- 1Hunan Provincial Key Laboratory of Phytohormones and Growth Development, College of Bioscience and Biotechnology, Hunan Agricultural University, Changsha, China
- 2Hengyang Medical College, University of South China, Hengyang, China
- 3Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China
- 4Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
Copy number variation (CNV) may have phenotypic effects by altering the expression level of the gene(s) or regulatory element(s) contained. It is believed that CNVs play pivotal roles in controlling plant architecture and other traits in plant. However, the effects of CNV contributing to special traits remain largely unknown. Here we report a CNV involved in rice architecture by modulating tiller number and leaf angle. In the genome of Oryza sativa ssp. japonica cv. Nipponbare, we found a locus Loc_Os08g34249 is derived from a 13,002-bp tandem duplication in the nearby region of OsMTD1, a gene regulating tillering in rice. Further survey of 230 rice cultivars showed that the duplication occurred in only 13 japonica rice cultivars. Phenotypic investigation indicated that this CNV region may contribute to tiller number. Moreover, we revealed that OsMTD1 not only influences rice tiller number and leaf angle, but also represses pri-miR156f transcription in the CNV region. Intriguingly, this CNV performs function through both the dosage and position effects on OsMTD1 and pri-miR156f. Thus, our work identified a CNV and revealed a molecular regulatory basis for its effects on plant architecture, implying this CNV may possess importance and application potential in molecular breeding in rice.
Introduction
Genomic rearrangements include duplications, deletions, and inversions of unique genomic segments at specific regions, as well as translocations, marker chromosomes, isochromosomes, and other complex rearrangements (Lupski, 1998; Feuk et al., 2006; Weckselblatt and Rudd, 2015). These rearrangements are not random events, but instead the reflection of higher-order architectural features of the genome (Lee and Lupski, 2006; Żmien’ko et al., 2014). Different from the whole genome duplication in a cell, the copy number variation (CNV) is the microduplication and deletion, which means an abnormal number of copies of one or more segments of DNA (Sebat et al., 2004). A CNV is commonly regarded as a DNA segment that has been deleted, inserted, or duplicated on certain chromosomes. The length of DNA is more than 1 kb and variable in copy number in comparison with a reference genome (Feuk et al., 2006). Previous studies indicated that CNVs not only involve in intraspecific genome variations, but also cause phenotypic differences. Thus, CNVs can be developed as markers for molecular identification. Genetic diversity can be differentiated by analyzing CNVs (Żmien’ko et al., 2014).
It was reported that CNVs are in variable linkage disequilibrium with flanking SNPs (Hinds et al., 2006; Locke et al., 2006; Yu et al., 2013). CNV could underlie a significant proportion of normal variation including differences in various features (Lee and Lupski, 2006). Known data suggest that CNV mainly affects the members of large families of functionally redundant genes, and the effects of individual CNV events on phenotype are usually modest (Żmien’ko et al., 2014). Altering copy number of a gene family member may only trigger quantitative rather than qualitative changes, making the CNV–phenotype association difficult to be detected. Increasing evidences showed that copy number polymorphisms contribute to natural genetic variation and adaptability in plants; some CNVs for specific genes have been linked to important traits such as flowering time, plant height, and stress resistance (Żmien’ko et al., 2014). A dramatic fruit size change due to a CNV with an insertion of 6–8 kb that affected gene regulation was described during tomato breeding (Cong et al., 2008). In wheat, a CNV has been found to determine the extreme dwarf phenotype by tandem segmental duplication of a region containing the green revolution gene Rht-D1b in the haploid genome (Li Y. et al., 2012).
Rice (Oryza sativa) is an important staple food crop in the world and a model plant of monocots; whether and how its CNVs are associated with specific traits have also been widely concerned. A CNV at the GL7 locus has been reported; a tandem duplication of a 17.1-kb segment leads to an increase in grain length (Wang Y. et al., 2015). A 1,212-bp deletion of qSW5 has been reported to be clearly associated with an increase in rice grain width (Shomura et al., 2008). It has been also reported that a natural tandem array of a 3,137-bp sequence in the upstream of IPA1 leads to superior yielding (Zhang et al., 2017). Although the knowledge of CNVs in higher plants is still poor, recent studies confirmed the prevalence of CNVs in the Oryza species and suggested that CNVs probably play a far more significant role in plant development than previously thought. High-level CNVs existing in different rice cultivars might associate to phenotypic diversity, yet how they affect yield, quality, resistance, and development processes is largely unknown (Li S. et al., 2012; Yu et al., 2013).
OsMTD1 is a tillering-related gene in rice (Liu et al., 2015). Here we describe a previously unknown transcriptional mechanism that OsMTD1 is able to repress pri-miR156f transcripts by the position effect. Furthermore, we provide evidences showing that OsMTD1-located region involved a CNV, a tandem segmental duplication resulting in the increasing expression of the OsMTD1 and reduction of tiller number. This CNV harbors a 13,002-bp region on the eighth chromosome, covering one protein-coding gene OsMTD1 and a microRNA precursor of osa-miR156f. The results by surveying a panel of 190 rice cultivars showed that 13 of 82 japonica cultivars harboring two copies of CNV corresponding sequence by segmental tandem duplication produce less tillers than the one-copy normal cultivars. Transgenic experiments indicated that the OsMTD1 not only influences tiller number and leaf angle, but also regulates pri-miR156f transcription in this CNV region.
Materials and Methods
Plant Materials, Field Trails, and Tiller Number Investigation
The mini-core collection accessions from the China National Crop Gene Bank in the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, as described in Supplementary Table 1, were used in our experiments. Another japonica cv. Kitaake was used for CRISPR/Cas9 editing and overexpression analysis. In addition, tobacco (Nicotiana benthamiana) leaves were used for Agrobacterium-mediated transient expression analysis.
Rice tiller number investigations were conducted in Beijing. Different rice cultivars were transplanted to a paddy field with single plant per hill. The tiller number was counted from three to six randomly chosen individual hills at heading stage in summer of 2011 and autumn of 2013, respectively (Supplementary Table 1).
Sequence Alignments and Comparisons
The bacterial artificial chromosome sequences from japonica cv. Nipponbare and indica cv. 93–11 were used to determine the start or the end point range in sequence of OsMTD1-located CNV. Then, the 13,002-bp reference genome sequence from Nipponbare was used in BLASTN (National Center for Biotechnology Information) searches against different rice databases for other cultivars, including japonica cv. Zhonghua 11 and indica cv. Zhenshan 97, Minghui 63, 93–11, Shuhui 498, and RP Bio-226, to determine their orthologous regions. The conserving segments, InDels, and substitution mutations in the orthologous regions of indica and japonica were identified by using the BLAST, MEGA, and DNAMAN programs.
Plasmid Construction and Plant Transformation
The vector constructions for the CRISPR/Cas9-mediated gene editing were performed as previously described (Miao et al., 2013). The vectors for OsMTD1 overexpression in which the OsMTD1 gene was driven by the CaMV 35S promoter were constructed as previously described (Liu et al., 2015). The constructs were transformed into ZH11 or Kitaake by Agrobacterium tumefaciens–mediated transformation (Hiei and Komari, 2008).
Expressions in tobacco leaves were performed in two different plasmids of pCAMBIA1301 and pSN1301 vectors using Golden Gate cloning strategy. The pSN1301 is an adapted form of pCAMBIA1301 in which a CaMV 35S promoter was added. The region containing the native sequence of OsMTD1 and pre-miR156f was amplified from a japonica cv. Nipponbare genomic DNA. The DNA fragment for pCAM1301::MTD1-OsmiR156f was amplified by primers 5′-gga tcc ccg ggt acc TGG CAG GTG TAA AGA GGT CA-3′ (prim-177) and 5′-tac gaa ttc gag ctc AAG GAG CAG TTA GAT AAT GGA G-3′ (prim-179) and the DNA fragment for pSN1301::MTD1-OsmiR156f was obtained by primers prim-177 and 5′-ggg aaa ttc gag ctc AAG GAG CAG TTA GAT AAT GGA G-3′ (prim-178) and then infused the fragment of interest with Kpn I-Sac I of pCAMBIA1301 and pSN1301 by using ClonExpress II one-step cloning kit (Vazyme, C112-01) to generate plasmid pCAM1301::MTD1-OsmiR156f and pSN1301::MTD1-OsmiR156f, respectively. The mutant form sequences were obtained by an overlap extension polymerase chain reaction (PCR) method. To generate the pCAM1301::△MTD1-OsmiR156f in which OsMTD1 gene sequence was deleted, primers 5′-gga tcc ccg ggt acc ctt aaa tgc tcc aat agc tag-3′ (prim-182) and prim-179 were used to amplify a fragment sequence from OsMTD1 gene stop codon to the 60-bp sequence downstream of pre-miR156f from genomic DNA, and then the DNA fragment was ligated into the binary vector pCAMBIA1301 for transformation. Similar strategies were carried out to construct pCAM1301::ATT-OsmiR156f in which the ATG start codon of OsMTD1 was mutated to ATT. The primers 5′-gga tcc ccg ggt acc aga tcg ccg gag atT agc cag aag tc-3′ (prim-183) and prim-179 were used in the ATT mutant fragment amplification. For pSN1301::△MTD1-OsmiR156f and the pSN1301::ATT-OsmiR156f, a CaMV 35S promoter was harbored at the upstream of pCAM1301::△MTD1-OsmiR156f and pCAM1301::ATT-OsmiR156f, respectively. The corresponding primers prim-182 and prim-178 were employed for pSN1301::△MTD1-OsmiR156f, and prim-183 and prim-178 for pSN1301::ATT-OsmiR156f. The constructed vectors were infiltrated into the tobacco leaves by Agrobacterium tumefaciens–mediated transformation.
PCR, Real-Time PCR, and Stem-Loop RT-PCR
Genomic DNA was extracted and purified from fresh young leaves of five plants using CTAB methods. PCR was carried out in a reaction system with a total volume of 20 μL. The primers 5′-ATG AGC CAG AAG TCG TCG TGG C-3′ and 5′-ACA CAT GAA CGT ACA CGG CGC C-3′ were used for OsMTD1 analysis. PCR validation for CNV was performed in all selected rice cultivars, and three independent experiments were performed for each cultivar. The primers were used as follows: primer64, 5′-AAA TGG CGG AAA CTT GAC AC-3′; primer65, 5′-TGA GCT AGC TGG ACA CAT GG-3′; primer66, 5′-CGG ACC TAA CCA CCG ATC TA-3′; primer67, 5′-ATC TTG GCG CTG CAA TTA TC-3′; inhF, 5′-ATG AGC CAG AAG TCG TCG TGG C-3′; inhR, 5′-ACA CAT GAA CGT ACA CGG CGC C-3′.
Total RNA was isolated from ∼100 mg leaves of five plants using a Trizol reagent (Invitrogen) and treated with RNase-free DNase I (Invitrogen) according to the manufacturer’s instructions. Approximately 5 μg of RNA was used to synthesize first-strand cDNA using poly (dT) oligo primer according to the manufacturer’s instructions in M-MLV kit (Invitrogen). Quantitative real-time reverse transcription PCR (RT-qPCR) was carried out in a reaction system with a total volume of 20 μL, which contained SYBR green I (Invitrogen) on a CFX96 system (BIO-RAD). The following programs were employed: predenaturing for 30 s at 95°C and then amplification for 40 cycles including denaturation for 10 s at 95°C, annealing for 30 s at 60°C, and extension at 72°C for 10 s. The pri-miR156f was normalized to the internal rice tubulinβ-4 gene, and the relative abundance was determined with 2–Δ Δ Ct method. The RT-qPCR analysis in different lines was repeated three independent times. The primers for testing pri-miR156f were 5′-CTT CCC TTC GAC AGG ATA GC-30 and 5′-AGC GGC AGC TGT ATC ATC A-3′.
Stem-loop RT-qPCR (Varkonyi-Gasic et al., 2007) was employed to detect the mature osa-miR156f. Relative expression levels of osa-miR156 were normalized to the internal control U6 in rice and NbEF1 in tobacco. PCR was carried out in a reaction system with a total volume of 20 μL, which contained SYBR green I (Invitrogen) on a CFX96 system (BIO-RAD). The following programs were employed: predenaturing for 30 s at 95°C and then amplification for 40 cycles including denaturation for 10 s at 95°C, annealing for 30 s at 60°C, and extension at 72°C for 10 s. The 2–ΔΔCt method was used to calculate the relative expression level of osa-miR156, and the analysis was repeated three independent times. The primers for U6 are 5′-TAC AGA TAA GAT TAG CAT GGC CCC-3′ and 5′-GGA CCA TTT CTC GAT TTG TAC GTG-3′, and primers for NbEF1 are 5′-GAT TGG TGG TAT TGG TAC TGT C-3′ and 5′-AGC TTC GTG GTG CAT CTC-3′.
Results
OsMTD1-Located Segment Involves a New CNV in Rice
OsMTD1 sequence was queried via BLAST against four databases: TIGR rice genome annotation1, Rice Information GateWay (RIGW2), National Center for Biotechnology Information3, and the Knowledge-Based Oryza Molecular Biological Encyclopedia (KOME4). In the reference genome sequence of O. sativa spp. japonica cv. Nipponbare, we found that another locus, Loc_Os08g34249, is identical in DNA sequence to OsMTD1, a gene previously reported responsible for tillering in rice. However, it is not the case in the genomes of indica cultivars such as 93–11 (Figure 1A) and Shuhui498. Further analysis showed that OsMTD1 and Loc_Os08g34249 genes located on rice chromosome 8 according to their positions given in the TIGR rice database. OsMTD1 and Loc_Os08g34249 genes coexisted in two overlapped PAC clone AP0082414 and clone AP004703, implying one segmental duplication event on OsMTD1. To detect the physical location whereby the CNV event began and ended, we mapped the DNA sequences with different lengths between OsMTD1 and Loc_Os08g34249 against the reference genome and found the Nipponbare harbored a 13,002-bp tandem segmental duplication on OsMTD1-located region on the eighth chromosome. OsMTD1 and Loc_Os08g34249 genes were reciprocal duplication, and each of them was encompassed in a 13,002-bp segment, respectively. Compared with the reference genome of Nipponbare, the tandem duplication in indica cultivar 93–11 is absent, and the varied length is more than 1 kb, so the 13,002-bp region encompassing OsMTD1 could be regarded as a CNV between different rice cultivars. Herein, this DNA segment (about 13,002-bp corresponding region) variation in different rice cultivars was designated as OsMTD1-located CNV.
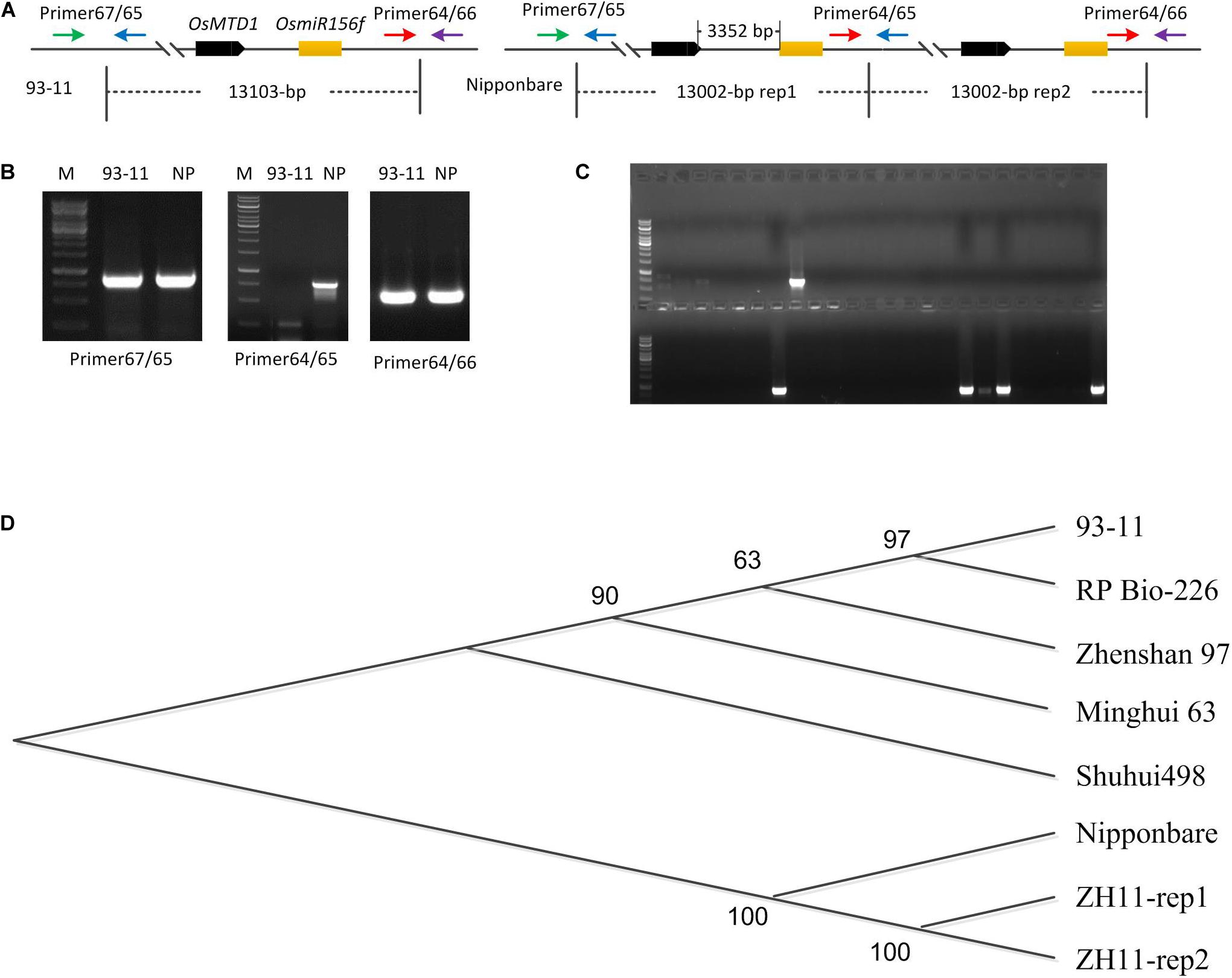
Figure 1. OsMTD1-located segment involves a CNV. (A) The schematic shows the fragment of OsMTD1 located on chromosome 8 in 93–11 and Nipponbare. Primer67/65, Primer64/66, and Primer64/65: primer pairs using for tandem duplication event analysis; OsMTD1 and OsmiR156f: the two comprising elements in the corresponding DNA sequence of the OsMTD1 located CNV. (B) Validity analysis on the primers detecting polymorphisms between 93–11 and Nipponbare. (C) Partial results from PCR amplification using different rice cultivars. (D) Phylogenetic analysis of CNV corresponding region’s sequences in different rice cultivars. Nipponbare: the first sequence of the two DNA segments in japonica cv. Nipponbare genome; ZH11-rep1, ZH11-rep2: the first and the second sequence of the two DNA segments in japonica cv. ZH11 genome; Shuhui498-rep, 93–11-rep, Minghui 63, RP Bio-226, and Zhanshan97-rep: the corresponding DNA sequence of the OsMTD1 located-CNV in different indica cultivars’ genome.
It was reported that the genome sizes of both indica and japonica subspecies have increased by greater than 2 and 6%, respectively, since their divergence from a common ancestor (Ma and Bennetzen, 2004). To find out whether this CNV contributes to intraspecific genome variations, PCR-amplified corresponding region was employed for a panel of 230 rice cultivars comprising both indica and japonica subspecies (Supplementary Table 1). The primers were designed according to the genomic sequence of both japonica cv. Nipponbare and indica cv. 93–11 to distinguish whether a tandem segmental duplication is harbored in the OsMTD1-located nearby region. A 754-bp fragment could be amplified from Nipponbare DNA with primer64 and primer65 but not from 93–11 (Figure 1A). The results showed that a clear band was obtained by two primer pairs (primer65 and primer67, primer64, and primer66) in all rice cultivars, representing the flanking sequences of the start or the end points of the corresponding 13,002-bp segment region in Nipponbare, respectively. However, a band was amplified with primer pair of primer64 and primer65 only in the ones whose genome harboring a tandem segmental duplication at the OsMTD1 gene locus nearby region (Figure 1B). After validation by PCR, only 13 japonica cultivars including Nipponbare were found to have a tandem duplication in the corresponding region of OsMTD1-located segment (Figure 1C and Supplementary Table 1).
The tandem duplication of OsMTD1-located CNV corresponding sequence only appears in some japonica cultivars, but not in all investigated indica cultivars (Figure 1C and Supplementary Table 1); thus, this CNV represents a large inserted region only in some japonica cultivars. We then used the corresponding sequence of OsMTD1-located CNV from Nipponbare as a query to search against rice database for other cultivars deposited in National Center for Biotechnology Information (see text footnote 3), including japonica cv. Zhonghua11 (ZH11) and indica cv. Zhenshan 97, Minghui 63, 93–11, Shuhui 498, and RP Bio-226. Comparative analysis showed that the sequences of OsMTD1-located CNV region in different rice cultivars were highly conserved, and the dramatic divergences were found between japonica and indica subspecies (Figure 1D and Supplementary Table 2). The corresponding fragment of OsMTD1-located CNV region includes 107 SNPs, 10 deletions, and 11 insertions, resulting in 111-bp increase in indica cv. Shuhui 498, compared with Nipponbare. However, those regions are highly conserved in indica cultivars; it reaches 99.96% identity with only a 10-bp deletion and three SNPs among all five indica cultivars. As in Nipponbare, a tandem segmental replication at the OsMTD1-located regions is found in japonica cv. Zhonghua11. However, different from the complete sequence identity of two replication regions in Nipponbare, the sequences of the two DNA segments (designed as rep1 and rep2 according to the order occurred in genome) harbor 35 SNPs or mutations in ZH11, and the identities with the sequence of Nipponbare in rep1 and rep2 are 99.74 and 99.92%, respectively. Distance and Homology matrix analysis using the sequences of OsMTD1-located CNV further showed distant evolutionary relationships among different cultivars (Supplementary Table 3).
Phenotypic Difference According to OsMTD1-Located CNV
Copy number variations locating regions that contain protein-coding genes or important regulatory elements often have phenotypic effects (Żmien’ko et al., 2014). Our previous report showed that a T-DNA insertion in OsMTD1 caused a dramatic change in tiller number (Liu et al., 2015). We therefore postulated this OsMTD1-located CNV has effects on a particular architecture trait, i.e., the tiller number. To investigate whether OsMTD1-located CNV affects tillering in rice, we performed phenotypic studies using 190 cultivars, including 108 indica and 82 japonica. Generally, indica and japonica cultivars show different tillering abilities, so the comparison between the two subspecies provided a reference to judge whether tiller number is a reliable trait for OsMTD1-located CNV conveying phenotype analysis. All cultivars were classified into four categories according to the tiller number: scale 1 (<10), scale 2 (10–20), scale 3 (21–30), and scale 4 (>30). Four indica while no japonica cultivars were classified into scale 4 (Figure 2A). On the contrary, more japonica cultivars were classified into scale 1 than indica cultivars, amounting to 55.5 and 22.5% of the investigated (Figures 2A,C,D), respectively. The average tiller number in indica was apparently higher than that in japonica cultivars (Figure 2B), indicating the selected 190 cultivars are a feasible representative group for tillering ability analysis. Further comparative analysis showed that the one-copy normal cultivars produced significantly increased tillers than the tandem duplicated cultivars (Figure 2B). In japonica cultivars, 6 of 55 one-copy normal cultivars (10.9%) showed tiller number of scale 3, whereas none was found in 13 two-copy cultivars (Figures 2C,D).
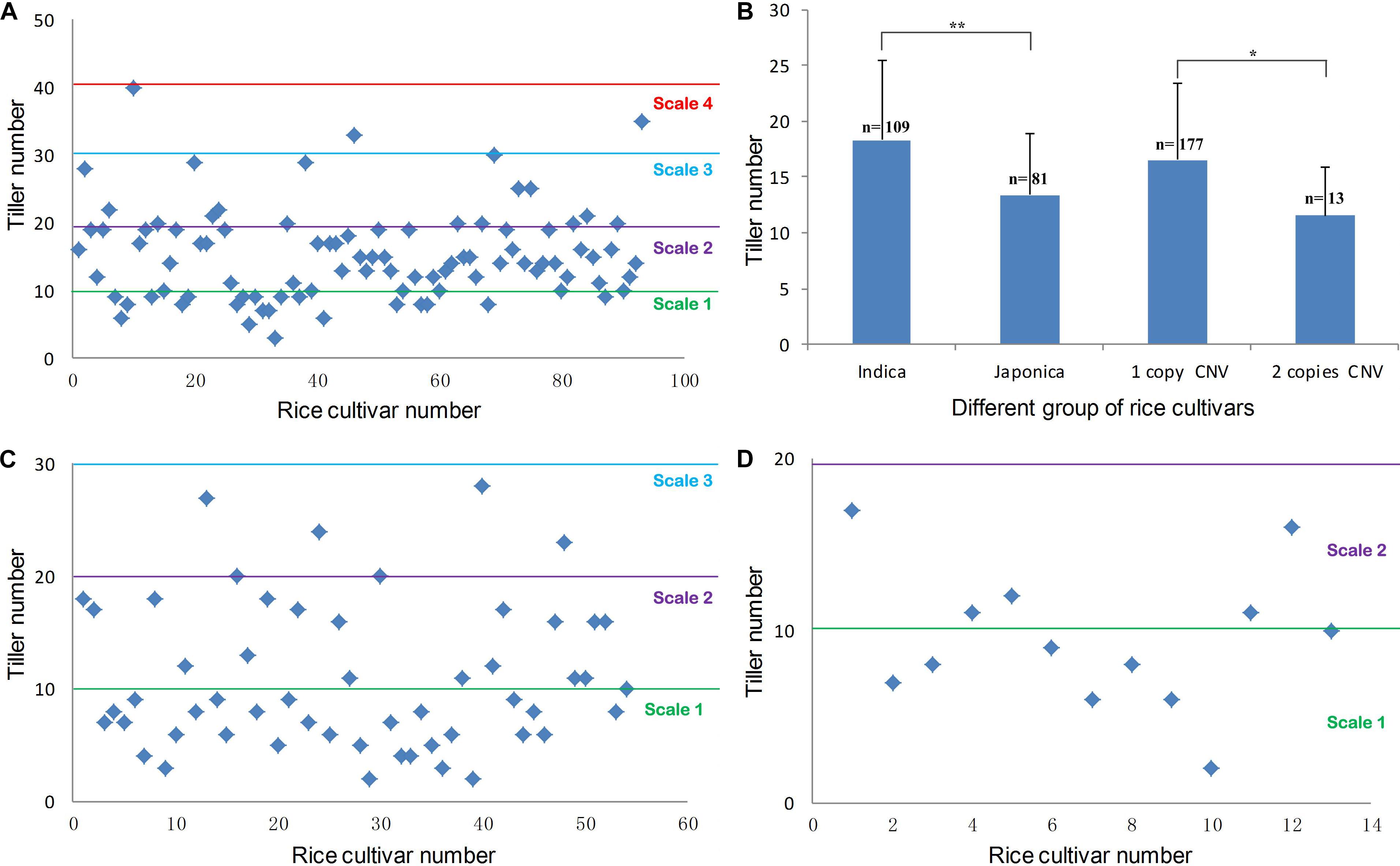
Figure 2. Phenotypic distribution of rice tiller number. (A) The maximum tiller number from 94 indica accessions. (B) The statistical result of the maximum tiller number between indica and japonica accessions, the maximum tiller number between one-copy and two-copy CNV cultivars. Statistical significance was estimated by Student t tests. *P < 0.05 and **P < 0.01. (C) The maximum tiller number in different japonica accessions with one copy of OsMTD1 nearby genome sequence. (D) The maximum tiller number in different japonica accessions with a tandem replication at OsMTD1 nearby genome sequence.
OsMTD1-Located CNV Involves in Rice Plant Architecture
The OsMTD1-located CNV region covers about 13,000 bp in different rice cultivars (Supplementary Table 2). GO analysis revealed that, apart from OsMTD1, an miR156 family member osa-miR156f was also contained in this CNV, and the pre-miR156f sequence located on downstream 3,352-bp away from OsMTD1 (Figure 1A). The comparative analysis indicated the sequences of OsMTD1 are identical in all cultivars, and pre-miR156f has identical sequences in all indica cultivars (Zhenshan 97, Minghui 63, 93–11, Shuhui 498, and RP Bio-226) but shows sequence differences in japonica cultivars (Nipponbare and ZH11); however, the final functional sequences of osa-miR156f and osa-miR156f∗ are completely identical in all investigated cultivars (Supplementary Figure 1). It is noteworthy that the osa-miR156 was confirmed to be positively correlated with rice tillering (Supplementary Figure 2; Schwab et al., 2005; Xie et al., 2006; Wang L. et al., 2015; Liu et al., 2019), whereas the tiller number comparison in cultivars with different CNV copies showed that the OsMTD1-located CNV region negatively affects rice tillering (Figures 2B,D). The osa-miR156f is aggressively antagonistic to OsMTD1-located CNV effect on tillering ability, suggesting that OsMTD1 plays a vital role in the CNV.
To better understand the role of OsMTD1 in the CNV, we further analyzed whether OsMTD1 is directly involved in tiller development; CRISPR/Cas9 genome-editing technology was employed to generate both Loc_Os08g34249 and OsMTD1 knockout lines under ZH11 background. In 34 independent T0 transgenic lines, sequence analysis revealed that each one belongs to heterogeneity accompanying an A/T/G/C insertion or deletion in OsMTD1 (Supplementary Figure 3). Surprisingly, no double-knockout mutant was obtained in the CRISPR/Cas9 editing line after self-crossing for four times, and all CRISPR/Cas9 editing lines displayed no obvious phenotypic change. We further carried out CRISPR/Cas9 and overexpression analysis in one copy japonica cv. Kitaake and obtained many independent single-base deletion or insertion transgenic lines. All of the single-base mutation lines, in which an A/T/G/C was inserted or deleted, resulting in a frame-shift mutation and the original stop codon of OsMTD1, were excluded (Supplementary Table 4). Some mutation lines (such as line A-8 and A-44) significantly increased tiller number, whereas others (such as A-3) showed no difference in tillering ability compared to the wild type. In fact, OsMTD1 overexpression significantly decreased tiller number (Figure 3), even though the lines showed different OsMTD1 increased levels (Supplementary Table 5). Intriguingly, the OsMTD1 overexpression caused multiple phenotypic defects, such as reduction in grain number and plant height, whereas the height of OsMTD1 CRISPR/Cas9 editing lines was comparable to that of the wild type plants (Supplementary Figure 4). These results under one-copy OsMTD1-located CNV background indicated that OsMTD1 plays a prominent role in the genetic control of tillering ability in rice.
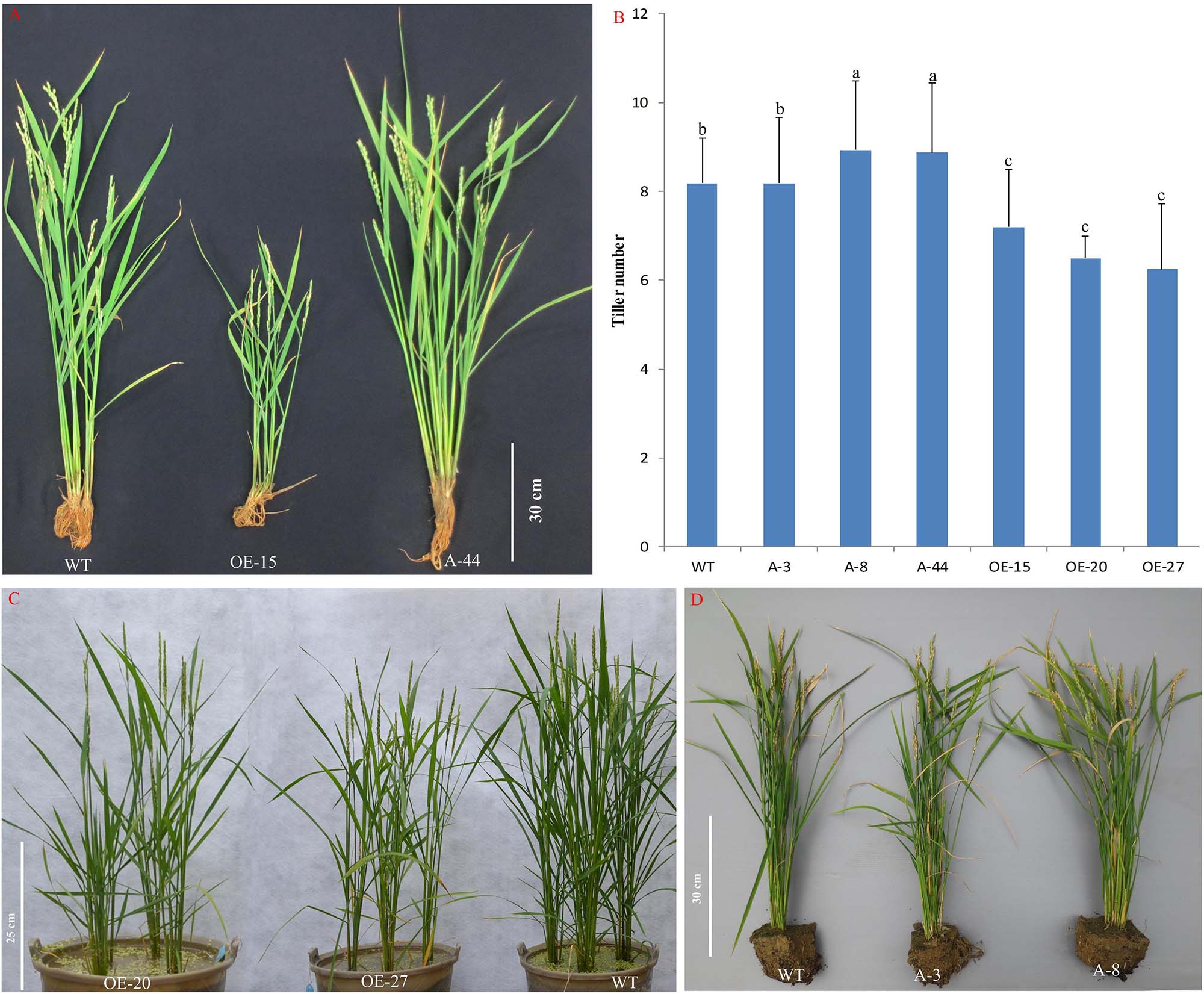
Figure 3. Effects of OsMTD1 on rice tillering. (A) Lines with different OsMTD1 expression levels and the tillering ability performed. (B) Comparative analysis of the tiller number in lines with different OsMTD1 expression levels. Statistical significance was estimated by Student t tests, and different letters indicate a significant difference (P < 0.05). (C) The OsMTD1 overexpression lines produce less tillers compared to wild type. (D) The OsMTD1 CRISPR/Cas9 editing lines show different tiller traits. WT: Kitaake; OE-15, OE-20, and OE-27: independent OsMTD1 overexpression line; A-3, A-8, and A-44: Independent OsMTD1 CRISPR/Cas9 editing line.
Leaf angle is an important agricultural trait determining rice plant architecture and ideotype (Zhou et al., 2017). In our experiments, the results suggest the pivotal role for OsMTD1 in leaf inclination. Compared with the wild type, some OsMTD1 CRISPR/Cas9 editing lines (such as A-8) showed no significant impact on leaf inclination at the mature stage, whereas overexpression of OsMTD1 significantly reduced flag leaf angle (Figure 4). Consistently, the OsMTD1 RNAi lines also presented increasing leaf inclination (Supplementary Figure 2). Conversely, overexpression of pre-miR156f increased leaf angle (Supplementary Figure 5). Meanwhile, leaf blades in miR156 knockout lines were found to be more erect than those of the wild type (Miao et al., 2019). These results also indicate that OsMTD1 and osa-miR156 play opposite roles in regulating leaf angle. Taken together, it could be concluded that OsMTD1-located CNV contributes to a compact plant architecture by influencing both tiller number and leaf angle in rice.

Figure 4. Effects of OsMTD1 on rice leaf angle. (A) Lines with different OsMTD1 expression levels show different flag leaf angles. (B) Comparative analysis the flag leaf angle in lines with different OsMTD1 expression levels. Statistical significance was estimated by Student t tests, and different letters indicate a significant difference (P < 0.05). WT: Kitaake; OE-15, OE-20, and OE-27: independent OsMTD1 overexpression line; A-3, A-8, and A-44: independent OsMTD1 CRISPR/Cas9 editing line.
OsMTD1 Inhibits the Transcript of MicroRNA156f
We next explored the underlying molecular mechanism of the OsMTD1-located CNV conveying phenotypes. Based on the experimental results mentioned above, an unexpected phenomenon is that the two elements or factors contained in the OsMTD1-located CNV region play opposite roles in controlling the architecture via tiller number and leaf angle: OsMTD1 alone negatively regulates while osa-miR156f alone positively modulates these traits.
The short miRNAs (19–23 nt in length) are processed from corresponding large pri-miRNAs. In the large pri-miRNAs containing short open reading frame sequences that encode regulatory peptides, this miRNA-encoded peptide (miPEP) increasing the transcription of the pri-miRNA was reported (Lauressergues et al., 2015). Because of the close position of the OsMTD1 to pri-miR156f, OsMTD1 might regulate pri-miR156f transcription. To test this hypothesis, we first analyzed the osa-miR156 levels in both the CRISPR/Cas9 editing and overexpression lines of OsMTD1 under Kitaake background. The results showed that the CRISPR/Cas9 editing lines produced more osa-miR156 than the wild type (Figure 5). Among the OsMTD1 overexpression plants, some lines (i.e., OE-15, OE-16, and OE-19) produced less osa-miR156, whereas some lines (i.e., OE-20, OE-26, and OE-27) produced comparable or more osa-miR156 than the wild type.
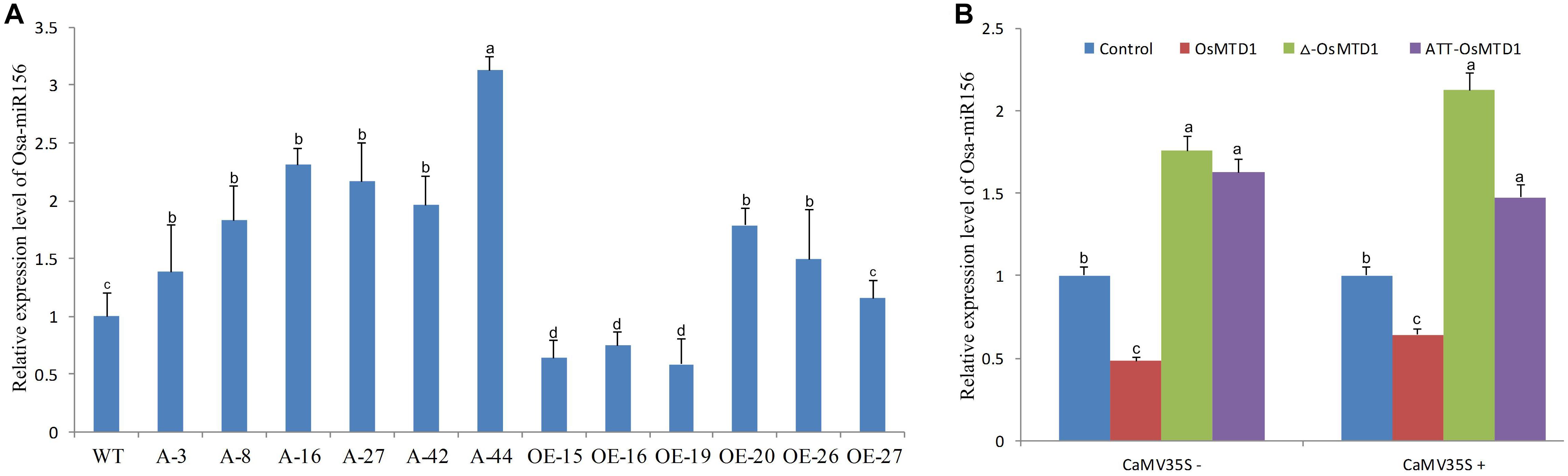
Figure 5. Effects of OsMTD1 on accumulation of miR156f. (A) The osa-miR156 relative levels in different rice lines. WT: Kitaake; A-3, A-8, A-16, A-27, A-42, and A-44: different OsMTD1 CRISPR/Cas9 editing lines; OE-15, OE-16, OE-19, OE-20, OE-26, OE-27: different OsMTD1 overexpression lines. (B) Quantification of miR156 in tobacco leaves expressing the pri-miR156f including OsMTD1 sequence (OsMTD1), or the pri-miR156f in which the OsMTD1 was deleted (Δ-OsMTD1), or in which the OsMTD1 start codon was mutated to ATT (ATT-OsMTD1). The empty vector was used as control. Statistical significance was estimated by Student t tests, and different letters indicate a significant difference (P < 0.05).
To further reveal regulatory role of OsMTD1 in osa-miR156f production, we used transformation of tobacco (Nicotiana tabacum) leaves to analyze the miR156f level by expressing both the native and mutant promoters of the pre-miR156f in pri-miR156f. Regardless of CaMV 35S promoter, compared with the amount of miR156f produced by expression of the native pri-miR156f, expression of an OsMTD1 deletion mutant showed higher miR156 abundance. Likewise, expression of a pri-miR56f in which the ATG start codon of OsMTD1 was mutated to ATT also produced higher miR156 level than expression of the native pri-miR156f (Figure 5), suggesting that OsMTD1 can inhibit the osa-miR156 accumulation when both are constructed in the same vector.
Discussion
Copy number variations are major sources of genetic variation influencing gene expression and eventually the phenotype. It is believed that there are more CNVs than chromosome structural variations among individuals, and the total number of nucleotides covered by CNVs is much larger than SNP number in the whole genome (Lupski, 2007; Yu et al., 2013). CNVs can create new genes, change gene dosage, reshape gene structures, and modify elements regulating gene expression (Henrichsen et al., 2009; Zhang et al., 2009). Here, we describe the identification of a new CNV, OsMTD1-located CNV, which involves an approximately 13,000-bp tandem duplication in DNA sequence on the eighth chromosome in different rice cultivars (Supplementary Tables 1, 2), and the corresponding sequences of OsMTD1-located CNV region in different cultivars are highly conserved, and the sequence includes two important regulator factors, i.e., OsMTD1 and pri-miR156f. The sequence of OsMTD1, osa-miR156f, and osa-miR156f∗ in all investigated cultivars is identified (Supplementary Figure 1).
Genome-scale studies indicated that CNVs significantly contribute to natural variation in plants (Yu et al., 2013; Żmien’ko et al., 2014; Bai et al., 2016). Changes in gene copy number provide the possibility to rapidly alter the dosage of a gene, which could directly cause a phenotypic variation, and as long as the new beneficial variation being selected over many generations under high selective pressure, the copy number alterations in a particular region may accumulate, and the phenotypic effects may intensify. Segmental duplications longer than 10 kb and of greater than ∼97% sequence identity can lead to local genomic instability (Stankiewicz and Lupski, 2010). As OsMTD1-located CNV covering approximately 13,000-bp DNA sequence is an evolutionarily recent duplication in some japonica cultivars after highly selective breeding programs, it is not surprising that OsMTD1-located CNV contributes to one or more currently advantageous traits in rice. In this article, we have investigated the tiller number on a panel of 190 rice cultivars, and results indicated that this CNV may have phenotypic effects on tiller development; for some, two-copy cultivars produced less tillers than one-copy cultivars (Figure 3). In the study, we also provided evidence that OsMTD1-located CNV contains two regulators, i.e., OsMTD1 and osa-miR156f, jointly regulating tillering and leaf angle (Figures 3, 4; Supplementary Figures 2, 4, 5). Together, these results indicate that OsMTD1-located CNV is important for rice plant architecture.
The essential role of a CNV depended on the genes or regulators contained in its region, so the roles of the OsMTD1-located CNV in rice phenotypes are determined by its two comprising elements: OsMTD1 and pri-miR156f. In order to reveal the function of OsMTD1, the first gene contained in OsMTD1-located CNV, CRISPR/Cas9-mediated gene editing technology was employed to knock out OsMTD1 in the one-copy Kitaake and two-copy ZH11. Some lines both with multiple tillers and large leaf inclination were found under the Kitaake background (Figures 3, 4). Although no obvious tillering phenotype change was observed in the CRISPR/Cas9 editing lines under the two-copy ZH11 background, the reason might be no knockout mutant was obtained in those experiments. Meanwhile, some OsMTD1 overexpression lines that produced less tiller and smaller leaf angle were obtained (Figures 3, 4). Combining the facts that both the OsMTD1 RNAi lines and mutant lines with a T-DNA insertion into the region of this CNV in two-copy ZH11 displayed multitillering phenotypes (Liu et al., 2015), it is clear that OsMTD1 alone could be regarded as an executive factor for tillering and leaf angle. For the second gene in OsMTD1-located CNV, i.e., pri-miR156f, we had demonstrated that the osa-miR156f plays crucial roles in rice tiller development (Liu et al., 2019), which is consistent with previous reports that high-level miR156 causes a bushy phenotype (Schwab et al., 2005; Xie et al., 2006). The effects of OsmiR156f on leaf angle are also verified (Supplementary Figure 5; Miao et al., 2019).
How OsMTD1-located CNVs regulate rice phenotype is another issue to be explored. It is believed that deletion and duplication can cause a phenotype change via several molecular mechanisms, and the commonly recognized mechanism is altering the copy number of a dosage-sensitive gene (or genes) (Lee and Lupski, 2006). The OsMTD1-located CNV enclosed two functional elements, OsMTD1 and pri-miR156f; either can act alone as a pleiotropic regulator to determine rice plant architecture in a dosage-dependent manner. However, it seems further explanation is needed for the joint regulation mechanism in plant architecture by OsMTD1, pri-miR156f, and OsMTD1-located CNV. If the CNV phenotype is conveyed by altering OsMTD1 and pri-miR156f dosage only, variation trends of the two contained genes should be the same – both increased or decreased along with the copy number change. In particular, transgenic experiments proved that changing two components of this CNV resulted in contradictory tillering phenotype: compared with the wild type, OsMTD1 overexpression lines produced less tillers (Figure 3), whereas pre-miR156f overexpression lines significantly increased tillers (Supplementary Figure 2). Similarly, OsMTD1 and pre-miR156f overexpressed lines also displayed opposite effects on leaf angle (Figure 4). All experimental data indicated that two genes contained in the CNV region, i.e., OsMTD1 and pri-miR156f, play opposite roles alone in tiller number and leaf angle. Finally, the role of OsMTD1-located CNV in rice tillering and leaf angle is apparently consistent with OsMTD1, whereas contradictory to osa-miR156f, it was implied that the transcript of OsMTD1 was more abundant in two-copy cultivars than in one-copy ones, whereas the opposite was true for pri-miR156f. Therefore, OsMTD1 exhibits the major effect and acts as a key factor in the OsMTD1-located CNV region and thus contributes to a compact architecture in rice. One possibility is the different extent of genetic buffering, as pri-miR156f belongs to a large functionally redundant gene family, and the duplication in the OsMTD1-located CNV has only minor effects compared with OsMTD1. An alternative explanation is that there might be else unknown factors that inhibit pri-miR156f transcription in the CNV.
In addition to changes in gene dosage, many other mechanisms are responsible for the potential effects of CNVs, including reshaping of the gene structure and modification of the elements that regulate gene expression (Henrichsen et al., 2009; Zhang et al., 2009). One possible mechanism is the position effect; i.e., a CNV encompassed regulatory elements might regulate a gene even if they are several Mbs away (Żmien’ko et al., 2014). In the corresponding region of OsMTD1-located CNV, OsMTD1 and pri-miR156f are neighboring genes approximately 3.3 kb apart (Figure 1). Hence, we reasoned that OsMTD1 can inhibit the transcripts of pri-miR156f via position effect. Validation for the unpredictable effects of the two distant components in the CNV region is informative. We hypothesized that OsMTD1 is a regulator repressing pri-miR156f transcription and provided some evidence. Compared with wild type, the OsMTD1 CRISPR/Cas9 editing lines showed higher osa-miR156 level, whereas some OsMTD1 overexpression lines showed lower osa-miR156 abundance (Figure 5A). Some OsMTD1 overexpression lines didn’t produce less osa-miR156 than wild type as expected, the reason might be that the insertion location is too far away from pri-miR156f in the genome. Furthermore, transformation results in tobacco leaves also showed that the native pri-miR156f vector produced less miR156 compared to the deleted and mutated types (Figure 5B). The above evidence implied that OsMTD1 can inhibit its neighboring pri-miR156f expression in vivo by the position effect. Different from previous report that miRNA-encoded peptide can enhance their corresponding pri-miRNA transcription (Lauressergues et al., 2015), OsMTD1 represses pri-miR156f transcription. Thus, our work revealed a novel regulatory mechanism for manipulating osa-miR156 level to control tiller number and leaf angle in rice.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
QL and LX conceived and designed the research. QL, JX, YZ, YM, and WK performed the experiments. QL, ZH, SX, YS, JT, CH, and WL analyzed the data. JX and X-FY investigated rice tiller number. C-ML provided technical support. QL, RW, KP, and LX wrote the manuscript. All authors read and approved the article.
Funding
This work was supported by the National Natural Science Foundation of China (91317312, 31900387, and 31570372), National Key Research and Development Program-Seven Major Crops Breeding Project (2016YFD0101803), and Scientific Research Fund of Hunan Provincial Education Department (16K042).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Li-Jia Qu and Prof. Genji Qin (Peking University) for their valuable suggestions. We also thank Dr. Jin Miao, Dr. Dongshu Guo, Dr. Xiaoru Wei, Dr. Tong Wei, and Dr. Qing Tao (Peking University) for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.620282/full#supplementary-material
Supplementary Figure 1 | The sequence comparison result of OsMTD1 and pre-OsmiR156f in different rice cultivars.
Supplementary Figure 2 | The CNV containing components OsMTD1 and OsmiR156f influenced rice tiller development. WT: wild type; OsMTD1-RNAi: OsMTD1 RNA interference line; OsmiR156f-OE: OsmiR156f overexpression line.
Supplementary Figure 3 | The OsMTD1 mutagenesis analysis in rice using CRISPR/Cas9 system under ZH11 background.
Supplementary Figure 4 | Effects of OsMTD1 on rice plant height. Statistical significance was estimated by Student t tests, and different letters indicate a significant difference (P < 0.05).
Supplementary Figure 5 | Effects of OsmiR156f on rice leaf angle.
Supplementary Table 1 | The surveying of CNV region and tiller number in different rice cultivars.
Supplementary Table 2 | DNA polymorphism analysis of OsMTD1-located CNV region in various rice cultivars. Nipponbare rep1, Nipponbare rep2: the first and the second sequence of the two DNA segments in Nipponbare genome; ZH11 rep1, ZH11 rep2: the first and the second sequence of the two DNA segments in ZH11 genome; Shuhui 498, 93–11, Minghui 63, RP Bio-226, and Zhenshan 97: the DNA sequence of the OsMTD1-located CNV in corresponding indica cultivar’s genome.
Supplementary Table 3 | The result of Distance and Homology matrix analysis. Evolutionary analysis were conducted in DNAMAN8, and the analysis involved eight sequences: Nipponbare rep1: the first sequence of the two DNA segments in Nipponbare genome; ZH11-rep1, ZH11-rep2: the first and the second sequence of the two DNA segments in ZH11 genome; Shuhui 498, 93–11, Minghui 63, RP Bio-226, and Zhenshan 97: the DNA sequence of the OsMTD1-located CNV in corresponding indica cultivar’s genome.
Supplementary Table 4 | The mutation information of OsMTD1 in different CRISPR/Cas9 editing lines.
Supplementary Table 5 | The OsMTD1 level in different overexpression lines.
Footnotes
- ^ http://rice.plantbiology.msu.edu/
- ^ http://rice.hzau.edu.cn/
- ^ http://www.ncbi.nlm.nih.gov/
- ^ http://www.cdna01.dna.affrc.go.jp/cDNA
References
Bai, Z., Chen, J., Liao, Y., Wang, M., Liu, R., Ge, S., et al. (2016). The impact and origin of copy number variations in the Oryza species. BMC Genom. 17:261. doi: 10.1186/s12864-016-2589-2
Cong, B., Barrero, L. S., and Tanksley, S. D. (2008). Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40, 800–804. doi: 10.1038/ng.144
Feuk, L., Carson, A. R., and Scherer, S. W. (2006). Structural variation in the human genome. Nat. Rev. Genet. 7, 85–97.
Henrichsen, C. N., Chaignat, E., and Reymond, A. (2009). Copy number variants, diseases and gene expression. Hum. Mol. Genet. 18, R1–R8.
Hiei, Y., and Komari, T. (2008). Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3, 824–834. doi: 10.1038/nprot.2008.46
Hinds, D. A., Kloek, A. P., Jen, M., Chen, X., and Frazer, K. A. (2006). Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 38, 82–85. doi: 10.1038/ng1695
Lauressergues, D., Couzigou, J.-M., Clemente, H. S., Martinez, Y., Dunand, C., Bécard, G., et al. (2015). Primary transcripts of microRNAs encode regulatory peptides. Nature 520, 90–93. doi: 10.1038/nature14346
Lee, J. A., and Lupski, J. R. (2006). Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron 52, 103–121. doi: 10.1016/j.neuron.2006.09.027
Li, S., Wang, S., Deng, Q., Zheng, A., Zhu, J., Liu, H., et al. (2012). Identification of genome-wide variations among three elite restorer lines for hybrid-Rice. PLoS One 7:e30952. doi: 10.1371/journal.pone.0030952
Li, Y., Xiao, J., Wu, J., Duan, J., Liu, Y., Ye, X., et al. (2012). A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol. 196, 281–291.
Liu, Q., Shen, G., Peng, K., Huang, Z., Tong, J., Kabir, M. H., et al. (2015). The alteration in the architecture of a T-DNA insertion rice mutant osmtd1 is caused by up-regulation of MicroRNA156f. J. Integrat. Plant Biol. 57, 819–829. doi: 10.1111/jipb.12340
Liu, Q., Su, Y., Zhu, Y., Peng, K., Hong, B., Wang, R., et al. (2019). Manipulating osa-MIR156f expression by D18 promoter to regulate plant architecture and yield traits both in seasonal and ratooning rice. Biol. Proced. 21:21.
Locke, D. P., Sharp, A. J., McCarroll, S. A., McGrath, S. D., Newman, T. L., Cheng, Z., et al. (2006). Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 79, 275–290. doi: 10.1086/505653
Lupski, J. R. (1998). Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14, 417–422. doi: 10.1016/s0168-9525(98)01555-8
Ma, J., and Bennetzen, J. L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. U.S.A. 101, 12404–12410. doi: 10.1073/pnas.0403715101
Miao, C., Wang, Z., Zhang, L., Yao, J., Hua, K., Liu, X., et al. (2019). The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat. Commun. 10:3822.
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236.
Schwab, R., Palatnik, J. F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527. doi: 10.1016/j.devcel.2005.01.018
Sebat, J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., et al. (2004). Large-scale copy number polymorphism in the human genome. Science 305, 525–528. doi: 10.1126/science.1098918
Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S., et al. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028. doi: 10.1038/ng.169
Stankiewicz, P., and Lupski, J. R. (2010). Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61, 437–455. doi: 10.1146/annurev-med-100708-204735
Varkonyi-Gasic, E., Wu, R., Wood, M., Walton, E. F., and Hellens, R. P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12. doi: 10.1186/1746-4811-3-12
Wang, L., Sun, S., Jiye, J., Fu, D., Yang, X., Weng, X., et al. (2015). Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 15504–15509. doi: 10.1073/pnas.1521949112
Wang, Y., Xiong, G., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. doi: 10.1038/ng.3346
Weckselblatt, B., and Rudd, M. K. (2015). Human structural variation: mechanisms of chromosome rearrangements. Trends Genet. 31, 587–599. doi: 10.1016/j.tig.2015.05.010
Xie, K., Wu, C., and Xiong, L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142, 280–293. doi: 10.1104/pp.106.084475
Yu, P., Wang, C., Xu, Q., Feng, Y., Yuan, X., Yu, H., et al. (2013). Genome-wide copy number variations in Oryza sativa L. BMC Genom. 14:649. doi: 10.1186/1471-2164-14-649
Zhang, F., Gu, W., Hurles, M. E., and Lupski, J. R. (2009). Copy number variation in human health, disease, and evolution. Annu. Rev. Genom. Hum. Genet. 10, 451–481.
Zhang, L., Yu, H., Ma, B., Liu, G., Wang, J., Wang, J., et al. (2017). A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 8:14789.
Zhou, L., Xiao, L., and Xue, H. (2017). Dynamic cytology and transcriptional regulation of rice lamina joint development. Plant Physiol. 174, 1728–1746. doi: 10.1104/pp.17.00413
Keywords: copy number variation, OsMTD1, rice, plant architecture, pri-miR156f
Citation: Liu Q, Xu J, Zhu Y, Mo Y, Yao X-F, Wang R, Ku W, Huang Z, Xia S, Tong J, Huang C, Su Y, Lin W, Peng K, Liu C-M and Xiao L (2021) The Copy Number Variation of OsMTD1 Regulates Rice Plant Architecture. Front. Plant Sci. 11:620282. doi: 10.3389/fpls.2020.620282
Received: 22 October 2020; Accepted: 31 December 2020;
Published: 11 February 2021.
Edited by:
Mingli Xu, University of South Carolina, United StatesReviewed by:
Gang Wu, Zhejiang Agriculture and Forestry University, ChinaShengben Li, Nanjing Agricultural University, China
Copyright © 2021 Liu, Xu, Zhu, Mo, Yao, Wang, Ku, Huang, Xia, Tong, Huang, Su, Lin, Peng, Liu and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Langtao Xiao, bHR4aWFvQGh1bmF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Qing Liu
Qing Liu Jinke Xu1†
Jinke Xu1† Ruozhong Wang
Ruozhong Wang Chao Huang
Chao Huang Chun-Ming Liu
Chun-Ming Liu Langtao Xiao
Langtao Xiao