- 1College of Life Sciences, Henan Agricultural University, Zhengzhou, China
- 2Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences, Shanghai, China
- 3Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN, United States
We have previously demonstrated that General Control Non-derepressible 1 (AtGCN1) is essential for translation inhibition under cold stress through interacting with GCN2 to phosphorylate eukaryotic translation initiation factor 2 (eIF2). Here, we report that the flower time of the atgcn1 mutant is later than that of the wild type (WT), and some siliques of atgcn1 cannot develop and produce seeds. Total and polysomal RNA of atgcn1-1 and wild type (WT) after cold treatments were sequenced. The sequencing results show that the mutation of atgcn1 selectively alters the expression of genes at both transcriptional and translational levels. The classification of AtGCN1 target genes reveals that AtGCN1 regulated gens are involved in flower development, seed dormancy and seed development, response to osmotic stress, amino acid biosynthesis, photosynthesis, cell wall organization, protein transport and localization, lipid biosynthesis, transcription, macroautophagy, proteolysis and cell death. Further analysis of AtGCN1 regulated genes at translational levels shows that the Kozak sequence and uORFs (upstream open reading frame) of transcripts affect translation selection. These results show that AtGCN1 is required for the expression of selective genes in Arabidopsis.
Introduction
Upon stress, the phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α) is catalyzed by eIF2α kinases to inhibit protein translation in mammals (Erickson and Hannig, 1996). There are four recognized eIF2α kinases in mammals, including a GCN2 (General Control Non-derepressible 2) which is induced under nutrition stress, a double-stranded-RNA-dependent protein kinase R (PKR) which participates in antiviral response, a PKR-like endoplasmic reticulum kinase (PERK) which responds to endoplasmic reticulum stress (ER stress) and a heme-regulated eIF2 inhibitor kinase (HRI) that is activated by heme deprivation. In yeast, however, only one GCN2 kinase has been reported and it is known to get activated by conditions of amino acid deficiency (Zhang et al., 2002; Zaborske et al., 2010), glucose deprivation (Yang et al., 2000), salt stress and UV irradiation (Narasimhan et al., 2004; Wu et al., 2004; Jiang and Wek, 2005).
Sensing amino acid starvation, yeast GCN2 kinase is activated to phosphorylate eIF2α, which results in the inhibition of protein translation (Zhang et al., 2002; Zaborske et al., 2010). Interestingly, eIF2α phosphorylation specifically enhances the translation of transcription factor (TF) GCN4 in yeast (Dever et al., 1992, 1993; Harding et al., 2000; Lu et al., 2004; Vattem and Wek, 2004), which in turn stimulates the expression of downstream genes to help yeast survive in stress conditions. The downstream genes have been found to involve in amino acid biosynthesis, purine biosynthesis, mitochondrial carrier, peroxisomes, energy metabolism and autophagy (Hinnebusch, 1988; Jia et al., 2000; Natarajan et al., 2001).
In Arabidopsis, GCN2 kinase, as the only recognized eIF2α kinase, participates in eIF2α phosphorylation and inhibits protein translation under amino acid deprivation (Lageix et al., 2008). In our previous research, we have found that Arabidopsis AtGCN1 interacts with GCN2 to phosphorylate eIF2α upon cold stress, which is essential for the cold-induced inhibition of protein translation and cold acclimation (Wang et al., 2017). However, genes that are regulated by AtGCN1/GCN2 in Arabidopsis have yet to be identified. To advance our understanding, we performed total RNA and polysomal RNA sequencing of mutant atgcn1-1 after cold treatments and found that AtGCN1 mediated eIF2α phosphorylation selectively regulates transcription and translation of genes. Our findings have moved one step closer into understanding the roles of AtGCN1 in Arabidopsis.
Materials and Methods
Plant Growth Conditions
Arabidopsis thaliana ecotype-Columbia, an ethyl methane sulfonate (EMS)-mutagenic mutant gl1 of Columbia (as the wild type of atgcn1-1, WT) and mutants were either grown in forest soil and vermiculite (1:1) or on a half-strength MS medium (M519, Sigma-Aldrich) containing 1% (w/v) of sucrose and 0.8% (w/v) agar at 22°C with a 16 h-light/8 h-dark cycle. To measure root length, fertilized seeds were sown in MS medium with 1% agar and grew for 8 days. For fresh weight measurement, the aboveground parts of four-week-old plants in the soil were cut to be weighed.
Protein Extraction and Western Blot Analysis
Ten-day-old seedlings were used for protein extraction and proteins were analyzed as described (Wang et al., 2017). The extraction buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.2% Triton X-100, 1 mM DTT, 1 mM PMSF) containing both Complete protease and PhosSTOP phosphatase inhibitor (Roche) was added into the grounded seedlings. The solutions were then centrifuged at 4°C for 5 min twice before extracting proteins from the upper layer. Extracted proteins were incubated at 95°C for 5 min, separated by SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) and transferred to PVDF (polyvinylidene fluoride) membranes for blotting. A rabbit monoclonal antibody of phospho-eIF2α (S51) (catalog no. 9721, Cell Signaling, 1/1,000 dilution) was probed and then reprobed for Actin (Abmart, 26F7, 1/10000 dilution) to ensure equal loading in each experiment.
Illumina Sequencing of Total and Polysomal RNA
Ten-day-old seedlings were transferred to cold conditions of 4°C temperature with 16 h-light/8 h-dark cycle for 24 h treatment and used to extract total RNA (TRIzol, Invitrogen) and polysomal RNA. Total and polysomal RNAs were respectively used to construct RNA library and sequenced by Illumina HiSeq2500 in Shanghai Center for Plant Stress Biology (PSC), following standard Illumina protocols (Chen et al., 2016). Two repeats were performed for each treatment.
For polysomal RNA sequencing, whole plants of 10-day-old seedlings after cold treatments were used to extract polysomes as previously reported (Sormani et al., 2011). 200 mg of 10-day-old seedlings was grounded in liquid nitrogen before placing into a 1 ml of lysis buffer (100 mM Tris–HCl, pH 8.4, 50 mM KCl, 25 mM MgCl2, 5 mM EGTA, 50 μg/ml cycloheximide, 50 μg/ml chloramphenicol, 0.5% Nonidet P40, Diethyl pyrocarbonate treated) for 10 min at 4°C. After centrifuging at 9,000 g for 15 min, the supernatant was loaded onto 12 mL of 0.8-1.5 M sucrose gradients. The gradients were then centrifuged at 175,000 g in a Beckman SW41 rotor for 150 min before getting the OD (260 nm) acquired with an ISCO gradient fractionator. Meanwhile, the gradients were fractionated into 21 tubes. According to the OD value at 260 nm, tubes 13-21 containing polysomes were collected together for polysomal RNA extraction using phenol extraction.
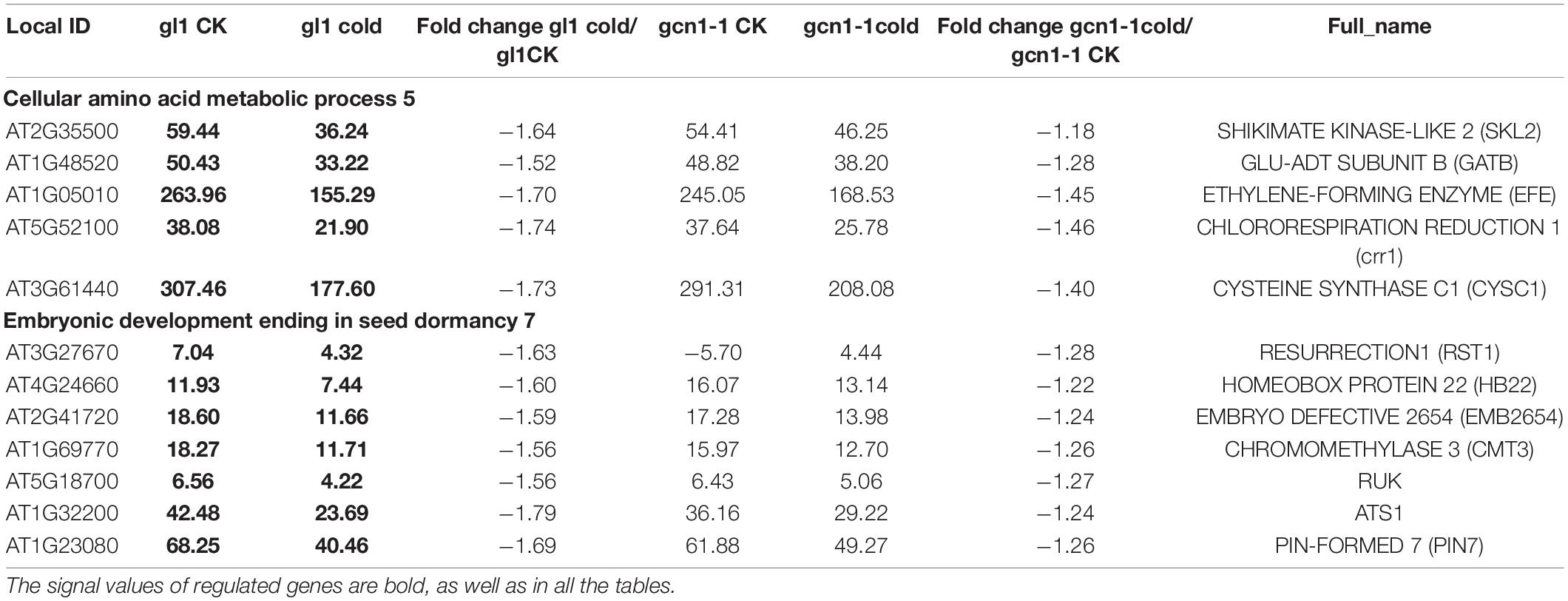
For the differentially expressed genes, a threshold of q-Value < 0.05, the sum of two signal values > 10 and at least 1.5-fold change were used in the study. Cold-AtGCN1 target genes were filtered out for hierarchical clustering analysis (Cluster 3.0). Functional classification of Cold-AtGCN1 target genes was carried out using DAVID (Database for Annotation, Visualization and Integrated Discovery1). The pie charts for Figures 3C,D, 4B,C were drawn in Excel after performing classification.
RNA Extraction and qPCR
Whole RNA of seedlings treated as the methods in RNA sequencing was extracted according to TRIzol method (Life, Invitrogen). Quantified RNAs were treated with DNase I (Turbo DNA-free kit, Ambion) before getting transcribed into cDNA using oligodT and reverse transcriptase (Goscript Reverse Transcription system;A5001). qPCR analysis was subsequently performed using an IQ5 Multicolor Real-Time PCR Detection system. The following parameters were used to perform qPCR: 95°C for 15 min, 44 cycles of 95°C for 15 s, 55°C for 15s, 72°C for 15 s, followed by a melting curve analysis. Actin was used for normalization and the primers used in qRT-PCR are listed in Supplementary Table 4.
Results
atgcn1 Mutants Flower Later Than the Wild Type
As showed in the previous results, atgcn1-1 is a point mutant from G to A, which is a truncated protein of AtGCN1 without the C-terminal, while atgcn1-2, on the other hand, was achieved through a T-DNA insertion that led to a truncated protein of AtGCN1 without the N-terminal (Wang et al., 2017). Here, we found that both atgcn1-1 and atgcn1-2 mutants flowered 2 days later than the WT under long-day conditions (16 h light/8 h dark) (Figures 1A,B). To exclude the germination difference, all the plants on soil were transferred from 5-day-old seedlings in 1/2 strong MS plates. Under short-day conditions (8 h light/16 h dark), mutants did not show to flower later than the WT (not shown). Moreover, we counted the number of rosette leaves at flower time. The result show that the atgcn1-1 has more rosette leaves than wild type at flower time (Figure 1C). Another mutant of atgcn1-2 has similar number of rosette leaves as wild type. FLOWERING LOCUS C (FLC) is a gene that negatively controls flowering (Michaels and Amasino, 1999),and we detected the expression of FLC in atgcn1. We found that FLC was up-regulated in atgcn1-1 and atgcn1-2, compared to that in WT (Figure 1D). These findings prove that atgcn1 mutations lead to a late flowering phenotype in Arabidopsis.
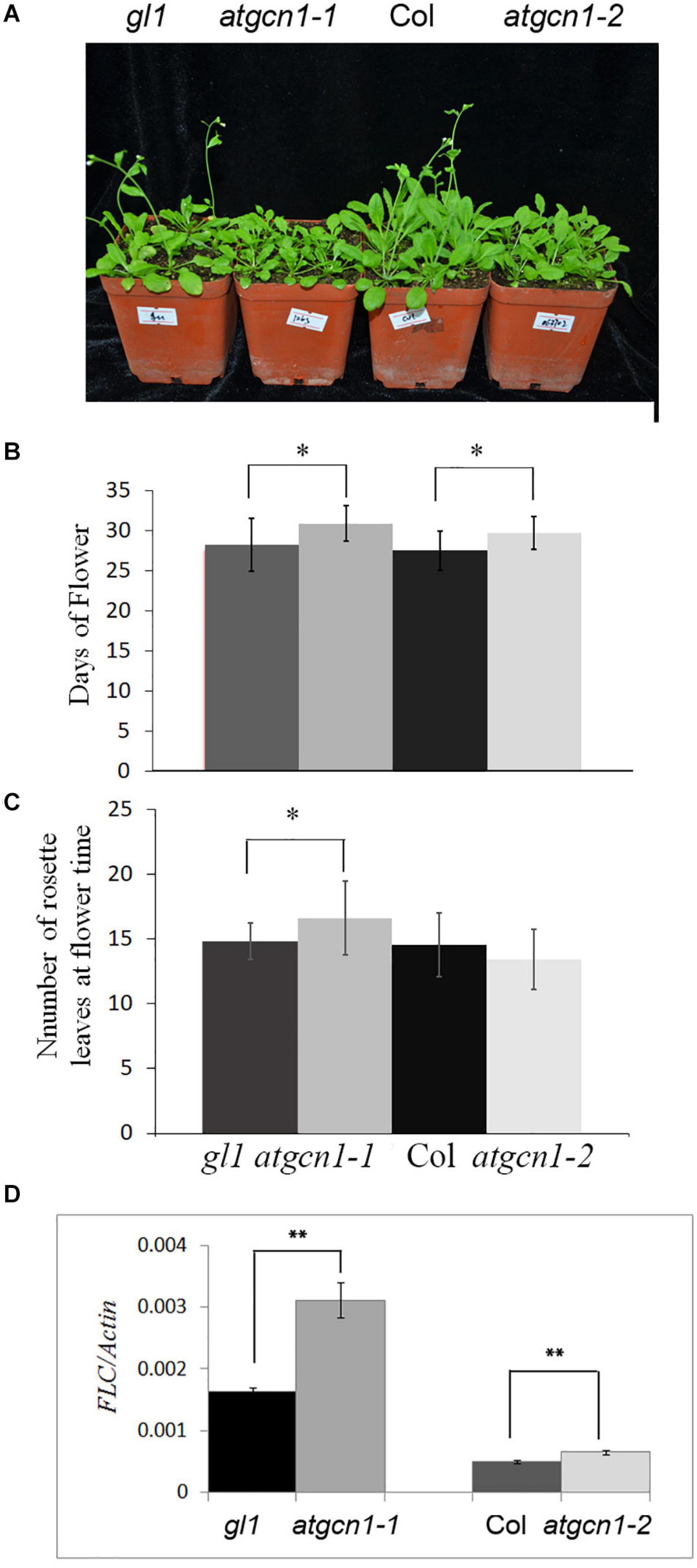
Figure 1. Early flowering of atgcn1-1 and atgcn1-2 in soil. (A) The flowering phenotype of atgcn1-1 and atgcn1-2 in soil. (B) The day statistics of flower time. (C) The number statistics of rosette leaves at flower time. (D) The expression of FLC is higher in atgcn1-1 and atgcn1-2 than in WT. Statistical t-tests were performed. Single asterisk indicates significant difference with a p-Value < 0.05. Double asterisks indicate a significant difference with a p-Value < 0.01. Experiments were repeated three times with similar results achieved.
Mutations of atgcn1 Influences Plant Growth and Seed Development
Since mutations of atgcn1 were reported to impair eIF2 phosphorylation, which has an effect on the translation of mRNAs (Wang et al., 2017), we checked for plant growth alteration in atgcn1. The root lengths of atgcn1 mutants were observed to be significantly shorter than that of WT (Figures 2A,B). Moreover, the weight of rosette leaves in atgcn1 was evidently lower than that in WT (Figure 2C). These data indicate that the mutation of atgcn1 inhibits growth in both root and shoot.
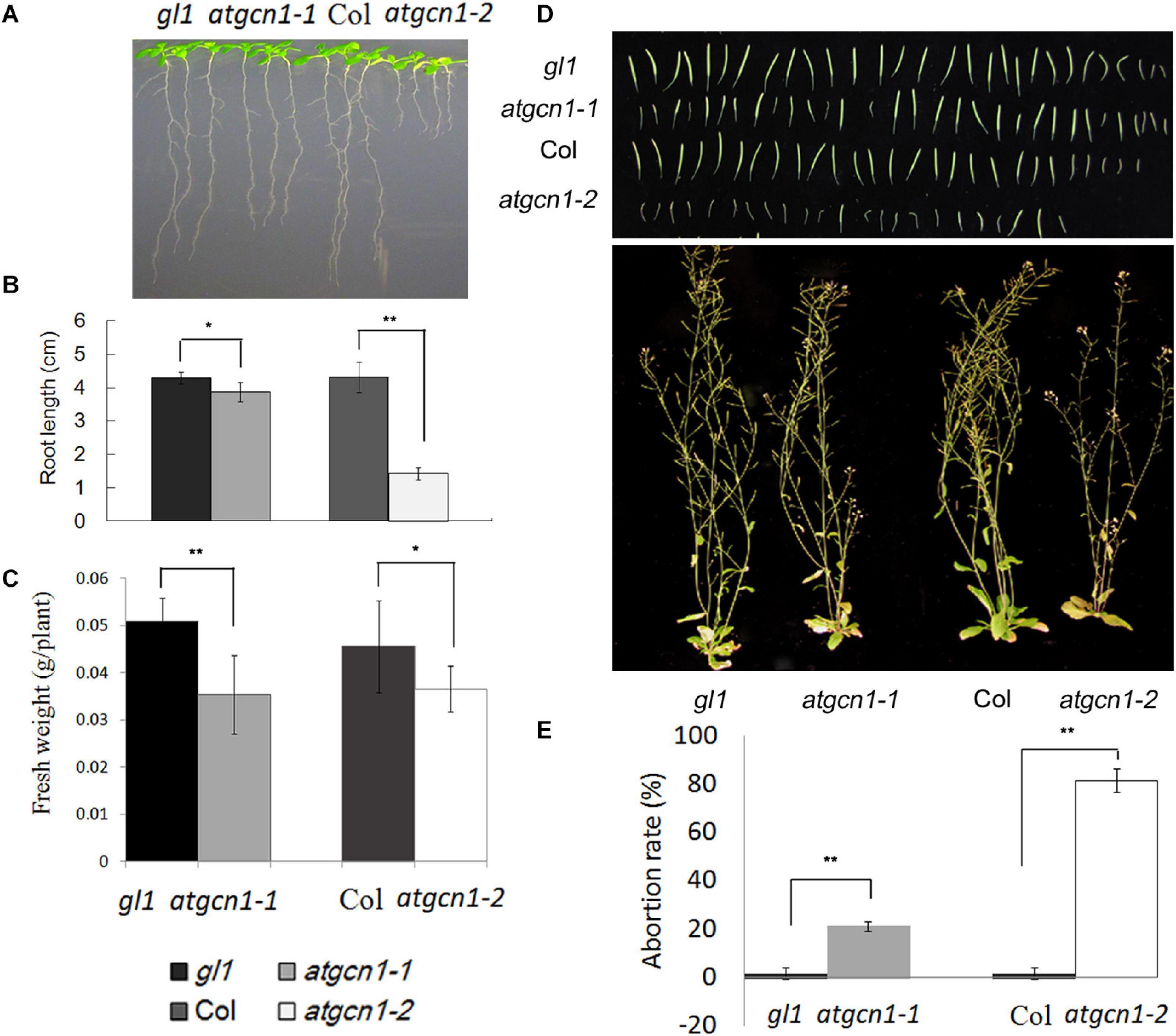
Figure 2. AtGCN1 is required for plant growth and seed development. (A) The root length of atgcn1-1 and atgcn1-2 is shorter than that of WT. (B) The measurement of root length. (C) The fresh weight of atgcn1-1 and atgcn1-2 is less than that of WT. (D) Seed development is impaired by atgcn1 mutations. (E) The statistical analysis of abortive siliques. Statistical t-tests were performed. Single asterisk indicates significant difference with a p-Value < 0.05. Double asterisks indicate significant difference with a p-Value < 0.01. Experiments were repeated three times with similar results attained.
In addition to the defect of vegetative growth, we also found that atgcn1 mutations impaired seed development as partial siliques of mutant plants were observed to be empty (Figures 2D,E). In the atgcn1-1 mutant line, the rate of aborted siliques is near 20%. In the atgcn1-2 mutant line, the rate of aborted siliques is around 80%. The results show that atgcn1 mutations impair the growth at both vegetative and reproductive stages.
As shown in Figure 2, the atgcn1-2 mutant line has more severe phenotype, with much shorter root length and higher rate of aborted siliques than atgcn1-1. The remarkable difference of the vegetative growth and seed production observed between atgcn1-1 and atgcn1-2 suggests that the N-terminal of AtGCN1 has a greater impact on plant functions than the C-terminal. The more severe atgcn1-2 allele has few seeds and it is not enough to be used, so the further experiments are conducted with the less severe atgcn1-1 allele.
The Phosphorylation of eIF2α Selectively Regulates Transcription in Cold Stress
AtGCN1 has been reported to interact with GCN2 to phosphorylate eIF2α, which results in translation inhibition under cold stress (Wang et al., 2017). As shown in Figure 3A, eIF2α phosphorylation was induced by cold stress in WT, while the phosphorylation disappeared in atgcn1-1. To explore how AtGCN1/GCN2-mediated eIF2α phosphorylation regulates gene expression in Arabidopsis, we extracted the total RNA of WT and atgcn1-1 after cold treatments for a global transcriptome analysis using RNA sequencing.
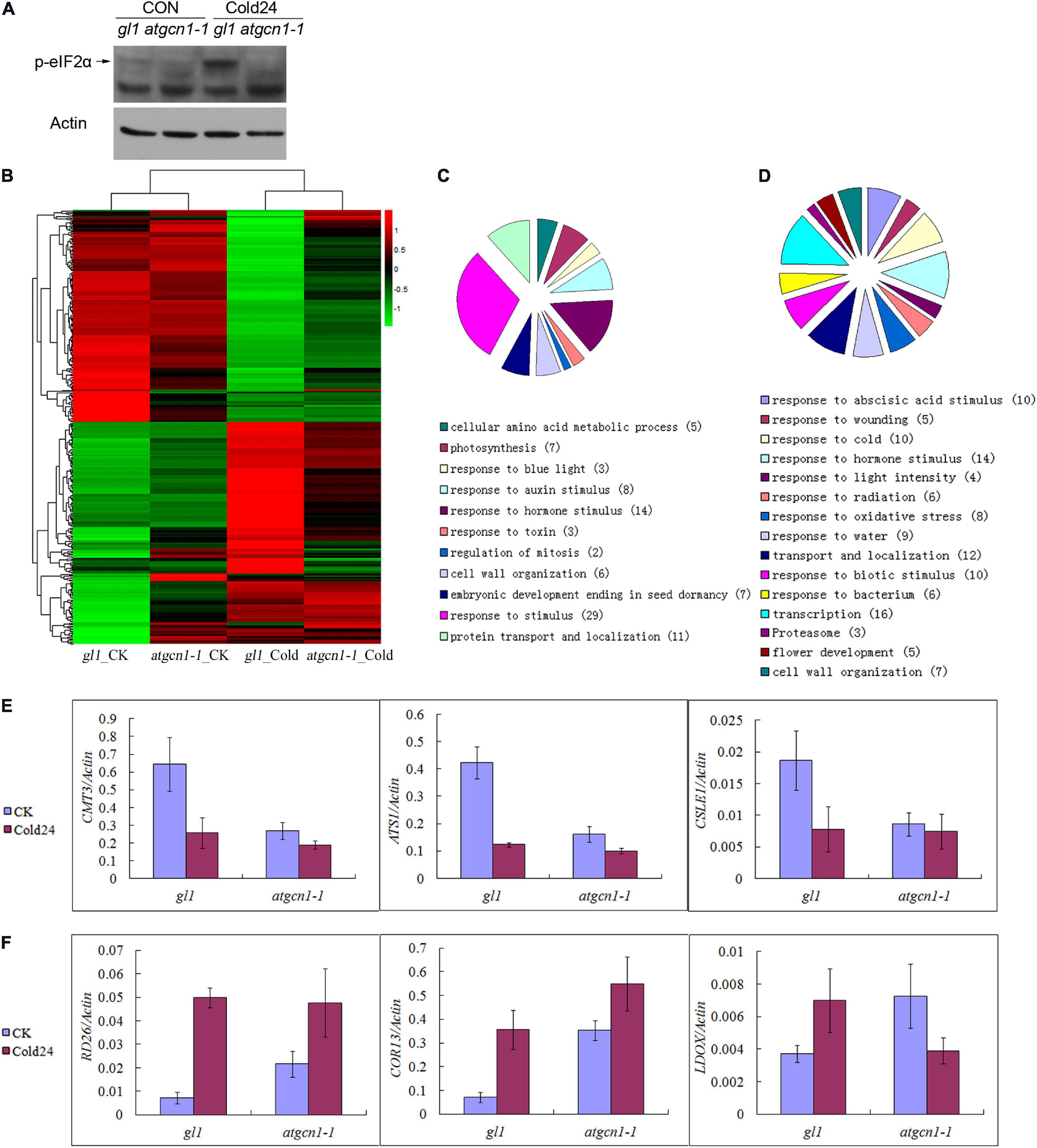
Figure 3. eIF2α phosphorylation selectively regulates gene transcription under cold stress conditions. (A) The immunoblotting assays of antibodies against phosphorylated eIF2α under normal temperature and cold stress conditions. Western blot results with Actin used as a control. (B) Hierarchical clustering analysis of cold-ATGCN1 genes at transcriptional levels. (C) GO terms analysis of cold-ATGCN1 down-regulated genes at transcriptional levels. (D) GO terms analysis of cold-ATGCN1 up-regulated genes at transcriptional levels. (E) Cold-ATGCN1 down-regulated genes selected for validation by using qRT-PCR. (F) Cold-ATGCN1 up-regulated genes selected for validation by using qRT-PCR. CK, grown in normal temperature conditions. Cold or Cold24, treated by cold stress conditions of 4°C temperature for 24 h treatment. qRT-PCR was repeated three times with similar results attained.
Based on statistical analysis and after filtering, 184 down-regulated and 179 up-regulated genes were identified and respectively named as cold-AtGCN1 down-regulated or up-regulated genes (Supplementary Table 1). Upon cold stress, these down-regulated or up-regulated genes were either down-regulated or up-regulated at transcriptional levels in WT (the sum of two signal values > 10; a signal ratio fold change ≥ 1.5; p-Value < 0.05), whereas the regulation of these genes remains unaltered in atgcn1-1 (the sum of two signal values > 10; a signal ratio fold change ≤ 1.5).
Cold-AtGCN1 regulated genes at transcriptional levels were then randomly selected for further validation by qRT-PCR. Genes such as CXG methyltransferase choromomethylase 3 (CMT3), chloroplast glycerol-3-phosphate acyltransferase (ATS1) and cellulose synthase like E1 (CSLE1) were discovered to be down-regulated by cold stress in WT, but the expression of these genes was noticeably less or lost in atgcn1-1 (Figure 3E and Supplementary Table 1). Additionally, TF in response to desiccation (RD26), cystine lyase involved in cysteine and ethylene biosynthesis (CORI3), leucoanthocyanidin dioxygenase involved in proanthocyanin biosynthesis (LDOX) were confirmed to be up-regulated by cold stress in WT, but the regulation was less or lost in atgcn1-1 (Figure 3F and Supplementary Table 1). The results from qRT-PCR are mostly consistent with those of RNA sequencing data, confirming the efficiency and reliability of our RNA sequencing results.
Our hierarchical clustering analysis of cold-AtGCN1 regulated genes showed that although the repression level was lower in atgcn1-1 than in WT (Figure 3B and Table 1), most cold-AtGCN1 down-regulated genes was repressed by cold stress in atgcn1-1. The result suggests that an alternative mechanism that is independent of eIF2α phosphorylation is involved to affect the transcription of cold-AtGCN1 down-regulated genes. Hierarchical clustering analysis of cold-AtGCN1 up-regulated genes also supports the existence of an alternative mechanism, apart from eIF2α phosphorylation, to induce the expression of cold-AtGCN1 up-regulated genes. As shown, most of the cold-AtGCN1 up-regulated genes were activated by cold stress in atgcn1-1, although the induced levels were lower than that in the WT (Figure 3B and Table 2).
We identified the cold-AtGCN1 regulated genes using DAVID (Database for Annotation, Visualization and Integrated Discovery, See Text footnote 1) (Figures 3C,D and Supplementary Table 1). Five enzymes that involve in amino acid synthesis were down-regulated in WT after cold treatment (Table 1), which was consistent with the fact that fewer amino acids were required for protein translation under cold stress. The five genes were not down-regulated in atgcn1-1 after cold stress treatment. In addition, seven genes taking part in embryonic development and seed dormancy were down-regulated in WT but not in atgcn1-1 after cold treatment. Cold-AtGCN1 down-regulated genes were also found to participate in photosynthesis (7), response to blue light (3), response to hormone stimulus (14), response to toxin (3), regulating mitosis (2), cell wall organization (6), and protein transport/localization (11) (Figure 3C and Supplementary Table 1, in the brackets is the number of regulated genes).
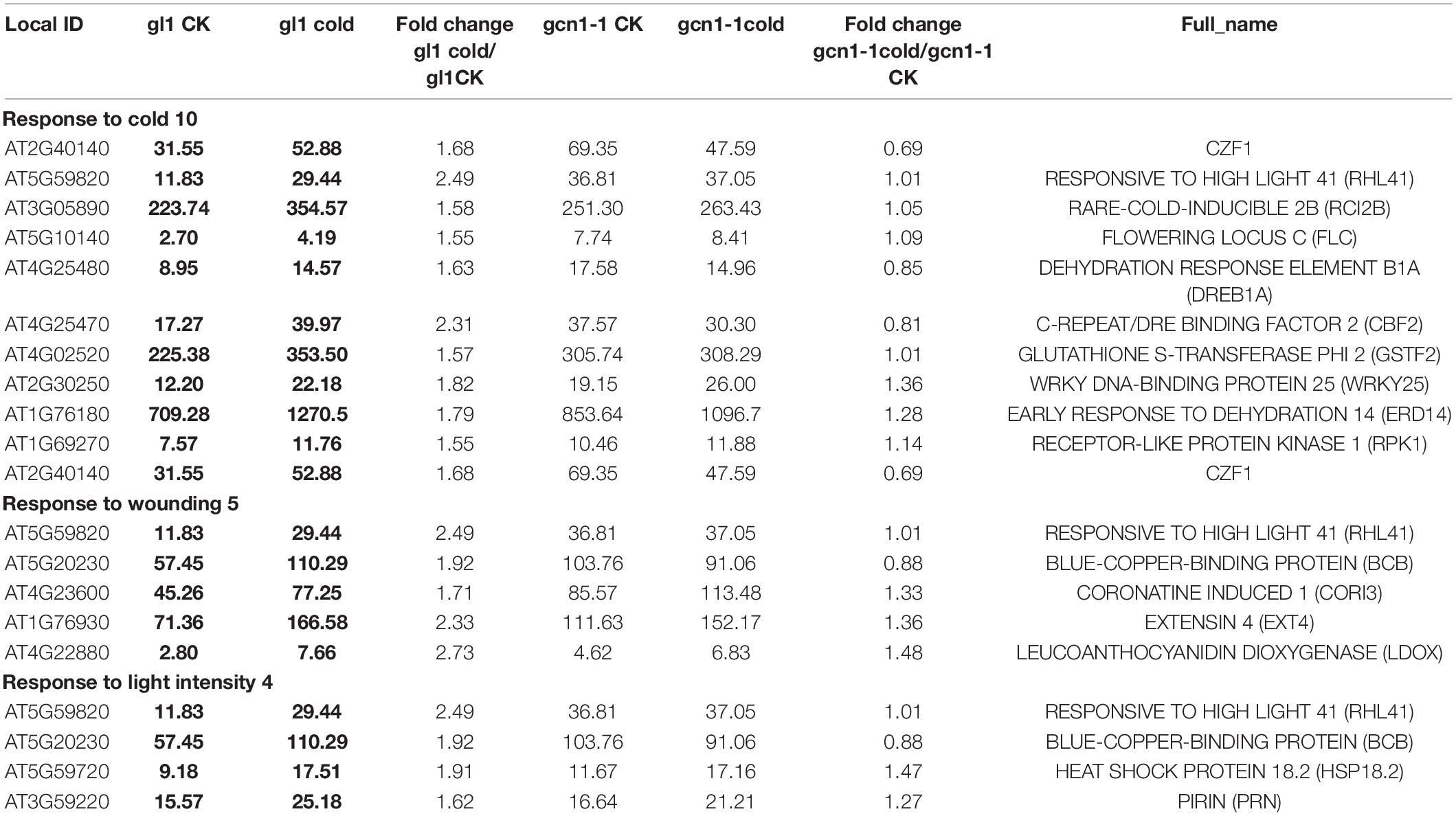
As for the cold-AtGCN1 up-regulated genes, we found that a lot of them were involved in response to abiotic stimuli, including abscisic acid, cold, light intensity, oxidative stress and water deprivation (Figure 3D, Table 2, and Supplementary Table 1). These results indicated that AtGCN1 may have potential functional roles in other abiotic stress conditions, in addition to cold stress. Cold-AtGCN1 up-regulated genes were also discovered to involve in protein transport and localization (12), response to biotic stimuli (10), gene transcription (16), proteasome (3), flower development (5) and cell wall organization (7) (Figure 3D and Supplementary Table 1).
AtGCN1 Selectively Regulates Protein Translation in Cold Stress
As previously mentioned, without eIF2α phosphorylation, polysomes in atgcn1-1 accumulated slightly higher than that in the WT under cold stress (Wang et al., 2017). To seek out the genes that are regulated by AtGCN1-mediated eIF2α phosphorylation at the translation level, we simultaneously sequenced the polysomal RNA of atgcn1-1 after cold treatments.
Based on statistical analysis and after filtering, 200 down-regulated and 205 up-regulated genes were identified and respectively named as cold-AtGCN1 down-translated or up-translated genes (Supplementary Table 2). Upon cold stress, the translation of cold-AtGCN1 down-translated or up-translated genes were either down-translated or up-translated in the WT (the sum of two signal values > 10; a fold change of ≥1.5; p-Value < 0.05), while the translational regulation of these proteins was not affected in atgcn1-1 (the sum of two signal values > 10; a fold change of ≤1.5). Additionally, to remove the affection of transcription, we excluded the regulated genes.
Hierarchical clustering analysis of cold-AtGCN1 regulated genes at translational levels demonstrated that most of cold-AtGCN1 inhibited or promoted genes were inhibited or promoted in atgcn1-1 after cold treatments (Figure 4A and Tables 3, 4), although the regulation level of translation was lower in atgcn1-1 than in WT. Consistent with the consequence reported for the transcription of regulated genes, the polysomal RNA sequencing data suggest that there are alternative mechanisms involved in regulating protein translation, independent of AtGCN1-mediated eIF2α phosphorylation.
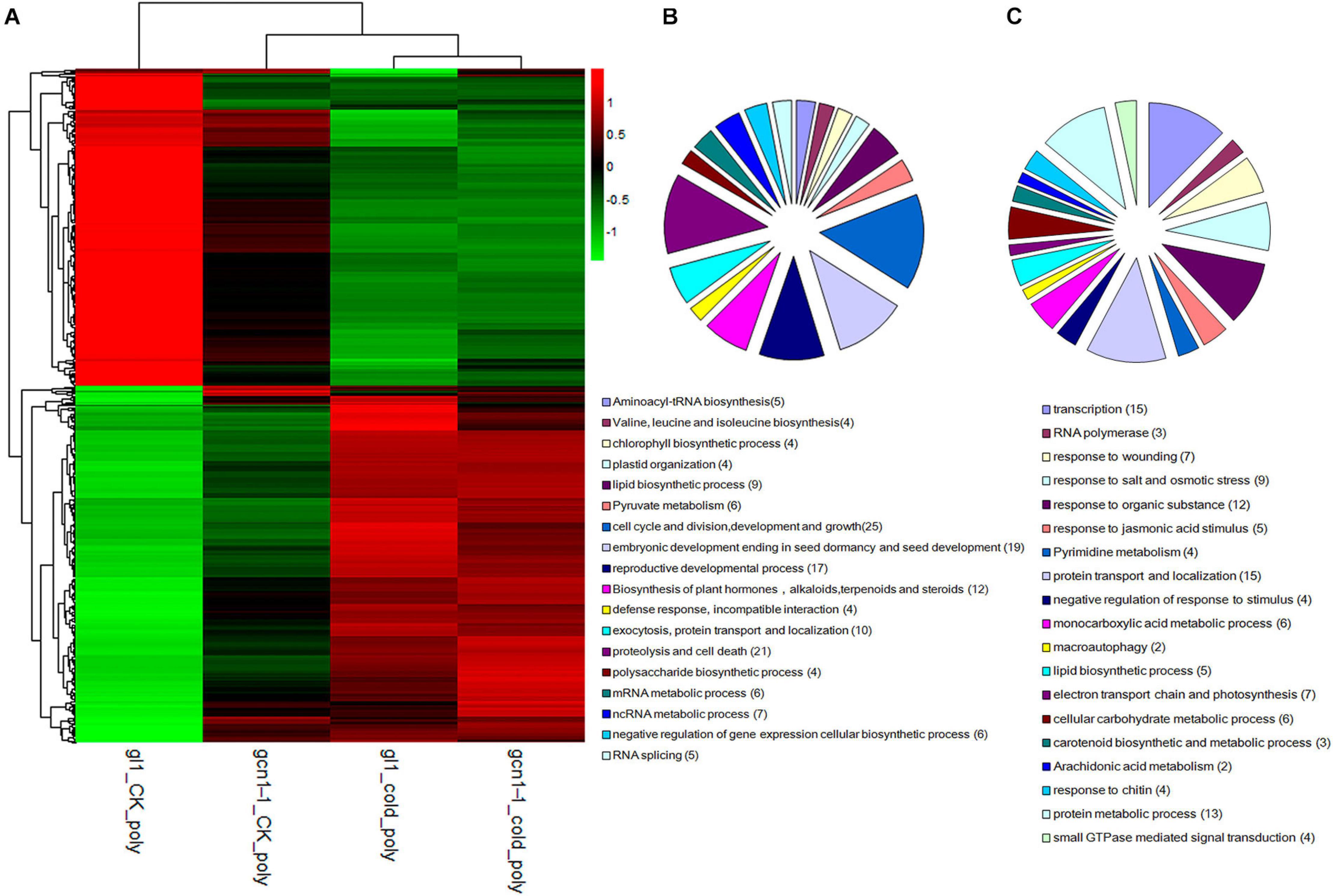
Figure 4. eIF2α phosphorylation selectively regulates protein translation under cold stress conditions. (A) Hierarchical clustering analysis of cold-ATGCN1 genes at translational levels. (B) GO terms analysis of cold-ATGCN1 genes that were down-regulated at translational levels. (C) GO terms analysis of cold-ATGCN1 genes that were up-regulated at translational levels.
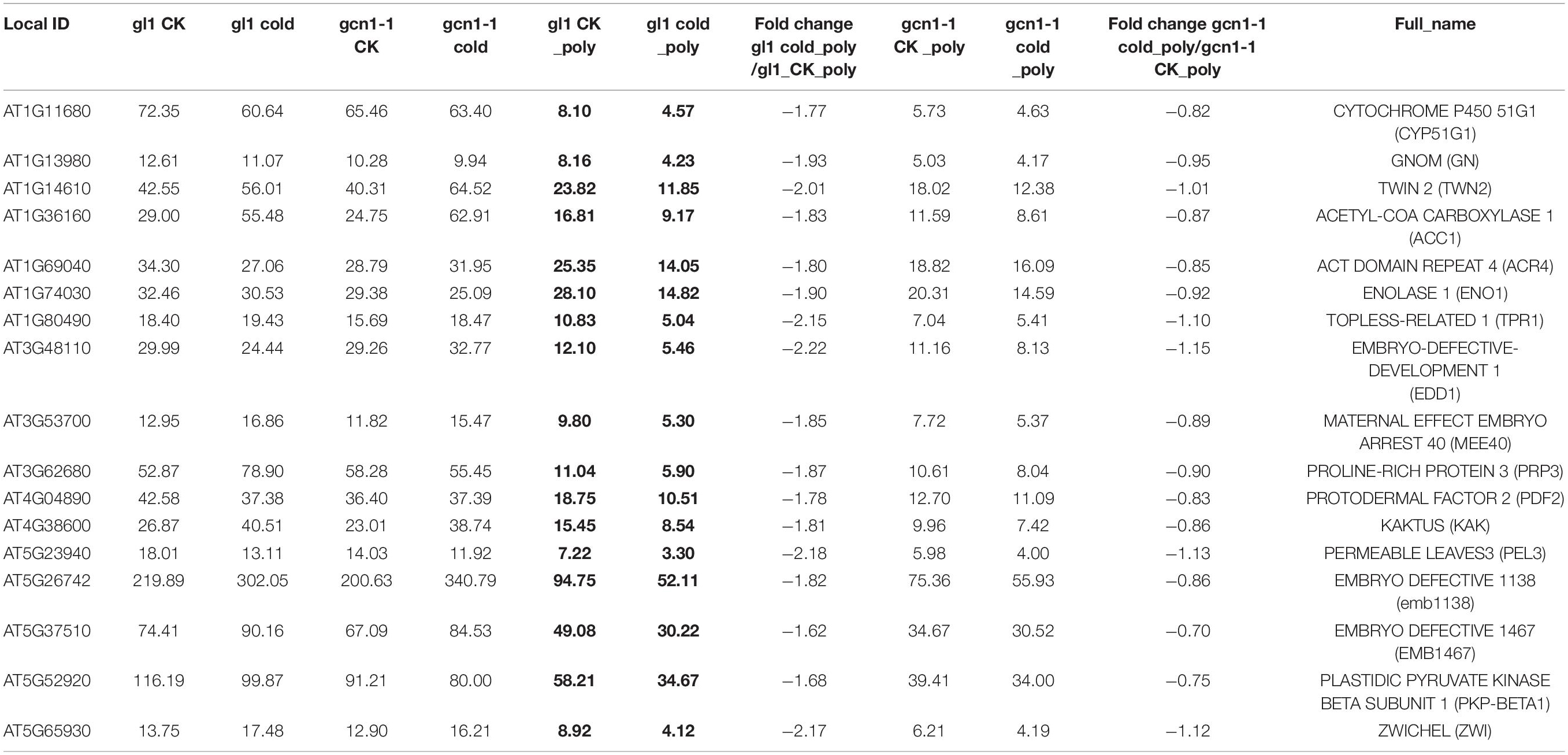
Table 3. Cold-ATGCN1 down-translated genes are involved in embryonic development ending in seed dormancy and seed development.
We classified the cold-AtGCN1 down-translated or up-translated genes using DAVID (Database for Annotation, Visualization and Integrated Discovery, see text footnote 1) (Figures 4B,C and Supplementary Table 2). 19 genes involved in embryonic development ending in seed dormancy and seed development were down-translated by cold stress in WT, due to cold-induced eIF2α phosphorylation (Table 3). On the other hand, they are not regulated in atgcn1-1, in which eIF2α phosphorylation is lost. Cold-AtGCN1 down-translated genes were also identified with roles in aminoacyl-tRNA biosynthesis (5), chlorophyll biosynthetic process (4), plastid organization (4), lipid biosynthetic process (9), pyruvate metabolism (6), cell cycle and division, development and growth (25), reproductive developmental process (17), exocytosis, protein transport and localization (10), proteolysis and cell death (21), polysaccharide biosynthetic process (4), mRNA metabolic process (6), ncRNA metabolic process (7), negative regulation of gene expression of cellular biosynthetic process (6), RNA splicing (5), and incompatible interaction with defense response(4) (Figure 4B and Supplementary Table 2).
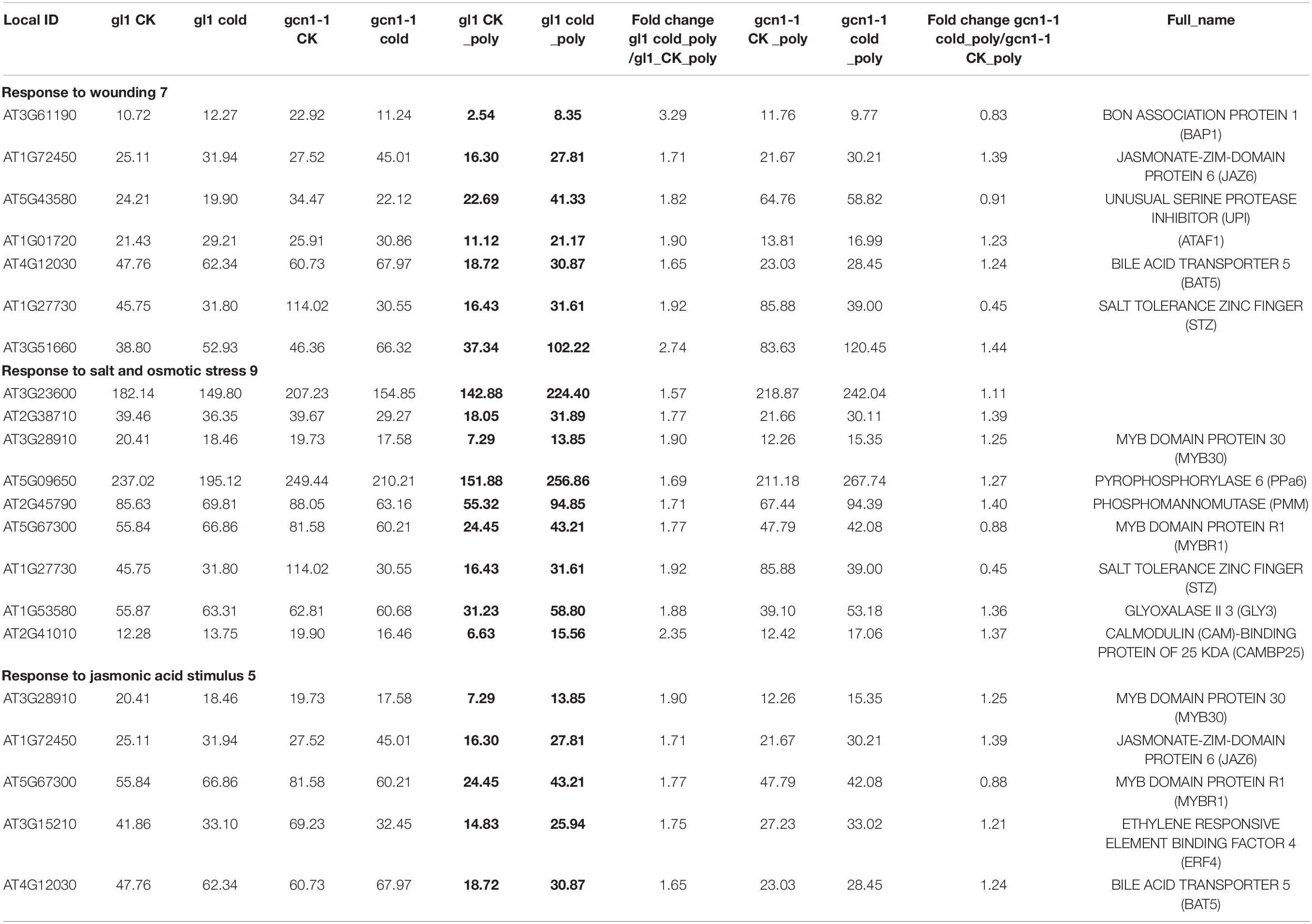
Lots of genes that were induced in the WT were noted to remain unchanged at translational levels in atgcn1-1 under cold stress, which are referred as cold-AtGCN1 up-translated genes. Classification results show that cold-AtGCN1 up-translated genes are involved in response to wounding, salt and osmotic stress, jasmonic acid (JA) stimulus (Table 4). Moreover, these genes take part in DNA-directed RNA polymerase (3), response to organic substance (12), pyrimidine metabolism (4), protein transport and localization (15), negative regulation of genes in response to stimulus (4), monocarboxylic acid metabolic process (6), macroautophagy (2), lipid biosynthetic process (5), electron transport chain and photosynthesis (7), cellular carbohydrate metabolic process (6), carotenoid biosynthetic and metabolic process (3), arachidonic acid metabolism (2), response to chitin (4), protein metabolic process (13), small GTPase mediated signal transduction (4), and gene transcription (15) (Figure 4C and Supplementary Table 2).
Kozak Sequence and uORFs Affect Translation Selection Under Cold Stress Conditions
The initiation codon context is known to be important for translation selection of a mRNA (Kozak, 1987; Hinnebusch, 2011), and the optimized sequence is (A/G)nnATG(G) (Kozak sequence), with the initiation codon (bold) and purine residues at −3 and +4. For a “strong” initiation codon, the nucleotides both at positions +4 (i.e., G in the consensus) and −3 (i.e., either A or G in the consensus) match the consensus; An “adequate” initiation codon match only 1 of 2 sites; a “weak” initiation codon match neither.
According to Araport11 on TAIR (https://www.arabidopsis.org), we analyzed the initiation codon of total genes, cold-AtGCN1 down-translated and up-translated genes (Table 5 and Supplementary Table 2). The rate of transcripts with strong, adequate and weak initiation codon of total genes is respectively 34.0%, 44.5%, and 10.6% in Arabidopsis. Among cold-AtGCN1 down-translated genes, 48.4% of the transcripts show a strong start codon and 41.8% of ones have an adequate initiation codon. Among cold-AtGCN1 up-translated genes, 41.3% of the transcripts show a strong start codon and 52.1% of ones have an adequate initiation codon. Thus, the percentage of cold-AtGCN1 down- and up-translated genes with a strong or adequate initiation codon is respectively 90.2 and 93.4%, which are both higher than that in total genes (78.5%). The results show that cold-AtGCN1 mediated eIF2α phosphorylation may regulate the translation preference to transcripts with strong or adequate initiation codon and transcripts with weak initiation codon or without 5′-UTR (untranslated regions, UTR) have low preference to be regulated by cold-AtGCN1 mediated eIF2α phosphorylation.
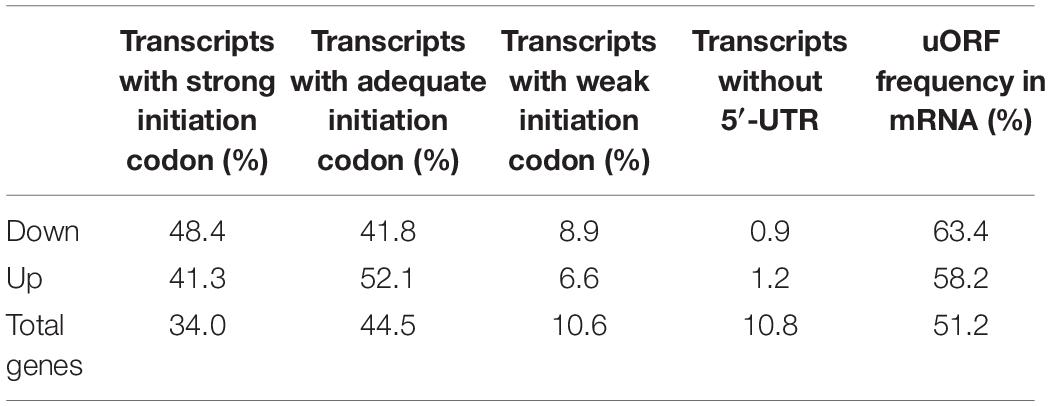
Table 5. The frequency of initiation codon and uORFs for cold-AtGCN1 down-translated or up-translated genes.
Yeast GCN4, whose translation is specifically increased in response to eIF2α phosphorylation, has four or two upstream open reading frames (uORFs) prior to the initiation codon (Mueller and Hinnebusch, 1986; Vattem and Wek, 2004). According to Araport11 on TAIR (https://www.arabidopsis.org), we analyzed the uORF of transcripts and the rate of transcripts with uORF was 51.2% (Supplementary Table 2). We hypothesized that eIF2α phosphorylation should promote the translation of transcripts with uORF in plants. However, as shown in Table 1, both 63.4% of down-translated transcripts and 58.2% of up-translated genes with uORFs are higher than 51.2% of total genes with uORFs. The results show that cold-AtGCN1 mediated eIF2α phosphorylation may selectively regulate the translation preference to transcripts with uORFs, which is different from the hypothesis we supposed.
AtGCN1 Selectively Regulates Gene Transcription and Protein Translation in Normal Temperature Conditions
Hierarchical clustering analysis of cold-AtGCN1 target genes showed that most of them were regulated in atgcn1-1 in normal temperature conditions (Figures 3B, 4A), suggesting that the low level of eIF2α phosphorylation mediated by AtGCN1 regulates the transcription or translation of specific genes. According to statistics and filtering (the sum of two signal values > 10; fold change ≥ 1.5; p-Value < 0.05.), we found 509 genes were up-translated and 249 genes were up-transcribed in atgcn1-1 in normal temperature conditions, compared to that in WT (Supplementary Table 3). On the other hand, 354 genes were down-translated and 47 genes were down-transcribed in atgcn1-1, compared to that in WT in normal temperature conditions. To exclude the effect of transcription, regulated genes at transcriptional levels were eliminated.
We respectively classified the up-translated and up-transcribed genes in atgcn1-1 using DAVID online. The up-translated genes in atgcn1-1 include genes responsive to not only abiotic stress including oxidation (16), cold (13), and salt (16), but also hormones including abscisic acid (ABA) (17), ethylene (9), salicylic acid (SA) (11), JA (9), auxin (11), brassinosteroid (BR) (4) and gibberellin (7) (Table 6 and Supplementary Table 3). The up-transcribed genes in atgcn1-1 are also found involved in abiotic stress and hormone signals including water deprivation (29), cold (19), wounding (24), salt (18), ABA (25), SA (12), JA (12) (Table 7 and Supplementary Table 3). The results show that AtGCN1 mediated eIF2α phosphorylation in normal temperature conditions inhibits the expression of genes involved in stress stimulus and hormone signals, at both transcriptional and translational levels. In addition, up-transcribed genes in atgcn1-1 are mostly up-regulated at translational levels, and we can’t infer whether the increased translation of these transcripts in atgcn1-1 contributes to the transcriptional increase or vice versa.
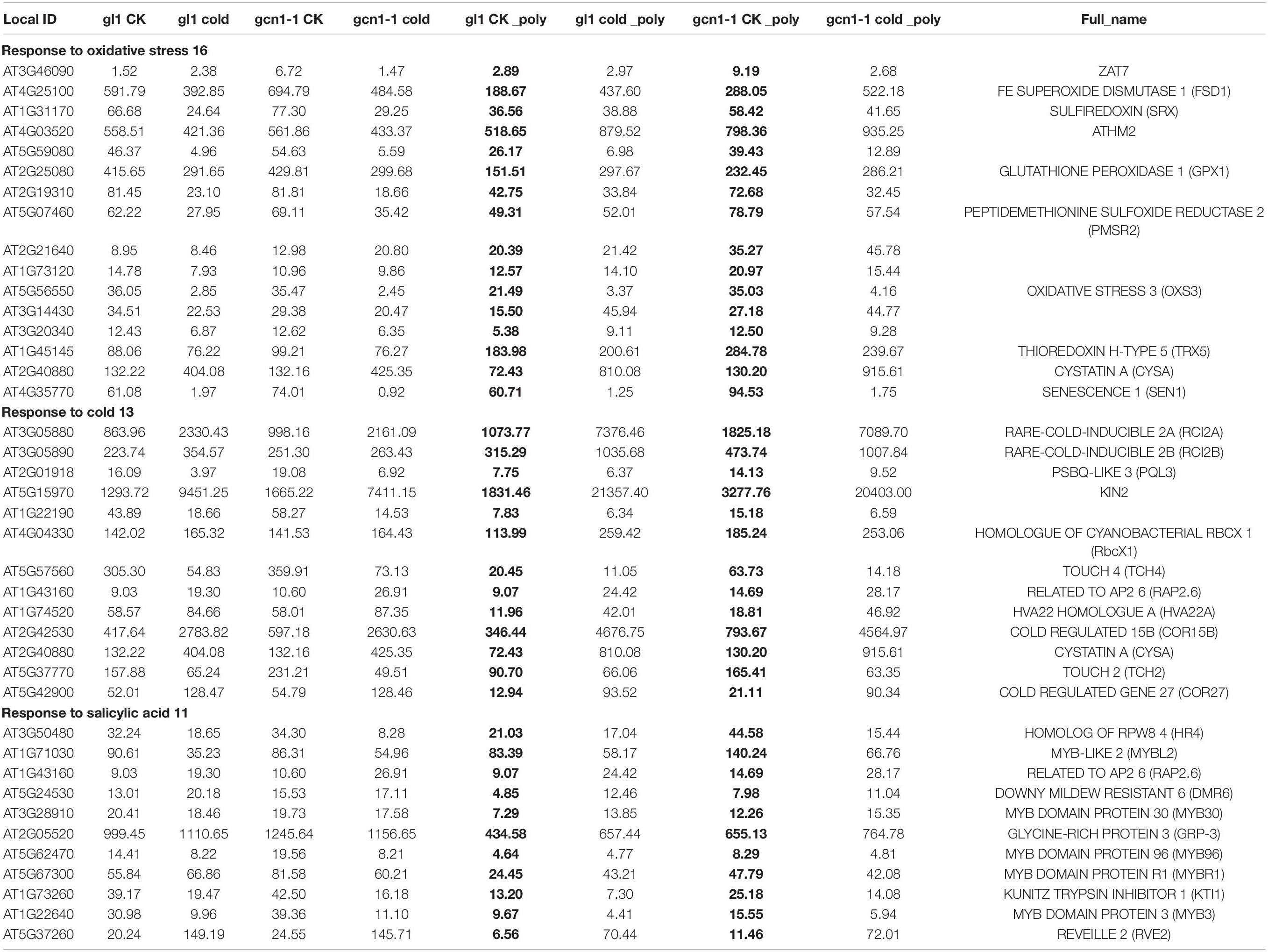
Table 6. Partial classification data of up-translated genes in atgcn1-1, compared with that in WT under normal temperature conditions.
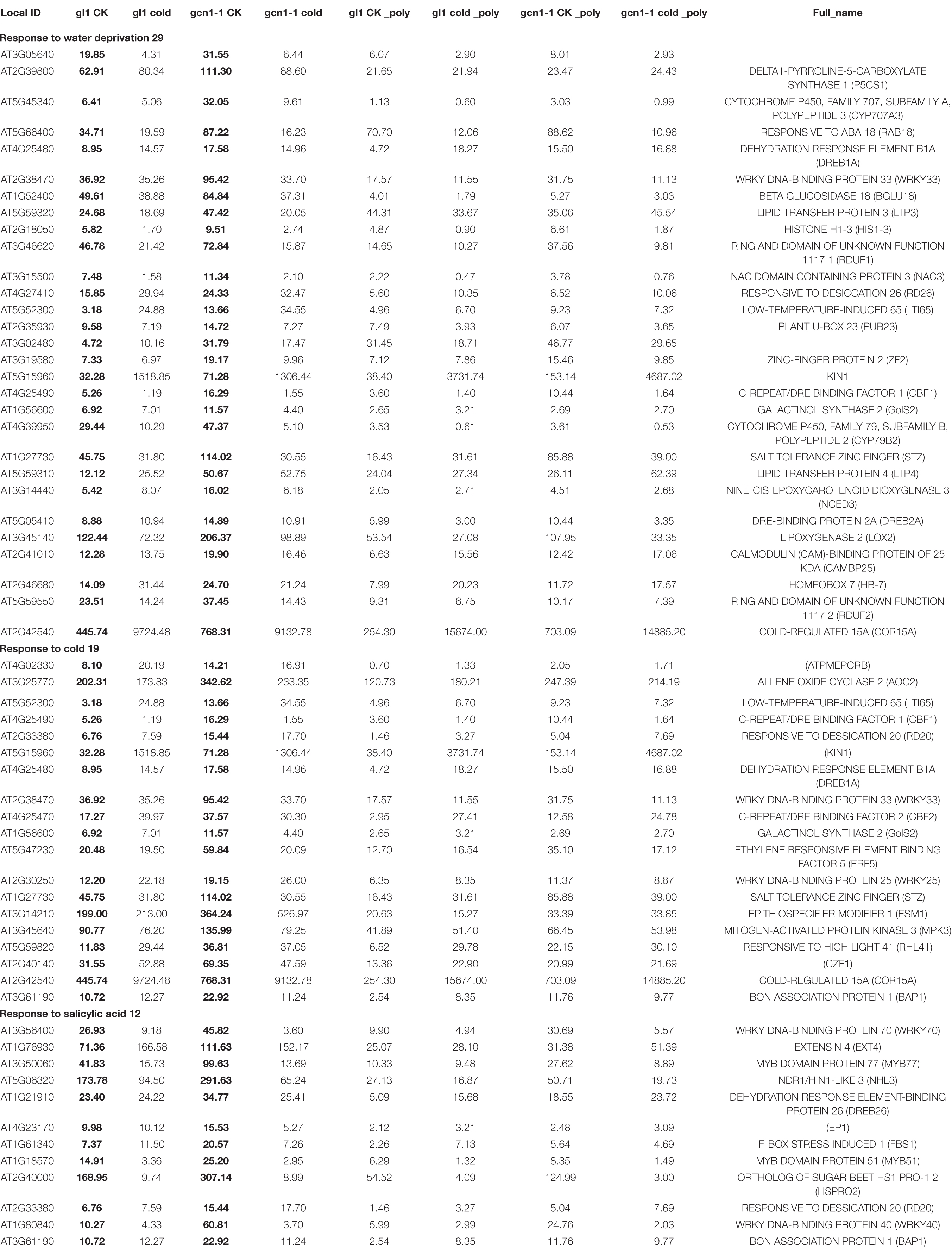
Table 7. Partial classification data of up-transcribed genes in atgcn1-1, compared with that in WT under normal temperature conditions.
Due to the limited number of down-regulated in atgcn1-1, the classification may not make sense. Down-translated genes in atgcn1-1 were found to be involved not only in plant development including flower development (11), post-embryonic development (5) and root hair elongation (5) (Table 8), but also in intracellular protein transport (14), protein folding (12), and endocytosis (8) (Supplementary Table 3).
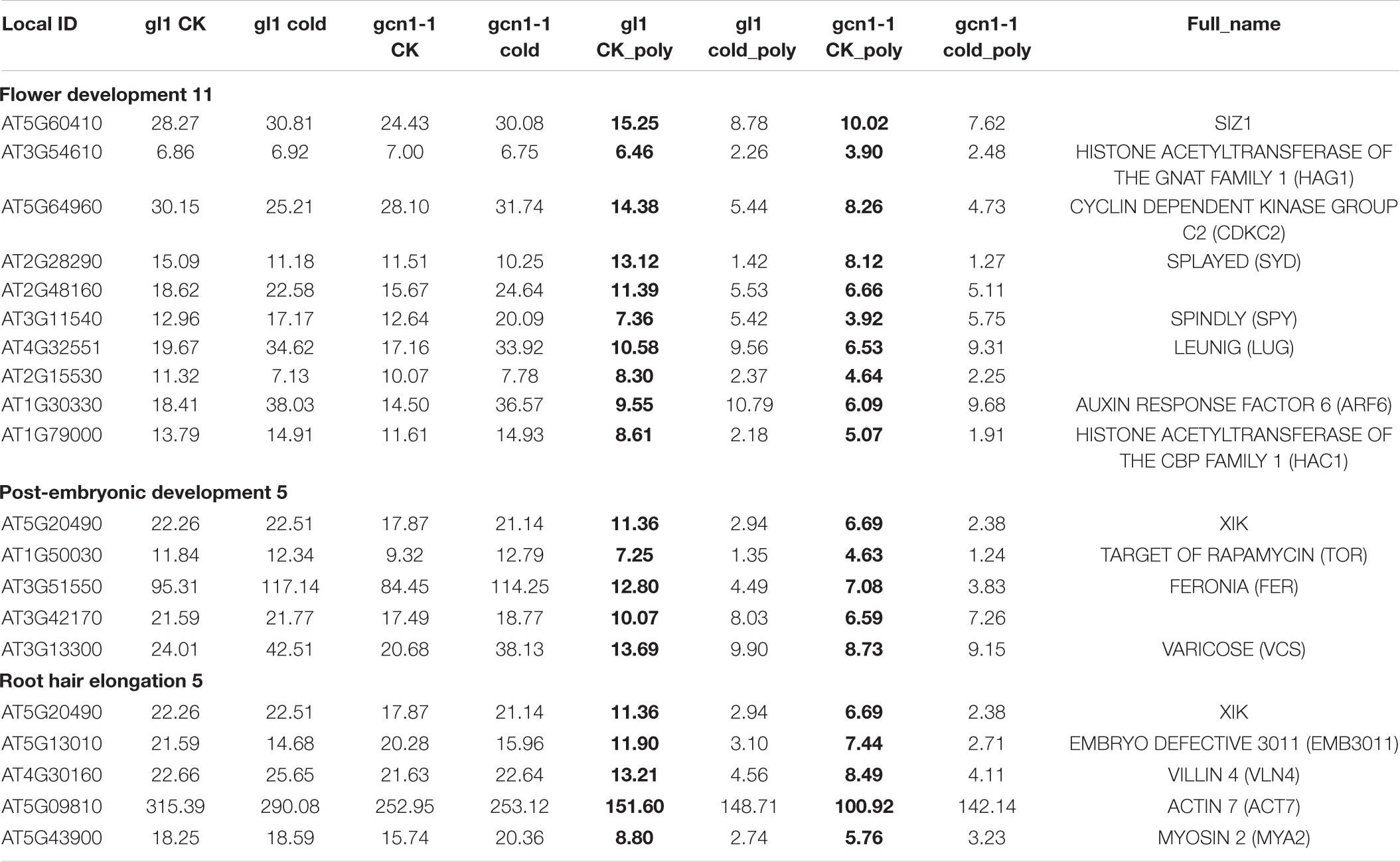
Table 8. Partial Classification data of down-translated genes in atgcn1-1, compared with that in WT under temperature conditions.
Kozak Sequence and uORFs Affect Translation Selection Under Normal Temperature Conditions
As shown in Figure 3A, WT plants have a low level of eIF2α phosphorylation in normal temperature conditions. The low level of eIF2α phosphorylation were showed to regulate expression of specific genes (Tables 6-8 and Supplementary Table 3). We wonder how the low level of eIF2α phosphorylation regulate protein translation, so we further analyzed the initiation codon and uORFs of the up-translated and down-translated genes in atgcn1-1 under normal temperature conditions.
The percentage of up-translated and down-translated transcripts with strong initiation codon in atgcn1-1 is respectively 45.6% and 41.8%, both are higher than 34% of total genes (Table 9). Meanwhile, the percentage of up-translated and down-translated transcripts with adequate initiation codon in atgcn1-1 are respectively 45.6% and 41.8%, both are higher than 34% of total genes. On the other hand, the percentage of up-translated and down-translated transcripts with weak initiation codon or without 5′-UTR are lower than that of total genes. The results show that the transcripts with strong or adequate initiation codon have the preference to be regulated by GCN1 mediated eIF2α phosphorylation, which is consistent with the preference of cold-AtGCN1 mediated eIF2α phosphorylation.
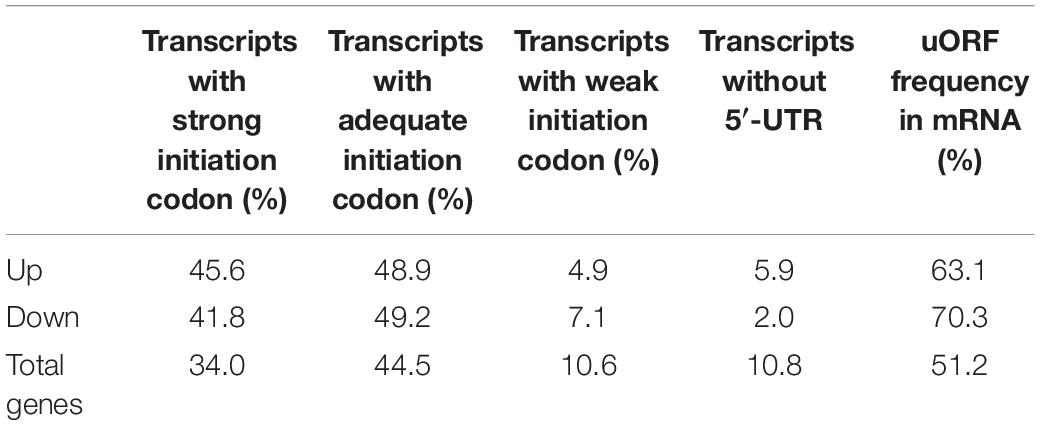
Table 9. The frequency of initiation codon and uORFs for up-translated and down-translated genes in atgcn1-1, compared with WT, under normal temperature conditions.
We next analyzed the uORFs of up-translated and down-translated genes in atgcn1-1, compared with WT. The uORF frequency of up-translated and down-translated transcripts is individually 63.1% and 70.3%, both are higher than that in total genes (51.2%). The results demonstrate that the transcripts with uORFs have the preference to be regulated by GCN1 mediated eIF2α phosphorylation in normal temperature conditions, which also coincides with the preference of cold-AtGCN1 up and down-translated genes by cold-induced eIF2α phosphorylation.
Discussion
In Arabidopsis, GCN2 is the only known eIF2α kinase to phosphorylate eIF2α. Additionally, eIF2α can’t be phosphorylated under amino acid starvation in gcn2-knockout mutants (Lageix et al., 2008). In our previous study, we have cloned AtGCN1 to demonstrate that AtGCN1 interacts with GCN2 to phosphorylate eIF2α upon cold stress (Wang et al., 2017). These evidence collectively indicate that the phosphorylation of eIF2α is defective in atgcn1 mutants and AtGCN1-GCN2 mediated eIF2α phosphorylation is essential for cold acclimation.
In this study, we further investigated the function of AtGCN1 in Arabidopsis. Firstly, we found that atgcn1 mutants flowered later than the WT (Figure 1). Secondly, we discovered that plant growth and seed development were arrested in atgcn1 mutants (Figure 2). FLC, a negative regulator of flower development, could be attributed to this observation as it was up-regulated in atgcn1-1 compared to the WT (Figure 2B and Supplementary Table 1).
Since the phosphorylation of eIF2α is defective in atgcn1, the translation of protein was proposed to be in an enhanced level in atgcn1. The defects noted in plant growth and seed development of mutant atgcn1, is opposite with the improved growth of AtTOR-overexpressing plants (Deprost et al., 2007), although the loss of eIF2α phosphorylation in atgcn1 as well as AtTOR-overexpression may promote protein translation (Wang et al., 2017). The results show that enhanced protein translation doesn’t associate with vigorous growth and AtGCN1 play roles in the growth at both vegetative and reproductive stages.
Thus far, AtGCN1 has been observed to interact with only eIF2 kinase GCN2 to phosphorylate eIF2α upon cold stress (Wang et al., 2017). However, it is still not clear how AtGCN1 mediated eIF2α phosphorylation upon cold stress affects transcription and protein translation in plants. Therefore, we performed sequencing on the total and polysomal RNA of atgcn1 mutants after cold treatments to further examine the function of eIF2α phosphorylation mediated by AtGCN1.
Hierarchical clustering analysis of cold-AtGCN1 regulated genes at both transcriptional and translational levels demonstrates that there are alternative pathways, independent of eIF2α phosphorylation, involved in regulating gene expression at both transcriptional and translational levels (Figures 3, 4). In mammals, concerted activation of AMPK (AMP-activated protein kinase), inhibition of TORC1 (TARGET OF RAPAMYCIN complex 1), phosphorylation of eIF2α and other signals may inhibit protein synthesis in parallel to ensure cellular survival under low temperature and endoplasmic reticulum (ER) stress (Hofmann et al., 2012; Guan et al., 2014). In Arabidopsis, there are at least two pathways required to suppress protein translation under cold stress, including the inhibition of TOR1 and eIF2α phosphorylation (Wang et al., 2017). The RNA sequencing results confirm that AtGCN1 mediated eIF2α phosphorylation is not the only way to monitor gene transcription and protein translation in cold stress.
Importantly, the RNA sequencing results demonstrated that AtGCN1-mediated eIF2α phosphorylation upon cold stress selectively regulates gene expression at both transcriptional and translation levels. The classifications of cold-AtGCN1 regulated genes show that AtGCN1-mediated eIF2α phosphorylation upon cold stress selectively regulate the transcription or translation of genes in response to blue light, hormone stimulus, toxin, wounding, salt stress, osmotic stress and organic substance; suggesting that AtGCN1 has a functional role in abiotic or biotic stress, in addition to cold stress. Moreover, AtGCN1-mediated eIF2α phosphorylation upon cold stress selectively regulates the gene expression involved in amino acid biosynthesis, photosynthesis, cell wall organization, protein transport and localization, lipid biosynthesis, gene transcription, macroautophagy, proteolysis and cell death.
In normal temperature conditions, the low level of eIF2α phosphorylation in WT is always detected (Figure 3A), so we infer that the low level of eIF2α phosphorylation may regulate gene expression. We compared the expression difference between atgcn1-1 and WT in normal temperature conditions and a lot of genes were filtered out (Supplementary Table 3). The results indicate that the low level of eIF2α phosphorylation in normal temperature conditions can selectively regulate gene expression at both transcriptional and translational level, as well as the cold-induced high level of eIF2α phosphorylation in cold stress conditions.
Interestingly, we found that cold-AtGCN1 induced or inhibited genes in WT at both transcriptional and translational levels were mostly induced or inhibited in atgcn1-1 in normal temperature conditions, compared with that in WT (Tables 1–4 and Supplementary Tables 1, 2). The results indicate that the high level of eIF2α phosphorylation in cold stress conditions and the low level of eIF2α phosphorylation in normal temperature conditions play opposite roles in the regulation of gene expression. In other words, the high level of eIF2α phosphorylation promotes or inhibits the expression of specific genes at both transcriptional and translational levels, while the low level of eIF2α phosphorylation oppositely regulates the expression of these genes.
The classifications of up-regulated genes in atgcn1-1 in normal temperature conditions show that AtGCN1 negatively regulates the transcription or translation of genes involved in biotic stress, abiotic stress and hormone signaling (Tables 6, 7). The results indicate that AtGCN1 mediated eIF2α phosphorylation should respond to a lot of stimulus, in addition to cold stress, which is consistent with the conclusion of cold-AtGCN1 up-regulated genes (Tables 2, 4). Moreover, a lot of genes in plant development were found to be down-translated in atgcn1-1 in normal temperature conditions (Table 8 and Supplementary Table 3), including flower development, post-embryonic development, cell wall organization, primary shoot apical meristem specification, root hair elongation, cell growth, cell division and seed germination.
The down-translated genes of atgcn1-1 in plant development suggest that eIF2alpha phosphorylation play an important role in plant growth. Firstly, we found that atgcn1 mutants flowered later than the WT (Figure 1). Meantime, we discovered that plant growth and seed development were arrested in atgcn1 mutants (Figure 2). The down-translated genes of atgcn1-1 in plant development may explain the late flowering, arrested growth and seed defect noticed in atgcn1 mutants.
We analyzed the initiation codon and uORFs of cold-AtGCN1 down-translated, cold-AtGCN1 up-translated genes and down/up-translated genes in atgcn1-1 in normal temperature conditions (Tables 5, 9). These results demonstrate that eIF2α phosphorylation results in the translation preference to transcripts with strong or adequate initiation codon. In other words, transcripts with a weak initiation codon or without 5′-UTR has less opportunity to be regulated by eIF2α phosphorylation than ones with a stronger initiation codon, which is the first report to our knowledge. Moreover, the uORF analysis of these genes shows that AtGCN1 mediated eIF2α phosphorylation may regulate the translation preference to transcripts with uORFs, which is different from the function of eIF2α phosphorylation in yeast and mammalians.
In yeast and mammalians, eIF2α phosphorylation specifically increases the translation of GCN4 or ATF4 with uORFs (Mueller and Hinnebusch, 1986; Vattem and Wek, 2004). Due to pathogen-triggered eIF2α phosphorylation, Arabidopsis TBF1 is supposed to be specifically translated through its two uORFs (Pajerowska-Mukhtar et al., 2012; Lokdarshi et al., 2020). However, TBF1 is not regulated in the data. The most likely cause is that TBF1 is specifically induced by pathogen infection, not by cold stress treatment used here.
In addition, among the down-translated genes in atgcn1-1 under normal temperature conditions, we found that lots of chloroplastic and mitochondrial genes were down-translated in atgcn1-1, compared with that in WT (Supplementary Table 3). The results suggest that the lack of eIF2α phosphorylation in atgcn1 may repress the translation of organellar genes, which need be explored in the future.
In this work, a further investigation of AtGCN1 was achieved. Total and polysomal RNA sequencing of atgcn1-1 shows that eIF2 phosphorylation mediated by AtGCN1 selectively regulates gene expression at both transcriptional and translational levels and Kozak sequence and uORFs of transcripts affect translation selection. Moreover, mutations of atgcn1 impairs flowering time, plant growth and seed development, which reflecting the affection of the expression alteration in atgcn1-1. All results show that AtGCN1 mediated eIF2 phosphorylation selectively regulates gene transcription and protein translation in Arabidopsis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences, https://bigd.big.ac.cn/gsa/, CRA003757.
Author Contributions
HZ designed the research. XC, KG, LW, ML, ZL, DZ, LD, and XL performed the experiments. HZ, WW, and WY analyzed the sequencing data. HZ and J-KZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We acknowledge the funding supported by the National Natural Science Foundation of China to HZ (31270309), and the Chinese Academy of Sciences to J-KZ. We also acknowledge the Arabidopsis Biological Resource Center (Columbus, OH, United States) for providing the T-DNA seeds of atgcn1-2.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.630311/full#supplementary-material
Supplementary Table 1 | Cold-AtGCN1 regulated genes at transcriptional levels.
Supplementary Table 2 | Cold-AtGCN1 regulated genes at translational levels.
Supplementary Table 3 | Regulated genes in atgcn1-1 in normal temperature conditions.
Supplementary Table 4 | Primers used for qRT-PCR.
Footnotes
References
Chen, Y., Lun, A. T., and Smyth, G. K. (2016). From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000 Res. 5:1438. doi: 10.12688/f1000research.8987.2
Deprost, D., Yao, L., Sormani, R., Moreau, M., Leterreux, G., Nicolaï, M., et al. (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8, 864–870. doi: 10.1038/sj.embor.7401043
Dever, T. E., Chen, J. J., Barber, G. N., Cigan, A. M., Feng, L., Donahue, T. F., et al. (1993). Mammalian eukaryotic initiation factor 2 alpha kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc. Natl. Acad. Sci. U.S.A. 90, 4616–4620. doi: 10.1073/pnas.90.10.4616
Dever, T. E., Feng, L., Wek, R. C., Cigan, A. M., Donahue, T. F., and Hinnebusch, A. G. (1992). Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68, 585–596. doi: 10.1016/0092-8674(92)90193-g
Erickson, F. L., and Hannig, E. M. (1996). Ligand interactions with eukaryotic translation initiation factor 2: role of the gamma-subunit. EMBO J. 15, 6311–6320. doi: 10.1002/j.1460-2075.1996.tb01021.x
Guan, B. J., Krokowski, D., Majumder, M., Schmotzer, C. L., Kimball, S. R., Merrick, W. C., et al. (2014). Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2alpha. J. Biol. Chem. 289, 12593–12611. doi: 10.1074/jbc.m113.543215
Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapir, M., et al. (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. doi: 10.1016/s1097-2765(00)00108-8
Hinnebusch, A. G. (1988). Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 52, 248–273. doi: 10.1128/mmbr.52.2.248-273.1988
Hinnebusch, A. G. (2011). Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467. doi: 10.1128/mmbr.00008-11
Hofmann, S., Cherkasova, V., Bankhead, P., Bukau, B., and Stoecklin, G. (2012). Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol. Biol. Cell 23, 3786–3800. doi: 10.1091/mbc.e12-04-0296
Jia, M. H., Larossa, R. A., Lee, J. M., Rafalski, A., Derose, E., Gonye, G., et al. (2000). Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol. Genomics 3, 83–92. doi: 10.1152/physiolgenomics.2000.3.2.83
Jiang, H. Y., and Wek, R. C. (2005). GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem. J. 385, 371–380. doi: 10.1042/bj20041164
Kozak, M. (1987). An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15, 8125–8148. doi: 10.1093/nar/15.20.8125
Lageix, S., Lanet, E., Pouch-Pelissier, M. N., Espagnol, M. C., Robaglia, C., Deragon, J. M., et al. (2008). Arabidopsis eIF2alpha kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 8:134. doi: 10.1186/1471-2229-8-134
Lokdarshi, A., Guan, J., Camacho, R. A. U., Cho, S. K., Morgan, P. W., Leonard, M., et al. (2020). Light activates the translational regulatory kinase GCN2 via reactive Oxygen Species Emanating from the Chloroplast. Plant Cell 32, 1161–1178. doi: 10.1105/tpc.19.00751
Lu, P. D., Harding, H. P., and Ron, D. (2004). Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33. doi: 10.1083/jcb.200408003
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.2307/3870827
Mueller, P. P., and Hinnebusch, A. G. (1986). Multiple upstream AUG codons mediate translational control of GCN4. Cell 45, 201–207. doi: 10.1016/0092-8674(86)90384-3
Narasimhan, J., Staschke, K. A., and Wek, R. C. (2004). Dimerization is required for activation of eIF2 kinase Gcn2 in response to diverse environmental stress conditions. J. Biol. Chem. 279, 22820–22832. doi: 10.1074/jbc.m402228200
Natarajan, K., Meyer, M. R., Jackson, B. M., Slade, D., Roberts, C., Hinnebusch, A. G., et al. (2001). Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell Biol. 21, 4347–4368. doi: 10.1128/mcb.21.13.4347-4368.2001
Pajerowska-Mukhtar, K. M., Wang, W., Tada, Y., Oka, N., Tucker, C. L., Fonseca, J. P., et al. (2012). The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 22, 103–112. doi: 10.1016/j.cub.2011.12.015
Sormani, R., Delannoy, E., Lageix, S., Bitton, F., Lanet, E., Saez-Vasquez, J., et al. (2011). Sublethal cadmium intoxication in Arabidopsis thaliana impacts translation at multiple levels. Plant Cell Physiol. 52, 436–447. doi: 10.1093/pcp/pcr001
Vattem, K. M., and Wek, R. C. (2004). Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274. doi: 10.1073/pnas.0400541101
Wang, L., Li, H., Zhao, C., Li, S., Kong, L., Wu, W., et al. (2017). The inhibition of protein translation mediated by AtGCN1 is essential for cold tolerance in Arabidopsis thaliana. Plant Cell Environ. 40, 56–68. doi: 10.1111/pce.12826
Wu, S., Tan, M., Hu, Y., Wang, J. L., Scheuner, D., and Kaufman, R. J. (2004). Ultraviolet light activates NFkappaB through translational inhibition of IkappaBalpha synthesis. J. Biol. Chem. 279, 34898–34902. doi: 10.1074/jbc.m405616200
Yang, R., Wek, S. A., and Wek, R. C. (2000). Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell Biol. 20, 2706–2717. doi: 10.1128/mcb.20.8.2706-2717.2000
Zaborske, J. M., Wu, X., Wek, R. C., and Pan, T. (2010). Selective control of amino acid metabolism by the GCN2 eIF2 kinase pathway in Saccharomyces cerevisiae. BMC Biochem. 11:29. doi: 10.1186/1471-2091-11-29
Keywords: protein translation, eIF2α phosphorylation, GCN2, AtGCN1, cold stress
Citation: Cui X, Gao K, Wang L, Lv M, Li Z, Zheng D, Wu W, Yao W, Ding L, Li X, Zhu J-K and Zhang H (2021) General Control Non-derepressible 1 (AtGCN1) Is Important for Flowering Time, Plant Growth, Seed Development, and the Transcription/Translation of Specific Genes in Arabidopsis. Front. Plant Sci. 12:630311. doi: 10.3389/fpls.2021.630311
Received: 18 November 2020; Accepted: 01 February 2021;
Published: 31 March 2021.
Edited by:
Raju Datla, Global Institute for Food Security, CanadaReviewed by:
Eiji Nambara, University of Toronto, CanadaDaoquan Xiang, National Research Council Canada (NRC-CNRC), Canada
Copyright © 2021 Cui, Gao, Wang, Lv, Li, Zheng, Wu, Yao, Ding, Li, Zhu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hairong Zhang, aHJ6aGFuZ0BoZW5hdS5lZHUuY24=
†These authors have contributed equally to this work
 Xiaona Cui1†
Xiaona Cui1† Linjuan Wang
Linjuan Wang Wen Yao
Wen Yao Jian-Kang Zhu
Jian-Kang Zhu Hairong Zhang
Hairong Zhang