- 1College of Life Sciences, Henan Normal University, Xinxiang, China
- 2Department of Plant Biology, University of Illinois at Urbana-Champaign, Champaign, IL, United States
Sex chromosome evolution has mostly been studied in species with heteromorphic sex chromosomes. The Spinacia genus serves as an ideal model for investigating evolutionary mechanisms underlying the transition from homomorphic to heteromorphic sex chromosomes. Among evolutionary factors, repetitive sequences play multiple roles in sex chromosome evolution while their forces have not been fully explored in Spinacia species. Here, we identified major repetitive sequence classes in male and female genomes of Spinacia species and their ancestral relative sugar beet to elucidate the evolutionary processes of sex chromosome evolution using next-generation sequencing (NGS) data. Comparative analysis revealed that the repeat elements of Spinacia species are considerably higher than of sugar beet, especially the Ty3/Gypsy and Ty1/Copia retrotransposons. The long terminal repeat retroelements (LTR) Angela, Athila, and Ogre may be accounted for the higher proportion of repeats in the spinach genome. Comparison of the repeats proportion between female and male genomes of three Spinacia species indicated the different representation in Spinacia tetrandra samples but not in the S. oleracea or S. turkestanica samples. From these results, we speculated that emergence of repetitive DNA sequences may correlate the formation of sex chromosome and the transition from homomorphic sex chromosomes to heteromorphic sex chromosomes as heteromorphic sex chromosomes exclusively existed in Spinacia tetrandra. Three novel sugar beet-specific satellites were identified and confirmed by fluorescence in situ hybridization (FISH); six out of eight new spinach-specific satellites were mapped to the short arm of sex chromosomes. A total of 141 copies of SolSat01-171-s were found in the sex determination region (SDR). Thus, the accumulation of satellite DNA on the short arm of chromosome 1 may be involved in the sex chromosome evolution in Spinacia species. Our study provides a fundamental resource for understanding repeat sequences in Spinacia species and their roles in sex chromosome evolution.
Introduction
The Spinacia genus belongs to family Chenopodiaceae. It includes cultivated spinach (S. oleracea L.) and two wild relatives (S. turkestanica Ilj. and S. tetrandra Stev.). The cultivated spinach genome was approximately 989 Mb (Arumuganathan and Earle, 1991). The direct progenitor of cultivated spinach is likely S. turkestanica, which was generally more closely related to the cultivated spinach than S. tetrandra (Xu et al., 2017). The three species of Spinacia are dioecious and occasionally monoecious, which is controlled by a pair of sex chromosomes X/Y (Onodera et al., 2008). The sex chromosomes of all S. oleracea accessions and all S. turkestanica accessions are homomorphic. However, sex chromosomes of two S. tetrandra accessions are homomorphic and the other three accessions are heteromorphic (Fujito et al., 2015). Both homomorphic and heteromorphic sex chromosomes evolved from a common ancestral homologous autosomes. For the homomorphic sex chromosomes in Spinacia genus, the Y chromosome was predicted to have diverged recently with its counterpart X chromosome (0.40 ± 0.08 Mya); the homomorphic sex chromosomes and heteromorphic sex chromosomes may have diverged approximately 5.7 Mya (Okazaki et al., 2019). The co-existence of two types of sex chromosomes under Spinacia makes it to be a good model for investigating the evolution of heteromorphic sex chromosomes from ancestral homomorphic sex chromosomes.
Species with nine pairs of chromosomes are prevalent in Chenopodiaceae. Molecular phylogenetic evidence revealed an independent dysploid chromosome loss in two lineages of Chenopodiaceae [one is Spinacia (2n = 2x = 12)]. The monoecious Blitum (2n = 2x = 18) is relative to Spinacia in Chenopodiaceae, indicating dysploidy loss happened during Spinacia evolution (Kühn et al., 1993; Fuentes-Bazan et al., 2012a, b). Spinacia diverged with Blitum was approximately 15.4 Mya (based on rbcL) and 17.4 Mya (based on atpB-rbcL) (Kadereit et al., 2010).
Sugar beet (Beta vulgaris, 2n = 2x = 18) is an important crop with hermaphroditic flowers in the Chenopodiaceae with an estimated genome size of 567 Mb, of which approximately 42.3% are repetitive sequences (Paesold et al., 2012; Dohm et al., 2014; Kowar et al., 2016). Many repetitive sequences, such as satellite DNAs, transposable elements, retrotransposons, and DNA transposons, have been fully investigated in the sugar beet genome (Weber and Schmidt, 2009; Wenke et al., 2009; Weber et al., 2010; Zakrzewski et al., 2010, 2014; Menzel et al., 2012; Wollrab et al., 2012; Heitkam et al., 2014; Schwichtenberg et al., 2016). The divergence between Arabidopsis and the common ancestor of spinach and sugar beet occurred following an ancient triplication event (g), while sugar beet may have diverged from spinach approximately 38.4 Mya (Takahata et al., 2016; Xu et al., 2017).
Eukaryotic genomes contain abundant repetitive DNA, but the relationship between repeat composition and genome size remains unclear. Research on plant sex chromosome evolution has mostly focused on species with heteromorphic sex chromosomes, such as Cannabis sativa (Sakamoto et al., 2000), Hippophae rhamnoides (Puterova et al., 2017), Coccinia grandis (Sousa et al., 2016), Silene latifolia, and Rumex acetosa (Vyskot and Hobza, 2004). Repetitive sequences are important for sex chromosome evolution, and are involved in recombination suppression, heterochromatization, sex chromosomes differentiation in structure, Y degeneration, and X dosage compensation (Hobza et al., 2015; Li et al., 2016). In spinach, 74.4% of the genome is repetitive sequences (Xu et al., 2017) and the region around the male-determining locus is highly repetitive (Kudoh et al., 2018). The repetitive components of the S. oleracea L genome was annotated in our previous study, where we found that Ogre/Tat lineage and two sex chromosome-specific satellites DNAs may be involved in sex chromosome evolution (Li et al., 2019). However, the role of repetitive sequences in sex chromosome evolution of Spinacia has not been fully investigated.
Graph-based clustering of sequence reads is a novel method of identifying repetitive elements following next-generation sequencing (NGS) (Novak et al., 2014). RepeatExplorer is a computational pipeline to perform de novo repeat elements identification using a graph-based sequence clustering algorithm (Novak et al., 2013). This pipeline has been used to perform repetitive sequence classification and estimate sequence similarity among individual genomes (Feng et al., 2017; Gaiero et al., 2019; Liu et al., 2019; Pamponet et al., 2019).
This study aimed to elucidate variations of repetitive sequences regarding distribution of male vs. female genomes of three Spinacia species, and to compare genomic repeat DNA sequences of the Spinacia species with their relative sugar beet; as such, the evolutionary processes shaping the sex chromosome. Global identification and classification of repeats elements from male and female Spinacia species were conducted to characterize the distribution of elements and relations to sex chromosomes evolution. We also surveyed the potential roles of repetitive sequences in genome expansion in Spinacia wild relatives, which might be related to the emergence of heteromorphic sex chromosomes. Our study provides a fundamental resource for understanding repeat sequences in Spinacia species and their roles in sex chromosome evolution.
Materials and Methods
Plant Materials
The seeds of three species of Spinacia, including S. oleracea L., S. turkestanica Ilj., and S. tetrandra Stev., were obtained from the U.S. National Plant Germplasm System (NPGS)1. Plant specimen information was deposited in NPGS. The accession numbers of the plant materials used in our study are PI 527332 (S. oleracea L.), PI 664498 (S. oleracea L.), PI 478393 (S. oleracea L.), PI 647862 (S. turkestanica Ilj.), PI 604792 (S. turkestanica Ilj.), PI 494751 (S. turkestanica Ilj.), PI 647861 (S. tetrandra Stev.), PI 677114 (S. tetrandra Stev.), and PI 608712 (S. tetrandra Stev.). The plant name and origin of plant specimen are listed in Supplementary Table 1.
Samples were planted in a garden field of Henan Normal University. The fresh leaves from male and female plants from each species were collected in flowering phase and DNA was extracted using traditional cetyl trimethylammonium bromide method (Rogers and Bendich, 1988).
Next-Generation Sequencing
We prepared three DNA aliquots (10 μL; ∼2 μg/μL) of each sample in parallel [samples 1–17; Sp-OL-M-1 (PI 527332), Sp-OL-F-1 (PI 527332), Sp-OL-M-2 (PI 664498), Sp-OL-F-2 (PI 664498), Sp-OL-M-3 (PI 478393), and Sp-OL-F-3 (PI 478393) from S. oleracea; Sp-TU-M-1 (PI 647862), Sp-TU-F-1 (PI 647862), Sp-TU-M-2 (PI 604792), Sp-TU-F-2 (PI 604792), Sp-TU-M-3 (PI 494751), and Sp-TU-F-3 (PI 494751) from S. turkestanica; Sp-TE-M-1 (PI 647861), Sp-TE-F-1 (PI 647861), Sp-TE-M-2 (PI 677114), Sp-TE-M-3 (PI 608712), and Sp-TE-F-3 (PI 608712) from S. tetrandra]. Paired-end libraries with insert sizes of 400 bp were constructed using TruSeq Library Construction Kit (Illumina®) and sequenced by an Illumina HiSeq4000 platform (LC⋅BioTech, Hangzhou, China). Then 20 Gb raw data sequencing reads were generated and trimmed using the FASTX Toolkit, and merged using the “fastqjoin” software with default Settings.
Repetitive DNA Characterization and Identification
RepeatExplorer pipeline2 (Novak et al., 2010, 2013) was used to detect and characterize the repetitive DNA families based on the NGS data of spinach and raw data (SRR952972) of the sugar beet genome from NCBI database (Dohm et al., 2014). Clustered read repeats were identified by De novo Cluster analysis using a graph-based approach.
Satellite DNA Identification
The TAREAN tool in RepeatExplorer was used to identify satellite DNA (Novak et al., 2017). Satellites were automatically detected based on the parameters “Connected component index (C)” and “Pair completeness index (P).” The identified satellites with C and P index close to one were defined as high-confidence satellites and used to develop FISH probes.
Fluorescence in situ Hybridization and Microscopy Imaging
Oligonucleotide probes were designed from DNA elements identified by TAREAN tool. For monomers of satellite DNA, 50 bp nucleotides was randomly selected and labeled directly with Texas Red-X (Invitrogen, Shanghai, China) during synthesis. 45S rDNA was labeled with Chroma Tide Alexa Fluor 488-5-dUTP (Invitrogen) to identify corresponded chromosomes.
Chromosome spreads from root tips were prepared as described from previous studies (Deng et al., 2012). Briefly, selected slides were cross-linked under UV for 2 min. The probe solution (20 ng/μL of each probe in 2 × saline-sodium citrate and 1 × Tris–ethylenediaminetetraacetic acid buffer) was denatured in boiling water for 5 min and kept on ice before use. Six μL of the denatured probe solution was added to each slide, heated for 5 min at 100°C, and left overnight at 55°C in a humid chamber. The slides were washed in 2 × saline-sodium citrate, mounted using the Vectashield mounting medium (containing 1.5 μg/mL 4’, 6-diamidino-2-phenylindole; Vector Laboratories, Burlingame, United States). FISH images were captured by ANDOR CCD under an Olympus BX63 fluorescence microscope.
Statistical Analysis
Each sample was validated with three replications. Multiple comparisons were performed to evaluate the difference between data groups by using SPSS software. Data were analyzed using least significance difference (LSD), and the significance level is 0.05.
Results
Repeat Proportion in Spinach and Sugar Beet Genome
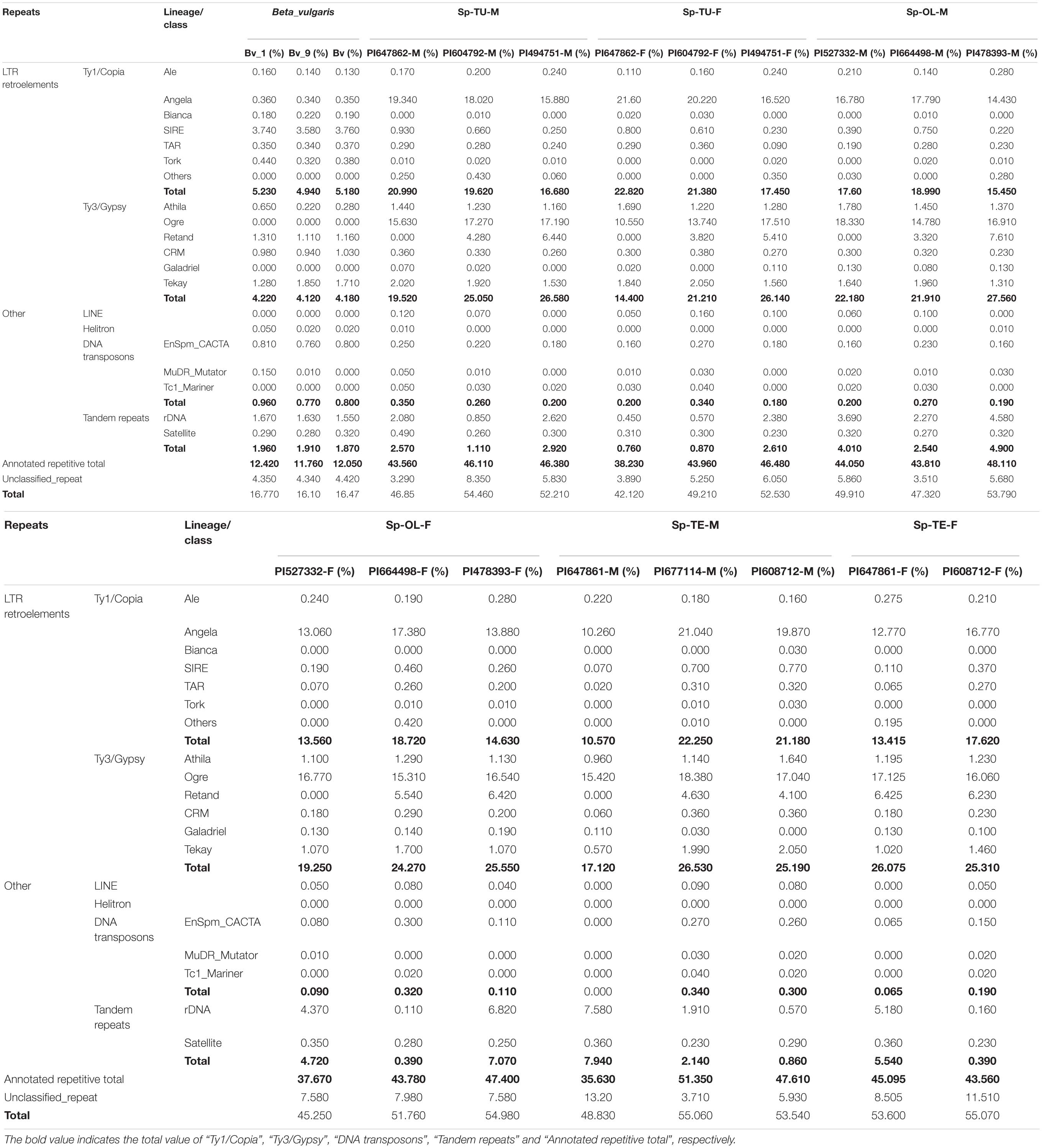
NGS generated 20 Gb of raw sequencing reads (Illumina 300 bp paired end) for each sample from S. oleracea, S. turkestanica, and S. tetrandra with an average 44.43% of guanine-cytosine (GC) content (Supplementary Table 2). The raw reads (SRR952972) of sugar beet genome were downloaded from NCBI3. For gragh-based clustering of reads, 800–1,000 Mb subsets were analyzed using RepeatExplorer. We estimated the repeat proportions of the genome of Spinacia species and sugar beet through comparative clustering in Repeat Explorer. As a result, the proportions of combined repeats identified in each species ranged from 16.45 to 54.34% of their respective estimated genome size (Table 1 and Supplementary Table 3). Interestingly, the proportion of repeat in the spinach genome is much higher than that in sugar beet (Table 1 and Figure 1A). The repeats identified were further classified by cluster shape and sequence similarities. As shown in Table 1 and Figure 1B, the long terminal repeat (LTR) retrotransposons Ty3/Gypsy and Ty1/Copia are the main forms of the repetitive elements in spinach (23.31 and 17.69%, respectively) and sugar beet (4.17 and 5.17%, respectively); tandem repeats, including rDNA and satellites, make up nearly 3.02 and 1.91% of the spinach genome and sugar beet genome, respectively. Nevertheless, DNA transposons only make up 2.12 and 0.84%, respectively. The repeat elements with the lowest proportions are LINE and Helitron, which are less than 0.1% of the spinach and sugar beet genomes. Among these repeats, the proportions of Ty3/Gypsy and Ty1/Copia were significantly enriched in the spinach genome, compared with that of sugar beet. Thus, Ty3/Gypsy and Ty1/Copia may be accounted for the increasing proportion of repeats in the spinach genome.
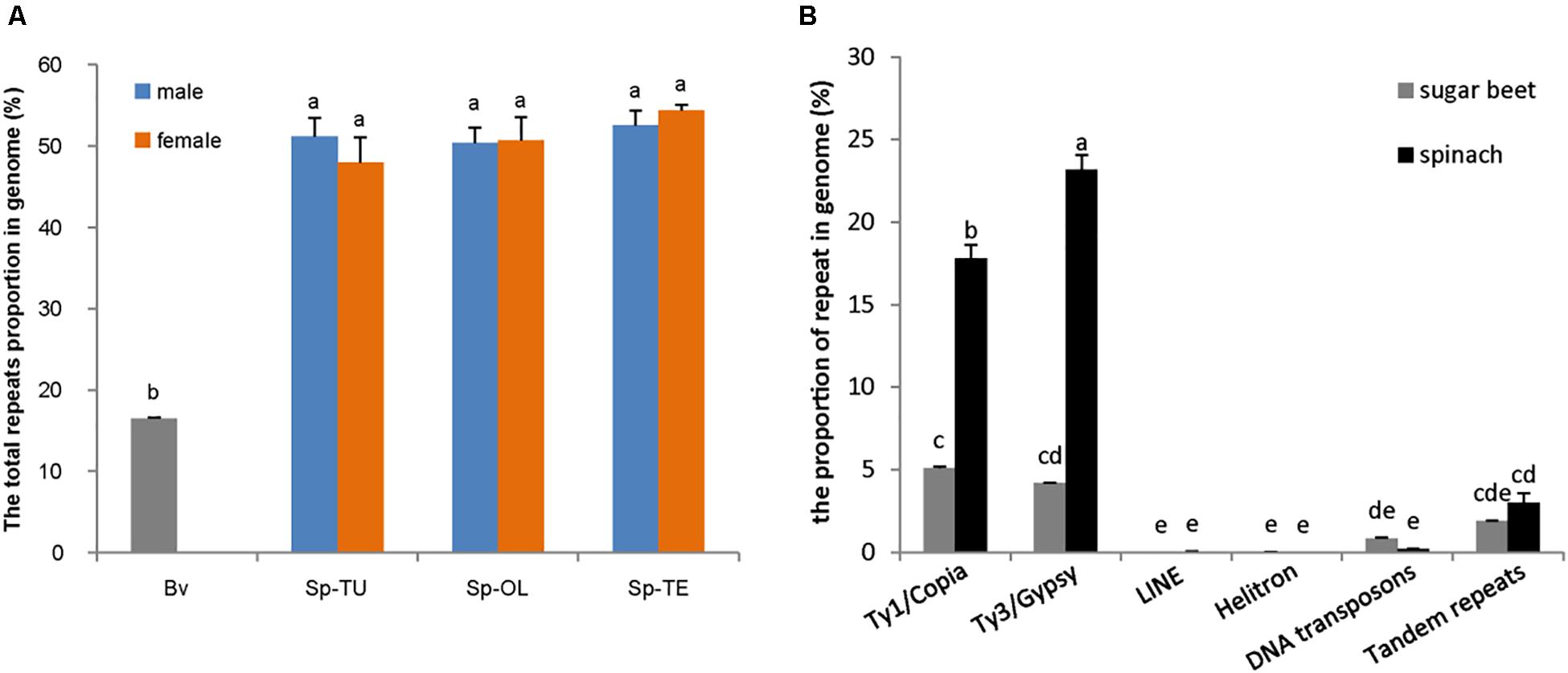
Figure 1. Comparison of repetitive sequence between spinach and sugar beet genome. (A) The total repeats proportion in spinach and sugar beet genome; Bv, sugar beet; Sp-TU, S. turkestanica; Sp-OL, S. oleracea; Sp-TE, S. tetrandra. (B) The repeat proportion in spinach and sugar beet genome. “a, b, c, d, e” means significance level by multiple comparison, p < 0.05.
For the Ty1/Copia repeats, the proportion of Angela was significantly higher in spinach than that in sugar beet, whereas the proportion of Bianca, SIRE, and TORK was significantly lower in spinach than that in sugar beet (Figure 2). For Ty3/Gypsy repeats, the proportion of Athila and Ogre are pronounced, while that in CRM was lower in spinach than that in sugar beet (Figure 2). Accordingly, Angela, Athila, and Ogre may contribute to the increasing proportion of repeats in the spinach genome.
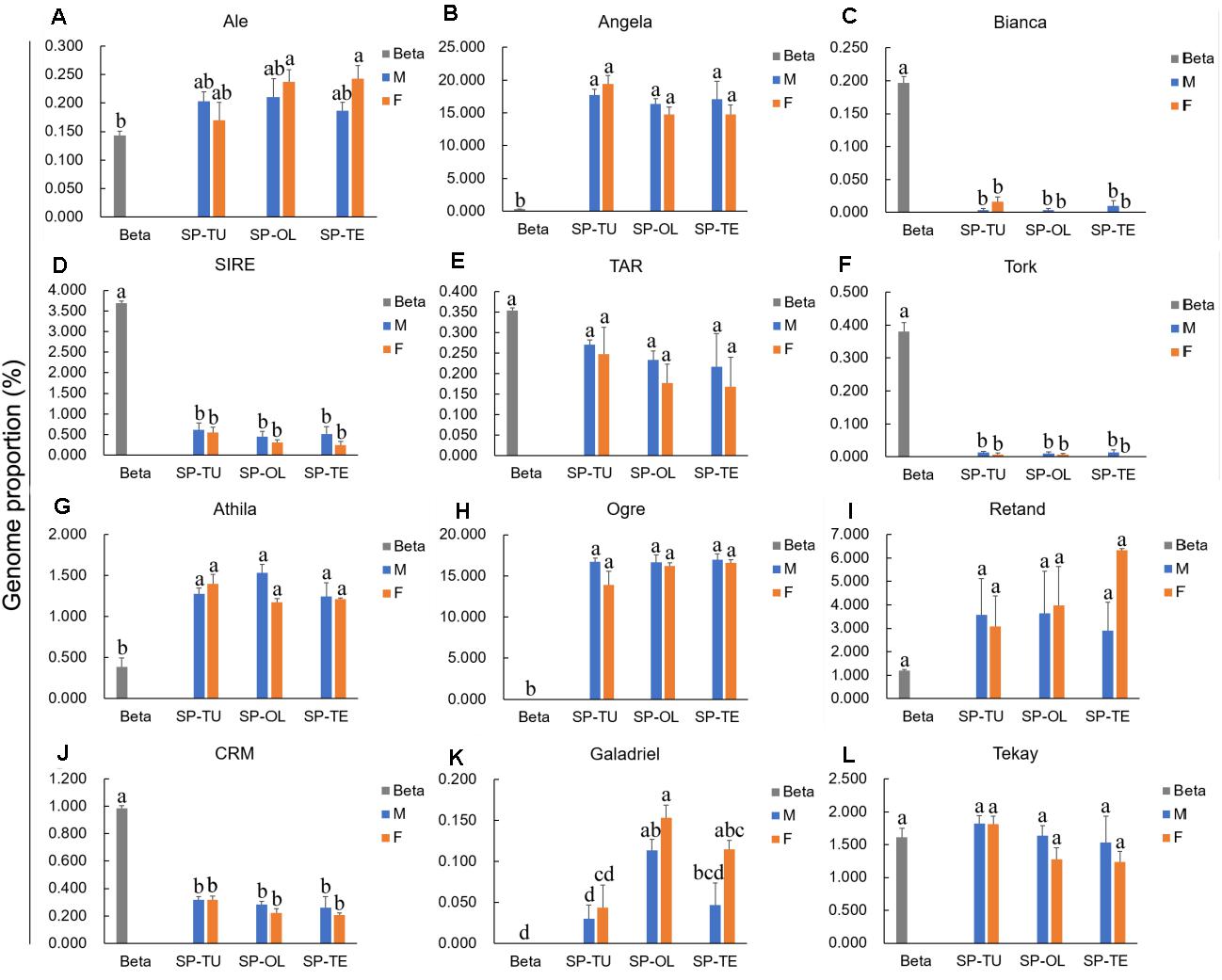
Figure 2. Comparison of Ty1/Copia and Ty3/Gypsy lineages proportion between spinach and sugar beet genome. (A–F) The proportion of Ty1/Copia repeats Ale, Angela, Bianca, SIRE, TAR and Tork in spinach and sugar beet genome. (G–L) The proportion of Ty3/Gypsy repeats Athila, Ogre, Retand, CRM, Galadriel and Tekay in spinach and sugar beet genome. “a, b, c, d,” means significance level by multiple comparison, p < 0.05.
Repeat Proportion in S. oleracea, S. turkestanica, and S. tetrandra Genome
Repetitive sequences are closely related to sex chromosome evolution. In the present, one S. tetrandra sample (PI647861) has heteromorphic sex chromosomes, while the sex chromosomes of the two other S. tetrandra samples and all S. oleracea and S. turkestanica samples are homomorphic. Assessment of the repeats proportions of genomes of Spinacia species revealed a conserved pattern between male and female genome of S. oleracea and S. turkestanica (Figure 2). However, in S. tetrandra, the proportion of several repeat lineages was significantly different between male and female genomes (Figure 3). For PI608712 with homomorphic sex chromosomes, the proportion of Ale and Retand was higher in the female genome than in the male genome, while the proportion of Angela, Bianca, CRM, Tekay, LINE, EnSpm_CACTA, and rDNA was higher in male genome than in the female genome (Figure 3). For PI647861 with heteromorphic sex chromosomes, the proportion of Angela, Athila, Ogre, CRM, Tekay, and EnSpm_CACTA was higher in the female genome than in the male genome, whereas the proportion of rDNA was higher in the male genome than in the female genome (Figure 4). Among these divergent repeat lineages, Angela, CRM, Tekay, EnSpm_CACTA, and rDNA are shared by PI608712 and PI647861, but an opposite trend of the shared repeats except rDNA is found between PI608712 and PI647861 (Figure 3). The proportion of rDNA is higher in male genome than in the female genome of PI608712 and PI647861. In addition, Ale, Retand, Bianca, and LINE are differentially abundant only in PI608712, while Athila and Ogre are differentially abundant only in PI647861.
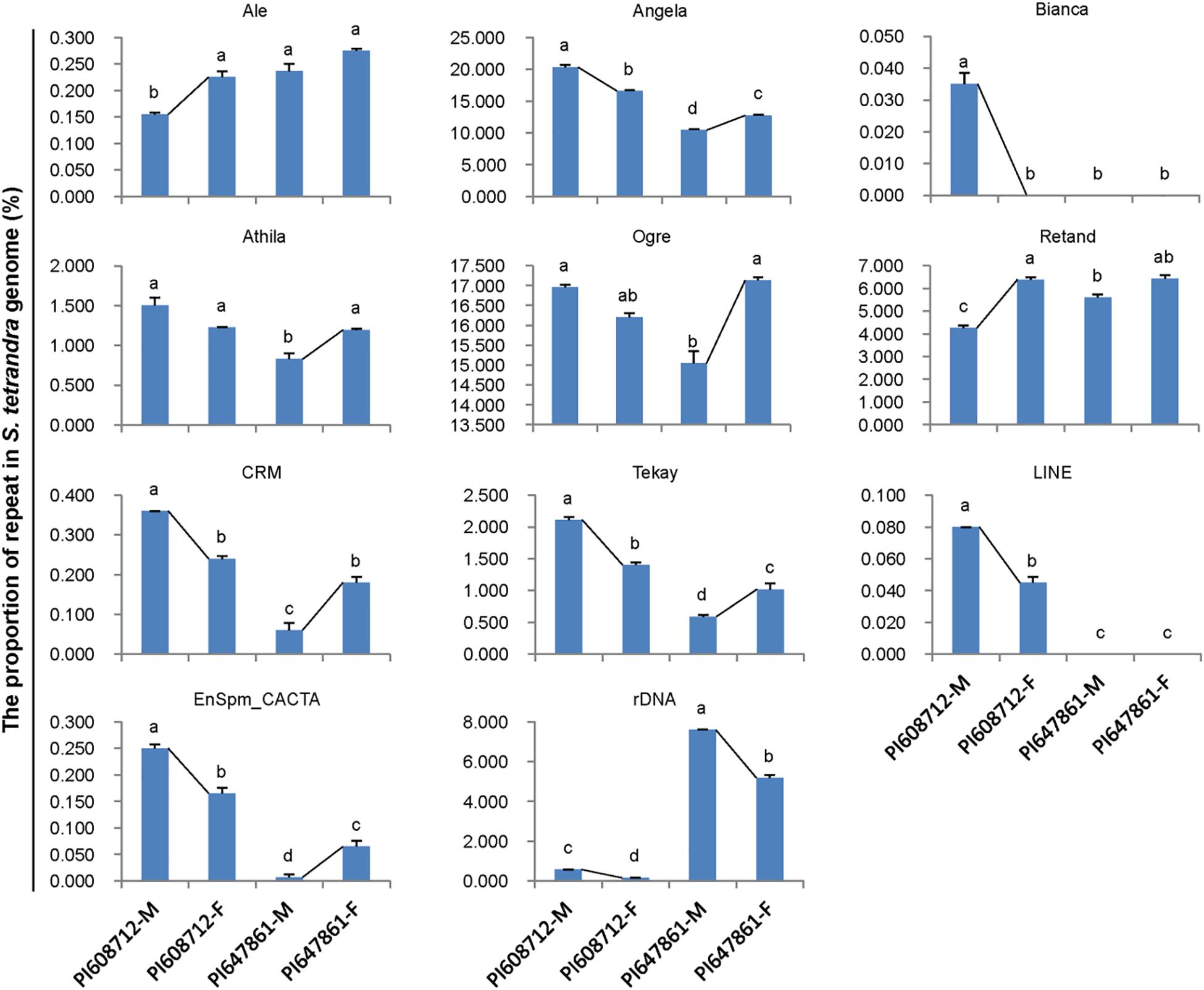
Figure 3. The differentially abundant repeats between male and female genome in two accessions of S. tetrandra. PI608712 with homomorphic sex chromosomes, PI647861 with heteromorphic sex chromosomes; PI608712-F, female genome of PI608712; PI608712-M, male genome of PI608712; PI647861-F, female genome of PI647861; PI647861-M, male genome of PI647861; the black line linking two bars indicates the variation trend; “a, b, c, d,” means significance level by multiple comparison, p < 0.05.
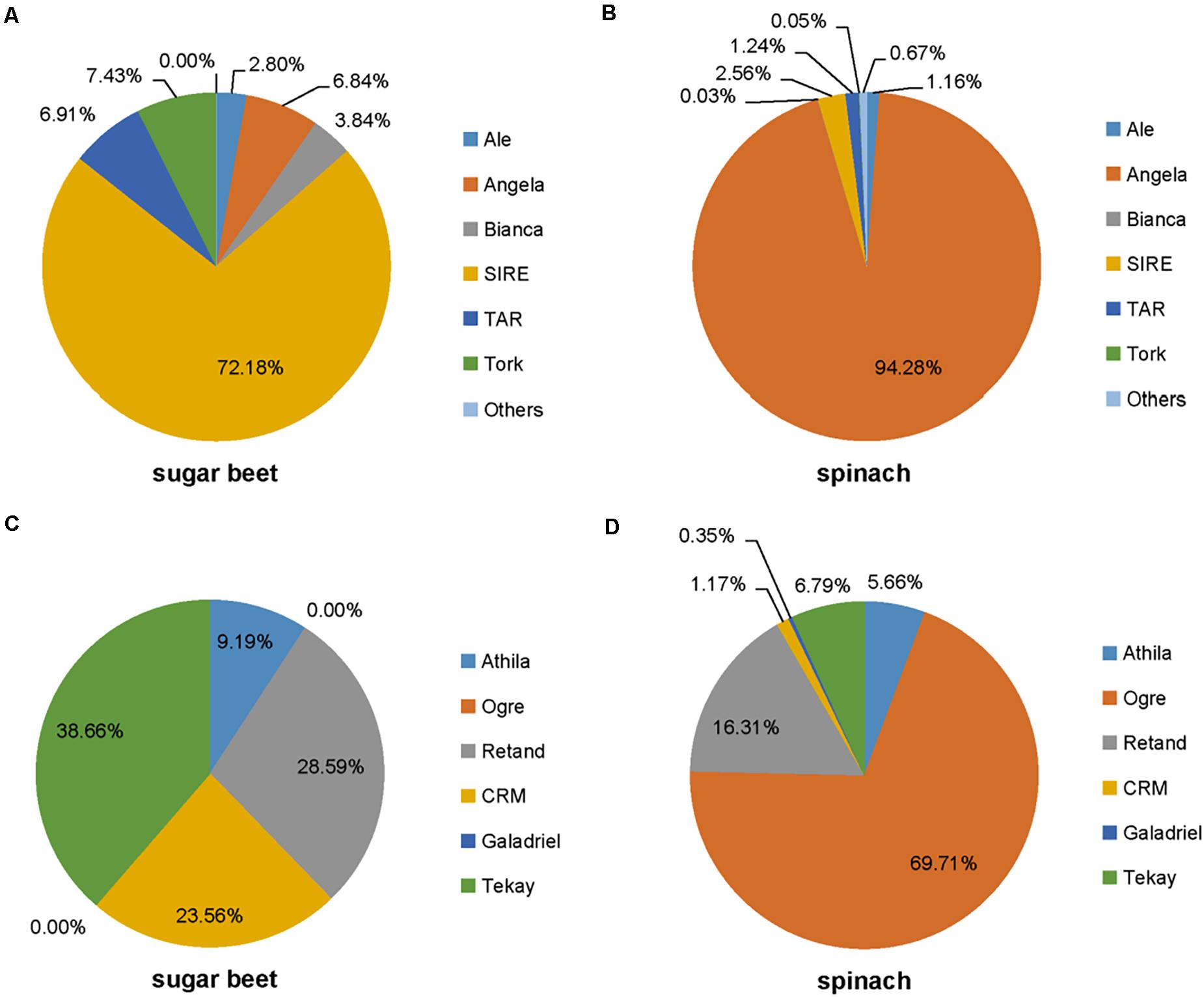
Figure 4. The abundance of each lineage in Ty1/Copia class (A,B) and Ty3/Gypsy class (C,D) in the spinach and sugar beet genomes.
Comparative Analysis of Interspersed Repeats
The repetitive element portions of genomes from sugar beet and spinach are mainly contributed by LTR retrotransposons (Table 1 and Figure 1B). In Spinacia species, Angela is the predominant lineage among Ty1/Copia elements, accounting for more than 94.28%; in B. vulgaris, SIRE lineage accounts for the majority (72.18%) of Ty1/Copia elements (Figure 5). For Ty3/Gypsy elements, Ogre is the most common type of lineages (69.71%) in Spinacia species; in B. vulgaris, Retand, and Tekay lineages form a relatively large proportion, accounting for 28.59 and 38.66%, respectively (Figure 4). Moreover, several LTR variants, namely, Ogre, Galadriel, LINE, and Tc1_Mariner exclusively appear in Spinacia species; however, Helitron only appears in B. vulgaris (Table 1 and Supplementary Table 4).
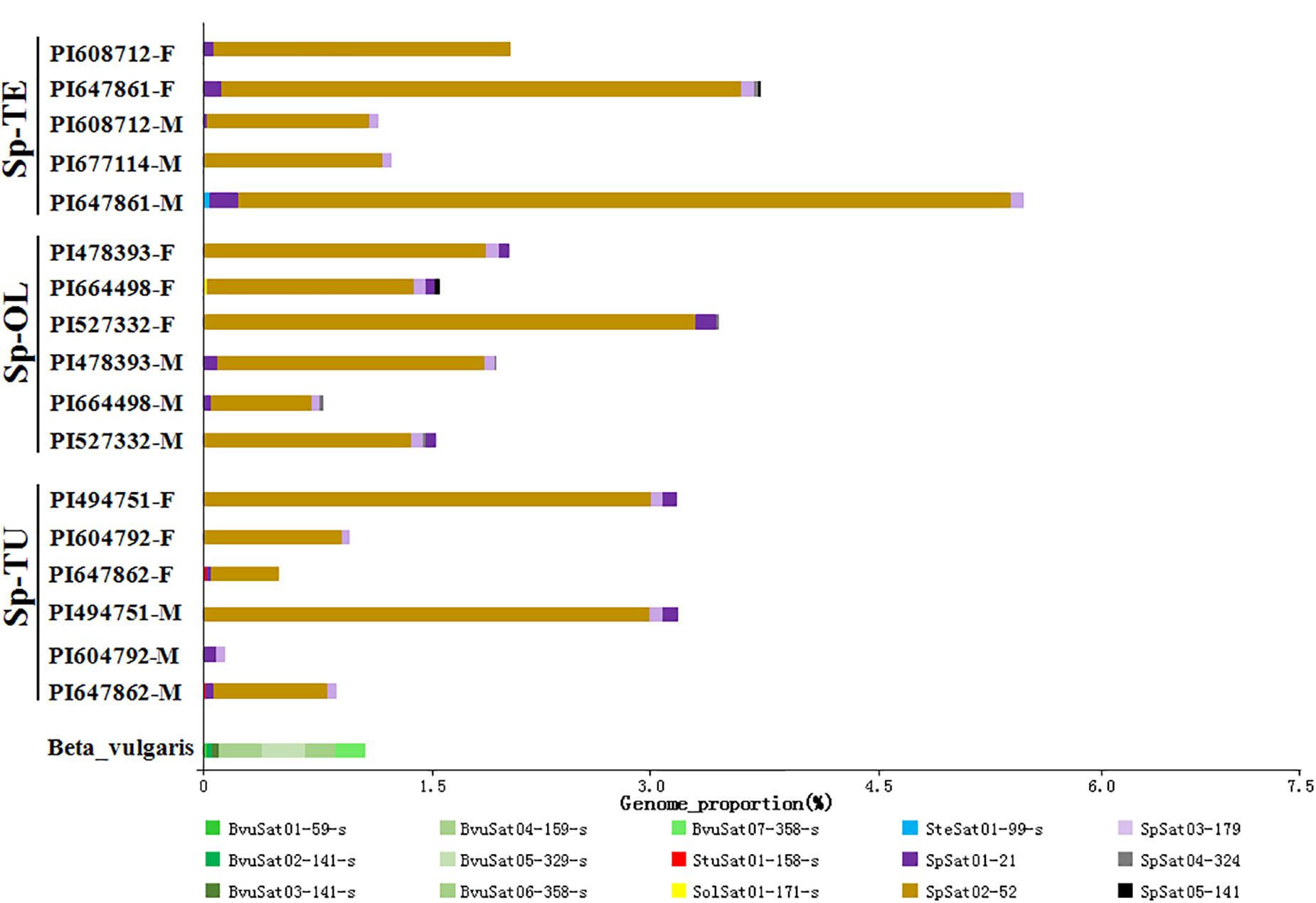
Figure 5. Estimated satellite abundance in spinach and sugar beet genome. The size of the rectangle is proportional to the number of reads in a cluster for each species. Colors of the rectangles correspond to repeat type. See Table 1 for species codes.
Comparative Analysis of Major Groups of Satellite
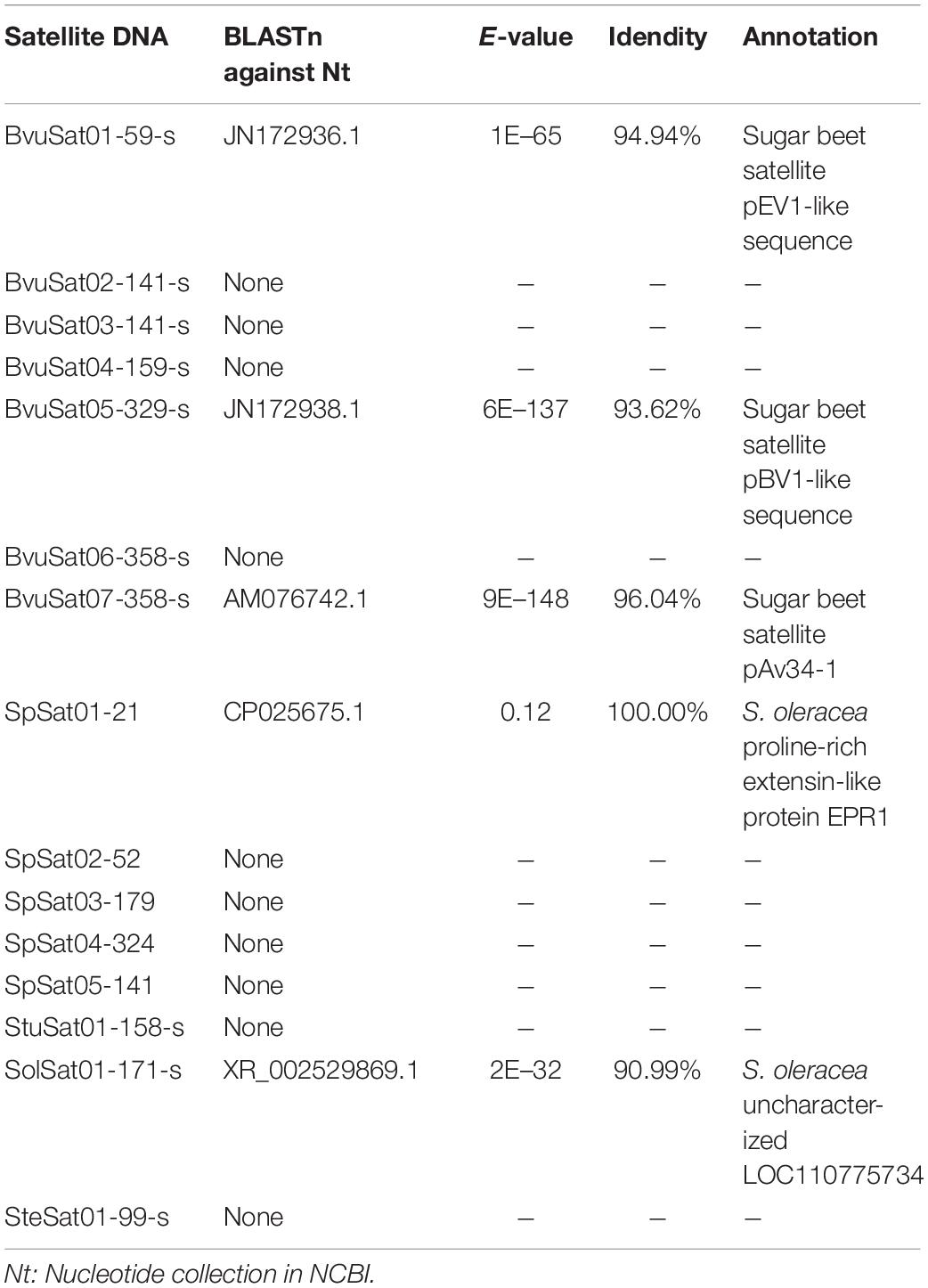
The proportion of satellite repeats was found approximately 0.3% in both the spinach and sugar beet genomes (Table 1), and the satellites which detected by RepeatExplorer and TAndem REpeat ANalyzer (TAREAN) are summarized in Supplementary Table 5. A total of 15 satellite DNA families were found (Figure 5 and Supplementary Table 5). The monomer size of the satellite DNAs is broad and ranged from 21 to 358 bp, among the four species (Supplementary Table 6). Subsequent analyses revealed that BvuSat01-59-s (MN918718), BvuSat02-141-s (MN918719), BvuSat03-141-s (MN918720), BvuSat04-159-s (MN918721), BvuSat05-329-s (MN918722), BvuSat06-358-s (MN918723), and BvuSat07-358-s (MN918724) were only present in the B. vulgaris genome. Previously, many satellite DNAs have been characterized in the sugar beet genome. Herein, we blasted the satellite DNAs identified in this work against these in GenBank (Table 2). BvuSat01-59-s, BvuSat02-141-s, BvuSat03-141-s, and BvuSat06-358-s had no similarities with other known sequences (Table 2). However, BvuSat04-159-s, BvuSat05-329-s, and BvuSat07-358-s shared a higher similarity (94.94, 93.62, and 96.04%) with sugar beet satellite pEV1-like sequence (JN172936.1), pBV1-like sequence (JN172938.1), and pAv34-1 (AM076742.1), respectively (Table 2). Hence, among these seven satellite DNAs identified in the sugar beet genome, four were novel DNAs in current work. In Spinacia genomes, eight satellite DNAs were identified (Figure 5 and Table 2). Five satellite DNAs, SpSat01–21 (5’-GCTATCGGCACCCGCCAACTA-3’), SpSat02-52 (MN918725), SpSat03-179 (MN918726), SpSat04-324 (MN918727), and SpSat05-141 (MN918728), were found in three Spinacia species (Figure 5). However, StuSat01-158-s (MN918729) was only found in the S. turkestanica genome; SolSat01-171-s (MN918730) was unique in the S. oleracea genome; SteSat01-99-s (MN918731) was only found in the S. tetrandra genome. Among the common satellite DNAs in Spinacia species, SpSat02-52 is the most abundant DNA.
Chromosomal Localization of Satellite DNAs
Fluorescence in situ hybridization (FISH) was used to analyze the chromosomal localization of the 15 satellites identified above in B. vulgaris, male S. oleracea and female S. oleracea mitotic metaphase chromosomes (Figures 6, 7). The hybridization revealed that BvuSat01-59-s, BvuSat02-141-s, and BvuSat03-141-s, are localized to the telomeric regions of a pair of chromosome (Figures 6A–C); BvuSat04-159-s is located in the intercalary positions of each chromosome-arm (Figure 6D); BvuSat05-329-s and BvuSat06-358-s are associated with the centromere (Figures 6E,F), and BvuSat07-358-s occurs on most chromosome ends with signals on the centromere of each chromosome (Figure 6G). However, all seven satellite DNA sequences identified in sugar beet did not show fluorescence signal when hybridized with the spinach mitotic metaphase chromosomes. For the eight spinach satellite DNA sequences (Figure 7), SpSat01-21 was localized to the telomeric regions of the sex chromosome (Chr 1) and the long arm of Chr 2, 3, 4, 5, and 6; SpSat02-52 was mostly concentrated in the telomeric regions of the Chr 3 and the long arm of Chr 4; hybridization signal of SpSat03-179 was mostly enriched on the telomeric regions of the sex chromosome (Chr 1) and the long arm of Chr 2, 3, and 6; SpSat04-324 and SpSat05-141 are featured by the distribution of hybridization signals on the telomeric region of the sex chromosome (Chr 1) short arm; the signals of StuSat01-158-s and SolSat01-171-s were found on the telomeric regions of the sex chromosome (Chr 1) and the long arm of Chr 2, 5, and 6; and the signals of SteSat01-99-s concentrated on the long arm of Chr 2 and 5. Signals of these eight satellites have notable difference between male and female S. oleracea mitotic metaphase. No fluorescence signals were found from hybridizations between eight satellite DNA sequences and B. vulgaris mitotic metaphase chromosomes, indicating that these repeats are Spinacia species specific. Satellite DNA sequences (SpSat01-21, SpSat02-52, SpSat03-179, SpSat04-324, and 45S rDNA) were used as probes, and sequential FISH was used to establish accurate spinach karyotype (Figure 8A). These four satellite clusters featured a star-like or circular graph topology (Supplementary Figures 1, 2). As shown in Figures 8B,C, Chr 1 pair: SpSat01-21 and SpSat03-179 on the telomeric region of both arms and SpSat04-324 on the telomeric region of short arm; Chr 2 pair: SpSat01-21 and SpSat03-179 on the telomeric region of long arm and 45S rDNA on the telomeric region of short arm; Chr 3 pair: SpSat02-52 on the telomeric region of both arms, SpSat01-21 and SpSat03-179 on the telomeric region of short arm; Chr 4 pair: SpSat01-21 and SpSat02-52 on the telomeric region of long arm; Chr 5 pair: SpSat01-21 on the telomeric region of long arm and 45S rDNA on the telomeric regions of short arm; Chr 6 pair: SpSat01-21 and SpSat03-179 on the telomeric region of long arm and 45S rDNA on the telomeric region of short arm. Thus, distribution characters of eight satellite DNAs are different among each chromosome pair, which can be used as DNA markers for accurate karyotype of spinach.
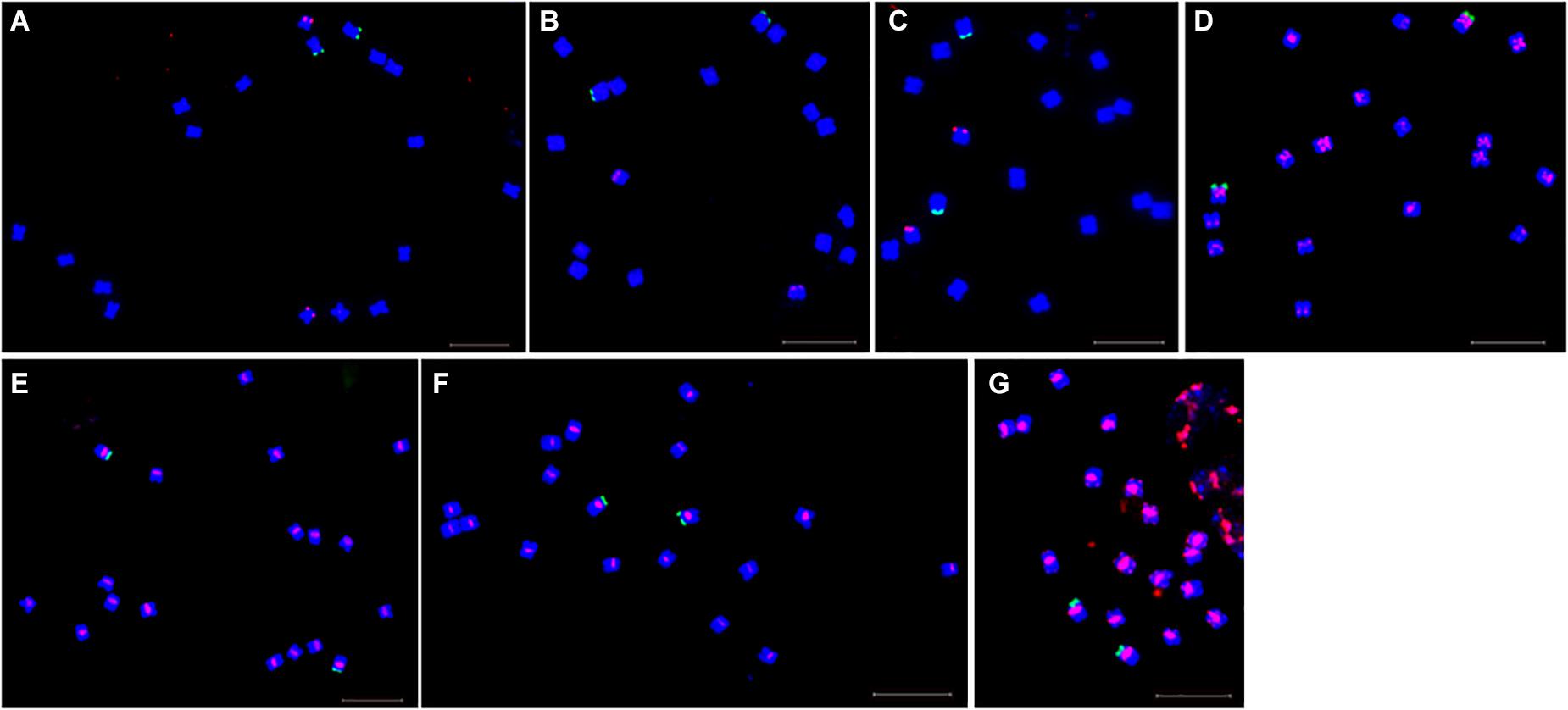
Figure 6. Localization of seven satellites on metaphase chromosomes of B. vulgaris using fluorescence in situ hybridization. Blue are DAPI stained chromosomes, green signals show chromosomal localization of 45 rDNA, The satellite DNAs were labeled with Texas red (red signal). (A) Probe BvuSat01-59-s (red). (B) Probe BvuSat02-141-s. (C) Probe BvuSat03-141-s. (D) Probe BvuSat04-159-s. (E) Probe BvuSat05-329-s. (F) Probe BvuSat06-358-s, (G) Probe BvuSat07-358-s. Bar indicates 10 μm.
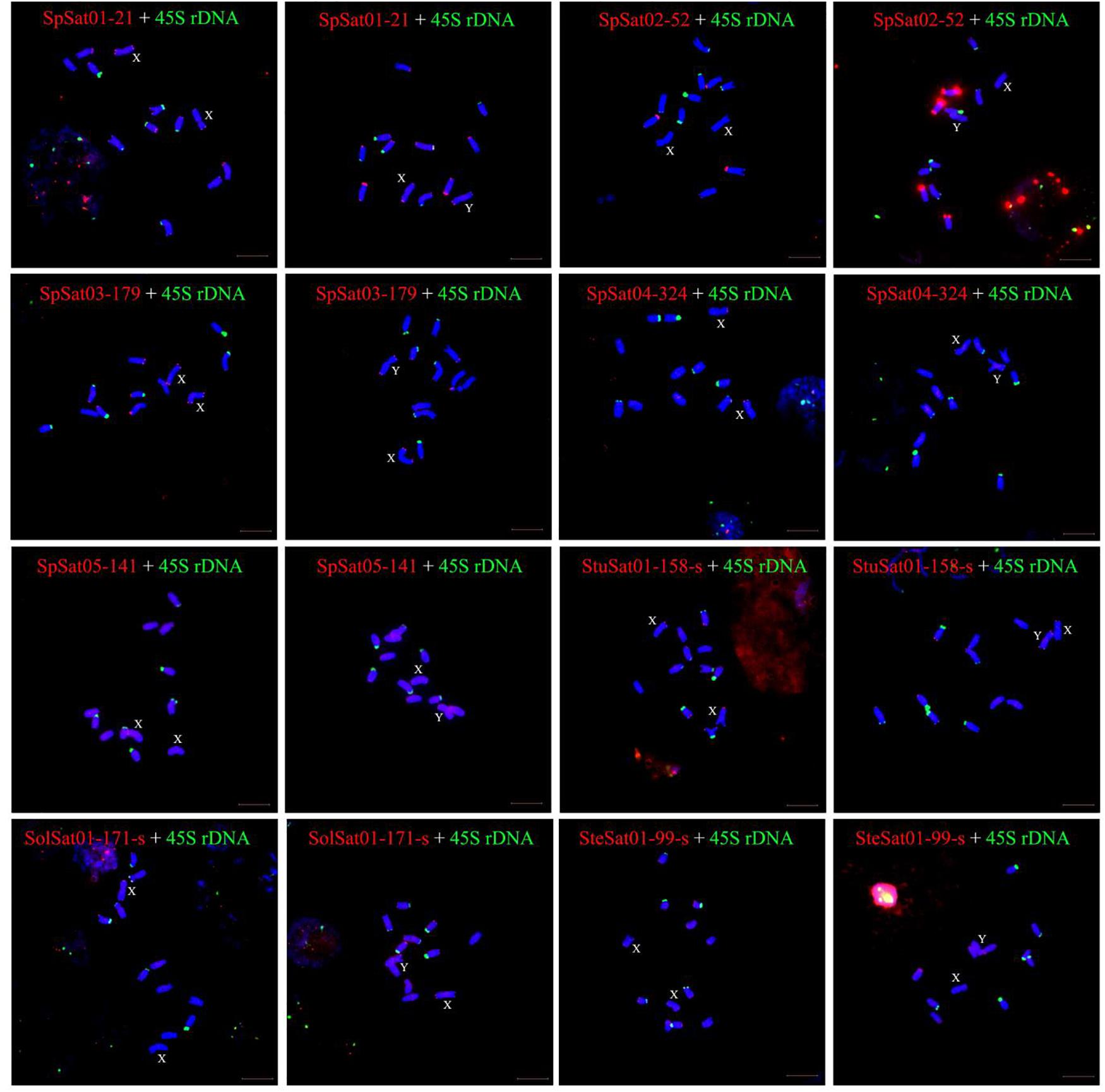
Figure 7. Localization of eight satellites on metaphase chromosomes of spinach using fluorescence in situ hybridization. The name of satellites and sex of individual are indicated inside each figure. Blue are DAPI stained chromosomes, the satellite DNAs were labeled with Texas red (red signal), and green signals show chromosomal localization of satellites. Bar indicates 10 μm.
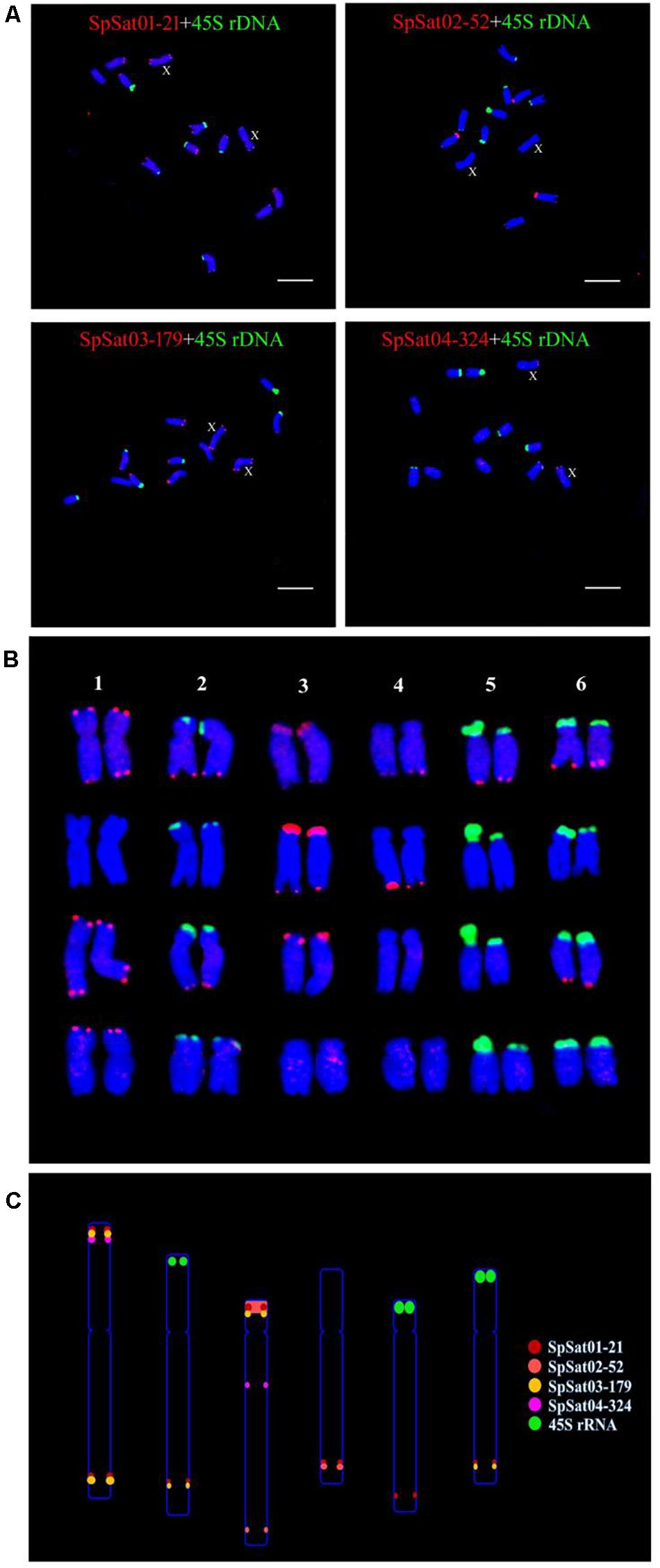
Figure 8. Karyotype and ideograph for spinach mitotic metaphase chromosomes. (A) FISH analysis of SpSat01-21, SpSat02-52, SpSat03-179, and SpSat04-324 (red), and 45S rDNA (green) on mitotic chromosomes. Arrows indicate the sex chromosomes. (B) Karyotype analysis of spinach based on the size and FISH signal pattern. (C) Ideogram shows the positions of SpSat01-21 (red), SpSat02-52 (yellow), SpSat03-179 (pink), SpSat04-324 (purple), and 45S rDNA (green). Bars = 10 μm.
Discussion
Genome Size Variation and Repeat Content
A recent sequenced draft sugar beet reference genome comprised 567 Mb, 42.3% of which were identified as repetitive sequences (Dohm et al., 2014). The spinach draft genome size is approximately 996 Mb, with 74.4% repetitive sequences (Xu et al., 2017). However, De novo sequencing only identified 16.45% of the total sugar beet genome (Organelle genome excluded) as repetitive DNA sequences; in Spinacia species genome, the repetitive sequences proportion is 49.56, 50.50, and 53.41%, respectively. The repeats proportion in our results is lower than that reported in previous studies, which might be attributed to the different methods for repeats annotation. Nevertheless, our results were consistent with Kudoh group’s report, which revealed a significantly higher proportion of repeats in Spinacia species compared with B. vulgaris (Kudoh et al., 2018). Moreover, the proportion of the LTR retrotransposon Ty3/Gypsy and Ty1/Copia was obviously higher in spinach genome than that in sugar beet genome (Figure 1B). This result is consistent with that LTR-retrotransposons may be related to genome size variation between the close plant relatives (Bennetzen et al., 2005; Kowar et al., 2016).
Sex Chromosome Evolution and Repeat Elements
In Spinacia, sex chromosomes among three of the S. tetrandra accessions are heteromorphic but the other Spinacia species are homomorphic s; thus, spinach provides a good opportunity for studying transition of homomorphic sex chromosomes to heteromorphic sex chromosomes The abundance of repetitive sequence around sex-determining loci can suppress the homologous recombination of sex-determining loci between X and Y chromosomes, and finally form the male-specific region (Li et al., 2016). Hence, variations of repeats proportions between female and male genomes can reflect differences between X and Y chromosomes. To understand the roles of repetitive DNA in spinach sex chromosome evolution, we characterized the differentially accumulated repeats elements in male and female spinach. Comparison of the repeats proportion between female and male genomes of three Spinacia species indicated that the difference was present in S. tetrandra samples but not in the S. oleracea or S. turkestanica samples. Herein, we analyzed two accessions of S. tetrandra, PI608712 with homomorphic sex chromosomes and PI647861 with heteromorphic sex chromosomes. S. oleracea and S. turkestanica samples with homomorphic sex chromosomes are at the early stage of sex chromosome evolution, which may result negligible differences of repeat proportion between female and male genome. However, in S. tetrandra, several repeats (Ale, Retand, Angela, Bianca, CRM, Tekay, LINE, EnSpm_CACTA, and rDNA) exhibit different abundances between the female and male genomes of PI647861 although its sex chromosomes are homomorphic (Figure 3). For PI647861, Angela, Athila, Ogre, CRM, Tekay, EnSpm_CACTA, and rDNA are differentially abundant repeats between female and male genomes (Figure 3). Among these differentially abundant repeats, only tandem repeat rDNA showed a higher proportion in male genome both of PI608712 and PI647861. We found that the proportions of Ale, Bianca, Athila, Ogre, Retand, and CRM were significantly different in the male genome but not in the female genome between PI608712 and PI647861 (Figure 4); compared with the male genome of PI608712, only the proportion of Ale and Retand was increased in the male genome of PI647861. We presumed that repetitive DNA sequences are correlated with the formation of sex chromosomes and the transition from homomorphic sex chromosomes to heteromorphic sex chromosomes in Spinacia species. The sex determination region (SDR) of spinach was identified (Yu et al., 2020). Meanwhile, k-mer analysis was useful in identifying the sex determination region in persimmons (Akagi et al., 2014) and date palm (Torres et al., 2018) efficiently. In the future, we will use this method to clone the SDR of male and female species of Spinacia and explore the function of repetitive DNA sequences on sex chromosome evolution in spinach.
Interspersed Repeats
The LTR retroelements, particularly the Ty3/Gypsy class, are the major contributions to the repetitive sequences in spinach genome (Xu et al., 2017). Based on the sequence composition of the largest 18 superclusters, superclusters 8 (Ogre), 11 (Galadriel), 13 (LINE), and 17 (MuDR_Mutator) only appear in Spinacia species and not in sugar beet. Thus, we proposed that insertion frequency of these elements, especially Ogre (abundant in female and specific to spinach), may be correlated with the mating system. Spinacia species are commonly dioecious and their offspring are obtained by outcrossing among different sexes. In B. vulgaris, diploid species are hermaphroditic. The single cluster Helitron was only found in B. vulgaris and not in Spinacia species. Helitron, a rolling-circle DNA transposon, plays an important role in plant evolution. As reported in Brassica napus, function loss of BnSP11-1, the self-incompatibility male determining gene, is due to the insertion of a 3.6 kb non-autonomous Helitron transposon into this gene promoter (Gao et al., 2016). Hence, further experiments will be needed to study the role of Ogre and Helitron transposon in spinach sex chromosome evolution.
Satellite Repeats and Sex Chromosome Evolution of Spinach
Satellite accumulation accompanies sex chromosomes evolution in some plant species with heteromorphic sex chromosomes. Compared with mammalian sex chromosomes that evolved 300 Mya, sex chromosomes in plants are incipient which ranges from 3 to 13 Mya (Navajas-Perez et al., 2005; Sousa et al., 2013; Kubat et al., 2014). In S. latifolia, TRAYC and STAR-Y satellites accumulated on the Y chromosome (Hobza et al., 2006, 2007). In Rumex acetosa, RA160 and RA690 satellites are enriched X and Y chromosomes, respectively (Steflova et al., 2013). Seabuckthorn (Hippophae rhamnoides) has a large X chromosome and a small Y chromosome; HRTR8 satellite is specific to the X chromosome, while HRTR12 satellite is Y-specific (Puterova et al., 2017). Satellites accumulate not only in the heterochromatic regions of the sex chromosomes but also in the euchromatic region. A model has revealed the accelerated spread of TEs by microsatellite targeting. However, the rate of satellites spreading during sex chromosome evolution and their roles in heterochromatinization process remain unclear. In Spinacia, the genome size and karyotype of S. tetrandra are clearly distinct from those of two Spinacia members. The sex chromosome pairs of three S. tetrandra accessions are heteromorphic, while sex chromosomes of the other Spinacia members are homomorphic; In the present study, the signals of six satellites (SpSat01-21, SpSat03-179, SpSat04-324, SpSat05-141, StuSat01-158-s, and SolSat01-171-s) were located on chromosome 1 (Figure 7). Our previous study showed that two satellites, namely, Spsat2 (has similar hybridization signal but different sequences with SpSat01-21) and Spsat3 provide hybridization signals on chromosome 1 (Li et al., 2019). Chromosome 1 is a sex chromosome of spinach (Deng et al., 2012, 2013; Okazaki et al., 2019). The SDR, which is approximately 18.4 Mb, is located on the short arm of Chromosome 1 (Yu et al., 2020), as consistent as signals of six satellites we developed in current study (SpSat01-21, SpSat03-179, SpSat04-324, SpSat05-141, StuSat01-158-s, and SolSat01-171-s) (Figure 7). Searching DNA sequences of the sex satellites against the DNA sequences of SDR achieved 141 copies of SolSat01-171-s in SDR, and the standard cutoff is Percentage of identical matches ≥ 90% and E-value ≤ 3.61E–61 (Supplementary Table 7). Thus, we speculated that the accumulation of satellite DNA on the short arm of chromosome 1 may be involved in the sex chromosome evolution in Spinacia species. However, the signals of the eight satellites between male and female S. oleracea mitotic metaphase have notable differences (Figure 7). This result may be related to the early stages of sex chromosome evolution in spinach. The sex chromosomes were accompanied by insufficient recombination suppression induced by repetitive sequence insertions. In our future work, we will use the eight satellite DNA sequences to hybridize with mitotic metaphase chromosomes of S. turkestanica, S. oleracea, and S. tetrandra to study the role of satellites in the sex chromosome evolution.
In conclusion, the global distribution of the major repetitive sequence landscape in the male and female genome of Spinacia species was characterized using next-generation sequencing data. Compared with that of sugar beet, the genome of spinach showed an increased total proportion, which is largely attributed to the accumulation of LTR retrotransposon Ty3/Gypsy (especially Athila and Ogre) and Ty1/Copia (especially Angela). Compared with B. vulgaris, Spinacia species had a higher percentage of repeat elements potentially derived from a recent lineage-specific repeat burst. Among Ty1/Copia retro-transposable elements, Angela lineage was predominant, accounting for more than 94.97% of the Ty1/Copia elements in Spinacia species; SIRE lineage was more popular (> 71.44%) among Ty1/Copia elements in sugar beet. Distribution of Ty3/Gypsy retro-transposable elements showed that Ogre was the most common type (69.28%) in Spinacia species; Retand and Tekay lineages are the main types among Ty3/Gypsy elements in sugar beet, accounting for 28.5 and 38.6%, respectively. Development of seven sugar beet-specific and eight spinach-specific satellite DNA probes provided evidence of repetitive sequence divergence between two genera; furthermore, the six satellite DNA probe signals from Spinacia species were identified on the short arm of the sex chromosome by FISH. A total of 141 copies of SolSat01-171-s were present in SDR. Thus, we proposed that the accumulation of satellite DNA on the short arm of chromosome 1 may be involved in the sex chromosome evolution of Spinacia species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA608209/.
Author Contributions
NL, XL, and CD designed the experiments. NL, XL, JZ, and CD conducted the study, processed the data, and wrote the manuscript. LY, SL, YZ, RQ, WG, and CD discussed the results and revised the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the grants from the National Natural Science foundation of China (31770346, 31970240, 31470334, and 32000243), Postgraduate research and innovation project of Henan Normal University (YL201828), and the key scientific research project plan for Henan provincial colleges and universities (20A180016). The funding bodies had no role in the design of the study, the collection, analysis, interpretation of data, and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members who participated in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.575462/full#supplementary-material
Abbreviations
NGS, Next-Generation Sequencing; FISH, fluorescence in situ hybridization; LTR, Long terminal repeat; TAREAN, TAndem REpeat Analyzer; SDR, Sex determination region.
Footnotes
- ^ https://npgsweb.ars-grin.gov/gringlobal/query/query.aspx
- ^ https://repeatexplorer-elixir.cerit-sc.cz/galaxy/
- ^ https://www.ncbi.nlm.nih.gov/sra/?term=SRR952972
References
Akagi, T., Henry, I. M., Tao, R., and Comai, L. (2014). A Y-chromosome–encoded small RNA acts as a sex determinant in persimmons. Science 346, 646–650. doi: 10.1126/science.1257225
Arumuganathan, K., and Earle, E. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218. doi: 10.1007/bf02672069
Bennetzen, J., Ma, J., and Devos, K. (2005). Mechanisms of recent genome size variation in flowering plants. Ann. Botany 95, 127–132. doi: 10.1093/aob/mci008
Deng, C., Qin, R., Cao, Y., Gao, J., Li, S., Gao, W., et al. (2013). Microdissection and painting of the Y chromosome in spinach (Spinacia oleracea). J. Plant Res. 126, 549–556. doi: 10.1007/s10265-013-0549-3
Deng, C., Qin, R., Gao, J., Cao, Y., Li, S., Gao, W., et al. (2012). Identification of sex chromosome of spinach by physical mapping of 45s rDNAs by FISH. Caryologia 65, 322–327. doi: 10.1080/00087114.2012.760879
Dohm, J., Minoche, A., Holtgräwe, D., Capella-Gutiérrez, S., Zakrzewski, F., Tafer, H., et al. (2014). The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505, 546–549. doi: 10.1038/nature12817
Feng, R. J., Wang, X., Tao, M., Du, G. C., and Wang, Q. S. (2017). Genome size and identification of abundant repetitive sequences in Vallisneria spinulosa. Peerj 5:e3982. doi: 10.7717/peerj.3982
Fuentes-Bazan, S., Mansion, G., and Borsch, T. (2012a). Towards a species level tree of the globally diverse genus Chenopodium (Chenopodiaceae). Mol. Phylogenet. Evol. 62, 359–374. doi: 10.1016/j.ympev.2011.10.006
Fuentes-Bazan, S., Uotila, P., and Borsch, T. (2012b). A novel phylogeny-based generic classification for Chenopodium sensu lato, and a tribal rearrangement of Chenopodioideae (Chenopodiaceae). Willdenowia 42, 5–24. doi: 10.3372/wi.42.42101
Fujito, S., Takahata, S., Suzuki, R., Hoshino, Y., Ohmido, N., and Onodera, Y. (2015). Evidence for a common origin of homomorphic and heteromorphic sex chromosomes in distinct spinacia species. G3-Genes Genom. Genet. 5, 1663–1673. doi: 10.1534/g3.115.018671
Gaiero, P., Vaio, M., Peters, S. A., Schranz, M. E., de Jong, H., and Speranza, P. R. (2019). Comparative analysis of repetitive sequences among species from the potato and the tomato clades. Ann. Bot-London 123, 521–532. doi: 10.1093/aob/mcy186
Gao, C., Zhou, G., Ma, C., Zhai, W., Zhang, T., Liu, Z., et al. (2016). Helitron-like transposons contributed to the mating system transition from out-crossing to self-fertilizing in polyploid Brassica napus L. Sci. Rep. 6:33785.
Heitkam, T., Holtgräwe, D., Dohm, J., Minoche, A., Himmelbauer, H., Weisshaar, B., et al. (2014). Profiling of extensively diversified plant LINEs reveals distinct plant-specific subclades. Plant J. 79, 385–397. doi: 10.1111/tpj.12565
Hobza, R., Kejnovsky, E., Vyskot, B., and Widmer, A. (2007). The role of chromosomal rearrangements in the evolution of Silene latifolia sex chromosomes. Mol. Genet. Genom. 278, 633–638. doi: 10.1007/s00438-007-0279-0
Hobza, R., Kubat, Z., Cegan, R., Jesionek, W., Vyskot, B., and Kejnovsky, E. (2015). Impact of repetitive DNA on sex chromosome evolution in plants. Chromosome Res. 23, 561–570. doi: 10.1007/s10577-015-9496-2
Hobza, R., Lengerova, M., Svoboda, J., Kubekova, H., Kejnovsky, E., and Vyskot, B. (2006). An accumulation of tandem DNA repeats on the Y chromosome in Silene latifolia during early stages of sex chromosome evolution. Chromosoma 115, 376–382. doi: 10.1007/s00412-006-0065-5
Kadereit, G., Mavrodiev, E. V., Zacharias, E. H., and Sukhorukov, A. P. (2010). Molecular phylogeny of Atripliceae (Chenopodioideae, Chenopodiaceae): implications for systematics, biogeography, flower and fruit evolution, and the origin of C-4 photosynthesis. Am. J. Botany 97, 1664–1687. doi: 10.3732/ajb.1000169
Kowar, T., Zakrzewski, F., Macas, J., Kobližková, A., Viehoever, P., Weisshaar, B., et al. (2016). Repeat composition of CenH3-chromatin and H3K9me2-marked heterochromatin in sugar beet (Beta vulgaris). BMC Plant Biol. 16:120. doi: 10.1186/s12870-016-0805-5
Kubat, Z., Zluvova, J., Vogel, I., Kovacova, V., Cermak, T., Cegan, R., et al. (2014). Possible mechanisms responsible for absence of a retrotransposon family on a plant Y chromosome. New Phytol. 202, 662–678. doi: 10.1111/nph.12669
Kudoh, T., Takahashi, M., Osabe, T., Toyoda, A., Hirakawa, H., Suzuki, Y., et al. (2018). Molecular insights into the non-recombining nature of the spinach male-determining region. Mol. Genet. Genom. 293, 557–568. doi: 10.1007/s00438-017-1405-2
Kühn, U., Bitteich, V., Carolin, R., Freitag, H., Hedge, I., Until, P., et al. (1993). “Chenopodiaceae,” in The Families and Genera of Vascular Plants, Vol. 2, eds K. Kubitzki, J. Rohwer, and V. Bittrich (Berlin: Springer), 253–281.
Li, S., Guo, Y., Li, J., Zhang, D., Wang, B., Li, N., et al. (2019). The landscape of transposable elements and satellite DNAs in the genome of a dioecious plant spinach (Spinacia oleracea L.). Mobile DNA 10:3.
Li, S., Zhang, G., Yuan, J., Deng, C., and Gao, W. (2016). Repetitive sequences and epigenetic modification: inseparable partners play important roles in the evolution of plant sex chromosomes. Planta 243, 1083–1095. doi: 10.1007/s00425-016-2485-7
Liu, Q., Li, X., Zhou, X., Li, M., Zhang, F., Schwarzacher, T., et al. (2019). The repetitive DNA landscape in Avena (Poaceae): chromosome and genome evolution defined by major repeat classes in whole-genome sequence reads. BMC Plant Biol. 19:226. doi: 10.1186/s12870-019-1769-z
Menzel, G., Krebs, C., Diez, M., Holtgräwe, D., Weisshaar, B., Minoche, A., et al. (2012). Survey of sugar beet (Beta vulgaris L.) hAT transposons and MITE-like hATpin derivatives. Plant Mol. Biol. 78, 393–405. doi: 10.1007/s11103-011-9872-z
Navajas-Perez, R., de la Herran, R., Gonzalez, G. L., Jamilena, M., Lozano, R., Rejon, C. R., et al. (2005). The evolution of reproductive systems and sex-determining mechanisms within Rumiex (Polygonaceae) inferred from nuclear and chloroplastidial sequence data. Mol. Biol. Evol. 22, 1929–1939. doi: 10.1093/molbev/msi186
Novak, P., Avila, R. L., Koblizkova, A., Vrbova, I., Neumann, P., and Macas, J. (2017). TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 5:e111. doi: 10.1093/nar/gkx257
Novak, P., Hribova, E., Neumann, P., Koblizkova, A., Dolezel, J., and Macas, J. (2014). Genome-Wide analysis of repeat diversity across the family musaceae. PLoS One 9:e98918. doi: 10.1371/journal.pone.0098918
Novak, P., Neumann, P., and Macas, J. (2010). Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinform. 11:378. doi: 10.1186/1471-2105-11-378
Novak, P., Neumann, P., Pech, J., Steinhaisl, J., and Macas, J. (2013). RepeatExplorera Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29, 792–793. doi: 10.1093/bioinformatics/btt054
Okazaki, Y., Takahata, S., Hirakawa, H., Suzuki, Y., and Onodera, Y. (2019). Molecular evidence for recent divergence of X- and Y-linked gene pairs in Spinacia oleracea L. PLoS One 14:e0214949. doi: 10.1371/journal.pone.0214949
Onodera, Y., Yonaha, I., Niikura, S., Yamazaki, S., and Mikami, T. (2008). Monoecy and gynomonoecy in Spinacia oleracea L.: morphological and genetic analyses. Scientia Horticult. 118, 266–269. doi: 10.1016/j.scienta.2008.06.008
Paesold, S., Borchardt, D., Schmidt, T., and Dechyeva, D. (2012). A sugar beet (Beta vulgaris L.) reference FISH karyotype for chromosome and chromosome-arm identification, integration of genetic linkage groups and analysis of major repeat family distribution. Plant J. 72, 600–611. doi: 10.1111/j.1365-313x.2012.05102.x
Pamponet, V. C. C., Souza, M. M., Silva, G. S., Micheli, F., de Melo, C. A. F., de Oliveira, S. G., et al. (2019). Low coverage sequencing for repetitive DNA analysis in Passiflora edulis sims: citogenomic characterization of transposable elements and satellite DNA. BMC Genom. 20:262. doi: 10.1186/s12864-019-5576-6
Puterova, J., Razumova, O., Martinek, T., Alexandrov, O., Divashuk, M., Kubat, Z., et al. (2017). Satellite DNA and transposable elements in seabuckthorn (Hippophae rhamnoides), a dioecious plant with small Y and large X chromosomes. Genome Biol. Evol. 9, 197–212.
Rogers, S. O., and Bendich, A. J. (1988). Extraction of DNA from plant tissues. Plant Mol. Biol. 6, 1–6.
Sakamoto, K., Ohmido, N., Fukui, K., Kamada, H., and Satoh, S. (2000). Site-specific accumulation of a LINE-like retrotransposon in a sex chromosome of the dioecious plant Cannabis sativa. Plant Mol. Biol. 44, 723–732.
Schwichtenberg, K., Wenke, T., Zakrzewski, F., Seibt, K., Minoche, A., Dohm, J., et al. (2016). Diversification, evolution and methylation of short interspersed nuclear element families in sugar beet and related amaranthaceae species. Plant J. 85, 229–244. doi: 10.1111/tpj.13103
Sousa, A., Bellot, S., Fuchs, J., Houben, A., and Renner, S. S. (2016). Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y chromosome reveals distinct Y centromeres. Plant J. 88, 387–396. doi: 10.1111/tpj.13254
Sousa, A., Fuchs, J., and Renner, S. S. (2013). Molecular cytogenetics (FISH, GISH) of Coccinia grandis: a ca. 3 myr-Old species of cucurbitaceae with the largest Y/Autosome divergence in flowering plants. Cytogenetic Genome Res. 139, 107–118. doi: 10.1159/000345370
Steflova, P., Tokan, V., Vogel, I., Lexa, M., Macas, J., Novak, P., et al. (2013). Contrasting patterns of transposable element and satellite distribution on sex chromosomes (XY1Y2) in the dioecious plant Rumex acetosa. Genome Biol. Evol. 5, 769–782. doi: 10.1093/gbe/evt049
Takahata, S., Yago, T., Iwabuchi, K., Hirakawa, H., Suzuki, Y., and Onodera, Y. (2016). Comparison of spinach sex chromosomes with sugar beet autosomes reveals extensive synteny and low recombination at the male-determining locus. J. Heredity 107, 679–685. doi: 10.1093/jhered/esw055
Torres, M. F., Mathew, L. S., Ahmed, I., Al-Azwani, I. K., Krueger, R., Rivera-Nuñez, D., et al. (2018). Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nat. Commun. 9:3969.
Vyskot, B., and Hobza, R. (2004). Gender in plants: sex chromosomes are emerging from the fog. Trends Genet. TIG 20, 432–438. doi: 10.1016/j.tig.2004.06.006
Weber, B., and Schmidt, T. (2009). Nested Ty3-gypsy retrotransposons of a single Beta procumbens centromere contain a putative chromodomain. Chromosome Res. 17, 379–396. doi: 10.1007/s10577-009-9029-y
Weber, B., Wenke, T., Frömmel, U., Schmidt, T., and Heitkam, T. (2010). The Ty1-copia families SALIRE and Cotzilla populating the Beta vulgaris genome show remarkable differences in abundance, chromosomal distribution, and age. Chromosome Res. 18, 247–263. doi: 10.1007/s10577-009-9104-4
Wenke, T., Holtgräwe, D., Horn, A., Weisshaar, B., and Schmidt, T. (2009). An abundant and heavily truncated non-LTR retrotransposon (LINE) family in Beta vulgaris. Plant Mol. Biol. 71, 585–597. doi: 10.1007/s11103-009-9542-6
Wollrab, C., Heitkam, T., Holtgräwe, D., Weisshaar, B., Minoche, A., Dohm, J., et al. (2012). Evolutionary reshuffling in the Errantivirus lineage elbe within the Beta vulgaris genome. Plant J. 72, 636–651. doi: 10.1111/j.1365-313x.2012.05107.x
Xu, C., Jiao, C., Sun, H., Cai, X., Wang, X., Ge, C., et al. (2017). Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat. Commun. 8:15275.
Yu, L., Ma, X. K., Deng, B., Yue, J. J., and Ming, R. (2020). Construction of high-density genetic maps defined sex determination region of the Y chromosome in spinach. Mol. Genet. Genom. doi: 10.1007/s00438-020-01723-1724 Online ahead of print.
Zakrzewski, F., Schubert, V., Viehoever, P., Minoche, A., Dohm, J., Himmelbauer, H., et al. (2014). The CHH motif in sugar beet satellite DNA: a modulator for cytosine methylation. Plant J. 78, 937–950. doi: 10.1111/tpj.12519
Keywords: repetitive DNA sequences, rDNA, homomorphic sex chromosome, heteromorphic sex chromosome, long terminal repeat retroelements, next generation sequencing
Citation: Li N, Li X, Zhou J, Yu L, Li S, Zhang Y, Qin R, Gao W and Deng C (2021) Genome-Wide Analysis of Transposable Elements and Satellite DNAs in Spinacia Species to Shed Light on Their Roles in Sex Chromosome Evolution. Front. Plant Sci. 11:575462. doi: 10.3389/fpls.2020.575462
Received: 23 June 2020; Accepted: 17 December 2020;
Published: 14 January 2021.
Edited by:
Lucia Natali, University of Pisa, ItalyReviewed by:
Amy Litt, University of California, Riverside, United States Alex Rajewski, University of California, Riverside, in collaboration with ALSara V. Good, University of Winnipeg, Canada
Copyright © 2021 Li, Li, Zhou, Yu, Li, Zhang, Qin, Gao and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanliang Deng, ZGVuZ2NodWFubGlhbmdAaHR1LmVkdS5jbg==
†These authors share first authorship
 Ning Li1†
Ning Li1† Xiaoyue Li
Xiaoyue Li Jian Zhou
Jian Zhou Yulan Zhang
Yulan Zhang Ruiyun Qin
Ruiyun Qin