- 1Collaborative Innovation Center for Efficient and Green Production of Agriculture in Mountainous Areas of Zhejiang Province, College of Horticulture Science, Zhejiang A&F University, Hangzhou, China
- 2Department of Biotechnology, University of Houston Clear Lake, Houston, TX, United States
MYC2/3/4, known as a basic helix–loop–helix (bHLH) transcription factor, directly activate the genes involved in diverse plant development and secondary metabolites biosynthesis. In this study, we identified and cloned five MYC paralogs (BrMYC2/3-1/3-2/4-1/4-2) from Chinese cabbage (Brassica rapa ssp. pekinensis). In-silico analyses for the physicochemical properties suggested that BrMYC2/3-1/3-2/4-2/4-3 are unstable hydrophobic and acidic proteins, while BrMYC4-1 is an unstable hydrophobic and basic protein. BrMYC2/3/4 belong to the bHLH superfamily and are closely related to AthMYC2/3/4 orthologs that mediate the regulation of various secondary metabolites. It was demonstrated that BrMYC2/3/4-GFP fusion protein localized in the nucleus and expression levels of five BrMYC2/3/4 homologous genes all elevated relative to control (Ctrl). When expressed in Arabidopsis under the control of 35S promoter, each of the BrMYC2/3-1/3-2/4-1/4-2 transgenes differentially influenced root and shoot elongation, vegetative phase change, flowering time, plant height and tiller number after flowering, and seed production. Despite the variation of phenotypes between the transgenic lines, all the lines except for BrMYC4-2 exhibited shorter seed length, less seed weight, higher accumulation of glucosinolates (GSs), and resistance to Sclerotinia sclerotiorum than Ctrl. Notably, BrMYC2 overexpression (OE) line significantly reduced the lengths of root and hypocotyl, seed length, and weight, along with faster bolting time and strikingly higher accumulation of total GSs. Accumulation of GSs at the highest levels in the BrMYC2OE line conferred the highest resistance to S. sclerotiorum. Unlike BrMYC3OE and BrMYC4OE, BrMYC2OE stimulated the growth of plant height after fluorescence. The results of this study point to the BrMYC2 overexpression that may provide a beneficial effect on plant growth and development via plant resistance to the fungal pathogen.
Introduction
Myelocytomatosis proteins (MYCs) are key transcription factors (TFs) in the response pathway of jasmonic acid (JA) in plants (Xie et al., 2020). Jasmonates (JAs) and its derivatives of JA methyl ester and amino acid conjugates (Per et al., 2018; Stella de Freitas et al., 2019) are well-known phytohormones connecting both developmental signals and environmental stresses to coordinately regulate diverse metabolic processes involved in plant growth and development (Song et al., 2017). Examples are seed development (Cao et al., 2016), root growth (To et al., 2019), shoot elongation (To et al., 2019), trichome development (del Rosario Cappellari et al., 2019), and fertility (Schubert et al., 2019). MYC2 (bHLH006), MYC3 (bHLH005), and MYC4 (bHLH004) belong to the family of the IIIe subgroup basic helix–loop–helix (bHLH) and contain a conserved domain of bHLH (Zhang et al., 2018, 2020; Wasternack and Strnad, 2019). MYC2/3/4 interact with F-box protein CORONATINE INSENSITIVE1 (COI1) (Major et al., 2017) and JA ZIM-domains (JAZs) (Garrido-Bigotes et al., 2018) to mediate diverse JA response, such as root growth inhibition (Yang et al., 2017), stamen development (Zhuo et al., 2020), leaf senescence (Xu et al., 2018), glucosinolates (GSs) biosynthesis (Costarelli et al., 2020), defenses against insect attack (Chen et al., 2021), and necrotrophic fungi (Tiwari, 2018).
Glucosinolates are a group of secondary metabolites found in Brassicaceae family and are composed of β-D-thioglucose and sulfonated oxime moieties (Tiwari, 2018). They have received considerable interest because of their commercial properties of anti-cancer agents (Arumugam and Razis, 2018), bio-pesticides (Malka and Cheng, 2017), and flavor condiment (Burow et al., 2015). They are typically classified as aliphatic GS (from alanine, valine, leucine, isoleucine, or methionine), aromatic GS (from phenylalanine or tyrosine), or indole GS (from tryptophan) on the basis of their modified side chains (R) derived from amino acids through a long chain lengthening process and hydroxylation or oxidation (Rahimi and Rahmanpour, 2020). Biosynthesis and metabolism of GSs are a complex process that is finely regulated by a variety of trans-acting regulatory factors; these include eight TFs of v-myb avian myeloblastosis viral oncogene homolog (MYB) family (Frerigmann and Gigolashvili, 2014; Zhang et al., 2015; Seo and Kim, 2017; Coleto et al., 2021), three TFs of MYC family (MYC2, MYC3, and MYC4) (Schweizer et al., 2013), and other TFs, such as Dof1.1, IQD1 (Kim et al., 2014), and ATR1 (Kai et al., 2011). MYC2, MYC3, and MYC4 are the main TFs regulating indole GSs (IGSs) biosynthesis, since those mutants significantly reduced levels of IGSs (Schweizer et al., 2013; Frerigmann et al., 2014). Non-methionine aliphatic GSs (AGSs) are present in all the Brassicaceae families, and the indole pathway is thought to have originated from a whole-genome duplication event (Mitreiter and Gigolashvili, 2021).
Chinese cabbage (Brassica rapa ssp. pekinensis) is an economically important leafy vegetable that is largely cultivated around the world, especially in Korea, Japan, and China, and its genome sequencing has been completed (Park et al., 2019). Due to the evolutionary relatedness, most of the genes in Chinese cabbage contain more than one ortholog of Arabidopsis (Wang et al., 2011). GSs and their hydrolysis products are known to play essential roles in plant–insects and plant–microbes' interactions (Pastorczyk et al., 2020; Ji et al., 2021). For example, isothiocyanates (ITCs), 4-hydroxyglucobrassicin (4-OHGBC), and 4-methoxyglucobrassicin (4MeGBC) are involved in plant resistance against pests (Pfalz et al., 2019; Döring and Ulber, 2020; Shukla and Beran, 2020), as well as plant innate immunity (Clay et al., 2009; Pangesti et al., 2016; Chen et al., 2020). ITCs are the main volatile compounds resulting from the hydrolysis of GSs; 3-butenyl-(55.6–71.8%) and 5-methylsulfinylpentyl-ITC (0–9.5%) originate from gluconapin (GNA) and glucoalyssin (GAS), respectively (BlaŽević et al., 2011). Anti-microbial activity of such volatile compounds was found to inhibit a wide range of bacteria and fungi (BlaŽević et al., 2019). Indole-3-carbinol (I3C), a hydrolysate of 4MeGBC, aides in guiding the oviposition of specific insects in Cruciferous vegetables (Katz and Chamovitz, 2017).
Evolutionary data suggest that Chinese cabbage is closely related to A. thaliana and that it underwent a whole-genome triplication since it has diverged from Arabidopsis thaliana (Wang et al., 2011). There are one, two, and three copies of BrMYC2/3/4 family genes in Chinese cabbage, respectively. Functions of MYC2/3/4 orthologs have been mainly investigated in A. thaliana, and they showed a functional divergence (Schweizer et al., 2013). However, the functions and regulatory roles of multiple paralogous copies of BrMYC2/3/4 family genes have remained unresolved.
In the current study, we identified, cloned five MYC2/3/4 paralogs (BrMYC2, BrMYC3-1, BrMYC3-2, BrMYC4-1, and BrMYC4-2) in Chinese cabbage, and conducted in-silico comparative studies on physicochemical properties, domains, and evolutionary relationships among the deduced BrMYC2/3/4 proteins. In order to study the phenotypes conferred by expression of BrMYC2/3/4, each of the cloned coding sequence of five MYC2/3/4 paralogs was introduced into a binary vector pCAMBIA2302, and each chimeric transgene expression construct was transformed into Arabidopsis. Subsequently, T2 lines showing 3:1 segregation ratio of selectable marker gene were selected, and T3 lines were used to analyze various phenotypes of plants at juvenile and adult stage, GSs contents, and resistance to white mold caused by necrotrophic Sclerotinia sclerotiorum.
Materials and Methods
Identification of B. Rapa MYC2/3/4 Genes
Gene IDs for BrMYC2/3/4 genes of Chinese cabbage, BraA05g023030.3C (BrMYC2), BraA09g022310.3C (BrMYC3-1), BraA06g041690.3C (BrMYC3-2), BraA01g009460.3C (BrMYC4-1), BraA01g009470.3C (BrMYC4-2), and BraA08g000150.3C (BrMYC4-3), were retrieved from Brassica database annotations (http://brassicadb.cn/#/Annotations/). Coding sequences of the retrieved BrMYC2/3/4 genes were obtained from Brassica database gene sequence (http://brassicadb.cn/#/GeneSequence/).
In-silico Analyses
The amino sequences deduced from coding sequences were used to predict the functional properties of BrMYC2/3/4 proteins. Physiochemical properties were inferred using ProtParam software (http://web.expacy.org/protparam/). The percentages of sequence identities among and between MYC2/3/4 in terms of coding sequence and deduced amino acid sequence were analyzed using DNAMAN software version 6.0 (Lynnon Biosoft, Montreal, Canada). The domains were predicted using the conserved domain database (CDD, https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), which has a domain prediction tool provided by the National Center for Biotechnology Information (NCBI, US National Library of Medicine). Structural and evolutionary analyses of MYC2/3/4 coding sequences in the U's triangle species were conducted with homologous sequences on Brassica database (http://brassicadb.cn/#/BLAST/) according to the neighbor-joining method (Tarahomi et al., 2020) using molecular evolutionary genetics analysis software (MEGA7 software) (Kumar et al., 2016). The main parameter settings were distance model, p-distance method (Sáez-López et al., 2019), gene tree robustness detection, bootstrap test (1,000 replicates) (Elateek et al., 2020), processing of gap missing data, and deletion between pairs.
Expression Constructs
All cloning experiments were carried out through homologous recombination using the One Step Cloning Kit (Vazyme, Nanjing, China). The coding sequences of BrMYC2, BrMYC3-1, BrMYC3-2, BrMYC4-1, and BrMYC4-2 were put under control of the constitutive CaMV 35S promoter in the binary vector pCAMBIA2302 with green fluorescent protein (GFP). The integrity of the cloned coding sequence was verified by colony PCR, followed by DNA sequencing.
Plant Materials and Growth Conditions
Arabidopsis ecotype Col-0 was used as the wild-type for all experiments. Plant seeds were sown in a mixture of grass charcoal soil and vermiculite as 1:1 ratio and cultivated in a greenhouse for 2 months under well-controlled conditions of 65% relative humidity and 600 μmol·m−2·s−1 maximum light intensity. The photoperiod and temperature were 16/8 h (light/dark) and 21/19°C, respectively, in day/night.
Plant Transformation and Transgenic Plants Selection
Recombinant plasmids (pCAMBIA2302, p2302MYC2, p2302MYC3-1, p2302MYC3-2, p2302MYC4-1, and p2302MYC4-2) were introduced into the Agrobacterium tumefaciens strain GV3101 (Shanghai Weidi Biotechnology Co, Ltd, China). Agrobacterium-mediated transformation was performed to produce T0 transgenic lines using the floral dip method (Van Eck, 2018). Overexpression (OE) transgenic lines obtained were named as Ctrl, BrMYC2OE, BrMYC3-1OE, BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE, respectively.
Arabidopsis seeds from T0 lines were first surface sterilized using 75% (v/v) ethanol for 90 s and then washed three times with double-distilled water (ddH2O). The seeds were further sterilized in 10% bleach for 10 min and washed three times with ddH2O. They were subsequently sown onto 1 × MS medium (Phyto Technology Laboratories, Shawnee Mission, KS, USA) supplemented with 3% sucrose (pH 5.8 and 0.8% (w/v) plant tissue culture agar) and 50 μg·ml−1 kanamycin (Phyto Technology Laboratories, Shawnee Mission, KS, USA). Seedlings were chilled at 4°C for 2 days and transferred to a growth room with controlled conditions (25°C, 16:8 h, light:dark regime) grown vertically for 4 days. Healthy seedlings were then transferred to the soil and cultivated in a greenhouse with controlled conditions as described above. Homozygous plants were selected at the T3 generation based on the 3:1 segregation ratio of T2 line in the presence of kanamycin.
In order to observe GFP fluorescence, 5-day-old kanamycin resistant seedlings were used under a confocal laser scanning microscope, Olympus FluoView FV10i (Olympus, Tokyo, Japan). Images were recorded and contrast-enhanced using ImageJ software (ImageJ, 1.47v, NIH, Bethesda, USA). Some seedlings were subjected to plasmolysis in 1 M mannitol for 5 min (Xiao et al., 2014). GFP fluorescence was detected using a 488-nm excitation laser and 525/50-nm emission filter.
Leaf samples obtained from different T3 transgenic lines were harvested, frozen in liquid nitrogen, and stored at −80°C until later use.
Semi-quantitative RT-PCR for Detection of Gene Expression Levels
Semi-quantitative RT-PCR to detect RNAs was carried out using total RNA isolated from 8-day-old seedlings. Total RNAs were isolated using the TRIzol reagent kit (Invitrogen) and treated with RNase-free DNase to remove any genomic DNA contaminants. All RNA samples were quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) and were adjusted to the same concentration with diethylpyrocarbonate-treated water. Complementary DNA (cDNA) was synthesized using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara Bio, Inc, Otsu, Japan) following the protocol of the manufacturer using an oligo dT primer and 1 μg of total RNA. PCR was then performed using gene-specific primers and KOD Plus Neo (Toyobo, Osaka, Japan). The reaction was initiated by predenaturating at 94°C for 2 min, followed by 35 cycles of denaturation (98°C for 10 s), annealing (50°C for 30 s), and extension (68°C for 30 s), and was terminated with a final extension of 10 min at 68°C. The amplification products were analyzed by 1.2% agarose gel electrophoresis.
AtActin2, a housekeeping gene of A. thaliana, was used as an internal control to adjust the amount of cDNA template for PCR. Stably expressed AtActin2 (AT3G18780) was used for normalization. MYC2/3/4 and Atactin2 primers are shown in Supplementary Table 1. Gel images were quantified using Gelpro 3.2 software (Media Cybernetics, Inc., Rockville, MD, USA). Three biological replicates were carried out for each sample.
Seed Production Evaluations
In order to determine the seed yield, 30 T3 lines were taken from each genotype. After the siliques matured, the first five incompletely developed siliques were removed from the main inflorescence axis as described before (Jiang et al., 2020). The first intact siliques were then used to assess the number of seeds per silique. The mature siliques were tiled on A4 paper, and the carpel wall was removed with a dissecting needle, photographed with a Leica stereomicroscope (MZ16FA, Leica, Germany) to count the seed number per silique. The remaining siliques were allowed to mature, and seeds from the siliques located on the basis of a major inflorescence were selected for observation. Approximately 2,000 mature seeds from Ctrl and BrMYC2/3-1/3-2/4-1/4-2OE of each line were randomly selected, observed, and photographed using a Leica stereomicroscope. The length and width of seeds were estimated using ImageJ software. Seed weight was measured using an electronic scale. Data points represent the average of 300 biological replicates.
Plant Phase Change Evaluations
To evaluate the phase change among different genotypes, ~30 seedlings were randomly selected from each T3 line, transferred to an autoclaved mixture of grass charcoal soil and vermiculite a 1:1 ratio, and cultivated in the greenhouse for about 2–3 weeks. In order to assess whether the juvenile seedlings have reached the mature stage, abaxial trichomes as a hallmark of vegetative phase change were scored with a Leica stereomicroscope. For leaf shape analysis, fully expanded leaves were removed, attached to a cardboard with double-sided tape, flattened with transparent tape, and then scanned using the Epson V700 Professional scanner (Epson, Suwa, Japan). The bolting time and rosette leaf number at bolting time were recorded to determine the flowering time in A. thaliana (Xing et al., 2018). Plant height and tiller number were determined just after flowering. Pictures were taken from the plant adjacent to a ruler that was used to calibrate the ImageJ software for precise measurement of distance. Maximum height (from the growing point to the highest leaf tip as depicted by the vertical line) was estimated from the silhouette of A. thaliana differing in size. Measurements were obtained from 12 independent biological replicates.
GSs Contents Determination
The leaves of 8-week-old homozygous T3 generation transgenic A. thaliana lines were used for determining GSs contents. Six seedlings were randomly selected from one transgenic line. GSs were essentially quantified as previously described (Zang et al., 2015). High-performance liquid chromatography (HPLC) analysis was performed using an Agilent1200 system (Agilent Technologies, Inc., Santa Clara, USA) with a C18 reverse-phase column (250 mm × 4 mm, 5 μm, Bischoff, Germany). Chromatography was performed over 60 min at a flow rate of 1 ml·min−1 in the following order: 100% H2O (2 min), a linear gradient of 0–20% ACN (32 min), 20% ACN (6 min), followed by 20–100% ACN (5 min), and 0% ACN before injecting the next sample. Eluents were monitored with a UV detector at 229 nm. Three biological and three technical replicates were performed.
Antifungal Activity Bioassay
Lyophilized leaf powder of each transgenic A. thaliana line was used to study the effect of different GSs on the visible growth of S. sclerotiorum. Anti-fungal activity was essentially assayed as previously described with minor modifications (Kelemu et al., 2004). S. sclerotiorum was preserved at 4°C and then reactivated in a Petri dish containing potato dextrose agar (PDA) medium (Becton Dickinson, Columbia, MD). The mycelium was inoculated into the center using a 5 mm puncher; the Petri dishes were then incubated at 22°C for 72 h to provide actively growing mycelium for subsequent experiments. New marginal hyphae were excised with a puncher from the Petri dishes. Filter paper disks of 10 mm in diameter were then placed onto the surface of PDA. Each filter-paper disk received 25 mg lyophilized powder and 100 μl ddH2O, along with 100 μl ddH2O only as a control. None of the three filter paper disks was a PDA Petri dish, and each of the three Petri dishes was a biological repeat. The filter-paper disks carrying lyophilized powder and S. sclerotiorum were incubated at 22°C. Fungal growth was assessed by observing visible mycelium growth and sclerotia number per disk after 72 h of incubation. Three technical replicates were performed.
Statistical Analysis
The data were evaluated using analysis of variance to determine statistical significance and were represented as means ± SD, as calculated by SPSS20.0 analysis software (IBM, Chicago, IL, USA). A difference was considered significant at the 95% confidence level (p < 0.05). All photographs were taken with a Nikon D5300 camera (Nikon Corporation, Tokyo, Japan) and edited in ImageJ. Graphs were plotted using Excel 2019 (Microsoft, Redmond, WA, USA) and figures were assembled using Microsoft PowerPoint.
Results
Structural Analyses of BrMYC2/3/4
Analysis of physicochemical properties showed that BrMYC2 consist of 605 amino acid residues, with a relative molecular mass of 65,848.54 and theoretical isoelectric point (pI) of 5.21, which suggests an acidic protein. Total average hydrophilicity and instability index are −0.548 and 45.84, respectively. Based on these values, BrMYC2 is an unstable hydrophobic and protein. BrMYC3-1 and BrMYC3-2 consist of 563 and 580 amino acid residues, with relative molecular masses of 61,940.76 and 63512.36, theoretical pI of 5.05 and 5.15, respectively. Total average hydrophilicity and instability index of BrMYC3-1 and BrMYC3-2 are −0.653 and −0.683, and 47.76 and 47.46, respectively. All these implicate that both BrMYC3-1 and BrMYC3-2 are unstable hydrophobic and acidic proteins. BrMYC4-1, BrMYC4-2, and BrMYC4-3 have 230, 297, and 60 amino acid residues with their relative molecular masses of 25,793.50, 31,956.93, and 6,509.31, respectively. Their theoretical pIs are 8.39, 4.66, and 5.05, respectively, suggesting that BrMYC4-1 is a basic protein, whereas BrMYC4-2 and BrMYC4-3 are acidic proteins. Total average hydrophilicity and instability index of BrMYC4-1, BrMYC4-2, and BrMYC4-3 are −0.647, −0.511, and −0.013 and 37.35, 46.81, and 39.05, respectively, indicating that BrMYC4-1, BrMYC4-2, and BrMYC4-3 are all hydrophobic and unstable proteins. In addition, the percentage of BrMYC2/3/4 coding sequence identities was 55.40%, the percentage of BrMYC2/3/4 deduced amino acid sequence identities was 47.40%. MYC2/3/4 coding sequence and deduced amino acid sequence in B. rapa share 18.69–81.53% and 5.74–84.82% sequence identity with the A. thaliana orthologs, respectively.
Domain Prediction of BrMYC2/3/4
The protein domains encoded by BrMYC2/3/4 coding sequences were predicted using CDD, an NCBI online domain analysis software. The results showed that BrMYC2, BrMYC3-1, BrMYC3-2, and BrMYC4-2 have a conserved domain named bHLH-MYC_N superfamily; BrMYC2, BrMYC3-1, BrMYC3-2, and BrMYC4-1 have a conserved domain named bHLH_AtABA-inducible bHLH-TYPE_like in the C-terminal region; and BrMYC4-3 contains a single domain named bHLH-MYC_N (Figure 1). A bHLH_AtAIB_like is the bHLH-type domain found in AIB and MYC proteins (MYC2, MYC3, and MYC4) of A. thaliana. AIB is an abscisic acid (ABA)-inducible transcriptional repressor that negatively regulates JA signaling (Nakata et al., 2013). These domain features implicate that BrMYC2/3/4 belongs to the bHLH superfamily of TFs mediating the positive and negative regulation of plant development and various secondary metabolites synthesis.
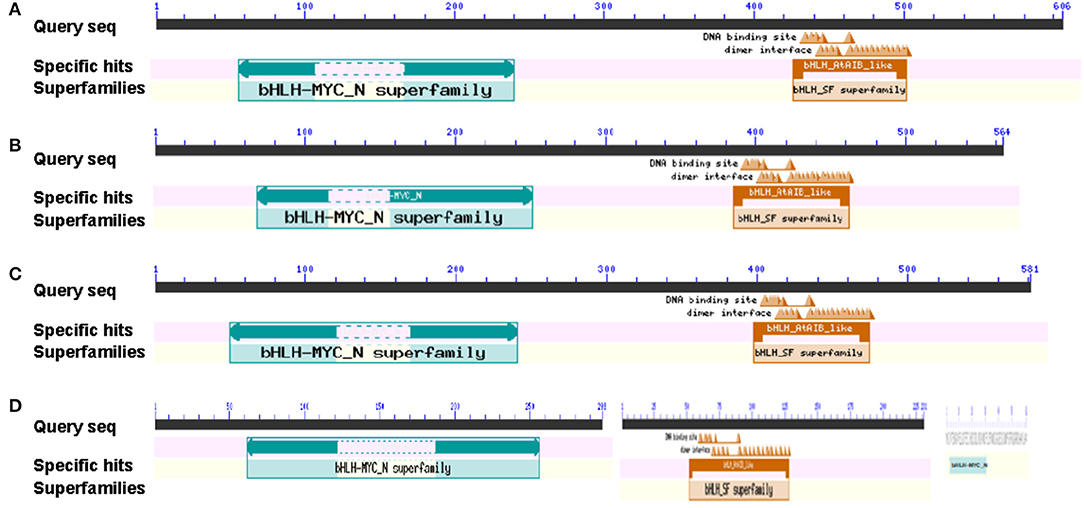
Figure 1. Putative BrMYC2/3/4 DNA-binding domains in Chinese cabbage. (A) BrMYC2 DNA-binding domains. (B) BrMYC3-1 DNA-binding domains. (C) BrMYC3-2 DNA-binding domains. (D) The DNA-binding domains illustrated; from left to right are BrMYC4-2, BrMYC4-1, and BrMYC4-3.
Evolutionary Analyses of BrMYC2/3/4
A cladogram of MYC2/3/4 in the U's triangle species genomes was constructed according to the neighbor-joining method. As shown in Figure 2, Brassicaceae MYC2/3/4 family members exhibited a divergent evolution. BraMYC2 (B. rapa) forms a clade with BnaMYC2 (B. napus), and they form a clade with AthMYC2 (A. thaliana) by a high nodal support value. BraMYC3-1 forms a clade with BnaMYC3-1, BjuMYC3-2 (B. juncea), and BjuMYC3-1. BraMYC3-2 forms a clade with BnaMYC3-2. BraMYC3-1 and BraMYC3-2 make up a clade with AthMYC3 by a high nodal support value. BraMYC4-1, 4-2, and 4-3 share an evolutionary lineage with the middle part, the first half, and a small part of the second half of AthMYC4, respectively. All these phylogenetic relationships are well in accordance with the classification based on morphological and biochemical characteristics of Brassicaceae plants.
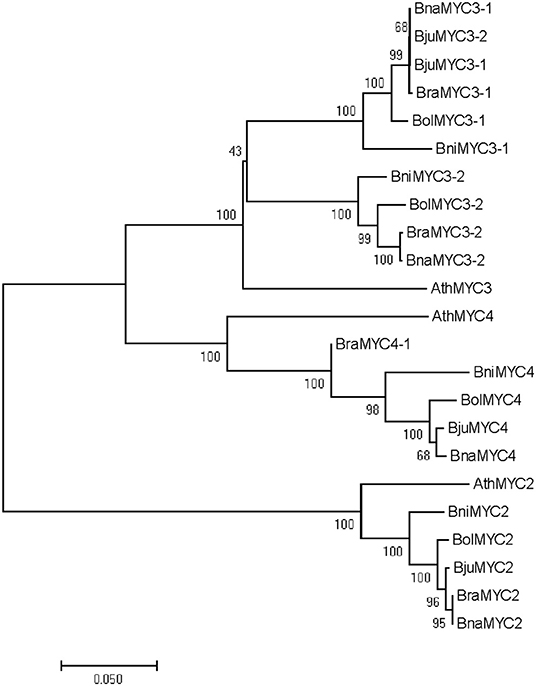
Figure 2. Phylogenetic tree of BrMYC2/3/4. Evolutionary analyses of full-length MYC2/3/4 coding sequences involved in the U's triangle of Brassica species. Numbers on the branches indicate the percentage of bootstrap support from 1,000 replicates; branch length represents the divergence distance. Ath, Arabidopsis thalinala; Bra, Brassica rapa; Bni, Brassica nigra; Bol, Brassica oleracea; Bna, Brassica napus; Bju, Brassica juncea.
Transgenic Plants Obtain and Differential Expression of BrMYC2/3/4 Genes
To determine the obtain of transgenic plants, the root tip of resistant Arabidopsis was observed using spinning disk confocal microscopy. BrMYC2/3/4-GFP fusion protein fluorescence signal was observed under the control of CaMV 35S promoter (Figure 3).
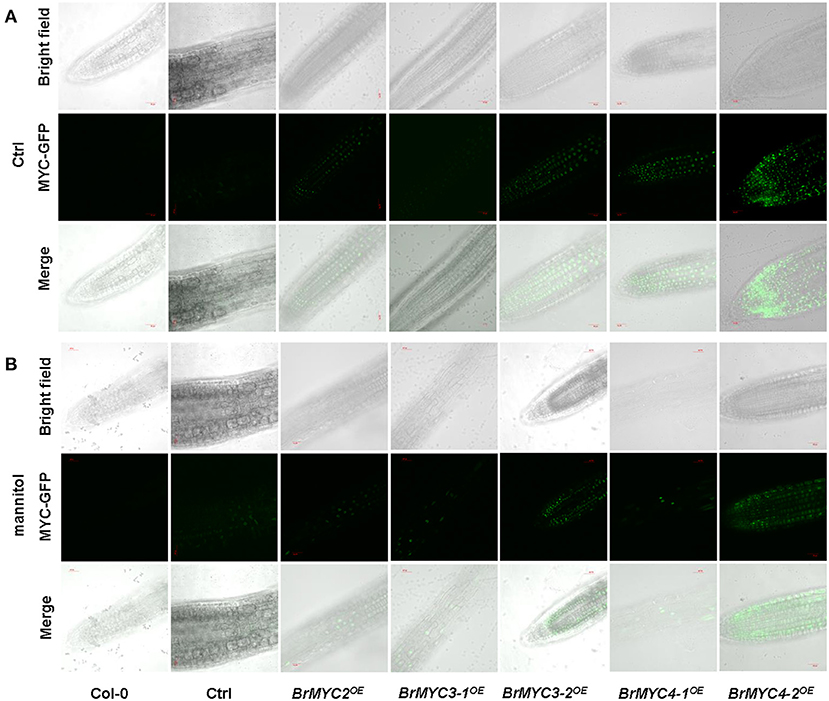
Figure 3. Green fluorescent protein (GFP) in the root tip of transgenic Arabidopsis, BrMYC2/3/4-GFP localizes to the nucleus. (A,B) Confocal images show the presence of BrMYC2/3/4-GFP fluorescence at the edges of root cells of 5-day-old transgenic Arabidopsis seedlings without (A) or with (B) plasmolysis induced by 1 M mannitol treatment for 5 min. Top panels, bright-field images; middle panels, fluorescence images of MYC-GFP; bottom panels, merged bright-field and fluorescence images. Bar = 30 μm.
Semi-quantitative RT-PCR was used to estimate the expression levels of five BrMYC2/3/4 homologous genes in BrMYCOE transgenic lines relative to Ctrl. Primers were designed from the gene-specific region (Supplementary Table 1). In the randomly selected three BrMYC2/3/4OE transgenic lines, BrMYC2/3/4 expression levels were all elevated (Supplementary Table 2). In BrMYC2OE, BrMYC2 expression levels elevated from 8.61 to 11.07 folds relative to Ctrl. In BrMYC3OE, BrMYC3-1 expression levels elevated from 5.72 to 7.50 folds, and BrMYC3-2 expression levels elevated from 2.97 to 4.43 folds. In BrMYC4OE, BrMYC4-1 expression levels elevated from 9.32 to 10.49 folds, and BrMYC4-2 expression levels elevated from 11.32 to 11.43 folds.
Effects of BrMYC2/3/4 Overexpression on Seed Production
Since BrMYC4-3 was only 63 bp and its predicted domain function was completely different from others, only BrMYC2/3-1/3-2/4-1/4-2 were cloned and transformed for further expression analysis. Transgenic Arabidopsis plants of Ctrl and BrMYC2/3-1/3-2/4-1/4-2OE were analyzed for seed yield in terms of seed size (length and width), 1,000 seed weight, and seed number per silique. In order to minimize the effects of environmental factors on seed development, all plants were kept under identical growing conditions, such as temperature, light, water, and nutrition. As shown in Figure 4, there was a significant difference in seed production among different transgenic plants. BrMYC2OE plant had the smallest seed length; there was no significant difference in seed length among BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE, all of which were significantly smaller than that of Ctrl (Figure 4C). BrMYC3-2OE and BrMYC4-1OE plants possessed the smallest seed width; there was no significant difference in seed width between BrMYC2OE and BrMYC4-2OE, as well as between BrMYC3-1OE and Ctrl (Figure 4D). Thousand seed weight significantly decreased in all the transgenic lines as compared with Ctrl (Figure 4E). BrMYC2OE line contained the smallest seed weight, followed by BrMYC3-2OE, BrMYC4-1OE, BrMYC4-2OE, and BrMYC3-1OE. Thousand seed weight of BrMYC3-1OE increased more than those of BrMYC3-2OE and BrMYC4-1OE, while there was no significant difference among those of BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE, as well as between BrMYC4-2OE and BrMYC3-1OE. BrMYC2OE line contained the lowest seed number per silique, while all other transgenic lines had significantly higher seed numbers than Ctrl (Figures 4B,F). BrMYC4-1OE line produced the highest seed number per silique followed by BrMYC3-2OE, BrMYC3-1OE, and BrMYC4-2OE. Taken the results together, constitutive expression of BrMYC2/3/4 TFs differentially affected the seed production with respect to the size, weight, and number of seed. Overexpression of BrMYC3-1/3-2/4-1/4-2 had a negative effect on both the seed length and seed weight and a positive effect on seed number per silique. In contrast, overexpression of BrMYC2 led to a negative effect on the seed length, seed weight, and seed number per silique.
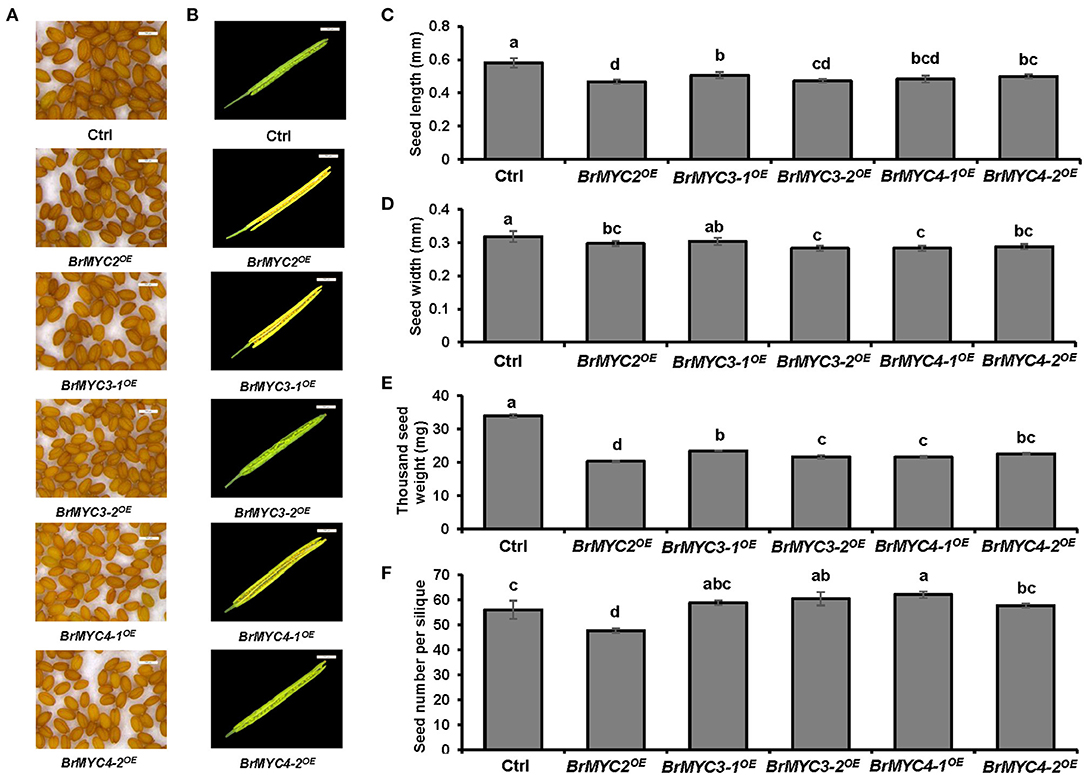
Figure 4. Seed production of T3 Arabidopsis lines transgenic for BrMYC2/3/4 genes. (A) Phenotype of the mature seed. (B) Phenotype of decapsulated mature siliques. (C) Comparison of the seed length. (D) Comparison of the seed width. (E) Comparison of 1,000 seed weight. (F) Comparison of seed number per silique. Tested transgenic lines are Ctrl, BrMYC2OE, BrMYC3-1OE, BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE. Seeds and siliques were randomly selected and pictures were taken under the same conditions. Data are shown as mean ± SD from three independent biological replicates. ANOVA was used for statistical analysis, followed by Tukey's multiple comparison test (p < 0.05). Bar = 500 μm.
Effects of BrMYC2/3/4 Overexpression on Plant Development
To determine if BrMYC2/3/4 expression influences vegetative and reproductive development, we examined the phenotypes of transgenic BrMYC2/3/4 plants at the seedling and bolting stage.
As for the vegetative stage, we studied the length of root and hypocotyl of seedlings. Significant differences in root and hypocotyl length were noted between Ctrl and BrMYC2/3-1/3-2/4-1/4-2OE lines (Figure 5). BrMYC2OE line exhibited the shortest root length, followed by BrMYC3-1OE, BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE (Figure 5B). Root length of BrMYC2OE and BrMYC3-1OE lines was shorter than that of Ctrl. Although there was no significant difference among BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE lines, their root length was significantly longer than that of Ctrl. BrMYC2OE had also the shortest length of hypocotyl, followed by BrMYC3-1OE, BrMYC4-2OE, BrMYC4-1OE, and BrMYC3-2OE (Figure 5C). BrMYC2OE line showed a shorter hypocotyl length than Ctrl, whereas transgenic BrMYC3-1OE, BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE lines exhibited longer hypocotyl lengths than Ctrl. Thus, overexpression of BrMYC2/3-1 and BrMYC3-2/4-1/4-2 appears to inhibit and promote root elongation in Arabidopsis, respectively. Overexpression of BrMYC2 and BrMYC3-1/3-2/4-1/4-2 seems to inhibit and foster hypocotyl elongation of Arabidopsis, respectively.
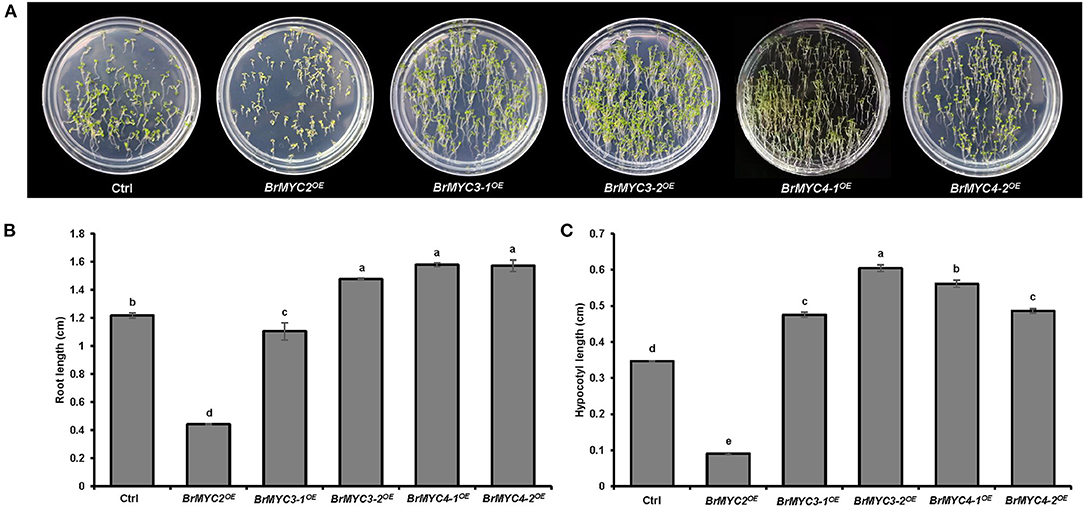
Figure 5. Root and hypocotyl length of T3 Arabidopsis lines transgenic for BrMYC2/3/4 genes. (A) Phenotype of 6-day old seedlings. (B) Comparison of the root length. (C) Comparison of the hypocotyl length. Transgenic lines are Ctrl, BrMYC2OE, BrMYC3-1OE, BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE. Data are shown as mean ± SD from three independent biological replicates. ANOVA was used for statistical analysis, followed by Tukey's multiple comparison test (p < 0.05). Bar = 1 cm.
About 3–4 weeks after seeds were sown, Arabidopsis seedlings transitioned from juvenile to adult vegetative growth phase (Figure 6). The leaf shape of transgenic plants was normal and indistinguishable from that of Ctrl. BrMYC2/3-1/3-2/4-1OE lines produced smaller number of the first leaves having trichomes than Ctrl, whereas BrMYC4-2OE produced a larger number of them on leaf than Ctrl (Figure 6B). For the leaf initiation rate, BrMYC2/3-1/4-1/4-2OE was significantly faster than Ctrl, but BrMYC3-2OE was similar to Ctrl (Figure 6C).
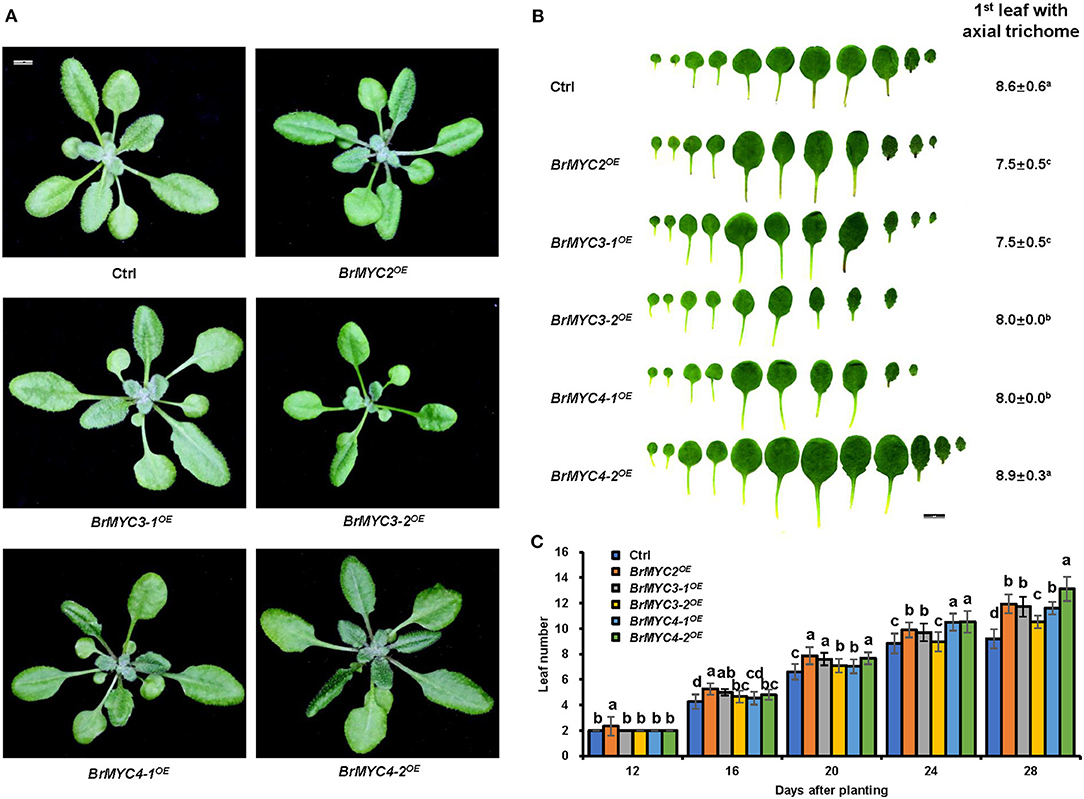
Figure 6. The effect of BrMYC2/3/4 expression on the growth of Arabidopsis. (A) Phenotype of 28-day old transgenic Arabidopsis. (B) Phenotype of leaf shape and abaxial trichome of fully expanded rosette leaves. The average numbers of the first leaf showing abaxial trichomes are presented. Different superscript letters on the numbers denote a significant difference between the transgenic lines. (C) Leaf initiation rate of transgenic Arabidopsis in short days. Leaf numbers were scored at 12, 16, 20, 24, and 28 days after planting. Transgenic lines are Ctrl, BrMYC2OE, BrMYC3-1OE, BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE. Data are shown as mean ± SD from three independent biological replicates. ANOVA was used for statistical analysis, followed by Tukey's multiple comparison test (p < 0.05). Bar = 1 cm.
As for the reproductive stage, we studied bolting time, rosette leaf number at the bolting time, plant height, and tiller number (Figure 7). BrMYC2OE line had significantly faster bolting time, while BrMYC3-2OE and BrMYC4-2OE lines had significantly slower bolting time than Ctrl. Both BrMYC3-1OE and BrMYC4-1OE were similar to Ctrl in bolting time (Figures 7A,B). The number of rosette leaves at bolting time significantly decreased in BrMYC2/3-1/4-1OE lines but increased in BrMYC3-2/4-2OE lines as compared with Ctrl (Figures 7A,C). Previously, flowering time was recorded as the number of rosette leaves at bolting, with the observation of late flowering plants having more rosette leaves (Lopez-Vernaza et al., 2012). Thus, it appears that BrMYC2OE, BrMYC3-1OE, and BrMYC4-1OE promoted flowering time, while BrMYC3-2OE and BrMYC4-2OE inhibited it in Arabidopsis. For the plant height, BrMYC2OE significantly increased, while BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE significantly decreased as compared with Ctrl. And there was no significant difference between BrMYC3-2OE and BrMYC4-2OE, as well as between BrMYC3-1OE and Ctrl (Figures 7A,E). Tiller number of BrMYC2OE and BrMYC3-1OE was significantly higher than Ctrl and no significant difference was noted among BrMYC3-2OE, BrMYC4-1OE, BrMYC4-2OE, and Ctrl (Figures 7A,F). Thus, expression of BrMYC2 only promoted the growth of both shoot elongation and plant height after fluorescence with faster bolting time.
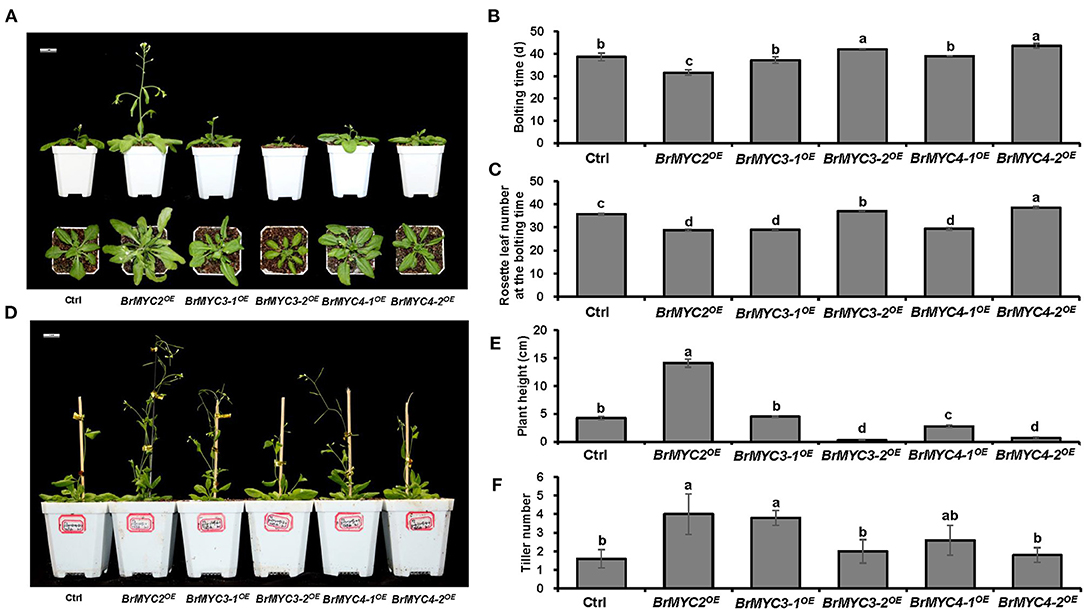
Figure 7. Plant height and flowering time of transgenic Arabidopsis. (A) Phenotype of 3-week-old Arabidopsis. (B) Bolting time. (C) Rosette leaf number at the bolting time. (D) Phenotype of 7-week-old Arabidopsis. (E) Plant height. (F) Tiller number. Transgenic lines are Ctrl, BrMYC2OE, BrMYC3-1OE, BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE. Data are shown as mean ± SD from three independent biological replicates. ANOVA was used for statistical analysis, followed by Tukey's multiple comparison test (p < 0.05). Bar = 1 cm.
Analysis of GSs Contents
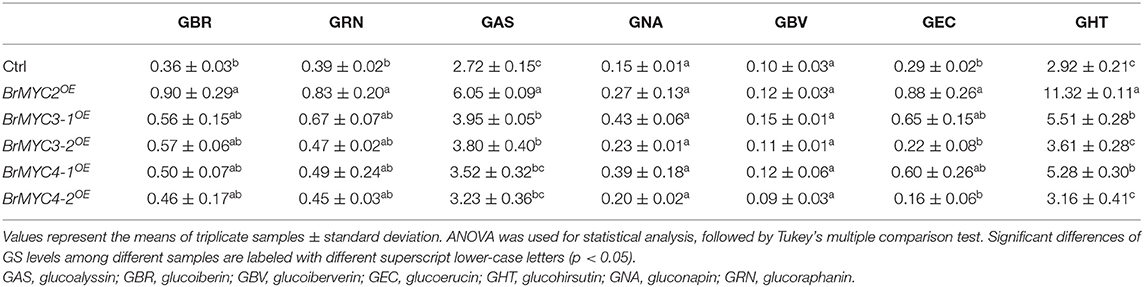
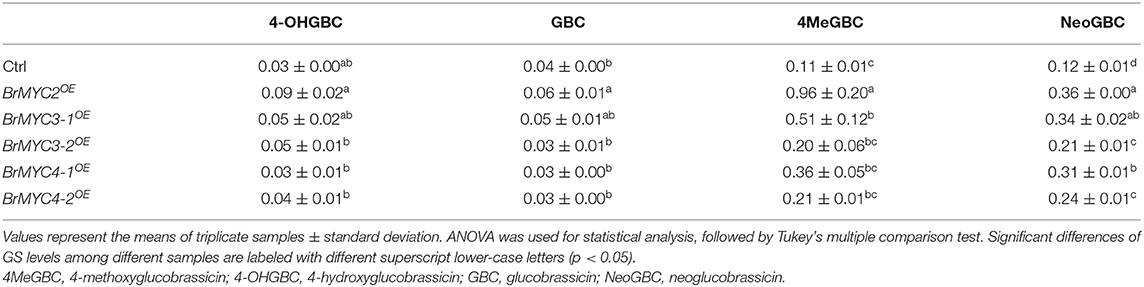
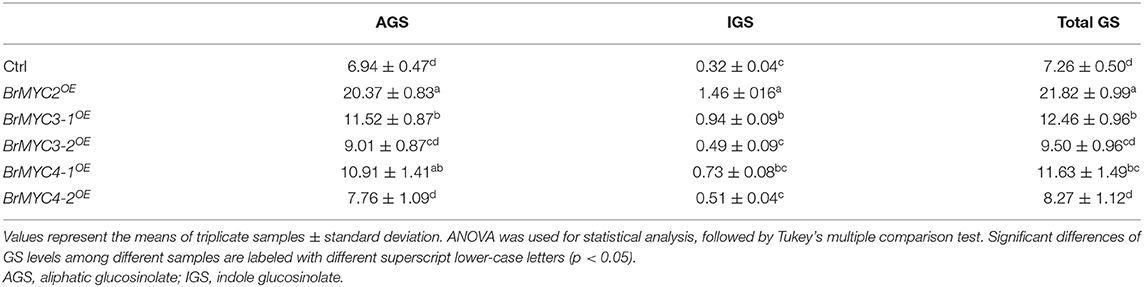
In order to investigate the effect of BrMYC2/3/4 transgene expression on GSs metabolism, GSs profiles in the leaves of T3 lines were determined using HPLC. Transgenic lines exhibited significantly increased levels of most of the short- and long-chain AGSs and IGSs when compared to those of Ctrl (Tables 1, 2). Glucoiberin (GBR), derived from homomethionine, increased by 2.5-fold in BrMYC2OE line. AGS derived from dihomomethionine, such as glucoerucin (GEC) and glucoraphanin (GRN), increased by 3.1- and 2.2-fold in BrMYC2OE line, respectively. Glucoalyssin (GAS), derived from trihomomethionine, increased by 2.2-fold in BrMYC2OE line. Glucohirsutin (GHT), derived from pentahomomethionine, increased by 3.9-fold in BrMYC2OE, 1.9-fold in BrMYC3-1OE, and 1.8-fold in BrMYC4-1OE lines. The major IGS such as glucobrassicin (GBC), 4MeGBC, and neoglucobrassicin (NeoGBC) increased by 1.6-, 8.9- and 1.2-fold in BrMYC2OE line, respectively. 4MeGBC increased by 2.9-fold in BrMYC3-1OE line. NeoGBC increased by 4.7-, 1.9-, 3.3-, and 1.9-fold in BrMYC3-1/3-2/4-1/4-2OE lines, respectively. As shown in Table 3, AGS increased by 2.9-, 1.7-, and 1.6-fold in BrMYC2/3-1/4-1OE lines, respectively. IGS increased by 4.5- and 2.9-fold in BrMYC2/3-1OE lines, respectively. Total GS increased by 3.0-, 1.7-, and 1.6-fold in BrMYC2/3-1/4-1OE lines, respectively. Hence, levels of most AGS and IGS significantly increased in all the lines except for BrMYC4-2OE.
Anti-fungal Activity
To study the extent of resistance of transgenic lines to S. sclerotiorum, 25 mg of the lyophilized powder of each transgenic line's leaf tissue was uniformly sprinkled around the hyphae. The visible growth of cottony mycelium of plaque was observed after 72 h of S. sclerotiorum incubation (Figure 8). The thinnest plaque was noted in BrMYC2OE line, followed by BrMYC3-1OE, BrMYC4-1OE, BrMYC3-2OE, and BrMYC4-2OE. Ctrl displayed much thinner plaque than water control, suggesting that endogenous basal levels of GSs present in the vector control line inhibited the growth of S. sclerotiorum to a certain degree. However, Ctrl showed thicker plaques than BrMYC2/3/4OE lines. This may result from the suppression of the mycelium growth by higher contents of GSs accumulated in BrMYC2/3/4OE lines.
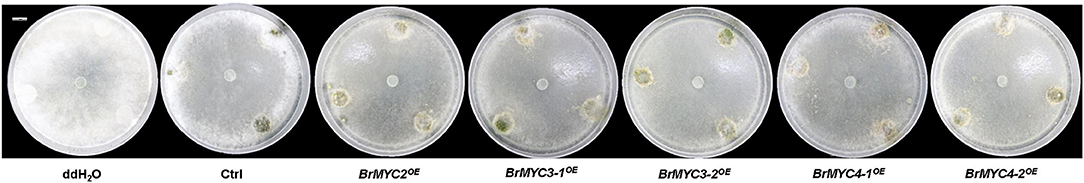
Figure 8. Inhibition of S. sclerotiorum growth by the lyophilized powder of each transgenic A. thaliana line on PDA. From left to right are: ddH2O, Ctrl, BrMYC2OE, BrMYC3-1OE, BrMYC3-2OE, BrMYC4-1OE, and BrMYC4-2OE. One sclerotium of S. sclerotiorum was placed in the center of each dish. Fungal hyphae growth was visible as white cottony mycelium plaque. Bar = 1 cm.
Discussion
Our in-silico analysis indicated that BrMYC2/3-1/3-2/4-2/4-3 are unstable hydrophobic and acidic proteins, whereas BrMYC4-1 is an unstable hydrophobic and acidic basic protein. BrMYC2/3/4 TFs were predicted to contain the bHLH AtAIB-like domain whose protein is involved in the positive regulation of ABA signaling in Arabidopsis (Figure 1). Notably, the molecular weight of BrMYC4-3 was far smaller and its domain function was very different from others. Phylogenetic relationship analyses indicated that BrMYC2/3/4 were closely related to AthMYC2/3/4 orthologs from the Brassicaceae family.
Transgenic Arabidopsis plants of BrMYC2/3/4 were verified by observation of GFP fluorescence (Figure 3). Expression levels of five BrMYC2/3/4 homologous genes, all elevated relative to Ctrl (Figure 9).
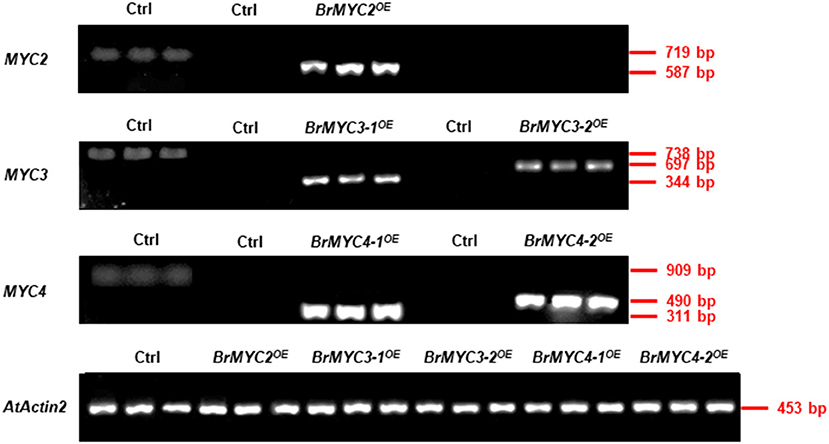
Figure 9. Semi-quantitative RT-PCR analysis of BrMYC2/3/4 genes in 8-day-old transgenic Arabidopsis. The left column of Ctrl with three lanes used primers from corresponding Arabidopsis genes, and the other's Ctrl used primers from corresponding Brassica rapa genes. AtActin2 serves as an internal control.
When expressed in Arabidopsis under the control of 35S promoter, each member of the BrMYC2/3-1/3-2/4-1/4-2 transgenes differentially influenced root and shoot elongation, vegetative phase change, flowering time, plant height and tiller number after fluorescence, and seed production. Seed size is a key agronomic trait that determines the grain yield and breeding of plants (Shirley et al., 2019). The number and weight of the seed ultimately determine seed yield (Jiang et al., 2020). In this study, we found that as a result of overexpression of each of BrMYC2/3-1/3-2/4-1/4-2, both the seed length and seed weight decreased (Figures 4C,E), while seed number per silique increased except for BrMYC2OE as compared to Ctrl (Figure 4F). The results appear to be consistent with a finding that MYC2, MYC3, and MYC4 act additively during seed development and that triple mutants produced the largest seeds with more seed storage proteins, while the seeds of single and double mutants were much larger than those of wild type (Gao et al., 2016). Hence, overexpression of the individual BrMYC2/3-1/3-2/4-1/4-2 family genes appears to have a negative effect on seed size and seed weight.
To examine the effect of BrMYC expression on plant development, the lengths of root and hypocotyl of seedlings were measured. We found that BrMYC2 expression significantly inhibited both root and hypocotyl elongation, whereas BrMYC3-2/4-1/4-2 expression significantly promoted both root and hypocotyl elongation when compared to Ctrl (Figure 5). In contrast, BrMYC3-1 expression significantly inhibited root elongation but promoted hypocotyl elongation. Thus, expression of the duplicated copies of BrMYC3, BrMYC3-1, and BrMYC3-2, did not lead to the same effect on the root length but on the hypocotyl length. This is an interesting observation, suggesting that they have divergent functions and regulatory roles in plant development. In A. thaliana, the transition from juvenile to adult vegetative phase referred to as “vegetative phase change” is signified by the formation of trichomes on the abaxial side of leaf blade and an increase in the leaf length/width ratio during shoot development (Xu et al., 2019). Our studies on abaxial trichome production and leaf initiation rate as shown in Figure 6 suggested that BrMYC2/3-1/4-1OE positively regulate the time of vegetative phase change in Arabidopsis, since the less number of the first-formed leaves having abaxial trichomes relative to control was correlated with promoting floral transition (Chien and Sussex, 1996). Previous studies showed that the plant height determines its ability to compete for light and therefore often correlates with the leaf mass and seed production, and that stem growth is initiated once the plant becomes reproductive and continues until termination of the inflorescence meristems (Serrano-Mislata et al., 2017; Moles et al., 2019). Based on the bolting time and rosette leaf number at bolting time, it appears that BrMYC2/3-1/4-1OE promoted flowering time, while BrMYC3-2/4-2OE inhibited it (Figure 7). Hence, expression of each set of the duplicated copies of BrMYC3 (3-1/3-2) and BrMYC4 (4-1/4-2) led to the opposite action on flowering time, suggesting that they have divergent function and regulatory roles in fluorescence. After flowering, BrMYC2OE promoted the growth of the plant height, whereas BrMYC3-2/4-1/4-2OE repressed it, suggesting that the three MYC TFs play different roles in signaling pathways. Notably, BrMYC2/3-1OE displayed a remarkable increase in tiller number after fluorescence. The earlier flowering phenotype of overexpressed BrMYC2 seems to be consistent with a previous finding that expression of all three MYC2/3/4 was required to inhibit flowering in Arabidopsis (Wang et al., 2017). Along this line, it was reported that MYC2 regulates diverse functions within the JA signaling pathway and that MYC2 forms homo- and/or heterodimers with MYC3 and MYC4 and binds to the conserved G-box present in the promoters of JA-responsive genes (Kazan and Manners, 2013). Moreover, Zhai et al. (2013) reported that high and low accumulation of the MYC2 protein correlated with positive regulation of early wound-responsive genes and negative regulation of late pathogen-responsive genes, respectively, and that MYC activity was further regulated by phosphorylation and the ubiquitin-proteasome system-mediated proteolysis. Accordingly, a wide range of differential effects of each BrMYC2/3/4 expression on the root and shoot elongation, vegetative phase change, flowering time, and seed production implicates MYC-mediated complex signaling networks for positive and negative regulation of plant growth and development to connect environmental stresses with developmental signals at both transcriptional and post-transcription levels.
It was reported that bHLH-type MYC TFs, participating in crosstalk among JA and other hormones as well as environmental signals, allow the coordinated and rapid regulation of GS genes (Mitreiter and Gigolashvili, 2021). MYC-interaction motif mediates interactions between MYC and MYB TFs in A. thaliana that are critical for constitutive and induced GS biosynthesis (Millard et al., 2021). MYC3/4 act additively with MYC2 to stimulate GS biosynthesis (Guo et al., 2013). In Arabidopsis, MYC2 binds directly to the promoters of several GS biosynthesis genes, and that MYC2/3/4 interact directly with GS-related MYBs (Schweizer et al., 2013). Therefore, MYC2/3/4 are involved in the regulation of GS biosynthesis. In line with these findings, our study showed that transgenic Arabidopsis plants overexpressing BrMYC2/3-13-2//4-1 accumulated significantly higher levels of GS relative to Ctrl (Tables 1–3). Levels of AGS remarkably increased in BrMYC2/3-1/4-1OE lines. GHT content also significantly rose in BrMYC3-1OE and BrMYC4-1OE lines. Levels of IGS were strikingly elevated in BrMYC2/3-1OE lines. NeoGBC content significantly increased in BrMYC2/3-1/3-2/4-1/4-2OE lines. Overall, total GS considerably increased in BrMYC2/3-1/3-2/4-1OE lines. BrMYC2 was far more effective to positively regulate the biosynthesis of both AGS and IGC than BrMYC3 and BrMYC4.
Modulation of GS content affected plant resistance to pathogenic infection in Arabidopsis (Madloo et al., 2019). Brassicaceae plants typically produce GSs which are hydrolyzed by myrosinases upon tissue damage to generate ITCs, toxic bioactive compounds acting against broad pathogens. 4MeGBC was activated by the atypical PEN2 myrosinase (a type of β-thioglucoside glucohydrolase) for antifungal defense (Bednarek et al., 2009). Fungal infection triggered the accumulation of both IGS and AGS in B. rapa (Abdel-Farid et al., 2010). In B. rapa, S. sclerotiorum inoculation in vivo induced GS accumulation and increased GS biosynthesis-related proteins (Teng et al., 2021). In our study, the lyophilized leaf powder of each Arabidopsis transgenic line was uniformly sprinkled around the hyphae in order to assess anti-fungal activity against S. sclerotiorum (Figure 8). PDA plate sprinkled with lyophilized powder of BrMYC2OE line showed the thinnest fungal hyphae plaque, followed by those of BrMYC3-1OE and BrMYC4-1OE lines. Such a varying degree of fungal resistance exhibited by the overexpression lines seemed to be in line with the accumulation levels of GS. IGSs such as GBC, 4MeGBC, and NeoGBC, as well as GHT accumulated at a higher level in the order of BrMYC2OE, BrMYC3-1OE, and BrMYC3-1OE lines (Table 2). GBC, 4MeGBC, and NeoGBC were not metabolized by S. sclerotiorum (Pedras and Hossain, 2011). Besides, a previous study showed that aliphatic ITCs inhibited the growth of S. sclerotiorum in vitro, and that anti-microbial activity of AGS-derived isothiocyanates was dependent on side chain elongation and modification, with glucohirsutin (GHT) being most toxic to S. sclerotiorum (Stotz et al., 2011).
Conclusions
In summary, Arabidopsis plants expressing each of the five MYC paralogous genes of Chinese cabbage exhibited a wide range of differing phenotypes with respect to the root and shoot elongation, vegetative phase change, flowering time, plant height and tiller number right after flowering, and seed production. Despite the wide variation of phenotypes between the transgenic lines, all of the lines except for BrMYC4-2OE exhibited shorter seed length, less seed weight, higher accumulation of GSs, and resistance to S. sclerotiorum than Ctrl. Notably, the highest GSs level accumulated in BrMYC2OE line was correlated with the highest extent of resistance to the necrotic fungal pathogen S. sclerotiorum. Unlike BrMYC3-1/3-2/4-/4-2, BrMYC2 expression stimulated the growth of plant height after fluorescence with a faster bolting time. The results presented here indicate that despite the complexity of GSs biosynthesis and metabolism regulated in the positive and negative manner by many other TFs, BrMYC2 expression alone was far more effective to positively regulate the biosynthesis of both AGS and IGC than BrMYC3 and BrMYC4, and thus may provide the beneficial effects on the plant growth and development via resistance to fungal pathogens.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
YZ conceived and designed the experiments and performed the analysis with discussions. ZT performed the experiment and data analysis. ZT, YY, and ZZ contributed to manuscript preparation. ZT and YZ wrote the original draft. S-BH, WZ, and ZZ edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was partially supported by the Zhejiang Provincial Natural Science Foundation of China (LY20C150001) and the National Natural Science Foundation of China (31572130).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.707054/full#supplementary-material
Abbreviations
4MeGBC, 4-methoxyglucobrassicin, 4-methoxy-3-indolylmethyl-glucosinolate; 4-OHGBC, 4-hydroxyglucobrassicin, 4-hydroxy-3-indolylmethyl-glucosinolate; ABA, abscisic acid; AGSs, aliphatic glucosinolates; AIB, ABA-inducible bHLH-TYPE; Ath, Arabidopsis thaliana; bHLH, basic helix–loop–helix; Bju, Brassica juncea; Bna, Brassica napus; Bni, Brassica nigra; Bol, Brassica oleracea; Bra, Brassica rapa; BrMYC2/3/4OE, transgenic Arabidopsis plants carrying BrMYC2/3/4 genes; CDD, conserved domain database; cDNA, complementary DNA; COI1, coronatine insensitive1; Ctrl, transgenic Arabidopsis plants carrying an empty vector pCAMBIA2302; ddH2O, double-distilled water; GAS, glucoalyssin, 5-methylsulphinylpentyl-glucosinolate; GBC, glucobrassicin, 3-indolylmethyl-glucosinolate; GBR, glucoiberin, 3-methylsulphinylpropyl-glucosinolate; GBV, glucoiberverin, 3-methylthiopropyl-glucosinolate; GEC, glucoerucin, 4-methylthiobutyl-glucosinolate; GFP, green fluorescent protein; GHT, glucohirsutin, 8-methylsulphinyloctyl-glucosinolate; GNA, gluconapin, 3-butenyl-glucosinolate; GRN, glucoraphanin, 4-methylsulphinylbutyl-glucosinolate; GSs, glucosinolates; HLH, helix–loop–helix; I3C, indole-3-carbinol; IGSs, indole glucosinolates; ITCs, isothiocyanates; JA, jasmonic acid; Jas, jasmonates; JAZs, JAZIM-domains; MEGA7 software, molecular evolutionary genetics analysis software version 7. 0; MYB, v-myb avian myeloblastosis viral oncogene homolog; MYCs, myelocytomatosis proteins; NCBI, National Center for Biotechnology Information; NeoGBC, neoglucobrassicin, 1-methoxy-3-indolylmethyl-glucosinolate; OE, overexpression; PDA, potato dextrose agar; pI, isoelectric point; TFs, transcription factors.
References
Abdel-Farid, I. B., Jahangir, M., Mustafa, N. R., Van Dam, N. M., Van den Hondel, C. A., Kim, H. K., et al. (2010). Glucosinolate profiling of Brassica rapa cultivars after infection by Leptosphaeria maculans and Fusarium oxysporum. Biochem. Syst. Ecol. 38, 612–620. doi: 10.1016/j.bse.2010.07.008
Arumugam, A., and Razis, A. F. A. (2018). Apoptosis as a mechanism of the cancer chemopreventive activity of glucosinolates: a review. Asian Pac. J. Cancer Prev. 19:1439.
Bednarek, P., Piślewska-Bednarek, M., Svatoš, A., Schneider, B., Doubský, J., Mansurova, M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106. doi: 10.1126/science.1163732
BlaŽević, I., Ðulović, A., Maravić, A., Cikeš Culić, V., Montaut, S., and Rollin, P. (2019). Antimicrobial and cytotoxic activities of Lepidium latifolium L. Hydrodistillate, extract and its major sulfur volatile allyl isothiocyanate. Chem. Biodivers. 16:e1800661. doi: 10.1002/cbdv.201800661
BlaŽević, I., Radonić, A., Skočibušić, M., De Nicola, G. R., Montaut, S., Iori, R., et al. (2011). Glucosinolate profiling and antimicrobial screening of Aurinia leucadea (Brassicaceae). Chem. Biodivers. 8, 2310–2321. doi: 10.1002/cbdv.201100169
Burow, M., Atwell, S., Francisco, M., Kerwin, R. E., Halkier, B. A., and Kliebenstein, D. J. (2015). The glucosinolate biosynthetic gene AOP2 mediates feed-back regulation of jasmonic acid signaling in Arabidopsis. Mol. Plant. 8, 1201–12012. doi: 10.1016/j.molp.2015.03.001
Cao, J., Li, M., Chen, J., Liu, P., and Li, Z. (2016). Effects of MeJA on Arabidopsis metabolome under endogenous JA deficiency. Sci. Rep. 6, 1–13. doi: 10.1038/srep37674
Chen, J., Ullah, C., Reichelt, M., Beran, F., Yang, Z. L., Gershenzon, J., et al. (2020). The phytopathogenic fungus Sclerotinia sclerotiorum detoxifies plant glucosinolate hydrolysis products via an isothiocyanate hydrolase. Nat. Commun. 11, 1–12. doi: 10.1038/s41467-020-16921-2
Chen, L., Zhang, L., Xiang, S., Chen, Y., Zhang, H., and Yu, D. (2021). The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 72, 1473–1489. doi: 10.1093/jxb/eraa529
Chien, J. C., and Sussex, I. M. (1996). Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiods in Arabidopsis thalina (L.) Heynh. Plant Physiol. 111, 1321–1328. doi: 10.1104/pp.111.4.1321
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101. doi: 10.1126/science.1164627
Coleto, I., Bejarano, I., Marín-Peña, A. J., Medina, J., Rioja, C., Burow, M., et al. (2021). Arabidopsis thaliana transcription factors MYB28 and MYB29 shape ammonium stress responses by regulating Fe homeostasis. New Phytol. 229, 1021–1035. doi: 10.1111/nph.16918
Costarelli, A., Bianchet, C., Ederli, L., Salerno, G., Piersanti, S., Rebora, M., et al. (2020). Salicylic acid induced by herbivore feeding antagonizes jasmonic acid mediated plant defenses against insect attack. Plant Signal. Behav. 15:1704517. doi: 10.1080/15592324.2019.1704517
del Rosario Cappellari, L., Santoro, M. V., Schmidt, A., Gershenzon, J., and Banchio, E. (2019). Induction of essential oil production in Mentha x piperita by plant growth promoting bacteria was correlated with an increase in jasmonate and salicylate levels and a higher density of glandular trichomes. Plant Physiol. Bioch. 141, 142–153. doi: 10.1016/j.plaphy.2019.05.030
Döring, A., and Ulber, B. (2020). Performance of cabbage stem flea beetle larvae (Psylliodes chrysocephala) in brassicaceous plants and the effect of glucosinolate profiles. Entomol. Exp. Appl. 168, 200–208. doi: 10.1111/eea.12891
Elateek, S. Y., Abdel-Razik, A. B., and Ibrahim, S. (2020). Initial clustering of some Egyptian Nematicides Serratia isolates based on their 16SrRNAs gene sequences. J. Genet. Genomics. 49.
Frerigmann, H., Berger, B., and Gigolashvili, T. (2014). bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 166, 349–369. doi: 10.1104/pp.114.240887
Frerigmann, H., and Gigolashvili, T. (2014). MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant. 7, 814–828. doi: 10.1093/mp/ssu004
Gao, C., Qi, S., Liu, K., Li, D., Jin, C., Li, Z., et al. (2016). MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis. Plant Physiol. Bioch. 108, 63–70. doi: 10.1016/j.plaphy.2016.07.004
Garrido-Bigotes, A., Figueroa, N. E., Figueroa, P. M., and Figueroa, C. R. (2018). Jasmonate signalling pathway in strawberry: genome-wide identification, molecular characterization and expression of JAZ s and MYC s during fruit development and ripening. PLoS ONE 13:e0197118. doi: 10.1371/journal.pone.0197118
Guo, J., Chen, Y. Z., Li, M. S., Shi, L., and Yan, X. F. (2013). Does MYC2 really play a negative role in jasmonic acid-induced indolic glucosinolate biosynthesis in Arabidopsis thaliana? Russ. J. Plant Physl. 60, 100–107. doi: 10.1134/S1021443713010020
Ji, R., Lei, J., Chen, I. W., Sang, W., Yang, S., Fang, J., et al. (2021). Cytochrome P450s CYP380C6 and CYP380C9 in green peach aphid facilitate its adaptation to indole glucosinolate–mediated plant defense. Pest Manag. Sci. 77, 148–158. doi: 10.1002/ps.6002
Jiang, H. L., Hong, J., Jiang, Y. T., Yu, S. X., Zhang, Y. J., Shi, J. X., et al. (2020). Genome-wide association analysis identifies candidate genes regulating seed number per silique in Arabidopsis thaliana. Plants 9:585. doi: 10.3390/plants9050585
Kai, K., Takahashi, H., Saga, H., Ogawa, T., Kanaya, S., and Ohta, D. (2011). Metabolomic characterization of the possible involvement of a Cytochrome P450, CYP81F4, in the biosynthesis of indolic glucosinolate in Arabidopsis. Plant Biotechnol. 28, 379–385. doi: 10.5511/plantbiotechnology.11.0704b
Katz, E., and Chamovitz, D. A. (2017). Wounding of Arabidopsis leaves induces indole-3-carbinol-dependent autophagy in roots of Arabidopsis thaliana. Plant J. 91, 779–787. doi: 10.1111/tpj.13610
Kazan, K., and Manners, J. M. (2013). MYC2: the master in action. Mol. Plant. 6, 686–703. doi: 10.1093/mp/sss128
Kelemu, S., Cardona, C., and Segura, G. (2004). Antimicrobial and insecticidal protein isolated from seeds of Clitoria ternatea, a tropical forage legume. Plant Physiol. Bioch. 42, 867–873. doi: 10.1016/j.plaphy.2004.10.013
Kim, Y. B., Chun, J. H., Kim, H. R., Kim, S. J., Lim, Y. P., and Park, S. U. (2014). Variation of glucosinolate accumulation and gene expression of transcription factors at different stages of Chinese cabbage seedlings under light and dark conditions. Nat. Prod. Commun. 9:1934578X1400900428. doi: 10.1177/1934578X1400900428
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33:1870–1874. doi: 10.1093/molbev/msw054
Lopez-Vernaza, M., Yang, S., M?ller, R., Thorpe, F., de Leau, E., and Goodrich, J. (2012). Antagonistic roles of SEPALLATA3, FT, and FLC genes as targets of the polycomb group gene curly leaf. PLoS ONE 7:e30715. doi: 10.1371/journal.pone.0030715
Madloo, P., Lema, M., Francisco, M., and Soengas, P. (2019). Role of major glucosinolates in the defense of kale against Sclerotinia sclerotiorum and Xanthomonas campestris pv. campestris. Phytopathology 109, 1246–1256. doi: 10.1094/PHYTO-09-18-0340-R
Major, I. T., Yoshida, Y., Campos, M. L., Kapali, G., Xin, X. F., Sugimoto, K., et al. (2017). Regulation of growth–defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 215, 1533–1547. doi: 10.1111/nph.14638
Malka, S. K., and Cheng, Y. (2017). Possible interactions between the biosynthetic pathways of indole glucosinolate and auxin. Front. Plant Sci. 8:2131. doi: 10.3389/fpls.2017.02131
Millard, P. S., Kragelund, B. B., and Burow, M. (2021). Evolution of A bHLH interaction motif. Int. J. Mol. Sci. 22:447. doi: 10.3390/ijms22010447
Mitreiter, S., and Gigolashvili, T. (2021). Regulation of glucosinolate biosynthesis. J. Exp. Bot. 72, 70–91. doi: 10.1093/jxb/eraa479
Moles, A. T., Warton, D. I., Warman, L., Swenson, N. G., Laffan, S. W., Zanne, A. E., et al. (2019). Global patterns in plant height. J. Ecol. 97, 923–932. doi: 10.1111/j.1365-2745.2009.01526.x
Nakata, M., Mitsuda, N., Herde, M., Koo, A. J., Moreno, J. E., Suzuki, K., et al. (2013). A bHLH-type transcription factor, ABA-inducible BHLH-type transcription factor/JA-associated MYC2-like1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25, 1641–1656. doi: 10.1105/tpc.113.111112
Pangesti, N., Reichelt, M., van de Mortel, J. E., Kapsomenou, E., Gershenzon, J., van Loon, J. J. A., et al. (2016). Jasmonic acid and ethylene signaling pathways regulate glucosinolate levels in plants during rhizobacteria-induced systemic resistance against a leaf-chewing herbivore. J. Chem. Ecol. 42, 1212–1225. doi: 10.1007/s10886-016-0787-7
Park, J. S., Park, J. H., and Park, Y. D. (2019). Construction of pseudomolecule sequences of Brassica rapa ssp. pekinensis inbred line CT001 and analysis of spontaneous mutations derived via sexual propagation. PLoS ONE 14:e0222283. doi: 10.1371/journal.pone.0222283
Pastorczyk, M., Kosaka, A., Piślewska-Bednarek, M., López, G., Frerigmann, H., Kułak, K., et al. (2020). The role of CYP71A12 monooxygenase in pathogen–triggered tryptophan metabolism and Arabidopsis immunity. New Phytol. 225, 400–412. doi: 10.1111/nph.16118
Pedras, M. S. C., and Hossain, S. (2011). Interaction of cruciferous phytoanticipins with plant fungal pathogens: indole glucosinolates are not metabolized but the corresponding desulfo-derivatives and nitriles are. Phytochemistry 72, 2308–2316. doi: 10.1016/j.phytochem.2011.08.018
Per, T. S., Khan, M. I. R., Anjum, N. A., Masood, A., Hussain, S. J., and Khan, N. A. (2018). Jasmonates in plants under abiotic stresses: crosstalk with other phytohormones matters. Environ. Exp. Bot. 145, 104–120. doi: 10.1016/j.envexpbot.2017.11.004
Pfalz, M., Vogel, H., and Kroymann, J. (2019). The gene controlling the indole glucosinolate modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21, 985–999. doi: 10.1105/tpc.108.063115
Rahimi, F., and Rahmanpour, S. (2020). Overcoming glucosinolate-myrosinase-isothiocyanate defense system by plant pathogenic fungi. Int. J. Endocrinol. 7, 19–27. doi: 10.21448/ijsm.697516
Sáez-López, E., Cristóvão, P., Costa, I., Pechirra, P., Conde, P., Guiomar, R., et al. (2019). Epidemiology and genetic variability of respiratory syncytial virus in portugal, 2014–2018. J. Clin. Virol. 121:104200. doi: 10.1016/j.jcv.2019.104200
Schubert, R., Dobritzsch, S., Gruber, C., Hause, G., Athmer, B., Schreiber, T., et al. (2019). Tomato MYB21 acts in ovules to mediate jasmonate-regulated fertility. Plant Cell 31, 1043–1062. doi: 10.1105/tpc.18.00978
Schweizer, F., Fernández-Calvo, P., Zander, M., Diez-Diaz, M., Fonseca, S., Glauser, G., et al. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25, 3117–3132. doi: 10.1105/tpc.113.115139
Seo, M. S., and Kim, J. S. (2017). Understanding of MYB transcription factors involved in glucosinolate biosynthesis in Brassicaceae. Molecules 22:1549. doi: 10.3390/molecules22091549
Serrano-Mislata, A., Bencivenga, S., Bush, M., Schiessl, K., Boden, S., and Sablowski, R. (2017). DELLA genes restrict inflorescence meristem function independently of plant height. Nat. Plants 3, 749–754. doi: 10.1038/s41477-017-0003-y
Shirley, N. J., Aubert, M. K., Wilkinson, L. G., Bird, D. C., Lora, J., Yang, X., et al. (2019). Translating auxin responses into ovules, seeds and yield: insight from Arabidopsis and the cereals. J. Integr. Plant Biol. 61, 310–336. doi: 10.1111/jipb.12747
Shukla, S. P., and Beran, F. (2020). Gut microbiota degrades toxic isothiocyanates in a flea beetle pest. Mol. Ecol. 29, 4692–4705. doi: 10.1111/mec.15657
Song, S., Huang, H., Wang, J., Liu, B., Qi, T., and Xie, D. (2017). MYC5 is involved in jasmonate-regulated plant growth, leaf senescence and defense responses. Plant Cell Physiol. 58, 1752–1763. doi: 10.1093/pcp/pcx112
Stella de Freitas, T. F., Stout, M. J., and Sant'Ana, J. (2019). Effects of exogenous methyl jasmonate and salicylic acid on rice resistance to Oebalus pugnax. Pest Manag. Sci. 75, 744–752. doi: 10.1002/ps.5174
Stotz, H. U., Sawada, Y., Shimada, Y., Hirai, M. Y., Sasaki, E., Krischke, M;, et al. (2011). Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant Cell Physiol. 67, 81–93. doi: 10.1111/j.1365-313X.2011.04578.x
Tarahomi, E., Fahimi, H., and Taghizadeh, M. (2020). Quantitative comparison of tree pairs resulted from gene and protein phylogenetic trees for sulfite reductase flavoprotein alpha-component and 5S rRNA and taxonomic trees in selected bacterial species. J. Biomed. Inform. 7, 326–336.
Teng, Z., Yu, Y., Zhu, Z., Hong, S. B., Yang, B., and Zang, Y. (2021). Melatonin elevated Sclerotinia sclerotiorum resistance via modulation of ATP and glucosinolate biosynthesis in Brassica rapa ssp. pekinensis. J. Proteomics 243:104264. doi: 10.1016/j.jprot.2021.104264
To, H. T. M., Nguyen, H. T., Dang, N. T. M., Nguyen, N. H., Bui, T. X., Lavarenne, J., et al. (2019). Unraveling the genetic elements involved in shoot and root growth regulation by jasmonate in rice using a genome-wide association study. Rice 12, 1–18. doi: 10.1186/s12284-019-0327-5
Van Eck, J. (2018). The status of Setaria viridis transformation: Agrobacterium-mediated to floral dip. Front. Plant Sci. 9:652. doi: 10.3389/fpls.2018.00652
Wang, H., Li, Y., Pan, J., Lou, D., Hu, Y., and Yu, D. (2017). The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in Arabidopsis. Mol. Plant. 10, 1461–1464. doi: 10.1016/j.molp.2017.08.007
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Wasternack, C., and Strnad, M. (2019). Jasmonates are signals in the biosynthesis of secondary metabolites—Pathways, transcription factors and applied aspects—a brief review. New biotechnol. 48, 1–11. doi: 10.1016/j.nbt.2017.09.007
Xiao, C., Somerville, C., and Anderson, C. T. (2014). Polygalacturonase involved in expansion1 functions in cell elongation and flower development in Arabidopsis. Plant Cell 26, 1018–1035. doi: 10.1105/tpc.114.123968
Xie, P. F., Zhu, L., Feng, L., Wu, J. C., and Liu, J. L. (2020). Research progress in transcription factor MYC2 mediating plant resistance to biological stress. J. Appl. Entomol. 57, 781.
Xing, L., Liu, Y., Xu, S., Xiao, J., Wang, B., Deng, H., et al. (2018). Arabidopsis O-GlcNAc transferase SEC activates histone methyltransferase ATX1 to regulate flowering. Embo. J. 37–19:e98115. doi: 10.15252/embj.201798115
Xu, L., Yang, H., Ren, L., Chen, W., Liu, L., Liu, F., et al. (2018). Jasmonic acid-mediated aliphatic glucosinolate metabolism is involved in clubroot disease development in Brassica napus L. Front. Plant Sci. 9:750. doi: 10.3389/fpls.2018.00750
Xu, Y., Qian, Z., Zhou, B., and Wu, G. (2019). Age dependent heteroblastic development of leaf hairs in Arabidopsis. New Phytol. 224, 741–748. doi: 10.1111/nph.16054
Yang, Z. B., He, C., Ma, Y., Herde, M., and Ding, Z. (2017). Jasmonic acid enhances Al-induced root growth inhibition. Plant Physiol. 173, 1420–1433. doi: 10.1104/pp.16.01756
Zang, Y., Zheng, W., He, Y., Hong, S. B., and Zhu, Z. (2015). Global analysis of transcriptional response of Chinese cabbage to methyl jasmonate reveals JA signaling on enhancement of secondary metabolism pathways. Sci. Hortic. 189, 159–167. doi: 10.1016/j.scienta.2015.04.008
Zhai, Q., Yan, L., Tan, D., Chen, R., Sun, J., Gao, L., et al. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9:e1003422. doi: 10.1371/journal.pgen.1003422
Zhang, C., Lei, Y., Lu, C., Wang, L., and Wu, J. (2020). MYC2, MYC3, and MYC4 function additively in wounding-induced jasmonic acid biosynthesis and catabolism. J. Integr. Plant Biol. 62, 1159–1175. doi: 10.1111/jipb.12902
Zhang, M., Jin, X., Chen, Y., Wei, M., Liao, W., Zhao, S., et al. (2018). TcMYC2a, a basic helix–loop–helix transcription factor, transduces JA-signals and regulates taxol biosynthesis in Taxus chinensis. Front. Plant Sci. 9:863. doi: 10.3389/fpls.2018.00863
Zhang, Y., Li, B., Huai, D., Zhou, Y., and Kliebenstein, D. (2015). The conserved transcription factors, MYB115 and MYB118, control expression of the newly evolved benzoyloxy glucosinolate pathway in Arabidopsis thaliana. Front. Plant Sci. 6:343. doi: 10.3389/fpls.2015.00343
Keywords: BrMYC2/3/4, plant development, glucosinolate, Sclerotinia sclerotiorum, disease resistance
Citation: Teng Z, Zheng W, Yu Y, Hong S-B, Zhu Z and Zang Y (2021) Effects of BrMYC2/3/4 on Plant Development, Glucosinolate Metabolism, and Sclerotinia sclerotiorum Resistance in Transgenic Arabidopsis thaliana. Front. Plant Sci. 12:707054. doi: 10.3389/fpls.2021.707054
Received: 08 May 2021; Accepted: 20 July 2021;
Published: 03 September 2021.
Edited by:
Stephan Pollmann, National Institute of Agricultural and Food Research and Technology, SpainReviewed by:
Branka Salopek Sondi, Rudjer Boskovic Institute, CroatiaArvind H. Hirani, Kemin Industries, Inc, United States
Copyright © 2021 Teng, Zheng, Yu, Hong, Zhu and Zang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhujun Zhu, emh1empAemFmdS5lZHUuY24=; Yunxiang Zang, eXh6YW5nQHphZnUuZWR1LmNu
†These authors have contributed equally to this work
 Zhiyan Teng
Zhiyan Teng Weiwei Zheng1†
Weiwei Zheng1† Seung-Beom Hong
Seung-Beom Hong Zhujun Zhu
Zhujun Zhu Yunxiang Zang
Yunxiang Zang