- 1Department of Biological Sciences, East Tennessee State University, Johnson City, TN, United States
- 2Department of Biological Systems Engineering, Virginia Tech, Blacksburg, VA, United States
- 3United States Department of Agriculture, Agricultural Research Service, New Orleans, LA, United States
WRINKLED1 (WRI1), a member of the APETALA2 (AP2) class of transcription factors regulates fatty acid biosynthesis and triacylglycerol (TAG) accumulation in plants. Among the four known Arabidopsis WRI1 paralogs, only WRI2 was unable to complement and restore fatty acid content in wri1-1 mutant seeds. Avocado (Persea americana) mesocarp, which accumulates 60-70% dry weight oil content, showed high expression levels for orthologs of WRI2, along with WRI1 and WRI3, during fruit development. While the role of WRI1 as a master regulator of oil biosynthesis is well-established, the function of WRI1 paralogs is poorly understood. Comprehensive and comparative in silico analyses of WRI1 paralogs from avocado (a basal angiosperm) with higher angiosperms Arabidopsis (dicot), maize (monocot) revealed distinct features. Predictive structural analyses of the WRI orthologs from these three species revealed the presence of AP2 domains and other highly conserved features, such as intrinsically disordered regions associated with predicted PEST motifs and phosphorylation sites. Additionally, avocado WRI proteins also contained distinct features that were absent in the nonfunctional Arabidopsis ortholog AtWRI2. Through transient expression assays, we demonstrated that both avocado WRI1 and WRI2 are functional and drive TAG accumulation in Nicotiana benthamiana leaves. We predict that the unique features and activities of ancestral PaWRI2 were likely lost in orthologous genes such as AtWRI2 during evolution and speciation, leading to at least partial loss of function in some higher eudicots. This study provides us with new targets to enhance oil biosynthesis in plants.
Introduction
WRINKLED1 (WRI1) belongs to the APETALA2/ethylene-responsive element binding protein (AP2/EREBP) class of transcription factors that play a master role in the developmental regulation of oil biosynthesis in plants (Cernac and Benning, 2004). The WRI1 transcription factor was first identified in Arabidopsis, where mutant seeds showed 80% reduction in triacylglycerol (TAG) content relative to wild-type (WT), 50% increase in sucrose levels, and a wrinkled seed coat phenotype. Subsequent studies revealed that WRI1 regulates a number of genes involved in controlling carbon allocation between sucrose and fatty acids in developing seeds (Focks and Benning, 1998). As such, Arabidopsis wri1 mutant seeds were unable to efficiently convert carbon from sucrose and glucose into fatty acids during seed development. The genes regulated by WRI1 include phosphoglycerate mutase, plastidial pyruvate kinase β-subunit 1 (PI-PKβ1), pyruvate dehydrogenase 1α (PDHE1α), biotin carboxyl carrier protein 2 (BCCP2), acyl carrier protein 1 (ACP1), and keto-ACP synthase 1 (KAS1) (Baud and Lepiniec, 2009; Maeo et al., 2009). Additionally, BIOTIN ATTACHMENT DOMAIN CONTAINING (BADC) proteins are conditional inhibitors of fatty acid biosynthesis; the genes encoding for BADC1, 2, and 3 were also positively regulated by WRI1 (Keereetaweep et al., 2018; Liu et al., 2019) Hence, fatty acid and storage lipid biosynthesis mediated through WRI1-induced genes is a complex and tightly regulated process.
DNA-binding studies and predictive structural analyses have shown that the regulation of glycolytic and fatty acid biosynthesis genes is achieved by the binding of AP2 domains in WRI1 with the AW-box, a 5′- upstream element ([CnTnG](n)7[CG]) in the promoter region of the target genes (Ruuska et al., 2002; Cernac and Benning, 2004; Baud et al., 2007, 2009; Maeo et al., 2009; Fukuda et al., 2013; Kuczynski et al., 2020). Specifically, in Arabidopsis WRI1 (AtWRI1), two AP2 domains (73 and 62 amino acid residues, respectively) are present in the N-terminal domain. A small exon coding for the predicted residues “VYL” (valine-tyrosine-leucine) is a functionally important element in the first AP2 domain and is widely conserved among many WRI1 orthologs. Mutation in any of these three residues rendered complementation constructs insufficient to rescue the low seed oil phenotype of retransformed wri1-1 plants (Ma et al., 2013). Furthermore, three intrinsically disordered regions (IDRs) were identified in AtWRI1, with one located near the N-terminus, preceding the first AP2 domain, and the other two towards the C-terminal region, after the second AP2 domain (Ma et al., 2015). While part of the C-terminal region from 307 to 397 in AtWRI1 is implicated in positive regulation of transcriptional activity (Ma et al., 2015), the occurrence of a PEST motif in nearby IDR3 likely acts as a recognition site for proteolytic degradation. The disordered state of IDR3 likely favors kinase access to key residues in this region and PEST residue phosphorylation serves as a signature that is recognized by key enzymes in the degradation pathway (Belizario et al., 2008). Since the PEST motif does not overlap with the transactivation domain (TAD), deletion or mutation of this motif increased the stability of WRI1 (Ma et al., 2015).
Since the discovery and characterization of WRI1 as a master regulator of fatty acid biosynthesis in seed tissues, its homologs have been identified in diverse plant species (Tang et al., 2019). In Arabidopsis, four paralogs of WRI1 (WRI1, WRI2, WRI3, and WRI4) were identified, all of which belong to the AP2/EREBP family of transcription factors. These genes, except AtWRI2, were also shown to play functional roles in fatty acid biosynthesis in various tissues (To et al., 2012). WRI1 is the dominant isoform that largely controls fatty acid metabolic gene expression during seed development and maturation. A basal level of transcriptional activity is however, maintained in wri1 mutant seeds, suggesting a redundant role played by functional paralogs (Baud et al., 2009). Both WRI3 and WRI4 are highly expressed in floral tissue and stems, and thus contribute to fatty acid production for sustained cutin synthesis (To et al., 2012). Conservation of functional homology of WRI1 genes from other species such as Zea mays (Pouvreau et al., 2011), Brassica napus (Liu et al., 2010), and Camelina sativa (An et al., 2017) was shown by the complementation of Arabidopsis wri1 mutants. In maize, there are two alternate splice forms of a WRI1 gene (ZmWRI1a and ZmWRI1b); ZmWRI1a is strongly expressed in endosperm and clearly regulates kernel oil accumulation. The tissue-specific and temporal regulation of ZmWRI1b expression is different and suggestive of an ongoing evolutionary functional specialization of these two isoforms (Pouvreau et al., 2011). Transgenic expression of two WRI1 isoforms from B. napus (BnWRI1-1 and BnWRI1-2) increased the seed oil content by 20% in Arabidopsis seeds (Liu et al., 2010). Similarly, CsWRI1 from Camelina sativa also partially restored the seed fatty acid levels in Arabidopsis wri1 mutants; its transient expression also increased the TAG content in tobacco leaves (An et al., 2017). More recently, various transcriptome studies have extended the role of WRI1 in regulation of oil biosynthesis beyond seed tissues. The function of WRI1 in regulation of fatty acid synthesis in oil palm (Elaeis guineensis) mesocarp, which accumulates about 90% oil by dry weight, was established unequivocally (Bourgis et al., 2011; Ma et al., 2013). Furthermore, EgWRI1 was able to rescue the low oil content in Atwri1-1 seeds suggesting that WRI1 is responsible for oil accumulation in both seed and non-seed tissues (Ma et al., 2013) in some plant species. Additional studies have identified high expression of tissue-specific WRI1 paralogs in diverse non-seed tissues such as Chinese tallow (Divi et al., 2016), potato, oat, and mesocarp of avocado (Kilaru et al., 2015; Rahman et al., 2016).
Avocado (Persea americana) is a basal angiosperm and occupies a useful evolutionary niche as one of the earliest lineages near the origin of flowering plants (Chanderbali et al., 2008). Avocado is also an ideal plant system to study oil biosynthesis for various reasons. The mesocarp of avocado accumulates 60–70% of oil by dry weight, of which, more than 80% is composed of nutritionally desirable oleic acid (Kikuta and Erickson, 1968; Kilaru et al., 2015). Additionally, transcriptome studies of developing mesocarp revealed that its oil accumulation is strongly correlated with the expression of fatty acid biosynthetic genes and WRI orthologs (Kilaru et al., 2015). Orthologs of all four members of the AtWRI gene family are present in avocado, referred to as putative PaWRI1 to PaWRI4, and all but the PaWRI4 are highly expressed in avocado mesocarp in correlation with oil accumulation. Avocado is unique in that it is the first plant system with confirmed concomitant expression of multiple WRI1 paralogs during oil accumulation, which has not been observed previously. PaWRI3 and PaWRI4 are likely the functionally diverged products of a relatively recent gene duplication event, given that PaWRI4 is not expressed in the presence of highly expressed PaWRI3 (Kilaru et al., 2015). Since AtWRI2 was unequivocally shown to lack any role in oil biosynthesis, high expression of its ortholog during non-seed oil accumulation in avocado was intriguing. Thus, we carried out comprehensive and comparative in silico analysis followed by transient expression studies to examine if PaWRI2 was functional and associated with TAG accumulation.
Materials and Methods
Predictive Structural Analyses
Sequences of the WRI homologs for different species were retrieved from the NCBI database based on the BLASTP search using corresponding WRI-paralogs from Arabidopsis as query. Putative transcript sequences of PaWRI1, 2, 3, and 4 were obtained from the previously published avocado transcriptome data (Ibarra-Laclette et al., 2015; Kilaru et al., 2015; Rendón-Anaya et al., 2019). Full-length sequences were subsequently confirmed by cDNA synthesis and PCR amplification using gene specific primers, and sequencing, and deposited in the GenBank. The accession numbers for all the sequences used in this study are provided in the Data Availability Statement. Various bioinformatics tools were utilized for the subsequent in silico analyses (Supplementary Table 1).
The predicted protein sequence of each avocado gene was generated with the ExPASy translation tool. Multiple Sequence Alignment was done using the PRALINE online tool (Simossis and Heringa, 2003; Supplementary Table 1). For sequence comparison, all three avocado WRI paralogs were compared to their orthologs from Arabidopsis and maize, with protein sequences in FASTA format was used as inputs. BLOSUM62 exchange weights matrix was used, with gap penalties value 12 for open and 1 for extension. Homology-extended alignment was performed keeping all other parameters at the default values provided by the software. The outputs were obtained in a color-coded conservation scoring format where the conservation at each position of amino acid was denoted by a score from 0 to 10; 0 being the least-conserved position and 10 (indicated by asterisk) being the highest. The information about the positioning of AP2 domains in each WRI protein sequence was obtained from the NCBI database.
The amino acid composition profile of each protein was determined using COMPOSITION PROFILER (Vacic et al., 2007; Supplementary Table 1). Each of the protein sequences was used as a query sample against the SwissProt 51 dataset. The graphical output was obtained in a bar chart format composed of twenty data points for each amino acid arranged in increasing order of hydrophobicity (Eisenberg), where bar heights indicate enrichment (upward) or depletion (downward). The software also performs statistical analyses to evaluate whether the enrichment and depletion of amino acids were significant at a (α) of 0.05. Only the significantly enriched amino acids are represented in green and the depleted amino acids in blue.
Composition of each of the secondary structures, such as α-helix, β-strand, β-turn, and random coil structures, were predicted using the Self-Optimized Prediction Method with Alignment (SOPMA) secondary structure prediction online tool (Combet et al., 2000; Supplementary Table 1) with default parameters. Percentages for each of the predicted secondary structure components are denoted in the tables and the respective positions of each are expressed in graphical format.
The prediction of IDRs was conducted using the Predictors of Natural Disordered Regions (PONDR-VL3) online tool (Peng et al., 2005; Supplementary Table 1). The disordered regions in the full-length protein sequences were identified and provided as graphic format outputs. A score above the threshold value of 0.5 indicates a propensity of that stretch of protein to be a part of an IDR. Only the stretches of >30 amino acid residues showing disordered tendency are considered as likely IDRs in the protein tertiary structure.
Kinase phosphorylation potential of each Ser (S), Thr (T), or Tyr (Y) residue was evaluated using the NetPhos 3.1 online tool (Blom et al., 1999). The likelihood threshold values were considered at 0.5. The positions of putative PEST motifs were predicted using the epestfind online tool (Supplementary Table 1).
Statistical Analysis
The transient expression assays were performed in triplicate and data were expressed as their mean value with standard deviation (SD). To test for significance amongst the dataset, one-way analysis of variance (ANOVA) followed by Tukey’s post-test was performed using Minitab software (version 18) at p-level of 0.05 (p < 0.05).
cDNA Synthesis and Cloning
Total RNA extract from avocado mesocarp was available from the previous study (Kilaru et al., 2015). From the total RNA, cDNA was synthesized using oligo dT primer and superscript reverse transcriptase (Promega). Full-length coding sequences of putative PaWRI1 and PaWRI2 were amplified by PCR using specific primers (PaWRI1-F: 5′-GCTCCCATGGACACATCTTCTCCCCTCTCCAATT-3′ and PaWRI1-R: 5′-CTATCCGCGGCTAAGAACATATGCTGATGG GAAGCGGAT-3′ for PaWRI1; PaWRI2-F : 5′-GCTCCC ATGGCTTCTTCTCCTTCGTCGTCG-3′ and PaWRI2-R: 5′-GCGGTCCGCGGTTATTCTTGACGGAAGAAAGTATATG TTG-3′ for PaWRI2) containing NcoI and SacII restriction sites (underlined) in their forward and reverse primers, respectively. Initiator methionine codons in the forward primers are shown in bold. The PCR products were digested with appropriate restriction endonucleases, and cloned into the corresponding sites in entry vector pK34 with a dual CaMV35S promoter and terminator (Shockey et al., 2015) using T4 DNA ligase (New England Biolabs). Ligation reactions were chemically transformed into Top10 E. coli competent cells, grown on solid media containing ampicillin, and insert-positive colonies were identified by colony PCR. Insert sequence accuracy was confirmed by sequencing. For expression in tobacco leaves, AscI fragments representing the promoter:gene:terminator cassettes from the K34-based entry plasmids were cloned into the binary expression vector pB110 (Shockey et al., 2015), followed by another round of E. coli transformation and colony selection, this time using kanamycin as the selective agent. The viral silencing suppressor protein gene P19 (Wood et al., 2009) was also cloned into pB110 behind the CaMV35S promoter using this strategy, and was later used for co-infiltration into N. benthamiana leaves.
Transient Expression
Wild type tobacco (Nicotiana benthamiana) was used for all transient expression experiments. All plants were grown in a growth chamber (Percival Scientific, Perry, IA, United States) under long day conditions (15 h L/9 h D) at 24°C and 60% relative humidity. Six weeks old plants were used for transient expression. For plant transformation, 100 μL of Agrobacterium LBA4404 competent cells were gently mixed with 100-1000 ng of binary plasmid DNA expressing the gene of interest, kept on ice for 5 min, then immediately transferred into liquid nitrogen. Subsequently, the flash-frozen mixture was incubated for 5 min at 37°C in a water bath and then transferred into 1 mL of LB media and incubated for 2-4 h at 28°C. After the incubation period, the bacterial cells were pelleted by gentle centrifugation and resuspended in 100 μl LB and immediately plated onto LB agar containing kanamycin (50 μg/mL) and rifampicin (50 μg/mL). Positive colonies were used for overnight culture in LB liquid media containing appropriate antibiotics at 28°C. The overnight culture was further supplemented with 100 μM acetosyringone and allowed to grow for additional 2 h. After incubation, the solution was centrifuged at 400 × g for 5 min at room temperature and the pellet was resuspended in infiltration buffer (5 mM MgSO4, pH 5.7, 5 mM methyl ethanesulfonate, and 100 mM acetosyringone). Finally, Agrobacterium constructs with an O.D. of 0.3 at 600 nm were prepared and infiltrated into N. benthamiana leaves. After infiltration, plants were maintained under ambient conditions in the growth chamber, to avoid any external stressors. Plants were allowed to grow for 6-7 days to express the recombinant protein. Subsequently, inoculated areas of leaves were harvested and either processed immediately or stored at −80°C for further analyses.
Nile Red Staining
After Agrobacterium infiltration, N. benthamiana plant leaf discs were collected and fixed with paraformaldehyde in 1X phosphate-buffered saline (PBS). Nile Red (in dimethyl sulfoxide) was diluted in 1X PBS (final concentration 4 μg/mL) was used for lipid droplet staining; leaf discs were immediately observed using a Leica TCS SP8 confocal fluorescence microscope. The excitation and emission wavelength for Nile Red staining were 488 nm and 560-620 nm, respectively. Images were acquired as a z-stack of 10 optical sections. The dimensions of each panel were 161.7 × 161.7 μm and the total number of LDs per view were quantified using ImageJ software the count was normalized to LD/mm2. Images were obtained from three biological replicates.
Extraction of Plant Lipids
Other infiltrated N. benthamiana leaf discs were collected at 6-7 days after Agrobacterium infiltration and used for total lipid extraction. Lipids were extracted using the hexane:isopropanol method (Hara and Radin, 1978). Briefly, 200 mg of plant tissue (fresh weight, FW) was ground in liquid nitrogen and 2 ml hot isopropanol was added, then heated to 70°C for 30 min in water to inactivate any internal lipase activity. After cooling to room temperature, one mL of chloroform and 250 μL water were added to achieve a final ratio of 2 mL isopropanol: 1 mL chloroform: 0.45 ml water. The samples were stored O/N at 4°C. The next day, the supernatant was collected from the sample after vortexing and centrifugation for 5 min at 5000 × g. One mL of chloroform and 2 mL of 1M KCl were added to the sample to achieve phase separation. After centrifugation, the upper aqueous phase and interphase were aspirated by using a Pasteur pipette. The sample was then washed twice with 2 mL of 1M KCl and the organic phase was collected and dried under nitrogen gas. Dried lipid samples were reconstituted in ∼1 mL of chloroform and transferred into a pre-weighed glass vial. After evaporating the chloroform in nitrogen gas, each glass vial was weighed again to obtain the total lipid weights. Before homogenization, an appropriate amount of heptadecanoic acid, 17:0 standard (100 μg; Sigma-Aldrich) was added to the plant tissue.
Fatty Acid Analysis by Gas Chromatography-Flame Ionization Detection (GC-FID)
Total lipids extracted from the infiltrated leaves were esterified to determine fatty acid composition. To perform fatty acid methyl esterification (FAME), HCl methanolic acid (1 N) was added to the samples and the mixture was heated for 2 h at 85°C in a water bath. After cooling to room temperature, 1 mL KCl and 1 mL hexane were added for phase separation. The hexane organic phase containing FAMEs was collected and dried under nitrogen gas at 40°C. Each FAME sample was resuspended in 1 μL of hexane and analyzed by gas chromatography coupled with a flame ionization detector (GC-FID, Varian). A capillary column (DB-23; 30 m × 0.32 mm I.D., 0.25 μm) with helium as carrier gas (flow rate of 1.5 mL/min) was used. The injection temperature was set at 150°C, which was ramped after 3 min to 240°C at 6°C/min; the detection temperature was 300°C. The retention time for each fatty was determined based on comparison to the Supelco FAME mix standards, and quantified relative to the C17:0 internal standards. Each sample was analyzed in triplicate.
Results and Discussion
Primary Sequence Features of WRI Paralogs in Avocado Differ From Their Dicot and Monocot Orthologs
To identify distinct structural characteristics among the WRI orthologs, comprehensive in silico analyses were conducted. Initially, phylogenetic analyses of WRI orthologs from select basal angiosperms (avocado and Amborella), monocots (rice, maize, and oil palm) and dicots (Arabidopsis, rapeseed, castor, grape, and poplar) was carried out (Supplementary Figure 1). These analyses revealed that WRI2-orthologs evolved independently and earlier than their corresponding paralogs in the respective plant species (Supplementary Figure 1). Further comprehensive analyses of structural features of PaWRI2, relative to its paralogs and orthologs specifically from maize and arabidopsis were determined by protein sequence comparison. Among the three species, the WRI1 protein sequences shared an overall identity of 50%, while WRI2, WRI3, and WRI4 showed 45, 57, and 59% identity, respectively (Figure 1). Additionally, phylogenetic analysis showed that avocado WRI3 and WRI4 share > 70% identity and are likely the result of a gene duplication event (Supplementary Figure 1). Duplicated genes may retain functional redundancy, but in some cases attain specialized temporal- or tissue-specific roles. PaWRI3 transcripts were abundant and PaWRI4 was poorly expressed in avocado mesocarp (Kilaru et al., 2015), suggestive of a larger role for PaWRI3 in mesocarp lipid metabolism. Whether the functional role of WRI2 orthologs in oil biosynthesis is conserved in any of the plant species has not been elucidated, except for AtWRI2, where it was shown to be non-functional based on its inability to complement Atwri1-1 mutant (To et al., 2012). As such, only WRI1, WRI2, and WRI3 orthologs were included in the subsequent in silico comparisons.

Figure 1. Primary sequence features of WRI homologs. Multiple sequence alignment of primary sequences of WRI1, 2, and 3 from P. americana (Pa), A. thaliana (At), and Z. mays (Zm) in a color-coded format; red and blue colors represent the most (10, indicated by asterisk) and least (0) conserved regions, respectively, on a scale of 0 to 10. Position of the AP2 domains and sequence length are indicated at the end of the alignment. The amino acid residues in the AP2 domain important for DNA interaction are represented by (▲).
Primary sequence comparisons revealed that avocado WRI1 and WRI3 proteins were generally shorter than their corresponding orthologs in Arabidopsis and maize (Figure 1) and also to some of other monocot and dicot orthologs examined (Supplementary Figure 2). Specifically, PaWRI1 is shorter than AtWRI1 and ZmWRI1a by 37 and 2 amino acids, respectively. PaWRI3 is only 10 amino acids shorter than AtWRI3, but 81 amino acid shorter than ZmWRI3/4b. PaWRI2 is 30 amino acids shorter than AtWRI2 and 16 amino acids longer than ZmWRI2. Interestingly, the relative position of the regions that contributed to shorter avocado WRI sequence among its paralogs varied. In the case of WRI1, both avocado and maize lacked several amino acids downstream of the second AP2 domain that are present in AtWRI1, while for WRI2, several amino acids in both N-terminal and C-terminal regions were absent. However, for WRI3, PaWRI3 and AtWRI3 lacked several N- and C-terminal amino acids that are present in ZmWRI3/4b. More than 45% of the shared identity is found in the N-terminal AP2 domains, which is a distinct feature of the APETALA protein family. As a result, the N-terminal region of WRI1 orthologs in different plant species is highly conserved, whereas the C-terminal region that harbors the TAD is variable (Kong et al., 2019; Tang et al., 2019; Fei et al., 2020).
Among the two highly conserved N-terminal AP2 domains, the first domain is typically more conserved than the second (Cernac and Benning, 2004), although both were similarly, conserved across the orthologs of the three species examined in detail here. Also, while both AP2 domains are present in the WRI1, WRI3, and WRI4 orthologs, the WRI2s, including PaWRI2, lacked the second AP2 domain (Figure 1 and Supplementary Figure 2). Moreover, unlike PaWRI2 or ZmWRI2, the single AP2 domain present in AtWRI2 was interrupted by a stretch of 35 additional amino acids (Figure 1 and Supplementary Figure 2). Plant AINTEGUMENTA transcription factors are able to bind DNA promoters despite having only a single AP2 domain (Krizek, 2003), but yeast-one hybrid studies have shown that AtWRI2 was unable to bind to the promoter of the BCCP2 gene (To et al., 2012), likely due to the interruption present in the AP2 domain. The need for an intact AP2 domain to bind DNA promoter motifs might explain the lack of function for AtWRI2; whether this function exists in ZmWRI2 remains unexplored.
The WRI AP2 domains interact with the DNA elements called the “AW-box” in target gene promoters (Cernac and Benning, 2004). While the identity of the protein residues that bind to the AW element is not yet known, based on the sequence and structural similarity to the GCC-box binding domain (GBD) of AtERF1 (1GCC; PDB database), 11 amino acid residues in the AP2 domain were predicted to interact with the target promoters (Okamuro et al., 1997; Allen et al., 1998). While these 11 amino acid residues are conserved among the WRI1 orthologs, AtWRI2 showed mutation in the first three residues (R→E, G→E, & T→S) and PaWRI3 had mutations in two residues (Q→K, Y→R) (Figure 1 and Supplementary Figure 2). The evolution of these three residues could also contribute to the loss of function in AtWRI2; DNA binding activity in PaWRI3 is yet to be explored.
Although the larger N-terminal region is highly conserved overall, the sequence upstream of the AP2 domains is variable among the orthologs. Particularly, the presence of repeat regions of serine residues in PaWRI1 and glycine in PaWRI2 is similar to their respective orthologs in Arabidopsis but not in maize. The serine-rich region in the N-terminus is thought to have gradually evolved among vascular plants (Tang et al., 2019), which may play a role in functional regulation and subcellular localization (Casal et al., 2002; Ma et al., 2015), whereas the role of glycine repeats in PaWRI2 is not known. The micro-exon encoding the “VYL” motif in the first AP2 domain is essential for AtWRI1 function (Ma et al., 2013). This domain is present in all WRI proteins here, except for PaWRI3 and PaWRI4 (Figure 1). Previous studies showed that site-directed mutagenesis of the amino acids “VYL” in AtWRI1 failed to complement Atwri1-1 mutant but the WRI1 isoform in castor bean (RcWRI1-B) and rice (OsWRI1-1) were functionally active despite lacking the motif (Ma et al., 2013; Ji et al., 2018; Mano et al., 2019). Thus, the mechanistic role of the “VYL” motif is not well-understood, and the functional importance appears to be species-dependent. High PaWRI3 expression during oil accumulation suggests that this protein might be functional, even in the absence of the motif. Additionally, two KIN10-mediated phosphorylation target residues (T70 and S166) identified in AtWRI1 are conserved in all the avocado paralogs, but S166 is absent in both AtWRI2 and ZmWRI2 (Supplementary Figure 3). Phosphorylation of these two residues by KIN10 triggers WRI1 degradation as a response to elevated sugar levels in the cell (Zhai et al., 2017). This suggests a possible PaWRI2-mediated, sugar-dependent regulation of avocado oil biosynthesis (that is likely aided by PaWRI1 and PaWRI3 as well), while such a mechanism is lost for WRI2 orthologs in at least some higher plants.
Additionally, other interesting protein structural variations were also observed. In general, the C-termini share very little sequence similarity. An acidic residue-rich region that overlaps with the putative TAD in Arabidopsis WRI1 was identified in monocots and other dicots (Ma et al., 2015; Tang et al., 2019). However, the TAD in AtWRI1 (307-397) is not conserved in other orthologs and about twenty six amino acid deletions were identified in that region of PaWRI1 (Figure 1). Among the three WRIs in avocado, PaWRI2 has the longest C-terminal region (sequences downstream of AP2 domain; 272 residues, compared to 169 and 134 in PaWRI1 and PaWRI3, respectively) although the length variation is commonly observed in other plant species (Tang et al., 2019). Also, both PaWRI3 and AtWRI3 lack several amino acids in their C-terminal domains, compared to their maize ortholog with an extended C-terminus (Figure 1).
Avocado WRI Sequences Include the Signature of a Structurally Disordered Protein Fold
Amino acid composition biases are important for protein function and are prominent among eukaryotic transcription factors. Particularly, enrichment with hydrophilic or depletion of hydrophobic amino acids leads to generation of a disordered protein fold that is associated with proteostasis (Dyson and Wright, 2005; Gao and Xu, 2012). The locally present unstructured regions due to amino acid composition biases are referred to as intrinsically disordered regions (IDRs) and the proteins as intrinsically disordered proteins (IDPs; Liu et al., 2006). The disordered value of AtWRI1 was ∼53% due to composition bias (Ma et al., 2015); similar analysis of other WRI homologs also revealed enriched polar amino acids and depleted hydrophobic amino acids (Supplementary Figure 4). Sequence alignment analysis has already indicated a great degree of dissimilarity in N- and C-terminal regions flanking the AP2 domains. To investigate compositional preference and to search for regions of significant enrichment, we used the SwissProt 51 database as a standard reference. The data showed that serine (S) is enriched in all the homologs, except ZmWRI3/4b. Hydrophobic amino acids like Ile (I), Val (V), and Leu (L) were significantly depleted in most of the WRI homologs (Supplementary Figure 4). For example, L and V were depleted in PaWRI1 and PaWRI2, respectively, while all the three residues (I, V, and L) were depleted in PaWRI3. However, unlike the other orthologs, the polar amino acid Lys (K) was significantly depleted only in PaWRI1 (Supplementary Figure 4). The tendency for enrichment of polar amino acids or depletion of hydrophobic amino acids was highest in AtWRI1 and lowest in PaWRI1, while the contrary was true for WRI3 orthologs. Among the WRI2 orthologs, however, the propensity was higher for ZmWRI2 followed by PaWRI2 and then AtWRI2. Overall, the degree of positive S bias and depletion of non-polar amino acids like I, V, and L suggests that WRI transcription factors in avocado have a characteristic feature of IDPs.
A disorder-promoting amino acid composition among the WRI homologs suggests that achieving secondary structure folds would be difficult. A hydrophobic core is important for nucleation of protein folding in an aqueous environment; depletion or low proportions of hydrophobic amino acids will typically lead to random coil (Eisenberg et al., 1986). Unstructured folds or random coil regions are common among eukaryotic proteins including transcription factors (Uversky and Dunker, 2010). Such features enable proteins to adopt different conformations to interact with multiple target partners and other coregulatory proteins (Khan and Kumar, 2009). The composition analyses of WRI homologs were used to predict the degree of secondary structure (α-helix, β-sheet, β- turn, and random coil) for each protein. In all the homologs, random coil structure was the highest in proportion, followed by α-helix, β-strand, and β-turn (Figure 2). All the avocado WRI paralogs have > 50% random coil region, which is greater than in their respective orthologs in Arabidopsis and maize. The random coil regions are mostly present in the N- and C-terminal regions, while the AP2 domain regions contain abundant α- helix and β-sheet content and a low proportion of random coils (Figure 2). The crystal structure of the GCC-box binding domain (similar to AP2 domain) of EREBP protein also has the same feature, where the β-sheets were particularly responsible for DNA binding (Allen et al., 1998). Although the 3D structure of the AP2 domains is yet to be resolved, based on the sequence homology, it is expected that their promoter binding ability is also achieved in a similar manner. Additionally, extensive random coil structure flanking the WRI AP2 domains can also contribute to effective binding to different promoters. Such functionality is evident from the demonstrated ability of AtWRI1 to bind to the promoter regions of various fatty acid synthesis genes (Baud and Lepiniec, 2009; Maeo et al., 2009). Overall, it can be inferred that a high proportion of random coil structure associated with WRI homologs, including those from avocado, contributes to high conformational flexibility to interact with their target genes resulting in a complex network of WRI-mediated regulation of oil biosynthesis. These findings support the possibility that PaWRI1, PaWRI2, and PaWRI3 have retained relatively more flexible and dynamic structures that allow for greater conformational promiscuity to interact with numerous target proteins.
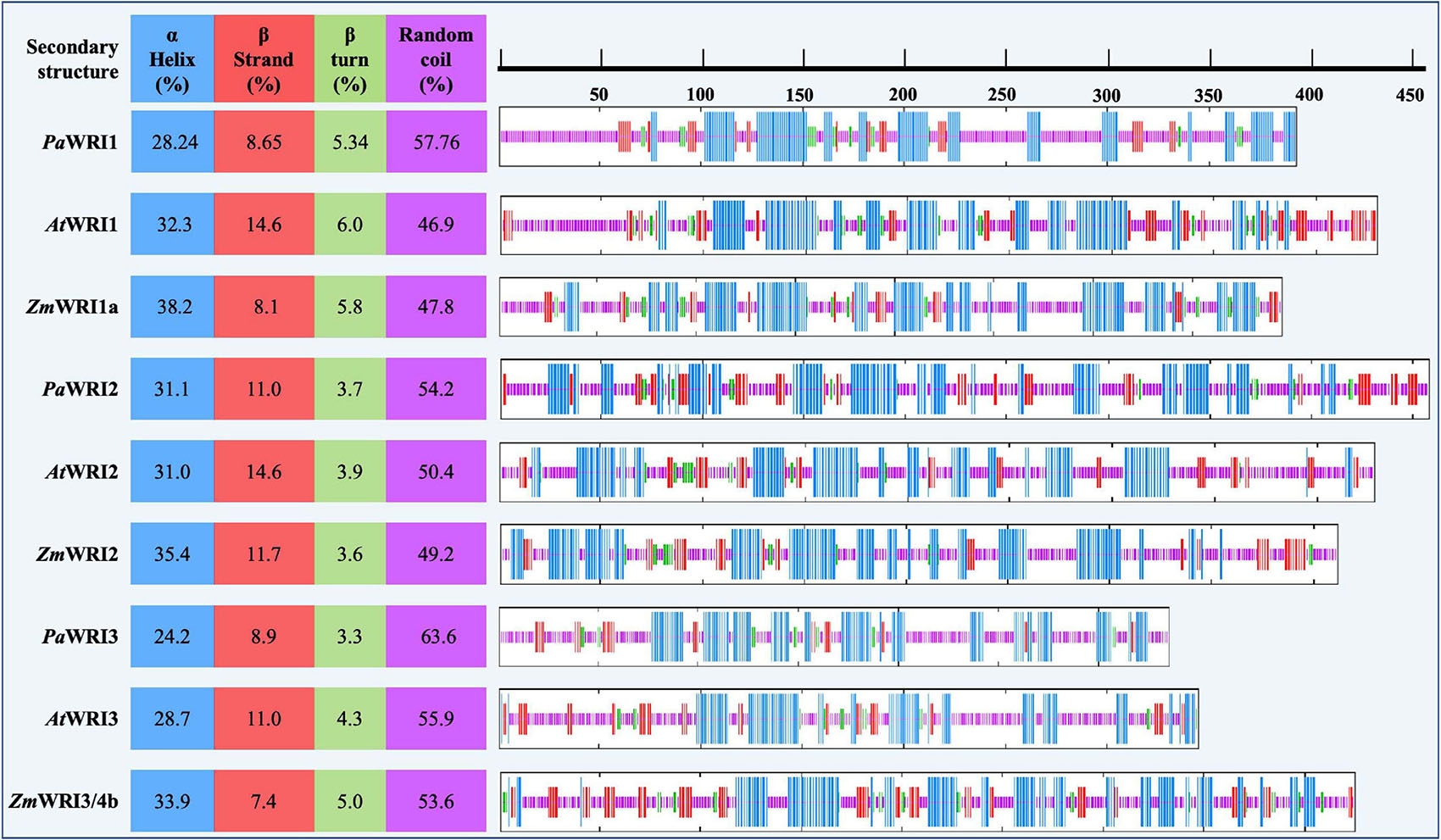
Figure 2. Proportion of the predicted secondary structures in WRI homologs. Position of each of the secondary structures α helix (blue), β strand (red), β turn (green), and random coil (pink) among all the WRI1 homologs are schematically represented on the right. The color-coded regions represent the position of the corresponding secondary structures. Proportion of each of the secondary structures is represented as a percentage value on the left, for each homolog.
Avocado WRI2 Lacks the C-Terminal IDR3 Region
Intrinsically disordered regions in the protein structure can prevent the formation of a hydrophobic core and result in improper folding (Uversky et al., 2000; Romero et al., 2001). In all three WRI homologs, in addition to the local random coil structures, amino acid bias also contributed to IDRs. The propensity for disorderliness was about two-fold higher among WRI homologs analyzed, compared to the mean value of 23% observed in the general Arabidopsis proteome (Oldfield et al., 2005). The overall disorder percentage among WRI1 orthologs was as follows: ZmWRI1a > AtWRI1 > PaWRI1, whereas for WRI2 orthologs, PaWRI2 > ZmWRI2 > AtWRI2 and for WRI3, ZmWRI3/4b > PaWRI3 > AtWRI3 (Table 1). While a higher percentage of disorderliness promotes flexibility and protein interaction, accessibility to proteases and the likelihood of protein degradation also increases. So, the relatively higher disordered values associated with both PaWRI2 and PaWRI3 and lower values with PaWRI1 might reflect a complex homeostasis, in which one protein could compensate for the degradation of the other to achieve sustained fatty acid biosynthesis during the prolonged period of TAG accumulation in avocado mesocarp.

Table 1. Details of the disorder value and the position of various intrinsically disordered regions (IDRs) and PEST motifs in WRI homologs.
The percentage disorder observed was reflected in the number of IDRs identified in each homolog. The protein sequences of PaWRI1, AtWRI1, and ZmWRI1 were predicted to contain three, three and four IDRs, respectively; all WRI2 and WRI3 proteins showed the presence of three IDRs (Figure 3). For further comparisons, the identified IDRs were named based on their position, similar to previously identified IDRs in AtWRI1, i.e., IDR1, IDR2, and IDR3. While IDR1 and IDR2 were unanimously predicted in all homologs, avocado WRI2 and WRI3 paralogs lacked the IDR3 in their C-terminal region. An additional short IDR (30-40 aa), which we referred to as IDR1a, was predicted in the linker region connecting the two AP2 domains (ZmWRI1a and all the three WRI3) and following the single AP2 domain in PaWRI2 proteins (Figure 3). Interestingly, the size of the IDRs was quite variable among the homologs (Table 1). Long IDRs (>30 aa) are common, comprising ∼33% of the eukaryotic proteome and affect transcription and translational activity, post-translational modifications, transportation and signal transduction activity, and also stability and degradation (Ward et al., 2004; Dyson and Wright, 2005; Van Der Lee et al., 2014). Shorter IDRs in the linker regions typically offer conformation flexibility that is needed for DNA-binding proteins (Tompa, 2005; Van Der Lee et al., 2014; Davey, 2019). The shorter IDR1a in the linker regions of some WRI orthologs, as well as the other longer IDRs around the AP2 domains are likely to provide conformational flexibility for an effective interaction with the target promoters while IDRs in the C-terminus might be associated with protein stability.

Figure 3. Graphical representation of the predicted IDRs in the WRI homologs. The predicted IDRs in all the WRI homologs are graphically plotted with the amino acid position on the x-axis and the PONDR score on the y-axis. Regions showing PONDR score > 0.5 are considered as IDRs.
Disordered N- and C-terminal regions are common in DNA-binding proteins; C-terminal IDR is especially prevalent in transcription factors (Van Der Lee et al., 2014). Previously characterized AtWRI1 IDR3 was shown to have a functional role in transactivation, although a three-fold increase in expression upon the truncation of IRD3 suggested that its presence likely also serves as a target for protein degradation pathways (Ma et al., 2015). IDR3 is absent in PaWRI2 although the other analyzed WRI2 orthologs have an IDR in their C-terminus (Figure 3 and Supplementary Figure 5). However, the PaWRI2 contains relatively long IDR1 and IDR2 (85 and 116 residues) compared to the respective orthologs in Arabidopsis and maize (57 aa and 100 aa, and 74 and 73 aa, respectively). These IDRs likely provide the necessary degree of transactivation capacity, while such function was lost in AtWRI2. The functional role of ZmWRI2 remains to be examined. Similarly, IDR3 is absent in yellow nutsedge (Cyperus esculentus) WRI1 (CeWRI1), although it retained its functional role in oil biosynthesis (Grimberg et al., 2015) and showed autoregulatory effect on the AtWRI1 promoter (Snell et al., 2019). Hence, it is possible that PaWRI2 is still able to activate the target genes in a similar manner to the transactivation mechanism shown by CeWRI1. On the other hand, long IDRs are also preferential targets for post-translational modification and protein binding that protect it from proteolytic cleavage (Dyson and Wright, 2005). The stability of AtWRI1 is enhanced upon interaction with either BTB/POZMATH1 (BPM1) or 14-3-3 proteins (Chen et al., 2013; Ma et al., 2016), although the complete interaction network is currently unknown. Hence, it is possible that the difference in functionality between PaWRI2 and AtWRI2 could be the result of distinct post-translational modifications and/or protein interactions.
The C-Terminal Region of Avocado WRI2 Lacks a PEST Motif
Among the WRI homologs examined, the C-terminal region downstream of the AP2 domain is highly variable (Figure 1). Nevertheless, this region does harbor some conserved features, without any particular trend in amino acid properties that are likely responsible for the regulation of the protein. Specifically, a region of the protein sequence that is enriched in proline (P), glutamic acid (E), serine (S), and threonine (T), referred to as PEST motifs occur among WRI homologs and were mostly associated with the IDRs (Ma et al., 2015). These PEST motifs are typically involved in proteasome-mediated protein turnover (García-Alai et al., 2006; Belizario et al., 2008). They are abundant among many key metabolic enzymes, transcription factors, protein kinases, and phosphatases in the cell, and function as a tool to maintain cellular homeostasis (Rechsteiner and Rogers, 1996). PEST-containing proteins make up a prevalent portion (∼25%) of the eukaryotic proteome and are enriched with disorder-promoting amino acids. The PEST motif associated with IDRs promotes proteolysis through ubiquitin-proteasome degradation or calpain cleavage (Gregory and Hann, 2000; Bordone and Campbell, 2002; Singh et al., 2006).
While all three WRI1 orthologs contained one PEST motif in their N-termini, AtWRI1 and PaWRI1 had one each, and ZmWRI1a had two, PEST motifs predicted in their respective C-terminal IDRs (Figure 4 and Table 1). However, the PEST motif in the C-terminal region of PaWRI1 has no potential phosphorylation site (Figure 4), and thus is an unlikely protease target. Interestingly, all WRI2 orthologs (including PaWRI2) and AtWRI3 lacked PEST motifs. Maize and avocado WRI3 orthologs, although lacking N-terminal PEST motifs, contained one and two each, respectively, embedded in their C-terminal IDRs (Figure 4 and Table 1). The association of PEST motifs with the IDR increases surface accessibility and favors it as a target for degradation signal attachment and phosphorylation (Singh et al., 2006). Additionally, while both N- and C-terminal PEST motifs are involved in proteolysis, a preference for C-terminal PEST motifs is noted in several proteins (Rechsteiner and Rogers, 1996; Singh et al., 2006). Among the WRI homologs, most PEST motifs were associated with the C-terminus and likely play a role in transcription factor stability control.
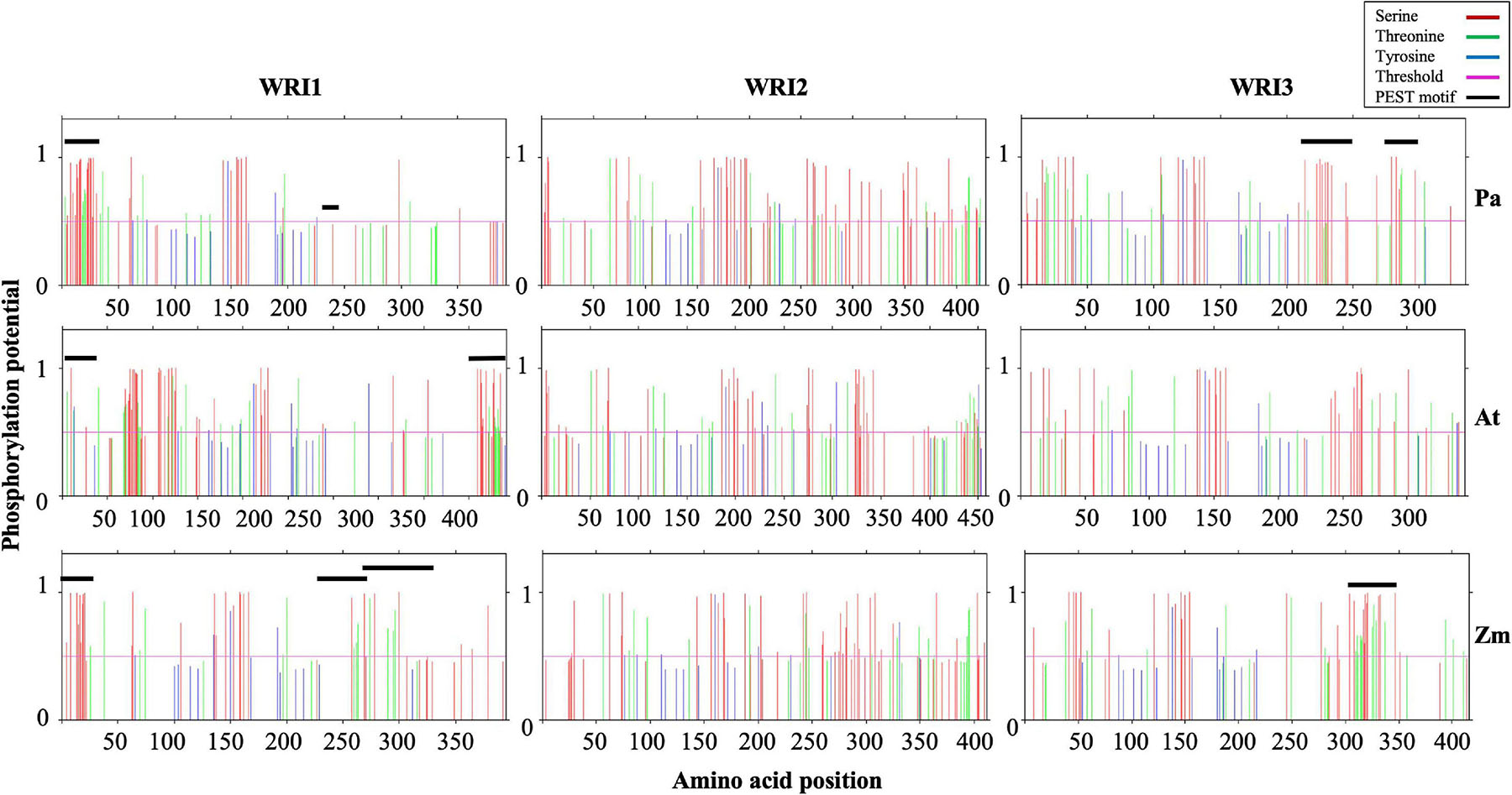
Figure 4. Schematic presentation of potential phosphorylation sites in the WRI homologs. Predicted phosphorylation potential for S (Red), T (green), and Y (blue) residue and their amino acid position are shown in the plots. The residues with potential higher than threshold value of 0.5 have higher probability of being phosphorylated. Residues that correspond to PEST motifs are also indicated by a black line.
The PEST motif associated with AtWRI1 IDR3 participates in phosphorylation-dependent protein degradation but was not critical for transactivation (Ma et al., 2015). Furthermore, transient expression of truncated AtWRI1 (1-397) in N. benthamiana leaves increased oil accumulation suggesting that the PEST motif plays a role in protein accumulation without affecting basic function (Ma et al., 2015). Hence, the absence of a C-terminal IDR-associated PEST motif in PaWRI2 might contribute to its increased stability and sustained activity, possibly contributing to high oil accumulation in avocado mesocarp. Although PaWRI3 transcript levels were high in avocado mesocarp during the period of oil accumulation, any effects of PEST motifs on transcript accumulation are currently unknown (Kilaru et al., 2015). The role of N-terminal PEST motifs in WRI homologs also has not been studied but is predicted to maintain homeostasis and protein interaction. Overall, our analyses suggest that lack of a PEST motif in the C-terminal region of PaWRI2, versus its presence in this region of PaWRI1 and PaWRI3, contributes to the mediation of the complex regulatory mechanism of homeostasis in avocado mesocarp tissue.
Both PaWRI1 and PaWRI2 Contain Phosphorylation Sites in the C-Terminal Region
Phosphorylation is among the major categories of post-translational modifications that control protein function. Phosphosites present in the long disordered regions in IDPs are preferentially phosphorylated by cellular kinases (Koike et al., 2020). To investigate the possible targets in our WRI protein set, we evaluated each of the putative residues Ser (S), Thr (T), and Tyr (Y) for phosphorylation modification potential using NetPhos 3.1 (Blom et al., 1999). The extensive S residues were most often identified as kinase target sites, followed by T and Y. Most of the identified phosphorylation sites overlapped with the predicted PEST motifs (Figure 5). Previous analysis of AtWRI1 identified four S residues in the IDR3-PEST
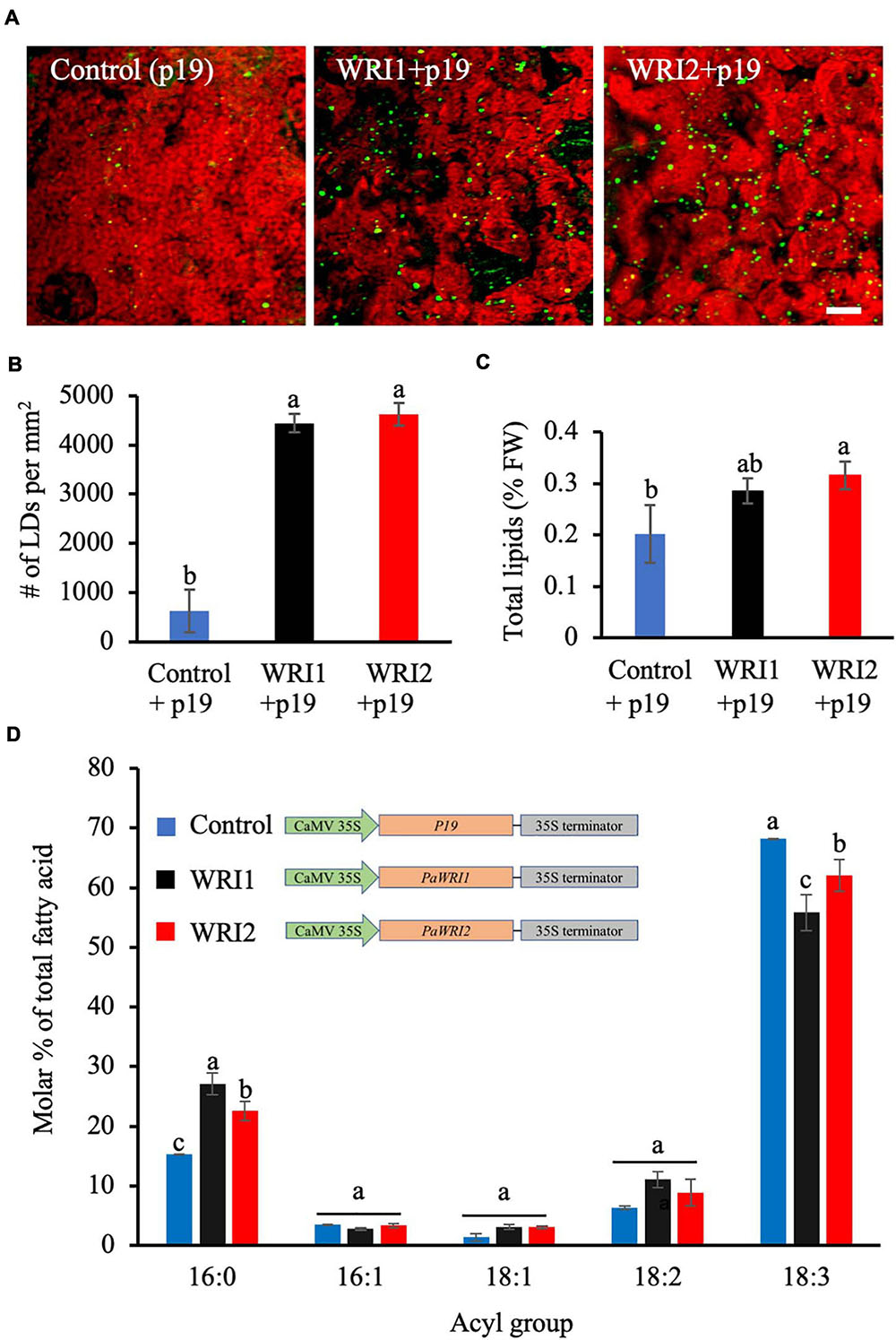
Figure 5. Lipid content and fatty acid composition of N. benthamiana leaves expressing PaWRI1 and PaWRI2. (A) Confocal images of accumulated lipid droplets (LDs) stained with Nile Red (green) in N. benthamiana leaves infected with Agrobacterium expressing control (p19 alone), PaWRI1 (+p19), and PaWRI2 (+p19) constructs. Scale bar corresponds to 20 μm. (B) Quantification of accumulated LDs per unit surface area of the leaf tissue. (C) Quantification of total lipid (TL) content. (D) fatty acid profile of total lipids extracted from the infiltrated leaves. Data represent mean ± sd (n = 3; p < 0.05).
motif as potential phosphorylation targets, and mutation of all these residues to Ala (A) (AtWRI14SA) resulted in increased stability and increased TAG accumulation in transiently expressed N. benthamiana leaves (Ma et al., 2015). Possibly, the Ser (S) residues are important and/or sufficient for the PEST motif recognition and phosphorylation by target enzymes to upregulate protein degradation. Since both PaWRI1 and PaWRI2 lack PEST motifs in their C-terminal ends, those regions did not show an enrichment of phosphorylation sites (Figure 4); these patterns thus differ significantly from those of AtWRI1 and AtWRI2. However, PaWRI2 was enriched with potential phosphorylation sites after amino acid position 350, which were absent in AtWRI2. Also, the region between the two AP2 domains in WRI1 and WRI3 orthologs and the region downstream of the AP2 domain in WRI2 orthologs were enriched with phosphorylation targets. Additionally, two KIN10-mediated phosphorylation target residues (T70 and S166, present in the first and second AP2 domains of AtWRI1) were also conserved in all WRI homologs except AtWRI2 and ZmWRI2 (Zhai et al., 2017). Although a large fraction of the identified phosphorylation sites remains unexplored, the high proportion of random coil structure and abundant IDR regions associated with each of the WRI proteins, including the avocado homologs, likely favor cellular protein kinase access, resulting in structural and/or functional modulation. Hence, phosphorylation, whether associated with residues in PEST domains or elsewhere, is also likely to play an important role in maintaining homeostasis of WRI paralogs in avocado mesocarp.
Distinct Features of PaWRI2 Are Likely Associated With Its Function
Previous studies reported the differentiation of avocado WRI2 from the other gene family members, likely due to an ancient gene duplication event. The loss of function in WRI2 from more recently evolved plant families such as Arabidopsis, suggests loss of selective pressure and enzyme function during evolution (Kilaru et al., 2015). We identified characteristic features of avocado WRI2 that might explain the retention of its functional role, through in silico analyses. All the WRI2 orthologs share only one AP2 domain, suggesting that it is functionally very important. Previous studies (Ma et al., 2013) suggested that the presence of a micro-exon, encoding the three residues “VYL,” is present in WRI1 orthologs from diverse plant species and is highly correlated with WRI protein functionality. PaWRI2 retains this exon, while AtWRI2 does not (Figure 1), suggesting that this element contributes to PaWRI2 protein function. Another unique difference between AtWRI2 and PaWRI2 is the absence of IDR3 but presence of IDR1a in PaWRI2 (Figure 3). Previous studies have reported that post-translational modifications in the C-terminal IDRs affect the stability of WRI1 proteins, for example, phosphorylation of the IDR3 PEST motif in AtWRI1 enhances protein degradation (Ma et al., 2015). As such, IDR3 in AtWRI2 might negatively affect its conformational flexibility and hence contribute to its loss of function. Conversely, in PaWRI2, the presence of IDR1a adjacent to the single AP2 domain (Figure 3) might contribute to productive interaction with target promoters. Furthermore, retention of function in PaWRI2
might be attained due to the presence of only one C-terminal IDR that does not contain PEST motifs (Figure 3 and Table 1), likely reducing the number of phosphorylation targets (Figure 4) and positively contributing to PaWRI2 protein stability.
These in silico analyses suggested many intriguing features that differentiate PaWRI2 from AtWRI2. Therefore, to confirm functionality in vivo, we performed transient expression assays in N. benthamiana leaves.
Unlike AtWRI2, the Expression of PaWRI2 Is Associated With Lipid Accumulation
Phylogenetic analyses of WRI1 homologs indicate that WRI2 has independently diverged from the remaining three orthologs, while WRI3 and WRI4 arose from a gene duplication event with WRI1 and evolved independently (Supplementary Figure 1; Kilaru et al., 2015). Among the four WRI1 orthologs in Arabidopsis, both AtWRI3 and AtWRI4, but not AtWRI2, shared functional similarity with AtWRI1 and were able to complement the wri1 mutant and restore seed oil biosynthesis to near-wild type levels (To et al., 2012). However, the uniquely high expression levels of PaWRI2 in avocado mesocarp during oil accumulation and distinct evolutionary separation from the remaining orthologs was intriguing (Kilaru et al., 2015). As such, to compare the functional role of PaWRI2 to that of PaWRI1 in oil accumulation, both genes were transiently expressed in N. benthamiana leaves. After seven days of co-infiltration of Agrobacterium expressing PaWRI1 or PaWRI2 and P19, a silencing−suppressor protein that enhances transgene expression (Naim et al., 2016), the leaves were harvested and analyzed for lipid accumulation. Interestingly, transient expression of both PaWRI1 and PaWRI2 resulted in a 7-fold increase in the number of LDs in N. benthamiana leaves (Figures 5A,B).
We further examined if these transcription factors affected fatty acid biosynthesis and thus lipid content in the leaf. Co-expression of PaWRI2/P19 generated a 1.5-fold increase in the total lipid content, relative to leaves expressing P19 alone (Figure 5C). The PaWRI2-mediated increase in lipid content, which was about 44% was greater than the 17-41% oil enhancement shown by the three transiently expressed isoforms of camelina CsWRI1 in N. benthamiana leaves (An et al., 2017). Typically, N. benthamiana leaves contain about 0.2 to 0.3% lipids by FW of which only 20-30% is TAG. Ectopic expression studies using WRI1 orthologs have reported increase in total lipids of which 80-90% was TAG (An et al., 2017). PaWRI1 expression resulted in higher LD numbers but only a smaller and statistically insignificant increase in total lipid content (Figure 5C).
Analysis of leaf total lipid fatty acid profile from samples expressing PaWRI1 or PaWRI2 further revealed a significant increase in 16:0 and decrease in 18:3 content, relative to the wild type leaves (Figure 5D). Although the role of WRI1 in oil accumulation has been unequivocally established, its ability to regulate fatty acid composition is not clear and is presumed to depend on the species, tissue type and its origin. For example, when camelina CsWRI1 was expressed in N. benthamiana leaves, the content of both 18:1 and 18:2 increased and 18:3 decreased, while in transgenic seeds of stably transformed Arabidopsis wri1 mutant plants, 18:3 content was highest and 18:1 remained low (An et al., 2017). Also, transient expression of WRI1 homologs from potato, poplar, nutsedge, oat, and Arabidopsis showed reduced 18:0 and higher proportions of 18:1, relative to the control leaves (Grimberg et al., 2015). Interestingly, although avocado mesocarp lipids are highly enriched in monounsaturated fatty acid (oleic acid, 18:1) (Kikuta and Erickson, 1968; Kilaru et al., 2015), transient expression of neither PaWRI nor PaWRI2 affected the oleic acid content in N. benthamiana leaves. Nevertheless, leaves expressing either PaWRI or PaWRI2 showed a ∼1.4 − 1.7-fold increase in 16:0 and a ∼0.8 − 0.9-fold decrease in 18:3 levels. With ∼20% reduction in 18:3, this effect was higher with PaWRI1 expression than with PaWRI2, where the decrease accounted for ∼10% (Figure 5D). Previous studies showed that transient expression of WRI1 homologs in N. benthamiana leaves led to downregulation of the transcripts encoding plastidial fatty acid desaturase 7 (FAD7) that is responsible for the desaturation of 18:2 to 18:3, and long chain acyl-CoA synthetase 1 and 2 (LACS 1, 2), which catalyze the synthesis of long chain acyl-CoA that serves as a substrate for TAG assembly (Lu et al., 2009; Zhang et al., 2012; Grimberg et al., 2015). Whether PaWRI1 and PaWRI2 transient expression caused increased 16:0 and decreased 18:3 content due to changes in FAD7, LACS1 and/or LACS2 expression remains to be examined. Together, the transient expression studies implicate that both PaWRI1 and PaWRI2 are functional and able to induce lipid accumulation in non-seed tissues and also affect fatty acid composition (Figures 5B,C). PaWRI2 enhanced lipid content and altered fatty acid composition more effectively than PaWRI1. These properties of PaWRI2 are functionally unique and contrast sharply to that of AtWRI2, which lacked any significant role in fatty acid biosynthesis (To et al., 2012). Considering the evolutionary position of avocado as a basal angiosperm, it is possible that PaWRI2 is structurally unique and evolved as an early regulator of fatty acid biosynthesis, while such ability was lost in AtWRI2 but retained by its paralogs.
Conclusion
Avocado mesocarp is one of the most oleate-rich sources of oil in the plant kingdom. Detailed molecular knowledge of the regulatory machinery responsible for such sustained fatty acid biosynthetic and TAG assembly activity during the long fruit development period remains unresolved for non-seed tissues. Additionally, while the role for WRI1 as a master regulator of oil biosynthesis in oil-rich seed tissues in several species is well established, its significance in non-seed tissues remains unclear. Although the three WRI2 orthologs analyzed in this study contained only one AP2 domain, reflecting an ancestral feature, an interruption in the domain by a 35 amino acid stretch in AtWRI2 was likely sufficient for loss of its function (Figure 1). Additionally, mutation in three amino acid residues in the AP2 domain of AtWRI2 that are important for DNA binding might have also affected its function (Figure 1). In contrast, structurally unique characteristics of PaWRI2, compared to its orthologs and paralogs, clarify its efficiency and functional novelty, despite containing only a single AP2 domain. PaWRI2 lacked IDR3 (Figure 4 and Table 1) and the C-terminal PEST motif (Figure 4), which is expected to increase protein stability and sustained activity. Additionally, while disorder-promoting amino acid composition bias resulting in random coil secondary structure was inherent in all the orthologs (Supplementary Figure 4), PaWRI2 displayed the highest disorder (Figure 2), suggesting maximum flexibility in interacting with target genes. Additionally, some conserved structural features such as the “VYL” micro exon and the KIN10 target residues (T70 and S166 identified in AtWRI1) suggest that PaWRI2 is involved in the maintenance of sugar-dependent lipid homeostasis in avocado mesocarp. Also, distinct phosphorylation targets that were predicted in the C-terminal region (Figure 4) might play a role in post-translational modifications, which can affect protein half-life, subcellular function, protein-protein interaction and homeostasis. Based on the previous RNA-seq analyses, it was clear that PaWRI1, 2 and 3 are highly expressed during the extended period of oil accumulation. In our study, we show that PaWRI2, unlike its ortholog AtWRI2, was functional and induced oil accumulation in and altered the fatty acid profile of transiently infected leaf tissue more effectively than PaWRI1 (Figure 5). Although the WRI2 orthologs formed a monophyletic clade as a result of independent divergence from their other paralogs, loss of function events among some orthologs were likely random. Bioinformatic analyses support the idea that all three mesocarp-expressed WRI paralogs, including PaWRI2, are functional and likely contribute to coordinated and sustained oil biosynthesis. Further studies are pertinent to establish more details of the underlying regulatory network in oil biosynthesis in non-seed tissues in plants.
Data Availability Statement
The original contributions presented in the study are available publicly at NCBI (https://www.ncbi.nlm.nih.gov/): PaWRI (MZ322905), PaWRI2 (MZ322906), PaWRI3 (MZ322907). The protein sequences used for data analyses are available publicly at NCBI (https://www.ncbi.nlm.nih.gov/): AtWRI1 (NP_001030857.1), AtWRI2 (NP_001189729.1), AtWRI3 (NP_563990.1), AtWRI4 (NP_178088.2), ZmWRI1a (NP_001137064.1), ZmWRI2 (NP_001145827.1), ZmWRI3/4b (XP_008651355.1), AtrWRI1 (XP_011620741.1), AtrWRI2 (XP_020526667.1), AtrWRI3 (XP_006845421.1), VvWRI1 (XP_010659009.1), VvWRI2 (XP_002284093.1), VvWRI3 (XP_010652396.1), RcWRI1 (NP_001310691.1), RcWRI2 (XP_015581065.1), RcWRI3 (NP_001310645.1), RcWRI4 (XP_015573687.1), OsWRI1 (BAD68218.1), OsWRI2 (XP_015638088.1), OsWRI3 (XP_015617951.1), OsWRI4 (XP_015619972.1), BrWRI1 (XP_009116120.1), BrWRI2 (XP_009133508.1), BrWRI3 (XP_009149048.1), BrWRI4 (XP_009106692.1), EgWRI1 (AHX71676.1), EgWRI2 (XP_010938537.1), EgWRI3 (XP_010912920.2), EgWRI4 (XP_010914036.1), PtWRI1 (XP_024465221.1), PtWRI2 (XP_024458881.1), PtWRI3 (XP_002297679.2), and PtWRI4 (XP_024467077.1).
Author Contributions
JB and SB conducted the bioinformatic analyses. SB and MR performed the cloning. MR conducted the transient expression assays. JB and MR performed the statistical analysis. JB, JS, and AK wrote the manuscript with assistance from MR and SB. All authors read and approved the final manuscript and contributed to the research design and data analyses.
Funding
This research was supported in part by minor and major grants from research and development committee, East Tennessee State University (ETSU) awarded to AK. Additionally, JB, MR, and SB received funds from School of Graduate Studies, ETSU. SB and MR received Sigma Xi GIAR Award. SB also received Fraley’s Memorial Research Award from the Department of Biological Sciences, ETSU.
Disclaimer
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement from the United States Department of Agriculture. The USDA is an equal opportunity provider and employer.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.648494/full#supplementary-material
Supplementary Figure 1 | Phylogenetic analysis and percentage identity among WRI1 homologs. (A) Phylogenetic analysis of WRI1 homologs in Arabidopsis thaliana (At), Zea mays (Zm), Amborella trichopoda (Atr), Populus trichocarpa (Pt), Ricinus communis (Rc), Brassica rapa (Br), Elaeis guineensis (Eg), Vitis vinifera (Vv), Oryza sativa (Os) and Persea americana (Pa) was carried out using UPGMA method (see Supplementary Table 1). An AP2-domain transcription factor from Chlamydomonas reinhardtii (CrAP2) was used as outgroup. Percentages indicate bootstrap values for 1000 replicates. The WRI-orthologs are shown in color; WRI1 (black), WRI2 (pink), WRI3 (orange), WRI4 (blue). (B) Percentage of amino acid sequence identity among the WRI1 homologs from avocado (Pa), Arabidopsis (At), and maize (Zm).
Supplementary Figure 2 | Sequence alignment of WRI1, WRI2, and WRI3 from various species showing conserved amino acids. Multiple sequence alignment of primary sequences of WRI1, 2, and 3 from species as described in Supplementary Figure 1 are shown in a color-coded format; red and blue colors represent the most (10, indicated by asterisk) and least (0) conserved regions, respectively on a scale of 0 to 10. Positions of the AP2 domains are shown as pink color bars under the sequences and the amino acid residues in the AP2 domain important for DNA interaction are represented by (▲).
Supplementary Figure 3 | Multiple sequence alignment of WRI1 homologs from the select three species. The alignment shows KIN10 target sites (red box) in the WRI1 homologs with reference to T70 and S166 identified in AtWRI1. The S166 residue is mutated in AtWRI2 and ZmWRI2. The highlighted amino acid residues correspond to the PEST motifs.
Supplementary Figure 4 | Amino acid composition profile of WRI1 homologs. Amino acid composition profile of WRI1 homologs showing the enriched and depleted amino acids as compared to SwissProt51 database. The amino acids are arranged in the increasing order of hydrophobic character on the x-axis and the y-axis represents the composition profile. The values >0 represents an enrichment and <0 represents a depletion of corresponding amino acids in the protein sequence. The significantly (p < 0.05) enriched (green) and depleted (blue) residues are denoted in color.
Supplementary Figure 5 | Graphical representation of the predicted IDRs. The predicted IDRs in all the WRI homologs are graphically plotted with the amino acid position on the x-axis and the PONDR score on the y-axis. Regions showing PONDR score > 0.5 are considered as IDRs. For AtrWRI1 and AtrWRI2, although the figures are not showing the PONDR score, the predicted IDRs by the same software are as follows: 30-53, 137-160, 234-348 for WRI1 and 12-84, 168-190 for WRI2. Both of them have the C-terminal IDR.
Supplementary Table 1 | List of online tools used for various predictive structural analysis of WRI1 orthologs.
References
Allen, M. D., Yamasaki, K., Ohme-takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a β -sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5484–5496. doi: 10.1093/emboj/17.18.5484
An, D., Kim, H., Ju, S., Go, Y. S., Kim, H. U., and Suh, M. C. (2017). Expression of camelina WRINKLED1 isoforms rescue the seed phenotype of the arabidopsis wri1 mutant and increase the triacylglycerol content in Tobacco leaves. Front. Plant Sci. 8, 1–13. doi: 10.3389/fpls.2017.00034
Baud, S., and Lepiniec, L. (2009). Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol. Biochem. 47, 448–455. doi: 10.1016/j.plaphy.2008.12.006
Baud, S., Mendoza, M. S., To, A., Harscoët, E., Lepiniec, L., and Dubreucq, B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50, 825–838. doi: 10.1111/j.1365-313X.2007.03092.x
Baud, S., Wuillème, S., To, A., Rochat, C., and Lepiniec, L. (2009). Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J. 60, 933–947. doi: 10.1111/j.1365-313X.2009.04011.x
Belizario, J., Alves, J., Garay-Malpartida, M., and Occhiucci, J. (2008). Coupling Caspase Cleavage and Proteasomal Degradation of Proteins Carrying PEST Motif. Curr. Protein Pept. Sci. 9, 210–220. doi: 10.2174/138920308784534023
Blom, N., Gammeltoft, S., and Brunak, S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362. doi: 10.1006/jmbi.1999.3310
Bordone, L., and Campbell, C. (2002). DNA ligase III is degraded by calpain during cell death induced by DNA-damaging agents. J. Biol. Chem. 277, 26673–26680. doi: 10.1074/jbc.M112037200
Bourgis, F., Kilaru, A., Cao, X., Ngando-Ebongue, G. F., Drira, N., Ohlrogge, J. B., et al. (2011). Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning (Proceedings of the National Academy of Sciences of the United States of America (2011) 108, 30 (12527-12532) DOI. Proc. Natl. Acad. Sci. U. S. A. 108:18186. doi: 10.1073/pnas.1115243108
Casal, J. J., Davis, S. J., Kirchenbauer, D., Viczian, A., Yanovsky, M. J., Clough, R. C., et al. (2002). The serine-rich N-terminal domain of oat phytochrome a helps regulate light responses and subnuclear localization of the photoreceptor. Plant Physiol. 129, 1127–1137. doi: 10.1104/pp.010977
Cernac, A., and Benning, C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40, 575–585. doi: 10.1111/j.1365-313X.2004.02235.x
Chanderbali, A. S., Albert, V. A., Ashworth, V. E. T. M., Clegg, M. T., Litz, R. E., Soltis, D. E., et al. (2008). Persea americana (avocado): Bringing ancient flowers to fruit in the genomics era. BioEssays 30, 386–396. doi: 10.1002/bies.20721
Chen, L., Lee, J. H., Weber, H., Tohge, T., Witt, S., Roje, S., et al. (2013). Arabidopsis BPM proteins function as substrate adaptors to a CULLIN3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 25, 2253–2264. doi: 10.1105/tpc.112.107292
Combet, C., Blanchet, C., Geourjon, C., and Deléage, G. (2000). NPS@: Network protein sequence analysis. Trends Biochem. Sci. 25, 147–150. doi: 10.1016/S0968-0004(99)01540-6
Davey, N. E. (2019). The functional importance of structure in unstructured protein regions. Curr. Opin. Struct. Biol. 56, 155–163. doi: 10.1016/j.sbi.2019.03.009
Divi, U. K., Zhou, X. R., Wang, P., Butlin, J., Zhang, D. M., Liu, Q., et al. (2016). Deep sequencing of the fruit transcriptome and lipid accumulation in a non-seed tissue of Chinese tallow, a potential biofuel crop. Plant Cell Physiol. 57, 125–137. doi: 10.1093/pcp/pcv181
Dyson, H. J., and Wright, P. E. (2005). Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208. doi: 10.1038/nrm1589
Eisenberg, D., Wilcox, W., and McLachlan, A. D. (1986). Hydrophobicity and amphiphilicity in protein structure. J. Cell. Biochem. 31, 11–17. doi: 10.1002/jcb.240310103
Fei, W., Yang, S., Hu, J., Yang, F., Qu, G., Peng, D., et al. (2020). Research advances of WRINKLED1 (WRI1) in plants. Funct. Plant Biol. 47, 185–194. doi: 10.1071/FP19225
Focks, N., and Benning, C. (1998). wrinkled1: A Novel, Low-Seed-Oil Mutant of Arabidopsis with a Deficiency in the Seed-Specific Regulation of Carbohydrate Metabolism. Plant Physiol. 118, 91–101. doi: 10.1104/pp.118.1.91
Fukuda, N., Ikawa, Y., Aoyagi, T., and Kozaki, A. (2013). Expression of the genes coding for plastidic acetyl-CoA carboxylase subunits is regulated by a location-sensitive transcription factor binding site. Plant Mol. Biol. 82, 473–483. doi: 10.1007/s11103-013-0075-7
Gao, J., and Xu, D. (2012). Correlation Between Posttranslational Modification and Intrinsic Disorder in Protein. Pac Symp Biocomput. 2012, 94–103.
García-Alai, M. M., Gallo, M., Salame, M., Wetzler, D. E., McBride, A. A., Paci, M., et al. (2006). Molecular basis for phosphorylation-dependent, PEST-mediated protein turnover. Structure 14, 309–319. doi: 10.1016/j.str.2005.11.012
Gregory, M. A., and Hann, S. R. (2000). c-Myc Proteolysis by the Ubiquitin-Proteasome Pathway: Stabilization of c-Myc in Burkitt’s Lymphoma Cells. Mol. Cell. Biol. 20, 2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000
Grimberg, Å, Carlsson, A. S., Marttila, S., Bhalerao, R., and Hofvander, P. (2015). Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 15:579. doi: 10.1186/s12870-015-0579-1
Hara, A., and Radin, N. S. (1978). Lipid Extraction of Tissues with a Low-Toxicity Solvent. Anal. Biochem. 90, 420–426. doi: 10.1016/0003-2697(78)90046-5
Ibarra-Laclette, E., Méndez-Bravo, A., Pérez-Torres, C. A., Albert, V. A., Mockaitis, K., Kilaru, A., et al. (2015). Deep sequencing of the Mexican avocado transcriptome, an ancient angiosperm with a high content of fatty acids. BMC Genomics 16, 1–18. doi: 10.1186/s12864-015-1775-y
Ji, X., Mao, X., Hao, Q., Liu, B., Xue, J., and Li, R. (2018). Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves. Int. J. Mol. Sci. 19:46. doi: 10.3390/ijms19010146
Keereetaweep, J., Liu, H., Zhai, Z., and Shanklin, J. (2018). Biotin Attachment Domain-Containing Proteins Irreversibly Inhibit Acetyl CoA Carboxylase. Plant Physiol. 177, 208–215. doi: 10.1104/pp.18.00216
Khan, S. H., and Kumar, R. (2009). An Overview of the Importance of Conformational Flexibility in Gene Regulation by the Transcription Factors. J. Biophys. 2009:210485. doi: 10.1155/2009/210485
Kikuta, Y., and Erickson, L. C. (1968). Seasonal Changes of Avocado Lipids During Fruit Development and Storage. Calif. Avocado Soc. 52, 102–108.
Kilaru, A., Cao, X., Dabbs, P. B., Sung, H. J., Rahman, M. M., Thrower, N., et al. (2015). Oil biosynthesis in a basal angiosperm: Transcriptome analysis of Persea Americana mesocarp. BMC Plant Biol. 15:586. doi: 10.1186/s12870-015-0586-2
Koike, R., Amano, M., Kaibuchi, K., and Ota, M. (2020). Protein kinases phosphorylate long disordered regions in intrinsically disordered proteins. Protein Sci. 29, 564–571. doi: 10.1002/pro.3789
Kong, Q., Yuan, L., and Ma, W. (2019). WRINKLED1, a “Master Regulator” in Transcriptional Control of Plant Oil Biosynthesis. Plants 8:238. doi: 10.3390/plants8070238
Krizek, B. A. (2003). AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain. Nucleic Acids Res. 31, 1859–1868. doi: 10.1093/nar/gkg292
Kuczynski, C., McCorkle, S., Keereetaweep, J., Shanklin, J., and Schwender, J. (2020). An Expanded Role for WRINKLED1 Metabolic Control Based on Combined Phylogenetic and Biochemical Analyses. bioRxiv, 2020.01.28.923292 [preprint] doi: 10.1101/2020.01.28.923292
Liu, H., Zhai, Z., Kuczynski, K., Keereetaweep, J., Schwender, J., and Shanklin, J. (2019). WRINKLED1 Regulates BIOTIN ATTACHMENT DOMAIN-CONTAINING Proteins that Inhibit Fatty Acid Synthesis. Plant Physiol. 181, 55–62. doi: 10.1104/pp.19.00587
Liu, J., Hua, W., Zhan, G., Wei, F., Wang, X., Liu, G., et al. (2010). Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 48, 9–15. doi: 10.1016/j.plaphy.2009.09.007
Liu, J., Perumal, N. B., Oldfield, C. J., Su, E. W., Uversky, V. N., and Dunker, A. K. (2006). Intrinsic disorder in transcription factors. Biochemistry 45, 6873–6888. doi: 10.1021/bi0602718
Lu, S., Song, T., Kosma, D. K., Parsons, E. P., Rowland, O., and Jenks, M. A. (2009). Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 59, 553–564. doi: 10.1111/j.1365-313X.2009.03892.x
Ma, W., Kong, Q., Arondel, V., Kilaru, A., Bates, P. D., Thrower, N. A., et al. (2013). WRINKLED1, A Ubiquitous Regulator in Oil Accumulating Tissues from Arabidopsis Embryos to Oil Palm Mesocarp. PLoS One 8, 1–13. doi: 10.1371/journal.pone.0068887
Ma, W., Kong, Q., Grix, M., Mantyla, J. J., Yang, Y., Benning, C., et al. (2015). Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 83, 864–874. doi: 10.1111/tpj.12933
Ma, W., Kong, Q., Mantyla, J. J., Yang, Y., Ohlrogge, J. B., and Benning, C. (2016). 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J. 88, 228–235. doi: 10.1111/tpj.13244
Maeo, K., Tokuda, T., Ayame, A., Mitsui, N., Kawai, T., Tsukagoshi, H., et al. (2009). An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 60, 476–487. doi: 10.1111/j.1365-313X.2009.03967.x
Mano, F., Aoyanagi, T., and Kozaki, A. (2019). Atypical Splicing Accompanied by Skipping Conserved Micro-exons Produces Unique WRINKLED1, An AP2 Domain Transcription Factor in Rice Plants. Plants 8:207. doi: 10.3390/plants8070207
Naim, F., Shrestha, P., Singh, S. P., Waterhouse, P. M., and Wood, C. C. (2016). Stable expression of silencing-suppressor protein enhances the performance and longevity of an engineered metabolic pathway. Plant Biotechnol. J. 14, 1418–1426. doi: 10.1111/pbi.12506
Okamuro, J. K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K. D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 94, 7076–7081. doi: 10.1073/pnas.94.13.7076
Oldfield, C. J., Cheng, Y., Cortese, M. S., Brown, C. J., Uversky, V. N., and Bunker, A. K. (2005). Comparing and combining predictors of mostly disordered proteins. Biochemistry 44, 1989–2000. doi: 10.1021/bi047993o
Peng, K., Vucetic, S., Radivojac, P., Brown, C. J., Dunker, A., and Obradovic, Z. (2005). Optimizing Long Intrinsic Disorder Predictors With Protein Evolutionary Information. J. Bioinform. Comput. Biol. 3, 35–60. doi: 10.1142/s0219720005000886
Pouvreau, B., Baud, S., Vernoud, V., Morin, V., Py, C., Gendrot, G., et al. (2011). Duplicate Maize Wrinkled1 Transcription Factors Activate Target Genes Involved in Seed Oil Biosynthesis. Plant Physiol. 156, 674–686. doi: 10.1104/pp.111.173641
Rahman, M., Liu, Q., Zhou, X.-R., and Singh, S. P. (2016). Oil-rich nonseed tissues for enhancing plant oil production. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 11, 1–11. doi: 10.1079/PAVSNNR201611021
Rechsteiner, M., and Rogers, S. W. (1996). PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21, 267–271. doi: 10.1016/S0968-0004(96)10031-1
Rendón-Anaya, M., Ibarra-Laclette, E., Méndez-Bravo, A., Lan, T., Zheng, C., Carretero-Paulet, L., et al. (2019). The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. U. S. A. 116, 17081–17089. doi: 10.1073/pnas.1822129116
Romero, P., Obradovic, Z., Li, X., Garner, E. C., Brown, C. J., and Dunker, A. K. (2001). Sequence complexity of disordered protein. Proteins 42, 38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3
Ruuska, S. A., Girke, T., Benning, C., and Ohlrogge, J. B. (2002). Contrapuntal Networks of Gene Expression during Arabidopsis Seed Filling. Plant Cell 14, 1191–1206. doi: 10.1105/tpc.000877
Shockey, J., Mason, C., Gilbert, M., Cao, H., Li, X., Cahoon, E., et al. (2015). Development and analysis of a highly flexible multi-gene expression system for metabolic engineering in Arabidopsis seeds and other plant tissues. Plant Mol. Biol. 89, 113–126. doi: 10.1007/s11103-015-0355-5
Simossis, V. A., and Heringa, J. (2003). The PRALINE online server: Optimising progressive multiple alignment on the web. Comput. Biol. Chem. 27, 511–519. doi: 10.1016/j.compbiolchem.2003.09.002
Singh, G. P., Ganapathi, M., Sandhu, K. S., and Dash, D. (2006). Intrinsic unstructuredness and abundance of PEST motifs in eukaryotic proteomes. Proteins Struct. Funct. Genet. 62, 309–315. doi: 10.1002/prot.20746
Snell, P., Grimberg, Carlsson, A. S., and Hofvander, P. (2019). WRINKLED1 is subject to evolutionary conserved negative autoregulation. Front. Plant Sci. 10:387. doi: 10.3389/fpls.2019.00387
Tang, T., Du, C., Song, H., Aziz, U., Wang, L., Zhao, C., et al. (2019). Genome-wide analysis reveals the evolution and structural features of WRINKLED1 in plants. Mol. Genet. Genomics 294, 329–341. doi: 10.1007/s00438-018-1512-8
To, A., Joubès, J., Barthole, G., Lécureuil, A., Scagnelli, A., Jasinski, S., et al. (2012). WRINKLED Transcription Factors Orchestrate Tissue-Specific Regulation of Fatty Acid Biosynthesis in Arabidopsis. Plant Cell 24, 5007–5023. doi: 10.1105/tpc.112.106120
Tompa, P. (2005). The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579, 3346–3354. doi: 10.1016/j.febslet.2005.03.072
Uversky, V. N., and Dunker, A. K. (2010). Understanding protein non-folding. Biochim. Biophys. Acta 1804, 1231–1264. doi: 10.1016/j.bbapap.2010.01.017
Uversky, V. N., Gillespie, J. R., and Fink, A. L. (2000). Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins Struct. Funct. Genet. 41, 415–427. doi: 10.1002/1097-0134(20001115)41:3
Vacic, V., Uversky, V. N., Dunker, A. K., and Lonardi, S. (2007). Composition Profiler: A tool for discovery and visualization of amino acid composition differences. BMC Bioinform. 8, 1–7. doi: 10.1186/1471-2105-8-211
Van Der Lee, R., Buljan, M., Lang, B., Weatheritt, R. J., Daughdrill, G. W., Dunker, A. K., et al. (2014). Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631. doi: 10.1021/cr400525m
Ward, J. J., Sodhi, J. S., McGuffin, L. J., Buxton, B. F., and Jones, D. T. (2004). Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J. Mol. Biol. 337, 635–645. doi: 10.1016/j.jmb.2004.02.002
Wood, C. C., Petrie, J. R., Shrestha, P., Mansour, M. P., Nichols, P. D., Green, A. G., et al. (2009). A leaf-based assay using interchangeable design principles to rapidly assemble multistep recombinant pathways. Plant Biotechnol. J. 7, 914–924. doi: 10.1111/j.1467-7652.2009.00453.x
Zhai, Z., Liu, H., and Shanklin, J. (2017). Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell 29, 871–889. doi: 10.1105/tpc.17.00019
Keywords: AP2 domain, Arabidopsis thaliana, fatty acids, mesocarp, Persea americana, transcription factor, triacylglycerol, WRINKLED1
Citation: Behera JR, Rahman MM, Bhatia S, Shockey J and Kilaru A (2021) Functional and Predictive Structural Characterization of WRINKLED2, A Unique Oil Biosynthesis Regulator in Avocado. Front. Plant Sci. 12:648494. doi: 10.3389/fpls.2021.648494
Received: 31 December 2020; Accepted: 03 May 2021;
Published: 08 June 2021.
Edited by:
Mie Shimojima, Tokyo Institute of Technology, JapanReviewed by:
Jin-Song Zhang, State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology (CAS), ChinaKoichi Kobayashi, Osaka Prefecture University, Japan
Copyright © 2021 Behera, Rahman, Bhatia, Shockey and Kilaru. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aruna Kilaru, a2lsYXJ1QGV0c3UuZWR1
†These authors have contributed equally to this work
 Jyoti R. Behera
Jyoti R. Behera Md. Mahbubur Rahman
Md. Mahbubur Rahman Shina Bhatia
Shina Bhatia Jay Shockey3
Jay Shockey3 Aruna Kilaru
Aruna Kilaru