- 1Key Laboratory of Plant Functional Genomics of the Ministry of Education/Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding, Yangzhou University, Yangzhou, China
- 2Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Yangzhou University, Yangzhou, China
Myeloblastosis (MYB)-related transcription factors comprise a large subfamily of the MYB family. They play significant roles in plant development and in stress responses. However, MYB-related proteins have not been comprehensively investigated in rapeseed (Brassica napus L.). In the present study, a genome-wide analysis of MYB-related transcription factors was performed in rapeseed. We identified 251 Brassica napus MYB (BnMYB)-related members, which were divided phylogenetically into five clades. Evolutionary analysis suggested that whole genome duplication and segmental duplication events have played a significant role in the expansion of BnMYB-related gene family. Selective pressure of BnMYB-related genes was estimated using the Ka/Ks ratio, which indicated that BnMYB-related genes underwent strong purifying selection during evolution. In silico analysis showed that various development-associated, phytohormone-responsive, and stress-related cis-acting regulatory elements were enriched in the promoter regions of BnMYB-related genes. Furthermore, MYB-related genes with tissue or organ-specific, stress-responsive expression patterns were identified in B. napus based on temporospatial and abiotic stress expression profiles. Among the stress-responsive MYB-related genes, BnMRD107 was strongly induced by drought stress, and was therefore selected for functional study. Rapeseed seedlings overexpressing BnMRD107 showed improved resistance to osmotic stress. Our findings not only lay a foundation for further functional characterization of BnMYB-related genes, but also provide valuable clues to determine candidate genes for future genetic improvement of B. napus.
Introduction
Transcription factors play a major role in the regulation of gene expression. Myeloblastosis (MYB) proteins represent a large family of transcription factors, members of which perform diverse biological functions in all eukaryotes. MYB transcription factors are characterized by a conserved MYB domain containing one to four imperfect repeats of ~52 amino acids (Dubos et al., 2010). Generally, each repeat can form three alpha helices, among which the second and third helices shape the helix-turn-helix structure via three consistently spaced tryptophan residues, which are essential to form the hydrophobic core of the amino acids, thereby stabilizing the three-dimensional structure of MYB repeat and promoting DNA-binding (Sue et al., 2006). In plants, MYB transcription factors can be roughly divided into four categories, designated as 1R-MYB, R2R3-MYB (2R-MYB), R1R2R3-MYB (3R-MYB), and 4R-MYB, according to the number of adjacent repeats (R) in MYB domain, which is located at the N-terminus (Dubos et al., 2010). R2R3-MYB is the predominant subgroup, with multiple regulatory functions covering almost every aspect of plant life-cycle, such as cell shape determination, secondary metabolism, plant organ patterning, and stress responses (Aydin et al., 2014). To date, studies have mostly concentrated on R2R3-MYB genes because of the large number of members of this subfamily. R1R2R3-MYB transcription factors bind to mitosis-specific activator (MSA) element, and modulate cytokinesis and cell cycle in plants (Haga et al., 2007). R4-type MYB proteins were identified later in plants, and little is known about their functions (Chen et al., 2006). 1R-MYB type proteins, also known as the MYB-related transcription factors, usually contain a single (sometimes only partial) binding repeat (Kirik and Baumlein, 1996; Li et al., 2019). MYB-related proteins act as pivotal regulatory components by activating or repressing gene transcription in plants (Andronis et al., 2008).
The first MYB-related gene (mybSt1) in plants was identified in potato and was demonstrated to function as a transcriptional activator (Baranowskij et al., 1994). In recent decades, a growing number of MYB-related genes were subsequently characterized in many plant species, including Arabidopsis thaliana (Yong et al., 2019), Oryza sativa (Piao et al., 2019), Vitis labruscana (Azuma et al., 2011), and Brassica napus (Li et al., 2020). In Arabidopsis, several MYB-related proteins were shown to be involved in the circadian clock (Nagel and Kay, 2012). Timing of CAB expression 1 (TOC1) is a key evening oscillator component of the transcriptional network that establishes the core mechanism of circadian rhythm in plants (Kolmos et al., 2008). REVEILLE8/LHY-CCA1-LIKE5 (RVE8/LCL5) was reported to be capable of binding to the promoter of TOC1 and promoted clock-controlled TOC1 gene expression to regulate circadian rhythms in Arabidopsis (Farinas and Mas, 2011; Rawat et al., 2011). As central components of the circadian oscillator, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) were indicated to function synergistically and redundantly in Arabidopsis circadian clock by negatively regulating TOC1 expression (Makino et al., 2002; Lu et al., 2009). Some MYB-related genes, such as EARLY-PHYTOCHROME-RESPONSIVE1 (EPR1) (Kuno et al., 2003), KUODA 1 (KUA1) (Lu et al., 2014), REVEILLE2 (RVE2) (Zhang et al., 2007), and MYB HYPOCOYL ELONGATION-RELATED (MYBH) (Kwon et al., 2013), were also revealed to participate in circadian clock regulation in Arabidopsis. REVEILLE1 (RVE1) is a critical node that integrates the circadian system and auxin signaling by regulating the expression of auxin biosynthetic gene YUCCA8 (YUC8, also known as CYTOKININ INDUCED ROOT CURLING2) (Rawat et al., 2009). A previous study suggested that MYBL2 could interact with MYB-bHLH-WDR (MBW) complexes to regulate flavonoid biosynthesis, and MYBD promoted anthocyanin accumulation by regulating MYBL2 expression in a circadian-dependent manner (Dubos et al., 2008; Nguyen and Lee, 2016). In addition to playing significant roles in circadian rhythm regulation, the MYB-related proteins also participate in the regulation of a broad range of developmental processes, such as seed germination (Zhao et al., 2019), trichome differentiation (Tominaga-Wada et al., 2017), leaf development and senescence (Piao et al., 2019), shoot apical meristem (SAM) formation (Lin et al., 2007), root development (Li et al., 2020), and flowering (Shibuta and Abe, 2017). A single-repeat MYB-related transcription factor TRIPTYCHON (TRY), was shown to be expressed in trichomes and mediated trichome patterning and position-dependent cell determination in root hair formation in Arabidopsis (Schellmann et al., 2002). MYB-related c1/pl1 genes were revealed to act as regulators of anthocyanin synthesis in maize in response to different light qualities and cytokinin (Piazza et al., 2002; Pilu et al., 2003). Grape VvmybA1-1, VvmybA1-2, and VvmybA2 were isolated from V. labruscana, and were indicated to manipulate the skin colors of grapes by regulating anthocyanin biosynthesis in grape berries (Azuma et al., 2008). The maize MYB-related protein-1 (ZmMRP-1), which is specifically expressed in the transfer cell layer of the endosperm, was reported to play a role in determining the differentiation of transfer cells by transcriptionally activating the expression of maize endosperm transfer cell-specific gene BETL1, and this process relied on the interaction between ZmMRP-1 and two C2H2 zinc finger proteins (ZmMRPI-1 and ZmMRPI-2) (Royo et al., 2009). Overexpression of the MYB-related gene, OsMYB102 in rice delayed leaf senescence, and OsMYB102 was suggested to modulate the expression of various kinds of senescence-associated genes, including genes participating in abscisic acid (ABA) metabolism and signaling (Piao et al., 2019). FE, which encodes a phloem-specific MYB-related protein, was documented to positively regulate the transcription level of FLOWERING LOCUS T (FT) and FLOWERING LOCUS T INTERACTING PROTEIN 1 (FTIP1), thereby promoting flowering in Arabidopsis (Abe et al., 2015). MYB-related AtCDC5 was proven to be critical for G2 to M transition of the cell cycle and controls the function of SAM in Arabidopsis (Lin et al., 2007). A wheat MYB-related gene, TaMYB72, was indicated to promote flowering in rice via upregulation of florigen genes, Hd3a and RFT1 (Zhang et al., 2016).
Notably, MYB-related genes were also demonstrated to play significant roles in improving abiotic stress tolerance in plants. A large number of MYB-related genes respond to diverse abiotic stresses and are crucial for plant resistance to adverse environmental conditions (Shi et al., 2015; Yong et al., 2019). A rice MYB-related gene, OsMYB48-1 was reported to regulate drought and salt tolerance positively by regulating ABA biosynthesis (Xiong et al., 2014). Overexpression of OsMYBR1 conferred improved drought tolerance and reduced ABA sensitivity to rice (Yin et al., 2017). Heterologous expression of a Fraxinus velutina gene, FvMYB1, in tobacco enhanced salt tolerance of the transgenic tobacco plants (Li et al., 2016). A recent study showed that RSM1 regulates seed germination and seedling development under ABA treatment and salt stress conditions by interacting with HY5/HYH (Yang et al., 2018). Overexpression of lily LlMYB3 in Arabidopsis increased cold, salt, and drought tolerance of the transgenic plants (Yong et al., 2019). Sheepgrass LcMYB2 was suggested to positively regulate drought tolerance by promoting root growth and osmotic adjustment (Zhao et al., 2019). Moreover, overexpression of GmMYB118 enhanced drought and salt tolerance in both transgenic Arabidopsis and soybean seedlings (Du et al., 2018). MYB-related genes were also revealed to be involved in cold tolerance. For example, the GARP-like transcription factor, VaAQUILO (VaAQ) was reported to dramatically improve cold tolerance by increasing the content of raffinose family oligosaccharides (RFOs) and osmoprotectants in transgenic Arabidopsis and in Amur grape calli (Sun et al., 2018).
Rapeseed is the third largest oil crop globally, and is a vital agricultural resource (Liao et al., 2019). Previous transcriptomic analysis revealed that MYB-related genes might be involved in the growth and development of B. napus (Hong et al., 2017; Li et al., 2020). However, systematic analysis of MYB-related proteins and the research demonstrating the roles of MYB-related genes in B. napus are still lacking. In the present study, various in silico approaches were applied to identify BnMYB-related genes, and 251 genes encoding BnMYB-related proteins were identified in B. napus. The phylogenetic relationships, gene structures, cis-elements, temporospatial expression patterns, and expression profiles of BnMYB-related genes under various stress conditions were studied comprehensively. Considering that little is known about the roles of MYB-related genes in B. napus, the results of this study will provide a useful reference for further understanding of the functions of BnMYB-related genes in response to abiotic stresses and in the regulation of developmental processes in B. napus.
Materials and Methods
Identification of MYB-Related Genes in the B. napus Genome
The genome and protein sequences of B. napus (Darmor-bzh) were obtained from the Genoscope database (http://www.genoscope.cns.fr/brassicanapus/). Conserved DNA binding domains of known MYB-related proteins were employed to search against the B. napus database (https://www.arabidopsis.org/) using the BLASTP program with an E value of less than 1e−10 to identify BnMYB-related proteins. Meanwhile, “MYB conserved domain” and “Myb_DNA-binding” were used as keywords to query the Pfam database (https://pfam.xfam.org/) (El-Gebali et al., 2019), and the corresponding Hidden Markov Model (HMM) profile (PF00249) was downloaded and employed to identify MYB-related proteins from B. napus using the HMMER tool (https://www.ebi.ac.uk/Tools/hmmer/). Moreover, NCBI-Conserved Domain Data (CDD) search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and InterproScan (http://www.ebi.ac.uk/interpro/search/sequence-search) were used to analyze and classify the BnMYB-related protein sequences. Then, the non-redundant sequences were compared with the MYB-related family in the plant transcription factor (TF) database version 4.0 (PlantTFDB 4.0, http://planttfdb.cbi.pku.edu.cn/) (Jin et al., 2017). Conserved domains of the retrieved putative MYB-related proteins were verified manually. Detailed information for the BnMYB-related genes is listed in Supplementary Table 1. ExPASy (https://web.expasy.org/protparam/) was used to calculate the number of amino acids, molecular weights (MW), and theoretical isoelectric points (pI) of BnMYB-related proteins.
Multiple Sequence Alignments and Phylogenetic Analysis
Multiple alignments of full-length BnMYB-related protein sequences were conducted using ClustalX (version 1.83) (Thompson et al., 1997). A phylogenetic tree was constructed using MEGA 7.0 software (https://www.megasoftware.net/) with the neighbor-joining (NJ) method, and bootstrap test was performed with 1,000 iterations, as described previously (Wang et al., 2017).
Gene Structure and Motif Analysis
Gene structure data were obtained from the B. napus genome annotation file on the Genoscope database. Exon-intron organizations of BnMYB-related genes were shown by the gene structure display server program (GSDS2.0, http://gsds.cbi.pku.edu.cn/) based on the alignment results of the complementary DNA (cDNA) sequences with the corresponding genomic DNA sequences of each gene. The MEME (http://meme-suite.org/tools/meme) program was used to identify the conserved motifs in BnMYB-related protein sequences. The motif distribution type was set as zero or one occurrence per sequence. The number of motifs was set as 20, and the motif width was between 6 and 50 amino acids. These data were integrated and visualized using TBtools (Chen et al., 2020).
Gene Location and Duplication
Location information of BnMYB-related genes was obtained from the B. napus genome annotation file on the Genoscope database. BnMYB-related genes were mapped to the B. napus genome using the MG2C (http://mg2c.iask.in/mg2c_v2.0/) online tools. Moreover, the whole-genome protein sequences from rapeseed were blasted against B. napus, B. rape, B. oleracea, and A. thaliana databases with an E value of less than 1e−10. MCScanX was used to detect the collinear blocks with default parameter settings (Wang et al., 2012). Ka/Ks ratios were calculated by ParaAT2.0 and KaKs_Calculator 2.0 programs (Wang et al., 2010; Zhang et al., 2012). The collinear blocks of BnMYB-related genes were obtained. In addition, the homolog pairs of MYB-related genes in eight rapeseed accessions of three ecotypes (winter-type, semi-winter-type, and spring-type) including ZS11, Gangan, No2127, Quinta, Shengli, Tapidor, Westar, and Zheyou7 were captured from the BnPIR database (Brassica napus pan-genome information resource, http://cbi.hzau.edu.cn/bnapus/) (Song et al., 2020). The Upset plot and colinearity between them were visualized using TBtools (Chen et al., 2020). The putative duplicated genes were linked with arcs in the relevant figure.
Regulatory Cis-Element Analysis
A 2-kb upstream sequence of coding regions of each BnMYB-related gene was selected as the promoter region for cis-element analysis. The Samtools program (http://www.htslib.org/) was used to obtain the promoter sequences from the B. napus genome, which were searched against the plant cis-acting regulatory DNA elements database (PlantCARE, http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to identify the putative cis-acting regulatory elements (Lescot et al., 2002).
Plant Materials, Growth Conditions, and Treatments
To examine the expression patterns of BnMYB-related genes in various B. napus tissues and organs, and under a variety of abiotic stress conditions, rapeseed seeds were germinated on wet filter papers. Germinated rapeseed seedlings were then transplanted to the field or a growth chamber with a 16 h light/8 h dark photocycle. For temporospatial expression analysis, the rapeseed plants were grown under field conditions over the whole growth period. Cotyledons, roots, stems, young leaves, SAM, mature leaves, flowers, ovaries, silique walls, and seeds at different stages [14 days, 24 days, 34 days, and 50 days after pollination (DAP)] were sampled. In order to establish the expression profile under various stress conditions, 4-week-old B. napus seedlings cultured in nutrient solutions in a glasshouse were subjected to various treatments, including drought, heat, salt, cold stress, and ABA treatment. For drought treatment, seedlings were taken out from the nutrient solution and dehydrated, and the leaves were sampled at 0.5 h after treatment. For salt treatment, seedlings were transferred to nutrient solutions containing 200 mM NaCl and sampled at 6 h after treatment. For cold and heat treatment, rapeseed seedlings were transferred to growth chambers at 4 and 42°C, and sampled at 6 and 1 h after treatment, respectively. For ABA treatment, seedlings were sprayed with 100 μM ABA and sampled at 6 h after treatment. Three biological replicates were performed for each experiment with five plants for each biological replicate.
RNA Isolation and Quantitative Real-Time Reverse Transcription PCR (qRT-PCR) Analysis
Plant tissues/organs were frozen in liquid nitrogen and ground into a fine powder using a mortar. Total RNA was extracted using an RNAprep pure plant kit (TIANGEN Biotech, Beijing, China) according to the instructions of the manufacturer. Then, 4 μg of total RNA of each sample was reverse transcribed using the HiScript® II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, Nanjing, China) to produce cDNA. For qPCR analysis, gene-specific primers were designed using PrimerExpress v3.0 (Applied Biosystems, Foster City, CA, USA). The qPCR assay was carried out using PowerUpTM SYBRTM Green Master Mix (Applied Biosystems, Waltham, MA, United States) according to the instructions of the manufacturer on StepOnePlusTM Real-time PCR instrument (Applied Biosystems, Waltham, MA, United States). The BnActin gene was used as an internal control. Three technical replicates were conducted for each sample. The reaction procedure was as follows: 50°C for 2 min and 95°C for 5 min; followed by 40 cycles of 95°C for 15 s, 58°C for 15 s, and 72°C for 30 s. Relative expression of BnMYB-related genes was calculated as described previously (Livak and Schmittgen, 2001). The primers used for PCR are listed in Supplementary Table 1.
Gene Cloning and Vector Construction
The full-length coding region of BnMRD107 (without its stop codon) was amplified and inserted into pMDC83 vector under the control of CaMV35S promoter to express a BnMRD107-GFP fusion protein, and the construct was designated as BnMRD107-OE. The primers used in this experiment are listed in Supplementary Table 1.
Generation of Transgenic B. napus Plants
Genetic transformation of B. napus was based on the Agrobacterium-mediated hypocotyl transformation method described previously (Dai et al., 2020), and a pure line semi-winter rapeseed J9712, was used as the transformation recipient. The regenerated B. napus plants were screened on a medium containing 25 mg/L hygromycin B.
Phenotyping of B. napus Plants for Osmotic and Salt Stress Tolerance at the Cotyledon Stage
The T2 generation of homozygous transgenic BnMRD107-OE lines in rapeseed “J9712” background was chosen for further analysis. For rapid identification of the performance of transgenic materials and control plants under various stress conditions, stress treatments on the culture medium at the cotyledon stage were carried out. Untransformed J9712 was used as the control. For osmotic and salt tolerance assays at the cotyledon stage, selfed seeds of BnMRD107-OE lines and J9712 were surface-sterilized with 70% ethyl alcohol and 10% sodium hypochlorite, and then germinated on plates comprising 1/2 Murashige and Skoog (MS) medium. The seeds were incubated at 22°C, 16 h light/20°C, 8 h dark cycle until germinated. Germinated seeds with the same growth vigor were then transferred to 1/2 MS medium, and 1/2 MS medium containing 150 mM mannitol (for osmotic stress) and 150 mM NaCl (for salt stress), respectively. The rapeseed materials were cultured in a 22°C, 16 h light/20°C, 8 h dark cycle for 5 days. Seedlings were photographed, and their hypocotyl length, root length, and fresh weight were measured. Three independent biological replicates were performed for each experiment with five seedlings.
3,3′-Diaminobenzidine (DAB) Staining
3,3′-diaminobenzidine (DAB) DAB staining of cotyledons of BnMRD107-OE and J9712 seedlings under normal, osmotic, and salt stress conditions was performed following a method described previously (Liu Y. et al., 2020).
Results
Identification of BnMYB-Related Proteins
Genome sequence of B. napus was downloaded from the GENOSCOPE database. BLASTP searching was applied to identify BnMYB-related proteins using the DNA-binding domain of known MYB-related proteins against the B. napus database. Putative MYB-related proteins in B. napus were also retrieved based on HMMER model using the PF00249 HMM profile. NCBI-CDD searching and InterproScan were used to test the dependability of the results. A total of 251 BnMYB-related genes were identified, with 120 and 131 genes from the AA- and CC-subgenomes, respectively (Supplementary Table 2). Validated BnMYB-related genes were named as BnMRD1-BnMRD251 based on their sequential locations on the B. napus chromosomes to keep the nomenclature consistent.
Molecular Characteristics of BnMYB-Related Proteins and Chromosomal Localization of BnMYB-Related Genes
Molecular characteristics, including the protein length, MW, and isoelectric point (pI) of BnMYB-related proteins were analyzed (Supplementary Table 2). The protein lengths of BnMYB-related proteins ranged from 51 (BnMRD54) to 1,885 amino acids (BnMRD27). The MWs of BnMYB-related proteins ranged from 5.67 (BnMRD54) to 208.31 kDa (BnMRD27). The pIs of 171 BnMYB-related proteins (68.92%) were >7, suggesting that these proteins were basic proteins, and the remaining 31.08% of the proteins were acidic proteins. Locations of all BnMYB-related genes on the B. napus chromosomes are shown in Supplementary Figure 1. Total of 251 BnMYB-related genes were distributed in 19 chromosomes (i.e., A01–A10, and C01–C09), whereas 34 members (e.g., BnMRD5, BnMRD111, BnMRD159, and BnMRD183) were not shown on the B. napus chromosomes because they were located on unanchored scaffolds, which were unable to map onto the specific chromosomes. As shown in Supplementary Figure 1, BnMYB-related genes were unevenly distributed on 19 chromosomes. The number of MYB-related genes distributed on C03 was the highest, whereas that distributed on A04 was the lowest.
Gene Structures, Motif Identification, and Phylogenetic Analysis
A neighbor-joining (NJ) phylogenetic tree of MYB-related family was constructed to investigate the evolutionary relationship of MYB-related proteins using the full-length sequences of MYB-related proteins. All the 251 MYB-related proteins were classified into five distinct clades, named as CCA1/R-R-like, I-box-like, TRF-like, CPC-like, and TBP-like, respectively (Figure 1). CCA1/R-R-like clade was the largest clade. To determine the structural conservation and diversification of MYB-related family, the exon-intron structures of each individual BnMYB-related gene were characterized, and an online MEME analysis tool was applied to identify the conserved motifs of BnMYB-related proteins.
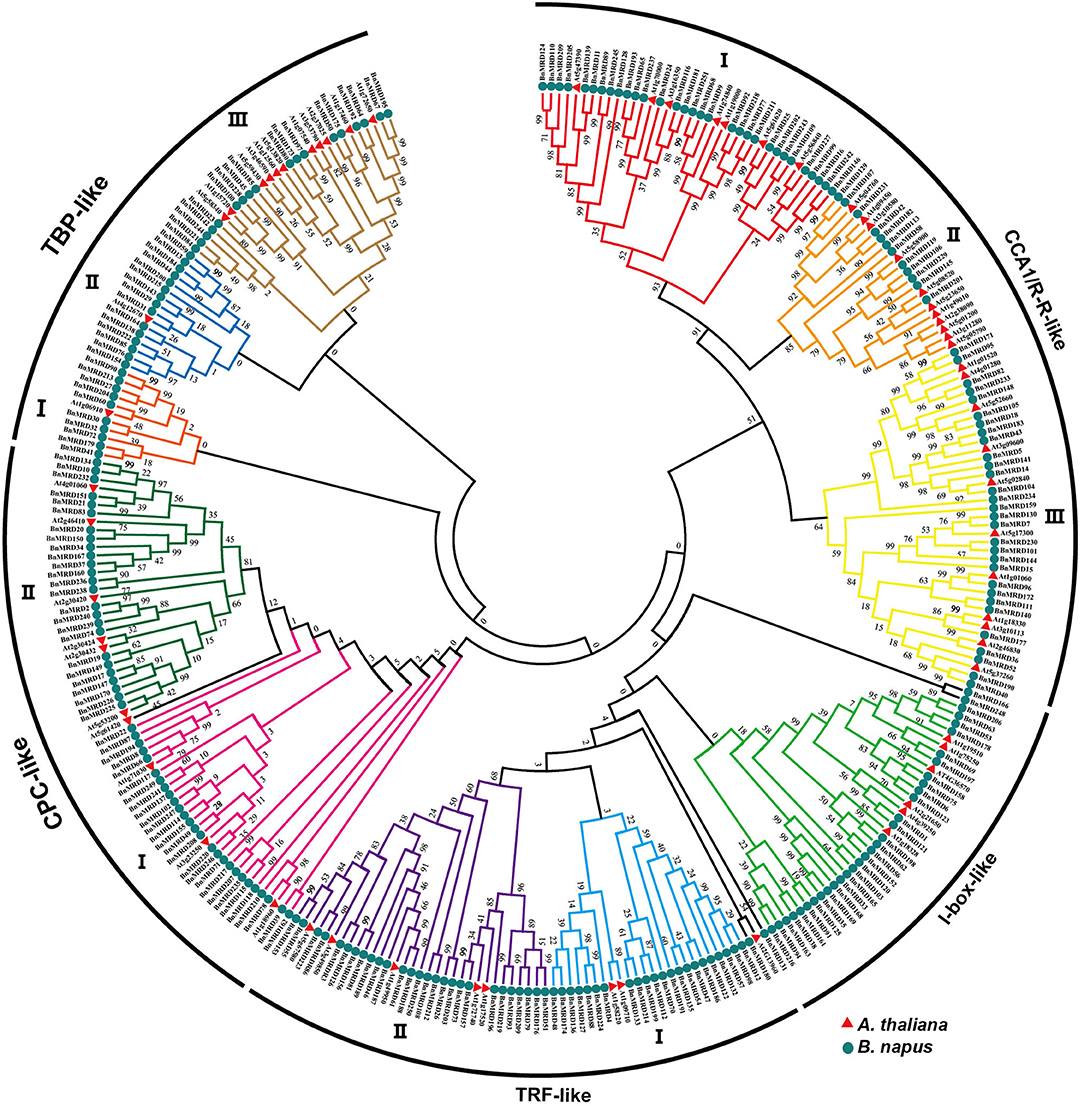
Figure 1. Phylogenetic analysis of MYB-related proteins using MEGA 7.0 with the neighbor-joining (NJ) method. Numbers at the nodes represent the reliability percentage of the bootstrap values based on 1,000 replications. Red triangles and green circles represent the MYB-related proteins in Arabidopsis thaliana (A. thaliana) and Brassica napus (B. napus), respectively.
The number of introns of MYB-related genes varied dramatically in B. napus (Figure 2A). BnMYB-related proteins were subdivided into smaller subgroups (I, II, III, and IV) according to the characteristics of motif composition and the exon-intron structures of their encoding genes. The numbers of exons ranged from 1 to 30 (Figure 2A). Nine genes of I-box-like clade had no introns. Most of the members of CCA1/R-R-like (I, II) group had 2–3 introns, while the members of CCA1/R-R-like (III) group had 8–15 introns. The majority of members of CPC-like clade had 2–3 introns. TBP-like group genes had the most complex exon-intron structure, with up to 24 introns. In general, there was no distinct regularity in the intron distribution pattern among the BnMYB-related genes; however, members belonging to the same clade shared identical or similar intron phases and motif compositions (Figure 2).
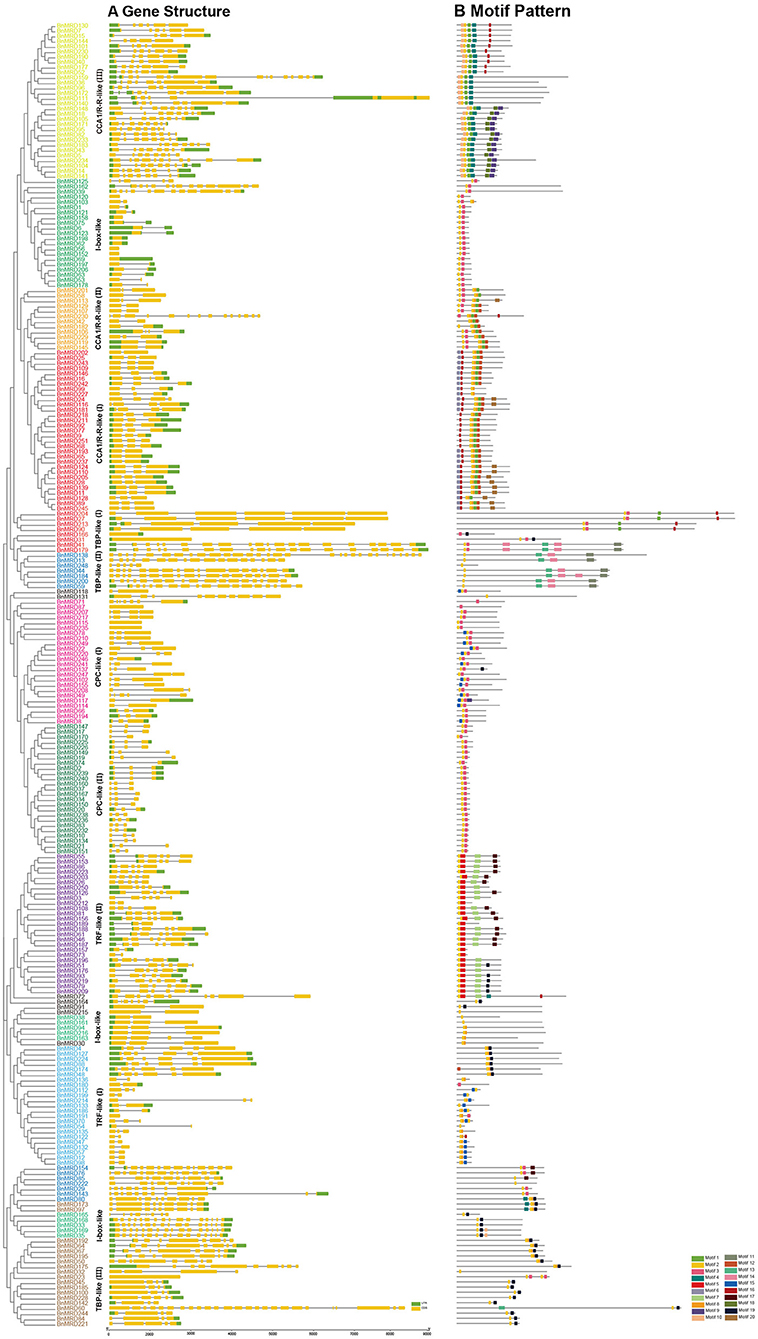
Figure 2. Gene structures and motif compositions of MYB-related proteins in B. napus. (A) Exon-intron structures of BnMYB-related proteins. Green boxes indicate 5′- and 3′-untranslated regions (UTR), respectively. Yellow boxes indicate exons, and black lines indicate introns. (B) Distribution of conserved motifs in BnMYB-related proteins. Motifs 1–20 identified by MEME analysis are displayed in different colored boxes.
Sequence alignment revealed that the conserved core sequence of MYB domains in MYB-related proteins belonging to different clades was remarkably divergent, whereas the tryptophan (W) residues were found to be evenly distributed in the conserved MYB repeat regions in most subgroups of MYB-related proteins. There were three W residues in MYB-domains of TRF-like (II) and CPC-like (II) members. Substitution or deletion at the first or the third W residue in MYB domain was observed in more than 20% of BnMYB-related proteins. For instance, members of CCA1/R-R-like subgroup had a conserved sequence of SHAQKY(N)F or SHAQK, in which the third W residue was substituted by Alanine (A). The third W residue was substituted by tyrosine (Y) in I-box-like clade proteins. The amino acid sequences of MYB domain were found to vary significantly in different subgroups in TBP-like clade members. However, a conserved sequence of DLKDKW(R/K) (N/T) was found in TBP-like (III) subgroup proteins. The conserved and subgroup-specific core sequences of MYB domains indicated that members in the same subgroup had a common ancestor during evolution (Figure 2). As shown in Figure 2B, 20 motifs were recognized in BnMYB-related proteins, with lengths of 11 to 50 amino acids. Motif 2 was the most conserved motif, being present in all the family members, which made up the core sequence of the single MYB domain. Motif 1, with the conserved SHAQKY sequence, was a distinct motif that existed in CCA1/R-R-like clade. CCA1/R-R-like (I) subgroup members generally harbored motif 1, motif 2, motif 6, motif 8, motif 12, and motif 16. Interestingly, N-termini of the proteins in this subgroup were relatively conserved. Motif 1, motif 2, motif 3, motif 8, and motif 12 were identified in CCA1/R-R-like (II) subgroup. Besides, motif 4 (QKSGTNIHIPPPRPKRKPAHPYPRKAGKN) and motif 10 (SGEDLAKKVRKPYTITKSRER) were considered as the basis to distinguish CCA1/R-R-like (III) members from members of the other two subgroups in CCA1/R-R-like clade. Proteins of I-box-like clade had the simplest motif composition with only two motifs (motif 2 and motif 3). TRF-like clade was divided into two subgroups. Motif 15 was found in 11 TRF-like (I) members and motif 19 was found in five TRF-like (II) members. Motif 3, motif 5, and motif 7 were the most common motifs in TRF-like (II) proteins. Motif 19 and motif 17 were also found in several members of TRF-like (II) subgroup. CPC-like clade also had two subgroups. CPC-like (I) proteins exhibited a similar motif composition (motif 2 and motif 3) to those of I-box-like proteins; however, the locations of conserved motifs in CPC-like (I) and I-box-like proteins were different. TBP-like clade proteins were basically classified into three subgroups. Multiple motifs were detected in each TBP-like subgroup, and the distribution of the motifs was relatively scattered in the proteins.
CCA1/R-R-like clade, with 73 members, was the largest of the five clades, and this clade could be further divided into three subgroups according to the exon-intron structure and motif characteristics. Among the 73 CCA1/R-R-like proteins, 31, 12, and 30 members belonged to group I, group II, and group III, respectively. All 73 members shared a highly conserved motif, SHAQKY(N)F or SHAQK, which is located in the third helix of MYB domain (Figure 3). Most of the MYB-related family members had a single MYB domain. However, CCA1/R-R-like (II) proteins in rapeseed contained two separate MYB domains, which are located in the N-terminus and the middle of the protein, respectively. This contrasted with the situation in 2R-MYB proteins, in which the two MYB domains are both distributed in the N-terminus of the proteins. TBP-like clade was the second largest, comprising 52 members. TBP-like clade had four subgroups with different exon-intron patterns and other conserved features. Ten, seven, 20, and 15 members fell into group I, group II, group III, and IV, respectively. Members of TBP-like (III) shared a consensus motif, DLKDKW(R/K) (N/T). Members of TBP-like clade had complicated exon-intron patterns (Figure 2). Both TRF-like and CPC-like clades comprised two subgroups. Most members of TBP-like (III) subgroup had two or three exons, while the majority of TBP-like (I) subgroup members had five to six exons (Figure 2). All of the individual MYB-related genes could be assigned into a clade based on phylogenetic analysis, except for BnMRD131. As shown in Figure 2, BnMRD131 had a complex exon-intron organization and a distinct motif composition, which supported the hypothesis that BnMRD131 was an orphan in BnMYB-related family. On the whole, most of the adjacent members in the phylogenetic tree exhibited similar exon-intron organizations and motif compositions.
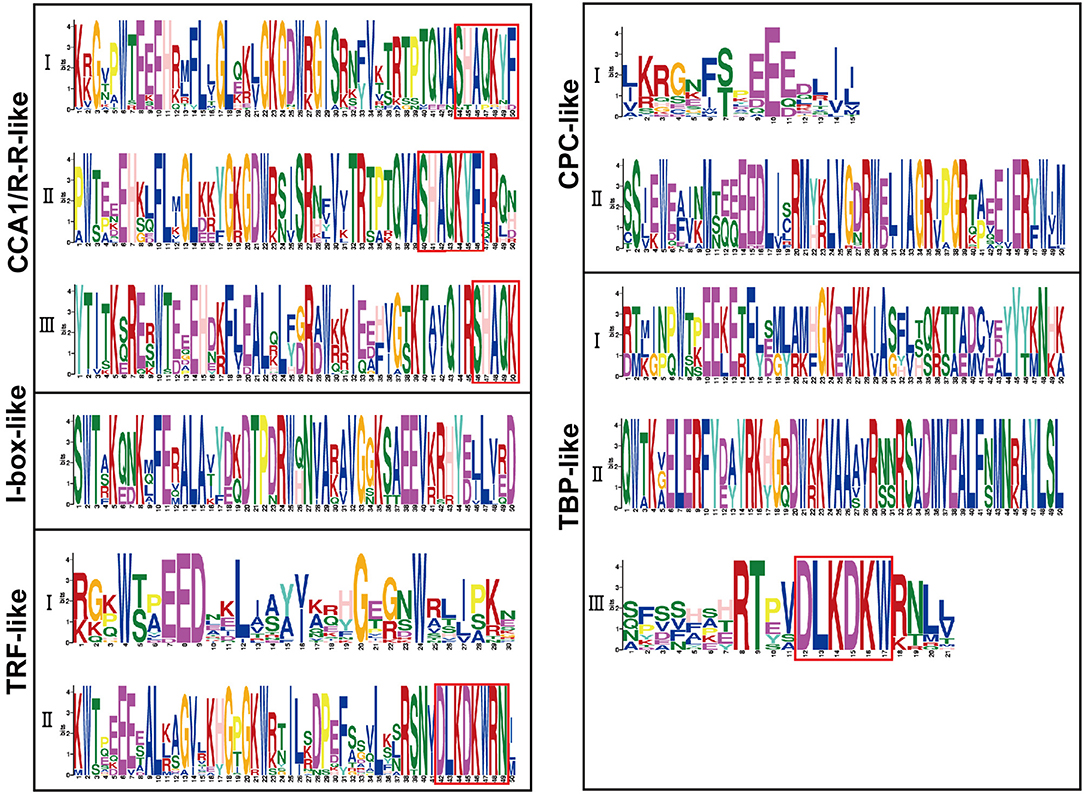
Figure 3. Amino acid sequence logos of the MYB domains in BnMYB-related proteins. Bit score indicates the information content for each position in the sequence. The conserved Trp (W) residues distributed in the amino acid sequence of MYB domains, and red boxes indicate the conserved motifs. The corresponding clades based on the phylogenetic tree (Figure 2) are indicated on the left for reference.
Synteny Analysis of MYB-Related Genes
The BLASTP and MCScanX programs were used to identify the homologous genes in BnMYB-related family based on synteny analysis. A total of 179 pairs of MYB-related paralogs were found in the B. napus genome (Supplementary Figure 2, Supplementary Table 3). Three tandemly duplicated genes, 194 pairs of whole genome duplication (WGD) or segmentally duplicated genes, 51 dispersedly duplicated gene pairs, and three proximal gene pairs were detected among BnMYB-related family (Supplementary Table 4). These results indicated that substantial numbers of BnMYB-related genes were generated by WGD or segmental duplication events, which played essential roles in the process of BnMYB-related family evolution.
To further demonstrate the expansion and clustering of MYB-related family members, chromosomal syntenic genes of BnMYB-related genes in Brassicaceae, including Arabidopsis, B. oleracea, and B. rapa were analyzed. About 45.42% (114/251) of BnMYB-related genes had a syntenic relationship with MYB-related genes in the other three Brassicaceae species. 64, 76, and 84 orthologous genes located in syntenic blocks were identified from Arabidopsis, B. oleracea, and B. rapa, respectively (Supplementary Figure 2, Supplementary Tables 5–7), indicating that the expansion of BnMYB-related family was accompanied by gene loss. However, 91 BnMYB-related genes might be newly generated, which suggested that WGD played an essential role in the expansion of BnMYB-related gene family. Moreover, the homolog pairs of MYB-related genes from eight rapeseed accessions were identified. The results suggested that a total of 178 MYB-related genes were conserved in all eight rapeseed accessions, while 54 MYB-related genes were specific to Darmor-bzh cultivar. Besides the above-mentioned 178 common members in eight accessions, 14 genes were shared between Darmor-bzh and ZS11, and 1–2 genes were shared in other accessions (Figure 4). The corresponding relationships of MYB-related homologs among the eight rapeseed accessions are listed in Supplementary Table 8. The homologs of 78.09% (196/251), 72.91% (183/251), 70.92% (178/251), 72.51% (182/251), 71.31% (179/251), 70.92% (178/251), 71.71% (180/251), and 70.92% (178/251) of MYB-related genes in Darmor-bzh were identified by alignment in ZS11, Gangan, No2127, Quinta, Shengli, Tapidor, Westar, and Zheyou7, respectively. Overall, these results demonstrated that WGD events were the major forces driving the expansion of MYB-related genes in B. napus and the MYB-related family is largely conserved in different rapeseed accessions.
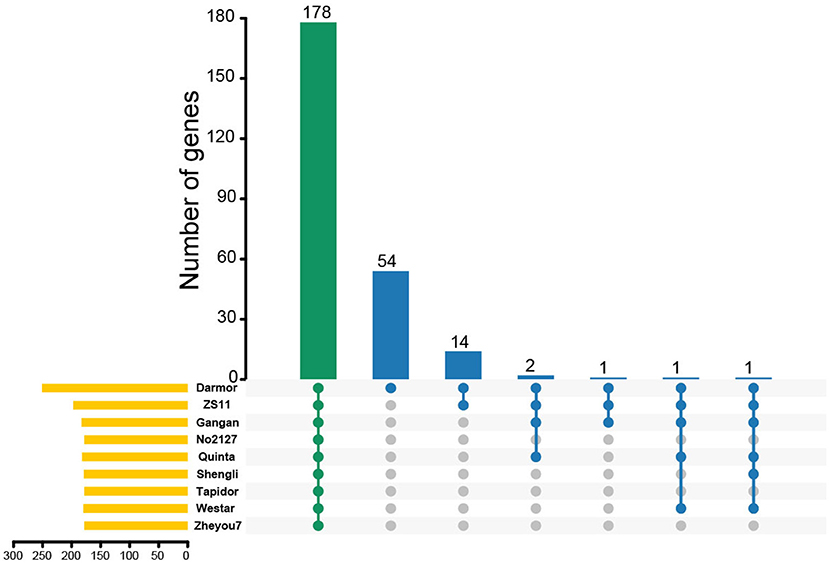
Figure 4. The Upset plot shows the distributions of homologous MYB-related genes in different rapeseed accessions. The bar chart above represents the number of common homologous genes shared among different rapeseed accessions. The left bar chart at the bottom represents the number of homologous genes in each accession. The right dotted line at the bottom shows the number of accessions contained in the group.
To gain a deeper insight into the evolutionary selective constraints on duplicated BnMYB-related genes, ParaAT2.0 (https://bigd.big.ac.cn/tools/paraat) was used to calculate non-synonymous (Ka) and synonymous (Ks) values, and the Ka/Ks substitution ratios. The Ka/Ks ratios were lower than 1 in all identified BnMYB-related segmental duplications, and the divergence time ranged from 0.515 (BnMRD215 and BnMRD32) to 0.0028 (BnMRD16 and BnMRD146) million years ago (Supplementary Table 3). This result provided evidence of strong purifying selection pressure on these duplicate genes in rapeseed. The Ka/Ks ratios were also calculated to evaluate the selection pressure among duplicated MYB-related gene pairs of A. thaliana vs. B. napus, B. oleracea vs. B. napus, and B. rapa vs. B. napus (Supplementary Tables 5–7). The Ka/Ks values from all the identified gene pairs were less than 1, indicating that MYB-related family in B. napus and A. thaliana, as well as in B. napus and its diploid progenitors, experienced purifying selection during family expansion.
Cis-Element Analysis of the Promoter Regions of BnMYB-Related Genes
A 2-kb genomic sequence upstream of the transcription start site (TSS) of each BnMYB-related gene was used to probe the regulatory cis-acting elements enriched in BnMYB-related genes using the plant CARE database. As shown in Figure 5 and Supplementary Table 9, 23 types of cis-acting elements were identified in the promoters of BnMYB-related genes. The existence of abundant cis-acting elements in the promoter regions of BnMYB-related genes might have implications for their transcriptional regulation and biological functions. The identified cis-acting elements were mainly divided into three types. Eleven elements including light responsive motif (GT1-motif), circadian control, zein metabolism regulation, meristem expression (CAT-box), endosperm expression (GCN4-motif), cell cycle regulation, endosperm specific negative expression (AACA-motif), meristem-specific activation (NON-box), differentiation of the palisade mesophyll cells, seed-specific regulation (RY-element), and root-specific cis-elements, were associated with plant growth and development. Light responsive elements appeared to be the most common cis-acting elements in BnMYB-related gene promoters, suggesting that BnMYB-related genes might play a vital role in the response of B. napus to variable light conditions. Several cis-elements, such as meristem-specific activation, seed-specific regulation, and root-specific elements, were considered to be associated with temporospatial-specific expression of the BnMYB-related genes. Five elements including gibberellin-responsive (GARE), ABA responsive (ABRE), auxin responsive (TGA), methyl jasmonate (MeJA)-responsive (TGACG-motif), and salicylic acid responsive elements (TCA) were responsible for phytohormone responses. Five elements including anaerobic induction (ARE), wound responsive (WUN-motif), defense and stress responsive (TC-rich repeats), low-temperature responsive (LTR), and dehydration, low-temperature, salt stress-responsive were involved in stress responses, implying that the BnMYB-related genes containing these cis-elements in their promoter regions might participate in the adaptation of B. napus to everchanging environmental conditions. Interestingly, an anaerobic induction element appeared as tandem repeats in some of the BnMYB-related gene promoters (BnMRD10, BnMRD40, BnMRD45, BnMRD47, BnMRD70, BnMRD71, BnMRD134, BnMRD118, BnMRD207, BnMRD225, BnMRD226, and BnMRD227), hinting that the regulatory proteins which bind to these regions might have special spatial structures. It is noteworthy that an MYB binding site was found in the promoter region of most BnMYB-related genes, revealing that the transcriptional abundance of BnMYB-related genes was largely modulated by MYB transcription factors.
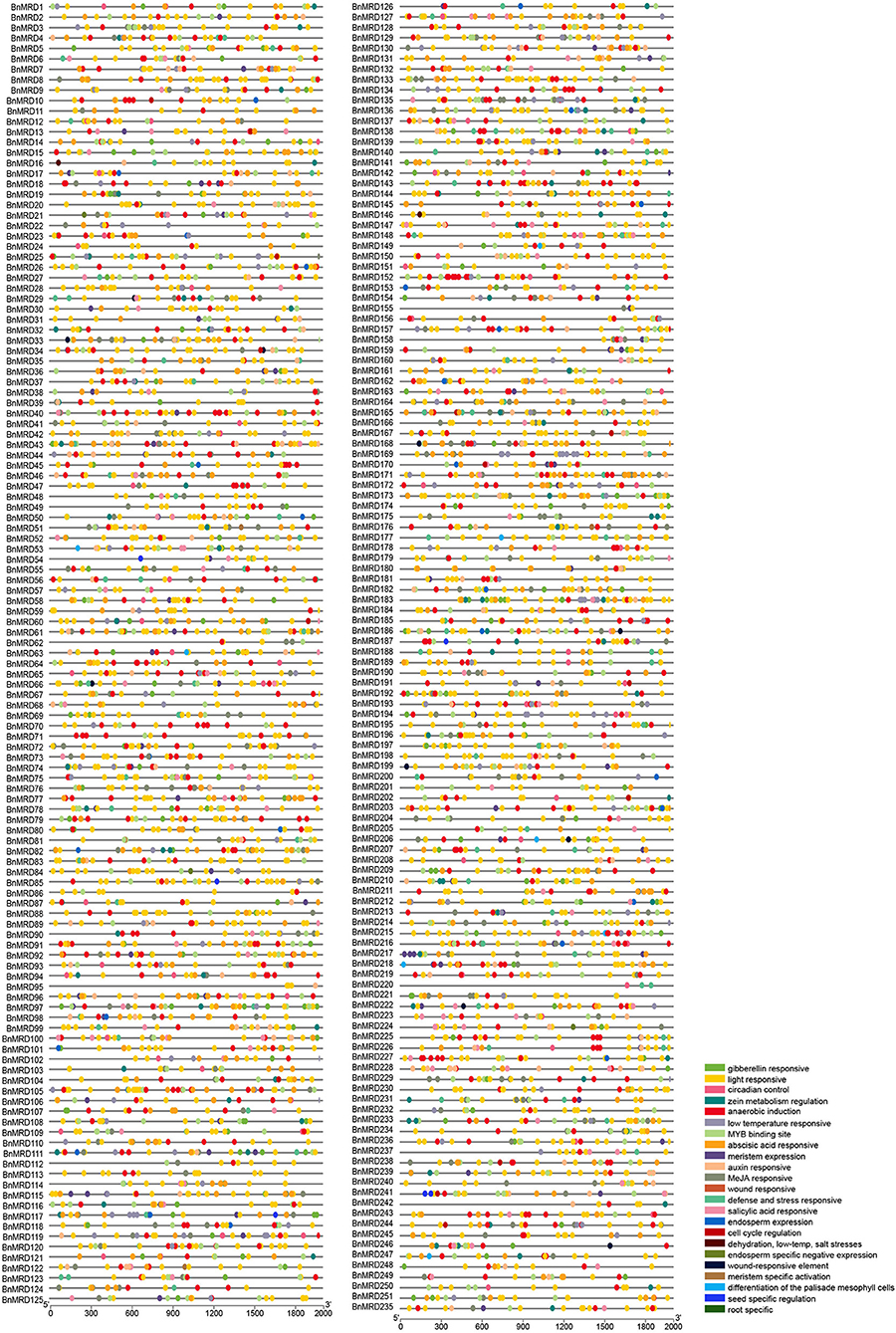
Figure 5. Regulatory cis-elements on the promoter regions of BnMYB-related genes. Locations of different cis-acting elements are arranged on the lines representing the 2 kb upstream region of TSS in the promoters of each BnMYB-related gene. Different colored ovals represent different types of cis-elements.
Temporospatial Expression Profiles of BnMYB-Related Genes
To dissect the expression characteristics of BnMYB-related genes, their transcript patterns in tissues and organs of the main developmental stages (cotyledon, root, stem, leaf, SAM, overleaf, flower, ovary, silique wall at 14, 24, 34, and 50 DAP, and seed at 14, 24, 34, 50 DAP) of B. napus were examined based on transcriptome data, and a visualization heatmap was generated (Figure 6, Supplementary Table 10). The results showed that BnMYB-related genes exhibited distinct expression patterns among the tissues and organs detected in this study, revealing their diverse functions during B. napus development. Among the BnMYB-related genes, 237 genes showed detectable expression in at least one tissue or organ. The remaining 14 BnMYB-related genes were not expressed in tested tissues or organs, 10 of which belonged to TRF-like clade, and four genes belonged to CPC-like clade. These genes might be non-functional, or else they have a highly specific expression pattern. Genes with undetectable expression are not shown in Figure 6. Transcripts of all CCA1/R-R-like genes were detectable in most tested tissues and organs, although their transcript abundance was relatively lower than that of genes from the other clades. It is important to note that 12 genes in CCA1/R-R-like clade (including BnMRD36, BnMRD96, BnMRD101, and BnMRD111) were preferentially expressed in SAM, suggesting that they might participate in SAM development. Ten genes including BnMRD95, BnMRD109, and BnMRD202 showed high and specific expression in seeds. BnMRD182 and BnMRD42 were predominantly expressed in flowers. Approximately half of the I-box-like clade genes were predominantly expressed in vegetative tissues and organs (especially cotyledon) at the seedling stage, indicating that they probably participate in vegetative growth. TRF-like genes displayed a diverse expression pattern in the samples tested. The majority of TRF-like genes showed higher expression in the reproductive tissues and organs than in the vegetative tissues and organs and had relatively low transcript abundance in cotyledons and leaves. Many genes of CPC-like clade had high expression in SAM, flowers, and ovaries. Transcripts of TBP-like clade genes were detected in almost all tested samples, and half of them showed a relatively high expression in flowers and ovaries. These findings supported the view that BnMYB-related genes might play fundamental roles in the entire developmental process in rapeseed.
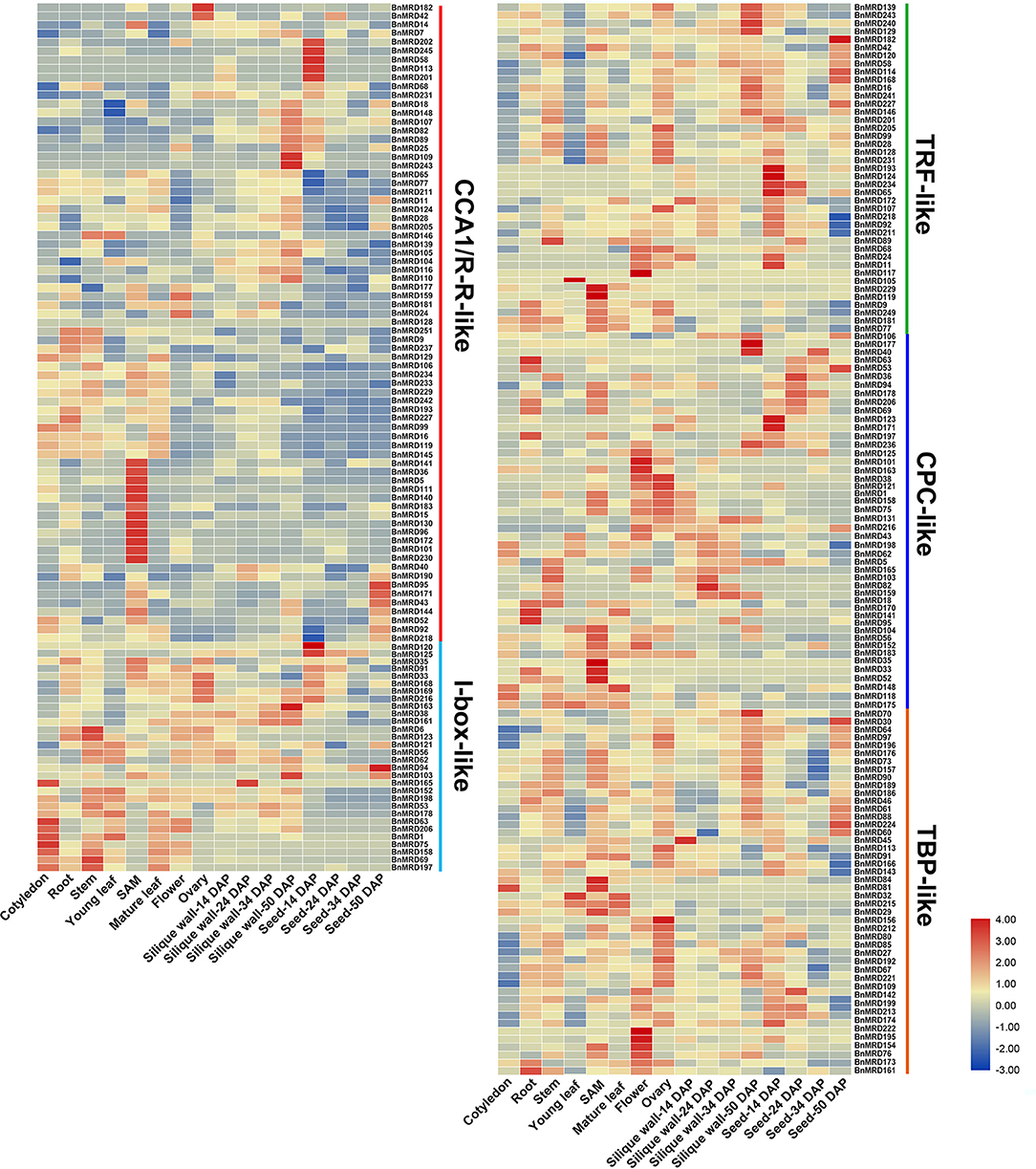
Figure 6. Temporospatial expression patterns of BnMYB-related genes. Fragments per kilobase of transcript per million mapped reads (FPKM) values of BnMYBR genes are log2 transformed and are used to represent the gene expression levels. The horizontal axis represents 16 tissues/organs of different developmental stages in rapeseed. The color scale represents gene expression levels with high transcript abundance (red) or low transcript abundance (blue).
Expression Profiles of BnMYB-Related Genes Under Abiotic Stress Conditions
Based upon the cis-element analysis, multiple abiotic stress response-associated elements were found in the promoter regions of BnMYB-related genes. To better understand the potential roles of BnMYB-related genes in responses to various abiotic stresses in B. napus, the expression patterns of individual members under drought, salt, cold, heat stress, and ABA treatment were investigated based on transcriptome data previously created by our lab (NCBI SRA BioProject ID: PRJNA687395 and PRJNA721476, Supplementary Table 11). A total of 195 MYB-related genes, more than half of all family members, displayed a stress-responsive expression pattern (Figure 7). The results showed that BnMYB-related genes had different expression patterns under four abiotic stresses and ABA treatment. Among them, 110 and 74 genes, 82 and 101 genes, 113 and 89 genes, 74 and 91 genes, were upregulated and downregulated by drought, salt, heat, and cold stress, respectively. The expression levels of 89 genes were elevated, whereas the expression levels of 96 genes were inhibited by ABA treatment. Further analysis showed that all four abiotic stresses and ABA treatment induced the expression of BnMRD40, BnMRD190, and BnMRD166. BnMRD17, BnMRD20, and BnMRD186 were upregulated, whereas BnMRD87, BnMRD78, BnMRD163, and BnMRD202 were downregulated by both drought stress and ABA treatment. Among MYB-related genes, 45.64% (89/195) (including BnMRD13, BnMRD116, and BnMRD126) showed increased expression under cold stress. Several BnMYB-related members exhibited an obvious response to heat stress. For instance, BnMRD96, BnMRD111, BnMRD172, and BnMRD140 were downregulated, whereas BnMRD52, BnMRD102, and BnMRD177 were upregulated by heat treatment. As shown in the heatmap (Figure 7), the expression patterns of BnMYB-related members varied greatly under abiotic stress conditions. These results implied that BnMYB-related genes contribute to a better adaptation of B. napus to the natural environment.
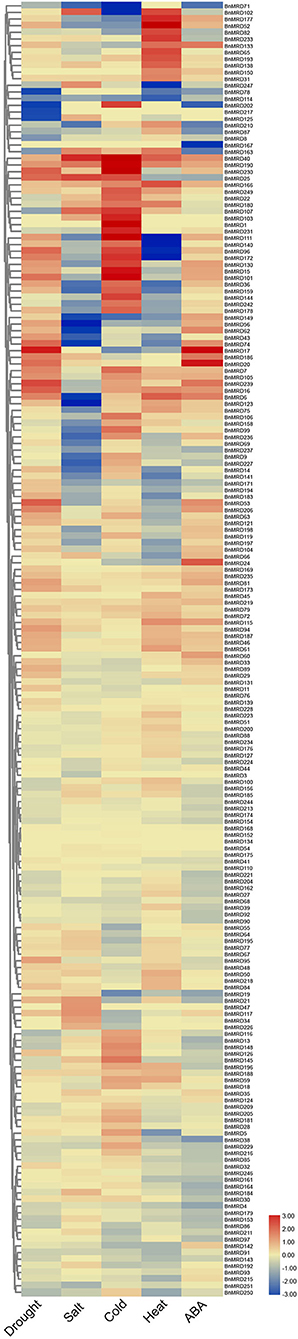
Figure 7. BnMYB-related genes in response to four abiotic stress conditions (drought, salt, cold, and heat) and ABA treatment. Ratios of FPKM values under stress conditions to FPKM values under normal conditions are log2 transformed and used to represent the gene expression levels.
Considering that quite a few BnMYB-related genes exhibited inducible expression under drought or cold stresses, the transcript abundance of nine genes under drought or cold treatment was determined using qPCR (Figure 8). Nine BnMYB-related genes including BnMRD15/25/105/107/148/173/186/250/251, displayed differential expression under normal and drought stress conditions. Notably, the expression levels of BnMRD15 and BnMRD107 were 150 times and 800 times higher after drought treatment for 3 and 6 h than that under normal conditions, respectively (Figure 8A). Moreover, nine genes, including BnMRD1/15/25/40/81/91/105/148/250, were upregulated under cold conditions. Except for BnMRD25, the transcript abundance of other eight genes elevated gradually from 0 to 12 h of cold treatment. The expression of BnMRD25 increased by ~7-fold at 3 h of cold treatments compared with that before stress, peaked at 6 h (by 8-fold), and then decreased at 12 h after cold treatment (Figure 8B). These findings provided insights into the potential roles of BnMYB-related genes in response to drought and cold stress, and will be helpful to explore candidate BnMYB-related genes for further functional determination and stress resistance improvement of B. napus.
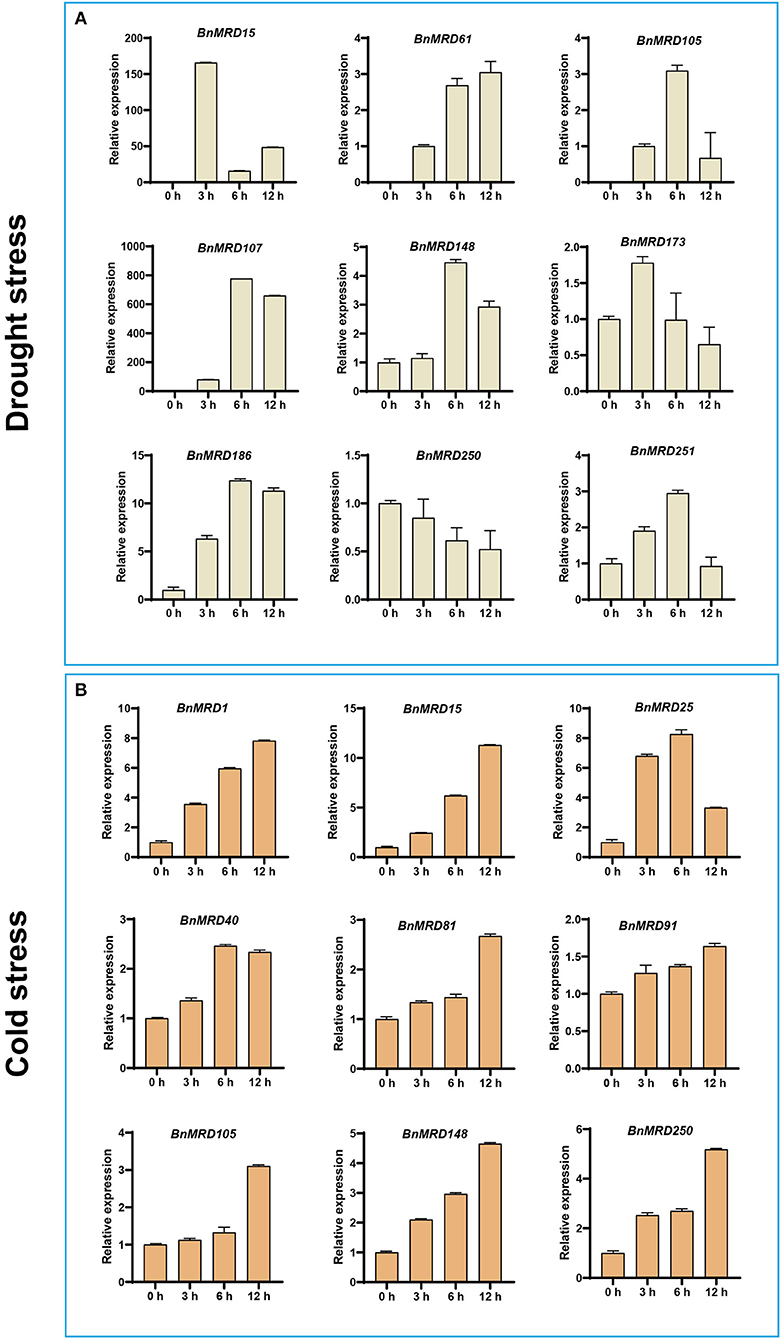
Figure 8. Expression analysis of 13 BnMYB-related genes under (A) drought treatment and (B) cold treatment by qPCR. The expression levels were normalized to BnActin. Data represented the mean ± SE (n =3).
BnMRD107 Enhances Osmotic and Salt Tolerance in Transgenic B. napus Seedlings
To further investigate the function of BnMYB-related genes under abiotic stress in B. napus, we carefully scanned the stress-responsive BnMYB-related genes. A member belonging to CCA1/R-R-like clade, BnMRD107, provoked our interest. BnMRD107 was significantly induced under drought stress, and its transcript abundance was elevated by almost 800-fold after 6 h of drought stress (Figure 8A). To reveal the potential role of BnMRD107 in osmotic response of B. napus, transgenic rapeseed plants overexpressing BnMRD107 were obtained by introducing a BnMRD107-OE construct into rapeseed using the Agrobacterium tumefaciens-mediated transformation approach (Supplementary Figure 3). Three T2 generations of homozygous transgenic lines (BnMRD107-OE-1, BnMRD107-OE-2, and BnMRD107-OE-3) were used to evaluate the performance of the plants under osmotic stress and salt treatments, and the wild-type seedlings (J9712) were used as controls (Figure 9). As shown in Figure 9A, no obvious differences were observed between J9712 and BnMRD107-OE lines under normal growth conditions. The BnMRD107-OE lines showed no significant difference compared with J9712 in hypocotyl length, root length, and fresh weight under normal condition (Figure 9B). However, the BnMRD107-OE lines (especially BnMRD107-OE-1 and BnMRD107-OE-2) showed apparently better growth performance than J9712 plants under both osmotic stress and salt treatments (Figure 9A), and the hypocotyl lengths of BnMRD107-OE plants were longer than those of J9712 plants under osmotic and salt stress conditions (Figures 9C,D). To examine the H2O2 accumulation in BnMRD107-OE and J9712 plants under normal, osmotic stress, and salt conditions, DAB-staining analysis of the plant cotyledons was performed. As shown in Figure 10, there was no visible difference in H2O2 accumulation between the BnMRD107-OE and J9712 plants under normal conditions, while BnMRD107-OE plants accumulated markedly less H2O2 than J9712 plants in their cotyledons after osmotic stress and salt treatments. These results indicated that BnMRD107 conferred osmotic and salt tolerance on transgenic rapeseed seedlings, and the rapeseed plants overexpressing BnMRD107 might possess stronger ROS-scavenging ability than wild-type J9712 plants.
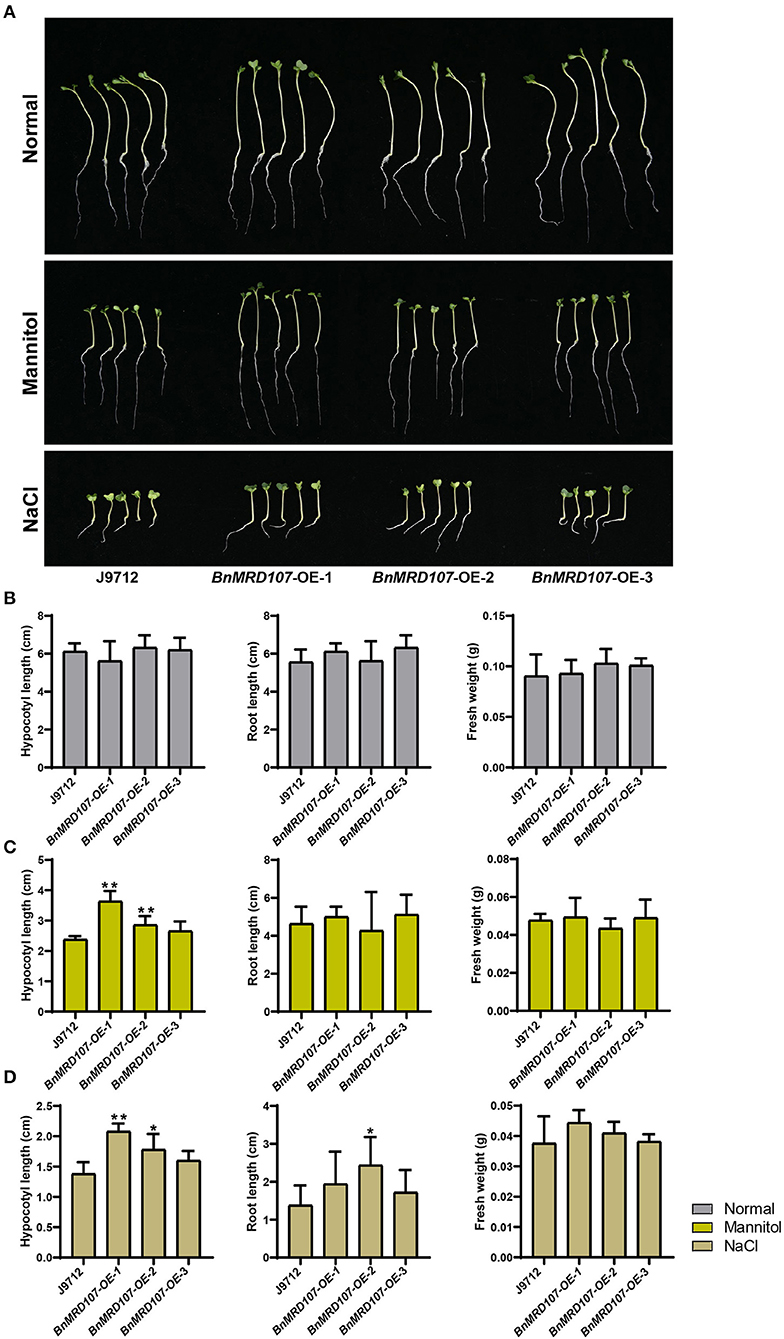
Figure 9. Phenotype of BnMYBR107-OE plants under normal conditions, osmotic stress, and salt treatments. (A) Growth performance of J9712 and BnMYBR107-OE plants under normal conditions, osmotic stress, and salt treatments. (B–D) Hypocotyl length, root length, and fresh weight under normal conditions, osmotic stress, and salt treatments. Data are presented as means ± SD (n = 5). Asterisks (*) indicate significant differences between the transgenic lines and J9712 based on Duncan's multiple range test (*p < 0.05, **p < 0.01).
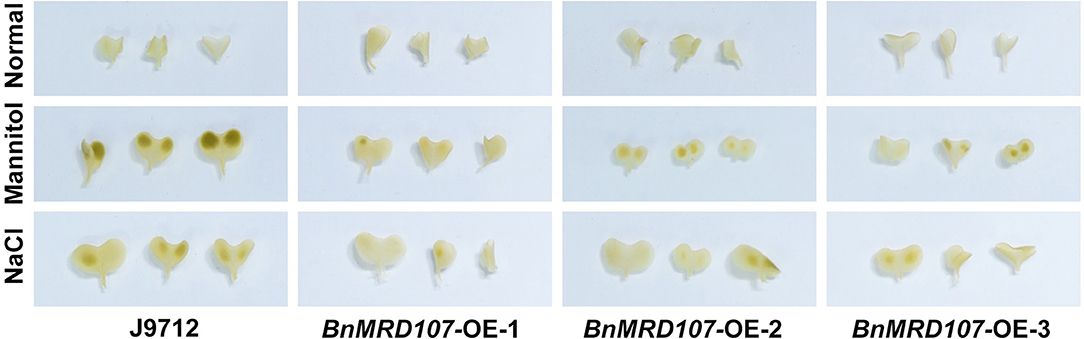
Figure 10. DAB-staining of cotyledons from J9712 and BnMYBR107-OE plants under normal conditions, osmotic stress, and salt treatments.
Discussion
MYB-related transcription factors comprise a prominent subtype of MYB superfamily in plants (Du et al., 2013). Based on the previous research, there are 64, and 223 MYB-related genes in the model plant Arabidopsis and rice, respectively (Chen et al., 2006, 2019). Recently, 62 genes in Physcomitrella patens (Pu et al., 2020), 138 genes in Solanum tuberosum (Liu Y. H. et al., 2020), 73 genes in Musa acuminate, and 59 genes in Musa balbisiana (Tan et al., 2020) were identified as MYB-related genes. Plant MYB-related genes have been reported to be involved in multiple processes, including cellular and organ morphogenesis, secondary metabolism, circadian rhythm, and drought tolerance (Baldoni et al., 2015). However, there has been little investigation of MYB-related genes in B. napus, and the functions of BnMYB-related family members are still largely unknown. In the present study, 251 MYB-related genes were identified in the B. napus genome. The phylogenetic relationships, gene structure, motif composition, syntenic analysis, cis-elements, and expression profiles of BnMYB-related family members were investigated systematically.
The present allotetraploid rapeseed (genome AACC) cultivars were generated by interspecific hybridization between its diploid progenitors B. rapa (genome AA) and B. oleracea (genome AACC) (Chalhoub et al., 2014). MYB-related genes, as a share of the entire MYB family genes, comprise 36.6% (97/265), 44.9% (106/236), 39.8% (202/508), 36.9% (171/464), and 33.9% (251/740) in Arabidopsis, rice, B. oleracea, B. rapa, and B. napus, respectively (Saha et al., 2016; Jin et al., 2017). In this study, 251 MYB-related genes were identified in B. napus, which showed a significantly higher number than that in the Arabidopsis genome (97 genes). In the diploid ancestors (B. oleracea and B. rapa), 76 and 84 orthologous genes were found, respectively. Four categories of gene duplication events including WGD/segmental duplication, dispersed duplication, proximal duplication, and tandem duplication were detected in BnMYB-related genes. Among the 251 sequences, 77.29% (194/251) were implicated in WGD/segmental duplication events, and 20.32% (51/251), 0.01% (3/251), and 0.01% (3/251) involved dispersed duplication, proximal duplication, and tandem duplication, respectively. These results indicated that WGD/segmental duplication and dispersed duplication were the vital driving force for MYB-related gene family expansion.
Similar to MYB-related proteins in other plants, BnMYB-related family members were characterized by a single or a partially conserved repeat. In this study, the 251 BnMYB-related proteins were classified phylogenetically into five major clades. CCA1/R-R-like clade is the largest clade, whose members have a common conserved motif SHAQKY(N)F or SHAQK, in which the third W residue was often substituted by A residue. The circadian-related cis-acting regulatory elements were found to be enriched in the promoter regions of CCA1/R-R-like clade genes, which were consistent with the result of previous studies, in which CCA1/R-R-like genes were identified as being involved in the circadian rhythm (Lu et al., 2009). A previous study on Arabidopsis illustrated that CCA1 protein can bind directly to the promoter of Light-harvesting chlorophyll a/b protein (Lhcb) to activate its transcription (Lu et al., 2009). Moreover, cis-elements associated with auxin response and cell cycle regulation were also found to be generally distributed in the promoter regions of CCA1/R-R-like genes. Cis-elements involved in meristem expression were detected in 12 genes including BnMRD36, BnMRD96, and BnMRD130, and expression analysis showed that these genes exhibited a strong and specific expression in SAM, suggesting their potential roles in meristem development. No consensus motifs were detected in the proteins of I-box-like, CPC-like, and TRF-like clades. However, W residues were evenly distributed in MYB repeats of the members of these clades. CPC-like (II) members contained three W residues in their MYB repeat regions. Investigations on tomato (Solanum lycopersicum) revealed that CPC-like members participated in anthocyanin biosynthesis (Ma and Constabel, 2019). Most of the TBP-like proteins had the consensus motif LKDKW(R/K) (N/T). Although the functional analysis of proteins of I-box-like, TRF-like, and TBP-like clades is relatively rare, their expression profile in various tissues and organs suggested that they might play important roles in B. napus development. Unlike members of other transcription factor families, the sequence identity of MYB-related proteins was relatively low, which might be relevant to their diverse functions in plants.
Although the MYB-related family widely exists in plant species, there has been much more research into 2R-MYB genes than into MYB-related genes in plants. In the present study, we observed that 77.69% (195/251) of MYB-related genes were responsive to at least one of the four tested abiotic stresses and ABA treatment, indicating that a large proportion of MYB-related genes are involved in the adaptation of B. napus to the environment. Cis-acting elements are determinate components of the specificity and expression level of genes (Lescot et al., 2002). Five elements associated with stress responsiveness (ARE, WUN-motif, TC-rich repeats, LTR, and DRE) and five hormonal responsive elements (GARE, ABRE, TGA, TGACG-motif, and TCA) are enriched in the promoters of MYB-related genes. Drought stress and cold stress are two principal natural disasters that occur during the lifespan of rapeseed. Based on the transcriptome expression data, 184 and 187 MYB-related genes showed a transcriptional response to drought and cold stress, respectively. The expression pattern of nine MYB-related genes under drought and cold stress, respectively, were verified using qPCR. Five genes, BnMRD15, BnMRD107, BnMRD148, BnMRD186, and BnMRD251, were strongly induced by drought, which was in accordance with the enrichment of cis-elements of MYB binding site involved in drought-inducibility in their promoters. In addition, certain genes (including BnMRD7, BnMRD9, BnMRD15, BnMRD81, and BnMRD102) that responded to cold stress were verified to contain cold-responsive cis-acting element LTRs in their promoters. These genes might be considered as potential targets to improve stress tolerance of B. napus.
In a previous study, the expansion of MYB superfamily and hormone-mediated expression of B. napus MYB genes were analyzed, and the functions of four 2R-MYB genes in root hair development were studied through ectopic expression in A. thaliana (Li et al., 2020). In the present study, comprehensive investigations of MYB-related family members in B. napus were performed. The homologs of MYB-related genes in eight rapeseed accessions were also identified for evolutionary conservation analysis. Combining cis-element analysis and temporospatial and abiotic stress expression profiles, we identified a number of tissue and organ-specific or stress-responsive MYB-related genes which might play major roles in B. napus development and response to external environmental stimuli. To confirm the function of the stress-responsive BnMYB-related genes, we chose BnMRD107, which was dramatically upregulated under drought stress, for cloning and transgenic functional verification. Overexpressing BnMRD107 led to significantly improved resistance to osmotic and salt stresses and enhanced ROS-scavenging ability in rapeseed seedlings. These results suggested that stress-responsive BnMYB-related genes, such as BnMRD107, can be considered as potential candidate genes for genetic improvement of B. napus. Detailed analysis of the candidate genes is required to gain insights into their biological functions and regulatory mechanisms in stress responses of B. napus.
Conclusion
In the current study, we performed a systematic genome-wide analysis of MYB-related family genes in B. napus. The 251 identified BnMYB-related genes are unevenly located on 19 chromosomes. Phylogenetic- and motif-based classification and diverse temporospatial expression patterns revealed that BnMYB-related genes are involved in modulating many growth and development processes in rapeseed. Furthermore, transcriptome data, together with qPCR analysis, illustrated that BnMYB-related genes might act as key transcription regulators of the response of B. napus to drought and cold stresses. In addition, we identified a gene, BnMRD107, which regulates osmotic and salt tolerance positively in B. napus. In summary, this study established a comprehensive picture of MYB-related gene family in B. napus. These findings will not only facilitate in-depth functional investigation of BnMYB-related genes but also offer candidate gene resources for improving the stress tolerance of B. napus.
Data Availability Statement
The original contributions generated for the study are publicly available. This data can be found at: NCBI Sequence Read Archive, and the BioProject ID are PRJNA687395 and PRJNA721476.
Author Contributions
JL and YF conceived the research, designed the experiments, and analyzed the data. JL, KL, SZ, and JW collected the sample materials and performed the experiments. JL prepared the original manuscript. YF and YW revised the manuscript and supervised the project. All authors have read and approved the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grant numbers U20A2028, 31501335, 31872874, and 31771824), the Natural Science Foundation of Jiangsu Province (Grant number BE2018356), the Undergraduate Training Program for Innovation and Entrepreneurship (Grant numbers XKYCX18_120 and XKYCX19_151), the Top Talent Support Program and the Qinglan Project of Yangzhou University to Yujie Fang, the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Project of Special Funding for Crop Science Discipline Development (yzuxk202006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely acknowledge Prof. Yongming Zhou (Huazhong Agricultural University) for providing the pMDC83 plasmid and seeds of J9712.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.678202/full#supplementary-material
Supplementary Figure 1. Distribution of MYB-related family genes on B. napus chromosomes. Several genes are not shown in this figure because they are located in unassembled scaffolds. Two hundred and thirty-one MYB-related genes were assigned to 19 chromosomes, and the names of the chromosomes are indicated at the top of each chromosome. The scale of the chromosomes was in megabases (Mb).
Supplementary Figure 2. Syntenic relationships of MYB-related genes between B. napus and three closely-allied plant species, Arabidopsis thaliana (A. thaliana), Brassica oleracea (B. oleracea), and Brassica rapa (B. rapa) genomes. Gray lines in the background indicate the collinear blocks within canola and other plant genomes, and the red lines highlight the MYB-related gene orthologs.
Supplementary Figure 3. Relative expression level of T0 generation of BnMRD107-OE transgenic plants.
Supplementary Table 1. List of the primers used in this study.
Supplementary Table 2. General information of BnMYB-related genes.
Supplementary Table 3. Ka/Ks ratios of the duplicated BnMYB-related genes pairs.
Supplementary Table 4. Duplication type of BnMYB-related genes.
Supplementary Table 5. Ka/Ks ratios of the duplicated MYB-related gene pairs of B. napus and Arabidopsis.
Supplementary Table 6. Ka/Ks calculation of the duplicated BnMYB-related gene pairs of B. napus and B. oleracea.
Supplementary Table 7. Ka/Ks ratios of the duplicated BnMYB-related gene pairs of B. napus and B. rapa.
Supplementary Table 8. Homologous members of MYB-related genes in eight rapeseed accessions.
Supplementary Table 9. Cis-elements in the promoter regions of BnMYB-related genes.
Supplementary Table 10. FPKM values of BnMYB-related genes in tissues/organs at different developmental stages.
Supplementary Table 11. Relative expression of BnMYB-related genes under abiotic stresses and ABA treatment.
References
Abe, M., Kaya, H., Watanabe-Taneda, A., Shibuta, M., Yamaguchi, A., Sakamoto, T., et al. (2015). FE, a phloem-specific myb-related protein, promotes flowering through transcriptional activation of FLOWERING LOCUS T and FLOWERING LOCUS T INTERACTING PROTEIN 1. Plant J. 83, 1059–1068. doi: 10.1111/tpj.12951
Andronis, C., Barak, S., Knowles, S. M., Sugano, S., and Tobin, E. M. (2008). The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol. Plant 1, 58–67. doi: 10.1093/mp/ssm005
Aydin, G., Yucel, M., Chan, M. T., and Oktem, H. A. (2014). Evaluation of abiotic stress tolerance and physiological characteristics of potato (Solanum tuberosum L. cv. Kennebec) that heterologously expresses the rice Osmyb4 gene. Plant Biotechnol. Rep. 8, 295–304. doi: 10.1007/s11816-014-0322-7
Azuma, A., Kobayashi, S., Mitani, N., Shiraishi, M., Yamada, M., Ueno, T., et al. (2008). Genomic and genetic analysis of myb-related genes that regulate anthocyanin biosynthesis in grape berry skin. Theor. Appl. Genet. 117, 1009–1019. doi: 10.1007/s00122-008-0840-1
Azuma, A., Udo, Y., Sato, A., Mitani, N., Kono, A., Ban, Y., et al. (2011). Haplotype composition at the color locus is a major genetic determinant of skin color variation in Vitis × labruscana grapes. Theor. Appl. Genet. 122, 1427–1438. doi: 10.1007/s00122-011-1542-7
Baldoni, E., Genga, A., and Cominelli, E. (2015). Plant MYB transcription factors: their role in drought response mechanisms. Int. J. Mol. Sci. 16, 15811–15851. doi: 10.3390/ijms160715811
Baranowskij, N., Frohberg, C., Prat, S., and Willmitzer, L. (1994). A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 13, 5383–5392. doi: 10.1002/j.1460-2075.1994.tb06873.x
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W., Chen, Z., Luo, F., Liao, M., Wei, S., Yang, Z., et al. (2019). RicetissueTFDB: a genome-wide identification of tissue-specific transcription factors in rice. Plant Genome 12:1. doi: 10.3835/plantgenome2017.09.0081
Chen, Y. H., Yang, X. Y., He, K., Liu, M. H., Li, J. G., Gao, Z. F., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Dai, C., Li, Y., Li, L., Du, Z., Lin, S., Tian, X., et al. (2020). An efficient Agrobacterium-mediated transformation method using hypocotyl as explants for Brassica napus. Mol. Breed. 40:96. doi: 10.1007/s11032-020-01174-0
Du, H., Wang, Y. B., Xie, Y., Liang, Z., Jiang, S. J., Zhang, S. S., et al. (2013). Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 20, 437–448. doi: 10.1093/dnares/dst021
Du, Y. T., Zhao, M. J., Wang, C. T., Gao, Y., Wang, Y. X., Liu, Y. W., et al. (2018). Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 18:320. doi: 10.1186/s12870-018-1551-7
Dubos, C., Le Gourrierec, J., Baudry, A., Huep, G., Lanet, E., Debeaujon, I., et al. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55, 940–953. doi: 10.1111/j.1365-313X.2008.03564.x
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Farinas, B., and Mas, P. (2011). Histone acetylation and the circadian clock: a role for the MYB transcription factor RVE8/LCL5. Plant Signal. Behav. 6, 541–543. doi: 10.4161/psb.6.4.14837
Haga, N., Kato, K., Murase, M., Araki, S., Kubo, M., Demura, T., et al. (2007). R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134, 1101–1110. doi: 10.1242/dev.02801
Hong, M. Y., Hu, K. N., Tian, T. T., Li, X., Chen, L., Zhang, Y., et al. (2017). Transcriptomic analysis of seed coats in yellow-seeded Brassica napus reveals novel genes that influence proanthocyanidin biosynthesis. Front. Plant Sci. 8:1674. doi: 10.3389/fpls.2017.01674
Jin, J. P., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J. C., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Kirik, V., and Baumlein, H. (1996). A novel leaf-specific myb-related protein with a single binding repeat. Gene 183, 109–113. doi: 10.1016/s0378-1119(96)00521-5
Kolmos, E., Schoof, H., Plümer, M., and Davis, S. J. (2008). Structural insights into the function of the core-circadian factor TIMING OF CAB2 EXPRESSION 1 (TOC1). J. Circadian Rhythms 6:3. doi: 10.1186/1740-3391-6-3
Kuno, N., Moller, S. G., Shinomura, T., Xu, X., Chua, N. H., and Furuya, M. (2003). The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 15, 2476–2488. doi: 10.1105/tpc.014217
Kwon, Y., Kim, J. H., Nguyen, H. N., Jikumaru, Y., Kamiya, Y., Hong, S. W., et al. (2013). A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J. Exp. Bot. 64, 3911–3922. doi: 10.1093/jxb/ert223
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, P. F., Wen, J., Chen, P., Guo, P. C., Ke, Y. Z., Wang, M. M., et al. (2020). MYB superfamily in Brassica napus: evidence for hormone-mediated expression profiles, large expansion, and functions in root hair development. Biomolecules 10:875. doi: 10.3390/biom10060875
Li, T., Sun, J. K., Bi, Y. P., and Peng, Z. Y. (2016). Overexpression of an MYB-related gene FvMYB1 from Fraxinus velutina increases tolerance to salt stress in transgenic tobacco. J. Plant. Growth. Regul. 35, 632–645. doi: 10.1007/s00344-015-9565-y
Li, X. X., Guo, C., Ahmad, S., Wang, Q., Yu, J., Liu, C., et al. (2019). Systematic analysis of MYB family genes in potato and their multiple roles in development and stress responses. Biomolecules 9:317. doi: 10.3390/biom9080317
Liao, P., Woodfield, H. K., Harwood, J. L., Chye, M. L., and Scofield, S. (2019). Comparative transcriptomics analysis of Brassica napus L. during seed maturation reveals dynamic changes in gene expression between embryos and seed coats and distinct expression profiles of acyl-CoA-binding proteins for lipid accumulation. Plant Cell Physiol. 60, 2812–2825. doi: 10.1093/pcp/pcz169
Lin, Z., Yin, K., Zhu, D., Chen, Z., Gu, H., and Qu, L. J. (2007). AtCDC5 regulates the G2 to M transition of the cell cycle and is critical for the function of Arabidopsis shoot apical meristem. Cell Res. 17, 815–828. doi: 10.1038/cr.2007.71
Liu, Y., Pei, L., Xiao, S., Peng, L., Liu, Z., Li, X., et al. (2020). AtPPRT1 negatively regulates salt stress response in Arabidopsis seedlings. Plant Signal. Behav. 15:1732103. doi: 10.1080/15592324.2020.1732103
Liu, Y. H., Zeng, Y. T., Li, Y. M., Liu, Z., Wang, K. L., Espley, R. V., et al. (2020). Genomic survey and gene expression analysis of the MYB-related transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 164, 2450–2464. doi: 10.1016/j.ijbiomac.2020.08.062
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, D. D., Wang, T., Persson, S., Mueller-Roeber, B., and Schippers, J. H. (2014). Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nat. Commun. 5:3767. doi: 10.1038/ncomms4767
Lu, S. X., Knowles, S. M., Andronis, C., Ong, M. S., and Tobin, E. M. (2009). CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 150, 834–843. doi: 10.1104/pp.108.133272
Ma, D., and Constabel, C. P. (2019). MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 24, 275–289. doi: 10.1016/j.tplants.2018.12.003
Makino, S., Matsushika, A., Kojima, M., Yamashino, T., and Mizuno, T. (2002). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 43, 58–69. doi: 10.1093/pcp/pcf005
Nagel, D. H., and Kay, S. A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22, R648–R657. doi: 10.1016/j.cub.2012.07.025
Nguyen, N. H., and Lee, H. (2016). MYB-related transcription factors function as regulators of the circadian clock and anthocyanin biosynthesis in Arabidopsis. Plant Signal. Behav. 11:e1139278. doi: 10.1080/15592324.2016.1139278
Piao, W., Kim, S. H., Lee, B. D., An, G., Sakuraba, Y., and Paek, N. C. (2019). Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 70, 2699–2715. doi: 10.1093/jxb/erz095
Piazza, P., Procissi, A., Jenkins, G. I., and Tonelli, C. (2002). Members of the c1/pl1 regulatory gene family mediate the response of maize aleurone and mesocotyl to different light qualities and cytokinins. Plant Physiol. 128, 1077–1086. doi: 10.1104/pp.010799
Pilu, R., Piazza, P., Petroni, K., Ronchi, A., Martin, C., and Tonelli, C. (2003). pl-bol3, a complex allele of the anthocyanin regulatory pl1 locus that arose in a naturally occurring maize population. Plant J. 36, 510–521. doi: 10.1046/j.1365-313x.2003.01898.x
Pu, X. J., Yang, L. X., Liu, L. N., Dong, X. M., Chen, S. L., Chen, Z. X., et al. (2020). Genome-wide analysis of the MYB transcription factor superfamily in physcomitrella patens. Int. J. Mol. Sci. 21:975. doi: 10.3390/ijms21030975
Rawat, R., Schwartz, J., Jones, M. A., Sairanen, I., Cheng, Y., Andersson, C. R., et al. (2009). REVEILLE1, a myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. U.S.A. 106, 16883–16888. doi: 10.1073/pnas.0813035106
Rawat, R., Takahashi, N., Hsu, P. Y., Jones, M. A., Schwartz, J., Salemi, M. R., et al. (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7:e1001350. doi: 10.1371/journal.pgen.1001350
Royo, J., Gomez, E., Barrero, C., Muniz, L. M., Sanz, Y., and Hueros, G. (2009). Transcriptional activation of the maize endosperm transfer cell-specific gene BETL1 by ZmMRP-1 is enhanced by two C2H2 zinc finger-containing proteins. Planta 230, 807–818. doi: 10.1007/s00425-009-0987-2
Saha, G., Park, J. I., Ahmed, N. U., Kayum, M. A., Kang, K. K., and Nou, I. S. (2016). Characterization and expression profiling of MYB transcription factors against stresses and during male organ development in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Physiol. Biochem. 104, 200–215. doi: 10.1016/j.plaphy.2016.03.021
Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. Embo J. 21, 5036–5046. doi: 10.1093/emboj/cdf524
Shi, G. Y., Guo, X. Y., Guo, J. Y., Liu, L. H., and Hua, J. P. (2015). Analyzing serial cDNA libraries revealed reactive oxygen species and gibberellins signaling pathways in the salt response of upland cotton (Gossypium hirsutum L.). Plant Cell Rep. 34, 1005–1023. doi: 10.1007/s00299-015-1761-5
Shibuta, M., and Abe, M. (2017). FE controls the transcription of downstream fowering regulators through two distinct mechanisms in leaf phloem companion cells. Plant Cell Physiol. 58, 2017–2025. doi: 10.1093/pcp/pcx133
Song, J. M., Guan, Z., Hu, J., Guo, C., Yang, Z., Wang, S., et al. (2020). Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 6, 34–45. doi: 10.1038/s41477-019-0577-7
Sue, S. C., Hsiao, H. H., Chung, B. C., Cheng, Y. H., Hsueh, K. L., Chen, C. M., et al. (2006). Solution structure of the Arabidopsis thaliana telomeric repeat-binding protein DNA binding domain: a new fold with an additional C-terminal helix. J. Mol. Biol. 356, 72–85. doi: 10.1016/j.jmb.2005.11.009
Sun, X. M., Matus, J. T., Wong, D. C. J., Wang, Z. M., Chai, F. M., Zhang, L. L., et al. (2018). The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J. Exp. Bot. 69, 1749–1764. doi: 10.1093/jxb/ery020
Tan, L., Ijaz, U., Salih, H., Cheng, Z., Ni Win Htet, N., Ge, Y., et al. (2020). Genome-wide identification and comparative analysis of MYB transcription factor family in Musa acuminata and Musa balbisiana. Plants 9:413. doi: 10.3390/plants9040413
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tominaga-Wada, R., Kurata, T., and Wada, T. (2017). Localization of the CAPRICE-ENHANCER OF TRY AND CPC1 chimera protein in Arabidopsis root epidermis. Biosci. Biotechnol. Biochem. 81, 1762–1767. doi: 10.1080/09168451.2017.1343120
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinf. 8, 77–80. doi: 10.1016/s1672-0229(10)60008-3
Wang, W., Zhang, X. P., Deng, F. N., Yuan, R., and Shen, F. F. (2017). Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genomics 18:376. doi: 10.1186/s12864-017-3768-5
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Xiong, H. Y., Li, J. Y., Liu, P. L., Duan, J. Z., Zhao, Y., Guo, X., et al. (2014). Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 9:e92913. doi: 10.1371/journal.pone.0092913
Yang, B., Song, Z., Li, C., Jiahao, J., Yangyang, Z., Wang, R., et al. (2018). RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet. 14:e1007839. doi: 10.1371/journal.pgen.1007839
Yin, X. M., Cui, Y. C., Wang, M. L., and Xia, X. J. (2017). Overexpression of a novel MYB-related transcription factor, OsMYBR1, confers improved drought tolerance and decreased ABA sensitivity in rice. Biochem. Biophys. Res. Commun. 490, 1355–1361. doi: 10.1016/j.bbrc.2017.07.029
Yong, Y. B., Zhang, Y., and Lyu, Y. M. (2019). A MYB-Related transcription factor from Lilium lancifolium L. (LlMYB3) is involved in anthocyanin biosynthesis pathway and enhances multiple abiotic stress tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 20:3195. doi: 10.3390/ijms20133195
Zhang, L. C., Liu, G. X., Jia, J. Z., Zhao, G. Y., Xia, C., Zhang, L. N., et al. (2016). The wheat MYB-related transcription factor TaMYB72 promotes flowering in rice. J. Integr. Plant Biol. 58, 701–704. doi: 10.1111/jipb.12461
Zhang, X. B., Chen, Y. H., Wang, Z. Y., Chen, Z. L., Gu, H. Y., and Qu, L. J. (2007). Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 51, 512–525. doi: 10.1111/j.1365-313X.2007.03156.x
Zhang, Z., Xiao, J., Wu, J., Zhang, H., Liu, G., Wang, X., et al. (2012). ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 419, 779–781. doi: 10.1016/j.bbrc.2012.02.101
Keywords: Brassica napus, MYB-related transcription factors, phylogenetic analysis, expression profiles, abiotic stress
Citation: Li J, Lin K, Zhang S, Wu J, Fang Y and Wang Y (2021) Genome-Wide Analysis of Myeloblastosis-Related Genes in Brassica napus L. and Positive Modulation of Osmotic Tolerance by BnMRD107. Front. Plant Sci. 12:678202. doi: 10.3389/fpls.2021.678202
Received: 09 March 2021; Accepted: 30 April 2021;
Published: 17 June 2021.
Edited by:
Meixue Zhou, University of Tasmania, AustraliaReviewed by:
Chengdao Li, Murdoch University, AustraliaMaoteng Li, Huazhong University of Science and Technology, China
Copyright © 2021 Li, Lin, Zhang, Wu, Fang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yujie Fang, eWpmYW5nQHl6dS5lZHUuY24=; Youping Wang, d2FuZ3lwQHl6dS5lZHUuY24=
 Jian Li
Jian Li Keyun Lin
Keyun Lin Shuai Zhang
Shuai Zhang Jian Wu
Jian Wu Yujie Fang
Yujie Fang Youping Wang
Youping Wang