- 1Biological Resources and Post-harvest Division, Japan International Research Center for Agricultural Sciences (JIRCAS), Tsukuba, Japan
- 2Technology Development Group, Actree Corporation, Hakusan, Japan
- 3Graduate School of Agriculture, Kyoto University, Kyoto, Japan
- 4Agri-Innovation Center, Iwate University, Morioka, Japan
- 5Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Japan
Quinoa (Chenopodium quinoa), native to the Andean region of South America, has been recognized as a potentially important crop in terms of global food and nutrition security since it can thrive in harsh environments and has an excellent nutritional profile. Even though challenges of analyzing the complex and heterogeneous allotetraploid genome of quinoa have recently been overcome, with the whole genome-sequencing of quinoa and the creation of genotyped inbred lines, the lack of technology to analyze gene function in planta is a major limiting factor in quinoa research. Here, we demonstrate that two virus-mediated transient expression techniques, virus-induced gene silencing (VIGS) and virus-mediated overexpression (VOX), can be used in quinoa. We show that apple latent spherical virus (ALSV) can induce gene silencing of quinoa phytoene desaturase (CqPDS1) in a broad range of quinoa inbred lines derived from the northern and southern highland and lowland sub-populations. In addition, we show that ALSV can be used as a VOX vector in roots. Our data also indicate that silencing a quinoa 3,4-dihydroxyphenylalanine 4,5-dioxygenase gene (CqDODA1) or a cytochrome P450 enzyme gene (CqCYP76AD1) inhibits betalain production and that knockdown of a reduced-height gene homolog (CqRHT1) causes an overgrowth phenotype in quinoa. Moreover, we show that ALSV can be transmitted to the progeny of quinoa plants. Thus, our findings enable functional genomics in quinoa, ushering in a new era of quinoa research.
Introduction
Rapid and large-scale changes in environmental and social conditions require rapid and fundamental changes in crop productivity and diversity (Eshed and Lippman, 2019). To withstand the effects of climate change and the population pressures on the global food system, the food supply will need to double by 2050 (Wheeler and Braun, 2013). Achieving global sustainable food production will require that people shift from a livestock-based diet to a plant-based diet (Shepon et al., 2018). This shift will need to be accompanied by an increase in the production of protein-rich crops (Eshed and Lippman, 2019). Of the tens of thousands of edible plants currently in existence, hundreds of species are cultivated around the world, but fewer than a dozen account for the majority of calories consumed (Eshed and Lippman, 2019). Diversifying staple crops is thus another requirement for achieving a food supply that is sustainable, resilient, and suited to local environments (Massawe et al., 2016).
In many parts of the world, underutilized, orphan, and neglected crops are cultivated in marginal lands (Massawe et al., 2016). Among these crops is quinoa (Chenopodium quinoa Willd.), an annual protein-rich pseudocereal with excellent nutritional properties and an ability to tolerate the stressful environments typical of the Andean region of South America. The seeds and leaves of quinoa contain a wide variety of minerals, vitamins, fats, dietary fiber, natural antioxidants, and high-quality proteins composed of high levels of essential amino acids (Vega-Gálvez et al., 2010; Nowak et al., 2016; Pathan et al., 2019; Rodríguez et al., 2020). Furthermore, quinoa can tolerate a wide spectrum of abiotic stresses such as drought, high salinity, frost, and low temperatures, making it a highly sustainable crop (Jacobsen et al., 2005; Hariadi et al., 2011; Yasui et al., 2016; Mizuno et al., 2020).
Quinoa was first domesticated at least 7,500 years ago (Dillehay et al., 2007). Although quinoa was a staple food for the indigenous people of the Andes in the Pre-Columbian era (González et al., 2015), the Spanish conquistadors banned quinoa production and consumption in the sixteenth century since it was revered as a sacred “mother grain” used in indigenous rituals (González et al., 2015). Unlike tomato (Solanum lycopersicum L.) and potato (Solanum tuberosum L.), which are also native to the Andes but spread throughout the world during the Spanish conquest, quinoa remained a neglected crop for almost 500 years, until the latter half of the twentieth century (Gomez-Pando, 2015). In the 1950s, plant virologists began to recognize quinoa as an important indicator plant for plant viruses (Uschdraweit, 1955; Hein, 1957). In the 1970s and 1980s, the National Academy of Sciences of the USA (NAS) and the US National Research Council (NRC) deemed quinoa as an underutilized plant with promising economic value for worldwide cultivation (National Academy of Sciences (NAS), 1975; National Research Council (NRC), 1989). In the 1990s, quinoa was considered as an important candidate crop to be grown as part of the USA’s National Aeronautics and Space Administration (NASA) Controlled Ecological Life Support System (CELSS) for long-term space missions due to its high productivity and desirable nutritional and growth features under controlled environments (Schlick and Bubenheim, 1993, 1996). In the twenty-first century, with the aim of raising public awareness of the nutritional benefits of this durable plant in the context of global food and nutrition security, the United Nations (UN) General Assembly designated 2013 as the “International Year of Quinoa,” in recognition of the ancestral practices of the Andean people, who have managed to preserve quinoa in its natural state as food for present and future generations (Bazile et al., 2016). Quinoa popularity is on the rise and is considered an ideal food source in many countries (Vega-Gálvez et al., 2010). However, while quinoa cultivation is currently being attempted in more than 95 countries around the world, the vast majority of commercial quinoa is produced in Bolivia and Peru (Bazile et al., 2016).
In many countries where quinoa is grown, stem lodging, low tolerance to high temperature stress, adverse effects of prolonged photoperiods, and high susceptibility to pests and diseases have been recognized as major limiting factors preventing the global expansion of quinoa cultivation and effective large-scale cultivation (Schmöckel et al., 2017; Gomez-Pando et al., 2019). Nonetheless, the Altiplano is a major production area supplying the world with high-quality quinoa, despite obstacles such as drought (rainfall below 200∼500 mm per year), high altitude (3,800 meters above sea level), frost, hail, and strong wind, which are only intensifying due climate change (Bonifacio, 2019). Addressing challenges in quinoa production and thereby contributing to local and global food and nutrition security will rely on the ability to genetically improve this crop.
Quinoa is an allotetraploid (2n = 4x = 36) species with an estimated genome size of approximately 1.5 Gbp (Palomino et al., 2008; Yangquanwei et al., 2013). For many years, molecular analysis of quinoa had been limited by its genome complexity, derived from allotetraploidy and its genetic heterogeneity due to partial outcrossing resulting from the existence of both hermaphrodite and female flowers on the same plant (Maughan et al., 2004; Christensen et al., 2007). Recently, our collaborative group (Yasui et al., 2016) and two other independent groups (Jarvis et al., 2017; Zou et al., 2017) have sequenced the quinoa genome. Subsequently, we have developed more than 130 genotyped quinoa inbred lines suitable for molecular analyses and illustrated the genotype–phenotype relationships among the inbred lines with respect to salt tolerance and key growth traits (Mizuno et al., 2020). Our integrative genome analyses using single-nucleotide polymorphisms (SNPs) indicated that the quinoa lines are divided into three genetic sub-populations, namely, the northern and southern highland and the lowland sub-populations (Mizuno et al., 2020). Nonetheless, since gain- and loss-of-function techniques have not been developed for quinoa, the research community does not have the resources needed to analyze the functions of endogenous genes in planta or to conduct functional genomics research in this important crop.
Virus-induced gene silencing (VIGS) is a powerful tool for functional genomics in plants, especially in plant species that are recalcitrant to stable transformation (Baulcombe, 1999; Burch-Smith et al., 2004; Senthil-Kumar and Mysore, 2011; Wang et al., 2016). VIGS is a post-transcriptional gene silencing-based technique used to knockdown the expression of endogenous target genes (Baulcombe, 1999; Burch-Smith et al., 2004; Senthil-Kumar and Mysore, 2011). Although no plant virus vectors have been shown to mediate VIGS in quinoa so far, quinoa has been used as a propagation host for apple latent spherical virus (ALSV), which in turn has been used as a VIGS vector for a wide range of dicot plants (Li et al., 2000; Igarashi et al., 2009; Yamagishi and Yoshikawa, 2009; Ogata et al., 2017). ALSV has icosahedral particles that harbor two single-stranded RNA species, RNA1 and RNA2 (Li et al., 2000). The ALSV-RNA2 VIGS vector can be engineered to carry additional DNA fragments for VIGS analysis (Li et al., 2004; Igarashi et al., 2009).
Here, we demonstrate that two virus-mediated transient expression techniques, VIGS and virus-mediated overexpression (VOX), can be used in quinoa. We show that ALSV induces gene silencing of quinoa phytoene desaturase (CqPDS1) in a broad range of quinoa inbred lines derived from the northern and southern highland and lowland sub-populations, and that ALSV can be used as a VOX vector in quinoa roots. Furthermore, we demonstrate that silencing a quinoa 3,4-dihydroxyphenylalanine (DOPA) 4,5-dioxygenase gene (CqDODA1) or a cytochrome P450 enzyme gene (CqCYP76AD1) inhibits betalain production and that knockdown of a reduced-height gene homolog (CqRHT1) causes an overgrowth phenotype in quinoa. In addition, we show that ALSV VIGS can be transmitted to the next plant generation in quinoa. Our findings present a strategy for conducting functional genomics in quinoa, opening the door to deciphering the molecular mechanisms of this promising crop with exceptional nutritional value and adaptability to various environments.
Results
ALSV Induces Silencing of CqPDS1 in Quinoa
ALSV can infect quinoa systemically (Li et al., 2000) and thus has been used as a propagation host for the ALSV VIGS vector system (Igarashi et al., 2009). We therefore wondered whether ALSV could be used as a vector to analyze endogenous genes in quinoa. To test whether ALSV could induce endogenous gene silencing in quinoa plants, we first used the PDS gene as a visible VIGS indicator. PDS encodes a key enzyme in carotenoid biosynthesis and has been widely used as a reporter gene, as silencing of PDS induces a characteristic photobleaching phenotype that can readily be detected (Ruiz et al., 1998). Based on the sequence data of putative PDS genes in quinoa obtained from public databases, we designed PCR primers in the conserved regions and then detected and named homoeolog CqPDS1 genes, CqPDS1A and CqPDS1B, in a standard quinoa inbred line, Kd (Yasui et al., 2016). The cloned coding sequence (CDS) of CqPDS1A shared 98.5% identity with that of CqPDS1B (Supplementary Figure 1). To construct the ALSV vectors harboring a specific trigger sequence required for VIGS, we then introduced two 300-bp fragments of the CqPDS1 homoeologs, CqPDSN and CqPDSC, from the highly conserved regions between these two genes (Supplementary Figure 1), into the ALSV-RNA2 vector. We obtained inocula for VIGS analyses, including the packaged viruses of ALSV-CqPDSN, ALSV-CqPDSC, and ALSV-WT, from the uninoculated upper leaves with chlorotic spots of quinoa plants (inbred Iw line) inoculated with ALSV-RNA1 cDNA plasmid combined with ALSV-RNA2 VIGS vectors either harboring or not a specific 300-bp trigger sequence required for VIGS (Supplementary Figure 1).
Photobleaching phenotypes first appeared in the uninoculated upper leaves of quinoa Iw plants infected with ALSV-CqPDSN or ALSV-CqPDSC at approximately 10 days post inoculation (dpi). By 14 dpi, photobleaching phenotypes were apparent in all upper leaves and stems from the sixth leaves upwards of Iw plants inoculated with ALSV-CqPDSN or ALSV-CqPDSC (Figures 1A,B). Consistently, reverse transcription (RT)-PCR analysis confirmed systemic infection of ALSV and showed that endogenous CqPDS1 expression was reduced in the uninoculated upper leaves (Figure 1C). Reverse transcription quantitative PCR (RT-qPCR) analysis showed that CqPDS1 was expressed at less than 20% of the level expressed in plants inoculated with ALSV-WT (Figure 1D). By 8 weeks post inoculation (wpi), photobleaching was observed even in the flower buds of Iw plants inoculated with ALSV-CqPDSN (Figure 1E). Thus, the ALSV vector can induce endogenous gene silencing in quinoa Iw line plants through systemic infection of ALSV.
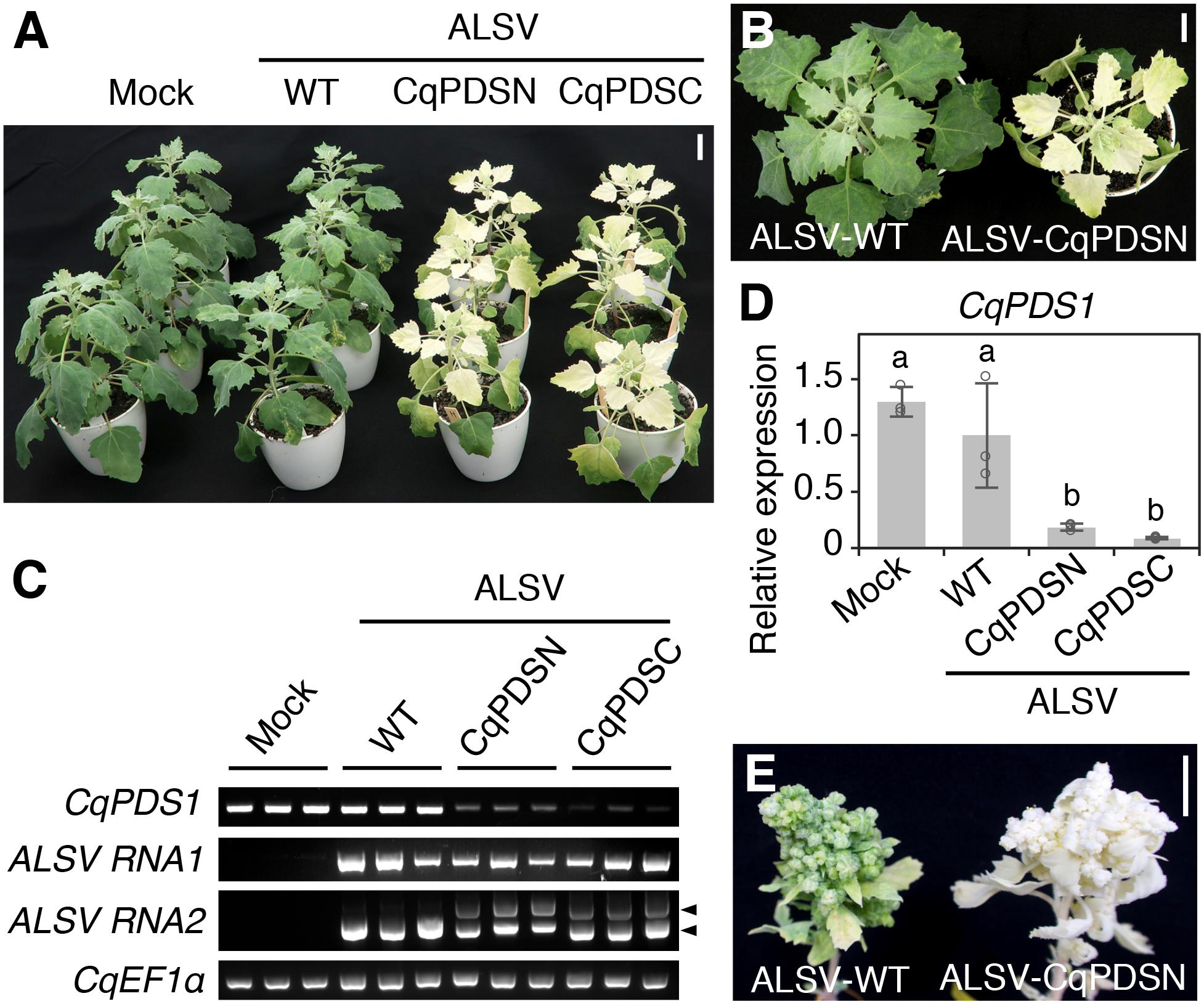
Figure 1. ALSV induces silencing of CqPDS1 genes in quinoa. (A) A representative image of plants (inbred Iw line) at 14 dpi with mock buffer, ALSV-WT, ALSV-CqPDSN, and ALSV-CqPDSC. Three plants were used per inoculum. Scale bar represents 2 cm. (B) Top view of representative plants at 14 dpi with ALSV-WT and ALSV-CqPDSN. Scale bar represents 2 cm. (C) Semi-quantitative RT-PCR of CqPDS1, ALSV RNA1, and RNA2 in the uninoculated upper leaves of plants at 14 dpi with mock, ALSV-WT, ALSV-CqPDSN, and ALSV-CqPDSC. Quinoa elongation factor 1α (CqEF1α) was used as an internal control. Arrowheads indicate the positions of recombinant ALSV RNA2 with the trigger sequence (top) and WT RNA2 without the trigger sequence (bottom). (D) RT-qPCR quantification of CqPDS1 transcripts in the uninoculated upper leaves of plants inoculated with the indicated inocula. Data were normalized to the 18S ribosomal RNA (18S rRNA) levels and are shown as means ± SD (n = 3). Different letters indicate significant differences by a Tukey’s HSD test (p < 0.05). (E) A representative image of flower buds of quinoa plants at 8 wpi with ALSV-WT and ALSV-CqPDSN. Scale bar represents 1 cm.
The ALSV Vector Is Suitable for Gene Function Analysis of a Variety of Quinoa Inbred Lines
Since quinoa line Iw, which we used in the VIGS of CqPDS1, belongs to the lowland sub-population, it was unclear whether ALSV could induce endogenous gene silencing in the other genotypes too. We thus tested whether ALSV-CqPDSC could induce silencing of CqPDS1 in 19 representative lines from the lowland (seven lines), southern highland (six lines), and northern highland (six lines) sub-populations (Table 1 and Figure 2A). Each representative line selected for this analysis contained 100% of the genetic background of each sub-population based on the results of our previous genotyping-by-sequencing (GBS) analysis of the inbred quinoa lines (Mizuno et al., 2020). For all 19 quinoa genotypes, entire photobleaching phenotypes were consistently observed by 16 dpi in the uninoculated upper leaves of quinoa plants inoculated with ALSV-CqPDSC (Figure 2B and Table 1). These findings suggest that ALSV induces gene silencing in a wide range of quinoa lines.
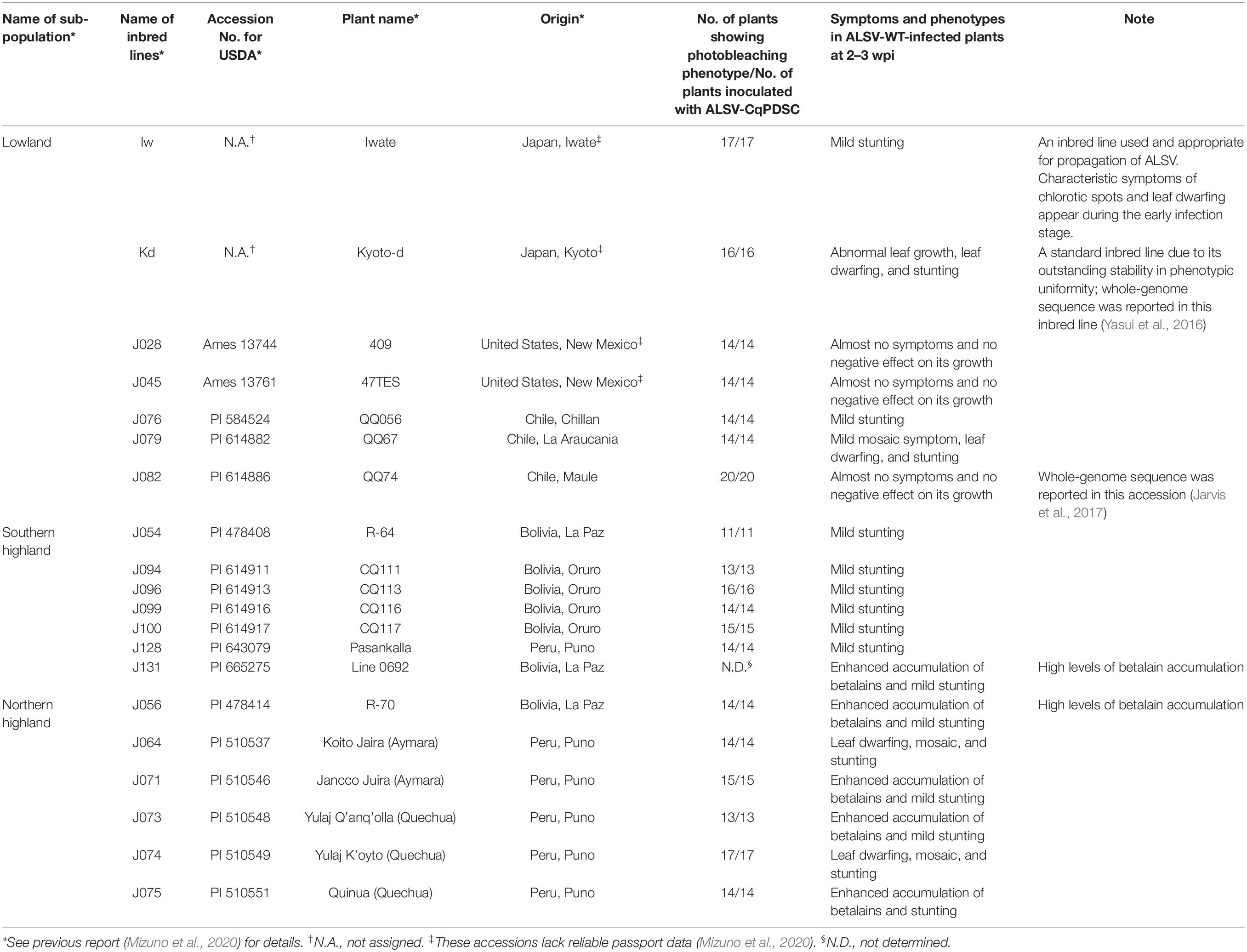
Table 1. List of quinoa inbred lines used in this study and summary of the occurrence of the photobleaching phenotype in ALSV-CqPDSC-infected plants.

Figure 2. The ALSV vector is suitable for gene function analysis in a variety of quinoa inbred lines. (A) Representative images of quinoa seeds of 19 representative inbred lines, including seven lowland, six southern, and six northern highland lines. Each representative inbred line selected in this analysis contains 100% of the genetic background of each sub-population based on the results of our previous GBS analysis of the inbred quinoa lines (Mizuno et al., 2020). Scale bar represents 5 mm. (B) Representative images of the 19 representative inbred line plants at 16 dpi with ALSV-CqPDSC. Scale bars represent 1 cm.
When analyzing the function of plant genes using VIGS, the effects of viral infection on the plant should be kept to a minimum. Therefore, we next inoculated the 19 representative quinoa lines with ALSV-WT and monitored the effect of ALSV infection on the quinoa plants. For most of the quinoa inbred lines examined, very mild mosaic symptoms were apparent in the uninoculated upper leaves of quinoa plants inoculated with ALSV-WT during the early stages of infection (at around 7–10 dpi) but the symptoms were nearly absent by 2–3 wpi (Table 1 and Supplementary Figures 2–4). ALSV RNA was detected in the uninoculated upper leaves of all of the quinoa inbred lines inoculated with ALSV-WT (Supplementary Figure 5), indicating that most of the genotypes have a latent ALSV infection. Three lowland inbred lines, J028, J045, and J082, displayed almost no symptoms, and showed no negative effect on their growth and plant size following inoculation with ALSV-WT (Table 1 and Supplementary Figure 2), suggesting that these three lines are well suited for ALSV VIGS analysis as they are not affected by ALSV infection. Mild stunted growth but no visible symptoms in the uninoculated upper leaves was observed in many quinoa plants of the northern and southern highland sub-populations inoculated with ALSV-WT (Table 1 and Supplementary Figures 3, 4). In addition, ALSV infection caused the red-violet color of the leaves and stems to become darker in the later stages of infection in four northern highland lines, J056, J071, J073, and J075 (Supplementary Figure 6A).
Unlike the other genotypes, the visible symptoms observed in J064, J074, J079, and Kd were sustained for several wpi (Table 1 and Supplementary Figures 2, 4). Particularly, ALSV infection induced stunting, leaf dwarfing, and visible mosaic symptoms in two northern highland inbred lines, J064 and J074 (Table 1 and Supplementary Figure 4), and also induced abnormal leaf growth and leaf dwarfing in the later stages of infection in Kd, a lowland inbred line (Table 1 and Supplementary Figure 2). However, considering that the induction of disease symptoms and VIGS in the leaves of quinoa plants inoculated with ALSV-WT and ALSV-CqPDSC, respectively, was accelerated and enhanced when plants were grown at 25°C compared to 22°C (Supplementary Figure 7), it may be possible to modulate the effects of ALSV infection by adjusting the growth conditions. Thus, these results demonstrate that the ALSV vector system is suitable for gene function analysis of a wide range of quinoa genotypes.
ALSV Can Be Used as a VOX Vector in Roots
The ALSV vector was used to express exogenous genes in quinoa leaves (Takahashi and Yoshikawa, 2008). To further expand the application of the ALSV vector in quinoa, we next examined whether ALSV could be used as a vector for virus-mediated overexpression (VOX) in roots as well as for VIGS in quinoa leaves. At 2 days post germination (dpg), the roots and hypocotyls of J082 plants, whose growth was not affected by ALSV infection during the vegetative stage, were inoculated with the packaged virus derived from recombinant ALSV harboring a full-length green fluorescence protein (GFP) CDS, using carborundum. GFP fluorescence and virus accumulation were observed in the roots of quinoa plants at 10 dpi (Supplementary Figure 8), indicating that ALSV can indeed be used as a VOX vector in roots. Hence, these collective results demonstrate that the ALSV could be used as a vector for both VIGS and VOX for gene function analysis in a whole quinoa plant.
Silencing of CqDODA1 and CqCYP76AD1 Inhibits Betalain Production in Quinoa
In plants, three major pigment classes, namely, carotenoids, anthocyanins, and betalains, are generally responsible for the attractive colors observed in fruits, flowers, and vegetative tissues (Tanaka et al., 2008). Most flowering plants produce anthocyanins, whereas functionally equivalent betalains are produced solely by Caryophyllales species such as quinoa (Stafford, 1994; Strack et al., 2003; Brockington et al., 2015). These two classes of pigments are known to be present in a mutually exclusive fashion (Stafford, 1994; Strack et al., 2003; Brockington et al., 2015). Although the biosynthesis of carotenoids and anthocyanins has been well characterized, betalain production and its regulation remain largely unknown (Polturak and Aharoni, 2018). Nevertheless, since betalains have been reported to have strong antioxidant and health-promoting properties, betalains are of scientific interest and may be of economic importance (Polturak and Aharoni, 2018).
Quinoa, beet (Beta vulgaris L.), and spinach (Spinacia oleracea L.) are Caryophyllales plant species that possess carotenoids and betalains in organs such as the leaves, stems, and seeds (Figure 2A and Supplementary Figures 2–4). To demonstrate that the ALSV VIGS vector induces silencing of genes in quinoa beyond those involved in carotenoid biosynthesis, we functionally analyzed genes involved in betalain biosynthesis using the ALSV-VIGS system. There are two structural groups of betalains: the red-violet betacyanins, and the yellow betaxanthins (Polturak and Aharoni, 2018). Currently, CYP76AD1 and DODA1 are considered to be involved in the key steps of betalain biosynthesis (Hatlestad et al., 2012; Polturak et al., 2016; Imamura et al., 2018; Sheehan et al., 2020). Although CqCYP76AD1 was previously implicated in betacyanin biosynthesis and shown to contribute to hypocotyl pigmentation (Imamura et al., 2018), it remains unknown whether CqCYP76AD1 and CqDODA1 are involved in betalain biosynthesis in whole quinoa plants.
To examine the possible roles of CqCYP76AD1 and CqDODA1 in the betalain biosynthesis pathway, VIGS was used to suppress gene expression in the quinoa inbred lines J056 and J131, which produce large amounts of betalains that can easily be seen under the experimental conditions (Figure 3 and Supplementary Figure 6). RT-qPCR analysis showed that the targeted genes were strongly silenced in the uninoculated upper leaves of both lines (Figure 3A). Silencing of CqCYP76AD1 resulted in the reduction of red-violet batalain pigments in the uninoculated upper leaves and stems of both inbred lines (Figures 3B–D), whereas silencing of CqDODA1 resulted in an almost complete loss of red-violet betalain pigments in the uninoculated upper leaves and stems in both lines (Figures 3B–D). By contrast, silencing of CqPDS1 resulted in photobleaching phenotypes in the uninoculated upper leaves of both lines and we were thus able to confirm the accumulation of red-violet batalain pigments in the leaves (Figure 3C). The observed changes in pigment phenotype in CqDODA1- or CqCYP76AD1-silenced J056 and J131 lines were verified through spectrophotometric measurements showing the reduction of red-violet betacyanins in the uninoculated upper leaves (Figure 3E) and high-performance liquid chromatography (HPLC) analysis indicating the reduction of betacyanins composed of amaranthin, celosianin II, and an unidentified compound (Imamura et al., 2019; Figure 3F). These observations demonstrate that CqCYP76AD1 and CqDODA1 are required for the production of betacyanin pigments in the leaves and stems of the inbred lines J056 and J131, suggesting that these genes function in key steps of betalain biosynthesis in whole quinoa plants. Notably, we also established that ALSV-WT infection significantly upregulated the expression of at least CqDODA1 in the inbred lines J056 and J131 (Figure 3A). This finding is consistent with the observation that ALSV-WT infection enhanced betalain accumulation in the leaves and stems of some northern highland lines such as J056, J071, J073, and J075, and the southern highland line J131 (Figures 3B–F and Supplementary Figure 6). These results suggest that ALSV infection induces defense responses, including the promotion of betalain accumulation, in these inbred lines.
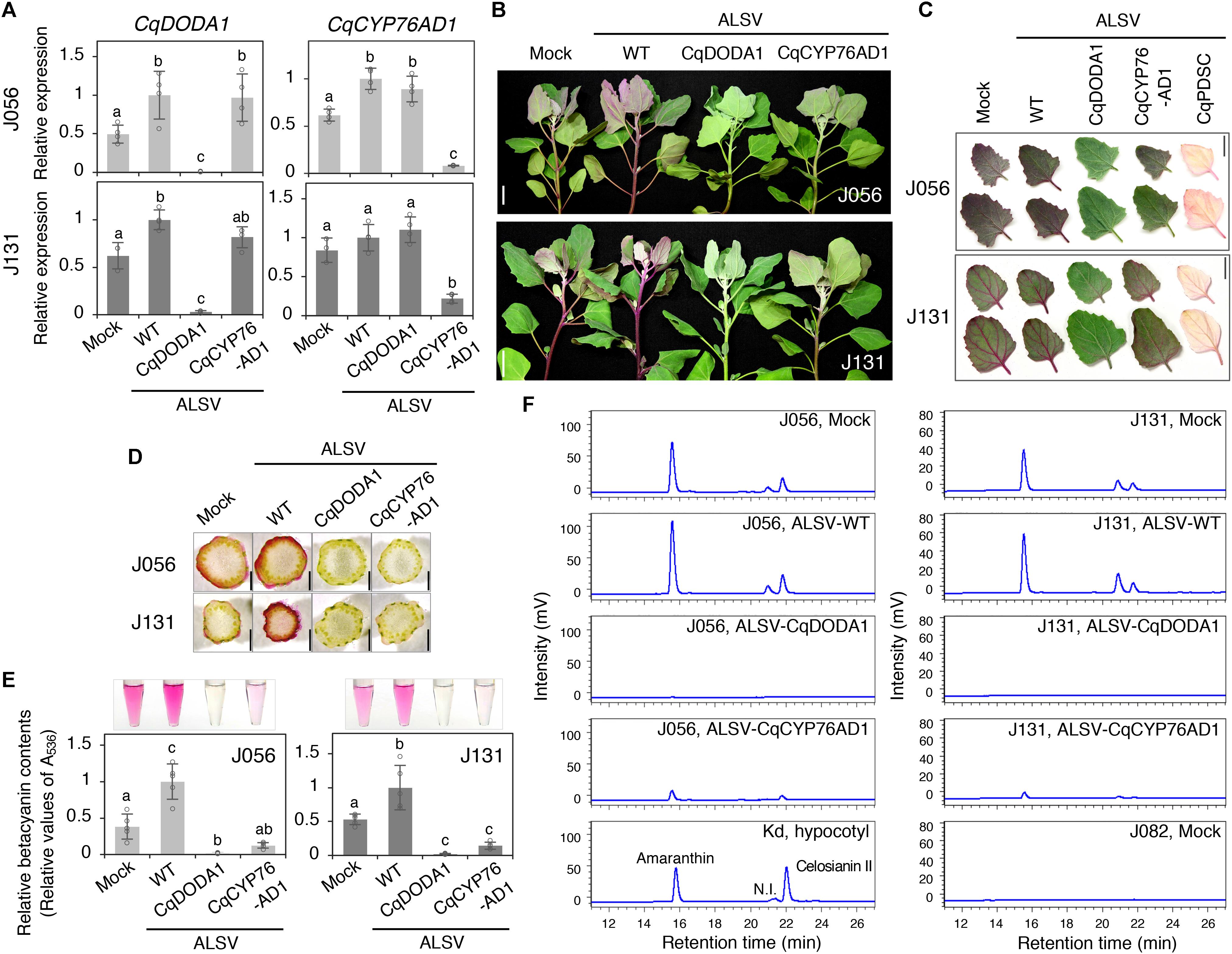
Figure 3. Silencing of CqDODA1 and CqCYP76AD1 inhibits betalain production in quinoa. (A) RT-qPCR analysis of CqDODA1 and CqCYP76AD1 transcripts in the uninoculated upper leaves of the inbred lines J056 and J131 inoculated with the indicated inocula. Data were normalized to quinoa polyubiquitin 10 (CqUBQ10) expression and are shown as means ± SD (n = 4). Different letters indicate significant differences by a Tukey’s HSD test (p < 0.05). (B) Representative images of quinoa plants at 28 dpi with mock buffer, ALSV-WT, ALSV-CqDODA1, and ALSV-CqCYP76AD1. Scale bars represent 2 cm. (C) Representative images of the uninoculated upper leaves of quinoa plants at 20 dpi with the indicated inocula. Scale bars represent 2 cm. (D) Representative images of stem cross-sections of plants at 28 dpi with the indicated inocula. Scale bars represent 1 mm. (E) Relative quantification of betacyanin content was assessed spectrophotometrically by light absorption measurements at 536 nm. Relative values are presented as means ± SD (n = 5). Betacyanin pigments were extracted from the uninoculated upper leaves of quinoa plants (inbred J056 and J131 lines) at 16 dpi inoculated with the indicated inocula. Representative images of aqueous extracts used for the measurements are shown. (F) High performance liquid chromatography analysis of betacyanin pigments in the aqueous extracts used in (E). The peak names of quinoa betacyanins composed of amaranthin, celosianin II, and a non-identified (N.I.) compound were identified based on the analysis of red-violet pigment extracted from hypocotyls of 14-day-old Kd plants (Imamura et al., 2019). No betacyanin peaks were detected in the uninoculated upper leaves of J082 inbred line plants inoculated with mock. The horizontal and vertical axes indicate the retention time (min) and signal intensity (mV), respectively.
Knockdown of CqRHT1 Causes an Overgrowth Phenotype in Quinoa
Plant morphology and architecture have been targeted as important agronomic traits for high productivity in crops (Boden and Østergaard, 2019; Eshed and Lippman, 2019). To illustrate that the ALSV VIGS vector can be used for functional genomics in quinoa beyond the genes involved in the production of secondary metabolites such as pigments, we examined the function of a height-regulating gene using the ALSV system. Height-regulating GAI/RGA/RHT/D8 genes, whose mutations enabled the “Green Revolution” in wheat (Triticum aestivum L.), have been identified as negative regulators of gibberellin (GA) signaling and belong to the GRAS family of transcriptional regulators with DELLA domains (Peng et al., 1999; Ikeda et al., 2001; Olszewski et al., 2002; Hedden, 2003).
Therefore, we sought to identify the GAI/RGA/RHT/D8 genes in quinoa. We obtained sequence data of quinoa orthologs of GAI/RGA/RHT/D8 genes, named CqRHT1, from public databases, and designed PCR primers based on the conserved regions for cloning the genes. We identified two CDS of CqRHT1 homoeologs, CqRHT1A and CqRHT1B (Supplementary Figure 9), in the standard quinoa inbred line Kd (Yasui et al., 2016). The cloned CDS sequence of CqRHT1A shared 97.9% identity with that of CqRHT1B (Supplementary Figure 9). A 300-bp fragment of the homoeolog CqRHT1 in the highly conserved regions between these two genes was introduced into the ALSV-RNA2 vector to construct a ALSV RNA2 VIGS vector. We obtained inocula for VIGS analyses, including the packaged viruses of ALSV-CqRHT1, from the uninoculated upper leaves with chlorotic spots of quinoa plants (Iw line) inoculated with ALSV-RNA1 cDNA plasmid combined with ALSV-RNA2 VIGS vectors either harboring or not a specific 300-bp trigger sequence required for VIGS. To test whether CqRHT1 influences plant morphology or architecture, we suppressed the expression of this gene using VIGS in the quinoa inbred line J082, which shows almost no symptoms and exhibited no negative effect on growth and plant size following inoculation with ALSV-WT (Table 1 and Supplementary Figure 2).
RT-qPCR analysis showed that CqRHT1 expression was significantly down-regulated in plants inoculated with ALSV-CqRHT1 compared with those inoculated with ALSV-WT (Figure 4A). Knockdown of CqRHT1 significantly enhanced plant height, stem diameter, and petiole length, indicating that CqRHT1 knockdown causes an overgrowth phenotype in quinoa (Figure 4 and Supplementary Figure 10). The enhanced plant height phenotype of plants inoculated with ALSV-CqRHT1 was due to the enhanced internode length rather than an increase in node number (Figures 4K,L and Supplementary Figures 10A,B). CqRHT1 knockdown resulted in an abnormal inflorescence size and architecture with elongated branches (Figures 4E–G). These findings are in accordance with the previous report that loss-of-function mutations in GAI/RGA/RHT/D8 genes caused an overgrowth phenotype in rice (Oryza sativa L.) and barley (Hordeum vulgare L.) (Ikeda et al., 2001; Chandler et al., 2002).
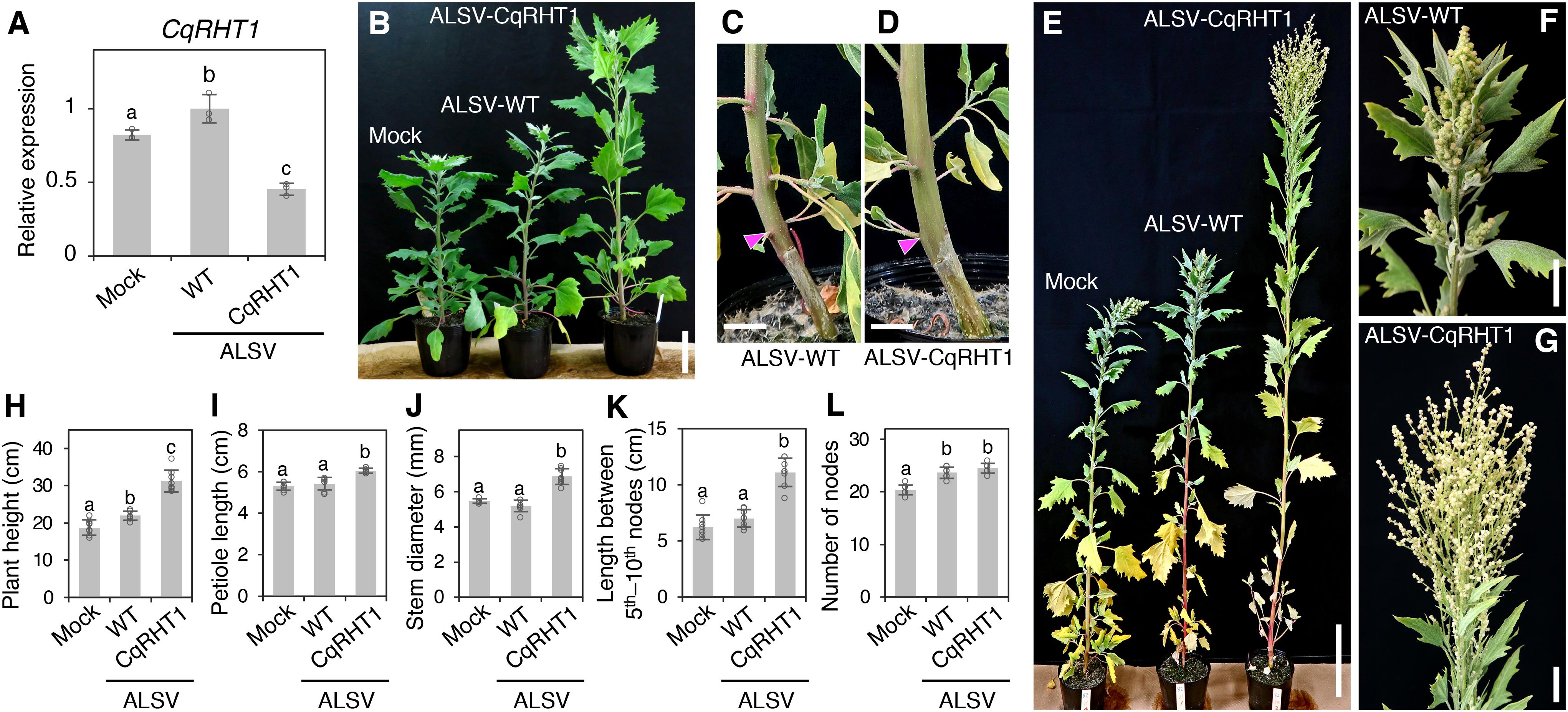
Figure 4. Knockdown of CqRHT1 causes an overgrowth phenotype in quinoa. (A) RT-qPCR quantification of CqRHT1 transcripts in the uninoculated upper leaves of the quinoa inbred line J082 inoculated with mock buffer, ALSV-WT, and ALSV-CqRHT1. Data were normalized to CqUBQ10 expression and are shown as means ± SD (n = 3). (B–G) Representative images of plants inoculated with mock buffer, ALSV-WT, and ALSV-CqRHT1 at 21 (B), 28 (C,D), and 37 (E–G) dpi. Whole plants (B,E), stem (C,D), and inflorescence (F,G) are shown. Arrowheads in (C) and (D) indicate the position at the first node. Scale bars represent 5 cm (B), 1 cm (C,D), 10 cm (E), or 2 cm (F,G). (H–L) Statistical analysis of the growth of quinoa plants inoculated with mock buffer, ALSV-WT, and ALSV-CqRHT1. Plant height (H), petiole length of the eighth leaf (I), stem diameter at the first node (J), internode length between the 5th and 10th nodes (K), and number of nodes (L) were measured at 20 dpi. The data are presented as means ± SD (n = 8). Different letters in (A,H–L) indicate significant differences by a Tukey’s HSD test (p < 0.05).
Furthermore, the CqRHT1-knockdown plants infected with ALSV-CqRHT1 showed enhanced cell division and elongation in the stem in comparison with the control plants infected with ALSV-WT (Supplementary Figures 10C–J). These results suggest that CqRHT1 functions as a negative regulator of GA signaling in quinoa. In support of this, we confirmed that putative GA-responsive genes were up-regulated in the quinoa plants inoculated with ALSV-CqRHT1 in comparison with the plants inoculated with ALSV-WT (Supplementary Figure 10K). Taken together, these observations show that ALSV VIGS vectors can be used to alter plant morphology and architecture, and demonstrate that CqRHT1 genes play a crucial role in controlling plant morphology and architecture through GA signaling in quinoa.
ALSV VIGS Can Be Transmitted to the Progeny Plants Through Seeds at a Low Frequency
As ALSV can be transmitted to progeny plants through seeds from infected apple (Malus domestica Borkh.), Nicotiana benthamiana L., and soybean (Glycine max L. Merr.) plants (Yamagishi and Yoshikawa, 2009; Nakamura et al., 2011; Kishigami et al., 2014; Kamada et al., 2018), we next examined whether ALSV VIGS was inherited by the progeny of infected quinoa plants. In the Iw line, 0% (0/169) and 3.7% (17/449) of the progeny from the quinoa plants inoculated with ALSV-CqPDSN and ALSV-CqPDSC, respectively, displayed photobleaching in cotyledons and thus retained silencing (Figure 5A and Table 2). RT-PCR analysis confirmed that viral RNAs were present in the seedlings with white cotyledons but not in those with green cotyledons (Figure 5B). We also found that 1.7% (2/120) and 0% (0/120) of progeny from the quinoa plants inoculated with ALSV-CqDODA1 did not exhibit betalain accumulation in whole plants and thus retained silencing in the inbred lines, J056 and J131, respectively (Figures 5C–E and Table 2). Viral RNAs were detected in the seedlings that lacked betalain accumulation but not in those that accumulated betalain (Figure 5F). Thus, the seed transmission rates of VIGS were very low (less than 5%). By contrast, 12.1% (19/176) of the progeny of the Iw plants inoculated with ALSV-WT showed characteristic chlorotic spots in the leaves (Figure 5G and Table 2). These observations suggest that the seed transmission rate of ALSV in quinoa depends on whether or not a trigger sequence has been inserted into the VIGS vector and on the genotype of quinoa used.

Figure 5. ALSV VIGS can be transmitted to progeny plants through seeds at a low frequency. (A) A representative image of progeny seedlings with the photobleaching phenotype at 7 dpg, derived from plants inoculated with ALSV-CqPDSC in the inbred Iw line. Arrowheads indicate progeny seedlings with the photobleaching phenotype. (B) Semi-quantitative RT-PCR of ALSV RNA1 and RNA2 in the progeny seedlings with (white) or without (green) the photobleaching phenotype shown in (A). CqEF1α was used as an internal control. Arrowheads indicate the positions of recombinant ALSV RNA2 with the trigger sequence (top) and WT RNA2 without the trigger sequence (bottom). (C) An arrowhead indicates a progeny seedling that has not accumulated betalain in the hypocotyl at 14 dpg, derived from J056 inbred line plants inoculated with ALSV-CqDODA1. (D,E) Representative images of a plant with a green hypocotyl (D) or red hypocotyl (E) at 4 wpg, derived from J056 inbred line plants inoculated with ALSV-CqDODA1. (F) Semi-quantitative RT-PCR of ALSV RNA1 and RNA2 in progeny seedlings with a green or red hypocotyl shown in (D,E). CqUBQ10 was used as an internal control. Arrowheads indicate positions of recombinant ALSV RNA2 with the trigger sequence (top) and WT RNA2 without the trigger sequence (bottom). (G) A representative image of a progeny plant with chlorotic spots in leaves at 20 dpg, derived from the Iw inbred line plants inoculated with ALSV-WT. Arrowheads point to chlorotic spots in leaves in progeny seedlings with ALSV-WT. Scale bars represent 1 cm.
Discussion
Here, we show that ALSV-mediated transient expression techniques enable gene function studies in a wide variety of quinoa lines spanning all three sub-populations and can be used to alter the production of secondary metabolites such as pigments, carotenoids, and betalains, and also the plant morphology and architecture of quinoa. Although quinoa gene functions have been evaluated by the ectopic transient overexpression of exogenous genes in quinoa hairy roots, Nicotiana benthamiana leaves, and tobacco BY-2 suspension cells (Imamura et al., 2018, 2019, 2020), our findings presented here provide the quinoa research community with the gain- and loss-of-function tools needed to analyze the functions of endogenous genes in planta and enable functional genomics in quinoa.
Our results illustrate that ALSV VOX as well as VIGS can be used in quinoa (Supplementary Figure 8). However, since viruses with icosahedral virions, including ALSV, allow just small inserts due to size constraints for virus genome packaging (Abrahamian et al., 2020), ALSV can only be used to overexpress relatively small proteins such as FLOWERING LOCUS T and GFP (Takahashi and Yoshikawa, 2008; Yamagishi et al., 2014). There is thus a growing need to develop transformation and genome editing techniques that allow for the dissection of gene function in quinoa. We were intrigued to note that ALSV could be transmitted to the progeny plants via seeds from infected quinoa plants, although the transmission efficiency was low (0–10%), depending on the presence or absence of the insertion sequence and other factors such as quinoa lines and insertion sequences (Figure 5 and Table 2). The mechanism by which ALSV is able to enter the quinoa meristem is unclear, but this property could be exploited in future studies. Although this method is not normally applicable to the analysis of young seedlings, it is possible to analyze young seedlings in which a specific gene is knocked down, by using the progeny plants to which ALSV has been transmitted (Figure 5). Since growth was affected to a lesser extent in CqDODA1-silenced seedlings than in CqPDS1-silenced seedlings (Figure 5), CqDODA1-silenced plants could serve as a useful visible marker to evaluate the efficiency with which ALSV VIGS is transmitted to progeny plants under different experimental conditions.
Although betalains have attracted attention in recent years as plant pigments with promising health-promoting potential and visible fluorescence (Gandía-Herrero et al., 2016; Guerrero-Rubio et al., 2020), research on quinoa betalains has only recently begun. A previous mutant analysis indicated that CqCYP76AD1 functions in betalain accumulation during the hypocotyl pigmentation process in quinoa (Imamura et al., 2018). Our VIGS results clearly extend the previous findings and show that CqDODA1 and CqCYP76AD1 play crucial roles in the production of betalain pigments accumulated in leaves and stems in quinoa (Figure 3). These findings support the notion that CqDODA1 and CqCYP76AD1 participate in key steps of betalain synthesis in whole quinoa plants. Recently, the function of quinoa amaranthin synthetase 1 (CqAMASY1), which is involved in the synthesis of amaranthin, a major component of quinoa red-violet betacyanins, has been evaluated using an overexpression approach in tobacco BY-2 cultured cells (Imamura et al., 2019). ALSV VIGS would be useful to establish whether CqAMASY1 and the other key genes involved in betalain synthesis function in allotetraploid quinoa as well. Plants of the Caryophyllales order, in which betalains occur in a mutually exclusive manner with anthocyanins (Polturak and Aharoni, 2018), are non-model plants and thus are not amenable to transformation in most cases. Therefore, viral vectors such as ALSV VIGS would be useful tools to decipher the biosynthesis pathways and evolution of betalain in Caryophyllales species.
The role of betalains in biotic stress responses in plants is poorly understood (Polturak and Aharoni, 2018). Our observations indicated that ALSV infection promotes betalain accumulation in quinoa (Figures 3B–F and Supplementary Figure 6), coupled with enhanced expression of betalain-related genes (Figure 3A). Based on previous reports of increased betalain synthesis induced by fungal and bacterial infections (Sepúlveda-Jiménez et al., 2004; Polturak et al., 2017), it is possible that the increase in betalain synthesis caused by ALSV infection may involve increased radical scavenging activity. The role of betalains in abiotic stress responses in quinoa is also not understood, even though quinoa is well adapted to harsh environments. A striking feature of the order Caryophyllales, to which quinoa belongs, is that its members are predominantly distributed in arid and semi-arid regions and in saline and alkaline soils (Polturak and Aharoni, 2018). The activity of betalains in defense against abiotic stress is likely mediated via their potent antioxidant capacity (Jain and Gould, 2015). With these considerations in mind, the role of betalains in abiotic and biotic stress responses in quinoa merits further investigation.
We show that CqRHT1 knockdown causes an overgrowth phenotype in quinoa, characterized by increased plant and inflorescence height, stem diameter, and internode length (Figure 4 and Supplementary Figure 10), demonstrating that the ALSV VIGS vector can be used to alter the plant morphology and architecture of quinoa. Considering that mutations in the flower-promoting protein florigen and its antagonist antiflorigen or in the growth stimulating phytohormone GA-DELLA system could be used to reduce plant height and extend the geographical distribution in quinoa (Eshed and Lippman, 2019), the ALSV vector could contribute to research aimed at radically improving the productivity of quinoa. Indeed, in recent years, quinoa has been used as a model plant to explore the molecular mechanisms underlying abiotic stress tolerance in plants. Salt tolerance has been extensively studied with respect to the roles and functions of epidermal bladder cells (Shabala et al., 2014; Kiani-Pouya et al., 2017, 2019, 2020; Zou et al., 2017; Böhm et al., 2018), xylem ion loading (Zarei et al., 2020), tonoplast channels (Bonales-Alatorre et al., 2013), and inorganic ions for osmotic adjustment (Hariadi et al., 2011). Unlike rice and maize (Zea mays L.), which require high-light conditions to keep plants healthy, quinoa plants grow well in the growth chambers normally used for the model plant Arabidopsis. Therefore, plant researchers could easily shift their focus from the weedy model plant Arabidopsis to the new model plant quinoa, which has great practical value, and utilize the research resources that are rapidly being developed for this species (Yasui et al., 2016; Jarvis et al., 2017; Zou et al., 2017; Mizuno et al., 2020).
In conclusion, we have demonstrated that the ALSV vector for VIGS and VOX can be used for gene function analyses in a wide range of quinoa inbred lines from all three sub-populations. We further showed that silencing of CqDODA1 and CqCYP76AD1 inhibits betalain biosynthesis, that knockdown of CqRHT1 results in an overgrowth phenotype in quinoa, and that ALSV is able to be transmitted to the progeny plants in quinoa. Thus, our findings enable functional genomics in quinoa, facilitating exploration of the molecular mechanisms of this mysterious plant with great environmental adaptability and outstanding nutritional profiles. Further, these techniques could be expected to contribute to the promotion of molecular breeding in quinoa.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of quinoa (Chenopodium quinoa Willd.) inbred lines were generated previously (Yasui et al., 2016; Mizuno et al., 2020) and are listed in Table 1. The inbred line Kd is more stable in terms of phenotypic uniformity than the other lines, indicating that Kd is a suitable standard inbred line for molecular genetics and analyses (Yasui et al., 2016). The Iw line is used for propagation of ALSV (Mizuno et al., 2020). Quinoa plants were grown in soil in pots as described previously (Yasui et al., 2016; Mizuno et al., 2020), with minor modifications. Quinoa seeds were sown in a peat moss mixture (Jiffy Mix, Sakata Seeds, Yokohama, Japan) in a cell tray and were allowed to germinate in a growth chamber (Biotron LH-350S/410S: Nippon Medical & Chemical Instruments, Osaka, Japan) under controlled conditions of a 16-h light/8-h dark photoperiod, 22°C day/20°C night cycles, and a light intensity of 150 μmol photons m–2 s–1. After 7 days, the seedlings were transferred to a standard potting mix (Tsuchitaro, Sumitomo Forestry, Tokyo, Japan) in pots.
Total RNA Isolation and RT-PCR Analysis
Total RNA isolation and complementary DNA (cDNA) synthesis were conducted essentially as described previously (Ogata et al., 2017). Total RNA was isolated from the uninoculated upper leaves of quinoa using RNAiso Plus (Takara Bio, Otsu, Shiga, Japan) according to the manufacturer’s instructions. Total RNA was pre-treated with RQ1 RNase-free DNase (Promega, Madison, WI, United States) and cDNA was synthesized using PrimeScript RT Master Mix (Takara Bio). Reverse transcription quantitative PCR (RT-qPCR) was performed using the QuantStudio 7 Flex real-time PCR system (Thermo Fisher Scientific, MA, United States) and TB Green Premix Ex Taq II (Takara Bio). Semiquantitative RT-PCR was performed using GoTaq Green Master Mix (Promega). The specific oligonucleotide primers used are listed in Supplementary Table 1.
Molecular Cloning and Plasmid Construction
Based on the CDS and genome sequence for putative CqPDS1 and CqRHT1 obtained from public databases such as Quinoa Genome DataBase1 and Phytozome database2, using the amino acid sequences of Arabidopsis PDS (At4g14210) and wheat Rht-A1a (KC767924) as queries in the BLASTP program, we designed PCR primers (Supplementary Table 1) to determine the full-length CDS of the quinoa homologs in the standard quinoa inbred line Kd (Yasui et al., 2016). RT-PCR was performed using cDNA prepared from the inbred line Kd, amplified DNA fragments were inserted into the pGEM-T/Easy vector (Promega), and the multiple cloned sequences were determined by Sanger sequencing. We thus identified the homoeolog CqPDS1 genes, CqPDS1A and CqPDS1B, and the homoeolog CqRHT1 genes, CqRHT1A and CqRHT1B, in the inbred Kd line. The sequence data were deposited in the DDBJ/EMBL/GenBank database under the following accession numbers: CqPDS1A (LC591855); CqPDS1B (LC591856); CqRHT1A (LC591857); and CqRHT1B (LC591858). We designed PCR primers (Supplementary Table 1) to amplify trigger regions for VIGS (Supplementary Table 2) in the determined CDSs.
In contrast to CqPDS1 and CqRHT1, multiple homologs of CqDODA1 and CqCYP76AD1 were identified in quinoa. We thus designed PCR primers that amplified trigger regions instead of sequencing the full-length CqDODA1 and CqCYP76AD1 genes. Based on the CDS of putative CqDODA1 and CqCYP76AD1 obtained from the public databases using Mirabilis jalapa DODA (AB435372) and Beta vulgaris CYP76AD1 (HQ656023) amino acid sequences as queries in the BLASTP program, we designed PCR primers (Supplementary Table 1) to amplify trigger regions for VIGS. RT-PCR was performed using cDNA from the Kd line and the amplified fragments were cloned into pEALSR2L5R5 (ALSV-RNA2 plasmid), which is the same as pER2L5R5XSB (Li et al., 2004; Supplementary Table 3). The cloned sequences were determined by Sanger sequencing (Supplementary Table 2). The cloned trigger sequences had 93% identity to CqCYP76AD1-1 (XM_021913610) and 95.8% identity to CqCYP76AD1-2 (XM_021876908), which were reported previously (Imamura et al., 2018).
ALSV-RNA2 vectors for VIGS analyses were constructed as described previously (Ogata et al., 2017), with minor modifications. The amplified DNA fragments containing trigger sequences of quinoa genes (Supplementary Table 2) for VIGS were cloned in-frame into the XhoI/BamHI site of pEALSR2L5R5, generating pEALSR2-CqPDSN, pEALSR2-CqPDSC, pEALSR2-CqDODA1, pEALSR2-CqCYP76AD1, and pEALSR2-CqRHT1, respectively. To construct the ALSV-RNA2 vector for VOX analyses, pGH-35S-sGFP (Fujita et al., 2009) was digested with SmaI/EcoRV and the CDS of the GFP fragment was cloned into the SmaI site of pEALSR2L5R5, generating pEALSR2-sGFP. The resulting viruses derived from the ALSV-RNA2 constructs were designated ALSV-CqPDSN, ALSV-CqPDSC, ALSV-CqDODA1, ALSV-CqCYP76AD1, ALSV-CqRHT1, and ALSV-GFP-OE, respectively.
Virus Inoculation
Quinoa plants were inoculated with ALSV as described previously (Ogata et al., 2017), with minor modifications. The plasmid DNAs (1–1.5 μg/μL) for the ALSV-RNA1 (pEALSR1) and each ALSV-RNA2 construct were mixed in equal amounts, and the DNA solution (5–10 μL) was mechanically rub-inoculated onto the true leaves of 12-day-old quinoa plants (inbred Iw line) using 600-mesh carborundum (Nakarai Tesque, Kyoto, Japan). Reverse osmosis (RO) water was used for mock inoculation. The inoculated quinoa plants were grown for 2–3 weeks and the uninoculated upper leaves showing chlorotic spots symptoms were harvested. The harvested leaves were grounded in extraction buffer (0.1 M Tris-HCl, pH 8.0, 0.1 M NaCl, 5 mM MgCl2) (Igarashi et al., 2009). Debris was precipitated by centrifugation at 18,800 g for 10 min at 4°C, and the supernatants were used as inocula for all quinoa inbred lines. The infected leaves or the inocula in extraction buffer were stored at –80°C before use. Seedlings (11–13 days old) of quinoa inbred lines of interest were mechanically rub-inoculated with the inocula (5–10 μL) using carborundum. ALSV derived from quinoa Iw plants inoculated with pEALSR1 and pEALSR2L5R5 was used as a control (ALSV-WT).
Pigment Chemical Analysis
Betalain pigments were extracted at 16 dpi from the uninoculated upper leaves of quinoa plants inoculated with mock buffer, ALSV-WT, ALSV-CqCYP76AD1, and ALSV-CqDODA1. Leaf samples were frozen in liquid nitrogen and then stored at –80°C before use. The samples were powdered using a ShakeMaster (BMS-A20TP, BioMedical Science, Tokyo, Japan) and extracted in 0.1% (w/v) ascorbic acid (Hatlestad et al., 2015) at a ratio of 0.1 g of fresh leaf per 1 mL buffer at 4°C overnight. After centrifugation at 18,800 g for 10 min at 4°C, the extracts were filtered through a 0.45 μm membrane filter (Shimadzu, Kyoto, Japan).
The relative betacyanin contents were estimated spectrophotometrically based on absorbance at 536 nm (Imamura et al., 2019; Guerrero-Rubio et al., 2020). UV–Vis spectra were acquired using a DeNovix DS-11 (DeNovix, Wilmington DE, United States) spectrophotometer.
HPLC separation of betalain pigments was performed essentially as described previously (Imamura et al., 2019). A JASCO HPLC system (Jasco, Tokyo, Japan) equipped with a pump (PU-4185-Binary), an auto-sampler (AS-4150), a column oven (CO-4060), and a photodiode array detector (MD-4010) was used for analytical HPLC separations. Samples were separated on a C18 column (4.6 × 250 mm, Wakopak Handy ODS, Fujifilm Wako Pure Chemicals, Osaka, Japan), and linear gradients were run from 0% B to 45% B over a 45 min period using 0.05% trifluoroacetic acid (TFA) in water (solvent A) and 0.05% TFA in acetonitrile (solvent B) at a flow rate of 0.5 mL/min at 25°C, with elution being monitored by absorbance at 536 nm.
VOX Analysis
Seeds of quinoa J082 plants, whose growth was not affected by ALSV infection, were surface-sterilized for 5 min in 1% (v/v) sodium hypochlorite solution and germinated on half-strength Murashige and Skoog (MS) salt medium supplemented with 1% (w/v) sucrose and 0.8% (w/v) agar at 22°C in the dark. At 2 dpg, the roots and hypocotyls of quinoa J082 plants were mechanically rub-inoculated with the packaged virus derived from recombinant ALSV harboring a full-length GFP CDS, using 600-mesh carborundum. The inoculated seedlings were incubated on MS agar plate at 22°C in the dark. GFP fluorescence was observed in the roots of quinoa plants at 10 dpi using a fluorescence stereoscopic microscope (SMZ25, Nikon, Tokyo, Japan) equipped with a dedicated digital camera DS-Fi3.
Statistical Analysis
One-way ANOVA with Tukey’s HSD tests were performed using R software version 3.6.3 (R Core Team, 2020).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
HM, TT, YY, and YF conceived a series of quinoa studies. NY conceived ALSV studies and provided the virus materials. TO designed and performed the most of experiments. MT produced and managed quinoa inbred lines and prepared the seed samples. CY-O and YK performed the HPLC experiments. KF assisted expression analysis. KT and YY performed database search. TO and YF wrote the manuscript. MT, YN, and NY revised and edited manuscript. YF supervised the research. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by cooperative research funds from Actree Co., the Science and Technology Research Partnership for Sustainable Development (SATREPS)—Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA) (Grant No. JPMJSA1907), Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant Nos. JP24510312 and JP16K07412 to YF), and the Ministry of Agriculture, Forestry and Fisheries (MAFF) of Japan.
Conflict of Interest
Conflict of Interest: KT, TT, and HM were employed by the company Actree Co.
The authors declare that this study received funding from the company Actree Co. The funder had the following involvement in the study: a series of quinoa research concepts and database searches.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank K. Shimizu, N. Hisatomi, Y. Masamura, I. Gejima, S. Shu, Y. Saito, K. Ozawa, J. Baba, A. Sugitani, M. Osaki, and Dr. Y. Murata (JIRCAS) for their excellent technical support, Drs. G. Almanza (UMSA), A. Bonifacio (PROINPA), and our other collaborators in Bolivia for helpful advice, and Plant Editors for skillful language-editing support of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.643499/full#supplementary-material
Supplementary Figure 1 | Alignment of nucleotide sequences of CqPDS1 homoeolog genes, CqPDS1A and CqPDS1B. Asterisks indicate conserved bases between the two homoeolog genes. Two 300-bp VIGS trigger sequences inserted into ALSV-CqPDSN and ALSV-CqPDSC vectors are highlighted in magenta and turquoise, respectively. The primer positions designed for RT-qPCR analysis are indicated with blue arrows.
Supplementary Figure 2 | Three lowland inbred lines of quinoa, J028, J045, and J082, display almost no symptoms and show no negative growth effects following ALSV infection. (A) Side (left panels) and top (right panels) views of representative images of lowland line plants at 16 dpi with mock buffer and ALSV-WT. (B) Representative images of enlarged views of the uninoculated upper leaves of Kd, J079, and J082 inbred line plants at 16 dpi with mock and ALSV-WT. Scale bars represent 2 cm.
Supplementary Figure 3 | Southern highland inbred lines of quinoa show mild growth retardation following ALSV infection. Side (left panels) and top (right panels) views of representative images of southern highland line plants at 16 dpi with mock buffer and ALSV-WT. Scale bars represent 2 cm.
Supplementary Figure 4 | Some northern highland inbred lines exhibit growth retardation and visible symptoms following ALSV infection. (A) Side (left panels) and top (right panels) views of representative northern highland line plants at 16 dpi with mock buffer and ALSV-WT. ALSV-infected plants showed some growth retardation to varying degrees in the northern highland lines used in this study. (B) Enlarged views of representative uninoculated upper leaves of J064 and J074 inbred line plants at 16 dpi with mock and ALSV-WT. Scale bars represent 2 cm.
Supplementary Figure 5 | ALSV can infect plants systemically in a variety of quinoa inbred lines. (A–C) Semi-quantitative RT-PCR of ALSV RNA1 in the uninoculated upper leaves of plants at 16 dpi with mock and ALSV-WT in 19 representative lines, including seven lowland (A), six southern (B), and six northern highland lines (C). CqUBQ10 was used as an internal control. No DNA indicates the absence of DNA template in PCR as a negative control. Two or three independent plants per inbred line were examined.
Supplementary Figure 6 | ALSV infection facilitates betalain accumulation in some highland inbred lines. (A,B) Top (left panels) and side (right panels) views of representative plants inoculated with mock buffer and ALSV-WT at 21 dpi for the northern highland lines (J056, J071, J073, and J075) (A) and at 28 dpi for the southern highland line (J131) (B). Scale bars represent 2 cm.
Supplementary Figure 7 | Temperature conditions can alter disease symptoms and VIGS induction in ALSV-infected quinoa plants. (A) Representative images of plants of the inbred Iw line at 7, 10, and 14 dpi with mock buffer, ALSV-CqPDSC, and ALSV-WT. The plants were grown at 22°C or 25°C from 0 dpi. (B) A representative image of 18-day-old plants at 0 dpi. Scale bars represent 2 cm.
Supplementary Figure 8 | ALSV can be used as a VOX vector in roots. (A) Representative images of GFP fluorescence in quinoa roots at 10 dpi with mock buffer and the packaged virus (ALSV-GFP-OE) derived from recombinant ALSV harboring a full-length GFP CDS. Scale bars represent 1 mm. TL indicates transmitted light microscopy images. (B) Semi-quantitative RT-PCR of ALSV RNA1 and RNA2 in roots at 3 wpi with the indicated inocula. 18S rRNA was used as an internal control.
Supplementary Figure 9 | Alignment of nucleotide sequences of CqRHT1 homoeolog genes, CqRHT1A and CqRHT1B. Asterisks indicate conserved bases between the two homoeologs. The VIGS trigger sequence that was inserted into the ALSV-CqRHT1 vector is highlighted in magenta. The positions of primers used for RT-qPCR analysis are indicated with blue arrows.
Supplementary Figure 10 | Knockdown of CqRHT1 results in cell expansion and elongation phenotypes. (A) A representative image of stems of J082 inbred line plants at 25 dpi with ALSV-WT and ALSV-CqRHT1. Blue arrows indicate internode regions between the second and fifth nodes of the quinoa plants, illustrating the location of the longitudinal sections shown in (B). Magenta arrows indicate the positions of the transverse sections (C–J) of the stems in the quinoa plants. (B) A representative image of longitudinal sections of the internode regions between the second and fifth nodes of the quinoa plants shown in (A). Scale bar represents 1 cm. (C–J) Representative microscopy images of transverse sections of the stems of the plants shown in (A), stained with 0.01% toluidine blue before observation. Scale bars represent 1 mm (C,D,G,H), 0.5 mm (E,I), and 0.1 mm (F,J). Pi, pith; Co, collenchyma; Ph, phloem; Xy, xylem. (K) Putative gibberellin-responsive genes were upregulated in the uninoculated upper leaves of J082 inbred line plants inoculated with ALSV-CqRHT1. The relative abundance of transcripts in the uninoculated upper leaves of plants at 15 dpi with mock, ALSV-WT, and ALSV-CqRHT1 was quantified by RT-qPCR analysis. Data were normalized to CqUBQ10 expression and are shown as means ± SD (n = 3). Different letters indicate significant differences by a Tukey’s HSD test (p < 0.05). To evaluate the expression of putative gibberellin-responsive genes in quinoa, CqACS9, CqAUX22B, CqEXPA11, CqEXPB3, CqGAST1, CqPIF3, CqPRE4, and CqSAUR21, primers for real time RT-qPCR were designed using the BLAST program in quinoa genome databases based on the nucleotide sequences of the corresponding Arabidopsis thaliana gibberellin-responsive gene homologs obtained from the NCBI database.
Supplementary Table 1 | Primers used in this study.
Supplementary Table 2 | Trigger sequences inserted into ALSV vectors.
Supplementary Table 3 | Nucleotide sequences of plasmids pEALSR1 and pEALSR2L5R5.
Footnotes
References
Abrahamian, P., Hammond, R. W., and Hammond, J. (2020). Plant virus–derived vectors: Applications in agricultural and medical biotechnology. Annu. Rev. Virol. 7, 513–535. doi: 10.1146/annurev-virology-010720-054958
Baulcombe, D. C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2, 109–113. doi: 10.1016/S1369-5266(99)80022-3
Bazile, D., Pulvento, C., Verniau, A., Al-Nusairi, M. S., Ba, D., Breidy, J., et al. (2016). Worldwide evaluations of quinoa: preliminary results from post international year of quinoa FAO projects in nine countries. Front. Plant Sci. 7:850. doi: 10.3389/fpls.2016.00850
Boden, S. A., and Østergaard, L. (2019). How can developmental biology help feed a growing population? Development 146:dev172965. doi: 10.1242/dev.172965
Böhm, J., Messerer, M., Müller, H. M., Scholz-Starke, J., Gradogna, A., Scherzer, S., et al. (2018). Understanding the molecular basis of salt sequestration in epidermal bladder cells of Chenopodium quinoa. Curr Biol. 28, 3075–3085.e7. doi: 10.1016/j.cub.2018.08.004
Bonales-Alatorre, E., Shabala, S., Chen, Z. H., and Pottosin, I. (2013). Reduced tonoplast fast-activating and slow-activating channel activity is essential for conferring salinity tolerance in a facultative halophyte, quinoa. Plant Physiol. 162, 940–952. doi: 10.1104/pp.113.216572
Bonifacio, A. (2019). Improvement of quinoa (Chenopodium quinoa Willd.) and Qañawa (Chenopodium pallidicaule Aellen) in the context of climate change in the high Andes. Cien. Inv. Agr. 46, 113–214. doi: 10.7764/rcia.v46i2.2146
Brockington, S. F., Yang, Y., Gandia-Herrero, F., Covshoff, S., Hibberd, J. M., Sage, R. F., et al. (2015). Lineage-specific gene radiations underlie the evolution of novel betalain pigmentation in Caryophyllales. New Phytol. 207, 1170–1180. doi: 10.1111/nph.13441
Burch-Smith, T. M., Anderson, J. C., Martin, G. B., and Dinesh-Kumar, S. P. (2004). Applications and advantages of virus−induced gene silencing for gene function studies in plants. Plant J. 39, 734–746. doi: 10.1111/j.1365-313X.2004.02158.x
Chandler, P. M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 129, 181–190. doi: 10.1104/pp.010917
Christensen, S. A., Pratt, D. B., Pratt, C., Nelson, P. T., Stevens, M. R., Jellen, E. N., et al. (2007). Assessment of genetic diversity in the USDA and CIP-FAO international nursery collections of quinoa (Chenopodium quinoa Willd.) using microsatellite markers. Plant Genet. Resources 5, 82–95. doi: 10.1017/S1479262107672293
Dillehay, T. D., Rossen, J., Andres, T. C., and Williams, D. E. (2007). Preceramic Adoption of peanut, squash, and cotton in northern peru. Science 316, 1890–1893. doi: 10.1126/science.1141395
Eshed, Y., and Lippman, Z. B. (2019). Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science 366:eaax0025. doi: 10.1126/science.aax0025
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132. doi: 10.1093/pcp/pcp147
Gandía-Herrero, F., Escribano, J., and García-Carmona, F. (2016). Biological activities of plant pigments betalains. Crit. Rev. Food Sci. Nutr. 56, 937–945. doi: 10.1080/10408398.2012.740103
Gomez-Pando, L. (2015). “Quinoa breeding,” in Quinoa: Improvement and Sustainable Production, eds K. Murphy and J. Matanguiban (Hoboken, NJ: Wiley Blackwell), 87–107.
Gomez-Pando, L. R., Aguilar-Castellanos, E., and Ibañez-Tremolada, M. (2019). “Quinoa (Chenopodium quinoa Willd.) breeding,” in Advances in Plant Breeding Strategies: Cereals, Vol. 5, ed. J. M. Al-Khayri et al. (Cham: Springer Nature), 259–313.
González, J. A., Eisa, S. S. S., Hussin, S. A. E. S., and Prado, F. E. (2015). “Quinoa: an incan crop to face global changes in agriculture,” in Quinoa: Improvement and Sustainable Production, eds K. Murphy and J. Matanguiban (Hoboken, NJ: Wiley Blackwell), 1–18.
Guerrero-Rubio, M. A., Escribano, J., García-Carmona, F., and Gandía-Herrero, F. (2020). Light emission in betalains: from fluorescent flowers to biotechnological applications. Trends Plant Sci. 25, 159–175. doi: 10.1016/j.tplants.2019.11.001
Hariadi, Y., Marandon, K., Tian, Y., Jacobsen, S. E., and Shabala, S. (2011). Ionic and osmotic relations in quinoa (Chenopodium quinoa Willd.) plants grown at various salinity levels. J. Exp. Bot. 62, 185–193. doi: 10.1093/jxb/erq257
Hatlestad, G. J., Akhavan, N. A., Sunnadeniya, R. M., Elam, L., Cargile, S., Hembd, A., et al. (2015). The beet Y locus encodes an anthocyanin MYB-like protein that activates the betalain red pigment pathway. Nat. Genet. 47, 92–96. doi: 10.1038/ng.3163
Hatlestad, G. J., Sunnadeniya, R. M., Akhavan, N. A., Gonzalez, A., Goldman, I. L., McGrath, J. M., et al. (2012). The beet R locus encodes a new cytochrome P450 required for red betalain production. Nat. Genet. 44, 816–820. doi: 10.1038/ng.2297
Hedden, P. (2003). The genes of the green revolution. Trends Genet. 19, 5–9. doi: 10.1016/s0168-9525(02)00009-4
Hein, A. (1957). Beitrage zur kenntnis der viruskrankheiten an unkrautern. II. Das Luzernmosaik-und das lamium-gelbmosaikvirus. Phytopath. Z. 29, 79–116.
Igarashi, A., Yamagata, K., Sugai, T., Takahashi, Y., Sugawara, E., Tamura, A., et al. (2009). Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology 386, 407–416. doi: 10.1016/j.virol.2009.01.039
Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., et al. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010. doi: 10.1105/tpc.13.5.999
Imamura, T., Isozumi, N., Higashimura, Y., Miyazato, A., Mizukoshi, H., Ohki, S., et al. (2019). Isolation of amaranthin synthetase from Chenopodium quinoa and construction of an amaranthin production system using suspension−cultured tobacco BY−2 cells. Plant Biotechnol. J. 17, 969–981. doi: 10.1111/pbi.13032
Imamura, T., Takagi, H., Miyazato, A., Ohki, S., Mizukoshi, H., and Mori, M. (2018). Isolation and characterization of the betalain biosynthesis gene involved in hypocotyl pigmentation of the allotetraploid Chenopodium quinoa. Biochem. Biophys. Res. Commun. 496, 280–286. doi: 10.1016/j.bbrc.2018.01.041
Imamura, T., Yasui, Y., Koga, H., Takagi, H., Abe, A., Nishizawa, K., et al. (2020). A novel WD40-repeat protein involved in formation of epidermal bladder cells in the halophyte quinoa. Commun. Biol. 3:513. doi: 10.1038/s42003-020-01249-w
Jacobsen, S. E., Monteros, C., Christiansen, J. L., Bravo, L. A., Corcuera, L. J., and Mujica, A. (2005). Plant responses of quinoa (Chenopodium quinoa Willd.) to frost at various phenological stages. Eur. J. Agron. 22, 131–139. doi: 10.1016/j.eja.2004.01.003
Jain, G., and Gould, K. S. (2015). Are betalain pigments the functional homologues of anthocyanins in plants? Environ. Exp. Bot. 119, 48–53. doi: 10.1016/j.envexpbot.2015.06.002
Jarvis, D. E., Ho, Y. S., Lightfoot, D. J., Schmöckel, S. M., Li, B., Borm, T. J. A., et al. (2017). The genome of Chenopodium quinoa. Nature 542, 307–312. doi: 10.1038/nature21370
Kamada, K., Omata, S., Yamagishi, N., Kasajima, I., and Yoshikawa, N. (2018). Gentian (Gentiana triflora) prevents transmission of apple latent spherical virus (ALSV) vector to progeny seeds. Planta 248, 1431–1441. doi: 10.1007/s00425-018-2992-9
Kiani-Pouya, A., Rasouli, F., Bazihizina, N., Zhang, H., Hedrich, R., and Shabala, S. (2019). A large-scale screening of quinoa accessions reveals an important role of epidermal bladder cells and stomatal patterning in salinity tolerance. Environ. Exp. Bot. 168:103885. doi: 10.1016/j.envexpbot.2019.103885
Kiani-Pouya, A., Rasouli, F., Shabala, L., Tahir, A. T., Zhou, M., and Shabala, S. (2020). Understanding the role of root-related traits in salinity tolerance of quinoa accessions with contrasting epidermal bladder cell patterning. Planta 251:103. doi: 10.1007/s00425-020-03395-1
Kiani-Pouya, A., Roessner, U., Jayasinghe, N. S., Lutz, A., Rupasinghe, T., Bazihizina, N., et al. (2017). Epidermal bladder cells confer salinity stress tolerance in the halophyte quinoa and Atriplex species. Plant Cell Environ. 40, 1900–1915. doi: 10.1111/pce.12995
Kishigami, R., Yamagishi, N., Ito, T., and Yoshikawa, N. (2014). Detection of apple latent spherical virus in seeds and seedlings from infected apple trees by reverse transcription quantitative PCR and deep sequencing: Evidence for lack of transmission of the virus to most progeny seedlings. J. Gen. Plant Pathol. 80, 490–498. doi: 10.1007/s10327-014-0541-3
Li, C., Sasaki, N., Isogai, M., and Yoshikawa, N. (2004). Stable expression of foreign proteins in herbaceous and apple plants using Apple latent spherical virus RNA2 vectors. Arch. Virol. 149, 1541–1558. doi: 10.1007/s00705-004-0310-2
Li, C., Yoshikawa, N., Takahashi, T., Ito, T., Yoshida, K., and Koganezawa, H. (2000). Nucleotide sequence and genome organization of apple latent spherical virus: a new virus classified into the family Comoviridae. J. Gen. Virol. 81, 541–547. doi: 10.1099/0022-1317-81-2-541
Massawe, F., Mayes, S., and Cheng, A. (2016). Crop diversity: an unexploited treasure trove for food security. Trends Plant Sci. 21, 365–368. doi: 10.1016/j.tplants.2016.02.006
Maughan, P. J., Bonifacio, A., Jellen, E. N., Stevens, M. R., Coleman, C. E., Ricks, M., et al. (2004). A genetic linkage map of quinoa (Chenopodium quinoa) based on AFLP, RAPD, and SSR markers. Theor. Appl. Genet. 109, 1188–1195. doi: 10.1007/s00122-004-1730-9
Mizuno, N., Toyoshima, M., Fujita, M., Fukuda, S., Kobayashi, Y., Ueno, M., et al. (2020). The genotype-dependent phenotypic landscape of quinoa in salt to- lerance and key growth traits. DNA Res. 27, 1–12. doi: 10.1093/dnares/dsaa022
Nakamura, K., Yamagishi, N., Isogai, M., Komori, S., Ito, T., and Yoshikawa, N. (2011). Seed and pollen transmission of Apple latent spherical virus in apple. J. Gen. Plant Pathol. 77, 48–53. doi: 10.1007/s10327-010-0275-9
National Academy of Sciences (NAS) (1975). Underexploited Tropical Plants with Promising Economic Value. Washington, DC: National Academy of Sciences.
National Research Council (NRC) (1989). Lost Crops of the Incas: Little-Known Plants of the Andes With Promise for Worldwide Cultivation. Washington, DC: The National Academies Press. doi: 10.17226/1398
Nowak, V., Du, J., and Charrondière, U. R. (2016). Assessment of the nutritional composition of quinoa (Chenopodium quinoa Willd.). Food Chem. 193, 47–54. doi: 10.1016/j.foodchem.2015.02.111
Ogata, T., Nagatoshi, Y., Yamagishi, N., Yoshikawa, N., and Fujita, Y. (2017). Virus-induced down-regulation of GmERA1A and GmERA1B genes enhances the stomatal response to abscisic acid and drought resistance in soybean. PLoS One 12:e0175650. doi: 10.1371/journal.pone.0175650
Olszewski, N., Sun, T. P., and Gubler, F. (2002). Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14, S61–S80. doi: 10.1105/tpc.010476
Palomino, G., Hernández, L. T., and de la Cruz Torres, E. (2008). Nuclear genome size and chromosome analysis in Chenopodium quinoa and C. berlandieri subsp. nuttalliae. Euphytica 164, 221–230. doi: 10.1007/s10681-008-9711-8
Pathan, S., Eivazi, F., Valliyodan, B., Paul, K., Ndunguru, G., and Clark, K. (2019). Nutritional composition of the green leaves of quinoa (Chenopodium quinoa Willd.). J. Food Res. 8, 55–65. doi: 10.5539/jfr.v8n6p55
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Polturak, G., and Aharoni, A. (2018). “La vie en rose”: biosynthesis, sources, and applications of betalain pigments. Mol. Plant 11, 7–22. doi: 10.1016/j.molp.2017.10.008
Polturak, G., Breitel, D., Grossman, N., Sarrion-Perdigones, A., Weithorn, E., Pliner, M., et al. (2016). Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytol. 210, 269–283. doi: 10.1111/nph.13796
Polturak, G., Grossman, N., Vela-Corcia, D., Dong, Y., Nudel, A., Pliner, M., et al. (2017). Engineered gray mold resistance, antioxidant capacity, and pigmentation in betalain-producing crops and ornamentals. Proc. Natl. Acad. Sci. U.S.A. 114, 9062–9067. doi: 10.1073/pnas.1707176114
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Rodríguez, J. P., Rahman, H., Thushar, S., and Singh, R. K. (2020). Healthy and resilient cereals and pseudo-cereals for marginal agriculture: Molecular advances for improving nutrient bioavailability. Front. Genet. 11:49. doi: 10.3389/fgene.2020.00049
Ruiz, M. T., Voinnet, O., and Baulcombe, D. C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946. doi: 10.1105/tpc.10.6.937
Schlick, G., and Bubenheim, D. L. (1993). Quinoa: an emerging “new” crop with potential for CELLS. NASA Tech. Pap. 3422, 1–9.
Schlick, G., and Bubenheim, D. L. (1996). “Quinoa: candidate crop for NASA’s controlled ecological life support systems,” in Progress in new crops, ed. J. Janick (Arlington, VA: ASHS Press), 632–640.
Schmöckel, S. M., Lightfoot, D. J., Razali, R., Tester, M., and Jarvis, D. E. (2017). Identification of putative transmembrane proteins involved in salinity tolerance in Chenopodium quinoa by integrating physiological data, RNAseq, and SNP analyses. Front. Plant Sci. 8:1023. doi: 10.3389/fpls.2017.01023
Senthil-Kumar, M., and Mysore, K. S. (2011). New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 16, 656–665. doi: 10.1016/j.tplants.2011.08.006
Sepúlveda-Jiménez, G., Rueda-Benítez, P., Porta, H., and Rocha-Sosa, M. (2004). Betacyanin synthesis in red beet (Beta vulgaris) leaves induced by wounding and bacterial infiltration is preceded by an oxidative burst. Physiol. Mol. Plant Pathol. 64, 125–133. doi: 10.1016/j.pmpp.2004.08.003
Shabala, S., Bose, J., and Hedrich, R. (2014). Salt bladders: do they matter? Trends Plant Sci. 19, 687–691. doi: 10.1016/j.tplants.2014.09.001
Sheehan, H., Feng, T., Walker-Hale, N., Lopez-Nieves, S., Pucker, B., Guo, R., et al. (2020). Evolution ofL-DOPA 4,5-dioxygenase activity allows for recurrent specialisation to betalain pigmentation in Caryophyllales. New Phytol. 227, 914–929. doi: 10.1111/nph.16089
Shepon, A., Eshel, G., Noor, E., and Milo, R. (2018). The opportunity cost of animal based diets exceeds all food losses. Proc. Natl. Acad. Sci. U.S.A. 115, 3804–3809. doi: 10.1073/pnas.1713820115
Stafford, H. A. (1994). Anthocyanins and betalains: evolution of the mutually exclusive pathways. Plant Sci. 101, 91–98. doi: 10.1016/0168-9452(94)90244-5
Strack, D., Vogt, T., and Schliemann, W. (2003). Recent advances in betalain research. Phytochemistry 62, 247–269. doi: 10.1016/s0031-9422(02)00564-2
Takahashi, T., and Yoshikawa, N. (2008). Analysis of cell-to-cell and long-distance movement of Apple latent spherical virus in infected plants using green, cyan, and yellow fluorescent proteins. Methods Mol. Biol. 451, 545–554. doi: 10.1007/978-1-59745-102-4_37
Tanaka, Y., Sasaki, N., and Ohmiya, A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749. doi: 10.1111/j.1365-313X.2008.03447.x
Uschdraweit, H. A. (1955). Chenopodium quinoa als Testpflanze für das Gurkenmosaik. Nachr. Bl. dtsch. Pfl. Sch. Dienst. (Braunschweig) 7, 151–152.
Vega-Gálvez, A., Miranda, M., Vergara, J., Uribe, E., Puente, L., and Martinez, E. A. (2010). Nutrition facts and functional potential of quinoa (Chenopodium quinoa willd.), an ancient Andean grain: a review. J. Sci. Food Agric. 90, 2541–2547. doi: 10.1002/jsfa.4158
Wang, R., Yang, X., Wang, N., Liu, X., Nelson, R. S., Li, W., et al. (2016). An efficient virus-induced gene silencing vector for maize functional genomics research. Plant J. 86, 102–115. doi: 10.1111/tpj.13142
Wheeler, T., and Braun, J. (2013). Climate change impacts on global food security. Science 341, 508–513. doi: 10.1126/science.1239402
Yamagishi, N., Kishigami, R., and Yoshikawa, N. (2014). Reduced generation time of apple seedlings to within a year by means of a plant virus vector: a new plant-breeding technique with no transmission of genetic modification to the next generation. Plant Biotechnol. J. 12, 60–68. doi: 10.1111/pbi.12116
Yamagishi, N., and Yoshikawa, N. (2009). Virus-induced gene silencing in soybean seeds and the emergence stage of soybean plants with Apple latent spherical virus vectors. Plant Mol. Biol. 71, 15–24. doi: 10.1007/s11103-009-9505-y
Yangquanwei, Z., Neethirajan, S., and Karunakaran, C. (2013). Cytogenetic analysis of quinoa chromosomes using nanoscale imaging and spectroscopy techniques. Nanoscale Res. Lett. 8:463. doi: 10.1186/1556-276X-8-463
Yasui, Y., Hirakawa, H., Oikawa, T., Toyoshima, M., Matsuzaki, C., Ueno, M., et al. (2016). Draft genome sequence of an inbred line of Chenopodium quinoa, an allotetraploid crop with great environmental adaptability and outstanding nutritional properties. DNA Res. 23, 535–546. doi: 10.1093/dnares/dsw037
Zarei, M., Shabala, S., Zeng, F., Chen, X., Zhang, S., Azizi, M., et al. (2020). Comparing kinetics of xylem ion loading and its regulation in halophytes and glycophytes. Plant Cell Physiol. 61, 403–415. doi: 10.1093/pcp/pcz205
Keywords: apple latent spherical virus, betalain, functional genomics, quinoa, virus-induced gene silencing, virus-mediated gene overexpression
Citation: Ogata T, Toyoshima M, Yamamizo-Oda C, Kobayashi Y, Fujii K, Tanaka K, Tanaka T, Mizukoshi H, Yasui Y, Nagatoshi Y, Yoshikawa N and Fujita Y (2021) Virus-Mediated Transient Expression Techniques Enable Functional Genomics Studies and Modulations of Betalain Biosynthesis and Plant Height in Quinoa. Front. Plant Sci. 12:643499. doi: 10.3389/fpls.2021.643499
Received: 18 December 2020; Accepted: 11 February 2021;
Published: 18 March 2021.
Edited by:
Joachim Hermann Schiemann, Julius Kühn-Institut, GermanyReviewed by:
Masaru Ohme-Takagi, Saitama University, JapanGuotian Li, Huazhong Agricultural University, China
Copyright © 2021 Ogata, Toyoshima, Yamamizo-Oda, Kobayashi, Fujii, Tanaka, Tanaka, Mizukoshi, Yasui, Nagatoshi, Yoshikawa and Fujita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasunari Fujita, eWFzdWZAYWZmcmMuZ28uanA=
†Present address: Chihiro Yamamizo-Oda, Plant and Microbial Research Unit, Division of Plant and Microbial Sciences, Institute of Agrobiological Sciences, National Agriculture and Food Research Organization (NARO), Tsukuba, Japan
 Takuya Ogata
Takuya Ogata Masami Toyoshima1
Masami Toyoshima1 Yasuo Yasui
Yasuo Yasui Nobuyuki Yoshikawa
Nobuyuki Yoshikawa Yasunari Fujita
Yasunari Fujita