- 1Joint International Research Laboratory of Metabolic & Developmental Sciences, Shanghai Jiao Tong University–University of Adelaide Joint Centre for Agriculture and Health, State Key Laboratory of Hybrid Rice, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
- 2Department of Genetics and Genome Biology, University of Leicester,Leicester, United Kingdom
The mammalian BREAST CANCER 2 (BRCA2) gene is a tumor suppressor that plays a crucial role in DNA repair and homologous recombination (HR). Here, we report the identification and characterization of OsBRCA2, the rice orthologue of human BRCA2. Osbrca2 mutant plants exhibit normal vegetative growth but experience complete male and female sterility as a consequence of severe meiotic defects. Pairing, synapsis and recombination are impaired in osbrca2 male meiocytes, leading to chromosome entanglements and fragmentation. In the absence of OsBRCA2, localization to the meiotic chromosome axes of the strand-invasion proteins OsRAD51 and OsDMC1 is severely reduced and in vitro OsBRCA2 directly interacts with OsRAD51 and OsDMC1. These results indicate that OsBRCA2 is essential for facilitating the loading of OsRAD51 and OsDMC1 onto resected ends of programmed double-strand breaks (DSB) during meiosis to promote single-end invasions of homologous chromosomes and accurate recombination. In addition, treatment of osbrca2-1 seedlings with mitomycin C (MMC) led to hypersensitivity. As MMC is a genotoxic agent that creates DNA lesions in the somatic cells that can only be repaired by HR, these results suggest that OsBRCA2 has a conserved role in DSB repair and HR in rice.
Introduction
Cellular DNA of living organisms experience DNA damage caused by exogenous and endogenous factors. DNA double-strand breaks (DSBs) are one of the most cytotoxic DNA lesions, as even one single unrepaired or misrepaired DSB will lead to genomic rearrangements and genome instability (Bennett et al., 1993; Sandell and Zakian, 1993). Non-homologous end joining (NHEJ) and homologous recombination (HR) are the two main pathways involved in the repair of DSBs (Bétermier et al., 2014; Guirouilh-Barbat et al., 2014). In the NHEJ pathway, DNA broken ends are repaired by rapidly ligating the two DNA ends with little to no DNA end processing. In contrast to the error-prone NHEJ pathway, the HR pathway uses an intact homologous DNA sequence as the repair template to ensure faithful repair. In addition to accidental mitotic DSBs, HR is indispensable for repairing programmed meiotic DSBs that are intentionally produced by a DNA topoisomerase VI-like complex at the beginning of meiotic prophase I (Fu et al., 2016; Fayos et al., 2020).
DSB repair via HR is initiated by 5' to 3' resection of the DNA ends with the creation of 3' single-stranded (ssDNA) overhangs, which are then coated by replication protein A (RPA) to prevent from degradation and forming secondary structures (Iftode et al., 1999). Subsequently, RPA proteins are replaced by recombinases, which are essential for strand invasion and DNA homology searching. In bacteria, RecA plays a pivotal role in strand exchange between homologous DNA molecules (Roca and Cox, 1997). In eukaryotic cells, two of RecA homologs, RAD51 and meiotic specific DMC1, possess the activity to catalyze the pairing of homologous DNA sequences and strand exchange reaction to fulfill the homology directed DSB repair (San Filippo et al., 2008). RAD51 uses the intact sister chromatid as the template to repair DSBs; while in meiotic cells, DMC1 employs homologous chromosomes as templates with the aid of RAD51 (Cloud et al., 2012; Kurzbauer et al., 2012; Da Ines et al., 2013), that may result in the formation of crossover (CO) between homologous chromosomes.
The formation of RAD51/DMC1 nucleoprotein filaments is considered as a rate-limiting process of HR and is mediated by the RAD51/DMC1 loader BRCA2. The BRCA2 gene was first identified in humans (Wooster et al., 1995), whose mutations have been reported to be the cause of hereditary breast cancers and confer a high risk to many other cancers (Hall et al., 1990). HsBRCA2 encodes a protein with 3418 amino acids, consisting of several functional domains, including eight BRC repeats, a phenylalanine-proline-proline (PhePP) motif, a DNA-binding domain (DBD) containing a helical rich region, three oligonucleotide/oligosaccharide binding (OB) folds, and a C-terminal TR2 domain (Dray et al., 2006; Thorslund et al., 2007; Seeliger et al., 2012). Biochemical studies revealed that BRCA2 is loaded onto both ends of the double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA), leading to the formation of ssDNA-dsDNA junctions (Yang et al., 2002, 2005). BRCA2 interacts with RAD51 and DMC1 via its BRC repeats and promotes their loading onto the ends of ssDNA covered by RPA (Wong et al., 1997; Chen et al., 1998). Besides, the C-terminal TR2 domain of BRCA2 interacts with multimeric RAD51 to stabilize the RAD51 nucleofilaments (Davies and Pellegrini, 2007; Esashi et al., 2007). Furthermore, BRCA2 interacts with multiple partner proteins to assist in RAD51/DMC1 loading and assembly, such as DSS1 (Marston et al., 1999), USP11 (Schoenfeld et al., 2004), BCCIP (Lu et al., 2005) FANCD2 (Hussain et al., 2003, 2004; Wang et al., 2004), FANCG (Hussain et al., 2003, 2004; Wang et al., 2004) and PALB2/FANCN (Xia et al., 2006; Sy et al., 2009; Zhang et al., 2009a,b; Orthwein et al., 2015). Thus, BRCA2 functions as a master regulator in promoting HR-mediated DSB repair to maintain genome integrity.
Homologs of tumor suppressor HsBRCA2 have been identified in various eukaryotes including plants but are absent from archaea, bacterial, and yeast. Although highly variable in protein size and the number of BRC repeats, the ability of BRCA2 homologs to associate with recombinases is conserved in several organisms. Arabidopsis is the only known organism that has two BRCA homologs (Pfeffer et al., 2017). In mammals, BRCA2 is essential for survival; its deficiency results in embryonic lethality. However, double knockouts of AtBRCA2A/B does not affect normal vegetative growth but causes severe abnormalities in meiosis, including defective homologous chromosome pairing and synapsis, chromosome entanglement, and fragmentation, leading to both male and female sterility (Seeliger et al., 2012). The fact that the brca2 phenotype is alleviated by the spo11 mutation indicates that its meiotic chromosomal instability is caused by failure in repairing programmed DSBs (Seeliger et al., 2012). AtBRCA2A/B has been proven to be able to interact with AtRAD51 and AtDMC1 in vitro and in vivo (Abe et al., 2009; Wang et al., 2010b). In atbrca2 double mutants, AtRAD51 and AtDMC1 foci could not be detected in meiotic cells (Seeliger et al., 2012). Additionally, the atbrac2a single mutant and atbrca2 double mutants display hypersensitivity to the genotoxic agent mitomycin C (MMC) and defects in somatic HR (Abe et al., 2009; Wang et al., 2010b). These data indicate that AtBRCA2 plays a conserved role in HR by recruiting RAD51 and DMC1, essential for HR mediated DSB repair in both meiotic and somatic cells.
Compared with the well-documented BRCA2 functions in animals and fungi, much less is known about the role of BRCA2 homologs in plants. Currently, only Arabidopsis BRCA2 has been functionally characterized and the role of the BRCA2 orthologs in other plants remains unknown. In this study, we identified and characterized the BRCA2 ortholog in Oryza sativa. Our results reveal that OsBRCA2 is essential for promoting HR and chromosome synapsis, as well as maintaining the genome stability in meiotic cells. Furthermore, osbrca2 is hypersensitive to the DNA damage agent MMC. Notably, we demonstrated that OsBRCA2 is able to facilitate OsRAD51 and OsDMC1 loading onto the chromosomes. This study provides further evidence to support the crucial function of the BRCA2 in HR-mediated DSBs repair pathway in plants.
Materials and Methods
Plant Materials, Growth Conditions, and Molecular Cloning of OsBRCA2
Rice (Oryza sativa) plants in the 9522 background (O. sativa ssp japonica) were grown in the paddy field of Shanghai Jiao Tong University under natural growth condition. The mapping population was collected from the F2 progenies that were generated from a cross between osbrca2-1 mutant and Guang-Lu-Ai4 (O. sativa ssp indica). Primary-mapping of OsBRCA2 was performed using bulked segregated analysis (Liu et al., 2005). Three BRCA2 alleles, including osbrca2-2, osbrca2-3, and osbrca2-4, were created using the CRISPR-Cas9 system kindly provided by Professor Jiankang Zhu. The sgRNA-Cas9 plant expression vectors were constructed as previously described (Wang et al., 2017). Primers used for fine mapping, constructing sgRNA-Cas9 plant expression vectors and verifying transgenic plants are listed in Supplementary Table S1.
Characterization of Mutant Plant Phenotypes
Rice plants, spikelets, and anthers at heading stage were photographed with digital camera (Nikon, E995) or under a stereoscope (Leica, M205A). The pollen viability assay was performed by 1% (w/v) I2-KI staining. The transverse sections of anthers were obtained according to the method previously published (Li et al., 2006).
Antibody Production
The OsBRCA2 and OsRAD51 polyclonal antibodies used in this study were prepared by Abclonal (Wuhan, China). A 600-bp DNA fragment encoding a 200-amino acid peptide of OsBRCA2 (residues 251-451) and a coding sequence of OsRAD51 encoding a 200-amino acid peptide of OsRAD51 (residues 1-200) were amplified from the rice anther cDNA and cloned into the protein expression vector pET-32a (GE) respectively. The recombinant proteins expressed in Escherichia coli BL21DE3 (Novagen) were purified and used to produce rabbit polyclonal antibodies. The polyclonal antibodies against OsREC8, γH2AX, COM1, RPA1c, RPA2c, RAD51C, DMC1, PAIR2, PAIR3, and ZEP1 have been described in previous studies (Fu et al., 2016; He et al., 2016).
FISH Analysis and Immunolocalization Assays
Fresh panicles containing male meiocytes were harvested and fixed with Carnoy’s solution (ethanol: glacial acetic 3:1, v/v). The preparation and DAPI staining of meiotic chromosomes were performed as previously described (He et al., 2016). Fluorescent in situ hybridization (FISH) assay was performed as described (Cheng, 2013). The sequences of the centromere and 5S rDNA FISH probes were designed and labeled as described (Mizuno et al., 2006; Zhang et al., 2019). Images were captured with an Eclipse Ni-E microscope and NIS elements software (Nikon). Immunolocalization assays were performed as described in previous studies (Cheng, 2013; He et al., 2016; Wang et al., 2017). Fresh panicles containing meiocytes were fixed in 4% (w/v) paraformaldehyde for 30 min at room temperature and then washed three times with 1 × PBS (pH 7.4). Anthers were squashed on a slide with 1 × PBS solution (pH 7.4) and soaked in liquid nitrogen. After removing the cover slips quickly with a blade, the slides were dehydrated through an ethanol series (70, 90, and 100%). Different antibody combinations diluted to 1:200 in TNB buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.5% blocking reagent) were added to the slides and then incubated in a humid chamber at 37°C for 2 h. After three rounds of washing in 1 × PBS, goat anti-rabbit antibody (Alexa Fluor® 555, Life Technologies, 1:500) and goat anti-mouse antibody (Dylight 488, Abbkine, 1:500) were added to the slides, and then incubated in a humid chamber at 37°C for 1 h. Finally, the slides were counterstained with DAPI after three rounds of washing in 1 × PBS. All fluorescence images were photographed and processed using an Eclipse Ni-E microscope (Nikon) with NIS-Elements Advanced Research software at the same parameter level to avoid the interference of multi-factor. Each channel keeps the same exposure values to capture red-green channel images as presented in visual results. For dot-like foci signals, image deconvolution will be further performed using “Mexican Hat” process to improve signal-to-noise ratio. The number of dot-like fluorescent foci signals was counted using ImageJ 1.52 software (Collins, 2007).
Yeast Two-Hybrid Assay
The full-length cDNAs of OsBRCA2 and a 2760-bp cDNA fragment encoding the six BRC domains of OsBRCA2 were amplified from rice anther cDNA and cloned into the pGBKT7 vector (Clontech) separately. The full-length cDNAs of OsDMC1A, OsDMC1B, OsRAD51A1, OsRAD51A2, was amplified from rice anther cDNA and cloned into the pGADT7 vector (Clontech), respectively. Subsequently, yeast two-hybrid (Y2H) assays were performed according to the manufacturer’s instructions (Clontech). Primers used for cDNA amplification and vector construction are listed in Supplementary Table S1.
qRT-PCR Assay
Total RNA from wild-type tissues was isolated using the Trizol Reagent Kit (Invitrogen) according to the manufacturer’s protocol. The rice anther development stage was defined as previously described (Zhang et al., 2011). Roots, shoots, and leaves were collected from 30-day-old seedlings and glumes were collected from stage 8 spikelets. An equal amount total RNA per sample was used to synthesize cDNA using Primescript™ RT reagent kit with genomic DNA eraser (Takara). The qRT-PCR analysis was performed according to the previous report (Fu et al., 2014). The rice Actin gene was used as the internal control, and primers used for qRT-PCR are listed in Supplementary Table S1. All reactions were performed in three independent biological replicates with three technical repeats each for statistical analysis. The gene expression was calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001).
MMS and MMC Sensitivity Test
The genotoxic agents sensitivity tests were performed using the methods as described by Chang et al. (2009) and modified as follows. Husked and surface-sterilized seeds of the heterozygous OsBRCA2-1+/− and wild type were germinated and grown on 1/2 MS (Murashige and Skoog) medium for 5 days. Subsequently, these seedlings were divided into three portions used for three independent biological repeats. Then seedlings were transferred to 1/2 MS (Murashige and Skoog) medium supplemented with 0.3% phytagel and with concentrations 0 μl/L to 150 μl/L of methylmethane sulphonate (MMS; Sigma-Aldrich, St. Louis, MO, United States) or 0–300 μg/ml MMC (Sangon, Shanghai, China). A total of 90 seedlings in each treatment arranged in six replications of 15 seedlings per plastic pot were placed in a light incubator. After 13 days treatment, the genotypes were verified by PCR and the height of seedlings was measured. Student’s t tests were performed for comparing the data differences, p < 0.05 was considered to be significant. * represented p < 0.05, and ** represented p < 0.01.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: OsBRCA2 (Os01g0164800, Os01g0164900), OsRAD51A1 (Os11g0615800), OsRAD51A2 (Os12g0497300), OsDMC1A (Os12g0143800), OsDMC1B (Os11g0146800), OsActin (Os03g0718100), AtBRCA2A (NP_191913.3), AtBRCA2B (NP_195783.3), and HsBRCA2 (CAA64484.1).
Results
Identification of the osbrca2-1 Mutant
To isolate genes essential for rice fertility, we screened for sterile mutants from a rice (O. sativa ssp japonica cultivar, 9522) mutant library (Chen et al., 2006) and identified a completely sterile mutant osbrca2-1. During vegetative developmental stage, the osbrca2-1 plant grew normally as wild-type plants (Figure 1A). However, during the reproductive stage, osbrca2-1 had small, pale-yellow stamens (Figures 1C,E) that could not produce mature pollen grains or seeds (Figures 1B,F,G). Female fertility of osbrca2-1 was also abnormal, shown by the smaller pistil (Figure 1D) and inability to set seeds when pollinated with wild-type pollen. Progeny from heterozygote plants segregated 298 fertile plants and 92 sterile plants (3:1, χ2 = 0.4137 < χ20.05, 1 = 3.84), indicating that osbrca2-1 was a single recessive mutation.

Figure 1. Phenotypic comparison between wild-type and osbrca2-1 mutant. (A) Wild-type and osbrca2-1 plants after heading. (B) Wild-type and osbrca2-1 panicles showing fertile and infertile grains, respectively. (C) Wild-type and osbrca2-1 spikelets after removing lemma. (D) Wild-type and osbrca2-1 pistils at stage 12. (E) Wild-type (left) and osbrca2-1 (right) anthers. (F) I2-KI staining of wild-type and (G) osbrca2-1 pollen grains at mature stage. Bars = 5 cm in (A) and (B), 5 mm in (C), 1 mm in (D) and (E), and 100 μm in (F) and (G).
A map-based cloning strategy was exploited to identify the mutated gene by using 298 mutants from an F2 mapping population. The mutated locus was located between two Indel molecular markers named SHY101-2-2 and SHY102-3-1 on chromosome 1 (Figure 2A). Whole-genome sequencing revealed a 5 bp deletion in the seventh exon of the candidate gene (LOC_Os01g07110), leading to a frame shift and a premature stop codon (Figure 2B). Sequence analysis showed that LOC_Os01g07110 encodes a protein sharing high sequence similarity with the human breast cancer susceptibility gene 2 (HsBRCA2; Thorslund and West, 2007), thus we named this gene as OsBRCA2. The predicted OsBRCA2 protein was 1575 amino acids in length and contained six putative BRC repeats, one OsBRCA2_helical, one OsBRCA2_OB1 domain, one OsBRCA2_OB2 domain containing one TOWER domain and one OsBRCA2_OB3 domain (Figure 2B).
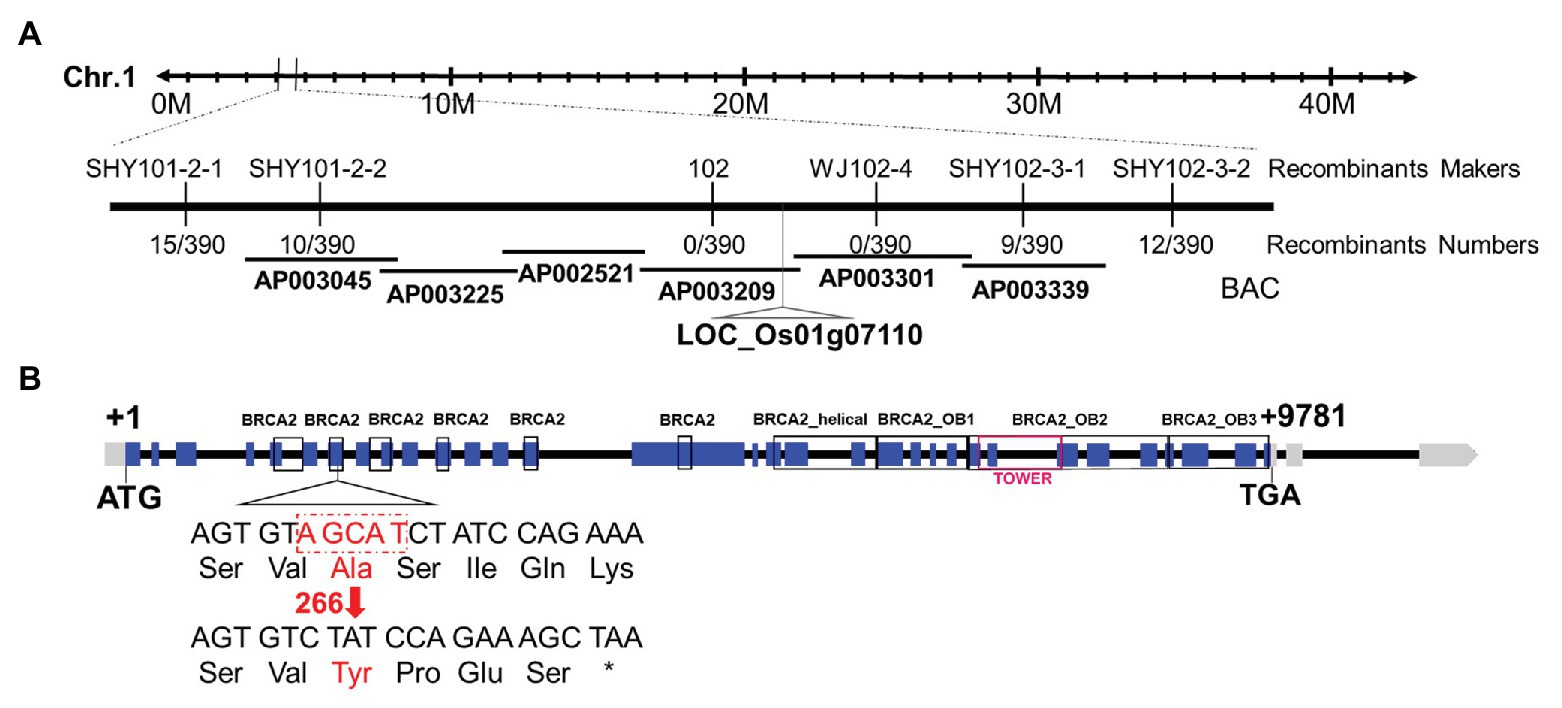
Figure 2. Molecular characterization of osbrca2-1. (A) Fine mapping of osbrca2-1 on chromosome 1. Names and positions of the markers are indicated. (B) A schematic representation of 32 exons and 31 introns of LOC_Os01g07110. The +1 indicates the putative starting nucleotide of translation, and the stop codon (TGA) is +9781. Blue boxes indicate exons, and intervening lines indicate introns. The deletion site in osbrca2-1 is shown (red arrow).
To confirm that the mutation in OsBRCA2 was responsible for the sterile phenotype of osbrca2-1 mutant, three independent alleles osbrca2-2, osbrca2-3, osbrca2-4 were generated using the CRISPR/Cas9 system. Osbrca2-2 and osbrca2-4 had a T insertion in the first BRC repeat and BRCA2_OB1 domain, which caused frame shift from 186th aa and 1153th aa respectively. Osbrca2-3 had a 2 bp (GT) deletion in the third BRC repeat that caused a frame shift from 332th aa and premature translation termination (Supplementary Figure S1A). All of the osbrca2-2, osbrca2-3, and osbrca2-4 homozygous plants were sterile (Supplementary Figures S1C,G,K) and displayed similar meiotic defects (see below), confirming that the sterility that occurred in osbrca2-1 was the consequence of mutation in OsBRCA2.
Expression Pattern of OsBRCA2
The spatial and temporal expression pattern of OsBRCA2 was investigated by qRT-PCR analysis. OsBRCA2 was highly expressed in the anther before entry into meiosis and then declined after meiosis. OsBRCA2 was also highly expressed in leaves and weakly expressed in shoot, glume, and root (Supplementary Figure S2). The results showed that the transcripts of OsBRCA2 were extensively expressed not only in reproductive organs but also in vegetative organs.
To further define the spatiotemporal localization of OsBRCA2 during meiosis, dual immunolocalization assays utilizing antibodies raised against OsREC8 and OsBRCA2 were performed. In wild type, OsBRCA2 foci were first observed at late leptotene (Figure 3A). The number of OsBRCA2 foci dramatically increased and peaked at zygotene (Figure 3B). After that, OsBRCA2 foci decreased and very few signals could be detected at late pachytene (Figures 3C–E). No OsBRCA2 signal can be detected in osbrca2-1 lines, indicating that the OsBRCA2 antibody is specific (Supplementary Figure S3).
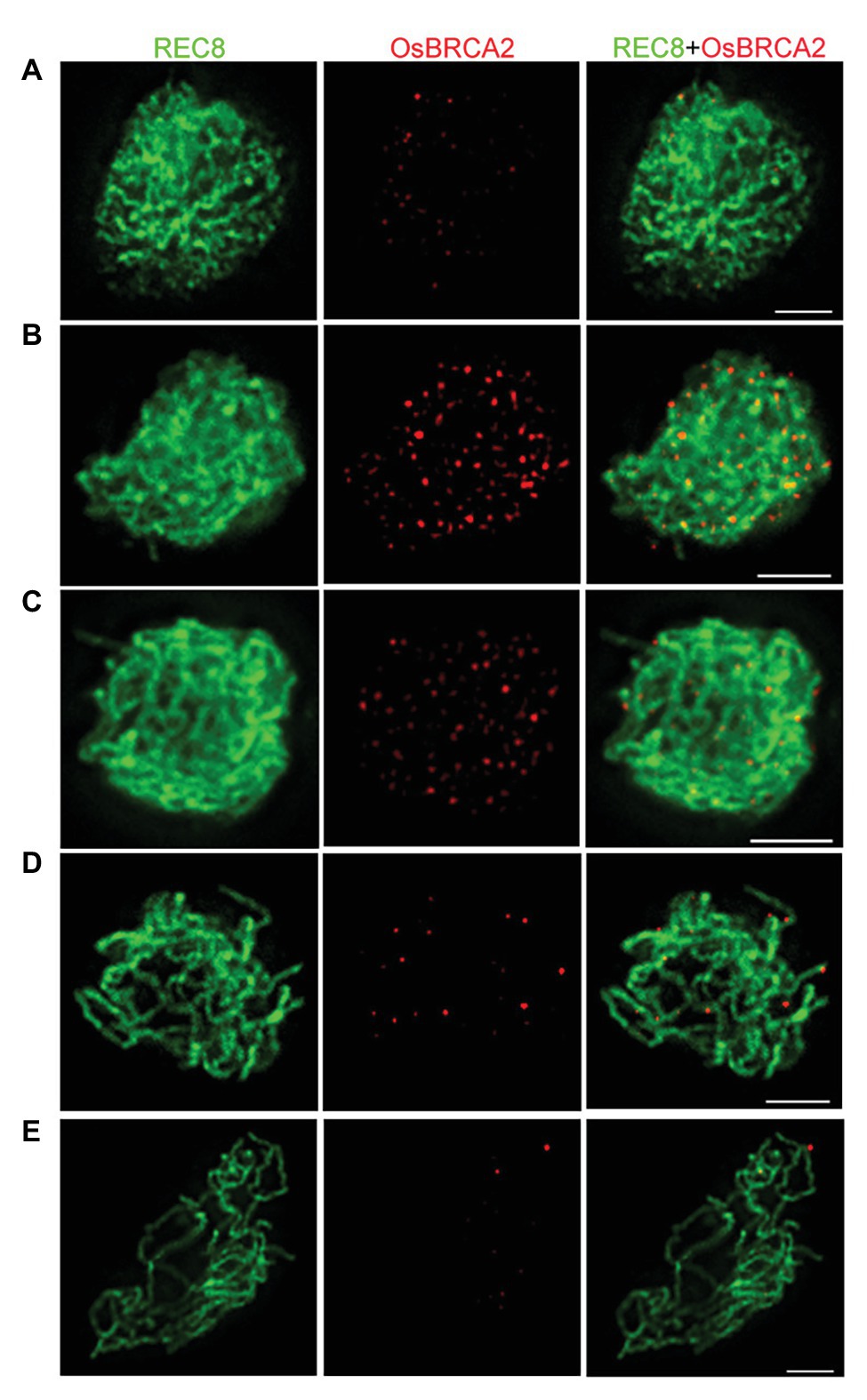
Figure 3. Dual immunolocalization assay of OsBRCA2 (red) and REC8 (green) in the wild-type meiocyte cells. (A) Late leptotene. (B) Zygotene. (C) Late zygotene. (D) Early pachytene. (E) Late pachytene. Bars = 5 μm.
Meiosis Is Impaired in osbrca2 Mutants
To identify the developmental defects responsible for osbrca2-1 male fertility, transverse sections of osbrca2-1 and wild-type anthers were performed. There were no distinguishable differences between wild type and osbrca2-1 before meiosis (Zhang et al., 2011). The pollen mother cells (PMCs) of wild type could produce dyads and tetrads, and released microspores normally. While PMCs of osbrca2-1 produced polyads at the end of meiosis, and generated severely distorted and shrunken microspores (Supplementary Figure S4).
Anther transverse sections indicated that the complete sterility of osbrca2-1 may be caused by meiotic defects. To investigate the roles of OsBRCA2 in meiosis, we performed 4', 6-diamidino-2-phenylindole (DAPI) staining to analyze the chromosome behavior in wild-type and osbrca2-1 male meiocytes. In wild type, the chromosomes completed the DNA replication process and began to condense into visible strands at leptotene (Figure 4A). During zygotene, the chromosomes continued to condense and pair as well as initiating synapsis (Figure 4B). At pachytene, synapsis and recombination occurred normally within homologous chromosomes (Figure 4C). At diplotene, the synaptonemal complexes (SCs) began to disassemble with the formation of chiasma that physically linked homologous chromosomes together. Twelve bivalents formed at diakinesis (Figure 4D) then aligned onto the equatorial plate at metaphase I (Figure 4E). At anaphase I, homologous chromosomes began to separate (Figure 4F) and migrated in opposite directions at telophase I (Figure 4G). Dyads and tetrads were formed at the end of meiosis I and II, respectively (Figures 4H,I). In osbrca2-1, chromosome behavior appeared to show no obvious differences from leptotene to zygotene when compared with wild type (Figures 4J,K). However, at pachytene, the chromosomes appeared only partially synapsed (Figure 4L). At diakinesis, irregularly shaped univalents, a small number of chromosome fragments and chromosome bridges were observed (Figure 4M). At metaphase I, chromosomes were entangled and could not align along the equatorial plate; chromosome bridges became more conspicuous (Figure 4N). At anaphase I, chromosomes asynchronously separated to the two opposite poles, as well as non-segregating chromosome fragments inside the nucleus (Figure 4O). At telophase I, lagging chromosome fragments could still be seen distributed between two newly formed nuclei (Figure 4P), leading to the formation of micronuclei in cells at the dyad and tetrad stage (Figures 4Q,R). We also monitored osbrca2-2, osbrca2-3, and osbrca2-4 meiotic progression by DAPI staining (Supplementary Figures S1D–F,H–J,L–N). The meiotic defects in these alleles were similar to those observed in osbrca2-1. These results indicate that the sterility of osbrca2 mutants was caused by the failure of DNA repair by HR and genome fragmentation during meiosis.
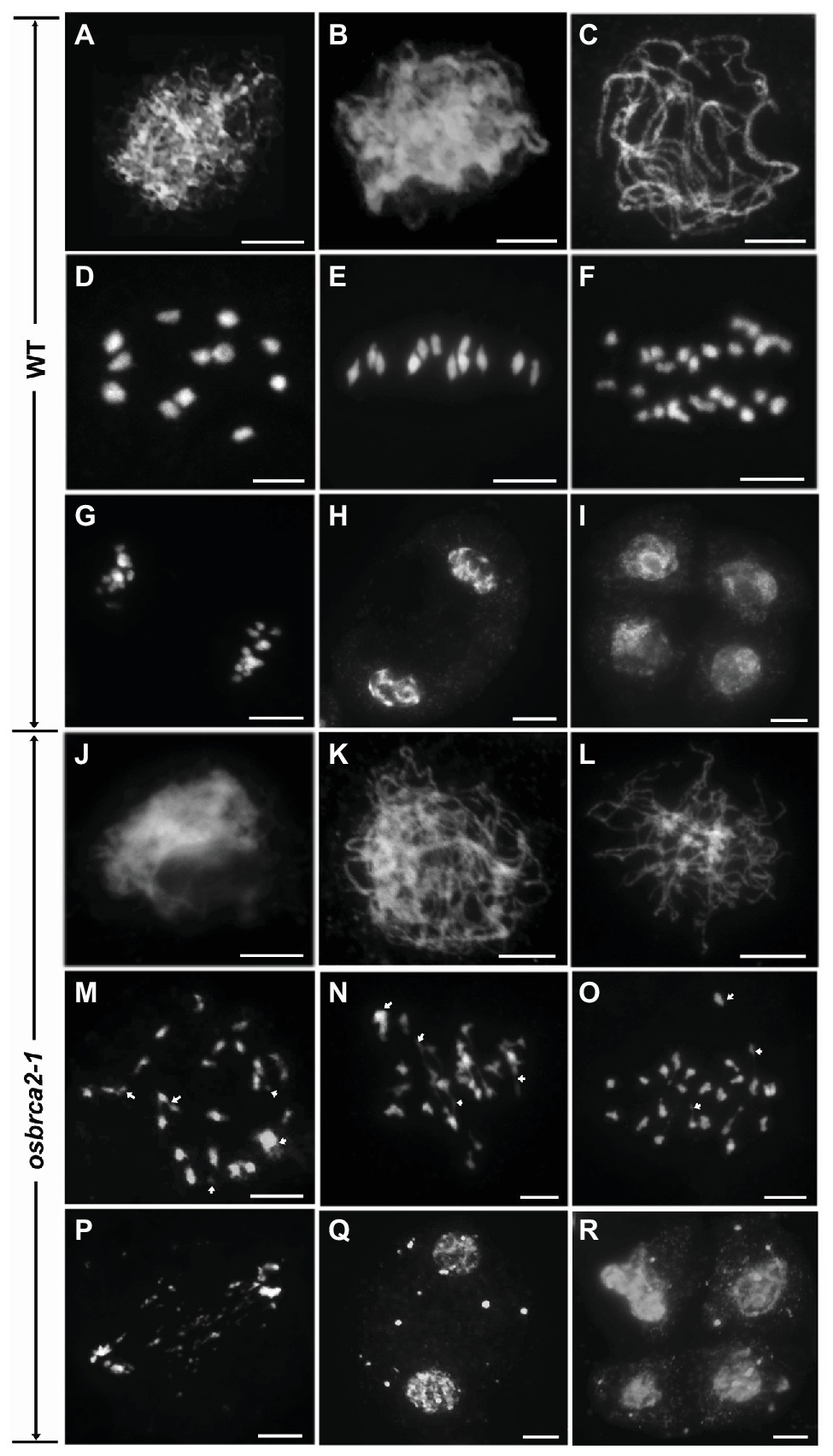
Figure 4. Meiotic chromosome behavior of male meiocytes in the wild-type and osbrca2-1. Chromosome behavior of male meiocytes of wild type (A–I) and osbrca2-1 (J–R) at various stages. (A,J) Leptotene. (B,K) Zygotene. (C,L) Pachytene. (D,M) Diakinesis. (E,N) Metaphase I. (F,O) Anaphase I. (G,P) Telophase I. (H,Q) Dyad. (I,R) Tetrad. Arrows indicated the abnormal chromosomes. Bars = 5 μm.
OsBRCA2 Is Essential for Homologous Chromosome Pairing and Synaptonemal Complex Formation
To investigate homologous chromosome pairing in the osbrca2-1 mutant, we performed fluorescent in situ hybridization (FISH) analysis using a centromere specific probe OsCenH3 and the 5S rDNA probe specifically distributed on the short arm of chromosome 11 (Zhang et al., 2005). In wild type, there were 12 OsCenH3 signals observed at pachytene (Figure 5A), but an average of 17 signals (n = 21, range 15–20) were detected in the osbrca2-1 mutant (Figure 5C). In the wild type, only one 5S rDNA signal can be detected (Figure 5B), which indicates full homologous chromosome pairing and synapsis. However, in the osbrca2-1 mutant, two totally separated 5S rDNA signals were observed in most nuclei (82.6%, n = 23; Figure 5D), suggesting that homologous chromosome pairing is defective.
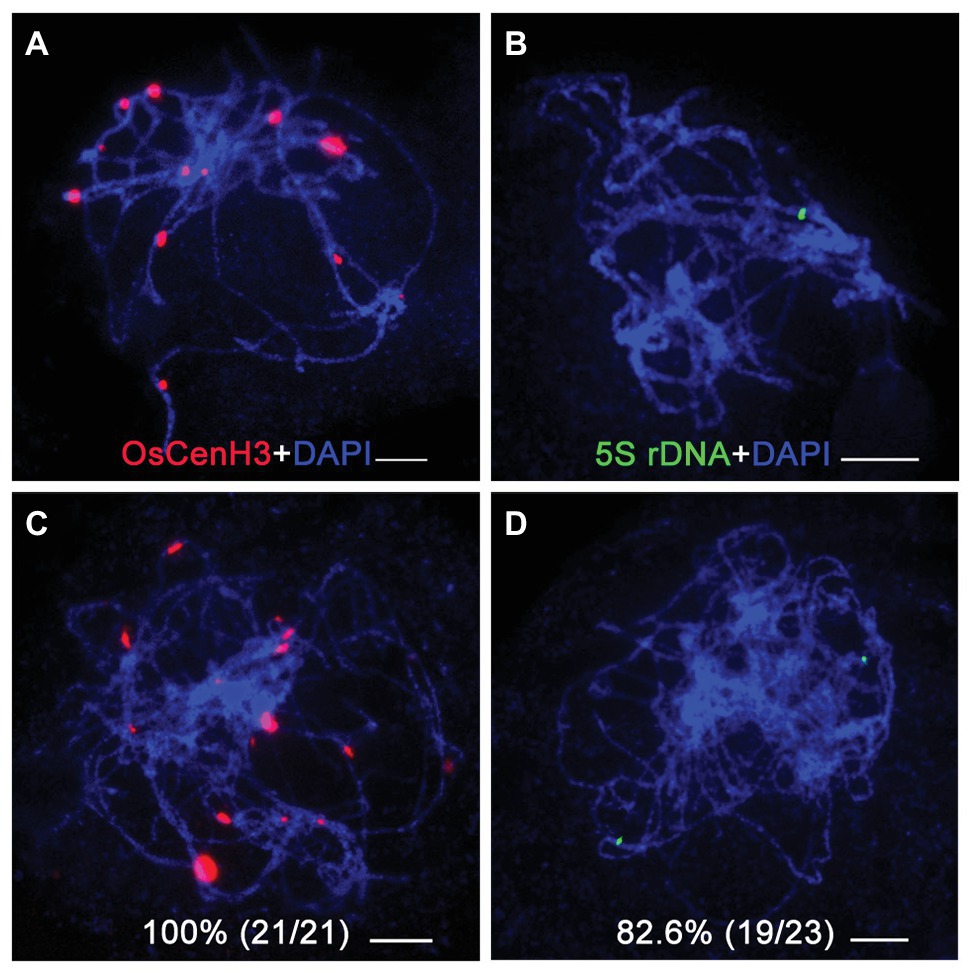
Figure 5. Homologous chromosomes paring is impaired in osbrca2-1 mutant. Homologous pairing analysis by fluorescent in situ hybridization (FISH) using OsCenH3 (red) and 5S rDNA (green) probes in wild-type (A,B) and osbrca2-1 mutant (C,D). Chromosomes (blue) are stained with DAPI. Percentage represents the number of abnormal cells, n = 21 and 23, respectively. Bars = 5 μm.
To further understand whether synaptonemal complex formation was defective in osbrca2-1, we conducted immunolocalization analysis using SC related proteins: OsREC8, PAIR2, PAIR3 and ZEP1 (Nonomura et al., 2006; Wang et al., 2010a, 2011; Shao et al., 2011). OsREC8 is a meiotic cohesin complex component and required for sister chromatid cohesion, axial element (AE) formation and homologous pairing (Shao et al., 2011). In osbrca2-1 male meiocytes, OsREC8 localized normally as that in wild-type male meiocytes and was used as a marker for further immunolocalization analysis (Figure 4). PAIR2 associated with axial elements (AEs) at leptotene and zygotene and then disassociated from the AEs of arm regions when homologous chromosomes fully synapsed (Nonomura et al., 2006). From zygotene to pachytene, PAIR2 signals became weaker and discontinuous on wild-type meiotic chromosomes (n = 10, Figure 6A). However, in osbrca2-1, PAIR2 foci did not disappear at pachytene (n = 13, Figure 6A). As an axis-associated protein, PAIR3 is also essential for SC assembly, which might provide a platform for other recombination elements, such as PAIR2 (Wang et al., 2011). By contrast, there was no obvious difference in PAIR3 localization between osbrca2-1 and wild type (n = 10, Supplementary Figure S5). ZEP1 encodes the transverse filament protein of the rice SC (Wang et al., 2010a). In wild type, ZEP1 formed punctate foci during zygotene and then elongated into continuous linear signals and aligned perfectly along the entire chromosome when homologous fully synapsed at pachytene (n = 11, Figure 6B). While in the osbrca2-1 mutant, only punctate or short discontinuous linear ZEP1 signal was observed even at late pachytene (n = 12, Figure 6B). These observations suggest that SC extension was severely interrupted in osbrca2-1 meiocytes.
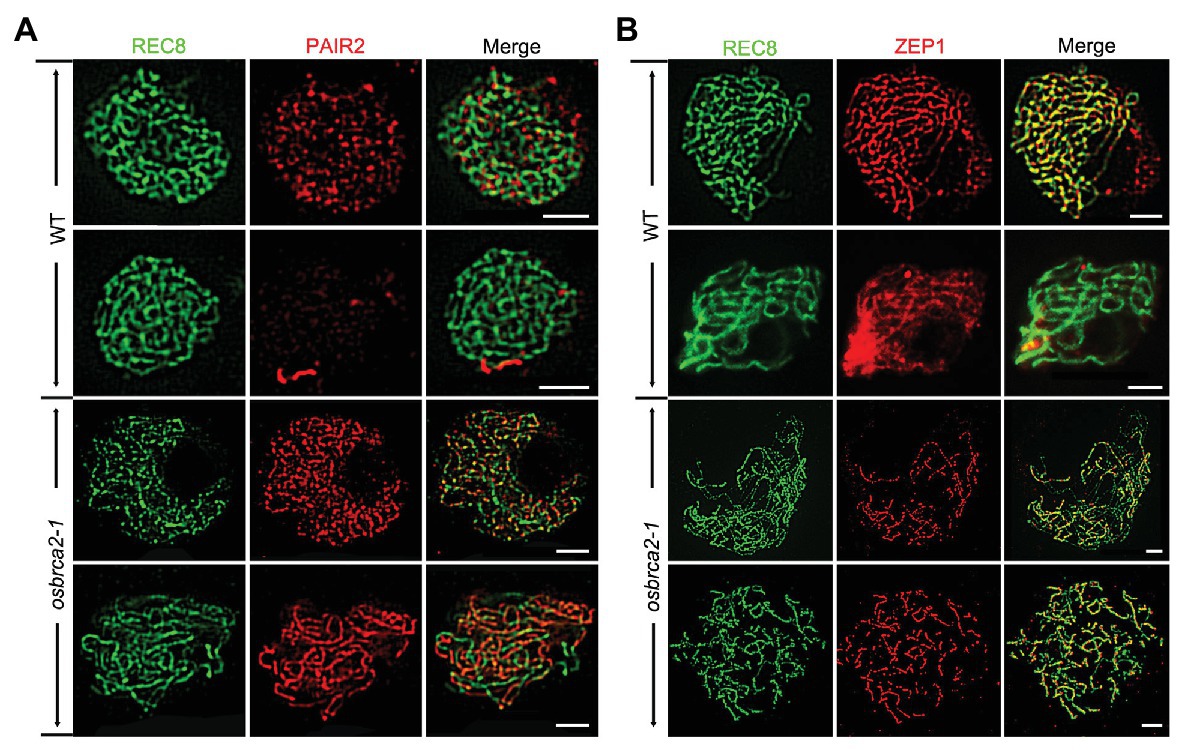
Figure 6. Synapsis complex is not correctly installed in osbrca2-1 mutant. Dual immunolocalization of PAIR2 (red) (A), ZEP1 (red) (B) and OsREC8 (green) in wild-type and osbrca2-1 PMCs. Bars = 5 μm.
OsBRCA2 Is Essential for Crossover Formation by Mediating Recruitment of OsRAD51 and OsDMC1
In many species, homologous chromosome recombination requires the formation and repair of double strand breaks (DSBs; Inagaki et al., 2010). Unrepaired or incorrectly repaired DSBs will lead to the formation of chromosomal aberrations such as broken chromosomes and/or gross chromosomal rearrangements (Pastink et al., 2001). In order to determine whether the chromosome fragments were caused by unrepaired or incorrectly repaired DSBs, we generated osbrca2-1 ossds double mutant. SDS is a meiosis specific cyclin-like protein, which is essential for meiotic DSB formation in rice (Wu et al., 2015). In an ossds single mutant, due to defective homologous chromosome pairing and synapsis, only univalents were detected at diakinesis and metaphase I (Supplementary Figures S6C1,C2), and no chromosome fragmentation was observed (Supplementary Figures S6C1–C6). In the osbrca2-1 ossds double mutants, no chromosome entanglements or fragments were observed, similar to that in the ossds single mutant (Supplementary Figures S6D1–D6). The chromosome abnormalities in osbrca2-1 were totally suppressed by loss of the OsSDS function. Therefore, we conclude that unrepaired DSBs are responsible for the chromosome entanglement and fragmentation generated in osbrca2-1.
To further determine whether OsBRCA2 is involved in DSB formation or processing, we applied antibodies raised against γH2AX, COM1, RPA1c, RPA2c, OsRAD51A, and OsDMC1, to perform an immunolocalization analysis on wild-type and osbrca2-1 male meiocytes. γH2AX is the phosphorylated form of Histone 2A.X and is a reliable marker for detection of DSB sites (Hunter et al., 2001; Mahadevaiah et al., 2001). γH2AX immunolocalization showed no significant difference between wild type (200.8, n = 35) and osbrca2-1 (212.2, n = 26; Supplementary Figure S7A), indicating that OsBRCA2 is dispensable for DSB initiation. It had been reported that COM1, RPA1c and RPA2c were required for DSB end-processing and 3' single strand invasion after DSB generation (Ji et al., 2012; Li et al., 2013; Tang et al., 2014). The localization of these proteins was also indistinguishable in osbrca2-1 (154.3, n = 30; 161.0, n = 22; and 211.8, n = 26, respectively) when compared with that in wild type (163.7, n = 34; 152.9, n = 22; and 211.2, n = 27, respectively; Supplementary Figures S7B–D). These results indicated that OsBRCA2 is not required for DSB production or resection end-processing.
During meiotic recombination, OsDMC1 and OsRAD51A are two key recombinases that play essential roles in crossover formation by promoting homology searches of the invading single strand and catalyzing strand exchange thereafter (Suwaki et al., 2011; Morozumi et al., 2013; Sansam and Pezza, 2015; Wang et al., 2016). In wild-type male meiocytes, OsDMC1 and OsRAD51A foci occurred at leptotene and peaked during zygotene (average 159.6, n = 28, and 140.8, n = 33, respectively). However, in osbrca2-1 zygotene meiocytes, the number of DMC1 and OsRAD51A foci was substantially decreased (average 3.5, n = 26, ***p < 0.0001, Figures 7A,C; and 18.2, n = 32, ***p < 0.0001, Figures 7B,D), indicating that functional OsBRCA2 is indispensable for the localization of OsDMC1 and OsRAD51A onto chromosomes during meiosis. These results suggest that disruption of crossover formation in osbrca2-1 is due to deficiency in OsDMC1 and OsRAD51A recruitment.
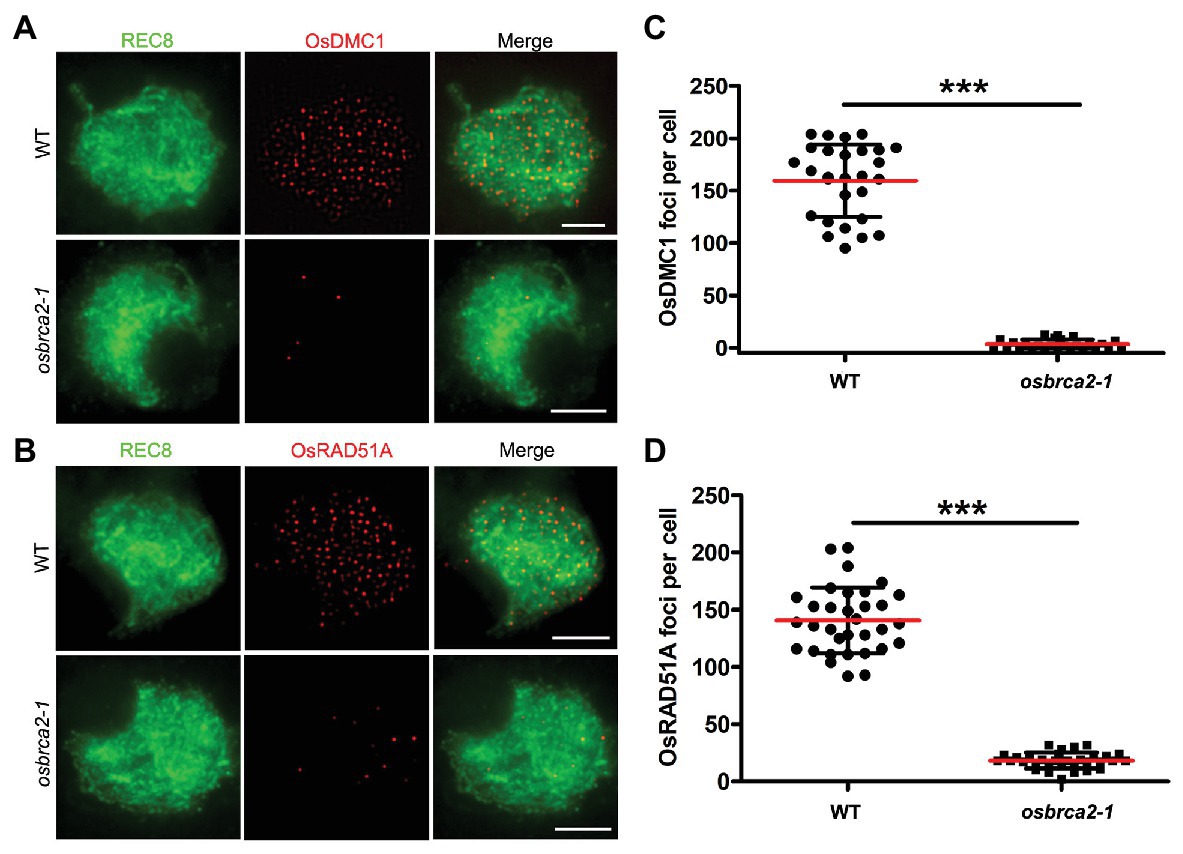
Figure 7. DMC1and RAD51A are not able to load on meiotic chromosomes in osbrca2-1. (A) Immunolocalization of DMC1 (red) and OsREC8 (green) in wild-type and osbrca2-1 meiocytes at zygotene. (B) Immunolocalization of RAD51A (red) and OsREC8 (green) in wild-type and osbrca2-1 meiocytes at zygotene. (C) Statistical analysis of the number of OsDMC1 foci per cell in wild type (n = 28) and osbrca2-1 mutants (n = 26). (D) Statistical analysis of the number of OsRAD51A foci per cell in wild type (n = 33) and osbrca2-1 mutants (n = 32). All values represent the mean ± SD, ***p < 0.0001, Student’s t tests. Bars = 2 μm.
It has been previously reported that BRCA2 acts as a universal recombinase regulator and interacts with RAD51 and DMC1 homologs via BRC domains in many species (Fradet-Turcotte et al., 2016). To investigate whether OsBRCA2 also directly interacts with OsRAD51 or OsDMC1 in rice, yeast two-hybrid (Y2H) was performed. Rice contains two RAD51 genes (RAD51A1 and RAD51A2) as well as two OsDMC1 genes (OsDMC1A and OsDMC1B). Y2H assays showed that both OsRAD51’s and OsDMC1’s interaction with the full length OsBRCA2 protein. Consistent with previous reports in other organisms, OsBRCA2 interacted with these proteins via its N-terminal OsBRC1-6 repeats (Figures 8A,B).
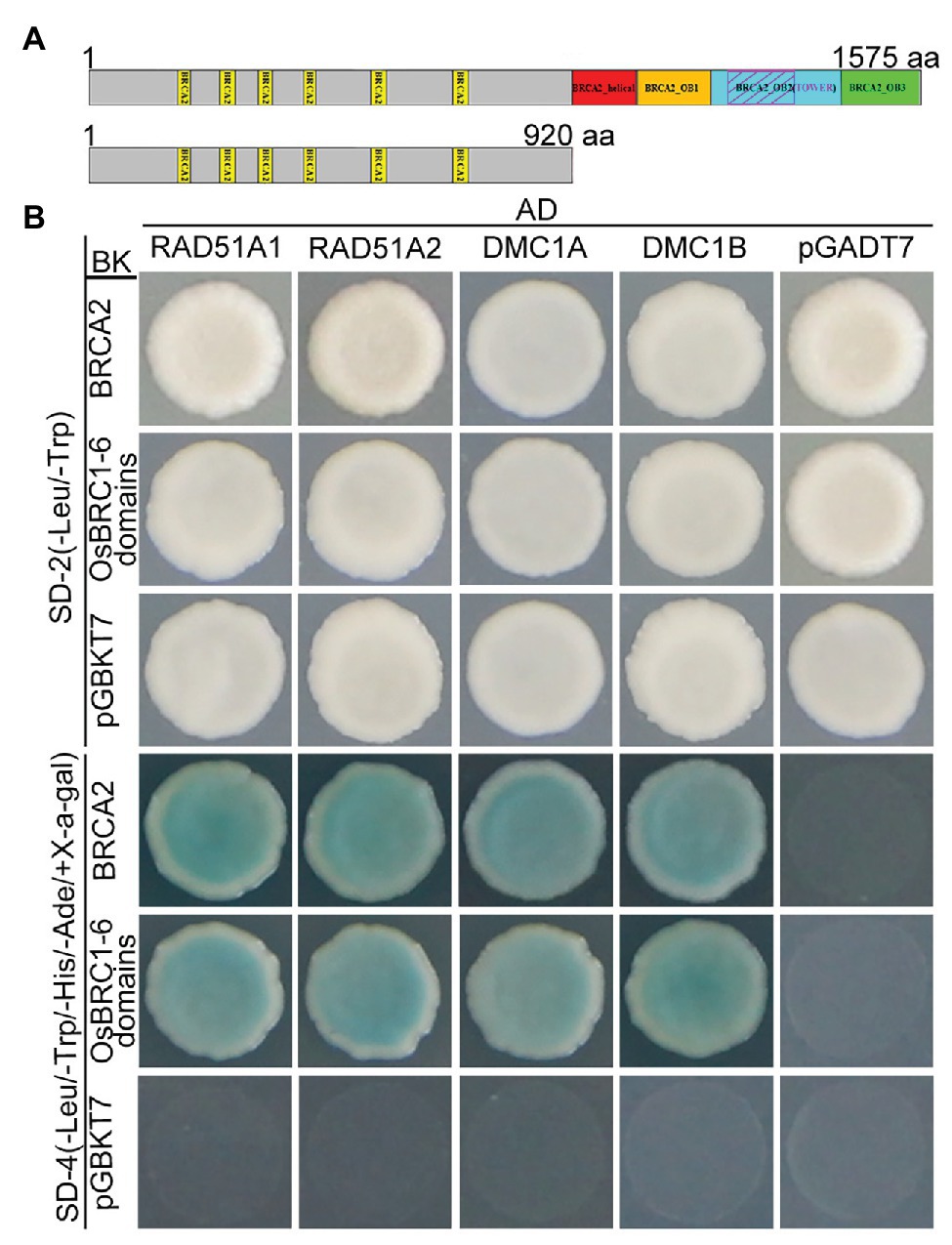
Figure 8. Interactions among OsBRCA2 and OsRAD51 paralogs. (A) A schematic diagram of OsBRCA2 and the truncation containing BRC1-6 motifs used in the following yeast two-hybrid assay is shown. (B) Yeast two-hybrid (Y2H) assay for interaction of OsBRCA2 with OsRAD51A1, OsRAD51A2, OsDMC1A, and OsDMC1B which verified on the SD-4 (-Leu/-Trp/-His/-Ade/+ X-α-gal) selection medium.
The osbrca2-1 Mutant Shows Hypersensitivity to Mitomycin C
Generally, mutants of genes involved in meiotic HR DSB repair show hypersensitivity to DNA mutagens, such as MMS, MMC, and UV irradiation (Waterworth et al., 2007; Chang et al., 2009; Kou et al., 2012; Zhang et al., 2015). Human pancreatic cancer cells expressing mutated BRCA2 lead to hypersensitivity from MMS treatment (Chen et al., 1998) and hsbrca2 mutant embryos are also hypersensitive to γ-irradiation (Sharan et al., 1997). Similarly, Arabidopsis brca2 double mutants are hypersensitive to the cross-linking agent MMC (Seeliger et al., 2012).
OsBRCA2 was broadly expressed not only in reproductive organs but also in vegetative organs, especially in leaves (Supplementary Figure S2). This suggests that although OsBRCA2 is not required for normal vegetative growth, it may be involved in HR mediated DNA damage repair in somatic cells. To test whether osbrca2-1 is sensitive to genotoxins like hsbrca2 or atbrca2, we treated 5-day-old seedlings of the heterozygous OsBRCA2-1+/− for 13 days with different concentrations of MMS or MMC. After treatment, the genotype of the seedlings was determined by PCR amplification and sequencing. There was no difference between osbrca2-1 and wild-type seedlings when grown on 1/2 MS medium without genotoxins (n = 15, p = 0.84, Figures 9A,E). While grown on a medium containing 100–300 μg/ml MMC, osbrca2-1 seedlings grew significantly slower than wild-type seedlings (n = 15, p < 0.05, Figures 9B,D,E; p < 0.01, Figures 9C,E) and after 20 days osbrca2-1 mutants could not survive both low and high MMC concentrations. Moreover, about 30% (n = 15) osbrca2-1+/− heterozygotes also died (Supplementary Figure S8). By contrast, the osbrca2-1 plants showed an undistinguishable level of suppressed growth to that of the wild-type plants on culture medium supplemented with 50 μl/L and 100 μl/L MMS (n = 15, p > 0.05, Supplementary Figures S9A–C). In addition, both wild-type plants and osbrca2-1 mutants could not survive after 150 μl/L MMS treatment (Supplementary Figure S9D). These results demonstrated that osbrca2-1 plants were hypersensitive to the DNA damaging agent that causes interstrand cross-linking, indicating that OsBRCA2 is essential for HR repair of DNA damage in somatic cells.
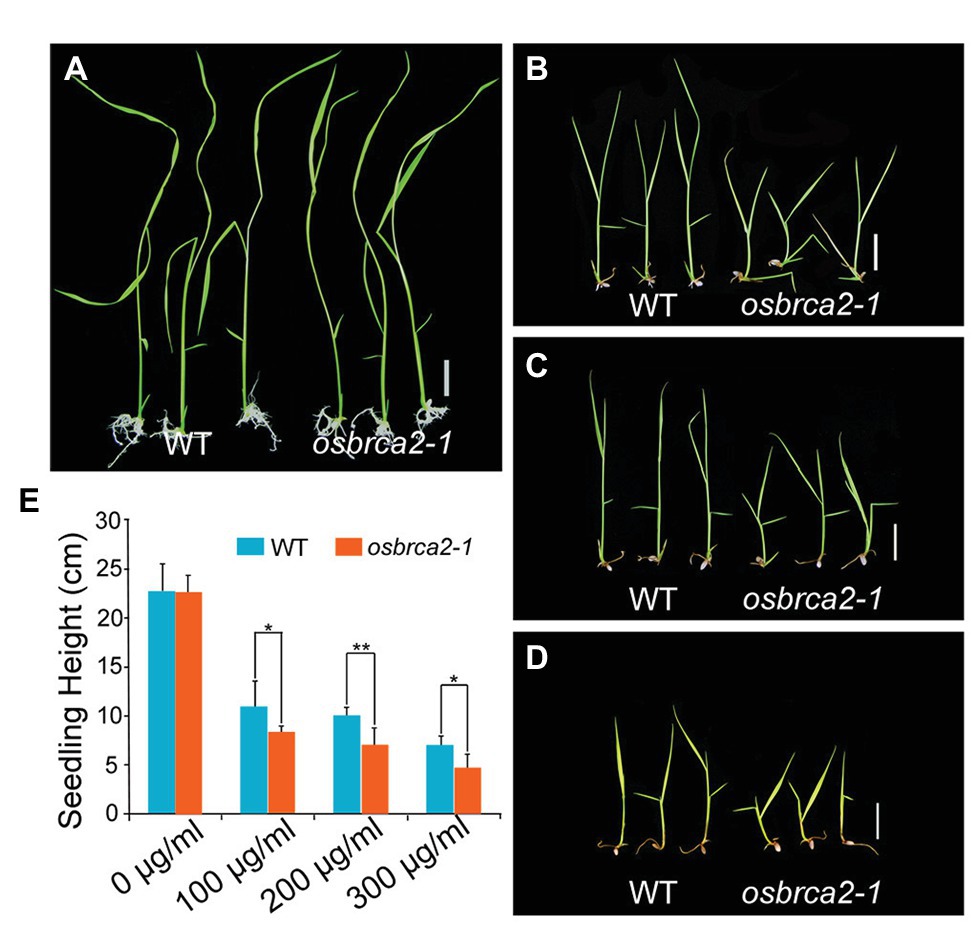
Figure 9. Osbrca2-1 mutants are sensitive to mitomycin C (MMC). Phenotypes of wild-type and osbrca2-1 seedlings growth in ½ MS medium at different MMC concentrations with 0 μg/ml (A), 100 μg/ml (B), 200 μg/ml (C), and 300 μg/ml (D). (E) Statistical analysis of the height of wild-type and osbrca2-1 seedlings (n = 15, *p < 0.05, **p < 0.01, Student’s t tests). All values represent the mean ± SD. Bar = 2 cm.
Discussion
OsBRCA2 Is Indispensable for Homologous Chromosome Pairing, Synapsis and DNA Repair in Rice Meiosis
In animals, BRCA2 plays essential roles in HR and HR-mediated DSB repair (O’Donovan and Livingston, 2010; Fradet-Turcotte et al., 2016). Compared with the extensive studies in animals, little is known about the function of BRCA2 in plants. Although orthologues of BRCA2 have been characterized in Arabidopsis (Siaud et al., 2004; Seeliger et al., 2012), it is yet unclear how widely these functions are conserved in other plants. In this study, our data reveals that mutations in rice BRCA2 lead to severe meiotic defects and infertility, but have no obvious effects on normal vegetative growth, unless exposed to genotoxic agent MMC.
Homologous pairing, synapsis, and recombination are interdependent processes during meiosis in many organisms (Wang and Copenhaver, 2018). HR is essential for facilitating the formation of crossovers (COs) and accurate chromosome segregation. FISH analysis using 5S rDNA and OsCenH3 probes shows that homologous pairing was compromised in osbrca2-1 (Figures 5C,D). Furthermore, the initiation of synapsis seemed normal but the synaptonemal complexes could not assemble successfully in the mutant, evidenced by the normal localization of PAIR3 while delayed depleting of PAIR2 and the failure of ZEP1 assembly (Figures 6A,B) at pachytene. At diakinesis, univalents instead of bivalents were observed in osbrca2, causing random segregation of homologous chromosomes at anaphase I. These results suggest that OsBRCA2 acts at an early stage of HR by mediating DSB dependent chromosome contacts, which is critical to establish physical linkage and ensure proper separation of homologous chromosomes.
The meiotic defects in osbrca2 are consistent with those observed in mammals and Arabidopsis for brca2 mutants, but in contrast to that of Caenorhabditis elegans, in which no homolog alignment or SC assembly aberrance is detected (Martin et al., 2005). The atbrca2a/b double mutant is able to set a few seeds when pollinated with wild type pollen grains (Seeliger et al., 2012), indicating that deletion of both AtBRCA2A/B does not lead to the complete loss of female gametophyte function. However, the male mice hypomorphic brca2 mutant is completely sterile, but in females some oocytes undergo meiosis and fertilization that finally develop into embryos (Sharan et al., 2004). By contrast, osbrca2-1 plants could not set seeds either used as maternal or paternal recipients. These observations suggest a functional divergence for the necessity of BRCA2 in different species and a different requirement of BRCA2 in male and female meiocyte development.
OsBRCA2 Is Essential for DNA Repair in Both Meiotic and Somatic Cells
Homologous recombination is one of the most effective mechanisms for DSB repair, which guarantees accurate repair of DSBs within homologous sequences (Cloud et al., 2012). BRCA2 plays a central role in HR-mediated DSB repair in animals and Arabidopsis (Gudmundsdottir and Ashworth, 2004; Thorslund and West, 2007). The normal localization of γH2AX, COM1, RPA1c and RPA2c in male meiocytes of osbrca2-1 suggests that OsBRCA2 is dispensable for DSB production and its end processing (Supplementary Figure S6). However, chromosome entanglements and fragments are observed at diakinesis and later stages (Figure 4). Furthermore, these abnormalities are completely eliminated by the mutation in OsSDS, a key factor required for meiotic DSB generation in rice (Supplementary Figure S5), indicating that OsBRCA2 plays a conserved role in programmed meiotic DSB repair.
BRCA2 is not only essential for meiotic DSB repair but also important for DSB repair in mitotic cell. Animals carrying a complete loss-of-function allele of BRCA2 are embryo lethal, suggesting cell cycle arrest and cell death triggered by the accumulating DNA damages and increased genomic instability. Hypomorphic brca2 mutants also showed hypersensitivity to DNA-damage agents, such as MMS, MMC, UV irradiation, which is a common feature of mutants defective in HR repair (Chen et al., 1998, 1999; Yuan et al., 1999; Yu et al., 2000; Masson et al., 2001; Wiese et al., 2002; Ohashi et al., 2005; Meyer et al., 2017). In Arabidopsis, both Atbrca2a and Atbrca2b single mutants and the double mutant are viable and appear healthy under normal growth conditions, while being hypersensitive to MMC (Wang et al., 2010b; Seeliger et al., 2012). Our study shows that osbrca2-1 seedlings are also hypersensitive to MMC (Figure 9), which introduces DNA interstrand cross-linking (Lehoczky et al., 2007). Interestingly, the heterozygotes of osbrca2-1 seedlings also exhibit higher ratio of death after treatment by low or high concentrations of MMC for 20 days (Supplementary Figure S7), indicating that adequate expression of BRCA2 is essential for DNA repair in rice somatic cells.
MMS causes damage by methylating DNA on the N7-deoxyguanine and N3-deoxyadenine (Vázquez et al., 2008). It has been shown that a truncated BRCA2 strain in mouse exhibits a striking increased sensitivity to MMS treatment. However, osbrca2-1 and wild-type seedlings displayed a similar sensitivity to MMS at a series of concentrations (Supplementary Figure S8). Different phenotypic outcomes of brca2 mutants to MMS in rice and mouse are probably due to DNA pathway choices. Another possibility is that there might be tissue or developmental stage specific sensitivity to genotoxic agents. For example, mouse embryo fibroblasts bearing the Brca2Tr/Tr mutation do not exhibit sensitivity to MMC, while Brca2Tr/Tr lymphoid cells are highly sensitive to MMC (Patel et al., 1998). These results confirmed the conserved function of BRCA2 in repairing DNA damage among different species.
The Role of OsBRCA2 in HR Mediated DNA Repair
In humans, BRCA2 interacts with RAD51 via BRC repeats and the TR2 domain located on the C-terminus. The BRC repeats of HsBRCA2 can also directly interact with DMC1 to stimulate single-strand invasion between homologous chromosomes. Besides, the PhePP domain of HsBRCA2 is specially bound by DMC1, but not for RAD51. (Davies and Pellegrini, 2007; Thorslund et al., 2007; Fradet-Turcotte et al., 2016; Martinez et al., 2016). The distinct interacting motifs on BRCA2 with RAD51 and DMC1 confer the universal functions of BRCA2 in regulating actions of two recombinases in both germinal and somatic cells. In Arabidopsis, there are two isoforms of BRCA2. Both AtBRCA2 can interact with AtRAD51 and AtDMC1 in vitro (Siaud et al., 2004; Dray et al., 2006) and are required for their recruitment to the chromosomes in meiocytes (Seeliger et al., 2012).
Our results show that both the full length and the truncated OsBRCA2 containing the six BRC repeats are capable of interacting with the two isoforms of OsRAD51A and OsDMC1 (Figure 8), suggesting a conserved function of the BRC repeats in OsBRCA2. Our results further reveal that both OsRAD51A and OsDMC1 foci are dramatically reduced in osbrca2-1 compared with wild type (Figure 7), confirming that the correct localization of OsRAD51As and OsDMC1 onto meiotic chromosomes also depends on a functional BRCA2 in rice. In contrast to Arabidopsis, OsDMC1 is not required for homologous chromosome pairing in rice (Wang et al., 2016). Previous reports have demonstrated the important role of RAD51 in homology searching and pairing in many organisms (Bishop, 1994; Pawlowski et al., 2003; Li et al., 2004). Thus, the abnormality in homologous pairing may mainly be due to the failure in recruiting OsRAD51A’s, although this is yet to be investigated in rice. Taken together, our study provides evidence for the conserved function of rice BRCA2 as a central regulator of RAD51 and DMC1 recruitment, to facilitate single-strand invasion during HR and HR-mediated DSB repair.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
WL conceived the project. RF, CW, HS, and JZ performed experiments. RF wrote the article. WL and JH supervised and complemented the writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the fund from the National Key Research and Development Program of China (2016YFD0100902), National Natural Science Foundation of China (U19A2031), China Innovative Research Team, Ministry of Education, and the Programme of Introducing Talents of Discipline to Universities (111 Project, B14016).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mingjiao Chen, Zhijin Luo, and Zibo Chen for screening the mutant and collecting the F2 population samples for mapping.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.600820/full#supplementary-material
References
Abe, K., Osakabe, K., Ishikawa, Y., Tagiri, A., Yamanouchi, H., Takyuu, T., et al. (2009). Inefficient double-strand DNA break repair is associated with increased fasciation in Arabidopsis BRCA2 mutants. J. Exp. Bot. 60, 2751–2761. doi: 10.1093/jxb/erp135
Bennett, C. B., Lewis, A. L., Baldwin, K. K., and Resnick, M. A. (1993). Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. U. S. A. 90, 5613–5617. doi: 10.1073/pnas.90.12.5613
Bétermier, M., Bertrand, P., and Lopez, B. S. (2014). Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 10:e1004086. doi: 10.1371/journal.pgen.1004086
Bishop, D. K. (1994). RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79, 1081–1092. doi: 10.1016/0092-8674(94)90038-8
Chang, Y., Gong, L., Yuan, W., Li, X., Chen, G., Li, X., et al. (2009). Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice. Plant Physiol. 151, 2162–2173. doi: 10.1104/pp.109.142877
Chen, P. L., Chen, C. F., Chen, Y., Xiao, J., Sharp, Z. D., and Lee, W. H. (1998). The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc. Natl. Acad. Sci. U. S. A. 95, 5287–5292. doi: 10.1073/pnas.95.9.5287
Chen, C. F., Chen, P. L., Zhong, Q., Sharp, Z. D., and Lee, W. H. (1999). Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J. Biol. Chem. 274, 32931–32935. doi: 10.1074/jbc.274.46.32931
Chen, L., Chu, H., Yuan, Z., Pan, A., Liang, W., Huang, H., et al. (2006). Isolation and genetic analysis for rice mutants treated with 60 co γ-ray. J. Xiamen Univ. 45, 82–85. doi: 10.3321/j.issn:0438-0479.2006.z1.021
Cheng, Z. (2013). Analyzing meiotic chromosomes in rice. Methods Mol. Biol. 990, 125–134. doi: 10.1007/978-1-62703-333-6_13
Cloud, V., Chan, Y. -L., Grubb, J., Budke, B., and Bishop, D. K. (2012). Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337, 1222–1225. doi: 10.1126/science.1219379
Da Ines, O., Degroote, F., Goubely, C., Amiard, S., Gallego, M. E., and White, C. I. (2013). Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet. 9, e1003787–e1003787. doi: 10.1371/journal.pgen.1003787
Davies, O. R., and Pellegrini, L. (2007). Interaction with the BRCA2 C terminus protects RAD51–DNA filaments from disassembly by BRC repeats. Nat. Struct. Mol. Biol. 14, 475–483. doi: 10.1038/nsmb1251
Dray, E., Siaud, N., Dubois, E., and Doutriaux, M. P. (2006). Interaction between Arabidopsis Brca2 and its partners Rad51, Dmc1, and Dss1. Plant Physiol. 140, 1059–1069. doi: 10.1104/pp.105.075838
Esashi, F., Galkin, V. E., Yu, X., Egelman, E. H., and West, S. C. (2007). Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 14, 468–474. doi: 10.1038/nsmb1245
Fayos, I., Meunier, A. C., Vernet, A., Sanz, S. N., Portefaix, M., Lartaud, M., et al. (2020). Assessment of the roles of OsSPO11-2 and OsSPO11-4 in rice meiosis using CRISPR/Cas9 mutagenesis. J. Exp. Bot. doi: 10.1093/jxb/eraa391 [Epub head of print]
Fradet-Turcotte, A., Sitz, J., Grapton, D., and Orthwein, A. (2016). BRCA2 functions: from DNA repair to replication fork stabilization. Endocr. Relat. Cancer 23, T1–T17. doi: 10.1530/ERC-16-0297
Fu, M., Wang, C., Xue, F., Higgins, J., Chen, M., Zhang, D., et al. (2016). The DNA topoisomerase VI-B subunit OsMTOPVIB is essential for meiotic recombination initiation in Rice. Mol. Plant 9, 1539–1541. doi: 10.1016/j.molp.2016.07.006
Fu, Z., Yu, J., Cheng, X., Zong, X., Xu, J., Chen, M., et al. (2014). The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26, 1512–1524. doi: 10.1105/tpc.114.123745
Gudmundsdottir, K., and Ashworth, A. (2004). BRCA2 in meiosis: turning over a new leaf. Trends Cell Biol. 14, 401–404. doi: 10.1016/j.tcb.2004.07.002
Guirouilh-Barbat, J., Lambert, S., Bertrand, P., and Lopez, B. S. (2014). Is homologous recombination really an error-free process? Front. Genet. 5:175. doi: 10.3389/fgene.2014.00175
Hall, J. M., Lee, M. K., Newman, B., Morrow, J. E., Anderson, L. A., Huey, B., et al. (1990). Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250, 1684–1689. doi: 10.1126/science.2270482
He, Y., Wang, C., Higgins, J. D., Yu, J., Zong, J., Lu, P., et al. (2016). MEIOTIC F-BOX is essential for male meiotic DNA double-Strand break repair in Rice. Plant Cell 28, 1879–1893. doi: 10.1105/tpc.16.00108
Hunter, N., Borner, G. V., Lichten, M., and Kleckner, N. (2001). Gamma-H2AX illuminates meiosis. Nat. Genet. 27, 236–238. doi: 10.1038/85781
Hussain, S., Wilson, J. B., Medhurst, A. L., Hejna, J., Witt, E., Ananth, S., et al. (2004). Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum. Mol. Genet. 13, 1241–1248. doi: 10.1093/hmg/ddh135
Hussain, S., Witt, E., Huber, P. A. J., Medhurst, A. L., Ashworth, A., and Mathew, C. G. (2003). Direct interaction of the Fanconi anaemia protein FANCG with BRCA2FANCD1. Hum. Mol. Genet. 12, 2503–2510. doi: 10.1093/hmg/ddg266
Iftode, C., Daniely, Y., and Borowiec, J. A. (1999). Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34, 141–180. doi: 10.1080/10409239991209255
Inagaki, A., Schoenmakers, S., and Baarends, W. M. (2010). DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis. Epigenetics 5, 255–266. doi: 10.4161/epi.5.4.11518
Ji, J., Tang, D., Wang, K., Wang, M., Che, L., Li, M., et al. (2012). The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J. 72, 18–30. doi: 10.1111/j.1365-313X.2012.05025.x
Kou, Y., Chang, Y., Li, X., Xiao, J., and Wang, S. (2012). The rice RAD51C gene is required for the meiosis of both female and male gametocytes and the DNA repair of somatic cells. J. Exp. Bot. 63, 5323–5335. doi: 10.1093/jxb/ers190
Kurzbauer, M. -T., Uanschou, C., Chen, D., and Schlögelhofer, P. (2012). The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell 24, 2058–2070. doi: 10.1105/tpc.112.098459
Lehoczky, P., McHugh, P. J., and Chovanec, M. (2007). DNA interstrand cross-link repair in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 31, 109–133. doi: 10.1111/j.1574-6976.2006.00046.x
Li, X., Chang, Y., Xin, X., Zhu, C., Li, X., Higgins, J. D., et al. (2013). Replication protein A2c coupled with replication protein A1c regulates crossover formation during meiosis in rice. Plant Cell 25, 3885–3899. doi: 10.1105/tpc.113.118042
Li, W., Chen, C., Markmann-Mulisch, U., Timofejeva, L., Schmelzer, E., Ma, H., et al. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. U. S. A. 101, 10596–10601. doi: 10.1073/pnas.0404110101
Li, N., Zhang, D. S., Liu, H. S., Yin, C. S., Li, X. X., Liang, W. Q., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999–3014. doi: 10.1105/tpc.106.044107
Liu, H., Chu, H., Li, H., Wang, H., Wei, J., Li, N., et al. (2005). Genetic analysis and mapping of rice (Oryza sativa L.) male-sterile (OsMS-L) mutant. Chin. Sci. Bull. 50, 122–125. doi: 10.1007/BF02897514
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, H., Guo, X., Meng, X., Liu, J., Allen, C., Wray, J., et al. (2005). The BRCA2-interacting protein BCCIP functions in RAD51 and BRCA2 focus formation and homologous Recombinational repair. Mol. Cell. Biol. 25, 1949–1957. doi: 10.1128/MCB.25.5.1949-1957.2005
Mahadevaiah, S. K., Turner, J. M., Baudat, F., Rogakou, E. P., de Boer, P., Blanco-Rodriguez, J., et al. (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27, 271–276. doi: 10.1038/85830
Marston, N. J., Richards, W. J., Hughes, D., Bertwistle, D., Marshall, C. J., and Ashworth, A. (1999). Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol. Cell. Biol. 19, 4633–4642. doi: 10.1128/MCB.19.7.4633
Martin, J. S., Winkelmann, N., Petalcorin, M. I. R., McIlwraith, M. J., and Boulton, S. J. (2005). RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-Strand break repair. Mol. Cell. Biol. 25, 3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005
Martinez, J. S., von Nicolai, C., Kim, T., Ehlén, Å., Mazin, A. V., Kowalczykowski, S. C., et al. (2016). BRCA2 regulates DMC1-mediated recombination through the BRC repeats. Proc. Natl. Acad. Sci. U. S. A. 113, 3515–3520. doi: 10.1073/pnas.1601691113
Masson, J. -Y., Stasiak, A. Z., Stasiak, A., Benson, F. E., and West, S. C. (2001). Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc. Natl. Acad. Sci. U. S. A. 98, 8440–8456. doi: 10.1073/pnas.111005698
Meyer, S., Stevens, A., Paredes, R., Schneider, M., Walker, M. J., Williamson, A. J. K., et al. (2017). Acquired cross-linker resistance associated with a novel spliced BRCA2 protein variant for molecular phenotyping of BRCA2 disruption. Cell Death Dis. 8:e2875. doi: 10.1038/cddis.2017.264
Mizuno, H., Wu, J., Kanamori, H., Fujisawa, M., Namiki, N., Saji, S., et al. (2006). Sequencing and characterization of telomere and subtelomere regions on rice chromosomes 1S, 2S, 2L, 6L, 7S, 7L and 8S. Plant J. 46, 206–217. doi: 10.1111/j.1365-313X.2006.02684.x
Morozumi, Y., Ino, R., Ikawa, S., Mimida, N., Shimizu, T., Toki, S., et al. (2013). Homologous pairing activities of two rice RAD51 proteins, RAD51A1 and RAD51A2. PLoS One 8:e75451. doi: 10.1371/journal.pone.0075451
Nonomura, K. -I., Nakano, M., Eiguchi, M., Suzuki, T., and Kurata, N. (2006). PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 119, 217–225. doi: 10.1242/jcs.02736
O’Donovan, P. J., and Livingston, D. M. (2010). BRCA1 and BRCA2: breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis 31, 961–967. doi: 10.1093/carcin/bgq069
Ohashi, A., Zdzienicka, M. Z., Chen, J., and Couch, F. J. (2005). Fanconi anemia complementation group D2 (FANCD2) functions independently of BRCA2- and RAD51-associated homologous recombination in response to DNA damage. J. Biol. Chem. 280, 14877–14883. doi: 10.1074/jbc.M414669200
Orthwein, A., Noordermeer, S. M., Wilson, M. D., Landry, S., Enchev, R. I., Sherker, A., et al. (2015). A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426. doi: 10.1038/nature16142
Pastink, A., Eeken, J. C., and Lohman, P. H. (2001). Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res. 480-481, 37–50. doi: 10.1016/s0027-5107(01)00167-1
Patel, K. J., Yu, V. P., Lee, H., Corcoran, A., Thistlethwaite, F. C., Evans, M. J., et al. (1998). Involvement of Brca2 in DNA repair. Mol. Cell 1, 347–357. doi: 10.1016/S1097-2765(00)80035-0
Pawlowski, W. P., Golubovskaya, I. N., and Cande, W. Z. (2003). Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15, 1807–1816. doi: 10.1105/tpc.012898
Pfeffer, C. M., Ho, B. N., and Singh, A. T. K. (2017). The evolution, functions and applications of the breast cancer genes BRCA1 and BRCA2. Cancer Genomics Proteomics 14, 293–298. doi: 10.21873/cgp.20040
Roca, A. I., and Cox, M. M. (1997). “RecA protein: structure, function, and role in recombinational DNA repair” in Progress in nucleic acid research and molecular biology. eds. W. E. Cohn and K. Moldave (New York, USA: Academic Press), 129–223.
Sandell, L. L., and Zakian, V. A. (1993). Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75, 729–739. doi: 10.1016/0092-8674(93)90493-A
San Filippo, J., Sung, P., and Klein, H. (2008). Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257. doi: 10.1146/annurev.biochem.77.061306.125255
Sansam, C. L., and Pezza, R. J. (2015). Connecting by breaking and repairing: mechanisms of DNA strand exchange in meiotic recombination. FEBS J. 282, 2444–2457. doi: 10.1111/febs.13317
Schoenfeld, A. R., Apgar, S., Dolios, G., Wang, R., and Aaronson, S. A. (2004). BRCA2 is Ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol. Cell. Biol. 24, 7444–7455. doi: 10.1128/MCB.24.17.7444-7455.2004
Seeliger, K., Dukowic-Schulze, S., Wurz-Wildersinn, R., Pacher, M., and Puchta, H. (2012). BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. New Phytol. 193, 364–375. doi: 10.1111/j.1469-8137.2011.03947.x
Shao, T., Tang, D., Wang, K., Wang, M., Che, L., Qin, B., et al. (2011). OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol. 156, 1386–1396. doi: 10.1104/pp.111.177428
Sharan, S. K., Morimatsu, M., Albrecht, U., Lim, D. S., Regel, E., Dinh, C., et al. (1997). Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386, 804–810. doi: 10.1038/386804a0
Sharan, S. K., Pyle, A., Coppola, V., Babus, J., Swaminathan, S., Benedict, J., et al. (2004). BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131, 131–142. doi: 10.1242/dev.00888
Siaud, N., Dray, E., Gy, I., Gerard, E., Takvorian, N., and Doutriaux, M. P. (2004). Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 23, 1392–1401. doi: 10.1038/sj.emboj.7600146
Suwaki, N., Klare, K., and Tarsounas, M. (2011). RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin. Cell Dev. Biol. 22, 898–905. doi: 10.1016/j.semcdb.2011.07.019
Sy, S. M. H., Huen, M. S. Y., and Chen, J. (2009). PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. U. S. A. 106, 7155–7160. doi: 10.1073/pnas.0811159106
Tang, D., Miao, C., Li, Y., Wang, H., Liu, X., Yu, H., et al. (2014). OsRAD51C is essential for double-strand break repair in rice meiosis. Front. Plant Sci. 5:167. doi: 10.3389/fpls.2014.00167
Thorslund, T., Esashi, F., and West, S. C. (2007). Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. EMBO J. 26, 2915–2922. doi: 10.1038/sj.emboj.7601739
Thorslund, T., and West, S. C. (2007). BRCA2: a universal recombinase regulator. Oncogene 26, 7720–7730. doi: 10.1038/sj.onc.1210870
Vázquez, M. V., Rojas, V., and Tercero, J. A. (2008). Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair 7, 1693–1704. doi: 10.1016/j.dnarep.2008.06.014
Wang, X., Andreassen, P. R., and D’Andrea, A. D. (2004). Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 24, 5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004
Wang, Y., and Copenhaver, G. P. (2018). Meiotic recombination: mixing it up in plants. Annu. Rev. Plant Biol. 69, 577–609. doi: 10.1146/annurev-arplant-042817-040431
Wang, S., Durrant, W. E., Song, J., Spivey, N. W., and Dong, X. (2010b). Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc. Natl. Acad. Sci. U. S. A. 107, 22716–22721. doi: 10.1073/pnas.1005978107
Wang, C., Higgins, J. D., He, Y., Lu, P., Zhang, D., and Liang, W. (2017). Resolvase OsGEN1 mediates DNA repair by homologous recombination. Plant Physiol. 173, 1316–1329. doi: 10.1104/pp.16.01726
Wang, H., Hu, Q., Tang, D., Liu, X., Du, G., Shen, Y., et al. (2016). OsDMC1 is not required for homologous pairing in Rice meiosis. Plant Physiol. 171, 230–241. doi: 10.1104/pp.16.00167
Wang, K., Wang, M., Tang, D., Shen, Y., Qin, B., Li, M., et al. (2011). PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol. Biol. Cell 22, 12–19. doi: 10.1091/mbc.e10-08-0667
Wang, M., Wang, K., Tang, D., Wei, C., Li, M., Shen, Y., et al. (2010a). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22, 417–430. doi: 10.1105/tpc.109.070789
Waterworth, W. M., Altun, C., Armstrong, S. J., Roberts, N., Dean, P. J., Young, K., et al. (2007). NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 52, 41–52. doi: 10.1111/j.1365-313X.2007.03220.x
Wiese, C., Collins, D. W., Albala, J. S., Thompson, L. H., Kronenberg, A., and Schild, D. (2002). Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 30, 1001–1008. doi: 10.1093/nar/30.4.1001
Wong, A. K., Pero, R., Ormonde, P. A., Tavtigian, S. V., and Bartel, P. L. (1997). RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 272, 31941–31944. doi: 10.1074/jbc.272.51.31941
Wooster, R., Bignell, G., Lancaster, J., Swift, S., Seal, S., Mangion, J., et al. (1995). Identification of the breast cancer susceptibility gene BRCA2. Nature 378, 789–792. doi: 10.1038/378789a0
Wu, Z., Ji, J., Tang, D., Wang, H., Shen, Y., Shi, W., et al. (2015). OsSDS is essential for DSB formation in rice meiosis. Front. Plant Sci. 6:21. doi: 10.3389/fpls.2015.00021
Xia, B., Sheng, Q., Nakanishi, K., Ohashi, A., Wu, J., Christ, N., et al. (2006) Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719–729. doi: 10.1016/j.molcel.2006.05.022
Yang, H., Jeffrey, P. D., Miller, J., Kinnucan, E., Sun, Y., Thoma, N. H., et al. (2002). BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297, 1837–1848. doi: 10.1126/science.297.5588.1837
Yang, H., Li, Q., Fan, J., Holloman, W. K., and Pavletich, N. P. (2005). The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433, 653–657. doi: 10.1038/nature03234
Yu, V. P. C. C., Koehler, M., Steinlein, C., Schmid, M., Hanakahi, L. A., van Gool, A. J., et al. (2000). Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 14, 1400–1406.
Yuan, S. S., Lee, S. Y., Chen, G., Song, M., Tomlinson, G. E., and Lee, E. Y. (1999). BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59, 3547–3551.
Zhang, F., Fan, Q., Ren, K., and Andreassen, P. R. (2009a). PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res. 7, 1110–1118. doi: 10.1158/1541-7786.MCR-09-0123
Zhang, D., Luo, X., and Zhu, L. (2011). Cytological analysis and genetic control of rice anther development. J. Genet. Genomics 38, 379–390. doi: 10.1016/j.jgg.2011.08.001
Zhang, F., Ma, J., Wu, J., Ye, L., Cai, H., Xia, B., et al. (2009b). PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 19, 524–529. doi: 10.1016/j.cub.2009.02.018
Zhang, J., Wang, C., Higgins, J. D., Kim, Y. -J., Moon, S., Jung, K. -H., et al. (2019). A multiprotein complex regulates interference-sensitive crossover formation in Rice. Plant Physiol. 181, 221–235. doi: 10.1104/pp.19.00082
Zhang, B., Wang, M., Tang, D., Li, Y., Xu, M., Gu, M., et al. (2015). XRCC3 is essential for proper double-strand break repair and homologous recombination in rice meiosis. J. Exp. Bot. 66, 5713–5725. doi: 10.1093/jxb/erv253
Keywords: meiosis, homologous recombination, BRCA2, DMC1, RAD51
Citation: Fu R, Wang C, Shen H, Zhang J, Higgins JD and Liang W (2020) Rice OsBRCA2 Is Required for DNA Double-Strand Break Repair in Meiotic Cells. Front. Plant Sci. 11:600820. doi: 10.3389/fpls.2020.600820
Edited by:
David Smyth, Monash University, AustraliaReviewed by:
Yiping Qi, University of Maryland, United StatesHua Jiang, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Germany
Copyright © 2020 Fu, Wang, Shen, Zhang, Higgins and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanqi Liang, d3FsaWFuZ0BzanR1LmVkdS5jbg==
 Ruifeng Fu
Ruifeng Fu Chong Wang
Chong Wang Hongyu Shen
Hongyu Shen Jie Zhang
Jie Zhang James D. Higgins
James D. Higgins Wanqi Liang
Wanqi Liang