- Regensburg Center for Biochemistry (RCB), Laboratory for RNA Biology, University of Regensburg, Regensburg, Germany
Short interfering RNAs (siRNAs) are processed from long double-stranded RNA (dsRNA), and a guide strand is selected and incorporated into the RNA-induced silencing complex (RISC). Within RISC, a member of the Argonaute protein family directly binds the guide strand and the siRNA guides RISC to fully complementary sites on-target RNAs, which are then sequence-specifically cleaved by the Argonaute protein—a process commonly referred to as RNA interference (RNAi). In animals, endogenous microRNAs (miRNAs) function similarly but do not lead to direct cleavage of the target RNA but to translational inhibition followed by exonucleolytic decay. This is due to only partial complementarity between the miRNA and the target RNA. SiRNAs, however, can function as miRNAs, and partial complementarity can lead to miRNA-like off-target effects in RNAi applications. Since siRNAs are widely used not only for screening but also for therapeutics as well as crop protection purposes, such miRNA-like off-target effects need to be minimized. Strategies such as RNA modifications or pooling of siRNAs have been developed and are used to reduce off-target effects.
Introduction
Double-stranded RNA (dsRNA) as trigger for RNA interference (RNAi) has been discovered decades ago in plants and nematodes (Baulcombe, 1996; Fire et al., 1998). Although these organisms are rather distant, the underlying mechanisms are remarkably conserved. dsRNA is generated by transcription or enzymes such as RNA-dependent RNA polymerases (RdRPs), which use single-stranded RNA as a template to generate long dsRNA (Meister and Tuschl, 2004; Mello and Conte, 2004; Sharp and Zamore, 2000). This RNA is further processed to short interfering RNAs (siRNAs), which serve as guides for the RNA-induced silencing complex (RISC) that binds and sequence-specifically cleaves complementary target RNAs (Zamore and Haley, 2005). This process is commonly referred to as RNAi. However, long dsRNA is toxic for animal organisms with more sophisticated immune systems that are capable of sensing long dsRNA as “foreign” as such RNAs could, for example, result from viral infections (Schlee and Hartmann, 2016). However, a breakthrough was reached when it was found that short siRNAs bypass immune sensing and can be used for gene knockdown also in higher organisms such as mammals (Elbashir et al., 2001). Besides broad usage in basic research, siRNAs have now been developed to target genes for therapy and indeed the first siRNAs reached the market (Dorsett and Tuschl, 2004; Sheridan, 2017). In addition to the therapeutic use in mammals, RNAi is also being explored as crop protection agent (Zhang et al., 2017). dsRNA directed against pests such as fungi, nematodes, or insects is sprayed onto the leaves of plants and upon uptake selectively affects growth of distinct target species. Since dsRNA is species specific, is a natural product, and with no genetically modified organism needs to be generated, such strategies are considered highly promising next-generation plant protection agents. Nevertheless, both for human disease and for plant protection purposes, high specificity is critical and off-target effects need to be minimized (Seok et al., 2018). The following chapters will summarize principles of small RNA functions and highlight strategies to reduce off-target effects in gene knockdown experiments.
RNAi Components in Animals and Plants
Both in animals and plants, long dsRNA is processed to double-stranded siRNAs by Dicer-like enzymes (Bernstein et al., 2001; Grishok et al., 2001; Ketting et al., 2001). In Arabidopsis thaliana, four Dicer-like enzymes exist (referred to as DCL1-4), which are specialized for the generation of different classes of small RNAs (Bologna and Voinnet, 2014). DCL1 processes primary microRNAs (miRNA) to 21-nt-long mature miRNAs. DCL2 is involved in antiviral strategies and cleave viral dsRNA to 21/22-nt-long siRNAs, which target viral RNAs. DCL3 functions in silencing processes targeting transposable elements and produces siRNAs of about 24 nt in length. Finally, DCL4 generates 21 nt transacting siRNAs (tasiRNAs), which silence specific endogenous genes. Except for DCL1 that engages already-folded and dsRNA precursors, DCL2-4 cooperate with specialized RdRPs that generate the dsRNA substrates from single-stranded transcripts [for more information on plant Dicer enzymes, see Bologna and Voinnet (2014), Fukudome and Fukuhara (2017)].
In animals, Dicer enzymes are typically less diverse (Meister and Tuschl, 2004). Dicer enzymes belong to the RNase III enzyme family, which recognizes the ends of long dsRNAs and cleave the RNA about 21 nt from the end (Treiber et al., 2019). Dicer-like enzymes possess two catalytic RNase III domains, which cleave both strands and due to their positioning on the dsRNA, leave two nucleotides 3′ overhangs (Filipowicz, 2005; Treiber et al., 2019). In subsequent steps, commonly referred to as RISC loading, a member of the Argonaute protein family recognizes particularly the 3′ overhangs and selects one strand of the duplex to become the guide strand (also referred to as the antisense strand). The other strand, referred to as passenger strand, is degraded (Dueck and Meister, 2014; Ipsaro and Joshua-Tor, 2015; Sheu-Gruttadauria and MacRae, 2017). While structural information for most Ago proteins is lacking (including all plant Ago proteins), human Ago2 is reasonably well understood and is presented as an example for general structural features of Ago proteins. Argonaute proteins are structurally highly conserved and typically contain four domains (Figures 1A,B), as follows: the N domain, which has been implicated in siRNA duplex unwinding (Kwak and Tomari, 2012), the PAZ domain that anchors the 3′ end of the selected guide strand (Lingel et al., 2003; Ma et al., 2004; Yan et al., 2003), the MID domain that binds the 5′ end of the guide strand (Ma et al., 2005; Parker et al., 2004), and the PIWI domain. The PIWI domain has structural similarities to RNase H, which cleaves RNA molecules in RNA-DNA hybrids (Song et al., 2004; Wang et al., 2009; Yuan et al., 2005). Thus, some, but not all Argonaute proteins are endonucleases that cleave the target RNA in siRNA-target RNA hybrids using a catalytic tetrad in their active centers (Nakanishi et al., 2012) (Figure 1C). These proteins are referred to as Slicer enzymes (Liu et al., 2004; Meister et al., 2004). Furthermore, structural work revealed that MID domains of animal and plant Ago proteins display a sequence bias regarding the 5′ terminal nucleotide, which is important for sorting specific classes of small RNAs into their correct silencing pathways (Frank et al., 2010; Frank et al., 2012; Mi et al., 2008).
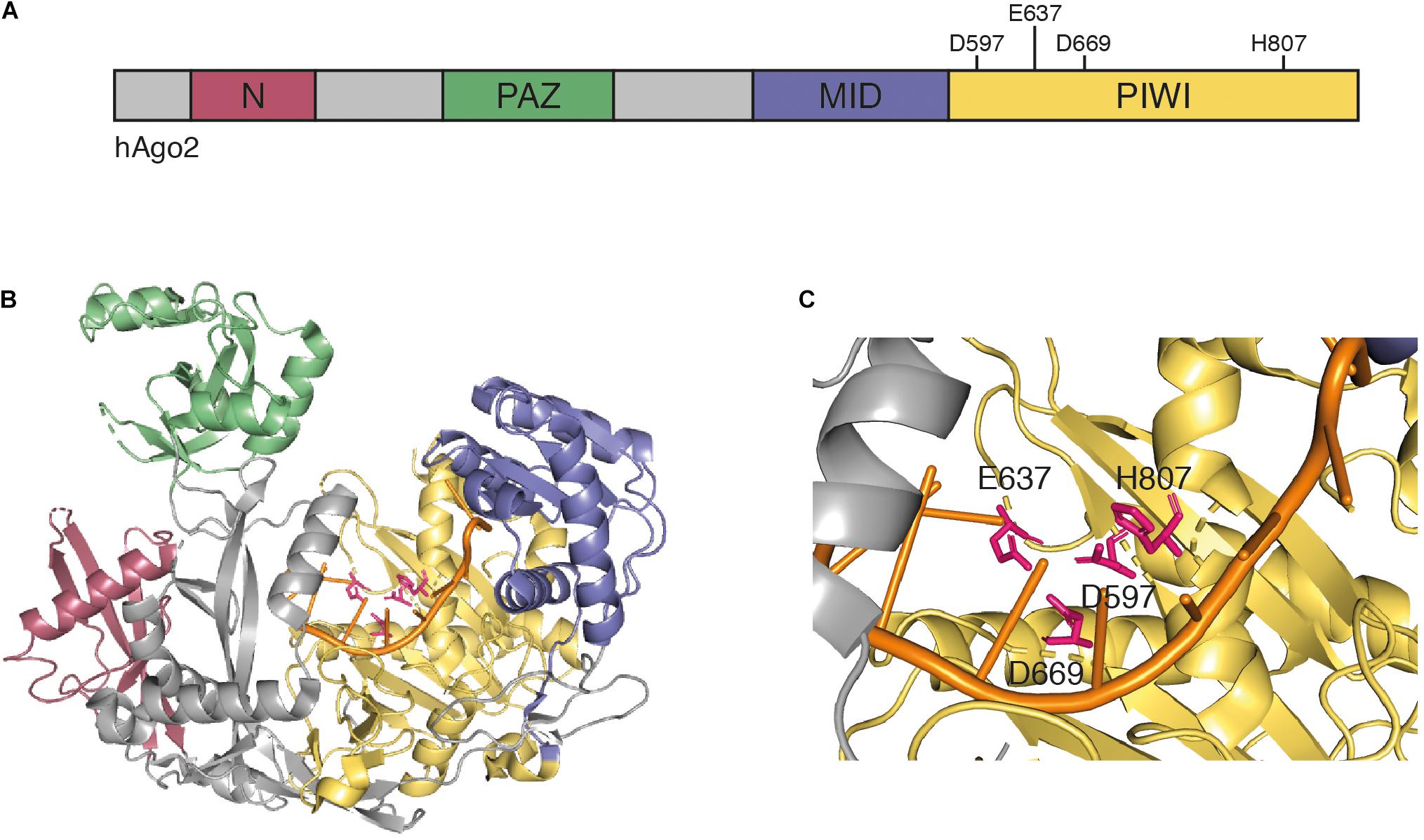
Figure 1. (A) Schematic representation of human Ago2. Argonaute proteins contain four conserved domains: the N domain (red), the PAZ domain (green), the MID domain (blue), and the PIWI domain (yellow). The PIWI domain contains the four catalytic residues D597, E637, D669, and H807 that are required for mRNA target cleavage. (B) Crystal structure of human Ago2 loaded with a siRNA (PDB ID 5JS1) (Schirle et al., 2016). The four domains are colored as in panel (A), the catalytic residues are highlighted in pink, and the 5′ region of the loaded siRNA is shown in orange. (C) Detailed view of catalytic center of the Ago2 structure from panel (B).
Plant Argonaute proteins are functionally diverse and are involved in various different gene silencing processes (Carbonell, 2017). In A. thaliana, Ago1, Ago7, and Ago10 bind to miRNAs and silence target genes. Ago4 has been implicated in RNA-directed DNA methylation and chromatin is epigenetically modified by this pathway. Generally, through their small RNA partner, plant Ago proteins are also involved in antiviral or bacterial defense mechanisms as well as responses to herbivore attack (Pradhan et al., 2017; Sibisi and Venter, 2020).
In plants as well as some animal species, which tolerate long dsRNA, the RNAi signal can be amplified by RdRPs (Maida et al., 2011). These enzymes use a siRNA strand bound to its target RNA as primer and synthesize the complementary strand to the target RNA resulting in a long dsRNA, which again enters Dicer processing and a second wave of siRNAs against a specific target is generated. Furthermore, in plants and also in some animal species, particularly nematodes such as Caenorhabditis elegans, siRNAs can be actively transported between cells and tissues [see, for example, (Das et al., 2019) for more information on this broad topic]. Remarkably, siRNA signals can also be inherited and target RNAs can be silenced over many generations. This process has been studied in C. elegans and is known as transgenerational gene silencing (TGS) [for more information on this exciting topic, please see, for example, (Iwasaki et al., 2015; Lev et al., 2019; Schraivogel and Meister, 2014)].
miRNA-Guided Gene Silencing
MiRNAs are found in almost all plants and animals and in contrast to siRNAs, are transcribed from distinct miRNA genes (Bartel, 2009; Kim et al., 2009). RNA polymerase II transcription results in capped and poly-adenylated primary miRNA transcripts (pri-miRNAs), which are recognized and processed by the nuclear microprocessor containing the RNase III enzyme Drosha and its interaction partner DGCR8 (Cai et al., 2004; Denli et al., 2004; Gregory et al., 2004; Han et al., 2004; Landthaler et al., 2004; Lee et al., 2003; Lee et al., 2004). Drosha/DGCR8 do not exist in plants and therefore DCL1 processes primary miRNA hairpins to mature miRNAs already in the nucleus of plant cells (Bologna and Voinnet, 2014; Bologna et al., 2018; Fukudome and Fukuhara, 2017).
Within pri-miRNAs, the miRNA strand itself is embedded in the stem of a local hairpin and the microprocessor cleaves the hairpin at the base of the stem. The resulting hairpin structure, referred to as miRNA precursor (pre-miRNA), is exported to the cytoplasm by the export receptor Exportin-5 in animals (Bohnsack et al., 2004; Lund et al., 2004; Yi et al., 2003). In the cytoplasm, Dicer binds to the end of the hairpin and cleaves off an approx. 21-nt-long dsmiRNA intermediate, which is reminiscent of a siRNA duplex described above (Zhang et al., 2002). Consistently, RISC loading processes are similar to siRNAs and both in plants and in animals require the action of heat shock protein 90 (HSP90) that holds Ago proteins in a loading competent open conformation (Dueck and Meister, 2014; Iki et al., 2012; Iwasaki et al., 2010; Miyoshi et al., 2010). Moreover, miRNAs function like siRNAs in case the miRNA and the target RNA are fully or almost fully complementary (Doench et al., 2003). This mechanism is predominant in plants (Song et al., 2019). In animals, however, target RNA binding as well as the mechanism of gene silencing is different. MiRNA-target sites are typically located in the 3′ untranslated region (UTR) of mRNAs (Bartel, 2009). Nucleotides 2–8 of the miRNA represent the seed sequence, which is generally fully complementary to the target site while the remaining sequence is often only partially paired (Rajewsky and Socci, 2004). This incomplete pairing prevents Slicer-mediated cleavage as it is observed also in siRNA-guided knockdown studies. Instead, Argonaute proteins recruit a member of the GW protein family, which coordinates the following steps in miRNA-guided gene silencing (Behm-Ansmant et al., 2006; Jakymiw et al., 2005; Liu et al., 2005; Meister et al., 2005). GW proteins are characterized by glycine-tryptophane repeats and are referred to as TNRC6 proteins in mammals (Pfaff and Meister, 2013; Pfaff et al., 2013). GW proteins establish interactions with the poly(A) tail of the mRNAs as well as deadenylase complexes including the CCR4/NOT complex or PAN2/3 leading to translational repression, deadenylation of the mRNA, and, finally, to the removal of the 5′ cap by decapping enzymes (Figure 2). The unprotected mRNA is then degraded by 5′–3′ exoribonucleases [for more details, see Braun et al. (2013), Jonas and Izaurralde (2015), Krol et al. (2010)]. Translation repression without site-specific cleavage has also been observed in plants of target sites that are fully complementary but located in the 3′ UTR of mRNAs (Brodersen et al., 2008). Since GW proteins are not conserved in plants, the extent of this type of miRNA action remains to be further investigated in plants (Song et al., 2019).
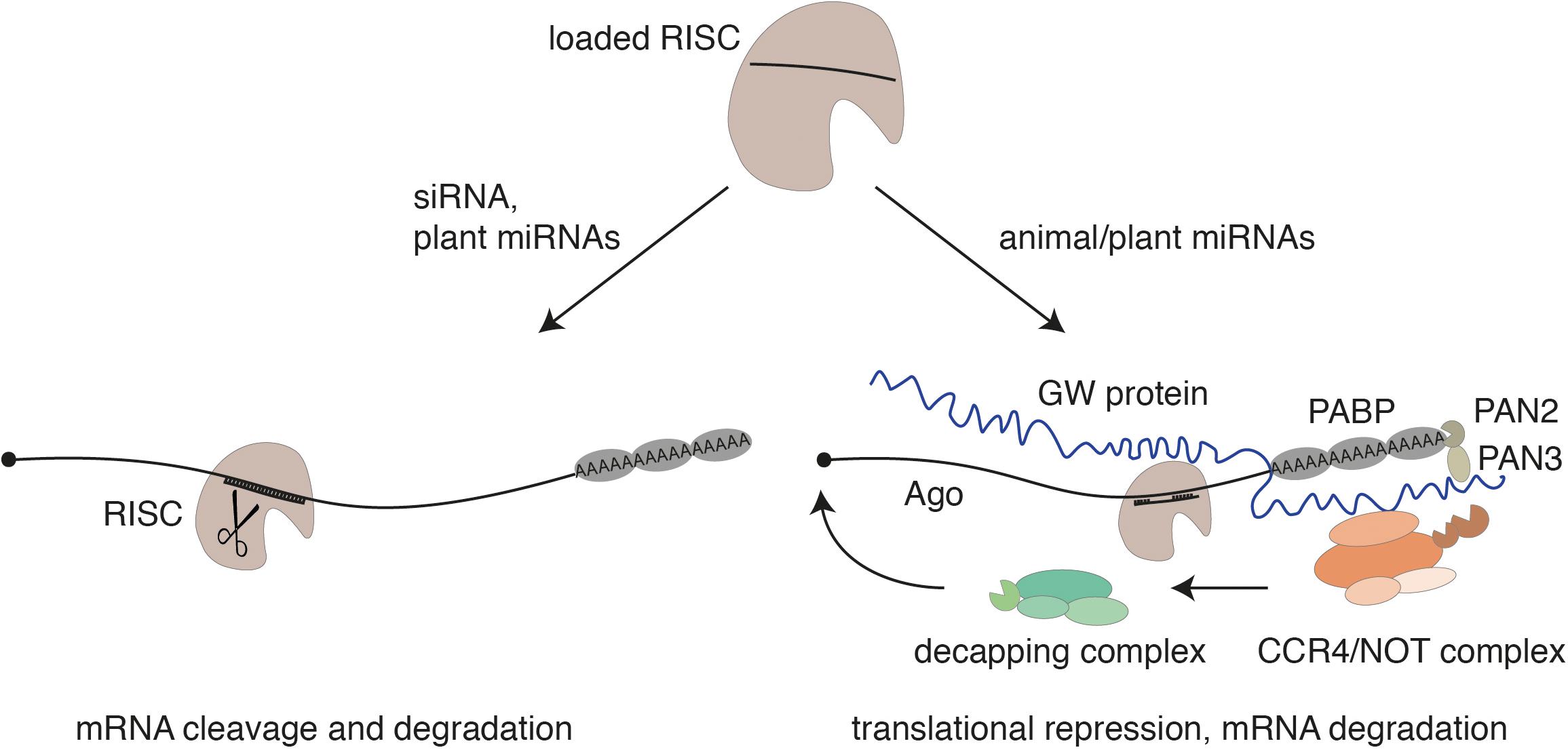
Figure 2. The guide strand of the siRNA or a miRNA is loaded into an Argonaute protein. In case of a perfect complementarity through siRNAs or miRNAs (which is often observed in plants), a catalytically active Argonaute protein cleaves the mRNA as part of RISC (left). As miRNAs in animals are only partially complementary to their target RNAs, Slicer-facilitated cleavage is impaired. In this case, Argonaute recruits a member of the GW protein family. These proteins mediate the interaction with further downstream acting factors like poly-(A)-binding proteins (PABPs) or the deadenylase complexes PAN2/PAN3 and CCR4/NOT. This leads to translational repression, deadenylation, decapping, and 5′–3′ exonucleolytic decay of the mRNA. Translational repression by miRNAs has also been observed for plant miRNAs.
miRNA-Like Off-Target Effects in RNAi Strategies
Although miRNA-guided gene silencing is distinct from siRNA-guided knockdown experiments, the pathways are intertwined and miRNAs can function as siRNAs and vice versa. This is particularly important for off-target effects observed in RNAi experiments (Seok et al., 2018). SiRNAs may, in addition to their fully complementary on-targets, bind to an undefined number of miRNA-like target sites in 3′ UTRs of mRNAs using their seed sequence. This will lead to silencing and unwanted off-target effects. Since such sequences are only 6–7 nt long, these unspecific target sites are hardly predictable and are thus very difficult to avoid. Indeed, miRNA-like off-target effects are highly problematic in large-scale RNAi screening approaches, and many hits are false positive and caused by off-target effects [e.g., (Birmingham et al., 2006; Buehler et al., 2012; Fedorov et al., 2006; Jackson et al., 2006b)]. Thus, strategies that control for or even reduce or eliminate such off-target effects are urgently needed.
In RNAi-mediated pest control, such off-target effects might not be predominantly problematic for the plant system since such a translational control system might be rather rare. However, in strategies, in which plants express si- or shRNAs that are taken up by animals and are toxic to defined species, off-target effects need to be considered. For example, non-target animals might incorporate these RNAs as well and, although perfect complementary target RNAs are absent, the expression of partially complementary sites could be affected through the endogenous miRNA system.
Strategies to Reduce miRNA-Like Off-Target Effects
SiRNAs are typically designed to avoid complementary sequences to other RNAs besides the on-target. However, miRNA-like seed matches are difficult to predict because they statistically occur very frequently on mRNAs and not all such matches are always leading to significant knockdown effects. A conclusive strategy to monitor such effects are whole transcriptome sequencing in case target organisms and cells are identifiable. However, molecules with strongly reduced off-target effects would be the most desirable approach. To reduce miRNA-like off-target activities, two main strategies have been developed (Jackson and Linsley, 2010; Seok et al., 2018). First, siRNA guide strands are chemically modified within their seed region particularly at position 2 from the 5′ end (Jackson et al., 2006a). Such modifications are either 2′-O-methylations or locked nucleic acid incorporations, in which the 2′-OH is chemically linked to the 4′ carbon of the ribose (Elmen et al., 2005). Both modifications weaken the interaction between the guide strand and the target. Since seed matches are short, such interactions are much stronger affected by this mild destabilization than siRNAs, which are typically fully complementary to their on-target. Thus, miRNA-like off-target interactions are reduced while on-target silencing is not compromised. In addition to the modification at position 2, other modifications have also been explored [for more details, please see Seok et al. (2018)]. A second approach to reduce off-target effects is pooling of multiple siRNAs. It is important to notice that miRNA-like off-target effects are specific to individual sequences. Thus, reducing the concentration of the applied siRNAs will also reduce miRNA-like off-target effects. This could be achieved by administration of very low concentrations (Persengiev et al., 2004). However, this would also directly affect on-target activity. An elegant way of lowering concentrations of siRNAs is siRNA pooling. Individual siRNAs within such a pool are directed against the same on-target at different positions, but each individual siRNA has a unique off-target signature. Consequently, all siRNAs act synergistically on the same on-target RNA. In complex pools, concentrations of individual siRNAs are very low and thus miRNA-like off-target effects are diluted out and cannot be measured anymore. Based on these ideas, three main pooling strategies are currently used. First, in the so-called smartPools, four individual siRNAs are combined. However, the complexity of such pools is low and thus the desired dilution effects are often not very pronounced. In contrast, endoribonuclease-produced siRNA pools (esiRNAs) are generated in vitro by recombinant RNase III digestion of long dsRNA (Kittler et al., 2005). These pools are then applied to cell cultures and, since these highly complex pools contain hundreds of different siRNAs, sequence-specific off-targets are not observed (Hannus et al., 2014; Kittler et al., 2005). A third strategy are so-called siPOOLs, which are highly complex but in contrast to esiRNAs, well defined. Up to 30 different siRNAs are designed and generated in vitro and such pools eliminate off-target effects even when a single siRNA with a pronounced off-target is included into the pool (Hannus et al., 2014).
Chemical modifications are the preferred choice when siRNAs are used for therapeutic purposes. For drug development, single and well-defined molecule species are preferred since broad toxicological validations are required during clinical trials and final approval. SiRNA pooling strategies are preferred in individual knockdown studies for research purposes or in genome-wide RNAi screening studies. Such pools are cost-efficient and thus genome-scale libraries are available.
Conclusion for RNA-Based Crop Protection and Outlook
Plants and animals with rather primitive immune systems tolerate long dsRNA and process it to siRNAs for gene silencing. One strategy in RNA-based crop protection is to spray dsRNA directed against pest-specific genes onto plants (Cai et al., 2018). Fungi or herbivores will take up these RNAs and process them to complex siRNA mixtures similar to esiRNA pools. This will kill or affect growth of the pathogens. Since such complex pools are naturally generated from dsRNAs in nematodes, insects, or fungi, miRNA-like off-target activity might be neglectable, when dsRNA is applied. In higher organisms such as mammals, the dsRNA will be fully degraded while transitioning through the digestive tract and only free nucleosides will be taken up. Thus, the administration of dsRNA to plants is an elegant and presumably very safe way of plant protection. SiRNAs are designed sequence specifically, and effects on other even highly related species could be minimized. Furthermore, since dsRNA is a natural product that is present in human diet, it might be better accepted by local communities than other plant protection strategies including the generation of genetically modified organisms (GMOs) or the use of conventional pesticides.
Author Contributions
GM structured and wrote the text. JN wrote the text and designed figures. Both authors contributed to the article and approved the submitted version.
Funding
The work in the Meister lab is supported by grants from the Deutsche Forschungsgemeinschaft (SFB 960) and the Bavarian Ministry for Education and Science (BioSysNet).
Conflict of Interest
GM is a co-founder of siTOOLs biotech.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript summarizes GM’s contribution during the OECD Conference on RNAi-based Pesticides, which was sponsored by the OECD Co-operative Research Programme: Biological Resource Management for Sustainable Agricultural Systems.

References
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Baulcombe, D. C. (1996). RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol. Biol. 32, 79–88. doi: 10.1007/978-94-009-0353-1_4
Behm-Ansmant, I., Rehwinkel, J., Doerks, T., Stark, A., Bork, P., and Izaurralde, E. (2006). mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20, 1885–1898. doi: 10.1101/gad.1424106
Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. doi: 10.1038/35053110
Birmingham, A., Anderson, E. M., Reynolds, A., Ilsley-Tyree, D., Leake, D., Fedorov, Y., et al. (2006). 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204. doi: 10.1038/nmeth854
Bohnsack, M. T., Czaplinski, K., and Gorlich, D. (2004). Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna 10, 185–191. doi: 10.1261/rna.5167604
Bologna, N. G., and Voinnet, O. (2014). The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev. Plant Biol. 65, 473–503. doi: 10.1146/annurev-arplant-050213-035728
Bologna, N. G., Iselin, R., Abriata, L. A., Sarazin, A., Pumplin, N., Jay, F., et al. (2018). Nucleo-cytosolic Shuttling of ARGONAUTE1 Prompts a Revised Model of the Plant MicroRNA Pathway. Mol. Cell 69, 709–719e5.
Braun, J. E., Huntzinger, E., and Izaurralde, E. (2013). The role of GW182 proteins in miRNA-mediated gene silencing. Adv. Exp. Med. Biol. 768, 147–163. doi: 10.1007/978-1-4614-5107-5_9
Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y. Y., Sieburth, L., et al. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190. doi: 10.1126/science.1159151
Buehler, E., Khan, A. A., Marine, S., Rajaram, M., Bahl, A., Burchard, J., et al. (2012). siRNA off-target effects in genome-wide screens identify signaling pathway members. Sci Rep. 2:428.
Cai, Q., He, B., Kogel, K. H., and Jin, H. (2018). Cross-kingdom RNA trafficking and environmental RNAi-nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 46, 58–64. doi: 10.1016/j.mib.2018.02.003
Cai, X., Hagedorn, C. H., and Cullen, B. R. (2004). Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna 10, 1957–1966. doi: 10.1261/rna.7135204
Carbonell, A. (2017). Plant ARGONAUTEs: Features, Functions, and Unknowns. Methods Mol. Biol. 1640, 1–21. doi: 10.1007/978-1-4939-7165-7_1
Das, S., Extracellular, R. N. A. C. C., Ansel, K. M., Bitzer, M., Breakefield, X. O., Charest, A., et al. (2019). The Extracellular RNA Communication Consortium: Establishing Foundational Knowledge and Technologies for Extracellular RNA Research. Cell 177, 231–242.
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F., and Hannon, G. J. (2004). Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235. doi: 10.1038/nature03049
Doench, J. G., Petersen, C. P., and Sharp, P. A. (2003). siRNAs can function as miRNAs. Genes Dev. 17, 438–442. doi: 10.1101/gad.1064703
Dorsett, Y., and Tuschl, T. (2004). siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 3, 318–329. doi: 10.1038/nrd1345
Dueck, A., and Meister, G. (2014). Assembly and function of small RNA - Argonaute protein complexes. Biol. Chem. 395, 611–629. doi: 10.1515/hsz-2014-0116
Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature 411, 494–498. doi: 10.1038/35078107
Elmen, J., Thonberg, H., Ljungberg, K., Frieden, M., Westergaard, M., Xu, Y., et al. (2005). Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 33, 439–447. doi: 10.1093/nar/gki193
Fedorov, Y., Anderson, E. M., Birmingham, A., Reynolds, A., Karpilow, J., Robinson, K., et al. (2006). Off-target effects by siRNA can induce toxic phenotype. Rna 12, 1188–1196. doi: 10.1261/rna.28106
Filipowicz, W. (2005). RNAi: the nuts and bolts of the RISC machine. Cell 122, 17–20. doi: 10.1016/j.cell.2005.06.023
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Frank, F., Hauver, J., Sonenberg, N., and Nagar, B. (2012). Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. Embo J. 31, 3588–3595. doi: 10.1038/emboj.2012.204
Frank, F., Sonenberg, N., and Nagar, B. (2010). Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465, 818–822. doi: 10.1038/nature09039
Fukudome, A., and Fukuhara, T. (2017). Plant dicer-like proteins: double-stranded RNA-cleaving enzymes for small RNA biogenesis. J. Plant Res. 130, 33–44. doi: 10.1007/s10265-016-0877-1
Gregory, R. I., Yan, K. P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., et al. (2004). The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240. doi: 10.1038/nature03120
Grishok, A., Pasquinelli, A. E., Conte, D., Li, N., Parrish, S., Ha, I., et al. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. doi: 10.1016/s0092-8674(01)00431-7
Han, J., Lee, Y., Yeom, K. H., Kim, Y. K., Jin, H., and Kim, V. N. (2004). The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027. doi: 10.1101/gad.1262504
Hannus, M., Beitzinger, M., Engelmann, J. C., Weickert, M. T., Spang, R., Hannus, S., et al. (2014). siPools: highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 42, 8049–8061. doi: 10.1093/nar/gku480
Iki, T., Yoshikawa, M., Meshi, T., and Ishikawa, M. (2012). Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. Embo J. 31, 267–278. doi: 10.1038/emboj.2011.395
Ipsaro, J. J., and Joshua-Tor, L. (2015). From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol. 22, 20–28. doi: 10.1038/nsmb.2931
Iwasaki, S., Kobayashi, M., Yoda, M., Sakaguchi, Y., Katsuma, S., Suzuki, T., et al. (2010). Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 39, 292–299. doi: 10.1016/j.molcel.2010.05.015
Iwasaki, Y. W., Siomi, M. C., and Siomi, H. (2015). PIWI-Interacting RNA: Its Biogenesis and Functions. Annu Rev. Biochem. 84, 405–433. doi: 10.1146/annurev-biochem-060614-034258
Jackson, A. L., and Linsley, P. S. (2010). Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 9, 57–67. doi: 10.1038/nrd3010
Jackson, A. L., Burchard, J., Leake, D., Reynolds, A., Schelter, J., Guo, J., et al. (2006a). Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. Rna 12, 1197–1205. doi: 10.1261/rna.30706
Jackson, A. L., Burchard, J., Schelter, J., Chau, B. N., Cleary, M., Lim, L., et al. (2006b). Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. Rna 12, 1179–1187. doi: 10.1261/rna.25706
Jakymiw, A., Lian, S., Eystathioy, T., Li, S., Satoh, M., Hamel, J. C., et al. (2005). Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 7, 1267–1274. doi: 10.1038/ncb1334
Jonas, S., and Izaurralde, E. (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433. doi: 10.1038/nrg3965
Ketting, R. F., Fischer, S. E., Bernstein, E., Sijen, T., Hannon, G. J., and Plasterk, R. H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659. doi: 10.1101/gad.927801
Kim, V. N., Han, J., and Siomi, M. C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139. doi: 10.1038/nrm2632
Kittler, R., Heninger, A. K., Franke, K., Habermann, B., and Buchholz, F. (2005). Production of endoribonuclease-prepared short interfering RNAs for gene silencing in mammalian cells. Nat. Methods 2, 779–784. doi: 10.1038/nmeth1005-779
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610. doi: 10.1038/nrg2843
Kwak, P. B., and Tomari, Y. (2012). The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 19, 145–151. doi: 10.1038/nsmb.2232
Landthaler, M., Yalcin, A., and Tuschl, T. (2004). The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 14, 2162–2167. doi: 10.1016/j.cub.2004.11.001
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419. doi: 10.1038/nature01957
Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. Embo J. 23, 4051–4060. doi: 10.1038/sj.emboj.7600385
Lev, I., Gingold, H., and Rechavi, O. (2019). H3K9me3 is required for inheritance of small RNAs that target a unique subset of newly evolved genes. Elife 8:e40448.
Lingel, A., Simon, B., Izaurralde, E., and Sattler, M. (2003). Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426, 465–469. doi: 10.1038/nature02123
Liu, J., Carmell, M. A., Rivas, F. V., Marsden, C. G., Thomson, J. M., Song, J. J., et al. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441. doi: 10.1126/science.1102513
Liu, J., Rivas, F. V., Wohlschlegel, J., Yates, J. R. III, Parker, R., and Hannon, G. J. (2005). A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7, 1161–1166.
Lund, E., Guttinger, S., Calado, A., Dahlberg, J. E., and Kutay, U. (2004). Nuclear export of microRNA precursors. Science 303, 95–98. doi: 10.1126/science.1090599
Ma, J. B., Ye, K., and Patel, D. J. (2004). Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429, 318–322. doi: 10.1038/nature02519
Ma, J. B., Yuan, Y. R., Meister, G., Pei, Y., Tuschl, T., and Patel, D. J. (2005). Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666–670. doi: 10.1038/nature03514
Maida, Y., Masutomi, K., and dependent, R. N. A. - (2011). RNA polymerases in RNA silencing. Biol Chem 392, 299–304.
Meister, G., and Tuschl, T. (2004). Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349. doi: 10.1038/nature02873
Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. (2004). Human Argonaute2 Mediates RNA Cleavage Targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197. doi: 10.1016/j.molcel.2004.07.007
Meister, G., Landthaler, M., Peters, L., Chen, P. Y., Urlaub, H., Luhrmann, R., et al. (2005). Identification of novel argonaute-associated proteins. Curr. Biol. 15, 2149–2155. doi: 10.1016/j.cub.2005.10.048
Mello, C. C., and Conte, D. Jr. (2004). Revealing the world of RNA interference. Nature 431, 338–342. doi: 10.1038/nature02872
Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., Ni, F., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127. doi: 10.1016/j.cell.2008.02.034
Miyoshi, T., Takeuchi, A., Siomi, H., and Siomi, M. C. (2010). A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat. Struct. Mol. Biol. 17, 1024–1026. doi: 10.1038/nsmb.1875
Nakanishi, K., Weinberg, D. E., Bartel, D. P., and Patel, D. J. (2012). Structure of yeast Argonaute with guide RNA. Nature 486, 368–374. doi: 10.1038/nature11211
Parker, J. S., Roe, S. M., and Barford, D. (2004). Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. Embo J. 23, 4727–4737. doi: 10.1038/sj.emboj.7600488
Persengiev, S. P., Zhu, X., and Green, M. R. (2004). Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). Rna 10, 12–18. doi: 10.1261/rna5160904
Pfaff, J., and Meister, G. (2013). Argonaute and GW182 proteins: an effective alliance in gene silencing. Biochem. Soc. Trans. 41, 855–860. doi: 10.1042/bst20130047
Pfaff, J., Hennig, J., Herzog, F., Aebersold, R., Sattler, M., Niessing, D., et al. (2013). Structural features of Argonaute-GW182 protein interactions. Proc. Natl. Acad. Sci. U S A 110, E3770–E3779.
Pradhan, M., Pandey, P., Gase, K., Sharaff, M., Singh, R. K., Sethi, A., et al. (2017). Argonaute 8 (AGO8) Mediates the Elicitation of Direct Defenses against Herbivory. Plant Physiol. 175, 927–946. doi: 10.1104/pp.17.00702
Rajewsky, N., and Socci, N. D. (2004). Computational identification of microRNA targets. Dev. Biol. 267, 529–535. doi: 10.1016/j.ydbio.2003.12.003
Schirle, N. T., Kinberger, G. A., Murray, H. F., Lima, W. F., Prakash, T. P., and MacRae, I. J. (2016). Structural Analysis of Human Argonaute-2 Bound to a Modified siRNA Guide. J. Am. Chem. Soc. 138, 8694–8697. doi: 10.1021/jacs.6b04454
Schlee, M., and Hartmann, G. (2016). Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 16, 566–580. doi: 10.1038/nri.2016.78
Schraivogel, D., and Meister, G. (2014). Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem. Sci. 39, 420–431. doi: 10.1016/j.tibs.2014.07.004
Seok, H., Lee, H., Jang, E. S., and Chi, S. W. (2018). Evaluation and control of miRNA-like off-target repression for RNA interference. Cell Mol. Life Sci. 75, 797–814. doi: 10.1007/s00018-017-2656-0
Sheridan, C. (2017). With Alnylam’s amyloidosis success, RNAi approval hopes soar. Nat. Biotechnol. 35, 995–997. doi: 10.1038/nbt1117-995
Sheu-Gruttadauria, J., and MacRae, I. J. (2017). Structural Foundations of RNA Silencing by Argonaute. J. Mol. Biol. 429, 2619–2639. doi: 10.1016/j.jmb.2017.07.018
Sibisi, P., and Venter, E. (2020). Wheat Argonaute 5 Functions in Aphid-Plant Interaction. Front. Plant Sci. 11:641.
Song, J. J., Smith, S. K., Hannon, G. J., and Joshua-Tor, L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437. doi: 10.1126/science.1102514
Song, X., Li, Y., Cao, X., and Qi, Y. (2019). MicroRNAs and Their Regulatory Roles in Plant-Environment Interactions. Annu Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Treiber, T., Treiber, N., and Meister, G. (2019). Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20, 5–20. doi: 10.1038/s41580-018-0059-1
Wang, Y., Juranek, S., Li, H., Sheng, G., Wardle, G. S., Tuschl, T., et al. (2009). Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461, 754–761. doi: 10.1038/nature08434
Yan, K. S., Yan, S., Farooq, A., Han, A., Zeng, L., and Zhou, M. M. (2003). Structure and conserved RNA binding of the PAZ domain. Nature 426, 468–474.
Yi, R., Qin, Y., Macara, I. G., and Cullen, B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016. doi: 10.1101/gad.1158803
Yuan, Y. R., Pei, Y., Ma, J. B., Kuryavyi, V., Zhadina, M., Meister, G., et al. (2005). Crystal Structure of A. aeolicus Argonaute, a Site-Specific DNA-Guided Endoribonuclease, Provides Insights into RISC-Mediated mRNA Cleavage. Mol. Cell 19, 405–419. doi: 10.1016/j.molcel.2005.07.011
Zamore, P. D., and Haley, B. (2005). Ribo-gnome: the big world of small RNAs. Science 309, 1519–1524. doi: 10.1126/science.1111444
Zhang, H., Kolb, F. A., Brondani, V., Billy, E., and Filipowicz, W. (2002). Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. Embo J. 21, 5875–5885. doi: 10.1093/emboj/cdf582
Keywords: siRNAs, microRNAs, off-target effects, RISC, RNAi
Citation: Neumeier J and Meister G (2021) siRNA Specificity: RNAi Mechanisms and Strategies to Reduce Off-Target Effects. Front. Plant Sci. 11:526455. doi: 10.3389/fpls.2020.526455
Received: 13 January 2020; Accepted: 15 December 2020;
Published: 28 January 2021.
Edited by:
Hailing Jin, University of California, Riverside, United StatesReviewed by:
Josh T. Cuperus, University of Washington, United StatesMatthew R. Willmann, Cornell University, United States
Copyright © 2021 Neumeier and Meister. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gunter Meister, Z3VudGVyLm1laXN0ZXJAdXIuZGU=
 Julia Neumeier
Julia Neumeier Gunter Meister
Gunter Meister