- Department of Biochemistry, University of Otago, Dunedin, New Zealand
Perennial ryegrass (Lolium perenne) is a temperate grass species commonly used as pasture for livestock. Flowering (heading) of ryegrass impacts metabolizable energy content and seed yield, therefore this trait is important for both farmers and seed producers. In related grass species, the VRN genes (VRN1-3) have been largely implicated in the determination of vernalization response and are responsible for much of the intra-species variation in this trait. Many other important flowering-time regulators have been cataloged in the model grass Brachypodium distachyon; however, in several cases, such as VRN2, their ryegrass homologs have not been well-characterized. Here, ryegrass homologs of important flowering time genes from B. distachyon were identified through available synteny data and sequence similarity. Phylogenetic analysis of VRN3/FT-like and VRN2-like genes was performed to elucidate these families further. The expression patterns of these genes were assessed during vernalization. This confirmed the key roles played by LpVRN1 and LpFT3 in the promotion of flowering. Furthermore, two orthologs of VRN2 identified here, as well as an ortholog of CO9, were expressed prior to vernalization, and were repressed in flowering plants, suggesting a role in floral repression. Significant variability in expression of these flowering pathway genes in diverse genotypes was detected and may underlie variation in flowering time and vernalization response.
Introduction
Perennial ryegrass (Lolium perenne) is a temperate grass, widely grown as pasture for livestock for its high levels of metabolizable energy. Ryegrass flowering (heading) occurs in the spring after a period of cold (i.e., winter) referred to as vernalization. As ryegrass is an obligate outcrosser, it is important that minimal variation in heading date occurs within a cultivar to ensure good seed production. Heading also impacts the availability of metabolizable energy, as plants switch from producing leaves toward less-nutritious stems and floral structures (Stockdale, 1999). Selection for different heading dates occurs during the breeding process, and many varieties with early and late heading dates are currently available. While some variation in heading date has been captured through introgression, a better understanding of the genetic mechanisms underlying heading date will enable breeders to develop cultivars with more specific and reliable heading dates.
The genetic mechanisms of flowering have been well studied in other model plants, such as Arabidopsis thaliana (Arabidopsis) and Brachypodium distachyon. In addition, a number of important flowering time genes have been identified in related crops such as wheat (Triticum sp.), barley (Hordeum vulgare) and rice (Oryza sativa). This intensive study has led to the description of a number of important genes and pathways controlling flowering time in grasses. The genes controlling vernalization response have been particularly well studied, as natural variation in vernalization requirement is widespread amongst different cultivars of wheat and barley. VRN1 is a MADS-box transcription factor which is induced by low temperatures (Yan et al., 2003). Extended periods of low temperatures result in mitotically stable expression of VRN1 through epigenetic marks (Oliver et al., 2009). Some varieties of spring wheat and barley have mutations at the VRN1 locus which result in constitutive expression of VRN1 and do not require vernalization in order to flower (Yan et al., 2003; Chu et al., 2011). Upon transition to long days, VRN1 is capable of activating transcription of the FLOWERING LOCUS T (FT) gene, VRN3, in leaves (Shimada et al., 2009). The VRN3/FT protein is transported from the leaves to the apical meristem, where it acts as a transcriptional co-activator (Tamaki et al., 2007). VRN1 is induced by FT in concert with a number of other proteins in the meristem, resulting in transcriptional activation of genes involved in floral meristem production (Li and Dubcovsky, 2008). In barley and B. distachyon natural variation exists in a relative of FT, HvFT3/BdFTL9, which acts during short days to promote flowering (Faure et al., 2007; Qin et al., 2019; Woods et al., 2019). In the case of BdFTL9, this is through a process called short day vernalization, and promotes floral competency upon return to long days (Woods et al., 2019).
Prior to vernalization, VRN2 is expressed in leaves, which represses VRN3 and possibly VRN1 (Yan et al., 2004; Dubcovsky et al., 2006). VRN2 is a zinc-finger CCT-domain protein and is down-regulated by short days and cold through the activity of VRN1 at the promoter (Chen and Dubcovsky, 2012; Deng et al., 2015). The CCT-domain is named after a 43-amino acid C-terminal domain found in three Arabidopsis proteins, CONSTANS (CO), CO-LIKE, and TIMING OF CAB1 (TOC1). Variations in the CCT domain of VRN2 have a profound impact on vernalization response in wheat, with non-functional variants abolishing vernalization requirement in spring varieties (Distelfeld et al., 2009). In rice, the CCT-domain protein Ghd7 acts to repress flowering in long days and is the ortholog of VRN2 from wheat (Griffiths et al., 2003; Xue et al., 2008; Woods et al., 2016). Other close homologs of VRN2/Ghd7 include the CO9-like genes, although these are less well-studied (Woods et al., 2016). The CO9-like gene from barley is expressed in short-day conditions and acts to repress flowering, suggesting that CO9-like genes also act as floral repressors (Kikuchi et al., 2012).
Ryegrass has been the subject of many flowering time studies, primarily using quantitative trait analysis, and a number of important flowering time genes have been identified (reviewed by Wang and Forster, 2017). VRN1 has been identified in ryegrass, and natural variation in the first intron affects vernalization response (Jensen et al., 2005; Andersen et al., 2006; Asp et al., 2011). A number of FT-like genes have also been described in ryegrass, with some of these homologs in close proximity to QTL affecting flowering time (Studer et al., 2009). Allelic variation at FT genes underlying these QTL has not yet been described. However, variation at the promoter of LpFT3, a putative ortholog of FT/VRN3 genes from other grasses, has been proposed to impact flowering time in a panel of different ryegrass germplasms (Skøt et al., 2011; Wang and Forster, 2017). No ortholog of VRN2 has been thoroughly described in ryegrass, although a locus on LG4 and a partial sequence have been reported (Paina et al., 2016; Woods et al., 2016). Two genes, named vrn2_2 and vrn2_3 were identified in a QTL analysis and corresponded to two CCT-domain proteins on LG7 (Andersen et al., 2006). Vrn2_2 was reported to be similar to VRN2 from wheat, while Vrn2_3 was a CO homolog (LpCO), although neither gene appeared to be regulated by vernalization.
Recently, a number of genomic resources have been developed for ryegrass, including two draft genomes, transcriptomic data, synteny data, and high-density mapping data (Studer et al., 2012; Paina et al., 2014; Byrne et al., 2015; Velmurugan et al., 2016). In this study, we aimed to use these resources to identify orthologs of important flowering time genes from B. distachyon. We further investigated FT-like genes and CCT-domain genes in an effort to find orthologous genes to BdFTL9, BdVRN3 and BdVRN2. We characterized homologs of VRN2 in a commercial cultivar during vernalization, flowering, and post-flowering. Finally, we looked for variation in the expression of these genes in a variety of different genotypes, which may underlie differences in their flowering habit.
Materials and Methods
Plant Materials and Growth Conditions
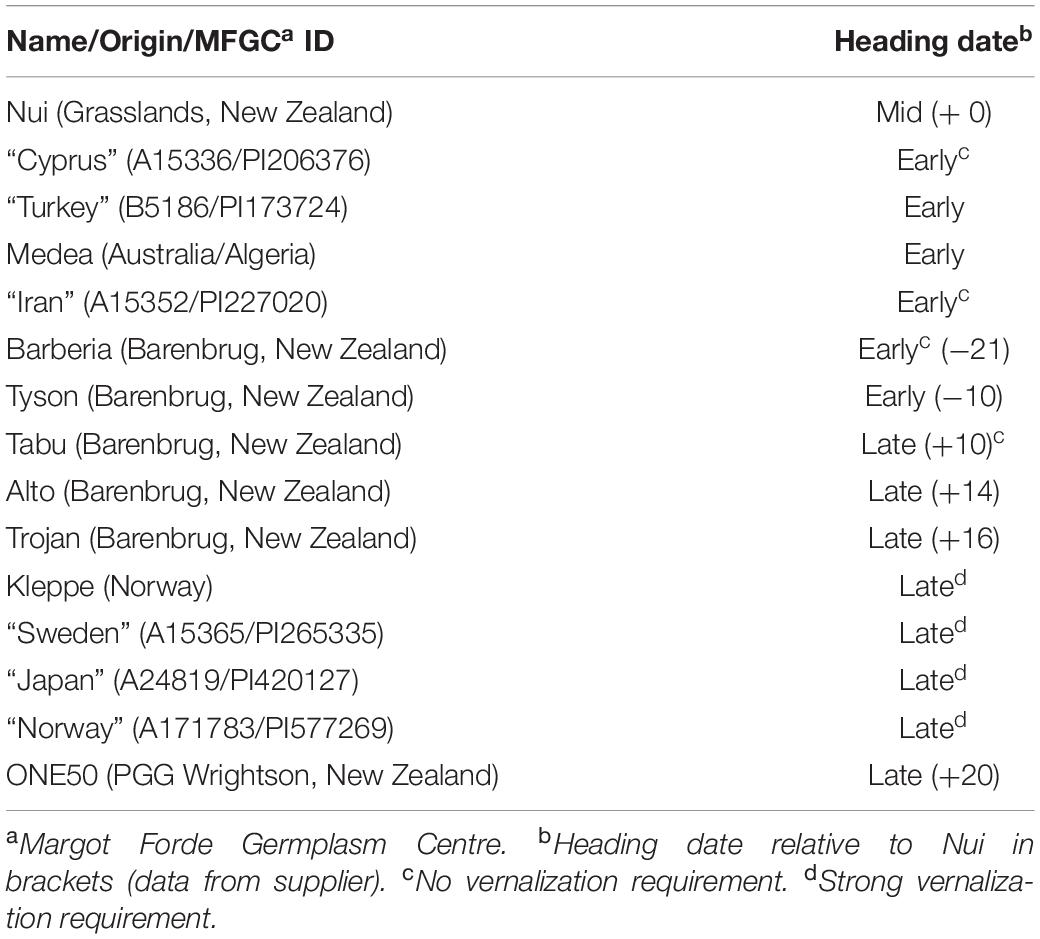
Lolium perenne plants were grown at 22°C 16 h light/8 h dark (Long Days; LD) or 8 h light/16 h dark (Short Days; SD) unless otherwise stated. A list of the ryegrass cultivars and accessions used in this study is provided in Table 1.
Flowering Time Gene Identification
tBLASTn was performed using B. distachyon peptides from B. distachyon predicted proteins (v1.2) against the L. perenne genome (The International Brachypodium Initiative, 2010; Byrne et al., 2015). For ease of identifying the matching annotations, predicted proteins from the ryegrass genome (Byrne et al., 2015) were also searched (BLASTp), and in cases where the top tBLASTn hit matched the top annotated protein hit, the annotation was noted and classified as the most-likely homolog. In cases where these did not match, the tBLASTn hit was examined on a genome browser (Integrative Genomics Viewer, IGV) to identify overlapping masked transcripts, or unannotated regions. The sequences of these genes/regions were aligned to their B. distachyon homologs to identify exons, and RNAseq reads (Paina et al., 2014) were simultaneously examined to determine splice sites where possible.
Phylogenetic Analysis of FT-Family and CCT-Domain Proteins
Homologous FT-family protein sequences in ryegrass were obtained from Veeckman et al. (2016). Homologous FT-like genes from B. distachyon, O. sativa and H. vulgare were also identified from the literature (Griffiths et al., 2003; Higgins et al., 2010). CCT-protein sequences were extracted from the L. perenne draft genome by searching predicted proteins (BLASTp) and a translated nucleotide database (tBLASTn) with the CCT domain of B. distachyon VRN2 (Byrne et al., 2015). Annotated CCT-domain proteins were identified in B. distachyon, O. sativa and H. vulgare genomes by searching for PFAM domain 06203 on Phytozome (B. distachyon and H. vulgare)1, or the rice genome2. All sequences were aligned using MUSCLE program followed by a gblock scan to identify conserved regions among the alignment which can later be used to infer phylogenetic relationship (Edgar, 2004). Phylogenetic trees were created using the CLC Genomics workbench (version 10) with the following settings: maximum likelihood method with JTT substitution matrix and 1000 bootstrap replication.
RNAseq Data Analysis
Previously published RNAseq data from two ecotypes (Falster and Veyo) was re-analzsed. Full details of the experimental conditions can be found in Paina et al. (2014). To summarize, leaf tissue from clonal Falster and Veyo plants was taken in short day conditions prior to vernalization in short days at 5°C. Leaf tissue was sampled after 2 days, 4 weeks, and 9 weeks of vernalization, and 7 days after returning to long days. Meristem tissue was taken after 9 weeks of vernalization, and 1 day, and 7 days after returning to long days. Raw data from Paina et al. (2014; EMBL-EBI ArrayExpress, accession no. E-MTAB-2623) was mapped to the L. perenne genome (Byrne et al., 2015) using STAR mapper (Dobin et al., 2013). Feature counts were extracted using HT-seq count (Anders et al., 2015) with the following settings: htseq-count -q -s no -f bam -t gene -i ID. A customized.gff file containing only the flowering genes of interest was used to avoid overlapping features preventing accurate quantification. Expression levels of each gene were normalized by the total number of mapped reads, and by the transcript length for plotting.
Sample Collection
For the 10 week vernalization time course, seeds of ONE50 were sterilized in a 1% bleach solution for 15 min and placed onto moist sterile filter paper in a petri dish and sealed with cling film. Seeds were chilled for 2 days in the dark at 4°C before transfer to LD conditions to germinate for 2 weeks, at which point seedlings are typically capable of responding to vernalization (Gangi et al., 1983). After 2 weeks in LD conditions, plates were transferred to 4°C in the dark for 10 weeks, and after 10 weeks of vernalization, seedlings were transferred to soil and grown in LD conditions at 22°C. Samples were collected prior to vernalization (0dV) and at 1 day, 1 week, 2 weeks, 4 weeks, 7 weeks, and 10 weeks vernalization (1dV, 1wV, 4wV, 7wV and 10wV), and at 7 days, 2 weeks and 4 weeks post-vernalization (7dLD, 2wLD, 4wLD). Each RNA sample from 0dV until 10wV was prepared by pooling tissue from the primary leaf of ∼10 seedlings. Samples after transfer to soil were prepared from a pool of single leaves from 3 plants. Samples were taken between 1–2 h after lights were turned on (ZT1- ZT2; 9am–10am), and at the same time each day in dark conditions. Leaf tissue was frozen in liquid nitrogen prior to RNA extraction.
For the comparison of flowering and non-flowering plants, each sample was prepared from leaves from several tillers from a single plant. Leaves were taken from three flowering and three non-flowering plants at ∼6 weeks after the transition to LD conditions when spikelets had clearly emerged in flowering plants. Further samples were taken from these plants 8 weeks later, and after 1 day or 1 week under vernalization conditions (SD + 5°C).
For comparison of gene expression between genotypes, seeds of the cultivars/accessions shown in Table 1 were sterilized and germinated as above for ONE50. Samples were taken after 2–3 weeks growth in long days (0dV) and after 2 days of cold (4°C)/dark treatment (2dV). Each RNA sample was prepared by pooling tissue from the primary leaf of ∼10 seedlings. Samples were taken between 1–2 h after lights were turned on (ZT1- ZT2; 9am–10am), and at the same time each day in dark conditions.
For analysis of LpFT08 expression in SD conditions, ONE50 plants were grown for 8 weeks in LD conditions (20 h light; 4 h dark). Leaf material from each plant was collected and pooled at T0 (prior to SD treatment), and 4 plants were moved to SD (8h light; 16 h dark) and 4 plants remained in LD (20 h light; 4 h dark). After 2.5 weeks in SD or LD, several leaves from each plant were pooled into a single sample (SD or LD) and frozen in liquid nitrogen prior to RNA extraction.
RNA Extraction, cDNA Synthesis and qPCR
Frozen leaf tissue (∼10 mg) was thawed in 300 μL Trizol (Invitrogen) prior to transfer to a ziplock bag. A dowel was rolled over the bag to homogenize the leaf tissue. Debris was removed by centrifugation at 16,000 × g for 5 min and discarding the pellet. RNA was extracted using Zymo Quickzol RNA microprep kit according to manufacturer’s instructions, eluting in 15 μL of water. cDNA was synthesized with 1 μg of RNA using SuperScript III Reverse transcriptase (Invitrogen) and oligo dT primers according to manufacturer’s instructions. qPCR was performed with SYBR 2x Mastermix (KAPA), using 0.2 μM each primer, in a final volume of 10 μL. Reactions were run in a Roche Lightcycler 480 with the following cycling conditions: a hold of 94 °C for 2 min followed by 50 cycles of 94 °C for 8 s, 58 °C for 10 s and 72 °C for 10 s. Tumor Homolog Protein (THP; Samarth and Jameson, 2019) and “expressed protein” (ExP; Narsai et al., 2010) were used as reference genes, and gene expression was calculated using the ΔCt method based on the geometric mean Ct of the two reference genes. Primers used for each gene are given in Supplementary Table 1. To compare variance of gene expression between different target genes in the array of genotypes, expression level of each sample was first normalized to the mean value, followed by pairwise F-tests to compare the variance of each target gene (LpVRN1, LpVRN2a/b and LpCO9). The log2 fold-change in expression level upon introduction to vernalization conditions was also calculated, and subsequent pairwise F-tests were performed on these values to determine significantly variable responses between target genes.
Results
Identification of Flowering Time Gene Homologs Based on Synteny and Sequence Similarity
To investigate the roles of known flowering time genes in vernalization response, we firstly identified likely orthologs of important flowering time genes from related grass species. A list of important flowering time proteins in B. distachyon identified by Higgins et al. (2010) was used to search for ryegrass homologs by searching a translated nucleotide database (tBLASTn) and this analysis was supplemented by identifying putative genomic locations using the L. perenne GenomeZipper (Byrne et al., 2015). In most cases, regions/genes identified in the GenomeZipper as homologous to B. distachyon flowering genes also contained the top tBLASTn hit. In some cases, no syntenic regions were identified in the GenomeZipper, and in these cases, the top BLAST hit was chosen as the most likely ortholog. In cases where the top BLAST hit was already associated with another B. distachyon gene, preference was given to the gene that was predicted to be syntenic (i.e., present in the GenomeZipper), and the non-syntenic gene was left without a likely homolog. Overall, from 158 B. distachyon genes, 89 had ryegrass homologs present in the GenomeZipper, while 56 had likely homologs identified via BLAST search, and 13 had no confidently predicted homologs. Five tBLASTn hits occurred in regions without an annotated transcript, in which case a note of the location of the BLAST hit was recorded, and the predicted protein was identified for future use (Supplementary Datasets 1, 2). A number of annotated ryegrass genes were previously omitted from the transcriptome/proteome (Byrne et al., 2015); the protein sequences of these genes have also been predicted and included in Supplementary Dataset 2. We also identified a small number of genes with truncated sequences in the proteome, which we manually re-annotated using homology to B. distachyon and RNAseq data to predict intron/exon boundaries and identify likely start and stop codons (Supplementary Dataset 2; Paina et al., 2014).
To validate these genes as orthologs of their respective B. distachyon genes, we performed a reciprocal BLAST search using their protein sequences against the B. distachyon proteome. Of the 89 homologs identified in the GenomeZipper, 82 were also the top hit to their respective B. distachyon counterpart, suggesting they are orthologous (Supplementary Dataset 1). A number of these genes have been previously identified via QTL analyses or other means, such as LpVRN1, LpCO and LpVRN3/FT3. Of particular interest are orthologs of important flowering time genes that have not been previously described. We discovered likely orthologs of B. distachyon VRN2 (discussed later), LEAFY (on scaffold_1714), and BdSOC1 and BdSOC1-like (on scaffold_156 and scaffold_10825). The expression of all identified homologs/orthologs during vernalization was assessed using available RNAseq data from leaves and meristems (Paina et al., 2014; Supplementary Dataset 3). This data was generated by sampling two genotypes (Falster and Veyo) with differing vernalization requirements at several time points during and after vernalization. Combining the expression pattern data for each gene, alongside its homology to known B. distachyon genes, unique identifiers from the draft genome, and putative genomic location, allows researchers to rapidly access useful information about these genes without the need to re-analyze next generation sequencing data.
Phylogeny and Expression Patterns Indicate LpFT3 and LpFT08 Likely Play Key Roles in Regulation of Ryegrass Flowering
FT-like genes are responsible for many aspects of flowering, and duplications and neo-functionalization of genes often occur in this family (Pin and Nilsson, 2012). For this reason, it is possible that there are some duplications or deletions of FT-like genes in L. perenne that confound the homology search performed above. For example, BdFTL7a and BdFTL7b both matched the same ryegrass gene using tBLASTn, which may indicate a duplication in B. distachyon or a deletion in L. perenne, and therefore requires further analysis. FT-like genes have been previously identified in ryegrass which we matched with annotated transcripts from the ryegrass proteome/transcriptome (Byrne et al., 2015; Veeckman et al., 2016; Supplementary Dataset 4). We constructed a phylogenetic tree of these FT genes, alongside those from B. distachyon, rice, and barley (Figure 1A). This analysis revealed that LpFT3 was most likely to be the ortholog of floral-inducers (e.g., BdFTL2/BdFT1 and OsFTL2 aka Hd3a; bootstrap support, 99) as has been previously proposed, and this was consistent with the synteny analysis, where it was predicted as the most likely ortholog of BdFTL2 (Supplementary Dataset 1). B. distachyon FTL9 has been implicated in a process known as short day vernalization, and grouped with LpFT08, as well as HvFT3 and BdFTL10 (Figure 1A; bootstrap support, 100; Qin et al., 2019; Woods et al., 2019). A ryegrass ortholog of TERMINAL FLOWER 1 (TFL1) has been previously identified, LpTFL1, which was closely grouped with BdTFL1 (bootstrap support, 91; Jensen et al., 2001). RCN2 and RCN4 are also important members of the FT family, controlling grain production in rice, and orthologs of these were detected in ryegrass – LpFT04 and LpFT06, respectively (bootstrap support, 68 and 46; Nakagawa et al., 2002; Ariyaratne et al., 2009). Despite the low bootstrap values of LpFT04 and LpFT06, these were also reciprocal BLAST hits of their B. distachyon orthologs (BdRCN2 and BdRCN4), although more comprehensive analysis may be required to fully elucidate the RCN2/RCN4 clade (Supplementary Dataset 1). In B. distachyon, FTL7 has three copies (BdFTL7a, b and c), while ryegrass has four copies (LpFT14-17; bootstrap support, 96), although these genes are yet to be characterized. Ryegrass also had two copies of MFT, one closer to BdMFT1a/b and one closer to BdMFT2 (LpFT05 and LpFT18, respectively; bootstrap support, 99 and 99).
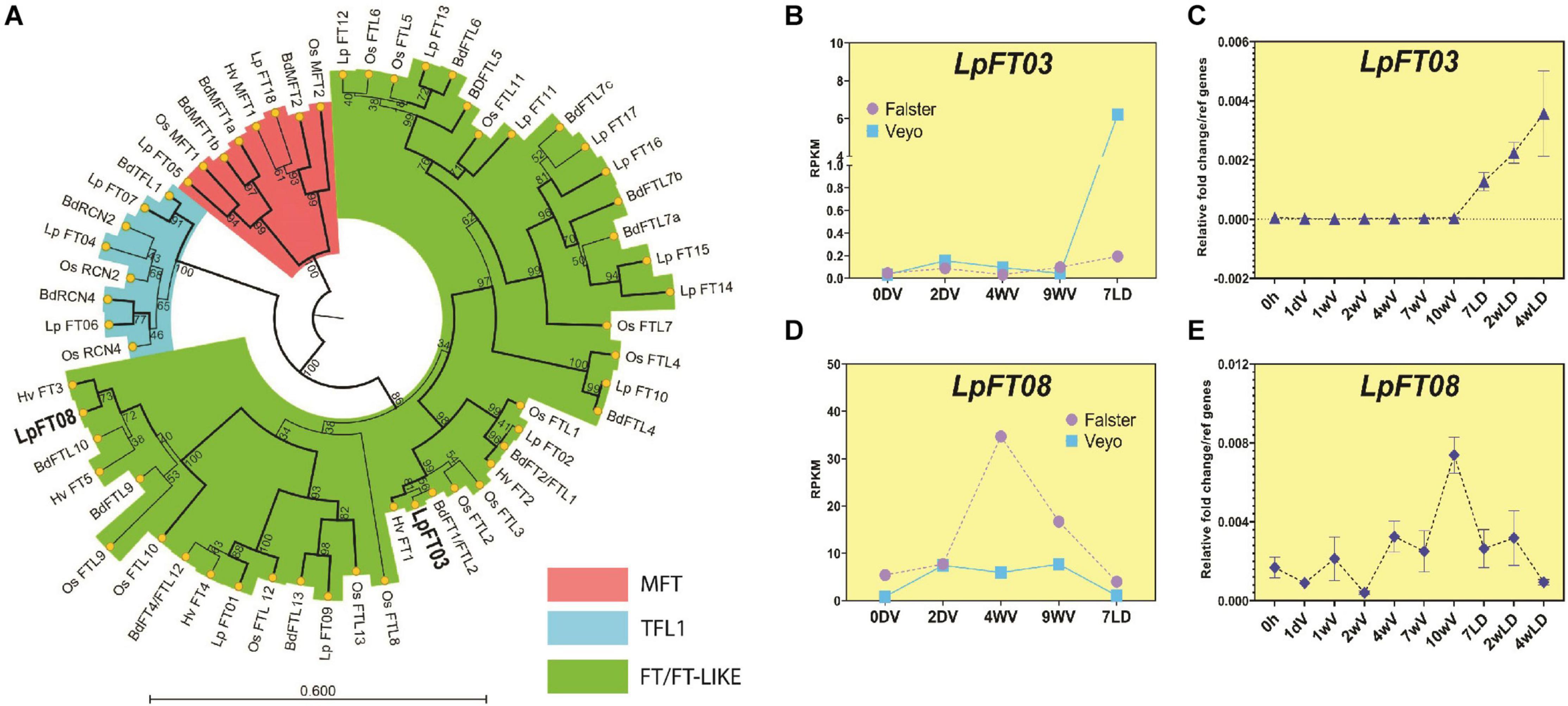
Figure 1. Phylogenetic relationships and expression patterns of FT-like proteins/genes from L. perenne. (A) Phylogenetic tree of LpFT01-LpFT18 proteins and their homologs from B. distachyon (Bd), H. vulgare (Hv) and O. sativa. (Os). The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed (scores represent the bootstrap support as a percentage). Branches with bootstrap support >70 are shown in bold. Branches in red represent the MFT clade, blue represent the TFL1 clade and green represent the FT clade. Bold highlights genes analyzed further in this study. MFT is the ancestral clade to TFL and FT clades and acts as an outgroup to root the tree. (B) Expression pattern of LpFT3 during and after vernalization in Falster and Veyo cultivars from RNAseq (data from Paina et al., 2014). (C) Expression pattern of LpFT3 during and after vernalization using qPCR in seedlings of the ONE50 cultivar. Expression was normalized to the geometric mean of two reference genes (ExP and THP), and shows the mean and standard deviation of two biological replicates of leaf tissue from ∼10 pooled seedlings. (D) Expression of LpFT08 during and after vernalization in Falster and Veyo from RNAseq (data from Paina et al., 2014). (E) Expression pattern of LpFT08 during and after vernalization using qPCR in seedlings of the ONE50 cultivar. Expression was normalized to the geometric mean of two reference genes (ExP and THP) and shows the mean and standard deviation of two biological replicates of leaf tissue from ∼10 pooled seedlings. RPKM, Reads Per Kilobase of transcript per Million mapped reads.
To determine which of the FT-like genes from ryegrass were responding to vernalization, we examined levels of FT-like genes during vernalization using available RNA-seq data (Paina et al., 2014). As expected of a floral inducer, LpFT3 expression occurred after vernalization when plants were returned to long days (Figure 1B). No other FT-like gene showed a similar expression pattern, suggesting that LpFT3 is likely to be the predominant FT-like gene acting as a floral inducer in these conditions (Supplementary Dataset 4). We confirmed the expression pattern of LpFT3 in a commercial cultivar (ONE50) during vernalization, which was similar to RNA seq data, despite differences in experimental approach (Figure 1C; see Methods). LpFT3 was also more highly expressed in flowering plants than non-flowering plants resulting from this experiment (discussed later; Supplementary Figure 1A). RNAseq data showed that LpFT08 was expressed during vernalization, and repressed upon introduction to long days, suggesting that it may act in a similar manner to BdFTL9 which is activated during short days (Figure 1D). LpFT08 expression in our vernalization conditions was not similar to the RNAseq data, as it initially decreased in expression, before increasing with prolonged vernalization (Figure 1E). This may be caused by differences in the genotype used for this study, or the difference in vernalization treatments. As the vernalization treatment in this experiment occurred under dark conditions, we tested LpFT08 expression in short days, which are known to induce the expression of BdFTL9 (Woods et al., 2019). LpFT08 expression increased in short days in the ONE50 cultivar in the absence of cold treatment, similarly to BdFTL9 (Supplementary Figure 1B). Initially we classified LpFT08 as an ortholog of BdFTL9 as these genes were linked in the GenomeZipper (Supplementary Dataset 1). However, LpFT08 grouped with BdFTL9 and BdFTL10 in the phylogenetic tree (Figure 1A) and we observed that BdFTL10 was absent from the GenomeZipper. To determine if LpFT08 was syntenic to BdFTL10 we examined the surrounding genes on scaffold_5827 and B. distachyon chromosome 2, revealing synteny between LpFT08 and BdFTL10 (Supplementary Figure 1C). LpFT08 is likely to be present on LG1 between markers PTA.648.C1 and P5G13, as that is the putative location of other genes on scaffold_5827.
Overall, the phylogenetic analysis of ryegrass FT-like genes has confirmed the identification of LpFT3 as a likely floral inducer, and LpFT08 as a potential inducer of floral competency similar to BdFTL9. We also grouped other known FT-like genes (such as LpTFL1) with their appropriate B. distachyon orthologs, and this has matched with the synteny data above. This suggests that our approaches to finding important flowering-time genes are appropriate and may be applied to less-well-known gene families.
Phylogenetic Analysis of CCT-Domain Proteins Revealed Candidates for VRN2 and CO9 in Ryegrass
To characterize the CCT-domain encoding genes from ryegrass, we queried predicted peptides from a translated nucleotide database based on the draft genome of L. perenne using the 43 amino acid CCT domain from BdVRN2 (Bradi3g10010) with tBLASTn. Simultaneously, we queried predicted proteins from the ryegrass transcriptome, to account for some cases where an intron interrupts the CCT domain (Cockram et al., 2012; Byrne et al., 2015). We identified annotated transcripts for each hit from which we derived the protein sequences (Supplementary Dataset 5). We also included the “vrn2_2” gene (present on scaffold_1808) which has been previously described but was not detected in our tBLASTn search as the CCT domain spans an intron, and is absent from the transcriptome (Andersen et al., 2006). Thirty-six CCT-domain proteins from ryegrass were aligned with CCT-domain proteins from B. distachyon, rice, and barley to generate a phylogenetic tree (Supplementary Figure 2). The ryegrass genes fell broadly into clades including PPD-like, CONSTANS-like, VRN2/Ghd7/CO9. From this tree we identified two putative VRN2-like genes, as well as a CO9-like gene in ryegrass. The gene annotated as “vrn2_2” present on scaffold_1808 was more similar to CO-like 14 from B. distachyon (Supplementary Figure 2; Andersen et al., 2006; Cockram et al., 2012). As no ryegrass homolog of VRN2 has been fully described to-date, we examined the VRN2/Ghd7/CO9-like clade more closely to identify genes which may act as floral repressors in a manner similar to VRN2 from wheat and barley (Figure 2A). From this tree, we could detect two possible homologs of VRN2 in ryegrass, present on scaffold_936 (LpVRN2a) and scaffold_1700 (LpVRN2b). These two genes had homologs in the L. multiflorum and F. pratensis genomes, suggesting conservation in the Lolium/Festuca complex (bootstrap support, 100; Knorst et al., 2019; Samy et al., 2020). A single ortholog of CO9/Ghd7 was also present on scaffold_10876 which was also present in the L. multiflorum and F. pratensis genomes (bootstrap support, 100).
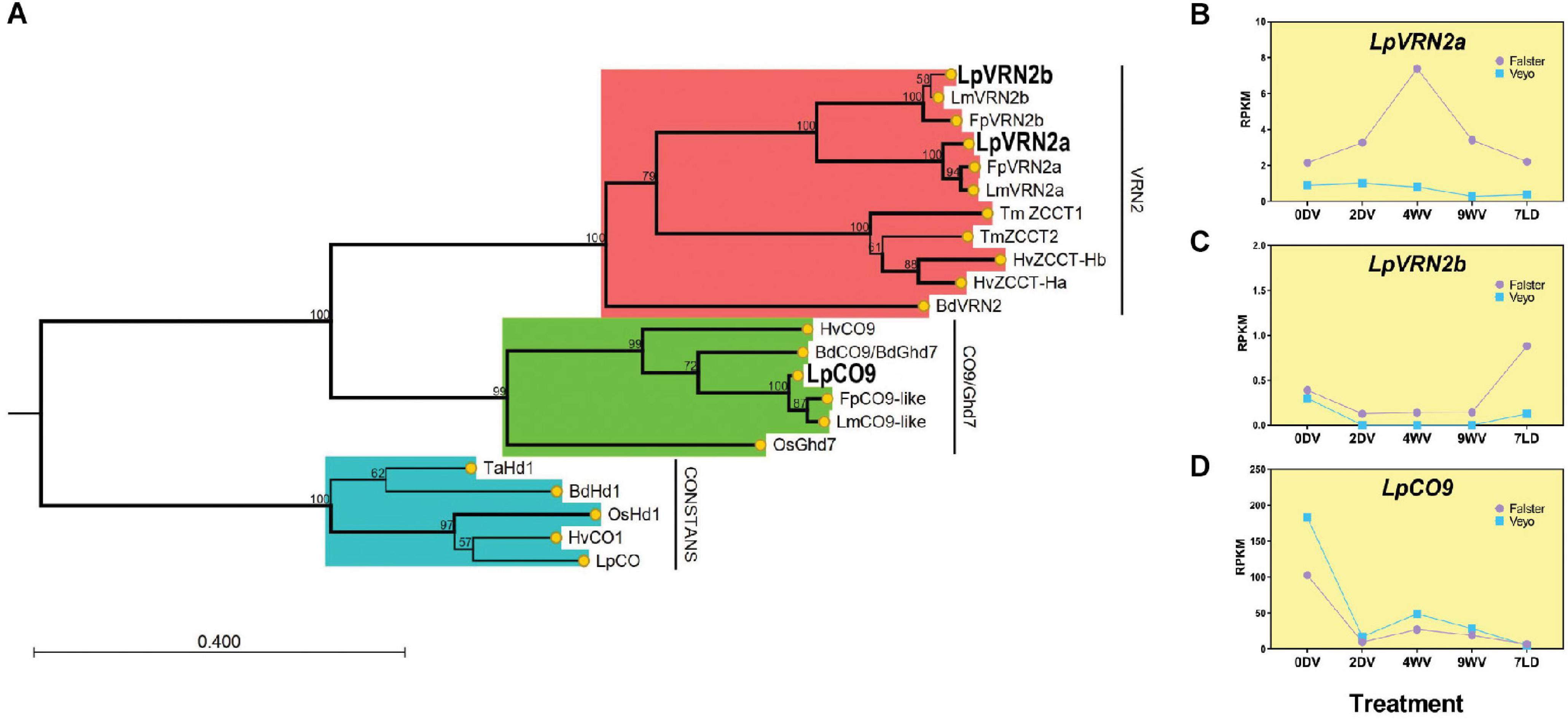
Figure 2. Phylogenetic relationships and expression patterns of VRN2/CO9-like proteins/genes from L. perenne. (A) Phylogenetic tree containing VRN2-like and CO9-like proteins from L. perenne (Lp), L. multiflorum (Lm), F. pratensis (Fp), B. distachyon (Bd), O. sativa (Os), T. monococcum (Tm), and H. vulgare (Hv). The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT + G matrix-based model. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed (scores represent the bootstrap support as a percentage). Branches with bootstrap support >70 are shown in bold. Branches in red represent the VRN2 clade, green represent the CO9/Ghd7 clade and blue represent the CONSTANS clade. CONSTANS acts as an outgroup to root the tree. (B–D) Expression patterns of LpVRN2a, LpVRN2b and LpCO9 during and after vernalization in Falster and Veyo cultivars from RNAseq (data from Paina et al., 2014). Expression was normalized to total mapped reads and transcript length. RPKM, Reads Per Kilobase of transcript per Million mapped reads.
Next, we aligned the CCT domains of LpVRN2a, LpVRN2b, and LpCO9 alongside homologs from wheat, barley, B. distachyon, L. multiflorum, F. pratensis and rice to determine whether any known functional residues were mutated in ryegrass species. Arginine residues 16, 35 and 39 have been reported to be mutated in non-functional versions of ZCCT in spring wheat, and these residues were unchanged in L. perenne or L. multiflorum, suggesting that the CCT domains of these proteins were likely to be functional (Distelfeld et al., 2009; Supplementary Figure 3A). In addition, residues conserved amongst a wide range of CCT-domain proteins, identified by Cockram et al. (2012), were unchanged in L. perenne, L. multiflorum and F. pratensis VRN2 and CO9-like proteins (Supplementary Figure 3A).
We also examined whether these three putative repressors were syntenic to known VRN2 genes in other grasses. Only LpVRN2a was present on a scaffold with more than one annotated gene, allowing us to easily determine syntenic regions from B. distachyon and wheat. The genes surrounding LpVRN2a were present on chromosome 1 from B. distachyon and 4B from durum wheat (Triticum turgidum; Supplementary Figure 3B). LpVRN2a was downstream of a norcoclaurine synthase gene; however, it was absent from B. distachyon and wheat. This suggests that LpVRN2a likely results from a novel insertion of a VRN2-like gene during the divergence of the Lolium genus.
Expression Analysis Indicates LpVRN2a, LpVRN2b and LpCO9 May Function as Floral Repressors
To gain a better understanding of the likely roles of LpVRN2a, LpVRN2b and LpCO9, we examined their expression pattern during vernalization in previously published RNAseq datasets from two ecotypes with differing vernalization requirements (Paina et al., 2014). LpVRN2a showed increased expression during vernalization in Falster compared to pre-vernalization levels, but was very lowly expressed in Veyo, while LpVRN2b showed very low expression in both varieties (Figures 2B,C). As the experiment had been conducted solely in short days, it is possible that both VRN homologs are repressed before cold treatment in these samples, as VRN2-like genes are repressed in short days in other species (Dubcovsky et al., 2006). LpCO9 was repressed during vernalization and stayed repressed on introduction to LD, similar to VRN2 from wheat as previously reported (Figure 2D; Paina et al., 2014).
We further characterized these expression patterns in the ONE50 cultivar, alongside the expression of LpVRN1, homologs of which act as repressors of VRN2 (Chen and Dubcovsky, 2012; Deng et al., 2015; Figure 3). LpVRN1 increased expression during vernalization, as previously demonstrated (Andersen et al., 2006; Figure 3A). LpVRN2a showed a similar expression pattern to the RNAseq data with a peak after several weeks of vernalization (Figure 3B). In contrast to the RNAseq data, LpVRN2b was expressed in ONE50 and showed a similar pattern to LpVRN2a (Figure 3C). LpCO9-like was also expressed; however, the pattern did not closely match that seen in the RNA seq data (Figure 3D). All three of these genes showed a reduction upon transfer from long days to vernalization conditions but unexpectedly increased later during vernalization, in contrast to similar experiments in wheat and barley (Figures 3B–D).
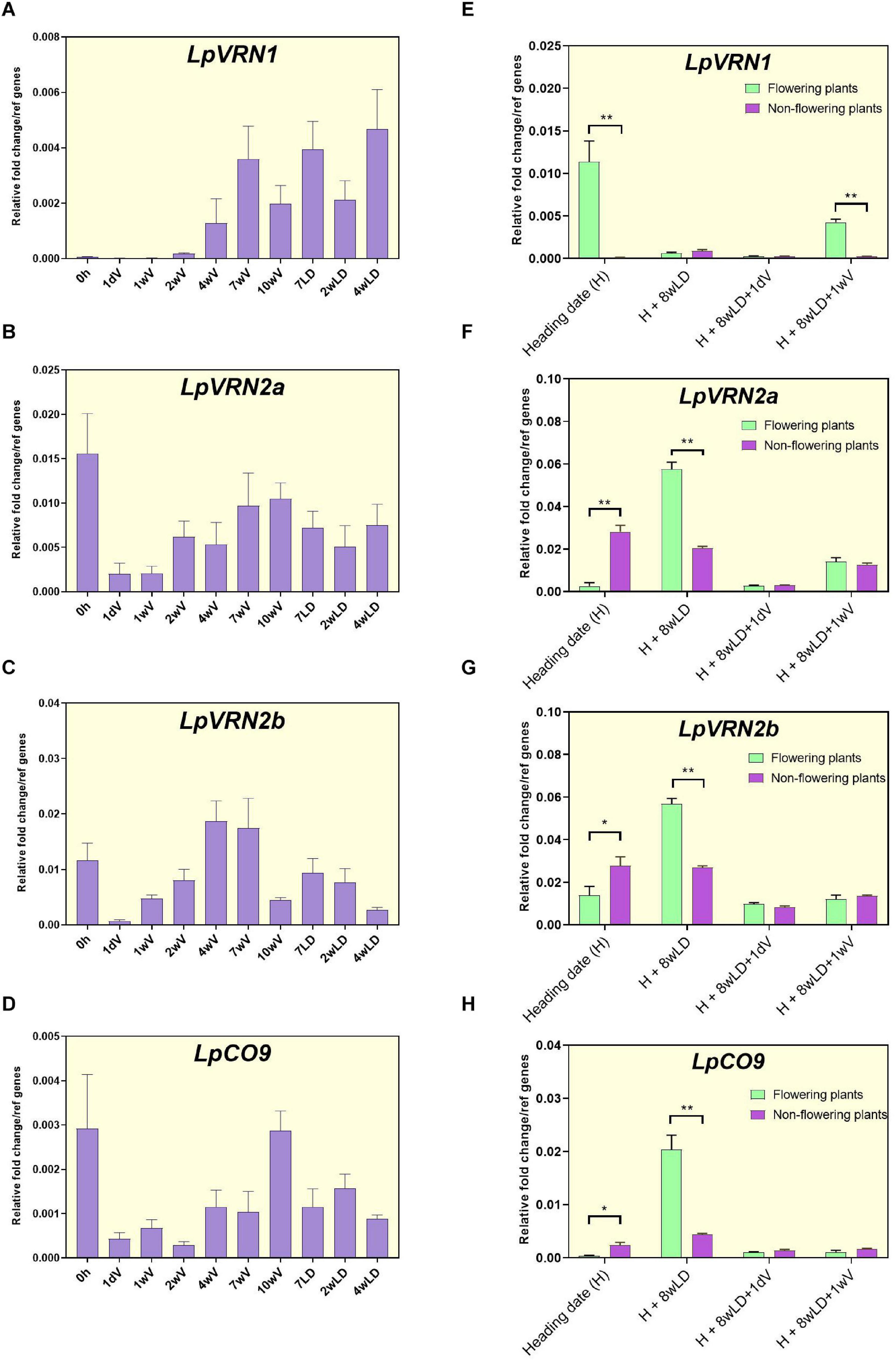
Figure 3. Expression patterns of VRN1 and VRN2-like genes from L. perenne during vernalization, heading, and re-vernalization. (A–D) Expression pattern of LpVRN1, LpVRN2a, LpVRN2b, and LpCO9 during and after vernalization using qPCR in seedlings of the ONE50 cultivar. Expression was normalized to the geometric mean of two reference genes (ExP and THP) and shows the mean and standard deviation of two biological replicates of leaf tissue from ∼10 pooled seedlings. (E–H) Expression pattern of LpVRN1, LpVRN2a, LpVRN2b, and LpCO9 at heading (H), 8 weeks after heading (H + 8wLD), and during reintroduction to vernalization conditions (H + 8wLD + 1dSDV and H + 8wLD + 1wSDV). Data is shown for plants that flowered (n = 3) and plants that did not flower (n = 3). Expression was normalized to the geometric mean of two reference genes (ExP and THP), the mean and standard deviation of three biological replicates is shown. Each biological replicate is from several leaf samples from a single plant. Asterisks represent the results of Student’s t-test between flowering and non-flowering results (*p < 0.1; **p < 0.05). LD, long days; SDV, short days with vernalization.
Samples used for qPCR time-course experiments were pooled from a number of related, but not identical ONE50 seedlings. Upon introduction to long days in our conditions, certain plants did not undergo flowering, suggesting that the pooled samples may contain induced and un-induced seedlings. We reasoned that the increase in expression of LpVRN2a, LpVRN2b and LpCO9 seen after ∼4 weeks of vernalization was due to expression in seedlings that were not responding to the vernalization treatment. To test this, we performed qPCR on flowering and non-flowering plants derived from this experiment (Figures 3E–H). As expected, VRN1 was highly expressed in flowering plants at heading (H), but not in non-flowering plants (Figure 3E). LpVRN2a, LpVRN2b and LpCO9 expression was low in flowering plants and high in non-flowering plants at heading (H; Figures 3F–H), suggesting that non-flowering seedlings may be the source of the observed increase in gene expression of these genes after ∼4 weeks of vernalization (Figures 3B–D).
As ryegrass is perennial, the vernalization response must be reset after flowering. We hypothesized that this would entail reversion of LpVRN1 and the floral repressors to their pre-vernalization levels, and that they would be able to respond to vernalization conditions again. We re-tested gene expression in the flowering and non-flowering plants from above 8 weeks after the completion of flowering (H + 8wLD), and also after 1 day (+ 1dV) and 1 week (+ 1wV) in vernalization conditions. Eight weeks after heading, LpVRN1 expression had returned to low pre-vernalization levels (Figure 3E), while LpVRN2a, LpVRN2b and LpCO9 had increased in plants that had previously flowered (Figures 3F–H). Upon re-introduction to vernalization conditions, LpVRN1 and the floral repressors responded as expected, suggesting that the plants were competent to undergo vernalization again (Figures 3E–H). Surprisingly, plants that had not initially flowered showed lower expression of floral repressors 8 weeks after heading (H + 8wLD) than plants that had flowered (Figures 3F–H). However, LpVRN2a, LpVRN2b and LpCO9 expression was still repressed upon introduction to vernalization conditions in these plants, suggesting that they were competent to respond to vernalization despite their already low levels of expression (Figures 3F–H). Interestingly, LpVRN1 did not strongly respond to vernalization conditions in the non-flowering plants, perhaps indicating that some genetic differences underlay the initial non-flowering phenotype (Figure 3E). In parallel, we assayed clones of these plants under short days (without cold; Supplementary Figure 4). LpVRN1 did not respond to short days, while the repressors responded in a similar manner in short days as they did to short days with cold (Supplementary Figure 4).
Variation in the Expression of Key Flowering-Time Genes Occurs in Ryegrass Genotypes With Different Vernalization Responses
Given the differences in LpVRN1 response seen in ONE50 plants above, we next aimed to determine whether the expression levels of LpVRN1, LpVRN2a, LpVRN2b and LpCO9 might contribute to the differences in vernalization response in different ryegrass genotypes (Table 1). Ryegrass genotypes were selected from a range of geographic locations, as well as a number of commercially grown cultivars with different flowering habits, in an effort to capture a wide array of genetic variation (Table 1; Faville et al., 2020). In wheat and barley, vernalization induces changes in gene expression of VRN1, firstly transiently, and then through stable epigenetic marks (Yan et al., 2003; Oliver et al., 2009; Diallo et al., 2012). Many spring varieties of wheat, as well as some ryegrass genotypes have high levels of VRN1 expression prior to vernalization which contributes to their lack of vernalization requirement (Andersen et al., 2006; Chu et al., 2011). We were interested in detecting variation in the level of LpVRN1 prior to vernalization and the transient response which may be indicative of genetic variation underlying different flowering habits. Overexpression of VRN2 and CO9-like genes also leads to delayed flowering, and may be another mechanism by which ryegrass can modulate its vernalization response (Kikuchi et al., 2012; Woods et al., 2016). As VRN1 regulates VRN2 expression at the promoter in other species, the absence of VRN2 repression upon transient VRN1 induction may indicate changes at the VRN2 promoter which could also contribute to varying vernalization responses (Deng et al., 2015). We predicted that the expression levels of LpVRN1 may be higher in earlier flowering varieties [similar to that seen in Veyo (early) compared to Falster (late); Andersen et al., 2006], and that the expression levels of the repressors may be lower. Alternatively, response to vernalization conditions may occur sooner in earlier flowering varieties, which would be illustrated by large changes in expression upon introduction to vernalization conditions. Genotypes with unusual expression levels or responses may contain useful genetic variation in the assayed genes that can be investigated further.
We examined the expression level of LpVRN1, LpVRN2a, LpVRN2b and LpCO9 prior to introduction to vernalization conditions in pooled 3-week-old seedlings to determine the pre-vernalization level of each gene (Supplementary Figure 5). To determine whether there was variation in the initial/transient response of these genes to vernalization conditions we also tested the expression of these genes after 2 days in cold (4°C) and dark and compared the values (Supplementary Figure 5). Gene expression was detected for all genes in all genotypes, except for LpCO9 in the Medea genotype (Supplementary Figure 5). In terms of their transient response to vernalization conditions, all plants had higher LpVRN1 expression after 2 days of cold, exemplified most clearly in the Iranian ecotype (Supplementary Figure 5E). LpVRN2a and LpVRN2b were repressed in all genotypes except Medea and Barberia, while LpCO9 varied greatly between genotypes upon transfer to vernalization conditions (Supplementary Figure 5F–H). Despite our earlier predictions, there was no association between early flowering and high LpVRN1 or low levels of repressors, and the transient response also did not appear to be significantly associated with flowering habit (Supplementary Figure 6). However, LpCO9 was more highly expressed in late-flowering genotypes prior to vernalization, which may implicate it in the late-flowering phenotype (Supplementary Figure 6).
Although we did not detect strong associations between flowering habit and gene expression in most cases, there was a high level of variation in expression and response between genotypes, particularly in LpVRN1 and LpCO9 which may indicate the presence of natural variation. We examined the variance between genotypes of LpVRN1, LpVRN2a/b and LpCO9 expression before and after 2 days in vernalization conditions, and also the variance in response (expressed as log2 fold change between conditions) to determine which genes are most likely to have underlying genetic variation (Figure 4). LpVRN1 and LpCO9 were highly variable, both in terms of expression level, and in terms of response, while LpVRN2a and LpVRN2b were significantly less variable (Figure 4). Overall, these results suggest that LpCO9 might hold more natural variation in expression than LpVRN2a and LpVRN2b. Natural variation in LpVRN1 has already been demonstrated and may exist in the genotypes examined here as well (Andersen et al., 2006).
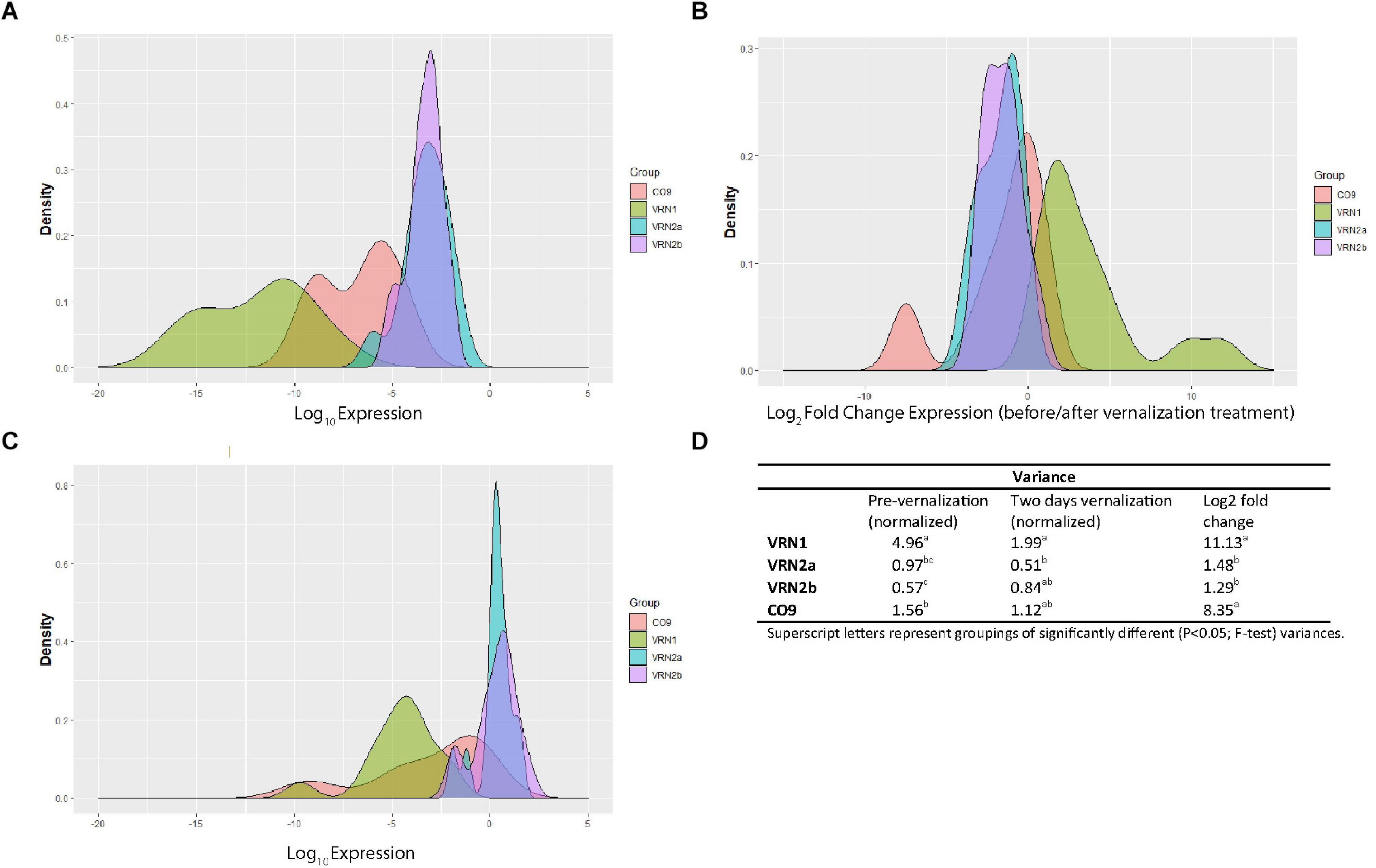
Figure 4. Analysis of the variability in expression and response to vernalization treatment of LpVRN1, LpVRN2a, LpVRN2b and LpCO9 in a variety of different ryegrass genotypes. (A) Density plot of log-transformed gene expression before transfer to vernalization conditions. (B) Density plot of log-transformed gene expression after 2 days in vernalization conditions. (C) Density plot of log2 fold change in gene expression after transfer to vernalization conditions. (D) Table showing significant differences in variances (p < 0.05; F-test) between each gene. LpVRN1 and LpCO9 were more variable than LpVRN2a and LpVRN2b.
Discussion
Ryegrass flowering impacts metabolizable energy and seed production, and is therefore an important trait for breeders to target. The advent of genomic selection now allows breeders to more accurately select for heading date (Hayes et al., 2013). However, understanding the genes and alleles responsible for variation in heading date is required to more easily identify useful germplasm, and for more targeted approaches, such as gene editing with CRISPR/Cas9. Translating knowledge from model species and well-studied crops is therefore an efficient way to further our understanding of heading date in ryegrass.
Here, we have used available genomic resources to compile a list of likely orthologs of important flowering-time genes from B. distachyon in L. perenne. We have further confirmed orthology for FT-like and VRN2-like genes using phylogeny (Figures 1, 2). Further phylogenetic analyses may confirm the orthology between other genes identified here (e.g., MADS-box domain genes, such as SOC1), and additional genomic sequences may enable the identification of more genes/paralogs in ryegrass. It is hoped that this thorough analysis can be used as a reference, particularly for determining the functions of FT-like and VRN2-like genes in ryegrass. Our phylogenetic analysis of FT-like genes in L. perenne was more complete than previous analyses (Arojju et al., 2016), and incorporated expression data to strengthen hypotheses regarding the function of each L. perenne FT-like gene. For example, it has been suggested that the FT-like gene present on scaffold_13332 (LpFT02) corresponds to the LpVRN3 marker (Arojju et al., 2016). However, the analysis by Arojju et al. (2016) did not include LpFT3 (likely because it was not included in the transcriptome), which perfectly matches the entire sequence of the LpVRN3 marker, and is supported to be the functional FT homolog in this (and other) studies (Skøt et al., 2011; Studer et al., 2012). Similarly, the gene associated with the vrn2_2 marker on scaffold_1808 matches more closely with CO-like 14 from B. distachyon (Andersen et al., 2006; Supplementary Figure 3). The two VRN2 homologs identified here are the closest match to VRN2 from B. distachyon and also ZCCT1 and ZCCT2 from wheat (Figure 3A). In addition, no closer genes were identified in F. pratensis or L. multiflorum, suggesting that it is unlikely that more VRN2-like genes exist in L. perenne than the two identified here.
In wheat, VRN2 is a key determinant of flowering habit between different cultivars. QTL analyses on ryegrass vernalization response identified LpVRN1 and LpCO as important determinants of the different responses between Falster and Veyo cultivars (Jensen et al., 2005; Andersen et al., 2006). LpVRN2a and LpVRN2b were not identified in this screen, although the genes underlying all QTL from this experiment are yet to be identified. LpVRN2a has been assigned to linkage group 4 and is included in a QTL relating to winter survival (Paina et al., 2016). LpVRN2b and LpCO9 have not been assigned to a linkage group in GenomeZipper or the recent high-density mapping population, making it difficult to determine whether these genes may correspond to a flowering QTL (Byrne et al., 2015; Velmurugan et al., 2016). Our data suggest that less variation in the expression levels (and response to vernalization conditions) of LpVRN2a and LpVRN2b exists in comparison to LpVRN1 and LpCO9 (Figure 4), which may explain why these genes have not been detected in previous QTL analyses; mutational studies or targeted F2 populations (e.g., utilizing Medea or Barberia individuals as parents, which lacked response in VRN2a and VRN2b, respectively) would be required to determine their function (Supplementary Figure 5). There may also be variation in LpVRN2a and LpVRN2b expression after long-term cold exposure/vernalization which was not represented in the initial response.
The high variability of LpCO9 response (Figure 4), and high expression levels of LpCO9 in late-flowering genotypes (Supplementary Figure 6), suggests that some genetic variation exists in the promoter of LpCO9, or in proteins that regulate its expression. How much variation in the expression levels of these genes exists between individuals of a cultivar/ecotype is also of interest – as we have pooled samples for our expression analysis (Supplementary Figure 5), it is possible that some individuals from each pool largely dictate the observed expression level of a gene. Whether the variability in LpCO9 expression observed after 2 days in vernalization conditions translates to variability at later stages remains to be seen.
VRN2 was also reported to be absent from the L. multiflorum transcriptome prior to vernalization, however, we have detected LpVRN2a and LpVRN2b transcripts in annual ryegrasses (Tabu, Barberia, and the Cypriot and Iranian ecotypes; Supplementary Figure 5; Czaban et al., 2015). This discrepancy may be due to differences in genotype, or to plant age, as we have used 3-week-old seedlings, compared to mature plants, which may indicate a role for LpVRN2a and LpVRN2b in repressing flowering in young annual ryegrass plants. The alleles present in L. multiflorum appear to have functional CCT domains (Supplementary Figure 3A), and the sequence of L. multiflorum VRN2a is present in the Falster transcriptome (data not shown; Paina et al., 2014). Alleles of LpVRN2a and LpVRN2b may play a more minor role in heading date and vernalization requirement compared to the large effect of different LpVRN1 alleles. We did not detect an association between early flowering and high LpVRN1 expression prior to vernalization (Supplementary Figure 5A), perhaps indicating that the early flowering (and annual) genotypes examined here contain novel variation affecting this trait, distinct from that which affects LpVRN1 expression (as seen in Falster and Veyo; Andersen et al., 2006). Alternatively, LpVRN1 expression may only be induced later in these genotypes, again suggesting that plant age plays a role in modulating expression of VRN genes in ryegrass.
How LpVRN1 impacts the expression level of LpVRN2a, LpVRN2b and LpCO9 in ryegrass remains to be determined. In the non-flowering ryegrass plants, and in ryegrass exposed to short days in the absence of cold, LpVRN1 was not induced, however, LpVRN2a/b and LpCO9 expression was reduced in these conditions, suggesting that regulators other than LpVRN1 are able to repress these genes (Figure 3 and Supplementary Figure 3). The alleles underlying LpVRN1, LpVRN2a/b and LpCO9 expression differences in the various genotypes we assayed here may also be of interest (Figure 4 and Supplementary Figure 5). Transgenic studies to test the function of LpFT08, LpVRN2a/b and LpCO9 will be critical to further our understanding of these genes in L. perenne.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
RH, S, LB, and RM designed the experiments and wrote and edited the manuscript. RH and S performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Ministry of Business Innovation and Employment (MBIE) funding grant number UOOX1911.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.640324/full#supplementary-material
Footnotes
References
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169.
Andersen, J. R., Jensen, L. B., Asp, T., and Lubberstedt, T. (2006). Vernalization response in perennial ryegrass (Lolium perenne L.) involves orthologues of diploid wheat (Triticum monococcum) VRN1 and rice (Oryza sativa) Hd1. Plant Mol. Biol. 60, 481–494. doi: 10.1007/s11103-005-4815-1
Ariyaratne, M., Takamure, I., and Kato, K. (2009). Shoot branching control by reduced culm number 4 in rice (Oryza sativa L.). Plant Sci. 176, 744–748.
Arojju, S. K., Barth, S., Milbourne, D., Conaghan, P., Velmurugan, J., Hodkinson, T. R., et al. (2016). Markers associated with heading and aftermath heading in perennial ryegrass full-sib families. BMC Plant Biol. 16:160. doi: 10.1186/s12870-016-0844-y
Asp, T., Byrne, S., Gundlach, H., Bruggmann, R., Mayer, K. F., Andersen, J. R., et al. (2011). Comparative sequence analysis of VRN1 alleles of Lolium perenne with the co-linear regions in barley, wheat, and rice. Mol. Genet. Genomics 286, 433–447. doi: 10.1007/s00438-011-0654-8
Byrne, S. L., Nagy, I., Pfeifer, M., Armstead, I., Swain, S., Studer, B., et al. (2015). A synteny-based draft genome sequence of the forage grass Lolium perenne. Plant J. 84, 816–826. doi: 10.1111/tpj.13037
Chen, A., and Dubcovsky, J. (2012). Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 8:e1003134. doi: 10.1371/journal.pgen.1003134
Chu, C.-G., Tan, C., Yu, G.-T., Zhong, S., Xu, S., and Yan, L. (2011). A novel retrotransposon inserted in the dominant Vrn-B1 allele confers spring growth habit in tetraploid wheat (Triticum turgidum L.). G3 Genes Genomes Genet. 1, 637–645.
Cockram, J., Thiel, T., Steuernagel, B., Stein, N., Taudien, S., Bailey, P. C., et al. (2012). Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS One 7:e45307. doi: 10.1371/journal.pone.0045307
Czaban, A., Sharma, S., Byrne, S. L., Spannagl, M., Mayer, K. F., and Asp, T. (2015). Comparative transcriptome analysis within the Lolium/Festuca species complex reveals high sequence conservation. BMC Genomics 16:249. doi: 10.1186/s12864-015-1447-y
Deng, W., Casao, M. C., Wang, P., Sato, K., Hayes, P. M., Finnegan, E. J., et al. (2015). Direct links between the vernalization response and other key traits of cereal crops. Nat. Commun. 6:5882. doi: 10.1038/ncomms6882
Diallo, A. O., Ali-Benali, M. A., Badawi, M., Houde, M., and Sarhan, F. (2012). Expression of vernalization responsive genes in wheat is associated with histone H3 trimethylation. Mol. Genet. Genomics 287, 575–590.
Distelfeld, A., Tranquilli, G., Li, C., Yan, L., and Dubcovsky, J. (2009). Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 149, 245–257. doi: 10.1104/pp.108.129353
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dubcovsky, J., Loukoianov, A., Fu, D., Valarik, M., Sanchez, A., and Yan, L. (2006). Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol. Biol. 60, 469–480. doi: 10.1007/s11103-005-4814-2
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Faure, S., Higgins, J., Turner, A., and Laurie, D. A. (2007). The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176, 599–609. doi: 10.1534/genetics.106.069500
Faville, M. J., Griffiths, A. G., Baten, A., Cao, M., Ashby, R., Ghamkhar, K., et al. (2020). Genomic assessment of white clover and perennial ryegrass genetic resources. J. N. Z. Grassl. 82, 27–34.
Gangi, A. S., Chilcote, D. O., and Frakes, R. V. (1983). Growth, floral induction and reproductive development in selected perennial ryegrass Lolium perenne L. Cultivars. Appl. Seed Prod. 1, 34–38.
Griffiths, S., Dunford, R. P., Coupland, G., and Laurie, D. A. (2003). The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 131, 1855–1867. doi: 10.1104/pp.102.016188
Hayes, B. J., Cogan, N. O., Pembleton, L. W., Goddard, M. E., Wang, J., Spangenberg, G. C., et al. (2013). Prospects for genomic selection in forage plant species. Plant Breed. 132, 133–143.
Higgins, J. A., Bailey, P. C., and Laurie, D. A. (2010). Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One 5:e10065. doi: 10.1371/journal.pone.0010065
Jensen, C. S., Salchert, K., and Nielsen, K. K. (2001). A TERMINAL FLOWER1-like gene from perennial ryegrass involved in floral transition and axillary meristem identity. Plant Physiol. 125, 1517–1528. doi: 10.1104/pp.125.3.1517
Jensen, L. B., Andersen, J. R., Frei, U., Xing, Y., Taylor, C., Holm, P. B., et al. (2005). QTL mapping of vernalization response in perennial ryegrass (Lolium perenne L.) reveals co-location with an orthologue of wheat VRN1. Theor. Appl. Genet. 110, 527–536. doi: 10.1007/s00122-004-1865-8
Kikuchi, R., Kawahigashi, H., Oshima, M., Ando, T., and Handa, H. (2012). The differential expression of HvCO9, a member of the CONSTANS-like gene family, contributes to the control of flowering under short-day conditions in barley. J. Exp. Bot. 63, 773–784. doi: 10.1093/jxb/err299
Knorst, V., Yates, S., Byrne, S., Asp, T., Widmer, F., Studer, B., et al. (2019). First assembly of the gene−space of Lolium multiflorum and comparison to other Poaceae genomes. Grassl. Sci. 65, 125–134.
Li, C., and Dubcovsky, J. (2008). Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 55, 543–554. doi: 10.1111/j.1365-313X.2008.03526.x
Nakagawa, M., Shimamoto, K., and Kyozuka, J. (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750. doi: 10.1046/j.1365-313x.2002.01255.x
Narsai, R., Ivanova, A., Ng, S., and Whelan, J. (2010). Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol. 10:56. doi: 10.1186/1471-2229-10-56
Oliver, S. N., Finnegan, E. J., Dennis, E. S., Peacock, W. J., and Trevaskis, B. (2009). Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc. Natl. Acad. Sci. U.S.A. 106, 8386–8391. doi: 10.1073/pnas.0903566106
Paina, C., Byrne, S. L., Domnisoru, C., and Asp, T. (2014). Vernalization mediated changes in the Lolium perenne transcriptome. PLoS One 9:e107365. doi: 10.1371/journal.pone.0107365
Paina, C., Byrne, S. L., Studer, B., Rognli, O. A., and Asp, T. (2016). Using a candidate gene-based genetic linkage map to Identify QTL for winter survival in perennial ryegrass. PLoS One 11:e0152004. doi: 10.1371/journal.pone.0152004
Pin, P. A., and Nilsson, O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35, 1742–1755.
Qin, Z., Bai, Y., Muhammad, S., Wu, X., Deng, P., Wu, J., et al. (2019). Divergent roles of FT-like 9 in flowering transition under different day lengths in Brachypodium distachyon. Nat. Commun. 10:812. doi: 10.1038/s41467-019-08785-y
Samarth and Jameson, P. E. (2019). Selection of reference genes for flowering pathway analysis in the masting plants, Celmisia lyallii and Chionochloa pallens, under variable environmental conditions. Sci. Rep. 9:9767.
Samy, J. K. A., Rognli, O. A., and Kovi, M. R. J. D. (2020). ForageGrassBase: molecular resource for the forage grass meadow fescue (Festuca pratensis Huds.). Database 2020:baaa046. doi: 10.1093/database/baaa046
Shimada, S., Ogawa, T., Kitagawa, S., Suzuki, T., Ikari, C., Shitsukawa, N., et al. (2009). A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene. VRN1, is upstream of FLOWERING LOCUS T. Plant J. 58, 668–681. doi: 10.1111/j.1365-313X.2009.03806.x
Skøt, L., Sanderson, R., Thomas, A., Skøt, K., Thorogood, D., Latypova, G., et al. (2011). Allelic variation in the perennial ryegrass FLOWERING LOCUS T gene is associated with changes in flowering time across a range of populations. Plant Physiol. 155, 1013–1022. doi: 10.1104/pp.110.169870
Stockdale, C. (1999). Effects of season and time since defoliation on the nutritive characteristics of three irrigated perennial pasture species in northern Victoria 1. Energy, protein and fibre. Aust. J. Exp. Agric. 39, 555–565.
Studer, B., Byrne, S., Nielsen, R. O., Panitz, F., Bendixen, C., Islam, M. S., et al. (2012). A transcriptome map of perennial ryegrass (Lolium perenne L.). BMC Genomics 13:140. doi: 10.1186/1471-2164-13-140
Studer, B., Jensen, L. B., Fiil, A., and Asp, T. (2009). “Blind” mapping of genic DNA sequence polymorphisms in Lolium perenne L. by high resolution melting curve analysis. Mol. Breed. 24, 191–199.
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
The International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. doi: 10.1038/nature08747
Veeckman, E., Vandepoele, K., Asp, T., Roldàn-Ruiz, I., and Ruttink, T. (2016). “Genomic variation in the FT gene family of perennial ryegrass (Lolium perenne),” in Breeding in a World of Scarcity, eds I. Roldán-Ruiz, J. Baert, and D. Reheul (Cham: Springer), 121–126.
Velmurugan, J., Mollison, E., Barth, S., Marshall, D., Milne, L., Creevey, C. J., et al. (2016). An ultra-high density genetic linkage map of perennial ryegrass (Lolium perenne) using genotyping by sequencing (GBS) based on a reference shotgun genome assembly. Ann. Bot. 118, 71–87. doi: 10.1093/aob/mcw081
Wang, J. P., and Forster, J. W. (2017). Flowering time regulation in perennial ryegrass. Euphytica 213:106.
Woods, D., Dong, Y., Bouche, F., Bednarek, R., Rowe, M., Ream, T., et al. (2019). A florigen paralog is required for short-day vernalization in a pooid grass. eLife 8:e42153. doi: 10.7554/eLife.42153
Woods, D. P., McKeown, M. A., Dong, Y., Preston, J. C., and Amasino, R. M. (2016). Evolution of VRN2/Ghd7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol. 170, 2124–2135. doi: 10.1104/pp.15.01279
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644. doi: 10.1126/science.1094305
Keywords: flowering time, vernalization, grasses, forage, transcriptome
Citation: Herridge R, Samarth, Brownfield L and Macknight R (2021) Identification and Characterization of Perennial Ryegrass (Lolium perenne) Vernalization Genes. Front. Plant Sci. 12:640324. doi: 10.3389/fpls.2021.640324
Received: 11 December 2020; Accepted: 15 February 2021;
Published: 05 March 2021.
Edited by:
Richard Amasino, University of Wisconsin-Madison, United StatesReviewed by:
Jill Christine Preston, University of Vermont, United StatesAna M. Casas, Aula Dei Experimental Station (EEAD), Spain
Copyright © 2021 Herridge, Samarth, Brownfield and Macknight. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rowan Herridge, Um93YW4uSGVycmlkZ2VAb3RhZ28uYWMubno=
 Rowan Herridge
Rowan Herridge Samarth
Samarth Lynette Brownfield
Lynette Brownfield Richard Macknight
Richard Macknight