- Floral and Nursery Plants Research Unit, United States National Arboretum, Agricultural Research Service, United States Department of Agriculture, Beltsville, MD, United States
Members of the Ralstonia solanacearum species complex cause a variety of wilting diseases across a wide range of hosts by colonizing and blocking xylem vessels. Of great concern are race 3 biovar 2 strains of R. solanacearum capable of causing brown rot of potato at cool temperatures, which are select agents in the United States. To gain a better understanding of cool-virulence mechanisms, we generated libraries of transposon mutants in the cool-virulent R. solanacearum strain UW551 and screened 10,000 mutants using our seedling assay for significantly reduced virulence at 20°C. We found several mutants that exhibited reduced virulence at 28 and 20°C and also mutants that were only affected at the cooler temperature. One mutant of the latter chosen for further study had the transposon inserted in an intergenic region between a type III secretion system effector gene ripS1 and a major facilitator superfamily (MFS) protein gene. Gene expression analysis showed that expression of ripS1 was altered by the transposon insertion, but not the MFS protein gene. An independent mutant with this insertion upstream of ripS1 was generated and used to confirm virulence and gene expression phenotypes. The effector, RipS1, has unknown function and is part of a family of effectors belonging to the largest known type III effectors. The functional connection between RipS1 and cool-virulence of R. solanacearum UW551 suggests that RipS1 (and/or its upstream promoter element) may serve as a potential target for development of cool-virulence-specific diagnostic tools to differentiate the highly regulated cool-virulent strains from non-cool-virulent strains of R. solanacearum. Our results provide important information for continued work toward a better understanding of cool-virulence of R. solanacearum and development of proper control strategies to combat this important plant pathogen.
Introduction
Collectively, the Ralstonia solanacearum species complex (Rssc) – a group of related Gram-negative beta-proteobacteria – is pathogenic to many plant species, including several crops important economically and to food security across the globe (Hayward, 1991). The Rssc persists in soil and invades host plant roots, ultimately building to high-density populations in xylem vessels, which causes a reduction in water flow from roots to aerial tissues and frequently results in wilt symptoms (Lowe-Power et al., 2018). Although many strains in the Rssc are highly effective pathogens in their native tropical climates, some strains are of great economic concern in more temperate settings because of the ability to effectively infect, colonize, and cause disease symptoms at cool temperatures (<25°C), a phenotype referred to as cool-virulence (Janse et al., 1998; Van Elsas et al., 2000). Due to changing availability of techniques to characterize the members of the Rssc, classification has also changed over time. Rssc strains that exhibit the cool-virulence phenotype with potato as the host were initially designated as race 3 biovar 2 strains on the basis of host range and biochemical properties and subsequently designated as phylotype IIB sequevars 1 and 2 based on molecular characters from multiplex PCR (Fegan and Prior, 2005). Subsequently, nomenclature was updated and these strains belong to the “true” R. solanacearum species (Safni et al., 2014). Whereas genetic markers and diagnostic assays have been developed for detection and classification of R. solanacearum based on phylogeny, diagnostic tools based on markers directly connected to the cool-virulence phenotype are lacking and the specific underlying mechanisms enabling cool-virulence remain poorly understood. Recently, we identified a DNA region associated with the cool-virulence of R. solanacearum strain UW551 (Stulberg et al., 2018), and we also developed the first PCR assay targeting the cool-virulence associated region for specific detection and identification of race 3 biovar 2 strains of R. solanacearum (Stulberg et al., 2018).
Many virulence-associated traits have been identified in the Rssc and some of the most important include exopolysaccharide production, motility, and type III secretion (Meng, 2013). These traits enable the Rssc to migrate to susceptible hosts, attach to and invade roots, finally entering and colonizing xylem vessels (Álvarez et al., 2010). Exopolysaccharides produced by Rssc members provide a matrix facilitating and supporting bacterial growth, especially in host vascular tissues (Araud-Razou et al., 1998). Rssc strains exhibit both flagellar and twitching motilities; however, evidence suggests that twitching motility may play a greater role in overall virulence (Kang et al., 2002; Corral et al., 2020). Twitching motility at cool temperatures has been associated with cool-virulent R. solanacearum and may have direct or indirect effects on the cool-virulence phenotype (Bocsanczy et al., 2012; Stulberg et al., 2018).
As for many plant pathogens, the Rssc type III secretion system plays a critical role in virulence as it enables direct delivery of effector proteins from the bacterial cytoplasm into host cytoplasm (van Gijsegem et al., 1995). Type III effector proteins are involved in host defense suppression and manipulation of host physiological processes such as ubiquitin ligases and acetyltransferases to favor bacterial proliferation (Deslandes and Genin, 2014). The Rssc pan-genome includes a large arsenal of type III effectors, and efforts to confirm and characterize these effectors have identified certain effectors with critical functions for virulence, including some responsible for host-specificity (Poueymiro et al., 2009; Mukaihara et al., 2010). Some effectors are conserved and present in single copies in nearly all Ralstonia, such as RipU or the pectate lyase, and RipW (Sabbagh et al., 2019). Many Rssc type III effectors are grouped into families of similarly structured effectors with the genome of each Rssc strain carrying many members of the effector family. The RipS or SKWP effector family is characterized by carrying an array of 10–20 repeats of a semi-conserved 42 amino-acid motif that begins with the consensus SKWP, making them the largest known type III effectors (Mukaihara and Tamura, 2009). A member of this family, RipS1, has been identified several times in various efforts to describe the type III effector suite of the Rssc (Mukaihara and Tamura, 2009; Mukaihara et al., 2010; Peeters et al., 2013). As a result, RipS1, SKWP1, rip27, and hpx37 are all synonymous appellations that all refer to this single gene and corresponding locus tags are Rsc3401 and RRSL_04182 in strains GMI1000 and UW551, respectively. In a screen of several Ralstonia pseudosolanacearum type III effectors, it was determined that RipS1 can be induced to be secreted in vitro, and the occurrence of such secretion is dependent on the HpaB chaperone (Mukaihara et al., 2010). Additional work determined that RipS1 is not only secreted, but also is actually translocated into plant cells, in an HrpB dependent manner (Mukaihara and Tamura, 2009). Homologous effectors from the RipS family are present not only in Ralstonia, but also in other plant associated bacteria from Xanthomonas, Rhizobium, and even a few Pseudomonads (Teper et al., 2016). Deletion mutants of RipS family effectors, including ripS1 in R. pseudosolanacearum and xopAD in Xanthomonas citri pv. citri have shown no virulence defect when this effector is lacking (Escalon et al., 2013; Lei et al., 2020). Indeed, the dispensability of these effectors is confirmed by the identification of natural isolates containing premature stop codons and insertion sequences likely resulting in loss of function (Escalon et al., 2013; Teper et al., 2016). An R. pseudosolanacearum strain lacking all RipS family effectors similarly had no observable virulence defect (Lei et al., 2020), and in plants inoculated with a 1:1 mix of wild-type and ripS1 mutant bacteria, both strains were recovered at the same rate indicating no competitive disadvantage from the loss of this effector (Chen et al., 2018).
Many virulence-associated traits are expressed under control of complex regulatory networks, including quorum sensing, and critical transcription factors such as PhcA and HrpG (Vasse et al., 2000; Genin et al., 2005). Genes encoding effector proteins and structural components of the type III secretion system are expressed at high levels under in planta conditions (Coll and Valls, 2013). These genes are often co-regulated through a shared promoter element called a plant-inducible-promoter (PIP), characterized by a conserved sequence motif termed a PIP-box that has a consensus sequence of TTCGC – N15 – TTCGC (Cunnac et al., 2004). Presence of a PIP-box element in the promoter region of a gene is often an indication of important virulence function.
In this study, we implemented a high-throughput seedling virulence assay to screen for random transposon mutagenesis-generated R. solanacearum mutant strains for significantly reduced virulence under cool (20°C) but not warm (28°C) temperature conditions. Through our screen, we identified a transposon insertion mutant of interest that implicates the type III secretion effector gene ripS1 in the cool-virulence phenotype. Expression of this type III effector gene is dysregulated in our transposon mutant, suggesting that appropriate expression of this effector is necessary for cool-virulence of R. solanacearum strain UW551.
Materials and Methods
Culture Conditions, Strains, and Plasmids
Ralstonia solanacearum strains from frozen stocks were routinely grown in casamino acid peptone glucose (CPG) culture medium (10 g L−1 peptone, 1 g L−1 yeast extract, 1 g L−1 casamino acids, and 5 g L−1 glucose) at 28°C and Escherichia coli strains were grown using LB culture medium (10 g L−1 tryptone, 5 g L−1 sodium chloride, and 5 g L−1 yeast extract) at 37°C, except where specified otherwise. The antibiotics kanamycin, ampicillin, and chloramphenicol were included in final concentrations of 30, 50, and 10 μg/ml, respectively, as appropriate. Supplementary Table S1 contains a list of strains and plasmids used in this study.
Transposon Mutagenesis and Selection for Mutants of Ralstonia solanacearum Strain UW551
Random transposon mutants were obtained through conjugation of a mini-Tn5 suicide-donor plasmid from E. coli into the cool-virulent R. solanacearum strain UW551. UW551 cells were transformed by electroporation with the low-copy, broad-host-range plasmid pSIM7 (Datta et al., 2006) to provide a means of selection against donor E. coli following conjugation. Our Nicotiana glutinosa seedling screen was used to verify that there was no significant difference in virulence between wild-type UW551 and UW551 carrying plasmid pSIM7. For conjugations, the recipient UW551 cells carrying plasmid pSIM7 were prepared by growing cultures for 48 h to stationary phase at 28°C. Donor E. coli, S17-1 carrying the mini-Tn5 plasmid pUT-miniTn5-luxCDABE (Weitz et al., 2001) or pBAMD1-2 (Martínez-García et al., 2014), was grown to exponential phase, and mixed in a 1:1 ratio with the recipient R. solanacearum cells. The pBAMD1-2 was a gift from Victor de Lorenzo (Addgene plasmid #61564; RRID::Addgene61564).1 The cell mix was collected by centrifugation, washed twice with sterile water, and then resuspended in 1/20th volume of sterile water. Washed cells were dripped onto the surface of a CPG plate lacking any glucose or antibiotics, allowed to dry, and incubated at 28°C for 18 h. Cells grown on the CPG plate were resuspended in 15% glycerol and the transposon mutant library was stored at −80°C. When ready for use, selections were made by plating onto CPG containing chloramphenicol and kanamycin to select for R. solanacearum receiving the transposon. Dilutions were made so that single colonies grew with separation and were incubated at 28°C until colonies grew. Individual colonies were picked from selection plates and used directly as distinct UW551 mutant isolates for virulence screen using N. glutinosa seedlings as described previously (Schachterle and Huang, 2021). Briefly, N. glutinosa seeds were sown quickly and easily on top of individual soft water agar wells of a 96-well microtiter plate by pipetting out desired number of seeds in an aqueous suspension. They were inoculated on the same day by first touching a bacterial colony with an autoclaved toothpick and then stabbing the toothpick into the center of the water agar well (Schachterle and Huang, 2021).
High-Throughput Virulence Screening of Transposon Mutants of R. solanacearum Strain UW551
Our recently developed N. glutinosa seedling screen (Schachterle and Huang, 2021) was used for high-throughput screening of R. solanacearum UW551 transposon mutants for significantly reduced virulence at 20°C. Each mutant colony was inoculated into an individual well containing N. glutinosa seeds as described above. Wild-type UW551 and sterile water were used for inoculation as positive and negative controls, respectively. The inoculated 96-well plates were incubated at 20°C for 18–21 days before evaluation of symptoms. Mutants in wells causing visibly reduced disease symptoms compared to the wild-type UW551 at 20°C were selected for additional testing. This was done by re-isolation of the selected mutant isolate from its water agar well by streaking from the water agar well onto solid CPG media, providing colonies to repeat the virulence assay using seedlings in three replicate wells at 20 and at 28°C to confirm its reduced virulence was specific only to the cool but not to the warm temperature.
Virulence Assay Using Tomato Seedlings
Virulence of R. solanacearum on tomato seedlings was tested using two approaches. In one approach, seedlings were not wounded, and seeds were sown in soft water agar and inoculated with the selected mutant isolate on the day of sowing, incubated at the indicated temperature and monitored for growth and symptom development. This approach mirrors what was done in the N. glutinosa seedling screen, except with a different host, and was used to confirm the temperature-specific virulence phenotype of the mutant isolate in N. glutinosa. This approach was also used to extract RNA from inoculated tomato seedlings at different temperatures for gene expression analysis described below. In the other approach (Khokhani et al., 2018), young tomato seedlings were grown in soil at 28°C. After wounding by removal of a leaf, and inoculation with 2 μl of a 105 bacterial cells/ml suspension on the petiole of the removed leaf to directly introduce the selected R. solanacearum strain to the xylem, the inoculated tomato seedlings were incubated at the indicated temperature and monitored daily for disease symptoms. This approach was used to determine whether delivering the mutant isolate directly into the plant has any effect on the temperature-specific virulence of the mutant isolate.
Phenotypic Trait Assays
Swimming motility was assayed as described (Corral et al., 2020). Briefly, overnight cultures of R. solanacearum were washed with sterile water and adjusted to an OD600 of 0.1, and 3 μl of this bacterial cell suspension were stab inoculated into the center of the soft-agar medium plate (10 g/L peptone, 2.5 g/L agar). The plate was then incubated at the indicated temperature for 24 h and imaged. The halo of cells swimming from the point of inoculation was quantified using ImageJ analysis software (Abràmoff et al., 2004). Susceptibility to exogenous hydrogen peroxide was conducted as described previously (Schachterle et al., 2019), with the modification that CPG medium was used instead of LB. Briefly, overnight cultures of R. solanacearum were washed with sterile water and diluted to an OD600 of 0.1. Then, 50 μl of the cell suspension was spread on a CPG plate. An 8 mm sterile filter paper disk was placed in the center of the plate on top of the CPG plate with cells and 10 μl of 1 M hydrogen peroxide was added to the filter paper, and plates were incubated at 20 or 28°C, as indicated. Plates were imaged after 48 h, and the zone of growth inhibition around the filter paper disk was quantified using ImageJ (Abràmoff et al., 2004). Twitching motility was assessed as described (Bocsanczy et al., 2012). Briefly, cells were diluted to grow individual colonies and grown on CPG medium at either 20°C for 48 h or 28°C for 24 h before analysis and imaging by microscopy. Each assay was repeated three times, each time with at least three replicates. For twitching motility, images shown are representative.
Transposon Insertion Site Sequencing and Identification
Transposon insertion locations were determined using arbitrary PCR amplification followed by Sanger sequencing. Arbitrary PCR was conducted in two amplification reactions as described previously (Miller-Williams et al., 2006; Mendez et al., 2018). In the first round of PCR, a primer specific to one side of the transposon (MCS-Arb Ext; Supplementary Table S2, or TDKm6; Mendez et al., 2018) is used with a degenerate primer (DGEN1; Miller-Williams et al., 2006) under a low annealing temperature to amplify the edge of the transposon. In the second round, a nested primer specific to the transposon edge MCS-Arb Int or TDKm-Int (Supplementary Table S2) is used with a primer specific to the degenerate primer (DGEN2; Miller-Williams et al., 2006) to provide amplification specificity to the transposon edge. All oligonucleotide sequences are included in Supplementary Table S2. Purified PCR products were submitted to Eurofins Genomics (Louisville, KY) for sequencing using the transposon-specific nested primer (MCS-Arb Int or TDKm-Int). Positions of flanking sequences were determined using blastn from the BLAST+ suite (Camacho et al., 2009) with the UW551 genome (Hayes et al., 2017) as database.
RNA Isolations
For RNA isolations from culture medium, R. solanacearum strains were grown overnight in CPG medium, washed, and resuspended to an OD600 of 1. For trials in CPG medium, 500 μl cells were resuspended in sterile water and diluted 100-fold into CPG and incubated at indicated temperature with 200 rpm agitation. For trials in minimal medium, 2 ml cells were resuspended in minimal medium and diluted 4-fold into minimal medium and incubated at the indicated temperature with 200 rpm agitation. After 24 h of growth, cells were collected by centrifugation and RNA was isolated using an approach as described (Rivas et al., 2001). For RNA isolations from tomato seedlings, seeds were sown in soft water agar (0.2% w/v) and the water agar was stab-inoculated on the same day as described above. Seven days post-sowing/inoculation, seedlings were collected and total RNA was isolated using the E.Z.N.A. Plant RNA kit (Omega Bio-tek, Norcross, GA, United States) according to manufacturer’s instructions. RNA quality and quantity were assessed using agarose gel electrophoresis and spectrophotometrically via NanoDrop (Thermo Scientific, Waltham, MA, United States).
qPCR and Gene Expression Analysis
RNA was reverse transcribed to cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, United States) following manufacturer’s protocol with random hexamers. Quantitative real-time PCR (qRT-PCR) was conducted using cDNA as template for reactions with iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, United States) and run on a C1000 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, United States). Three biological replicate cultures were used for each strain-temperature-medium combination for RNA isolation and cDNA synthesis. Each cDNA sample was in turn tested with three technical replicates per primer set in qPCR reactions. Gene expression was calculated using the ddCt method (Livak and Schmittgen, 2001) with an established 16S rRNA primer set as endogenous control (Addy et al., 2012).
Generation and Testing of Independent Mutant UW551-TnR1-ck
To confirm that the observed phenotypes in the transposon mutant UW551-TnR1 were caused by insertion of the transposon at the determined insertion site, not spontaneous mutations at other genomic positions, an independent mutant containing the exact transposon insertion at the same genomic site in UW551 was generated by site-directed double homologous recombination. Briefly, the transposon and flanking genomic DNA were amplified from UW551-TnR1 genomic DNA by high-fidelity PCR with Phusion polymerase (New England Biolabs, Ipswich, MA, United States) using the RRSL04180 forward primer and the ripS1 qPCR reverse primer. The resulting PCR fragment contained the transposon insertion site – which carries kanamycin resistance – with 688 nt and 1,391 nt of DNA flanking the transposon on the side of ripS1 or RRSL_04180, respectively, allowing for homologous recombination to independently introduce the mutation to wild-type strain UW551. To prepare electrocompetent cells, overnight cultures of strain UW551 grown in CPG medium were iced, washed five times with ice-cold sterile water, and resuspended in a 0.1X volume, concentrating the cells 10-fold from the original culture density. PCR products were purified using the QIAquick PCR purification kit according to manufacturer’s instructions (Qiagen, Germantown, MD, United States), and 10 μg of PCR product was added to 50 μl of competent UW551 in a pre-chilled electroporation cuvette (2 mm gap width). Electroporation was conducted using a MicroPulser (BioRad, Hercules, CA, United States) on setting EC2. Following electroporation, 1 ml of CPG medium was added to the cuvette and recovery of cells was conducted at 28°C for 4 h. Following recovery, cells were plated onto CPG plates containing kanamycin to select for recombinant mutants and incubated at 28°C until colonies grew. A colony that was confirmed by PCR and Sanger sequencing to contain the transposon insertion at the same flanking sequences as UW551-TnR1 was selected and designated UW551-TnR1-ck. Virulence testing of this strain was conducted using seedlings of N. glutinosa and tomato grown in soft water agar as described above. RNA isolation, cDNA synthesis, and qPCR for gene expression analysis of UW551-TnR1-ck was conducted as described above.
Results
Generation of Transposon Mutants of R. solanacearum UW551 and Screen for Mutants With Reduced Virulence
We generated libraries of random mini-Tn5 transposon insertion mutants by conjugating transposon donor plasmid pUT-miniTn5-luxCDABE or pBAMD1-2 into the cool virulent R. solanacearum strain UW551 (Supplementary Table S1). We then employed our recently developed N. glutinosa seedling virulence assay (Schachterle and Huang, 2021) to screen 10,000 isolates from our mutant libraries. The screen was done at 20°C first and 302 isolates exhibiting reduced virulence were chosen for further testing. These 302 isolates were tested with additional replicates at both 20 and 28°C. Of these, 16 had reduced virulence at both temperatures tested and 10 isolates had reduced virulence only at 20°C, but not 28°C. Of the 26 mutants of interest collected from our screen, arbitrary PCR followed by Sanger sequencing was successful for 20 mutants, indicating a single transposon insertion. Of particular interest, we obtained three mutant isolates that all had an identical transposon insertion, potentially arising from a single transposon insertion event followed by cell division before selection pressure was imposed. This transposon insertion was located on the UW551 chromosome at nucleotide 3,408,846 (assembly GCA_ 002251655.1) in an intergenic region between a major facilitator superfamily (MFS) protein gene (RRSL_04180 in assembly GCA_000167955.1; B7R79_15985 in assembly GCA_002251655.1) and a type III secretion system effector gene, ripS1 (RRSL_04182 in assembly GCA_000167955.1; B7R79_15995 in assembly GCA_002251655.1) 339 bases upstream of the ripS1 start codon (Figure 1A). We designated this mutant strain as UW551-TnR1 and chose this mutant for further study because of its proximity to a type III effector.
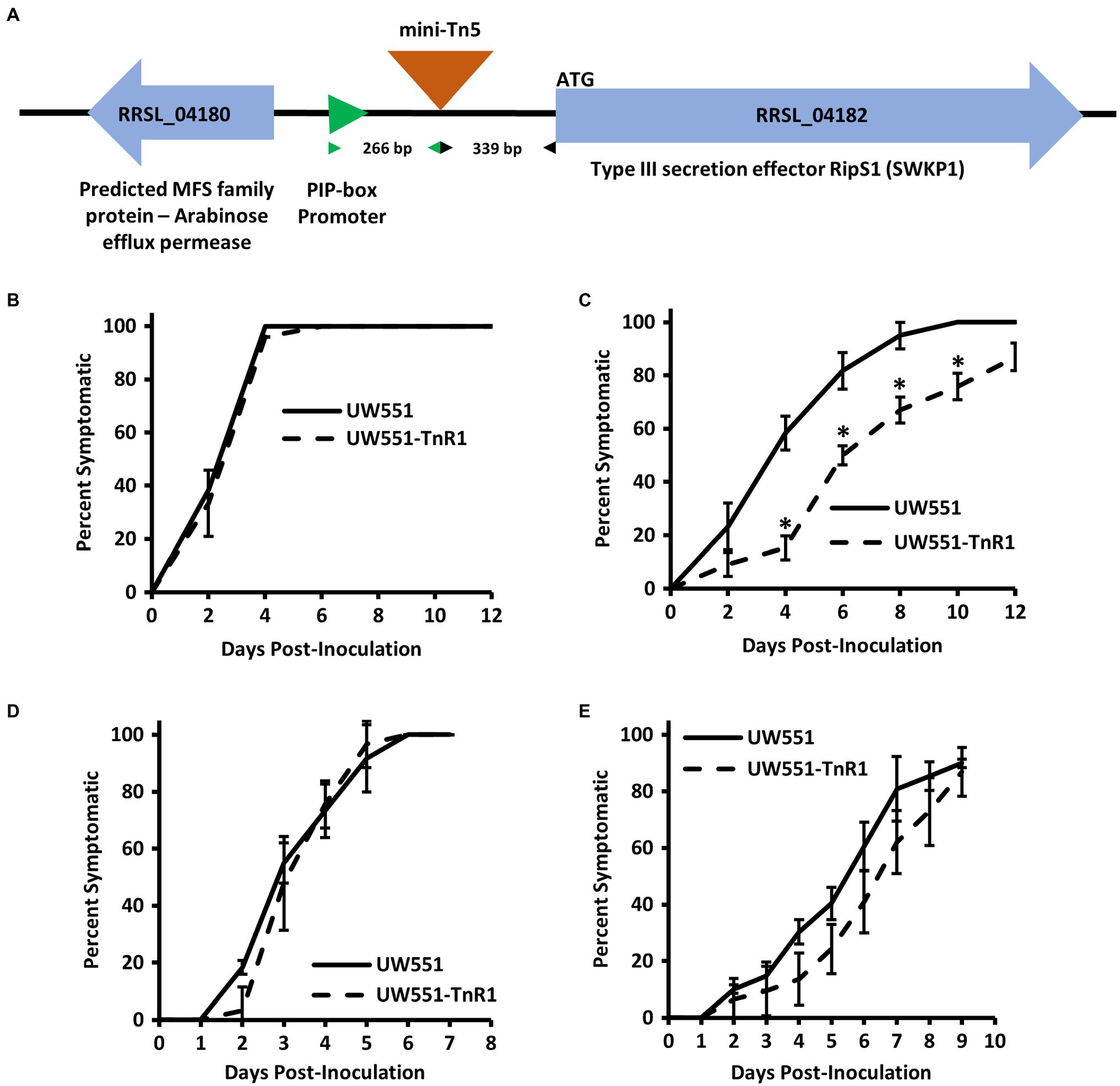
Figure 1. Intergenic transposon insertion in mutant strain UW551-TnR1 reduces virulence at 20°C in tomato seedlings, but not when the mutant was introduced directly to the xylem. Schematic diagram showing the insertion site of the mini-Tn5 transposon in UW551-TnR1 between the major facilitator superfamily (MFS) protein gene RRSL_04180 and ripS1 in parental UW551, as well as the distance between the transposon insertion and associated plant-inducible-promoter (PIP) element and ripS1, respectively (A). Confirmation that mutant UW551-TnR1 recovered from Nicotiana glutinosa seedling screen exhibits full virulence at 28°C (B) and reduced virulence at 20°C (C) in tomato seedlings. No significant difference in virulence was observed between wild-type UW551 and mutant UW551-TnR1 at individual time points when each bacterium was inoculated directly to tomato xylem through a wounded petiole regardless of whether grown at 28 (D) or 20°C (E). Experiments were repeated four times, each time with at least five seedlings across two wells of a 12-well plate (B,C) or five seedlings grown in soil (D,E). Each individual seedling was assessed for any symptoms, including wilt, water soaking of leaves, collapse of stem, or brown discolored lesions on stem, and those showing symptoms were calculated as percent of all seedlings in that treatment group. Values show average of all four experiments and error bars represent SE. Asterisks denote statistically significant difference (p < 0.05) from wild-type strain UW551 at the indicated timepoint by Student’s t-test.
To confirm the temperature dependent effects on virulence of the transposon insertion in UW551-TnR1, we assessed virulence of the mutant strain using seedlings of tomato, a commonly and widely used host of R. solanacearum, grown on water agar at 20 or 28°C. Similar to N. glutinosa seedling results, we observed that in tomato seedlings grown at 28°C, the mutant displayed full virulence compared to wild-type UW551, but at 20°C, seedlings inoculated with the mutant showed delayed appearance of disease symptoms such as stem discoloration and lesions, water soaking, and wilt compared to the wild-type strain (Figures 1B,C).
Direct Introduction of Mutant UW551-TnR1 Into Tomato Xylem Overcomes Reduced Cool Virulence Phenotype in the Mutant
Because infection and symptom development in our seedling assay requires bacterial movement to the seedling root and invasion of growing plant tissues, it is possible that the cool virulence defect observed in UW551-TnR1 is due to some defect in an early-stage infection process. To determine whether the UW551-TnR1 cool-virulence defect occurs before or after bacterial entry into the host plant, we introduced the bacterial inoculum directly to the xylem of tomato plants via a wounded petiole. We observed no significant differences in virulence between the wild-type strain UW551 and the mutant strain UW551-TnR1 at individual timepoints regardless of whether the virulence assay was done at 20 or 28°C (Figures 1D,E).
We further assessed the UW551-TnR1 mutants for various traits potentially involved in cool-virulence in early stages of infection. We found that the mutant strain was unaltered compared to the wild-type strain for flagellar motility, twitching motility, or susceptibility to exogenous hydrogen peroxide, regardless of whether grown at 20 or 28°C (Supplementary Figure S1).
Transposon Insertion in R. solanacearum Strain UW551 Alters Expression of ripS1 but Not RRSL_04180
To determine any effect of the transposon insertion in the mutant strain UW551-TnR1 on expression of the neighboring genes, we assessed their expression by qRT-PCR. We found that in cells grown in CPG medium at 28°C, expression of the ripS1 effector gene was increased by about 10-fold in the UW551-TnR1 mutant compared to the wild-type strain UW551 (Figure 2A). When compared to cells grown in CPG medium at 20°C, the decrease in temperature resulted in decreased abundance or ripS1 mRNA by a factor of 2–3-fold in both the UW551-TnR1 mutant and wild-type UW551 cells, with the UW551-TnR1 still about 10-fold higher than UW551 (Figure 2A). Expression of the MFS gene (RRSL_04180) on the other side of the transposon insertion was unaffected between wild-type UW551 and mutant UW551-TnR1 (Figure 2B). To ensure that no systematic bias was introduced in our sample treatment methods, we tested two other type III effector genes, one located on the R. solanacearum chromosome and one located on the megaplasmid, ripW, and ripU, respectively. Neither ripW nor ripU showed temperature dependent expression, and their expression was not significantly altered by the transposon insertion (Supplementary Figure S2).
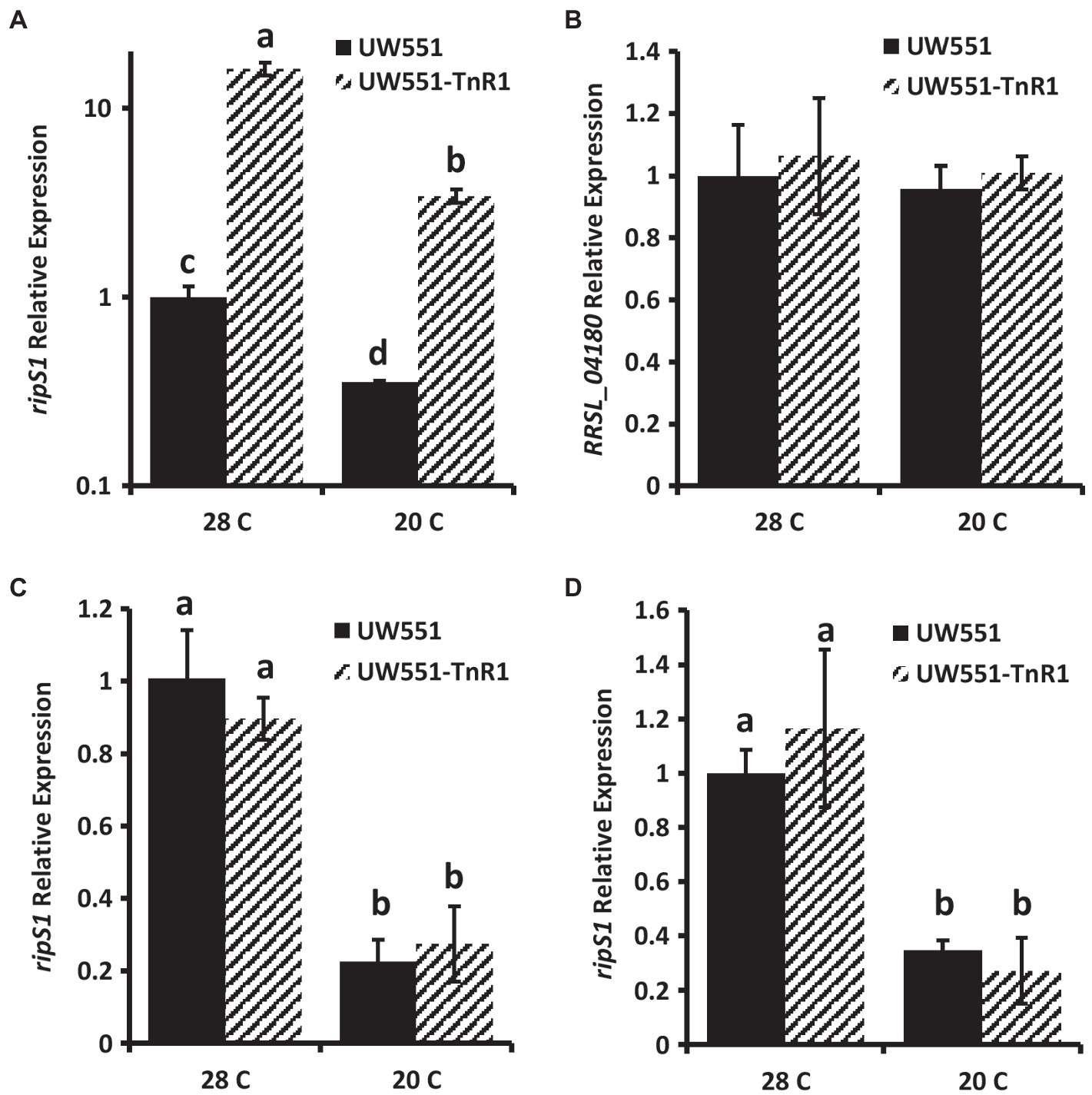
Figure 2. Relative mRNA abundance in wild-type UW551 or transposon mutant UW551-TnR1 of ripS1 (A,C,D) or RRSL_04180 (B). cDNA from RNA isolated from bacterial cells grown for 24 h in CPG medium (A,B), in minimal medium (C) or using RNA from infected tomato seedlings (D) grown at 28 or 20°C served as template for quantitative real-time PCR (qRT-PCR) reactions. Three independent samples were collected for each strain-growth combination, and each sample was tested in triplicate. Values represent average of all samples and error bars represent SD. Samples with different letters are significantly different (p < 0.05, Student’s t-test, all combinations).
To confirm that the phenotypes of the UW551-TnR1 mutant were due to single transposon insertion and not due to mutations at other locations in the genome, we generated an independent mutant containing the same transposon insertion at nucleotide 3,408,846 in UW551 using homologous recombination methods. This mutant, designated UW551-TnR1-ck did not differ significantly in growth from UW551-TnR1, and neither of these mutant strains differed significantly from wild-type UW551 in growth (Figure 3A). The virulence patterns observed in the independent mutant UW551-TnR1-ck did not differ significantly from those of UW551-TnR1 (Figure 3B), and similar gene expression patterns for ripS1 and RRSL_04180 were also observed in UW551-TnR1-ck as in UW551-TnR1 (Figures 3C,D).
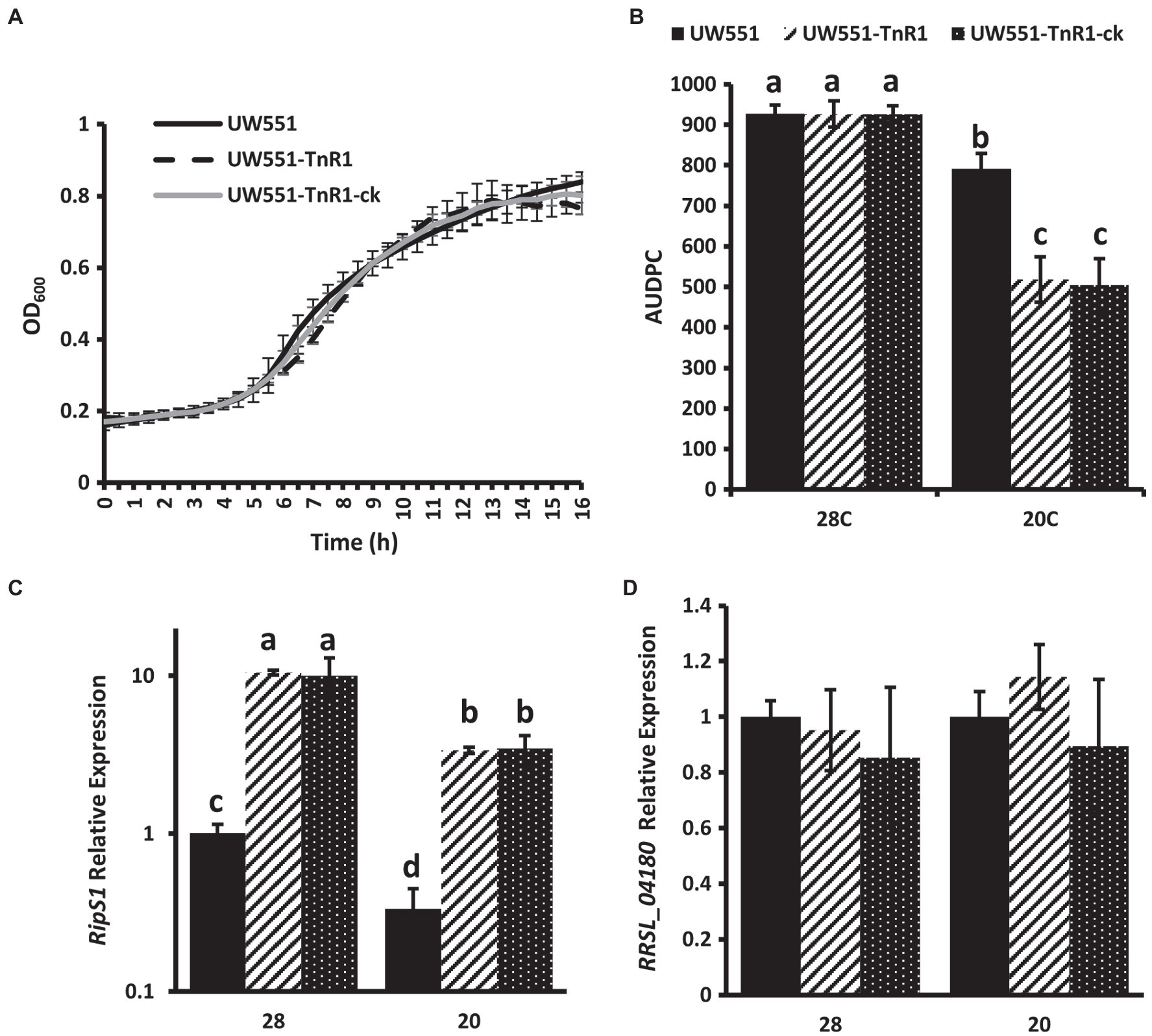
Figure 3. Independent mutant UW551-TnR1-ck confirms phenotypes of random transposon mutant UW551-TnR1. No significant differences observed in growth in CPG medium (A). Area under the disease progress curve (AUDPC) for virulence assays in tomato seedling assays shows no difference between transposon mutant UW551-TnR1 and independent mutant UW551-TnR1-ck, as compared to the wild-type strain UW551 (B). Similar relative mRNA abundance of ripS1 (C) or RRSL_04180 (D) in transposon mutant UW551-TnR1 and independent mutant UW551-TnR1-ck, as compared to the wild-type strain UW551. cDNA from RNA isolated from bacterial cells grown for 24 h in CPG medium at the indicated temperature served as template for qRT-PCR reactions. Three independent samples were collected for each strain-growth combination, and each sample was tested in triplicate. Values represent average of all samples and error bars represent SD. Samples with different letters are significantly different (p < 0.05, Student’s t-test, all combinations).
A PIP-box upstream of ripS1 was predicted in strain GMI1000 (Mukaihara and Tamura, 2009), suggesting that ripS1 is likely co-regulated with type III secretion system genes. We generated a multiple sequence alignment of the ripS1 PIP-box from several R. solanacearum and R. pseudosolanacearum strains and found that it is conserved and that the sequence for R. solanacearum strains are a better PIP consensus match than for R. pseudosolanacearum strains (Figure 4A). We also performed a multiple sequence alignment of the entire intergenic region between RRSL_04180 and ripS1 from a variety of Ralstonia strains and found clear separation between R. solanacearum and R. pseudosolanacearum (Figure 4B). Ralstonia solanacearum strain clustering further separated phylotype IIB sequevar 1 from sequevar 4 and both were separated from phylotype IIA strains (Figure 4B). Strain Rs489, which has previously proven difficult to classify (Stulberg et al., 2018), grouped near the phylotype IIB sequevar 1 strains, but does not contain identical sequence (Figure 4B). Grouping based on multiple sequence alignments of the amino acid sequence of effector RipS1 shows similar patterns but with phylotype IIB sequevar 4 grouping closer to the phylotype IIB sequevar 1 strains than strain Rs489 (Supplementary Figure S3).
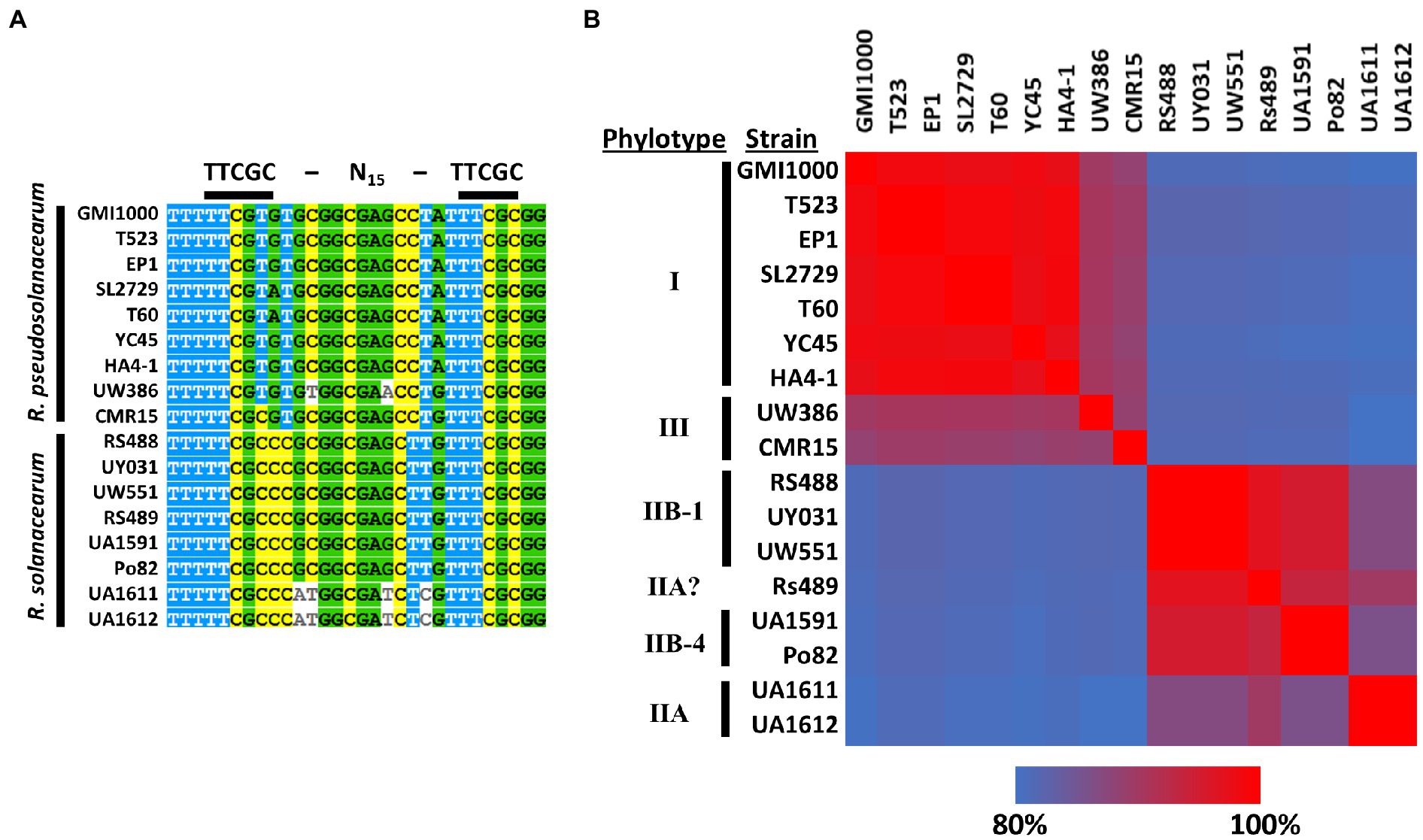
Figure 4. Multiple sequence alignment of PIP-box located about 600 bases upstream of RipS1 in various Ralstonia solanacearum species complex (Rssc) strains shows that phylotype II strains harbor a perfect PIP-box consensus match at this site (A). Heatmap showing percent identity for the intergenic region between RRSL_04180 and ripS1 in various R. solanacearum species complex strains indicates that phylotype II strains clearly separate from phylotypes I and III in this region and IIB-1 strains can be separated from all other strains (B). Multiple sequence alignments were generated and percent identity calculated using MAFFT.
Transposon Insertion Effect Lost Under Type III Secretion Inducing Conditions
To test if the transposon insertion in UW551-TnR1 altered ripS1 expression under conditions in which the PIP-box is predicted to be active, we grew cells for RNA extraction and qPCR analysis using a minimal medium in which R. solanacearum has been shown to express type III secretion system genes at high levels (Arlat et al., 1992). We found no significant difference in ripS1 expression between wild-type and mutant cells grown at the same temperature (Figure 2C). However, the temperature dependent difference in expression was still manifest, since for both strains, expression of ripS1 was higher at 28°C, compared to at 20°C (Figure 2C). We further tested ripS1 expression during infection of tomato seedlings to determine whether host-specific factors would have a different effect. In these samples, we again observed temperature dependent differences between growth at 20 and 28°C but like in minimal medium there was no difference between wild-type and mutant infected samples under the same temperature (Figure 2D).
Discussion
Using a newly developed high-throughput virulence screening method (Schachterle and Huang, 2021) to screen 10,000 mutants of R. solanacearum strain UW551 in N. glutinosa grown in water agar wells of 96-well plates led to the discovery of 26 mutants with reduced virulence, suggesting the effectiveness of the screening method. One of the mutants, UW551-TnR1, was chosen for further study and displayed significantly reduced virulence under cool temperatures but maintained full virulence under warm temperature conditions in the screen assay, and such virulence phenotype was confirmed by subsequent tomato seedling assays.
The mutant UW551-TnR1 contains a transposon insertion between the gene encoding the type III effector RipS1 and its predicted PIP promoter. This insertion resulted in increased expression of ripS1 when grown in rich medium, but no difference under type III secretion inducing conditions. This suggests that the transposon insertion is causing a constitutive activation of ripS1. Our data indicate that this dysregulation, or inability of R. solanacearum to shut down expression of this gene, is leading to a reduction in virulence at 20°C, but that these bacteria are fully virulent at 28°C. Significant reduction in virulence was not observed when a high dose of bacterial cells were directly introduced to tomato xylem, suggesting the role of RipS1 during early infection processes. Indeed, recent work provides evidence that certain RipS effectors, including RipS1 may play more important virulence roles than other members of this family during infection of eggplant (Chen et al., 2021). However, we observed no change in the virulence traits of swimming or twitching motility, and no change in susceptibility to exogenous oxidative stress from hydrogen peroxide in mutant UW551-TnR1 compared to wild-type UW551. While other early-stage infection virulence traits may play a role, such as ability to spread between xylem vessels, our data suggest that the specific function of RipS1 as an effector may be responsible for the reduction in cool virulence and not pleiotropic effects on other virulence traits tested in this study. Our further work to confirm virulence and gene expression phenotypes using the independently generated strain UW551-TnR1-ck provides additional evidence that this effect appears to be through RipS1, whether directly or indirectly, and not any other spontaneous mutations that may have arisen during transposon mutagenesis.
Because several deletion mutants lacking RipS family effectors have no virulence defect (Lei et al., 2020), our data suggest that the significantly reduced virulence, we observed in mutant UW551-TnR1 is not because of any loss of function of RipS1, but is more likely due to an inappropriate expression at some critical stage of cool-virulence or in some subpopulation of cells. However, it is noteworthy that the studies conducted with ripS deletion mutants were in non-cool virulent R. pseudosolanacearum and were not tested at cool temperature, and functions of ripS at different temperature conditions in different Ralstonia species may be different.
Our results indicate that ripS1 is expressed at lower levels at 20°C compared to 28°C. A transcriptomic study of strain UW551 grown in CPG medium also found a 2.33-fold decrease in ripS1 expression at 20°C compared to 28°C (Meng et al., 2015). However, other transcriptomic and genomic studies did not associate ripS1 with differential expression based on temperature or in planta growth or differential presence between strains (Jacobs et al., 2012; Bocsanczy et al., 2017). Similarly, RipS1 was not detected in a proteomic study of R. solanacearum at different temperatures (Bocsanczy et al., 2014), but this is not surprising given its nature as a secreted protein, that other type III effectors were not detected, and the difficulty of consistent detection of low-abundance proteins in shotgun proteomic analyses. The difficulties of these other genome-scale approach to converge on factors associated with the cool-virulence magnifies an advantage of our use of a forward genetic screen to specifically test for the cool-virulence phenotype.
A PIP-box was predicted 683 bases upstream from the ripS1 start codon in strain GMI1000 (Mukaihara and Tamura, 2009), and we found that PIP-box is conserved in the promoter region of ripS1 in R. solanacearum and R. pseudosolanacearum, with R. solanacearum strains perfectly matching the PIP consensus sequence (Figure 4A). Although this promoter, and associated transcriptional start site suggested by the PIP-box have not been experimentally validated, this would represent a very long 5´UTR that could be subject to many types of regulation, including post-transcriptional regulation (Oliva et al., 2015). Indeed, it is in this region that the transposon inserted in UW551-TnR1. The increased mRNA abundance due to uncoupling of the PIP-box from ripS1 suggests that there are likely binding sites for other transcription factors or other regulatory elements in the ripS1 promoter and 5´UTR region. Our data support this because regardless of the presence of the transposon insertion or cell growth conditions, we observed temperature dependence in ripS1 mRNA abundance but no temperature dependence in other type III effectors ripW and ripU. Consistent with other findings, our results do not point to the well-characterized type III secretion regulatory systems as temperature dependent, but suggest another mechanism exists regulating temperature dependence. Additional work is needed to uncover the mechanism of temperature dependence in ripS1 expression in cool-virulent strains of R. solanacearum.
Although the function of RipS family effectors remains unknown, a recent study predicted RipS1 and several other R. solanacearum type III effectors to carry nuclear localization signals (NLSs) that would enable them to localize to the plant host nucleus following translocation (Jeon et al., 2020). RipS1 and three other members of the RipS effector family were predicted to carry one or more NLSs. Although shown for other effectors with predicted NLSs, functionality was not determined for any members of the RipS effector family. Previous work has suggested that the cool-virulence phenotype of some R. solanacearum is dependent on interaction with temperature dependent host-derived factors (Milling et al., 2009). The significantly reduced virulence due to increased expression of ripS1 at cool temperatures could fit this hypothesis, especially when no reduction in virulence was observed, while this effector was also overexpressed at warm temperature. Although not a well understood phenomenon, similar observations have been made in which overexpression of specific type III effectors in planta reduces pathogen fitness (Macho et al., 2012). However, we found no difference in ripS1 mRNA abundance between UW551 and UW551-TnR1 under hrp-inducing conditions, such as in minimal medium or in tomato seedlings, regardless of temperature. Because of the possibility that ripS1 expression changes over time and in different niche environments presented by the host, expression differences between UW551 and UW551-TnR1 may exist that were not observed due to our limited testing, which occurred at 7 days post-inoculation and after symptom appearance. Thus, unobserved differences in expression may still exist. Nonetheless, our data suggest that additional complexities must be involved to explain these apparently contradictory results. Several possibilities exist that could cause such a contradiction. These include post-transcriptional or post-translational regulations altering RipS1 activity without affecting mRNA abundance. Another possibility is localized expression of ripS1 by a subpopulation of cells in different in planta environments, or only during specific stages of disease development such as during attachment, invasion, colonization, or systemic spread. Furthermore, epistatic interactions with additional genes could result in behaviors that are inexplicable when the expression of only ripS1 is considered. Indeed, each of these potential complexities has been observed in plant pathogenic bacteria (Cui et al., 2018; Yuan et al., 2019), and support an abundance of evidence that indicates that cool-virulence is complex and likely controlled by a suite of genes, rather than a single actor.
Deletion of a previously identified genomic region that resulted in reduced cool-virulence also resulted in cool temperature reduction in twitching motility (Stulberg et al., 2018). Although associations between cool-virulent R. solanacearum and twitching motility have been established (Bocsanczy et al., 2012; Stulberg et al., 2018), our transposon mutant UW551-TnR1 did not exhibit any qualitative reduction in twitching motility at cool or warm temperatures indicating that likely multiple virulence factors contribute to the cool-irulence phenotype in R. solanacearum.
Our data implicate the type III effector RipS1 as playing an important role in cool-virulence of R. solanacearum. Additional work is needed to determine whether RipS1 has a direct role such as interacting with a specific host factor or protein at cool temperatures, or if it only has an indirect role in cool-virulence, such as overexpression of ripS1 in the UW551-TnR1 mutant prevents efficient secretion of another type III effector that in turn has a direct role. Multiple sequence alignments indicate that the RipS1 protein and its associated promoter region can separate cool-virulent R. solanacearum (phylotype IIB sequevar 1) strains from others and may prove useful for development of diagnostic tools specific to cool-virulent R. solanacearum. Better understanding of the mechanisms underlying the cool-virulence phenotype will facilitate accurate definition, detection, and control of the select agent strains of R. solanacearum to safeguard United States Agriculture.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
JS and QH conceived and designed the study, analyzed the data, and wrote the manuscript. JS conducted the experiments. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Hedda J. Weitz from University of Aberdeen, Scotland, United Kingdom and Donald L. Court from National Cancer Institute, United States for providing plasmids pUT-miniTn5-luxCDABE and pSIM7, respectively, for our study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.705717/full#supplementary-material
Footnotes
References
Abràmoff, M. D., Magalhães, P. J., and Ram, S. J. (2004). Image processing with imageJ. Biophoton. Int. 11, 36–42.
Addy, H. S., Askora, A., Kawasaki, T., Fujie, M., and Yamada, T. (2012). Loss of virulence of the phytopathogen Ralstonia solanacearum through infection by φRSM filamentous phages. Phytopathology 102, 469–477. doi: 10.1094/PHYTO-11-11-0319-R
Álvarez, B., Biosca, E. G., and López, M. M. (2010). On the life of Ralstonia solanacearum, a destructive bacterial plant pathogen. Technol. Educ. Top. Appl. Microbiol. Microb. Biotechnol. 1, 267–279.
Araud-Razou, I., Vasse, J., Montrozier, H., Etchebar, C., and Trigalet, A. (1998). Detection and visualization of the major acidic exopolysaccharide of Ralstonia solanacearum and its role in tomato root infection and vascular colonization. Eur. J. Plant Pathol. 104, 795–809. doi: 10.1023/A:1008690712318
Arlat, M., Gough, C. L., Zischek, C., Barberis, P. A., Trigalet, A., and Boucher, C. A. (1992). Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 5, 187–193. doi: 10.1094/MPMI-5-187
Bocsanczy, A. M., Achenbach, U. C. M., Mangravita-Novo, A., Chow, M., and Norman, D. J. (2014). Proteomic comparison of Ralstonia solanacearum strains reveals temperature dependent virulence factors. BMC Genomics 15:280. doi: 10.1186/1471-2164-15-280
Bocsanczy, A. M., Achenbach, U. C. M., Mangravita-Novo, A., Yuen, J. M. F., and Norman, D. J. (2012). Comparative effect of low temperature on virulence and twitching motility of Ralstonia solanacearum strains present in Florida. Phytopathology 102, 185–194. doi: 10.1094/PHYTO-05-11-0145
Bocsanczy, A. M., Huguet-Tapia, J. C., and Norman, D. J. (2017). Comparative genomics of Ralstonia solanacearum identifies candidate genes associated with cool virulence. Front. Plant Sci. 8:1565. doi: 10.3389/fpls.2017.01565
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chen, L., Lei, N., Kiba, A., Hikichi, Y., and Ohnishi, K. (2021). Contribution of RipS type III effector family of Ralstonia solanacearum Japanese strain OE1-1 to disease development in eggplant. J. Gen. Plant Pathol. 87, 77–82. doi: 10.1007/s10327-020-00977-5
Chen, L., Li, J., Shirota, M., and Ohnishi, K. (2018). Analysis of the SKWP effectors to bacterial fitness in host plant by a novel competition assay. Acta Microbiol. Sin. 58, 432–442. doi: 10.13343/j.cnki.wsxb.20170221
Coll, N. S., and Valls, M. (2013). Current knowledge on the Ralstonia solanacearum type III secretion system. Microb. Biotechnol. 6, 614–620. doi: 10.1111/1751-7915.12056
Corral, J., Sebastià, P., Coll, N. S., Barbé, J., Aranda, J., and Valls, M. (2020). Twitching and swimming motility play a role in Ralstonia solanacearum pathogenicity. mSphere 5, e00740–e00819. doi: 10.1128/msphere.00740-19
Cui, Z., Yuan, X., Yang, C. H., Huntley, R. B., Sun, W., Wang, J., et al. (2018). Development of a method to monitor gene expression in single bacterial cells during the interaction with plants and use to study the expression of the type III secretion system in single cells of Dickeya dadantii in potato. Front. Microbiol. 9:1429. doi: 10.3389/fmicb.2018.01429
Cunnac, S., Boucher, C., and Genin, S. (2004). Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 186, 2309–2318. doi: 10.1128/JB.186.8.2309-2318.2004
Datta, S., Costantino, N., and Court, D. L. (2006). A set of recombineering plasmids for gram-negative bacteria. Gene 379, 109–115. doi: 10.1016/j.gene.2006.04.018
Deslandes, L., and Genin, S. (2014). Opening the Ralstonia solanacearum type III effector tool box: insights into host cell subversion mechanisms. Curr. Opin. Plant Biol. 20, 110–117. doi: 10.1016/j.pbi.2014.05.002
Escalon, A., Javegny, S., Vernière, C., Noël, L. D., Vital, K., Poussier, S., et al. (2013). Variations in type III effector repertoires, pathological phenotypes and host range of Xanthomonas citri pv. Citri pathotypes. Mol. Plant Pathol. 14, 483–496. doi: 10.1111/mpp.12019
Fegan, M., and Prior, P. (2005). “How complex is the Ralstonia solanacearum species complex,” in Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex. eds. C. Allen, P. Prior, and A. C. Hayward (Minnesota: APS Press), 449–461.
Genin, S., Brito, B., Denny, T. P., and Boucher, C. (2005). Control of the Ralstonia solanacearum type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 579, 2077–2081. doi: 10.1016/j.febslet.2005.02.058
Hayes, M. M., MacIntyre, A. M., and Allen, C. (2017). Complete genome sequences of the plant pathogens Ralstonia solanacearum type strain K60 and R. solanacearum race 3 biovar 2 strain UW551. Genome Announc. 5, e01088–e01117. doi: 10.1128/genomeA.01088-17
Hayward, A. C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29, 65–87. doi: 10.1146/annurev.py.29.090191.000433
Jacobs, J. M., Babujee, L., Meng, F., Milling, A., and Allen, C. (2012). The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. mBio 3, e00114–e00212. doi: 10.1128/mBio.00114-12
Janse, J. D., Araluppan, F. A. X., Schans, J., Wenneker, M., and Westerhuis, W. (1998). “Experiences with brown rot Ralstonia solanacearum biovar 2, race 3 in the Netherlands,” in Bacterial Wilt Disease. eds. P. Prior, C. Allen, and J. Elphinstone (Berlin: Springer), 146–152.
Jeon, H., Kim, W., Kim, B., Lee, S., Jayaraman, J., Jung, G., et al. (2020). Ralstonia solanacearum type III effectors with predicted nuclear localization signal localize to various cell compartments and modulate immune responses in Nicotiana spp. Plant Pathol. J. 36:43. doi: 10.5423/PPJ.OA.08.2019.0227
Kang, Y., Liu, H., Genin, S., Schell, M. A., and Denny, T. P. (2002). Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46, 427–437. doi: 10.1046/j.1365-2958.2002.03187.x
Khokhani, D., Tuan, T., Lowe-Power, T., and Allen, C. (2018). Plant assays for quantifying Ralstonia solanacearum virulence. Bio-Protocol 8:e3028. doi: 10.21769/bioprotoc.3028
Lei, N., Chen, L., Kiba, A., Hikichi, Y., Zhang, Y., and Ohnishi, K. (2020). Super-multiple deletion analysis of type III effectors in Ralstonia solanacearum OE1-1 for full virulence toward host plants. Front. Microbiol. 11:1683. doi: 10.3389/fmicb.2020.01683
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lowe-Power, T. M., Khokhani, D., and Allen, C. (2018). How Ralstonia solanacearum exploits and thrives in the flowing plant xylem environment. Trends Microbiol. 26, 929–942. doi: 10.1016/j.tim.2018.06.002
Macho, A. P., Zumaquero, A., Gonzalez-Plaza, J. J., Ortiz-Martín, I., Rufián, J. S., and Beuzón, C. R. (2012). Genetic analysis of the individual contribution to virulence of the type III effector inventory of Pseudomonas syringae pv. Phaseolicola. PLoS One 7:e35871. doi: 10.1371/journal.pone.0035871
Martínez-García, E., Aparicio, T., de Lorenzo, V., and Nikel, P. I. (2014). New transposon tools tailored for metabolic engineering of gram-negative microbial cell factories. Front. Bioeng. Biotechnol. 2:46. doi: 10.3389/fbioe.2014.00046
Mendez, J., Cascales, D., Torrico, A. I. G., and Guijarro, J. A. (2018). Temperature-dependent gene expression in Yersinia ruckeri: tracking specific genes by bioluminescence during in vivo colonization. Front. Microbiol. 9:1098. doi: 10.3389/fmicb.2018.01098
Meng, F. (2013). The virulence factors of the bacterial wilt pathogen Ralstonia solanacearum. J. Plant Pathol. Microbiol. 4:168. doi: 10.4172/2157-7471.1000168
Meng, F., Babujee, L., Jacobs, J. M., and Allen, C. (2015). Comparative transcriptome analysis reveals cool virulence factors of Ralstonia solanacearum race 3 biovar 2. PLoS One 10:e0139090. doi: 10.1371/journal.pone.0139090
Miller-Williams, M., Loewen, P. C., and Oresnik, I. J. (2006). Isolation of salt-sensitive mutants of Sinorhizobium meliloti strain Rm1021. Microbiology 152, 2049–2059. doi: 10.1099/mic.0.28937-0
Milling, A., Meng, F., Denny, T. P., and Allen, C. (2009). Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2. Phytopathology 99, 1127–1134. doi: 10.1094/PHYTO-99-10-1127
Mukaihara, T., and Tamura, N. (2009). Identification of novel Ralstonia solanacearum type III effector proteins through translocation analysis of hrp B-regulated gene products. Microbioloy 155, 2235–2244. doi: 10.1099/mic.0.027763-0
Mukaihara, T., Tamura, N., and Iwabuchi, M. (2010). Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant-Microbe Interact. 23, 251–262. doi: 10.1094/MPMI-23-3-0251
Oliva, G., Sahr, T., and Buchrieser, C. (2015). Small RNAs, 5’ UTR elements and RNA-binding proteins in intracellular bacteria: impact on metabolism and virulence. FEMS Microbiol. Rev. 39, 331–349. doi: 10.1093/femsre/fuv022
Peeters, N., Carrère, S., Anisimova, M., Plener, L., Cazalé, A. C., and Genin, S. (2013). Repertoire, unified nomenclature and evolution of the type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics 14:859. doi: 10.1186/1471-2164-14-859
Poueymiro, M., Cunnac, S., Barberis, P., Deslandes, L., Peeters, N., Cazale-Noel, A. C., et al. (2009). Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host-range specificity on tobacco. Mol. Plant-Microbe Interact. 22, 538–550. doi: 10.1094/MPMI-22-5-0538
Rivas, R., Vizcaíno, N., Buey, R. M., Mateos, P. F., Martínez-Molina, E., and Velázquez, E. (2001). An effective, rapid and simple method for total RNA extraction from bacteria and yeast. J. Microbiol. Methods 47, 59–63. doi: 10.1016/S0167-7012(01)00292-5
Sabbagh, C. R. R., Carrere, S., Lonjon, F., Vailleau, F., Macho, A. P., Genin, S., et al. (2019). Pangenomic type III effector database of the plant pathogenic Ralstonia spp. PeerJ 7:e7346. doi: 10.7717/peerj.7346
Safni, I., Cleenwerck, I., De Vos, P., Fegan, M., Sly, L., and Kappler, U. (2014). Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int. J. Syst. Evol. Microbiol. 64, 3087–3103. doi: 10.1099/ijs.0.066712-0
Schachterle, J. K., and Huang, Q. (2021). A high-throughput virulence screening method for the Ralstonia solanacearum species complex. J. Microbiol. Methods 187:106270. doi: 10.1016/j.mimet.2021.106270
Schachterle, J. K., Onsay, D. M., and Sundin, G. W. (2019). Small RNA ArcZ regulates oxidative stress response genes and regulons in Erwinia amylovora. Front. Microbiol. 10:2775. doi: 10.3389/fmicb.2019.02775
Stulberg, M. J., Cai, X., Ahmad, A. A., and Huang, Q. (2018). Identification of a DNA region associated with the cool virulence of Ralstonia solancearum strain UW551 and its utilization for specific detection of the bacterium’s race 3 biovar 2 strains. PLoS One 13:e0207280. doi: 10.1371/journal.pone.0207280
Teper, D., Burstein, D., Salomon, D., Gershovitz, M., Pupko, T., and Sessa, G. (2016). Identification of novel Xanthomonas euvesicatoria type III effector proteins by a machine-learning approach. Mol. Plant Pathol. 17, 398–411. doi: 10.1111/mpp.12288
Van Elsas, J. D., Kastelein, P., Van Bekkum, P., Van der Wolf, J. M., De Vries, P. M., and Van Overbeek, L. S. (2000). Survival of Ralstonia solanacearum biovar 2, the causative agent of potato brown rot, in field and microcosm soils in temperate climates. Phytopathology 90, 1358–1366. doi: 10.1094/PHYTO.2000.90.12.1358
van Gijsegem, F., Gough, C., Zischek, C., Niqueux, E., Arlat, M., Genin, S., et al. (1995). The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol. Microbiol. 15, 1095–1114. doi: 10.1111/j.1365-2958.1995.tb02284.x
Vasse, J., Genin, S., Frey, P., Boucher, C., and Brito, B. (2000). The hrpB and hrpG regulatory genes of Ralstonia solanacearum are required for different stages of the tomato root infection process. Mol. Plant-Microbe Interact. 13, 259–267. doi: 10.1094/MPMI.2000.13.3.259
Weitz, H. J., Ritchie, J. M., Bailey, D. A., Horsburgh, A. M., Killham, K., and Glover, L. A. (2001). Construction of a modified mini-Tn5 luxCDABE transposon for the development of bacterial biosensors for ecotoxicity testing. FEMS Microbiol. Lett. 197, 159–165. doi: 10.1111/j.1574-6968.2001.tb10598.x
Yuan, X., Zeng, Q., Khokhani, D., Tian, F., Severin, G. B., Waters, C. M., et al. (2019). A feed-forward signalling circuit controls bacterial virulence through linking cyclic di-GMP and two mechanistically distinct sRNAs, ArcZ and RsmB. Environ. Microbiol. 21, 2755–2771. doi: 10.1111/1462-2920.14603
Keywords: Ralstonia solanacearum, cool-virulence, virulence, seedling screen, type III effector, RipS1
Citation: Schachterle JK and Huang Q (2021) Implication of the Type III Effector RipS1 in the Cool-Virulence of Ralstonia solanacearum Strain UW551. Front. Plant Sci. 12:705717. doi: 10.3389/fpls.2021.705717
Edited by:
Youfu “Frank” Zhao, University of Illinois at Urbana-Champaign, United StatesReviewed by:
Gang Yu, Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences (CAS), ChinaTuan Minh Tran, Nanyang Technological University, Singapore
Copyright © 2021 Schachterle and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Huang, cWkuaHVhbmdAdXNkYS5nb3Y=
 Jeffrey K. Schachterle
Jeffrey K. Schachterle Qi Huang
Qi Huang