- 1Fujian Agriculture and Forestry University, Fuzhou, China
- 2College of Forestry, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Department of Chemistry, Birendra Multiple Campus, Tribhuvan University, Chitwan, Nepal
- 4Key Laboratory of Crop Ecology and Molecular Physiology, Fujian Agriculture and Forestry University, Fuzhou, China
- 5Fujian Provincial Key Laboratory of Agroecological Processing and Safety Monitoring, School of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
The growth and productivity of Casuarina equisetifolia is negatively impacted by planting sickness under long-term monoculture regimes. In this study, Illumina MiSeq sequencing targeting nifH genes was used to assess variations in the rhizospheric soil diazotrophic community under long-term monoculture rotations. Principal component analysis and unweighted pair-group method with arithmetic means (UPGMA) clustering demonstrated distinct differences in diazotrophic community structure between uncultivated soil (CK), the first rotation plantation (FCP), the second rotation plantation (SCP), and the third rotation plantation (TCP). Taxonomic analysis showed that the phyla Proteobacteria increased while Verrucomicrobia decreased under the consecutive monoculture (SCP and TCP). The relative abundance of Paraburkholderia, Rhodopseudomonas, Bradyrhizobium, Geobacter, Pseudodesulfovibrio, and Frankia increased significantly while Burkholderia, Rubrivivax, and Chlorobaculum declined significantly at the genus level under consecutive monoculture (SCP and TCP). Redundancy analysis (RDA) showed that Burkholderia, Rubrivivax, and Chlorobaculum were positively correlated with total nitrogen and available nitrogen. In conclusion, continuous C. equisetifolia monoculture could change the structure of diazotrophic microbes in the rhizosphere, resulting in the imbalance of the diazotrophic bacteria population, which might be a crucial factor related to replanting disease in this cultivated tree species.
Introduction
Casuarina equisetifolia Forst has covers over 300,000 ha in southeastern China. It is one of the preferred trees for forming shelterbelts in coastal areas, as it vegetates the coasts and contributes to stabilizing coastal sand and protecting against storms (Zhong et al., 2005; Karthikeyan et al., 2013). However, continuous monocultures of C. equisetifolia result in productivity decline and regeneration failure. This phenomenon is referred to as the consecutive monoculture problem (CMP) (Wardle et al., 2004). Replanting diseases are observed in many cultivated tree species, including Chinese fir, Pinus elliottii, Picea abies, and Eucalyptus spp. (Wu et al., 2017). The depletion in soil nutrients (Bennett et al., 2012), the autointoxication of root exudations (Araya et al., 2012), and the imbalance of rhizospheric microflora (Wu et al., 2016a) are thought to be the main reasons for CMP. Increasing the amount of chemical fertilizer does not alleviate this problem (Wardle et al., 2004). Root exudates not only directly inhibit the growth and reproduction of plants but also restrain rhizosphere microorganisms (Li et al., 2014; Arafat et al., 2017). Great attention has been paid to rhizospheric biological processes in recent years. Lu et al. (2017) indicated that soil microbial community compositions and theirs structures had distinct variations under Poplar monocultures. Similar results were found in Chinese fir, Eucalyptus, and Pinus halepensis (Fernandez et al., 2008; Jie et al., 2014; Wu et al., 2017).
Biological nitrogen fixation has attracted widespread attention due to its influential role in the nitrogen cycle. Hsu and Buckley (2009) investigated the diversity of nitrogen-fixing genes and their functional microbial communities through high-throughput MiSeq amplicon sequencing of nifH gene, which encodes a subunit of the nitrogenase enzyme complex related to nitrogen fixation efficiency. Penton et al. (2016) amplified nifH gene sequencing and found that permafrost thawing in Alaskan soil altered the N-fixing microbial community composition in soil as the depth of groundwater changed. Wang et al. (2017) found that long-term inorganic fertilization in acid soil altered the diazotrophic community structure and decreased the abundance of operational taxonomic units (OTUs). Biological nitrogen fixation has been recognized as a crucial source of nitrogen to support certain primary production (Zehr et al., 1998). N-fixation by diazotrophs is an important strategy by which most organisms regulate biological productivity (Li et al., 2018). Nevertheless, the relationship between the diazotrophic microbial community and monocultures of C. equisetifolia has received little attention. On the basis of the above facts, we hypothesized that diazotrophic microbial community structure might be altered by C. equisetifolia monocultures (Wu et al., 2016b; Chen et al., 2017; Zhou et al., 2019).
In this study, quantitative PCR (qPCR) assays and the MiSeq high-throughput sequencing technique were applied to assess the shifts in the abundance of nifH gene and diazotrophic community composition in rhizosphere soil, respectively, after C. equisetifolia monoculture. Meanwhile, correlation analyses were used to resolve the relationships between the diazotrophic microbial community and environmental parameters, including total nitrogen (TN), total phosphorus (TP), total potassium (TK), available nitrogen (AN), available phosphorus (AP), available potassium (AK), and pH. This research will help improve our understanding of the nitrogen-fixing microbial community structure in soil under CMP. It will provide effective scientific-based references on the molecular ecological mechanisms of soil restoration and improvement.
Materials and Methods
Research Plot and Sample Collection
In this study, the plot was located at Chihu National Forest Center in Fujian Province, China (24°54′N and 118°55′E). Since the 1960s, the Chihu Forestry Farm cultivated a large-scale coastal windbreak and gradually established a settled coastal shelterbelt system. The annual mean temperature is 19.8°C (the extreme high and low temperatures are 35°C and 1°C, respectively), with a mean annual precipitation of 1,029 mm and a mean annual evaporation of 2,000 mm. There are three generations of C. equisetifolia plantation in the forest farm that were planted in 1987 (first rotation plantation, FCP), 2011 (second rotation plantation, SCP), and 2014 (third rotation plantation, TCP). Due to the long growth period of the arbor and field condition, the method of “space replacing time” is often applied in forestry research (Shi et al., 2018; Deng et al., 2019).
Three sampling positions (20 m × 20 m) were established at the FCP, SCP, and TCP on January 6, 2019. At the same time, a vacant area of soil in the forest farm with no C. equisetifolia cultivation was selected as a blank control, or CK. Each sampling position was set with three duplicate quadrats for a total of 12 quadrats. Soil samples were randomly collected from a depth of 0–20 cm at 12 quadrats. Twenty random replicated samples were taken from each quadrat. Soil samples from the FCP, SCP, and TCP were collected from the rhizosphere of C. equisetifolia. The field samplings were performed according to the method described by Zhou et al. (2019).
Determination of Soil Nutrients
Soil pH was measured using a glass electrode pH meter (1:2.5 soil–water suspensions) (Zhao et al., 2017). The available and total amounts of NPK were determined referring to the methods by Jackson (2005).
DNA Extraction
Soil DNA extractions were completed using a Power Soil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, United States) following the manufacturer’s specifications. The genomic DNA was confirmed using 1.2% agarose gels.
qPCR for nifH
Quantitative PCR was performed to quantify the relative abundance of the nifH gene in four individual rhizospheric soils with the primers nifH-F (AAAGGYGGWATCGGYAART CCACCAC) and nifH-R (TTGTTSGCSGCRTACATSGCCAT CAT) (Zhang et al., 2016). The reaction mixture (15 μl) for qPCR consisted of 7.5 μl 2 × SYBR green I Super Real Premix (TIANGEN, Beijing, China), 10 μM of each primer and 40 ng template DNA. The amplification conditions were 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 40 s. The standard curve equation was Y = −4.083X + 46.196 (R2 = 0.99).
PCR Amplification and nifH Sequencing
High-throughput sequencing was used to explore the influences of successive monocultures of C. equisetifolia on rhizospheric soil diazotrophic microbial communities. For nifH gene sequencing, the primers nifH-F and nifH-R (the same as above) were applied to amplify the nifH gene. The PCR amplification was implemented on a Mastercycler Gradient (Eppendorf, Germany) with a 25 μl volume, including 12.5 μl 2 × Taq PCR MasterMix, 3 μl BSA (2 ng/μl), 2 μl Primer (5 μM), 2 μl template DNA, and 5.5 μl ddH2O. Cycling parameters were 95°C for 5 min, followed by 32 cycles of 95°C for 45 s, 55°C for 50 s, and 72°C for 45 s with a final extension at 72°C for 10 min. The amplicons were purified with a QIAquick Gel Extraction Kit (QIAGEN, Germany), sequenced on the MiSeq PE300 platform at Allwegene Company, Beijing, China.
Data Analyses
The low-quality (q < 20) and short sequences (<200 bp) were removed via the Pear (v. 0.9.6). The final length of sequences was 400–420. The datasets were then analyzed using QIIME (v. 1.8.0, Caporaso et al., 2010). The qualified sequences were clustered into OTUs at a 97% identity threshold (Edgar, 2013). All effective tags were classified into different taxonomic groups using the Ribosomal Database Project (RDP) classifier (Cole et al., 2009). Alpha diversity indices were employed to identify the richness and diversity within samples (Xiong et al., 2015). In order to make the sequencing depth consistent, the abundances of OTUs were normalized. Alpha diversity indices were calculated by R software (v. 2.15.3, Caporaso et al., 2010) based on the normalized data (21,778 taxa). The principal component analysis (PCA), clustering analyses, and non-metric multidimensional scaling (NMDS) were carried out to assess the similarity and difference between individual samples (Wang et al., 2012). PCA and NMDS were implemented based on the R with vegan package (v. 3.0.2, Oksanen et al., 2009; Goossen et al., 2010). To compare the diazotrophic community compositions and structures in the rhizosphere, heat maps were applied within the top 20 most abundant OTUs using Mothur (Jami et al., 2013). Redundancy analysis (RDA) was performed to establish which environmental parameters played an important role in the variation of top 12 most abundant OTUs using Canoco 5.0. Statistical analysis was carried out by DPS 7.05 software, and analysis of variance (ANOVA) was applied to identify significance difference by LSD’s test (P < 0.05).
Results
Soil Physicochemical Data
The results of soil nutrient analysis showed that replanting soils had a lower level of pH under the extended C. equisetifolia monoculture regime. The TN content was higher in the FCP than in the SCP (significant, P < 0.05) and TCP (not significant, P > 0.05). AN was higher in the FCP than in the SCP and TCP (P > 0.05). In the FCP and SCP, TP was significantly higher than in the TCP (P < 0.05). No significant differences were observed in AP, TK, and AK (Supplementary Table 1).
Abundance of the nifH Gene
Figure 1 indicates that the quantity of the nifH gene changed significantly under monoculture regime (p < 0.05). The number of copies of the nifH gene was between 0.746 × 106 and 3.647 × 106/g soil, and the highest and lowest numbers were found in the SCP and CK, respectively. Compared with the CK, the number of copies in the FCP, SCP, and TCP were increased by 54.4, 388.9, and 127.6%, respectively.
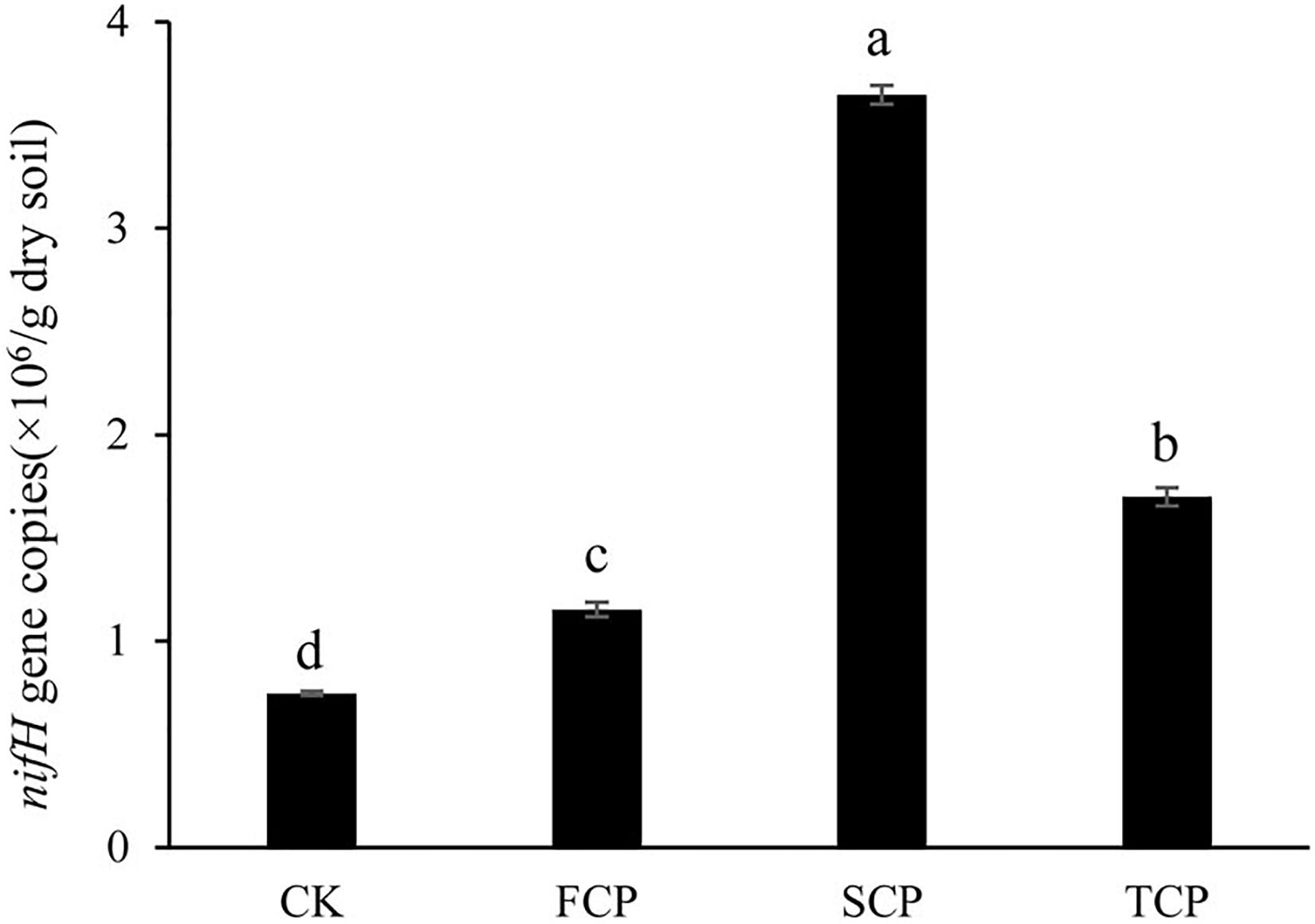
Figure 1. Quantification of nifH gene in four different soil samples. Vertical bars show standard deviations. CK, FCP, SCP, and TCP represent the control with no C. equisetifolia cultivation, the first rotation plantation, the second rotation plantation and the third rotation plantation.
OTU Clusters and Species Annotation
After filtration analysis, a sum of 656,599 high-quality tags (clean tags) was acquired from the CK, FCP, SCP, and TCP, with a mean of 54,717 effective sequences (Supplementary Table 2). Rarefaction curves indicated that the number of observed species was stable over 20,000 tags (Supplementary Figure 1). At the 97% similarity cut-off, effective tags from 12 soil samples were clustered into 4,610 OTUs. The average OTU numbers in the CK, FCP, SCP, and TCP were 272, 332, 375, and 559, respectively (Supplementary Table 2). The raw sequences data was deposited in NCBI (PRJNA666767).
Alpha Diversity Indices
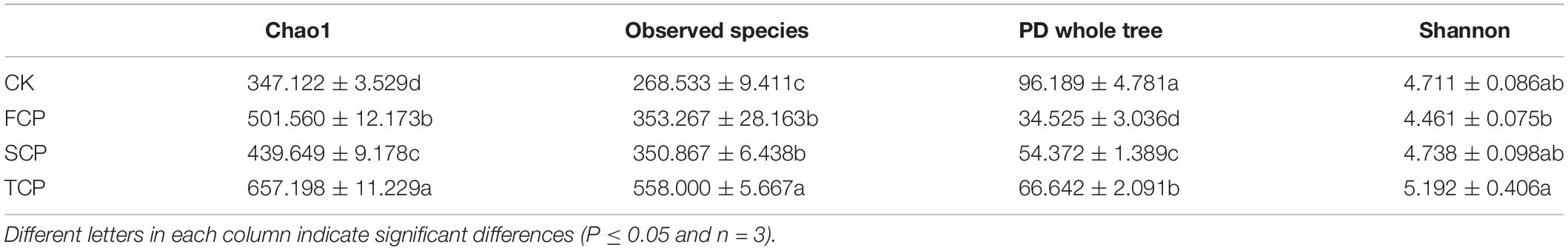
In this research, the richness and diversity of diazotrophic microbial communities were obtained according to 21,778 taxa. Table 1 exhibits the Chao1, Observed species, PD whole tree, and Shannon index of nitrogen-fixing microbial communities. The observed number of species was dramatically higher in the TCP than in the FCP and SCP (p < 0.05). The Shannon index was significantly higher in the TCP than in the FCP (p < 0.05) and it was also higher than in the SCP (not significant, p > 0.05). In the TCP, the soil nitrogen-fixing microbial communities showed significantly higher Chao1 and PD whole tree values than in the FCP and SCP (p < 0.05) (Table 1).
PCA, UPGMA Clustering, NMDS, and PERMANOVA
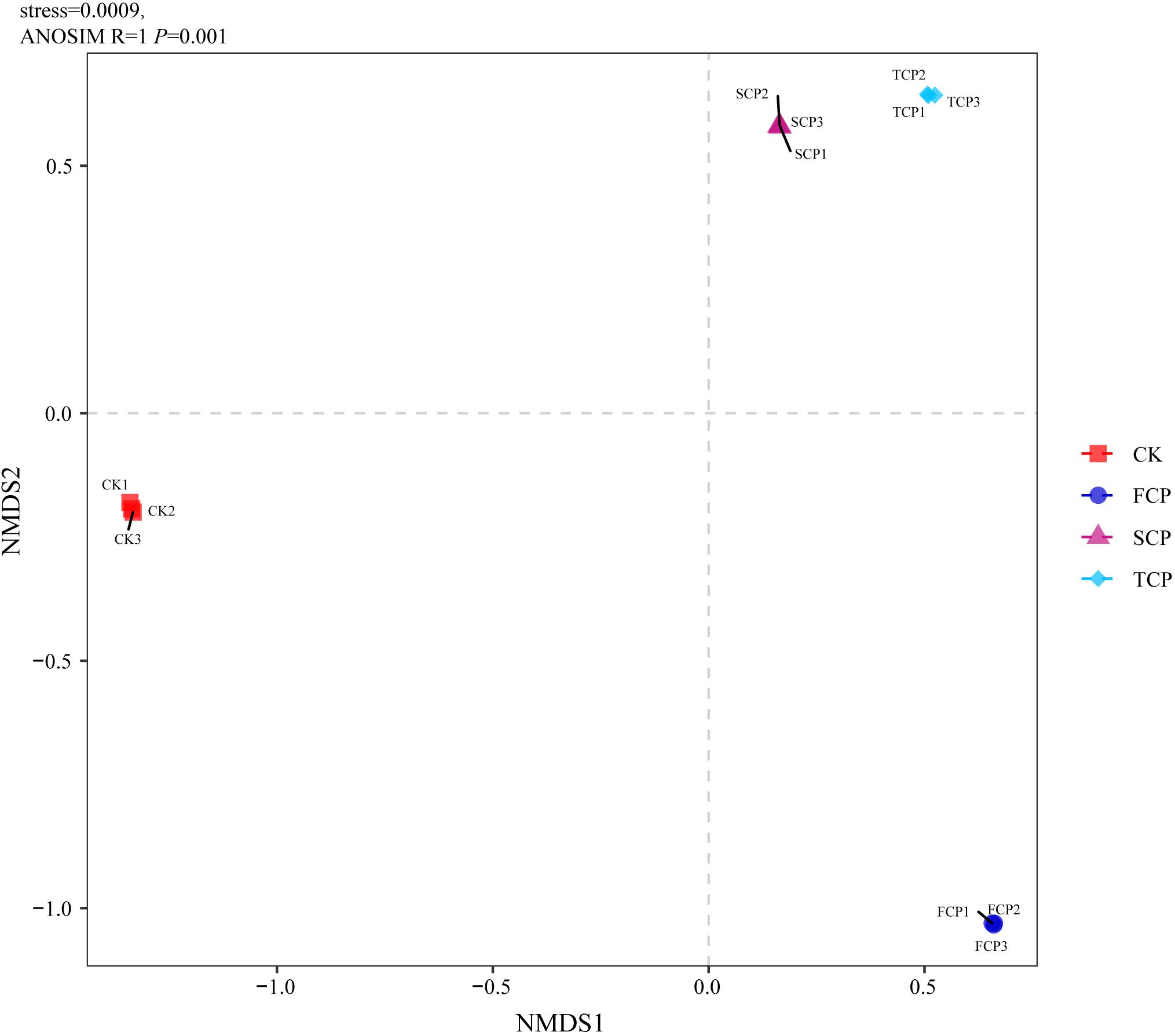
The PCA analysis indicated obvious dissimilarities in the soil diazotrophic community structures among the four soil samples. The PCA plot completely accounted for 72.56% of the overall variation in the soil nitrogen-fixing microbial community. The PC1 explained 47.85% and the PC2 explained 24.71% of the microbial variation (Supplementary Figure 2A). When observing the unweighted pair-group method with arithmetic means (UPGMA) clustering, the figure showed that diazotrophic community structures clustered together from the SCP and TCP, whereas they were separated from the FCP, and the CK formed a separate group (Supplementary Figure 2B). The results of PCA and UPGMA clustering revealed that diazotrophic microbial community composition is different under monoculture regimes. Non-metric multidimensional scaling (NMDS) was applied to illustrate the differences in diazotrophic community composition and structure (Tibbits, 2008). Pairwise contrasts demonstrated that the CK, FCP, SCP, and TCP plots were significantly (P < 0.05) separated (Figure 2). The PERMANOVA analysis showed significant differences in the composition and distribution of the diazotrophic microbial community (F = 26.7519 and P = 0.001).
Venn Diagram Analysis
The shared and exclusive OTUs among the FCP, SCP, and TCP were explored through Venn analysis. Supplementary Figure 3 indicates that the proportion of OTUs shared in the FCP, SCP and TCP was 30.9% (323 species). The proportion of OTUs found only in the FCP was 5.4% (57 species). The abundance of OTUs exclusively constructed in the SCP accounted for up to 13.4% (140 species). The percentage of OTUs constructed only in the TCP was 23.8% (249 species).
Abundance Change in the Diazotrophic Microbial Communities
Diazotrophic microbial OTUs consisted mostly of five phyla: Proteobacteria, Verrucomicrobia, Chlorobi, unidentified, and Actinobacteria. Among them, Proteobacteria was the dominant phylum, accounting for 85.7, 65.7, 94.4, and 85.5% of the total species in the CK, FCP, SCP, and TCP, respectively (Supplementary Figure 4).
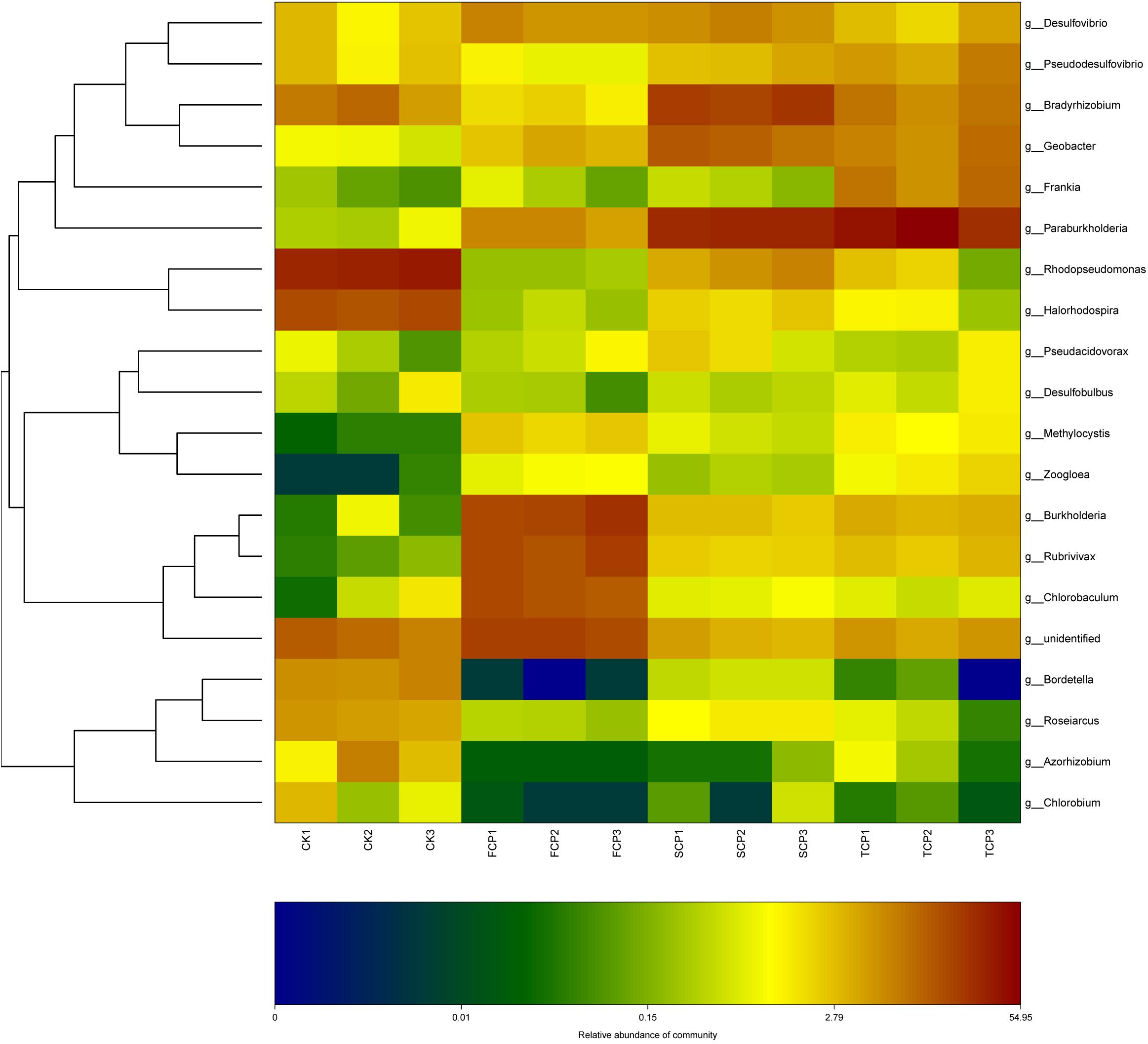
At genus level, the C. equisetifolia monoculture regime generated a distinct enhancement in the relative abundance of Paraburkholderia, Rhodopseudomonas, Bradyrhizobium, Geobacter, Pseudodesulfovibrio, and Frankia and a distinct reduction in Burkholderia, Rubrivivax, and Chlorobaculum (Table 2). Compared with the FCP, the genus Desulfovibrio was slightly higher in the SCP but decreased in the TCP. The most common diazotrophic microbial communities of C. equisetifolia are listed in Table 2, including Paraburkholderia, Bradyrhizobium, Burkholderia, and Frankia.
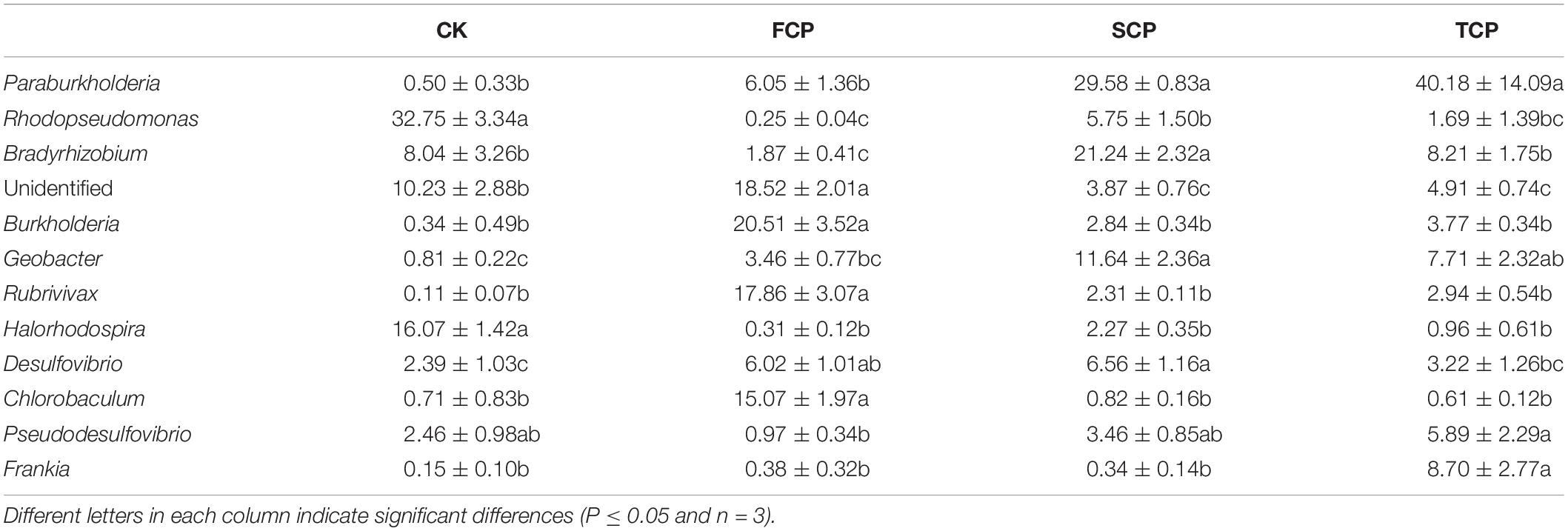
Table 2. Calculations of the top diazotrophic bacteria for four different soil samples at the genus level.
It can be seen from the heat map analysis of the top 20 diazotrophic genera that distinct variations in diazotrophic community structure arose with the increase of CMP. Compared with the FCP, the discrepancy of community structures of diazotrophic bacteria increased within the extended monoculture regime, implying that diazotrophic community structure gradually changed under the CMP (Figure 3).
Linear discriminant analysis effect size (LEfSe) was applied to identify biomarkers (potential discriminating species in relative abundance between groups) from metagenome taxa (Xia et al., 2018). The LEfSe analysis revealed that there were 30 potential bacterial markers distinguishing the CK, FCP, SCP, and TCP with linear discriminant analysis (LDA) scores more than 3. In these tests, one phylum (unidentified), three classes (Alphaproteobacteria, Gammaproteobacteria, and unidentified), three orders (Rhizobiales, Chromatiales, and unidentified), and six families (Bradyrhizobiaceae, Roseiarcaceae, Xanthobacteraceae, Alcaligenaceae, Ectothiorhodospiraceae, and unidentified) were biomarkers for CK. Two phyla (Chlorobi and Verrucomicrobia), two classes (Chlorobia and Opitutae), two orders (Chlorobiales and Opitutales), and four families (Chlorobiaceae, Methylocystaceae, Opitutaceae, and unidentified) were significantly different in the FCP. In the SCP, biomarkers were mostly clustered in Proteobacteria, including Deltaproteobacteria, Desulfuromonadales, and Geobacteraceae. In the TCP, biomarkers were Betaproteobacteria, Burkholderiales, and Burkholderiaceae (Figure 4).
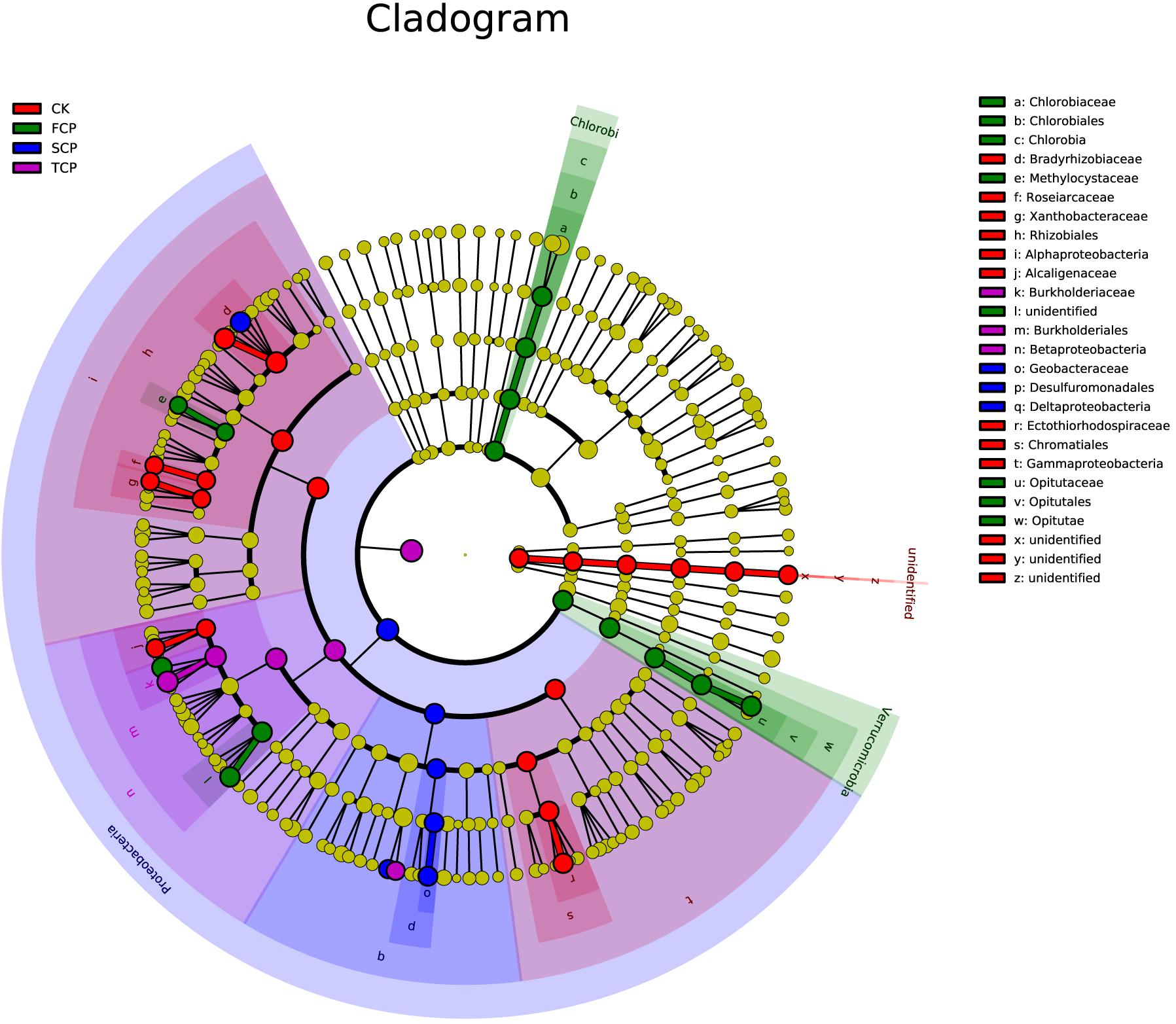
Figure 4. Phylogenetic dendrogram of biomarkers in C. equisetifolia rhizospheric soil diazotrophic bacteria in CK, FCP, SCP, and TCP. The circles from inside to outside exhibit diazotrophic taxonomic levels from phylum to genus. Yellow dots mean that there is no significant difference in diazotrophic abundance among individual soil samples. The colors of the biomarkers are based on their corresponding class colors on the right.
Correlation Analysis Between Diazotrophic Microbial Communities and Environmental Parameters
To detect the relationships between diazotrophic microbial communities and environmental parameters, RDA was performed based on the top 12 species of the diazotrophic microbial community and environmental parameters (Figure 5). The influence of environmental factors on the diazotrophic microbial community can be seen from the length of the arrow and the angle between the arrow and the microbe. RDA canonical axes 1 and 2, respectively, described 58.40 and 33.52% of the variation in the diazotrophic microbial community, suggesting a remarkable correlation between microbial communities and environmental parameters. As shown in Figure 5, the SCP and TCP were clustered together and distributed at the negative end of RDA1, away from the CK and FCP, reflecting the significant shift of the diazotrophic microbial community under a monoculture regime. The results of RDA were consistent with the PCA results (Supplementary Figure 2B). In addition, from the results in Figure 5, it can be seen that Burkholderia, Rubrivivax and Chlorobaculum were positively correlated with the environmental parameters of TN, AN and TK, but negatively correlated with AK. However, the opposite was true for the Bradyrhizobium which was positively correlated with AK.
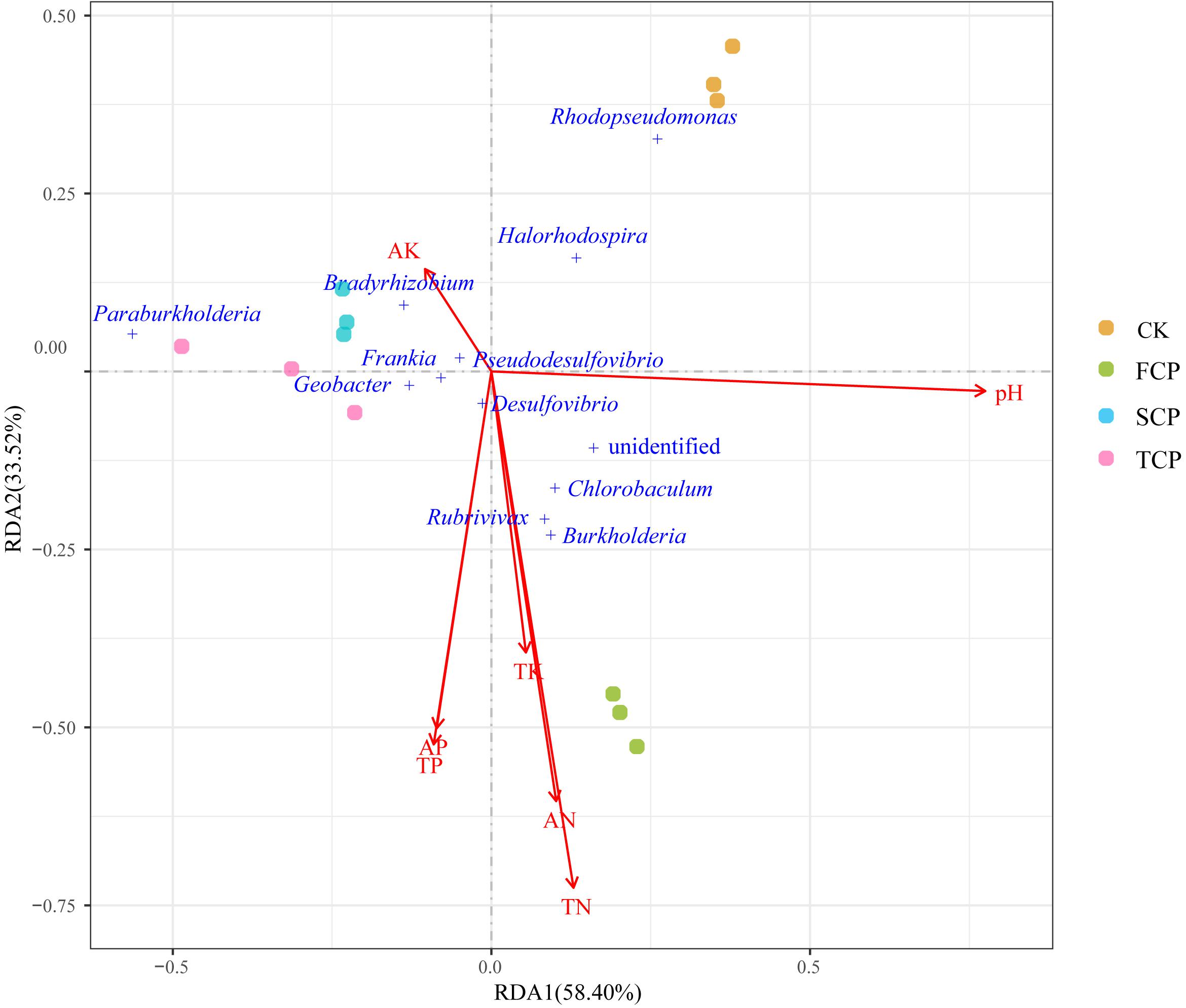
Figure 5. RDA of soil physicochemical properties and predominant nitrogen-fixing microbial communities in four individual soil samples.
Discussion
Consecutive monoculture problem, also known as soil sickness, has a severe impact on the growth and health of plants. It is a widespread and complex phenomenon in numerous cultivated tree species (Huang et al., 2013). Many factors are thought to contribute to the continuous cropping problem, including the physical and chemical properties of soil, the accumulation of allelochemicals, and changes in the soil microbial community (Wu et al., 2016a; Chen et al., 2018). In this study, we found that the contents of soil nitrogen (AN and TN) under monoculture regimes (SCP and TCP) were significantly lower than that in the FCP (Supplementary Table 1). The results indicated that soil physical and chemical properties may indirectly affect the healthy growth of C. equisetifolia through regulating the structure of the soil microbial community (Berendsen et al., 2012).
Recently, many studies have revealed that shifts in the soil microbial community are related to this soil sickness (Li et al., 2014; Liu et al., 2017). Soil microbes play a vital role in the growth and health of plants. Soil microbes are also known as the second genome of plants (Berendsen et al., 2012; Berg et al., 2017). N-fixing organisms play a profound part in nitrogen cycling in forest ecosystems (Yousuf et al., 2014). In the present study, the nifH gene abundance in C. equisetifolia increased with the extension of monoculture. These results are contradictory to those of a previous study on a Eucalyptus successive plantation (Monteiro et al., 2020). Species diversity is a crucial factor for soil health (Chaparro et al., 2012). Wu et al. (2017) reported that the rhizosphere microbial diversity in Chinese fir was decreased after long-term successive rotations. In this study, most of the diversity indices were significantly lower in the FCP and in the SCP than TCP (Table 1). The differing results among these studies may be due to differences in factors including plant types, soil environments, and root chambers. The number of copies of the nifH gene increased under monoculture regime, while diversity indices were increased or decreased with monoculture. That is the diazotrophic community compositions and structures were changed under long-term monoculture C. equisetifolia plantations, indicating that our hypothesis was correct.
In recent years, more attention has been paid to the relationship between key rhizosphere microbes and plants, which is central to plant growth and development (Mendes et al., 2011; Chen et al., 2017). The Proteobacteria phylum is commonly explored in soil systems (Gaby and Buckley, 2011). In this study, the most predominant phylum present in the rhizosphere was Proteobacteria. The diazotrophic bacterial populations in the C. equisetifolia rhizospheric soil (Supplementary Figure 4) accounted for 85.69, 65.75, 94.40, and 85.54% of the total in the CK, FCP, SCP, and TCP, respectively, which was in accordance with the results found by Bazylinski et al. (2013). Taxonomic analysis (genus level) showed that continuous monoculture of C. equisetifolia increased the relative abundance of Paraburkholderia, Rhodopseudomonas, Bradyrhizobium, Geobacter, Pseudodesulfovibrio, and Frankia, whereas the abundance of Burkholderia, Rubrivivax, and Chlorobaculum decreased significantly (Table 2). Among these diazotrophs, compared with the FCP, the relative abundance of Bradyrhizobium increased by 1,135.83 and 439.04% in the SCP and TCP, respectively. Bradyrhizobium was the dominant diazotroph in soil systems due to strong persistence of diazotrophic bacteria across varying soil environmental conditions (Pereira et al., 2013). Relative to the FCP, the abundance of Burkholderia decreased by 722.18 and 544.03% in the SCP and TCP, respectively. Burkholderia can be used for biological control through such methods as promoting plant growth and bio-remediation. It has been reported that Burkholderia has the ability of nodulation and N-fixation in certain legume plants including Burkholderia vietnamiensis, Burkholderia phymatum, and Burkholderia tuberum (Coenye and Vandamme, 2003). Wu et al. (2016a) reported that monoculture of Pseudostellaria heterophylla led to a significant decline in the abundance of Burkholderia. Mendes et al. (2011) showed that the relative abundances of Burkholderiaceae was positively associated with disease suppression of sugar beet. Thus, certain species of diazotrophic bacteria found in this study were inhibited under C. equisetifolia monocultures (SCP and TCP) that might be key rhizosphere microbes or plant growth promoters. This issue should be studied in the future.
Many reports have indicated that soil microbial community composition was indirectly influenced by environmental parameters (Li and Wu, 2018; Zhang et al., 2019). In our study, RDA results showed that key rhizosphere microbes (Burkholderia, Rubrivivax and Chlorobaculum) were positively correlated with soil nitrogen (AN and TN). Therefore, based on their abundance (a significant decrease in SCP and TCP), soil nitrogen seems to play an important role in C. equisetifolia monocultures. The decline of Burkholderia, Rubrivivax and Chlorobaculum may be related to soil nitrogen under continuous planting. Their abundance may indicate an important role in plant-microbial interactions and soil function as well as nitrogen fixation. In addition, Burkholderia is considered a rhizospheric-plant promoting group (Zhao K. et al., 2014). It could play an important role in maintaining the stability of soil communities (Wakelin et al., 2017). The results of RDA suggested that the diazotrophic microbial community may respond differently to environmental parameters, but this hypothesis should be further tested.
A growing number of researchers have reported that the imbalance of key rhizosphere microflora is considered to be the main cause of CMP. Zhao J. et al. (2014) revealed that Eucalyptus monocultures increased the abundance of fungi in rhizospheric soil. Successive rotations of Cunninghamia lanceolata generated imbalance in the soil microbial community. Most studies have reported that this phenomenon of imbalance resulted from alterations of the rhizospheric microflora induced by plant root exudations rather than direct allelopathy (Li et al., 2014; Wu et al., 2015). In this study, the continuous monoculture of C. equisetifolia shifted the structure of the diazotrophic population in rhizospheric soils. This shift may result in the imbalance of diazotrophic bacteria population structure, which might be a crucial factor in replanting disease of this cultivated tree species. Many previous studies have shown that root exudates restructure the plant-associated rhizospheric microbes and these microbes impact plants (Paterson et al., 2007; Haichar et al., 2008; Michalet et al., 2013). Therefore, further studies are required to analyze the effects of the C. equisetifolia root exudates to obtain precise knowledge of the soil nitrogen-fixing microorganism community structure.
Conclusion
Our result indicates that the continuous monoculture of C. equisetifolia distinctly influenced the diazotrophic community compositions and structures in the rhizosphere. The phyla Proteobacteria increased, whereas Verrucomicrobia decreased with increasing continuous monoculture. At the genus level, the relative abundance of Paraburkholderia, Rhodopseudomonas, Bradyrhizobium, Geobacter, Pseudodesulfovibrio, and Frankia increased significantly, while Burkholderia, Rubrivivax, and Chlorobaculum declined significantly. To more efficiently explore the causes of obstacles to continuous monoculture, the isolation of specific N-fixing organisms (i.e., Bradyrhizobium and Burkholderia) and their functions, which may be correlated with CMP, should be further studied. It is noteworthy that most specific N-fixing organisms appear to be highly sensitive to changes in the soil under monoculture regimes. This should be investigated further.
Data Availability Statement
The data presented in the study are deposited in the (NCBI) repository, accession number (PRJNA666767; SRP286063).
Author Contributions
ZW, WL, and LZ conceived the study. ZW, YZ, LZ, and WL designed all the experiments. JL, JC, YB, and CZ performed the experiments. LZ, CW, GP, and JL performed the statistical analyses. LZ and ZW wrote the manuscript with the assistance and approval of all authors.
Funding
This work was supported by the Chinese National Natural Science Foundation (31500443), Natural Science Foundation of Fujian Province, China (Grant No. 2018J01617), the Scientific Research Foundation of Fujian Agriculture and Forestry University (XJQ201718), the Fujian-Taiwan Joint Innovative Centre for Germplasm Resources and Cultivation of Crops (Grant No. 2015-75. FJ 2011 Program, China), and the Special Foundation of Science and Technology Innovation of Fujian Agriculture and Forestry University (CXZX2019051G).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.578812/full#supplementary-material
References
Arafat, Y., Wei, X. Y., Jiang, Y. H., Chen, T., Saqib, H. S. A., Lin, S., et al. (2017). Spatial distribution patterns of Root-Associated bacterial communities mediated by root exudates in different aged ratooning tea monoculture systems. Int. J. Mol. Sci. 18:1727. doi: 10.3390/ijms18081727
Araya, H., Otaka, J., Nishihara, E., and Fujii, Y. (2012). First isolation and identification of salicylate from Betula grossa var. Ulmifolia - a potent root growth inhibitor. Allelopathy J. 30, 153–158.
Bazylinski, D. A., Williams, T. J., Lefevre, C. T., Berg, R. J., Zhang, C. L., et al. (2013). Magnetococcus marinus gen. Nov., Sp nov., A marine, magnetotactic bacterium that represents a novel lineage (Magnetococcaceae fam. Nov., Magnetococcales ord. Nov.) at the base of the Alphaproteobacteria. Int. J. Syst. Evol. Micr. 63, 801–808. doi: 10.1099/ijs.0.038927-0
Bennett, A. J., Bending, G. D., Chandler, D., Hilton, S., and Mills, P. (2012). Meeting the demand for crop production: The challenge of yield decline in crops grown in short rotations. Biol. Rev. 87, 52–71. doi: 10.1111/j.1469-185X.2011.00184.x
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., Koeberl, M., Rybakova, D., Mueller, H., Grosch, R., Smalla, K., et al. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:fix050. doi: 10.1093/femsec/fix050
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., and Bushman, F. D. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chaparro, J. M., Sheflin, A. M., Manter, D. K., and Vivanco, J. M. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fert. Soils 48, 489–499. doi: 10.1007/s00374-012-0691-4
Chen, J., Wu, L. K., Xiao, Z. G., Wu, Y. H., Wu, H. M., Qin, X. J., et al. (2017). Assessment of the diversity of Pseudomonas spp. and Fusarium spp. in Radix pseudostellariae rhizosphere under monoculture by combining DGGE and quantitative PCR. Front. Microbiol. 8:1748. doi: 10.3389/fmicb.2017.01748
Chen, S. C., Yu, H. J., Zhou, X. G., and Wu, F. Z. (2018). Cucumber (Cucumis sativus L.) Seedling Rhizosphere Trichoderma and Fusarium spp. Communities Altered by Vanillic Acid. Front. Microbiol. 9:2195. doi: 10.3389/fmicb.2018.02195
Coenye, T., and Vandamme, P. (2003). Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5, 719–729. doi: 10.1046/j.1462-2920.2003.00471.x
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145. doi: 10.1093/nar/gkn879
Deng, C. H., Wu, L. L., Zhang, Y. T., Qiao, H., Liu, X. Y., Hu, Y. J., et al. (2019). The stoichiometry characteristics of soil and plant carbon, nitrogen, and phosphorus in different stand ages in Camellia oleifera plantation. Acta Ecol. Sin. 39, 9152–9161. doi: 10.5846/stxb201809202055
Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10:996. doi: 10.1038/NMETH.2604
Fernandez, C., Voiriot, S., Mévy, J., Vila, B., Orme, O. E., Dupouyet, S., et al. (2008). Regeneration failure of Pinus halepensis Mill.: The role of autotoxicity and some abiotic environmental parameters. Forest Ecol. Manag. 255, 2928–2936. doi: 10.1016/j.foreco.2008.01.072
Gaby, J. C., and Buckley, D. H. (2011). A global census of nitrogenase diversity. Environ. Microbiol. 13, 1790–1799. doi: 10.1111/j.1462-2920.2011.02488.x
Goossen, W. T. F., Epping, P. J. M. M., Heuvel, W. J. V. D., Feuth, T., and Hasman, A. (2010). Development of the Nursing Minimum Data Set for the Netherlands (NMDSN): Identification of categories and items. J. Adv. Nurs. 31, 536–547. doi: 10.1046/j.1365-2648.2000.01308.x
Haichar, F. Z., Marol, C., Berge, O., Rangel-Castro, J. I., and Prosser, J. I. (2008). Plant host habitat and root exudates shape soil bacterial community structure. Isme. J. 2, 1221–1230. doi: 10.1038/ismej.2008.80
Hsu, S., and Buckley, D. H. (2009). Evidence for the functional significance of diazotroph community structure in soil. ISME J. 3, 124–136. doi: 10.1038/ismej.2008.82
Huang, L. F., Song, L. X., Xia, X. J., Mao, W. H., Shi, K., Zhou, Y. H., et al. (2013). Plant-Soil feedbacks and soil sickness: From mechanisms to application in agriculture. J. Chem. Ecol. 39, 232–242. doi: 10.1007/s10886-013-0244-9
Jackson, M. L. (2005). Soil Chemical Analysis: Advanced Course. Madison, UW: Libraries Parallel Press, 432–433. doi: 10.1097/01.ss.0000223482.86664.14
Jami, E., Israel, A., Kotser, A., and Mizrahi, I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Jie, Z., Songze, W., Chenlu, Z., Zhanfeng, L., and Lixia, Z. (2014). Contributions of understory and/or overstory vegetations to soil microbial PLFA and nematode diversities in eucalyptus monocultures. Plos One 9:e85513.
Karthikeyan, A., Chandrasekaran, K., Geetha, M., and Kalaiselvi, R. (2013). Growth response of Casuarina equisetifolia Forst. Rooted stem cuttings;to Frankia in nursery and field conditions. J. Biosci. 38, 741–747. doi: 10.1007/s12038-013-9362-3
Li, S., and Wu, F. Z. (2018). Diversity and co-occurrence patterns of soil bacterial and fungal communities in seven intercropping systems. Front. Microbiol. 9:1521. doi: 10.3389/fmicb.2018.01521
Li, X. G., Ding, C. F., Hua, K., Zhang, T. L., Zhang, Y. N., Zhao, L., et al. (2014). Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 78, 149–159. doi: 10.1016/j.soilbio.2014.07.019
Li, Y. Y., Chen, X. H., Xie, Z. X., Li, D. X., Wu, P. F., Kong, L. F., et al. (2018). Bacterial diversity and nitrogen utilization strategies in the upper layer of the northwestern pacific ocean. Front. Microbiol. 9:797. doi: 10.3389/fmicb.2018.00797
Liu, J. G., Li, X. G., Jia, Z. J., Zhang, T. L., and Wang, X. X. (2017). Effect of benzoic acid on soil microbial communities associated with soilborne peanut diseases. Appl. Soil Ecol. 110, 34–42. doi: 10.1016/j.apsoil.2016.11.001
Lu, Q., Zhang, J. C., and Chen, L. S. (2017). Impact of monoculture of poplar on the rhizosphere microbial communities over time. Pedosphere 30, 487–495. doi: 10.1016/S1002-0160(17)60416-8
Mendes, R., Kruijt, M., Bruijn, I., Dekkers, E., Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Michalet, S., Rohr, J., Warshan, D., Bardon, C., Roggy, J. C., Domenach, A. M., et al. (2013). Phytochemical analysis of mature tree root exudates in situ and their role in shaping soil microbial communities in relation to tree N-acquisition strategy. Plant Physiol. Bioch. 72, 169–177. doi: 10.1016/j.plaphy.2013.05.003
Monteiro, D. A., Fonseca, E. S., Rodrigues, R. D. A. R., Silva, J. J. N., Silva, E. P., Balieiro, F. C., et al. (2020). Structural and functional shifts of soil prokaryotic community due to Eucalyptus plantation and rotation phase. Scient. Rep. 10, 1–14.
Oksanen, J., Kindt, R., Legendre, P., O’Hara, B., Simpson, G. L., et al. (2009). Vegan: Community Ecology Package. R. package. Version. 15–12.
Paterson, E., Gebbing, T., Abel, C., Sim, A., and Telfer, G. (2007). Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 173, 600–610. doi: 10.1111/j.1469-8137.2006.01931.x
Penton, C. R., Yang, C. Y., Wu, L. Y., Wang, Q., Zhang, J., Liu, F. F., et al. (2016). nifH-Harboring Bacterial Community Composition across an Alaskan Permafrost Thaw Gradient. Front. Microbiol. 7:1894. doi: 10.3389/fmicb.2016.01894
Pereira, E., Silva, M. C., Schloter-Hai, B., Schloter, M., van Elsas, J. D., and Salles, J. F. (2013). Temporal dynamics of abundance and composition of Nitrogen-Fixing communities across agricultural soils. PLoS One. 8:e74500. doi: 10.1371/journal.pone.0074500
Shi, D., Ni, J. P., Ni, C. S., and Liu, J. C. (2018). The effects of different restoration years on plant diversity in Wushan’s alpine emigrant region. Acta Ecol. Sin. 39, 5584–5593. doi: 10.5846/stxb201802260392
Tibbits, K. J. (2008). Non-parametric multivariate comparisons of soil fungal composition: Sensitivity to thresholds and indications of structural redundancy in T-RFLP data. Soil Biol. Biochem. 40, 1601–1611. doi: 10.1016/j.soilbio.2008.01.008
Wakelin, S., Young, S., Gerard, E., Mander, C., and O’Callaghan, M. (2017). Isolation of root-associated Pseudomonas and Burkholderia spp. with biocontrol and plant growth-promoting traits. Biocontrol. Sci. Techn. 27, 139–143. doi: 10.1080/09583157.2016.1248899
Wang, C., Zheng, M., Song, W., Wen, S., and Wang, B. (2017). Impact of 25 years of inorganic fertilization on diazotrophic abundance and community structure in an acidic soil in southern China. Soil Biol. Biochem. 113, 240–249. doi: 10.1016/j.soilbio.2017.06.019
Wang, Y., Sheng, H. F., He, Y., Wu, J. Y., Jiang, Y. X., et al. (2012). Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microb. 78, 8264–8271. doi: 10.1128/AEM.01821-12
Wardle, D. A., Bardgett, R. D., Klironomos, J. N., Setala, H., Putten, W. H., and Wall, D. H. (2004). Ecological linkages between aboveground and belowground biota. Science 304, 1629–1633. doi: 10.1126/science.1094875
Wu, L. K., Chen, J., Wu, H. M., Qin, X. J., Wang, J. Y., Wu, Y. H., et al. (2016a). Insights into the Regulation of Rhizosphere Bacterial Communities by Application of Bio-organic Fertilizer in Pseudostellaria heterophylla Monoculture Regime. Front. Microbiol. 7:1788. doi: 10.3389/fmicb.2016.01788
Wu, L. K., Wang, J. Y., Huang, W. M., Wu, H. M., Chen, J., Yang, Y. Q., et al. (2015). Plant-microbe rhizosphere interactions mediated by Rehmannia glutinosa root exudates under consecutive monoculture. Sci. Rep. 5:15871. doi: 10.1038/srep15871
Wu, L. K., Wu, H. M., Chen, J., Wang, J. Y., and Lin, W. X. (2016b). Microbial community structure and its temporal changes in Rehmannia glutinosa rhizospheric soils monocultured for different years. Eur. J. Soil Biol. 72, 1–5. doi: 10.1016/j.ejsobi.2015.12.002
Wu, Z. Y., Li, J. J., Zheng, J., Liu, J. F., Liu, S. Y., Lin, W. X., et al. (2017). Soil microbial community structure and catabolic activity are significantly degenerated in successive rotations of Chinese fir plantations. Sci. Rep. 7:6691. doi: 10.1038/s41598-017-06768-x
Xia, Y., Lu, M. X., Chen, G., Cao, J. M., Gao, F. Y., Wang, M., et al. (2018). Effects of dietary Lactobacillus rhamnosus JCM1136 and Lactococcus lactis subsp lactis JCM5805 on the growth, intestinal microbiota, morphology, immune response and disease resistance of juvenile Nile tilapia, Oreochromis niloticus. Fish Shellfish Immun. 76, 368–379. doi: 10.1016/j.fsi.2018.03.020
Xiong, W., Zhao, Q. Y., Zhao, J., Xun, W. B., Li, R., Zhang, R. F., et al. (2015). Different continuous cropping spans significantly affect microbial community membership and structure in a vanilla-grown soil as revealed by deep pyrosequencing. Microbiol. Ecol. 70, 1–10. doi: 10.1007/s00248-014-0516-0
Yousuf, B., Kumar, R., Mishra, A., and Jha, B. (2014). Differential distribution and abundance of diazotrophic bacterial communities across different soil niches using a gene-targeted clone library approach. FEMS Microbiol. Lett. 360, 117–125. doi: 10.1111/1574-6968.12593
Zehr, J. P., Mellon, M. T., and Zani, S. (1998). New Nitrogen-Fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444. doi: 10.1002/(SICI)1097-0290(19980905)59:5
Zhang, L. N., Wang, D. C., Hu, Q., Dai, X. Q., Xie, Y. S., Li, Q., et al. (2019). Consortium of plant growth-promoting rhizobacteria strains suppresses sweet pepper disease by altering the rhizosphere microbiota. Front. Microbiol. 10:1668. doi: 10.3389/fmicb.2019.01668
Zhang, M. Y., Xu, Z. H., Teng, Y., Christie, P., Wang, J., Ren, W. J., et al. (2016). Non-target effects of repeated chlorothalonil application on soil nitrogen cycling: The key functional gene study. Sci. Tot. Environ. 543, 636–643. doi: 10.1016/j.scitotenv.2015.11.053
Zhao, J., Mei, Z., Zhang, X., Xue, C., Zhang, C. Z., Ma, T. F., et al. (2017). Suppression of Fusarium wilt of cucumber by ammonia gas fumigation via reduction of Fusarium population in the field. Sci. Rep. 7:43103. doi: 10.1038/srep43103
Zhao, J., Wan, S. Z., Zhang, C. L., Liu, Z. F., Zhou, L. X., Fu, S. L., et al. (2014). Contributions of understory and/or overstory vegetations to soil microbial PLFA and nematode diversities in Eucalyptus monocultures. PLoS One. 9:e85513. doi: 10.1371/journal.pone.0085513
Zhao, K., Penttinen, P., Zhang, X. P., Ao, X. L., Liu, M. K., Yu, X. M., et al. (2014). Maize rhizosphere in Sichuan, China, hosts plant growth promoting Burkholderia cepacia with phosphate solubilizing and antifungal abilities. Microbiol. Res. 169, 76–82. doi: 10.1016/j.micres.2013.07.003
Zhong, C. L., Bai, J. Y., and Zhang, Y. (2005). Introduction and conservation of casuarina trees in china. Forest Res. 18, 345–350. doi: 10.1360/biodiv.050121
Zhou, L. T., Li, J. J., Luo, Y., Liu, S. Y., Chen, J., Wang, J. Y., et al. (2019). Variation in soil fungal community structure during successive rotations of Casuarina equisetifolia plantations as determined by high-throughput sequencing analysis. Plant Growth Regul. 87, 445–453. doi: 10.1007/s10725-019-00483-5
Keywords: Casuarina equisetifolia, monoculture rotations, nifH gene sequencing, diazotrophic microbial community, soil
Citation: Zhou L, Li J, Pokhrel GR, Chen J, Zhao Y, Bai Y, Zhang C, Lin W, Wu Z and Wu C (2021) nifH Gene Sequencing Reveals the Effects of Successive Monoculture on the Soil Diazotrophic Microbial Community in Casuarina equisetifolia Plantations. Front. Plant Sci. 11:578812. doi: 10.3389/fpls.2020.578812
Received: 01 July 2020; Accepted: 21 December 2020;
Published: 25 January 2021.
Edited by:
Sanushka Naidoo, University of Pretoria, South AfricaReviewed by:
Bing Yang, Chengdu Institute of Biology, Chinese Academy of Sciences, ChinaAmit K. Jaiswal, Purdue University, United States
Copyright © 2021 Zhou, Li, Pokhrel, Chen, Zhao, Bai, Zhang, Lin, Wu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zeyan Wu, d3V6ZXlhbjA5NzdAMTI2LmNvbQ==
†These authors share first authorship
 Liuting Zhou
Liuting Zhou Jianjuan Li2†
Jianjuan Li2† Ganga Raj Pokhrel
Ganga Raj Pokhrel Jun Chen
Jun Chen Yanlin Zhao
Yanlin Zhao Wenxiong Lin
Wenxiong Lin