- 1College of Life Sciences, Capital Normal University, and Beijing Key Laboratory of Plant Gene Resources and Biotechnology for Carbon Reduction and Environmental Improvement, Beijing, China
- 2State Key Laboratory of Plant Genomics, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
In higher-plant reproduction, the compatibility of pollen tube germination in the pistil is essential for successful double fertilization. It has been reported that Mildew Locus O (MLO) family gene NTA (MLO7), expressing in synergid cells, can correctly guide pollen tubes. However, the molecular mechanism underlying the interacting partners to MLOs in the fertilization is still unknown. In our study, we identified the direct protein interaction between CML9 and MLO10 within a non-canonical CaMBD. In GUS reporter assays, CML9 expresses in a high level in pollens, whereas MLO10 can be specifically detected in stigma which reaches up to a peaking level before fertilization. Therefore, the spatio-temporal expression patterns of MLO10 and CML9 are required for the time-window of pollination. When we observed the pollen germination in vitro, two cml9 mutant alleles dramatically reduced germination rate by 15% compared to wild-type. Consistently, the elongation rate of pollen tubes in planta was obviously slow while manually pollinating cml9-1 pollens to mlo10-1 stigmas. Additionally, cml9-1 mlo10-1 double mutant alleles had relatively lower rate of seed setting. Taken together, protein interaction between MLO10 and CML9 is supposed to affect pollen tube elongation and further affect seed development.
Introduction
Reproductive process is critical for flowering plants. After physically attaching to the stigma epidermal, pollen grains successively adhere, hydrate, and germinate. Then pollen tubes penetrating along the style are guided to the ovary to release the sperms (Zheng et al., 2018). Those steps require the matured male and female organs to communicate and recognize each other, and any molecule inhibiting male–female recognition leads to incompatibility. During pollination, pollen–stigma recognition usually depends on specific protein–protein interactions, which is classified into several types of compatibilities (Knox et al., 1976; Dumas and Knox, 1983). For example, when expressing the SRK (S-locus receptor kinase) and SCR (S-locus cysteine rich protein) genes in the stigma and pollen respectively, SCR–SRK protein interaction leads to unsuccessful germination of stigma-attached pollens (Schopfer et al., 1999; Takasaki et al., 2000; Nasrallah et al., 2002). In the GSI (Gametophytic Self-incompatibility) type, while S-RNase in the stigma recognizes and interacts with SFB (S-locus F-Box) from the germinated pollen, it results in defective of pollen tube elongation (McClure and Franklin-Tong, 2006; Hiscock and Allen, 2008). In pollination stages, specific protein interactions determine the compatibility or incompatibility of pollens with stigma and other pistil parts.
When a pollen tube is guided to the ovule, several recognition combinations of ligand-receptors like LURE1-MIK/MDIS1, RALF34-BUPS-ANX have been characterized (Wang et al., 2016; Ge et al., 2017). In Mildew Locus O (MLO) gene family, recent studies have also identified a fertilization related gene MLO7 (also named NOTIA) localized in synergid cells. In mlo7 mutant, the pollen tube cannot be correctly guided to the embryo sac, which further fails to burst the tip and release sperm cells to continue the double fertilization (Kessler et al., 2010; Jones and Kessler, 2017). Based on phylogenetic analysis, MLO10, expressed in flower organs, is considered to be a MLO7-related gene. However, it remains unclear whether MLO10 is involved in fertilization (Wuest et al., 2010; Jones et al., 2017). Therefore, we aim to investigate MLO10 as well as its interaction partners that may mediate recognition between the pollen tube and stigma or embryo sac through direct protein–protein interaction.
Based on bioinformatic analysis, it has been found that one of the domains called CaMBD (Calmodulin binding domain) is significantly enriched in the C-terminal of 15 MLO proteins (Knox et al., 1976; Feechan et al., 2008; Appiano et al., 2015). These indicate that MLOs may interact with calmodulins to regulate the reproduction process. Accumulated pieces evidence proves that calcium plays a critical role in plant fertilization (Iwano et al., 2009; Qin and Yang, 2011; Hepler et al., 2012; Wudick et al., 2018; Zhang et al., 2020). Calmodulins (CaMs) or Calmodulins like (CMLs) proteins have been identified to regulate plant development and stress mediated reactions (Cheval et al., 2013; Zhu et al., 2015). Several CaMs and CMLs have been found to be involved in pollen germination and tube elongation. For example, CaM2 forms a complex with CNGC8/18 to control pollen tube elongation through modulating calcium oscillations at the tips (Pan et al., 2019). Meanwhile, mutation of CaM2 significantly inhibits pollen germination (Landoni et al., 2010; Leba et al., 2012). CML25 was also reported to affect pollen tube development and seed setting (Wang et al., 2015).
In conclusion, although some components of MLOs or CaM/CMLs have been reported to be involved in plant reproduction, the direct link between these two-family proteins is missing. In our study, we have identified the new protein interacting complex MLO10–CML9 which is involved in pollen germination, pollen tube elongation as well as seed setting. We also investigate and speculate how the direct interaction between stigma-expressed MLO10 and pollen-expressed CML9 regulates reproductive physiology.
Materials and Methods
Plant Materials and Growth Conditions
Experiments with Arabidopsis thaliana were performed on ecotype Columbia-0. The knock-out mutant lines cml9-1 (SALK_126787C) and cml9-2 (SALK_006380C), were obtained from the ABRC (https://www.arabidopsis.org/abrc/) as described (Heyer et al., 2018). Seeds were stratified for 2 d at 4°C in darkness and then grown under a 16-h-light/8-h-dark cycle at a photon fluence rate of approximately 120 μmol·m−2·s−1 during the day. Plants were kept at 22°C with approximately 60% humidity. The plates containing 0.5 strength Murashige and Skoog media and 1% (w/v) sucrose solidified with 0.6% (w/v) Agar. After 7 days, the seedlings were transplanted into the soil under the same light cycle, temperature, and humidity.
CRISPR-Cas9 Mediated Gene Editing Events of MLO10 in Arabidopsis
The method for MLO10 editing was performed as Gao et al. (2016) described. PHEN401 is used as MLO10 gene-editing vector. The CRISPR/Cas9 constructs were transformed into Arabidopsis wild-type Columbia-0 through floral dipping. T1 plants were selected on 25 µg·L−1 hygromycin B. Genomic DNA samples extracted from leaf tissues of 2-week-old T1 plants were used as templates for PCR. The PCR product amplified with targeting site-specific primers was digested using restriction enzyme XhoI. Putative mutations should produce XhoI-resistant band. Then results were verified by sequencing the PCR products. Cas9-free T3 seeds were isolated. Our experiments were performed on the Cas9-free lines mlo10-1 and mlo10-2 (Supplementary Figure 3A). The primers used in these cloning procedures are listed in Table 1.
Yeast Two-Hybrid Assays
Coding regions of MLO10-CT-(P1-P5) and CaM2, CaM4, CaM6, CaM7, CML8, CML9, CML10 sequences were cloned into the pCBKT7 vector and pGADT7 vector respectively. Primers are listed in Table 1. Then the plasmids (MLO10-CT::BD and CaMs/CMLs::AD, MLO10-CT::BD fragments P1–P5 and CML9::AD, respectively) were co-transformed into the yeast AH109 strain, using the method of PEG/LiAc. Transformants were selected on the SD medium lacking Leu and Trp. After 3 days of growth, the haploid cells were transferred to a selected medium containing 1.5 mM 3-AT but lacking Leu, Trp, His. The X-Gal filter assay was done as described earlier and modified (Moeini-Naghani and Navaratnam, 2016). The primers used in these cloning procedures are listed in Table 1.
Subcellular Localization and BiFC Assay
We used Arabidopsis mesophyll protoplasts for the GFP and BiFC transient expression assay. To generate the 35S::MLO10-GFP, 35S::CML9-GFP (Green Fluorescent Protein) plasmid, CDS sequences (without stop codon) were inserted into the pCAMBIA1302 vector. For BiFC vectors, sequences of MLO10 and CML9 were inserted into pSAT1-nVenus-C (pE3242) and pSAT1-cCFP-C (pE3228) respectively. Primers are listed in Table 1. Plasmids were transferred into Arabidopsis thaliana mesophyll protoplasts as described (Yoo et al., 2007). After 16 h of incubation at 23°C in the dark, the GFP or CFP, YFP signals were detected using a confocal laser scanning microscopy (LSM 780; Carl Zeiss).
Construction of Transgenic Lines and Analyses of GUS Activity
About 5-week-old Col wild type plants with several mature flowers were transformed with the MLO10pro::GUS and CML9pro::GUS constructs in PBI101 respectively, via the Agrobacterium tumefaciens strain GV3101 by using the floral dip method (Bent and Clough, 1998). The primers used in these cloning procedures are listed in Table 1. T1 transgenic plant seedlings were selected on 1/2 MS plates containing 50 μg/L kanamycin. Examination of GUS activity in transgenic seedlings was performed as described (Rosado et al., 2012). Samples were fixed for 20 min in ice-cold 90% (vol/vol) acetone, were washed three times (5 min per wash) with ice-cold phosphate buffer [100 mM sodium phosphate (pH 7), 10 μM EDTA, 0.1% Triton X-100, 2 mM K4Fe(CN)6.3H2O, 2 mM K3Fe(CN)6], and were stained in X-gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid) solution (2 mM X-gluc in the same phosphate buffer) at 37°C in darkness, check the GUS activity every 1 h. After the assay these tissues were washed in 70% ethanol, 80% ethanol, 90% ethanol, and finally in 100% ethanol. The plant tissues after GUS staining were photographed with microscope (Discovery.V20, ZEISS).
Aniline Blue Staining Assay
Aniline blue staining of pollen tubes in pistils was performed as described (Jiang et al., 2005) and modified. The pre-emasculated mature wild-type and mlo10-1 stigma were pollinated either with wild-type or cml9-1 pollen respectively. The pollinated pistils were collected 4 hap (hour after pollination) and briefly fixed in a fixing solution of ethanol:acetic acid (3:1) for 2 h at room temperature. The fixed pistils were washed three times with distilled water and treated in softening solution of 8 M NaOH overnight. Then, the pistil tissues were washed in distilled water and stained in aniline blue solution (0.1% aniline blue in 0.1 M K4Fe(CN)6•3H2O buffer, pH 11) for 3 to 5 h in the dark. The stained pistils were observed and photographed with a Leica DMRA fluorescence microscope. The images were digitized, and the pollen tube lengths were measured using ImageJ 1.52a software.
Analysis of Pollen Germination and Pollen Tubes Length
In vitro germination assays were modified as described (Fan et al., 2001; Mouline et al., 2002). Pollens were collected by tapping the open anthers on solid agar and then incubated in the dark at 28°C. The solid pollen germination medium consisted of 10 μg ·ml−1 inositol, 1.5 mM H3BO3, 5 mM MES, 10 mM CaCl2, 1 mM KCl, 0.8 mM MgSO4, 1% agar, and 20% sucrose, pH 5.8. For analysis of pollen germination and pollen tube growth in vitro, samples were incubated for 6 h. Pollen tubes were photographed with microscope (Discovery.V20, ZEISS). The images were digitized, and the pollen tube lengths were measured using ImageJ 1.52a software.
Results
Physical Interaction Between CML9 and MLO10
MLO family proteins contain one CaM-binding domain (CaMBD) in most higher plants. It has been reported that HvMLO1 can bind to CaMs via CaMBDs at the C-terminal (CT) cytosolic regions. We focused on the possible interacting partners especially the calcium sensor proteins to MLO10 in Arabidopsis thaliana. However, CaM binding has not been studied well with MLO10, and the specific binding sites between CaMs and MLO10 remain unknown. First, based on the bioinformatic prediction by TMHMM 2.0, MLO10-CT is supposed to localize at the cytoplasm. Thus, taking MLO10-CT as bait, we conducted a yeast two hybrid (Y2H) screen with Arabidopsis CaMs and CMLs. Interestingly, we identified the interaction event between MLO10 and CML9. When co-expressing MLO10-CT-BD and CML9-AD, the yeast colony became blue by adding X-gal into the selection media (Figure 1A), which indeed confirms the direction interaction of MLO10-CML9. However, no obvious colony was observed when combining MLO10 and CML9 isoforms like CaM2, CaM4, CaM6, CaM7 and CML8, CML10 (Figure 1A, Supplementary Figure 1), which suggests the MLO10–CML9 interaction could be specific. Next, we wanted to map the specific interaction domain of MLO10. When aligning amino acids of several MLOs, generally the divergent domain ranges from 430 to 590 aa (MLO C-terminal), and the putative CaMBD, which was marked with arrows (Supplementary Figures 2A, C), may be involved in determining the specificity of protein interactions. Therefore, we divided MLO10-CT which has 136 aa (434–569 aa) into five truncation fragments P1–5 (Figure 1B). Surprisingly, through the X-gal based Y2H assay, the results showed that P1, P3, and P5 can interact with CML9 (Figure 1C), which suggests the shared P5 fragment (536–569 aa) is the critical domain of the C-terminal of MLO10. We concluded that the P5 domain on the C-terminal of MLO10, but not the canonical CaMBD, is responsible for specific interaction with CML9.
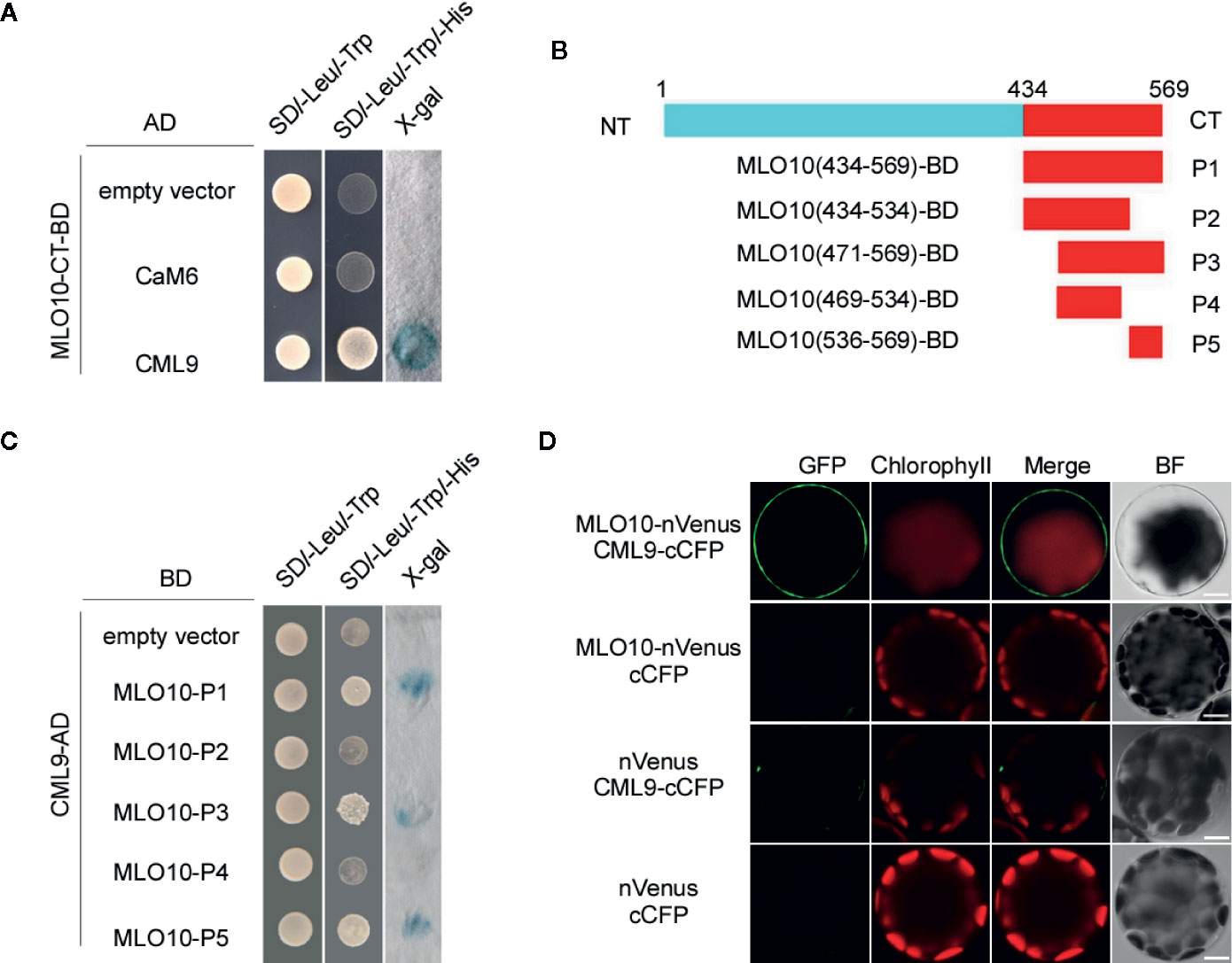
Figure 1 Protein Interaction of CML9 and MLO10. (A) Yeast two-hybrid assay of calmodulins (CaMs)/CaM-like proteins (CMLs) and the C-terminal of MLO10 (MLO10-CT). The combination of pGBKT7-MLO10-CT and empty pGADT7 vectors was used as a negative control. SD/-Trp/-Leu represents synthetic dextrose minimal medium without tryptophan and leucine; SD/-Trp/-Leu/-His indicates synthetic dextrose minimal medium without tryptophan, leucine, and histidine. Growth of yeast on SD/-Trp/-Leu/-His plate indicates interaction between the two tested proteins. (B) Diagram representing various domains of the MLO10-CT (P1–P5), which were cloned and used as prey during yeast two-hybrid screening. (C) Yeast two-hybrid analysis of CaM1 with various fragments of MLO10-CT. (D) Bimolecular fluorescence complementation (BiFC) analysis between MLO10 and CML9 in Arabidopsis mesophyll protoplasts. Vectors encoding MLO10-nVenus, CML9-cCFP, and nVenus were co-expressed in various combinations in Arabidopsis protoplasts. Green fluorescence (GFP), red fluorescence (Chlorophyll), merged fluorescence of green and red (Merge), and bright field (BF) were detected, respectively. Scale bars, 10 μm.
We further confirmed MLO10–CML9 interaction in BiFC (Bimolecular Fluorescence Complementation) assay. When co-expressing MLO10-nVenus and CML9-cCFP, green fluorescence signal was detected significantly in the plasma membrane of Arabidopsis protoplasts (Figure 1D), which was obviously different compared to the controls like nVenus + cCFP, nVenus + CML9-cCFP and MLO10-nVenus + cCFP. It indicates that MLO10 recruits and physically interacts with CML9 in the plasma membrane (Figure 1D). Taken together, we have identified the direct interaction of MLO10–CML9 in both Y2H and BiFC systems.
Subcellular Localization of CML9 and MLO10
To investigate the subcellular localization of CML9 and MLO10, we generated 35S:MLO10-GFP and 35S:CML9-GFP vectors which were individually introduced into Arabidopsis protoplasts. Only the GFP protein as control is localized in the nucleus, cytoplasm, and membrane (Figure 2). We can observe the obvious signal of the fusion protein MLO10-GFP mainly in the cell membrane and very weak signal in some other membrane-attached parts (Figure 2). However, the CML9-GFP expressed alone is mainly localized in the cytoplasm (Figure 2). Therefore, combined with the observation of BiFC results above, we concluded that the protein interaction between membrane-localized MLO10 and cytoplasm-localized CML9 occurs in the plant cell (Figure 1D).
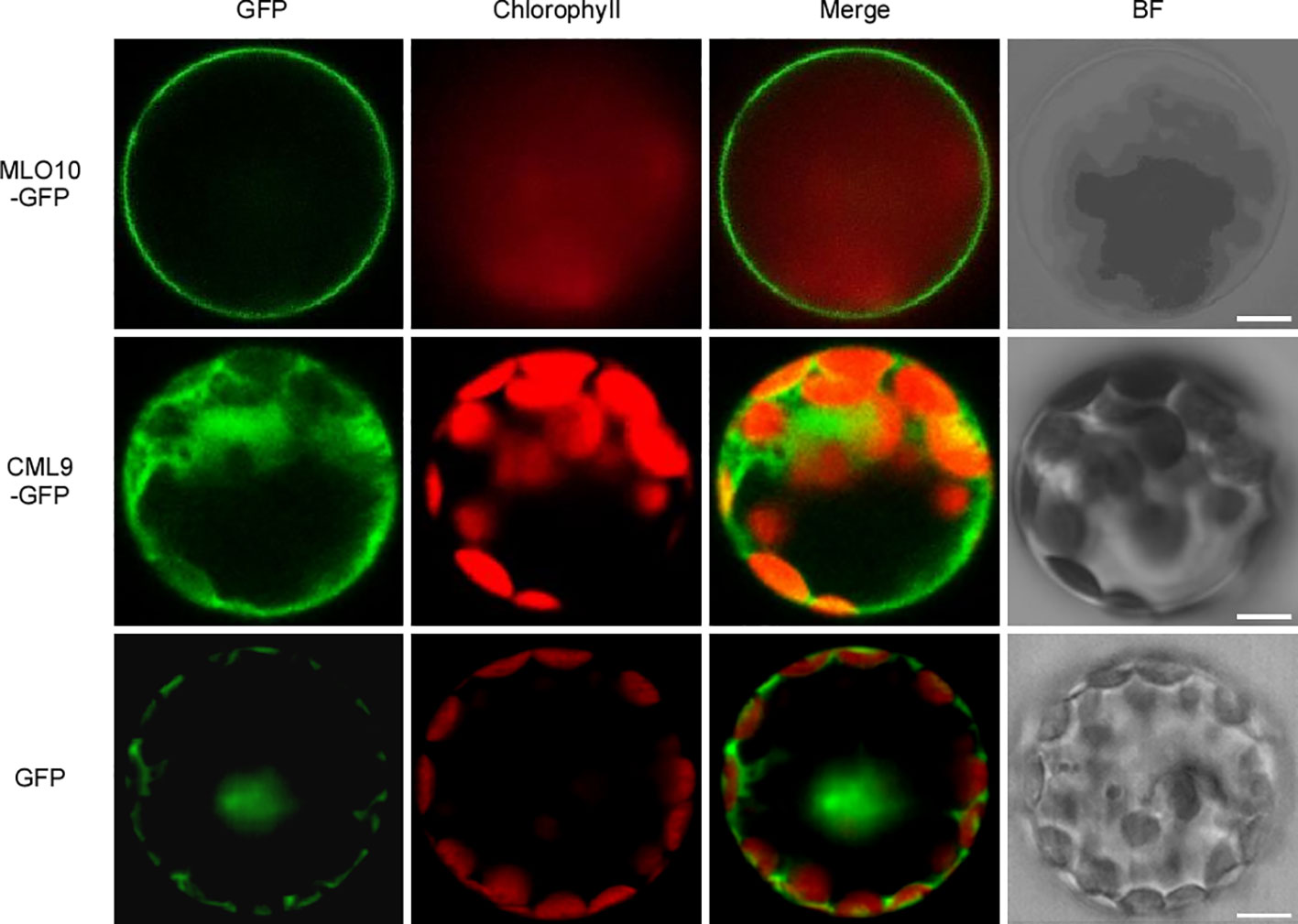
Figure 2 Subcellular localization of MLO10 and CML9 in Arabidopsis mesophyll protoplast. AtMLO10-GFP and AtCML9-GFP proteins were transiently expressed in protoplasts under the control of the cauliflower mosaic virus 35S promoter. Green fluorescence (GFP), red fluorescence (Chlorophyll), yellow fluorescence (merged fluorescence of green and red), and bright field (BF) were detected, respectively. Scale bars, 10 μm.
MLO10 and CML9 Are Co-Expressed in Flowers Within a Spatio-Temporal Specific Manner
In order to test the tissue specific expression pattern of CML9 and MLO10, we fused CML9 and MLO10 promoters with GUS reporter gene respectively. The results showed that CML9 is generally expressed in the root tips and leaf vasculature bundle, especially in the filament and anther of flowers (Figure 3A). The reporter GUS signal driven by CML9 promoter was stained not only in matured pollens released from the opening anther, but also highly in the germinated pollens and pollen tubes (Figures 3A-ii, -iv). This observation demonstrated that CML9 probably plays an important role in the pollen and pollen tube development during pollination.
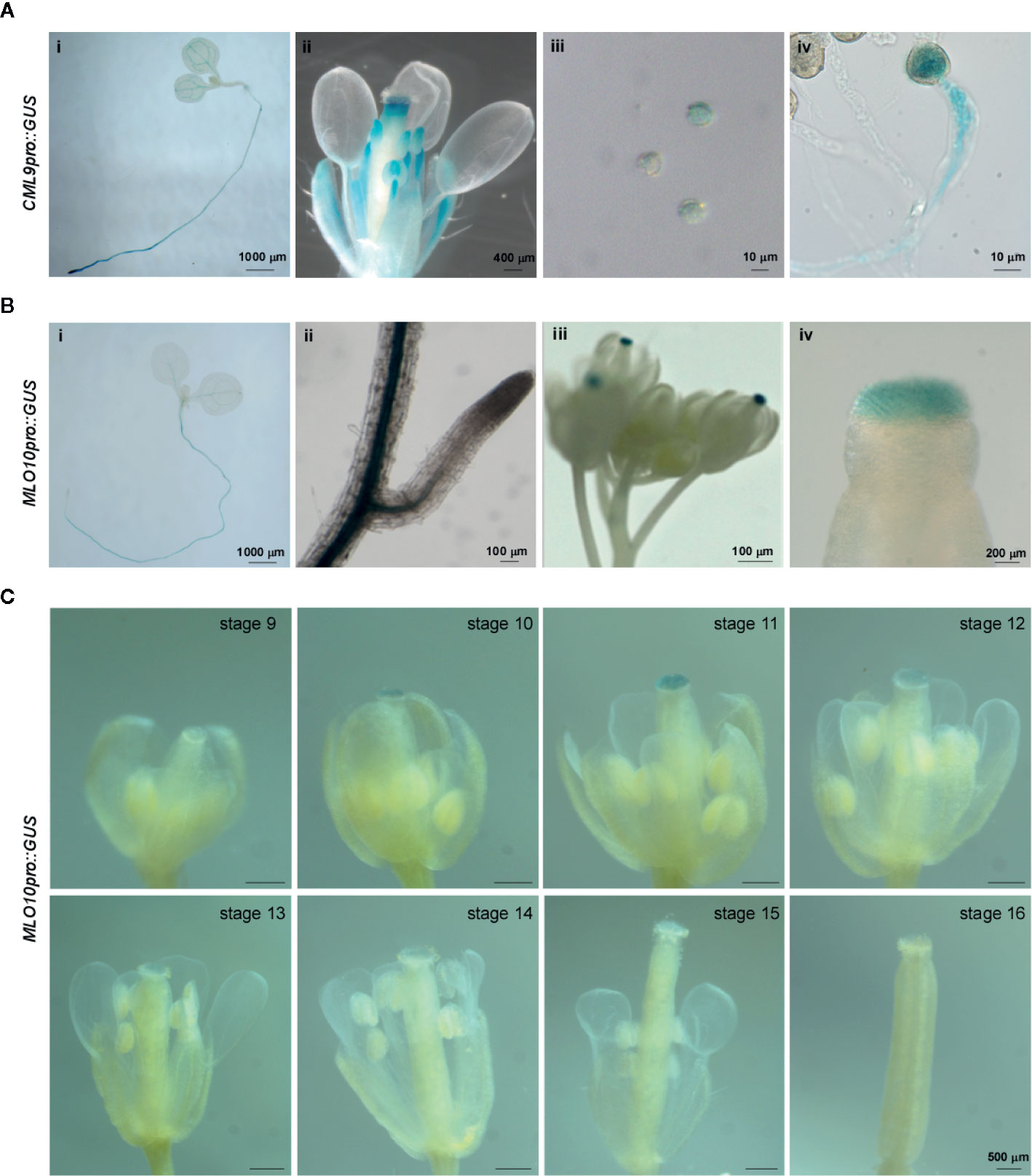
Figure 3 Tissue specific expression pattern of MLO10 and CML9. (A) GUS staining of CML9pro::GUS transgenic Arabidopsis lines in whole seedling (i), lateral root (ii), inflorescence (iii). and stigma (iv). (B) GUS staining of MLO10pro::GUS transgenic Arabidopsis lines in whole seedling (i), inflorescence (ii), pollen (iii), and pollen tube (iv). (C) Expression pattern of MLO10pro::GUS in flower development stages 9–16. Scale bars, 500 μm.
According to the GUS staining assay, MLO10 staining signals were restricted to the root, leaf vasculature bundle, and stigma (Figure 3B). When we further analyzed GUS signal in different developmental stages of flowers, the results suggest that MLO10 expression gets to a peaking level at stage 11 and displays relatively low expression levels in other flower developmental stages (Figure 3C). Flower developmental stages 13–14 are considered as the time-window of pollination recognition between matured pollen and stigma papilla, which is in line with the equal height of stamen and carpel [Figure 1C and (Alvarez-Buylla et al., 2010)]. In addition, MLO10 expression was also observed in siliques previously (Chen et al., 2006).
Taken together, the specific spatio-temporal expression pattern led us to hypothesize that MLO10, expressing slightly early before pollinaton, may get ready to interact with later expressed CML9.
Mutation of CML9 Reduced the Rates of Pollen Germination and Pollen Tube Elongation
Next, we investigated the physiological function of the MLO10–CML9 protein-protein interaction on affecting reproductive development. Because CML9 was highly expressed in the pollen and pollen tube (Figure 1B), we firstly measured the germination rates of two cml9 mutant alleles and Col-0 pollens. The results showed that after 4 h in vitro germination assay the rate of Col-0 was about 75%, but cml9-1 and cml9-2 mutants significantly reduced to 60% (Figures 4A, B). Consistently, at 4 hap (hour after pollination), the pollen tube lengths in cml9-1 and cml9-2 were severely shorter than that in Col-0 (Figure 4C). Collectively, the mutation of CML9 affects the germination rate of pollens and elongation rate of pollen tubes. To further test whether it is also true in planta, we carried out the manual pollination through four combinations that take cml9-1 and Col-0 as male-parent, mlo10-1 and Col-0 as female-parent. In line with the observation in vitro germination, the pollen tube penetration rate of Col-0 ♀ × cml9-1 ♂ was obliviously reduced by about 14% as compared with Col-0 ♀ × Col-0 ♂ (Figures 4D, E). Interestingly, mlo10-1 ♀ × cml9-1 ♂ showed lowest pollen tube penetration rate among the four tested combinations (Figures 4D, E), suggesting that both CML9 and MLO10 contribute to pollen tube elongation in planta, which is possibly mediated through MLO10–CML9 interacting module.
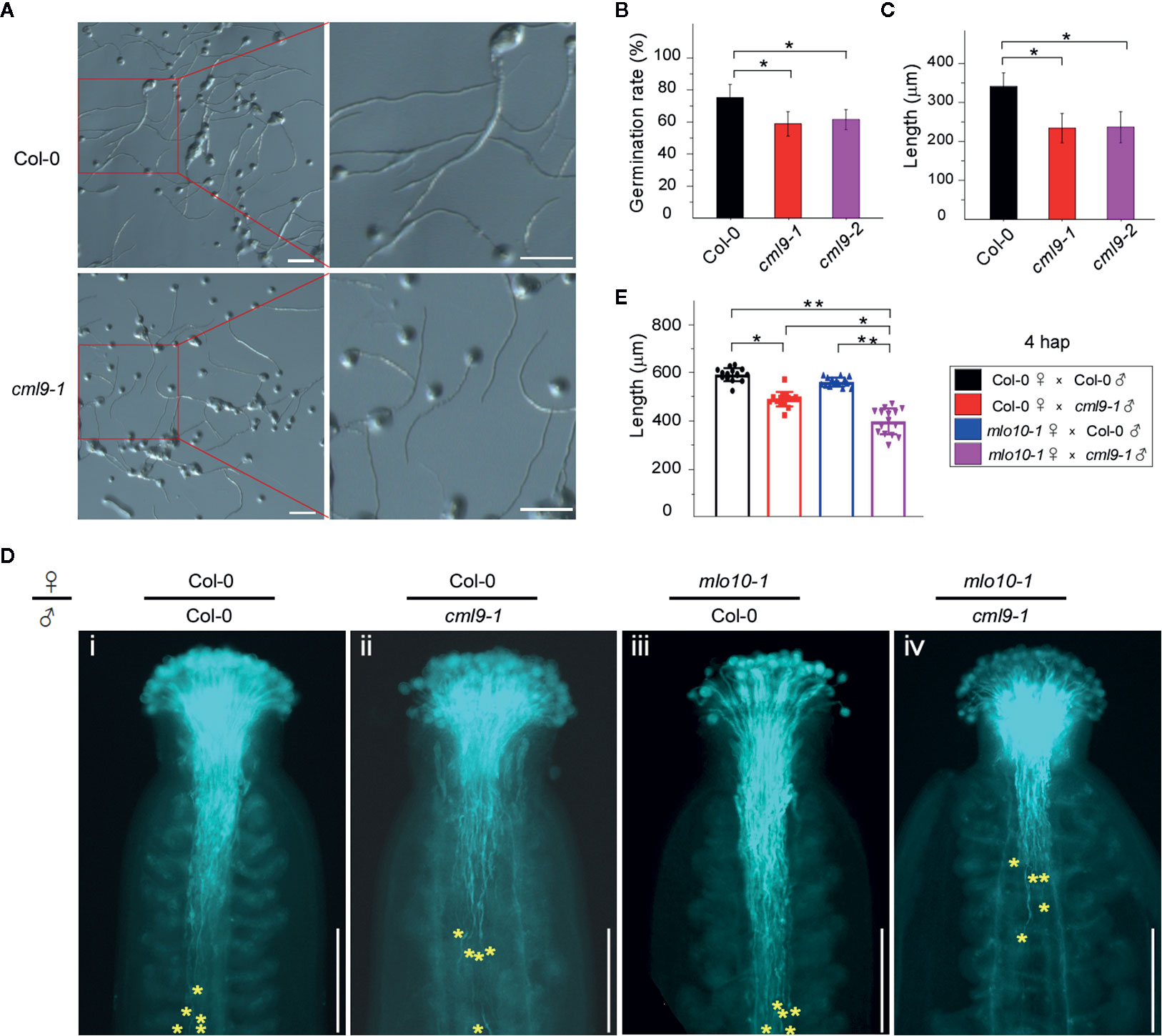
Figure 4 Mutation of CML9 affect pollen germination and pollen tube elongation. (A) In vitro test of pollen germination in wild-type and cml9-1 (4hap). Scale bars, 100 μm. (B) Quantification of pollen germination rate of different genotypes from (A). Data are shown as the mean ± s.e.m. n = 20. Two-tailed Students’ t-test. (C) Quantification of pollen tube length of Col-0, cml9-1, and cml9-2. Data are shown as the mean ± s.e.m. n = 100. Two-tailed Students’ t-test. (D) Manual pollination assay showed different pollen tube lengths in Col-0 ♀ × Col-0 ♂, Col-0 ♀ × cml9-1 ♂, mlo10-1 ♀ × Col-0 ♂, mlo10-1 ♀ × cml9-1 ♂ (4 hap). Yellow stars show top 5 of the furthest pollen tubes. Scale bars, 250 μm. (E). Statistics of D. Data are shown as the mean ± s.e.m. n = 15. Two-tailed Students’ t-test. *p < 0.05, **p < 0.01.
Genetic Interaction of MLO10 and CML9 Affects Seed Setting Rate
We further asked whether the pollen tube elongation mediated by MLO10–CML9 may affect later developmental stages like silique development in Arabidopsis. The self-pollination siliques were checked among the materials of Col-0, cml9-1, cml9-2, mlo10-1, mlo10-2, and cml9-1 mlo10-1. Phenotypically, a few seeds were obviously defective in cml9-1, cml9-2, and cml9-1 mlo10-1 (Figure 5A). We found that seed setting rates in cml9-1, cml9-2, and cml9-1 mlo10-1 were less by a reduction rate around 5% compared to Col-0 (Figures 5A, B). Taken together, MLO10–CML9 module could affect the developmental process of seed setting.
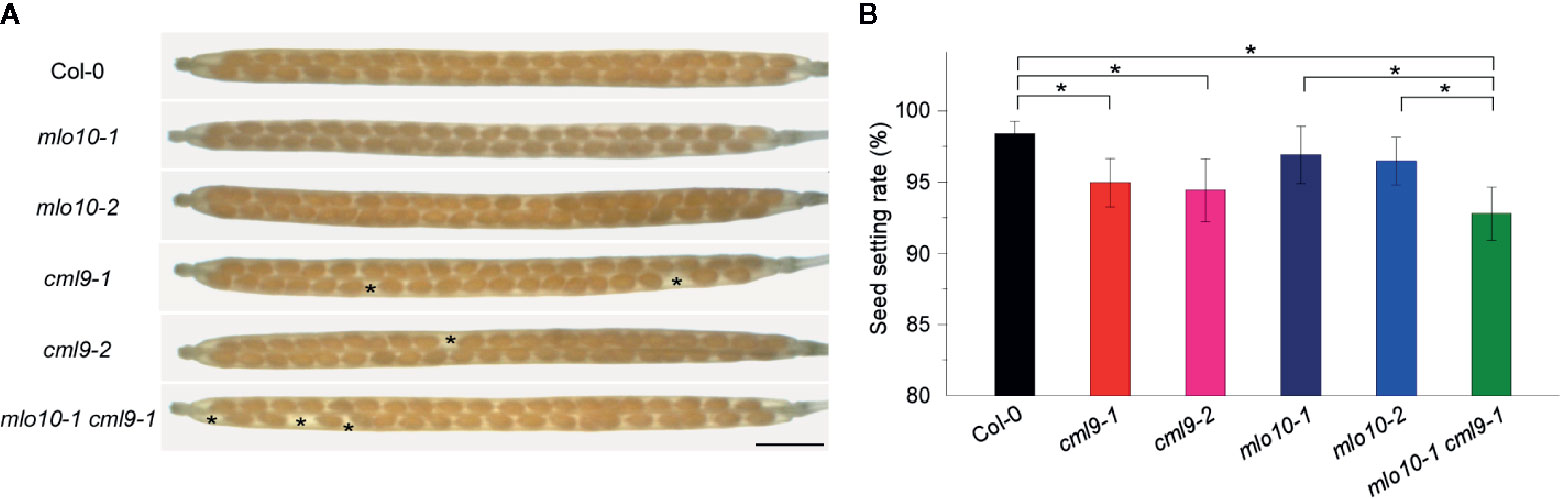
Figure 5 cml9-1 mlo10-1 double mutant reduced seed setting. (A) Seed setting rates in Col-0, cml9-1, cml9-2, mlo10-1, mlo10-2, and cml9-1 mlo10-1. Scale bars, 1,500 μm. (B) Statistics of (A). Data are shown as the mean ± s.e.m. n = 40. Two-tailed Students’ t-test. p < 0.05.
Discussion
In this study, we identified a protein complex MLO10–CML9 which may be involved in pollen–stigma recognition and affect pollen tube elongation and seed setting. These two proteins display the maternal- and paternal-specific expression manner during pollination period. Their genetic interaction further demonstrates the function of MLO10–CML9 in plant reproduction physiology.
MLOs May Have General Function for Recognition
MLO family members have divergent biological functions which mediate many communications, such as the touching effect between roots and environmental materials (Chen et al., 2009; Bidzinski et al., 2014), pathogen attack to leaves (Consonni et al., 2006; Acevedo-Garcia et al., 2017), recognition between pollen tube and synergid cells (Kessler et al., 2010; Jones and Kessler, 2017), as well as pollen–stigma communication in this study. In brief, we may consider MLOs as plant endogenous monitors to various environmental or internal stimuli. Therefore, mutation of MLOs may block or inhibit the recognition process in plant.
In Barley, mutation of HvMLOs inhibits the pathogen attack of powdery mildew by inducing local leaf neurosis (Consonni et al., 2006). In Arabidopsis mlo7 mutant, pollen tubes cannot be correctly guided to the embryo sac (Kessler et al., 2010), indicating that this recognition may be mediated by the interaction of MLO7 and some unknown protein. Interestingly, in this study, we identified a new interacting module MLO10–CML9 which could be related to pollen–stigma recognition. Especially, genetic mutations of MLO10 in stigma as well as CML9 in pollen result in significant reduction of pollen germination and pollen tube elongation rates, suggesting MLO10–CML9 contributes to fertilization. Our future work will figure out the detailed molecular interactions of MLO10 and CML9 in planta, as well as the possibility of redundancy modules to MLO10-CML9.
Taken together, MLO members interacting with their partners may function in sensing environmental stimuli and cell–cell communications. The previous and our studies will provide a new insight to deeply understand the functions of MLO family.
Interaction of Calmodulins and MLOs Provide a New Module in Regulating Reproductive Physiology and Stress Response
Calcium oscillations at pollen tube tips are important for maintaining its polarity growth (Iwano et al., 2009; Qin and Yang, 2011; Hepler et al., 2012; Wudick et al., 2018). CNGC8/18 as calcium channels interact with CaM2 to form a complex to encode calcium oscillations at pollen tube tips (Pan et al., 2019). Mutation of CaM2 leads to low rate of pollen germination (Landoni et al., 2010), and cml25 mutant was also reported to reduce rates of pollen germination in vitro and in planta (Wang et al., 2015). Those observations are compatible with our results in cml9-1 and cml9-2 mutants. Therefore, we conclude that CaM/CMLs may have an important and basic role in pollen–stigma recognition as well as polarity growth of pollen tubes. MLO10–CML9 complex may further create a new connection among the membrane protein, calcium sensor, and calcium oscillations.
Considering the stress responses, CML37 positively promotes ABA synthesis in response to drought stress (Wang et al., 2015), whereas CML42 negatively regulates plant defense by downregulation of JA response genes (Vadassery et al., 2012). CML9 acts as a positive player in response to bacterial effector triggered immunity (ETI) (Leba et al., 2012). A recent study identified that a protein complex CaM–CNGC2–CNGC4 mediated the early event of calcium-dependent ETI process (Tian et al., 2019).
In barley, calmodulin can interact with the C-terminal domain of MLO protein to regulate pathogen defense (Kim et al., 2002b). Especially, extracellar calmodulin can bind to a plant cell membrane (Wang et al., 2009), suggesting that camodulin could function as a mobile signal across cells. In our study, we found CML9 interacts with MLO10 which is involved in fertilization and seed setting. Therefore, in future work it will be worth to investigate the direct connection between MLO10–CML9 complex mediated calcium and ROS signaling (Zhang et al., 2020), which can further lead to deeply understanding the physiology of double fertilization.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material; further inquiries can be directed to the corresponding authors.
Author Contributions
LeL, QZ and LiL conceived and designed the project. QZ and CH performed most of the research and data analysis, and YT, MT, ZR, CF, JS, XW, TL, ML and WT prepared the genetic materials and observed the phenotype and collected data. LeL and LiL drafted the manuscript and LeL, LiL, and JQ revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from by the key program of the National Natural Science Foundation of China (No. 31930010 to LeL), the National Key Research and Development Program of China (Grant YFD0300102-3 to LeL), the general program of National Natural Science Foundation of China (No. 31270297 and No. 31470356 to LeL and No. 31571258 and No. 31800224 to LiL), and the Capacity Building for Sci-Tech Innovation-Fundamental Scientific Research Funds (No. 19530050165 to LeL).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01119/full#supplementary-material
References
Acevedo-Garcia, J., Gruner, K., Reinstadler, A., Kemen, A., Kemen, E., Cao, L., et al. (2017). The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Sci. Rep. 7 (1), 9319. doi: 10.1038/s41598-017-07188-7
Alvarez-Buylla, E. R., Benitez, M., Corvera-Poire, A., Chaos Cador, A., de Folter, S., Gamboa de Buen, A., et al. (2010). Flower development. Arabidopsis Book 8, e0127. doi: 10.1199/tab.0127
Appiano, M., Catalano, D., Santillan Martinez, M., Lotti, C., Zheng, Z., Visser, R. G., et al. (2015). Monocot and dicot MLO powdery mildew susceptibility factors are functionally conserved in spite of the evolution of class-specific molecular features. BMC Plant Biol. 15, 257. doi: 10.1186/s12870-015-0639-6
Bent, A. F., Clough, S. J. (1998). “Agrobacterium Germ-Line Transformation: Transformation of Arabidopsis without Tissue Culture,” in Plant Molecular Biology Manual. Eds. Gelvin, S. B., Schilperoort, R. A. (Dordrecht: Springer Netherlands), 17–30.
Bhat, R. A., Miklis, M., Schmelzer, E., Schulze-Lefert, P., Panstruga, R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. U.S.A. 102 (8), 3135–3140. doi: 10.1073/pnas.0500012102
Bhat, R. A., Lahaye, T., Panstruga, R. (2006). The visible touch: in planta visualization of protein-protein interactions by fluorophore-based methods. Plant Methods 2:12. doi: 10.1186/1746-4811-2-12
Bidzinski, P., Noir, S., Shahi, S., Reinstadler, A., Gratkowska, D. M., Panstruga, R. (2014). Physiological characterization and genetic modifiers of aberrant root thigmomorphogenesis in mutants of Arabidopsis thaliana MILDEW LOCUS O genes. Plant Cell Environ. 37 (12), 2738–2753. doi: 10.1111/pce.12353
Chen, Z., Hartmann, H. A., Wu, M. J., Friedman, E. J., Chen, J. G., Pulley, M., et al. (2006). Expression analysis of the AtMLO gene family encoding plant-specific seven-transmembrane domain proteins. Plant Mol. Biol. 60 (4), 583–597. doi: 10.1007/s11103-005-5082-x
Chen, Z., Noir, S., Kwaaitaal, M., Hartmann, H. A., Wu, M. J., Mudgil, Y., et al. (2009). Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell 21 (7), 1972–1991. doi: 10.1105/tpc.108.062653
Cheval, C., Aldon, D., Galaud, J. P., Ranty, B. (2013). Calcium/calmodulin-mediated regulation of plant immunity. Biochim. Biophys. Acta 1833 (7), 1766–1771. doi: 10.1016/j.bbamcr.2013.01.031
Consonni, C., Humphry, M. E., Hartmann, H. A., Livaja, M., Durner, J., Westphal, L., et al. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38 (6), 716–720. doi: 10.1038/ng1806
Dumas, C., Knox, R. B. (1983). Callose and determination of pistil viability and incompatibility. Theor. Appl. Genet. 67 (1), 1–10. doi: 10.1007/BF00303914
Elliott, C., Muller, J., Miklis, M., Bhat, R. A., Schulze-Lefert, P., Panstruga, R. (2005). Conserved extracellular cysteine residues and cytoplasmic loop-loop interplay are required for functionality of the heptahelical MLO protein. Biochem. J. 385 (Pt 1), 243–254. doi: 10.1042/BJ20040993
Fan, L. M., Wang, Y. F., Wang, H., Wu, W. H. (2001). In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. J. Exp. Bot. 52 (361), 1603–1614. doi: 10.1093/jexbot/52.361.1603
Feechan, A., Jermakow, A. M., Torregrosa, L., Panstruga, R., Dry, I. B. (2008). Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct. Plant Biol. 35 (12), 1255–1266. doi: 10.1071/FP08173
Gao, X., Chen, J., Dai, X., Zhang, D., Zhao, Y. (2016). An Effective Strategy for Reliably Isolating Heritable and Cas9-Free Arabidopsis Mutants Generated by CRISPR/Cas9-Mediated Genome Editing. Plant Physiol. 171 (3), 1794–1800. doi: 10.1104/pp.16.00663
Ge, Z., Bergonci, T., Zhao, Y., Zou, Y., Du, S., Liu, M. C., et al. (2017). Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358 (6370), 1596–1600. doi: 10.1126/science.aao3642
Hepler, P. K., Kunkel, J. G., Rounds, C. M., Winship, L. J. (2012). Calcium entry into pollen tubes. Trends Plant Sci. 17 (1), 32–38. doi: 10.1016/j.tplants.2011.10.007
Heyer, M., Scholz, S. S., Voigt, D., Reichelt, M., Aldon, D., Oelmuller, R., et al. (2018). Herbivory-responsive calmodulin-like protein CML9 does not guide jasmonate-mediated defenses in Arabidopsis thaliana. PloS One 13 (5), e0197633. doi: 10.1371/journal.pone.0197633
Hiscock, S. J., Allen, A. M. (2008). Diverse cell signalling pathways regulate pollen-stigma interactions: the search for consensus. New Phytol. 179 (2), 286–317. doi: 10.1111/j.1469-8137.2008.02457.x
Iwano, M., Entani, T., Shiba, H., Kakita, M., Nagai, T., Mizuno, H., et al. (2009). Fine-tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiol. 150 (3), 1322–1334. doi: 10.1104/pp.109.139329
Jiang, L., Yang, S. L., Xie, L. F., Puah, C. S., Zhang, X. Q., Yang, W. C., et al. (2005). VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17 (2), 584–596. doi: 10.1105/tpc.104.027631
Jones, D. S., Kessler, S. A. (2017). Cell type-dependent localization of MLO proteins. Plant Signal Behav. 12 (11), e1393135. doi: 10.1080/15592324.2017.1393135
Jones, D. S., Yuan, J., Smith, B. E., Willoughby, A. C., Kumimoto, E. L., Kessler, S. A. (2017). MILDEW RESISTANCE LOCUS O Function in Pollen Tube Reception Is Linked to Its Oligomerization and Subcellular Distribution. Plant Physiol. 175 (1), 172–185. doi: 10.1104/pp.17.00523
Kessler, S. A., Shimosato-Asano, H., Keinath, N. F., Wuest, S. E., Ingram, G., Panstruga, R., et al. (2010). Conserved molecular components for pollen tube reception and fungal invasion. Science 330 (6006), 968–971. doi: 10.1126/science.1195211
Kim, M. C., Lee, S. H., Kim, J. K., Chun, H. J., Choi, M. S., Chung, W. S., et al. (2002a). Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein. Isolation and characterization of a rice Mlo homologue. J. Biol. Chem. 277 (22), 19304–19314. doi: 10.1074/jbc.M108478200
Kim, M. C., Panstruga, R., Elliott, C., Muller, J., Devoto, A., Yoon, H. W., et al. (2002b). Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416 (6879), 447–451. doi: 10.1038/416447a
Knox, R. B., Clarke, A., Harrison, S., Smith, P., Marchalonis, J. J. (1976). Cell recognition in plants: Determinants of the stigma surface and their pollen interactions. Proc. Natl. Acad. Sci. U.S.A. 73 (8), 2788–2792. doi: 10.1073/pnas.73.8.2788
Landoni, M., De Francesco, A., Galbiati, M., Tonelli, C. (2010). A loss-of-function mutation in Calmodulin2 gene affects pollen germination in Arabidopsis thaliana. Plant Mol. Biol. 74 (3), 235–247. doi: 10.1007/s11103-010-9669-5
Leba, L. J., Cheval, C., Ortiz-Martin, I., Ranty, B., Beuzon, C. R., Galaud, J. P., et al. (2012). CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway. Plant J. 71 (6), 976–989. doi: 10.1111/j.1365-313X.2012.05045.x
McClure, B. A., Franklin-Tong, V. (2006). Gametophytic self-incompatibility: understanding the cellular mechanisms involved in “self” pollen tube inhibition. Planta 224 (2), 233–245. doi: 10.1007/s00425-006-0284-2
Moeini-Naghani, I., Navaratnam, D. S. (2016). Yeast Two-Hybrid Screening to Test for Protein-Protein Interactions in the Auditory System. Methods Mol. Biol. 1427, 95–107. doi: 10.1007/978-1-4939-3615-1_6
Mouline, K., Very, A. A., Gaymard, F., Boucherez, J., Pilot, G., Devic, M., et al. (2002). Pollen tube development and competitive ability are impaired by disruption of a Shaker K+ channel in Arabidopsis. Genes Dev. 16 (3), 339–350. doi: 10.1101/gad.213902
Nasrallah, M. E., Liu, P., Nasrallah, J. B. (2002). Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297 (5579), 247–249. doi: 10.1126/science.1072205
Pan, Y., Chai, X., Gao, Q., Zhou, L., Zhang, S., Li, L., et al. (2019). Dynamic Interactions of Plant CNGC Subunits and Calmodulins Drive Oscillatory Ca2+ Channel Activities. Dev. Cell 48 (5), 710–725 e715. . doi: 10.1016/j.devcel.2018.12.025
Qin, Y., Yang, Z. (2011). Rapid tip growth: insights from pollen tubes. Semin. Cell Dev. Biol. 22 (8), 816–824. doi: 10.1016/j.semcdb.2011.06.004
Rosado, A., Li, R., van de Ven, W., Hsu, E., Raikhel, N. V. (2012). Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc. Natl. Acad. Sci. U.S.A. 109 (48), 19537–19544. doi: 10.1073/pnas.1214774109
Schopfer, C. R., Nasrallah, M. E., Nasrallah, J. B. (1999). The male determinant of self-incompatibility in Brassica. Science 286 (5445), 1697–1700. doi: 10.1126/science.286.5445.1697
Takasaki, T., Hatakeyama, K., Suzuki, G., Watanabe, M., Isogai, A., Hinata, K. (2000). The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403 (6772), 913–916. doi: 10.1038/35002628
Tian, W., Hou, C., Ren, Z., Wang, C., Zhao, F., Dahlbeck, D., et al. (2019). A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572 (7767), 131–135. doi: 10.1038/s41586-019-1413-y
Vadassery, J., Reichelt, M., Hause, B., Gershenzon, J., Boland, W., Mithofer, A. (2012). CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 159 (3), 1159–1175. doi: 10.1104/pp.112.198150
Wang, Q., Chen, B., Liu, P., Zheng, M., Wang, Y., Cui, S., et al. (2009). Calmodulin binds to extracellular sites on the plasma membrane of plant cells and elicits a rise in intracellular calcium concentration. J. Biol. Chem. 284 (18), 12000–12007. doi: 10.1074/jbc.M808028200
Wang, S. S., Diao, W. Z., Yang, X., Qiao, Z., Wang, M., Acharya, B. R., et al. (2015). Arabidopsis thaliana CML25 mediates the Ca2+ regulation of K+ transmembrane trafficking during pollen germination and tube elongation. Plant Cell Environ. 38 (11), 2372–2386. doi: 10.1111/pce.12559
Wang, T., Liang, L., Xue, Y., Jia, P. F., Chen, W., Zhang, M. X., et al. (2016). A receptor heteromer mediates the male perception of female attractants in plants. Nature 531 (7593), 241–244. doi: 10.1038/nature16975
Wudick, M. M., Portes, M. T., Michard, E., Rosas-Santiago, P., Lizzio, M. A., Nunes, C. O., et al. (2018). CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca2+ homeostasis. Science 360 (6388), 533–536. doi: 10.1126/science.aar6464
Wuest, S. E., Vijverberg, K., Schmidt, A., Weiss, M., Gheyselinck, J., Lohr, M., et al. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20 (6), 506–512. doi: 10.1016/j.cub.2010.01.051
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 (7), 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, M. J., Zhang, X. S., Gao, X. Q. (2020). ROS in the Male-Female Interactions During Pollination: Function and Regulation. Front. Plant Sci. 11:177. doi: 10.3389/fpls.2020.00177
Zheng, Y. Y., Lin, X. J., Liang, H. M., Wang, F. F., Chen, L. Y. (2018). The Long Journey of Pollen Tube in the Pistil. Int. J. Mol. Sci. 19 (11), 3529 doi: 10.3390/ijms19113529
Keywords: CML9, MLO10, pollination, pollen tube, stigma, recognition
Citation: Zhang Q, Hou C, Tian Y, Tang M, Feng C, Ren Z, Song J, Wang X, Li T, Li M, Tian W, Qiu J, Liu L and Li L (2020) Interaction Between AtCML9 and AtMLO10 Regulates Pollen Tube Development and Seed Setting. Front. Plant Sci. 11:1119. doi: 10.3389/fpls.2020.01119
Received: 08 April 2020; Accepted: 07 July 2020;
Published: 23 July 2020.
Edited by:
Stefan de Folter, Instituto Politécnico Nacional de México (CINVESTAV), MexicoReviewed by:
Ravishankar Palanivelu, University of Arizona, United StatesXian Sheng Zhang, Shandong Agricultural University, China
Copyright © 2020 Zhang, Hou, Tian, Tang, Feng, Ren, Song, Wang, Li, Li, Tian, Qiu, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangyu Liu, bGlhbmd5dS5saXVAY251LmVkdS5jbg==; Legong Li, bGdsaUBjbnUuZWR1LmNu
†These authors have contributed equally to this work
 Qian Zhang
Qian Zhang Congcong Hou
Congcong Hou Yudan Tian1
Yudan Tian1 Mitianguo Tang
Mitianguo Tang Zhijie Ren
Zhijie Ren Xiaohan Wang
Xiaohan Wang Liangyu Liu
Liangyu Liu Legong Li
Legong Li