- 1Zhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Department of Horticulture, Zhejiang University, Hangzhou, China
- 2Key Laboratory of Horticultural Plants Growth, Development and Quality Improvement, Agricultural Ministry of China, Hangzhou, China
Autophagy is a highly conserved cellular process for the degradation and recycling of unnecessary cytoplasmic components in eukaryotes. Various studies have shown that autophagy plays a crucial role in plant growth, productivity, and survival. The extensive functions of plant autophagy have been revealed in numerous frontier studies, particularly those regarding growth adjustment, stress tolerance, the identification of related genes, and the involvement of metabolic pathways. However, elucidation of the molecular regulation of plant autophagy, particularly the upstream signaling elements, is still lagging. In this review, we summarize recent progress in research on the molecular mechanisms of autophagy regulation, including the roles of protein kinases, phytohormones, second messengers, and transcriptional and epigenetic control, as well as the relationship between autophagy and the 26S proteasome in model plants and crop species. We also discuss future research directions for the potential application of autophagy in agriculture.
Plain Language Summary
In the last two decades, numerous studies have reported that the autophagy pathway is precisely regulated in plants. We summarize recent progress on the molecular mechanisms of autophagy regulation in plants and discuss future research directions for the potential application of autophagy in agriculture.
Introduction
Autophagy, literally meaning “self-eating,” is a highly conserved cellular process for the degradation and recycling of unnecessary cytoplasmic components, including unnecessary proteins, damaged nuclear fragments, dysfunctional complexes, and even whole organelles, in eukaryotes. Three distinct but not mutually exclusive types of autophagy have been reported in plants, including macroautophagy, microautophagy, and mega-autophagy (Marshall and Vierstra, 2018). Macroautophagy is characterized by the sequestration of cellular cargos by double-membrane structures called autophagosomes, which fuse with the vacuole for digestion and recycling. Macroautophagy is the best-characterized form of autophagy; therefore, it is simply regarded as autophagy. Macroautophagy can be either nonselective or selective. Nonselective macroautophagy, stimulated by nutritional deficiency, normally refers to random bulk protein degradation, whereas selective macroautophagy specifically removes specific components and involves the recognition of autophagy substrates by dedicated receptors (Johansen and Lamark, 2020). Based on the specific recognition and degradation of organelles or pathogens, the forms of selective autophagy are named mitophagy for mitochondria degradation, chlorophagy for chloroplast degradation, reticulophagy for endoplasmic reticulum (ER) degradation, and xenophagy for intracellular pathogen degradation (Abdrakhmanov et al., 2020). In contrast, microautophagy is the direct uptake of cytoplasmic materials into the vacuole by invagination or protrusion of the tonoplast, such as anthocyanin aggregate transport from the cytosol to the vacuole, and these materials are directly engulfed by the vacuolar membrane, eventually becoming free in the vacuolar lumen (Chanoca et al., 2015). Mega-autophagy is an extreme autophagic process accomplished by permeabilization or rupture of the vacuolar membrane (van Doorn and Woltering, 2005). Mega-autophagy appears to be the most common type during programmed cell death (PCD), which occurs during development or in response to pathogenic invasion as in the case of xylem formation in Arabidopsis (Kwon et al., 2010), senescence (Liu and Bassham, 2012), and plant–pathogen interactions (Leary et al., 2019). Additionally, chaperone-mediated autophagy (CMA), which is a selective form of autophagy, occurs in most mammalian cells through cytosolic chaperone proteins that target substrates, but no functional counterparts have been identified in plants.
The genetic machinery of macroautophagy (hereafter termed as autophagy) has been systematically deciphered by the identification and functional analysis of over 40 autophagy-related (ATG) genes in eukaryotes (Furukawa et al., 2019). Plant genomes encode multiple orthologs of identified ATG members in yeast and mammals. These ATG proteins are traditionally divided into four protein complexes, including the ATG1 complex with scaffold protein ATG11/17 for the initiation of autophagy, transmembrane core protein ATG9 with ATG2/ATG18 for nucleation and phagophore expansion, the phosphatidylinositol 3-kinase (PI3K) complex for phagophore decoration, and ATG8/12 conjugation systems for autophagosome maturation (Tang and Bassham, 2018; Zhuang et al., 2018). Many excellent reviews have discussed the functions and categories of ATG genes (Bassham et al., 2006; Vanhee and Batoko, 2011; Michaeli et al., 2016; Antonioli et al., 2017; Galluzzi et al., 2017); thus, these topics are not covered here in detail.
In the first two decades of the current century, research on autophagy in plants expanded rapidly and explored the elements and the molecular mechanisms of autophagy, multiple ultrastructures involved in autophagy, and significant roles of autophagy in plant development and environmental responses (Figure 1). Compared to previous extensive functional research on autophagy, research on the regulatory mechanisms of the autophagy pathway is still lagging. Hence, a comprehensive review outlining the recent research on autophagy regulators in plants is needed. In the present review, we summarize recent advances in research on the molecular regulation of autophagy in plants, including the roles of protein kinases, phytohormones, second messengers, and transcriptional and epigenetic regulators. We also discuss the connection and distinction between autophagy and the 26S proteasome and the applications and prospects of autophagy in agriculture.
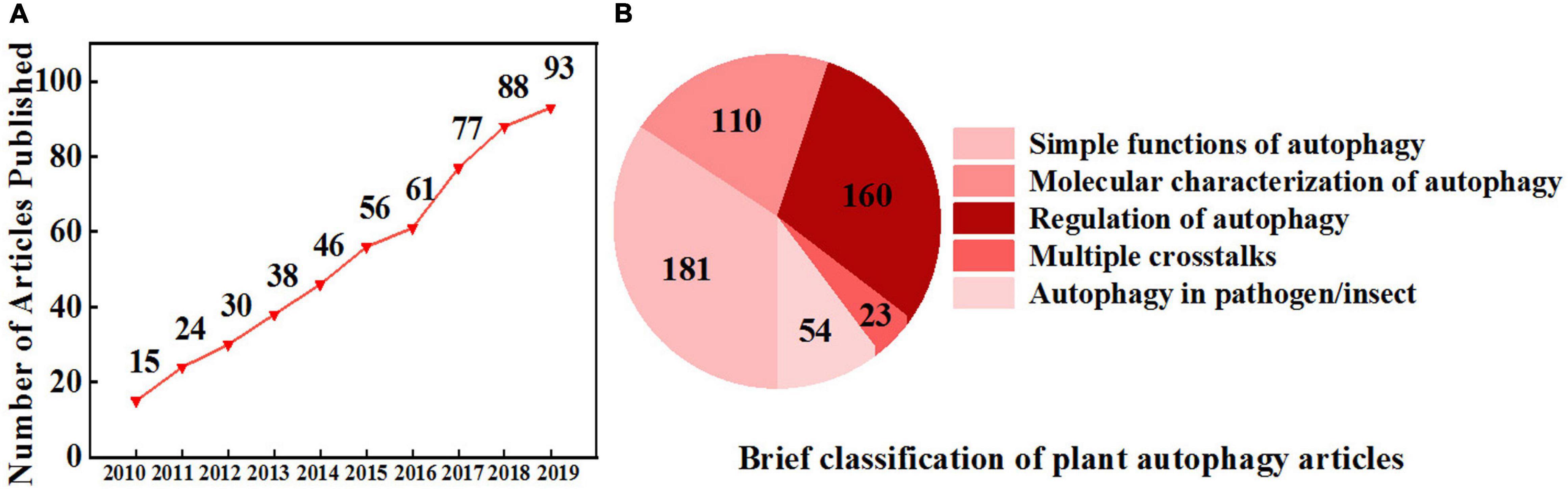
Figure 1. Increasing interest in plant autophagy. (A) The number of research articles published on autophagy in plants from 2010 to 2019. Information was retrieved from the Web of Science with the topic “plant autophagy” refined by the database “Web of Science Core Collection.” (B) A brief classification of plant autophagy articles from 2010 to 2019 based on (A) by manual division.
Regulators of Autophagy Induction
The conserved hierarchical steps of autophagosome formation include the following two major initiation complexes in plants (Figure 2A): (1) the ATG1 kinase complex, including ATG1, ATG13, ATG11, and ATG101, which responds to nutritional signals, and (2) the PI3K complex, which is characterized by two heterotetramers, namely, complex I and complex II, and remodels autophagic membranes (Zhuang et al., 2018). Complex I includes vacuolar protein sorting 34 (VPS34) and the accessory proteins VPS15, ATG6, and ATG14, whereas in complex II, ATG14 is replaced with VPS38 (Liu et al., 2020). ATG1 is the main switch activating autophagy, and it is normally regulated by upstream kinases (Hurley and Young, 2017). Target of rapamycin (TOR) and sucrose nonfermenting-1-related protein kinase 1 (SnRK1) are two evolutionarily conserved protein kinase complexes that play central and antagonistic roles in the initiation of autophagy (Rodriguez et al., 2019). TOR inhibits and SnRK1 activates ATG1 kinase under nutrient starvation and stress conditions, respectively. Mammals and yeast have TOR complex 1 (TORC1) and TORC2; however, only TORC1 has been identified in plants. Plant TORC1 includes the central element TOR kinase, two regulatory-associated protein of TOR (RAPTOR) subunits and lethal with sec thirteen 8 (LST8). TOR-dependent control of autophagy was first studied in animals. It has been reported that TOR promotes hyperphosphorylation of ATG13 to decrease its affinity to ATG1 and represses unc-51-like autophagy activating kinase 1 (ULK1, ATG1 homolog) activity through direct dephosphorylation of Ser757 under nutrient sufficiency (Kamada et al., 2010; Kim et al., 2011). Similar to the regulation in animals, the autophagy-initiating ATG1/ATG13 kinase complex is negatively regulated by the TOR complex in plants (Suttangkakul et al., 2011). During nutrition deprivation, inhibition of TOR signaling leads to dephosphorylation of ATG13 and hyperphosphorylation of ATG1a to activate the autophagy pathway (Li and Vierstra, 2012). Moreover, the mammalian homolog of SnRK1, AMP-activated protein kinase (AMPK), was previously reported to promote autophagy by directly activating ULK1 through phosphorylation of Ser317 and Ser777 (Kim et al., 2011). It has been recently reported that plant SnRK1 activates autophagy via inhibition of the TOR signaling pathway or direct activation of ATG proteins. For instance, Arabidopsis SnRK1 subunit KIN10 has been shown to interact with RAPTOR in the cytosol and to phosphorylate RAPTOR by kinase assays, suggesting that SnRK1 phosphorylation of RAPTOR represses TOR complex activity to activate autophagy in plants (Nukarinen et al., 2016; Pu et al., 2017b). KIN10 has also been shown to directly phosphorylate ATG1, and overexpression of Arabidopsis KIN10 enhanced the phosphorylation of ATG1 under carbon starvation and activated the autophagy signaling pathway (Chen L. et al., 2017). Furthermore, in addition to regulating the ATG1 complex, SnRK1 can also directly phosphorylate ATG6 to activate the PI3K complex by sensing nutritional status (Huang X. et al., 2019). Thus, SnRK1-mediated activation of the PI3K complex is a possible alternative route of autophagy initiation when the ATG1 initial complex is under prolonged fixed-carbon starvation in plants.
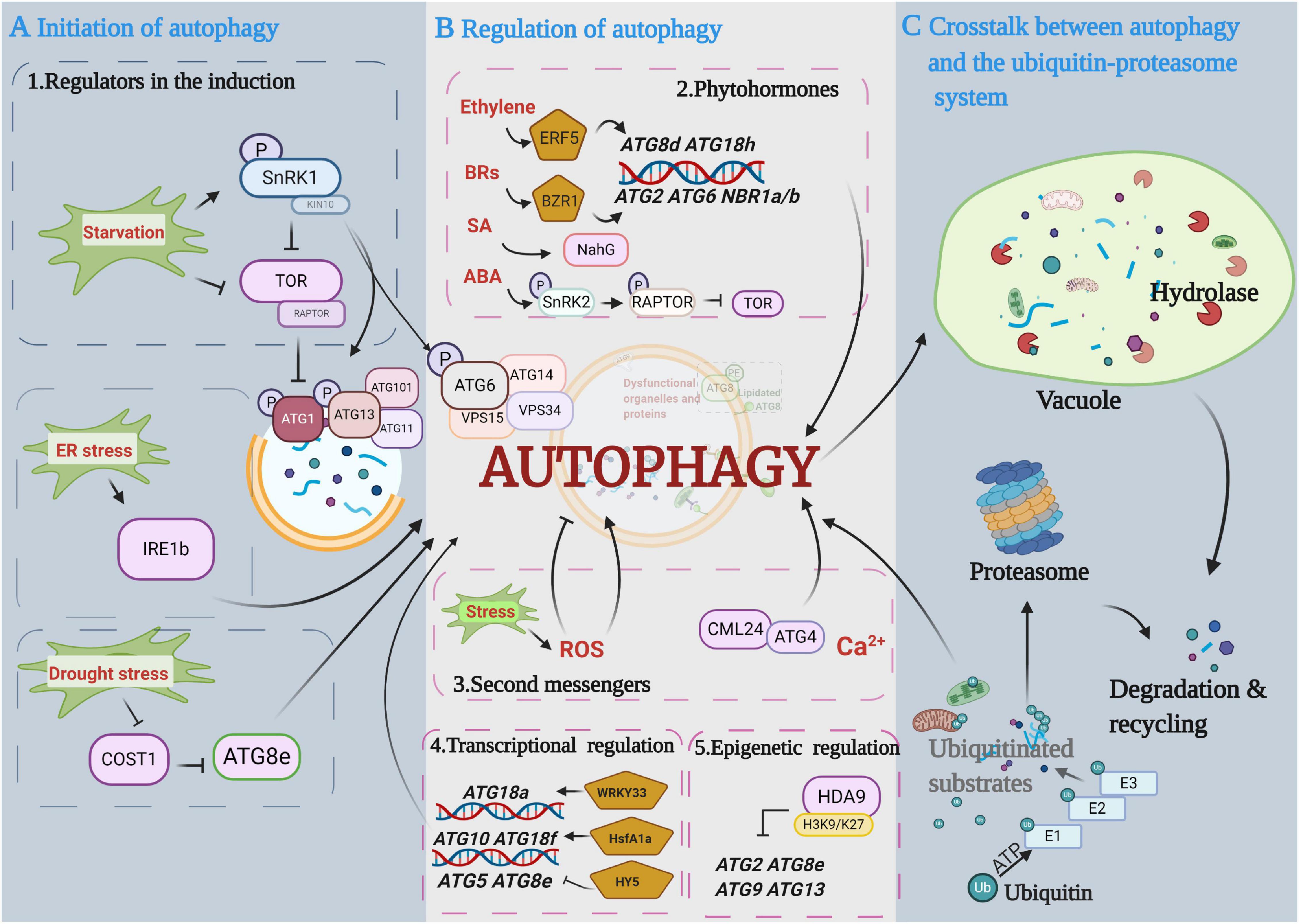
Figure 2. Diagram depicting the main proposed events in the autophagy pathway. (A) Initiation of autophagy. Autophagy is initiated by protein kinases and developmental and environmental signals. Autophagy induction and nucleation involve the autophagy-related gene 1 (ATG1) complex that is negatively controlled by target of rapamycin (TOR) kinase and activated by sucrose nonfermenting-1-related protein kinase 1 (SnRK1) kinase. (B) Regulation of autophagy in autophagosome expansion and maturation. Multiple regulatory mechanisms are involved in autophagy processes, including phytohormones, secondary messengers, and transcriptional and epigenetic regulators. (C) Crosstalk between autophagy and the ubiquitin-proteasome pathway (UPS). Ubiquitinated substrates are precisely identified and degraded by the UPS and autophagy pathways, respectively. This figure was created by BioRender (https://biorender.com/).
In addition to nutrient starvation-induced autophagy, multiple types of stress-induced autophagy are initiated through TOR/SnRK1-independent signaling pathways in plants. For instance, one potential regulator is inositol-requiring enzyme 1b (IRE1b), a dual protein kinase and ribonuclease, which indirectly activates autophagy by degrading the RNA transcripts of factors that interfere with the induction of autophagy under ER stress (Bao et al., 2018). Constitutively stressed 1 (COST1) has been reported as a possible negative regulator of autophagy through direct interaction with ATG8e in plants. Arabidopsis cost1 mutants exhibited strong drought tolerance with constitutive induction of broad expression of typical stress-responsive genes and autophagy initiation (Bao and Bassham, 2020). Furthermore, the stress-responsive protein SnRK2, a core abscisic acid (ABA) signaling kinase, appears to inhibit TOR activity and thus indirectly induces autophagy (Wang et al., 2018). However, whether SnRK2 can directly interact with ATG proteins remains to be further explored.
The regulation of selective autophagy initiation is seldom studied in plants. In mammals, it has been previously demonstrated that autophagic processes could be mediated independently by cargo receptors, such as nuclear dot protein 52 and TANK-binding kinase 1 (NDP52/TBK1), which initiate autophagy by recruiting ULK1 to cargo in the absence of microtubule-associated protein 1 light chain 3 (LC3, the mammalian homolog of ATG8) in HeLa cells (Vargas et al., 2019). A major cargo receptor p62/Sequestosome-1 (SQSTM1) binding to the scaffolding protein FIP200 (homolog of yeast ATG17) can promote autophagosome formation via the interaction between disordered residues 326–380 in p62 and the C-terminal region of FIP200 in HAP1 cells (Turco et al., 2019).
Phytohormones
Phytohormones are critical endogenous molecules that regulate physiological and molecular reactions during plant growth and development and during the stress response. A growing body of evidence suggests that plant autophagy is regulated by phytohormones (Figure 2B). The application of exogenous hormones can directly regulate autophagy initiation. For instance, benzothiadiazole (BTH), a functional analog of salicylic acid (SA), can induce autophagy through the SA signal transducer Nonexpresser of PR genes 1 (NPR1). BTH-induced rapid accumulation of autophagosomes was compromised in npr1 mutants (Yoshimoto, 2010; Munch et al., 2014). Overexpression of the NahG gene, which encodes a bacterial SA hydroxylase that converts SA to an inactive form, clearly suppressed SA-mediated early senescence in atg mutants (Yoshimoto, 2010). Furthermore, zeatin, a natural cytokinin (CTK), inhibited Arabidopsis primary root growth and reduced autophagy in root epidermis cells (Slavikova et al., 2008). Phytohormone response factors also participate in the regulation of autophagy. Ethylene response factor 5 (ERF5) directly binds to the ATG8d and ATG18h gene promoters and induces the transcription of both genes and autophagy in tomato under drought stress (Zhu et al., 2018). Meanwhile, the tomato brassinosteroid (BR) signaling transcription factor (TF) brassinazole resistance 1 (BZR1) also directly binds to the ATG2 and ATG6 gene promoters, and BZR1-overexpressing plants showed enhanced tolerance to nitrogen starvation along with an increase in ATG gene expression and autophagy (Wang et al., 2019). In addition to direct regulation, there are some indirect connections between autophagy and phytohormones. An increase in intercellular ABA can reduce the persulfidation of ATG4 by hydrogen sulfide and subsequently increase ATG4 protease activity and the formation of autophagosomes (Laureano-Marin et al., 2020). Other connections exist between autophagy and ABA as mentioned above; for example, ABA activates SnRK2 kinases, which phosphorylates RAPTOR and represses TOR activity to induce autophagy under osmotic stress (Wang et al., 2018). In addition, it has been reported that SnRK2 kinases have dual roles in the regulation of SnRK1 during plant growth and stress (Belda-Palazon et al., 2020; Laureano-Marin et al., 2020). Under optimal conditions, SnRK2s, together with the harbored type 2C phosphatases (PP2Cs), form “repressor complexes” that sequester SnRK1 to promote plant growth. Under stress conditions, stress-induced ABA helps disassemble SnRK2s and PP2C-containing SnRK1 repressor complexes, and thus, the released SnRK2s and SnRK1α trigger stress responses (Belda-Palazon et al., 2020). These results led us to hypothesize that SnRK2 not only directly represses TOR activity but also regulates SnRK1 activation to induce autophagy under stress conditions. In addition, mounting evidence also suggests that phytohormone biosynthesis and signal transduction pathways are affected by autophagy. The expression of phytohormone biosynthetic genes and the levels of endogenous phytohormones are changed in atg mutants. Higher auxin levels were observed in Arabidopsis atg5 and atg7 root tips, with enhanced root meristem activities on 3% glucose-based media. Moreover, significantly lower auxin levels were observed in atg mutants on media lacking glucose than in wild-type seedlings, indicating that auxin biosynthesis is changed in atg mutants under different growth conditions (Huang L. et al., 2019). Endogenous levels of active forms of gibberellins (GAs, including GA1, GA4, and GA7) and CTK (trans-zeatin) were significantly lower in the anthers of rice atg7 mutants, which show limited dehiscence and a sterility phenotype (Kurusu et al., 2017). Thus, the loss of autophagy function results in phytohormone and cellular metabolism disorders. Furthermore, phytohormone signal transduction is also connected to autophagy. The BR core signaling element BZR1 can be degraded through an autophagy-dependent pathway under sugar starvation. Treatment with the autophagy inhibitor 3-methyladenine (3MA) prevents estradiol-induced BZR1 degradation (Zhang et al., 2016). The Arabidopsis BR master regulator BRI1-EMS suppressor 1 (BES1) interacts with the ubiquitin receptor protein dominate suppressor of KAR2 (DSK2) and is targeted by autophagy for degradation during stress via the interaction of DSK2 with ATG8 (Nolan et al., 2017). EXO70D-mediated selective autophagy can target the negative regulators of CTK signaling, type-A response regulators (type-A ARR), for degradation in Arabidopsis roots (Acheampong et al., 2020). All of these results demonstrate that selective receptor-mediated autophagy could precisely modulate phytohormone signaling.
Moreover, several hormone-related proteins, including indole-3-acetic acid inducible 17 (IAA17), IAA27, polar auxin transport inhibitor sensitive 1 (PIS1), and ABI5-binding protein 3 (AFP3), contain an ATG8-interacting motif (AIM) or ubiquitin-interacting motif (UIM) and interacted with ATG8 in a yeast two-hybrid assay in vitro; thus, they are potential substrates for autophagy (Marshall et al., 2019). Therefore, autophagy may precisely regulate multiple phytohormone signaling pathways by degrading the signal components.
Second Messengers
Autophagy, as a process of cytoplasmic component recycling and reuse, is also regulated by second messengers, such as reactive oxygen species (ROS), nitric oxide (NO), Ca2+, and the cyclic nucleotides cAMP and cGMP (Figure 2B).
Under abiotic and biotic stresses, ROS, including superoxide anions (O2–), hydrogen peroxide (H2O2), singlet oxygen (1O2), and hydroxyl radical (OH), have been thought to play a dual role in plant biology (Mittler, 2017), as they can operate as important second messengers that trigger several signaling cascades at low levels and cause severe oxidative damage to DNA, RNA, proteins, and cellular membranes at high levels (Medeiros et al., 2020). ROS can modulate autophagy by targeting upstream factors or key autophagy genes. Redox signals directly modulate the kinase activity of the autophagy upstream regulator SnRK1; for example, it has been demonstrated that Arabidopsis KIN10 activity is strongly dependent on the redox status in vitro and that this redox sensitivity is conferred by a single cysteine residue (Wurzinger et al., 2017). Furthermore, ATG4 proteases were inhibited by ROS to ensure lipidation of ATG8 and autophagy progression in Arabidopsis and Chlamydomonas reinhardtii under stress conditions (Woo et al., 2014; Perez-Perez et al., 2016). NO signaling is related to autophagy through its master regulator S-nitrosoglutathione reductase 1 (GSNOR1). The conformation of GSNOR1 can be changed to expose its AIM by S-nitrosylation at the Cys10 residue, after which it is bound by ATG8 and degraded in an AIM-dependent manner during hypoxia responses in Arabidopsis (Zhan et al., 2018). Moreover, highly reactive and toxic oxidative species cause oxidation and denaturation of cellular proteins, which are specific substrates for autophagic degradation. For instance, more oxidative proteins were aggregated in autophagy-impaired plants under oxidative stress (Xiong et al., 2007). Exogenous H2O2 application-damaged peroxisomes were selectively degraded by autophagy in Arabidopsis (Shibata et al., 2013). Unnecessary or damaged peroxisomes can be degraded by selective autophagy, called pexophagy, which is a crucial quality control system of peroxisomes in plant cells (Borek et al., 2019; Su et al., 2020). However, how peroxisomes are marked for degradation in plants is not yet clear (Su et al., 2020). Moreover, aggregation of peroxisomes and high levels of ROS accumulation are observed in Arabidopsis atg mutants, which leads to disorders of guard cell ROS homeostasis and stomatal defects (Yamauchi et al., 2019). In addition, autophagy was decreased in mitochondrial alternative oxidase 1a (AOX1a) RNAi tomato plants with increased levels of H2O2 (Zhu et al., 2018), and increased catalase aggregation occurred in Arabidopsis selective autophagy cargo receptor next to BRCA1 gene 1 (nbr1) mutants under heat stress (Zhou et al., 2014b).
Unlike the large amounts of ROS and autophagy research, reports on the connection between calcium signaling and autophagy in plants are limited, though this connection has been extensively reported in animals. Previous results from an animal study showed that intracellularly sequestered calcium could induce autophagy in hepatocytes (Gordon et al., 1993). Subsequent studies showed that Ca2+ signaling is an essential component of the AMPK-dependent autophagy pathway. AMPK could be activated by Ca2+/calmodulin-dependent protein kinase kinase-β (CaMKKβ) in insulinoma cells, providing a further association between Ca2+ signaling and autophagy (Witters et al., 2006). In Arabidopsis, calmodulin-related protein 24 (CML24) could affect autophagy progression and the resistance of darkness-induced starvation through interacting with ATG4 (Tsai et al., 2013). However, mechanistic details of the regulation between calcium signaling and autophagy in plants are not fully known. Additionally, early insightful studies have shown that cyclic nucleotide second messengers (cAMP and cGMP) regulate cellular autophagic capacity by directly affecting autophagy genes or indirect regulation in animals. The first insight comes from mammalian systems, where cAMP or dibutyryl cAMP injections produced a wave of autophagy in the rat liver (Shelburne et al., 1973). A number of subsequent studies have further explored the mechanism; for example, cyclic GMP-AMP (cGAMP) induced robust LC3 lipidation through WIPI2 (WD-repeat phosphatidylinositol-3-phosphate effector proteins) and ATG5-dependent pathways in human fibroblast cells (Gui et al., 2019).
Transcriptional and Epigenetic Regulation of Autophagy
Recently, a growing body of research revealed that the transcriptional regulation of ATG genes is an important mechanism for autophagy to maintain cellular homeostasis under nutrient starvation and stress conditions. Furthermore, a growing number of studies also suggest that epigenetic changes, such as histone modification and DNA methylation, influence the expression of ATG genes and subsequent autophagic processes (Figure 2B).
Transcription Factors
Transcription factors are important players controlling various processes of plant development and responses to different external stimuli. Increasing evidence in the last decade clearly indicates that nuclear transcriptional events play major roles in autophagy regulation under adverse environmental conditions in plants. Arabidopsis WRKY33 is the first reported TF that interacts with ATG18a and is required for resistance to necrotrophic pathogens (Lai et al., 2011). Likewise, its tomato homologs WRKY33a/b also play key roles in heat tolerance and regulation of stress-induced autophagy. Silencing SlWRKY33s reduced the expression of heat-induced ATG genes and the formation of autophagosomes (Zhou et al., 2014a). The first reported plant TF to transcriptionally regulate ATG genes was tomato HsfA1a, which directly binds to the promoters of ATG10 and ATG18f and enhances their transcript levels under drought stress (Wang Y. et al., 2015). Subsequently, more TFs that regulate autophagy genes have been discovered, such as Arabidopsis elongated hypocotyl 5 (HY5), which directly binds to the promoters of ATG5 and ATG8e to suppress their gene expression and thus negatively modulates autophagy (Yang et al., 2020b). Furthermore, key downstream signaling elements of phytohormones have also been reported to transcriptionally regulate autophagy genes. Tomato ERF5, a typical drought-responsive TF, is involved in ethylene-mediated autophagy through binding to the promoters of ATG8d and ATG18h via the DRE-binding site (ACCGAC) and promoting the expression of both genes (Zhu et al., 2018). BRs and their signaling element BZR1 can also transcriptionally upregulate ATG genes and the selective autophagy receptor NBR1 and induce accumulation of NBR1 proteins and autophagosome formation in tomato under nitrogen starvation and chilling stress (Wang et al., 2019; Chi et al., 2020). These results indicate that TFs regulate ATG gene expression and promote autophagosome formation. Moreover, they are also involved in the regulation of selective autophagy receptors responsible for the recognition of damaged proteins. Furthermore, using a yeast one-hybrid library screening system, 225 TFs from 35 families were identified to bind to the promoters of ATG8s. These TFs are generally involved in plant development processes and environmental stress response (Wang P. et al., 2020). Whether more potential TFs are involved in plant autophagy signaling pathways remains to be demonstrated.
DNA Methylation
DNA methylation is a major epigenetic modification that occurs in eukaryotes ranging from fungi to mammals. In plants, DNA methylation occurs in CG, CHG, and CHH (where H represents A, T, or C) sequence contexts and is mediated by DNA methyltransferases, like methyltransferase (MET), chromomethylase (CMT), and domain-rearranged methylase (DRM) (Qi et al., 2020). Several expression profiles have indicated that DNA methylation regulates autophagy genes in plants. For example, almost all ATG loci are enriched in different cytosine sequence contexts across gene regions in tomato (Zhong et al., 2013). Consistent with this, one genome-wide analysis of Arabidopsis DNA methylation also uncovered almost every ATG gene with a methylated modification (Zhong et al., 2015). Moreover, the methylation profiling of the DNA methyltransferase mutant drm2 showed that the levels of methylated CG in ATG6 and ATG7 were lower than those in wild-type Arabidopsis (Zhong et al., 2015). Furthermore, the ATG8f promoter region was hypomethylated when evaluating global DNA methylation, and ATG8f expression was induced under TOR inhibition in Arabidopsis (Zhu et al., 2020). These studies allowed us to propose that DNA methylation plays a critical role in the regulation of autophagy in plants.
Histone Modification
Histone modifications are essential transcriptional regulators that adjust chromatin structure and recruit histone modifiers. Histones contain five major subtypes (H1, H2A, H2B, H3, and H4) and include at least eight types of modifications: acetylation, methylation, phosphorylation, ubiquitylation, sumoylation, ADP ribosylation, deamination, and proline isomerization (Kouzarides, 2007). Accumulating evidence indicates that some histone modifications are related to the regulation of autophagy in animals. For instance, the methyltransferase enhancer of zeste homolog 2 (EZH2), which di- and trimethylates Lys27 of histone H3 (H3K27me2/3), can repress ATG5 and ATG7 protein levels in vascular smooth muscle cells. Moreover, inhibition or knockdown of EZH2 induced the accumulation of ATG5 and ATG7 and autophagosome formation (Li et al., 2018). Plants generally possess histone modifications similar to those in animals. Arabidopsis histone deacetylase 9 (HDA9) can directly bind and repress the expression of the ATG2, ATG9, ATG8e, and ATG13 genes. Moreover, HDA9 can be recruited by HY5 to ATG5 and ATG8e loci to repress their expression through the deacetylation of H3K9 and H3K27 under nitrogen-sufficient or light conditions (Yang et al., 2020b). According to the enrichment analysis of essential histone markers on ATG genes in Arabidopsis, most ATG loci display high accumulation of the active markers H3K9/27/56Ac and H3K4/36me3, while only ATG18e shows enrichment of the repressive marker H3K27me3, which is closely related to the low expression of ATG18e in all Arabidopsis organs (Yang et al., 2020a).
Noncoding RNAs
Noncoding RNAs (ncRNAs) have been shown to regulate a variety of cellular processes and functions by controlling gene expression. MicroRNAs (miRNAs) refer to a class of ncRNAs comprising 21–25 nucleotides that target multiple genes to regulate their expression. Long ncRNAs (lncRNAs) are noncoding transcripts of more than 200 nucleotides and have complex secondary structures to bind proteins, RNA, and DNA, thus endowing them with a variety of regulatory capabilities. Abundant studies in animals have shown that miRNAs and lncRNAs are broadly involved in the core pathways of autophagy, including vesicle nucleation, elongation, retrieval, and fusion. For instance, MIR223 restrains autophagy and promotes central nervous system inflammation by targeting ATG16L1, and MIR223 deficiency increases ATG16L1 expression in murine cells (Li et al., 2019). In plants, one possible miRNA involved in the regulation of autophagy is MIR447a.2, which is highly expressed in Arabidopsis pollen and is predicted to target ATG18h (Borges et al., 2011). MIR447a.2 could be induced by Pseudomonas syringae pv. tomato (Pst avrRpt2) infection in Arabidopsis, while the expression of its target gene ATG18h was reduced under Pst avrRpt2 treatment (Zhang et al., 2010). Compared with miRNAs, lncRNAs are larger and act through diverse sets of mechanisms to regulate autophagy in animals. For instance, the lncRNA maternally expressed gene 3 (MEG3) upregulates LC3 and ATG3 expression levels, leading to autophagosome formation in epithelial ovarian cancer (Xiu et al., 2017). In plants, the roles of lncRNAs in the regulation of autophagy remain to be elucidated. Genome-wide analysis of 200 Arabidopsis transcriptome data sets successfully uncovered 6,480 lncRNAs, and the expression of 1,832 lncRNAs was significantly altered after drought, cold, high-salt, and ABA treatments (Matsui et al., 2008; Liu et al., 2012). Moreover, ATG genes were shown to be transcriptionally upregulated under these conditions (Liu et al., 2009; Chi et al., 2020; Wang M. et al., 2020). However, whether these ATG genes are modulated by lncRNAs remains unknown. Furthermore, the lncRNA-miRNA interaction also regulates autophagy at the molecular level in animals. MIR188-3p inhibits autophagy and cell death by targeting ATG7, while lncRNA autophagy promoting factor (APF) targets MIR188-3p and inhibits its activity; therefore, APF promotes autophagy signaling through targeting the MIR188-3p/ATG7 axis in cardiomyocytes (Wang K. et al., 2015).
Crosstalk Between Autophagy and the Ubiquitin–Proteasome System
Autophagy and the ubiquitin-26S proteasome pathway (UPS) constitute two major mechanisms of cellular protein degradation in eukaryotes, and they coordinately enable nutrient recycling, ensure cellular well-being and regulate growth (Pohl and Dikic, 2019). Unlike autophagy as an intracellular vesicle transport system, the UPS uses its own protease activity to degrade target proteins. UPS mediates the ubiquitination of target proteins by a three-step cascade of the E1 (activation), E2 (conjugation), and E3 (ligation) enzymes and then promotes the degradation of ubiquitinated proteins through the 26S proteasome in an ATP-dependent manner (Figure 2C). The 26S proteasome is a barrel-shaped organelle that is composed of the 20S core protease (CP) and lid 19S regulatory particles (RP) (Nam et al., 2017). It is now recognized that UPS normally aims to remove single, unfolded substrate polypeptides because the narrow entrance of the pore loops is only a 30- to 40-Å gap (Bard et al., 2018), while autophagy can degrade intact protein complexes, protein aggregates, or even organelles (Schreiber and Peter, 2014). Proteomic analyses have revealed that the ratio of protein degradation by proteasome or autophagy depends on cell types and states (Mathew et al., 2014; Braten et al., 2016). In addition, Arabidopsis autophagy receptor nbr1 mutants and chaperone-associated E3 ubiquitin ligase Hsc70-interacting protein (chip) mutants both accumulated a large number of unfolded proteins under heat stress, and unfolded proteins were further increased in the chip nbr1 double mutant (Zhou et al., 2014b), suggesting that both UPS and autophagy pathways collaboratively degrade aggregated proteins, and either pathway can be functionally compensated by the other when one is dysfunctional. Moreover, during plant aging, UPS mainly impacts the timing and onset of senescence, but autophagy is closely related to the degradation of bulk proteins during aging (Wang and Schippers, 2019).
Accumulating evidence has shown that crosstalk exists between autophagy and the proteasomal degradation pathway. Some autophagy components are directly regulated via ubiquitin-mediated UPS in plant cells. ATG6 can be ubiquitinated by the E3 ligases SINAT1 and SINAT2, leading to its degradation by the 26S proteasome in the presence of tumor necrosis factor receptor-associated factor 1a (TRAF1a) and TRAF1b in Arabidopsis (Qi et al., 2017). Moreover, inactivated proteasomes can be degraded by autophagy. For instance, the Arabidopsis 26S proteasome is the degradation substrate of ATG8-mediated autophagy (proteaphagy) under nitrogen starvation, and the RP non-ATPase subunit (RPN10) acts as a selective autophagy receptor in this process by targeting inactive 26S proteasomes and tethering them to autophagic vesicles. Furthermore, plant RPN10 serves as a dual receptor in both autophagy and the 26S proteasome pathway. RPN10 recognizes ubiquitylated targets when integrated into the 19S RP lid and has a specific UIM to bind to ATG8 docking sites (Marshall et al., 2015). Additionally, both Arabidopsis and tomato autophagy-deficient mutants hyperaccumulated ubiquitylated protein aggregates in response to heat and oxidative stresses, indicating that autophagy also recognizes ubiquitin and degrades ubiquitinylated substrates similar to the UPS (Zhou et al., 2013, 2014b). The mutual compensation mechanisms between autophagy and UPS are complex in plants, and further research is needed to integrate cellular protein quality control systems under different conditions.
Agricultural Applications
The functions of autophagy in growth, development, and stress responses have been deciphered in various crop species. Therefore, how to use autophagy to improve agricultural benefits, such as high yield, quality, and multifaceted resistance, is an important research direction. Here, we highlight the roles of autophagy in crop growth, yield, and stress tolerance and discuss future research directions for potential applications in agriculture (Figure 3).
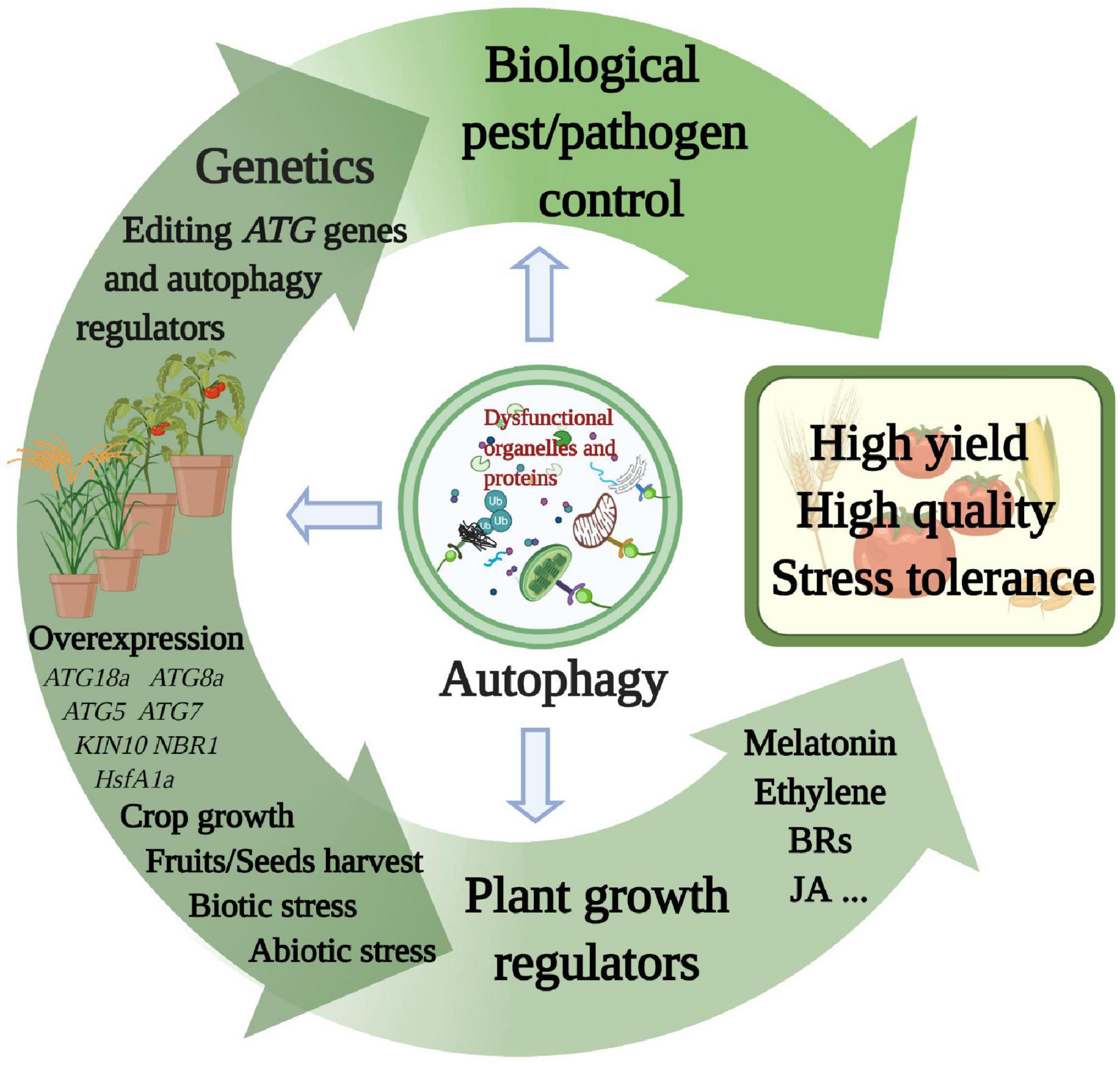
Figure 3. Multiple proposed agricultural applications related to autophagy. Each of the methods described in the main text explores a component of autophagy function. Superior yield, quality, and stress resistance characteristics will emerge in various crop species from autophagy applications via genetic research, plant growth regulators, and biological pest and pathogen control. This figure was created by BioRender (https://biorender.com/).
Utilizing Plant Growth Regulators and Biological Pest/Pathogen Control
As discussed above, some plant growth regulators can regulate autophagy; thus, the application of growth regulators may be an effective way to activate autophagy for agricultural benefits. For instance, elevation of both exogenous and endogenous melatonin results in enhanced thermotolerance in tomato mediated by ATG expression and autophagy, which are related to the degradation of aggregated proteins under heat stress (Xu et al., 2016). Exogenous BRs can be used to increase crop resistance to nitrogen starvation and chilling stress through BZR1-mediated autophagy (Wang et al., 2019; Chi et al., 2020). Thus, melatonin and BRs are possibly utilized as plant growth regulators to increase crop resistance through autophagy signaling. Ethylene has also been suggested to mediate autophagy, contributing to enhanced survival during flooding, hypoxia, and reoxygenation stress through the induction of ATG genes and ROS levels in soybean and tomato (Hartman et al., 2019). Considering its function in fruit ripening and leaf senescence, the manipulation of ethylene might be an effective means to regulate plant growth and fruit quality via autophagy. Moreover, plant resistance against necrotrophic phytopathogens is mediated by autophagy via modulation of hormone homeostasis. For example, autophagy apparently plays a positive role in the induction of jasmonic acid (JA)-regulated plant defensin 1.2 (PDF1.2) expression as a defense against Botrytis cinerea in Arabidopsis (Lai et al., 2011). In addition, we can exploit the corresponding biological pesticides to manipulate autophagic activity in pathogens and pests. For instance, Tomato yellow leaf curl virus (TYLCV) is a whitefly-transmitted geminivirus that causes severe yield losses in tomato production. Activating autophagy in whitefly inhibits the transmission efficiency of TYLCV by reducing the amount of viral coat protein and genomic DNA transmitted to tomato plants. Feeding with rapamycin activates whitefly autophagy to prevent TYLCV transmission to solanaceous plants (Wang et al., 2016). Interestingly, autophagy induced by different pathogens in insect vectors causes different results; for example, the Rice gall dwarf virus (RGDV)-induced autophagy pathway promotes viral replication in the leafhopper Recilia dorsalis, causing severe viral infection and transmission to rice plants. Moreover, leafhopper-borne viral spread was decreased by the autophagy inhibitor 3MA (Chen Y. et al., 2017). Thus, by exploring the function and the regulation of autophagy in insect-borne pathogens, we can exploit pesticides that can activate/inhibit the autophagy pathway in insect vectors and block vector-borne plant viruses. With ongoing autophagy research, plant growth regulators and pesticides will be developed in agricultural production based on autophagy signaling.
Exploring Potential Applications of Functional Genes
The evolution and practical breeding of crops essentially depend on genetic variation. With the development of genomics and genome-editing techniques, we are able to select excellent agronomic traits and increase yields by editing ATG genes and autophagy regulators. For example, overexpression of foxtail millet SiATG8a in Arabidopsis and rice OsATG8a conferred tolerance to nitrogen starvation, with an increase in root and leaf areas and increased nitrogen absorption (Izumi et al., 2015; Li et al., 2015). Overexpression of apple MdATG18a also improved nitrogen absorption by upregulating nitrate uptake genes and the accumulation of anthocyanins (Sun et al., 2018), indicating that the application of ATG genes contributes to crop adaptability to low-nitrogen environments and improves crop growth. Moreover, autophagy also affects pollen growth, development, yield, and fruit ripening. For example, rice mutants defective in autophagy showed sporophytic male sterility and immature pollen (Kurusu and Kuchitsu, 2017). Ripened pepper fruits exhibited increases in the expression of ATG4, ATG8a, and ATG9 (Lopez-Vidal et al., 2020), and postharvest fruit senescence of Ziziphus jujube was delayed by the inhibition of autophagy (Deng et al., 2019). Constitutive overexpression of ATG5 or ATG7 increased seed yields and the levels of fatty acids in Arabidopsis seeds (Minina et al., 2018), suggesting that upregulated autophagy has a positive effect on increasing crop fitness and oil accumulation in the breeding of high-yield oil crops (Ortiz et al., 2020). However, these overexpressing transgenic plants are generated by increasing protein expression from cloned transgenes with special enhancers. Considering the biosafety of agricultural products, transgene-free genome editing might be a better way to expedite crop improvement by enhancing ATG gene expression. Manipulation of upstream open reading frames (uORFs) by genome editing can fine-tune mRNA translation and thereby increase the amounts of protein synthesized (Zhang et al., 2018).
Autophagy is widely regarded to enhance stress resistance. Overexpression of ATG8 conferred tolerance to low-nitrate conditions and led to an increase in yield and nitrogen remobilization efficiency in Arabidopsis (Chen et al., 2019). Overexpression of ATG5 and ATG7 increased ATG8 protein lipidation and autophagic flux, thereby exhibiting increased resistance to necrotrophic pathogens and oxidative stress in Arabidopsis (Minina et al., 2018). Overexpression of Joka2 (homolog of NBR1) significantly restricted the size of disease lesions caused by Phytophthora infestans in potato, but virus-induced gene silencing of Joka2 resulted in increased disease lesions, indicating that Joka2-mediated selective autophagy contributes to defense against P. infestans (Dagdas et al., 2016). Therefore, manipulation of ATG transcriptional changes by genetic stimulation seems to be an effective approach for enhancing plant resistance.
In addition to directly focusing on ATG genes in various stages of autophagy, multiplex gene editing, knockouts, and regulation of gene transcription can be used to regulate upstream signaling pathways of autophagy. For example, overexpressing the tomato TF HsfA1a, which activates ATG genes, would enhance plant drought tolerance (Wang Y. et al., 2015). TOR and SnRK1 are essential upstream regulators of autophagy. Downregulation of TOR expression or kinase activity led to constitutive activation of autophagy, while the overexpression of TOR was sufficient to block starvation-, salt-, and drought-induced autophagy in Arabidopsis (Liu and Bassham, 2010; Pu et al., 2017a). Arabidopsis KIN10-overexpressing lines exhibited enhanced tolerance to hypoxia with increasing autophagy (Chen L. et al., 2017; Janse van Rensburg et al., 2019).
Conclusion and Future Perspectives
The core mechanisms of autophagy are conserved in all eukaryotes and have been well-studied in plants. However, the functions and regulatory networks of autophagy are still not fully understood. First, although many ATG genes have been identified, their multiple functions beyond self-eating are still unknown, and some ATG proteins may play multiple roles and have pleiotropic effects during numerous processes or pathways. For instance, ATG1 is an essential protein for the formation of autophagy vesicles and can be phosphorylated by the upstream kinase TOR; thus, is there a possibility that ATG1, as a kinase, regulates other signaling pathways besides autophagy? ATG10 acts as an E2-like enzyme to help ATG5 conjugate with ATG12; is there a possibility that ATG10, as an E2-like enzyme, has other substrates besides autophagy? Therefore, the multiple functions of ATG are interesting research topics that warrant further investigation in plants. Second, the regulatory networks of autophagy are intricate, and different levels of autophagy regulation might play complex and ambiguous roles. Different phytohormones are mutually antagonistic and synergistic during plant growth and development or under stress. Phytohormones regulate autophagy, and in turn, autophagy feedback influences the balance of phytohormones. ATG-overexpressing and atg mutant plants can both exhibit high levels of the same phytohormones, suggesting that crosstalk interactions between hormonal signals and autophagy are more complicated. Third, the role of autophagy remains to be further explored, and more precise approaches are needed to provide clearer insights into the agricultural applications of autophagy, such as artificial evolution of critical autophagy genes by base editing. Overall, in the coming years, more interesting and fundamental research will likely emerge to answer existing questions regarding plant autophagy and will shine light on agricultural applications.
Author Contributions
JZ planned the review manuscript. J-JC and C-XL analyzed the data. J-JC, JZ, and S-JS wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFD1000300) and the National Natural Science Foundation of China (31922078 and 31872089).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.618944/full#supplementary-material
Supplementary Figure 1 | DNA methylation in plants.
Supplementary Table 1 | DNA methylation of Arabidopsis ATG genes.
References
Abdrakhmanov, A., Gogvadze, V., and Zhivotovsky, B. (2020). To eat or to die: deciphering selective forms of autophagy. Trends Biochem. Sci. 45, 347–364. doi: 10.1016/j.tibs.2019.11.006
Acheampong, A. K., Shanks, C., Cheng, C. Y., Schaller, G. E., Dagdas, Y., and Kieber, J. J. (2020). EXO70D isoforms mediate selective autophagic degradation of type-A ARR proteins to regulate cytokinin sensitivity. Proc. Natl. Acad. Sci. U.S.A. 117, 27034–27043. doi: 10.1073/pnas.2013161117
Antonioli, M., Di Rienzo, M., Piacentini, M., and Fimia, G. M. (2017). Emerging mechanisms in initiating and terminating autophagy. Trends Biochem. Sci. 42, 28–41. doi: 10.1016/j.tibs.2016.09.008
Bao, Y., and Bassham, D. C. (2020). COST1 balances plant growth and stress tolerance via attenuation of autophagy. Autophagy 16, 1157–1158. doi: 10.1080/15548627.2020.1752981
Bao, Y., Pu, Y., Yu, X., Gregory, B. D., Srivastava, R., Howell, S. H., et al. (2018). IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana. Autophagy 14, 1562–1573. doi: 10.1080/15548627.2018.1462426
Bard, J. A. M., Goodall, E. A., Greene, E. R., Jonsson, E., Dong, K. C., and Martin, A. (2018). Structure and function of the 26S proteasome. Annu. Rev. Biochem. 87, 697–724. doi: 10.1146/annurev-biochem-062917-011931
Bassham, D. C., Laporte, M., Marty, F., Moriyasu, Y., Ohsumi, Y., Olsen, L. J., et al. (2006). Autophagy in development and stress responses of plants. Autophagy 2, 2–11. doi: 10.4161/auto.2092
Belda-Palazon, B., Adamo, M., Valerio, C., Ferreira, L. J., Confraria, A., Reis-Barata, D., et al. (2020). A dual function of SnRK2 kinases in the regulation of SnRK1 and plant growth. Nat. Plants 6, 1345–1353. doi: 10.1038/s41477-020-00778-w
Borek, S., Stefaniak, S., Sliwinski, J., Garnczarska, M., and Pietrowska-Borek, M. (2019). Autophagic machinery of plant peroxisomes. Int. J. Mol. Sci. 20, 4754. doi: 10.3390/ijms20194754
Borges, F., Pereira, P. A., Slotkin, R. K., Martienssen, R. A., and Becker, J. D. (2011). MicroRNA activity in the Arabidopsis male germline. J. Exp. Bot. 62, 1611–1620. doi: 10.1093/jxb/erq452
Braten, O., Livneh, I., Ziv, T., Admon, A., Kehat, I., Caspi, L. H., et al. (2016). Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc. Natl. Acad. Sci. U.S.A. 113, e4639–e4647. doi: 10.1073/pnas.1608644113
Chanoca, A., Kovinich, N., Burkel, B., Stecha, S., Bohorquez-Restrepo, A., Ueda, T., et al. (2015). Anthocyanin vacuolar inclusions form by a microautophagy mechanism. Plant Cell 27, 2545–2559. doi: 10.1105/tpc.15.00589
Chen, L., Su, Z. Z., Huang, L., Xia, F. N., Qi, H., Xie, L. J., et al. (2017). The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front. Plant Sci. 8:1201. doi: 10.3389/fpls.2017.01201
Chen, Q., Soulay, F., Saudemont, B., Elmayan, T., Marmagne, A., and Masclaux-Daubresse, C. (2019). Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 60, 343–352. doi: 10.1093/pcp/pcy214
Chen, Y., Chen, Q., Li, M., Mao, Q., Chen, H., Wu, W., et al. (2017). Autophagy pathway induced by a plant virus facilitates viral spread and transmission by its insect vector. PLoS Pathog. 13:e1006727. doi: 10.1371/journal.ppat.1006727
Chi, C., Li, X., Fang, P., Xia, X., Shi, K., Zhou, Y., et al. (2020). Brassinosteroids act as a positive regulator of NBR1-dependent selective autophagy in response to chilling stress in tomato. J. Exp. Bot. 71, 1092–1106. doi: 10.1093/jxb/erz466
Dagdas, Y. F., Belhaj, K., Maqbool, A., Chaparro-Garcia, A., Pandey, P., Petre, B., et al. (2016). An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 5, e10856. doi: 10.7554/eLife.10856
Deng, B., Guo, M., Liu, H., Tian, S., and Zhao, X. (2019). Inhibition of autophagy by hydroxychloroquine enhances antioxidant nutrients and delays postharvest fruit senescence of Ziziphus jujuba. Food Chem. 296, 56–62. doi: 10.1016/j.foodchem.2019.05.189
Furukawa, K., Innokentev, A., and Kanki, T. (2019). Regulatory mechanisms of mitochondrial autophagy: lessons from yeast. Front. Plant Sci. 10:1479. doi: 10.3389/fpls.2019.01479
Galluzzi, L., Baehrecke, E. H., Ballabio, A., Boya, P., Pedro, J. M. B.-S., Cecconi, F., et al. (2017). Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836. doi: 10.15252/embj.201796697
Gordon, P. B., Holen, I., Fosse, M., Rotnes, J. S., and Seglen, P. O. (1993). Dependence of hepatocytic autophagy on intracellularly sequestered calcium. J. Biol. Chem. 268, 26107–26112. doi: 10.1016/s0021-9258(19)74287-2
Gui, X., Yang, H., Li, T., Tan, X., Shi, P., Li, M., et al. (2019). Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266. doi: 10.1038/s41586-019-1006-9
Hartman, S., Sasidharan, R., and Voesenek, L. (2019). The role of ethylene in metabolic acclimations to low oxygen. New Phytol. 229, 64–70. doi: 10.1111/nph.16378
Huang, L., Yu, L. J., Zhang, X., Fan, B., Wang, F. Z., Dai, Y. S., et al. (2019). Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 15, 407–422. doi: 10.1080/15548627.2018.1520547
Huang, X., Zheng, C., Liu, F., Yang, C., Zheng, P., Lu, X., et al. (2019). Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell 31, 2973–2995. doi: 10.1105/tpc.19.00066
Hurley, J. H., and Young, L. N. (2017). Mechanisms of autophagy initiation. Annu. Rev. Biochem. 86, 225–244. doi: 10.1146/annurev-biochem-061516-044820
Izumi, M., Hidema, J., Wada, S., Kondo, E., Kurusu, T., Kuchitsu, K., et al. (2015). Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol. 167, 1307–1320. doi: 10.1104/pp.114.254078
Janse van Rensburg, H. C., Van den Ende, W., and Signorelli, S. (2019). Autophagy in plants: both a puppet and a puppet master of sugars. Front. Plant Sci. 10:14. doi: 10.3389/fpls.2019.00014
Johansen, T., and Lamark, T. (2020). Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432, 80–103. doi: 10.1016/j.jmb.2019.07.016
Kamada, Y., Yoshino, K., Kondo, C., Kawamata, T., Oshiro, N., Yonezawa, K., et al. (2010). TOR directly controls the ATG1 kinase complex to regulate autophagy. Mol. Cell Biol. 30, 1049–1058. doi: 10.1128/MCB.01344-09
Kim, J., Kundu, M., Viollet, B., and Guan, K. L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141. doi: 10.1038/ncb2152
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Kurusu, T., Koyano, T., Kitahata, N., Kojima, M., Hanamata, S., Sakakibara, H., et al. (2017). Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant Signal. Behav. 12, e1365211. doi: 10.1080/15592324.2017.1365211
Kurusu, T., and Kuchitsu, K. (2017). Autophagy, programmed cell death and reactive oxygen species in sexual reproduction in plants. J. Plant Res. 130, 491–499. doi: 10.1007/s10265-017-0934-4
Kwon, S. I., Cho, H. J., Jung, J. H., Yoshimoto, K., Shirasu, K., and Park, O. K. (2010). The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 64, 151–164. doi: 10.1111/j.1365-313X.2010.04315.x
Lai, Z., Wang, F., Zheng, Z., Fan, B., and Chen, Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66, 953–968. doi: 10.1111/j.1365-313X.2011.04553.x
Laureano-Marin, A. M., Aroca, A., Perez-Perez, M. E., Yruela, I., Jurado-Flores, A., Moreno, I., et al. (2020). Abscisic acid-triggered persulfidation of cysteine protease ATG4 mediates regulation of autophagy by sulfide. Plant Cell 32, 3902–3920. doi: 10.1105/tpc.20.00766
Leary, A. Y., Savage, Z., Tumtas, Y., and Bozkurt, T. O. (2019). Contrasting and emerging roles of autophagy in plant immunity. Curr. Opin. Plant Biol. 52, 46–53. doi: 10.1016/j.pbi.2019.07.002
Li, F., and Vierstra, R. D. (2012). Regulator and substrate: dual roles for the ATG1-ATG13 kinase complex during autophagic recycling in Arabidopsis. Autophagy 8, 982–984. doi: 10.4161/auto.20240
Li, R., Yi, X., Wei, X., Huo, B., Guo, X., Cheng, C., et al. (2018). EZH2 inhibits autophagic cell death of aortic vascular smooth muscle cells to affect aortic dissection. Cell Death Dis. 9, 180. doi: 10.1038/s41419-017-0213-2
Li, W. W., Chen, M., Zhong, L., Liu, J. M., Xu, Z. S., Li, L. C., et al. (2015). Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 468, 800–806. doi: 10.1016/j.bbrc.2015.11.035
Li, Y., Zhou, D., Ren, Y., Zhang, Z., Guo, X., Ma, M., et al. (2019). Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy 15, 478–492. doi: 10.1080/15548627.2018.1522467
Liu, F., Hu, W., Li, F., Marshall, R. S., Zarza, X., Munnik, T., et al. (2020). AUTOPHAGY-RELATED14 and its associated phosphatidylinositol 3-kinase complex promotes autophagy in Arabidopsis. Plant Cell 32, 3939–3960. doi: 10.1105/tpc.20.00285
Liu, J., Jung, C., Xu, J., Wang, H., Deng, S., Bernad, L., et al. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24, 4333–4345. doi: 10.1105/tpc.112.102855
Liu, Y., and Bassham, D. C. (2010). TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS One 5:e11883. doi: 10.1371/journal.pone.0011883
Liu, Y., and Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. doi: 10.1146/annurev-arplant-042811-105441
Liu, Y., Xiong, Y., and Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963. doi: 10.4161/auto.5.7.9290
Lopez-Vidal, O., Olmedilla, A., Sandalio, L. M., Sevilla, F., and Jimenez, A. (2020). Is autophagy involved in pepper fruit ripening? Cells 9, 106. doi: 10.3390/cells9010106
Marshall, R. S., Hua, Z., Mali, S., McLoughlin, F., and Vierstra, R. D. (2019). ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 177, 766–781. doi: 10.1016/j.cell.2019.02.009
Marshall, R. S., Li, F., Gemperline, D. C., Book, A. J., and Vierstra, R. D. (2015). Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 58, 1053–1066. doi: 10.1016/j.molcel.2015.04.023
Marshall, R. S., and Vierstra, R. D. (2018). Autophagy: the master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208. doi: 10.1146/annurev-arplant-042817-040606
Mathew, R., Khor, S., Hackett, S. R., Rabinowitz, J. D., Perlman, D. H., and White, E. (2014). Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Mol. Cell 55, 916–930. doi: 10.1016/j.molcel.2014.07.019
Matsui, A., Ishida, J., Morosawa, T., Mochizuki, Y., Kaminuma, E., Endo, T. A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149. doi: 10.1093/pcp/pcn101
Medeiros, D. B., Barros, J. A. S., Fernie, A. R., and Araujo, W. L. (2020). Eating away at ROS to regulate stomatal opening. Trends Plant Sci. 25, 220–223. doi: 10.1016/j.tplants.2019.12.023
Michaeli, S., Galili, G., Genschik, P., Fernie, A. R., and Avin-Wittenberg, T. (2016). Autophagy in plants–What’s new on the menu? Trends Plant Sci. 21, 134–144. doi: 10.1016/j.tplants.2015.10.008
Minina, E. A., Moschou, P. N., Vetukuri, R. R., Sanchez-Vera, V., Cardoso, C., Liu, Q., et al. (2018). Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 69, 1415–1432. doi: 10.1093/jxb/ery010
Munch, D., Rodriguez, E., Bressendorff, S., Park, O. K., Hofius, D., and Petersen, M. (2014). Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy 10, 1579–1587. doi: 10.4161/auto.29406
Nam, T., Han, J. H., Devkota, S., and Lee, H. W. (2017). Emerging paradigm of crosstalk between autophagy and the ubiquitin-proteasome system. Mol. Cells 40, 897–905. doi: 10.14348/molcells.2017.0226
Nolan, T. M., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z., et al. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41, e37. doi: 10.1016/j.devcel.2017.03.013
Nukarinen, E., Nagele, T., Pedrotti, L., Wurzinger, B., Mair, A., Landgraf, R., et al. (2016). Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 6, 31697. doi: 10.1038/srep31697
Ortiz, R., Geleta, M., Gustafsson, C., Lager, I., Hofvander, P., Lofstedt, C., et al. (2020). Oil crops for the future. Curr. Opin. Plant Biol. 56, 181–189. doi: 10.1016/j.pbi.2019.12.003
Perez-Perez, M. E., Lemaire, S. D., and Crespo, J. L. (2016). Control of autophagy in Chlamydomonas is mediated through redox-dependent inactivation of the ATG4 protease. Plant Physiol. 172, 2219–2234. doi: 10.1104/pp.16.01582
Pohl, C., and Dikic, I. (2019). Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 366, 818–822. doi: 10.1126/science.aax3769
Pu, Y., Luo, X., and Bassham, D. C. (2017a). TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front. Plant Sci. 8:1204. doi: 10.3389/fpls.2017.01204
Pu, Y., Soto-Burgos, J., and Bassham, D. C. (2017b). Regulation of autophagy through SnRK1 and TOR signaling pathways. Plant Signal. Behav. 12, e1395128. doi: 10.1080/15592324.2017.1395128
Qi, H., Xia, F. N., and Xiao, S. (2020). Autophagy in plants: physiological roles and post-translational regulation. J. Integr. Plant Biol. doi: 10.1111/jipb.12941 [Epub ahead of print].
Qi, H., Xia, F. N., Xie, L. J., Yu, L. J., Chen, Q. F., Zhuang, X. H., et al. (2017). TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell 29, 890–911. doi: 10.1105/tpc.17.00056
Rodriguez, M., Parola, R., Andreola, S., Pereyra, C., and Martinez-Noel, G. (2019). TOR and SnRK1 signaling pathways in plant response to abiotic stresses: do they always act according to the “yin-yang” model? Plant Sci. 288, 110220. doi: 10.1016/j.plantsci.2019.110220
Schreiber, A., and Peter, M. (2014). Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim. Biophys. Acta 1843, 163–181. doi: 10.1016/j.bbamcr.2013.03.019
Shelburne, J. D., Arstila, A. U., and Trump, B. F. (1973). Studies on cellular autophagocytosis: cyclic Amp-stimulated and dibutyryl cyclic Amp-stimulated autophagy in rat liver. Am. J. Pathol. 72, 521–539.
Shibata, M., Oikawa, K., Yoshimoto, K., Kondo, M., Mano, S., Yamada, K., et al. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25, 4967–4983. doi: 10.1105/tpc.113.116947
Slavikova, S., Ufaz, S., Avin-Wittenberg, T., Levanony, H., and Galili, G. (2008). An autophagy-associated ATG8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59, 4029–4043. doi: 10.1093/jxb/ern244
Su, T., Li, X., Yang, M., Shao, Q., Zhao, Y., Ma, C., et al. (2020). Autophagy: an intracellular degradation pathway regulating plant survival and stress response. Front. Plant Sci. 11:164. doi: 10.3389/fpls.2020.00164
Sun, X., Jia, X., Huo, L., Che, R., Gong, X., Wang, P., et al. (2018). MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 41, 469–480. doi: 10.1111/pce.13110
Suttangkakul, A., Li, F., Chung, T., and Vierstra, R. D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23, 3761–3779. doi: 10.1105/tpc.111.090993
Tang, J., and Bassham, D. C. (2018). Autophagy in crop plants: what’s new beyond Arabidopsis? Open Biol. 8, 180162. doi: 10.1098/rsob.180162
Tsai, Y. C., Koo, Y., Delk, N. A., Gehl, B., and Braam, J. (2013). Calmodulin-related CML24 interacts with ATG4b and affects autophagy progression in Arabidopsis. Plant J. 73, 325–335. doi: 10.1111/tpj.12043
Turco, E., Witt, M., Abert, C., Bock-Bierbaum, T., Su, M. Y., Trapannone, R., et al. (2019). FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol. Cell 74, 330–346. doi: 10.1016/j.molcel.2019.01.035
van Doorn, W. G., and Woltering, E. J. (2005). Many ways to exit? Cell death categories in plants. Trends Plant Sci. 10, 117–122. doi: 10.1016/j.tplants.2005.01.006
Vanhee, C., and Batoko, H. (2011). Autophagy involvement in responses to abscisic acid by plant cells. Autophagy 7, 655–656. doi: 10.4161/auto.7.6.15307
Vargas, J. N. S., Wang, C., Bunker, E., Hao, L., Maric, D., Schiavo, G., et al. (2019). Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol. Cell 74, 347–362. doi: 10.1016/j.molcel.2019.02.010
Wang, H., and Schippers, J. H. M. (2019). The role and regulation of autophagy and the proteasome during aging and senescence in plants. Genes 10, 267. doi: 10.3390/genes10040267
Wang, K., Liu, C. Y., Zhou, L. Y., Wang, J. X., Wang, M., Zhao, B., et al. (2015). APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 6, 6779. doi: 10.1038/ncomms7779
Wang, L. L., Wang, X. R., Wei, X. M., Huang, H., Wu, J. X., Chen, X. X., et al. (2016). The autophagy pathway participates in resistance to tomato yellow leaf curl virus infection in whiteflies. Autophagy 12, 1560–1574. doi: 10.1080/15548627.2016.1192749
Wang, M., Li, X., Luo, S., Fan, B., Zhu, C., and Chen, Z. (2020). Coordination and crosstalk between autophagosome and multivesicular body pathways in plant stress responses. Cells 9, 119. doi: 10.3390/cells9010119
Wang, P., Nolan, T. M., Yin, Y., and Bassham, D. C. (2020). Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 16, 123–139. doi: 10.1080/15548627.2019.1598753
Wang, P., Zhao, Y., Li, Z., Hsu, C. C., Liu, X., Fu, L., et al. (2018). Reciprocal eregulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 69, 100–112. doi: 10.1016/j.molcel.2017.12.002
Wang, Y., Cai, S. Y., Yin, L. L., Shi, K., Xia, X., Zhou, Y. H., et al. (2015). Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 11, 2033–2047. doi: 10.1080/15548627.2015.1098798
Wang, Y., Cao, J. J., Wang, K. X., Xia, X. J., Shi, K., Zhou, Y. H., et al. (2019). BZR1 mediates brassinosteroid-induced autophagy and nitrogen starvation in tomato. Plant Physiol. 179, 671–685. doi: 10.1104/pp.18.01028
Witters, L. A., Kemp, B. E., and Means, A. R. (2006). Chutes and ladders: the search for protein kinases that act on AMPK. Trends Biochem. Sci. 31, 13–16. doi: 10.1016/j.tibs.2005.11.009
Woo, J., Park, E., and Dinesh-Kumar, S. P. (2014). Differential processing of Arabidopsis ubiquitin-like ATG8 autophagy proteins by ATG4 cysteine proteases. Proc. Natl. Acad. Sci. U.S.A. 111, 863–868. doi: 10.1073/pnas.1318207111
Wurzinger, B., Mair, A., Fischer-Schrader, K., Nukarinen, E., Roustan, V., Weckwerth, W., et al. (2017). Redox state-dependent modulation of plant SnRK1 kinase activity differs from AMPK regulation in animals. FEBS Lett. 591, 3625–3636. doi: 10.1002/1873-3468.12852
Xiong, Y., Contento, A. L., Nguyen, P. Q., and Bassham, D. C. (2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143, 291–299. doi: 10.1104/pp.106.092106
Xiu, Y. L., Sun, K. X., Chen, X., Chen, S., Zhao, Y., Guo, Q. G., et al. (2017). Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget 8, 31714–31725. doi: 10.18632/oncotarget.15955
Xu, W., Cai, S. Y., Zhang, Y., Wang, Y., Ahammed, G. J., Xia, X. J., et al. (2016). Melatonin enhances thermotolerance by promoting cellular protein protection in tomato plants. J. Pineal Res. 61, 457–469. doi: 10.1111/jpi.12359
Yamauchi, S., Mano, S., Oikawa, K., Hikino, K., Teshima, K. M., Kimori, Y., et al. (2019). Autophagy controls reactive oxygen species homeostasis in guard cells that is essential for stomatal opening. Proc. Natl. Acad. Sci. U.S.A. 116, 19187–19192. doi: 10.1073/pnas.1910886116
Yang, C., Luo, M., Zhuang, X., Li, F., and Gao, C. (2020a). Transcriptional and epigenetic regulation of autophagy in plants. Trends Genet. 36, 676–688. doi: 10.1016/j.tig.2020.06.013
Yang, C., Shen, W., Yang, L., Sun, Y., Li, X., Lai, M., et al. (2020b). HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol. Plant 13, 515–531. doi: 10.1016/j.molp.2020.02.011
Yoshimoto, K. (2010). Plant autophagy puts the brakes on cell death by controlling salicylic acid signaling. Autophagy 6, 192–193. doi: 10.4161/auto.6.1.10843
Zhan, N., Wang, C., Chen, L., Yang, H., Feng, J., Gong, X., et al. (2018). S-Nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol. Cell 71, 142–154. doi: 10.1016/j.molcel.2018.05.024
Zhang, H., Si, X., Ji, X., Fan, R., Liu, J., Chen, K., et al. (2018). Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36, 894–898. doi: 10.1038/nbt.4202
Zhang, W., Gao, S., Zhou, X., Xia, J., Chellappan, P., Zhou, X., et al. (2010). Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol. 11, R81. doi: 10.1186/gb-2010-11-8-r81
Zhang, Z., Zhu, J. Y., Roh, J., Marchive, C., Kim, S. K., Meyer, C., et al. (2016). TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr. Biol. 26, 1854–1860. doi: 10.1016/j.cub.2016.05.005
Zhong, S., Fei, Z., Chen, Y. R., Zheng, Y., Huang, M., Vrebalov, J., et al. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31, 154–159. doi: 10.1038/nbt.2462
Zhong, X., Hale, C. J., Nguyen, M., Ausin, I., Groth, M., Hetzel, J., et al. (2015). Domains rearranged methyltransferase3 controls DNA methylation and regulates RNA polymerase V transcript abundance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, 911–916. doi: 10.1073/pnas.1423603112
Zhou, J., Wang, J., Cheng, Y., Chi, Y. J., Fan, B., Yu, J. Q., et al. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9:e1003196. doi: 10.1371/journal.pgen.1003196
Zhou, J., Wang, J., Yu, J. Q., and Chen, Z. (2014a). Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 5:174. doi: 10.3389/fpls.2014.00174
Zhou, J., Zhang, Y., Qi, J., Chi, Y., Fan, B., Yu, J. Q., et al. (2014b). E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 10:e1004116. doi: 10.1371/journal.pgen.1004116
Zhu, T., Li, L., Feng, L., Mo, H., and Ren, M. (2020). Target of rapamycin regulates genome methylation reprogramming to control plant growth in Arabidopsis. Front. Genet. 11:186. doi: 10.3389/fgene.2020.00186
Zhu, T., Zou, L., Li, Y., Yao, X., Xu, F., Deng, X., et al. (2018). Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol. J. 16, 2063–2076. doi: 10.1111/pbi.12939
Keywords: agricultural applications, autophagy, autophagy-related genes, epigenetic regulation, phytohormone, transcription factor, ubiquitin–proteasome system
Citation: Cao J-J, Liu C-X, Shao S-J and Zhou J (2021) Molecular Mechanisms of Autophagy Regulation in Plants and Their Applications in Agriculture. Front. Plant Sci. 11:618944. doi: 10.3389/fpls.2020.618944
Received: 19 October 2020; Accepted: 28 December 2020;
Published: 16 February 2021.
Edited by:
Vasileios Fotopoulos, Cyprus University of Technology, CyprusReviewed by:
Farida Minibayeva, Kazan Institute of Biochemistry and Biophysics (RAS), RussiaChristian Meyer, INRA UMR 1318 Institut Jean Pierre Bourgin, France
Copyright © 2021 Cao, Liu, Shao and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Zhou, amllQHpqdS5lZHUuY24=
 Jia-Jian Cao1
Jia-Jian Cao1 Chen-Xu Liu
Chen-Xu Liu Jie Zhou
Jie Zhou