- 1Rice Breeding Platform, International Rice Research Institute, Metro Manila, Philippines
- 2School of Agricultural Biotechnology, Punjab Agricultural University, Ludhiana, India
- 3International Rice Research Institute South Asia Regional Centre, Varanasi, India
There is an urgent need to breed dry direct-seeded adapted rice varieties in order to address the emerging scenario of water–labor shortage. The aim of this study was to develop high-yielding, direct-seeded adapted varieties utilizing biparental to multiparental crosses involving as many as six different parents in conventional breeding programs and 12 parents in genomics-assisted breeding programs. The rigorous single plant selections were followed from the F2 generation onwards utilizing phenotypic selection and quantitative trait locus (QTL)/gene-based/linked markers for tracking the presence of desirable alleles of targeted QTL/genes. In conventional breeding, multiparent lines had significantly higher yields (2,072–6,569 kg ha−1) than the biparental lines (1,493–6,326 kg ha−1). GAB lines derived from multiparent crosses had significantly higher (3,293–6,719 kg ha−1) yields than the multiparent lines from conventional breeding (2,072–6,569 kg ha−1). Eleven promising lines from genomics-assisted breeding carrying 7–11 QTL/genes and eight lines from conventional breeding with grain-yield improvement from 727 to 1,705 kg ha−1 and 68 to 902 kg ha−1, respectively, over the best check were selected. The developed lines may be released as varieties/parental lines to develop better rice varieties for direct-seeded situations or as novel breeding material to study genetic interactions.
Introduction
Rice is mainly cultivated under anaerobic conditions and primarily adapted and evolved under these conditions (Kumar and Ladha, 2011). A shortage of water and labor input makes rice production through transplanted rice (TPR) more expensive, less profitable, and unsustainable (Farooq et al., 2011). The estimates specify that the water requirement for irrigation by 2025 could be approximately 561 km3 for a low-demand scenario and 611 km3 for a high-demand scenario (Kumar et al., 2005). The demand for water in various non-agricultural sectors in the country reduces its availability for agriculture; as a result, the agricultural sector is likely to face another 10–15% reduction in irrigation water by 2025 (Boretti and Rosa, 2019). With 90% of the world's rice grown and consumed in Asia and the large water requirements of conventional transplanted rice cultivation systems, it is evident that this water scarcity will severely affect rice production in this continent. The water scarcity situation will lead to rice cultivation with less water (Wang et al., 2002).
Cultivation of dry direct-seeded rice (DDSR) helps in avoiding three basic operations: puddling, transplanting, and maintaining standing water. DDSR cultivation in saturated soil has been widely adopted in southern Brazil, Chile, Cuba, Venezuela, some of the Caribbean countries, and in certain areas of Colombia (Fischer and Antigua, 1996). Currently, DDSR is becoming more popular in South Asia, South East Asia, and, to some extent, in West Africa. The cultivation of DDSR is being practiced with several modifications of tillage or land preparation and crop establishment (CE) with a site-specific package, but has not gained the required popularity because of one or two unsolved deficiencies. In addition to higher economic returns, DDSR crops vis-à-vis the transplanted (TPR) system are faster and easier to plant, have a shorter duration, are less labor intensive, and consume less water (Bhushan et al., 2007). The mechanized DDSR cultivation system has been estimated to provide potential irrigation water savings of 40 cm ha−1, labor savings of 25 person-days ha−1, energy savings of 1,500 MJ ha−1, a reduction of GHG (green house gas) emissions of 1,500 kg CO2 equivalent ha−1, and a yield increase of 0.5 t ha−1 with an increased net economic return of USD 50 ha−1 in most rice-growing countries (Kumar and Ladha, 2011).
The development of DDSR varieties depends on several factors, such as selection of traits, identification, and introgression of genomic regions associated with those particular traits of interest. The traits that are expected to play an important role in providing yield stability and adaptability under direct-seeded conditions include anaerobic germination (ability to germinate under water, Ghosal et al., 2019), early uniform seedling emergence (Dixit et al., 2015; Sandhu et al., 2015), vegetative vigor, root phenotypic plasticity, proper nutrient uptake (Sandhu et al., 2015), and lodging resistance (Yadav et al., 2017; Sandhu et al., 2019b; Subedi et al., 2019). The biotic stress (disease and insect) resistance includes blast (Qu et al., 2006), brown spot (Sato et al., 2008), bacterial blight (Ullah et al., 2012), sheath blight, brown planthopper (Jairin et al., 2007), gall midges (Nair et al., 1996), and nematode resistance (Das et al., 2011).
Existing rice varieties are not specifically developed for direct-seeded ecosystems. However, some earlier developed rice varieties for transplanted conditions have been adapted to direct-seeded conditions exhibiting a yield decline under direct-seeded conditions. The newly developed drought-submergence-tolerant, rainfed-adapted rice varieties have shown good performance in India and Nepal, providing a yield advantage of 1.0 t ha−1 over the currently grown drought-submergence susceptible varieties (Sandhu et al., 2019a). Varieties such as Sahbhagi dhan, DRR 42, DRR 44, CR dhan 201, CR dhan 203, and Swarna Shreya (Sandhu and Kumar, 2017) developed by the International Rice Research Institute (IRRI) and genomics-assisted derived CR dhan 801 varieties have become highly popular in India and are reported to be cultivated on at least an area of 0.8 m ha. Similarly, in Nepal, the rice varieties Sukha dhan 3 and Sukha dhan 5 (Sandhu and Kumar, 2017), and the genomics-assisted derived lines Bahuguni dhan-1 and Bahuguni dhan-2 (Sandhu et al., 2019a) have become highly popular and contributed to the increased rice production in Nepal. Based on these achievements, the development of direct-seeded adapted rice varieties could provide opportunities for a significant increase in rice productivity in rice-growing countries.
Breeding methods such as QTL/gene pyramiding and multiparent application have been reported to be effective in developing resistant/tolerant varieties against biotic and abiotic stresses (Koide et al., 2009; Bandillo et al., 2013; Sandhu et al., 2019b). Conventional breeding is an efficient approach for the development of novel genetic variants (Breseghello and Coelho, 2013). The conventional breeding approach suffers from the problem of linkage drag, leading to the transfer of undesired traits closely linked with traits of interest. The use of molecular markers in breeding programs improves the efficiency of traditional breeding by enabling breeders to select trait-linked molecular markers (Collard and Mackill, 2008). The existence of abundant genetic diversity, genomic variation in the rice gene pool, and availability of modern genomic tools/techniques provides opportunities to choose suitable donors that are free from undesirable linkage and to dissect the quantitative nature of associated traits of interest. Most of the rice landraces perform better under various nutrient-deficient conditions (Wissuwa and Ae, 2001). Screening of diverse genotypes under direct-seeded conditions by assessing various biometrical traits, such as root architecture, plant height, and yield-related traits, will help to identify genotypes with improved yield, better root traits, higher nutrient uptake, higher nutrient utilization efficiency, and better plant type traits.
However, identification of donors, traits, and QTL for traits increasing rice yield under direct-seeded conditions and their introgression in elite genetic backgrounds using advances in genomics-assisted breeding (GAB) present a unique scenario to achieve significant yield advantages under direct-seeded conditions. The present study was undertaken to develop new direct-seeded adapted rice varieties utilizing the identified donors, genes/QTL, and their introgression in elite genetic backgrounds, compare the performance of bi-parental and multi-parental developed lines under dry direct-seeded conditions, and compare the performance of multiparent developed lines through conventional breeding and GAB approaches.
Materials and Methods
The study on the development of direct-seeded adapted rice lines through conventional and GAB approaches began at the International Rice Research Institute (IRRI), Los Baños, Laguna (Philippines) in 2009, with the identification of suitable traits and genomic regions associated with the traits improving grain yield and adaptability of rice under dry direct-seeded cultivation conditions.
Selection of Parents for the Conventional Breeding Program
The parents used in the conventional breeding program included widely grown high-yielding rice varieties under the transplanted system, drought-tolerant lines adapted to upland and shallow lowland conditions, and direct-seeded adapted breeding lines possessing traits required to increase rice adaptability to direct-seeded conditions. The conventional breeding program involved two, three, four, five, and six parent crosses. Detailed information on breeding lines/varieties used and the traits they possessed is presented in Table 1.
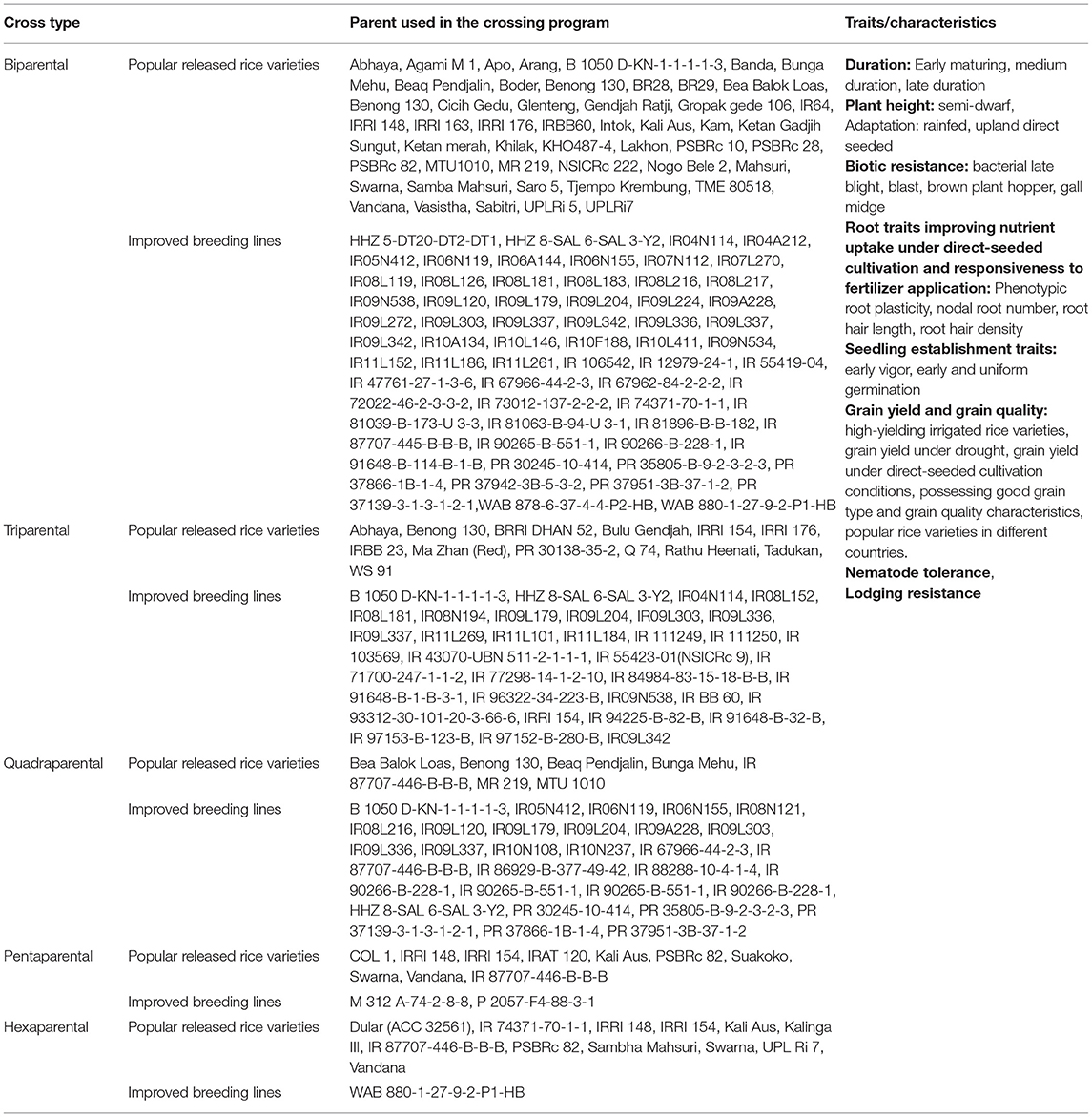
Table 1. Detailed information on trait characteristics and parents used to develop breeding lines in conventional breeding program.
Conventional Breeding Program
The conventional breeding hybridization program to develop suitable adapted breeding lines involving the parents presented in Table 1 was initiated in the dry season (DS) of 2009. New crosses were included in each successive season. The F1s were self-pollinated and phenotypic selection of F2s involving selection for bacterial late blight, blast, plant height, and visual grain yield was conducted under direct-seeded conditions. The best plants with enhanced plant type, disease resistance, grain type, and grain yield were carried forward in successive generations until the F4 generation. The plant-to-progeny testing of selected F4 plants was attempted in the F5 and F6 generation followed by an observational yield trial (OYT) and advanced yield trial (AYT) (Figure 1). The selection of disease resistance (blast and bacterial late blight) and grain quality was conducted in AYTs. The disease screening protocol followed was as described by Sandhu et al. (2018). The advanced breeding lines were analyzed for grain quality parameters at the IRRI GQNC (Grain Quality and Nutrition Center) laboratory facility. The detailed information on the grain quality testing is presented in Yadav et al. (2020). In the F5 and F6 generations, the progenies were tested in replicated yield trials with a plot size of 1.6 m2, whereas the plot sizes in OYT and AYT were 3.2 and 4.8 m2, respectively. In each generation, the plants were grown maintaining 20 cm (hill to hill) × 20 cm (row to row) distance. Detailed information on soil characteristics of upland fields, land preparation, field management, and phenotypic screening was provided by Sandhu et al. (2019b).
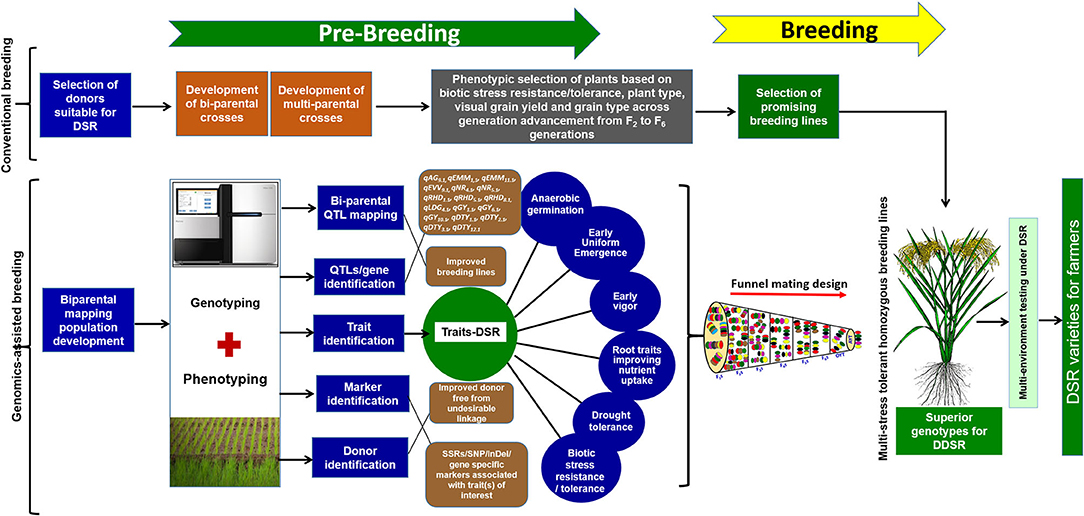
Figure 1. Detailed schematic representation of the pre-breeding steps, use of genomics-assisted breeding following funnel mating design to assemble the targated QTL/genes to develop multi-stress-tolerant homozygous breeding lines suitable for direct-seeded cultivation conditions.
Selection of Traits, Donors, and QTL for the Genomics-Assisted Introgression Program
Seedling emergence and establishment traits selected for the development of direct-seeded adapted rice varieties included early uniform emergence (germination rate, uniformity, and the percentage of seedling emergence; Dixit et al., 2015) and early vegetative vigor (higher relative growth rate and biomass accumulation, Sandhu et al., 2015). The abiotic stress tolerance traits included anaerobic germination (germination under flooding conditions; Ismail et al., 2009; Angaji et al., 2010) and drought tolerance (Vikram et al., 2011; Mishra et al., 2013). Root traits improving nutrient uptake, adaptability, and grain yield under direct-seeded cultivation conditions included nodal root, root hair length, root hair density, and root plasticity (Sandhu et al., 2015, 2016). The biotic stress tolerance/resistance traits included resistance to blast (Fjellstrom et al., 2004; Koide et al., 2011; Shikari et al., 2013), brown plant hopper (Sun et al., 2005; Jairin et al., 2007), gall midges (Nair et al., 1996), and bacterial blight (Song et al., 1997; Chu et al., 2006; Perumalsamy et al., 2010; Ullah et al., 2012). The other traits included lodging resistance (Dixit et al., 2015) and nematode tolerance (Galeng-Lawilao et al., 2018).
During the last 10 years of research at IRRI, the donors and QTL associated with these traits of interest were identified. At the IRRI, QTL for anaerobic germination (qAG9.1, Angaji et al., 2010), early uniform emergence (qEMM1, 1, qEMM11.1; Dixit et al., 2015), early vigor (qEVV9.1; Sandhu et al., 2015), higher root length density (qNR4.1, qNR5.1, qRHD1.1, qRHD5.1, qRHD8.1; Sandhu et al., 2015), facilitating higher N, P, and K uptake (qN5.1) under variable anaerobicaerobic soil conditions, lodging resistance (qLDG4.1, Dixit et al., 2015), and grain yield under dry direct-seeded conditions (qGY1.1, qGY6.1, qGY10.1; Sandhu et al., 2015) were identified. In addition to the above-mentioned QTL, those for increased yield under drought conditions (qDTY1.1, qDTY2.1, qDTY3.1, qDTY12.1) (Vikram et al., 2011; Mishra et al., 2013) have been identified. Previously reported genes for biotic stress resistance were used in the genomics-assisted introgression program. Detailed information on the donors and QTL selected and used in the present genomics-assisted introgression study is presented in Table 2.
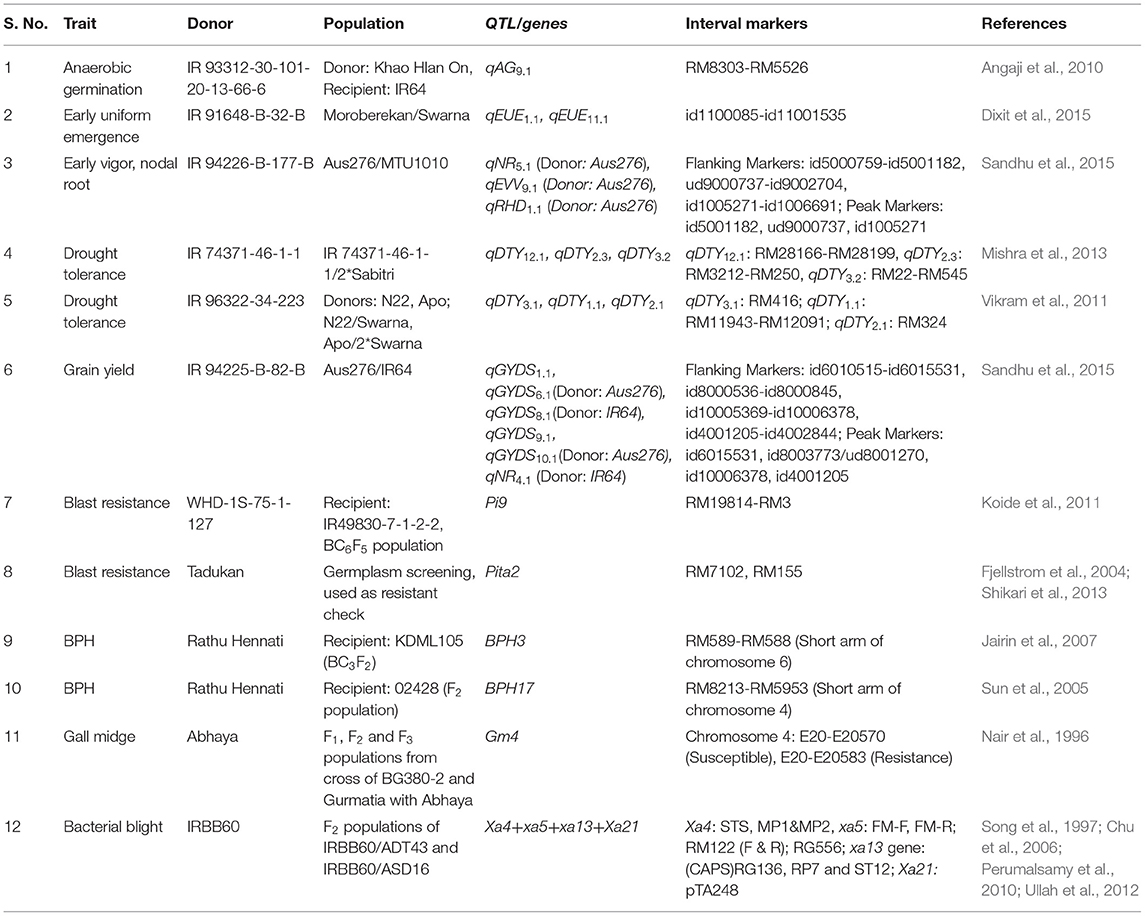
Table 2. Detailed information on the donors and QTL selected and used in the genomics-assisted introgression study.
Genomics-Assisted Introgression Program
To combine anaerobic germination, blast resistance, brown plant hopper resistance, bacterial blight resistance, gall midge resistance, grain yield under direct-seeded cultivation conditions, early vigor, nodal roots, and early and uniform emergence in the background of high-yielding irrigated rice cultivar IR09N538 (IRRI 132/PR 30138-35-2//IR04N114) with preferable grain type, a complex crossing program began in the 2014 DS with 12 donors, including donors for biotic and abiotic stress tolerance and DSR-adapted traits. Based on the synchronization of flowering and availability of a true F1 generation with desired trait/QTL combinations, different cross combinations were attempted to achieve success in the breeding program. A detailed schematic representation of the pre-breeding steps and the genomics-assisted breeding strategy followed for the introgression of QTL/genes to develop a multi-stress-tolerant, direct-seeded, adapted superior rice genotype suitable for DDSR cultivation is presented in Figure 1.
Genotyping
In the genomics-assisted introgression program, the F1s from biparental and multiparental crosses were tested in each season to check for true F1s using rice microsatellite (SSR; simple sequence repeats) markers. The SSR markers were identified in the previously reported QTL associated with traits, such as early uniform germination, early vegetative vigor, nodal roots, root hair length, root hair density, grain yield under direct-seeded conditions, and drought stress tolerance. A total of 1420 SSR markers in the QTL regions were used for parental polymorphism. Eighty-one SSR markers, one indel, and eight gene-specific polymorphic markers were used to screen the F1s and the progenies. A detailed description of the markers used to check the introgressed region is presented in Table 3.
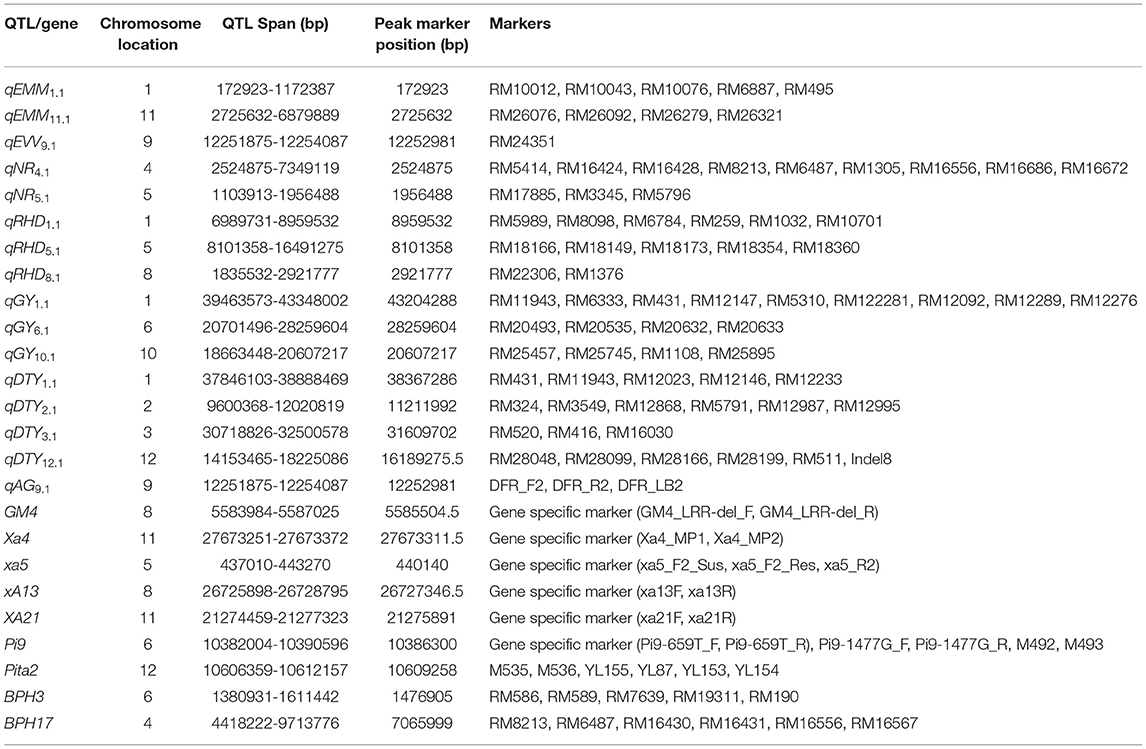
Table 3. Detailed information on the SSR/InDel/gene-specific markers used in the genomics-assisted introgression study.
Fresh, young leaves from the 15-day-old rice seedlings were collected from each F1 plant and the respective parents. Genomic DNA was extracted from F1s using a modified CTAB protocol (Murray and Thompson, 1980). The DNA from the phenotypically selected plants from generations F2 onwards was used for further studies. The amplification of genomic region using PCR (polymerase chain reaction), separation of bands on agarose, and 6–8% (v/v) PAGE (polyacrylamide gel electrophoresis (CBS scientific model MGV-202-33); depending on the SSR, indel, and gene-specific marker product size was performed. After staining with SYBR Safe™, the separated genomic fragments were visualized under a UV trans-illuminator (AlphaImager™ System). The associations of the polymorphic SSRs, indel, and gene-specific markers with the introgressed genomic regions were exploited for the accurate detection of the F1s and the progenies. Because there are many introgressed genomic regions/genes and many are not fine mapped, a sequential two-stage genotyping approach was used to make the genotyping labor and cost effective. The first stage selection was performed on phenotypically selected plants using the peak markers associated with the introgressed genomic region. The second-stage selection was performed on the selected plants harboring the selected donor allele using flanking and all other markers underlying the QTL regions.
Phenotyping
To combine traits of true F1s to increase rice adaptation under direct-seeded cultivation conditions, –early uniform emergence, early vigor, high-nutrient uptake through improved root traits, tolerance to drought, and grain yield under conditions together with resistance to biotic stresses were selected using trait-linked markers. The number of F1s developed per cross per season is listed in Supplementary Table 1. At each generation, phenotypic plant selection was conducted initially based on plant type, duration, plant height, number of tillers, grain type, and visual yield, and then selected plants were evaluated genotypically with the trait associated markers. A single plant selection strategy was followed from the F2 to F6 generation for the presence of QTL/gene combinations together with phenotypic selection for yield to reject plants showing negative interactions. A plot size of 8 m2 was used for the screening of each breeding line at the F4, F5, and F6 generation. At each advancing generation step, the plants were grown maintaining 20 cm (hill to hill) × 20 cm (row to row) distance. Field management was conducted following the procedure described by Sandhu et al. (2019b). The data on plant height (cm) and grain yield of single plant selection (g) and plot yield (kg ha−1) were collected. Plant height was measured from the root-shoot junction to the tip of the uppermost panicle on the main tiller. After harvesting at maturity, the grains were first dried to 14% moisture, and then weighed to record GY (g, kg ha−1).
Statistical Analysis
The agronomic data collected from all the conventional and GAB experiments were analyzed using statistical tools and software. The experimental means and standard error of difference of the progeny testing experiments were calculated using IRRI PBTools v1.4. The least significant difference (LSD) at P = 1 and 5% levels of significance were used to compare the means of test entries and to estimate the significant variations existing between parents and the breeding lines for the particular trait of interest. An ANOVA (analysis of variance) was estimated using following mixed linear model:
where μ, Gi, Rj, BK(Rj), and eijk are the overall mean, effect of ith breeding line, effect of jth replicate, block effect of the jth replicate, and error, respectively. While estimating the entry mean, the genotypes and replication block effects were kept as fixed and random, respectively.
Mean Comparison of Different Parental Classes
The hypothesis regarding mean differences among the breeding lines developed involving different numbers of parental lines under direct-seeded cultivation conditions was performed using the following linear model in SAS v9.2 (SAS Institute Inc. 2009).
where μ, rk, b(r)kl, qi, g(q)ij, and eijkl symbolize the population mean, effect of the kth replicate, effect of the lth block within the kth replicate, effect of the ith parental class, effect of the jth breeding line nested within the ith parental class, and the error, respectively (Knapp, 2001). The effects of parental class and the breeding lines within the parental class were considered as fixed effects, and the replicates and blocks effects within replicates were set as random.
Results
Conventional Breeding Program
A total of 184 crosses involving 137 biparent crosses, 21 triparent crosses, 16 quadraparent, 5 pentaparent, and 5 hexaparent crosses were attempted and evaluated from the F2 generation for advanced yield trials. A highly stringent phenotypic selection based on plant type, duration, plant height, number of tillers, grain type, and visual yield across generations was made. The stringency of selection can be estimated from the number of plants/breeding lines selected across generation advancement, as represented in Supplementary Table 2.
The crosses involving three or more parents showed grain yield improvement over the biparental lines and the upland adapted check varieties (Table 4). The high mean grain performance of the parental class involving more than two parents in the F4 generation was consistent in the F5 and F6 generations and also in observational and advanced yield trials across seasons. The highest average grain yield was observed in the hexaparent class across generations and seasons (Table 4). The average grain yield of the biparent class ranged from 3,517 to 4,589 kg ha−1 in the WS (wet season) and from 5,010 to 5,796 kg ha−1 in the DS across generations. The parental class involving three, four, and five parents showed average grain yields ranging from 4,434 to 5,904 kg ha−1, 4,532 to 6,002 kg ha−1, and 4,566 to 6,124 kg ha−1, respectively, across generations. The grain yield of the hexaparent class ranged from 4,680 to 5,603 kg ha−1 in the WS and from 5,505 to 6,292 kg ha−1 in the DS across generations. Across season and generation advancement, the grain yield advantage of the biparent class ranged from 100 to 540 kg ha−1, triparent class from 35 to 1,008 kg ha−1, and quadraparent class from 352 to 1,095 kg ha−1 over the check varieties. The parental classes involving five and six parents consistently outperformed the check varieties in terms of average grain yield advantage (Table 4). The multi-parent lines had significantly higher yield (2,072–6,569 kg ha−1) than the bi-parental lines (1,493–6,326 kg ha−1) at the F6 generation in the DS.
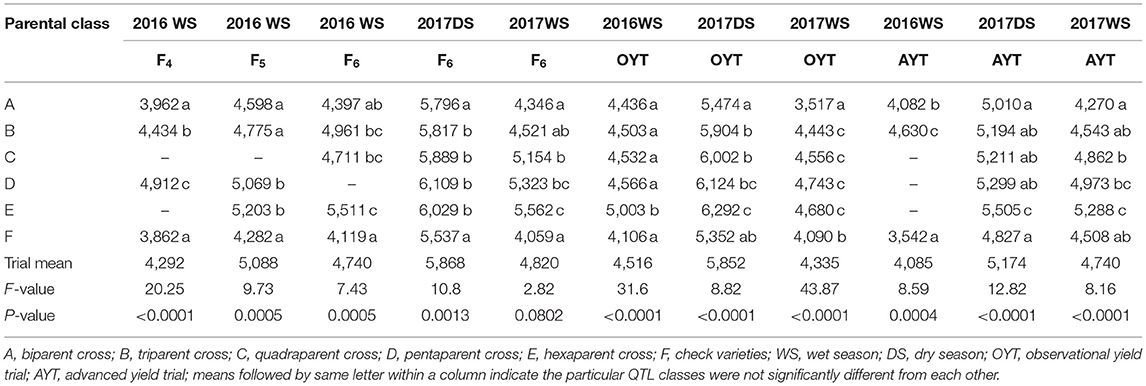
Table 4. Mean comparison of QTL classes for grain yield (kg ha−1) from the F4 to F6 generations in observational and advanced yield trials under direct-seeded cultivation conditions.
The breeding lines developed involving more than two parents could ensure 0.3–1.5 kg ha−1 improvement in grain yield over the check varieties and 0.1–1.3 kg ha−1 grain yield improvement over the breeding lines involving two parents. DTF of the different classes were comparable to the check varieties, except in some seasons where 1 or 2 d earliness was observed (Table 5). Most of the classes showed comparable or slightly higher PHT than did the check varieties (Table 5).
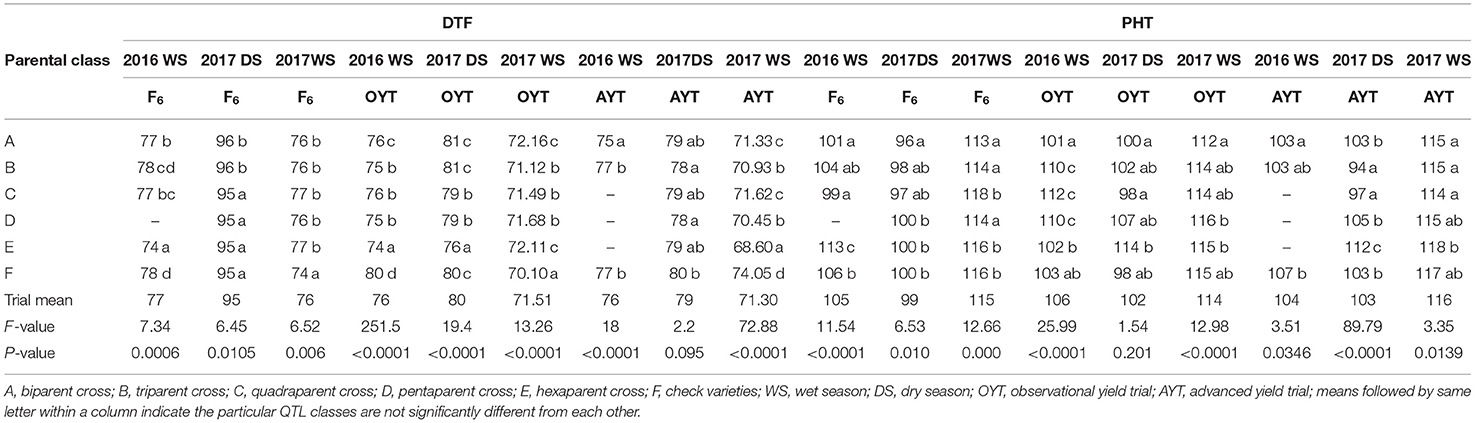
Table 5. Mean comparison of QTL classes for DTF (d) and PHT (cm) from the F4 to F6 generations in observational and advanced yield trials under direct-seeded cultivation conditions.
Genomics-Assisted Breeding Program
The GAB program involving 12 donors and IR09N538 as the recipient was initiated in the 2014 DS. The final multiparent F1s were generated in the 2016DS Figure 2. A single plant selection strategy was followed across generations. The multiparental 4,200 F1s were grown in an F1 nursery at IRRI under puddled transplantation conditions in the 2016WS. The number of QTL/genes varied from two to 15 and grain yield ranged from 1.0 to 58.8 g/plant. Thirty-five F2s derived F1s were selected based on different QTL/gene combinations. The number of QTL/genes in the selected plants ranged from 8 to 15 and the grain yield varied from 8.0 to 58.8 g/plant. An F2 population of approximately 40,000 plants, F3 population of 13,780 plants, F4 population of 144 breeding lines, F5 population of 243 breeding lines, and F6 population of 70 breeding lines were raised and screened under DDSR cultivation conditions. A total of 13,468 F2 plants of required plant type, duration, plant height, number of tillers, grain type, and visual yield were screened genotypically with the trait-associated markers, and a total of 153 F2 derived F3 plants were selected. The number of QTL/genes in the selected plants varied from 3 to 12, plant height from 80 to 130 cm, and grain yield from 5.9 to 68 g/plant. The 1,177 F3 derived F4 plants and 1,411 F4 derived F5 plants were selected and advanced by combining phenotypic genotypic selection in the 2017WS and the 2018DS, respectively. More than 30 genomics-assisted derived F5 breeding lines with different combinations of QTL/genes yielded more than check varieties MTU1010, IRRI 155, UPLRi7, and the recipient parent, IR09N538. F4 derived F5 breeding lines with 223 different combinations of QTL/genes were evaluated in the 2018WS. Significant variability in grain yield, ranging from 1,026 to 5,956 kg ha−1, was observed (data not shown). After genotypic selection at the F5 level, a total of 427 single plants were selected phenotypically, and then 70 plants to progeny were carried forward to the F6 generation. The grain yield variability in the selected F6 breeding lines varied from 3,293 to 6,719 kg ha−1, plant height from 65 to 115 cm, and days to 50% flowering from 77 to 104 d (data not shown). The QTL/genes in selected F6 breeding lines varied from 6 to 12, and a total of 55 different combinations of QTL/genes with acceptable phenotype and grain characteristics were selected.
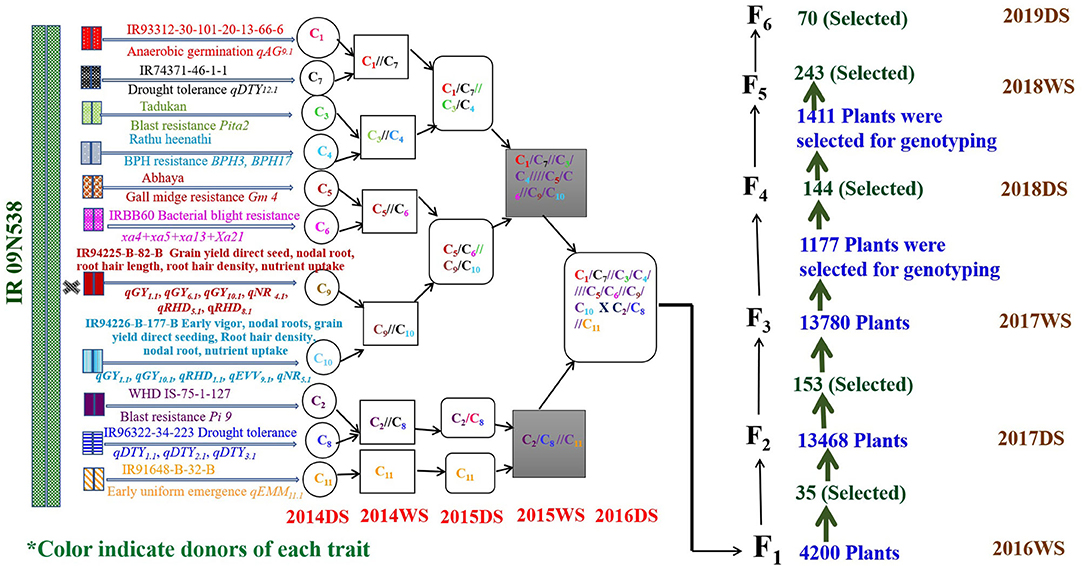
Figure 2. Detailed schematic representation of crossing strategy followed for the introgression of QTL/genes from 12 donors in the background of IR09N538 and the number of plants selected across successive generations. #C: represents the cross.
Comparison of Conventional and Genomics-Assisted Derived Breeding Lines
The analysis of the differences in mean grain yield of genomics-assisted derived breeding lines was significantly higher than that of breeding lines derived from conventional breeding programs Table 6. The grain yield of GAB lines derived from multi-parent crosses ranged from 3,293 to 6,719 kg ha−1; however, the grain yield of multi-parent lines from conventional breeding ranged from 2,072 to 6,569 kg ha−1 at the F6 generation in the DS. The GAB lines IR 129477-2815-41-4-7-1, IR 129477-3343-500-36-5-1, IR 129477-4139-439-1-1-2, and IR 129477-1629-14-1-2-2 with eight, nine, nine, and 11 QTL/genes, respectively, showed grain yield of more than 5,500 kg ha−1 in the WS at the F5 generation advancement stage and more than 6,000 kg ha−1 in the DS at the F6 generation Table 7. The breeding lines IR 115844-B-B-475-1-2, IR 107891-B-B-1253-1-1, IR 115845-B-B-296-2-1, and IR 115846-B-B-197-1-1 from four different crosses of the conventional breeding program involving five/six parents yielded more than 4,500 kg ha−1 in the WS at the F5 generation advancement stage and more than 5,500 kg ha−1 in the DS at the F6 generation (Table 7). The genomics-assisted derived breeding lines showed better yield performance (5,100–7,004 kg ha−1) when tested at Varanasi, India, under observational yield trials (Table 7). The breeding lines from the conventional breeding program showed delayed flowering compared to the genomics-assisted breeding lines, except the lines derived from the cross Dular (acc. 32561)/IRRI 148///IRRI 154/UPLRi 7//IR87707-446-B-B-B/Kali Aus (Table 7). The genomics-assisted derived breeding lines were more dwarf than the breeding lines developed from the conventional breeding program (Table 7). Further, the breeding lines developed from the crosses (NSIC Rc 222/IR87707-446-B-B-B)/IR87707-446-B-B-B/IRRI148//Vandana/Kali Aus, and Dular (acc. 32561)/IRRI 148///IRRI 154/UPLRi 7//IR87707-446-B-B-B/Kali Aus showed better performance across generation advancement followed by (Sahbhagi dhan/UPLRi 7)/(PSBRc 82/Kali Aus)//(Vandana/IRRI148) and (Sahbhagi dhan/IR87707-446-B-B)/(PSBRc 82/IRRI148)//(Kali Aus/Kalinga 3) (Table 7).
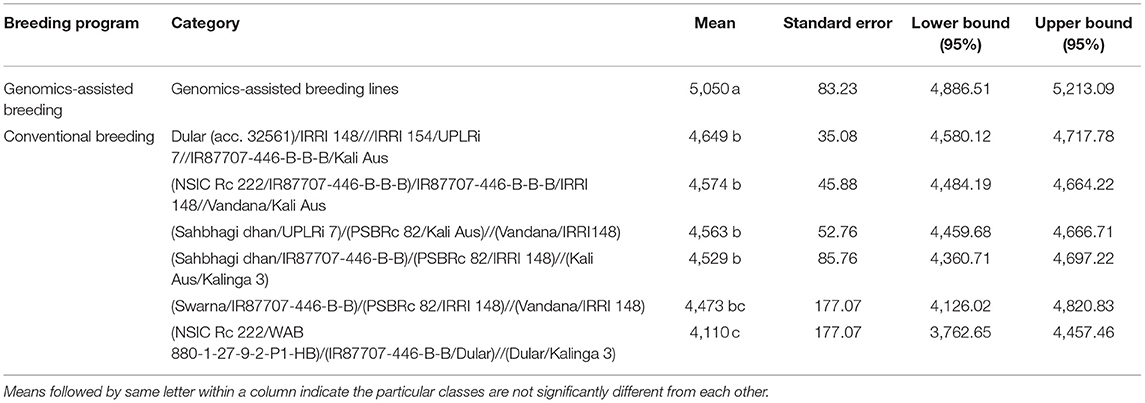
Table 6. Analysis of the differences for grain yield (kg ha−1) at the F6 generation in the dry season between the categories of breeding lines (with a confidence interval of 95%) developed following genomic assisted breeding and conventional breeding approaches.
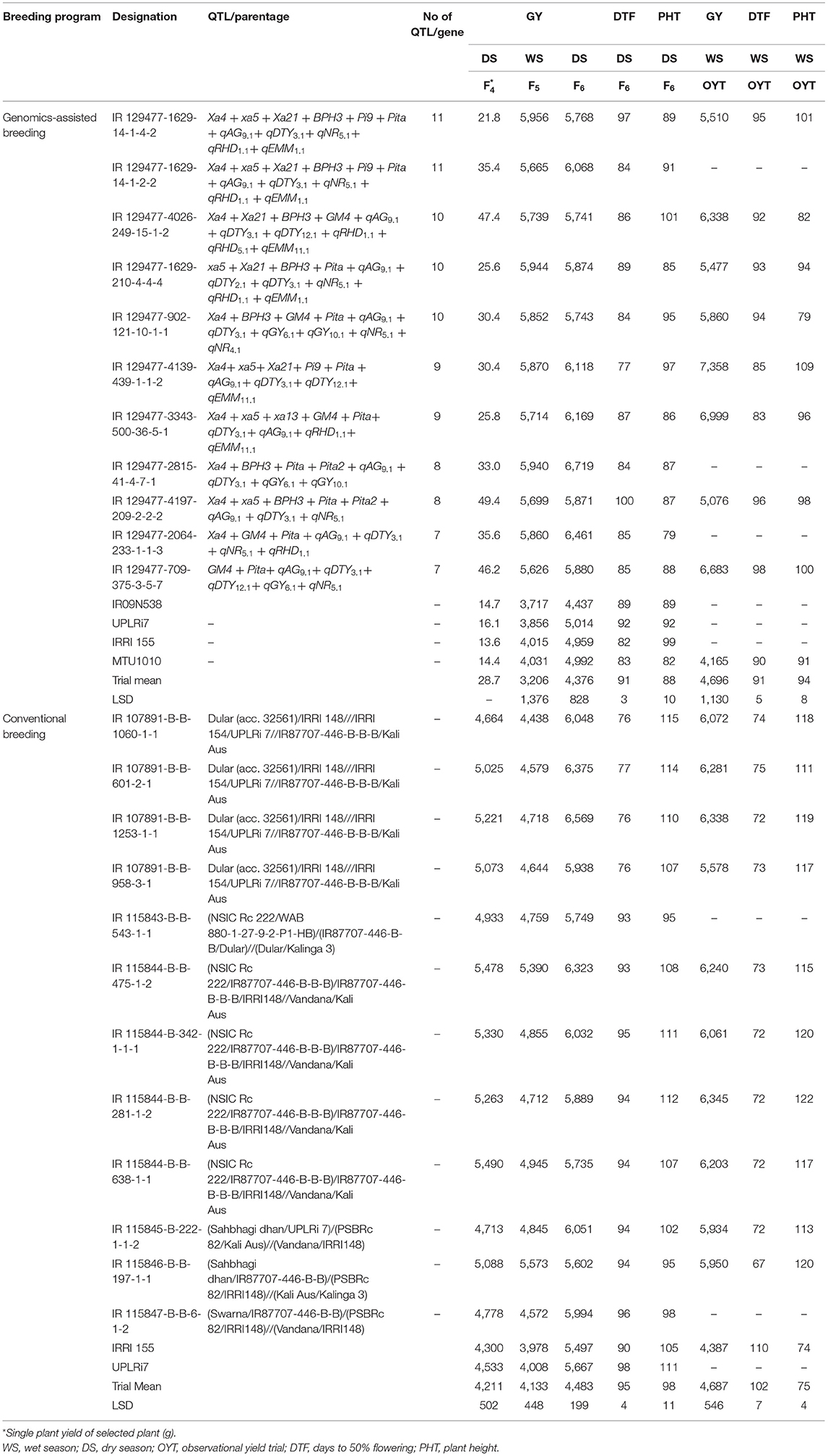
Table 7. Mean comparison of grain yield (kg ha−1), DTF (days), and PHT (cm) of breeding lines developed under the genomics-assisted breeding program and conventional breeding program.
Selection of Promising Lines/Donors
The breeding lines carrying multiple QTL/genes from GAB programs and multiparent lines from conventional breeding programs performed well in terms of grain yield and adaptability under DDSR and also exhibited phenotypically high levels of tolerance/resistance to abiotic and biotic stresses with desired grain quality characteristics. A total of 11 promising lines carrying 7–11 QTL/genes for various DDSR adaptable traits, as well as biotic stresses tolerance/resistance (Table 7, Figures 3a–c), and eight promising lines from the multiparental conventional breeding program were selected (Table 8, Figures 3d,e) for further evaluation in multiple environments for varietal release. The recipient variety IR 09N538 is presented in Figure 3f and the upland adapted check varieties, UPLRi7 and Katihan 1 are presented in Figures 3g,h, respectively. It is interesting to note that the genomics-assisted breeding lines with a combination of DDSR adaptable traits and abiotic stress tolerance/resistance QTL/genes showed better yield advantages, viz. IR 129477-2815-41-4-7-1 (Xa4 + BPH3 + Pita + Pita2 + qAG9.1 + qDTY3.1 + qGY6.1 + qGY10.1) showed ~33% and IR 129477-1629-14-1-2-2 (Xa4 + xa5 + Xa21 + BPH3 + Pita + qAG9.1+ Pi9 + qDTY3.1 + qNR5.1 + qRHD1.1 + qEMM1.1) showed 22% yield improvement over the best performing check variety. These lines were 6–7 d earlier than the other lines. The breeding lines IR 115844-B-B-281-1-2 and IR 115844-B-342-1-1-1 from five parent cross (NSIC Rc 222/IR87707-446-B-B-B)/IR87707-446-B-B-B/IRRI148//Vandana/Kali Aus yielded more than 7,000 kg ha−1 under direct-seeded cultivation conditions in an advanced yield trial (Table 8). Most of the selected breeding lines possessed less chalkiness, medium amylose content, and intermediate GT content (Table 8). Lines with chalkiness >10% and amylose content <10% were not carried forward in the breeding program. The promising breeding lines from both the conventional and GAB programs can be further tested for adaptability and yield stability by conducting multi-location trials in their targeted environments. These pyramided lines may serve as novel donors for the development of lines with multiple abiotic and biotic stress tolerance/resistance traits.
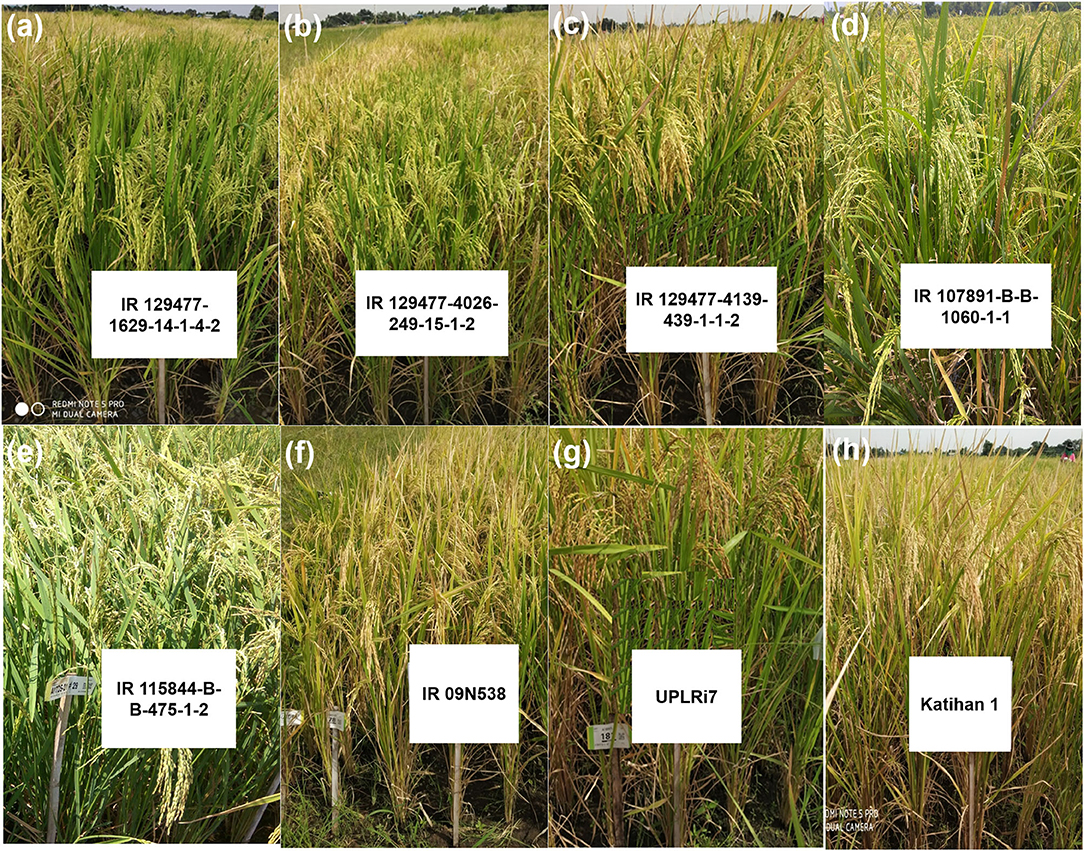
Figure 3. Selected promising breeding lines from (a–c) genomics-assisted breeding program (d,e), conventional breeding program, and (f) recipient variety used in genomics-assisted breeding program (g,h) for upland adapted check varieties.
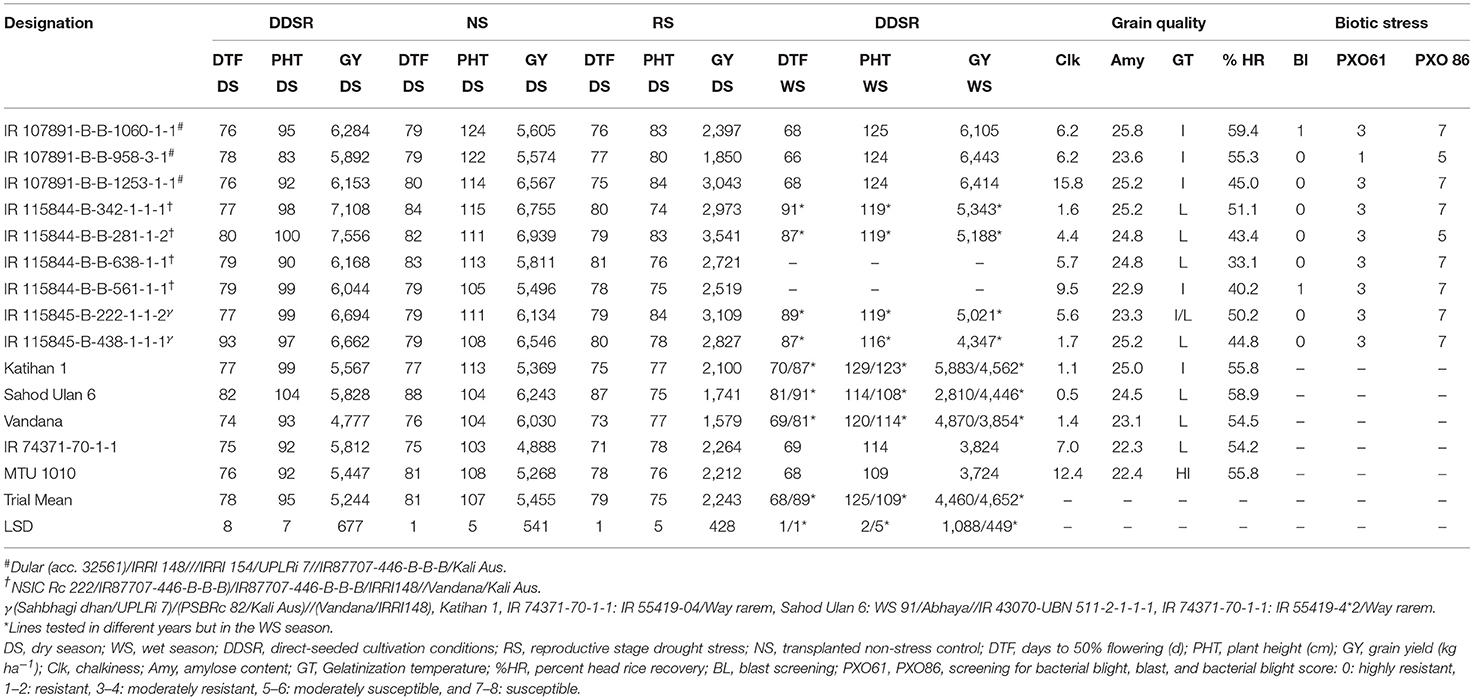
Table 8. Performance of selected promising multi-parent breeding lines over check varieties from a conventional breeding program in advanced yield trials at IRRI, Philippines.
Discussion
To meet the gaps between rice crop yields and global rice consumption, rice breeders need to continuously release new rice varieties with better yield potential, adaptability under DDSR, acceptable grain quality traits, high nutrient-use efficiency, and resistance to various biotic/abiotic stresses (Leng et al., 2017). The longstanding idea behind using an insecticide mixture to broaden the insect resistance spectrum laid the foundation of gene pyramiding (Ye and Smith, 2008). The nature of donors/traits selected for introgression, number of genes transferred, distance between the introgressed genes and flanking markers, and the number of plants selected with different gene combinations across each breeding generation, are critical for an effective QTL/genes pyramiding program.
To the best of our knowledge, the present study is the first to report the pyramiding of 7–11 QTL/genes associated with both traits, providing better adaptability and biotic stress tolerance/resistance under DSSR, exploiting genomics-assisted foreground selection. Previous studies reported the marker-assisted pyramiding of QTL for either tolerance to abiotic stress traits, such as drought and submergence (Swamy et al., 2013; Septiningsih et al., 2015; Shamsudin et al., 2016; Dixit et al., 2017; Sandhu et al., 2019a), or resistance to blast (Singh et al., 2013; Fukuoka et al., 2015), bacterial blight (Suh et al., 2013; Pradhan et al., 2015; Das et al., 2018), brown plant hoppers (Wang et al., 2015; Jena et al., 2017), and gall midges (Divya et al., 2015) in rice.
Various high-yielding rice varieties from the conventional breeding program involving phenotypic selection (Sandhu and Kumar, 2017) and genomics-assisted selection (Sandhu et al., 2019b) have been released. However, labor and time use, linkage drag, and low efficiency are some of the major barriers impeding conventional breeding (Prohens, 2011). Advances in molecular mapping, precise phenotyping, novel marker development, and multi-parent population development have paved the way for genomics-assisted breeding. These advances have gradually shifted the focus of traditional plant breeding from phenotype-based selection to genotype-based selection (Xu and Crouch, 2008). The GAB strategies involve backcrossing or introgression of QTL/genes, enrichment of multiple favorable alleles, and selection for introgressed traits/QTL/genes using trait-linked markers (Hospital et al., 1992; Eathington et al., 2007; Gupta et al., 2010).
A GAB program combining 25 QTL/genes and a conventional breeding program involving multi-parent cross-governing better adaptation of rice to direct-seeded cultivation conditions was undertaken at IRRI. The final step of the introgression program was to fix the targeted QTL/genes into a homozygous state across successive generations. Generally, recombinant inbred lines (RILs) and double haploid (DH) production techniques are used for the development of homozygous lines. We did not succeed in combining all 25 QTL/genes in one breeding line because: (i) some with large introgressed regions were lost during successive recombination events, (ii) some QTL/genes combination altered the flowering cycle and led to the failure of planned crossing strategy in successive seasons because of non-synchronization of flowering, (iii) some of the complex F1s crosses with possible negative interactions among the introgressed QTL/genes were rejected and not utilized for further crossing, and (iv) some of the QTL that were in a heterozygous state were lost in succeeding generations. We succeeded in pyramiding 15 QTL/genes in the F2/F3 generation in the background of IR09N538 but some QTL/genes, which were in a heterozygous state, were lost in succeeding generations. In this regard, DH production has been reported to provide satisfactory results in becoming fully homozygous in a very short time (Dunwell, 2010; Mishra and Rao, 2016), which was not used in the present study. Additionally, genetic engineering (GE) also aids the precise transfer of genes of interest to crop plants (Lemaux, 2008) to generate crops with desired trait/s. Although DH and GE are precise, universal, and fast methods to transfer the desired gene/s into different crop plants (Nicholl, 2008), it will not replace GAB, but it will definitely add to the efficiency of rice crop improvement. Some QTL/genes combination that showed negative interactions leading to poor grain yield and adaptability under DDSR with unacceptable plant and grain type were rejected in successive generations. Finally, the pyramided lines possessing 7–11 QTL/genes under homozygous conditions were selected, indicating the existence of positive interactions among different biotic and abiotic stress-related QTL/genes.
The DDSR lines developed through conventional and GAB strategies were evaluated at an advanced stage under dry direct-seeding conditions. The increased yield of breeding lines developed under conventional breeding programs involving multi-parents performed over the bi-parental lines across generations could result from the accumulation of alleles for traits that increase rice adaptability to direct-seeded situations (Huang et al., 2012). It can be predicted that, with the use of a greater number of parents, there will be greater identification of the complex traits (Huang et al., 2012), a greater number of recombinations, and greater chances of accumulation of favorable alleles. Nevertheless, multi-parent crosses require more time and higher costs for the development of the population, but increased genetic variations may provide opportunities to improve grain yield and adaptability under DDSR. In the present study, we had succeeded in breaking the unfavorable linkages, elimination of the inferior plants with poor plant and grain type during the selection process. The better grain yield performance of GAB-derived multi-parental breeding lines compared to the breeding lines developed from conventional breeding programs could be the result of precise marker-based selection that helps reduce undesirable linkage drags. GAB aids in obtaining the desirable QTL/genes combination without any unwanted genes, minimizing the linkage drag around the target QTL/genes. GAB has been proven effective for transferring QTL/genes from pyramided lines into improved varieties and new plants (Magar et al., 2014).
A stepwise hybridization and selection strategy in the GAB program led to the selection of the most suitable plants with desired QTL/gene combinations. Arbelaez et al. (2019) reported that an average of 800 F2 individuals plants were required to obtain 50 lines with the desired genotype fixed at two loci. Compared to the F2 generation, the frequency of desired homozygotes for two linked genomic loci will decrease nearly three times in the F6 generation (Arbelaez et al., 2019). Keeping the above points in view, a large population size of 40,000 (F2) and 13,780 (F3) plants capturing the hidden genetic variations were maintained in the early generations to select appropriate plants possessing positive interactions among targeted QTL/gene combinations, with desired plant type, grain type, better grain yield, and adaptability under DDSR. The increased yield of the GAB-derived lines from multiparent crosses has been achieved, keeping the plant height and days to 50% flowering, similar to check varieties, also indicates yield increase as a result of accumulation of favorable alleles, increasing rice adaptability to direct-seeded situations caused by introgression of genes/QTL for such traits. Notably, stepwise phenotypic and genotypic selection in the early generations has reduced the genotypic efforts, finally reducing the genotyping cost. The effectiveness of the above-mentioned strategy targeting the pyramiding of various drought QTL has been successfully demonstrated in various genetic backgrounds by Kumar et al. (2018).
Most of the GAB-improved lines developed in the present study carry the xa5 gene in combination with xa13 and Xa21 genes. Based on previous reports, xa5 (Jiang et al., 2006; Huang et al., 2016), xa13 (Ogawa et al., 1987; Chu et al., 2006), and Xa21 (Huang et al., 1997; Cao et al., 2005) genes are considered as the most effective R genes, providing broader levels of resistance, and have been widely used for introgression programs against BLB in most rice growing countries in Asia. The selected promising lines carrying either the Pita, Pita2, or Pi9 gene alone showed stable resistance to blast disease. Similarly, Qu et al. (2006) reported that the Pi9 gene can provide broader and durable resistance against rice blast disease. Additionally, the selected breeding lines possessed a genomic region providing resistance to gall midge, tolerance to reproductive stage drought stress, adaptability, and improved yield under DDSR. The selected improved breeding lines carrying the genomic region contributing to biotic stress resistance will further be evaluated phenotypically. The better performance of breeding lines under transplanted control non-stress and reproductive stage drought stress conditions (Table 8), in addition to DDSR, indicated their phenotypic plasticity. Enhanced seedling establishment, improved and stable yield, and phenotypic plasticity of the root system have been reported to result in improved adaptability of selected promising lines across variable growing conditions (Sandhu et al., 2016, 2017). Grain quality parameters play a crucial role in varietal release and wider acceptance of the released varieties (Rani et al., 2006). Thus, our field selection criteria also involved selection for grain type across each successive generation. The breeding lines from the conventional and GAB programs were selected considering the preferences of various countries in South and South East Asia. The developed breeding lines possessing medium amylose content are suitable for India and Nepal, those with higher amylose content for Bangladesh, and those with lower amylose content for South East Asia.
Conclusions
The study showed GAB was a better strategy for improving yield and adaptation of rice under DDSR utilizing multiple QTL/genes compared to conventional breeding. A higher number of parents and more recombination events resulted in higher grain yield advantage compared to the classical bi-parental population. These findings are vital for researchers working on multi-parental populations, because this will remarkably reduce the efforts of developing large numbers of bi-parental populations. The rigorous selection from the F2 generation onwards resulted in the development of promising breeding lines carrying 7–11 QTL/genes for different traits. Promising GAB lines carrying multiple QTL/genes under homozygous conditions with preferable grain type and quality traits can address the constraints faced by farmers caused by both biotic and abiotic stresses simultaneously under DDSR in addition to the labor-water shortage. The GAB-derived breeding lines and multi-parent conventional breeding lines could further be recommended for varietal release after multi-location evaluation in national and provincial coordinated trials in different countries. These breeding lines may serve as a novel genetic resource that could aid breeders and molecular biologists to further conduct genetic and interaction studies for a wide range of breeder-relevant parameters.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
NS conducted the experiment, analysis, data interpretation, and drafting of the manuscript. SY assisted in conducting experiments. MCa assisted in genotyping. MCr performed management of field experiments. AK conceived the study and was involved in critical revision and final approval of the manuscript. All authors thoroughly discussed the results.
Funding
The authors thank the Asian Development Bank (ADB), Manila, and Government of Finland for the financial support to conduct the experiments and publish this study through the project grant TA8441-Development and dissemination of climate resilient rice varieties for water short areas of South Asia and Southeast Asia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.637488/full#supplementary-material
References
Angaji, S. A., Septiningsih, E. M., Mackill, D. J., and Ismail, A. M. (2010). QTLs associated with tolerance of flooding during germination in rice (Oryza sativa L.). Euphytica 172, 159–168. doi: 10.1007/s10681-009-0014-5
Arbelaez, J. D., Dwiyanti, M. S., Tandayu, E., Llantada, K., Jarana, A., Ignacio, J. C., et al. (2019). 1k-RiCA (1K-Rice Custom Amplicon) a novel genotyping amplicon-based SNP assay for: genetics and breeding applications in rice. Rice 12:55. doi: 10.1186/s12284-019-0311-0
Bandillo, N., Raghavan, C., Muyco, P. A., Sevilla, M. A. L., Lobina, I. T., Dilla-Ermita, C. J., et al. (2013). Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6:11. doi: 10.1186/1939-8433-6-11
Bhushan, L., Ladha, J. K., Gupta, R. K., Singh, S., Tirol-Padre, A., Saharawat, Y. S., et al. (2007). Saving of water and labor in a rice–wheat system with no-tillage and direct seeding technologies. Agron. J. 99, 1288–1296. doi: 10.2134/agronj2006.0227
Boretti, A., and Rosa, L. (2019). Reassessing the projections of the world water development report. npj Clean Water 2:15. doi: 10.1038/s41545-019-0039-9
Breseghello, F., and Coelho, A. S. G. (2013). Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.). J. Agric. Food Chem. 61, 8277–8286. doi: 10.1021/jf305531j
Cao, L. Y., Zhan, X. D., Zhuang, J., and Cheng, S. H. (2005). Breeding of indica hybrid rice Guodao 1 with good quality, high yield and resistance to bacterial leaf blight by marker-assisted selection technique. Hybrid Rice 20, 16–18.
Chu, Z., Fu, B., Yang, H., Xu, C., Li, Z., Sanchez, A., et al. (2006). Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. App. Genet. 112, 455–461. doi: 10.1007/s00122-005-0145-6
Collard, B. C., and Mackill, D. J. (2008). Marker-assisted selection, an approach for precision plant breeding in the twenty-first century. Phil. Trans. Royal Soc. B Bio. Sci. 363, 557–572. doi: 10.1098/rstb.2007.2170
Das, G., Rao, G. J., Varier, M., Prakash, A., and Prasad, D. (2018). Improved Tapaswini having four BB resistance genes pyramided with six genes/QTLs, resistance/tolerance to biotic and abiotic stresses in rice. Sci. Rep. 8, 1–16. doi: 10.1038/s41598-018-20495-x
Das, K., Zhao, D., De Waele, D., Tiwari, R. K. S., Shrivastava, D. K., and Kumar, A. (2011). Reactions of traditional upland and aerobic rice genotypes to rice root knot nematode (Meloidogyne graminicola). J. Plant Breed. Crop Sci. 3, 131–137.
Divya, D., Himabindu, K., Nair, S., and Bentur, J. S. (2015). Cloning of a gene encoding LRR protein and its validation as candidate gall midge resistance gene, Gm4, in rice. Euphytica 203, 185–195. doi: 10.1007/s10681-014-1302-2
Dixit, S., Grondin, A., Lee, C. R., Henry, A., Olds, T. M., and Kumar, A. (2015). Understanding rice adaptation to varying agro-ecosystems, trait interactions and quantitative trait loci. BMC Genet. 16:86. doi: 10.1186/s12863-015-0249-1
Dixit, S., Singh, A., Sandhu, N., Bhandari, A., Vikram, P., and Kumar, A. (2017). Combining drought and submergence tolerance in rice, marker-assisted breeding and QTL combination effects. Mol. Breed. 37:143. doi: 10.1007/s11032-017-0737-2
Dunwell, J. M. (2010). Haploids in flowering plants, origins and exploitation. Plant Biotech. J. 8, 377–424. doi: 10.1111/j.1467-7652.2009.00498.x
Eathington, S. R., Crosbie, T. M., Edwards, M. D., Reiter, R. S., and Bull, J. K. (2007). Molecular markers in a commercial breeding program. Crop Sci. 47:S-154. doi: 10.2135/cropsci2007.04.0015IPBS
Farooq, M., Siddique, K. H., Rehman, H., Aziz, T., Lee, D. J., and Wahid, A. (2011). Rice direct seeding, experiences, challenges and opportunities. Soil Till. Res. 111, 87–98. doi: 10.1016/j.still.2010.10.008
Fischer, A. J., and Antigua, G. (1996). “Weed management for rice in Latin America and the Caribbean,” in Weed Management in Rice, eds B. A. Auld and K. U. Kim. FAO Plant Production and Protection Paper 139. (Rome: FAO), 159–179.
Fjellstrom, R., Conaway-Bormans, C. A., McClung, A. M., Marchetti, M. A., Shank, A. R., and Park, W. D. (2004). Development of DNA markers suitable for marker assisted selection of three Pi genes conferring resistance to multiple Pyricularia grisea pathotypes. Crop Sci. 44, 1790–1798. doi: 10.2135/cropsci2004.1790
Fukuoka, S., Saka, N., Mizukami, Y., Koga, H., Yamanouchi, U., Yoshioka, Y., et al. (2015). Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 5, 1–7. doi: 10.1038/srep07773
Galeng-Lawilao, J., Kumar, A., and De Waele, D. (2018). QTL mapping for resistance to and tolerance for the rice root-knot nematode, Meloidogyne graminicola. BMC Genet. 19:53. doi: 10.1186/s12863-018-0656-1
Ghosal, S., Casal, C., Quilloy, F. A., Septiningsih, E. M., Mendioro, M. S., and Dixit, S. (2019). Deciphering Genetics Underlying stable anaerobic germination in rice, phenotyping, QTL identification, and interaction analysis. Rice 12:50. doi: 10.1186/s12284-019-0305-y
Gupta, P. K., Langridge, P., and Mir, R. R. (2010). Marker-assisted wheat breeding, present status and future possibilities. Mol. Breed. 26, 145–161. doi: 10.1007/s11032-009-9359-7
Hospital, F., Chevalet, C., and Mulsant, P. (1992). Using markers in gene introgression breeding programs. Genetics 132, 1199–1210. doi: 10.1093/genetics/132.4.1199
Huang, B. E., George, A. W., Forrest, K. L., Kilian, A., Hayden, M. J., Morell, M. K., et al. (2012). A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotech. J. 10, 826–839. doi: 10.1111/j.1467-7652.2012.00702.x
Huang, N., Angeles, E. R., Domingo, J., Magpantay, G., Singh, S., Zhang, G., et al. (1997). Pyramiding of bacterial blight resistance genes in rice, marker-assisted selection using RFLP and PCR. Theor. App. Genet. 95, 313–320. doi: 10.1007/s001220050565
Huang, S., Antony, G., Li, T., Liu, B., Obasa, K., Yang, B., et al. (2016). The broadly effective recessive resistance gene xa5 of rice is a virulence effector-dependent quantitative trait for bacterial blight. Plant J. 86, 186–194. doi: 10.1111/tpj.13164
Ismail, A. M., Ella, E. S., Vergara, G. V., and Mackill, D. J. (2009). Mechanisms associated with tolerance to flooding during germination and early seedling growth in rice (Oryza sativa). Ann. Bot. 103, 197–209. doi: 10.1093/aob/mcn211
Jairin, J., Phengrat, K., Teangdeerith, S., Vanavichit, A., and Toojinda, T. (2007). Mapping of a broad-spectrum brown planthopper resistance gene, Bph3, on rice chromosome 6. Mol. Breed. 19, 35–44. doi: 10.1007/s11032-006-9040-3
Jena, K. K., Hechanova, S. L., Verdeprado, H., Prahalada, G. D., and Kim, S. R. (2017). Development of 25 near-isogenic lines (NILs) with ten BPH resistance genes in rice (Oryza sativa L.), production, resistance spectrum, and molecular analysis. Theor. App. Genet. 130, 2345–2360. doi: 10.1007/s00122-017-2963-8
Jiang, G. H., Xia, Z. H., Zhou, Y. L., Wan, J., Li, D. Y., Chen, R. S., et al. (2006). Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAgamma1. Mol. Genet. Genom. 275, 354–366. doi: 10.1007/s00438-005-0091-7
Knapp, S. J. (2001). “Mapping quantitative trait loci,” in DNA Based Markers in Plants, eds R. L. Philipps and I. K. Vasil (Springer), 58–96.
Koide, Y., Ebron, L. A., Kato, H., Tsunematsu, H., Telebanco-Yanoria, M. J., Kobayashi, N., et al. (2011). A set of near-isogenic lines for blast resistance genes with an Indica-type rainfed lowland elite rice (Oryza sativa L.) genetic background. Field Crops Res. 123, 19–27. doi: 10.1016/j.fcr.2011.04.005
Koide, Y., Kobayashi, N., Xu, D., and Fukuta, Y. (2009). Resistance genes and selection DNA markers for blast disease in rice (Oryza sativa L.). Jpn. Agric. Res. 43, 255–280. doi: 10.6090/jarq.43.255
Kumar, A., Sandhu, N., Dixit, S., Yadav, S., Swamy, B. P. M., and Shamsudin, N. A. A. (2018). Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice 11:35. doi: 10.1186/s12284-018-0227-0
Kumar, R., Singh, R. D., and Sharma, K. D. (2005). Water resources of India. Curr. Sci. 89, 794–811.
Kumar, V., and Ladha, J. K. (2011). Direct seeding of rice, recent developments and future research needs. Adv. Agron. 111, 297–413. doi: 10.1016/B978-0-12-387689-8.00001-1
Lemaux, P. G. (2008). Genetically engineered plants and foods, a scientist's analysis of the issues (Part I). Ann. Review Plant Bio. 59, 771–812. doi: 10.1146/annurev.arplant.58.032806.103840
Leng, P. F., Lübberstedt, T., and Xu, M. L. (2017). Genomics-assisted breeding-A revolutionary strategy for crop improvement. J. Integ. Agric. 16, 2674–2685. doi: 10.1016/S2095-3119(17)61813-6
Magar, M. M., Rani, C. V. D., and Anuradha, G. (2014). Marker assisted selection for bacterial leaf blight resistance in segregating populations of Cottondora sannalu. Int. J. Appl. Sci. Biotechnol. 2, 229–237. doi: 10.3126/ijasbt.v2i3.10570
Mishra, K. K., Vikram, P., Yadaw, R. B., Swamy, B. M., Dixit, S., Cruz, M. T. S., et al. (2013). qDTY12.1, a locus with a consistent effect on grain yield under drought in rice. BMC Genet. 14:12. doi: 10.1186/1471-2156-14-12
Mishra, R., and Rao, G. J. N. (2016). In-vitro androgenesis in rice, advantages, constraints and future prospects. Rice Sci. 23, 57–68. doi: 10.1016/j.rsci.2016.02.001
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucl. Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nair, S., Kumar, A., Srivastava, M. N., and Mohan, M. (1996). PCR-based DNA markers linked to a gall midge resistance gene, Gm4t, has potential for marker-aided selection in rice. Theor. App. Genet. 92, 660–665. doi: 10.1007/BF00226086
Nicholl, D. (2008). An Introduction to Genetic Engineering. Cambridge, UK, Cambridge University Press.
Ogawa, T., Lin, L., Tabien, R. E., and Khush, G. S. (1987). A new recessive gene for resistance to bacterial blight of rice. Rice Genetic Newsl. 4, 98–100.
Perumalsamy, S., Bharani, M., Sudha, M., Nagarajan, P., Arul, L., Saraswathi, R., et al. (2010). Functional marker-assisted selection for bacterial leaf blight resistance genes in rice (Oryza sativa L.). Plant Breed. 129, 400–406. doi: 10.1111/j.1439-0523.2009.01705.x
Pradhan, S. K., Nayak, D. K., Mohanty, S., Behera, L., Barik, S. R., Pandit, E., et al. (2015). Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety. Jalmagna. Rice 8:51. doi: 10.1186/s12284-015-0051-8
Prohens, J. (2011). Plant breeding, a success story to be continued thanks to the advances in genomics. Front. Plant Sci. 2:51. doi: 10.3389/fpls.2011.00051
Qu, S., Liu, G., Zhou, B., Bellizzi, M., Zeng, L., Dai, L., et al. (2006). The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172, 1901–1914. doi: 10.1534/genetics.105.044891
Rani, N. S., Pandey, M. K., Prasad, G. S. V., and Sudharshan, I. (2006). Historical significance, grain quality features and precision breeding for improvement of export quality basmati varieties in India. Ind. J. Crop Sci. 1, 29–41.
Sandhu, N., Dixit, S., Swamy, B. M., Vikram, P., Venkateshwarlu, C., Catolos, M., et al. (2018). Positive interactions of major-effect QTLs with genetic background that enhances rice yield under drought. Sci. Rep. 8:1626. doi: 10.1038/s41598-018-20116-7
Sandhu, N., Dixit, S., Swamy, B. P. M., Raman, A., Kumar, S., Singh, S. P., et al. (2019a). Marker assisted breeding to develop multiple stress tolerant varieties for flood and drought prone areas. Rice 12:8. doi: 10.1186/s12284-019-0269-y
Sandhu, N., and Kumar, A. (2017). Bridging the rice yield gaps under drought, QTLs, genes, and their use in breeding programs. Agronomy 7:27. doi: 10.3390/agronomy7020027
Sandhu, N., Raman, K. A., Torres, R. O., Audebert, A., Dardou, A., Kumar, A., et al. (2016). Rice root architectural plasticity traits and genetic regions for adaptability to variable cultivation and stress conditions. Plant Physio. 171, 2562–2576. doi: 10.1104/pp.16.00705
Sandhu, N., Subedi, S. R., Singh, V. K., Sinha, P., Kumar, S., Singh, S. P., et al. (2019b). Deciphering the genetic basis of root morphology, nutrient uptake, yield, and yield-related traits in rice under dry direct-seeded cultivation systems. Sci. Rep. 9, 1–16. doi: 10.1038/s41598-019-45770-3
Sandhu, N., Subedi, S. R., Yadaw, R. B., Chaudhary, B., Prasai, H., Iftekharuddaula, K., et al. (2017). Root traits enhancing rice grain yield under alternate wetting and drying condition. Front. Plant Sci. 8:1879. doi: 10.3389/fpls.2017.01879
Sandhu, N., Torres, R. O., Sta Cruz, M. T., Maturan, P. C., Jain, R., Kumar, A., et al. (2015). Traits and QTLs for development of dry direct-seeded rainfed rice varieties. J. Exp. Bot. 66, 225–244. doi: 10.1093/jxb/eru413
Sato, H., ando, I., Hirabayashi, H., Takeuchi, Y., Arase, S., Kihara, J., et al. (2008). QTL analysis of brown spot resistance in rice (Oryza sativa L.). Breed. Sci. 58, 93–96. doi: 10.1270/jsbbs.58.93
Septiningsih, E. M., Hidayatun, N., Sanchez, D. L., Nugraha, Y., Carandang, J., Pamplona, A. M., et al. (2015). Accelerating the development of new submergence tolerant rice varieties, the case of Ciherang-Sub1 and PSB Rc18-Sub1. Euphytica 202, 259–268. doi: 10.1007/s10681-014-1287-x
Shamsudin, N. A. A., Swamy, B. M., Ratnam, W., Cruz, M. T. S., Sandhu, N., Raman, A. K., et al. (2016). Pyramiding of drought yield QTLs into a high quality Malaysian rice cultivar MRQ74 improves yield under reproductive stage drought. Rice 9:21. doi: 10.1186/s12284-016-0093-6
Shikari, A. B., Khanna, A., Krishnan, S. G., Singh, U. D., Rathour, R., Tonapi, V., et al. (2013). Molecular analysis and phenotypic validation of blast resistance genes Pita and Pita2 in landraces of rice (Oryza sativa L.). Indian J. Genet. 73, 131–141. doi: 10.5958/j.0975-6906.73.2.020
Singh, V. K., Singh, A., Singh, S. P., Ellur, R. K., Singh, D., Gopala Krishnan, S., et al. (2013). Marker-assisted simultaneous but stepwise backcross breeding for pyramiding blast resistance genes Piz5 and Pi54 into an elite Basmati rice restorer line ‘PRR 78'. Plant Breed. 132, 486–495. doi: 10.1111/pbr.12077
Song, W. Y., Pi, L. Y., Wang, G. L., Gardner, J., Holsten, T., and Ronald, P. C. (1997). Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9, 1279–1287. doi: 10.1105/tpc.9.8.1279
Subedi, S. R., Sandhu, N., Singh, V. K., Sinha, P., Kumar, S., Singh, S. P., et al. (2019). Genome-wide association study reveals significant genomic regions for improving yield, adaptability of rice under dry direct seeded cultivation condition. BMC Genom. 20:471. doi: 10.1186/s12864-019-5840-9
Suh, J. P., Jeung, J. U., Noh, T. H., Cho, Y. C., Park, S. H., Park, H. S., et al. (2013). Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 6:5. doi: 10.1186/1939-8433-6-5
Sun, L., Su, C., Wang, C., Zhai, H., and Wan, J. (2005). Mapping of a major resistance gene to the brown planthopper in the rice cultivar Rathu Heenati. Breed. Sci. 55, 391–396. doi: 10.1270/jsbbs.55.391
Swamy, B. P. M., Ahmed, H. U., Henry, A., Mauleon, R., Dixit, S., Vikram, P., et al. (2013). Genetic, physiological, and gene expression analyses reveal that multiple QTL enhance yield of rice mega-variety IR64 under drought. PLoS ONE 8:e62795. doi: 10.1371/journal.pone.0062795
Ullah, I., Jamil, S., Iqbal, M. Z., Shaheen, H. L., Hasni, S. M., Jabeen, S., et al. (2012). Detection of bacterial blight resistance genes in basmati rice landraces. Genet. Mol. Res. 11, 1960–1966. doi: 10.4238/2012.July.20.1
Vikram, P., Swamy, B. M., Dixit, S., Ahmed, H. U., Cruz, M. T. S., Singh, A. K., et al. (2011). qDTY1.1, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet. 12:89. doi: 10.1186/1471-2156-12-89
Wang, H., Bouman, B. A. M., Zhao, D., Wang, C., and Moya, P. F. (2002). “Aerobic rice in northern China, opportunities and challenges,” in Water-Wise Rice Production. Proceedings of the International Workshop on Water-Wise Rice Production, 8-11–April 2002, Los Baños, Philippines, eds B. A. M. Bouman, H. Hengsdijk, B. Hardy, P. S. Bindraban, T. P. Tuong, J. K. Ladha (Los Baños, Philippines: International Rice Research Institute), 143–154.
Wang, Y., Cao, L., Zhang, Y., Cao, C., Liu, F., Huang, F., et al. (2015). Map-based cloning and characterization of BPH29, a B3 domain-containing recessive gene conferring brown planthopper resistance in rice. J. Exp. Bot. 66, 6035–6045. doi: 10.1093/jxb/erv318
Wissuwa, M., and Ae, N. (2001). Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breed. 120, 43–48. doi: 10.1046/j.1439-0523.2001.00561.x
Xu, Y., and Crouch, J. H. (2008). Marker-assisted selection in plant breeding from publications to practice. Crop Sci. 48, 391–407. doi: 10.2135/cropsci2007.04.0191
Yadav, S., Sandhu, N., Dixit, S., Singh, V. K., Catolos, M., Mazumder, R. R., et al. (2020). Genomics-assisted breeding for successful development of multiple-stress-tolerant, climate-smart rice for southern and southeastern Asia. Plant Genome 14:e20074. doi: 10.1002/tpg2.20074
Yadav, S., Singh, U. M., Naik, S. M., Venkateshwarlu, C., Ramayya, P. J., Raman, K. A., et al. (2017). Molecular mapping of QTLs associated with lodging resistance in dry direct-seeded rice (Oryza sativa L.). Front. Plant Sci. 8:1431. doi: 10.3389/fpls.2017.01431
Keywords: direct seeded, genomics, multiparent, QTL/genes, pyramiding, Oryza sativa
Citation: Sandhu N, Yadav S, Catolos M, Cruz MTS and Kumar A (2021) Developing Climate-Resilient, Direct-Seeded, Adapted Multiple-Stress-Tolerant Rice Applying Genomics-Assisted Breeding. Front. Plant Sci. 12:637488. doi: 10.3389/fpls.2021.637488
Received: 03 December 2020; Accepted: 11 March 2021;
Published: 15 April 2021.
Edited by:
Hanwei Mei, Shanghai Agrobiological Gene Center, ChinaReviewed by:
Weiya Xue, Pennsylvania State University (PSU), United StatesShouvik Das, Indian Agricultural Research Institute (ICAR), India
Copyright © 2021 Sandhu, Yadav, Catolos, Cruz and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arvind Kumar, YXJ2aW5kLmt1bWFyQGNnaWFyLm9yZw==
 Nitika Sandhu1,2
Nitika Sandhu1,2 Shailesh Yadav
Shailesh Yadav Ma Teresa Sta Cruz
Ma Teresa Sta Cruz Arvind Kumar
Arvind Kumar