- 1Key Laboratory of Forestry Genetics & Biotechnology of Ministry of Education, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
- 2Experimental Center of Desert Forestry, Chinese Academy of Forestry, Dengkou, China
- 3Research Center of Saline and Alkali Land of National Forestry and Grassland Administration, China Academy of Forestry, Beijing, China
- 4State Forestry Administration Key Laboratory of Biodiversity Conservation in Karst Mountainous Areas of Southwestern China, Guizhou Normal University, Guiyang, China
- 5Guizhou Provincial Key Laboratory of Plant Physiology and Developmental Regulation, Guizhou Normal University, Guiyang, China
Calcineurin B-like protein-interacting protein kinases (CIPKs) play essential roles in plant abiotic stress response. In order to better understand salt tolerance, we cloned and analyzed the NtCIPK9 gene from the halophyte Nitraria tangutorum. Phylogenetic analysis shows that NtCIPK9 belongs to a sister clade with the Arabidopsis AtCIPK9 gene and is thought to localize to the plasma membrane. NtCIPK9 shows the highest expression level in the Nitraria tangutorum root under normal growth conditions, whereas after NaCl treatment, the highest expression was found in the blade. NtCIPK9-overexpressing Arabidopsis plants have a higher seed germination rate, longer root length, and displayed higher salt tolerance than wild type seedlings under salt stress conditions. Furthermore, NtCIPK9 overexpression might enhance the expression of genes related to K+ transportation after NaCl treatment. Thus, we conclude that NtCIPK9 increases transgenic plant salt tolerance and reduces damage associated with salt stress by promoting the expression of genes controlling ion homeostasis. Our results suggest that NtCIPK9 could serve as an ideal candidate gene to genetically engineer salt-tolerant plants.
Introduction
Soil salinity gradually accumulates along with global environmental degradation and limits the quality and productivity of most important agricultural crops and trees worldwide (Mahajan and Tuteja, 2005; Zhou et al., 2014; Guo et al., 2018). Currently, soil salinization is estimated to result in one-third of the world’s limited arable land loss, and will increase to 50% by the middle of 21st century (Zhu et al., 1998; Wang et al., 2003). Thus, improving the adaptability of plants to salinized soil is crucial not only for plant survival, but also for effective soil utilization. Genetic engineering to improve the salt tolerance of crops has been actively investigated by plant scientists worldwide (Wang et al., 2003; Hu et al., 2005; Ashraf and Akram, 2009).
Calcineurin B-like proteins (CBLs) and their targets CBL-interacting protein kinases (CIPKs) are involved in the Ca2+ signal pathway that functions during stress response (Kudla et al., 1999; Shi et al., 1999; Kim et al., 2000); they play important roles in maintaining cytoplasm ion homeostasis and improving salt tolerance. CIPKs contain a typical protein kinase domain with a putative activation loop and a unique C-terminal regulatory region with a conserved NAF/FISL motif, both of which are necessary and sufficient for the function of these genes (Shi et al., 1999; Albrecht et al., 2001; Guo et al., 2001). CIPKs have been identified in many plant species (Albrecht et al., 2001; Kolukisaoglu et al., 2004; Lyzenga et al., 2013; Zhang et al., 2014), and remarkable progress has been made in exploring the functions of CIPKs (Zhu et al., 1998; Halfter et al., 2000; Shi et al., 2000; Qiu et al., 2002; Cheong et al., 2003; Chinnusamy et al., 2004; Sánchez-Barrena et al., 2005; Luan, 2009). In Arabidopsis, CIPKs affect cellular ion homeostasis under saline conditions by regulating ion transporters, such as HKT1 (HIGH-AFFINITY K+ TRANSPORTER1). HKT1 improves salinity tolerance by removing Na+ from the transpiration stream and promoting the absorption of K+ in Arabidopsis (Kato et al., 2001; Cheong et al., 2003; Li et al., 2006; Xu et al., 2006; Grefen and Blatt, 2012). In addition, CIPKs also regulate the expression of stress-responsive genes mediated by the abscisic acid pathway during seed germination and at the seedling stage (Kim et al., 2003; Pandey et al., 2008a; Piao et al., 2010). CIPK function is highly conserved across plant families, shown by studies in apple and tomato (Hu et al., 2016), Cicer arietinum (Tripathi et al., 2009), Hordeum brevisubulatum (Li et al., 2012), and Nitraria tangutorum (Himabindu et al., 2016). However, most studies have been performed in salinity sensitive plants (glycophytes), such as Arabidopsis, rice, and maize (Kolukisaoglu et al., 2004; Zhao et al., 2009; Rigó et al., 2016), limiting our understanding of how plants may adapt to saline environments. Therefore, studying how halophyte CIPKs homologs function might provide crucial perspective in addressing this question, as these genes could be functionally more efficient than their glycophyte counterparts (Himabindu et al., 2016).
Nitraria tangutorum Bobr. (a halophyte), is a shrub that belongs to the family Nitrariaceae Nitraria in Sapindales (Zhao et al., 2002; Chase and Reveal, 2009; Group, 2016; Lu et al., 2018), and is widely distributed in China’s northwestern region (Wang et al., 2014). This typical plant has a strong adaptability to high salinity, arid or semiarid environments. It can efficiently alleviate the degree of soil salinity–alkalinity, which could improve the utilization of saline areas and prevent soil desertification (Zhao et al., 2002; Yang et al., 2010a; Jie et al., 2011). Due to its ecological effect, studies on Nitraria tangutorum have mainly focused on the physiological and biochemical aspects of its adaptive mechanisms to abiotic stresses (Yang et al., 2010a; Yang et al., 2010b; Yang et al., 2012; Yang et al., 2013). Although NtCIPK2 and NtP5CS from Nitraria tangutorum have been cloned and analyzed to a certain extent (Zheng L. et al., 2014; Zheng L. L. et al., 2014), there is a current lack of knowledge on how Nitraria tangutorum responds molecularly to salt stress. In order to reveal the functional genes supporting Nitraria tangutorum to deal with high salinity and promote the application of these functional genes from halophyte to glycophyte, in our study, we used rapid amplification of cDNA end (RACE) cloning to identify a novel Nitraria tangutorum CIPK gene, which shows significant homology to Arabidopsis CIPK9. Therefore, we named it NtCIPK9 (Nitraria tangutorum CIPK9). Quantitative PCR analysis showed that NtCIPK9 positively responds to 500 mM NaCl treatment in both the root and leaf of Nitraria tangutorum. We overexpressed NtCIPK9 in Arabidopsis and compared the different abilities of salt resistance between transgenic plants and wild type plants. NtCIPK9 overexpressing-plants displayed a higher germination efficiency, longer root length, more leaves, and a lower death rate than the wild type under salt stresses. The high K+ content and AtHKT1 expression level in transgenic seedlings suggest that NtCIPK9 enhanced salt tolerance by regulating expression of genes controlling ion homeostasis.
Materials and Methods
Plant Materials and Treatments
Nitraria tangutorum
Nitraria tangutorum seeds, provided by the Experimental Center for Desert Forestry of the Chinese Academy of forestry, were kept in sand with around 7% water at 4℃ for four weeks. Seeds were germinated in pots containing a mixture of soil and sand (1:1 ratio) at 26°C to 28°C and 16-h light/8-h dark cycle condition. The humidity of the chamber for plant growth was 55% to 60%. 6-month-old Nitraria tangutorum plants were irrigated with 500 mM NaCl and harvested at 0, 1, and 2 h after treatment for RNA extraction.
Arabidopsis thaliana
The Arabidopsis thaliana Columbia ecotype was used for NtCIPK9 gene transformation through the floral dip method (Clough and Bent, 1998). Positive transgenic plants were selected using 50 mg/L Kanamycin and confirmed by PCR. Wild type and transgenic plants were grown under identical growth conditions in parallel. Seeds were harvested at the same time for phenotypical analysis. Arabidopsis seeds were sown on ½MS with 0, 100, and 150 mM NaCl for germination rate analysis, phenotypical observation, and ion content measurements. Three biological replicates were used for germination rate analysis. Each biological replicate included three technical replicates. At least 200 seeds per line have been used. WT and transgenic plants were sown on the same plates. 10-day-old seedlings were transferred to ½MS with 0, 100, and 150 NaCl for another 10 days to analyze survival rate of transgenic plants, three biological replicates for each experiment. Seedlings from 0 and 100 mM NaCl treatment were frozen in liquid nitrogen and stored at −80°C for qPCR experiments. NtCIPK9 transgenic T2 heterozygous seeds have been sown in the pots. After four weeks, seedlings in pots were watered by 200 mM NaCl for 4 days. Three biological replicates and four experimental repeats have been conducted.
Gene Cloning
Total RNA was extracted from Nitraria tangutorum leaves using a Total RNA Purification Kit (NORGEN, Thorold, ON, Canada), followed by removal of genomic DNA contamination using DNaseI (TaKaRa, Japan). Total RNA concentration and integrity were quantified by ultraviolet spectrophotometry and electrophoresis, respectively. First-strand cDNA was synthesized using reverse transcriptase (Invitrogen, Carlsbad, USA). Degenerate primers to amplify the CIPK fragment were designed based on the homeodomain of CIPK from poplar (Chen et al., 2011) and are listed in Supplementary Table 1. The full length sequence was obtained by 5’ and 3’-RACE (rapid amplification of cDNA ends) according to the SMARTer™ RACE cDNA Amplification Kit User manual (BD Bioscience Clontech, USA). Primers for RACE are listed in Supplementary Table 2. The amplified PCR product was purified and cloned into pMD19-T (TaKaRa, Japan) and sequenced (GenScript, Nanjing, China). After assembly, the complete NtCIPK9 sequence was amplified from cDNA using the primers mentioned in Supplementary Table 3. To confirm whether the NtCIPK9 genomic region also contains introns, we amplified NtCIPK9 from Nitraria tangutorum genomic DNA using the same primers (Supplementary Table 3).
Bioinformatics Analysis
The NtCIPK9 homolog was identified by using NCBI blastp. Multisequence alignment was performed using DNAMAN 6.0 software (Lynnon Biosoft, Quebec, Canada). Conserved domains of NtCIPK9 were predicted using InterProScan online software (http://www.ebi.ac.uk/InterProScan/). Phylogenetic trees were constructed with amino acid sequences of NtCIPK9 and 26 Arabidopsis CIPK proteins using the Neighbor-joining method with 1,000 bootstrap replications and the Jones-Taylor-Thornton model in Mega 6 software. Sequence accession numbers are listed in Supplementary Table 4. Hydrophobic analysis and transmembrane domain prediction of the NtCIPK9 protein were performed using ProtScale (http://ca.expasy.org/tools/protscale.html) and the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/).
56 protein-coding genes from 46 chloroplast genomes (Supplementary Table 5) were selected for phylogenetic analysis of Nitraria tangutorum, Vitis vinifera as outgroup. Sequences alignment was performed using ClustalW. Each orthologous gene was trimmed with trimAl version 1.2 (Capella-Gutiérrez et al., 2009). The trimmed alignments were concatenated using SequenceMatriX version 1.7.8 (Vaidya et al., 2011). A nucleotide matrix of 44780 sites was then constituted for Maximum parsimony analysis by PAUP 4.0 (Swofford, 2002).
Subcellular Localization Assay
The full-length coding region of NtCIPK9 was cloned into vector pJIT166:GFP for subcellular localization analysis. The recombinant plasmids were bombarded into onion epidermal cells according to a previously described method (Yokoi et al., 2002) and followed by fluorescence detection using a ZEISS X-cite 120Q fluorescence microscope (ZEISS, Germany). Three technical replicates have been performed. The primers for construction of GFP tagged NtCIPK9 are listed in Supplementary Table 6.
Quantitative Real-Time PCR Analysis
Total RNA isolation and reverse transcription were performed as mentioned above. Quantitative real-time PCR was performed using a SYBR-Green PCR Mastermix on a LightCycler®480 real-time PCR detection system (Roche, Basel, Switzerland) according to the manufacturer’s instruction. Expression levels of target genes were normalized using the housekeeping gene actin in Nitraria tangutorum (Wang et al., 2012) and ubiquitin10 (UBQ10) in Arabidopsis (Geldner et al., 2009). Three technical replicates for three independent transgenic lines were carried out for real-time PCR. Sequence-specific primers were designed using Primer 3.0 and Oligo 7 and are listed in Supplementary Table 7.
Cation Content Measurements
Twenty-day-old seedlings grown on ½MS with 0, 100, and l50 mM NaCl were collected, respectively, and washed three times with ddH2O, then dried at 80°C for 3 day. Harvested samples were digested with the HNO3-HClO4 method (Zhao et al., 1994). After acid digestion, samples were diluted to a total volume of 50 mL with ddH2O and kept in new tubes before analysis using flame atomic absorption spectrophotometry (FAAS) (Karpiuk et al., 2016). Three biological replicates were performed for each ion content test experiment. Three technical replicates were repeated for each biological replicate.
Results
Conserved Domain of NtCIPK9
To start, we analyzed the basic properties of NtCIPK9 sequence. Full length NtCIPK9 is 1735 bp with a predicted open reading frame of 1332 bp nucleotides, a 5′UTR of 239 bp and a 3′UTR of 164 bp in length, encoding 443 amino acids with an estimated molecular weight 50.52 kDa. The acquired coding sequence shares high similarity with CIPKs from different plant species. The deduced NtCIPK9 protein sequence showed 82.18% identity with Theobroma cacao CIPK9 (TcCIPK9), 80% identity with Populus trichocarpa CIPK9 (PtCIPK9) and 77.78% identity with Arabidopsis thaliana CIPK9 (AtCIPK9) (Figure 1A). Consistent with other CIPKs, NtCIPK9 possesses an N-terminal SNF-1–related serine/threonine protein kinase domain (14–268 aa) and a C-terminal regulatory domain (305–421 aa) with a CBL-interacting NAF/FISL module (Figure 1B). Thus, this gene was designated as NtCIPK9, a novel member of the plant CIPK gene family.
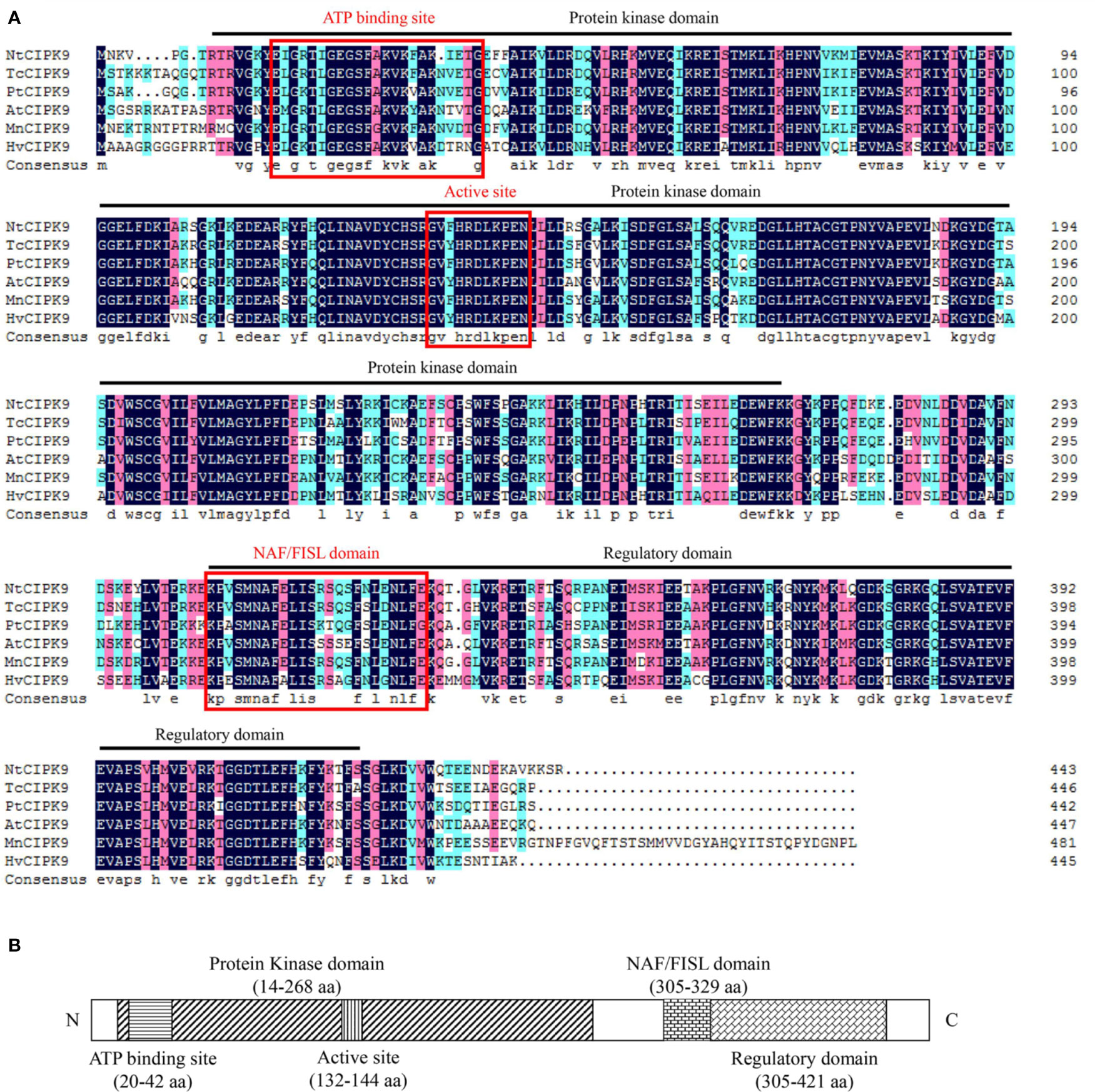
Figure 1 Multiple alignment and domain prediction of NtCIPK9. (A) Amino acid sequence alignment of NtCIPK9 with homologs from Theobroma cacao (EOX95608.1), Populus trichocarpa (XP_024450759.1), Arabidopsis thaliana (NP_171622.1), Morus notabilis (EXB83832.1), Oryza sativa (XP_015630713.1), Hordeum vulgare (AEZ51503.1), and Zea mays (PWZ05904.1). Dark blue shading indicates identical residues, and pink shading indicates similar residues. Dark lines demarcate the N-terminal kinase and C-terminal regulatory domains, while their active sites and NAF/FISH domain are marked with a red lines, the sequence identities between NtCIPK9 and others were shown at the end of the Multiple Alignment. (B) Schematic diagram of the domains of the NtCIPK9 protein. The amino acid position of the NtCIPK9 domain borders was predicted by InterProScan online software.
Phylogenetic comparison of NtCIPK9 with the Arabidopsis CIPK family clustered NtCIPK9 as a sister branch of AtCIPK9 to the intron-rich subgroup (Yu et al., 2007) (Figure 2). To analyze whether the NtCIPK9 gene contains introns, we amplified the genomic NtCIPK9 sequence from genomic DNA. The result show genomic NtCIPK9 harbors introns by DNA electrophoresis and sequencing (Supplementary Figure 1A), which is consistent with the results of our phylogenetic analysis. Besides, evolutionary study showed the Nitraria tangutorum was claded with Sapindus mukorossi, Azadirachta indica, Zanthoxylum piperitum, and Citrus sinensis in Sapindales (Supplementary Figure 2). Citrus sinensis is one of most important commercial fruit crops (Bausher et al., 2006).
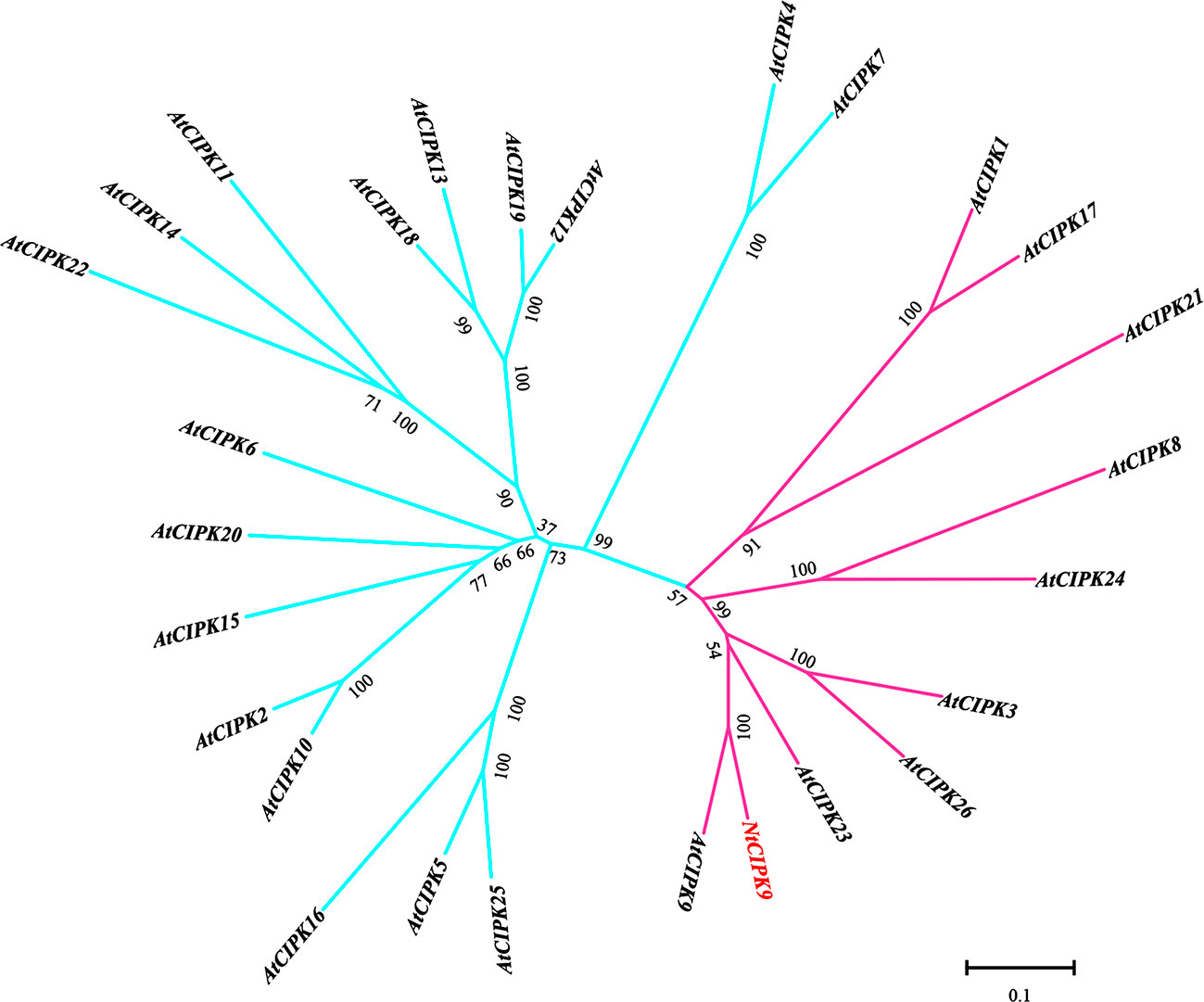
Figure 2 Phylogenetic analysis of NtCIPK9 with Arabidopsis CIPKs. The pink branch represents the subgroup of CIPKs with introns. The blue branch represents the clusters without intron.
Subcellular Location of NtCIPK9
A hydrophobicity blot indicated that the most hydrophobic segment of NtCIPK9 was located between amino acid residues 196 to 211 (Figure 3A), corresponding to the transmembrane domain predicted by the TMHMM Server 2.0 (Figure 3B). To further confirm the subcellular localization of NtCIPK9 in plant cells, a 35S:NtCIPK9-GFP translational fusion was constructed with GFP tagged to the C-terminus of NtCIPK9 and 35S:GFP was used as control (Figure 3C). The two vectors were bombarded into onion epidermal cells and transient expression of NtCIPK9-GFP was detected by epi-fluorescence. 35S:GFP fluorescence was detected in the membrane and cytoplasm (Figures 3D, E), similar to the localization of NtCIPK9-GFP (Figures 3F, G). The hydrophobicity and subcellular location analysis suggest that NtCIPK9 might be one of membrane-bound proteins.
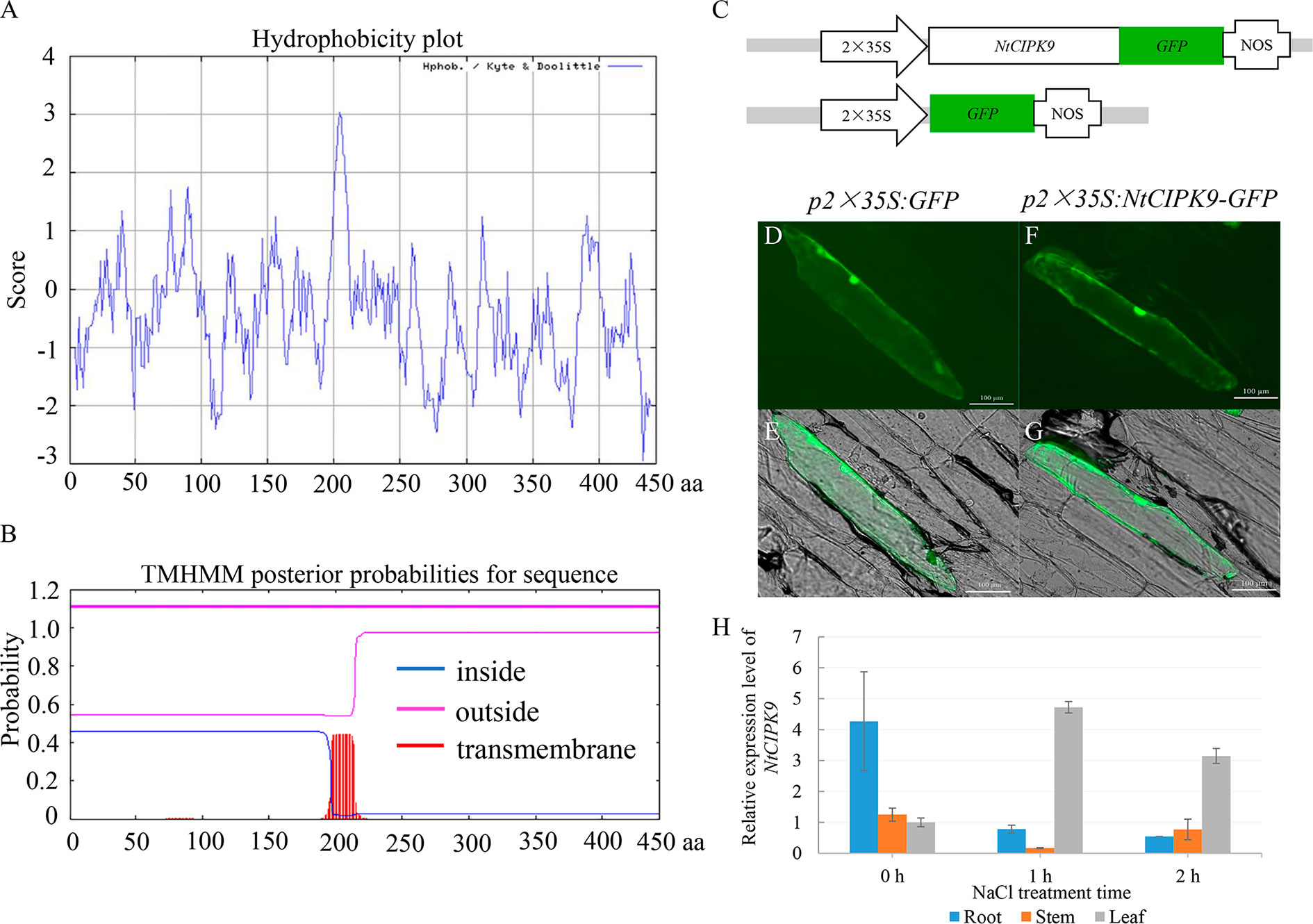
Figure 3 Structure analysis and subcellular localization of NtCIPK9. (A) Hydrophobicity plot of NtCIPK9. (B) The predicted transmembrane helix domain of NtCIPK9. (C) Schematic of the vectors used for analysis of NtCIPK9 subcellular localization. (D–G) NtCIPK9 subcellular localization. p35S:GFP serves as the control. (H) Relative expression level of NtCIPK9 in different tissues of Nitraria tangutorum. Data represent means ± SD from three biological replicates.
NtCIPK9 Responds to Salt Treatment in Nitraria tangutorum
To assess the expression of NtCIPK9 in Nitraria tangutorum under salt stress conditions, we isolated total mRNA from different tissues (including root, stem, and leaf) after 2 h 500 mM NaCl treatment. qPCR results revealed that NtCIPK9 showed relatively higher expression levels in the root than in the leaf and stem before salt treatment (Figure 3H). However, NtCIPK9 transcription was upregulated in leaves after a 500 mM NaCl treatment (Figure 3H). Besides, the expression patterns in whole plants also showed the positive response of NtCIPK9 to salt stress (Supplementary Figure 1B and Supplementary Figure 3A).
Ectopic Expression of NtCIPK9 in Arabidopsis Promotes Seed Germination Under Salt Stress
To further investigate how NtCIPK9 affects salt tolerance, we overexpressed (35S:NtCIPK9) it in Arabidopsis. Seeds from three individual homozygous lines and WT were sown on ½ MS-agar plates to test their germination rate. On ½ MS without added NaCl, wild type and transgenic seeds showed no difference; yet on ½ MS with 100 and 150 mM added NaCl, 98.18% and 65.91% of 35S:NtCIPK9 seeds germinated, respectively, while only 54.39% and 7.42% of WT seeds germinated under the same conditions (Figures 4A–L). Therefore, we conclude that NtCIPK9 significantly promotes seed germination under salt stress conditions (Figure 4M).
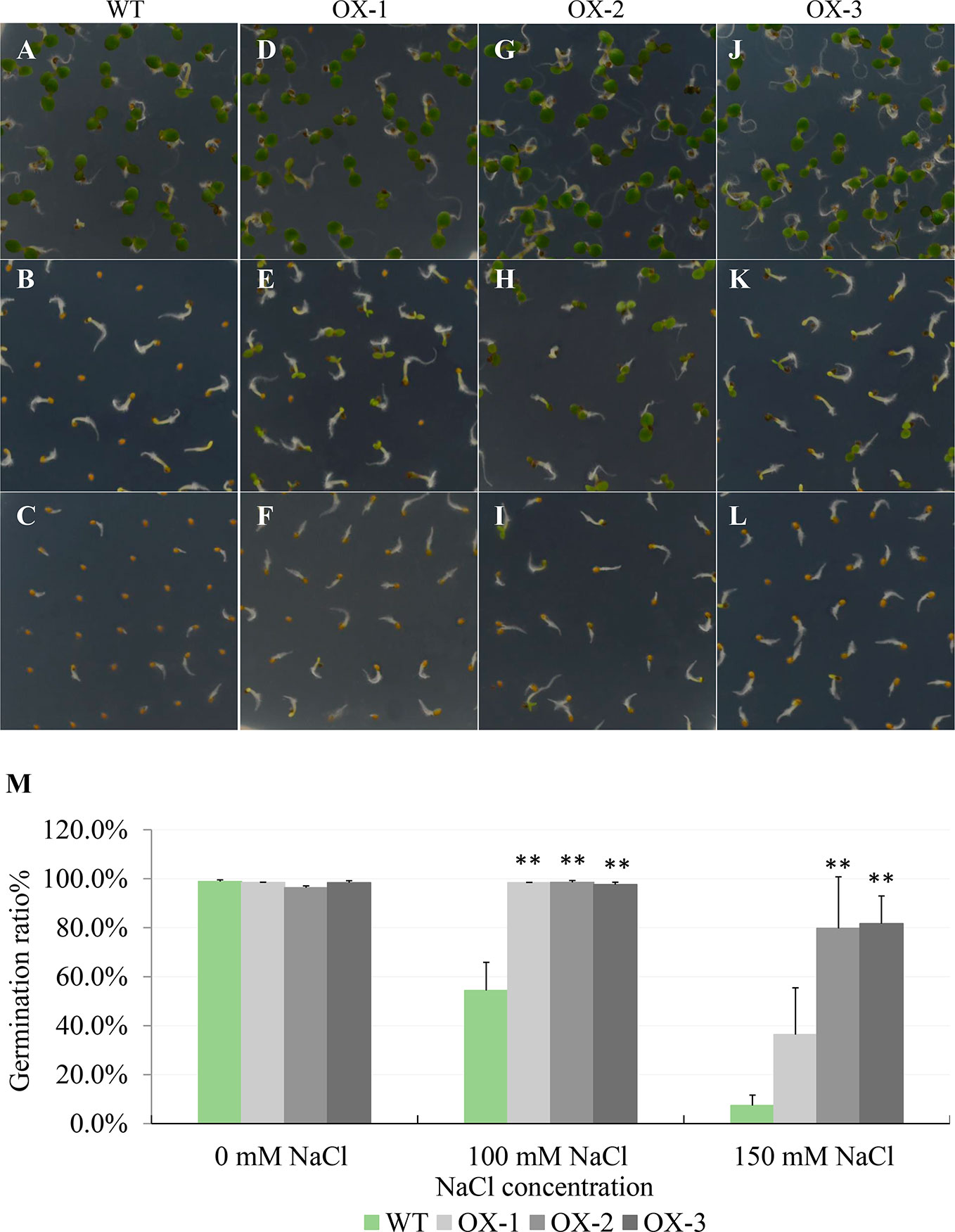
Figure 4 NtCIPK9 promotes seed germination rate of Arabidopsis. (A–C) Wild type seeds germinated on ½ MS medium with 0, 100, and 150 mM NaCl respectively after 4 days. (D–L) Three independent transgenic lines of 35S:NtCIPK9, depicted as OX-1, OX-2, and OX-3, germinated on different salt media as indicated. (M) Graphic of germination rate. Six technical replicates for each line were used in three biological replicates. **P < 0.01, ANOVA test was used for statistical analysis.
Ectopic Expressing NtCIPK9 Enhances Salt Tolerance in Arabidopsis
To address whether ectopic expression of NtCIPK9 could influence salt tolerance of plants, we grew 35S:NtCIPK9 and WT seeds on salt-rich media with 100 and 150 mM NaCl. 35S:NtCIPK9 seedlings showed better growth with more leaves and longer primary root on both media compared to WT plants, 20 days after germination (Figures 5A–C). This effect is more clear when plants grew on medium with a higher salt concentration (Figures 5B, C). To further assess salt-tolerance of the transgenic plants, 10-day-old seedlings were treated with 150 mM NaCl. 10 days after treatment, plants grown on medium without salt (Figure 6A) displayed no different phenotype. However, the number of whitening leaves and the mortality rate in WT were significantly higher than that of three transgenic lines grown on media with 150 mM NaCl (Figures 6B, C). In addition, enhanced tolerance to salt was also observed in plants grown in pots. Four weeks-old plants of WT and T2 heterozygous transgenic lines, four in a pot in duplicate, were irrigated with 200 mM NaCl for 4 days. All plants displayed withering blades from first day after salt treatment (Figures 7A–D). But the plants overexpressing NtCIPK9 showed a lower percentage of withering leaves than WT under salt stress (Figure 7E). Similarly, four-week-old T3 homozygous transgenic plants in pots also showed a higher salt tolerance than WT under 200 mM NaCl treatment for 4 days (Figures 8A, B).
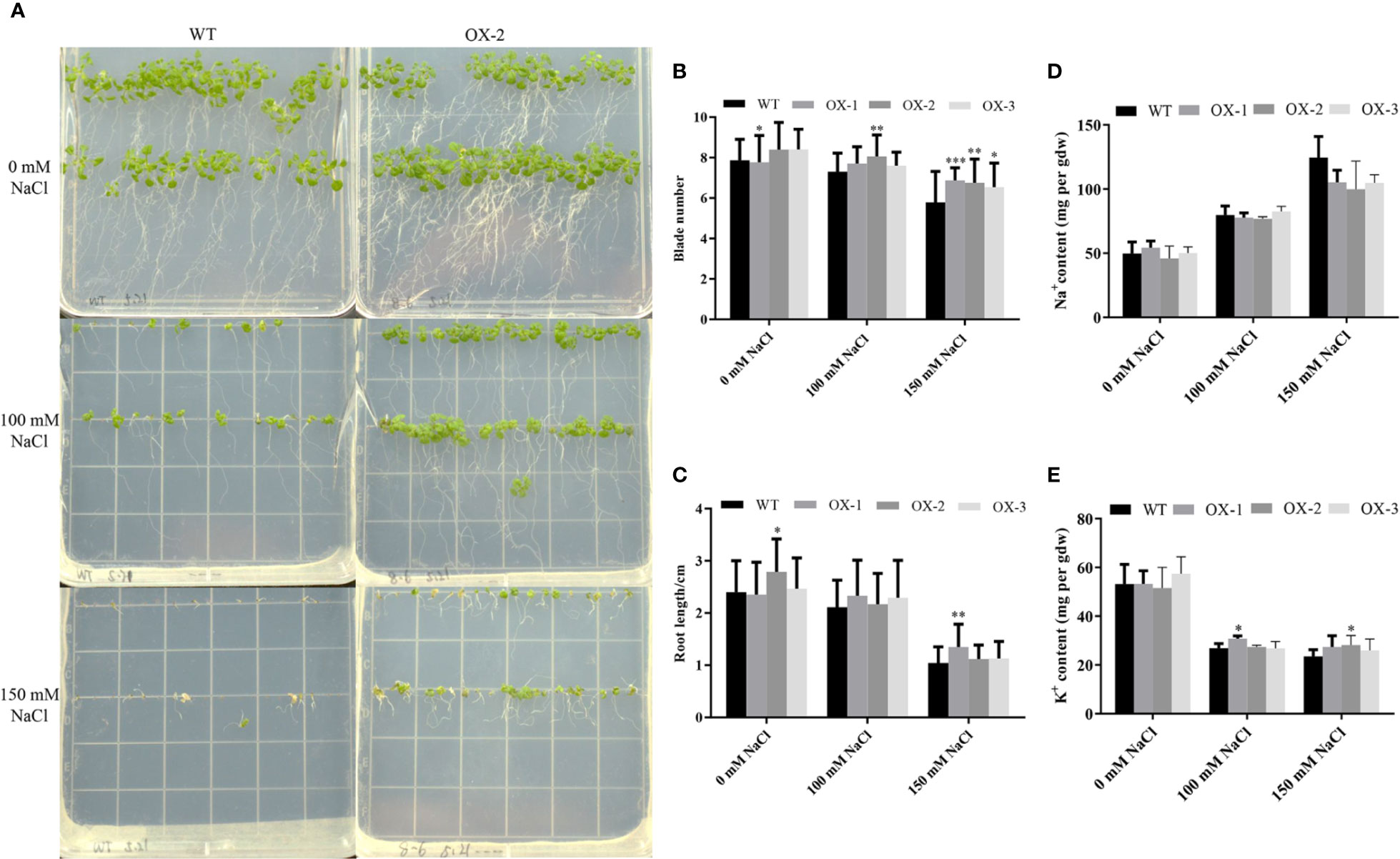
Figure 5 NtCIPK9 transgenic plants show more vigorous growth than WT. (A) Transgenic plants grew faster 20 days post germination under different NaCl treatment conditions. (B) Quantification of plant leaf number. (C) Plant primary root length. At least 90 plants were analyzed for each line. (D) Na+ content of whole seedlings treated with 100 mM NaCl. (E) K+ content of whole seedlings treated with 100 mM NaCl. Three biological replicates and three technical replicates were performed for each ion content test experiment. ***P < 0.001; **P < 0.01; *P < 0.05. ANOVA test was performed for statistical analysis. Data represent means ± SD from three technical replicates.
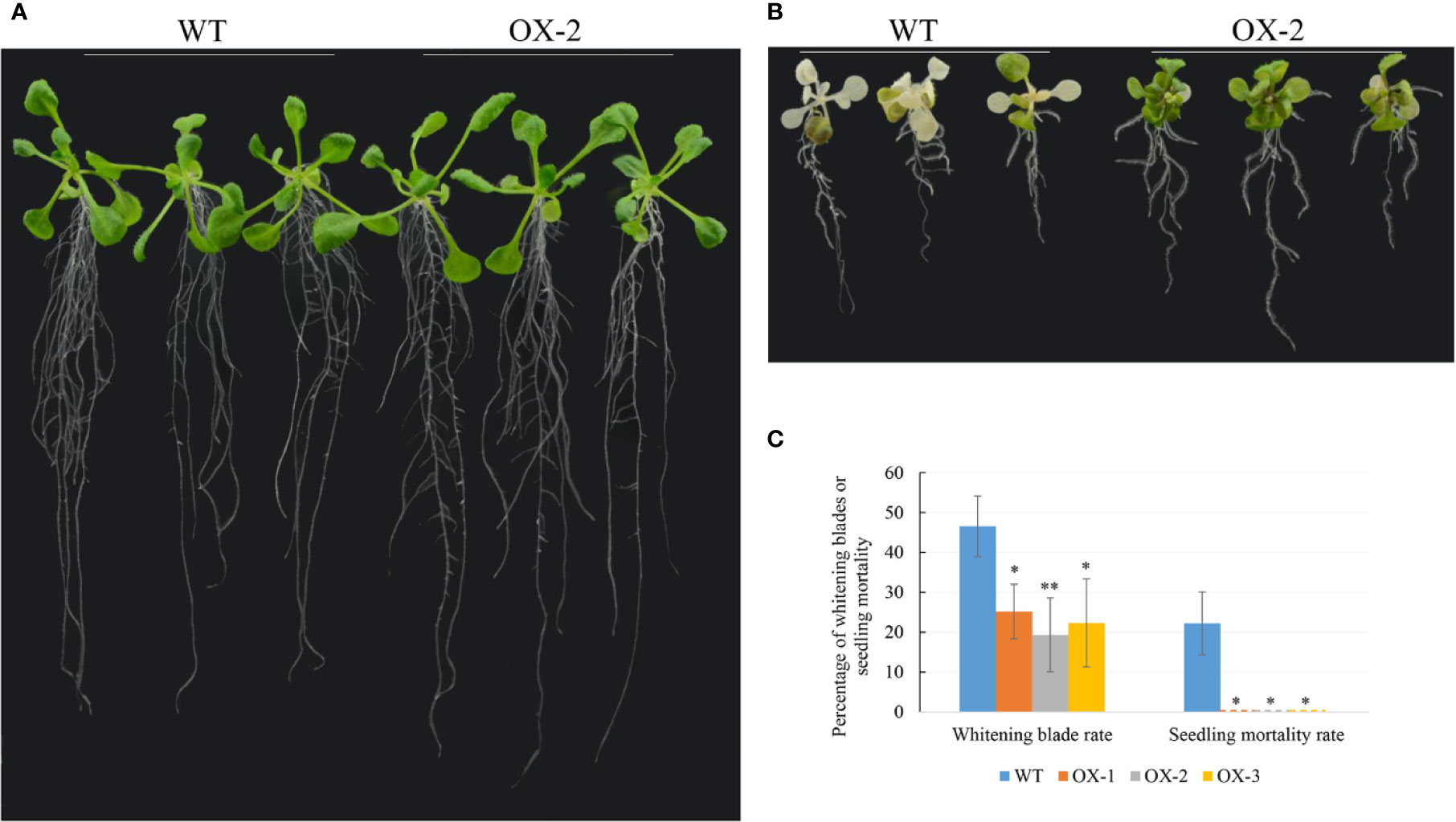
Figure 6 35S:NtCIPK9 transgenic plants show a lower mortality rate than WT. (A) WT and transgenic plants grown on media without added NaCl for 20 days. (B) The phenotypes of WT and transgenic plants grown on media supplemented with 150 mM NaCl for 10 days. (C) The percentage of chlorotic leaves and mortality of plants. 18 plants were used for statistics by ANOVA, three biological replicates for each experiment. ANOVA test was used for statistical analysis. **P < 0.01; *P < 0.05.
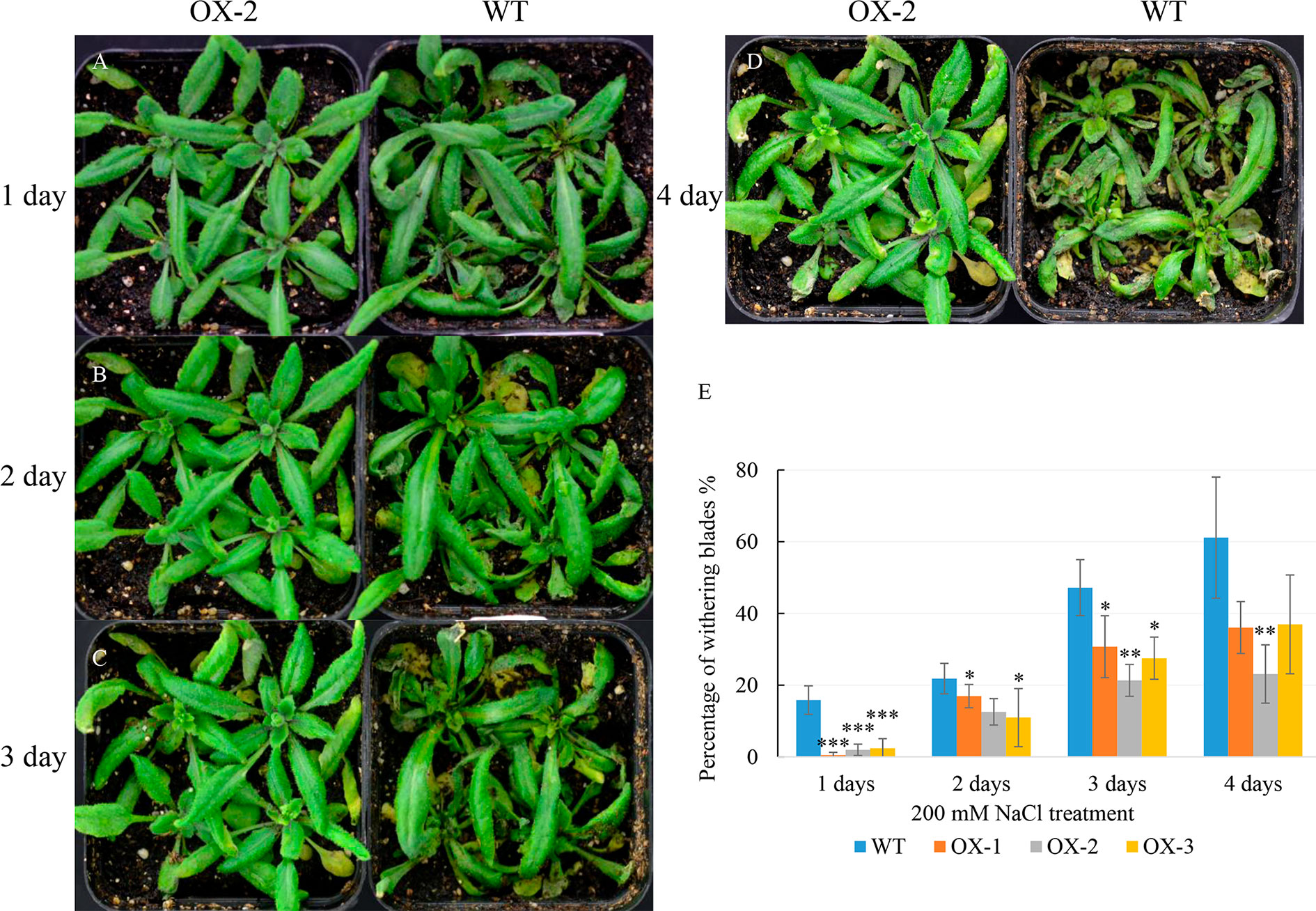
Figure 7 Improved salinity tolerance in heterozygous plants with NtCIPK9 overexpression. (A–D) 200 mM NaCl treated 4-week-old T2 transgenic plants and WT in pots for four days respectively. (E) Percentage of withering leaves during salt treatment in pots. ANOVA test was used for statistical analysis. ***P < 0.001; **P < 0.01; *P < 0.05. Four experimental repeats have been used in three independent replicates.
Ectopic Expression of NtCIPK9 in Arabidopsis Elevates K+ Accumulation
To investigate how ectopic expression of NtCIPK9 causes increased salt tolerance, we measured the Na+ and K+ content of 35S:NtCIPK9 transgenic plants under salt stress. Under normal conditions, the ion content of these transgenic lines has no difference with WT. By contrast, although salt stress increased the Na+ content of both WT and transgenic plants, transgenic plants do show a slightly lower Na+ content than WT (Figure 5D). More importantly, salt treatment reduced the K+ content of transgenic plants to a lesser extent than that of WT (Figure 5E).
To figure out what might be causing the difference in Na+ and K+ content, we analyzed five genes which are known to be involved in Na+ or/and K+ transportation. We found that AtHKT1 was around 2-fold upregulated in at least two NtCIPK9 transgenic plants, compared to wildtype plants treated in pots (Figure 8C). Similarly, the AtHKT1 expression level was also significantly upregulated in the transgenic plants treated on petri dishes (Figure 9A). Besides, the expression level of the other four genes (AtNHX1, AtNHX7, AtTRH1, and AtAKT2) was higher in the transgenic plants than in WT seedlings under salt stress (Figures 9B–E). To further ensure the function of CIPKs in salt stress, we checked the transcription of other known CIPKs in Nitraria tangutorum (NtCIPK2) (Zheng L. L. et al., 2014). The results showed that the CIPKs were also positively response to salt stress in both Nitraria tangutorum and Arabidopsis thaliana (Supplementary Figure 3).
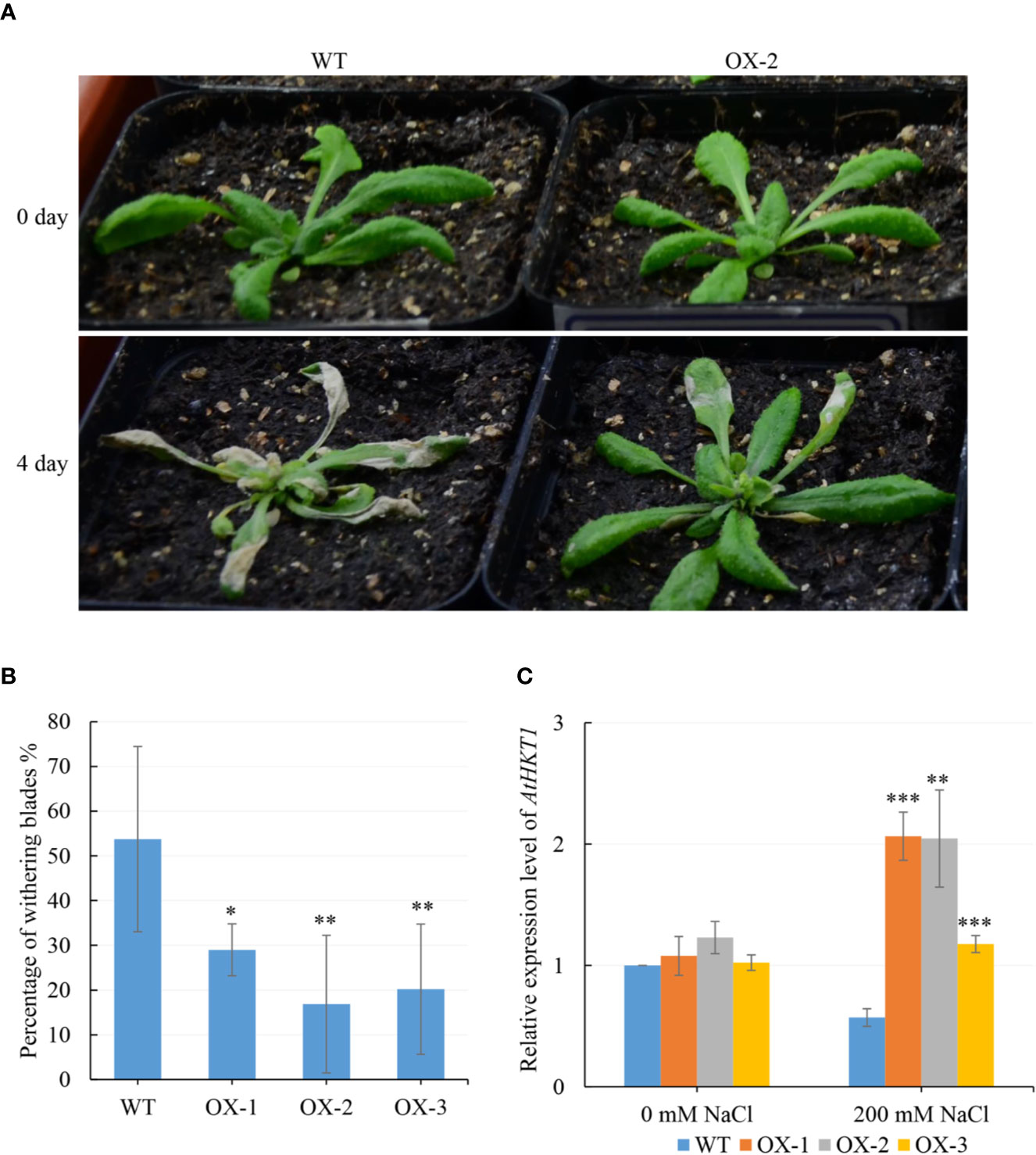
Figure 8 NtCIPK9 overexpressing homozygous Arabidopsis showed a higher salt tolerance than WT. (A) 200 mM NaCl treated T3 transgenic plants and WT for 4 days. (B) Percentage of withering blades. (C) AtHKT1 expression of transgenic plants (OX-1, OX-2, OX-3) and WT in pots treated by salt stress. ANOVA test was conducted to determine statistical significance of the results. ***P < 0.001; **P < 0.01; *P < 0.05.
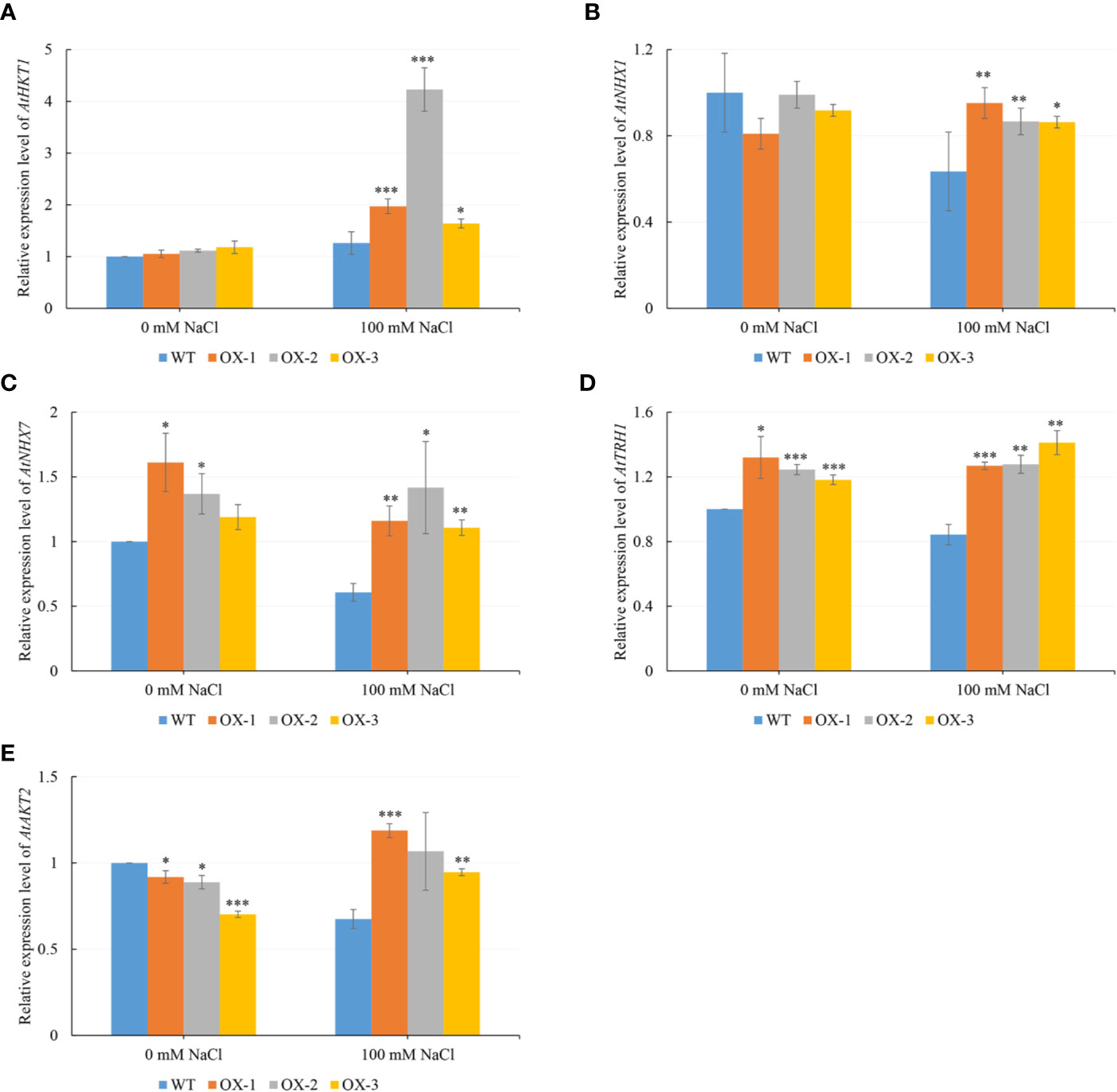
Figure 9 NtCIPK9 promoted the expression of Na+ or/and K+ transporter genes. (A–E) Gene expression level of transgenic plants (OX-1, OX-2, OX-3) and WT treated by 100 mM NaCl on petri dishes for 20 days. Data represent means ± SD from three technical replicates. ANOVA test was used for statistical analysis. ***P < 0.001; **P < 0.01; *P < 0.05.
Discussion
Salinization of arable land is a serious threat to agricultural and ecological stability. Therefore, halophytes have become promising candidates for further management of salinized areas. In order to adapt to their stressful environment, these plants developed a series of regulatory mechanisms during evolution. At the same time, genetic engineering of glycophytes by transforming genes from halophytes has been widely used to improve their salt resistance (Himabindu et al., 2016). In our study, we found that overexpression of NtCIPK9 from Nitraria tangutorum in Arabidopsis increased seed germination rate under salt stress. There’s one possible reason that could cause the higher seed germination of transgenic plants. AtCIPK3 from Arabidopsis has been reported to be involved in the phytohormone abscisic acid (ABA) response, which plays a vital role in seed maturation, dormancy, and seed germination (Finkelstein et al., 2002; Pandey et al., 2008b). Therefore, we thought NtCIPK9 increasing seed germination rate could also be related with plant endogenous ABA. On the media without NaCl, we found the WT has a high germination rate as the NtCIPK9-overexpressing seeds. NtCIPK9 here didn’t show a positive function on seed germination because the germination rate of WT was close to 100% at normal condition. However, NtCIPK9 effectively enhanced the seed germination under salt treatment, that reflected the function of NtCIPK9 on coping with salt stress.
BnCIPK9 from Brassica napus L. was reported to regulate seed oil content, a different function than was reported for Arabidopsis AtCIPK9 (Guo et al., 2018), suggesting that CIPK orthologs from different species can also have roles other than only being involved in salt tolerance. However, we found that NtCIPK9 may share a similar role in regulating ion homeostasis with the ortholog of Arabidopsis (Pandey et al., 2007; Li-Li et al., 2013). AtCIPK9 has been identified as a critical regulator of potassium transporters in Arabidopsis, that are involved in potassium acquisition, with some of them being critical for potassium nutrition under low potassium conditions (Pandey et al., 2007). In our study, the results also suggested that the overexpression of NtCIPK9 might regulate the expression of potassium transporter AtHKT1 to promote the homeostasis of Na+ and K+ in Arabidopsis resistance for salt stress. This revealed the common identity of orthologs from different species. However, other genes for cation transportation (AtNHX1, AtNHX7, AtTRH1 and AtAKT2), which have been reported to respond to salinity, were not upregulated even in transgenic plants after salt treatment. But compared with WT, the transgenic plants have a relative higher expression level (Figures 9B–E). The possible reason could be that these gene are not the key factors regulated by NtCIPK9; or these genes responding to the stress are asynchrony because of the necessity for long-time stress resistance.
Conclusion
In our research, we identified a novel CIPK gene, NtCIPK9, which positively responds to salt stress in Nitraria tangutorum. Overexpression of NtCIPK9 in Arabidopsis plants increases seed germination rate, root length, leaf number, and reduces mortality rate under salt stress. Furthermore, NtCIPK9 may enhance the tolerance of transgenic plants to salinity by increasing the expression level of genes in balancing ion homeostasis after the salt treatment. Altogether, our study revealed that NtCIPK9 from the halophyte Nitraria tangutorum could improve the salt tolerance of Arabidopsis, which would further contribute to the genetic engineering of other glycophytes for stronger salt resistance and sheds light on the molecular mechanism causing the enhanced resistance. However, more practical application of halophytes facing various degree of stresses need to be further investigated.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/nuccore/MN852853.
Author Contributions
JC and JS contributed conception and design of the study. LZ, ML, JZ, XY, PW, YL, TC, and YY performed the experiments and carried out the statistical analysis. LL and XC wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Key research and development plan of Jiangsu Province (BE2017376), Foundation of Jiangsu forestry bureau (LYKJ[2017]42), the Nature Science Foundation of China (31770715), the Qinglan project of Jiangsu province, A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Insitutions (PAPD), Natural Science Foundation of Jiangsu Province (BK20181176) and the Joint Fund of the Natural Science Foundation of China and the Karst Science Research Center of Guizhou Province (Grant No. U1812401).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01112/full#supplementary-material
References
Albrecht, V., Ritz, O., Linder, S., Harter, K., Kudla, J. (2001). The NAF domain defines a novel protein–protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 20, 1051–1063. doi: 10.1093/emboj/20.5.1051
Ashraf, M., Akram, N. A. (2009). Improving salinity tolerance of plants through conventional breeding and genetic engineering: an analytical comparison. Biotechnol. Adv. 27, 744–752. doi: 10.1016/j.biotechadv.2009.05.026
Bausher, M. G., Singh, N. D., Lee, S.-B., Jansen, R. K., Daniell, H. (2006). The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var’Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 6, 1. doi: 10.1186/1471-2229-6-21
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chase, M. W., Reveal, J. L. (2009). A phylogenetic classification of the land plants to accompany APG III. Bot. J. Linn. Soc. 161, 122–127. doi: 10.1111/j.1095-8339.2009.01002.x
Chen, X., Gu, Z., Xin, D., Hao, L., Liu, C., Huang, J., et al. (2011). Identification and characterization of putative CIPK genes in maize. J. Genet. Genomics 38, 77–87. doi: 10.1016/j.jcg.2011.01.005
Cheong, Y. H., Kim, K.-N., Pandey, G. K., Gupta, R., Grant, J. J., Luan, S. (2003). CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15, 1833–1845. doi: 10.1105/tpc.012393
Chinnusamy, V., Schumaker, K., Zhu, J. K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 55, 225–236. doi: 10.1093/jxb/erh005
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Finkelstein, R. R., Gampala, S. S. L., Rock, C. D. (2002). Abscisic Acid Signaling in Seeds and Seedlings. Plant Cell 14, S15. doi: 10.1105/tpc.010441
Geldner, N., Dénervaud-Tendon, V., Hyman, D. L., Mayer, U., Stierhof, Y. D., Chory, J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59, 169–178. doi: 10.1111/j.1365-313X.2009.03851.x
Grefen, C., Blatt, M. R. (2012). Do calcineurin B-like proteins interact independently of the serine threonine kinase CIPK23 with the K+ channel AKT1? Lessons learned from a menage a trois. Plant Physiol. 159, 915–919. doi: 10.1104/pp.112.198051
Group, T. (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 161, 105–121. doi: 10.1111/boj.12385
Guo, Y., Halfter, U., Ishitani, M., Zhu, J.-K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13, 1383–1400. doi: 10.1105/TPC.010021
Guo, Y., Huang, Y., Gao, J., Pu, Y., Wang, N., Shen, W., et al. (2018). CIPK9 is involved in seed oil regulation in Brassica napus L. and Arabidopsis thaliana (L.) Heynh. Biotechnol. Biofuels 11, 124. doi: 10.1186/s13068-018-1122-z
Halfter, U., Ishitani, M., Zhu, J.-K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. 97, 3735–3740. doi: 10.1073/pnas.97.7.3735
Himabindu, Y., Chakradhar, T., Reddy, M. C., Kanygin, A., Redding, K. E., Chandrasekhar, T. (2016). Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 124, 39–63. doi: 10.1016/j.envexpbot.2015.11.010
Hu, L., Lu, H., Liu, Q., Chen, X., Jiang, X. (2005). Overexpression of mtlD gene in transgenic Populus tomentosa improves salt tolerance through accumulation of mannitol. Tree Physiol. 23, 1273–1281. doi: 10.1093/treephys/25.10.1273
Hu, D. G., Ma, Q. J., Sun, C. H., Sun, M. H., You, C. X., Hao, Y. J. (2016). Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol. Plant. 156, 201–214. doi: 10.1111/ppl.12354
Jie, Z., Hui, L., Ding, C. X., Suo, Y. R., Wang, L. S., Wang, H. L. (2011). Anthocyanins composition and antioxidant activity of two major wild Nitraria tangutorun Bobr. variations from Qinghai–Tibet Plateau. Food Res. Int. 44, 2041–2046. doi: 10.1016/j.foodres.2010.07.008
Karpiuk, U. V., Azzam, K. M. A., Abudayeh, Z. H. M., Kislichenko, V., Naddaf, A., Cholak, I., et al. (2016). Qualitative and quantitative content determination of macro-minor elements in Bryonia Alba L. roots using flame atomic absorption spectroscopy technique. Adv. Pharm. Bull. 6, 285. doi: 10.15171/apb.2016.040
Kato, Y., Sakaguchi, M., Mori, Y., Saito, K., Nakamura, T., Bakker, E. P., et al. (2001). Evidence in support of a four transmembrane-pore-transmembrane topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proc. Natl. Acad. Sci. 98, 6488–6493. doi: 10.1073/pnas.101556598
Kim, K.-N., Cheong, Y. H., Gupta, R., Luan, S. (2000). Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 124, 1844–1853. doi: 10.1104/pp.124.4.1844
Kim, K.-N., Cheong, Y. H., Grant, J. J., Pandey, G. K., Luan, S. (2003). CIPK3, a calcium sensor–associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15, 411–423. doi: 10.1105/tpc.006858
Kolukisaoglu, Weinl, S., Blazevic, D., Batistic, O., Kudla, J. (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58. doi: 10.1104/pp.103.033068
Kudla, J., Xu, Q., Harter, K., Gruissem, W., Luan, S. (1999). Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. 96, 4718–4723. doi: 10.1073/pnas.96.8.4718
Li, L., Kim, B.-G., Cheong, Y. H., Pandey, G. K., Luan, S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. 103, 12625–12630. doi: 10.1073/pnas.0605129103
Li, R., Zhang, J., Wu, G., Wang, H., Chen, Y., Wei, J. (2012). HbCIPK2, a novel CBL-interacting protein kinase from halophyte Hordeum brevisubulatum, confers salt and osmotic stress tolerance. Plant Cell Environ. 35, 1582–1600. doi: 10.1111/j.1365-3040.2012.02511.x
Li-Li, L., Hui-Min, R., Li-Qing, C., Yi, W., Wei-Hua, W. (2013). A protein kinase, calcineurin B-like protein-interacting protein Kinase9, interacts with calcium sensor calcineurin B-like Protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant Physiol. 161, 266–277. doi: 10.1104/pp.112.206896
Lu, L., Li, X., Hao, Z., Yang, L., Zhang, J., Peng, Y., et al. (2018). Phylogenetic studies and comparative chloroplast genome analyses elucidate the basal position of halophyte Nitraria sibirica (Nitrariaceae) in the Sapindales. Mitochondrial DNA Part A 29, 745–755. doi: 10.1080/24701394.2017.1350954
Luan, S. (2009). The CBL–CIPK network in plant calcium signaling. Trends Plant Sci. 14, 37–42. doi: 10.1016/j.tplants.2008.10.005
Lyzenga, W. J., Liu, H., Schofield, A., Muise-Hennessey, A., Stone, S. L. (2013). Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin–proteasome system. J. Exp. Bot. 64, 2779–2791. doi: 10.1093/jxb/ert123
Mahajan, S., Tuteja, N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444, 139–158. doi: 10.1016/j.abb.2005.10.018
Pandey, G. K., Yong, H. C., Kim, B. G., Grant, J. J., Li, L. (2007). CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell Res. 17, 411–421. doi: 10.1038/cr.2007.39
Pandey, G., Cheong, Y., Li, L., Luan, S. (2008a). The calcineurin B-like protein CBL9 and its interacting kinase CIPK3 functions in ABA-regulated seed germination. Mol. Plant 1, 238–248. doi: 10.1093/mp/ssn003
Pandey, G. K., Grant, J. J., Hwa, C. Y., Beom-Gi, K., Gong, L. L., Sheng, L. (2008b). Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 1, 238–248. doi: 10.1093/mp/ssn003
Piao, H.-L., Xuan, Y.-H., Park, S. H., Je, B. I., Park, S. J., Park, S. H., et al. (2010). OsCIPK31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants. Mol. Cells 30, 19–27. doi: 10.1007/s10059-010-0084-1
Qiu, Q.-S., Guo, Y., Dietrich, M. A., Schumaker, K. S., Zhu, J.-K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. 99, 8436–8441. doi: 10.1073/pnas.122224699
Rigó, G., Valkai, I., Faragó, D., Kiss, E., Van, H. S., Van, D. S. N., et al. (2016). Gene mining in halophytes: functional identification of stress tolerance genes in Lepidium crassifolium. Plant Cell Environ. 39, 2074–2084. doi: 10.1111/pce.12768
Sánchez-Barrena, M. J., Martínez-Ripoll, M., Zhu, J.-K., Albert, A. (2005). The structure of the Arabidopsis thaliana SOS3: molecular mechanism of sensing calcium for salt stress response. J. Mol. Biol. 345, 1253–1264. doi: 10.1016/j.jmb.2004.11.025
Shi, J., Kim, K.-N., Ritz, O., Albrecht, V., Gupta, R., Harter, K., et al. (1999). Novel protein kinases associated with calcineurin B–like calcium sensors in Arabidopsis. Plant Cell 11, 2393–2405. doi: 10.1105/tpc.11.12.2393
Shi, H., Ishitani, M., Kim, C., Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. 97, 6896–6901. doi: 10.1073/pnas.120170197
Swofford, D. L. (2002). “PAUP: phylogenetic analysis using parsimony, version 4.0 b10” (Sunderland, MA: Sinauer Associates).
Tripathi, V., Parasuraman, B., Laxmi, A., Chattopadhyay, D. (2018). CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 58 (5), 778–790. doi: 10.1111/j.1365-313X.2009.03812.x
Vaidya, G., Lohman, D. J., Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Wang, W., Vinocur, B., Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Wang, L., Fei-Feng, L. I., Zhang, W. B., Chen, G. L., Lin, X. F. (2012). Isolation and characterization of Nitraria sibirica actin gene. Acta Prataculturae Sin. 21, 151–158.
Wang, N., Gao, J., Zhang, S. Q., Wang, G. X. (2014). Variations in leaf and root stoichiometry of Nitraria tangutorum along aridity gradients in the Hexi Corridor, northwest China. Contemp. Probl. Ecol. 7, 308–314. doi: 10.1134/S1995425514030123
Xu, J., Li, H.-D., Chen, L.-Q., Wang, Y., Liu, L.-L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. doi: 10.1016/j.cell.2006.06.011
Yang, Y., Shi, R., Wei, X. (2010a). Effect of salinity on antioxidant enzymes in calli of the halophyte Nitraria tangutorum Bobr. Plant Cell Tissue Organ Cult. (PCTOC) 102, 387–395. doi: 10.1007/s11240-010-9745-1
Yang, Y., Wei, X., Shi, R., Fan, Q. (2010b). Salinity-induced physiological modification in the callus from Halophyte nitraria tangutorum Bobr. J. Plant Growth Regul. 29, 465–476. doi: 10.1007/s00344-010-9158-8
Yang, Y. L., Zhang, Y. Y., Lu, J., Zhang, H., Liu, Y., Jiang, Y., et al. (2012). Exogenous H2O2 increased catalase and peroxidase activities and proline content in Nitraria tangutorum callus. Biol. Plant. 56, 330–336. doi: 10.1007/s10535-012-0094-2
Yang, Y., Yang, F., Li, X. (2013). Signal regulation of proline metabolism in callus of the halophyte Nitraria tangutorum Bobr. grown under salinity stress. Plant Cell Tissue Organ Cult. (PCTOC) 112, 33–42. doi: 10.1007/s11240-012-0209-7
Yokoi, S., Quintero, F. J., Cubero, B., Ruiz, M. T., Bressan, R. A., Hasegawa, P. M., et al. (2002). Differential expression and function of Arabidopsis thaliana NHX Na + /H + antiporters in the salt stress response. Plant J. 30, 529–539. doi: 10.1046/j.1365-313X.2002.01309.x
Yu, Y., Xia, X., Yin, W., Zhang, H. (2007). Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul. 52, 101–110. doi: 10.1007/s10725-007-9165-3
Zhang, H., Yang, B., Liu, W.-Z., Li, H., Wang, L., Wang, B., et al. (2014). Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 14, 1. doi: 10.1186/1471-2229-14-8
Zhao, F., Mcgrath, S., Crosland, A. (1994). Comparison of three wet digestion methods for the determination of plant sulphur by inductively coupled plasma atomic emission spectroscopy (ICP-AES). Commun. Soil Sci. Plant Anal. 25, 407–418. doi: 10.1080/00103629409369047
Zhao, K., Hai, F., Ungar, I. A. (2002). Survey of halophyte species in China. Plant Sci. 163, 491–498. doi: 10.1016/S0168-9452(02)00160-7
Zhao, J., Sun, Z., Zheng, J., Guo, X., Dong, Z., Huai, J., et al. (2009). Cloning and characterization of a novel CBL-interacting protein kinase from maize. Plant Mol. Biol. 69, 661–674. doi: 10.1007/s11103-008-9445-y
Zheng, L., Dang, Z., Li, H., Zhang, H. (2014). Isolation and characterization of a Δ1-pyrroline-5-carboxylate synthetase (NtP5CS) from Nitraria tangutorum Bobr. and functional comparison with its Arabidopsis homologue. Mol. Biol. Rep. 41, 563–572. doi: 10.1007/s11033-013-2893-8
Zheng, L. L., Gao, Z., Wang, J., Zhang, H. R., Wang, Y. C. (2014). Molecular cloning and functional characterization of a novel CBL-interacting protein kinase NtCIPK2 in the halophyte Nitraria tangutorum. Genet. Mol. Res. GMR 13, 4716–4728. doi: 10.4238/2014.July.2.1
Zhou, J., Wang, J., Bi, Y., Wang, L., Tang, L., Yu, X., et al. (2014). Overexpression of PtSOS2 enhances salt tolerance in transgenic poplars. Plant Mol. Biol. Rep. 32, 185–197. doi: 10.1007/s11105-013-0640-x
Keywords: halophyte, Nitraria tangutorum, CIPK9, ion homeostasis, salt tolerance
Citation: Lu L, Chen X, Zhu L, Li M, Zhang J, Yang X, Wang P, Lu Y, Cheng T, Shi J, Yi Y and Chen J (2020) NtCIPK9: A Calcineurin B-Like Protein-Interacting Protein Kinase From the Halophyte Nitraria tangutorum, Enhances Arabidopsis Salt Tolerance. Front. Plant Sci. 11:1112. doi: 10.3389/fpls.2020.01112
Received: 14 October 2019; Accepted: 06 July 2020;
Published: 21 August 2020.
Edited by:
Jayakumar Bose, University of Adelaide, AustraliaReviewed by:
Dong-Ha Oh, Louisiana State University, United StatesSalman Gulzar, University of Karachi, Pakistan
Copyright © 2020 Lu, Chen, Zhu, Li, Zhang, Yang, Wang, Lu, Cheng, Shi, Yi and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinhui Chen, Y2hlbmpoQG5qZnUuZWR1LmNu
†These authors have contributed equally to this work
 Lu Lu
Lu Lu Xinying Chen
Xinying Chen Liming Zhu1
Liming Zhu1 Jingbo Zhang
Jingbo Zhang Xiuyan Yang
Xiuyan Yang Pengkai Wang
Pengkai Wang Ye Lu
Ye Lu Tielong Cheng
Tielong Cheng Jisen Shi
Jisen Shi Yin Yi
Yin Yi Jinhui Chen
Jinhui Chen