- 1Zhengzhou Research Base, State Key Laboratory of Cotton Biology, Zhengzhou University, Zhengzhou, China
- 2State Key Laboratory of Cotton Biology, Institute of Cotton Research of Chinese Academy of Agricultural Sciences, Key Laboratory of Cotton Genetic Improvement, Ministry of Agriculture and Rural Affairs, Anyang, China
- 3Department of Plant and Environmental Sciences, New Mexico State University, Las Cruces, NM, United States
Cotton (Gossypium spp.) is an economically important crop grown for natural fiber and seed oil production. DA1 is a ubiquitin receptor that determines final seed and organ size by restricting the period of cell proliferation. In the present study, we identified 7 DA1-like genes each in cultivated tetraploid (AADD) G. hirsutum and G. barbadense, and 4 and 3 DA1-like genes in their ancestral diploid G. arboreum (A2A2) and G. raimondii (D5D5), respectively. The 7 GhDA1 genes were confirmed to be distributed on four At and three Dt subgenome chromosomes in G. hirsutum. GhDA1-1A showed a high sequence similarity to AtDA1 in Arabidopsis, and they possessed the same functional domains, suggesting conserved functions. The overexpression of GhDA1-1AR301K in Arabidopsis significantly increased seed size and seed weight, indicating that GhDA1-1A is a promising target for cotton improvement. This study provides information on the molecular evolutionary properties of DA1-like genes in cotton, which will be useful for the genetic improvement of cotton.
Introduction
Organ size is one of the most important features and is regulated by complex developmental processes involving both internal and external signals (Cai et al., 2020). Seeds represent the core of plant life cycle traits and are involved in the mechanisms of plant diffusion, germination, seedling survival and overall reproductive success (Cardinal-McTeague et al., 2019; Keren et al., 2020). In contrast to the reproductive advantage of small-seeded species, the key advantage of larger seeds appears to be their tolerance to abiotic stresses such as shade or drought, and seed size is also an important agronomic trait that greatly affects crop yield (Liu et al., 2020b). The signaling pathways that affect endosperm and/or maternal tissue growth to determine seed size include the HAIKU1 (IKU), ubiquitin-proteasome, G-protein signaling, and mitogen-activated protein kinase, plant hormone, and transcription regulator pathways (Li and Li, 2016).
Cotton is an economically important crop. In cotton breeding, focusing on a higher lint percentage (a ratio between lint weight and total seed cotton weight from seed and lint) inadvertently leads to the reduction in seed size or weight (i.e., the seed index, or the weight in g of 100 cotton seeds), which is an indicator of the quality of seeds (Zhao et al., 2015a). Generally, cottonseeds with larger volume and mass tend to content more storage material and have higher vigor (Zhao et al., 2019). Previous studies have shown that plants with large seeds exhibit better traits than those with small seeds on the basis of testing the effect of seed size on cotton seedling growth. Large seeds exhibit more nutrient accumulation than small seeds, which may affect seed germination and even the growth and development of plants (Wang et al., 2008).
There are many factors affecting the size of a seed, including genetic factors, environment (including pests and diseases) and genotype-by-environment interactions (Wang et al., 2016). In recent years, the completion of plant genome sequencing and the construction of plant transcription factor databases have greatly advanced research progress related to transcription factors involved in seed development and their molecular regulation mechanisms (Wang et al., 2013). It has been shown that multiple components of the ubiquitin pathway are involved in the regulation of seed size: for example, E3 ubiquitin ligases, the proteasome, and ubiquitination modification play important roles in regulating seed size. Several genes related to plant organ size have been cloned and functionally verified, such as EBPI (Horváth et al., 2006), DA1 (Li et al., 2008) and DA2 (Xia et al., 2013). There are three different protein domains in AtDA1, including two UIMs proximal to the N-terminus, one zinc-binding LIM domain and one DA1-like functional domain next to the C-terminus (Peng et al., 2015). The DA1 genes of Arabidopsis thaliana encode a ubiquitin receptor. When the conserved arginine (R) at the 358th position in the AtDA1 protein sequence is mutated to lysine (K), the resulted mutant produces larger seeds than wild-type plants, indicating that the DA1 gene negatively regulates both seed and tissue size (Li et al., 2008). The overexpression of AtDA1R358K can increase the rapeseed yield in Brassica napus (Wang et al., 2017). In addition, the overexpression of the mutant ZmDA1 (Zmda1) or ZmDAR1 (Zmdar1) gene improves sugar import in sink organs and starch synthesis in maize kernels (Xie et al., 2018).
However, DA1-like proteins without conserved mutation may play different roles in different plant species. Different cis-acting regulatory elements in the promoter sequences of Arabidopsis thaliana and rice respond to different hormones (such as abscisic acid and salicylic acid) and stress signals (such as heat stress and drought stress) (Li et al., 2009). As a receptor for E3 ubiquitin ligase, DA1-like genes also play important roles in regulating ABA signaling pathways to participate in drought stress (Li et al., 2008). In Glycine soja, constitutive GsoDA1 expression can improve salt resistance with no effect on seed size (Zhao et al., 2015b). The overexpression of TaDA1 decreased the size and weight, while the downregulation of TaDA1 might be effective in improving grain yields in wheat (Liu et al., 2020a).
Till now, there is no report whether DA1-like genes from cotton regulate seed size. In the present study, sequence characteristics and expression patterns of DA1-like genes were analyzed in cotton. GhDA1-1A is the homologous gene of AtDA1 in cotton. Previous studies had demonstrated that seed size of Atda1-1 mutant increased (Li et al., 2008). We wanted to know whether over-expression of mutated GhDA1-1A would have a similar phenotype. Then, GhDA1-1AR301K sequence was designed containing a single-nucleotide G-to-A transition at 902nd nucleotide site of GhDA1-1A (GH_A01G1154), which was predicted to cause an arginine-to-lysine change at the 301st amino acid site. GhDA1-1AR301K was transformed into Arabidopsis thaliana ecotype Col-0. The relationship between GhDA1-1AR301K and seed size was elaborated.
Materials and Methods
Sequence Retrieval and Identification of DA1-Like Genes in Gossypium
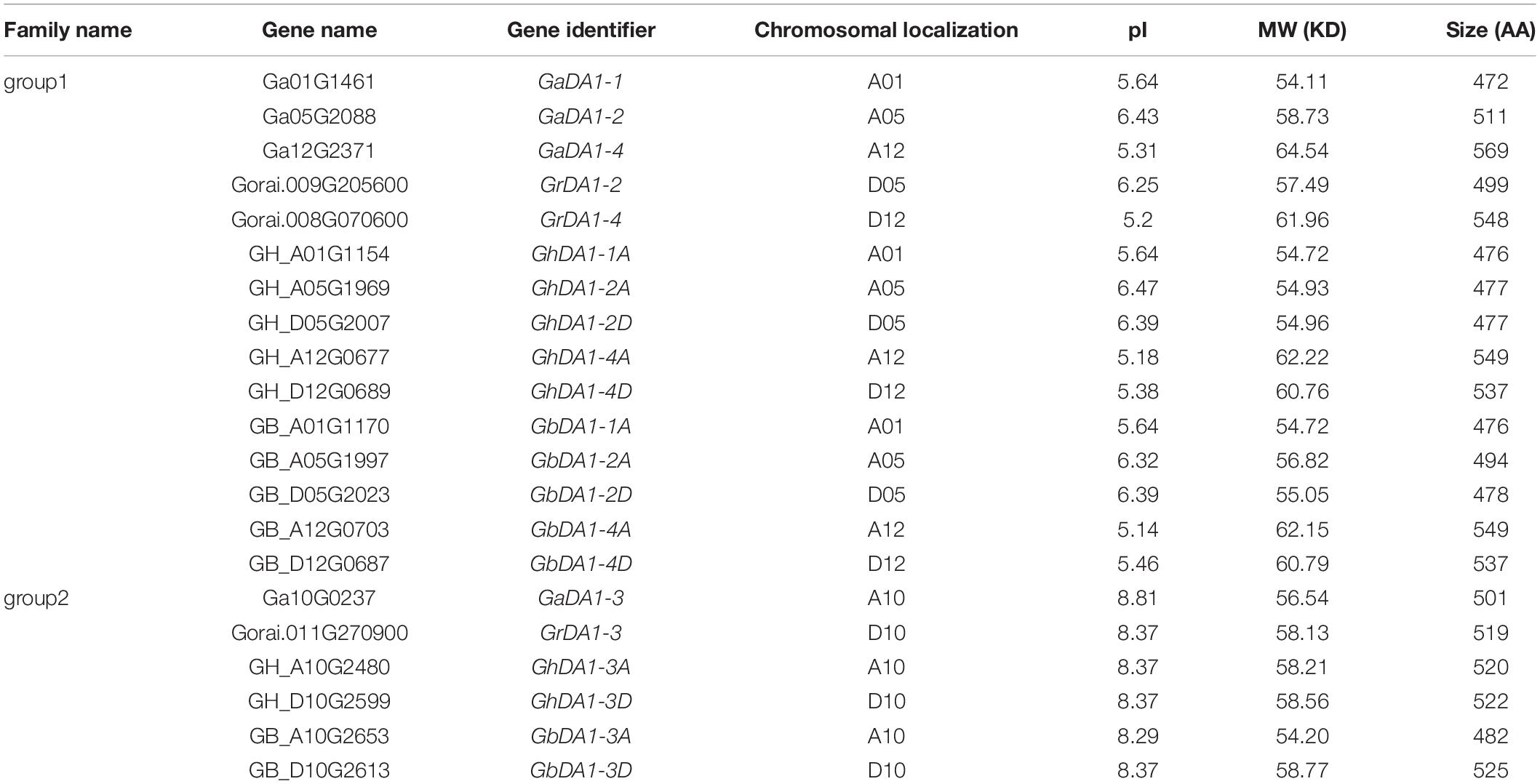
The DA1 genome sequences and protein sequences of Arabidopsis and Glycine max were retrieved from The Arabidopsis Information Resource (TAIR release 10)1 and SoyBase2, respectively. At the CottonGEN website3, we downloaded the genome sequences of G. arboreum (A2, CRI_V1.0) (Du et al., 2018), G. raimondii (D5, JGI v2_a2.1) (Paterson et al., 2012), G. hirsutum acc. TM-1 (AD1, ZJU) (Hu et al., 2019), and G. barbadense acc.3-79 (AD2, ZJU) (Hu et al., 2019). The candidate DA1 protein sequences were used as the query sequences, and BlastP (E-value = 10 × 10–5) searches were performed in the above genome databases. The default parameter settings were used. Then, the candidate sequences were submitted to Pfam4 (Finn et al., 2014) and further verified in the SMART5 (Letunic et al., 2015) database to determine whether the candidate sequence contained one zinc-binding LIM domain and one DA1-like functional domain next to the C-terminus. Multiple sequence alignments of all DA1 full-length protein sequences were performed using Clustal X2.0 software (Larkin et al., 2007) with the default values. Subsequently, the neighbor-joining (NJ) method was employed to construct phylogenetic trees by using MEGA v7.0 software (Tamura et al., 2013) with the pairwise deletion option, Poisson correction model and uniform rates. The statistical reliability of the phylogenetic tree was evaluated using the bootstrap method with 1000 repeats. Furthermore, the theoretical molecular weight (MW) and isoelectric point (pI) of the DA1-like proteins were predicted using the online ExPASy tool6 (Bjellqvist et al., 1994).
Chromosomal Location
All DA1-like genes of G. raimondii, G. arboreum, G. hirsutum, and G. barbadense were mapped on the corresponding chromosomes according to their positional information provided in the genome annotation document. The chromosome location of the cotton DA1-like genes was illustrated with MapChart v2.2 software (Voorrips, 2002).
Genetic Structure Analysis and Protein Domain Detection
Tbtools software (Chen et al., 2020) was used to predict DA1-like gene structure. The NCBI database7 was used for the identification of DA1-like protein domains.
Plant Materials and Growth Conditions
Upland cotton TM-1 was used for gene cloning and spatiotemporal quantitative real-time PCR (qRT-PCR) analysis and was grown at Anyang (AY), Henan, China. Roots, stems and leaves were collected at the seedling stage, and fibre and ovule samples were collected at 0, 5, 10, 20 and 30 days post-anthesis (DPA) for RNA extraction. Each experiment was independently repeated in triplicate. Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as the wild-type line. The RNA-seq data of G. hirsutum acc. TM-1 (Hu et al., 2019) were used to identify the expression levels of GhDA1-like genes. The expression profiles of AtDA1 and AtDAR1-7 genes were extracted from the Arabidopsis eFP Browser database8 were used to identify the expression levels of AtDA1 and AtDAR1-7 genes. Seeds were surface-sterilized with 10% (v/v) household bleach for 10 min, washed at least three times with sterile water, stratified at 4°C for 3 days in the dark, dispersed on Murashige and Skoog (MS) medium with 0.88% agar, and then grown at 22°C. Plants were grown under long-day conditions (16-h light/8-h dark) at 22°C.
Constructs and Transformation
The complete coding sequence of GhDA1-1A was amplified with gene-specific primers (Supplementary Table 1). GhDA1-1AR301K DNA sequencing revealed a single-nucleotide G-to-A transition in the GhDA1-1A gene (GH_A01G1154), which was predicted to cause an arginine-to-lysine change at the 301st amino acid site. GhDA1-1A-F and mGhDA1-1Ar as well as mGhDA1-1Af and GhDA1-1A-R bridge PCR primer pairs were used in the first PCR amplification to introduce a G to A single-base mutant in two fragments. GhDA1-1A-F and GhDA1-1A-R were used to join the two fragments together. The complete coding sequence of GhDA2 was amplified with gene-specific primers (Supplementary Table 1).
The 35S:GhDA1-1AR301K construct was generated using a PCR-based system. The specific primers used to produce the 35S:GhDA1-1AR301K construct were 35S:GhDA1-1AR301K-F and 35S:GhDA1-1AR301K-R (Supplementary Table 1). The PCR products were subcloned into a pBI121 vector digested with BamHI and SacI using the ClonExpress® II One Step Cloning Kit (Vazyme, Nanjing, China). The 35S:GhDA1-1 plasmid was introduced into Col-0 plants using Agrobacterium GV3101, and transformants were selected on medium containing kanamycin (50 mg/L). The progeny of the transformants showed approximately 3:1 segregation of live and dead phenotypes, and homozygous lines of the T3 generation were used for further analysis.
Subcellular Localization Analysis
The CDS of GhDA1-1A (1,431bp) and GhDA2 (1,272 bp) was cloned into the KpnI and SmaI sites of the 35S-GFP vector to generate 35S-GhDA1-1-GFP and 35S-GhDA2-GFP with a ClonExpress II One Step Cloning Kit (Vazyme, C112-01). The 35S-GhDA1-1-GFP and 35S-GhDA2-GFP construct was introduced into tobacco (N. benthamiana) leaves, respectively. GFP fluorescence in tobacco leaves were observed by confocal microscopy.
Bimolecular Fluorescence Complementation Assays
The CDS of GhDA1-1A was cloned into the BamHI and SalI sites of the pSPYNE vector and the CDS of GhDA2 was cloned into the BamHI and SalI sites of the pSPYCE vector with a ClonExpress II One Step Cloning Kit (Vazyme, C112-01), respectively. The constructs were transferred into Agrobacterium GV3101 cells. Agrobacterium cells were grown in LB medium containing 1% (m/v) peptone, 0.5% (m/v) yeast extract, and 1% (m/v) NaCl (pH 7) at 28°C to an OD600 of 1.2. The bacteria were pelleted and resuspended at a concentration corresponding to an OD600 of 1.2 in a solution containing 10mM MES (pH 5.8), 10mM MgCl2, and 150mM acetosyringone. Different combinations of pSPYNE-GhDA1-1A/pSPYCE-GhDA2, pSPYNE/pSPYCE-GhDA2, pSPYNE-GhDA1-1A/pSPYCE, and pSPYNE/pSPYCE were infiltrated into N. benthamiana leaves. The YFP fluorescence was detected 2 days after infiltration by confocal microscopy.
Luciferase Complementation Imaging Assay
The CDS of GhDA1-1A was cloned into the SacI and SalI sites of the pCAMBIA1300-nLUC vector with a ClonExpress II One Step Cloning Kit (Vazyme, C112-01). The CDS of GhDA2 was cloned into the KpnI and SalI sites of the pCAMBIA1300-cLUC vector with a ClonExpress II One Step Cloning Kit (Vazyme, C112-01). The constructs were transferred into Agrobacterium GV3101 cells. Agrobacterium cells were grown in LB medium containing 1% (m/v) peptone, 0.5% (m/v) yeast extract, and 1% (m/v) NaCl (pH 7) at 28°C to an OD600 of 1.2. The bacteria were pelleted and resuspended at a concentration corresponding to an OD600 of 0.6 in a solution containing 10mM MES (pH 5.8), 10mM MgCl2, and 150mM acetosyringone. Different combinations of GhDA1-1A-nLuc/c-Luc-GhDA2, nLuc/c-Luc-GhDA2, and GhDA1-1A-nLuc/c-Luc were introduced into N. benthamiana leaves by Agrobacterium tumefaciens-mediated transformation. Luciferase activity was detected 2 days after infiltration. Luciferin (Promega, e1601) at a 1 mM concentration was sprayed onto leaves, and the materials were kept in the dark for 10 min. Images were obtained with a charge-coupled device (CCD) imaging apparatus (Tanon-5200 Multl, Shanghai China).
Results
Genome-Wide Identification and Phylogenetic Analysis of DA1-Like Genes in Gossypium
To identify all the DA1-like proteins in cotton, BLASTP searches were performed against the diploid cotton (G. raimondii and G. arboreum) and tetraploid cotton (G. hirsutum and G. barbadense) protein databases using the AtDA1 and AtDAR1-7 protein sequences of Arabidopsis as queries. The candidate genes were further subjected to analysis in the NCBI database to identify their protein domains. After a strict two-step selection process, 4 deduced DA1-like genes were identified in G. arboreum, along with 3 in G. raimondii, 7 in G. barbadense, and 7 in G. hirsutum. More information about DA1-like genes, such as identifiers and predicted properties of DA1-like proteins, is listed in Table 1.
To assess the phylogenetic relationships of DA1-like genes among four cotton species, Arabidopsis and soybean, a comprehensive phylogenetic tree was constructed using the NJ method (Figure 1). In accordance with previous studies (Zhao et al., 2015b), the DA1-like genes could be divided into two groups. There were 15 members in DA1-like group 1: 5 from G. hirsutum, 5 from G. barbadense, 3 from G. arboreum, and 2 from G. raimondii. In addition, DA1-like group 2 consisted of 6 members: 2, 2, 1, and 1 from G. hirsutum, G. barbadense, G. arboretum, and G. raimondii, respectively.
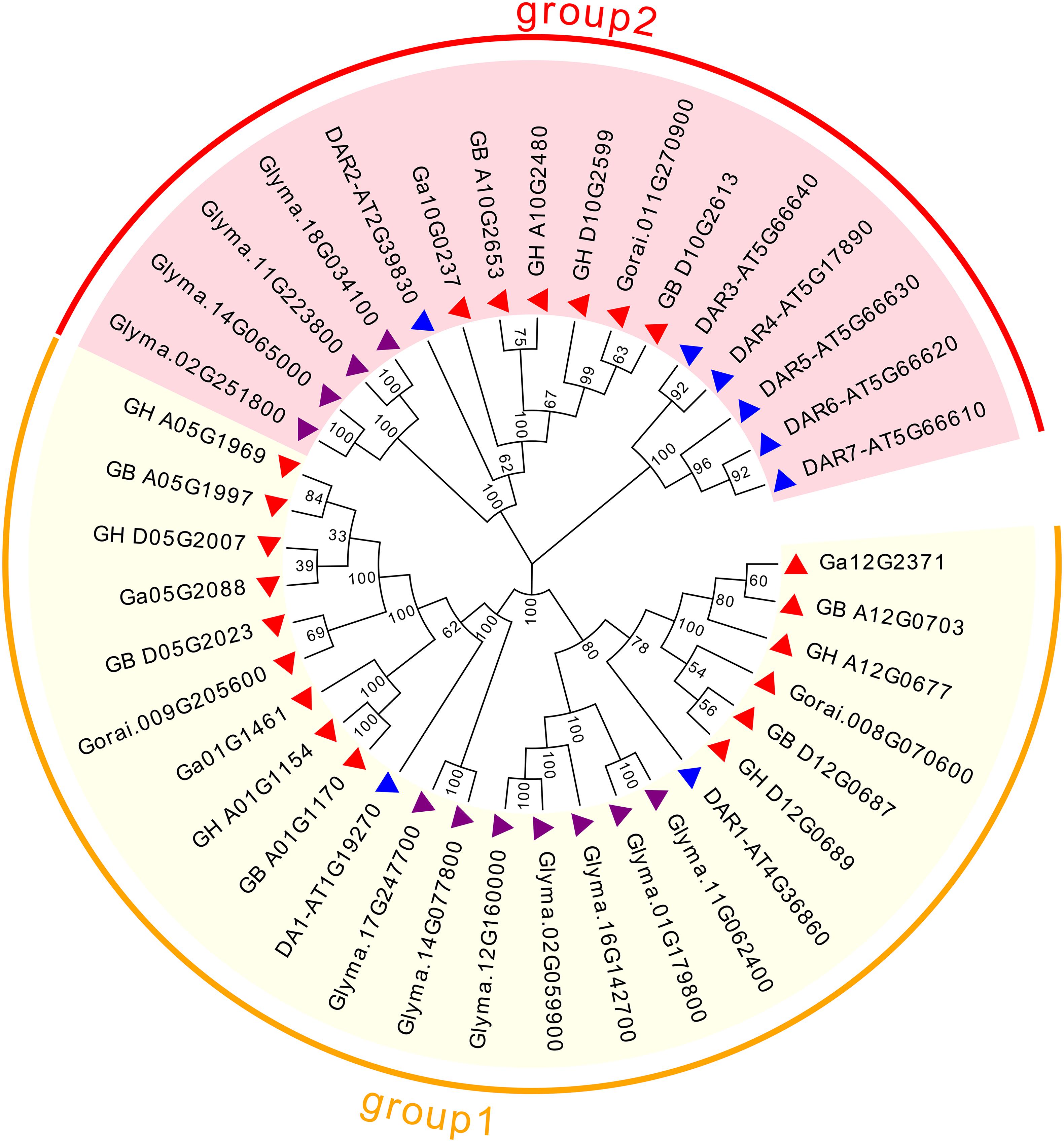
Figure 1. Phylogenetic relationships of DA1-like genes from Gossypium, Arabidopsis, and soybean. Phylogenetic analysis was performed using the neighbor-joining method with 1,000 replicates. The DA1-like genes from Gossypium, Arabidopsis, and soybean are marked with red, blue, and purple triangles, respectively. Each group is indicated with a specific color.
Chromosomal Distribution of DA1-Like Genes
The mapping of 21 DA1-like genes to chromosomes based on the available genomic information on the four cotton species revealed that all the DA1-like genes were evenly distributed on chromosomes. In the G. arboreum genome, 4 GaDA1s were distributed on four chromosomes (A01, A05, A10, and A12) (Table 1 and Supplementary Figure 1A). Three GrDA1 genes were mapped to 3 chromosomes of G. raimondii: chromosome 09 (D05), chromosome 11 (D10), and chromosome 08 (D12) (Table 1 and Supplementary Figure 1B). In the G. hirsutum genome, we found that four GhDA1s were located on At subgenome chromosomes (A01, A05, A10, and A12), while three GhDA1s genes were located on three Dt subgenome chromosomes (D05, D10, and D12) (Table 1 and Supplementary Figure 1C). In the G. barbadense genome, we also found that four GbDA1s were located on the four At subgenome chromosomes (A01, A05, A10, and A12). The other three GbDA1s were located on Dt subgenome chromosomes (D05, D10, and D12) (Table 1 and Supplementary Figure 1D). We speculate that DA1-like genes were conserved during diploid to tetraploid evolution.
Gene Structure and Protein Domain Analyses of DA1-Like Genes
The analysis of gene structure is a very effective method for determining gene function and can reflect the phylogenetic relationships among DA1-like genes. By comparing the GFF3 files of each DA1-like gene family member in G. hirsutum, G. barbadense, G. raimondii, and G. arboreum with their corresponding coding sequences, we evaluated the gene structure of DA1-like gene family members (Figure 2A). A common feature that can be observed is that DA1-like genes may contain more than ten exons: eleven DA1-like genes contain 11 exons, eight genes contain 12 exons and two genes contain 13 exons (Figure 2A). To better understand the similarity and diversity of DA1 proteins, their putative protein domains were predicted using the NCBI database. Previous studies have shown that UIM domains are absent in some DA1-like proteins (Li et al., 2008). Our results showed that most of the DA1-like proteins contained two UIM domains, one LIM domain, and one conserved DA1-like protein domain at the C-terminus. However, GrDA1-1, GhDA1-3D, GbDA1-3D, GaDA1-4, GhDA1-4A, and GbDA1-4A exhibited only one LIM domain and one DA1-like protein domain (Figure 2B). In addition, ten different conserved motifs were indicated by the MEME online program9 (Bailey and Elkan, 1994). It was demonstrated that most of these motif sare conserved in DA1-like proteins, except in GbDA1-3A, GhDA1-3A, GaDA1-3, GrDA1-3, GbDA1-2D, and GhDA1-2D, in which motif-10 was absent. It remains to be determined whether the lack of motif-10 alters protein function (Figure 2C).
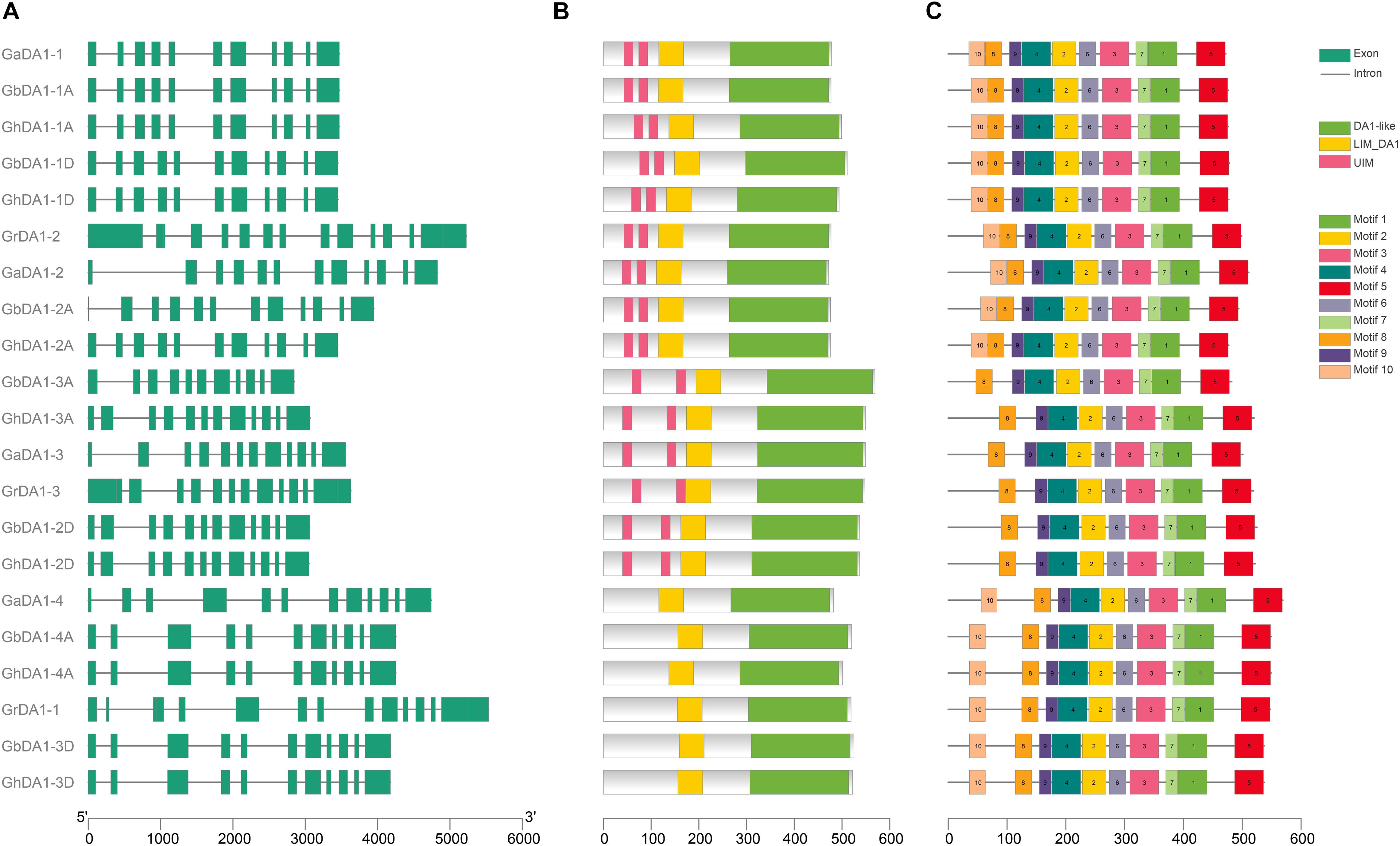
Figure 2. Gene structure and protein domain analyses of DA1-like genes in Gossypium. (A) The exon-intron structure of DA1-like genes in G. raimondii, G. arboreum, G. hirsutum, and G. barbadense. (B) DA1-like protein domain prediction. (C) The motifs of DA1-like genes in G. raimondii, G. arboreum, G. hirsutum, and G. barbadense, respectively.
Tissue-Specific Expression Profiles of GhDA1 Genes
To reveal the tissue-specific expression profiles of DA1-like genes in cotton and Arabidopsis, published TM-1 expression data (Hu et al., 2019) and the public Arabidopsis expression data (See Text Footnote 8) were used for analysis (Supplementary Figure 2A,B). GhDA1-like genes exhibited different expression patterns in different tissues of TM-1 as that in Arabidopsis, indicating that GhDA1-like genes have multiple biological functions in cotton growth and development. qRT-PCR results also showed that GhDA1-1A was highly expressed in roots, stems, and leaves but presented almost no expression in the early stage of ovule development (Figure 3A). GhDA1-2 (including the expression of GhDA1-2A and GhDA1-2D) was most highly expressed in roots (Figure 3B). GhDA1-3 (including the expression of GhDA1-3A and GhDA1-3D) and GhDA1-4 (including the expression of GhDA1-4A and GhDA1-4D) expression was highest in 30 DPA fibers, suggesting the involvement of these genes in the late period of cotton fiber development (Figures 3C,D).
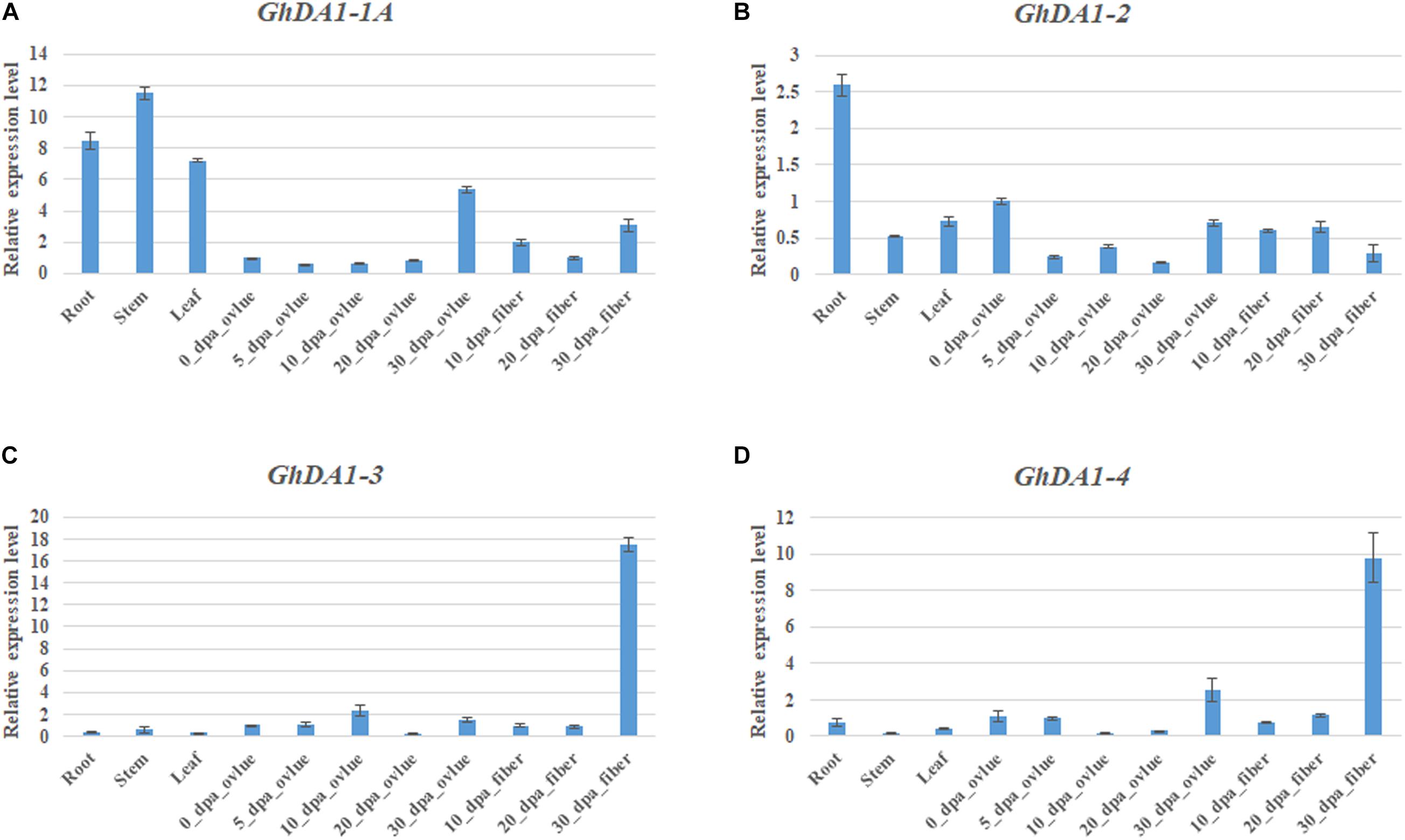
Figure 3. Tissue-specific expression profiles of GhDA1s in different tissues of G. hirsutum accession TM-1. (A) Relative expression level of GhDA1-1A. (B) Relative expression level of GhDA1-2A and GhDA1-2D. (C) Relative expression level of GhDA1-3A and GhDA1-3D. (D) Relative expression level of GhDA1-4A and GhDA1-4D. The ΔCt value of GhDA1 in 0-DPA-ovules was set as the control. The data presented are the means ± SD of three replicates.
GhDA1-1A and AtDA1 showed similar expression patterns and were widely expressed, indicating that GhDA1-1A has similar biological functions during cotton growth and development.
Generation of GhDA1-1AR301K-Overexpressing Arabidopsis Lines
AtDA1, GhDA1-1A, GhDA1-2A, GhDA1-3A, GhDA1-4A, GhDA1-2D, GhDA1-3D, and GhDA1-4D contain 532, 476, 477, 520, 549, 477, 522, and 537 amino acids, respectively, and they share 44.31% to 63.79% identity, indicating the high conservation of these homologs. The amino acid sequence of GhDA1-1A showed the closest similarity to that of AtDA1 and contained the same functional domain (Figure 4). The overexpression of AtDA1R358K results in large seeds and organs (Li et al., 2008; Weng et al., 2008), therefore, we were interested in whether GhDA1-1A could exhibit similar functions. Hence, GhDA1-1A was selected for further functional analysis.
Based on sequence alignment, mutation site 358 in AtDA1 is equivalent to conserved amino acid 301 in the DA1-like functional domain of GhDA1-1A (Figure 4). A corresponding single-nucleotide mutation was designed as a G-to-A transition in the GhDA1-1A gene to cause an arginine-to-lysine change in the conserved amino acid at position 301 (Figure 4). Arabidopsis plants overexpressing the sequence were generated and preliminarily identified by PCR (Figure 5A). qRT-PCR was performed to further assess relative expression levels using cDNA from three different transgenic lines and WT plants as templates. A total of three lines with high expression levels were obtained and used for further studies (Figure 5B).
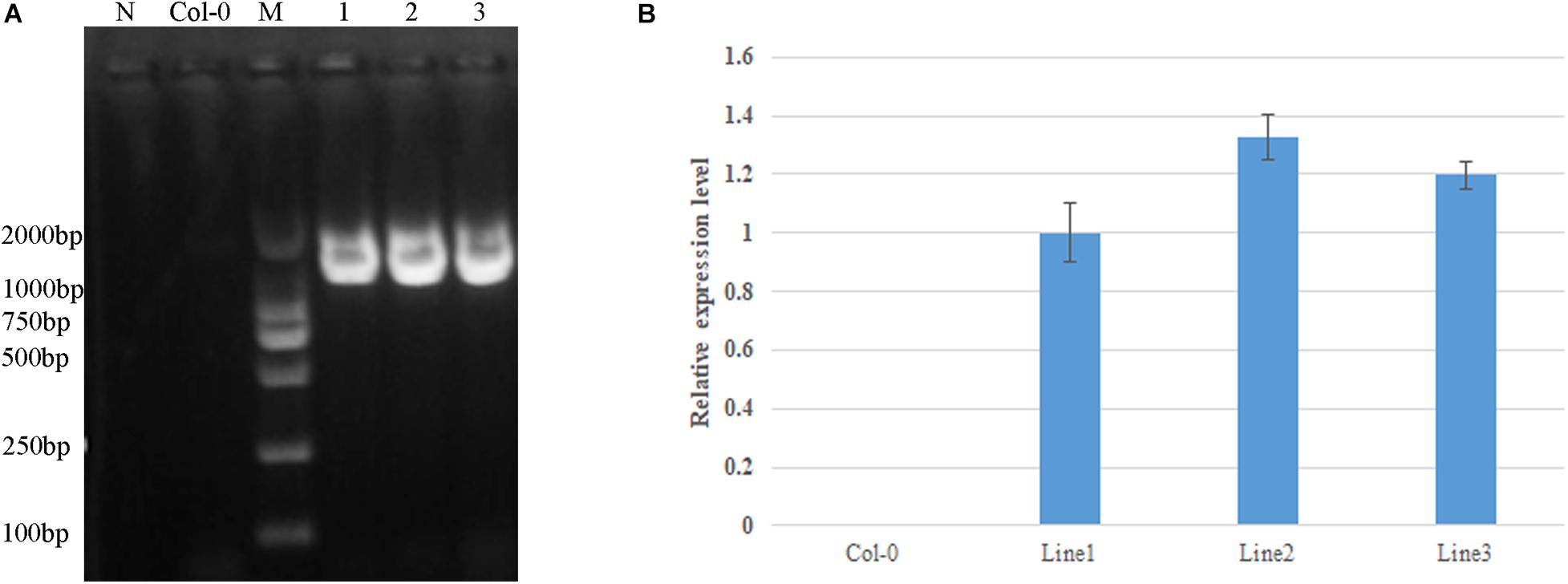
Figure 5. Identification of GhDA1-1AR301K transgenic plants by PCR. (A) The primers used were 35S-F in the 35S promotor and GhDA1-1A-R in the GhDA1-1A gene (Supplementary Table 1). The “N” is the negative control without any DNA. (B) Relative expression level of GhDA1-1A in three transgenic Arabidopsis lines. The ΔCt value of GhDA1-1A in transgenic line 1 was set as the control. The data presented are the means ± SD of three biological replicates.
Overexpression of GhDA1-1AR301K Increases Seed Size and Seed Weight
To evaluate the applicability of GhDA1-1AR301K to transgenic breeding for seed size, we characterized the phenotypes of GhDA1-1AR301K transgenic Arabidopsis at different developmental stages. The seed size of the transgenic lines was examined and was shown to be significantly increased compared to that of Col-0 (Figures 6A–D). After the seeds germinated, the cotyledon area of 9-day-old seedlings was further measured. The results showed that the seedling size of the transgenic lines was greater than that of Col-0 (Figures 6E,F). Moreover, the transgenic lines produced large flowers (Figures 6G,H), indicating that the overexpression of GhDA1-1AR301K influenced flower development and the seed mass of line 2 was increased to 128% of the Col-0 seed mass (Figures 6I–K). However, there were no differences in flowering timing, frequency, or duration (data not shown). Therefore, the overexpression of GhDA1-1AR301K increased seed size, seed weight, cotyledon size and flower size.
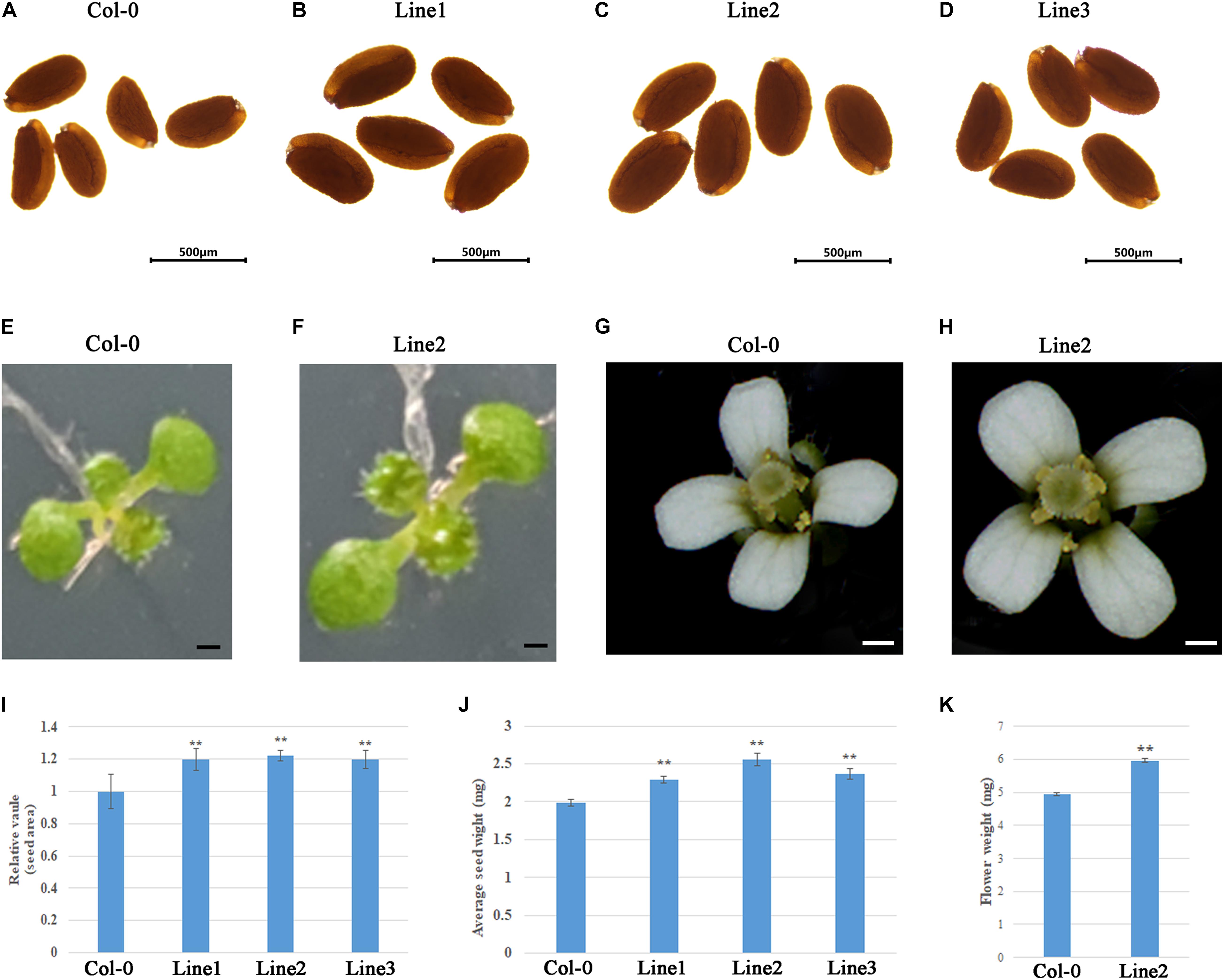
Figure 6. The overexpression of GhDA1-1AR301K increases the weight and size of the seeds. (A-D) The seeds from transgenic plant lines were compared to seeds from CK. Bar = 500 μm. (E) Nine-day-old seedlings of Col-0 (E) and transgenic plant lines 2 (F). Line2 has larger cotyledons than Col-0. ((G,H)) Flowers of Col-0 (G) and Line2 (H). (I) The relative area of seeds from Col-0 and three transgenic plant lines.(J) Average 1000-seed weights of the CK and transgenic plant lines. Values represent the mean ± standard error from three independent samples. (K) Mass of five fresh flowers. **represents significant differences at the P < 0.01 level.
GhDA1-1A Interacts With GhDA2
DA1 is a ubiquitin receptor that interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis and maize (Xia et al., 2013; Liu et al., 2020a). We were interested in the relationship between GhDA1-1A and GhDA2 (GH_D05G3532) in G. hirsutum.
To determine the subcellular localization of GhDA1-1A and GhDA2, the GhDA1-1A and GhDA2 construct fused with green fluorescent protein (GFP) were infiltrated into tobacco leaves, respectively. The epidermal cells of tobacco leaves were observed under a confocal microscope (Olympus). In contrast to the pattern observed in plants carrying the empty vector, the GFP signal of GhDA1-1A was found in membrane. Besides, the 35S-GhDA2-GFP signal was not only detected in the nucleus but also found in the membrane (Figure 7A).
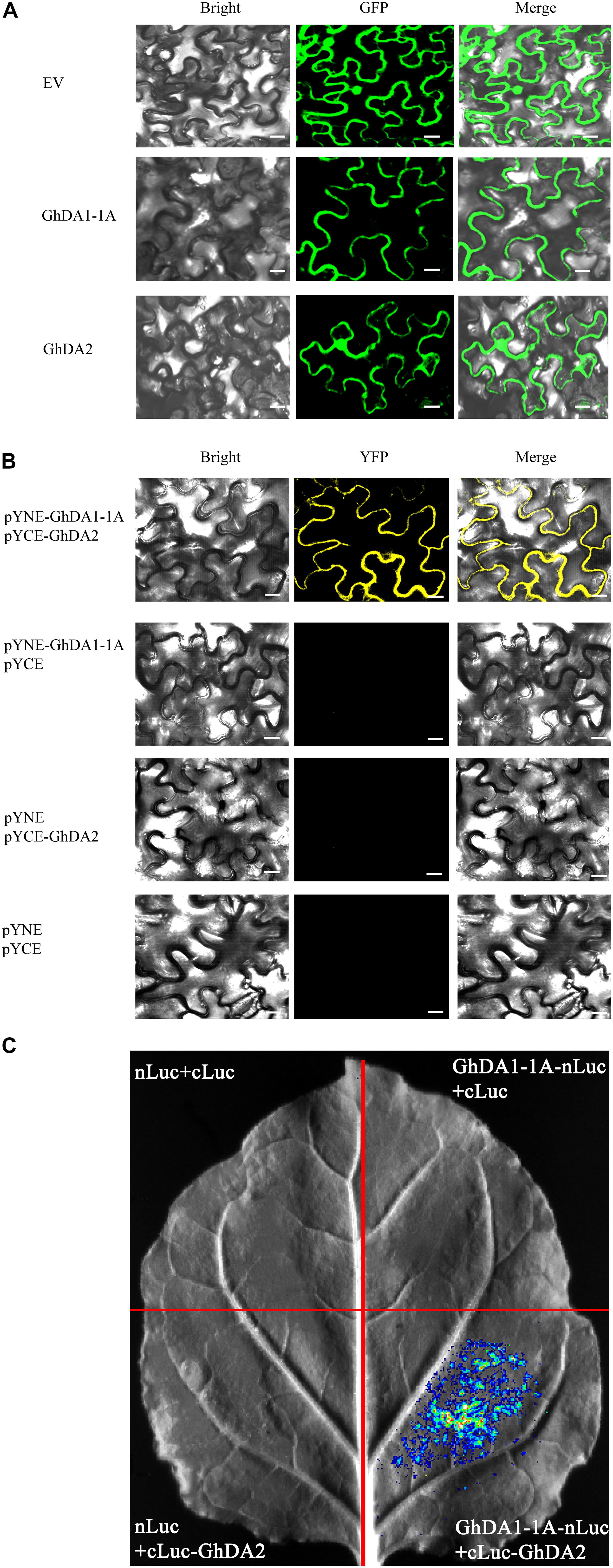
Figure 7. The subcellular localization and interaction of GhDA1-1A and GhDA2 (A) Subcellular localization of 35S-GhDA1-1A-GFP and 35S-GhDA2-GFP. (B) BiFC analysis of the interactions between GhDA1-1A and GhDA2. The pYNE-GhDA1-1A and the pYCE-GhDA2 were co-expressed into N. benthamiana leaf cells. The yellow fluorescence was detected 2 days later. (C) Firefly luciferase complementation imaging assay showing the interactions between GhDA1-1A and GhDA2. GhDA1-1A-nLUC and cLUC-GhDA2 were co-expressed in N. benthamiana leaf cells. Luciferase activity was detected 2 days after infiltration.
BiFC assays were performed in tobacco leaves to test whether GhDA1-1A could interact with GhDA2. The results showed the fusion proteins of pSPYNE-GhDA1-1A and pSPYCE-GhDA2 co-localized in the membrane (Figure 7B). Then, the firefly luciferase complementation imaging assay (Chen et al., 2008) was used to provide further evidence for the interactions between GhDA1-1A and GhDA2 using agroinfiltration (Figure 7C). The results indicated that GhDA1-1A may interact with GhDA2.
Discussion
Seed size is a key agronomic trait that strongly affects the grain yield of plants (Liu et al., 2020b). However, in cotton, fibers are the main economic product. Given that fiber cells develop from the cotton seed epidermis, seed development strongly influences fiber growth, yield, and quality (Ruan, 2013). Cotton seeds are also the sixth-largest source of vegetable oil worldwide (Liu et al., 2009). Large-seeded plants accumulate abundant nutrients to increase stress tolerance, whereas small-seeded plants flourish via dispersal and colonization (Moles et al., 2005). The mechanisms controlling seed size are thus of interest in both agriculture and biology.
In recent years, with the completion of various plant genome sequences, DA1-like genes in Arabidopsis, Brassica napus, Zea mays, and Triticum aestivum have been cloned and functionally verified (Li et al., 2008; Wang et al., 2017; Xie et al., 2018; Liu et al., 2020a). The overexpression of AtDA1R358K increases seed size in Arabidopsis and B. napus. In addition, the expression of the Zmda1 or Zmdar1 mutant gene improves grain yield in maize. The overexpression of TaDA1 decreases the size and weight of wheat kernels, while RNA interference (RNAi) has the opposite effect. The above results may suggest that mutation is necessary to increase seed size.
In the present study, the DA1-like genes of four typical Gossypium cotton species were identified. Three, four, seven, and seven DA1 genes were identified in G. raimondii, G. arboreum, G. hirsutum, and G. barbadense. Previous studies have shown that DA1 proteins without the UIM and LIM domains exist in crop plants, including rice and maize (Li et al., 2008). Our studies also identified some UIM domain-lacking DA1-like genes, such as GrDA1-1, GhDA1-3D, GbDA1-3D, GaDA1-4, GhDA1-4A, and GbDA1-4A, as typical representatives of DA1-like genes.
DA1-like genes play important roles in increasing seed yield and biomass during plant growth and development (Li et al., 2008; Wang et al., 2017; Liu et al., 2020a). Therefore, sequence alignment was used to identify protein sequence similarities to AtDA1. All GhDA1-like genes were shown to be conserved, and the GhDA1-1A homolog shared 63.79% identity to AtDA1. AtDA1 and GhDA1-1A contain the same functional domains, including two UIM domains, a LIM domain, and a DA1-like functional domain, suggesting their similar functions. Furthermore, GhDA1-1A and AtDA1 show similar expression patterns and are both widely expressed. Therefore, GhDA1-1A was cloned from G. hirsutum for further research. Previous studies have shown that OsDA1 is detectable around the plasma membrane of tobacco epidermal cells, while the fused TaDA1-A, -B, or -D proteins are distributed throughout the cytoplasm and nucleus (Liu et al., 2020a).
Because the overexpression of AtDA1R358K was shown to increase seed size and seed weight (Li et al., 2008; Wang et al., 2017), the GhDA1-1AR301K mutant was transformed into Arabidopsis, which increased seed size and seed weight. Moreover, the transgenic plants produced larger seedlings and larger flowers than the wild-type plants, indicating that GhDA1-1A plays important roles in regulating seed and organ size. Researches had showed that the DA1 can interacts with DA2 in Arabidopsis and maize (Xia et al., 2013; Liu et al., 2020a). However, the relationship between DA1 and DA2 was unknow in cotton. Therefore, the BiFC assay and dual-luciferase reporter assay were performed and verified that GhDA1-1A has an interaction with GhDA2, which showed that GhDA2 may function synergistically with GhDA1-1A to regulate seed size in cotton.
Previous studies showed that the seedling vigor and oil content are related to seed size. Seedlings from large seeds were more vigorous than the small ones in Brassica napus and cotton (Elliott et al., 2008; Pahlavani et al., 2009). In addition, there was a positive correlation between oil content and seed size in cotton (Pahlavani et al., 2009; Snider et al., 2014). Cotton ovules are composed of developing seeds and fibers. Developing seeds and fibers assimilate photosynthetic products through the same ovules. However, the relationship between seed size and fiber is complex and needs to be studied further. In our future work, we will generate transgenic cotton lines to explore the relationship between seed size and fiber.
In summary, we performed a genome-wide comprehensive analysis of DA1-like genes in the four sequenced Gossypium species and identified a total of 21 DA1-like genes. Larger seeds and organs present potential economic value in oil crop improvement and biomass energy production (Wang et al., 2016). The overexpression of GhDA1-1AR301K in Arabidopsis increased seed size and size weight, which may be useful in the improvement of seed size in cotton. Our detailed analysis of DA1-like genes in cotton has far-reaching significance for breeding work.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JY and XZ directed the experiments. LH, JS, LL, YB, BJ, LW, YX, MW, and JZ participated in the study. SY conceived the study, performed the experiments, and wrote the manuscript. JY, XZ, and JZ revised the manuscript. All authors read and approved the final manuscript.
Funding
The research was supported by grants from the National Natural Science Foundation of China (Grant No. 31801415), and the Natural Science Foundation of Xinjiang Uygur Autonomous Region of China (Grant No. 2020D01A135), and the Central Public-interest Scientific Institution Basal Research Fund (No. 1610162021016), and Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.647091/full#supplementary-material
Supplementary Figure 1 | Chromosomal localization of DA1-like genes in four Gossypium species. A total of 21 DA1-like genes were mapped onto different chromosomes of G. arboreum (A), G. raimondii (B), G. hirsutum (C), and G. barbadense (D). The scale represents megabases (Mb).
Supplementary Figure 2 | Expression level of DA1-like genes in G. hirsutum and Arabidopsis. (A) Expression patterns of 7 GhDA1-like genes. (B) Expression patterns of AtDA1 and AtDAR1-7 genes.
Supplementary Table 1 | Primers used in this study.
Supplementary File 2 | Coding sequence of GhDA1-1A, GhDA1-1AR301K and GhDA2.
Footnotes
- ^ http://www.arabidopsis.org
- ^ https://www.soybase.org
- ^ https://www.cottongen.org/
- ^ http://pfam.sanger.ac.uk/
- ^ http://smart.emblheidelberg.de/
- ^ http://web.expasy.org/
- ^ https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi
- ^ http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi
- ^ http://meme.nbcr.net/meme/
References
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Bjellqvist, B., Basse, B., Olsen, E., and Celis, J. E. (1994). Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15, 529–539. doi: 10.1002/elps.1150150171
Cai, Y., Cai, X., Wang, Q., Wang, P., Zhang, Y., Cai, C., et al. (2020). Genome sequencing of the Australian wild diploid species Gossypium australe highlights disease resistance and delayed gland morphogenesis. Plant Biotechnol. J. 18, 814–828. doi: 10.1111/pbi.13249
Cardinal-McTeague, W. M., Wurdack, K. J., Sigel, E. M., and Gillespie, L. J. (2019). Seed size evolution and biogeography of Plukenetia (Euphorbiaceae), a pantropical genus with traditionally cultivated oilseed species. BMC Evol. Biol. 19:29. doi: 10.1186/s12862-018-1308-9
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Du, X., Huang, G., He, S., Yang, Z., Sun, G., Ma, X., et al. (2018). Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50, 796–802.
Elliott, R. H., Franke, C., and Rakow, G. F. W. (2008). Effects of seed size and seed weight on seedling establishment, vigour and tolerance of Argentine canola (Brassica napus) to flea beetles. Phyllotreta spp 88, 207–217.
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Horváth, B. M., Magyar, Z., Zhang, Y., Hamburger, A. W., Bakó, L., Visser, R. G., et al. (2006). EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J 25, 4909–4920. doi: 10.1038/sj.emboj.7601362
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371-5
Keren, I., Lacroix, B., Kohrman, A., and Citovsky, V. (2020). Histone deubiquitinase OTU1 epigenetically regulates DA1 and DA2, which control arabidopsis seed and organ size. Science 23:100948. doi: 10.1016/j.isci.2020.100948
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, A., Li, L., Fang, Z., and Liu, G. (2009). Bioinformatic analysis of rice DA1 gene. Shangdong Agric. Sci. 8, 5–8. doi: 10.3969/j.issn.1001-4942.2009.08.002
Li, N., and Li, Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Li, Y., Zheng, L., Corke, F., Smith, C., and Bevan, M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Liu, H., Li, H., Hao, C., Wang, K., Wang, Y., Qin, L., et al. (2020a). TaDA1, a conserved negative regulator of kernel size, has an additive effect with TaGW2 in common wheat (Triticum aestivum L.). Plant Biotechnol. J. 18, 1330–1342. doi: 10.1111/pbi.13298
Liu, Q., Singh, S., Chapman, K., and Green, A. (2009). “Bridging Traditional and molecular genetics in modifying cottonseed oil,” in Genetics and Genomics of Cotton. Plant Genetics and Genomics: Crops and Models, Vol. 3, ed. A. H. Paterson (New York, NY: Springer).
Liu, Z., Li, N., Zhang, Y., and Li, Y. (2020b). Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis. Nat. Commun. 11:1846. doi: 10.1038/s41467-020-15603-3
Moles, A. T., Ackerly, D. D., Webb, C. O., Tweddle, J. C., Dickie, J. B., and Westoby, M. (2005). A brief history of seed size. Science 307, 576–580. doi: 10.1126/science.1104863
Pahlavani, M., Miri, A., Kazemi, G. J. P. B., and Science, S. (2009). Response of oil and protein content to seed size in cotton(Gossypium hirsutum L., cv. Sahel). Plant Breed. Seed Sci. 59, 53.
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Peng, Y., Chen, L., Lu, Y., Wu, Y., Dumenil, J., Zhu, Z., et al. (2015). The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 27, 649–662.
Ruan, Y. L. (2013). Boosting seed development as a new strategy to increase cotton fiber yield and quality. J. Integr. Plant Biol. 55, 572–575. doi: 10.1111/jipb.12074
Snider, J., Collins, G., Whitaker, J., Chapman, K., and Grey, T. L. (2014). Seed size and oil content are key determinants of seedling vigor in Gossypium hirsutum. J. Cotton Sci. 18, 1–9.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, C., Kong, W., and Li, J. (2013). Current research method of transcription factors in plants. Lett. Biotech. 24, 118–123. doi: 10.3969/j.issn.1009-0002.2013.01.028
Wang, J. L., Tang, M. Q., Chen, S., Zheng, X. F., Mo, H. X., Li, S. J., et al. (2017). Down-regulation of BnDA1, whose gene locus is associated with the seeds weight, improves the seeds weight and organ size in Brassica napus. Plant Biotechnol. J. 15, 1024–1033. doi: 10.1111/pbi.12696
Wang, J., Chen, S., Fu, Z., Liu, R., Chen, S., Wang, Z., et al. (2016). Seed size of lateral branches-removed Brassica napus and its cytological mechanism. Jiangsu J. Agric. Sci. 32, 759–762. doi: 10.3969/j.issn.1000-4440.2016.04.007
Wang, X., Lei, B., and Li, R. (2008). Effect of seed size on the plant growth at seedling stage in cotton. J. Anhui Agric. Sci. 36, 9939–9940. doi: 10.3969/j.issn.0517-6611.2008.23.052
Weng, J., Gu, S., Wan, X., Gao, H., Guo, T., Su, N., et al. (2008). Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18, 1199–1209. doi: 10.1038/cr.2008.307
Xia, T., Li, N., Dumenil, J., Li, J., Kamenski, A., Bevan, M. W., et al. (2013). The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25, 3347–3359. doi: 10.1105/tpc.113.115063
Xie, G., Li, Z., Ran, Q., Wang, H., and Zhang, J. (2018). Over-expression of mutated ZmDA1 or ZmDAR1 gene improves maize kernel yield by enhancing starch synthesis. Plant Biotechnol. J. 16, 234–244. doi: 10.1111/pbi.12763
Zhao, J., Bai, W., Zeng, Q., Song, S., Zhang, M., Li, X., et al. (2015a). Moderately enhancing cytokinin level by down-regulation of GhCKX expression in cotton concurrently increases fiber and seed yield. Mol. Breed. 35:60. doi: 10.1007/s11032-015-0232-6
Zhao, M., Gu, Y., He, L., Chen, Q., and He, C. (2015b). Sequence and expression variations suggest an adaptive role for the DA1-like gene family in the evolution of soybeans. BMC Plant Biol. 15:120. doi: 10.1186/s12870-015-0519-0
Keywords: cotton, seed, DA1-like, expression pattern, GhDA1-1A
Citation: Yang S, Huang L, Song J, Liu L, Bian Y, Jia B, Wu L, Xin Y, Wu M, Zhang J, Yu J and Zang X (2021) Genome-Wide Analysis of DA1-Like Genes in Gossypium and Functional Characterization of GhDA1-1A Controlling Seed Size. Front. Plant Sci. 12:647091. doi: 10.3389/fpls.2021.647091
Received: 29 December 2020; Accepted: 26 April 2021;
Published: 20 May 2021.
Edited by:
Goetz Hensel, Heinrich Heine University Düsseldorf, GermanyReviewed by:
Khurram Bashir, Lahore University of Management Sciences, PakistanQian-Hao Zhu, Plant Industry (CSIRO), Australia
Copyright © 2021 Yang, Huang, Song, Liu, Bian, Jia, Wu, Xin, Wu, Zhang, Yu and Zang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiwen Yu, eXVqdzY2NkBob3RtYWlsLmNvbQ==; Xinshan Zang, emFuZ3hpbnNoYW5AMTYzLmNvbQ==
 Shuxian Yang
Shuxian Yang Li Huang2
Li Huang2 Lisen Liu
Lisen Liu Man Wu
Man Wu Jiwen Yu
Jiwen Yu Xinshan Zang
Xinshan Zang