Abstract
Marine diatoms, the most successful photoautotrophs in the ocean, efficiently sequester a significant part of atmospheric CO2 to the ocean interior through their participation in the biological carbon pump. However, it is poorly understood how marine diatoms fix such a considerable amount of CO2, which is vital information toward modeling their response to future CO2 levels. The Tara Oceans expeditions generated molecular data coupled with in situ biogeochemical measurements across the main ocean regions, and thus provides a framework to compare diatom genetic and transcriptional flexibility under natural CO2 variability. The current study investigates the interlink between the environmental variability of CO2 and other physicochemical parameters with the gene and transcript copy numbers of five key enzymes of diatom CO2 concentration mechanisms (CCMs): Rubisco activase and carbonic anhydrase (CA) as part of the physical pathway, together with phosphoenolpyruvate carboxylase, phosphoenolpyruvate carboxykinase, and malic enzyme as part of the potential C4 biochemical pathway. Toward this aim, we mined >200 metagenomes and >220 metatranscriptomes generated from samples of the surface layer of 66 globally distributed sampling sites and corresponding to the four main size fractions in which diatoms can be found: 0.8–5 μm, 5–20 μm, 20–180 μm, and 180–2,000 μm. Our analyses revealed that the transcripts for the enzymes of the putative C4 biochemical CCM did not in general display co-occurring profiles. The transcripts for CAs were the most abundant, with an order of magnitude higher values than the other enzymes, thus implying the importance of physical CCMs in diatom natural communities. Among the different classes of this enzyme, the most prevalent was the recently characterized iota class. Consequently, very little information is available from natural diatom assemblages about the distribution of this class. Biogeographic distributions for all the enzymes show different abundance hotspots according to the size fraction, pointing to the influence of cell size and aggregation in CCMs. Environmental correlations showed a complex pattern of responses to CO2 levels, total phytoplankton biomass, temperature, and nutrient concentrations. In conclusion, we propose that biophysical CCMs are prevalent in natural diatom communities.
Introduction
Diatoms are among the most successful and diversified eukaryotic photoautotrophs in the present day ocean (Armbrust, 2009; Malviya et al., 2016; Pierella Karlusich et al., 2020). Their fast growth rates in high-nutrient environments and comparatively large sizes make them important contributors to organic carbon production. On an annual scale, marine diatoms fix 10–20 billion metric tons of inorganic carbon (comparable to all global rainforests combined), corresponding to up to 40% of the total marine primary production and as much as 20% of the total primary production on Earth (Field et al., 1998; Smetacek, 1999; Granum et al., 2005; Jin et al., 2006; Falkowski and Raven, 2013; Tréguer et al., 2018). Thus, diatoms are main contributors to marine food chains and in sequestering atmospheric CO2 to the ocean interior through gravitational sinking of particles (biological carbon pump) (Figure 1A), and hence have high biogeochemical significance (Tréguer et al., 2018; Boyd et al., 2019). Diatoms possess a peculiar gene complement derived from green and red algal sources, and have many genes in common with animals and bacteria (Bowler et al., 2008; Moustafa et al., 2009; Dorrell et al., 2017, 2021), mostly owing to their chimeric evolutionary origins as well as to horizontal gene transfer events (Armbrust, 2009; Dorrell et al., 2021). It is believed that these genes have enabled them to develop unique and highly efficient carbon (Schoefs et al., 2017) and nitrogen metabolism pathways (Wilhelm et al., 2006; Busseni et al., 2019). In the present context of climate change and the substantial anthropogenic perturbations in the ocean (increasing CO2 and temperature, acidification, disturbances in nutrient cycles, etc.), a key question is how marine diatoms will respond. In order to do this, a clear understanding of diatom carbon metabolism is required.
FIGURE 1
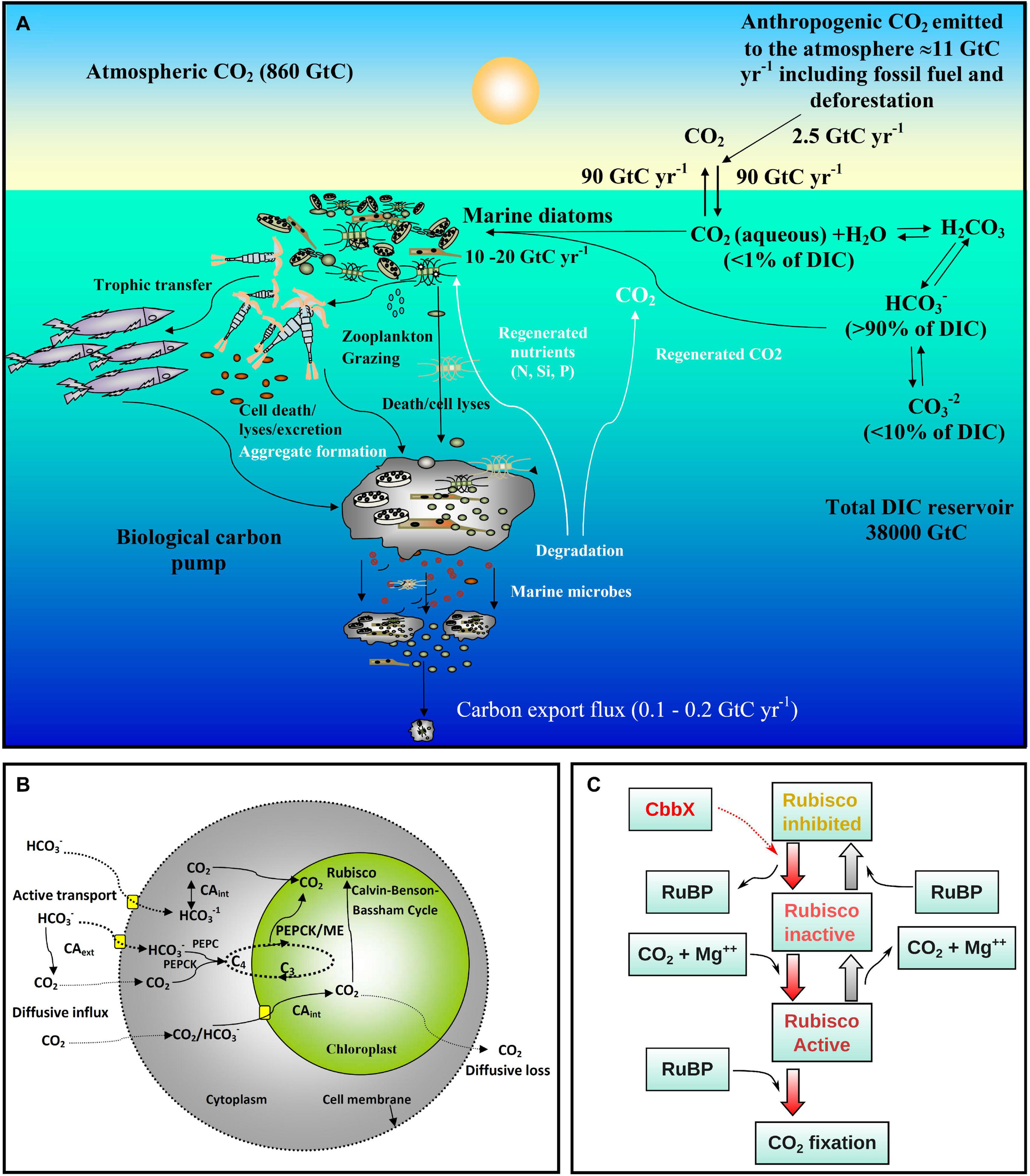
Overview of ocean carbon cycle and diatom carbon dioxide concentration mechanisms. (A) Schematic representation of the ocean carbon cycle depicting the role of marine diatoms in the biological carbon pump. The anthropogenic CO2 emission to the atmosphere (mainly generated by fossil fuel burning and deforestation) is nearly 11 Gigaton carbon (GtC) per year, of which almost 2.5 GtC is taken up by the surface ocean. In surface seawater (pH 8.1–8.4), bicarbonate (HCO3–) and carbonate ions (CO32–) constitute nearly 90 and <10% of dissolved inorganic carbon (DIC) respectively, while dissolved CO2 (CO2 aqueous) contributes <1%. Despite this low level of CO2 in the ocean and its slow diffusion rate in water, diatoms fix 10–20 GtC annually via photosynthesis thanks to their carbon dioxide concentration mechanisms (CCMs), allowing them to sustain food chains. In addition, 0.1–1% of this organic material produced in the euphotic layer sinks down as particles, thus transferring the surface carbon toward the deep ocean and sequestering atmospheric CO2 for thousands of years or longer. The remaining organic matter is remineralized through respiration. Thus, diatoms are one of the main players in this biological carbon pump, which is arguably the most important biological mechanism in the Earth System allowing CO2 to be removed from the carbon cycle for very long period. Based on data from Friedlingstein et al., 2020. (B) Schematic representation of the CCMs in diatoms. The low levels of CO2 in the ocean and its slow diffusion rate in water have led diatoms and other photosynthetic organisms to evolve CCMs that utilize the higher concentrations of HCO3–. The biophysical CCM consists of various bicarbonate transporters and carbonic anhydrases (CAs) that serve to increase the CO2 flux balance toward the pyrenoid, a low CO2-permeable subcellular compartment in the chloroplast containing most of the Rubisco. In addition, some diatoms may also have a biochemical (C4-like) CCM involving phosphoenolpyruvate carboxylase (PEPC), phosphoenolpyruvate carboxykinase (PEPCK) and/or malic enzyme (ME). (C) Schematic presentation of Rubisco activation by CbbX in diatoms and other phototrophs with red-type Rubisco. CbbX functions as a mechanochemical motor protein and uses the energy from ATP hydrolysis to modify the structure of Rubisco. This process facilitates the dissociation of inhibitory sugar phosphates [ribulose-1,5-bisphosphate (RuBP) and others] from the active site of Rubisco.
The key carbon fixing enzyme, ribulose-1,5-bisphosphate carboxylase/oxygenase, Rubisco, is one of the most abundant proteins on Earth and is responsible for 100 billion tons of carbon fixation annually (Erb and Zarzycki, 2018; Bar-On and Milo, 2019). Remarkably however, Rubisco is highly inefficient because it shows specificity toward O2 which competes with CO2. This is believed to be a remnant of its evolution at a time when oxygen levels were minimal, and leads to a wasteful process called photorespiration (Poudel et al., 2020). Diatoms possess a red algal type Rubisco (type ID) which is one of the most efficient Rubisco forms, with the highest preference toward carboxylation over oxygenation (Young et al., 2016). However, CO2 concentration in the present day surface oceans is on average 10–12 μmol kg–1 [<1% of the dissolved inorganic carbon (DIC) pool] (Figure 1A), which is well below the half saturation constant of Rubisco for CO2 (Badger et al., 1998). Instead, the DIC pool is >90% bicarbonate ions (HCO3–) (Figure 1A). To optimize carboxylation even at present CO2 levels, most photoautotrophs have developed active carbon dioxide concentration mechanisms (CCMs) which can be biophysical or biochemical (Figure 1B). Such CCMs aim to maintain a higher CO2 concentration over O2 in the vicinity of Rubisco (Reinfelder, 2011). In a biophysical CCM, the cells actively pump bicarbonate ions (HCO3–) inside the cell followed by the conversion of HCO3– to CO2 (Hopkinson et al., 2011; Reinfelder, 2011) via the metalloenzyme carbonic anhydrase (CA; Morel et al., 1994; Badger, 2003). CO2 molecules enter the cell through the lipid bilayer membrane (Gutknecht et al., 1977) and can easily diffuse out due to the high concentration gradient. It has been proposed that in diatoms cytoplasmic CA continuously maintains low CO2 levels by converting CO2 to HCO3– for facilitating a CO2 diffusive influx (Matsuda et al., 2017). Hence, CA plays a key role in carbon acquisition in diatoms. The functioning of biophysical CCMs in marine diatoms has been well studied in laboratory conditions (Hopkinson et al., 2016; reviewed by Matsuda et al., 2017) and was shown to be highly diverse and more efficient than in C4 plants (Young et al., 2016). Down regulation of CCM/photorespiratory genes under elevated CO2 levels in model marine diatoms has been observed in experimental studies (Ohno et al., 2012; Hennon et al., 2015; Li et al., 2015).
In biochemical CCMs, the enzyme phosphoenolpyruvate carboxylase (PEPC) works as a primary carboxylase in the cytoplasm, forming oxaloacetate (C4) from phosphoenolpyruvate (C3) and HCO3– (Figure 1B). This C4 acid is then transported into the chloroplast and releases CO2 in the vicinity of Rubisco by action of the enzyme phosphoenolpyruvate carboxykinase (PEPCK) (Reinfelder et al., 2000, 2004; Roberts et al., 2007a,b; and references therein). The process of decarboxylation can also be performed by the malic enzyme (ME). Oxaloacetate (OAA) is converted to malate via malate dehydrogenase which is then transferred to another compartment (likely mitochondria) and forms pyruvate and CO2 via ME (Kustka et al., 2014). Co-occurrences of PEPCK and ME driven decarboxylation mechanisms have been reported in C4 plants (Cacefo et al., 2019) and marine diatoms (Kroth et al., 2008). It has been proposed that ME may not be actively involved in CCMs and probably plays a role in photorespiration and mitochondrial metabolism in marine diatoms (Davis et al., 2017). The study by Kroth et al. (2008) stated that in case of a C4 pathway in diatoms, the processes of decarboxylation of OAA as well as malate and carboxylation by Rubisco may take place separately in mitochondria and plastids, respectively. In such a case the CO2 molecule released in mitochondria via decarboxylation needs to be transferred to Rubisco. It is possible that the CO2 is then converted to HCO3– again via CA and a further conversion to CO2 takes place within the plastid in the vicinity of Rubisco before carboxylation. These double conversions involve a considerable amount of energy and diatoms may use a C4 CCM for dissipating extra energy which they acquire via the light reactions. Thus, diatoms living under optimum light conditions might actively use a C4 CCM, whereas the diatoms from light limited areas and in deep chlorophyll maxima may down-regulate this process to avoid energy loss.
However, the existence of a fully functional biochemical CCM (C4 pathway) in marine diatoms is not yet proven (Tanaka et al., 2014; Clement et al., 2016) despite some experimental studies (Roberts et al., 2007b; Kustka et al., 2014). A short term metabolic C14 labeling study of two model marine diatoms (Thalassiosira pseudonana and Thalassiosira weissflogii) showed that the initial labeled products in T. pseudonana were mostly C3 and C6, whereas T. weissflogii produced a mixture of C3 and C4 acids (Roberts et al., 2007b). Notwithstanding, C4 enzymes were documented in both species. This suggests that some diatoms may operate a mixture of C3 and C4 CCMs. A significant increase in expression of genes encoding C4 enzymes under low CO2 acclimatized cells was reported in model marine diatoms (Kroth et al., 2008; Saade and Bowler, 2009). However, the evidence for an active biochemical CCM in natural communities of marine diatoms has remained inconclusive.
Another inefficient feature of Rubisco in green algae and land plants is its deactivation by sugar phosphates (ribulose-1,5-bisphosphate and others). To perform optimum photosynthesis, Rubisco is usually reactivated by a motor protein, named Rubisco activase (RCA), by binding to the inactive Rubisco via ATP hydrolysis (Shivhare and Mueller-Cajar, 2017; Figure 1C). The gradient of pH and Mg++ concentrations are two key factors that control RCA activity. A non-substrate CO2 and a Mg++ ion need to bind to Rubisco before carboxylation and therefore the concentration of CO2 is also important for activation of Rubisco prior to carboxylation (Pollock et al., 2003). In the study by Young et al. (2016) it was noticed that the activation levels of Rubisco in eleven experimental diatom species were quite low suggesting a strong possibility for the presence of a RCA type of enzyme. Surprisingly, no structural homolog of RCA has been reported in diatoms. Instead, a functional homolog of RCA, denoted Calvin-Benson-Bassham protein (CbbX) complex, was identified from a red type Rubisco in proteobacteria (Mueller-Cajar et al., 2011) and red algae (Loganathan et al., 2016). Jensen et al. (2017) reported a BLAST search that revealed the presence of CbbX homologs in almost 100 stramenopiles including some model diatoms. The authors also established that CbbX is encoded in the plastid genome unlike in green plants where the RCA gene is encoded in the nucleus. However, it was subsequently found that in red algae and diatoms, another CbbX gene is also encoded in the nucleus (Bhat et al., 2017). Jensen et al. (2017) also argued in favor of the existence of an allosteric control of Rubisco by CbbX in diatoms (Figure 1C). Other than this, to our knowledge there have been no other studies of the abundance and functioning of CbbX in diatoms, neither in lab studies nor in natural populations. Conversely, only a few discrete studies have reported gene expression within phytoplankton communities as a function of changing ocean carbon chemistry (Endo et al., 2015; Hennon et al., 2015; Hopkinson et al., 2016). Moreover, most of the studies are based on model diatoms and hence there is a strong need to study natural diatom assemblages.
Therefore, we deemed it important to characterize the diatom CCM in the environment under natural CO2 variability. With this motivation, the present study investigates the interlink between the abundance and expression of the genes encoding five key enzymes (CbbX, CA, PEPC, PEPCK, and ME) involved in diatom CCMs under variable CO2 levels. We did so by mining the Tara Oceans datasets (Figure 2), which were generated from samples across the global ocean in a standardized manner, including the measurement of carbonate chemistry and other physicochemical parameters and the generation of >200 metagenomes and >220 metatranscriptomes (Carradec et al., 2018).
FIGURE 2
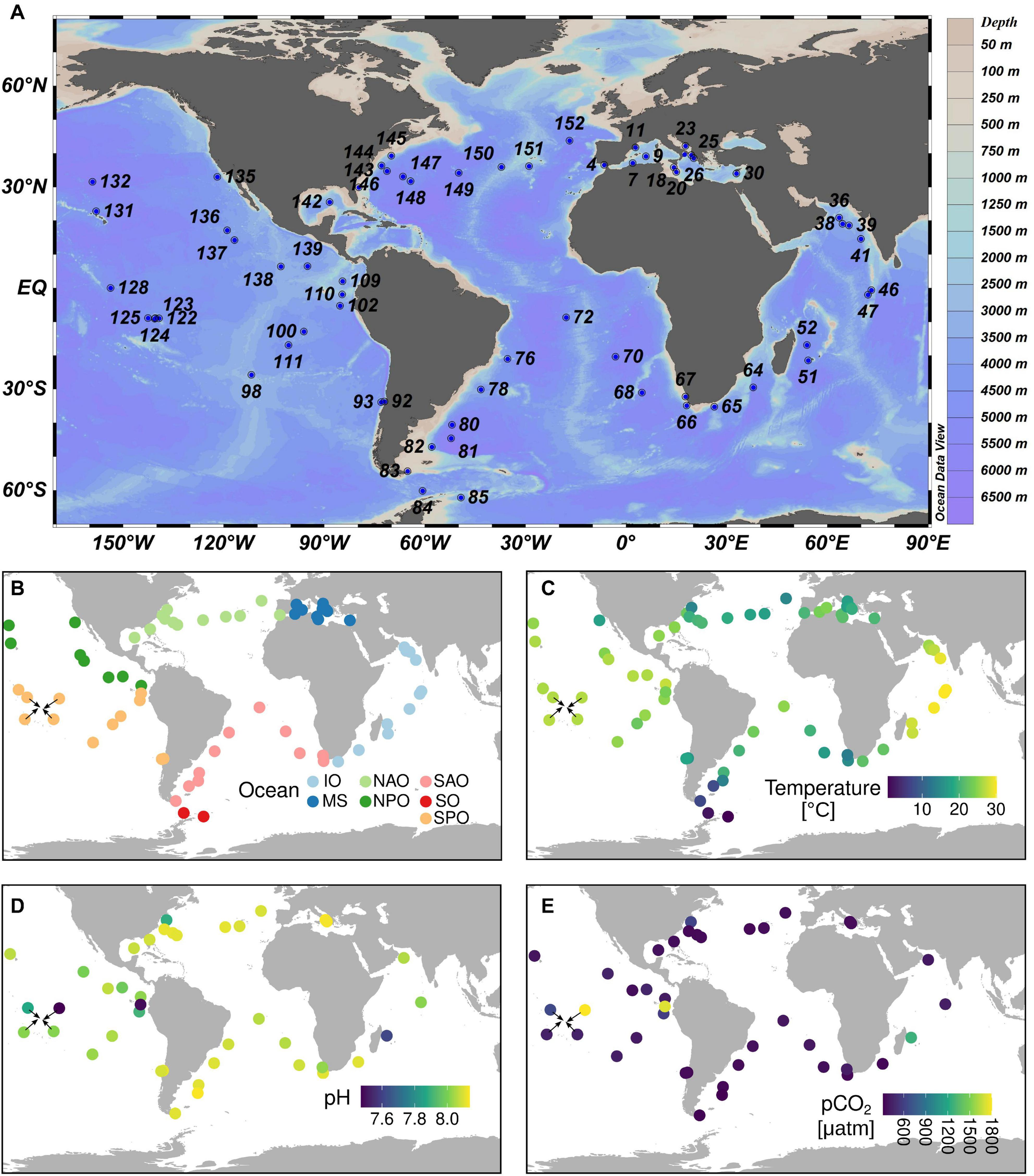
Tara Oceans sampling sites relevant to the current study. (A) Station labels. (B) Ocean regions. (C) Temperature measurements. (D) pH measurements. (E) CO2 partial pressure measurements. The sampling covers almost all main ecogeographic locations. Complete contextual data is available in Supplementary Table 1. IO, Indian Ocean; MS, Mediterranean Sea; NAO, North Atlantic Ocean; NPO, North Pacific Ocean; SAO, South Atlantic Ocean; SO, Southern Ocean; SPO, South Pacific Ocean.
Materials and Methods
Sequence Search and Analysis in the Tara Oceans Eukaryotic Gene Catalog
We searched for sequences of interest in version 1 of the Marine Atlas of Tara Oceans Unigenes (MATOU.v1; Carradec et al., 2018). It consists of 116 million transcribed sequences mainly from eukaryotic plankton in size fractions ranging from 0.8 to 2,000 μm. It was generated by assembling 441 poly-A + metatranscriptomes from samples across the main ocean basins (with the exception of the Arctic Ocean) and then clustered at 95% identity to define a non-redundant catalog (Carradec et al., 2018).
A HMMer search (version 3.2.1 with gathering threshold option)1 was performed in the translated version of MATOU.v1 using the following Pfam models: PF00004 (AAA; ATPase family associated with various cellular activities) for detecting CbbX, PF00311 (PEPcase; Phosphoenolpyruvate carboxylase) for PEPC, PF01293 (PEPCK_ATP; Phosphoenolpyruvate carboxykinase) for PEPCK, PF00390 (malic; Malic enzyme N-terminal domain) and PF03949 (Malic_M; Malic enzyme NAD binding domain) for ME, PF00194 (Carb_anhydrase; Eukaryotic-type carbonic anhydrase) for alpha-CA, PF00484 (Pro_CA; Carbonic anhydrase) for beta-CA, PF00132 (Hexapep; Bacterial transferase hexapeptide) for gamma-CA, PF10563 (CA_like; Putative carbonic anhydrase) for delta-CA, PF18484 (CDCA; Cadmium carbonic anhydrase repeat) for zeta-CA, PF18599 (LCIB_C_CA; Limiting CO2-inducible proteins B/C beta carbonic anhydrases) for theta-CA, and PF08332 (CaMKII_AD; Calcium/calmodulin dependent protein kinase II association domain) for iota-CA. To compare with primary and housekeeping pathways, we also retrieved the sequences coding for the nuclear-encoded subunits of photosystem II (PF05151, PsbM; PF01716, MSP; PF05757, PsbQ; PF06514, PsbU; PF18240, PSII_Pbs31) and for ribosomal proteins (112 Pfam models listed in Supplementary Table 1). Taxonomic assignment of MATOU.v1 is already available based on sequence similarity against a reference database containing UniRef90, MMETSP, and other sources (see Carradec et al., 2018). Based on this assignment, we only kept with those sequences assigned as diatoms for further analysis.
In order to discard homologous proteins of interest, we carried out a combination of sequence similarity network and phylogeny approaches for functional assignment. Briefly, we carried out a HMMer v3.2.1 search (as previously mentioned) for sequences containing the Pfam domains of interest among the sequenced genomes available in the Integrated Microbial Genome (IMG) database2 (Chen et al., 2018) and the sequenced transcriptomes from MMETSP (Keeling et al., 2014). The retrieved sequences were translated in the correct frame and the Pfam domain region was extracted. These sequences were used for building a protein similarity network using EFI-EST tool (Zallot et al., 2019) and Cytoscape visualization (Shannon et al., 2003), which allowed us to inspect the different protein clusters varying the score cut-off. By this step, we found that most Pfams were specific to the enzymes of interest (at least in diatoms) with the only exceptions of CbbX and iota CA (Supplementary Figures 1, 2). The final list of MATOU.v1 sequences used in the current work is displayed in Supplementary Table 1.
In the case of CbbX, it is part of one of the many clusters detected in the sequence similarity network of the AAA domain sequences (Supplementary Figure 1A). This network was built using a score cut-off of 40 after a previous step of reducing sequence redundancy to 80% identity with CD-HIT version 4.6.4 (Li and Godzik, 2006). Therefore, we then built a phylogeny for all the AAA domain sequences of this cluster (Supplementary Figures 1B,C). For this, we aligned the sequences with MAFFT version 6 using the G-INS-I strategy (Katoh and Toh, 2008) and used the resulting alignment to generate the tree with PhyML version 3.0 (Guindon et al., 2010). Four categories of rate variation were used. The starting tree was a BIONJ tree and the type of tree improvement was subtree pruning and regrafting. Branch support was calculated using the approximate likelihood ratio test (aLRT) with a Shimodaira–Hasegawa-like (SH-like) procedure. CbbX sequences formed a distinctive branch (Supplementary Figure 1B), which included the experimentally validated sequences from the proteobacterium Rhodobacter sphaeroides and the nuclear- and plastid-encoded versions from the red alga Cyanidioschyzon merolae (Loganathan et al., 2016). The remaining branches of the tree are annotated as stage V sporulation protein K (KEEG id: K06413) by BlastKOALA (Kanehisa et al., 2016). Therefore, the sequence similarity network and the phylogenies were used as references for the selection of Tara Oceans unigenes coding for diatom CbbX.
In the case of iota-CA, it forms one of the two main clusters in the sequence similarity network of CaMKII_AD domain sequences (Supplementary Figure 2), which was built using a score cut-off of 18 and a previous step of reducing redundancy at 90% identity with CD-HIT version 4.6.4 (Li and Godzik, 2006). The iota-CA cluster contains sequences from bacteria and eukaryotes, including the experimentally validated iota-CA from T. pseudonana (Jensen et al., 2019) as well as orthologous sequences from other species (Jensen et al., 2019; Nonoyama et al., 2019). The other subfamily contains eukaryotic sequences annotated as canonical Calcium/calmodulin dependent protein kinases. Therefore, we used the protein similarity network to keep exclusively with the iota-CAs among those MATOU-v1 sequences with the CaMKII_AD domain.
Analysis of Biogeographical and Environmental Patterns of Gene and Transcript Abundances
Tara Oceans performed a worldwide sampling of plankton between 2009 and 2013 (Figure 2 and Supplementary Table 2) using a serial filtration system for separating the plankton into discrete size fractions (Pesant et al., 2015). In the current work, we analyzed a total of 203 metagenomes and 224 metatranscriptomes generated from samples of the surface layer (5 m depth) of 66 globally distributed stations (Figure 2) and corresponding to the four main size fractions enriched in protists: 0.8–5 μm, 5–20 μm, 20–180 μm, and 180–2,000 μm (Carradec et al., 2018). Thus, we retrieved the metagenomic and metatranscriptomic read abundances of the selected MATOU.v1 sequences (described in the section “Sequence Search and Analysis in the Tara Oceans Eukaryotic Gene Catalog”) and normalized them by the total read abundance for genes or transcripts of the whole diatom community of the corresponding sample. Results are displayed in Supplementary Table 3.
We compared the metagenomic and metatranscriptomic abundance patterns with the environmental data collected during Tara Oceans expeditions.3 The contextual data used in the current work is displayed in Supplementary Table 2. Carbonate chemistry was determined in 40 stations.4 Total alkalinity and DIC were measured potentiometrically (Edmond, 1970), and other carbonate chemistry parameters (pH on total scale, CO2, pCO2, HCO3–, CO3–) were calculated using seacarb (Nisumaa et al., 2010). The average CO2 values were 12 ± 2μmol kg–1 which is very common for present day surface seawater values. However, there were four stations (TARA_110, TARA_122, TARA_052, TARA_145) with more than double these CO2 levels. The station locations were highly diverse; from tropical, subtropical and higher latitude locations (from 54.37°S to 43.67°N) including upwelling, shallow lagoon and deep sea stations (Figure 2 and Supplementary Table 2).
Measurements of temperature, conductivity, salinity, depth, pressure, and oxygen were carried out at each station with a vertical profile sampling system (CTD-rosette) and Niskin bottles (Picheral et al., 2014). Chlorophyll a concentrations were measured using high-performance liquid chromatography (Van Heukelem and Thomas, 2001; Ras et al., 2008). Phosphate and silicate concentrations were determined using segmented flow analysis (Aminot et al., 2009). Iron concentrations were derived from the biogeochemical model PISCES2 (Aumont et al., 2015). Monthly average estimates of photosynthetically active radiation (PAR) were derived from satellite data5.
Plotting and Statistical Analysis
All analyses were carried out in R language6. Correlation matrices were generated with the rcorr function of the Hmisc package and plotted using the corrplot library. Other graphs were plotted with R library ggplot2 (Wickham, 2009). Spearman rho correlation analysis were carried out with cor.test function.
Results
Diversity and Abundance of Sequences Coding for Diatom Carbon Dioxide Concentration Enzymes
To investigate the diversity and environmental distribution of CCMs in natural populations of diatoms, we searched for sequences coding for CbbX, CA, PEPC, PEPCK, and ME in the eukaryotic unigene catalog of Tara Oceans (Carradec et al., 2018) using profile hidden Markov models and sequence similarity networks (see section “Materials and Methods”). The total number of retrieved distinct diatom sequences was: 40 for CbbX, 4,860 for CAs, 943 for PEPC, 488 for PEPCK, and 336 for ME. The obtained CA sequences corresponded to the following classes: 434 alpha, 39 beta, 1,231 delta, 895 gamma, 1,477 iota, 637 theta, and 147 zeta (Supplementary Table 1).
We then retrieved the metagenomic and metatranscriptomic read abundances of these sequences across the four main eukaryotic size fractions (Figure 3A and Supplementary Figure 3 and Supplementary Table 3). CAs were dominant both in gene number and transcript abundance, with almost one order of magnitude higher levels than the other enzymes under study (Figure 3A). CAs comprise on average the 0.2% of the total diatom metatranscriptomic reads, which is similar to the values of all nuclear-encoded subunits of photosystem II (Supplementary Figure 3B). These results emphasize the importance of CAs in diatom CCMs. For the five enzymes, we found differences between size fractions, probably related with differential needs for maintaining CCMs according to cell sizes and/or aggregation forms: while CbbX gene and transcript abundance increases when moving toward the bigger size fractions, the opposite is observed for the other enzymes (Figure 3A and Supplementary Figure 3).
FIGURE 3
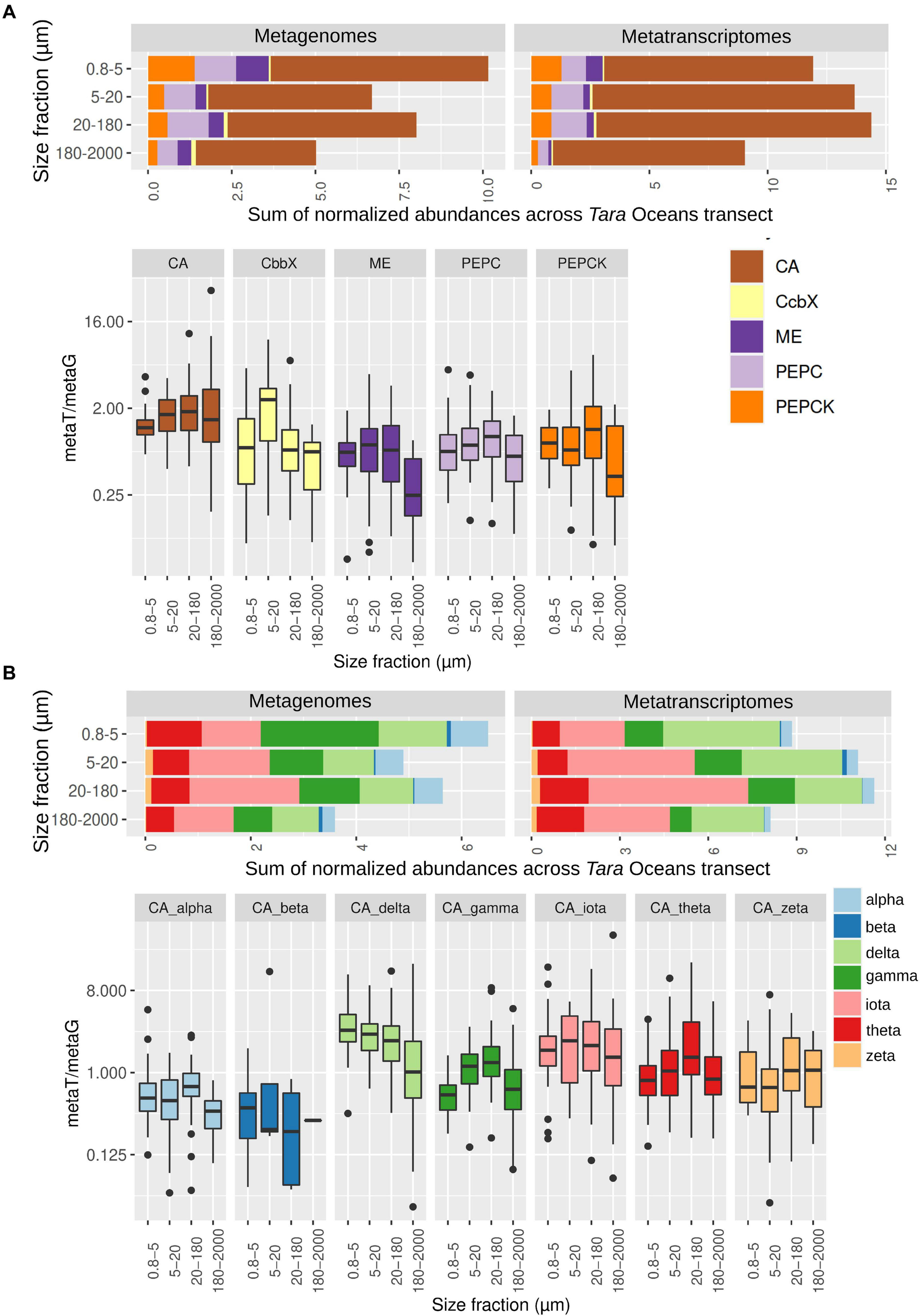
Abundance of genes and transcripts potentially involved in diatom carbon dioxide concentration mechanisms across the different size-fractionated seawater samples collected during the Tara Oceans transect. (A) Gene and transcript abundances for the five enzymes under study. Barplots show the sum of normalized abundances for all samples in a given size fraction. Boxplots show the gene expression levels based on the abundance ratio in metatranscriptomes and metagenomes (metaT/metaG), and are displayed in logarithmic scale. Abbreviations: carbonic anhydrase (CA), Rubisco activase (CbbX), malic enzyme (ME), phosphoenolpyruvate carboxylase (PEPC), and phosphoenolpyruvate carboxykinase (PEPCK). (B) Gene and transcript abundances for the different types of CAs. Barplots and boxplots are displayed as indicated in panel (A).
Among the different classes of CAs (Figure 3B and Supplementary Figure 4), delta, gamma and iota are the most abundant (18–37% and 9–47% of the total CA gene and transcript abundance, respectively, with the percentage range corresponding to the minimum and maximum values depending on the size fraction), followed by theta (13–16% and 9–10%) and alpha (7–11% and 2–4%), whereas zeta and beta represent <2% of gene or transcript abundance. Iota-CA showed the highest gene abundances, and the highest transcript abundances together with delta-CA. The CA classes show differences in abundance between metagenomes and metatranscriptomes, reflecting differences in the expression levels of their genes (Figure 3B). Delta CA is the most expressed and shows a clear expression increase toward the smaller size classes. It is followed by iota, whose expression does not vary between size fractions. On the opposite, alpha and beta are the least expressed classes.
We also analyzed the correlations between the transcript abundances of the different enzymes (Figure 4). In general, we did not find strong correlations in expression of the potential components of a biochemical CCM: ME, PEPC, and PEPCK (Figure 4). An exception was nonetheless noted in the largest size fraction (180–2,000 μm) (Figure 4), where epizoic and large chain-forming diatoms are found. Thus, this pathway cannot be discarded, but it seems clear that it would not be universal in diatom communities.
FIGURE 4
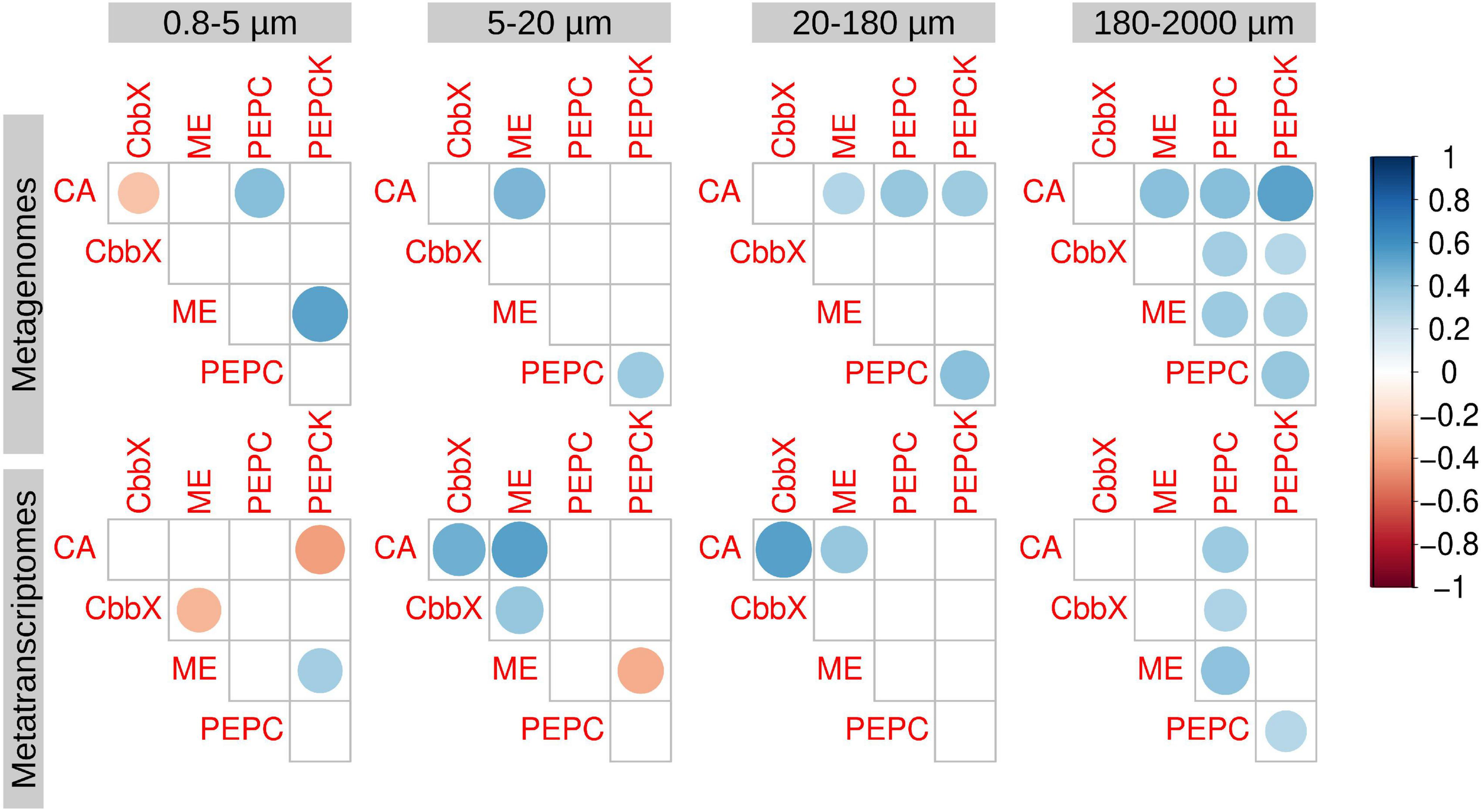
Correlation analysis between the diatom genes and transcripts potentially involved in carbon dioxide concentration mechanisms. Circle size and color intensity are proportional to the Spearman’s rho correlation coefficients. Empty spaces refer to non-significant correlation values (two.tailed p-value > 0.05). carbonic anhydrase (CA), Rubisco activase (CbbX), malic enzyme (ME), phosphoenolpyruvate carboxylase (PEPC), and phosphoenolpyruvate carboxykinase (PEPCK).
Biogeographical Distribution of Genes and Transcripts of Diatom Carbon Dioxide Concentration Enzymes Show Abundance Hotspots
We plotted the biogeographical abundance distributions of the genes and transcripts under study (Figure 5 and Supplementary Figures 5, 6). All enzymes show a widespread occurrence, but with some regional patterns in abundance. A clear regional pattern is found for PEPCK, which shows its lowest gene and transcript abundances in the Southern Ocean (SO) across all size fractions. In addition, we detected several stations that can be considered abundance hotspots for the genes and/or the transcripts coding for carbon concentrating enzymes, but showing divergence between size fractions, pointing to the effect of cell size and/or aggregation.
FIGURE 5
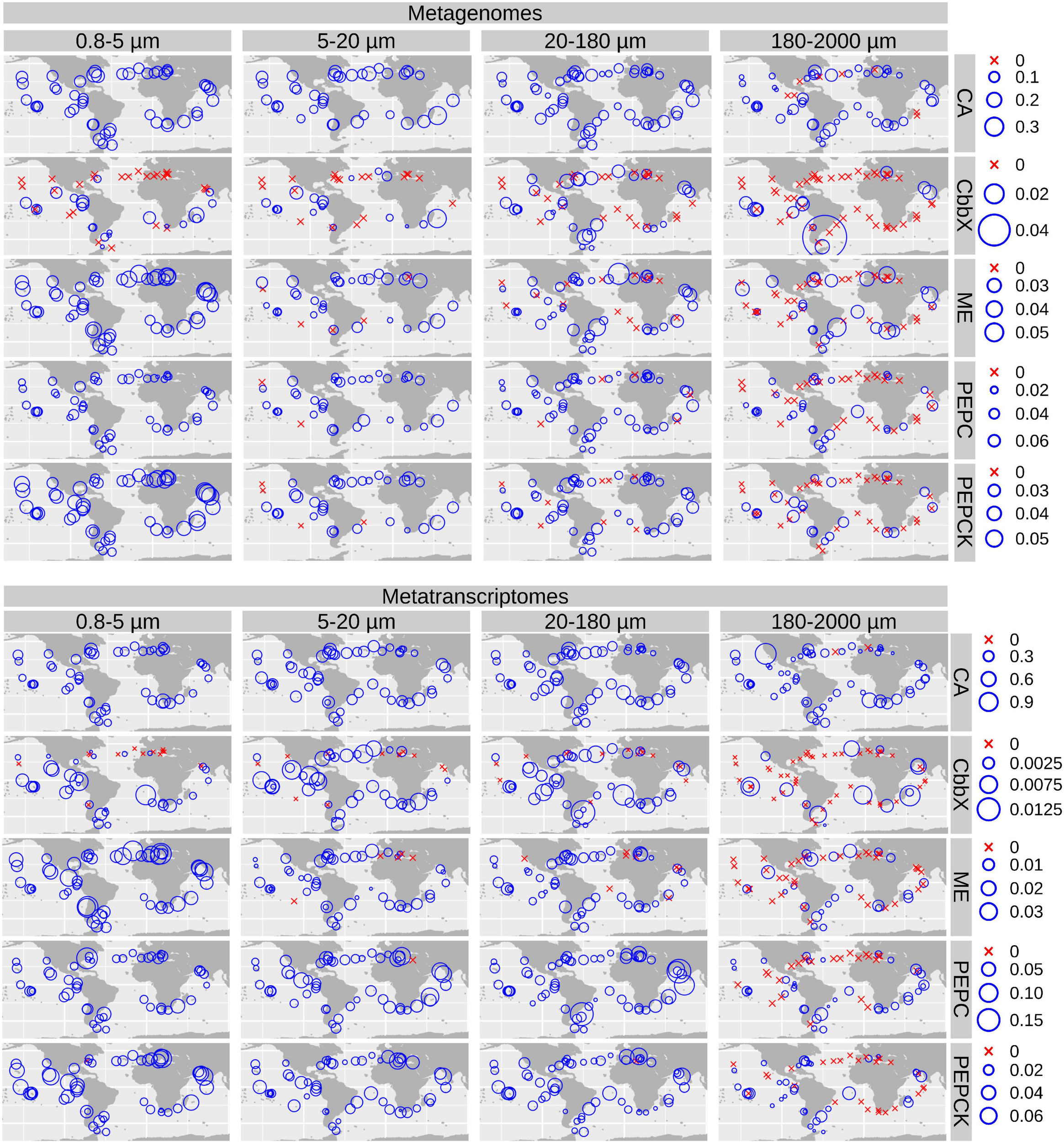
Biogeography of genes and transcripts potentially involved in diatom carbon dioxide concentration mechanisms. Circle sizes are proportional to the gene or transcript abundance (% of total diatom gene or transcript read abundance), while crosses indicate absence of detection. Color code varies according to the size fractions. CA, carbonic anhydrase; CbbX, Rubisco activase; ME, malic enzyme; PEPC, phosphoenolpyruvate carboxylase, PEPCK, phosphoenolpyruvate carboxykinase.
For CA, the highest gene and transcript abundances were detected in the Indian and North Atlantic Oceans (IO and NAO, respectively) as well as in a few stations in the South Atlantic Ocean (SAO; Figure 6 and Supplementary Figures 7, 8). The most abundant CA classes are widespread in the global ocean (but with some differences in their abundances). On the contrary, the low-abundant zeta and beta classes are mainly detected outside the equatorial region (Figure 6).
FIGURE 6
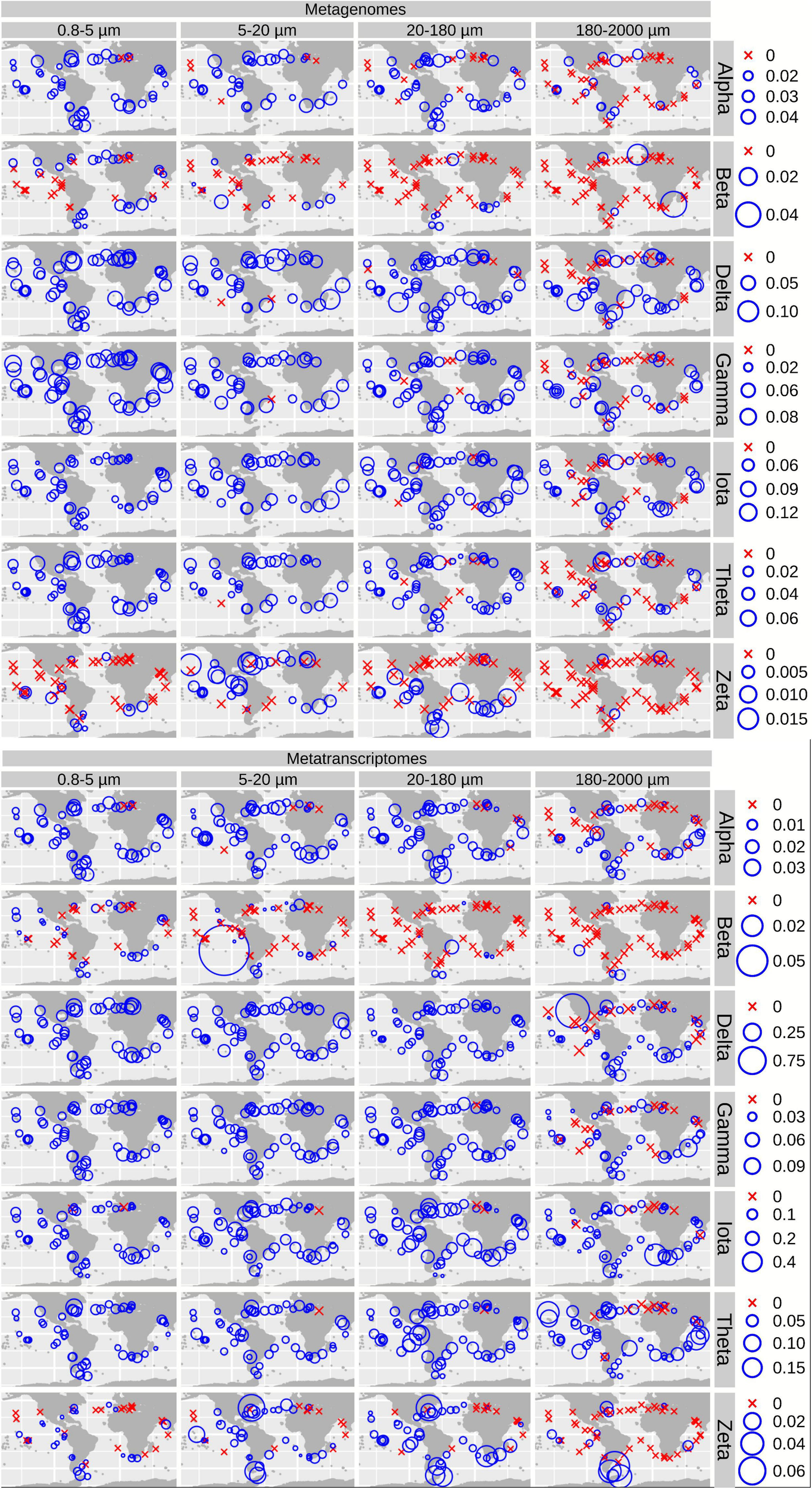
Biogeographical patterns for the diatom genes and transcripts encoding carbonic anhydrases. Circle sizes are proportional to the gene or transcript abundance (% of total diatom gene or transcript abundance), while crosses indicate absence of detection.
Correlations Between the Environmental Variables and Genes Encoding Diatom Carbon Dioxide Concentration Enzymes
We carried out a correlation analysis between gene and transcript abundances of the enzymes under study and the physicochemical and carbon chemistry variables (Figure 7). Many of these variables are correlated among each other (Figure 7A), which has to be taken into account when interpreting the patterns.
FIGURE 7
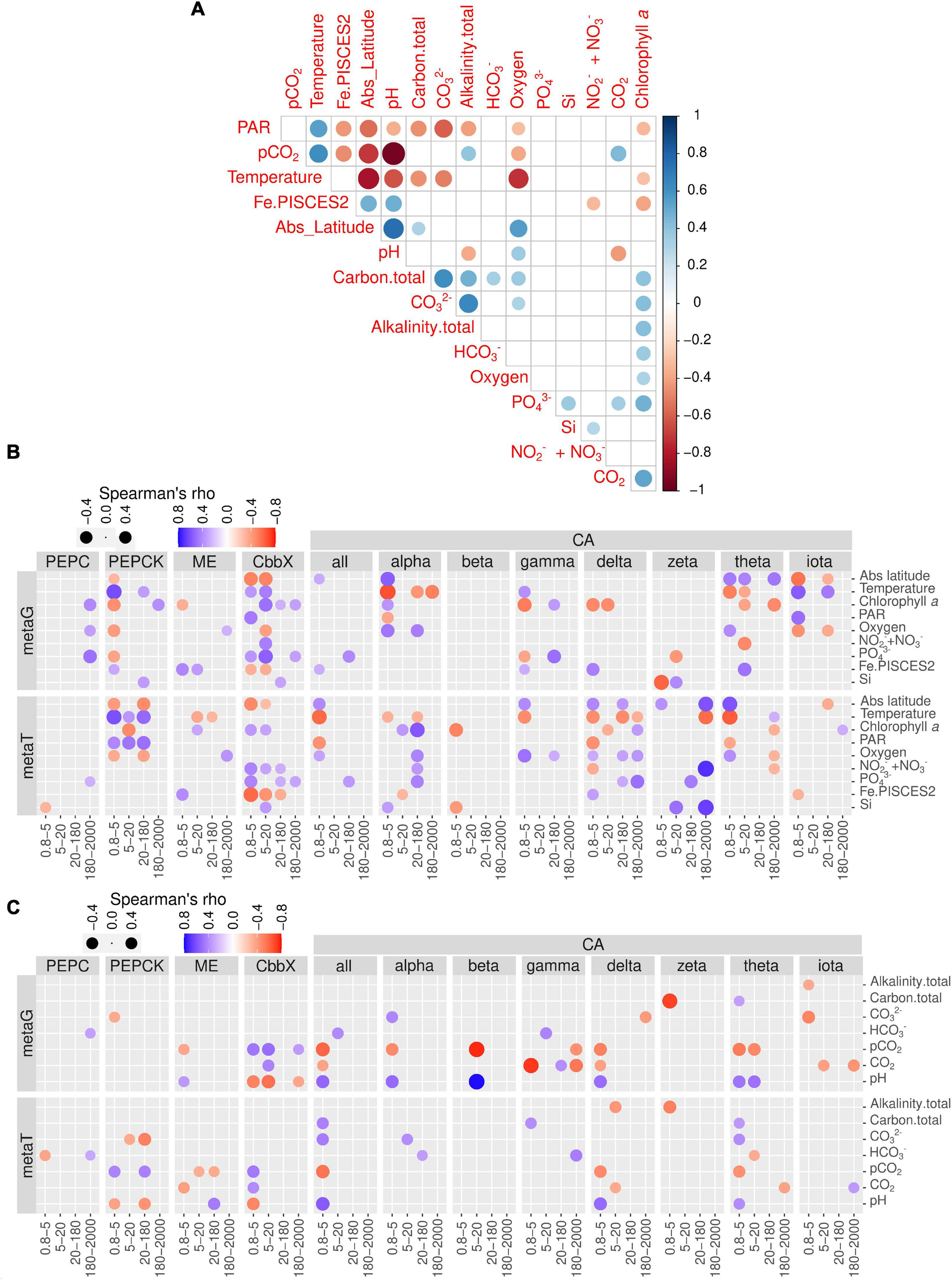
Environmental distribution of genes and transcript potentially involved in diatom carbon dioxide concentration mechanisms. (A) Pairwise correlation of the matrix of contextual parameters. (B) Correlations of nutrients, chlorophyll a and temperature with gene and transcript abundances for the enzymes under study. (C) Correlations of carbonate chemistry measurements with gene and transcript abundances for the enzymes under study. Circle color varies according to Spearman rho’s correlation coefficient, while size varies according to the absolute value of the coefficient. Only statistically significant correlations are displayed (two-tailed test, p < 0.05). PAR, photosynthetically active radiation; CA, carbonic anhydrase; CbbX, Rubisco activase; ME, malic enzyme; PEPC, phosphoenolpyruvate carboxylase, PEPCK, phosphoenolpyruvate carboxykinase.
When focusing on transcript abundances, PEPCK and CbbX displayed an anticorrelation with absolute latitude, whereas ME and most of the CA classes showed the opposite (Figure 7B). These patterns can be related to the effect of temperature and/or PAR, or the fact that in the current dataset the absolute latitude is linked to nutrient and carbon chemistry variables (Figure 7A). CbbX is correlated with phosphate, as are many CA classes.
The correlation matrix with the carbon chemistry variables and CCM enzymes are displayed in Figure 7C. The trends revealed that the partial pressure of CO2 displayed no correlation with PEPC transcript abundance in any size fraction. By contrast, PEPCK showed strong positive correlations in two size fractions (0.8–5 and 20–180 μm) with the partial pressure of CO2 and strong negative correlations with pH. On the contrary, ME was significantly negatively correlated with CO2 and positively with pH. Interestingly, CbbX, the least expressed enzyme of diatom CCM, showed significant positive correlations with CO2 (partial pressure and concentrations) and negatively varied with pH only in the smallest size fractions.
CAs in general displayed strong positive correlations with bicarbonate, carbonate ion concentrations, as well as total alkalinity, and negatively correlated with the partial pressure of CO2 only in the smallest size class. Specifically, delta and theta classes show strong negative correlations with the partial pressure of CO2 for smaller size groups. Surprisingly, iota-CA, one of the most abundant CAs, was generally not well correlated with the carbon chemistry variables. Similarly, beta, gamma and zeta-CA did not show any clear trends with carbon chemistry parameters. Zeta-CA gene expression levels for the largest size class exhibited strong positive correlations with absolute latitude, Si and NO3- + NO2- levels and varied inversely with temperature (Figure 7B). The expression levels of alpha, delta, gama, and theta for the smallest size class were negatively correlated with temperature and hence the average expression level for all CAs also indicated a similar trend.
Discussion
CbbX
The identification of CbbX and its functional role as a Rubisco activation system in diatoms were reported less than a decade ago (Mueller-Cajar et al., 2011) and very little information is available from natural diatom assemblages. We present here the first baseline data regarding the natural variability of this important protein.
The number of CbbX sequences was very low compared with the other sequences. This can be related to the fact that the Tara Oceans gene catalog corresponds to assembled sequences from transcriptomes of polyadenylated RNA (Alberti et al., 2017; Carradec et al., 2018), thus minimizing the detection of plastid-encoded versions of CbbX. In addition chloroplast sequences were removed from the final catalog (Carradec et al., 2018), which might also filter the nuclear-encoded versions of CbbX due to its similarity to the plastid encoded versions (Bhat et al., 2017).
The metatranscriptomic read abundance for the sequences coding for CbbX was also very low. A priori, a high expression would be expected if we consider the ability of marine diatoms to fix one fifth of global carbon fixation per year and that Rubisco is the most abundant protein on the planet. However, this low total metatranscriptomic read abundance is probably an underestimation due to the low number of retrieved sequences, as the expression of these genes (based on the abundance ratio between metranscriptomes and metagenomes) is similar to those of the other enzymes under study (Figure 3A). In addition, low transcript abundances do not necessarily imply a low enzymatic activity. It can be possible that the CbbX function in marine diatoms is controlled by both nuclear and plastid-encoded CbbX versions. Moreover, the gene expression for both CbbX and Rubisco can be linearly varied, and hence a low transcript abundance for CbbX would indicate low transcript abundance for Rubisco. Indeed, diatoms possess an efficient CCM, thus they do not require a high Rubisco concentration: the amount is <6% of the total cellular protein according to both field and culture experiments (Losh et al., 2013), much less than in land plants. All this information may justify the low transcript levels for CbbX in the current work. This must be particularly true in the oligotrophic open ocean where nitrogen can be limiting because Rubisco plays a role as a nitrogen reservoir (Herrig and Falkowski, 1989). Under nitrogen limitation, the nuclear-encoded proteins are synthesized preferentially over those proteins that are encoded in the plastid (Herrig and Falkowski, 1989). In this contest, it is worth mentioning the significant positive correlation between transcript levels for CbbX and NO3– + NO2– concentrations. The strongest correlation was observed for the smallest size fraction while no correlation was detected for the largest size range, which can be related to the fact that smaller diatoms allocate lesser nitrogen resources to build Rubisco than the large centric diatoms (Wu et al., 2014). Finally, the negative correlation of gene and transcript abundance for CbbX against Fe concentrations may indicate its higher activity in the open ocean iron-limited areas. Under nitrogen limitation, the cellular demand for Fe can be significantly low since Fe is essential for nitrogen metabolism. Small-sized diatoms growing under low nitrogen and Fe limited area can allocate less amount of nitrogen resource to synthesize Rubisco and this could be an evolutionary strategy for the open ocean diatoms.
Our analysis shows that the gene abundance and expression levels of CbbX were positively correlated with pCO2 and negatively related with pH in the smallest size fraction (0.8–5 μm). This trend suggests that within the smallest diatoms from the global ocean, CbbX activation is likely to be a prominent feature. CbbX homologs have been detected in the model diatoms T. pseudonana and Phaeodactylum tricornutum, as well as Asterionella formosa and other stramenopiles (Jensen et al., 2017). Nevertheless, it has been already shown that the quantitative level of Rubisco protein does not represent the rate of carboxylation (Raines, 2003; Gontero and Salvucci, 2014). This is likely because the rate of carboxylation is controlled principally through the activation of Rubisco by a motor protein like CbbX (Jensen et al., 2017). However, such a conformational change of Rubisco from an inactive protein to its active form involves several factors and is more complicated than was initially presumed (Gontero and Salvucci, 2014). Therefore, our observation reveals the likely significance of CbbX protein in diatoms. Furthermore, the structure and activation mechanism of RCA in higher plants or green algal lineages are considerably different from the CbbX protein found in red algal lineage taxa characterized by ID type Rubisco. Based on such information, it has been hypothesized that these two different types of motor proteins for Rubisco activation probably resulted from convergent evolution coupled with changing atmospheric CO2/O2 levels (Mueller-Cajar et al., 2014).
Based on the recently reported high diversity of diatom CCMs (Young et al., 2016; Iñiguez et al., 2020) and the efficiency of the Rubisco 1D type, it has been postulated that diatom CCMs and Rubisco might have co-evolved (Young and Hopkinson, 2017) with changing environmental variables like decreasing CO2 and increasing O2 levels (Reinfelder, 2011; Clement et al., 2017). The photorespiratory energy loss is relatively lower in marine diatoms than in other phytoplankton (Rech et al., 2008), while the specificity factor (τ) of Rubisco for CO2 relative to O2 is considerably higher (Tortell, 2000). This strengthens the fact that diatoms are capable of maintaining a high CO2:O2 ratio in the vicinity of Rubisco through active DIC pumping systems (Reinfelder, 2011). The main evolutionary diversification in marine diatoms took place during the time when atmospheric CO2 levels dropped significantly (Reinfelder et al., 2000) and therefore diatoms among the other phytoplankton groups are likely to have developed the most efficient CCMs and Rubisco type (Young et al., 2012). This type ID Rubisco from red algal lineage can perform its highest activity under low CO2:O2 ratio and demands low nutrients as well as energy investment in a CCM; this was likely to be the key factor for mass expansion of diatoms and coccolithophores in the Phanerozoic oceans under very high O2 and low CO2 levels (Rickaby and Hubbard, 2019).
At the heart of the CCM of diatoms and other algae is the pyrenoid (Badger et al., 1998), a spherical structure in the chloroplast stroma consisting of a matrix of tightly packed Rubisco and RCA. The molecular mechanism by which Rubisco aggregates to form the pyrenoid matrix was recently resolved in the model green alga Chlamydomonas reinhardtii, where a low-complexity repeat protein, Essential Pyrenoid Component 1 (EPYC1), links Rubisco to form the pyrenoid (Mackinder et al., 2016). The primary sequences of disordered proteins like EPYC1 are known to evolve rapidly compared with those of structured proteins, but their physicochemical properties are under selective pressure and are evolutionarily maintained. Therefore, Mackinder et al., 2016 searched for proteins with similar physicochemical properties (i.e., repeat number, length, high isoelectric point, disorder profile, and absence of transmembrane domains) across a broad range of algae. They found potential EPYC1-like proteins in the diatoms T. pseudonana and P. tricornutum, which do not exhibit sequence conservation between them. Expectelly, a BLAST search using these sequences against the MATOU-v1 catalog did not retrieve any similar sequences (data not shown).
Carbonic Anhydrases
Carbonic anhydrases are one of the highest upregulated CCM enzymes in diatom cells grown in CO2 limited conditions (Clement et al., 2017), however, CAs also play several other physiological roles apart from photosynthesis (Raven, 1995). Out of eight different types of CAs (Jensen et al., 2020), seven subclasses of CAs are found to be constitutively expressed in diatoms (Samukawa et al., 2014; Jensen et al., 2019). The present study also noticed the presence of the expressed genes of all eight types of CAs in the natural diatom populations from the surface ocean. Such high variability and abundance of CAs in diatoms are quite exclusive relative to other organisms and could be due to their evolutionary complexity. The fact that CA transcript levels are the highest in the Tara Oceans dataset also explains its profound role in CCMs in marine natural populations of diatoms and indicates that diatoms in the global oceans are likely to be operating a biophysical CCM. Marine diatoms usually show very high intercellular conversion of bicarbonate to CO2 and vice-versa to maximize CO2 levels in the vicinity of Rubisco and reduce the diffusive loss of CO2 from the cell (Matsuda and Kroth, 2014) and hence the significance of CAs are eminently important. Zeng et al. (2019) noticed a strong correlation between Rubisco and CA activities in the model marine diatom P. tricornutum and suggests that the rate of carboxylation is directly dependent on the rate of DIC supply which is mediated by CA.
The subcellular location of different CAs can be directly linked to CO2 acquisition. There are some isoforms which are found in the diatom chloroplast, such as iota-CA, beta-CA and theta-CA (Tanaka et al., 2005; Kikutani et al., 2016). The proximity of such CAs to Rubisco probably results in a more efficient CO2 acquisition. Consistent with this view, our observation of a significant negative correlation between gene and transcript abundances of theta-CA against pCO2 for the smallest size fraction also points to an upregulated function of this enzyme at low pCO2 levels. The presence of the chloroplast-targeted theta-CA in some haptophyte species suggest that the diatom ancestor might have acquired this CA gene via horizontal gene transfer (Nonoyama et al., 2019). Regarding iota-CA, there are many gene copies coding for chloroplast-targeted iota-CAs in common marine diatoms like Odontella, but in a few other species the gene is absent (Nonoyama et al., 2019).
The recent research by Clement et al. (2017) reported the regulation of the latest type of CA, known as “Low CO2 inducible protein of 63kDa” or LCIP63 in the marine diatom T. pseudonana. Later Jensen et al. (2019) confirmed its biochemical function as a CA and renamed it as iota-CA, also showing its widespread occurrence in the Tara Oceans dataset. Most importantly, the authors also reported that this type of CA showed its highest expression in surface waters and decreased with increasing depths. It should therefore also be noted that CCMs are likely to play a role in energy dissipation to remove extra energy from the cells and hence, under light inhibition in surface waters, CCMs involving this iota-CA might be used both for carbon acquisition as well as energy dissipation (Kroth et al., 2008). With increasing depth, light stress reduces and CO2 levels increase and therefore, the need for running a CCM involving iota-CA may be reduced. Our results also found that the highest abundant and expressed gene was iota-CA within marine diatoms from surface waters of the global ocean.
The absence of any significant correlation between iota-CA and carbon chemistry in general probably suggests that this enzyme functions despite CO2 variability in surface waters. This shows that the expression levels of CAs may not necessarily be coupled with CO2 levels. For example, in the coccolithophore Emiliania huxleyi the transcript of a delta-CA can exhibit high levels of expression irrespective of CO2 variability (Soto et al., 2006).
Carbonic anhydrase-zeta showed its highest expression at high latitudes for 180–2,000 μm size and seemed to be associated with larger diatoms. The positive correlations with NO2– + NO3– and Si levels also support this view since the large-celled diatoms in high latitude regions are usually found within eutrophic waters because they have very low surface area to volume ratios.
Within the smallest size fraction (0.8–5 μm) the positive correlation between CA gene expression and pH (coupled with negative correlation with pCO2) indicates that under high pH smaller diatoms use CA in their CCMs.
Our results also show that CAs are ubiquitous among all size classes of diatoms, and display high diversity. The abundance and expression of different types of CAs can largely be impacted by trace metal availability in the sea. Importantly, marine diatoms showed the ability to replace a specific metal ion with other more available forms under metal limited conditions (Lane et al., 2005). These metalloenzymes mostly use zinc (Zn) as a cofactor, but other metals such as cadmium (Cd), cobalt (Co), iron (Fe), and manganese (Mn) have also been reported to be associated with different CAs (Morel et al., 2020). In fact the Zn-CAs have been identified to substitute Zn with Co and Cd in surface waters (Morel et al., 2020). In the present study, out of these seven CAs detected, alpha, beta and theta-CAs use Zn ions, whereas, gama, delta and zeta showed the ability to substitute Zn with other metal ions including Cd, Co, and even Fe (Jensen et al., 2020). The highest expression (i.e., metatranscriptomic to metagenomic abundance ratios) were seen in those CAs which are capable of replacing Zn with other metals (Figure 3B). The recently identified iota-CA contains Mn and the availability of Mn can be much higher than Zn, particularly in coastal regions. Hence, marine diatoms might have selectively used this particular Mn-containing CA to cope with the available metal ions. However, this will remain a topic for future research to correlate different CA abundance and expression with trace metal concentrations in the global oceans.
The C4 Enzymes
Our analyses revealed that the transcripts for the enzymes of the putative C4 biochemical CCM did not display co-occurring profiles, with the exeption of the largest size fraction (180–2,000 μm). It has to be noted that this size fraction has a prevalence of copepods, considered one of the most abundant multicellular organisms on the planet, and thus the sequencing signal from diatoms is weaker than in the other size fractions. This can be reflected by the higher variability in this size fraction with respect to the absence/presence of diatom genes and transcripts in the different sampling sites. Therefore, we cannot extend so far the speculations about this biochemical pathway, but it seems clear that the process is unlikely to be prevalent in natural communities, as the transcript levels for the three enzymes of a potential biochemical CCM were significantly lower than CA.
There are many experimental studies on marine diatoms showing the expression of all C4 enzymes (Reinfelder et al., 2000, 2004, Reinfelder, 2011; Roberts et al., 2007b), however their active functioning was not confirmed. The negative correlation between gene expression levels of ME and pCO2/fugacity (as well as the positive correlation with pH) suggests that under CO2 limitation the diatoms are likely to use this enzyme (except in the largest size fraction). Clement et al. (2017) observed that ME showed the lowest activity among all C4 enzymes and the ratio of Rubisco to PEPC was persistently >1 in the experimental marine diatoms. Our results are also consistent with this observation. Haimovich-Dayan et al. (2013) conducted an experiment by genetically silencing an essential C4 enzyme (pyruvate−orthophosphate dikinase, PPDK) in P. tricornutum and observed no major reduction in carboxylation rate. The authors concluded that marine diatoms are likely to use a C4 CCM for dissipating extra light energy. In another study by McGinn and Morel (2008), it was noticed that inhibition of two C4 enzymes (PEPC and PEPCK) resulted in significant reduction in photosynthetic activity in three model marine diatoms. There was almost no study available from any natural diatom population on this aspect and therefore, this study confirms that the relative contribution of C4 CCMs in surface water diatoms is significantly lower than C3 CCM. Moreover, a detailed investigation on deep chlorophyll maxima diatoms is essential to have a clearer picture about functioning of C4 CCM in marine diatoms. Furthermore, additional information on bicarbonate transporter proteins would also shed more light on this topic.
There is some experimental evidence showing higher resilience of phytoplankton communities to increasing CO2 levels from the oceanic region within the “subtropical north and above” (Schulz et al., 2013; Holding et al., 2015; Hoppe et al., 2017 and references therein). Diatoms from the Arctic and other high latitude seas showed high resilience to variable CO2 levels (Feng et al., 2009; Hoppe et al., 2018a,b; Sett et al., 2018; Wolf et al., 2018, 2019). This suggests that certain diatom species have high physiological plasticity to tackle the problem of increasing CO2 levels and therefore, no alteration in photosynthetic performance or growth rate was noticed in relation to changing CO2 levels in the experimental simulations (Hoppe et al., 2018a; Hoppe et al., 2018b; Wolf et al., 2018; Biswas et al., unpublished data). The diatoms from this region are likely to possess a constitutive CCM and therefore variable CO2 levels did not reveal any correlation with the gene expression of CbbX protein and other enzymes.
Hoppe et al. (2018a, b) and Wolf et al. (2018), and Biswas et al. (unpublished data) showed that Arctic diatoms are also highly resilient to the combined stress of irradiance and CO2 levels. This suggests that they have highly evolved cellular mechanisms to counteract photo-inhibition mechanisms. Unpublished data from Biswas et al. showed that an Arctic diatom has high plasticity to control pigment synthesis to combat light limitation/inhibition. Likewise, active functioning of CCM in the surface waters also could be used for these diatoms and the expression levels of C4 enzymes as well as CA can be high. Low latitude phytoplankton may face a stronger impact of photo-inhibition, particularly in the surface waters than the high latitude groups (Tortell, 2000). Hence, the cells living in surface waters may trade off cellular energy between photo-protection and carboxylation. In that case, CA gene expression may be high on the surface. Light is never limiting in this region and hence light dependent DIC uptake can never be hampered. In an experimental study by Biswas et al. (2017) on a tropical coastal diatom community, it was noticed that when light and CO2 both became limiting, carboxylation significantly hampered and resulted in low organic carbon accumulation. On the other hand, under saturated light the signature of non-photochemical quenching was noticed, even though carbon biomass accumulation was higher. Moreover, there is a continuous need of photosystem repair in the surface water due to the breakdown of the D1 protein of pigment system-II (Lavaud et al., 2016). A CCM, either C3- or C4-like mechanism could also be used for dissipating extra light energy in the surface waters (Haimovich-Dayan et al., 2013). It is also possible that a functional CCM in diatom cells from this region may help alleviate light stress and allow photosynthetic performance to remain unaffected. The recent study by Jensen et al. (2019) showed that iota-CA showed the highest expression in surface waters and decreased with increasing depth. Light/energy limitation in the subsurface water may be the reason for such down regulation (Kroth et al., 2008).
Conclusion
This is the first attempt to assess the diversity, abundance, and distribution of CCMs in natural diatom assemblies at a global ocean scale. We carried out paired metagenomic and metatranscriptomic analyses, targeting five key enzymes, including components of the physical pathway as well as components associated with the putative biochemical mechanism.
We observed changes in transcript abundances in the different size fractions depending on the enzymes, pointing to the effect of different cell sizes and/or aggregation forms, such as chains.
CA was the most abundant and highly expressed gene with almost an order of magnitude higher values than the remaining enzymes, thus confirming the importance of biophysical CCM in natural diatom communities. Among the different classes of this enzyme, the most prevalent was the iota class, which was only recently characterized as a CA (Jensen et al., 2019) and so the information presented here represents the first data on its abundance in natural diatom assemblages.
Biogeographical and environmental distributions showed a complex pattern of responses to CO2 levels, total phytoplankton biomass, temperature and nutrient concentrations. This is in part due to the current limitations in the dataset, such as the correlations between different environmental variables or the poor representation of certain conditions. The future generation of data from new regions (e.g., Arctic Ocean) can ameliorate these limitations. It is nevertheless expected to obtain complex patterns when assessing the bulk responses of natural diatom populations, since species can differ in their physiological and molecular responses to the environment.
The transcript levels for the three enzymes of a potential biochemical CCM were significantly lower than CA. In addition, we did not find strong correlations among them, except in the largest size fraction (180–2,000 μm), where epizoic and large chain-forming diatoms are found. Thus, while the biochemical pathway cannot be excluded, it seems clear that the process is unlikely to be prevalent in natural communities.
Overall, this work provides a snapshot of diatom CCMs in the global ocean, providing valuable information toward the prediction of diatom responses in an ocean under anthropogenic change.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
HB and CB designed the project. JJPK carried out the bioinformatic analysis. JJPK, CB, and HB analyzed the results and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the FFEM—French Facility for Global Environment, French Government ‘Investissements d’Avenir’ programmes OCEANOMICS (ANR-11-BTBR-0008), FRANCE GENOMIQUE (ANR-10-INBS-09-08), MEMO LIFE (ANR-10-LABX-54), PSL Research University (ANR-11-IDEX-0001-02), the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Diatomic; grant agreement No. 835067), and Agence Nationale de la Recherche “Phytomet” (ANR-16-CE01-0008), and BrownCut (ANR-19-CE20-0020) projects. JJPK acknowledges postdoctoral funding from the Fonds Français pour l’Environnement Mondial. This article is contribution number 115 of Tara Oceans.
Acknowledgments
We would like to thank all colleagues from the Tara Oceans consortium as well as the Tara Ocean Foundation for their inspirational vision. We also acknowledge Dr. Shruti Malviya and Dr. Federico Ibarbalz for their scientific input during the development of this project and Ms. Saumya Silori for helping with the station location map. We are also grateful to the two reviewers for their useful comments. HB is also thankful to the Director NIO, for his kind support to carry out this research. NIO contribution number is 6711.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer YM declared a past co-authorship with one of the authors CB to the handling editor.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.657821/full#supplementary-material
Supplementary Figure 1Sequence analysis of CbbX and homologs. (A) Protein similarity network for the Pfam domain AAA (PF00004). Each node represents a given sequence and those sequences with similarity higher than a score cutoff are linked (score cut-off of 40 in blast alignment). The network was built with sequences retrieved from the literature and from reference genomes and transcriptomes. Nodes are colored according to their taxonomy. The cluster containing CbbX sequences and close homologs is circled. (B) Phylogeny of the Pfam domain AAA from the sequences belonging to the cluster highlighted in panel (A). The branch for CbbX is colored in yellow, whereas the remaining back branches are annotated as stage V sporulation protein K. (C) Phylogeny of the Pfam domain AAA from CbbX sequences, corresponding to the branch highlighted in panel (B). Color code varies according to the taxonomy. The sequence similarity network and the phylogenies were used as references for the selection of Tara Oceans unigenes encoding diatom CcbX. The list of sequences and the alignment are available in Supplementary Table 1.
Supplementary Figure 2Sequence analysis of iota carbonic anhydrase and homologs. Protein similarity network for the Pfam domain CaMKII_AD (PF08332). Each node represents a given sequence and those sequences with similarity higher than a score cutoff are linked (score cut-off of 18 in blast alignment). The network was built with sequences retrieved from the literature and from reference genomes and transcriptomes, as well as Tara Oceans unigenes. Nodes are colored according to their taxonomy. The cluster containing the iota carbonic anhydrase is encircled, as well as the cluster containing calcium/calmodulin-dependent protein kinase II. The list of sequences is available in Supplementary Table 1.
Supplementary Figure 3Relative abundance of genes and transcripts potentially involved in diatom carbon dioxide concentration mechanisms in comparison to genes involved in other metabolisms. (A) Sum of normalized abundances for all samples in a given size fraction. (B) Gene and transcript abundances. Values in the box plots correspond to the% of total diatom gene or transcript abundance in the corresponding sample, and are displayed in log2 scale. In order to compare with other pathways, we also show the abundances for ribosomal proteins and for the nuclear-encoded subunits of photosystem II. Abbreviations: PSII, photosystem II; CA, carbonic anhydrase; CcbX, Rubisco activase; ME, malic enzyme; PEPC, phosphoenolpyruvate carboxylase; PEPCK, phosphoenolpyruvate carboxykinase.
Supplementary Figure 4Relative abundance of genes and transcripts coding for the different classes of diatom carbonic anhydrases across the size-fractionated seawater samples collected during the Tara Oceans transect. (A) Sum of normalized abundances for all samples in a given size fraction. (B) Gene and transcript abundances. Values in the box plots correspond to the% of total diatom gene or transcript abundance, and are displayed in log2 scale.
Supplementary Figure 5Biogeographical distribution of genes potentially involved in diatom carbon dioxide concentration mechanisms. Barplots are proportional to the gene abundance (% of the total diatom gene read abundance), while color indicates the enzyme: CA, carbonic anhydrase; CbbX, Rubisco activase; ME, malic enzyme; PEPC, phosphoenolpyruvate carboxylase, PEPCK, phosphoenolpyruvate carboxykinase. The Y axis shows the Tara Oceans stations and the ocean regions: MS, Mediterranean Sea; IO, Indian Ocean; SAO, South Atlantic Ocean; SO, Southern Ocean; SPO, South Pacific Ocean; NPO, North Pacific Ocean; NAO, North Atlantic Ocean.
Supplementary Figure 6Biogeographical distribution of transcripts potentially involved in diatom carbon dioxide concentration mechanisms. Barplots are proportional to the transcript abundance (% of the total diatom transcript read abundance), while color indicates the enzyme. Abbreviations: CA, carbonic anhydrase; CbbX, Rubisco activase; ME, malic enzyme; PEPC, phosphoenolpyruvate carboxylase, PEPCK, phosphoenolpyruvate carboxykinase.
Supplementary Figure 7Biogeographical distribution of genes coding for the different classes of diatom carbonic anhydrases. Barplots are proportional to the gene abundance (% of the total diatom gene read abundance), while color indicates the carbonic anhydrase class. The Y axis shows the Tara Oceans stations and the ocean regions. Abbreviations: MS, Mediterranean Sea; IO, Indian Ocean; SAO, South Atlantic Ocean; SO, Southern Ocean; SPO, South Pacific Ocean; NPO, North Pacific Ocean; NAO, North Atlantic Ocean.
Supplementary Figure 8Biogeographical distribution of transcripts coding for the different classes of diatom carbonic anhydrases. Barplots are proportional to the transcript abundance (% of the total diatom transcript read abundance), while color indicates the carbonic anhydrase class. The Y axis shows the Tara Oceans stations and the ocean regions. Abbreviations: MS, Mediterranean Sea; IO, Indian Ocean; SAO, South Atlantic Ocean; SO, Southern Ocean; SPO, South Pacific Ocean; NPO, North Pacific Ocean; NAO, North Atlantic Ocean.
Supplementary Table 1Sequences used in the current work. (A) List of Pfam models. (B) MATOU-v1 unigenes from diatoms coding for the five analyzed enzymes. (C) Sequences used for Supplementary Figure 1A. (D) Sequences and alignments used for Supplementary Figure 1B. (E) Sequences and alignments used for Supplementary Figure 1C. (F) Sequences used for Supplementary Figure 2.
Supplementary Table 2Contextual data for the Tara Oceans samples used in the current work. Original source: https://doi.org/10.1594/PANGAEA.875582 and https://doi.pangaea.de/10.1594/PANGAEA.875567.
Supplementary Table 3Abundance of genes and transcripts potentially involved in diatom carbon dioxide concentration mechanisms across the different Tara Oceans samples. Values correspond to the% of total diatom read abundance (in rpkm). carbonic anhydrase (CA), Rubisco activase (CbbX), malic enzyme (ME), phosphoenolpyruvate carboxylase (PEPC), and phosphoenolpyruvate carboxykinase (PEPCK).
Footnotes
3.^https://doi.org/10.1594/PANGAEA.875582
4.^https://doi.pangaea.de/10.1594/PANGAEA.875567
References
1
AlbertiA.PoulainJ.EngelenS.LabadieK.RomacS.FerreraI.et al (2017). Viral to metazoan marine plankton nucleotide sequences from the Tara Oceans expedition.Sci. Data4:170093.
2
AminotA.KérouelR.CoverlyS. C. (2009). “Nutrients in seawater using segmented flow analysis,” in Practical Guidelines for the Analysis of Seawater, ed.WurlO. (Boca Raton, FL: CRC Press), 143–178.
3
ArmbrustE. V. (2009). The life of diatoms in the world’s oceans.Nature459185–192. 10.1038/nature08057
4
AumontO.EthéC.TagliabueA.BoppL.GehlenM. (2015). PISCES-v2: an ocean biogeochemical model for carbon and ecosystem studies.Geosci. Model Dev.82465–2513. 10.5194/gmd-8-2465-2015
5
BadgerM. (2003). The roles of carbonic anhydrases in photosynthetic CO2 concentrating mechanisms.Photosynth. Res.77:83.
6
BadgerM. R.AndrewsT. J.WhitneyS. M.LudwigM.YellowleesD. C.LeggatW.et al (1998). The diversity and coevolution of rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae.Can. J. Bot.761052–1071. 10.1139/cjb-76-6-1052
7
Bar-OnY. M.MiloR. (2019). The global mass and average rate of rubisco.Proc. Natl. Acad. Sci. U.S.A.1164738–4743. 10.1073/pnas.1816654116
8
BhatJ. Y.Thieulin-PardoG.HartlF. U.Hayer-HartlM. (2017). Rubisco activases: AAA+ chaperones adapted to enzyme repair.Front. Mol. Biosci.4:20. 10.3389/fmolb.2017.00020
9
BiswasH.ShaikA. U. R.BandyopadhyayD.ChowdhuryN. (2017). CO2 induced growth response in a diatom dominated phytoplankton community from SW Bay of Bengal coastal water.Estuar. Coast. Shelf Sci.19829–42. 10.1016/j.ecss.2017.07.022
10
BowlerC.AllenA. E.BadgerJ. H.GrimwoodJ.JabbariK.KuoA.et al (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes.Nature456239–244.
11
BoydP. W.ClaustreH.LevyM.SiegelD. A.WeberT. (2019). Multi-faceted particle pumps drive carbon sequestration in the ocean.Nature568327–335. 10.1038/s41586-019-1098-2
12
BusseniG.Rocha Jimenez VieiraF.AmatoA.PelletierE.Pierella KarlusichJ. J.FerranteM. I.et al (2019). Meta-omics reveals genetic flexibility of diatom nitrogen transporters in response to environmental changes.Mol. Biol. Evol.362522–2535. 10.1093/molbev/msz157
13
CacefoV.RibasA. F.ZillianiR. R.NerisD. M.DominguesD. S.MoroA. L.et al (2019). Decarboxylation mechanisms of C4 photosynthesis in Saccharum spp. increased PEPCK activity under water-limiting conditions.BMC Plant Biol.19:144. 10.1186/s12870-019-1745-7
14
CarradecQ.PelletierE.Da SilvaC.AlbertiA.SeeleuthnerY.Blanc-MathieuR.et al (2018). A global ocean atlas of eukaryotic genes.Nat. Commun.9:373.
15
ChenI. M. A.ChuK.PalaniappanK.PillayM.RatnerA.HuangJ.et al (2018). IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes.Nucleic Acids Res.47666–677.
16
ClementR.DimnetL.MaberlyS. C.GonteroB. (2016). The nature of the CO2−concentrating mechanisms in a marine diatom, Thalassiosira pseudonana.New Phytol.2091417–1427. 10.1111/nph.13728
17
ClementR.LignonS.MansuelleP.JensenE.PophillatM.LebrunR.et al (2017). Responses of the marine diatom Thalassiosira pseudonana to changes in CO2 concentration: a proteomic approach.Sci. Rep.7:42333.
18
DavisA.AbbrianoR.SmithS. R.HildebrandM. (2017). Clarification of photorespiratory processes and the role of malic enzymes in diatoms.Protist168134–153. 10.1016/j.protis.2016.10.005
19
DorrellR. G.GileG.MccallumG.MéheustR.BaptesteE. P.KlingerC. M.et al (2017). Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome.eLife6:e23717.
20
DorrellR. G.VillainA.Perez-LamarqueB.de KerdrelG. A.McCallumG.WatsonA. K.et al (2021). Phylogenomic fingerprinting of tempo and functions of horizontal gene transfer within ochrophytes.Proc. Natl. Acad. Sci. U.S.A.118:e2009974118. 10.1073/pnas.2009974118
21
EdmondJ. M. (1970). High precision determination of titration alkalinity and total carbon dioxide content of sea water by potentiometric titration.Deep-Sea Res. Oceanogr. Abstr.17737–750. 10.1016/0011-7471(70)90038-0
22
EndoH.SugieK.YoshimuraT.SuzukiK. (2015). Effects of CO2 and iron availability on rbcL gene expression in Bering sea diatoms.Biogeosciences122247–2259. 10.5194/bg-12-2247-2015
23
ErbT. J.ZarzyckiJ. (2018). A short history of RubisCO: the rise and fall (?) of Nature’s predominant CO2 fixing enzyme.Curr. Opin. Biotechnol.49100–107. 10.1016/j.copbio.2017.07.017
24
FalkowskiP. G.RavenJ. A. (2013). Aquatic Photosynthesis.Princeton, NJ: Princeton University Press.
25
FengY.HareC. E.LeblancK.RoseJ. M.ZhangY.DiTullioG. R.et al (2009). Effects of increased pCO2 and temperature on the North Atlantic spring bloom. I. The phytoplankton community and biogeochemical response.Mar. Ecol. Prog. Ser.38813–25. 10.3354/meps08133
26
FieldC. B.BehrenfeldM. J.RandersonJ. T.FalkowskiP. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components.Science281237–240. 10.1126/science.281.5374.237
27
FriedlingsteinP.O’SullivanM.JonesM. W.AndrewR. M.HauckJ.OlsenA.et al (2020). Global carbon budget 2020.Earth Syst. Sci. Data123269–3340.
28
GonteroB.SalvucciM. E. (2014). Regulation of photosynthetic carbon metabolism in aquatic and terrestrial organisms by rubisco activase, redox-modulation and CP12.Aquat. Bot.11814–23. 10.1016/j.aquabot.2014.05.011
29
GranumE.RavenJ. A.LeegoodR. C. (2005). How do marine diatoms fix 10 billion tonnes of inorganic carbon per year?Can. J. Bot.83898–908. 10.1139/b05-077
30
GuindonS.DufayardJ.-F.LefortV.AnisimovaM.HordijkW.GascuelO. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0.Syst. Biol.59307–321. 10.1093/sysbio/syq010
31
GutknechtJ.BissonM. A.TostesonF. C. (1977). Diffusion of carbon dioxide through lipid bilayer membranes. Effects of carbonic anhydrase, bicarbonate, and unstirred layers.J. Gen. Physiol.69:779. 10.1085/jgp.69.6.779
32
Haimovich-DayanM.GarfinkelN.EweD.MarcusY.GruberA.WagnerH.et al (2013). The role of C4 metabolism in the marine diatom Phaeodactylum tricornutum.New Phytol.197177–185.
33
HennonG. M.AshworthJ.GroussmanR. D.BerthiaumeC.MoralesR. L.BaligaN. S.et al (2015). Diatom acclimation to elevated CO2 via cAMP signalling and coordinated gene expression.Nat. Clim. Chang.5:761. 10.1038/nclimate2683
34
HerrigR.FalkowskiP. G. (1989). Nitrogen limitation in Isochrysis galbana (haptophyceae). I. Photosynthetic energy conversion and growth efficiencies.I.J. Phycol.25462–471. 10.1111/j.1529-8817.1989.tb00251.x
35
HoldingJ. M.DuarteC. M.Sanz-MartínM.MesaE.ArrietaJ. M.ChiericiM.et al (2015). Temperature dependence of CO2-enhanced primary production in the European Arctic Ocean.Nat. Clim. Chang.51079–1082. 10.1038/nclimate2768
36
HopkinsonB. M.DupontC. L.AllenA. E.MorelF. M. (2011). Efficiency of the CO2-concentrating mechanism of diatoms.Proc. Natl. Acad. Sci. U.S.A.1083830–3837. 10.1073/pnas.1018062108
37
HopkinsonB. M.DupontC. L.MatsudaY. (2016). The physiology and genetics of CO2 concentrating mechanisms in model diatoms.Curr. Opin. Plant Biol.3151–57. 10.1016/j.pbi.2016.03.013
38
HoppeC. J.SchubackN.SemeniukD. M.MaldonadoM. T.RostB. (2017). Functional redundancy facilitates resilience of subarctic phytoplankton assemblages toward ocean acidification and high irradiance.Front. Mar. Sci.4:229. 10.3389/fmars.2017.00229
39
HoppeC. J. M.SchubackN.SemeniukD.GiesbrechtK.MolJ.ThomasH.et al (2018a). Resistance of arctic phytoplankton to ocean acidification and enhanced irradiance.Polar Biol.41399–413. 10.1007/s00300-017-2186-0
40
HoppeC. J. M.WolfK. K.SchubackN.TortellP. D.RostB. (2018b). Compensation of ocean acidification effects in arctic phytoplankton assemblages.Nat. Clim. Change8529–533. 10.1038/s41558-018-0142-9
41
IñiguezC.Capó−BauçàS.NiinemetsÜStollH.Aguiló−NicolauP.GalmésJ. (2020). Evolutionary trends in rubisco kinetics and their co−evolution with CO2 concentrating mechanisms.Plant J.101897–918. 10.1111/tpj.14643
42
JensenE.ClémentR.MaberlyS. C.GonteroB. (2017). Regulation of the calvin–benson–bassham cycle in the enigmatic diatoms: biochemical and evolutionary variations on an original theme.Philos. Trans. R. Soc. B372:20160401. 10.1098/rstb.2016.0401
43
JensenE. L.ClementR.KostaA.MaberlyS. C.GonteroB. (2019). A new widespread subclass of carbonic anhydrase in marine phytoplankton.ISME J.132094–2106. 10.1038/s41396-019-0426-8
44
JensenE. L.MaberlyS. C.GonteroB. (2020). Insights on the functions and ecophysiological relevance of the diverse carbonic anhydrases in microalgae.Int. J. Mol. Sci.21:2922. 10.3390/ijms21082922
45
JinX.GruberN.DunneJ. P.SarmientoJ. L.ArmstrongR. A. (2006). Diagnosing the contribution of phytoplankton functional groups to the production and export of particulate organic carbon, CaCO3, and opal from global nutrient and alkalinity distributions.Global Biogeochem. Cycles20:GB2015.
46
KanehisaM.SatoY.MorishimaK. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences.J. Mol. Biol.428726–731. 10.1016/j.jmb.2015.11.006
47
KatohK.TohH. (2008). Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework.BMC Bioinformatics9:212. 10.1186/1471-2105-9-212
48
KeelingP. J.BurkiF.WilcoxH. M.AllamB.AllenE. E.Amaral-ZettlerL. A.et al (2014). The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing.PLoS Biol.12:e1001889. 10.1371/journal.pbio.1001889
49
KikutaniS.NakajimaK.NagasatoC.TsujiY.MiyatakeA.MatsudaY. (2016). Thylakoid luminal θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum.Proc. Natl. Acad. Sci. U.S.A.1139828–9833. 10.1073/pnas.1603112113
50
KrothP. G.ChiovittiA.GruberA.Martin-JezequelV.MockT.ParkerM. S.et al (2008). A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis.PLoS One3:e1426. 10.1371/journal.pone.0001426
51
KustkaA. B.MilliganA. J.ZhengH.NewA. M.GatesC.BidleK. D.et al (2014). Low CO2 results in a rearrangement of carbon metabolism to support C4 photosynthetic carbon assimilation in Thalassiosira pseudonana.New Phytol.204507–520. 10.1111/nph.12926
52
LaneT. W.SaitoM. A.GeorgeN. G.PickeringI. J.PrinceR. C.MorelF. M. M. (2005). A cadmium enzyme from a marine diatom.Nature43542–42. 10.1038/435042a
53
LavaudJ.SixC.CampbellD. A. (2016). Photosystem II repair in marine diatoms with contrasting photophysiologies.Photosynth. Res.127189–199. 10.1007/s11120-015-0172-3x
54
LiG.BrownC. M.JeansJ. A.DonaherN. A.McCarthyA.CampbellD. A. (2015). The nitrogen costs of photosynthesis in a diatom under current and future pCO2.New Phytol.205533–543. 10.1111/nph.13037
55
LiW.GodzikA. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences.Bioinformatics221658–1659. 10.1093/bioinformatics/btl158
56
LoganathanN.TsaiY. C. C.Mueller-CajarO. (2016). Characterization of the heterooligomeric red-type rubisco activase from red algae.Proc. Natl. Acad. Sci. U.S.A.11314019–14024. 10.1073/pnas.1610758113
57
LoshJ. L.YoungJ. N.MorelF. M. (2013). Rubisco is a small fraction of total protein in marine phytoplankton.New Phytol.19852–58. 10.1111/nph.12143
58
MackinderL. C.MeyerM. T.Mettler-AltmannT.ChenV. K.MitchellM. C.CaspariO.et al (2016). A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle.Proc. Natl. Acad. Sci. U.S.A.1135958–5963. 10.1073/pnas.1522866113
59
MalviyaS.ScalcoE.AudicS.VincentF.VeluchamyA.PoulainJ.et al (2016). Insights into global diatom distribution and diversity in the world’s ocean.Proc. Natl. Acad. Sci. U.S.A.113E1516–E1525.
60
MatsudaY.HopkinsonB. M.NakajimaK.DupontC. L.TsujiY. (2017). Mechanisms of carbon dioxide acquisition and CO2 sensing in marine diatoms: a gateway to carbon metabolism.Philos. Trans. R. Soc. Lond B.372:20160403. 10.1098/rstb.2016.0403
61
MatsudaY.KrothP. G. (2014). “Carbon fixation in diatoms,” in The Structural Basis of Biological Energy Generation, ed.Hohmann-MarriottM. (Dordrecht: Springer), 335–362. 10.1007/978-94-017-8742-0_18
62
McGinnP. J.MorelF. M. (2008). Expression and inhibition of the carboxylating and decarboxylating enzymes in the photosynthetic C4 pathway of marine diatoms.Plant Physiol.146300–309. 10.1104/pp.107.110569
63
MorelF. M.LamP. J.SaitoM. A. (2020). Trace metal substitution in marine phytoplankton.Annu. Rev. Earth Planet. Sci.48491–517. 10.1146/annurev-earth-053018-060108
64
MorelF. M. M.ReinfelderJ. R.RobertsS. B.ChamberlainC. P.LeeJ. G.YeeD. (1994). Zinc and carbon co-limitation of marine phytoplankton.Nature369740–742. 10.1038/369740a0
65
MoustafaA.BeszteriB.MaierU. G.BowlerC.ValentinK.BhattacharyaD. (2009). Genomic footprints of a cryptic plastid endosymbiosis in diatoms.Science3241724–1726. 10.1126/science.1172983
66
Mueller-CajarO.StotzM.BracherA. (2014). Maintaining photosynthetic CO2 fixation via protein remodelling: the Rubisco activases.Photosynth. Res.119191–201. 10.1007/s11120-013-9819-0
67
Mueller-CajarO.StotzM.WendlerP.HartlF. U.BracherA.Hayer-HartlM. (2011). Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase.Nature479:194. 10.1038/nature10568
68
NisumaaA. M.PesantS.BellerbyR. G. J.DelilleB.MiddelburgJ. J.OrrJ. C.et al (2010). EPOCA/EUR-OCEANS data compilation on the biological and biogeochemical responses to ocean acidification.Earth Syst. Sci. Data2167–175. 10.5194/essd-2-167-2010
69
NonoyamaT.KazamiaE.NawalyH.GaoX.TsujiY.MatsudaY.et al (2019). Metabolic innovations underpinning the origin and diversification of the diatom chloroplast.Biomolecules9:322. 10.3390/biom9080322
70
OhnoN.InoueT.YamashikiR.NakajimaK.KitaharaY.IshibashiM.et al (2012). CO2-cAMP-responsive cis-elements targeted by a transcription factor with CREB/ATF-like basic zipper domain in the marine diatom Phaeodactylum tricornutum.Plant Physiol.158499–513. 10.1104/pp.111.190249
71
PesantS.NotF.PicheralM.Kandels-LewisS.Le BescotN.GorskyG.et al (2015). Open science resources for the discovery and analysis of Tara Oceans data.Sci. Data2:150023.
72
PicheralM.SearsonS.TaillandierV.BricaudA.BossE.RasJ.et al (2014). Vertical profiles of environmental parameters measured on discrete water samples collected with Niskin bottles during the Tara Oceans expedition 2009-2013.PANGAEA10.1594/PANGAEA.836319
73
Pierella KarlusichJ. J.IbarbalzF. M.BowlerC. (2020). Phytoplankton in the Tara Ocean.Ann. Rev. Mar. Sci.12233–265. 10.1146/annurev-marine-010419-010706
74
PollockS. V.ColomboS. L.ProutD. L.GodfreyA. C.MoroneyJ. V. (2003). Rubisco activase is required for optimal photosynthesis in the green alga Chlamydomonas reinhardtii in a low-CO2 atmosphere.Plant Physiol.1331854–1861. 10.1104/pp.103.032078
75
PoudelS.PikeD. H.RaananH.ManciniJ. A.NandaV.RickabyR. E. M.et al (2020). Biophysical analysis of the structural evolution of substrate specificity in RuBisCO.Proc. Natl. Acad. Sci. U.S.A.11730451–30457. 10.1073/pnas.2018939117
76
RainesC. A. (2003). The Calvin cycle revisited.Photosynth. Res.751–10. 10.1163/9789004244672_002
77
RasJ.ClaustreH.UitzJ. (2008). Spatial variability of phytoplankton pigment distributions in the Subtropical South Pacific Ocean: comparison between in situ and predicted data.Biogeosciences5353–369. 10.5194/bg-5-353-2008
78
RavenA. (1995). Photosynthetic and non-photosynthetic roles of carbonic anhydrase in algae and cyanobacteria.Phycologia3493–101. 10.2216/i0031-8884-34-2-93.1
79
RechM.Morant-ManceauA.TremblinG. (2008). Carbon fixation and carbonic anhydrase activity in Haslea ostrearia (Bacillariophyceae) in relation to growth irradiance.Phtosynthetica4656–62. 10.1007/s11099-008-0011-2
80
ReinfelderJ. R. (2011). Carbon concentrating mechanisms in eukaryotic marine phytoplankton.Ann. Rev. Mar. Sci.3291–315. 10.1146/annurev-marine-120709-142720
81
ReinfelderJ. R.KraepielA. M.MorelF. M. (2000). Unicellular C4 photosynthesis in a marine diatom.Nature407996–999. 10.1038/35039612
82
ReinfelderJ. R.MilliganA. J.MorelF. M. (2004). The role of the C4 pathway in carbon accumulation and fixation in a marine diatom.Plant Physiol.1352106–2111. 10.1104/pp.104.041319
83
RickabyR. E.HubbardM. E. (2019). Upper ocean oxygenation, evolution of RuBisCO and the Phanerozoic succession of phytoplankton.Free Radic. Biol. Med.140295–304. 10.1016/j.freeradbiomed.2019.05.006
84
RobertsK.GranumE.LeegoodR.RavenJ. A. (2007a). Carbon acquisition by diatoms.Photosynth. Res.9379–88. 10.1007/s11120-007-9172-2
85
RobertsK.GranumE.LeegoodR. C.RavenJ. A. (2007b). C3 and C4 pathways of photosynthetic carbon assimilation in marine diatoms are under genetic, not environmental, control.Plant Physiol.145230–235. 10.1104/pp.107.102616
86
SaadeA.BowlerC. (2009). Molecular tools for discovering the secrets of diatoms.BioScience59757–765. 10.1525/bio.2009.59.9.7
87
SamukawaM.ShenC.HopkinsonB. M.MatsudaY. (2014). Localization of putative carbonic anhydrases in the marine diatom, Thalassiosira pseudonana.Photosynth. Res.121235–249. 10.1007/s11120-014-9967-x
88
SchoefsB.HuH.KrothP. G. (2017). The peculiar carbon metabolism in diatoms.Phil. Trans. R. Soc. B372:20160405. 10.1098/rstb.2016.0405
89
SchulzK. G.BellerbyR. G. J.BrussaardC. P.BüdenbenderJ.CzernyJ.EngelA.et al (2013). Temporal biomass dynamics of an Arctic plankton bloom in response to increasing levels of atmospheric carbon dioxide.Biogeosciences10161–180. 10.5194/bg-10-161-2013
90
SettS.SchulzK. G.BachL. T.RiebesellU. (2018). Shift towards larger diatoms in a natural phytoplankton assemblage under combined high-CO2 and warming conditions.J. Plankton Res.40391–406. 10.1093/plankt/fby018
91
ShannonP.MarkielA.OzierO.BaligaN. S.WangJ. T.RamageD.et al (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks.Genome Res.132498–2504. 10.1101/gr.1239303
92
ShivhareD.Mueller-CajarO. (2017). “Rubisco Activase: the molecular chiropractor of the world’s most abundant protein,” in Photosynthesis And Bioenergetics, edsBarberJ.RubanA. V. (Singapore: World Scientific), 159–187. 10.1142/9789813230309_0008
93
SmetacekV. (1999). Diatoms and the ocean carbon cycle.Protist15025–32. 10.1016/s1434-4610(99)70006-4
94
SotoA. R.ZhengH.ShoemakerD.RodriguezJ.ReadB. A.WahlundT. M. (2006). Identification and preliminary characterization of two cDNAs encoding unique carbonic anhydrases from the marine alga Emiliania huxleyi.Appl. Environ. Microbiol.725500–5511. 10.1128/AEM.00237-06
95
TanakaR.KikutaniS.MahardikaA.MatsudaY. (2014). Localization of enzymes relating to C4 organic acid metabolisms in the marine diatom, Thalassiosira pseudonana.Photosynth. Res.121251–263. 10.1007/s11120-014-9968-9
96
TanakaY.NakatsumaD.HaradaH.IshidaM.MatsudaY. (2005). Localization of soluble beta-carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Sorting to the chloroplast and cluster formation on the girdle lamellae.Plant Physiol.138207–217. 10.1104/pp.104.058982
97
TortellP. D. (2000). Evolutionary and ecological perspectives on carbon acquisition in phytoplankton.Limnol. Oceanogr.45744–750. 10.4319/lo.2000.45.3.0744
98
TréguerP.BowlerC.MoriceauB.DutkiewiczS.GehlenM.AumontO.et al (2018). Influence of diatom diversity on the ocean biological carbon pump.Nat. Geosci.1127–37. 10.1038/s41561-017-0028-x
99
Van HeukelemL.ThomasC. S. (2001). Computer-assisted high-performance liquid chromatography method development with applications to the isolation and analysis of phytoplankton pigments.J. Chromatogr. A.91031–49. 10.1016/s0378-4347(00)00603-4
100
WickhamH. (2009). ggplot2: Elegant Graphics for Data Analysis.New York: Springer Science & Business Media.
101
WilhelmC.BüchelC.FisahnJ.GossR.JakobT.LaRocheJ.et al (2006). The regulation of carbon and nutrient assimilation in diatoms is significantly diff- erent from green algae.Protist15791–124. 10.1016/j.protis.2006.02.003
102
WolfK. K.HoppeC. J.RostB. (2018). Resilience by diversity: large intraspecific differences in climate change responses of an Arctic diatom.Limnol. Oceanogr.63397–411. 10.1002/lno.10639
103
WolfK. K.RomanelliE.RostB.JohnU.CollinsS.WeigandH.et al (2019). Company matters: the presence of other genotypes alters traits and intraspecific selection in an Arctic diatom under climate change.Glob. Chang. Biol.252869–2884. 10.1111/gcb.14675
104
WuY.JeansJ.SuggettD. J.FinkelZ. V.CampbellD. A. (2014). Large centric diatoms allocate more cellular nitrogen to photosynthesis to counter slower Rubisco turnover rates.Front. Mar. Sci.1:68. 10.3389/fmars.2014.00068
105
YoungJ. N.HeureuxA. M.SharwoodR. E.RickabyR. E.MorelF. M.WhitneyS. M. (2016). Large variation in the Rubisco kinetics of diatoms reveals diversity among their carbon-concentrating mechanisms.J. Exp. Bot.673445–3456. 10.1093/jxb/erw163
106
YoungJ. N.HopkinsonB. M. (2017). The potential for co-evolution of CO2-concentrating mechanisms and Rubisco in diatoms.J. Exp. Bot.683751–3762. 10.1093/jxb/erx130
107
YoungJ. N.RickabyR. E. M.KapralovM. V.FilatovD. A. (2012). Adaptive signals in algal Rubisco reveal a history of ancient atmospheric carbon dioxide.Philos. Trans. R. Soc. Lond. B367483–492. 10.1098/rstb.2011.0145
108
ZallotR.ObergN.GerltJ. A. (2019). The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways.Biochemistry584169–4182. 10.1021/acs.biochem.9b00735
109
ZengX.JinP.XiaJ.LiuY. (2019). Correlation of carbonic anhydrase and Rubisco in the growth and photosynthesis in the diatom Phaeodactylum tricornutum.J. Appl. Phycol.31123–129. 10.1007/s10811-018-1537-8
Summary
Keywords
Tara Oceans, diatoms, carbon metabolism, carbon dioxide concentration mechanisms, metagenomics, metatranscriptomics
Citation
Pierella Karlusich JJ, Bowler C and Biswas H (2021) Carbon Dioxide Concentration Mechanisms in Natural Populations of Marine Diatoms: Insights From Tara Oceans. Front. Plant Sci. 12:657821. doi: 10.3389/fpls.2021.657821
Received
24 January 2021
Accepted
23 March 2021
Published
30 April 2021
Volume
12 - 2021
Edited by
Benoit Schoefs, Le Mans Université, France
Reviewed by
Ansgar Gruber, Institute of Parasitology, Academy of Sciences of the Czech Republic (ASCR), Czechia; Yusuke Matsuda, Kwansei Gakuin University, Japan
Updates
Copyright
© 2021 Pierella Karlusich, Bowler and Biswas.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haimanti Biswas, haimanti.biswas@nio.org
This article was submitted to Marine and Freshwater Plants, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.