- 1Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
- 2College of Biology and the Environment, Nanjing Forestry University, Nanjing, China
The Salt Overly Sensitive (SOS) signaling pathway is key in responding to salt stress in plants. SOS2, a central factor in this pathway, has been studied in non-halophytes such as Arabidopsis and rice, but has so far not been reported in the halophyte Nitraria tangutorum. In order to better understand how Nitraria tangutorum acquires its tolerance for a high salt environment, here, the NtSOS2 was cloned from Nitraria tangutorum, phylogenetic analyses showed that NtSOS2 is homologous to the SOS2 of Arabidopsis and rice. Gene expression profile analysis showed that NtSOS2 localizes to the cytoplasm and cell membrane and it can be induced by salt stress. Transgenesis experiments showed that exogenous expression of NtSOS2 reduces leaf mortality and improves the germination rate, biomass and root growth of Arabidopsis under salt stress. Also, exogenous expression of NtSOS2 affected the expression of ion transporter-related genes and can rescue the phenotype of sos2-1 under salt stress. All these results revealed that NtSOS2 plays an important role in plant salt stress tolerance. Our findings will be of great significance to further understand the mechanism of salt tolerance and to develop and utilize molecular knowledge gained from halophytes to improve the ecological environment.
Introduction
Soil salinization has become a major constraint to the sustainable development of the ecological environment, more than 20% of arable land suffered from soil salinization (Shrivastava and Kumar, 2015; Rui and Ricardo, 2017) and a trend for ever increasing salinization has been observed (Cuevas et al., 2019; Singh, 2021). The area of salinized soil in the world is gradually expanding due to human activity and natural phenomena. Soil salinization seriously inhibits plant growth (Tian et al., 2020) through soil hardening and decreases in soil total porosity and fertility, in general lowering a plant’s chances of survival (Jez et al., 2016; Van Zelm et al., 2020). Excessively high salt concentrations make it difficult for plants to germinate, causing plant biomass to decline or even to die, hindering the comprehensive utilization and sustainable development of soil.
The main reason why plants have difficulty in living in salinized soil is that they are subjected to abiotic stress, which will lead to osmotic stress and ion toxicity under salt stress (Liang et al., 2018; Arif et al., 2020). When plants subjected to stress, Ca2+ concentration increases and activates protein kinases by calmodulin then induce a series of cellular reactions to make plants adapt to or reduce the damage caused by salt stress (Villalobo and Berchtold, 2020). Plant protein kinases (a kind of phosphotransferase) participate in the process of plant stress resistance signaling and play an important role when plants suffer from salt damage and drought stress (Yip Delormel and Boudsocq, 2019; Chen et al., 2021). Calcineurin B-like proteins-interacting protein kinases (CIPKs) are plant-specific protein kinases, which contain conserved Protein Kinase and NAF/FLSL domains (Tang et al., 2020). CIPKs are involved in a variety of abiotic stresses such as salt stress and drought stress, and have been identified and functionally validated in plant (Ma et al., 2019, 2020). CIPK24 is also known as SOS2 in Arabidopsis and encodes a serine/threonine protein kinase with two important functional domains: an N-terminal catalytic domain and a C-terminal regulatory domain. SOS2 was first identified and cloned from Arabidopsis using bitmap cloning methods and was found to play an important role in the SOS signaling pathway, which maintaining the ion balance within plant cells (Liu et al., 2000). The SOS2 C-terminal regulatory region contains a conserved NAF/FISL motif that interacts with the Ca2+ binding protein SOS3/CBL4 to form a binary complex, after which they integrate into the plasma membrane and activate the SOS1 gene (Quintero et al., 2002; Qiu et al., 2004; Yang et al., 2009).
Nitraria tangutorum is a species within the Nitraria Linnaeus genus, which is widely distributed in arid saline-alkali areas within northwest China (Zhou et al., 2015). Nitraria tangutorum has a strongly developed root system which can withstand sand burial, and has excellent tolerance to drought and salinity. It is one of the outstanding pioneer species in a saline-alkali environment, making it an excellent model species for research into salt/drought stress resistance (Liu et al., 2016). So, the use of Nitraria tangutorum to reduce the accumulation of salt on the soil surface and can repair saline-alkali land up to a point. At present, research on Nitraria tangutorum has mainly focused on the determination of physiological and biochemical indices of stress resistance and fruit composition (Liu et al., 2014), with research into the molecular mechanism of salt tolerance being quite scarce.
Furthermore, SOS2 has been primarily studied more in well-known plant model species such as Arabidopsis and rice (Batelli et al., 2007; Martínez-Atienza et al., 2007; Fujii and Zhu, 2009), with such studies indicating that SOS2 plays a key role in plant saline-alkali resistance. Summarizing, few molecular studies on Nitraria tangutorum, such as on the NtCIPK2 and NtCIPK9 genes, have been reported (Zheng et al., 2014; Lu et al., 2020), while any study on the Nitraria tangutorum SOS2 gene is still lacking.
Salt tolerant plants can grow in salinized soil. Therefore, it is a great significance to understand the mechanism of salt tolerance in halophytes in order to alleviate and improve soil salinization. Studies on model species have shown that SOS2 plays an important role in the process of plant stress tolerance, so a key question is how does this gene respond to salt stress in the halophyte Nitraria tangutorum? To explore the role of SOS2 in Nitraria tangutorum, the NtSOS2 gene was cloned and analyzed its expression pattern under salt stress. Meanwhile, to explore whether the exogenous expression of NtSOS2 may affect Arabidopsis salt resistance, the germination rate, leaf white blade rate, fresh weight, root length and phenotype of transgenic Arabidopsis were analyzed and compared it to the wild type. Our results indicated that exogenous expression of NtSOS2 enhances Arabidopsis salt tolerance. These experiments indicated that NtSOS2 plays a key role in plant salinity tolerance.
Materials and Methods
Plant Materials and Growth Conditions
Nitraria tangutorum seeds were collected from Dengkou county, Inner Mongolia, China. The seeds were mixed with wet sand at 4°C for ∼2 months to vernalize, then placed on germination box. After germination, seedlings were planted in a matrix with a peat soil: perlite ratio of 4:1 and grown at 23°C with a 16-h light/8-h dark light cycle and 60% air humidity. Seedlings were grown for 3 months, after which they were treated with a 500 mM NaCl solution, collected at 0, 1, 6, 12, and 24 h intervals, then submerged in liquid nitrogen to quickly freeze for subsequent RNA extraction.
The Arabidopsis thaliana Columbia ecotype (Col-0) used in this study were provided by Prof. Thomas Laux (Signalling Research Centres BIOSS and CIBSS, Faculty of Biology, University of Freiburg, Germany); The mutant sos2-1 obtained from Nottingham Arabidopsis Stock Centre (NASC ID: N3863). These seeds were sown on 1/2 MS medium after disinfection, stored at 4°C for 3 days to stratify, after which they were cultivated in a light incubator having identical culture conditions as Nitraria tangutorum above. When seedlings reached the four leaves stage, they were transferred to a substrate of peat soil: vermiculite: perlite with 5:1:1 ratio for further cultivation.
Cloning and Sequence Analysis of NtSOS2
Total ribonucleic acid was isolated from Nitraria tangutorum leaves with an RNA extraction kit (Bioteke, China) and treated with DNase I (Takara, Japan) to remove contaminating desoxyribonucleic acid. cDNA was synthesized with 1st strand cDNA synthesis Kit (Vazyme, China). The NtSOS2 was amplified from Nitraria tangutorum’s cDNA using specific primers which designed by Oligo 7.60 (Cascade, CO 80809, United States) that were listed in Supplementary Table 1. Homologous proteins were searched for using the NCBI blastp program and multiple alignments were performed using DNAMAN v9.0 (Lynnon Corporation, San Ramon, CA, United States) with the deduced amino acid sequences. A phylogenetic tree was constructed through the Neighbor-Joining method, and the bootstrap value was calculated according to 1,000 repetitions using MEGA-X v10.1 (Temple, Philadelphia, PA, United States).
Quantitative Real-Time PCR Analyses
Quantitative real-time PCR (qRT-PCR) was performed using the AceQ qPCR SYBR Green Master Mix (Vazyme, China). Cycling conditions were 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Each reaction was performed in triplicate. Relative expression values were calculated through the relative quantization method (2–ΔΔCT) (Livak and Schmittgen, 2001), primer and actin sequences for qRT-PCR are listed in Supplementary Table 2.
Subcellular Localization
The NtSOS2 open reading frame (ORF) was fused in-frame upstream to GFP in pJIT166 vector using XbaI and BamHI restriction sites. The primers used to construct this vector were listed in Supplementary Table 3. Then the constructive fusion plasmid was transfected into onion epidermal cells by microprojectile bombardment (Nebenführ, 2014) and incubated for 24 h in the dark. The cells were subsequently observed using Zeiss fluorescence microscope (X-cite 120Q, Carl Zeiss).
Arabidopsis Transformation
A pair of primers (listed in Supplementary Table 4) was designed to amplify the ORF of NtSOS2 and clone it into the pBI121 vector using XbaI and BamHI restriction sites. The vector was then transformed into Arabidopsis columbia and sos2-1 mutant using the method reported by Clough and Bent (1998), after which positive plants were screened using kana-containing 1/2MS medium. Transformed plants were confirmed using PCR amplification.
Analysis of Salt Tolerance of Transgenic Arabidopsis
To analyze the germination rate, transgenic Arabidopsis of the T3 generation and Col-0 were seeded on 9 × 9 cm square dishes containing 1/2 MS medium supplemented with 0, 100, or 150 mM NaCl. ∼70 seeds were placed on each dish for each strain, plants were then cultured for 7 days in the light incubator, after which the germination rate was determined. For phenotypical observation (root length, leaf blade whitening ratio and fresh weight), seedlings were grown on 1/2 MS culture medium containing 0, 100, or 150 mM NaCl for 10 days of vertical culture. They were then transferred to soil for another 2 weeks of growth, after which 200 mM NaCl was used to induce salt stress and surface appearance and leaf mortality rate were scored. For rescue phenotype of sos2-1, the root length and fresh weight are observated seedlings were grown on 1/2 MS culture medium containing 0, 60, or 120 mM NaCl for 7 days of vertical culture and then transferred to soil culture are the same as above conditions, after which 150 mM NaCl was used to induce salt stress and surface appearance and leaf mortality rate were scored. Each experiment was performed using three biological replicates.
Analyze Physiological Changes of Salt Stress
The Arabidopsis growing in soil were exposed to salt solution for 3 days for physiological determination. The Soluble protein, Superoxide Dismutase (SOD), Malondialdehyde (MDA) were determinated and analyzed by the soluble protein, superoxide dismutase and malondialdehyde assay kits, respectively (Jiancheng Bioengineering Institute, Nanjing, China).
For the determination of chlorophyll, the protocol referred to the previously reported method (Lichtenthaler and Wellburn, 1983), 0.1 g fresh leaves were taken and cut into 10 ml 96% ethanol. The absorbance was measured at 665 and 649 nm under dark conditions at room temperature until the leaves turned white completely. Each experiment was performed using three biological replicates.
Results
Cloning and Sequence Analysis of NtSOS2
In order to clone SOS2 of Nitraria tangutorum, the SOS2 protein sequences from rice and Arabidopsis were compared to our unpublished transcriptome data using the NCBI blastp program. A pair of specific primers were designed to obtain the SOS2 target sequence by PCR amplification (Supplementary Figure 1). Gene sequence analysis showed that NtSOS2 coding sequence (CDS) length is 1,332 bp which encodes a protein 443 amino acids in length, with molecular weight of 50.34 kDa.
Then DNAMAN software was used to compare the NtSOS2 protein sequence to homologous sequences from seven additional plant species and found the amino acid sequence homology of NtSOS2 to the other seven species to be average of 82.01% (Figure 1), indicating that NtSOS2 has a relatively high sequence similarity to homologs from other species. In addition, the NCBI conserved domain program was used to identify conserved Protein Kinase (PF00069) and NAF/FLSL (PF03822) domains (Supplementary Figure 2), showing that NtSOS2 is a typical CIPK gene as these domains are essential for CIPK function.

Figure 1. Multiple amino acid sequence alignment of NtSOS2. The amino acid from Arachis hypogaea (HG797656.1), Arabidopsis thaliana (NM_122932.5), Gossypium hirsutum (GU_188961.1), Populus alba (KT_875331.1), Sorghum bicolor (KY_202762.1), Solanum lycopersicum (NM_001247281.2), Oryza sativa (XP_015643084.1), the blue lines represent the Protein Kinase and NAF/FLSL domain, respectively.
Furthermore, the NtSOS2 protein sequence was aligned with the CIPK gene family of Arabidopsis and rice to construct a phylogenetic tree, and the results showed that NtSOS2, AtCIPK24, and OsCIPK24 are distributed on one branch (Figure 2). This showed that NtSOS2 has a high homology to AtCIPK24 and OsCIPK24 that have been previously described as SOS2 genes, which indicated that NtSOS2 may have similar functions compared with SOS2 of Arabidopsis and rice.
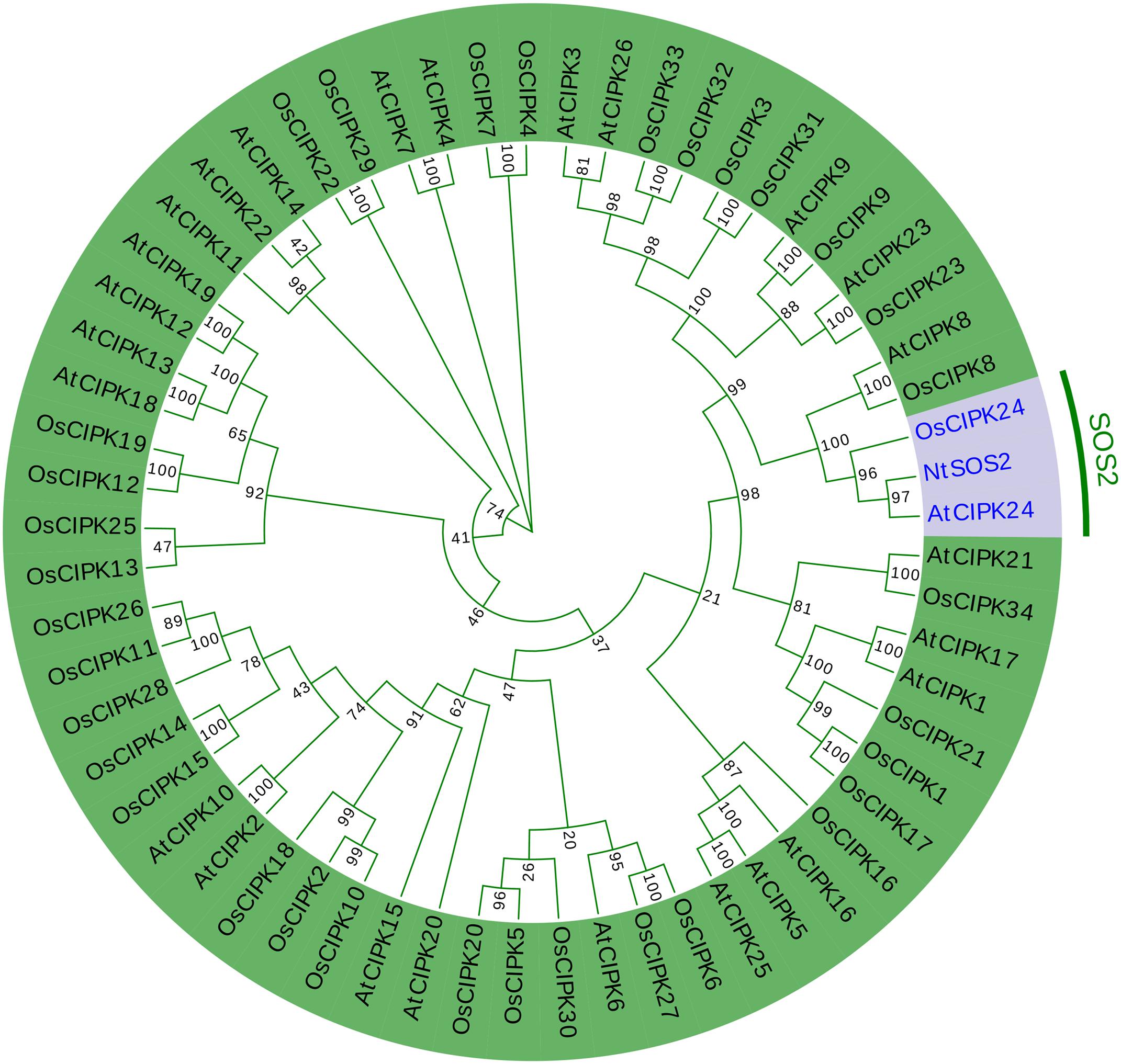
Figure 2. Phylogenetic analysis of NtSOS2 with Arabidopsis CIPKs and Rice CIPKs. The red section highlights the SOS2 gene of all three species, while the green section represents all other CIPK gene family members from Arabidopsis and rice.
Subcellular Localization of NtSOS2
To understand the expression pattern of NtSOS2, the ProtComp 9.0 tools was used to predict and analyze the subcellular localization of NtSOS2, and the result showed that the probability of NtSOS2 localizing to the cytoplasm and cell membrane is relatively high (Supplementary Figure 1). To further determine the subcellular localization of NtSOS2, the coding region of NtSOS2 was combined into a pJIT166-GFP vector. Then, this plasmid was instantly expressed in onion epidermal cells via microparticle bombardment and 35S:GFP was used as a control. As shown in Figure 3, 35S:GFP could be detected in the nucleus and cytosol, while 35S:NtSOS2-GFP was detected in the cytoplasm and plasma membrane, consistent with the subcellular localization prediction of ProtComp.
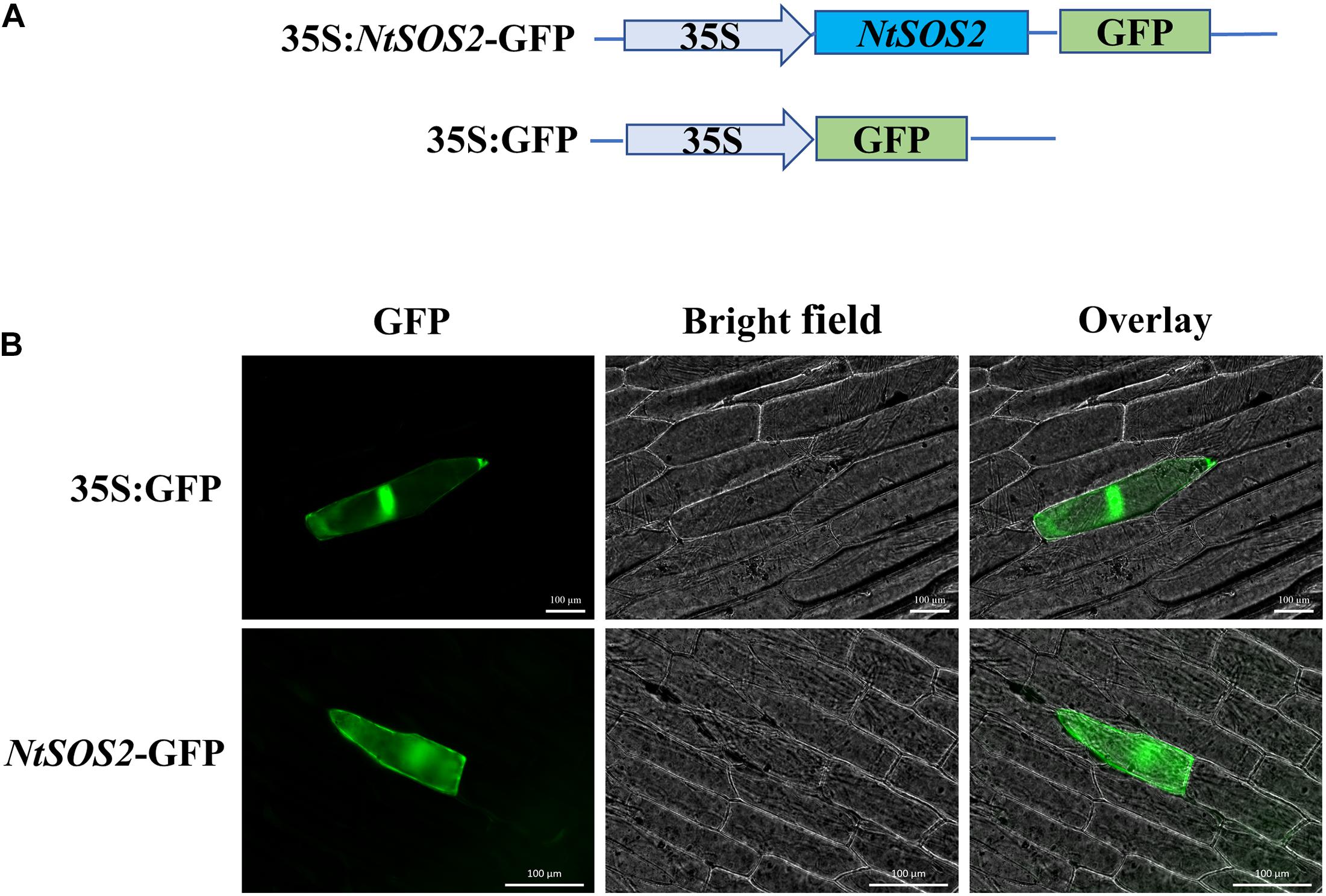
Figure 3. Subcellular localization of NtSOS2-GFP. (A) Schematic diagram of the SOS2 subcellular localization and GFP control vectors. (B) Fluorescence microscopy analysis of the GFP signal from onion epidermal cells transiently expressing GFP and NtSOS2-GFP fusions as indicated. Scale bar = 100 μm.
Expression Analysis of NtSOS2 Under Salt Stress
Abiotic stress can induce changes in the expression of SOS2 (Kaur et al., 2015), the Nitraria tangutorum seedlings were stressed in solution containing 500 mM NaCl to investigate whether SOS2 had a similar expression pattern, the root, stem and leaf tissue were collected at 0, 1, 6, 12, and 24 h, respectively, for real-time quantitative PCR analysis of NtSOS2 expression. NtSOS2 expression in the root continuously increased in response to 500 mM NaCl between 0 and 12 h, followed by downregulation at 24 h, while NtSOS2 expression in the stem reached its maximum 1 h after stress exposure (Figure 4). In the leaf, the expression of NtSOS2 was observed a more wave-like pattern, with an expression peak observed after 6 h. Overall, NtSOS2 expression under NaCl stress tended to be up-regulated first, then down-regulated.
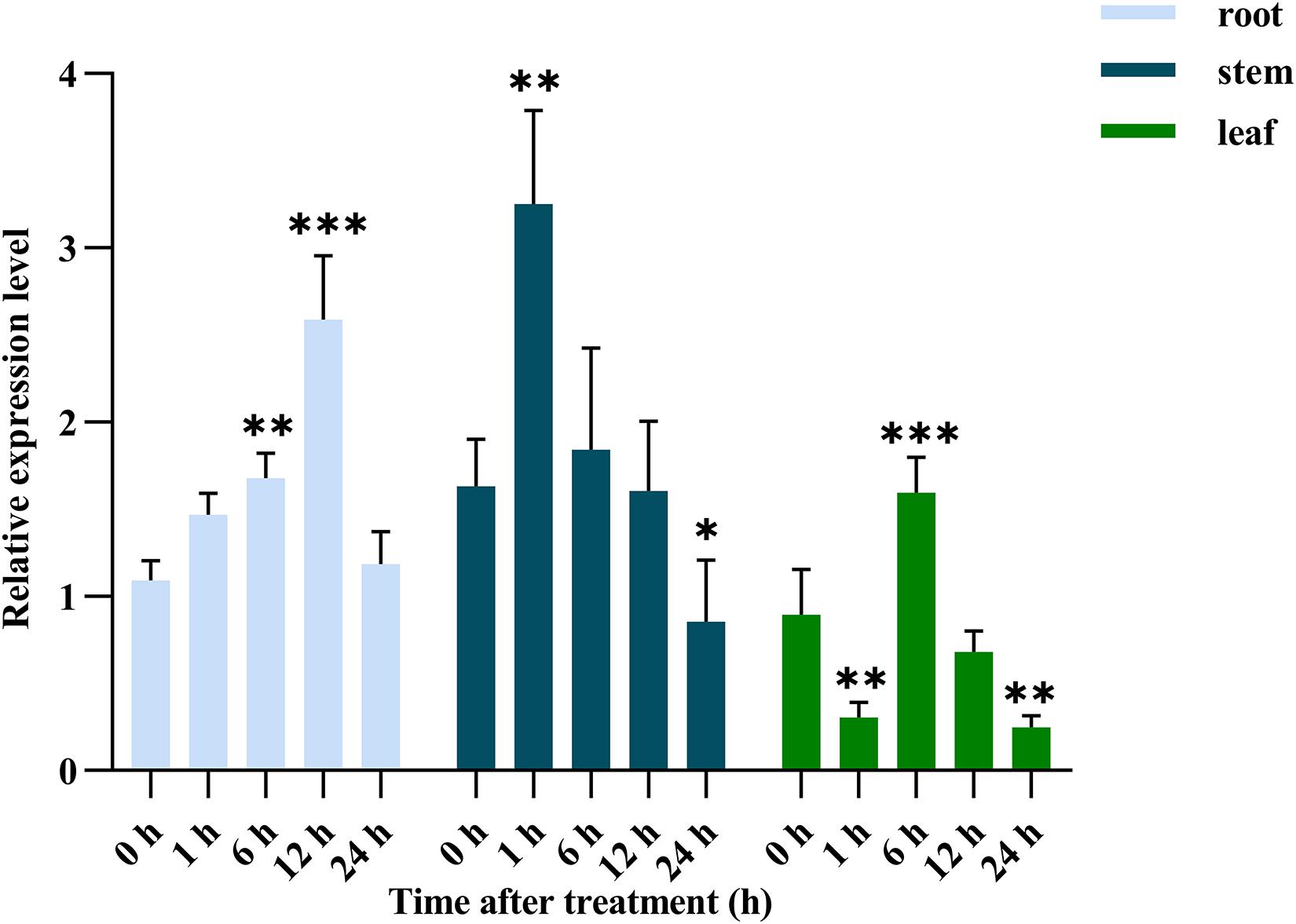
Figure 4. Relative expression of NtSOS2 in roots, stems and leaves under 500 mM NaCl salt stress. Displayed is the mean ± SD. Error bars indicate the standard deviation of three biological replicates. Statistically significant differences were assessed using ANOVA test (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). The significant differences were analyzed in root, stem and leaf, respectively transgenic lines.
Exogenous Expression of NtSOS2 Enhances Germination Under Salt Stress in Arabidopsis
In order to verify the potential function of NtSOS2, an overexpression vector of NtSOS2 was constructed and transformed into Arabidopsis, three of transgenic lines with relatively high expression of NtSOS2 were selected through semi-quantitative experiments (Supplementary Figure 3).
Salt stress can significantly inhibit the germination of seeds (Zhang et al., 2019). Since NtSOS2 expression increases in response to salt stress, whether exogenous NtSOS2 expression may increase germination rate under salt stress conditions, so wild type and transgenic Arabidopsis over expressing NtSOS2 were grown on 1/2 MS medium containing 0, 100, and 150 mM NaCl for 7 days. The result showed that the germination rate of transgenic Arabidopsis was not affected under conditions without salt stress (Figures 5A,D). However, in the 1/2MS medium containing 100 mM NaCl, the germination rate of wild-type seeds decreased significantly, which may be due to the significant inhibition of NtSOS2 over expression (Figures 5B,D).
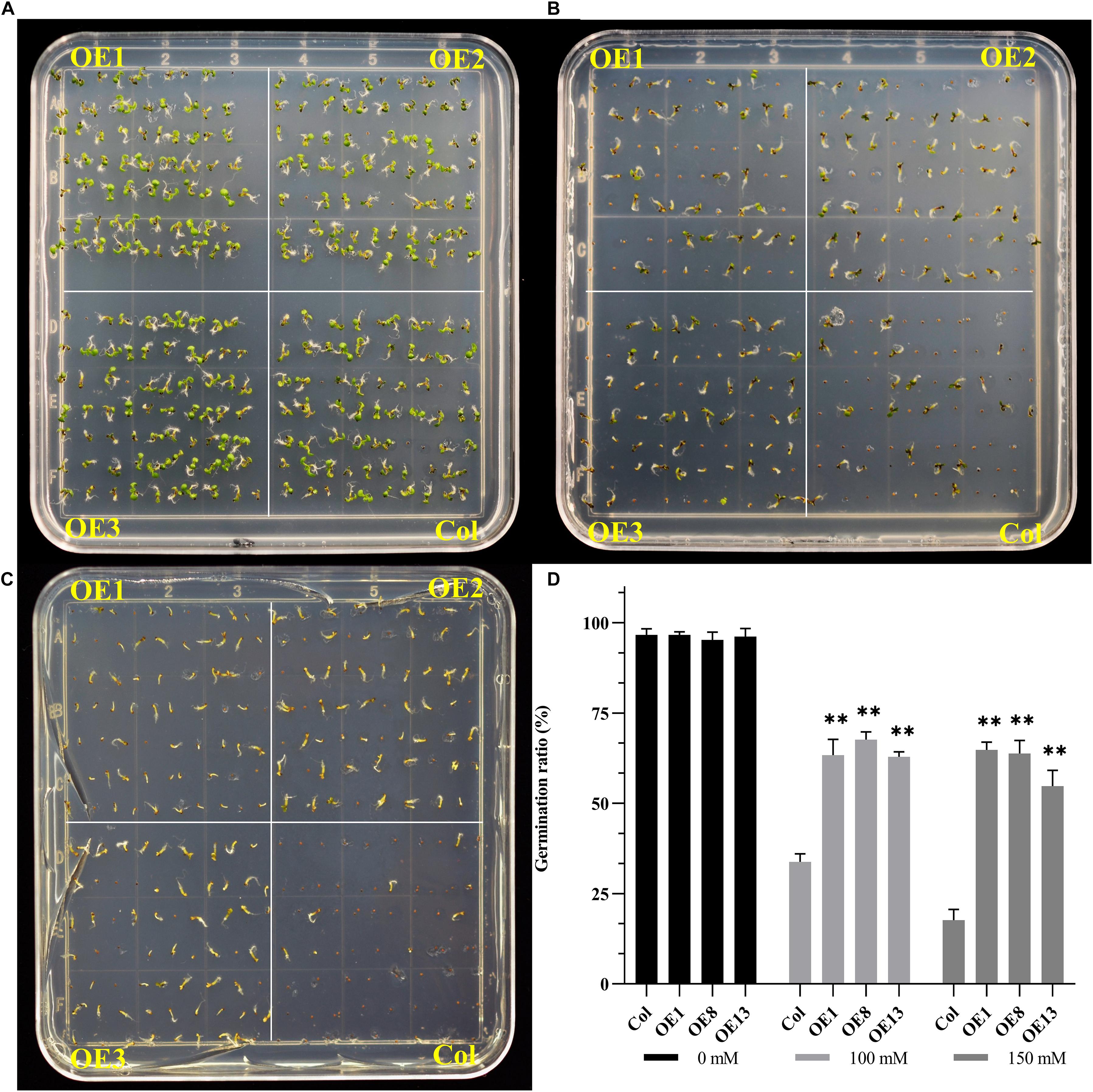
Figure 5. Exogenous expression of NtSOS2 increases germination rate under salt stress. (A–C) Wild-type (Col) and transgenic (OE1-3) Arabidopsis seeds germinating on 1/2 MS medium supplemented with NaCl (0, 100 mM, 150 mM) 7 day after being transferred to the light. (D) Quantification of germinate rate. n = 70 seeds for each condition (genotype/NaCl conc.). The germination rate is shown as the mean ± SD (n = 3). Statistically significant differences were assessed using an ANOVA test (∗∗P < 0.01). OE1-3 represent three independent transgenic lines.
The decline in germination rate of wild type seeds was even more pronounced when applying 150 mM salt stress, while germination of transgenic seeds did not decrease further (Figures 5C,D). In wild type Arabidopsis the germination rate decreased from 96.66 to 17.14% under heavy salt stress on average, in transgenic seeds the germination rate decreased to 62.38% on average under heavy salt stress, a much less dramatic effect. Therefore, these data demonstrate that overexpression of NtSOS2 can significantly decrease the effect that salt stress has on seed germination rate in Arabidopsis.
Exogenous Expression of NtSOS2 Enhances Salt Tolerance in Arabidopsis
To verify whether NtSOS2 overexpression can improve the tolerance of Arabidopsis seedlings to salt stress, wild type and transgenic Arabidopsis were grown on vertically placed plates containing 1/2 MS medium supplemented with 0, 100, and 150 mM NaCl for 10 days. On medium without added NaCl, wild type and transgenic Arabidopsis showed no obvious differences in growth, while on 100 or 150 mM NaCl, growth of wild type Arabidopsis was clearly reduced, with wild type plants growing on 150 mM NaCl clearly showing stronger whitening and anthocyanin accumulation than NtSOS2 overexpressing plants (Figure 6A). In addition, NtSOS2 overexpression significantly reduces loss of fresh weight, leaf whitening and inhibition of root growth under high salt stress conditions (Figures 6B–D). To continue observing plant growth past the seed stage, four-leaved seedlings were transferred to soil, growing them for 2 weeks with 200 mM NaCl irrigation. After 1 day of growth, no differences could be observed, but starting from 3 days of growth, wild-type Arabidopsis showed significantly more leaf mortality than NtSOS2 overexpressing lines (Figures 7A,B).
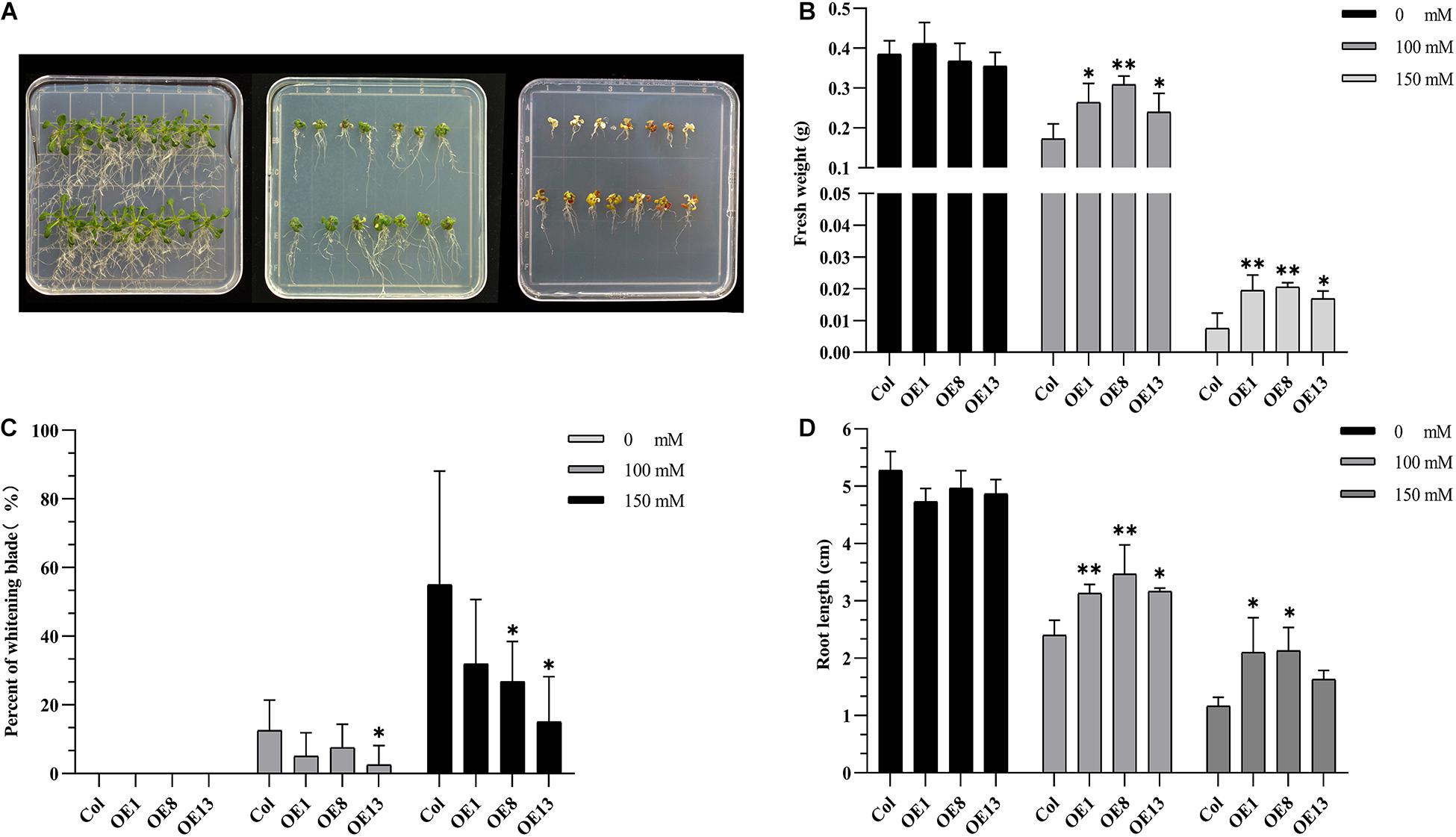
Figure 6. Exogenous expression of NtSOS2 enhances salt tolerance in Arabidopsis. (A) Wild-type (top-rows) and NtSOS2 overexpressing (bottom rows) Arabidopsis were grown on 1/2 MS medium containing 0 (left), 100 (middle) and 150 mM (right) NaCl for 10 days. (B–D) Fresh weight (g) (B), Leaf blade whitening (%) (C) and Main root length (cm) (D) of wild-type and NtSOS2 overexpressing Arabidopsis under salt stress. See legend for NaCl concentrations used. Each experiment was performed with three biological replicates. An ANOVA test was used for statistical analysis. ∗P < 0.05, ∗∗P < 0.01. Scale bar = 1 cm.
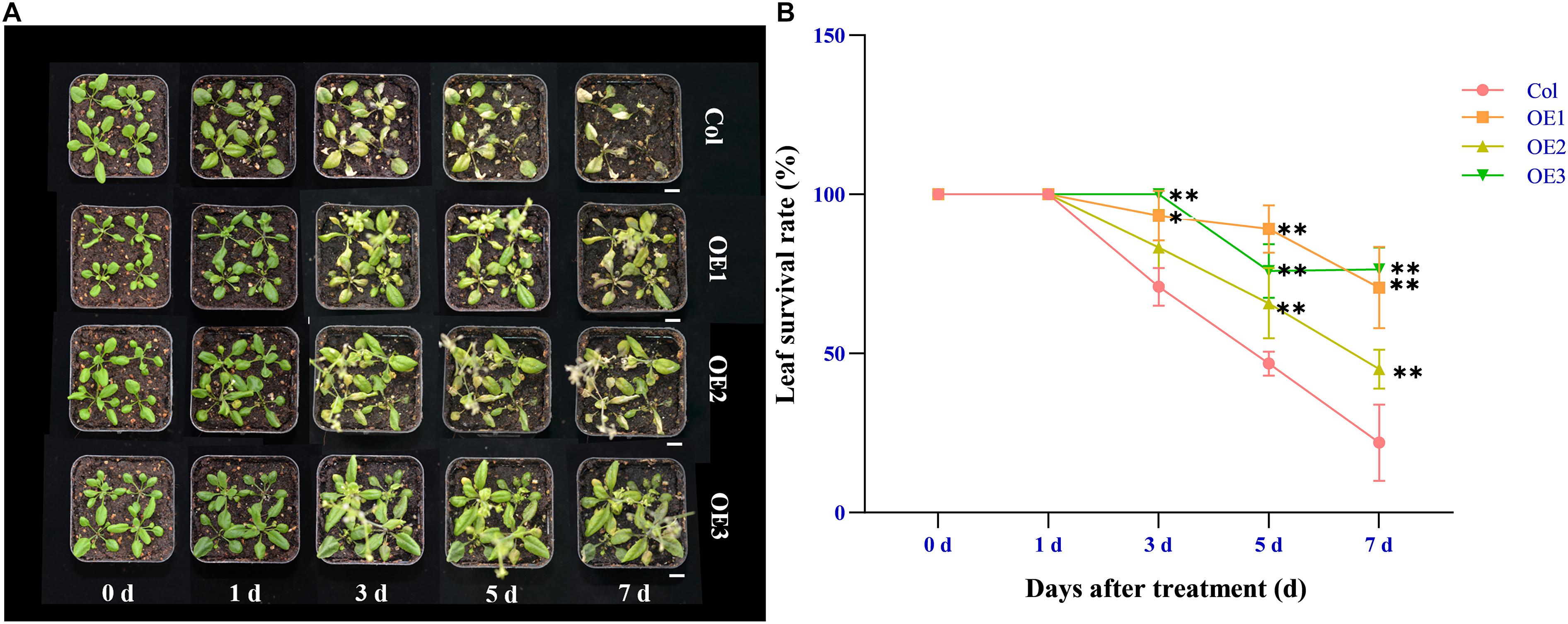
Figure 7. Ectopic expression of NtSOS2 reduces the leaf mortality rate. (A) Representative photograph of wild-type and NtSOS2 overexpressing lines under 200 mM NaCl stress. (B) The leaf survival rate under salt stress of wild-type (red) and NtSOS2 overexpression (orange, green, blue) lines. ANOVA test was used for statistical analysis, ∗P < 0.05, ∗∗P < 0.01. Scale bar = 1 cm.
Judging from the results, there were more obvious phenotypic differences after 3 days of salt stress for wild type and transgenic type. To characterize their change in physiological state, wild-type and transgenic Arabidopsis that had been subjected to salt irrigation for 3 days were chosen to detected the chlorophyll, soluble protein, MDA and SOD content. The results showed that in the absence of salt stress, there was no significant difference in chlorophyll content between transgenic lines and wild-type lines (Figure 8). However, there were significant differences in chlorophyll, soluble protein, MDA and SOD content between transgenic and wild-type strains under salt stress. The content of chlorophyll, soluble protein and SOD in the transgenic plants was higher than that in the wild-type, while the content of MDA was the opposite (Figure 8), which may indicate that the wild-type plants showed higher salt intolerance. Taken together, these results show that NtSOS2 can counteract the physiological effects that high salt stress has on growth of Arabidopsis, thereby endowing plants with increased salt tolerance.
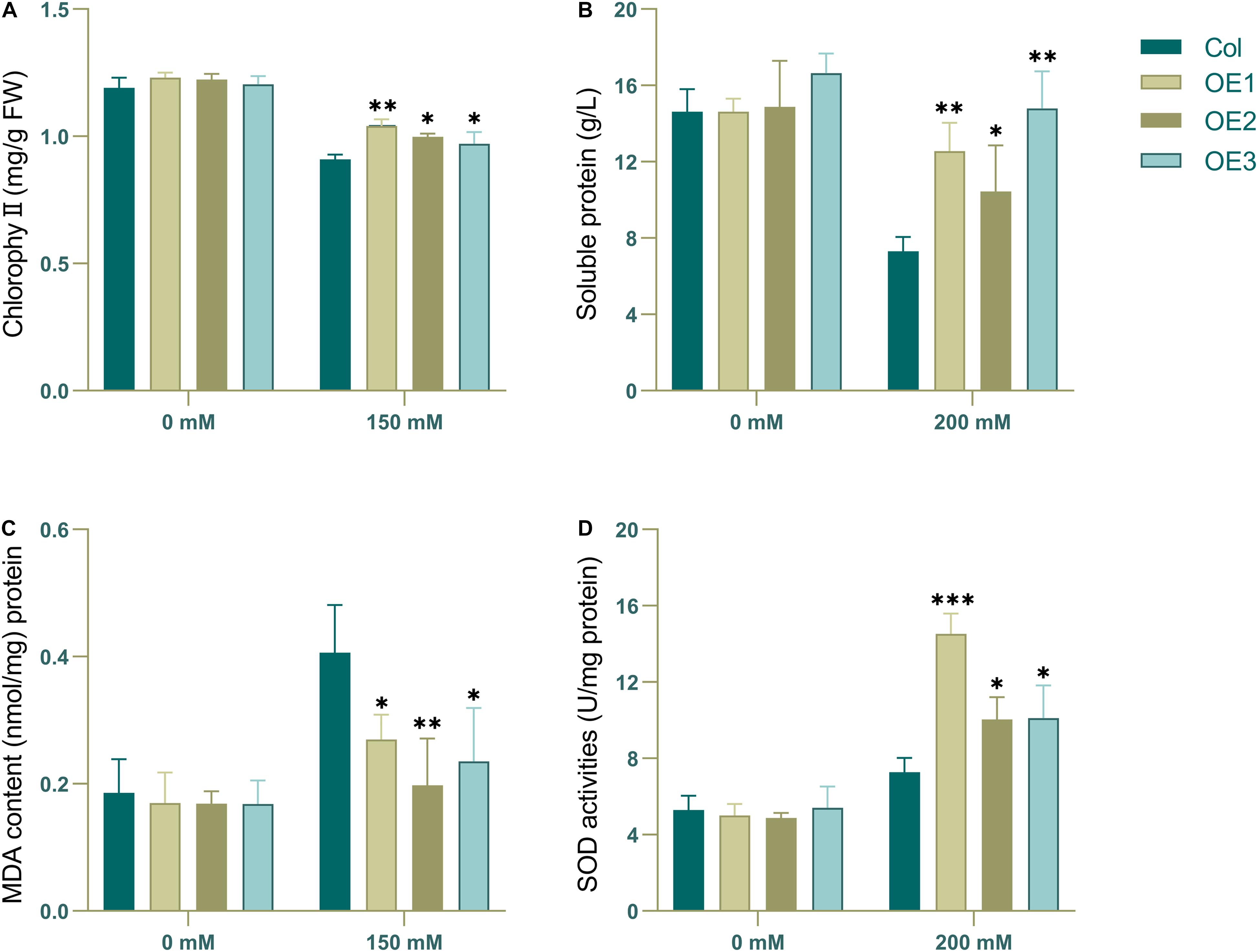
Figure 8. The chlorophyll, MDA and antioxidative enzyme activity analyses. 2-week-old wild type and transgenic Arabidopsis were treated with 0 or 200 mM NaCl for 3 days. (A) Chlorophyll content. (B) Soluble protein content. (C) MDA level. (D) SOD activity. ANOVA test was used for statistical analysis. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Exogenous Expression of NtSOS2 Can Rescue Mutant sos2-1 Phenotype
In Arabidposis, mutates SOS2 leads to salt stress sensitive phenotype (Liu et al., 2000). Exogenous expression of CIPK24 in canola and foxtail millet can complement the phenotype of sos2-1 (Zhang et al., 2014, 2017). In order to verify whether NtSOS2 has a similar function with other species, the NtSOS2 gene were exogenously expressed into sos2-1. Three of transgenic lines were used for functional analysis (Supplementary Figure 3), on medium without added NaCl, sos2-1 and transgenic Arabidopsis showed no obvious differences in growth, while on 60 or 120 mM NaCl, growth of mutant Arabidopsis was clearly reduced, with sos2-1 plants growing on 120 mM NaCl clearly showing stronger whitening and anthocyanin accumulation than NtSOS2 overexpressing plants (Figure 9A).
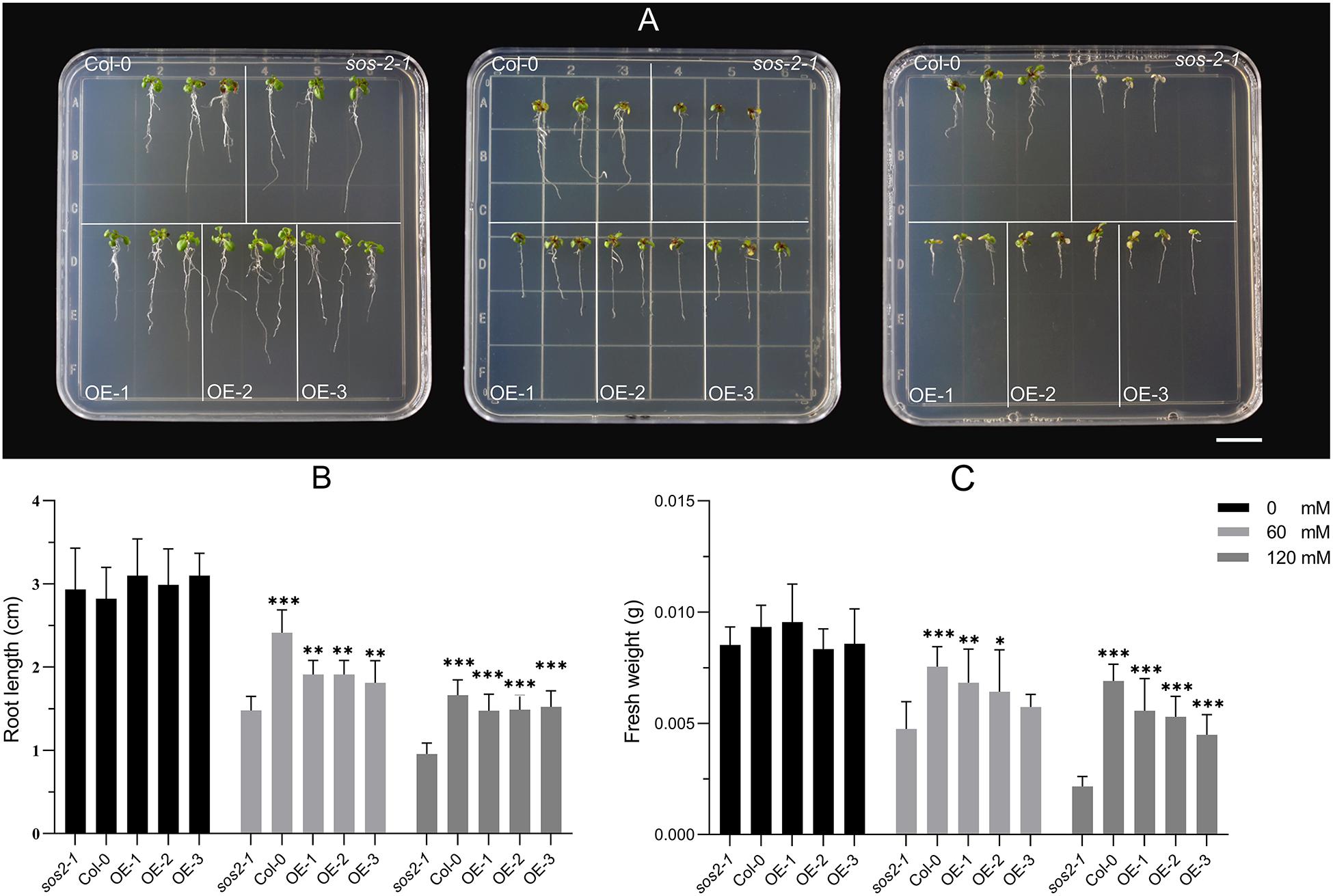
Figure 9. Exogenous expression of NtSOS2 could rescue sos2-1 phenotype on salt stress. (A) Col, mutant sos2-1 and transgenic Arabidopsis were treated at 0, 60, 120 mM NaCl for 7 days. (B) The root length (cm) of sos2-1 and transgenic Arabidopsis on salt stress. (C) The fresh weight (g) of sos2-1 and transgenic Arabidopsis on salt stress. Three biological replicates for each experiment. ANOVA test was used for statistical analysis. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.01.
In addition, under high salt stress conditions, the seedings that overexpress the NtSOS2 gene significantly reduces loss of fresh weight and inhibition of root growth under high salt stress conditions (Figures 9B,C). To continue observing plant growth past the seed stage, four-leaved seedlings were transferred to soil, when growing for 2 weeks with 150 mM NaCl irrigation. After 1 day of growth, no differences could be observed, but starting from 3 days of growth, sos2-1 showed significantly more leaf mortality than NtSOS2 overexpressing lines (Figure 10).
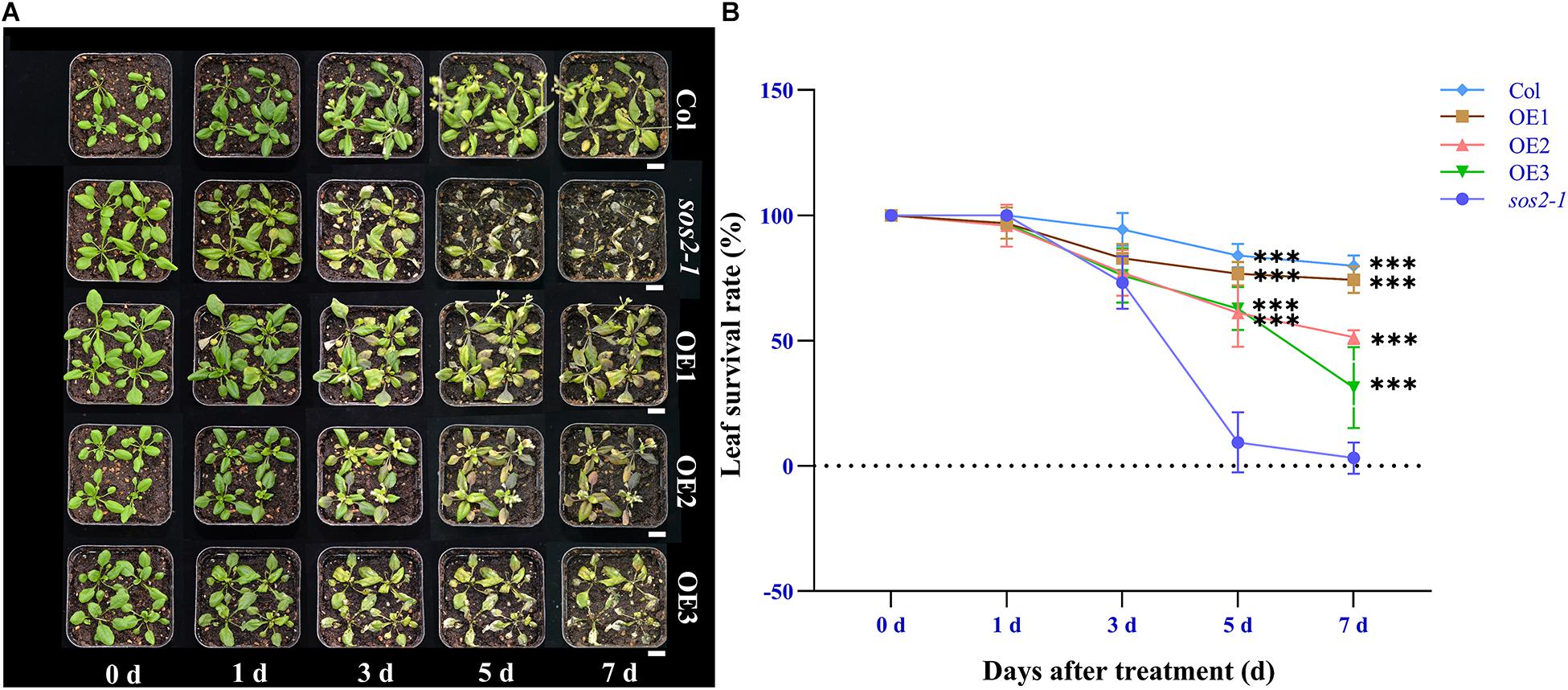
Figure 10. Ectopic expression of NtSOS2 can rescue sos2-1 phenotype. (A) Phenotype observation of sos2-1 and overexpression lines under 150 mM NaCl stress. (B) Statistics of leaf survival rate under salt stress in sos2-1, wild type and overexpression lines. ANOVA test was used for statistical analysis. Scale bar = 1 cm. ANOVA test was used for statistical analysis. ∗∗∗P < 0.001.
To characterize their change in physiological state, these plants were taken to detected the chlorophyll content, soluble protein, MDA and SOD content (Figure 11), and the results showed that the contents of chlorophyll, soluble protein, MDA and POD were not significantly different between the wild types and transgenic types cultured under normal conditions. However, under salt stress, the contents of chlorophyll, MDA and soluble protein of the mutant were significantly different from those of the transgenic plants. At the same time, the SOD content of the transgenic lines was generally higher than that of the mutant sos2-1, although the SOD content did not reach a statistically significant difference (P<0.05).
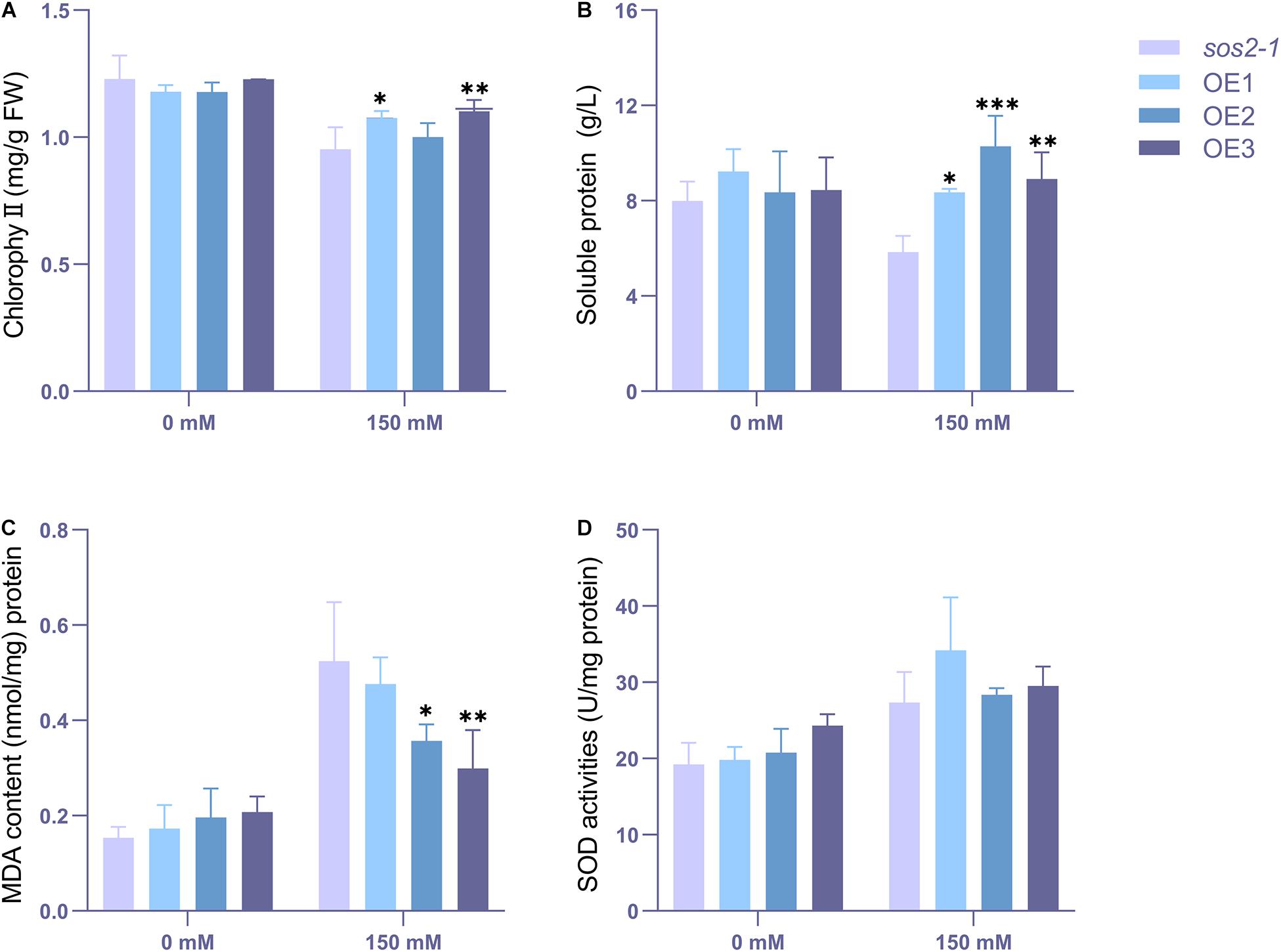
Figure 11. The chlorophyll, MDA and antioxidative enzyme activity analyses. 2-week-old mutant and transgenic Arabidopsis were treated with 0 or 150 mM NaCl for 3 days. (A) Chlorophyll content. (B) Soluble protein content. (C) MDA level. (D) SOD activity. ANOVA test was used for statistical analysis. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
NtSOS2 Affects the Expression of Ion Transporter-Related Genes
In Arabidopsis, the AtSOS2 gene plays an important role in maintaining cellular ion balance. In order to explore whether exogenous expression of NtSOS2 affects the expression of Arabidopsis ion transport-related genes, 10-day-old wild-type and transgenic Arabidopsis plants were placed on MS medium contains 100 mM NaCl for 7 days of continuous salt stress, after which were collected for fluorescence quantitative PCR experiments. Four genes which reported to be related to ion transport: AtSOS1, AtHKT1, AtCAX1, and AtNHX1 were selected to detected the change of expression, and the results showed that under salt stress, the expression levels of these genes were significantly increased in both wild-type and transgenic Arabidopsis (Figure 12). Furthermore, the expression of AtSOS1, AtCAX1, and AtNHX1 under salt stress was higher in transgenic than in wild-type plants, while the expression level of AtHKT1 was slightly decreased in transgenic plants under these conditions (Figure 12).
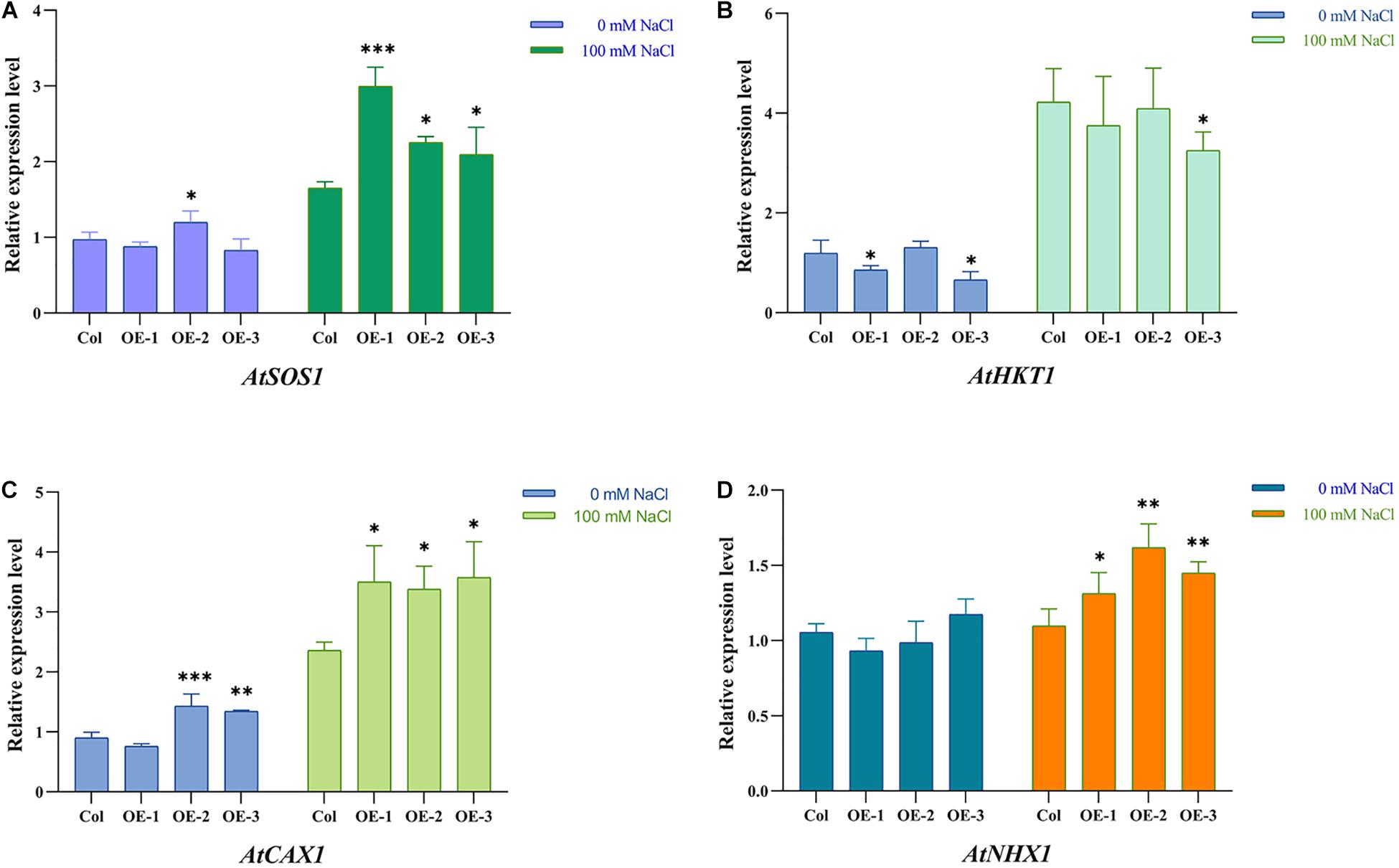
Figure 12. Ectopic expression of NtSOS2 affected Ion transporter-related genes expression. (A–D) Ion transporter-related genes relative quantification. ANOVA test was used for statistical analysis. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Discussion
Soil salinization is a common environmental stress factor that causes soil hardening and soil fertility decline, seriously affecting the germination and growth of plants (Litalien and Zeeb, 2020). More and more cultivated land is suffering from saline-alkali harm, making crop yields decrease (Xun et al., 2015) and reducing the grain surplus. At present, there are several known methods to reduce soil saline-alkalization, among which phytoremediation is one of the better methods with a low cost and no side effects to the environment. Plant mulching is beneficial to slow down the evaporation rate of soil water in saline-alkali land and reduces the accumulation of salt in the soil surface layer. Meanwhile, some saline-living plants can also accumulate salt to slow down or treat salinized soil. Therefore, plant remediation has a good prospect for saline-alkali land remediation.
Long-term saline-alkali stress causes halophytes to adapt and change so that they can grow and reproduce in the harsh saline-alkali land. Therefore, halophytes can be used in saline-alkali land restoration, and are suitable to research the plant salt tolerance mechanism. More and more studies focus on the mechanism of plant salt tolerance from the molecular perspective, but few reports on Nitraria tangutorum have been published.
Genetic engineering is widely used in the study of plant salt tolerance and plays an important role in the study of its molecular mechanism (Wei et al., 2017). Recent studies have shown that SOS2 is a key gene controlling plant salt resistance: SlSOS2 in tomato increases salt tolerance (Baghour et al., 2019), overexpression of PtSOS2 improves tolerance to salt stress in poplar (Yang et al., 2015) and the MdSOS2 gene contributes to the salt tolerance of apple callus and improves the salt tolerance of transgenic Arabidopsis (Hu et al., 2012).
The germination rate of seeds under salt stress is related to the concerning plant’s salt tolerance. Salt stress can increase the seed coat’s permeability, and salt ions may damage the seed’s embryo, one of the main reasons for decreases in germination rate (Deng et al., 2014). In our study, overexpressing NtSOS2 increases germination rate (Figure 5) under salt stress, indicating that ectopic expression of NtSOS2 increases salt tolerance in Arabidopsis.
Salt stress restricts the growth and development of plants and may even cause plant death (Acosta-Motos et al., 2017). Here, we found that exogenous expression of NtSOS2 improves salt stress tolerance in Arabidopsis, as it increases biomass and root growth and reduces leaf mortality (Figures 6, 7). These findings are analogous to and corroborate previous studies on SOS2 gene activity (Wang et al., 2004).
The stress of adversity often leads to the changes of physiological state in plants.
Plants under salt stress often show the phenotype of leaf albinism and reduced chlorophyll (Taïbi et al., 2016). The content of MDA often reflects the degree of lipid peroxidation in the body and indirectly reflects the degree of cell damage (Jan et al., 2017). And also the improvement of antioxidant enzyme activity is an important factor to protect cells under stress (Farhangi-Abriz and Torabian, 2017). In this experiment, the transgenic Arabidopsis showed less chlorophyll loss, higher soluble protein and SOD accumulation under salt stress, and showed lower MDA accumulation (Figures 8, 11). All these indicated that the transgenic lines had better physiological state and stronger salt tolerance ability under salt stress, indicated that exogenous expression of SOS2 could improve salt tolerance of plants under stress.
Such studies have shown that SOS2 plays an important role in maintaining a plant cell’s ion balance. In this study, exogenous expression of NtSOS2 affects the expression of AtSOS1, AtCAX1, and AtNHX1 genes, increasing it significantly in comparison to wild-type Arabidopsis (Figure 12). SOS2 can form a complex with SOS3 to assist the Na+/H+ antiporter SOS1, promoting the efflux of Na+ from plant cells to maintain a balance between ions inside and outside the cell, reducing ion toxicity (Ji et al., 2013). In non-halophytic plants such as Arabidopsis, the SOS2 complex activates the expression of SOS1 to enhance the salt tolerance (Quintero et al., 2002; Lin et al., 2009), and the same effect has been found in halophytic plants such as Thellungiella salsuginea (Oh et al., 2009, 2010). Similarly, the result observed in this experiment are similar to them (Figure 12A) which indicated that SOS pathway is conserved response to salt stress in plant. In addition, the SOS2/SOS3 complex can inhibit the high-affinity K+ transporter HKT1 to reduce the inward transport of Na+ and maintain Na+ homeostasis (Mahajan et al., 2008). Vacuoles can maintain the stability of cellular morphology and are important organelles in the process of plant stress resistance. Under salt stress, CAX1 can promote Ca2+ transport into the vacuole as well as outward H+ transport, while SOS2 can further stimulate this process (Cheng et al., 2004). In addition, SOS2 may interact with CBL10 to activate NHX1 and promote Na+ influx in vacuoles, maintaining their ion balance (Quan et al., 2007). It’s possible that the improved salt tolerance that observed in Arabidopsis overexpressing NtSOS2 exogenously is due to NtSOS2 similarly affecting the activity of ion transporters and maintaining the K+ and Na+ balance in plant cells under salt stress. Further studies will be required to confirm these hypotheses.
Conclusion
In this study, a new gene NtSOS2 was obtained from the halophyte Nitraria tangutorum and some functional verifications were done. Our findings suggest that NtSOS2, like its homologs from other plant species, stimulates salt tolerance. First, NtSOS2 responds to salt stress in Nitraria tangutorum by increasing its expression. Second, overexpression of NtSOS2 in Arabidopsis improves its salt tolerance and increases survival under salt stress. These results indicated that NtSOS2 plays an important role in saline-alkali stress resistance, which is also of considerable significance to further understand Nitraria tangutorum’s mechanism of salt-tolerance. However, more research is needed to determine the molecular mechanism of NtSOS2 function and to fully understand how exactly Nitraria tangutorum copes with salt stress. Such findings should eventually aid in the discovery of new methods to restore overtly saline soil.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JC and TC contributed to the design of this research. LL, YLu, ZH, and JS carried out the statistical analysis. ZL, ML, JH, LZ, and YLi performed the experiments. LZ wrote sections of the manuscript. All authors contributed to manuscript revision and approved the submitted version.
Funding
This research was supported by the Nature Science Foundation of China (Nos. 31770715 and 32071784), Distinguished Professor Project of Jiangsu Province, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). Funding didn’t play a role in the design of the study, nor collection, analysis, interpretation of data, nor in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We wish to thank the editor and reviewers for their helpful comments and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.716855/full#supplementary-material
Supplementary Figure 1 | Agarose gel electrophoresis showing NtSOS2 PCR. The first lanes is 2,000 DNA marker and the second to fifth lanes are NtSOS2 genes from four separate seedings.
Supplementary Figure 2 | Protein domains within the NtSOS2 protein as shown by NCBI CDD (https://www.ncbi.nlm.nih.gov/cdd).
Supplementary Figure 3 | Semi-quantification of NtSOS2 in transgenic Arabidopsis.
Supplementary Figure 4 | Prediction of NtSOS2 subcellular localization by ProtComp9.0 (http://linux1.softberry.com/berry.phtml).
Supplementary Table 1 | Primers for Quantitative RT-PCR.
Supplementary Table 2 | Primers for Subcellular localization. The underlined bases indicated the location of the restriction sites.
Supplementary Table 3 | Primers for constructing overexpression gene vector. The underlined bases indicated the location of the restriction sites.
Supplementary Table 4 | SOS2 gene and its sequence number in different species used in DNAMAN.
Supplementary Table 5 | The CIPK genes used to construct phylogenetic tree in Arabidopsis and rice.
Supplementary Table 6 | Primers for isolation of NtSOS2 fragment.
Abbreviations
SOS, Salt Overly Sensitive; CIPKs, Calcineurin B-like proteins-Interacting protein kinases; NAF, Asn-Ala-Phe; FISL, Phe-Ile-Ser-Leu; ORF, Open Reading Frame; CDS, Coding sequence; Cdna, Complementary DNA; qPCR, Quantitative real-time PCR; OE, Overexpress; HKT1, high-affinity K+ transporter 1 gene; NHX1, Na+/H+ exchange 1 gene; SOS1, Salt Overly Sensitive 1 gene; CAX1, Ca2+/H+ Cation Exchanger 1 gene.
References
Acosta-Motos, J. R., Ortuño, M. F., Bernal-Vicente, A., Diaz-Vivancos, P., Sanchez-Blanco, M. J., and Hernandez, J. A. (2017). Plant responses to salt stress: adaptive mechanisms. Agronomy 7:18. doi: 10.3390/agronomy7010018
Arif, Y., Singh, P., Siddiqui, H., Bajguz, A., and Hayat, S. (2020). Salinity induced physiological and biochemical changes in plants: an omic approach towards salt stress tolerance. Plant Physiol. Biochem. 156, 64–77. doi: 10.1016/j.plaphy.2020.08.042
Baghour, M., Gálvez, F. J., Sánchez, M. E., Aranda, M. N., Venema, K., and Rodríguez-Rosales, M. P. (2019). Overexpression of LeNHX2 and SlSOS2 increases salt tolerance and fruit production in double transgenic tomato plants. Plant Physiol. Biochem. 135, 77–86. doi: 10.1016/j.plaphy.2018.11.028
Batelli, G., Verslues, P. E., Agius, F., Qiu, Q., Fujii, H., Pan, S., et al. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell. Biol. 27, 7781–7790. doi: 10.1128/MCB.00430-07
Chen, X., Ding, Y., Yang, Y., Song, C., Wang, B., Yang, S., et al. (2021). Protein kinases in plant responses to drought, salt, and cold stress. J. Integrat. Plant Biol. 63, 53–78. doi: 10.1111/jipb.13061
Cheng, N., Pittman, J. K., Zhu, J., and Hirschi, K. D. (2004). The protein kinase SOS2 activates the Arabidopsis H+/Ca2 + antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 279, 2922–2926. doi: 10.1074/jbc.M309084200
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cuevas, J., Daliakopoulos, I. N., Del Moral, F., Hueso, J. J., and Tsanis, I. K. (2019). A review of soil-improving cropping systems for soil salinization. Agronomy 9:295. doi: 10.3390/agronomy9060295
Deng, Y., Yuan, F., Feng, Z., Ding, T., Song, J., and Wang, B. (2014). Comparative study on seed germination characteristics of two species of Australia saltbush under salt stress. Acta Ecol. Sin. 34, 337–341. doi: 10.1016/j.chnaes.2013.07.011
Farhangi-Abriz, S., and Torabian, S. (2017). Antioxidant enzyme and osmotic adjustment changes in bean seedlings as affected by biochar under salt stress. Ecotoxicol. Environ. Safety 137, 64–70. doi: 10.1016/j.ecoenv.2016.11.029
Fujii, H., and Zhu, J. K. (2009). An autophosphorylation site of the protein kinase SOS2 is important for salt tolerance in arabidopsis. Mol. Plant 2, 183–190. doi: 10.1093/mp/ssn087
Hu, D., Li, M., Luo, H., Dong, Q., Yao, Y., You, C., et al. (2012). Molecular cloning and functional characterization of MdSOS2 reveals its involvement in salt tolerance in apple callus and Arabidopsis. Plant Cell Rep. 31, 713–722. doi: 10.1007/s00299-011-1189-5
Jan, A. U., Hadi, F., Nawaz, M. A., and Rahman, K. (2017). Potassium and zinc increase tolerance to salt stress in wheat (Triticum aestivum L.). Plant Physiol. Biochem. 116, 139–149. doi: 10.1016/j.plaphy.2017.05.008
Jez, J. M., Lee, S. G., and Sherp, A. M. (2016). The next green movement: plant biology for the environment and sustainability. Science 353, 1241–1244. doi: 10.1126/science.aag1698
Ji, H., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., and Li, X. (2013). The salt overly sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6, 275–286. doi: 10.1093/mp/sst017
Kaur, C., Kumar, G., Kaur, S., Ansari, M. W., Pareek, A., Sopory, S. K., et al. (2015). Molecular cloning and characterization of salt overly sensitive gene promoter from Brassica juncea (BjSOS2). Mol. Biol. Rep. 42, 1139–1148. doi: 10.1007/s11033-015-3851-4
Liang, W., Ma, X., Wan, P., and Liu, L. (2018). Plant salt-tolerance mechanism: a review. Biochem. Biophys. Res. Commun. 495, 286–291. doi: 10.1016/j.bbrc.2017.11.043
Lichtenthaler, H. K., and Wellburn, A. R. (1983). Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 11, 591–592. doi: 10.1042/bst0110591
Lin, H., Yang, Y., Quan, R., Mendoza, I., Wu, Y., Du, W., et al. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21, 1607–1619. doi: 10.1105/tpc.109.066217
Litalien, A., and Zeeb, B. (2020). Curing the earth: a review of anthropogenic soil salinization and plant-based strategies for sustainable mitigation. Sci. Total Environ. 698:134235. doi: 10.1016/j.scitotenv.2019.134235
Liu, J., Ishitani, M., Halfter, U., Kim, C., and Zhu, J. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 97, 3730–3734. doi: 10.1073/pnas.060034197
Liu, W., Zhang, Y., Yuan, X., Xuan, Y., Gao, Y., and Yan, Y. (2016). Exogenous salicylic acid improves salinity tolerance of Nitraria tangutorum. Russ. J. Plant Physiol. 63, 132–142. doi: 10.1134/S1021443716010118
Liu, Z., Mei, L., Wang, Q., Shao, Y., and Tao, Y. (2014). Optimization of subcritical fluid extraction of seed oil from Nitraria tangutorum using response surface methodology. LWT Food Sci. Technol. 56, 168–174. doi: 10.1016/j.lwt.2013.10.048
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, L., Chen, X., Zhu, L., Li, M., Zhang, J., Yang, X., et al. (2020). NtCIPK9: a Calcineurin B-like protein-interacting protein kinase from the halophyte Nitraria tangutorum, enhances Arabidopsis salt tolerance. Front. Plant Sci. 11:1112. doi: 10.3389/fpls.2020.01112
Ma, X., Li, Q., Yu, Y., Qiao, Y., and Gong, Z. (2020). The CBL–CIPK pathway in plant response to stress signals. Int. J. Mol. Sci. 21:5668. doi: 10.3390/ijms21165668
Ma, Y., Cao, J., Chen, Q., He, J., Liu, Z., Wang, J., et al. (2019). The kinase CIPK11 functions as a negative regulator in drought stress response in Arabidopsis. Int. J. Mol. Sci. 20:2422. doi: 10.3390/ijms20102422
Mahajan, S., Pandey, G. K., and Tuteja, N. (2008). Calcium- and salt-stress signaling in plants: shedding light on SOS pathway. Arch. Biochem. Biophys. 471, 146–158. doi: 10.1016/j.abb.2008.01.010
Martínez-Atienza, J., Jiang, X., Garciadeblas, B., Mendoza, I., Zhu, J., Pardo, J. M., et al. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143, 1001–1012. doi: 10.1104/pp.106.092635
Nebenführ, A. (2014). Identifying subcellular protein localization with fluorescent protein fusions after transient expression in onion epidermal cells. Methods Mol. Biol. 1080, 77–85. doi: 10.1007/978-1-62703-643-6_6
Oh, D., Dassanayake, M., Haas, J. S., Kropornika, A., Wright, C., d’Urzo, M. P., et al. (2010). Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiol. 154, 1040–1052. doi: 10.1104/pp.110.163923
Oh, D., Zahir, A., Yun, D., Bressan, R. A., and Bohnert, H. J. (2009). SOS1 and halophytism. Plant Signal. Behav. 4, 1081–1083. doi: 10.4161/psb.4.11.9786
Qiu, Q., Guo, Y., Quintero, F. J., Pardo, J. M., Schumaker, K. S., and Zhu, J. K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279, 207–215. doi: 10.1074/jbc.M307982200
Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., et al. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. The Plant Cell 19, 1415–1431. doi: 10.1105/tpc.106.042291
Quintero, F. J., Ohta, M., Shi, H., Zhu, J., and Pardo, J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 99, 9061–9066. doi: 10.1073/pnas.132092099
Rui, M., and Ricardo, S. (2017). Soil salinity: effect on vegetable crop growth. management practices to prevent and mitigate soil salinization. Horticulturae 3:13. doi: 10.3390/horticulturae3020030
Shrivastava, P., and Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Singh, A. (2021). Soil salinization management for sustainable development: a review. J. Environ. Manag. 277:111383. doi: 10.1016/j.jenvman.2020.111383
Taïbi, K., Taïbi, F., Abderrahim, L. A., Ennajah, A., Belkhodja, M., and Mulet, J. M. (2016). Effect of salt stress on growth, chlorophyll content, lipid peroxidation and antioxidant defence systems in Phaseolus vulgaris L. South Afr. J. Bot. 105, 306–312. doi: 10.1016/j.sajb.2016.03.011
Tang, R., Wang, C., Li, K., and Luan, S. (2020). The CBL–CIPK calcium signaling network: unified paradigm from 20 years of discoveries. Trends Plant Sci. 25, 604–617. doi: 10.1016/j.tplants.2020.01.009
Tian, F., Hou, M., Qiu, Y., Zhang, T., and Yuan, Y. (2020). Salinity stress effects on transpiration and plant growth under different salinity soil levels based on thermal infrared remote (TIR) technique. Geoderma 357:113961. doi: 10.1016/j.geoderma.2019.113961
Van Zelm, E., Zhang, Y., and Testerink, C. (2020). Salt tolerance mechanisms of plants. Ann. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Villalobo, A., and Berchtold, M. W. (2020). The role of calmodulin in tumor cell migration, invasiveness, and metastasis. Int. J. Mol. Sci. 21:765. doi: 10.3390/ijms21030765
Wang, J., Wu, W., Zuo, K., Fei, J., Sun, X., Lin, J., et al. (2004). Isolation and characterization of a serine/threonine protein kinase SOS2 gene from Brassica napus. Cell. Mol. Biol. Lett. 9, 465–473. doi: 10.1007/s00018-003-3367-2
Wei, D., Zhang, W., Wang, C., Meng, Q., Li, G., Chen, T. H. H., et al. (2017). Genetic engineering of the biosynthesis of glycinebetaine leads to alleviate salt-induced potassium efflux and enhances salt tolerance in tomato plants. Plant Sci. 257, 74–83. doi: 10.1016/j.plantsci.2017.01.012
Xun, F., Xie, B., Liu, S., and Guo, C. (2015). Effect of plant growth-promoting bacteria (PGPR) and arbuscular mycorrhizal fungi (AMF) inoculation on oats in saline-alkali soil contaminated by petroleum to enhance phytoremediation. Environ. Sci. Poll. Res. 22, 598–608. doi: 10.1007/s11356-014-3396-4
Yang, Q., Chen, Z., Zhou, X., Yin, H., Li, X., Xin, X. F., et al. (2009). Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2, 22–31. doi: 10.1093/mp/ssn058
Yang, Y., Tang, R. J., Jiang, C. M., Li, B., Kang, T., Liu, H., et al. (2015). Overexpression of the P t SOS 2 gene improves tolerance to salt stress in transgenic poplar plants. Plant Biotechnol. J. 13, 962–973. doi: 10.1111/pbi.12335
Yip Delormel, T., and Boudsocq, M. (2019). Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 224, 585–604. doi: 10.1111/nph.16088
Zhang, C., Luo, W., Li, Y., Zhang, X., Bai, X., Niu, Z., et al. (2019). Transcriptomic analysis of seed germination under salt stress in two desert sister species (Populus euphratica and P. Pruinosa). Front. Genet. 10:231. doi: 10.3389/fgene.2019.00231
Zhang, H., Yang, B., Liu, W., Li, H., Wang, L., Wang, B., et al. (2014). Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 14:8. doi: 10.1186/1471-2229-14-8
Zhang, Y., Linghu, J., Wang, D., Liu, X., Yu, A., Li, F. T., et al. (2017). Foxtail millet CBL4 (SiCBL4) interacts with SiCIPK24, modulates plant salt stress tolerance. Plant Mol. Biol. Rep. 35, 634–646. doi: 10.1007/s11105-017-1051-1
Zheng, L. L., Gao, Z., Wang, J., Zhang, H. R., and Wang, Y. C. (2014). Molecular cloning and functional characterization of a novel CBL-interacting protein kinase NtCIPK2 in the halophyte Nitraria tangutorum. Genet. Mol. Res. 13, 4716–4728. doi: 10.4238/2014.July.2.1
Keywords: salt stress, SOS2, salt overly sensitive pathway, Nitraria tangutorum, halophyte plant
Citation: Zhu L, Li M, Huo J, Lian Z, Liu Y, Lu L, Lu Y, Hao Z, Shi J, Cheng T and Chen J (2021) Overexpression of NtSOS2 From Halophyte Plant N. tangutorum Enhances Tolerance to Salt Stress in Arabidopsis. Front. Plant Sci. 12:716855. doi: 10.3389/fpls.2021.716855
Received: 29 May 2021; Accepted: 09 August 2021;
Published: 06 September 2021.
Edited by:
Crystal Sweetman, Flinders University, AustraliaReviewed by:
Akhtar Ali, Konkuk University, South KoreaRatna Karan, University of Florida, United States
Copyright © 2021 Zhu, Li, Huo, Lian, Liu, Lu, Lu, Hao, Shi, Cheng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tielong Cheng, Y2hlbmd0bEBuamZ1LmVkdS5jbg==; Jinhui Chen, Y2hlbmpoQG5qZnUuZWR1LmNu
 Liming Zhu
Liming Zhu Mengjuan Li
Mengjuan Li Junnan Huo
Junnan Huo Ziming Lian
Ziming Lian Yuxin Liu
Yuxin Liu Lu Lu
Lu Lu Ye Lu
Ye Lu Zhaodong Hao
Zhaodong Hao Jisen Shi
Jisen Shi Tielong Cheng
Tielong Cheng Jinhui Chen
Jinhui Chen