- 1Interdepartmental Program in Plant Biology, Iowa State University, Ames, IA, United States
- 2Department of Horticulture, Iowa State University, Ames, IA, United States
- 3Innovation Center of Rice Cultivation Technology in Yangtze River Valley, Ministry of Agriculture/Key Laboratory of Crop Genetics and Physiology of Jiangsu Province, Yangzhou University, Yangzhou, China
- 4Christopher S. Bond Life Sciences Center, Division of Plant Sciences, University of Missouri, Columbia, MO, United States
- 5Donald Danforth Plant Science Center, St. Louis, MO, United States
Tillering is an important biomass yield component trait in switchgrass (Panicum virgatum L.). Teosinte branched 1 (tb1)/Branched 1 (BRC1) gene is a known regulator for tillering/branching in several plant species; however, its role on tillering in switchgrass remains unknown. Here, we report physiological and molecular characterization of mutants created by CRISPR/Cas9. We successfully obtained nonchimeric Pvtb1a and Pvtb1b mutants from chimeric T0 mutants using nodal culture. The biallelic Pvtb1a-Pvtb1b mutant plants produced significantly more tillers and higher fresh weight biomass than the wild-type plants. The increased tiller number in the mutant plants resulted primarily from hastened outgrowth of lower axillary buds. Increased tillers were also observed in transgene-free BC1 monoallelic mutants for either Pvtb1a-Pvtb1b or Pvtb1b gene alone, suggesting Pvtb1 genes act in a dosage-dependent manner. Transcriptome analysis showed 831 genes were differentially expressed in the Pvtb1a-Pvtb1b double knockdown mutant. Gene Ontology analysis revealed downregulation of Pvtb1 genes affected multiple biological processes, including transcription, flower development, cell differentiation, and stress/defense responses in edited plants. This study demonstrates that Pvtb1 genes play a pivotal role in tiller production as a negative regulator in switchgrass and provides opportunities for further research aiming to elucidate the molecular pathway regulating tillering in switchgrass.
Introduction
Switchgrass (Panicum virgatum), a C4 perennial grass with demonstrated high biomass yield is native to North America and is well adapted to marginal land not suitable for food crops (Mitchell et al., 2008; Narasimhamoorthy et al., 2008). The low production cost and high lignocellulose-based biofuel potential make it well-suited for bioenergy crop (McLaughlin and Kszos, 2005). It was named the model species for herbaceous bioenergy crop by the U.S. Department of Energy in 1991 following more than a decade of research (Wright and Turhollow, 2010). Switchgrass is an out-crossing species with varied ploidy levels and is self-incompatible with individual plants being highly heterozygous (Narasimhamoorthy et al., 2008). Upland and lowland ecotypes are the two major representative taxa of switchgrass (Zhang et al., 2011). Most of the low-tillering lowland ecotypes are tetraploid (2n=4x=36), while the high-tillering upland ecotypes contain both tetraploids and octoploids (2n=8x=72) with hexaploids (2n=6x=54) being reported rarely (Zhang et al., 2011). Unlike the lowland ecotype, which has a caespitose growth habit and poor freezing tolerance, upland ecotypes are highly rhizomatous and cold hardy (Mitchell and Schmer, 2012).
High biomass yield is a high priority for switchgrass breeding (Casler, 2010; Okada et al., 2010; Lu et al., 2013). Tiller number is positively correlated with biomass yield (Das et al., 2003; Boe, 2007; Boe and Beck, 2008). Therefore, understanding the molecular mechanism regulating tillering in switchgrass can facilitate development of high-yielding cultivars. Tillers result from the outgrowth of axillary buds at nodes in grass species and the number of tillers produced from a single plant varies greatly by species and genotypes. Tillering or branching is regulated by numerous endogenous and environmental factors in both eudicots and monocots (Kebrom et al., 2010; Whipple et al., 2011; Reddy and Finlayson, 2014; Gonzalez-Grandio et al., 2017; Holalu and Finlayson, 2017). For instance, auxin is a well-known contributor to apical dominance, which inhibits axillary bud outgrowth, while cytokinins (CKs) promote bud outgrowth (Thimann and Skoog, 1933; Morris, 1977; Minakuchi et al., 2010; Braun et al., 2012). Strigolactones (SLs), another key plant hormone, represses branching or tillering in both monocots and eudicots (Sorefan et al., 2003; Zou et al., 2006; Arite et al., 2007). Additionally, tiller production is often inhibited under severe shade (Kebrom et al., 2006; Finlayson et al., 2010; Kebrom et al., 2010), insufficient mineral nutrients or photosynthates (Wang et al., 2019).
The teosinte branched 1 (tb1) gene is an important tillering/branching-related transcription factor gene that regulates tillering by integrating environmental and developmental cues (Doebley et al., 1997; Whipple et al., 2011; Seale et al., 2017). In cultivated maize (Zea mays L.), an insertion in the regulatory region of the tb1 gene elevates its expression at leaf axils where axillary buds are located and greatly suppresses tillering, while its ancestor teosinte lacking the insertional sequence is highly branched (Studer et al., 2011). tb1 belongs to the TCP (TEOSINTE BRANCHED1, CYCLOIDEA, PCF) gene family that all encode proteins with a 59-amino acid, noncanonical basic helix-loop-helix (bHLH) motif that allows DNA binding and protein-protein interactions (Martin-Trillo and Cubas, 2010). Closely related tb1 genes with similar functions have been identified in both monocots and dicots (Kebrom et al., 2006; Aguilar-Martinez et al., 2007; Martin-Trillo et al., 2011; Choi et al., 2012; Nicolas et al., 2015). For example, the OsTB1 gene functions as a negative regulator for lateral branching in rice (Oryza sativa L.) (Takeda et al., 2003; Choi et al., 2012). The ortholog of tb1 in bread wheat (Triticum aestivum L.) regulates inflorescence architecture and outgrowth of axillary buds (Dixon et al., 2018). In Arabidopsis, two orthologs of tb1, BRANCHED 1 (BRC1) and BRANCHED 2 (BRC2), regulate branching with BRC1 having the major effect (Aguilar-Martinez et al., 2007). Many studies demonstrate tb1 genes integrate multiple signaling pathways to regulate bud outgrowth and tillering (Kebrom et al., 2013; Rameau et al., 2015). Low ratio of red light to far-red light (R/FR), as a result of shade, inhibits the outgrowth of axillary buds in sorghum (Sorghum bicolor (L.) Moench) by upregulating the expression of SbTB1, an ortholog of tb1 (Kebrom et al., 2006; Kebrom et al., 2010). In Arabidopsis (Arabidopsis thaliana (L.) heynh.), BRC1 suppresses lateral bud growth in response to shade by promoting abscisic acid accumulation inside axillary buds (Gonzalez-Grandio et al., 2017). Cytokinins activate axillary bud outgrowth by inhibiting localized PsBRC1 expression in pea (Pisum sativum L.) (Braun et al., 2012) or OsTB1 in rice (Minakuchi et al., 2010). These studies all demonstrate that BRC1/TB1 is a common target for hormonal and environmental signals that regulate tillering/branching across species. Hence, understanding the function of switchgrass tb1 genes can provide valuable information to the understanding of the tillering mechanisms in switchgrass.
The lowland switchgrass cv. Alamo from which the reference genome was produced is an allotetraploid (2n = 4x = 36, NNKK). It is therefore hypothesized that the majority of genes are present as homoeologs. There are two Pvtb1 genes (Pvtb1a and Pvtb1b) in ‘Alamo’ with 89% DNA sequence identity between them (Panicum virgatum v4.1, DOE-JGI, http://phytozome.jgi.doe.gov/). Because of its self-incompatibility, it is difficult to obtain homozygous mutants in switchgrass by inbreeding hemizygous mutants as is done for self-pollinating transgenic plants. The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 nuclease (Cas9) based genome editing tools have become a powerful genetic tool for gene function analysis or crop improvement (Zhao et al., 2016; Wang et al., 2018; Yang et al., 2018). A multiplex CRISPR/Cas9 platform can be used to simultaneously edit multiple genes, which is particularly advantageous for a polyploid species such as switchgrass in which homeologs of a same gene exist, or for members of a gene family that are present in tandem (Zhao et al., 2016).
We previously created switchgrass tb1 mutant plants by using CRISPR/Cas9 (Liu et al., 2018). However, the allelic composition of Pvtb1 genes in these T0 mutant plants was not fully characterized. In addition, it remained possible that these primary mutants are chimeric, which prevented us from accurately assessing the function of tb1 genes. In plant species that can be vegetatively propagated, nonchimeric, solid mutants can be successfully isolated from chimeric mutants (Maluszynski et al., 1995). For example, separating mutated sectors from chimeric mutants by in vitro multiplication has been achieved in banana, cassava and other vegetatively propagated crops (Harten et al., 1981; Novák et al., 1990; Maluszynski et al., 1995; Das et al., 2000). Switchgrass can be readily propagated using node culture (Alexandrova et al., 1996). Hence, generation of primary mutants with CRISPR/Cas9, followed by nodal culture would allow us to generate nonchimeric mutants without the need of producing progeny.
The relative ease with which transgene-free mutant plants can be obtained is a unique advantage for CRISPR/Cas9-based genome editing. Transgene-free mutants have been obtained in multiple crops using CRISPR/Cas9-based genome editing tools and stable inheritance of CRISPR/Cas9 induced mutations has been demonstrated in other plant species, (Pyott et al., 2016; Zhang et al., 2016; He et al., 2018) but not yet in switchgrass. Germplasm generated through CRISPR/Cas9-based genome editing method without foreign DNAs may bypass or simplify the regulatory process required for crops created with traditional transgenic approaches (Waltz, 2018).
Here, we report the successful use of micropropagation to generate nonchimeric mutants from chimeric primary mutants induced by CRISPR/Cas9, eliminating the need of obtaining progeny mutants for gene function analysis. Moreover, the transmission of CRISPR/Cas9-induced mutations in switchgrass is demonstrated in this study. Transgene-free progeny mutants, preserving the phenotypic effect of Pvtb1 mutations similar to that exhibited in T0 mutants, were generated in this study. We showed that double biallelic mutant for Pvtb1a and 1b genes enhanced tiller production and increased biomass yield, indicating that Pvtb1 genes negatively regulate tillering in switchgrass. Transcriptome analysis of a Pvtb1 knockdown mutant 52-1 and wild-type (WT) plant WT-1 suggest that Pvtb1 genes are involved in multiple pathways to regulate tillering in switchgrass.
Materials and Methods
Micropropagation and Seed Propagation of CRISPR/Cas9-Induced Pvtb1 Mutants
Primary mutant plants were generated previously (Liu et al., 2018). Briefly, the CRISPR/Cas9 construct containing two gRNAs targeting the conserved regions of Pvtb1a and Pvtb1b genes was delivered into the genome of switchgrass through Agrobacterium-mediated transformation of caryopsis-derived embryogenic callus from which primary mutant plants were obtained. To produce nonchimeric mutants from chimeric primary mutants, we micropropagated Pvtb1 mutants by culturing nodes containing axillary buds according to (Harten et al., 1981; Alexandrova et al., 1996). Each of the two basal nodes from a stem containing an axillary bud with a portion of (~1.5-cm long) internode above and below the node was selected for in vitro culture on the MS-0 medium with 3 gL-1 Phytagel (Sigma Chemical Co., St. Louis) and no plant growth regulator. These nodal segments were surface sterilized in commercial bleach (5% aqueous solution of sodium hypochlorite) for 30 min, and then rinsed three times with sterile distilled deionized water. Three experiments were conducted for the 52-1 and 35-2 mutants and their corresponding WT (WT-1) plants with 20 nodes for each experiment. After 8 weeks of culture in a growth chamber with a light intensity of 140 µmol m-2 s-1 at a photoperiod of 16/8 h light/dark and a temperature of 25°C, regeneration efficiency (regenerated plantlets/total nodes number) were calculated. After roots emerged, plantlets were grown in 6-inch diameter (1 L) pots using commercial soil mix (Sunshine soil mix #1, Sun Gro) of peat moss and perlite and moved into a mist room for 7–10 days before they were placed in a greenhouse at 26°C with a 16/8 h (day/night) photoperiod and a light intensity of approximately 400 µmol m-2 s-1.
To determine the inheritance of CRISPR/Cas9 induced mutations in switchgrass, BC1 progeny were obtained by crossing mutants with genetically compatible WT plants. Seeds were harvested from the mutant parents and were sowed in a 1,020 tray (54.5 cm L × 27.8 cm W × 6.2 cm H) with the same soil as described previously that was maintained at a mist room until germination. Seedlings were transferred to the same greenhouse as described earlier. The presence of the transgene was analyzed using PCR with gRNA/Cas9-specific primers (Table S6).
Genotyping of CRISPR/Cas9-Induced Pvtb1 Mutants by Next Generation Sequencing
NGS was used to genotype the Pvtb1 mutant plants generated from micropropagation of primary mutants and BC1 Pvtb1 mutant progeny. To amplify the target fragments of each Pvtb1 gene, gene-specific primers were designed for Pvtb1a and Pvtb1b respectively. The amplicon size for Pvtb1a is 250 bp, while that of Pvtb1b is 288 bp, both are sufficiently long to cover the two target sequences. The Illumina overhang adapter sequences were added to the gene-specific primers (Table S6). A second round of PCR was used to add the dual indices with the Nextra XT Index Kit (Illumina, San Diego, CA, USA). The quality of these sequencing libraries was determined by the Qubit® 2.0 Fluorometer. The 150-cycle HiSeq sequencing was done for all the amplicon libraries at the DNA Facility at Iowa State University (ISU). For each library, at least 5,000 reads were generated to determine the sequence of the respective amplicons. The results were analyzed by CRISPR-DAV pipeline (Wang et al., 2017).
Sequence Alignment and Phylogenetic Analysis
The sequence of maize tb1 gene was used to blast the sequences of Pvtb1 genes from the P. virgatum genome project v4.1 (Panicum virgatum v1.0, DOE-JGI, http://phytozome.jgi.doe.gov/). The Pvtb1 genes were subsequently isolated and sequenced with the Pvtb1 gene-specific primers (Table S6). Full-length putative amino acid sequences of tb1 gene orthologs were obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) (Table S7). Alignments of the putative protein sequences of TB1 were performed using Clustal Omega (Sievers et al., 2011) (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Phenotypic Characterization of Pvtb1 Mutant Plants With Hydroponic Culture
To investigate the effects of Pvtb1 genes on phenotype of switchgrass, we established a hydroponic system to observe plant growth. Nodal segments were cut from the wild-type plant (WT, genotype AABB) and mutant 52-1-3 (genotype aabb), 52-1-1 (genotype Aabb) plants, and cultured in vitro as described previously. Plants of similar height that were grown in 6-inch pots for 10 days were transferred into a hydroponic device in a greenhouse where temperature was maintained at 26°C with a 16h/8h (day/night) photoperiod with a light intensity of approximately 400 μmol m-2 s-1. For hydroponic culture, the stem base of each plant was wrapped around by sponge and placed into a small basket which was then inserted into a hole on the lid of a plastic container (51 cm L × 43 cm W × 15 cm H) that was filled with a nutrient solution (16 - 4 - 17 Oasis Hydroponic, JR Peters Inc., PA, USA). Electrical conductivity (EC) and pH of the nutrient solution were measured by a portable pH/EC/Temperature meter (HI9813-6, Hanna Instruments, Inc., RI, USA) daily and maintained at 1.35 mS cm-1 and 6.0, respectively. The pH was lowered or raised by phosphoric acid or sodium hydroxide, respectively, while the EC was maintained by adding concentrated fertilizer stock solution or clear water. The nutrient solution was aerated by an aquarium air pump (ActiveAQUA, Hydrofarm, CA, USA) to ensure adequate oxygen supply.
Statistical Analysis of Morphological Traits
Primary, secondary, tertiary or quaternary tillers were visually determined according to Davidson and Chevalier (1987). Primary tillers were defined as those which arise from leaf axils in nodes of the main stem whereas secondary tillers arise from leaf axils in nodes of the primary tillers. By the same rule, tertiary and quaternary tillers were defined as those arise from leaf axils of nodes of secondary and tertiary tillers, respectively. Adventitious roots of newly formed tillers tend to have creamy white color whereas roots of older tillers appear in darker color. This color variation was used to assist classification of tillers. Plant height was determined by measuring the tallest tiller for each plant. Root length was determined by measuring the length of the longest root for each plant. Stem diameter was determined by the average diameter of the middle internode of five largest tillers for each plant. The number of tillers and roots (longer than 2 cm) were also counted. To determine biomass yield, pot-grown plants were manually cut at the ground level using a pruning shears and weighted for fresh weight. Samples were then oven-dried at 70°C for 4 days when they reached a constant weight to obtain the dry weight. Three to six biological replicates were used in each measurement. Student’s t-test was used to determine the significance of difference between the mutant plants and the WT plants.
Transcriptome Sequencing and Analysis of the mRNA-Seq Data
It was shown in the expression database that Pvtb1 genes in switchgrass expressed highly in axillary buds (Panicum virgatum v1.0, DOE-JGI, http://phytozome.jgi.doe.gov/). Thus, we extracted RNAs from axillary buds of tillers. The mutant 52-1 with highly enhanced tiller production and the WT plant (WT-1) generated from the same callus line from which the mutant 52-1 was obtained were chosen for mRNA-seq. Three biological replications were sampled for each plant. All visible axillary buds collected from two to three tillers are treated as one biological replicate. Total RNA was isolated from tissue samples using the Qiagen RNeasy Plant Isolation kit according to the manufacturer’s protocol (Qiagen Inc., Valencia, CA, USA) and the RNA quality was checked using BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). cDNA libraries were constructed by the ISU DNA Facility according to the instructions in the Illumina sequencing manual. The Illumina HiSeq 3000 150 paired-end platform was used for sequencing by the DNA Facility at Iowa State University (http://www.dna.iastate.edu/).
Library adapter and low quality nucleotides were trimmed off using Trimmomatic (Bolger et al., 2014). Then, STAR was applied to align the trimmed reads to the reference genome V4.1 of switchgrass (Dobin et al., 2013). The number of reads mapped to each gene was calculated using HTseq-count (Anders et al., 2015). Normalization was done using the Upper Quartile method and gene expression differences were analyzed with package edgeR (Robinson et al., 2010) in the statistical software ‘R’ (Version 3.4.3) (The R FAQ, https://CRAN.R-project.org/doc/FAQ/). Genes are classified as differentially expressed when an absolute log2-fold change value is ≥1 and a false discovery rate is ≤ 0.05 (Benjamini and Hochberg, 1995). Gene Ontology (GO) analysis for DEGs were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al., 2009a; Huang et al., 2009b). ReviGO (Supek et al., 2011) and Cytoscape (Shannon et al., 2003) were applied to visualize enriched GOs. Six genes were selected to validate the expression patterns determined by RNA-seq. Primers were listed in Table S6.
Results
Micropropagation Can Effectively Generate Nonchimeric Mutants From Chimeric Primary (T0) CRISPR/Cas9-Induced Mutants
We micropropagated the primary (T0) Pvtb1 mutants (52-1 and 35-2) using in vitro node culture (Figure S1). After about 8 weeks of culture, 48 and 20 plants were successfully regenerated from the primary mutants 35-2 and 52-1, respectively.
To determine the allelic compositions of Pvtb1a and Pvtb1b genes in 10 randomly selected micropropagated plants including 9 plants generated from primary mutants and 1 plant from the wild-type, sequencing libraries for the tb1a amplicons with an insert size of 250 bp and tb1b amplicons with an insert size of 288 bp were constructed for each individual plant. For each library, at least 5,000 reads were generated from Illumina HiSeq3000 to determine the sequence of the respective amplicons. Three distinct genotypes were found among the five micropropagated plants (52-1-1, -2, -3, -4, -5) derived from 52-1, while four distinct genotypes were observed among the four micropropagated plants (35-2-1, -2, -3, -4) derived from 35-2, clearly indicating that the two primary Pvtb1 mutants from which micropropagated plants were obtained are chimeric (Table 1).
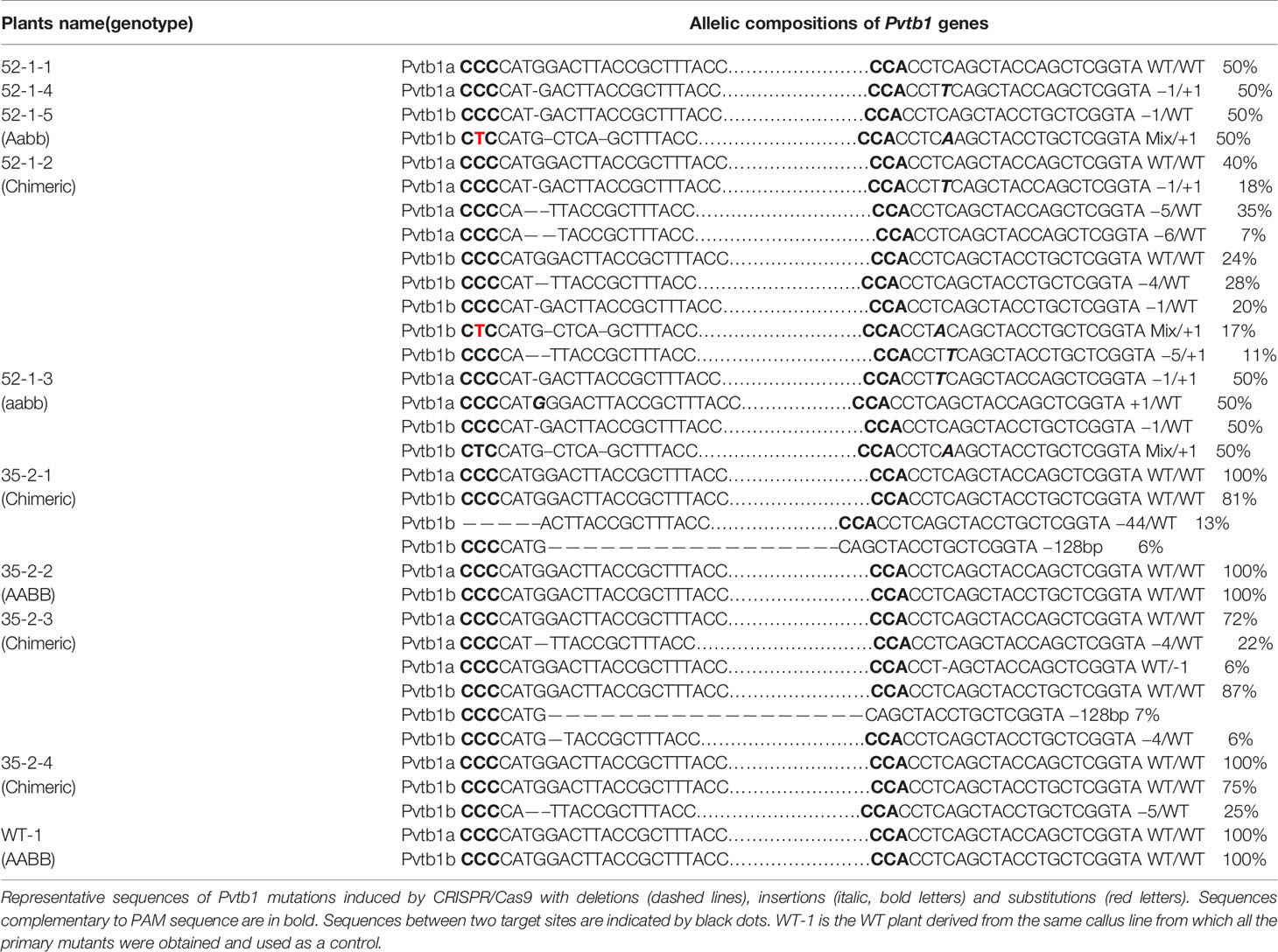
Table 1 Estimation of allelic composition of Pvtb1 genes in plants regenerated from node culture of the primary mutants 52-1 and 35-2.
Four nonchimeric, solid mutants, 52-1-1, 52-1-3, 52-1-4, and 52-1-5 were obtained from the T0 mutant 52-1 with 52-1-1, -4, and -5 all having the same genotype, i.e. 50% of WT Pvtb1a allele and 50% Pvtb1a allele with a G deletion at the first target site and a T insertion at the second target site (Table 1), suggesting these plants were likely originated from a single progenitor mutant cell carrying a monoallelic Pvtb1a mutation. In the other solid mutant 52-1-3, all alleles of the Pvtb1a were mutated with half of the Pvtb1a alleles contains a G deletion at the first target site and a T insertion at the second target site, while the remaining half of Pvtb1a alleles contains only one change, an insertion of G at the first target site (Table 1), suggesting 52-1-3 was likely derived from a single progenitor mutant cell carrying biallelic mutations of Pvtb1a gene. These four plants all contain the same biallelic mutations for Pvtb1b with no WT alleles (Table 1). Fifty percent of the amplicons from each of the four solid mutants contained one C deletion at the first target site, while the remaining half amplicons contained the same mixed mutation (deletions, insertions and substitution) at the first target site and one A insertion at the second target site (Table 1), suggesting these four plants were developed from a single progenitor cell or mutant sector carrying biallelic mutations of the Pvtb1b gene.
Some of the micropropagated plants remained chimeric following micropropagation. In 52-1-2, four types of Pvtb1a amplicons including 3 different mutant alleles accounting for 60% of the reads and 40% WT reads were found (Table 1). In 35-2-3, there were three types of Pvtb1a amplicons including 28% mutated amplicons and 72% WT (Table 1). For Pvtb1b, in 52-1-2, 35-2-1, 35-2-3, and 35-2-4, the percentage of mutant reads are 76%, 19%, 13%, and 25%, respectively (Table 1). The makeup of the WT and mutant alleles is not proportional to the expected allelic composition for a tetraploid species, strongly indicating these four micropropagated plants (52-1-2, and 35-2-1, -3, -4) remain chimeric mutants comprising a mixture of mutant cells carrying different mutations induced by CRISPR/Cas9. These results indicate the axillary bud contained in the explants used for node culture were likely chimeric.
Knockout of Pvtb1 Genes Leads to Increased Tillering in Switchgrass
Based on the most recent annotated switchgrass genome in Phytozome (Goodstein et al., 2012) (https://phytozome.jgi.doe.gov/pz/portal.html), two tb1-like genes, Pvtb1a (Pavir.Ia00838/Pavir.9NG142700) and Pvtb1b (Pavir.Ib04362/Pavir.9KG031700), were isolated from switchgrass using gene-specific primers. These two Pvtb1 genes reside on homeologous chromosomes 9N and 9K, thus are likely homeologous to each other. Amino acid sequence alignment of Pvtb1 gene products and other tb1 orthologs of related species showed that both PvTB1 proteins have the conserved TCP domain and R domain (Figure 1). To determine the function of Pvtb1 genes, tiller numbers of the Pvtb1a-Pvtb1b double biallelic mutants (aabb), Pvtb1a monoallelic and Pvtb1b biallelic mutant plants (Aabb) and WT (AABB) plants were compared. Because switchgrass tillers develop from nodes near or below soil surface, we grew plants in a hydroponic system, which allows close monitoring of tiller and root growth without the destructive excavation of the underground parts of these plants. As shown in Figure 2, the first primary tiller of WT plants did not develop until the end of the fourth week following establishment in the hydroponic system, while most of the mutant plants developed their first primary tiller by the end of the first week. After 4 weeks, the average tiller number of aabb mutants is 5.7 which is about two times the tiller number of WT plants, while the average tiller number of Aabb mutants is 3.7 which is about 1.5 times the tiller number of WT plants.

Figure 1 Multiple sequence alignment of Teosinte branched 1 (TB1) proteins from various species. The black line indicates the TCP domain, while the grey line indicates the R domain. Protein names are shown before each sequence.
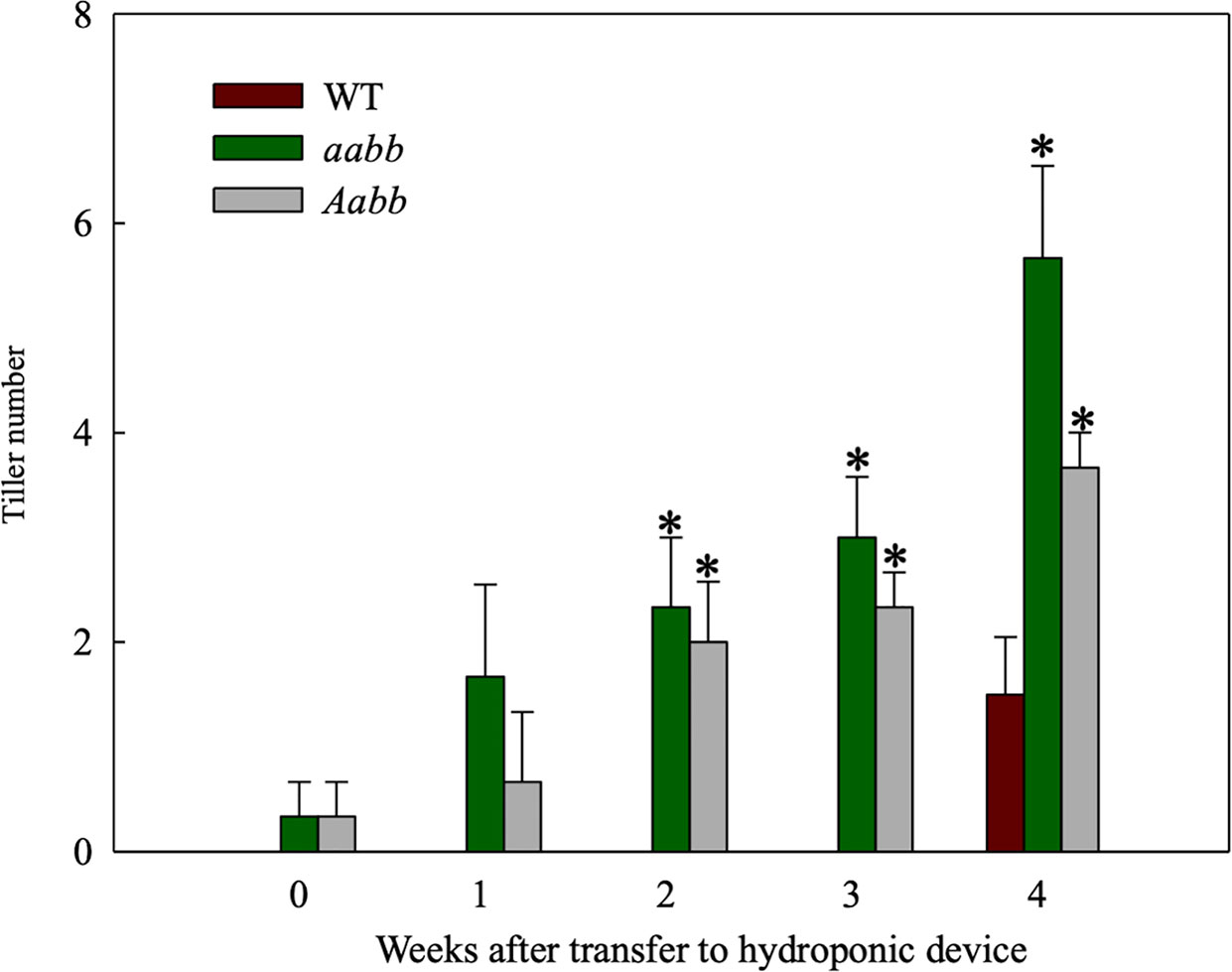
Figure 2 Tiller development in the double biallelic mutant 52-1-3 (aabb) and 52-1-1 mutant (monoallelic for Pvtb1a and biallelic for Pvtb1b, Aabb) and the wild type (WT) at various times after transfer to hydroponic devices. Values are means ± s.d. (mutants, n = 3 plants; WT, n = 6 plants). * indicated significance differences between mutants and WT at P < 0.05.
To further investigate where the increased tiller originated from mutants, development of tillers of different orders in the aabb mutant plants and WT plants were closely monitored for 8 weeks in a separate hydroponic experiment. As shown in Figure 3A, the pvtb1a-pvtb1b (aabb) mutant plants had significantly more tillers than WT plants. The change of tiller numbers over the course of 8 weeks show the tiller numbers of mutant plants are significantly higher than WT plants at all weekly sampling times (Figure 3B). The WT plants did not generate new tillers until the end of the fourth week after establishment in the hydroponic system, while the mutant plants started to produce new tillers at the end of the first week, which is similar to the results from the shorter-term study described above. However, the pvtb1a-pvtb1b mutant plants had no significant effect on the number of primary tillers (1°) compared with WT plants (Figures 3C, D), which suggested that Pvtb1 genes do not regulate the formation of primary axillary buds in the main stem, and the significant increase in secondary (2°), tertiary (3°), or quaternary (4°) tillers in the mutant plants is the result of hastened outgrowth of primary axillary buds. To further confirm the effect of Pvtb1 on the outgrowth of axillary buds and eliminate possible effect of physiological conditions of the starting materials, primary tillers developed from the pvtb1a-pvtb1b double biallelic mutant plants and the WT plants were grown as starting material in a separate hydroponic study. The total number of new tillers developed from the primary tillers of the mutant plants was three-fold of that produced by the primary tillers of the WT plants (Figures 4A, B). Similarly, plants established from the primary tillers of the pvtb1a-pvtb1b mutant had similar number of the first order tillers (1°) to that of the WT, but had greatly increased number of secondary (2°) and tertiary tillers (3°) than those from the primary tillers of the WT. These results strongly suggest that, regardless of the origin of the starting materials, the pvtb1a-pvtb1b mutant plants consistently promote axillary bud outgrowth, increasing tiller number. Aerial tillers, however, were not observed in either the mutant plants or the WT plants, suggesting that Pvtb1 genes did not affect the outgrowth of the upper axillary buds. Interestingly, while the first axillary bud (the lowest in the stem base) in the WT was generally dormant, it was frequently elongated to eventually become a tiller in the pvtb1a-pvtb1b mutant (Figure S2). These results further demonstrate that Pvtb1 genes affect the outgrowth of the axillary bud rather than the formation of new axillary buds.
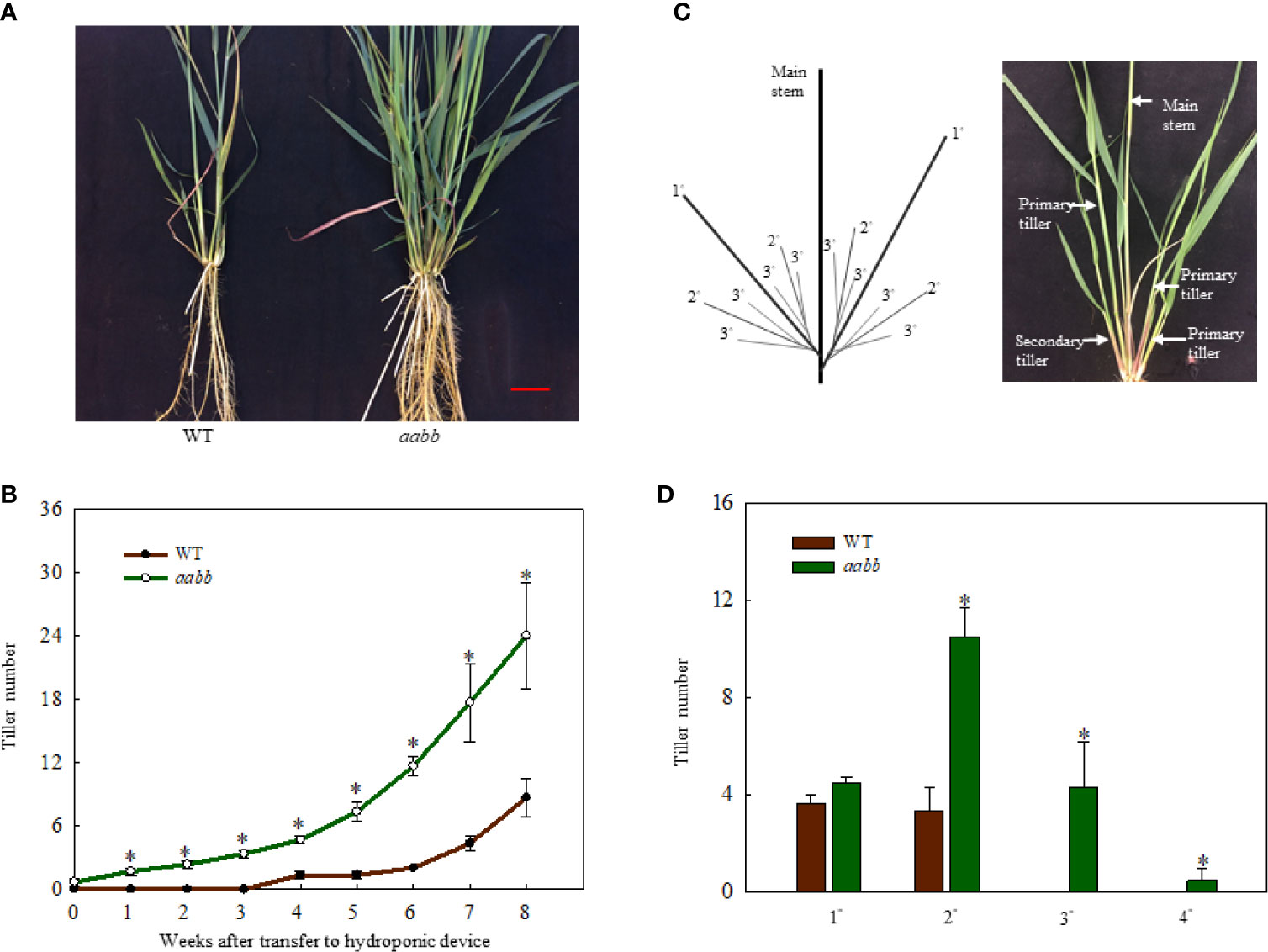
Figure 3 Phenotypic characterization of the Pvtb1a-Pvtb1b biallelic mutant (52-1-3, aabb) and the WT. (A) Phenotype of the Pvtb1a-Pvtb1b biallelic mutant and the WT after 8 weeks of growth in a hydroponic device. Bar = 3 cm. (B) Weekly changes in tiller numbers in the 52-1-3 mutant and the WT after transfer to a hydroponic device. (C) Schematic diagram for tiller ordering in switchgrass. 1° denotes primary tillers, 2° denotes secondary tillers, 3° denotes tertiary tillers. (D) Number of tillers of different orders in the aabb mutant and the wild type (WT) after 8 weeks of growth in hydroponic devices. Values are means ± s.d. [(B), n = 3 plants; (D), n = 6 plants]. * indicated significant differences at P < 0.05.
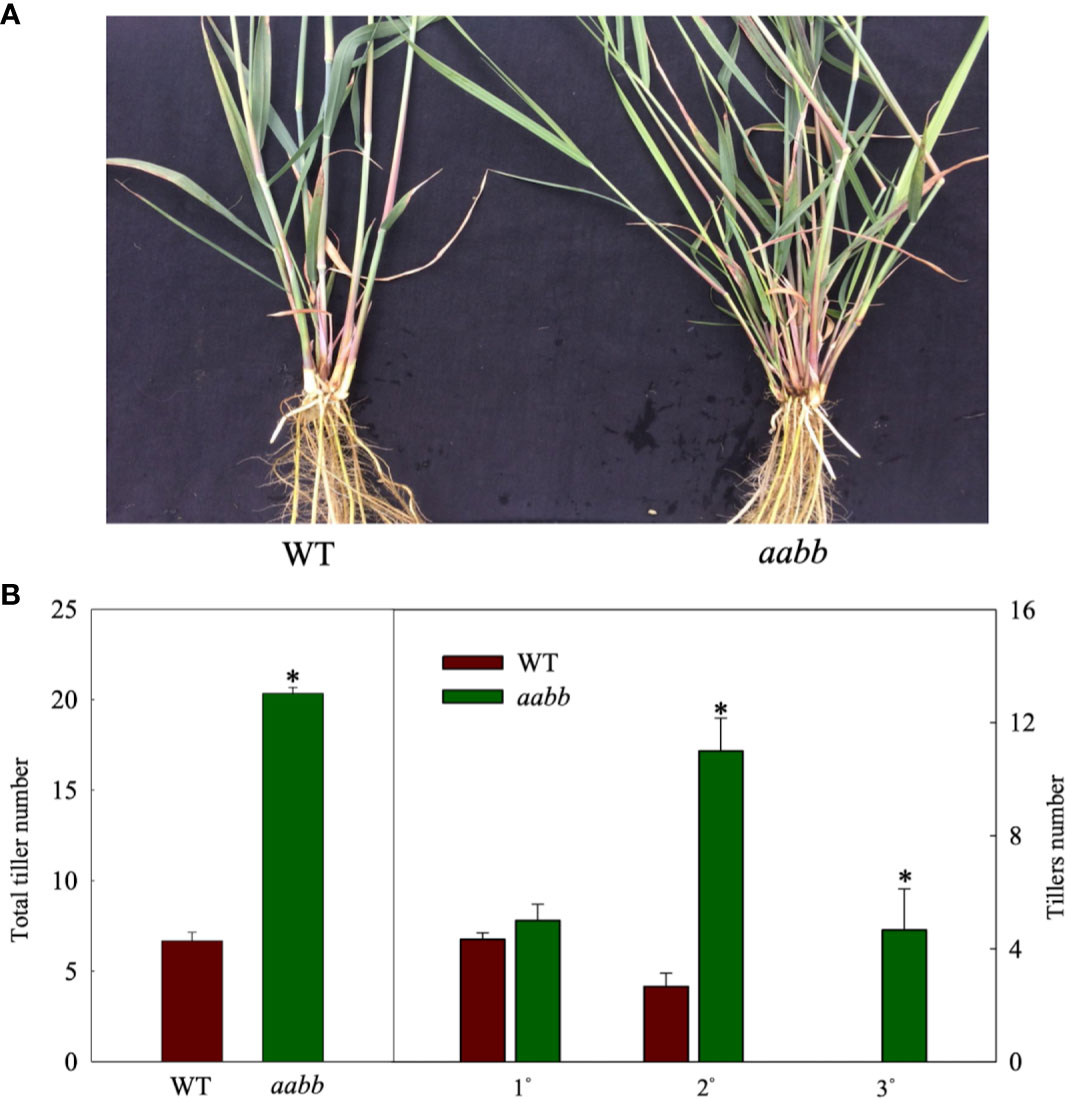
Figure 4 Phenotypic characterization of tiller production with primary tillers developed from main stems of the Pvtb1a-Pvtb1b double biallelic mutant (52-1-3, aabb) as starting materials and cultured in a hydroponic devise for 8 weeks. (A) Visual appearance of the wild type (WT) (left) and the mutant (right) after 8 weeks of culture. (B) Total number of tillers and tiller number for each class of tillers after 8 weeks of culture. 1° denotes primary tillers, 2° denotes secondary tillers, 3° denotes tertiary tillers. Values are means ± s.d. (n = 3 plants). * indicated significant differences at P < 0.05.
Pvtb1 Knock-out Mutant Plants Exhibit Increased Root Production and Biomass Yield
The average root number in mutant plants was similar to that of the WT plants at the beginning of the hydroponic culture (Figure 5). Starting from the second week, the difference between the average root number in mutant plants and WT plants started to increase as the experiment progressed (Figure 5). At the end of the experiment, the average root number of the mutant plants was 28, twice the average root number of the WT plants (Figure 5). Hence, disrupting Pvtb1 gene function in switchgrass enhances root production.
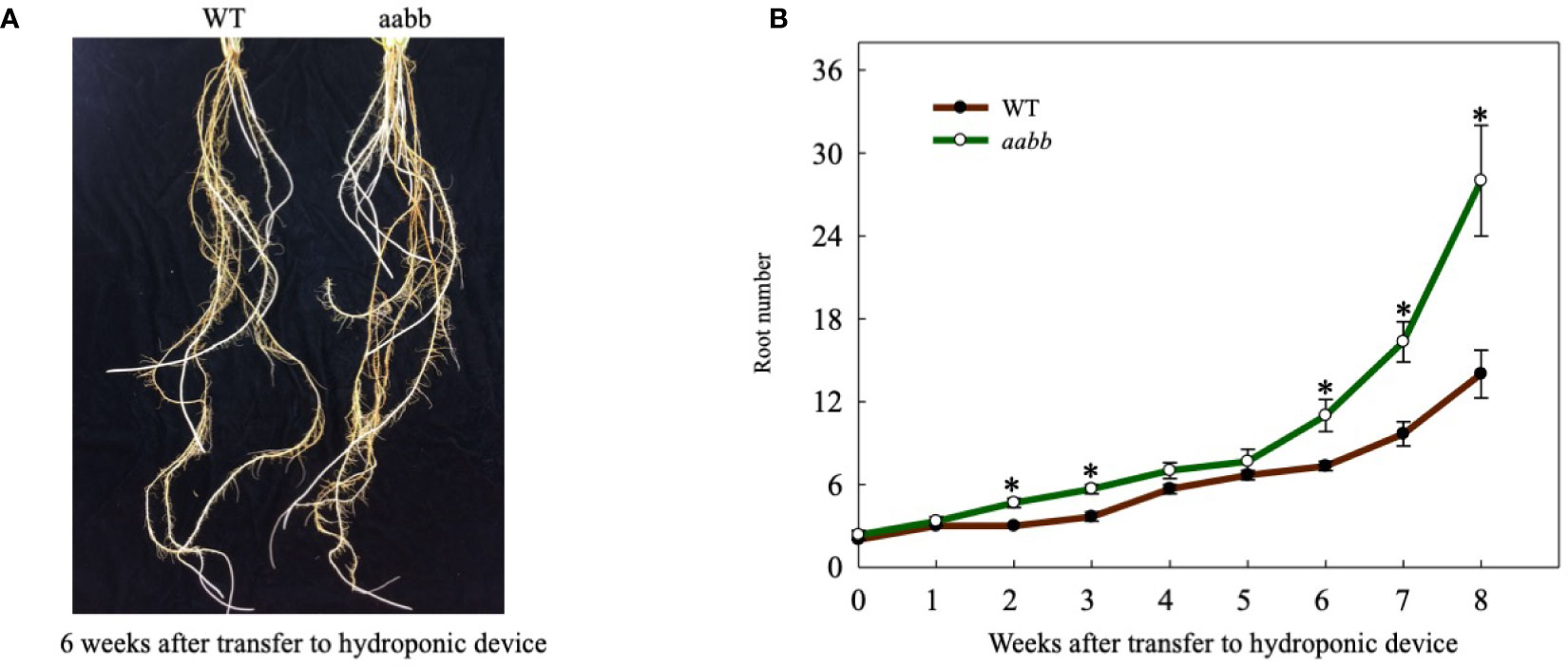
Figure 5 Phenotypic characterization of root development for the Pvtb1a-Pvtb1b biallelic mutant (52-1-3, aabb) and the wild type (WT). (A) Representative plants of WT and a mutant (aabb) with increased root numbers are shown. (B) Weekly changes in root number in the Pvtb1a-Pvtb1b double biallelic mutant (52-1-3, aabb) and the WT after transfer to hydroponic devices. Values are means ± s.d. (n = 3 plants). * indicated significant differences at P < 0.05.
To eliminate the potential unintended effect of the hydroponic growth environment on growth and development, we also evaluated tiller number, stem diameter, plant height, fresh weight and dry weight of pvtb1a-pvtb1b mutant plants and WT plants grown in the soil. Similar to the hydroponic experiments, pvtb1a-pvtb1b mutant plants produced significantly more tillers (2.53-fold) compared with the WT plants (Table 2). Plant height of the mutant plants was similar to that of the WT, while stem diameter of the mutant plants was 13.6% smaller than that of the WT (Table 2). The fresh and dry biomass of the mutant plants increased 29.6% and 15.5% over these of the WT plants, respectively, suggesting the enhanced numbers of tillers of mutants were more than sufficient to compensate for the loss in biomass from decreased stem diameter (Table 2).
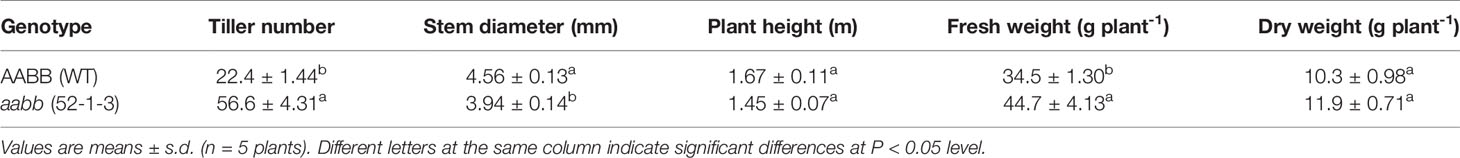
Table 2 Phenotype of the wild-type (WT) and the micropropagated biallelic mutant plants, after 12 weeks of growth in pots filled with soil.
CRISPR/Cas9-Induced Mutations Are Transmitted to Progeny
Only transgene-free BC1 plants, derived from crossing the T0 mutants as maternal parents with a compatible WT plant, were examined to avoid complications that could have resulted from the continuing action of the CRISPR/Cas9 transgene. Among the 30 progeny of the primary mutant 52-1, 12 were transgene-free with ten of them carrying mutations. Meanwhile three of six progeny of the cross between mutant 35-1 and a WT plant carried no transgenes and of these, only one carried mutations (Table 3). Additionally, 18 of 20 progeny from the mutant 35-2 were transgene-free (Table 3), but only two carried mutation.
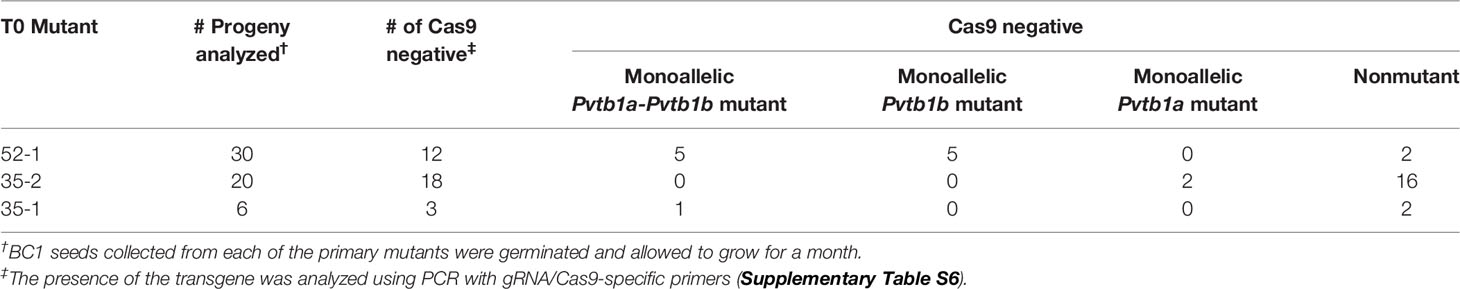
Table 3 Transgene-free BC1 mutants obtained from crossing each of the primary mutants with genetically compatible wild-type plants.
The Pvtb1 mutations observed in all three primary mutants were also observed in their progeny, indicating a stable transmission of the original mutations to the progeny (Table S1). Seven BC1 plants of the 52-1 had both Pvtb1 genes mutated (Table S1), while the other five plants, in BC1 generation, only contained mutations of Pvtb1b (Table S1). All mutations in these BC1 plants were present in the primary mutant 52-1 except for the 52-1-BC1-24, which has a mutated Pvtb1a allele carrying a 128 bp deletion which was not observed in the primary 52-1 mutant (Table S1). Two monoallelic Pvtb1a mutants (35-2-BC1-1 and -6) were obtained from the progeny of the 35-2 (Table 3). Both carried identical mutations observed in the primary mutant 35-2 (Table S1). Furthermore, one doubly monoallelic Pvtb1a-tb1b mutant with big deletions for both tb1 genes was found in the progeny of 35-1 (Table S1).
To test if mutant progeny still retains the phenotypic effects of Pvtb1 mutations, tiller numbers of Pvtb1b monoallelic mutants (genotype AABb with A represents the WT and a represents the mutant allele of Pvtb1a and B represents the WT and b represents the mutant allele of Pvtb1b), and Pvtb1a-Pvtb1b doubly monoallelic mutant plants (genotype AaBb) in the BC1 generation were investigated. Tiller numbers of doubly Pvtb1a-Pvtb1b monoallelic mutants and single Pvtb1b monoallelic mutants were significantly higher than the siblings carrying no mutations (null segregants), suggesting that Pvtb1 genes act in dosage-dependent manner. The average percentage increase in tiller number was 45% for doubly Pvtb1a-Pvtb1b monoallelic mutant plants and 30% for the Pvtb1b monoallelic mutant plants (Table 4).
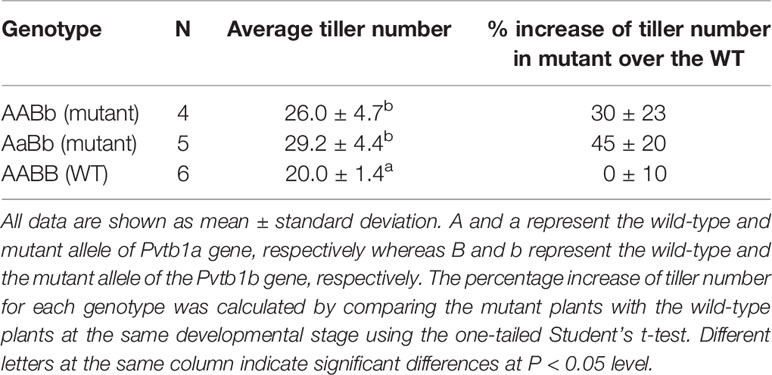
Table 4 The average tiller number of transgene-free BC1 mutants and that of the siblings carrying no mutations (null segregants) and the percentage increase of tiller number for mutants over nonmutant siblings.
mRNA-Seq of Pvtb1 Mutants Suggests Pvtb1 Genes Are Involved in Multiple Pathways That Regulate Tillering in Switchgrass
Although the primary mutant 52-1 was later found to be chimeric, its enhanced tiller production (Liu et al., 2018) with 30%–50% of the tb1a alleles and 61%–94% of the tb1b alleles mutated are sufficient to be treated as a knockdown mutant of Pvtb1 gene (Figure S3). Using a generalized linear model with the R package edgeR (Robinson et al., 2010), 831 genes showed significantly differential expression between the WT and 52-1 when the false discovery rate (FDR) was controlled at 0.05 (Benjamini and Hochberg, 1995) (Table S2). Among them, 364 genes were significantly upregulated while 467 genes showed significantly down regulation in the mutant 52-1 (Figure S4). To validate the RNA-seq data, qRT-PCR was performed on 4 differentially expressed genes and 2 genes with similar expression level between the WT and 52-1. As shown in Figure S5, the expression trends of these 6 genes observed in qRT-PCR and RNA-seq results were similar in WT_vs_52-1 and a high expression correlation was observed between fold changes of 6 genes in RNA-seq and qPCR experiments using linear regression analysis (R2 = 0.871, p=0.0065), indicating the results of RNA-seq are reliable.
Orthologous Arabidopsis genes for these DEGs were subjected to Gene Ontology (GO) analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al., 2009a; Huang et al., 2009b). Regarding the biological process, 27 terms (P<0.05, Table S4) were enriched for downregulated genes in the mutant, and 12 terms (P<0.05, Table S3) for the upregulated genes. A major GO term for these DEGs was DNA-templated transcription (GO: 0006351) (Figures 6A, B). Among the genes enriched in this GO term, transcript levels for 34 genes increased significantly in the Pvtb1 gene knockdown mutant 52-1 while the remaining 43 genes were downregulated (Table S5). The majority of the genes in this GO term encode transcription factors (TFs). For instance, genes for 3 TCP family TFs (TCP2, 4, and 5) and 4 MADS-box family TFs (AP1) were upregulated in the mutant 52-1, while genes for 8 WRKY family TFs (WRKY30, 33, 40, 41, 46, and 53) and 3 NAC family TFs (NAC02, 036, and 102) were down- regulated. GO terms associated with cell differentiation and positive regulation of development were also enriched among the upregulated genes in the mutant. Other GOs enriched in both up- and downregulated genes were involved in response to various phytohormones suggesting Pvtb1 genes are, as in other species involved in various hormonal signaling pathways in regulating tillering in switchgrass (Tables S3, S4). For example, BRH1, a brassinosteroid-responsive RING-H2 gene, was downregulated in the Pvtb1 gene knockdown mutant (Figure 7). In contrast, genes for auxin-responsive factor and ABA-induced proteins were both upregulated in the mutant (Figure 7). In addition, GO terms involved in negative regulation of flower development and heterochromatin maintenance were overrepresented among the downregulated genes (Figure 6B). For example, Arabidopsis DRD1 and EDM2 both regulate DNA methylation patterns (Cho et al., 2016; Duan et al., 2017) and the orthologs of these two were downregulated in the present study. Hence, we speculate that epigenetic modification plays a role in Pvtb1-mediated regulation in tillering in switchgrass. Intriguingly, GOs associated with stress/defense responses were significantly enriched in downregulated genes, indicating Pvtb1 genes might function in maintaining homeostasis in the regulation of growth and defense (Figures 6B, 7). These data suggest the gene regulatory network of tillering in switchgrass is complex and Pvtb1 genes are important hubs in this network.
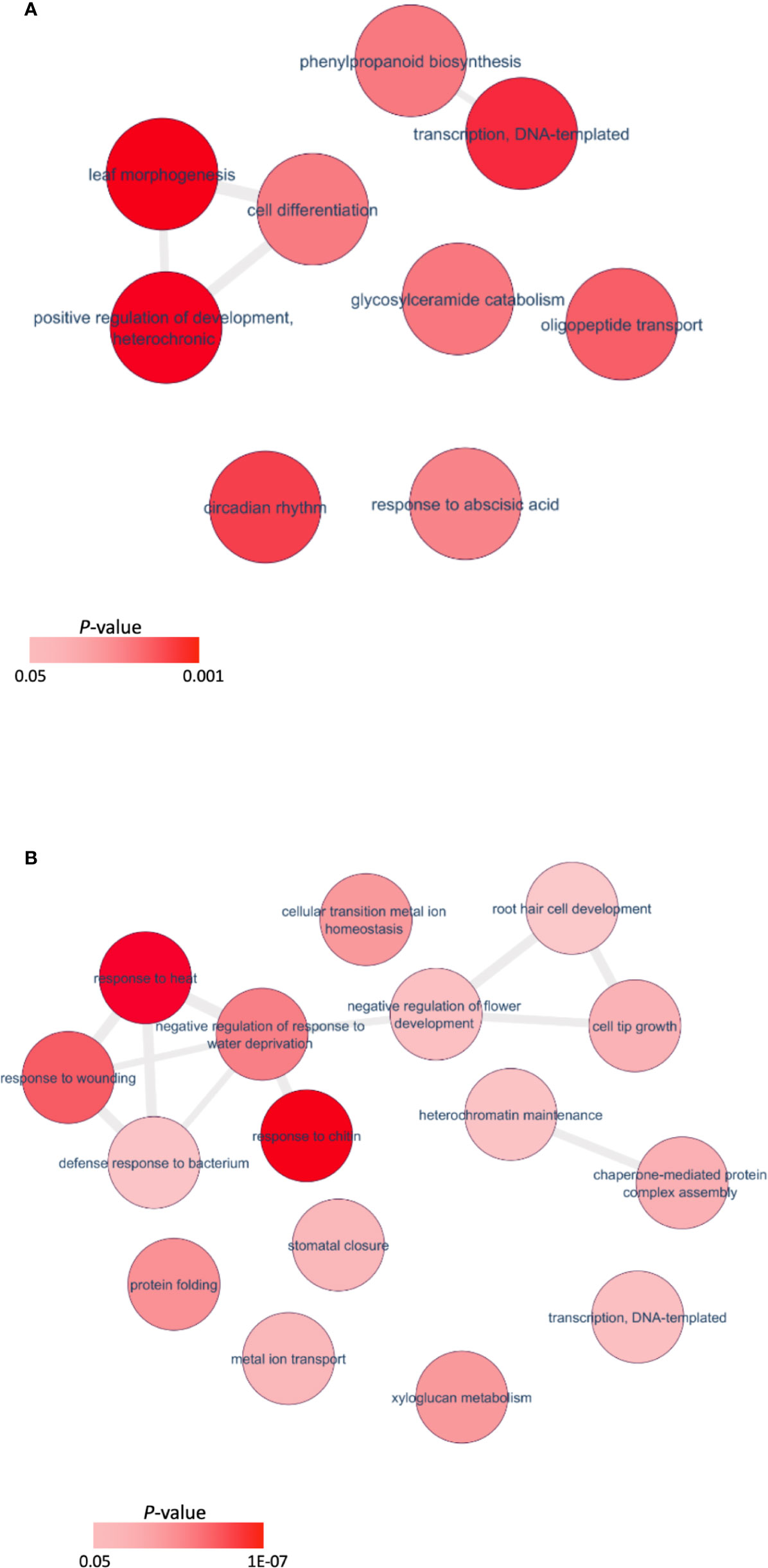
Figure 6 Enriched Gene Ontology (GO) terms and differentially expressed genes of the Pvtb1 genes knockdown mutant. (A) Gene set enrichment analysis of upregulated genes; (B) Gene set enrichment analysis of downregulated genes. Each red circle represents a GO term. Two color bars indicate P-value ranging from 0.05 to 0.001 and 1.0 × 10−7, respectively.
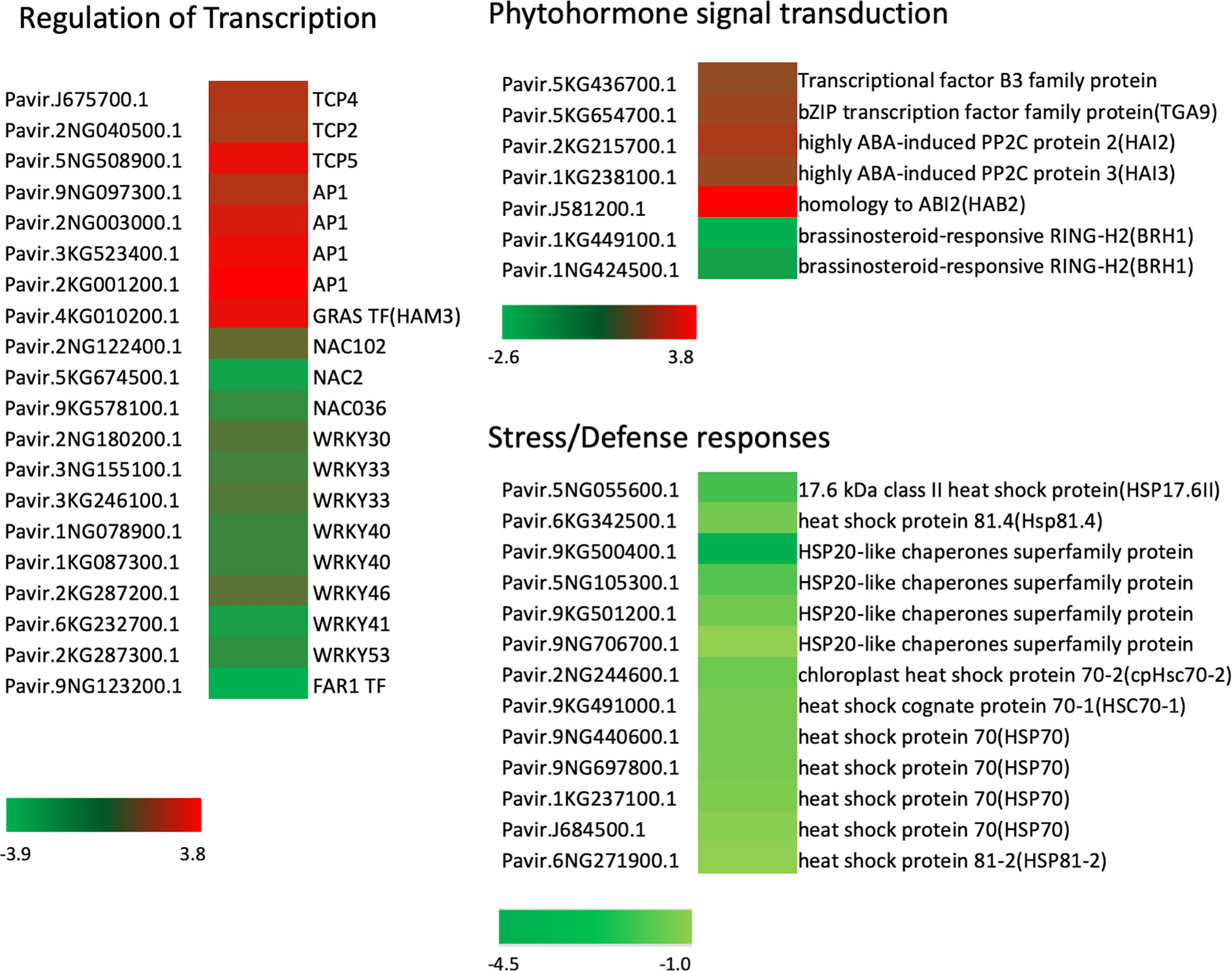
Figure 7 Expression patterns of representative differentially expressed genes (DEGs) involved in transcription regulation, phytohormone signal transduction and stress/defense responses previously characterized in Arabidopsis. Three color bars represent the range of fold change for DEGs, where red indicates upregulation while green indicates downregulation.
Discussion
CRISPR/Cas9-based genome editing tool affords new possibilities for reverse genetics-based studies in switchgrass (Liu et al., 2018). In the present study, through the node culture of chimeric primary mutants induced by the CRISPR/Cas9 system, we successfully obtained a nonchimeric biallelic Pvtb1a-Pvtb1b mutant, and two nonchimeric mutants carrying biallelic Pvtb1b mutations and one Pvtb1a mutated allele. These results demonstrate that micropropagation is an effective way of isolating mutant sectors from chimeric mutants in switchgrass and obtaining nonchimeric mutants. Solid nonchimeric mutants were isolated from the chimeric primary mutant, 52-1. Among five plants regenerated from nodes culture of 52-1, four plants are nonchimeric mutants, suggesting the primary mutant plant 52-1 is composed primarily of mutant cells. However, for the primary mutant 35-2, none of the regenerated plants are nonchimeric mutants, indicating this plant, or regions where axillary buds arise are composed primarily of nonmutant cells. Overall, we demonstrated that nonchimeric mutants can be successfully isolated from chimeric mutants using micropropagation, but the chimeric pattern of chimeric mutants is a determining factor on the efficiency of isolating nonchimeric mutants, and varies greatly between mutants. Strong genetic-incompatibility of switchgrass is an obstacle to generate homozygous mutants. To set seeds carrying homozygous mutations, individual mutants used for crossing must have different alleles of S and Z genes (Martinez-Reyna and Vogel, 2002), which is difficult to determine because the molecular basis of the self-incompatibility in switchgrass has not been studied. Also, it takes over 6 months for switchgrass seedlings to reach reproductive stage (Hopkins et al., 1995; VanEsbroeck et al., 1997) for crossing. Given all these obstacles, the successful purification of chimeric mutants using micropropagation clearly has advantages. In addition, nonchimeric homozygous mutants can be obtained from T0 chimeric mutants within one month of using the micropropagation method, which is time-saving compared to traditional breeding method.
Switchgrass cv. Alamo is an allotetraploid with two homeologous subgenomes, but detailed information about chromosome pairing, whole or partial genome duplications, and allelic diversity of specific genes is lacking (Missaoui et al., 2005; Okada et al., 2010). With limited information gained by sequencing clones of PCR amplicons spanning the target regions, the nature of mutants was not resolved unequivocally in our previous study (Liu et al., 2018). Using the Next Generation Sequencing (NGS) technology, genotypes of micropropagated mutants were fully characterized. The two primary mutants, 52-1 and 35-2 were both revealed by NGS to be chimeric mutants, which were likely the results of continuous action of CRISPR/Cas9 in somatic cells. Indeed, chimeric mutations induced by CRISPR/Cas9 have been reported in different species (Feng et al., 2014; Pan et al., 2017). For instance, Pan et al. (2017) showed that 63.9% T0 transgenic plants carried chimeric mutations in tomato (Solanum lycopersicum L.). In rice, the segregation ratio of CRISPR/Cas9-induced mutations in BC1 generation did not follow the expected segregation ratio, indicating the chimeric mutations in T0 plants (Xu et al., 2015).
Transgene-free mutants were successfully obtained by crossing primary mutant plants with WT plants. The majority of the mutations observed in progeny were identical to the mutations in the primary mutants. For example, BC1 progeny of the 52-1 carried the Pvtb1b mutations that were present in all five tillers of the primary mutant 52-1. These results demonstrate mutations induced by the CRISPR/Cas9 in T0 mutants were stably transmitted to the BC1 generation without alteration. However, a mutation (128bp deletion) of Pvtb1a, not detected in the primary mutant 52-1, was observed in the BC1 transgene-free mutant plant 52-1-BC1-24. This deletion likely escaped detection in T0 plants due to the chimeric nature. The similar phenomenon has also been observed in maize (Lee et al., 2019).
Pvtb1 genes are closely related to the tb1 genes in other monocots. The Pvtb1a-Pvtb1b double biallelic mutants produced significantly higher tiller number than the WT plants under both hydroponic and soil culture conditions. Results from the hydroponic experiments in which starting plant materials were primary tillers validated the observed differences between the mutants and the WT plants were the result of genetic alteration instead of the existing physiological difference between the WT and the mutant. Our results strongly indicate Pvtb1 genes play an important role in regulating tillering in switchgrass. There was no significant difference in the number of primary tillers between the WT and 52-1-3, suggesting the function of Pvtb1 genes in switchgrass is only to regulate the rate of outgrowth of axillary buds that are destined to become tillers with one possible exception, i.e., the release of the outgrowth of the bud at the lowest node (Figure S2). This is similar to orthologs of tb1 in other species, reflecting the functional conservation of tb1 genes across species (Takeda et al., 2003; Kebrom et al., 2006; Aguilar-Martinez et al., 2007; Braun et al., 2012).
The Pvtb1a-Pvtb1b double biallelic mutant plants also produced more roots compared with WT plants. This result is consistent with Gaudin et al. (2014) who reports a decrease in tb1 function in maize resulted in a larger root system. The increased root growth and development is likely the indirect result of increased tiller production, as each tiller normally develops its own adventitious roots. Increased tiller number has the potential to increase biomass yield in switchgrass (Chuck et al., 2011; Fu et al., 2012). The Pvtb1a-Pvtb1b mutants produced 29.6% and 15.5% more fresh and dry biomass, respectively. Although the difference on dry biomass between the wild-type and mutant plants is not statistically significant, this may change if mutant plants with more tillers and increased root mass are grown in the field where more resources are available.
In maize, studies have shown that tb1 regulates branching in a dosage-dependent manner (Doebley et al., 1995; Hubbard et al., 2002). Maize heterozygous tb1 mutants had slightly more tillers than WT plants, while homozygous tb1 mutants produced more tillers than heterozygous tb1 mutants (Doebley et al., 1995). In this study, we noticed the tiller number in monoallelic heterozygous Pvtb1b mutants (AABb) was significantly higher than WT plants, suggesting that Pvtb1b functions in a dosage-dependent manner. In addition, due to the high amino acid sequence identities between PvTB1A and PvTB1B, it is reasonable to expect they regulate tillering of switchgrass redundantly or additively. However, comparing monoallelic Pvtb1b mutant plants (AABb) with the doubly monoallelic Pvtb1a-Pvtb1b mutant plants (AaBb) did not show significant differences on tiller numbers, suggesting there is no significant additive effect between Pvtb1a and Pvtb1b. Therefore, Pvtb1a might have a minor effect on tillering in switchgrass. This is similar to Arabidopsis in which only BRC1 regulates branching, despite both BRC1 and BRC2 have the conserved TCP and R domains (Aguilar-Martinez et al., 2007; Gonzalez-Grandio et al., 2013; Seale et al., 2017).
To have a better understanding of the functions of Pvtb1 genes, we examined global transcriptional changes caused by the downregulation of Pvtb1 genes. Increased expression level of genes for TCP TFs that are associated with cell differentiation and positive regulation of development in the mutant suggested that Pvtb1 genes inhibit the tiller production through deactivating cell differentiation. In addition, HAIRY MERISTEM 3 (HAM3), the gene for GRAS family TF that interacts with WUSCHEL (WUS) TF to promote shoot meristem development (Zhou et al., 2015), was upregulated in the mutant, suggesting increased shoot stem cell proliferation in the Pvtb1 gene knockdown mutant.
Our transcriptomic analysis results suggest PvTB1a and PvTB1b regulate tillering in switchgrass by interacting with complex hormonal signaling pathways. Six cytochrome P450 genes were up regulated in the mutant (Table S5). The members of cytochrome P450 family catalyze the biosynthesis of several phytohormones including auxin, brassinosteroids, and strigolactones which regulate branching across various plants species (Zhao, 2008; Kebrom et al., 2013). In addition, increased expression of ABA-responsive genes, PROTEIN PHOSPHATASE 2C (PP2C) genes, was observed in the mutant. These results suggest that Pvtb1 genes regulate bud development by modulating phytohormone biosynthesis and signaling. In Arabidopsis and maize, it has been shown TB1/BRC1 promotes ABA accumulation and the expression of ABA response factors to inhibit bud outgrowth (Gonzalez-Grandio et al., 2013; Yao and Finlayson, 2015; Gonzalez-Grandio et al., 2017; Holalu and Finlayson, 2017). Although it is well-known that TB1/BRC1 are involved in hormonal signaling pathways in different plant species, these regulation pathways are not conserved across various species (Kebrom et al., 2013). For instance, cytokinins repress the expression of TB1 in rice, while they act in a pathway independent of BRC1 in Arabidopsis (Aguilar-Martinez et al., 2007; Minakuchi et al., 2010). The timing of hormonal signaling in tillering has not been decided in switchgrass. Hence, more studies are needed to understand how the hormonal signals are involved in the Pvtb1-mediated regulation of branching.
Altered expression of genes in response to red or far-red light in the mutant suggest that Pvtb1 genes integrate the light signal to regulate tillering in switchgrass (Figure 7). FAR-RED-IMPAIRED RESPONSE1 (FAR1)-related sequence (FRS) family of transcription factors regulate plant growth and development in response to far-red light in Arabidopsis (Wang and Wang, 2015). The homolog of FAR1, FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) promotes shoot branching in Arabidopsis (Stirnberg et al., 2012). Further, FAR1/FHY3 promotes FHY1/FHL gene expression to facilitate phyA nuclear accumulation under far-red light condition. The downregulation of FRS TF genes in the switchgrass mutant suggested that Pvtb1 genes may be involved in promotion of phyA nuclear accumulation to inhibit the axillary bud initiation or outgrowth. Additionally, knockdown of Pvtb1 genes increased the expression level of the Phytochrome interacting factor 4 (PIF4) gene that has been shown to regulate expression of genes involved in cell expansion (Huq and Quail, 2002). Because PIF4 is a TF regulated by phyB-mediated signaling, its activity is regulated by the red light signal (Xu, 2018). These results suggest Pvtb1 genes regulate tillering through light signaling pathways. It has been reported that tb1 genes inhibit bud outgrowth in the process of shade-avoidance-syndrome (SAS) (Kebrom et al., 2013). In Arabidopsis and sorghum, the expression levels of BRC1 and SbTB1 were both upregulated under shade (Kebrom et al., 2006; Kebrom et al., 2010; Gonzalez-Grandio et al., 2013). Therefore, Pvtb1 genes might also sense the low R:FR ratio to inhibit bud outgrowth in switchgrass.
Several genes associated with stress/defense responses were significantly downregulated in the mutant. For example, 13 genes for Heat-shock proteins (Hsps)/chaperones which assist in protein refolding under stress conditions (Wang et al., 2004) were downregulated in the mutant. It has been shown overexpression of alfalfa (Medicago sativa L.) MsHSP70 gene could enhance Arabidopsis drought and heat stress tolerance (Li et al., 2017). Soybean (Glycine max (L.) Merril) Hsp 90 family members respond differentially to abiotic stresses and reduce the damage caused by abiotic stresses in Arabidopsis (Xu et al., 2013). Hence, the downregulation of Hsp genes in the switchgrass mutant suggest hastened outgrowth of axillary buds of the mutant might trigger increased expression of stress/defense responsive genes. Further, the expression levels of several WRKY TF genes responsive to chitin elicitation functioning in plant defense to fungal pathogens (Libault et al., 2007) also decreased in the mutant. In Arabidopsis, WRKY TFs are necessary for resistance to pathogen infection (Zheng et al., 2006) or resistance to abiotic stresses (Chen et al., 2010). It is well-known tolerance-growth trade-offs occur in plants under the low-resource conditions (Koziol et al., 2012; Bristiel et al., 2018). It has been shown that TB1 may influences sucrose levels and energy balance within dormancy buds in maize (Dong et al., 2019). Exploration of the mechanism of PvTB1s controlling energy balance in switchgrass would provide valuable information for the improvement of switchgrass for biomass production and development of enhanced stress-tolerant cultivars.
Conclusions
We successfully isolated mutated segments from chimeric mutants using micropropagation. This method overcomes the difficulties of obtaining nonchimeric mutants in self-incompatible species. Further, transgene-free mutants were obtained in this research, which provided valuable germplasm for switchgrass genetic research and breeding. More importantly, we proved the stable transmission of mutations induced by the CRISPR/Cas9 system in switchgrass. We propose that Pvtb1 genes negatively regulates tillering in switchgrass. RNA-seq analysis revealed a complex regulatory network potentially regulating tillering in switchgrass and provided some clues to the pathways of Pvtb1 genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories: The repository name is Sequence Read Archive (SRA). The project name is' Transcriptome sequencing of axillary buds of switchgrass Pvtb1 mutant and WT plant' The accession number is PRJNA660286. To access the data, please use the link: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA660286.
Author Contributions
S-zF and YL conceived the original research plans. YL conducted the experiments on gene editing, micropropagation, and sequencing and analyzed relevant data. YB provided technical assistance to YL. WW conducted the hydroponic experiment. CC provided technical assistance to WW. YL and S-zF wrote the article with contributions from all authors. S-zF agrees to serve as the author responsible for contact and ensures communication.
Funding
This work was partially supported by the National Institute of Food and Agriculture of the US Department of Agriculture (2013-33522-21091 to BY and S-zF) and the Crop Bioengineering Center of Iowa State University (S-zF).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors declare no competing financial interests and no conflict-of-interest. The authors also wish to thank Michael Baker at the ISU DNA Facility for his assistance on NGS experiment and Zhaoxia Li and Xianran Li for their valuable suggestions on NGS data analysis. This manuscript has been released as a pre-print at bioRXiv (Liu et al., 2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.572193/full#supplementary-material
Supplementary Figure 1 | Micropropagation of switchgrass. Longitudinally split nodal segments were cultured on the MS-0 medium without plant growth regulators.
Supplementary Figure 2 | Outgrowth of the lowest axillary bud in mutant plants and the wild-type plants. Arrow indicates a tiller developing from the lowest node in the Pvtb1a-Pvtb1b mutant (52-1-3, aabb), which is usually absent in the WT. The WT plant was about 2 weeks older than the mutant plant. Other tillers were removed for better view.
Supplementary Figure 3 | Estimation of allelic composition of Pvtb1 genes in cDNA samples of the mutant 52-1.
Supplementary Figure 4 | Mean-Difference plot showing the log-fold change and average abundance of each gene. Significantly up- and down-regulated genes in the mutant are highlighted in red and blue, respectively.
Supplementary Figure 5 | Expression patterns of six genes in both qRT-PCR and RNA-seq results. Three independent biological replicates and technique replicates were applied for each qRT-PCR assay.
References
Aguilar-Martinez, J., Poza-Carrion, C., Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19 (2), 458–472. doi: 10.1105/tpc.106.048934
Alexandrova, K., Denchev, P., Conger, B. (1996). Micropropagation of switchgrass by node culture. Crop Sci. 36 (6), 1709–1711. doi: 10.2135/cropsci1996.0011183X003600060049x
Anders, S., Pyl, P., Huber, W. (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31 (2), 166–169. doi: 10.1093/bioinformatics/btu638
Arite, T., Iwata, H., Ohshima, K., Maekawa, M., Nakajima, M., Kojima, M., et al. (2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51 (6), 1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x
Benjamini, Y., Hochberg, Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B-Methodol. 57 (1), 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Boe, A., Beck, D. (2008). Yield components of biomass in switchgrass. Crop Sci. 48 (4), 1306–1311. doi: 10.2135/cropsci2007.08.0482
Boe, A. (2007). Variation between two switchgrass cultivars for components of vegetative and seed biomass. Crop Sci. 47 (2), 636–642. doi: 10.2135/cropsci2006.04.0260
Bolger, A., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Braun, N., de Saint Germain, A., Pillot, J., Boutet-Mercey, S., Dalmais, M., Antoniadi, I., et al. (2012). The Pea TCP Transcription Factor PsBRC1 Acts Downstream of Strigolactones to Control Shoot Branching. Plant Physiol. 158 (1), 225–238. doi: 10.1104/pp.111.182725
Bristiel, P., Gillespie, L., Ostrem, L., Balachowski, J., Violle, C., Volaire, F. (2018). Experimental evaluation of the robustness of the growth-stress tolerance trade-off within the perennial grass Dactylis glomerata. Funct. Ecol. 32 (8), 1944–1958. doi: 10.1111/1365-2435.13112
Casler, M. (2010). Changes in Mean and Genetic Variance During Two Cycles of Within- family Selection in Switchgrass. Bioenergy Res. 3 (1), 47–54. doi: 10.1007/s12155-009-9071-9
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., Xu, X. (2010). Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10, 1–5. doi: 10.1186/1471-2229-10-281
Cho, E. J., Choi, S. H., Kim, J. H., Kim, J. E., Lee, M. H., Chung, B. Y., et al. (2016). A mutation in plant-specific SWI2/SNF2-like chromatin-remodeling proteins, DRD1 and DDM1, delays leaf senescence in Arabidopsis thaliana. PloS One 11 (1), e0146826. doi: 10.1371/journal.pone.0146826
Choi, M., Woo, M., Koh, E., Lee, J., Ham, T., Seo, H., et al. (2012). Teosinte Branched 1 modulates tillering in rice plants. Plant Cell Rep. 31 (1), 57–65. doi: 10.1007/s00299-011-1139-2
Chuck, G., Tobias, C., Sun, L., Kraemer, F., Li, C., Dibble, D., et al. (2011). Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc. Natl. Acad. Sci. United States America 108 (42), 17550–17555. doi: 10.1073/pnas.1113971108
Das, A., Gosal, S., Sidhu, J., Dhaliwal, H. (2000). Induction of mutations for heat tolerance in potato by using in vitro culture and radiation. Euphytica 114 (3), 205–209. doi: 10.1023/A:1003965724880
Das, M., Fuentes, R., Taliaferro, C. (2003). Genetic variability and trait relationships in switchgrass. Crop Sci. 44 (2), 443–448. doi: 10.2135/cropsci2004.4430
Davidson, D., Chevalier, P. (1987). Influence of Polyethylene Glycol-Induced Water Deficits on Tiller Production in Spring Wheat 1. Crop Sci. 27 (6), 1185–1187. doi: 10.2135/cropsci1987.0011183X002700060019x
Dixon, L., Greenwood, J., Bencivenga, S., Zhang, P., Cockram, J., Mellers, G., et al. (2018). TEOSINTE BRANCHED1 Regulates Inflorescence Architecture and Development in Bread Wheat (Triticum aestivum). Plant Cell 30 (3), 563–581. doi: 10.1105/tpc.17.00961
Dobin, A., Davis, C., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 (1), 15–21. doi: 10.1093/bioinformatics/bts635
Doebley, J., Stec, A., Gustus, C. (1995). teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141 (1), 333–346.
Doebley, J., Stec, A., Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386 (6624), 485–488. doi: 10.1038/386485a0
Dong, Z., Xiao, Y., Govindarajulu, R., Feil, R., Siddoway, M. L., Nielsen, T., et al. (2019). The regulatory landscape of a core maize domestication module controlling bud dormancy and growth repression. Nat. Commun. 10 (1), 1–15. doi: 10.1038/s41467-019-11774-w
Duan, C.-G., Wang, X., Zhang, L., Xiong, X., Zhang, Z., Tang, K., et al. (2017). A protein complex regulates RNA processing of intronic heterochromatin-containing genes in Arabidopsis. Proc. Natl. Acad. Sci. 114 (35), E7377–E7384. doi: 10.1073/pnas.1710683114
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. United States America 111 (12), 4632–4637. doi: 10.1073/pnas.1400822111
Finlayson, S., Krishnareddy, S., Kebrom, T., Casal, J. (2010). Phytochrome Regulation of Branching in Arabidopsis. Plant Physiol. 152 (4), 1914–1927. doi: 10.1104/pp.109.148833
Fu, C., Sunkar, R., Zhou, C., Shen, H., Zhang, J., Matts, J., et al. (2012). Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 10 (4), 443–452. doi: 10.1111/j.1467-7652.2011.00677.x
Gaudin, A. C., McClymont, S. A., Soliman, S. S., Raizada, M. N. (2014). The effect of altered dosage of a mutant allele of Teosinte branched 1 (tb1-ref) on the root system of modern maize. BMC Genet. 15 (1), 23. doi: 10.1186/1471-2156-15-23
Gonzalez-Grandio, E., Poza-Carrion, C., Sorzano, C., Cubas, P. (2013). BRANCHED1 Promotes Axillary Bud Dormancy in Response to Shade in Arabidopsis. Plant Cell 25 (3), 834–850. doi: 10.1105/tpc.112.108480
Gonzalez-Grandio, E., Pajoro, A., Franco-Zorrilla, J., Tarancon, C., Immink, R., Cubas, P. (2017). Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. United States America 114 (2), E245–E254. doi: 10.1073/pnas.1613199114
Goodstein, D., Shu, S., Howson, R., Neupane, R., Hayes, R., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40 (D1), D1178–D1186. doi: 10.1093/nar/gkr944
Harten, A. M., Bouter, H., Broertjes, C. (1981). In vitro adventitious bud techniques for vegetative propagation and mutation breeding of potato (Solanum tuberosum L.). II. Significance for mutation breeding. Euphytica 30 (1), 1–8. doi: 10.1007/BF00033653
He, Y., Zhu, M., Wang, L., Wu, J., Wang, Q., Wang, R., et al. (2018). Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol. Plant 11 (9), 1210–1213. doi: 10.1016/j.molp.2018.05.005
Holalu, S., Finlayson, S. (2017). The ratio of red light to far red light alters Arabidopsis axillary bud growth and abscisic acid signalling before stem auxin changes. J. Exp. Bot. 68 (5), 943–952. doi: 10.1093/jxb/erw479
Hopkins, A. A., Vogel, K. P., Moore, K. J., Johnson, K. D., Carlson, I. T. (1995). Genotype Effects and Genotype by Environment Interactions for Traits of Elite Switchgrass Populations. Crop Sci. 35 (1), 125–132. doi: 10.2135/cropsci1995.0011183X003500010023x
Huang, D., Sherman, B., Lempicki, R. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 (1), 1–13. doi: 10.1093/nar/gkn923
Huang, D., Sherman, B., Lempicki, R. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 (1), 44–57. doi: 10.1038/nprot.2008.211
Hubbard, L., McSteen, P., Doebley, J., Hake, S. (2002). Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162 (4), 1927–1935.
Huq, E., Quail, P. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21 (10), 2441–2450. doi: 10.1093/emboj/21.10.2441
Kebrom, T., Burson, B., Finlayson, S. (2006). Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 140 (3), 1109–1117. doi: 10.1104/pp.105.074856
Kebrom, T., Brutnell, T., Finlayson, S. (2010). Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 33 (1), 48–58. doi: 10.1111/j.1365-3040.2009.02050.x
Kebrom, T., Spielmeyer, W., Finnegan, E. (2013). Grasses provide new insights into regulation of shoot branching. Trends Plant Sci. 18 (1), 41–48. doi: 10.1016/j.tplants.2012.07.001
Koziol, L., Rieseberg, L. H., Kane, N., Bever, J. D. (2012). Reduced drought tolerance during domestication and the evolution of weediness results from tolerance–growth trade-offs. Evol.: Int. J. Organic Evol. 66 (12), 3803–3814. doi: 10.1111/j.1558-5646.2012.01718.x
Lee, K., Zhang, Y., Kleinstiver, B. P., Guo, J. A., Aryee, M. J., Miller, J., et al. (2019). Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 17; 362–372. doi: 10.1111/pbi.12982
Li, Z., Long, R., Zhang, T., Wang, Z., Zhang, F., Yang, Q., et al. (2017). Molecular cloning and functional analysis of the drought tolerance gene MsHSP70 from alfalfa (Medicago sativa L.). J. Plant Res. 130 (2), 387–396. doi: 10.1007/s10265-017-0905-9
Libault, M., Wan, J., Czechowski, T., Udvardi, M., Stacey, G. (2007). Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant-Microbe Interact. 20 (8), 900–911. doi: 10.1094/MPMI-20-8-0900
Liu, Y., Merrick, P., Zhang, Z., Ji, C., Yang, B., Fei, S. Z. (2018). Targeted mutagenesis in tetraploid switchgrass (Panicum virgatum L.) using CRISPR/Cas9. Plant Biotechnol. J. 16 (2), 381–393. doi: 10.1111/pbi.12778
Liu, Y., Wang, W., Yang, B., Currey, C., Fei, S. Z. (2020). Functional analysis of the teosinte branched 1 gene in the tetraploid switchgrass (Panicum virgatum L.) by CRISPR/Cas9-directed mutagenesis. bioRxiv. doi: 10.1101/2020.05.23.112961
Lu, F., Lipka, A., Glaubitz, J., Elshire, R., Cherney, J., Casler, M., et al. (2013). Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol. PloS Genet. 9 (1), e1003215. doi: 10.1371/journal.pgen.1003215
Maluszynski, M., Ahloowalia, B. S., Sigurbjörnsson, B. (1995). Application of in vivo and in vitro mutation techniques for crop improvement. Euphytica 85 (1-3), 303–315. doi: 10.1007/BF00023960
Martinez-Reyna, J., Vogel, K. (2002). Incompatibility systems in switchgrass. Crop Sci. 42 (6), 1800–1805. doi: 10.2135/cropsci2002.1800
Martin-Trillo, M., Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15 (1), 31–39. doi: 10.1016/j.tplants.2009.11.003
Martin-Trillo, M., Grandio, E., Serra, F., Marcel, F., Rodriguez-Buey, M., Schmitz, G., et al. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 67 (4), 701–714. doi: 10.1111/j.1365-313X.2011.04629.x
McLaughlin, S., Kszos, L. (2005). Development of switchgrass (Panicum virgatum) as a bioenergy feedstock in the United States. Biomass Bioenergy 28 (6), 515–535. doi: 10.1016/j.biombioe.2004.05.006
Minakuchi, K., Kameoka, H., Yasuno, N., Umehara, M., Luo, L., Kobayashi, K., et al. (2010). FINE CULM1 (FC1) Works Downstream of Strigolactones to Inhibit the Outgrowth of Axillary Buds in Rice. Plant Cell Physiol. 51 (7), 1127–1135. doi: 10.1093/pcp/pcq083
Missaoui, A., Paterson, A., Bouton, J. (2005). Investigation of genomic organization in switchgrass (Panicum virgatum L.) using DNA markers. Theor. Appl. Genet. 110 (8), 1372–1383. doi: 10.1007/s00122-005-1935-6
Mitchell, R., Schmer, M. (2012). “Switchgrass harvest and storage,” in Switchgrass (London: Springer), 113–127.
Mitchell, R., Vogel, K., Sarath, G. (2008). Managing and enhancing switchgrass as a bioenergy feedstock. Biofuels Bioprod. Biorefining-Biofpr. 2 (6), 530–539. doi: 10.1002/bbb.106
Morris, D. (1977). Transport of exogenous auxin in two-branched dwarf pea seedlings (Pisum sativum L.). Planta 136 (1), 91–96. doi: 10.1007/BF00387930
Narasimhamoorthy, B., Saha, M., Swaller, T., Bouton, J. (2008). Genetic Diversity in Switchgrass Collections Assessed by EST-SSR Markers. Bioenergy Res. 1 (2), 136–146. doi: 10.1007/s12155-008-9011-0
Nicolas, M., Rodriguez-Buey, M., Franco-Zorrilla, J., Cubas, P. (2015). A Recently Evolved Alternative Splice Site in the BRANCHED1a Gene Controls Potato Plant Architecture. Curr. Biol. 25 (14), 1799–1809. doi: 10.1016/j.cub.2015.05.053
Novák, F. J., Afza, R., Vanduren, M., Omar, M. (1990). Mutation induction by gamma irradiation of in vitro cultured shoot-tips of banana and plantain (Musa cvs). Trop. Agric. 67 (1), 21–28.
Okada, M., Lanzatella, C., Saha, M., Bouton, J., Wu, R., Tobias, C. (2010). Complete Switchgrass Genetic Maps Reveal Subgenome Collinearity, Preferential Pairing and Multilocus Interactions. Genetics 185 (3), 745–760. doi: 10.1534/genetics.110.113910
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2017). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations (vol 6, 24765, 2016). Sci. Rep. 7, 24765. doi: 10.1038/srep46916
Pyott, D. E., Sheehan, E., Molnar, A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 17 (8), 1276–1288. doi: 10.1111/mpp.12417
Rameau, C., Bertheloot, J., Leduc, N., Andrieu, B., Foucher, F., Sakr, S. (2015). Multiple pathways regulate shoot branching. Front. Plant Sci. 5, 741. doi: 10.3389/fpls.2014.00741
Reddy, S., Finlayson, S. (2014). Phytochrome B Promotes Branching in Arabidopsis by Suppressing Auxin Signaling. Plant Physiol. 164 (3), 1542–1550. doi: 10.1104/pp.113.234021
Robinson, M., McCarthy, D., Smyth, G. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1), 139–140. doi: 10.1093/bioinformatics/btp616
Seale, M., Bennett, T., Leyser, O. (2017). BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 144 (9), 1661–1673. doi: 10.1242/dev.145649
Shannon, P., Markiel, A., Ozier, O., Baliga, N., Wang, J., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi: 10.1101/gr.1239303
Sievers, F., Wilm, A., Dineen, D., Gibson, T., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Sorefan, K., Booker, J., Haurogne, K., Goussot, M., Bainbridge, K., Foo, E., et al. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17 (12), 1469–1474. doi: 10.1101/gad.256603
Stirnberg, P., Zhao, S., Williamson, L., Ward, S., Leyser, O. (2012). FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J. 71 (6), 907–920. doi: 10.1111/j.1365-313X.2012.05038.x
Studer, A., Zhao, Q., Ross-Ibarra, J., Doebley, J. (2011). Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43 (11), 1160. doi: 10.1038/ng.942
Supek, F., Bosnjak, M., Skunca, N., Smuc, T. (2011). REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PloS One 6 (7), e21800. doi: 10.1371/journal.pone.0021800
Takeda, T., Suwa, Y., Suzuki, M., Kitano, H., Ueguchi-Tanaka, M., Ashikari, M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33 (3), 513–520. doi: 10.1046/j.1365-313X.2003.01648.x
Thimann, K., Skoog, F. (1933). Studies on the growth hormone of plants III The inhibiting action of the growth substance on bud development. Proc. Natl. Acad. Sci. United States America 19, 714–716. doi: 10.1073/pnas.19.7.714
VanEsbroeck, G., Hussey, M., Sanderson, M. (1997). Leaf appearance rate and final leaf number of switchgrass cultivars. Crop Sci. 37 (3), 864–870. doi: 10.2135/cropsci1997.0011183X003700030028x
Waltz, E. (2018). With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 36 (1), 6–7. doi: 10.1038/nbt0118-6b
Wang, H., Wang, H. (2015). Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci. 20 (7), 453–461. doi: 10.1016/j.tplants.2015.04.003
Wang, W., Vinocur, B., Shoseyov, O., Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9 (5), 244–252. doi: 10.1016/j.tplants.2004.03.006
Wang, X., Tilford, C., Neuhaus, I., Mintier, G., Guo, Q., Feder, J., et al. (2017). CRISPR-DAV: CRISPR NGS data analysis and visualization pipeline. Bioinformatics 33 (23), 3811–3812. doi: 10.1093/bioinformatics/btx518
Wang, W., Simmonds, J., Pan, Q., Davidson, D., He, F., Battal, A., et al. (2018). Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat. Theo. Appl. Genet. 131 (11), 2463–2475. doi: 10.1007/s00122-018-3166-7
Wang, M., Le Moigne, M.-A., Bertheloot, J., Crespel, L., Perez-Garcia, M.-D., Ogé, L., et al. (2019). BRANCHED1: a key hub of shoot branching. Front. Plant Sci. 10, 76. doi: 10.3389/fpls.2019.00076
Whipple, C., Kebrom, T., Weber, A., Yang, F., Hall, D., Meeley, R., et al. (2011). grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. United States America 108 (33), E506–E512. doi: 10.1073/pnas.1102819108
Wright, L., Turhollow, A. (2010). Switchgrass selection as a “model” bioenergy crop: A history of the process. Biomass Bioenergy 34 (6), 851–868. doi: 10.1016/j.biombioe.2010.01.030
Xu, J., Xue, C., Xue, D., Zhao, J., Gai, J., Guo, N., et al. (2013). Overexpression of GmHsp90s, a Heat Shock Protein 90 (Hsp90) Gene Family Cloning from Soybean, Decrease Damage of Abiotic Stresses in Arabidopsis thaliana. PloS One 8 (7), e69810. doi: 10.1371/journal.pone.0069810
Xu, R., Li, H., Qin, R., Li, J., Qiu, C., Yang, Y., et al. (2015). Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5, 11491. doi: 10.1038/srep11491
Xu, D. (2018). Multifaceted Roles of PIF4 in Plants. Trends Plant Sci. 23 (9), 749–751. doi: 10.1016/j.tplants.2018.07.003
Yang, J., Luo, D., Yang, B., Frommer, W., Eom, J. (2018). SWEET11 and 15 as key players in seed filling in rice. New Phytol. 218 (2), 604–615. doi: 10.1111/nph.15004
Yao, C., Finlayson, S. (2015). Abscisic Acid Is a General Negative Regulator of Arabidopsis Axillary Bud Growth. Plant Physiol. 169 (1), 611–626. doi: 10.1104/pp.15.00682
Zhang, Y., Zalapa, J., Jakubowski, A., Price, D., Acharya, A., Wei, Y., et al. (2011). Natural Hybrids and Gene Flow between Upland and Lowland Switchgrass. Crop Sci. 51 (6), 2626–2641. doi: 10.2135/cropsci2011.02.0104
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. doi: 10.1038/ncomms12617
Zhao, C., Zhang, Z., Xie, S., Si, T., Li, Y., Zhu, J. (2016). Mutational Evidence for the Critical Role of CBF Transcription Factors in Cold Acclimation in Arabidopsis. Plant Physiol. 171 (4), 2744–2759. doi: 10.1104/pp.16.00533
Zhao, Y. (2008). The role of local biosynthesis of auxin and cytokinin in plant development. Curr. Opin. Plant Biol. 11 (1), 16–22. doi: 10.1016/j.pbi.2007.10.008
Zheng, Z., Abu Qamar, S., Chen, Z., Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48 (4), 592–605. doi: 10.1111/j.1365-313X.2006.02901.x
Zhou, Y., Liu, X., Engstrom, E., Nimchuk, Z., Pruneda-Paz, J., Tarr, P., et al. (2015). Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517 (7534), 377–U528. doi: 10.1038/nature13853
Keywords: CRISPR/Cas9, gene editing, Teosinte branched 1, tillering, micropropagation, switchgrass
Citation: Liu Y, Wang W, Yang B, Currey C and Fei S-z (2020) Functional Analysis of the teosinte branched 1 Gene in the Tetraploid Switchgrass (Panicum virgatum L.) by CRISPR/Cas9-Directed Mutagenesis. Front. Plant Sci. 11:572193. doi: 10.3389/fpls.2020.572193
Received: 13 June 2020; Accepted: 03 September 2020;
Published: 23 September 2020.
Edited by:
Zeng-Yu Wang, Qingdao Agricultural University, ChinaReviewed by:
Jiyi Zhang, BASF, United StatesSalim Al-Babili, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2020 Liu, Wang, Yang, Currey and Fei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shui-zhang Fei, c2ZlaUBpYXN0YXRlLmVkdQ==
†Present address: Yang Liu, Department of Genetics, Cell Biology, and Development, University of Minnesota, St Paul, MN, United States
 Yang Liu
Yang Liu Weiling Wang2,3
Weiling Wang2,3 Bing Yang
Bing Yang Christopher Currey
Christopher Currey Shui-zhang Fei
Shui-zhang Fei