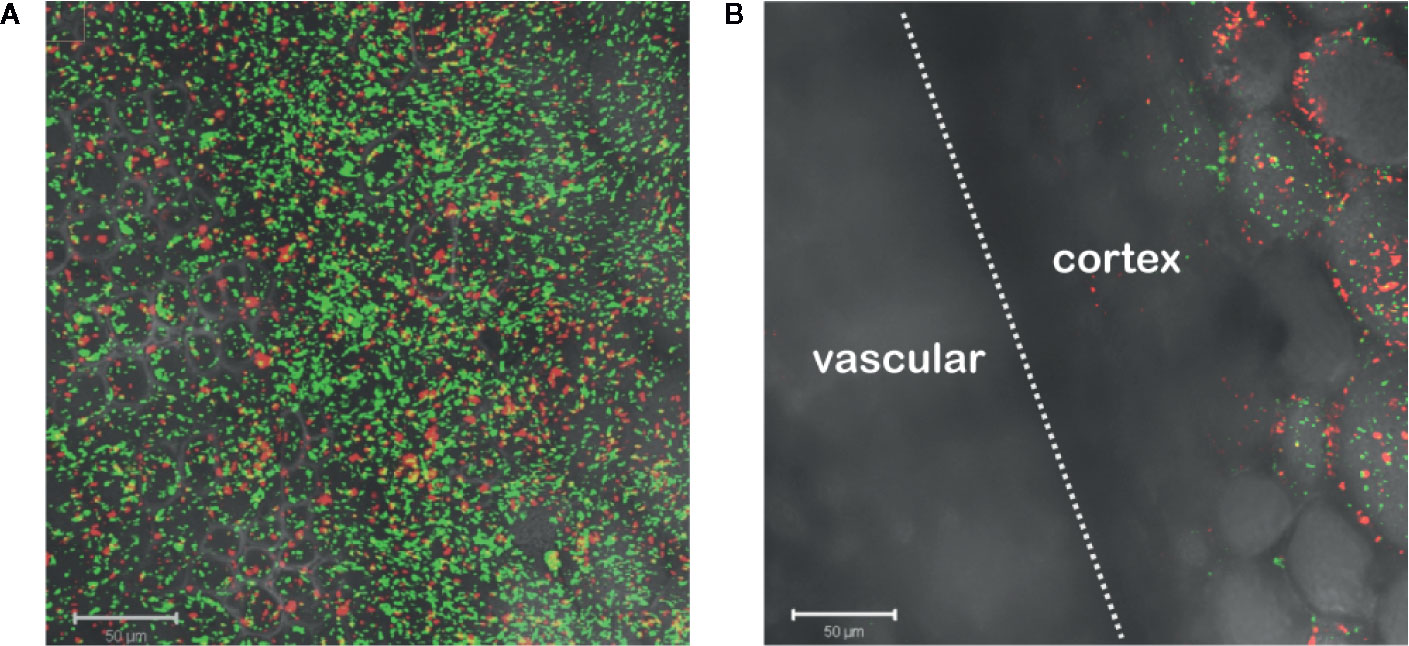
Figure 3 Agrobacterium colonize into the apoplast of tobacco explants within two days of co-cultivation. YHL324s+ colonize on the surfaces (A) and 50 µm below the surface (B) of the stem explants. The white dashed line indicates the junction between vascular tissue and the cortex tissue.
Agrobacterium Behaviors at Monocot and Dicot Wound Sites Differ
Monocots initially appeared resistant to Agrobacterium pathogenesis and indeed the plants lack sufficient amounts of one or more vir inducers upon wounding (Hooykaas, 1989; Smith and Hood, 1995). The successful transformation of rice enhanced with exogenous AS supported that assertion (Hiei et al., 1994; Vijayachandra et al., 1995; Nishimura, 2020). Wall phenol structures from monocots differ widely from those of dicots and even among monocot species (Usami et al., 1988; Messens et al., 1990). Additionally, monocot plant growth regulators or secondary metabolites have been reported to inhibit the process of vir gene induction (Sahi et al., 1990; Zhang et al., 2000; Maresh et al., 2006). Given these results, we were interested in the behavior of our three engineered strains of Agrobacterium at monocot wound sites. Using tobacco as a dicot control and maize as a monocot non-host, midrib explants were inoculated with each of these bacterial strains. AB650wt was induced by the dicot (Figure 4A) but not the monocot (Figure 4B); YHL301ss was strongly induced by the dicot (Figure 4C), but not the monocot (Figure 4D); and YHL324s+ was induced by both (Figures 4E, F), suggesting that both tobacco and maize wound sites contain sufficient sugar to induce the sugar responsive YHL324s+ strain. It is important to note that the YHL324s+ strain does retain virulence while the AB650wt and YHL301ss strains do not (see Table 2), which could contribute to differential induction seen in these experiments.
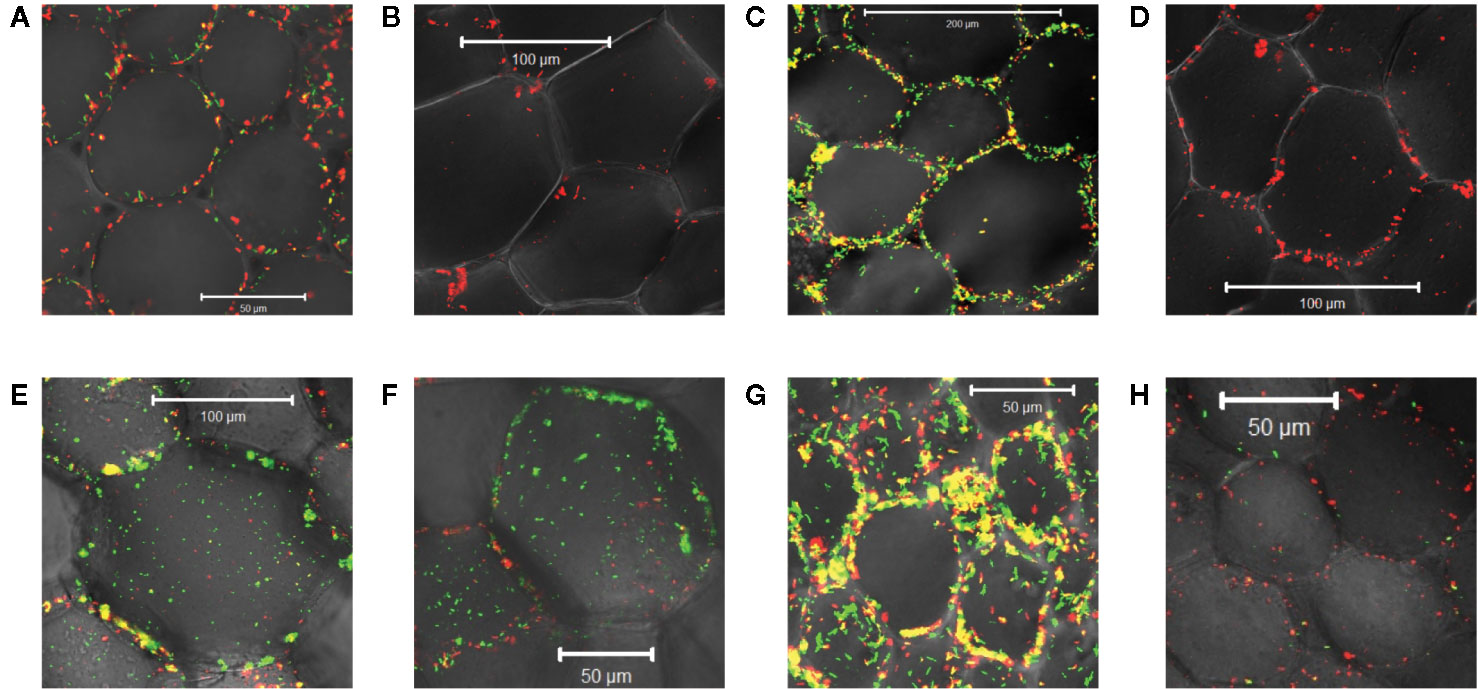
Figure 4 Virulence induction events in tobacco vs. maize midrib explants. AB650wt with explants of tobacco (A) and maize (B) on glycerol plates; YHL301ss with explants of tobacco (C) and maize (D) on glycerol plates; YHL324s+ with explants of tobacco (E) and maize (F) on glycerol plates; AB650wt with tobacco explants on glucose plates (G), and AB650wt with tobacco explants on AS plates (H).
To test whether sugar might limit induction in the tobacco wound site, we co-cultivated the mid-rib explants with AB650wt on 0.5X MS medium supplemented with 56 mM glucose, a saturating sugar concentration for virulence induction. These conditions strongly induced AB650wt (Figure 4G), but when supplemented with 100 μM AS and no sugar, AB650wt was only minimally induced (Figure 4H). These results suggest that these wounded tobacco midrib explants fail to induce high virulence expression (as in Figure 4A) due to low levels of the xenognostic sugar in the wound, as opposed to low phenol levels, and motivated our efforts to quantify GFP expression in each Agrobacterium strain via flow cytometry.
Cell Counts of Agrobacterium at Wound Sites
Bacterial cells harvested from co-cultivation with plant tissues were analyzed via flow cytometry. While absolute quantification is limited by washing efficiency, we sought a relative comparison between similar bacterial strains. Our engineered strains vary in sensitivity to phenol and sugar concentrations as defined in Figure 1 (Liu, 2012). As in Figure 1, we did not include YHL324s+ because the production of GFP in these cells was from a different source than AB650wt and YHL301ss. We compared host vs nonhost plants conditioned by wounding, organ/tissue type, and age (Table 4) with the AS and glucose equivalents estimated according to the ratio of YHL301ss induction to AB650wt induction. To determine whether phenols are limiting for vir-induction in a particular plant tissue, co-cultivations with different bacterial strains were performed in the presence of saturating glucose concentrations. Similarly, to determine whether vir-inducing sugars are limiting in plant tissues, co-cultivations were conducted in the presence of saturating levels of the inducing phenol, AS. The complete comparisons are available in supplemental information (Figures S4–6), including how YHL324s+ responds in this assay, and a typical example in each test group contained in Figure 5 is discussed in the sections below.
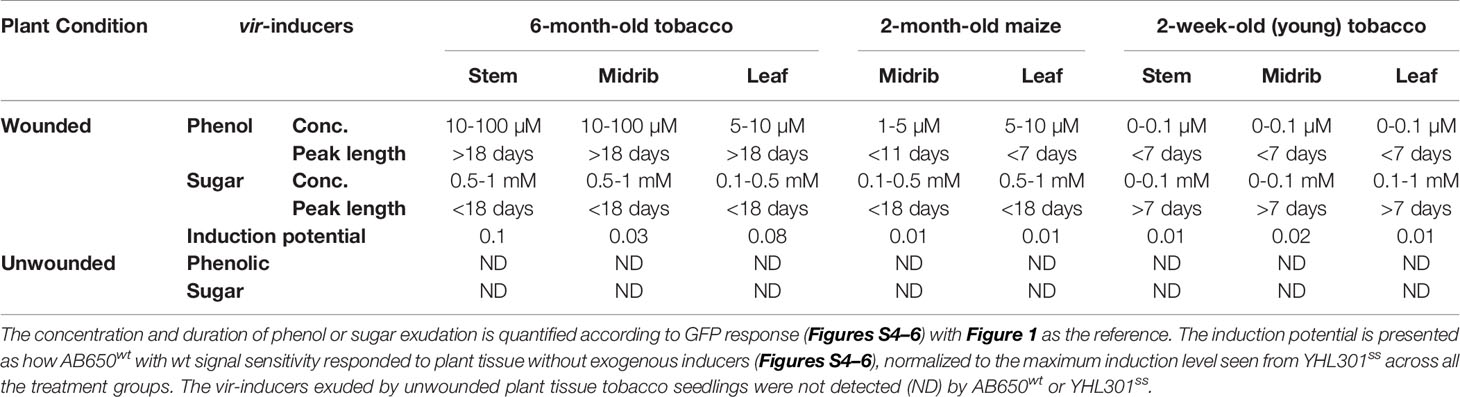
Table 4 Summary of the quantified phenolic, sugar, or combined effect of both inducers, released from tobacco and maize as conditioned by wounding, organ/tissue types, or age.
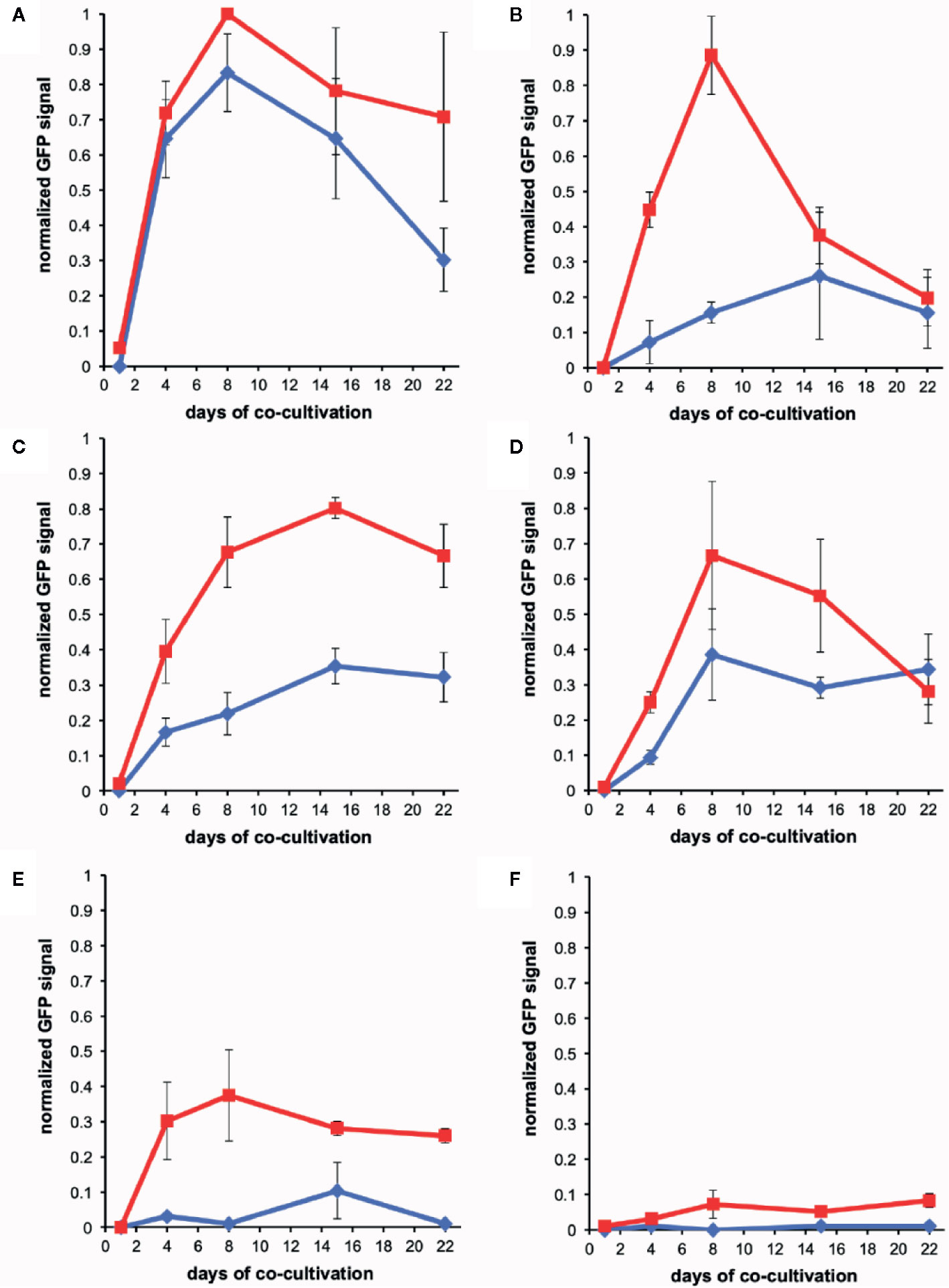
Figure 5 Quantification of wound-induced virulence inducers by co-cultivation. Bacteria were co-incubated with plant explants, vortexed, and GFP intensity was determined using flow cytometry (see Methods). Experiments were performed in triplicate and error bars represent ± SE. AB650wt (blue diamonds) or YHL301ss (red squares) with tobacco midrib explants on glucose plates (A), maize midrib explants on glucose plates (B), tobacco midrib explants on acetosyringone (AS) plates (C), maize midrib explants on AS plates (D), tobacco midrib explants on glycerol plates (E), and maize midrib explants on glycerol plates (F).
Xenognostic Phenols in Wounded Hosts vs. Nonhosts
The wound-released active phenols were compared according to the virulence expression on glucose plates where glucose concentrations are saturating. For these experiments, explants were obtained from tobacco leaves on the midrib near the petiole and always incubated in the same orientation on plates. Similarly, maize explants were excised from the midrib about halfway between the plant stem and leaf tip. On the tobacco midrib explants (Figures 5A, B), YHL301ss and AB650wt responded at similar rates and to similar levels, indicating the explants released saturating concentrations of xenognostic phenols (Figure 1A). The maize midrib explants (Figure 5B) induced YHL301ss four times higher (0.8 vs. 0.2) and twice as fast as AB650wt (8 days vs. 15 days), confirming that the maize explants release lower concentrations of active phenols. The observation that the peak of induction on maize explants disappeared within 4-7 days is consistent with previous reports that, in contrast to dicots, monocots have weaker short-lived wounding responses (Savatin et al., 2014; Hayta et al., 2019). Initial data with AB650wt cultures pre-induced with 100 µM AS on glycerol 0.5X MS plates suggest that it takes up to 8 days after removal of AS for the pre-existing GFP to be completely turned over (Figure S8). Therefore, the duration of phenols released was estimated by measuring for how many days the YHL301ss induction was above the threshold of 0.2 (Table 4).
Xenognostic Sugars in Wounded Hosts vs. Nonhosts
On plates containing 100 µM AS, the ratio of responses between the two Agrobacterium strains is very similar between wounded tobacco and maize explants (Figures 5C, D), suggesting that tobacco and maize release similar amount of sugars at wound sites. This result suggests that the different susceptibility to Agrobacterium infection between host dicots and non-host monocots rests with wound-induced phenols rather than sugars. YHL324s+ however responded very differently on these AS plates relative to the glucose plates, with induction higher than YHL301ss in all cases (Figures S4–S6). Given that it only takes 10–20 mM glucose for complete induction of YHL324s+ (see Figure S2), the AS plates may allow YHL324s+ to perform normally because of the enhanced glucose sensitivity. Finally, testing each of these strains on glycerol plates confirmed both the designation of the YHL301ss strain as sugar sensitive and the observation that dicots can induce a virulence response at conditions where monocots cannot (Figures 5E, F).
Induction by Young Nicotiana tabacum Seedlings Mirrors That of the Parasitic Plant Striga Asiatica
The process of signal generation, or semagenesis, in parasitic plants depends on low cell wall phenol content in the parasite. Here the production of H2O2 at the parasite root tip is proposed to create a mild wound response via peroxidase oxidation of wall phenols in the host in order to generate the haustorial inducing benzoquinone at the host/parasite interface (Chang and Lynn, 1986; Keyes et al., 2007). The parasite has been shown to have low phenol content in its own cell wall to avoid self-response, and this concept has been generalized to root meristematic tissue of other dicots including tobacco (Fuller et al., 2017). In contrast to mature tobacco tissue, two-week-old tobacco seedlings failed to induce AB650wt or YHL301ss (data not shown). These young plants weakly induced YHL324s+ when unwounded (Figure 6A) but do show greater induction upon wounding (Figure 6B) (Liu, 2012). Consistent with the hypothesis that the parasite appears to have weaker phenol content, seedlings of the dicot Striga asiatica do not induce expression in any of the three strains when unwounded (Figure 6C), wounded (Figure 6D), following haustorium induction (Figure 6E), or even following wounding of the haustorium (Figure 6F) on glycerol plates. These results are consistent with a critical role for wall phenols in the host age dependence in Agrobacterium pathogenesis.
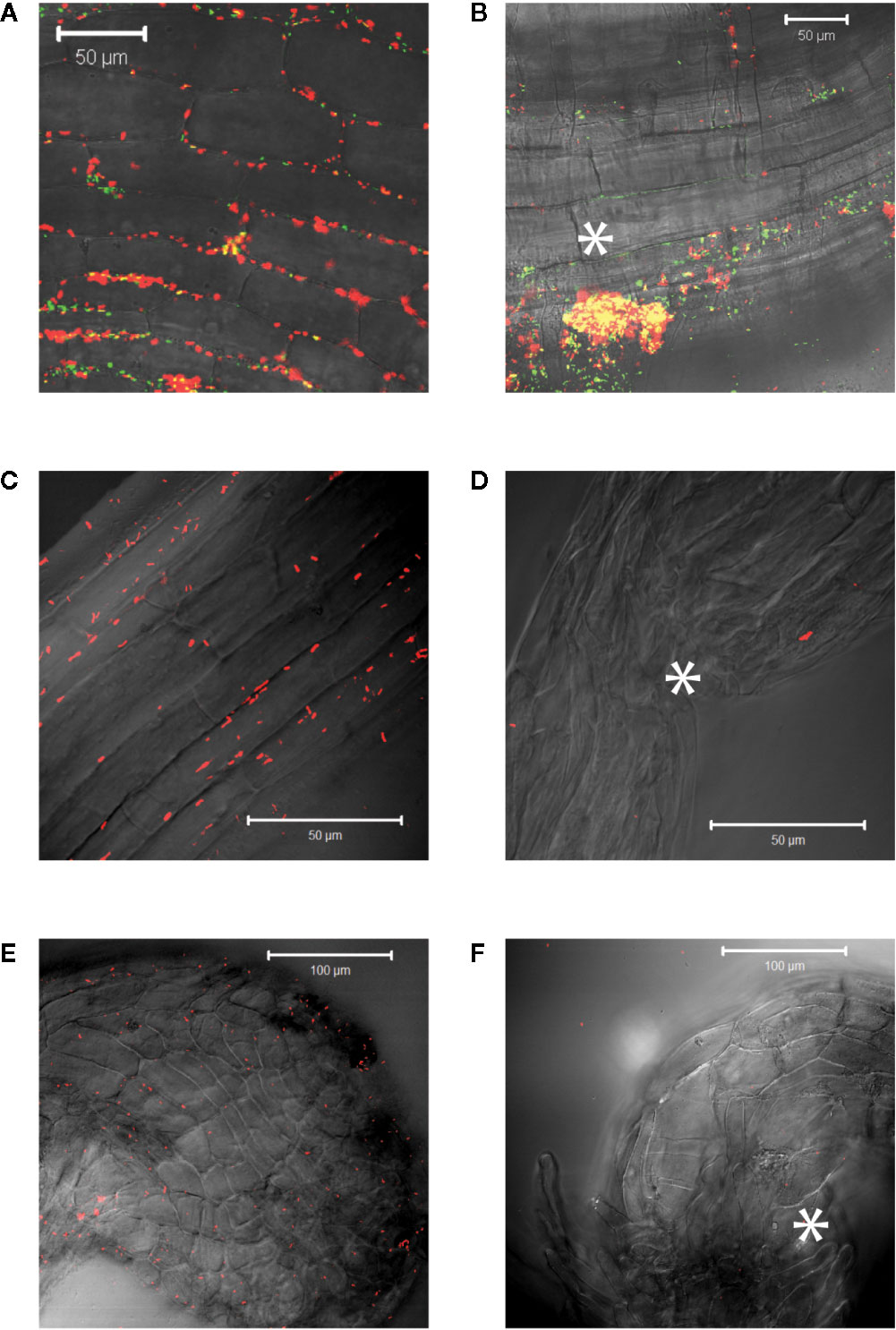
Figure 6 Virulence induction events reveal similarities between young seedlings of different species. Strain YHL324s+ was co-cultivated with two-week old tobacco seedlings on glycerol plates and an unwounded root (A) and a wounded root (B) are shown. A similar experiment was performed with five-day-old S. asiatica seedlings and YHL324s+ on glycerol plates and representative images are shown: unwounded root of a five-day-old S. asiatica seedling (C), a wounded root of a S. asiatica seedling (D), a haustorium of a S. asiatica seedling (E), and a wounded haustorium of a S. asiatica seedling (F). White asterisks indicate approximate wound locations.
Hypersenstive Agrobacterium Strains Differentially Infect Dicots
While the hypersensitive strain YHL324s+ is strongly induced by maize explants, published data indicate that while T-DNA transfer can occur in maize leaf tissues, tumorigenesis was not observed (Shen et al., 1993). These authors suggest this result could arise from the absence of cell division in maize leaf tissues even after transformation. After two weeks of co-cultivation on tobacco explants, however, YHL320s+ (YHL324s+ without the GFP containing plasmid) or wild type strain A348wt initiated tumor growths on the leaves and midribs of tobacco (Figures 7A–H) (Liu, 2012). Interestingly, YHL320s+ caused the stem explants to display necrosis, including a reduction in pith tissue and, possibly for that reason, no tumor production (Figures 7I, J). In contrast, A348wt bacteria did not cause such severe necrosis and these stem segments were able to form tumors (Figures 7K, L). Including glucose in the media aided tumor formation with A348wt in tobacco, but not for YHL320s+ (Table 4).
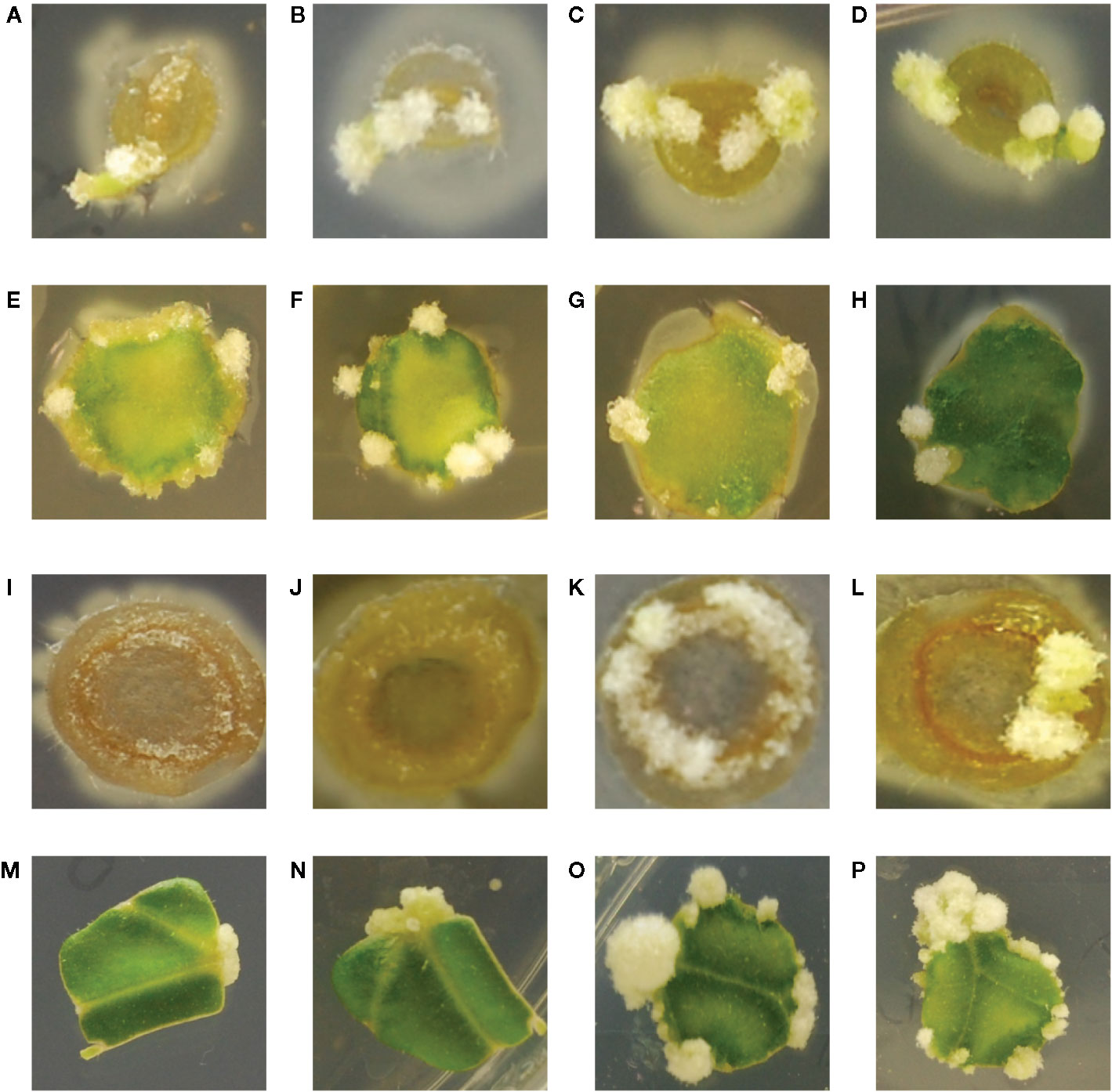
Figure 7 Crown gall tumor induction on tobacco explants. Using mature plants started out in tobacco midrib explants were co-cultivated with YHL320s+ on glucose plates (A), and glycerol plates (B), or with A348wt on glucose plates (C) and glycerol plates (D); tobacco leaf explants with YHL320 s+ on glucose plates (E) and glycerol plates (F), or with A348wt on glucose plates (G) and glycerol plates (H); tobacco stem explants with YHL320 s+ on glucose plates (I) and glycerol plates (J), or with A348wt on glucose plates (K) and glycerol plates (L) for 51 days on 0.5X MS plates. Additionally, tobacco leaf explants are shown co-cultivated with A348wt for 20 days (M), 34 days (N), or 78 days (O, P). Representative photos are shown and quantification of tumor induction rate is found in Table 5.
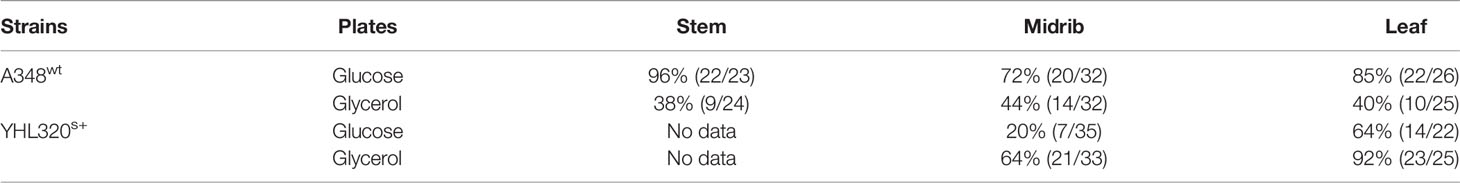
Table 5 Tumor induction rate on tobacco explants co-cultivated with A348wt and hypersensitive YHL320s+.
To further evaluate the relationship between increased sugar sensitivity and successful tumorigenesis, we inoculated Kalanchoe diagremontiana with A348wt and YHL320s+ strains (Figure 8). While the wild type A. tumefaciens produced typical tumors (Figure 8A), the tumors produced by strains containing the VirAY293F mutation appeared small and irregular (Figure 8B), indicating a further phenotypic difference between the two strains. The preliminary location of the tumors is also informative. We observed that the tumors in tobacco formed preferentially at cambium cells in the junction between two tissue types (Figures 7I–L), possibly due to the presence of stem cells (Figures 7C–H). These tumors appeared to arise in a polar fashion (Figures 7M, N) and only when the co-cultivation lasted for more than 35 days did the opposite cut end and additional spots around the cut edge of the leaf disks initiate tumor growth (Figures 7O, P). While this localization pattern supports the hypothesis that it is potentially the rapidly dividing cells generated at a wound site that are susceptible to Agrobacterium-mediated transformation, further experimentation probing the precise locations of the cell divisions will be necessary.
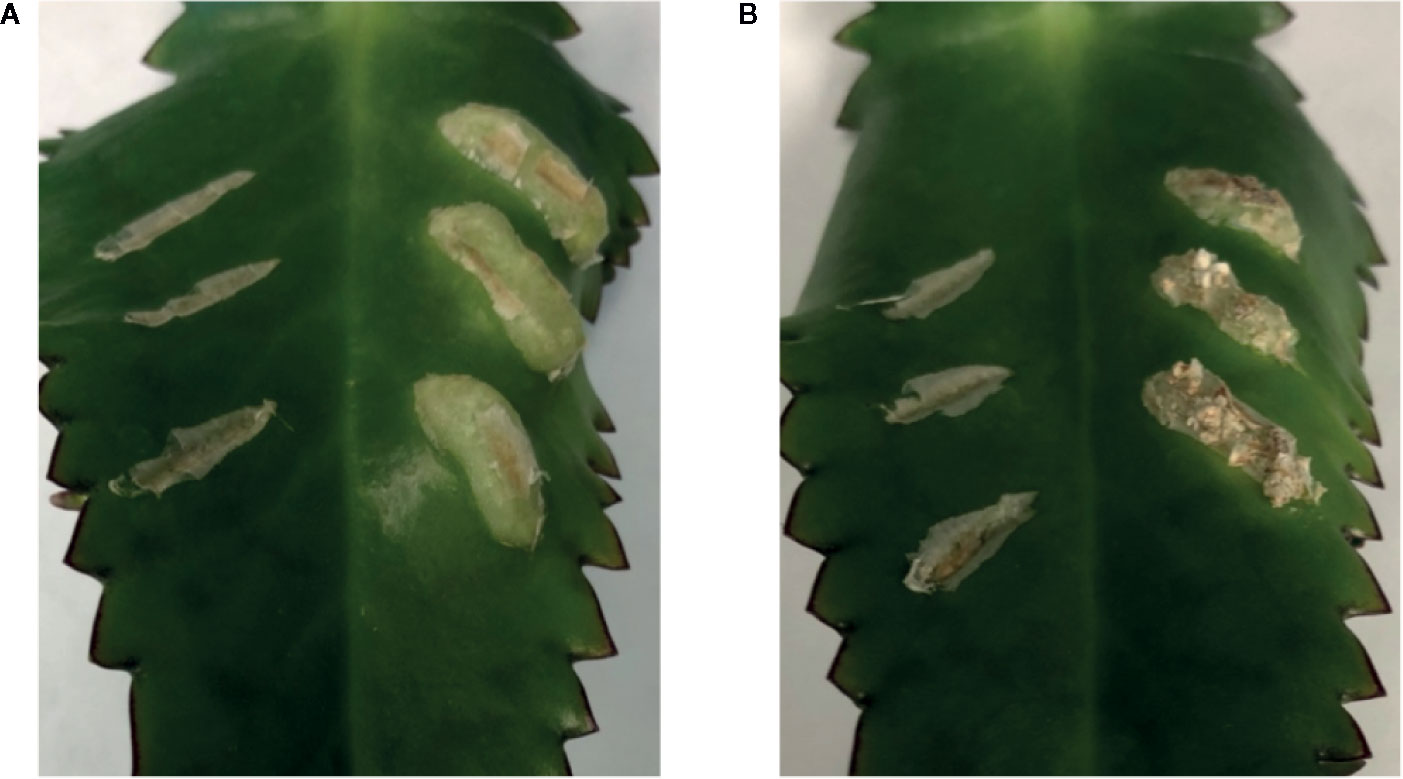
Figure 8 Both VirAwt and VirAY293F Agrobacterium induce crown gall tumors. Kalanchoe diagremontiana leaves were inoculated with A348wt (A) and YHL320s+ (B) in triplicate on the right of each leaf, while the wounds to the left were left as controls. Tumors for A348wt appeared larger and more uniform.
Discussion
Mapping the behavior of Agrobacterium within hosts tissues has the potential to correlate models for plant wounding (Savatin et al., 2014) with the evolution of plant pathogenesis (Lin et al., 2014; Hayta et al., 2019). The xenognosin receptor of A. tumefaciens functions as an AND gate, requiring both simple sugars and phenols to initiate pathogenesis (Fang et al., 2015). We show that while the exudation of sugars is similar between certain tissues of host (tobacco leaves) and non-host (maize leaves) plants, wound-localized xenognostic phenols in maize are orders of magnitude lower and occur as a transient spike rather than the sustained exudation observed in tobacco. In situ imaging identifies the accumulation of A. tumefaciens cells around viable host tissues. Accumulation is localized below the wound surface, rather than the adjacent vascular cells at the same depth, and is consistent with the pathogen targeting viable cells (Dubravina et al., 2005). This contrasting behavior highlights the differing wounding profiles between host and non-host and the well-known weak wound responses (Hiei et al., 1994) and low levels of vir-inducing exudates from monocots (Hooykaas, 1989).
Several strains of A. tumefaciens were constructed to further understand the molecular origins of these pathogen behaviors. YHL301ss contains a mutation within the xenognostic VirA receptor, allowing it to function as an OR gate where either sugar or phenol alone induce virulence gene expression (Fang et al., 2015). YHL324s+ contains the same mutation as well as the deletion of the multiple monosaccharide transporter B (δmmsB), a deletion which confers hypersensitivity (Zhao and Binns, 2011). Virulence gene expression is induced in both of these strains by dicot explants, and YHL324s+ is also induced by monocot tissues. However, this hypersensitive strain did not induce tumors in wounded maize plants, suggesting that pathogen incompatibility in monocots may be limited by other intrinsic factors than those responsible for the initial stages of virulence gene induction (Maresh et al., 2006; Maresh et al., 2007). As outlined in Figure 9, wound healing in dicots involves dedifferentiation and proliferation to repair the wound, but monocot cells lose the ability to dedifferentiate very early in development (Graves et al., 1988). Instead, monocots form a lignified ring of hardened cells that quickly seals the wound from invading microbes (Sood et al., 2011; Miedes et al., 2014; Lee et al., 2019), a process that likely limits plant transformation(Graves et al., 1988). Indeed, Agrobacterium-mediated transformation of monocot cells has been more successful in embryonic calli and immature embryos (Chan et al., 1993; Ishida et al., 1996; Cheng et al., 1997), while wounding in dicots appears to generate significant numbers of competent cells at the wound site (Binns, 1990; Ikeuchi et al., 2013).
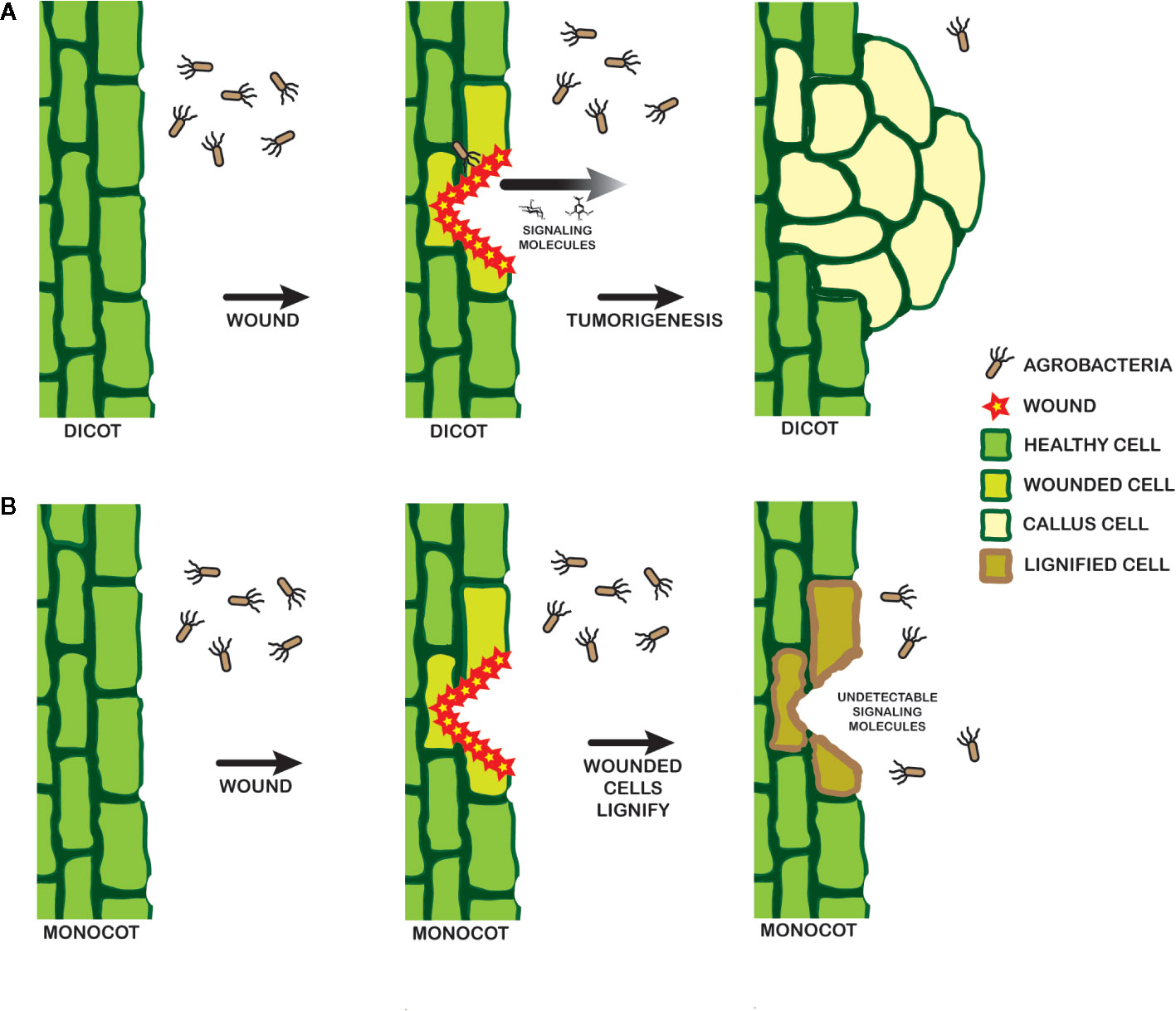
Figure 9 Model for signal landscape of monocots and dicots. In dicots (A), wounding events release detectable signals into the rhizosphere, which can be recognized by a pathogenic Agrobacterium. Chemotaxis follows, leading to DNA transfer and tumorigenesis. Wounding in monocots (B), in contrast, leads to lignification in the cell walls and a lack of strong signals by a pathogenic Agrobacterium.
Spatiotemporal mapping with Agrobacterium then opens a window into the complex and dynamic behaviors of the communities that naturally inhabit the plant wound site. More generally, the bacterial strains reported here may be extended to probe the reaction-diffusion networks that contribute to specifying the complex cellular order of the rhizosphere (Taran et al., 2019). Such methods for mapping the dynamic behaviors of the epiphytes, symbionts, pathogens, and other integral members of this complex multicellular community will become increasingly important for achieving a more sustainable agriculture and addressing the food challenges we will face in our changing global climate.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
SL and BP initiated this project in the lab of DL, and BP developed new aspects of this research in his own lab. Y-HL significantly contributed to the creation of the constructs. BP, SL, NW, AM, and JA performed the experiments reported. BP and DL wrote the manuscript with contributions from SL, AM, and AB.
Funding
This work was funded by the University of Richmond School of Arts & Sciences which provided startup funds for BP in addition to his National Institute of Health K12 GM000680 Fellowship in Research and Science Teaching (FIRST) Institutional Research and Academic Career Development Award (IRACDA). The undergraduate students AM, NW, and JA were all funded through the University of Richmond research grants. The work of DL, SL, and Y-HL were funded through The National Science Foundation award IOS # 1423862.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge the contribution of John Hayden, University of Richmond, to the creation of our model in Figure 9. We also would like to recognize that a portion of the experiments in this article appeared in the thesis of SL, and would like to thank her thesis committee for their careful readings and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01074/full#supplementary-material
References
Akinsulire, O., Aibin, I., Adenipekun, T., Adelowotan, T., Odugbemi, T. (2008). In Vitro Antimicrobial Activity Of Crude Extracts From Plants Bryophyllum pinnatum And Kalanchoe crenata. Afr. J. Tradit. Complement. Altern. Medicines 4, 338–344. doi: 10.4314/ajtcam.v4i3.31227
Ashby, A. M., Watson, M. D., Loake, G. J., Shaw, C. H. (1988). Ti plasmid-specified chemotaxis of Agrobacterium tumefaciens C58C1 toward vir-inducing phenolic compounds and soluble factors from monocotyledonous and dicotyledonous plants. J. Bacteriol. 170, 4181–4187. doi: 10.1128/jb.170.9.4181-4187.1988
Badri, D. V., Vivanco, J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32, 666–681. doi: 10.1111/j.1365-3040.2009.01926.x
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., Vivanco, J. M. (2006). The Role Of Root Exudates In Rhizosphere Interactions With Plants And Other Organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Binns, A. N., Zhao, J. (2020). The MexE/MexF/AmeC efflux pump of Agrobacterium tumefaciens and its role in Ti plasmid virulence gene expression. J. Bacteriol. 202. doi: 10.1128/jb.00609-19
Binns, A. N. (1990). Agrobacterium-mediated gene delivery and the biology of hostrange limitations. Physiol. Plantarum 79, 135–139. doi: 10.1111/j.1399-3054.1990.tb05875.x
Chan, M.-T., Chang, H.-H., Ho, S.-L., Tong, W.-F., Yu, S.-M. (1993). Agrobacterium-mediated production of transgenic rice plants expressing a chimeric ?-amylase promoter/?-glucuronidase gene. Plant Mol. Biol. 22, 491–506. doi: 10.1007/bf00015978
Chang, M., Lynn, D. G. (1986). The haustorium and the chemistry of host recognition in parasitic angiosperms. J. Chem. Ecol. 12, 561–579. doi: 10.1007/bf01020572
Chang, M., Netzly, D. H., Butler, L. G., Lynn, D. G. (1986). Chemical regulation of distance. Characterization of the first natural host germination stimulant for Striga asiatica. J. Am. Chem. Soc. 108, 7858–7860. doi: 10.1021/ja00284a074
Cheng, M., Fry, J. E., Pang, S., Zhou, H., Hironaka, C. M., Duncan, D. R., et al. (1997). Genetic Transformation of Wheat Mediated by Agrobacterium tumefaciens. Plant Physiol. 115, 971–980. doi: 10.1104/pp.115.3.971
Chesnokova, O., Coutinho, J. B., Khan, I. H., Mikhail, M. S., Kado, C., II (1997). Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol. Microbiol. 23, 579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x
Dietrich, L. E. P., Teal, T. K., Price-Whelan, A., Newman, D. K. (2008). Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Sci. New Y. N. Y. 321, 1203–1206. doi: 10.1126/science.1160619
Dubravina, G. A., Zaytseva, S. M., Zagoskina, N. V. (2005). Changes in Formation and Localization of Phenolic Compounds in the Tissues of European and Canadian Yew during Dedifferentiation In Vitro. Russ J. Plant Physl+ 52, 672–678. doi: 10.1007/s11183-005-0100-z
Fang, F., Lin, Y.-H., Pierce, B. D., Lynn, D. G. (2015). A Rhizobium radiobacter Histidine Kinase Can Employ Both Boolean AND and OR Logic Gates to Initiate Pathogenesis. Chembiochem. Eur. J. Chem. Biol. 16, 2183–2190. doi: 10.1002/cbic.201500334
Fate, G., Chang, M., Lynn, D. G. (1990). Control of Germination in Striga asiatica: Chemistry of Spatial Definition. Plant Physiol. 93, 201–207. doi: 10.1104/pp.93.1.201
Fuller, A. W., Young, P., Pierce, B. D., Kitson-Finuff, J., Jain, P., Schneider, K., et al. (2017). Redox-mediated quorum sensing in plants. PloS One 12, e0182655. doi: 10.1371/journal.pone.0182655
Garfinkel, D. J., Simpson, R. B., Ream, L. W., White, F. F., Gordon, M. P., Nester, E. W. (1981). Genetic analysis of crown gall: Fine structure map of the T-DNA by site-directed mutagenesis. Cell 27, 143–153. doi: 10.1016/0092-8674(81)90368-8
Glasser, N. R., Saunders, S. H., Newman, D. K. (2017). The Colorful World of Extracellular Electron Shuttles. Annu. Rev. Microbiol. 71, 731–751. doi: 10.1146/annurev-micro-090816-093913
Goulian, M., van der Woude, M. (2006). A simple system for converting lacZ to gfp reporter fusions in diverse bacteria. Gene 372, 219–226. doi: 10.1016/j.gene.2006.01.004
Graves, A. E., Goldman, S. L., Banks, S. W., Graves, A. C. (1988). Scanning electron microscope studies of Agrobacterium tumefaciens attachment to Zea mays, Gladiolus sp., and Triticum aestivum. J. Bacteriol. 170, 2395–2400. doi: 10.1128/jb.170.5.2395-2400.1988
Hayta, S., Smedley, M. A., Demir, S. U., Blundell, R., Hinchliffe, A., Atkinson, N., et al. (2019). An efficient and reproducible Agrobacterium-mediated transformation method for hexaploid wheat (Triticum aestivum L.). Plant Methods 15, 121. doi: 10.1186/s13007-019-0503-z
Heindl, J. E., Wang, Y., Heckel, B. C., Mohari, B., Feirer, N., Fuqua, C. (2014). Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium. Front. Plant Sci. 5, 176. doi: 10.3389/fpls.2014.00176
Hiei, Y., Ohta, S., Komari, T., Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313x.1994.6020271.x
Hooykaas, P. J. J. (1989). Transformation of plant cells via Agrobacterium. Plant Mol. Biol. 13, 327–336. doi: 10.1007/bf00025321
Hu, X., Zhao, J., DeGrado, W. F., Binns, A. N. (2012). Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals. P. Natl. Acad. Sci. U.S.A. 110, 678–683. doi: 10.1073/pnas.1215033110
Huang, X.-F., Chaparro, J. M., Reardon, K. F., Zhang, R., Shen, Q., Vivanco, J. M. (2014). Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92, 267–275. doi: 10.1139/cjb-2013-0225
Ikeuchi, M., Sugimoto, K., Iwase, A. (2013). Plant callus: mechanisms of induction and repression. Plant Cell 25, 3159–3173. doi: 10.1105/tpc.113.116053
Ishida, Y., Saito, H., Ohta, S., Hiei, Y., Komari, T., Kumashiro, T. (1996). High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 14, 745–750. doi: 10.1038/nbt0696-745
Keyes, W. J., Palmer, A. G., Erbil, W. K., Taylor, J. V., Apkarian, R. P., Weeks, E. R., et al. (2007). Semagenesis and the parasitic angiosperm Striga asiatica. Plant J. 51, 707–716. doi: 10.1111/j.1365-313x.2007.03171.x
Lagendijk, E. L., Validov, S., Lamers, G. E. M., Weert, S. D., Bloemberg, G. V. (2010). Genetic tools for tagging Gram-negative bacteria with mCherry for visualization in vitro and in natural habitats, biofilm and pathogenicity studies. FEMS Microbiol. Lett. 305, 81–90. doi: 10.1111/j.1574-6968.2010.01916.x
Lee, K., Dudley, M. W., Hess, K. M., Lynn, D. G., Joerger, R. D., Binns, A. N. (1992). Mechanism of activation of Agrobacterium virulence genes: identification of phenol-binding proteins. Proc. Natl. Acad. Sci. 89, 8666–8670. doi: 10.1073/pnas.89.18.8666
Lee, Y.-W., Ha, U.-H., Sim, W.-S., Nester, E. W. (1998). Characterization of an unusual sensor gene (virA) of Agrobacterium. Gene 210, 307–314. doi: 10.1016/s0378-1119(98)00082-1
Lee, M., Jeon, H. S., Kim, S. H., Chung, J. H., Roppolo, D., Lee, H., et al. (2019). Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J. 38, e101948. doi: 10.15252/embj.2019101948
Liang, L., Liu, Y., Jariwala, J., Lynn, D. G., Palmer, A. G. (2016). Detection and Adaptation in Parasitic Angiosperm Host Selection. Am. J. Plant Sci. 07, 1275–1290. doi: 10.4236/ajps.2016.78123
Lin, Y.-H., Pierce, B. D., Fang, F., Wise, A., Binns, A. N., Lynn, D. G. (2014). Role of the VirA histidine autokinase of Agrobacterium tumefaciens in the initial steps of pathogenesis. Front. Plant Sci. 5, 195. doi: 10.3389/fpls.2014.00195
Liu, Y. (2012). Comparing and contrasting two plant pathogens provide a unique window into the differences in the innate immune responses between dicotsand monocots. Unpublished doctoral dissertation. Atlanta (GA): Emory University.
Loake, G. J., Ashby, A. M., Shaw, C. H. (1988). Attraction of Agrobacterium tumefaciens C58C1 towards Sugars Involves a Highly Sensitive Chemotaxis System. Microbiology+ 134, 1427–1432. doi: 10.1099/00221287-134-6-1427
Maresh, J., Zhang, J., Lynn, D. G. (2006). The Innate Immunity of Maize and the Dynamic Chemical Strategies Regulating Two-Component Signal Transduction in Agrobacterium tumefaciens. ACS Chem. Biol. 1, 165–175. doi: 10.1021/cb600051w
Maresh, J., Zhang, J., Tzeng, Y.-L., Goodman, N. A., Lynn, D. G. (2007). Rational design of inhibitors of VirA–VirG two-component signal transduction. Bioorg. Med. Chem. Lett. 17, 3281–3286. doi: 10.1016/j.bmcl.2007.04.018
McCullen, C. A., Binns, A. N. (2006). Agrobacterium tumefaciens and Plant Cell Interactions and Activities Required for Interkingdom Macromolecular Transfer. Annu. Rev. Cell Dev. Bi 22, 101–127. doi: 10.1146/annurev.cellbio.22.011105.102022
Meij, A., van der, Willemse, J., Schneijderberg, M. A., Geurts, R., Raaijmakers, J. M., et al. (2018). Inter- and intracellular colonization of Arabidopsis roots by endophytic actinobacteria and the impact of plant hormones on their antimicrobial activity. Antonie Van Leeuwenhoek 111, 679–690. doi: 10.1007/s10482-018-1014-z
Mendes, R., Kruijt, M., Bruijn, I., Dekkers, E., Voort, M., van der, et al. (2011). Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Merritt, P. M., Danhorn, T., Fuqua, C. (2007). Motility and Chemotaxis in Agrobacterium tumefaciens Surface Attachment and Biofilm Formation. J. Bacteriol. 189, 8005–8014. doi: 10.1128/jb.00566-07
Messens, E., Dekeyser, R., Stachel, S. E. (1990). A nontransformable Triticum monococcum monocotyledonous culture produces the potent Agrobacterium vir-inducing compound ethyl ferulate. Proc. Natl. Acad. Sci. 87, 4368–4372. doi: 10.1073/pnas.87.11.4368
Miedes, E., Vanholme, R., Boerjan, W., Molina, A. (2014). The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 5, 358. doi: 10.3389/fpls.2014.00358
Nester, E. W. (2014). Agrobacterium: nature’s genetic engineer. Front. Plant Sci. 5, 730. doi: 10.3389/fpls.2014.00730
Nishimura, A. (2020). Agrobacterium Transformation in the Rice Genome. Methods Mol. Biol. Clifton N. J. 2072, 207–216. doi: 10.1007/978-1-4939-9865-4_17
Philippot, L., Raaijmakers, J. M., Lemanceau, P., Putten, W. H., van der (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Rasmann, S., Turlings, T. C. (2016). Root signals that mediate mutualistic interactions in the rhizosphere. Curr. Opin. Plant Biol. 32, 62–68. doi: 10.1016/j.pbi.2016.06.017
Robbs, S. L., Hawes, M. C., Lin, H.-J., Pueppke, S. G., Smith, L. Y. (1991). Inheritance of Resistance to Crown Gall in Pisum sativum. Plant Physiol. 95, 52–57. doi: 10.1104/pp.95.1.52
Sahi, S. V., Chilton, M. D., Chilton, W. S. (1990). Corn metabolites affect growth and virulence of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. 87, 3879–3883. doi: 10.1073/pnas.87.10.3879
Savatin, D. V., Gramegna, G., Modesti, V., Cervone, F. (2014). Wounding in the plant tissue: the defense of a dangerous passage. Front. Plant Sci. 5, 470. doi: 10.3389/fpls.2014.00470
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., Pühler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Shen, W. H., Escudero, J., Schlappi, M., Ramos, C., Hohn, B., Koukolikova-Nicola, Z. (1993). T-DNA transfer to maize cells: histochemical investigation of beta-glucuronidase activity in maize tissues. Proc. Natl. Acad. Sci. 90, 1488–1492. doi: 10.1073/pnas.90.4.1488
Smith, R. H., Hood, E. E. (1995). Agrobacterium tumefaciens Transformation of Monocotyledons. Crop Sci. 35, 301. doi: 10.2135/cropsci1995.0011183x003500020001x
Smith, C. E., Dudley, M. W., Lynn, D. G. (1990). Vegetative/Parasitic Transition: Control and Plasticity in Striga Development. Plant Physiol. 93, 208–215. doi: 10.1104/pp.93.1.208
Sood, P., Bhattacharya, A., Sood, A. (2011). Problems and possibilities of monocot transformation. Biol. Plantarum 55, 1–15. doi: 10.1007/s10535-011-0001-2
Stachel, S. E., Nester, E. W. (1986). The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 5, 1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x
Stacy, A., Everett, J., Jorth, P., Trivedi, U., Rumbaugh, K. P., Whiteley, M. (2014). Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. P. Natl. Acad. Sci. U.S.A. 111, 7819–7824. doi: 10.1073/pnas.1400586111
Stacy, A., McNally, L., Darch, S. E., Brown, S. P., Whiteley, M. (2015). The biogeography of polymicrobial infection. Nat. Rev. Microbiol. 14, 93–105. doi: 10.1038/nrmicro.2015.8
Taran, O., Patel, V., Lynn, D. G. (2019). Small molecule reaction networks that model the ROS dynamics of the rhizosphere. Chem. Commun. Camb. Engl. 55, 3602–3605. doi: 10.1039/c8cc08940j
Taye, B., Giday, M., Animut, A., Seid, J. (2011). Antibacterial activities of selected medicinal plants in traditional treatment of human wounds in Ethiopia. Asian Pac. J. Trop. BioMed. 1, 370–375. doi: 10.1016/s2221-1691(11)60082-8
Tomilov, A., Tomilova, N., Shin, D. H., Jamison, D., Torres, M., Reagan, R., et al. (2006). “Chemical signaling between plants: mechanistic similarities between phytotoxic allelopathy and host recognition by parasitic plants,” in Chemical Ecology: From Gene to Ecosystem. Ed. Dicke, M. (Dordrecht, Netherlands: Springer), 55–69. doi: 10.1007/978-1-4020-5369-6_5
Usami, S., Okamoto, S., Takebe, I., Machida, Y. (1988). Factor inducing Agrobacterium tumefaciens vir gene expression is present in monocotyledonous plants. Proc. Natl. Acad. Sci. 85, 3748–3752. doi: 10.1073/pnas.85.11.3748
Uteau, D., Hafner, S., Pagenkemper, S. K., Peth, S., Wiesenberg, G. L. B., Kuzyakov, Y., et al. (2015). Oxygen and redox potential gradients in the rhizosphere of alfalfa grown on a loamy soil. J. Plant Nutr. Soil Sc. 178, 278–287. doi: 10.1002/jpln.201300624
Vijayachandra, K., Palanichelvam, K., Veluthambi, K. (1995). Rice scutellum induces Agrobacterium tumefaciens vir genes and T-strand generation. Plant Mol. Biol. 29, 125–133. doi: 10.1007/bf00019124
Walker, T. S., Bais, H. P., Grotewold, E., Vivanco, J. M. (2003). Root Exudation and Rhizosphere Biology: Fig. 1. Plant Physiol. 132, 44–51. doi: 10.1104/pp.102.019661
Whiteley, M., Diggle, S. P., Greenberg, E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. doi: 10.1038/nature24624
Winans, S. C. (1992). Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 56, 12–31. doi: 10.1128/MMBR.56.1.12-31.1992
Zhang, J., Boone, L., Kocz, R., Zhang, C., Binns, A. N., Lynn, D. G. (2000). At the maize/Agrobacterium interface: natural factors limiting host transformation. Chem. Biol. 7, 611–621. doi: 10.1016/s1074-5521(00)00007-7
Keywords: rhizosphere, Agrobacterium tumefaciens, confocal microscopy, spatiotemporal mapping, fluorescent biosensor, chemical network
Citation: Liu S, Lin Y-H, Murphy A, Anderson J, Walker N, Lynn DG, Binns AN and Pierce BD (2020) Mapping Reaction-Diffusion Networks at the Plant Wound Site With Pathogens. Front. Plant Sci. 11:1074. doi: 10.3389/fpls.2020.01074
Received: 31 March 2020; Accepted: 30 June 2020;
Published: 16 July 2020.
Edited by:
Claudia Knief, University of Bonn, GermanyReviewed by:
Anton Hartmann, Ludwig-Maximilian-Universität München, GermanyRyohei Thomas Nakano, Max Planck Institute for Plant Breeding Research, Germany
Copyright © 2020 Liu, Lin, Murphy, Anderson, Walker, Lynn, Binns and Pierce. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: B. Daniel Pierce, YnBpZXJjZUByaWNobW9uZC5lZHU=
 Stephanie Liu
Stephanie Liu Yi-Han Lin
Yi-Han Lin Aidan Murphy
Aidan Murphy Josh Anderson3
Josh Anderson3 Nicole Walker
Nicole Walker David G. Lynn
David G. Lynn Andrew N. Binns
Andrew N. Binns B. Daniel Pierce
B. Daniel Pierce

