- 1College of Tobacco Science, Guizhou University, Guiyang, China
- 2The Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region, Ministry of Education, Institute of Agro-Bioengineering, Guizhou University, Guiyang, China
- 3College of Tea Science, Guizhou University, Guiyang, China
Drought is the primary disaster that endangers agricultural production, including animal husbandry, and affects the distribution, growth, yield, and quality of crops. Previous study had revealed that DIP, as a potential regulator of DBF activity, played an important role in response to drought stress in maize. In this study, a total of 67 DIPs were identified from seventeen land plants, including six tobacco DIPs (NtDIPs). NtDIP6 gene was further selected as a candidate gene for subsequent experiments based on the phylogenetic analysis and structural analysis. The transgenic tobacco and poplar plants over-expressing NtDIP6 gene were generated using the Agrobacterium- mediated method. Although there was not phenotypic difference between transgenic plants and wild-type plants under normal conditions, overexpression of the NtDIP6 gene in transgenic tobacco and poplar plants enhanced the drought tolerance under drought treatments in comparison with the wild type. The content of antioxidant defense enzymes peroxidase (POD), catalase (CAT), and the photosynthetic rate increased in NtDIP6-Ox transgenic tobacco and poplar plants, while the content of malondialdehyde decreased, suggesting that the overexpression of NtDIP6 enhances the antioxidant capacity of transgenic poplar. Furthermore, the results of qRT-PCR showed that the level of expression of drought-related response genes significantly increased in the NtDIP6-Ox transgenic plants. These results indicated that NtDIP6, as a positive response regulator, improves drought stress tolerance by scavenging superoxide via the accumulation of antioxidant defense enzymes.
Introduction
As sessile organisms, the growth and development of plant is severely restricted by environmental stresses, such as drought, high salinity, and high and low temperatures. Thus, plants have evolved complex mechanisms to respond and adapt to different environmental stresses at the physiological and biochemical levels (Figueiredo et al., 2012). Among the various abiotic stresses, drought stress is the major factor that hinders the growth and development of crops throughout the world (Farooq et al., 2009). Recent studies revealed that gene expression, transcriptional regulation, and signal transduction are involved in the regulation of responses of plants to drought (Zhu et al., 2010).
DBF (Dehydration responsive element binding factor), as a transcription factor, was introduced by Kizis and Pages (2002), activates drought stress tolerance genes in many plants. Moreover, the DBF is also a part of the Apetala 2/Ethylene Response Factor (AP2/ERF) transcription factor family and induces the rab17 (responsive to abscisic acid) gene expression under drought stress conditions (Kizis and Pages, 2002). In maize, DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Xu et al. (2008) identified three new DBF genes in T. aestivum (named TaAIDFs, T. aestivum abiotic stress-induced DBFs) by screening a wheat cDNA library after drought treatment.
A previous study found that DBF1-interactor protein 1 (DIP1) that contained two conserced core domains (R3H and SUZ) was localized in the cytoplasm and regulates the activity of DBF1 in stress responses (Saleh et al., 2006). The R3H domain is highly conserved and widely distributed in many organisms, including eubacteria, plants, fungi, and metazoans (Saleh et al., 2006). This domain is involved in the binding of polynucleotides, including DNA, RNA, and single stranded DNA (Liepinsh et al., 2003). Moreover, the SUZ domain is a conserved RNA-binding domain found in eukaryotes and enriched in positively charged amino acids. Although Saleh et al. (2006) had revealed that DIP protein was interacted with DBF protein using yeast two-hybrid analyses, the gene function of DIP1 has not been further evaluated.
In this study, we identified DIP family genes in tobacco and other land plants. We constructed a phylogenetic tree and performed protein and gene structure analyses. We also generated transgenic tobacco and poplar plants overexpressing NtDIP6 under the control of the 35S–CaMV promoter to explore the phenotypic changes and drought resistance of transgenic plants compared with wild type (WT). In addition, we determined the activities of peroxidase (POD) and catalase (CAT) and the content of malondialdehyde (MDA) in transgenic tobacco and poplar plants to confirm the capacity of antioxidation. We further investigated the expression pattern of drought response genes (PtDBF1, PtWRKY1, PtWRKY3, and PtNCED1) using RT-qPCR. The results provide valuable information on the roles of NtDIP6 in the regulation of drought tolerance.
Materials and Methods
Homolog Identification
To explore the evolution relationship of DIP genes in land plants, seventeen sequenced species, representing the major lineage of land plants (Supplementary Table S1), were selected and analyzed. The complete genome sequences and corresponding annotation information for seventeen land plants were downloaded from the JGI and NCBI databases. The hidden Markov model profiles of the R3H (PF01424) and SUZ (PF12752) from the Pfam database was used as the queries to search for homologous sequences in the proteome data sets. Sequences with an E-value of 10–4 were considered candidates. After removing the redundant sequences and short proteins (lengths < 100 aa), the candidates were also confirmed the presence of the R3H and SUZ domains in each candidate using the SMART databases1 with an E-value cut off 10–10.
Phylogenetic Trees, Motif Distribution, and Gene Structure
The full-length amino acid sequences of DIPs from seventeen land plants were aligned using MAFFT with default parameters (Katoh and Standley, 2013). A maximum-likelihood phylogeny based on the MAFFT alignment was constructed using the PhyML software under the WAG evolution model (v. 3.0, Guindon et al., 2010). Bootstrapping with 100 replicates was used to test the reliability of trees obtained (Wang et al., 2019). A phylogenetic tree was visualized using FIGTREE2. MEME software was used to identify conserved motifs with the default parameters3. Exon/intron information for the DIP genes from six flowering plants was extracted from the corresponding genome annotation database. The data were then plotted using MapInspect software4.
Construction of Plasmids and Transformation of Plants
The complete coding sequence of NtDIP6 was amplified using primers (forward primer: 5′-CAGTTTGAGTTCCCA CATTTCC-3′ and reverse primer: 5′-CTGACCATCCA CCACATTATCC-3′) from the cDNA of N. tabacum and cloned into the HindIII and XbaI sites of the expression vector pSH737. Transgenic tobacco and poplar plants were generated by the Agrobacterium tumefaciens-mediated transformation of strain LBA4404 using methods described previously (Fillatti et al., 1987). The transgenic tobacco and poplar plants were planted in pots with nutrient soil in soil at 25°C under 120 μmol m–2 s–1 irradiance, 50% relative humidity and a 16 h light/8 h dark photoperiod in a growth chamber.
Drought Stress Treatment
After 1 month of growth in Murashige and Skoog (MS) media, WT and transgenic tobacco and poplar plants constitutively overexpressing NtDIP6 were transferred into pots as described in the previous section under a 16 h light/8 h dark photoperiod at a constant temperature of 25 °C, 50% relative humidity, and light intensity of 120 μmol photons m–2 s–1 and fertilized once a week. After 1 month, three transgenic tobacco and poplar lines (each line contained three plants) and three wild type (WT) plants were used for the drought treatment experiment. This experiment was repeated three times. Similar results were observed in all the replicates. Photos of the drought-stressed plants (one wild type plants and three transgenic lines) were taken after 2 weeks of drought treatment. The collected samples were immediately frozen in liquid nitrogen, and their RNA was extracted for subsequent experiments.
qRT-PCR Analyses
Total RNA of transgenic poplar plants was extracted using a Quick RNA Isolation Kit (Huayueyang, Beijing, China). The total RNA was treated with DNaseI according to the manufacturer’s instructions. One microgram of total RNA of each sample was used as a template for cDNA synthesis using a SuperScript gDNA Removal cDNA Synthesis Kit (Huayueyang). For quantitative real-time reverse transcriptase–PCR (RT-qPCR), SYBR Premix Ex Tag was used on a CFX Connect Real-Time PCR system (Bio-Rad, Shanghai, China).
The qRT-PCR reactions were conducted in 96-well plates. The melting temperature of the products was determined to verify the specificity of the amplified fragments. The primers used for qRT-PCR are listed in Supplementary Table S2. The results were analyzed by the ΔΔCT method using PtActin as the reference gene. All the qRT-PCR data points were established as three biological replicates, and three technical replicates were conducted.
Determination of Enzyme Activities
Transgenic tobacco lines, poplar lines and wild type (WT) seedlings were treated with PEG 6000 to simulate drought stress for 15 d, and the treated plant leaves (the fifth leaf from the base to the top) were used to detect the activities of POD and CAT enzymes using a kit (Beijing Suolaibao Biotechnology Co., Ltd., Beijing, China). Each lines contained three individuals.
Results
Genome-Wide Identification and Phylogenetic Analysis of DIP Proteins in N. tabacum
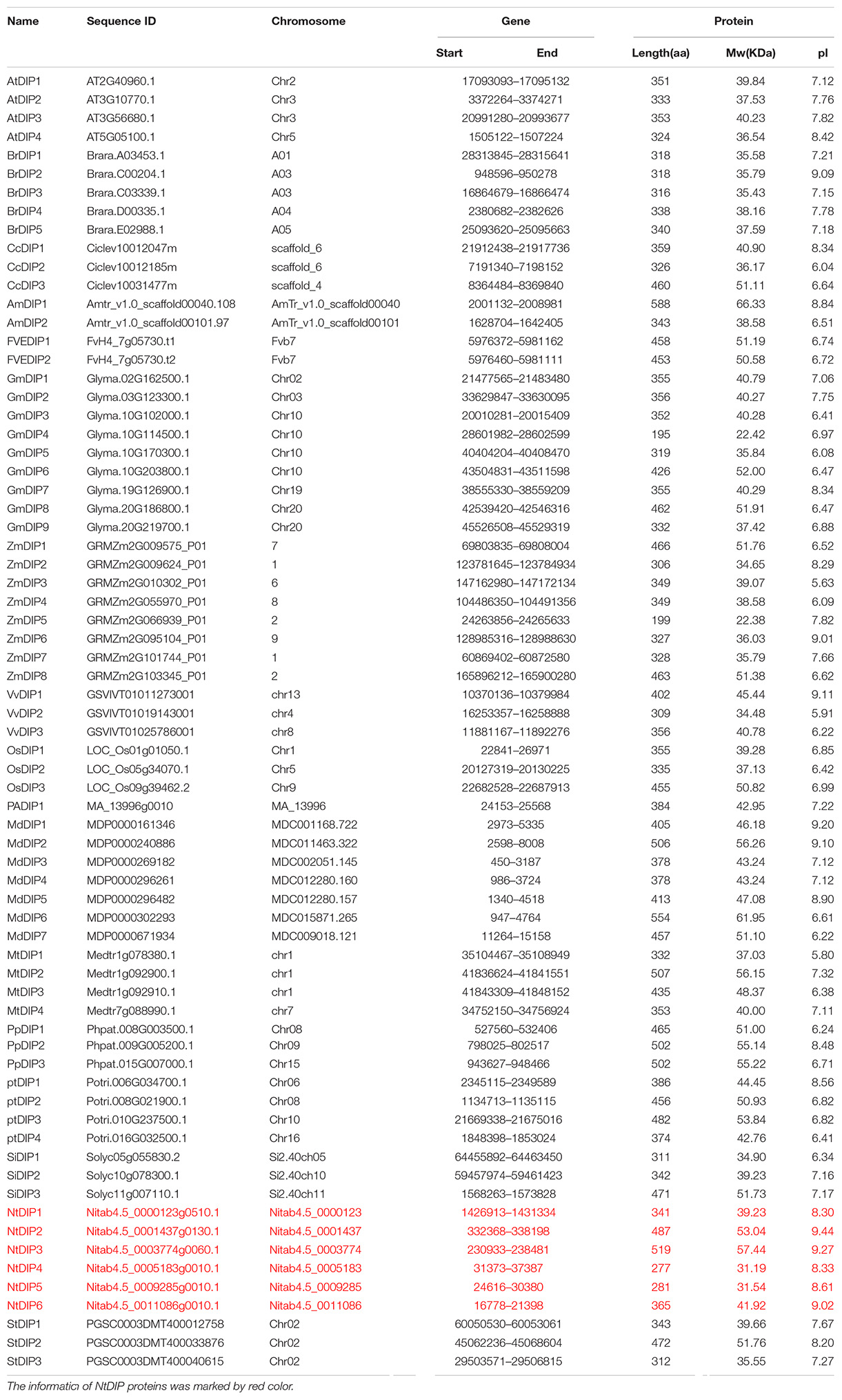
Two conserved domains [R3H (PF01424) and SUZ (PF12752)] of the DIP family proteins were downloaded from the Pfam database (v. 33.1) and used as queries to search the seventeen land plant genomes using the HMMER package (Supplementary Table S1). A total of 67 proteins were considered as candidates, and those proteins contained R3H and SUZ domains. Basic information of the DIP proteins was listed in Table 1, including the protein ID in NCBI, chromosomal distribution, sequence length, isoelectric point, and protein molecular mass, and gene length. In tobacco, NtDIP3 and NtDIP4 were on chromosome 11; NtDIP1 was on chromosome 24, and NtDIP2, 5, 6 were on three scaffolds (Nitab4.5_0001437, Nitab4.5_0009285, and Nitab4.5_0011086), respectively. The length of DIP proteins ranged from 281 to 589 amino acids; the isoelectric point was between 8.3 and 9.27, and the molecular mass was 31.19∼57.44 kDa (Table 1).
To study the deeper relationships among the DIP family members in tobacco, a maximum-likelihood phylogenetic tree was constructed based on the MAFFT alignment of these DIP proteins from seventeen land plants. According to the phylogeny, the DIP proteins could be divided into three groups: I, II, and III (Supplementary Figure S1). The group I was mainly composed of the members of flowering plants except A. trichopoda, group II includes almost all the species investigated except the members of f A. thaliana and B. rapa. The group III contained the members of twelve species investigated except the members of A. thaliana, B. rapa, P. trichocarpa, F. vesca, and P. abies. This result suggested that the members of groups II and III was more ancient than the members of goup I. In addition, the topological structure of the phylogenetic tree is further verified by the motif analysis of DIPs (Supplementary Figure S2). In addition, the two tobacco genes (NtDIP1 and NtDIP6) were clustered with the maize DIP1 and with putative proteins [AtDIP3 (AtNP_191227) and AtDIP1 (AtNP_565947)] from Arabidopsis (Figure 1 and Supplementary Figure S1).
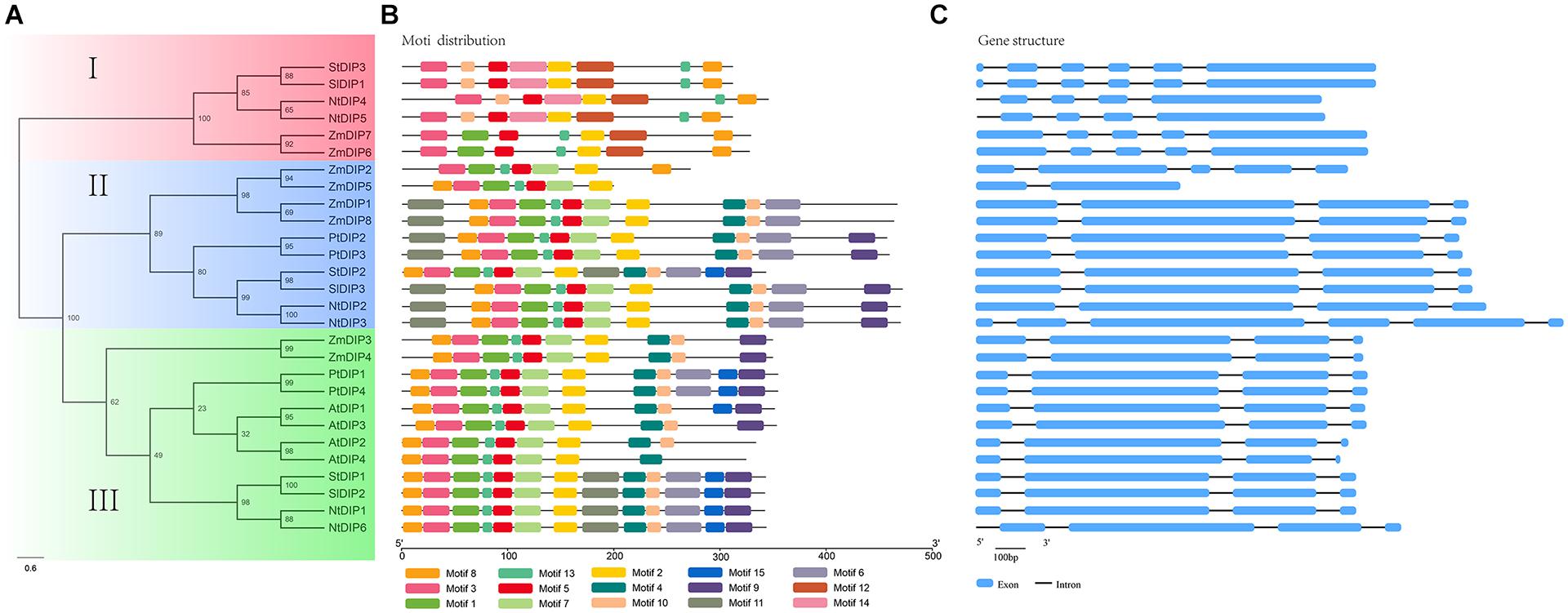
Figure 1. Maximum-likelihood phylogeny (Left), protein motif (Middle), and gene structure (Right) analyses of the DIP proteins from tobacco and five other flowering plants. Left: the phylogeny was constructed based on the amino acid sequences of full-length DIP proteins with 100 bootstrapping replicates. Rectangular color blocks represent different groups (Left). Middle: the motif analysis was performed using PhyML. Right: The gene structure analysis was conducted based on genome annotation. Blue boxes represent exons, and Dashes represent introns.
Motif and Gene Structures of the DIP Genes
The phylogenetic analysis, protein motif, and gene structures of the DIP family genes from six representative flowering plants were further analyzed (Figure 1). A total of 15 conserved motifs (motifs 1–15) were identified using the MEME suite (Figure 1B and Supplementary Figure S3). Motifs 1, 3, 5, 8, and 13 correspond to the R3H domain, and motifs 2 and 7 correspond to the SUZ domain and have been identified in nearly all DIP proteins. Motif 12 and 14 were specifically distributed in group I, while motif 5 was primarily distributed in group III. Motifs 4, 6, 7, 9, and 11 were commonly distributed in groups II and III. In addition, motif 10 was only present in the DIP family proteins in dicots. The analysis of gene structure indicated that 21 (75%) of the 28 DIP family genes possess three introns (Figure 1C). The average number of introns per intron-containing DIP genes was 3.25. Only one gene contained a single intron, while the others contained four to five introns. The DIP genes in same group displayed a similar gene structure (Figure 1).
The Overexpression of NtDIP6 in Tobacco and Poplar Enhanced Drought and Oxidative Stress Tolerance
We further investigate the expression pattern of NtDIP1 and NtDIP6 genes under drought stress, found that the expression level of these two genes was increased after drought treatment, and the expression level of NtDIP6 is 1.3 times the expression level of NtDIP1 (Supplementary Figure S4). Thus, we select NtDIP6 as a candidate gene for functional verification. Transgenic tobacco and poplar plants was generated overexpressing NtDIP6 gene under the control of 35S promoter after PCR detection and GUS staining (Supplementary Figure S5). Transgenic plants overexpressing NtDIP6 displayed the same phenotypes in comparison with the WT plants under the normal conditions (data not shown). Drought stress is one of the limiting factors that inhibits the yields of crops throughout the world. Among various techniques that enhance the drought resistance of plants, the use of genetic modification technology has proven to be promising (Hervé and Serraj, 2009). Thus, the drought stress tolerance was examined on WT and transgenic plants after 15 days of drought treatment (Figures 2, 3). Compared with the initial treatment, the growth and development of wild type tobacco was severely inhibited after 15 days of drought treatment, and the leaves displayed some degree of wilting, while there is no significant change in the transgenic tobacco plants (Figure 2). In addition, the leaves of wild type poplars had begun to wilt after 10 days of drought treatment in comparison with initial treatment, while no change was found in the leaves of transgenic poplar plants. Although the WT and transgenic poplars wilted, the wilted phenotype of wild type after drought treatment for 20 days was more severe than that of transgenic poplar plants (Figure 3).

Figure 2. Phenotypic differences between the WT and transgenic tobacco plants under drought stress. The WT (Left) and 35S:NtDIP6 overexpressing (L3, 7 and 10, right) tobacco plants after 2 weeks of drought treatment.
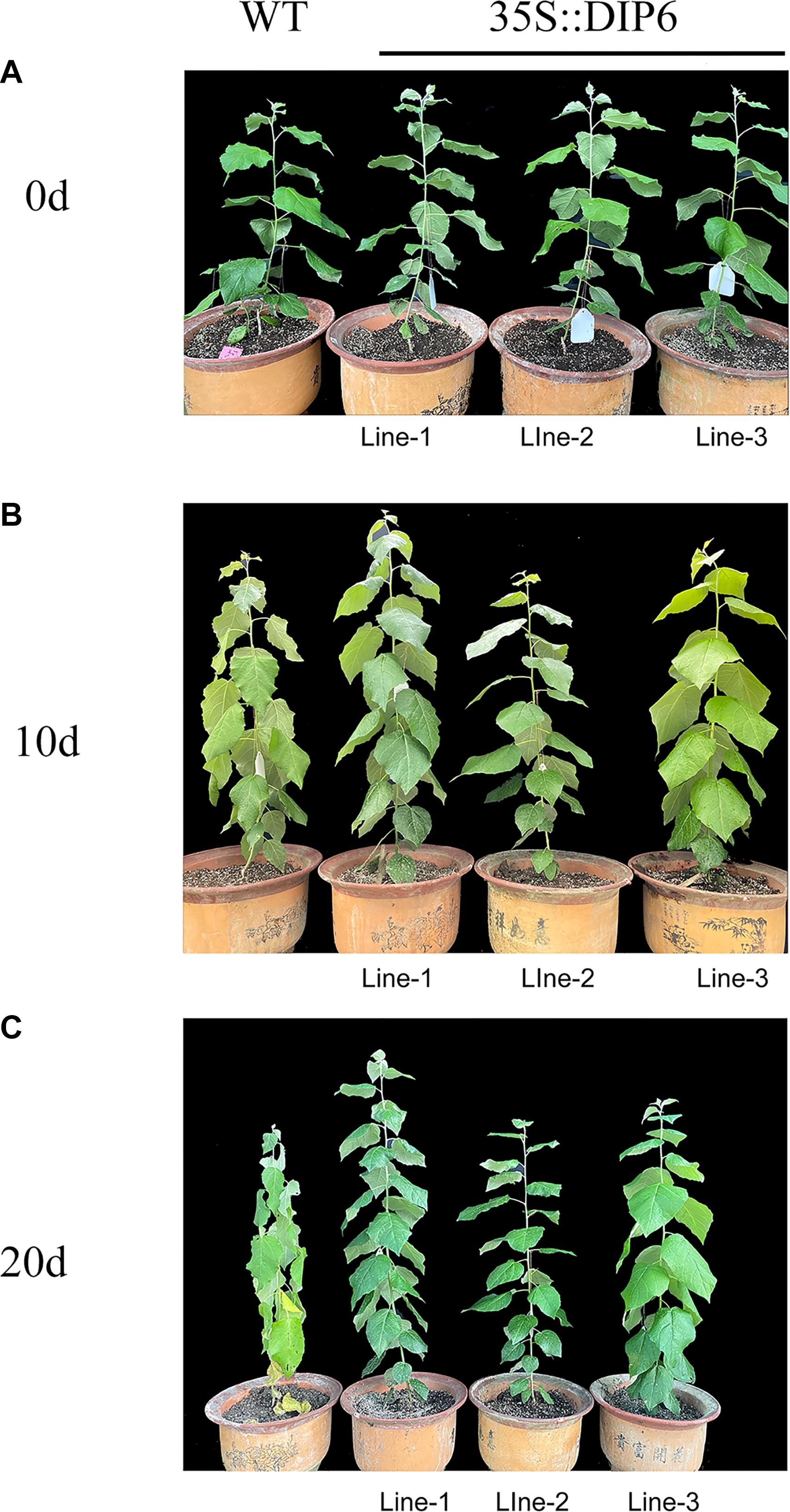
Figure 3. Phenotypic differences between the WT and transgenic poplar plants under drought stress. (A) The WT (left) and 35S:NtDIP6 overexpressing (L1–3, right) poplar plants before drought treatment, (B) 10 days after the stop of watering, and (C) 20 days after the stop of watering.
Oxidative stress tolerance is a basis of tolerance to many abiotic stresses, such as drought and extreme temperatures. To further compare the difference in antioxidant capacity between wild type and transgenic tobacco and poplar plants overexpressing NtDIP6, we measured the content of MDA and the activities of POD and CAT in WT and transgenic tobacco and poplar plants after PEG treatment. Compared with wild type tobacco, the content of CAT in transgenic tobacco lines 7 and 10 was increased by 2.07 and 2.61 times (Figure 4A), and the content of POD in transgenic tobacco lines 7 and 10 was increased by 1.63 and 1.69 times in comparison after PEG treatment, respectively (Figure 4B). In addition, the content of MDA in transgenic tobacco lines 7 and 10 was decreased by 0.34 and 0.47, respectively (Figure 4C). The content of CAT and POD in the transgenic poplars were higher than those in the WT plants (Figures 5A,B). In addition, the content of MDA in the transgenic poplar was significantly lower than that in the wild type (Figure 5C). These results indicated that the transgenic plants were more tolerant to drought than the wild type.
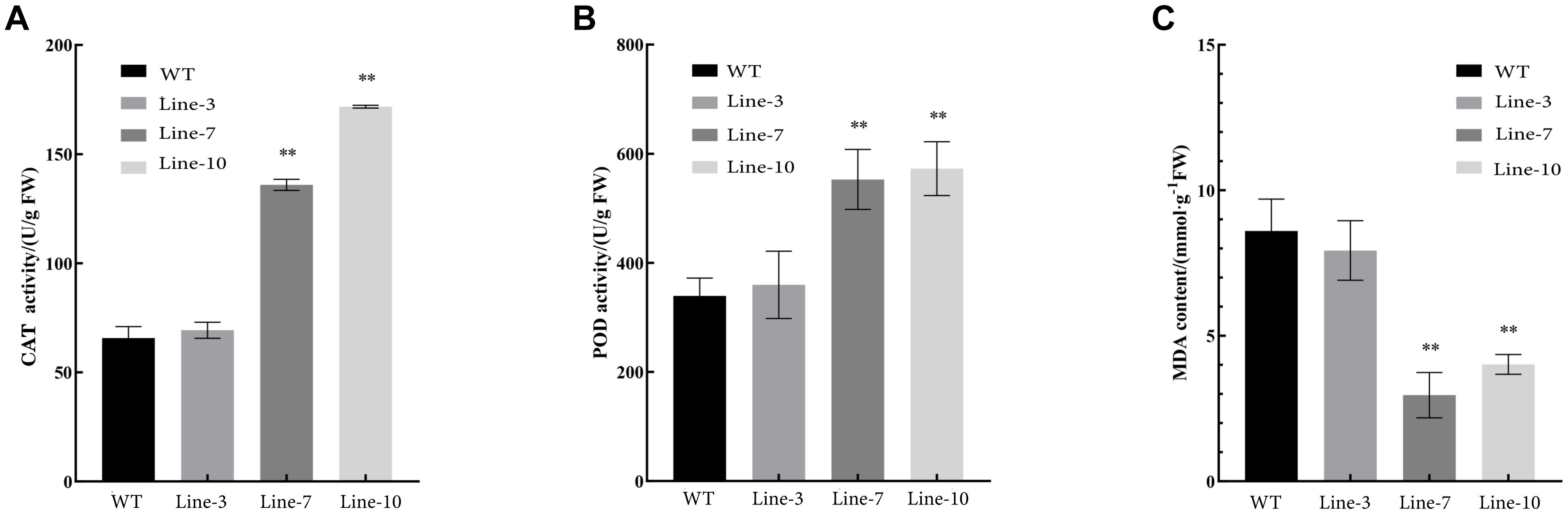
Figure 4. The activities of POD and CAT and the content of MDA in transgenic and wild type tobacco plants subjected to drought. (A,B) Higher activities of catalase (CAT) and peroxidase (POD) were observed in the leaves of transgenic tobaccos than in the leaves of WT plants after PEG treatment. (C) A lower content of malondialdehyde (MDA) in transgenic tobacco than in the leaves in WT after PEG treatment. Asterisks above the error bars indicate significant differences between transgenic tobacco lines and WT (∗p < 0.05; ∗∗p < 0.01). WT, wild type; Lines 3, 7, and 10, transgenic poplar lines 3, 7, and 10.
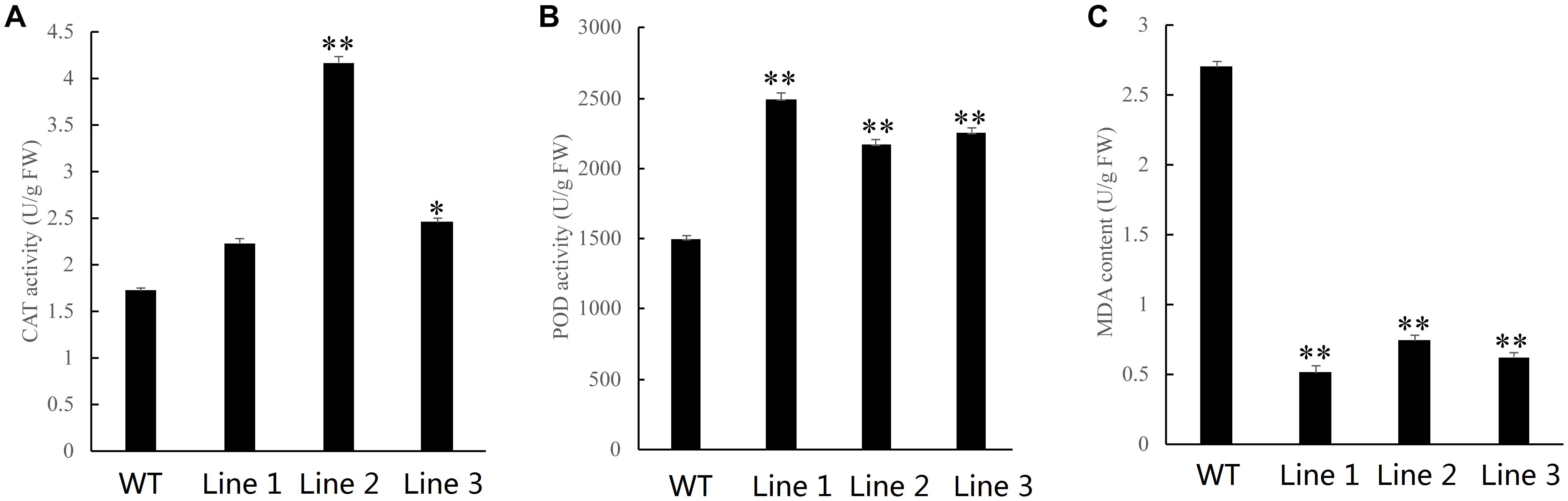
Figure 5. The activities of POD and CAT and the content of MDA in transgenic and wild type poplar plants subjected to drought. (A,B) Higher activities of catalase (CAT) and peroxidase (POD) were observed in the leaves of transgenic poplars than in the leaves of WT plants after PEG treatment. (C) A lower content of malondialdehyde (MDA) in transgenic poplars than in the leaves in WT after PEG treatment. Asterisks above the error bars indicate significant differences between transgenic poplar lines and WT (*p < 0.05; **p < 0.01). WT, wild type; Lines 1–3, transgenic poplar lines 1, 2, and 3.
Improved Photosynthetic Capacity in Transgenic Poplars That Overexpress NtDIP6
Photosynthesis is the complex process by which plants use solar energy to produce glucose from carbon dioxide and water. Improving the water use efficiency of plants under drought stress is one of the ways to enhance plant drought resistance. Previous studies have shown that a high photosynthetic capability decreases the stomatal conductance and closes stomata, which increase the intrinsic and instantaneous water-use efficiency under short term drought stress (Guo et al., 2014; Bian et al., 2019). We compared the net photosynthetic rate, CO2 concentration, stomatal conductance, and transpiration rate between transgenic poplars and the WT and found that these values in the transgenic plants were higher than those of the WT poplars after 2 weeks of drought treatment. The results of photosynthetic analysis showed that this photosynthetic rate was higher in the transgenic poplar compared with that in the WT poplar (average over this time period: 10:00–17:00 sunny day) (Figure 6).
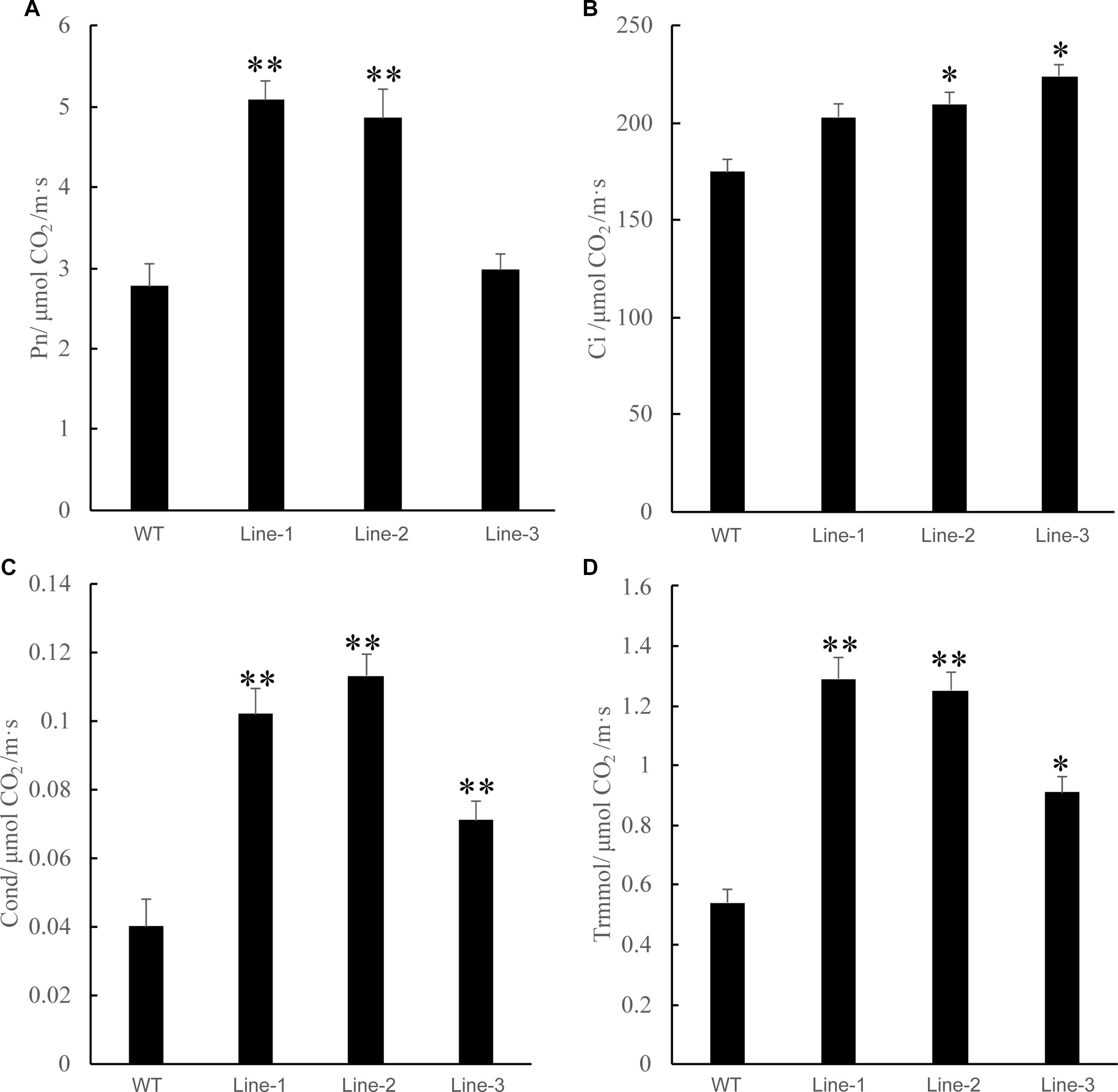
Figure 6. Analysis of the net photosynthetic rate and the intercellular CO2 concentration (Ci) in WT and transgenic poplars (Lines 1–3) in response to drought stress. (A) Net photosynthetic rate. (B) Carbon dioxide concentration. (C) Cond stomatal conductance. (D) Trmmol transpiration rate. Asterisks above the error bars indicate significant differences between trangenic poplar lines and WT (∗p < 0.05; ∗∗p < 0.01). WT, wild type; Lines 1–3, transgenic poplar lines 1, 2, and 3.
Effects of the Overexpression of NtDIP6 on the Levels of Transcripts of Drought-Related Response Genes
To further verify the drought resistance of transgenic plants, we identified the expression level of homologous sequences of four representative drought-responsive genes derived from the previous studies (Saleh et al., 2006; Zhu et al., 2016; Hwang et al., 2018; Rongrong et al., 2018), named PtDBF1, PtWRKY1, PtWRKY3, and PtNCED. The expression level of four genes was investigated in both the transgenic and WT plants under drought treatment using qRT-PCR (Figure 7). All the selected drought-related genes were highly expressed in NtDIP6-overexpressed poplar compared with the WT poplar, and the transgenic lines seemed to possess a higher level of selected drought-related genes. This suggested that NtDIP6 was involved in the regulation of the expression of genes related to drought stress.
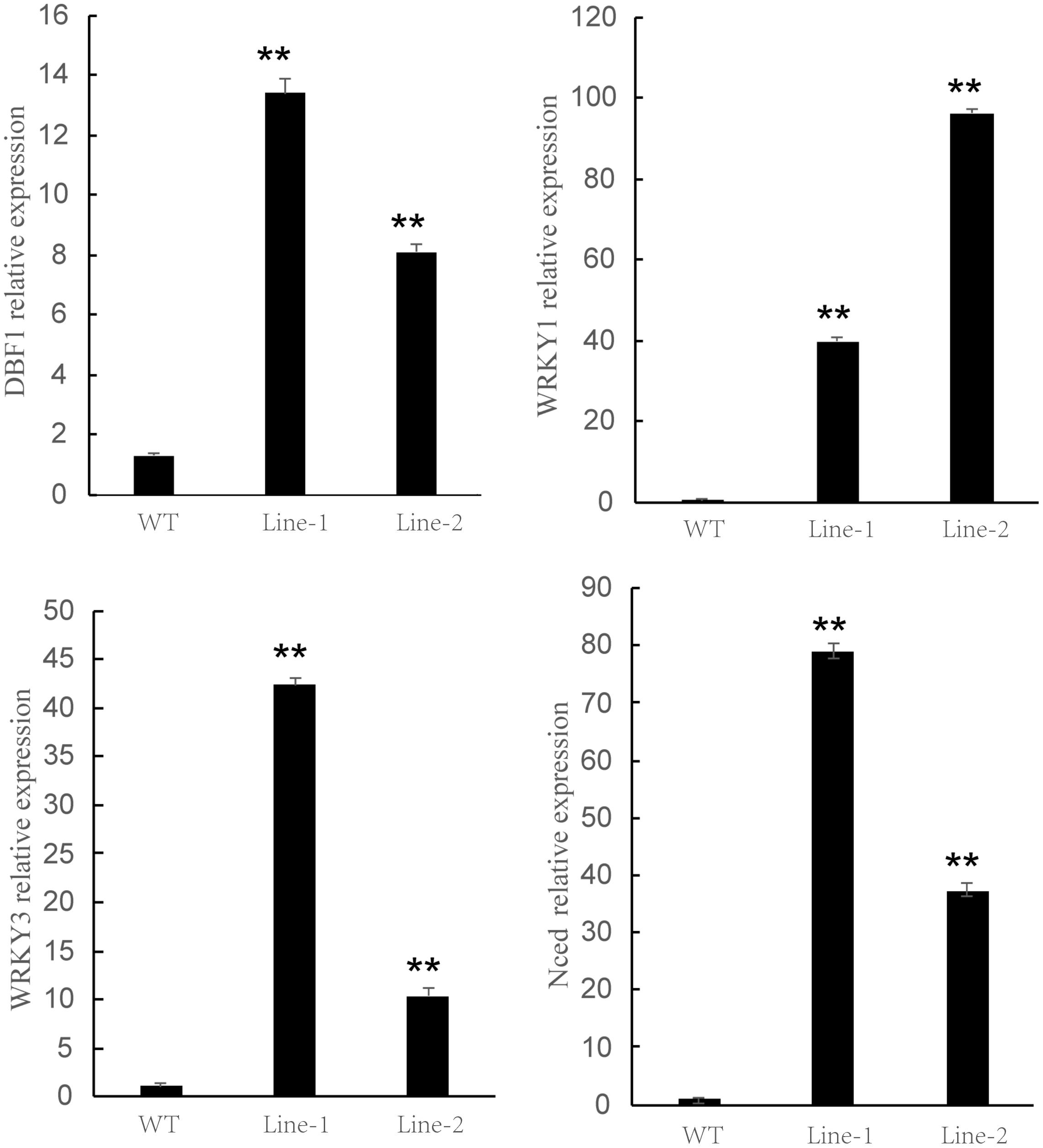
Figure 7. The expression pattern of drought response genes in NtDIP6-Ox transgenic poplars subjected to drought treatment. The expression levels relative to Actin were measured by quantitative RT-qPCR. Three biological replicates and three technical replicates were obtained for each data point. Asterisks above the error bars indicate significant differences between trangenic poplar lines and WT (∗∗p < 0.01). WT, wild type; Lines 1, 2, transgenic poplar lines 1, 2.
Discussion
Plants, as sessile organisms, cannot escape from the environmental stresses that can negatively impact their survival, development, and productivity. Plants have evolved complex mechanisms at the physiological and biochemical levels to adapt to different stresses (Figueiredo et al., 2012). Drought stress is one of the major environmental factors that affects the growth and development of plants. Previous studies have revealed that TFs are involved in binding the promoters of drought response genes to enhance the tolerance of plants. This is the first study to identify DIP family genes in seventeen land plants. A total of 67 DIP genes have been identified and divided into three groups based on phylogenetic, motif, and gene structure analyses (Supplementary Figure S1 and Table 1). Furthermore, two tobacco NtDIP genes (NtDIP1 and NtDIP6) and Arabidopsis DIP3 identified were clustered together and distributed in group III, suggesting that these genes may have similar functions (Figure 1).
The transgenic plants overexpressing NtDIP6 gene under the control of 35S promoter were generated. No significant difference in phenotype between transgenic plants and wild type was detected under normal conditions. Overexpression of NtDIP6 enhanced the drought resistance of transgenic plants (Figures 2, 3). These results were consistent with a previous study that DIP1 is a potential regulator of DBF1 activity in stress responses (Saleh et al., 2006). The DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an abscisic acid-dependent pathway in plants (Kizis and Pages, 2002; Xu et al., 2008). Oxidative stress tolerance is a basis of tolerance to many abiotic stresses, such as drought and extreme temperatures. The accumulation of reactive oxygen species, such as O–2 and H2O2, results in cell membrane peroxidation and degreasing, and increased permeability and ion outflow (Liu et al., 2005; Yao et al., 2017). Endogenous protective enzyme systems in the plants removed reactive oxygen free radicals and avoided the toxicity of free radicals to enhance the adaptability of plants. POD and CAT, as the protective enzyme systems in plants, are mostly involved in repairing the damage to cell membrane and signal transduction that results from drought stress (Wang et al., 2005; Liu et al., 2016). MDA, one of the final products of lipid peroxidation of plant cell membranes, which represents the degree of damage to plants: an increase in the content of MDA results in greater damage to plant cells (Pinhero et al., 1997; Shu et al., 2011). NtDIP6-overexpressing tobacco and poplar plants exhibited enhanced drought resistance with higher antioxidant enzyme activities (CAT and POD), a lower MDA content (Figures 4, 5), which is consistent with the findings of the previous studies (Saleh et al., 2006; Ning et al., 2017). In addition, NtDIP6-overexpressing poplar improved photosynthetic capacity with high net photosynthetic rate, CO2 concentration, stomatal conductance, and transpiration rate. Several studies have reported the existence of a positive correlation between photosynthetic efficiency maintenance and tolerance to drought stress in plants amended with compost and/or inoculated with AMF/PGPR (Wu et al., 2006; Sandhya et al., 2010; Tartoura, 2010; Abd El-Mageed et al., 2018, 2019; Duo et al., 2018; Khosravi Shakib et al., 2019). In this study, the expression level of four drought-responsive genes (PtDBF1, PtWRKY1, PtWRKY3, and PtNCED1) were highly expressed in NtDIP6-overexpressed poplar compared with the WT poplar. Previous studies had revealed that four genes (DBF1, WRKY1, WRKY3, and NCED) plays a positive regulatory role in drought stress (Saleh et al., 2006; Zhu et al., 2016; Hwang et al., 2018; Rongrong et al., 2018), which is similarly to our results of qRT-PCR.
In summary, we first identified NtDIP genes and randomly screened NtDIP6 as a candidate gene for transgenic functionality. We found that transgenic plants that expressed NtDIP6 displayed greater resistance to drought and oxidation. The level of expression of five drought-related response genes increased in NtDIP6-Ox transgenic poplars in comparison with the WT. Therefore, NtDIP6 can be used as a candidate gene for the molecular breeding of drought-tolerant varieties of poplar and has potential economic value in improving drought tolerance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YL and XY conceived and designed the project. XY and LZ grew the plant material and conducted the experiments. YL contributed analytical tools and analyzed data. XY wrote the manuscript. LL and RL oversaw the experiments and revised the manuscript. All the authors read and approved the manuscript.
Funding
This research was supported by the Construction and Application of Gene Editing Technology System, project number: Qianyanke No. 201602 and Guizhou Provincial High-level Innovative Talents Training Project Hundred Talents Program, project number: No. 20165663.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.601585/full#supplementary-material
Supplementary Figure 1 | Maximum-likelihood phylogeny of the DIP proteins from seventeen land plants. The phylogeny was constructed based on the amino acid sequences of full-length DIP proteins with 100 bootstrapping replicates. Green, black and orange arcs indicate different groups of DIP proteins.
Supplementary Figure 2 | The logo sequence in Supplementary Figure S1.
Supplementary Figure 3 | The logo sequence in Figure 1B.
Supplementary Figure 4 | The expression pattern of NtDIP1 and NtDIP6 after 3 h of drought treatment. The expression levels relative to Actin were measured by quantitative RT-qPCR. Three biological replicates and three technical replicates were obtained for each data point. Asterisks above the error bars indicate significant differences between control and treated tobacco plants (∗p < 0.05).
Supplementary Figure 5 | Molecular analyses of NtDIP 6 transgenic poplars. (a) Schematic map of construct. Expression of NtDIP6 is driven by the 35S promoter. (b) PCR verification of different independently regenerated transgenic lines. L1–3: different transgenic poplar lines. (c) GUS staining in leaves of NtDIP6 transgenic poplars.
Supplementary Table 1 | Distribution of Land-plant DIP genes among every subgroup of the phylogeny in Supplementary Figure S1.
Supplementary Table 2 | The primers used in this study.
Footnotes
- ^ http://smart.embl-heidelberg.de/
- ^ http://tree.bio.ed.ac.uk/software/fifigtree/
- ^ http://meme-suite.org/
- ^ http://mapinspect.software.informer.com/
References
Abd El-Mageed, T. A., El-Samnoudi, I. M., Ibrahim, A. E. A. M., and Abd El Tawwab, A. R. (2018). Compost and mulching modulates morphological, physiological responses and water use efficiency in Sorghum bicolor L. (Moench) under low moisture regime. Agric. Water Manag. 208, 431–439. doi: 10.1016/j.agwat.2018.06.042
Abd El-Mageed, T. A., El-Sherif, A. M. A., Abd El-Mageed, S. A., and Abdou, N. M. (2019). A novel compost alleviate drought stress for sugar beet production grown in Cd-contaminated saline soil. Agric. Water Manag. 226:105831. doi: 10.1016/j.agwat.2019.105831
Bian, Z., Zhang, X., Wang, Y., and Lu, C. (2019). Improving drought tolerance by altering the photosynthetic rate and stomatal aperture via green light in tomato (solanum lycopersicum) seedlings under drought conditions. Environ. Exp. Bot. 167:103844. doi: 10.1016/j.envexpbot.2019.103844
Duo, L. A., Liu, C. X., and Zhao, S. L. (2018). Alleviation of drought stress in turfgrass by the combined application of nano-compost and microbes from compost. Russ. J. Plant Physiol. 65, 419–426. doi: 10.1134/S102144371803010X
Farooq, M., Wahid, A., Kobayashi, N., Fujita, D., and Basra, S. M. A. (2009). Plant drought stress: effects, mechanisms and management. Agron. Sustain. 29, 185–212. doi: 10.1051/agro:2008021
Figueiredo, D. D., Barros, P. M., Cordeiro, A. M., Serra, T. S., Lourenço, T., Chander, S., et al. (2012). Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J. Exp. Bot. 63, 3643–3656. doi: 10.1093/jxb/ers035
Fillatti, J. J., Kiser, J., Rose, R., and Comai, L. (1987). Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium Tumefaciens vector. Nat. Biotechnol. 5, 726–730. doi: 10.1038/nbt0787-726
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guo, M., Pingyi, G., Xianguang, Y., Hong, G., Zenpan, G., Feng, L. et al. (2014). Effects of foliar application of Na2 SeO3 on photosynthetic characteristics and yield of foxtail millet. Journal of Nuclear Agricultural Sciences. 28, 1099–1107. doi: 10.11869/j.issn.100-8551.2014.06.1099
Hervé, P., and Serraj, R. (2009). “GM technology for drought resistance,” in Drought Frontiers In Rice: Crop Improvement for Increased Rainfed Production, eds R. Serraj, J. Bennett, and B. Hardy (Toh Tuck Link: World Scientific Publishing Co), 333–350. doi: 10.1142/9789814280013_0019
Hwang, S. G., Lee, C. Y., and Tseng, C. S. (2018). Heterologous expression of rice 9-cis-epoxycarotenoid dioxygenase 4 (OsNCED4) in Arabidopsis confers sugar oversensitivity and drought tolerance. Bot. Stud. 59, 1–12. doi: 10.1186/s40529-018-0219-9
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Khosravi Shakib, A., Rezaei Nejad, A., Khandan Mirkohi, A., and Kalate Jari, S. (2019). Vermicompost and manure compost reduce water-deficit stress in pot marigold (Calendula officinalis L. cv. Candyman Orange). Compost. Sci. Util. 27, 61–68. doi: 10.1080/1065657X.2019.1602489
Kizis, D., and Pages, M. (2002). Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J. 30, 679–689. doi: 10.1046/j.1365-313X.2002.01325.x
Liepinsh, E., Leonchiks, A., Sharipo, A., Guignard, L., and Otting, G. (2003). Solution structure of the R3H domain from human Smubp-2. J. Mol. Biol. 326, 217–223. doi: 10.1016/s0022-2836(02)01381-5
Liu, M. Q., Liu, Q. Y., and Ding, X. W. (2005). Heat resistance increases in seedlings of transgenic tobacco lines with SOD, POD genes. J. Yunnan Agric. Univ. 20, 620–623.
Liu, Y., Yao, X. Z., and Lv, L. T. (2016). Cloning of SbSKIP gene from sorghum (Sorghum bicolor) and analysis of drought-resistant function in tobacco (Nicotiana tabacum). J. Agric. Biotechnol. 24, 1500–1511.
Ning, W., Zhai, H., Yu, J., Liang, S., Yang, X., Xing, X., et al. (2017). Overexpression of Glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol. Breeding 37:19. doi: 10.1007/s11032-016-0614-4
Pinhero, R. G., Rao, M. V., Paliyath, G., Murr, D. P., and Fletcher, R. A. (1997). Changes in activities of antioxidant enzymes and their relationship to genetic and paclobutrazol-induced chilling tolerance of maize seedlings. Plant Physiol. 114, 695–704. doi: 10.1104/pp.114.2.695
Rongrong, G., Hengbo, Q., Jiao, Z., Xianhang, W., Mingxing, T., Chunlei, G., et al. (2018). The Grape VlWRKY3 gene promotes abiotic and biotic stress tolerance in transgenic Arabidopsis thaliana. Front. Plant Sci. 9:545. doi: 10.3389/fpls.2018.00545
Saleh, A., Lumbreras, V., Lopez, C., Dominguez-Puigjaner, E., Kizis, D., and Pagès, M. (2006). Maize DBF1-interactor protein 1 containing an R3H domain is a potential regulator of DBF1 activity in stress responses. Plant J. 46, 747–757. doi: 10.1111/j.1365-313X.2006.02742.x
Sandhya, V., Ali, S. Z., Grover, M., Reddy, G., and Venkateswarlu, B. (2010). Effect of plant growth promoting Pseudomonas spp. on compatible solutes, antioxidant status and plant growth of maize under drought stress. Plant Growth Regul. 62, 21–30. doi: 10.1007/s10725-010-9479-4
Shu, H. M., Ni, W. C., and Guo, S. Q. (2011). Brassinosteroid: Biosynthesis, metabolism and its regulationon plant salt tolerance. M. P. B. 9, 1261–1266.
Tartoura, K. A. H. (2010). Alleviation of oxidative-stress induced by drought through application of compost in wheat (Triticum aestivum L.) plants. Am. J. Agric. Environ. Sci. 9, 208–216.
Wang, H., Zhao, Z., Xu, C., and Liu, J. (2005). Nanometric La1–xKx MnO3 Perovskite-type oxides – highly active catalysts for the combustion of diesel soot particle under loose contact conditions. Catal. Lett. 102, 251–256. doi: 10.1007/s10562-005-5864-4
Wang, X., Lin, S., Liu, D., Wang, Q., McAvoy, R., Ding, J., et al. (2019). Characterization and expression analysis of ERF genes in Fragaria vesca suggest different divergences of tandem ERF duplicates. Front. Genet. 280:497–508. doi: 10.3389/fgene.2019.00805
Wu, Q., Xia, R., and Hu, Z. (2006). Effect of arbuscular mycorrhiza on the drought tolerance of Poncirus trifoliata seedlings. Front. Forst. Chn. 1:100–104. doi: 10.1007/s11461-005-0007-z
Xu, Z. S., Ni, Z. Y., Liu, L., Nie, L. N., Li, L. C., Chen, M., et al. (2008). Characterization of the TaAIDFa gene encoding a crt/dre-binding factor responsive to drought, high-salt, and cold stress in wheat. Mol. Genet. Genomics 280, 497–508. doi: 10.1007/s00438-008-0382-x
Yao, Y., Liu, H., Zhang, H., Wang, H., Zhang, C., Zhang, Z., et al. (2017). Risk factors for recurrent herniation after percutaneous endoscopic lumbar discectomy. World Neurosurg. 100, 1–6. doi: 10.1016/j.wneu.2016.12.089
Zhu, Q., Chun, L. L., and Wei, Z. (2016). WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana. Plant Mol. Biol. 91, 53–65. doi: 10.1007/s11103-016-0441-3
Keywords: NtDIP6, POD, CAT, drought stress, RT-qPCR analysis, transgenic poplars
Citation: Liu Y, Yao X, Zhang L, Lu L and Liu R (2021) Overexpression of DBF-Interactor Protein 6 Containing an R3H Domain Enhances Drought Tolerance in Populus L. (Populus tomentosa). Front. Plant Sci. 12:601585. doi: 10.3389/fpls.2021.601585
Received: 01 September 2020; Accepted: 13 January 2021;
Published: 04 February 2021.
Edited by:
Honghong Wu, Huazhong Agricultural University, ChinaReviewed by:
Vivek Dogra, Institute of Himalayan Bioresource Technology (CSIR), IndiaFernanda Lazzarotto, Federal University of Rio Grande do Sul, Brazil
Copyright © 2021 Liu, Yao, Zhang, Lu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Litang Lu, bHRsdkBnenUuZWR1LmNu; Renxiang Liu, cnhsaXVAZ3p1LmVkdS5jbg==
 Yang Liu1,2
Yang Liu1,2 Xinzhuan Yao
Xinzhuan Yao Litang Lu
Litang Lu Renxiang Liu
Renxiang Liu