- 1Institute of Horticulture, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2Institute of Crop Science, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 3Key Laboratory of Microbiol Technology and Bioinformatics of Zhejiang Province, Zhejiang Institute of Microbiology, Hangzhou, China
Saline stress is a significant factor that caused crop growth inhibition and yield decline. SHORT INTERNODES/STYLISH (SHI/STY) and SHI-RELATED SEQUENCE (SRS) transcription factors are specific to plants and share a conserved RING-like zinc-finger domain (CX2CX7CX4CX2C2X6C). However, the functions of SHI/STY and SRS genes in cotton responses to salt stress remain unclear. In this study, 26 GhSRSs were identified in Gossypium hirsutum, which further divided into three subgroups. Phylogenetic analysis of 88 SRSs from8 plant species revealed independent evolutionary pattern in some of SRSs derived from monocots. Conserved domain and subcellular location predication of GhSRSs suggested all of them only contained the conserved RING-like zinc-finger domain (DUF702) domain and belonged to nucleus-localized transcription factors except for the GhSRS22. Furthermore, synteny analysis showed structural variation on chromosomes during the process of cotton polyploidization. Subsequently, expression patterns of GhSRS family members in response to salt and drought stress were analyzed in G. hirsutum and identified a salt stress-inducible gene GhSRS21. The GhSRS21 was proved to localize in the nuclear and silencing it in G. hirsutum increased the cotton resistance to salt using the virus-induced gene silencing (VIGS) system. Finally, our transcriptomic data revealed that GhSRS21 negatively controlled cotton salt tolerance by regulating the balance between ROS production and scavenging. These results will increase our understanding of the SRS gene family in cotton and provide the candidate resistant gene for cotton breeding.
1 Introduction
SHI/STY and SRS family members, a plant-specific transcription factors, are defined by the presence of a conserved RING-like zinc-finger domain (CX2CX7CX4CX2C2X6C), the vast majority of which also contain the IXGH domain (Kuusk et al., 2006; Zhao et al., 2020). The RING finger domain is believed to confer E3 ubiquitin ligase activity and mediate the ubiquitination and proteasome-dependent degradation of target protein or confer the DNA and RNA binding activity (Fridborg et al., 2001; Zhao et al., 2020). The IXGH domain is rich in acidic amino acid residues, which are considered as transcriptional activators (Singh et al., 2020).
It has been shown that SRS transcription factors are involved in gibberellin (GA) and auxin signaling pathways (Fridborg et al., 1999; Fridborg et al., 2001; Singh et al., 2020; Yuan et al., 2020) and diverse growth and development processes in plant such as lateral root development and floral organ morphogenesis (Kuusk et al., 2002; Sohlberg et al., 2006; Staldal et al., 2012; Singh et al., 2020; Yuan et al., 2020). In Arabidopsis, there are 9 described members in the SHI/STY and SRS family (Eklund et al., 2011; Zhao et al., 2020). The first SHORT INTERNODES/STYLISH (SHI/STY) and SRS family gene to be identified was AtSHI and its transposon insertion mutants displayed a dwarf phenotype similar to the mutants defective in the biosynthesis of gibberellin (Fridborg et al., 1999). However, the application of GA could not rescue the phenotype the dwarf phenotype of atshi, indicating the potential role of AtSHI in GA response (Fridborg et al., 1999). LATERAL ROOT PRIMORDIUM1 (LRP1), another SHORT INTERNODES/STYLISH (SHI/STY) and SRS member, has been reported to interact with SHI, STY1, SRS3, SRS6 and SRS7 and affect the homeostasis and biosynthesis of auxin through the regulation of several YUCCA (YUC) genes during lateral root development (Singh et al., 2020). Meanwhile, the expression of LRP1 is subject to feedback regulation by auxin (Singh et al., 2020). Interestingly, the crosstalk between auxin and another SRS member SRS5 has been characterized and indicates that SRS5 negatively regulates lateral root formation by repressing the expression of LBD16 and LBD29 (Yuan et al., 2020). Besides, SRS5 promotes photomorphogenesis activating the expression of HY5, BBX21, and BBX22 upon exposure to light, whereas it undergoes COP1-mediated degradation via the 26S proteasome system in darkness (Yuan et al., 2018). In addition, SHI/STY and SRS proteins also play vital roles in floral organ development. In Arabidopsis, STY1 promotes stamen and gynoecium development while STY2 promotes gynoecium development (Kuusk et al., 2002; Sohlberg et al., 2006; Staldal et al., 2012).
Cotton is an important cash crop and provides raw material for textiles producing. Saline stress is a significant factor limiting crop productivity and survival (Deinlein et al., 2014; Chen et al., 2018a). In cotton, some QTLs (quantitative trait loci) and genes related to salt tolerance have been identified through either forward genetic or reverse genetic studies in recent years (Jia et al., 2016; Sun et al., 2018; Ullah et al., 2018; Dilnur et al., 2019; Li et al., 2019; Mu et al., 2019; Yasir et al., 2019; Yuan et al., 2019; Long et al., 2020). Yasir et al. identified two salt tolerance-related genes located on chromosome A10 and D10 by genome-wide association study and expression pattern analysis in upland cotton (Gossypium hirsutum) (Yasir et al., 2019). Dilnur et al. found two SNP loci associated with salt-stress tolerance on chromosome 7 in G. arboretum (Dilnur et al., 2019). Yuan et al. detected 13 QTLs using genome-wide association study and further identified 35 candidate genes responsible for cotton salt tolerance at the germination stage by RNA-seq analysis (Yuan et al., 2019). In addition, reverse genetic studies focused on cotton resistance to saline stress have made some progress in recent years. GhRaf19, a member of MAPKKK family in G. hirsutum, negatively controlled the salt tolerance by regulating the accumulation of endogenous reactive oxygen species (ROSs) in G. hirsutum (Jia et al., 2016). Similarly, GhWRKY6, a salt-induced gene, has proved to be a negative regulators of salt resistance using VIGS system (Li et al., 2019). Interestingly, another WRKY family member GhWRKY6-like had the opposite effect on cotton resistance to salt stress, which improved salt tolerance in G. hirsutum by activating the ABA signaling pathway and scavenging of ROSs (Ullah et al., 2018). Furthermore, protein phosphatase GhDsPTP3a interacted with a membrane protein GhANN8b and inhibited GhANN8b phosphorylation, resulting in changes of the salt induced calcium influx, the expression of GhSOS1, the outflow of sodium ions and decreased salt tolerance in G. hirsutum (Mu et al., 2019). In addition to regulators of cotton tolerance to salt stress, the gene structures, evolutionary relationships and expression patterns of Na+/H+ antiporters (NHXs) members in G. arboreum, G. raimondii and G. hirsutum were identified by Long et al. (Long et al., 2020). Then GhNHX1 was further proved to be located in the vacuolar system and played a crucial role in salt tolerance using VIGS system (Long et al., 2020). However, little is known about the functions of SHI/STY and SRS family genes responses to abiotic stresses in cotton.
In this study, we systematically identified 26 SRS family members in Gossypium hirsutum and analyze their phylogenetic relationships, protein structures, chromosomal locations, conserved motif distribution patterns, gene collinearity and expression pattern. Then, we further identified the function of a salt-inducible protein GhSRS21 under salt stress. Finally, we revealed that GhSRS21 played a negative role in salt tolerance of Gossypium hirsutum by controlled the balance of ROS production and scavenging. Our results will helpful to elucidate the salt response and regulation mechanism in Gossypium hirsutum and provide theoretical support for further in-depth research of GhSRSs.
2 Material and methods
2.1 Plant materials and treatment
All G. hirsutum plant materials used in the research were TM-1 (Texas Marker-1, the upland cotton genetic standard line) background. For subsequent quantitative reverse transcription (qRT)-PCR experiments, seeds of TM-1 were germinated and planted in soil under the following conditions: 12000 Lux light 16 h at 25 °C/dark 8 h at 23 °C, 80% humidity for 14 days (the first true leaf appeared). The seedlings above were divided into two groups and watered by 1/2 MS nutrient solution as the control or by 1/2 MS nutrient solution with 500 mM NaCl for 12 h.
2.2 Identification and property analysis of GhSRS genes
The genome datasets of G. hirsutum (ZJU, version 2.1) and G. barbadense (ZJU, version 1.1) were downloaded from COTTONOMICS (http://cotton.zju.edu.cn/index.htm), G. arboreum (WHU, version 3.0) and G. raimondii (NSF, version 1.0) from CottonGen (https://www.cottongen.org) and Arabidopsis from TAIR 10 (http://www.arabidopsis.org/). Other plant species genome datasets were downloaded from Phytozome v12.1 (https://phytozome.jgi.doe.gov/). The databases of PlantTFDB (http://planttfdb.cbi.pku.edu.cn/) and SMART (http://smart.embl.de/) were used to confirm the conserved RING-linke zinc-finger domain (DUF702). The molecular weight (MW), isoelectric point (pI) of each GhSRS were calculated using the ExPASy (https://web.expasy.org/) compute pI/Mw tool, Plant-Ploc (http://www.csbio.sjtu.edu.cn/bioinf/plant/) was for subcellular localizational prediction (Chou and Shen, 2007). All gene names and their IDs were listed in Supplementary Table 1.
2.3 Multiple alignments and phylogenetic analysis
SRS family amino acid sequences were used to perform multiple alignments by MEGA X software (Kumar et al., 2018) with MUSCLE default parameters, and then visualized using DNAMAN v7. Furthermore, the rooted and unrooted phylogenetic trees were constructed using MEGA X with Neighbor-Joining (NJ) methods, and 1000 bootstrap replicates were used to test reliability in each node and the maximum likelihood (ML) tree was constructed using “One Step Build a ML Tree” plugin in Tbtool software (Chen et al., 2020) with 5000 bootstrap replicates.
2.4 Gene structure, conserved motifs and synteny analysis
The gene structures of GhSRS genes were inferred by corresponding coding sequences. The MEME (https://meme-suite.org/meme/) program was used to identify conserved motifs in GhSRS proteins (Bailey et al., 2009). Tbtools Gene Structure View was used to draw the exon-intron structure, conserved motifs, and DUF702 domain distribution (Chen et al., 2020). The MCScanX software was used to analyze SRS protein sequences synteny and collinearity relationship between G. hirsutum, G. arboreum, and G. raimondii.
2.5 Collinearity analysis of SRS genes in G. hirsutum, G. arboreum and G. raimondii
Chromosomal positions of GhSRS genes were obtained from gff annotation files for G. hirsutum (ZJU, version 2.1). The synteny and collinearity analysis between G. hirsutum, G. arboreum and G. raimondii were employed by the MCScanX software (Wang et al., 2012). Tbtools Gene Location Visualize and Advanced Circos were used to draw the distribution of GhSRS genes chromosomal mapping and synteny relationships (Chen et al., 2020).
2.6 SRS genes expression patterns under biotic stress
To analysis the expression patterns of SRS genes under abiotic stress, a high-through RNA-seq datasets of leaf tissue under control and two types of stress (NaCl and PEG) treatments were obtained from COTTONOMICS (http://cotton.zju.edu.cn/) (Hu et al., 2019). Fragments per kilobase of exon per million fragments mapped (FPKM) was used for the quantification of gene expression. The clustered heatmap was drawn and normalized by the average expression levels (log2) based on FPKM values.
2.7 Virus-induced gene silencing assay and abiotic stress treatment
349 bp fragment of coding DNA sequence (CDS) of GhSRS21 was amplified from G. hirsutum (TM-1) cDNA and constructed into pTRV2. Then, the recombinant vector above, pTRV1 vector and CLA-pTRV2 were transferred into Agrobacterium strain GV3101, respectively, for subsequent experiments.
Seeds of TM-1 were germinated and planted in soil under the following conditions: 12000 Lux light 16 h at 25 °C/dark 8 h at 23 °C, 80% humidity, until the cotyledons were fully opened (about 10 d after seeds germinated). The Agrobacterium strains containing the GhSRS21-pTRV2 and pTRV1 or CLA-pTRV2 and pTRV1 or pTRV2 and pTRV1 plasmids were mixed and injected into TM-1 leaves as experimental groups, positive and negative controls, respectively. Cotton infected by Agrobacterium tumefaciens was cultured under the conditions above until the white striped leaf or albino phenotype appeared in the positive control (about 10 d after infected). Half of the experimental and negative control groups was watered by 1/2 MS nutrient solution as the control while the other half was watered by 500 mM NaCl regularly after every 3 days until the phenotypes appeared (about 14 d after treatment).
2.8 RNA isolation and qRT-PCR
Total RNA was isolated from leaf tissues of TM-1 under salt stress treatment at 0 h, 1 h, 3 h, 6 h and 12 h using RNAprep pure Plant Kit (code: DP432, Tiangen, Beijing, China). First-strand cDNA was synthesized using Hifair 1st Strand cDNA Synthesis SuperMix for qPCR (YEASEN Biotech, Shanghai, China). The qRT-PCR was performed using Hieff qPCR SYBR GreenMaster Mix (YEASEN Biotech, Shanghai, China). The cotton ubiquitin gene UBQ7 was used as the internal control for the relative expression calculation. RNA isolation and qRT-PCR was manipulated based on the manufacturer’s instructions. The primers used for qRT-PCR were listed in Supplementary Table 2.
2.9 Measurement of chlorophyll content and determination of malondialdehyde, H2O2, O2·- and antioxidant enzymes activities
The samples of cotton leaves obtained from negative control groups and GhSRS21 VIGS lines (experimental groups) treated with or without salt and PEG6000 in method 2.6 were used for measurement of chlorophyll content and determination of malondialdehyde, H2O2, O2·- and antioxidant enzymes activities. The detail method for determination of malondialdehyde, H2O2, O2·- and antioxidant enzymes activities was performed as described by Zhan et al. (Zhan et al., 2021), and method for measurement of chlorophyll contents were performed according to our previously described methods (Liu et al., 2019).
2.10 Subcellular localization
The CDS of GhSRS21 fused with GFP (green fluorescent protein) tag at the 3′-end was ligated into the pCAMBIA1300 vector. The recombinant plasmid above was mixed with the nuclear marker NLS-mCherry and co-transformed into Arabidopsis mesophyll protoplasts as described (Yoo et al., 2007). The protoplasts were observed and photographed by a fluorescence microscopy (Zeiss Imager.A2, Germany). The primers used above were listed in Supplementary Table 2.
2.11 RNA-seq and KEGG analysis
Total RNA was isolated from leaf tissues of negative control groups and GhSRS21 VIGS lines when the albino-like appearance on the leaves of positive control was observed. The fragments were purified by agarose gel electrophoresis and sequenced with NovaSeq 6000 Sequencer (Illumina Inc., San Diego, CA, USA) with a read length of 150 bp. Three biological replicates were performed separately. The raw data were filtered with fastp (https://github.com/OpenGene/fastp) (Chen et al., 2018b). The reads filtered above were then mapped to the cotton reference genome using HISAT2 software (http://ccb.jhu.edu/software/hisat2) (Kim et al., 2015; Zhang et al., 2015). Transcript analysis was performed using StringTie (https://ccb.jhu.edu/software/stringtie/) (Pertea et al., 2015), and differential expression genes (DEGs) analysis was performed by DEseq2 (Love et al., 2014), FPKM > 1.0 (FPKM, Fragments Per Kilobase of exon per Million mapped reads) were regarded as valid DEGs. Subsequently, KEGG annotations were performed using the online software EggNOG-Mapper (http://eggnog-mapper.embl.de/) and KEGG enrichment analysis were performed using Tbtools software (Chen et al., 2020).
2.12 Data processing and analysis
Statistical analysis was performed using SPSS version 23.0 statistical software (SPSS, Inc., Chicago, IL, USA). All data were subjected to analysis of variance (One-way ANOVA) and mean comparisons were carried out by Ducan’s multiple range test (p < 0.05).
3 Results
3.1 Identification of SRS genes in three cotton species
To identify all members of the SRS gene family in cotton, the conserved RING-like zinc-finger domain (DUF702) (Pfam ID: PF05142) from the Pfam databases (http://pfam.xfam.org/) were employed as queries to search against three main representative cotton species. Then, the puatative protein sequences using the PlantTFDB (http://planttfdb.cbi.pku.edu.cn/) and SMART (http://smart.embl.de/) databases to confirm the predicted functional domains contained DUF702 (PF05142) families. A total of 53 SRS members were identified in G. arboreum, G. raimondii, and G. hirsutum, of which 14 were GaSRS genes, 13 were GrSRS genes, and 26 were GhSRS genes (13 SRSs from At subgenome, 13 SRSs from Dt subgenome). Therefore, we named them GhSRS1~GhSRS26 based on their gene ID number and genomic distribution. The encoded protein length of GhSRS genes ranging from 195 (GhSRS3) aa to 434 (GhSRS22) aa, and molecular weight (MW) from 22116.24 Da to 45074.19 Da, isoelectric point (pI) varying from 5.49 (GhSRS25) to 9.22 (GhSRS26), in addition to predicting the subcellular location of all GhSRS members, 16 of GhSRS proteins were nucleus-localized. Other basic information for all SRS members in three cotton species were listed in Supplementary Table 3.
To better understand the phylogenetic and evolution relationships of SRS genes in cotton, the unrooted phylogenetic tree constructed by MEGA X revealed the SRS family genes can be divided into three subgroups (Figure 1A). The number of SRS genes in G. hirsutum was almost the sum of the number of those in G. arboreum and G. raimondii, which was consistent with polyploidy and whole-genome duplication (WGD) events during hybridization.
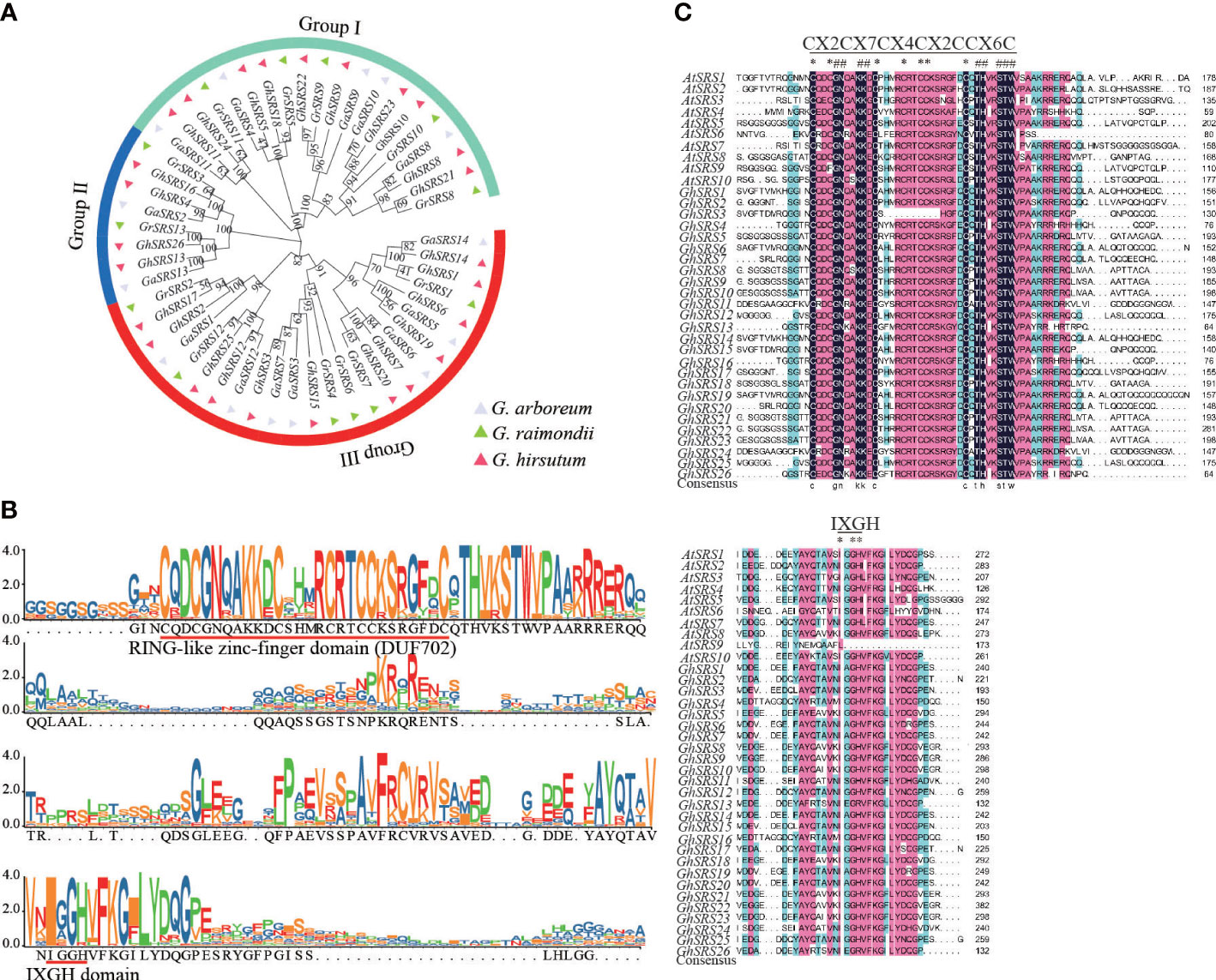
Figure 1 Phylogenetic and domain structure analysis of the SRS family proteins in three cotton species. (A) the unrooted phylogenetic tree of the SRS family in G arboreum, G raimondii, and G hirsutum at the amino acid sequences level using Neighbor-Joining method in MEGA X with 1000 bootstrap. The subgroup was divided according to the value on the node at the root of the evolutionary tree (the value>=80: all genes under this node belong to one subgroup; the value<80: all genes under this node can be further divided into two or more subgroups). (B) The conserved RING-like zinc-finger domain (CX2CX7CX4CX2C2X6C). The alignment domain sequence was obtained from CottonGen (https://www.cottongen.org/). (C) Alignment of conserved RING-like zinc-finger domain and the IXGH domain in A thaliana and G hirsutum.
We found that almost all members of this family contained a RING-like zinc-finger domain (CX2CX7CX4CX2C2X6C) through sequence alignment of amino acid residues. But lacking part of the RING domain in GhSRS3 may leading to a decrease in the binding ability to DNA, RNA, protein, and lipid substrates (Figure 1B). Moreover, GhSRSs also share a IXGH domain except GhSRS13/26, and this conserved region longer than IXGH domain in GmSRSs (Figure 1C) (Zhao et al., 2020).
3.2 Phylogenetic analysis of SRS genes
To investigate the evolutionary relationships of SRS gene family, we constructed a separate rooted phylogenetic tree using 13 plant species genome datasets from lower aquatic to higher terrestrial plants. We totally identified 115 genes in different moss (2 in P. patens), fern (4 in S. moellendorffii), monocotyledons (5 in O. sativa, 10 in Z. mays), and dicotyledons (5 in T. cacao, 10 in A. thaliana, 14 in G. arboreum, 13 in G. raimondii, 26 in G. barbadense and 26 in G. hirsutum), while no SRS gene was found in picophytoplankton (M. pusilla) and algae (Os. tauri, V. carteri) (Figure 2A, B). Results revealed that SRS gene first appeared in moss (P. patens), and the number of SRS gene increased dramatically in G. barbadense and G. hirsutum (Figure 2B).
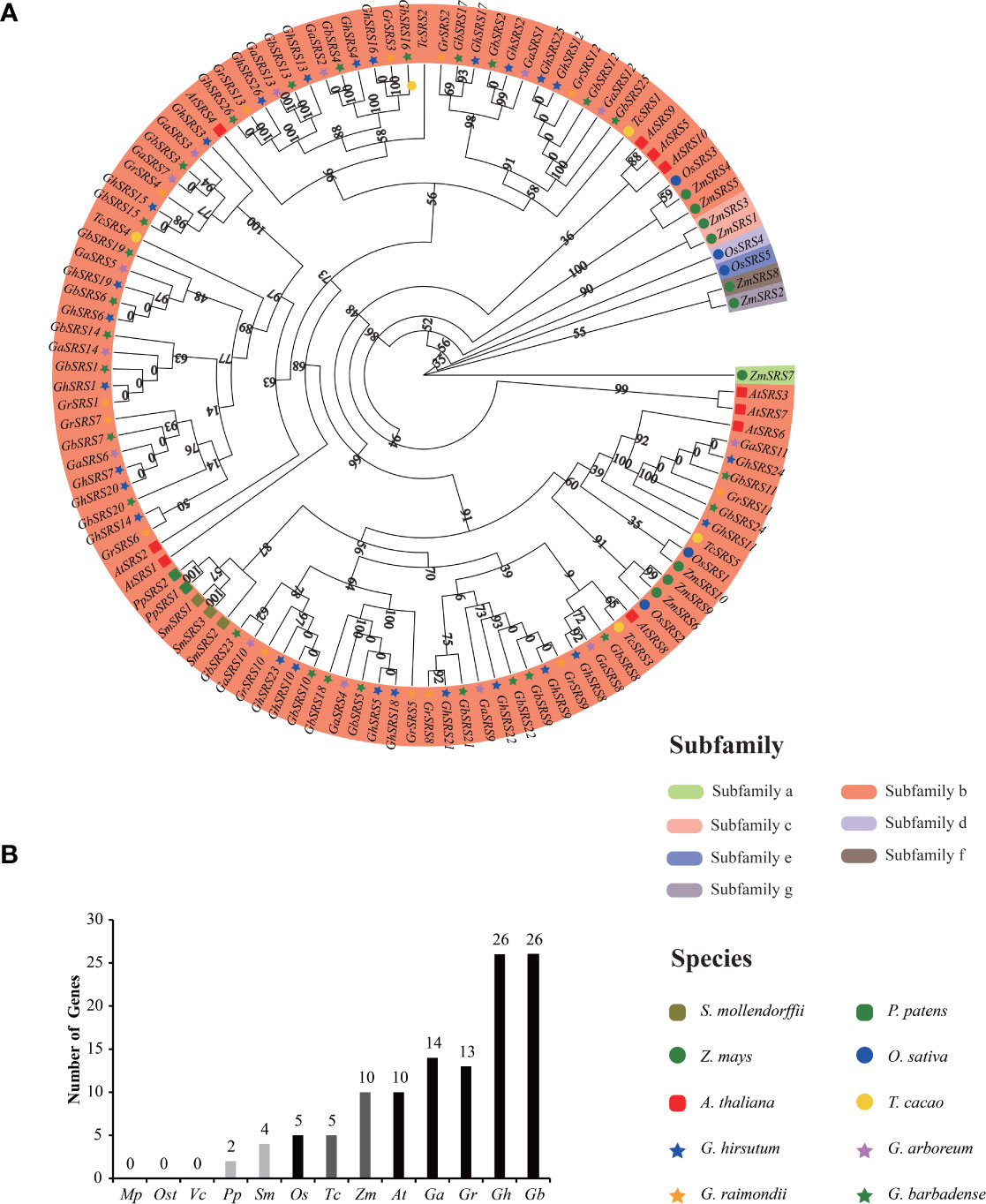
Figure 2 Phylogenetic and evolution relationships of the SRS gene family among different organisms (A) the rooted phylogenetic tree of the SRS gene family at the amino acid sequences level using the Maximum Likelihood method in Tbtools with 5000 bootstrap. The subgroup was divided according to the value on the node at the root of the evolutionary tree (the value>=80: all genes under this node belong to one subgroup; the value<80: all genes under this node can be further divided into two or more subgroups). (B) Comparisons of SRS gene numbers across a wide range of organisms. The prefix Mp, Ost, Vc, Pp, Sm, Os, Tc, Zm, At, Ga, Gr, Gh were used to describe the names of M. pusilla, Os. tauri, V. carteri, P. patens, S. moellemdorffii, O. sativa, T. cacao, Z. mays, A thaliana, G arboreum, G raimondii, G barbadense, G hirsutum, respectively.
The SRS genes in multiple plant species can be divided into 7 clades and named a to g subfamilies. The SRS subfamiliy a and c-gonly existed in monocotyledon and each subfamiliy above contained only one or two members. However, SRS subfamily b was the largest subfamily and contained 108 SRS members including all SRS genes derived from the above dicotyledons and some SRS genes derived from the above monocotyledons (Figure 2A). Furthermore, the number of GhSRS genes in G. barbadense or G. hirsutum were almost the sum of SRS genes in G. arboreum and G. raimondii (Figure 2B), which confirmed the ideas that allotretraploid cotton (G. barbadense and G. hirsutum) evolved from hybridization and polyploidization between two diploid cotton species (G. arboreum and G. raimondii).
3.3 Gene structure and domain analysis
To figure out the structure similarity of the SRS family in cotton, the full-length protein sequence of 26 GhSRSs were aligned to display phylogenetic tree with conserved motifs, exon-intron, and domain structure. Conserved motifs in the SRS protein sequences were performed by MEME online service (https://meme-suite.org/meme/), 12 different motifs were identified and distributed on the GhSRS protein sequences. GhSRSs in the same cluster shared similar conserved motif composition, especially all GhSRS members contained some conserved motifs (motif 1, motif 2, and motif 4) (Figure 3A). And the motif number of each protein ranging from five to eleven (Figure 3A). Then, we performed the exon-intron gene structure analysis by comparing genomic sequence to the extended cDNA sequence (CDS) of GhSRSs. The number of coding exons of GhSRSs in Gossypium hirsutum was conserved, as they all contained two exons (Figure 3B). However, the length of the introns of the GhSRS genes was largely variable, ranging from 73 bp to 497 bp (Figure 3B). The domain structure of SRS proteins was analyzed using the SMART protein-domain search interface (http://smart.embl.de/), and the results showed all GhSRSs shared a conserve domain named DUF702, containing the RING-like zinc-finger domain (CX2CX7CX4CX2C2X6C) (Figure 3C). Interestingly, only the GhSRS22 possessed a transmembrane domain. The results above indicated that the GhSRS22 was likely to be a membrane-bound protein, which was consistent with the result listed in Supplementary Table 3.
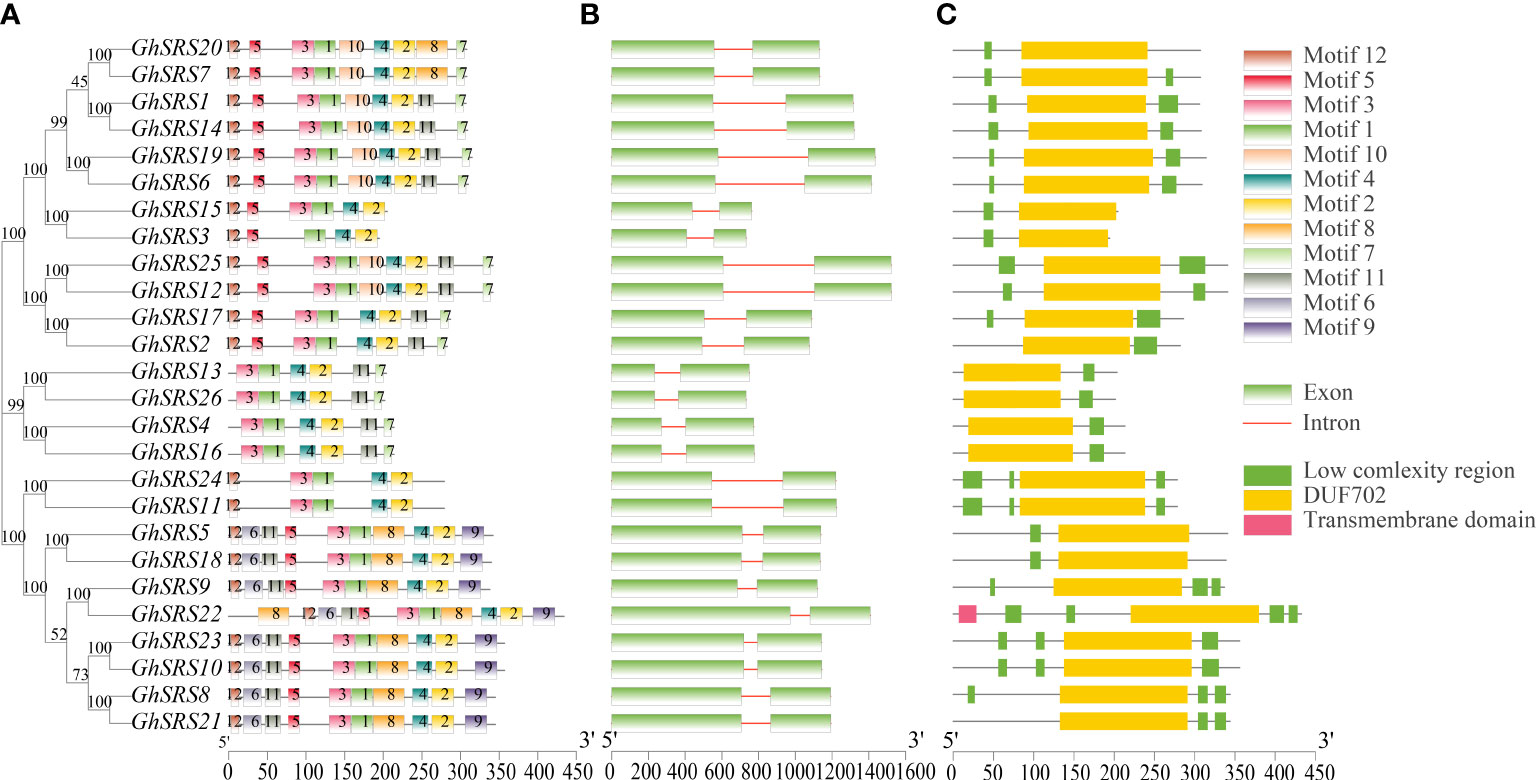
Figure 3 Conserved motifs, gene structures and conserved domains of SRS family members (A) Conserved motifs identification through Multiple EM for Motif Elicitation (MEME) (B) Gene structures of SRS proteins, green rectangle represents the exon of SRS genes and line represents intron (C) Conserved motifs of SRS proteins, the blue rectangle represents the SRS conserved domain containing the RING-like zinc-finger domain (CX2CX7CX4CX2C2X6C) and named DUF702.
3.4 Genomic distribution and synteny analysis
To investigate the chromosomal distribution and the duplication events of the SRS family in cotton, chromosomal distribution and collinearity analysis was performed. In G. arboreum, 13 SRS family members were distributed on chromosomes At02, At03, At05, At06, At07, At08, At09, At010, At11, At13. Meanwhile, a total of 13 SRSs were unevenly distributed on chromosomes Dt01, Dt02, Dt03, Dt05, Dt06, Dt07, Dt08, Dt09, Dt10, Dt11, Dt13 (Figure 4). Interestingly, although the number of SRS genes in G. hirsutum were not altered, their distributions on chromosomes in G. hirsutum displayed differences compared with those in G. arboreum and G. raimondii, implying structural variation on chromosomes during the process of cotton polyploidization. For instance, GrSRS1 was located on chromosome Dt01 in G. raimondii while there was no SRS gene on chromosome At01 in G. arboreum (Figure 4). However, in G. hirsutum, GhSRS1 was located in chromosome At01 and exhibited high gene collinearity with GrSRS1, probably resulting from the gene duplication and interchromosomal translocation. In addition, one SRS gene loss occurred in chromosome Dt03 (Figure 4). In summary, the SRS genes in G. hirsutum were unevenly distributed on all chromosomes except for the chromosomes At04, At12, Dt04, Dt12. Besides, SRS gene duplication and loss events occurred during ancestral allopolyploidization of G. hirsutum.
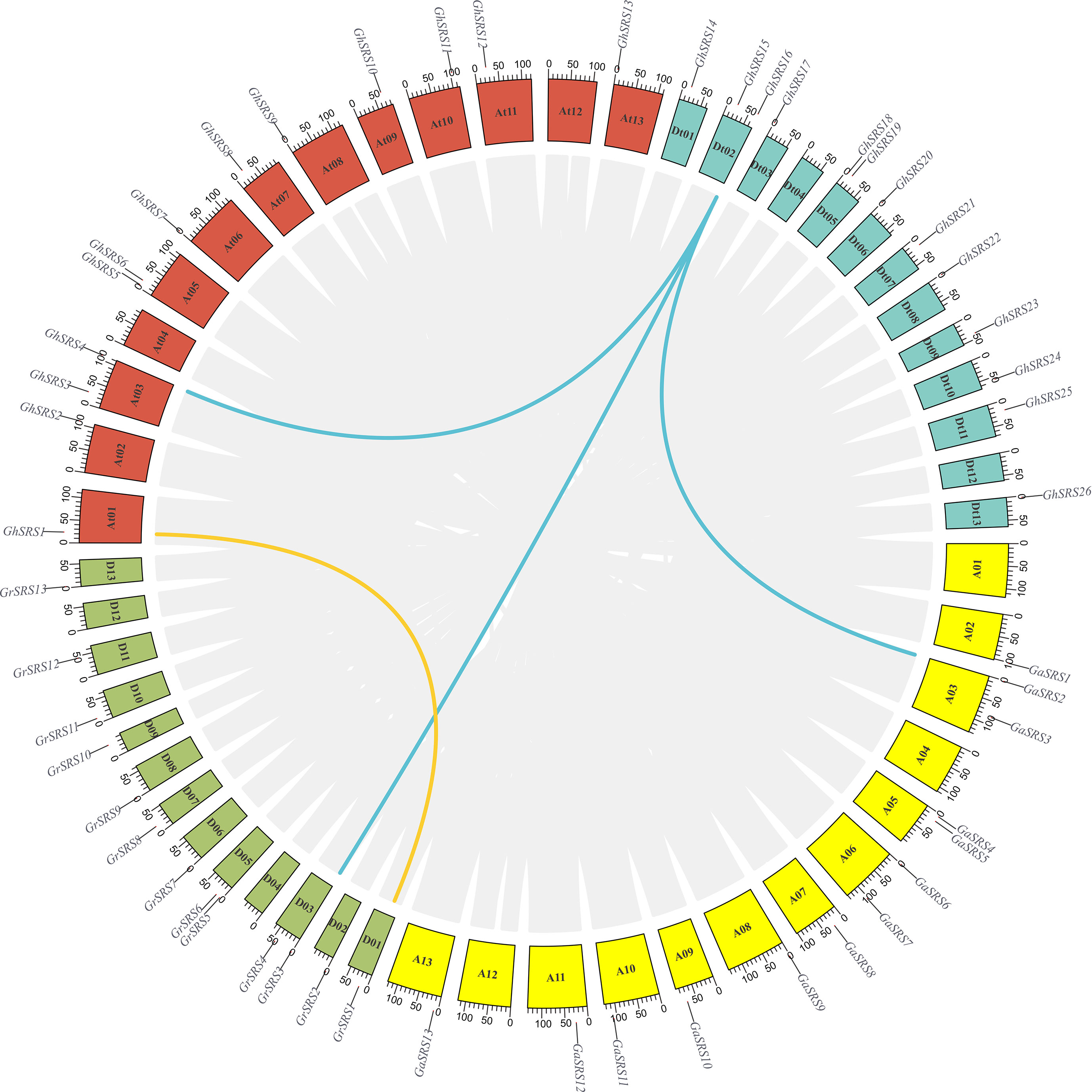
Figure 4 Collinearity analysis of SRS genes between G. hirsutum, G. arboreum and G. raimondii. A01 – A13 represent 13 chromosomes in G. arboreum, D01 – D13 represent 13 chromosomes in G. raimondii, At01 – At13, Dt01 – Dt13 represent 13 At subgenome chromosomes and 13 Dt subgenome chromosomes in G. hirsutum. Gene joined by yellow and blue lines are the product of gene duplication.
3.5 Expression patterns of GhSRSs under salt and drought stresses
To better understand the function of GhSRS genes under salt and drought stresses in Gossypium hirsutum, the expression pattern of GhSRSs in response to salt and drought stress was examined using the FPKM values of GhSRSs extracted from COTTONOMICS (http://cotton.zju.edu.cn/index.htm) (Figure 5). The results showed that most GhSRSs, such as GhSRS1, GhSRS2, GhSRS3, GhSRS4, GhSRS6, GhSRS7, GhSRS10, GhSRS12, GhSRS13, GhSRS14, GhSRS15, GhSRS16, GhSRS17, GhSRS18, GhSRS19, GhSRS20, GhSRS23, GhSRS24, GhSRS25, GhSRS26, expressed at a very low level (average FPKM<5) under control, salt and drought stress (Figure 5). Of the remaining 6 genes, GhSRS21 was induced by salt stress (3h, 12h and 24h after salt treatment) while the transcripts of GhSRS5, GhSRS22 showed high level accumulation after drought treatment (Figure 5), indicating their potential roles in salt or drought tolerance.
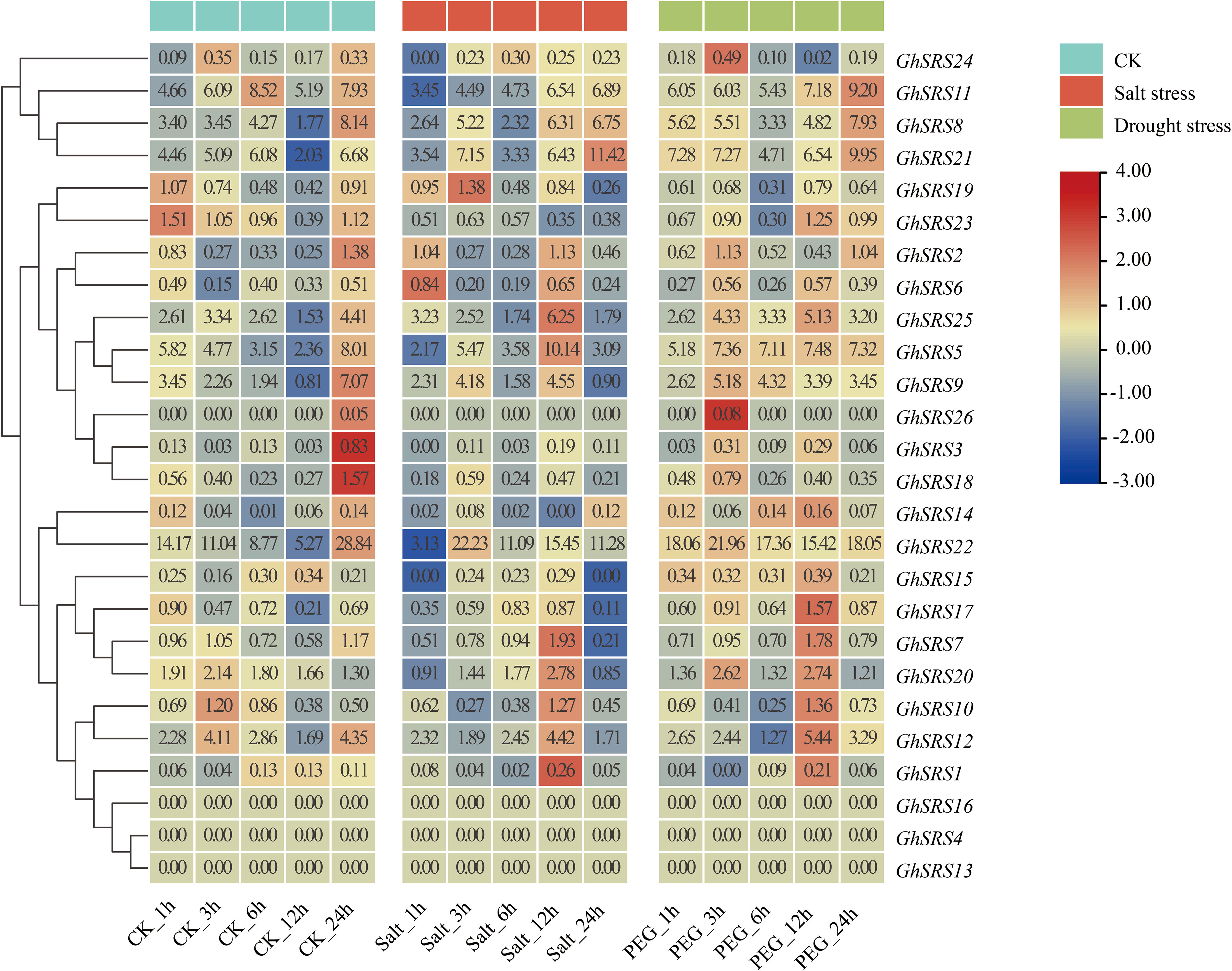
Figure 5 Expression patterns of GhSRS family members under salt and drought stress. Color intensity displayed in the heatmap are the Log2 transformed RPKM gene expression value. The origin FPKM values were showed in the squares. CK represents the control group.
3.6 GhSRS21 subcellular localization
To further confirm the nuclear localization of GhSRS21 and presume its potential role in regulation of the expression of eukaryotic genes, a nuclear localization sequence fused with the mCherry (NLS- mCherry) was used as nuclear localization marker and cotransformed with the plasmid (GhSRS21 ORF fused with GFP) to the Arabidopsis protoplasts. There was overlapping between green fluorescence and red fluorescence, indicating GhSRS21 located in the nucleus (Figure 6).
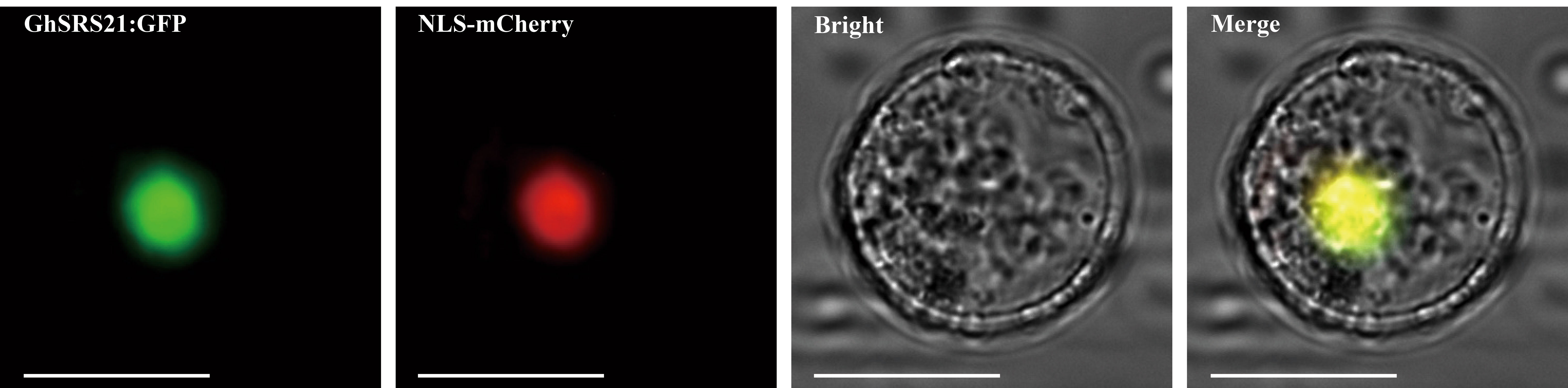
Figure 6 Subcellular location of GhSRS21. NLS-mCherry: nuclear localization sequence fused with a reporter gene mCherry, which is used as nuclear localization marker. Bar=10 μm.
3.7 GhSRS21 negatively regulates salt tolerance in a manner dependent on reactive oxygen species metabolic process in G. hirsutum
The expression pattern of GhSRS21 under salt stress has been described previously using the COTTONOMICS public database ((http://cotton.zju.edu.cn/index.htm)) (Figure 5). To further confirm the expression pattern of GhSRS21 in response to salt stress, RT-qPCR was performed and the results revealed that the GhSRS21 gene expression was highly induced by salt stress (Figure 7A). Subsequently, the function of GhSRS21 under salt stress conditions was determined using the VIGS (virus-induced gene silencing) system. When the albino-like appearance on the leaves of positive control was observed (Supplementary Figure 1), we examined the expression of GhSRS21 in negative control and GhSRS21 VIGS lines. The data showed that the expression levels of GhSRS21 were significantly decreased in GhSRS21 VIGS lines compared with those in negative control (Figure 7B). To further understand the role of GhSRS21 in G. hirsutum under salt stress, salt treatment was performed and silenced GhSRS21 in TM-1 resulted in enhanced salt tolerance (Figure 7C). In addition, we further detected the activities of antioxidant enzymes and relevant physiological indicators of negative control and VIGS plant under control and salt treatment. The results exhibited the increased activity of catalase (CAT), peroxidase (POD) and increased content of hydrogen peroxide (H2O2), malondialdehyde (MDA) significantly in both GhSRS21 VIGS cottons and negative control after salt treatment (Figures 7D–G). However, a significantly higher CAT activity was observed in the GhSRS21 VIGS lines compared with that of the negative control after salt treatment (Figure 7D). Meanwhile, the content of H2O2 and MDA in GhSRS21 VIGS lines was significantly lower than that in the negative control after salt treatment (Figures 7E, G). Taken together, GhSRS21 negatively regulates salt tolerance in G. hirsutum through increased antioxidant capacity of cotton.
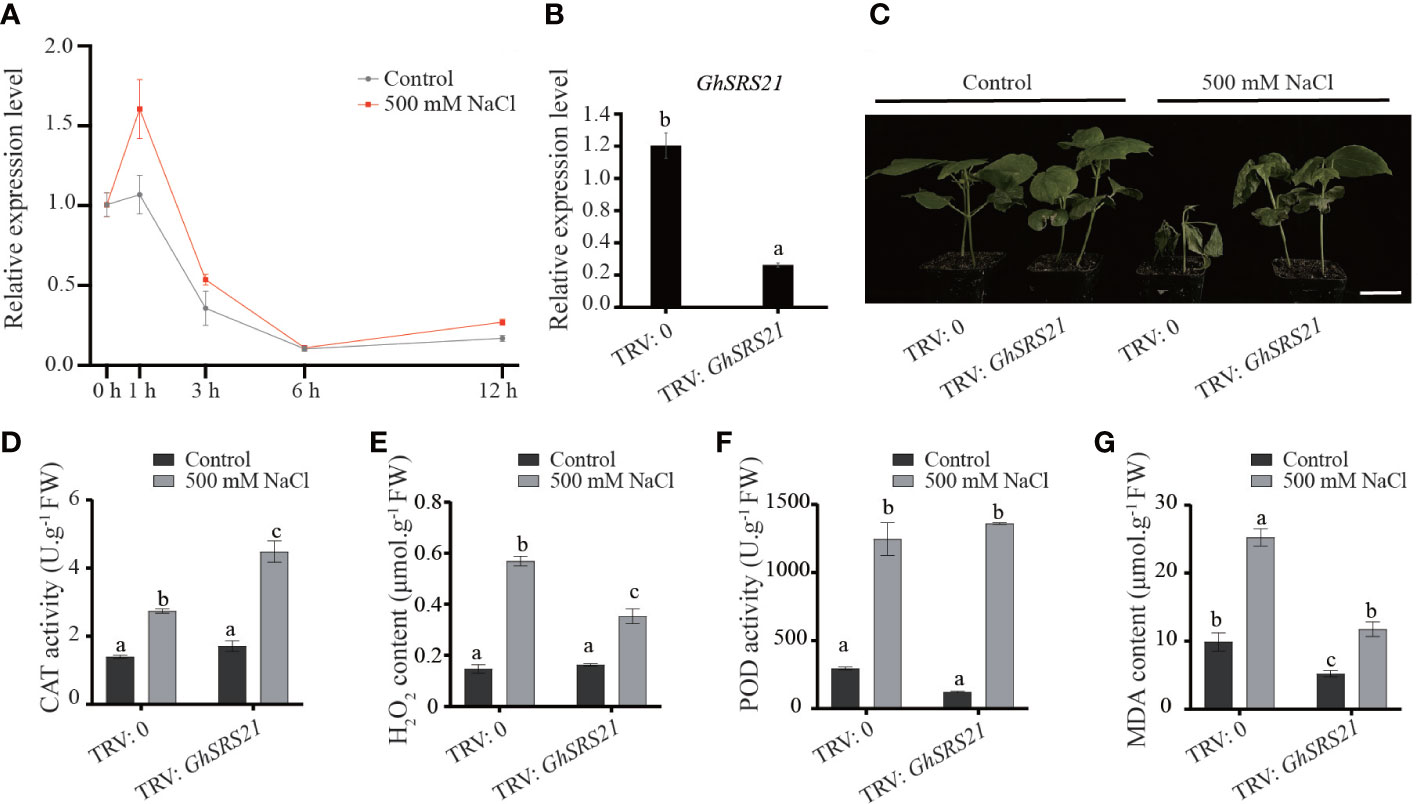
Figure 7 Phenotypic identification of GhSRS21 VIGS line under salt stress. (A) The expression levels of GhSRS21 in the negative control and GhSRS21 VIGS lines under control and 500 mM NaCl at 0 h, 1 h, 3 h, 6 h, 12 h. (B) The expression levels of GhSRS21 in the negative control and GhSRS21 VIGS lines. Each qPCR reaction was performed with three technical replicates. (C) Phenotypic identification of GhSRS21 VIGS line under control and 500 mM NaCl treatment. TRV2: 0 and TRV2: GhSRS21 represent the negative control and GhSRS21 VIGS line, respectively. Bar = 4 cm (D–G)SOD activity, POD activity and H2O2, MDA content in the negative control and GhSRS21 VIGS lines under control and 500 mM NaCl treatment. FW indicates fresh weight. Significant differences are determined using one-way ANOVA and Ducan’s Multiple Range Test, as indicated with different letters at P < 0.05 significance level.
To explore the potential mechanism of GhSRS21 in regulation of H2O2 production, genes differentially expressed in the negative control and GhSRS21 silenced plants were analyzed via the transcriptome data. The results indicated that the number of down-regulated genes (306) was more than that of up-regulated genes (71) in GhSRS21 silenced lines compared with the negative control (Figure 8A). Furthermore, the enrichment of the differentially expressed genes (DEGs) in the KEGG pathway was analyzed. Most up-regulated genes were enriched in the pathways of genetic information processing, carbohydrate metabolism, transcription, translation, peroxisome, etc, while the down-regulated genes showed significant enrichment on metabolism, transporters, flavonoid biosynthesis, etc (Figures 8B, C). There were four up-regulated genes (GH_A05G0875, GH_A09G1066, GH_A13G1914, GH_D09G1018), enriched in peroxisome KEGG pathway, belong to copper/zinc superoxide dismutase (SODC), and shared potential biological functions in ROS (reactive oxygen species) scavenging. To further figure out their roles in salt resistance of the upland cotton, the expression pattern of the mentioned SODCs above were analyzed using the public COTTONOMICS Database. The results showed that the four SODCs above were significantly suppressed by salt stress while GhSRS21 exhibited the opposite expression pattern under salt treatment (Figure 8D). Moreover, the four SODCs were induced when the GhSRS21 was repressed in the VIGS lines (Figure 8E). In summary, the GhSRS21 negatively regulates salt tolerance in a manner dependent on reactive oxygen species metabolic process and probably by negative regulation of SODCs expression.
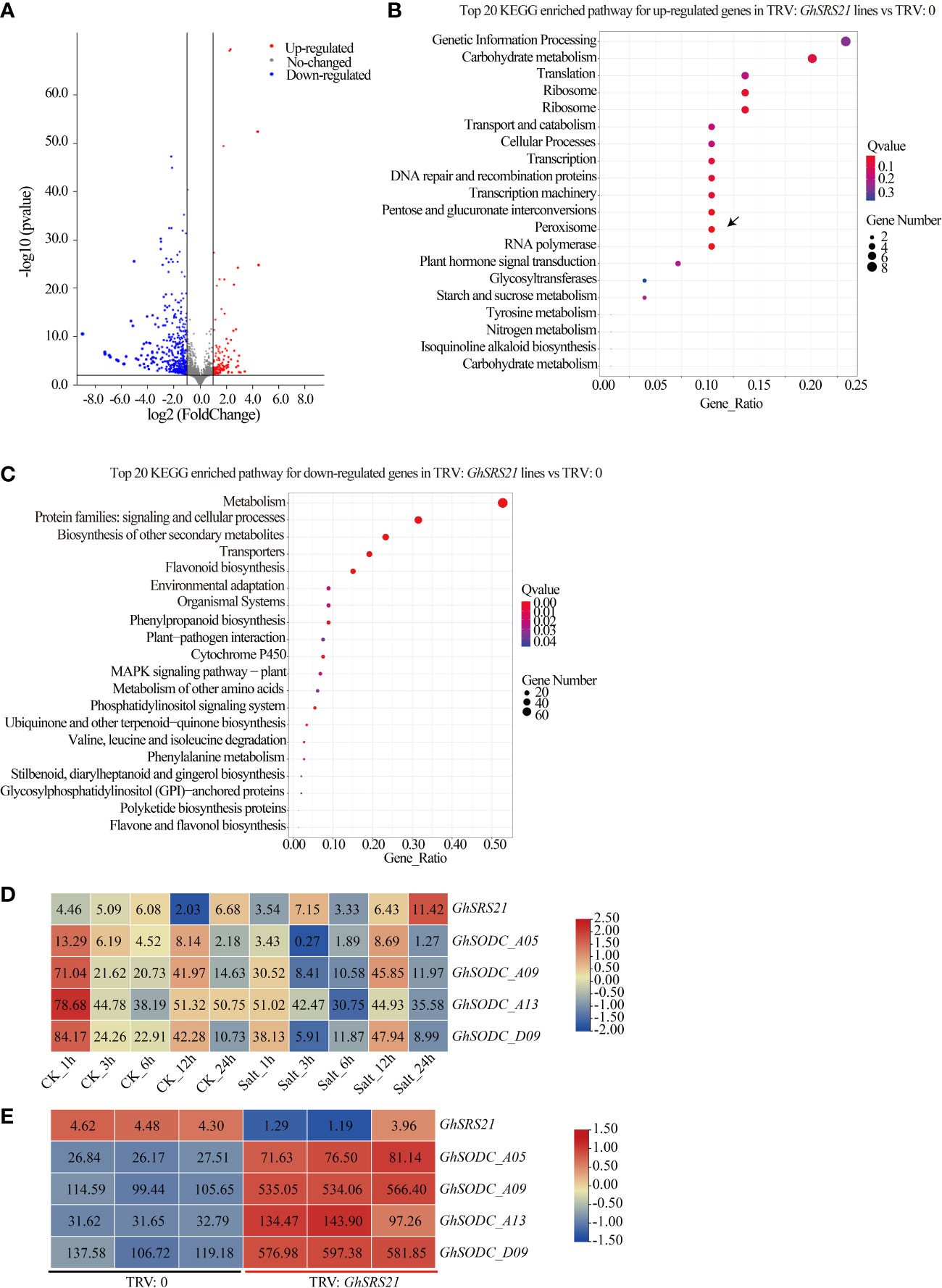
Figure 8 Transcriptome analysis of GhSRS21 VIGS lines.(A) Volcano map of DEGs (GhSRS21 VIGS lines vs negative control). (B, C) KEGG pathway analysis for up-regulated genes and down-regulated genes in GhSRS21 VIGS lines vs negative control (TRV: GhSRS21 lines represent the GhSRS21 VIGS lines and TRV: 0 represent the negative control), respectively. (D) Expression pattern of GhSRS21 and SODCs under control and salt treatment. (E) Expression pattern of GhSRS21 and SODCs in GhSRS21 VIGS lines vs negative control. Heatmap in (D, E) was normalized by the average log2 of FPKM values and the numbers in rectangle indicate FPKM values.
4 Discussion
The global soil salinization is major and growing ecological problems due to the rising sea level from global climate warming and inappropriate irrigation practice (Munns and Gilliham, 2015; Zhou et al., 2017). Salinity stress is one of the most important constraints on crop yield (Deinlein et al., 2014). Salt stress includes three types: osmotic stress, ionic stress, and oxidative damage (Ismail and Horie, 2017; Yang and Guo, 2018). Plants produce compatible osmolytes such as proline and soluble sugars and increase the cellular osmolarity to maintain the capability to absorb water under salt stress, which help the plants under stress in osmotic adjustment (Deinlein et al., 2014; Park et al., 2016; Yang and Guo, 2018). On the other hand, Plants exposure to salt stress induces overproduction of reactive oxygen species (ROS), which results in membrane injury and increased MDA production (Choudhury et al., 2017). The antioxidant enzymatic system, such as superoxide dismutase (SOD), ascorbate peroxidase (APX), CAT, guaiacolperoxidase (GPX) and POD, is responsible for scavenging of ROS induced by salt stress (Yang and Guo, 2018).
The SHI/STY and SRS members are plant-specific transcription factors (Zhao et al., 2020). Available researches of SRS transcription factors have focused on their role in regulation of plant growth and development (Zhao et al., 2020). However, the role of SRS transcription factors participated in plant’s resistance to abiotic stress is few reported. Besides, the function of SRS members in Gossypium hirsutum is largely unclear. In this study, we identified 26 GhSRSs in G. hirsutum and most of them were predicted to located in the nucleus (Supplementary Table 3), indicating their crucial roles in regulating nuclear gene expression. Then the conserved domain of the GhSRSs was analyzed and the result was consistent with the reported literatures (Fridborg et al., 2001; Zhao et al., 2020). Most SRS members in Gossypium hirsutum share a RING-like zinc-finger domain (CX2CX7CX4CX2C2X6C) and a IXGH domain (Figures 1B, C), which is responsible for biological macromolecules binding (DNA, RNA, protein and/or lipid substrates) (Klug, 1999) and transcriptional activation (Fridborg et al., 2001; Singh et al., 2020), respectively. Nevertheless, defective in RING-like zinc-finger domain of GhSRS3 and defective in IXGH domain of GhSRS13/26 most likely result in loss of function (Figures 1B, C). Furthermore, we identified the SRS genes in M. pusilla, Os. tauri, V. carteri, P. patens, S. moellemdorffii, O. sativa, T. cacao, Z. mays, A. thaliana, G. arboreum, G. raimondii, G. barbadense, G. hirsutum and investigated their evolutionary relationships. Interestingly, the SRS genes were missing in some algae (M. pusilla, Os. Tauri and V. carteri) but appeared in the land plants (P. patens, S. moellemdorffii, O. sativa, T. cacao, Z. mays, A. thaliana, G. arboreum, G. raimondii, G. barbadense and G. hirsutum) (Figures 2A, B). The results above indicated the potential role of SRS genes in the transition of plant from the water to the land. Besides, all members in SRS subfamily a and c-g were derived from monocots (O. sativa and Z. mays) (Figure 2A), indicating that these genes might evolve separately and played a specific role in monocots grown and stress defense. Our result above is also in agreement with the work of Yang et al. (Yang et al., 2020). Besides, Tetraploid cotton (represented by G. barbadense and G. hirsutum; AD1) originated from an allopolyploidization event between an A-genome (G. herbaceum- or G. arboreum-like) and a D-genome (G. raimondii-like) diploid species circa 1 to 2 million years ago (Wendel and Grover, 2015; Hu et al., 2019). The number of GhSRS genes in G. barbadense or G. hirsutum were almost the sum of SRS genes in G. arboreum and G. raimondii (Figure 2B), which confirmed the ideas that allotretraploid cotton (G. barbadense and G. hirsutum) evolved from hybridization and polyploidization between two diploid cotton species (G. arboreum and G. raimondii). Subsequently, we analyzed the gene structures and protein domains of GhSRSs. The gene structures are well conserved in cotton genome (all contained only one intron) (Figure 3B) in contrast to those in soybean, maize and alfalfa (He et al., 2020; Zhao et al., 2020; Yang et al., 2021). Besides, GhSRS22 is a unique member of the GhSRSs, containing the transmembrane domain (Figure 3C) and belonging to plant membrane-bound transcription factors. The plant membrane-bound transcription factors are usually located in cellular membranes and represented in inactive state (Liu et al., 2018; De Backer et al., 2022). However, the plant membrane-bound transcription factors are activated and relocate to the nucleus by protease cleavage of themselves in response to an intra- or extra-cellular trigger (Kim et al., 2010; De Backer et al., 2022). Consequently, GhSRS22 could play a crucial role in the regulation of the gene expression process under specific conditions.
To further explore biological functions of GhSRSs under multiple abiotic stresses, we analyzed their expression patterns in response to salt and drought stress in G. hirsutum and GhSRS21, a salt-inducible gene, was identified (Figure 5). The results of subcellular localization showed GhSRS21 belonged to the nuclear transcription factors (Figure 6), which was consistent with our predictions (Supplementary Table 3). The results above also implied its biological functions involved in regulation of nuclear gene expression. Furthermore, GhSRS21 silenced in Gossypium hirsutum L. increased cotton resistance to salt (Figures 7A–C), further conforming the negative regulatory role of GhSRS21 in cotton tolerance to salt. Similarly, Zhao et al. demonstrated that GmSRS18 negatively controlled drought and salt resistance in transgenic Arabidopsis (Zhao et al., 2020). Therefore, the SRSs tend to be negative regulators when plants are subjected to various abiotic stresses. H2O2, MDA and ROS are important markers reflecting the severity of the salinity stress while CAT and POD are responsible for ROS and H2O2 scavenging (Wang et al., 2013; Yang and Guo, 2018). Our further researches indicated that GhSRS21 modulated salt stress tolerance of cotton through negative regulation of CAT activity and H2O2 scavenging (Figures 7E, F). Nevertheless, the mechanism by which GhSRS21 negatively regulated H2O2 scavenging is unclear. Thus, the transcriptome sequencing was performed using the negative control and GhSRS21 VIGS lines and the results reveal that the up-regulated DEGs (GhSRS21 VIGS lines compared with negative control) were enriched in peroxisome pathway, containing four copper/zinc superoxide dismutase (SODC) genes (Figure 8B). Meanwhile, the four SODC genes above and GhSRS21 showed opposite expression pattern when cotton were subjected to salt stress (Figure 8D). SODC, responsible for ROS scavenging, play a crucial role in plant salt tolerance (Sunkar et al., 2006). It follows that GhSRS21 controlled salt sensitivity of cotton by regulation of the balance between ROS production and scavenging. In general, we identified a negative regulatory transcription factor GhSRS21 involving in cotton salt tolerance may provide valuable candidates for efforts toward the genetic improvement of cotton.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA893181.
Author contributions
CS conceived and designed the research. CS, LY, and SZ performed the experiments. CS, LY, and QG analyzed the data. CS, QG and MW contributed to writing the manuscript. CS, LY, QG and MW modified and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (32201710), Natural Science Foundation of Zhejiang Province, China (LQ19C020006) and Key Research Program of Zhejiang Province, China (2020C02001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1078083/full#supplementary-material
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Liu, A., Ren, H., Yu, Y., Duanmu, H., Duan, X., et al. (2018a). Genome-wide analysis of glycine soja response regulator GsRR genes under alkali and salt stresses. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01306
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018b). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Choudhury, F. K., Rivero, R. M., Blumwald, E., Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. doi: 10.1111/tpj.13299
Chou, K. C., Shen, H. B. (2007). Large-Scale plant protein subcellular location prediction. J. Cell Biochem. 100, 665–678. doi: 10.1002/jcb.21096
De Backer, J., Van Breusegem, F., De Clercq, I. (2022). Proteolytic activation of plant membrane-bound transcription factors. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.927746
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Dilnur, T., Peng, Z., Pan, Z., Palanga, K. K., Jia, Y., Gong, W., et al. (2019). Association analysis of salt tolerance in Asiatic cotton (Gossypium arboreum) with SNP markers. Int. J. Mol. Sci. 20, 2168. doi: 10.3390/ijms20092168
Eklund, D. M., Cierlik, I., Staldal, V., Claes, A. R., Vestman, D., Chandler, J., et al. (2011). Expression of arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiol. 157, 2069–2080. doi: 10.1104/pp.111.182253
Fridborg, I., Kuusk, S., Moritz, T., Sundberg, E. (1999). The arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 11, 1019–1032. doi: 10.1105/tpc.11.6.1019
Fridborg, I., Kuusk, S., Robertson, M., Sundberg, E. (2001). The arabidopsis protein SHI represses gibberellin responses in arabidopsis and barley. Plant Physiol. 127, 937–948. doi: 10.1104/pp.010388
He, B., Shi, P., Lv, Y., Gao, Z., Chen, G. (2020). Gene coexpression network analysis reveals the role of SRS genes in senescence leaf of maize (Zea mays l.). J. Genet. 99, 3. doi: 10.1007/s12041-019-1162-6
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371-5
Ismail, A. M., Horie, T. (2017). Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 68, 405–434. doi: 10.1146/annurev-arplant-042916-040936
Jia, H., Hao, L., Guo, X., Liu, S., Yan, Y., Guo, X. (2016). A raf-like MAPKKK gene, GhRaf19, negatively regulates tolerance to drought and salt and positively regulates resistance to cold stress by modulating reactive oxygen species in cotton. Plant Sci. 252, 267–281. doi: 10.1016/j.plantsci.2016.07.014
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, S. G., Lee, S., Seo, P. J., Kim, S. K., Kim, J. K., Park, C. M. (2010). Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95, 56–65. doi: 10.1016/j.ygeno.2009.09.003
Klug, A. (1999). Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 293, 215–218. doi: 10.1006/jmbi.1999.3007
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kuusk, S., Sohlberg, J. J., Long, J. A., Fridborg, I., Sundberg, E. (2002). STY1 and STY2 promote the formation of apical tissues during arabidopsis gynoecium development. Development 129, 4707–4717. doi: 10.1242/dev.129.20.4707
Kuusk, S., Sohlberg, J. J., Magnus Eklund, D., Sundberg, E. (2006). Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J. 47, 99–111. doi: 10.1111/j.1365-313X.2006.02774.x
Li, Z., Li, L., Zhou, K., Zhang, Y., Han, X., Din, Y., et al. (2019). GhWRKY6 acts as a negative regulator in both transgenic Arabidopsis and cotton during drought and salt stress. Front. Genet. 10. doi: 10.3389/fgene.2019.00392
Liu, Y., Li, P., Fan, L., Wu, M. (2018). The nuclear transportation routes of membrane-bound transcription factors. Cell Commun. Signal 16, 12. doi: 10.1186/s12964-018-0224-3
Liu, X., Wu, X., Sun, C., Rong, J. (2019). Identification and expression profiling of the regulator of chromosome condensation 1 (RCC1) gene family in Gossypium hirsutum l. under abiotic stress and hormone treatments. Int. J. Mol. Sci. 20, 1727. doi: 10.3390/ijms20071727
Long, L., Zhao, J. R., Guo, D. D., Ma, X. N., Xu, F. C., Yang, W. W., et al. (2020). Identification of NHXs in Gossypium species and the positive role of GhNHX1 in salt tolerance. BMC Plant Biol. 20, 147. doi: 10.1186/s12870-020-02345-z
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Munns, R., Gilliham, M. (2015). Salinity tolerance of crops - what is the cost? New Phytol. 208, 668–673. doi: 10.1111/nph.13519
Mu, C., Zhou, L., Shan, L., Li, F., Li, Z. (2019). Phosphatase GhDsPTP3a interacts with annexin protein GhANN8b to reversely regulate salt tolerance in cotton (Gossypium spp.). New Phytol. 223, 1856–1872. doi: 10.1111/nph.15850
Park, H. J., Kim, W. Y., Yun, D. J. (2016). A new insight of salt stress signaling in plant. Mol. Cells 39, 447–459. doi: 10.14348/molcells.2016.0083
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Singh, S., Yadav, S., Singh, A., Mahima, M., Singh, A., Gautam, V., et al. (2020). Auxin signaling modulates LATERAL ROOT PRIMORDIUM1 (LRP1) expression during lateral root development in arabidopsis. Plant J. 101, 87–100. doi: 10.1111/tpj.14520
Sohlberg, J. J., Myrenas, M., Kuusk, S., Lagercrantz, U., Kowalczyk, M., Sandberg, G., et al. (2006). STY1 regulates auxin homeostasis and affects apical-basal patterning of the arabidopsis gynoecium. Plant J. 47, 112–123. doi: 10.1111/j.1365-313X.2006.02775.x
Staldal, V., Cierlik, I., Chen, S., Landberg, K., Baylis, T., Myrenas, M., et al. (2012). The Arabidopsis thaliana transcriptional activator STYLISH1 regulates genes affecting stamen development, cell expansion and timing of flowering. Plant Mol. Biol. 78, 545–559. doi: 10.1007/s11103-012-9888-z
Sunkar, R., Kapoor, A., Zhu, J. K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065. doi: 10.1105/tpc.106.041673
Sun, Z., Li, H., Zhang, Y., Li, Z., Ke, H., Wu, L., et al. (2018). Identification of SNPs and candidate genes associated with salt tolerance at the seedling stage in cotton (Gossypium hirsutum l.). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01011
Ullah, A., Hakim, S. H., Yang, X., Zhang, X. (2018). A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant 162, 439–454. doi: 10.1111/ppl.12651
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, Z., Wang, M., Liu, L., Meng, F. (2013). Physiological and proteomic responses of diploid and tetraploid black locust (Robinia pseudoacacia l.) subjected to salt stress. Int. J. Mol. Sci. 14, 20299–20325. doi: 10.3390/ijms141020299
Wendel, J. F., Grover, C. E. (2015). “Taxonomy and evolution of the cotton genus, gossypium,” in Cotton John Wiley & Sons 2015, 25–44.
Yang, Y., Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Y., Qi, L., Nian, L., Zhu, X., Yi, X., Jiyu, Z., et al. (2021). Genome-wide identification and expression analysis of the SRS gene family in medicago sativa. DNA Cell Biol 40 (12), 1539–1553. doi: 10.1089/dna.2021.0462
Yang, J., Xu, P., Yu, D. (2020). Genome-wide identification and characterization of the SHI-related sequence gene family in rice. Evol. Bioinform. Online 16, 1176934320941495. doi: 10.1177/1176934320941495
Yasir, M., He, S., Sun, G., Geng, X., Pan, Z., Gong, W., et al. (2019). A genome-wide association study revealed key SNPs/Genes associated with salinity stress tolerance in upland cotton. Genes (Basel) 10, 829. doi: 10.3390/genes10100829
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yuan, Y., Xing, H., Zeng, W., Xu, J., Mao, L., Wang, L., et al. (2019). Genome-wide association and differential expression analysis of salt tolerance in Gossypium hirsutum l at the germination stage. BMC Plant Biol. 19, 394. doi: 10.1186/s12870-019-1989-2
Yuan, T. T., Xu, H. H., Li, J., Lu, Y. T. (2020). Auxin abolishes SHI-RELATED SEQUENCE5-mediated inhibition of lateral root development in Arabidopsis. New Phytol. 225, 297–309. doi: 10.1111/nph.16115
Yuan, T. T., Xu, H. H., Zhang, Q., Zhang, L. Y., Lu, Y. T. (2018). The COP1 target SHI-RELATED SEQUENCE5 directly activates photomorphogenesis-promoting genes. Plant Cell 30, 2368–2382. doi: 10.1105/tpc.18.00455
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum l. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhan, Y., Wu, T., Zhao, X., Wang, Z., Chen, Y. (2021). Comparative physiological and full-length transcriptome analyses reveal the molecular mechanism of melatonin-mediated salt tolerance in okra (Abelmoschus esculentus l.). BMC Plant Biol. 21, 180. doi: 10.1186/s12870-021-02957-z
Zhao, S. P., Song, X. Y., Guo, L. L., Zhang, X. Z., Zheng, W. J. (2020). Genome-wide analysis of the shi-related sequence family and functional identification of GmSRS18 involving in drought and salt stresses in soybean. Int. J. Mol. Sci. 21 (5), 1810. doi: 10.3390/ijms21051810
Keywords: genome-wide characterization, SRS family, Gossypium hirsutum, salt stress, regulation of gene expression
Citation: Sun C, Yu L, Zhang S, Gu Q and Wang M (2023) Genome-wide characterization of the SHORT INTER-NODES/STYLISH and Shi-Related Sequence family in Gossypium hirsutum and functional identification of GhSRS21 under salt stress. Front. Plant Sci. 13:1078083. doi: 10.3389/fpls.2022.1078083
Received: 24 October 2022; Accepted: 12 December 2022;
Published: 04 January 2023.
Edited by:
Libei Li, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Abdul Qayyum, The University of Haripur, PakistanGuoyuan Liu, Nantong University, China
Dr. Abdul Rehman, Zhengzhou University, China
Copyright © 2023 Sun, Yu, Zhang, Gu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chendong Sun, MTAwNTUwOTkxOUBxcS5jb20=; Qijuan Gu, Z3FqNjU5MEB0a2dlbmVjbHViLmNvbQ==; Mei Wang, d2FuZ21laUB6YWFzLmFjLmNu
†These authors have contributed equally to this work and share first authorship
 Chendong Sun
Chendong Sun Li Yu2†
Li Yu2† Shuojun Zhang
Shuojun Zhang