Abstract
The salt overly sensitive (SOS) signal transduction pathway is one of the most highly studied salt tolerance pathways in plants. However, the molecular mechanism of the salt stress response in Tamarix hispida has remained largely unclear. In this study, five SOS genes (ThSOS1–ThSOS5) from T. hispida were cloned and characterized. The expression levels of most ThSOS genes significantly changed after NaCl, PEG6000, and abscisic acid (ABA) treatment in at least one organ. Notably, the expression of ThSOS3 was significantly downregulated after 6 h under salt stress. To further analyze ThSOS3 function, ThSOS3 overexpression and RNAi-mediated silencing were performed using a transient transformation system. Compared with controls, ThSOS3-overexpressing transgenic T. hispida plants exhibited greater reactive oxygen species (ROS)-scavenging capability and antioxidant enzyme activity, lower malondialdehyde (MDA) and H2O2 levels, and lower electrolyte leakage rates under salt stress. Similar results were obtained for physiological parameters in transgenic Arabidopsis, including H2O2 and MDA accumulation, superoxide dismutase (SOD) and peroxidase (POD) activity, and electrolyte leakage. In addition, transgenic Arabidopsis plants overexpressing ThSOS3 displayed increased root growth and fresh weight gain under salt stress. Together, these data suggest that overexpression of ThSOS3 confers salt stress tolerance on plants by enhancing antioxidant enzyme activity, improving ROS-scavenging capability, and decreasing the MDA content and lipid peroxidation of cell membranes. These results suggest that ThSOS3 might play an important physiological role in salt tolerance in transgenic T. hispida plants. This study provides a foundation for further elucidation of salt tolerance mechanisms involving ThSOSs in T. hispida.
Introduction
High salinity is a major adverse environmental factor affecting plant growth and development due to osmotic and ionic stress (El Mahi et al., 2019; Yang et al., 2019). Plants have evolved several mechanisms to respond to harsh environments and adjust their growth under high-salt conditions (Munns and Tester, 2008). Ca2+ is a ubiquitous secondary messenger that is involved in the signaling of various environmental and developmental stimuli (Hilleary et al., 2018; Srinivasan and Roberto, 2018). Rapid and significant changes in intracellular Ca2+ concentrations can occur in response to such stimuli, and these changes are sensed and decoded by Ca2+ sensors, including calmodulins, calmodulin-like proteins, calcineurin B-like proteins (CBLs), and Ca2+-dependent protein kinases (Kudla et al., 2010). Ca2+ sensors or Ca2+-binding proteins can sense transient Ca2+ changes and alter the protein phosphorylation and gene expression of related proteins or genes, thus enabling plants to survive stress conditions (Luan, 2009).
The salt overly sensitive (SOS) 3 gene acts as a Ca2+ receptor in plants and is involved in Ca2+ signal-mediated stress responses. This gene, which encodes a calcineurin-like protein, belongs to the CBL gene family and is also known as CBL4. The SOS3 gene was originally identified in the model plant Arabidopsis (Kamei et al., 2010). The SOS3-encoded protein contains EF-hand domains in its C-terminus and a myristylation site in its N-terminal region. Myristylation is important for recruitment of SOS3 to the plasma membrane and for salt tolerance in plants (Quintero et al., 2002). SOS3 physically interacts with SOS2, a serine/threonine protein kinase. In the presence of Ca2+, SOS3 activates the substrate phosphorylation activity of SOS2. The binding of SOS3 to SOS2 is mediated by the 21-amino acid (aa) FISL motif in SOS2 and activates SOS1, a Na+/H+ antiporter, leading to Na+ efflux from the cytosol (Quintero et al., 2002; Nutan et al., 2018). Moreover, an SOS2–SOS3 interaction in the SOS pathway has also been demonstrated using sos3/sos2 double-mutant Arabidopsis (Guo et al., 2001). In addition to interacting with SOS3 at the plasma membrane, SOS2 has also been reported to interact with and thereby regulate the activity of several tonoplast-localized transporters, such as the Ca2+/H+ antiporter, vacuolar V-ATPase, and the Na+/H+ exchanger (Cheng et al., 2004; Batelli et al., 2007; Huertas et al., 2012).
SOS3s and CIPKs from different plant species have been found to function in different tissues in response to abiotic stress. For example, SOS2 and SOS3 specifically mediate salt stress signal transduction in Arabidopsis roots (Qiu et al., 2002; Zhang et al., 2017). OsSOS2 and OsSOS3 can coordinate to activate OsSOS1 in yeast cells and can be exchanged with their Arabidopsis counterparts to form heterologous protein kinase modules that activate both OsSOS1 and AtSOS1 and suppress the salt sensitivity of sos2 and sos3 Arabidopsis mutants (Martínez-Atienza et al., 2007).
The SOS3 gene has also been reported to be involved in plant salt stress responses. For example, in Arabidopsis, SOS3 has been shown to play a unique role in these responses (Kamei et al., 2010). Notably, duplication of SOS3 increases the Ca2+-mediated signaling capacity in Eutrema and confers increased salt tolerance on salt-sensitive Arabidopsis (Monihan et al., 2019). Overexpression of the SOS3 gene in tobacco increases salt stress by causing exclusion of Na+ from the cytosol and retention of high K+ levels in the cytosol to re-establish ion homeostasis (Li et al., 2013). In Populus trichocarpa, PtSOS1, PtSOS2, and PtSOS3 have been identified to cooperate in the activation of PtSOS1, thus conferring salt tolerance on P. trichocarpa (Tang et al., 2010).
Tamarix hispida is a woody halophyte species with excellent stress resistance. It can form natural forests in soil with a 1% salt content and is thus an excellent material for research on salt tolerance mechanisms and for cloning of salt tolerance genes. A search of T. hispida transcriptome libraries yielded 5 ThSOS genes. In this study, the 5 ThSOS genes (ThSOS1–ThSOS5) were cloned and characterized, and their expression levels under different abiotic stress conditions were analyzed by qRT-PCR. To further study ThSOS3 function, appropriate vectors for ThSOS3 overexpression and RNAi-mediated silencing were generated and transformed into T. hispida. The results showed that ThSOS3 significantly improved salt tolerance in transgenic plants. The results of this investigation thus provide a candidate salt tolerance gene for forest genetic breeding.
Materials and Methods
Plant Growth and Stress Treatments
T. hispida seedlings (Turpan Desert Botanical Garden) were planted in pots containing a perlite/vermiculite/soil mixture (1:1:4 v/v) in a greenhouse (70–75% relative humidity; 14 h light/10 h dark photocycle, approximately 600 mmol m–2 s–1; 24°C). Uniformly developed 2 months old T. hispida seedlings were irrigated with a solution of 0.4 M NaCl, 20% (w/v) PEG6000 or 100 μM abscisic acid (ABA), and the tissues were harvested at 6, 12, 24, 48, and 72 h. Seedlings irrigated with fresh water were harvested at the corresponding time points as controls. Each treatment was repeated three times.
Arabidopsis thaliana Columbia (wild-type, WT) plants were used in this study. After vernalization, Arabidopsis seeds were sterilized for 3–5 min with 2.5% (v/v) sodium hypochlorite and washed three times with distilled water. The seeds were soaked on 1/2-strength Murashige and Skoog (MS) solid medium plates containing 0.6% agar. One-week-old seedlings were transferred from the plates to pots filled with vermiculite/soil/perlite (1:3:1) and grown in a greenhouse under a 16 h light/8 h dark photocycle, 70–75% relative humidity, 500 μmol m–2 s–1 light intensity, and a stable temperature of 22°C.
Cloning and Sequence Analysis of ThSOSs
ThSOS genes were searched in the transcriptome library database and were further identified using conserved domain BLAST1. A phylogenetic tree was constructed with T. hispida SOS proteins and SOS homologs from other species using the neighbor-joining method in MEGA 5.0 via the bootstrap method with 1,000 replications (Tamura et al., 2011). Multiple sequence alignment was performed using ClustalX with a gap extension and gap opening penalties of 10 and 0.1, respectively (Thompson et al., 1997). The theoretical isoelectric point (pI) and molecular weight (MW) of each ThSOS protein were studied with the Expert Protein Analysis System (ExPASy) “compute pI/Mw” tool2. ThSOS subcellular localization was predicted using CELLO v.2.53. The ∼2 kb upstream promoter region of each ThSOS gene was searched with the T. hispida genome database. The putative cis-acting elements of 3 ThSOS genes were analyzed with the Plant Cis-Acting Regulatory DNA Elements (PLACE) database4.
RNA Extraction and qRT-PCR Analyses
Total RNA was extracted from T. hispida plants with a Plant RNeasy Extraction Kit (BioTeke, China), and first-strand cDNA was synthesized from 1 μg of purified RNA using a PrimeScriptTM RT Reagent Kit (TaKaRa, Beijing, China). qRT-PCR was performed on a qTOWER3 G (Analytik Jene AG, Germany) with the Actin (FJ618517) and β-tubulin (FJ618519) genes as internal controls. Each 20 μL reaction mixture contained 10 μL of SYBR-Green Real-time PCR Master Mix (Toyobo, Shanghai, China), specific primers (0.5 μM each), and 2 μL of cDNA template. Amplification was performed using the following cycling parameters: 94°C for 30 s; 45 cycles of 94°C for 12 s, 58°C for 30 s, and 72°C for 45 s; and then 82°C for 1 s for plate reading (Liu et al., 2018). The relative abundance was determined by the 2–ΔΔCt method (Livak and Schmittgen, 2001). Three replicates were included for each sample (the primers used for qRT-PCR are shown in Table 1).
TABLE 1
| Constructs | Forward and reverse primers (5′–3′) | ||
|
|
|||
| Genes | Primers used in real-time RT-PCR analysis | ||
| ThSOS1 | CTGATGCTGATCTGGATCCTAT | ATGCTAGACTGAAGAAATCGGT | |
| ThSOS2 | AGTAGAGGCCTTGTACGAGCT | ACCCAGTATGCCTCAGATCAT | |
| ThSOS3 | TGACGTTGATCCGATCAATTC | CCATAACAGGATCACATGCATAT | |
| ThSOS4 | CAATTTGCCTTATTCAGGAAT | CCACTATGCTCCCAACGATCT | |
| ThSOS5 | ATAGCCCACCATGGACGGCTT | ACCCTTGTGACTGAGAACCT | |
| Actin (FJ618517) | AAACAATGGCTGATGCTG | ACAATACCGTGCTCAATAGG | |
| β-tubulin (FJ618519) | GGAAGCCATAGAAAGACC | CAACAAATGTGGGATGCT | |
|
|
|||
| Primers used in constructing plant plasmids | |||
|
|
|||
| pROKII-ThSOS3 | GCTCTAGAATGGGCTGCTTCCATTCAAAG | GGTACCCCGTACTTCTGAATCTTCAACTT | |
| pFGC5941-ThSOS3 | ThSOS3-Sense-F: | ThSOS3-Sense-R: | |
| CATGCCATGGATGGGCTGCTTCCATTCAAAG | GCTCTAGATTATACTTCTGAATCTTCAACTTCCG | ||
| ThSOS3-Anti-F: GCTCTAGAATGGGCTGCTTCCATTCAAAG | ThSOS3-Anti-R: CATGCCATGGTTATACTTCTGAATCTTCAACTTCCG | ||
Primers sequences used in this study.
Transient Expression of the Thsos3 Gene in T. hispida
A 642 bp cDNA sequence of ThSOS3 was amplified, cloned into the prokII vector with the CaMV 35S promoter, and named 35S::ThSOS3. A 200 bp truncated inverted-repeat cDNA sequence of ThSOS3 was cloned into pFGC5941 flanking the CHSA intron to generate pFGC5941::ThSOS3 and used to silence the expression of ThSOS3 (the primers for vector construction are listed in Table 1).
T. hispida plants were transiently transformed according to the methods of Zhang et al. (2018). Three groups of transgenic T. hispida plants were generated by transient transformation: 35S::ThSOS3 plants were generated to overexpress ThSOS3 (OE), pFGC5941::ThSOS3 plants were generated to silence the expression of ThSOS3 (SE), and empty pROKII vector-transformed plants were generated as controls (Con). After growth for 12, 24, and 36 h under normal conditions or salt treatment, the relative abundance of ThSOS3 in these transformed T. hispida plants was studied using qRT-PCR. Three biological replicates were included that contained at least 60 transformed seedlings.
Stress Tolerance Analysis
Stably transformed Arabidopsis plants were generated by the floral dip method (Clough and Bent, 1998). Two T3 generation homozygous ThSOS3 transgenic lines (OE1 and OE2) were selected to further evaluate stress tolerance. After vernalization, Arabidopsis seeds were sown on 1/2-strength MS medium and grown for 5 days prior to being transferred to 1/2-strength MS medium with or without 120 mM NaCl for 10–14 days. The root length and fresh weight were measured, and the seedlings were imaged. To assess salt tolerance in soil, seeds of two T3 generation homozygous transgenic lines (OE1 and OE2) of ThSOS3 and the Con (WT) line were sown on 1/2 MS solid medium for 5–7 days and then transferred to soil. After 3 weeks of growth, the seedlings were irrigated with a 150 mM NaCl solution for 5 days and then imaged. The treatments were independently repeated at least three times. Each sample was analyzed and contained at least 15 transformed seedlings.
Biochemical Staining
Hydrogen peroxide (H2O2) and superoxide (O2–) staining were performed by infiltration with 3-3-diaminobenzidine (DAB) or nitro blue tetrazolium (NBT) following the procedures described by Zhang et al. (2011). Evans blue staining was performed to investigate cell death, as described by Liu et al. (2018).
Physiological Index Measurement of Transformed Plants
Transformed T. hispida plantlets were grown on 1/2 MS solid medium supplemented with 150 mM NaCl for 12–36 h. Four-week-old Arabidopsis seedlings were subjected to 150 mM NaCl for 5 days. After treatment, the seedlings were collected and subjected to physiological index analysis. Superoxide dismutase (SOD) and peroxidase (POD) activity and H2O2 content were measured using corresponding reagent kits (Nanjing Jiancheng Bioengineering Institute, China) according to the manufacturer’s instructions. The MDA content was determined according to the methods of Dhindsa et al. (1981). Electrolyte leakage was analyzed as described by Ben-Amor et al. (1999). Each sample contained at least nine harvested seedlings, and all of the experiments were repeated at least three times.
Statistical Analyses
Statistical analyses were carried out using Excel software. The data were compared using Student’s t-test, and differences were considered significant if P < 0.05. ∗Represents a significant difference (P < 0.05), and ∗∗represents a very significant difference (P < 0.01).
Results
Gene Identification and Sequence Analysis of ThSOS Genes
In total, five candidate SOS genes were selected and identified. The 5 ThSOS proteins ranged from 213 to 1,165 aa in length (Table 2). Large variations were found in the theoretical pI values (ranging from 4.76 to 6.42) and the MW values (ranging from 22.42 to 128.83 kDa) of the proteins encoded by the five ThSOS genes. The prediction results showed that the five ThSOS genes were localized in the plasma membrane, cytoplasm, extracellular space, or chloroplasts (Table 2).
TABLE 2
| Name | Locus | ORF (bp) | Introns | Protein length | Theoretical pI | Aliphatic index | Molecular weight (kD) | Localization predictions |
| ThSOS1 | Unigene22889 | 3,498 | 11 | 1,165 | 6.42 | 103.24 | 128.83 | Plasma membrane |
| ThSOS2 | Unigene13265 | 1,371 | 2 | 456 | 6.29 | 92.08 | 51.44 | Cytoplasmic |
| ThSOS3 | Unigene1212 | 642 | 6 | 213 | 4.76 | 96.53 | 22.42 | Cytoplasmic |
| ThSOS4 | Unigene24293 | 927 | 11 | 308 | 6.22 | 105.1 | 33.58 | Extracellular or Chloroplast |
| ThSOS5 | Unigene1744 | 675 | 0 | 224 | 5.00 | 88.39 | 25.57 | Cytoplasmic |
Features of ThSOS genes in T. hispida.
To determine the subclasses of the ThSOS genes, phylogenetic analysis was performed using the sequences of the ThSOS proteins and SOS proteins from other species (Figure 1A and Supplementary Table S1). The results revealed that ThSOS1, ThSOS2, and ThSOS3 genes were closely related to the SOS1, SOS2, and SOS3 subfamilies in Arabidopsis. ThSOS3 belonged to the CBL4 subfamily and clustered into the same clade as PtrSOS3, MnSOS3, and AtSOS3. Multiple sequence alignment analysis showed that ThSOS3 was closely related to PtrSOS3 (XP-002318422.1) from P. trichocarpa, MnSOS3 (XP-010100753.1) from Morus notabilis, and AtSOS3-1 (AT5G24270) from A. thaliana (Figure 1B).
FIGURE 1

Phylogenetic and sequence analysis of ThSOS proteins. (A) Phylogenetic analysis of ThSOSs and other SOS proteins from different plant species. (B) Alignment of the ThSOS3 protein sequence with other plant SOS3 protein sequences. The sequences of SOS proteins were downloaded from the GenBank database, and their GenBank accession numbers are listed in Supplementary Table S1.
Characterization of Cis-Elements in ThSOS Gene Promoters
Numerous stress-related consensus cis-acting elements were detected, including an ABA-responsive element (ABRE), an antioxidant response element (ARE), and TC-rich repeats. Moreover, ThSOS1 included 1 abiotic stress-related element, the MYB-binding site (MBS); nine hormone stress-related elements, such as the salicylic acid-responsive element (TCA element), methyl jasmonate (MeJA)-responsive element (CGTCA motif or TGACG motif), gibberellin-responsive element (TATC-box), and auxin-responsive element (TGA element); and 1 development-related endosperm expression element (GCN4 motif). ThSOS3 contained an MBS and low temperature response (LTR) response elements consisting of five hormone stress-related elements, auxin-responsive elements (TGA elements or AuxRR core elements), and salicylic acid-responsive elements (TCA elements). Similarly, ThSOS4 contained many abiotic and hormone stress-related elements, as shown in Supplementary Figure S1.
Expression of ThSOS Genes Under Abiotic Stress and ABA Treatment
To analyze the relative abundance of ThSOS genes, the expression profiles of the 5 ThSOS genes were measured under different stresses (NaCl or PEG6000 exposure) or hormone treatment (ABA) using qRT-PCR.
In roots under NaCl stress, the expression of most ThSOS genes was upregulated. Notably, ThSOS4 and ThSOS5 exhibited upregulated expression at all stress time points. The highest expression levels of ThSOS4 and ThSOS5 were 8.02- and 4.86-fold higher than control levels, respectively. The other three ThSOS genes, ThSOS1, ThSOS2, and ThSOS3, were downregulated at the initial stress time point and upregulated at later stages. The lowest expression levels of the three ThSOS genes in the roots all occurred at 6 h. The expression levels of ThSOS1, ThSOS2, and ThSOS3 at this time point were 3.55, 6.11, and 0.20% of control levels, respectively. These results indicate that these 3 genes can respond rapidly to salt stress in T. hispida roots. In leaves, ThSOS gene expression was mainly downregulated during the stress period. ThSOS2 and ThSOS3 reached their lowest expression levels (3.78 and 1.56% of baseline levels, respectively) in the control plants at 6 h. The relative abundance of ThSOS1, ThSOS4, and ThSOS5 was similar to that of ThSOS2 and ThSOS3, but the lowest expression levels were achieved at 24 h (Figure 2A).
FIGURE 2
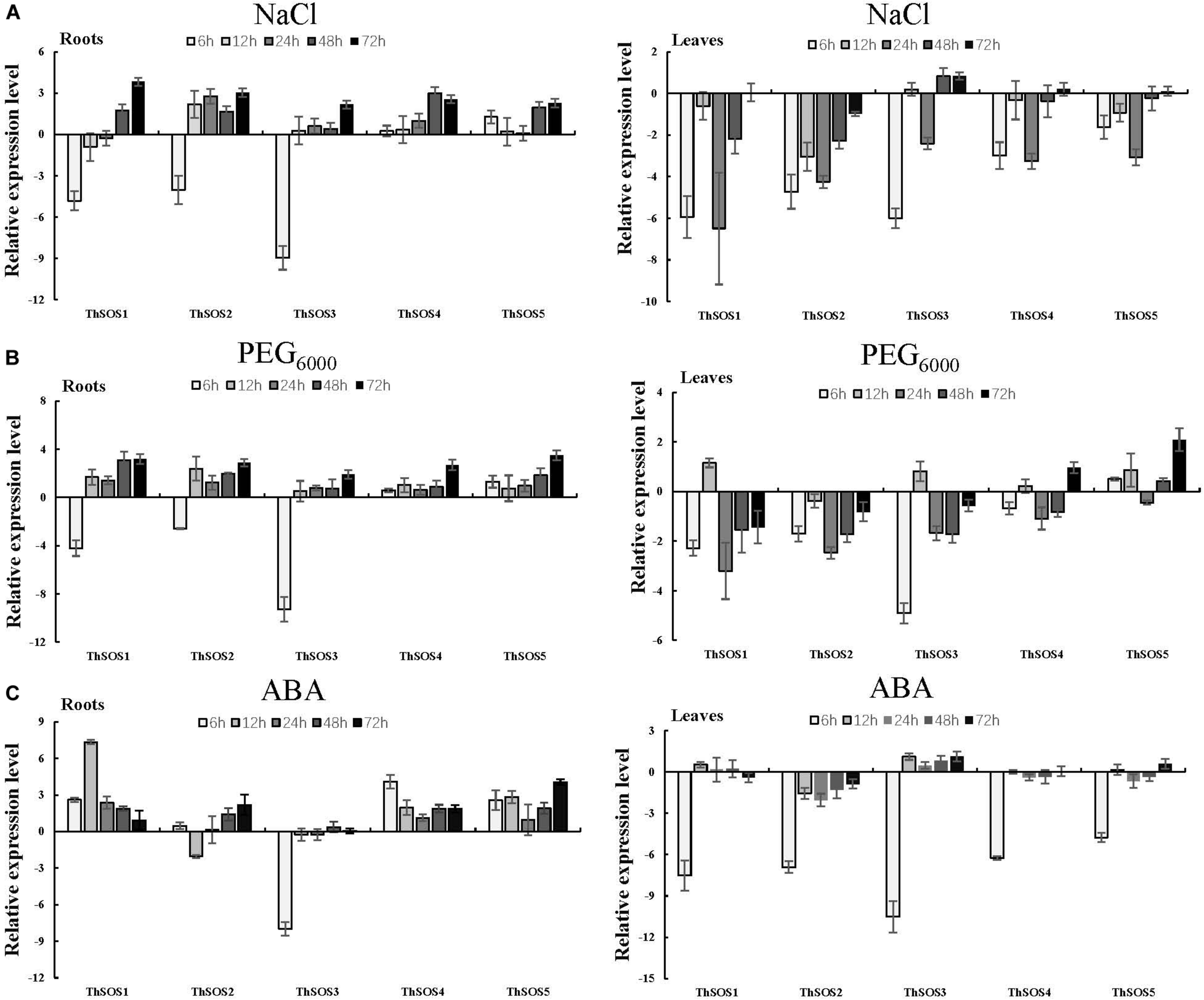
Expression analysis of the 5 ThSOS genes in the roots and leaves in response to abiotic stresses (NaCl and PEG6000 exposure) and hormone (ABA) treatment. (A) 0.4 M NaCl. (B) 20% (w/v) PEG6000. (C) 100 μM ABA. All relative transcription levels were log2-transformed. The error bars were obtained from multiple replicates of qRT-PCR.
Under PEG6000 stress, most of the ThSOS genes were significantly upregulated, and all ThSOS genes achieved their highest expression levels at 72 h. Interestingly, the expression levels of ThSOS1, ThSOS2, and ThSOS3 in the roots were significantly downregulated after 6 h of PEG6000 stress (5.39, 16.67, and 0.16% of the control levels, respectively). In leaves, the expression levels of the ThSOS1, ThSOS2, and ThSOS3 genes were mainly downregulated throughout the stress period, and ThSOS1 and ThSOS2 achieved their lowest expression levels at 24 h. However, ThSOS3 reached its lowest expression level during the early stage of stress (6 h). In contrast to the gene expression patterns of these three ThSOS genes, the relative expression of ThSOS5 was significantly upregulated at almost all stress points (in addition to 24 h) and peaked at 72 h. The expression of ThSOS4 did not change significantly under PEG6000 stress (Figure 2B).
Under ABA stress, the relative abundance of ThSOS1, ThSOS4, and ThSOS5 was significantly upregulated in the roots. The most strongly upregulated gene was ThSOS1; its expression peaked at levels 161.28-fold higher than control levels at 12 h. The relative expression of ThSOS2 was mainly upregulated except at 12 h, when its expression was only 24.3% of the control level. However, ThSOS3 expression was clearly downregulated at 6 h (0.4% of the control level) and showed no significant changes at any other stress time points. In leaves, no significant changes were found in the expression of any of the ThSOS genes except at 6 h. All the ThSOS genes (ThSOS1, ThSOS2, ThSOS3, ThSOS4, and ThSOS5) reached their lowest expression levels under ABA stress at 6 h (0.55, 0.83, 0.07, 1.31, and 3.17% of the control levels, respectively) (Figure 2C).
Transient Expression of ThSOS3 in T. hispida
To ascertain whether the ThSOS3 gene was successfully transiently overexpressed and suppressed in T. hispida, the ThSOS3 transcript levels in Con, OE, and SE plants were examined by qRT-PCR. Compared with that in Con plants, the ThSOS3 expression in OE plants was significantly increased under salt stress conditions, while that in SE plants was significantly decreased (Figure 3), indicating that the gain and loss of function of ThSOS3 in T. hispida plants were successfully achieved.
FIGURE 3
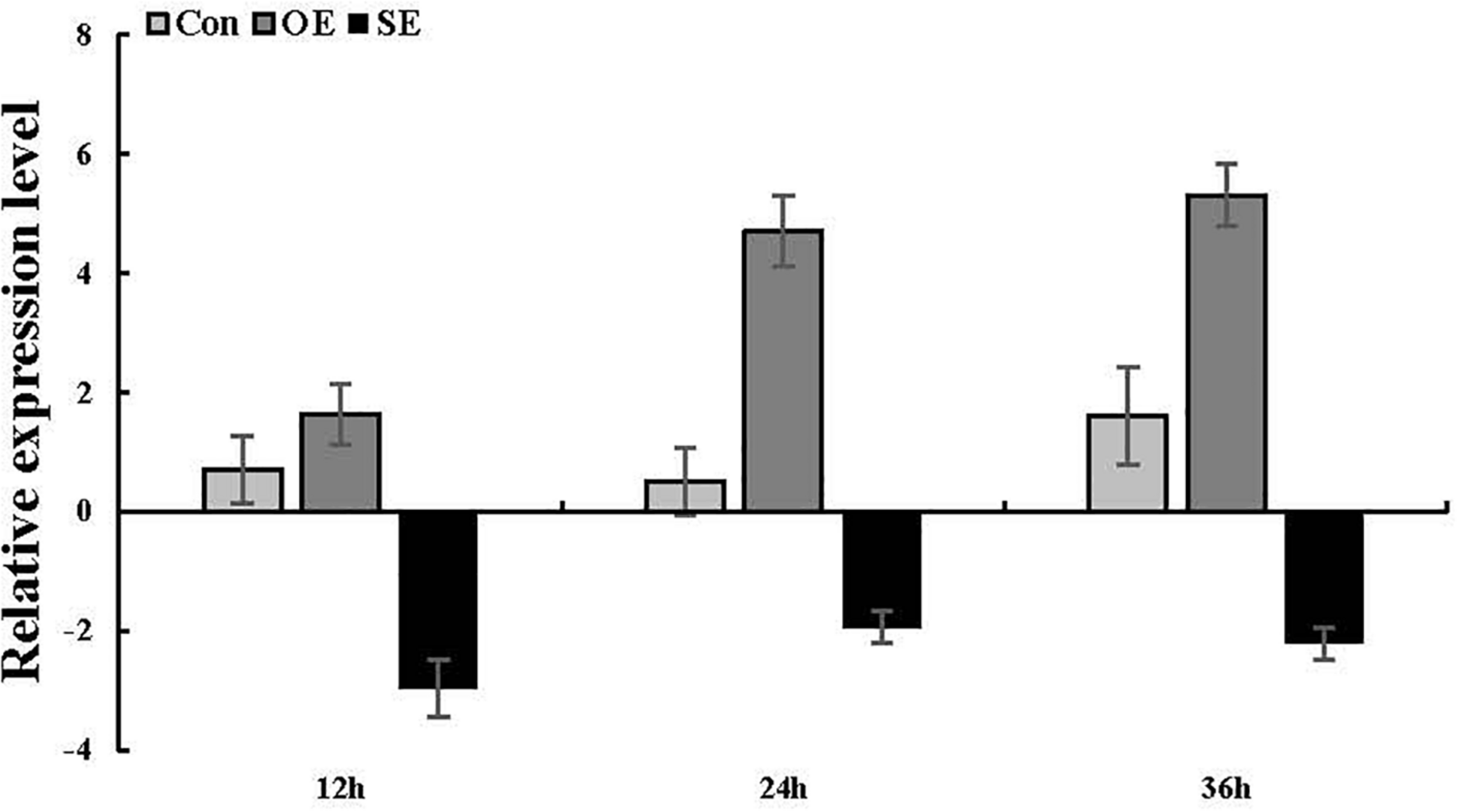
ThSOS3 transcript levels in T. hispida plants with transient overexpression or knockdown of ThSOS3. The expression data were log2-transformed. Two-month-old T. hispida plants were transiently transformed with empty pROKII, 35S::SOS3 or pFGC::SOS3. After transformation for 36 h, T. hispida plants were treated with 150 mM NaCl for 12, 24, or 36 h, and the expression of ThSOS3 was determined. OE, ThSOS3 overexpression; SE, ThSOS3 RNAi; Con, pROKII vector control.
ThSOS3 Confers Salt Stress Tolerance on Transgenic Plants
To preliminarily explore the function of the ThSOS3 gene, DAB staining and NBT staining were performed, and the related physiological indexes of three differently transformed T. hispida plants were studied. The reactive oxygen species (ROS) accumulation levels in OE, SE, and Con plants before and after abiotic stress were determined by DAB and NBT staining. Under salt stress, the staining intensity in OE plants was lower than that in Con plants, while that in SE plants was higher than that in Con plants (Figures 4A,B). Additionally, H2O2 and MDA levels were measured in different transgenic T. hispida. The results failed to demonstrate differences in H2O2 and MDA levels among the three transiently transgenic plants under normal conditions. However, under salt stress, SE plants showed the highest H2O2 and MDA levels, followed by Con plants; the OE plants had the lowest H2O2 and MDA levels. The levels of H2O2 and MDA in SE plants were 1.27 and 1.53 times those in Con plants, respectively. However, the H2O2 and MDA levels in OE plants were the lowest at only 82.02 and 85.2% of those in Con plants, respectively, at 24 h (Figures 4D,E).
FIGURE 4
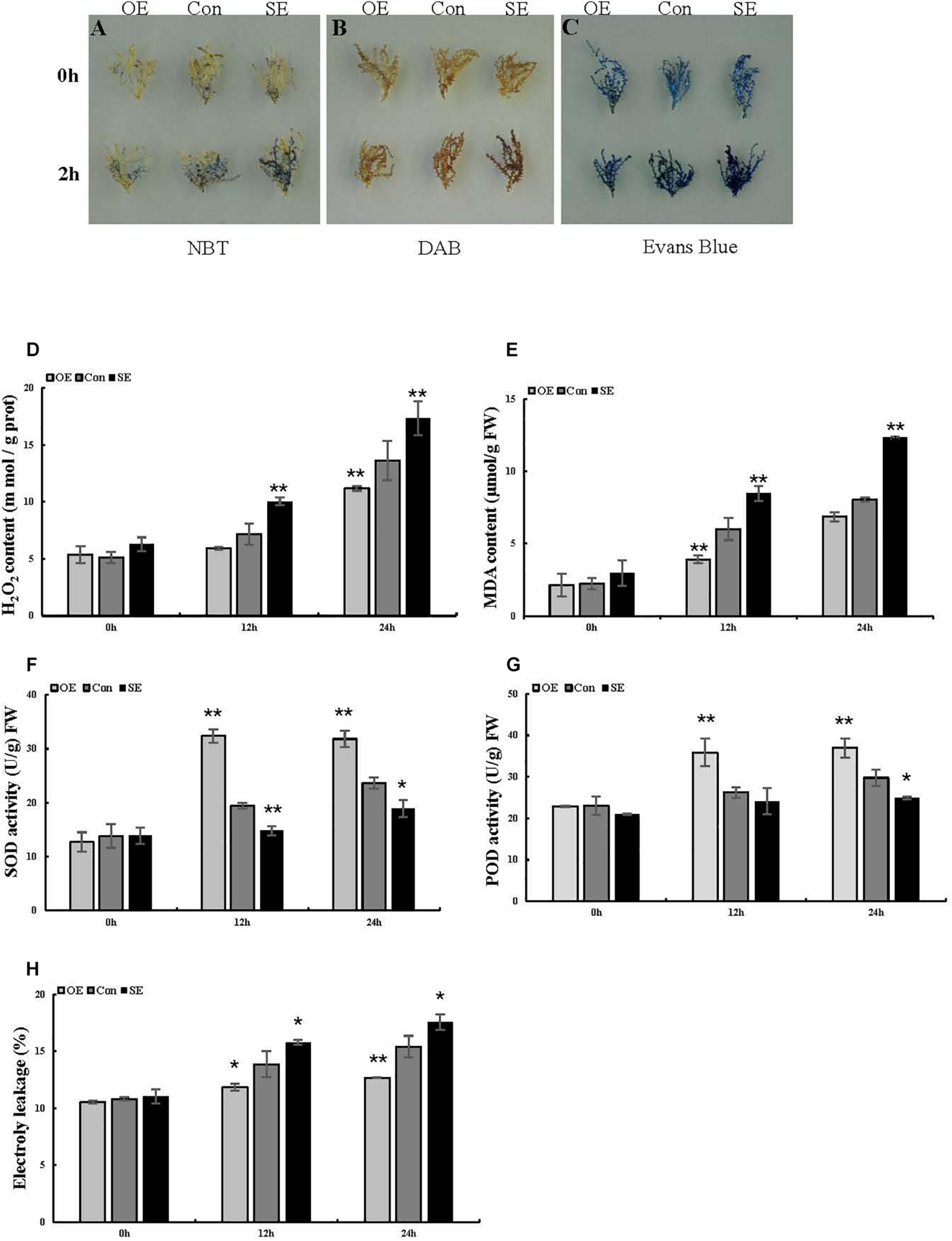
Analysis of ROS-scavenging capability and cell death in T. hispida plants with overexpression or RNAi-mediated knockdown of ThSOS3. (A) NBT and (B) DAB staining were performed to detect O2– and H2O2, respectively. (C) Evans blue staining was used to analyze cell death. Young branches from transformed T. hispida plants treated with 150 mM NaCl for 2 h were used for DAB, NBT, and Evans blue staining. (D–H) Analysis of H2O2 and MDA content, SOD and POD activity, and electrolyte leakage in three different transgenic T. hispida plants. Transformed T. hispida plantlets grown on 1/2 MS solid medium supplemented with 150 mM NaCl for 24 h were used to measure the H2O2(D) and MDA (E) content, SOD (F), and POD (G) activity, and electrolyte leakage (H). *Represents a significant difference (P < 0.05). **Represents a very significant difference (P < 0.01).
To better interpret the results of transient expression of ThSOS3 in T. hispida, ThSOS3 was overexpressed in Arabidopsis. Two independent T3 homozygous transgenic lines (OE1 and OE2) overexpressing ThSOS3 were selected and studied. Under salt treatment, the H2O2 and MDA levels in both OE lines were lower than those in the WT line, although the lines had similar H2O2 and MDA levels under normal conditions (Figures 5A,B,D). In addition, the phenotype, fresh weight, and root length were not different between the transgenic and WT plants under normal conditions. However, under salt treatment, the OE1 and OE2 plants presented significantly higher fresh weights and root lengths than the WT plants (Figures 6A–D).
FIGURE 5
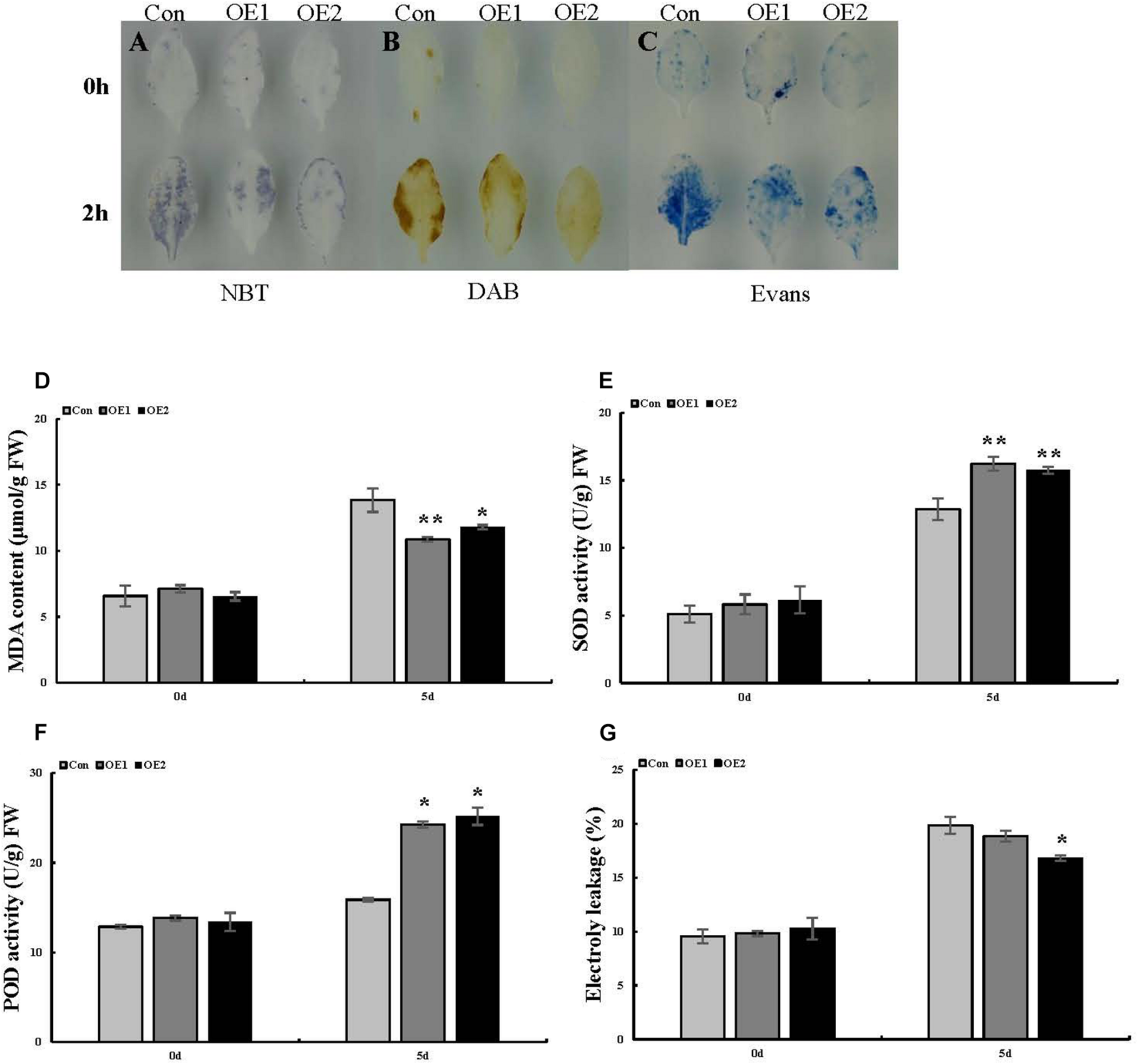
Analysis of ROS-scavenging capability and cell death in Arabidopsis plants overexpressing ThSOS3. (A) NBT, (B) DAB, and (C) Evans blue staining were performed. Leaves from ThSOS3-transformed and Con (WT Arabidopsis) plants treated with 150 mM NaCl for 24 h were used for histochemical staining. (D–F) Analysis of MDA content, SOD and POD activity, and electrolyte leakage in transgenic and Con (WT Arabidopsis) plants. Four-week-old Arabidopsis seedlings subjected to 150 mM NaCl for 24 h were used to detect the MDA (D) content, SOD (E) and POD (F) activity, and electrolyte leakage (G). *Represents a significant difference (P < 0.05). **Represents a very significant difference (P < 0.01).
FIGURE 6
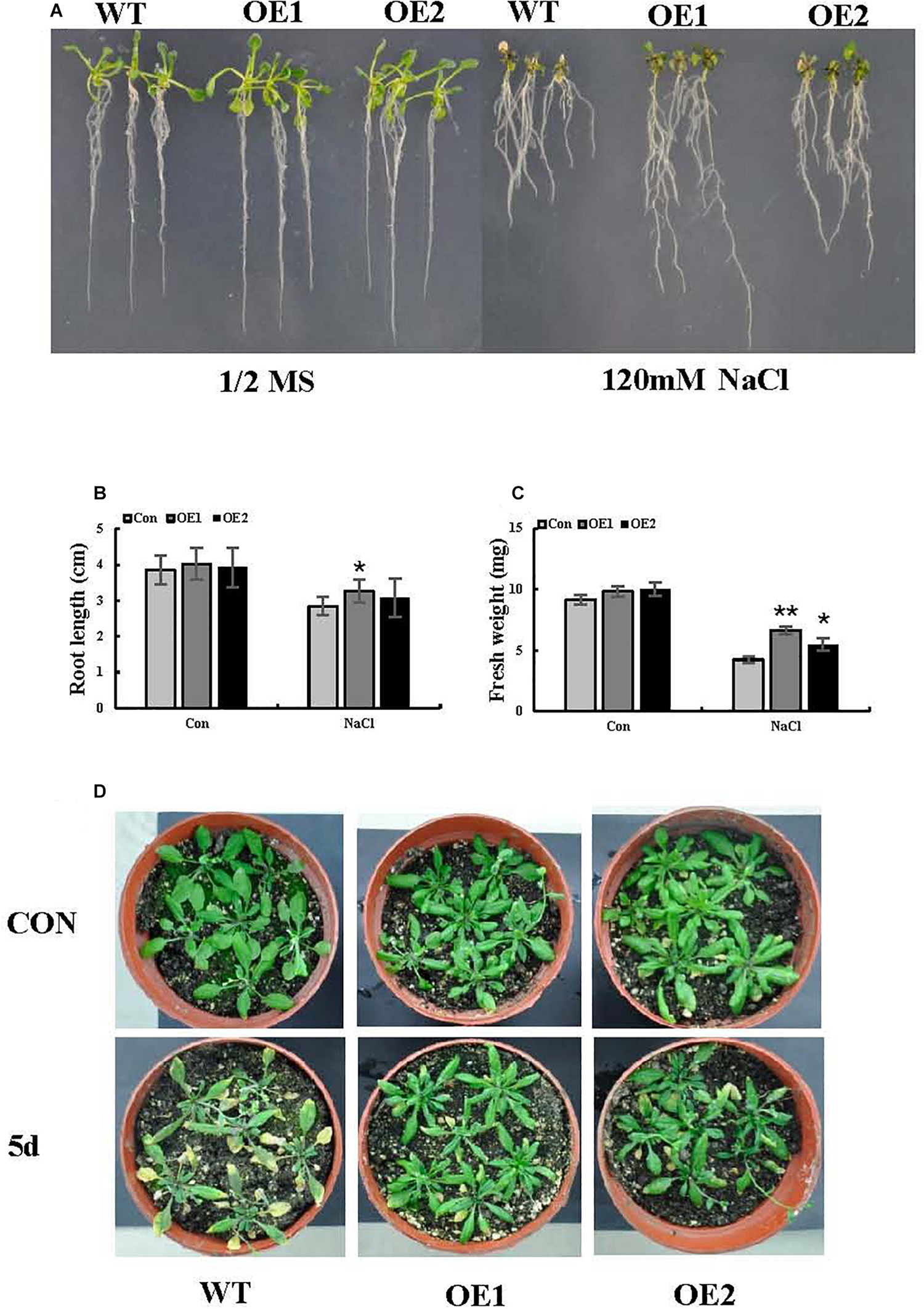
Salt stress tolerance associated with ThSOS3. (A) Growth comparison between OE and WT plants. Arabidopsis plants grown on 1/2-strength MS medium (control) and 1/2-strength MS medium supplied with 120 mM NaCl were used for growth analysis. (B) Root length and (C) fresh weight were also analyzed in at least 30 seedlings under each treatment. (D) Comparison of growth phenotypes between OE and WT Arabidopsis lines grown in soil. The plants were treated with 150 mM NaCl for 5 days for analysis. Plants grown under normal conditions were used as controls. *Represents a significant difference (P < 0.05). **Represents a very significant difference (P < 0.01).
ThSOS3 Improves ROS-Scavenging Capability
The antioxidant enzymes SOD and POD are the two most important ROS-scavenging enzymes influencing cellular ROS levels. Thus, we further studied POD and SOD activity. Under normal growth conditions, there were no obvious differences in activity levels between Con and transgenic T. hispida. However, SOD and POD activity levels were significantly increased in OE plants under salt stress. At 24 h, the activity levels of SOD and POD in OE plants were 1.35 and 1.24 times those in Con plants, while the activity levels in SE plants were only 80 and 83.73% of those in Con plants, respectively (Figures 4F,G).
Similarly, in Arabidopsis, SOD and POD activity did not obviously differ among the studied lines in the absence of stress. Under salt stress, the two OE lines showed higher antioxidant enzyme (SOD and POD) activity than the WT line (Figures 5E,F), consistent with the results obtained in T. hispida.
Cell Death and Electrolyte Leakage Analysis
Evans blue staining was used to assess cell membrane damage on the basis of the intensity of the staining under salt stress. Compared with the Con plants, the OE plants presented light-blue puncta with smaller areas under salt stress, while SE plants presented larger staining areas (Figure 4C). We then measured electrolyte leakage. Electrolyte leakage did not significantly differ among the three differently transformed T. hispida plants under normal conditions. Under salt stress, the relative electrolyte leakage rates of SE plants were the highest at 24 h at 1.14 times those of Con plants, while the rates of the OE plants were 0.82 times those of the Con plants (Figure 4H). Moreover, we detected changes in the levels of corresponding physiological indicators in Arabidopsis, which were consistent with the changes in T. hispida (Figures 5C,G).
Discussion
The SOS gene plays an important role in plants, and its function has been studied in A. thaliana (Qiu et al., 2004; Yang et al., 2009; Wang et al., 2019), Nicotiana tabacum (Yue et al., 2012), Oryza sativa (El Mahi et al., 2019), Gossypium raimondii (Che et al., 2019), Lycopersicon esculentum (Olias et al., 2009), Zea mays (Zörb et al., 2005), and P. trichocarpa (Tang et al., 2010; Zhou et al., 2014). However, few studies have investigated the salt tolerance function of ThSOS in T. hispida.
In our study, five monomorphic and intact ThSOS genes were selected. An unrooted phylogenetic tree and multiple sequence alignment analysis showed that ThSOS3 shared 85.92, 84, and 70% identity with PtrSOS3, MnSOS3, and AtSOS3-1, respectively. It has been reported that AtSOS3 and PtrSOS3 enhance salt tolerance in Arabidopsis and P. trichocarpa (Quan et al., 2007; Tang et al., 2010). The relative abundance of most ThSOS genes in T. hispida was significantly changed under NaCl, PEG6000, and ABA stresses. Notably, ThSOS3 expression was significantly downregulated under salt stress at 6 h. SOS3 is a Ca2+-regulated upstream regulatory protein of the SOS pathway and plays important roles in plant salt stress response pathways (Yang et al., 2009). Kim et al. (2013) confirmed that AtSOS3 expression is strongly induced by NaCl treatment. In addition, overexpression of LeSOS3-1 enhances salt stress tolerance in tobacco by regulating stress-associated physiological changes, such as by enhancing ROS-scavenging capability and maintaining K+/Na+ homeostasis.
Regulatory elements in promoter sequences are essential for the temporal, spatial, and cell type-specific control of gene expression (Jiang et al., 2014). Previous studies have shown that many abiotic and hormone stress-related elements are present in ThSOS gene promoters. As shown in Supplementary Figure S1, ABREs, AREs, and TC-rich repeats were found in the promoters of three ThSOS genes. The LTR element and MBS element were found in the promoter of the ThSOS3 gene. ThSOS3 also contained 4 hormone stress-related elements (a TGA element, a TCA element, an ABRE, and an AuxRR-core element). This result indicates that ThSOS might be involved in stress responses (to abiotic stress and hormone treatment) as well as in plant development. Moreover, previous studies have shown that OSBZ8 mediates salt and dehydration stress tolerance by binding to the ABRE motif (Narusaka et al., 2003), and AtMYB44 inhibits oxidative damage and hypersensitivity to abiotic stresses by binding to MBSs to activate the expression of related downstream genes (Persak and Pitzschke, 2014).
When the results of the analysis of cis-elements in ThSOS gene promoters and the phylogenetic analysis were combined, ThSOS3 was shown to contain abundant abiotic and hormone stress-related elements and to be closely related to AtSOS3 and PtrSOS3. Therefore, we predict that ThSOS3 might also play roles in responses to salt stress.
Plants produce high levels of ROS in adverse environments. ROS act as signaling molecules to control several physiological processes (Liu et al., 2018). Baxter et al. (2013) found that H2O2 and O2– signaling networks are involved in responses to abiotic stimuli. Under salt stress, the staining intensity was lower in OE plants and higher in SE plants than in Con plants. Furthermore, the H2O2 levels were consistent with the DAB and NBT staining results in transgenic T. hispida plants. The results showed that overexpression of ThSOS3 resulted in the lowest H2O2 and MDA accumulation. Conversely, compared to ThSOS3 overexpression, RNAi silencing induced the opposite physiological changes among the three differently transformed T. hispida plants. Next, Evans blue staining was performed to assess cell death in T. hispida plants under salt stress. The results indicated that compared with Con plants under salt stress, OE plants under salt stress presented light blue puncta with smaller areas, while SE plants showed the opposite results. We then measured electrolyte leakage, which did not significantly differ among the three differently transformed T. hispida plants under normal conditions. Under salt stress, the relative electrolyte leakage rates of SE plants were the highest at 24 h at 1.14 times those of Con plants; moreover, those of the OE plants were 0.82-fold those of the Con plants. The electrolyte leakage assay further confirmed the Evans blue staining results. Similar to our study, a previous study revealed that overexpression of the wheat TaAQP8 gene can confer salt tolerance on transgenic tobacco plants by maintaining ionic balance, reducing H2O2 accumulation and reducing membrane damage (Hu et al., 2012). In addition, ThWRKY4 can improve tolerance to salt and ABA treatment by increasing SOD and POD activity, decreasing O2 and H2O2 levels, reducing electrolyte leakage, preventing chlorophyll loss, and protecting cells from death (Zheng et al., 2013).
As ROS levels were significantly altered, we further studied the activity of POD and SOD, which are the two most important ROS-scavenging enzymes. In the absence of stress conditions, the activity levels of these enzymes did not significantly differ among the Con, OE, and SE plants. However, SOD and POD activity levels were significantly increased in OE plants under salt stress. At 24 h, the activity levels of SOD and POD in OE plants were 1.35 and 1.24 times those in Con plants, while the levels in SE plants were 80 and 83.73% of Con plants, respectively. As in our study, ThZFP1 was shown to enhance salt and osmotic stress tolerance in a previous study by positively regulating proline accumulation and SOD and POD activity (Zang et al., 2015). In addition, ThNAC7 has been found to induce the transcription of genes associated with stress tolerance to enhance salt and osmotic stress tolerance by increasing osmotic potential and enhancing ROS scavenging (He et al., 2019). In summary, the physiological indicator results suggest that ThSOS3 confers salt stress tolerance by increasing the activity of antioxidant enzymes (SOD and POD), reducing ROS accumulation, and decreasing the MDA content and lipid peroxidation of cell membranes.
Conclusion
The SOS gene plays important roles in responses to salt stress. However, few studies have evaluated the roles of ThSOSs in salt tolerance in T. hispida. In this study, five ThSOS genes were cloned and identified. Their expression patterns in response to different abiotic stresses (NaCl and PEG6000 exposure) and hormone (ABA) stress were analyzed using qRT-PCR. The expression levels of most ThSOS genes were significantly altered under NaCl, PEG6000, and ABA treatment in at least one organ. Notably, ThSOS3 expression was significantly downregulated under salt stress at 6 h. Furthermore, the role of ThSOS3 in salt tolerance was studied. The results showed that overexpression of ThSOS3 confers salt stress tolerance on T. hispida by enhancing antioxidant enzyme activity, improving ROS-scavenging capability and decreasing the MDA content and lipid peroxidation of cell membranes. This study provides a foundation for further elucidation of salt tolerance mechanisms involving ThSOSs in T. hispida. However, the molecular mechanism by which ThSOS3 confers salt stress tolerance on T. hispida is unclear. Future studies should focus on ThSOS3 mechanisms under salt stress.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
CG designed the research. ZL and FT conducted the experiments and performed data analysis. ZL and QX wrote the manuscript. JW and WD performed data analysis. CW and CG revised the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by grants from the Province in Heilongjiang Outstanding Youth Science Fund (JC2017004), the National Natural Science Foundation of China (31670679), the Overseas Expertise Introduction Project for Discipline Innovation (B16010), and the Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team) to GC.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.597480/full#supplementary-material
Supplementary Figure 1(A) Locations of cis-elements in the ThSOS gene promoters. (B) Descriptions of the cis-elements.
Supplementary Table 1Classification of SOS genes in different species.
Footnotes
1.^ https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome
2.^ http://www.expasy.org/tools/protparam.html
3.^ http://cello.life.nctu.edu.tw/
4.^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
1
Batelli G. Verslues P. E. Agius F. Qiu Q. Fujii H. Pan S. et al (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity.Mol. Cell. Biol.277781–7790. 10.1128/MCB.00430-07
2
Baxter A. Mittler R. Suzuki N. (2013). ROS as key players in plant stress signalling.J. Exp. Bot.651229–1240. 10.1093/jxb/ert375
3
Ben-Amor M. Flores B. Latché A. Bouzayen M. Pech J. C. Fomojaro F. (1999). Inhibition of ethylene biosynthesis by antisense acc oxidase RNA prevents chilling injury in charentais cantaloupe melons.Plant Cell Environ.221579–1586. 10.1046/j.1365-3040.1999.00509.x
4
Che B. Cheng C. Fang J. Liu Y. Jiang L. Yu B. (2019). The recretohalophyte Tamarix TrSOS1 gene confers enhanced salt tolerance to transgenic hairy root composite cotton seedlings exhibiting virus-induced gene silencing of GhSOS1.Intern. J. Mol. Sci.20:2930. 10.3390/ijms20122930
5
Cheng N. H. Pittman J. K. Zhu J. K. Hirschi K. D. (2004). The protein kinase SOS2 activates the Arabidopsis H+/Ca2 + antiporter CAX1 to integrate calcium transport and salt tolerance.J. Biol. Chem.2792922–2926. 10.1074/jbc.M309084200
6
Clough S. J. Bent A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana.Plant J.16735–743. 10.1046/j.1365-313x.1998.00343.x
7
Dhindsa R. S. Plumb-Dhindsa P. Thorpe T. A. (1981). Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase.J. Exp. Bot.3293–101. 10.1093/jxb/32.1.93
8
El Mahi H. Perez-Hormaeche J. De Luca A. Villalta I. Espartero J. Gamez-Arjona F. et al (2019). A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice.Plant Physiol.1801046–1065. 10.1104/pp.19.00324
9
Guo Y. Halfter U. Ishitani M. Zhu J. K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance.Plant Cell131383–1400. 10.1105/TPC.010021
10
He Z. Li Z. Lu H. Huo L. Wang Z. Wang Y. et al (2019). The NAC protein from Tamarix hispida, ThNAC7, confers salt and osmotic stress tolerance by increasing reactive oxygen species scavenging capability.Plants8:221. 10.3389/fpls.2017.00635
11
Hilleary R. Choi W. G. Kim S. H. Lim S. D. Gilroy S. (2018). Sense and sensibility: the use of fluorescent protein-based genetically encoded biosensors in plants.Curr. Opin. Plant Biol.4632–38. 10.1016/j.pbi.2018.07.004
12
Hu W. Yuan Q. Wang Y. Cai R. Deng X. Wang J. et al (2012). Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco.Plant Cell Physiol.532127–2141. 10.1093/pcp/pcs154
13
Huertas R. Olias R. Eljakaoui Z. Gálvez F. J. Li J. U. N. De M. et al (2012). Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato.Plant Cell Environ.351467–1482. 10.1111/j.1365-3040.2012.02504.x
14
Jiang Y. Duan Y. Yin J. Ye S. Zhu J. Zhang F. et al (2014). Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses.J. Exp. Bot.226629–6644. 10.1093/jxb/eru381
15
Kamei A. Seki M. Umezawa T. Ishida J. Satou M. Akiyama K. et al (2010). Analysis of gene expression profiles in Arabidopsis salt overly sensitive mutants sos2-1 and sos3-1.Plant Cell Environ.281267–1275. 10.1111/j.1365-3040.2005.01363.x
16
Kim J. H. Nguyen N. H. Jeong C. Y. Nguyen N. T. Hong S. W. Lee H. (2013). Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis.J. Plant Physiol.1701461–1465. 10.1016/j.jplph.2013.05.011
17
Kudla J. Batistiè O. Hashimoto K. (2010). Calcium signals: the lead currency of plant information processing.Plant Cell22541–563. 10.1105/tpc.109.072686
18
Li D. Ma N. N. Wang J. R. Yang D. Y. Zhao S. J. Meng Q. W. (2013). Overexpression of tomato enhancer of SOS3-1 (LeENH1) in tobacco enhanced salinity tolerance by excluding Na+ from the cytosol.Plant Physiol. Biochem.70150–158. 10.1016/j.plaphy.2013.05.014
19
Liu Z. Wang P. Zhang T. Li Y. Wang Y. Gao C. (2018). Comprehensive analysis of BpHSP genes and their expression under heat stresses in Betula platyphylla.Environ. Exp. Bot.152167–176. 10.1016/j.envexpbot.2018.04.011
20
Livak K. J. Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method.Methods25402–408. 10.1006/meth.2001.1262
21
Luan S. (2009). The CBL-CIPK network in plant calcium signaling.Trends Plant Sci.1437–42. 10.1016/j.tplants.2008.10.005
22
Martínez-Atienza J. Jiang X. Garciadeblas B. Mendoza I. Zhu J. K. Pardo J. M. et al (2007). Conservation of the salt overly sensitive pathway in rice.Plant Physiol.1431001–1012. 10.1104/pp.106.092635
23
Monihan S. M. Ryu C. H. Magness C. A. Schumaker K. S. (2019). Linking duplication of a calcium sensor to salt tolerance in Eutrema salsugineum.Plant Physiol.1791176–1192. 10.1104/pp.18.01400
24
Munns R. Tester M. (2008). Mechanisms of salinity tolerance.Annu. Rev. Plant Biol.59651–681. 10.1146/annurev.arplant.59.032607.092911
25
Narusaka Y. Nakashima K. Shinwari Z. K. Sakuma Y. Furihata T. Abe H. et al (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis RD29A gene in response to dehydration and high-salinity stresses.Plant J.34137–148. 10.1046/j.1365-313X.2003.01708.x
26
Nutan K. K. Kumar G. Singla-Pareek S. L. Pareek A. (2018). A salt overly sensitive pathway member from Brassica juncea BjSOS3 can functionally complement ΔAtsos3 in Arabidopsis.Curr. Genom.1960–69. 10.2174/1389202918666170228133621
27
Olias R. Eljakaoui Z. Li J. U. N. De M. P. A. Marín-Manzano M. C. Pardo J. M. et al (2009). The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs.Plant Cell Environ.32904–916. 10.1111/j.1365-3040.2009.01971.x
28
Persak H. Pitzschke A. (2014). Dominant repression by Arabidopsis transcription factor MYB44 causes oxidative damage and hypersensitivity to abiotic stress.Intern. J. Mol. Sci.152517–2537. 10.3390/ijms15022517
29
Qiu Q. S. Guo Y. Dietrich M. A. Schumaker K. S. Zhu J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3.Proc. Natl. Acad. Sci. U.S.A.998436–8441. 10.1073/pnas.122224699
30
Qiu Q. S. Guo Y. Quintero F. J. Pardo J. M. Schumaker K. S. Zhu J. K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279, 207–215. 10.1074/jbc.M307982200
31
Quan R. Lin H. Mendoza I. Zhang Y. Cao W. Yang Y. et al (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress.Plant Cell191415–1431. 10.1105/tpc.106.042291
32
Quintero F. J. Ohta M. Shi H. Zhu J. K. Pardo J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis.Proc. Natl. Acad. Sci. U.S.A.999061–9066. 10.1073/pnas.132092099
33
Srinivasan R. Roberto D. (2018). Membrane proteins in trypanosomatids involved in Ca2 + homeostasis and signaling.Genes9304–322. 10.3390/genes9060304
34
Tamura K. Peterson D. Peterson N. Stecher G. Nei M. Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.Mol. Biol. Evol.282731–2739. 10.1093/molbev/msr121
35
Tang R. J. Liu H. Bao Y. Lv Q. D. Yang L. Zhang H. X. (2010). The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress.Plant Mol. Biol.74367–380. 10.1007/s11103-010-9680-x
36
Thompson J. D. Gibson T. J. Plewniak F. Jeanmougin F. Higgins D. G. (1997). The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools.Nucleic Acids Res.254876–4882. 10.1093/nar/25.24.4876
37
Wang J. Xu X. Ma D. (2019). Establishment of an efficient alfalfa regeneration system via organogenesis and the co-expression of Arabidopsis SOS genes improves salt tolerance in transgenic alfalfa (Medicago sativa L.).Can. J. Plant Sci.99348–363. 10.1139/CJPS-2018-0105
38
Yang Q. Chen Z. Z. Zhou X. F. Yin H. B. Li X. Xin X. F. et al (2009). Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis.Mol. Plant222–31. 10.1093/mp/ssn058
39
Yang Y. Zhang C. Tang R. J. Xu H. X. Lan W. Z. Zhao F. et al (2019). Calcineurin B-Like proteins CBL4 and CBL10 mediate two independent salt tolerance pathways in Arabidopsis.Intern. J. Mol. Sci.20:2421. 10.3390/ijms20102421
40
Yue Y. Zhang M. Zhang J. Duan L. Li Z. (2012). SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio.J. Plant Physiol.169255–261. 10.1016/j.jplph.2011.10.007
41
Zang D. Wang C. Ji X. Wang Y. (2015). Tamarix hispida zinc finger protein ThZFP1 participates in salt and osmotic stress tolerance by increasing proline content and SOD and POD activities.Plant Sci.235111–121. 10.1016/j.plantsci.2015.02.016
42
Zhang T. Zhao Y. Wang Y. Liu Z. Gao C. (2018). Comprehensive analysis of MYB gene family and their expressions under abiotic stresses and hormone treatments in Tamarix hispida.Front. Plant Sci.9:1303. 10.3389/fpls.2018.01303
43
Zhang X. Wang L. Meng H. Wen H. Fan Y. Zhao J. (2011). Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species.Plant Mol. Biol.75365–378. 10.1007/s11103-011-9732-x
44
Zhang Y. Linghu J. Wang D. Liu X. Yu A. Li F. et al (2017). Foxtail millet CBL4 (SiCBL4) interacts with SiCIPK24, modulates plant salt stress tolerance.Plant Mol. Biol. Rep.35634–646. 10.1007/s11105-017-1051-1
45
Zheng L. Liu G. Meng X. Liu Y. Ji X. Li Y. et al (2013). A WRKY gene from Tamarix hispida, ThWRKY4, mediates abiotic stress responses by modulating reactive oxygen species and expression of stress-responsive genes.Plant Mol. Biol.82303–320. 10.1007/s11103-013-0063-y
46
Zhou J. Wang J. Bi Y. Wang L. Tang L. Yu X. et al (2014). Overexpression of PtSOS2 enhances salt tolerance in transgenic poplars.Plant Mol. Biol. Rep.32185–197. 10.1007/s11105-013-0640-x
47
Zörb C. Noll A. Karl S. Leib K. Yan F. Schubert S. (2005). Molecular characterization of Na+/H+ antiporters (ZmNHX) of maize (Zea mays L.) and their expression under salt stress.J. Plant Physiol.16255–66. 10.1016/j.jplph.2004.03.010
Summary
Keywords
ROS-scavenging capability, salt stress, Tamarix hispida , Arabidopsis thaliana , ThSOS
Citation
Liu Z, Xie Q, Tang F, Wu J, Dong W, Wang C and Gao C (2021) The ThSOS3 Gene Improves the Salt Tolerance of Transgenic Tamarix hispida and Arabidopsis thaliana. Front. Plant Sci. 11:597480. doi: 10.3389/fpls.2020.597480
Received
21 August 2020
Accepted
09 December 2020
Published
15 January 2021
Volume
11 - 2020
Edited by
Giovanni Stefano, University of Florence, Italy
Reviewed by
Yucheng Wang, Shenyang Agricultural University, China; Iris Aloisi, University of Bologna, Italy
Updates
Copyright
© 2021 Liu, Xie, Tang, Wu, Dong, Wang and Gao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caiqiu Gao, gaocaiqiu@nefu.edu.cn
†These authors have contributed equally to this work
This article was submitted to Plant Abiotic Stress, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.