- Agronomy, Horticulture, and Plant Science Department, South Dakota State University, Brookings, SD, United States
Own-rooted grapevines and grapevine rootstocks are vegetatively propagated from cuttings and have an adventitious root system. Unraveling the genetic underpinnings of the adventitious root system architecture (RSA) is important for improving own-rooted and grafted grapevine sustainability for a changing climate. Grapevine RSA genetic analysis was conducted in an Vitis sp. ‘VRS-F2’ population. Nine root morphology, three total root system morphology, and two biomass traits that contribute to root anchorage and water and nutrient uptake were phenotyped. Quantitative trait loci (QTL) analysis was performed using a high density integrated GBS and rhAmpSeq genetic map. Thirty-one QTL were detected for eleven of the RSA traits (surface area, root volume, total root length, fresh weight, number of tips, forks or links, longest root and average root diameter, link length, and link surface area) revealing many small effects. Several QTL were colocated on chromosomes 1, 9, 13, 18, and 19. QTL with identical peak positions on chromosomes 1 or 13 were enriched for AP2-EREBP, AS2, C2C2-CO, HMG, and MYB transcription factors, and QTL on chromosomes 9 or 13 were enriched for the ALFIN-LIKE transcription factor and regulation of autophagy pathways. QTL modeling for individual root traits identified eight models explaining 13.2 to 31.8% of the phenotypic variation. ‘Seyval blanc’ was the grandparent contributing to the allele models that included a greater surface area, total root length, and branching (number of forks and links) traits promoting a greater root density. In contrast, V. riparia ‘Manitoba 37’ contributed the allele for greater average branch length (link length) and diameter, promoting a less dense elongated root system with thicker roots. LATERAL ORGAN BOUNDARY DOMAIN (LBD or AS2/LOB) and the PROTODERMAL FACTOR (PFD2 and ANL2) were identified as important candidate genes in the enriched pathways underlying the hotspots for grapevine adventitious RSA. The combined QTL hotspot and trait modeling identified transcription factors, cell cycle and circadian rhythm genes with a known role in root cell and epidermal layer differentiation, lateral root development and cortex thickness. These genes are candidates for tailoring grapevine root system texture, density and length in breeding programs.
1 Introduction
Grapevine (Vitis sp.) is one of the most economically important fruit crops cultivated in the USA and the world (Nass, 2019). The increased temperature, water, and pest stresses associated with climate change frequently exceeds practical viticulture solutions. Therefore, to maintain an ecologically sound production system, development of cultivars with improved biotic and abiotic stress tolerance, water use, fruit quality, yield, and manageability is necessary (Limera et al., 2017). Grapevines are vegetatively propagated and either grown own-rooted or grafted to a rootstock. Adventitious roots provide anchoring and mechanical support, absorb water and nutrients from the soil, store carbohydrates, and exposure to beneficial soil microorganisms (Bellini et al., 2014). They serve as the only interface to sense and respond to changing soil environments, enabling plants to overcome abiotic stress challenges. The grape phylloxera epidemic of the late 1800s in Europe spurred the development of phylloxera resistant rootstocks and hybrid own-rooted grapevines. A large portion of the resulting rootstock cultivars are closely related and selected for phylloxera resistance and graft compatibility rather than stress tolerance and there has been limited genetic analysis of grapevine root systems (Walker et al., 2014; Ollat et al., 2016; Dalbó and Souza, 2019; Riaz et al., 2019). In contrast, genetic analysis of grapevine scion cultivars for improved fruit quality, abiotic and biotic resistance, cold tolerance, seedlessness, and other enological and phenological traits have proceeded rapidly (Correa et al., 2014; Delrot et al., 2020; Gautier et al., 2020; Zinelabidine et al., 2021; Vezzulli et al., 2022). The need for improved stress tolerance and pest resistance in changing climatic conditions highlights the need for a greater understanding the genetic basis of root system architecture (RSA) to continue development of improved own-rooted cultivars and rootstocks (Serra et al., 2014; Hugalde et al., 2021b).
The RSA is the spatial distribution of roots and reflects the shape, three-dimensional distribution, and branching pattern of post-embryonically generated roots or adventitious roots (Lynch, 1995; Satbhai et al., 2015). Root architecture can be described by morphology, topology, geometry, and growth dynamics (Lynch, 1995). Morphology refers to root shape, diameter, length, and orientation. The topology is the connection of roots through branching such as primary or secondary order roots. The positional gradient of roots or root biomass/length and their soil depth contribute to root geometry. Root growth rate and lateral emergence rate are examples of root dynamics. Roots have a seasonal growth pattern that optimizes nutrient and water uptake and anchorage. The main environmental factor that impacts root system growth and development is the heterogeneity of the soil properties such as bulk density, texture, water availability, and nutrient content (Smart et al., 2006; Comas et al., 2013; Serra et al., 2014; Yildirim et al., 2018). RSA shows considerable variation among species, genotypes within a species, and cultivars (Lynch, 1995). Root trait mapping studies for the annual crops rice (Uga et al., 2013), soybean (Valliyodan et al., 2017), barley (Robinson et al., 2018), and wheat (Zhang et al., 2019) have identified colocated quantitative trait loci (QTL) for root branching, morphology, biomass, and yield. In the deciduous tree Populus, adventitious root QTL are colocated with related shoot traits indicating the importance of the root system on shoot development (Han et al., 1994; Sun et al., 2019). Scion transpiration rate and acclimation to water deficit are controlled by rootstock genetics (Marguerit et al., 2012). An interspecific V. vinifera L. × V. riparia Michx. rootstock population study indicates that scion transpiration is controlled by a small number of loci each responsible for<10% of the phenotypic variance and suggests that hormonal (abscisic acid) and hydraulic (aquaporins) signaling genes play a role in the rootstock genotype response to water deficit (Marguerit et al., 2012). Studies of V. riparia × V. labrusca and ‘Riparia Glorie’ drought sensitive rootstocks have shallower root distribution than the drought-tolerant rootstocks ‘110R’ (Vitis berlandieri × Vitis rupestris) and ‘Ramsey’ (Vitis champinii) (Fort et al., 2017). The interspecific rootstocks V. riparia × V. rupestris Scheele. and V. vinifera × V. berlandieri Planch. had different capacity to withstand water and sodium chloride stress indicating that genotype variation contributes to root physiological and functional differences in response to the environment (Meggio et al., 2014). Petiolar nutrient concentration is influenced by rootstock and V. riparia contributes to lower petiolar magnesium and phosphorus and higher sulphur concentration in the scions in comparison to rootstocks with V. rupestris or V. berlandieri (Gautier et al., 2020).
The adventitious root system and rootstocks contribute to the sustainability of the grapevine; however, most reported root traits are controlled by multiple genes, each governing small effects and often changing with environmental conditions (De Dorlodot et al, 2007; Cooper et al., 2009). Marker assisted selection for efficient grapevine root systems has been limited due to lack of rapid and accurate phenotyping methods for roots and linkage of root phenotype to crop productivity (Lynch, 1995). The degree of diversity and associated variation in root traits, their complex genetic control, and the strong environmental effect on morphological traits inhibits traditional genetic studies in grapevine root systems (De Dorlodot et al, 2007). These difficulties result in a gap in our understanding of the genetic control of RSA and contribute to the lack of markers needed to assist in selection for improved root systems in own-rooted or rootstock cultivars. Therefore, this study was undertaken to explore the genetic architecture of root morphology in an interspecific F2 grapevine population and identify loci and candidate gene influencing adventitious root system morphology.
2 Materials and methods
2.1 Plant material and growing conditions
The VRS-F2 diploid population was produced by selfing a single F1 (V. sp. ‘16_9_2’) developed from a cross between V. riparia Michx. (seed parent, ‘Manitoba 37’, PI#588289) and V. sp. ‘Seyval blanc’ (pollen parent, VIVC#11558) (Fennell et al., 2005). Six-year-old potted VRS-F2 vines for this study were cycled annually through the greenhouse and cold storage in South Dakota State University, Brookings, SD (44.31°C N, 96.80°C W). The ecodormant vines were root pruned and repotted in soil, perlite, and peat growing medium (1:2:2 by volume), grown five months, induced into dormancy by natural short daylengths, and returned to cold storage after harvesting canes. Dormant replicate canes were collected from 266 F2 individuals, the population parent, and grandparents in early November and stored at 4°C as three-node cuttings (nodes 3-5 from the cane base) keeping genotype identity. For this study, chilling fulfilled canes were placed in a container with 10 cm water layer for three days to ensure uniform hydration. After three days of hydration, single node cuttings with swollen buds (six cm cane sections) were selected from the center of the 3-node cutting and placed randomly in a rooting box (60×45×15 cm (length x width x depth)) of perlite. Six replicate cuttings were used for each VRS-F2 genotype, 28 replicates for the F1 parent, and 18 replicates for each grandparent. Cuttings were placed randomly (maintaining identity) 7.5 cm apart within row and between row spacing. The root boxes were flooded and drained daily to maintain uniform moisture content. The rooting study was conducted in the greenhouse with >14-hour natural daylength at 26 ± 3°C and 80% relative humidity.
2.2 Trait measurements
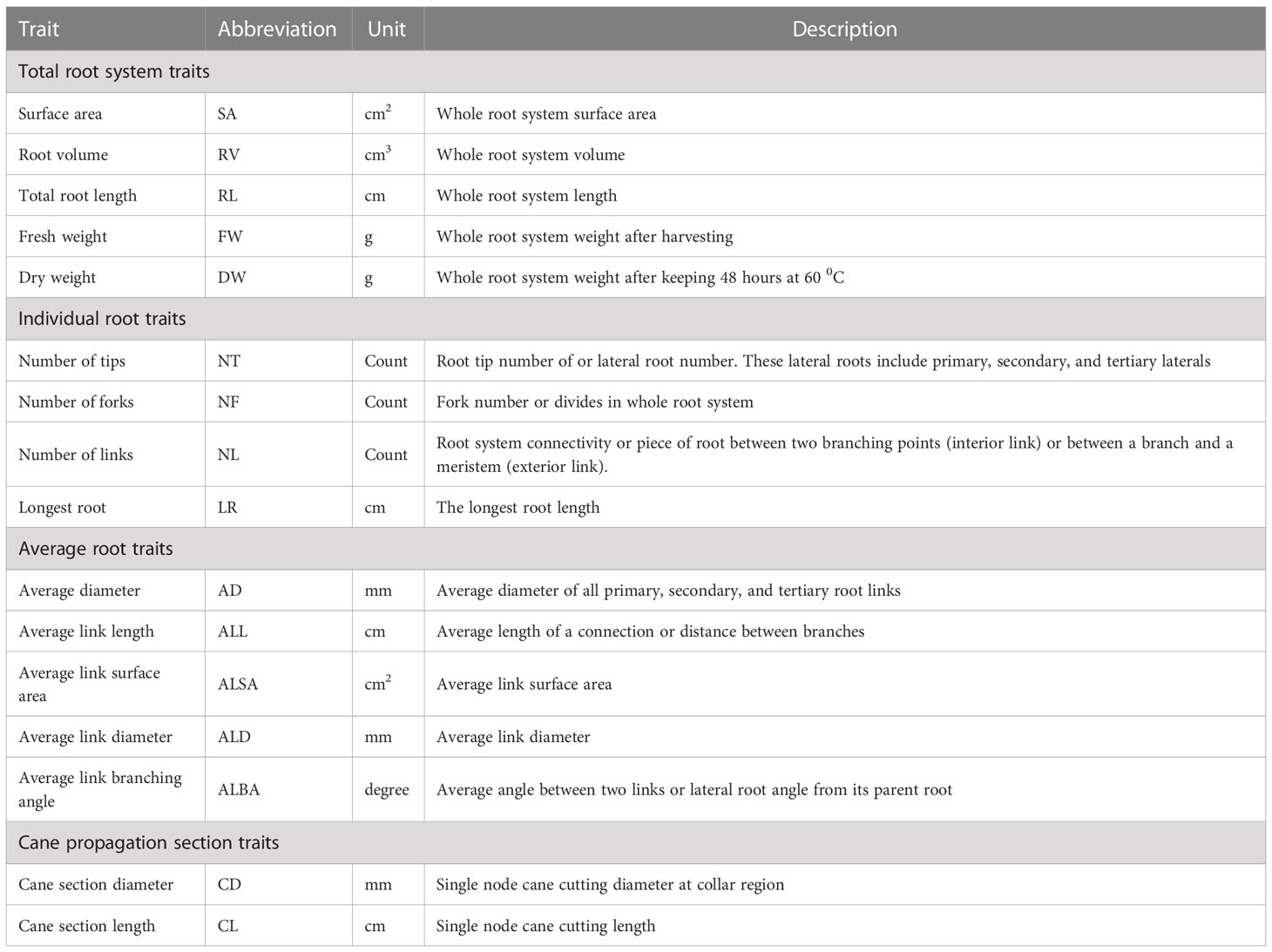
After 35 days, plants were harvested and cleaned using tap water to remove all perlite particles. Samples were stored (in plastic bags with 1 ml of water) in a 4°C cooler until they were scanned. Genotypes and their RSA replicate-identity were maintained throughout the experiment. Each root system was scanned using Epson scanner (PERFECTION V700PHOTO, Seiko Epson Corporation, Tokyo, Japan). Each root system was scanned twice (two different faces on scanner) to get the mean traits. After scanning, the length of the longest root was measured with a ruler. All the roots were then cut off at the collar region. The root fresh weight (FW) was measured, and the roots were dried at 60°C for 48 hours and the dry weight (DW) measured. Finally, the diameter (CD) and length (CL) of the single node cane section were measured manually. Eleven traits related to RSA were measured using WinRhizo software Reg 2016a (Regent Instruments Inc, Quebec, Canada). The root and propagule traits were categorized into four groups (total root system characters, individual root characters, average root characters, and stem characters) based on their morphology (Table 1). These root trait category names and trait name abbreviations are used throughout the results and discussion.
2.3 Statistical analysis
For each trait, the mean, median, minimum, maximum, and standard error were calculated. The grandparent’s trait means were tested for significant differences using a t-test in R (R Core Team, 2019). Only VRS- F2 genotypes (239) with no missing replicates were used for analysis. The genotype mean trait values for 16 traits were explored for their correlation, major trait contribution by principal component (PCA), and genotype relationship to grandparents or parent by cluster analysis. The data were analyzed using different packages in R statistical software (R Core Team, 2019). Descriptive data analyses were performed using dplyr (Wickham et al., 2019) and psych (Revelle, 2018) packages. The Hmisc package (Harrell, 2019) was used to calculate significant trait Pearson correlation coefficients. Principal component analysis was conducted for 16 traits (including 14 RSA traits and 2 stem traits) to identify trait contribution to RSA using the factoextra package (Kassambara and Mundt, 2017). Unsupervised k-means clustering method was used to categorize the 239 VRS-F2 genotypes with zero missing trait values using cluster package (Maechler et al., 2019).
2.4 Integrated VRS-F2 map
An integrated VRS-F2 GBS-rhAmpSeq genetic map using 1449 GBS markers as described in Yang et al. (2016) and 1970 rhAmpSeq markers developed from a genus-wide core genome and described in Zou et al. (2020). Genotyping and SNP calling of GBS markers were performed as described in Yang et al. (2016). rhAmpSeq marker development including DNA processing and genome assembly, core-genome construction, genus-wide variant calling, marker design pipeline, rhAmpSeq sequencing and genotyping, and quality control were detailed in Zou et al. (2020). As described in Alahakoon et al. (2022) distorted GBS and rhAmpSeq markers were tested using a threshold 1 × 10-21 chi-square p-value. Linkage groups containing a total of 2519 markers across 19 chromosomes with LOD = 5 were established using JoinMap (version 5, Kyazma B. V., Wageningen, Netherlands) (Alahakoon et al., 2022). Quantitative mapping was carried out for each of the 14 root traits, using the VRS-F2 GBS-rhAmpSeq integrated genetic map and R/qtl software (Broman et al., 2003; Alahakoon et al., 2022). Normal distribution of RSA phenotypes was measured using Shapiro-Wilk test and those not normally distributed were transformed to achieve normality. QTL were determined using scanone function in R/qtl. A permutation test was performed to identify 5% genome-wide log10 likelihood ratio (LOD) threshold (1,000 permutations). Eight traits had multiple significant QTL and QTL modeling was performed for those eight traits. All significant QTL used to build an additive model (y~x1+⋯+xn+∈, where y is the trait, x is the QTL, n is the number of significant QTL, and ∈ is the error term). Modeling of multiple QTL for a given trait was conducted according to the R/qtl package and QTL contributing to the model were tested for significant interactions and no interactions were significant. The QTL peak position marker, LOD score, percent of variation explained by individual and modeled QTL were evaluated by analysis of variance. Using Bayesian method, 95% confidence intervals were calculated. Dominant allele and the contributing grandparent for each of the modeled trait were identified.
2.5 Pathway analysis
Pathway enrichment analysis of individual traits was conducted by extracting the genes in the Vitis vinifera ‘PN40024 12X.v2 genome using 700 Kb either side of peak position considering that candidate genes should be within 3-4 cM of peak position or 1.4 Mb total based on grape genome size (Cipriani et al., 2011; Hugalde et al., 2021a). This 1.4 Mb region was used to identify enriched VitisNet pathways and candidate genes (Cipriani et al., 2011; Grimplet et al., 2012). The pathways were analyzed using the VitisNet functional annotation of the Vitis vinifera ‘PN40024 12X.v2’ genome and Fisher’s test (p-value< 0.05 for enrichment (Grimplet et al., 2012; Osier, 2016). VitisNet pathway enrichment analysis was also conducted for QTL that had three or more traits with the same peak position to identify RSA candidate genes.
3 Results
3.1 RSA phenotypic variation among grandparents, the parent and F2 population
Grapevine adventitious roots that developed under well-watered conditions were analyzed for 239 VRS-F2 genotypes. The RSA phenotypic variation for the grandparents and the parent of the VRS-F2 population is shown in Figure 1. Significant differences between means of the grandparents were detected for all measured RSA traits except RV, FW, LR, and ALBA (Table 2). SA, TRL, DW, NT, NF, and NL values were greater for ‘Seyval blanc’ than for V. riparia ‘Manitoba 37’ and AD, ALL, ALSA, and ALD were greater for V. riparia than for ‘Seyval blanc’. The values for the VRS-F2 population showed wide variation for all traits with values ranging from less to greater than the means of the parent and grandparents (Table 2). Original trait distributions showed deviation from the normality (Supplementary Figure 1). Traits were normalized and a Shapiro-Wilk normality test p-values revealed that eleven traits were successfully transformed with p-value > 0.05.

Figure 1 Representative adventitious root systems for the grandparents and parent of the VRS-F2 population. V. riparia ‘Manitoba 37’ (left) and ‘Seyval blanc’ (middle), and the VRS-F2 population parent 16-9-2 (F1) (right).
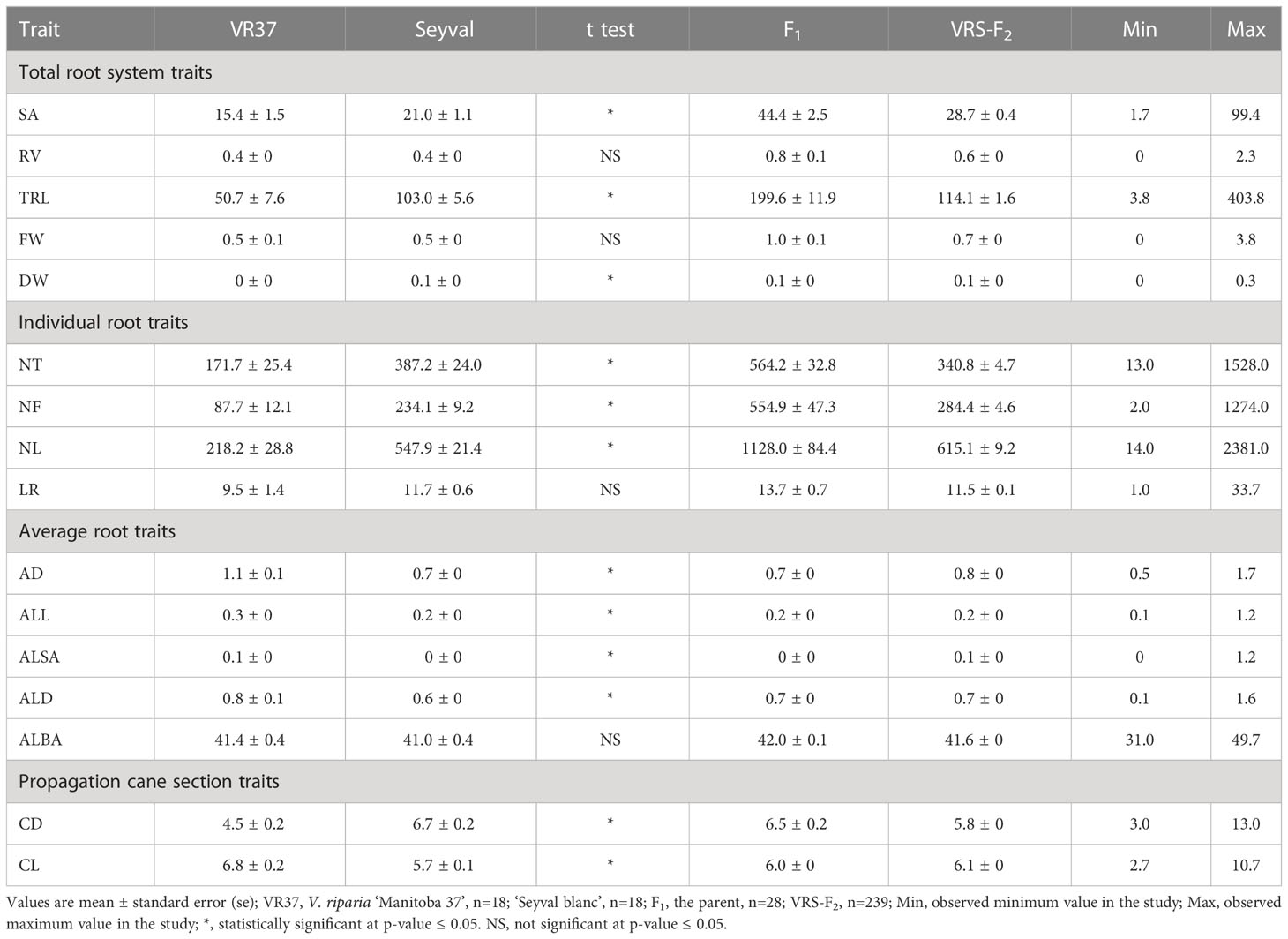
Table 2 Descriptive RSA trait statistics for V. riparia ‘Manitoba 37’ and ‘Seyval blanc’ (grandparents), F1 (parent), and the F2 population.
3.2 Trait correlations
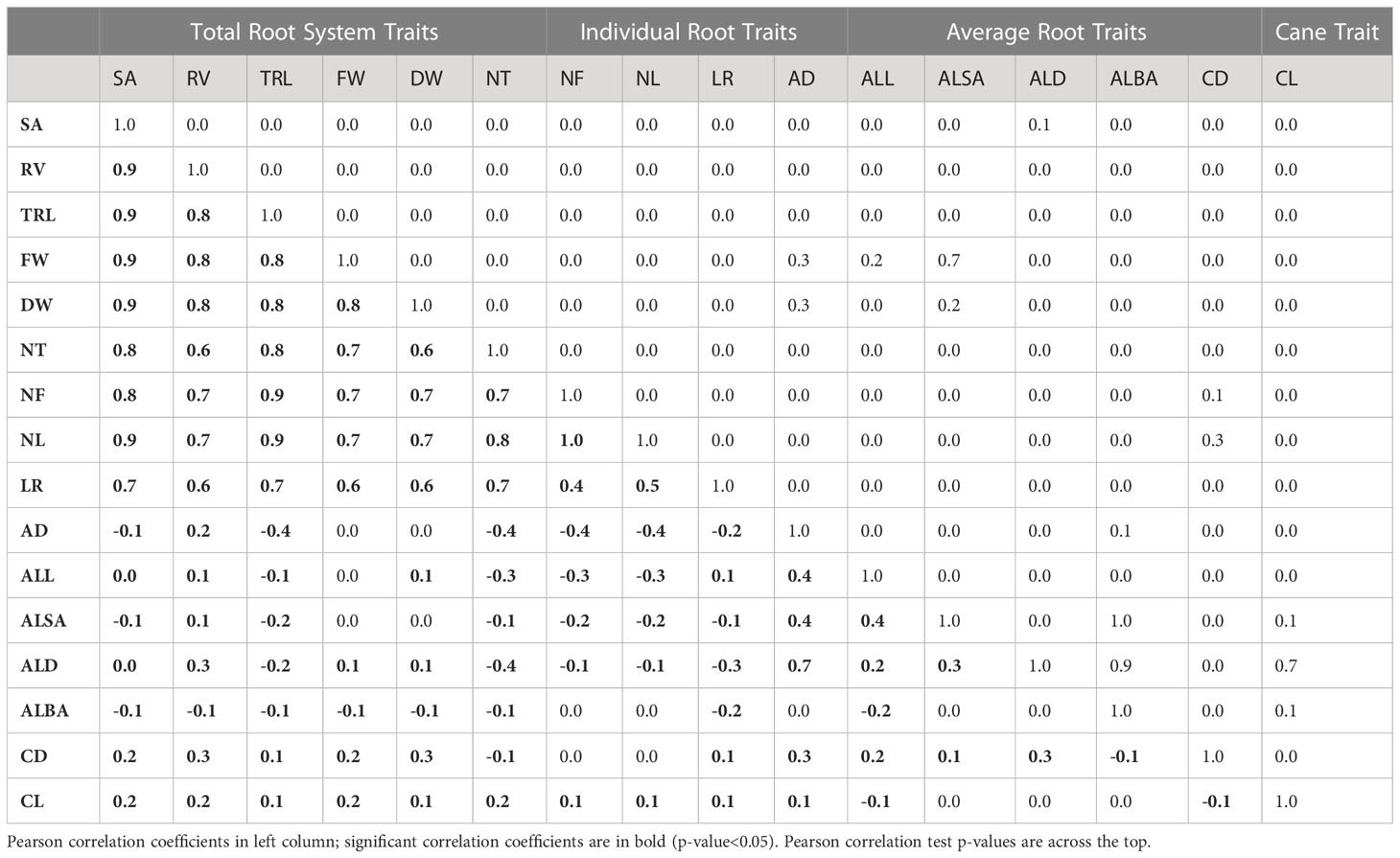
The total root system traits and individual root characteristics traits showed significant (p-value<0.05) and strong positive correlations (Table 3). Average root characteristic traits did not display a strong correlation with total root system traits or individual root characteristics traits. The AD and total root system characteristics and individual root characteristics traits that would contribute to root length and number had a negative correlation. AD had a positive relationship with the other average traits (ALL, ALSA, and ALD). The cane propagule section diameter and length (CD and CL) did not show a strong correlation with any RSA traits.
3.3 Morphological traits explain major part of the RSA
The principal component analysis was performed for all RSA traits measured to identify the trait contribution to RSA. The first principal component (Dim1), and second (Dim2), and third (Dim3) explained 45.9%, 18.8%, and 10.3% of the root system variation, respectively. The first dimension was predominately explained by total root system characteristics and individual root characteristics traits. Average root characteristics traits contributed to the second dimension. ALBA mainly characterized the third dimension. Traits that contributed to first and second dimensions clustered separately in the PCA biplot (Figure 2).
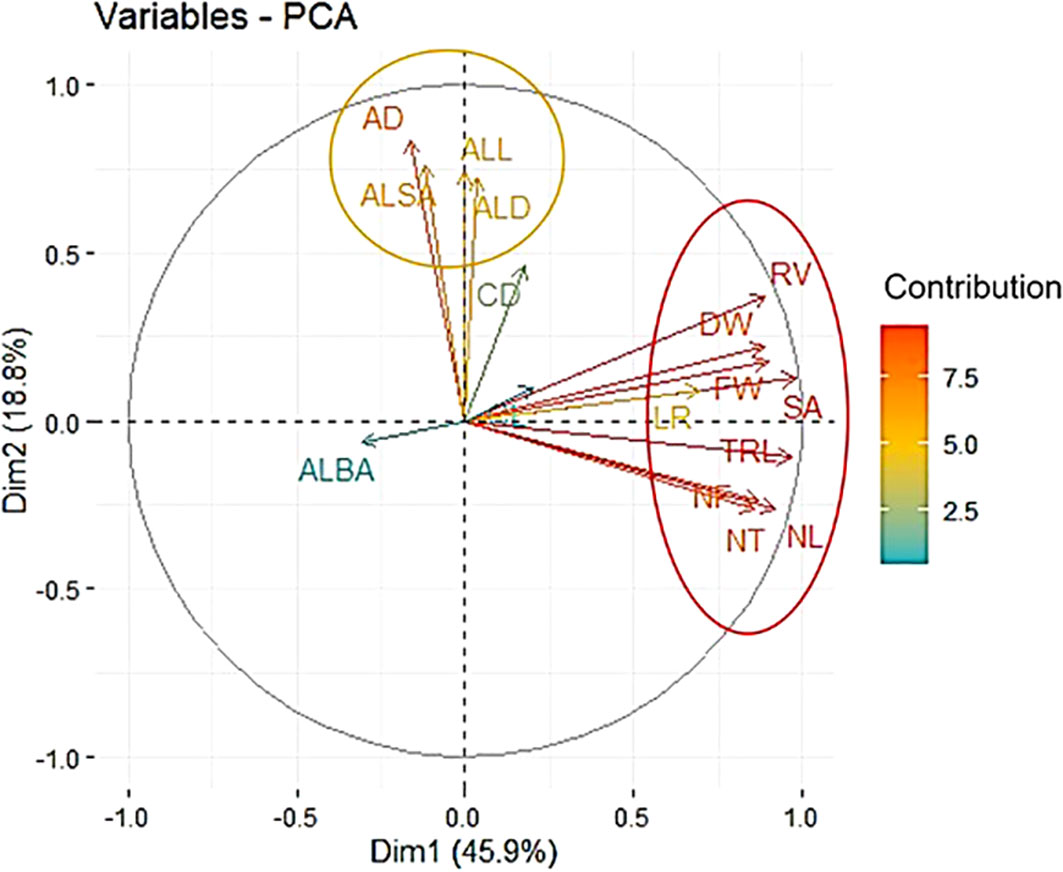
Figure 2 Root system architecture trait principal component biplot. The first (Dim1) and second (Dim2) principal component dimensions are in x and y-axis respectively. Red and orange ovals represent clusters formed. Vector color represents the percent contribution of each individual trait to the first and second principal components.
3.4 Genetic diversity in VRS-F2 population
Cluster analysis of the 14 RSA traits and the 2 propagule traits classified 239 genotypes into three groups (Figure 3). Cluster 1 comprised 55 genotypes plus the female grandparent, V. riparia ‘Manitoba 37’. A second cluster contained ‘Seyval blanc’ and 120 genotypes, while the third cluster included 66 genotypes plus the F1 parent.
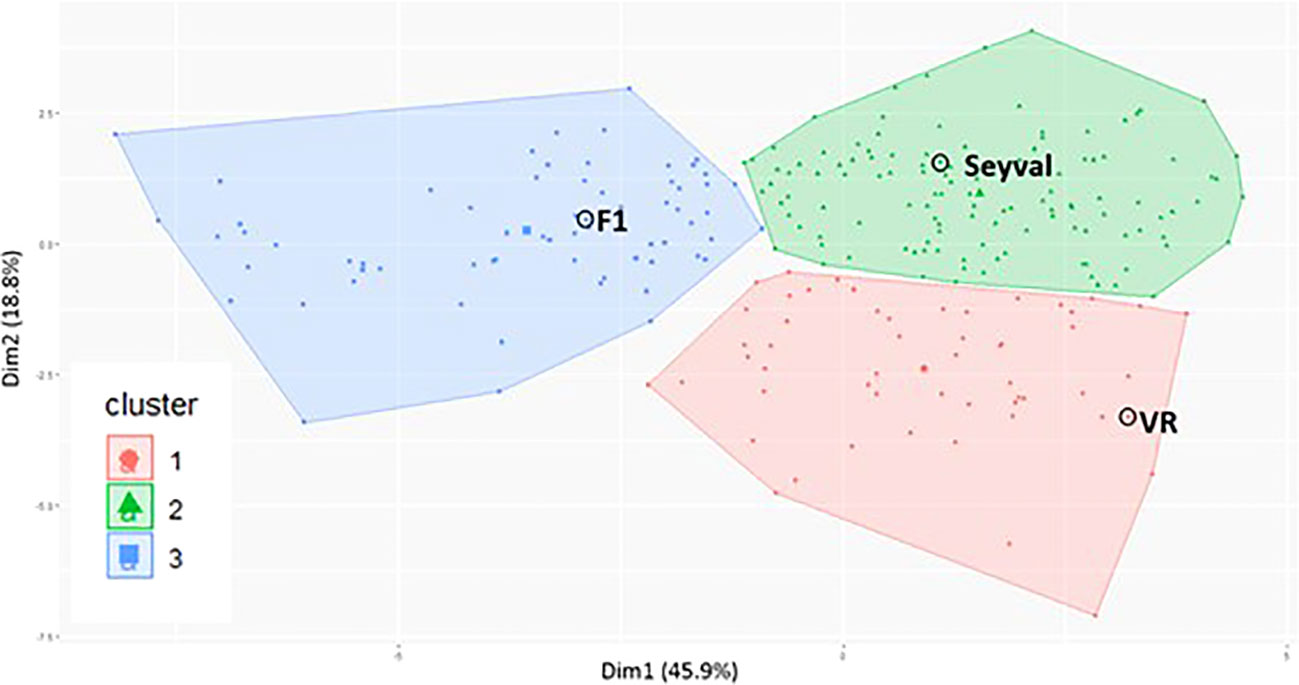
Figure 3 VRS-F2 population cluster plot for 14 root system traits and 2 propagule traits. Cluster plot was generated by using k-means clustering method and principal component analysis. VR indicates V. riparia ‘Manitoba 37’, Seyval indicates ‘Seyval blanc’ and F1 indicates the VRS-F2 population parent. Dots not circled represent VRS-F2 genotypes.
3.5 QTL detection and colocalization
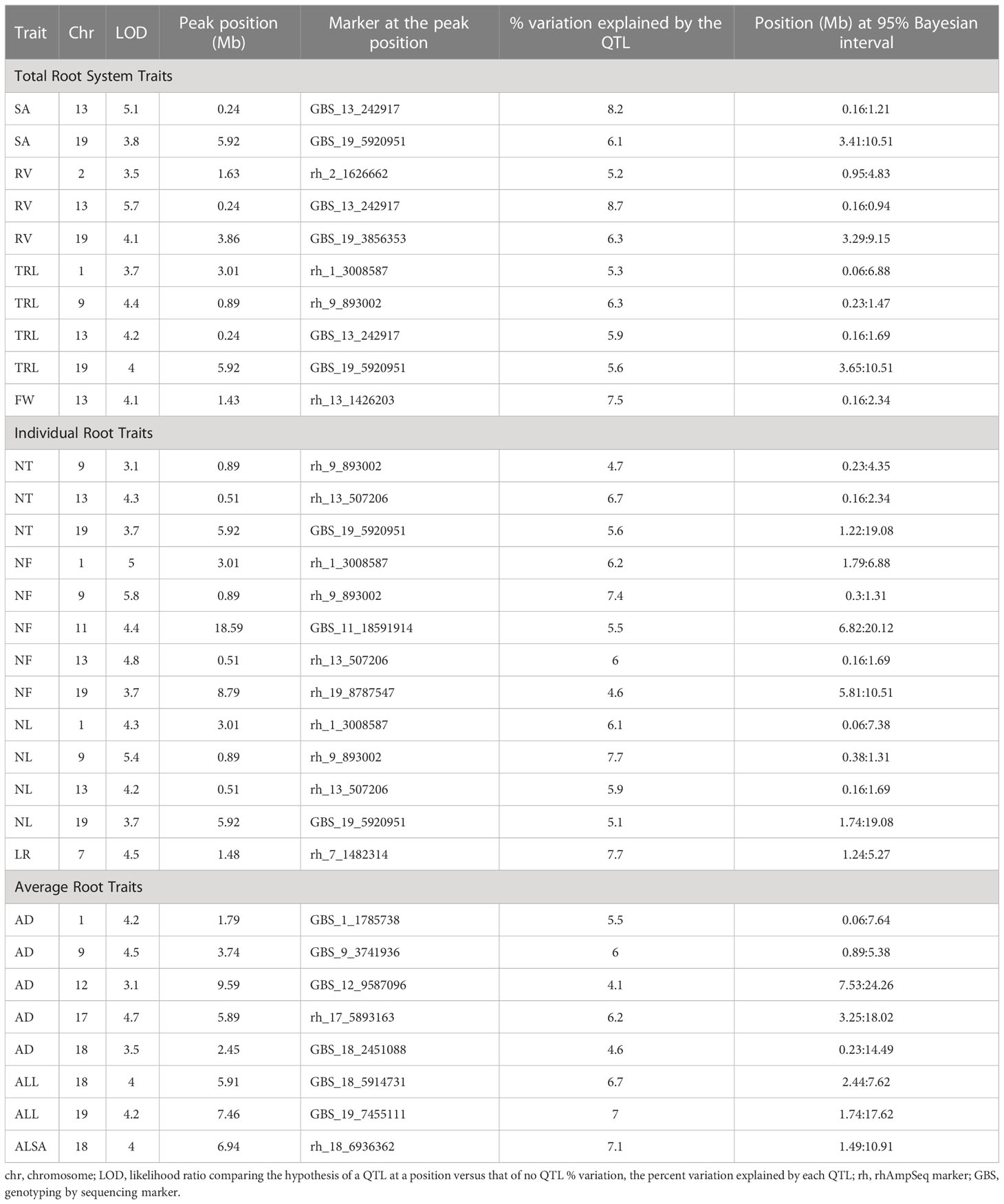
Thirty-one QTL were detected for 11 RSA traits on chromosomes 1, 2, 7, 9, 11, 12, 13, 17, 18, and 19 (Table 4, Figure 4). No QTL were observed for the DW biomass trait, ALBA, and ALD average morphology traits or the CD and CL propagule traits. Multiple QTL were closely located on chromosomes 1, 9, 13, 18, and 19. In chromosomes 1, 9, 13, and 19 there were five identical peak positions for three or more trait QTL on chromosome 1 (NF, NL, and TRL; 3.01 Mb), 9 (TRL, NT, NF, and NL; 0.89 Mb), 13 (SA, RV, and TRL; 0.24 Mb) and (NT, NF, and NL; 0.51 Mb), and 19 (SA, TRL, NT, and NL; 5.92 Mb) (Table 4). The two separate peak positions for multiple traits on chromosome 13 were close to the end of the chromosome and had overlapping and small confidence intervals; therefore, for enrichment analysis they were analyzed as one position. None of the QTL identified on chromosome 18 had identical peak positions (Table 4). Single QTL were found on chromosomes 2 (RV), 7 (LR), 11 (NF), 12 (AD), and 17 (AD) (Table 4).
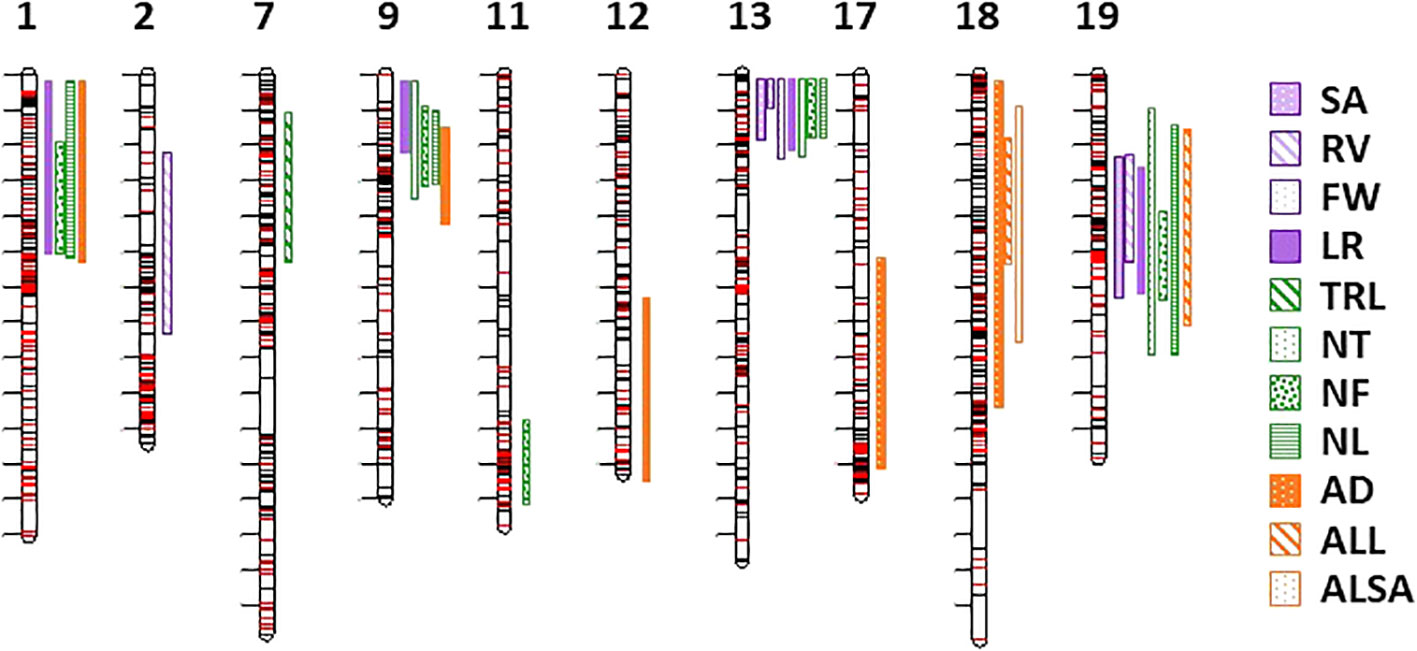
Figure 4 Chromosomal location of 31 QTL for eleven RSA traits. Chromosome number is at the top of the image. Legend on right of image identifies RSA traits with QTL (surface area (SA); root volume (RV); fresh weight (FW); total root length (TRL); longest root (LR); number of tips, forks, and links (NT, NF, NL); and average diameter, link length, and link surface area (AD, ALL, ALSA). Length of the band identifies the confidence interval.
3.6 Identical QTL peak positions identify inter-relationship of root traits
Chromosomes 1, 9, 13, and 19 had QTL peak positions that were identical for three to six root traits on each chromosome (Figure 5). These QTL had narrower confidence intervals than most of the single QTL. Examination of enriched VitisNet pathways underlying 700 kb either side of the peak position on each chromosome identified 17 enriched pathways that occurred in at least 2 of these loci and showed the interplay between individual and total root traits (Figure 5). All enriched pathways for each QTL are noted for the chromosome hotspot in Supplementary Table 1. In chromosomes 1 and 13 the enriched transcription factor pathway contained LATERAL ORGAN BOUNDARIES DOMAIN genes (LBD4 and ASYMETRIC LEAVES 2, vv60007AS2) close to the QTL peak position. Enriched ALFIN-LIKE transcription factor and auxin signaling pathways on chromosome 9 contained an ALFIN-LIKE PHD FINGER PROTEIN (ALFINDOM8, vv60002ALFIN) and six auxin signaling genes near the QTL peak position. On chromosome 13, which had 6 traits mapping with an identical peak QTL, there were cell cycle and AS2 transcription factor pathways enriched and these included several cell cycle genes and two additional LBD/LOB genes in the loci. The QTL on chromosome 19 contained enriched actin cytoskeleton and HOMEO BOX DOMAIN (HB) transcription factor pathways which included TORTIFOLIA1 (TOR1, which regulates cortical microtubules) and PROTODERMAL FACTOR 2 (PDF2) near the peak position. On chromosome 1 and 19, circadian rhythm pathway was enriched with the presence of CONSTANS-LIKE 11 and 16 and an ALTERED RESPONSE TO GRAVITY (ARG1) from the primary transporter enriched pathway were within the QTL confidence interval. Regulation of autophagy pathway was enriched (AUTOPHAGY, APG 12 and 18) for QTL on chromosome 9 and 13.
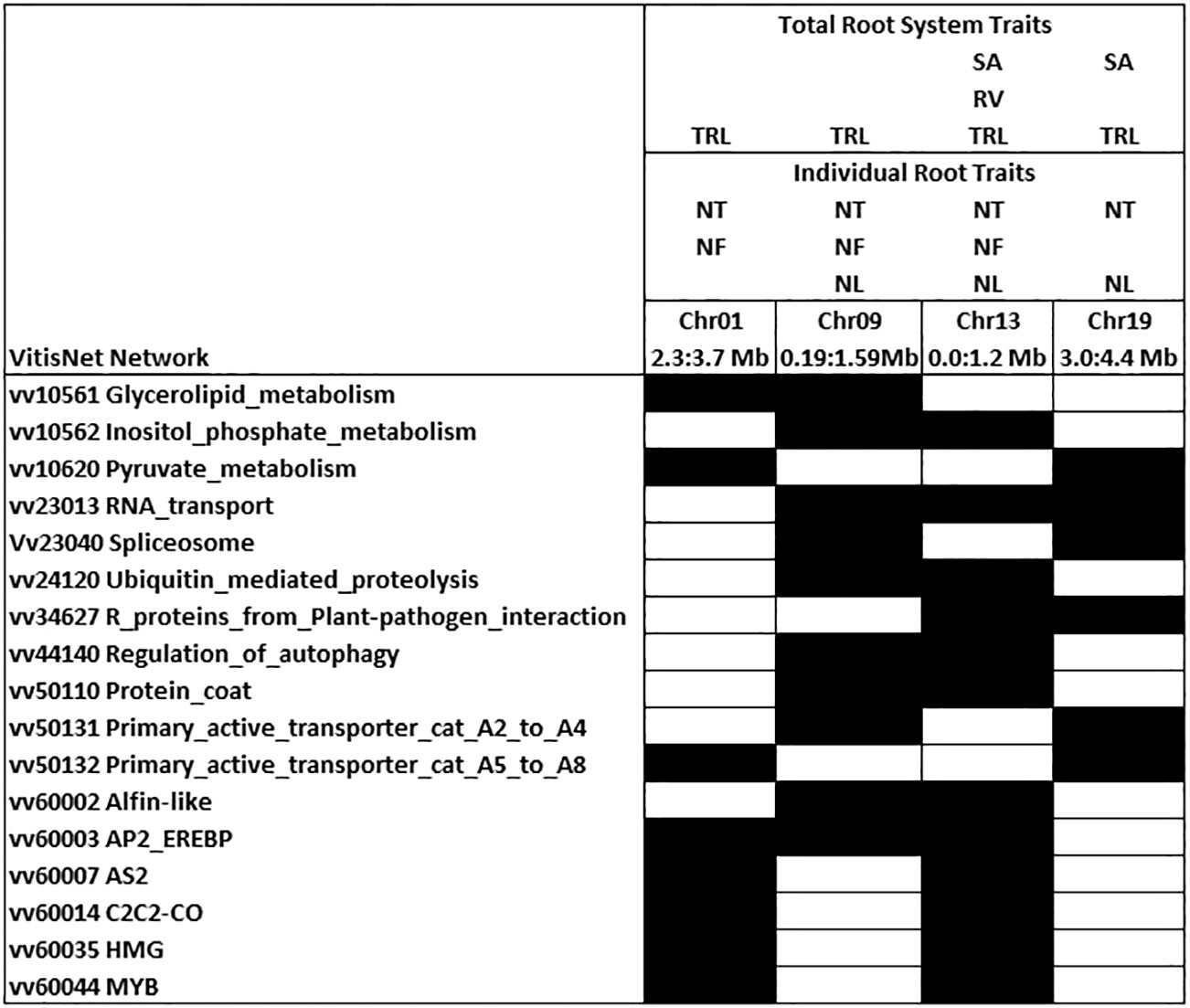
Figure 5 Enriched VitisNet networks for traits with identical QTL peak positions. Shading indicates the significantly enriched pathway (Fisher test p-value 0.05) in at least two of the loci. Trait abbreviations are noted in Table 1. All enriched pathways and contributing genes for each hotspot are identified in Supplementary Table 1. The two peak positions on chromosome 13 were merged into one hotspot as the 700 kb regions overlapped.
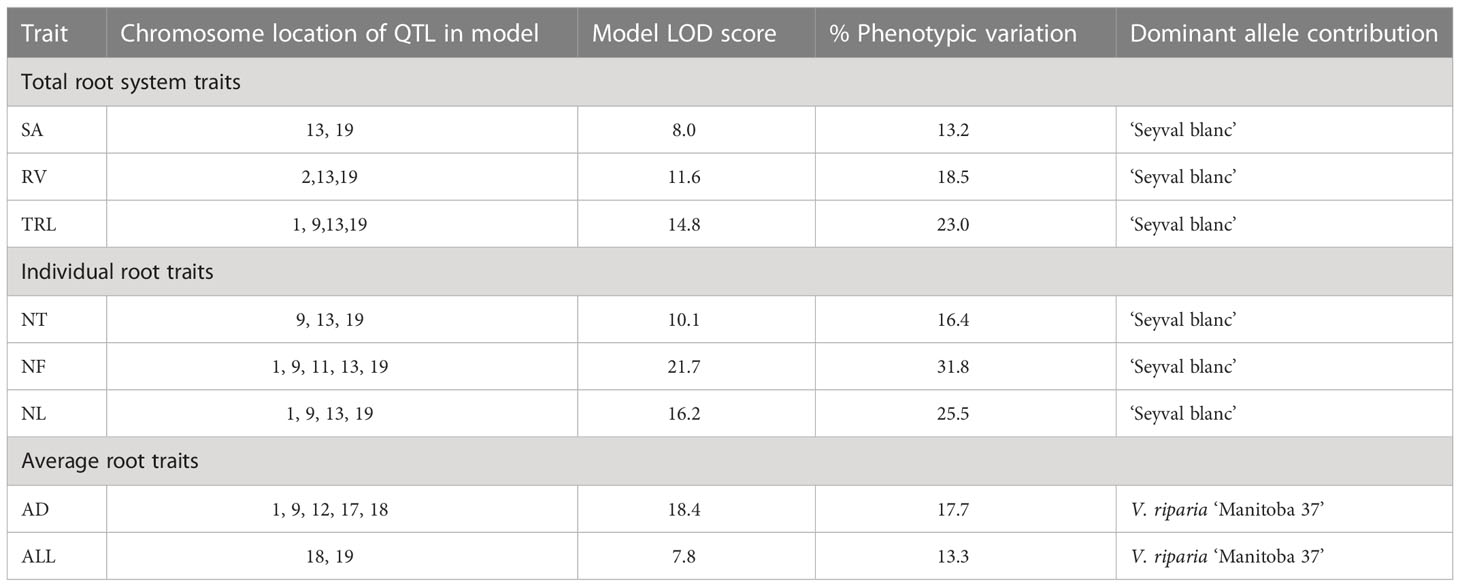
The variation explained by each QTL was small; therefore, models were tested for the individual root traits with more than one QTL. All the trait models were additive and no significant interactions were dectedted for this study. Modeling increased the explained phenotypic variation two- to five-fold for each trait to individual QTL (Tables 4 and 5). Both grandparents contributed dominant alleles for different RSA traits. ‘Seyval blanc’ grandparent contributed dominant allele for modeled traits (TRL, SA, RV, NT, NF, and NL). Three pathways were uniquely enriched for the individual NT, NF, and NL QTL; specifically, the regulation of autophagy (VV44140), the APETALA2-ETHYLENE- RESPONSIVE ELEMENT BINDING PROTEIN (VV60003 AP2-EREBP) and ASYMMETRIC LEAVES2 (VV60007 AS2) transcriptions factors pathways (Supplementary Table 1). Exploration of the genotype effect for NF using two markers near the LATERAL ORGAN BINDING DOMAIN (LBD) candidate genes showed the dominant A allele in homozygous or heterozygous genotypes (A contributed by ‘Seyval blanc’) had a greater number of forks than the homozygous B genotypes (Table 5, Figure 6). A negative correlation existed between the NF and AD traits. Examination of the genotype effect for the same markers relative to the AD phenotype indicated that the dominant allele B is associated with greater AD. V. riparia contributed dominant allele for modeled traits AD and ALL (Table 5). The dominant allele contributor for the single QTL traits FW and LR was ‘Seyval blanc’ and for the ALSA trait it was V. riparia.
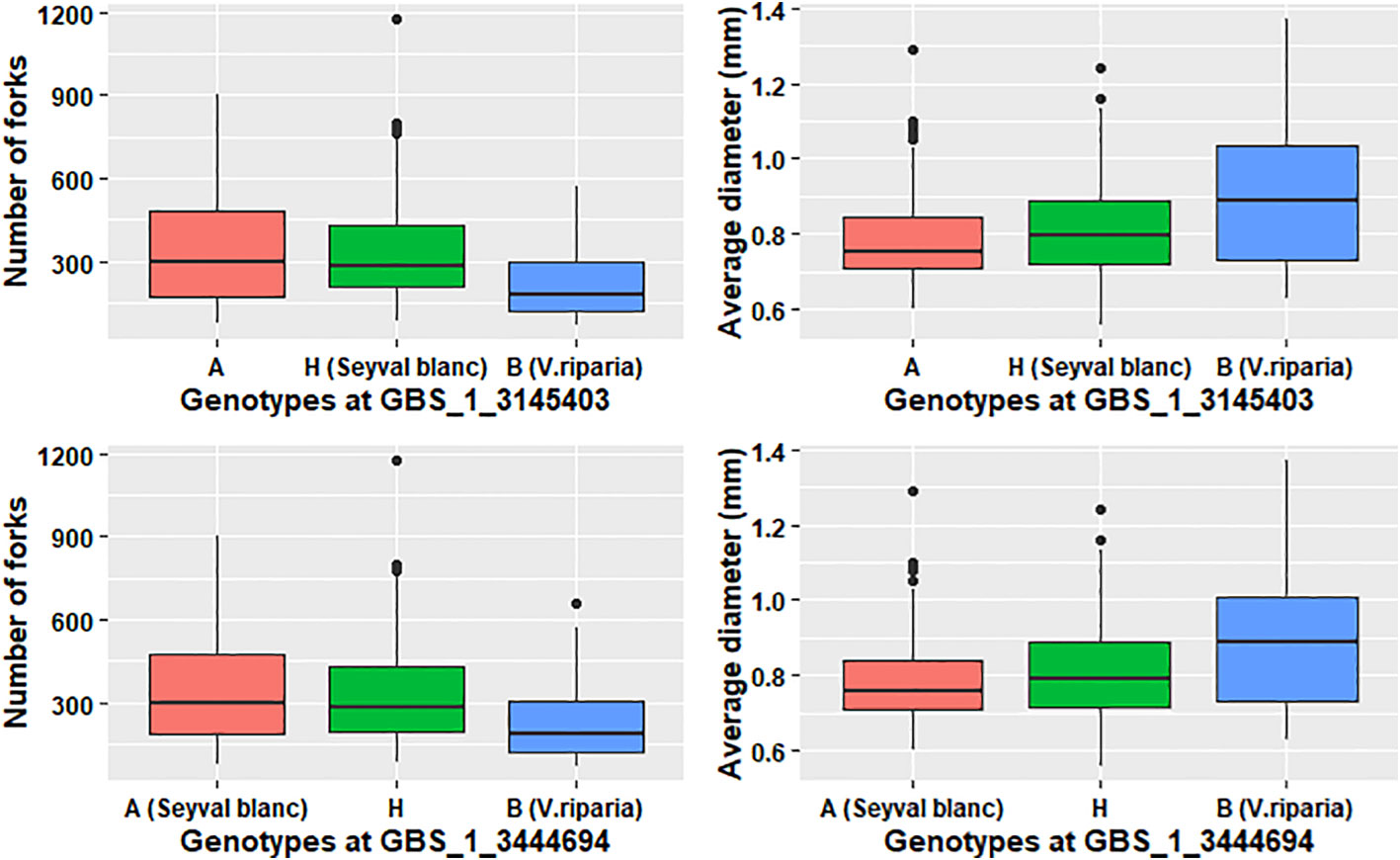
Figure 6 Genotype effect for markers surrounding LATERAL ORGAN BOUNDARIES DOMAIN candidate genes. The grandparent genotype is indicated in parentheses.
3.7 VitisNet Pathway enrichment analysis in modeled traits
An analysis of all genes underlying the 700 Kb region either side of each QTL peak position for modeled traits indicated > 2200 unique genes for the regions underlying modeled trait QTL. Enriched VitisNet pathways and their contributing genes for all modeled trait QTL are indicated in Supplementary Table 2. The number of enriched pathways varied by trait and there were 27 pathways enriched for five or more of the eight modeled traits (Figure 7). TRL and individual root traits (NT, NF, and NL) showed the most enriched pathways in-common (Figure 7), similarly they had the greatest co-localization of QTL (Table 4). The individual root traits (NT, NF, and NL) showed 25 in-common enriched pathways underlying their QTL, with ABA and Auxin signaling, circadian rhythm, cell cycle, Regulation of autophagy, AP2_EREBP and BASIC HELIX-SPAN-LOOP-HELIX BHSH transcription factors (Supplementary Table 2). The modeled average traits AD and ALL had 12 enriched pathways in common and nine of these were in common with other root traits (Supplementary Table 2; Figure 7). Of the average root traits, ALL had the most enriched pathways in common with the individual root traits and TRL; notably circadian rhythm, cell cycle and HB and MYB transcriptions factors (Supplementary Table 2).
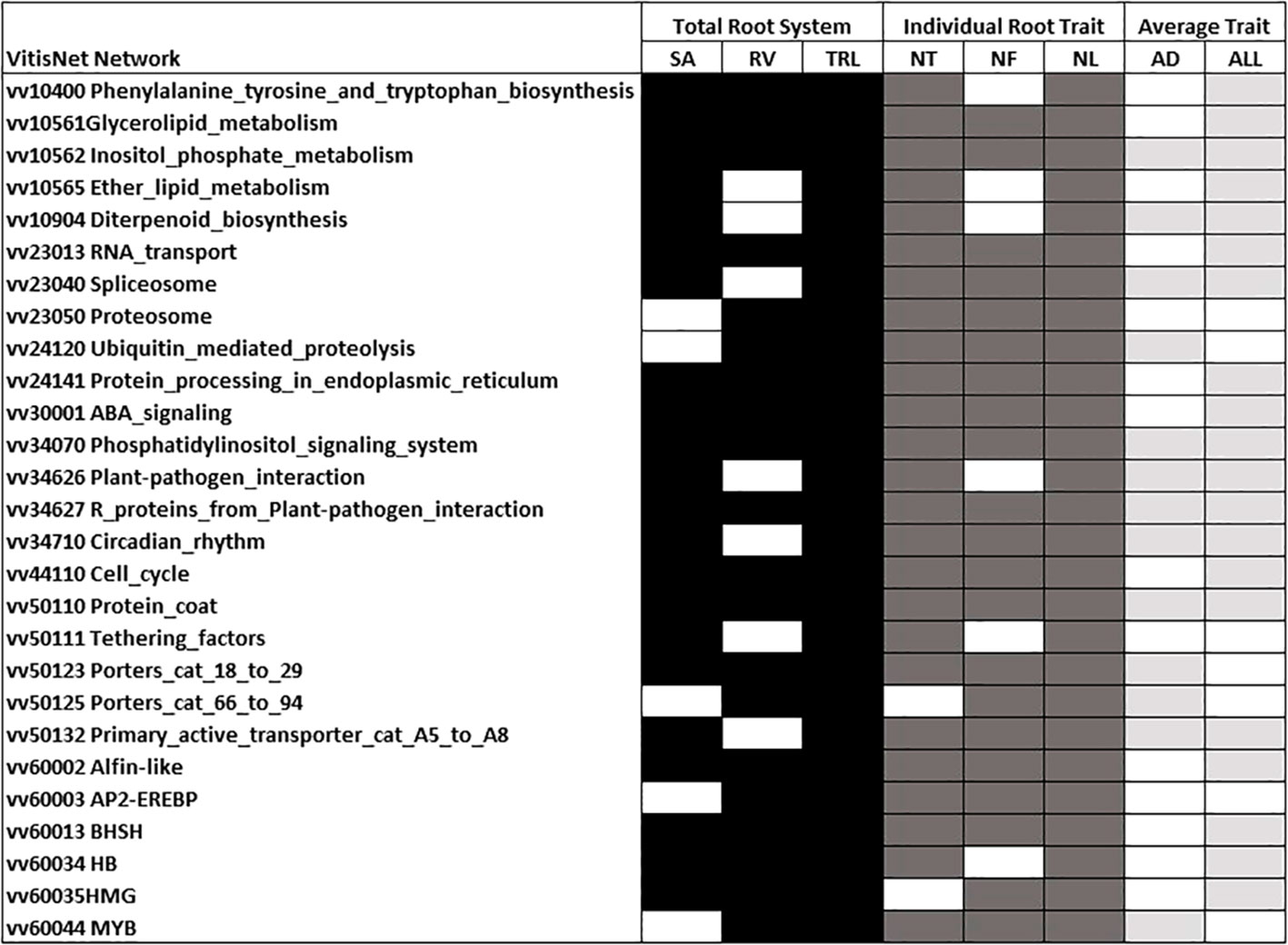
Figure 7 Enriched VitisNet pathways for modeled total root system, individual root and average traits. Shading indicates the significantly enriched pathway for total root system (black), individual root (dark gray), and average root (light gray/) traits (Fisher test p-value 0.05) White indicates not enriched. Only pathways enriched in at least five of the modeled traits are presented. All enriched pathways for an individual modeled trait are in Supplementary Table 2. Trait abbreviations are noted in Table 1.
4 Discussion
4.1 VRS-F2 grandparents differ in trait contribution to RSA
The challenge of increased summer temperatures and decreased water availability associated with climate change emphasizes the need for improved own-rooted grapevine or rootstock cultivars to maintain globally sustainable production (Smart et al., 2006; Delrot et al., 2020; Schmitz et al., 2021). While significant effort is paid to the genetic analysis of cluster and canopy architecture, less information is available for the grapevine root architecture as this hidden portion of the plant is more difficult to phenotype (Iandolino et al., 2013; Correa et al., 2014; Li et al., 2019; Krzyzaniak et al., 2021; Zinelabidine et al., 2021). The highly heterozygous grapevines are vegetatively propagated as own-rooted or grafted on rootstock to maintain their unique traits; thus, the entire root system of grapevines in production is formed of adventitious roots (Smart et al., 2002). In typical vine propagation, the cane or stem size impacts the available carbon and vascular development for root development in cuttings (Smart et al., 2002). In this study there was a very low correlation between RSA and cutting size suggesting that the cutting size was uniform and resulted in negligible variation across the genotypes. For multigenic traits like root system architecture, selection of parents that show genetic and phenotypic divergence increases the power of QTL detection (Hung et al., 2012). In this study, a VRS-F2 mapping population known to segregate for cold hardiness and berry quality traits was used to explore the genetic characteristics of a young grapevine adventitious root system (Fennell et al., 2005; Yang et al., 2016; Fennell et al., 2019; Alahakoon et al., 2022). VRS-F2 progeny showed morphological variation across all traits and progeny clustered with the parent or one of the grandparents, indicating that genetic contribution from both grandparents were important in determining the RSA. The SA, TRL, NT, NF, NL, and LR traits provide measures of the total root system and are key factors influencing soil exploration (vertical and horizontal) and play a role in anchorage and water and mineral nutrient uptake (Comas et al., 2013; Yildirim et al., 2018). Exploration of the LBD candidate gene markers identified in QTL hotspot showed that the grandparent ‘Seyval blanc’ contributed the dominant allele for the NF trait. In contrast, the cold-hardy grandparent V. riparia contributed the dominant allele for AD the trait which impact root thickness/diameter. Similarly in tropical and temperate tree species AD correlates with cortical thickness and plays a major role in increasing absorptive capacity and isolating the stele from environmental stress especially in cold climate (Gu et al., 2014; Wang et al., 2019a). Several studies have shown that RSA traits work together to ensure resource uptake and plant survival under environmental stress (Comas et al., 2013; Uga et al., 2013; Koevoets et al., 2016). The SA, TRL, RV, NT, NF, and NL are all strongly correlated and can contribute to increased root system size, which would be beneficial for increased soil exploration, water and nutrient uptake, and strong plant anchorage as shown under drought conditions in Juglans and maize (Sun et al., 2011; Comas et al., 2013; Lynch, 2013). In contrast, root diameter impacts cortical thickness and thicker roots support increased absorptive capacity, thus acting as a buffer that protects the stele from environmental stress and contributes to better anchorage (Smart et al., 2002; Aloni et al., 2006; Alvarez-Uria and Körner, 2007; De Dorlodot et al, 2007; Gu et al., 2014; Wang et al., 2019a). The negative correlation between AD (V. riparia) contributing the dominant allele and NF (‘Seyval’ contributing dominant allele) provide a point of genetic interaction that may control the overall differences root system morphology in population for the more ‘Seyval’-like and V. riparia-like progeny.
4.2 QTL hotspots identify functional genomic units of RSA
Adventitious RSA and soil layer exploration result from an interplay of root traits. Root studies in several grain crops have shown that individual RSA trait QTL co-locate creating hotspots. In 24 rice QTL studies, many QTL for maximum root length co-located creating QTL hotspots (Comas et al., 2013). In Populus, 150 QTLs were associated with adventitious root traits and 25 hotspots were identified for these traits (Sun et al., 2019). In a grapevine rootstock population, developed to explore pH related nutrient deficiencies (Bert et al., 2013), 30 QTL for shoot and root traits show root biomass, root section and coarse root number co-localizing in field grown vines using an SSR map (Tandonnet et al., 2018). Similarly, in the VRS-F2 population there were seven areas of co-localization encompassing three to seven trait QTLs for chromosomes 1, 9, 13, 18, and 19. Although different traits were phenotyped in field vines with mature root systems, hotspots were identified in similar regions of chromosome 1 and 9 for traits that could contribute to root biomass in either study, providing further confidence in QTL identified in newly rooted cuttings. Genes found to regulate Arabidopsis root development such as auxin signaling were identified in the confidence interval of Tandonnet et al. (2018) and the current study. However, this study identified LATERAL ORGAN BOUNDARIES DOMAIN genes (LBD4 and ASYMETRIC LEAVES 2) and ALFIN-LIKE transcription factors near the peak positions in the hotspots which have also been shown to play a role in root development. Pathway enrichment was conducted for genes located 700 kb up and downstream of the QTL peak position based on the recommendation that candidate genes be within 3-4 cM of the peak (Cipriani et al., 2011; Hugalde et al., 2021). The AP2_EREBP enriched pathway on chromosomes 1 and 9 included the ETHYLENE-RESPONSIVE transcription factor ERF118, a gene associated with xylem cell expansion in Populus gene co-expression analysis (Seyfferth et al., 2018). In addition, there were two AP2 DOMAIN CONTAINING RAP2 genes that are known to regulate shoot development and are expressed in root vasculature during root development (Che et al., 2006). Another AP2_EREBP gene AINTEGUMENTA (ANT1) often regulating floral organogenesis was found in the chromosome 9 hotspot. This was interesting as a GROWTH REGULATING FACTOR (GRF) transcription factor gene is a potential target of ANT1 regulation and this may play a role in the regulation of meristematic competence (Krizek et al., 2020). In Arabidopsis ANT1 binds to genes similar to another related transcription factor PLT2 which is known to function in primarily in roots (Krizek et al., 2020). The transport related gene ALTERED RESPONSE TO GRAVITY 1 (ARG1) gene known to interact with PIN-FORMED auxin transport proteins regulating the distribution of auxin in response to gravitropic signals is also found in the chromosome 1 hotspot of this study (Konstantinova et al., 2021). In addition, four lateral organ boundary domain genes (LBD41-3, LBD41-4, LBD21, and ASL5) were found near the peak position for TRL, NT, NF and NL on chromosomes 1 and 13. LBD genes are known to have a role in lateral root development in grapevine and other species (Grimplet et al., 2017; Trinh, 2019). The enriched C2C2-CO transcription factors are CONSTANS-LIKE (COL11 and COL16) genes known to be involved in floral development. Several MYB transcription factor genes with potential roles in root function and development contributed to the enrichment of the MYB pathway on chromosomes 1 and 9 (MYB3R1, MYB62, WEREWOLF-5, MYB7-1, TT2, and MYB26) (Baudry et al., 2004; Gu et al., 2017; Wang et al., 2018; Wang et al., 2019b; Van Den Broeck et al., 2021). MYB3R1 is a cell cycle related gene that forms a regulatory hub with the TSO1 transcription factor to coordinate cell division in root and shoot (Wang et al., 2018). WEREWOLF-5 has a role in regulating root epidermal cell root-hair and nonhair cell types (Wang et al., 2019b). MYB7 is thought to have a role, in Arabidopsis roots, as a repressor regulating CYC6 a cortex/endodermis asymmetric stem cell division gene (Wang et al., 2019b). Overexpression of MYB26 in Populus promotes secondary wall deposition and is a potential master switch controlling secondary-wall biosynthesis (Xiao et al., 2021). Thus, these genes have the potential to be involved in regulating cell division, root hair and lateral root development. Autophagy, a cytoplasmic degradation pathway, has recently been shown to be involved in glucose-mediated root meristem activity by peroxisome regulation of the production of reactive oxygen species and auxin biosynthesis (Huang et al., 2019; Su et al., 2020). In Populus, expression level of several autophagy (ATG) genes alternated during the differentiation of xylem and phloem tissues and different ATG genes were specific to primary and secondary root tissue development (Wojciechowska et al., 2019). Over-expression of ATG in Arabidopsis resulted in fewer lateral roots (Su et al., 2020). In the current study, two AUTOPHAGY genes (APG12a and ATG18) were identified underlying the hotspots on chromosome 9 and 13 for NT, NF, and NL traits, suggesting a role in root branching. ALFIN-like transcription factors have a role in root hair elongation, meristem development, root development, and abiotic stress and have been shown to enhance drought and salt tolerance (Kayum et al., 2015; Tao et al., 2018). Therefore, the ALFINDOM1 and ALFINDOM8 which are identified in SA, RV, TRL, NT, NF, and NL may play a role in the adventitious root morphology. Taken together the enriched suite of transcription factor pathways and the autophagy pathway genes underlying the hotspots for the morphological traits related to root system size and branching patterns present strong candidate genes for further analysis in the genetic control and development of RSA.
4.3 Modeling RSA QTL reveals the inter-relationship of small effect traits underlying the RSA complexity
In the present study, all QTL had minor effects that explained less than 10% of phenotypic variance. This is similar to other reports that the RSA is a multigenic trait and often shows 3-6 QTL and a low percentage of variability explanation for individual root traits (Smith, 2010; Bert et al., 2013; Tandonnet et al., 2018). De Dorlodot et al. (2007) suggests that modeling of RSA traits may address the complexity of RSA and reveal interesting relationships between the traits (Comas et al., 2013). They indicate that the possibility of identifying QTL loci in related species using comparative QTL mapping is as an advantage of modeling (Comas et al., 2013). Enriched pathway analysis by trait, in contrast to the analysis of the individual hotspots provided a global perspective of the genes related to each of the root traitsIt is well known that adventitious root initiation is regulated by hormone signaling and a central role for auxin biosynthesis and signaling in lateral root initiation; however, when both the hotspots and modeled QTL were interrogated, several other enriched pathways were identified. Therefore, emphasis here is placed on the less frequently described cell cycle, circadian rhythm, and the HB transcription factor family enriched pathways underlying root trait specific QTL (Trinh, 2019).
Unique to modeled traits were distinct cell cycle genes, two CYCLINA1 and two CYCLINB1 (CYCA1-1, CYCA1-2, and CYCB1-2) which are expressed in root tips, dividing root cells, lateral roots and root epidermis in Arabidopsis, thus also implicating a role in the grapevine RSA (Masucci et al., 1996; Himanen et al., 2002). Clock related genes were found in the enriched circadian rhythm pathway in this study (Figure 7, Supplementary Table 2). The circadian rhythm pathway had seven genes contributing to its pathway enrichment with five of these having a function in circadian clock signaling (two EARLY FLOWERING 4 (ELF4), PHYTOCHROME C (PHYC), PHYTOCHROME-ASSOCIATED PROTEIN 1 (PAP1), and ADAGIO PROTEIN 1 (ZTL)) and the signaling gene CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1). Light modulates primary root elongation and lateral root development and elongation (Yang and Liu, 2020). In grapevine roots there is a strong network of ABA, cytokinin, and circadian rhythm gene expression during water stress induced growth reduction (Khadka et al., 2019). In roots, the circadian clock is simpler than in the shoot, runs faster in the root tip than shoot, and rephases in roots to controls levels of auxin and auxin related genes during lateral root emergence (Voß et al., 2015; Greenwood et al., 2022). PHYC is active in root tips and has a role in gravitropism in Arabidopsis hypocotyls but a limited role in root gravitropism (Tóth et al., 2001; Salisbury et al., 2007; Kumar et al., 2008). ELF4 is a mobile shoot to root signal that promotes regulation of the root clock speed in response to temperature conditions (Chen et al., 2020). The enrichment of the circadian rhythm pathway in several root traits in this study suggest further investigation is warranted as there is limited literature on its function in adventitious RSA. There were six HB transcription factors contributing to the enrichment of this pathway for six traits. Of these PROTODERMAL FACTOR2 (PDF2) and ANTHOCYANINLESS2 (ANL20) are potential candidate genes in RSA as they play a role in regulating epidermal layer and root cell differentiation and root development (Kubo et al., 1999; Kubo and Hayashi, 2011; Ogawa et al., 2015). These genes regulate cellular organization and the identification of these genes near peak position on chromosomes 13 and 19 support a role in RSA.
5 Conclusion
The variation and genetic architecture of own-rooted VRS-F2 population are reported. VRS-F2 genotypes that had similar root morphology to either grandparent or the parent were identified. ‘Seyval blanc’ contributed to dominant allele for SA, RV, FW, TRL, LR, NT, NF, and NL traits that maximize resource uptake and anchorage. V. riparia contributed dominant allele to greater root thickness and link length in the AD, ALSA, and ALL traits which has been noted in species with environmental stress tolerance. QTL hotspots with identical peak positions were identified on chromosomes 1, 9, 13, and 19. These hotspots were underlain by AUTOPHAGY genes and LBD, MYB, and ALFINDOM transcription factor genes that have been shown to have a role in root development in other species. QTL modeling and candidate gene identification revealed interrelatedness of small effect traits causing the RSA complexity. Enriched pathways underline QTL confidence intervals identified genes involved lateral root growth (LBD and PDF) and cell cycle and circadian clock genes which have previously been shown to have a role in Arabidopsis and alfalfa root development. The combined analysis of QTL hotspots and modeled root trait QTLs in a grapevine F2 population with grandparents of differing RSA has provided a suite of candidate genes that can be explored for selection of improved adventitious root system architecture in grapevine and other woody species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://openprairie.sdstate.edu/, https://openprairie.sdstate.edu/ahps_datasets/1, https://www.try-db.org/TryWeb/Home.php, DOI: 10.17871/TRY.92.
Author contributions
AF and DA conceptualization, DA acquired data and conducted QTL analysis, AF conducted pathway analysis. DA and AF prepared and finalized the manuscript.
Funding
This research was partially funded by the South Dakota Agricultural Experiment Station, Hatch Project No. SD00H63318 and the National Institute of Food and Agriculture of USDA (USDA-NIFA) Specialty Crop Research Initiative Award No. 2017-51181-26829.
Acknowledgments
We acknowledge Ramandeep Kuar for running script for gene set enrichment analysis and Yajun Wu for reviewing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1083374/full#supplementary-material
Supplementary Table 1 | Enriched pathways underlying QTL for identical peak position root traits on chromosomes 1, 9, 13, 19 and the genes contributing to the enrichment.
Supplementary Table 2 | Enriched pathways underlying modeled QTL traits and genes contributing to the enrichment.
References
Alahakoon, D., Fennell, A., Helget, Z., Bates, T., Karn, A., Manns, D., et al. (2022). Berry anthocyanin, acid, and volatile trait analyses in a grapevine-interspecific F2 population using an integrated GBS and rhAmpSeq genetic map. Plants 11, 696. doi: 10.3390/plants11050696
Aloni, R., Aloni, E., Langhans, M., Ullrich, C. I. (2006). Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 97, 883–893. doi: 10.1093/aob/mcl027
Alvarez-Uria, P., Körner, C. (2007). Low temperature limits of root growth in deciduous and evergreen temperate tree species. Funct. Ecol. 21, 211–218. doi: 10.1111/j.1365-2435.2007.01231.x
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Bellini, C., Pacurar, D. I., Perrone, I. (2014). Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 65, 639–666. doi: 10.1146/annurev-arplant-050213-035645
Bert, P. F., Bordenave, L., Donnart, M., Hevin, C., Ollat, N., Decroocq, S. (2013). Mapping genetic loci for tolerance to lime-induced iron deficiency chlorosis in grapevine rootstocks (Vitis sp.). Theor. Appl. Genet. 126, 451–473. doi: 10.1007/s00122-012-1993-5
Broman, K. W., Wu, H., Sen, S., Churchill, G. A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. doi: 10.1093/bioinformatics/btg112
Che, P., Lall, S., Nettleton, D., Howell, S. H. (2006). Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 141, 620–637. doi: 10.1104/pp.106.081240
Chen, W. W., Takahashi, N., Hirata, Y., Ronald, J., Porco, S., Davis, S. J., et al. (2020). A mobile ELF4 delivers circadian temperature information from shoots to roots. Nat. Plants 6, 416–426. doi: 10.1101/612671
Cipriani, G., Di Gaspero, G., Canaguier, A., Jusseaume, J., Tassin, J., Lemainque, A., et al. (2011). Molecular linkage maps: strategies, resources and achievements (A.-F. Adam-Blondon, J.M. Martinez-Zapater, Chittaranjan Kole: Science Publishers and CRC Press), 111–136.
Comas, L. H., Becker, S. R., Cruz, V. M., Byrne, P. F., Dierig, D. A. (2013). Root traits contributing to plant productivity under drought. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00442
Cooper, M., Van Eeuwijk, F. A., Hammer, G. L., Podlich, D. W., Messina, C. (2009). Modeling QTL for complex traits: detection and context for plant breeding. curr opin. Plant Biol. 12, 231–240. doi: 10.1016/j.pbi.2009.01.006
Correa, J., Mamani, M., Munoz-Espinoza, C., Laborie, D., Munoz, C., Pinto, M., et al. (2014). Heritability and identification of QTLs and underlying candidate genes associated with the architecture of the grapevine cluster (Vitis vinifera l.). Theor. Appl. Genet. 127, 1143–1162. doi: 10.1007/s00122-014-2286-y
Dalbó, M., Souza, A. (2019). Rootstock breeding for resistance to grapevine decline and dieback in southern Brazil. Acta Hortic. 1248, 123–128. doi: 10.17660/ActaHortic.2019.1248.18
De Dorlodot, S., Forster, B., Pages, L., Price, A., Tuberosa, R., Draye, X. (2007). Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 12, 474–481. doi: 10.1016/j.tplants.2007.08.012
Delrot, S., Grimplet, J., Carbonell-Bejerano, P., Schwandner, A., Bert, P.-F., Bavaresco, L., et al. (2020). Genetic and genomic approaches for adaptation of grapevine to climate change, in genomic designing of climate-smart fruit crops. Springer 157–270. doi: 10.1007/978-3-319-97946-5_7
Fennell, A., Alahakoon, D., Luby, J., Clark, M. (2019). Mapping the genetic architecture of grapevine bud growth-cycling quantitative traits. Acta Hortic. 1248, 337–344. doi: 10.17660/ActaHortic.2019.1248.49
Fennell, A., Mathiason, K., Luby, J. (2005). Genetic segregation for indicators of photoperiod control of dormancy induction in Vitis species. Acta Hort. 689, 533–539. doi: 10.17660/ActaHortic.2005.689.66
Fort, K., Fraga, J., Grossi, D., Wlaker, M. A. (2017). Early measures of drought tolereance in four grape rootstocks. J. Am. Soc. Horticulture Sci. 142, 36–46. doi: 10.21273/JASH0391916
Gautier, A., Cookson, S. J., Lagalle, L., Ollat, N., Marguerit, E. (2020). Influence of the three main genetic backgrounds of grapevine rootstocks on petiolar nutrient concentrations of the scion, with a focus on phosphorus. OENO One 54, 1–13. doi: 10.20870/oeno-one.2020.54.1.2458
Greenwood, M., Tokuda, I. T., Locke, J. C. (2022). A spatial model of the plant circadian clock reveals design principles for coordinated timing. Mol. Syst. Biol. 18, e10140. doi: 10.15252/msb.202010140
Grimplet, J., Pimentel, D., Agudelo-Romero, P., Martinez-Zapater, J. M., Fortes, A. M. (2017). The LATERAL ORGAN BOUNDARIES domain gene family in grapevine: genome-wide characterization and expression analyses during developmental processes and stress responses. Sci. Rep. 7, 1–18. doi: 10.1038/s41598-017-16240-5
Grimplet, J., Van Hemert, J., Carbonell-Bejerano, P., Diaz-Riquelme, J., Dickerson, J., Fennell, A., et al. (2012). Comparative analysis of grapevine whole-genome gene predictions, functional annotation, categorization and integration of the predicted gene sequences. BMC Res. Notes 5, 213. doi: 10.1186/1756-0500-5-213
Gu, J., Xu, Y., Dong, X., Wang, H., Wang, Z. (2014). Root diameter variations explained by anatomy and phylogeny of 50 tropical and temperate tree species. Tree Physiol. 34, 415–425. doi: 10.1093/treephys/tpu019
Gu, M., Zhang, J., Li, H., Meng, D., Li, R., Dai, X., et al. (2017). Maintenance of phosphate homeostasis and root development are coordinately regulated by MYB1, an R2R3-type MYB transcription factor in rice. J. Exp. Bot. 68, 3603–3615. doi: 10.1093/jxb/erx174
Han, K.-H., Bradshaw, H., Gordon, M. (1994). Adventitious root and shoot regeneration in vitro is under major gene control in an F2 family of hybrid poplar (Populus trichocarpa x deltoides). Int. J. For. Genet 1, 139–146.
Himanen, K., Boucheron, E., Vanneste, S., De Almeida Engler, J., Inzé, D., Beeckman, T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14, 2339–2351. doi: 10.1105/tpc.004960
Huang, L., Yu, L.-J., Zhang, X., Fan, B., Wang, F.-Z., Dai, Y.-S., et al. (2019). Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in arabidopsis. Autophagy 15, 407–422. doi: 10.1080/15548627.2018.1520547
Hugalde, I., Paolinelli, M., Agüero, C. B., Riaz, S., Gomez Talquenca, S., Walker, M. A., et al. (2021). Prioritization of vigor QTL-associated genes for future genome-directed vitis breeding: Priorización de genes relacionados a vigor en Vitis. Revista De La Facultad De Ciencias Agrarias UNCuyo. 53, 27–35. doi: 10.48162/rev.39.036
Hung, H. Y., Browne, C., Guill, K., Coles, N., Eller, M., Garcia, A., et al. (2012). The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity (Edinb) 108, 490–499. doi: 10.1038/hdy.2011.103
Iandolino, A., Pearcy, R., Williams, L. E. (2013). Simulating three-dimensional grapevine canopies and modelling their light interception characteristics. Aust. J. Grape Wine Res. 19, 388–400. doi: 10.1111/ajgw.12036
Kayum, M., Park, J.-I., Ahmed, N. U., Jung, H.-J., Saha, G., Kang, J.-G., et al. (2015). Characterization and stress-induced expression analysis of alfin-like transcription factors in Brassica rapa. Mol. Genet. Genomics 290, 1299–1311. doi: 10.1007/s00438-015-0993-y
Kasssambara, A., Mundt, F. (2017). Factoextra: extract and visualize the results of multivariate data analyses. R package version 1.0.5. Available at: https://CRAN.R-project.org/package=factoextra.
Khadka, V. S., Vaughn, K., Xie, J., Swaminathan, P., Ma, Q., Cramer, G. R., et al. (2019). Transcriptomic response is more sensitive to water deficit in shoots than roots of Vitis riparia (Michx.). BMC Plant Biol. 19, 1–20. doi: 10.1186/s12870-019-1664-7
Koevoets, I. T., Venema, J. H., Elzenga, J. T., Testerink, C. (2016). Roots withstanding their environment: exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01335
Konstantinova, N., Korbei, B., Luschnig, C. (2021). Auxin and root gravitropism: addressing basic cellular processes by exploiting a defined growth response. Int. J. Mol. Sci. 22. doi: 10.3390/ijms22052749
Krizek, B. A., Blakley, I. C., Ho, Y. Y., Freese, N., Loraine, A. E. (2020). The arabidopsis transcription factor AINTEGUMENTA orchestrates patterning genes and auxin signaling in the establishment of floral growth and form. Plant J. 103, 752–768. doi: 10.1111/tpj.14769
Krzyzaniak, Y., Cointault, F., Loupiac, C., Bernaud, E., Ott, F., Salon, C., et al. (2021). In situ phenotyping of grapevine root system architecture by 2D or 3D imaging: Advantages and limits of three cultivation methods. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.638688
Kubo, H., Hayashi, K. (2011). Characterization of root cells of anl2 mutant in Arabidopsis thaliana. Plant Sci. 180, 679–685. doi: 10.1016/j.plantsci.2011.01.012
Kubo, H., Peeters, A. J., Aarts, M. G., Pereira, A., Koornneef, M. (1999). ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11, 1217–1226. doi: 10.1105/tpc.11.7.1217
Kumar, P., Montgomery, C. E., Kiss, J. Z. (2008). The role of phytochrome c in gravitropism and phototropism in Arabidopsis thaliana. Funct. Plant Biol. 35, 298–305. doi: 10.1071/FP08013
Li, M., Klein, L. L., Duncan, K. E., Jiang, N., Chitwood, D. H., Londo, J. P., et al. (2019). Characterizing 3D inflorescence architecture in grapevine using X-ray imaging and advanced morphometrics: implications for understanding cluster density. J. Exp. Bot. 70, 6261–6276. doi: 10.1093/jxb/erz394
Limera, C., Sabbadini, S., Sweet, J. B., Mezzetti, B. (2017). New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01418
Lynch, J. (1995). Root architecture and plant productivity. Plant Physiol. 109, 7–13. doi: 10.1104/pp.109.1.7
Lynch, J. P. (2013). Steep, cheap and deep: an ideotype to optimize water and n acquisition by maize root systems. Ann. Bot. 112, 347–357. doi: 10.1093/aob/mcs293
Maechler, M., Rousseeuw, P., Struyf, A., Hubert, M., Hornik, K. (2019). Cluster: Cluster analysis basics and extensions. R version 2, 0. ed, 1.
Marguerit, E., Brendel, O., Lebon, E., Van Leeuwen, C., Ollat, N. (2012). Rootstock control of scion transpiration and its acclimation to water deficit are controlled by different genes. New Phytol. 194, 416–429. doi: 10.1111/j.1469-8137.2012.04059.x
Masucci, J. D., Rerie, W. G., Foreman, D. R., Zhang, M., Galway, M. E., Marks, M. D., et al. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of. Arabidopsis thaliana. Dev. 122, 1253–1260. doi: 10.1242/dev.122.4.1253
Meggio, F., Prinsi, B., Negri, A. S., Simone Di Lorenzo, G., Lucchini, G., Pitacco, A., et al. (2014). Biochemical and physiological responses of two grapevine rootstock genotypes to drought and salt treatments. Aust. J. Grape Wine Res. 20, 310–323. doi: 10.1111/ajgw.12071
Nass (2019). National statistics for grapes (USDA). Available at: https://www.nass.usda.gov/Statistics_by_State/California.
Ogawa, E., Yamada, Y., Sezaki, N., Kosaka, S., Kondo, H., Kamata, N., et al. (2015). ATML1 and PDF2 play a redundant and essential role in arabidopsis embryo development. Plant Cell Physiol. 56, 1183–1192. doi: 10.1093/pcp/pcv045
Ollat, N., Bordenave, L., Tandonnet, J., Boursiquot, J., Marguerit, E. (2016). Grapevine rootstocks: origins and perspectives. Acta Hortic. 1136, 11–22. doi: 10.17660/ActaHortic.2016.1136.2
Osier, M. (2016). VitisPathways: gene pathway analysis for V. vinifera. VITIS-Journal Grapevine Res. 55, 129–133. doi: 10.5073/vitis.2016.55.129-133
R Core Team (2019). A language and environment for statistical computing. Vienna Austria: R Foundation for Statistical Computing. Available at: https://www.R-project.org.
Revelle, W. (2018). Psych: procedures for psychological, psychometric, and personality research (Evanston, Illinois: Northwestern University).
Riaz, S., Pap, D., Uretsky, J., Laucou, V., Boursiquot, J.-M., Kocsis, L., et al. (2019). Genetic diversity and parentage analysis of grape rootstocks. Theor. Appl. Genet. 132, 1847–1860. doi: 10.1007/s00122-019-03320-5
Robinson, H., Kelly, A., Fox, G., Franckowiak, J., Borrell, A., Hickey, L. (2018). Root architectural traits and yield: exploring the relationship in barley breeding trials. Euphytica 214, 1–16. doi: 10.1007/s10681-018-2219-y
Salisbury, F. J., Hall, A., Grierson, C. S., Halliday, K. J. (2007). Phytochrome coordinates arabidopsis shoot and root development. Plant J. 50, 429–438. doi: 10.1111/j.1365-313X.2007.03059.x
Satbhai, S. B., Ristova, D., Busch, W. (2015). Underground tuning: quantitative regulation of root growth. J. Exp. Bot. 66, 1099–1112. doi: 10.1093/jxb/eru529
Schmitz, R., Atkinson, B., Sturrock, C., Hausmann, L., Toepfer, R., Herzog, K. (2021). High-resolution 3D phenotyping of the grapevine root system using X-ray computed tomography. VITIS 60, 21–27. doi: 10.5073/vitis.2021.60.21-27
Serra, I., Strever, A., Myburgh, P., Deloire, A. (2014). The interaction between rootstocks and cultivars (Vitis vinifera l.) to enhance drought tolerance in grapevine. Aust. J. Grape Wine Res. 20, 1–14. doi: 10.1111/ajgw.12054
Seyfferth, C., Wessels, B., Jokipii-Lukkari, S., Sundberg, B., Delhomme, N., Felten, J., et al. (2018). Ethylene-related gene expression networks in wood formation. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00272
Smart, D. R., Kocsis, L., Walker, M. A., Stockert, C. (2002). Dormant buds and adventitious root formation by vitis and other woody plants. J. Plant Growth Regul. 21, 296–314. doi: 10.1007/s00344-003-0001-3
Smart, D. R., Schwass, E., Lakso, A., Morano, L. (2006). Grapevine rooting patterns: a comprehensive analysis and a review. Am. J. Enology Viticulture 57, 89–104. doi: 10.5344/ajev.2006.57.1.89
Smith, B. P. (2010). Genetic and molecular mapping studies on a population derived from vitis vinifera x muscadinia rotundifolia and genetic diversity of wild muscadinia rotundifolia (Davis: University of California).
Su, T., Li, X., Yang, M., Shao, Q., Zhao, Y., Ma, C., et al. (2020). Autophagy: an intracellular degradation pathway regulating plant survival and stress response. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00164
Sun, P., Jia, H., Zhang, Y., Li, J., Lu, M., Hu, J. (2019). Deciphering genetic architecture of adventitious root and related shoot traits in populus using QTL mapping and RNA-seq data. Int. J. Mol. Sci. 20. doi: 10.3390/genes10120973
Sun, S. J., Meng, P., Zhang, J. S., Wan, X. (2011). Variation in soil water uptake and its effect on plant water status in Juglans regia l. during dry and wet seasons. Tree Physiol. 31, 1378–1389. doi: 10.1093/treephys/tpr116
Tandonnet, J.-P., Marguerit, E., Cookson, S. J., Ollat, N. (2018). Genetic architecture of aerial and root traits in field-grown grafted grapevines is largely independent. Theor. Appl. Genet. 131, 903–915. doi: 10.1007/s00122-017-3046-6
Tao, J.-J., Wei, W., Pan, W.-J., Lu, L., Li, Q.-T., Ma, J.-B., et al. (2018). An alfin-like gene from atriplex hortensis enhances salt and drought tolerance and abscisic acid response in transgenic arabidopsis. Sci. Rep. 8, 1–13. doi: 10.1038/s41598-018-21148-9
Tóth, R., Kevei, E., Hall, A., Millar, A. J., Nagy, F., Kozma-Bognár, L. (2001). Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 127, 1607–1616. doi: 10.1104/pp.010467
Trinh, D. C. (2019). “Gene regulatory network for lateral root formation,” in Arabidopsis thaliana (France: Université Montpellier).
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Valliyodan, B., Ye, H., Song, L., Murphy, M., Shannon, J. G., Nguyen, H. T. (2017). Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J. Exp. Bot. 68, 1835–1849. doi: 10.1093/jxb/erw433
Van Den Broeck, L., Spurney, R. J., Fisher, A. P., Schwartz, M., Clark, N. M., Nguyen, T. T., et al. (2021). A hybrid model connecting regulatory interactions with stem cell divisions in the root. quantitative. Plant Biol. 2, E2. doi: 10.1017/qpb.2021.1
Vezzulli, S., Gramaje, D., Tello, J., Gambino, G., Bettinelli, P., Pirrello, C., et al. (2022). Genomic Designing for Biotic Stress Resistant Grapevine." In Genomic Designing for Biotic Stress Resistant Fruit Crops. C, Kole. (eds). (Cham: Springer). doi: 10.1007/978-3-030-91802-6_4
Voß, U., Wilson, M. H., Kenobi, K., Gould, P. D., Robertson, F. C., Peer, W. A., et al. (2015). The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat. Commun. 6, 1–9. doi: 10.17863/CAM.26324
Walker, M., Lund, K., Agüero, C., Riaz, S., Fort, K., Heinitz, C., et al. (2014). Breeding grape rootstocks for resistance to phylloxera and nematodes – it’s not always easy. Acta Hortic. 1045, 89–98. doi: 10.17660/ActaHortic.2014.1045.12
Wang, W., Ryu, K. H., Barron, C., Schiefelbein, J. (2019b). Root epidermal cell patterning is modulated by a critical residue in the WEREWOLF transcription factor. Plant Physiol. 181, 1239–1256. doi: 10.1104/pp.19.00458
Wang, W., Sijacic, P., Xu, P., Lian, H., Liu, Z. (2018). Arabidopsis TSO1 and MYB3R1 form a regulatory module to coordinate cell proliferation with differentiation in shoot and root. Proc. Natl. Acad. Sci. 115, E3045–E3054. doi: 10.1073/pnas.1715903115
Wang, H., Wang, Z., Dong, X. (2019a). Anatomical structures of fine roots of 91 vascular plant species from four groups in a temperate forest in northeast China. PloS One 14, e0215126. doi: 10.1371/journal.pone.0215126
Wickham, H., François, R., Henry, L., Müller, K. (2022). dplyr: A Grammar of Data Manipulation. Available at: https://github.com/tidyverse/dplyr.
Wojciechowska, N., Smugarzewska, I., Marzec-Schmidt, K., Zarzyńska-Nowak, A., Bagniewska-Zadworna, A. (2019). Occurrence of autophagy during pioneer root and stem development in Populus trichocarpa. Planta 250, 1789–1801. doi: 10.1007/s00425-019-03265-5
Xiao, R., Zhang, C., Guo, X., Li, H., Lu, H. (2021). MYB transcription factors and its regulation in secondary cell wall formation and lignin biosynthesis during xylem development. Int. J. Mol. Sci. 22. doi: 10.3390/ijms22073560
Yang, S., Fresnedo-Ramirez, J., Sun, Q., Manns, D. C., Sacks, G. L., Mansfield, A. K., et al. (2016). Next generation mapping of enological traits in an F2 interspecific grapevine hybrid family. PloS One 11, e0149560. doi: 10.1371/journal.pone.0149560
Yang, Y., Liu, H. (2020). Coordinated shoot and root responses to light signaling in arabidopsis. Plant Commun. 1, 100026. doi: 10.1016/j.xplc.2020.100026
Yildirim, K., Yagci, A., Sucu, S., Tunc, S. (2018). Responses of grapevine rootstocks to drought through altered root system architecture and root transcriptomic regulations. Plant Physiol. Biochem. 127, 256–268. doi: 10.1016/j.plaphy.2018.03.034
Zhang, M., Gao, M., Zheng, H., Yuan, Y., Zhou, X., Guo, Y., et al. (2019). QTL mapping for nitrogen use efficiency and agronomic traits at the seedling and maturity stages in wheat. Mol. Breed. 39, 1–17. doi: 10.1007/s11032-019-0965-8
Zinelabidine, L. H., Torres-Perez, R., Grimplet, J., Baroja, E., Ibanez, S., Carbonell-Bejerano, P., et al. (2021). Genetic variation and association analyses identify genes linked to fruit set-related traits in grapevine. Plant Sci. 306, 110875. doi: 10.1016/j.plantsci.2021.110875
Keywords: LATERAL ORGAN BOUNDARY DOMAIN, protodermal factor, circadian rhythm, autophagy, grapevine, adventitious roots
Citation: Alahakoon D and Fennell A (2023) Genetic analysis of grapevine root system architecture and loci associated gene networks. Front. Plant Sci. 13:1083374. doi: 10.3389/fpls.2022.1083374
Received: 28 October 2022; Accepted: 29 December 2022;
Published: 02 February 2023.
Edited by:
Jianfu Jiang, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Antonio Lupini, Mediterranea University of Reggio Calabria, ItalyOlfa Zarrouk, Smart Farm CoLab (SFCOLAB), Portugal
Lei Sun, Beijing Academy of Agricultural and Forestry Sciences, China
Copyright © 2023 Alahakoon and Fennell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anne Fennell, YW5uZS5mZW5uZWxsQHNkc3RhdGUuZWR1
 Dilmini Alahakoon
Dilmini Alahakoon Anne Fennell
Anne Fennell