- 1National Key Laboratory of Plant Molecular Genetics, Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China
- 2Department of Microbiology and Immunology, School of Medicine & Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, China
Phenolamides represent one of the largest classes of plant-specialized secondary metabolites and function in diverse physiological processes, including defense responses and development. The biosynthesis of phenolamides requires the BAHD-family acyltransferases, which transfer acyl-groups from different acyl-donors specifically to amines, the acyl-group acceptors. However, the mechanisms of substrate specificity and multisite-acylation of the BAHD-family acyltransferases remain poorly understood. In this study, we provide a structural and biochemical analysis of AtSHT and AtSDT, two representative BAHD-family members that catalyze the multisite acylation of spermidine but show different product profiles. By determining the structures of AtSHT and AtSDT and using structure-based mutagenesis, we identified the residues important for substrate recognition in AtSHT and AtSDT and hypothesized that the acyl acceptor spermidine might adopt a free-rotating conformation in AtSHT, which can undergo mono-, di-, or tri-acylation; while the spermidine molecule in AtSDT might adopt a linear conformation, which only allows mono- or di-acylation to take place. In addition, through sequence similarity network (SSN) and structural modeling analysis, we successfully predicted and verified the functions of two uncharacterized Arabidopsis BAHD acyltransferases, OAO95042.1 and NP_190301.2, which use putrescine as the main acyl-acceptor. Our work provides not only an excellent starting point for understanding multisite acylation in BAHD-family enzymes, but also a feasible methodology for predicting possible acyl acceptor specificity of uncharacterized BAHD-family acyltransferases.
Introduction
Phenolamides, also known as phenylamides or hydroxycinnamic acid amides, are ubiquitous secondary metabolites in plants. They are produced by mono-conjugation of aromatic monoamines (tyramine, tryptamine, and dopamine) or poly-conjugation of aliphatic polyamines (putrescine, spermidine, spermine, and agmatine) with phenolic acids (coumaric, caffeic, ferulic, and sinapoyl acids; Edreva et al., 2007). Numerous studies have suggested that phenolamides play critical roles in plant biotic or abiotic stress responses (Keller et al., 1996; Von Ropenack et al., 1998; Walters et al., 2001; Galis et al., 2006; Edreva et al., 2007; Muroi et al., 2009; Kaur et al., 2010; Park et al., 2014; Cho and Lee, 2015; Schafer et al., 2017). The accumulation of p-coumaroyl/feruloyl-agmatine and p-coumaroyl/feruloyl-putrescine was induced in leaves by pathogenic infections of Solanum tuberosum and Arabidopsis thaliana (Keller et al., 1996; Von Ropenack et al., 1998; Muroi et al., 2009). Two major phenolamides of Nicotinana attenuate plants, caffeoylputrescine and dicaffeoylspermidine, increased dramatically in local and systemic tissues after an herbivore attack (Keller et al., 1996; Galis et al., 2006; Schafer et al., 2017). N-trans-cinnamoyltryptamine and N-p-coumaroylserotonin could be isolated from UV-treated rice leaves, and the genes for arylamine biosynthesis of them were also up-regulated by UV irradiation (Park et al., 2014).
In recent years, enzymes and genes accounting for the biosynthesis of mono- or di- substituted phenolamides in plants have been identified through biochemical and metabolome-based genome-wide association study (GWAS) approaches, and their expression has been analyzed (Grienenberger et al., 2009; Luo et al., 2009; Onkokesung et al., 2012; Wen et al., 2014; Dong et al., 2015; Elejalde-Palmett et al., 2015; Delporte et al., 2018). Agmatine coumaroyltransferase (ACT) synthesizes p-coumaroyl/feruloyl-agmatine and p-coumaroyl/feruloyl-putrescine in the Arabidopsis leaves and barley seedlings (Burhenne et al., 2003; Muroi et al., 2009). Spermidine disinapoyl/dicoumaroyl transferases AtSDT and AtSCT were expressed mainly in the seed and root of Arabidopsis (Luo et al., 2009). Putrescine hydroxycinnamoyl transferase (AT1) and spermidine hydroxycinnamoyl acyltransferase (DH29) were found to be expressed in the leaves of native tobacco (Onkokesung et al., 2012). Two spermidine hydroxycinnamoyl transferases were identified in leaves, while four putrescine hydroxycinnamoyl transferases were found to be constitutively expressed in Oryza sativa flowers, roots, and leaves (Wen et al., 2014; Tanabe et al., 2016). In contrast to the mono- or di- substituted phenolamides, which seem to be widely distributed in different organs, the poly-substituted phenolamides, tri-substituted spermidine, and tetra-substituted spermidine were found to be restricted to the flower pollen coat. Spermidine/spermine hydroxycinnamoyl transferases (SHT), accounting for the synthesis of tri- or tetra- substituted spermidine/spermidine, have been identified and studied in Arabidopsis thaliana (AtSHT), Malus domestica (MdSHT), and Cichorium intybus (CiSHT; Grienenberger et al., 2009; Elejalde-Palmett et al., 2015; Delporte et al., 2018).
All these phenolamide synthesis enzymes belong to the acyl-coenzyme A (CoA)-dependent BAHD acyltransferases family, which was named according to the first letter of each of its first four biochemically characterized enzymes: Benzylalcohol O-acetyltransferase, Anthocyanin O-HCT, HCT of anthranilate, and Deacetylvindoline 4-O-acetyltransferase (D’Auria, 2006). It has been established that BAHD acyltransferases catalyze the transfer of acyl groups from acyl-donors (Acyl-CoA) to various acyl-acceptors. Several structural studies of BAHD acyltransferases have been also reported (Ma et al., 2005; Unno et al., 2007; Garvey et al., 2008, 2009; Lallemand et al., 2012; Chiang et al., 2018; Levsh et al., 2019). However, the BAHD family enzymes in plants have complicated substrates (various acyl-donors and acceptors) and diverse products, while only sharing low sequence similarities, which makes it extremely challenging to predict the possible substrate specificity of uncharacterized BAHD family enzymes from their amino acid sequences. In addition, the multisite acylation mechanisms of BAHD-family acyltransferases also remain poorly understood until now.
In this study, we first performed structural analyses on two representative BAHD family enzymes, the Arabidopsis mono-/di-acyltransferase-SDT (AtSDT) and poly acyltransferase-SHT (AtSHT), then explored the acceptor-molecule specificity and potential molecular mechanisms of multisite acylation by structure-based mutagenesis approaches, and, finally, successfully predicted and varified the acyl acceptor substrates of two uncharacterized BAHD family transferases in Arabidopsis using our prediction methods on the basis of a sequence similarity network (SSN) and of structure modeling. All these results here could not only could deepen our understanding of substrate specificity and multisite acylation mechanisms of this large enzyme family, but also provide important insights on the function of many diverse and so far, uncharacterized BAHD-family proteins in plants.
Materials and Methods
Gene Cloning, Site-Directed Mutagenesis, Expression, and Protein Purification
Wild type SDT (AT2G19070), SHT (AT2G23510), OAO95042.1 (AT5G07080), and NP_190301.2 (AT3G47170) genes from Arabidopsis thaliana were cloned into the pET28a vector under control of the bacteriophage T7 gene promoters using NdeI and XhoI. The resulting plasmids were transformed into Escherichia coli strain BL21(DE3; Invitrogen). Single colonies of the resulting transformants were used to inoculate 50 ml LB broth containing 50 μg/ml kanamycin, and cultures were incubated 16 h at 37°C with shaking. Aliquots (10 ml) were used to inoculate 1 L LB broth containing 50 μg/ml kanamycin, cultures were incubated at 37°C with shaking until OD600 = 0.8, cultures were induced by addition of isopropyl-β-D-thiogalactoside to 1 mM, and cultures were incubated 16 h at 16°C. Cells were harvested by centrifugation (4,000 × g; 15 min at 4°C), re-suspended in buffer A (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 5 mM DTT, and 5% glycerol), and were lysed using an EmulsiFlex-C5 cell disruptor (Avestin). The lysate was centrifuged (20,000 × g; 30 min at 4°C) and the cell debris was removed. The supernatant was loaded onto a 5 ml column of Ni2+-NTA-agarose (Qiagen) pre-equilibrated in buffer A, and the column was washed with 10 × 5 ml buffer A containing 25 mM imidazole and eluted with 50 ml buffer A containing 250 mM imidazole. The sample was concentrated to around 10.0 mg/ml and further purified by gel filtration chromatography on a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) in 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, and 1 mM β-mercaptoethanol; The peak fractions were collected and concentrated to 10 mg/ml in the same buffer using 30 kDa MWCO Amicon Ultra-15 centrifugal ultrafilters (EMD Millipore); and stored in aliquots at −80°C. Yields were ~5 mg/l, and purities were ~95%. Site-directed mutations were prepared using one step PCR method, and proteins were expressed and purified following similar protocols as wild type.
In vitro Activity Assays
The in vitro activity assays of the purified recombinant protein were performed at 30°C for 30 min in 100 μl 100 mM Tris-HCl buffer (pH 7.5) containing 60 μM acyl donor (caffeoyl-CoA or feruloyl-CoA or sinapoyl-CoA) and 200 μM acyl acceptor (spermidine or spermine or putrescine) and 10 μg purified protein. The reactions were terminated by adding 20 μl ice-cold 0.5% trifluoroacetic acid and directly subjected to liquid chromatography-mass spectrometry (LC-MS) on an Agilent 1260 system (Agilent Technologies, CA), equipped with electron spray ionization mass spectrometer 6125B. The temperature of column oven is 30°C; Electron Spray Ionization (ESI) is in the positive mode; capillary voltage is at 3 Kv; and for full-scan mode, the wavelength range is from 190 to 600 nm. Samples were separated on a reverse-phase C18 column [Thermo Syncronis C18 analytical column (150 mm × 4.6 mm, 5 μm)] at a flow rate of 0.8 ml/min and a gradient mobile phase as follows: 0–5 min, 15% solvent B (0.2% acetic acid in acetonitrile) in solvent A (0.2% acetic acid in water); 5–25 min, 15–100% solvent B; 25–35 min, 100% solvent B; 35–40 min, 100 to 15% B (Grienenberger et al., 2009; Luo et al., 2009). CoA esters were synthesized according to published methods (Semler et al., 1987) and were identified and quantified by spectrophotometry (Stockigt and Zenk, 1974). All of the reactions were run for two technical replicates, and each assay was repeated for at least three independent experiments.
Crystallization, Data Collection and Structure Determination
Robotic crystallization trials were performed for AtSDT co-crystallized with spermidine and CoA-HS for AtSHT co-crystallized with spermdine and CoA-HS by using a Gryphon liquid handling system (Art Robbins Instruments), commercial screening solutions (Emerald Biosystems, Hampton Research, and Qiagen), and the sitting-drop vapor-diffusion technique (drop: 0.2 μl protein plus 0.2 μl screening solution; reservoir: 60 μl screening solution; 20°C). About 900 conditions were screened. Under several conditions, AtSDT and AtSHT crystals appeared within 2 weeks. Conditions were optimized using the hanging-drop vapor-diffusion technique at 20°C. The optimized crystallization condition for AtSDT was 0.1 M MES (pH 6.5), 25% W/V PEG 4000 at 20°C; the optimized crystallization condition for AtSHT was 0.2 M ammonium sulfate, 0.1 M Tris-HCl (pH 8.5), 25% W/V PEG 3350 at 20°C. Crystals were transferred to a reservoir solution containing 20% (v/v) glycerol and flash-cooled with liquid nitrogen.
Diffraction data were collected from cryo-cooled crystals at SSRF BL17U. Data were processed using HKL2000 (Otwinowski and Minor, 1997). The resolution cut-off criteria were: (i) I/σ < =2.0 and (ii) CC1/2 (highest resolution shell) < 0.5.
The structures of AtSDT and AtSHT were solved by molecular replacement with MOLREP (Collaborative Computational Project Number 4, 1994) using the structure of native HCT from Coffea canephora (PDB 4G0B) as a starting model. The molecular replacement solution was outstanding, and an automatic model building was performed with Phenix (Adams et al., 2010). Additional model building was done manually with Coot (Emsley et al., 2010) and refined with Phenix. The final model of AtSDT and AtSHT was refined to 2.4 Å and 2.3 Å resolution, respectively. The final models for AtSDT and AtSHT were refined to Rwork and Rfree of 0.19/0.24 and 0.19/0.25, respectively (Table 1).
Molecular Docking Studies
All molecular docking studies were performed using Autodock4.2 package (Morris et al., 2009). Briefly, the crystal structure of AtSHT or AtSDT was docked with their potential products (CoA-HS for AtSHT/AtSDT when docking with their final products or intermediate products). The molecule was added with non-polar hydrogens and assigned partial atomic charges using AutoDockTools (ADT; Morris et al., 2009). The coordinates of feruloyl-CoA and spermidine in AtSHT structure were generated based on the coordinates of p-coumaroyl shikimate from the crystal structure of AtHCT (PDB 5KJT) and of HS-CoA from the crystal structure of AtSHT (PDB 6LPV) in combination with the CORINA Classic online service. A grid box with 40 × 40 × 40 grid points and 0.2 Å grid spacing centered roughly at the feruloyl-CoA binding position was used as the searching space. 100 runs of Larmarckian Genetic Algorithm were performed to search the protein-ligand interactions. The results were clustered and ranked. Result analyses and figure rendering were performed using PyMOL.
Sequence Similarity Network and Phylogenetic Analysis
The sequence datasets of the BAHD family were gathered by using AtSDT, AtSHT, and AtHCT as searching the template for Blast e-value cut-off at 1 × 10−25. All the sequences were filtered by a redundancy check and a conserve motif search. The final number of 12,768 non-redundant BAHD proteins were further used to generate the sequence similarity network (SSN) by using Pythoscape (Barber and Babbitt, 2012) and visualized at e-value cut-off 1 × 10–51.5 in Cytoscape. Two hundred and twenty-seven protein sequences from each of the clusters in the SSN, including AtSCT, AtSHT, AtHCT, AtSDT, were used to generate the neighbor-joining tree by MEGA8 software (Tamura et al., 2013) and draw the final maps using the iTOL online software.
Data Availability
The crystal structures of AtSHT and AtSDT have been deposited into Protein Data Bank under accession numbers 6LPV and 6LPW. GenBank accession numbers: AT5G07080 for OAO95042.1 and AT3G47170 for NP_190301.2.
Results
Structural and Biochemical Characterization of AtSHT Enzyme
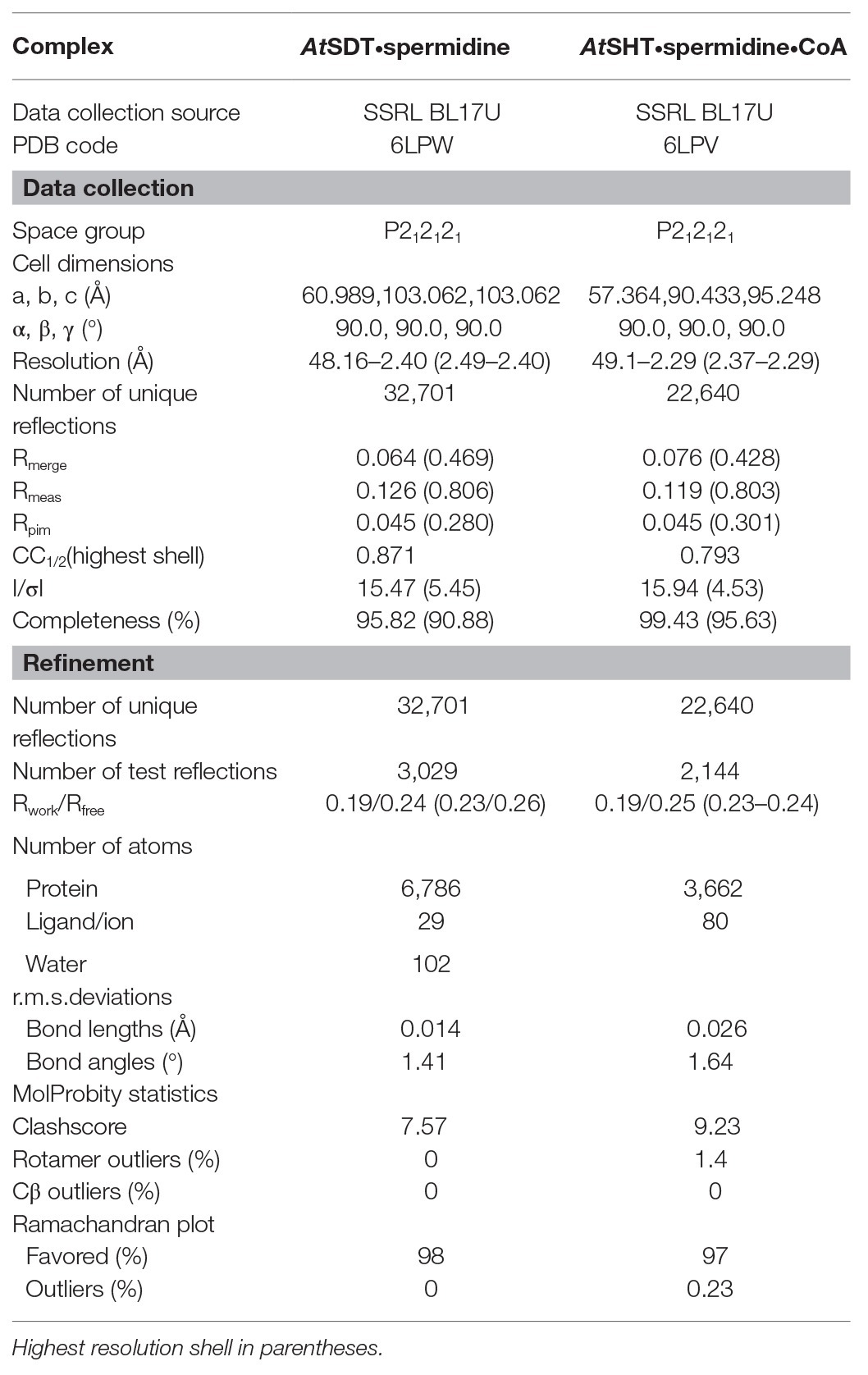
BAHD family acyltransferases, AtSHT and AtSDT, both catalyze the spermidine acylation. AtSHT uses hydroxycinnamoyl CoAs, including caffeoyl/feruoyl/p-coumaroyl/sinapoyl-CoA as acyl donors to fully substitute the N1, N5, and N10 positions of spermidine (Grienenberger et al., 2009), while AtSDT uses caffeoyl/feruoyl/sinapoyl-CoA with the N1 and N10 positions of spermidine (Figure 1A; Supplementary Figure 1; Luo et al., 2009). Despite the similar substrates’ specificity, they differ greatly in sequence identity (only sharing 22.2%) and the final products’ acyl-group substitution sites (Supplementary Figures 2–4). In addition, the expression pattern and distribution of these two enzymes in Arabidopsis thaliana are different (Grienenberger et al., 2009; Luo et al., 2009; Dong et al., 2015; Elejalde-Palmett et al., 2015; Delporte et al., 2018). All of this suggests that AtSHT and AtSDT may gain similar enzyme activities through a possible convergent-evolutionary pathway. To elucidate the molecular basis underlying differences in the acceptor-molecule specificity and possible molecular multisite acylation mechanisms of AtSHT and AtSDT, we expressed the AtSHT and AtSDT enzymes, and determined the crystal structure of AtSHT in complex CoA-HS at 2.2 Å resolution, as well as the crystal structure of AtSDT in the apo form at 2.4 Å resolution. Statistics of data collection and model refinement are summarized in Table 1. The overall structures of both AtSHT and AtSDT are similar to known BAHD family acyltransferases. The structures consist of two pseudo-symmetric N-terminal and C-terminal domains that are connected by a long loop. The N-terminal (residues 1–173 and 391–411) and C-terminal (residues 230–390 and 412–451) domains of AtSHT feature a similar spatial arrangement with a β-sheet core flanked by α-helices (Figure 1B). The active site is located at the interface of the two domains with a residue His155 in between. The extra density near His155 was determined as CoA-HS (Figure 1B). The 3'-phosphoadenosine group of CoA-HS binds with residues Arg246, Arg263, Ser387, and Thr390 in the structure, while residues Glu265, Thr262, and Arg298 interact with the diphosphate group of CoA-HS (Supplementary Figure 5), and these residues are conserved in both AtSDT and AtSHT (Supplementary Figure 4). To view the relative positions of the acyl acceptor and donor, we modeled the potential acyl donor feruloyl-CoA and the acyl-acceptor spermidine into the AtSHT structure by using AtHCT/p-coumaroyl-CoA (PDB ID 5KJT) and AtHCT/p-coumaroylshikimate (PDB ID 5KJU) structures as references (Figure 1C).
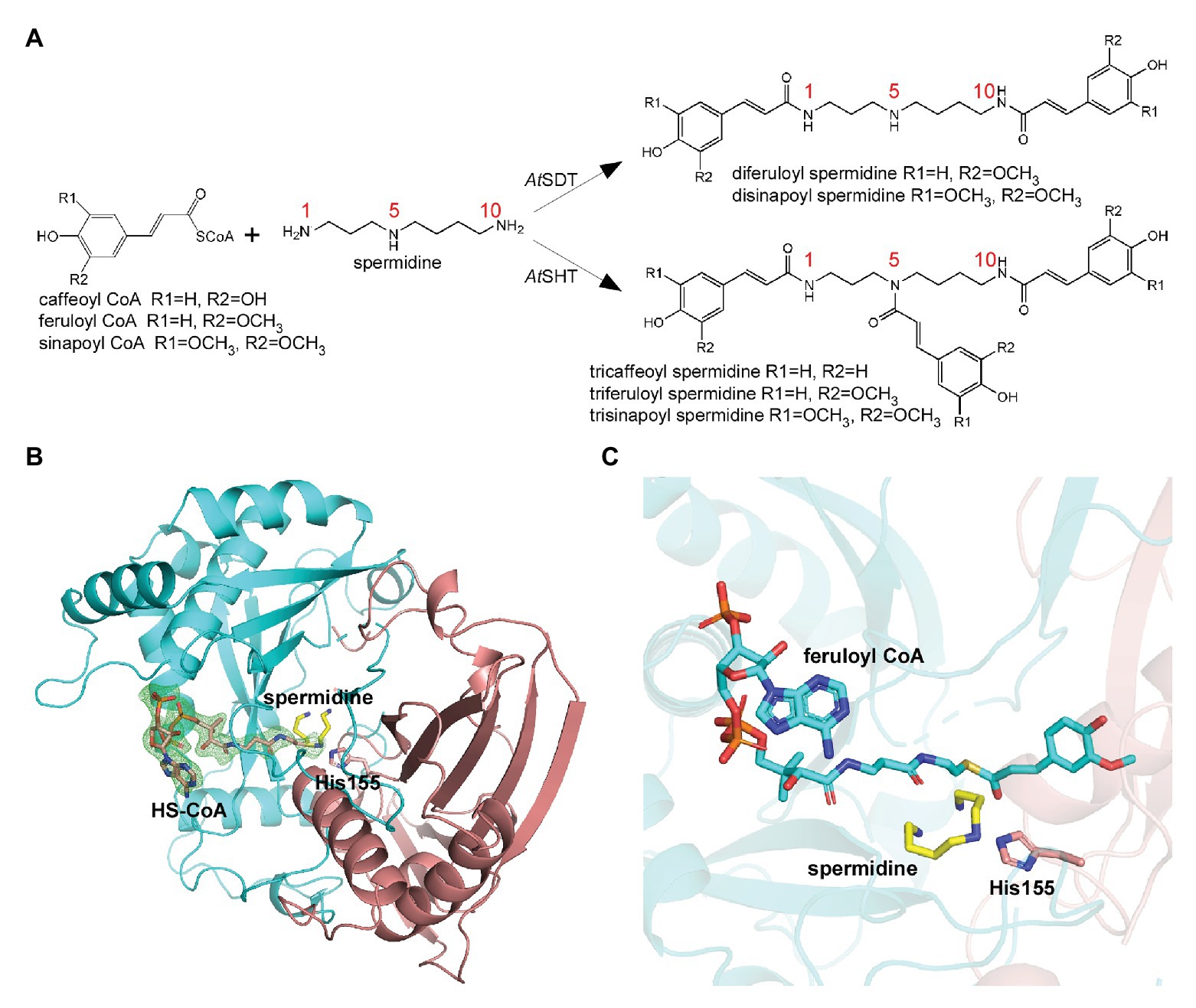
Figure 1. Overall structure of AtSHT. (A) Spermidine conjugates by AtSHT and AtSDT in Arabidopsis. AtSDT catalyzes the di- acylation of spermidine N1 and N10, while AtSHT catalyzes the tri- acylation of spermidine N1, N5, and N10. (B) Overall structure of AtSHT bound with spermidine and HS-CoA. Residues 1–201 are colored red and residues 202–451 are colored cyan. Spermidine and HS-CoA are shown with yellow and pink stick models, respectively. The conserved catalytic residue His155 is also shown with sticks; the Fo-Fc electron density omits the map for HS-CoA (contoured at 2.5σ), is shown with green mesh. (C) The modeled structure of AtSHT showing the binding site of feruloyl-CoA and spermidine. The feruloyl CoA is shown with cyan sticks.
In the modeled structure, several hydrophobic residues were found to surround the feruloyl head, which could serve to improve the affinity-to-fit the phenolic acid group and is consistent with the previous study (Levsh et al., 2016; Supplementary Figure 6). Interestingly, we observed a possible electron density in the modeled position of spermidine from the AtSHT map. The residues around the position form a C-shape channel, which is highly negative charged (Figures 2A,B). We further used spermidine and N1, N5,N10-trihydroxyferuloyl spermidine as ligands to dock in the position, and the results shows both of them fit well in the channel (Supplementary Figure 7). Based on our docking results, spermidine seems to be able to adopt three differently rational conformations that each fits the electron density, and interactions with the surrounding protein residues make a head-to-tail “C” shape (Figures 2A,B). In all three possible conformations, spermidine can form hydrogen bonds with Asp314, Thr33, and the conserved catalytic residue His155. In addition, residues Asp416 and His411, as well as a water molecule, form a hydrogen-bonding network stabilizing spermidine; while residues Cys292, Gly290, Thr312, Val386, and Ile37 further stabilize spermidine via van der Waals interactions (Figures 2A,B); all of these interaction residues form a possible spermidine binding pocket in AtSHT.
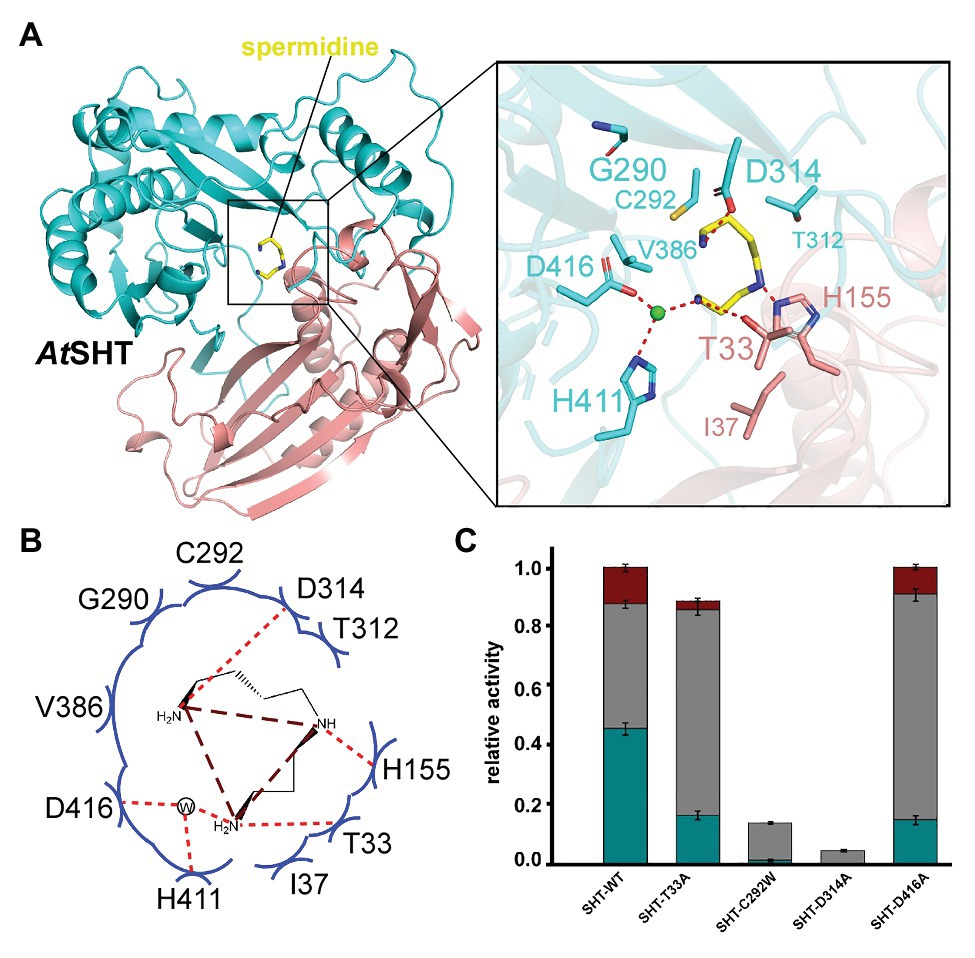
Figure 2. The acyl acceptor-spermidine-binding site of AtSHT. (A) Overall structure of AtSHT bound with spermidine. Zoom-in view shows the residues interacting with spermidine. Color coding is the same as in Figure 1B. The light green sphere indicates a water molecule. (B) Summary of residue interactions with spermidine. Red dashed lines indicate H-bonds; blue arcs indicate van der Waals interactions. (C) A histogram showing the relative activity of wide-type and mutant AtSHTs. The mono-, di-, and tri- acylated products are shown with dark-cyan, gray, and dark-red colors, respectively. Values are means ± S.D., and error bars indicate the S.D. for three biological replicates.
To verify this potential spermidine binding pocket in the AtSHT structure, we mutated the relevant residues within the spermidine binding pockets of AtSHT to assess the effects on enzyme activity and reaction products. The experimental results clearly showed that wild-type AtSHT can efficiently convert spermidine and feruloyl-CoA to monoferuloyl spermidine (46%), diferuloyl spermidine (42%), and triferuloyl spermidine (12%; Figure 2C; Supplementary Figure 2A). AtSHT mutation D314A absolutely abolishes the hydrogen-bonding interaction with the amino group of spermidine, and mutation C292W disrupts the binding of spermidine, thereby resulting in substantially decreased AtSHT enzymatic activity (Figure 2C). In addition, the T33A mutation destabilizes the binding of spermidine and impairs the enzyme activity (fully substituted spermidine was reduced from 12 to 3%), while D416A had less effect on the enzymes as it interacts with spermidine via a water molecule. These results confirmed the potential interactions within the proposed spermidine binding pocket of the AtSHT crystal structure (Figure 2).
Structural and Biochemical Characterization of AtSDT Enzyme
AtSDT adopts an overall structure that is similar to AtSHT (RMSD = 1.68 Å), which also contains two pseudo-symmetric N-terminal (residues 1–187 and 384–407) and C-terminal (residues 233–383 and 408–451) domains that are connected by a long loop (residues 188–232; Figure 3A). No extra electron density for the acyl donor or the reaction product CoA-HS was seen in the AtSDT map (in contrast with AtSHT), which may be due to the crystal packing since the crystallography symmetry related AtSDT molecule blocks the entry binding tunnel, thereby preventing acyl donor and putative reaction product entry. Unexpectedly, after superimposed with the AtSHT structure, we observed a possible electron density for spermidine in the corresponding modeled acyl-acceptor position from the AtSDT map. The residues around the position form a linear-shape channel, which is highly negative charged (Figures 3A,B). We further used spermidine, N1,N10-dihydroxysinapoyl spermidine, N1,N5,N10-trihydroxysinapoyl spermidine as ligands to dock in the position and the results show spermidine and N1,N10-dihydroxysinapoyl spermdine fit well in the channel but crash with N1,N5,N10-trihydroxysinapoyl spermidine (Supplementary Figures 7B,C). According to the docking results, the spermidine in AtSDT seems be able to adopt a linear rational conformation to fit that electron density and forms a hydrogen bond through its τ-nitrogen with the conserved catalytic residue His169. This spatial arrangement is consistent with the role of His169 as a general base that deprotonates the acyl-acceptor spermidine, priming it for the nucleophilic attack of the carbonyl carbon of the hydroxycinnamoyl-CoA acyl donor. Specificity-determining residues, Asp316 and Ser294, form hydrogen bonds with the N1 and N5 groups of spermidine. In addition, Tyr47, Trp381, and Thr379 form hydrogen bonds through a water molecule with the spermidine N10 group. These interactions stabilize the binding of spermidine. Furthermore, N-terminal domain residues Tyr314, Tyr318, Cys377, Glu354, Thr358, and Gly292, and C-terminal domain residues, Asp40 and Asn43, contact spermidine via van der Waals interactions (Figures 3A,B). All of these surrounding residues will also form a possible spermidine binding pocket in AtSDT, and spermidine may adopt a totally different conformation from that in AtSHT (Figures 3A,B).
Figure 3. The acyl acceptor spermidine-binding site of AtSDT. (A) Overall structure of AtSDT bound with spermidine. Residues 1–200 are colored light orange, residues 201–449 are colored light blue; spermidine is shown as a magenta stick model. Zoom-in view shows the residues interacting with spermidine. The light green sphere indicates a water molecule. (B) Summary of residue interactions with spermidine. Red dashed lines indicate H-bonds; blue arcs indicate van der Waals interactions. (C) A histogram showing the relative activity of wide-type and mutant AtSDTs. The mono- and di- acylated products are shown with dark cyan and gray colors, respectively. Values are means ± S.D., and error bars indicate the S.D. for three biological replicates.
To confirm this potential spermidine binding pocket in the AtSDT structure, we mutated the relevant residues within the spermidine-binding pockets of AtSDT to assess the effects on enzyme activity and reaction products. The results showed that wild-type AtSDT can efficiently couple spermidine with sinapoyl CoA to yield monosinapoyl (2%) spermidine and disinapoyl spermidine (98%; Figure 3C; Supplementary Figure 3). Meanwhile, eight single or double AtSDT mutants that potentially affect the binding of spermidine were generated. The results showed that the D316A mutation absolutely abolishes the hydrogen bonding interaction with the N5 group of spermidine, and mutant S294W not only disrupts the potential hydrogen bonding interaction with the N1 group, but also prevents spermidine binding, thereby substantially decreasing AtSDT enzymatic activities. The S294I mutation impairs the enzyme activity (compared with wild-type AtSDT, the production of disinapoyl spermidine was reduced from 98 to 63%), suggesting that disruption of the hydrogen bond interaction destabilizes spermidine binding. In addition, the Y47A and W381A mutations seem to have a greater effect on the enzyme activity than the S294I mutation (the product disinapoyl spermidine was reduced from 98 to 28% or 49%, respectively), as these two residues also form the hydrogen bond interactions with spermidine via a water molecule, and these results indicated that the hydrogen bond interaction between S294 has less effect on the enzyme activity than that of Y47 or W384 and spermidine, while the N43A and Y314A mutations disrupt the van der Waals interactions with spermidine, slightly impairing enzyme activities (Figure 3C). Furthermore, the double mutant E354A/T358A, which changes residues located at the predicted spermidine entry channel in the AtSDT structure (Supplementary Figure 8A), showed decreased activity compared with wild-type AtSDT (Figure 3C). Interestingly, these two residues are also close to a loop (Met364-Leu375 in AtSDT) which is unstable compared with other parts of the structure and shows a huge movement when aligned with AtSHT and AtHCT structures (Supplementary Figures 8B,C). As the loop is located near the active center, its movement might lead the active center open/close to the outside environment, which thus may help in maintaining the catalytic environment when it is in a close state or releasing the products when it is an open state. Since its proposed function is like a lid, we here named this loop as “lid-loop.”
These results are fully consistent with the interactions revealed from the modeled structures and suggest that key interactions from the proposed spermidine-binding pockets of AtSDT and AtSHT are critical for acyl acceptor substrate recognition and binding.
Comparison on Spermidine Binding Pockets of AtSDT and AtSHT
Guided by structural information of AtSDT and AtSHT, we performed structure-based sequence alignments by using AtSHT, AtSDT, and other representative BAHD-transferase family homologues (Supplementary Figure 4) and found that residues in the proposed acyl-acceptor spermidine-binding pocket of AtSDT were conserved with SDT homologues and were variable from SHT homologues (Supplementary Figure 4, shown as blue stars). Meanwhile, the residues from the AtSHT acyl-acceptor spermidine-binding pocket were conserved among SHT homologues and were variable in SDT homologues (Supplementary Figure 4, shown as green circles). These findings further confirmed that different evolution strategies may be adopted for acyl acceptor spermidine binding in AtSHT and AtSDT.
In addition, we also compared the proposed acyl-acceptor binding pocket of AtSHT and AtSDT with that of AtHCT. Although the overall structures of AtSHT and AtSDT are very similar to that of AtHCT (the RMSDs of AtHCT with AtSHT and AtSDT are 1.70 Å and 2.03 Å, respectively; Figure 4A), the interior features and the electrostatic potential of the acyl acceptor binding pockets of AtSHT and AtSDT differ significantly from those of AtHCT. The formers are composed of negatively-charged residues to accommodate spermidine, while the latter is mainly comprised of positively-charged residues suitable for attracting and binding shikimate (Figure 4B). This suggests that it may be possible for us to predict and evaluate the acyl-acceptor substrate preference of unknown BAHD transferase family proteins based on the charge distributions of the residues comprising the acyl acceptor binding pocket.
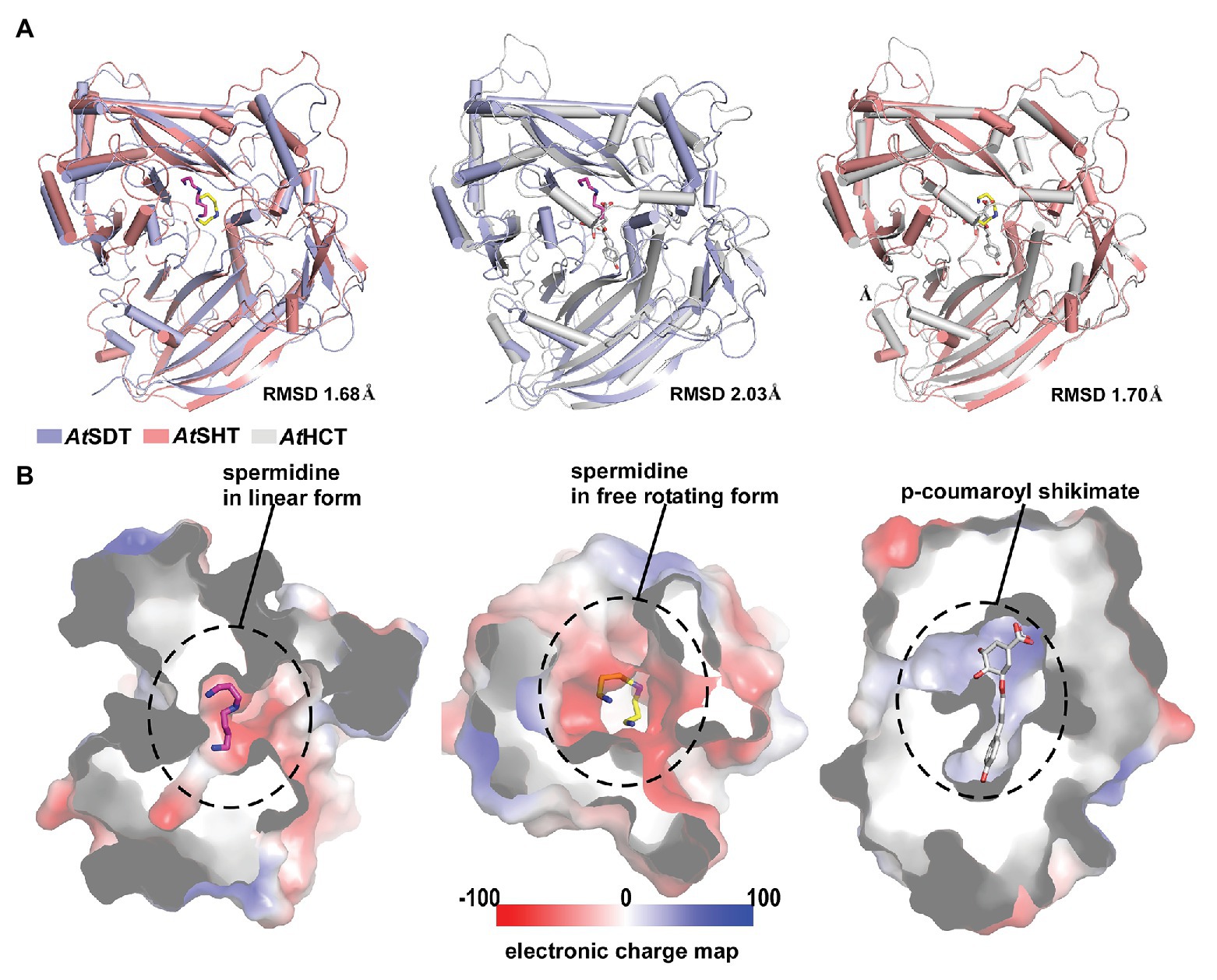
Figure 4. Overall structure and acyl-acceptor binding site comparison between AtSDT, AtSHT, and AtHCT. (A) Structure superimpositions of AtSDT and AtSHT (left), AtSDT, and AtSHCT (middle), and AtSHT and AtHCT(right). Structures of AtSDT, AtSHT, and AtHCT are shown with ribbons colored slate-blue, pink, and gray, respectively. Their respective acyl-acceptors are shown as stick models colored magenta, yellow, and gray. (B) Electron static surface models of AtSDT(left), AtSHT(middle) and AtHCT(right) show the charge distributions in their acyl-acceptor binding sites.
Prediction of Unknown BAHD Family Proteins
With the structural information (especially the acyl-acceptor interaction residues’ distributions) from AtSDT and AtSHT, we expanded our analysis on other unknown BAHD-family proteins. A total number of 12,768 non-redundant protein sequences from the National Center for Biotechnology Information (NCBI) protein database was identified, including 49 A. thaliana BAHD transferase proteins (Dataset 1). Since the SSN can not only can display the same topology compared with phylogenetic tree (Davidson et al., 2018; Burroughs et al., 2019), but also have an advantage dealing with a large sequence dataset and a better global overview, we applied the SSN analysis in studying the BAHD-protein family sequences. The SSN map was generated using Pythoscape (Barber and Babbitt, 2012), where nodes represent sequences, and edges represent pairwise local alignments with e-values cut-off of 1e-51.5 (Supplementary Figure 9). Forty-nine A. thaliana BAHD transferase proteins were assigned to the SSN map where 18 functionally characterized sequences (Supplementary Table S1) were marked as purple triangle and 31 functionally unknown sequences were marked as blue circles (Supplementary Figure 9). The AtSDT, AtSCT, AtSHT, and AtHCT proteins were also highlighted in this map. Interestingly, AtSDT and AtSHT were present in two different clusters in the SSN map. The AtSDT cluster contains 62 nodes, including 437 sequences in which AtSCT, the spermidine dicoumaroyl transferase, is also present (sequence identity between AtSDT and AtSCT is 52.5%). The AtSHT belongs to a more complicated cluster which contains several sub-clusters. The AtSHT sub-cluster contains 96 nodes including 592 sequences and MdSHT, and CiSHT are in the same node with AtSHT. The sub-cluster containing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase (HCT) connected with AtSHT sub-cluster showing the close relationship with AtSHT. Indeed, the sequence of AtHCT in this sub-cluster shows higher sequence identity with AtSHT (sequence identity 36.4%) rather than with AtSDT (sequence identity 22.2%) or AtSCT (sequence identity 20.5%). To our surprise, there are two uncharacterized A. thaliana BAHD proteins (OAO95042.1 and NP_190301.2) in the same cluster with AtSDT and AtSCT. We then further evaluate their acyl-acceptor substrate preference on the basis of the charge distributions of the residues comprising the acyl-acceptor binding pocket. The structures of OAO95042.1 and NP_190301.2 were modeled using the AtSDT structure as a template through the online Swiss-modeling program (Arnold et al., 2006), and negatively charged acyl-acceptor substrate binding pockets were found (Supplementary Figure 11), suggesting a basic substrate with characteristics similar to polyamines. For verification, the corresponding genes, AT5G07080 and AT3G47170 for OAO95042.1 and NP_190301.2, respectively, were cloned from A. thaliana and recombinant proteins were expressed in E. coli. The potential activity was tested using caffeoyl/feruloyl/sinapoyl-CoA as acyl donors and positively charged molecules putrescine/spermidine/spermine as acyl acceptors. The HPLC-MS results showed that both AT5G07080 and AT3G47170 encoded enzymes that could efficiently convert putrescine and caffeoyl-CoA to di-caffeoyl putrescine (Figures 5, 6 and Supplementary Figure 12). Further, OAO95042.1 enzyme can even convert spermidine/spermine and feruloyl CoA to mono-feruloyl spermidine/spermine (Figure 6 and Supplementary Figure 12). Results also suggested that the enzyme encoded by AT5G07080 has a preference for feruloyl-CoA binding, but little acyl-acceptor specificity, while the enzyme encoded by AT3G47170 has a preference for caffeoyl CoA and putrescine.
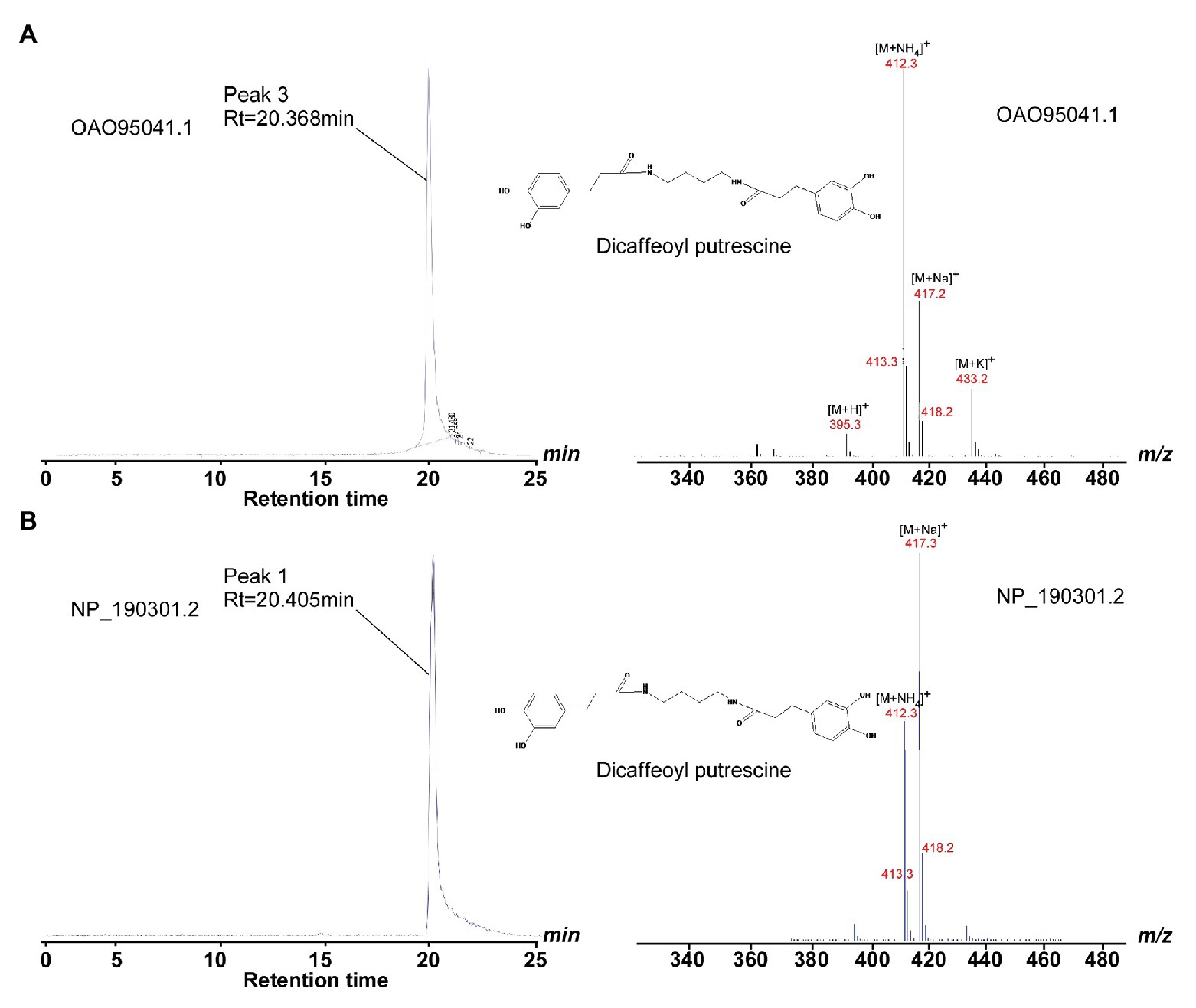
Figure 5. HPLC and LC/MS profiles show the catalytic activity of OAO95042.1 and NP_190301.2. (A) Left panel, HPLC profile of methanolic extracts showing products of wild-type OAO95042.1 reaction with caffeoyl-CoA and putrescine substrates; right panel, LC/MS fragmentation of di-caffeoyl putrescine (SIM mode, m/z 412). (B) Left panel, HPLC profile of methanolic extracts showing products of wild-type NP_190301.2 reaction with caffeoyl CoA and putrescine substrates; right panel, LC/MS fragmentation of di-caffeoyl putrescine (SIM mode, m/z 412). The di-caffeoyl putrescine (MW 412) perhaps easily formed the dehydration product (MW 394) in the reaction or detection conditions, and the peaks of 395.3, 412.3, 417.3, and 433.2 are [M+ H]+, [M+ NH4]+, [M + Na]+, and [M + K]+ of its dehydration products, respectively.
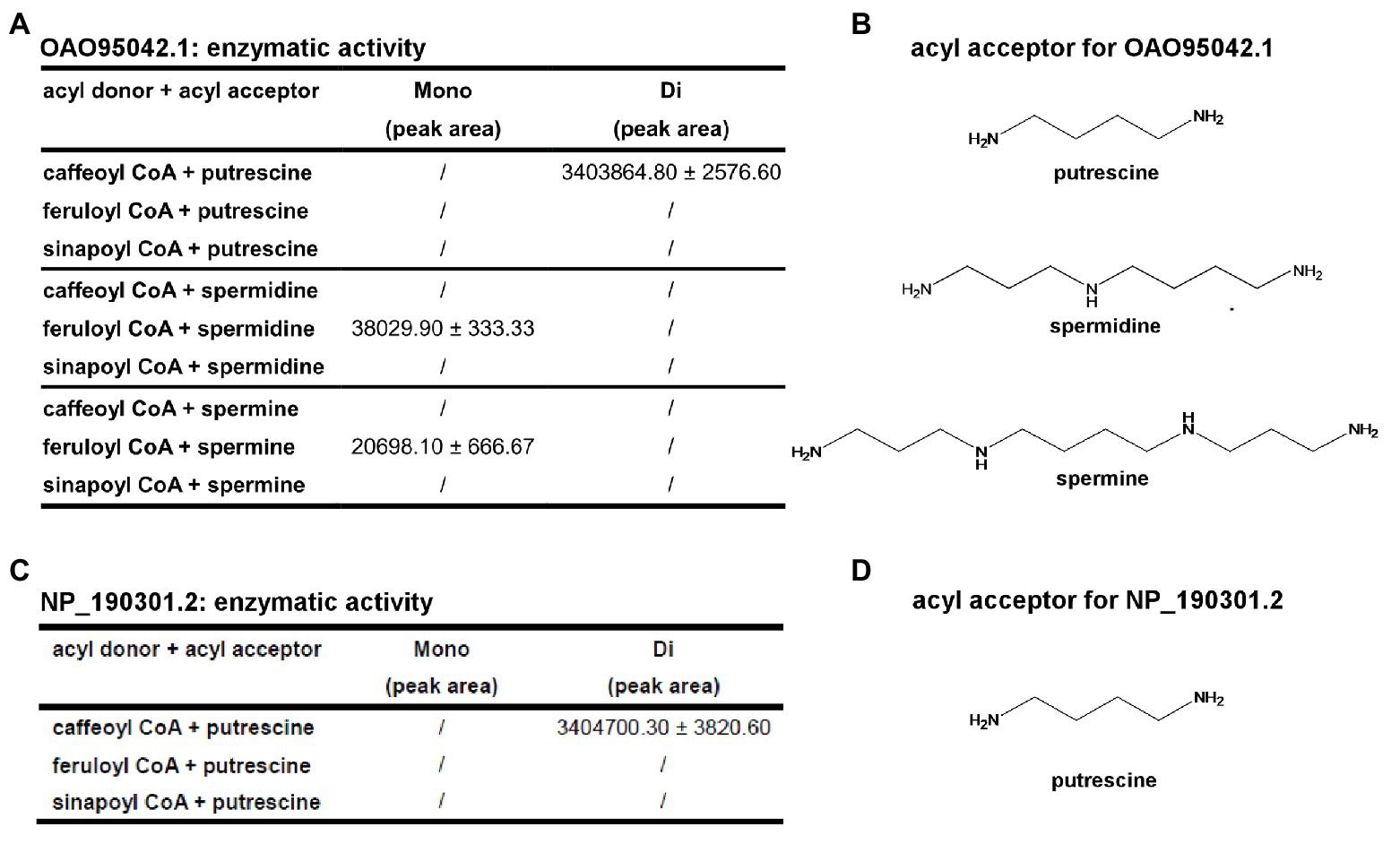
Figure 6. Summary of the catalytic activity of OAO95042.1 and NP_190301.2 with different acyl donors and acceptors. (A) Identification and quantification of products from different acyl acceptors and acyl donors for OAO95042.1(AT5G07080.1). (B) Acyl acceptors utilized by OAO95042.1. (C) Identification and quantification of products from different acyl acceptors and acyl donors for NP_190301.2(AT3G47170). (D) Acyl acceptor utilized by NP_190301.2. Values are means ± S.D. and error bars indicate the S.D. for three biological replicates.
The successful prediction on substrate specificity of AT5G07080- and AT3G47170 encoded BAHD enzymes demonstrated the feasibility of predicting and identifying possible substrates for uncharacterized BAHD-family transferases on the basis of the charge distributions of the residues comprising the acyl-acceptor binding pocket. It would provide great possibilities for us to gain a better understanding of the functionally uncharacterized BAHD-family proteins, and the potential strategy to predict substrates would be very useful for uncharacterized plant metabolite enzymes.
Discussion
Phenolamides are ubiquitous secondary metabolites in plants. They are produced by BAHD-family acyltransferases that mono-conjugation of aromatic monoamines or poly-conjugation of aliphatic polyamines with phenolic acids. More and more research findings have highlighted the importance of phenolamides in diverse plant physiological processes, including defense responses and development. However, BAHD-family acyltransferases usually only share low sequence similarities among them, which makes it extremely challenging to predict the substrate specificity of uncharacterized BAHD family enzymes from their amino acid sequences. To solve this remaining question, our work here provides a new feasible methodology for the substrate specificity prediction of unknown BAHD family transferases in plants. The phylogenetic tree has been used for a long time on the function analysis of the unknown genes; however, with the limitation of gene numbers, the results are less reliable and irreproducible (Shen et al., 2020). By using the SSN, we could use all the information from the BAHD family sequences and have a global view on the classification of each gene, which would greatly accelerate the discovery of target genes. By combining the SSN analysis and structural information, we further clarified the potential target and validated them by more expensive in vitro experiment. Just based on this important finding, we successfully predicted the possible substrates of two uncharacterized BAHD-family enzyme OAO95042.1 and NP_190301.2, which turned out encoding the putrescine hydroxycinnamoyl transferases.
Our study also highlights the potential convergent-evolutionary way of AtSDT and AtSHT genes. Although both AtSDT and AtSHT use spermidine as an acyl-acceptor, the expression pattern and distribution of these two enzymes are different (Grienenberger et al., 2009; Luo et al., 2009). AtSHT were mainly expressed in the tapetum of Arabidopsis anthers and synthiszedsynthesized fully substituted products. The other enzymes that synthesize the fully substituted products also have the similar expression pattern, such as CiSHT, which promotes tetrahydroxycinnamoyl spermine accumulation in the pollen coat of the Asteraceae family, and MdSHT which synthesizes the trihydroxycinnamoyl spermidines in the pollen coat of core Eudicotyledons. Disrupting the function of AtSHT would lead to abnormal formation of pollengrains in the sht mutant of Arabidopsis, indicating the probable function of trihydroxycinnamoyl spermidine derivatives in sporopollenin ultrastructure that the fully substituted products may provide for a barrier for pollen or may function as a supporting structure. Interestingly, according to our SSN map and phylogenetic tree, all of all these SHTs genes are close to HCTs, which are the key enzymes in lignin metabolism, and that both HCTs and SHTs may evolute from a same ancestor. On the other hand, the AtSDT is mainly expressed in the seed and the root of Arabidopsis and synthesizes the mono- or di- substituted phenolamides. Unlike the SHT, the enzymes that synthesis the mono- or di- substituted phenolamides seem to be widely distributed in different organs and functions as plant biotic or abiotic stress responses. In our SSN map and phylogenetic tree, AtSDT is far away from HCT, but close to our newly discovered putrescine transferases OAO95042.1 and NP_190301.2. Furthermore, according to our structures, residues in the proposed acyl-acceptor spermidine binding pocket of AtSDT were conserved with SDT homologues and were variable from SHT homologues (Supplementary Figure 4, shown as blue stars). Meanwhile, the residues from the AtSHT acyl acceptor spermidine-binding pocket were conserved among SHT homologues and were variable in SDT homologues (Supplementary Figure 4, shown as green circles). Taken together, we suggest that the AtSHT and AtSDT may undergo the convergent-evolutionary way and thus gain similar spermidine transferase activity.
The molecular mechanisms of multisite acylation of BAHD-family acyltransferases remain poorly understood so far. In this study, we tried to answer this tough question by determining the crystal structures of AtSHT and AtSDT, two BAHD family members catalyzing the multisite acylation of spermidine, and but showing different product profiles in Arabidopsis thaliana. We closely compared the differences in their potential spermidine binding pockets. The possible electron density shape for spermidine in the AtSHT structure and our molecular docking results suggests that it may adopt a freely-rotating conformation in the center of the binding pocket (Supplementary Figure 13A) by interacting with residues Thr33, Asp314, Asp416, and His155, establishing equal probabilities for acylation of N1, N5, and N10 atoms in spermidine. In contrast, the possible electron density shape for spermidine in the AtSDT structure, in combination with the molecular docking results, suggests a linear conformation at the center of the binding pocket (Supplementary Figure 13B). Therefore, spermidine in AtSDT could only be docked into the binding pocket in two different orientations (“N1 to N10” or “N10 to N1”). In view of these spermidine conformation differences in AtSHT and AtSDT, we propose a “linear/rotation” model here, which may be able to clarify the potential mechanism of the different acylation activities of AtSHT and AtSDT (Supplementary Figure 14). The full acylation activity of AtSHT is enabled by the “freely-rotating” conformation adopted by the acyl-acceptor spermidine in the binding pocket, while AtSDT only binds in a linear conformation that is limited to the “head-tail” acylation (Supplementary Figure 14). That is, the acyl-acceptor spermidine adopts a free-rotating conformation in AtSHT and can undergo mono-, di- or tri-acylation; while the spermidine molecule in AtSDT adopts a linear conformation, which only allows mono- or di-acylation to take place. Our biochemical results all support this proposal that changing the spermidine binding pattern will decrease or abolish the production of fully acylated products, and thus match our proposed “linear/rotation” model (Supplementary Figure 14). Meanwhile, by superpositioning AtSDT and AtSHT structure with AtHCT, we observe a potential movement of the “lid-loop,” which is located near the active center and may function in the maintenance of catalytic environments and the release of products (Supplementary Figure 8).
In summary, our extensive structural and biochemical analyses on AtSDT and AtSHT in this study provides an excellent starting point for predicting the biochemical functions of uncharacterized BAHD-family enzymes and understanding multisite acylation in BAHD-family enzymes. However, to further elucidate the potential molecular mechanism underlying the differing acylation activities of AtSHT and AtSDT, crystal structures of AtSDT and AtSHT in complex with its acyl donor and acceptor are still anticipated in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: http://www.wwpdb.org/, 6LPV and 6LPW.
Author Contributions
CW, JL, and WL designed experiments. CW and JL performed the bulk of the experiments. MM contributed to protein expression, purification, and crystallization. ZL and WH contributed to enzymatic assay experiments. PZ, CW, and WL analyzed the data and wrote the manuscript. PZ conceived the project. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Key R&D Program of China (2018YFA0900600), the National Natural Science Foundation of China (31870720, 81903526, 31700263, 81991523, and 82072240), the Strategic Priority Research Program of CAS (XDB27020103), grants from the Shanghai Science and Technology Commission (19XD1424500), and grants from Jiangsu Province of China (BK20190798 to WL). WL is supported by Jiangsu Specially-appointed Professor Talent Program. JL is supported by the Foundation of Youth Innovation Promotion Association of the Chinese Academy of Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff at BL19U1/BL17U1-SSRF for technical assistance in diffraction data collection, and the staff Dr. Shanshan Wang at the core facility center of Institute of Plant Physiology and Ecology for X-ray diffraction analysis and Mass Spectrometry analysis. We also thank Dr. Jie Luo from Huazhong Agriculture University for providing us the AtSHT, AtSDT, and AtSCT genes.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.610118/full#supplementary-material
References
Adams, P., Afonine, P., Bunkóczi, G., Chen, V., Davis, I., Echols, N., et al. (2010). PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. doi: 10.1107/S0907444909052925
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: a web-based enviroment for protein structure homology modelling. Bioinformatics 22, 195–201. doi: 10.1093/bioinformatics/bti770
Barber, A. E. 2nd, and Babbitt, P. C. (2012). Pythoscape: a framework for generation of large protein similarity networks. Bioinformatics 28, 2845–2846. doi: 10.1093/bioinformatics/bts532
Burhenne, K., Kristensen, B. K., and Rasmussen, S. K. (2003). A new class of N-hydroxycinnamoyltransferases. Purification, cloning, and expression of a barley agmatine coumaroyltransferase (EC 2.3.1.64). J. Biol. Chem. 278, 13919–13927. doi: 10.1074/jbc.M213041200
Burroughs, A. M., Glasner, M. E., Barry, K. P., Taylor, E. A., and Aravind, L. (2019). Oxidative opening of the aromatic ring: tracing the natural history of a large superfamily of dioxygenase domains and their relatives. J. Biol. Chem. 294, 10211–10235. doi: 10.1074/jbc.RA119.007595
Chiang, Y. C., Levsh, O., Lam, C. K., Weng, J. K., and Wang, Y. (2018). Structural and dynamic basis of substrate permissiveness in hydroxycinnamoyltransferase (HCT). PLoS Comput. Biol. 14:e1006511. doi: 10.1371/journal.pcbi.1006511
Cho, M. H., and Lee, S. W. (2015). Phenolic Phytoalexins in Rice: biological functions and biosynthesis. Int. J. Mol. Sci. 16, 29120–29133. doi: 10.3390/ijms161226152
Collaborative Computational Project Number 4 (1994). The CCP4 suite: programs for protein crystallography. Acta Cryst. D. 54, 760–763. doi: 10.1107/S0907444994003112
D’Auria, J. C. (2006). Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9, 331–340. doi: 10.1016/j.pbi.2006.03.016
Davidson, R., Baas, B. -J., Akiva, E., Holliday, G. L., Polacco, B. J., Levieux, J. A., et al. (2018). A global view of structure-function relationships in the tautomerase superfamily. J. Biol. Chem. 293, 2342–2357. doi: 10.1074/jbc.M117.815340
Delporte, M., Bernard, G., Legrand, G., Hielscher, B., Lanoue, A., Molinie, R., et al. (2018). A BAHD neofunctionalization promotes tetrahydroxycinnamoyl spermine accumulation in the pollen coat of the Asteraceae family. J. Exp. Bot. 69, 5355–5371. doi: 10.1093/jxb/ery320
Dong, X., Gao, Y., Chen, W., Wang, W., Gong, L., Liu, X., et al. (2015). Spatiotemporal distribution of phenolamides and the genetics of natural variation of hydroxycinnamoyl spermidine in rice. Mol. Plant 8, 111–121. doi: 10.1016/j.molp.2014.11.003
Edreva, A. M., Velikova, V. B., and Tsonev, T. D. (2007). Phenylamides in plants. Russ. J. Plant Physl. 54, 287–301. doi: 10.1134/S1021443707030016
Elejalde-Palmett, C., de Bernonville, T. D., Glevarec, G., Pichon, O., Papon, N., Courdavault, V., et al. (2015). Characterization of a spermidine hydroxycinnamoyltransferase in Malus domestica highlights the evolutionary conservation of trihydroxycinnamoyl spermidines in pollen coat of core Eudicotyledons. J. Exp. Bot. 66, 7271–7285. doi: 10.1093/jxb/erv423
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66:486. doi: 10.1107/S0907444910007493
Galis, I., Simek, P., Narisawa, T., Sasaki, M., Horiguchi, T., Fukuda, H., et al. (2006). A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant J. 46, 573–592. doi: 10.1111/j.1365-313X.2006.02719.x
Garvey, G. S., McCormick, S. P., Alexander, N. J., and Rayment, I. (2009). Structural and functional characterization of TRI3 trichothecene 15-o-acetyltransferase from Fusarium sporotrichioides. Protein Sci. 18, 747–761. doi: 10.1002/pro.80
Garvey, G. S., McCormick, S. P., and Rayment, I. (2008). Structural and functional characterization of the TRI101 trichothecene 3-O-acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: kinetic insights to combating Fusarium head blight. J. Biol. Chem. 283, 1660–1669. doi: 10.1074/jbc.M705752200
Grienenberger, E., Besseau, S., Geoffroy, P., Debayle, D., Heintz, D., Lapierre, C., et al. (2009). A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermdines. Plant J. 58, 246–259. doi: 10.1111/j.1365-313X.2008.03773.x
Kaur, H., Heinzel, N., Schottner, M., Baldwin, I. T., and Galis, I. (2010). R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiol. 152, 1731–1747. doi: 10.1104/pp.109.151738
Keller, H., Hohlfeld, H., Wray, V., Hahlbrock, K., Scheel, D., and Strack, D. (1996). Changes in the accumulation of soluble and cell wall-bound phenolics in elicitor-treated cell suspension cultures and fungus-infected leaves of Solanum tuberosum. Phytochemistry 42, 389–396. doi: 10.1016/0031-9422(95)00866-7
Lallemand, L. A., Zubieta, C., Lee, S. G., Wang, Y., Acajjaoui, S., Timmins, J., et al. (2012). A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiol. 160, 249–260. doi: 10.1104/pp.112.202051
Levsh, O., Chiang, Y. C., Tung, C. F., Noel, J. P., Wang, Y., and Weng, J. K. (2016). Dynamic conformational states dictate selectivity toward the native substrate in a substrate-permissive Acyltransferase. Biochemistry 55, 6314–6326. doi: 10.1021/acs.biochem.6b00887
Levsh, O., Pluskal, T., Carballo, V., Mitchell, A. J., and Weng, J. K. (2019). Independent evolution of rosmarinic acid biosynthesis in two sister families under the Lamiids clade of flowering plants. J. Biol. Chem. 294, 15193–15205. doi: 10.1074/jbc.RA119.010454
Luo, J., Fuell, C., Parr, A., Hill, L., Bailey, P., Elliott, K., et al. (2009). A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell 21, 318–333. doi: 10.1105/tpc.108.063511
Ma, X., Koepke, J., Panjikar, S., Fritzsch, G., and Stockigt, J. (2005). Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J. Biol. Chem. 280, 13576–13583. doi: 10.1074/jbc.M414508200
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi: 10.1002/jcc.21256
Muroi, A., Ishihara, A., Tanaka, C., Ishizuka, A., Takabayashi, J., Miyoshi, H., et al. (2009). Accumulation of hydroxycinnamic acid amides induced by pathogen infection and identification of agmatine coumaroyltransferase in Arabidopsis thaliana. Planta 230, 517–527. doi: 10.1007/s00425-009-0960-0
Onkokesung, N., Gaquerel, E., Kotkar, H., Kaur, H., Baldwin, I. T., and Galis, I. (2012). MYB8 controls inducible phenolamide levels by activating three novel hydroxycinnamoyl-coenzyme a: polyamine transferases in Nicotiana attenuata. Plant Physiol. 158, 389–407. doi: 10.1104/pp.111.187229
Otwinowski, Z., and Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Meths. Enzymol. 276, 307–326. doi: 10.1016/S0076-6879(97)76066-X
Park, H. L., Yoo, Y., Hahn, T. R., Bhoo, S. H., Lee, S. W., and Cho, M. H. (2014). Antimicrobial activity of UV-induced phenylamides from rice leaves. Molecules 19, 18139–18151. doi: 10.3390/molecules191118139
Schafer, M., Brutting, C., Xu, S., Ling, Z., Steppuhn, A., Baldwin, I. T., et al. (2017). NaMYB8 regulates distinct, optimally distributed herbivore defense traits. J. Integr. Plant Biol. 59, 844–850. doi: 10.1111/jipb.12593
Semler, U., Schmidtberg, G., and Gross, G. G. (1987). Synthesis of piperoyl coenzyme-a thioester. Z. Naturforsch. C 42, 1070–1074. doi: 10.1515/znc-1987-9-1011
Shen, X. X., Li, Y., Hittinger, C. T., Chen, X. X., and Rokas, A. (2020). An investigation of irreproducibility in maximum likelihood phylogenetic inference. Nat. Commun. 11:6096. doi: 10.1038/s41467-020-20005-6
Stockigt, J., and Zenk, M. H. (1974). Enzymatic synthesis of chlorogenic acid from caffeoyl coenzyme a and quinic acid. FEBS Lett. 42, 131–134. doi: 10.1016/0014-5793(74)80769-6
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanabe, K., Hojo, Y., Shinya, T., and Galis, I. (2016). Molecular evidence for biochemical diversification of phenolamide biosynthesis in rice plants. J. Integr. Plant Biol. 58, 903–913. doi: 10.1111/jipb.12480
Unno, H., Ichimaida, F., Suzuki, H., Takahashi, S., Tanaka, Y., Saito, A., et al. (2007). Structural and mutational studies of anthocyanin malonyltransferases establish the features of BAHD enzyme catalysis. J. Biol. Chem. 282, 15812–15822. doi: 10.1074/jbc.M700638200
Von Ropenack, E., Parr, A., and Schulze-Lefert, P. (1998). Structural analyses and dynamics of soluble and cell wall-bound phenolics in a broad spectrum resistance to the powdery mildew fungus in barley. J. Biol. Chem. 273, 9013–9022. doi: 10.1074/jbc.273.15.9013
Walters, D., Meurer-Grimes, B., and Rovira, I. (2001). Antifungal activity of three spermidine conjugates. FEMS Microbiol. Lett. 201, 255–258. doi: 10.1016/S0378-1097(01)00278-6
Keywords: BAHD family acyltransferases, spermidine, acyl acceptor, phenolamides, multisite-acylation, crystal structure
Citation: Wang C, Li J, Ma M, Lin Z, Hu W, Lin W and Zhang P (2021) Structural and Biochemical Insights Into Two BAHD Acyltransferases (AtSHT and AtSDT) Involved in Phenolamide Biosynthesis. Front. Plant Sci. 11:610118. doi: 10.3389/fpls.2020.610118
Edited by:
Chang-Jun Liu, Brookhaven National Laboratory (DOE), United StatesReviewed by:
Desigan Kumaran, Brookhaven National Laboratory (DOE), United StatesGoro Taguchi, Shinshu University, Japan
Copyright © 2021 Wang, Li, Ma, Lin, Hu, Lin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Zhang, cGVuZ3poYW5nMDFAY2VtcHMuYWMuY24=; Wei Lin, d2VpbGluQG5qdWNtLmVkdS5jbg==
†These authors have contributed equally to this work
‡Present address: Chengyuan Wang, Waksman Institute and Department of Chemistry, Rutgers University, Piscataway, NJ, United States
 Chengyuan Wang
Chengyuan Wang Jianxu Li1†
Jianxu Li1† Wenli Hu
Wenli Hu Wei Lin
Wei Lin Peng Zhang
Peng Zhang