- 1Faculty of Science and Natural Resources, Universiti Malaysia Sabah, Sabah, Malaysia
- 2Horticulture Research Centre, Malaysian Agricultural Research and Development Institute (MARDI), Selangor, Malaysia
The microbial diseases cause significant damage in agriculture, resulting in major yield and quality losses. To control microbiological damage and promote plant growth, a number of chemical control agents such as pesticides, herbicides, and insecticides are available. However, the rising prevalence of chemical control agents has led to unintended consequences for agricultural quality, environmental devastation, and human health. Chemical agents are not naturally broken down by microbes and can be found in the soil and environment long after natural decomposition has occurred. As an alternative to chemical agents, biocontrol agents are employed to manage phytopathogens. Interest in lactic acid bacteria (LAB) research as another class of potentially useful bacteria against phytopathogens has increased in recent years. Due to the high level of biosafety, they possess and the processes they employ to stimulate plant growth, LAB is increasingly being recognized as a viable option. This paper will review the available information on the antagonistic and plant-promoting capabilities of LAB and its mechanisms of action as well as its limitation as BCA. This review aimed at underlining the benefits and inputs from LAB as potential alternatives to chemical usage in sustaining crop productivity.
1. Introduction
According to the Food and Agriculture Organization (FAO) of the United Nations (UN), farmers will need to produce 70 to 100 percent more food to meet the demand of the predicted growing population of 9.3 billion people by 2050 (UNDP, 2021). Apart from the growing population, depletion of natural resources, climate change, and the emergence of new pests and diseases are among the several factors that have a negative impact on agricultural production and productivity. Various pests and diseases contribute 20 to 40 percent yearly economic losses in agricultural products by reducing crop output, degrading its quality, and contaminating food with hazardous compounds (Guo et al., 2013). Plant diseases caused by phytopathogens can limit crop production in a vast array of plant species worldwide, resulting in significant annual losses on a global scale. The most common infectious plant diseases are caused by pathogenic organisms such as fungi, bacteria, viruses, protozoa, insects, and parasitic plants. Wilting, spotting (necrosis), mold, pustules, rot, overgrowth (hypertrophy), distortion (mummification), staining (discoloration), and destruction of afflicted tissue are the symptoms of plant diseases (Nazarov et al., 2020).
Plant diseases can be mitigated in a variety of ways, including cultural, physical, chemical, and behavioral practices. Growers typically rely on the use of chemical treatments, such as fungicides and bactericides, which have been in widespread use for more than a century but none of them are sustainable (Malik et al., 2021). On the other hand, the use of these synthetic chemical treatments in agriculture has been linked to causing numerous issues such as the development of resistance in pathogen populations, detrimental impacts on human health, loss of beneficial soil microbes, the introduction of leftover harmful material into the food chain, and decreased in biodiversity of microorganisms (Sindhu et al., 2016). Efforts to find alternatives to chemical treatments have been bolstered by the understanding that this method of treating plant diseases is suboptimal at best or prohibited by regulation. In addition, due to the growing demand for safety and quality of food production, the search to find alternatives with an eco-friendlier approach has become a priority. The widespread use of agrochemicals can be replaced with a method that is less hazardous. Thanks to the development of microbial biocontrol agents. The term “biological control” refers to the practice of reducing the prevalence of plant disease through the application of naturally occurring organisms, such as beneficial microorganisms or their by-products or extracts from plants or animals (Sundin et al., 2016). Their mechanisms of action toward the target pathogen can be varying either directly or indirectly, such as competition, predation, antibiosis, induced host resistance, or by the activity of lytic enzymes. Over the past decades, numerous research has been published on the potential application of beneficial microorganisms as biological control agents (BCAs) against plant pathogenic bacteria, and strains from the genera Pseudomonas, Burkholderia, Streptomyces, Bacillus, and Trichoderma are very well-known for their antimicrobial capability and synthesis of a wide range of bioactive compounds (Alexander and Phin, 2014; Haryadi et al., 2019; Lim et al., 2019; Alexander et al., 2021).
Interest in lactic acid bacteria (LAB) research as another class of potentially useful microorganisms against phytopathogens has increased in recent years. The application of LAB for plant protection and plant growth stimulation first appeared in the 1980s by Visser et al. (1986) and Higa and Kinjo (1989). It has been demonstrated that LAB has the ability to produce compounds that are effective in the management of a wide range of bacterial and fungal phytopathogens (Gajbhiye and Kapadnis, 2016; Daranas et al., 2019; Duha and Abdullah, 2021). Moreover, its long and widespread use history in food processing allows researchers to understand the physiological processes and bioactive substances being produced. This resulted in its being given the generally regarded as safe (GRAS) status with few exceptions (Goldstein et al., 2015), meaning that its use in edible crop cultivation poses no health concerns to humans. This paper will review the available information on the antagonistic and plant-promoting capabilities of LAB and its mechanisms of action as well as its limitation as BCA. This review aimed at underlining the benefits and inputs from LAB as potential alternatives to chemical usage in sustaining crop productivity.
2. Lactic acid bacteria
LAB is a group of Gram-positive, catalase-negative, non-sporulating, facultatively anaerobic, rod-shaped (bacilli) or spherical (cocci) bacteria that include 6 families and 38 genera in the Lactobacillales order (Holzapfel and Wood, 2014). It produces lactic acid (LA) as the primary end product of saccharolytic metabolism (Hayek and Ibrahim, 2013) and has been classified into homofermentative and heterofermentative strains based on their lactic acid (LA) production. Homofermentative LAB converts sugars to lactic acid, while heterofermentative LAB produces lactic acid, ethanol or acetic acid, and carbon dioxide. The most prevalent species include Lactobacillus, Lactococcus, Enterococcus, Streptococcus, Pediococcus, Leuconostoc, Weissella, and Bifidobacterium (Abubakr and Al-Adiwish, 2017). Studies on LAB identification was done long time ago because it was important to know their properties and ensure that the strains were safe to use. Phenotypic and chemical approaches were formerly used to identify LAB. Different types of LAB activity, such as carbohydrate fermentation, hetero- or homofermentative, gas production, motility, and spore production, form the basis for these techniques (Ashmaig et al., 2009). However, LAB identification based on carbohydrate fermentation profiles is imprecise and insufficient to identify closely related strains due to their similar nutritional requirements (Perricone et al., 2014). Therefore, genomic sequencing is the most reliable way to accurately identify bacteria.
Rapid advances in molecular biology in recent years have had far-reaching effects on the discipline of microbiology allowing the use of 16S rDNA gene sequencing techniques for the identification of bacteria, including LAB. These conserved genes exhibit sufficient variation to be regarded as excellent phylogenetic markers, which can be used for identifying organisms down to the genus and species level (Chong et al., 2013; Lo and Chong, 2020). The development of whole genome sequencing (WGS) technology in recent years has also substantially accelerated the development and application of LAB resources. It enables researchers to systematically and comprehensively understand the metabolic characteristics, potential beneficial functions, and application directions of the strains based on the information contained in the whole genomes of LAB (Buron-Moles et al., 2019). In addition, WGS allows the determination of the genetic evolution and classification of LAB in a more accurate manner (Huang et al., 2020). The first complete LAB genome sequence was published in 2001 for the species Lactococcus lactis IL1403 (Bolotin et al., 2001). Since then, 7,055 species of LAB that includes Lactobacillus, Lactococcus, Bifidobacterium, and Streptococcus thermophilus have had genomic data (comprising whole genomes and framework genomes of varying degrees) deposited in the GenBank (Yanwei et al., 2022) enabling for a comprehensive understanding of the industrial application and metabolic characteristics of LAB. Whole genome information can reflect the safety of LAB strains by assessing genes related to drug resistance, virulence, and pathogenicity and determining whether the related genes can be transmitted horizontally (Rodríguez-Serrano et al., 2018; Toropov et al., 2020; Zhou et al., 2021). Furthermore, the genome-scale metabolic model (GSMM) can be reconstructed from whole-genome data to simulate and anticipate how bacteria will behave in each environment and to systematically direct metabolic engineering efforts. The first GSMM of LAB was generated using the genome sequence of Lactococcus lactis IL1403, which effectively predicted and confirmed the minimum medium for strain development and steered the metabolism to enhance diacetyl production (Oliveira et al., 2005). Other successful examples of GSMM in guiding metabolic engineering of microbial improvement in LAB have also been documented (Vinay-Lara et al., 2014; Xu et al., 2015; Kristjansdottir et al., 2019).
2.1. Diversity, abundance, and plant colonization attributes of LAB in plant microbiome
Lactic acid bacteria (LAB) are found virtually everywhere in the natural world, including in the phyllosphere, endosphere, and rhizosphere (Liu et al., 2014). Their abundance varies greatly from one environment to another. The genomic variation in LAB strains is largely attributable to the selective pressure exerted by these settings (McAuliffe, 2018). LAB in the phyllosphere microbiota was discovered to make up a relatively small percentage of the total bacteria detected on plant tissues, ranging between 102 and 104 CFU/g (di Cagno et al., 2013). It has been acknowledged that LAB may live as an endophyte in a variety of crop plants (Suzzi et al., 2018; Filannino et al., 2019) and that it can survive in seeds (Taha et al., 2019). In the rhizosphere, several LAB strains were discovered to exhibit antimicrobial properties (Fhoula et al., 2013). Carbon-richness is a major determinant of LAB abundance and variety in soils (Yanagida et al., 2006; Reyes-Escogido et al., 2010; Ain et al., 2017). Soil acidity may also play a role in the recovery of several halotolerant LAB, which are known to survive and even thrives in arid settings. Among the commonly found genera of the LAB families from plant microbiomes includes Enterococcus, Lactococcus, Lactobacillus, Leuconostoc, Streptococcus, and Weissella (Yu et al., 2020).
Plants are usually thought to be difficult settings for many microbes. The phyllosphere, for instance, is subject to fast shifts in water supply, UV radiation, oxidative stress, and temperature, and can have low nutrient contents. However, many types of plant-associated bacteria such as LAB have adapted to live and thrive on plant microbiomes despite these stressors. Some properties of LAB suggest that these bacteria could also be successful plant colonists. LAB prefers glucose, fructose, and sucrose as carbon sources for fermentative development. These sugars are found in the phyllosphere and can amount up to 12.5 μg g−1 leaf or 38 g/100 ml of floral nectar (Canto and Herrera, 2012). LAB are not known to metabolize hemicellulose, yet genes associated in hemicellulose breakdown such as α-glucuronidase (algA), polysaccharide deacetylase (pda), endoxylanase/endoglucanase (llkf_1370), and acetylesterase (llkf_1374), were found to be upregulated in L. lactis KF147 grown in a lysate of Arabidopsis thaliana leaves (Golomb and Marco, 2015). LAB also produces metabolites that have a direct impact on plant development (e.g. 2,3-butanediol and acetoin) (Sharifi and Ryu, 2018). This was demonstrated for L. plantarum WCFS1, which was found to produce plant hormone-like compounds that interfered with plant root development when incubated under slow growth, substrate-limited conditions (Goffin et al., 2010). These findings imply that, even in the absence of measurable levels of plant growth, LAB may be an important modifier of plant physiology.
A great number of LAB are also able to survive in aerobic (aerotolerant) and low water activity (osmotolerant) environments. They have the ability to eliminate reactive oxygen species (ROS) through the synthesis of NADH oxidases, superoxide dismutase, cytochrome d oxidase or nonenzymatic dismutation of hydrogen peroxide (H2O2) by Mn2+ (Papadimitriou et al., 2016). Osmotolerance in LAB is mostly connected with enhanced transport of compatible solutes and elevated synthesis of protease, chaperones, and peptidoglycan. Exopolysaccharides (EPS) synthesis is also linked to LAB’s ability to survive in environments with limited water (Nwodo et al., 2012). In addition, the ability of LAB to generate antimicrobial substances may also aid in its colonization in the plant microbiomes. Intra-specific genetic and phenotypic variations among LAB also provide more evidence that certain LAB strains are better adapted to plant environments than others (Strafella et al., 2021). Ayad et al. (1999) reported that plant-isolated Lactococcus lactis was discovered to have lower amino acid needs for growth compared to L. lactis isolated from other sources. Comparative genomic analysis of the L. lactis strains KF147 (isolated from mung bean sprouts) and IL1403 (isolated from cheese) revealed that the KF147 genome contains more genes encoding for carbohydrate metabolism (xylan and arabinose degradation), EPS biosynthesis, bacterial defense (nisin biosynthesis), and stress response (Siezen et al., 2010). L. lactis KF147 also grew faster and had a greater final cell density than IL1403 when cultured in Arabidopsis thaliana leaf lysate (Golomb and Marco, 2015).
3. Mechanism of action of LAB in controlling disease and stimulating plant growth
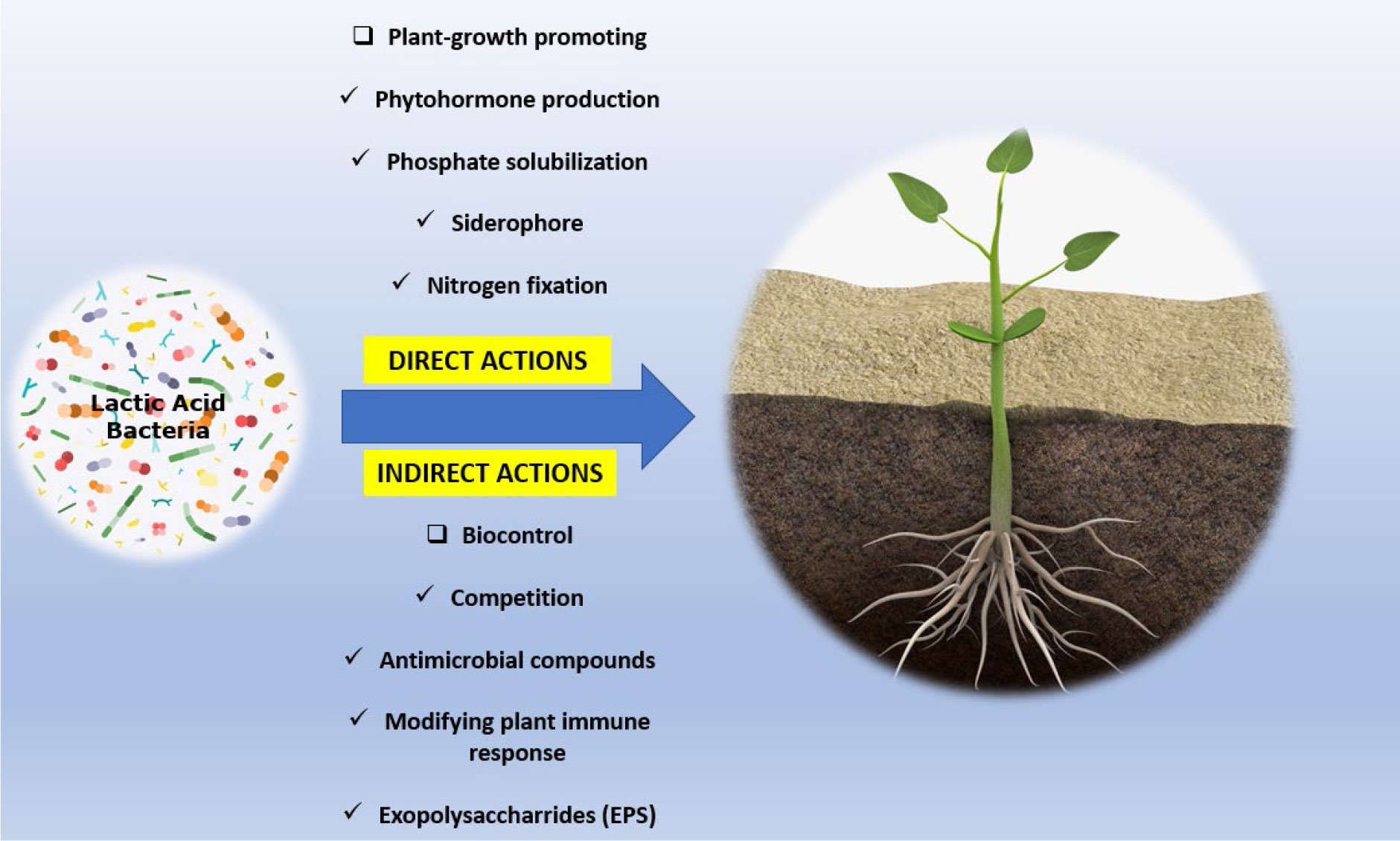
The properties of some LAB species made them the potential candidate for biological control agent (BCA) (Tsuda et al., 2016; Daranas et al., 2019). Despite not having the same reputation as other groups of BCA like Pseudomonas, Burkholderia, Streptomyces, Bacillus, and Trichoderma, several LAB species were reported for the ability to suppress the action of phytopathogens and were also found to stimulate plant growth (Jaini et al., 2022). As illustrated in Figure 1, LAB can help directly with plant disease control and plant growth by modulating the uptake of important nutrients like phosphorus and potassium, fixing nitrogen, and the production of plant hormones and siderophores. Indirectly, LAB could aid in the biocontrol of phytopathogens through the production of a wide range of antimicrobial compounds including diketopiperazines, hydroxy derivatives of fatty acids, 3-phenyllactate; antibacterial bacteriocins and bacteriocin-like inhibitory substances (BLIS) organic acids, hydrogen peroxide, pyrrolidone-5-carboxylic acid, diacetyl, and reuterin (Lamont et al., 2017), modulating defense mechanism by creating systemic resistance, and decreasing pathogen iron availability. It has also been proposed that multiple mechanisms of action might be involved in the attack of LAB against phytopathogens (Sangmanee and Hongpattarakere, 2014).
3.1. Direct mechanisms
3.1.1. Modulating the intake of nutrients and the fixation of nitrogen
Some strains of LAB can boost the availability of nutrients derived from compost and other forms of organic or inorganic matter to plants (Lamont et al., 2017). Phosphorus (P), the major macronutrient for plant growth, is mostly stored in the soil either as an organic compound or as an inorganic precipitate. Similarly, a deficit in potassium (K) which is predominantly contained in fixed form, has detrimental effects on the overall growth and yield of a plant. It was suggested by de Lacerda et al. (2016) that the presence of gene sequences encoding for two types of alkaline phosphatase—the enzymes that catalyze phosphate mineralization—allows L. lactis to solubilize several sources of phosphorus compound. The acidity induced by LAB, which is caused by the synthesis of organic acids, is also responsible for the solubilization of P and K, which then makes these elements available for the plant to absorb. Apart from LAB capabilities to dissolve phosphate, Giassi et al. (2016) reported that some strains of the LAB were also able to fix atmospheric nitrogen for plant consumption. Biological nitrogen fixation (BNF) is a process in which atmospheric N2 is transformed into ammonia and nitrate with the help of the nitrogenase enzyme complex. Higdon et al. (2020) reported that L. lactis isolated from mucilage microbiota of Sierra Mixe maize were recently found as diazotrophs capable of BNF. Molecular functions associated with polysaccharide catabolism, glycan-mediated host adhesion, iron/siderophore utilisation, FeMo cofactor biosynthesis (NifB), and novel oxidoreductase activities were discovered through protein domain analysis of the identified unknown genes in L. lactis, indicating their importance for the BNF trait.
3.1.2. Phytohormones production
Plants and bacteria both generate phytohormones in very low concentrations that can influence plant growth. These phytohormones enhance root hair length and surface area, which improves plant root nutrition and water uptake (Kumar et al., 2022). The increased metabolic activity caused by phytohormones production aids in defence, normal cell function, and abiotic stress management (Khan et al., 2020). Several LAB species are capable of secreting phytohormones such as gibberellin (GA) and auxins such as indole-3-acetic acid (IAA) which play various functions in plant growth promotion (Lamont et al., 2017). According to Turaeva et al. (2021), GA4 and GA7 were detected from the culture fluid of L. plantarum which enhances the plant growth and development of wheat coleoptiles through the usage of HPLC-MS. However, there is still a lack of clarity behind the mechanisms of action.
3.2. Indirect mechanisms
3.2.1. Organic acids
Organic acids have been implicated in several studies as a major mechanism by which LAB exerts its antimicrobial activity against a wide variety of target microorganisms (Tofalo et al., 2016). Lactic acid is the most common LAB metabolite, however other acids such as acetic, propionic, formic, benzoic, and PLA acids are also produced. The antibacterial impact of lactic acid is commonly believed by interfering the membrane functions of the pathogen, inhibition of active transport, reduction of intracellular pH, and inhibition of several metabolic activities, thus killing the target microorganism (Rattanachaikunsopon and Phumkhachorn, 2010). However, the generation of lactic acid and its pH lowering effect are affected by species or strain, culture mix, and growth conditions (Olaoye and Onilude, 2011). Many bacteria, fungi, and yeasts are killed off by the presence of lactic acid in its undissociated form at low pH. The degree to which various microbes are affected by lactic acid can vary widely.
3.2.2. Hydrogen peroxide
Hydrogen peroxide (H2O2) is a reactive oxygen species that is also generated by LAB in the presence of oxygen. Hydrogen peroxide has a powerful oxidizing effect on microbial cells, causing irreparable damage to the fundamental molecular structures of protein involved in cellular metabolism (Sunil and Narayana, 2008). H2O2, once generated, can inhibit the development of both psychotropic and pathogenic microorganisms. Newer research, however, suggests the antimicrobial activity of H2O2 is probably limited, as bacteria only create a small amount, and that its effects are largely in conjunction with other antifungals substances (le Lay et al., 2016).
3.2.3. Bacteriocin
Bacteriocins are ribosomal generated antimicrobial peptides produced by bacteria, can kill or inhibit related or unrelated bacterial strains without harming the bacteria themselves (Yang et al., 2012). Their antimicrobial modes of action are multiples including interference with cell wall development, disruption of the cytoplasmic membrane, suppression of protein synthesis, interference with the replication and transcription of DNA, and interference with the septum formation (Ahmad et al., 2017). Certain members of LAB produce bacteriocins and bacteriocin-like inhibitory substances (BLIS). The majority of LAB bacteriocins are small thermostable or large thermolabile proteins or protein complexes with antibacterial activity against other microbes, although producer cells are immune to their own bacteriocin(s) (Zacharof and Lovitt, 2012). The pH, nutrition sources, and incubation temperature have a significant impact on bacteriocin synthesis. Based on biochemical and genetic characterization, four distinct classes of LAB bacteriocins have been identified: lantibiotics (class 1); small, heat-stable nonlanthionine peptides (class 2); large heat-labile proteins (class 3) and complex bacteriocins with chemical moieties such as lipid and carbohydrate (class 4) (Hernández et al., 2005). The management of bacterial infections in economically important crops may be possible via bacteriocin-mediated resistance in plants, according to recent research by Rooney et al. (2020a); Rooney et al. (2020b).
3.2.4. Reuterin
Several lactobacilli have been demonstrated to produce the glycerol-derived antimicrobial compound reuterin, and its production is stimulated directly or indirectly by the presence of glycerol in anaerobic conditions. LAB do not have an oxidative pathway that would allow them to use glycerol as their primary carbon source. Consequently, LAB must utilize an alternate carbon source in order to breakdown glycerol (Bergsma et al., 2022). Reuterin is a strong inhibitory chemical with broad spectrum activity that is pH independent. It is resistant to proteolytic and lipolytic enzymes and inhibits DNA replication (Singh, 2018). Reuterin has been proven to be effective against a variety of fungus, including several species of Fusarium, Penicillium, and Aspergillus (Vimont et al., 2019), and has been linked to the prevention of mycotoxin development in fermented foods. It also inhibits the growth of gram-positive and gram-negative bacteria, as well as enteropathogens, yeast, fungi, protozoa, and viruses. (Nes et al., 2011).
3.2.5. Cyclic dipeptides
Cyclic dipeptides or cyclodipeptides (CDPs), also known as 2, 5-diketopiperazines, are the smallest cyclic peptides, and existing data indicate that bacteria produce nearly 90% of CDPs (Mishra et al., 2017). Among the cyclic dipeptides extracted from LAB with known antimicrobial properties are cyclo(Gly-Leu), cyclo(Phe-Pro), cyclo(Phe-OH-Pro), and cyclo(Phe-OH-Pro) (Leu-Leu) (Silpa and Rupachandra, 2022). Due to their stability in many environments (pH, heat, and enzymes), cyclic peptides have received a lot of interest. Cyclo(Gly-Leu) from Lb. plantarum VTT E-78076 was discovered to be an antifungal chemical with antifungal activity against plant fungal pathogens Fusarium avenaceum (Zhao et al., 2017). In spite of its antimicrobial potential, significant research is necessary to know its mode of action and range of uses.
3.2.6. Fatty acids
Antimicrobial effects of hydroxy fatty acids (FAs) have been observed by (Hou and Forman, 2000; Granér et al., 2003), and they are found in a wide variety of organisms, including mammals and plants. 3-OH-FAs are found in bacteria as lipopolysaccharides or poly-hydroxyalkanoic acids. The lipopolysaccharides and polyhydroxyalkanoic acids of LAB have not been reported. Over 90% of all cellular FAs in LAB are saturated and monounsaturated FAs, and these are the ones that have been utilized for classification of distinct LAB. However, Lee et al. (1996) found that various Leuconostoc strains contained 2-hydroxyhexadecanoic acid and 3-hydroxyheptadecanoic acid. Unsaturated FAs can be converted into OH-FAs by LAB (Wanikawa et al., 2002), suggesting metabolic pathways for hydroxylation of FAs, albeit the precise function of 3-OH-FAs in LAB metabolism has yet to be determined. A study by (Sjögren et al., 2003) suggested that the antimicrobial effect of 4 OH-FAs extracted from L. plantarum MiLAB 14 is owing to the compounds’ detergent-like characteristics, which disrupt the structure of the target organisms’ cell membranes. A molecule comparable to the 3-OH-FAs discovered here, cis-9-heptadecenoic acid, partitions efficiently into the lipid bilayers of fungal membranes. This finally causes the cytoplasmic disintegration of fungal cells by increasing membrane permeability and the release of intracellular electrolytes and proteins.
4. Potential role of LAB in plant stress tolerance
4.1. Defending plant against biotic stress
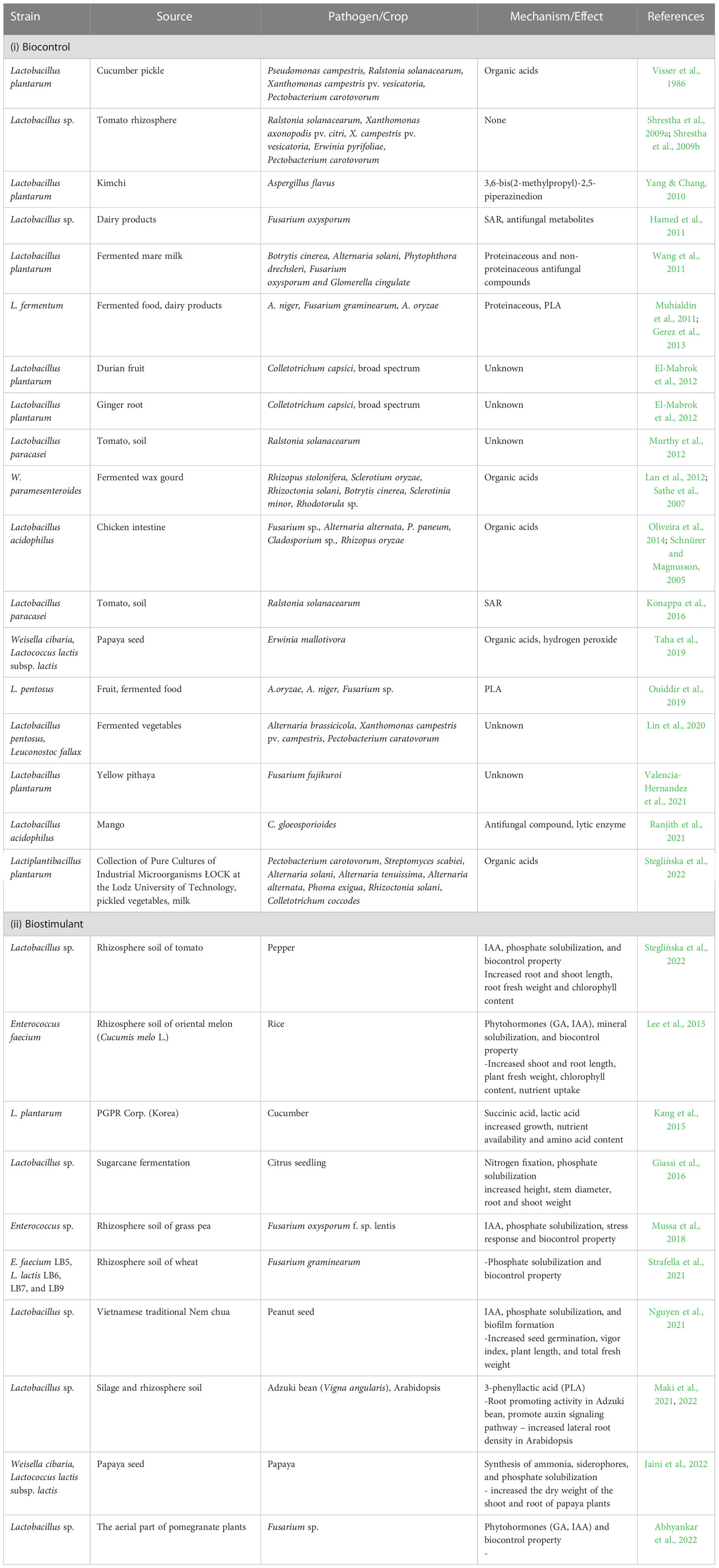
The increase in biotic and abiotic stressors poses a threat to the productivity of crops. Extreme occurrences such as the advent of plant diseases and pests and the effect of climate change are becoming more common around the world. LABs have demonstrated the ability to improve crop development and productivity by developing tolerance traits to various types of stress. These bacteria have a wide range of functional characteristics and can colonize themselves in plant tissues, positively influencing plant development and survival. Numerous studies have looked into the potentiality of LAB in controlling bacterial and fungal phytopathogens from causing destruction to crops (Table 1). For instance, Lactobacillus plantarum and Leuconostoc mesenteroides strains were successfully screened by using in vitro and in planta assays against three bacterial pathogens affecting three different crops, namely Pseudomonas syringae in kiwifruit, Xanthomonas arboricola in Prunus, and Xanthomonas fragariae in strawberry (Daranas et al., 2019). They were selected due to their broad-spectrum activity in preventing all 3 pathogens from infecting their plant hosts. Also, in both semi-field and field studies, the biocontrol performance of the L. plantarum strains was on par with reference controls. The generation of lactic acid and a decrease in pH was partially responsible for the inhibitory mechanism observed in vitro and both strains had comparable rates of survival when placed on leaf surfaces. Similar broad-spectrum inhibition was observed by the species L. paracasei and L. plantarum isolated from wine fermentations (López-Seijas et al., 2019). The LAB strains were not only able to inhibit several food spoilage Gram-positive bacteria but in vitro studies also showed that the LAB strains had a 55-76% effect in preventing the growth of Fusarium oxysporum sp. lycopersici, a phytopathogenic fungus that causes disease in tomatoes. The efficacy of these malolactic LAB strains was very competitive when compared to the previous studies of L. plantarum isolated from different sources (Rouse et al., 2008; Dalié et al., 2010). It has also been shown that the plant-derived Weissella confusa and Pediococcus pentosaceous strains both have broad-range inhibitory action against fungal diseases of fruit crops. (Crowley et al., 2012a; Crowley et al., 2012b).
Steglińska et al. (2022) reported LAB screening of ten phytopathogens related to potato including Pectobacterium carotovorum, Fusarium oxysporum and Rhizoctonia solani, showed a 40-90% disease reduction except for Fusarium oxysporum and Fusarium sambucinum which were not inhibited by the LAB, Lactiplantibacillus plantarum KB2 LAB 03. In the metabolic profiles of the LAB strains, the most abundant compounds were found to be from organic acids and ethanol. Zebboudj et al. (2020) indicated that L. lactis subsp. diacetylactis were able to inhibit Fusarium species of tomato crown and root rot up to 62.42% on MRS agar medium. Another in vitro assessment done by Valencia-Hernandez et al. (2021) showed biomass fraction of Lactobacillus plantarum isolated from yellow pithaya inhibit Fusarium fujikuroi growth by 100% over 48 hr of fermentation. In another finding by Lin et al. (2020), Lactobacillus pentosus and Leuconostoc fallax recovered from fermented vegetables in combination with chitosan present a powerful inhibitory effect against three cruciferous vegetable diseases, including cabbage black spot caused by Alternaria brassicicola, black rot caused by Xanthomonas campestris, and soft rot caused by Pectobacterium caratovorum. The LAB/chitosan mixture is also antagonistic against Colletotrichum higginsianum, Sclerotium rolfsii, and Fusarium oxysporum f. rapae, suggesting a broad-spectrum activity of LAB/chitosan. Futhemore, as indicating by the experiment numerous applications are more successful than a single application. A considerable reduction in the severity of the papaya dieback disease was seen after the application of the LAB combination, Weisella cibaria and Lactococcus lactis in nurseries (Taha et al., 2019). Lactobacillus acidophilus which was isolated from mango (Mangifera indica L.) had an inhibitory action of more than 40% against post-harvest anthracnose caused by C. gloeosporioides. Evaluation in vitro demonstrated that the isolates produced antifungal chemicals as well as lytic enzymes as a mechanism of antagonism against the fungus (Fenta and Kibret, 2021).
LAB has been demonstrated in studies to have a variety of plant growth-stimulating properties to enhance the availability of nutrients to its host plants (Table 1), allowing them to deal with stress and inhibit plant nematodes (Amprayn et al., 2016; Ibrahim et al., 2022). According to research by Strafella et al. (2021), sixteen LAB strains evaluated in experimental settings were able to solubilize a significant amount of phosphate, and the findings corresponded to strains of Enterococcus sp. isolated by Mussa et al. (2018). Plant development can be promoted directly by increasing mineral and nutrient intake or indirectly by modulating plant hormones such as indole-3-acetic acid (IAA), cytokinin, and ethylene. In this case, LAB has also been demonstrated to produce indole-3-acetic acid (IAA), plant hormones that stimulate the rapid development of plants (Mohite, 2013). Three isolates from the aerial sections of the pomegranate plant that were identified as Leuconostoc sp. and Lactobacillus sp. were tested for plant growth-boosting properties by Abhyankar et al. (2022). Besides demonstrating antifungal activity against Fusarium sp., isolates of Lactobacillus sp. also exhibited 1-Aminocyclopropane 1-carboxylic acid (ACC) deaminase activity, with LAB isolate GYP3 exhibiting the highest level. This enzyme is necessary to reduce ethylene to non-toxic levels in order to protect the plants. It was also discovered that the isolate GYP3 produced indole-3-acetic acid (IAA) and Gibberellin, both of which aid in root elongation and flowering. Exopolysaccharide (EPS) was also produced by all three isolates. An oriental melon (Cucumis melo L.) rhizosphere LAB strain, Enterococcus faecium LKE12, was investigated in gibberellin (GA)-deficient rice dwarf mutant (waito-C) and a normal GA biosynthetic rice cultivar for its plant growth-promoting capacity (Hwayongbyeo) (Lee et al., 2015). Both regular and dwarf rice cultivars benefited greatly from E. faecium LKE12’s ability to secrete a wide variety of GA and IAA, which increased the shoot length and biomass of the plants, indicating a beneficial interaction between E. faecium LKE12 and plants. Isolates of Lactobacillus spp. L5, L3, and L2N found in traditional Vietnamese Nem chua exhibited Indole-acetic acid (IAA) synthesis, P-solubilization, and biofilm development (Nguyen et al., 2021). Peanut seed treatment with the same mixed bacterial cultures improved seed germination and vigor index when compared to untreated control seeds and those treated with fungicide. Those that were treated with LAB grew in both height and total fresh weight by 22.4% and 99.6%. Greenhouse and field evaluation by Shrestha et al. (2014) reported that due to its ability to secrete a significant amount of IAA, LAB strains KLF01 and KPD03 outperformed LAB strain KLC02 in terms of growth promotion, whereas KLC02 outperformed KLF01 and KPD03 in the field. Environmental conditions, root colonization, competition, and the synthesis of antagonistic metabolites are just some of the abiotic and biotic elements that could explain why greenhouse and field testing produce different results. Growth-promoting effects of several other LABs were also observed on cucumber and tomato seedlings (Lutz et al., 2012).
Effective Microorganisms (EM) consortiums are known to consist yeast, mould fungus, LAB, photosynthetic bacteria, actinomycetes, and other microorganisms. Compost incorporated with EM has been found to boost yields and nutrient uptake in a variety of crops (Javaid, 2011; Lamont et al., 2017). Fermented compost products based on lactic acid bacteria also improve soil fertility, soil structure, and aeration, neutralize alkalinity, and enhance moisture retention. Bokashi fertilizer, a traditional type of fertilizer often used in Japan, is an example of this EM-inoculated compost. Maki et al. (2021) identified 3-phenyllactic acid (PLA), a root-promoting compound from Bokashi fertilizer. PLA is a significant organic acid generated by many bacteria, particularly LAB, as a catabolic result of phenylalanine via phenylpyruvic acid (PPA) and has been shown to be biologically active as a plant root promoter. Recent study by Maki et al. (2022) found that PLA stimulated the auxin signalling system and influenced root development in Arabidopsis. PLA promoted lateral root density while decreasing primary root growth in Arabidopsis and elevated the expression of the auxin response marker gene IAA19 in roots. PLA’s auxin-like activity was clearly reduced in the auxin signalling mutant, tir1-1 afb2, indicating that PLA regulates root development via the auxin signalling pathway. In a pot experiment by Javaid (2011), adding lactic acid bacteria to farmyard manure boosted rice (Oryza sativa L.) root and shoot growth, but not in NPK-amended soil. Lactococcus lactis isolated from organic soil was also found to promote plant growth in cabbage. (Somers et al., 2007). Previously, it was believed that LAB had almost minimal iron (Fe) requirement and did not produce siderophores. However, the genomes of two vegetable-isolated Lactococcus lactis strains isolated by Shrestha et al. (2014) revealed non-ribosomal peptide pathways, indicating the ability of LABs to produce siderophores. Further research by Jaini et al. (2022) revealed the synthesis of ammonia and siderophores, as well as the solubilization of phosphate, resulting in an increase in the dry weight of the shoot and root of papaya plants by the endophytic LAB identified in the work of Taha et al. (2019).
4.2. Alleviating abiotic stress in plant
Plant development can be stunted by a variety of abiotic stresses, such as flooding, dehydration, high temperatures or salt levels, the presence of toxic metals, and exposure to organic pollutants. Under abiotic stimuli, the intracellular redox balance of plants is upset which results in the production of reactive oxygen species (ROS). As a result, the plant’s enzymatic and non-enzymatic antioxidants are activated to withdraw the effects of ROS. In a condition of drought or dehydration, plant biosynthesis of nitric oxide (NO) increases in order to reduce the effects of oxidative stress. It has been observed that root treatment of wheat seedlings with L. plantarum 8P-A3 managed to alleviate oxidative stress during dehydration (Yarullina et al., 2014). Increases in total integral antioxidant capacity (IAC) and catalase activity indicate that NO has a role in the stress-limiting activities of lactobacilli by mitigating the deleterious effects of dehydration. Excessive salt in the soil causes ion imbalance and toxicity in plants. Plants respond to salinity stress by synthesizing polyamines and osmolytes, activating defense systems, blocking the deposition of reactive oxygen species, and controlling the transfer of ions. To counter salt-induced oxidative stress, Swertia chirayita inoculated with L. plantarum demonstrates better salinity stress tolerance by adopting more energy-efficient defensive mechanisms and efficiently partitioning carbon flow between primary and secondary metabolism (Phoboo et al., 2016). Despite the complexity of plant stress response networks still not being fully understood, LAB treatment can somehow manage to improve the stress response of plant.
5. Limitations, challenges, and the way forward
Similar to other types of BCAs, LAB also has its limitations and challenges when it comes to application. Currently, evidence linking LAB antagonism in vitro to actual pathogen control in the field is still scarce. Its basic limitation of use in agricultural applications, as with other kinds of BCA, is the capacity to survive and produce sufficient amounts of bioactive compounds in suitable circumstances. This could be overcome by selecting or designing strains through biotechnology that can flourish in the phytomicrobiome, enhacing cultures with necessary nutrients or protective carriers, and reapplying cultures to maintain a large number of viable cells. Anyhow, these methods are complicated and will take a long time. Transgenic strains with diverse modes of action can be developed using biotechnology to improve strains with desirable features such as simplicity of formulation, stability, or extraordinary suitability for plant colonization. Another alternative is to use a LAB strain more often in places that are better for its growth, like fruits, flowers, and soils with a lot of organic matter. This strategy has been effective in preventing and eradicating floral diseases that affect rosaceous tree crops (Bonaterra et al., 2014) and has promising results against postharvest infections as well (Trias et al., 2008).
The production of bioactive substances could also be accomplished through the use of LAB that has been grown in bioreactors under optimum conditions. Previous studies by Omer et al. (2009) and Limanska et al. (2015a); Limanska et al., 2015b) have shown that the metabolites produced by LABs are responsible for their activity, and the method of isolating and purifying this metabolite has been successfully applied (Maki et al., 2021). Even though LAB can endure a wide range of environmental stresses, they have specific dietary needs to thrive. Researchers have looked into how sugar beet and sweet potato processing wastes can be used to make industrial LAB media (Krzywonos and Eberhard, 2011; Hayek et al., 2013), but more consistent LAB medium is still needed to make industrial LAB culture last longer. It is also important to take precautions when planning the establishment of mixed consortia LAB with other PGPM groups to prevent incompatibilities. Nanomaterials, which have been effectively used in industries like energy, medicine, and electronics, are a newer avenue of nanotechnology being investigated and implemented in agriculture (Cruz-Luna et al., 2021). Successful applications of metal nanoparticles (M-NPs) such as silver (Ag), iron (Fe), copper (Cu), zinc (Zn), and selenium (Se) have been reported in the suppression of several phytopathogens as well as promoting plant growth in agriculture (Consolo et al., 2020; Akpinar et al., 2021). However, the chemical and physical processes utilized to create M-NPs can be both expensive and potentially hazardous to human health and the environment. As a result, ‘green’ synthesis is leading the way in this emerging discipline by exploring the viability of microorganisms and plants as nanofactories. Green synthesis of M-NPs has the benefits of being environmentally friendly, cost-effective, non-toxic, quick and reliable, stable and sustainable, with low polydispersity, scalability, and biocompatibility. Several studies have recently brought attention to the promising nanobiotechnological applications of LAB in the synthesis of intracellular and extracellular M-NPs (Alam et al., 2020; Aziz Mousavi et al., 2020; Ghosh et al., 2022). This will pave the way for further research into the role of this bacterial group in facilitating plant growth and controlling phytopathogens.
Despite its history of safe usage and “GRAS” status, the safety of the chosen LAB must be assured before industrial application to prevent having an impact on the biodiversity of the ecosystem or causing diseases in humans, animals, or plants. For example, Linares-Morales et al. (2020) reported encouraging results of E. faecium against post-harvest pathogens, however, further evaluation of the strains’ safety is necessary as some of the Enterococcus strains can potentially carry harmful genes (Venegas-Ortega et al., 2020). The increased efficiency of genome analysis over the past decade allows screening on the safety of LAB strains by assessing genes related to drug resistance, virulence, and pathogenicity and determining whether the related genes can be transmitted horizontally (Rodríguez-Serrano et al., 2018; Toropov et al., 2020; Zhou et al., 2021).
6. Conclusion
LAB strains can stimulate crop production in a number of ways, including functioning as a BCA, increasing the availability of nutrients, mitigating the effects of biotic and abiotic stressors, and stimulating plant growth directly. Its GRAS status and extensive history in food research make them ideal for use in crop protection. Although LABs are ubiquitous in the phytomicrobiome, their potential roles as BCAs and promoters of plant development have been generally disregarded. Evidence from the past and present points to the fact that LAB has the ability to serve as renewable and safe agricultural inputs that can aid in the control of plant diseases and the promotion of plant growth. However, more LAB studies are needed and should focus on its biocontrol efficiency under field conditions as well as LAB bioproduction and formulations. The integration of LAB as biocontrol agents that could be used with other biocontrol techniques in an integrated control program would be a viable way to increase efficacy against phytopathogens and help solve the challenges to achieving sustainable food security.
Author contributions
Conceptualization, NJ and KC. Methodology, NJ and KC. Validation, RJ and KP. formal analysis, NJ. Investigation, NJ. Resources, KC. Writing—original draft NJ. Writing—review and editing, RJ and KC. Visualization, NJ & KC. Supervision, RJ & KC. All authors contributed to the article and approved the submitted version.
Funding
This work is financially supported by Malaysian the Ministry of Higher Education (MoHE) under the Fundamental Research Grant Scheme (FRGS) FRGS/1/2022/WAB04/UMS/01/1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abhyankar, P. S., Gunjal, A. B., Kapadnis, B. P., Ambade, S. V. (2022). Potential of lactic acid bacteria in plant growth promotion. Bhartiya Krishi Anusandhan Patrika 36, 326–329. doi: 10.11648/j.ijmb.20170202.12
Abubakr, M. A. S., Al-Adiwish, W. M. (2017). Isolation and identification of lactic acid bacteria from different fruits with proteolytic activity. Int. J. Microbiol. Biotechnol. 2, 58–64. doi: 10.11648/j.ijmb.20170202.12
Ahmad, V., Khan, M. S., Jamal, Q. M. S., Alzohairy, M. A., al Karaawi, M. A., Siddiqui, M. U. (2017). Antimicrobial potential of bacteriocins: in therapy, agriculture and food preservation. Int. J. Antimicrob. Agents 49, 1–11. doi: 10.1016/J.IJANTIMICAG.2016.08.016
Ain, N., Bakar, A., Hariz, M., Rahman, A., Shakri, N. A. (2017). Malaysian Journal of microbiology identification and characterization of lactic acid bacteria isolated from fruit tree rhizosphere in MARDI, Malaysia. Malays J. Microbiol. 13, 61–66 doi: 10.21161/mjm.79715
Akpinar, I., Unal, M., Sar, T. (2021). Potential antifungal effects of silver nanoparticles (AgNPs) of different sizes against phytopathogenic fusarium oxysporum f. sp. radicis-lycopersici (FORL) strains. SN Appl. Sci. 3, 1–9. doi: 10.1007/S42452-021-04524-5/TABLES/2
Alam, H., Khatoon, N., Khan, M. A., Husain, S. A., Saravanan, M., Sardar, M. (2020). Synthesis of selenium nanoparticles using probiotic bacteria lactobacillus acidophilus and their enhanced antimicrobial activity against resistant bacteria. J. Clust Sci. 31, 1003–1011. doi: 10.1007/s10876-019-01705-6
Alexander, A., Khai, R., Lo, S., Chong, K. P. (2021). The effectiveness of selected biological control agents in controlling ganoderma boninense. Penerbit UMT J. Sustainability Sci. Manage. 16, 128–137. doi: 10.46754/jssm.2021.08.011
Alexander, A., Phin, C. K. (2014). Combination of biological agents in suppressing colonization of ganoderma boninense of basal stem rot. American-Eurasian J. Sustain. Agric. 8, 1–7.
Amprayn, K.-O., Supawong, V., Kengkwasingh, P., Getmala, A. (2016). “Plant growth promoting traits of lactic acid bacterium isolated from rice rhizosphere and its effect on rice growth,” in Proceedings of the Burapha University International Conference 2016, 28-29 July 2016, Bangsaen, Chonburi, Thailand, 28–29.
Ashmaig, A., Hasan, A., Gaali, E. (2009). Identification of lactic acid bacteria isolated from traditional Sudanese fermented camel’s milk (Gariss). Afr. J. Microbiol. Res. 3, 451–457.
Ayad, E. H. E., Verheul, A., de Jong, C., Wouters, J. T. M., Smit, G. (1999). Flavour forming abilities and amino acid requirements of lactococcus lactis strains isolated from artisanal and non-dairy origin. Int. Dairy J. 9, 725–735. doi: 10.1016/S0958-6946(99)00140-5
Aziz Mousavi, S. M. A., Mirhosseini, S. A., Rastegar Shariat Panahi, M., Mahmoodzadeh Hosseini, H. (2020). Characterization of biosynthesized silver nanoparticles using lactobacillus rhamnosus GG and its In vitro assessment against colorectal cancer cells. Probiotics Antimicrob. Proteins 12, 740–746. doi: 10.1007/s12602-019-09530-z
Bergsma, S., Jan, G., Euverink, W., Charalampogiannis, N., Poulios, E., Janssens, T. K. S., et al. (2022). Biotechnological and medical aspects of lactic acid bacteria used for plant protection: A comprehensive review. BioTech 11, 40. doi: 10.3390/BIOTECH11030040
Bolotin, A., Wincker, P., Mauger, S., Jaillon, O., Malarme, K., Weissenbach, J., et al. (2001). The complete genome sequence of the lactic acid bacterium lactococcus lactis ssp. lactis IL1403. Genome Res. 11, 731–753. doi: 10.1101/GR.169701
Bonaterra, A., Cabrefiga, J., Mora, I., Roselló, G., Francés, J., Montesinos, E. (2014). Gram-positive bacteria producing antimicrobial peptides as efficient biocontrol agents of fire blight. Acta Hortic. 1056, 117–122. doi: 10.17660/ACTAHORTIC.2014.1056.1
Buron-Moles, G., Chailyan, A., Dolejs, I., Forster, J., Mikš, M. H. (2019). Uncovering carbohydrate metabolism through a genotype-phenotype association study of 56 lactic acid bacteria genomes. Appl. Microbiol. Biotechnol. 103, 3135–3152. doi: 10.1007/S00253-019-09701-6/FIGURES/5
Canto, A., Herrera, C. M. (2012). Micro-organisms behind the pollination scenes: microbial imprint on floral nectar sugar variation in a tropical plant community. Ann. Bot. 110, 1173–1183. doi: 10.1093/AOB/MCS183
Chong, K. P., Abdullah, S., Ng, T. L. (2013). Molecular fingerprint of ganoderma spp. from sabah, Malaysia. Int. J. Agric. Biol. 15, 1112–1118.
Consolo, V. F., Torres-Nicolini, A., Alvarez, V. A. (2020). Mycosinthetized Ag, CuO and ZnO nanoparticles from a promising trichoderma harzianum strain and their antifungal potential against important phytopathogens. Sci. Rep. 1 (10), 1–9. doi: 10.1038/s41598-020-77294-6
Crowley, S., Mahony, J., van Sinderen, D. (2012a). Broad-spectrum antifungal-producing lactic acid bacteria and their application in fruit models. Folia Microbiologica 4 (58), 291–299. doi: 10.1007/S12223-012-0209-3
Crowley, S., Mahony, J., van Sinderen, D. (2012b). Comparative analysis of two antifungal lactobacillus plantarum isolates and their application as bioprotectants in refrigerated foods. J. Appl. Microbiol. 113, 1417–1427. doi: 10.1111/JAM.12012
Cruz-Luna, A. R., Cruz-Martínez, H., Vásquez-López, A., Medina, D. I. (2021). Metal nanoparticles as novel antifungal agents for sustainable agriculture: Current advances and future directions. J. Fungi 7, 1033. doi: 10.3390/jof7121033
Dalié, D. K. D., Deschamps, A. M., Richard-Forget, F. (2010). Lactic acid bacteria – potential for control of mould growth and mycotoxins: A review. Food Control 21, 370–380. doi: 10.1016/J.FOODCONT.2009.07.011
Daranas, N., Roselló, G., Cabrefiga, J., Donati, I., Francés, J., Badosa, E., et al. (2019). Biological control of bacterial plant diseases with lactobacillus plantarum strains selected for their broad-spectrum activity. Ann. Appl. Biol. 174, 92–105. doi: 10.1111/aab.12476
de Lacerda, J. R. M., da Silva, T. F., Vollú, R. E., Marques, J. M., Seldin, L. (2016). Generally recognized as safe (GRAS) lactococcus lactis strains associated with lippia sidoides cham. are able to solubilize/mineralize phosphate. Springerplus 5, 1–7. doi: 10.1186/s40064-016-2596-4
di Cagno, R., Coda, R., de Angelis, M., Gobbetti, M. (2013). Exploitation of vegetables and fruits through lactic acid fermentation. Food Microbiol. 33, 1–10. doi: 10.1016/j.fm.2012.09.003
Duha, F. A., Abdullah, A. H. (2021). “Isolation and identification of ssome lactobacillus spp. bacteria and evaluation their efficacy in the management of damping off disease on peas,” in Proceedings of IOP Conf. Series: Earth and Environmental Science, Fourth International Conference for Agricultural and Sustainability Sciences 4-5 October 2021, Babil, Iraq. 910. doi: 10.1088/1755-1315/910/1/012106
El-Mabrok, A. S. W., Hassan, Z., Mokhtar, A. M., Hussain, K. M. A., Kahar, F. K. S. B. A. (2012). Screening of lactic acid bacteria as biocontrol against (Colletotrichum capsici) on chilli bangi. Res. J. Appl. Sci. 7, 466–473. doi: 10.3923/RJASCI.2012.466.473
Fenta, L., Kibret, M. (2021). Biocontrol potential of lactobacillus spp. against post-harvest mango (Mangifera indica l.) anthracnose disease caused by colletotrichum gloeosporioides. Res. Crops 22, 858–867. doi: 10.31830/2348-7542.2021.141
Fhoula, I., Najjari, A., Turki, Y., Jaballah, S., Boudabous, A., Ouzari, H. (2013). Diversity and antimicrobial properties of lactic acid bacteria isolated from rhizosphere of olive trees and desert truffles of Tunisia. BioMed. Res. Int. 14, 1–14. doi: 10.1155/2013/405708
Filannino, P., di Cagno, R., Tlais, A. Z. A., Cantatore, V., Gobbetti, M. (2019). Fructose-rich niches traced the evolution of lactic acid bacteria toward fructophilic species. Crit. Rev. Microbiol. 45, 65–81. doi: 10.1080/1040841X.2018.1543649
Gajbhiye, M. H., Kapadnis, B. P. (2016). Antifungal-activity-producing lactic acid bacteria as biocontrol agents in plants. Biocontrol Sci. Technol. 26, 1451–1470. doi: 10.1080/09583157.2016.1213793
Gerez, C. L., Torres, M. J., Font de Valdez, G., Rollán, G. (2013). Control of spoilage fungi by lactic acid bacteria. Biol. Control 64, 231–237. doi: 10.1016/J.BIOCONTROL.2012.10.009
Ghosh, S., Sarkar, B., Kaushik, A., Mostafavi, E. (2022). Nanobiotechnological prospects of probiotic microflora: Synthesis, mechanism, and applications. Sci. Total Environ. 838, 156212. doi: 10.1016/J.SCITOTENV.2022.156212
Giassi, V., Kiritani, C., Kupper, K. C. (2016). Bacteria as growth-promoting agents for citrus rootstocks. Microbiol. Res. 190, 46–54. doi: 10.1016/J.MICRES.2015.12.006
Goffin, P., van de Bunt, B., Giovane, M., Leveau, J. H. J., Höppener-Ogawa, S., Teusink, B., et al. (2010). Understanding the physiology of lactobacillus plantarum at zero growth. Mol. Syst. Biol. 6, 413. doi: 10.1038/MSB.2010.67
Goldstein, E. J. C., Tyrrell, K. L., Citron, D. M. (2015). Lactobacillus species: Taxonomic complexity and controversial susceptibilities. Clin. Infect. Dis. 60, S98–S107. doi: 10.1093/cid/civ072
Golomb, B. L., Marco, M. L. (2015). Lactococcus lactis metabolism and gene expression during growth on plant tissues. J. Bacteriol 197, 371. doi: 10.1128/JB.02193-14
Granér, G., Hamberg, M., Meijer, J. (2003). Screening of oxylipins for control of oilseed rape (Brassica napus) fungal pathogens. Phytochemistry 63, 89–95. doi: 10.1016/S0031-9422(02)00724-0
Guo, L., Rasool, A., Li, C. (2013). Antifungal substances of bacterial origin and plant disease management. Bacteria Agrobiol: Dis. Manage. 9783642336393, 473–485. doi: 10.1007/978-3-642-33639-3_18
Hamed, H., Moustafa, Y., Journal, S. A.-A.-L. S. (2011). In vivo efficacy of lactic acid bacteria in biological control against fusarium oxysporum for protection of tomato plant. Life Sci. J. 8, 1097–8135.
Haryadi, D., Sidhu, S. M., Panjaitan, T., Hadi, H., Chong, K. P. (2019). The potential of endophytic trichoderma from oil palm (Elaeis guineensis jacq.) roots of north Sumatra, Indonesia against ganoderma boninense. J. Oil Palm Res. 31, 592–603. doi: 10.21894/jopr.2019.0049
Hayek, S. A., Ibrahim, S. A. (2013). Current limitations and challenges with lactic acid bacteria: A review. Food Nutr. Sci. 04, 73–87. doi: 10.4236/fns.2013.411a010
Hayek, S. A., Shahbazi, A., Awaisheh, S. S., Shah, N. P., Ibrahim, S. A. (2013). Sweet potatoes as a basic component in developing a medium for the cultivation of lactobacilli. Biosci. Biotechnol. Biochem. 77, 2248–2254. doi: 10.1271/BBB.130508
Hernández, D., Cardell, E., Zárate, V. (2005). Antimicrobial activity of lactic acid bacteria isolated from tenerife cheese: initial characterization of plantaricin TF711, a bacteriocin-like substance produced by lactobacillus plantarum TF711. J. Appl. Microbiol. 99, 77–84. doi: 10.1111/J.1365-2672.2005.02576.X
Higa, T., Kinjo, S. (1989). Effect of lactic acid fermentation bacteria on plant growth and soil humus formation. (Okinawa, Japan: University of the Ryukyus)
Higdon, S. M., Huang, B. C., Bennett, A. B., Weimer, B. C. (2020). Identification of nitrogen fixation genes in lactococcus isolated from maize using population genomics and machine learning. Microorganisms 8, 1–25. doi: 10.3390/MICROORGANISMS8122043
Holzapfel, W. H., Wood, B. J. B. (2014). “Introduction to the LAB,” in Lactic acid bacteria (Chichester, UK: John Wiley & Sons, Ltd), 1–12. doi: 10.1002/9781118655252.ch1
Hou, C. T., Forman, R. J. (2000). Growth inhibition of plant pathogenic fungi by hydroxy fatty acids. J. Ind. Microbiol. Biotechnol. 24, 275–276. doi: 10.1038/SJ.JIM.2900816
Huang, C. H., Chen, C. C., Liou, J. S., Lee, A. Y., Blom, J., Lin, Y. C., et al. (2020). Genome-based reclassification of lactobacillus casei: Emended classification and description of the species lactobacillus zeae. Int. J. Syst. Evol. Microbiol. 70, 3755–3762. doi: 10.1099/IJSEM.0.003969/CITE/REFWORKS
Ibrahim, D., Amer, M., El-Shishtawy, H., Elmaghraby, M., Abdellatif, A., El-Deriny, M. (2022). Nematicidal activity of lactic acid bacteria against root-knot nematodes nematicidal activity of lactic acid bacteria against root-knot nematodes. J. Microbiol. Biotechnol. 7, 1–12. doi: 10.23880/oajmb-16000216
Jaini, M. F. M., Roslan, N. F., Yusof, M. T., Saidi, N. B., Ramli, N., Zainudin, N. A. I. M., et al. (2022). Investigating the potential of endophytic lactic acid bacteria isolated from papaya seeds as plant growth promoter and antifungal agent. Pertanika J. Trop. Agric. Sci. 45, 207–233. doi: 10.47836/PJTAS.45.1.12
Javaid, A. (2011). Effects of biofertilizers combined with different soil amendments on potted rice plants. Chil J. Agric. Res. 71, 157. doi: 10.4067/S0718-58392011000100019
Kang, S. M., Radhakrishnan, R., You, Y. H., Khan, A. L., Park, J. M., Lee, S. M., et al. (2015). Cucumber performance is improved by inoculation with plant growth-promoting microorganisms. Acta Agric. Scand. B Soil Plant Sci. 65, 36–44. doi: 10.1080/09064710.2014.960889
Khan, N., Bano, A., Ali, S., Babar, M. A. (2020). Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 2 (90), 189–203. doi: 10.1007/S10725-020-00571-X
Konappa, N. M., Maria, M., Uzma, F., Krishnamurthy, S., Nayaka, S. C., Niranjana, S. R., et al. (2016). Lactic acid bacteria mediated induction of defense enzymes to enhance the resistance in tomato against ralstonia solanacearum causing bacterial wilt. Sci. Hortic. 207, 183–192. doi: 10.1016/j.scienta.2016.05.029
Kristjansdottir, T., Bosma, E. F., Branco Dos Santos, F., Özdemir, E., Herrgård, M. J., França, L., et al. (2019). A metabolic reconstruction of lactobacillus reuteri JCM 1112 and analysis of its potential as a cell factory. Microb. Cell Fact 18, 1–19. doi: 10.1186/S12934-019-1229-3/TABLES/5
Krzywonos, M., Eberhard, T. (2011). Electronic journal of biotechnology high density process to cultivate lactobacillus plantarum biomass using wheat stillage and sugar beet molasses. J. Biotechnol. 14, 1–9. doi: 10.2225/vol14-issue2-fulltext-10
Kumar, S., Diksha, Sindhu, S. S., Kumar, R. (2022). Biofertilizers: An ecofriendly technology for nutrient recycling and environmental sustainability. Curr. Res. Microb. Sci. 3, 100094. doi: 10.1016/J.CRMICR.2021.100094
Lamont, J. R., Wilkins, O., Bywater-Ekegärd, M., Smith, D. L. (2017). From yogurt to yield: Potential applications of lactic acid bacteria in plant production. Soil Biol. Biochem. 111, 1–9. doi: 10.1016/j.soilbio.2017.03.015
Lan, W.-T., Chen, Y.-S., Wu, H.-C., Yanagida, F. (2012). Bio-protective potential of lactic acid bacteria isolated from fermented wax gourd. Folia Microbiologica 2 (57), 99–105. doi: 10.1007/S12223-012-0101-1
Lee, J.-S., Chun, C.-O., Kim, H.-J., Joo, Y.-J., Lee, H.-J., Park, C.-S., et al. (1996). Analysis of cellular fatty acid methyl esters (FAMEs) for the identification of leuconostoc strains isolated from kimchi. J. Microbiol. 34, 225–228.
Lee, K. E., Radhakrishnan, R., Kang, S. M., You, Y. H., Joo, G. J., Lee, I. J., et al. (2015). Enterococcus faecium LKE12 cell-free extract accelerates host plant growth via gibberellin and indole-3-Acetic acid secretion. J. Microbiol. Biotechnol. 25, 1467–1475. doi: 10.4014/JMB.1502.02011
le Lay, C., Coton, E., le Blay, G., Chobert, J. M., Haertlé, T., Choiset, Y., et al. (2016). Identification and quantification of antifungal compounds produced by lactic acid bacteria and propionibacteria. Int. J. Food Microbiol. 239, 79–85. doi: 10.1016/J.IJFOODMICRO.2016.06.020
Limanska, N. V., Babenko, D. O., Yamborko, V. O., Ivanytsia (2015b). Detection of plantaricin genes in strains of lactobacillus plantarum – antagonists of phytopathogenic bacteria. Microbiology&Biotechnology 0, 27–33. doi: 10.18524/2307-4663.2015.2(30).48071
Limanska, N., Korotaeva, N., Ivanytsia, T., Biscola, V., Franco, B. D. G. M., Merlich, A. (2015a). Study of the potential application of lactic acid bacteria in the control of infection caused by agrobacterium tumefaciens. J. Plant Pathol. Microbiol. 06, 1–9. doi: 10.4172/2157-7471.1000292
Lim, P. H., Gansau, J. A., Chong, K. P. (2019). Biocontrol of basal stem rot pathogen ganoderma boninense by pseudomonas aeruginosa. Bangladesh J. Bot. 48, 209–215. doi: 10.3329/BJB.V48I2.47494
Linares-Morales, J. R., Cuellar-Nevárez, G. E., Rivera-Chavira, B. E., Gutiérrez-Méndez, N., Pérez-Vega, S. B., Nevárez-Moorillón, G. V. (2020). Selection of lactic acid bacteria isolated from fresh fruits and vegetables based on their antimicrobial and enzymatic activities. Foods 9, 1399. doi: 10.3390/FOODS9101399
Lin, Y. C., Chung, K. R., Huang, J. W. (2020). A synergistic effect of chitosan and lactic acid bacteria on the control of cruciferous vegetable diseases. Plant Pathol. J. 36, 157–169. doi: 10.5423/PPJ.OA.01.2020.0004
Liu, W., Pang, H., Zhang, H., Cai, Y. (2014). “Biodiversity of lactic acid bacteria,” in Lactic acid bacteria (Dordrecht: Springer Netherlands).
Lo, R. K. S., Chong, K. P. (2020). Metagenomic data of soil microbial community in relation to basal stem rot disease. Data Brief 31, 106030. doi: 10.1016/J.DIB.2020.106030
López-Seijas, J., García-Fraga, B., da Silva, A. F., Sieiro, C. (2019). Wine lactic acid bacteria with antimicrobial activity as potential biocontrol agents against fusarium oxysporum f. sp. lycopersici. Agronomy 10, 31. doi: 10.3390/AGRONOMY10010031
Lutz, M. P., Michel, V., Martinez, C., Camps, C. (2012). Lactic acid bacteria as biocontrol agents of soil-borne pathogens. Biol. Control Fungal Bacterial Plant Pathog. IOBC-WPRS Bull. 78, 285–288.
Maki, Y., Soejima, H., Kitamura, T., Sugiyama, T., Sato, T., Watahiki, M. K., et al. (2021). 3-phenyllactic acid, a root-promoting substance isolated from bokashi fertilizer, exhibits synergistic effects with tryptophan. Plant Biotechnol. (Tokyo) 38, 9–16. doi: 10.5511/PLANTBIOTECHNOLOGY.20.0727A
Maki, Y., Soejima, H., Sugiyama, T., Watahiki, M. K., Sato, T., Yamaguchi, J. (2022). 3-phenyllactic acid is converted to phenylacetic acid and induces auxin-responsive root growth in arabidopsis plants. Plant Biotechnol. (Tokyo) 39, 111–117. doi: 10.5511/PLANTBIOTECHNOLOGY.21.1216A
Malik, M. T., Rehman, A. U., Naqvi, S. A. H., Hasnain, A., Umar, U. U. D., Azeem, H., et al. (2021). Biological mediated management of bacterial diseases in crop-plants: A review. Pakistan J. Phytopathol. 33, 217–232. doi: 10.33866/PHYTOPATHOL.033.01.0669
McAuliffe, O. (2018). Symposium review: Lactococcus lactis from nondairy sources: Their genetic and metabolic diversity and potential applications in cheese1. J. Dairy Sci. 101, 3597–3610. doi: 10.3168/jds.2017-13331
Mishra, A. K., Choi, J., Choi, S. J., Baek, K. H. (2017). Cyclodipeptides: An overview of their biosynthesis and biological activity. Molecules 22, 1796. doi: 10.3390/MOLECULES22101796
Mohite, B. (2013). Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 13, 638–649. doi: 10.4067/S0718-95162013005000051
Muhialdin, B. J., Zaiton, H., Sadon, S., Zulkifli, N. A., Azfar, A. A. (2011). Effect of pH and heat treatment on antifungal activity of lactobacillus fermentum Te007, lactobacillus pentosus G004 and pediococcus pentosaceus Te010. Innov. Rom Food Biotechnol. 8, 41–53.
Murthy, K., Malini, M., Savitha, J., Srinivas, C. (2012). Lactic acid bacteria (LAB) as plant growth promoting bacteria (PGPB) for the control of wilt of tomato caused by ralstonia solanacearum. Pest Manage. Hortic. Ecosyst. 161, 60–65.
Mussa, A., Million, T., Assefa, F. (2018). Rhizospheric bacterial isolates of grass pea (Lathyrus sativus l.) endowed with multiple plant growth promoting traits. J. Appl. Microbiol. 125, 1786–1801. doi: 10.1111/JAM.13942
Nazarov, P. A., Baleev, D. N., Ivanova, M. I., Sokolova, L. M., Karakozova, M. V. (2020). Infectious plant diseases: Etiology, current status, problems and prospects in plant protection. Acta Naturae 12, 46. doi: 10.32607/ACTANATURAE.11026
Nes, I. F., Kjos, M., Diep, D. B. (2011). “Antimicrobial components of lactic acid bacteria,” in Lactic acid bacteria: Microbiological and functional aspects, fourth edition (Boca Raton: CRC Press), 285–329.
Nguyen, H. H., Tram, B., Le, T. (2021). Use of lactic acid bacteria in peanut seed treatment. J. Technol. Innovation (JTIN) 1, 20–22. doi: 10.26480/jtin.01.2021.20.22
Nwodo, U. U., Green, E., Okoh, A. I. (2012). Bacterial exopolysaccharides: Functionality and prospects. Int. J. Mol. Sci. 13, 14002–14015. doi: 10.3390/IJMS131114002
Olaoye, O. A., Onilude, A. A. (2011). Quantitative estimation of antimicrobials produced by lactic acid bacteria isolated from Nigerian beef. Int. Food Res. J. 18, 1155–1161.
Oliveira, A. P., Nielsen, J., Förster, J. (2005). Modeling lactococcus lactis using a genome-scale flux model. BMC Microbiol. 5, 1–15. doi: 10.1186/1471-2180-5-39/TABLES/4
Oliveira, P. M., Zannini, E., Arendt, E. K. (2014). Cereal fungal infection, mycotoxins, and lactic acid bacteria mediated bioprotection: From crop farming to cereal products. Food Microbiol. 37, 78–95. doi: 10.1016/J.FM.2013.06.003
Omer, Z. S., Karin, J., Eberhard, T. H., Johansson, L. K. H. (2009). Bacteria considered as biocontrol agents to control growth of white clover on golf courses. Acta Agric. Scand. B Soil Plant Sci. 60, 193–198. doi: 10.1080/09064710902773637
Ouiddir, M., Bettache, G., Lyva Salas, M., Pawtowski, A., Donot, C., Brahimi, S., et al. (2019). Selection of Algerian lactic acid bacteria for use as antifungal bioprotective cultures and application in dairy and bakery products. Food Microbiol. 82, 160–170. doi: 10.1016/j.fm.2019.01.020
Papadimitriou, K., Alegría, Á., Bron, P. A., de Angelis, M., Gobbetti, M., Kleerebezem, M., et al. (2016). Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 80, 837–890. doi: 10.1128/MMBR.00076-15
Perricone, M., Bevilacqua, A., Corbo, M. R., Sinigaglia, M. (2014). Technological characterization and probiotic traits of yeasts isolated from altamura sourdough to select promising microorganisms as functional starter cultures for cereal-based products. Food Microbiol. 38, 26–35. doi: 10.1016/J.FM.2013.08.006
Phoboo, S., Sarkar, D., Bhowmik, P. C., Jha, P. K., Shetty, K. (2016). Improving salinity resilience in swertia chirayita clonal line with lactobacillus plantarum. Can. J. Plant Sci. 96, 117–127. doi: 10.1139/CJPS-2015-0178/ASSET/IMAGES/LARGE/CJPS-2015-0178F9.JPEG
Ranjith, F. H., Muhialdin, B. J., Yusof, N. L., Mohammed, N. K., Miskandar, M. H., Shobirin, A., et al. (2021). Effects of lacto-fermented agricultural by-products as a natural disinfectant against post-harvest diseases of mango (Mangifera indica l.). Plants 10, 285. doi: 10.3390/plants10020285
Rattanachaikunsopon, P., Phumkhachorn, P. (2010). Lactic acid bacteria: their antimicrobial compounds and their uses in food production. Ann. Biol. Res. 1, 218–228.
Reyes-Escogido, L., Balam-Chi, M., Rodríguez-Buenfil, I., Valdés, J., Kameyama, L., Martínez-Pérez, F. (2010). Purification of bacterial genomic DNA in less than 20 min using chelex-100 microwave: examples from strains of lactic acid bacteria isolated from soil samples. Antonie van Leeuwenhoek 98, 465–474. doi: 10.1007/S10482-010-9462-0
Rodríguez-Serrano, G. M., García-Garibay, J. M., Cruz-Guerrero, A. E., Gómez-Ruiz, L. D. C., Ayala-Niño, A., Castañeda-Ovando, A., et al. (2018). Proteolytic system of streptococcus thermophilus. J. Microbiol. Biotechnol. 28, 1581–1588. doi: 10.4014/JMB.1807.07017
Rooney, W. M., Chai, R., Milner, J. J., Walker, D. (2020a). Bacteriocins targeting gram-negative phytopathogenic bacteria: Plantibiotics of the future. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.575981
Rooney, W. M., Grinter, R. W., Correia, A., Parkhill, J., Walker, D. C., Milner, J. J. (2020b). Engineering bacteriocin-mediated resistance against the plant pathogen pseudomonas syringae. Plant Biotechnol. J. 18, 1296–1306. doi: 10.1111/PBI.13294
Rouse, S., Harnett, D., Vaughan, A., Sinderen, D.v. (2008). Lactic acid bacteria with potential to eliminate fungal spoilage in foods. J. Appl. Microbiol. 104, 915–923. doi: 10.1111/J.1365-2672.2007.03619.X
Sangmanee, P., Hongpattarakere, T. (2014). Inhibitory of multiple antifungal components produced by lactobacillus plantarum K35 on growth, aflatoxin production and ultrastructure alterations of aspergillus flavus and aspergillus parasiticus. Food Control 40, 224–233. doi: 10.1016/J.FOODCONT.2013.12.005
Sathe, S. J., Nawani, N. N., Dhakephalkar, P. K., Kapadnis, B. P. (2007). Antifungal lactic acid bacteria with potential to prolong shelf-life of fresh vegetables. J. Appl. Microbiol. 103, 2622–2628. doi: 10.1111/j.1365-2672.2007.03525.x
Schnürer, J., Magnusson, J. (2005). Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 16, 70–78. doi: 10.1016/J.TIFS.2004.02.014
Sharifi, R., Ryu, C. M. (2018). Revisiting bacterial volatile-mediated plant growth promotion: lessons from the past and objectives for the future. Ann. Bot. 122, 349–358. doi: 10.1093/AOB/MCY108
Shrestha, A., Choi, K.-U., Lim, C.-K., Hur, J.-H., Cho, S.-Y. (2009a). Antagonistic effect of lactobacillus sp. strain KLF01 against plant pathogenic bacteria ralstonia solanacearum. Korean J. Pesticide Sci. 13, 45–53.
Shrestha, A., Kim, E.-C., Lim, C.-K., Cho, S.-Y., Hur, J.-H., Park, D.-H. (2009b). Biological control of soft rot on Chinese cabbage using beneficial bacterial agents in greenhouse and field. Korean J. Pesticide Sci. 13, 325–331.
Shrestha, A., Kim, B. S., Park, D. H. (2014). Biological control of bacterial spot disease and plant growth-promoting effects of lactic acid bacteria on pepper. Biocontrol Sci. Technol. 24, 763–779. doi: 10.1080/09583157.2014.894495
Siezen, R. J., Tzeneva, V. A., Castioni, A., Wels, M., Phan, H. T. K., Rademaker, J. L. W., et al. (2010). Phenotypic and genomic diversity of lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 12, 758–773. doi: 10.1111/J.1462-2920.2009.02119.X
Silpa, S., Rupachandra, S. (2022). Cyclic peptide production from lactic acid bacteria (LAB) and their diverse applications. Crit. Rev. Food Sci. Nutr. 62, 2909–2927. doi: 10.1080/10408398.2020.1860900
Sindhu, S. S., Sehrawat, A., Sharma, R., Dahiya, A. (2016). Biopesticides: Use of rhizosphere bacteria for biological control of plant pathogens. Def Life Sci. J. 1, 135. doi: 10.14429/DLSJ.1.10747
Singh, V. P. (2018). Recent approaches in food bio-preservation - a review. Open Vet. J. 8, 104–111. doi: 10.4314/OVJ.V8I1.16
Sjögren, J., Magnusson, J., Broberg, A., Schnürer, J., Kenne, L. (2003). Antifungal 3-hydroxy fatty acids from lactobacillus plantarum MiLAB 14. Appl. Environ. Microbiol. 69, 7554. doi: 10.1128/AEM.69.12.7554-7557.2003
Somers, E., Amake, A., Croonenborghs, A., Overbeek, L. S., Vanderleyden, J. (2007). Lactic acid bacterial in organic agricultural soil. Proc. Rhizosphere 2 Montpellier France.
Steglińska, A., Kołtuniak, A., Motyl, I., Berłowska, J., Czyżowska, A., Cieciura-Włoch, W., et al. (2022). Lactic acid bacteria as biocontrol agents against potato (Solanum tuberosum l.) pathogens. Appl. Sci. 12, 7763. doi: 10.3390/app12157763
Strafella, S., Simpson, D. J., Khanghahi, M. Y., de Angelis, M., Gänzle, M., Minervini, F., et al. (2021). Comparative genomics and In vitro plant growth promotion and biocontrol traits of lactic acid bacteria from the wheat rhizosphere. Microorganisms 9, 78. doi: 10.3390/microorganisms
Sundin, G. W., Castiblanco, L. F., Yuan, X., Zeng, Q., Yang, C. H. (2016). Bacterial disease management: Challenges, experience, innovation and future prospects: Challenges in bacterial molecular plant pathology. Mol. Plant Pathol. 17, 1506–1518. doi: 10.1111/mpp.12436
Sunil, K., Narayana, B. (2008). Spectrophotometric determination of hydrogen peroxide in water and cream samples. Bull. Environ. Contamination Toxicol. 4 (81), 422–426. doi: 10.1007/S00128-008-9477-7
Suzzi, G., Francesca, N., di Cagno, R., Pontonio, E., di Cagno, R., Tarraf, W., et al. (2018). Dynamic and assembly of epiphyte and endophyte lactic acid bacteria during the life cycle of origanum vulgare l. Front. Microbiol. 9, 1372. doi: 10.3389/fmicb.2018.01372
Taha, M. D. M., Jaini, M. F. M., Saidi, N. B., Rahim, R. A., Shah, U. K. M., Hashim, A. M. (2019). Biological control of erwinia mallotivora, the causal agent of papaya dieback disease by indigenous seed-borne endophytic lactic acid bacteria consortium. PloS One 14, 1–20. doi: 10.1371/journal.pone.0224431
Tofalo, R., Hadavi, E., Spano, G., Arena, M. P., Silvain, A., Normanno, G., et al. (2016). Use of lactobacillus plantarum strains as a bio-control strategy against food-borne pathogenic microorganisms. Front. Microbiol. 1. doi: 10.3389/fmicb.2016.00464
Toropov, V., Demyanova, E., Shalaeva, O., Sitkin, S., Vakhitov, T. (2020). Whole-genome sequencing of lactobacillus helveticus D75 and D76 confirms safety and probiotic potential. Microorganisms 8, 329. doi: 10.3390/MICROORGANISMS8030329
Trias, R., Bañeras, L., Montesinos, E., Badosa, E. (2008). Lactic acid bacteria from fresh fruit and vegetables as biocontrol agents of phytopathogenic bacteria and fungi. Int. Microbiol. 11, 231–236. doi: 10.2436/20.1501.01.66
Tsuda, K., Tsuji, G., Higashiyama, M., Ogiyama, H., Umemura, K., Mitomi, M., et al. (2016). Biological control of bacterial soft rot in Chinese cabbage by lactobacillus plantarum strain BY under field conditions. Biol. Control 100, 63–69. doi: 10.1016/J.BIOCONTROL.2016.05.010
Turaeva, B. I., Qizi, K. K. F., Soliev, A. B., Kutlieva, G. J. (2021). Gibberellic and indole acetic acids producing features of bacteria from the genus lactobacillus and their effect on plant development. Asian J. Biol. Life Sci. 10, 681–686. doi: 10.5530/AJBLS.2021.10.91
UNDP (2021). United nations development programme, precision agriculture for smallholder farmers (UNDP global centre for technology, innovation and sustainable development (Singapore: UNDP Global Centre for Technology, Innovation and Sustainable Development).
Valencia-Hernandez, L. J., Lopez-Lopez, K., Gomez-Lopez, E. D., Sernacock, L., Aguilar, C. N. (2021). In-vitro assessment for the control of fusarium species using a lactic acid bacterium isolated from yellow pitahaya (Selenicereus megalanthus (K. schum. ex vaupel moran)). J. Integr. Agric. 20, 159–167. doi: 10.1016/S2095-3119(20)63284-1
Venegas-Ortega, M. G., Flores-Gallegos, A. C., Aguilar, C. N., Rodríguez-Herrera, R., Martínez-Hernández, J. L., Nevárez-Moorillón, G. V. (2020). Multi-functional potential of presumptive lactic acid bacteria isolated from chihuahua cheese. Foods 9, 276. doi: 10.3390/FOODS9030276
Vimont, A., Fernandez, B., Ahmed, G., Fortin, H. P., Fliss, I. (2019). Quantitative antifungal activity of reuterin against food isolates of yeasts and moulds and its potential application in yogurt. Int. J. Food Microbiol. 289, 182–188. doi: 10.1016/J.IJFOODMICRO.2018.09.005
Vinay-Lara, E., Hamilton, J. J., Stahl, B., Broadbent, J. R., Reed, J. L., Steele, J. L. (2014). Genome –scale reconstruction of metabolic networks of lactobacillus casei ATCC 334 and 12A. PloS One 9, e110785. doi: 10.1371/JOURNAL.PONE.0110785
Visser, R., Holzapfel, W. H., Bezuidenhout, J. J., Kotze, J. M. (1986) Antagonism of lactic acid bacteria against phytopathogenic bacteria. Available at: https://journals.asm.org/journal/aem.
Wang, H., Yan, H., Shin, J., Huang, L., Zhang, H., Qi, W. (2011). Activity against plant pathogenic fungi of lactobacillus plantarum IMAU10014 isolated from xinjiang koumiss in China. Ann. Microbiol. 61, 879–885. doi: 10.1007/S13213-011-0209-6
Wanikawa, A., Shoji, H., Hosoi, K., Nakagawa, K. I. (2002). Stereospecificity of 10-hydroxystearic acid and formation of 10-ketostearic acid by lactic acid bacteria. J. Am. Soc. Brewing Chem. 60, 14–20. doi: 10.1094/ASBCJ-60-0014
Xu, N., Liu, J., Ai, L., Liu, L. (2015). Reconstruction and analysis of the genome-scale metabolic model of lactobacillus casei LC2W. Gene 554, 140–147. doi: 10.1016/J.GENE.2014.10.034
Yanagida, F., Chen, Y. S., Shinohara, T. (2006). Searching for bacteriocin-producing lactic acid bacteria in soil. J. Gen. Appl. Microbiol. 52, 21–28. doi: 10.2323/JGAM.52.21
Yang, E. J., Chang, H. C. (2010). Purification of a new antifungal compound produced by lactobacillus plantarum AF1 isolated from kimchi. Int. J. Food Microbiol. 139, 56–63. doi: 10.1016/J.IJFOODMICRO.2010.02.012
Yang, E., Fan, L., Jiang, Y., Doucette, C., Fillmore, S. (2012). Antimicrobial activity of bacteriocin-producing lactic acid bacteria isolated from cheeses and yogurts. AMB Express 2, 1–12. doi: 10.1186/2191-0855-2-48/FIGURES/4
Yanwei, Z., Ling, Y., Jianghao, L., Chao, Y., Hongyan, W., Ling, L., et al. (2022). Application progress of lactic acid bacteria whole genome sequencing. Sci. Technol. Food Industry 43 (15), 444–450. doi: 10.13386/J.ISSN1002-0306.2021080128
Yarullina, D. R., Asafova, E. v., Kartunova, J. E., Ziyatdinova, G. K., Ilinskaya, O. N. (2014). Probiotics for plants: NO-producing lactobacilli protect plants from drought. Appl. Biochem. Microbiol. 50, 166–168. doi: 10.1134/S0003683814020197
Yu, A. O., Leveau, J. H. J., Marco, M. L. (2020). Abundance, diversity and plant-specific adaptations of plant-associated lactic acid bacteria. Environ. Microbiol. Rep. 12, 16–29. doi: 10.1111/1758-2229.12794
Zacharof, M. P., Lovitt, R. W. (2012). Bacteriocins produced by lactic acid bacteria a review article. APCBEE Proc. 2, 50–56. doi: 10.1016/J.APCBEE.2012.06.010
Zebboudj, N., Yezli, W., Hamini-Kadar, N., Kihal, M. (2020). Antifungal activity of lactic acid bacteria against fusarium species responsible for tomato crown and root rots. Environ. Exp. Biol. 18, 7–13. doi: 10.22364/eeb.18.02
Zhao, H., Vegi, A., Wolf-Hall, C. (2017). Screening of lactic acid bacteria for anti-fusarium activity and optimization of incubation conditions. J. Food Prot 80, 1648–1656. doi: 10.4315/0362-028X.JFP-17-100
Keywords: lactic acid bacteria, phytopathogens, biological control agent, plant disease management, plant growth
Citation: Jaffar NS, Jawan R and Chong KP (2023) The potential of lactic acid bacteria in mediating the control of plant diseases and plant growth stimulation in crop production - A mini review. Front. Plant Sci. 13:1047945. doi: 10.3389/fpls.2022.1047945
Received: 19 September 2022; Accepted: 27 December 2022;
Published: 13 January 2023.
Edited by:
Islam Hamim, Bangladesh Agricultural University, BangladeshReviewed by:
Deepa N., University of Mysore, IndiaBartholomew Saanu Adeleke, Olusegun Agagu University of Science and Technology, Nigeria
Abhay K. Pandey, Tea Research Association, India
Elsherbiny A. Elsherbiny, Mansoura University, Egypt
Copyright © 2023 Jaffar, Jawan and Chong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Khim Phin Chong, Y2hvbmdrcEB1bXMuZWR1Lm15
 Nur Sulastri Jaffar
Nur Sulastri Jaffar Roslina Jawan
Roslina Jawan Khim Phin Chong
Khim Phin Chong