- 1Shandong Collaborative Innovation Center of Fruit & Vegetable Quality and Efficient Production, College of Horticulture, Qingdao Agricultural University, Qingdao, China
- 2Editorial Office of YanTai Fruits, Yantai Academy of Agricultural Sciences, Yantai, China
- 3College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, China
Drought is a major environmental factor that significantly limits crop yield and quality worldwide. Basic helix-loop-helix (bHLH) transcription factors have been reported to participate in the regulation of various abiotic stresses. In this study, a bHLH transcription factor in apple, MdbHLH130, which contains a highly conserved bHLH domain, was isolated and characterized. qRT-PCR and PMdbHLH130::GUS analyses showed that MdbHLH130 was notably induced in response to dehydration stress. Compared with the wild-type (WT), transgenic apple calli overexpressing MdbHLH130 displayed greater resistance to PEG6000 treatment. In contrast, the MdbHLH130-Anti lines were more sensitive to PEG6000 treatment than WT. Moreover, ectopic expression of MdbHLH130 in tobacco improved tolerance to water deficit stress, and plants exhibited higher germination rates and survival rates, longer roots, and lower ABA-induced stomatal closure and leaf water loss than the WT control. Furthermore, overexpression of MdbHLH130 in tobacco also led to lower electrolyte leakage, malondialdehyde contents, and reactive oxygen species (ROS) accumulation and upregulation of the expression of some ROS-scavenging and stress-responsive genes under water deficit stress. In addition, MdbHLH130 transgenic tobacco plants exhibited improved tolerance to oxidative stress compared with WT. In conclusion, these results indicate that MdbHLH130 acts as a positive regulator of water stress responses through modulating stomatal closure and ROS-scavenging in tobacco.
Introduction
Plants are inevitably challenged by environmental factors during their life cycle (including biotic and abiotic stresses), and drought is a major stressor because it limits plant growth and crop quality and productivity (Umezawa et al., 2006; Singh and Laxmi, 2015). To survive, plants must evolve a wide range of physiological and biochemical processes for reducing water loss or increasing tolerance under drought stress (Shinozaki et al., 2003; Basu et al., 2016; Rasheed et al., 2016).
Previous studies in Arabidopsis, rice, and other plants have shown that the transcript levels of many drought-inducible genes are significantly increased or decreased in response to drought stress (Shinozaki and Yamaguchi-Shinozaki, 2007; Buscaill and Rivas, 2014). In Arabidopsis, drought-inducible genes can be classified into two groups. The first group consists of functional proteins, including water channel proteins, detoxification enzymes, key osmolyte biosynthesis enzymes, and proteases. The second group consists of regulatory proteins, including transcription factors (TFs), protein kinases, and protein phosphatases (Liu et al., 2014; Joshi et al., 2016). Among the various regulatory genes, TFs act as key regulators to transduce stress signals and regulate stress-associated target gene expression to protect plants from drought stress damage (Singh and Laxmi, 2015). In plants, many TFs, including APETALA2/ethylene-responsive element binding protein (AP2/EREBP), basic leucine zipper (bZIP), NAM/ATAF1, 2/CUC2 (NAC), nuclear factor-Y (NF-Y), basic helix-loop-helix (bHLH), dehydration-responsive element-binding (DREBs), ABA-responsive element-binding (AREBs), myeloblastosis (MYB), and WRKY, were identified in response to drought stress via microarray analyses (Tran et al., 2004; Sakuma et al., 2006; Nakashima et al., 2009; Ishida et al., 2012; Nuruzzaman et al., 2013; Osakabe et al., 2014; Kudo et al., 2017). The bHLH protein family, one of the largest families of TFs, is present in all eukaryotic organisms. The family is defined by the bHLH domain, which contains 50–60 amino acids and two functionally distinct regions: the 13–17 basic region located at the N-terminus for binding to specific E-box (CANNTG) motifs, and the HLH region, which forms two amphipathic α-helices separated by a loop of variable length for forming homodimers or heterodimers with its partners (Toledo-Ortiz et al., 2003; Feller et al., 2011).
bHLH proteins are one of the important families of conserved TFs that regulate many cellular processes in both plants and animals, and they are also involved in responses to biotic and abiotic stresses (Toledo-Ortiz et al., 2003; Castilhos et al., 2014; Sun et al., 2018; Zhao et al., 2018). In animals, bHLHs are involved in many physiological and developmental processes, such as responses to environmental signals, neurogenesis, myogenesis, sex differentiation, and cell differentiation (Atchley and Fitch, 1997; Tomita et al., 2000; Nieto et al., 2001; Stevens et al., 2008). In plants, bHLH TFs play key roles in the regulation of seed germination (Penfield et al., 2005); flowering time (Kumar et al., 2012); hypocotyl growth (Lorrain et al., 2008); trichome initiation (Qi et al., 2011); anthocyanin biosynthesis, flavonoid synthesis (Ramsay and Glover, 2005); light signaling (Leivar et al., 2008); hormone signaling (Abe et al., 2003; Fernández-Calvo et al., 2011); responses to wounding, drought, salt, and low temperatures (Sun et al., 2018); iron deficiency, and phosphate starvation (Ling et al., 2002; Zhao et al., 2016). In Arabidopsis, there are 162 genes encoding bHLHs, and 80% have not yet been functionally characterized (Pires and Dolan, 2010; Feller et al., 2011). Recently, mounting evidence has shown that bHLH TFs are important for plant responses to abiotic stresses. For example, AtICE1 and MdCIbHLH1 increase cold stress tolerance in Arabidopsis and apple (Feng et al., 2012); bHLH122 regulates resistance to drought, salt, and osmotic stress, and increases cellular ABA levels in Arabidopsis (Liu et al., 2014); bHLH104 and MdbHLH104 increase iron deficiency tolerance in Arabidopsis and apple (Zhang et al., 2015; Zhao et al., 2016); AtMYC2 increases drought resistance by positively regulating the transcription of the drought-responsive gene RD22 in Arabidopsis (Abe et al., 2003); AtbHLH112 is a transcriptional activator that mediates proline biosynthesis and reactive oxygen species (ROS)-scavenging pathways to enhance abiotic stresses tolerance; OsbHLH148, an AtMYC2 homolog, improves drought tolerance in rice (Seo et al., 2011); NtbHLH123 increases cold tolerance by regulating the NtCBF genes and ROS-scavenging in Nicotiana tabacum (Zhao et al., 2018); and PebHLH35 improves plant tolerance to drought by regulating stomatal density and stomatal pore size (Dong et al., 2014).
Apples constitute an important part of the daily human diet and are one of the best-loved fruit in many parts of the world. The trees are most affected by drought stress, which significantly decreases fruit yield and quality. Thus, they have developed a number of sophisticated mechanisms to adapt to the surrounding conditions. Recently, some bHLH TFs have been involved in response to drought stress in plants (Singh and Laxmi, 2015). However, only a few apple bHLH genes have been functioned in drought response. Mao et al. (2017) identified 188 MdbHLH proteins in apple, and some of them regulated responses to drought stress. Here, we further identified a bHLH gene, MdbHLH130, which is highly induced by dehydration treatment, and selected it for further study. Transgenic apple calli overexpressing MdbHLH130 displayed greater resistance to PEG6000 treatment and were more sensitive to the suppression of MdbHLH130 compared with that of the wild type (WT). The overexpression of MdbHLH130 in tobacco increased water deficit tolerance, reduced electrolyte leakage and malondialdehyde (MDA) levels, elevated antioxidant enzyme activity, and increased the expression of ROS-scavenging genes and stress-related genes compared with the WT plants. Moreover, MdbHLH130-overexpressing plants showed decrease sensitivity to oxidative stress. Overall, these data might provide new insights into the mechanism of bHLH proteins in response to water deficit stress.
Materials and Methods
Plant Materials and Treatments
Apple (Malus × domestic ‘Royal Gala’) seedlings were grown at 23°C under 8/16-h dark/light photoperiod. The method of dehydration treatment was performed according to Liao et al., 2017. The seedlings were transferred to filter paper and dried at 23°C for the indicated times (0, 1, 3, 6, 12, and 24 h), and then immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
‘Orin’ apple calli (wild-type) were used for genetic transformation in this study. The calli were cultivated on medium as previously described (Zhao et al., 2016).
Tobacco cultivars (N. tabacum L. “NC89”) were used for genetic transformation. The seeds were surface-sterilized with 2.6% NaClO for 10 min and then germinated on 1/2 MS medium containing 2.5% sucrose and 1.0% agar at 25 ± 1°C with a 16-h light/8-h dark photoperiod.
Real-Time Quantitative PCR
Total RNA was extracted from the apple seedlings, apple calli, and tobacco whole seedlings using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions, and then reverse transcripts were synthesized with a PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, China). Real-time quantitative PCR was carried out on an Applied Biosystem 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using the SYBR Premix Ex Taq (TaKaRa, Dalian, China) in a 20-μl volume. The PCR reaction conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. The MdActin (GenBank: CN938023) was used as an internal reference for apple, while NtActin (GenBank: U60495) was used for tobacco. Relative gene expression analyses were conducted using the cycle threshold (Ct) 2−Δ;Δ;CT method (Livak and Schmittgen, 2001). Each sample at each time point was performed in three independent replicates. The primer sequences in this study are shown in Table S1.
Plasmid Construction and Genetic Transformation
To generate the PMdbHLH130::GUS construct, a 2.0 kb promoter fragment was amplified and inserted into the pCXGUS-P vector containing the GUS reporter gene. For the overexpression vector, the full-length cDNA of MdbHLH130 was cloned into pRI-GFP vectors under the control of a CaMV 35S promoter. For the antisense suppression vector, a 280-bp specific fragment of MdbHLH130 was amplified and linked into the pRI vector. Then, the recombinant plasmid PMdbHLH130::GUS, pRI-MdbHLH130-GFP and pRI-MdbHLH130-Anti were introduced into the A. tumefaciens strain EHA105. The primer sequences are shown in Table S2.
Transgenic apple calli were obtained as described previously (Zhao et al., 2016). ‘Orin’ apple calli were immersed into Agrobacterium suspension cultures for 10-15 min. Then, the calli were co-cultivated in MS medium with 1.5 mg−1 L 2,4-D and 0.4 mg−1 L 6-BA at 23 ± 1°C in the dark for 2–3 days. After co-cultivation, the calli were transferred to the screening medium containing MS + 1.5 mg−1 L 2,4-D + 0.4 mg−1 L 6-BA + 100 mg−1 L kanamycin + 250 mg−1 L carbenicillin to obtain transgenic calli.
For transgenic tobacco, the A. tumefaciens strain EHA105 containing binary constructs were transformed into tobacco by leaf disc transformation (Horsch et al., 1985). The leaves were cut into small strips and immersed into A. tumefaciens suspension cultures for 15 min. Then, the pieces were dried with sterile filter paper and screened onto selection medium (containing 100 mg−1 L kanamycin). T3 seedlings from three independent transgenic lines were used in further investigations.
GUS Staining and Activity Analyses
GUS staining of ProMdbHLH130::GUS transgenic apple calli and the WT was performed according to Jefferson et al. (1987). The apple calli were transferred to a filter paper and dried at 23°C for 4 h and then immersed into GUS staining buffer at 37°C in the dark overnight. After staining, the samples were de-stained and photographed.
GUS activity was detected as described previously (Jefferson et al., 1987). The total proteins from the samples were extracted with extraction buffer and reacted with 4-MUG (Sigma-Aldrich, St. Louis, MO, USA) at 37°C. The GUS activity was determined using a VersaFlour spectrofluorometer (Bio Rad Laboratory Inc., Hercules, CA) at an Ex = 365 nm and Em = 450 nm.
Subcellular Localization and Transactivation Assay
The construct 35S::MdbHLH130-GFP (pRI-MdbHLH130-GFP) was transferred into the A. tumefaciens GV3101 strain. Then, the Agrobacterium-harboring 35S::MdbHLH130-GFP construct was infiltrated into the epidermal cells of Nicotiana benthamiana leaves at 24°C for 2–3 days. 4′,6-diamidino-2-phenylindole (DAPI) was used to stain the nuclei, and the fluorescence was photographed with a confocal laser scanning microscope (Ex = 488 nm by argon laser, Em = 505–530 nm by BP filter) (Zeiss LSM510 Meta, Germany).
The open reading frames (ORFs) of MdbHLH130 were cloned into the pGBKT7 vector (Clontech) to generate pGBKT7-MdbHLH130. Then, the pGBKT7-MdbHLH130 + pGADT7 was transformed into the yeast strain Y2H Gold. The transformants were spotted on SD/-Trp and SD/-Trp/-His/-Ade medium.
Protein Extraction and Western Blotting
Samples of tobacco whole seedlings were ground in a protein extraction buffer. The proteins were separated by SDS-PAGE gel and then transferred onto polyvinylidene fluoride (PVDF) membrane (Roche, USA). The signals were visualized using the ECL plus kit (Millipore, Bedford, MA) and the MdbHLH130 protein levels were detected with anti-GFP antibody as described previously (Burnette, 1981). ACTIN served as a protein-loading control.
Stress Tolerance Assay
For the PEG 6000 treatment and germination assay, the seeds of transgenic lines and WT were sterilized by 2.6% NaClO and planted on 1/2 MS medium with or without 10% PEG6000, and then the germination rates were measured daily (Quan et al., 2004). For the root length assay, the seeds were germinated on 1/2 MS medium for 10 d. Seedlings with the same root length were then transferred to 1/2 MS medium supplemented with 10% PEG6000 and placed upright in the chamber. The root lengths were measured after the treatment.
For the drought treatment, 35-day-old MdbHLH130-overexpression and WT plants were grown in a greenhouse and adequately watered. Subsequently, water was withheld for 15 d, and the survival rates were recorded after re-watering for 7 d. To estimate the water loss, fully expanded leaves were detached from 35-day-old plants and weighed immediately. Then, the leaves were placed on filter paper in the ambient environment and weighed at the designated times, and the water loss rate was calculated relative to that of the initial fresh weights.
For the oxidative stress treatment, the leaf discs were detached from the fully expanded leaves of 35-day-old plants and incubated in a methyl viologen (MV) solution (0 or 100 μM) for 48 h, and then the Chl contents were measured. All the results are based on the average of three independent biological replicates.
Stomatal Aperture Ratio Analysis
Leaves of the WT and transgenic lines were incubated in an opening solution (10 mM KCl, 50 mM CaCl2, and 10 mM MES-Tris, pH 6.15) for 4 h under light conditions. To observe the stomatal aperture ratio under dehydration stress conditions, the leaves were placed on filter paper for 4 h. To investigate the ABA response, the leaves were treated with 10 µM ABA solution for 4 h. Subsequently, the stomata were imaged using a microscope (Olympus ix71, Tokyo, Japan). The widths and lengths of the stomata were determined with ImageJ software. At least 100 stomata measurements were performed for each treatment.
ROS Staining and Physiological Measurements
ROS levels were monitored using the fluorescent probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFH2-DA, Sigma-Aldrich, USA). The leaves were treated with 10 µM DCFH2-DA for 30 min and then washed with distilled H2O. The images were obtained using the confocal microscope (Ex = 488 nm; Em = 522 nm) (Leica Microsystems, Wetzlar, Germany). Fluorescence intensity of leaves was analyzed using ImageJ software.
The electrolyte leakage (EL) measurement in leaves was examined according to the method of Lurie and Ben-Arie (1983). The leaves were placed in distilled water for 3–4 h at room temperature and measured with electric conductivity meter (E1). Then the samples were boiled for 20 min and cooled to room temperature, the electrolyte conductivity (E2) was measured, and the Electrolyte leakage (%) = 100 × E1/E2. The malondialdehyde (MDA) contents were measured using the thiobarbituric acid-based method with a MDA assay kit (A003-3, Jiancheng, Nanjing, China). For the total chlorophyll content (a + b), the young leaves were collected and immersed in 80% acetone and then centrifuged at 10,000 g for 5 min. The total chlorophyll concentrations were measured from the spectroscopy absorbance measurements at 663, 645, and 480 nm.
For the enzyme assays, the SOD activity was measured following the manufacturer’s instructions of the SOD Detection Kit (A001, Jiancheng, Nanjing, China). The CAT activity was measured using a CAT Detection Kit by detecting the degradation of H2O2 at 405 nm (A007, Jiancheng, Nanjing, China). The POD activity was detected in the leaves according to POD Detection Kit for plant (A084-3, Jiancheng, Nanjing, China). Each assay was replicated at least three times per sample.
Statistical Analysis
The SPSS 16.0 software (SPSS Inc., Chicago, IL, USA) was used for data analyses. Experimental data were analyzed with Student’s t-test, taking P < 0.01 (*) as significant. All experiments were repeated at least three replicates, and sample variability is given as the standard deviation (SD) of the mean.
Results
Phylogenetic Tree and Sequence Analysis of MdbHLH130
Studies have reported that bHLH TFs have important functions in the regulation of plant stress responses. Previous studies have shown that a bHLH TF (MDP0000581816) is upregulated by drought stress treatment (Mao et al., 2017; Benny et al., 2019), suggesting that it may be involved in drought stress tolerance; thus, it was selected for further study. The phylogenetic tree of all bHLH genes of apple has been analyzed by Mao et al. (2017). The MDP0000581816 protein clustered within the same clade as MDP0000307685 (similarity 75.37%), so the MDP0000581816 gene is not duplicated. The open reading frame of MDP0000581816 is 1236 bp in length and encodes proteins with 411 amino acids. The phylogenetic tree showed that MDP0000581816 shared most sequence similarities with the uncharacterized Arabidopsis gene AtbHLH130 (AT2G42280), which is named MdbHLH130 (Figure 1A). Multiple sequence alignment showed that MdbHLH130 had a single bHLH conserved domain in the C-terminal (Figure 1B).
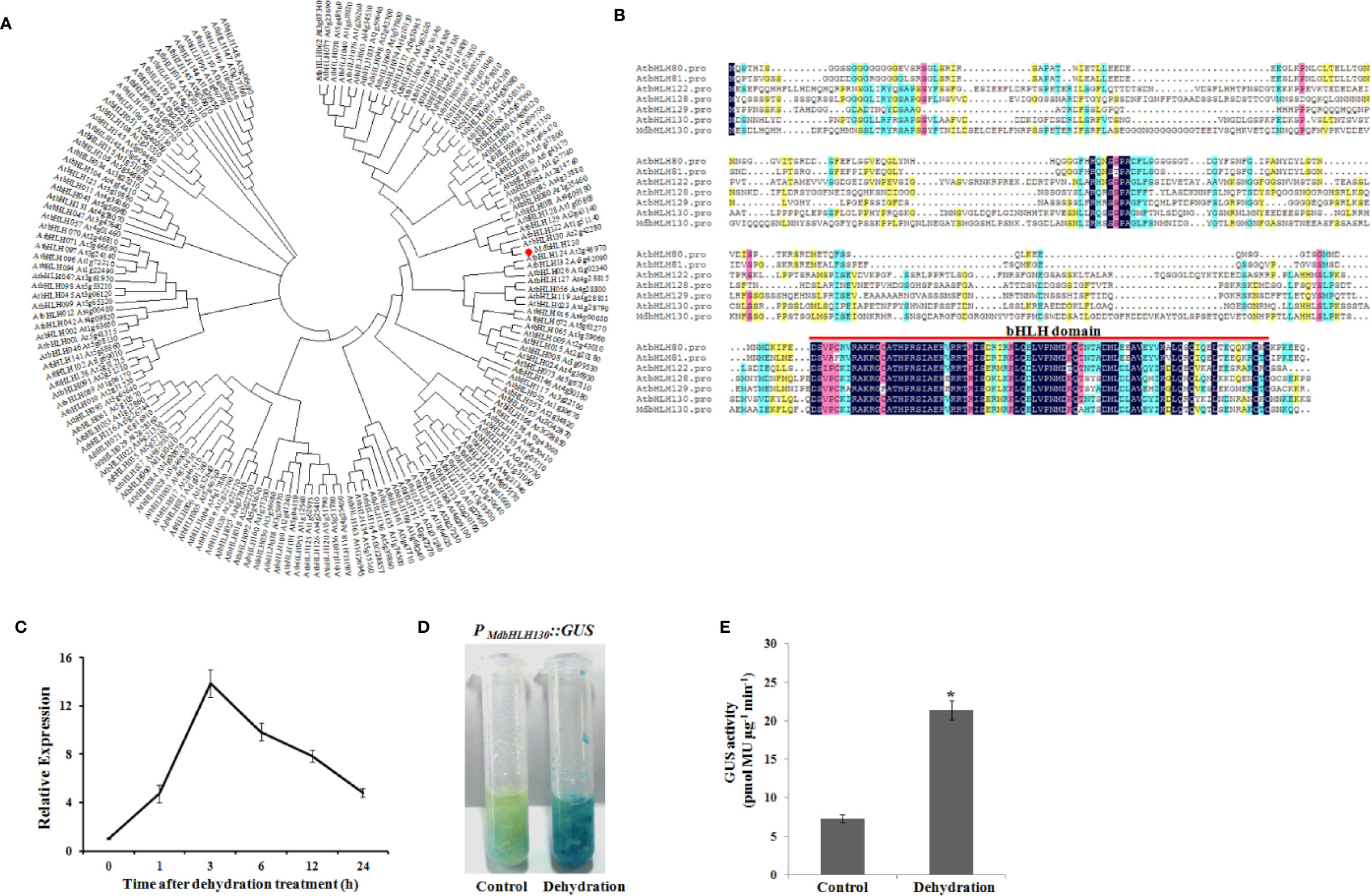
Figure 1 Phylogenetic tree, sequence alignment and transcript levels of the MdbHLH130. (A) Phylogenetic analysis of bHLH proteins between 166 AtbHLH proteins and the MdbHLH130 protein via MEGA4.0 using the neighbor-joining method. MdbHLH130 is in the red box. (B) Multiple sequence alignment of the MdbHLH130 protein with its known Arabidopsis homologs. Identical amino acids are shaded in black. The conserved bHLH motif is indicated with a red line. (C) qRT-PCR analysis of MdbHLH130 observed in 30-day-old apple seedlings after the dehydration treatment applied for 0, 1, 3, 6, 12, and 24 h. Data are the means ± SD of three independent biological replicates. (D) and (E) GUS staining and activity analysis of PMdbHLH130::GUS transgenic apple calli after the dehydration treatment. Error bars indicate the means ± SD from three independent biological replicates. Asterisks indicate significant differences relative to the control (*P < 0.01).
MdbHLH130 Is Induced by Dehydration Stress
The expression pattern of MdbHLH130 was examined in response to dehydration stress via a quantitative real-time (qRT) PCR analysis, and the results showed that MdbHLH130 expression levels were greatly induced and gradually reached a peak at 3 h (12.6-fold) and then decreased (Figure 1C). To further confirm the expression patterns of MdbHLH130, the promoter region (2.0 kb) was cloned and fused to a β-glucuronidase (GUS) reporter gene to achieve PMdbHLH130::GUS transgenic apple calli. Consistent with the qRT-PCR analysis results, the GUS staining and activity results revealed that PMdbHLH130::GUS transgenic apple calli had higher GUS activity compared with the control under dehydration stress treatment (Figures 1D, E). These results indicated that the expression of MdbHLH130 is induced by dehydration stress.
MdbHLH130 Is a Nuclear Protein That Function as a TF
The bioinformatic prediction analysis implied that MdbHLH130 might be targeted to the nucleus (PredictNLS software). To determine the subcellular localization of MdbHLH130, the CDS region of MdbHLH130 was fused with the green fluorescent protein (GFP) reporter gene under the control of the CaMV 35S promoter. Because the Agrobacterium-harboring 35S::MdbHLH130-GFP construct had difficulty infiltrating into the epidermal cells of apple leaves, the sub-localization of apple genes were usually done in Nicotiana benthamiana leaves. The construct was then transiently injected into the epidermal cells in N. benthamiana leaves using A. tumefaciens-mediated transformation. After 2–3 days, the GFP signal was detected only in the nucleus of epidermal cells (Figure 2A), indicating that MdbHLH130 was localized to the nucleus. Nuclear localization was confirmed by staining with DAPI.
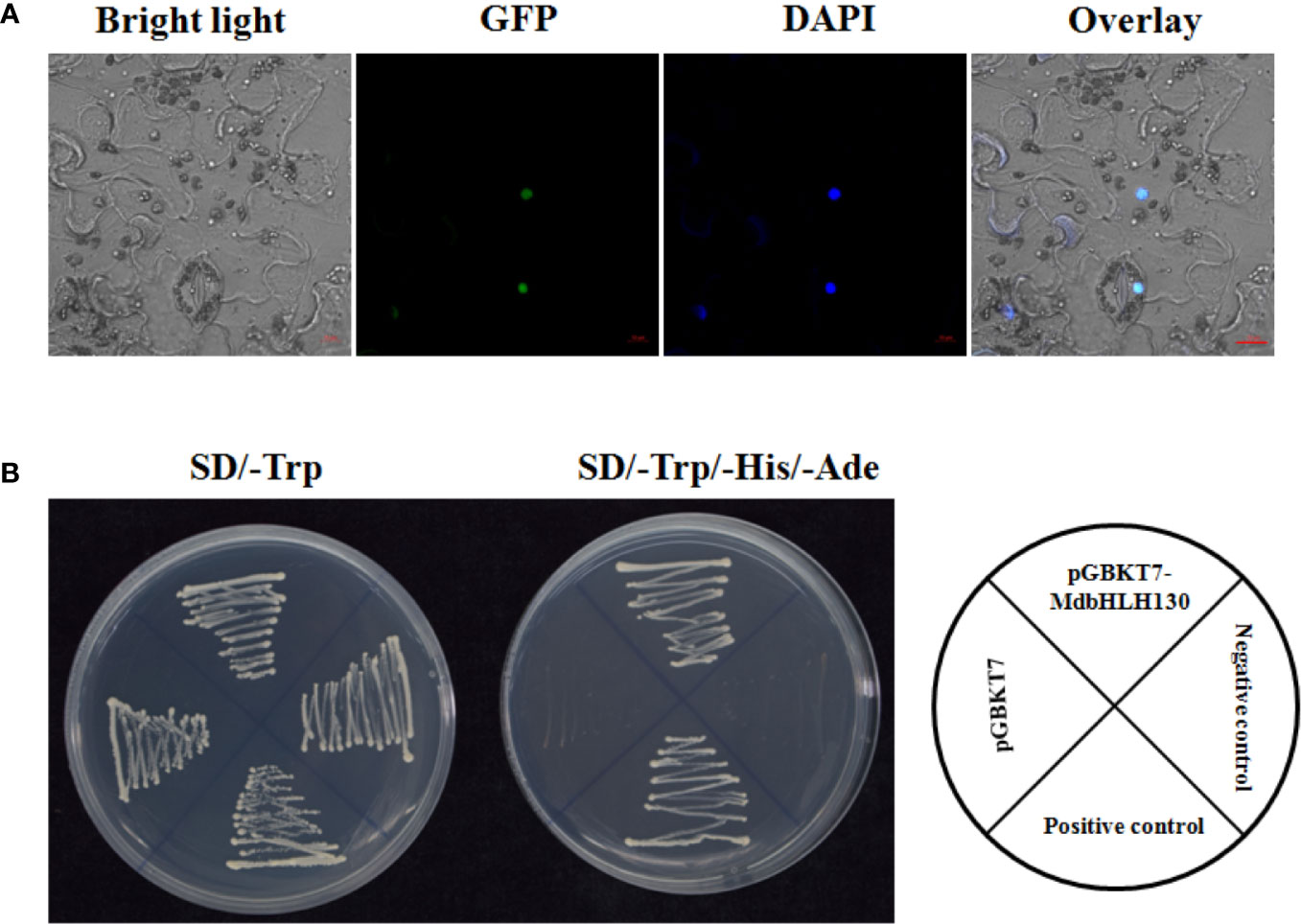
Figure 2 Subcellular localization and transcriptional activity of the MdbHLH130 protein. (A) Subcellular localization of MdbHLH130. The 35S::MdbHLH130-GFP fusion proteins were transiently transferred into epidermal cells of N. benthamiana leaves. Green fluorescence was visualized using confocal microscopy. Scale bar, 10 μm. (B) Transactivation assay of MdbHLH130 in yeast. The yeast cells transformed with different constructs on SD/-Trp or SD/-Trp/-His/-Ade medium for 3–5 days.
bHLH TFs have been reported to possess transcriptional activity (Toledo-Ortiz et al., 2003). To determine whether MdbHLH130 had transcriptional activity, the full-length MdbHLH130 coding sequence (CDS) was fused with a GAL4 DNA-binding domain (GDBD) in the yeast expression vector pGBKT7. The different combinations were transformed separately into the yeast strain Y2H Gold. The different transformants were transformed, and all grew well on synthetic dextrose synthetic dropout (SD)/-Trp medium. However, the yeast cells transformed with the control vector did not survive on selective SD/-Trp/-His/-Ade media. In contrast, yeast cells harboring pGBKT7-MdbHLH130 grew normally on the same medium (Figure 2B), indicating that MdbHLH130 exhibited transcriptional activity in yeast cells.
Stress Tolerance Assay of MdbHLH130 in Transgenic Apple Calli
Because it is difficult to obtain transgenic apple plants using the apple calli “Orin”, so the apple calli were thereafter used for function analysis. As a model system, the apple transgenic calli were used to characterize the function of some genes in modulating stresses tolerance (Zhao et al., 2016; Ma et al., 2019; Ren et al., 2019). To characterize the functions of MdbHLH130, the ORF sequence and specific cDNA fragments were cloned and fused into the PRI vector to generate 35S::MdbHLH130 (MdbHLH130-ox) and 35S::MdbHLH130-Anti (MdbHLH130-Anti) constructs, respectively. Then, the constructs were transformed into apple calli (‘Orin’). qRT-PCR analysis indicated that the MdbHLH130-ox line had higher transcript levels relative to the WT control while the MdbHLH130-Anti calli produced lower levels (Figure 3A), indicating that transgenic apple calli were obtained. Subsequently, the 10-day-old MdbHLH130-ox, MdbHLH130-Anti, and WT calli were grown on MS medium with or without 6% PEG6000 for another 20 days. Compared with the control, MdbHLH130-ox calli exhibited faster growth than the WT control, while the MdbHLH130-Anti calli had the opposite phenotype (Figures 3B, C). Consistent with the phenotype, the MDA content levels in the MdbHLH130-ox calli were lower than that of the WT lines, while the MdbHLH130-Anti calli showed increased levels (Figure 3D). Overall, MdbHLH130 enhances the tolerance to PEG6000 in transgenic apple calli.
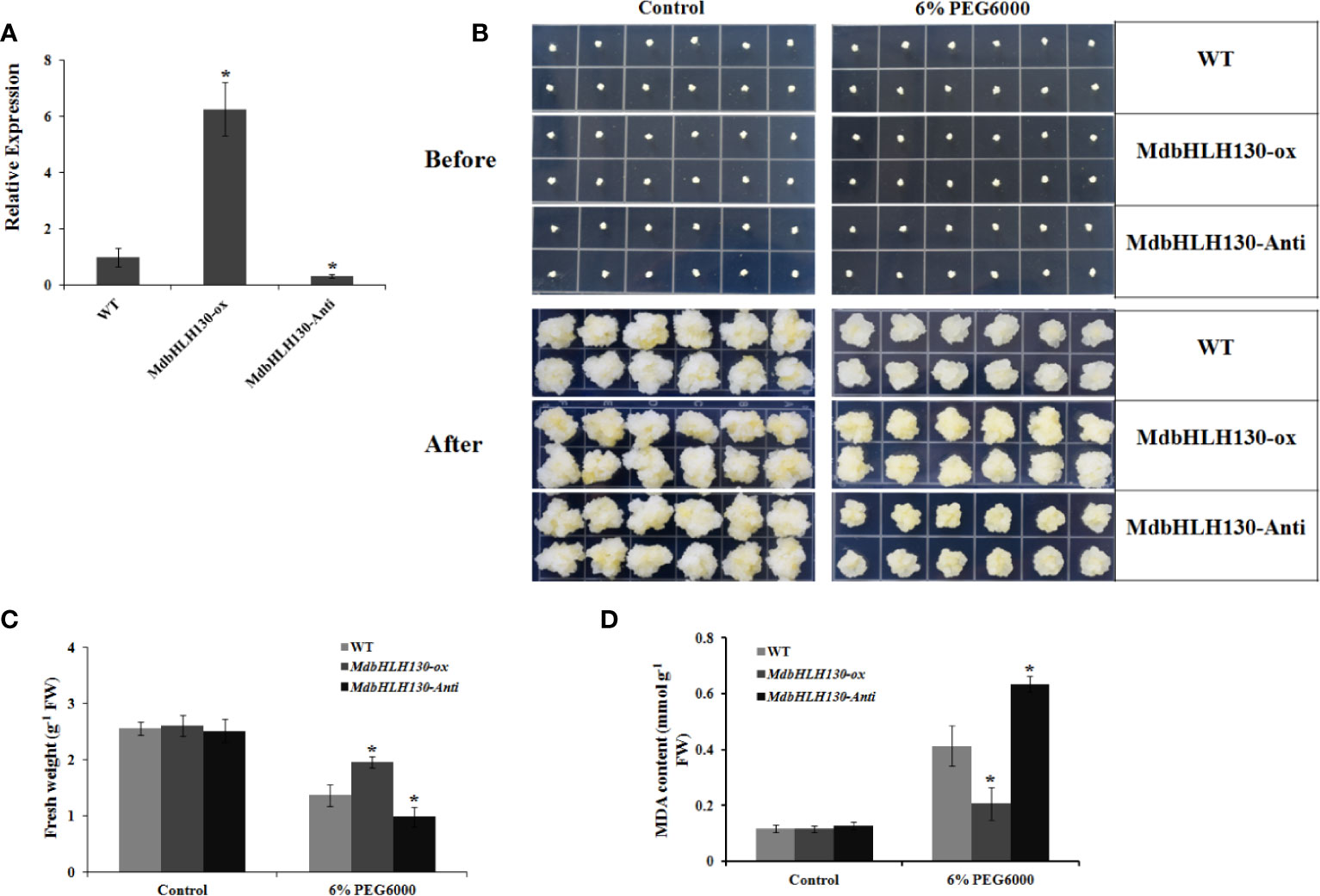
Figure 3 Effect of MdbHLH130 on PEG6000 tolerance in transgenic apple calli. (A) Expression analysis of MdbHLH130 in WT, MdbHLH130-ox and MdbHLH130-Anti transgenic calli by RT-PCR. The results were normalized using the internal control MdActin. Data are expressed as the mean ± SD as determined from three independent biological replicates. (B) Growth phenotypes of WT, MdbHLH130-ox and MdbHLH130-Anti transgenic apple calli. The WT and transgenic apple calli were grown on medium at 23°C for 7 days and then treated with 6% PEG6000 for another 20 days. (C, D) Fresh weights and MDA contents of WT, MdbHLH130-ox and MdbHLH130-Anti transgenic apple calli under the control or PEG6000 treatment conditions for another 20 days. Error bars represent the means ± SD taken from three independent biological replicates. Asterisks indicate significant differences relative to the WT (*P < 0.01).
Next, we examined the transcript abundances of ROS-scavenging and stress-related genes. qRT-PCR results indicated that the gene expression in MdbHLH130-OX lines was slightly higher than in the MdbHLH130-Anti lines and wild-type under control conditions. However, the transcript levels were significantly higher in the MdbHLH130-OX lines than in wild-type, whereas suppression of MdbHLH130 had the opposite results (Supplementary Figure S1).
Overexpression of MdbHLH130 in Tobacco Enhances Water Deficit Tolerance
Transgenic tobacco plants overexpressing MdbHLH130 were generated to further investigate the role of MdbHLH130 in water deficit stress tolerance. A total of eight independent transgenic lines were obtained using PCR and western blotting analyses (Supplementary Figures S2A–C). Three T3 homozygous lines (L1, L2, and L3) were selected, and they had higher MdbHLH130 expression levels; thus, they were selected for further analyses using semi-quantitative RT-PCR and western blotting (Supplementary Figures S3A, B).
First, the germination rates of the WT and MdbHLH130-ox plants on MS medium with or without 10% PEG6000 were explored. On MS medium, the MdbHLH130-ox lines and WT plants showed similar germination rates (Supplementary Figure S4). However, supplementation with 10% PEG6000 significantly inhibited the germination rates of the WT but had a limited effect on the MdbHLH130-ox transgenic plants (Supplementary Figure S4). To further evaluate the effects of drought stress on the transgenic plants, 10-day-old WT and MdbHLH130-ox seedlings were placed on MS medium containing 10% PEG6000 for 8 days. The root elongation was significantly higher for the MdbHLH130-ox (3.7 to 4.6 cm) transgenic plants than for the WT (3.1 cm) plants (Figures 4A, B), indicating that MdbHLH130 is involved in the response to PEG6000 treatment at the germination and root elongation stages.
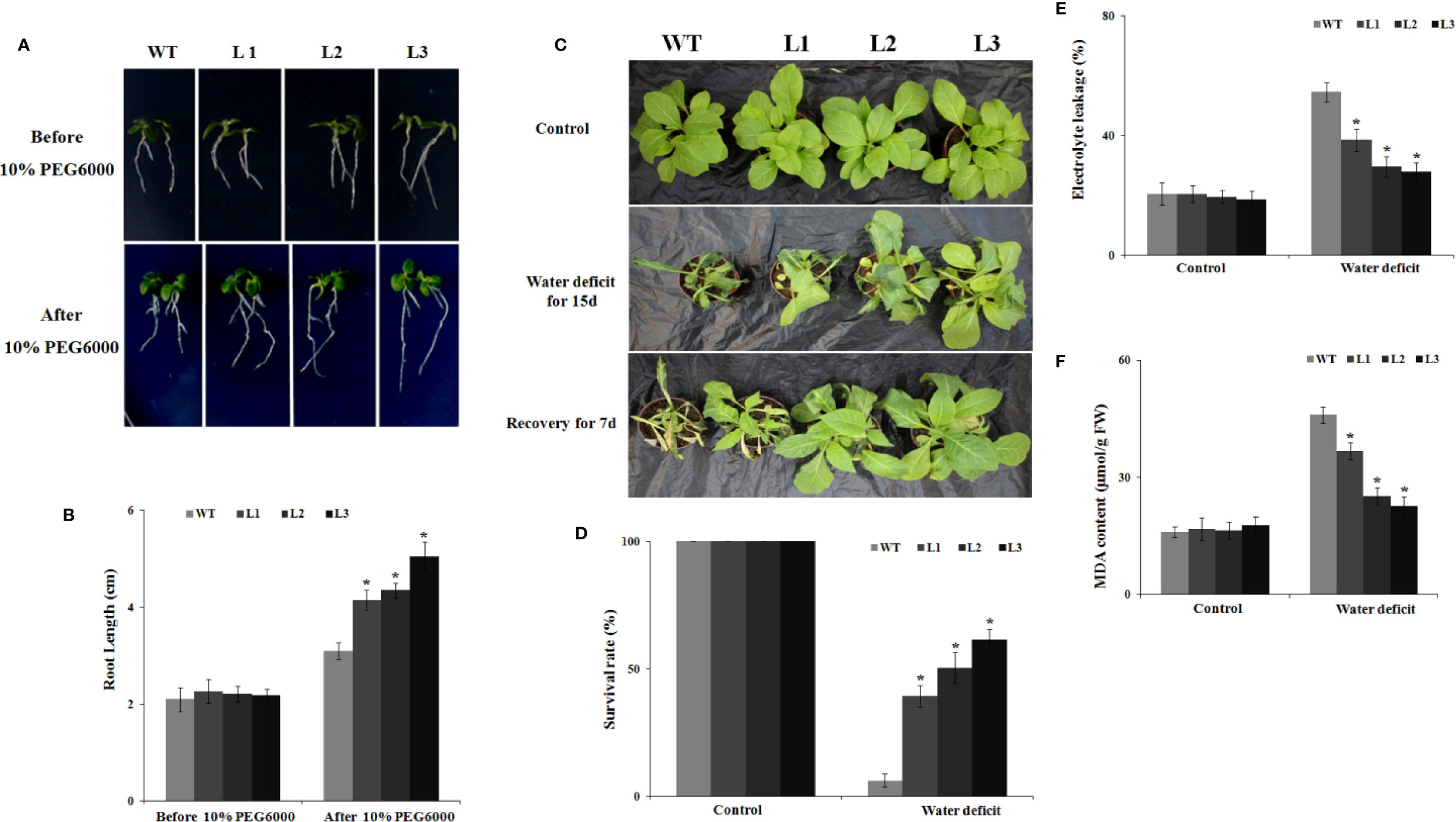
Figure 4 Water deficit stress tolerance to tobacco plants overexpressing MdbHLH130. (A, B) Primary root growth of the WT and MdbHLH130-ox (L1, L2, and L3) seedlings before and after 10% PEG6000 treatment. The seedlings were grown on 1/2 MS medium supplemented with 10% PEG6000 and placed upright in the chamber for 8 days. (C) Water deficit stress phenotypes of the WT and MdbHLH130 transgenic lines. The WT and three transgenic tobacco lines (L1, L2, and L3) were grown at 24°C and then withheld water for 15 days, and photos were taken 7 days after the plants were re-watered. (D, E) Statistical analysis of the survival rates (D), electrolyte leakage (E) and MDA contents (F) in the WT and transgenic lines after water deficit treatment. FW, Fresh weight. Error bars represent the means ± SD of more than 30 plants from three independent biological replicates. Asterisks indicate significant differences relative to the WT (*P < 0.01).
Subsequently, 35-day-old seedlings of the WT and transgenic plants were exposed to water deficit for 15 days in soil. Most of the WT plants wilted, whereas the MdbHLH130-ox transgenic plants remained turgid and green (Figure 4C). Moreover, 45–63% of the MdbHLH130-ox transgenic plants survived after re-watering, which was significantly higher than for the WT (11%) plants (Figure 4D). These results suggested that MdbHLH130-ox plants may have depleted their water resources more slowly than the WT transgenic plants and thus grew well. Additional indicators of stress are the malondialdehyde (MDA) content and electrolyte leakage (EL), which can reflect the degree of damage to cell membranes (Moore and Roberts, 1998). MDA and EL accumulation were detected in the transgenic lines and WT under water deficit stress. The transgenic lines exhibited significantly less MDA and EL accumulation compared to the WT (Figures 4E, F), confirming that the overexpression of MdbHLH130 reduced the damage and maintained the membrane integrity during water deficit stress. In addition, the total chlorophyll levels in the leaves of the WT plants (0.22 mg/g FW) were also lower than those of the transgenic plants (0.37–0.53 mg/g FW) after the water deficit treatment (Supplementary Figure S5A). Consistent with the total chlorophyll levels, the PN and Fv/Fm values were decreased in both the WT and transgenic plants under water deficit stress; however, their values remained higher in the transgenic plants compared with the WT (Supplementary Figures S5B, C).
To investigate whether there was a difference in water loss rate, we measured the water loss rates from the leaves of the WT and MdbHLH130-ox plants under normal and drought conditions. As shown in Figure 5A, the water loss rate of the leaves from the transgenic MdbHLH130-ox plants was much lower compared with that of the WT plants after the dehydration treatment, indicating that MdbHLH130 overexpression in tobacco leads to superior water status. Consistent with these results, the stomatal aperture ratio indices of the MdbHLH130-ox plants decreased to 0.32–0.24, which was smaller than that of the WT after 4 h of dehydration (Figures 5B, C). A previous study reported that ABA-mediated stomatal closure is crucial to reducing transpiration under drought stress (Castilhos et al., 2014). We checked the leaf stomatal aperture ratios in the presence of exogenous ABA, and the stomatal aperture ratio index was 0.40 for the WT plants and 0.28–0.17 for the MdbHLH130-ox plants (Figures 5B, C). These results suggested that MdbHLH130 improved the water deficit tolerance at least partly by promoting the plants’ ability to close their stomata and reduce transpiration.
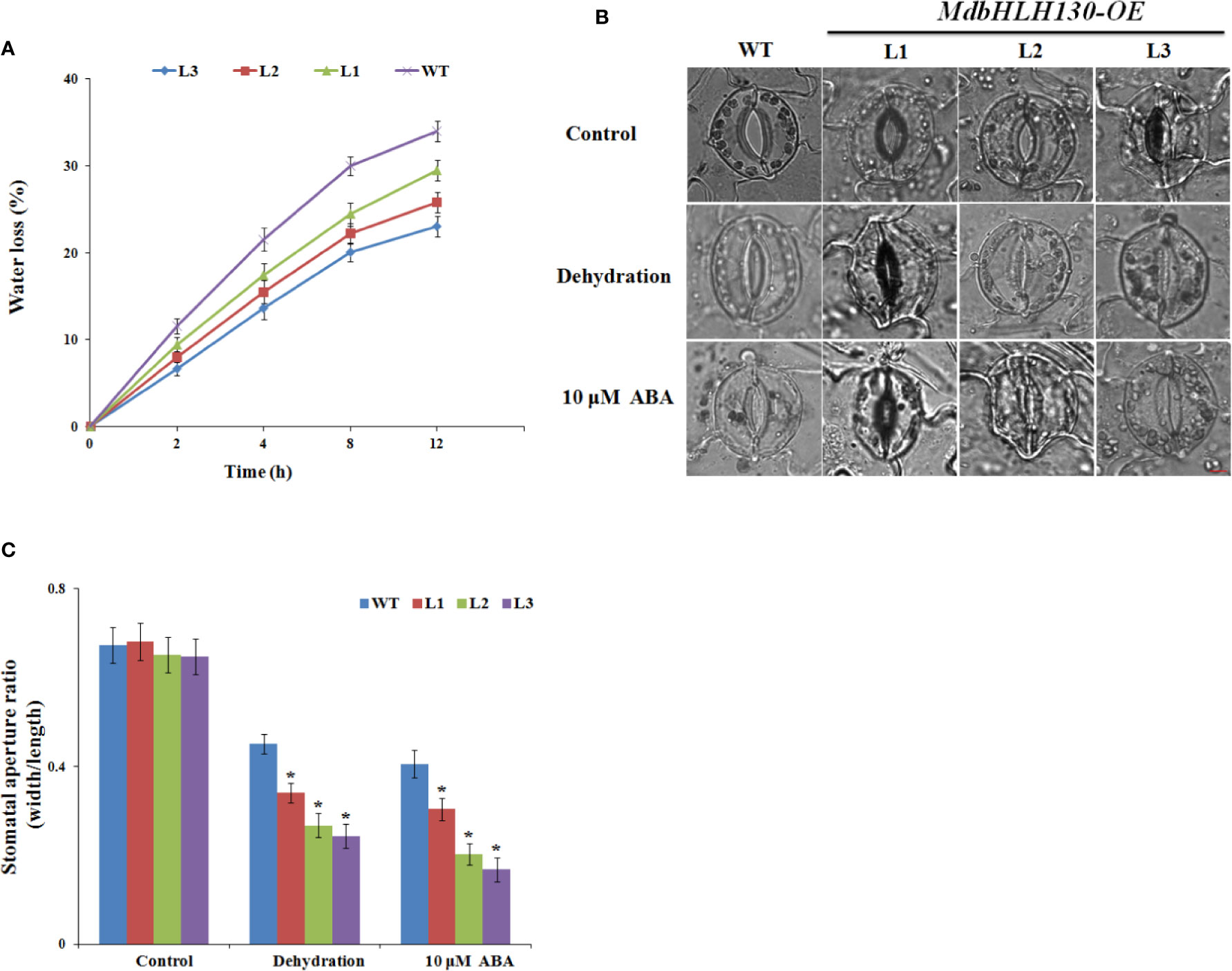
Figure 5 Water loss rate and stomatal aperture ratios from WT and MdbHLH130-ox plants after treatments. (A) Water loss of the detached leaves from the WT and MdbHLH130-ox plants. (B, C) Images of stomata and measurement of stomatal aperture ratios (width/length ratio) of the WT and MdbHLH130-ox plant leaves in response to dehydration or 10 μM ABA. Scale bar, 10 μm. Error bars represent the means ± SD of 100 stomata from three independent biological replicates. Asterisks indicate significant differences between MdbHLH130-ox plants and WT (*P < 0.01).
Overall, these results indicate that MdbHLH130 functions as a positive regulator in plant responses to water deficit stress.
ROS Accumulation and Antioxidant Enzyme Activities in MdbHLH130-Overexpressing Plants Under Water Deficit Stress Conditions
Water deficit leads to the production of ROS, and excess ROS levels cause damage to cell growth following oxidative stress (Choudhury et al., 2017). To test whether MdbHLH130 regulates ROS levels in response to water deficit stress, leaves from the WT plants and MdbHLH130-ox plants were stained with 2′,7′-dichlorofluorescin diacetate (H2DCFDA) after water deficit treatment for 10 days. ROS staining in MdbHLH130-ox plants was lower than that of the WT plants under water deficit treatment (Figure 6A), showing that the WT plants accumulated more ROS. Consistent with these results, the H2O2 content levels for the transgenic plants were lower than those of the WT plants under water deficit stress (Figure 6B).
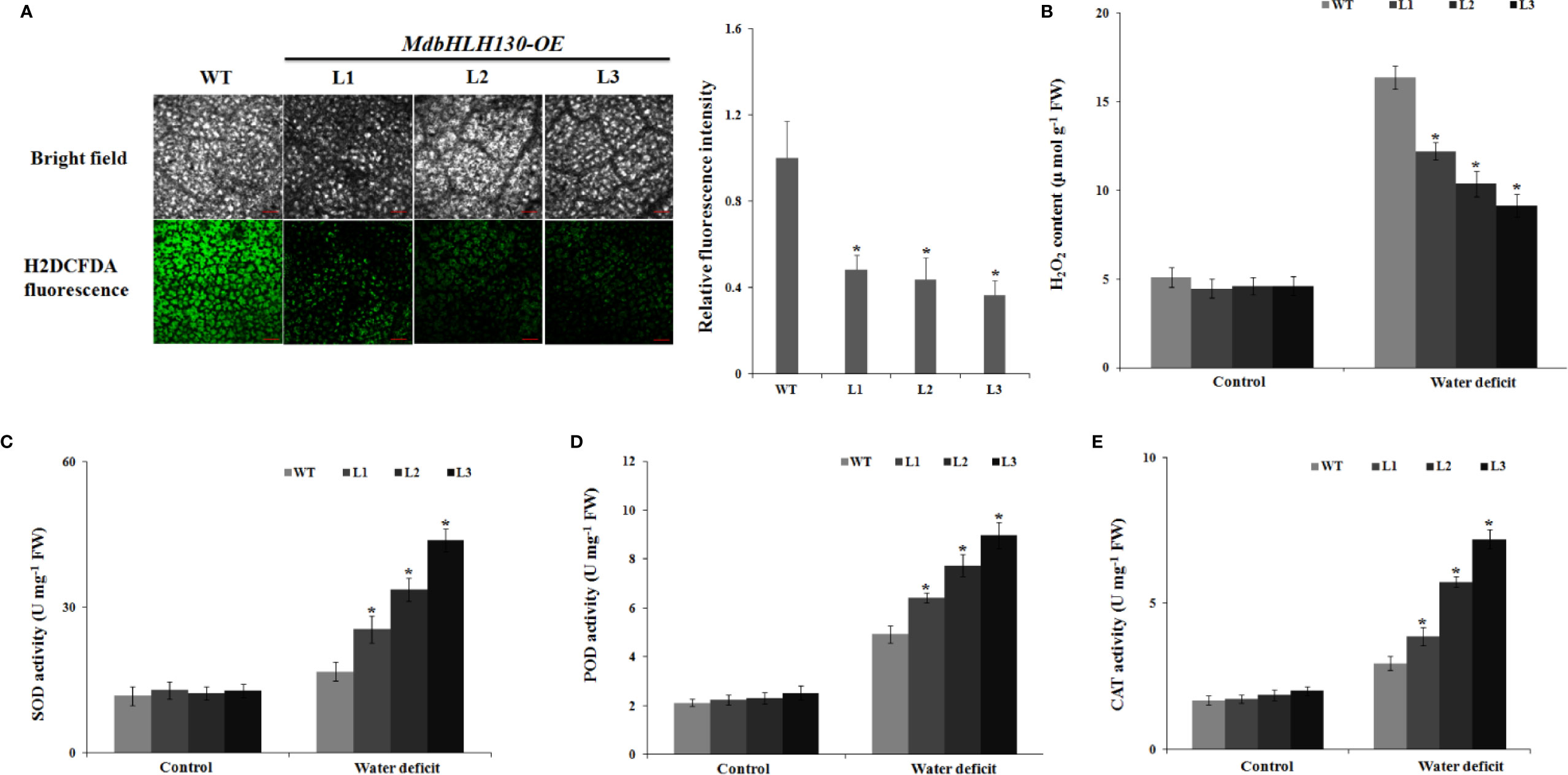
Figure 6 ROS accumulation and activity of antioxidant enzymes in MdbHLH130-ox plants and WT after water deficit treatment. (A) Fluorescence detection and quantification of ROS by H2DCFDA staining in MdbHLH130-ox plants and WT leaves after water deficit treatment. Bar = 250 µm. (B–E) Statistical analysis of the H2O2 content (B), SOD activity (C), POD activity (D), and CAT activity (E) in the WT and three MdbHLH130-ox plants after water deficit treatment. Error bars represent the means ± SD taken from three independent biological replicates. Asterisks indicate significant differences relative to the WT (*P < 0.01).
Superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) are important antioxidant enzymes responsible for scavenging ROS in response to various stresses (Choudhury et al., 2017; Mittler, 2017). Thus, we confirmed the activities of these enzymes. Under normal conditions, the SOD, POD, and CAT activities in the transgenic lines showed no differences compared with that of the WT (Figures 6C–E). However, after the water deficit stress treatment, the transgenic lines presented significantly higher SOD, POD, and CAT activities than the WT (Figures 6C–E), which is consistent with ROS staining. These results suggest that the overexpression of MdbHLH130 reduced ROS accumulation under water deficit stress by enhancing the activities of ROS-scavenging enzymes.
MdbHLH130 Positively Regulates Genes Related to ROS Scavenging and Stress Responses Under Water Deficit Stress Conditions
To obtain additional insights into the molecular mechanisms underlying the MdbHLH130-mediated signaling pathway in water deficit resistance, we examined the transcript abundance of genes related to ROS scavenging (NtSOD, NtPOD, and NtCAT) and stress responses (NtDREB3, NtERD10C, NtERD10D, NtNCED1, NtLEA5, and NtLTP1) by qRT-PCR. Under normal conditions, the mRNA abundance of genes related to ROS scavenging and stress responses did not differ between the WT and MdbHLH130-ox plants (Figure 7). Upon exposure to water deficit conditions, however, the transcript levels of these genes were significantly higher in the MdbHLH130-ox plants compared with the WT plants (Figure 7). These data suggest that MdbHLH130 increases water deficit tolerance in tobacco plants by upregulating the transcript levels of genes related to ROS scavenging and stress responses.
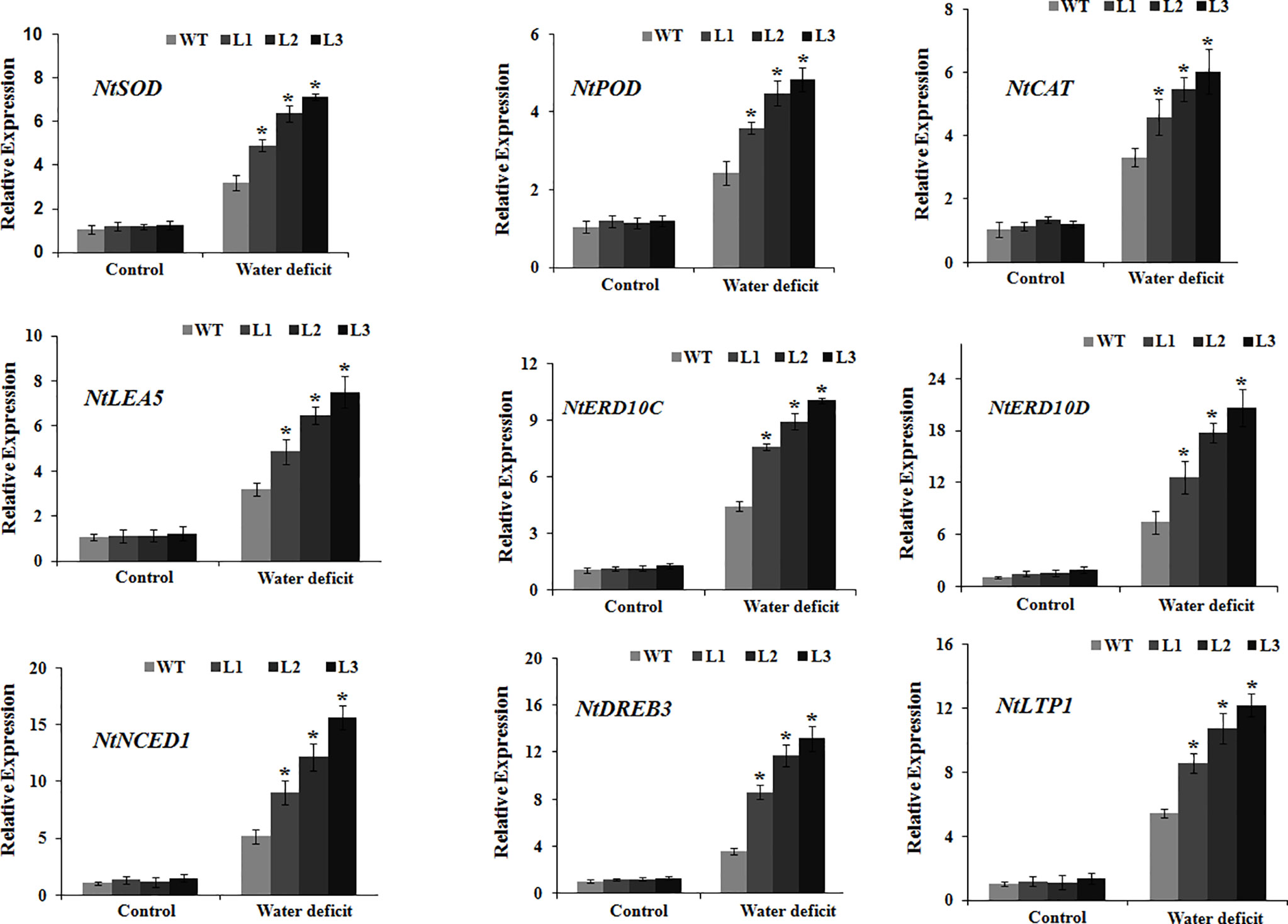
Figure 7 Expression of the genes involved in ROS scavenging and stress responses in the WT and transgenic plants. The expression levels were analyzed by qRT-PCR in tobacco seedlings under the control or water deficit treatment conditions. Error bars represent the means ± SD taken from three independent biological replicates. Asterisks indicate significant differences from the WT (*P < 0.01).
MdbHLH130 Overexpression Enhances Oxidative Stress Tolerance
To assess the role of MdbHLH130 in regulating antioxidant mechanisms, we investigated the relationship between MdbHLH130 and oxidative stress. Methyl viologen (MV) is a herbicide that causes Chl degradation and cell membrane leakage through ROS production (Kurepa et al., 1998). Leaf discs were incubated in distilled water or 100 μM methyl viologen (MV) solution. There were no conspicuous differences between the WT and transgenic lines incubated in distilled water, whereas after 100 μM MV treatment, the WT exhibited more browned and necrotic leaf discs than did the transgenic lines (Figure 8A). The Chl content was higher in the transgenic lines than in the WT (Figure 8B). These data indicate that ectopic expression of MdbHLH130 in tobacco decreased the sensitivity to oxidative stress.
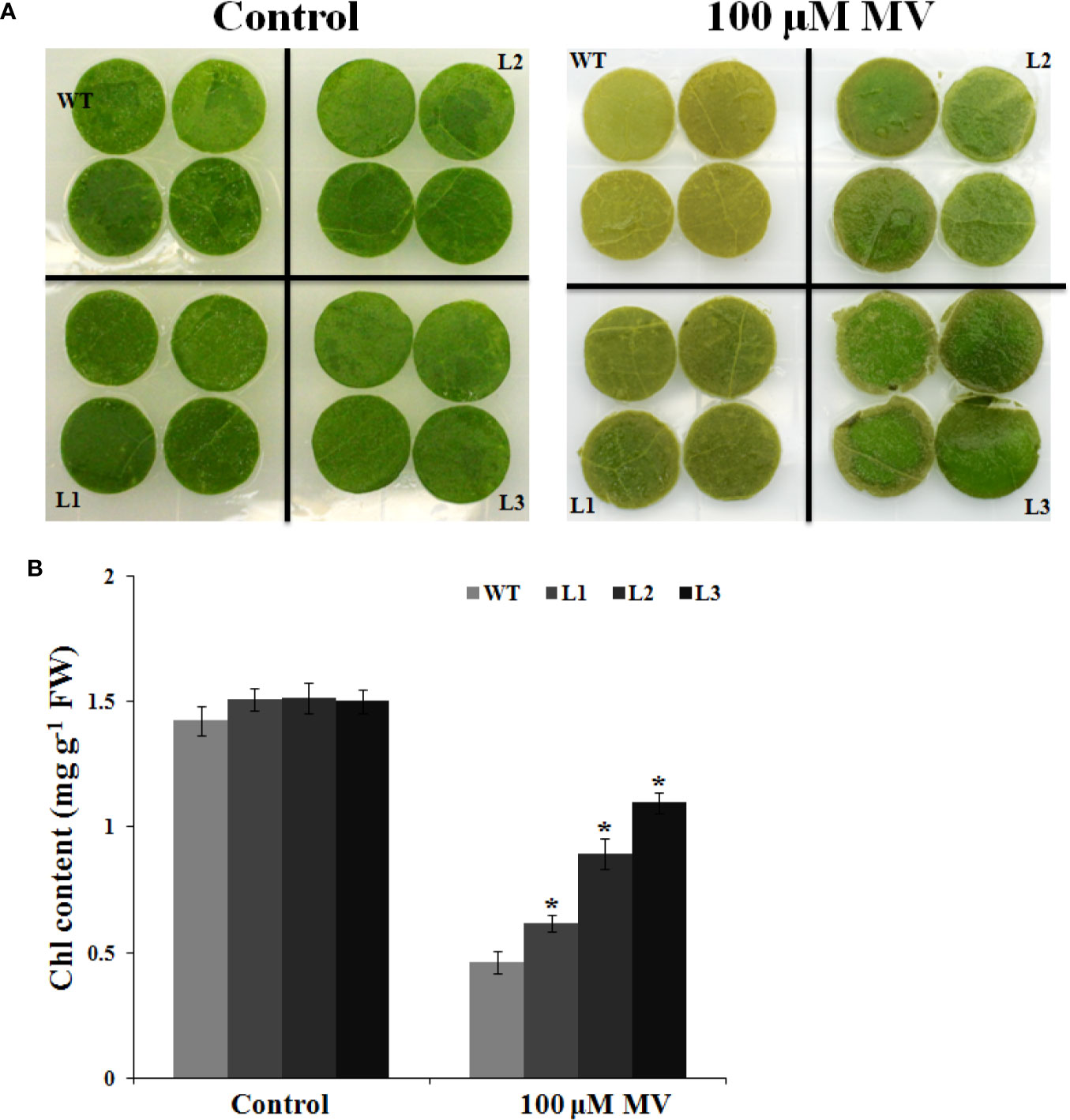
Figure 8 Oxidative stress tolerance to MdbHLH130-ox plants. (A) Phenotype of the leaf discs from the WT and three MdbHLH130-ox plants after incubation with or without 100 μM MV solution for 48 h. (B) Relative total chlorophyll content in the leaf discs after 100 μM MV treatment. Three independent biological replicates were performed. Vertical bars refer to means ± SD of at least three independent biological replicates. Asterisks indicate a significant difference between the WT and MdbHLH130-ox lines (*P < 0.01).
Discussion
In nature, plant growth and development and crop productivity are inevitably affected by many abiotic stresses, such as drought stress (Bray et al., 2000; Shinozaki and Yamaguchi-Shinozaki, 2000; Zhu, 2016). To survive, plants have evolved a wide range of mechanisms to respond to drought stress (Singh and Laxmi, 2015). Among the numerous stress-related genes, transcription factor (TF) genes are modulated in plants under drought stress to help the plants maintain normal growth and development (Zhu, 2016). bHLH proteins are the second largest subfamily of TFs in plants and participate in various biological processes, including drought stress response, by binding to the G-box/E-box cis-elements in the promoter regions of its target genes (Robinson and Lopes, 2000; Toledo-Ortiz et al., 2003; Song et al., 2016). However, only a few apple bHLH genes have been reported to play essential roles in drought stress tolerance. In this study, we isolated a bHLH gene named MdbHLH130 from apple (Figure 1). The overexpression of MdbHLH130 in apple calli and tobacco transgenic plants showed enhanced resistance to water stress (Figures 3, 4), indicating that MdbHLH130 is a positive regulatory factor involved in the water stress tolerance of plants.
Previous studies showed that bHLH genes played important roles in response to abiotic stresses (Seo et al., 2011). For instance, the PtrbHLH gene is upregulated by various stresses, particularly cold stress (Huang et al., 2013); and the OrbHLH2 gene is upregulated by salt and osmotic stress but not by cold stress (Zhou et al., 2009). In our work, the MdbHLH130 transcript level was induced by dehydration treatment (Figures 1C–E), indicating that it possibly plays a role in regulating the response to water stress. bHLH TFs are involved in several processes, including stomatal development, root hair formation, root meristem size, and hormone metabolism in response to drought stress (Abe et al., 2003; Pillitteri and Torii, 2007; Makkena and Lamb, 2013; Castilhos et al., 2014). Stomata represent a key determinant for transpirational water loss, and stomatal closure results in reduced water loss, and this behavior is critical for maintaining a high water potential under drought stress conditions in plants (Nilson and Assmann, 2007). In Arabidopsis, the ABA levels were markedly induced under drought conditions, resulting in stomatal closure (Hu et al., 2013). Drought stress has been reported to trigger the ABA-dependent signaling pathway (Zhu, 2002). Certain bHLH genes have been reported to be induced in response to drought stress dependent on the ABA-mediated pathway. For example, AtMYC2 functions as a transcriptional activator in the ABA-mediated drought stress signaling pathway (Abe et al., 2003). Overexpression of the bHLH122 gene increased the resistance of transgenic plants to drought and osmotic stresses by increasing the cellular ABA levels (Liu et al., 2014). In this study, the stomatal aperture ratio indices of the MdbHLH130 transgenic tobacco lines were lower than those observed of the WT plants after the dehydration treatment and ABA treatments (Figure 5). In accordance with these results, the leaf water loss rate of the transgenic tobacco plants was low (Figure 5). Moreover, the gene encoding NtNCED1, a key enzyme of ABA biosynthesis, is a known participant in ABA-mediated responses (Shinozaki et al., 2003; Hu et al., 2013). The expression of NtNCED1 was upregulated in the MdbHLH130-overexpressing plants compared to the WT plants, implying that ABA biosynthesis may be promoted in transgenic plants (Figure 7). Therefore, these results indicate that MdbHLH130 functions as a positive regulator of water deficit stress responses by partly involving ABA-mediated stomatal aperture ratio and transpiration. In addition, photosynthesis is an essential process for maintaining plant growth and development, and total chlorophyll fluorescence parameters (e.g., Fv/Fm) represent a rapid and accurate approach for detecting and quantifying water stress tolerance in plants (Percival and Sheriffs, 2002; Anjum et al., 2011). Under the water deficit stress condition, the transgenic plants exhibited higher survival rates and better total chlorophyll contents based on phenotypic observations, suggesting that MdbHLH130 conferred tolerance to water deficit stress by regulating different pathways (Figure S5).
EL is an indicator of the severity of a membrane injury, and MDA is a product of oxidative attack on membrane functionality and integrity (Moore and Roberts, 1998). Under water deficit stress conditions, the transgenic tobacco plants exhibited decreased IL and MDA contents (Figures 4E, F). NtERD10C, NtERD10D, and NtLEA5 encode the late embryogenesis abundant (LEA) proteins that protect cellular and macromolecular structures during dehydration tolerance (Hundertmark and Hincha, 2008). The higher expression of NtERD10C, NtERD10D, and NtLEA5 in transgenic tobacco plants resulted in greater water retention and less membrane damage compared to the WT plants under water deficit stress (Figures 4, 7). ROS, which mainly include O2− and H2O2, cause damage to cell growth by oxidizing proteins, lipids, and DNA or acting as signal molecules that mediate tolerance to various stresses (Choudhury et al., 2017; Mittler, 2017). Previous studies have shown that the overexpression of PtrbHLH or AtbHLH112 increases SOD or POD activity and reduces ROS accumulation under stress conditions (Huang et al., 2013). Given the importance of ROS in stress tolerance, we investigated whether ROS-scavenging ability is regulated by MdbHLH130. The staining and measurements results revealed that ROS accumulated at lower levels in the MdbHLH130-ox tobacco plants compared with the WT plants (Figures 6A, B). Furthermore, the activities of three important ROS-scavenging enzymes were higher in the MdbHLH130-expressing tobacco plants (Figures 6C–E). Moreover, the transcripts of the ROS-scavenging genes were positively correlated with the transcript levels of MdbHLH130 (Figure 7). In addition, the MdbHLH130-ox transgenic plants were damaged to a less serious degree under the MV treatment, suggesting that they were more resistant to oxidative stresses than the WT (Figure 8). Taken together, these results indicate that MdbHLH130 might function to mediate the transcriptional upregulation of ROS-scavenging genes to enhance the ROS-scavenging ability and improved water deficit stress tolerance.
Plants have evolved a variety of mechanisms to improve drought tolerance (Miller et al., 2010). Dehydration-responsive element-binding proteins (DREBs) also play important roles in regulating water deficit stress responses in ABA-independent pathways (Umezawa et al., 2006). In this study, the DREB family member NtDREB3 was induced in transgenic tobacco plants under drought stress, which indicated that MdbHLH130 is involved in water stress through ABA-dependent and ABA-independent pathways (Figure 7). NtLTP1 belongs to the lipid transfer protein family, which is universally involved in the response to ABA and drought stress (Hu et al., 2013; Wang et al., 2015). In the present study, MdbHLH130-ox transgenic tobacco plants showed enhanced NtLTP1 expression under drought stress, suggesting that MdbHLH130 affects the expression of some lipid-transfer protein genes that are responsive to water stress (Figure 7).
Water deficit stress often affects plant growth and development (Zhu, 2016). Apple trees are among the crops most affected by water stress, which significantly decreases fruit yield and quality (Reid and Kalcsits, 2020). It is well known that numerous genes are involved in drought tolerance. However, just few of them are characterized and used for genetic improvement in fruit tree. In summary, we isolated and characterized a dehydration stress-responsive bHLH TF (MdbHLH130) from apple, which acted as a positive regulator of water stress tolerance. We concluded that MdbHLH130 exhibits important physiological functions in the water stress response through ABA signaling and antioxidant system regulation, thus preventing plants from oxidative damage. Our findings provide a deeper understanding of the regulatory mechanism of MdbHLH130 in response to water deficit stress and may have candidate application for drought improvement in horticultural crop breeding. Further analyses of the direct downstream target genes of MdbHLH130 can help clarify its mechanisms in the enhanced tolerance to water stress.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding authors.
Author Contributions
QZ and YW conceived and designed the research. QZ, ZF, LQ, QC, TW, and YL provided experimental materials. YW and QZ analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFD1000300), Shandong Province Government (SDAIT-06-05), Breeding Plan of Shandong Provincial Qingchuang Research Team (2019), the Talents of High Level Scientific Research Foundation of Qingdao Agricultural University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.543696/full#supplementary-material
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Anjum, S., Xie, X. Y., Wang, L. C., Saleem, M. F., Man, C., Wang, L. (2011). Morphological, physiological and biochemical responses of plants to drought stress. Afr. J. Agr. Res. 6, 2026–2032. doi: 10.5897/AJAR10.027
Atchley, W. R., Fitch, W. M. (1997). A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. U. S. A. 94, 5172–5176. doi: 10.1073/pnas.94.10.5172
Basu, S., Ramegowda, V., Kumar, A., Pereira, A. (2016). Plant adaptation to drought stress. F1000Research 5 (F1000 Faculty Rev), 1554. doi: 10.12688/f1000research.7678.1
Benny, J., Pisciotta, A., Caruso, T., Martinelli, F. (2019). Identification of key genes and its chromosome regions linked to drought responses in leaves across different crops through meta-analysis of RNA-Seq data. BMC Plant Biol. 19, 194. doi: 10.1186/s12870-019-1794-y
Bray, E. A., Bailey-Serres, J., Weretilnyk, E. (2000). “Responses to abiotic stresses,” in Biochemistry and Molecular Biology of Plants. Eds. Buchanan, B. B., Gruissem, W., Jones, R. L. (Rockville, MD: American Society of Plant Physiologists), 1158–1203.
Burnette, W. N. (1981). “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate–polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112, 195–203. doi: 10.1016/0003-2697(81)90281-5
Buscaill, P., Rivas, S. (2014). Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 20, 35–46. doi: 10.1016/j.pbi.2014.04.004
Castilhos, G., Lazzarotto, F., Spagnolo-Fonini, L., Bodanese-Zanettini, M. H., Margis-Pinheiro, M. (2014). Possible roles of basic helix-loop-helix transcription factors in adaptation to drought. Plant Sci. 223, 1–7. doi: 10.1016/j.plantsci.2014.02.010
Choudhury, F. K., Rivero, R. M., Blumwald, E., Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. doi: 10.1111/tpj.13299
Dong, Y., Wang, C., Han, X., Tang, S., Liu, S., Xia, X., et al. (2014). A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis. Biochem. Bioph. Res. Co. 450, 453–458. doi: 10.1016/j.bbrc.2014.05.139
Feller, A., Machemer, K., Braun, E. L., Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Feng, X. M., Zhao, Q., Zhao, L. L., Qiao, Y., Xie, X. B., Li, H. F., et al. (2012). The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Bio. 12, 22. doi: 10.1186/1471-2229-12-22
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Horsch, R. B., Fry, J. E., Hoffman, N. L., Eichholtz, D., Rogers, S. A., Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1232. doi: 10.1126/science.227.4691.1229
Hu, W., Huang, C., Deng, X., Zhou, S., Chen, L., Li, Y., et al. (2013). TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ. 36, 1449–1464. doi: 10.1111/pce.12074
Huang, X. S., Wang, W., Zhang, Q., Liu, J. H. (2013). A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol. 162, 1178–1194. doi: 10.1104/pp.112.210740
Hundertmark, M., Hincha, D. K. (2008). LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9, 118.
Ishida, T., Osakabe, Y., Yanagisawa, S. (2012). “Transcription factors: improving abiotic stress tolerance in plants,” in Improving Stress Resistance to Abiotic Stress. Ed. Tuteja, N. (Berlin, Germany: Wiley-Blackwell), 589–619.
Jefferson, R. A., Kavanagh, T. A., Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901. doi: 10.1002/j.1460-2075.1987.tb02730.x
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7, 1029. doi: 10.3389/fpls.2016.01029
Kudo, M., Kidokoro, S., Yoshida, T., Mizoi, J., Todaka, D., Fernie, A. R., et al. (2017). Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants. Plant Biotechnol. J. 15, 458–471. doi: 10.1111/pbi.12644
Kumar, S. V., Lucyshyn, D., Jaeger, K. E., Alós, E., Alvey, E., Harberd, N. P., et al. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242–245. doi: 10.1038/nature10928
Kurepa, J., Smalle, J., Montagu, M. V., Inzé, D. (1998). Polyamines and paraquat toxicity in Arabidopsis thaliana. Plant Cell Physiol. 39 (9), 987–992. doi: 10.1093/oxfordjournals.pcp.a029463
Leivar, P., Monte, E., Oka, Y., Liu, T., Carle, C., Castillon, A., et al (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823.
Liao, X., Guo, X., Wang, Q., Wang, Y., Zhao, D., Yao, L., et al. (2017). Overexpression of MsDREB 6.2 results in cytokinin-deficient developmental phenotypes and enhances drought tolerance in transgenic apple plants. Plant J. 89, 510–526. doi: 10.1111/tpj.13401
Ling, H. Q., Bauer, P., Bereczky, Z., Keller, B., Ganal, M. (2002). The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc. Natl. Acad. Sci. U. S. A. 99, 13938–13943. doi: 10.1073/pnas.212448699
Liu, W., Tai, H., Li, S., Gao, W., Zhao, M., Xie, C., et al. (2014). bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 201, 1192–1204. doi: 10.1111/nph.12607
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C., Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. doi: 10.1111/j.1365-313X.2007.03341.x
Lurie, S., Ben-Arie, R. (1983). Microsomal membrane changes during the ripening of apple fruit. Plant Physiol. 73, 636–638. doi: 10.1104/pp.73.3.636
Ma, Q. J., Sun, M. H., Lu, J., Kang, H., You, C. X., Hao, Y. J. (2019). An apple sucrose transporter MdSUT2. 2 is a phosphorylation target for protein kinase MdCIPK22 in response to drought. Plant Biotechnol. J. 17, 625–637. doi: 10.1111/pbi.13003
Makkena, S., Lamb, R. S. (2013). The bHLH transcription factor SPATULA regulates root growth by controlling the size of the root meristem. BMC Plant Biol. 13, 1. doi: 10.1186/1471-2229-13-1
Mao, K., Dong, Q., Li, C., Liu, C., Ma, F. (2017). Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant Sci. 8, 480. doi: 10.3389/fpls.2017.00480
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Moore, K., Roberts, L. J. (1998). Measurement of lipid peroxidation. Free Radical Res. 28, 659–671. doi: 10.3109/10715769809065821
Nakashima, K., Ito, Y., Yamaguchi-Shinozaki, K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Nieto, M., Schuurmans, C., Britz, O., Guillemot, F. (2001). Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron 29, 401–413. doi: 10.1016/S0896-6273(01)00214-8
Nilson, S. E., Assmann, S. M. (2007). The control of transpiration. Insights from Arabidopsis. Plant Physiol. 14, 19–27. doi: 10.1104/pp.106.093161
Nuruzzaman, M., Sharoni, A. M., Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 248. doi: 10.3389/fmicb.2013.00248
Osakabe, Y., Osakabe, K., Shinozaki, K., Tran, L. S. P. (2014). Response of plants to water stress. Front. Plant Sci. 5, 86. doi: 10.3389/fpls.2014.00086
Penfield, S., Josse, E. M., Kannangara, R., Gilday, A. D., Halliday, K. J., Graham, I. A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15, 1998–2006. doi: 10.1016/j.cub.2005.11.010
Percival, G. C., Sheriffs, C. N. (2002). Identification of drought-tolerant woody periennials using chlorophyll fluorescence. J. Arboricul. 28, 215–223.
Pillitteri, L. J., Torii, K. U. (2007). Breaking the silence: three bHLH proteins direct cell-fate decisions during stomatal development. Bioessays 29, 861–870. doi: 10.1002/bies.20625
Pires, N., Dolan, L. (2010). Early evolution of bHLH proteins in plants. Plant Signal. Behave 5, 911–912. doi: 10.4161/psb.5.7.12100
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., et al. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814. doi: 10.1105/tpc.111.083261
Quan, R., Shang, M., Zhang, H., Zhao, Y., Zhang, J. (2004). Engineering of enhanced glycine betaine synthesis improves drought tolerance in maize. Plant Biotechnol. J. 2, 477–486. doi: 10.1111/j.1467-7652.2004.00093.x
Ramsay, N. A., Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Rasheed, S., Bashir, K., Matsui, A., Tanaka, M., Seki, M. (2016). Transcriptomic analysis of soil-grown Arabidopsis thaliana roots and shoots in response to a drought stress. Front. Plant Sci. 7, 180. doi: 10.3389/fpls.2016.00180
Reid, M., Kalcsits, L. (2020). Water Deficit Timing Affects Physiological Drought Response, Fruit Size, and Bitter Pit Development for ‘Honeycrisp’ Apple. Plants (Basel) 9, 874.
Ren, Y. R., Yang, Y. Y., Zhang, R., You, C. X., Zhao, Q., Hao, Y. J. (2019). MdGRF11, an apple 14-3-3 protein, acts as a positive regulator of drought and salt tolerance. Plant Sci. 288, 110219. doi: 10.1016/j.plantsci.2019.110219
Robinson, K. A., Lopes, J. M. (2000). Survey and summary: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res. 28, 1499–1505. doi: 10.1093/nar/28.7.1499
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65, 907–921. doi: 10.1111/j.1365-313X.2010.04477.x
Shinozaki, K., Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. doi: 10.1016/S1369-5266(00)00067-4
Shinozaki, K., Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Shinozaki, K., Yamaguchi-Shinozaki, K., Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Singh, D., Laxmi, A. (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6, 895. doi: 10.3389/fpls.2015.00895
Song, L., Huang, S. S. C., Wise, A., Castanon, R., Nery, J. R., Chen, H., et al. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354, 1550. doi: 10.1126/science.aag1550
Stevens, J. D., Roalson, E. H., Skinner, M. K. (2008). Phylogenetic and expression analysis of the basic helix-loop-helix transcription factor gene family: genomic approach to cellular differentiation. Differentiation 76, 1006–1042. doi: 10.1111/j.1432-0436.2008.00285.x
Sun, X., Wang, Y., Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Bioph. Res. Co. 503, 397–401. doi: 10.1016/j.bbrc.2018.07.123
Toledo-Ortiz, G., Huq, E., Quail, P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
Tomita, K., Moriyoshi, K., Nakanishi, S., Guillemot, F., Kageyama, R. (2000). Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 19, 5460–5472. doi: 10.1093/emboj/19.20.5460
Tran, L. S. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Umezawa, T., Fujita, M., Fujita, Y., Yamaguchi-Shinozaki, K., Shinozaki, K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes unlock the future. Curr. Opin. Biotech. 17, 113–122. doi: 10.1016/j.copbio.2006.02.002
Wang, X., Zeng, J., Li, Y., Rong, X., Sun, J., Sun, T., et al. (2015). Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 6, 615. doi: 10.3389/fpls.2015.00615
Zhang, J., Liu, B., Li, M., Feng, D., Jin, H., Wang, P., et al. (2015). The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27, 787–805. doi: 10.1105/tpc.114.132704
Zhao, Q., Ren, Y. R., Wang, Q. J., Yao, Y. X., You, C. X., Hao, Y. J. (2016). Overexpression of MdbHLH 104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnol. J. 14, 1633–1645. doi: 10.1111/pbi.12526
Zhao, Q., Xiang, X., Liu, D., Yang, A., Wang, Y. (2018). Tobacco transcription factor NtbHLH123 confers tolerance to cold stress by regulating the NtCBF pathway and reactive oxygen species homeostasis. Front. Plant Sci. 9, 381. doi: 10.3389/fpls.2018.00381
Zhou, J., Li, F., Wang, J. L., Ma, Y., Chong, K., Xu, Y. Y. (2009). Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt-and osmotic stress in Arabidopsis. J. Plant Physiol. 166, 1296–1306. doi: 10.1016/j.jplph.2009.02.007
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Keywords: apple, bHLH transcription factor, MdbHLH130, water stress, stomatal closure, ROS-scavenging
Citation: Zhao Q, Fan Z, Qiu L, Che Q, Wang T, Li Y and Wang Y (2020) MdbHLH130, an Apple bHLH Transcription Factor, Confers Water Stress Resistance by Regulating Stomatal Closure and ROS Homeostasis in Transgenic Tobacco. Front. Plant Sci. 11:543696. doi: 10.3389/fpls.2020.543696
Received: 18 March 2020; Accepted: 31 August 2020;
Published: 09 October 2020.
Edited by:
Woo Taek Kim, Yonsei University, South KoreaReviewed by:
Xin Deng, Chinese Academy of Sciences, ChinaChan Yul Yoo, Oklahoma State University, United States
Copyright © 2020 Zhao, Fan, Qiu, Che, Wang, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Zhao, emhhb3FpYW5nMDAwNjY2QDE2My5jb20=; Yongzhang Wang, cWF1d3l6QDE2My5jb20=
 Qiang Zhao
Qiang Zhao Zihao Fan1
Zihao Fan1 Yuanyuan Li
Yuanyuan Li